Abstract
Primary bone tumor, also known as osteosarcoma (OS), is the most common primary malignancy of bone in children and young adults. Current treatment protocols yield a 5-year survival rate of near 70% although approximately 80% of patients have metastatic disease at the time of diagnosis. However, long-term survival rates have remained virtually unchanged for nearly four decades, largely due to our limited understanding of the disease process. One major signaling pathway that has been implicated in human OS tumorigenesis is the insulin-like growth factor (IGF)/insulin-like growth factor-1 receptor (IGF1R) signaling axis. IGF1R is a heterotetrameric α2β2 receptor, in which the α subunits comprise the ligand binding site, whereas the β subunits are transmembrane proteins containing intracellular tyrosine kinase domains. Although numerous strategies have been devised to target IGF/IGF1R axis, most of them have failed in clinical trials due to the lack of specificity and/or limited efficacy. Here, we investigated whether a more effective and specific blockade of IGF1R activity in human OS cells can be accomplished by employing dominant-negative IGF1R (dnIGF1R) mutants. We engineered the recombinant adenoviruses expressing two IGF1R mutants derived from the α (aa 1-524) and β (aa 741-936) subunits, and found that either dnIGF1Rα and/or dnIGF1Rβ effectively inhibited cell migration, colony formation, and cell cycle progression of human OS cells, which could be reversed by exogenous IGF1. Furthermore, dnIGF1Rα and/or dnIGF1Rβ inhibited OS xenograft tumor growth in vivo, with the greatest inhibition of tumor growth shown by dnIGF1Rα. Mechanistically, the dnIGF1R mutants down-regulated the expression of PI3K/AKT and RAS/RAF/MAPK, BCL2, Cyclin D1 and most EMT regulators, while up-regulating pro-apoptotic genes in human OS cells. Collectively, these findings strongly suggest that the dnIGF1R mutants, especially dnIGF1Rα, may be further developed as novel anticancer agents that target IGF signaling axis with high specificity and efficacy.
Keywords: Osteosarcoma, bone tumor, IGF-1R, IGF signaling, dominant-negative mutants, targeted therapy
Introduction
Primary bone tumor, also known as osteogenic sarcoma or osteosarcoma (OS), is the most common primary malignancy of bone in children and young adults, with a peak incidence in the second decade of life [1-7]. Even though the current treatment protocols, including wide margin local resection, pre-operative and/or post-operative chemotherapy, yield a 5-year disease-free survival rate of near 70% [1,2,6-9], approximately 80% of patients have either macro- or micro-metastatic disease at the time of diagnosis [7-9]. Furthermore, despite significant improvement in surgical techniques and chemotherapeutic regimens, long-term survival rates have remained virtually unchanged for nearly four decades, largely due to our limited understanding of the disease processes [6-9]. Therefore, there is an unmet critical need to understand the pathogenesis and precise molecular mechanisms leading to the development of OS [1-7].
Like other human tumors, many genetic and epigenetic alterations have been implicated in the development and/or progression of human OS, and these alterations include mutations in oncogenes, tumor suppressor genes, numerous epigenetic modifications, oncometabolic changes and noncoding RNAs [1,6,7,10-27]. One of the major signaling pathways shown to play a critical role in cancer development and progression, including OS tumorigenesis, is insulin like growth factor (IGF) signaling axis [28-32]. In fact, the local expression of IGF-1 is implicated in OS development since the highest levels of IGF-1 usually coincide with the peak incidence of OS; and the increased levels of IGF-1 and IGF-1R have been detected in OS samples [30,33]. Furthermore, overexpression of IGF-1/IGF-1R signaling contributed to tumor cell survival, metastasis, and resistance to chemotherapeutic drugs; and IGF-1R expression was correlated with a poor prognosis of OS [30,33,34].
The IGF signaling axis plays critical roles in many biological processes, including normal growth and development, and cell proliferation and survival, as well as in numerous pathological processes, such as the cancer development and progression, when dysregulated [28-32]. IGFs are members of a ligand family that includes IGF1, IGF2 and insulin. The functions of IGF1, IGF2 and insulin are mediated through interaction with their cell surface receptor tyrosine kinases (RTKs) type 1 IGF receptor, called IGF-1R, or IGF1R, and insulin receptor (INSR) [28,29,32]. IGF1R is a heterotetrameric α2β2 receptor, in which the two α subunits are extracellular and together comprise the binding site for the ligand, whereas the two β subunits are transmembrane proteins that include the intracellular tyrosine kinase domains [32]. IGF1R binds IGF1 with high affinity (~1-5 nM) and with significantly lower affinity for IGF2 and insulin (4-5 fold and >100-fold lower, respectively) [32].
While IGF ligand binding to IGF1R leads to the activation of multiple signaling pathways, two of the best-characterized downstream pathways are Phosphoinositide 3-kinase-Protein kinase B (PI3K-AKT) and RAS-Mitogen-activated protein kinase (MAPK), which are also activated by cell signals, such as epidermal growth factor (EGF), fibroblast growth factors (FGFs), and hepatocyte growth factor (HGF) [28-32]. Since IGF signaling is widely up-regulated in human cancer, targeting IGF1R has long been attractive in the field of cancer research. Numerous strategies including therapeutic antibodies, small molecule inhibitors and shRNA reagents have been devised in the past decades [32]. We previously demonstrated that insulin growth factor binding protein 5 (IGFBP5), a member of the naturally occurring IGF-binding antagonist IGFBP family, can inhibit OS tumorigenesis and metastasis [30,35-37]. Nonetheless, most of the current IGF1R targeting strategies have been hampered in clinical trials due to the lack of specificity and/or limited efficacy.
In this study, we investigated whether effective and specific blockade of IGF1R activity can block the proliferation and growth of human OS cells. To accomplish this objective, we engineered two dominant-negative IGF1R mutants that express the ligand binding region of the α subunit (i.e., dnIGF1Rα) and the extracellular domain of the β subunit (i.e., dnIGF1Rβ), and investigated whether these soluble decoy receptors could specifically suppress IGF signaling in human OS cells in a dominant-negative inhibition fashion. We found that the exogenous expression of either dnIGF1Rα and/or dnIGF1Rβ effectively inhibited cell migration, colony formation, and cell cycle progression of human OS cells. The inhibitory effect on cell proliferation by the dnIGF1R mutants could be relieved by exogenous IGF1 protein. Furthermore, exogenous expression of dnIGF1Rα and/or dnIGF1Rβ significantly inhibited OS xenograft tumor growth in vivo, with the greatest inhibition of tumor growth shown by dnIGF1Rα. Mechanistically, the dnIGF1R mutants were shown to effectively down-regulate PI3K/AKT and RAS/RAF/MAPK signaling pathways, as well as the expression of BCL2, Cyclin D1 and most EMT regulators, while up-regulating the expression of pro-apoptotic genes in human OS cells. Collectively, these findings strongly suggest that the dominant-negative IGF1R mutants, in particular dnIGF1Rα, may be further developed as novel anticancer agents that target IGF signaling pathway with high specificity and efficacy.
Materials and methods
Cell culture
Human osteosarcoma cell line 143B and HEK-293 cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). The 293pTP and RAPA cells were HEK293 derivative lines that were previously characterized [38,39]. All cell lines were maintained in DMEM containing 10% fetal bovine serum (FBS, Gemini Bio-Products, West Sacramento, CA), supplemented with 100 U/ml penicillin and 100 mg/ml streptomycin at 37°C in 5% CO2 as described [40-44]. Human IGF-1 was purchased from PEPRO TECH (Cat# G502522, Cranbury, NJ). Unless indicated otherwise, other chemicals/reagents were purchased from Thermo Fisher Scientific (Waltham, MA) or Sigma-Aldrich (St. Louis, MO).
Construction and generation of recombinant adenoviruses expressing the dominant-negative IGF1R mutants, dnIGF-1Rα and dnIGF-1Rβ, or GFP
Recombinant adenoviruses were generated by using the AdEasy technology as described [45-48]. Briefly, for the construction of the dnIGF-1Rα, the coding region for amino acids 1-524 of the human IGF1R (NM_000875.3) was PCR amplified and cloned into the adenoviral shuttle vector, pAdTrace-CMV as described [40,49-51], yielding pAdTrace-dnIGF1Rα. For the construction of the dnIGF-1Rβ, the coding region for amino acids 741-936 of the human IGF1R was PCR amplified and cloned into the pDisplay vector (Invitrogen, Carlsbad, CA) and maintained in-frame with the Igκ signal peptide. The fragment containing the coding sequences of Igκ signal peptide-IGF1R AA 741-936 was then cloned into the pAdTrace-CMV vector, yielding pAdTrace-dnIGF1Rβ. The transgenes are driven by the CMV promoter as independent expression cassettes. All PCR amplified sequences and cloning junctions were verified by DNA sequencing. Detailed information about vector construction is available upon request.
Both pAdTrace-dnIGF1Rα and pAdTrace-dnIGF1Rβ were used for homologous recombination with the pAdEasy1 backbone vector in BJ5183 bacterial cells, generating the recombinant adenoviral plasmids pAdR-dnIGF1Rα and pAdR-dnIGF1Rβ, which were subsequently used to generate adenoviruses in 293pTP and/or RAPA cells as reported [46,52-56]. The resultant adenoviruses were designated as AdR-dnIGF1Rα and AdR-dnIGF1Rβ, both of which also express red fluorescent protein (RFP) as a tracking marker of infection efficiency. An adenovirus that expresses GFP only was used a mock virus control [57-60]. Polybrene (final concentration at 4 µg/mL) was added to all adenovirus infections to enhance adenoviral transduction efficiency [61-63].
RNA isolation and touchdown real-time quantitative PCR (TqPCR) analysis
RNA was isolated with the TRIzol Reagent (Invitrogen) by following the manufacturer’s instructions and subjected to reverse transcription reactions using random hexamer and M-MuLV Reverse Transcriptase (New England Biolabs, Ipswich, MA). The resultant cDNA products were diluted and used as qPCR templates. PCR primers were designed by using the Primer3 Plus program [64]. The quantitative PCR analysis was carried out using our previously optimized TqPCR protocol [44,65,66]. Briefly, the SYBR Green qPCR reactions (Bimake, Houston, TX) were set up according to manufacturer’s instructions. The cycling program was modified by incorporating 4 cycles of touchdown steps prior to the regular cycling program as described [67-72]. GAPDH was used as a reference gene. All sample values were normalized to GAPDH expression by using the 2-ΔΔCt method. The qPCR primer sequences are shown in the Supplementary Table 1.
Wound healing/cell migration assay
Subconfluent 143B cells were seeded in 6-well cell culture plates, infected with the indicated adenoviruses, and cultured to confluence (usually within 24 h). Wound injuries were introduced by scratching the monolayer cells with 10 µL pipette tips, and floater cells were removed by washing. The wound areas were monitored under bright field and photographed at 0, 12, 24, and 36 hours post wounding as described [73-76]. Each assay condition was set up in triplicate.
Colony formation assay
Approximately 3,000 of 143B cells infected with different adenoviruses were seeded into 6-well culture plates in triplicate, and cultured for 8 days. The cells were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet. The stained colonies were recorded photographically and quantitatively determined as described [77-79].
WST-1 cell proliferation analysis
Exponentially growing cells were first infected with adenoviruses for 16 h, replated into 96-well plate at 30% confluence in triplicate, and treated with or without human IGF-1 at the pre-determined optimal concentration 30 ng/mL. At 0 h, 24 h, 48 h, 72 h after IGF1 treatment, WST-1 substrate working mix was added to the wells and incubated for 1 h, and then subjected to absorbance readings at 450 nm with the use of a microplate reader as described [54,73,79-84].
