Abstract
Objective
The Unified Medical Language System (UMLS) is 1 of the most successful, collaborative efforts of terminology resource development in biomedicine. The present study aims to 1) survey historical footprints, emerging technologies, and the existing challenges in the use of UMLS resources and tools, and 2) present potential future directions.
Materials and Methods
We collected 10 469 bibliographic records published between 1986 and 2019, using a Web of Science database. graph analysis, data visualization, and text mining to analyze domain-level citations, subject categories, keyword co-occurrence and bursts, document co-citation networks, and landmark papers.
Results
The findings show that the development of UMLS resources and tools have been led by interdisciplinary collaboration among medicine, biology, and computer science. Efforts encompassing multiple disciplines, such as medical informatics, biochemical sciences, and genetics, were the driving forces behind the domain’s growth. The following topics were found to be the dominant research themes from the early phases to mid-phases: 1) development and extension of ontologies and 2) enhancing the integrity and accessibility of these resources. Knowledge discovery using machine learning and natural language processing and applications in broader contexts such as drug safety surveillance have recently been receiving increasing attention.
Discussion
Our analysis confirms that while reaching its scientific maturity, UMLS research aims to boundary-span to more variety in the biomedical context. We also made some recommendations for editorship and authorship in the domain.
Conclusion
The present study provides a systematic approach to map the intellectual growth of science, as well as a self-explanatory bibliometric profile of the published UMLS literature. It also suggests potential future directions. Using the findings of this study, the scientific community can better align the studies within the emerging agenda and current challenges.
Keywords: unified medical language system, science mapping, visual analytics, text mining, content analysis
INTRODUCTION
The Unified Medical Language System (UMLS) is 1 of the most impactful multidisciplinary projects initiated to create a set of interoperable terminologies and applications in biomedicine.1 Developed and maintained by the National Library of Medicine, UMLS aims to capture a variety of medical entities and relationships that reference the same concepts, but are often expressed in very idiosyncratic forms. It also facilitates the interoperation between different medical systems with reduced barriers. Finally, it serves as a compendium of knowledge bases in biomedicine as well as comprehensive thesaurus and ontology. Over the past 3 decades, there have been considerable collaborative, multisite efforts for designing, developing, and tooling these resources.
Facing the monumental 30-year anniversary and scientific maturity of UMLS resources, tools, and applications, it is important to highlight current uses and impacts as well as technical milestones of UMLS. Based on a thorough survey of where it has been, where it is, and where it may go, we can properly identify future directions as well as better position our work with respect to emerging trends and current challenges. As the volume of literature in UMLS research has enormously increased, this study aims to systematically conduct a holistic review of the domain’s intellectual landscapes. We explore the epistemological characteristics, historical developments, emerging technologies, and current challenges of the domain. The diffusion of knowledge is also investigated to understand the intellectual growth in much broader contexts. We employ a scientometrics review2–6 using a set of quantitative and visual analytics. Compared to the conventional reviews, this approach offers the following advantages: 1) a more diverse range of bibliographic entities can be analyzed; 2) this type of domain analysis can be conducted as frequently as needed without prior experience in a target domain; and 3) citation analysis used in this work provides topically relevant, influential references which, otherwise, can be chosen less objectively. The following research questions guide the remainder of the present study:
RQ1: What are the intellectual driving forces of UMLS resources and projects?
RQ2: What thematic patterns characterize the domain’s historic footprint?
RQ3: What are the emerging tools, applications, and current challenges?
MATERIALS AND METHODS
Data collection
We collected topically relevant articles to UMLS from the Web of Science (WoS). The WoS was chosen as our primary data source because it is known as an authoritative source of scientific literature, and our study leveraged scientometrics tool kits that use bibliographic records retrieved from the WoS. After determining that the WoS topic search on “unified medical language system” OR “umls” retrieved many irrelevant records (eg, with UMLs as an acronym for “upper mixed layers,” we devised a 2-step approach: First, the following query was run on PubMed, which resulted in 1228 PMIDs (PubMed identifiers): “unified medical language system” [Text Word]. Given the retrieved PMIDs, we conducted the PubMed ID search on the WoS and retrieved 906 articles, proceedings, and reviews written in English between 1986 and 2019, as of December 31, 2019. This still limited the inclusiveness of records compared to the initial search on PubMed, but we decided to balance the trade-off between higher relevancy and less noise. The vocabulary mismatch has been often reported to be a challenge for keyword-based searches.7 This data set was labeled “Core.” Second, we used the citation report to retrieve a broader context of UMLS research via citation indexing.8 A total of 9563 records that cited the core data were collected. We named this set “Expanded.” In the remainder of the study, Core was used to map the scientific profile of the UMLS literature while Expanded was considered as evidence representing the diffusion of knowledge. A statistical summary of the collected data sets is presented in Table 1. As rendered in Figure 1, the number of records in Core is consistent over time although it has received increasing attention in recent years.
