Abstract
Nicotinamide adenine dinucleotide (NAD+) is an essential cofactor metabolite and is the currency of metabolic transactions critical for cell survival. Depending on tissue context and genotype, cancer cells have unique dependencies on NAD+ metabolic pathways. Poly(ADP-ribose) polymerases (PARPs) catalyze oligomerization of NAD+ monomers into poly(ADP-ribose) (PAR) chains during cellular response to alkylating chemotherapeutics, including procarbazine or temozolomide. Here, we find that, in endogenous IDH1 mutant tumor models, alkylator-induced cytotoxicity is markedly augmented by pharmacologic inhibition or genetic knockout of the PAR breakdown enzyme poly(ADP-ribose) glycohydrolase (PARG). Both in vitro and in vivo, we observe that concurrent alkylator and PARG inhibition depletes freely available NAD+ by preventing PAR breakdown, resulting in NAD+ sequestration and collapse of metabolic homeostasis. This effect reversed with NAD+ rescue supplementation, confirming the mechanistic basis of cytotoxicity. Thus, alkylating chemotherapy exposes a genotype-specific metabolic weakness in tumor cells that can be exploited by PARG inactivation.
Keywords: Glioma, temozolomide, PARG inhibitor, NAD metabolism, IDH1 mutation
Introduction
Gliomas characterized by mutations in the isocitrate dehydrogenase (IDH) genes IDH1 and IDH2 are the most common primary brain cancer of adults in the 3rd, 4th and 5th decades of life (1,2). Typically presenting as lower grade lesions within the World Health Organization diagnostic classification (grades II and III) (3,4), these infiltrative tumors are slow-growing but cause progressive neurological morbidity in patients. Alkylating chemotherapy has been proven effective for patients with IDH mutant gliomas, with extended survival demonstrated in international randomized clinical trials combining radiation with the procarbazine, lomustine and vincristine (PCV) regimen (5,6) or the oral chemotherapeutic temozolomide (TMZ) (7). However, while effective, these therapies are not curative and strategies to improve treatment are needed.
Mutant IDH enzymes catalyze the abnormal overproduction of D-2-hydroxyglutarate (2-HG) from alpha-ketoglutarate, resulting in substantial metabolic derangements in cancer cells, including low basal levels of NAD+(8). Many studies have shown that these metabolic derangements have created selective susceptibilities in IDH mutant cancers (9–14). In addition, recent work has shown that DNA damage response and PARP signaling is a susceptibility in IDH mutant cancer cells (15–18), which display sensitivity to PARP inhibitors (PARPi) (19).
As a key component of the cellular metabolic response to alkylating chemotherapy, poly(ADP-ribose) polymerase (PARP) activity is acutely upregulated upon chemotherapeutic exposure (20), transiently consuming nicotinamide adenine dinucleotide (NAD+) through enzymatic polymerization of monomeric NAD+ into poly(ADP-ribose) (PAR) chains (21). This PARylation signal then recruits DNA repair machinery to the sites of chemotherapeutic-induced DNA damage (22–24). Unique to IDH mutant gliomas, which have low basal levels of NAD+, widespread activation of PARP causes available NAD+ to be critically depleted after alkylating chemotherapy, exposing a transient metabolic vulnerability (25).
Importantly, PARylation is regulated not only by PARPs, but also by poly(ADP-ribose) glycohydrolase (PARG). PARG is the primary enzyme that mediates PAR breakdown, by cleaving the glycoside bonds of PAR to release mono ADP-ribose (26). Therefore, PARG inhibition after alkylating chemotherapy could, in theory, result in hyper-accumulation of PAR by blocking enzymatic degradation, with the simultaneous arrest of PAR recycling to NAD+ leading to catastrophic collapse of free NAD+ levels (27). We hypothesized that, for IDH mutant gliomas, a therapeutic effect could be maximized by combining alkylating chemotherapy with PARG inhibition, causing both DNA damage and scarcity of available monomeric NAD+.
Here, we report that the combination of TMZ and PARG inhibitors is highly effective against IDH mutant gliomas. We show that, as hypothesized, TMZ treatment promotes PARP activation and outflow consumption of cellular NAD+ pools, while PARG inactivation then freezes this NAD+ as polymerized PAR, by blocking subsequent breakdown. This resulting state is lethal in IDH mutant tumor cells in both in vitro and in vivo models.
Results
PARP inhibitors block NAD+ consumption induced by TMZ in IDH mutant patient-derived cancer cell lines
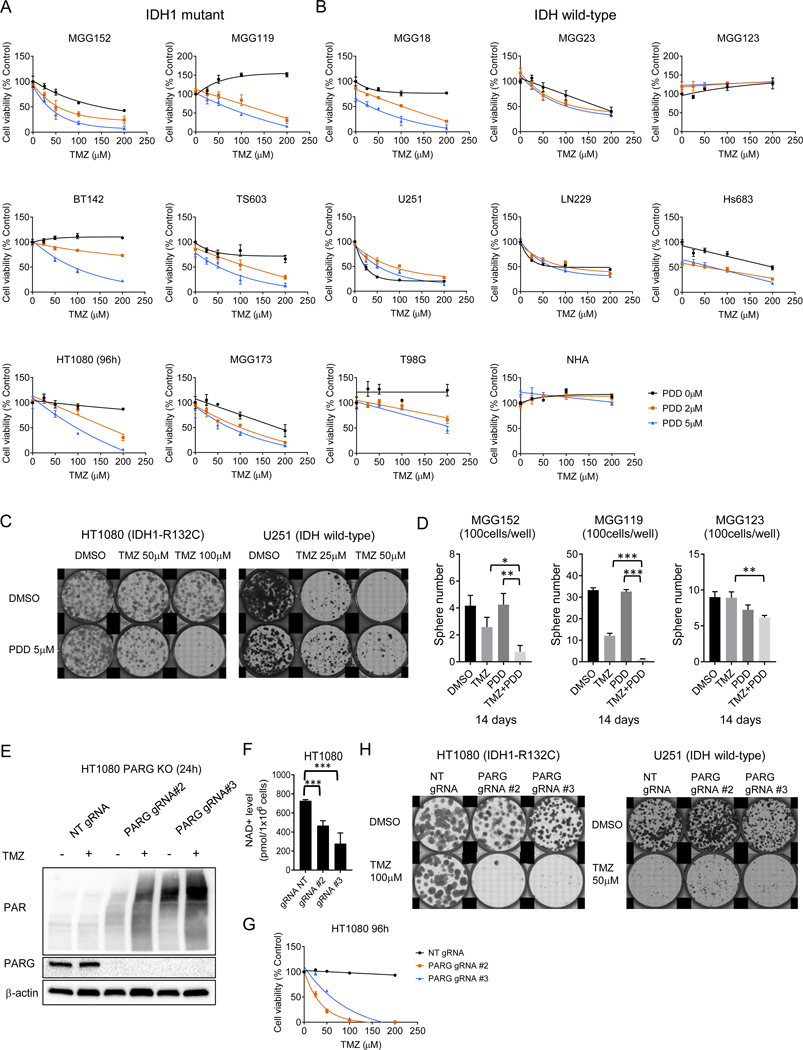
We first treated a panel of patient-derived endogenous IDH1 mutant and wild-type glioma lines, as well as the IDH1 mutant fibrosarcoma line HT1080, with the small molecule PARP inhibitor olaparib. PARP inhibition (PARPi) displayed a substantial effect as monotherapy in IDH1 mutant lines HT1080 and TS603 (Figure 1A, Supplementary Fig. S1A), and this cytotoxic effect was further augmented by the addition of TMZ. Both findings are consistent with prior reports (16,17). In several patient-derived IDH1 mutant glioma lines, however, including MGG152, MGG119, and BT142, the effect of PARPi monotherapy was weaker, and augmentation of that effect with TMZ was muted. As a control, we tested a panel of IDH wild-type glioma lines and, consistent with recent reports (28,29), also observed a range of response to PARPi monotherapy and combination with TMZ (Figure 1A, Supplementary Fig. S1A). While some wild-type glioma lines like Hs683 and T98G were sensitive to PARPi monotherapy and PARPi+TMZ (Supplementary Figs. S1A, S1B), normal human astrocytes (NHA) cells used as controls were relatively insensitive to either treatment, at doses across the physiologic range (Supplementary Fig. S1C). These data suggest that the association between IDH mutation and PARP-mediated synthetic lethality in cancer cells may be context specific, depending on additional genomic or metabolite differences in different cancer lines.
Figure 1. PARP inhibitors block NAD+ consumption induced by TMZ in IDH mutant patient-derived cancer cell lines.
A. Relative cell viability of IDH1-mutant and wild-type cancer cells after exposure to 0, 2 and 5 μM of olaparib (Ola) with temozolomide (TMZ) at indicated concentrations. Drug exposure was 120 h except 96 h for HT1080. Upper row, IDH1-mutant cells; Lower row, IDH1 wild-type cells.
B. Mutation profiles and MGMT methylation status of IDH1 mutant and wild-type cells.
C. Relative NAD+ levels in IDH1-mutant (upper row) and IDH wild-type lines (lower row) after 6 and 24-hour treatment with TMZ (200 μM), Ola (5 μM), or TMZ plus Ola. Bars, ± SEM; *p < 0.05, **p < 0.01, ***p < 0.001.
D. Western blot analysis of PAR, PARP and cleaved PARP (cPARP) expression in HT1080 transfected with cDNA carrying no (control) and PARP1 cDNA (PARP OE), at 72-hour. PARP OE, DNA 2.5 μg/well. β− actin is loading control.
E. Relative cell viability after 96-hour treatment with DMSO, Ola (1 μM) or NMN (1 mM) to HT1080 transfected with cDNA carrying no (control) and PARP1 cDNA (PARP OE). Bars, ± SEM; ***p < 0.001. NS, non-significant.
F. elative NAD+ levels after 6 and 24-hour treatment with DMSO or Ola (1 μM) to HT1080 transfected with cDNA carrying no (control) and PARP1 cDNA (PARP OE). Bars, ± SEM; **p < 0.01, ***p < 0.001.
G. Relative cell viability of IDH1-mutant cancer cells after 48-hour exposure to Ola (0 or 5 μM) with FK866 at indicated concentrations.