Cell cycle/flow cytometry analysis
Exponentially growing 143B cells were infected with adenoviruses for 16 h, replated into 60 mm cell culture dishes in triplcate, and treated with or without human IGF-1 (30 ng/mL) for 24 hours. The cells were fixed with 70% ethanol, washed with PBS and stained with the PI and RNase stain solution (BD Biosciences Pharmingen, Cat # 550825). The stained cells were then subjected to FACS using the BD™ LSR II Flow Cytometer. The acquired flow cytometry data were analyzed with the ModFit Lt software as described [57,66,69,79].
Subcutaneous tumor cell implantation, xenograft tumor formation, and Xenogen bioluminescence imaging
The animal use and care in this study was approved by the Institutional Animal Care and Use Committee, and all animal experimental procedures were carried out in accordance with the approved guidelines. Subcutaneous cancer cell implantation procedure was performed as described [35,36,85-89]. Briefly, 143B cells were first retrovirally tagged with firefly luciferase, yielding 143B-FLuc. Subconfluent 143B-FLuc cells were infected with different adenoviruses. After 24 h post infection, the cells were collected, resuspended in sterile PBS, and injected subcutaneously into the flanks of athymic nude mice (Envigo/Harlan Research Laboratories, n=5/group, female, 6-8 week old, 2×106 cells per injection site). The animals were maintained ad lib in the biosafety barrier facility. Tumor volumes were measured with a digital clipper at days 7, 10, 13 and 16. The mice were also subjected to Xenogen IVIS 200 imaging at days 7, 10, 13 and 16. The pseudoimages were obtained by superimposing the emitted light over the grayscale photographs of the animals. Quantitative analysis was done with Xenogen’s Living Image V2.50.1 software as described [73,79,85]. At 16 days after implantation, the mice were euthanized, the tumors were retrieved for volume and weight measure, and also histological evaluation.
Hematoxylin and Eosin (H&E) and immunohistochemistry staining
The retrieved tumor masses were fixed with 10% buffered formalin and embedded in paraffin. Serial sections at 5 μm of the embedded specimens were carried out, and subjected to H & E staining as described [49,56,90-94]. The immunohistochemical staining was carried out as described [37,88,95-97]. Briefly, slides were deparaffinized and rehydrated, followed by antigen retrieval treatment through boiling the slides for 5 min in 0.01 M citrate buffer. Endogenous peroxidase was blocked using 3% hydrogen peroxide for 15 min. Non-specific binding was blocked with 10% donkey serum (Jackson ImmunoResearch Laboratories) for 60 min at room temperature. The slides were incubated at 4°C with primary antibody against PNCA (sc-56, 1:300 dilution, Santa Cruz Biotechnology) overnight, and then with donkey anti-mouse IgG secondary antibody conjugated with HRP (Jackson ImmunoResearch Laboratories) for 30 min at room temperature. The staining was visualized with 5% 3, 3-diaminobenzidine tetrachloride (DAB), and counterstained with light green. The minus anti-PNCA and mouse IgG staining groups were used negative controls. The staining results were recorded under a bright field microscope.
Statistical analysis
All quantitative assays were performed in triplicate and/or repeated in three independent batches. Data were expressed as mean ± standard deviation. Statistical significances were determined by one-way analysis of variance and the Student t-test. Statistical analysis was performed using the SPSS software version 19.0. A p-value less than 0.05 was considered statistically significant.
Results
The soluble forms of IGF-1R decoy receptor function as dominant-negative inhibitors of IGF-1R activity by suppressing cell migration and colony formation of human osteosarcoma (OS) cells
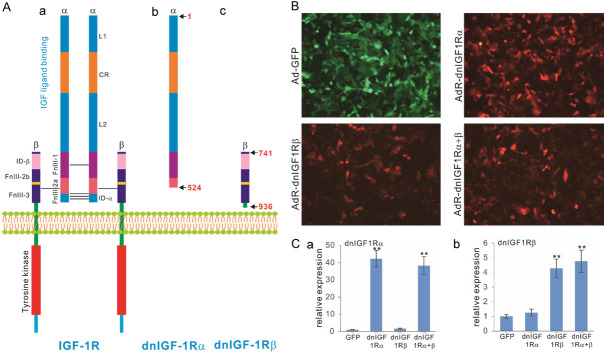
We sought to construct soluble forms of the α and β subunits of the human IGF-1R receptor. As shown in Figure 1A, human IGF-1R is a tetramer consisting of two α subunits and two β subunits, in which the α subunits are located in the extracellular space and mainly bind to IGF ligands, whereas the β subunits contain a small extracellular domain, a transmembrane span and a large cytoplasmic domain with tyrosine kinase activity (Figure 1Aa). To engineer soluble dominant-negative mutants that may effectively block IGF-1R activity in human OS cells, we constructed two mutants that express the large ligand-binding portion of the α subunits (aa 1-524; aka, dnIGF1Rα) and the extracellular portion of the β subunits that interact with the α subunits (aa 741-936; aka, dnIGF1Rβ), respectively (Figure 1Ab, 1Ac).
Figure 1.
Construction of dominant-negative IGF-1R (dnIGF1R) mutants as decoy receptors to specifically target IGF-1R signaling activity. A. Schematic depictions of the hetero-tetrameric structure and domains of the wild type IGF-1R and two dnIGF1R mutants, dnIGF-1Rα and dnIGF-1Rβ. L1, ligand binding domain 1; L2, ligand binding domain 2; CR, cysteine rich domain; ID-a, insert domain a; ID-b, insert domain b; FnIII-1, fibronectin type III domain 1; FnIII-2a, fibronectin type III domain 2a; FnIII-2b, fibronectin type III domain 2b; FnIII-3, fibronectin type III domain 3. The schematic diagrams are not drawn to scale. B. The dnIGF1R mutant expressing adenoviral vectors effectively transduce human osteosarcoma (OS) cells. Subconfluent OS line 143B cells were infected with Ad-GFP control virus, AdR-IGF1Rα, or AdR-IGF1Rβ, or co-infected with AdR-IGF1Rα and AdR-IGF1Rβ (AdR-dnIGF1Rα+β). Fluorescent signals were recorded at 48 h post infection. Representative images are shown. C. Recombinant adenovirus-mediated expression of the dnIGF1R mutants in human OS cells. Subconfluent OS line 143B cells were infected with Ad-GFP control virus, AdR-IGF1Rα, or AdR-IGF1Rβ, or co-infected with AdR-dnIGF1Rα+β. Total RNA was isolated at 48 h post infection and subjected to reverse transcription and qPCR analyses with the primers for the coding regions of the dnIGF1R mutants. GAPDH was used as a reference gene. “**”, P<0.01 compared with that of the GFP group’s.
In order to effectively express these mutants in OS cells, we generated recombinant adenoviruses that express dnIGF1Rα and dnIGF1Rβ, designated as AdR-dnIGF1Rα and AdR-dnIGF-1Rβ, respectively. These adenoviruses were shown to effectively infect human OS line 143B cells (Figure 1B). Co-infection of AdR-dnIGF1Rα and AdR-dnIGF1Rβ (aka, AdR-dnIGF1Rα+β) was utilized to achieve simultaneous inhibition of both α and β subunits. High levels of the adenovirus-mediated expression of the mutants, either individually or in co-infection, were confirmed by qPCR analysis (Figure 1Ca, 1Cb).
When the human OS cells were infected with AdR-dnIGF1Rα, AdR-dnIGF1Rβ, or AdR-dnIGF1Rα+β, the cell wounding assay revealed that the adenovirus-mediated expression of dnIGF-1Rα, dnIGF1Rβ, or dnIGF1Rα+β significantly inhibited the wound closure at 24 h and 36 h after monolayer cell injury, compared with the GFP control (Figure 2A). Colony formation analysis further revealed that dnIGF1Rα, dnIGF1Rβ, or dnIGF1Rα+β effectively suppressed the numbers of colonies formed in human OS cells, compared with the GFP control (Figure 2Ba). Quantitatively, the expression of dnIGF1Rα and dnIGF1Rβ decreased the numbers of colonies formed to 58.2% and 61.6% of the GFP control numbers, while the colony numbers decreased to 25.8% of the GFP control (Figure 2Bb). These in vitro results indicate that expression of dnIGF1Rα, and/or dnIGF1Rβ effectively suppressed wounding monolayer cell healing and colony formation in human OS 143B cells.
Figure 2.
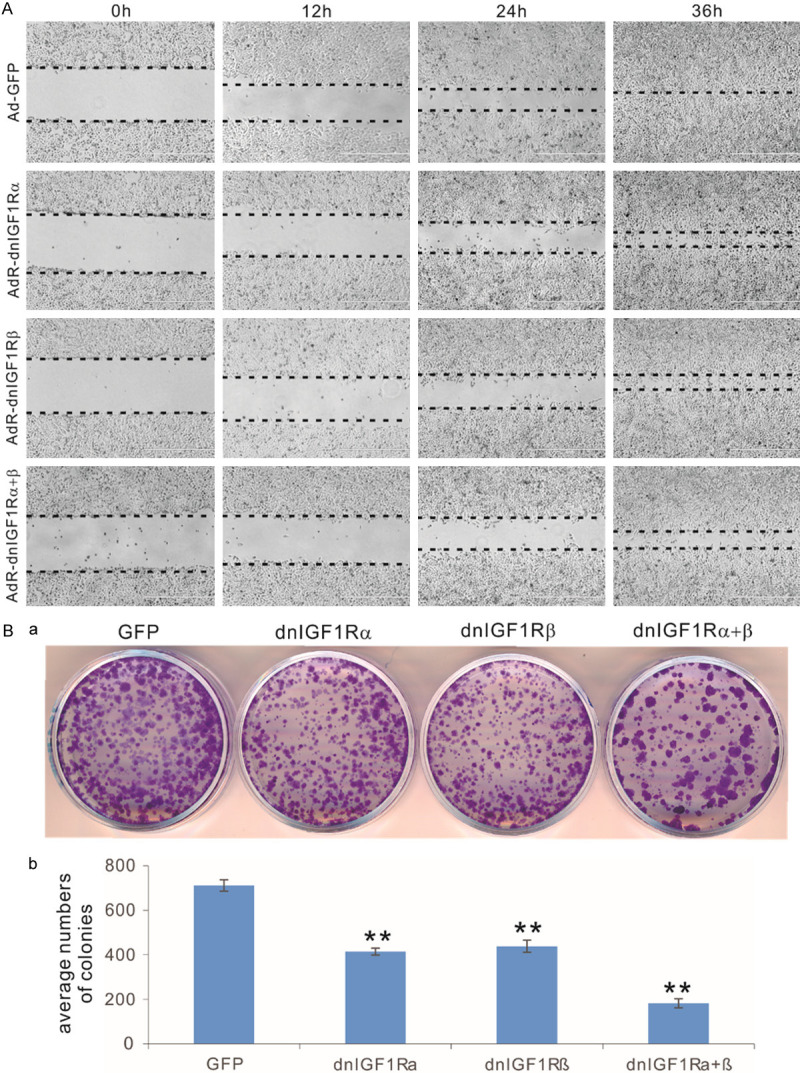
The dominant-negative IGF-1R (dnIGF1R) mutants effectively inhibit cell migration and colony formation of human OS cells. (A) The dnIGF1R mutants inhibit cell wound healing in OS cells. Subconfluent 143B cells were infected with the indicated adenoviral vectors for 16 h and replated at near confluence. The well attached cells were injured with fine pipette tips, and the injured gaps were recorded and monitored for 36 h. Images at approximately same locations were taken under a bright field microscope. The wounding front lines are indicated with the dotted lines. (B) The dnIGF1R mutants inhibit colony formation from human OS cells. Subconfluent 143B cells were infected with the indicated adenoviral vectors for 16 h. The same numbers of infected OS cells were reseeded in 12-well cell culture plates in triplicate, and cultured for 10 days with medium changes every 2-3 days. At 10 days post replating, the cells were fixed and stained with crystal violet. Representative images are shown (a), and the established colonies were further quantitatively determined (b). “**”, P<0.01 compared with that of the GFP group’s.
