Highlights
-
•
Bacterial nucleomodulins are proteins targeting and influencing host nucleus.
-
•
Bacterial nucleomodulins are involved in plant tumor development.
-
•
Their role is still debated in animal tumors.
-
•
Nucleomodulins are also found in bacteria suspiciously involved with human cancer.
-
•
Study of nucleomodulins can provide ways to manage infection associated cancer.
Keywords: Oncogenesis, Host-pathogen interaction, Infection, Pathogens, Protein targeting
Abstract
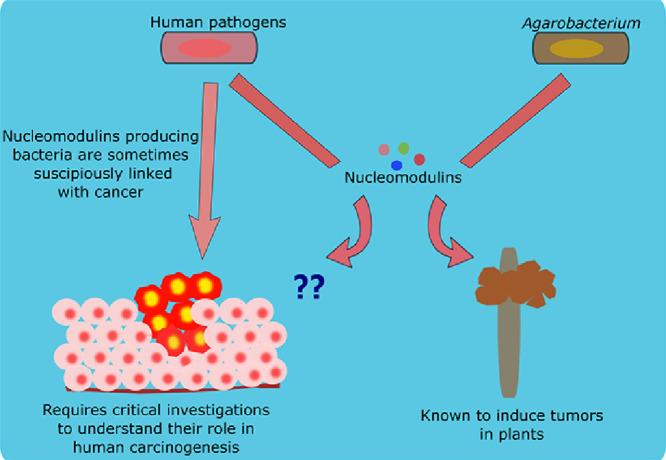
Recent studies in microbial pathogenesis have identified several bacterial proteins with the potential to influence host cell nuclei. This field of research is in its infancy, however it is rapidly growing. In particular, the role of bacterial nucleomodulins in animal oncogenesis is an area that requires attention. Earlier research has suggested the role of nucleomodulins in plant tumor development and these findings may provide us with a better understanding of the role of these proteins in human cancer development. This proposition is further supported by previous identification of nucleomodulins present in bacteria that have been associated with cancer development, but their role in human cancer is unclear. In this article, we provide an update on the status of these nucleomodulins and their role in cancer etiology. We collected information about known bacterial nucleomodulins and tried to relate their mechanistic implication with already known plant tumor development model. The present research indicates that bacterial nucleomodulins may be an important target in cancer etiology and knowledge of their role in human oncogenesis may help us to create suitable alternative cancer management strategies.
Graphical abstract
Introduction
While pathogens may be small, they have the superb ability to influence comparatively more complex, larger host cells. Evolutionary advantage made the pathogens advanced and efficient with multiple strategies to target host cells. They can secrete certain effectors to hijack host cellular and subcellular machinery for their survival. Bacteria are also known to release proteins that target important subcellular organelles including the nucleus, mitochondria, endoplasmic reticulum, Golgi body etc. Several reviews concerning the regulation of host cellular machinery by pathogen's proteins through subcellular targeting are available and the list of such proteins is increasing rapidly [23,51]. Among these proteins, some have the ability to target and modulate nuclear function specifically. These proteins are known as nucleomodulins and have been shown to regulate nuclear function from inside to outside nucleus [9].
The nucleus serves as the core of the cell machinery and controls several important biological processes. It harbors genetic information and regulates expression of genes. The modulation of the nucleus by bacterial regulators, such as nucleomodulins, is important for controlling host cell machinery for bacterial advantage. Although, a deeper understanding of these nucleomodulins is needed and the detection of them in different bacteria is ongoing, several articles on the role of nucleomodulins in mediating host cell regulation arose recently [10]. The majority of literature surrounding bacterial nucleomodulins focuses on their role in host cell regulation and consequent contribution in disease pathogenesis, rather than their link to other mechanisms such as cancer development.
In fact, several bacterial infections are linked with cancer. It has been estimated that around 20% of cancer are attributed to infections [98]. There is substantial mechanistic involvement of microbial infections in the development of cancer, but exact mechanisms behind these findings are still disputed and studies investigating the mechanistic links between infection and cancer development are still ongoing.
Bacterial nucleomodulins may be an important player in cancer development due to their ability to alter nuclear function. Many reviews detail the role of bacterial nucleomodulins in host cell modulation, but their subsequent contributions in cancer etiology remains unexplored. Therefore, this article attempts to provide a glimpse into current status of bacterial nucleomodulins and their subsequent effects on cancer development through microbial involvement. Strong evidence of plant tumor development by bacterial nucleomodulins are currently available, but the similar role in human cancer development remains unclear.
Bacterial nucleomodulins and their consequences on host cell
As the nucleus plays a central role in cellular functions, modulation of nuclear function will ultimately lead to substantial effects on the host cell. Nucleomodulins are considered to be bacterial factors that target the nucleus [9,10]. Several articles are already available on the regulation of host cell by nucleomodulins so we briefly covered this aspect in order to provide necessary background. Our main intention is to discuss the possible role of bacterial nucleomodulins in human oncogenesis and a representative group of nucleomodulins are presented in Table 1 Nucleomodulins can act in a variety of ways in order to alter host cellular functions and some of these are presented below.
Table 1.
A list of representative bacterial nucleomodulins and their potential mode of action inside host cell nuclei.
| Sr. No. | Name of Protein | Organism | Function (TSS) |
|---|---|---|---|
| Plant Host | |||
| 1 | VIRE2 [97] | A. tumefaciens | It is involved in host nuclear uptake of bacterial ssDNA element known as transferred DNA and results in DNA transformation (T4SS). |
| 2 | VirE3 [27] | A. tumefaciens | Plant transcriptional activator (T4SS). |
| 3 | VirD5 [86] | A. tumefaciens | VirD5 is known to have a dual function. It acts as a transcriptional activator and regulates host gene expression, it also prevents the degradation of coat proteins of the bacterial T-complex through the host's ubiquitin proteasome system [86] (T4SS). |
| 4 | T-DNA border endonuclease virD2 [60] | A. tumefaciens | It is involved in the cleavage of unique sites within T-DNA of bacteria and involves the transfer and integration of particular segments (T-DNA) of the Ti plasmid DNA into host. T4SS effector (T4SS). |
| 5 | Protein VirF [80] | A. tumefaciens | In the host cell, VirF is involved in ubiquitination and subsequent proteoasomal degradation of target proteins. It is also involved in T-DNA translocation and is essential for tumor formation in some plant species (T4SS). |
| 6 | Transcription Activator-Like (TAL) effectors [38] | Xanthomonas oryzae pv. oryzae | Functions as a transcription factor in rice plant cells (T3SS). |
| 7 | XopD [36] | X. campestris | Interferes with host proteins regulation by mimicking host SUMO isopeptidase (T3SS). |
| 8 | AvrXa7 [89] | Xanthomonas oryzae pv. oryzae | Bind to host double-stranded DNA and affects transcriptional machinery (T3SS). |
| 9 | HsvG [14] | Pantoea agglomerans pv. gypsophilae (Pag) | DNA binding protein that controls expression of host gene HSVGT (T3SS). |
| 10 | HsvB [14] | Pantoea agglomerans pv. gypsophilae (Pag) | Act as a transcriptional activator (T3SS). |
| 11 | HopAI1 [14] | Pseudomonas syringae | Proposed to Affect host gene expression by modulating post-translational modification of histones (T3SS). |
| 12 | PopP2 [14] | Ralstonia solanacearum | Modulates host cell gene transcription (T3SS) |
| Animal Host | |||
| 13 | SET Domain protein (The Chlamydia nuclear effector (NUE)) [65] | Chlamydia trachomatis | Leads to host cell chromatin modification (T3SS). |
| 14 | hypothetical protein CT311 [53] | Chlamydia trachomatis | ?? (sec‑dependent pathway) |
| 15 | Nuclear Effector AnkA [26] | Anaplasma phagocytophilum | Binds to host AT rich DNA and affects transcription of antimicrobial defense genes in granulocytes [71] (T4SS) |
| 16 | p200 [95] | Ehrlichia spp. | Binds to adenine-rich DNA and affects host cell gene transcription (T1SS). |
| 17 | TRP32 [25] | E. chaffeensis | Binds to G rich motif of host DNA and modulates gene transcription (T1SS). |
| 18 | TRP47 [46] | E. chaffeensis | Enters host nucleus through MYND binding domain and regulates transcription of essential genes (T1SS). |
| 19 | TRP120 [94] | E. chaffeensis | Binds to GC-rich motif and shows transcriptional activator activity (T1SS) |
| 16 | OspF [4] | Shigella flexneri | Affects histone protein phosphorylation and host immunity-related genes expression (T3SS). |
| 17 | OspB [99] | Shigella flexneri | Modulates MAP Kinase pathway and affects host inflammatory response (T3SS). |
| 18 | E3 ubiquitin-protein ligase ipaH9.8 [5] | S. flexneri | Affect Nf-kB-mediated inflammatory response (T3SS). |
| 19 | E3 ubiquitin-protein ligase SspH1 [31] | Salmonella enterica serovar typhimurium | Inhibits Nf-kB associated genes (T3SS). |