Table 1.
Bibliographic records statistics
| Context | Duration | Total | Articles | Proceedings | Reviews | Authors | Keywords | References |
|---|---|---|---|---|---|---|---|---|
| Core | 1986–2019 | 906 | 646 | 374 | 19 | 3724 | 9764 | 23 185 |
| Expanded | 1989–2019 | 9563 | 6706 | 2971 | 618 | 45 467 | 110 676 | 391 569 |
Figure 1.
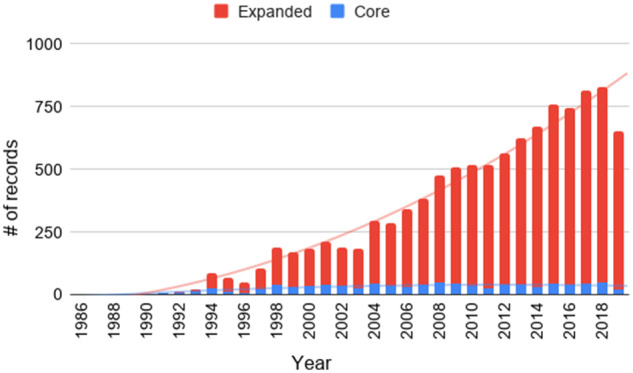
Data distribution over time.
Scientific mapping and text mining
We represented the intellectual landscapes of the domain with a variety of bibliographic entities such as publications, author and indexer keywords (keyword and keyword plus), cited references, and textual content. In interpreting such representations, we took a deductive approach moving from publication-level citations, subject category assignment, keyword co-occurrence and bursts, and document co-citation networks, to content analysis of landmark articles. This enabled our findings to be triangulated with different levels of granularity with consistent, richer meanings as we moved on to the next subsections. Our work leveraged CiteSpace v5.5.R2,9,10 VOSviewer v. 1.6.14,11 and Gephi v0.9.2, which are widely used scientific mapping tool kits. The followings describe structural measures and machine learning frameworks to communicate the present study’s analytical approaches:
Citation analysis: Citation analysis is the study of frequency, patterns, and interconnections of references in literature.8 Citation analysis draws intellectual graphs of the data sets.
Dual-map overlay: A dual-map overlay is a publication-level citation pattern visualization technique.12 This representation was used to depict the domain-level growth of knowledge in the literature where a base map consists of the inter-citations among over 10 000 journals and conferences.
Graph reduction: Drawing the entirety of nodes and edges on a graph is computationally costly and less likely to deliver an important structure. To remedy this, we selected the top 10% most occurring entities per year.
Topological metrics: A graph density is defined as the number of actual links divided by the number of possible links. The higher, the more interconnections among the vertices.13 Betweenness centrality is a measure based on the shortest paths passing through a node.14 A vertex with a high betweenness value has an influence over the flow of information on a network. PageRank is an algorithm that measures the quantity and quality of links to a node.15 The higher, the more important links the node receives.
Burst detection: Burst detection is an algorithm that models the periods and strengths in which certain features rise sharply in frequency.16 This technique identifies the keywords showing the surging frequencies.
Community detection: Community detection is a clustering approach to identify the latent sets of densely connected nodes on a network. To group the cited references, we employed a model where global modularity ranges between 0 and 1,17 meaning that the higher, the higher the number of communities.
