Abstract
Sarcomas are heterogeneous and clinically challenging soft tissue and bone cancers. Although constituting only 1% of all human malignancies, sarcomas represent the second most common type of solid tumors in children and adolescents and comprise an important group of secondary malignancies. More than 100 histological subtypes have been characterized to date, and many more are being discovered due to molecular profiling. Owing to their mostly aggressive biological behavior, relative rarity, and occurrence at virtually every anatomical site, many sarcoma subtypes are in particular difficult‐to‐treat categories. Current multimodal treatment concepts combine surgery, polychemotherapy (with/without local hyperthermia), irradiation, immunotherapy, and/or targeted therapeutics. Recent scientific advancements have enabled a more precise molecular characterization of sarcoma subtypes and revealed novel therapeutic targets and prognostic/predictive biomarkers. This review aims at providing a comprehensive overview of the latest advances in the molecular biology of sarcomas and their effects on clinical oncology; it is meant for a broad readership ranging from novices to experts in the field of sarcoma.
Keywords: bone sarcoma, molecular diagnostics, molecular medicine, soft tissue sarcoma, targeted therapy
Subject Categories: Cancer, Molecular Biology of Disease, Musculoskeletal System
Sarcomas are heterogeneous and clinically challenging soft tissue and bone cancers. The current article comprehensively reviews recent advances in the molecular characterization of sarcoma subtypes, and describes novel therapeutic targets and biomarkers in this field.
Glossary
- Cancer stem cells (CSCs)
Cells within the tumor found in very small fractions that are thought to be responsible for resistance to cancer treatments and thus relapse.
- Cell dormancy
Stage in cancer progression during which tumor cells cease dividing but survive in a quiescent state while waiting for appropriate environmental conditions.
- Chorioallantoic Membrane (CAM) models
Chick embryo CAM models used to study tumor formation, angiogenesis, and metastasis.
- Circulating tumor cells (CTCs)
Cells that leak into the vasculature or lymphatics from a primary tumor and are carried around the body in the blood circulation.
- Epigenomic alterations
Heritable change that does not affect the DNA sequence but results in a change in gene expression.
- Extracellular vesicles (EVs)
Heterogeneous family of vesicles generated from different subcellular compartments and released into the extracellular space or the blood circulation.
- Genomic alterations
Permanent modifications in the DNA sequence including somatic mutations, copy‐number variations (CNVs), and gene fusions.
- Immunotherapy
Type of cancer treatment that aids the immune system to fight tumors.
- Oncolytic viruses
Viruses that, by their intrinsic properties or through genetic engineering, specifically replicate in and kill cancer cells.
- Orthotopic xenografts
Animal models based on the injection of tumor cell lines in the location where the tumors typically appear in humans.
- Patient‐derived xenografts (PDXs)
Animal model based on transplantation of human tumor biopsies that encompass tumor cells and the TME in immunodeficient animals.
- Pediatric tumors
Tumors that typically arise between 0–14 years of age.
- Precision medicine
Approach to patient care that allows physicians to select the treatments that are most likely to help patients based on a molecular understanding of their disease.
- Sarcomas
Malignant neoplasms that originate from the skeleton or soft tissues.
- Tumor microenvironment (TME)
Cellular environment in which cancer cells reside encompassing the extracellular matrix and stromal cells (endothelial cells, fibroblasts, and immune cells)
Epidemiology of sarcoma
Although sarcomas are rare among adult malignancies, they represent 12–15% of all pediatric tumors (Stiller et al, 2013). Despite the implementation and continuous optimization of multimodal therapies, around one‐third of sarcoma patients still succumb to the disease. Historically, sarcomas have been clustered in two large subgroups, according to the anatomical site of occurrence—sarcomas of the skeleton and sarcomas of the soft tissues (hereafter referred to as “bone sarcomas” or “soft tissue sarcomas” [STSs], respectively). Both subgroups comprise a variety of histological subtypes, and recent technological advances have enabled to decipher a constantly increasing number of subtypes at the molecular level (Fig 1; Baldauf et al, 2018a; Koelsche et al, 2018a; Watson et al, 2018; Weidema et al, 2020). Table 1 summarizes the major sarcoma subtypes discussed in this review and their main features.
Figure 1. Diversity of sarcomas as highlighted by DNA methylation profiling.
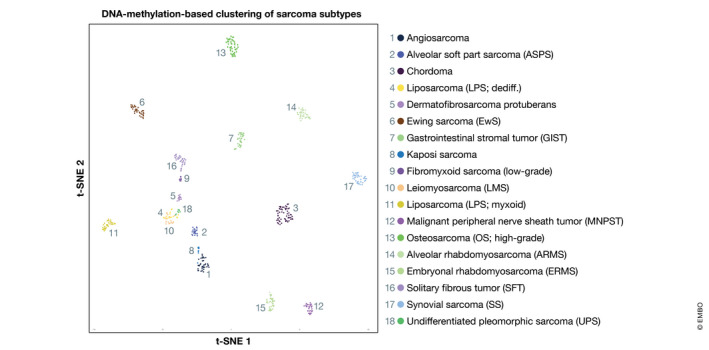
t‐distributed stochastic neighbor embedding (t‐SNE) plot of n = 18 major sarcoma and soft tissue tumor subtypes based on genome‐wide DNA methylation profiling on Illumina EPIC arrays (Koelsche et al, 2018a,b). Web‐link to classifier: www.molecularsarcomapathology.org.
Table 1.
Main sarcoma subtypes discussed in this review and their characteristics
| Sarcoma subtype | Abbreviation | Main features |
|---|---|---|
| Bone sarcomas | ||
| Chondrosarcomaa | CHS |
|
| Ewing sarcomaa | EwS |
|
| Osteosarcomaa | OS |
|
| Soft Tissue Sarcomas (STSs) | ||
| Fibrosarcomaa |
|
|
| GastroIntestinal Stromal Tumors | GIST |
|
| Leiomyosarcoma | LMS |
|
| Liposarcomaa | LPS |
|
| Rhabdomyosarcoma | RMS |
|
| Undifferentiated pleomorphic sarcomaa | UPS |
|
| Synovial sarcoma | SS |
|
The most common bone sarcoma and STS subtypes (WHO Classification of Tumours: Soft Tissue and Bone Tumours, 2020).
Among bone sarcomas, osteosarcoma (OS) is the most frequent subtype (Heymann, 2014). OS primarily affects adolescents and young adults, with the first and largest peak of incidence at age ~10–14 years. Coinciding with the pubertal growth spurt, the incidence rate of OS is 4 (3.5–4.6) for the range 0–14 years and 5 (4.6–5.6) for the range of 0–19 years per year per million persons (Ottaviani & Jaffe, 2009). The current standard of care was first introduced in the late 1970s and remains largely unaltered despite numerous efforts to improve outcomes (Rosen et al, 1976). Nowadays, patients with localized disease still face 5‐year overall survival rates < 70%, and < 20% of patients who develop metastatic disease or relapse survive > 3 years (Roberts et al, 2019). Ewing sarcoma (EwS) is included in the group of bone sarcomas because it is an aggressive sarcoma of both bone (~85% of cases) and soft tissue (~15% of cases), and because it has an incidence and survival rate similar to OS.
The STS subgroup comprises ~70–80% of all sarcomas with > 70 heterogenous histological subtypes (WHO Classification of Tumours: Soft Tissue and Bone Tumours, 2020). Although STSs represent < 1% of all cancers, they have the highest incidence among rare malignancies. Overall, the 5‐year survival for STS is estimated at ~57–62% and can vary widely depending on the disease stage and the complex interplay between anatomical site and STS subtype (Lyu et al, 2019). Unfortunately, the epidemiological data on specific STS subtypes are limited and frequently incomplete. National initiatives are ongoing to improve the databases, and they likely will benefit from the use of “big data” approaches. A recent review on the epidemiology of STSs in Italy and other European countries stated that they generally have an incidence of 6.27 and 4.71 cases per 100,000 inhabitants per year in Italy and Europe, respectively (Trama et al, 2019), with median ages at diagnosis of 58 and 63 years, respectively. Leiomyosarcoma (LMS), liposarcoma (LPS), and undifferentiated pleomorphic sarcoma (UPS; previously termed malignant fibrous histiocytoma) are the most common STS subtypes (WHO Classification of Tumours: Soft Tissue and Bone Tumours, 2020). A recent study in the Australian population reported that the incidence rate has almost doubled in the last 30 years (Bessen et al, 2019), which could be related to improved diagnostics or molecular pathology sub‐classification.
The complex biology of sarcoma: How current knowledge may affect therapy
To date, targeted therapy of sarcomas has only been partially effective, possibly due to the existence of compensatory pathways, the intrinsically heterogeneous nature of sarcomas, and the complex interplay with the tumor microenvironment (TME; Brown et al, 2018). In the TME, multiple intermingled cell types coexist through complex heterotypic cellular interactions and communicate via a large array of paracrine signals. The heterogeneity of different cancer cell subpopulations is further modulated by the extracellular matrix, admixed with intra‐ and extracellular reactive elements, such as metabolites, oxygen tension, and pH.
Impact of the tumor microenvironment on the stemness and behavior of sarcoma cells
Similar to the “seed and soil” theory described for other malignancies, sarcoma cells evolve in a permissive milieu favoring their quiescence and drug resistance or their proliferation and aggressiveness. Sarcoma cells are embedded in a highly heterogeneous tissue context composed of immune cells, endothelial cells, pericytes, mesenchymal stem cells (MSCs), cancer‐associated fibroblasts (CAFs), and nerve fibers, all of which may influence their behavior and favor “stemness” properties. Cancer stem cells (CSCs) usually represent only a very small fraction of the tumor cell mass, yet their eradication is critical for improving drug response. Indeed, CSCs have a great potential for self‐renewal and develop protective mechanisms against conventional anti‐tumor treatments, thereby causing sarcoma relapse and metastasis (Abarrategi et al, 2016; Brown et al, 2017a; Fourneaux et al, 2019; Hatina et al, 2019). Common methods of isolating/enriching CSCs to model sarcoma heterogeneity in vitro include culturing floating three‐dimensional (3D)‐colonies (tumorspheres), cell sorting based on the expression of specific markers (i.e., CD133, ABCG2, CD44, CD184, STRO1, CD117, CD271, or aldehyde dehydrogenase 1), the ability to extrude fluorescent dyes (side populations), or the selective pressure induced by long‐term culturing with chemotherapeutic drugs. CSCs have been extensively characterized in both bone sarcomas and STSs (Salerno et al, 2013; Abarrategi et al, 2016; Brown et al, 2018; Genadry et al, 2018; Skoda & Veselska, 2018; Hatina et al, 2019; Schiavone et al, 2019; Fig 2).
Figure 2. Biological features of sarcomas and therapeutic approaches.
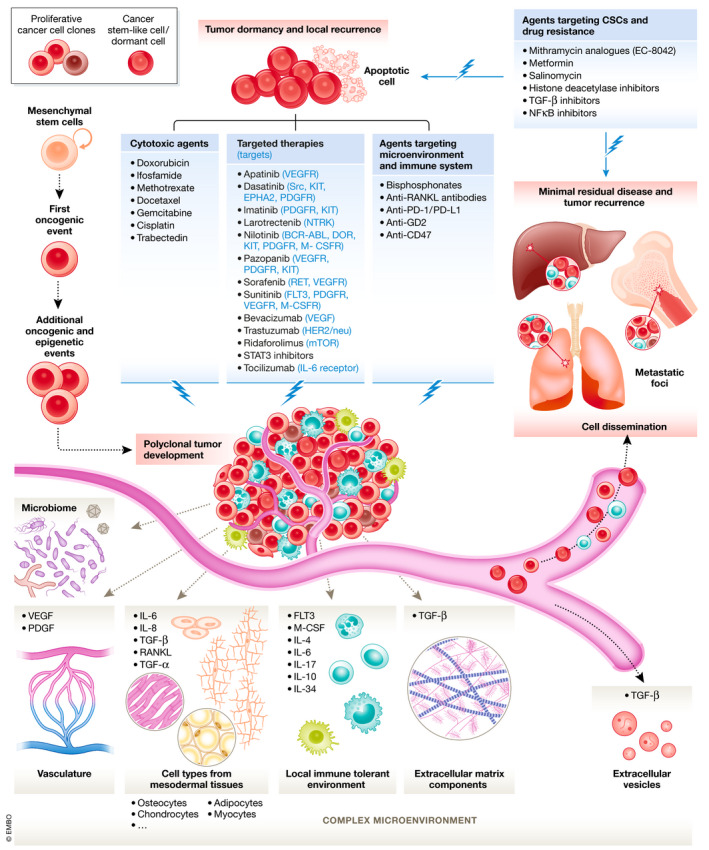
Sarcoma development results from a complex biological process. Their natural history combines the emergence of a first oncogenic hit followed by secondary oncogenic and epigenetic events with a conjuncture of a permissive microenvironment composed by cell types from mesodermal tissues, immune infiltrate, vascular, and extracellular matrix components. Sarcoma cells interact with their close environment through direct contact, enhanced cytokine/growth factors/miRNA signaling under a soluble form or encapsulated in extracellular vesicles. Sarcoma cells are characterized by a phenotypic and genetic heterogeneity coming from the successive oncogenic/epigenetic events occurring during tumor development and by cancer cells acquiring stemness properties that become progressively quiescent. Sarcomas are prone to induce distant metastatic foci spread by circulating tumor cells and invading after extravasation appropriate metastatic niches. Cancer cells installed in distant organs can spread again and enrich other metastatic sites increasing the tumor heterogeneity and potentially drug resistances. Persisting cells after resection of the primary tumor or dormant cancer cells located in distant organs characterize the minimal residual disease and are responsible of tumor recurrences. A selection of approved and experimental treatments aimed to prevent tumor growth and/or dissemination is shown.
Stemness in sarcoma is a fluctuating functional state orchestrated by the expression of pluripotency factors, such as OCT3/4, NANOG, KLF4, and, especially, SOX2 (Basu‐Roy et al, 2012; Maurizi et al, 2018; Skoda & Veselska, 2018; Sannino et al, 2019). The expression of these factors in sarcomas is oncogene‐driven and triggered by a combination of mutational and epigenetic events or by developmental programs (Rodriguez et al, 2012; Xiao et al, 2013). These events ultimately result in the deregulation of pathways that control stemness and differentiation, such as Hedgehog, Notch, Wnt/β‐Catenin, Hippo, or ALK (Graf Finckenstein et al, 2008; Naka et al, 2010; Riggi et al, 2010; Rodriguez et al, 2013; Basu‐Roy et al, 2015, 2016; Eid & Garcia, 2015; Tamaki et al, 2015; Abarrategi et al, 2016; Almazán‐Moga et al, 2017; Slemmons et al, 2017; Deel et al, 2018; Genadry et al, 2018; Hatina et al, 2019; Rodríguez‐Núñez et al, 2019; Schiavone et al, 2019; Trautmann et al, 2019). Alternatively, both stemness and aggressiveness can be regulated by the interaction with cells in the TME (Alfranca et al, 2015; Schiavone et al, 2019), or physical and chemical properties of the TME (i.e., hypoxia and extracellular acidosis) (Zeng et al, 2011; Alfranca et al, 2015; Avnet & Cortini, 2016; Avnet et al, 2017).
Several recent studies have focused on characterizing the sarcoma‐associated stroma and its effect on drug response (Tarnowski et al, 2010; Ehnman et al, 2013; Baglio et al, 2017; Cortini et al, 2017, 2019; Avnet et al, 2019). OS cells interact closely with MSCs, CAFs, osteoblasts, osteocytes, osteoclasts, chondrocytes, immune infiltrates, or components of the extracellular matrix to drive stemness‐promoting signaling (Avnet et al, 2008; Basu‐Roy et al, 2012; Zhang et al, 2013; Alfranca et al, 2015; Avnet & Cortini, 2016; Heymann et al, 2019). Moreover, MSCs/CAFs regulate tumor growth and metastasis through PDGFRα/β and MIF‐CXCR4/7 signaling, enhancing sarcoma aggressiveness via the secretion of inflammatory cytokines, exosomes (Miller et al, 2013; Cortini et al, 2016; Avnet et al, 2017; Baglio et al, 2017; preprint: Evdokimova et al, 2019), or metabolites that can fuel the oxidative metabolism of tumor cells (Bonuccelli et al, 2014). Metabolic fueling of sarcoma cells by stromal cells may be particularly relevant to sustain the energy demand of uncontrolled tumor growth and progression (Zhang et al, 2010; Ren et al, 2017; Gaude et al, 2018; Zhu et al, 2019). Consequently, the composition of the local TME has direct influence on the histological response to chemotherapy (Crenn et al, 2017). In addition, although axonogenesis has largely been neglected in sarcoma preclinical modeling so far, increasing evidence suggests that nerves in the TME may contribute to tumorigenesis, progression, and cancer‐associated pain in several sarcoma subtypes, such as fibrosarcoma, OS, EwS, LPS, and extraskeletal myxoid chondrosarcoma (CHS; Cain et al, 2001; Wacnik et al, 2005; Endo et al, 2008; Ghilardi et al, 2010; Kanojia et al, 2015; Moriarity et al, 2015; Shor et al, 2015; Brenca et al, 2019).
Moreover, the sarcoma TME may contain a specific microbiome (Nejman et al, 2020): A recent study described that bacterial DNA can be found in most CHSs. Bacteria were mostly intracellular and were detectable in immune and tumor cells (Nejman et al, 2020). Interestingly, metabolic functions related to intratumoral bacteria appeared tumor type‐specific; that is, degradation of hydroxyprolines by bacteria was enriched in CHSs (Nejman et al, 2020). Although more work is needed to decipher the precise role(s) of this symbiotic microenvironment, it is tempting to speculate that it could affect the stemness/differentiation and metabolic state of CHSs and possibly other sarcomas.
To date, several clinical and preclinical studies have reported treatments able to target the TME and/or CSCs in sarcomas (Abarrategi et al, 2016; Genadry et al, 2018; Schiavone et al, 2019) (Fig 2). The advent of techniques for single‐cell analysis, such as single‐cell DNA/RNA sequencing and spatial transcriptomics, will accelerate studying and modeling of sarcoma tissue heterogeneity and possibly lead to the identification of novel biomarkers and/or therapeutic targets.
The immune infiltrate in sarcoma as a source of new therapeutic targets
The TME of sarcoma cells is infiltrated by different immune cell populations (Fig 3). For example, OS tumor tissues are infiltrated by T lymphocytes (tumor‐infiltrating lymphocytes, TILs) in a very high percentage of patients, mainly expressing CD8+ (Théoleyre et al, 2005; Palmerini et al, 2017), and both TILs and tumor cells showed a high expression of HLA‐DR compared with other, non‐malignant bone tumors (Trieb et al, 1998). In preclinical models, CD8+ TILs are cytotoxic against allogeneic tumor cells (Théoleyre et al, 2005), and the number of CD8+ or CD8+/TIA1+ TILs correlates positively with longer survival in patients (van Erp et al, 2017; Gomez‐Brouchet et al, 2017; Palmerini et al, 2017). Similarly, in a small percentage of tumors, FOXP3+ (regulatory T cells, Tregs), and Arginase+ (myeloid‐derived suppressor cells, MDSCs), immune‐suppressive infiltrating cells were detected (Fritzsching et al, 2015; Palmerini et al, 2017). Notably, the CD8+/FOXP3+ ratio had a positive prognostic value (Fritzsching et al, 2015). Furthermore, a high pretreatment ratio of infiltrating neutrophils to lymphocytes, high levels of C‐reactive protein, Glasgow prognostic score, platelet–lymphocyte ratio score, and lymphocyte‐monocyte ratio or systemic absolute leukocyte counts in post‐therapeutic early recovery are independent prognostic markers (Moore et al, 2010; Liu et al, 2016; Vasquez et al, 2017).
Figure 3. Sarcomas are characterized by an immune oasis.
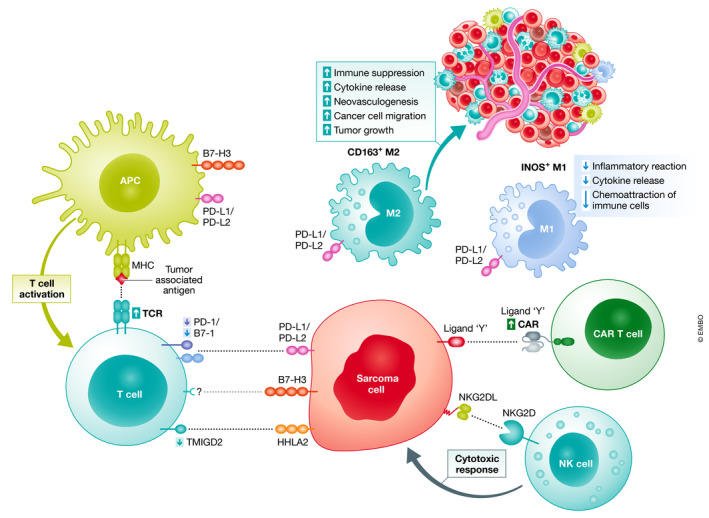
Sarcomas are infiltrated by numerous immune cells, which are in some sarcoma subtypes deleterious by establishing an immune tolerant microenvironment that can be at the origin of innovative therapeutic approaches. In physiological condition, the adaptive immune system is activated by exogenous antigens leading to initiation of an effective immune response against the host at the origin of these antigens. Unfortunately, in most cases immune activation by tumor‐associated antigen is counterbalanced by inhibitory signals transmitted after the binding of immune checkpoint molecules (e.g., PD‐1) expressed by immune effectors to their ligands expressed by cancer cells such as PD‐L1. Macrophages also contribute to the immune surveillance in sarcomas with two main distinct subsets: M1 macrophages with pro‐tumor activities and M2 macrophages with anti‐tumor and immunosuppressive functions. This immune landscape has led to the development of immunotherapies including immune checkpoint inhibitors, activated NK cells, or genetically modified T lymphocytes (CAR T cells) in order to reactivate the tumor immune surveillance.
Sarcomas are also frequently infiltrated by macrophages, which represent the main immune infiltrate and a highly heterogeneous population (Toulmonde et al, 2018; Mu et al, 2019; Stahl et al, 2019). Macrophage subpopulations are composed of a balance between immune‐stimulatory M1 and immune‐suppressive M2 macrophages that can be dysregulated in sarcomas. Both subpopulations are CD68+ and can be distinguished by the INOS and CD163 expression in M1 and M2 macrophages, respectively (Jayasingam et al, 2020). However, their roles are complex, as revealed by the functional discrepancy observed according to the given sarcoma subtype. Indeed, CD163+ is required for their protumoral activities (Shiraishi et al, 2018) and is a prognostic marker for specific sarcoma subtypes such as embryonal rhabdomyosarcoma (ERMS; Kather et al, 2019), whereas in OSs, CD163+ M2 macrophages are proangiogenic, facilitating cancer cell extravasation and promoting the metastatic process (Dumars et al, 2016; Han et al, 2016; Gomez‐Brouchet et al, 2017). Conflicting results showed a positive association of tumor‐associated dendritic cells (CD1a+) and macrophages with either a worse disease‐free survival (Koirala et al, 2016) or inhibition of metastases (Buddingh et al, 2011). However, their phenotype has not been fully characterized.
Sarcomas driven by reciprocal fusion oncoproteins, such as EwS, generally exhibit a low immune infiltrate, constituting so‐called “cold” tumors. Few available studies have demonstrated that TILs and dendritic cells are quite rare (immune desert) and that programmed death‐ligand 1 (PD‐L1) expression is usually low (Spurny et al, 2018). The presence of infiltrating macrophages has been associated with poorer overall survival (Vakkila et al, 2006), and elevated levels of circulating proinflammatory factors (e.g., interleukin 6, IL‐6) correlate with tumor‐associated fever at advanced stages (Lissat et al, 2015), implying the recruitment of immunosuppressing myeloid dendritic cells, macrophages, and other inflammatory cells at the tumor site (preprint: Evdokimova et al, 2019).
For STSs, only a very few recent reports have aimed to determine the “hot” or “cold” tumor immunophenotypes and their potential as biomarkers for response to therapy (Galon & Bruni, 2019). Kim et al reported the presence of PD‐1+ and PD‐L1+ TILs at rates of 65% and 58%, respectively, in various STS subtypes (Kim et al, 2013a). Similarly, the infiltrations of PD‐L1‐expressing macrophages and lymphocytes were observed in 58% and 30%, respectively, of 50 analyzed STS samples (D'Angelo et al, 2015), and the PD‐L1 expression was associated with a higher density of CD3+ PD‐1+ TILs, a higher tumor grading, and a lower overall survival (Orth et al, 2020). PD‐L1 was also expressed by tumor cells in 12% of cases, with the highest prevalence in gastroIntestinal stromal tumors (GISTs). Finally, the detection of low CD3+ or CD4+ TILs was significantly correlated with better overall survival by a univariate analysis (D'Angelo et al, 2015). However, recent reports have provided a more panoramic view of PD‐1 and PD‐L1 expression in larger series of STS and revealed that most STS subtypes show expression of both factors (Dancsok et al, 2019; Orth et al, 2020). However, the bioclinical relevance of PD‐1 and PD‐L1 (e.g., prognostic value) remains controversial in sarcomas, mainly due to their high heterogeneity (Fujii et al, 2014; Nduom et al, 2016; Nowicki et al, 2017).
Collectively, the immune infiltrates observed in sarcomas offer a rich opportunity for implementation of immunotherapeutic approaches in sarcomas. Yet, a complete and more standardized immune score may help to better understand the different immunophenotypes related to each sarcoma subtype and to improve immunotherapeutic approaches.
Models for studying the biology of sarcomas
Human cancer cell lines have become the cornerstone of cancer research. However, the accumulation of (epi‐)genetic mutations over time and across laboratories can have crucial implications when investigating new treatments as shown in carcinoma cell lines (Liu et al, 2019), since they affect drug response (Ben‐David et al, 2018). Whether this holds true for translocation‐driven sarcomas, such as alveolar rhabdomyosarcoma (ARMS), EwS, myxoid LPS, and SS, which display rather “silent” genomes, remains to be determined. Yet, the use of low‐passaged primary cell lines can prevent accumulation of mutations: A recent study of CHS patient samples and their derived cell lines characterized the genetic drift process of primary cell lines after 20–34 in vitro cell culture passages (Rey et al, 2019). Although the adaptation of tumor cells to in vitro cell culture is accompanied by additional genetic mutations, these rather low‐passaged CHS cell lines retained the most relevant mutations of the patient's founder clone (Rey et al, 2019).
For preclinical modeling of sarcoma, 3D culture has recently emerged as a tool for better prediction of drug efficacy and development of precision medicine approaches (Vaira et al, 2010; Santoro et al, 2015; Bregenzer et al, 2019). These 3D models include microfluidic devices, bioprinted cell‐enriched structures with tailorable biomechanical properties, and well‐defined tumoroids (Murphy & Atala, 2014), which contain different cell types, defined gradients of bioactive factors, and “physiological” biomaterials to precisely recapitulate the natural TME (Ma et al, 2018). This will help to elucidate the mechanical cross‐talk between sarcoma cells and “normal” cells (including vasculature and immune cells) (Huang et al, 2014; Datta et al, 2017), as well as components of the extracellular matrix (Doraiswamy et al, 2007; Pavlou et al, 2019). However, although a recent study has successfully employed a mineralized 3D bone model to evaluate the effect of the small‐molecule elesclomol on EwS cells (Marchetto et al, 2020), 3D models for the study of sarcoma are still in their infancy (Barron et al, 2004, 2005).
In vivo, the chick chorioallantoic membrane (CAM) assay is a valuable option due to its low costs and relatively easy implementation. CAM assays have been employed to study sarcoma angiogenesis, fibroblast infiltration, tumorigenesis, tumor invasion, and metastasis in CHS, EwS, fibrosarcoma, LPS, and OS (Sys et al, 2013; Patil et al, 2014; Manjunathan & Ragunathan, 2015; Cimpean et al, 2018; Kunz et al, 2019; Perut et al, 2019; Steinestel et al, 2020). Numerous additional in vivo models of inducible or spontaneous sarcomas have been described in non‐mammalian vertebrates (e.g., zebrafish; Leacock et al, 2012; Mohseny et al, 2012; Brown et al, 2017b; Hayes & Langenau, 2017; Ignatius et al, 2018; Fleming et al, 2019) and in mammalians (e.g., mouse, rat, and dog; Cannon, 2015; Jacques et al, 2018; Castillo‐Tandazo et al, 2019; Pomella & Rota, 2020). Genetically modified zebrafish and xenotransplantation of human sarcoma cells in fish were simultaneously proposed. Their main advantages are (i) their small size, allowing the maintenance of many animals at low costs; (ii) their high rate of proliferation (> 200 embryos per pairing); (iii) ex utero development of embryos, facilitating cell transplantations; (iv) their transparency, which facilitates non‐invasive and repeated imaging; (v) the possibility of imaging at the single‐cell level; (vi) studies of human cells and host factors facilitated by the use of transgenic lines; (vii) no immune rejection in early cell transplantation; and (viii) facilitation of high‐throughput drug screening due the animals’ permeability to small molecules through diffusion. Yet, the lack of specific organs (e.g., lungs) and the difference with human TME are two major limitations of zebrafish models (Mohseny et al, 2012; Brown et al, 2017b; Hayes & Langenau, 2017).
