Abstract
Serotyping has traditionally been used for subtyping of non-typhoidal Salmonella (NTS) isolates. However, its discriminatory power is limited, which impairs its use for epidemiological investigations of source attribution. Whole-genome sequencing (WGS) analysis allows more accurate subtyping of strains. However, because of the relative newness and cost of routine WGS, large-scale studies involving NTS WGS are still rare. We aimed to revisit the big picture of subtyping NTS with a public health impact by using traditional serotyping (i.e. reaction between antisera and surface antigens) and comparing the results with those obtained using WGS. For this purpose, we analysed 18 282 sequences of isolates belonging to 37 serotypes with a public health impact that were recovered in the USA between 2006 and 2017 from multiple sources, and were available at the National Center for Biotechnology Information (NCBI). Phylogenetic trees were reconstructed for each serotype using the core genome for the identification of genetic subpopulations. We demonstrated that WGS-based subtyping allows better identification of sources potentially linked with human infection and emerging subpopulations, along with providing information on the risk of dissemination of plasmids and acquired antimicrobial resistance genes (AARGs). In addition, by reconstructing a phylogenetic tree with representative isolates from all serotypes (n=370), we demonstrated genetic variability within and between serotypes, which formed monophyletic, polyphyletic and paraphyletic clades. Moreover, we found (in the entire data set) an increased detection rate for AARGs linked to key antimicrobials (such as quinolones and extended-spectrum cephalosporins) over time. The outputs of this large-scale analysis reveal new insights into the genetic diversity within and between serotypes; the polyphyly and paraphyly of certain serotypes may suggest that the subtyping of NTS to serotypes may not be sufficient. Moreover, the results and the methods presented here, leading to differentiation between genetic subpopulations based on their potential risk to public health, as well as narrowing down the possible sources of these infections, may be used as a baseline for subtyping of future NTS infections and help efforts to mitigate and prevent infections in the USA and globally.
Keywords: antimicrobial resistance, foodborne infections, non-typhoidal Salmonella, Salmonella subtyping, source attribution
Data Summary
A list of the Sequence Read Archive (SRA) numbers of all NTS isolate sequences included in the final analysis (n=18282), their metadata (collection period, source, etc.) and genetic attributes are included in Table S1 (available in the online version of this article). All sequences were downloaded from the National Center for Biotechnology Information (NCBI) SRA repository.
Impact Statement.
Isolates of non-typhoidal Salmonella (NTS), a major foodborne pathogen, are traditionally subtyped into serotypes. However, serotyping has low discriminatory power, especially in comparison to the increasingly available whole-genome sequencing (WGS) technology. Thus far, the use of WGS in NTS studies has been limited to small-scale studies on certain serotypes, mainly as part of outbreak investigations. Here, in one of the largest analysis of NTS WGS published in the peer-reviewed literature, we analysed 18 282 sequences of isolates belonging to 37 serotypes with a public health impact that were recovered in the USA between 2006 and 2017 from multiple sources. Serotype and WGS-based subtyping were compared, and the advantages of the latter were demonstrated. In addition, we demonstrated that high variability within certain serotypes might result in the misclassification of distinct bacterial subpopulations into a single serotype. We found an increase in the detection of resistance genes to key antimicrobials over time, and that certain genetic subpopulations may present higher risk for their horizontal dissemination. Overall, this analysis provides a bird’s eye view of the genetic variation within and between NTS serotypes with a public health impact, and outputs may set the ground for future studies focusing on specific genetic subpopulations with a public health impact.
Introduction
Every year, approximately 1 million people fall ill, 20 000 are hospitalized and 400 die due to foodborne zoonotic salmonellosis in the USA [1]. The annual economic burden of the disease has been estimated to reach USD $3.7 billion (ranging between $193 million and $9.5 billion) in the country [2].
Serotyping has been used for many years for the subtyping of Salmonella isolates based on the immunological variability of two main surface structures, namely the O (somatic) and the H (flagellar) antigens [3]. Based on to the Kauffmann–White–Le Minor scheme, more than 2600 serotypes had been identified up to 2019; of these, approximately 1600 belong to the subspecies enterica (subspecies I), which is the subspecies involved in the vast majority of the foodborne zoonotic salmonellosis caused by non-typhoidal Salmonella (NTS) [4]. Even though many serotypes may contribute to human illness, the 20 most prevalent serotypes account for almost 70 % of the reported human cases in the USA according to a Centers for Disease Control and Prevention (CDC) report [5]. Serotyping is a useful and efficient way to subtype NTS infections, but it has low discriminatory power for differentiating between similar isolates belonging to the same serovar, which limits its use for epidemiological purposes, including tracing sources of infections [6]. Therefore, over the years, multiple molecular methods, such as pulsed-field gel electrophoresis (PFGE), multilocus sequence typing (MLST) and whole-genome sequencing (WGS), have been developed and increasingly applied to subtype Salmonella strains accurately, as reviewed by Tang et al. [6]. WGS is the most recent of these molecular methods, and can provide the highest subtyping resolution if used, for example, for single-nucleotide polymorphism (SNP) typing or core genome multilocus sequence typing (cgMLST) [7, 8]. This method is also gradually being adopted as the main method for pathogen subtyping by public health surveillance systems in different countries, including the USA [6].
The high discriminatory power of methods such as WGS to identify sources of infection can be particularly useful in the case of foodborne zoonoses, due to the complexity of the food processing chain, which includes multiple steps from farm to fork, in which cross-contamination may occur. Successful traceback of the sources of NTS outbreaks has been achieved in the past [9–11], allowing the implementation of control measures to reduce the risk of infection. However, the outputs of such studies may be limited to the specific outbreak being investigated. Moreover, outbreaks only account for a small portion of all Salmonella infections in humans, and most (60–80 %) cases are considered to be sporadic infections [12] for which no specific source is attributed. Pires et al. have reviewed the available approaches (other than WGS) for source attribution in the case of sporadic NTS infections [13]. These approaches provided an important contribution to our understanding of Salmonella epidemiology. However, the increasing availability of WGS opens up new opportunities for better identification of sources through finer resolution in strain subtyping [14, 15].
Here we used sequences of NTS isolates collected from various sources between 2006 and 2017 in the USA to generate WGS-based subtyping in comparison with the classic serotype-based approach. We grouped strains based on their serotype and based on the genetic subpopulations identified in the phylogenetic tree of each serotype, and compared the usefulness of each technique to identify isolates belonging to groups not typically associated with human infections and those belonging to emerging subpopulations that are associated with human infections and linked to one or more specific sources. Furthermore, we identified the presence of acquired antimicrobial resistance genes (AARGs), usually harboured by plasmids, in the sequences to estimate their potential risk for dissemination through these emerging subpopulations. The outputs of this large-scale analysis provide insights into the genetic diversity within and between serotypes and may be used as a baseline for subtyping of future NTS with a public health impact infections and estimating their potential source and public health risk.
Methods
Study population
The National Center for Biotechnology Information (NCBI) Pathogen Detection repository (https://www.ncbi.nlm.nih.gov/pathogens) Salmonella metadata were downloaded (13 October 2017) and explored to identify NTS isolates recovered in the USA between 2006 and 2017 from multiple sources (see below). We defined serotypes with a public health impact as those including metadata for at least 100 isolates, of which 1 or more were retrieved from humans. Based on this definition, the sequences of 25 897 isolates belonging to 37 serotypes were considered for inclusion in the analysis.
Data analysis
Data quality
For each serotype, paired-end Illumina reads were downloaded from the NCBI’s ftp server and assembled using SPAdes assembler v3.12.0 [16]. QUAST v4.6.3 [17] and Salmonella In Silico Typing Resource (SISTR) v1.0.2 [18] outputs were used for the filtration of assemblies with an N50 value <30 000 base pairs (bp) and mismatched serotypes, respectively (overall 1 685 isolates were excluded from the analysis; see the Supplementary Material for details). In addition, the average coverage (depth) of 99.6 % of the assemblies was at least 20 (see the Supplementary Material for details).
Genetic analyses
The assembled contigs were used as follows:
For the detection of AARGs, MLST and plasmid replicon types using ResFinder v2.1 [19], MLST v1.6 [20] and PlasmidFinder v1.2 [20], respectively. These bioinformatic tools and their databases [included in the ‘bacterial analysis pipeline’ at the Center for Genomic Epidemiology server (https://cge.cbs.dtu.dk/services/cge/)] were downloaded and used on a local server (with a local blast v2.4.0+ [21]) using the ‘bacterial analysis pipeline’ default settings.