| 20 | Cycle inhibiting factor (Cif) [41] | E. coli | Induces accumulation of cyclin-dependent kinase inhibitors p21 and p27, and inhibits cell cycle at the both G1/S, G2/M phases (T3SS). |
| 21 | EspF [21] | Entero-pathogenic E. coli | Disrupts host cell nucleolus and many other cellular events (T3SS). |
| 22 | YopM [7] | Y. pestis | Interacts with p90 ribosomal S6 kinase 1 (RSK1) and helps in virulence (T3SS). |
| 23 | Uncharacterized protein (CifYp) [19] | Y. pseudotuberculosis | Cell Cycle inhibiting factors (T3SS). |
| 24 | CifBp [68] | Burkholderia pseudomallei | Cell Cycle inhibiting factors (T3SS). |
| 25 | BtSET [23] | Burkholderia thailandensis | Induces methylation of host cell chromatin H3K4 protein and activates ribosomal DNA transcription (T3SS). |
| 26 | CifPl [10] | Photorhabdus luminescens | Cell cycle inhibiting factors (T3SS). |
| 27 | BaSET [23] | B. anthracis | Targets host cell histone protein H1 |
| 28 | RomA [23] | Legionella pneumophila | Leads to methylation of histone H3 Lys 14 (T4SS). |
| 29 | AnkH [84] | L. pneumophila | Interferes with host cell transcription (T4SS). |
| 30 | AnkX [91] | L. pneumophila | Interacts with host nuclear protein PLEKHN1, manipulating inflammation (T4SS). |
| 31 | SnpL [74] | L. pneumophila | Targets host RNA polymerase II and regulates host gene expression (T4SS) |
| 32 | LntA [50] | Listeria monocytogenes | Targets host cell chromatin repressor BAHD1 and stimulates interferon genes. |
| 33 | SuAT1 [77] | Theileria annulata (Protozoa) | Function as a DNA binding protein with the ability to alter host cell phenotype. |
| 34 | Rv2966c [76] | Mycobacterium tuberculosis | Interacts with host chromatin and affects gene expression. |
| 35 | Rv1988 [90] | Mycobacterium tuberculosis | Interacts with host chromatin and affects histone methylation. |
| 36 | serine/threonine phosphatase (SP-STP) [1] | Streptococcus pyogenes | Induces apoptosis in infected cells. |
| 37 | HP0425 [47] | Helicobacter pylori | DNAse activity. |
Bacterial proteins acting as transcription factors
A number of articles are already available on the regulation of host cells by nucleomodulins through various mechanisms. Several bacteria are known to regulate host cell transcription through the secretion of a variety of proteins that either directly or indirectly affect nuclear functions. Some bacterial nucleomodulins can even act as transcription activators. For example, the VirE3 protein of Agrobacterium can bind to the plants cell-specific transcription factor, pBrp, which belongs to the TF2B family. VirE3 is also able to induce transcription in yeast after binding to DNA [27]. The same category of proteins was found in another plant pathogen Pantoea agglomerans. It is known to secrete HsvG and HsvB, two proteins that have two nuclear localization signal to target host nuclei and are known to induce transcription of certain genes in host cells [87]. In addition, there is a specific category of bacterial nucleomodulins known as Transcription Activator-Like (TAL) effectors, function as transcription factors for host cells and induce expression of certain genes (Table 1) [38].
Histone modification and chromatin regulation
Nucleomodulins can act as transcriptional regulators granting them with exclusive unhindered access to host genetic material. Eukaryotic genetic material is larger than prokaryotes and it is usually packed in a smaller, more limited nuclear space. The packing of DNA is done in order to ensure no disturbance to the vital properties of genetic material- like replication, transcription, repair, and chromosomal segregation. Histone and chromatin remodeling proteins maintain this tight packaging of nuclear DNA, known as chromatin. Several microorganisms, including bacteria, have the ability to regulate chromatin in their host cells in order to regulate transcriptional control of their desired genes. The modification of the chromatin structure by bacteria has been reviewed already and many bacterial nucleomodulins are known to target this process [29]. Numerous bacteria are known to secrete special SET domain containing proteins, that possess the ability to modify chromatin structure [2]. Table 1 also lists some bacterial nucleomodulins known to alter chromatin structure.
Effects on cell cycle and DNA integrity
In addition to the role of bacteria in chromatin regulation and subsequent control on transcriptional machinery, several bacterial nucleomodulins are able to damage DNA, affect DNA repair, as well as alter the cell cycle. Some bacteria are also known to secrete DNA damaging bacterial effectors. For example, E. coli is known to secrete a bacterial toxin colibactin, with the ability to cause DNA damage [20]. Another bacterial protein, cytolethal distending toxin (CDT) is known to have DNA damaging properties that induce cell cycle arrest. Many Gram negative bacteria are known to produce CDT. The role of bacterial proteins in host DNA damage has been further reviewed in detail in other articles [28].
Nucleus invaders bacteria
Some bacteria themselves are known to directly invade host cell nuclei and influence their morphology and functions. These bacteria are often protected from host cytoplasmic defense mechanisms due to this unusual target location. Intranuclear bacteria are largely found to be associated with Protists, but their association with animals has also been observed. Holospora spp. are one of the more well-known intranuclear bacteria, and infect Paramecia. However, this infection does not always cause problems for Paramecia, instead it provides a selective advantage as Holospora induces expression of heart shock proteins, which results in increased cell survival. Much remains uncertain about intranuclear bacteria, but the current status has been reviewed in a recent article [75]. Intranuclear bacteria are also found to be associated with mammalian cells. For example Rickettsia rickettsia has occasionally been observed to undergo intranuclear growth in host cells [13]. Moreover, another bacterium associated with Rickettsia is found within the nuclei of human liver cell and is known as Orientia tsutsugamushi [66]. While it has been suggested that this unusual intranuclear colonization may be accidental [75], this bacteria is known to cause various nuclear injuries, degeneration, and in some instances clumping of nucleoproteins. This suggests their presence in the nucleus may not be completely unintentional since there are direct effects observed on nuclear morphology and its functions [66]. Another bacterium known as CC99 is known for its ability to infect Amoeba and is often known as Amoeba resistant bacteria. CC99 is thought to infect nuclei of human HeLa and U937 macrophage like cells [24]. The knowledge about intranuclear bacteria is still in its infancy and has not yet been identified properly. Future research directed towards such bacteria will shed more light on the role of these bacteria in their contribution to disease pathogenesis.
Potential of same category proteins in oncogenesis
Primarily cancer involves the dysregulation of cellular homeostasis caused by altered cell growth control. This altered cell growth is associated with several peculiar hallmarks, including constant proliferative stimuli, evasion of growth suppression mechanisms and cell death resistance, increased immortality, angiogenesis, invasion, and metastasis activity. It is considered that these hallmarks of cancer are as a result of genomic instability and inflammation [30]. The involvement of bacteria in genomic instability and inflammation is widely discussed but their exact role in cancer etiology remains under investigation. Bacterial nucleomodulins are thought to have significant potential to alter genetic regulation since bacteria themselves are a great contributor to inflammatory mechanisms inside host cells.
It is firmly believed that targets of nucleomodulins in host cells are directly involved in cancer etiology, and many studies are ongoing to utilize these targets and associated pathways for cancer management (Fig. 1). The role of bacterial nucleomodulins in cancer etiology is yet to be established, but the cancer causing ability of the targets of nucleomodulins provides several caveats. Regulation of transcription factors is associated with many diseases including cancer and the role of transcription factors in cancer etiology has been substantially studied. For example many cancer cells depend on the transcription factor c-Myc for growth, proliferation, cancer severity and their derived clinical effects. The role of transcription factors in cancer etiology has been reviewed numerous times [52]. Recently, the targeting of transcription factors in cancer management has also been reviewed [8,57,70].