RESULTS
Disciplinary-level research trends
Dual-map overlays
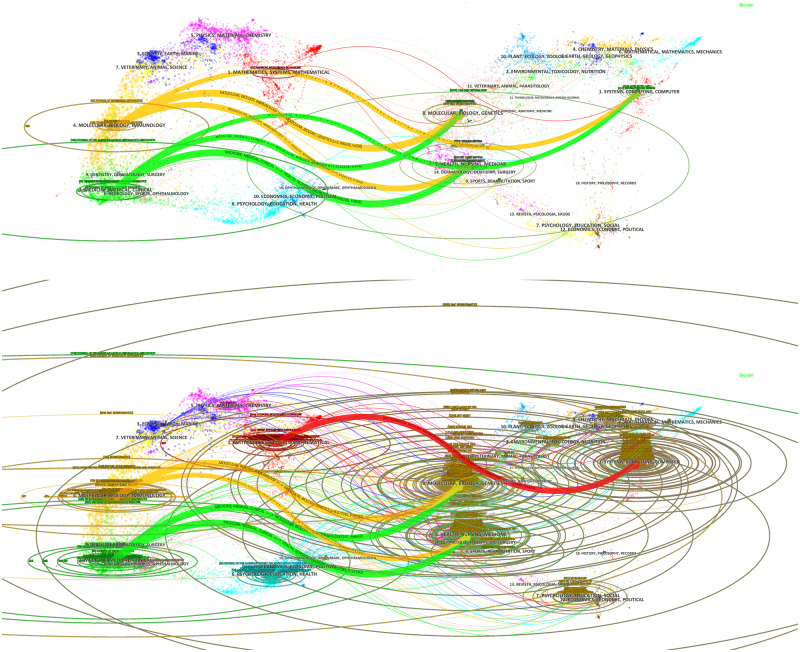
Figure 2 displays the dual-map overlays rendering domain-level citation patterns in Core (upper) and Expanded (lower), respectively. The visuals consist of 2 groups of domains: 1) publication domains (on the left) representing the scientific domains where the data sets are published and 2) reference domains (on the right) showing the domains from which the published articles cited their references. In these domains, each subregion is labeled with terms commonly found in the journal/conference titles in that subregion. The citation paths between the publication and reference domains are colored based on the publication domains’ colors; the width of a path is proportional to the frequency of citations. Table 2 describes the paths with the publication and reference domains in descending order of frequency in z-score.
Figure 2.
Dual-map overlays: Core (upper) and Expanded (lower).
Table 2.
Disciplinary-level citation trajectories (rows colored with corresponding paths in Figure 2 upper and lower)
| Context | Publication domains | Reference domains | Z-score |
|---|---|---|---|
| Core | Medicine, medical, clinical | Health, nursing, medicine | 5.049 |
| Molecular, biology, immunology | Molecular, biology, genetics | 3.786 | |
| Medicine, medical, clinical | Molecular, biology, genetics | 2.613 | |
| Molecular, biology, immunology | Health, nursing, medicine | 2.342 | |
| Medicine, medical, clinical | Systems, computing, computer | 1.917 | |
| Molecular, biology, immunology | Systems, computing, computer | 1.750 | |
| Expanded | Molecular, biology, immunology | Molecular, biology, genetics | 8.678 |
| Medicine, medical, clinical | Health, nursing, medicine | 5.118 | |
| Medicine, medical, clinical | Molecular, biology, genetics | 3.825 | |
| Mathematics, systems, mathematical | Systems, computing, computer | 3.332 | |
| Molecular, biology, immunology | Health, nursing, medicine | 2.155 |
In the remainder of this section, “interdisciplinarity” is used for the combination of more than 1 branch of knowledge into a synthesis of approaches while “multidisciplinarity” draws on their disciplinary knowledge. In Core, the literature published in medical (medicine, medical, clinical) and biological domains (molecular, biology, immunology) heavily cites from healthcare (health, nursing, medicine), biological (molecular, biology, genetics), and computational (systems, computing, computer) domains. The citations in the core literature show an interdisciplinary pattern where the publication domains have been created by combining 3 reference domains. The publication domains are also partially multidisciplinary; 2 publication domains have led the creation of knowledge. In the Expanded data set, the published research in biological (molecular, biology, immunology) and medical (medicine, medical, clinical) domains mainly cites from biological (molecular, biology, genetics) and healthcare (health, nursing, medicine) domains. The other set of research published in mathematical domains (mathematics, systems, mathematical) cites from computational (systems, computing, computer) domains. The expanded literature in biological and medical domains cites the computer science domain to a lesser degree, whereas only the literature in mathematical domains references the computer domains. The diffusion of knowledge shows a less interdisciplinary but more multidisciplinary pattern.
Subject category assignment
Table 3 lists the top 20 subject categories most frequently assigned to the records in Core and Expanded, showing a high-level thematic concentration. Exclusive categories in each data set were colored in gray. In Core, medical informatics is the most dominant category followed by computer science and related fields (computer science, interdisciplinary applications, and computer science information systems) and healthcare sciences. The expanded data set shows similar trends: medical informatics is the top category and there are 17 overlapping subject categories. In contrast to the number of records being highly concentrated to a very few subject categories in Core, the expanded data set is more evenly distributed across multiple domains such as biochemical technologies and genetics. Along with the findings from the dual-map overlays showing Core being more interdisciplinary and Expanded being more multidisciplinary, the subject category assignment suggests that the UMLS research has been broadened from the interdisciplinary concentrated efforts toward the applications in more diverse contexts.