For annotation using Prokka v1.13.3 [22] and reconstruction of a pan and core genomes. Pan and core genome reconstruction was conducted for each serotype separately (with two outgroup sequences in each analysis; see below) using the Prokka GFF format output in Roary v3.12.0 [23]. Core genes (genes present in at least 99 % of the genomes) were used to construct a multiple FASTA alignment file, from which the SNPs were extracted using SNP-sites v2.4.0 [24]. An approximately maximum-likelihood phylogenetic tree of each serotype was reconstructed with FastTree v2.1.10 [25] using the general time-reversible substitution evolutionary model with gamma correction (GTR+Γ). Trees were rooted using S. Paratyphi type A outgroup (SRR3033248, SRR3277289) and support for the tree branches was assessed using 5000 bootstrap replicates (see the Supplementary Material for additional details). The packages ape v5.0 [26] and ggtree v1.10.5 [27] in R software v3.4.3 [28] were used for visualization. For each serotype, the phylogenetic tree was visualized, and genetically distinct subpopulations were defined as clusters including more than 10 genetically similar sequences (with branch bootstrap support higher than 70 %; not necessarily a monophyletic group). Sequences not included in these subpopulations were defined as ‘not grouped’. These were excluded from the analyses when only the subpopulations were analysed or when comparing the subtyping by serotype and genetic subpopulations.
Moreover, the phylogenetic trees were scanned visually and 10 sequences were selected to represent each of the serotypes (and their distinct genetic subpopulations), and a core genome of these sequences (n=370) was created (as described above). The SNP alignment was then used for reconstruction of a maximum-likelihood (ML) phylogenetic tree with RAxML v8.2.10 [29]. The tree was rooted using the S. Paratyphi type A outgroup strain (see above) and 5000 bootstrap replicates were used for branch support.
The data quality and genetic analyses are further described in the Supplementary Material and the analysis pipeline is illustrated in Fig. S1.
Data interpretation
Comparison between subtyping by serotypes and genetic subpopulations
The sources of the sequenced isolates were categorized as either human, bovine, poultry or porcine (including isolates from both animals and food products), and ‘others’ (including isolates from food products not originated from livestock, environment, wildlife, domestic animals and with no information; see the Supplementary Material for additional details). In addition, the study period was divided into three equal time intervals: 2006–2009, 2010–2013 and 2014–2017.
The traditional serotype-based subtyping approach and the WGS approach (genetic subpopulations) were compared. For this purpose, we evaluated which of the sources (i.e. human, bovine, poultry, porcine and others) were found at least once in each serotype and subpopulation. Then, Venn diagrams (obtained through the package VennDiagram v1.6.18 [30] in R [28]) were used to illustrate the number of serotypes and genetic subpopulations for which a specific source and/or combination of sources were found. Furthermore, to allow a better identification of potentially emerging subpopulations and their sources, the proportion of isolates from each period and source in the subpopulations (and serotypes) were visualized using bar plots.
The genetic characteristics (i.e. presence of AARGs, MLST and plasmid replicon types) found in the genomes were summarized by serotype and genetic subpopulation, and the percentage of sequences in which AARGs conferring resistance to key antimicrobials were found was calculated for each subpopulation (see the Supplementary Material for additional details on data summarization). The risk for the dissemination of key antimicrobials was defined as follows: ‘no current risk’ when no AARGs were found in the genomes; ‘low’ for 1–10 % AARG prevalence; ‘moderate’ for 11–50 % AARG prevalence; and ‘high’ for >50 % AARG prevalence.
For genomes harbouring plasmid replicons, an estimate of their size was obtained using the average size of the replicon type, which was calculated based on the plasmid sizes described by Carattoli et al. [20]. Plasmids were then categorized into groups: (1) ‘small’, up to 6 kbp; (2) ‘intermediate’, between 6 and 100 kbp; and (3) ‘large’, more than 100 kbp. The percentage of sequences harbouring the different plasmid size groups was summarized for each genetic subpopulation. In addition, we evaluated which of the plasmid size categories (i.e. small, intermediate and large) were found at least once in each serotype and subpopulation, and summarized and compared the number of serotypes and genetic subpopulations for which a specific plasmid size category and/or combination of plasmid size categories were found.
General trends in the presence of AARGs and plasmid replicons
The entire data set was used to identify temporal trends of the following:
The presence of AARGs conferring antimicrobial resistance to ampicillin, streptomycin, sulfonamides and tetracycline, with/without genes conferring resistance to chloramphenicol – ACSSuT and ASSuT profiles, respectively. These profiles were defined, based on their definition in emerging serotypes (see the Discussion for further details), by the simultaneous presence of the genes blaCARB-2 (formerly named blaPSE-1), floR, aadA2, sul1 and tetG [31] for the ACSSuT profile, and the simultaneous presence of the genes blaTEM-1B, strA and strB, sul2 and tetB [32, 33] for the ASSuT profile.
The presence of AARGs conferring resistance to key antimicrobials including quinolones (aac(6′)Ib-cr, qnr and oqx genes), ESCs (including bla CMY, bla SHV, bla CTX-M and bla OXA genes) and colistin (mcr genes).
The presence of plasmid size groups.
Results
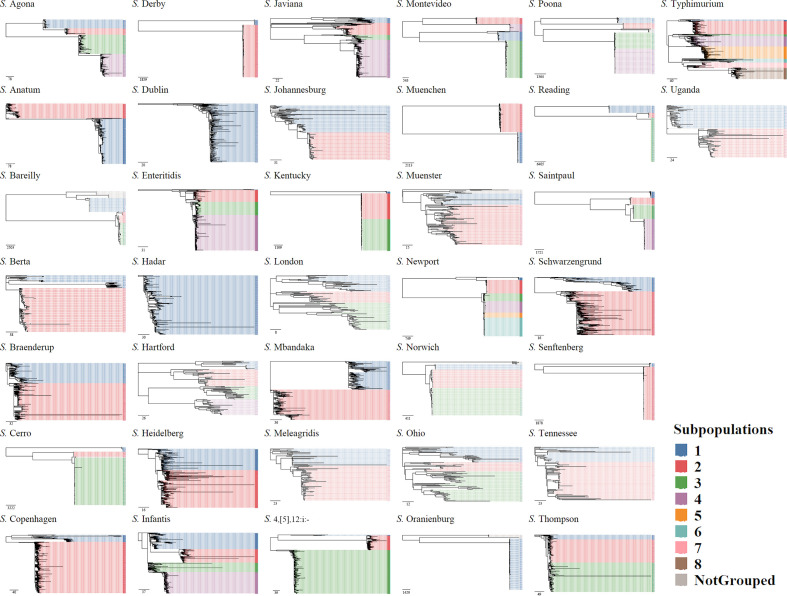
Overall the whole-genome sequences of 1282 NTS isolates belonging to 37 serotypes that were collected between 2006 and 2017 in the USA from multiple sources [human (n=6 180), bovine (n=1 702), poultry (n=6 129), swine (n=2 071) and others (n=2 200)] were included in the analysis. In the final analysis (after performing a quality control check and removing genetically identical duplicates; see below) the number of isolates per serotype varied between 94 and 2 113 (median=304), and the five predominant serotypes were S. Enteritidis (n=2 113), S. Typhimurium (n=1 926), S. Kentucky (n=1 635), S. Newport (n=1 159) and S. 4,[5],12:i:- (n=1 015). The number of core genes (found in at least 99 % of the isolates, including the S. Paratyphi A outgroup strains) in each serotype varied between 3 195 and 4 109 (median=3 793). The serotypes’ core genome alignment lengths varied between 2861875 and 3911965 bp (median=3561091 bp) and the SNPs’ variable site alignment lengths varied between 15663 and 71013 bp (median=40915 bp). The maximal difference between two isolates within a serotype (excluding the S. Paratyphi A outgroup strains) ranged between 151 (S. London; core genome alignment length=3611490 bp) and 38 926 (S. Reading; core genome alignment length=3561091 bp) SNPs, and between one and eight distinct genetic subpopulations were identified within each serotype (Fig. 1).
Fig. 1.