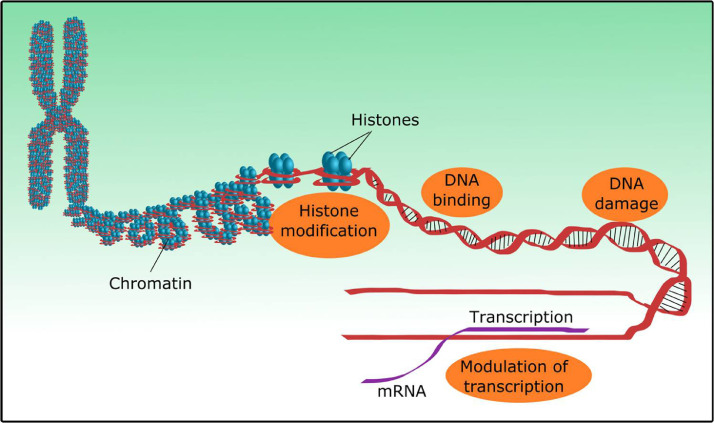
Fig. 1.
Possible strategies for bacterial nucleomodulin mediated alteration of cellular functions that may contribute to cancer development. The orange circles represent possible molecular targets of bacterial nucleomodulins.
The role of histone modification in cancer etiology has also been reviewed in many recent articles [6,73]. Post-translational modifications in histone proteins including methylation, phosphorylation and acetylation are found to be linked with cancer etiology. This modulation in epigenetic mechanisms controlling gene expression can lead to the development of cancer [85].
The effects of cell cycle modulation in oncogenesis are also known. Briefly, cancer is a disease involving uncontrolled cell division. The cell cycle is a multi-step process and each step is controlled by certain check points. The well-known protein p53, is mainly involved in blocking the progression of the cell cycle through the G1 step by activating production of protein p21 which inhibits cyclin dependent kinase. Generally p53 prevents cell cycle progression before DNA repair. Even in some instances, the p53 induces cell apoptosis, if the DNA damage is severe. Mutations in p53 genes are evident in many cancers and its effects have been reviewed extensively [58]. For example, Li-Fraumeni syndrome is associated with germline mutations in p53 gene, and leads to inherited susceptibility to multiple cancers and also linked with altered cell cycle control [54]. In addition, the OrfX protein produced by L. monocytogenes interacts with the RybP gene in the nucleus. This RybP gene has the ability to regulate p53 and its expression is decreased in human cancer tissues. Therefore this suggests the potential role of bacterial nucleomodulins in p53 regulation [17,67]. Alongside cell cycle control modulation and its role in carcinogenesis, repair mechanisms as a result of DNA damage are also altered during the development of cancer. Several anti-cancer treatment strategies target these repair pathways for cancer management [34].
Bacterial nucleomodulins have great potential to contribute to cancer etiology by affecting several cancer targets, nevertheless their conclusive role in mammalian oncogenesis is yet to be established. Scattered evidence infers the carcinogenic potential of bacterial nucleomodulins, but the overall mechanistic link leading to oncogenesis is much awaited. The following section will attempt to gather pieces of literature concerning the role of bacterial nucleomodulins in mammalian carcinogenesis.
Bacterial nucleomodulins and cancer etiology
Agrobacterium tumefaciens: a model for the involvement of bacterial nucleomodulins in tumorigenesis
It was understood over 100 years ago by Smith and Townsend that Gram-negative soil bacterium Agrobacterium tumefaciens, induces tumors in plants. Later it was found that a large Ti plasmid is required for Agrobacterium to induce tumors [11]. Now, it has been widely established that Agrobacterium have a complete set of machinery that induces tumors in host cells. This machinery includes several proteins including nucleomodulins. In addition to nucleomodulins, it has a specific sequence of DNA known as T-DNA, which gets transformed into the host cell. T-DNA transformation in host cells acts as a transfer of machinery for generation of bacterial nucleomodulins inside host nucleus. T-DNA induces the production of excessive amounts of growth hormones inside the host cell, including auxin and cytokinin, and leads to the induction of tumors by the uncontrolled proliferation of host cells [72].
Agrobacterium is known to produce two categories of proteins, one is known as Ti-plasmid encoded virulence gene products (vir), and the second is the chromosomally-encoded virulence proteins (chv). Several articles are available on Agrobacterium pathogenesis and consequent tumor formation in host plants. Fig. 2 gives an overview of some nucleomodulins involved in the process, in order to get an idea behind role of bacterial nucleomodulins in plant tumorigenesis.
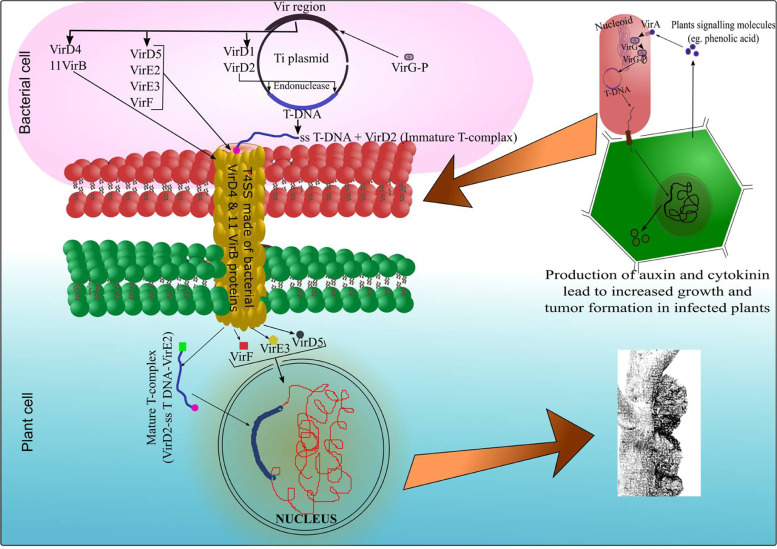
Fig. 2.
Schematic representation of the tumor development process in plants by Agrobacterium-mediated transformation. The process involves several bacterial nucleomodulins and the transfer of nucleomodulins producing machinery in the host cell nucleus as T-DNA. The bacterial VirA senses host plants and activates the VirG protein through phosphorylation, which, in turn, activates the expression of proteins from the vir region of T-DNA. Vir D1 and VirD2 proteins act as endonucleases to release ss T-DNA from Ti-plasmid by strand replacement.VirD4, along with other 11 VirB proteins, make T4SS to transfer bacterial molecules into the host cell. VirD2 binds with T-DNA to make an immature T-complex. Other molecules including VirD5, VirE2, VirE3, and VirF are also translocated into the host cell. The VirD2-T DNA complex binds with VirE2 in the host cell to make a mature T-complex. VirD2 and VirE2 contain nuclear localization signals and transport the mature T-complex into the host cell nucleus [64]. VirD5, VirE3, and VirF are also translocated into the host cell nucleus. VirF is supposed to help in the uncoating of the mature T-complex and facilitate its integration into host cell genetic material [49]. VirD5 localizes to the host's centrosome/kinetochore [93] while VirE3 act as a transcriptional activator for the induction of the expression of certain genes [63]. The whole machinery involves several nucleomodulins that lead to increased growth and production of tumor in the host cell. Other membranes between bacterial and plant cells are not shown for the ease of simplicity in the figure.
Nucleomodulins and cancer associated bacterial infection
Nucleomodulins are known to have the potential to alter normal cellular functions, which can contribute to cancer development. The possible strategies of bacterial nucleomodulins in mediating tumor development are shown in Fig. 1. In addition, Table 2 identifies nucleomodulins found in bacteria that are associated with cancer and therefore with the potential role in tumorigenesis. As the list of nucleomodulins is still in its infancy, the complete picture of nucleomodulin mediated effects on tumorigenesis can only be inferred for now, Fig. 3 provides mechanisms for the contributions of bacterial nucleomodulins to cancer etiology based on current evidences. The next section is intended to cover a few case studies of bacterial nucleomodulins that originate from cancer associated bacteria.
Table 2.