The dnIGF1R mutants competitively inhibit IGF1-induced cell proliferative activity in human OS cells
Mechanistically, we next tested whether these IGF1R mutants would suppress the IGF signaling activity in dominant-negative inhibition fashion by assessing whether the dnIGF1R mutant-mediated inhibition of cell proliferation could be overcome by exogenous IGF ligands. Using the human IGF1 recombinant protein, we demonstrated that the human OS line 143B cells were highly responsive to IGF1 stimulation in low FBS growth condition in a dose-dependent fashion (in the 0 to 50 ng/mL range) (data not shown). When the OS line 143B cells were infected with AdR-dnIGF1Rα, AdR-dnIGF1Rβ, AdR-dnIGF1Rα+β, or AdGFP, we found that exogenous expression of dnIGF1Rα, dnIGF1Rβ, or dnIGF1Rα+β significantly inhibited cell proliferation, which could be effectively reversed by the addition of IGF1 (at 30 ng/mL) at as early as 24 h post stimulation, and becoming more pronounced at 72 h (Figure 3A). In fact, the addition of exogenous IGF1 in the cells expressing dnIGF1Rα, dnIGF1Rβ, or dnIGF1Rα+β effectively restored the cell proliferative activity close to the level of the GFP control’s (Figure 3B). These results strongly suggest that the dnIGF1Rα and dnIGF1Rβ may function as dominant-negative mutants to suppress the IGF1R signaling activity in human OS cells.
Figure 3.
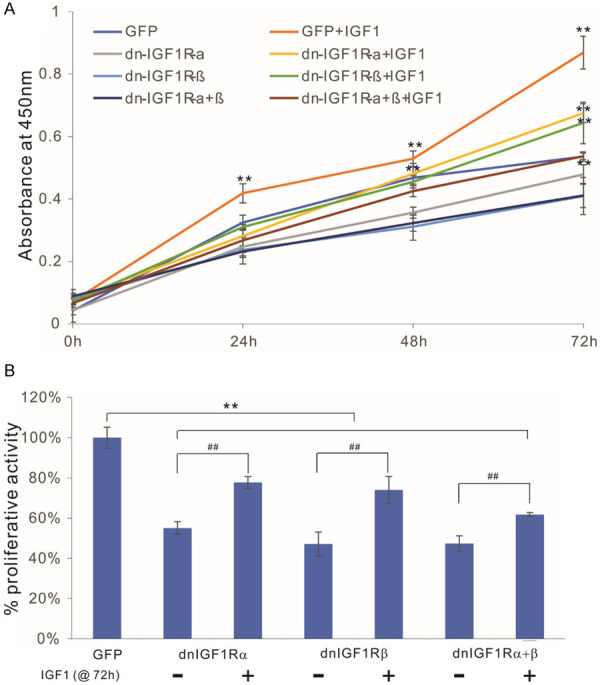
The dominant-negative IGF-1R (dnIGF1R) mutants compete with IGF1-induced cell proliferative activity in human OS cells. Exponentially growing 143B cells were infected with the indicated adenoviral vectors for 16 h and replated into 96-well cell culture plates, with or without IGF1 stimulation (30 ng/mL). At the indicated time points, the cells were subjected to WST-1 assays (A), and the WST-1 assay results at 72 h were further quantitatively analyzed (B). “**”, P<0.01 compared with that of the GFP group’s; “##”, P<0.01 compared with that of the -IGF1 group’s.
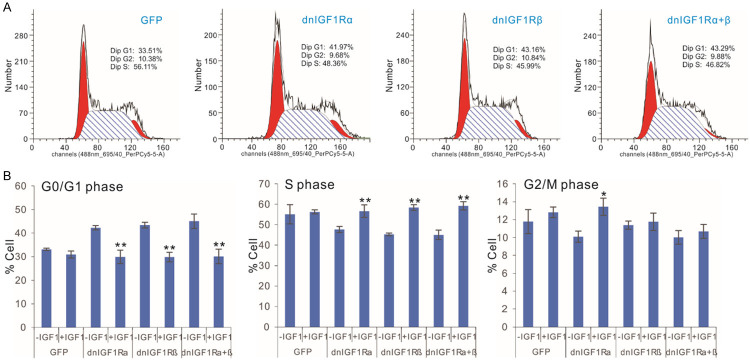
The dnIGF1R mutants suppress OS cell cycle progression by increasing G0/G1 phase and reducing S phase, and effectively inhibit tumor growth in the xenograft OS tumor model
We infected the subconfluent 143B cells with AdR-dnIGF1Rα, AdR-dnIGF1Rβ, AdR-dnIGF1Rα+β, or AdGFP, in the presence or absence of human IGF1 stimulation (30 ng/mL) for 48 h and conducted flow cytometry analysis. We found that exogenous expression of dnIGF1Rα, dnIGF1Rβ, or dnIGF1Rα+β significantly inhibited cell cycle progression by increasing numbers of cells in G0/G1 phase, while decreasing the numbers of cells in S phase (Figure 4A). As expected, the addition of IGF1 (at 30 ng/mL) effectively reversed the inhibition of cell cycle progression caused by the expression of dnIGF1Rα, dnIGF1Rβ, or dnIGF1Rα+β in OS line 143B cells (Figure 4B). However, it seems that these mutants had little impact on G2/M phase in the OS cells expressing dnIGF1Rα, dnIGF1Rβ, or dnIGF1Rα+β mutant.
Figure 4.
The dominant-negative IGF-1R (dnIGF1R) mutants inhibit OS cell cycle progression by increasing G0/G1 phase and reducing S phase. A. Subconfluent 143B cells were infected with the indicated adenoviral vectors for 48 h and subjected to FACS analysis. The assays were done in triplicate, and representative results are shown. B. Subconfluent 143B cells were infected with the indicated adenoviral vectors and cultured with or without IGF1 (30 ng/mL). At 48 h post infection, the cells were collected and subjected to FACS analysis. “**”, P<0.01 compared with that of the -IGF1 group’s.
We further investigated the effect of these mutants on tumor growth in vivo. We infected the subconfluent Firefly luciferase-tagged 143B cells with AdR-dnIGF1Rα, AdR-dnIGF1Rβ, AdR-dnIGF1Rα+β, or AdGFP, and the infected cells were injected subcutaneously into the flanks of athymic nude mice. Whole body bioluminescence imaging analysis indicated that the tumor growth was significantly inhibited in the groups injected with OS tumor cells infected with AdR-dnIGF1Rα, AdR-dnIGF1Rβ, or AdR-dnIGF1Rα+β, compared with that of the AdGFP group (Figure 5A). Interestingly, the AdR-dnIGF1Rα group exhibited the greatest reduction of tumor growth. These results were further confirmed by the quantitative analysis of the bioluminescence imaging data during the course of tumor growth (Figure 5B). The tumor volume changes were in general correlated well with the bioluminescence imaging results and the AdR-dnIGF1Rα group showed the greatest reduction of tumor volume growth (Figure 5C).
Figure 5.
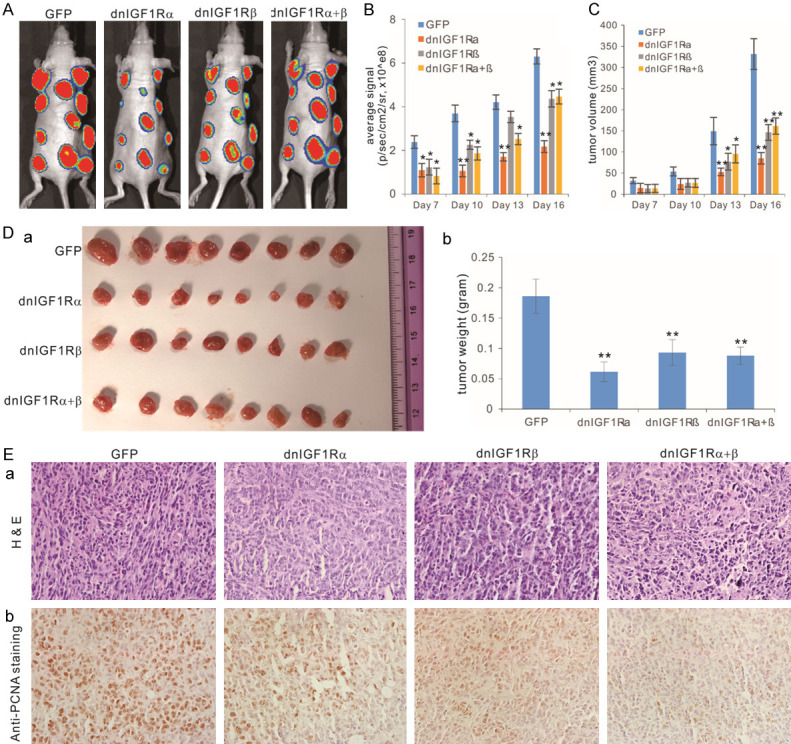
The dominant-negative IGF-1R (dnIGF1R) mutants suppress tumor growth in the xenograft tumor model of human OS. (A) Subconfluent Firefly luciferase-tagged 143B cells were infected with the indicated adenoviral vectors for 24 h, and collected for subcutaneous injection into the flanks of athymic nude mice. The mice were subjected to Xenogen imaging at days 7, 10, 13 and 16. The representative results for day 16 are shown. (B) Quantitative analysis of the average signal from the xenogeny imaging. “*”, P<0.05, “**”, P<0.01, compared with that of the GFP group’s. (C) Tumor volumes monitored with clipper measurement. “*”, P<0.05, “**”, P<0.01, compared with that of the GFP group’s. (D) Gross images of the retrieved tumor masses. Representative images of the retrieved tumor masses (a) and average tumor weight of the retrieved masses (b). (E) Histologic and immunohistochemical staining. The retrieved tumor samples were paraffin-embedded and subjected to H & E staining (a) and immunohistochemical staining using a PCNA antibody (b). Minus primary antibody and control IgG were used as negative controls (data not shown). Representative images are shown.
At the endpoint of day 16, the xenograft tumor masses were retrieved from the athymic nude mice, and the tumor sizes in the AdR-dnIGF1Rα, AdR-dnIGF1Rβ, and AdR-dnIGF1Rα+β groups were smaller than that of the AdGFP group, while the AdR-dnIGF1Rα group had the smallest tumor size (Figure 5Da), which was further confirmed by the quantitative measurement of average tumor weight (Figure 5Db). H&E histologic evaluation indicates that the tumor masses retrieved from the AdR-dnIGF1Rα, AdR-dnIGF1Rβ, and AdR-dnIGF1Rα+β groups exhibited significantly lower proliferating cells, compared with the AdGFP group (Figure 5Ea). Immunohistochemical staining analysis revealed that PNCA-positive cells significantly decreased in the tumor masses retrieved from the AdR-dnIGF1Rα, AdR-dnIGF1Rβ, and AdR-dnIGF1Rα+β groups, compared with that in the Ad-GFP group (Figure 5Eb). Collectively, the above in vivo results strongly demonstrate that the dnIGF1R mutants could effectively inhibit tumor growth and reduce tumor cell proliferative activity in the xenograft model of human osteosarcoma.