H. A schematic model of NAD+ metabolic and DNA repair cycle.
To explore this finding further, we performed genomic profiling of our cell line panel; however, these analyses did not reveal an alteration that consistently predicted PARPi responsiveness (Figure 1B). MGMT is methylated in the majority of lines, although cases where it is unmethylated (ie. HT1080, TS603) could potentially account for differences in the effect of combination TMZ and olaparib treatment compared to relative insensitivity to TMZ monotherapy. Given the utilization of NAD+ as a PARP substrate, we then considered the metabolic effects of NAD+ levels on PARPi monotherapy across our panel of cancer cell lines. To assess the relationship between PARP inhibition and NAD+ consumption, we noted that, at transient time points (6 hours), TMZ treatment often caused detectable NAD+ depletion in IDH mutant cell lines (Figure 1C), an effect not seen in wild-type lines which have higher levels of basal NAD+ (8). This depletion was reversed by PARPi (Figure 1C), recovering at 24hrs in all cases. Due to the lower basal levels of NAD+, IDH mutant cells are vulnerable to treatment with inhibitors of the NAD+ biosynthesis enzyme NAMPT (8). Indeed, in prior studies we have observed that NAMPT inhibitor plus TMZ had an additive growth inhibitory effect in IDH mutant cells (25). Interestingly however, it has been noted that PARPi synthetic lethality is mediated through amplification of DNA damage response, as a selective vulnerability in IDH mutant cancer cells, and not dependent upon NAD+ (16). To assess this molecular mechanism, we therefore tested the effect of PARP1 overexpression. We found that PARP1 overexpression does lead to PAR hyperaccumulation and cytotoxicity in IDH1 mutant cells (Figure 1D, E). Not surprisingly, measurement of NAD+ levels revealed they were markedly depleted. Furthermore, attempted rescue with the NAD+ precursor nicotinamide mononucleotide (NMN) was only minimally effective when compared to NMN rescue of NAMPT inhibitor (8) (Figure 1F), consistent with robust PARylation signaling as a primary mechanism of effect.
We also tested the potential contribution of the lowered availability of substrate NAD+ for PARP consumption, which could render a pass-through mechanism of “pseudo-PARP inhibition” by NAMPT inhibition. We therefore considered whether PARPi would additively promote the cytotoxicity of NAMPT inhibitors in IDH mutant gliomas. Notably, we observed the opposite -- PARPi treatment consistently reversed the cytotoxic effect of NAMPT inhibition at 24 and 48 hours (Figure 1G, Supplementary Fig. S1D), potentially highlighting a distinct metabolic weakness of depleted basal NAD+ in these tumors, separable from PARP-mediated DNA damage signaling.
Thus, our working model for understanding PARPi-mediated cytotoxicity in susceptible cancer cells is centered on the amplification of DNA damage response by PARP enzymes through PARylation trapping of available NAD+ (Figure 1H). The complex interplay between PARP inhibition, PAR recycling and NAD+ equilibrium led us to consider whether this repair cycle could be intercepted in IDH mutant gliomas at a different point, maximally exploiting both vulnerabilities.
Inhibition of PARG augments treatment with TMZ
We hypothesized that a therapeutic effect could be optimized by targeting PAR breakdown via PARG inhibition, thereby combining the effect of PAR-mediated damage signaling with an arrest of NAD+ captured within PAR to simultaneously drive metabolic stress. In a test of this hypothesis, across our panel of IDH mutant cancer cell lines, we observed that cytotoxicity was significantly and consistently induced by a combination of TMZ with the selective small molecule PARG inhibitor (PARGi) PDD00017273 (hereafter abbreviated as PDD), at doses of PDD that were only modestly cytotoxic as monotherapy (Figure 2A, Supplementary Fig. S2A). Conversely, we did not observe augmentation of TMZ cytotoxicity by PDD in 5 of 7 IDH wild-type lines, nor did we observe this effect in NHA cells (Figure 2B). We further observed that treatment with PDD augmented the TMZ-mediated inhibition of clone formation of IDH mutant HT1080 cells, but not IDH wild-type U251 cells (Figure 2C). Analogously, the TMZ and PDD combination suppressed sphere formation in IDH mutant neurosphere lines MGG152 and MGG119, compared to an IDH wild-type control line (Figure 2D). We used a live cell protease activity assay as a control in parallel with an ATP-based cell viability assay to demonstrate consistency across assays confirming cell death from combined TMZ and PDD treatment (Supplementary Fig. S2B).
Figure 2. Inhibition of PARG augments treatment with TMZ.
A. B.
Relative cell viability of IDH1-mutant (A) and wild-type cancer cells or normal human astrocyte (NHA) (B) after exposure to 0, 2 and 5 μM of PDD00017273 (PDD) with temozolomide (TMZ) at indicated concentrations. Drug exposure was 120-hour except 96-hour for HT1080.
C. Clonogenic assay of HT1080 (IDH1 R132C) and U251 (IDH wild-type) treated with DMSO or PDD (5 μM) with TMZ (25, 50 or 100 μM) after 7–10-day exposure.
D. Sphere formation assay of MGG152 and MGG119 (IDH1 R132H) and MGG123 (IDH wild-type) treated with DMSO, TMZ (100 μM), PDD (5 μM), and TMZ plus PDD after 14 days exposure. Bars, ± SEM; *p < 0.05, **p < 0.01, ***p < 0.001.
E. Western blot analysis of PAR and PARG in non-targeting (NT), PARG gRNA #2 and #3-transduced HT1080 cells. β-Actin, loading control.
F. Quantitation of basal NAD+ levels in NT, PARG gRNA #2 and #3-transduced HT1080 cells. Bars, ± SEM; ***p < 0.001.
G. Relative cell viability after 96-hour exposure to TMZ at indicated concentrations in NT, PARG gRNA #2 and #3-transduced HT1080 cells.
H. Clonogenic assay of NT, PARG gRNA #2 and #3-transduced HT1080 and U251 cells after 7 days exposure to DMSO or TMZ (100 μM for HT1080, 50 μM for U251).
To assess whether this PDD cytotoxic effect was “on-target”, we engineered IDH mutant HT1080 and TS603 and IDH wild-type U251 PARG knock-out (KO) cells using two independent CRISPR guide RNAs (Genscript gRNA #2 and #3) (Figure 2E, Supplementary S2C). Baseline PAR levels were increased, and TMZ treatment strongly induced PAR accumulation in PARG KO HT1080 cells, compared to non-targeting control gRNA transduced cells (Figure 2E). We also observed lower basal NAD+ and NADH levels in HT1080 PARG KO lines (Figure 2F, Supplementary Fig. S2D). TMZ treatment potently inhibited cell viability in both PARG KO IDH mutant cell lines, but not in control PARG KO IDH wild-type cell line (Figure 2G, Supplementary Fig S2E), an effect analogous to those seen above with combined TMZ and PDD. Of note, baseline cell growth was somewhat slower in HT1080 PARG KO than control (Supplementary Fig. S2F), suggestive of lowered fitness for PARG KO in this genetic background. We therefore compared clonogenicity after TMZ treatment, confirming marked sensitization in PARG KO compared to wild-type control (Figure 2H)
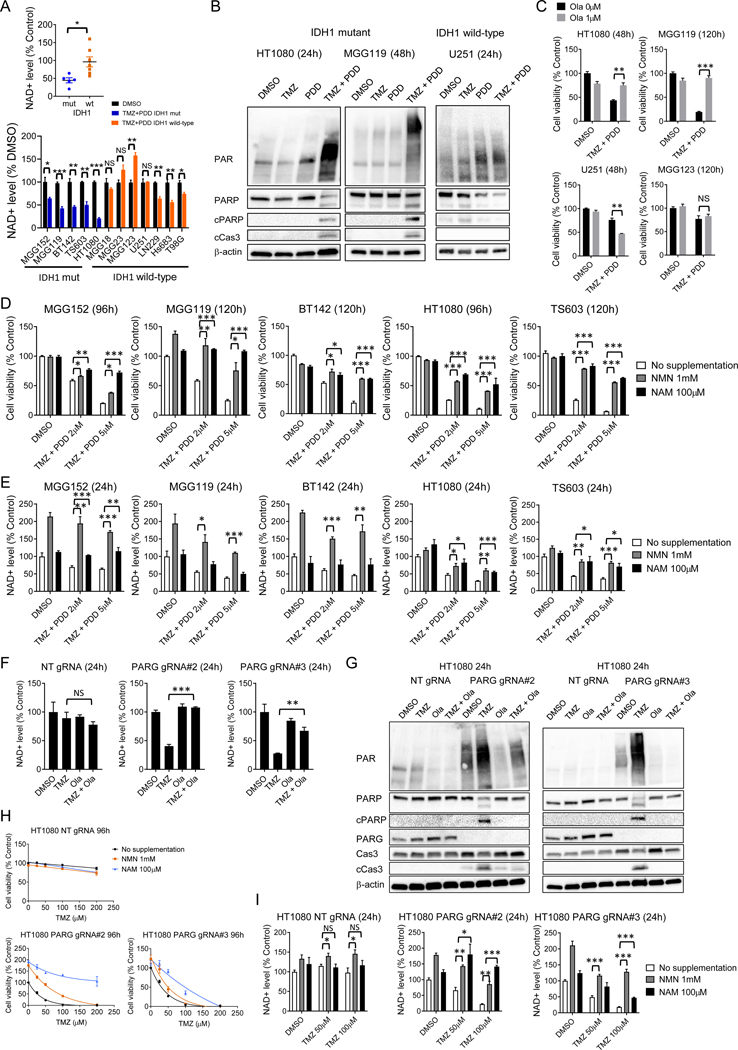
Metabolic depletion of NAD+ contributes to combined TMZ and PARG inhibition toxicity
After exposure to TMZ only, the decrease of NAD+ and upregulation of PAR is transient in IDH mutant gliomas (25), with a measurable PAR increase in the 1–6 hours following treatment that resolves within 24 hours. The rapid return to baseline levels suggests that enzymatic PAR breakdown by PARG may be contributing to the restoration of NAD+ homeostasis. To test whether PARG inhibition after TMZ was sequestering NAD+ as PAR, we next measured NAD+ levels in cells treated with combined TMZ + PDD, and observed significant and sustained depletion in five endogenous IDH-mutant cancer lines compared to IDH wild-type cancer lines (Figure 3A, Supplementary Fig. S3A), a finding that supports our proposed mechanism. In addition, in these IDH mutant cell lines, TMZ + PDD, but not TMZ alone, led to marked PAR accumulation and concomitant apoptosis (Figure 3B, Supplementary Fig. S3B), compared to IDH wild-type glioma U251 control. Olaparib potently suppressed TMZ-induced PAR expression (Supplementary Fig. S3C). Strikingly, olaparib also reversed the cell viability reduction induced by the TMZ + PDD combination in IDH mutant cells (Figure 3C, Supplementary Fig. S3D). Taken together with the previous results that PARP inhibition alone did not decrease NAD+ (Figure 1C), and that TMZ alone caused only transient PAR accumulation (25), these data suggest that the TMZ + PDD combination induces cytotoxicity by a distinct mechanism. We considered that the aberrant metabolism of NAD+ depletion could be the primary cause of the observed effect, in place of (or in addition to) intensified alkylating DNA damage. Indeed, during the short-term timeframe of our in vitro experiments, an effective dose of the TMZ + PDD combination elicited no, or only minimal, activation of gamma H2AX, a DNA damage marker, or LC3, an autophagy marker, in HT1080 and MGG119. (Supplementary Fig. S3E). Interestingly, addition of PDD seems to suppress TMZ-induced gamma-H2AX activation, suggesting possible secondary effects leading to alterations of DNA damage signaling pathways.