Approximate maximum-likelihood phylogenetic trees were reconstructed with FastTree using SNPs found in the core genomes of the 37 Salmonella serotypes. For each serotype, a core genome alignment was created including two S. Paratyphi type A outgroup strains (SRR3033248, SRR3277289; not included in the figure). Bootstrap replicates (n=5000) were used for branch support. Tree tips were coloured according to the identified genetic subpopulations. The scale bar indicates SNP difference.
Comparison between subtyping by serotypes and by genetic subpopulations
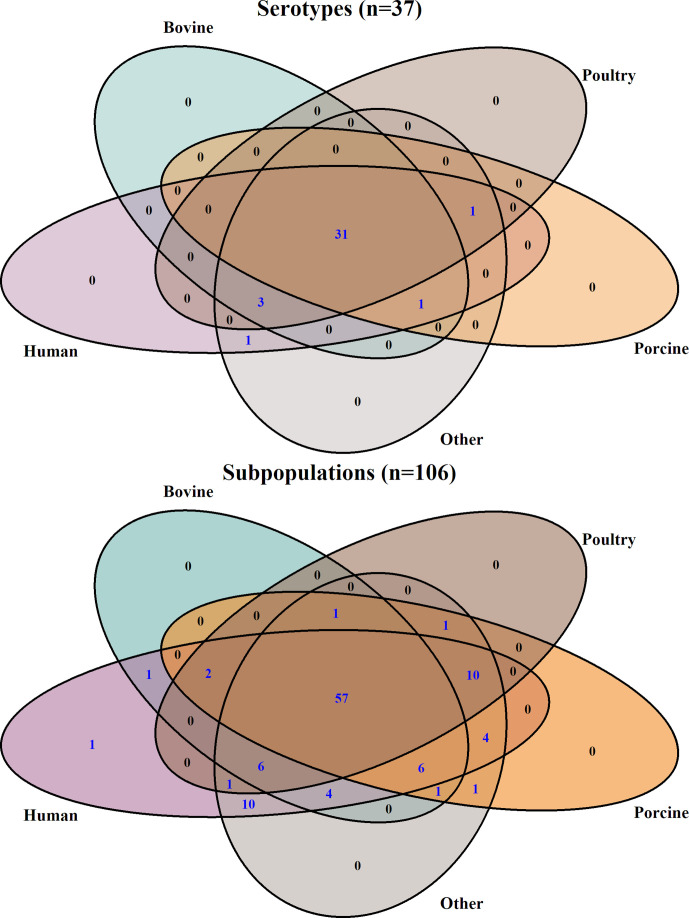
While subtyping the data by serotypes, 31/37 (84 %) of the serotypes included isolates collected from all sources considered, resulting in limited ability to identify a unique source for human infections. However, when using the genetic subpopulations for subtyping, only 57/106 (54 %) of the genetic subpopulations included all possible sources (Fig. 2; see also Fig. S2 for the proportions of different sources within the genetic subpopulations). Moreover, changes in the proportions of the different subpopulations within a serotype over time were demonstrated using WGS subtyping (Fig. S3). For example, the total number of S. 4,[5],12:i:- sequences increased over time. However, when examining the proportion of sequences from each genetic subpopulation, it was apparent that subpopulation 3 of this serotype, which was first detected during 2010–2013, was the main contributor to the dramatic increase of this serotype in 2014–2017 (an increase was also evident in subpopulations 1 and 2. However, subpopulation 3 consisted almost 75 % of the sequences of this serotype). Large variability in genetic resistance (based on presence of AARGs) to antimicrobial classes was found across serotypes (Table 1). Furthermore, the genetic characteristics (AARGs, MLST and predominant plasmid replicons) varied between subpopulations within serotypes. For example, resistance to phenicols (mostly due to the presence of floR gene) was mainly found in serotypes Dublin (85.19 % of the sequences within the serotype), Infantis (28.22 %), Typhimurium (21.81 %), Agona (15.67 %) and Newport (13.29 %). However, in all but serotype Dublin, in which only one subpopulation was defined, certain subpopulations were the main contributors for this resistance: S. Infantis subpopulation 4 [244/336 (72.62 %)]; S. Typhimurium subpopulations 3 [35/67 (71.43 %)] and 5 [328/400 (82 %)]; S. Agona subpopulations 1 [23/46 (50 %)] and 2 [14/34 (41.18 %)]; and S. Newport subpopulation 2 [146/262 (55.73 %)] (Table S2).
Fig. 2.
Venn diagrams demonstrating the degree of overlap between sources when data were subtyped by serotypes (upper inset) or genetic subpopulations (lower inset). The number of serotypes/subpopulations is indicated within each category (values higher than zero are highlighted in blue). Sources were coloured as follows: human (purple), bovine (blue), poultry (brown), porcine (orange) and other (grey).
Table 1.
Genotypic resistance to different antimicrobial classes in non-typhoid Salmonella sequences of isolates belonging to 37 serotypes with a public health impact that were collected in the USA between 2006 and 2017. Resistance was predicted based on the presence of AARGs. The number of sequences harbouring AARGs linked to each antimicrobial class and their percentage within a serotype are presented for each serotype
|
Serotype |
n |
Beta-lactams |
Aminoglycosides |
Folate pathway inhibitors |
Tetracyclines |
Macrolides and lincosamides |
Quinolones |
Phenicols |
Others |
|---|---|---|---|---|---|---|---|---|---|
|
Agona |
300 |
75 (25 %) |
124 (41.33 %) |
122 (40.67 %) |
130 (43.33 %) |
3 (1 %) |
13 (4.33 %) |
47 (15.67 %) |
0 (0 %) |
|
Anatum |
646 |
26 (4.02 %) |
27 (4.18 %) |
17 (2.63 %) |
217 (33.59 %) |
2 (0.31 %) |
20 (3.1 %) |
3 (0.46 %) |
0 (0 %) |
|
Bareilly |
128 |
9 (7.03 %) |
1 (0.78 %) |
1 (0.78 %) |
1 (0.78 %) |
0 (0 %) |
2 (1.56 %) |
0 (0 %) |
0 (0 %) |
|
Berta |
179 |
26 (14.53 %) |
28 (15.64 %) |
18 (10.06 %) |
50 (27.93 %) |
0 (0 %) |
1 (0.56 %) |
0 (0 %) |
0 (0 %) |
|
Braenderup |
345 |
12 (3.48 %) |
10 (2.9 %) |
5 (1.45 %) |
9 (2.61 %) |
0 (0 %) |
4 (1.16 %) |
1 (0.29 %) |
0 (0 %) |
|
Cerro |
250 |
1 (0.4 %) |
7 (2.8 %) |
2 (0.8 %) |
9 (3.6 %) |
0 (0 %) |
0 (0 %) |
0 (0 %) |
0 (0 %) |
|
Copenhagen |
587 |
362 (61.67 %) |
198 (33.73 %) |
551 (93.87 %) |
546 (93.02 %) |
1 (0.17 %) |
1 (0.17 %) |
31 (5.28 %) |
0 (0 %) |
|
Derby |
407 |
30 (7.37 %) |
196 (48.16 %) |
183 (44.96 %) |
321 (78.87 %) |
5 (1.23 %) |
14 (3.44 %) |
12 (2.95 %) |
0 (0 %) |
|
Dublin |
297 |
251 (84.51 %) |
268 (90.24 %) |
262 (88.22 %) |
263 (88.55 %) |
2 (0.67 %) |
4 (1.35 %) |
253 (85.19 %) |
0 (0 %) |
|
Enteritidis |
2113 |
109 (5.16 %) |
59 (2.79 %) |
