Abstract
The beautiful interplay between light and matter can give rise to many striking physical phenomena, surface plasmon resonance (SPR) being one of them. Plasmonic immunosensors monitor refractive index changes that occur as a result of specific ligand–analyte or antibody–antigen interactions taking place on the sensor surface. The coronavirus disease (COVID‐19) pandemic has jeopardized the entire world and has resulted in economic slowdown of most countries. In this work, a model of a sandwich plasmonic biosensor that utilizes gold nanorods (Au NRs) for the detection of COVID‐19 SARS‐CoV‐2 spike protein is presented. Simulation results for different prismatic configurations for the basic Kretschmann layout are presented. It is found that a BK7 glass prism‐based SPR sensor has an incremental sensitivity of 111.11 deg RIU−1. Additionally, using Comsol Multiphysics the electric field enhancement observed for various aspect ratios and layouts of Au NRs are discussed in depth.
Keywords: COVID‐19, gold nanorods, kretschmann layouts, plasmonic immunosensors
The coronavirus disease (COVID‐19) pandemic has adversely affected the entire world, where apart from stressing the medical fraternity, it has resulted in a major economic slowdown. In this paper, a unique detection scheme for quantifying SARS‐CoV‐2 virus using a plasmonic immunosensor that uses gold nanorods for signal amplification is presented.
1. Introduction
A sudden outbreak of several unexplained instances of pneumonia in Wuhan, China in the last week of December 2019 alarmed the medical community. Later, a few months down the line, the scenario changed completely and now the outbreak has spread globally and paralyzed 200 countries which included United States of America and many European countries as the ones who were badly affected. The outbreak was found to be caused by a unique strain of coronavirus (CoV) that was named as the novel coronavirus disease (COVID‐19).[ 1 , 2 , 3 , 4 , 5 ] The World Health Organization has declared the situation to be a pandemic where it is still evolving in many countries like India. The COVID‐19 virus has symptoms similar to earlier outbreaks caused by the severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS) CoVs. Both COVID‐19 and SARS‐CoV viruses have the same structure with 70% match in the sequence of their constituent amino acids. Also, their mode of entry into the human body is by binding with the same receptor called angiotensin‐converting enzyme 2. The COVID‐19 virus was thus named as SARS‐CoV‐2.[ 6 , 7 , 8 , 9 , 10 ] All the three viruses (SARS‐CoV, MERS‐CoV, and SARS‐CoV‐2) are positive stranded RNA viruses that belong to the betacoronoviride family. The virus can cause mild to acute respiratory infections and has varied symptoms like fever, cough, difficulty breathing, loss of taste, and smell. Due to its ability to mutate into different forms, its detection has not been easy. Elderly people, infants, adolescent children, and people with compromised immune system are the ones at maximum risk of contracting the virus.[ 11 , 12 , 13 , 14 , 15 ]
The SARS‐CoV‐2 genome comprises of 27 proteins that includes four structural and other nonstructural proteins. The four structural proteins are the spike (S), envelope (E), nucleocapsid (N), and matrix (M) proteins. Being a surface protein, the S protein is primarily responsible for entry into the host cell. The virus was initially diagnosed using molecular techniques like reverse transcription polymerase chain reaction (RT‐PCR). However, considering the increasing number of cases, the PCR kits have not been able to meet the testing demand.[ 16 , 17 , 18 , 19 , 20 ] Also, underdeveloped and developing countries where there are many people who still live in rural areas, lack proper PCR infrastructure in rural and suburban areas. Finally, since the technique detects the genome structure of the live virus present in the body, it cannot identify recovered asymptomatic patients. Thus, there has been ongoing research on finding easier and reliable detection methods. Serological testing has long been used for the detection of many viruses like human immunodeficiency virus, SARS‐CoV, and MERS‐CoV. This testing scheme mainly utilizes the reactivity between specific antibodies developed by the human immune system in response to the presence of viral antigens.[ 21 , 22 , 23 , 24 , 25 , 26 , 27 , 28 , 29 , 30 ] Many researchers have come up with unique schemes for detecting the spike protein of the SARS‐CoV‐2 virus. Mahari et al. used fluorine doped tin oxide electrode along with Au nanoparticles as a detection scheme.[ 26 ] Zhang et al. designed a graphene field effect transistor (FET) that was highly sensitive to the SARS‐CoV‐2 virus spike protein antibody–antigen interaction.[ 27 ]
