We establish a new pathway of sulfide oxidation with thiosulfate as a key intermediate in Cupriavidus pinatubonensis JMP134. The bacterium mainly oxidizes sulfide by using sulfide:quinone oxidoreductase, persulfide dioxygenase, and the Sox system with thiosulfate as a key intermediate. Although the purified and reconstituted Sox system oxidizes sulfide, its rate of sulfide oxidation in C. pinatubonensis JMP134 is too low to be physiologically relevant. The findings reveal how these sulfur-oxidizing enzymes participate in sulfide oxidation in a single bacterium.
KEYWORDS: sulfide, sulfane sulfur, thiosulfate, sulfate, heterotrophic bacteria, sulfur oxidation pathway, sulfate reduction
ABSTRACT
Heterotrophic bacteria actively participate in the biogeochemical cycle of sulfur on Earth. The heterotrophic bacterium Cupriavidus pinatubonensis JMP134 contains several enzymes involved in sulfur oxidation, but how these enzymes work together to oxidize sulfide in the bacterium has not been studied. Using gene-deletion and whole-cell assays, we determined that the bacterium uses sulfide:quinone oxidoreductase to oxidize sulfide to polysulfide, which is further oxidized to sulfite by persulfide dioxygenase. Sulfite spontaneously reacts with polysulfide to produce thiosulfate. The sulfur-oxidizing (Sox) system oxidizes thiosulfate to sulfate. Flavocytochrome c sulfide dehydrogenase enhances thiosulfate oxidation by the Sox system but couples with the Sox system for sulfide oxidation to sulfate in the absence of sulfide:quinone oxidoreductase. Thus, C. pinatubonensis JMP134 contains a main pathway and a contingent pathway for sulfide oxidation.
IMPORTANCE We establish a new pathway of sulfide oxidation with thiosulfate as a key intermediate in Cupriavidus pinatubonensis JMP134. The bacterium mainly oxidizes sulfide by using sulfide:quinone oxidoreductase, persulfide dioxygenase, and the Sox system with thiosulfate as a key intermediate. Although the purified and reconstituted Sox system oxidizes sulfide, its rate of sulfide oxidation in C. pinatubonensis JMP134 is too low to be physiologically relevant. The findings reveal how these sulfur-oxidizing enzymes participate in sulfide oxidation in a single bacterium.
INTRODUCTION
Sulfur oxidation is a key step in the biogeochemical cycling of sulfur on Earth. Under anoxic conditions, sulfur-reducing bacteria use sulfate as the terminal electron acceptor for the oxidation of organic compounds, generating hydrogen sulfide (H2S), which can be oxidized back to sulfate by chemolithoautotrophic bacteria under oxic conditions or used as the reducing power for photosynthesis by phototrophic bacteria under anoxic conditions (1, 2). Further, many heterotrophic bacteria are also capable of oxidizing H2S (3, 4). H2S is a common product of the microbial metabolism of l-cysteine, and heterotrophic bacteria carrying sulfide:quinone oxidoreductase (SQR) and persulfide dioxygenase (PDO) oxidize self-produced H2S or exogenously introduced H2S to sulfite and thiosulfate (3, 5, 6). Pseudomonas putida oxidizes thiosulfate to tetrathionate (7, 8), Hyphomicrobium denitrificans oxidizes thiosulfate to tetrathionate and sulfate (9), and other bacteria, including bacteria isolated from marine sediments and hydrothermal vents, oxidize thiosulfate to sulfate (10). These reports suggest that different heterotrophic bacteria may work together to oxidize sulfide to sulfate.
The soil bacterium Cupriavidus pinatubonensis (formerly Ralstonia eutropha and Alcaligenes eutrophus) JMP134 has been widely used to study the microbial degradation of aromatic compounds (11, 12). It also oxidizes H2S during normal growth on organic compounds (3), offering an opportunity to investigate how the heterotrophic bacterium oxidizes H2S. When its SQR and PDO genes are introduced into Escherichia coli, the recombinant cells oxidize H2S to sulfite with polysulfide as an intermediate, and the sulfite spontaneously reacts with polysulfide to produce thiosulfate (5). From genome sequence analysis, C. pinatubonensis JMP134 also contains genes coding for the flavocytochrome c sulfide dehydrogenase (FCSD) system, the sulfur-oxidizing (Sox) system, and sulfite:cytochrome c oxidoreductase (SOR).
The FCSD system consists of a flavin adenine dinucleotide-containing protein (FccB) and a cytochrome c (FccA), which are soluble proteins in the periplasmic space. FccB and FccA are also called SoxF and SoxE, respectively, in chemolithoautotrophic bacteria, as their genes are often clustered with other sox genes (13). FCSD uses cytochrome c as the electronic acceptor, while SQR uses ubiquinone. FCSD was first identified in the purple photosynthetic bacterium Allochromatium vinosum (14), and it has subsequently been found to be widely present in autotrophic bacteria (15) and in heterotrophic bacteria (16). FCSD from Cupriavidus necator H16 oxidizes sulfide to polysulfide, which is further oxidized by PDO to sulfite and thiosulfate when both are cloned in Pseudomonas aeruginosa (16). Since C. pinatubonensis JMP134 contains both SQR and FCSD, it is unclear which one is preferentially used to oxidize H2S.
The Sox system is a versatile enzyme system for the oxidation of reduced sulfur species, and the Sox proteins are soluble in the periplasmic space (17). The most extensively studied Sox system is from Paracoccus pantotrophus GB17 (ATCC 35512T or DSM 2944), a facultative chemolithotroph able to growth on thiosulfate (17). The core enzyme of the Sox system consists of four enzymes, SoxYZ (a heterodimer of SoxY and SoxZ), SoxXA, SoxCD, and SoxB, encoded by seven sox genes. How the Sox system oxidizes thiosulfate has been characterized. SoxYZ is a carrier protein with the sulfur molecule to be oxidized being covalently linked to a conserved cysteine residue of SoxY (18, 19). SoxXA catalyzes the loading of thiosulfate to SoxYZ-SH, producing SoxYZ-S-thiosulfate (20). SoxB hydrolytically removes the terminal sulfonate group, producing SoxYZ-SSH and sulfate (21). SoxYZ-SSH is oxidized by SoxCD to SoxYZ-S-SO3− (22, 23), and the sulfonate group is again released by SoxB. The electrons from sulfur oxidation enter the electron transfer chain via a cytochrome c-type cytochrome (24). The purified and reconstituted Sox system also oxidizes sulfide, elemental sulfur, and sulfite to sulfate (24), but it has not been confirmed whether P. pantotrophus or any other bacteria use the Sox system to oxidize these sulfur species.
SOR consists of two proteins, SorA and SorB. SorA is a large protein containing molybdopterin, and SorB is a small protein containing cytochrome c. SOR, located in the periplasmic space, oxidizes sulfite to sulfate (25). It is unknown if SOR oxidizes the sulfite generated by PDO to sulfate in C. pinatubonensis JMP134.
In this study, we investigated how C. pinatubonensis JMP134 uses SQR/PDO, FCSD, the Sox system, and SOR to oxidize sulfide to sulfate. On the basis of genomic analysis and our experimental data, we identified a new pathway of H2S oxidation in which SQR and PDO collectively oxidize sulfide to thiosulfate and then the Sox system oxidizes thiosulfate to sulfate. FCSD’s main function was to enhance thiosulfate oxidation by the Sox system; however, FCSD oxidized H2S to zero-valent sulfur, part of which was directly oxidized by the Sox system when sqr was deleted in C. pinatubonensis JMP134.
RESULTS
Sulfur-oxidizing genes in C. pinatubonensis JMP134.
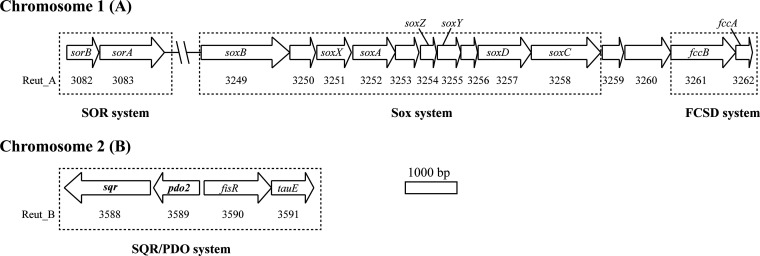
The genome of C. pinatubonensis JMP134 consists of two chromosomes, chromosome A (3.8 Mb) and chromosome B (2.72 Mb), and two large plasmids (11). The sqr-pdo2 gene cluster, located on chromosome B (Fig. 1; see also Table S1 in the supplemental material), codes for four possible proteins, SQR, PDO, a possible sulfite exporter protein (TauE), and a gene regulator (FisR) (3, 26). Another pdo gene (pdo1) is located on chromosome A (Table S1) and encodes a type I PDO (27). The sox genes on chromosome A consist of seven sox genes (soxB, soxX, soxA, soxZ, soxY, soxD, soxC), encoding the core components of the Sox system (Fig. 1; Table S1). Downstream (locus tags, Reut_A3261 and Reut_A3262) of the sox genes are soxF and soxE, which are also known as the fccB and fccA genes, respectively, coding for FccB and FccA of the FCSD system. The sequence similarities of FccB and FccA in C. pinatubonensis JMP134 to FccB (GenBank accession number CAJ94633.1) and FccA (GenBank accession number CAJ94634.1) in C. necator H16 were 85% (query cover, 99%) and 79% (query cover, 84%), respectively. In C. necator H16, which does not have SQR, the FCSD system oxidizes H2S (16). SOR has two subunits, SorA and SorB, whose genes are also located on chromosome A (Fig. 1; Table S1). Genes coding for thiosulfate dehydrogenases, oxidizing thiosulfate to tetrathionate (28, 29), and tetrathionate hydrolase, converting tetrathionate to sulfate, sulfur, and thiosulfate (30), were not found in C. pinatubonensis JMP134. Genes involved in dissimilatory sulfur reduction, such as qmoABC, aprAB, and sat (31), were not found in the bacterium. The gene coding for a ribulose bisphosphate carboxylase, necessary for carbon fixation and chemolithoautotrophic growth, was not found in the bacterium either.