Figure 3. Metabolic depletion of NAD+ contributes to combined TMZ and PARG inhibition cytotoxicity.
A. Scatter plot and bar graph of relative NAD+ levels in IDH1-mutant and IDH wild-type cells treated with DMSO or temozolomide (TMZ, 200 μM) plus PDD00017273 (PDD, 5 μM) at 24 hours. *p < 0.05 for difference from DMSO. Bars, ± SEM; *p < 0.05, **p < 0.01, ***p < 0.001
B. Western blot analysis of PAR, PARP, cleaved PARP (cPARP), and cleaved caspase3 (cCas3) in HT1080, MGG119 (IDH mutant) and U251 (IDH wild-type) cells treated with DMSO, TMZ (200 μM), PDD (5 μM), and TMZ plus PDD for 24 (for HT1080 and U251) or 48 hours (for MGG119). β-actin is loading control.
C. Relative cell viability of IDH mutant cells (HT1080 and MGG119) and IDH wild-type cells (U251 and MGG123) after 48 or 120 h exposure to DMSO or TMZ (200 μM) plus PDD (5 μM), either with or without 1 μM olaparib (Ola). Bars, ± SEM; **p < 0.01, ***p < 0.001.
D. Relative cell viability of IDH mutant cells after 96–120h exposure to DMSO, TMZ (200 μM) plus PDD (2 μM), or TMZ (200 μM) plus PDD (5 μM) with no supplementation, NMN (1 mM), or NAM (100 μM). Bars, ± SEM; *p < 0.05, **p < 0.01, ***p < 0.001.
E. Relative NAD+ level of IDH mutant cells after 24h exposure to DMSO, TMZ (200 μM) plus PDD (2 μM), or TMZ (200 μM) plus PDD (5 μM) with no supplementation, NMN (1 mM), or NAM (100 μM). Bars, ± SEM; *p < 0.05, **p < 0.01, ***p<0.001.
F. Relative NAD+ level in non-targeting (NT), PARG gRNA #2 and #3-transduced HT1080 cells with DMSO, TMZ (100 μM), Ola (1 μM), or TMZ plus Ola at 24 hours. Bars, ± SEM; **p < 0.01, ***p < 0.001. NS, non-significant.
G. Western blot analysis of PAR, PARP, cPARP, caspase 3, and cCas3 after treatment with DMSO, TMZ (200 μM), Ola (1 μM) or TMZ plus Ola for 24 hours in NT, PARG gRNA #2 and #3-transduced HT1080 cells. β-actin, loading control.
H. Relative cell viability of HT1080 NT gRNA, PARG gRNA #2 and #3 cells after 96h-exposure to TMZ at indicated dose with no supplementation, NMN (1 mM) and NAM (100 μM).
I. Relative NAD+ level of HT1080 NT gRNA, PARG gRNA #2 and #3 cells after 24h-exposure to DMSO, TMZ (50 μM or 100 μM) with no supplementation, NMN (1 mM), or NAM (100 μM). Bars, ± SEM; *p < 0.05, **p < 0.01, ***p < 0.001.
Strongly supporting our hypothesis, we were able to partially or fully reverse the effects of TMZ + PDD on both NAD+ levels and cell viability by rescue supplementation with NAD+ precursors nicotinamide (NAM) or nicotinamide mononucleotide (NMN) (Figure 3D, E, Supplementary Fig. S3F). The lesser magnitude of effect on NAD+ level, when compared to cell viability, that we observed with NAM rescue suggests that cytotoxicity may be offset with NAD+ levels recovering above an appropriate threshold in a critical timeframe. Furthermore, PAR levels decreased with NAM supplementation (Supplementary Fig. S3G), possibly due to the established mechanism by which NAM acts as a feedback negative regulator on PARPs, suppressing PARylation (30,31) and impacting the synthetic lethal DNA damage signaling pathways.
To further evaluate our hypothesized metabolic mechanism, we then tested NAD+ alterations in our independent PARG KO lines. TMZ treatment induced marked NAD+ depletion in both PARG KO HT1080 lines, compared to non-targeting control (Figure 3F). TMZ treatment also induced PAR upregulation and apoptosis, as evidenced by cleaved caspase3 induction, in both PARG KO HT1080 lines (Figure 3G, Supplementary Fig. S4A), findings fully consistent with those observed after PARG inhibition by PDD. Conversely, PARP inhibition with olaparib abrogated TMZ-induced PAR accumulation and NAD+ depletion in PARG KO cells, rescuing them from early (24h) apoptosis (Figure 3F, 3G). These results constitute independent evidence that inactivation of PARG activity, and resulting PAR accumulation, is required for cell death after TMZ treatment in PARG KO cells, in contrast to the control cells. Furthermore, we could reverse the NAD+ level and cell viability inhibition by rescue supplementation with NMN or NAM in PARG KO cells (Figure 3H, 3I, Supplementary Fig. S4B, S4C), a result consistent with the effect seen with TMZ + PDD treatment. All told, these results indicate that metabolic depletion of NAD+ contributes mechanistically to combined TMZ and PARGi toxicity.
Next, we assessed the relationship between PARG inhibition and metabolic enhancement with other alkylating regents that are clinically-effective in glioma patients, such as procarbazine (PCZ), carmustine (BCNU), and lomustine (CCNU) (Supplementary Fig. S5A–C). Interestingly, only monofunctional DNA alkylating agents TMZ and PCZ activated PAR in PARG KO lines (Supplementary Fig. S5D). These monofunctional alkylators produce DNA adducts primarily at O6- and N7-methyl guanine (O6meG and 7meG) and N3-methyl adenine (3meA). On the other hand, nitrosoureas such as BCNU and CCNU, which react with adjacent cytosine bases to generate guanine-cytosine (G-C) inter-strand crosslinks, did not activate PAR in PARG KO lines. These results highlight the differential role of PARPs in response to various DNA damage adducts, supporting the specificity of our hypothesized mechanism (32). Thus, we conclude that PARG inhibition after monofunctional alkylators sequesters NAD+ as PAR, both prolonging the hyperactive PAR-mediated signaling state and critically depleting NAD+ levels, resulting in increased cytotoxicity.
The effect of combined TMZ and PARGi in an IDH mutant background
We then considered the genomic determinant of sensitivity to alkylating chemotherapy and PARGi in IDH mutant cancer cells, given the possibility that the combination could be toxic in normal cells. Given our prior evidence of the selectivity of NAD+ depletion-mediated toxicity for IDH mutant cells (8), we used an isogenic model to directly test the causal effect of IDH mutation on the combination-responsive phenotype.
First, we tested TMZ + PARGi in IDH wild-type MGG18 cells which were engineered to express a doxycycline-inducible IDH1 mutant protein (Figure 4A). Altered NAD+ equilibrium in IDH mutant cells is related to downregulation of the biosynthesis enzyme NAPRT1 (8,33). After 3 months of exposure to doxycycline, a time course consistent with previous findings (8,34), we observed an induction of mutant IDH1 expression, a high concentration of 2-HG, NAPRT1 inactivation and decreased NAD+ and NADH (Figure 4B–D). In this model system, we observed that TMZ + PARGi had a greater and more rapid cell inhibitory effect after mutant IDH1 induction, compared to the non-induced state (Figure 4E). These results were also recapitulated in an additional IDH wild-type line MGG123 engineered to express IDH1-R132H (Supplementary Fig S6A–D).
Figure 4. The effect of combined TMZ and PARG inhibitor is exposed in an IDH mutant background.
A. Western blot analysis of IDH1R132H in tet-inducible MGG18-IDH1-R132H cells with or without 3-month exposure to 1 μg/mL doxycycline (Tet). Vinculin is loading control.
B. Quantification of 2-HG levels with or without 3-month exposure to Tet in MGG18-IDH1-R132H cells. ***p < 0.001.
C. Western blot analysis of NAPRT1 in MGG18-IDH1-R132H cells with or without 3-monthTet. Vinculin is loading control.
D. Quantitative NAD+ and NADH levels in MGG18-IDH1-R132H cells with or without 3-month Tet. ***p < 0.001.
E. Relative cell viability of MGG18-IDH1-R132H cells after 48 or 96-hour exposure to 2 μM PDD00017273 (PDD) with temozolomide (TMZ) at indicated concentrations. Tet was used for 3 months in the MGG18-IDH1-R132H Tet+ line. Tet+ and Tet- were each compared to PDD untreated. *p < 0.05, **p < 0.01 for difference between Tet+ and Tet-.
F. Relative NAD+ levels in Tet- and Tet+ MGG18-IDH1-R132H cells treated with DMSO, TMZ (200 μM), PDD (5 μM), or TMZ plus PDD for 6 and 24 hours. Bars, ± SEM; **p < 0.01, ***p < 0.001.
G. Western blot analysis of PAR, PARP, cleaved PARP (cPARP), caspase3 (Cas3) and cleaved caspase 3 (cCas3) in Tet- and Tet+ MGG18-IDH1-R132H cells treated with DMSO, TMZ (200 μM), PDD (5 μM), and TMZ plus PDD for 24 hours. β-actin is loading control.
H. Relative cell viability of endogenous IDH1 mutant MGG152 glioma cells ± long term (3 months) IDH1 inhibitor (IDHi) after 120-hour exposure to 0, 2 or 5 μM PDD with TMZ at indicated concentrations. Bars, ± SEM; **p < 0.01, ***p < 0.001.