Plasmons are generated as a result of electromagnetic light wave interacting with the free surface electrons on nanosized metals. Surface plasmon polaritons (SPP) are electromagnetic (EM) waves that travel along the surface of the metals. The excitation of plasmons requires proper structural arrangement and can only be achieved by incorporating special techniques like the attenuated total reflection configuration. Direct coupling of incident light wave (LW) on the metal surface cannot excite the plasmons as the incident light wave vector is lower than the propagation constant of the surface plasmon wave (SPW). By allowing the incident light to be passed through a high refractive index (R.I.) prism, the wavevector of the incident LW can be enhanced and matched to the propagation constant of the SPW.[ 31 , 32 , 33 , 34 , 35 ] Thus, the standard Kretschmann model utilizes a high R.I. prism like SF10, SF11 or BK7. A nanosized plasmonic metal film, usually made of 50 nm Au is coated on the prism. For the standard configuration, there are three different media. Medium 1 is the high R.I. prism. Medium 2 is that of the plasmonic metal and medium 3 is the analyte. For most commercial sensors, the analyte comprises of the running buffer like the Hepes buffered saline used by Biacore SPR systems. The matching of the propagation constant takes place only at a particular incident angle known as the SPR angle. Since this angle is highly sensitive to R.I. changes of the analyte, any perturbation in the composition of the analyte like ligand–analyte interaction or antibody–antigen binding can result in the movement of the SPR angle and a corresponding change in the real‐time response curve, also known as sensorgram, in case of commercial sensors. If used carefully with proper and timely maintenance of commercial sensors, the plasmonic sensing approach can offer many benefits like real‐time and label‐free detection, reusability of sensor chips and high repeatability of results. Many SPR‐based detection schemes have been used for detecting different kinds of viruses.[ 36 , 37 ]
Localized surface plasmon resonance (LSPR) is a unique phenomenon observed in noble metal nanoparticles (MNPs). When the incident EM light wave falls on the MNPs, the electric field interacts with the free electrons and causes them separate from the metal core. The repulsive force acting among the electrons causes them to move in opposite directions that results them to oscillate and thus excite the LSPR. The excitation of LSPR can induce strong light absorption. Au NRs have been enormously studied for many decades and have been used in varied applications like plasmonic photothermal therapy and cancer imaging, and even enhanced sensing and detection. Au NRs generally have two absorption bands namely longitudinal plasmon band (LPB) and transverse plasmon band (TPB) where the electrons oscillate along the long and short axes of the NR, respectively.[ 38 , 39 , 40 , 41 , 42 ] The aspect ratio (AR) has almost no impact on the TPB, whereas the increase of AR redshifts the LPB. Additionally, the LPB is also highly sensitive to R.I. changes. Experimental studies also reveal the linear relationship between the AR and the peak wavelength of LPB. In our current work, with the help of simulation, we demonstrate how Au NR can be utilized for signal amplification of conventional SPR sensors.[ 43 , 44 , 45 , 46 , 47 , 48 , 49 , 50 , 51 , 52 , 53 , 54 , 55 ] Thus, using the antigen–antibody interaction principle, we describe a sandwich‐type plasmonic immunoassay model that uses Au NR for enhancement of output signal. In Sections 2 and 3, we describe the structure, simulation technique along with relevant mathematical equations in Section 2. Section 3 outlines the results and discussion and finally we end with a concluding note in Section 4.
2. Structure, Equation, and Simulation Method
2.1. Sensor Structure
The conventional SPR scheme consists of a plasmonic metal, mainly gold, coated on a high R.I. prism. The structure of our Au NR modified layout is shown in Figure 1 . In this sandwich‐type structure, the gold nanosheet is functionalized with SARS‐CoV‐2 spike (S) protein antibody (Ab). Additionally, Au NRs are conjugated with the same Ab. The antigen is the SARS‐CoV‐2 virus in serum/nasopharyngeal swab samples. The procedure of immobilization starts with functionalization of the Au nanosheet with the S protein Ab. The serum samples that contain the antigen are allowed to flow as an analyte. And finally, the Ab conjugated Au NR is applied for sensitivity enhancement. The electric field enhancement and consequently the improvement in sensitivity depend on many factors like the Au NR AR, the placement of the NR, or the distance between the nanosheet and the NR and also the working wavelength of the sensor. Choosing a proper Au NR that has the peak absorbance near the working wavelength can ensure maximum coupling of local electric fields of Au NR and the evanescent field at the sensor surface. In Section 3, we discuss in depth about how the placement of Au NR can affect the field enhancement and thus it constitutes an important parameter that needs to be assessed while designing immunosensors.
Figure 1.
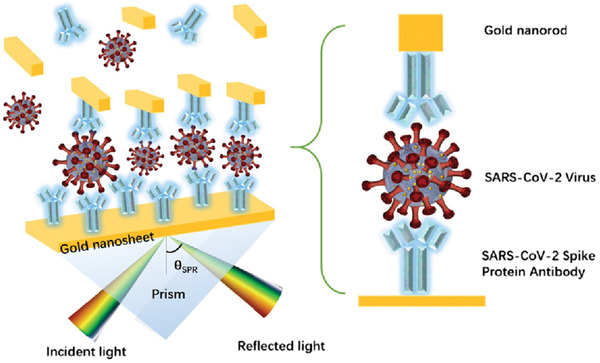
Structure of the plasmonic immunoassay.
2.2. Mathematical Equations
We use angular interrogation technique for our simulation study and utilize Fresnel's Equations (transfer matrix method (TMM)) for arriving at the reflectivity of the multilayer SPR structure
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
| (6) |
Elucidating the variables used in Equations (1) through (6), the total number of layers of the sensor structure is represented by N. For our conventional scheme N = 3. λ is the excitation wavelength. The R.I. and dielectric constant of the k‐th layer are represented by n k and εk, respectively. Additionally, the thickness of the k‐th layer is d k and θ1 is the angle of incidence. The reflectivity, SPR angle and R.I. of the analyte layer are denoted by R p, θSPR, and n bio, respectively. M is th characteristic matrix of the N‐layer model and it is the main matrix of the TMM.