FIG 1.
Schematic overview of the sulfur-oxidizing genes in C. pinatubonensis JMP134. The genome of C. pinatubonensis JMP134 includes two chromosomes, A and B. Genes encoding the SOR, Sox, and FCSD systems are located on chromosome A, and the sqr-pdo operon is on chromosome B. The SOR system is encoded by sorA (GenBank accession number AAZ62443.1) and sorB (GenBank accession number AAZ62442.1). The Sox system genes include soxB (GenBank accession number AAZ62608.1), soxX (GenBank accession number AAZ62610.1), soxA (GenBank accession number AAZ62611.1), soxZ (GenBank accession number AAZ62613.1), soxY (GenBank accession number AAZ62614.1), soxD (GenBank accession number AAZ62616.1), and soxC (GenBank accession number AAZ62617.1). The FCSD system is encoded by fccB (GenBank accession number AAZ62620.1) and fccA (a possible cytochrome c; GenBank accession number AAZ62621.1). An operon coding for the SQR/PDO system genes consists of sqr (GenBank accession number AAZ62946.1), pdo2 (GenBank accession number AAZ62947.1), fisR (GenBank accession number AAZ62948.1), and tauE (GenBank accession number AAZ62949.1). The locus tag of each gene is given below the gene (e.g., Reut_A3252 is the tag for soxA).
Deletion of genes involved in sulfur oxidation.
Selected genes were deleted via homologous recombination by using a suicide plasmid carrying the DNA fragments before and after the gene. All the deletions were in frame, with about 0 to 10 amino acid residues at the N terminus and about 0 to 36 residues at the C terminus remaining in the mutants to avoid affecting downstream and upstream genes. All mutants grew similarly to the wild type when growing in Luria-Bertani (LB) medium or in a mineral medium (MM) with sulfate, sulfite, or thiosulfate as the sole sulfur source (data not shown), suggesting that these genes associated with sulfur oxidation are not essential for the bacterium during heterotrophic growth.
SQR was the primary enzyme responsible for sulfide oxidation.
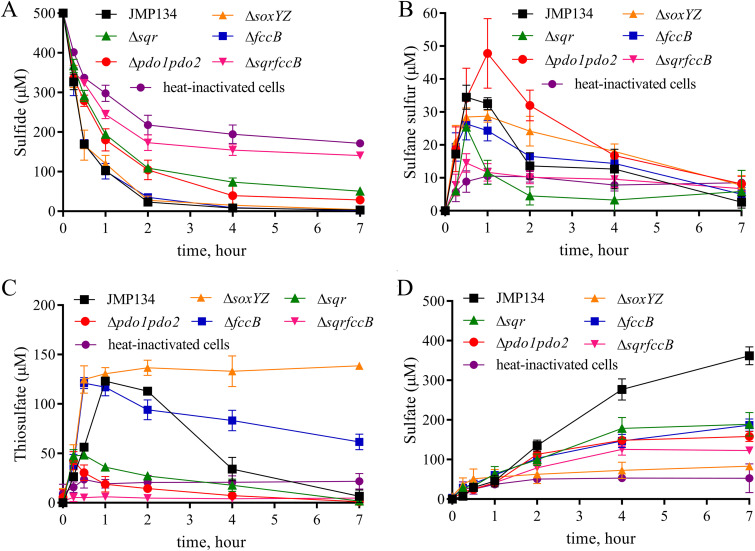
To investigate whether SQR, FCSD, or the Sox system was mainly responsible for sulfide oxidation in C. pinatubonensis JMP134, we tested the rate of sulfide oxidation by sulfide-induced cells of the wild type and the sqr, fccB, or soxY-soxZ deletion mutants. The ΔsoxYZ mutant oxidized sulfide at a rate similar to that for the wild type (Table 1; Fig. 2), the ΔfccB, Δsqr, Δsqr ΔfccB, and Δsqr ΔfccB ΔsoxYZ strains oxidized sulfide at 98%, 27%, 11%, and 8% of the wild-type rate, respectively (Table 1; Fig. 2). The slow sulfide oxidation by the Δsqr ΔfccB ΔsoxYZ mutant is likely due to nonspecific oxidation by other enzymes, as superoxide dismutase and catalase are able to oxidize sulfide (32, 33). The reduced rates of sulfide oxidation in the mutants were recovered when the deleted genes were complemented in trans on a plasmid (Table 1; Fig. S1). These results indicate that for sulfide oxidation, SQR is the main enzyme, FCSD plays a supplemental role, and the Sox system is only marginally active.
TABLE 1.
Rates of sulfur oxidation by JMP134 and its mutant cellsa
| Strain | Oxidation rate (nmol/mg/min)b
|
||
|---|---|---|---|
| HS− | HSn− | S2O32− | |
| JMP134 | 6.2 ± 0.1 | 11.8 ± 0.4 | 1.7 ± 0.1 |
| Δsqr | 1.7 ± 0.4** | — | — |
| ΔfccB | 6.1 ± 0.2 | — | 0.4 ± 0.1* |
| ΔfccB/fccB | 6.1 ± 0.5 | — | 0.9 ± 0.1* |
| Δsqr Δpdo1 Δpdo2 | 1.2 ± 0.2** | — | — |
| Δsqr Δpdo1 Δpdo2/sqr pdo2 | 6.1 ± 1.0 | — | — |
| Δsqr Δpdo1 Δpdo2 ΔsoxYZ | 1.6 ± 0.7** | — | — |
| Δsqr ΔfccB | 0.7 ± 0.1** | — | — |
| Δsqr ΔfccB ΔsoxYZ | 0.5 ± 0.4** | — | — |
| ΔsoxYZ | 6.3 ± 1.0 | 10.8 ± 1.3 | 0.2 ± 0.1* |
| ΔsoxYZ/soxYZ | 6.4 ± 0.2 | — | 1.2 ± 0.3 |
| Δpdo1 | 6.1 ± 0.5 | 11.0 ± 0.6 | — |
| Δpdo2 | 3.5 ± 0.2** | 2.6 ± 0.5** | — |
| Δpdo1 Δpdo2 | 2.2 ± 0.4** | 1.3 ± 0.2** | — |
| Δpdo1 Δpdo2/pdo2 | 5.7 ± 0.2 | 8.1 ± 0.4* | — |
| Δpdo1 Δpdo2 ΔsoxYZ | 1.9 ± 0.3** | 0.7 ± 0.1** | — |
| ΔsorA | 6.0 ± 0.9 | — | — |
| ΔtauE | 6.1 ± 0.2 | — | — |
| Buffer | 0.6 ± 0.3** | — | — |
Cells were induced, harvested, and resuspended in 100 mM HEPES buffer (pH 7.4) at an OD of 2. The cell suspensions were used to oxidize 500 μM sulfide, 800 μM polysulfide, or 600 μM thiosulfate at 30°C. The induction was with the corresponding substrate before harvesting.
For HS− and HSn−, the rates were calculated with data obtained at 0.5 h by using the cell dry weight. For S2O32−, the rates were calculated with data obtained at 2 h by using the cell dry weight. When calculating all rates, the rate for the heat-inactivated cell as a control was deducted. The rates of sulfide, polysulfide (HSn−), and thiosulfate oxidation of heat-inactivated cells were 6.0 ± 0.1, 0.7 ± 0.1, and 0 nmol/mg/min, respectively. —, not measured; *, the result for the mutant was different (P < 0.05) from that for the wild type; **, the result for the mutant was significantly different (P < 0.01) from that for the wild type. The data are the averages for at least three samples with standard deviations.
FIG 2.
The function of SQR, PDO, the Sox system, and FccAB in C. pinatubonensis JMP134. Cells were harvested, washed, and resuspended in 100 mM HEPES buffer, pH 7.4, at an OD600 of 2.0. Sulfide was added to 500 μM to initiate the reaction. Sulfide (A), sulfane sulfur (B), thiosulfate (C), and sulfate (D) concentrations were determined. There was no apparent difference in the decrease in sulfide levels in HEPES buffer with or without heat-inactivated cells, but the heat-inactivated cells produced more sulfate (53 ± 13 μM) than the buffer (5 ± 3 μM) at 7 h (see Fig. S1 in the supplemental material). The apparent decrease in the buffer with heat-inactivated cells was likely due to evaporation via shaking and autoxidation. All data are averages for at least three samples with standard deviations (error bars).
