Abstract
Radiologic techniques remain the main method for early detection for breast cancer and are critical to achieve a favorable outcome from cancer. However, more sensitive detection methods to complement radiologic techniques are needed to enhance early detection and treatment strategies. Using our recently established culturing method that allows propagation of normal and cancerous breast epithelial cells of luminal origin, flow cytometry characterization, and genomic sequencing, we show that cancer cells can be detected in breast milk. Cells derived from milk from the breast with cancer were enriched for CD49f+/EpCAM−, CD44+/CD24− and CD271+ cancer stem-like cells (CSC). These CSC carried mutations within the cytoplasmic retention domain of HDAC6, stop/gain insertion in MORF4L1, and deletion mutations within SWI/SNF complex component SMARCC2. CSC were sensitive to HDAC6 inhibitors, BET bromodomain inhibitors, and EZH2 inhibitors, as mutations in SWI/SNF complex components are known to increase sensitivity to these drugs. Among cells derived from breast milk of additional 10 women not known to have breast cancer, two of them contained cells that were enriched for the CSC phenotype and carried mutations in NF1 or KMT2D, which are frequently mutated in breast cancer. Breast milk-derived cells with NF1 mutations also carried copy number variations in CDKN2C, PTEN, and REL genes. The approach described here may enable rapid cancer cell characterization including driver mutation detection and therapeutic screening for pregnancy/post-partum breast cancers. Furthermore, this method can be developed as a surveillance or early detection tool for women at high risk for developing breast cancer.
Introduction:
Detection and treatment prior to lymphatic spread is associated with favorable outcomes in breast cancer (1). For example, in the case of women who have estrogen receptor alpha-positive (ERα+) breast cancer and are treated with anti-estrogens, recurrences occur at a steady rate from years 5 to 20 after diagnosis. However, this recurrence is dependent on the status of lymph nodes at the time of diagnosis. Distal recurrence is 13% in women without lymph node involvement but 34% in women with four or more lymph nodes involved (1). Radiologic techniques remain the mainstay of breast cancer detection but these methods detect tumors above 5 mm in diameter (2). Unfortunately, these methods are still not always reliable, with high rate of false positives and false negatives. An examination of the Dutch national registry for the frequency of missed breast cancers in women participating in a high-risk MRI screening program has revealed the limitations of using only radiological techniques for diagnosis. Among 131 breast cancer cases with a negative MRI 0-24 months before cancer detection, 31% of cases had MRI detectable cancers that were missed, whereas 34% of cases showed minimal signs (3). Therefore, complementary molecular assays are needed to improve earlier cancer detection.
Breast milk can be one source of cancer cells as it contains a variety of cell types (4). Cells from breast milk have been cultured and characterized for stemness activity and have been suggested to be a source of cells for regenerative medicine (4,5). Earlier culturing methods favored the propagation of stem/basal and mesenchymal stem cells from the breast milk (5). Since the majority of breast cancers likely originate from luminal progenitor cells (6), earlier methods are not ideal for the expansion of luminal and cancer cells from breast milk for robust characterization. We recently reported a breast epithelial culturing method that did not require feeder layer cells and allowed for the propagation of breast epithelial cells that phenotypically correspond to stem, luminal progenitor and mature/differentiated luminal cells (7).
We initiated this study upon the contribution of breast milk from a patient who was diagnosed with ER+/Progesterone Receptor (PR)-positive (ER+/PR+) cancer in her right breast while breast feeding. We determined whether the above technique can be used to propagate and characterize cancer cells in breast milk and show that breast milk contains cells that display cancer stem cell (CSC) properties and carry mutations that are enriched in breast cancer. Since incidence of breast cancer during pregnancy is 1 in 3000 (8), is expected to increase due to delay in childbearing (9), and breast cancers diagnosed during pregnancy or postpartum tend to be highly metastatic (10), the method described here is useful for rapid characterization of cancer cells in these patients including identification of driver mutations and screening for drugs that may potentially target these mutations.
Materials and Methods:
Breast milk-derived cell culture and donor information.
Breast milk was collected in sterile cups and cultured within an hour of donation directly on 60-mm tissue culture plates pre-coated with conditioned media from 804G cells overnight in 50% culture media along with ROCK, TGFβ and BMP inhibitors as previously described with further addition of Cipro (7). In total, breast milk from 11 women were cultured and characterized. Five of them (all Caucasian) completed a questionnaire, which requested information on the number of pregnancies, the duration of breast-feeding, smoking, and contraceptive use etc. Time of milk collection varied between 1-5 months after delivery. Four women were nursing from their first pregnancy and one was nursing from her second pregnancy. None of the women had a breast cancer diagnosis or known BRCA mutations. Two women had a family history of breast cancer. All of the women had used birth control pills. There were no discernable differences between donors and the age range of donors was 29-37 years.
Cell culture plates with breast milk were washed extensively with PBS the next day and cells were passaged once plates became confluent. We usually observed faster growth of cells from milk from the breasts with abnormalities. Flow cytometry characterization and antibodies used have been described previously (11).
Mammosphere culture, cell proliferation assays, and drug sensitivity studies:
Mammosphere assays were performed using MammoCult media (Stem Cell Technologies) as described previously (12). Cell proliferation rates were measured using Bromodeoxyuridine ELISA by plating 1000 cells in 96 well plates precoated with conditioned media from 804G cells overnight as previously described (12). Sensitivity to drugs was measured after five days of treatment with one media and drug exchange. HDAC6 inhibitor CAY10603 (#S7596), EZH2 inhibitor GSK126 (#S7061), and the BET bromodomain inhibitor JQ1 (#S7110) were purchased from Selleckchem.com. We used an unpaired t test (Graphpad.com) to determine statistical differences in proliferation rate and data from technical replicates are presented although drug sensitivity studies were performed thrice.
RNA and DNA preparation for sequencing:
RNA from biologic triplicates was prepared using the RNAeasy kit from Qiagen and. Total DNA was prepared using the Qiagen DNA mini kit (Cat #51304).
Sequence alignment and gene counts for RNA-seq:
RNA sequencing was performed as previously described (11). The sequencing data were first assessed using FastQC (v.0.11.5, Babraham Bioinformatics, Cambridge, UK) for quality control. All sequenced libraries were mapped to the human genome (UCSC hg38) using the STAR RNA-seq aligner (v.2.5) with the following parameter: “--outSAMmapqUnique 60” (13). The read distribution across the genome was assessed using bamutils (from ngsutils v.0.5.9) (14). Uniquely mapped sequencing reads were assigned to hg38 refGene genes using featureCounts (subread v.1.5.1) with the following parameters: “-s 2 –p –Q 10” (15). Each sample was analyzed independently and genes with read count per million (CPM) < 0.5 in more than 3 replicate samples were removed from the comparisons. The data was normalized using the TMM (trimmed mean of M values) method. Multi-dimensional scaling analysis was done with limma (v.3.38.3) (16). Differential expression analyses were performed using edgeR (v.3.12.1) (16). The false discovery rate (FDR) was computed from p-values using the Benjamini-Hochberg procedure. Table S1 provides detailed RNA-seq data. RNA sequencing results have been deposited to GEO with the accession number GSE157785.
DNA sequencing, alignment, and mutation detection:
Whole genome sequencing libraries were generated with Illumina Nextera DNA Flex Library Prep Kit according to the manufacturer’s instruction. The libraries were sequenced on Illumina NovaSeq 6000 S4 flow cell with 150 bp paired-end reads. Breast milk-derived cell DNA and blood samples were targeted for ~80X and 40X coverage, respectively.