2.3. Simulation Technique
Most commercial SPR sensors work in the NIR spectrum of the electromagnetic light wave. Spreeta SPR sensors manufactured by Texas Instruments are miniaturized form of SPR sensors and are widely used for research purposes. A wide variety of commercial SPR sensors are available in the market like Biacore manufactured by GE, SPRm 200 manufactured by Biosensing Instrument, Carterra LSA SPR platform, IBIS‐MX96 by IBIS Technologies, SensiQ pioneer by SensiQ technologies. However, all these sensors are bulky and are not designed to meet the needs of point‐of‐care (POC) diagnostic technique. Spreeta SPR sensors are low‐cost portable sensors that can be widely deployed for designing POC SPR detection kits. Spangler et al. used these sensors and found that it could detect reasonable quantity of 70 pmol of Escherichia Coli (E. Coli).[ 51 ] Additionally, Chinowsky et al. designed a portable sensing unit using Spreeta sensors and could detect low concentrations of multiple analytes like small molecules, proteins, viruses, and bacteria.[ 52 ] The sensor can provide detection results within a few minutes compared to other laborious methods like enzyme‐linked immunosorbent assay that can take several hours. Soelberg et al. also used these miniaturized SPR sensors for real‐time monitoring of environmental toxins.[ 53 ] Additionally, they have also been used for the development of a portable biosensor system for the detection of Staphylococcal enterotoxin B (SEB).[ 54 ] Hence, we perform the simulation at the working wavelength of these sensors λ = 830 nm. The working R.I. range of these miniature sensors are from 1.32 to 1.368 with a precision of 5E‐6 RIU. Thus, we vary the R.I. of the analyte layer n bio from 5E‐6 to 4.8E‐2 in steps of 5E‐6 and calculate the change in SPR angle . The thickness of the gold nanosheet is 50 nm and we carry out the simulation for three kinds of prism namely SF10, SF11, and BK7. The R.I. of the three prisms at 830 nm are 1.7099, 1.7630, and 1.5102, respectively. Additionally, the R.I. of Au at the working wavelength is 0.2328 + 4.8144i. The first layer of the sensor is the high R.I. prism followed by the second layer of Au, and the third layer is the analyte which is considered as deionized water whose R.I. is chosen to be 1.33.
Since SPPs are electron density waves, the evanescent electric field simulation can provide us a good insight into the sensitivity. The normal component of the electric field decays exponentially as one moves away from the sensing surface toward the analyte. The penetration depth is defined as the distance at which the field becomes 1/e of its value at the sensing surface. Biomolecular interactions that take place in this depth can result in RI changes of the medium. Increased electric field can enhance the penetration depth and thus the probability of the interaction getting detected increases. Hence, by the aid of finite‐difference time domain simulation, we can have an idea of the electric field coupling of the SPPs at the Au film sensor surface and LSPR waves oscillating along the longitudinal axis of the Au NR. For the electric field simulation, we use Comsol Multiphysics. The diameter of the Au NR is chosen to be 10 nm. We vary the AR from 1 to 4 and the distance of the Au NR from the Au nanosheet from 2 to 14 nm in steps of 3 nm. Therefore, in totality we evaluate 20 different scenarios by taking all the combinations of ARs and Au NR distance from nanosheet.
3. Results and Discussion
We perform the simulation at 830 nm. The SPR reflectivity, which is the ratio between incident and reflected light intensities, has been shown in Figure 2 for different scenarios. After crossing the critical angle of incidence, the reflected light undergoes total internal reflection. At a particular incident angle known as the SPR angle, the reflectivity is minimum. Beyond this angle, the reflectivity again starts to increase. Figure 2a shows the SPR reflectivity curve for Au NR with an AR of 1, whereas Figure 2b shows the same for AR = 4. Figure S1 (Supporting Information) shows the same curve for ARs 2 and 3.
Figure 2.
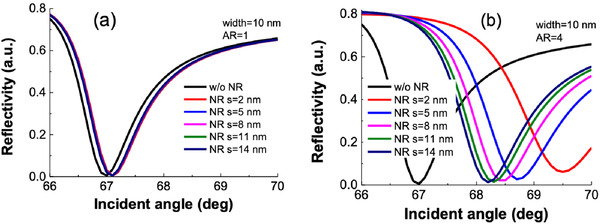
SPR reflectivity curve.
For the angular interrogation technique, we evaluate three different prismatic configurations. By varying the R.I. of the analyte from 5E‐6 to 4.8E‐2 in steps of 5E‐6, we observe the corresponding change in SPR angle. Additionally, for most commercial sensors, the change in R.I. that occurs due to ligand–analyte interaction is represented in the sensorgram, a real‐time response curve, in the form of change in sensor response. A change in 0.1° in the SPR angle results in a change of 1000 RU in the sensorgram response.[ 34 ] Thus, Figure 3 shows the respective SPR angle and sensorgram response changes for the three prism types, namely SF10, SF11, and BK7. From the simulation results, we observe that the SPR angle changes in the form of steps. This is not very evident from the curve which shows a linear variation since the step size is very small. However, the figure inset shows the magnified step variation. The corresponding step sizes for SF10, SF11, and BK7 prism‐based sensors are 1.5E‐4, 1.65E‐4, and 9E‐5 RIU, respectively. This means that only when the R.I. changes by these amounts, the respective sensors can detect a change in the SPR angle. Hence, the lower the step size, the better the sensor resolution. Hence, BK7 prism‐based sensor gives the best sensor resolution among the three prism types. The limit of detection (LOD), which is the minimum R.I. change the sensor can detect, for the SF10, SF11, and BK7‐prism based sensors are 4E‐5, 8.5E‐5, and 7E‐5 RIU, respectively. Thus, in terms of LOD, SF10 prism‐based sensor gives the best result. If we talk about incremental sensitivity, defined as the ratio of change in SPR angle to the step‐size change in RI, the values for the SF10, SF11, and BK7 prism‐based sensors are 66.67, 60.60, and 111.11 deg RIU−1, respectively. Hence, in terms of incremental sensitivity, BK7 prism‐based sensor gives the ideal performance. Thus, designing sensors based on BK7 prism can detect small RI changes easily and hence help in detecting lower concentrations of SARS‐CoV‐2 S protein antigen.