53 (2.51 %) |
68 (3.22 %) |
0 (0 %) |
10 (0.47 %) |
14 (0.66 %) |
0 (0 %) |
|
Hadar |
381 |
123 (32.28 %) |
345 (90.55 %) |
46 (12.07 %) |
338 (88.71 %) |
0 (0 %) |
2 (0.52 %) |
0 (0 %) |
0 (0 %) |
|
Hartford |
110 |
4 (3.64 %) |
0 (0 %) |
0 (0 %) |
0 (0 %) |
0 (0 %) |
1 (0.91 %) |
0 (0 %) |
0 (0 %) |
|
Heidelberg |
930 |
281 (30.22 %) |
375 (40.32 %) |
234 (25.16 %) |
295 (31.72 %) |
4 (0.43 %) |
55 (5.91 %) |
73 (7.85 %) |
0 (0 %) |
|
Infantis |
964 |
288 (29.88 %) |
393 (40.77 %) |
376 (39 %) |
378 (39.21 %) |
2 (0.21 %) |
8 (0.83 %) |
272 (28.22 %) |
0 (0 %) |
|
Javiana |
558 |
34 (6.09 %) |
12 (2.15 %) |
6 (1.08 %) |
3 (0.54 %) |
0 (0 %) |
9 (1.61 %) |
0 (0 %) |
0 (0 %) |
|
Johannesburg |
147 |
8 (5.44 %) |
10 (6.8 %) |
8 (5.44 %) |
25 (17.01 %) |
0 (0 %) |
6 (4.08 %) |
1 (0.68 %) |
0 (0 %) |
|
Kentucky |
1635 |
162 (9.91 %) |
1216 (74.37 %) |
63 (3.85 %) |
911 (55.72 %) |
2 (0.12 %) |
10 (0.61 %) |
4 (0.24 %) |
1 (0.06 %) |
|
London |
109 |
11 (10.09 %) |
19 (17.43 %) |
11 (10.09 %) |
31 (28.44 %) |
0 (0 %) |
5 (4.59 %) |
0 (0 %) |
0 (0 %) |
|
Mbandaka |
304 |
9 (2.96 %) |
22 (7.24 %) |
21 (6.91 %) |
46 (15.13 %) |
0 (0 %) |
6 (1.97 %) |
1 (0.33 %) |
0 (0 %) |
|
Meleagridis |
94 |
5 (5.32 %) |
11 (11.7 %) |
11 (11.7 %) |
20 (21.28 %) |
1 (1.06 %) |
0 (0 %) |
8 (8.51 %) |
0 (0 %) |
|
4,[5],12:i:- |
1015 |
674 (66.4 %) |
681 (67.09 %) |
662 (65.22 %) |
782 (77.04 %) |
36 (3.55 %) |
87 (8.57 %) |
58 (5.71 %) |
2 (0.2 %) |
|
Montevideo |
594 |
21 (3.54 %) |
51 (8.59 %) |
31 (5.22 %) |
65 (10.94 %) |
1 (0.17 %) |
19 (3.2 %) |
9 (1.52 %) |
0 (0 %) |
|
Muenchen |
382 |
13 (3.4 %) |
58 (15.18 %) |
54 (14.14 %) |
52 (13.61 %) |
0 (0 %) |
5 (1.31 %) |
0 (0 %) |
0 (0 %) |
|
Muenster |
135 |
9 (6.67 %) |
40 (29.63 %) |
36 (26.67 %) |
40 (29.63 %) |
0 (0 %) |
10 (7.41 %) |
8 (5.93 %) |
0 (0 %) |
|
Newport |
1159 |
192 (16.57 %) |
178 (15.36 %) |
174 (15.01 %) |
181 (15.62 %) |
7 (0.6 %) |
19 (1.64 %) |
154 (13.29 %) |
0 (0 %) |
|
Norwich |
110 |
9 (8.18 %) |
1 (0.91 %) |
0 (0 %) |
0 (0 %) |
0 (0 %) |
0 (0 %) |
0 (0 %) |
0 (0 %) |
|
Ohio |
113 |
15 (13.27 %) |
15 (13.27 %) |
15 (13.27 %) |
16 (14.16 %) |
1 (0.88 %) |
2 (1.77 %) |
8 (7.08 %) |
0 (0 %) |
|
Oranienburg |
172 |
3 (1.74 %) |
1 (0.58 %) |
0 (0 %) |
1 (0.58 %) |
0 (0 %) |
3 (1.74 %) |
0 (0 %) |
0 (0 %) |
|
Poona |
154 |
3 (1.95 %) |
4 (2.6 %) |
1 (0.65 %) |
2 (1.3 %) |
0 (0 %) |
3 (1.95 %) |
1 (0.65 %) |
0 (0 %) |
|
Reading |
224 |
48 (21.43 %) |
74 (33.04 %) |
62 (27.68 %) |
59 (26.34 %) |
0 (0 %) |
1 (0.45 %) |
3 (1.34 %) |
0 (0 %) |
|
Saintpaul |
541 |
220 (40.67 %) |
113 (20.89 %) |
51 (9.43 %) |
256 (47.32 %) |
1 (0.18 %) |
9 (1.66 %) |
2 (0.37 %) |
1 (0.18 %) |
|
Schwarzengrund |
457 |
26 (5.69 %) |
278 (60.83 %) |
37 (8.1 %) |
50 (10.94 %) |
9 (1.97 %) |
6 (1.31 %) |
1 (0.22 %) |
0 (0 %) |
|
Senftenberg |
302 |
59 (19.54 %) |
68 (22.52 %) |
50 (16.56 %) |
44 (14.57 %) |
1 (0.33 %) |
13 (4.3 %) |
14 (4.64 %) |
0 (0 %) |
|
Tennessee |
96 |
1 (1.04 %) |
2 (2.08 %) |
0 (0 %) |
2 (2.08 %) |
0 (0 %) |
0 (0 %) |
0 (0 %) |
0 (0 %) |
|
Thompson |
317 |
7 (2.21 %) |
5 (1.58 %) |
7 (2.21 %) |
3 (0.95 %) |
1 (0.32 %) |
3 (0.95 %) |
2 (0.63 %) |
1 (0.32 %) |
|
Typhimurium |
1926 |
779 (40.45 %) |
729 (37.85 %) |
1042 (54.1 %) |
1015 (52.7 %) |
39 (2.02 %) |
70 (3.63 %) |
420 (21.81 %) |
2 (0.1 %) |
|
Uganda |
101 |
7 (6.93 %) |
16 (15.84 %) |
14 (13.86 %) |
17 (16.83 %) |
1 (0.99 %) |
1 (0.99 %) |
9 (8.91 %) |
0 (0 %) |
|
Total |
18 282 |
3912 (21.4 %) |
5635 (30.82 %) |
4226 (23.12 %) |
6246 (34.16 %) |
119 (0.65 %) |
422 (2.31 %) |
1409 (7.71 %) |
7 (0.04 %) |
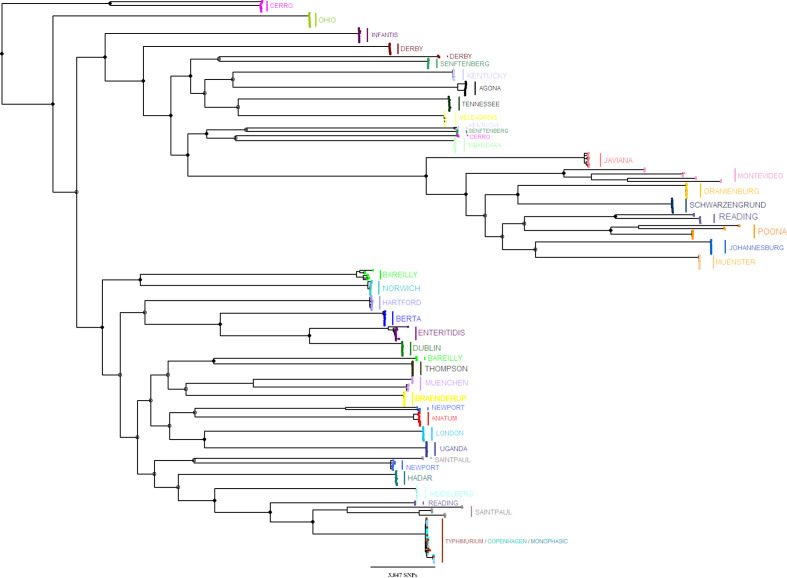
The reconstruction of the ML phylogenetic tree of all 37 serotypes (n=370) was conducted using 128 217 SNP variable sites that were found in 2965 core genes (including the S. Paratyphi A outgroup strains; Fig. 3). Different levels of genetic variation within serotypes were observed, and the selected sequences (n=10 from each serotype) formed monophyletic, polyphyletic and paraphyletic clades in different serotypes (Figs 3 and S4; see the Discussion for further details).
Fig. 3.
An ML phylogenetic tree was reconstructed with RAxML using SNPs found in the core genome of representative sequences from all 37 serotypes (n=370). Ten sequences were selected from each serotype phylogeny to represent the diversity of the genetic subpopulations. The tree was rooted using S. Paratyphi type A outgroup strains (SRR3033248, SRR3277289; not included in the figure). Bootstrap replicates (n=5000) were used for branch support. Full and empty circles indicate ≥70 and <70 % bootstrap support for major branches, respectively. Tree tips were coloured according to the serotype (Fig. S3 includes the same tree, annotated by the serotype and the genetic subpopulations).
