Abstract
In free B-DNA, slow (microsecond-to-millisecond) motions that involve equilibrium between Watson–Crick (WC) and Hoogsteen (HG) base-pairing expand the DNA dynamic repertoire that could mediate DNA–protein assemblies. R1ρ relaxation dispersion NMR methods are powerful tools to capture such slow conformational exchanges in solution using 13C/15 N labelled DNA. Here, these approaches were applied to a dodecamer containing a TTAAA element that was assumed to facilitate nucleosome formation. NMR data and inferred exchange parameters assign HG base pairs as the minor, transient conformers specifically observed in three successive A·T base pairs forming the TAA·TTA segment. The abundance of these HG A·T base pairs can be up to 1.2% which is high compared to what has previously been observed. Data analyses support a scenario in which the three adenines undergo non-simultaneous motions despite their spatial proximity, thus optimising the probability of having one HG base pair in the TAA·TTA segment. Finally, revisiting previous NMR data on H2 resonance linewidths on the basis of our results promotes the idea of there being a special propensity of A·T base pairs in TAA·TTA tracts to adopt HG pairing. In summary, this study provides an example of a DNA functional element submitted to slow conformational exchange. More generally, it strengthens the importance of the role of the DNA sequence in modulating its dynamics, over a nano- to milli-second time scale.
Subject terms: Biophysics, Structural biology
Introduction
DNA–protein recognition processes occur through so-called direct and indirect readout of DNA by proteins. The formation of nucleoprotein complexes requires in particular, recognition of DNA chemical patterns specific to each base, and DNA structural and dynamic features that are sequence dependent. Deciphering the dynamics of DNA is not experimentally easy. Nuclear Magnetic Resonance (NMR) has long been, and remains a powerful technique for capturing picosecond dynamics at atomic resolution; more recent developments of relaxation dispersion experiments extend the timescale up to milliseconds and quantitatively investigate slow conformational exchange processes1–4.
Initially, the existence of slow dynamic movements in DNA emerged from early NMR data collected on adenines of TpA steps, by detecting an excess linewidth of adenine H2-resonance protons in biologically active DNA sequences5,6. Further investigations showed that this linewidth broadening was sensitive to the TpA tetranucleotide sequence context6–8. The observation of these resonance experiments was interpreted as being due to a slow exchange between two conformational states7,9,10, which could arise because of poor TpA stacking9,10.
R1ρ relaxation dispersion NMR experiments have led to major advances in this field. These experiments were used to reveal and characterize slow conformational equilibria between major and minor conformational states, assimilated to ground and excited states in analogy with spectroscopy. The use of double 13C and 15N labelled molecules allowed the detection and analysis of excited conformer populations of less than 1%. This approach initially provided insights on various topics essentially related to proteins such as folding, enzymatic catalysis, ligand binding and recognition1,2,11, but now also gives information about intrinsic slow motions of nucleic acids3,12–16. Applied to B-DNA double helices, this methodology revealed an unexpected equilibrium between two schemes of base-pairing: standard Watson–Crick (WC) base pairs being able to transiently adopt the Hoogsteen (HG) configuration15,17–20. Prior to these NMR studies, the first experimental evidence of HG pairing in DNA was obtained in 2002 using a DNA containing 6 A·T base pairs that all crystallized in the HG mode21. Both experimental22 and theoretical23 approaches showed that d(AT)n sequences were globally more stable in the classical WC B-DNA than in a fully HG double helix, at least in dilute aqueous solutions and in the absence of cofactors. However, earlier models24 as well as more sophisticated MD structures15,25,26 demonstrated that HG pairing can coexist with WC base pairs in the same DNA without generating prohibitive energy cost. Thus, the presence of transient HG base pairs in solution, as shown by NMR, is strongly supported by these previous reports.
Relaxation dispersion NMR studies provided quantitative information that covered three different aspects of the WC ↔ HG exchange process, thermodynamic (conformer populations), kinetic (exchange rates) and structural (chemical shifts of the minor conformer). Modulation of this equilibrium by the base pair type (A·T versus G·C) and the sequence surrounding the base pair15,17,27 was established, in agreement with, and as a complement to, analyses of H2-linewidth broadening.
That HG or HG-like T·A in TpA steps are found relatively frequently in X-ray structures of DNA duplexes suggests a biological role for the WC ↔ HG equilibrium27. In addition to their possible involvement through base-pairing, TpA steps also play a role in nucleoprotein complexes as illustrated by the case of nucleosomes whose positioning along eukaryotic genomes is biased by the DNA sequence. Thus, favourable sequences for forming nucleosomes in vivo as well as in vitro are composed in such a way that A·T-rich and G·C-rich minor grooves generally tend to face towards and away from the histone core, respectively28–32. This is particularly the case for the 601 sequence, also called the “Widom sequence”, which is widely used for positioning nucleosomes because of its high affinity for the histone octamer33, further enhanced by an enrichment of additional strategically located TpA steps34–37. To better understand the properties of the 601 sequence we carried out classic NMR experiments37 and modelling38 studying four dodecamers that together cover 39 base pairs of the 5′ half of the 601 sequence. One of these dodecamers contains the TTAAA fragment that, in its ground state, shows a remarkable narrowing of its minor groove37. This structural characteristic is thought to be associated with an enhanced electronegative potential39 that is especially attractive for the histone arginines anchoring the DNA. Given the above, we decided to further extend the exploration of the properties of the dodecamer whose sequence and numbering is given below:
5′- C1 C2 G3 C4 T5 T6 A7 A8 A9 C10G11C12 -3′
3′- G24G23C22G21A20A19T18T17T16G15C14G13 -5′
Here, we will describe NMR experiments, in particular R1ρ relaxation dispersion experiments, that were used to study this dodecamer. Careful analysis and interpretation of the NMR data led to the detection and characterization of slow motions on a patch of 3 successive A·T base pairs in TpA·TpA and ApA·TpT contexts, which participate in WC ↔ HG equilibria. These findings were put into perspective with regard to previous studies in order to discuss the sequence effect on slow motion.
Results
1D spectra of H2 protons
In 1D (Supplementary Fig. S1) and 1D-T1 inversion-recovery (Fig. 1a) spectra, the five H2 protons of A7, A8, A9, A19 and A20 show a very large spectral dispersion compared to those of other non-exchangeable protons. Such dispersion is expected because H2 chemical shifts are extremely sensitive to their sequence environment6–8, notably with high-field shifts specific to TpA adenines9,10.
Figure 1.

Chemical shifts and linewidth of H2 adenine proton resonances. (a) 1D-T1 inversion recovery spectrum of the aromatic region of the unlabelled oligomer at 25 °C in 2H2O, after having selected the inversion recovery delay to only obtain the H2 resonances of adenines. (b) Linewidths (measured at half-width) of H2 adenine proton resonances as a function of temperature. The numbering of the DNA segment of interest is given on the right of the Figure.
Here, the H2 chemical shifts of A7 and A19 are in fact strongly high-field shifted (Fig. 1a) in the unique TpA7·TpA19 complementary dinucleotide of the studied dodecamer (see the sequence in above “Introduction” section). Although these adenines share the same tetranucleotide context (TTA7A·TTA19A), their H2 chemical shifts are clearly different (Fig. 1a). Thus, the sensitivity of H2 chemical shifts to the sequence depends on both 5′ and 3′ nearest neighbours and not only on the 3′ base as previously postulated10,40.
A key point is that the H2 resonances of A7 and A19 are severely broadened compared to those of A8, A9 and A20 (Fig. 1a). These A7 and A19 H2 broadenings are detected below the Tm of 57 °C (Supplementary Fig. S2) and their amplitudes are maximal between 25 and 30 °C (Fig. 1b). Such behaviour of H2 adenine resonances resembles other TpA results that were interpreted as being the signature for conformational motion5–9 occurring at the microsecond-millisecond range41.
C2–H2 cross-peaks were then identified from a constant-time 1H–13C spectrum. Weak intensity and significant broadenings are observed for only A19 and A7 cross-peaks. 1D H2 inversion-recovery and 1D dispersion R1ρ relaxation spectra show that both H2 and C2 resonances of A19 and A7 are involved in these specific cross-peak particularities. So, the motions detected here influence the NMR behaviour of at least these two adenine atoms.