Figure 3.
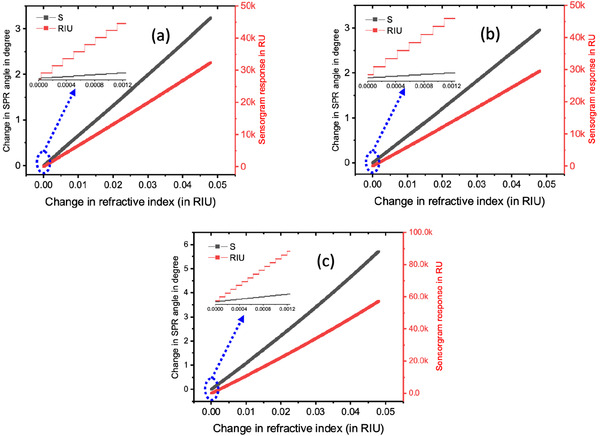
Variation of SPR angle and sensorgram response as a function of change in R.I. of the analyte for a) SF10, b) SF11, and c) BK7 prisms.
For simulation of electric field, we used Comsol Multiphysics. Briefly, the Au NR is represented as a 2D rectangular structure with a width of 10 nm and varying heights depending on the AR. The simulation is carried out at 830 nm. We consider the effect of different ARs from AR = 1 to 4 and varying distances of 2, 5, 8, 11, and 14 nm between the NR and the Au nanosheet. Figure 4 below shows the evanescent field variation as a function of distance from the plasmonic nanosheet. As can be seen the field gradually decreases as one moves away from the metal nanosheet and hence the name evanescent. The penetration depth of this field is defined as the distance at which the field is 1/e times its value at the metal surface. The field normally vanishes and becomes negligible at one‐fourth to one‐third wavelength distance. All ligand–analyte interactions that take place in between this zone can cause R.I. changes of the analyte. Hence, it is essential to keep the entire sensing system compact with minimum distance between the different layers.
Figure 4.
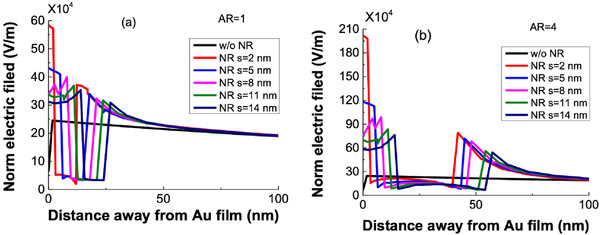
Evanescent decay of the electric field penetrating into the analyte solution from the surface of Au nanosheet along the central line of Au NR at the resonance condition for a) AR = 1 and b) AR = 4.
For the simple Au nanosheet configuration, the electric field is 2.45E5 V m−1. The phenomenon of the plasmonic effect in the nanosheet and the NR are different in nature. While Au NR experiences LSPR, it is the SPPs that travel along the plasmonic metal nanosheet. The LSPR field is concentrated around the nanostructure and decays within 10–30 nm and is thus quite sensitive to distance from the metal sheet and the RI changes of the surrounding area. In contrast to this, SPPs are evanescent in nature that progressively reduce to 0 within 100–200 nm distance. The confined electron cloud oscillation in the Au NRs can produce intense local EM fields that are several orders higher than the incident field. Upon the introduction of Au NR into the plasmonic nanosheet‐based system, the SPPs travelling along the Au nanosheet can couple with the local EM oscillations at the Au NR tips that can lead to resonance and boost the decaying evanescent field by several orders of magnitude. Hence, the evanescent field perturbation caused by the NRs can lead to signal augmentation and corresponding enhancement in the sensitivity.[ 32 , 46 ] Thus, incorporating Au NR can allow the SPPs to couple with LSPR EM field, leading to enhanced electric field and lower detection limits. We observe from Figure 4a,b that as we increase the distance between the plasmonic metal nanosheet and the Au NR, the enhancement in local field reduces. This can be attributed to reduced evanescent field coupling between the Au nanosheet and the Au NR. Thus, the field enhancement is highest for a distance of 2 nm between the sheet and the NR and lowest for 14 nm. Figure S2 (Supporting Information) shows the evanescent field variation for AR = 2 and 3.