The paired-end sequence reads were first processed to remove Illumine adapter sequences and low-quality base calls with Trim Galore (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/). The resulting high-quality reads were aligned to human reference genome hg38 using BWA-MEM and BWAKIT (v0.7.15, (17) (18). Sentieon version 201911 (Sentieon, Inc, https://www.sentieon.com/) (19) was applied in the following step to identify somatic variants. The sentieon analysis included duplicated reads marking, indel realignment, base quality recalibration (BQSR), and co-realignment of tumor and normal samples. Quality metrics including alignment stats, whole genome sequence metrics and insert size metrics were generated during the process. Somatic variants were detected with the Sentieon TNSeq algorithm. The variants passed filters were next functionally annotated with ANNOVAR using various databases (20).
Copy number variation (CNV) measurement using nanoString:
We used nCounter® v2 Cancer Copy number assay (115000112: XT_PCN_HuV2_Cancercopy-CSO from Nanostring) to determine CNVs in 87 genes commonly deleted or amplified in cancer. The assay was done as per manufacturer recommendations using 300 ngs of genomic DNA. Values 1.6-2.4 are considered normal, 2.6-3.4 considered amplification, whereas 0.4 to 1.4 are considered deletions. For M1 and M2, DNA from the right breast milk-derived cells was compared to DNA from the left breast milk-derived cells, whereas for M10, DNA from the left breast milk-derived cells was compared to DNA from the right breast milk-derived cells. Thus, controls for the assay is from the same individual. Raw numbers of triplicate assays are presented in Table S2.
Study approval:
Breast milk was obtained with informed written consent from the subjects. All experiments were carried out in accordance with the approved guidelines of the Indiana University Institutional Review Board. International Ethical Guidelines for Biomedical Research Involving Human Subjects were followed. All donors consented for genomic sequencing.
Results:
Breast milk from the breast diagnosed with cancer is enriched for cells with CSC properties:
Breast milk was collected from both the breasts prior to initiating any treatment and was cultured using the protocol previously described (7) with the addition of Cipro at 10 μgs/ml to reduce growth of bacteria in milk. Breast epithelial cells grew within a day (Fig. 1A) and cells from both breasts were subjected to flow cytometry using various markers that discriminate stem, luminal progenitor and mature/differentiated cells (11). A schematic view of the study design is shown in Fig. S1A. CD49f+/EpCAM−, CD49f+/EpCAM+ and CD49f−/EpCAM+ cells are considered to enrich for basal/stem, luminal progenitors, and mature/differentiated cells, respectively (21). The CD49f/EpCAM staining pattern revealed the presence of higher levels of CD49f+/EpCAM− basal/stem cells in milk of the breast with cancer (right breast) compared to the milk of the breast without cancer (left breast) (Fig. 1B). Cancer stem cells (CSCs), which are defined based on CD44+/CD24− characteristics (22) and whose presence in breast tumors is associated with unfavorable outcomes (23), were also enriched among cells propagated from milk of the right breast with cancer, although statistical significance was not reached (p=0.07). In general, there was a trend of elevated CSC/progenitor-like features (CD201+/EpCAM+, and CD271+/EpCAM± (24,25)) in cells derived from milk of the breast with cancer compared to milk of the non-cancerous breast. For example, CD271−/EpCAM+ differentiated cells were lower in the right breast milk-derived cells compared to the left milk-derived cells (p=0.0085). Cells grown from milk from the cancerous breast contained a distinct population of CD10+/EpCAM+ cells. While CD10+/EpCAM− and CD10−/EpCAM+ cells are characterized as basal/myoepithelial and luminal cells, respectively (26), characteristics of CD10+/EpCAM+ cells are unknown. There was limited fibroblast growth under our culturing condition as cells were negative for CD140b. Additionally, we noted that while cryopreserved cells from the right breast milk could be grown for more than 15 passages, cells from the left breast milk did not survive for long for additional characterization.
Fig. 1: Cells derived from milk of the breast with cancer show enrichment of cells with CSC phenotype.
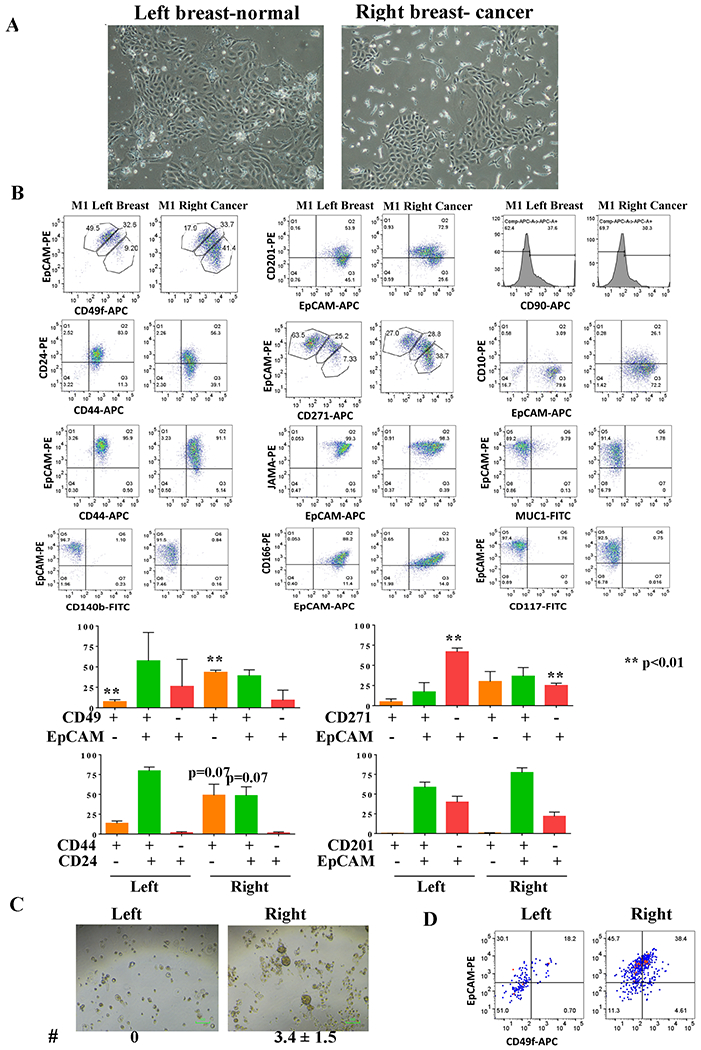
A) Phase contrast images of cells derived from the breast milk. B) Cell surface marker profiles of cells from breast milk without (left) and with (right) cancer. Numerical values of different cell populations from two biological replicates are shown at the bottom. C) Cells from the right breast milk generated larger size mammospheres. D) Mammospheres of cells from the right breast milk were enriched for luminal progenitors (CD49f+/EpCAM+) compared to mammospheres of cells from the left breast milk.
We subjected cells from milk from both breasts to a mammosphere assay to determine whether cells derived from milk from the right breast with cancer show unique properties compared to cells derived from milk from the left breast without cancer. While cells from the left breast milk generated only very small mammospheres (<30 micrometers), larger mammospheres (>50 micrometers) were detected in the right breast milk derived cells (Fig. 1C). Composition of mammospheres was also different as CD49f+/EpCAM+ luminal progenitor cells were enriched in mammospheres of the right breast milk-derived cells compared to mammospheres from cells derived from the left breast milk (Fig. 1D).
