Abstract
MicroRNAs (miRNAs) are a group of small noncoding RNA molecules with significant capacity to regulate the gene expression at the post-transcriptional level in a sequence-specific manner either through translation repression or mRNA degradation triggering a fine-tuning biological impact. They have been implicated in several processes, including cell growth and development, signal transduction, cell proliferation and differentiation, metabolism, apoptosis, inflammation, and immune response modulation. However, over the last few years, extensive studies have shown the relevance of miRNAs in human pathophysiology. Common human parasitic diseases, such as Malaria, Leishmaniasis, Amoebiasis, Chagas disease, Schistosomiasis, Toxoplasmosis, Cryptosporidiosis, Clonorchiasis, and Echinococcosis are the leading cause of death worldwide. Thus, identifying and characterizing parasite-specific miRNAs and their host targets, as well as host-related miRNAs, are important for a deeper understanding of the pathophysiology of parasite-specific diseases at the molecular level. In this review, we have demonstrated the impact of human microRNAs during host−parasite interaction as well as their potential to be used for diagnosis and prognosis purposes.
Keywords: microRNAs, Human parasitic diseases, Pathophysiology, Prognosis, Biomarker
Introduction
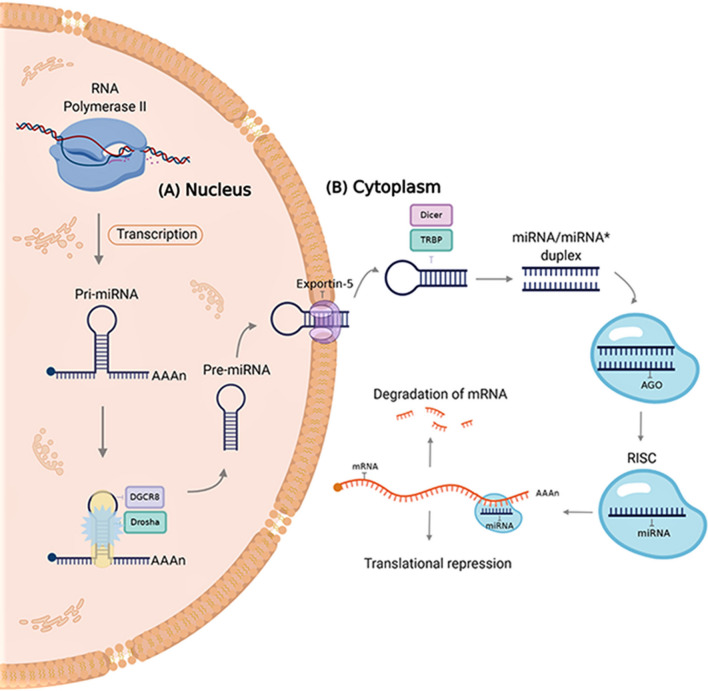
miRNAs are small (~ 22 nucleotides in length), endogenous, evolutionarily conserved regulatory ncRNAs that are implicated in the post-transcriptional regulation of cellular signaling pathways in both animals and plants (Paul et al. 2018, 2020a, d; De la Fuente et al. 2020). Because the discovery of these molecules in Caenorhabditis elegans by Lee et al. (1993) it has been shown that they are widely distributed in most eukaryotes, including humans (Felden and Gilot 2019; Paul et al. 2020b, c). Biogenesis of miRNAs consists of sequential events occurring in the cell nucleus and cytoplasm. In the nucleus, miRNA genes are first transcribed by the RNA polymerase II and fold into long double-strand primary miRNA transcripts (pri-miRNA). Then, the RNase type III Drosha and DGCR8 (also known as Pasha) complex processes the pri-miRNA molecule to form miRNA precursor (pre-miRNA) (Fig. 1a). The resulted pre-miRNA is then translocated into the cytoplasm by Exportin 5 where it is processed one more time by the complex composed of the RNase III Dicer and the Trans-Activation Responsive RNA-Binding Protein (TRBP) to form the mature miRNA/miRNA* duplex (Treiber et al. 2019). To be functional, the resulted miRNA/miRNA* duplex is then separated by a helicase and the single miRNA strand is incorporated into the RNA-induced silencing complex (RISC) coupled with the Argonaute (AGO) protein family which guides to interact with the target mRNA. Binding of miRNAs to the mRNAs (partial or full complementarity) leads to the regulation of their expression either by the degradation of the mRNA or by inhibiting its translation (Fig. 1b) (Wang 2014).
Fig. 1.
The canonical pathway of miRNA biogenesis. a In the nucleus, the miRNA gene is transcribed by RNA polymerase II and then fold into a long pri-miRNA with a hairpin structure. The long pri-miRNA is then cleaved by the microprocessor complex made up of Drosha and DGCR8 (Pasha) proteins, generating a precursor miRNA (pre-miRNA). b Exportin-5 binds to the pre-miRNA and facilitates its export to the cytoplasm. In the cytoplasm, the type III RNase Dicer complex with the double-stranded RNA binding protein TRBP and PACT cleaves the precursor’s hairpin and the resulting duplex is isolated by a helicase enzyme. Finally, the functional strand is loaded together with the Argonaute (AGO) protein into the RNA-induced silencing complex (RISC) to target mRNAs by sequence complementary binding and mediates gene suppression through mechanisms of either translational repression or mRNA degradation
Evidence has shown that miRNAs influence numerous cell biological processes including growth and development, signal transduction, cell proliferation and differentiation, metabolism, cell death, and immune regulation (Miska 2005; Wu and Lu 2017; Pockar et al. 2019). It has been reported that miRNAs participate in the regulation of nearly 60% of human protein-coding genes (Friedman et al. 2009) and 2654 distinct human mature miRNAs available in miRbase (https://www.mirbase.org/) so far supports the idea that miRNAs are involved in complex regulatory networks. Their role has been established in several pathophysiologies including cancer, autoimmune, and metabolic disorders (Garo and Murugaiyan 2016; Naveed et al. 2017). However, in the past ten years, deep sequencing and miRNA microarray technology have evidenced that during the onset of pathogenic infection the host miRNAs regulate cellular responses (Bruscella et al. 2017; Zhou et al. 2018) resulting in signaling and physiological modifications. Parasites are known to have complex interactions with their specific hosts and these interactions are becoming a leading research field for infectious diseases. miRNAs have been involved in both the inflammatory response during the induction of the immune response and the modulation of innate and adaptive immune responses in infectious diseases (Pockar et al. 2019). Moreover, miRNAs also participate in mediating intercellular communications as they are secreted into vesicles or circulating extracellular fluids as exosomes, emphasizing its potential role as biomarkers for a variety of disorders, including parasitic diseases (Makarova et al. 2016). Although studies have highlighted the important role of miRNAs as regulators of gene expression related to the pathogenesis of numerous human diseases the specific functions of miRNAs in human parasitic infections are still not very clear. Thus, unraveling the regulatory roles of miRNAs in host−parasite interactions will not only provide new insights into our understanding of parasite disease pathogenesis but will also offer a foundation for new therapeutic approaches to be established. In this review, we demonstrate our current understanding of the influence of miRNAs in the development and progression of common parasitic diseases in humans, such as Malaria, Leishmaniasis, Amoebiasis, Chagas disease, Schistosomiasis, Toxoplasmosis, Cryptosporidiosis, Clonorchiasis, and Echinococcosis, as well as their possible use as clinically meaningful diagnostic biomarkers.