I. Quantitative 2-HG levels in MGG152 cells ± IDHi exposure for 3 months. Bars, ± SEM; ***p < 0.001.
J. Quantitative NAD+ and NADH levels in MGG152 cells ± IDH1i exposure for 3 months. ***p < 0.001.
In the IDH1 mutant induced state, as compared to non-induced MGG18, NAD+ was depleted to a greater degree and for a longer time after treatment with the combination, inducing apoptotic cell death (Figure 4F, G). In the MGG18-IDH1 mutant line, transient high expression of gamma H2AX was observed at 24 hours, but it was repaired at 48 hours (Supplementary Fig. S6E) and no significant change occurred in the autophagy marker LC3. On the other hand, short-term incubation of doxycycline (6 days) caused sufficient expression of IDH1-R132H to increase 2-HG level but did not result in changes in NAPRT1 expression or NAD+/NADH levels (Supplementary Fig. S6F–I). Consistent with the lack of difference in NAD+ level, no significant change occurred in the sensitivity to TMZ + PARGi in this cellular context (Supplementary Fig. S6J). The addition of octyl-2HG, a cell-permeable form of 2-HG, similarly increased the intracellular 2-HG concentration, but did not change the sensitivity to TMZ + PARGi (Supplementary Fig. S6K, L). This time course suggests that long-term 2-HG exposure is required for NAPRT1 suppression, which leads to altered NAD+ metabolism and TMZ + PARGi sensitivity.
Also supportive of this mechanism, the patient-derived endogenous IDH mutant astrocytoma line MGG152, tested with and without prolonged exposure (>3 month) to the IDH inhibitor AGI5198, displayed a reversal of the combination TMZ + PDD cytotoxic effect after inhibition of mutant IDH1 (Figure 4H). Long-term IDH inhibitor treatment suppressed 2-HG and was associated with greater steady-state NAD+ and NADH accumulation (Figure 4I, J).
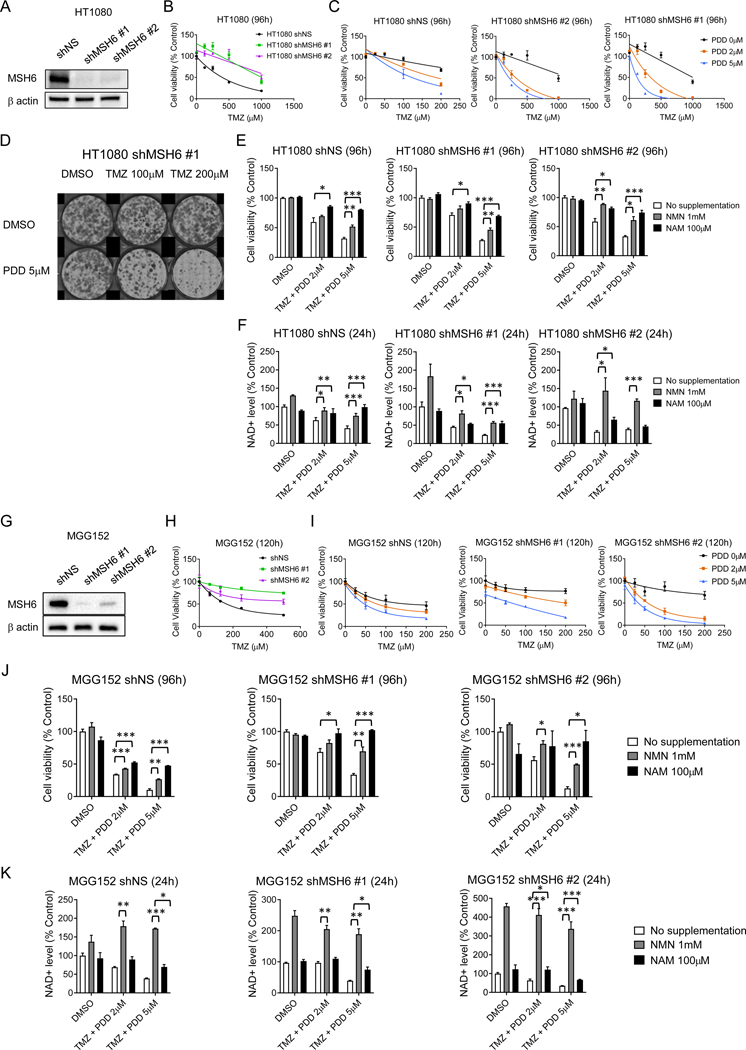
Metabolic depletion of NAD+ achieved by combined TMZ and PARGi treatment can overcome TMZ monotherapy resistance mediated by deficiencies in DNA mismatch repair
Genetic inactivation of the mismatch repair system (MMR) leads to the emergence of clones resistant to alkylating chemotherapy in cancer cell populations (35), thus demonstrating the key role MMR signaling plays in mediating the cytotoxicity of alkylating DNA damage. In recurrent gliomas, somatic acquisition of MMR pathway deficiency confers resistance to TMZ, albeit at the cost of a hypermutator phenotype in the post-TMZ setting (36,37). MMR deficiency is particularly enriched in post-treatment recurrent IDH mutant gliomas (38,39). To experimentally isolate the impact of MMR on the combined effect of TMZ and PARGi in the context of endogenous IDH mutation, we established stable MSH6 knockdown lines of IDH1 mutant HT1080 (Figure 5A), which is MGMT unmethylated (Figure 1B) (25). In this model system, we confirmed the expected acquired resistance to the cytotoxic effects of TMZ monotherapy (Figure 5B). Notably however, MSH6 knockdown lines retain sensitivity to the TMZ + PDD combination in cell viability and clonogenicity assays (Figure 5C, D). In addition, in experiments mirroring those above, we could rescue the cytotoxic and NAD+ depletion effect by supplementation with NMN or NAM (Figure 5E, F, Supplementary Fig. S7A).
Figure 5. Metabolic depletion of NAD+ achieved by combined TMZ and PARGi can overcome TMZ monotherapy resistance mediated by deficiencies in DNA mismatch repair.
A. Western blot analysis of MSH6 in non-silencing (NS) and MSH6 shRNA (#1, 2)-transduced HT1080 cells. Vinculin, loading control.
B. Relative cell viability of shNS-transduced and MSH6 shRNA-transduced HT1080 cells with 96-hour exposure to temozolomide (TMZ) at indicated concentrations.
C. Relative cell viability of shNS, MSH6 sh1 and 2-transduced HT1080 cells after 96-hour exposure to 0, 2 and 5 μM PDD00017273 (PDD) with TMZ at indicated concentrations.
D. Clonogenic assay of MSH6 shRNA #1-transduced HT1080 cells treated with DMSO or PDD (5 μM) with TMZ (0, 100 or 200 μM) for 7 days.
E. Relative cell viability of HT1080 shNS, MSH6 shRNA #1 and #2 cells after 96h-exposure to DMSO, TMZ (200 μM) plus PDD (2 μM), or TMZ (200 μM) plus PDD (5 μM) with no supplementation, NMN (1 mM), or NAM (100 μM). Bars, ± SEM; *p < 0.05, **p < 0.01, ***p < 0.001.
F. Relative NAD+ level in HT1080 shNS, MSH6 shRNA #1 and #2 cells after 24h exposure to DMSO, TMZ (200 μM) plus PDD (2 μM), or TMZ (200 μM) plus PDD (5 μM) with no supplementation, NMN (1 mM), or NAM (100 μM). Bars, ± SEM; *p < 0.05, **p < 0.01, ***p < 0.001.
G. Western blot analysis of MSH6 in NS and MSH6 shRNA-transduced MGG152. β-actin, loading control.
H. Relative cell viability of shNS and MSH6 shRNA #1 and #2 transduced MGG152 cells after 120h TMZ treatment at indicated concentrations.
I. Relative cell viability of shNS and MSH6 shRNA #1 and #2 transduced MGG152 cells after 120-hour exposure to 0, 2 or 5 μM PDD with TMZ at indicated concentrations.
J. Relative cell viability of MGG152 shNS, MSH6 shRNA #1 and #2 transduced cells after 96h-exposure to DMSO, TMZ (100 μM) plus PDD (2 μM), or TMZ (100 μM) plus PDD (5 μM) with no supplementation, NMN (1 mM), or NAM (100 μM). Bars, ± SEM; *p < 0.05, **p < 0.01, ***p < 0.001.
K. Relative NAD+ level in MGG152 shNS, MSH6 shRNA #1 and #2 transduced cells after 24h exposure to DMSO, TMZ (100 μM) plus PDD (2 μM), or TMZ (100 μM) plus PDD (5 μM) with no supplementation, NMN (1 mM), or NAM (100 μM). Bars, ± SEM; *p < 0.05, **p < 0.01, ***p < 0.001.
Furthermore, in MGG152 glioma cells, which are MGMT methylated (Figure 1B), we established two independent stable MSH6 knockdown cells and confirmed MSH6 knockdown increased resistance to TMZ (Figure 5G, H). Here again, the TMZ plus PDD combination displayed equal efficacy even in MSH6 knockdown cells (Figure 5I). The cytotoxic and NAD+ depletion effect was again rescued by supplementation with NMN or NAM (Figure 5J, K, Supplementary Fig. S7B). As a control to test whether this finding was specific for IDH mutant cells, we tested the MGMT methylated IDH wild-type glioblastoma, LN229, utilizing engineered MSH6 knockdown lines (Supplementary Fig. S7C). In this IDH wild-type glioma, TMZ + PDD did not show any benefit, irrespective of the MSH6 status (Supplementary Fig. S7D). Therefore, combining TMZ and PARGi is effective in IDH1-mutant cells notwithstanding the MMR-mediated resistance mechanism.
PARG inactivation markedly enhances the efficacy of TMZ in IDH mutant xenograft model in vivo
Given our observations pointing towards the key role of NAD+ in mediating TMZ + PARGi cytotoxicity, and the potential for variation in metabolism when comparing in vitro and in vivo environments, we tested whether PARG inactivation in vivo would produce the same results. We established a paired test, injecting the left flank of an individual mouse with non-targeting control sgRNA HT1080 and the right flank of the same mouse with PARG-targeting sgRNA #3 HT1080 (Figure 6A). Pairing tumors in the same animal provides the experimental advantage of an internal control for TMZ dosing, as each animal harbors an isogenic comparison for the same systemic TMZ administration. We noted a slowing of tumor growth in PARG KO lines even without chemotherapeutic treatment, a result likely indicative of more stringent growth conditions in vivo (40,41), compared to in vitro. Importantly, the tumor-specific effect of TMZ was augmented in the PARG KO line, which displayed markedly smaller tumor size and weight across treated cohorts (Figure 6B, C, Supplementary Fig. S8A). Confirming the metabolic mechanism in vivo, we observed significantly lower basal NAD+ levels in the PARG KO tumors, in which NAD+ and NADH levels were further decreased after 4 days (one cycle) of TMZ treatment (Figure 6D), with a concomitant increase in PAR (Figure 6E). In immunohistochemical analysis of the treated tumors, we observed evidence of increased apoptosis in the PARG KO tumors (Figure 6F, G). PARG KO tumors had a decreased Ki-67 index, indicating suppression of proliferation, consistent with a recent report (41). The animals displayed no significant systemic toxicity during treatment (Figure 6H). We also tested the orthotopic flank model using a second independent clone of HT1080 with PARG-targeting sgRNA #2 (Supplementary Fig. S8B–E) and observed findings mirroring those seen with PARG sgRNA #3. Thus, PARG inactivation potentiates the efficacy of TMZ in an IDH mutant xenograft model in vivo.