The tumor in the right breast of this donor was a 2.1 cm, moderately differentiated, ER+/PR+ (80% nuclear positivity for both), HER2−, grade 2/3, 2/6 lymph node positive, E-cadherin-positive pT2, pN1a invasive ductal carcinoma. E-cadherin positivity indicated that it is not a lobular carcinoma. Treatment consisted of a lumpectomy, followed by chemotherapy, a mastectomy, and radiation. We propagated cells from mastectomy breast tissues. At the time of surgery, there was no evidence of cancer. Flow cytometry showed distinct patterns between the right and left breasts. Cells from the breast with cancer (right) were negative for EpCAM+ epithelial cells but were likely enriched for mesenchymal stem cells as all cells were CD44+ and >75% of cells were CD90+/CD73+. The remaining cells were CD90+/CD73− (Fig. 2). These cells could also be myoepithelial/basal cells as 100% of the cells were CD10+/EpCAM− (26). At the beginning of culturing, there were small patch of epithelial cells, which were rapidly overtaken by mesenchymal stem-like cells. Cells from the left breast milk and tissue-derived cells from the same breast showed some similarities with respect to CD49f/EpCAM, CD271/EpCAM, Jam-A/EpCAM and MUC1/EpCAM staining patterns but displayed uniqueness with respect to CD44/CD24, CD201/EpCAM and CD10/EpCAM staining patterns. These results suggest that not all cell types of the breast are released into breast milk.
Fig. 2: Cells grown from tissues of the breast with cancer after lumpectomy and chemotherapy treatment are enriched for mesenchymal stem-like cells.
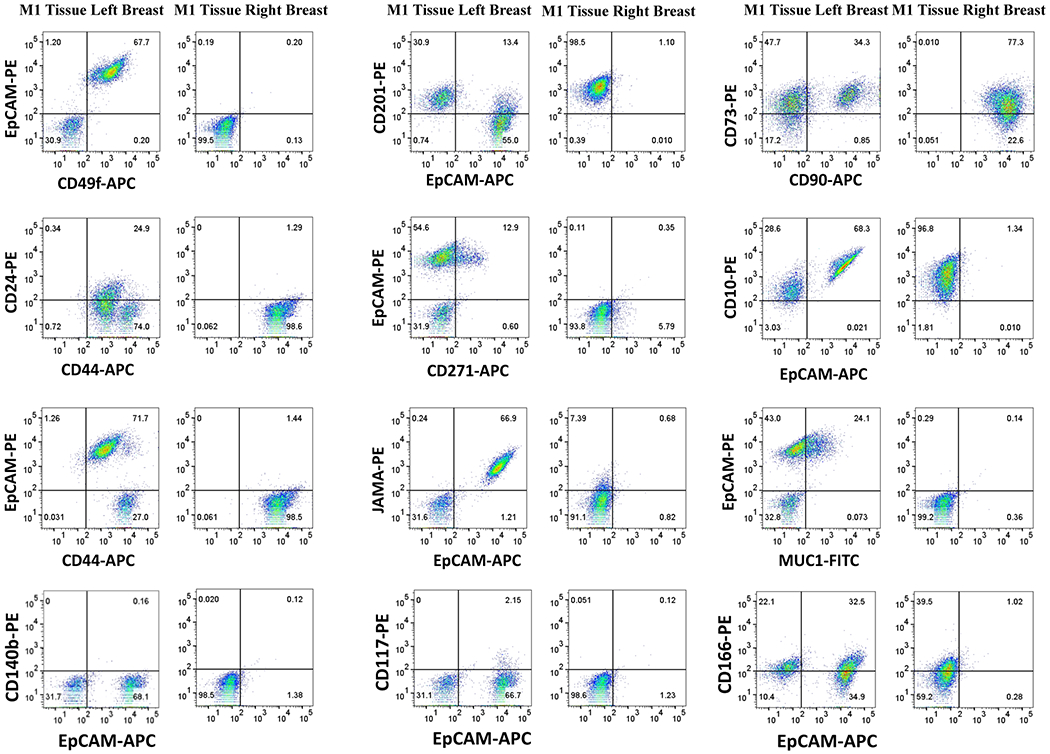
Tissues from mastectomy after successful treatment were cultured under the same condition as milk and resulting cells were characterized for cell surface markers. Cells derived from the right breast had no EpCAM+ cells and likely enriched for mesenchymal stem cells (CD44+, CD90+/CD73+) and/or basal/myoepithelial cells (CD10+/EpCAM−), whereas cells from the left breast had significant levels of CD49f+/EpCAM+ luminal progenitor cells.
In a select few cases, breast milk from women with no known cancer have abnormal cells:
We obtained breast milk from 10 nursing women, propagated cells and characterized cells using flow cytometry. Note that cells from both breasts were cultured for a similar duration and the analyses were done within five passages. Profiles of milk-derived cells from these women are presented in Fig. 3A, Fig. 3B, Fig. 4 and Fig. S1A–S1C, Fig. S2 and Fig. S3. Fig. S1D and S3 show representative isotype controls. Since right breast milk-derived cells from donor 1 showed differences in CD49f/EpCAM, CD44/CD24, CD201/EpCAM, CD271/EpCAM and CD10/EpCAM staining patterns compared to left breast milk-derived cells, we paid particular attention to the presence of these differences among breast milk-derived cells from these 10 donors. Breast milk-derived cells from two donors (M2 and M10) showed differences between the right and the left breast with cells from one of the breasts showing similarity to donor 1 right breast milk-derived cells with cancer. For example, cells derived from the right breast milk of M2 compared to cells from the left breast milk contained significantly higher numbers of CD49f+/EpCAM+ luminal progenitors (68% versus 23%, p=0.0061), CD44+/CD24− (52% versus 21%, p=0.02), and CD271+/EpCAM+ (69% versus 27%), subpopulations (Fig. 3). Although mammospheres generated from cells of both breasts milk of M2 did not show any differences in size, mammospheres generated by cells from the right breast contained higher levels of luminal progenitor cells. By contrast, mammospheres generated from breast milk-derived cells of donors M3 and M5 did not show any differences in luminal progenitor populations between breasts (Fig. S4A). With respect to proliferation rate, cells from the right breast milk of M2 proliferated at a faster rate compared to the left breast milk-derived cells (Fig. S4B). In fact, cells from the right breast milk grew past 15 passages like an established cell line, while cells from the left breast milk ceased to grow by 10 passages.
Fig. 3: Presence of cells enriched for CSC-like properties in breast milk of two donors.