C. pinatubonensis JMP134 oxidizes sulfide to sulfate with sulfane sulfur and thiosulfate as detectable intermediates.
To understand the pathway, we monitored the intermediates and products during sulfide oxidation by C. pinatubonensis JMP134. When 500 μM NaHS was added to a suspension of sulfide-induced C. pinatubonensis JMP134 cells at an optical density at 600 nm (OD600) of 2, about 100 μM sulfide remained at 1 h (Fig. 2A) and the cells produced about 32 μM sulfane sulfur (S0) (including polysulfide and elemental sulfur), 120 μM thiosulfate, and 45 μM sulfate (Fig. 2B to D). Sulfane and thiosulfate were further consumed and gradually decreased to almost zero (Fig. 2B and C), and the cell suspension produced 362 μM sulfate at 7 h (Table 2; Fig. 2D). Sulfite was not detected during sulfide oxidation by the bacterium. The rate of sulfide oxidation was lower than that of polysulfide oxidation but higher than that of thiosulfate oxidation by C. pinatubonensis JMP134 (Table 1), reflecting the increased accumulation of thiosulfate during sulfide oxidation (Fig. 2).
TABLE 2.
Substrates and products at 7 h of sulfide oxidation by whole cellsa
| Strain | Concn (μM) |
Recovery of sulfur (%) | |||
|---|---|---|---|---|---|
| HS− | Sulfane sulfur | S2O32− | SO42− | ||
| JMP134 | 3 ± 1 | 3 ± 1 | 7 ± 5 | 362 ± 16 | 76 |
| Δsqr | 51 ± 6 | 6 ± 5 | 2 ± 0 | 189 ± 21 | 44 |
| ΔfccB | 2 ± 2 | 5 ± 2 | 62 ± 6 | 187 ± 11 | 63 |
| ΔfccB/fccB | 4 ± 1 | 2 ± 1 | 1 ± 0 | 252 ± 17 | 52 |
| Δsqr Δpdo1 Δpdo2 | 92 ± 8 | 6 ± 3 | 1 ± 0 | 200 ± 11 | 51 |
| Δsqr Δpdo1 Δpdo2/sqr pdo2 | 5 ± 2 | 3 ± 1 | 0 ± 0 | 436 ± 38 | 89 |
| Δsqr Δpdo1 Δpdo2 ΔsoxYZ | 100 ± 5 | 24 ± 1 | 40 ± 1 | 33 ± 2 | 34 |
| Δsqr ΔfccB | 141 ± 5 | 7 ± 1 | 5 ± 2 | 123 ± 4 | 39 |
| Δsqr ΔfccB ΔsoxYZ | 143 ± 15 | 10 ± 1 | 69 ± 3 | 28 ± 1 | 49 |
| ΔsoxYZ | 4 ± 2 | 8 ± 2 | 139 ± 2 | 83 ± 6 | 74 |
| ΔsoxYZ/soxYZ | 20 ± 4 | 3 ± 3 | 1 ± 0 | 448 ± 17 | 94 |
| Δpdo1 Δpdo2 | 29 ± 7 | 8 ± 2 | 1 ± 0 | 158 ± 9 | 36 |
| Δpdo1 Δpdo2/pdo2 | 7 ± 2 | 3 ± 1 | 8 ± 4 | 327 ± 18 | 70 |
| Δpdo1 Δpdo2 ΔsoxYZ | 65 ± 9 | 4 ± 1 | 38 ± 4 | 30 ± 1 | 25 |
| ΔsorA | 3 ± 1 | 3 ± 0 | 0 ± 0 | 357 ± 32 | 72 |
| ΔtauE | 7 ± 1 | 5 ± 3 | 7 ± 4 | 363 ± 5 | 77 |
| Buffer | 176 ± 10 | 3 ± 2 | 16 ± 5 | 5 ± 3 | 12 |
| Heat-inactivated cell | 172 ± 7 | 8 ± 2 | 22 ± 6 | 53 ± 13 | 32 |
The oxidation of 500 μM sulfide by cell suspensions at an OD of 2 in 100 mM HEPES buffer, pH 7.4, at 30°C was used. Sulfur compounds were analyzed at 7 h. All data are the averages for at least three samples with standard deviations.
SQR, PDO, and the Sox system sequentially oxidized sulfide to sulfate.
Whether C. pinatubonensis JMP134 used PDO or the Sox system to oxidize the sulfane sulfur produced by SQR was further investigated by the construction of mutants with deletions of the corresponding genes. The deletion of pdo1 did not affect sulfide oxidation, but the deletion of pdo2 significantly slowed sulfide oxidation (Table 1). The Δpdo1 Δpdo2 mutant showed a further reduction in the sulfide oxidation rate (Table 1; Fig. 2A), with the elevated production of sulfane sulfur, the decreased production of thiosulfate, as well as the decreased production of sulfate (Table 2; Fig. 2B to D). This observation implies product inhibition: SQR oxidizes sulfide to sulfane sulfur, which accumulates and inhibits SQR activity in the absence of PDO, which oxidizes sulfane sulfur (5). The resting state of cells of the ΔsoxYZ mutant did not affect sulfide oxidation or the transitory accumulation of sulfane sulfur but largely abolished thiosulfate oxidation and sulfate production (Table 2; Fig. 2A to D); complementation of soxYZ partially restored thiosulfate oxidation and sulfate production (Table 2; Fig. S1). The partial recovery seen after complementation could have been due to the level of gene expression or an imbalanced ratio of the proteins that make up the Sox system. A downstream effect is also possible, as it is not clear whether there are internal promoters within the deleted gene, albeit the deletion was in frame.
Cells in the polysulfide-induced resting state were also used to determine the rates of polysulfide oxidation by C. pinatubonensis JMP134 and its mutants (Table 1). The ΔsoxYZ, Δpdo1 Δpdo2, and Δpdo1 Δpdo2 ΔsoxYZ mutants oxidized polysulfide at rates of 91%, 11%, and 6% of the wild-type rate, respectively, suggesting that the two PDOs are the primary enzymes for the oxidation of added polysulfide, which is a specific form of sulfane sulfur and which is likely the direct product of SQR (5, 34). The Δpdo1 mutant and the Δpdo2 mutant oxidized polysulfide at rates of 93% and 22% of the rate of the wild type, respectively (Table 1), suggesting that PDO2 is the main PDO in the bacterium. The complementation of the Δpdo1 Δpdo2 mutant with pdo2 led to recovery of the rate of polysulfide oxidation to 69% of the wild-type rate (Table 1).
Cells in the thiosulfate-induced resting state were used to determine the rates of thiosulfate oxidation by C. pinatubonensis JMP134 and its mutants (Table 1), as thiosulfate induces the expression of sox genes in P. pantotrophus GB17 (17). The ΔsoxYZ cells oxidized thiosulfate at a rate of 12% of the wild-type rate. Complementation with soxYZ led to the partial recovery of the rate of thiosulfate oxidation to 71% of the wild-type rate (Table 1).
Collectively, the data suggest that SQR oxidizes sulfide to sulfane sulfur; PDO oxidizes sulfane sulfur to sulfite, which reacts with sulfane sulfur to generate thiosulfate in C. pinatubonensis JMP134 (5); and the Sox system further oxidizes thiosulfate to sulfate.
SQR did not couple with the Sox system for sulfide oxidation.
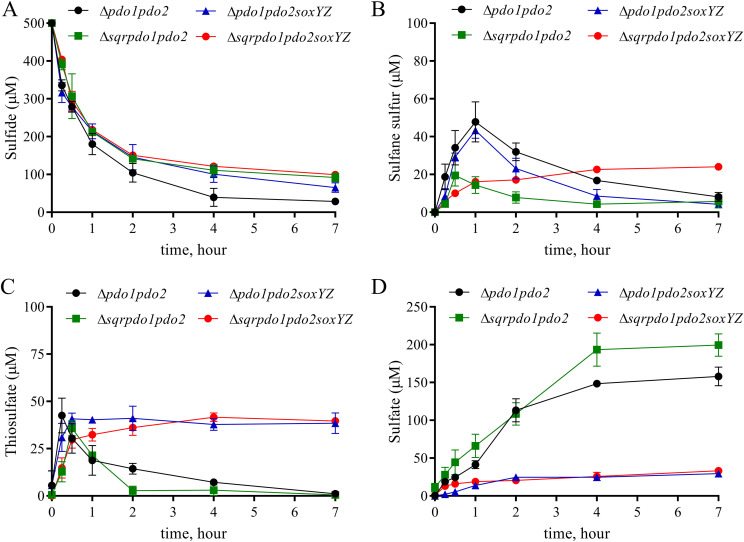
The direct coupling of SQR and the Sox system was not apparent, as the Δpdo1 Δpdo2 and Δpdo1 Δpdo2 ΔsoxYZ mutants oxidized sulfide at similar rates (Table 1; Fig. 3A), suggesting that the product (sulfane sulfur) inhibition in SQR was not alleviated by the Sox system in the Δpdo1 Δpdo2 mutant. The level of sulfane sulfur was higher for both the Δpdo1 Δpdo2 and the Δpdo1 Δpdo2 ΔsoxYZ mutants than for the wild type (Fig. 2B and 3B). Thiosulfate was also produced by the Δpdo1 Δpdo2 and Δpdo1 Δpdo2 ΔsoxYZ mutants (Fig. 2C and 3C), suggesting that sulfane sulfur is slowly oxidized to thiosulfate by other enzymes, like catalase (33), or by autoxidation (35). The Δpdo1 Δpdo2 mutant with the Sox system further oxidized thiosulfate to sulfate, while the Δpdo1 Δpdo2 ΔsoxYZ mutant accumulated thiosulfate (Fig. 3C and D). However, the Δpdo1 Δpdo2 ΔsoxYZ mutant also produced 30 μM sulfate (Table 2; Fig. 3D), suggesting nonspecific oxidation of either sulfane sulfur or thiosulfate at a very low rate by the mutant. Thus, the sulfane sulfur produced by SQR is mainly oxidized by PDO to thiosulfate, which is oxidized by the Sox system to sulfate.