miRNAs: fine modulators of parasitic infections
Malaria and miRNAs
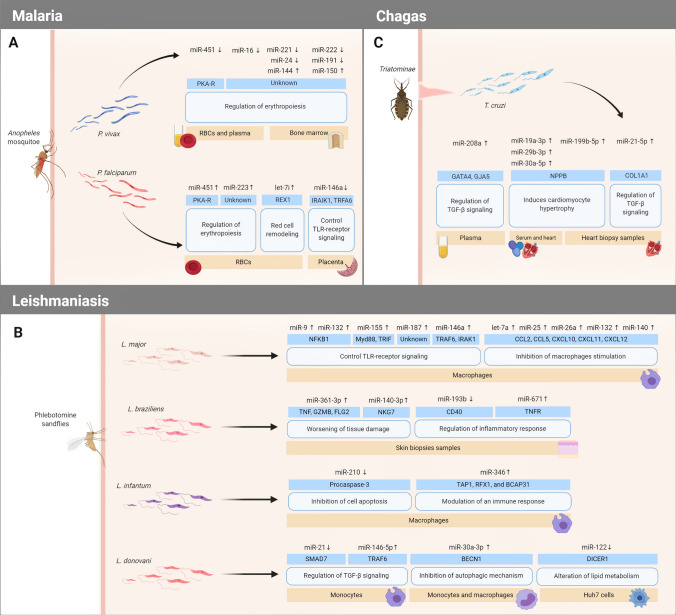
Plasmodium is a genus of protist parasites that is transmitted by female Anopheles mosquitoes which injects sporozoites to vertebrate hosts (including humans) and quickly invades liver cells undergoing rapid multiplication causing malaria (Fig. 2a) (White 2017). There are six known Plasmodium species that infect humans, such as P. vivax, P. ovale curtisi, P. ovale wallikeri, P. malariae, P. knowlesi, and P. falciparum (Singh et al. 2017), and among them, P. falciparum is considered the deadliest since it is responsible for driving the most severe forms of the disease (Phillips et al. 2017; Garrido-Cárdenas et al. 2019). Malaria is one of the foremost infectious illnesses in the world (generally in low-income countries) affecting around 228 million people per year (World Health Organization 2019). Although it is widely known that P. falciparum lacks the classical functional RNAi machinery, accumulating evidence point to the possible exploitation of the host RNAi machinery or by employing a novel mechanism unique to Plasmodium, to manipulate host miRNA expression favoring their growth and survival leading to a potential alteration in the expression of erythrocytic miRNAs (Rathjen et al. 2006; Hakimi and Ménard 2010). It has been shown that Plasmodium parasite principally upregulates those host miRNAs whose target proteins are involved in immune response and downregulates those miRNAs that participate in the inhibition of parasitic translation, host cell proliferation, metabolism, and survival (Table 1) suggesting a high probability to be involved in the manipulation of both MAPK/ERK (Paroo et al. 2009) and Transforming Growth Factor-β (TGF-β) signaling pathways (Lourembam et al. 2013). Increasing evidence suggests that miRNA-451, miR-223, and let-7i are significantly upregulated in Plasmodium-infected red blood cells (RBCs) (Xue et al. 2008; Lamonte et al. 2013) targeting genes, such as Protein Kinase A Regulatory (PKA-R) and Reduced Expression protein 1 (REX1) involved in the regulation of erythropoiesis (the process of RBCs production) and red cells remodeling (Lamonte et al. 2013). Moreover, few studies indicated that host miRNAs may participate in the prognosis of the disease after the Plasmodium infection, supporting the idea that they could be considered as biomarkers for the diagnosis of parasite−host response and the disease progression. For example, miR-146a rs2910164 polymorphism, which could affect the expression level of mature miR-146a (downregulation), has been associated with increased susceptibility to P. falciparum infection in placental samples of pregnant women (Van Loon et al. 2019). Taganov and colleagues (2006) demonstrated that miR-146a is involved in the regulation of important immune response genes, such as tumor necrosis factor receptor-associated factor 6 (TRAF6) and IL-1 receptor-associated kinase 1 genes (IRAK1). On the other hand, P. vivax infection triggers a downregulation of miR-451 and miR-16 in plasma and RBCs of malaria patients and those have been also involved in the regulation of erythropoiesis by targeting PKA-R (Chamnanchanunt et al. 2015). In addition, it has been demonstrated that miR-221, miR-222, miR-24, and miR-191 were downregulated in the bone marrow of P. vivax infected patients, while miR-144, which is generally upregulated during erythropoiesis, and miR-150, which drives megakaryocyte formation while inhibiting erythropoiesis, were overexpressed, respectively (Baro et al. 2017). To date, a few reports have been reviewed to understand the consequence of Plasmodium infection on the host's miRNA expression profile and the disturbance of cellular homeostasis (Shrivastava and Rajasubramaniam 2018), however, the specific interaction between human miRNAs and malaria infection is still unknown.
Fig. 2.
A graphical illustration of human microRNAs and their targets profile during host−parasite interaction of the most important insect vector transmitted parasitic diseases discussed in this review: (a) Malaria (b) Leishmaniasis, and (c) Chagas disease
Table 1.
Relevant miRNAs involved in different parasitic diseasesa
| Disease | Parasite | miRNAs | Target genes | Biological mechanism | Sources | Ref |
|---|---|---|---|---|---|---|
| Malaria | P. falciparum | miR-451 ↑ | PKA-R | Regulation of erythropoiesis | RBCs | (Xue et al. 2008; LaMonte et al. 2013; Chamnanchanunt et al. 2015) |
| miR-223 ↑ | Unknown | |||||
| let-7i ↑ | REX1 | Red cell remodeling | ||||
| miR-146a ↓ | IRAK1 and TRAF6 | Control TLR-receptor signaling | Placental samples | (Van Loon et al. 2019; Taganov et al. 2006) | ||
| P. vivax | miR-451 ↓ | PKA-R | Regulation of erythropoiesis | RBCs and plasma | (Chamnanchanunt et al. 2015) | |
| miR-16 ↓ | Unknown | |||||
| miR-221 ↓ | Bone marrow | (Baro et al. 2017) | ||||
| miR-222 ↓ | ||||||
| miR-24 ↓ | ||||||
| miR-191 ↓ | ||||||
| miR-144 ↑ | ||||||
| miR-150 ↑ | ||||||
| Leishmaniasis | L. major | miR-9 ↑ | NFKB1 | Control TLR-receptor signaling | Macrophages | (Guerfali et al. 2008; Bazzoni et al. 2009; Lemaire et al. 2013) |
| miR-132 ↑ | ||||||
| miR-155 ↑ | Myd88 and TRIF | |||||
| miR-187 ↑ | Unknown | |||||
| miR-146a ↑ | TRAF6, IRAK1 | |||||
| let-7a ↑ | CCL2, CCL5, CXCL10, CXCL11, and CXCL12 | Inhibits macrophages stimulation | ||||
| miR-25 ↑ | ||||||
| miR-26a ↑ | ||||||
| miR-132 ↑ | ||||||
| miR-140 ↑ | ||||||
| L. braziliens | miR-361-3p ↑ | TNF, GZMB, and FLG2 | Worsening of tissue damage | Skin biopsies sample | (Lago et al. 2018) | |
| miR-140-3p ↑ | NKG7 | |||||
| miR-193b ↓ | CD40 | Regulation of inflammatory response | (Nunes et al. 2018) | |||
| miR-671 ↑ | TNFR | |||||
| L. infantum | miR-210 ↓ | Procaspase-3 | Inhibition of cell apoptosis | Macrophages | (Lemaire et al. 2013) | |
| miR-346 ↑ | TAP1, RFX1, and BCAP31 | Modulation of an immune response | (Diotavelli et al. 2018) | |||
| L.donovani | miR-21 ↓ | SMAD7 | Regulation of TGF-β signaling | Monocytes | (Geraci et al. 2015) | |
| miR-146b-5p ↑ | TRAF6 | |||||
| miR-30a-3p ↑ | BECN1 | Inhibition of autophagic mechanism | Monocytes and macrophages | (Singh et al. 2016) | ||
| miR-122 ↓ | DICER1 | Alteration of lipid metabolism | Huh7 cells | (Ghosh et al. 2013) | ||
| Amoebiasis | E. hystolytica | miR-526b-5p ↑ | XIAP, BAK1, BNIP3L | Induces cell cycle arrest, tumor suppression, and apoptosis | Epithelial colon cells | (Lopez-Rosas et al. 2019) |
| miR-643 ↑ | XIAP, BCL2L1, BCL2L10, BCL2L11, BCL2L14, API5 | |||||
| miR-615-5p ↑ | BCL2L1, BCLAF1, BMF, AIFM3, AATK | |||||
| miR-525 ↑ |
BCL2L1, BLC2L13, BAG3, BAG1, XIAP, CASP7, BMF |
|||||
| miR-150 ↑ | BCL2L2, NAIF1, CASP7, CASP8, AIFM3, AIFM2, CFLAR | |||||
| miR-409-3p ↓ | BCL2, BCL2L11, BCL2L15 | |||||
| Chagas Disease | T.cruzi | miR-208a ↑ | GATA4 and GJA5 | Regulation of TGF-β signaling | Plasma | (Linhares-Lacerda et al. 2018) |
| miR-19a-3p ↑ | NPPB | Induces cardiomyocyte hypertrophy | Serum and heart biopsy sample | (Nonaka et al. 2019) | ||
| miR-29b-3p ↑ | ||||||
| miR-30a-5p ↑ | ||||||
| miR-199b-5p ↑ | Heart biopsy sample | |||||
| miR-21-5p ↑ | COL1A1 | Regulation of TGF-β signaling | ||||
| Schistosomiasis | S. japonicum | miR-150-5p ↑ | KANK4, DRD1, and MT1H | Actin reorganization and cell contractility | Liver sample | (Cabantous et al. 2017) |
| miR-146b-5p ↑ | Unknown | Regulation of lipid metabolism | ||||
| miR-143-3p ↑ | PL1N1 and VNN1 | Induces late fibrosis | ||||