Genetically engineered mouse models (GEMMs) are considered reliable models for studying cancer development. Indeed, by inducing the formation of spontaneous tumors mimicking the natural history of human pathologies, GEMMs are privileged models to functionally identify and characterize molecular drivers or genetic initiator events of the disease (Kersten et al, 2017). While EwS, for which no bona fide GEMMs have been developed to date, is an exception among sarcomas, numerous GEMMs of bone sarcomas (for reviews, see ref. Jacques et al, 2018, 2019) and STSs (for review, see ref. Dodd et al, 2015) have been described. The first GEMM overexpressed the AP‐1 transcription factor c‐Fos in murine osteoblasts, which led to the development of OS without inducing metastatic foci (Grigoriadis et al, 1993). More recent models include deletion of Tp53, Rb, Prx‐1, or Prkar1a; overexpression of Sonic Hedgehog signaling components; or targeting Apc and Twist, and lead to the formation of metastatic OS (Jacques et al, 2018). Similarly, conditional loss of Tp53 or Ink4a/Arf in an Ext1‐driven GEMM results in the formation of CHS (de Andrea et al, 2015).
GEMMs of STSs were also developed (Dodd et al, 2015). For example, the conditional Pax3‐Fkhr knock‐in allele is associated with the development of ARMS with a frequency that can be increased by the loss of function of Ink4a/ARF and Tp53 (Keller et al, 2004). In addition, ERMS can be induced from the adipocyte lineage by adipocyte‐restricted activation of Hedgehog signaling through constitutive expression of an active Smoothened allele (Hatley et al, 2012). The latter model has also helped to demonstrate that Hedgehog signaling drives aberrant expression of myogenic specification factors, which may induce ERMS from non‐myogenic endothelial progenitors (Drummond et al, 2018). More recently, GEMMs for sarcomas have been obtained by CRISPR‐Cas9 technology (Huang et al, 2017).
Xenografts are alternatives to GEMMs and can be obtained by injection of tumor cells into immunodeficient mice. Xenografts are relatively easy to generate and highly reproducible (Picarda et al, 2010; Gambera et al, 2018; Jacques et al, 2018), but cannot fully recapitulate the TME of many sarcoma subtypes, and only rarely give rise to spontaneous metastases (Jacques et al, 2018). In this context, orthotopic xenografts obtained through injection of a suspension of tumor cells into the para‐ or intraosseous site for OS and EwS modeling (Hauer et al, 2013; Lamora et al, 2014; Stewart et al, 2014; Ségaliny et al, 2015; Baglio et al, 2017), or through intramuscular injection for the modeling of “soft tissue EwS” (Jaboin et al, 2002; Merchant et al, 2004), more closely recapitulated the TME of the respective tumor histotype. Similarly, early passage patient‐derived xenografts (PDXs) constitute a powerful tool for preserving the TME, histology, and genetic profiles of sarcomas (Hoffman, 2015; Stewart et al, 2017). PDXs are obtained through subcutaneous or orthotopic implantation of small fragments of tumors isolated from patients in immunodeficient mice. However, so far, only few studies have been published on PDXs in sarcoma due to the low success rate of the engraftment, the complex implantation procedure (Stewart et al, 2017; Nanni et al, 2019; Rainusso et al, 2019), and the costs required for the stabilization of the model, which may require up to a year (Nanni et al, 2019).
Current standard therapies for sarcomas
The therapeutic care of bone sarcoma and STS patients requires specialized sarcoma units. In fact, treatment in such specialized centers has been shown to result in improved surgical and oncologic outcomes (Blay et al, 2017). In addition, due to the potentially devastating consequences that can arise from poorly performed biopsies, biopsies of lesions suspected of being a sarcoma should be carried out in (or directed by) a specialized center (Mankin et al, 1982; Potter et al, 2008; Pretell‐Mazzini et al, 2015; Traub et al, 2018). The cornerstone of bone sarcoma and STS management is surgical resection of the primary tumor, which is typically accompanied by neoadjuvant and/or adjuvant chemotherapy and/or irradiation. Radiation therapy contributes to local control of tumor growth with positive margins or high‐grade STS (Kim et al, 2008). Chemotherapy regimens of bone sarcomas (e.g., OS, EwS) combine doxorubicin, cisplatin, methotrexate, and ifosfamide administered before and/after surgery for 6–12 months (Brown et al, 2018). Similarly, systemic treatments of STSs are mainly based on anthracyclines (e.g., doxorubicin) alone or in combination with an alkylating agent (e.g., ifosfamide) (Judson et al, 2014; Gómez & Tsagozis, 2020; Smrke et al, 2020). Interestingly, the use of adjuvant chemotherapy or radiotherapy may be defined by biological risk factors in high‐risk STSs (Sundby Hall et al, 2018). Although systemic therapy is the treatment of choice in metastatic disease (Meyers, 2015), resection of the primary tumor may still be performed with palliative intent, or rarely, in combination with resection of oligometastatic disease (Blakely et al, 2015). Wide margin surgery then remains the crucial technical approach in sarcoma treatment (Patrikidou et al, 2011).
For bone sarcomas, studies have demonstrated that oncologic outcomes of OS and EwS are similar between limb salvage and amputation when wide margins are achieved (Simon et al, 1986; Rougraff et al, 1994; Alamanda et al, 2012; Jauregui et al, 2018). Thus, the current standard of care is limb salvage surgery if preservation of neurovascular structures allows reconstruction of a functional extremity (Yang et al, 2017). Special considerations are made for limb reconstruction in the growing child, such as the use of growing prostheses, vascularized autografts, or van Nes rotationplasty. The choice of (neo)adjuvant treatment modalities is largely driven by the histological subtype: For instance, OS and EwS are usually chemosensitive and treated with neoadjuvant and adjuvant chemotherapy to decrease the risk of systemic disease progression, while STSs are frequently treated with neoadjuvant radiation therapy to decrease the risk of local recurrence (Gaspar et al, 2015; Brown et al, 2018; Le Cesne, 2018; Ray‐Coquard et al, 2018; Fig 2). In contrast, high‐grade CHS is largely resistant to existing chemo‐ and radiotherapies; thus, achieving a wide margin resection is currently the best option for prevention of disease progression (Reed et al, 2017; Brown et al, 2018; Whelan & Davis, 2018).
GIST is one of the STS subtypes for which the therapeutic development has been the most spectacular (Farag et al, 2020). For instance, up to 85% of patients with advanced GIST benefit from imatinib treatment (Blay, 2011). In fact, 90% of GISTs harbor driver mutations in the KIT proto‐oncogene receptor tyrosine kinase (KIT) and platelet‐derived growth factor receptor alpha (PDGFRA), which can be targeted by tyrosine kinase inhibitors (TKIs). Their therapeutic efficacy is directly linked to the type of mutation, and consequently, the acquisition of secondary mutations can result in drug resistance (see section “Resistance to targeted therapies”), which remains the most significant challenge in the treatment of locally advanced and metastatic GIST (Li & Raut, 2019). However, even fourth‐line therapy with TKIs may still be effective in advanced GIST (Blay et al, 2020).
Yet, the mostly moderate efficacy of any second‐line treatment for the majority of relapsed bone sarcomas and STSs highlights the need for intensified research to identify novel targets and improved preclinical models to predict drug response in molecularly defined cohorts of patients suffering from refractory and/or recurrent disease.
Mechanisms of drug resistance
Chemoresistance has been largely associated with the expression of specific detoxifying molecules, such as efflux pumps (ATP‐binding cassette (ABC) family proteins or ALDH enzymes), as it has also been recently demonstrated for CSCs (Lohberger et al, 2012). In particular, P‐glycoprotein is a 170 kDa transmembrane energy‐dependent efflux pump encoded by the MDR1 gene. Its expression leads to a multidrug resistance phenotype rather than an increased biological aggressiveness (Scotlandi et al, 1996; Baldini, 1997), which is associated with decreased event‐free survival in OS patients (Baldini et al, 1995) and in a small percentage of STS patients (Serra et al, 1996), and has also recently been found in bone sarcoma PDXs (Nanni et al, 2019).
Besides P‐glycoprotein, additional drug resistance mechanisms are caused by tumor heterogeneity arising from high DNA repair capacity, deregulation of apoptotic factors, adoption of a quiescent state (Honoki et al, 2010; Abarrategi et al, 2016; Martinez‐Cruzado et al, 2016; Roundhill et al, 2019; Vallette et al, 2019), drug delivery failure, the epithelial–mesenchymal transition (EMT) (Sannino et al, 2017), increased autophagy (Xiao et al, 2018), enrichment of CSCs (Eyler & Rich, 2008), protective signaling traits after chemotherapeutic treatment (Martins‐Neves et al, 2016; Yu et al, 2016), and immune evasion (Vasan et al, 2019).
In addition, resistance to conventional TKIs (e.g., imatinib) is associated with secondary mutations of KIT or PDGFRA in GIST (see section “Resistance to targeted therapies”). To overcome such acquired resistance, “switch pocket inhibitors” have been developed (Blay et al, 2020). A switch pocket inhibitor has the same target as the conventional inhibitors but acts like a light switch that deactivates cell signaling associated with the targeted receptor via blocking conformational activation of the kinase. For example, ripretinib targets KIT, PDGFRα/β, kinase insert domain receptor (KDR), and colony‐stimulating factor 1 receptor (CSF1R alias C‐FMS) and has been developed to overcome the TKI resistance occurring in GIST patients. The Asp842Val (D842V) mutation of PDGFRA was identified as the primary driver mutation in 5–6% of GISTs, which are refractory to all currently approved TKIs (Corless et al, 2005). The D842V mutation is located in the exon 18 encoding the PDGFRA activation loop and modifies the protein conformation to a “constitutive” active form.
Avapritinib is a new TKI designed on the base of its selectively property to target the active conformation of KIT and PDGFRA. A phase I clinical trial (ClinicalTrials.gov No. NCT02508532) has recently assessed its safety, tolerability, and anti‐tumor activity (Heinrich et al, 2020). Interestingly, 9% of complete response and 79% a partial response was observed. Ripretinib—an inhibitor of all known KIT and PDGFRA mutations—forces the switching of the mutated receptors to assume the “off” position. A recent double‐blind, randomized, placebo‐controlled, phase 3 clinical trial (ClinicalTrials.gov No. NCT03353753) showed that ripretinib significantly improved the progression‐free survival with an acceptable safety profile in patients suffering from advanced GIST resistant to approved treatment (Blay et al, 2020).
Similarly, the classification of BRAF mutations, the knowledge about dysregulated signaling pathways and dysregulated circuitries related to these mutations, and the function of BRAF in sarcoma led to the development of new therapeutic options to overcome resistance to conventional chemotherapy. For instance, the BRAF V600E mutation was recently identified as a potential therapeutic target in a small subset of SS (Watanabe et al, 2020). It is interesting to note that resistance to BRAF mutation inhibitors may be overcome by combining BRAF inhibitors with EGFR, PI3K, mTOR, MEK, RTK, HGF, and MET inhibitors, leading to the targeting of the MAPK and PI3K‐AKT‐mTOR signaling pathways (Liu et al, 2020). CX‐6258 is a pan‐Pim kinase inhibitor selected for its potent activity against sensitive and resistant cancer cells to RAF/MEK inhibitor (Haddach et al, 2011).
Using a KINOMEscan assay platform, haspin kinase was identified as a target of CX‐6258. The inhibition of haspin reduced cancer cell proliferation and regulated the immune system by increasing the frequency of interferon γ (IFNγ)‐producing CD8+ T cells and reducing the number of Tregs in vivo (Melms et al, 2019). Interestingly, the haspin kinase inhibitor can overcome RAF/MEK inhibitor‐resistant cancer cells and shows anti‐tumor effects in EwS (Melms et al, 2019). Acquired resistance to cisplatin observed in OS patients is associated with a poor prognosis (Higuchi et al, 2019). Peroxisome proliferator‐activated receptor gamma (PPARγ) was reported to enhance the efficacy and overcome resistance to cisplatin in various oncological entities and exhibits similar properties in OS (Higuchi et al, 2019).
The cell differentiation state also affects drug sensitivity (Dawson et al, 2020). A subpopulation of RMS cells that expressed MYOD1 and NOG exhibited primary resistance to vincristine and doxorubicin, which can be partly overcome by the combination of 12‐O‐tetradecanoylphorbol‐13‐acetate (TPA) and an enhancer of zeste homolog 2 (EZH2) inhibitor (GSK126) (Dawson et al, 2020). EZH2 is an epigenetic drug acting as a histone methyltransferase inhibitor that has been recently approved for metastatic or locally advanced epithelioid sarcoma (Rugo et al, 2020). The elimination and recycling of damaged proteins and organelles are driven by autophagy, which provides energy to the cells. Autophagy can be activated by chemotherapy and can promote increased chemosensitivity, as well as drug resistance in OS (Camuzard et al, 2019; Liao et al, 2019). Thus, drugs regulating autophagy may be an option to overcome drug resistance in the future.
Cell dormancy and recurrence
The risk of recurrence in oncology is associated with the persistence of cancer cells, which are not clinically/biologically detectable after resection of the primary tumor (Arlt et al, 2013). The latency without any detectable disease varies according to the clinical condition (e.g., histological grade and subtype) and depends on cancer cells characterized by slow cycling, low metabolism and fitness, and consequently, long‐term survival mechanisms (Vallette et al, 2019). Awakened cancer cells re‐acquire an active state, with capacities of proliferation and spreading to distant sites, and they define the minimal residual disease (Riethmüller & Klein, 2001). Dormant cells have been identified in several sarcoma subtypes, including fibrosarcoma (Dobson & Dickey, 1956; Varani et al, 1981; Cao et al, 1998), LPS (Almog et al, 2006; Rogers et al, 2014), RMS (Kimura et al, 2002), Kaposi sarcoma (Indraccolo et al, 2006), and OS (Naumov et al, 2006; Shimizu et al, 2014; Avril et al, 2016a,b; Guo et al, 2017). These rare dormant cells exhibit stemness properties (Visvader, 2011), and they have been related to drug resistance (De Angelis et al, 2019; Smith & Macleod, 2019; Vallette et al, 2019). The emergence of dormant cells is a conserved biological process linked to cell survival and controlled by multiple parameters, including genetic and epigenetic alterations, clonal cell evolution, cell–matrix interactions within the TME (e.g., immune tolerance), and diversity/heterogeneity. No specific molecular signature of dormant sarcoma cells has yet been identified. The most recent molecular approaches (e.g., single‐cell RNA sequencing, RNA/DNA methylation profiling) should lead to the identification of their specific molecular profile and of the molecular drivers of this state. For instance, myeloma dormant cells are switched “on” by engagement with osteoblastic cells and switched “off” by active osteoclasts (Lawson et al, 2015), which illustrates the clinical interest of targeting cell dormancy also in the context of bone sarcomas and STSs (Endo & Inoue, 2019; Recasens & Munoz, 2019; Tellez‐Gabriel et al, 2019).
Resistance to targeted therapies
TKIs are the largest class of targeted therapies approved by the Food and Drug Administration (FDA). In particular, GIST commonly shows activating mutations in the receptor tyrosine kinases KIT and PDGFRA. While physiological KIT or PDGFRα signaling are involved in cell differentiation and survival, activating mutations in both genes results in constitutive ligand‐independent receptor activation, leading to GIST tumorigenesis. TKIs are the standard of care in the primary treatment of GIST, and imatinib is the most commonly used compound (Casali et al, 2018). The resistance toward TKIs in GIST is mainly related to secondary mutations of KIT (Li & Raut, 2019; Napolitano & Vincenzi, 2019), but can also be triggered by PDGFRA mutations (Lim et al, 2008; Kalfusova et al, 2019).
In non‐GIST STSs, the currently approved targeted therapies are limited to the multi‐target TKI pazopanib, which targets VEGFR‐1, VEGFR‐2, and VEGFR‐3, PDGFRα and PDGFR‐β; and KIT (Lee et al, 2019). It has been demonstrated that anti‐angiogenic TKIs, including pazopanib, do not succeed in targeting sarcoma stem cells (Canter et al, 2014), whereas treatment with pazopanib in a human SS model promotes the development of resistance (Lanzi et al, 2019). Despite a strong inhibition of the main target of pazopanib, PDGFRα/β, the activation of the AKT and ERK signaling pathways was only partially impaired, possibly due to the over activation of other tyrosine kinase receptors, including the insulin‐like growth factor receptor type 1 (IGF1R) and insulin receptor (IR). Similarly, in another SS cell line, the presence of an NRAS mutation sustained ERK activation and caused resistance to pazopanib treatment (Lanzi et al, 2019). Thus, a combination treatment with either an IGF1R/IR inhibitor or a MEK inhibitor has been suggested to restore the inhibition of the PDGFRα/β pathways and effectively promote apoptosis (Lanzi et al, 2019). Phosphoproteomic profiling of pazopanib‐resistant cells identified the inhibition of HSP90 as a therapeutic route to overcome resistance (Vyse et al, 2018).
These findings highlight the importance of patient‐specific tumor profiling to identify the underlying activated signaling pathways, thereby avoiding the “one‐size‐fits‐all” paradigm and moving toward personalized, multi‐line, and patient‐specific treatment regimens (Wilding et al, 2019). Biomarker‐guided basket trials, such as the CREATE trial, which evaluates multiple disease types with a common oncogenic driver matched to a specific targeted therapy, may be considered in this respect (Péron et al, 2019). Moreover, characterization of interpatient pharmacokinetic variability will be a valuable tool to predict and overcome the development of resistance (Cardoso et al, 2020).
Other types of resistances
Several other indirect mechanisms of drug resistance in sarcoma have been identified, such as the formation of abnormal TME, hypoxia, and acidosis. Elevated levels of hypoxia and hypoxia‐inducible factor 1α (HIF1α) in human sarcomas correlate with tumor progression and radiation resistance (Kim et al, 2013b). In particular, in STS, HIF1α expression was found in 25.5% of tumors and was associated with both shorter overall survival and progression‐free survival (Kim et al, 2015). Moreover, translational activation of HIF1α by YB‐1 was found to promote metastasis in preclinical models of EwS, OS, and RMS (El‐Naggar et al, 2015). Similarly, in OS, hypoxia was responsible for the induction of the Wnt/β‐catenin signaling pathway and resulted in 6–13 times more cell resistance to doxorubicin‐mediated toxicity than under normoxic conditions (Roncuzzi et al, 2014; Scholten et al, 2014). In EwS, hypoxia has been found to protect tumor cells against anticancer drugs, while suppression of HIF1α enhanced drug‐induced apoptosis (Kilic et al, 2007). Accordingly, metabolic characterization, including hypoxic phenotypes, may help to identify specific treatment modalities in OS, other bone sarcomas, and STSs (Eary et al, 2011; Campanile et al, 2013). Along these lines, a recent pilot study characterized different metabolic parameters in a small group of STS patients using specific positron emission tomography (PET) agents to assess the individual risk associated with biological characteristics of the tumors (Wolsztynski et al, 2018).
Tumor acidosis is a metabolic adaptation observed in cancers and characterized by the fermentation of glucose to lactic acid. This process occurs in the presence of oxygen and is called aerobic glycolysis or Warburg effect. This adaptative mechanism modulates the drug sensitivity and leads to drug resistances by intrinsic (e.g., modulation of the mutational profile driven by a cell adaptation to stress) or extrinsic (e.g., structural/functional modulation of drugs induced by the local pH modifications) mechanisms (Kolosenko et al, 2017). Indeed, the pH of the local microenvironment regulates the passive diffusion of small molecules such as cancer drugs across biological barriers by modulating charged components of cell membranes, process named ion trapping or pH‐partitioning (Scott et al, 2017). Many cancer drugs are ionizable molecules containing weak bases or acids in their structure and are subjected to pH‐partitioning resistance (Zhitomirsky & Assaraf, 2016). That is the case for doxorubicin (weak base compound) in OS, which is trapped in the acidic extracellular microenvironment and consequently cannot target cancer cells (Avnet et al, 2016). On the contrary, the cytotoxic effects of cisplatin (weakly acidic drug) are increased in OS by the local tumor acidosis, which favors its neutral form and then facilitates its passive diffusion across the cell membranes (Avnet et al, 2016). In the cytoplasm, cisplatin is ionized by the low alkaline pH and trapped in the cell. A similar phenomenon was described in RMS, and the diffusion of weak base drugs across cell membranes and their sequestration in the lysosomal compartment are facilitated by ion trapping (Salerno et al, 2014; Zhitomirsky & Assaraf, 2016).
Molecular signatures of sarcomas: Effects on diagnosis and prognosis
In past decades, an unbiased and systematic search for gene fusions combined with unsupervised gene expression and (epi)genetic analyses of different sarcoma subtypes led to better classification systems (WHO Classification of Tumours: Soft Tissue and Bone Tumours, 2020). In addition, these molecular signatures provide information about the biology of these tumors, reflecting both the characteristics of the sarcoma's cell of origin and the activated pathways driving the malignant phenotype (Taylor et al, 2011).
Genomic and transcriptomic alterations
The Cancer Genome Atlas (TCGA) Research Network reported a recent analysis of 206 adult STSs representing six major subtypes (Cancer Genome Atlas Research Network, 2017). Here, the authors showed that common sarcomas (except for SS) are characterized by a high number of copy‐number variations (CNVs) and recurrent point mutations in relatively few genes, such as TP53, ATRX, and RB1. Importantly, specific genomic and transcriptomic alterations also define molecular subtypes, which are associated with patient outcome (Cancer Genome Atlas Research Network, 2017). Other studies have identified whole‐genome duplication as a cause of the structural complexity of UPS (Steele et al, 2019), and CDKN2A alterations as a predictor of worse overall survival across sarcoma subtypes (Bui et al, 2019). Integrated analysis of genomic and transcriptomic data confirmed the mutational profiles of STSs and identified PDGFRα as a putative target in complex karyotype STSs (Kim et al, 2018). Indeed, a PDGFRα‐blocking antibody (olaratumab) in combination with doxorubicin showed promising results for non‐GIST STS treatment (Klug & Heinrich, 2017). Given the widespread presence of CDK4‐amplification/high expression and CDKN2A loss across sarcomas subtypes, CDK4 inhibitors such as palbociclib are also a promising strategy in RB‐positive tumors (Dickson et al, 2013). It is noteworthy that ATRX has been shown to be required for response to CDK4 inhibitors in LPS, providing a potential biomarker for upcoming clinical trials (Kovatcheva et al, 2015; Cancer Genome Atlas Research Network, 2017). Integration of genomic and transcriptome analysis has also uncovered a “BRCAness” mutational signature in LMS, which confers sensitivity to DNA double‐strand break‐inducing drugs (Helleday, 2011; Chudasama et al, 2018) and sensitivity toward the combination of the poly(ADP‐ribose) polymerase (PARP) inhibitor olaparib and cisplatin (Chudasama et al, 2018). Olaparib combined with trabectedin (an alkylating drug) showed manageable toxicities at active dose levels and encouraging anti‐tumor activity in STS (Grignani et al, 2018). A phase 2 study on this topic is ongoing (ClinicalTrials.gov No. NCT04076579).
Exome sequencing has revealed a combination of single‐base substitutions, loss of heterozygosity events, and/or large‐scale genome instability involving 14 driver genes (ATM, ATRX, BAP1, BRCA2, FANCA, MDC1, MUTYH, NUMA1, PTEN, RB1, RECQL4, RET, TP53, and WRN) and many additional genes that define a “BRCAness” signature in > 80% of OS (Kovac et al, 2015). In fact, OS is characterized by a very complex altered genomic landscape explained by chromothripsis‐generating driver mutations and multiple genomic rearrangements (Behjati et al, 2017). However, in some cases, OS tumorigenesis is associated with germline alterations in TP53, RB1, and RECQL1/2/3 predisposing patients to the accumulation of high numbers of somatic mutations (Smida et al, 2017; Baumhoer et al, 2019; Sayles et al, 2019). In addition, two recent publications hypothesized that specific somatic CNV profiles of OS can be used for outcome prediction and for identification of altered genes and associated pathways as potential therapeutic targets (Smida et al, 2017; Sayles et al, 2019). Similar preliminary findings have been reported for EwS and RMS (Cheng et al, 2019). Olaparib combined with ceritinib (ALK inhibitor) in OS showed limited toxicity and should be further evaluated (Beck et al, 2020). A clinical trial assessing olaparib combined with ceralasertib (ATR inhibitor) is currently in progress in the context of OS (ClinicalTrials.gov No. NCT04417062).
In contrast to OS and most sarcomas of adulthood, translocation‐driven pediatric sarcomas, such as EwS, SS, or fusion‐positive ARMS, exhibit much lower rates of single‐nucleotide variants and CNVs, and, instead, appear to be driven by marked epigenetic and transcriptomic perturbations induced by the fusion oncoproteins (Shern et al, 2014; Tirode et al, 2014; Cancer Genome Atlas Research Network, 2017). In fact, through the integration of transcriptomic and genetic data, a recent study found that EWSR1‐FLI1 hijacks the developmental transcription factor SOX6 and thus promotes proliferation of EwS cells, which provides opportunities for targeted therapeutic intervention for the oxidative stress inducer elesclomol (Marchetto et al, 2020). New molecular studies have also shed light on the role of the interplay between germline variants and somatic mutations in interindividual tumor heterogeneity in EwS (Musa & Grünewald, 2020). Musa et al recently showed that EWSR1‐FLI1 binds to a polymorphic enhancer‐like GGAA‐microsatellite, through which it regulates the expression of the oncogenic transcription factor MYBL2 (Musa et al, 2019). Importantly, variability at this MYBL2‐associated GGAA‐microsatellite is inherited via the germline and linked to intertumoral variation in MYBL2 expression (Musa et al, 2019). As MYBL2 is phosphorylated and activated by CDK2 (Musa et al, 2017), high MYBL2 expression sensitizes EwS cells to CDK2 inhibition, indicating the potential for using MYBL2 as a biomarker in anti‐CDK2 therapy (Musa et al, 2019).
While oncogenic gene fusions involving transcription factors remain largely undruggable (Knott et al, 2019), clinical trials using larotrectinib, a kinase inhibitor targeting gene fusions involving NTRK1/2/3, have shown promising results and could offer a strategy for the treatment of NTRK‐fusion‐positive sarcomas (Doebele et al, 2015; Fig 3). In addition, DNA minor groove‐binding agents in DNA, such as trabectedin or mithramycin, have been described as potent inhibitors of EWSR1‐FLI1‐mediated transcription with anti‐tumor potential (Bailey et al, 2019; Harlow et al, 2019). A recent clinical trial showed that mithramycin was too toxic at the dose required to inhibit EWSR1‐FLI1 (Grohar et al, 2017). However, the development of less toxic second‐generation mithramycin analogs, such as EC‐8042, opens the possibility of using this compound clinically (Osgood et al, 2016; Tornin et al, 2016; Fig 3).
Epigenetic alterations
Mutations in chromatin remodeler components have recently been recognized as oncogenic drivers in adult and pediatric sarcomas (Nacev et al, 2019). Recurrent somatic missense mutations in histone H3 at lysine 36 impair the mesenchymal differentiation program and promote the initiation of UPS (Fang et al, 2016; Lu et al, 2016). These mutations result in hypomethylation of H3K36 and a gain in H3K27 methylation that leads to the de‐repression and redistribution of polycomb repressive complex 1 (PRC1) associated with a blockade of mesenchymal differentiation. K36M mutations in H3F3B have also been detected in most chondroblastomas (Behjati et al, 2013). The detection of histone mutations could help in therapeutic choices as recently evidenced by an instructive case of a patient diagnosed with a histiocytic neoplasm harboring a histone H3K36I mutation. This patient did not respond to multiple histiocytosis treatments, but showed a stable therapeutic response after chemotherapy and radiation therapy used for STS (Snuderl et al, 2019). Similarly, mutations in chromatin remodeling genes, including ATRX, DOT1L, and H3F3A, have been identified in 14 UPS cases highlighting the potential involvement of deregulated chromatin remodeling pathways in tumorigenesis (Ali et al, 2019).