The dnIGF1R mutants inhibit the expression of major downstream mediators of the IGF-1R signaling pathway in OS cells
Lastly, we investigated the impact of the dnIGF1R mutants on the major signaling mediators of the IGF-1R pathway. IGF1R is activated by IGF1 and IGF2, which triggers the auto-phosphorylation of IGF1R and subsequently activates the downstream PI3K/AKT and RAS/RAF/MAPK signaling pathways, promoting proliferation, growth and survival [28,29,31]. When the subconfluent 143B cells were infected with AdR-dnIGF1Rα, AdR-dnIGF1Rβ, AdR-dnIGF1Rα+β, or AdGFP for 48 h, qPCR analysis revealed that the expression of PI3KCA, AKT1, BRAF, KRAS, NRAS, ERK1, and MEK1 was significantly inhibited in the OS cells infected with AdR-dnIGF1Rα, AdR-dnIGF1Rβ, or AdR-dnIGF1Rα+β, although the decreased expression of MTOR was less pronounced, compared with that infected with AdGFP (Figure 6A). These results indicated the dnIGF1R mutants can effectively suppress important downstream mediators of the IGF-1R pathway, including PI3K/AKT and RAS/RAF/MAPK signaling pathways.
Figure 6.
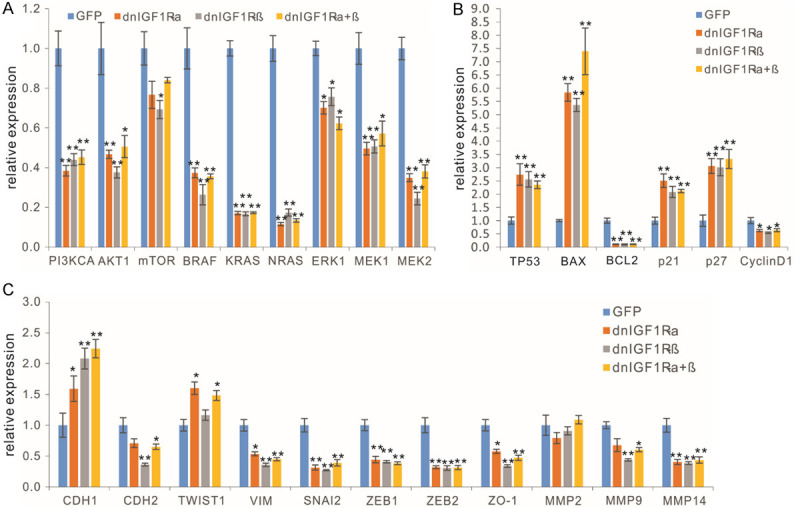
The dominant-negative IGF-1R (dnIGF1R) mutants effectively inhibit the downstream mediators of the IGF-1R signaling pathway in OS cells. Subconfluent 143B cells were infected with the indicated adenoviral vectors for 48 h. Total RNA was isolated and subjected to qPCR analyses of the IGF-1R signal downstream mediators (A), the apoptosis and cell cycle related genes (B), and EMT related genes (C). “*”, P<0.05, “**”, P<0.01, compared with that of the GFP group’s.
We further examined the effect of the dnIGF1R mutants on apoptosis and cell cycle regulators. The qPCR results revealed that the expression of pro-apoptotic regulators and/or cell cycle inhibitors TP53, BAX, p21 and p27 was significantly up-regulated in the OS cells infected with AdR-dnIGF1Rα, AdR-dnIGF1Rβ, or AdR-dnIGF1Rα+β, while anti-apoptotic factor BCL2 and cell cycle promoter Cyclin D1 were down-regulated in the OS cells infected with AdR-dnIGF1Rα, AdR-dnIGF1Rβ, or AdR-dnIGF1Rα+β (Figure 6B). We also analyzed the effect of the dnIGF1R mutants on epithelial-mesenchymal transition (EMT) regulators and found that AdR-dnIGF1Rα, AdR-dnIGF1Rβ, or AdR-dnIGF1Rα+β inhibited the expression of EMT regulators including CDH2, VIM, SNAI2, ZEB1, ZEB2, ZO-1, MMP9 and MMP14, while up-regulating CDH1, although MMP2 expression was not affected and TWIST1 was slightly upregulated by AdR-dnIGF1Rα, or AdR-dnIGF1Rα+β, compared with that of the GFP group (Figure 6C). Collectively, these results demonstrate that the dnIGF1R mutants can effectively up-regulate the expression of pro-apoptotic regulators while suppressing the expression of anti-apoptotic BCL2, Cyclin D1, and most of the EMT regulators in human OS cells.
Discussion
Since aberrant activation of the IGF/IGF-1R axis has been associated with many pathogenic processes including cancer, this signal axis has long been targeted for therapeutic intervention [32]. IGF1R was indeed the first tyrosine kinase receptor to be targeted by a monoclonal antibody in preclinical studies, which was shown to inhibit breast cancer cell proliferation in vitro and xenograft tumor growth in nude mice [98]. Several anti-IGF-1R monoclonal antibodies that bind to the IGF1R α subunit and neutralize IGF binding have been evaluated in clinical trials in cancer patients [32]. However, some of these antibodies bind IGF1R: INSR-A/B hybrid receptors, causing dose-limiting hyperglycemia in patients treated with IGF-1R antibody, or other cases causing endocrine response to IGF-1R blockade and hepatic secretion of growth hormone and IGFBPs that can impair glucose tolerance [32].
Small molecule tyrosine kinase inhibitors (TKIs) of IGF1R represent another major strategy to inhibit IGF1R kinase activity [32,99]. While numerous experimental IGF1R TKIs, such as OSI-906, have shown strong anti-proliferative effects in preclinical studies [99], very few have undergone clinical evaluation and results have been disappointing [32]. A major obstacle is to overcome high levels of sequence homology (~85%) between IGF-1R and INSR-A/B kinase domains, which contain 100% identity in the ATP-binding cleft [100]. Therefore, ATP-competitive IGF1R TKIs also inhibit INSR [101], leading to impairment of metabolic insulin signaling and hyperinsulinemia and dose-limiting hyperglycemia disappointing [32]. Non-ATP competitive IGF1R inhibitors, such as AXL1717 and INSM-18, may overcome the above adverse effects and inhibit IGF-1R without INSR blockade [32,102]. Thus, some of this class of inhibitors have been moved to various stages of clinical trials.
Interestingly, IGF binding proteins (IGFBPs) are a family of naturally occurring IGF inhibitors and have been identified as a focus for drug development. Recombinant human IGFBP3 (rhIGFBP3) was shown to have anticancer activity in vitro and in vivo [103]. We previously demonstrated IGFBP5 is a critical inhibitor of OS tumorigenesis and metastasis [36]. IGFBP5 expression was also decreased in metastatic OS lesions when compared to primary tumors in patient samples [35]. Furthermore, we found that IGFBP5 acted as a potent tumor suppressor, and over-expression of IGFBP5 inhibited OS cell proliferation, migration and invasion in vitro; while inducing apoptosis while siRNA knockdown of IGFBP5 promoted OS growth and lung metastasis in vivo [36,37]. While it is not clear whether IGFBPs have comparable or more potent inhibitory effect than that of dnIGF1R mutant’s, it is conceivable that a combination of IGFBPs and dnIGF1R mutants may act synergistically in blocking IGF signaling in cancer cells. An alternative therapeutic strategy is to specifically target the IGF ligands with neutralizing antibodies, such as Dusigitumab (MEDI-573, Medimmune) and xentuzumab (BI 836845), both of which entered clinical trials [104,105].
Previously, dnIGF1Rs expressing IGF1R residues 1~486, 1~482, 1~950, and 1~976, were constructed and shown to induce apoptosis and inhibit xenograft tumor growth in breast cancer, lung cancer and gastric cancer [106-108]. Furthermore, an IGF-TRAP was generated by fusing the IGF1R extracellular domain (aa 1-933) to the Fc region of human IgG1, which was shown to bind IGF ligands and suppress growth of breast cancer xenografts and colon and lung cancer liver metastases in vivo [109]. However, the Fc-fusion protein tends to form high molecular weight complexes due to the propensity of cysteine to form disulfides between Fc fragments, and there are currently no IGF-TRAPS in clinical use. In this study, unlike the previous reports we engineered two distinct dominant-negative IGF1R mutants that were derived from the α (aa 1-524) and β (aa 741-936) subunits, respectively, and demonstrated the dnIGF1R mutants could effectively suppress OS cell proliferation and tumor growth in vitro and in vivo. Our reported work was the first to demonstrate that the 196-aa fragment containing the extracellular domain of the IGF1R β subunit is sufficient and effective to block IGF1R signaling activity. It is conceivable our dnIGF1R mutants, in particular dnIGF1Rβ, provide more specific interactions with IGF ligands and thus minimize potential interference with insulin functions.
In summary, in order to overcome the lack of specificity and/or limited efficacy of the current IGF1R targeting strategies, we engineered two dominant-negative IGF1R mutants dnIGF1Rα and dnIGF1Rβ, and demonstrated that these soluble decoy receptors could specifically suppress IGF signaling in human OS cells in a dominant-negative inhibition fashion. Specifically, we found that the exogenous expression of either dnIGF1Rα and/or dnIGF1Rβ effectively inhibited cell migration, colony formation, and cell cycle progression of human OS cells, which could be relieved by exogenous IGF1 protein. Furthermore, exogenous expression of dnIGF1Rα and/or dnIGF1Rβ, especially dnIGF1Rα alone, significantly inhibited OS xenograft tumor growth in vivo. Mechanistically, the dnIGF1R mutants were shown effectively down-regulate PI3K/AKT and RAS/RAF/MAPK signaling pathways, as well as the expression of BCL2, Cyclin D1 and most EMT regulators, while up-regulate the expression of pro-apoptotic genes in human OS cells. Therefore, these findings strongly suggest that these dominant-negative IGF1R mutants, in particular dnIGF1Rα, may be further developed as novel anticancer agents in the clinical arena that target IGF signaling pathway with high specificity and efficacy.