The next step was to apply R1ρ relaxation dispersion experiments to the 13C/15N labelled dodecamer, as a means to follow and describe slow conformational exchanges. The 1D selective R1ρ spectrum of the A9 resonance illustrates the good selectivity of this type of experiment, ensuring that the excitation of a given carbon does not affect its spectrum neighbours (Fig. 2).
Figure 2.
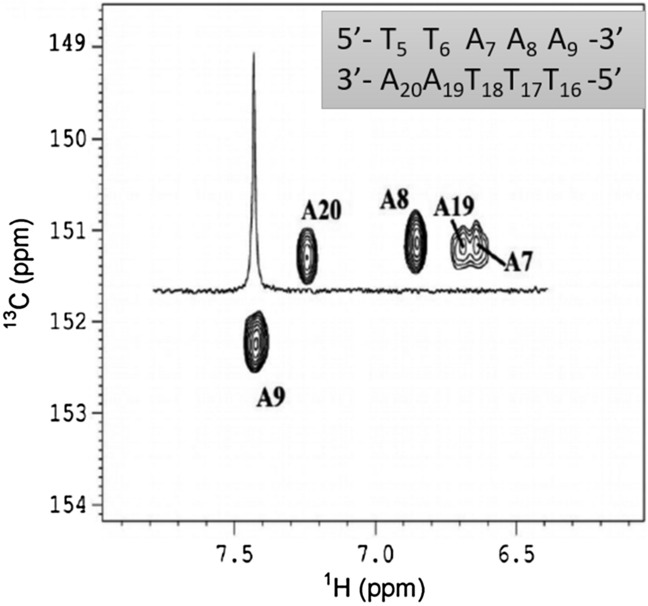
H2–C2 region of adenines of a constant-time HSQC spectrum. The H2–C2 region of adenines of a 13C–1H constant-time HSQC spectrum (600 MHz) was obtained on the labelled oligomer at 25 °C. The 1H 1D spectrum was superposed on the proton frequencies from 1D 13C R1ρ dispersion experiment at the 1H and 13C frequency of A9 signal.
On-resonance 13C R1ρ dispersion relaxation experiments: evidence of slow conformational exchanges
R1ρ relaxation dispersion experiments14 in the on-resonance version constituted a first approach for identifying those carbon atoms of nucleotides submitted to slow conformational exchange. The R1ρ rates were carefully measured to obtain the best estimate of the exponential decreasing 13C magnetization. Typical plots of mono-exponential decays of the type shown in Supplementary Fig. S3 illustrate the quality of the data. The R1ρ profiles measured as a function of effective spin-lock field power were fitted using a two-state model and two variables, the relaxation rate R2 and the exchange rate Rex (see “Materials and Methods”); 500 runs were performed per fit, to obtain R2 and Rex standard deviations.
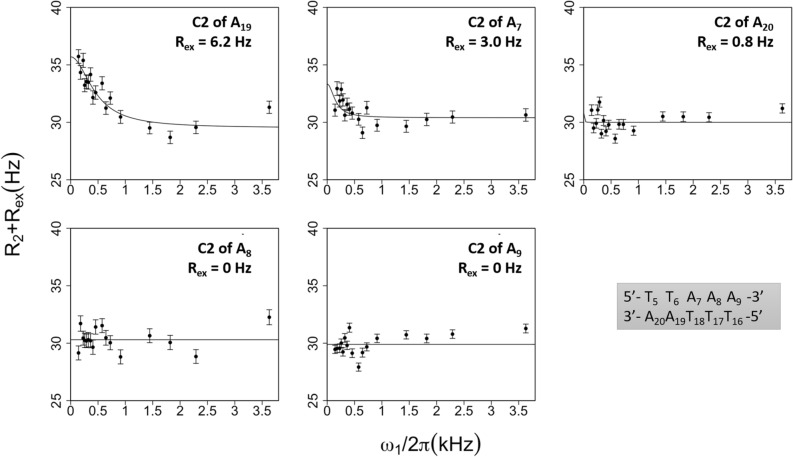
C2 atoms of adenines
The C2 atoms of A7, A8, A9, A19 and A20 (Fig. 3) were studied first. Evidence of a slow conformational exchange on A7 and A19 emerges from the profiles of their R1ρ (= R2 + Rex) rates measured as a function of effective spin-lock field power (Fig. 3). These profiles and their fits show that C2-Rex is higher for A19 than for A7 (Fig. 3 and Supplementary Table S2-1 for both R2 and Rex values). The consistency between these results and those obtained on H2 adenine atoms (Figs. 1 and 2) argues in favour of a major effect of slow motion on H2 and C2 linewidths. The on-resonance data collected on A8, A9 and A20 C2 atoms reveal invariant R1ρ values (Fig. 3) and thus null Rex values (Supplementary Table S2-1).
Figure 3.
On-resonance R1ρ relaxation dispersion profiles of C2 atoms of adenines. R1ρ (= R2 + Rex) rates of the C2 atoms of the five adenines were plotted as a function of the effective spin-lock field power (ω1/2π). The experiments were performed at 25 °C. The two-state model fits (solid lines) were obtained using the protocol described in “Materials and Methods”; (R2 + Rex) standard deviations were calculated from the 500 runs carried out for each fit. The averaged Rex values are specified in each panel. Top: Rex > 0; bottom: Rex ~ 0.
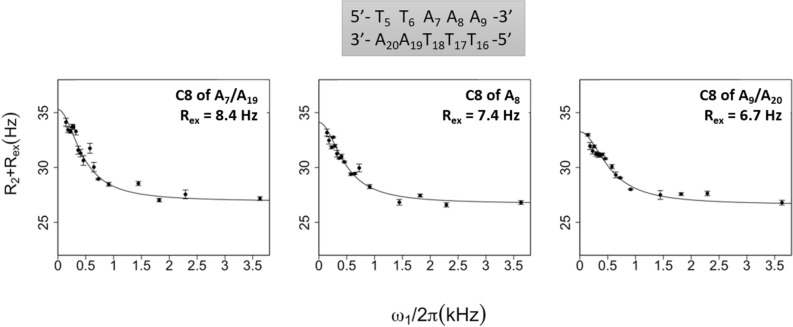
C6 or C8 atoms
The C6/C8 on-resonance NMR signals were resolved for over half of the dodecamer nucleotides, comprising A9, but severe spectral overlaps occurred for several residues, preventing in particular the distinction of A7 from A19, and A9 from A20. The fitting R2 and Rex values for the profiles of R1ρ rates as a function of effective spin-lock field power are given in Supplementary Table S2-2. Thus, the existence of a conformational exchange is attested for A7 and/or A19, A9 and/or A20, and A8 (Fig. 4), with Rex values similar to the highest ones obtained on previous DNA slow motion studies performed in appropriate pH and temperature conditions15,17,18. Such events are excluded or at least much more questionable for the remaining nucleotides that display flat R1ρ profiles (examples in Supplementary Fig. S4) and therefore null or low Rex values (< 2.3 Hz) (Supplementary Table S2-2).
Figure 4.
On-resonance R1ρ relaxation dispersion profiles of adenine C8 atoms. R1ρ (= R2 + Rex) rates of the C8 atoms of adenines were plotted as a function of the effective spin-lock field power (ω1/2π). The experiments were performed at 25 °C. The two-state model fits (solid lines) were obtained using the protocol described in “Materials and Methods”. (R2 + Rex) standard deviations were calculated from the 500 runs carried out for each fit. The averaged Rex values are specified in each panel.
C1′ atoms
A third series of experiments was carried out on the anomeric C1′ atoms. The (R2 + Rex) profiles (Fig. 5) and the R2 and Rex values (Fig. 5 and Supplementary Table S2-3) show that the data related to the C1′ atoms of A7, A8, A9 and A19 are compatible with a slow conformational equilibrium. Among these four adenines, A7 and A19 are associated with especially high Rex values (> 15 Hz) that exceed available measurements15,17 and reflect a relative abundance of excited state3. The flat R1ρ profiles obtained for the C1′ atoms of other nucleotides, comprising thymine partners of adenines, correspond to much lower or null Rex values (Supplementary Table S2-3).