The predicted risk for dissemination of AARGs varied between subpopulations (Tables 2 and S3). In 57/106 (54 %), 9/106 (8 %) and 4/106 (4 %) of the subpopulations, AARGs conferring resistance to extended-spectrum cephalosporins (ESCs) by means of AmpC, extended-spectrum beta-lactamases (ESBLs) and carbapenemases were found, respectively. These subpopulations were estimated to have the potential to disseminate resistance to ESCs with a level of risk ranging between low and high (1–10 and >50 % of the sequences within the subpopulation harbouring resistance genes to ESCs, respectively). In 68/106 (64 %) and 54/106 (51 %) of the subpopulations one or more AARGs conferring resistance to quinolones or only the predominant qnrB19 gene were found, respectively, and it was estimated that the risk for dissemination was up to moderate (11–50 % of the sequences within the subpopulations harbouring resistance genes to quinolones).
Table 2.
The estimated level of risk for resistance dissemination from genetic subpopulations of non-typhoid Salmonella
|
Resistance dissemination risk level* |
# of subpopulations in the category (% out of all 106 subpopulations) |
||||
|---|---|---|---|---|---|
|
ESC classes |
Quinolones |
||||
|
AmpC† |
ESBL‡ |
Carbapenems§ |
All|| |
qnrB19 |
|
|
No current |
49 (46.23 %) |
97 (91.51 %) |
102 (96.23 %) |
38 (35.85 %) |
52 (49.06 %) |
|
Low |
46 (43.4 %) |
8 (7.55 %) |
3 (2.83 %) |
63 (59.43 %) |
52 (49.06 %) |
|
Moderate |
6 (5.66 %) |
– |
1 (0.94 %) |
5 (4.72 %) |
2 (1.89 %) |
|
High |
5 (4.72 %) |
1 (0.94 %) |
– |
– |
– |
*Genetic subpopulations were categorized according the percentage of sequences that harboured the AARGs. The categories were defined as follows: ‘no current’ – none were found; ‘low’ – between 1 and 10 % harboured the AARGs; ‘moderate’ – between 11 and 50 % harboured the AARGs; and ‘high’ – above 50 % harboured the AARGs.
†Presence of bla CMY genes.
‡Presence of bla SHV and/or bla CTX-M genes.
§Presence of bla OXA genes.
||Presence of qnr and/or aac(6')-Ib-cr and/or oqx genes.
Plasmid replicons indicated the presence of small (<6000 bp), intermediate (≥6 000 bp, <100000 bp) and large (≥100000 bp) plasmids in sequences from 36, 37 and 34 of the serotypes respectively, with all serotypes but 1 harbouring plasmids of more than 1 size category. When genetic subpopulations were considered, plasmids belonging to only one category size were found in 11/94, 20/103 and 11/77 of the subpopulations harbouring small, intermediate and large plasmids, respectively (Table 3). Moreover, the percentage of isolates harbouring plasmids varied within serotypes and subpopulations: 0–98 % (median=9 %), 0–100 % (median=21.5 %) and 0–100 % (median=5.5 %) of the isolates within subpopulation were harbouring small, intermediate and large plasmids, respectively (Table S3, Fig. S5).
Table 3.
The presence of plasmids (based on the identification of plasmid replicons) in serotypes and genetic subpopulations of non-typhoid Salmonella , categorized according to the plasmids’ estimated size into ‘small’ (<6000 bp), ‘intermediate’ (≥6 000 bp, <100000 bp) and ‘large’ (≥100000 bp). The total number and the extent of overlap between plasmid size groups within serotype and genetic subpopulations are presented
|
Plasmid size* category |
Grouping method |
Found in (n) |
Alone |
Overlap with other plasmid size categories |
|---|---|---|---|---|
|
Small (<6000 bp) |
Serotype |
36 |
0 (0 %) |
36 (100 %) |
|
Subpopulation |
94 |
11 (11.7 %) |
83 (88.3 %) |
|
|
Intermediate (≥6000 bp, <100000 bp) |
Serotype |
37 |
0 (0 %) |
37 (100 %) |
|
Subpopulation |
103 |
20 (19.4 %) |
83 (80.6 %) |
|
|
Large (≥100000 bp) |
Serotype |
34 |
1 (2.9 %) |
33 (97.1 %) |
|
Subpopulation |
77 |
11 (14.3 %) |
66 (85.7 %) |
*Approximate average sizes for Col/Inc plasmid groups were determined using Carattoli et al. [19] (as detailed in the the Methods section).
General trends in the presence of AARGs and plasmid replicons
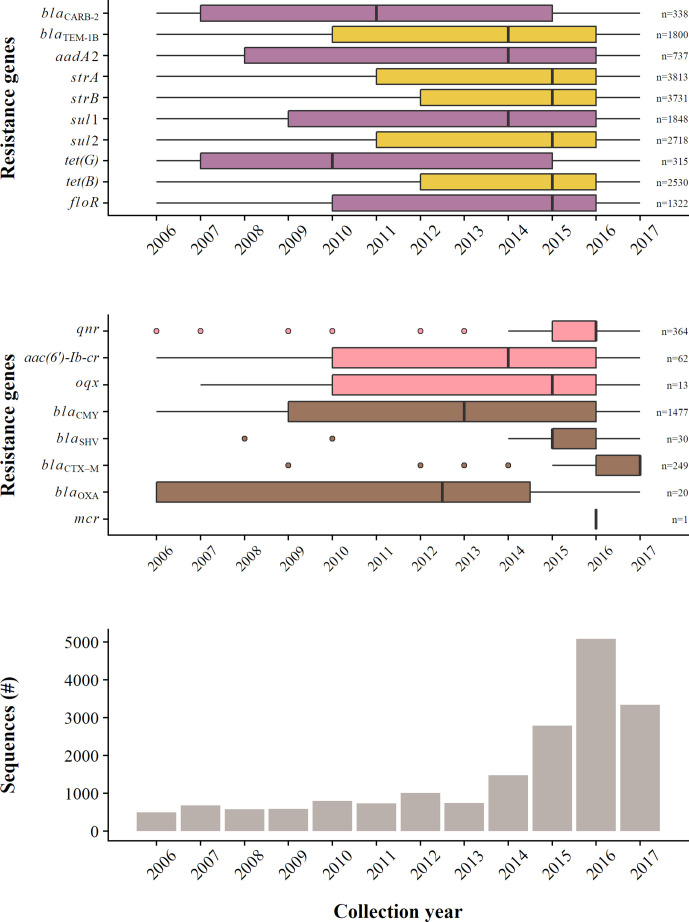
The AARGs conferring the ACSSuT and ASSuT resistance profiles (see above) were found in isolates recovered throughout the period between 2006 and 2017, regardless of the sequencing intensity (Fig. 4, upper and lower insets, respectively). However, the frequency of the AARGs conferring ASSuT were higher than those conferring ACSSuT (e.g. 1800 bla TEM-1B as opposed to only 338 bla CARB-2 genes being detected between 2006 and 2017). Moreover, while some of the AARGs conferring the ACSSuT profile [i.e. bla CARB-2 and tet(G)] have been detected less frequently since 2015 (despite a larger number of sequences being available between 2015 and 2017), the majority (75 %) of the isolates harbouring the AARGs conferring the ASSuT genetic profile were detected starting 2010 and 2012 period.
Fig. 4.
General trends found in the data during the period between 2006 and 2017. (i) The presence of different AARGs sets conferring resistance to ampicillin, streptomycin, sulfonamides, tetracycline and chloramphenicol (i.e. ACSSuT; purple) or without chloramphenicol (i.e. ASSuT; yellow) (see text for additional details) (upper inset). The number of genes detected is indicated on the right. (ii) The presence of selected acquired antimicrobial resistance genes (AARGs) conferring resistance to ESCs (brown) or quinolones (pink) (middle inset). The number of genes detected is indicated on the right. (iii) The number of available whole-genome sequences (as raw reads) of NTS isolates in the NCBI SRA (lower inset).