Acknowledgements
The authors wish to thank the technical support provided by the Integrated Small Animal Imaging Research Resource (iSAIRR) Faculty at The University of Chicago. The reported work was supported in part by a research grant from the Natural National Science Foundation of China (grant# 81672656 to ZL). WW was supported by the Medical Scientist Training Program of the National Institutes of Health (T32 GM007281). This project was also supported in part by The University of Chicago Cancer Center Support Grant (P30CA014599) and the National Center for Advancing Translational Sciences (NCATS) of the National Institutes of Health (NIH) through Grant Number 5UL1TR002389-02 that funds the Institute for Translational Medicine (ITM) although the contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH. TCH was also supported by the Mabel Green Myers Research Endowment Fund and The University of Chicago Orthopaedics Alumni Fund. Funding sources were not involved in the study design; in the collection, analysis and interpretation of data; in the writing of the report; and in the decision to submit the paper for publication.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Tang N, Song WX, Luo J, Haydon RC, He TC. Osteosarcoma development and stem cell differentiation. Clin Orthop Relat Res. 2008;466:2114–2130. doi: 10.1007/s11999-008-0335-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haydon RC, Luu HH, He TC. Osteosarcoma and osteoblastic differentiation: a new perspective on oncogenesis. Clin Orthop Relat Res. 2007;454:237–246. doi: 10.1097/BLO.0b013e31802b683c. [DOI] [PubMed] [Google Scholar]
- 3.Wagner ER, He BC, Chen L, Zuo GW, Zhang W, Shi Q, Luo Q, Luo X, Liu B, Luo J, Rastegar F, He CJ, Hu Y, Boody B, Luu HH, He TC, Deng ZL, Haydon RC. Therapeutic implications of PPARgamma in human osteosarcoma. PPAR Res. 2010;2010:956427. doi: 10.1155/2010/956427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wagner ER, Luther G, Zhu G, Luo Q, Shi Q, Kim SH, Gao JL, Huang E, Gao Y, Yang K, Wang L, Teven C, Luo X, Liu X, Li M, Hu N, Su Y, Bi Y, He BC, Tang N, Luo J, Chen L, Zuo G, Rames R, Haydon RC, Luu HH, He TC. Defective osteogenic differentiation in the development of osteosarcoma. Sarcoma. 2011;2011:325238. doi: 10.1155/2011/325238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whelan JS. Osteosarcoma. Eur J Cancer. 1997;33:1611–1618. doi: 10.1016/s0959-8049(97)00251-7. discussion 1618-1619. [DOI] [PubMed] [Google Scholar]
- 6.Lamplot JD, Denduluri S, Qin J, Li R, Liu X, Zhang H, Chen X, Wang N, Pratt A, Shui W, Luo X, Nan G, Deng ZL, Luo J, Haydon RC, He TC, Luu HH. The current and future therapies for human osteosarcoma. Curr Cancer Ther Rev. 2013;9:55–77. doi: 10.2174/1573394711309010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Denduluri SK, Wang Z, Yan Z, Wang J, Wei Q, Mohammed MK, Haydon RC, Luu HH, He TC. Molecular pathogenesis and therapeutic strategies of human osteosarcoma. J Biomed Res. 2015:30. doi: 10.7555/JBR.29.20150075. [DOI] [PubMed] [Google Scholar]
- 8.Davis AM, Bell RS, Goodwin PJ. Prognostic factors in osteosarcoma: a critical review. J. Clin. Oncol. 1994;12:423–431. doi: 10.1200/JCO.1994.12.2.423. [DOI] [PubMed] [Google Scholar]
- 9.Mankin HJ, Hornicek FJ, Rosenberg AE, Harmon DC, Gebhardt MC. Survival data for 648 patients with osteosarcoma treated at one institution. Clin Orthop Relat Res. 2004:286–291. doi: 10.1097/01.blo.0000145991.65770.e6. [DOI] [PubMed] [Google Scholar]
- 10.Mostafa S, Pakvasa M, Coalson E, Zhu A, Alverdy A, Castillo H, Fan J, Li A, Feng Y, Wu D, Bishop E, Du S, Spezia M, Li A, Hagag O, Deng A, Liu W, Li M, Ho SS, Athiviraham A, Lee MJ, Wolf JM, Ameer GA, Luu HH, Haydon RC, Strelzow J, Hynes K, He TC, Reid RR. The wonders of BMP9: from mesenchymal stem cell differentiation, angiogenesis, neurogenesis, tumorigenesis, and metabolism to regenerative medicine. Genes Dis. 2019;6:201–223. doi: 10.1016/j.gendis.2019.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin Z, Liu Y. DNA methylation in human diseases. Genes Dis. 2018;5:1–8. doi: 10.1016/j.gendis.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Letson GD, Muro-Cacho CA. Genetic and molecular abnormalities in tumors of the bone and soft tissues. Cancer Control. 2001;8:239–251. doi: 10.1177/107327480100800304. [DOI] [PubMed] [Google Scholar]
- 13.Li S, Li J, Chen C, Zhang R, Wang K. Pan-cancer analysis of long non-coding RNA NEAT1 in various cancers. Genes Dis. 2018;5:27–35. doi: 10.1016/j.gendis.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng Y, Spezia M, Huang S, Yuan C, Zeng Z, Zhang L, Ji X, Liu W, Huang B, Luo W, Liu B, Lei Y, Du S, Vuppalapati A, Luu HH, Haydon RC, He TC, Ren G. Breast cancer development and progression: risk factors, cancer stem cells, signaling pathways, genomics, and molecular pathogenesis. Genes Dis. 2018;5:77–106. doi: 10.1016/j.gendis.2018.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qin JJ, Li X, Hunt C, Wang W, Wang H, Zhang R. Natural products targeting the p53-MDM2 pathway and mutant p53: recent advances and implications in cancer medicine. Genes Dis. 2018;5:204–219. doi: 10.1016/j.gendis.2018.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fardi M, Solali S, Farshdousti Hagh M. Epigenetic mechanisms as a new approach in cancer treatment: an updated review. Genes Dis. 2018;5:304–311. doi: 10.1016/j.gendis.2018.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chawsheen HA, Ying Q, Jiang H, Wei Q. A critical role of the thioredoxin domain containing protein 5 (TXNDC5) in redox homeostasis and cancer development. Genes Dis. 2018;5:312–322. doi: 10.1016/j.gendis.2018.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He RZ, Luo DX, Mo YY. Emerging roles of lncRNAs in the post-transcriptional regulation in cancer. Genes Dis. 2019;6:6–15. doi: 10.1016/j.gendis.2019.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adorno-Cruz V, Liu H. Regulation and functions of integrin alpha2 in cell adhesion and disease. Genes Dis. 2019;6:16–24. doi: 10.1016/j.gendis.2018.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie M, Bu Y. SKA2/FAM33A: a novel gene implicated in cell cycle, tumorigenesis, and psychiatric disorders. Genes Dis. 2019;6:25–30. doi: 10.1016/j.gendis.2018.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mussell A, Frangou C, Zhang J. Regulation of the Hippo signaling pathway by deubiquitinating enzymes in cancer. Genes Dis. 2019;6:335–341. doi: 10.1016/j.gendis.2019.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Y, Sun XX, Sears RC, Dai MS. Writing and erasing MYC ubiquitination and SUMOylation. Genes Dis. 2019;6:359–371. doi: 10.1016/j.gendis.2019.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ragland BD, Bell WC, Lopez RR, Siegal GP. Cytogenetics and molecular biology of osteosarcoma. Lab Invest. 2002;82:365–373. doi: 10.1038/labinvest.3780431. [DOI] [PubMed] [Google Scholar]
- 24.Zinatizadeh MR, Momeni SA, Zarandi PK, Chalbatani GM, Dana H, Mirzaei HR, Akbari ME, Miri SR. The role and function of ras-association domain family in cancer: a review. Genes Dis. 2019;6:378–384. doi: 10.1016/j.gendis.2019.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu H, Li X, Li H. Gene fusions and chimeric RNAs, and their implications in cancer. Genes Dis. 2019;6:385–390. doi: 10.1016/j.gendis.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fuchs B, Pritchard DJ. Etiology of osteosarcoma. Clin Orthop Relat Res. 2002:40–52. doi: 10.1097/00003086-200204000-00007. [DOI] [PubMed] [Google Scholar]
- 27.Yang S, Li X. Recent advances in extracellular vesicles enriched with non-coding RNAs related to cancers. Genes Dis. 2018;5:36–42. doi: 10.1016/j.gendis.2017.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pollak M. The insulin and insulin-like growth factor receptor family in neoplasia: an update. Nat Rev Cancer. 2012;12:159–169. doi: 10.1038/nrc3215. [DOI] [PubMed] [Google Scholar]
- 29.Pollak M. The insulin receptor/insulin-like growth factor receptor family as a therapeutic target in oncology. Clin Cancer Res. 2012;18:40–50. doi: 10.1158/1078-0432.CCR-11-0998. [DOI] [PubMed] [Google Scholar]
- 30.Denduluri SK, Idowu O, Wang Z, Liao Z, Yan Z, Mohammed MK, Ye J, Wei Q, Wang J, Zhao L, Luu HH. Insulin-like growth factor (IGF) signaling in tumorigenesis and the development of cancer drug resistance. Genes Dis. 2015;2:13–25. doi: 10.1016/j.gendis.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chitnis MM, Yuen JS, Protheroe AS, Pollak M, Macaulay VM. The type 1 insulin-like growth factor receptor pathway. Clin Cancer Res. 2008;14:6364–6370. doi: 10.1158/1078-0432.CCR-07-4879. [DOI] [PubMed] [Google Scholar]
- 32.Osher E, Macaulay VM. Therapeutic targeting of the IGF axis. Cells. 2019;8:895. doi: 10.3390/cells8080895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hong SH, Briggs J, Newman R, Hoffman K, Mendoza A, LeRoith D, Helman L, Yakar S, Khanna C. Murine osteosarcoma primary tumour growth and metastatic progression is maintained after marked suppression of serum insulin-like growth factor I. Int J Cancer. 2009;124:2042–2049. doi: 10.1002/ijc.24169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.MacEwen EG, Pastor J, Kutzke J, Tsan R, Kurzman ID, Thamm DH, Wilson M, Radinsky R. IGF-1 receptor contributes to the malignant phenotype in human and canine osteosarcoma. J Cell Biochem. 2004;92:77–91. doi: 10.1002/jcb.20046. [DOI] [PubMed] [Google Scholar]
- 35.Su Y, Luo X, He BC, Wang Y, Chen L, Zuo GW, Liu B, Bi Y, Huang J, Zhu GH, He Y, Kang Q, Luo J, Shen J, Chen J, Jin X, Haydon RC, He TC, Luu HH. Establishment and characterization of a new highly metastatic human osteosarcoma cell line. Clin Exp Metastasis. 2009;26:599–610. doi: 10.1007/s10585-009-9259-6. [DOI] [PubMed] [Google Scholar]