| miR-199a-3p ↑ | Accumulation of triglycerides | |||||
| miR-10a-5p ↑ | KANK4 and VNN1 | Regulation of TGF-β signaling | ||||
| miR-4521 ↑ | CTNNA3 | Reorganization of extracellular matrix | ||||
| miR-31-5p ↑ | Unknown | Live fibrosis progression | ||||
| miR-222-3p ↑ | KANK4 and SLC39A8 | |||||
| miR-221-3p ↑ | DRD1 | |||||
| miR-663b ↓ | GBP5 | Oxidative stress damage | ||||
| Bantam* | Unknown | Unknown | Serum | (Meningher et al. 2016) | ||
| miR-2c-3p* | ||||||
| miR-3488* | ||||||
| miR-2a-5p* | ||||||
| Toxoplasmosis | T. gondii | miR-125 ↑ | Unknown | Inhibition of apoptosis | Macrophages | (Cai et al. 2013, 2014) |
| miR-27b ↑ | ||||||
| miR-30c ↑ | ||||||
| miR19a ↑ | BIM | |||||
| miR-19b ↑ | ||||||
| miR-20a ↑ | ||||||
| miR-17 ~ 92 cluster ↑ | HFF | (Zeiner et al. 2010) | ||||
| miR-106b ~ 25 cluster ↑ | Unknown | |||||
| miR-146a ↑ | IRAK1 and TRAF6 | Control TLR-receptor signaling | HFF-Me49 infected cells | (Cannella et al. 2014) | ||
| miR-146a ↓ | HHF-RH infected cells | |||||
| miR-132 ↑ | APAF1, KRAS, MAPK3, and PPP2R5E | Regulation of cell apoptosis and immune response | Neuroepithelial cells | (Ngô et al. 2017; Xiao et al. 2014) | ||
| Cryptosporidiosis | C. parvum | let-7 family ↓ | SNAP23 and SOCS4 | Activation of TLR4/ NF-kB signaling | Biliary epithelial cells | (Hu et al. 2010, 2013) |
| miR-27b ↑ | KSRP | (Zhou et al. 2012) | ||||
| miR-98 ↓ | SOCS4 | Regulation of cytokine signaling | (Hu et al. 2010; Sato et al. 2017) | |||
| miR-34b-5p ↓ | ELAVL1, RAB10, RAB14 | Regulation of cell apoptosis and immune response | HCT-8 cells | (Wang et al. 2019) | ||
| miR-3591-3p ↓ | ELK4, SOS2, TAB2, DAXX, FGF14, MAPK3 | |||||
| miR-18b-3p ↓ | Unknown | |||||
| miR-3976 ↓ | ||||||
| Clonorchiasis | C. sinensis | miR373 ↑ | MMP9 | Modulation of cell adhesion, migration, invasion, and metastasis | HuCCT1 cells and H69 cells | (Pak et al. 2014) |
| miR24 ↑ | PTP | |||||
| miR342-5p ↑ | AKT1 | Activation of proinflammatory mediators | ||||
| miR181d ↑ | CDH13 and RASSF1 | Inactivation of tumor suppressor genes via hypermethylation | ||||
| miR31 ↑ | LAST2 and PPP2R2A | Inhibition of cancer prevention pathways | ||||
| miR185 ↑ | PTEN and PTPN13 | |||||
| miR136 ↑ | Unknown | Cell proliferation and inhibition of tumor suppression | ||||
| miR-95 ↑ | ||||||
| miR-93 ↑ | ||||||
| miR-153 ↑ | ||||||
| miR-16-2 ↑ | ||||||
| miR-195 ↑ | Suppression of cell growth | |||||
| miR-199a-3p ↑ | CAV2 | Increases the proliferative and survival cell activities | ||||
| let7i ↓ | TLR4, RAS, MYC, and HMGA2 | Inhibition of proliferation and differentiation of tumor cells | ||||
| let7a ↓ | ||||||
| miR-124 ↓ | STAT3 and EZH2 | Inhibition of cell proliferation. Induction of apoptosis, and suppression of tumor growth | HuCCT1, H69, HepG-2, and gastric cancer cells | |||
| Echinococcosis | E. granulosus | let-7g-5p ↑ | IL-13, IL-10, and IL-6 | Proliferation and activation of macrophages, inflammation, apoptosis and oxidative damage | Whole blood samples | (Mariconti et al. 2019) |
| let-7a-5p ↑ | ||||||
| miR-26a-5p ↑ | TMEM184B | Increases the expression of type I IFN | ||||
| miR-26b-5p ↑ | PTEN | Modulation of NF-κB pathway | ||||
| miR-195-5p ↑ | BCL2 | Promotion of apoptosis | ||||
| miR-16-5p ↑ | ||||||
| miR-30c-5p ↑ | Unknown | Regulation of the innate immunity, type I IFN signaling | ||||
| miR-223-3p ↑ | GZMB and HDAC2 | |||||
| egr-miR-71* | Unknown | Unknown | Plasma | (Alizadeh et al. 2020) | ||
| egr-let-7* | ||||||
| miR-19↓ | COL1A1 and COL3A | Suppression of cell proliferation by blocking signal transmission in the TGF-β pathway | LX-2 cells and liver tissue | (Zhang et al. 2016) | ||
| E. multilocularis | miR-483-3p ↑ | LBR | Promotes wound healing and cancer progression | Plasma | (Ren et al. 2019) |
*Parasite derived miRNAs
↑Indicates upregulated
↓Indicates downregulated
Leishmaniasis and miRNAs
The genus Leishmania was first described in 1903 for the highly pathogenic species L. donovani, but since then several pathogenic species of the genus Leishmania have been reported. Leishmania flagellates are transmitted to vertebrates by the bite of infected female phlebotomine sandflies generating leishmaniasis with symptoms ranging from skin lesions to fatal leishmaniasis (Fig. 2b) (Akhoundi et al. 2017; Borghi et al. 2017; Derici et al. 2018). It has been estimated up to 1 million new cases every year, resulting in 26,000–65,000 deaths worldwide (World Health Organization 2020). Over the past decade, miRNAs have been shown to be related to the pathogenesis of leishmaniasis (Table 1). It has been seen that at the first hours of the L. major infection upregulation of miR9, miR132, miR-155, miR-187, and miR-146a occur which is related with the control of TLR-receptor signaling and targeting transcripts, such as Nuclear Factor NF-κ-B p105 (NFKB1), Myeloid differentiation primary response 88 (Myd88), TIR domain-containing adaptor protein-inducing Interferon β (TRIF), TRAF6, and IRAK1 in macrophages, suggesting that these miRNAs are negative regulators of fine-tuned inflammatory reactions. Likewise, an upregulation of let-7a, miR-25, miR-26a, miR-132, and miR-140 in L. major-infected human macrophages led to a negative correlation on the expression of their specific chemokine targets CCL2, CCL5, CXCL10, CXCL11, and CXCL12 inhibiting macrophage stimulation (Guerfali et al. 2008; Bazzoni et al. 2009; Lemaire et al. 2013). On the other hand, it has been stated that miR-361-3p and miR-140-3p were significantly overexpressed in cutaneous leishmaniasis lesions (CL) generated by L. braziliensis infection as compared to normal skin samples targeting genes involved in worsening of tissue damage such as TNF, Granzyme B (GZMB), Filaggrin-2 (FLG2) and Natural Killer cell Granule protein 7 (NKG7) (Lago et al. 2018). While downregulation of miR-193b and upregulation of miR-671 are correlated with their respective target genes CD40 and TNF receptors (TNFR) modulating the inflammatory response in lesions caused by this parasite (Nunes et al. 2018). In addition, Lemaire and colleagues (2013) highlighted the downregulation of miR-210 during L. infantum infection which led to inhibition of cell apoptosis by targeting procaspase-3 in monocyte-derived macrophages. Likewise, Diotavelli et al. (2018) described the upregulation of miR-346 during L. infantum infection of human U937 and THP-1-derived macrophages decreasing the mRNA level of major histocompatibility complex- or interferon-associated genes, such as antigen peptide transporter 1 (TAP1), regulatory factor X1 (RFX1) and B-cell receptor-associated protein 31 (BCAP31) involved in both immune response regulation and cell survival under endoplasmic reticulum stress during infection; thus, miR-346 could be considered as an enticing target for anti-Leishmania approaches. Ghosh et al. (2013) assessed an interesting connection between altered lipid metabolism during L. donovani infection and Huh7 cells miR-122 levels (downregulation) by targeting DICER1. Moreover, Geraci and colleagues (2015) demonstrated significant correlations between miR-21 (downregulation) and miR-146b-5p (upregulation) in L. donovani infected dendritic cells and specific members of the TGF-β signaling pathway SMAD7 and TRAF6. Furthermore, a decrease infectivity of L. donovani due to the inhibition of the autophagic mechanism via negative regulation of Beclin 1 (BECN1) in THP-1 and human monocyte-derived macrophages has been reported to be triggered by the upregulation of miR-30a-3p during infection (Singh et al. 2016).