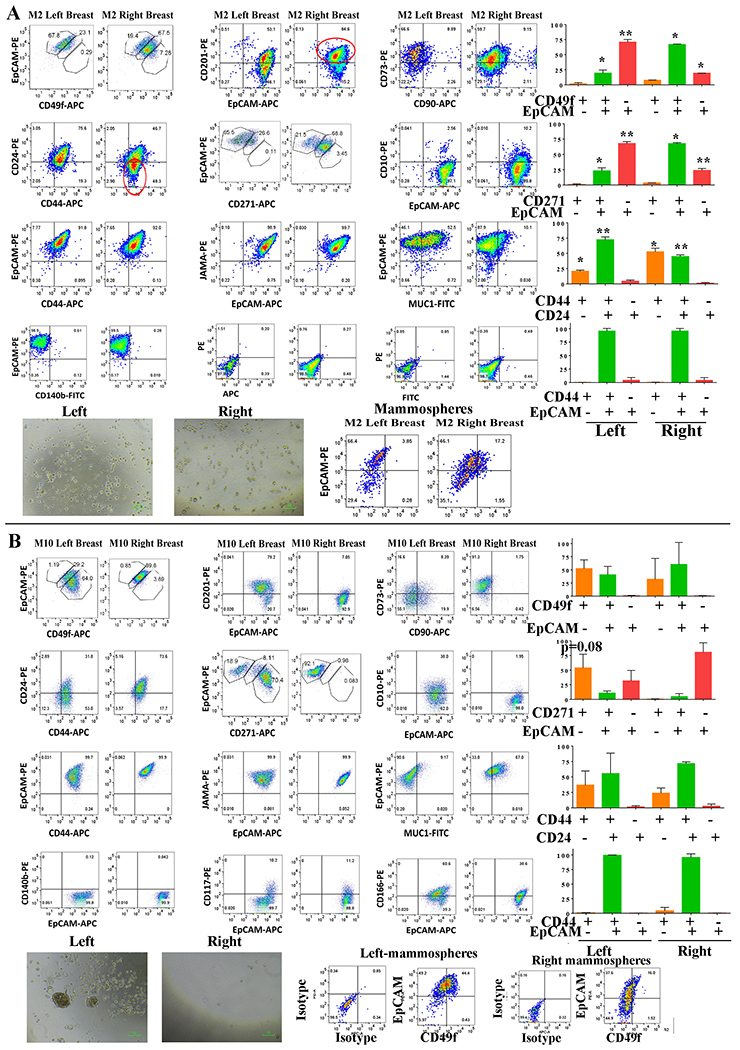
A) Cell surface marker profiles of the left and the right breast milk-derived cells from the donor M2. Although this donor is clinically not known to have breast cancer, cells derived from the right breast milk were enriched for CD49f+/EpCAM+, CD271+/EpCAM+, and CD44+/CD24− cells compared to cells from the left breast milk. Mammospheres generated by cells from the right breast milk showed elevated luminal progenitor cells (CD49f+/EpCAM+) compared to cells from the left breast milk, although there were no differences in size of mammospheres. B) Left breast milk-derived cells of donor M10 display CSC-like properties. Cell surface marker profiling showed a trend towards enrichment of CD271+/EpCAM− subpopulation in the left breast milk-derived cells compared to the right breast milk-derived cells of this donor. Left breast milk-derived cells also generated larger mammospheres and these mammospheres were enriched for luminal progenitor cells (CD49f+/EpCAM+) cells compared to cells from the right breast milk.
Fig. 4: Breast milk-derived cells from the left and the right breasts of not all donors show phenotypic differences.
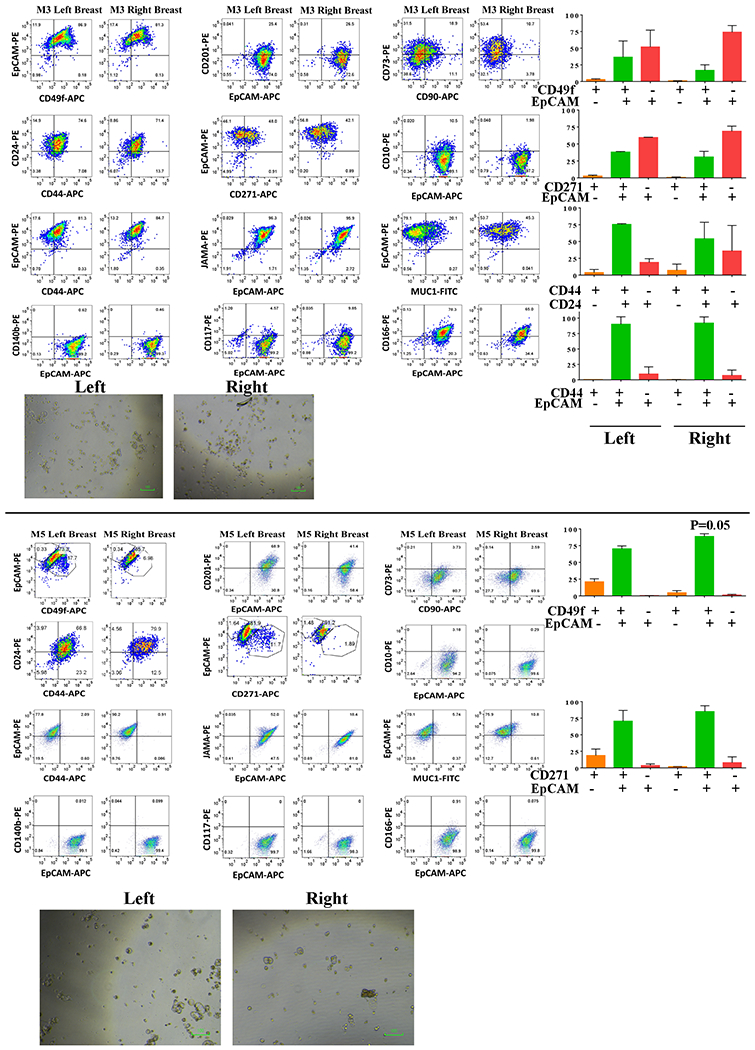
Phenotype of breast epithelial cells derived from breast milk of donors M3 and M5. There were only marginal differences in phenotype and in both cases, the mammosphere size was <30 micrometer.
Cells from the left breast milk and the right breast milk of M10 showed differences in subpopulation of CD49f+/EpCAM−, CD44+/CD24− and CD271+/EpCAM− cells, although none of the differences reached statistical difference (Fig. 3). However, similar to cells derived from the right breast of donor 1, cells from the left breast milk in this donor generated larger mammospheres and these mammospheres were enriched for CD49f+/EpCAM+ luminal progenitor cells compared to cells from the right breast. Thus, a phenotypically different population of cells can be obtained from milk of two breasts of the same woman, which may provide the first indication of abnormalities in one of the breasts. We also noted inter-individual differences in cell subpopulations in breast milk-derived cells, similar to differences we reported with cells propagated from the breast tissue of healthy women (27). For example, while CD271+/EpCAM+ and CD201+/EpCAM+ subpopulations of cells were present at a higher level in milk-derived cells of M5 and M7 (Fig. S2), these subpopulations were lower in M9 (Fig. S3).
Luminal characteristics and differential p63 expression in breast milk-derived cells:
We employed RNA sequencing to determine whether phenotypic changes between breast milk-derived cells of the two breasts of the same individual could be aligned with transcriptomic changes to obtain evidence for the presence of abnormal cells in one of the breasts. RNA sequencing was uninformative as breast milk-derived cells from the right and the left breasts with and without cancer or aberrant cells showed significant differences in gene expression patterns. Supplementary Table S1 provides details of gene expression patterns in the right and the left breast milk-derived cells of six women. MDS plots of all samples are shown in Fig. 5A. Nonetheless, RNA sequencing reconfirmed abundant expression of luminal-differentiated cell enriched keratins such as KRT8, KRT18 and KRT19 in milk-derived cells, which suggests that the method of propagating cells enriches for luminal cells including luminal cancer cells and is not biased towards basal/stem cells (Table S1). Furthermore, breast milk-derived cells expressed abundant levels of luminal markers FOXA1 and GATA3 (Fig. 5B, Ct values of <25 in qRT-PCR). These cells also expressed low levels of ER (Ct value range 29-34), further confirming that our culturing method favors the growth of luminal cells.