- 36.Su Y, Wagner ER, Luo Q, Huang J, Chen L, He BC, Zuo GW, Shi Q, Zhang BQ, Zhu G, Bi Y, Luo J, Luo X, Kim SH, Shen J, Rastegar F, Huang E, Gao Y, Gao JL, Yang K, Wietholt C, Li M, Qin J, Haydon RC, He TC, Luu HH. Insulin-like growth factor binding protein 5 suppresses tumor growth and metastasis of human osteosarcoma. Oncogene. 2011;30:3907–3917. doi: 10.1038/onc.2011.97. [DOI] [PubMed] [Google Scholar]
- 37.Luther GA, Lamplot J, Chen X, Rames R, Wagner ER, Liu X, Parekh A, Huang E, Kim SH, Shen J, Haydon RC, He TC, Luu HH. IGFBP5 domains exert distinct inhibitory effects on the tumorigenicity and metastasis of human osteosarcoma. Cancer Lett. 2013;336:222–230. doi: 10.1016/j.canlet.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu N, Zhang H, Deng F, Li R, Zhang W, Chen X, Wen S, Wang N, Zhang J, Yin L, Liao Z, Zhang Z, Zhang Q, Yan Z, Liu W, Wu D, Ye J, Deng Y, Yang K, Luu HH, Haydon RC, He TC. Overexpression of Ad5 precursor terminal protein accelerates recombinant adenovirus packaging and amplification in HEK-293 packaging cells. Gene Ther. 2014;21:629–637. doi: 10.1038/gt.2014.40. [DOI] [PubMed] [Google Scholar]
- 39.Wei Q, Fan J, Liao J, Zou Y, Song D, Liu J, Cui J, Liu F, Ma C, Hu X, Li L, Yu Y, Qu X, Chen L, Yu X, Zhang Z, Zhao C, Zeng Z, Zhang R, Yan S, Wu X, Shu Y, Reid RR, Lee MJ, Wolf JM, He TC. Engineering the Rapid Adenovirus Production and Amplification (RAPA) cell line to expedite the generation of recombinant adenoviruses. Cell Physiol Biochem. 2017;41:2383–2398. doi: 10.1159/000475909. [DOI] [PubMed] [Google Scholar]
- 40.Zeng Z, Huang B, Huang S, Zhang R, Yan S, Yu X, Shu Y, Zhao C, Lei J, Zhang W, Yang C, Wu K, Wu Y, An L, Ji X, Gong C, Yuan C, Zhang L, Liu W, Feng Y, Zhang B, Dai Z, Shen Y, Wang X, Luo W, Haydon RC, Luu HH, Zhou L, Reid RR, He TC, Wu X. The development of a sensitive fluorescent protein-based transcript reporter for high throughput screening of negative modulators of lncRNAs. Genes Dis. 2018;5:62–74. doi: 10.1016/j.gendis.2018.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu X, Chen L, Wu K, Yan S, Zhang R, Zhao C, Zeng Z, Shu Y, Huang S, Lei J, Ji X, Yuan C, Zhang L, Feng Y, Liu W, Huang B, Zhang B, Luo W, Wang X, Liu B, Haydon RC, Luu HH, He TC, Gan H. Establishment and functional characterization of the reversibly immortalized mouse glomerular podocytes (imPODs) Genes Dis. 2018;5:137–149. doi: 10.1016/j.gendis.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang B, Huang LF, Zhao L, Zeng Z, Wang X, Cao D, Yang L, Ye Z, Chen X, Liu B, He TC, Wang X. Microvesicles (MIVs) secreted from adipose-derived stem cells (ADSCs) contain multiple microRNAs and promote the migration and invasion of endothelial cells. Genes Dis. 2019;7:225–234. doi: 10.1016/j.gendis.2019.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang LH, Luo Q, Shu Y, Zeng ZY, Huang B, Feng YX, Zhang B, Wang X, Lei Y, Ye ZY, Zhao L, Cao DG, Yang LJ, Chen X, Liu B, Wagstaff W, Reid RR, Luu HH, Haydon RC, Lee MJ, Wolf JM, Fu Z, He TC, Kang Q. Transcriptomic landscape regulated by the 14 types of bone morphogenetic proteins (BMPs) in lineage commitment and differentiation of mesenchymal stem cells (MSCs) Genes Dis. 2019;6:258–275. doi: 10.1016/j.gendis.2019.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shu Y, Wu K, Zeng Z, Huang S, Ji X, Yuan C, Zhang L, Liu W, Huang B, Feng Y, Zhang B, Dai Z, Shen Y, Luo W, Wang X, Liu B, Lei Y, Ye Z, Zhao L, Cao D, Yang L, Chen X, Luu HH, Reid RR, Wolf JM, Lee MJ, He TC. A simplified system to express circularized inhibitors of miRNA for stable and potent suppression of miRNA functions. Mol Ther Nucleic Acids. 2018;13:556–567. doi: 10.1016/j.omtn.2018.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.He TC, Zhou S, da Costa LT, Yu J, Kinzler KW, Vogelstein B. A simplified system for generating recombinant adenoviruses. Proc Natl Acad Sci U S A. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luo J, Deng ZL, Luo X, Tang N, Song WX, Chen J, Sharff KA, Luu HH, Haydon RC, Kinzler KW, Vogelstein B, He TC. A protocol for rapid generation of recombinant adenoviruses using the AdEasy system. Nat Protoc. 2007;2:1236–1247. doi: 10.1038/nprot.2007.135. [DOI] [PubMed] [Google Scholar]
- 47.Lee CS, Bishop ES, Zhang R, Yu X, Farina EM, Yan S, Zhao C, Zheng Z, Shu Y, Wu X, Lei J, Li Y, Zhang W, Yang C, Wu K, Wu Y, Ho S, Athiviraham A, Lee MJ, Wolf JM, Reid RR, He TC. Adenovirus-mediated gene delivery: potential applications for gene and cell-based therapies in the new era of personalized medicine. Genes Dis. 2017;4:43–63. doi: 10.1016/j.gendis.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yan S, Zhang R, Wu K, Cui J, Huang S, Ji X, An L, Yuan C, Gong C, Zhang L, Liu W, Feng Y, Zhang B, Dai Z, Shen Y, Wang X, Luo W, Liu B, Haydon RC, Lee MJ, Reid RR, Wolf JM, Shi Q, Luu HH, He TC, Weng Y. Characterization of the essential role of bone morphogenetic protein 9 (BMP9) in osteogenic differentiation of mesenchymal stem cells (MSCs) through RNA interference. Genes Dis. 2018;5:172–184. doi: 10.1016/j.gendis.2018.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang B, Yang L, Zeng Z, Feng Y, Wang X, Wu X, Luo H, Zhang J, Zhang M, Pakvasa M, Wagstaff W, He F, Mao Y, Qin K, Ding H, Zhang Y, Niu C, Wu M, Zhao X, Wang H, Huang L, Shi D, Liu Q, Ni N, Fu K, Athiviraham A, Moriatis Wolf J, Lee MJ, Hynes K, Strelzow J, El Dafrawy M, Xia Y, He TC. Leptin potentiates BMP9-induced osteogenic differentiation of mesenchymal stem cells through the activation of JAK/STAT signaling. Stem Cells Dev. 2020;29:498–510. doi: 10.1089/scd.2019.0292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao C, Jiang W, Zhou N, Liao J, Yang M, Hu N, Liang X, Xu W, Chen H, Liu W, Shi LL, Oliveira L, Wolf JM, Ho S, Athiviraham A, Tsai HM, He TC, Huang W. Sox9 augments BMP2-induced chondrogenic differentiation by downregulating Smad7 in mesenchymal stem cells (MSCs) Genes Dis. 2017;4:229–239. doi: 10.1016/j.gendis.2017.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao C, Zeng Z, Qazvini NT, Yu X, Zhang R, Yan S, Shu Y, Zhu Y, Duan C, Bishop E, Lei J, Zhang W, Yang C, Wu K, Wu Y, An L, Huang S, Ji X, Gong C, Yuan C, Zhang L, Liu W, Huang B, Feng Y, Zhang B, Dai Z, Shen Y, Wang X, Luo W, Oliveira L, Athiviraham A, Lee MJ, Wolf JM, Ameer GA, Reid RR, He TC, Huang W. Thermoresponsive citrate-based graphene oxide scaffold enhances bone regeneration from BMP9-stimulated adipose-derived mesenchymal stem cells. ACS Biomater Sci Eng. 2018;4:2943–2955. doi: 10.1021/acsbiomaterials.8b00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang X, Wang F, Zhao C, Yang S, Cheng Q, Tang Y, Zhang F, Zhang Y, Luo W, Wang C, Zhou P, Kim S, Zuo G, Hu N, Li R, He TC, Zhang H. Dentinogenesis and tooth-alveolar bone complex defects in BMP9/GDF2 knockout mice. Stem Cells Dev. 2019;28:683–694. doi: 10.1089/scd.2018.0230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Song D, Huang S, Zhang L, Liu W, Huang B, Feng Y, Liu B, He TC, Huang D, Reid RR. Differential responsiveness to BMP9 between patent and fused suture progenitor cells from craniosynostosis patients. Plast Reconstr Surg. 2020;145:552e–562e. doi: 10.1097/PRS.0000000000006597. [DOI] [PubMed] [Google Scholar]
- 54.Huang E, Bi Y, Jiang W, Luo X, Yang K, Gao JL, Gao Y, Luo Q, Shi Q, Kim SH, Liu X, Li M, Hu N, Liu H, Cui J, Zhang W, Li R, Chen X, Shen J, Kong Y, Zhang J, Wang J, Luo J, He BC, Wang H, Reid RR, Luu HH, Haydon RC, Yang L, He TC. Conditionally immortalized mouse embryonic fibroblasts retain proliferative activity without compromising multipotent differentiation potential. PLoS One. 2012;7:e32428. doi: 10.1371/journal.pone.0032428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang J, Bi Y, Zhu GH, He Y, Su Y, He BC, Wang Y, Kang Q, Chen L, Zuo GW, Luo Q, Shi Q, Zhang BQ, Huang A, Zhou L, Feng T, Luu HH, Haydon RC, He TC, Tang N. Retinoic acid signalling induces the differentiation of mouse fetal liver-derived hepatic progenitor cells. Liver Int. 2009;29:1569–1581. doi: 10.1111/j.1478-3231.2009.02111.x. [DOI] [PubMed] [Google Scholar]
- 56.Wang H, Cao Y, Shu L, Zhu Y, Peng Q, Ran L, Wu J, Luo Y, Zuo G, Luo J, Zhou L, Shi Q, Weng Y, Huang A, He TC, Fan J. Long non-coding RNA (lncRNA) H19 induces hepatic steatosis through activating MLXIPL and mTORC1 networks in hepatocytes. J Cell Mol Med. 2020;24:1399–1412. doi: 10.1111/jcmm.14818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shu Y, Yang C, Ji X, Zhang L, Bi Y, Yang K, Gong M, Liu X, Guo Q, Su Y, Qu X, Nan G, Zhao C, Zeng Z, Yu X, Zhang R, Yan S, Lei J, Wu K, Wu Y, An L, Huang S, Gong C, Yuan C, Liu W, Huang B, Feng Y, Zhang B, Dai Z, Shen Y, Luo W, Wang X, Haydon RC, Luu HH, Reid RR, Wolf JM, Lee MJ, He TC, Li Y. Reversibly immortalized human umbilical cord-derived mesenchymal stem cells (UC-MSCs) are responsive to BMP9-induced osteogenic and adipogenic differentiation. J Cell Biochem. 2018;119:8872–8886. doi: 10.1002/jcb.27140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hu X, Li L, Yu X, Zhang R, Yan S, Zeng Z, Shu Y, Zhao C, Wu X, Lei J, Li Y, Zhang W, Yang C, Wu K, Wu Y, An L, Huang S, Ji X, Gong C, Yuan C, Zhang L, Liu W, Huang B, Feng Y, Zhang B, Haydon RC, Luu HH, Reid RR, Lee MJ, Wolf JM, Yu Z, He TC. CRISPR/Cas9-mediated reversibly immortalized mouse bone marrow stromal stem cells (BMSCs) retain multipotent features of mesenchymal stem cells (MSCs) Oncotarget. 2017;8:111847–111865. doi: 10.18632/oncotarget.22915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liao J, Wei Q, Zou Y, Fan J, Song D, Cui J, Zhang W, Zhu Y, Ma C, Hu X, Qu X, Chen L, Yu X, Zhang Z, Wang C, Zhao C, Zeng Z, Zhang R, Yan S, Wu T, Wu X, Shu Y, Lei J, Li Y, Luu HH, Lee MJ, Reid RR, Ameer GA, Wolf JM, He TC, Huang W. Notch signaling augments BMP9-induced bone formation by promoting the osteogenesis-angiogenesis coupling process in mesenchymal stem cells (MSCs) Cell Physiol Biochem. 2017;41:1905–1923. doi: 10.1159/000471945. [DOI] [PubMed] [Google Scholar]