Amoebiasis and miRNAs
Entamoeba histolytica is a single-celled anaerobic protozoan parasite that causes human amoebiasis. It is spread by fecal–oral transmission and is most prevalent in areas plagued by overcrowding, poverty, and poor sanitation (Nourollahpour Shiadeh et al. 2016; Pineda and Perdomo 2017; Deere et al. 2019). Amoebiasis affects around 50 million people globally of which approximately 10% of infected individuals are at risk of contracting invasive amoebiasis that includes amoebic colitis and amoebic liver abscesses. Invasive amoebiasis kills up to 100,000 people worldwide annually, mainly in tropical countries (Saidin et al. 2019). The effects of parasites on host miRNAs expression have been described in a few species of protozoans and nematodes, but very little is known about E. histolytica. The contribution of miRNAs in amoebiasis has been described to act in the modulation of gene expression of physiological and pathophysiological factors (Table 1). It has been suggested that miRNA-controlled pathways such as clathrin receptor-mediated internalization of lipid and protein molecules, as well as gene regulation and signal transduction of the Ras family GTPase are disrupted during E. hystolytica infection (Mar-Aguilar et al. 2013). Interestingly, evidence indicates that the components of miRNA biogenesis machinery, such as Argonaute (AGO) are present in E. histolytica while Dicer protein is still elusive, suggesting the presence of some unknown mechanisms to regulate gene expression without a Dicer enzyme (Mar-Aguilar et al. 2013). However, recent studies have shown an important dysregulation of miRNAs repertoire in epithelial colon cells, specifically, López-Rosas and coworkers (2019) evidenced that after 45 min of E. histolytica infection, there is an upregulation of miR-526b-5p, miR-643, miR-615-5p, miR-525, and miR150, and a downregulation of miR-409-3p, which altogether may impact in the expression of genes involved in at least five major biochemical pathways in SW-480 cells, including biosynthesis of unsaturated fatty acids, phosphatidylinositol 3-kinase/Protein kinase B (PI3K/AKT) signaling pathway, ubiquitin-mediated proteolysis, mRNA surveillance pathways, and apoptosis. Interestingly, the amoeba can induce apoptosis in host cells by partially altering miRNAs that regulate genes involved in lipid metabolism, the PI3K/AKT signaling pathway, and apoptosis. Of particular note, the aforementioned six modulated microRNAs potentially target apoptosis-related genes. Thus, the expression of B Cell Lymphoma 2 (BCL-2) protein family, BCL2 Antagonist/Killer 1 (BAK1), BCL2 Interacting Protein 3 Like (BNIP3L), X-linked Inhibitor of Apoptosis Protein (XIAP), Apoptosis Inducing Factor Mitochondria associated 2 and 3(AIFM 2 and AIFM3), Apoptosis Inhibitor 5 (API5), BCL2 Associated transcription Factor 1 (BCLAF1), Apoptosis-Associated Tyrosine Kinase (AATK), BAG1, BAG3, BMF, Nuclear Apoptosis Inducing Factor 1 (NAIF1), CFLAR, and Caspase 7 and 8 (CASP7 and CASP8) can potentially be regulated by the aforesaid miRNAs suggesting their possible usage in the clinical diagnosis of amoebiasis (Lopez-Rosa et al. 2019).
Chagas disease and miRNAs
Chagas disease (CD) is an anthropozoonosis caused by the protozoan parasite Trypanosoma cruzi (Fig. 2c) which is transmitted by insect vector Triatominae or kissing bugs and affects about 10–12 million people just in America leading to approximately 50,000 deaths per year (Corti and Villafañe 2017; Pérez-Molina and Molina 2018; De Oliveira et al. 2020). The infection has two successive phases. The acute phase is characterized by a high parasitemia, usually asymptomatic or oligosymptomatic, while it may progress to the chronic phase with neurological, cardiac, digestive, or cardiodigestive clinical complaints (Pérez-Molina and Molina 2018; De Souza 2019). Among chronic Chagas patients, modulation of gene expression in myocardial tissue is mostly associated with immune response, metabolism, and cell stress response (Ferreira 2014). Over the past decade, the central role of miRNAs has been established to impact the resistance to infection and the pathogenesis of CD (Table 1). Linhares-Lacerda et al. (2018) detected higher levels of miR-208a in plasma samples from patients with chronic CD and they suggested that this might be correlated with TGF-β stimulation and regulation of genes involved in cardiac hypertrophy and fibrosis, such as GATA binding 4 (GATA4) and Gap Junction Alpha-5 (GJA5). More recently, Nonaka et al. (2019) reported an elevated level of miR-19a-3p, miR-29b-3p, and miR-30a-5p in serum, and miR-19a-3p, miR-21-5p, miR-29b-3p, miR-30a-5p, and miR-199b-5p in heart samples from chronic CD patients suggesting that some of them might be correlated with cardiac injury and disease severity, targeting Natriuretic Peptide B (NPPB) and Collagen type I Alpha 1 chain (COL1A1). However, the relationship of miRNAs with CD pathogenesis could be further explored in order to validate new biomarkers or molecular targets for therapeutic intervention.
Schistosomiasis and miRNAs
Schistosomiasis (also known as Bilharziasis) is a parasitic infection caused by several species of blood-flukes of the genus Schistosoma and it is one of the most prevalent zoonotic diseases, affecting over 258 million individuals in 54 countries (McManus et al. 2018; Chuah et al. 2019; Salari et al. 2020). Trematode parasites S. mansoni (mainly distributed in Africa, South America, Caribbean, and the Middle East), S. haematobium (Africa and the Middle East), and S. japonicum (China and Southeast Asia) are the main species that cause this human disease (Meningher et al. 2020). Infections take place in freshwater bodies, where schistosomes penetrate human skin; followed by penetration, schistosome cercariae migrate to the host portal−mesenteric vein system where the female worm lay a large number of eggs that are either discharged into the environment through feces or urine or are retained in host tissues where they induce inflammation (Colley et al. 2014; Wu et al. 2015). Several lines of evidence have been demonstrated that miRNAs can modulate schistosomiasis pathogenesis (Table 1). For example, Cabantous et al. (2017) showed that in the liver of S. japonicum infected patients the levels of miR-150-5p, miR-146b-5p, miR-143-3p, miR-199a-3p, miR-10a-5p, miR-4521, miR-31-5p, miR-222-3p, and miR-221-3p were elevated, while miR-663b was present in low level. Furthermore, they reported that the predicted target genes of the aforesaid miRNAs, such as KANK4, Dopamine Receptor D1 (DRD1), Metallothionein-1H (MT1H), PL1N1, Vanin 1 (VNN1), Catenin Alpha-3 (CTNNA3), SLC39A8, and Guanylate-Binding Protein 5 (GBP5) are involved in crucial processes implicated in hepatic fibrosis progressions, such as cellular proliferation and differentiation, reorganization of the extracellular matrix, lipolysis, and cellular detoxification. To date, four schistosomal miRNAs have been isolated from extracellular vesicles in sera from Schistosoma infected individuals, such as Bantam, miR-2c-3p, miR-3488, and miR-2a-5p suggesting that can be used both as a diagnostic tool for infection and to monitor treatment effectiveness (Meningher et al. 2016). More recently, the role of miRNAs in the pathogenesis of hepatic fibrosis in schistosomiasis caused by both S. japonicum and S. mansoni has been reviewed, highlighting their role in the regulation of antifibrosis and profibrosis mechanisms (Chen et al. 2019). Moreover, the recent advances in characterizing miRNA profiles in extracellular vesicles secreted by Schistosoma species have upstretched the possibility for validating more parasite-derived miRNAs as potential biomarkers for schistosomiasis detection (Cabantous et al. 2017).