FIG 3.
Sulfide oxidation by the Δpdo1 Δpdo2 mutant and its derivatives. Cells were harvested, washed, and resuspended at an OD600 of 2.0 in 100 mM HEPES buffer, pH 7.4. Sulfide was added to 500 μM to initiate the reaction. Sulfide (A), polysulfides (B), thiosulfate (C), and sulfate (D) levels were determined at different times. All data are averages for at least three samples with standard deviations (error bars).
FccAB coupled with the Sox system for sulfide oxidation.
The Δsqr Δpdo1 Δpdo2 mutant produced some thiosulfate during sulfide oxidation, suggesting that the sulfane sulfur produced by FccAB is oxidized to thiosulfate by other enzymes, as catalase slowly oxidizes polysulfide (33). Further, three lines of evidence supported the suggestion that the sulfane sulfur produced by FCSD was directly oxidized to sulfate by the Sox system in the absence of SQR. First, the Δpdo1 Δpdo2 mutant produced less sulfate than the Δsqr Δpdo1 Δpdo2 mutant did (Table 2; Fig. 3D), even though it oxidized sulfide faster (Table 1; Fig. 3A) and transitorily accumulated more sulfane sulfur and thiosulfate (Fig. 3C). Here (Fig. 3C), the reduced production of thiosulfate by the Δsqr Δpdo1 Δpdo2 mutant in comparison with that by the Δpdo1 Δpdo2 mutant suggests the direct coupling of FCSD with the Sox system, in which thiosulfate is not produced. Second, the rates of sulfide oxidation and the final levels of production of sulfate by the Δsqr and the Δsqr Δpdo1 pdo2 mutants were similar (Tables 1 and 2), indicating that PDO is not necessary during sulfide oxidation in that Δsqr mutant and that FCSD can couple with the Sox system for sulfide oxidation. Third, the Δsqr Δpdo1 Δpdo2 ΔsoxYZ mutant accumulated more sulfane sulfur than the Δsqr Δpdo1 Δpdo2 mutant did during sulfide oxidation (Table 2; Fig. 3B), suggesting that the Sox system oxidizes sulfane sulfur in the Δsqr Δpdo1 Δpdo2 mutant. Thus, in the sqr deletion mutants, the sulfane sulfur produced by FCSD is oxidized either by nonspecific enzymes, like catalase, or by the Sox system. When FCSD couples with the Sox system, thiosulfate is not an intermediate during sulfide oxidation to sulfate.
The sulfite produced during sulfide oxidation is not released into the medium.
Recombinant E. coli with cloned sqr and pdo2 from C. pinatubonensis JMP134 oxidizes sulfide to sulfite and thiosulfate (5), but we did not detect a transitory accumulation of sulfite in C. pinatubonensis JMP134 or its mutants during sulfide oxidation. C. pinatubonensis JMP134 contains a SOR system, encoded by sorA and sorB; however, the ΔsorA mutant metabolized sulfide in a similar way as the wild type did (Fig. 2; Fig. S2), and sulfite was still undetectable. The results indicate that C. pinatubonensis JMP134 produces sulfite during sulfide oxidation and that sulfite reacts with sulfane sulfur to generate thiosulfate inside the cell without being released.
Sulfite was oxidized to sulfate by the SOR system and the Sox system.
The sulfite-induced resting cells of C. pinatubonensis JMP134 quickly consumed 500 μM sulfite within 2 h, producing about 500 μM sulfate; the ΔsorA mutant completely lost the ability to oxidize sulfite; sulfite oxidation was restored in the ΔsorA/sorA strain. The ΔsoxYZ mutant oxidized sulfite as fast as the wild type did (Fig. S3), suggesting that the Sox system is not involved in sulfite oxidation in the wild type. When the cells were induced with thiosulfate and then used to oxidize sulfite, the ΔsorA mutant oxidized sulfite at a significantly lower rate (0.6 ± 0.3 nmol/mg [dry weight]/min) than the wild type did (5.4 ± 0.2 nmol/mg [dry weight]/min), and the ΔsorA ΔsoxYZ double mutant completely lost the ability to oxidize sulfite (Fig. S4). Thus, the Sox system also oxidized sulfite but did so at a rate of 11% of the rate that SOR did in C. pinatubonensis JMP134.
DISCUSSION
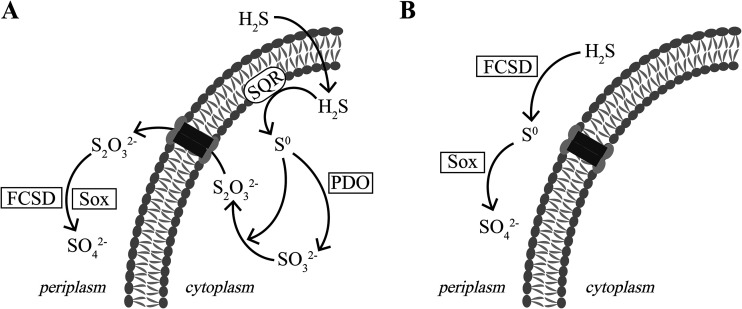
Our results indicate that C. pinatubonensis JMP134 oxidizes sulfide to sulfate via a new pathway (Fig. 4A). The pathway has not been previously observed in any bacteria. First, H2S enters the cytoplasm via diffusion, as membrane transporters are not required for the process (36). Second, SQR oxidizes H2S to sulfane sulfur (S0) in the cytoplasm (37). Third, PDO oxidizes sulfane sulfur to sulfite, which spontaneously reacts with sulfane sulfur to generate thiosulfate (5). The reaction between sulfane sulfur and sulfite likely occurs inside the cytoplasm, as both reactants are produced in the cytoplasm (37). Fourth, the produced thiosulfate is transported to the periplasmic space by an unknown transporter. Although tauE, encoding a hypothetical sulfite exporter (see Table S1 in the supplemental material), is next to the sqr-pdo2 operon, its role is not essential, as its deletion did not have detectable effects on sulfide oxidation and sulfate production (Tables 1 and 2). Fifth, the Sox system oxidizes thiosulfate to sulfate in the periplasmic space (Fig. 4A). In C. pinatubonensis JMP134, FCSD does not play a significant role in sulfide oxidation but enhances thiosulfate oxidation by the Sox system, as the ΔfccB mutant oxidized sulfide at a rate similar to that for the wild type but oxidized thiosulfate at a significantly reduced rate (Table 1). The role of FCSD in enhancing thiosulfate oxidation by the Sox system also occurs in P. pantotrophus GB17 (38). In sqr deletion mutants, such as the Δsqr and the Δsqr Δpdo1 Δpdo2 mutants, FCSD is likely able to direct couple with the Sox system for sulfide oxidation to sulfate (Fig. 4B), but at a significantly reduced rate (Table 1). Thus, C. pinatubonensis JMP134 contains a main pathway (Fig. 4A) and a contingent pathway (Fig. 4B) for sulfide oxidation.
FIG 4.
Proposed pathways of sulfide oxidation in C. pinatubonensis JMP134 and its Δsqr Δpdo1 Δpdo2 mutant. (A) Sulfide oxidation in the wild type. Sulfide is oxidized to sulfane sulfur (S0) by SQR; PDO oxidizes S0 to sulfite, which spontaneously reacts with S0 to generate thiosulfate in the cytoplasm. Thiosulfate is transported to the periplasmic space and is oxidized by the Sox system to sulfate. (B) Sulfide oxidation by the Δsqr Δpdo1 Δpdo2 mutant without SQR and PDO. FCSD oxidizes sulfide to S0, which is then oxidized by the Sox system to sulfate. This pathway is marginal in the wild type.
The main pathway is different from other documented pathways in bacteria. The anoxygenic purple sulfur bacterium Allochromatium vinosum uses H2S as the electron donor for photosynthesis, producing sulfate. It oxidizes sulfide to polysulfide by SQR and then to sulfite by reverse dissimilatory sulfite reductase (34). Sulfite is mainly oxidized to sulfate by a polysulfide reductase-like iron–sulfur molybdoprotein (SoeABC) (39).
The dimethylsulfide-degrading bacterium Hyphomicrobium denitrificans degrades dimethylsulfide to sulfide and then further oxidizes sulfide to sulfate under oxic conditions. Interestingly, the bacterium also produces thiosulfate as a metabolic intermediate. Although it contains PDO, its role in sulfane sulfur oxidation during dimethylsulfide oxidation is not observed. It employs heterodisulfide reductase to oxidize sulfane sulfur to sulfite, which reacts with sulfane sulfur to generate thiosulfate (9). Since the bacterium contains the incomplete Sox system without SoxCD, the system cannot completely oxidize thiosulfate to sulfate. The incomplete Sox system works with heterodisulfide reductase to oxidize thiosulfate to sulfate and sulfite (9). Sulfite reacts with sulfane sulfur to generate thiosulfate. The net process leads to the oxidation of reduced sulfur compounds to sulfate (40).