Figure 5.
On-resonance R1ρ relaxation dispersion profiles of adenines C1′ atoms. R1ρ (= R2 + Rex) rates of the C1′ atoms of adenines were plotted as a function of the effective spin-lock field power (ω1/2π). The experiments were performed at 25 °C. The two-state model fits (solid lines) were obtained using the protocol described in “Materials and Methods”. (R2 + Rex) standard deviations were calculated from the 500 runs carried out for each fit. The averaged Rex values are specified in each panel.
When C1′ and C6–C8 relaxation dispersion experiments are available for the same nucleotide, R2 and Rex values are consistent (Supplementary Table S2-2 vs Supplementary Table S2-3). Assuming that this property is true for all the nucleotides, the present relaxation dispersion experiments help to interpret the data subject to C8 atom resonance overlaps which concern in particular the nucleotide couples A7/A19 and A9/A20 (Fig. 4). Thus, both A7 and A19 likely contribute to C8 relaxation dispersion; the Rex values, clearly higher for C1′ of A9 than for C1′ of A20, advocate for a major contribution of A9 to C8-Rex.
In sum
Null Rex values show that there is no slow exchange on C·G base pairs, which is the norm at our pH conditions (pH 6.5)42. Also, the Rex values calculated for the thymines paired with the five adenines do not furnish any robust evidence for motions (Supplementary Table S2). In contrast, non-null Rex values appear on the five dodecamer adenines (Table 1). It is clear that A7 and A19 in T6pA7·T18pA19 are submitted to a slow conformational equilibrium, according to consistent, high Rex values from C2, C8 and C1′ on-resonance experiments and to H2 and C2 linewidth broadenings. Among the three neighbouring adenines, A8 and A9 exhibit signs of dynamic events although the behaviour of their H2 and C2 atoms differ from those of A7 and A19; the case of A20 is much more disputable, given the low Rex value associated with its C1′ atom.
Table 1.
Adenines in T5T6A7A8A9·T16T17T18A19A20 associated with non-null Rex value according to on-resonance relaxation dispersion experiments.
| Atom type | High Rex value | Low Rex value |
|---|---|---|
| C2 | Rex ≥ 2.5 s−1: A7 A19 | – |
| C8 | Rex ≥ 6.7 s−1: (A7 and/or A19) A8 (A9 and/or A20) | – |
| C1’ | Rex ≥ 6.0 s−1: A7 A8 A9 A19 | Rex ≤ 2.5 s−1: A20 |
On-resonance experiments are commonly used to provide information about the presence or absence of slow motion but by themselves they are insufficiently accurate for a viable quantification of the exchange parameters. Consequently, more sophisticated off-resonance experiments were also carried out to extract significant information about the conformational exchange process.
Off-resonance R1ρ dispersion relaxation experiments: characteristics of slow conformational exchange
Off-resonance dispersion relaxation experiments performed at a single magnetic field were used to obtain thermodynamic, kinetic and structural information about conformational exchange. This type of experiment was applied to the C1′ resonances of A7, A8, A9 and A19; the four adenines for which signs of exchange arise from on-resonance dispersion relaxation experiments. Additional investigations focused on T6, C4, G15 and G21, nucleotides associated to very low or null Rex (Supplementary Table S2). C1′ resonances were chosen because they showed large signal to noise ratios and were subject to only a few overlaps so that data could be collected for most nucleotides of the dodecamer (Supplementary Tables S2, S3).
The off-resonance data are compatible with slow motions for A7, A8, and A19 alone (Fig. 6). A9, as well as the other nucleotides are associated with flat (R2 + Rex) profiles (examples in Supplementary Fig. S5). The A7, A8, and A19 (R2 + Rex) profiles were fitted using two distinct methods, an approach that enables to assess the robustness of the resulting parameters. We implemented first a classical analytic, algebraic method15,43–45, described as Method 1 in “Materials and Methods”, performing 1000 runs of calculations for each fit. The same data were also analysed using a completely different method recently published3, called here Method 2, which is based on the numerical integration of the Bloch-McConnell equations. Basically, both methods rely on a two-state exchange model and three variables, the rate exchange (kex), the population of the minor conformer (pE) and the difference between the chemical shifts of major and minor conformers (Δω). Examples of fits with Method 2 are given in Fig. 6.
Figure 6.
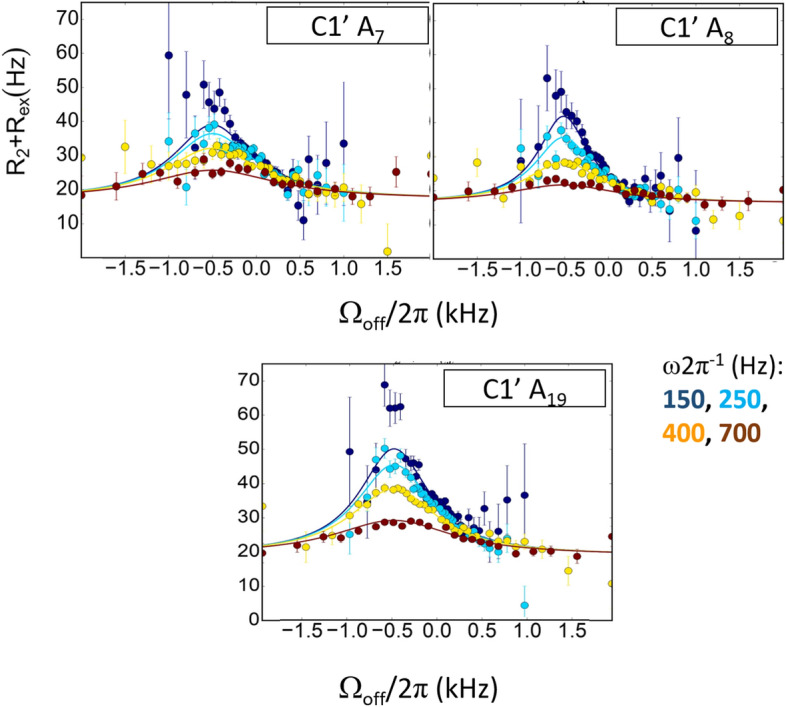
Off-resonance R1ρ relaxation dispersion profiles for C1′ atoms of A7, A8 and A19. R2 + Rex (= R1ρ) values are given as a function of the resonance offset from the major state (Ωoff/2π). Error bars represent experimental uncertainties. The experiments were carried out at four different spin-lock powers (From 150 to 700 Hz, colour code given in the bottom right of the Figure). The fits (solid lines) were performed using Method 2, described in “Materials and Methods”. The resulting exchange parameters are reported in Tables 2 and S3.
Applied to A7, A8 and A19 data, the exchange parameters from either Methods 1 or 2 are remarkably consistent (Tables 2 and S3), in the range of those previously published15,17–20. The rather modest standard deviations associated with the parameter values from Method 1 show the reliability of each run series. Nevertheless, some subtle differences are observed for the A7 exchange obtained from the two methods. Although it is clear that this nucleotide is submitted to a slow exchange, the A7-Δω value and standard deviation (4.16 ± 1.06 ppm) obtained from Method 1 appear too high compared to published data15,17–20. In addition, one expects a substantial pE value, similar to that of A19, given their comparable high Rex values that are primarily sensitive to the excited state population3. For these reasons, we prefer to give more weight to the A7-parameters from Method 2.
Table 2.