Toxoplasmosis and miRNAs
Toxoplasma gondii, one of the most common human parasites in the world, is a ubiquitous pathogen that is the causative agent of toxoplasmosis and can infect a large range of hosts including humans. The transmission begins with the ingestion of contaminated raw meat and then the parasite starts to infect as many cells as possible. Manifestations are highly variable, ranging from asymptomatic to severe, especially in cases of brain and eye infection (Parlog et al. 2015; Assolini et al. 2017). Nearly all infections are silent, and it has been shown that this parasite specifically modulates the expression of essential miRNAs in the host, altering their response to the infection (Table 1). The first study in this subject demonstrated that Toxoplasma effectors are responsible for the alterations in host miR-17 ~ 92 and miR-106b ~ 25 family expression that are upregulated after infection with RH toxoplasma strain in primary human foreskin fibroblasts (HFFs) (Zeiner et al. 2010). Similarly, Cai and colleagues (2013) found an increased expression of miR-20a, miR-125, miR-19a, miR-19b, miR-27b, and miR-30c in human macrophage at 6 h and 12 h postinfection. Interestingly, some of those miRNAs (miR-19a, miR-19b, and miR-20a) are part of the miR-17 ~ 92 cluster and have been associated with a novel mechanism for the regulation of apoptosis by inhibiting proapoptotic molecule BIM allowing T. gondii to evade immune responses through this mechanism (Cai et al. 2014). On the other hand, Cannella and colleagues (2014) used an infected HFF with two different Toxoplasma strains (Me49 and RH) and reported an upregulation of miR-146a in HFF infected with Me49 and a downregulation in the RH infected cells, targeting IRAK and TRAF which are fine modulator of TLR-receptor signaling pathway. Furthermore, it is also known that T. gondii affects the human brain in many pathways related to epilepsy, neurodegeneration, and cancer. A positive regulation of miR-132 has been reported in the human neuroepithelial cell line infected with RH-2F, PRU, and CTG strains which have been related to a modulation in the expression of genes involved in the metabolism of dopamine, such as apoptotic protease-activating F-factor 1 (APAF1), Kirsten Rat Sarcoma (KRAS), MAPK3 and PPP2R5E (Xiao et al. 2014; Ngô et al. 2017). Finally, it has been recently reviewed that this parasite also features its own miRNA processing system and possesses the capacity to secret exosomes that contain miRNAs (Menard et al. 2019). Changes in the miRNA profiles of the host in T. gondii infection represent a powerful mechanism for a better understanding of this pathology.
Cryptosporidiosis and miRNAs
The genus Cryptosporidium is composed of protozoan parasites that infect the gastrointestinal epithelium and other mucosal surfaces of their host, including humans (Vanathy et al. 2017). Transmission occurs through the fecal−oral path, and sources of infection with Cryptosporidium include contaminated food or water (Dumaine et al. 2019). Human cryptosporidial infections have been related mainly to C. parvum and the severity of infection may range from an asymptomatic shedding of ingested oocysts to a serious life-threatening, and prolonged disease (Ryan et al. 2014; Lender et al. 2015). The exact molecular pathogenic mechanisms of cryptosporidial infection are still not completely known. However, a growing number of functional studies have documented the role of miRNAs in the response of human hosts to Cryptosporidium (Table 1). For example. increasing evidence suggests that host miRNAs help in the elimination of the parasites by regulating TLR4 and NF-κB signaling pathways and with the regulation of the release of antimicrobial peptides (Chen et al. 2007; Hu et al. 2013). Specifically, it has been demonstrated that C. parvum infection reduces the expression of the let-7 family miRNAs in biliary epithelial cells, which lead to an increase of synaptosome-associated protein 23 (SNAP23) expression, coordinating the subsequent release of exosomes carrying antimicrobial peptides (Hu et al. 2013). In addition, Zhou and colleagues (2012) demonstrated the role of miR-27b (upregulation) in the modulation of TLR4/NF-κB-mediated epithelial anti-C. parvum responses in human biliary epithelial cells, targeting KSRP. On the other hand, a couple of studies suggest that host miRNAs are also used by C. parvum to strengthen its own survival. Specifically, C. parvum infection downregulates the expression of miR-98 and let-7 family to induce the suppressor of cytokine signaling (SOCS4) proteins that are negative regulators of cytokine signaling (Hu et al. 2010; Sato et al. 2017). Recently, Wang and collaborators (2019) reported for the first time the miRNA expression profile of human intestinal epithelial cells infected with C. parvum and they evidenced that most miRNAs were not significantly differentially expressed in the infected HCT-8 cells as compared to uninfected cells. Nonetheless, they reported that the miR-34b-5p, miR-18b-3p, miR-3976, and miR-3591-3p were downregulated after C. parvum infection and those have been associated with the regulation of both apoptotic processes and epithelial immune responses, targeting genes, such as ELAVL1, RAB10, RAB14, ELK4, SOS2, TAB2, DAXX, fibroblast growth factor 14 (FGF14), and MAPK3.
Clonorchiasis and miRNAs
Clonorchiasis is a food-borne parasitic disease caused by the fluke Clonorchis sinensis through the consumption of raw or undercooked freshwater fish, containing metacercariae of C. sinensis. Clonorchiasis generally appears as bile duct obstruction, biliary inflammation, liver cirrhosis, hepatic carcinoma, and cholangiocarcinoma (Wu et al. 2012; Tang et al. 2016). This parasite infects about 35 million people globally, mainly distributed in Asian countries, such as China, Japan, Vietnam, and Korea (Han et al. 2016). Although molecular mechanisms of carcinogenesis associated with hepatic fluke infestation are not completely understood, some studies have investigated variations in miRNA expression patterns and associations with specific biological functions in C. sinensis infection indicating a possible association between miRNA and cholangiocarcinogenesis (Table 1). Specifically, in vitro experiments using human cholangiocarcinoma cells (HuCCT1) treated with excretory-secretory protein (ESP) of C. sinensis have demonstrated an upregulation in expression levels of miR373, miR342-5p, miR-199a-3p, miR-195, miR185, miR181d, miR-153, miR136, miR-95, miR-93, miR31, miR24, and miR-16-2 along with the downregulation of miR-124a, let7i, and let7a, in a time-dependent manner compared with untreated controls. Similarly, ESP-treated normal cholangiocytes (H69) revealed that the expressions of nine miRNAs (miR-16-2, miR-93, miR-95, miR-153, miR-195, miR-199-3P, let7a, let7i, and miR-124a) were similarly regulated, showing that the cell proliferation and inhibition of tumor suppression mediated by these miRNAs are common to both cancerous and noncancerous cells (Pak et al. 2014). Functional clustering of these dysregulated miRNAs revealed their involvement in cell differentiation/proliferation, inflammation, metastasis, oncogene regulation, and DNA methylation by regulating various cancer-related signaling pathways, such as TGF-β, MAPK, TLR, and PI3K/AKT, and by targeting several genes, such as matrix metalloproteinase-9 (MMP9), Tyrosine-protein phosphatase (PTP), AKT serine/threonine kinase 1 (AKT1), CDH12M, Ras Association domain family member 1 (RASSF1), LAST2, PPP2R2A, Phosphatase and Tensin homolog (PTEN), PTPN13, Caveolin-2 (CAV2), TLR4, RAS, MYC, HMGA2, STAT3, and EZH2 (Pak et al. 2014; Yan et al. 2016). This information could allow the identification of potential targets and miRNA-associated genes involved in multiple oncogenic pathways during C. sinensis infection and establishing new tools that may function as indicators for the diagnosis and prognosis of the disease.
Echinococcosis and miRNAs
Cystic echinococcosis (CE) and alveolar echinococcosis (AE), two severe zoonotic tapeworm diseases, are triggered by Echinococcus granulosus sensu lato (s.l.) and Echinococcus multilocularis, respectively (Wang et al. 2013; Wen et al. 2019). CE (considered as a neglected disease by the WHO) infects mainly the liver and lungs and it is categorized as a major public health problem resulting in 1.2 million cases per year (Mariconti et al. 2019; Alizadeh et al. 2020). On the other hand, AE, one of the most dangerous human parasitic zoonosis in the northern hemisphere, is mainly characterized by a tumor-like development of metacestodes in human livers causing around 200 cases per year (Geramizadeh and Baghernezhad 2016). MicroRNA-based diagnostics have attracted great interest in biomarker research for clinical diagnosis and monitoring of echinococcosis (Table 1). The recent studies show a substantial upregulation of eight miRNAs (let-7g-5p, let-7a-5p, miR-26a-5p, miR-26b-5p, miR-195-5p, miR-16-5p, miR-30c-5p, and miR-223-3p) in whole blood samples of patients with active larval cysts as compared to those with inactive cysts. These upregulated miRNAs have been involved in a variety of biological processes, such as macrophages proliferation and activation, inflammation, apoptosis, oxidative damage, targeting, modulation of the NF-κB pathway and type I interferon signaling by targeting genes, such as Interleukin 6, 10, and 13 (IL-6, IL-10, and IL-13), Transmembrane protein 184B (TMEM184B), PTEN, BCL2, GZMB, and Histone Deacetylase 2 (HDAC2) (Mariconti et al. 2019). Furthermore, Zhang and colleagues (2016) reported that the expression of miR-19 showed a significant reduction in hepatic stellate cells (LX-2 cells) treated with hydatid cyst fluid (HCF) in pericystic collagen-rich liver tissue of CE patients, as compared to normal liver, leading to a significant suppression of cell proliferation by blocking signal transmission in the TGF-β pathway, decreasing COL1A1 and COL3A protein expression rates, suggesting that miR-19 could be an important biomarker in humans infected with E. granulosus (s.l.). Likewise, Alizadeh and colleagues (2020) reported that egr-miR-71 and egr-let-7 can be detected in human plasma during hydatid cyst infection and can be used as possible biomarkers for the early diagnosis and monitoring of CE. Similarly, miR-483-3p that targets Lamin-B receptor (LBR) and has been associated with cancer progression, is assessed as a potential marker due to substantial upregulation in the plasma of AD patients as compared to normal controls that provide a new approach to the clinical diagnosis of AE (Ren et al. 2019).