Figure 6. PARG inactivation enhances the efficacy of TMZ in IDH1 mutant xenograft models in vivo.
A. Experimental schema of HT1080 flank xenograft model.
B. Tumor growth curves in non-targeting (NT) gRNA and PARG knockout (gRNA #3, PARG) HT1080 flank xenograft model. Animals were treated with DMSO (n = 7) or temozolomide (TMZ) (50 mg/kg i.p., 5 days/week x 2 cycles) (n = 7). Data are presented as mean tumor volume and SEM in each group. Arrows, time points when treatment doses were given. *p = 0.0003, DMSO vs. TMZ in PARG KO tumors on day 18, unpaired t test.
C. Weight of NT gRNA and PARG KO (gRNA #3) HT1080 tumors on day 18. Data are presented as mean tumor volume +/− SEM in each group. *p = 0.0281, DMSO vs TMZ in NT tumors; ***p = 0.0009, DMSO vs TMZ in PARG KO tumors (unpaired t test).
D. Mean absolute NAD+ and NADH levels in HT1080 xenograft specimens treated with DMSO or TMZ (50 mg/kg for 5 days). **p < 0.01.
E. Western blot analysis of PAR and PARG expression in NT and PARG KO tumors treated with DMSO or TMZ (50 mg/kg). β-actin is loading control.
F. Representative microscopic pictures of hematoxylin and eosin (H&E), Ki-67, and TUNEL staining of HT1080 xenograft tissues after treatment. Scale bars, 100 μm.
G. Bar graphs showing percent positively staining cells of Ki-67 and TUNEL. Six pictures were captured with 20X magnification per section and used for quantitative analysis of immunopositivity. Bars, ± SEM; *p < 0.05 for difference between NT gRNA TMZ vs PARG KO TMZ. NS, non-significant.
H. Mean body weight of mice with flank HT1080 tumors for each treatment groups (same experiment as B). Bars indicate +/− SEM. Mouse weight was measured three days a week. Arrows, the time points when treatment doses were given.
Discussion
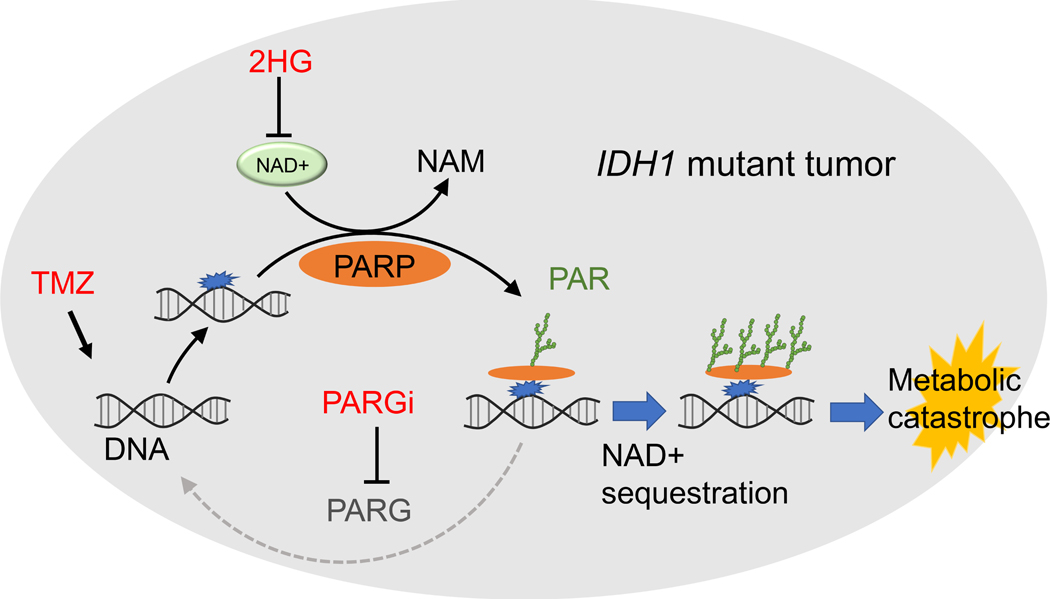
Here, we report a unique metabolic consequence of PARG inactivation after TMZ treatment, namely PAR hyper-accumulation and concurrent sequestration of NAD+ (Figure 7). This combination of alkylating chemotherapy and PARG inhibition is highly effective against IDH mutant cancer cells, as confirmed by isogenic experiments. PARGi prolongs and deepens the TMZ-induced depletion of free NAD+ levels, a metabolic state which is ultimately lethal in these vulnerable cells (8). The cytotoxic effect of the combined regimen is reversed by NAD+ supplementation, experimentally demonstrating the key contribution of metabolic stress, and mechanistically consistent with prior studies of NAD+ trafficking in IDH mutant and wild-type cells (25,27).
Figure 7. Proposed model of PARG inhibition combined with TMZ in IDH mutant cancer cells.
Low basal NAD+ levels in IDH mutant cancer cells, combined with sequestration of NAD+ as PAR by temozolomide (TMZ) + PARG inhibitor (PARGi), lead to metabolic catastrophe and cell death.
A similar mechanism was observed with the monofunctional alkylator PCZ when combined with PARG inhibition, highlighting the potential translational relevance of this finding to clinically-utilized glioma treatment regimens. The most widely-used alkylating chemotherapeutic regimens, which include either TMZ or PCZ, lack durability in treating IDH mutant glioma patients. Recurrences emerge in a meaningful subset of patients within a decade after diagnosis (42), in many cases recurring as malignant tumor subclones that drive lethal disease progression (43,44). Post-alkylator recurrences of IDH mutant glioma often display the mutational signature of an on-target selective pressure of chemotherapy, with escape from DNA damage surveillance mediated by MMR-deficient hypermutant genomic evolution (38,45). This partial effectiveness of chemotherapy has motivated research efforts to identify additional adjuvant agents that are specific for IDH mutant gliomas and can be combined with standard-of-care alkylators. Encouragingly, we demonstrate that a combination of a monofunctional alkylator plus PARG inhibition can be effective in MMR-deficient IDH mutant glioma, potentially allowing for more effective treatment outcomes.
From a therapeutic translational standpoint, this combination may prove effective in other cancer cells with metabolic alterations in NAD+ trafficking (33,46,47). Nevertheless, one hurdle still to overcome is the lack of a suitable PARGi for in vivo combination with TMZ, and studies on PARGi to date have primarily explored only in vitro models. The cell permeable PARGi PDD is not amenable for in vivo use, despite its potency, due to its short half-life in circulation (48–50). A more recently available PARGi, COH34, has improved stability in vitro and in vivo, but its potency is limited in otherwise PARGi-sensitive cell lines (51). This situation, however, is rapidly changing, as increasingly stable new agents enter active preclinical development. An important consideration when testing potential combination regimens is that the toxicity of these agents in normal tissues is currently unknown. Reassuringly however, we did not detect a measurably significant effect of combination treatment in either normal control NHA cells or most IDH wild-type tumor lines. Although further translational studies are needed, a key strength of our work is the support provided to the proposed combination of alkylating chemotherapy and PARG inhibition in IDH mutant cancers by independent and convergent lines of genetic and pharmacologic evidence in multiple patient-derived models, both in vitro and in vivo.
We also observed a sensitivity to PARP inhibition as monotherapy in our endogenous IDH mutant glioma lines, consistent with prior reports (16,17). However, we noted variability in the responsiveness of IDH mutant lines, suggesting that important modifiers of this effect are yet to be discovered. Notably, we found that PARP inhibition counteracted NAD+ biosynthesis inhibition, a metabolic perturbation that could potentially inform the combination therapy of TMZ and PARPi in IDH mutant tumor cells. The complexity of cancer PARylation metabolism is further highlighted by the disparate effects observed in other studies of PARG (50), inhibitors of which can, in varying contexts, alternatively antagonize (49) or phenocopy (52) the HRD-associated synthetic lethal effects of PARPi, or can mediate differential killing via HRD-independent mechanisms altogether (40,53). Additionally, the hyper-PARylation we observed after alkylating chemotherapy and PARGi likely magnifies PAR-mediated inhibition of hexokinase activity and glycolysis (54,55), independently of metabolic effects on NAD+. Our findings of a PARGi-mediated metabolic effect therefore suggest that a TMZ + PARGi combination could represent another therapeutic option to counter clinical PARPi resistance across multiple cancer types.
In conclusion, the combination treatment we propose uses a two-hit disruption of NAD+ homeostasis to achieve targeting of an intrinsic IDH mutant metabolic weakness, while limiting the escape avenues available for the emergence of sub-clonal resistance mutations. Our proposed mechanism extends and unifies several prior observations of selective vulnerability in IDH mutant gliomas to chemotherapy, NAD+ biosynthesis inhibition and isolated PARPi monotherapy sensitivity. Importantly, the observed non-overlapping mechanisms of DNA damage and metabolic cytotoxicity may allow for a treatment strategy that could improve IDH mutant glioma treatment.
METHODS
Cell Lines
The patient-derived glioma lines used in this study (MGG18, MGG23, MGG119, MGG123, MGG152, and MGG173) were obtained from 2008 to 2018 under IRB-approved protocols. The TS603 (IDH1-R132H, WHO grade III anaplastic oligodendroglioma) gliomasphere was derived from a patient who underwent tumor resection at Memorial Sloan-Kettering Cancer Center. The BT142 (IDH1R132H), HT1080 (IDH1R132C), T98G (IDH wild-type), U251 (IDH wild-type), LN229 (IDH wild-type), and Hs683 (IDH1 wild-type) lines were authenticated in 2019 by comparing their STR profiles to those of the ATCC public dataset. Normal human astrocytes (NHAs), purchased from ScienCell, were cryopreserved at passage number 3 or lower before their in vitro use. Patient-derived glioma neurosphere lines (BT142, TS603, MGG18, MGG23, MGG119, MGG123, MGG152, and MGG173) were cultured in serum-free Neurobasal medium (Gibco) as described previously (43,56,57). NHA, T98G, LN229, Hs683, and U251 cells were cultured in DMEM. HT1080 cells were cultured in EMEM. All standard growth media were supplemented with 10% fetal bovine serum and penicillin, streptomycin, and amphotericin B. All glioma cell lines were determined to be mycoplasma free using a LookOut Mycoplasma PCR Detection Kit (Sigma) in 2018 and 2019. All cells were maintained at 37°C in a humidified atmosphere of 5% CO2/95% air.