A7, A8 and A19 conformational exchange parameters from C1′ off-resonance R1ρ relaxation dispersion experiments.
| C1′ nucleotide | kex (s−1) | pE (%) | Δω (ppm) | |||
|---|---|---|---|---|---|---|
| Method 1 | Method 2 | Method 1 | Method 2 | Method 1 | Method 2 | |
| A7 | 2991 ± 693 | 3309 ± 357 | 0.62 ± 0.19 | 0.80 ± 0.08 | 4.16 ± 1.06 | 3.32 ± 0.16 |
| A8 | 1580 ± 306 | 1994 ± 199 | 0.58 ± 0.09 | 0.59 ± 0.02 | 3.53 ± 0.51 | 3.42 ± 0.1 |
| A19 | 2680 ± 456 | 2872 ± 205 | 1.20 ± 0.01 | 1.00 ± 0.05 | 3.31 ± 0.32 | 3.29 ± 0.09 |
The exchange parameters presented in this Table correspond to the population of the minor conformer (pE), the rate exchange (kex) and the difference between the chemical shifts of major and minor conformers (Δω); they were inferred from individual fits of C1′ off-resonance relaxation dispersion data according to a classical, home-made protocol (Method 1, see “Materials and Methods”) or an approach developed by Al Hashimi’s group (Method 2). Experimental data were collected at 600 MHz, 25 °C and pH 6.5.
The data related to A7 and A19 strengthen the interpretation of H2 and C2 linewidth broadening proposed above from on-resonance results. Indeed, there is now a clear parallel between H2 and C2 broadenings (Figs. 1 and 2), Rex values (Fig. 3) and pE (Table 2), all of these parameters being more accentuated for A19 than for A7. We therefore postulate that H2 and C2 broadenings not only reveal slow motions, but are also correlated with the excited state population. We will see below that this conclusion is very pertinent in the reconsideration of the results of previous 1D NMR experiments.
The case of A9 is intriguing and deserves a short comment. The Rex profiles are either incompatible (off-resonance experiments, Supplementary Fig. S5) or compatible (on-resonance experiments, Fig. 5) with a conformational exchange. This unexpected situation has already been encountered46 and may correspond to a scenario in which the typical limits of slow exchange are no longer valid; in particular, A9 exchange rate could be faster than those of A7, A8 or A19.
The results above were obtained with NMR datasets from only C1′ sugar resonances. Because of numerous resonance overlaps, off-resonance data could only be collected for C8 and C2 atoms of A19, which allowed the constitution of a new extended dataset, involving sugar and base atoms. The fits of this dataset were performed assuming that C1′, C8 and C2 atoms are subjected to the same conformational exchange, with one unique value for kex and one unique value for pE. The resulting values of kex, pE and Δω are remarkably coherent with those obtained from C1′ resonances alone (Table 3), confirming in particular that the population of A19 minor conformer reaches 1.2/1.1%. From a methodological point of view, this approach shows that an off-resonance dataset from only one atom type is sufficient to produce reliable exchange parameters, with the obvious condition that the atom under consideration is implicated in the conformational transition of interest.
Table 3.
A19 conformational exchange parameters from off-resonance R1ρ relaxation dispersion experiments.
| A19 atom(s) | kex (s−1) | pE (%) | Δω (ppm) for C1’ | |||
|---|---|---|---|---|---|---|
| Method 1 | Method 2 | Method 1 | Method 2 | Method 1 | Method 2 | |
| C1′ alone | 2680 ± 456 | 2872 ± 205 | 1.20 ± 0.01 | 1.00 ± 0.05 | 3.31 ± 0.32 | 3.29 ± 0.09 |
| C1′, C8, C2 together | 2671 ± 459 | 3138 ± 228 | 1.20 ± 0.20 | 1.10 ± 0.08 | 3.31 ± 0.31 | 3.31 ± 0.09 |
The exchange parameters presented in this Table correspond to the population of the minor conformer (pE), the rate exchange (kex) and the difference between the chemical shifts of major and minor conformers (Δω); they were inferred from fits of off-resonance relaxation dispersion data according to Methods 1 or 2; the data are those collected from resonances of either C1′ alone or C1′, C8 and C2 together, at 600 MHz, 25 °C and pH 6.5.
The similarities between the two-state exchange characteristics of A7, A8 and A19 (Table 2) could be compatible with simultaneous transitions of the three nucleotides between ground and excited states, implying the transient co-existence of three successive base pair minor conformers. Thus, we completed our analyses by testing this hypothesis of synchronous motions. A collective transition implies that kex and pE are identical for the three nucleotides and these conditions were therefore integrated as restraints in the fit calculations. This model provides reasonable values of conformational exchange parameters (Table 4) that, at first sight, are comparable to those obtained for individual motions (Table 2). Fisher tests were thus performed to evaluate which of the two hypotheses, non-simultaneous or simultaneous motions, led to the best result in terms of χ2. With both Methods 1 and 2, the F-values largely exceed the F-table value (Table 5). In accordance with the p-values that show the statistical significance of the tests, this means without ambiguity that the model assuming individual motions of each adenine is more effective at best representing the off-resonance data.
Table 4.
Conformational exchange parameters from off-resonance R1ρ relaxation dispersion experiments: model of coordinated motions of A7, A8 and A19.
| kex (s−1) | pE (%) | Δω (ppm) | ||||
|---|---|---|---|---|---|---|
| Method 1 | Method 2 | Method 1 | Method 2 | Method 1 | Method 2 | |
| C1′ of A7, A8 and A19 | 2391 ± 299 | 2678 ± 153 | 0.85 ± 0.53 | 0.85 ± 0.11 | 3.32 ± 0.43 | 2.83 ± 0.11 |
| 3.09 ± 0.33 | 2.59 ± 0.07 | |||||
| 3.71 ± 0.26 | 3.70 ± 0.09 | |||||
The exchange parameters presented in this Table were produced by fitting a model in which the three nucleotides undergo coordinated motions; this hypothesis implies identical kex and pE for the three nucleotides.
Table 5.
Comparison between fits assuming coordinated or uncoordinated motion models for A7, A8 and A19.
| Degree of freedom | F-table value | F-value | p-value | |
|---|---|---|---|---|
| Method 1 | 417 | 2.39 | 126 | 1.7 10–70 |
| Method 2 | 48 | 2 10–33 |
Statistical F-test using χ2 and F-distribution analysis was performed to compare the fits based on models where A7, A8 and A19 undergo either coordinated, collective or uncoordinated, individual transitions; p-values validate the results.
Thus, the interpretation of the off-resonance dispersion relaxation experiments draws a picture of a block of three specific adenines that undergo slow motions between two states, the excited one representing from 0.6 to 1.2% of the conformers. While the three adenines are clustered in the T6A7A8·T17T18A19 segment, they adopt a non-synchronous regime of motions.
Nature of the excited conformer
As mentioned above, both on- and off-resonance dispersion relaxation experiments and the inferred exchange parameters agree remarkably well with those obtained by Al Hashimi’s group within B-DNA sequences15,17–20. The asymmetry of the NMR data in A·T base pairs, in which evidence of slow dynamics is observable on adenines but not on their thymine partners, is also in line with these earlier results. It is now accepted that these NMR-based characteristics correspond to a dynamic equilibrium of particular base pairs between the canonical Watson–Crick (WC) conformation and an excited, short-lived state of low abundance, which is the Hoogsteen (HG) pairing3,15,18,19. HG base-pairing corresponds to N7purine − N3pyrimidine and N6/O6purine − O4/N4pyrimidine hydrogen bonds, which imply purine and pyrimidine nucleotides in syn- and anti-configurations, respectively. Thus, the WC → HG transition does not dramatically affect the pyrimidine but requires one major change, i.e. the slow motion of the purine around the glycosidic angle χ. Here, we can confidently attribute a WC ↔ HG equilibrium to the three successive A·T base pairs composing the T6A7A8·T17T18A19 segment. It should be underlined that the HG percentages calculated here for A7, A8 and A19 are among the highest measured populations15,17–20.
In addition to the purine rotation, the WC → HG transition is accompanied by some changes in sugar pucker and backbone torsion angles, which likely optimize hydrogen-bonding and stacking with neighbours19,27,42. In particular, a study of either WC or HG A·T containing oligomers showed that 31P signals are up-field shifted in and around HG A·T base pairs compared to their WC counterparts42. Such shifts are usually interpreted as being due to the presence of more BI conformers47,48.