- 60.Fan J, Feng Y, Zhang R, Zhang W, Shu Y, Zeng Z, Huang S, Zhang L, Huang B, Wu D, Zhang B, Wang X, Lei Y, Ye Z, Zhao L, Cao D, Yang L, Chen X, Liu B, Wagstaff W, He F, Wu X, Zhang J, Moriatis Wolf J, Lee MJ, Haydon RC, Luu HH, Huang A, He TC, Yan S. A simplified system for the effective expression and delivery of functional mature microRNAs in mammalian cells. Cancer Gene Ther. 2020;27:424–437. doi: 10.1038/s41417-019-0113-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhao C, Wu N, Deng F, Zhang H, Wang N, Zhang W, Chen X, Wen S, Zhang J, Yin L, Liao Z, Zhang Z, Zhang Q, Yan Z, Liu W, Wu D, Ye J, Deng Y, Zhou G, Luu HH, Haydon RC, Si W, He TC. Adenovirus-mediated gene transfer in mesenchymal stem cells can be significantly enhanced by the cationic polymer polybrene. PLoS One. 2014;9:e92908. doi: 10.1371/journal.pone.0092908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liao J, Yu X, Hu X, Fan J, Wang J, Zhang Z, Zhao C, Zeng Z, Shu Y, Zhang R, Yan S, Li Y, Zhang W, Cui J, Ma C, Li L, Yu Y, Wu T, Wu X, Lei J, Wang J, Yang C, Wu K, Wu Y, Tang J, He BC, Deng ZL, Luu HH, Haydon RC, Reid RR, Lee MJ, Wolf JM, Huang W, He TC. lncRNA H19 mediates BMP9-induced osteogenic differentiation of mesenchymal stem cells (MSCs) through Notch signaling. Oncotarget. 2017;8:53581–53601. doi: 10.18632/oncotarget.18655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fan J, Wei Q, Liao J, Zou Y, Song D, Xiong D, Ma C, Hu X, Qu X, Chen L, Li L, Yu Y, Yu X, Zhang Z, Zhao C, Zeng Z, Zhang R, Yan S, Wu T, Wu X, Shu Y, Lei J, Li Y, Zhang W, Haydon RC, Luu HH, Huang A, He TC, Tang H. Noncanonical Wnt signaling plays an important role in modulating canonical Wnt-regulated stemness, proliferation and terminal differentiation of hepatic progenitors. Oncotarget. 2017;8:27105–27119. doi: 10.18632/oncotarget.15637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG. Primer3--new capabilities and interfaces. Nucleic Acids Res. 2012;40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Q, Wang J, Deng F, Yan Z, Xia Y, Wang Z, Ye J, Deng Y, Zhang Z, Qiao M, Li R, Denduluri SK, Wei Q, Zhao L, Lu S, Wang X, Tang S, Liu H, Luu HH, Haydon RC, He TC, Jiang L. TqPCR: a touchdown qPCR assay with significantly improved detection sensitivity and amplification efficiency of SYBR green qPCR. PLoS One. 2015;10:e0132666. doi: 10.1371/journal.pone.0132666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shu Y, Wang Y, Lv WQ, Peng DY, Li J, Zhang H, Jiang GJ, Yang BJ, Liu S, Zhang J, Chen YH, Tang S, Wan KX, Yuan JT, Guo W, Fu G, Qi XK, Liu ZD, Liu HY, Yang C, Zhang LH, Liu FJ, Yu J, Zhang PH, Qu B, Zhao H, He TC, Zou L. ARRB1-promoted NOTCH1 degradation is suppressed by oncomiR miR-223 in T-cell acute lymphoblastic leukemia. Cancer Res. 2020;80:988–998. doi: 10.1158/0008-5472.CAN-19-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liao J, Wei Q, Fan J, Zou Y, Song D, Liu J, Liu F, Ma C, Hu X, Li L, Yu Y, Qu X, Chen L, Yu X, Zhang Z, Zhao C, Zeng Z, Zhang R, Yan S, Wu T, Wu X, Shu Y, Lei J, Li Y, Zhang W, Wang J, Reid RR, Lee MJ, Huang W, Wolf JM, He TC, Wang J. Characterization of retroviral infectivity and superinfection resistance during retrovirus-mediated transduction of mammalian cells. Gene Ther. 2017;24:333–341. doi: 10.1038/gt.2017.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu W, Deng Z, Zeng Z, Fan J, Feng Y, Wang X, Cao D, Zhang B, Yang L, Liu B, Pakvasa M, Wagstaff W, Wu X, Luo H, Zhang J, Zhang M, He F, Mao Y, Ding H, Zhang Y, Niu C, Haydon RC, Luu HH, Wolf JM, Lee MJ, Huang W, He TC, Zou Y. Highly expressed BMP9/GDF2 in postnatal mouse liver and lungs may account for its pleiotropic effects on stem cell differentiation, angiogenesis, tumor growth and metabolism. Genes Dis. 2020;7:235–244. doi: 10.1016/j.gendis.2019.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li Y, Wagner ER, Yan Z, Wang Z, Luther G, Jiang W, Ye J, Wei Q, Wang J, Zhao L, Lu S, Wang X, Mohammed MK, Tang S, Liu H, Fan J, Zhang F, Zou Y, Song D, Liao J, Haydon RC, Luu HH, He TC. The calcium-binding protein S100A6 accelerates human osteosarcoma growth by promoting cell proliferation and inhibiting osteogenic differentiation. Cell Physiol Biochem. 2015;37:2375–2392. doi: 10.1159/000438591. [DOI] [PubMed] [Google Scholar]
- 70.Zeng Z, Huang B, Wang X, Fan J, Zhang B, Yang L, Feng Y, Wu X, Luo H, Zhang J, Zhang M, He F, Mao Y, Pakvasa M, Wagstaff W, Li AJ, Liu B, Ding H, Zhang Y, Niu C, Wu M, Zhao X, Wolf JM, Lee MJ, Huang A, Luu HH, Haydon RC, He TC. A reverse transcriptase-mediated ribosomal RNA depletion (RTR2D) strategy for the cost-effective construction of RNA sequencing libraries. J Adv Res. 2020;24:239–250. doi: 10.1016/j.jare.2019.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang X, Zhao L, Wu X, Luo H, Wu D, Zhang M, Zhang J, Pakvasa M, Wagstaff W, He F, Mao Y, Zhang Y, Niu C, Wu M, Zhao X, Wang H, Huang L, Shi D, Liu Q, Ni N, Fu K, Hynes K, Strelzow J, El Dafrawy M, He TC, Qi H, Zeng Z. Development of a simplified and inexpensive RNA depletion method for plasmid DNA purification using size selection magnetic beads (SSMBs) Genes Dis. 2020 doi: 10.1016/j.gendis.2020.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang X, Yuan C, Huang B, Fan J, Feng Y, Li AJ, Zhang B, Lei Y, Ye Z, Zhao L, Cao D, Yang L, Wu D, Chen X, Liu B, Wagstaff W, He F, Wu X, Luo H, Zhang J, Zhang M, Haydon RC, Luu HH, Lee MJ, Moriatis Wolf J, Huang A, He TC, Zeng Z. Developing a versatile shotgun cloning strategy for single-vector-based multiplex expression of short interfering RNAs (siRNAs) in mammalian cells. ACS Synth Biol. 2019;8:2092–2105. doi: 10.1021/acssynbio.9b00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang X, Wu X, Zhang Z, Ma C, Wu T, Tang S, Zeng Z, Huang S, Gong C, Yuan C, Zhang L, Feng Y, Huang B, Liu W, Zhang B, Shen Y, Luo W, Wang X, Liu B, Lei Y, Ye Z, Zhao L, Cao D, Yang L, Chen X, Haydon RC, Luu HH, Peng B, Liu X, He TC. Monensin inhibits cell proliferation and tumor growth of chemo-resistant pancreatic cancer cells by targeting the EGFR signaling pathway. Sci Rep. 2018;8:17914. doi: 10.1038/s41598-018-36214-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang F, Li Y, Zhang H, Huang E, Gao L, Luo W, Wei Q, Fan J, Song D, Liao J, Zou Y, Liu F, Liu J, Huang J, Guo D, Ma C, Hu X, Li L, Qu X, Chen L, Yu X, Zhang Z, Wu T, Luu HH, Haydon RC, Song J, He TC, Ji P. Anthelmintic mebendazole enhances cisplatin’s effect on suppressing cell proliferation and promotes differentiation of head and neck squamous cell carcinoma (HNSCC) Oncotarget. 2017;8:12968–12982. doi: 10.18632/oncotarget.14673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liao Z, Nan G, Yan Z, Zeng L, Deng Y, Ye J, Zhang Z, Qiao M, Li R, Denduluri S, Wang J, Wei Q, Geng N, Zhao L, Lu S, Wang X, Zhou G, Luu HH, Haydon RC, He TC, Wang Z. The anthelmintic drug niclosamide inhibits the proliferative activity of human osteosarcoma cells by targeting multiple signal pathways. Curr Cancer Drug Targets. 2015;15:726–738. doi: 10.2174/1568009615666150629132157. [DOI] [PubMed] [Google Scholar]
- 76.Zhang W, Zhang H, Wang N, Zhao C, Zhang H, Deng F, Wu N, He Y, Chen X, Zhang J, Wen S, Liao Z, Zhang Q, Zhang Z, Liu W, Yan Z, Luu HH, Haydon RC, Zhou L, He TC. Modulation of beta-catenin signaling by the inhibitors of MAP kinase, tyrosine kinase, and PI3-kinase pathways. Int J Med Sci. 2013;10:1888–1898. doi: 10.7150/ijms.6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li R, Zhang W, Cui J, Shui W, Yin L, Wang Y, Zhang H, Wang N, Wu N, Nan G, Chen X, Wen S, Deng F, Zhang H, Zhou G, Liao Z, Zhang J, Zhang Q, Yan Z, Liu W, Zhang Z, Ye J, Deng Y, Luu HH, Haydon RC, He TC, Deng ZL. Targeting BMP9-promoted human osteosarcoma growth by inactivation of notch signaling. Curr Cancer Drug Targets. 2014;14:274–285. doi: 10.2174/1568009614666140305105805. [DOI] [PubMed] [Google Scholar]
- 78.Deng Y, Wang Z, Zhang F, Qiao M, Yan Z, Wei Q, Wang J, Liu H, Fan J, Zou Y, Liao J, Hu X, Chen L, Yu X, Haydon RC, Luu HH, Qi H, He TC, Zhang J. A blockade of IGF signaling sensitizes human ovarian cancer cells to the anthelmintic niclosamide-induced anti-proliferative and anticancer activities. Cell Physiol Biochem. 2016;39:871–888. doi: 10.1159/000447797. [DOI] [PubMed] [Google Scholar]
- 79.Deng Y, Zhang J, Wang Z, Yan Z, Qiao M, Ye J, Wei Q, Wang J, Wang X, Zhao L, Lu S, Tang S, Mohammed MK, Liu H, Fan J, Zhang F, Zou Y, Liao J, Qi H, Haydon RC, Luu HH, He TC, Tang L. Antibiotic monensin synergizes with EGFR inhibitors and oxaliplatin to suppress the proliferation of human ovarian cancer cells. Sci Rep. 2015;5:17523. doi: 10.1038/srep17523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bi Y, He Y, Huang J, Su Y, Zhu GH, Wang Y, Qiao M, Zhang BQ, Zhang H, Wang Z, Liu W, Cui J, Kang Q, Zhang Z, Deng Y, Li R, Zhang Q, Yang K, Luu HH, Haydon RC, He TC, Tang N. Functional characteristics of reversibly immortalized hepatic progenitor cells derived from mouse embryonic liver. Cell Physiol Biochem. 2014;34:1318–1338. doi: 10.1159/000366340. [DOI] [PubMed] [Google Scholar]
- 81.Wang N, Zhang W, Cui J, Zhang H, Chen X, Li R, Wu N, Chen X, Wen S, Zhang J, Yin L, Deng F, Liao Z, Zhang Z, Zhang Q, Yan Z, Liu W, Ye J, Deng Y, Wang Z, Qiao M, Luu HH, Haydon RC, Shi LL, Liang H, He TC. The piggyBac transposon-mediated expression of SV40 T antigen efficiently immortalizes mouse embryonic fibroblasts (MEFs) PLoS One. 2014;9:e97316. doi: 10.1371/journal.pone.0097316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lamplot JD, Liu B, Yin L, Zhang W, Wang Z, Luther G, Wagner E, Li R, Nan G, Shui W, Yan Z, Rames R, Deng F, Zhang H, Liao Z, Liu W, Zhang J, Zhang Z, Zhang Q, Ye J, Deng Y, Qiao M, Haydon RC, Luu HH, Angeles J, Shi LL, He TC, Ho SH. Reversibly immortalized mouse articular chondrocytes acquire long-term proliferative capability while retaining chondrogenic phenotype. Cell Transplant. 2015;24:1053–1066. doi: 10.3727/096368914X681054. [DOI] [PubMed] [Google Scholar]