The detection of AARGs conferring resistance to key antimicrobial classes was variable (Fig. 4, middle inset). bla CMY genes (n=1477), of which 1348 (91 %) were bla CMY-2, the predominant gene conferring resistance to ESCs, were detected in sequences of isolates collected throughout the period between 2006 and 2017, while the increasingly abundant bla CTX-M (n=249) had mainly been found since 2015, with 50 % of genes detected in isolates collected during 2017. qnr genes (n=364) were the predominant AARGs associated with quinolone resistance. These had already been detected in isolates collected in 2006, but most genes were found after 2014, with 50 % of the genes detected in sequences of isolates collected since 2016. mcr gene (mcr-1) was only detected once in a sequence of an isolate collected in 2016.
Small (<6 000 bp), intermediate (≥6 000 bp, <100000 bp) and large (≥100000 bp) plasmids were identified in sequences of isolates collected throughout the period between 2006 and 2017, with the majority (75 %) of plasmids detected since 2011, 2012 and 2012, respectively (data not shown).
Discussion
We have analysed a large data set of 18 282 whole-genome sequences of NTS isolates that were collected in the USA between 2006 and 2017 belonging to 37 serotypes with a public health impact. By aligning the core genes of isolates within each serotype, we were able to reconstruct the phylogenetic trees and identify genetic subpopulations within serotypes. We used the assembled contigs to identify unique genetic characteristics, including AARGs, and this information was combined with the source and collection time. By comparing the subtyping of isolates based on traditional serotyping and the one based on the phylogenies, we demonstrated the usefulness of the latter for the detection of specific subpopulations that may be associated with a limited number of sources and/or might be increasing in prevalence over time. For example, eggs and poultry meat are regarded as the main source of S. Enteritidis [34]. However, when the WGS information for S. Enteritidis from the USA is considered, this association with poultry and its products is reflected in subpopulations 3 and 4, but not in subpopulations 1 and 2 of this serotype (Fig. S2). The latter are mainly associated with the ‘other’ category, which includes various possible sources. It is possible that isolates in these subpopulations ultimately originated from poultry and ended up being recovered from other sources due to cross-contamination, but it is also possible that non-poultry sources are the main origin of a proportion of human cases. This is important when considering disease control and mitigation and, in fact, the increasing availability of WGS data through surveillance programmes for foodborne infections [35] has allowed the identification of the sources of infections due to certain Salmonella serovars through machine learning. These approaches may not always be straightforward, however: Wheeler [15] summarized the findings of two studies that applied machine learning to track the sources of S. Typhimurium infections in the USA and found contradictory results regarding the source of this pathogen, possibly due to differences in the data selected for use in the models. This is, however, another example of the potential usefulness of using the discriminatory power of WGS for subtyping of Salmonella and source tracking.
Given the high number of the NTS sequences that were considered for inclusion in this analysis (above 25000; Fig. S1), filtration of poor quality and/or potentially misclassified sequences was required to avoid errors that might require repetition of the analyses. Among other measures to ensure the quality of the data, in silico serotyping was used as an initial filtration step and isolates for which the predicted serotype was in disagreement with the metadata were excluded. High accuracy (approximately 95 %) of SISTR was estimated previously, as described in a review by Tang et al. [6]. Accordingly, here in most serotypes the predicted serotypes of only few isolates (up to 6.7 % of the sequences within serotype) were found to disagree with the metadata. However, in S. Enteritidis 959/6225 (15.41 %) of the sequences were excluded. This may be a result of low coverage (depth) of the specific genes detected by SISTR or it may indicate of a limitation of SISTR to identify this serotype. However, it is also possible that errors in traditional serotyping occurred, as this technique can be error-prone [6, 36].
While reconstructing the phylogenetic trees of different serotypes, a relatively large genetic heterogeneity was observed between subpopulations in certain serotypes (S. Bareilly, S. Cerro, S. Derby, S. Kentucky, S. Montevideo, S. Newport, S. Reading, S. Saintpaul, S. Senftenberg and S. Poona), but not in others (e.g. S. Hadar and S. Dublin). This variability was also evident in the phylogenetic tree that included all 37 serotypes: for most serotypes, all genetic subpopulations were included in a monophyletic clade, but isolates from serotypes with large genetic variability (except S. Derby, S. Montevideo and S. Poona) constructed polyphyletic clades with genetic subpopulations divided into different tree branches. The existence of polyphyletic serotypes has been reported before [37–39], including when comparing between subtyping of Salmonella by serotyping and sequence types (STs) and/or by genetically closely related clusters called eBurstGroups (eBGs) [36], but never in the context of an analysis including such a large number of isolates of multiple serotypes with public health relevance, as in this study. This within-serotype genetic variability further supports the need to use techniques with high discriminatory power to study the epidemiology of these important pathogens. This is particularly evident in the case of S. Typhimurium and its two variants, serotypes 4,[5],12:i:- and Copenhagen [40, 41]. In the phylogenetic tree of all serotypes, S. Typhimurium formed a paraphyletic clade and its variants (S. 4,[5],12:i:- and S. Copenhagen) formed polyphyletic clades with the genetic subpopulations divided into different branches. A similar structure was also evident in a previous study we conducted [32], in which a phylogenetic tree of S. Typhimurium and S. 4,[5],12:i:- sequences was divided into two main clades according to two genetic subpopulations of S. 4,[5],12:i:-, with S. Typhimurium isolates included in both. Therefore, our findings suggest that differentiation of isolates belonging to these three serotypes based on their antigenic formulas may be misleading, and that genetic characterization will give a more accurate picture of their epidemiological relationship. However, this study was designed to provide a bird’s eye view of the genetic structure of multiple NTS serotypes with a public health impact, and for this reason a single genetically related outgroup (S. Paratyphi A) was included in all of the phylogeny reconstructions. This decision allowed standardization of the analyses, but inevitable variation in the genetic similarity between different serotypes and the selected outgroup may have resulted in reduced ability to identify minor changes within those. Additional focused studies that may also include the use of models to estimate the evolution, such as Bayesian evolutionary analysis sampling trees (beast [42]), of S. Typhimurium and its variants are required for finer estimation of the epidemiology of these serotypes.
We identified specific genetic characteristics (such as the presence of AARGs and plasmids) linked to certain genetic subpopulations, which may pose a risk to public health. Plasmids play an important role in horizontal transmission of genetic material, which may also include AARGs, between isolates [20, 43]. When subtyping sequences by serotypes, all serotypes were found to harbour plasmids. However, subtyping by genetic subpopulations revealed that not all genetic subpopulations within a certain serotype were equally likely to harbour plasmids or to harbour plasmids of a specific size category (e.g. only 73 % of the subpopulations harboured large plasmids). Conjugation, the main mechanism for horizontal transmission of plasmids, requires the presence of both mobility (MOB) and mating pair formation (MPF) complexes that involve multiple genes [43, 44]. In the presence of the MOB complex only, plasmids may be mobilized (horizontally) by using MPF (from another plasmid) or another genetic element present in the host cell [44]. Furthermore, Smillie et al. [44] estimated that mobilizable plasmids (the size distribution of mobilizable plasmids peaked at 5 kbp with an additional secondary peak at around 150 kbp) were generally smaller than conjugable plasmids (average size of approximately 100 kbp). We found that in 11.7 and 19.4 % of the genetic subpopulations only small and intermediate plasmids were found, respectively. This finding may suggest that in these genetic subpopulations horizontal transmission may be limited as, even though plasmids may have the potential to be mobilized horizontally, in certain cases, especially for small plasmids, this ability may be limited due to the lack of MPF complexes in the host bacterial cells.
With the constant increase in antimicrobial resistance over time and the predictions for further increases in the future, the importance of reserving efficient antimicrobials is further aggravated [45]. We found a relatively high number of sequences encoding the bla CMY-2 (n=1348), a gene commonly found in ESC-resistant NTS isolates from multiple sources in the USA [46–48]. bla CTX-M genes, which were previously rarely found in the USA [49], were identified in 249 sequences (1.36 %) here. Similarly, qnr genes were rarely found in the USA before 2007 [50], but their presence has been increasing in recent years [51] and 364 sequences (1.99 %) harboured these genes. In addition, based on the percentage of sequences in which AARGs linked to quinolones and ESCs were found, we estimated that at least 55 and 4 % of the subpopulations could be a source for the dissemination of AARGs conferring resistance to quinolones and ESCs, respectively, assuming that these genes are still harboured by plasmids and were not incorporated in the bacterial chromosome.