Epigenetic alterations and signatures have also been extensively explored in EwS. In fact, EwS has been defined as an “enhancer disease” with substantial levels of epigenetic heterogeneity (Tomazou et al, 2015; Sheffield et al, 2017). In contrast to many other cancers, inter‐tumor epigenetic heterogeneity did not uncover discrete subgroups in EwS, but, rather, defined a continuous spectrum along two distinct and biologically interpretable dimensions (“Ewing‐like” and “mesenchymal versus stem‐like”; Sheffield et al, 2017). Although the clinical relevance of this epigenetic heterogeneity in sarcoma remains to be clarified, recent studies have highlighted the potential of epigenetic therapies in OS and EwS: Selective inhibition of BET bromodomain epigenetic signaling interferes with the bone‐associated tumor's vicious cycle in OS and inhibits the oncogenic transcription factor EWSR1‐FLI1 in EwS (Lamoureux et al, 2014; Jacques et al, 2016; Baud'huin et al, 2017). Super‐enhancers (SEs), which are large genomic regions enriched in active enhancers, have been identified as regulators of cellular identity (Whyte et al, 2013). In pediatric fusion‐positive ARMS, PAX3‐FOXO1 was shown to establish a miswired myoblastic SE landscape, creating a dependency on BET bromodomains (Gryder et al, 2017, 2019, 2020). BET inhibitors ablate PAX3‐FOXO1 function, providing a rationale for their use in the treatment of fusion‐positive ARMS patients (Gryder et al, 2017, 2019, 2020).
Deregulation of epigenetic programs also plays key roles in other sarcoma subtypes, such as SS, an STS that often occurs in young adults. The defining genetic event present in all histological variants of SS is the translocation of the SS18 gene on chromosome 18q11 to an SSX gene (mainly SSX1 or SSX2) located on chrXp11 (Clark et al, 1994). A recent RNA interference screen to find specific epigenetic vulnerabilities created by the SS18‐SSX oncoprotein identified a critical role for KDM2B, a member of the non‐canonical polycomb repressive complex 1 (PRC1.1) in sustaining SS cell proliferation (Banito et al, 2018). PRC1.1 is required for the recruitment of SS18‐SSX and the mSWI/SNF complex to unmethylated CpG islands, which enables the fusion to activate genes that would otherwise be repressed (Banito et al, 2018). In addition, two recent studies found a dependency of SS on the mSWI/SNF subunit BRD9 (Brien et al, 2018; Michel et al, 2018). However, further work should determine whether these results pinpoint a requirement of BRD9 for the SS18‐SSX‐driven expression program (Brien et al, 2018) and whether this constitutes a synthetic lethal interaction by regulation of fusion‐independent genomic sites (Michel et al, 2018).
Apart from their roles in sarcomagenesis, specific epigenetic alterations can be used to improve bone sarcoma and STS classification, diagnosis, and patient stratification (Fig 1; Koelsche et al, 2018a; Weidema et al, 2020). The promising results of brain tumor DNA methylation‐based classification (Capper et al, 2018) fostered adaptation of this principle to the decision‐making process in sarcoma diagnostics, which is often clinically equally challenging (Koelsche et al, 2018a). Analyses of more than 1,000 mesenchymal tumor samples comprising more than 50 STS and bone sarcoma subtypes of pediatric and adult patients by array‐based methylation profiling suggested that methylation signatures can be used to accurately predict sarcoma entities such as “small round blue” cell tumors (Koelsche et al, 2018a). Furthermore, this allows for defining novel subgroups within the sarcoma subtypes, for example, in angiosarcoma (Weidema et al, 2020). Methylation profiling also provides evidence for defining novel entities, such as the recently described primary intracranial sarcoma subtype with highly recurrent DICER1 mutations (Koelsche et al, 2018b). Thus, array‐based DNA methylation analysis will be a major step forward to quickly and reliably discriminate between mesenchymal tumor subtypes, thus increasing diagnostic accuracy. A free access classifier tool currently under development will allow sarcoma subtypes to be predicted using array‐generated DNA methylation data (www.molecularsarcomapathology.org). These molecular signatures will continue to improve the knowledge and classification of mesenchymal tumors, as well as patient outcome through more personalized therapies.
Recent developments in functional assessment of sarcoma biology through imaging
Imaging plays a critical role in the diagnosis, staging, and monitoring of therapeutic response in sarcomas as well as in assessment of recurrence. Routine imaging modalities include plain radiography; despite limitations in contrast resolution, this modality is low cost, widely available, and useful in detecting mineralization and distinguishing ossification from calcification for diagnostic purposes (Kransdorf & Meis, 1993). Computed tomography (CT) is of limited utility in evaluating STSs due to radiation concerns and poor contrast resolution, but the ability to provide three‐dimensional information is mainly exploited to guide biopsy procedures and detect lung metastases (Casali et al, 2018). Magnetic resonance imaging (MRI) is the modality of choice for evaluating sarcomas, given its excellent tissue contrast and lack of ionizing radiations, particularly to determine tumor size and delineation of mass extent and to identify invasion of the compartments and occasionally for histological classification using conventional T1‐weighted, T2‐weighted, and fluid‐sensitive sequences (Fayad et al, 2012).
In addition to these common imaging modalities, novel techniques are emerging for the functional characterization of tumors, including metabolism and the microenvironment, and for a reliable estimation of treatment response by complementing functional assessments with anatomical evaluation. PET, in combination with 18F 2‐fluoro‐2‐deoxy‐D‐glucose (FDG), is a valuable tool for the characterization of cancer metabolism, since the uptake of FDG—a non‐metabolizable derivative of native glucose—correlates with the pathological grade and can be used to discriminate between benign lesions and STSs (Ioannidis & Lau, 2003). Moreover, it can be used to detect metastases for the follow‐up of treatments and to identify the target regions for biopsy (Kubo et al, 2016; Harrison et al, 2017).
Magnetic resonance imaging has taken a lead in the functional characterization of tumors, since it has the capability to provide multiparametric analysis of biological features of sarcoma by exploiting a variety of approaches, including chemical shift imaging (CSI), diffusion‐weighted imaging (DWI), magnetic resonance spectroscopy (MRS), and quantitative dynamic contrast‐enhanced (DCE)‐MRI (Subhawong & Wilky, 2015). DCE‐MRI provides information on tissue vascularization, perfusion, and permeability that can be exploited for differentiating STS from benign soft tissue tumors (Tuncbilek et al, 2005; Pepin et al, in press), or in monitoring tumor response by revealing early perfusion changes (Amit et al, 2014; Crombé et al, 2019), or in cell proliferation assessment (Lee et al, 2020). DWI provides measurements of tissue cellularity and membrane integrity by assessing the Brownian motion of water molecules in tissues. Malignant lesions are usually more cellular than benign lesions, leading to modified Brownian motion (Amit et al, 2014). DWI may be particularly suited for assessing treatment response, with an increase in water diffusion that is usually associated with a positive therapeutic response (Dudeck et al, 2008). MRS can provide the metabolic profile of tumors and is frequently used in sarcoma to evaluate the concentration of the membrane phospholipid choline, which may serve as a marker of malignancy in musculoskeletal STSs (Fayad et al, 2007, 2012). The quantitative parameters of CSI, DWI, MRS, and DCE‐MRI have also shown promising potential as biomarkers for osseous tumors (e.g., differentiation of tumor from edema, determination of biological aggressiveness) (Fukuda et al, 2019).
Tumor acidosis is considered a major player in promoting tumor angiogenesis, progression, invasion, and resistance to chemo‐radiotherapy (Pillai et al, 2019). In OS, the acidic microenvironment strongly affects the activation of MSCs by inducing clonogenicity and invasion, in addition to promoting multidrug resistance (described above) (Avnet et al, 2016, 2017). Indirect measurements of acidic regions in the TME have been obtained in canine OS samples by immunohistochemistry (IHC) analysis (Avnet et al, 2017). Consequently, non‐invasive imaging approaches are needed to provide accurate in vivo measurements of tumor acidosis (Anemone et al, 2019; Consolino et al, 2020). Previous MRS approaches reported intratumoral acidosis in murine fibrosarcoma models, but lacked the ability to assess the spatial distribution (Vaupel et al, 1989, 1994). Recently, a novel MRI‐based approach has been proposed for in vivo imaging of extracellular tumor pH with high accuracy and spatial resolution by exploiting iopamidol, an FDA‐approved X‐ray contrast medium that allows potential clinical translation (Longo et al, 2014; Anemone et al, 2019). Preclinical studies have shown the capability of this pH mapping method to assess the correlation between dysregulated glycolysis and tumor acidosis (Longo et al, 2016) and monitor the treatment response to anticancer therapies targeting glycolysis (Anemone et al, 2017). This novel tumor pH imaging approach may be of particular importance for investigating tumor acidosis in the field of sarcomas.
It is interesting to note that advances in imaging technology have paved the way for imaging modalities that are capable of defining drug response at earlier stages of treatment. As an example, the use of FDG‐PET after 2 weeks of treatment with pazopanib was able to correctly classify 42% of STS patients as non‐responders (Vlenterie et al, 2019).
Novel biomarkers of sarcomas
Traditionally, histomorphological assessment of sarcoma samples in conjunction with clinical and imaging features (See section “Recent developments in functional assessment of sarcoma biology through imaging”) has led to the establishment of diagnosis. In addition, the identification of fusion gene products or overexpressed oncogenes by IHC has enriched the clinical practice (Heymann, 2014; WHO Classification of Tumours: Soft Tissue and Bone Tumours, 2020). However, sarcomas often do not express specific IHC markers. In contrast to studies on tumor biopsies, the discovery of circulating tumor cells (CTCs), cell‐free circulating tumor DNA (cfDNA), and tumor‐derived extracellular vesicles (EVs), as well as the advent of new technologies to detect, quantify, and analyze these biological entities in peripheral blood, hold great promise for developing minimally invasive methods to improve patient care. Indeed, liquid biopsies may enable longitudinal monitoring of treatment response, early detection of relapse, and the identification of druggable driver mutations. Although IHC markers remain important tools for diagnostics in sarcomas (as reviewed in ref. Wei et al, 2017), the aim of this section is to focus on recent advances in the field of liquid biopsies in sarcoma.
Circulating cytokines as markers associated with prognosis
Deregulated levels of cytokines and their receptors can be detected in cancer patients both locally and systemically, and they may be of a high prognostic value in several tumor types (Kumar et al, 1998; Belluco et al, 2000; Kawashima et al, 2000), including sarcomas. Increased serum levels of cytokines and their soluble receptors that are involved in bone degradation (e.g., IL‐6 and IL‐8) and bone formation (e.g., tumor necrosis factor receptor I [TNFRI]) are positively correlated with tumor size and local tumor extent, which is associated with worse overall survival in adult bone sarcoma patients (Rutkowski et al, 2003). Several studies have recognized the negative prognostic significance of various chemokines or cytokines, such as CXCL4/CXCL6 (Li et al, 2011), CXCL10 (Flores et al, 2017), IL‐17A (Wang et al, 2013), IL‐6, IL‐8, and TNF‐α (Xiao et al, 2014) in OS patients. IL‐6 levels were also elevated in serum of a subgroup of EwS patients with poor prognosis (Lissat et al, 2015) and constitute an indicator of poor overall survival and event‐free survival in STS, suggesting a possible association with aggressive tumor behavior (Hagi et al, 2017). Besides IL‐6, other cytokine signaling components including IL‐8, TNF‐R, sIL‐2R, and M‐CSF have been shown to correlate with tumor grade and size in STS patients, and the serum levels of some of these proteins were associated with the prognosis (Rutkowski et al, 2003). To date, the identification of specific cytokine components involved in sarcoma progression is far from being complete, and future studies are essential for generating innovative prognostic tools and facilitating therapy and risk‐stratification.
Extracellular vesicles (EVs) and micro RNAs (miRNAs)
EVs are intercellular messengers where cargo (nucleic acids, proteins, lipids, and metabolites) can be characterized and potentially used as new or supplementary biomarkers in liquid biopsy approaches (Mader & Pantel, 2017). EVs isolated from peripheral blood samples derive not only from tumor cells but also from cells of the TME (See section “The complex biology of sarcoma: How current knowledge may affect therapy”). Thus, EVs can be representative of the interaction between cells in the TME and may bring useful information to follow disease progression (Baglio et al, 2017; Mannerström et al, 2019). One major advantage of EVs in the liquid biopsy approaches is their membranous structure that protects their cargo and gives them enough stability to allow EV sample storage before analysis, which facilitates their clinical use (Jeyaram & Jay, 2017).
In 2013, Miller et al initiated the study of EVs’ diagnostic potential for sarcoma by demonstrating the efficient isolation of EVs derived from EwS and containing EwS‐specific transcripts, including EWSR1‐FLI1, in a pre‐clinical model for patient plasma (Miller et al, 2013). Since then, only few clinical studies have been conducted in limited patient cohorts exploring sarcoma‐derived EVs as biomarkers. Circulating EV‐associated transforming growth factor β (TGF‐β) levels were elevated in OS patients compared with healthy individuals (Baglio et al, 2017), and circulating vesicular miR‐25-3p and miR‐92a-3p were elevated in LPS patients (Casadei et al, 2017). Moreover, miR‐25-3p and miR‐92a-3p modulated macrophages in the local TME, which in turn released IL‐6, increasing the proliferation, migration, and invasion of cancer cells. EVs secreted by dedifferentiated LPSs were also carriers of MDM2 DNA transferable to preadipocytes, which acquired oncogenic properties (e.g., impaired TP53) (Casadei et al, 2019). In addition, miR‐642a, miR‐1260b, and miR‐4286 were significantly higher in serum collected from myxofibrosarcoma patients compared with healthy controls, and miR‐1260b expression was associated with tumor burden and the infiltrative nature of sarcoma (Morita et al, 2020). Moreover, EVs derived from the plasma of GIST patients expressed activated KIT, which was undetectable in samples from healthy donors (Atay & Godwin, 2014). Promising data were also obtained for SS, where serum miR‐92b-3p constituted a robust marker for discriminating patients with SS from other STS patients and was elevated in EVs compared with AGO2‐positive fractions (Uotani et al, 2017). miR‐761 released in EVs enhanced pazopanib resistance in SS (Shiozawa et al, 2018) and correlated with increased resistance. Such resistance may be explained by the modulation of NAD‐dependent protein deacetylase sirtuin‐3 (SIRT3) expression. Interestingly, pazopanib regulated the protein contents of EVs released by SS (Shiozawa et al, 2018), more specifically proteins from the Wnt pathway, which is crucial for SS (Baird et al, 2005). RMS also secreted EVs, which upregulated the proliferation of RMS cells and fibroblasts of the TME, and initiated the migration/invasion of tumor‐associated fibroblasts through promotion of angiogenesis (Ghayad et al, 2016). EVs secreted by cancer cells appeared as key regulators of bone sarcoma biology. A pilot study analyzing RNA isolated from plasma‐derived EVs of OS patients found a higher tumor mutational burden in patients with metastatic disease than in OS patients without metastases (Bao et al, 2018). The response to chemotherapy can be monitored by the identification of dysregulated levels of miRNAs (miR‐124, miR‐133a, miR‐135b, miR‐148a, miR‐199a-3p, miR‐27a, miR‐385, and miR‐9) and mRNAs (ANNEXIN2, CDC5L, CDKN1B, CIP4, MTAP, PEDF, SMAD2, and WWOX) in EVs isolated from the serum of OS patients with a poor chemotherapeutic response when compared with good responders (Xu et al, 2017). However, before being incorporated into routine clinical practice, a careful optimization and standardization of EVs isolation protocols from blood samples and validation studies in larger patient cohorts are required. In particular, the position paper recently published by the International Society for Extracellular Vesicles stresses the importance of a variety of critical parameters (pre‐analytical parameters, such as time to processing, type of container(s), and choice of anticoagulant) (Théry et al, 2018).
Circulating tumor cells (CTCs)
Circulating tumor cells are cells released from primary and metastatic tumor foci and migrating in secondary organs through the peripheral blood. The biological value of CTCs was assessed by comparing the molecular profiles of CTCs and primary tumors (Keller & Pantel, 2019). Controversial conclusions showed that CTCs only partly reflect the spectrum of mutations in the primary and metastatic tumors (Paoletti et al, 2018; Wu et al, 2018; Brown et al, 2019; Keller & Pantel, 2019). CTCs may be considered a snapshot of tumor tissue heterogeneity at a given time and could have strong implications for longitudinal patient monitoring (Brown et al, 2019; Tellez‐Gabriel et al, 2019). In contrast to studies in carcinomas (Pantel & Alix‐Panabières, 2019), studies of CTCs in sarcomas are currently limited (Tellez‐Gabriel et al, 2016). The restricted number of patients, the high heterogeneity of sarcoma subtypes, and the absence of specific markers expressed by most sarcoma cells contribute to the limited advances in this field. Despite the absence of specific markers, various methods of cell isolation based on physical specificity (e.g., higher size and higher cell deformability of tumor cells) or biological properties (e.g., immunomagnetic isolation) have been proposed with success (Gabriel et al, 2016; Hayashi et al, 2017; Li et al, 2017). CTCs are detectable in bone sarcomas (Chinen et al, 2014; Benini et al, 2018) and STS patients (Braun et al, 2018; Mihály et al, 2018; Przybyl et al, 2019). To improve the sensitivity and specificity of detection and isolation of CTCs across sarcoma subtypes, investigators have been looking for universal sarcoma markers (Satelli et al, 2014; Li et al, 2018). Cell‐surface Vimentin was expressed in CTCs isolated from 24 sarcoma patients comprising OS, EwS, angiosarcoma, LMS, and UPS (Satelli et al, 2014). More recently, a new class of CD45− CTCs expressing macrophage markers CD14 and CD68, cell‐surface Vimentin, and specific GIST markers (DOG1 and KIT) have been identified (Li et al, 2018). This CTC subset was more abundant in patients with metastatic disease than with localized GIST. In contrast, cell‐surface Vimentin‐positive cells that did not express macrophage markers failed to predict GIST metastasis (Li et al, 2018). These studies underlined the potential clinical interest in CTCs as prognostic or predictive markers, although longitudinal clinical trials with a large series of patients may be required.
Cell‐free circulating tumor DNA (cfDNA)
cfDNA is composed of DNA fragments released into the bloodstream by healthy and cancer tissues alike, as a result of cell death (e.g., apoptosis, necrosis) or active release (Volckmar et al, 2018; Chen & Zhao, 2019). The cfDNA fraction released from tumor tissues, called circulating tumor DNA (ctDNA), may reflect the genetic aberrations of cancer cells at a given time. cfDNA was recently detected in plasma of bone sarcoma (Gutteridge et al, 2017; Shukla et al, 2017; Barris et al, 2018) and STS patients (Boonstra et al, 2018; Eastley et al, 2018; Namløs et al, 2018; Ogino et al, 2018; Shulman et al, 2018). In these studies, total cfDNA levels were frequently increased in the plasma of sarcoma patients compared with the cancer‐free controls. Cancer‐associated mutations, such as in TP53, PIK3CA, and IDH1 or fusion oncogenes (e.g., SS18‐SSX1/2), were also detected. In patients affected by GIST, mutations of KIT and PDGFRA were detected, and the amount of mutant cfDNA correlated with clinical progression (Maier et al, 2013). Interestingly, the usefulness of cfDNA analysis was demonstrated to identify TKI‐resistant mutations (Yoo et al, 2014). In a series of CHSs, ctDNA levels detected by mutated IDH1 correlated with tumor grade and prognosis (Gutteridge et al, 2017). Patient‐specific somatic alterations in cfDNA were observed in OS (Barris et al, 2018) and were associated with inferior outcomes in EwS and OS patients (Shulman et al, 2018). Individual genomic EWSR1‐ETS fusion sequences can be quantified from cfDNA in EwS patients’ plasma, and as such represent suitable serum markers for therapy assessment (Krumbholz et al, 2016). Indeed, copy numbers of cell‐free EWSR1‐ETS fusion sequences correlate with patients’ risk factors such as tumor volume, pelvic tumor, and metastatic status, and most EwS patients show a fast reduction of cfDNA levels during treatment, while recurrence of increasing cfDNA levels indicates relapse (Krumbholz et al, 2016). In addition to somatic mutations and DNA methylation, recent studies have reported the detection of circulating nucleosomes in blood, showing that cfDNA retains at least some features of nuclear chromatin. Most importantly, whole‐genome sequencing of cfDNA was shown to yield a dense, genome‐wide map of nucleosome occupancy that enables identification of the cell types that contribute to circulating cfDNA (Snyder et al, 2016; Ulz et al, 2016). This is highly relevant to EwS as it supports the idea of monitoring the chromatin state of EwS‐specific enhancer elements (Riggi et al, 2014; Tomazou et al, 2015; Sheffield et al, 2017) over time and during the treatment course, enabling the development of enhancer‐based minimally invasive assays for live monitoring of therapy response.
Overall, the detection and characterization of cfDNA and ctDNA in sarcomas show promising results, and efforts are now needed to profile larger biological cohorts with complete clinical annotations to validate their clinical value.
Recent therapeutic developments
Precision medicine in sarcoma: General considerations
The ultimate goal of personalized medicine is to be able to integrate clinical, genomic, transcriptomic, and epigenomic data to increase the accuracy of diagnosis and prognosis, and to identify the most effective therapy for treatment (Burdach et al, 2018; Salgado et al, 2018; Gargallo et al, 2020). Recent advances in machine learning‐based methods for analysis of histology and radiography imaging may also play an increasingly important role (Blackledge et al, 2019; Wang et al, 2019; Malinauskaite et al, 2020). For instance, clinical investigations into immune checkpoint therapy have designated UPS, myxofibrosarcoma, and similar genomically complex histotypes as “UPS” (Que et al, 2017), making comparisons with other studies difficult. However, the inclusion of genomic analyses led to the re‐classification of 13% of sarcoma cases and would have resulted in changes to the clinical treatment pathway or prognosis in 11% of cases, demonstrating the importance of including molecular and computational tools for classification and risk‐stratification of sarcomas (Italiano et al, 2016).
Several recent studies have identified therapeutically targetable mutations in sarcoma patients and have used this knowledge to guide treatment (Groisberg et al, 2017). Yet, not all attempts were successful (Demetri et al, 2013; Perry et al, 2014), indicating that genomic data alone are not sufficient for the accurate prediction of response to therapy.
The clinical trial MULTISARC (ClinicalTrials.gov No. NCT03784014) should provide the first glimpse into the successes and potential pitfalls of personalized medicine in sarcoma. Based on a retrospective survey of genomic alterations that could be therapeutically actionable (Lucchesi et al, 2018), MULTISARC is a two‐arm, randomized trial aiming to prospectively evaluate their potential as predicative biomarkers for response to therapy. STS patients will be randomized to receive standard therapy or undergo genomic profiling for suitability for therapy with 16 different agents. Sarcomas were identified as a priority for the 100,000 genomes project in the United Kingdom with 500 to be sequenced as part of the study, although it will focus on LMS, myxofibrosarcoma, SS, and rare histotypes such as alveolar soft part sarcoma (ASPS). In addition to collecting both genomic and clinical data from patients, the project's Genomics England Clinical Interpretation Partnerships (GeCIPs), including the Sarcoma GeCIP, will also identify training and standardization of practice needed to bring personalized medicine toward routine clinical practice.
Likewise, genomic analyses in combination with screening cancer cell lines against libraries of drugs have the potential to improve the correlation between genomic biomarkers and response to therapy. Such an approach has been used to identify biomarkers for response to therapy of several sarcomas using cell lines, patient‐derived samples, and canine sarcoma as proof of principle (Berlow et al, 2019). This approach is challenging for studying sarcoma, due to the limited number of cell lines available, although isolation of new cell lines (Salawu et al, 2016) and sarcoma PDX models is improving (Stebbing et al, 2014). The next step will be to take advantage of combining molecular information gained through next‐generation sequencing (NGS) technologies with functional drug screening using primary organoid cultures that include both stromal cells and cancerous cells to improve prediction of response to therapy, as observed in other cancers (Tiriac et al, 2018; Vlachogiannis et al, 2018).
Photodynamic therapy
An interesting approach, based on photo‐ and radiodynamic therapy following acridine orange administration, has been extensively investigated and successfully applied for the treatment of sarcomas (Matsubara et al, 2013; Kusuzaki et al, 2018; Martano et al, 2019). Photodynamic therapy with hematoporphyrin prevented local recurrence following minimally invasive surgery in preclinical models (Duchi et al, 2016) as well as in clinical settings (Hourigan et al, 1993). Acridine orange has the advantage of selectively binding to tumor tissue due to the acidic microenvironment specific to malignant cells (Matsubara et al, 2006) and to specifically exert a strong cytotoxic activity on tumor cells, which is further enhanced by photo‐ and radioactivation (Matsubara et al, 2013; Kusuzaki et al, 2018). Therefore, following marginal or even intralesional gross removal of the tumor, it is possible to selectively target residual sarcoma and spare the surrounding normal tissues, with a satisfactory functional result (Martano et al, 2019). The procedure is safe, without local or systemic complications (Martano et al, 2019). Systemic administration of acridine orange with low‐dose radiation therapy is currently under evaluation in Japan for non‐resectable sarcomas (Kusuzaki et al, 2018). This procedure appears to be safe, and the preliminary results are encouraging.
Immune‐based therapies
Sarcomas are highly heterogeneous, including the TME, which might dictate their heterogeneous response to different immunotherapeutic approaches (section “The complex biology of sarcoma: How current knowledge may affect therapy”, Figs 2 and 3). While checkpoint inhibitor immunotherapies have already been introduced for the first‐/second‐line treatment of several carcinomas, their efficacy in sarcoma treatment is currently unclear, and clinical trials are ongoing (Thanindratarn et al, 2019). Unfortunately, the first results showed only sporadic therapeutic responses in STSs and bone sarcomas, highlighting the need for further investigations (Merchant et al, 2016).
Some STS subtypes (e.g., myxofibrosarcoma and UPS) are characterized by a high mutational burden, which may constitute a biomarker for response to immune checkpoint blockade (Pollack et al, 2017). In addition, recent profiling studies of immune checkpoints expression in STSs and bone sarcomas revealed their correlation with poor clinical outcomes and provide rationales for their targeting (Dancsok et al, 2019; Orth et al, 2020). In fact, a new study revealed a positive correlation between immune infiltration and response to anti‐PD‐L1 therapy in sarcoma (Keung et al, 2020). Similarly, a gene expression study in 608 tumors across STS subtypes established a classification between immune‐low, immune‐high, and vascularized phenotypes (Petitprez et al, 2020). The phenotype with the highest immune cell infiltration featured tertiary lymphoid structures with T cells, dendritic cells, and B cells. Interestingly, B cells were the strongest prognostic factor, and they were associated with improved survival and high response rates to PD‐1 blockade (Petitprez et al, 2020).