- 83.Yang K, Chen J, Jiang W, Huang E, Cui J, Kim SH, Hu N, Liu H, Zhang W, Li R, Chen X, Kong Y, Zhang J, Wang J, Wang L, Shen J, Luu HH, Haydon RC, Lian X, Yang T, He TC. Conditional immortalization establishes a repertoire of mouse melanocyte progenitors with distinct melanogenic differentiation potential. J Invest Dermatol. 2012;132:2479–2483. doi: 10.1038/jid.2012.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li M, Chen Y, Bi Y, Jiang W, Luo Q, He Y, Su Y, Liu X, Cui J, Zhang W, Li R, Kong Y, Zhang J, Wang J, Zhang H, Shui W, Wu N, Zhu J, Tian J, Yi QJ, Luu HH, Haydon RC, He TC, Zhu GH. Establishment and characterization of the reversibly immortalized mouse fetal heart progenitors. Int J Med Sci. 2013;10:1035–1046. doi: 10.7150/ijms.6639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Luo X, Chen J, Song WX, Tang N, Luo J, Deng ZL, Sharff KA, He G, Bi Y, He BC, Bennett E, Huang J, Kang Q, Jiang W, Su Y, Zhu GH, Yin H, He Y, Wang Y, Souris JS, Chen L, Zuo GW, Montag AG, Reid RR, Haydon RC, Luu HH, He TC. Osteogenic BMPs promote tumor growth of human osteosarcomas that harbor differentiation defects. Lab Invest. 2008;88:1264–1277. doi: 10.1038/labinvest.2008.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Luo X, Sharff KA, Chen J, He TC, Luu HH. S100A6 expression and function in human osteosarcoma. Clin Orthop Relat Res. 2008;466:2060–2070. doi: 10.1007/s11999-008-0361-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Luo X, Wang CZ, Chen J, Song WX, Luo J, Tang N, He BC, Kang Q, Wang Y, Du W, He TC, Yuan CS. Characterization of gene expression regulated by American ginseng and ginsenoside Rg3 in human colorectal cancer cells. Int J Oncol. 2008;32:975–983. [PMC free article] [PubMed] [Google Scholar]
- 88.He BC, Chen L, Zuo GW, Zhang W, Bi Y, Huang J, Wang Y, Jiang W, Luo Q, Shi Q, Zhang BQ, Liu B, Lei X, Luo J, Luo X, Wagner ER, Kim SH, He CJ, Hu Y, Shen J, Zhou Q, Rastegar F, Deng ZL, Luu HH, He TC, Haydon RC. Synergistic antitumor effect of the activated PPARgamma and retinoid receptors on human osteosarcoma. Clin Cancer Res. 2010;16:2235–2245. doi: 10.1158/1078-0432.CCR-09-2499. [DOI] [PubMed] [Google Scholar]
- 89.He BC, Gao JL, Zhang BQ, Luo Q, Shi Q, Kim SH, Huang E, Gao Y, Yang K, Wagner ER, Wang L, Tang N, Luo J, Liu X, Li M, Bi Y, Shen J, Luther G, Hu N, Zhou Q, Luu HH, Haydon RC, Zhao Y, He TC. Tetrandrine inhibits Wnt/beta-catenin signaling and suppresses tumor growth of human colorectal cancer. Mol Pharmacol. 2011;79:211–219. doi: 10.1124/mol.110.068668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Song D, Zhang F, Reid RR, Ye J, Wei Q, Liao J, Zou Y, Fan J, Ma C, Hu X, Qu X, Chen L, Li L, Yu Y, Yu X, Zhang Z, Zhao C, Zeng Z, Zhang R, Yan S, Wu T, Wu X, Shu Y, Lei J, Li Y, Zhang W, Wang J, Lee MJ, Wolf JM, Huang D, He TC. BMP9 induces osteogenesis and adipogenesis in the immortalized human cranial suture progenitors from the patent sutures of craniosynostosis patients. J Cell Mol Med. 2017;21:2782–2795. doi: 10.1111/jcmm.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ye J, Wang J, Zhu Y, Wei Q, Wang X, Yang J, Tang S, Liu H, Fan J, Zhang F, Farina EM, Mohammed MK, Zou Y, Song D, Liao J, Huang J, Guo D, Lu M, Liu F, Liu J, Li L, Ma C, Hu X, Haydon RC, Lee MJ, Reid RR, Ameer GA, Yang L, He TC. A thermoresponsive polydiolcitrate-gelatin scaffold and delivery system mediates effective bone formation from BMP9-transduced mesenchymal stem cells. Biomed Mater. 2016;11:025021. doi: 10.1088/1748-6041/11/2/025021. [DOI] [PubMed] [Google Scholar]
- 92.Lu S, Wang J, Ye J, Zou Y, Zhu Y, Wei Q, Wang X, Tang S, Liu H, Fan J, Zhang F, Farina EM, Mohammed MM, Song D, Liao J, Huang J, Guo D, Lu M, Liu F, Liu J, Li L, Ma C, Hu X, Lee MJ, Reid RR, Ameer GA, Zhou D, He T. Bone morphogenetic protein 9 (BMP9) induces effective bone formation from reversibly immortalized multipotent adipose-derived (iMAD) mesenchymal stem cells. Am J Transl Res. 2016;8:3710–3730. [PMC free article] [PubMed] [Google Scholar]
- 93.Yan Z, Yin L, Wang Z, Ye J, Zhang Z, Li R, Denduluri SK, Wang J, Wei Q, Zhao L, Lu S, Wang X, Tang S, Shi LL, Lee MJ, He TC, Deng ZL. A novel organ culture model of mouse intervertebral Disc tissues. Cells Tissues Organs. 2016;201:38–50. doi: 10.1159/000439268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang Y, Hong S, Li M, Zhang J, Bi Y, He Y, Liu X, Nan G, Su Y, Zhu G, Li R, Zhang W, Wang J, Zhang H, Kong Y, Shui W, Wu N, He Y, Chen X, Luu HH, Haydon RC, Shi LL, He TC, Qin J. Noggin resistance contributes to the potent osteogenic capability of BMP9 in mesenchymal stem cells. J Orthop Res. 2013;31:1796–1803. doi: 10.1002/jor.22427. [DOI] [PubMed] [Google Scholar]
- 95.Haydon RC, Deyrup A, Ishikawa A, Heck R, Jiang W, Zhou L, Feng T, King D, Cheng H, Breyer B, Peabody T, Simon MA, Montag AG, He TC. Cytoplasmic and/or nuclear accumulation of the beta-catenin protein is a frequent event in human osteosarcoma. Int J Cancer. 2002;102:338–342. doi: 10.1002/ijc.10719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Luu HH, Kang Q, Park JK, Si W, Luo Q, Jiang W, Yin H, Montag AG, Simon MA, Peabody TD, Haydon RC, Rinker-Schaeffer CW, He TC. An orthotopic model of human osteosarcoma growth and spontaneous pulmonary metastasis. Clin Exp Metastasis. 2005;22:319–329. doi: 10.1007/s10585-005-0365-9. [DOI] [PubMed] [Google Scholar]
- 97.Luu HH, Zhou L, Haydon RC, Deyrup AT, Montag AG, Huo D, Heck R, Heizmann CW, Peabody TD, Simon MA, He TC. Increased expression of S100A6 is associated with decreased metastasis and inhibition of cell migration and anchorage independent growth in human osteosarcoma. Cancer Lett. 2005;229:135–148. doi: 10.1016/j.canlet.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 98.Arteaga CL, Kitten LJ, Coronado EB, Jacobs S, Kull FC Jr, Allred DC, Osborne CK. Blockade of the type I somatomedin receptor inhibits growth of human breast cancer cells in athymic mice. J Clin Invest. 1989;84:1418–1423. doi: 10.1172/JCI114315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.García-Echeverría C, Pearson MA, Marti A, Meyer T, Mestan J, Zimmermann J, Gao J, Brueggen J, Capraro HG, Cozens R, Evans DB, Fabbro D, Furet P, Porta DG, Liebetanz J, Martiny-Baron G, Ruetz S, Hofmann F. In vivo antitumor activity of NVP-AEW541-A novel, potent, and selective inhibitor of the IGF-IR kinase. Cancer Cell. 2004;5:231–239. doi: 10.1016/s1535-6108(04)00051-0. [DOI] [PubMed] [Google Scholar]
- 100.Favelyukis S, Till JH, Hubbard SR, Miller WT. Structure and autoregulation of the insulin-like growth factor 1 receptor kinase. Nat Struct Biol. 2001;8:1058–1063. doi: 10.1038/nsb721. [DOI] [PubMed] [Google Scholar]
- 101.Mulvihill MJ, Cooke A, Rosenfeld-Franklin M, Buck E, Foreman K, Landfair D, O’Connor M, Pirritt C, Sun Y, Yao Y, Arnold LD, Gibson NW, Ji QS. Discovery of OSI-906: a selective and orally efficacious dual inhibitor of the IGF-1 receptor and insulin receptor. Future Med Chem. 2009;1:1153–1171. doi: 10.4155/fmc.09.89. [DOI] [PubMed] [Google Scholar]
- 102.Aiken R, Axelson M, Harmenberg J, Klockare M, Larsson O, Wassberg C. Phase I clinical trial of AXL1717 for treatment of relapsed malignant astrocytomas: analysis of dose and response. Oncotarget. 2017;8:81501–81510. doi: 10.18632/oncotarget.20662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jerome L, Alami N, Belanger S, Page V, Yu Q, Paterson J, Shiry L, Pegram M, Leyland-Jones B. Recombinant human insulin-like growth factor binding protein 3 inhibits growth of human epidermal growth factor receptor-2-overexpressing breast tumors and potentiates herceptin activity in vivo. Cancer Res. 2006;66:7245–7252. doi: 10.1158/0008-5472.CAN-05-3555. [DOI] [PubMed] [Google Scholar]
- 104.Haluska P, Menefee M, Plimack ER, Rosenberg J, Northfelt D, LaVallee T, Shi L, Yu XQ, Burke P, Huang J, Viner J, McDevitt J, LoRusso P. Phase I dose-escalation study of MEDI-573, a bispecific, antiligand monoclonal antibody against IGFI and IGFII, in patients with advanced solid tumors. Clin Cancer Res. 2014;20:4747–4757. doi: 10.1158/1078-0432.CCR-14-0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hussain SA, Maroto P, Climent MA, Bianchini D, Jones RH, Lin CC, Wang SS, Dean E, Crossley K, Schlieker L, Bogenrieder T, De Bono JS. Targeting IGF-1/2 with xentuzumab (Xe) plus enzalutamide (En) in metastatic castration-resistant prostate cancer (mCRPC) after progression on docetaxel chemotherapy (DCt) and abiraterone (Abi): randomized phase II trial results. J. Clin. Oncol. 2019;37:5030–5030. [Google Scholar]
- 106.Ambrosio C, Ferber A, Resnicoff M, Baserga R. A soluble insulin-like growth factor I receptor that induces apoptosis of tumor cells in vivo and inhibits tumorigenesis. Cancer Res. 1996;56:4013. [PubMed] [Google Scholar]
- 107.Samani AA, Chevet E, Fallavollita L, Galipeau J, Brodt P. Loss of tumorigenicity and metastatic potential in carcinoma cells expressing the extracellular domain of the type 1 insulin-like growth factor receptor. Cancer Res. 2004;64:3380–3385. doi: 10.1158/0008-5472.CAN-03-3780. [DOI] [PubMed] [Google Scholar]
- 108.Sachdev D, Hartell JS, Lee AV, Zhang X, Yee D. A dominant negative type I insulin-like growth factor receptor inhibits metastasis of human cancer cells. J Biol Chem. 2004;279:5017–5024. doi: 10.1074/jbc.M305403200. [DOI] [PubMed] [Google Scholar]
- 109.Wang N, Rayes RF, Elahi SM, Lu Y, Hancock MA, Massie B, Rowe GE, Aomari H, Hossain S, Durocher Y, Pinard M, Tabaries S, Siegel PM, Brodt P. The IGF-Trap: novel inhibitor of carcinoma growth and metastasis. Mol Cancer Ther. 2015;14:982–993. doi: 10.1158/1535-7163.MCT-14-0751. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.