Conclusion
Identifying and characterizing parasite-specific miRNAs and their targets in hosts, as well as miRNAs interfering with host pathology, are crucial for a better understanding of the pathophysiology of parasitic diseases at the molecular level. The biological knowledge acquired about miRNAs, especially in biomedical research, is expected to be widely translated in the field of parasitology in the coming years since miRNAs have great potential to lead to a transition to a novel class of theranostic tool. Thus, we can expect the discovery of more and more specific miRNAs with highly specialized functions linked to cellular processes in parasites that can provide novel guidelines for the management of parasitic diseases.
Code availability
The figures were exported under a paid subscription. Created with BioRender.com.
Compliance with ethical standards
Conflicts of interest
The authors declare that they have no conflict of interest.
References
- Akhoundi M, Downing T, Votýpka J, et al. Leishmania infections: molecular targets and diagnosis. Mol Aspects Med. 2017;57:1–29. doi: 10.1016/j.mam.2016.11.012. [DOI] [PubMed] [Google Scholar]
- Alizadeh Z, Mahami-Oskouei M, Spotin A, et al. Parasite-derived microRNAs in plasma as novel promising biomarkers for the early detection of hydatid cyst infection and post-surgery follow-up. Acta Trop. 2020;202:105255. doi: 10.1016/j.actatropica.2019.105255. [DOI] [PubMed] [Google Scholar]
- Assolini JP, Concato VM, Gonçalves MD, et al. Nanomedicine advances in toxoplasmosis: diagnostic, treatment, and vaccine applications. Parasitol Res. 2017;116:1603–1615. doi: 10.1007/s00436-017-5458-2. [DOI] [PubMed] [Google Scholar]
- Baro B, Deroost K, Raiol T, et al. Plasmodium vivax gametocytes in the bone marrow of an acute malaria patient and changes in the erythroid miRNA profile. PLoS Negl Trop Dis. 2017;11:6–13. doi: 10.1371/journal.pntd.0005365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzoni F, Rossato M, Fabbri M, et al. Induction and regulatory function of miR-9 in human monocytes and neutrophils exposed to proinflammatory signals. Proc Natl Acad Sci USA. 2009;106:5282–5287. doi: 10.1073/pnas.0810909106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghi SM, Fattori V, Conchon-Costa I, et al. Leishmania infection: painful or painless? Parasitol Res. 2017;116:465–475. doi: 10.1007/s00436-016-5340-7. [DOI] [PubMed] [Google Scholar]
- Bruscella P, Bottini S, Baudesson C, et al. Viruses and miRNAs: more friends than foes. Front Microbiol. 2017;8:824. doi: 10.3389/fmicb.2017.00824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabantous S, Hou X, Louis L, et al. Evidence for an important role of host microRNAs in regulating hepatic fibrosis in humans infected with Schistosoma japonicum. Int J Parasitol. 2017;47:823–830. doi: 10.1016/j.ijpara.2017.05.007. [DOI] [PubMed] [Google Scholar]
- Cai Y, Chen H, Jin L, et al. STAT3-dependent transactivation of miRNA genes following Toxoplasma gondii infection in macrophage. Parasites and Vectors. 2013;6:1–9. doi: 10.1186/1756-3305-6-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Chen H, Mo X, et al. Toxoplasma gondii inhibits apoptosis via a novel STAT3-miR-17-92-Bim pathway in macrophages. Cell Signal. 2014;26:1204–1212. doi: 10.1016/j.cellsig.2014.02.013. [DOI] [PubMed] [Google Scholar]
- Cannella D, Brenier-Pinchart MP, Braun L, VanRooyen JM, et al. miR-146a and miR-155 delineate a microRNA fingerprint associated with toxoplasma persistence in the host brain. Cell Rep. 2014;6:928–937. doi: 10.1016/j.celrep.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamnanchanunt S, Kuroki C, Desakorn V, et al. Downregulation of plasma miR-451 and miR-16 in Plasmodium vivax infection. Exp Parasitol. 2015;155:19–25. doi: 10.1016/j.exppara.2015.04.013. [DOI] [PubMed] [Google Scholar]
- Chen XM, Splinter PL, O’Hara SP, LaRusso NF. A cellular micro-RNA, let-7i, regulates toll-like receptor 4 expression and contributes to cholangiocyte immune responses against Cryptosporidium parvum infection. J Biol Chem. 2007;282:28929–28938. doi: 10.1074/jbc.M702633200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Zhang J, Zheng T, et al. The role of microRNAs in the pathogenesis, grading and treatment of hepatic fibrosis in schistosomiasis. Parasit Vectors. 2019;12:1–10. doi: 10.1186/s13071-019-3866-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuah C, Gobert GN, Latif B, et al. Schistosomiasis in Malaysia: a review. Acta Trop. 2019;190:137–143. doi: 10.1016/j.actatropica.2018.11.012. [DOI] [PubMed] [Google Scholar]
- Colley DG, Bustinduy AL, Secor WE, King CH. Human schistosomiasis. Lancet. 2014;383:2253–2264. doi: 10.1016/S0140-6736(13)61949-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti M, Villafañe MF. AIDS and Chagas’ disease. Am Trypanos Chagas Dis One Hundred Years Res Second Ed. 2017 doi: 10.1016/B978-0-12-801029-7.00031-9. [DOI] [Google Scholar]
- De la Fuente JL, Sharma A, Paul S. Characterization of miRNAs from sardine (Sardina pilchardus Walbaum, 1792) and their tissue-specific expression analysis in brain and liver. 3 Biotech. 2020;10:318. doi: 10.1007/s13205-020-02298-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza W. Biology of Trypanosoma cruzi. Rio de Janeiro: Intechopen; 2019. Introductory chapter: biology of Trypanosoma cruzi; pp. 1–4. [Google Scholar]
- De Oliveira ABB, Tedeschi BBB, De Oliveira J, et al. Parasite x vector relationship in Chagas disease: does Trypanosoma cruzi (Chagas, 1909) infection affect the spermatogenesis of Triatoma infestans (Klug, 1834)? Parasitol Res. 2020 doi: 10.1007/s00436-020-06788-z. [DOI] [PubMed] [Google Scholar]
- Deere JR, Parsons MB, Lonsdorf EV, et al. Entamoeba histolytica infection in humans, chimpanzees and baboons in the Greater Gombe Ecosystem, Tanzania. Parasitology. 2019;146:1116–1122. doi: 10.1017/S0031182018001397. [DOI] [PubMed] [Google Scholar]
- Derici MK, Cansaran-Duman D, Taylan-Özkan A. Usnic acid causes apoptotic-like death in Leishmania major, L.infantum and L. tropica. 3 Biotech. 2018;8:384. doi: 10.1007/s13205-018-1409-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diotavelli A, De Santi M, Buffi G, Ceccarelli M, Vitale F, Galluzzi L, Magnani M. Leishmania infection induces microRNA hsa-miR-346 in human cell line-derived macrophages. Front Microbiol. 2018;9:1019. doi: 10.3389/fmicb.2018.01019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumaine JE, Tandel J, Striepen B. Cryptosporidium parvum. Trends Parasitol. 2019 doi: 10.1016/j.pt.2019.11.003. [DOI] [PubMed] [Google Scholar]
- Felden B, Gilot D. Modulation of bacterial sRNAs activity by epigenetic modifications: inputs from the eukaryotic miRNAs. Genes (Basel) 2019;10:1–16. doi: 10.3390/genes10010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira LRP. Interferon-γ and other inflammatory mediators in cardiomyocyte signaling during Chagas disease cardiomyopathy. World J Cardiol. 2014;6:782. doi: 10.4330/wjc.v6.i8.782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RC, Farh KKH, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garo LP, Murugaiyan G. Contribution of microRNAs to autoimmune diseases. Cell Mol Life Sci. 2016;73:2041–2051. doi: 10.1007/s00018-016-2167-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido-Cárdenas JA, González-Cerón L, Manzano-Agugliario F, Mesa-Valle C. Plasmodium genomics: an approach for learning about and ending human malaria. Parasitol Res. 2019;118:1–27. doi: 10.1007/s00436-018-6127-9. [DOI] [PubMed] [Google Scholar]