Nucleomodulins from bacteria either known or suspiciously involved in induction of tumor.
| Sr. No | Name of nucleomodulins | Bacteria | Function related to carcinogenesis | Type of cancer |
|---|---|---|---|---|
| 1 | VirE3 | Agrobacterium | Transcription induction [27] | Plant Tumor |
| VirF | Agrobacterium tumefaciens | T-DNA translocation for tumor formation | Plant Tumor | |
| 2 | HsvG | Pantoea agglomerans | DNA binding protein, transcriptional activator [87] | Plant Tumor |
| 3 | HsvB | |||
| 4 | SET Domain protein (NUE) | Chlamydia trachomatis | Chromatin modification [65] | Cervical cancer |
| 5 | cpnSET | Chlamydophila pneumoniae | Chromatin modification [59] | Lung cancer |
| 6 | Colibactin | E. coli | DNA damage [83] | Colorectal cancer |
| 7 | Cytolethal distending toxin (CDT) | E. coli | DNA damage, cell cycle arrest [39] | Colorectal cancer |
| 8 | Cif | E. coli | Migrates to host cell nucleus and affects NEDD8 [41] | Colorectal cancer |
| 9 | HP0425 | H. pylori | Translocates to nucleus DNase I-like enzymatic activity [47] | Gastric cancer |
| 10 | HP0059 | H. pylori | Translocates to nucleus DNase I-like enzymatic activity [48] | Gastric cancer |
| 11 | SspH1 | Salmonella enterica serovar Typhimurium | Translocates to nucleus and inhibits NF-kB dependent genes [31] | Hepatobiliary cancer |
| 12 | Rv3423.1 | Mycobacterium tuberculosis | A 8 Kda proteins acetylating histone H3 at the K9/K14 positions [40]. | Lung cancer |
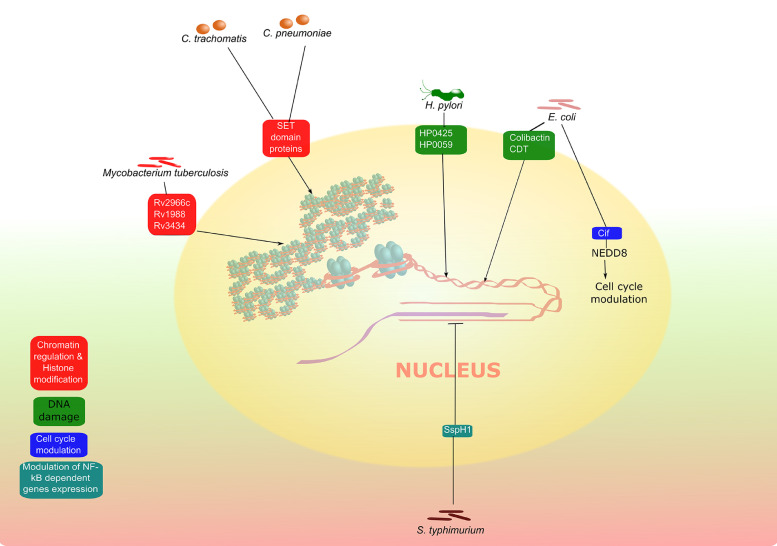
Fig. 3.
Nucleomodulins from cancer-associated bacteria and their subsequent effects on host cell nuclei. These are just a few out of a larger, more complete list which is beyond the coverage of this figure.The nucleomodulins targeting different nuclear aspects are shown in different colors. The nucleomodulins can have variety of other consequences on the host cells, but only primary effects mentioned in the literature are shown here.
Helicobacter pylori
H. pylori is a proven example of bacterial involvement in cancer etiology. Epidemiological evidence suggests the involvement of this bacterium in the etiology of gastric carcinoma specifically [82] . It is known to produce several proteins with the ability to alter nuclear function and some of these proteins are briefly discussed in Table 2. Among these proteins, some are directly translocated to the nucleus while some indirectly influence nuclear function. Recent studies found some H. pylori proteins directly localized in the host cell nucleus and contributing to gastric cancer development. For example, H. pylori proteins, HP0425 and HP0059 contain nuclear localization signal allowing them to translocate to the host cell nucleus. These proteins have DNAse activity which is an important factor for tumor development [47,48]. In addition to these nucleomodulins, CagA and VacA are the most widely acclaimed proteins found in H. pylori known to induce a variety of effects on the host cell leading to its suggested carcinogenic potential. CagA is encoded by CagA pathogenicity island, which also encodes the T4SS secretion system similar to vir genes found in Agrobacterium. The CagA pathogenicity island is present in highly virulent strains of H. pylori and is assumed to have been acquired by horizontal gene transfer events [18]. CagA is also involved in a variety of events not restricted to its host nucleus, including tyrosine phosphorylation by Src family kinase and induction of changes in cell structure leading to a hummingbird phenotype in cell culture [33]. Furthermore, CagA is known to localize to the inner membrane of host cell surface, but it can activate the ERK signaling pathway which influences the expression of several genes in the host nucleus, inducing cell proliferation and gastric cancer. CagA is also involved in nuclear accumulation of β-catenin, induction of the transcription factor NF-kB, which contributes to cancer progression [62,78]. In addition, CagA also induces DNA damage by increasing the expression of spermine oxidase [16]. It has been found that CagA has ability to target nucleus and small fraction of CagA is also detected in host cell nuclei [81] making it an important component of bacterial nucleomodulins involved in cancer. Another widely acclaimed H. pylori protein VacA localizes to cell membrane in order to form anion conducting channels. It causes the release of cytochrome C from mitochondria, leading to apoptosis. In contrast, some forms of this protein are strongly associated with gastric cancer and it is assumed that all H. pylori virulence factors act together to drive cancerous transformation. Further, VacA is involved in the translocation of β-catenin into nucleus resulting in cell proliferation [61], their direct targeting to host nucleus is not known. The role of CagA in the induction of gastric cancer has been reviewed extensively and therefore, in this article we only specify the aspects involved in modulation of nuclear function.
Escherichia coli
The role of E. coli in colorectal cancer has been suggested in a number of sources including epidemiological, experimental and computational studies. However, the exact reason behind the involvement of E. coli in colorectal cancer etiology is still awaited [42,44,45,79]. Several nucleomodulins have been detected in E. coli and noted for their potential involvement in CRC etiology. Some of these nucleomodulins are discussed in Table 2. E. coli is known to produce a toxin named Colibactin, which has the ability to cause DNA damage in host cells [83]. In addition, the contributions of other nucleomodulins like cytotoxic necrotizing factors (CNF) and cytolethal distending toxins (CDT) are also subject to current research. An in silico study predicted several E. coli proteins with the ability to localize in their host cell's nucleus [42], but the exact role of this category of proteins in E. coli mediated colorectal cancer etiology is still unknown.
Chlamydia
Chlamydia is another bacterial genus which has been noted for its involvement in cancer. While, contradictory evidence exists, the role of C. trachomtis, C. psittaci, C. pneumonie is suggested in cervical cancer [96], ocular adnexal lymphoma [15] and in lung cancer [92], respectively. The exact role of Chlamydia in the etiology of cancer is still a matter of debate, but the presence of nucleomodulins in these organisms provides support for their etiological potential. Some Chlamydial spp. are known to have SET domain proteins with the ability to alter chromatin structure. The role of SET domain proteins in cancer etiology has already reviewed recently [37], and Table 1 and 2 cover a few SET domain proteins identified in a few Chlamydia spp. with the ability to alter host chromatin. In addition, some Chlamydia spp. are known to have several other proteins targeting the host nucleus during pathogenesis. CT621 is a C. trachomatis protein, which is involved in translocation into the host nucleus and cytoplasm through the type 3 secretion system [35]. SINC is a T3SS protein from C. psittaci, which translocates to the inner nuclear membrane of host cell [56]. Nevertheless, there are many other Chlamydial proteins that are yet to be investigated for their potential to target host nuclei. Perhaps future research will unveil the reasoning behind the intriguing involvement of Chlamydia in cancer etiology and these nucleomodulins will serve as a link to fill the gap between the role of bacteria in cancer induction.
Salmonella
Salmonella is another debated bacterium suspected to be involved in cancer etiology. Many studies have linked the infection of Salmonella with hepatobiliary cancer risk [55]. Salmonella is also known to have proteins with the ability to target and alter host cell nuclear function. It has been reported that Salmonella also produces cytolethal distending toxin (CDT), which translocate into the host nucleus in order to cause DNA damage and chromatin fragmentation [69]. SspH1 is another Salmonella effector which translocate into the host nucleus and suppress NF-kB dependent gene expression [31], however its exact role in Salmonella mediated cancer risk must be evaluated as the role of several other Salmonella effectors is known in modulating cancer related events. For example, SopE is a T3SS protein from S. typhimurium, which has the ability to activate Rho GTpase (CDC42 & RAC1) resulting in cytoskeletal rearrangement and ultimately leading to the entry of bacteria into non phagocytic cells. In addition, SopE is involved in the activation of MAP kinase resulting in the activation of signaling pathways leading to altered nuclear function [32]. Similarly, another protein SopE2, is thought to have same ability of modulating nuclear function, but it has also ability to activate NF-kB. Moreover, SopB is involved in protection of host cell from apoptosis.