Examining high-resolution X-ray structures of free DNA suggests that another backbone alteration could also occur on α/γ angles, as briefly previously mentioned27. Indeed, structures containing exclusively HG pairings (PDB codes 1GQU and 1RSB, of 2.5 and 2.2 Å of resolution, respectively) enclose TpA junctions adopting unusual α/γ:g+ /g− angles, instead of the canonical g-/g+ configuration. Unusual α/γ angles are infrequent in WC B-DNA X-ray structures49,50 because they are energetically very costly to generate50. Nevertheless, as rare as they are, these atypical α/γ conformations are mainly encountered in ApA steps or A·T-rich contexts (Supplementary Table S4). So, the potential ability of A·T-rich patches to adopt unusual α/γ conformations could be a factor promoting the emergence of HG base pairs.
Sequence effect on TpA steps
The results presented above demonstrate that adenine H2 resonances associated with linewidth broadenings reflect the existence of a slow conformational exchange, as previously postulated7,9,10. However, any adenine submitted to slow motions is not associated with such anomalies that exclusively arise on TpA adenines. Accordingly, H2 linewidth broadenings are observed in the dodecamer on only those adenines belonging to TpA, A7 and A19 (Fig. 1). This specific phenomenon may be related to the especially high-field shifted H2 resonance of the major conformer (Fig. 1) that likely maximizes Δω, and consequently Rex—recalling that a high Rex broadens the resonance.
This intimate connection between H2 linewidth broadening and slow motion allows the re-examination of earlier NMR data. A study reported measurements of H2 linewidth broadenings on 14 oligomers containing the 16 possible immediate sequence contexts of TpA steps (i.e. NTAN)7,8. This systematic approach showed that broadening of H2 occurs in diverse TpA environments, the maximal values corresponding to the central adenines of (Y/R)TAA tetramer fragments (Table 6).
Table 6.
TpA steps associated with large H2 linewidth broadenings in the literature.
| References | Oligomer sequence | A in TpA with max H2 linewidth > 15 Hz |
|---|---|---|
| McAteer et al. 1995 | CTTTA1AATTTA2AAG | TTA1A (~ 15 Hz) and TTA2A (~ 19 Hz) |
| CTTTACATGTAAAG | GTAA | |
| CTTTAGATCTAAAG | CTAA | |
| CTTTATATATAAAG | ATAA | |
| McAteer et al. 2000 | GCTTATATAAGC | ATAA |
| GCATACGTATGC | GTAT | |
| GCTTAGCTAAGC | CTAA | |
| GCTTA1ATTA2AGC | TTA1A (~ 18 Hz) and TTA2A (~ 22 Hz) | |
| GCTTACGTAAGC | GTAA |
This table reports the sequences studied by Kennedy’s group in which the H2 linewidth broadening of one or two adenines (in bold, underlined) in TpA steps is equal or exceeds 15 Hz at 25 °C. The immediate environment of such TpA is reported in the last column. Note that all sequences are palindromic.
However, considering the nearest neighbour is insufficient to explain the slight but significant disparity of H2 linewidth broadenings of the TpA adenines that are in the same tetrameric environment in CTTTAAATTTAAAG8 and GCTTAATTAAGC7 (Table 6). Similarly, H2 broadening (Fig. 1) and slow motion characteristics differ between A7 and A19 in TTA7AA·TTA19A, for instance the population of the excited state (pE) (Table 2). These differences suggest a subtle, long-range influence of the sequence on the DNA’s ability to undergo WC ↔ HG transitions. At this stage, the only certitude is that slow DNA motions are modulated at dinucleotide and tetranucleotide levels, as previously established in solution for nano-second dynamics37,47,51.
Discussion
This NMR study focused on a dodecamer containing the TTAAA·TTTAA fragment that was assumed to be an element facilitating histone anchoring upon nucleosome formation34–37. Slow conformational exchanges in the dodecamer were revealed and characterized from R1ρ relaxation dispersion experiments. These NMR approaches also allowed a re-examination of classical 1D and 1D-T1 inversion-recovery experiments and to demonstrate the relationship between H2 linewidth broadenings and slow motions. Given that linewidth is easily observable and quantifiable, it represents an interesting means of detecting nucleotide slow dynamics, even if it appears to be limited to TpA adenine H2 protons.
As expected, most nucleotides of the dodecamer do not show consistent, conclusive signs of any slow motion. However, A7, A8, and A19 in the TA7A8·TTA19 fragment are specifically subjected to conformational exchange in the milli-second time range. The corresponding exchange parameters perfectly match the signature of WC ↔ HG base pair equilibrium as published by Al-Hashimi’s group15,17–20. Thus, we can confidently postulate that the TA7A8·TTA19 fragment is composed of three base pairs that transiently flip toward the HG conformation.
Although the crystallographic form of the ATTAAT·ATTAAT hexamer52 attests for the co-existence of three or more successive HG base pairs, our analyses agree with non-simultaneous WC ↔ HG transitions of the three A·T base pairs, which does not exclude a certain degree of cooperation between the motions. For instance, that one HG base pair locally destabilizes the double helix42 could favour a WC → HG flip of its closest neighbours. In any case, individual motions have the effect of magnifying the time during which one HG base pair is present in the TAA·TTA tract. Thus, according to the individual HG populations calculated here (~ 1% for each of the three HG base pairs), and given the non-simultaneous character of the motions highlighted by our fits, 3% of TAA·TTA fragment contains one HG A·T base pair.
This relatively high occurrence underlines the relevance of the question of an eventual function of transient HG base pairs in the nucleosome context, remembering that the TTAAA element likely helps histone anchoring. The first idea is that DNA wrapping could involve HG base pairs but our analyses failed to detect such pairing in high-resolution X-ray nucleosome structures, conversely to what was observed on several other types of DNA–protein complexes27. Another proposal relates to the exploitation of shape features specific to HG base pairs upon nucleosome formation. To our knowledge, there is only one structure ensemble based on NMR data that was collected on an oligomer containing one HG m1A·T base pair in the Am1ACC·GGTT environment42. This oligomer has as a main characteristic a major-groove kink. Indeed, analyses of X-ray structures of DNA bound to proteins or small molecules suggested that such major-groove curvature is induced by any HG base pair in any context27,42. This structural particularity cannot be considered as a favourable pre-organization for the nucleosome since TTAAA bound to the histone octamer presents a minor groove curvature34,53. In addition to the determination of the structure of our HG oligomer itself, another factor such as long-range consequence of HG base pair could be investigated. It has to be borne in mind that, at this point in time, there is no clear indication of how those HG base pairs present in the TAA element impact on nucleosome structure or dynamics. Indeed, it is now widely accepted that nucleosome assembly is primarily modulated by sequences that introduce structural variability for helical parameters such as roll or slide along the free WC B-DNA.
The percentage of HG conformers of the three A·T base pairs in T6A7A8·T17T18A19 is unusually high (~ 1%) for such minor, short-lived states that very rarely exceed 0.5%15,17–20. Multiple examples of enhanced dynamics on TA·TA adenines in the same TAA·TTA context emerge when one considers studies reporting linewidth broadenings (Table 4)7,8. However, the structural foundation for this specific, sequence dependent, stabilisation of HG base pairs remains unclear and poorly documented. One can only formulate hypotheses, being aware that such speculations will require future investigation. A first point concerns atypical features observed in A·T containing sequences. Thus, the photo reactivities of TT steps in TTAA·TTAA leading to T^T dimers are clearly outsized compared to those of any other dinucleotides, comprising other TT steps53; this particularity is added to (but cannot be totally explained by) the TTAA·TTAA marked positive rolls and low twists attested by both NMR37,54 and simulation55 data. Also, as highlighted in one of the above sections, A·T rich sequences show a specific propensity to adopt unusual α/γ backbone angle conformations (Supplementary Table S4). Such unusual structural elements could destabilize the WC form of A·T, favour the flipping-out of adenine from the double helix via the major groove15,56, which preludes WC ↔ HG conversions, and, in the case of α/γ angles, stabilize the HG base pair. Another suggestion emerges from the X-ray structure of the DNA bound to the MATα2 homeodomain57. In this complex, the T6A7A8·T36T37A38 fragment contains one HG base pair, A7·T37, in which A7 engages two hydrogen bonds with T37 and T36. A similar way to increase the stability of HG conformers may occur in any TAA·TTA fragment, in the present case via hydrogen bonds between A7A8 and T16T17T18 on the one hand and between A19 and T5T6 on the other.