The chemolithotroph Acidithiobacillus caldus, a bioleaching agent, and members of the genus Thioalkalivibrio, a genus of haloalkaliphilic sulfur-oxidizing bacteria, contain genes similar to those seen in H. denitrificans (41, 42), and they may produce thiosulfate as a key intermediate during sulfur oxidation. However, Acidithiobacillus spp. and Thioalkalivibrio spp. also possess sulfite:quinone oxidoreductase, which may directly oxidize sulfite to sulfate (41, 42).
Both SQR and FCSD oxidize sulfide to sulfane sulfur (Fig. 2 and 3). Sequence analysis of 4,929 bacterial genomes showed that 1,014 bacteria contain SQR (3) and 190 bacteria contain FccB (16). The popularity of SQR may suggest its significance in H2S oxidation. Among the 190 bacteria with FccB, 121 bacteria (63.7%) also carried SQR (16). The frequent co-occurrence of FccB and SQR within a single bacterium indicates that they may have different physiological roles. In C. pinatubonensis JMP134, SQR is the primary sulfide oxidase, and the deletion of fccB did not reduce the sulfide oxidation rate but significantly reduced the rate of thiosulfate oxidation (Table 1). This finding is similar to that for A. vinosum, which contains both sqr and fccAB, and the inactivation of fccAB does not affect its rate of sulfide oxidation (43). The enhancement of the Sox activity by FCSD has previously been reported (38). Further, FCSD oxidizes sulfide at a lower rate in the Δsqr mutant (Table 1). Thus, FCSD oxidizes sulfide in bacteria that do not contain SQR, as previously reported (16). The choice of SQR over FCSD is likely due to energy conservation, as SQR uses ubiquinone as its electron acceptor (44), producing more membrane potential than FCSD, which uses cytochrome c as its electron acceptor (16).
The Sox system was extensively studied in P. pantotrophus, which is able to grow on organic compounds as well as on thiosulfate (45). Because the purified and reconstituted Sox system from P. pantotrophus oxidizes sulfide, thiosulfate, sulfur, and sulfite, the Sox system has been proposed to be a general sulfur-oxidizing system (24, 46). Early genetic analysis further supported the involvement of the Sox system in the oxidation of sulfide and thiosulfate in the anaerobic phototrophic bacterium Rhodovulum sulfidophilum (47). However, the role of Sox in sulfide oxidation should be revisited, as it should be confirmed whether R. sulfidophilum contains SQR and FCSD. Our data support the suggestion that the physiological substrate of the Sox system is thiosulfate in C. pinatubonensis JMP134 (Table 1). In the Δsqr Δpdo1 Δpdo2 mutant (Fig. 3) and the ΔsorA mutant (Fig. S2), the Sox system oxidizes S0 and sulfite at significantly reduced rates, suggesting that the Sox system can oxidize S0 and sulfite but that the efficiency is much lower than that for PDO and SOR in C. pinatubonensis JMP134.
The Sox system oxidized the sulfane sulfur generated by FCSD, but not by SQR or added polysulfide (Table 1). Since both FCSD and the Sox system are in the periplasmic space, it is possible that FccB directly transfers sulfane sulfur to SoxY, producing SoxYZ-SSH, which can be further oxidized by the Sox system to sulfate (Fig. 4B). When polysulfide is added to neutral solutions, it is rapidly converted to elemental sulfur in the form of S8 (48); perhaps the reaction of S8 with SoxYZ-SH is kinetically slow or needs additional reducing power. Since the active site of SQR in C. pinatubonensis JMP134 is in the cytoplasm (37), the membrane separation of SQR-produced sulfane sulfur and the Sox system may contribute to the uncoupling of SQR and the Sox system.
Our evidence does not support the suggestion that the bacterium uses the Sox system for the efficient oxidation of sulfide. First, the ΔsoxYZ mutant oxidized sulfide at the same rate as the wild type (Table 1; Fig. 2). A. vinosum also does not use the Sox system for sulfide oxidation, as the deletion of the sox genes in A. vinosum does not affect its sulfide oxidation (29). Second, the Δsqr ΔfccB mutant with the Sox system did not show any meaningful rate of sulfide oxidation (Table 1; Fig. 2), suggesting that C. pinatubonensis JMP134 does not use the Sox system for H2S oxidation. Since P. pantotrophus contains SQR (GenBank accession number RKS43125.1) and FCSD (49), it should be investigated whether it uses the Sox system to oxidize sulfide.
In C. pinatubonensis JMP134, SQR prefers to work with PDO, and the Sox system may oxidize the sulfane sulfur produced by FCSD. The preference may be related to the subcellular localization of the enzymes (Fig. 4). SQR is a membrane protein with its active site in the cytoplasm, and PDO is also in the cytoplasm (37), whereas FCSD and the Sox system are soluble proteins in the periplasmic space (16, 46, 50). The preference is also reflected in the linkages of these genes on the chromosome, with sqr and pdo2 being organized in an operon on chromosome B and the sox genes and fccAB being adjacently located on chromosome A (Fig. 1).
The H2S oxidation network of SQR-PDO, FCSD, and the Sox system of C. pinatubonensis JMP134 may also be present in other bacteria, including P. pantotrophus GB17, Roseobacter denitrificans OCh 114, and Ruegeria pomeroyi DSS-3, as they all contain these genes (Table S2). These bacteria may use the same pathway to oxidize H2S. C. pinatubonensis JMP134 is a soil bacterium (12); P. pantotrophus GB17 is from a denitrifying, sulfide-oxidizing effluent treatment plant in the Netherlands (51); R. denitrificans OCh 114 and R. pomeroyi DSS-3 are the model bacteria of the Roseobacter clade, dominant in coastal seawater and surface sediments (52). These bacteria all belong to the Proteobacteria and are capable of heterotrophic growth. Their wide distribution in soil, wastewater treatment plants, and marine waters implies the potential role of these bacteria in H2S oxidation in the natural environment.
MATERIALS AND METHODS
Strains, plasmids, and primers.
The strains and plasmids used in this study are listed in Table 3. All the primers are listed in Table 4.
TABLE 3.
Strains and plasmids used in this study
| Strain or plasmid | Characteristic or target protein | Source |
|---|---|---|
| Strains | ||
| E. coli S17-1 | recA pro thi hsdS, RP4 tra functions, supE44 | Invitrogen |
| C. pinatubonensis JMP134 | Wild type | Our labs |
| Plasmids | Characteristics | Source |
| pBBR1MCS2 | Kanamycin resistance, mob+, pBBR1 replicon, cloning vector | Qi Qingsheng |
| pK18mobsacB | Widely used gene-knockout vector, kanamycin resistance | Our labs |
| pBBR-soxYZ | pBBR1MCS2 containing soxY and soxZ | This study |
| pBBR-pdo2-sqr | pBBR1MCS2 containing pdo2 and sqr | This study |
| pBBR-fccB | pBBR1MCS2 containing fccB | This study |
| pBBR-pdo2 | pBBR1MCS2 containing pdo2 | This study |
| pBBR-sorA | pBBR1MCS2 containing sorA | This study |
| Mutants of JMP134 | ||
| Δsqr | sqr deleted | This study |
| ΔfccB | fccB deleted | This study |
| Δsqr Δpdo1 Δpdo2 | sqr, pdo2, and pdo1 deleted | This study |
| Δsqr Δpdo1 Δpdo2 ΔsoxYZ | sqr, pdo2, pdo1, soxY, and soxZ deleted | This study |
| Δsqr ΔfccB | sqr and fccB deleted | This study |
| Δsqr ΔfccB ΔsoxYZ | sqr, fccB, soxY, and soxZ deleted | This study |
| ΔsoxYZ | soxY and soxZ deleted | This study |
| Δpdo1 | pdo1 deleted | This study |
| Δpdo2 | pdo2 deleted | This study |
| Δpdo1 Δpdo2 | pdo1 and pdo2 deleted | This study |
| Δpdo1 Δpdo2 ΔsoxYZ | pdo1, pdo2, soxY, and soxZ deleted | This study |
| ΔsorA | sorA deleted | This study |
| ΔtauE | tauE deleted | This study |
| Complemented strains | ||
| ΔfccB/fccB | ΔfccB mutant with plasmid pBBR-fccB | This study |
| Δsqr Δpdo1 Δpdo2/sqr pdo2 | Δsqr Δpdo1 Δpdo2 mutant with plasmid pBBR-pdo2-sqr | This study |
| ΔsoxYZ/soxYZ | ΔsoxYZ mutant with plasmid pBBR-soxYZ | This study |
| Δpdo1 Δpdo2/pdo2 | Δpdo1 Δpdo2 mutant with plasmid pBBR-pdo2 | This study |
| ΔsorA/sorA | ΔsorA mutant with plasmid pBBR-sorA | This study |
| Δsqr Δpdo1 Δpdo2 ΔsoxYZ/soxYZ | Δsqr Δpdo1 Δpdo2 ΔsoxYZ mutant with plasmid pBBR-soxYZ | This study |
TABLE 4.