Fig. 5: Milk-derived cells express luminal-differentiated cell enriched genes.
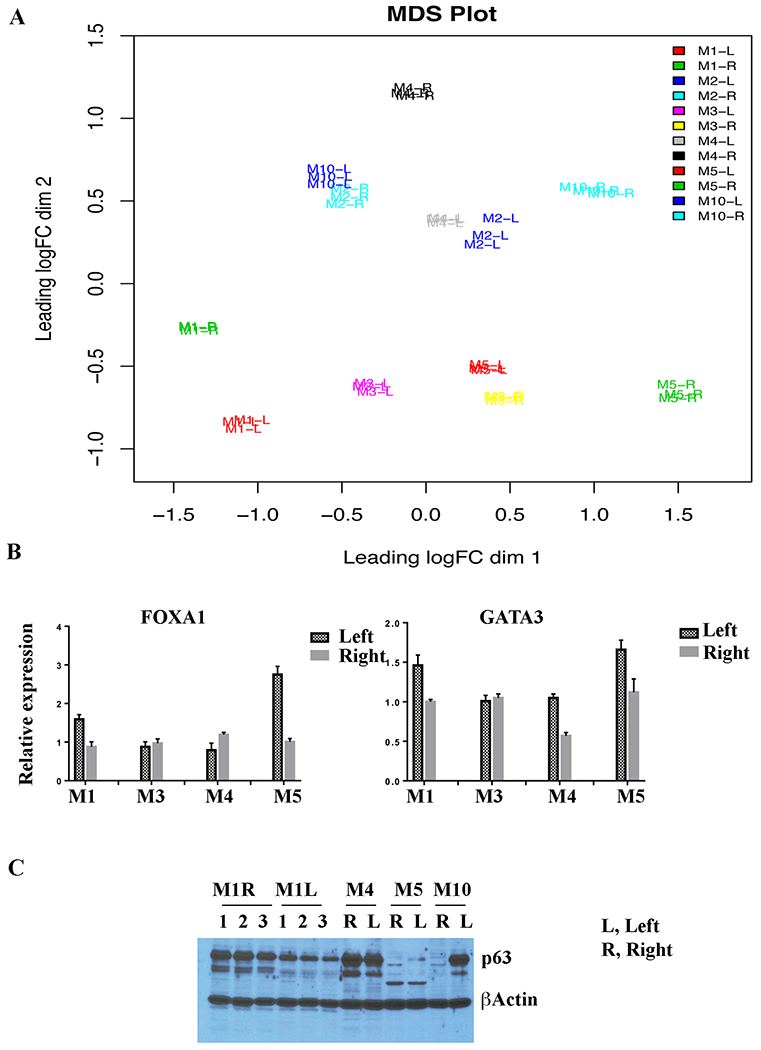
A) MDS plot of RNA-seq data of cells derived from breast milk of six donors. Note RNA expression differences between cells of the right and the left breast milk irrespective of whether one of the breasts had cancer (M1) or phenotypically aberrant cells (M2 and M10). B) qRT-PCR analyses of RNA from milk-derived cells for the expression of FOXA1 and GATA3. RNAs from milk-derived cells were analyzed in biologic triplicates. C) Left breast milk-derived cells of M10 express high levels of stemness-associated protein TP63 compared to the right breast milk-derived cells. TP63 expression was also higher in the right breast milk-derived cells of M1. Cell extracts obtained from three independent batch of cells were analyzed in case of M1.
Others and we had previously demonstrated a close relationship between cancer stem cell phenotype and epithelial to mesenchymal transition (EMT) (28,29). We examined RNA-seq data for differences in the expression of EMT-associated genes between the right and the left breast milk-derived cells. Expression levels of SNAI1, ZEB1, ZEB2, and SOX2 were extremely low, which is consistent with luminal characteristics of cells (Table S1). SNAI2 (Slug) showed two-fold expression differences between the right breast milk and the left breast milk-derived cells in few cases. The major difference was noted with TP63, which controls self-renewal of breast cancer stem cells through sonic hedgehog pathway (30). While two-fold differences in expression between the right and the left breast milk-derived cells were common across all samples, the difference was 36-fold in case of M10 (Table S1). We verified these differences at protein levels. Consistent with mRNA data, M1 right breast milk-derived cells contained 2-fold higher levels of p63 protein (Fig. 5C). In case of M10, cells from the left breast milk, which are phenotypically enriched for cancer stem cells, expressed substantially elevated levels of p63 protein compared to cells from the right breast milk. Thus, phenotypic changes in the left breast milk-derived cells in this donor correlates with increased expression of p63.
Detection of driver mutations in breast milk-derived cells of M1, M2 and M10:
We performed whole genome sequencing of breast milk-derived cell DNA and compared the DNA sequencing results with sequencing data of the corresponding germ line DNA from blood. Summary of the results are presented Fig. 6A. Consistent with a recent report of high level somatic mutations in benign breast tissues with and without proliferative changes (31), significant number of mutations in exonic, intronic, downstream, untranslated regions as well as intergenic regions were noted in all samples, irrespective of whether these samples are enriched for phenotypically cancer stem-like cells (Fig. 6A). However, a closer look at the exonic mutations revealed the presence of mutations that are considered driver mutations in M1-right, M2-right, and M10-left breast milk-derived cells. Schematic view of these mutations is shown in Fig. 6B. For example, M1-right breast milk-derived cells contained deletion mutation in the SE14 domain of HDAC6. SE14 domain of HDAC6 is required for cytoplasmic retention of this protein (32). These cells also contained insertion leading to premature termination of MORF4L1, an epigenetic regulator and a member of the BRCA multiprotein complex involved in DNA repair and tumor suppressor activity (33). A deletion mutation was also observed in SMARCC2 (BAF170), a component of SWI/SNF complex that is defective in 20% of human tumors (34). Cells from the right breast milk of M2 contained frameshift mutations in NF1 gene, a tumor suppressor that controls ERα activity and mutated in breast cancer (35). Cells from the left breast milk of M10 contained an amino acid deletion in KMT2D gene, which is also frequently mutated in breast cancer and is a regulator of ERα activity (36). Analysis of cBioportal database further showed significant alterations of these genes in breast cancer (Fig. 6C) (37).
Fig. 6: Milk-derived cells of M1-right, M2-right and M10-left breasts contain driver mutations.