Figure 5 shows the electric field distribution across the sensor surface at the resonance condition. The local field enhancement at the tip of the Au NRs can be easily seen as red‐hot spots. Figure S3 (Supporting Information) shows the electric field distribution for AR = 2 and 3. The electric field and the corresponding enhancements for all the 20 combinations (4 different ARs X 5 different distances between the sheet and the NR) of Au NR and Au nanosheet arrangement have been summarized in Table 1 . We observe that as we increase the AR, the local field enhancement increases. For our sandwich‐type assay, the immobilization with S protein Ab and the flow of antigen containing serum samples can take up about 10–15 nm distance on top of the sensing film. With compact packing of the different layers, this can be reduced to 10 nm. Thus, to counteract the reduced enhancement of the local electric field due to increased distance between the Au nanosheet and the Au NR, we need to choose higher aspect ratios. Hence, for our case, we observe that by choosing an Au NR with AR = 4 that is located 14 nm above the Au nanosheet, we can get an enhancement of 376.33%. For the graphene‐based FET SARS‐CoV‐2 S spike protein detector, the researchers could detect a minimum concentration of 0.2 × 10−12 m.[ 27 ] Additionally, the SARS‐CoV‐2 spike protein sensor developed by Mahari et al. had a LOD of 90 × 10−15 m and it displayed high linearity in the range of 1 × 10−15 m to 10 × 10−6 m concentration of the virus.[ 26 ] Since no particular benchmark has been arrived regarding the LOD, we believe that by incorporating properly customized Au NRs the LOD can be lowered down by several orders of magnitude.
Figure 5.
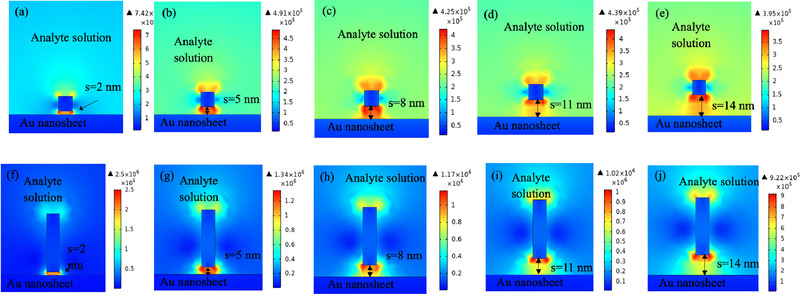
Normal electric field distributions at the resonance condition for Au NR with different aspect ratios and located at various distances from the Au nanosheet for AR = 1 a–e) and AR = 4 f–j).
Table 1.
Enhancement of electric field for various arrangements of Au NR as compared to the standard Au nanosheet (without Au NR)
| Distance from nanosheet | 2 nm | Enhancement [%] | 5 nm | Enhancement [%] | 8 nm | Enhancement [%] | 11 nm | Enhancement [%] | 14 nm | Enhancement [%] |
|---|---|---|---|---|---|---|---|---|---|---|
| Au nanosheet (without NR) | 2.45 × 105 | — | 2.45 × 105 | — | 2.45 × 105 | — | 2.45 × 105 | — | 2.45 × 105 | — |
| Au nanosheet + Au NR with AR = 1 | 7.42 × 105 | 302.86 | 4.91 × 105 | 200.41 | 4.25 × 105 | 173.47 | 4.39 × 105 | 179.18 | 3.95 × 105 | 161.22 |
| Au nanosheet + Au NR with AR = 2 | 1.28 × 106 | 522.45 | 7.68 × 105 | 313.47 | 6.51 × 105 | 265.71 | 6.16 × 105 | 251.43 | 5.39 × 105 | 220 |
| Au nanosheet + Au NR with AR = 3 | 1.86 × 106 | 759.18 | 1.03 × 106 | 420.41 | 8.96 × 105 | 365.71 | 7.98 × 105 | 325.71 | 7.23 × 105 | 295.10 |
| Au nanosheet + Au NR with AR = 4 | 2.5 × 106 | 1020.41 | 1.34 × 106 | 546.94 | 1.17 × 106 | 477.55 | 1.02 × 106 | 416.33 | 9.22 × 105 | 376.33 |
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Au nanoparticles have been used for many decades for enhancing the sensing properties of not only plasmonic sensors but also other sensors that work on different techniques. Lepinay et al. designed a tilted fiber Bragg grating optical fiber sensor that used Au nanocages (Au NCs)/Au nanospheres (Au NSs) for detection of various target proteins.[ 55 ] With the help of Au NPs, the researchers were able to lower the detection limit from the nanomolar range to the picomolar range. Additionally, Le et al. used Au NSs that were stabilized with acetylated chitosan, and that showed plasmonic properties, for the detection of melamine, bacteria, and uric acid.[ 56 ] Moreover, many SPR‐based schemes have been used in the past that have incorporated Au NPs for the detection of DNA hybridization, oligonucleotide and p53 cDNA determination and interaction assay of Carbamate‐Acetylcholinesterase.[ 57 , 58 , 59 ] Thus, we believe that Au NRs can be highly effective in enhancing the sensitivity and lowering the detection limit of conventional SPR scheme. Additionally, since NRs have a higher surface volume as compared to NSs, they can bind to more antibodies and hence it will lower the concentration of NR required.