Therapeutic strategies based on (genetically modified) T cells are currently underway. Their main objectives are to enhance T‐cell infiltration into tumor tissues and identify specific tumor target antigens only expressed by malignant cells (Baldauf et al, 2018b). Some encouraging results have been described, such as the therapeutic benefit observed in SS upon inoculation of autologous T cells engineered to express an affinity‐enhanced T‐cell receptor (TCR) recognizing the NY‐ESO‐1‐derived peptide (D'Angelo et al, 2018). Similarly, chimeric antigen receptor (CAR) T cells characterized by the expression of a chimeric receptor (fusion of specific antibody‐derived single‐chain variable fragments with the signaling domain of a T‐cell receptor) are capable of inducing conventional activation signals from TCRs in a non‐MHC restricted manner (Majzner & Mackall, 2018; Pollack et al, 2018). Although some sarcomas subtypes express tumor epitopes, such as HER2, GD2, ROR2, or EGFRvIII, B7‐H3 (Majzner et al, 2019), or oncofetal glycosaminoglycans (Salanti et al, 2015), these tumor epitopes are often only expressed at low levels. CAR T cells may overcome the low levels of tumor antigen expression, and several clinical trials are currently in progress to evaluate their therapeutic benefit (Majzner & Mackall, 2018; Pollack et al, 2018). Interestingly, a first completed phase I/II trial with HER2‐CAR T cells showed that cells can persist for 6 weeks without evident toxicities, setting the stage for studies that combine CAR T cells with other immunomodulatory approaches to enhance their expansion and persistence (ClinicalTrials.gov No. NCT00902044; Ahmed et al, 2015). OS (Théoleyre et al, 2005; Koirala et al, 2016), EwS (Machado et al, 2018), and CHS (Simard et al, 2017; Richert et al, 2019) are moderately infiltrated by lymphocytes with moderate functional impact (Heymann et al, in press). However, the number of T lymphocytes appeared to be significantly higher in metastatic foci than in primary tumors and in local relapses, suggesting the potential benefit of TIL‐based immunotherapy in metastatic clinical situation (Sundara et al, 2017; Shi et al, 2020). T lymphocyte infiltration has also been described in STS (Dancsok et al, 2019; Que et al, 2019; Shi et al, 2020). Two phase 2 clinical trials have recently been set up for treating sarcoma patients with autologous TIL expanded ex vivo (ClinicalTrials.gov No. NCT03449108 & NCT03935893). Similarly, adoptive immune cell therapy options based on infusion of NK cells were assessed in preclinical models of bone sarcomas and STS (Thiel et al, 2013; Fernández et al, 2015). Case reports including ERMS and EwS showed a beneficial anti‐tumor activity of allogeneic hematopoietic stem cell transplantation (Pérez‐Martínez et al, 2009). A pilot phase 1/2 clinical study named “NKEXPSARC” will assess the clinical potential of activated haploidentical natural killer cell infusions in sarcomas (ClinicalTrial.gov No. NCT02409576).
Oncolytic viruses
The approval of the Herpes virus Talimogene Laherparepvec (T‐VEC; Imlygic) by the FDA and EMA for recurrent melanoma confirms that virotherapy has emerged as a feasible therapeutic strategy in oncology (Andtbacka et al, 2015; Ribas et al, 2017). Oncolytic viruses have been assessed in bone sarcomas and STSs (Lacroix et al, 2018; MacNeill et al, 2018; Smith et al, 2019; Tazawa et al, 2020). They are tumor selective, destroy cancer cells, and trigger an anti‐tumor immune response (Garcia‐Moure et al, 2017; Varela‐Guruceaga et al, 2018). Table 2 summarizes the main potential therapeutic viruses for the treatment of sarcomas.
Table 2.
Summary of main oncolytic viruses applied in sarcoma treatment
| Virus | Disease | Trial |
|---|---|---|
| DNA | ||
| Adenovirus (Ad) | Respiratory and gastrointestinal infections | Preclinicalphase I |
| Herpes simplex virus (HSV) | Oral and genital ulcerations | Preclinicalphase I |
| Vaccinia virus | Flu | Preclinical |
| RNA | ||
| Reovirus | Respiratory and gastrointestinal infections | Preclinicalphase I |
| Semliki forest virus (SFV) | Non‐pathogenic in humans / encephalitis in mice | Preclinical |
| Vesicular stomatitis virus (VSV) | Non‐pathogenic | Preclinical |
| Measles virus (MeV) | Measles | Preclinical |
| Poliovirus | Neurological disorders (poliomyelitis) | Preclinical |
| Newcastle disease virus (NDV | Respiratory and gastrointestinal infections | Preclinicalphase I/II |
In the group of DNA viruses, Adenovirus, Herpes virus, and Vaccinia virus are commonly employed. These three types of viruses have advanced to clinical trials. For example, Telomelysin, a human telomerase reverse transcriptase (hTERT) promoter‐driven modified oncolytic Adenovirus, was tested in a phase I clinical trial to assess its clinical safety in patients with advanced solid tumors (Nemunaitis et al, 2010). Herpes virus HSV1716 was tested in pediatric patients with non‐central nervous system solid tumors (ClinicalTrials.gov No. NCT00931931), including two patients with OS (Streby et al, 2017). This virus was delivered as a single dose of 105–107 infectious units via CT‐guided intratumoral injection, and tumor response was measured by imaging. HSV1716 was safe in the pediatric population, with minimal toxicities reported; however, no clinical responses were observed in this phase I trial (Streby et al, 2017). Finally, the Vaccinia virus, armed with GM‐CSF (JX‐594), has also been tested in a phase I clinical trial in pediatric solid tumors (ClinicalTrials.gov No. NTC01169584) but did not include sarcomas. This virus did not show toxicity, but exhibited biological activity in the pediatric population (Cripe et al, 2015). The group of RNA viruses, including Semliki Forest Virus, Poliovirus, Newcastle Disease Virus, Measles, or Reovirus (Table 2), have also transitioned to clinical trials (Schneider et al, 2018). However, only the Reovirus Reolysin has been tested in OS (Kolb et al, 2015). Twenty‐four patients were treated in this trial, including OS and other extracranial pediatric tumors, to establish virus safety. The virus was well tolerated and showed a safe profile, but no response was observed (Kolb et al, 2015).
The therapeutic effect of several oncolytic viruses in STSs (Leddon et al, 2015; Siurala et al, 2015; Wilkinson et al, 2016; Chen et al, 2017) and bone sarcomas (Witlox et al, 2004; Graat et al, 2006; Hingorani et al, 2014; Martínez‐Vélez et al, 2016; Martinez‐Velez et al, 2014) was tested in various preclinical studies. Due to their versatility and lack of toxicity, oncolytic Adenoviruses are commonly used (Fig 4). Because the Rb pathway is frequently mutated in sarcomas, oncolytic Adenoviruses based on selective replication conditional to Rb pathway deregulation have been developed. VCN‐01 (Martínez‐Vélez et al, 2016) and Delta‐24-RGD (Martinez‐Velez et al, 2014) are Adenoviruses that showed in vitro and in vivo anti‐sarcoma activity. Delta‐24-RGD is a replication‐competent Adenovirus that harbors a 24‐bp deletion in the E1A region (responsible for binding Rb protein) that triggers tumor selectivity. The addition of an RGD‐4C motif in the fiber H1 loop allows enhanced infectivity through integrins that are widely expressed in cancer cells (Suzuki et al, 2001). VCN‐01 is an oncolytic Adenovirus where the E1A gene also contains deletions in the pRb binding site, thus rendering its selective replication in Rb‐deficient tumor cells (Rodríguez‐García et al, 2015). Importantly, both viruses have shown efficacy not only against the primary tumor but also against lung metastases (Martinez‐Velez et al, 2014; Martínez‐Vélez et al, 2016). It should be noted that most of the oncolytic Adenoviruses are amenable to be used in combination with standard chemotherapy, small molecules, nanoparticles, immunotherapy with immune checkpoint inhibitors, and CAR T cells.
Figure 4. Main features and functional aspects of oncolytic virus.
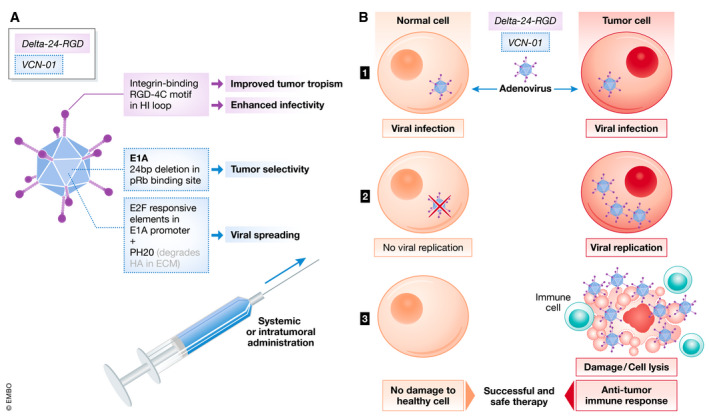
(A) Characteristics of oncolytic Adenoviruses Delta‐24-RGD and VCN‐01. These two Adenoviruses harbor different modifications (black for Delta‐24-RG (D24‐RGD) and dashed blue for VCN‐01) that render them with tumor specificity and enhanced infectivity. (B) Schematic representation of the virus’ mechanism of action. (1) The viruses are able to infect both normal and tumor cells. (2) However, due to their tumor specificity they only replicate and lyse the tumor cells. (3) They exert a potent cytolytic effect, and they are able to trigger an anti‐tumor immune response, which is crucial to successfully eliminate the tumors.
Conclusions
Sarcomas comprise relatively rare but diverse cancer entities affecting patients of all ages. Bone sarcomas are more frequent in adolescents and young adults, and the frequency of STS increases with age. Most sarcomas exhibit a high cellular, molecular, and genetic/epigenetic heterogeneity, which makes identification of single therapeutic targets more difficult. Fortunately, in some instances, identification of new targets has revolutionized the therapeutic management of sarcoma patients, as illustrated by the use of imatinib mesylate targeting receptor tyrosine kinases in GIST even if secondary resistance is observed (Napolitano & Vincenzi, 2019), which can be overcome with other, rationally designed TKIs (Blay et al, 2020). The TME plays a key role in the pathogenesis of sarcomas, not only for tumor initiation but also in the metastatic process. Like other cancers, sarcomas are now in the era of immunotherapy (e.g., PD‐L1 inhibitors, CAR T‐cell therapy) and numerous clinical trials are currently ongoing. Epigenetic profiles emerge as useful tools to improve diagnostic accuracy in sarcomas and to discover or better delineate new sarcoma subtypes. In addition, epigenetic events occurring during sarcomagenesis have been identified as new, promising opportunities for treating sarcomas. Innate or acquired resistances of sarcomas are the principal obstacles to treatment efficacy, and a better understanding of these cellular/molecular processes will help to define better therapeutic lines. Tackling MDR, CSCs, and/or cell dormancy are all tracks for progress. Finally, the high heterogeneity of sarcoma requires better classification of sarcoma subtypes based on (epi)genetic characteristics (e.g., CTCs, circulating RNA/DNA, immune infiltrates) to identify the best therapeutic option for each patient. Thus, future advances in the field of molecular biology related to sarcomas hold great promise to overcome treatment resistance and treatment‐related toxicity through individualized precision medicine approaches.
Conflict of interest
Marta Alonso has obtained research grant from DNAtrix. Stefan Budach has an ownership interest in PDL BioPharma and has had US and EU intellectual properties in gene expression analysis. He served as consultant to EOS Biotechnology Inc. and serves as advisor to Bayer AG and Swedish Orphan Biovitrum AB. Dominique Heymann has an ownership interest in Atlanta SAS (Saint‐Herblain, France).
Pending issues.
-
•
Identification of unknown extrinsic factors that may have a role in sarcoma progression and response to therapy and that may derive from the following: (i) the tumor microbiome, (ii) immune infiltrates, and (iii) other cells of the tumor‐associated stroma (including neurons).
-
•
Development of novel and more representative 3D preclinical models to be used in place of animal models to develop new therapeutic options.
-
•
Further generation of immunocompetent and bona fide GEMMs for all sarcoma subtypes for a better understanding of sarcomagenesis.
-
•
Elucidation of the mechanisms that lead to resistance toward TKIs in non‐GIST STS.
-
•
Elaboration of non‐invasive assays for the monitoring of drug response and for early detection of drug resistance.
-
•
Development of compounds that enhance tumor antigen presentation and of therapeutic protocols based on immunotherapies for the treatment of sarcoma.
-
•
Investigation of the use of photodynamic therapies for limb‐preserving surgery.
-
•
Optimization and clinical translation of oncolytic virus therapies for sarcomas.
For more information
Societies and Network for health scientists and professionals
-
•
EuSARC (European network for SARComa): https://eusarc.com/
-
•
NIH website for information to health professional, related to bone cancer: https://www.cancer.gov/types/bone/hp
-
•
World sarcoma network: http://www.worldsarcomanetwork.com/
Patient associations
-
•
UK patient association on sarcoma: https://sarcoma.org.uk/about-sarcoma/understanding-sarcoma-0
-
•
The Liddy Shriver sarcoma initiative: http://sarcomahelp.org/sarcoma-centers.html#tpm1_1
-
•
Sarcoma patients Euronet: https://www.sarcoma-patients.eu/it/sarcoma-research/research-networks
OMIM site
-
•
Ewing sarcoma: https://www.omim.org/entry/612219?search=sarcoma&highlight=sarcoma
-
•
GastroIntestinal Stromal Tumor: https://www.omim.org/entry/606764?search=GIST&highlight=gist
-
•
Kaposi sarcoma: https://www.omim.org/entry/148000?search=sarcoma&highlight=sarcoma
-
•
Osteosarcoma: https://www.omim.org/entry/259500?search=osteosarcoma&highlight=osteosarcoma
-
•
Synovial sarcoma: https://www.omim.org/entry/300813?search=sarcoma&highlight=sarcoma
Database
-
•
Surveillance, Epidemiology, and End Results (SEER) database: https://seer.cancer.gov/statfacts/html/bones.html
-
•
National Program of Cancer Registries (NPCR): https://www.cdc.gov/cancer/npcr/index.htm
-
•
National Cancer Database (NCDB): https://www.facs.org/quality-programs/cancer/ncdb
-
•
ClinicalTrials.gov: https://clinicaltrials.gov/
Reference book
-
•
WHO Classification of Tumours, 5th Edition, Volume 3. Soft Tissue and Bone Tumours WHO Classification of Tumours Editorial Board. IARC publication Ed. (Lyon, FR) 2020: https://www.iarc.fr/news-events/publication-of-the-who-classification-of-tumours-5th-edition-volume-3-soft-tissue-and-bone-tumours/
Diagnostic sarcoma classifier
-
•
DNA methylation‐based classification: https://www.molecularsarcomapathology.org
Acknowledgments
The authors thank the European EuSARC initiative, which addresses the clinical challenges posed by sarcomas and aims at accelerating the translation of new molecular findings into clinical practice through the organization of annual conferences that foster interdisciplinary and international collaboration in the field of sarcomas (www.eusarc.com).
EMBO Mol Med (2020) 12: e11131
See the Glossary for abbreviations used in this article.
Contributor Information
Thomas GP Grünewald, Email: t.gruenewald@dkfz-heidelberg.de.
Nicola Baldini, Email: nicola.baldini@ior.it.
Dominique Heymann, Email: dominique.heymann@univ-nantes.fr.
References
- Abarrategi A, Tornin J, Martinez‐Cruzado L, Hamilton A, Martinez‐Campos E, Rodrigo JP, González MV, Baldini N, Garcia‐Castro J, Rodriguez R (2016) Osteosarcoma: cells‐of‐origin, cancer stem cells, and targeted therapies. Stem Cells Int 2016: 3631764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed N, Brawley VS, Hegde M, Robertson C, Ghazi A, Gerken C, Liu E, Dakhova O, Ashoori A, Corder A et al (2015) Human epidermal growth factor receptor 2 (HER2) ‐specific chimeric antigen receptor‐modified T cells for the immunotherapy of HER2‐positive sarcoma. J Clin Oncol 33: 1688–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alamanda VK, Crosby SN, Archer KR, Song Y, Schwartz HS, Holt GE (2012) Amputation for extremity soft tissue sarcoma does not increase overall survival: a retrospective cohort study. Eur J Surg Oncol 38: 1178–1183 [DOI] [PubMed] [Google Scholar]
- Alfranca A, Martinez‐Cruzado L, Tornin J, Abarrategi A, Amaral T, de Alava E, Menendez P, Garcia‐Castro J, Rodriguez R (2015) Bone microenvironment signals in osteosarcoma development. Cell Mol Life Sci 72: 3097–3113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali NM, Niada S, Brini AT, Morris MR, Kurusamy S, Alholle A, Huen D, Antonescu CR, Tirode F, Sumathi V et al (2019) Genomic and transcriptomic characterisation of undifferentiated pleomorphic sarcoma of bone. J Pathol 247: 166–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almazán‐Moga A, Zarzosa P, Vidal I, Molist C, Giralt I, Navarro N, Soriano A, Segura MF, Alfranca A, Garcia‐Castro J et al (2017) Hedgehog pathway inhibition hampers sphere and holoclone formation in rhabdomyosarcoma. Stem Cells Int 2017: 7507380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almog N, Henke V, Flores L, Hlatky L, Kung AL, Wright RD, Berger R, Hutchinson L, Naumov GN, Bender E et al (2006) Prolonged dormancy of human liposarcoma is associated with impaired tumor angiogenesis. FASEB J 20: 947–949 [DOI] [PubMed] [Google Scholar]
- Amit P, Patro DK, Basu D, Elangovan S, Parathasarathy V (2014) Role of dynamic MRI and clinical assessment in predicting histologic response to neoadjuvant chemotherapy in bone sarcomas. Am J Clin Oncol 37: 384–390 [DOI] [PubMed] [Google Scholar]
- de Andrea CE, Zhu J‐F, Jin H, Bovée JVMG, Jones KB (2015) Cell cycle deregulation and mosaic loss of Ext1 drive peripheral chondrosarcomagenesis in the mouse and reveal an intrinsic cilia deficiency. J Pathol 236: 210–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andtbacka RHI, Kaufman HL, Collichio F, Amatruda T, Senzer N, Chesney J, Delman KA, Spitler LE, Puzanov I, Agarwala SS et al (2015) Talimogene laherparepvec improves durable response rate in patients with advanced melanoma. J Clin Oncol 33: 2780–2788 [DOI] [PubMed] [Google Scholar]
- Anemone A, Consolino L, Conti L, Reineri F, Cavallo F, Aime S, Longo DL (2017) In vivo evaluation of tumour acidosis for assessing the early metabolic response and onset of resistance to dichloroacetate by using magnetic resonance pH imaging. Int J Oncol 51: 498–506 [DOI] [PubMed] [Google Scholar]
- Anemone A, Consolino L, Arena F, Capozza M, Longo DL (2019) Imaging tumor acidosis: a survey of the available techniques for mapping in vivo tumor pH. Cancer Metastasis Rev 38: 25–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlt MJE, Banke IJ, Bertz J, Ram Kumar RM, Muff R, Born W, Fuchs B (2013) Reduced latency in the metastatic niche contributes to the more aggressive phenotype of LM8 compared to dunn osteosarcoma cells. Sarcoma 2013: 404962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atay S, Godwin AK (2014) Tumor‐derived exosomes: a message delivery system for tumor progression. Commun Integr Biol 7: e28231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avnet S, Longhi A, Salerno M, Halleen JM, Perut F, Granchi D, Ferrari S, Bertoni F, Giunti A, Baldini N (2008) Increased osteoclast activity is associated with aggressiveness of osteosarcoma. Int J Oncol 33: 1231–1238 [PubMed] [Google Scholar]
- Avnet S, Cortini M (2016) Role of pericellular matrix in the regulation of cancer stemness. Stem Cell Rev Rep 12: 464–475 [DOI] [PubMed] [Google Scholar]
- Avnet S, Lemma S, Cortini M, Pellegrini P, Perut F, Zini N, Kusuzaki K, Chano T, Grisendi G, Dominici M et al (2016) Altered pH gradient at the plasma membrane of osteosarcoma cells is a key mechanism of drug resistance. Oncotarget 7: 63408–63423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avril P, Duteille F, Ridel P, Heymann M‐F, De Pinieux G, Rédini F, Blanchard F, Heymann D, Trichet V, Perrot P (2016b) Opposite effects of soluble factors secreted by adipose tissue on proliferating and quiescent osteosarcoma cells. Plast Reconstr Surg 137: 865–875 [DOI] [PubMed] [Google Scholar]
- Avnet S, Di Pompo G, Chano T, Errani C, Ibrahim‐Hashim A, Gillies RJ, Donati DM, Baldini N (2017) Cancer‐associated mesenchymal stroma fosters the stemness of osteosarcoma cells in response to intratumoral acidosis via NF‐κB activation. Int J Cancer 140: 1331–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avnet S, Lemma S, Cortini M, Di Pompo G, Perut F, Baldini N (2019) Pre‐clinical models for studying the interaction between mesenchymal stromal cells and cancer cells and the induction of stemness. Front Oncol 9: 305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avril P, Le Nail L‐R, Brennan MÁ, Rosset P, De Pinieux G, Layrolle P, Heymann D, Perrot P, Trichet V (2016a) Mesenchymal stem cells increase proliferation but do not change quiescent state of osteosarcoma cells: potential implications according to the tumor resection status. J Bone Oncol 5: 5–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baglio SR, Lagerweij T, Pérez‐Lanzón M, Ho XD, Léveillé N, Melo SA, Cleton‐Jansen A‐M, Jordanova ES, Roncuzzi L, Greco M et al (2017) Blocking tumor‐educated msc paracrine activity halts osteosarcoma progression. Clin Cancer Res 23: 3721–3733 [DOI] [PubMed] [Google Scholar]
- Bailey K, Cost C, Davis I, Glade‐Bender J, Grohar P, Houghton P, Isakoff M, Stewart E, Laack N, Yustein J et al (2019) Emerging novel agents for patients with advanced Ewing sarcoma: a report from the Children's Oncology Group (COG) New Agents for Ewing Sarcoma Task Force, F1000Res 8: F1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird K, Davis S, Antonescu CR, Harper UL, Walker RL, Chen Y, Glatfelter AA, Duray PH, Meltzer PS (2005) Gene expression profiling of human sarcomas: insights into sarcoma biology. Cancer Res 65: 9226–9235 [DOI] [PubMed] [Google Scholar]
- Baldauf MC, Gerke JS, Orth MF, Dallmayer M, Baumhoer D, de Alava E, Hartmann W, Kirchner T, Grünewald TGP (2018a) Are EWSR1‐NFATc2‐positive sarcomas really Ewing sarcomas? Mod Pathol 31: 997–999 [DOI] [PubMed] [Google Scholar]
- Baldauf MC, Gerke JS, Kirschner A, Blaeschke F, Effenberger M, Schober K, Rubio RA, Kanaseki T, Kiran MM, Dallmayer M et al (2018b) Systematic identification of cancer‐specific MHC‐binding peptides with RAVEN. Oncoimmunology 7: e1481558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldini N, Scotlandi K, Barbanti‐Bròdano G, Manara MC, Maurici D, Bacci G, Bertoni F, Picci P, Sottili S, Campanacci M (1995) Expression of P‐glycoprotein in high‐grade osteosarcomas in relation to clinical outcome. N Engl J Med 333: 1380–1385 [DOI] [PubMed] [Google Scholar]
- Baldini N (1997) Multidrug resistance–a multiplex phenomenon. Nat Med 3: 378–380 [DOI] [PubMed] [Google Scholar]
- Banito A, Li X, Laporte AN, Roe J‐S, Sanchez‐Vega F, Huang C‐H, Dancsok AR, Hatzi K, Chen C‐C, Tschaharganeh DF et al (2018) The SS18‐SSX oncoprotein hijacks KDM2B‐PRC1.1 to drive synovial sarcoma. Cancer Cell 33: 527–541.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Q, Gong L, Wang J, Wen J, Shen Y, Zhang W (2018) Extracellular vesicle RNA sequencing reveals dramatic transcriptomic alterations between metastatic and primary osteosarcoma in a liquid biopsy approach. Ann Surg Oncol 25: 2642–2651 [DOI] [PubMed] [Google Scholar]
- Barris DM, Weiner SB, Dubin RA, Fremed M, Zhang X, Piperdi S, Zhang W, Maqbool S, Gill J, Roth M et al (2018) Detection of circulating tumor DNA in patients with osteosarcoma. Oncotarget 9: 12695–12704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barron JA, Wu P, Ladouceur HD, Ringeisen BR (2004) Biological laser printing: a novel technique for creating heterogeneous 3‐dimensional cell patterns. Biomed Microdevices 6: 139–147 [DOI] [PubMed] [Google Scholar]
- Barron JA, Krizman DB, Ringeisen BR (2005) Laser printing of single cells: statistical analysis, cell viability, and stress. Ann Biomed Eng 33: 121–130 [DOI] [PubMed] [Google Scholar]
- Basu‐Roy U, Seo E, Ramanathapuram L, Rapp TB, Perry JA, Orkin SH, Mansukhani A, Basilico C (2012) Sox2 maintains self renewal of tumor‐initiating cells in osteosarcomas. Oncogene 31: 2270–2282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu‐Roy U, Bayin NS, Rattanakorn K, Han E, Placantonakis DG, Mansukhani A, Basilico C (2015) Sox2 antagonizes the Hippo pathway to maintain stemness in cancer cells. Nat Commun 6: 6411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu‐Roy U, Han E, Rattanakorn K, Gadi A, Verma N, Maurizi G, Gunaratne PH, Coarfa C, Kennedy OD, Garabedian MJ et al (2016) PPARγ agonists promote differentiation of cancer stem cells by restraining YAP transcriptional activity. Oncotarget 7: 60954–60970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud'huin M, Lamoureux F, Jacques C, Rodriguez Calleja L, Quillard T, Charrier C, Amiaud J, Berreur M, Brounais‐LeRoyer B, Owen R et al (2017) Inhibition of BET proteins and epigenetic signaling as a potential treatment for osteoporosis. Bone 94: 10–21 [DOI] [PubMed] [Google Scholar]
- Baumhoer D, Amary F, Flanagan AM (2019) An update of molecular pathology of bone tumors. Lessons learned from investigating samples by next generation sequencing. Genes Chromosom Cancer 58: 88–99 [DOI] [PubMed] [Google Scholar]
- Beck O, Paret C, Russo A, Burhenne J, Fresnais M, Steimel K, Seidmann L, Wagner D‐C, Vewinger N, Lehmann N et al (2020) Safety and activity of the combination of ceritinib and dasatinib in osteosarcoma. Cancers (Basel) 12: 793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behjati S, Tarpey PS, Presneau N, Scheipl S, Pillay N, Van Loo P, Wedge DC, Cooke SL, Gundem G, Davies H et al (2013) Distinct H3F3A and H3F3B driver mutations define chondroblastoma and giant cell tumor of bone. Nat Genet 45: 1479–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behjati S, Tarpey PS, Haase K, Ye H, Young MD, Alexandrov LB, Farndon SJ, Collord G, Wedge DC, Martincorena I et al (2017) Recurrent mutation of IGF signalling genes and distinct patterns of genomic rearrangement in osteosarcoma. Nat Commun 8: 15936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belluco C, Nitti D, Frantz M, Toppan P, Basso D, Plebani M, Lise M, Jessup JM (2000) Interleukin‐6 blood level is associated with circulating carcinoembryonic antigen and prognosis in patients with colorectal cancer. Ann Surg Oncol 7: 133–138 [DOI] [PubMed] [Google Scholar]