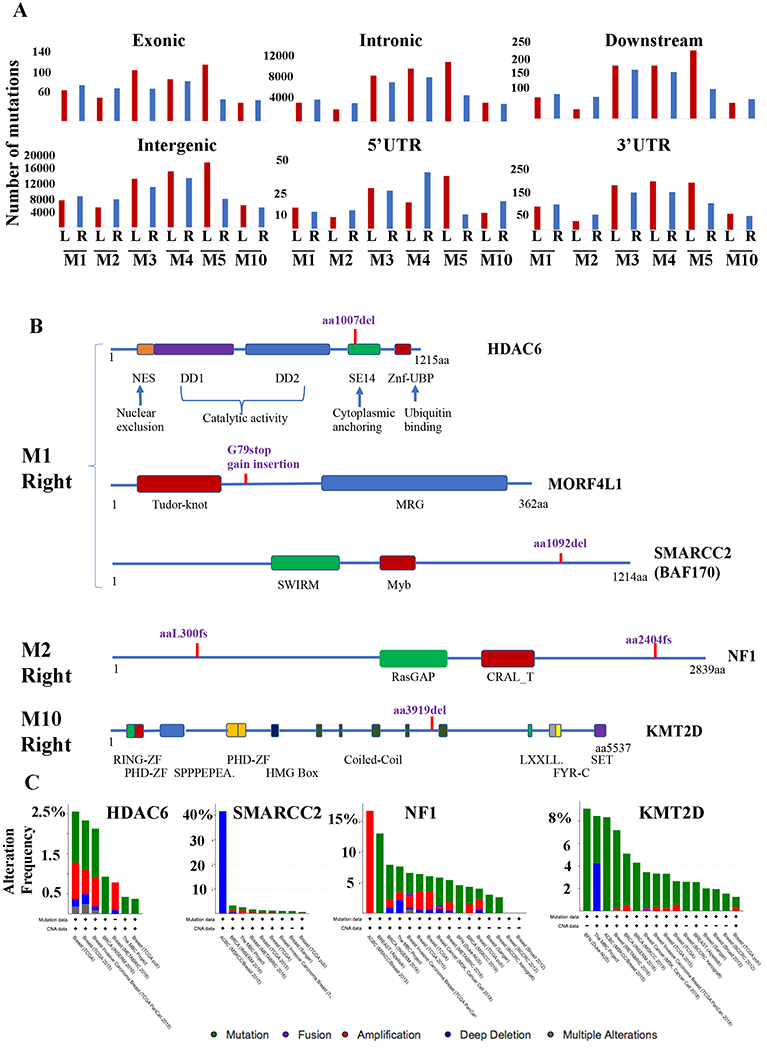
A) Summary of somatic mutation patterns revealed through whole genome sequencing of DNA from indicated samples. DNA from blood of individual donor was sequenced for comparison. Mutation frequency in DNA was similar in breast milk-derived cells with and without enrichment of cells with cancer stem cell phenotype. B) Schematic view of few of the driver mutations observed in cells from M1-right, M2-right and M10-left breast milk-derived cells. Various characterized domains of the corresponding proteins are indicated. C) Frequency of mutations in HDAC6, SMARCC2, NF1 and KMT2D in breast cancer. Data were generated using the cBioportal database.
We performed CNV analysis of 87 cancer-relevant genes using DNA from breast milk-derived cells of donors M1. M2 and M10. Breast milk-derived cell DNA from the breast without aberrant cells from the same donor was used as control for comparison. While the right breast milk-derived cells of M1 and the left breast milk-derived cells of M10 did not carry any CNVs in these 87 genes, the right breast milk-derived cells of M2 showed amplification in CDKN2C, PTEN, and REL (Fig. 7A and Table S2). cBioportal database analysis showed amplification of these genes in 1-3% of breast cancers. Collectively, data presented here demonstrate feasibility of using breast milk-derived cells to identify driver mutations, which can be correlated with phenotypic changes in cells.
Fig. 7: Sensitivity of right breast milk-derived cells of donors M1 and M2 to targeted therapies.
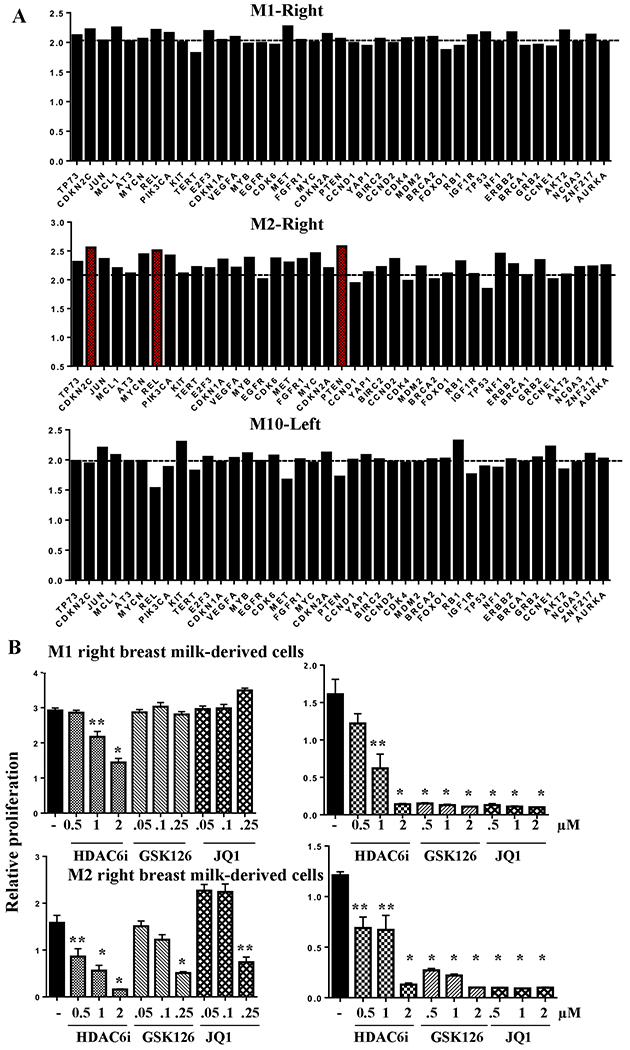
A) CNV analysis of DNA from breast milk-derived cells of M1, M2 and M10. CNVs in DNA from breast milk-derived cells of right of M1, right of M2 and left of M10 were determined by comparing to DNAs from breast milk-derived cells of left of M1, left of M2 and right of M10. Horizontal line indicates normal copy number value of 2. Genes with numbers close to 2.6 or above are considered amplified (indicated in red in case of M2-Right). Raw values for all 87 genes are provided in Table S2. B) Cells were treated with indicated drugs for five days and cell proliferation was measured using bromodeoxyuridine incorporation-ELISA. Results of two independent experiments at different concentrations of GSK126 and JQ1 are shown. Data shown are average and standard errors of six technical replicates. *p<0.001. **<0.02, untreated versus drug treated cells.
Sensitivity of milk-derived cells with driver mutations to targeted therapies.
Prior studies have demonstrated synthetic lethality of SWI/SNF mutated cancers to inhibitors of EZH2 and bromodomains (38,39). Similarly, NF1 mutated tumors are susceptible to bromodomain inhibitors (40). Since cells from the right breast milk of M1 contained mutations in SWI/SNF complex and the right breast milk of M2 had a mutation in NF1 gene, we examined the sensitivity of these cells to EZH2 inhibitor GSK126 and bromodomain inhibitor JQ1. Since cells from right breast milk of M1 contained a mutation in HDAC6 gene, we also tested the effects of the HDAC6 inhibitor CAY10603. Unfortunately, we were unable to use cells from the other breast of the same donors as controls as these cells ceased to proliferate by the time we reached this stage of studies. Cells from the donor M10 also could not be tested because antibiotics used in the culture media failed to suppress milk-resident bacterial growth in this sample. While cells from both donors showed modest sensitivity to the HDAC6 inhibitor, they were extremely sensitive to GSK126 and JQ1 (Fig. 7B). Thus, the assays presented in this study not only allow detection of driver mutations in breast milk-derived cells but also demonstrate the feasibility of using these cells for in vitro screening of potential targeted therapies.