The AuNR‐based SPR model present here can be effectively realized and designed using commercial Spreeta SPR sensors. With features like real‐time sensing, quantitative analysis, internal fault detection mechanism, robust design, small, and lightweight, these sensors can be efficiently used for diagnostics. With a volumetric dimension of 1.63 × 1.139 × 0.531 cm3, and an in‐built integrated circuit chip system, they are quite compatible for the design of POC devices where they can be merged together with other software and hardware platforms to design a complete wireless plasmonic sensing scheme that encompasses an electronic wireless transmission module that can store the test results in the data cloud for global data monitoring and analysis. The reason behind choosing gold nanorods is due to its easy and comparatively simple method of preparation as compared to other nanomaterials. Additionally, commercially prepared Au NRs with different aspect ratios having a particular LSPR wavelength can be procured easily as the synthesis method is quite mature and many chemical companies can actually prepare customized Au NRs. Also, there have been established methods of synthesizing customized Au NRs. The seed‐mediated growth method forms the basis of many improved methods. By tweaking various parameters, many researchers have come up with different techniques of preparing fine‐tuned Au NRs where they can control the AR by varying the reaction conditions like temperature or the concentration of a particular solvent.[ 60 , 61 , 62 , 63 , 64 ] Given the high biocompatibility of these Au NRs, they can be conjugated to Abs in a facile and efficient way. For instance, they can attach themselves to the Abs by various mechanisms like Abs being electrostatically adsorbed on the cetyltrimethylammonim bromide (CTAB) layer on the Au NRs; coating the NR CTAB layer with charged polymers and following it with electrostatic adsorption of Abs; replacing the CTAB layer with biofunctional ligand to which the Abs can attach through covalent binding; and replacing the CTAB layer with methoxypoly(ethyleneglycol)(mPEG)‐thiol molecules and following it with directional conjugation that takes place with the help of the carbohydrate portion situated on the heavy chains of the Fc region of antibodies.[ 65 ] The chemical binding of the various constituents can affect the R.I. of the analyte. In a qualitative way, the R.I. change affects the change in SPR angle and thus the change in sensorgram response. Thus, due to experimental errors if the binding is not ideal and results in a decreased R.I. change, the sensorgram response will be reduced. The virus can still be detected but the concentration deduced from the response will be lower than the actual. Considering all these aspects, we believe that the actual model of the proposed plasmonic immunosensor can be fruitfully realized. We would also like to emphasize that the use of Au NRs can not only help this particular miniaturized version of the sensing system but can also immensely improve the sensitivity of commercial SPR systems.
4. Conclusion
The rapidly rising cases of COVID‐19 have placed enormous stress on the medical fraternity. The easy transmittivity of the COVID‐19 virus has led to an exponential increase in the number of infected people. Thus, faster and efficient diagnostic techniques are required that can detect even trace amounts of the SARS‐CoV‐2 virus. Plasmonic sensing scheme offers many advantages like label‐free and real‐time detection technique. Thus, in this work we present a sandwich‐type plasmonic model that makes use of Au NRs to enhance the sensitivity by virtue of magnification of electric field. We present simulation results for various ARs and arrangements of the Au NRs. We find that increasing the AR can effectively lead to improved electric field. Alternatively, increasing the distance between the Au NR and the plasmonic Au nanosheet can reduce the evanescent field. For a typical 10 nm diameter Au NR‐based configuration (Au nanosheet + Au NR with AR = 4), we observe an evanescent field improvement of 376.33% as compared to the basic Au nanosheet‐based Kretschmann model.
Conflict of Interest
The authors declare no conflict of interest.
Supporting information
Supporting Information
Acknowledgements
C.M.D. and Y.G. contributed equally to this work. This work was supported by the National Research Foundation (NRF) Singapore and French National Research Agency (ANR), Grant No. (NRF2017–ANR002 2DPS) and Provost's Chair in Electrical and Electronic Engineering Award (002354–00001).
References
- 1. Yadav S. R., Kumar R., Gupta N., Ish P., Chakrabarti S., Kumar A., Monaldi Arch. Chest Dis. 2020, 90. [Google Scholar]