Indeed, the effect of sequence in WC ↔ HG transition whilst incontestable remains complex and thus so far is only partially elucidated as already noted17. Further investigations are clearly required to clarify in particular, the effect of the tetra or even hexanucleotide contexts on the HG population. This topic is in fact essential for capturing the multiple aspects of DNA double helix functional versatility. A lot of information about nano-second dynamics keeping WC pairing intact is already available; these are primarily sensitive to the dinucleotide sequence while modulated at the tetramer level, as shown by experimental X-ray49,58,59 or NMR data47,48,51,54 and modelling38,55,60. NMR data in particular revealed that such rapid motions are especially enhanced in G·C rich elements47,51,54. Several examples demonstrated their importance in the structural adjustment of DNA to its protein partners, transcription factors61–63 or other proteins37,51,64. Beyond what occurs on WC G·C rich sequences at short timescales, A·T rich elements could be more specialized in milli-second conformational transitions, with the possibility of generating regions of Hoogsteen base-pair hot-spots that could play a topological role in genomic DNA27.
Materials and methods
Samples and resonance assignments
Samples were purchased as single-stranded oligonucleotides 5′-CCGCTTAAACGC-3′ and 5′-GCGTTTAAGCGG-3′ from Eurogentec (Belgium) for unlabelled DNAs or from Eurisotop (France) for fully 15N/13C–labeled DNAs. The two complementary strands were resuspended in 66 mM sodium phosphate buffer with 0.1 mM EDTA, for a total ionic strength of 0.1 M at pH 6.5; they were then mixed with a 1:1 ratio in 450 μL H2O. In a next step, the samples were lyophilized three times in 99.99% 2H2O. The final concentration of the unlabelled and labelled duplexes were 1.2 mM and 0.85 mM respectively in volumes of 500 μL and 180 μL respectively.
The full assignment of 1H and 13C resonances of the dodecamer were previously described37,54. To complete the carbon assignments and notably to obtain those of the quaternary carbons of bases, we performed additional experiments on a Bruker Avance spectrometer equipped with a 5-mm triple-resonance cryogenic probe, at 600 MHz frequency or 150 MHz for 13C experiments: (i) constant time 1H-13C HSQC optimized separately for aromatic and aliphatic carbons, C2 of adenine, C8 of purines, C5 and C6 of pyrimidines and C1′ of any base and, (ii) for the quaternary carbons C4, C5, C6 of adenines, 3D TROSY relayed HCCH-COSY65 according to Hansen et al.14.
R1ρ relaxation dispersion experiments
All experiments were performed on the same spectrometer as used for assignments, at pH 6.5 and 25 °C. As specified in the Results section, the linewidth broadening of H2-proton resonances is maximal at this temperature. 1D selective 13C R1ρ pulse was applied as previously described14,16. The spin-lock powers (ω) needed to be carefully controlled, and were calibrated accordingly14. On-and off-resonance experiments were performed on C2, C6/C8 and C1′ atoms of the 13C/15N-labeled DNA. On-resonance data were recorded at various ω, from ~ 100 to 3,500 Hz; off-resonance data were collected at various spin-lock offset frequencies (Ω) and at three or four different spin-lock powers (ω). Details of these experiments are given in Supplementary Table S1. Ten delays were used to determine Trelax, the monoexponential decrease of the magnetization: 0, 4, 8, 12, 12, 16, 20, 24, 32, 32 ms for C6, C8 and C2 carbons and 0, 4, 8, 12, 12, 18, 26, 34, 42 and 42 ms for C1′ carbons.
Note that the duplication of two delays in each experiment allowed an evaluation of errors in the measurement of the peak heights. These errors were subsequently used in Monte Carlo simulations to determine R1ρ uncertainties. The data corresponding to Hartmann-Hahn matching were omitted from the fits of on-and off-resonance measurements as previously described14. The 1D dispersion data were processed using NMRPipe66.
Equilibrium parameters from R1ρ relaxation dispersion experiments
Our approach assumed a two–state equilibrium between a ground state G and a minor, excited state E in which pG and pE, the G and E populations, are strongly asymmetric: pG ≫ pE. The equilibrium parameters that can be extracted from relaxation dispersion experiments are pG, pE, kex the exchange rate, ΔωGE the difference between the frequencies of G and E, and Rex, the quantity added to the relaxation rate due to exchange, which depends on pG, pE, kex and ΔωGE.
On-resonance experiments
Rex was obtained by fitting the profiles of data from on-resonance experiments carried out in function of spin-lock offset powers (ω). In these experiments, θ, the angle between the effective field and the z axis, has the particular value of π/2. The fit is based on Eq. (1), a version of Eq. (2) that is simplified by using θ = π/2 to represent the condition of on-resonance experiments. For each fit, 500 runs were performed using R1ρ values.
| 1 |
R1 and R2 are the intrinsic longitudinal and transverse relaxation rates, respectively, which are supposed to be identical for the two-states G and E; ω1 is the spin-lock power.
Off-resonance experiments
The profiles of the off-resonance relaxation dispersion data measured in function of spin-lock offset frequencies (Ω) and power (ω) were fitted by two methods described below.
Method 1 is based on the Eq. (2)15,43–45.
| 2 |
In this case, restraints were applied on R2: R2 ≥ 16 s−1 and R2 ≤ 19 s−1, according to the range inferred from on-resonance experiments; ΩE and ΩG are the resonance offsets for the excited and the ground states E and G, respectively; ΔωGE = ΩE − ΩG;
ω1 is the spin-lock power related to by the relation
kex = kG + kE is the exchange rate with kG = pEkex and kE = pGkex, kG and kE representing the forward and reverse rate constants, respectively.
For each fit, 1000 runs were performed using R1ρ values. The R1ρ values from off-resonance experiments were systematically and randomly varied using standard deviations derived from the fits of the exponentially decreasing intensities of the peaks as function of the relaxation time. This protocol implemented using home-based Python and R scripts enables to derive the errors associated to each exchange parameter according to a strategy described previously67.
Method 2 used the “Bloch-McConnell Numerical Simulator” (BMNS) developed by Al-Hashimi’s group3, which is exclusively based on a two-state exchange model. The calculations were performed with the same constraints on the R2 relaxation rates than for the fits described above.
Supplementary information
Acknowledgements
Dr Al-Hashimi and Dr Kimsey are warmly thanked for having helped in the development of dispersion relaxation methods in our laboratory. The authors also thank the NMR team in Gif/Yvette, in particular Carine van Heijenoort and Nelly Morellet, for instructive discussions.
Author contributions
A.B.I. and L.Z. performed the NMR experiments under the supervision of O.M., the leader project. A.B.I. and O.M. carried out most analyses and wrote the original draft. A.B.I., O.M. and B.H. prepared all the figures. B.H. participated to some analyses and wrote the final manuscript with M.B. All authors reviewed the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-75645-x.