Discussion:
This study, initiated due to foresight of a patient, has established a mechanism to identify and characterize aberrant cells in the breast potentially before a radiologic and/or routine breast exam can detect suspicious abnormalities. Cells from breast milk of a donor with known cancer were enriched with CSCs and contained mutations in genes that are linked to breast cancer initiation and/or progression. In 82% of studies that correlated CSC biomarker expression in tumors with outcomes showed an association of CSC marker enrichment with poor overall survival (23). Since there are no universal CSC biomarkers and characterized biomarkers are associated with different aspects of cancer progression such as drug resistance and metastasis, the method described here would allow for the comprehensive characterization of CSC marker enrichment during early stage of the disease and may potentially aid in monitoring disease progression and making treatment decisions. These phenotypic characterizations coupled with DNA sequencing provided details of earliest lesions associated with breast cancer. For example, in addition to a mutation in HDAC6, which itself is an ERα-regulated gene and mediates estradiol-induced cell migration (41), DNA from breast milk with ER+/PR+ cancer cells showed mutations in SMARCC2 (BAF170). BAF170 is part of the SWI/SNF complex and the activity of ARID1A-SWI/SNF is essential for luminal cell identity and the control of ER activity and anti-estrogen response in breast cancer (42,43). Thus, two major players in ERα functions are aberrant in this cancer.
One in 3000 women are diagnosed with breast cancer during pregnancy or post-partum (8). These cancers tend to be highly metastatic because remodeling of organs such as liver during lactation provides fertile niche for metastatic cells (44). In certain instances, as in the case of donor 1, it is possible to propagate cancer cells from breast milk for genomic sequencing and focused screening of drugs that may target driver mutations. Thus, the method described here is beneficial for characterizing at least pregnancy and post-partum breast cancers.
We observed phenotypic, proliferative and molecular defects in breast milk-derived cells of two other donors. Data presented here are insufficient to draw clinical conclusions and, since this is a research study, it is not designed to go back to clinical evaluation. Since benign breast diseases with and without proliferative changes show driver mutations, including the well-studied PIK3CA-H1047R mutation (31), changes we observed in breast milk-derived cells of these two donors could simply reflect the existence of benign breast disease. A large clinical study in future will likely address this issue.
The method presented here to detect cancer cells is non-invasive, which increases the feasibility of using this method in a surveillance setting. Our previous studies have shown that culturing of primary cells does not introduce mutations (45); therefore, any mutations detected in milk-derived cells are not likely due to culturing artifacts. This method has an inbuilt control (milk from the other breast without aberrations from the same woman), which should help to eliminate any culture-induced artifacts corrupting cancer-specific mutation detection as well as bias attributable to inter-individual variations in the genotype and phenotype of breast epithelial cells.
One minor caveat of the method is the recent observations of somatic mutations in normal tissues, although the mutation load is lower in breast tissue compared to tissues such as lung, thyroid, and esophagus mucosa (46), which can lead to false positive mutation detection. Further refinement of the method such as direct sequencing of DNA from milk similar to ctDNA sequencing in cancer patients (47) and targeted sequencing of catalogued breast cancer mutations used in a precision genomics clinic (48) should further improve accuracy and speed of cancer detection. In this respect, DNA from breast milk has previously been used to determine race- and lactation-duration dependent differences in DNA methylation patterns in the breast (49).
The method described here is particularly useful for screening women who are considered to be at a higher risk of developing breast cancer due to family history, germ line breast cancer risk alleles, and dense breasts that hinder radiologic detection. Additionally, since breast milk is enriched for cells with proliferative capacity, the method would help in cataloguing somatic mutation rates in BRCA1/BRCA2 mutation carriers or any other risk alleles associated with impaired DNA repair pathways.
Supplementary Material
Significance:
Findings describe how a simple method for characterization of cancer cells in pregnancy and postpartum breast cancer can be exploited as a surveillance tool for women at risk of developing breast cancer.
Acknowledgements:
We thank Ms. Emily Nelson for organizing breast milk collection and women who donated breast milk for this research study. We also thank members of the IUSCC flow cytometry core, Medical Genomics core, IUSCC Clinical Trial Office lab and Immunohistochemistry cores for their services. In addition, we thank Drs. Steven D Rhodes and Abbi Elise Smith of Wells Center for Pediatrics Research for assistance in CNV analysis and Mr. Jerid Robinson of Nanostring technology for blinded analysis of CNV data. Zeta Tau Sorority and Susan G. Komen for the Cure (SAC110025) funded this study (to H. Nakshatri). Vera Bradley Foundation Center for Breast Cancer Research supports breast cancer research at Indiana University School of Medicine.
Footnotes
Conflict of Interest: Indiana University is in the process of submitting a patent application
References:
- 1.Pan H, Gray R, Braybrooke J, Davies C, Taylor C, McGale P, et al. 20-Year Risks of Breast-Cancer Recurrence after Stopping Endocrine Therapy at 5 Years. N Engl J Med 2017;377(19):1836–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shaevitch D, Taghipour S, Miller AB, Montgomery N, Harvey B. Tumor size distribution of invasive breast cancers and the sensitivity of screening methods in the Canadian National Breast Screening Study. J Cancer Res Ther 2017;13(3):562–69. [DOI] [PubMed] [Google Scholar]
- 3.Vreemann S, Gubern-Merida A, Lardenoije S, Bult P, Karssemeijer N, Pinker K, et al. The frequency of missed breast cancers in women participating in a high-risk MRI screening program. Breast Cancer Res Treat 2018;169(2):323–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ninkina N, Kukharsky MS, Hewitt MV, Lysikova EA, Skuratovska LN, Deykin AV, et al. Stem cells in human breast milk. Hum Cell 2019;32(3):223–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sani M, Hosseini SM, Salmannejad M, Aleahmad F, Ebrahimi S, Jahanshahi S, et al. Origins of the breast milk-derived cells; an endeavor to find the cell sources. Cell Biol Int 2015;39(5):611–8. [DOI] [PubMed] [Google Scholar]
- 6.Chaffer CL, Weinberg RA. Cancer cell of origin: spotlight on luminal progenitors. Cell Stem Cell 2010;7(3):271–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prasad MS KB, Bhat-Nakshatri P, Anjanappa M, Sandusky G, Miller KD, Storniolo AM, and Nakshatri H. Dual TGFbeta/BMP pathway inhibition enables expansion and characterization of multiple epithelial cell types of the normal and cancerous breast. Molecular Cancer Research 2019;17(7):1556–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee GE, Rosenberg SM, Mayer EL, Borges V, Meyer ME, Schapira L, et al. Contemporary management of breast cancer during pregnancy and subsequent lactation in a multicenter cohort of young women with breast cancer. Breast J 2019;25(6):1104–10. [DOI] [PubMed] [Google Scholar]