Other bacteria involved in cancer etiology or cancer-associated infections
Mycobacterium tuberculosis infection is common among lung cancer patients. Some studies suggest that pulmonary tuberculosis is associated with an increased risk for lung cancer [88], while some reports indicate that the immunocompromised status of cancer patients makes them more susceptible to tuberculosis infection [3]. Under both circumstances, tuberculosis is strongly associated with lung cancer. The protein Rv3423.1 found in M. tuberculosis is isolated from chromatin of human macrophages infected with these bacteria. It was found that this protein has ability to acetylate histone H3 [40]. The involvement of Streptococcus in the etiology of colorectal cancer is also a controversial point, but several studies have found an association between Streptococcus and colorectal cancer. It has been found that cell wall proteins of S. bovis can induce oncogenic transformation in the colon [22]. It is found to have a histone like protein HlpA, although its role has been proposed in the adherence to colorectal cancer cells [12]. Nevertheless, their histone like structure leaves scope for their inclusion in the category of nucleomodulins. Future research may reveal various novel nucleomodulins- like properties of HlpA or even in other proteins found in S. bovis, as this bacterium has been identified for its ability to modulate nuclear function and its carcinogenic potential without identifying exact bacterial proteins (Table 2).
Concluding remarks
Host pathogen interactions span a very complex set of mechanisms involving a variety of molecules including nucleomodulins. Nucleus is the core of a cell and plays a very important role during oncogenesis. Hijacking of cell organelles, such as the nucleus, by pathogens is a very important aspect for understanding the role of bacteria in cancer etiology. While, many methods are available to detect nuclear localization of bacterial proteins and their subsequent effects on nuclear function of a certain proteins, they are extremely arduous for analyzing whole bacterial proteomes for their potential to contain nucleomodulins. Some computational tools are also available to alleviate this difficulty by predicting the nuclear localization of certain protein. However, while this process may be fast and convenient, recent research indicate that these tools also have some limitations when predicting bacterial protein localization in the nucleus of human cell [43]. In addition, some proteins can modulate nuclear function without translocating into the host cell nucleus making the situation more complex. Perhaps future research will identify alternative fast track methods to detect entire bacterial proteomes for their nucleomodulins potential and their subsequent impact on cancer etiology. Moreover, among Cif family, E. coli effector is known to target host nucleus, but much is not known about Cif from other bacteria and they are mentioned in text due to their homology with E. coli Cif and possible similar effects. Similarly, colibactin is known to cause DNA damage and nucleomodulation, but its physical nuclear targeting needs investigation. We included here several bacterial proteins with the ability to modulate nuclear functions, but their direct nuclear targeting needs more investigations. Therefore, more studies are needed to understand mechanisms of nucleomodulins homologs and other nuclear function modulators.
Despite these limitations, various findings are beginning to be made on the modulation of nuclear function by bacteria. A similar category of molecules is also frequently detected in cancer-associated bacteria and provides several implications. Bacterial nucleomodulins may be the perfect candidate for the study of the bacterial role in cancer etiology since the role of similar nucleomodulins found in plants in the induction of plant tumors has already been proven. Increased understanding of this mechanism and its applicability to human tumor development may open up many ways to manage cancer by controlling certain risk factors. Future research in this direction is needed to reveal the association between bacterial nucleomodulins and animal oncogenesis which will ultimately aid in the study of cancer associated microbes and their etiologic potential.
Declaration of Competing Interest
Authors declare no potential conflicts of interest related to text of this manuscript.
Acknowledgments
Acknowledgment
Authors are thankful to Jade Thurnham, Cedars-Sinai Medical Center for critical reading and valuable suggestions during preparation of this manuscript.
Author contribution statement
Abdul Arif Khan: Formal Analysis, Investigation, Resources, Data Curation, Writing Original Draft, Project Administration; Zakir Khan: Formal Analysis, Investigation, Writing-Review and Editing.
Funding
None.
References
- 1.Agarwal S., Jin H., Pancholi P., Pancholi V. Serine/threonine phosphatase (SP-STP), secreted from Streptococcus pyogenes, is a pro-apoptotic protein. J. Biol. Chem. 2012;287:9147–9167. doi: 10.1074/jbc.M111.316554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alvarez-Venegas R. Bacterial SET domain proteins and their role in eukaryotic chromatin modification. Front. Genet. 2014;5:65. doi: 10.3389/fgene.2014.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anibarro L., Pena A. Tuberculosis in patients with haematological malignancies. Mediterr. J. Hematol. Infect. Dis. 2014;6 doi: 10.4084/MJHID.2014.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arbibe L., Kim D.W., Batsche E., Pedron T., Mateescu B., Muchardt C., Parsot C., Sansonetti P.J. An injected bacterial effector targets chromatin access for transcription factor NF-kappaB to alter transcription of host genes involved in immune responses. Nat. Immunol. 2007;8:47–56. doi: 10.1038/ni1423. [DOI] [PubMed] [Google Scholar]
- 5.Ashida H., Kim M., Schmidt-Supprian M., Ma A., Ogawa M., Sasakawa C. A bacterial E3 ubiquitin ligase IpaH9.8 targets NEMO/IKKgamma to dampen the host NF-kappaB-mediated inflammatory response. Nat. Cell Biol. 2010;12:66–73. doi: 10.1038/ncb2006. sup pp 61-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Audia J.E., Campbell R.M. Histone modifications and cancer. Cold Spring Harb. Perspect. Biol. 2016;8 doi: 10.1101/cshperspect.a019521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benabdillah R., Mota L.J., Lutzelschwab S., Demoinet E., Cornelis G.R. Identification of a nuclear targeting signal in YopM from Yersinia spp. Microb. Pathog. 2004;36:247–261. doi: 10.1016/j.micpath.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 8.Bhagwat A.S., Vakoc C.R. Targeting transcription factors in cancer. Trends Cancer. 2015;1:53–65. doi: 10.1016/j.trecan.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bierne H., Cossart P. When bacteria target the nucleus: the emerging family of nucleomodulins. Cell Microbiol. 2012;14:622–633. doi: 10.1111/j.1462-5822.2012.01758.x. [DOI] [PubMed] [Google Scholar]
- 10.Bierne H., Pourpre R. Bacterial factors targeting the nucleus: the growing family of nucleomodulins. Toxins (Basel) 2020;12:220. doi: 10.3390/toxins12040220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Binns A., Campbell A. eLS, John Wiley & Sons, Ltd; 2001. Agrobacterium Tumefaciens-Mediated Transformation of Plant Cells. [Google Scholar]
- 12.Boleij A., Schaeps R.M., de Kleijn S., Hermans P.W., Glaser P., Pancholi V., Swinkels D.W., Tjalsma H. Surface-exposed histone-like protein a modulates adherence of Streptococcus gallolyticus to colon adenocarcinoma cells. Infect. Immun. 2009;77:5519–5527. doi: 10.1128/IAI.00384-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burgdorfer W., Anacker R.L., Bird R.G., Bertram D.S. Intranuclear growth of Rickettsia rickettsii. J. Bacteriol. 1968;96:1415–1418. doi: 10.1128/jb.96.4.1415-1418.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Canonne J., Rivas S. Bacterial effectors target the plant cell nucleus to subvert host transcription. Plant Sig. Behav. 2012;7:217–221. doi: 10.4161/psb.18885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chanudet E., Zhou Y., Bacon C.M., Wotherspoon A.C., Muller-Hermelink H.K., Adam P., Dong H.Y., de Jong D., Li Y., Wei R., Gong X., Wu Q., Ranaldi R., Goteri G., Pileri S.A., Ye H., Hamoudi R.A., Liu H., Radford J., Du M.Q. Chlamydia psittaci is variably associated with ocular adnexal MALT lymphoma in different geographical regions. J. Pathol. 2006;209:344–351. doi: 10.1002/path.1984. [DOI] [PubMed] [Google Scholar]
- 16.Chaturvedi R., de Sablet T., Peek R.M., Wilson K.T. Spermine oxidase, a polyamine catabolic enzyme that links Helicobacter pylori CagA and gastric cancer risk. Gut Microbes. 2012;3:48–56. doi: 10.4161/gmic.19345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen D., Zhang J., Li M., Rayburn E.R., Wang H., Zhang R. RYBP stabilizes p53 by modulating MDM2. EMBO Rep. 2009;10:166–172. doi: 10.1038/embor.2008.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Covacci A., Telford J.L., Del Giudice G., Parsonnet J., Rappuoli R. Helicobacter pylori virulence and genetic geography. Science. 1999;284:1328–1333. doi: 10.1126/science.284.5418.1328. [DOI] [PubMed] [Google Scholar]
- 19.Crow A., Hughes R.K., Taieb F., Oswald E., Banfield M.J. The molecular basis of ubiquitin-like protein NEDD8 deamidation by the bacterial effector protein Cif. Proc. Natl. Acad. Sci. U. S. A. 2012;109:E1830–E1838. doi: 10.1073/pnas.1112107109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cuevas-Ramos G., Petit C.R., Marcq I., Boury M., Oswald E., Nougayrède J.-.P. Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proceed. Natl. Acad. Sci. 2010;107:11537. doi: 10.1073/pnas.1001261107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dean P., Scott J.A., Knox A.A., Quitard S., Watkins N.J., Kenny B. The enteropathogenic E. coli effector EspF targets and disrupts the nucleolus by a process regulated by mitochondrial dysfunction. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ellmerich S., Scholler M., Duranton B., Gosse F., Galluser M., Klein J.P., Raul F. Promotion of intestinal carcinogenesis by Streptococcus bovis. Carcinogenesis. 2000;21:753–756. doi: 10.1093/carcin/21.4.753. [DOI] [PubMed] [Google Scholar]
- 23.Escoll P., Mondino S., Rolando M., Buchrieser C. Targeting of host organelles by pathogenic bacteria: a sophisticated subversion strategy. Nat. Rev. Microbiol. 2016;14:5–19. doi: 10.1038/nrmicro.2015.1. [DOI] [PubMed] [Google Scholar]
- 24.A.L. Farone, S.G. Berk, M.B. Farone, J.H. Gunderson, The isolation and characterization of naturally-occurring amoeba-resistant bacteria from water samples, Final report R833102, US Environmental Protection Agency, 2010, USA.