Compounds and Chemicals
The following chemical compounds were used in culture: AGI-5198 (Selleckchem), FK866 (Cayman Chemical), Dimethyl Sulfoxide (Sigma-Aldrich), Doxycycline hyclate (Sigma-Aldrich), Nicotinamide (Sigma-Aldrich), Nicotinamide mononucleotide (Sigma-Aldrich), Octyl-(R)-2HG (Sigma-Aldrich), Olaparib (Selleckchem), PDD00017273 (Tocris), and Temozolomide (Cayman Chemical).
IDH1-R132H Cell Line Generation
MGG18 cells expressing tetracycline-inducible IDH1-R132H, MGG18-IDH1-R132H, were generated with lentiviral transduction and described previously (8). To induce expression of mutant IDH1, MGG18-IDH1-R132H cells were cultured with doxcycline (1 μg/mL; Sigma-Aldrich) for 3 days to 3 months. GBM cell line MGG123 overexpressing IDH1R132H was generated by lentivirus infection using pLenti6.3/TO/V5 containing IDH1R132H, pCMV-psPAX2 (Addgene), and pCMV-VSVG (Addgene), followed by selection with blasticidin (0.5 μg/ml).
PARP overexpression
Cells were seeded in 6 well plate in antibiotic free medium. 70–80% confluent cells were subjected to transfection. PARP1 cDNA ORF clone (Cat #: OHu2551) and control vector (pcDNA3.1+/C-(K)DYK, Cat # SC1849) were purchased from GenScript. Transfection was carried out using Opti-MEM medium (Thermo Fisher Scientific) and Lipofectamine 3000 reagent (Invitrogen). After 72h, PARP overexpression was confirmed by Western blot.
shRNA and Control shRNA Cell Lines
MSH6 knockdown HT1080 and MGG152 cells were generated with lentiviral transduction as described previously (25). Detailed protocols are found in the Supplementary Methods.
CRISPR/Cas9 Genome Editing
Lentivirus vector plasmids containing gRNA sequences for PARG were pLentiCrispr v2-PARG gRNA from Genscript (gRNA2 and 3). Non-targeting gRNA lentivirus vector was from Genscript. To generate lentiviral particles, 293T cells were transfected with lentiviral plasmid, packaging plasmid (pCMV-psPAX2), and envelope plasmid (pCMV-VSV-G) with Fugene HD (Promega). Cells were infected with lentivirus in the presence of polybrene (8 μg/mL) for 8 h. After 3 days, the cells were selected with puromycin (0.6 μg/mL for HT1080) for another 3 days before use. Single cells were isolated in a clear 96-well plate, and once single colonies were identified, they were scaled up to 12-well plates and plated in 10-cm dishes. Knockout was confirmed by western blot.
gRNA sequences were as follows:
Plasmid: pLentiCrispr v2 non-targeting gRNA: ATCTACGGGTTTATGCCAAT
Plasmid: pLentiCrispr v2-PARG gRNA2: TGCTATTCTGAAATACAATG
Plasmid: pLentiCrispr v2-PARG gRNA3: CAACACATTATAAAGATTTG
Molecular Analyses
Genomic and bisulfite-modified DNA was extracted using AllPrep DNA/RNA/miRNA Universal Kit and EpiTect Bisulfite kits (Qiagen) per the manufacturer’s protocol. Detailed protocols are described in the Supplementary Methods.
To assess MGMT promoter methylation status, methylation-specific PCR was performed in a two-step approach as described previously (8). MGMT protein expression was performed by western blot analysis. Tumors with mixed results (ie. Evidence of partially MGMT methylated MSP, but detectable expression of MGMT protein), were designed “unmethylated” to signify the presence of TMZ-resistant subclones.
Cell Viability Assay
Cells were seeded in 96-well plates at 1,000–3,000 cells per well. After a 4-h incubation, compounds were serially diluted and added to wells, as indicated in the figure legends. Cell viability was evaluated by Cell Titer-Glo (Promega) and CellTiter-Fluor Cell Viability Assay (Promega) according to the manufacturer’s protocol at the indicated time points. Cell viability was evaluated on a daily basis after drug exposure to determine the time course of treatment effects and was plotted as % cell viability relative to DMSO control.
Clonogenic Assay
To assess the clonogenicity of attached cells, 100 HT1080 and U251 cells were seeded on 6-well plates. Cells were exposed to DMSO, PDD00017273 (5 μM), TMZ (50–200 μM), or PDD00017273 plus TMZ for 8–14 days. After 1 × PBS washes, cells were fixed in 6% glutaraldehyde and stained with crystal violet for 30 min. Images were captured with the EVOS™ FL Auto 2 Imaging System (Thermo Fisher Scientific).
Sphere Formation Assay
To assess clonogenicity in gliosphere cells (MGG123, MGG152 and MGG119), 100 cells were seeded per well in clear 96-well plates. Cells were exposed to DMSO, PDD00017273 (5 μM), TMZ (100 μM), or PDD00017273 plus TMZ. Fresh medium was added every week. The sphere number was counted directly under a microscope at 14 days. To exclude bias, spheres were counted in a blinded manner by SR.
Cell Growth Assay
To test cell growth, 4,000 cells of HT1080 NT gRNA, PARG gRNA2, and PARG gRNA3, were seeded on 6-well plates. Cell numbers were counted 0, 2, and 5 days afterwards using a Luna-FL™ Automated Cell Counter (Logos Biosystems).
Apoptosis Assay
Caspase3/7 activities were evaluated with a Caspase-Glo 3/7 assay (Promega) according to the manufacturer’s recommendations. GBM tumor spheres and HT1080 cells were dissociated into single cells and seeded in 96-well plates at 1,000–3,000 cells/well. After 4 h, serially diluted DMSO, TMZ, and TMZ with or without PDD00017273 were added to the wells at the indicated concentrations. After 24–72 h, an equal volume of caspase 3/7 reagent was added to the samples and mixed. After a 30-min incubation, luminescence was recorded with a Synergy™ HT Multi-Mode Microplate Reader (BioTek).
NAD+ Quantitation
To obtain qualitative values of NAD+ and NADH, the NAD/NADH-Glo Assay (Promega) was used according to the manufacturer’s recommendations. Briefly, 1 × 105 cells were lysed with 200 μL of PBS and then 200 μL of 0.2 N NaOH with 1% dodecyltrimethylammonium bromide (DTAB; Sigma-Aldrich) was added. To measure NAD+, 50 μL of 0.4 N HCl was added to lysed cell samples (100 μL), which were then heated at 60°C for 15 min. After incubation at room temperature for 10 min, 0.5 M Trizma base buffer (50 μL; Sigma-Aldrich) was added. To measure NADH, lysed cell samples (100 μL) were heated at 60°C for 15 min and then incubated at room temperature for 10 min, followed by the addition of an equal volume of HCl/Trizma solution. Finally, samples were seeded in 96-well plates and incubated with NAD/NADH-Glo detection reagent for 60 min. Data were compared with DMSO-treated cells and expressed as % control.
To measure NAD+ quantitatively, NAD+/NADH Quantification Colorimetric Kit (BioVision Incorporated) was used according to the manufacturer’s recommendations. For each experiment, 2 × 105 cells, as indicated, were washed with cold PBS and extracted in NADH/NAD extraction buffer via 2 freeze/thaw cycles.
To measure NAD+ in tumor tissue, 4 × 106 HT1080/NT and PARG gRNA3 cells were subcutaneously transplanted into the left and right flank of athymic nude mice, respectively. Four days after transplantation, the mice were randomized to treatment with vehicle (n = 3) or TMZ (50 mg/kg, n = 3) given daily for 5 days. Six h after the last treatment, the mice were euthanized and the tumor tissues collected, washed with PBS and homogenized in NADH/NAD extraction buffer. Protein concentrations were measured using a BCA assay and used for normalization. After vortex extraction for 10 s, samples were centrifuged at 14,000 rpm for 5 min and the supernatant was transferred to a new tube. In order to detect total NADt (NADH and NAD), 50 μL of the extracted samples were transferred into a 96-well plate. To detect NADH, NAD+ was decomposed by heating at 60°C for 30 min. Then, 50 μL of NAD-decomposed samples were transferred into a 96-well plate. The NADH/NAD supernatant was transferred into a tube and NADt (NADH and NAD) and NADH signals were measured at OD 450 nm (BioTek). NAD+ concentrations were calculated by subtracting NADH from NADt. Data are expressed as pmol/1 × 106 cells or pmol/mg protein.
2-HG Quantitation
To obtain qualitative values of 2-HG, a D-2-Hydroxyglutarate (D2HG) Assay Kit (Sigma-Aldrich) was used according to the manufacturer’s recommendations. Detailed protocols are found in the Supplementary Methods.
Western Blot Analysis
Primary antibodies for blotting were as follows: IDH1 R132H (Dianova, #DIA-H09), NAPRT1 (Sigma-Aldrich, #HPA023739), PAR (Trevigen, #4336-BPC), Beta-Actin (Cell Signaling Technology, #3700), Cleaved Caspase-3 (Cell Signaling Technology, #9661), Cleaved PARP (Cell Signaling Technology, #5625), LC3B antibody (Cell Signaling Technology, #2775), MGMT(Cell Signaling Technology, #2739), MSH6 (Cell Signaling Technology, #5424), PARG (Cell Signaling Technology, #66564), PARP (Cell Signaling Technology, #9542), Phospho-Histone H2A.X (Cell Signaling Technology, #2577), and Vinculin (Thermo Fisher Scientific, # 700062). Detailed protocols are described in the Supplementary Methods.
Immunohistochemistry
Tumor tissue sections were incubated with anti-Ki-67 (1:200; Dako, # M7240) overnight at 4°C and then with secondary antibody (ImmPRESS HRP Horse Anti-Rabbit IgG [Peroxidase] Polymer Detection Kit; Vector Laboratories) for 30 min at room temperature. Three slides were stained per tumor. Six pictures were captured with 20X magnification per section and used for quantitative analysis of immunopositivity. Detailed protocols are described in the Supplementary Methods.
TUNEL assay
In situ detection of apoptotic cells was carried out using the TUNEL assay kit according to the manufacturer’s protocol (Millipore).