- Ben‐David U, Siranosian B, Ha G, Tang H, Oren Y, Hinohara K, Strathdee CA, Dempster J, Lyons NJ, Burns R et al (2018) Genetic and transcriptional evolution alters cancer cell line drug response. Nature 560: 325–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benini S, Gamberi G, Cocchi S, Garbetta J, Alberti L, Righi A, Gambarotti M, Picci P, Ferrari S (2018) Detection of circulating tumor cells in liquid biopsy from Ewing sarcoma patients. Cancer Manag Res 10: 49–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlow NE, Rikhi R, Geltzeiler M, Abraham J, Svalina MN, Davis LE, Wise E, Mancini M, Noujaim J, Mansoor A et al (2019) Probabilistic modeling of personalized drug combinations from integrated chemical screen and molecular data in sarcoma. BMC Cancer 19: 593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessen T, Caughey GE, Shakib S, Potter JA, Reid J, Farshid G, Roder D, Neuhaus SJ (2019) A population‐based study of soft tissue sarcoma incidence and survival in Australia: an analysis of 26,970 cases. Cancer Epidemiol 63: 101590 [DOI] [PubMed] [Google Scholar]
- Blackledge MD, Winfield JM, Miah A, Strauss D, Thway K, Morgan VA, Collins DJ, Koh D‐M, Leach MO, Messiou C (2019) Supervised machine‐learning enables segmentation and evaluation of heterogeneous post‐treatment changes in multi‐parametric MRI of soft‐tissue sarcoma. Front Oncol 9: 941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakely AM, McPhillips J, Miner TJ (2015) Surgical palliation for malignant disease requiring locoregional control. Ann Palliat Med 4: 48–53 [DOI] [PubMed] [Google Scholar]
- Blay J‐Y (2011) A decade of tyrosine kinase inhibitor therapy: historical and current perspectives on targeted therapy for GIST. Cancer Treat Rev 37: 373–384 [DOI] [PubMed] [Google Scholar]
- Blay J‐Y, Soibinet P, Penel N, Bompas E, Duffaud F, Stoeckle E, Mir O, Adam J, Chevreau C, Bonvalot S et al (2017) Improved survival using specialized multidisciplinary board in sarcoma patients. Ann Oncol 28: 2852–2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blay J‐Y, Serrano C, Heinrich MC, Zalcberg J, Bauer S, Gelderblom H, Schöffski P, Jones RL, Attia S, D'Amato G et al (2020) Ripretinib in patients with advanced gastrointestinal stromal tumours (INVICTUS): a double‐blind, randomised, placebo‐controlled, phase 3 trial. Lancet Oncol 21: 923–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonuccelli G, Avnet S, Grisendi G, Salerno M, Granchi D, Dominici M, Kusuzaki K, Baldini N (2014) Role of mesenchymal stem cells in osteosarcoma and metabolic reprogramming of tumor cells. Oncotarget 5: 7575–7588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonstra PA, Ter Elst A, Tibbesma M, Bosman LJ, Mathijssen R, Atrafi F, van Coevorden F, Steeghs N, Farag S, Gelderblom H et al (2018) A single digital droplet PCR assay to detect multiple KIT exon 11 mutations in tumor and plasma from patients with gastrointestinal stromal tumors. Oncotarget 9: 13870–13883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun AC, de Mello CAL, Corassa M, Abdallah EA, Urvanegia AC, Alves VS, Flores BCTCP, Díaz M, Nicolau UR, Silva VSE et al (2018) EGFR expression in circulating tumor cells from high‐grade metastatic soft tissue sarcomas. Cancer Biol Ther 19: 454–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bregenzer ME, Horst EN, Mehta P, Novak CM, Raghavan S, Snyder CS, Mehta G (2019) Integrated cancer tissue engineering models for precision medicine. PLoS ONE 14: e0216564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenca M, Stacchiotti S, Fassetta K, Sbaraglia M, Janjusevic M, Racanelli D, Polano M, Rossi S, Brich S, Dagrada GP et al (2019) NR4A3 fusion proteins trigger an axon guidance switch that marks the difference between EWSR1 and TAF15 translocated extraskeletal myxoid chondrosarcomas. J Pathol 249: 90–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brien GL, Remillard D, Shi J, Hemming ML, Chabon J, Wynne K, Dillon ET, Cagney G, Van Mierlo G, Baltissen MP et al (2018) Targeted degradation of BRD9 reverses oncogenic gene expression in synovial sarcoma. Elife 7: e41305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown HK, Tellez‐Gabriel M, Heymann D (2017a) Cancer stem cells in osteosarcoma. Cancer Lett 386: 189–195 [DOI] [PubMed] [Google Scholar]
- Brown HK, Schiavone K, Tazzyman S, Heymann D, Chico TJ (2017b) Zebrafish xenograft models of cancer and metastasis for drug discovery. Expert Opin Drug Discov 12: 379–389 [DOI] [PubMed] [Google Scholar]
- Brown HK, Schiavone K, Gouin F, Heymann M‐F, Heymann D (2018) Biology of bone sarcomas and new therapeutic developments. Calcif Tissue Int 102: 174–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown HK, Tellez‐Gabriel M, Cartron P‐F, Vallette FM, Heymann M‐F, Heymann D (2019) Characterization of circulating tumor cells as a reflection of the tumor heterogeneity: myth or reality? Drug Discov. Today 24: 763–772 [DOI] [PubMed] [Google Scholar]
- Buddingh EP, Kuijjer ML, Duim RAJ, Bürger H, Agelopoulos K, Myklebost O, Serra M, Mertens F, Hogendoorn PCW, Lankester AC et al (2011) Tumor‐infiltrating macrophages are associated with metastasis suppression in high‐grade osteosarcoma: a rationale for treatment with macrophage activating agents. Clin Cancer Res 17: 2110–2119 [DOI] [PubMed] [Google Scholar]
- Bui NQ, Przybyl J, Trabucco SE, Frampton G, Hastie T, van de Rijn M, Ganjoo KN (2019) A clinico‐genomic analysis of soft tissue sarcoma patients reveals CDKN2A deletion as a biomarker for poor prognosis. Clin Sarcoma Res 9: 12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdach SEG, Westhoff M‐A, Steinhauser MF, Debatin K‐M (2018) Precision medicine in pediatric oncology. Mol Cell Pediatr 5: 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cain DM, Wacnik PW, Turner M, Wendelschafer‐Crabb G, Kennedy WR, Wilcox GL, Simone DA (2001) Functional interactions between tumor and peripheral nerve: changes in excitability and morphology of primary afferent fibers in a murine model of cancer pain. J Neurosci 21: 9367–9376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campanile C, Arlt MJE, Krämer SD, Honer M, Gvozdenovic A, Brennecke P, Fischer CR, Sabile AA, Müller A, Ametamey SM et al (2013) Characterization of different osteosarcoma phenotypes by PET imaging in preclinical animal models. J Nucl Med 54: 1362–1368 [DOI] [PubMed] [Google Scholar]
- Camuzard O, Santucci‐Darmanin S, Carle GF, Pierrefite‐Carle V (2019) Role of autophagy in osteosarcoma. J Bone Oncol 16: 100235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network (2017) Comprehensive and integrated genomic characterization of adult soft tissue sarcomas. Cell 171: 950–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon CM (2015) Cats, cancer and comparative oncology. Vet Sci 2: 111–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canter RJ, Ames E, Mac S, Grossenbacher SK, Chen M, Li C‐S, Borys D, Smith RC, Tellez J, Sayers TJ et al (2014) Anti‐proliferative but not anti‐angiogenic tyrosine kinase inhibitors enrich for cancer stem cells in soft tissue sarcoma. BMC Cancer 14: 756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, O'Reilly MS, Marshall B, Flynn E, Ji RW, Folkman J (1998) Expression of angiostatin cDNA in a murine fibrosarcoma suppresses primary tumor growth and produces long‐term dormancy of metastases. J Clin Invest 101: 1055–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capper D, Jones DTW, Sill M, Hovestadt V, Schrimpf D, Sturm D, Koelsche C, Sahm F, Chavez L, Reuss DE et al (2018) DNA methylation‐based classification of central nervous system tumours. Nature 555: 469–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso E, Guidi M, Blanchet B, Schneider MP, Decosterd LA, Buclin T, Csajka C, Widmer N (2020) Therapeutic drug monitoring of targeted anticancer protein kinase inhibitors in routine clinical use: a critical review. Ther Drug Monit 42: 33–44 [DOI] [PubMed] [Google Scholar]
- Casadei L, Calore F, Creighton CJ, Guescini M, Batte K, Iwenofu OH, Zewdu A, Braggio DA, Bill KL, Fadda P et al (2017) Exosome‐derived miR‐25‐3p and miR‐92a‐3p stimulate liposarcoma progression. Cancer Res 77: 3846–3856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casali PG, Abecassis N, Aro HT, Bauer S, Biagini R, Bielack S, Bonvalot S, Boukovinas I, Bovee JVMG, Brodowicz T et al (2018) Gastrointestinal stromal tumours: ESMO‐EURACAN Clinical Practice Guidelines for diagnosis, treatment and follow‐up. Ann Oncol 29: iv68–iv78 [DOI] [PubMed] [Google Scholar]
- Casadei L, Calore F, Braggio DA, Zewdu A, Deshmukh AA, Fadda P, Lopez G, Wabitsch M, Song C, Leight JL et al (2019) MDM2 derived from dedifferentiated liposarcoma extracellular vesicles induces MMP2 production from preadipocytes. Cancer Res 79: 4911–4922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo‐Tandazo W, Mutsaers AJ, Walkley CR (2019) Osteosarcoma in the post genome era: preclinical models and approaches to identify tractable therapeutic targets. Curr Osteoporos Rep 17: 343–352 [DOI] [PubMed] [Google Scholar]
- Chen C‐Y, Wang P‐Y, Hutzen B, Sprague L, Swain HM, Love JK, Stanek JR, Boon L, Conner J, Cripe TP (2017) Cooperation of oncolytic herpes virotherapy and PD‐1 blockade in murine rhabdomyosarcoma models. Sci Rep 7: 2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Zhao H (2019) Next‐generation sequencing in liquid biopsy: cancer screening and early detection. Hum Genomics 13: 34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Pandya PH, Liu E, Chandra P, Wang L, Murray ME, Carter J, Ferguson M, Saadatzadeh MR, Bijangi‐Visheshsaraei K et al (2019) Integration of genomic copy number variations and chemotherapy‐response biomarkers in pediatric sarcoma. BMC Med Genomics 12: 23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinen LTD, Mello CAL, Abdallah EA, Ocea LM, Buim ME, Breve NM, Gasparini JL, Fanelli MF, Paterlini‐Bréchot P (2014) Isolation, detection, and immunomorphological characterization of circulating tumor cells (CTCs) from patients with different types of sarcoma using isolation by size of tumor cells: a window on sarcoma‐cell invasion. Onco Targets Ther 7: 1609–1617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chudasama P, Mughal SS, Sanders MA, Hübschmann D, Chung I, Deeg KI, Wong S‐H, Rabe S, Hlevnjak M, Zapatka M et al (2018) Integrative genomic and transcriptomic analysis of leiomyosarcoma. Nat Commun 9: 144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimpean AM, Lalošević D, Lalošević V, Banović P, Raica M, Mederle OA (2018) Disodium cromolyn and anti‐podoplanin antibodies strongly inhibit growth of BHK 21/C13‐derived fibrosarcoma in a chick embryo chorioallantoic membrane model. In Vivo 32: 791–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark J, Rocques PJ, Crew AJ, Gill S, Shipley J, Chan AM, Gusterson BA, Cooper CS (1994) Identification of novel genes, SYT and SSX, involved in the t(X;18)(p11.2;q11.2) translocation found in human synovial sarcoma. Nat Genet 7: 502–508 [DOI] [PubMed] [Google Scholar]
- Consolino L, Anemone A, Capozza M, Carella A, Irrera P, Corrado A, Dhakan C, Bracesco M, Longo DL (2020) Non‐invasive investigation of tumor metabolism and acidosis by MRI‐CEST imaging. Front Oncol 10: 161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corless CL, Schroeder A, Griffith D, Town A, McGreevey L, Harrell P, Shiraga S, Bainbridge T, Morich J, Heinrich MC (2005) PDGFRA mutations in gastrointestinal stromal tumors: frequency, spectrum and in vitro sensitivity to imatinib. J Clin Oncol 23: 5357–5364 [DOI] [PubMed] [Google Scholar]
- Cortini M, Massa A, Avnet S, Bonuccelli G, Baldini N (2016) Tumor‐activated mesenchymal stromal cells promote osteosarcoma stemness and migratory potential via IL‐6 secretion. PLoS ONE 11: e0166500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortini M, Avnet S, Baldini N (2017) Mesenchymal stroma: role in osteosarcoma progression. Cancer Lett 405: 90–99 [DOI] [PubMed] [Google Scholar]
- Cortini M, Baldini N, Avnet S (2019) New advances in the study of bone tumors: a lesson from the 3D environment. Front Physiol 10: 814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crenn V, Biteau K, Amiaud J, Dumars C, Guiho R, Vidal L, Nail L‐RL, Heymann D, Moreau A, Gouin F et al (2017) Bone microenvironment has an influence on the histological response of osteosarcoma to chemotherapy: retrospective analysis and preclinical modeling. Am J Cancer Res 7: 2333–2349 [PMC free article] [PubMed] [Google Scholar]
- Cripe TP, Ngo MC, Geller JI, Louis CU, Currier MA, Racadio JM, Towbin AJ, Rooney CM, Pelusio A, Moon A et al (2015) Phase 1 study of intratumoral Pexa‐Vec (JX‐594), an oncolytic and immunotherapeutic vaccinia virus, in pediatric cancer patients. Mol Ther 23: 602–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crombé A, Saut O, Guigui J, Italiano A, Buy X, Kind M (2019) Influence of temporal parameters of DCE‐MRI on the quantification of heterogeneity in tumor vascularization. J Magn Reson Imaging 50: 1773–1788 [DOI] [PubMed] [Google Scholar]
- Dancsok AR, Setsu N, Gao D, Blay J‐Y, Thomas D, Maki RG, Nielsen TO, Demicco EG (2019) Expression of lymphocyte immunoregulatory biomarkers in bone and soft‐tissue sarcomas. Mod Pathol 32: 1772–1785 [DOI] [PubMed] [Google Scholar]
- D'Angelo SP, Shoushtari AN, Agaram NP, Kuk D, Qin L‐X, Carvajal RD, Dickson MA, Gounder M, Keohan ML, Schwartz GK et al (2015) Prevalence of tumor‐infiltrating lymphocytes and PD‐L1 expression in the soft tissue sarcoma microenvironment. Hum Pathol 46: 357–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Angelo SP, Melchiori L, Merchant MS, Bernstein D, Glod J, Kaplan R, Grupp S, Tap WD, Chagin K, Binder GK et al (2018) Antitumor activity associated with prolonged persistence of adoptively transferred NY‐ESO‐1 c259T cells in synovial sarcoma. Cancer Discov 8: 944–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta P, Ayan B, Ozbolat IT (2017) Bioprinting for vascular and vascularized tissue biofabrication. Acta Biomater 51: 1–20 [DOI] [PubMed] [Google Scholar]
- Dawson LE, D'Agostino L, Hakim AA, Lackman RD, Brown SA, Sensenig RB, Antonello ZA, Kuzin II (2020) Induction of myogenic differentiation improves chemosensitivity of chemoresistant cells in soft‐tissue sarcoma cell lines. Sarcoma 2020: 8647981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Angelis ML, Francescangeli F, La Torre F, Zeuner A (2019) Stem cell plasticity and dormancy in the development of cancer therapy resistance. Front Oncol 9: 626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deel MD, Slemmons KK, Hinson AR, Genadry KC, Burgess BA, Crose LES, Kuprasertkul N, Oristian KM, Bentley RC, Linardic CM (2018) The transcriptional coactivator TAZ is a potent mediator of alveolar rhabdomyosarcoma tumorigenesis. Clin Cancer Res 24: 2616–2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demetri GD, Chawla SP, Ray‐Coquard I, Le Cesne A, Staddon AP, Milhem MM, Penel N, Riedel RF, Bui‐Nguyen B, Cranmer LD et al (2013) Results of an international randomized phase III trial of the mammalian target of rapamycin inhibitor ridaforolimus versus placebo to control metastatic sarcomas in patients after benefit from prior chemotherapy. J Clin Oncol 31: 2485–2492 [DOI] [PubMed] [Google Scholar]
- Dickson MA, Tap WD, Keohan ML, D'Angelo SP, Gounder MM, Antonescu CR, Landa J, Qin L‐X, Rathbone DD, Condy MM et al (2013) Phase II trial of the CDK4 inhibitor PD0332991 in patients with advanced CDK4‐amplified well‐differentiated or dedifferentiated liposarcoma. J Clin Oncol 31: 2024–2028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson L, Dickey LB (1956) Spontaneous regression of malignant tumors; report of a twelve‐year spontaneous complete regression of an extensive fibrosarcoma, with speculations about regression and dormancy. Am J Surg 92: 162–173 [DOI] [PubMed] [Google Scholar]
- Dodd RD, Añó L, Blum JM, Li Z, Van Mater D, Kirsch DG (2015) Methods to generate genetically engineered mouse models of soft tissue sarcoma. Methods Mol Biol 1267: 283–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebele RC, Davis LE, Vaishnavi A, Le AT, Estrada‐Bernal A, Keysar S, Jimeno A, Varella‐Garcia M, Aisner DL, Li Y et al (2015) An oncogenic NTRK fusion in a patient with soft‐tissue sarcoma with response to the tropomyosin‐related kinase inhibitor LOXO‐101. Cancer Discov 5: 1049–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doraiswamy A, Narayan RJ, Harris ML, Qadri SB, Modi R, Chrisey DB (2007) Laser microfabrication of hydroxyapatite‐osteoblast‐like cell composites. J Biomed Mater Res A 80: 635–643 [DOI] [PubMed] [Google Scholar]
- Drummond CJ, Hanna JA, Garcia MR, Devine DJ, Heyrana AJ, Finkelstein D, Rehg JE, Hatley ME (2018) Hedgehog pathway drives fusion‐negative rhabdomyosarcoma initiated from non‐myogenic endothelial progenitors. Cancer Cell 33: 108–124.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchi S, Ramos‐Romero S, Dozza B, Guerra‐Rebollo M, Cattini L, Ballestri M, Dambruoso P, Guerrini A, Sotgiu G, Varchi G et al (2016) Development of near‐infrared photoactivable phthalocyanine‐loaded nanoparticles to kill tumor cells: An improved tool for photodynamic therapy of solid cancers. Nanomedicine 12: 1885–1897 [DOI] [PubMed] [Google Scholar]
- Dudeck O, Zeile M, Pink D, Pech M, Tunn P‐U, Reichardt P, Ludwig W‐D, Hamm B (2008) Diffusion‐weighted magnetic resonance imaging allows monitoring of anticancer treatment effects in patients with soft‐tissue sarcomas. J Magn Reson Imaging 27: 1109–1113 [DOI] [PubMed] [Google Scholar]
- Dumars C, Ngyuen J‐M, Gaultier A, Lanel R, Corradini N, Gouin F, Heymann D, Heymann M‐F (2016) Dysregulation of macrophage polarization is associated with the metastatic process in osteosarcoma. Oncotarget 7: 78343–78354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eary JF, Link JM, Muzi M, Conrad EU, Mankoff DA, White JK, Krohn KA (2011) Multiagent PET for risk characterization in sarcoma. J Nucl Med 52: 541–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastley NC, Ottolini B, Neumann R, Luo J‐L, Hastings RK, Khan I, Moore DA, Esler CP, Shaw JA, Royle NJ et al (2018) Circulating tumour‐derived DNA in metastatic soft tissue sarcoma. Oncotarget 9: 10549–10560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehnman M, Missiaglia E, Folestad E, Selfe J, Strell C, Thway K, Brodin B, Pietras K, Shipley J, Östman A et al (2013) Distinct effects of ligand‐induced PDGFRα and PDGFRβ signaling in the human rhabdomyosarcoma tumor cell and stroma cell compartments. Cancer Res 73: 2139–2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eid JE, Garcia CB (2015) Reprogramming of mesenchymal stem cells by oncogenes. Semin Cancer Biol 32: 18–31 [DOI] [PubMed] [Google Scholar]
- El‐Naggar AM, Veinotte CJ, Cheng H, Grunewald TGP, Negri GL, Somasekharan SP, Corkery DP, Tirode F, Mathers J, Khan D et al (2015) Translational activation of HIF1α by YB‐1 promotes sarcoma metastasis. Cancer Cell 27: 682–697 [DOI] [PubMed] [Google Scholar]
- Endo Y, Beauchamp E, Woods D, Taylor WG, Toretsky JA, Uren A, Rubin JS (2008) Wnt‐3a and Dickkopf‐1 stimulate neurite outgrowth in Ewing tumor cells via a Frizzled3‐ and c‐Jun N‐terminal kinase‐dependent mechanism. Mol Cell Biol 28: 2368–2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo H, Inoue M (2019) Dormancy in cancer. Cancer Sci 110: 474–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Erp AEM, Versleijen‐Jonkers YMH, Hillebrandt‐Roeffen MHS, van Houdt L, Gorris MAJ, van Dam LS, Mentzel T, Weidema ME, Savci‐Heijink CD, Desar IME et al (2017) Expression and clinical association of programmed cell death‐1, programmed death‐ligand‐1 and CD8+ lymphocytes in primary sarcomas is subtype dependent. Oncotarget 8: 71371–71384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evdokimova V, Ruzanov P, Gassmann H, Zaidi SH, Peltekova V, Heisler LE, McPherson JD, Orlic‐Milacic M, Specht K, Steiger K et al (2019) Exosomes transmit retroelement RNAs to drive inflammation and immunosuppression in Ewing Sarcoma. bioRxiv 10.1101/806851 [PREPRINT] [DOI] [Google Scholar]
- Eyler CE, Rich JN (2008) Survival of the fittest: cancer stem cells in therapeutic resistance and angiogenesis. J Clin Oncol 26: 2839–2845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang D, Gan H, Lee J‐H, Han J, Wang Z, Riester SM, Jin L, Chen J, Zhou H, Wang J et al (2016) The histone H3.3K36M mutation reprograms the epigenome of chondroblastomas. Science 352: 1344–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farag S, Smith MJ, Fotiadis N, Constantinidou A, Jones RL (2020) Revolutions in treatment options in gastrointestinal stromal tumours (GISTs): the latest updates. Curr Treat Options Oncol 21: 55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fayad LM, Barker PB, Bluemke DA (2007) Molecular characterization of musculoskeletal tumors by proton MR spectroscopy. Semin Musculoskelet Radiol 11: 240–245 [DOI] [PubMed] [Google Scholar]
- Fayad LM, Jacobs MA, Wang X, Carrino JA, Bluemke DA (2012) Musculoskeletal tumors: how to use anatomic, functional, and metabolic MR techniques. Radiology 265: 340–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández L, Valentín J, Zalacain M, Leung W, Patiño‐García A, Pérez‐Martínez A (2015) Activated and expanded natural killer cells target osteosarcoma tumor initiating cells in an NKG2D‐NKG2DL dependent manner. Cancer Lett 368: 54–63 [DOI] [PubMed] [Google Scholar]
- Fleming JT, Brignola E, Chen L, Guo Y, Zhao S, Wang Q, Li B, Correa H, Ermilov AN, Dlugosz AA et al (2019) Insight into the etiology of undifferentiated soft tissue sarcomas from a novel mouse model. Mol Cancer Res 17: 1024–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores RJ, Kelly AJ, Li Y, Nakka M, Barkauskas DA, Krailo M, Wang LL, Perlaky L, Lau CC, Hicks MJ et al (2017) A novel prognostic model for osteosarcoma using circulating CXCL10 and FLT3LG. Cancer 123: 144–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourneaux B, Bourdon A, Dadone B, Lucchesi C, Daigle SR, Richard E, Laroche‐Clary A, Le Loarer F, Italiano A (2019) Identifying and targeting cancer stem cells in leiomyosarcoma: prognostic impact and role to overcome secondary resistance to PI3K/mTOR inhibition. J Hematol Oncol 12: 11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsching B, Fellenberg J, Moskovszky L, Sápi Z, Krenacs T, Machado I, Poeschl J, Lehner B, Szendrõi M, Bosch AL et al (2015) CD8+/FOXP3+‐ratio in osteosarcoma microenvironment separates survivors from non‐survivors: a multicenter validated retrospective study. Oncoimmunology 4: e990800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Arakawa A, Utsumi D, Sumiyoshi S, Yamamoto Y, Kitoh A, Ono M, Matsumura Y, Kato M, Konishi K et al (2014) CD8+ tumor‐infiltrating lymphocytes at primary sites as a possible prognostic factor of cutaneous angiosarcoma. Int J Cancer 134: 2393–2402 [DOI] [PubMed] [Google Scholar]
- Fukuda T, Wengler K, de Carvalho R, Boonsri P, Schweitzer ME (2019) MRI biomarkers in osseous tumors. J Magn Reson Imaging 50: 702–718 [DOI] [PubMed] [Google Scholar]
- Gabriel MT, Calleja LR, Chalopin A, Ory B, Heymann D (2016) Circulating tumor cells: a review of non‐EpCAM‐based approaches for cell enrichment and isolation. Clin Chem 62: 571–581 [DOI] [PubMed] [Google Scholar]
- Galon J, Bruni D (2019) Approaches to treat immune hot, altered and cold tumours with combination immunotherapies. Nat Rev Drug Discov 18: 197–218 [DOI] [PubMed] [Google Scholar]
- Gambera S, Abarrategi A, González‐Camacho F, Morales‐Molina Á, Roma J, Alfranca A, García‐Castro J (2018) Clonal dynamics in osteosarcoma defined by RGB marking. Nat Commun 9: 3994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia‐Moure M, Martinez‐Vélez N, Patiño‐García A, Alonso MM (2017) Oncolytic adenoviruses as a therapeutic approach for osteosarcoma: a new hope. J Bone Oncol 9: 41–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gargallo P, Juan A, Yáñez Y, Dolz S, Segura V, Castel V, Cañete A (2020) Precision medicine in Ewing sarcoma: a translational point of view. Clin Transl Oncol 22: 1440–1454 [DOI] [PubMed] [Google Scholar]
- Gaspar N, Hawkins DS, Dirksen U, Lewis IJ, Ferrari S, Le Deley M‐C, Kovar H, Grimer R, Whelan J, Claude L et al (2015) Ewing sarcoma: current management and future approaches through collaboration. J Clin Oncol 33: 3036–3046 [DOI] [PubMed] [Google Scholar]
- Gaude E, Schmidt C, Gammage PA, Dugourd A, Blacker T, Chew SP, Saez‐Rodriguez J, O'Neill JS, Szabadkai G, Minczuk M et al (2018) NADH shuttling couples cytosolic reductive carboxylation of glutamine with glycolysis in cells with mitochondrial dysfunction. Mol Cell 69: 581–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genadry KC, Pietrobono S, Rota R, Linardic CM (2018) Soft tissue sarcoma cancer stem cells: an overview. Front Oncol 8: 475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghayad SE, Rammal G, Ghamloush F, Basma H, Nasr R, Diab‐Assaf M, Chelala C, Saab R (2016) Exosomes derived from embryonal and alveolar rhabdomyosarcoma carry differential miRNA cargo and promote invasion of recipient fibroblasts. Sci Rep 6: 37088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghilardi JR, Freeman KT, Jimenez‐Andrade JM, Mantyh WG, Bloom AP, Kuskowski MA, Mantyh PW (2010) Administration of a tropomyosin receptor kinase inhibitor attenuates sarcoma‐induced nerve sprouting, neuroma formation and bone cancer pain. Mol Pain 6: 87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez J, Tsagozis P (2020) Multidisciplinary treatment of soft tissue sarcomas: an update. World J Clin Oncol 11: 180–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez‐Brouchet A, Illac C, Gilhodes J, Bouvier C, Aubert S, Guinebretiere J‐M, Marie B, Larousserie F, Entz‐Werlé N, de Pinieux G et al (2017) CD163‐positive tumor‐associated macrophages and CD8‐positive cytotoxic lymphocytes are powerful diagnostic markers for the therapeutic stratification of osteosarcoma patients: an immunohistochemical analysis of the biopsies fromthe French OS2006 phase 3 trial. Oncoimmunology 6: e1331193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graat HCA, Witlox MA, Schagen FHE, Kaspers GJL, Helder MN, Bras J, Schaap GR, Gerritsen WR, Wuisman PIJM, van Beusechem VW (2006) Different susceptibility of osteosarcoma cell lines and primary cells to treatment with oncolytic adenovirus and doxorubicin or cisplatin. Br J Cancer 94: 1837–1844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf Finckenstein F, Shahbazian V, Davicioni E, Ren Y‐X, Anderson MJ (2008) PAX‐FKHR function as pangenes by simultaneously inducing and inhibiting myogenesis. Oncogene 27: 2004–2014 [DOI] [PubMed] [Google Scholar]