- Geraci NS, Tan JC, McDowell MA. Characterization of microRNA expression profiles in leishmania infected human phagocytes. Parasite Immunol. 2015;37:43–51. doi: 10.1111/pim.12156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geramizadeh B, Baghernezhad M. Hepatic alveolar hydatid cyst: a brief review of published cases from Iran in the last 20 years. Hepat Mon. 2016;16:10–13. doi: 10.5812/hepatmon.38920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh J, Bose M, Roy S, Bhattacharyya SN. Leishmania donovani targets dicer1 to downregulate mir-122, lower serum cholesterol, and facilitate murine liver infection. Cell Host Microbe. 2013;13:277–288. doi: 10.1016/j.chom.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerfali FZ, Laouini D, Guizani-Tabbane L, et al. Simultaneous gene expression profiling in human macrophages infected with Leishmania major parasites using SAGE. BMC Genomics. 2008;9:1–18. doi: 10.1186/1471-2164-9-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakimi MA, Ménard R. Do apicomplexan parasites hijack the host cell microRNA pathway for their intracellular development? F1000 Biol Rep. 2010;2:16–18. doi: 10.3410/B2-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Tang Q, Lu X, et al. Dysregulation of hepatic microRNA expression profiles with Clonorchis sinensis infection. BMC Infect Dis. 2016;16:1–8. doi: 10.1186/s12879-016-2058-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Zhou R, Liu J, et al. MicroRNA-98 and let-7 regulate expression of suppressor of cytokine signaling 4 in biliary epithelial cells in response to Cryptosporidium parvum infection. J Infect Dis. 2010;202:125–135. doi: 10.1086/653212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Gong AY, Roth AL, et al. Release of luminal exosomes contributes to TLR4-mediated epithelial antimicrobial defense. PLoS Pathog. 2013 doi: 10.1371/journal.ppat.1003261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lago TS, Silva JA, Lago EL, et al. The miRNA 361-3p, a Regulator of GZMB and TNF is associated with therapeutic failure and longer time healing of cutaneous Leishmaniasis caused by L. (viannia) braziliensis. Front Immunol. 2018;9:2621. doi: 10.3389/fimmu.2018.02621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaMonte G, Philip N, Reardon J, et al. Contributes to malaria resistance. Cell Host Microbe. 2013;12:187–199. doi: 10.1016/j.chom.2012.06.007.Translocation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementary to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- Lemaire J, Mkannez G, Guerfali FZ, et al. microRNA Expression profile in human macrophages in response to leishmania major infection. PLoS Negl Trop Dis. 2013;7:e2478. doi: 10.1371/journal.pntd.0002478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lender M, Böttcher D, Delling C, et al. A novel CDPK1 inhibitor—a potential treatment for cryptosporidiosis in calves? Parasitol Res. 2015;114:335–336. doi: 10.1007/s00436-014-4228-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linhares-Lacerda L, Granato A, Gomes-Neto JF, Luciana C, et al. Circulating plasma microRNA-208a as potential biomarker of chronic indeterminate phase of chagas disease. Front Microbiol. 2018;9:1–9. doi: 10.3389/fmicb.2018.00269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Rosas I, Lopez-Camarillo C, Salinas-Vera YM, et al. Entamoeba histolytica up-regulates microRNA-643 to promote apoptosis by targeting XIAP in human epithelial colon cells. Front Cell Infect Microbiol. 2019;9:1–15. doi: 10.3389/fcimb.2018.00437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lourembam SD, Sawian CE, Baruah S. Dysregulation of cytokines expression in complicated falciparum malaria with increased TGF-β and IFN-γ and decreased IL-2 and IL-12. Cytokine. 2013;64:503–508. doi: 10.1016/j.cyto.2013.08.007. [DOI] [PubMed] [Google Scholar]
- Makarova JA, Shkurnikov MU, Wicklein D, et al. Intracellular and extracellular microRNA: an update on localization and biological role. Prog Histochem Cytochem. 2016;51:33–49. doi: 10.1016/j.proghi.2016.06.001. [DOI] [PubMed] [Google Scholar]
- Mar-Aguilar F, Trevino V, Salinas-Hernández JE, et al. Identification and characterization of microRNAS from Entamoeba histolytica HM1-IMSS. PLoS ONE. 2013;8:e68202. doi: 10.1371/journal.pone.0068202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariconti M, Vola A, Manciulli T, et al. Role of microRNAs in host defense against Echinococcus granulosus infection: a preliminary assessment. Immunol Res. 2019 doi: 10.1007/s12026-018-9041-4. [DOI] [PubMed] [Google Scholar]
- McManus DP, Dunne DW, Sacko M, et al. Schistosomiasis. Nat Rev Dis Prim. 2018;4:1–19. doi: 10.1038/s41572-018-0013-8. [DOI] [PubMed] [Google Scholar]
- Menard KL, Haskins BE, Denkers EY. Impact of Toxoplasma gondii infection on host non-coding RNA responses. Front Cell Infect Microbiol. 2019;9:1–8. doi: 10.3389/fcimb.2019.00132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meningher T, Lerman G, Regev-Rudzki N, Gold D, et al. Schistosomal microRNAs isolated from extracellular vesicles in sera of infected patients: a new tool for diagnosis and follow-up of human schistosomiasis. J Infect Dis. 2016;215(378):386. doi: 10.1093/infdis/jiw539. [DOI] [PubMed] [Google Scholar]
- Meningher T, Barsheshet Y, Ofir-Birin Y, et al. Schistosomal extracellular vesicle-enclosed miRNAs modulate host T helper cell differentiation. EMBO Rep. 2020;21:1–17. doi: 10.15252/embr.201947882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miska EA. How microRNAs control cell division, differentiation and death. Curr Opin Genet Dev. 2005;15:563–568. doi: 10.1016/j.gde.2005.08.005. [DOI] [PubMed] [Google Scholar]
- Naveed A, Ur-Rahman S, Abdullah S, Naveed MA. A concise review of MicroRNA exploring the insights of microRNA regulations in bacterial, viral and metabolic diseases. Mol Biotechnol. 2017;59:518–529. doi: 10.1007/s12033-017-0034-7. [DOI] [PubMed] [Google Scholar]
- Ngô HM, Zhou Y, Lorenzi H, et al. Toxoplasma modulates signature pathways of human epilepsy, neurodegeneration and cancer. Sci Rep. 2017;7:1–32. doi: 10.1038/s41598-017-10675-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka CKV, Macêdo CT, Cavaicante BRR, de Alcântara AC, et al. Circulating miRNAs as potential biomarkers associated with cardiac remodeling and fibrosis in chagas disease cardiomyopathy. J Mol Sci. 2019;20:1–16. doi: 10.3390/ijms20164064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nourollahpour Shiadeh M, Niyyati M, Fallahi S, Rostami A. Human parasitic protozoan infection to infertility: a systematic review. Parasitol Res. 2016;115:469–477. doi: 10.1007/s00436-015-4827-y. [DOI] [PubMed] [Google Scholar]
- Nunes S, Silva IB, Ampuero MR, et al. Integrated analysis reveals that miR-193b, miR-671, and TREM-1 correlate with a good response to treatment of human localized cutaneous leishmaniasis caused by Leishmania braziliensis. Front Immunol. 2018;9:1–13. doi: 10.3389/fimmu.2018.00640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak JH, Kim IK, Kim SM, et al. Induction of cancer-related microRNA expression profiling using excretory-secretory products of Clonorchis sinensis. Parasitol Res. 2014;113:4447–4455. doi: 10.1007/s00436-014-4127-y. [DOI] [PubMed] [Google Scholar]
- Parlog A, Schlüter D, Dunay IR. Toxoplasma gondii-induced neuronal alterations. Parasite Immunol. 2015;37:159–170. doi: 10.1111/pim.12157. [DOI] [PubMed] [Google Scholar]
- Paroo Z, Ye X, Chen S, Liu Q. Phosphorylation of the human microRNA-generating complex mediates MAPK/Erk signaling. Cell. 2009;139:112–122. doi: 10.1016/j.cell.2009.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul SC, Sharma A, Mehta R, Paul S. genome wide computational identification of Tuna (Thunnus orientalis) microRNAs and their targets. Ocean Sci J. 2018;53:727–734. doi: 10.1007/s12601-018-0041-z. [DOI] [Google Scholar]
- Paul S, de la Fuente-Jiménez JL, Manriquez CG, Sharma A. Identification, characterization and expression analysis of passion fruit (Passiflora edulis) microRNAs. 3 Biotech. 2020;10:25. doi: 10.1007/s13205-019-2000-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S, Reyes PR, Garza BS, Sharma A. MicroRNAs and child neuropsychiatric disorders: a brief review. Neurochem Res. 2020;45:323–240. doi: 10.1007/s11064-019-02917-y. [DOI] [PubMed] [Google Scholar]