References
- 1.Mittermaier AK, Kay LE. Observing biological dynamics at atomic resolution using NMR. Trends Biochem. Sci. 2009;34:601–611. doi: 10.1016/j.tibs.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 2.Palmer AG. Chemical exchange in biomacromolecules: past, present, and future. J. Magn. Reson. 2014;241:3–17. doi: 10.1016/j.jmr.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rangadurai A, Szymaski ES, Kimsey IJ, Shi H, Al-Hashimi HM. Characterizing micro-to-millisecond chemical exchange in nucleic acids using off-resonance R1ρ relaxation dispersion. Prog. Nucl. Magn. Reson. Spectrosc. 2019;112–113:55–102. doi: 10.1016/j.pnmrs.2019.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walinda E, Morimoto D, Sugase K. Overview of relaxation dispersion NMR spectroscopy to study protein dynamics and protein-ligand interactions. Curr. Protoc. Protein Sci. 2018;92:e57. doi: 10.1002/cpps.57. [DOI] [PubMed] [Google Scholar]
- 5.Lefevre JF, Lane AN, Jardetzky O. A temperature dependent transition in the Pribnow box of the trp promoter. FEBS Lett. 1985;190:37–40. doi: 10.1016/0014-5793(85)80422-1. [DOI] [PubMed] [Google Scholar]
- 6.Lefevre JF, Lane AN, Jardetzky O. A description of conformational transitions in the Pribnow box of the trp promoter of Escherichia coli. Biochemistry. 1988;27:1086–1094. doi: 10.1021/bi00404a002. [DOI] [PubMed] [Google Scholar]
- 7.McAteer K, Kennedy MA. NMR evidence for base dynamics at all TpA steps in DNA. J. Biomol. Struct. Dyn. 2000;17:1001–1009. doi: 10.1080/07391102.2000.10506588. [DOI] [PubMed] [Google Scholar]
- 8.McAteer K, Ellis PD, Kennedy MA. The effects of sequence context on base dynamics at TpA steps in DNA studied by NMR. Nucleic Acids Res. 1995;23:3962–3966. doi: 10.1093/nar/23.19.3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kennedy MA, Nuutero ST, Davis JT, Drobny GP, Reid BR. Mobility at the TpA cleavage site in the T3A3-containing AhaIII and PmeI restriction sequences. Biochemistry. 1993;32:8022–8035. doi: 10.1021/bi00082a025. [DOI] [PubMed] [Google Scholar]
- 10.Lingbeck J, et al. Effect of adenine methylation on the structure and dynamics of TpA steps in DNA: NMR structure determination of [d(CGAGGTTTAAACCTCG)]2 and its A9-methylated derivative at 750 MHz. Biochemistry. 1996;35:719–734. doi: 10.1021/bi951364k. [DOI] [PubMed] [Google Scholar]
- 11.Palmer AG. Enzyme dynamics from NMR spectroscopy. Acc. Chem. Res. 2015;48:457–465. doi: 10.1021/ar500340a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baronti L, et al. Base-pair conformational switch modulates miR-34a targeting of Sirt1 mRNA. Nature. 2020;583:139–144. doi: 10.1038/s41586-020-2336-3. [DOI] [PubMed] [Google Scholar]
- 13.Bothe JR, et al. Characterizing RNA dynamics at atomic resolution using solution-state NMR spectroscopy. Nat. Methods. 2011;8:919–931. doi: 10.1038/nmeth.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hansen AL, Nikolova EN, Casiano-Negroni A, Al-Hashimi HM. Extending the range of microsecond-to-millisecond chemical exchange detected in labeled and unlabeled nucleic acids by selective carbon R(1rho) NMR spectroscopy. J. Am. Chem. Soc. 2009;131:3818–3819. doi: 10.1021/ja8091399. [DOI] [PubMed] [Google Scholar]
- 15.Nikolova EN, et al. Transient Hoogsteen base pairs in canonical duplex DNA. Nature. 2011;470:498–502. doi: 10.1038/nature09775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xue Y, et al. Characterizing RNA excited states using NMR relaxation dispersion. Meth. Enzymol. 2015;558:39–73. doi: 10.1016/bs.mie.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alvey HS, Gottardo FL, Nikolova EN, Al-Hashimi HM. Widespread transient Hoogsteen base pairs in canonical duplex DNA with variable energetics. Nat Commun. 2014;5:4786. doi: 10.1038/ncomms5786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nikolova EN, Gottardo FL, Al-Hashimi HM. Probing transient Hoogsteen hydrogen bonds in canonical duplex DNA using NMR relaxation dispersion and single-atom substitution. J. Am. Chem. Soc. 2012;134:3667–3670. doi: 10.1021/ja2117816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi H, et al. Atomic structures of excited state A-T Hoogsteen base pairs in duplex DNA by combining NMR relaxation dispersion, mutagenesis, and chemical shift calculations. J. Biomol. NMR. 2018;70:229–244. doi: 10.1007/s10858-018-0177-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu Y, McSally J, Andricioaei I, Al-Hashimi HM. Modulation of Hoogsteen dynamics on DNA recognition. Nat. Commun. 2018;9:1473. doi: 10.1038/s41467-018-03516-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abrescia NGA, Thompson A, Huynh-Dinh T, Subirana JA. Crystal structure of an antiparallel DNA fragment with Hoogsteen base pairing. Proc. Natl. Acad. Sci. USA. 2002;99:2806–2811. doi: 10.1073/pnas.052675499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abrescia NGA, González C, Gouyette C, Subirana JA. X-ray and NMR studies of the DNA oligomer d(ATATAT): Hoogsteen base pairing in duplex DNA. Biochemistry. 2004;43:4092–4100. doi: 10.1021/bi0355140. [DOI] [PubMed] [Google Scholar]
- 23.Cubero E, Abrescia NGA, Subirana JA, Luque FJ, Orozco M. Theoretical study of a new DNA structure: the antiparallel Hoogsteen duplex. J. Am. Chem. Soc. 2003;125:14603–14612. doi: 10.1021/ja035918f. [DOI] [PubMed] [Google Scholar]
- 24.Singh UC, Pattabiraman N, Langridge R, Kollman PA. Molecular mechanical studies of d(CGTACG)2: complex of triostin A with the middle A–T base pairs in either Hoogsteen or Watson-Crick pairing. Proc. Natl. Acad. Sci. U.S.A. 1986;83:6402–6406. doi: 10.1073/pnas.83.17.6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chakraborty D, Wales DJ. Energy Landscape and Pathways for Transitions between Watson-Crick and Hoogsteen Base Pairing in DNA. J Phys Chem Lett. 2018;9:229–241. doi: 10.1021/acs.jpclett.7b01933. [DOI] [PubMed] [Google Scholar]
- 26.Cubero E, Luque FJ, Orozco M. Theoretical study of the Hoogsteen-Watson-Crick junctions in DNA. Biophys. J. 2006;90:1000–1008. doi: 10.1529/biophysj.105.059535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou H, et al. New insights into Hoogsteen base pairs in DNA duplexes from a structure-based survey. Nucleic Acids Res. 2015;43:3420–3433. doi: 10.1093/nar/gkv241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brogaard K, Xi L, Wang J-P, Widom J. A map of nucleosome positions in yeast at base-pair resolution. Nature. 2012;486:496–501. doi: 10.1038/nature11142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Langley SA, Karpen GH, Langley CH. Nucleosomes shape DNA polymorphism and divergence. PLoS Genet. 2014;10:e1004457. doi: 10.1371/journal.pgen.1004457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lieleg C, Krietenstein N, Walker M, Korber P. Nucleosome positioning in yeasts: methods, maps, and mechanisms. Chromosoma. 2015;124:131–151. doi: 10.1007/s00412-014-0501-x. [DOI] [PubMed] [Google Scholar]
- 31.Nalabothula N, et al. Archaeal nucleosome positioning in vivo and in vitro is directed by primary sequence motifs. BMC Genom. 2013;14:391. doi: 10.1186/1471-2164-14-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Struhl K, Segal E. Determinants of nucleosome positioning. Nat. Struct. Mol. Biol. 2013;20:267–273. doi: 10.1038/nsmb.2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thåström A, Bingham LM, Widom J. Nucleosomal locations of dominant DNA sequence motifs for histone-DNA interactions and nucleosome positioning. J. Mol. Biol. 2004;338:695–709. doi: 10.1016/j.jmb.2004.03.032. [DOI] [PubMed] [Google Scholar]