- Grignani G, D'Ambrosio L, Pignochino Y, Palmerini E, Zucchetti M, Boccone P, Aliberti S, Stacchiotti S, Bertulli R, Piana R et al (2018) Trabectedin and olaparib in patients with advanced and non‐resectable bone and soft‐tissue sarcomas (TOMAS): an open‐label, phase 1b study from the Italian Sarcoma Group. Lancet Oncol 19: 1360–1371 [DOI] [PubMed] [Google Scholar]
- Grigoriadis AE, Schellander K, Wang ZQ, Wagner EF (1993) Osteoblasts are target cells for transformation in c‐fos transgenic mice. J Cell Biol 122: 685–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grohar PJ, Glod J, Peer CJ, Sissung TM, Arnaldez FI, Long L, Figg WD, Whitcomb P, Helman LJ, Widemann BC (2017) A phase I/II trial and pharmacokinetic study of mithramycin in children and adults with refractory Ewing sarcoma and EWS‐FLI1 fusion transcript. Cancer Chemother Pharmacol 80: 645–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisberg R, Hong DS, Holla V, Janku F, Piha‐Paul S, Ravi V, Benjamin R, Kumar Patel S, Somaiah N, Conley A et al (2017) Clinical genomic profiling to identify actionable alterations for investigational therapies in patients with diverse sarcomas. Oncotarget 8: 39254–39267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryder BE, Yohe ME, Chou H‐C, Zhang X, Marques J, Wachtel M, Schaefer B, Sen N, Song Y, Gualtieri A et al (2017) PAX3‐FOXO1 establishes myogenic super enhancers and confers BET bromodomain vulnerability. Cancer Discov 7: 884–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryder BE, Pomella S, Sayers C, Wu XS, Song Y, Chiarella AM, Bagchi S, Chou H‐C, Sinniah RS, Walton A et al (2019) Histone hyperacetylation disrupts core gene regulatory architecture in rhabdomyosarcoma. Nat Genet 51: 1714–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryder BE, Wachtel M, Chang K, El Demerdash O, Aboreden NG, Mohammed W, Ewert W, Pomella S, Rota R, Wei JS et al (2020) Miswired enhancer logic drives a cancer of the muscle lineage. IScience 23: 101103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P, Huang J, Moses MA (2017) Characterization of dormant and active human cancer cells by quantitative phase imaging. Cytometry A 91: 424–432 [DOI] [PubMed] [Google Scholar]
- Gutteridge A, Rathbone VM, Gibbons R, Bi M, Archard N, Davies KEJ, Brown J, Plagnol V, Pillay N, Amary F et al (2017) Digital PCR analysis of circulating tumor DNA: a biomarker for chondrosarcoma diagnosis, prognostication, and residual disease detection. Cancer Med 6: 2194–2202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddach M, Michaux J, Schwaebe MK, Pierre F, O'Brien SE, Borsan C, Tran J, Raffaele N, Ravula S, Drygin D et al (2011) Discovery of CX‐6258. A potent, selective, and orally efficacious pan‐pim kinases inhibitor. ACS Med Chem Lett 3: 135–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagi T, Nakamura T, Iino T, Matsubara T, Asanuma K, Matsumine A, Sudo A (2017) The diagnostic and prognostic value of interleukin‐6 in patients with soft tissue sarcomas. Sci Rep 7: 9640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Q, Shi H, Liu F (2016) CD163(+) M2‐type tumor‐associated macrophage support the suppression of tumor‐infiltrating T cells in osteosarcoma. Int Immunopharmacol 34: 101–106 [DOI] [PubMed] [Google Scholar]
- Harlow ML, Chasse MH, Boguslawski EA, Sorensen KM, Gedminas JM, Kitchen‐Goosen SM, Rothbart SB, Taslim C, Lessnick SL, Peck AS et al (2019) Trabectedin inhibits EWS‐FLI1 and evicts SWI/SNF from chromatin in a schedule‐dependent manner. Clin Cancer Res 25: 3417–3429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison DJ, Parisi MT, Shulkin BL (2017) The role of 18F‐FDG‐PET/CT in pediatric sarcoma. Semin Nucl Med 47: 229–241 [DOI] [PubMed] [Google Scholar]
- Hatina J, Kripnerova M, Houfkova K, Pesta M, Kuncova J, Sana J, Slaby O, Rodríguez R (2019) Sarcoma stem cell heterogeneity. Adv Exp Med Biol 1123: 95–118 [DOI] [PubMed] [Google Scholar]
- Hatley ME, Tang W, Garcia MR, Finkelstein D, Millay DP, Liu N, Graff J, Galindo RL, Olson EN (2012) A mouse model of rhabdomyosarcoma originating from the adipocyte lineage. Cancer Cell 22: 536–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauer K, Calzada‐Wack J, Steiger K, Grunewald TGP, Baumhoer D, Plehm S, Buch T, Prazeres da Costa O, Esposito I, Burdach S et al (2013) DKK2 mediates osteolysis, invasiveness, and metastatic spread in Ewing sarcoma. Cancer Res 73: 967–977 [DOI] [PubMed] [Google Scholar]
- Hayashi M, Zhu P, McCarty G, Meyer CF, Pratilas CA, Levin A, Morris CD, Albert CM, Jackson KW, Tang C‐M et al (2017) Size‐based detection of sarcoma circulating tumor cells and cell clusters. Oncotarget 8: 78965–78977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes MN, Langenau DM (2017) Discovering novel oncogenic pathways and new therapies using zebrafish models of sarcoma. Methods Cell Biol 138: 525–561 [DOI] [PubMed] [Google Scholar]
- Heinrich MC, Jones RL, von Mehren M, Schöffski P, Serrano C, Kang Y‐K, Cassier PA, Mir O, Eskens F, Tap WD et al (2020) Avapritinib in advanced PDGFRA D842V‐mutant gastrointestinal stromal tumour (NAVIGATOR): a multicentre, open‐label, phase 1 trial. Lancet Oncol 21: 935–946 [DOI] [PubMed] [Google Scholar]
- Helleday T (2011) The underlying mechanism for the PARP and BRCA synthetic lethality: clearing up the misunderstandings. Mol Oncol 5: 387–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heymann D (2014) Bone Cancer. Academic Press, San Diego, California, USA; [Google Scholar]
- Heymann M‐F, Lézot F, Heymann D (2019) The contribution of immune infiltrates and the local microenvironment in the pathogenesis of osteosarcoma. Cell Immunol 343: 103711 [DOI] [PubMed] [Google Scholar]
- Heymann M‐F, Schiavone K, Heymann D (2020) Bone sarcomas in the immunotherapy era. Br J Pharmacol 10.1111/bph.14999 [DOI] [PubMed] [Google Scholar]
- Higuchi T, Sugisawa N, Miyake K, Oshiro H, Yamamoto N, Hayashi K, Kimura H, Miwa S, Igarashi K, Kline Z et al (2019) Pioglitazone, an agonist of PPARγ, reverses doxorubicin‐resistance in an osteosarcoma patient‐derived orthotopic xenograft model by downregulating P‐glycoprotein expression. Biomed Pharmacother 118: 109356 [DOI] [PubMed] [Google Scholar]
- Hingorani P, Sampson V, Lettieri C, Kolb EA (2014) Oncolytic viruses for potential osteosarcoma therapy. Adv Exp Med Biol 804: 259–283 [DOI] [PubMed] [Google Scholar]
- Hoffman RM (2015) Patient‐derived orthotopic xenografts: better mimic of metastasis than subcutaneous xenografts. Nat Rev Cancer 15: 451–452 [DOI] [PubMed] [Google Scholar]
- Honoki K, Fujii H, Kubo A, Kido A, Mori T, Tanaka Y, Tsujiuchi T (2010) Possible involvement of stem‐like populations with elevated ALDH1 in sarcomas for chemotherapeutic drug resistance. Oncol Rep 24: 501–505 [DOI] [PubMed] [Google Scholar]
- Hourigan AJ, Kells AF, Schwartz HS (1993) In vitro photodynamic therapy of musculoskeletal neoplasms. J Orthop Res 11: 633–637 [DOI] [PubMed] [Google Scholar]
- Huang TQ, Qu X, Liu J, Chen S (2014) 3D printing of biomimetic microstructures for cancer cell migration. Biomed Microdevices 16: 127–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Chen M, Whitley MJ, Kuo H‐C, Xu ES, Walens A, Mowery YM, Van Mater D, Eward WC, Cardona DM et al (2017) Generation and comparison of CRISPR‐Cas9 and Cre‐mediated genetically engineered mouse models of sarcoma. Nat Commun 8: 15999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignatius MS, Hayes MN, Moore FE, Tang Q, Garcia SP, Blackburn PR, Baxi K, Wang L, Jin A, Ramakrishnan A et al (2018) tp53 deficiency causes a wide tumor spectrum and increases embryonal rhabdomyosarcoma metastasis in zebrafish. Elife 7: e37202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indraccolo S, Stievano L, Minuzzo S, Tosello V, Esposito G, Piovan E, Zamarchi R, Chieco‐Bianchi L, Amadori A (2006) Interruption of tumor dormancy by a transient angiogenic burst within the tumor microenvironment. Proc Natl Acad Sci USA 103: 4216–4221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannidis JPA, Lau J (2003) 18F‐FDG PET for the diagnosis and grading of soft‐tissue sarcoma: a meta‐analysis. J Nucl Med 44: 717–724 [PubMed] [Google Scholar]
- Italiano A, Di Mauro I, Rapp J, Pierron G, Auger N, Alberti L, Chibon F, Escande F, Voegeli A‐C, Ghnassia J‐P et al (2016) Clinical effect of molecular methods in sarcoma diagnosis (GENSARC): a prospective, multicentre, observational study. Lancet Oncol 17: 532–538 [DOI] [PubMed] [Google Scholar]
- Jaboin J, Wild J, Hamidi H, Khanna C, Kim CJ, Robey R, Bates SE, Thiele CJ (2002) MS‐27‐275, an inhibitor of histone deacetylase, has marked in vitro and in vivo antitumor activity against pediatric solid tumors. Cancer Res 62: 6108–6115 [PubMed] [Google Scholar]
- Jacques C, Lamoureux F, Baud'huin M, Rodriguez Calleja L, Quillard T, Amiaud J, Tirode F, Rédini F, Bradner JE, Heymann D et al (2016) Targeting the epigenetic readers in Ewing sarcoma inhibits the oncogenic transcription factor EWS/Fli1. Oncotarget, 7: 24125–24140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques C, Renema N, Lezot F, Ory B, Walkley CR, Grigoriadis AE, Heymann D (2018) Small animal models for the study of bone sarcoma pathogenesis:characteristics, therapeutic interests and limitations. J Bone Oncol 12: 7–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques C, Renema N, Ory B, Walkley CR, Grigoriadis AE, Heymann D (2019) Murine models of bone sarcomas. Methods Mol Biol 1914: 331–342 [DOI] [PubMed] [Google Scholar]
- Jauregui JJ, Nadarajah V, Munn J, Pivec R, Kapadia BH, Lerman DM, Maheshwari AV (2018) Limb salvage versus amputation in conventional appendicular osteosarcoma: a systematic review. Indian J Surg Oncol 9: 232–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayasingam SD, Citartan M, Thang TH, Mat Zin AA, Ang KC, Ch'ng ES (2020) Evaluating the polarization of tumor‐associated macrophages into M1 and M2 phenotypes in human cancer tissue: technicalities and challenges in routine clinical practice. Front Oncol 9: 1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeyaram A, Jay SM (2017) Preservation and storage stability of extracellular vesicles for therapeutic applications. AAPS J 20: 1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson I, Verweij J, Gelderblom H, Hartmann JT, Schöffski P, Blay J‐Y, Kerst JM, Sufliarsky J, Whelan J, Hohenberger P et al (2014) Doxorubicin alone versus intensified doxorubicin plus ifosfamide for first‐line treatment of advanced or metastatic soft‐tissue sarcoma: a randomised controlled phase 3 trial. Lancet Oncol 15: 415–423 [DOI] [PubMed] [Google Scholar]
- Kalfusova A, Linke Z, Kalinova M, Krskova L, Hilska I, Szabova J, Vicha A, Kodet R (2019) Gastrointestinal stromal tumors – Summary of mutational status of the primary/secondary KIT/PDGFRA mutations, BRAF mutations and SDH defects. Pathol Res Pract 215: 152708 [DOI] [PubMed] [Google Scholar]
- Kanojia D, Nagata Y, Garg M, Lee DH, Sato A, Yoshida K, Sato Y, Sanada M, Mayakonda A, Bartenhagen C et al (2015) Genomic landscape of liposarcoma. Oncotarget 6: 42429–42444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kather JN, Hörner C, Weis C‐A, Aung T, Vokuhl C, Weiss C, Scheer M, Marx A, Simon‐Keller K (2019) CD163+ immune cell infiltrates and presence of CD54+ microvessels are prognostic markers for patients with embryonal rhabdomyosarcoma. Sci Rep 9: 9211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima O, Kamiyoshihara M, Sakata S, Endo K, Saito R, Morishita Y (2000) The clinicopathological significance of preoperative serum‐soluble interleukin‐2 receptor concentrations in operable non‐small‐cell lung cancer patients. Ann Surg Oncol 7: 239–245 [DOI] [PubMed] [Google Scholar]
- Keller C, Arenkiel BR, Coffin CM, El‐Bardeesy N, DePinho RA, Capecchi MR (2004) Alveolar rhabdomyosarcomas in conditional Pax3: Fkhr mice: cooperativity of Ink4a/ARF and Trp53 loss of function. Genes Dev 18: 2614–2626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller L, Pantel K (2019) Unravelling tumour heterogeneity by single‐cell profiling of circulating tumour cells. Nat Rev Cancer 19: 553–567 [DOI] [PubMed] [Google Scholar]
- Kersten K, de Visser KE, van Miltenburg MH, Jonkers J (2017) Genetically engineered mouse models in oncology research and cancer medicine. EMBO Mol Med 9: 137–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keung EZ, Burgess M, Salazar R, Parra ER, Rodrigues‐Canales J, Bolejack V, Van Tine BA, Schuetze SM, Attia S, Riedel RF et al (2020) Correlative analyses of the SARC028 trial reveal an association between sarcoma‐associated immune infiltrate and response to pembrolizumab. Clin Cancer Res 26: 1258–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilic M, Kasperczyk H, Fulda S, Debatin K‐M (2007) Role of hypoxia inducible factor‐1 alpha in modulation of apoptosis resistance. Oncogene 26: 2027–2038 [DOI] [PubMed] [Google Scholar]
- Kim J, Kim JH, Kang HG, Park SY, Yu JY, Lee EY, Oh SE, Kim YH, Yun T, Park C et al (2018) Integrated molecular characterization of adult soft tissue sarcoma for therapeutic targets. BMC Med Genet 19: 216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JI, Choi KU, Lee IS, Choi YJ, Kim WT, Shin DH, Kim K, Lee JH, Kim JY, Sol MY (2015) Expression of hypoxic markers and their prognostic significance in soft tissue sarcoma. Oncol Lett 9: 1699–1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JR, Moon YJ, Kwon KS, Bae JS, Wagle S, Kim KM, Park HS, Lee H, Moon WS, Chung MJ et al (2013a) Tumor infiltrating PD1‐positive lymphocytes and the expression of PD‐L1 predict poor prognosis of soft tissue sarcomas. PLoS ONE 8: e82870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YB, Shin KH, Seong J, Roh JK, Kim GE, Hahn SB, Suh CO (2008) Clinical significance of margin status in postoperative radiotherapy for extremity and truncal soft‐tissue sarcoma. Int J Radiat Oncol Biol Phys 70: 139–144 [DOI] [PubMed] [Google Scholar]
- Kim Y‐J, Lee H‐J, Kim T‐M, Eisinger‐Mathason TK, Zhang AY, Schmidt B, Karl DL, Nakazawa MS, Park PJ, Simon MC et al (2013b) Overcoming evasive resistance from vascular endothelial growth factor a inhibition in sarcomas by genetic or pharmacologic targeting of hypoxia‐inducible factor 1α. Int J Cancer 132: 29–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S, Kawaguchi S, Wada T, Nagoya S, Yamashita T, Kikuchi K (2002) Rhabdomyosarcoma arising from a dormant dumbbell ganglioneuroma of the lumbar spine: a case report. Spine 27: E513–E517 [DOI] [PubMed] [Google Scholar]
- Klug LR, Heinrich MC (2017) PDGFRA antibody for soft tissue sarcoma. Cell 168: 555 [DOI] [PubMed] [Google Scholar]
- Knott MML, Hölting TLB, Ohmura S, Kirchner T, Cidre‐Aranaz F, Grünewald TGP (2019) Targeting the undruggable: exploiting neomorphic features of fusion oncoproteins in childhood sarcomas for innovative therapies. Cancer Metastasis Rev 38: 625–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koelsche C, Hartmann W, Schrimpf D, Stichel D, Jabar S, Ranft A, Reuss DE, Sahm F, Jones DTW, Bewerunge‐Hudler M et al (2018a) Array‐based DNA‐methylation profiling in sarcomas with small blue round cell histology provides valuable diagnostic information. Mod Pathol 31: 1246–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koelsche C, Mynarek M, Schrimpf D, Bertero L, Serrano J, Sahm F, Reuss DE, Hou Y, Baumhoer D, Vokuhl C et al (2018b) Primary intracranial spindle cell sarcoma with rhabdomyosarcoma‐like features share a highly distinct methylation profile and DICER1 mutations. Acta Neuropathol 136: 327–337 [DOI] [PubMed] [Google Scholar]
- Koirala P, Roth ME, Gill J, Piperdi S, Chinai JM, Geller DS, Hoang BH, Park A, Fremed MA, Zang X et al (2016) Immune infiltration and PD‐L1 expression in the tumor microenvironment are prognostic in osteosarcoma. Sci Rep 6: 30093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb EA, Sampson V, Stabley D, Walter A, Sol‐Church K, Cripe T, Hingorani P, Ahern CH, Weigel BJ, Zwiebel J et al (2015) A phase I trial and viral clearance study of reovirus (Reolysin) in children with relapsed or refractory extra‐cranial solid tumors: a Children's Oncology Group Phase I Consortium report. Pediatr Blood Cancer 62: 751–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolosenko I, Avnet S, Baldini N, Viklund J, De Milito A (2017) Therapeutic implications of tumor interstitial acidification. Semin Cancer Biol 43: 119–133 [DOI] [PubMed] [Google Scholar]
- Kovac M, Blattmann C, Ribi S, Smida J, Mueller NS, Engert F, Castro‐Giner F, Weischenfeldt J, Kovacova M, Krieg A et al (2015) Exome sequencing of osteosarcoma reveals mutation signatures reminiscent of BRCA deficiency. Nat Commun 6: 8940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovatcheva M, Liu DD, Dickson MA, Klein ME, O'Connor R, Wilder FO, Socci ND, Tap WD, Schwartz GK, Singer S et al (2015) MDM2 turnover and expression of ATRX determine the choice between quiescence and senescence in response to CDK4 inhibition. Oncotarget 6: 8226–8243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kransdorf MJ, Meis JM (1993) From the archives of the AFIP. Extraskeletal osseous and cartilaginous tumors of the extremities. Radiographics 13: 853–884 [DOI] [PubMed] [Google Scholar]
- Krumbholz M, Hellberg J, Steif B, Bäuerle T, Gillmann C, Fritscher T, Agaimy A, Frey B, Juengert J, Wardelmann E et al (2016) Genomic EWSR1 fusion sequence as highly sensitive and dynamic plasma tumor marker in ewing sarcoma. Clin Cancer Res 22: 4356–4365 [DOI] [PubMed] [Google Scholar]
- Kubo T, Furuta T, Johan MP, Ochi M (2016) Prognostic significance of (18)F‐FDG PET at diagnosis in patients with soft tissue sarcoma and bone sarcoma; systematic review and meta‐analysis. Eur J Cancer 58: 104–111 [DOI] [PubMed] [Google Scholar]
- Kumar H, Heer K, Lee PW, Duthie GS, MacDonald AW, Greenman J, Kerin MJ, Monson JR (1998) Preoperative serum vascular endothelial growth factor can predict stage in colorectal cancer. Clin Cancer Res 4: 1279–1285 [PubMed] [Google Scholar]
- Kunz P, Schenker A, Sähr H, Lehner B, Fellenberg J (2019) Optimization of the chicken chorioallantoic membrane assay as reliable in vivo model for the analysis of osteosarcoma. PLoS ONE 14: e0215312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusuzaki K, Takai T, Yoshimura H, Inoue K, Takai S, Baldini N (2018) Clinical trial of radiotherapy after intravenous injection of acridine orange for patients with cancer. Anticancer Res 38: 481–489 [DOI] [PubMed] [Google Scholar]
- Lacroix J, Kis Z, Josupeit R, Schlund F, Stroh‐Dege A, Frank‐Stöhr M, Leuchs B, Schlehofer JR, Rommelaere J, Dinsart C (2018) Preclinical testing of an oncolytic parvovirus in ewing sarcoma: protoparvovirus H‐1 induces apoptosis and lytic infection in vitro but fails to improve survival in vivo . Viruses 10: 302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamora A, Talbot J, Bougras G, Amiaud J, Leduc M, Chesneau J, Taurelle J, Stresing V, Le Deley MC, Heymann MF et al (2014) Overexpression of smad7 blocks primary tumor growth and lung metastasis development in osteosarcoma. Clin Cancer Res 20: 5097–5112 [DOI] [PubMed] [Google Scholar]
- Lamoureux F, Baud'huin M, Rodriguez Calleja L, Jacques C, Berreur M, Rédini F, Lecanda F, Bradner JE, Heymann D, Ory B (2014) Selective inhibition of BET bromodomain epigenetic signalling interferes with the bone‐associated tumour vicious cycle. Nat Commun 5: 3511. [DOI] [PubMed] [Google Scholar]
- Lanzi C, Dal Bo L, Favini E, Tortoreto M, Beretta GL, Arrighetti N, Zaffaroni N, Cassinelli G (2019) Overactive IGF1/insulin receptors and NRASQ61R mutation drive mechanisms of resistance to pazopanib and define rational combination strategies to treat synovial sarcoma. Cancers (Basel) 11: 408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson MA, McDonald MM, Kovacic N, Hua Khoo W, Terry RL, Down J, Kaplan W, Paton‐Hough J, Fellows C, Pettitt JA et al (2015) Osteoclasts control reactivation of dormant myeloma cells by remodelling the endosteal niche. Nat Commun 6: 8983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Cesne A (2018) Making the best of available options for optimal sarcoma treatment. Oncology 95(Suppl 1): 11–20 [DOI] [PubMed] [Google Scholar]
- Leacock SW, Basse AN, Chandler GL, Kirk AM, Rakheja D, Amatruda JF (2012) A zebrafish transgenic model of Ewing's sarcoma reveals conserved mediators of EWS‐FLI1 tumorigenesis. Dis Model Mech 5: 95–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leddon JL, Chen C‐Y, Currier MA, Wang P‐Y, Jung FA, Denton NL, Cripe KM, Haworth KB, Arnold MA, Gross AC et al (2015) Oncolytic HSV virotherapy in murine sarcomas differentially triggers an antitumor T‐cell response in the absence of virus permissivity. Mol Ther Oncolytics 1: 14010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ATJ, Jones RL, Huang PH (2019) Pazopanib in advanced soft tissue sarcomas. Signal Transduct Target Ther 4: 16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Yoon YC, Seo SW, Choi Y‐L, Kim HS (2020) Soft tissue sarcoma: DWI and DCE‐MRI parameters correlate with Ki‐67 labeling index. Eur Radiol 30: 914–924 [DOI] [PubMed] [Google Scholar]
- Li GZ, Raut CP (2019) Targeted therapy and personalized medicine in gastrointestinal stromal tumors: drug resistance, mechanisms, and treatment strategies. Onco Targets Ther 12: 5123–5133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Meng QH, Noh H, Batth IS, Somaiah N, Torres KE, Xia X, Wang R, Li S (2017) Detection of circulating tumor cells from cryopreserved human sarcoma peripheral blood mononuclear cells. Cancer Lett 403: 216–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Meng QH, Noh H, Somaiah N, Torres KE, Xia X, Batth IS, Joseph CP, Liu M, Wang R et al (2018) Cell‐surface vimentin‐positive macrophage‐like circulating tumor cells as a novel biomarker of metastatic gastrointestinal stromal tumors. Oncoimmunology 7: e1420450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Flores R, Yu A, Okcu MF, Murray J, Chintagumpala M, Hicks J, Lau CC, Man T‐K (2011) Elevated expression of CXC chemokines in pediatric osteosarcoma patients. Cancer 117: 207–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y‐X, Yu H‐Y, Lv J‐Y, Cai Y‐R, Liu F, He Z‐M, He S‐S (2019) Targeting autophagy is a promising therapeutic strategy to overcome chemoresistance and reduce metastasis in osteosarcoma. Int J Oncol 55: 1213–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim K‐H, Huang M‐J, Chen L‐T, Wang T‐E, Liu C‐L, Chang C‐S, Liu M‐C, Hsieh R‐K, Tzen C‐Y (2008) Molecular analysis of secondary kinase mutations in imatinib‐resistant gastrointestinal stromal tumors. Med Oncol 25: 207–213 [DOI] [PubMed] [Google Scholar]
- Lissat A, Joerschke M, Shinde DA, Braunschweig T, Meier A, Makowska A, Bortnick R, Henneke P, Herget G, Gorr TA et al (2015) IL6 secreted by Ewing sarcoma tumor microenvironment confers anti‐apoptotic and cell‐disseminating paracrine responses in Ewing sarcoma cells. BMC Cancer 15: 552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Huang Y, Sun Y, Zhang J, Yao Y, Shen Z, Xiang D, He A (2016) Prognostic value of inflammation‐based scores in patients with osteosarcoma. Sci Rep 6: 39862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Nazmun N, Hassan S, Liu X, Yang J (2020) BRAF mutation and its inhibitors in sarcoma treatment. Cancer Med 10.1002/cam4.3103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Mi Y, Mueller T, Kreibich S, Williams EG, Van Drogen A, Borel C, Frank M, Germain P‐L, Bludau I et al (2019) Multi‐omic measurements of heterogeneity in HeLa cells across laboratories. Nat Biotechnol 37: 314–322 [DOI] [PubMed] [Google Scholar]
- Lohberger B, Rinner B, Stuendl N, Absenger M, Liegl‐Atzwanger B, Walzer SM, Windhager R, Leithner A (2012) Aldehyde dehydrogenase 1, a potential marker for cancer stem cells in human sarcoma. PLoS ONE 7: e43664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo DL, Bartoli A, Consolino L, Bardini P, Arena F, Schwaiger M, Aime S (2016) In vivo imaging of tumor metabolism and acidosis by combining PET and MRI‐CEST pH imaging. Cancer Res 76: 6463–6470 [DOI] [PubMed] [Google Scholar]
- Longo DL, Sun PZ, Consolino L, Michelotti FC, Uggeri F, Aime S (2014) A general MRI‐CEST ratiometric approach for pH imaging: demonstration of in vivo pH mapping with iobitridol. J Am Chem Soc 136: 14333–14336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Jain SU, Hoelper D, Bechet D, Molden RC, Ran L, Murphy D, Venneti S, Hameed M, Pawel BR et al (2016) Histone H3K36 mutations promote sarcomagenesis through altered histone methylation landscape. Science 352: 844–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchesi C, Khalifa E, Laizet Y, Soubeyran I, Mathoulin‐Pelissier S, Chomienne C, Italiano A (2018) Targetable alterations in adult patients with soft‐tissue sarcomas: insights for personalized therapy. JAMA Oncol 4: 1398–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyu HG, Haider AH, Landman AB, Raut CP (2019) The opportunities and shortcomings of using big data and national databases for sarcoma research. Cancer 125: 2926–2934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Liu J, Zhu W, Tang M, Lawrence N, Yu C, Gou M, Chen S (2018) 3D bioprinting of functional tissue models for personalized drug screening and in vitro disease modeling. Adv Drug Deliv Rev 132: 235–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado I, López‐Guerrero JA, Scotlandi K, Picci P, Llombart‐Bosch A (2018) Immunohistochemical analysis and prognostic significance of PD‐L1, PD‐1, and CD8+ tumor‐infiltrating lymphocytes in Ewing's sarcoma family of tumors (ESFT). Virchows Arch 472: 815–824 [DOI] [PubMed] [Google Scholar]
- MacNeill AL, Weishaar KM, Séguin B, Powers BE (2018) Safety of an oncolytic myxoma virus in dogs with soft tissue sarcoma. Viruses 10: 398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mader S, Pantel K (2017) Liquid biopsy: current status and future perspectives. Oncol Res Treat 40: 404–408 [DOI] [PubMed] [Google Scholar]