Primers used in this study
| Target gene | Primera | Sequence (5′–3′)b |
|---|---|---|
| Deletion | ||
| soxY and soxZ | Up-f | CAGGAAACAGCTATGACATGATTACGAATTCCGTCATTTTCGGATATCGC |
| Up-r | TGGTCGCTTCCCGCAGTACTTCTCTTCGTTTC | |
| Down-f | AGTACTGCGGGAAGCGACCATCGCCTGATT | |
| Down-r | TTCAGGATCCCCGGGTACCGAGCTCGAATTCGCTGTTCGAGATCCATGAC | |
| V-f | GATGGTAGGGTGGATTCTTGAGGC | |
| V-r | CTACGGGGCCTTCAAAGGTGTT | |
| fccB | UP-f | CAGGAAACAGCTATGACATGATTACGAATTGTCAACGAACTGGAAATCACGTC |
| UP-r | TAGCTCAGCACTTCAGAAAGTTGCGTCGTTGC | |
| Down-f | CTTTCTGAAGTGCTGAGCTAGCCGTTCCGCAT | |
| Down-r | TTCAGGATCCCCGGGTACCGAGCTCGAATTAACCGGCCCCACATGGTGTAG | |
| V-f | GATCAAGCCTCCACCTCGCAGAAC | |
| V-r | CGCAAAGCGTCAACAGAAACCCG | |
| sqr | Up-f | CAGGAAACAGCTATGACATGATTACGAATTGACGGGGGCCTTGAACTTTTATC |
| Up-r | GACCATGCAATTGCAGGGATAACACCCGAAG | |
| Down-f | ATCCCTGCAATTGCATGGTCGTCTGTTCCTTG | |
| Down-r | TTCAGGATCCCCGGGTACCGAGCTCGAATTAATCGCAACGCTCTGCTAACC | |
| V-f | GCACCGGTGCCTTTGTATTG | |
| V-r | TCCTGTACATGTGCCACGAC | |
| pdo1 | Up-f | CAGGAAACAGCTATGACATGATTACGAATTAGACGATTACCTGGTCTACACCTTC |
| Up-r | CAGCTGTTCGTACAGGCGCGTCAAATCCTTCTAT | |
| Down-f | CGCGCCTGTACGAACAGCTGATAGAAGGTTTGCAT | |
| Down-r | TTCAGGATCCCCGGGTACCGAGCTCGAATTGGCTGATGATGGAGAACGAAC | |
| V-f | TATTGGCTGCCATCTGCT | |
| V-r | GCTCTACAAGCTCAATGCG | |
| pdo2 | Up-f | CAGGAAACAGCTATGACATGATTACGAATTCGAGGTCGTAGCGGTAGTTG |
| Up-r | ACACACATGAGCTATCTGAAGATTCCCCTCAAC | |
| Down-f | TTCAGATAGCTCATGTGTGTCTATCCGTGGTTAGC | |
| Down-r | TTCAGGATCCCCGGGTACCGAGCTCGAATTCCATTTCATCGAGGAATAGCGT | |
| V-f | ATGGCGTCCCAATCCAGCTT | |
| V-r | TTGCCTGGAGAGTGGCTTTG | |
| sorA | Up-f | CAGGAAACAGCTATGACATGATTACGAATTATCCGTCGGCAGGTAACAGC |
| Up-r | CATATAGCCGATTGTCAGGCGCATTCATGG | |
| Down-f | GCCTGACAATCGGCTATATGCGCAACGTGG | |
| Down-r | TTCAGGATCCCCGGGTACCGAGCTCGAATTGCGCGTGGAGGAAGTGATCC | |
| V-f | TCTTTCGCTTGCGTCTGATG | |
| V-r | CAGCGGATCTTGCGATCTAC | |
| tauE | Up-f | CAGGAAACAGCTATGACATGATTACGAATTGCTGTCGATGCAGGTCAAATT |
| Up-r | ACGTTCAGCTCAATGTCTTGCATCTGTCATTCC | |
| Down-f | CAAGACATTGAGCTGAACGTCAGGGTGAAATC | |
| Down-r | TTCAGGATCCCCGGGTACCGAGCTCGAATTCACCTCTGCGAGTGTTTCATTAG | |
| V-f | TGATGGTCAGTCAAGGGCTGTT | |
| V-r | CTGTTGAAGGGACAGGGATAGG | |
| Complementation | ||
| soxY and soxZ | Forward | CACACAGGAAACAGCTATGAATTCGAAACGAAGAGAAG |
| Reverse | TTCCATTCGCCATTCATCAGGCGATGGTCGCT | |
| fccB | Forward | CACACAGGAAACAGCTATGCAACGACGCAACTTTCTGAAG |
| Reverse | TAACAAAATATTAACGCTTAGCTCAGCATGTCGGC | |
| sorA | Forward | CACACAGGAAACAGCTATGAATGCGCCTGACAATCCCC |
| Reverse | TTCCATTCGCCATTCAACTGCGGTAACGCGCGTGGA | |
| sqr and pdo2 | Forward | CACACAGGAAACAGCTATGACACCGACCATGCCAAGC |
| Reverse | TTCCATTCGCCATTCATTATCCCTGCAACTCGGGTGTCG | |
| pdo2 | Forward | CACACAGGAAACAGCTATGACACCGACCATGCCAAG |
| Reverse | TTCCATTCGCCATTCATCAGAGGGCGTTGAGGGG | |
| Linearization of pBBR1MCS2 | Forward | TGAATGGCGAATGGAAATTGTAAG |
| Reverse | AGCTGTTTCCTGTGTGAAATTGTTATC |
Up, primers used to clone the upstream sequence of the target gene; Down, primers used to clone the downstream sequence of the target gene; V, primers used to verify the mutants.
For the deletion mutants, underlining represents the sequences overlapping the plasmid pK18mobsacB sequence. For the complemented strains, underlining represents the sequences overlapping the plasmid pBBR1MCS2 sequence.
Culture conditions.
C. pinatubonensis JMP134 and its mutants were grown at 30°C in Luria-Bertani (LB) medium containing 10 g of NaCl per liter or in a mineral medium (MM) (53). MM consisted of 0.58 g of K2HPO4, 0.19 g of KH2PO4, 0.25 g of NaNO3, 0.5 g of MgCl2, and 1 ml of a trace element solution per liter of deionized water, and its pH was adjusted to 7.0. The carbon source was 0.5% (wt/vol) monosodium glutamate. NaSO4, Na2SO3, or Na2S2O3 (1 mM) was used as the sole sulfur source. The trace element solution contained 10 ml of concentrated HCl, 4.74 mg of ZnCl2, 2.53 mg of MnCl2·4H2O, 30 mg of H3BO3, 20 mg of CoCl2·6H2O, 1 mg of CuCl2·2H2O, 2 mg of NiCl2·6H2O, 3.24 mg of Na2MoO4·2H2O per liter.
Sulfur compound preparation.
NaHS, Na2SO3, or Na2S2O3 was freshly prepared in 100 mM HEPES buffer (pH 7.4). The polysulfide was prepared according to a published method by mixing sulfur power and sodium sulfide in anoxic distilled water under argon gas (5).
Gene deletion and complementation.
The method used to delete the sulfur-related genes in C. pinatubonensis JMP134 is essentially similar to that previously reported (3). The primers used in the deletion process are shown in Table 4. After the upstream and downstream fragments of the target gene were obtained by PCR, these two fragments were ligated with the linearized plasmid pK18mobsacB by a modified In-Fusion method (54) to construct a deletion plasmid, and the deletion plasmid was transformed into E. coli S17-1 and then transferred to C. pinatubonensis JMP134 by conjugation. After two rounds of screening by using colony PCR, the correct deletion strain was obtained.
The complementation strain was generated by transforming a recombinant plasmid into the corresponding mutant. The recombinant plasmid was constructed by assembling the PCR-amplified gene in the broad-host-range plasmid pBBR1MCS2 (linearized via PCR) by using a modified In-Fusion method (54). The primers used to amplify the target gene and linearized plasmid pBBR1MCS2 are listed in Table 4.
Whole-cell assay.
A single colony of the wild type, deletion strains, or complementation strains was inoculated in LB medium (with antibiotics, as necessary) and incubated at 30°C overnight (200 rpm). The culture was transferred to 100 ml fresh medium (1% inoculation) and incubated at 30°C with shaking (200 rpm) to an OD600 of 1.0 to 1.5, and then 20 μM NaHS, 20 μM polysulfides, 100 μM Na2SO3, or 100 μM Na2S2O3 was added for induction with continuous incubation to an OD600 of 2.5 (about 1 h). Cells were collected by centrifugation (4,000 × g, 5 min) and resuspended in HEPES buffer (pH 7.4, 100 mM) at an OD600 of 2.0. The heat-inactivated cells were prepared by incubating 10 ml of cell suspension in a boiling water bath for 15 min.
Ten milliliters of the suspension was transferred to a 50-ml large plastic centrifuge tube (Labcon, USA); NaHS, polysulfide, Na2SO3, or Na2S2O3 was added; and the tube was covered with a silicone stopper. The sample was mixed and incubated with shaking (30°C, 100 rpm). One milliliter of sample was taken at 15 min, 30 min, 1 h, 2 h, 4 h, and 7 h, and the concentrations of the sulfur compounds were measured. Suspensions were used directly for the detection of sulfide and sulfane sulfur. When sulfite, thiosulfate, and sulfate were tested, the suspension was centrifuged (13,000 × g, 3 min) and the supernatant was taken for detection.
Sulfur compound detection.