- 2. Jasti M., Nalleballe K., Dandu V., Onteddu S., J. Neurol. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Smith M. L., Gandolfi S., Coshall P. M., Rahman P. K. S. M., Front. Microbiol. 2020, 11, 1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jindal V., Sahu K. K., Gaikazian S., Siddiqui A. D., Jaiyesimi I., Med. Oncol. 2020, 37, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gupta N., Agrawal S., Ish P., Monaldi Arch. Chest Dis. 2020, 90. [DOI] [PubMed] [Google Scholar]
- 6. Kadkhoda K., mSphere 2020, 5, e00344‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bergquist R., Rinaldi L., Geospat Health 2020, 15. [DOI] [PubMed] [Google Scholar]
- 8. Dowd J. B., Andriano L., Brazel D. M., Rotondi V., Block P., Ding X., Liu Y., Mills M. C., Proc. Natl. Acad. Sci. USA 2020, 117, 9696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wu Y., Jing W., Liu J., Ma Q., Yuan J., Wang Y., Du M., Liu M., Sci. Total Environ. 2020, 729, 139051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bulut C., Kato Y., Turk. J. Med. Sci. 2020, 50, 563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ambrosino I., Barbagelata E., Ortona E., Ruggieri A., Massiah G., Giannico O. V., Politi C., Moretti A. M., Monaldi Arch. Chest Dis. 2020, 90. [DOI] [PubMed] [Google Scholar]
- 12. Lau H., Khosrawipour V., Kocbach P., Mikolajczyk A., Ichii H., Schubert J., Bania J., Khosrawipour T., J. Microbiol., Immunol. Infect. 2020, 53, 454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kebede Y., Yitayih Y., Birhanu Z., Mekonen S., Ambelu A., PLoS One 2020, 15, e0233744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Das G., Mukherjee N., Ghosh S., ACS Chem. Neurosci. 2020, 11, 1206. [DOI] [PubMed] [Google Scholar]
- 15. Tezer H., Bedir Demirdag T., Turk. J. Med. Sci. 2020, 50, 592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nguyen T., Duong Bang D., Wolff A., Micromachines 2020, 11, 306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. van Kasteren P. B., van der Veer B., van den Brink S., Wijsman L., de Jonge J., van den Brandt A., Molenkamp R., Reusken C., Meijer A., J. Clin. Virol. 2020, 128, 104412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Somvanshi S. B., Kharat P. B., Saraf T. S., Somwanshi S. B., Shejul S. B., Jadhav K. M., Mater. Res. Innovations 2020, 1. [Google Scholar]
- 19. Yang T., Wang Y. C., Shen C. F., Cheng C. M., Diagnostics 2020, 10, 165. [Google Scholar]
- 20. Tahamtan A., Ardebili A., Expert Rev. Mol. Diagn. 2020, 20, 453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. de Assis R. R., Jain A., Nakajima R., Jasinskas A., Felgner J., Obiero J. M., Adenaiye O., Tai S., Hong F., Norris P. J., Stone M., Simmons G., Bagri A., Schreiber M., Buser A., Holbro A., Battegay M., Hosimer P., Noesen C., Milton D. K., Prometheus Study Group , Davies D. H., Contestable P., Corash L. M., Busch M. P., Felgner P. L., Khan S., bioRxiv 2020. 10.1101/2020.04.15.043364. [DOI] [Google Scholar]
- 22. Zhang Y., Xiao M., Zhang S., Xia P., Cao W., Jiang W., Chen H., Ding X., Zhao H., Zhang H., Wang C., Zhao J., Sun X., Tian R., Wu W., Wu D., Ma J., Chen Y., Zhang D., Xie J., Yan X., Zhou X., Liu Z., Wang J., Du B., Qin Y., Gao P., Qin X., Xu Y., Zhang W., Li T., Zhang F., Zhao Y., Li Y., Zhang S., N. Engl. J. Med. 2020, 382, e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Petherick A., Lancet 2020, 395, 1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Udugama B., Kadhiresan P., Kozlowski H. N., Malekjahani A., Osborne M., Li V. Y. C., Chen H., Mubareka S., Gubbay J. B., Chan W. C. W., ACS Nano 2020, 14, 3822. [DOI] [PubMed] [Google Scholar]
- 25. Qiu G., Gai Z., Tao Y., Schmitt J., Kullak‐Ublick G. A., Wang J., ACS Nano 2020, 14, 5268. [DOI] [PubMed] [Google Scholar]
- 26. Mahari S., Roberts A., Shahdeo D., Gandhi S., bioRxiv 2020. 10.1101/2020.04.24.059204. [DOI] [Google Scholar]
- 27. Zhang X., Qi Q., Jing Q., Ao S., Zhang Z., Ding M., Wu M., Liu K., Wang W., Ling Y., Zhang Z., Fu W., preprint arXiv:2003.12529 2020.
- 28. Yi J., Mudumbai R., Xu W., preprint arXiv:2004.05759 2020.
- 29. Poh C. M., Carissimo G., Wang B., Amrun S. N., Lee C. Y.‐P., Chee R. S.‐L., Yeo N. K.‐W., Lee W.‐H., Leo Y.‐S., Chen M. I.‐C., Tan S.‐Y., Chai L. Y. A., Kalimuddin S., Thien S.‐Y., Young B. E., Lye D. C., Wang C. I., Renia L., Ng L. F. P., Nat. Commun. 2020, 11, 2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Seo G., Lee G., Kim M. J., Baek S.‐H., Choi M., Ku K. B., Lee C.‐S., Jun S., Park D., Kim H. G., Kim S.‐J., Lee J.‐O., Kim B. T., Park E. C., Kim S. I., ACS Nano 2020, 14, 5135. [DOI] [PubMed] [Google Scholar]
- 31. Sharma A. K., Gupta B. D., J. Appl. Phys. 2007, 101, 093111. [Google Scholar]