- Paul S, Bravo Vázquez LA, Pérez Uribe S, Roxana Reyes-Pérez P, Sharma A. Current status of microRNA-based therapeutic approaches in neurodegenerative disorders. Cells. 2020;9:1698. doi: 10.3390/cells9071698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul SC, Sharma A, Mehta R, Paul S. In silico characterization of microRNAs and their target transcripts form cranberry (Vaccinium macrocarpon) Cytol Genet. 2020;54:82–90. doi: 10.3103/S0095452720010120. [DOI] [Google Scholar]
- Pérez-Molina JA, Molina I. Chagas disease. Lancet. 2018;391:82–94. doi: 10.1016/S0140-6736(17)31612-4. [DOI] [PubMed] [Google Scholar]
- Phillips MA, Burrows JN, Manyando C, et al. Malaria. Nat Rev Dis Prim. 2017;3:1–24. doi: 10.1038/nrdp.2017.50. [DOI] [PubMed] [Google Scholar]
- Pineda E, Perdomo D. Entamoeba histolytica under oxidative stress: what countermeasure mechanisms are in place? Cells. 2017;6:1–14. doi: 10.3390/cells6040044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pockar S, Globocnik Petrovic M, Peterlin B, Vidovic Valentincic N. MiRNA as biomarker for uveitis—a systematic review of the literature. Gene. 2019;696:162–175. doi: 10.1016/j.gene.2019.02.004. [DOI] [PubMed] [Google Scholar]
- Rathjen T, Nicol C, McConkey G, Dalmay T. Analysis of short RNAs in the malaria parasite and its red blood cell host. FEBS Lett. 2006;580:5185–5188. doi: 10.1016/j.febslet.2006.08.063. [DOI] [PubMed] [Google Scholar]
- Ren B, Wang H, Ren L, et al. Screening for microRNA-based diagnostic markers in hepatic alveolar echinococcosis. Med (United States) 2019;98:1–7. doi: 10.1097/MD.0000000000017156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan U, Fayer R, Xiao L. Cryptosporidium species in humans and animals: current understanding and research needs. Parasitology. 2014;141:1667–1685. doi: 10.1017/S0031182014001085. [DOI] [PubMed] [Google Scholar]
- Saidin S, Othman N, Noordin R. Update on laboratory diagnosis of amoebiasis. Eur J Clin Microbiol Infect Dis. 2019;38:15–38. doi: 10.1007/s10096-018-3379-3. [DOI] [PubMed] [Google Scholar]
- Salari P, Fürst T, Knopp S, et al. Cost of interventions to control schistosomiasis: a systematic review of the literature. PLoS Negl Trop Dis. 2020;14:1–23. doi: 10.1371/journal.pntd.0008098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Koshizuka T, Ishibashi K, et al. Involvement of herpes simplex virus type 1 UL13 protein kinase in the induction of SOCS genes, the negative regulator of cytokine signaling running. Microbiol Immunol. 2017;61:159–167. doi: 10.1111/1348-0421.12483. [DOI] [PubMed] [Google Scholar]
- Shrivastava R, Rajasubramaniam S. Host−plasmodium interaction: role of RNAi. Curr Sci. 2018;115:2219–2231. doi: 10.18520/cs/v115/i12/2219-2231. [DOI] [Google Scholar]
- Singh AK, Pandey RK, Shaha C, Madhubala R. MicroRNA expression profiling of Leishmania donovani-infected host cells uncovers the regulatory role of MIR30A-3p in host autophagy. Autophagy. 2016;12:1817–1831. doi: 10.1080/15548627.2016.1203500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SP, Srivastava D, Mishra BN. Genome-wide identification of novel vaccine candidates for Plasmodium falciparum malaria using integrative bioinformatics approaches. 3 Biotech. 2017;7:318. doi: 10.1007/s13205-017-0947-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-κB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. PNAS. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang ZL, Huang Y, Yu XB. Current status and perspectives of Clonorchis sinensis and clonorchiasis: epidemiology, pathogenesis, omics, prevention, and control. Infect Dis Poverty. 2016;5:1–12. doi: 10.1186/s40249-016-0166-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treiber T, Treiber N, Meister G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat Rev Mol Cell Biol. 2019;20:5–20. doi: 10.1038/s41580-018-0059-1. [DOI] [PubMed] [Google Scholar]
- Van Loon W, Gai PP, Hamann L, et al. MiRNA-146a polymorphism increases the odds of malaria in pregnancy 11 medical and health sciences 1107 immunology 11 medical and health sciences 1108 medical microbiology. Malar J. 2019;18:1–7. doi: 10.1186/s12936-019-2643-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanathy K, Parija SC, Mandal J, Hamide A, Krishnamurthy S. Cryptosporidiosis: a mini review. Trop Parasitol. 2017;7:72–80. doi: 10.4103/tp.TP_25_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. Composition of seed sequence is a major determinant of microRNA targeting patterns. Bioinformatics. 2014;30:1377–1383. doi: 10.1093/bioinformatics/btu045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Gao C, Steverding D, et al. Differential diagnosis of cystic and alveolar echinococcosis using an immunochromatographic test based on the detection of specific antibodies. Parasitol Res. 2013;112:3627–3633. doi: 10.1007/s00436-013-3550-9. [DOI] [PubMed] [Google Scholar]
- Wang C, Liu L, Zhu H, Zhang L, et al. MicroRNA expression profile of HTC-8 cells in the early phase of Cryptosporidium parvum infection. BMC Genomics. 2019;20:37. doi: 10.1186/s12864-018-5410-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen H, Vuitton L, Tuxun T, Li J, Vuitton DA, Zhang W, McManus DP. Echinococcosis: advances in the 21st century. Clin Microbiol Rev. 2019;32:e00075–18. doi: 10.1128/CMR.00075-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White NJ. Malaria parasite clearance. Malar J. 2017;16:1–14. doi: 10.1186/s12936-017-1731-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (2019) World malaria report 2019. WHO
- World Health Organization (2020) Leishmaniasis. WHO
- Wu C-J, Lu L-F. MicroRNA in immune regulation. In: Yoshimura A, editor. Emerging concepts targeting immune checkpoints in cancer and autoimmunity. Cham: Springer International Publishing; 2017. pp. 249–267. [Google Scholar]
- Wu W, Qian X, Huang Y, Hong Q. A review of the control of Clonorchiasis sinensis and Taenia solium taeniasis/cysticercosis in China. Parasitol Res. 2012;111:1879–1884. doi: 10.1007/s00436-012-3152-y. [DOI] [PubMed] [Google Scholar]
- Wu W, Feng A, Huang Y. Research and control of advanced schistosomiasis japonica in China. Parasitol Res. 2015;114:17–27. doi: 10.1007/s00436-014-4225-x. [DOI] [PubMed] [Google Scholar]
- Xiao J, Li Y, Prandovszky E, et al. MicroRNA-132 dysregulation in Toxoplasma gondii infection has implications for dopamine signaling pathway. Neuroscience. 2014;268:128–138. doi: 10.1016/j.neuroscience.2014.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue X, Zhang Q, Huang Y, et al. No miRNA were found in Plasmodium and the ones identified in erythrocytes could not be correlated with infection. Malar J. 2008;7:1–6. doi: 10.1186/1475-2875-7-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C, Shen LP, Ma R, et al. Characterization and identification of differentially expressed microRNAs during the process of the peribiliary fibrosis induced by Clonorchis sinensis. Infect Genet Evol. 2016;43:321. doi: 10.1016/j.meegid.2016.06.009. [DOI] [PubMed] [Google Scholar]
- Zeiner GM, Norman KL, Thomson JM, et al. Toxoplasma gondii infection specifically increases the levels of key host microRNAs. PLoS ONE. 2010;5:e8741. doi: 10.1371/journal.pone.0008742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Wang L, Ali T, Li L, et al. Hydatid cyst fluid promotes peri-cystic fibrosis in cystic echinococcosis by suppressing miR-19 expression. Parasit Vectors. 2016;9:1–9. doi: 10.1186/s13071-016-1562-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Gong AY, Eischeid AN, Chen XM. mir-27b Targets KSRP to coordinate TLR4-mediated epithelial defense against Cryptosporidium parvum Infection. PLoS Pathog. 2012;8:e1002702. doi: 10.1371/journal.ppat.1002702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Li X, Wu M. miRNAs reshape immunity and inflammatory responses in bacterial infection. Signal Transduct Target Ther. 2018;3:1–13. doi: 10.1038/s41392-018-0006-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The figures were exported under a paid subscription. Created with BioRender.com.