Sulfide was detected by using the methylene blue method, and sulfane sulfur (including polysulfide, persulfide, and elemental sulfur) was detected by using the cyanide method (5). The detection of sulfite, thiosulfate, and sulfate in the supernatant was carried out by using an ion chromatograph system (ICS; model ICS-1100; Dionex, USA) in the anion detection mode with an Ion Pac AS19 column, an eluent automatic generation device (RFC-30), a column temperature of 30°C, an ASRS_4 mm suppressor, and an eluent of 22 mM KOH with isocratic elution (1 ml/min KOH). Under these conditions, the elution times of sulfite, sulfate, and thiosulfate were 7.9 min, 8.5 min, and 27.5 min, respectively. This method had a sulfite detection limit of 10 μM.
Bioinformatics.
The basic sulfur metabolic pathway was derived from the analysis of C. pinatubonensis JMP134 at the KEGG website (55). The query sequences were from A. vinosum DSM 180T (locus tag, Alvin_0091; GenBank accession number ADC61061) and Cupriavidus metallidurans CH34 (locus tag, Rmet_5347; GenBank accession number ABF12206) for thiosulfate dehydrogenases, from Acidithiobacillus ferrooxidans (GenBank accession number AAB93983) for tetrathionate hydrolase, and from Thiobacillus denitrificans ATCC 25259 (GenBank accession number Q60028.3) for ribulose bisphosphate carboxylase. Similar proteins of each sulfur-related enzyme of C. pinatubonensis JMP134 were searched for in selected microbial genomes by using the TBLASTN program (56).
Data availability.
Data and NCBI accession numbers related to sulfide oxidation by additional mutants and complementation strains and sulfur-metabolizing enzymes in C. pinatubonensis JMP134 and other bacteria are given in the supplemental material.
Supplementary Material
ACKNOWLEDGMENTS
The work was financially supported by grants from the National Natural Science Foundation of China (31870097 and 91951202).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Dahl C. 2015. Cytoplasmic sulfur trafficking in sulfur-oxidizing prokaryotes. IUBMB Life 67:268–274. doi: 10.1002/iub.1371. [DOI] [PubMed] [Google Scholar]
- 2.Wang R, Lin JQ, Liu XM, Pang X, Zhang CJ, Yang CL, Gao XY, Lin CM, Li YQ, Li Y, Lin JQ, Chen LX. 2018. Sulfur oxidation in the acidophilic autotrophic Acidithiobacillus spp. Front Microbiol 9:3290. doi: 10.3389/fmicb.2018.03290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xia Y, Lu C, Hou N, Xin Y, Liu J, Liu H, Xun L. 2017. Sulfide production and oxidation by heterotrophic bacteria under aerobic conditions. ISME J 11:2754–2766. doi: 10.1038/ismej.2017.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luebke JL, Shen J, Bruce KE, Kehl-Fie TE, Peng H, Skaar EP, Giedroc DP. 2014. The CsoR-like sulfurtransferase repressor (CstR) is a persulfide sensor in Staphylococcus aureus. Mol Microbiol 94:1343–1360. doi: 10.1111/mmi.12835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xin Y, Liu H, Cui F, Liu H, Xun L. 2016. Recombinant Escherichia coli with sulfide:quinone oxidoreductase and persulfide dioxygenase rapidly oxidises sulfide to sulfite and thiosulfate via a new pathway. Environ Microbiol 18:5123–5136. doi: 10.1111/1462-2920.13511. [DOI] [PubMed] [Google Scholar]
- 6.Hou N, Xia Y, Wang X, Liu H, Liu H, Xun L. 2018. H2S biotreatment with sulfide-oxidizing heterotrophic bacteria. Biodegradation 29:511–524. doi: 10.1007/s10532-018-9849-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mason J, Kelly DP. 1988. Thiosulfate oxidation by obligately heterotrophic bacteria. Microb Ecol 15:123–134. doi: 10.1007/BF02011707. [DOI] [PubMed] [Google Scholar]
- 8.Sorokin DY, Tourova TP, Muyzer G. 2005. Oxidation of thiosulfate to tetrathionate by an haloarchaeon isolated from hypersaline habitat. Extremophiles 9:501–504. doi: 10.1007/s00792-005-0465-0. [DOI] [PubMed] [Google Scholar]
- 9.Koch T, Dahl C. 2018. A novel bacterial sulfur oxidation pathway provides a new link between the cycles of organic and inorganic sulfur compounds. ISME J 12:2479–2491. doi: 10.1038/s41396-018-0209-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teske A, Brinkhoff T, Muyzer G, Moser DP, Rethmeier J, Jannasch HW. 2000. Diversity of thiosulfate-oxidizing bacteria from marine sediments and hydrothermal vents. Appl Environ Microbiol 66:3125–3133. doi: 10.1128/aem.66.8.3125-3133.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lykidis A, Perez-Pantoja D, Ledger T, Mavromatis K, Anderson IJ, Ivanova NN, Hooper SD, Lapidus A, Lucas S, Gonzalez B, Kyrpides NC. 2010. The complete multipartite genome sequence of Cupriavidus necator JMP134, a versatile pollutant degrader. PLoS One 5:e9729. doi: 10.1371/journal.pone.0009729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Don RH, Pemberton JM. 1981. Properties of six pesticide degradation plasmids isolated from Alcaligenes paradoxus and Alcaligenes eutrophus. J Bacteriol 145:681–686. doi: 10.1128/JB.145.2.681-686.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gregersen LH, Bryant DA, Frigaard NU. 2011. Mechanisms and evolution of oxidative sulfur metabolism in green sulfur bacteria. Front Microbiol 2:116. doi: 10.3389/fmicb.2011.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bartsch RG, Kamen MD. 1960. Isolation and properties of two soluble heme proteins in extracts of the photoanaerobe Chromatium. J Biol Chem 235:825–831. [PubMed] [Google Scholar]
- 15.Kostanjevecki V, Brige A, Meyer TE, Cusanovich MA, Guisez Y, van Beeumen J. 2000. A membrane-bound flavocytochrome c-sulfide dehydrogenase from the purple phototrophic sulfur bacterium Ectothiorhodospira vacuolata. J Bacteriol 182:3097–3103. doi: 10.1128/jb.182.11.3097-3103.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lü C, Xia Y, Liu D, Zhao R, Gao R, Liu H, Xun L. 2017. Cupriavidus necator H16 uses flavocytochrome c sulfide dehydrogenase to oxidize self-produced and added sulfide. Appl Environ Microbiol 83:e01610-17. doi: 10.1128/AEM.01610-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedrich CG, Rother D, Bardischewsky F, Quentmeier A, Fischer J. 2001. Oxidation of reduced inorganic sulfur compounds by bacteria: emergence of a common mechanism? Appl Environ Microbiol 67:2873–2882. doi: 10.1128/AEM.67.7.2873-2882.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grabarczyk DB, Berks BC. 2017. Intermediates in the Sox sulfur oxidation pathway are bound to a sulfane conjugate of the carrier protein SoxYZ. PLoS One 12:e0173395. doi: 10.1371/journal.pone.0173395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quentmeier A, Friedrich CG. 2001. The cysteine residue of the SoxY protein as the active site of protein-bound sulfur oxidation of Paracoccus pantotrophus GB17. FEBS Lett 503:168–172. doi: 10.1016/s0014-5793(01)02727-2. [DOI] [PubMed] [Google Scholar]
- 20.Grabarczyk DB, Chappell PE, Johnson S, Stelzl LS, Lea SM, Berks BC. 2015. Structural basis for specificity and promiscuity in a carrier protein/enzyme system from the sulfur cycle. Proc Natl Acad Sci U S A 112:E7166–E7175. doi: 10.1073/pnas.1506386112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sauve V, Roversi P, Leath KJ, Garman EF, Antrobus R, Lea SM, Berks BC. 2009. Mechanism for the hydrolysis of a sulfur-sulfur bond based on the crystal structure of the thiosulfohydrolase SoxB. J Biol Chem 284:21707–21718. doi: 10.1074/jbc.M109.002709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu WP, Kelly DP. 1984. Properties and role of sulphite: cytochrome c oxidoreductase purified from Thiobacillus versutus (A2). Microbiology 130:1683–1692. doi: 10.1099/00221287-130-7-1683. [DOI] [Google Scholar]
- 23.Zander U, Faust A, Klink BU, de Sanctis D, Panjikar S, Quentmeier A, Bardischewsky F, Friedrich CG, Scheidig AJ. 2011. Structural basis for the oxidation of protein-bound sulfur by the sulfur cycle molybdohemo-enzyme sulfane dehydrogenase SoxCD. J Biol Chem 286:8349–8360. doi: 10.1074/jbc.M110.193631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rother D, Henrich HJ, Quentmeier A, Bardischewsky F, Friedrich CG. 2001. Novel genes of the sox gene cluster, mutagenesis of the flavoprotein SoxF, and evidence for a general sulfur-oxidizing system in Paracoccus pantotrophus GB17. J Bacteriol 183:4499–4508. doi: 10.1128/JB.183.15.4499-4508.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robin S, Arese M, Forte E, Sarti P, Giuffre A, Soulimane T. 2011. A sulfite respiration pathway from Thermus thermophilus and the key role of newly identified cytochrome c550. J Bacteriol 193:3988–3997. doi: 10.1128/JB.05186-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H, Li J, Lu C, Xia Y, Xin Y, Liu H, Xun L, Liu H. 2017. FisR activates σ54-dependent transcription of sulfide-oxidizing genes in Cupriavidus pinatubonensis JMP134. Mol Microbiol 105:373–384. doi: 10.1111/mmi.13725. [DOI] [PubMed] [Google Scholar]