In the historic S. Typhimurium DT-104, phenotypic resistance to ACSSuT was conferred by the genes blaCARB-2 (formerly named blaPSE-1), floR, aadA2, sul1 and tetG, respectively [31]. These were localized on a 13 kb multidrug resistance region on Salmonella genomic island (SGI)-1 [52]. In the recently emerging S. 4,[5],12:i:- ST34, a similar resistance profile, excluding the resistance to chloramphenicol (ASSuT), was conferred by the genes blaTEM-1B, strA and strB, sul2 and tetB, respectively [32, 33]. The resistance array that includes this cassette of genes was previously described as RR3 [53] and was found in a large data set of S. 4,[5],12:i:- ST34 sequences from Europe and the USA [32]. Our findings here suggest that the gene cassettes conferring the ACSSuT and ASSuT phenotypic profiles can be found in multiple genetic subpopulations of several NTS serotypes. The dissemination of these genetic resistance determinants between serotypes may be attributed to recent insertions of mobile transposons, as suggested by Petrovska et al. [33], and may also reflect the risk of plasmid dissemination among Enterobacteriaceae [54]. However, insertion of integrative and conjugative elements (ICEs) [55] in the bacterial chromosome and vertical transmission of genes cassettes between evolving serotypes with/without partial loss of genes may serve as an alternative hypothesis of ancestral origin for the dissemination of the resistance gene cassettes in NTS. Moreover, the higher frequency of detection of genes conferring resistance to ASSuT in comparison to genes conferring ACCSuT over the study period may suggest that the former provides an evolutionary advantage for isolates harbouring it.
The inherent bias caused by the increasing availability of sequencing data over time might have influenced the observed trends in detection of certain genetic components, such as plasmids. However, the increase of qnr genes in sequenced isolates is likely to represent a true rise in their frequency, as reflected by the increase in genotypic and phenotypic resistance to quinolones in NTS in the USA in recent years that was described in a National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS) report [51]. Potential additional biases of a study such as this, relying on publicly available data submitted to the NCBI, would be the over-representation of specific sources that could be a result of: (a) the nature of the data submitted to NCBI, representing more clinical cases of human and samples from poultry than other sources; and/or (b) the submission of multiple samples with the same or an epidemiologically linked origin, as may occur during an outbreak. However, the approach taken here included removal of sequences with genetically identical core genomes (duplicates) and therefore such cases are less likely to affect the outputs of this analysis. In addition, the majority of the duplicates found in this study belonged to S. Enteritidis, which may be a consequence of the low genetic variability within this serotype, as demonstrated here and elsewhere [8].
In conclusion, we performed a large-scale analysis of multiple NTS serotypes with a public health impact and demonstrated the usefulness of using WGS to reconstruct phylogeny trees for isolate subtyping. As opposed to using WGS in small-scale analyses for outbreak investigations (as reviewed by Tang et al. [6]), here we aimed to provide a broad view on multiple serotypes with a public health impact and the study outputs may establish the basis for further focused analyses of specific emerging genetic subpopulations and/or genetic subpopulations demonstrating higher risk for antimicrobial resistance spread. Identifying these genetic subpopulations and understanding their epidemiology may contribute to efforts invested in the prevention and mitigation of these important foodborne pathogens. Moreover, the outcomes of the analysis here may help in the identification of potential sources/potential risk due to the carriage of resistance determinants of future Salmonella outbreaks in the USA based on genetic similarity to certain genetic subpopulations.
Supplementary Data
Funding information
This work was supported by the Global Food Venture–MnDrive Initiative, the National Institute of Food and Agriculture (Animal Health Formula Fund project MIN-62–091) of the USDA, the Rapid Agricultural Response Fund (RARF) and the Swine Disease Eradication Center (SDEC) at the University of Minnesota. In addition, E.E. was supported by BARD, the United States–Israel Binational Agricultural Research and Development Fund (Vaadia–BARD Postdoctoral Fellowship award no. FI-565–17) and J.A. was supported by the Ramón y Cajal postdoctoral contract from the Spanish Ministry of Economy, Industry and Competitiveness (MINECO) (RYC-2016–20422).
Acknowledgements
We want to thank Belinda Befort for her help with the initial data analysis.
Conflicts of interest
The authors declare that there are no conflicts of interest
Author contributions
E. E. and J. A. designed the study. S. L. H., S. L. and E. E. contributed to data processing and analysis. E. E. (with scientific advice from T. J. J., A. P. and J. A.) interpreted the results, and drafted and edited the paper. All co-authors contributed to revising and structuring the paper, and all approved the final draft.
Footnotes
Abbreviations: AARG, acquired antimicrobial resistance genes; ACSSuT, ampicillin, chloramphenicol, streptomycin, sulfonamides, and tetracycline; ASSuT, ampicillin, streptomycin, sulfonamides, and tetracycline; ESBL, extended spectrum beta lactamases; ESC, extended spectrum cephalosporins; MLST, multilocus sequence typing; NCBI, National Center for Biotechnology Information; NTS, non-typhoidal Salmonella; PFGE, pulsed-field gel electrophoresis; SISTR, Salmonella In Silico Typing Resource; SRA, Sequence Read Archive; WGS, whole genome sequencing.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Two supplementary tables and five supplementary figures are available with the online version of this article.
References
- 1.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, et al. Foodborne illness acquired in the United States-major pathogens. Emerg Infect Dis. 2011;17:7–15. doi: 10.3201/eid1701.p11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoffmann S, Maculloch B, Batz M. Economic burden of major foodborne illnesses acquired in the United States. economic information Bulletin -140: U.S. Department of agriculture, economic research service. 2015.
- 3.Brenner FW, Villar RG, Angulo FJ, Tauxe R, Swaminathan B. Salmonella nomenclature. J Clin Microbiol. 2000;38:2465–2467. doi: 10.1128/JCM.38.7.2465-2467.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng RA, Eade CR, Wiedmann M. Embracing diversity: differences in virulence mechanisms, disease severity, and host adaptations contribute to the success of nontyphoidal Salmonella as a foodborne pathogen. Front Microbiol. 2019;10:1368. doi: 10.3389/fmicb.2019.01368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention (CDC) National Salmonella Surveillance Annual Report, 2016. Atlanta, Georgia: National Salmonella Surveillance Annual Report, 2016: US Department of Health and Human Services CDC2018; 2018. [Google Scholar]
- 6.Tang S, Orsi RH, Luo H, Ge C, Zhang G, et al. Assessment and comparison of molecular subtyping and characterization methods for Salmonella . Front Microbiol. 2019;10:1591. doi: 10.3389/fmicb.2019.01591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barco L, Barrucci F, Olsen JE, Ricci A. Salmonella source attribution based on microbial subtyping. Int J Food Microbiol. 2013;163:193–203. doi: 10.1016/j.ijfoodmicro.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 8.Taylor AJ, Lappi V, Wolfgang WJ, Lapierre P, Palumbo MJ, et al. Characterization of foodborne outbreaks of Salmonella enterica serovar enteritidis with whole-genome sequencing single nucleotide polymorphism-based analysis for surveillance and outbreak detection. J Clin Microbiol. 2015;53:3334–3340. doi: 10.1128/JCM.01280-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cito F, Baldinelli F, Calistri P, Di Giannatale E, Scavia G, et al. Outbreak of unusual Salmonella enterica serovar Typhimurium monophasic variant 1,4,[5],12:i:-, Italy, June 2013 to September 2014. Euro Surveill. 2016;21 doi: 10.2807/1560-7917.ES.2016.21.15.30194. [DOI] [PubMed] [Google Scholar]
- 10.Mossong J, Marques P, Ragimbeau C, Huberty-Krau P, Losch S, et al. Outbreaks of monophasic Salmonella enterica serovar 4,[5],12:i:- in Luxembourg, 2006. Euro Surveill. 2007;12:E11–12. doi: 10.2807/esm.12.06.00719-en. [DOI] [PubMed] [Google Scholar]
- 11.Gieraltowski L, Higa J, Peralta V, Green A, Schwensohn C, et al. National outbreak of multidrug resistant Salmonella heidelberg infections linked to a single poultry company. PLoS One. 2016;11:e0162369. doi: 10.1371/journal.pone.0162369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heyman DL. Control of Communicable Diseases Manual. American Public Health Association APHA; 2008. Salmonellosis; pp. 534–540. [Google Scholar]