Three slides were stained per tumor. Six pictures were captured with 20X magnification per section and used for quantitative analysis of immunopositivity. Detailed protocols are described in the Supplementary Methods.
In Vivo Studies
All animal experiments were approved by the Institutional Animal Care and Use Committee of Massachusetts General Hospital (Boston, MA). Non-targeting (NT) gRNA HT1080 cells and PARG gRNA3 HT1080 cells (4 × 106 each) were subcutaneously transplanted into the left and right flank of 7- to 10-week-old female athymic nude mice (weight, 21–26 g; Charles River), respectively. Mice were housed under a 12-h light: dark cycle in single cages (21–24°C with 45% humidity) with standard bedding and enrichment and ad libitum access to food and water. When the maximum tumor diameter reached 5 mm, the mice were randomized to a vehicle group (PBS, i.p., n = 7) and a temozolomide group (TMZ; 50 mg/kg, i.p., n = 7). PBS with 10% DMSO was used as vehicle control. Treatments were administered 5 times a week for 2 weeks with a 1-week off period. A digital caliper was used to measure tumor diameters 3 times a week. The volume (mm3) was calculated as length (mm) × width (mm)2 × 0.5. Tumor volume was normalized to that of day 0.
To evaluate changes in NAD+ in tumor tissue, 4 × 106 HT1080/NT cells and HT1080/PARG gRNA3 cells were subcutaneously transplanted into the left and right flank of athymic nude mice, respectively. When the maximum tumor diameter reached 5 mm, the mice were randomized to treatment with vehicle (n = 3) or TMZ (50 mg/kg, n = 3). Treatment was administered 5 times daily. Six hours after the last treatment, the mice were sacrificed and tumor tissues collected. Tumor tissue was washed with PBS and homogenized in NADH/NAD extraction buffer, followed by NAD+ quantitation. For western blot analysis, a separate set of mice with bilateral flank tumors and the same vehicle or TMZ (50 mg/kg) treatment were used. Tumor tissues were collected 6 hours after the last treatment, washed with PBS and homogenized in RIPA buffer for tissue lysates. To examine apoptosis and cell proliferating in vivo, the same bilateral tumor model was set up with vehicle or TMZ (50 mg/kg) treatment in 5 consecutive days. Six hours after the last treatment, the mice were sacrificed and the tumor tissues collected, fixed in neutral-buffered formalin and embedded in paraffin, followed by immunohistochemistry.
Statistical Analysis
Prism 8 software (GraphPad) was used for all statistical analyses. Data are presented as mean ± standard error of the mean and were analyzed using a Student t-test (unpaired) for 2 group comparisons. Details of statistical analyses are provided in the figure legends. Key in vitro experiments were performed at least three times with consonant results. Key in vivo experiments were performed at least two times with consonant results. P values less than 0.05 were considered statistically significant.
Supplementary Material
Statement of Significance.
Oncogenic mutations in the isocitrate dehydrogenase (IDH) genes IDH1 or IDH2 initiate diffuse gliomas of younger adulthood. Strategies to maximize the effectiveness of chemotherapy in these tumors are needed. We discover alkylating chemotherapy and concurrent PARG inhibition exploits an intrinsic metabolic weakness within these cancer cells to provide genotype-specific benefit.
Acknowledgements
We thank the patients for their donation of critical research materials, and Donald Glazer, Fred Barker, and members of the Translational Neuro-Oncology Laboratory for helpful discussions and input.
Financial Support: This work was supported by the NIH R01CA227821 (HW/DPC), P50CA165962 (DPC), NCI Paul Calabresi Career Development Award in Clinical Oncology for Nervous System Tumors K12CA090354 (JJM). We also acknowledge the Tawingo Fund (DPC), Loglio Foundation (DPC), Richard B. Simches Scholars Award (JJM), a Seeman Family MGH Scholar in Neuro-Oncology Award (JJM), and the Overseas Research Fellowship of the Uehara Memorial Foundation (HN).
Footnotes
Conflict of Interest: All authors have no conflicts of interest to report with regard to this manuscript.
Disclosures: D.P.C. has received honoraria and travel reimbursement from Merck, and has served as a consultant for Lilly.
Declaration of Interest: All authors have no conflicts of interest to report with regard to this manuscript.
REFERENCES
- 1.Waitkus MS, Diplas BH, Yan H. Biological Role and Therapeutic Potential of IDH Mutations in Cancer. Cancer Cell 2018;34(2):186–95 doi 10.1016/j.ccell.2018.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller JJ, Shih HA, Andronesi OC, Cahill DP. Isocitrate dehydrogenase-mutant glioma: Evolving clinical and therapeutic implications. Cancer 2017;123(23):4535–46 doi 10.1002/cncr.31039. [DOI] [PubMed] [Google Scholar]
- 3.Louis DN OH, Wiestler OD, Cavenee WK (eds.). World Health Organization Histological Classification of Tumours of the Central Nervous System. France: International Agency for Research on Cancer; 2016. [Google Scholar]
- 4.Ostrom QT, Gittleman H, Xu J, Kromer C, Wolinsky Y, Kruchko C, et al. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2009–2013. Neuro Oncol 2016;18(suppl_5):v1–v75 doi 10.1093/neuonc/now207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cairncross JG, Wang M, Jenkins RB, Shaw EG, Giannini C, Brachman DG, et al. Benefit from procarbazine, lomustine, and vincristine in oligodendroglial tumors is associated with mutation of IDH. J Clin Oncol 2014;32(8):783–90 doi 10.1200/JCO.2013.49.3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buckner JC, Shaw EG, Pugh SL, Chakravarti A, Gilbert MR, Barger GR, et al. Radiation plus Procarbazine, CCNU, and Vincristine in Low-Grade Glioma. N Engl J Med 2016;374(14):1344–55 doi 10.1056/NEJMoa1500925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van den Bent MJ, Baumert B, Erridge SC, Vogelbaum MA, Nowak AK, Sanson M, et al. Interim results from the CATNON trial (EORTC study 26053–22054) of treatment with concurrent and adjuvant temozolomide for 1p/19q non-co-deleted anaplastic glioma: a phase 3, randomised, open-label intergroup study. Lancet 2017;390(10103):1645–53 doi 10.1016/S0140-6736(17)31442-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tateishi K, Wakimoto H, Iafrate AJ, Tanaka S, Loebel F, Lelic N, et al. Extreme Vulnerability of IDH1 Mutant Cancers to NAD+ Depletion. Cancer Cell 2015;28(6):773–84 doi 10.1016/j.ccell.2015.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seltzer MJ, Bennett BD, Joshi AD, Gao P, Thomas AG, Ferraris DV, et al. Inhibition of glutaminase preferentially slows growth of glioma cells with mutant IDH1. Cancer Res 2010;70(22):8981–7 doi 10.1158/0008–5472.CAN-10–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grassian AR, Parker SJ, Davidson SM, Divakaruni AS, Green CR, Zhang X, et al. IDH1 Mutations Alter Citric Acid Cycle Metabolism and Increase Dependence on Oxidative Mitochondrial Metabolism. Cancer Res 2014. doi 10.1158/0008-5472.CAN-14-0772-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reitman ZJ, Duncan CG, Poteet E, Winters A, Yan LJ, Gooden DM, et al. Cancer-associated isocitrate dehydrogenase 1 (IDH1) R132H mutation and d-2-hydroxyglutarate stimulate glutamine metabolism under hypoxia. J Biol Chem 2014;289(34):23318–28 doi 10.1074/jbc.M114.575183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karpel-Massler G, Ishida CT, Bianchetti E, Zhang Y, Shu C, Tsujiuchi T, et al. Induction of synthetic lethality in IDH1-mutated gliomas through inhibition of Bcl-xL. Nat Commun 2017;8(1):1067 doi 10.1038/s41467-017-00984-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McBrayer SK, Mayers JR, DiNatale GJ, Shi DD, Khanal J, Chakraborty AA, et al. Transaminase Inhibition by 2-Hydroxyglutarate Impairs Glutamate Biosynthesis and Redox Homeostasis in Glioma. Cell 2018. doi doi.org/10.1016/j.cell.2018.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Izquierdo-Garcia JL, Viswanath P, Eriksson P, Cai L, Radoul M, Chaumeil MM, et al. IDH1 Mutation Induces Reprogramming of Pyruvate Metabolism. Cancer Res 2015;75(15):2999–3009 doi 10.1158/0008-5472.CAN-15-0840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohba S, Mukherjee J, See WL, Pieper RO. Mutant IDH1-driven cellular transformation increases RAD51-mediated homologous recombination and temozolomide resistance. Cancer Res 2014;74(17):4836–44 doi 10.1158/0008-5472.CAN-14-0924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sulkowski PL, Corso CD, Robinson ND, Scanlon SE, Purshouse KR, Bai H, et al. 2-Hydroxyglutarate produced by neomorphic IDH mutations suppresses homologous recombination and induces PARP inhibitor sensitivity. Science translational medicine 2017;9(375) doi 10.1126/scitranslmed.aal2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu Y, Kwintkiewicz J, Liu Y, Tech K, Frady LN, Su YT, et al. Chemosensitivity of IDH1-Mutated Gliomas Due to an Impairment in PARP1-Mediated DNA Repair. Cancer Res 2017;77(7):1709–18 doi 10.1158/0008-5472.CAN-16-2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nunez FJ, Mendez FM, Kadiyala P, Alghamri MS, Savelieff MG, Garcia-Fabiani MB, et al. IDH1-R132H acts as a tumor suppressor in glioma via epigenetic up-regulation of the DNA damage response. Science translational medicine 2019;11(479) doi 10.1126/scitranslmed.aaq1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pommier Y, O’Connor MJ, de Bono J. Laying a trap to kill cancer cells: PARP inhibitors and their mechanisms of action. Science translational medicine 2016;8(362):362ps17 doi 10.1126/scitranslmed.aaf9246. [DOI] [PubMed] [Google Scholar]
- 20.Zong WX, Ditsworth D, Bauer DE, Wang ZQ, Thompson CB. Alkylating DNA damage stimulates a regulated form of necrotic cell death. Genes & development 2004;18(11):1272–82 doi 10.1101/gad.1199904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Curtin NJ. PARP inhibitors for cancer therapy. Expert reviews in molecular medicine 2005;7(4):1–20. [DOI] [PubMed] [Google Scholar]