- 27.Liu H, Xin Y, Xun L. 2014. Distribution, diversity, and activities of sulfur dioxygenases in heterotrophic bacteria. Appl Environ Microbiol 80:1799–1806. doi: 10.1128/AEM.03281-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Denkmann K, Grein F, Zigann R, Siemen A, Bergmann J, van Helmont S, Nicolai A, Pereira IA, Dahl C. 2012. Thiosulfate dehydrogenase: a widespread unusual acidophilic c-type cytochrome. Environ Microbiol 14:2673–2688. doi: 10.1111/j.1462-2920.2012.02820.x. [DOI] [PubMed] [Google Scholar]
- 29.Hensen D, Sperling D, Truper HG, Brune DC, Dahl C. 2006. Thiosulphate oxidation in the phototrophic sulphur bacterium Allochromatium vinosum. Mol Microbiol 62:794–810. doi: 10.1111/j.1365-2958.2006.05408.x. [DOI] [PubMed] [Google Scholar]
- 30.De Jong GA, Hazeu W, Bos P, Kuenen JG. 1997. Isolation of the tetrathionate hydrolase from Thiobacillus acidophilus. Eur J Biochem 243:678–683. doi: 10.1111/j.1432-1033.1997.00678.x. [DOI] [PubMed] [Google Scholar]
- 31.Ramos AR, Keller KL, Wall JD, Pereira IA. 2012. The membrane QmoABC complex interacts directly with the dissimilatory adenosine 5′-phosphosulfate reductase in sulfate reducing bacteria. Front Microbiol 3:137. doi: 10.3389/fmicb.2012.00137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olson KR, Gao Y, Arif F, Arora K, Patel S, DeLeon ER, Sutton TR, Feelisch M, Cortese-Krott MM, Straub KD. 2018. Metabolism of hydrogen sulfide (H(2)S) and production of reactive sulfur species (RSS) by superoxide dismutase. Redox Biol 15:74–85. doi: 10.1016/j.redox.2017.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olson KR, Gao Y, DeLeon ER, Arif M, Arif F, Arora N, Straub KD. 2017. Catalase as a sulfide-sulfur oxido-reductase: an ancient (and modern?) regulator of reactive sulfur species (RSS). Redox Biol 12:325–339. doi: 10.1016/j.redox.2017.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prange A, Engelhardt H, Truper HG, Dahl C. 2004. The role of the sulfur globule proteins of Allochromatium vinosum: mutagenesis of the sulfur globule protein genes and expression studies by real-time RT-PCR. Arch Microbiol 182:165–174. doi: 10.1007/s00203-004-0683-3. [DOI] [PubMed] [Google Scholar]
- 35.Kleinjan WE, de Keizer A, Janssen AJ. 2005. Kinetics of the chemical oxidation of polysulfide anions in aqueous solution. Water Res 39:4093–4100. doi: 10.1016/j.watres.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 36.Mathai JC, Missner A, Kugler P, Saparov SM, Zeidel ML, Lee JK, Pohl P. 2009. No facilitator required for membrane transport of hydrogen sulfide. Proc Natl Acad Sci U S A 106:16633–16638. doi: 10.1073/pnas.0902952106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao R, Liu H, Xun L. 2017. Cytoplasmic localization of sulfide:quinone oxidoreductase and persulfide dioxygenase of Cupriavidus pinatubonensis JMP134. Appl Environ Microbiol 83:e01820-17. doi: 10.1128/AEM.01820-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bardischewsky F, Quentmeier A, Friedrich CG. 2006. The flavoprotein SoxF functions in chemotrophic thiosulfate oxidation of Paracoccus pantotrophus in vivo and in vitro. FEMS Microbiol Lett 258:121–126. doi: 10.1111/j.1574-6968.2006.00210.x. [DOI] [PubMed] [Google Scholar]
- 39.Dahl C, Franz B, Hensen D, Kesselheim A, Zigann R. 2013. Sulfite oxidation in the purple sulfur bacterium Allochromatium vinosum: identification of SoeABC as a major player and relevance of SoxYZ in the process. Microbiology (Reading) 159:2626–2638. doi: 10.1099/mic.0.071019-0. [DOI] [PubMed] [Google Scholar]
- 40.Tanabe TS, Leimkuhler S, Dahl C. 2019. The functional diversity of the prokaryotic sulfur carrier protein TusA. Adv Microb Physiol 75:233–277. doi: 10.1016/bs.ampbs.2019.07.004. [DOI] [PubMed] [Google Scholar]
- 41.Chen L, Ren Y, Lin J, Liu X, Pang X, Lin J. 2012. Acidithiobacillus caldus sulfur oxidation model based on transcriptome analysis between the wild type and sulfur oxygenase reductase defective mutant. PLoS One 7:e39470. doi: 10.1371/journal.pone.0039470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berben T, Overmars L, Sorokin DY, Muyzer G. 2019. Diversity and distribution of sulfur oxidation-related genes in Thioalkalivibrio, a genus of chemolithoautotrophic and haloalkaliphilic sulfur-oxidizing bacteria. Front Microbiol 10:160. doi: 10.3389/fmicb.2019.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reinartz M, Tschape J, Bruser T, Truper HG, Dahl C. 1998. Sulfide oxidation in the phototrophic sulfur bacterium Chromatium vinosum. Arch Microbiol 170:59–68. doi: 10.1007/s002030050615. [DOI] [PubMed] [Google Scholar]
- 44.Cherney MM, Zhang Y, Solomonson M, Weiner JH, James MN. 2010. Crystal structure of sulfide:quinone oxidoreductase from Acidithiobacillus ferrooxidans: insights into sulfidotrophic respiration and detoxification. J Mol Biol 398:292–305. doi: 10.1016/j.jmb.2010.03.018. [DOI] [PubMed] [Google Scholar]
- 45.Rainey FA, Kelly DP, Stackebrandt E, Burghardt J, Hiraishi A, Katayama Y, Wood AP. 1999. A re-evaluation of the taxonomy of Paracoccus denitrificans and a proposal for the combination Paracoccus pantotrophus comb. nov. Int J Syst Bacteriol 49:645–651. doi: 10.1099/00207713-49-2-645. [DOI] [PubMed] [Google Scholar]
- 46.Friedrich CG, Quentmeier A, Bardischewsky F, Rother D, Kraft R, Kostka S, Prinz H. 2000. Novel genes coding for lithotrophic sulfur oxidation of Paracoccus pantotrophus GB17. J Bacteriol 182:4677–4687. doi: 10.1128/jb.182.17.4677-4687.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Appia-Ayme C, Little PJ, Matsumoto Y, Leech AP, Berks BC. 2001. Cytochrome complex essential for photosynthetic oxidation of both thiosulfate and sulfide in Rhodovulum sulfidophilum. J Bacteriol 183:6107–6118. doi: 10.1128/JB.183.20.6107-6118.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Toohey JI. 2011. Sulfur signaling: is the agent sulfide or sulfane? Anal Biochem 413:1–7. doi: 10.1016/j.ab.2011.01.044. [DOI] [PubMed] [Google Scholar]
- 49.Quentmeier A, Hellwig P, Bardischewsky F, Wichmann R, Friedrich CG. 2004. Sulfide dehydrogenase activity of the monomeric flavoprotein SoxF of Paracoccus pantotrophus. Biochemistry 43:14696–14703. doi: 10.1021/bi048568y. [DOI] [PubMed] [Google Scholar]
- 50.Frigaard NU, Dahl C. 2009. Sulfur metabolism in phototrophic sulfur bacteria. Adv Microb Physiol 54:103–200. doi: 10.1016/S0065-2911(08)00002-7. [DOI] [PubMed] [Google Scholar]
- 51.Robertson LA, Kuenen JG. 1983. Thiosphaera pantotropha gen. nov. sp. nov., a facultatively anaerobic, facultatively autotrophic sulphur bacterium. J Gen Microbiol 129:2847–2855. [Google Scholar]
- 52.Lenk S, Moraru C, Hahnke S, Arnds J, Richter M, Kube M, Reinhardt R, Brinkhoff T, Harder J, Amann R, Mußmann M. 2012. Roseobacter clade bacteria are abundant in coastal sediments and encode a novel combination of sulfur oxidation genes. ISME J 6:2178–2187. doi: 10.1038/ismej.2012.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Louie TM, Webster CM, Xun L. 2002. Genetic and biochemical characterization of a 2,4,6-trichlorophenol degradation pathway in Ralstonia eutropha JMP134. J Bacteriol 184:3492–3500. doi: 10.1128/jb.184.13.3492-3500.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xia Y, Li K, Li J, Wang T, Gu L, Xun L. 2019. T5 exonuclease-dependent assembly offers a low-cost method for efficient cloning and site-directed mutagenesis. Nucleic Acids Res 47:e15. doi: 10.1093/nar/gky1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kanehisa M, Goto S. 2000. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data and NCBI accession numbers related to sulfide oxidation by additional mutants and complementation strains and sulfur-metabolizing enzymes in C. pinatubonensis JMP134 and other bacteria are given in the supplemental material.