- 25.Farris T.R., Dunphy P.S., Zhu B., Kibler C.E., McBride J.W. Ehrlichia chaffeensis TRP32 is a nucleomodulin that directly regulates expression of host genes governing differentiation and proliferation. Infect. Immun. 2016;84:3182–3194. doi: 10.1128/IAI.00657-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garcia-Garcia J.C., Rennoll-Bankert K.E., Pelly S., Milstone A.M., Dumler J.S. Silencing of host cell CYBB gene expression by the nuclear effector AnkA of the intracellular pathogen Anaplasma phagocytophilum. Infect. Immun. 2009;77:2385–2391. doi: 10.1128/IAI.00023-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garcia-Rodriguez F.M., Schrammeijer B., Hooykaas P.J. The Agrobacterium VirE3 effector protein: a potential plant transcriptional activator. Nucleic Acids Res. 2006;34:6496–6504. doi: 10.1093/nar/gkl877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guerra L., Guidi R., Frisan T. Do bacterial genotoxins contribute to chronic inflammation, genomic instability and tumor progression? Febs J. 2011;278:4577–4588. doi: 10.1111/j.1742-4658.2011.08125.x. [DOI] [PubMed] [Google Scholar]
- 29.Hamon M.A., Cossart P. Histone modifications and chromatin remodeling during bacterial infections. Cell Host Microbe. 2008;4:100–109. doi: 10.1016/j.chom.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 30.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 31.Haraga A., Miller S.I. A Salmonella enterica serovar typhimurium translocated leucine-rich repeat effector protein inhibits NF-kappa B-dependent gene expression. Infect. Immun. 2003;71:4052–4058. doi: 10.1128/IAI.71.7.4052-4058.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hardt W.D., Chen L.M., Schuebel K.E., Bustelo X.R., Galan J.E. S. typhimurium encodes an activator of Rho GTPases that induces membrane ruffling and nuclear responses in host cells. Cell. 1998;93:815–826. doi: 10.1016/s0092-8674(00)81442-7. [DOI] [PubMed] [Google Scholar]
- 33.Hatakeyama M., Higashi H. Helicobacter pylori CagA: a new paradigm for bacterial carcinogenesis. Cancer Sci. 2005;96:835–843. doi: 10.1111/j.1349-7006.2005.00130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Helleday T., Petermann E., Lundin C., Hodgson B., Sharma R.A. DNA repair pathways as targets for cancer therapy. Nat. Rev. Cancer. 2008;8:193–204. doi: 10.1038/nrc2342. [DOI] [PubMed] [Google Scholar]
- 35.Hobolt-Pedersen A.-.S., Christiansen G., Timmerman E., Gevaert K., Birkelund S. Identification of Chlamydia trachomatis CT621, a protein delivered through the type III secretion system to the host cell cytoplasm and nucleus. FEMS Immunol. Med. Microbiol. 2009;57:46–58. doi: 10.1111/j.1574-695X.2009.00581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hotson A., Chosed R., Shu H., Orth K., Mudgett M.B. Xanthomonas type III effector XopD targets SUMO-conjugated proteins in planta. Mol. Microbiol. 2003;50:377–389. doi: 10.1046/j.1365-2958.2003.03730.x. [DOI] [PubMed] [Google Scholar]
- 37.Huang L., Xu A.M. SET and MYND domain containing protein 3 in cancer. Am. J. Transl. Res. 2017;9:1–14. [PMC free article] [PubMed] [Google Scholar]
- 38.Hummel A.W., Wilkins K.E., Wang L., Cernadas R.A., Bogdanove A.J. A transcription activator-like effector from Xanthomonas oryzae pv. oryzicola elicits dose-dependent resistance in rice. Mol. Plant Pathol. 2016;18:55–66. doi: 10.1111/mpp.12377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jinadasa R.N., Bloom S.E., Weiss R.S., Duhamel G.E. Cytolethal distending toxin: a conserved bacterial genotoxin that blocks cell cycle progression, leading to apoptosis of a broad range of mammalian cell lineages. Microbiology. 2011;157:1851–1875. doi: 10.1099/mic.0.049536-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jose L., Ramachandran R., Bhagavat R., Gomez R.L., Chandran A., Raghunandanan S., Omkumar R.V., Chandra N., Mundayoor S., Kumar R.A. Hypothetical protein Rv3423.1 of Mycobacterium tuberculosis is a histone acetyltransferase. FEBS J. 2016;283:265–281. doi: 10.1111/febs.13566. [DOI] [PubMed] [Google Scholar]
- 41.Jubelin G., Taieb F., Duda D.M., Hsu Y., Samba-Louaka A., Nobe R., Penary M., Watrin C., Nougayrede J.P., Schulman B.A., Stebbins C.E., Oswald E. Pathogenic bacteria target NEDD8-conjugated cullins to hijack host-cell signaling pathways. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khan A.A. In silico prediction of escherichia coli proteins targeting the host cell nucleus, with special reference to their role in colon cancer etiology. J. Comput. Biol.: J. Comput. Mol. Cell Biol. 2014;21:466–475. doi: 10.1089/cmb.2014.0001. [DOI] [PubMed] [Google Scholar]
- 43.Khan A.A., Khan Z., Kalam M.A., Khan A.A. Inter-kingdom prediction certainty evaluation of protein subcellular localization tools: microbial pathogenesis approach for deciphering host microbe interaction. Brief. Bioinf. 2018;19:12–22. [Google Scholar]
- 44.Khan A.A., Khan Z., Malik A., Kalam M.A., Cash P., Ashraf M.T., Alshamsan A. Colorectal cancer-inflammatory bowel disease nexus and felony of Escherichia coli. Life Sci. 2017;180:60–67. doi: 10.1016/j.lfs.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 45.Khan A.A., Khan Z., Malik A., Shrivastava A., Jain S.K., Alshamsan A. Computational prediction of Escherichia coli proteins host subcellular targeting and their implications in colorectal cancer etiology. Cancer Lett. 2015;364:25–32. doi: 10.1016/j.canlet.2015.04.024. [DOI] [PubMed] [Google Scholar]
- 46.Kibler C.E., Milligan S.L., Farris T.R., Zhu B., Mitra S., McBride J.W. Ehrlichia chaffeensis TRP47 enters the nucleus via a MYND-binding domain-dependent mechanism and predominantly binds enhancers of host genes associated with signal transduction, cytoskeletal organization, and immune response. PLoS ONE. 2018;13 doi: 10.1371/journal.pone.0205983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim J.M., Choe M.H., Asaithambi K., Song J.Y., Lee Y.S., Lee J.C., Seo J.H., Kang H.L., Lee K.H., Lee W.K., Cho M.J., Rhee K.H., Youn H.S., Baik S.C. Helicobacter pylori HP0425 targets the nucleus with DNase I-like activity. Helicobacter. 2016;21:218–225. doi: 10.1111/hel.12271. [DOI] [PubMed] [Google Scholar]
- 48.Kwon Y.C., Kim S., Lee Y.S., Lee J.C., Cho M.-.J., Lee W.-.K., Kang H.-.L., Song J.-.Y., Baik S.C., Ro H.S. Novel nuclear targeting coiled-coil protein of Helicobacter pylori showing Ca2+-independent, Mg2+-dependent DNase I activity. J. Microbiol. 2016;54:387–395. doi: 10.1007/s12275-016-5631-9. [DOI] [PubMed] [Google Scholar]
- 49.B. Lacroix, V. Citovsky, Nopaline-type Ti plasmid of Agrobacterium encodes a VirF-like functional F-box protein, 5 (2015) 16610. [DOI] [PMC free article] [PubMed]