- 22.Trivedi RN, Almeida KH, Fornsaglio JL, Schamus S, Sobol RW. The role of base excision repair in the sensitivity and resistance to temozolomide-mediated cell death. Cancer Research 2005;65(14):6394–400. [DOI] [PubMed] [Google Scholar]
- 23.Irshad S, Ashworth A, Tutt A. Therapeutic potential of PARP inhibitors for metastatic breast cancer. Expert Rev Anticancer Ther 2011;11(8):1243–51 doi 10.1586/era.11.52. [DOI] [PubMed] [Google Scholar]
- 24.Goellner EM, Grimme B, Brown AR, Lin YC, Wang XH, Sugrue KF, et al. Overcoming temozolomide resistance in glioblastoma via dual inhibition of NAD+ biosynthesis and base excision repair. Cancer Res 2011;71(6):2308–17 doi 10.1158/0008-5472.CAN-10-3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tateishi K, Higuchi F, Miller JJ, Koerner MVA, Lelic N, Shankar GM, et al. The Alkylating Chemotherapeutic Temozolomide Induces Metabolic Stress in IDH1-Mutant Cancers and Potentiates NAD(+) Depletion-Mediated Cytotoxicity. Cancer Res 2017;77(15):4102–15 doi 10.1158/0008-5472.CAN-16-2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wright RH, Lioutas A, Le Dily F, Soronellas D, Pohl A, Bonet J, et al. ADP-ribose-derived nuclear ATP synthesis by NUDIX5 is required for chromatin remodeling. Science 2016;352(6290):1221–5 doi 10.1126/science.aad9335. [DOI] [PubMed] [Google Scholar]
- 27.Shirai H, Poetsch AR, Gunji A, Maeda D, Fujimori H, Fujihara H, et al. PARG dysfunction enhances DNA double strand break formation in S-phase after alkylation DNA damage and augments different cell death pathways. Cell Death Dis 2013;4:e656 doi 10.1038/cddis.2013.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ning JF, Stanciu M, Humphrey MR, Gorham J, Wakimoto H, Nishihara R, et al. Myc targeted CDK18 promotes ATR and homologous recombination to mediate PARP inhibitor resistance in glioblastoma. Nat Commun 2019;10(1):2910 doi 10.1038/s41467-019-10993-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Higuchi F, Nagashima H, Ning J, Koerner MVA, Wakimoto H, Cahill DP. Restoration of Temozolomide Sensitivity by Poly(ADP-Ribose) Polymerase inhibitors in Mismatch Repair Deficient Glioblastoma is Independent of Base Excision Repair. Clin Cancer Res 2020. doi 10.1158/1078-0432.CCR-19-2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hwang ES, Song SB. Nicotinamide is an inhibitor of SIRT1 in vitro, but can be a stimulator in cells. Cell Mol Life Sci 2017;74(18):3347–62 doi 10.1007/s00018-017-2527-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dominguez-Gomez G, Diaz-Chavez J, Chavez-Blanco A, Gonzalez-Fierro A, Jimenez-Salazar JE, Damian-Matsumura P, et al. Nicotinamide sensitizes human breast cancer cells to the cytotoxic effects of radiation and cisplatin. Oncol Rep 2015;33(2):721–8 doi 10.3892/or.2014.3661. [DOI] [PubMed] [Google Scholar]
- 32.Fu D, Calvo JA, Samson LD. Balancing repair and tolerance of DNA damage caused by alkylating agents. Nat Rev Cancer 2012;12(2):104–20 doi 10.1038/nrc3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chowdhry S, Zanca C, Rajkumar U, Koga T, Diao Y, Raviram R, et al. NAD metabolic dependency in cancer is shaped by gene amplification and enhancer remodelling. Nature 2019;569(7757):570–5 doi 10.1038/s41586-019-1150-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turcan S, Makarov V, Taranda J, Wang Y, Fabius AWM, Wu W, et al. Mutant-IDH1-dependent chromatin state reprogramming, reversibility, and persistence. Nat Genet 2018;50(1):62–72 doi 10.1038/s41588-017-0001-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Branch P, Aquilina G, Bignami M, Karran P. Defective Mismatch Binding and a Mutator Phenotype in Cells Tolerant to DNA Damage. Nature 1993;362(6421):652–4. [DOI] [PubMed] [Google Scholar]
- 36.Cahill DP, Levine KK, Betensky RA, Codd PJ, Romany CA, Reavie LB, et al. Loss of the mismatch repair protein MSH6 in human glioblastomas is associated with tumor progression during temozolomide treatment. Clin Cancer Res 2007;13(7):2038–45 doi 13/7/2038 [pii] 10.1158/1078-0432.CCR-06-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choi S, Yu Y, Grimmer MR, Wahl M, Chang SM, Costello JF. Temozolomide-associated hypermutation in gliomas. Neuro Oncol 2018;20(10):1300–9 doi 10.1093/neuonc/noy016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson BE, Mazor T, Hong C, Barnes M, Aihara K, McLean CY, et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science 2014;343(6167):189–93 doi 10.1126/science.1239947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barthel FP, Johnson KC, Varn FS, Moskalik AD, Tanner G, Kocakavuk E, et al. Longitudinal molecular trajectories of diffuse glioma in adults. Nature 2019;576(7785):112–20 doi 10.1038/s41586-019-1775-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jain A, Agostini LC, McCarthy GA, Chand SN, Ramirez A, Nevler A, et al. Poly (ADP) Ribose Glycohydrolase Can Be Effectively Targeted in Pancreatic Cancer. Cancer Res 2019;79(17):4491–502 doi 10.1158/0008-5472.CAN-18-3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marques M, Jangal M, Wang LC, Kazanets A, da Silva SD, Zhao T, et al. Oncogenic activity of poly (ADP-ribose) glycohydrolase. Oncogene 2019;38(12):2177–91 doi 10.1038/s41388-018-0568-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller JJ, Loebel F, Juratli TA, Tummala SS, Williams EA, Batchelor TT, et al. Accelerated progression of IDH mutant glioma after first recurrence. Neuro Oncol 2019;21(5):669–77 doi 10.1093/neuonc/noz016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wakimoto H, Tanaka S, Curry WT, Loebel F, Zhao D, Tateishi K, et al. Targetable Signaling Pathway Mutations Are Associated with Malignant Phenotype in IDH-Mutant Gliomas. Clin Cancer Res 2014;20(11):2898–909 doi 10.1158/1078-0432.CCR-13-3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tateishi K, Nakamura T, Juratli TA, Williams EA, Matsushita Y, Miyake S, et al. PI3K/AKT/mTOR Pathway Alterations Promote Malignant Progression and Xenograft Formation in Oligodendroglial Tumors. Clin Cancer Res 2019;25(14):4375–87 doi 10.1158/1078-0432.CCR-18-4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hunter C, Smith R, Cahill DP, Stephens P, Stevens C, Teague J, et al. A hypermutation phenotype and somatic MSH6 mutations in recurrent human malignant gliomas after alkylator chemotherapy. Cancer Res 2006;66(8):3987–91 doi 66/8/3987 [pii] 10.1158/0008-5472.CAN-06-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tateishi K, Iafrate AJ, Ho Q, Curry WT, Batchelor TT, Flaherty KT, et al. Myc-Driven Glycolysis Is a Therapeutic Target in Glioblastoma. Clin Cancer Res 2016;22(17):4452–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fons NR, Sundaram RK, Breuer GA, Peng S, McLean RL, Kalathil AN, et al. PPM1D mutations silence NAPRT gene expression and confer NAMPT inhibitor sensitivity in glioma. Nat Commun 2019;10(1):3790 doi 10.1038/s41467-019-11732-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.James DI, Smith KM, Jordan AM, Fairweather EE, Griffiths LA, Hamilton NS, et al. First-in-Class Chemical Probes against Poly(ADP-ribose) Glycohydrolase (PARG) Inhibit DNA Repair with Differential Pharmacology to Olaparib. ACS Chem Biol 2016;11(11):3179–90 doi 10.1021/acschembio.6b00609. [DOI] [PubMed] [Google Scholar]
- 49.Gogola E, Duarte AA, de Ruiter JR, Wiegant WW, Schmid JA, de Bruijn R, et al. Selective Loss of PARG Restores PARylation and Counteracts PARP Inhibitor-Mediated Synthetic Lethality. Cancer Cell 2018;33(6):1078–93 e12 doi 10.1016/j.ccell.2018.05.008. [DOI] [PubMed] [Google Scholar]
- 50.O’Sullivan J, Tedim Ferreira M, Gagne JP, Sharma AK, Hendzel MJ, Masson JY, et al. Emerging roles of eraser enzymes in the dynamic control of protein ADP-ribosylation. Nat Commun 2019;10(1):1182 doi 10.1038/s41467-019-08859-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen SH, Yu X. Targeting dePARylation selectively suppresses DNA repair-defective and PARP inhibitor-resistant malignancies. Sci Adv 2019;5(4):eaav4340 doi 10.1126/sciadv.aav4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gravells P, Grant E, Smith KM, James DI, Bryant HE. Specific killing of DNA damage-response deficient cells with inhibitors of poly(ADP-ribose) glycohydrolase. DNA Repair (Amst) 2017;52:81–91 doi 10.1016/j.dnarep.2017.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pillay N, Tighe A, Nelson L, Littler S, Coulson-Gilmer C, Bah N, et al. DNA Replication Vulnerabilities Render Ovarian Cancer Cells Sensitive to Poly(ADP-Ribose) Glycohydrolase Inhibitors. Cancer Cell 2019;35(3):519–33 e8 doi 10.1016/j.ccell.2019.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Andrabi SA, Umanah GK, Chang C, Stevens DA, Karuppagounder SS, Gagne JP, et al. Poly(ADP-ribose) polymerase-dependent energy depletion occurs through inhibition of glycolysis. Proc Natl Acad Sci U S A 2014;111(28):10209–14 doi 10.1073/pnas.1405158111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fouquerel E, Goellner EM, Yu Z, Gagne JP, Barbi de Moura M, Feinstein T, et al. ARTD1/PARP1 negatively regulates glycolysis by inhibiting hexokinase 1 independent of NAD+ depletion. Cell Rep 2014;8(6):1819–31 doi 10.1016/j.celrep.2014.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wakimoto H, Kesari S, Farrell CJ, Curry WT Jr., Zaupa C, Aghi M, et al. Human glioblastoma-derived cancer stem cells: establishment of invasive glioma models and treatment with oncolytic herpes simplex virus vectors. Cancer Res 2009;69(8):3472–81 doi 10.1158/0008-5472.CAN-08-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wakimoto H, Mohapatra G, Kanai R, Curry WT Jr., Yip S, Nitta M, et al. Maintenance of primary tumor phenotype and genotype in glioblastoma stem cells. Neuro Oncol 2012;14(2):132–44 doi 10.1093/neuonc/nor195. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.