- 32. Li M., Cushing S. K., Wu N., Analyst 2015, 140, 386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kaushik S., Tiwari U. K., Pal S. S., Sinha R. K., Biosens. Bioelectron. 2019, 126, 501. [DOI] [PubMed] [Google Scholar]
- 34. Ravi Shankaran D., Miura N., J. Phys. D: Appl. Phys. 2007, 40, 7187. [Google Scholar]
- 35. Xue T., Liang W., Li Y., Sun Y., Xiang Y., Zhang Y., Dai Z., Duo Y., Wu L., Qi K., Shivananju B. N., Zhang L., Cui X., Zhang H., Bao Q., Nat. Commun. 2019, 10, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nasrin F., Chowdhury A. D., Takemura K., Lee J., Adegoke O., Deo V. K., Abe F., Suzuki T., Park E. Y., Biosens. Bioelectron. 2018, 122, 16. [DOI] [PubMed] [Google Scholar]
- 37. Lee T., Kim G. H., Kim S. M., Hong K., Kim Y., Park C., Sohn H., Min J., Colloids Surf., B 2019, 182, 110341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang J., Sun Y., Xu B., Zhang H., Gao Y., Zhang H., Song D., Biosens. Bioelectron. 2013, 45, 230. [DOI] [PubMed] [Google Scholar]
- 39. Cao J., Galbraith E. K., Sun T., Grattan K. T. V., Sens. Actuators, B 2012, 169, 360. [Google Scholar]
- 40. Gormley A. J., Greish K., Ray A., Robinson R., Gustafson J. A., Ghandehari H., Int. J. Pharm. 2011, 415, 315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cao J., Sun T., Grattan K. T. V., Sens. Actuators, B 2014, 195, 332. [Google Scholar]
- 42. Castellana E. T., Gamez R. C., Gomez M. E., Russell D. H., Langmuir 2010, 26, 6066. [DOI] [PubMed] [Google Scholar]
- 43. Liu Y., Zhao Y., Wang Y., Li C. M., J. Colloid Interface Sci. 2015, 439, 7. [DOI] [PubMed] [Google Scholar]
- 44. Wu Q., Chen L., Huang L., Wang J., Liu J., Hu C., Han H., Biosens. Bioelectron. 2015, 74, 16. [DOI] [PubMed] [Google Scholar]
- 45. Zhan P., Dutta P. K., Wang P., Song G., Dai M., Zhao S.‐X., Wang Z.‐G., Yin P., Zhang W., Ding B., Ke Y., ACS Nano 2017, 11, 1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Law W.‐C., Yong K.‐T., Baev A., Prasad P. N., ACS Nano 2011, 5, 4858. [DOI] [PubMed] [Google Scholar]
- 47. Mackey M. A., Ali M. R., Austin L. A., Near R. D., El‐Sayed M. A., J. Phys. Chem. B 2014, 118, 1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ma X., Gao L., Tang Y., Miao P., Part. Part. Syst. Charact. 2018, 35, 1700326. [Google Scholar]
- 49. Jinwoo J., Saji U., Jiyoung L., Hyejin H., Gibum K., Pil J. Y., Bruce D. H., Christine S. K., Yeon‐Su P., In‐Kyu P., Sens. Actuators, B 2018, 266, 710. [Google Scholar]
- 50. Bobin L., Jin‐Ho P., Ju‐Young B., Joon Heon K., Min‐Gon K., Biosens. Bioelectron. 2018, 102, 504.29197812 [Google Scholar]
- 51. Spangler B. D., Wilkinson E. A., Murphy J. T., Tyler B. J., Anal. Chim. Acta 2001, 444, 149. [Google Scholar]
- 52. Chinowsky T. M., Soelberg S. D., Baker P., Swanson N. R., Kauffman P., Mactutis A., Grow M. S., Atmar R., Yee S. S., Furlong C. E., Biosens. Bioelectron. 2007, 22, 2268. [DOI] [PubMed] [Google Scholar]
- 53. Soelberg S. D., Chinowsky T., Geiss G., Spinelli C. B., Stevens R., Near S., Kauffman P., Yee S., Furlong C. E., J. Ind. Microbiol. Biotechnol. 2005, 32, 669. [DOI] [PubMed] [Google Scholar]
- 54. Soelberg S. D., Stevens R. C., Limaye A. P., Furlong C. E., Anal. Chem. 2009, 81, 2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lepinay S., Staff A., Ianoul A., Albert J., Biosens. Bioelectron. 2014, 52, 337. [DOI] [PubMed] [Google Scholar]
- 56. Te L. T., Nguyen H.‐P., Dinh Q.‐K., Hoang T.‐L., Nguyen Q.‐H., Tran T.‐H., Nguyen T.‐D., Mater. Chem. Phys. 2015, 149–150, 324. [Google Scholar]
- 57. He L., Musick M. D., Nicewarner S. R., Salinas F. G., Benkovic S. J., Natan M. J., Keating C. D., J. Am. Chem. Soc. 2000, 122, 9071. [Google Scholar]
- 58. Yao X., Li X., Toledo F., Zurita‐Lopez C., Gutova M., Momand J., Zhou F., Anal. Biochem. 2006, 354, 220. [DOI] [PubMed] [Google Scholar]
- 59. Huang X., Tu H., Zhu D., Du D., Zhang A., Talanta 2009, 78, 1036. [DOI] [PubMed] [Google Scholar]
- 60. Huang S., Neretina S., El‐Sayed M. A., Adv. Mater. 2009, 21, 4880. [DOI] [PubMed] [Google Scholar]
- 61. Mackey M. A., Ali M. R. K., Austin L. A., Near R. D., El‐Sayed M. A., J. Phys. Chem. B 2014, 118, 1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Nikoobakht B., El‐Sayed M. A., Chem. Mater. 2003, 15, 1957. [Google Scholar]
- 63. Yin F., Yang C., Wang Q., Zeng S., Hu R., Lin G., Tian J., Hu S., Lan R. F., Yoon H. S., Lu F., Wang K., Yong K.‐T., Theranostics 2015, 5, 818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yong K.‐T., Sahoo Y., Swihart M. T., Schneeberger P. M., Prasad P. N., Top. Catal. 2008, 47, 49. [Google Scholar]
- 65. Joshi P. P., Yoon S. J., Hardin W. G., Emelianov S., Sokolov K. V., Bioconjugate Chem. 2013, 24, 878. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information