- 13.Pires SM, Vieira AR, Hald T, Cole D. Source attribution of human salmonellosis: an overview of methods and estimates. Foodborne Pathog Dis. 2014;11:667–676. doi: 10.1089/fpd.2014.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mather AE, Vaughan TG, French NP. Molecular approaches to understanding transmission and source attribution in nontyphoidal Salmonella and their application in Africa. Clin Infect Dis. 2015;61:S259–265. doi: 10.1093/cid/civ727. [DOI] [PubMed] [Google Scholar]
- 15.Wheeler NE. Tracing outbreaks with machine learning. Nat Rev Microbiol. 2019;17:269. doi: 10.1038/s41579-019-0153-1. [DOI] [PubMed] [Google Scholar]
- 16.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoshida CE, Kruczkiewicz P, Laing CR, Lingohr EJ, Gannon VPJ, et al. The Salmonella In Silico Typing Resource (SISTR): an open web-accessible tool for rapidly typing and subtyping draft Salmonella genome assemblies. PLoS One. 2016;11:e0147101. doi: 10.1371/journal.pone.0147101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014;58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 22.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 23.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Page AJ, Taylor B, Delaney AJ, Soares J, Seemann T, et al. SNP-sites: rapid efficient extraction of SNPs from multi-FASTA alignments. Microb Genom. 2016;2:e000056. doi: 10.1099/mgen.0.000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Price MN, Dehal PS, Arkin AP. FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paradis E, Claude J, Strimmer K. Ape: analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 27.Yu G, Smith DK, Zhu H, Guan Y, TTY L. ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol. 2017;8:28–36. [Google Scholar]
- 28.R Core Team R: A Language and Environment for Statistical Computing. Vienna, Austria: 2016. [Google Scholar]
- 29.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen H, Boutros PC. VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics. 2011;12:35. doi: 10.1186/1471-2105-12-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mather AE, Reid SWJ, Maskell DJ, Parkhill J, Fookes MC, et al. Distinguishable epidemics of multidrug-resistant Salmonella Typhimurium DT104 in different hosts. Science. 2013;341:1514–1517. doi: 10.1126/science.1240578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Elnekave E, Hong S, Mather AE, Boxrud D, Taylor AJ, et al. Salmonella enterica Serotype 4,[5],12:i:- in Swine in the United States Midwest: an emerging multidrug-resistant clade. Clin Infect Dis. 2018;66:877–885. doi: 10.1093/cid/cix909. [DOI] [PubMed] [Google Scholar]
- 33.Petrovska L, Mather AE, AbuOun M, Branchu P, Harris SR, et al. Microevolution of Monophasic Salmonella Typhimurium during Epidemic, United Kingdom, 2005-2010. Emerg Infect Dis. 2016;22:617–624. doi: 10.3201/eid2204.150531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tack DM, Marder EP, Griffin PM, Cieslak PR, Dunn J, et al. Preliminary Incidence and Trends of Infections with Pathogens Transmitted Commonly Through Food - Foodborne Diseases Active Surveillance Network, 10 U.S. Sites, 2015-2018. MMWR Morb Mortal Wkly Rep. 2019;68:369–373. doi: 10.15585/mmwr.mm6816a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Allard MW, Strain E, Melka D, Bunning K, Musser SM, et al. Practical value of food pathogen traceability through building a whole-genome sequencing network and database. J Clin Microbiol. 2016;54:1975–1983. doi: 10.1128/JCM.00081-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Achtman M, Wain J, Weill F-X, Nair S, Zhou Z, et al. Multilocus sequence typing as a replacement for serotyping in Salmonella enterica . PLoS Pathog. 2012;8:e1002776. doi: 10.1371/journal.ppat.1002776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Worley J, Meng J, Allard MW, Brown EW, Timme RE. Salmonella enterica phylogeny based on whole-genome sequencing reveals two new clades and novel patterns of horizontally acquired genetic elements. mBio. 2018;9:e02303-18. doi: 10.1128/mBio.02303-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sangal V, Harbottle H, Mazzoni CJ, Helmuth R, Guerra B, et al. Evolution and population structure of Salmonella enterica serovar Newport. J Bacteriol. 2010;192:6465–6476. doi: 10.1128/JB.00969-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Timme RE, Pettengill JB, Allard MW, Strain E, Barrangou R, et al. Phylogenetic diversity of the enteric pathogen Salmonella enterica subsp. enterica inferred from genome-wide reference-free SNP characters. Genome Biol Evol. 2013;5:2109–2123. doi: 10.1093/gbe/evt159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rabsch W, Andrews HL, Kingsley RA, Prager R, Tschäpe H, et al. Salmonella enterica serotype Typhimurium and its host-adapted variants. Infect Immun. 2002;70:2249–2255. doi: 10.1128/iai.70.5.2249-2255.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Soyer Y, Moreno Switt A, Davis MA, Maurer J, McDonough PL, et al. Salmonella enterica serotype 4,5,12:i:-, an emerging Salmonella serotype that represents multiple distinct clones. J Clin Microbiol. 2009;47:3546–3556. doi: 10.1128/JCM.00546-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.San Millan A, MacLean RC. Fitness costs of plasmids: a limit to plasmid transmission. Microbiol Spectr. 2017;5 doi: 10.1128/microbiolspec.MTBP-0016-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smillie C, Garcillán-Barcia MP, Francia MV, Rocha EPC, de la Cruz F. Mobility of plasmids. Microbiol Mol Biol Rev. 2010;74:434–452. doi: 10.1128/MMBR.00020-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Laxminarayan R, Duse A, Wattal C, Zaidi AKM, Wertheim HFL, et al. Antibiotic resistance-the need for global solutions. Lancet Infect Dis. 2013;13:1057–1098. doi: 10.1016/S1473-3099(13)70318-9. [DOI] [PubMed] [Google Scholar]
- 46.Centers for Disease Control and Prevention (CDC) National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS): Human Isolates Surveillance Report for 2014 (Final Report) Atlanta, Georgia: U.S: Department of Health and Human Services, CDC; 2016. [Google Scholar]
- 47.Alcaine SD, Sukhnanand SS, Warnick LD, Su W-L, McGann P, et al. Ceftiofur-resistant Salmonella strains isolated from dairy farms represent multiple widely distributed subtypes that evolved by independent horizontal gene transfer. Antimicrob Agents Chemother. 2005;49:4061–4067. doi: 10.1128/AAC.49.10.4061-4067.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao S, White DG, Friedman SL, Glenn A, Blickenstaff K, et al. Antimicrobial resistance in Salmonella enterica serovar Heidelberg isolates from retail meats, including poultry, from 2002 to 2006. Appl Environ Microbiol. 2008;74:6656–6662. doi: 10.1128/AEM.01249-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McDermott PF, Tyson GH, Kabera C, Chen Y, Li C, et al. Whole-Genome Sequencing for Detecting Antimicrobial Resistance in Nontyphoidal Salmonella . Antimicrob Agents Chemother. 2016;60:5515–5520. doi: 10.1128/AAC.01030-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sjölund-Karlsson M, Howie R, Rickert R, Krueger A, Tran T-T, et al. Plasmid-mediated quinolone resistance among non-Typhi Salmonella enterica isolates, USA. Emerg Infect Dis. 2010;16:1789–1791. doi: 10.3201/eid1611.100464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.The National Antimicrobial Resistance Monitoring System The National Antimicrobial Resistance Monitoring System. NARMS Integrated Report, 2015. Laurel, MD: U.S: Department of Health and Human Services, FDA2017; 2017. [Google Scholar]
- 52.Boyd D, Peters GA, Cloeckaert A, Boumedine KS, Chaslus-Dancla E, et al. Complete nucleotide sequence of a 43-kilobase genomic island associated with the multidrug resistance region of Salmonella enterica serovar Typhimurium DT104 and its identification in phage type DT120 and serovar Agona. J Bacteriol. 2001;183:5725–5732. doi: 10.1128/JB.183.19.5725-5732.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.García P, Malorny B, Rodicio MR, Stephan R, Hächler H, et al. Horizontal Acquisition of a Multidrug-Resistance Module (R-type ASSuT) Is Responsible for the Monophasic Phenotype in a Widespread Clone of Salmonella Serovar 4,[5],12:i: Front Microbiol. 2016;7:680. doi: 10.3389/fmicb.2016.00680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carattoli A. Resistance plasmid families in Enterobacteriaceae . Antimicrob Agents Chemother. 2009;53:2227–2238. doi: 10.1128/AAC.01707-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Johnson CM, Grossman AD. Integrative and conjugative elements (ICEs): what they do and how they work. Annu Rev Genet. 2015;49:577–601. doi: 10.1146/annurev-genet-112414-055018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.