- Maier J, Lange T, Kerle I, Specht K, Bruegel M, Wickenhauser C, Jost P, Niederwieser D, Peschel C, Duyster J et al (2013) Detection of mutant free circulating tumor DNA in the plasma of patients with gastrointestinal stromal tumor harboring activating mutations of CKIT or PDGFRA. Clin Cancer Res 19: 4854–4867 [DOI] [PubMed] [Google Scholar]
- Majzner RG, Mackall CL (2018) Tumor antigen escape from CAR T‐cell therapy. Cancer Discov 8: 1219–1226 [DOI] [PubMed] [Google Scholar]
- Majzner RG, Theruvath JL, Nellan A, Heitzeneder S, Cui Y, Mount CW, Rietberg SP, Linde MH, Xu P, Rota C et al (2019) CAR T cells targeting B7‐H3, a pan‐cancer antigen, demonstrate potent preclinical activity against pediatric solid tumors and brain tumors. Clin Cancer Res 25: 2560–2574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinauskaite I, Hofmeister J, Burgermeister S, Neroladaki A, Hamard M, Montet X, Boudabbous S (2020) Radiomics and machine learning differentiate soft‐tissue lipoma and liposarcoma better than musculoskeletal radiologists. Sarcoma 2020: 7163453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manjunathan R, Ragunathan M (2015) Chicken chorioallantoic membrane as a reliable model to evaluate osteosarcoma‐an experimental approach using SaOS2 cell line. Biol Proced Online 17: 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankin HJ, Lange TA, Spanier SS (1982) The hazards of biopsy in patients with malignant primary bone and soft‐tissue tumors. J Bone Joint Surg Am 64: 1121–1127 [PubMed] [Google Scholar]
- Mannerström B, Kornilov R, Abu‐Shahba AG, Chowdhury IM, Sinha S, Seppänen‐Kaijansinkko R, Kaur S (2019) Epigenetic alterations in mesenchymal stem cells by osteosarcoma‐derived extracellular vesicles. Epigenetics 14: 352–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetto A, Ohmura S, Orth MF, Knott MML, Colombo MV, Arrigoni C, Bardinet V, Saucier D, Wehweck FS, Li J et al (2020) Oncogenic hijacking of a developmental transcription factor evokes vulnerability toward oxidative stress in Ewing sarcoma. Nat Commun 11: 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martano M, Morello E, Avnet S, Costa F, Sammartano F, Kusuzaki K, Baldini N (2019) Photodynamic surgery for feline injection‐site sarcoma. Biomed Res Int 2019: 8275935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez‐Cruzado L, Tornin J, Santos L, Rodriguez A, García‐Castro J, Morís F, Rodriguez R (2016) Aldh1 expression and activity increase during tumor evolution in sarcoma cancer stem cell populations. Sci Rep 6: 27878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez‐Velez N, Xipell E, Jauregui P, Zalacain M, Marrodan L, Zandueta C, Vera B, Urquiza L, Sierrasesúmaga L, Julián MS et al (2014) The oncolytic adenovirus Δ24‐RGD in combination with cisplatin exerts a potent anti‐osteosarcoma activity. J Bone Miner Res 29: 2287–2296 [DOI] [PubMed] [Google Scholar]
- Martínez‐Vélez N, Xipell E, Vera B, Acanda de la Rocha A, Zalacain M, Marrodán L, Gonzalez‐Huarriz M, Toledo G, Cascallo M, Alemany R et al (2016) The oncolytic adenovirus VCN‐01 as therapeutic approach against pediatric osteosarcoma. Clin Cancer Res 22: 2217–2225 [DOI] [PubMed] [Google Scholar]
- Martins‐Neves SR, Paiva‐Oliveira DI, Wijers‐Koster PM, Abrunhosa AJ, Fontes‐Ribeiro C, Bovée JVMG, Cleton‐Jansen A‐M, Gomes CMF (2016) Chemotherapy induces stemness in osteosarcoma cells through activation of Wnt/β‐catenin signaling. Cancer Lett 370: 286–295 [DOI] [PubMed] [Google Scholar]
- Matsubara T, Kusuzaki K, Matsumine A, Nakamura T, Sudo A (2013) Can a less radical surgery using photodynamic therapy with acridine orange be equal to a wide‐margin resection? Clin Orthop Relat Res 471: 792–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara T, Kusuzaki K, Matsumine A, Shintani K, Satonaka H, Uchida A (2006) Acridine orange used for photodynamic therapy accumulates in malignant musculoskeletal tumors depending on pH gradient. Anticancer Res 26: 187–193 [PubMed] [Google Scholar]
- Maurizi G, Verma N, Gadi A, Mansukhani A, Basilico C (2018) Sox2 is required for tumor development and cancer cell proliferation in osteosarcoma. Oncogene 37: 4626–4632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melms JC, Vallabhaneni S, Mills CE, Yapp C, Chen J‐Y, Morelli E, Waszyk P, Kumar S, Deming D, Moret N et al (2019) Inhibition of Haspin kinase promotes cell‐intrinsic and extrinsic anti‐tumor activity. Cancer Res 80: 798–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant MS, Wright M, Baird K, Wexler LH, Rodriguez‐Galindo C, Bernstein D, Delbrook C, Lodish M, Bishop R, Wolchok JD et al (2016) Phase I clinical trial of ipilimumab in pediatric patients with advanced solid tumors. Clin Cancer Res 22: 1364–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant MS, Yang X, Melchionda F, Romero M, Klein R, Thiele CJ, Tsokos M, Kontny HU, Mackall CL (2004) Interferon gamma enhances the effectiveness of tumor necrosis factor‐related apoptosis‐inducing ligand receptor agonists in a xenograft model of Ewing's sarcoma. Cancer Res 64: 8349–8356 [DOI] [PubMed] [Google Scholar]
- Meyers PA (2015) Systemic therapy for osteosarcoma and Ewing sarcoma. Am Soc Clin Oncol Educ Book e644–e647 [DOI] [PubMed] [Google Scholar]
- Michel BC, D'Avino AR, Cassel SH, Mashtalir N, McKenzie ZM, McBride MJ, Valencia AM, Zhou Q, Bocker M, Soares LMM et al (2018) A non‐canonical SWI/SNF complex is a synthetic lethal target in cancers driven by BAF complex perturbation. Nat Cell Biol 20: 1410–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihály D, Nagy N, Papp G, Pápai Z, Sápi Z (2018) Release of circulating tumor cells and cell‐free nucleic acids is an infrequent event in synovial sarcoma: Liquid biopsy analysis of 15 patients diagnosed with synovial sarcoma. Diagn Pathol 13: 81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller IV, Raposo G, Welsch U, Prazeres da Costa O, Thiel U, Lebar M, Maurer M, Bender H‐U, von Luettichau I, Richter GHS et al (2013) First identification of Ewing's sarcoma‐derived extracellular vesicles and exploration of their biological and potential diagnostic implications. Biol Cell 105: 289–303 [DOI] [PubMed] [Google Scholar]
- Mohseny AB, Hogendoorn PCW, Cleton‐Jansen A‐M (2012) Osteosarcoma models: from cell lines to zebrafish. Sarcoma 2012: 417271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore C, Eslin D, Levy A, Roberson J, Giusti V, Sutphin R (2010) Prognostic significance of early lymphocyte recovery in pediatric osteosarcoma. Pediatr Blood Cancer 55: 1096–1102 [DOI] [PubMed] [Google Scholar]
- Moriarity BS, Otto GM, Rahrmann EP, Rathe SK, Wolf NK, Weg MT, Manlove LA, LaRue RS, Temiz NA, Molyneux SD et al (2015) A Sleeping Beauty forward genetic screen identifies new genes and pathways driving osteosarcoma development and metastasis. Nat Genet 47: 615–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita T, Fujiwara T, Yoshida A, Uotani K, Kiyono M, Yokoo S, Hasei J, Kunisada T, Ozaki T (2020) Clinical relevance and functional significance of cell‐free microRNA‐1260b expression profiles in infiltrative myxofibrosarcoma. Sci Rep 10: 9414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu J, Sun P, Ma Z, Sun P (2019) BRD4 promotes tumor progression and NF‐κB/CCL2‐dependent tumor‐associated macrophage recruitment in GIST. Cell Death Dis 10: 935 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Murphy SV, Atala A (2014) 3D bioprinting of tissues and organs. Nat Biotechnol 32: 773–785 [DOI] [PubMed] [Google Scholar]
- Musa J, Aynaud M‐M, Mirabeau O, Delattre O, Grünewald TG (2017) MYBL2 (B‐Myb): a central regulator of cell proliferation, cell survival and differentiation involved in tumorigenesis. Cell Death Dis 8: e2895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musa J, Cidre‐Aranaz F, Aynaud M‐M, Orth MF, Knott MML, Mirabeau O, Mazor G, Varon M, Hölting TLB, Grossetête S et al (2019) Cooperation of cancer drivers with regulatory germline variants shapes clinical outcomes. Nat Commun 10: 4128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musa J, Grünewald TGP (2020) Interaction between somatic mutations and germline variants contributes to clinical heterogeneity in cancer. Mol Cell Oncol 7: 1682924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nacev BA, Feng L, Bagert JD, Lemiesz AE, Gao J, Soshnev AA, Kundra R, Schultz N, Muir TW, Allis CD (2019) The expanding landscape of ‘oncohistone’ mutations in human cancers. Nature 567: 473–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naka N, Takenaka S, Araki N, Miwa T, Hashimoto N, Yoshioka K, Joyama S, Hamada K‐I, Tsukamoto Y, Tomita Y et al (2010) Synovial sarcoma is a stem cell malignancy. Stem Cells 28: 1119–1131 [DOI] [PubMed] [Google Scholar]
- Namløs HM, Boye K, Mishkin SJ, Barøy T, Lorenz S, Bjerkehagen B, Stratford EW, Munthe E, Kudlow BA, Myklebost O et al (2018) Noninvasive detection of ctDNA reveals intratumor heterogeneity and is associated with tumor burden in gastrointestinal stromal tumor. Mol Cancer Ther 17: 2473–2480 [DOI] [PubMed] [Google Scholar]
- Nanni P, Landuzzi L, Manara MC, Righi A, Nicoletti G, Cristalli C, Pasello M, Parra A, Carrabotta M, Ferracin M et al (2019) Bone sarcoma patient‐derived xenografts are faithful and stable preclinical models for molecular and therapeutic investigations. Sci Rep 9: 12174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napolitano A, Vincenzi B (2019) Secondary KIT mutations: the GIST of drug resistance and sensitivity. Br J Cancer 120: 577–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumov GN, Bender E, Zurakowski D, Kang S‐Y, Sampson D, Flynn E, Watnick RS, Straume O, Akslen LA, Folkman J et al (2006) A model of human tumor dormancy: an angiogenic switch from the nonangiogenic phenotype. J Natl Cancer Inst 98: 316–325 [DOI] [PubMed] [Google Scholar]
- Nduom EK, Wei J, Yaghi NK, Huang N, Kong L‐Y, Gabrusiewicz K, Ling X, Zhou S, Ivan C, Chen JQ et al (2016) PD‐L1 expression and prognostic impact in glioblastoma. Neuro‐Oncology 18: 195–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nejman D, Livyatan I, Fuks G, Gavert N, Zwang Y, Geller LT, Rotter‐Maskowitz A, Weiser R, Mallel G, Gigi E et al (2020) The human tumor microbiome is composed of tumor type–specific intracellular bacteria. Science 368: 973–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemunaitis J, Tong AW, Nemunaitis M, Senzer N, Phadke AP, Bedell C, Adams N, Zhang Y‐A, Maples PB, Chen S et al (2010) A phase I study of telomerase‐specific replication competent oncolytic adenovirus (telomelysin) for various solid tumors. Mol Ther 18: 429–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowicki TS, Akiyama R, Huang RR, Shintaku IP, Wang X, Tumeh PC, Singh A, Chmielowski B, Denny C, Federman N et al (2017) Infiltration of CD8 T Cells and Expression of PD‐1 and PD‐L1 in Synovial Sarcoma. Cancer Immunol Res 5: 118–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino S, Konishi H, Ichikawa D, Hamada J, Shoda K, Arita T, Komatsu S, Shiozaki A, Okamoto K, Yamazaki S et al (2018) Detection of fusion gene in cell‐free DNA of a gastric synovial sarcoma. World J Gastroenterol 24: 949–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth MF, Buecklein VL, Kampmann E, Subklewe M, Noessner E, Cidre‐Aranaz F, Romero‐Pérez L, Wehweck FS, Lindner L, Issels R et al (2020) A comparative view on the expression patterns of PD‐L1 and PD‐1 in soft tissue sarcomas. Cancer Immunol Immunother 69: 1353–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osgood CL, Maloney N, Kidd CG, Kitchen‐Goosen S, Segars L, Gebregiorgis M, Woldemichael GM, He M, Sankar S, Lessnick SL et al (2016) Identification of mithramycin analogues with improved targeting of the EWS‐FLI1 transcription factor. Clin Cancer Res 22: 4105–4118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottaviani G, Jaffe N (2009) The epidemiology of osteosarcoma. Cancer Treat Res 152: 3–13 [DOI] [PubMed] [Google Scholar]
- Palmerini E, Agostinelli C, Picci P, Pileri S, Marafioti T, Lollini P‐L, Scotlandi K, Longhi A, Benassi MS, Ferrari S (2017) Tumoral immune‐infiltrate (IF), PD‐L1 expression and role of CD8/TIA‐1 lymphocytes in localized osteosarcoma patients treated within protocol ISG‐OS1. Oncotarget 8: 111836–111846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantel K, Alix‐Panabières C (2019) Liquid biopsy and minimal residual disease – latest advances and implications for cure. Nat Rev Clin Oncol 16: 409–424 [DOI] [PubMed] [Google Scholar]
- Paoletti C, Schiavon G, Dolce EM, Darga EP, Carr TH, Geradts J, Hoch M, Klinowska T, Lindemann J, Marshall G et al (2018) Circulating biomarkers and resistance to endocrine therapy in metastatic breast cancers: correlative results from AZD9496 Oral SERD Phase I Trial. Clin Cancer Res 24: 5860–5872 [DOI] [PubMed] [Google Scholar]
- Patil N, Ahmed Kabeer Rasheed S, Abba M, Hendrik Leupold J, Schwarzbach M, Allgayer H (2014) A mechanistic study on the metastasis inducing function of FUS‐CHOP fusion protein in liposarcoma. Int J Cancer 134: 2808–2819 [DOI] [PubMed] [Google Scholar]
- Patrikidou A, Domont J, Cioffi A, Le Cesne A (2011) Treating soft tissue sarcomas with adjuvant chemotherapy. Curr Treat Options Oncol 12: 21–31 [DOI] [PubMed] [Google Scholar]
- Pavlou M, Shah M, Gikas P, Briggs T, Roberts SJ, Cheema U (2019) Osteomimetic matrix components alter cell migration and drug response in a 3D tumour‐engineered osteosarcoma model. Acta Biomater 96: 247–257 [DOI] [PubMed] [Google Scholar]
- Pepin K, Grimm R, Kargar S, Howe BM, Fritchie K, Frick M, Wenger D, Okuno S, Ehman R, McGee K et al (2019) Soft tissue sarcoma stiffness and perfusion evaluation by MRE and DCE‐MRI for radiation therapy response assessment: a technical feasibility study. Biomed Phys Eng Express 10.1088/2057-1976/ab2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez‐Martínez A, Leung W, Muñoz E, Iyengar R, Ramírez M, Vicario JL, Lassaletta A, Sevilla J, González‐Vicent M, Madero L et al (2009) KIR‐HLA receptor‐ligand mismatch associated with a graft‐versus‐tumor effect in haploidentical stem cell transplantation for pediatric metastatic solid tumors. Pediatr Blood Cancer 53: 120–124 [DOI] [PubMed] [Google Scholar]
- Péron J, Marreaud S, Staelens D, Raveloarivahy T, Nzokirantevye A, Flament J, Steuve J, Lia M, Collette L, Schöffski P (2019) A multinational, multi‐tumour basket study in very rare cancer types: the European Organization for Research and Treatment of Cancer phase II 90101 “CREATE” trial. Eur J Cancer 109: 192–195 [DOI] [PubMed] [Google Scholar]
- Perry JA, Kiezun A, Tonzi P, Van Allen EM, Carter SL, Baca SC, Cowley GS, Bhatt AS, Rheinbay E, Pedamallu CS et al (2014) Complementary genomic approaches highlight the PI3K/mTOR pathway as a common vulnerability in osteosarcoma. Proc Natl Acad Sci USA 111: E5564–E5573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perut F, Roncuzzi L, Zini N, Massa A, Baldini N (2019) Extracellular nanovesicles secreted by human osteosarcoma cells promote angiogenesis. Cancers (Basel) 11: 779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petitprez F, de Reyniès A, Keung EZ, Chen TW‐W, Sun C‐M, Calderaro J, Jeng Y‐M, Hsiao L‐P, Lacroix L, Bougoüin A et al (2020) B cells are associated with survival and immunotherapy response in sarcoma. Nature 577: 556–560 [DOI] [PubMed] [Google Scholar]
- Picarda G, Lamoureux F, Geffroy L, Delepine P, Montier T, Laud K, Tirode F, Delattre O, Heymann D, Rédini F (2010) Preclinical evidence that use of TRAIL in Ewing's sarcoma and osteosarcoma therapy inhibits tumor growth, prevents osteolysis, and increases animal survival. Clin Cancer Res 16: 2363–2374 [DOI] [PubMed] [Google Scholar]
- Pillai SR, Damaghi M, Marunaka Y, Spugnini EP, Fais S, Gillies RJ (2019) Causes, consequences, and therapy of tumors acidosis. Cancer Metastasis Rev 38: 205–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollack SM, He Q, Yearley JH, Emerson R, Vignali M, Zhang Y, Redman MW, Baker KK, Cooper S, Donahue B et al (2017) T‐cell infiltration and clonality correlate with programmed cell death protein 1 and programmed death‐ligand 1 expression in patients with soft tissue sarcomas. Cancer 123: 3291–3304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollack SM, Ingham M, Spraker MB, Schwartz GK (2018) Emerging targeted and immune‐based therapies in sarcoma. J Clin Oncol 36: 125–135 [DOI] [PubMed] [Google Scholar]
- Pomella S, Rota R (2020) The CRISP(Y) future of pediatric soft tissue sarcomas. Front Chem 8: 178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter BK, Adams SC, Pitcher JD, Temple HT (2008) Local recurrence of disease after unplanned excisions of high‐grade soft tissue sarcomas. Clin Orthop Relat Res 466: 3093–3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pretell‐Mazzini J, Barton MD, Conway SA, Temple HT (2015) Unplanned excision of soft‐tissue sarcomas: Current concepts for management and prognosis. J Bone Joint Surg Am 97: 597–603 [DOI] [PubMed] [Google Scholar]
- Przybyl J, van de Rijn M, Rutkowski P (2019) Detection of SS18‐SSX1/2 fusion transcripts in circulating tumor cells of patients with synovial sarcoma. Diagn Pathol 14: 24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Que Y, Fang Z, Guan Y, Xiao W, Xu B, Zhao J, Chen H, Zhang X, Zeng M, Liang Y et al (2019) LAG‐3 expression on tumor‐infiltrating T cells in soft tissue sarcoma correlates with poor survival. Cancer Biol Med 16: 331–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Que Y, Xiao W, Guan Y‐X, Liang Y, Yan S‐M, Chen H‐Y, Li Q‐Q, Xu B‐S, Zhou Z‐W, Zhang X (2017) PD‐L1 expression is associated with FOXP3+ regulatory T‐cell infiltration of soft tissue sarcoma and poor patient prognosis. J Cancer 8: 2018–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainusso N, Cleveland H, Hernandez JA, Quintanilla NM, Hicks J, Vasudevan S, Marco RAW, Allen‐Rhoades W, Wang LL, Yustein JT (2019) Generation of patient‐derived tumor xenografts from percutaneous tumor biopsies in children with bone sarcomas. Pediatr Blood Cancer 66: e27579 [DOI] [PubMed] [Google Scholar]
- Ray‐Coquard I, Serre D, Reichardt P, Martín‐Broto J, Bauer S (2018) Options for treating different soft tissue sarcoma subtypes. Future Oncol 14: 25–49 [DOI] [PubMed] [Google Scholar]
- Recasens A, Munoz L (2019) Targeting cancer cell dormancy. Trends Pharmacol Sci 40: 128–141 [DOI] [PubMed] [Google Scholar]
- Reed DR, Hayashi M, Wagner L, Binitie O, Steppan DA, Brohl AS, Shinohara ET, Bridge JA, Loeb DM, Borinstein SC et al (2017) Treatment pathway of bone sarcoma in children, adolescents, and young adults. Cancer 123: 2206–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren L, Hong ES, Mendoza A, Issaq S, Tran Hoang C, Lizardo M, LeBlanc A, Khanna C (2017) Metabolomics uncovers a link between inositol metabolism and osteosarcoma metastasis. Oncotarget 8: 38541–38553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey V, Menendez ST, Estupiñan O, Rodriguez A, Santos L, Tornin J, Martinez‐Cruzado L, Castillo D, Ordoñez GR, Costilla S et al (2019) New chondrosarcoma cell lines with preserved stem cell properties to study the genomic drift during in vitro/in vivo growth. J Clin Med 8: 455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribas A, Dummer R, Puzanov I, VanderWalde A, Andtbacka RHI, Michielin O, Olszanski AJ, Malvehy J, Cebon J, Fernandez E et al (2017) Oncolytic virotherapy promotes intratumoral T cell infiltration and improves anti‐PD‐1 immunotherapy. Cell 170: 1109–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richert I, Gomez‐Brouchet A, Bouvier C, Du Bouexic De Pinieux G, Karanian MA, Blay J‐Y, Dutour A (2019) The immune landscape of chondrosarcoma reveals an immunosuppressive environment in the dedifferentiated subtypes and exposes CSFR1+ macrophages as a promising therapeutic target. J Bone Oncol 20: 100271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riethmüller G, Klein CA (2001) Early cancer cell dissemination and late metastatic relapse: Clinical reflections and biological approaches to the dormancy problem in patients. Semin Cancer Biol 11: 307–311 [DOI] [PubMed] [Google Scholar]
- Riggi N, Knoechel B, Gillespie SM, Rheinbay E, Boulay G, Suvà ML, Rossetti NE, Boonseng WE, Oksuz O, Cook EB et al (2014) EWS‐FLI1 utilizes divergent chromatin remodeling mechanisms to directly activate or repress enhancer elements in Ewing sarcoma. Cancer Cell 26: 668–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggi N, Suvà M‐L, De Vito C, Provero P, Stehle J‐C, Baumer K, Cironi L, Janiszewska M, Petricevic T, Suvà D et al (2010) EWS‐FLI‐1 modulates miRNA145 and SOX2 expression to initiate mesenchymal stem cell reprogramming toward Ewing sarcoma cancer stem cells. Genes Dev 24: 916–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts RD, Lizardo MM, Reed DR, Hingorani P, Glover J, Allen‐Rhoades W, Fan T, Khanna C, Sweet‐Cordero EA, Cash T et al (2019) Provocative questions in osteosarcoma basic and translational biology: a report from the Children's Oncology Group. Cancer 125: 3514–3525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez R, Rubio R, Menendez P (2012) Modeling sarcomagenesis using multipotent mesenchymal stem cells. Cell Res 22: 62–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez R, Tornin J, Suarez C, Astudillo A, Rubio R, Yauk C, Williams A, Rosu‐Myles M, Funes JM, Boshoff C et al (2013) Expression of FUS‐CHOP fusion protein in immortalized/transformed human mesenchymal stem cells drives mixoid liposarcoma formation. Stem Cells 31: 2061–2072 [DOI] [PubMed] [Google Scholar]
- Rodríguez‐García A, Giménez‐Alejandre M, Rojas JJ, Moreno R, Bazan‐Peregrino M, Cascalló M, Alemany R (2015) Safety and efficacy of VCN‐01, an oncolytic adenovirus combining fiber HSG‐binding domain replacement with RGD and hyaluronidase expression. Clin Cancer Res 21: 1406–1418 [DOI] [PubMed] [Google Scholar]
- Rodríguez‐Núñez P, Romero‐Pérez L, Amaral AT, Puerto‐Camacho P, Jordán C, Marcilla D, Grünewald TGP, Alonso J, de Alava E, Díaz‐Martín J (2019) Hippo pathway effectors YAP1/TAZ induce an EWS‐FLI1‐opposing gene signature and associate with disease progression in Ewing sarcoma. J Pathol 250: 374–386 [DOI] [PubMed] [Google Scholar]
- Rogers MS, Novak K, Zurakowski D, Cryan LM, Blois A, Lifshits E, Bø TH, Oyan AM, Bender ER, Lampa M et al (2014) Spontaneous reversion of the angiogenic phenotype to a nonangiogenic and dormant state in human tumors. Mol Cancer Res 12: 754–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roncuzzi L, Pancotti F, Baldini N (2014) Involvement of HIF‐1α activation in the doxorubicin resistance of human osteosarcoma cells. Oncol Rep 32: 389–394 [DOI] [PubMed] [Google Scholar]
- Rosen G, Murphy ML, Huvos AG, Gutierrez M, Marcove RC (1976) Chemotherapy, en bloc resection, and prosthetic bone replacement in the treatment of osteogenic sarcoma. Cancer 37: 1–11 [DOI] [PubMed] [Google Scholar]
- Rougraff BT, Simon MA, Kneisl JS, Greenberg DB, Mankin HJ (1994) Limb salvage compared with amputation for osteosarcoma of the distal end of the femur. A long‐term oncological, functional, and quality‐of‐life study. J Bone Joint Surg Am 76: 649–656 [DOI] [PubMed] [Google Scholar]
- Roundhill EA, Jabri S, Burchill SA (2019) ABCG1 and Pgp identify drug resistant, self‐renewing osteosarcoma cells. Cancer Lett 453: 142–157 [DOI] [PubMed] [Google Scholar]
- Rugo HS, Jacobs I, Sharma S, Scappaticci F, Paul TA, Jensen‐Pergakes K, Malouf GG (2020) The promise for histone methyltransferase inhibitors for epigenetic therapy in clinical oncology: a narrative review. Adv Ther 37: 3059–3082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkowski P, Kamińska J, Kowalska M, Ruka W, Steffen J (2003) Cytokine and cytokine receptor serum levels in adult bone sarcoma patients: correlations with local tumor extent and prognosis. J Surg Oncol 84: 151–159 [DOI] [PubMed] [Google Scholar]
- Salanti A, Clausen TM, Agerbæk MØ, Al Nakouzi N, Dahlbäck M, Oo HZ, Lee S, Gustavsson T, Rich JR, Hedberg BJ et al (2015) Targeting human cancer by a glycosaminoglycan binding malaria protein. Cancer Cell 28: 500–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salawu A, Fernando M, Hughes D, Reed MWR, Woll P, Greaves C, Day C, Alhajimohammed M, Sisley K (2016) Establishment and molecular characterisation of seven novel soft‐tissue sarcoma cell lines. Br J Cancer 115: 1058–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salerno M, Avnet S, Bonuccelli G, Eramo A, De Maria R, Gambarotti M, Gamberi G, Baldini N (2013) Sphere‐forming cell subsets with cancer stem cell properties in human musculoskeletal sarcomas. Int J Oncol 43: 95–102 [DOI] [PubMed] [Google Scholar]
- Salerno M, Avnet S, Bonuccelli G, Hosogi S, Granchi D, Baldini N (2014) Impairment of lysosomal activity as a therapeutic modality targeting cancer stem cells of embryonal rhabdomyosarcoma cell line RD. PLoS ONE 9: e110340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgado R, Moore H, Martens JWM, Lively T, Malik S, McDermott U, Michiels S, Moscow JA, Tejpar S, McKee T et al (2018) Steps forward for cancer precision medicine. Nat Rev Drug Discov 17: 1–2 [DOI] [PubMed] [Google Scholar]
- Sannino G, Marchetto A, Kirchner T, Grünewald TGP (2017) Epithelial‐to‐mesenchymal and mesenchymal‐to‐epithelial transition in mesenchymal tumors: a paradox in sarcomas? Cancer Res 77: 4556–4561 [DOI] [PubMed] [Google Scholar]
- Sannino G, Marchetto A, Ranft A, Jabar S, Zacherl C, Alba‐Rubio R, Stein S, Wehweck FS, Kiran MM, Hölting TLB et al (2019) Gene expression and immunohistochemical analyses identify SOX2 as major risk factor for overall survival and relapse in Ewing sarcoma patients. EBioMedicine 47: 156–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro M, Lamhamedi‐Cherradi S‐E, Menegaz BA, Ludwig JA, Mikos AG (2015) Flow perfusion effects on three‐dimensional culture and drug sensitivity of Ewing sarcoma. Proc Natl Acad Sci USA 112: 10304–10309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satelli A, Mitra A, Cutrera JJ, Devarie M, Xia X, Ingram DR, Dibra D, Somaiah N, Torres KE, Ravi V et al (2014) Universal marker and detection tool for human sarcoma circulating tumor cells. Cancer Res 74: 1645–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayles LC, Breese MR, Koehne AL, Leung SG, Lee AG, Liu H‐Y, Spillinger A, Shah AT, Tanasa B, Straessler K et al (2019) Genome‐informed targeted therapy for osteosarcoma. Cancer Discov 9: 46–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiavone K, Garnier D, Heymann M‐F, Heymann D (2019) The heterogeneity of osteosarcoma: the role played by cancer stem cells. Adv Exp Med Biol 1139: 187–200 [DOI] [PubMed] [Google Scholar]