- 50.Lebreton A., Lakisic G., Job V., Fritsch L., To N.T., Camejo A., Mattei P.J., Regnault B., Nahori M.A., Cabanes D., Gautreau A., Ait-Si-Ali S., Dessen A., Cossart P., Bierne H. A bacterial protein targets the BAHD1 chromatin complex to stimulate type III interferon response. Science. 2011;331:1319–1321. doi: 10.1126/science.1200120. [DOI] [PubMed] [Google Scholar]
- 51.Lebreton A., Stavru F., Cossart P. Organelle targeting during bacterial infection: insights from Listeria. Trends Cell Biol. 2015;25:330–338. doi: 10.1016/j.tcb.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 52.Lee T.I., Young R.A. Transcriptional regulation and its misregulation in disease. Cell. 2013;152:1237–1251. doi: 10.1016/j.cell.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lei L., Dong X., Li Z., Zhong G. Identification of a novel nuclear localization signal sequence in Chlamydia trachomatis-secreted hypothetical protein CT311. PLoS ONE. 2013;8:e64529. doi: 10.1371/journal.pone.0064529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Malkin D. Li-Fraumeni syndrome. Genes Cancer. 2011;2:475–484. doi: 10.1177/1947601911413466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mellemgaard A., Gaarslev K. Risk of hepatobiliary cancer in carriers of Salmonella typhi. JNCI: J. Natl. Cancer Inst. 1988;80:288. doi: 10.1093/jnci/80.4.288. -288. [DOI] [PubMed] [Google Scholar]
- 56.Mojica S.A., Hovis K.M., Frieman M.B., Tran B., Hsia R.C., Ravel J., Jenkins-Houk C., Wilson K.L., Bavoil P.M. SINC, a type III secreted protein of Chlamydia psittaci, targets the inner nuclear membrane of infected cells and uninfected neighbors. Mol. Biol. Cell. 2015;26:1918–1934. doi: 10.1091/mbc.E14-11-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Morgan R., Boxall A., Harrington K.J., Simpson G.R., Michael A., Pandha H.S. Targeting HOX transcription factors in prostate cancer. BMC Urol. 2014;14:17. doi: 10.1186/1471-2490-14-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Muller P.A., Vousden K.H. p53 mutations in cancer. Nat. Cell Biol. 2013;15:2–8. doi: 10.1038/ncb2641. [DOI] [PubMed] [Google Scholar]
- 59.Murata M., Azuma Y., Miura K., Rahman M.A., Matsutani M., Aoyama M., Suzuki H., Sugi K., Shirai M. Chlamydial SET domain protein functions as a histone methyltransferase. Microbiology. 2007;153:585–592. doi: 10.1099/mic.0.29213-0. [DOI] [PubMed] [Google Scholar]
- 60.Mysore K.S., Bassuner B., Deng X.B., Darbinian N.S., Motchoulski A., Ream W., Gelvin S.B. Role of the Agrobacterium tumefaciens VirD2 protein in T-DNA transfer and integration. Mol. Plant Microbe In. 1998;11:668–683. doi: 10.1094/MPMI.1998.11.7.668. [DOI] [PubMed] [Google Scholar]
- 61.Nakayama M., Hisatsune J., Yamasaki E., Isomoto H., Kurazono H., Hatakeyama M., Azuma T., Yamaoka Y., Yahiro K., Moss J., Hirayama T. Helicobacter pylori VacA-induced inhibition of GSK3 through the PI3K/Akt signaling pathway. J. Biol. Chem. 2009;284:1612–1619. doi: 10.1074/jbc.M806981200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Neal J.T., Peterson T.S., Kent M.L., Guillemin K. H. pylori virulence factor CagA increases intestinal cell proliferation by Wnt pathway activation in a transgenic zebrafish model. Dis. Models Mech. 2013;6:802–810. doi: 10.1242/dmm.011163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Niu X., Zhou M., Henkel C.V., van Heusden G.P., Hooykaas P.J. The Agrobacterium tumefaciens virulence protein VirE3 is a transcriptional activator of the F-box gene VBF. Plant J.: Cell Mol. Biol. 2015;84:914–924. doi: 10.1111/tpj.13048. [DOI] [PubMed] [Google Scholar]
- 64.Păcurar D.I., Thordal-Christensen H., Păcurar M.L., Pamfil D., Botez C., Bellini C. Agrobacterium tumefaciens: from crown gall tumors to genetic transformation. Physiol. Mol. Plant Pathol. 2011;76:76–81. [Google Scholar]
- 65.Pennini M.E., Perrinet S., Dautry-Varsat A., Subtil A. Histone methylation by NUE, a novel nuclear effector of the intracellular pathogen Chlamydia trachomatis. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pongponratn E., Maneerat Y., Chaisri U., Wilairatana P., Punpoowong B., Viriyavejakul P., Riganti M. Electron-microscopic examination of Rickettsia tsutsugamushi-infected human liver. Trop. Med. Int. Health. 1998;3:242–248. doi: 10.1046/j.1365-3156.1998.00231.x. [DOI] [PubMed] [Google Scholar]
- 67.Prokop A., Gouin E., Villiers V., Nahori M.A., Vincentelli R., Duval M., Cossart P., Dussurget O. OrfX, a Nucleomodulin required for Listeria monocytogenes Virulence. MBio. 2017;8 doi: 10.1128/mBio.01550-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pumirat P., Broek C.V., Juntawieng N., Muangsombut V., Kiratisin P., Pattanapanyasat K., Stevens J.M., Stevens M.P., Korbsrisate S. Analysis of the prevalence, secretion and function of a cell cycle-inhibiting factor in the melioidosis pathogen Burkholderia pseudomallei. PLoS ONE. 2014;9:e96298. doi: 10.1371/journal.pone.0096298. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 69.Puneet G.Nath, Shukla V.K. Possible strategies of bacterial involvement in cancer development. In: Khan A.A., editor. Bacteria and Cancer. Springer; Netherlands, Dordrecht: 2012. pp. 165–184. [Google Scholar]
- 70.Redmond A.M., Carroll J.S. Defining and targeting transcription factors in cancer. Genome Biol. 2009;10:311. doi: 10.1186/gb-2009-10-7-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rennoll-Bankert K.E., Garcia-Garcia J.C., Sinclair S.H., Dumler J.S. Chromatin-bound bacterial effector ankyrin A recruits histone deacetylase 1 and modifies host gene expression. Cell Microbiol. 2015;17:1640–1652. doi: 10.1111/cmi.12461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sakakibara H., Kasahara H., Ueda N., Kojima M., Takei K., Hishiyama S., Asami T., Okada K., Kamiya Y., Yamaya T., Yamaguchi S. Agrobacterium tumefaciens increases cytokinin production in plastids by modifying the biosynthetic pathway in the host plant. Proc. Natl. Acad. Sci. U. S. A. 2005;102:9972–9977. doi: 10.1073/pnas.0500793102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sawan C., Herceg Z. Histone modifications and cancer. Adv. Genet. 2010;70:57–85. doi: 10.1016/B978-0-12-380866-0.60003-4. [DOI] [PubMed] [Google Scholar]
- 74.Schuelein R., Spencer H., Dagley L.F., Li P.F., Luo L., Stow J.L., Abraham G., Naderer T., Gomez-Valero L., Buchrieser C., Sugimoto C., Yamagishi J., Webb A.I., Pasricha S., Hartland E.L. Targeting of RNA Polymerase II by a nuclear Legionella pneumophila Dot/Icm effector SnpL. Cell Microbiol. 2018;20:e12852. doi: 10.1111/cmi.12852. [DOI] [PubMed] [Google Scholar]