- 34.Chua EYD, Vasudevan D, Davey GE, Wu B, Davey CA. The mechanics behind DNA sequence-dependent properties of the nucleosome. Nucleic Acids Res. 2012;40:6338–6352. doi: 10.1093/nar/gks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elbahnsi A, Retureau R, Baaden M, Hartmann B, Oguey C. Holding the nucleosome together: a quantitative description of the DNA-histone interface in Solution. J. Chem. Theory Comput. 2018;14:1045–1058. doi: 10.1021/acs.jctc.7b00936. [DOI] [PubMed] [Google Scholar]
- 36.Wu B, Mohideen K, Vasudevan D, Davey CA. Structural insight into the sequence dependence of nucleosome positioning. Structure. 2010;18:528–536. doi: 10.1016/j.str.2010.01.015. [DOI] [PubMed] [Google Scholar]
- 37.Xu X, et al. NMR studies of DNA support the role of pre-existing minor groove variations in nucleosome indirect readout. Biochemistry. 2014;53:5601–5612. doi: 10.1021/bi500504y. [DOI] [PubMed] [Google Scholar]
- 38.Ben Imeddourene A, et al. Simulations meet experiment to reveal new insights into DNA intrinsic mechanics. PLoS Comput. Biol. 2015;11:e1004631. doi: 10.1371/journal.pcbi.1004631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rohs R, West SM, Liu P, Honig B. Nuance in the double-helix and its role in protein-DNA recognition. Curr. Opin. Struct. Biol. 2009;19:171–177. doi: 10.1016/j.sbi.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wijmenga SS, Kruithof M, Hilbers CW. Analysis of (1)H chemical shifts in DNA: Assessment of the reliability of (1)H chemical shift calculations for use in structure refinement. J. Biomol. NMR. 1997;10:337–350. doi: 10.1023/A:1018348123074. [DOI] [PubMed] [Google Scholar]
- 41.Palmer AG. NMR characterization of the dynamics of biomacromolecules. Chem. Rev. 2004;104:3623–3640. doi: 10.1021/cr030413t. [DOI] [PubMed] [Google Scholar]
- 42.Sathyamoorthy B, et al. Insights into Watson-Crick/Hoogsteen breathing dynamics and damage repair from the solution structure and dynamic ensemble of DNA duplexes containing m1A. Nucleic Acids Res. 2017;45:5586–5601. doi: 10.1093/nar/gkx186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Korzhnev DM, Orekhov VY, Kay LE. Off-resonance R(1rho) NMR studies of exchange dynamics in proteins with low spin-lock fields: an application to a Fyn SH3 domain. J. Am. Chem. Soc. 2005;127:713–721. doi: 10.1021/ja0446855. [DOI] [PubMed] [Google Scholar]
- 44.Palmer AG, Massi F. Characterization of the dynamics of biomacromolecules using rotating-frame spin relaxation NMR spectroscopy. Chem. Rev. 2006;106:1700–1719. doi: 10.1021/cr0404287. [DOI] [PubMed] [Google Scholar]
- 45.Trott O, Palmer AG. R1rho relaxation outside of the fast-exchange limit. J. Magn. Reson. 2002;154:157–160. doi: 10.1006/jmre.2001.2466. [DOI] [PubMed] [Google Scholar]
- 46.Dethoff EA, Chugh J, Mustoe AM, Al-Hashimi HM. Functional complexity and regulation through RNA dynamics. Nature. 2012;482:322–330. doi: 10.1038/nature10885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heddi B, Foloppe N, Bouchemal N, Hantz E, Hartmann B. Quantification of DNA BI/BII backbone states in solution. Implications for DNA overall structure and recognition. J. Am. Chem. Soc. 2006;128:9170–9177. doi: 10.1021/ja061686j. [DOI] [PubMed] [Google Scholar]
- 48.Tian Y, et al. 31P NMR investigation of backbone dynamics in DNA binding sites. J. Phys. Chem. B. 2009;113:2596–2603. doi: 10.1021/jp711203m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Djuranovic D, Hartmann B. Conformational characteristics and correlations in crystal structures of nucleic acid oligonucleotides: evidence for sub-states. J. Biomol. Struct. Dyn. 2003;20:771–788. doi: 10.1080/07391102.2003.10506894. [DOI] [PubMed] [Google Scholar]
- 50.Várnai P, Djuranovic D, Lavery R, Hartmann B. Alpha/gamma transitions in the B-DNA backbone. Nucleic Acids Res. 2002;30:5398–5406. doi: 10.1093/nar/gkf680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Heddi B, Oguey C, Lavelle C, Foloppe N, Hartmann B. Intrinsic flexibility of B-DNA: the experimental TRX scale. Nucleic Acids Res. 2010;38:1034–1047. doi: 10.1093/nar/gkp962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Acosta-Reyes FJ, Alechaga E, Subirana JA, Campos JL. Structure of the DNA duplex d(ATTAAT)2 with Hoogsteen hydrogen bonds. PLoS ONE. 2015;10:e0120241. doi: 10.1371/journal.pone.0120241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hatakeyama A, Hartmann B, Travers A, Nogues C, Buckle M. High-resolution biophysical analysis of the dynamics of nucleosome formation. Sci. Rep. 2016;6:27337. doi: 10.1038/srep27337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Imeddourene AB, et al. The intrinsic mechanics of B-DNA in solution characterized by NMR. Nucleic Acids Res. 2016;44:3432–3447. doi: 10.1093/nar/gkw084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dans PD, et al. The static and dynamic structural heterogeneities of B-DNA: extending Calladine-Dickerson rules. Nucleic Acids Res. 2019 doi: 10.1093/nar/gkz905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lindahl V, Villa A, Hess B. Sequence dependency of canonical base pair opening in the DNA double helix. PLoS Comput. Biol. 2017;13:e1005463. doi: 10.1371/journal.pcbi.1005463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aishima J, et al. A Hoogsteen base pair embedded in undistorted B-DNA. Nucleic Acids Res. 2002;30:5244–5252. doi: 10.1093/nar/gkf661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Djuranovic D, Hartmann B. DNA fine structure and dynamics in crystals and in solution: the impact of BI/BII backbone conformations. Biopolymers. 2004;73:356–368. doi: 10.1002/bip.10528. [DOI] [PubMed] [Google Scholar]
- 59.Schneider B, et al. A DNA structural alphabet provides new insight into DNA flexibility. Acta Crystallogr D Struct Biol. 2018;74:52–64. doi: 10.1107/S2059798318000050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zgarbová M, et al. Influence of BII backbone substates on DNA twist: a unified view and comparison of simulation and experiment for all 136 distinct tetranucleotide sequences. J. Chem. Inf. Model. 2017;57:275–287. doi: 10.1021/acs.jcim.6b00621. [DOI] [PubMed] [Google Scholar]
- 61.Djuranovic D, Oguey C, Hartmann B. The role of DNA structure and dynamics in the recognition of bovine papillomavirus E2 protein target sequences. J. Mol. Biol. 2004;339:785–796. doi: 10.1016/j.jmb.2004.03.078. [DOI] [PubMed] [Google Scholar]
- 62.Heddi B, Foloppe N, Oguey C, Hartmann B. Importance of accurate DNA structures in solution: the Jun-Fos model. J. Mol. Biol. 2008;382:956–970. doi: 10.1016/j.jmb.2008.07.047. [DOI] [PubMed] [Google Scholar]
- 63.Tisné C, Delepierre M, Hartmann B. How NF-kappaB can be attracted by its cognate DNA. J. Mol. Biol. 1999;293:139–150. doi: 10.1006/jmbi.1999.3157. [DOI] [PubMed] [Google Scholar]
- 64.Heddi B, Abi-Ghanem J, Lavigne M, Hartmann B. Sequence-dependent DNA flexibility mediates DNase I cleavage. J. Mol. Biol. 2010;395:123–133. doi: 10.1016/j.jmb.2009.10.023. [DOI] [PubMed] [Google Scholar]
- 65.Simon B, Zanier K, Sattler M. A TROSY relayed HCCH-COSY experiment for correlating adenine H2/H8 resonances in uniformly 13C-labeled RNA molecules. J. Biomol. NMR. 2001;20:173–176. doi: 10.1023/A:1011214914452. [DOI] [PubMed] [Google Scholar]
- 66.Delaglio F, et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 67.Bothe JR, Stein ZW, Al-Hashimi HM. Evaluating the uncertainty in exchange parameters determined from off-resonance R1ρ relaxation dispersion for systems in fast exchange. J. Magn. Reson. 2014;244:18–29. doi: 10.1016/j.jmr.2014.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.