- 9.Cottreau CM, Dashevsky I, Andrade SE, Li DK, Nekhlyudov L, Raebel MA, et al. Pregnancy-Associated Cancer: A U.S. Population-Based Study. J Womens Health (Larchmt) 2019;28(2):250–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Callihan EB, Gao D, Jindal S, Lyons TR, Manthey E, Edgerton S, et al. Postpartum diagnosis demonstrates a high risk for metastasis and merits an expanded definition of pregnancy-associated breast cancer. Breast Cancer Res Treat 2013;138(2):549–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumar B, Prasad MS, Bhat-Nakshatri P, Anjanappa M, Kalra M, Marino N, et al. Normal breast-derived epithelial cells with luminal and intrinsic subtype-enriched gene expression document inter-individual differences in their differentiation cascade. Cancer Res 2018;78(17):5107–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhat-Nakshatri P, Goswami CP, Badve S, Sledge GW Jr., Nakshatri H. Identification of FDA-approved drugs targeting breast cancer stem cells along with biomarkers of sensitivity. Scientific reports 2013;3:2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Breese MR, Liu Y. NGSUtils: a software suite for analyzing and manipulating next-generation sequencing datasets. Bioinformatics 2013;29(4):494–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014;30(7):923–30. [DOI] [PubMed] [Google Scholar]
- 16.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43(7):e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25(14):1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.H L. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:13033997v1 q-bioGN 2013. [Google Scholar]
- 19.Kendig KI, Baheti S, Bockol MA, Drucker TM, Hart SN, Heldenbrand JR, et al. Sentieon DNASeq Variant Calling Workflow Demonstrates Strong Computational Performance and Accuracy. Front Genet 2019;10:736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;38(16):e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lim E, Wu D, Pal B, Bouras T, Asselin-Labat ML, Vaillant F, et al. Transcriptome analyses of mouse and human mammary cell subpopulations reveal multiple conserved genes and pathways. Breast Cancer Res 2010;12(2):R21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A 2003;100(7):3983–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lathia J, Liu H, Matei D. The Clinical Impact of Cancer Stem Cells. Oncologist 2020;25(2):123–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang D, Hu X, Liu C, Jia Y, Bai Y, Cai C, et al. Protein C receptor is a therapeutic stem cell target in a distinct group of breast cancers. Cell Res 2019;29(10):832–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim J, Villadsen R, Sorlie T, Fogh L, Gronlund SZ, Fridriksdottir AJ, et al. Tumor initiating but differentiated luminal-like breast cancer cells are highly invasive in the absence of basal-like activity. Proc Natl Acad Sci U S A 2012;109(16):6124–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keller PJ, Arendt LM, Skibinski A, Logvinenko T, Klebba I, Dong S, et al. Defining the cellular precursors to human breast cancer. Proc Natl Acad Sci U S A 2012;109(8):2772–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakshatri H, Anjanappa M, Bhat-Nakshatri P. Ethnicity-Dependent and -Independent Heterogeneity in Healthy Normal Breast Hierarchy Impacts Tumor Characterization. Scientific reports 2015;5:13526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sheridan C, Kishimoto H, Fuchs RK, Mehrotra S, Bhat-Nakshatri P, Turner CH, et al. CD44+/CD24− breast cancer cells exhibit enhanced invasive properties: an early step necessary for metastasis. Breast Cancer Res 2006;8(5):R59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 2008;133(4):704–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Memmi EM, Sanarico AG, Giacobbe A, Peschiaroli A, Frezza V, Cicalese A, et al. p63 Sustains self-renewal of mammary cancer stem cells through regulation of Sonic Hedgehog signaling. Proc Natl Acad Sci U S A 2015;112(11):3499–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zeng Z, Vo A, Li X, Shidfar A, Saldana P, Blanco L, et al. Somatic genetic aberrations in benign breast disease and the risk of subsequent breast cancer. NPJ Breast Cancer 2020;6:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boyault C, Sadoul K, Pabion M, Khochbin S. HDAC6, at the crossroads between cytoskeleton and cell signaling by acetylation and ubiquitination. Oncogene 2007;26(37):5468–76. [DOI] [PubMed] [Google Scholar]
- 33.Martrat G, Maxwell CM, Tominaga E, Porta-de-la-Riva M, Bonifaci N, Gomez-Baldo L, et al. Exploring the link between MORF4L1 and risk of breast cancer. Breast Cancer Res 2011;13(2):R40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chabanon RM, Morel D, Postel-Vinay S. Exploiting epigenetic vulnerabilities in solid tumors: Novel therapeutic opportunities in the treatment of SWI/SNF-defective cancers. Semin Cancer Biol 2020;61:180–98. [DOI] [PubMed] [Google Scholar]
- 35.Zheng ZY, Anurag M, Lei JT, Cao J, Singh P, Peng J, et al. Neurofibromin Is an Estrogen Receptor-alpha Transcriptional Co-repressor in Breast Cancer. Cancer Cell 2020;37(3):387–402 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Toska E, Osmanbeyoglu HU, Castel P, Chan C, Hendrickson RC, Elkabets M, et al. PI3K pathway regulates ER-dependent transcription in breast cancer through the epigenetic regulator KMT2D. Science 2017;355(6331):1324–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer discovery 2012;2(5):401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim KH, Kim W, Howard TP, Vazquez F, Tsherniak A, Wu JN, et al. SWI/SNF-mutant cancers depend on catalytic and non-catalytic activity of EZH2. Nat Med 2015;21(12):1491–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shorstova T, Marques M, Su J, Johnston J, Kleinman CL, Hamel N, et al. SWI/SNF-Compromised Cancers Are Susceptible to Bromodomain Inhibitors. Cancer Res 2019;79(10):2761–74. [DOI] [PubMed] [Google Scholar]
- 40.Patel AJ, Liao CP, Chen Z, Liu C, Wang Y, Le LQ. BET bromodomain inhibition triggers apoptosis of NF1-associated malignant peripheral nerve sheath tumors through Bim induction. Cell reports 2014;6(1):81–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saji S, Kawakami M, Hayashi S, Yoshida N, Hirose M, Horiguchi S, et al. Significance of HDAC6 regulation via estrogen signaling for cell motility and prognosis in estrogen receptor-positive breast cancer. Oncogene 2005;24(28):4531–9. [DOI] [PubMed] [Google Scholar]
- 42.Xu G, Chhangawala S, Cocco E, Razavi P, Cai Y, Otto JE, et al. ARID1A determines luminal identity and therapeutic response in estrogen-receptor-positive breast cancer. Nat Genet 2020;52(2):198–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nagarajan S, Rao SV, Sutton J, Cheeseman D, Dunn S, Papachristou EK, et al. ARID1A influences HDAC1/BRD4 activity, intrinsic proliferative capacity and breast cancer treatment response. Nat Genet 2020;52(2):187–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Borges VF, Lyons TR, Germain D, Schedin P. Postpartum Involution and Cancer: An Opportunity for Targeted Breast Cancer Prevention and Treatments? Cancer Res 2020;80(9):1790–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anjanappa M, Hao Y, Simpson ER, Bhat-Nakshatri P, Nelson JB, Tersey SA, et al. A system for detecting high impact-low frequency mutations in primary tumors and metastases. Oncogene 2018:185–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garcia-Nieto PE, Morrison AJ, Fraser HB. The somatic mutation landscape of the human body. Genome Biol 2019;20(1):298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krebs MG, Metcalf RL, Carter L, Brady G, Blackhall FH, Dive C. Molecular analysis of circulating tumour cells-biology and biomarkers. Nature reviews Clinical oncology 2014;11(3):129–44. [DOI] [PubMed] [Google Scholar]
- 48.Buono G, Gerratana L, Bulfoni M, Provinciali N, Basile D, Giuliano M, et al. Circulating tumor DNA analysis in breast cancer: Is it ready for prime-time? Cancer Treat Rev 2019;73:73–83. [DOI] [PubMed] [Google Scholar]
- 49.Davis Lynn BC, Bodelon C, Pfeiffer RM, Yang HP, Yang HH, Lee M, et al. Differences in Genome-wide DNA Methylation Profiles in Breast Milk by Race and Lactation Duration. Cancer Prev Res (Phila) 2019;12(11):781–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