- 75.Schulz F., Horn M. Intranuclear bacteria: inside the cellular control center of eukaryotes. Trends Cell Biol. 2015;25:339–346. doi: 10.1016/j.tcb.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 76.Sharma G., Upadhyay S., Srilalitha M., Nandicoori V.K., Khosla S. The interaction of mycobacterial protein Rv2966c with host chromatin is mediated through non-CpG methylation and histone H3/H4 binding. Nucleic Acids Res. 2015;43:3922–3937. doi: 10.1093/nar/gkv261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shiels B.R., McKellar S., Katzer F., Lyons K., Kinnaird J., Ward C., Wastling J.M., Swan D. A Theileria annulata DNA binding protein localized to the host cell nucleus alters the phenotype of a bovine macrophage cell line. Eukaryot. Cell. 2004;3:495–505. doi: 10.1128/EC.3.2.495-505.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sokolova O., Naumann M. NF-kappaB signaling in gastric cancer. Toxins (Basel) 2017;9:119. doi: 10.3390/toxins9040119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Swidsinski A., Khilkin M., Kerjaschki D., Schreiber S., Ortner M., Weber J., Lochs H. Association between intraepithelial Escherichia coli and colorectal cancer. Gastroenterology. 1998;115:281–286. doi: 10.1016/s0016-5085(98)70194-5. [DOI] [PubMed] [Google Scholar]
- 80.Tzfira T., Vaidya M., Citovsky V. Involvement of targeted proteolysis in plant genetic transformation by Agrobacterium. Nature. 2004;431:87–92. doi: 10.1038/nature02857. [DOI] [PubMed] [Google Scholar]
- 81.Uchida T., Kanada R., Tsukamoto Y., Hijiya N., Matsuura K., Yano S., Yokoyama S., Kishida T., Kodama M., Murakami K., Fujioka T., Moriyama M. Immunohistochemical diagnosis of the cagA-gene genotype of Helicobacter pylori with anti-East Asian CagA-specific antibody. Cancer Sci. 2007;98:521–528. doi: 10.1111/j.1349-7006.2007.00415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Uemura N., Okamoto S., Yamamoto S., Matsumura N., Yamaguchi S., Yamakido M., Taniyama K., Sasaki N., Schlemper R.J. Helicobacter pylori infection and the development of gastric cancer. N. Engl. J. Med. 2001;345:784–789. doi: 10.1056/NEJMoa001999. [DOI] [PubMed] [Google Scholar]
- 83.van Ooij C. Bacterial toxins: escherichia coli damages host DNA. Nat. Rev. Microbiol. 2010;8:534. doi: 10.1038/nrmicro2414. [DOI] [PubMed] [Google Scholar]
- 84.Von Dwingelo J., Chung I.Y.W., Price C.T., Li L., Jones S., Cygler M., Abu Kwaik Y. Interaction of the Ankyrin H core effector of legionella with the host LARP7 component of the 7SK snRNP complex. MBio. 2019:10. doi: 10.1128/mBio.01942-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Waldmann T., Schneider R. Targeting histone modifications-epigenetics in cancer. Curr. Opin. Cell Biol. 2013;25:184–189. doi: 10.1016/j.ceb.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 86.Wang Y., Peng W., Zhou X., Huang F., Shao L., Luo M. The putative Agrobacterium transcriptional activator-like virulence protein VirD5 may target T-complex to prevent the degradation of coat proteins in the plant cell nucleus. New Phytol. 2014;203:1266–1281. doi: 10.1111/nph.12866. [DOI] [PubMed] [Google Scholar]
- 87.Weinthal D.M., Barash I., Tzfira T., Gaba V., Teper D., Sessa G., Manulis-Sasson S. Characterization of nuclear localization signals in the type III effectors HsvG and HsvB of the gall-forming bacterium Pantoea agglomerans. Microbiology. 2011;157:1500–1508. doi: 10.1099/mic.0.047118-0. [DOI] [PubMed] [Google Scholar]
- 88.Wu C.Y., Hu H.Y., Pu C.Y., Huang N., Shen H.C., Li C.P., Chou Y.J. Pulmonary tuberculosis increases the risk of lung cancer: a population-based cohort study. Cancer. 2011;117:618–624. doi: 10.1002/cncr.25616. [DOI] [PubMed] [Google Scholar]
- 89.Yang B., Zhu W., Johnson L.B., White F.F. The virulence factor AvrXa7 of Xanthomonas oryzae pv. oryzae is a type III secretion pathway-dependent nuclear-localized double-stranded DNA-binding protein. Proc. Natl. Acad. Sci. U. S. A. 2000;97:9807–9812. doi: 10.1073/pnas.170286897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yaseen I., Kaur P., Nandicoori V.K., Khosla S. Mycobacteria modulate host epigenetic machinery by Rv1988 methylation of a non-tail arginine of histone H3. Nat. Commun. 2015;6:8922. doi: 10.1038/ncomms9922. [DOI] [PubMed] [Google Scholar]
- 91.Yu X., Noll R.R., Romero Duenas B.P., Allgood S.C., Barker K., Caplan J.L., Machner M.P., LaBaer J., Qiu J., Neunuebel M.R. Legionella effector AnkX interacts with host nuclear protein PLEKHN1. BMC Microbiol. 2018;18:5. doi: 10.1186/s12866-017-1147-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhan P., Suo L.J., Qian Q., Shen X.K., Qiu L.X., Yu L.K., Song Y. Chlamydia pneumoniae infection and lung cancer risk: a meta-analysis. Eur. J. Cancer. 2011;47:742–747. doi: 10.1016/j.ejca.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 93.Zhang X., van Heusden G.P.H., Hooykaas P.J.J. Virulence protein VirD5 of Agrobacterium tumefaciens binds to kinetochores in host cells via an interaction with Spt4. Proceed. Natl. Acad. Sci. 2017;114:10238–10243. doi: 10.1073/pnas.1706166114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhu B., Kuriakose J.A., Luo T., Ballesteros E., Gupta S., Fofanov Y., McBride J.W. Ehrlichia chaffeensis TRP120 binds a G+C-rich motif in host cell DNA and exhibits eukaryotic transcriptional activator function. Infect. Immun. 2011;79:4370. doi: 10.1128/IAI.05422-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhu B., Nethery K.A., Kuriakose J.A., Wakeel A., Zhang X.F., McBride J.W. Nuclear translocated Ehrlichia chaffeensis ankyrin protein interacts with a specific adenine-rich motif of host promoter and intronic Alu elements. Infect. Immun. 2009;77:4243–4255. doi: 10.1128/IAI.00376-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhu H., Shen Z., Luo H., Zhang W., Zhu X. Chlamydia trachomatis infection-associated risk of cervical cancer: a meta-analysis. Medicine (Baltimore) 2016;95:e3077. doi: 10.1097/MD.0000000000003077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zupan J.R., Citovsky V., Zambryski P. Agrobacterium VirE2 protein mediates nuclear uptake of single-stranded DNA in plant cells. Proc. Natl. Acad. Sci. U. S. A. 1996;93:2392–2397. doi: 10.1073/pnas.93.6.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zur Hausen H. The search for infectious causes of human cancers: where and why. Virology. 2009;392:1–10. doi: 10.1016/j.virol.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 99.Zurawski D.V., Mumy K.L., Faherty C.S., McCormick B.A., Maurelli A.T. Shigella flexneri type III secretion system effectors OspB and OspF target the nucleus to downregulate the host inflammatory response via interactions with retinoblastoma protein. Mol. Microbiol. 2009;71:350–368. doi: 10.1111/j.1365-2958.2008.06524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]