- Schneider JR, Patel NV, Kwan K, Boockvar JA (2018) Recurrent glioblastoma treated with recombinant poliovirus. Neurosurgery 83: E200 [DOI] [PubMed] [Google Scholar]
- Scholten DJ, Timmer CM, Peacock JD, Pelle DW, Williams BO, Steensma MR (2014) Down regulation of wnt signaling mitigates hypoxia‐induced chemoresistance in human osteosarcoma cells. PLoS ONE 9: e111431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotlandi K, Serra M, Nicoletti G, Vaccari M, Manara MC, Nini G, Landuzzi L, Colacci A, Bacci G, Bertoni F et al (1996) Multidrug resistance and malignancy in human osteosarcoma. Cancer Res 56: 2434–2439 [PubMed] [Google Scholar]
- Scott DO, Ghosh A, Di L, Maurer TS (2017) Passive drug permeation through membranes and cellular distribution. Pharmacol Res 117: 94–102 [DOI] [PubMed] [Google Scholar]
- Ségaliny AI, Mohamadi A, Dizier B, Lokajczyk A, Brion R, Lanel R, Amiaud J, Charrier C, Boisson‐Vidal C, Heymann D (2015) Interleukin‐34 promotes tumor progression and metastatic process in osteosarcoma through induction of angiogenesis and macrophage recruitment. Int J Cancer 137: 73–85 [DOI] [PubMed] [Google Scholar]
- Serra M, Scotlandi K, Manara MC, Maurici D, Benini S, Sarti M, Nini G, Barbanti‐Brodano G, Baldini N (1996) Evaluation of P‐glycoprotein expression in soft tissue sarcomas of the extremities. Cytotechnology 19: 253–256 [DOI] [PubMed] [Google Scholar]
- Sheffield NC, Pierron G, Klughammer J, Datlinger P, Schönegger A, Schuster M, Hadler J, Surdez D, Guillemot D, Lapouble E et al (2017) DNA methylation heterogeneity defines a disease spectrum in Ewing sarcoma. Nat Med 23: 386–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shern JF, Chen L, Chmielecki J, Wei JS, Patidar R, Rosenberg M, Ambrogio L, Auclair D, Wang J, Song YK et al (2014) Comprehensive genomic analysis of rhabdomyosarcoma reveals a landscape of alterations affecting a common genetic axis in fusion‐positive and fusion‐negative tumors. Cancer Discov 4: 216–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Li M, Yang R (2020) Tumor‐infiltrating lymphocytes as a feasible adjuvant immunotherapy for osteosarcoma with a poor response to neoadjuvant chemotherapy. Immunotherapy 12: 641–652 [DOI] [PubMed] [Google Scholar]
- Shimizu T, Sugihara E, Yamaguchi‐Iwai S, Tamaki S, Koyama Y, Kamel W, Ueki A, Ishikawa T, Chiyoda T, Osuka S et al (2014) IGF2 preserves osteosarcoma cell survival by creating an autophagic state of dormancy that protects cells against chemotherapeutic stress. Cancer Res 74: 6531–6541 [DOI] [PubMed] [Google Scholar]
- Shiozawa K, Shuting J, Yoshioka Y, Ochiya T, Kondo T (2018) Extracellular vesicle‐encapsulated microRNA‐761 enhances pazopanib resistance in synovial sarcoma. Biochem Biophys Res Commun 495: 1322–1327 [DOI] [PubMed] [Google Scholar]
- Shiraishi D, Fujiwara Y, Horlad H, Saito Y, Iriki T, Tsuboki J, Cheng P, Nakagata N, Mizuta H, Bekki H et al (2018) CD163 is required for protumoral activation of macrophages in human and murine sarcoma. Cancer Res 78: 3255–3266 [DOI] [PubMed] [Google Scholar]
- Shor S, Fadl‐Alla BA, Pondenis HC, Zhang X, Wycislo KL, Lezmi S, Fan TM (2015) Expression of nociceptive ligands in canine osteosarcoma. J Vet Intern Med 29: 268–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla NN, Patel JA, Magnan H, Zehir A, You D, Tang J, Meng F, Samoila A, Slotkin EK, Ambati SR et al (2017) Plasma DNA‐based molecular diagnosis, prognostication, and monitoring of patients with EWSR1 fusion‐positive sarcomas. JCO Precis Oncol 10.1200/PO.16.00028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulman DS, Klega K, Imamovic‐Tuco A, Clapp A, Nag A, Thorner AR, Van Allen E, Ha G, Lessnick SL, Gorlick R et al (2018) Detection of circulating tumour DNA is associated with inferior outcomes in Ewing sarcoma and osteosarcoma: a report from the Children's Oncology Group. Br J Cancer 119: 615–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simard FA, Richert I, Vandermoeten A, Decouvelaere A‐V, Michot J‐P, Caux C, Blay J‐Y, Dutour A (2017) Description of the immune microenvironment of chondrosarcoma and contribution to progression. Oncoimmunology 6: e1265716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon MA, Aschliman MA, Thomas N, Mankin HJ (1986) Limb‐salvage treatment versus amputation for osteosarcoma of the distal end of the femur. J Bone Joint Surg Am 68: 1331–1337 [PubMed] [Google Scholar]
- Siurala M, Bramante S, Vassilev L, Hirvinen M, Parviainen S, Tähtinen S, Guse K, Cerullo V, Kanerva A, Kipar A et al (2015) Oncolytic adenovirus and doxorubicin‐based chemotherapy results in synergistic antitumor activity against soft‐tissue sarcoma. Int J Cancer 136: 945–954 [DOI] [PubMed] [Google Scholar]
- Skoda J, Veselska R (2018) Cancer stem cells in sarcomas: getting to the stemness core. Biochim Biophys Acta Gen Subj 1862: 2134–2139 [DOI] [PubMed] [Google Scholar]
- Slemmons KK, Crose LES, Riedel S, Sushnitha M, Belyea B, Linardic CM (2017) A novel notch‐YAP circuit drives stemness and tumorigenesis in embryonal rhabdomyosarcoma. Mol Cancer Res 15: 1777–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smida J, Xu H, Zhang Y, Baumhoer D, Ribi S, Kovac M, von Luettichau I, Bielack S, O'Leary VB, Leib‐Mösch C et al (2017) Genome‐wide analysis of somatic copy number alterations and chromosomal breakages in osteosarcoma. Int J Cancer 141: 816–828 [DOI] [PubMed] [Google Scholar]
- Smith AG, Macleod KF (2019) Autophagy, cancer stem cells and drug resistance. J Pathol 247: 708–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith HG, Mansfield D, Roulstone V, Kyula‐Currie JN, McLaughlin M, Patel RR, Bergerhoff KF, Paget JT, Dillon MT, Khan A et al (2019) PD‐1 blockade following isolated limb perfusion with vaccinia virus prevents local and distant relapse of soft‐tissue sarcoma. Clin Cancer Res 25: 3443–3454 [DOI] [PubMed] [Google Scholar]
- Smrke A, Wang Y, Simmons C (2020) Update on systemic therapy for advanced soft‐tissue sarcoma. Curr Oncol 27: 25–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snuderl M, Dolgalev I, Heguy A, Walsh MF, Benayed R, Jungbluth AA, Ladanyi M, Karajannis MA (2019) Histone H3K36I mutation in a metastatic histiocytic tumor of the skull and response to sarcoma chemotherapy. Cold Spring Harb Mol Case Stud 5: a004606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder MW, Kircher M, Hill AJ, Daza RM, Shendure J (2016) Cell‐free DNA comprises an in vivo nucleosome footprint that informs its tissues‐of‐origin. Cell 164: 57–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurny C, Kailayangiri S, Jamitzky S, Altvater B, Wardelmann E, Dirksen U, Hardes J, Hartmann W, Rossig C (2018) Programmed cell death ligand 1 (PD‐L1) expression is not a predominant feature in Ewing sarcomas. Pediatr Blood Cancer 10.1002/pbc.26719 [DOI] [PubMed] [Google Scholar]
- Stahl D, Gentles AJ, Thiele R, Gütgemann I (2019) Prognostic profiling of the immune cell microenvironment in Ewing′s Sarcoma Family of Tumors. Oncoimmunology 8: e1674113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbing J, Paz K, Schwartz GK, Wexler LH, Maki R, Pollock RE, Morris R, Cohen R, Shankar A, Blackman G et al (2014) Patient‐derived xenografts for individualized care in advanced sarcoma. Cancer 120: 2006–2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele CD, Tarabichi M, Oukrif D, Webster AP, Ye H, Fittall M, Lombard P, Martincorena I, Tarpey PS, Collord G et al (2019) Undifferentiated sarcomas develop through distinct evolutionary pathways. Cancer Cell 35: 441–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinestel K, Trautmann M, Jansen E‐P, Dirksen U, Rehkämper J, Mikesch J‐H, Gerke JS, Orth MF, Sannino G, Arteaga M‐F et al (2020) Focal adhesion kinase confers pro‐migratory and antiapoptotic properties and is a potential therapeutic target in Ewing sarcoma. Mol Oncol 14: 248–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart E, Federico SM, Chen X, Shelat AA, Bradley C, Gordon B, Karlstrom A, Twarog NR, Clay MR, Bahrami A et al (2017) Orthotopic patient‐derived xenografts of paediatric solid tumours. Nature 549: 96–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart E, Goshorn R, Bradley C, Griffiths LM, Benavente C, Twarog NR, Miller GM, Caufield W, Freeman BB, Bahrami A et al (2014) Targeting the DNA repair pathway in Ewing sarcoma. Cell Rep 9: 829–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiller CA, Trama A, Serraino D, Rossi S, Navarro C, Chirlaque MD, Casali PG, RARECARE Working Group (2013) Descriptive epidemiology of sarcomas in Europe: report from the RARECARE project. Eur J Cancer 49: 684–695 [DOI] [PubMed] [Google Scholar]
- Streby KA, Geller JI, Currier MA, Warren PS, Racadio JM, Towbin AJ, Vaughan MR, Triplet M, Ott‐Napier K, Dishman DJ et al (2017) Intratumoral injection of HSV1716, an oncolytic herpes virus, is safe and shows evidence of immune response and viral replication in young cancer patients. Clin Cancer Res 23: 3566–3574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subhawong TK, Wilky BA (2015) Value added: functional MR imaging in management of bone and soft tissue sarcomas. Curr Opin Oncol 27: 323–331 [DOI] [PubMed] [Google Scholar]
- Sundara YT, Kostine M, Cleven AHG, Bovée JVMG, Schilham MW, Cleton‐Jansen A‐M (2017) Increased PD‐L1 and T‐cell infiltration in the presence of HLA class I expression in metastatic high‐grade osteosarcoma: a rationale for T‐cell‐based immunotherapy. Cancer Immunol Immunother 66: 119–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundby Hall K, Bruland ØS, Bjerkehagen B, Zaikova O, Engellau J, Hagberg O, Hansson L, Hagberg H, Ahlström M, Knobel H et al (2018) Adjuvant chemotherapy and postoperative radiotherapy in high‐risk soft tissue sarcoma patients defined by biological risk factors‐A Scandinavian Sarcoma Group study (SSG XX). Eur J Cancer 99: 78–85 [DOI] [PubMed] [Google Scholar]
- Suzuki K, Fueyo J, Krasnykh V, Reynolds PN, Curiel DT, Alemany R (2001) A conditionally replicative adenovirus with enhanced infectivity shows improved oncolytic potency. Clin Cancer Res 7: 120–126 [PubMed] [Google Scholar]
- Sys GML, Lapeire L, Stevens N, Favoreel H, Forsyth R, Bracke M, De Wever O (2013) The in ovo CAM‐assay as a xenograft model for sarcoma. J Vis Exp 77: e50522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki S, Fukuta M, Sekiguchi K, Jin Y, Nagata S, Hayakawa K, Hineno S, Okamoto T, Watanabe M, Woltjen K et al (2015) SS18‐SSX, the oncogenic fusion protein in synovial sarcoma, is a cellular context‐dependent epigenetic modifier. PLoS ONE 10: e0142991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarnowski M, Grymula K, Liu R, Tarnowska J, Drukala J, Ratajczak J, Mitchell RA, Ratajczak MZ, Kucia M (2010) Macrophage migration inhibitory factor is secreted by rhabdomyosarcoma cells, modulates tumor metastasis by binding to CXCR4 and CXCR7 receptors and inhibits recruitment of cancer‐associated fibroblasts. Mol Cancer Res 8: 1328–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor BS, Barretina J, Maki RG, Antonescu CR, Singer S, Ladanyi M (2011) Advances in sarcoma genomics and new therapeutic targets. Nat Rev Cancer 11: 541–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tazawa H, Hasei J, Yano S, Kagawa S, Ozaki T, Fujiwara T (2020) Bone and soft‐tissue sarcoma: a new target for telomerase‐specific oncolytic virotherapy. Cancers (Basel) 12: 478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tellez‐Gabriel M, Brown HK, Young R, Heymann M‐F, Heymann D (2016) The challenges of detecting circulating tumor cells in sarcoma. Front Oncol 6: 202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tellez‐Gabriel M, Heymann M‐F, Heymann D (2019) Circulating tumor cells as a tool for assessing tumor heterogeneity. Theranostics 9: 4580–4594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanindratarn P, Dean DC, Nelson SD, Hornicek FJ, Duan Z (2019) Advances in immune checkpoint inhibitors for bone sarcoma therapy. J Bone Oncol 15: 100221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théoleyre S, Mori K, Cherrier B, Passuti N, Gouin F, Rédini F, Heymann D (2005) Phenotypic and functional analysis of lymphocytes infiltrating osteolytic tumors: use as a possible therapeutic approach of osteosarcoma. BMC Cancer 5: 123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, Antoniou A, Arab T, Archer F, Atkin‐Smith GK et al (2018) Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles 7: 1535750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel U, Koscielniak E, Blaeschke F, Grunewald TGP, Badoglio M, Diaz MA, Paillard C, Prete A, Ussowicz M, Lang P et al (2013) Allogeneic stem cell transplantation for patients with advanced rhabdomyosarcoma: a retrospective assessment. Br J Cancer 109: 2523–2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiriac H, Belleau P, Engle DD, Plenker D, Deschênes A, Somerville TDD, Froeling FEM, Burkhart RA, Denroche RE, Jang G‐H et al (2018) Organoid profiling identifies common responders to chemotherapy in pancreatic cancer. Cancer Discov 8: 1112–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirode F, Surdez D, Ma X, Parker M, Le Deley MC, Bahrami A, Zhang Z, Lapouble E, Grossetête‐Lalami S, Rusch M et al (2014) Genomic landscape of Ewing sarcoma defines an aggressive subtype with co‐association of STAG2 and TP53 mutations. Cancer Discov 4: 1342–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomazou EM, Sheffield NC, Schmidl C, Schuster M, Schönegger A, Datlinger P, Kubicek S, Bock C, Kovar H (2015) Epigenome mapping reveals distinct modes of gene regulation and widespread enhancer reprogramming by the oncogenic fusion protein EWS‐FLI1. Cell Rep 10: 1082–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornin J, Martinez‐Cruzado L, Santos L, Rodriguez A, Núñez L‐E, Oro P, Hermosilla MA, Allonca E, Fernández‐García MT, Astudillo A et al (2016) Inhibition of SP1 by the mithramycin analog EC‐8042 efficiently targets tumor initiating cells in sarcoma. Oncotarget 7: 30935–30950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toulmonde M, Penel N, Adam J, Chevreau C, Blay J‐Y, Le Cesne A, Bompas E, Piperno‐Neumann S, Cousin S, Grellety T et al (2018) Use of PD‐1 targeting, macrophage infiltration, and IDO pathway activation in sarcomas: a phase 2 clinical trial. JAMA Oncol 4: 93–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trama A, Badalamenti G, Baldi GG, Brunello A, Caira M, Drove N, Marrari A, Palmerini E, Vincenzi B, Dei Tos AP et al (2019) Soft tissue sarcoma in Italy: from epidemiological data to clinical networking to improve patient care and outcomes. Cancer Epidemiol 59: 258–264 [DOI] [PubMed] [Google Scholar]
- Traub F, Griffin AM, Wunder JS, Ferguson PC (2018) Influence of unplanned excisions on the outcomes of patients with stage III extremity soft‐tissue sarcoma. Cancer 124: 3868–3875 [DOI] [PubMed] [Google Scholar]
- Trautmann M, Cheng Y‐Y, Jensen P, Azoitei N, Brunner I, Hüllein J, Slabicki M, Isfort I, Cyra M, Berthold R et al (2019) Requirement for YAP1 signaling in myxoid liposarcoma. EMBO Mol Med 11: e9889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trieb K, Lechleitner T, Lang S, Windhager R, Kotz R, Dirnhofer S (1998) Evaluation of HLA‐DR expression and T‐lymphocyte infiltration in osteosarcoma. Pathol Res Pract 194: 679–684 [DOI] [PubMed] [Google Scholar]
- Tuncbilek N, Karakas HM, Okten OO (2005) Dynamic contrast enhanced MRI in the differential diagnosis of soft tissue tumors. Eur J Radiol 53: 500–505 [DOI] [PubMed] [Google Scholar]
- Ulz P, Thallinger GG, Auer M, Graf R, Kashofer K, Jahn SW, Abete L, Pristauz G, Petru E, Geigl JB et al (2016) Inferring expressed genes by whole‐genome sequencing of plasma DNA. Nat Genet 48: 1273–1278 [DOI] [PubMed] [Google Scholar]
- Uotani K, Fujiwara T, Yoshida A, Iwata S, Morita T, Kiyono M, Yokoo S, Kunisada T, Takeda K, Hasei J et al (2017) Circulating MicroRNA‐92b‐3p as a novel biomarker for monitoring of synovial sarcoma. Sci Rep 7: 14634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaira V, Fedele G, Pyne S, Fasoli E, Zadra G, Bailey D, Snyder E, Faversani A, Coggi G, Flavin R et al (2010) Preclinical model of organotypic culture for pharmacodynamic profiling of human tumors. Proc Natl Acad Sci USA 107: 8352–8356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakkila J, Jaffe R, Michelow M, Lotze MT (2006) Pediatric cancers are infiltrated predominantly by macrophages and contain a paucity of dendritic cells: a major nosologic difference with adult tumors. Clin Cancer Res 12: 2049–2054 [DOI] [PubMed] [Google Scholar]
- Vallette FM, Olivier C, Lézot F, Oliver L, Cochonneau D, Lalier L, Cartron P‐F, Heymann D (2019) Dormant, quiescent, tolerant and persister cells: four synonyms for the same target in cancer. Biochem Pharmacol 162: 169–176 [DOI] [PubMed] [Google Scholar]
- Varani J, Lovett EJ, Lundy J (1981) A model of tumor cell dormancy: effects of anesthesia and surgery. J Surg Oncol 17: 9–14 [DOI] [PubMed] [Google Scholar]
- Varela‐Guruceaga M, Tejada‐Solís S, García‐Moure M, Fueyo J, Gomez‐Manzano C, Patiño‐García A, Alonso MM (2018) Oncolytic viruses as therapeutic tools for pediatric brain tumors. Cancers (Basel) 10: 226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasan N, Baselga J, Hyman DM (2019) A view on drug resistance in cancer. Nature 575: 299–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasquez L, León E, Beltran B, Maza I, Oscanoa M, Geronimo J (2017) Pretreatment neutrophil‐to‐lymphocyte ratio and lymphocyte recovery: independent prognostic factors for survival in pediatric sarcomas. J Pediatr Hematol Oncol 39: 538–546 [DOI] [PubMed] [Google Scholar]
- Vaupel P, Okunieff P, Kallinowski F, Neuringer LJ (1989) Correlations between 31P‐NMR spectroscopy and tissue O2 tension measurements in a murine fibrosarcoma. Radiat Res 120: 477–493 [PubMed] [Google Scholar]
- Vaupel P, Schaefer C, Okunieff P (1994) Intracellular acidosis in murine fibrosarcomas coincides with ATP depletion, hypoxia, and high levels of lactate and total Pi. NMR Biomed 7: 128–136 [DOI] [PubMed] [Google Scholar]
- Visvader JE (2011) Cells of origin in cancer. Nature 469: 314–322 [DOI] [PubMed] [Google Scholar]
- Vlachogiannis G, Hedayat S, Vatsiou A, Jamin Y, Fernández‐Mateos J, Khan K, Lampis A, Eason K, Huntingford I, Burke R et al (2018) Patient‐derived organoids model treatment response of metastatic gastrointestinal cancers. Science 359: 920–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlenterie M, Oyen WJ, Steeghs N, Desar IME, Verheijen RB, Koenen AM, Grootjans W, DE Geus‐Oei L‐F, Van Erp NP, Van der Graaf WT (2019) Early metabolic response as a predictor of treatment outcome in patients with metastatic soft tissue sarcomas. Anticancer Res 39: 1309–1316 [DOI] [PubMed] [Google Scholar]
- Volckmar A‐L, Sültmann H, Riediger A, Fioretos T, Schirmacher P, Endris V, Stenzinger A, Dietz S (2018) A field guide for cancer diagnostics using cell‐free DNA: from principles to practice and clinical applications. Genes Chromosom Cancer 57: 123–139 [DOI] [PubMed] [Google Scholar]
- Vyse S, McCarthy F, Broncel M, Paul A, Wong JP, Bhamra A, Huang PH (2018) Quantitative phosphoproteomic analysis of acquired cancer drug resistance to pazopanib and dasatinib. J Proteomics 170: 130–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacnik PW, Baker CM, Herron MJ, Kren BT, Blazar BR, Wilcox GL, Hordinsky MK, Beitz AJ, Ericson ME (2005) Tumor‐induced mechanical hyperalgesia involves CGRP receptors and altered innervation and vascularization of DsRed2 fluorescent hindpaw tumors. Pain 115: 95–106 [DOI] [PubMed] [Google Scholar]
- Wang M, Wang L, Ren T, Xu L, Wen Z (2013) IL‐17A/IL‐17RA interaction promoted metastasis of osteosarcoma cells. Cancer Biol Ther 14: 155–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Yang DM, Rong R, Zhan X, Xiao G (2019) Pathology image analysis using segmentation deep learning algorithms. Am J Pathol 189: 1686–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S, Shimomura A, Kubo T, Sekimizu M, Seo T, Watanabe S‐I, Kawai A, Yamamoto N, Tamura K, Kohno T et al (2020) BRAF V600E mutation is a potential therapeutic target for a small subset of synovial sarcoma. Mod Pathol 33: 1660–1668 [DOI] [PubMed] [Google Scholar]
- Watson S, Perrin V, Guillemot D, Reynaud S, Coindre J‐M, Karanian M, Guinebretière J‐M, Freneaux P, Loarer FL, Bouvet M et al (2018) Transcriptomic definition of molecular subgroups of small round cell sarcomas. J Pathol 245: 29–40 [DOI] [PubMed] [Google Scholar]
- Wei S, Henderson‐Jackson E, Qian X, Bui MM (2017) Soft tissue tumor immunohistochemistry update: illustrative examples of diagnostic pearls to avoid pitfalls. Arch Pathol Lab Med 141: 1072–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidema ME, van de Geer E, Koelsche C, Desar IME, Kemmeren P, Hillebrandt‐Roeffen MHS, Ho VKY, van der Graaf WTA, Versleijen‐Jonkers YMH, von Deimling A et al (2020) DNA methylation profiling identifies distinct clusters in angiosarcomas. Clin Cancer Res 26: 93–100 [DOI] [PubMed] [Google Scholar]
- Whelan JS, Davis LE (2018) Osteosarcoma, chondrosarcoma, and chordoma. J Clin Oncol 36: 188–193 [DOI] [PubMed] [Google Scholar]
- WHO Classification of Tumours: Soft Tissue and Bone Tumours (2020). WHO Editorial Board WHO Classification of Tumours: Soft Tissue and Bone Tumours, 5th Ed. 978‐92-8324502‐5 (IARC).
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA (2013) Master transcription factors and mediator establish super‐enhancers at key cell identity genes. Cell 153: 307–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilding CP, Elms ML, Judson I, Tan A‐C, Jones RL, Huang PH (2019) The landscape of tyrosine kinase inhibitors in sarcomas: looking beyond pazopanib. Expert Rev Anticancer Ther 19: 971–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson MJ, Smith HG, Pencavel TD, Mansfield DC, Kyula‐Currie J, Khan AA, McEntee G, Roulstone V, Hayes AJ, Harrington KJ (2016) Isolated limb perfusion with biochemotherapy and oncolytic virotherapy combines with radiotherapy and surgery to overcome treatment resistance in an animal model of extremity soft tissue sarcoma. Int J Cancer 139: 1414–1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witlox AM, Van Beusechem VW, Molenaar B, Bras H, Schaap GR, Alemany R, Curiel DT, Pinedo HM, Wuisman PIJM, Gerritsen WR (2004) Conditionally replicative adenovirus with tropism expanded towards integrins inhibits osteosarcoma tumor growth in vitro and in vivo . Clin Cancer Res 10: 61–67 [DOI] [PubMed] [Google Scholar]
- Wolsztynski E, O'Sullivan F, Keyes E, O'Sullivan J, Eary JF (2018) Positron emission tomography‐based assessment of metabolic gradient and other prognostic features in sarcoma. J Med Imaging (Bellingham) 5: 024502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L‐L, Tang M, Zhang Z‐L, Qi C‐B, Hu J, Ma X‐Y, Pang D‐W (2018) Chip‐assisted single‐cell biomarker profiling of heterogeneous circulating tumor cells using multifunctional nanospheres. Anal Chem 90: 10518–10526 [DOI] [PubMed] [Google Scholar]
- Xiao H, Chen L, Luo G, Son H, Prectoni JH, Zheng W (2014) Effect of the cytokine levels in serum on osteosarcoma. Tumour Biol 35: 1023–1028 [DOI] [PubMed] [Google Scholar]
- Xiao W, Mohseny AB, Hogendoorn PCW, Cleton‐Jansen A‐M (2013) Mesenchymal stem cell transformation and sarcoma genesis. Clin Sarcoma Res 3: 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X, Wang W, Li Y, Yang D, Li X, Shen C, Liu Y, Ke X, Guo S, Guo Z (2018) HSP90AA1‐mediated autophagy promotes drug resistance in osteosarcoma. J Exp Clin Cancer Res 37: 201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J‐F, Wang Y‐P, Zhang S‐J, Chen Y, Gu H‐F, Dou X‐F, Xia B, Bi Q, Fan S‐W (2017) Exosomes containing differential expression of microRNA and mRNA in osteosarcoma that can predict response to chemotherapy. Oncotarget 8: 75968–75978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Han L, He Z, Li X, Yang S, Yang J, Zhang Y, Li D, Yang Y, Yang Z (2017) Advances in limb salvage treatment of osteosarcoma. J Bone Oncol 10: 36–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo C, Ryu M‐H, Na YS, Ryoo B‐Y, Park SR, Kang Y‐K (2014) Analysis of serum protein biomarkers, circulating tumor DNA, and dovitinib activity in patients with tyrosine kinase inhibitor‐refractory gastrointestinal stromal tumors. Ann Oncol 25: 2272–2277 [DOI] [PubMed] [Google Scholar]
- Yu L, Fan Z, Fang S, Yang J, Gao T, Simões BM, Eyre R, Guo W, Clarke RB (2016) Cisplatin selects for stem‐like cells in osteosarcoma by activating Notch signaling. Oncotarget 7: 33055–33068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W, Wan R, Zheng Y, Singh SR, Wei Y (2011) Hypoxia, stem cells and bone tumor. Cancer Lett 313: 129–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Wu H, Zheng J, Yu P, Xu L, Jiang P, Gao J, Wang H, Zhang Y (2013) Transforming growth factor β1 signal is crucial for dedifferentiation of cancer cells to cancer stem cells in osteosarcoma. Stem Cells 31: 433–446 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Qiu Y, Hua Y, Wang Y, Chen T, Zhao A, Chi Y, Pan L, Hu S, Li J et al (2010) Serum and urinary metabonomic study of human osteosarcoma. J Proteome Res 9: 4861–4868 [DOI] [PubMed] [Google Scholar]
- Zhitomirsky B, Assaraf YG (2016) Lysosomes as mediators of drug resistance in cancer. Drug Resist Updat 24: 23–33 [DOI] [PubMed] [Google Scholar]
- Zhu R, Li X, Ma Y (2019) miR‐23b‐3p suppressing PGC1α promotes proliferation through reprogramming metabolism in osteosarcoma. Cell Death Dis 10: 381 [DOI] [PMC free article] [PubMed] [Google Scholar]