Table 3.
Top 20 WoS subject categories
| Context | WoS Categories | Records | % of 906 |
|---|---|---|---|
| Core | Medical informatics | 583 | 64.349 |
| Computer science interdisciplinary applications | 395 | 43.598 | |
| Computer science information systems | 386 | 42.605 | |
| Health care sciences services | 354 | 39.073 | |
| Information science library science | 223 | 24.614 | |
| Mathematical computational biology | 103 | 11.369 | |
| Computer science artificial intelligence | 72 | 7.947 | |
| Biochemical research methods | 65 | 7.174 | |
| Engineering biomedical | 63 | 6.954 | |
| Biotechnology applied microbiology | 58 | 6.402 | |
| Computer science theory methods | 56 | 6.181 | |
| Engineering electrical electronic | 23 | 2.539 | |
| Statistics probability | 19 | 2.097 | |
| Biochemistry molecular biology | 13 | 1.435 | |
| Biology | 11 | 1.214 | |
| Radiology nuclear medicine medical imaging | 10 | 1.104 | |
| Genetics heredity | 9 | 0.993 | |
| Medicine research experimental | 9 | 0.993 | |
| Public environmental occupational health | 9 | 0.993 | |
| Emergency medicine | 6 | 0.662 | |
| Expanded | Medical informatics | 3124 | 32.668 |
| Computer science information systems | 2731 | 28.558 | |
| Computer science interdisciplinary applications | 2445 | 25.567 | |
| Health care sciences services | 1903 | 19.900 | |
| Computer science artificial intelligence | 1307 | 13.667 | |
| Mathematical computational biology | 1283 | 13.416 | |
| Information science library science | 1110 | 11.607 | |
| Computer science theory methods | 932 | 9.746 | |
| Biochemical research methods | 711 | 7.435 | |
| Biotechnology applied microbiology | 620 | 6.483 | |
| Engineering electrical electronic | 520 | 5.438 | |
| Engineering biomedical | 452 | 4.727 | |
| Multidisciplinary sciences | 308 | 3.221 | |
| Biochemistry molecular biology | 290 | 3.033 | |
| Computer science software engineering | 240 | 2.510 | |
| Genetics heredity | 215 | 2.248 | |
| Radiology nuclear medicine medical imaging | 201 | 2.102 | |
| Pharmacology pharmacy | 194 | 2.029 | |
| Statistics probability | 190 | 1.987 | |
| Public environmental occupational health | 161 | 1.684 |
Keywords as evidence of emerging technologies
Keyword co-occurrence
In this section, we investigated the keywords, considering them as important yet mid-high-level indicators of the underlying concepts in the UMLS research. Table 4 describes the top 20 keywords that most frequently occur in Core and Expanded. The “Year” column indicates the year a keyword first appears and the “Density” column indicates the average Count from its first appearance to 2019 (ie, Count/(2019 − Year + 1). Density describes a keyword’s distributed impact over multiple years. The “Between” column shows the betweenness centrality of the keyword on the co-word networks. Exclusive keywords in each data set are colored in gray. The rows are sorted in ascending order of Year and descending order of Count within Year. For the remainder of the article, we divided the entire study duration into 3 phases: 1) P1 (1986–1997), 2) P2 (1998–2008), and 3) P3 (2009–2019).
Table 4.
Top 20 most frequent keywords (sorted in ascending order of Year and descending order of Count)
| Context | Phase | Keyword | Year | Count | Density | Between |
|---|---|---|---|---|---|---|
| Core | P1 | unified medical language system | 1991 | 63 | 2.172 | 0.120 |
| Medline | 1993 | 19 | 0.704 | 0.010 | ||
| System | 1994 | 87 | 3.346 | 0.140 | ||
| Information | 1994 | 62 | 2.385 | 0.240 | ||
| Vocabulary | 1994 | 22 | 0.846 | 0.050 | ||
| Umls | 1995 | 160 | 6.400 | 0.280 | ||
| P2 | Terminology | 1998 | 62 | 2.818 | 0.170 | |
| Knowledge | 1998 | 42 | 1.909 | 0.050 | ||
| Database | 1998 | 35 | 1.591 | 0.040 | ||
| information retrieval | 1998 | 29 | 1.318 | 0.060 | ||
| medical language system | 1999 | 49 | 2.333 | 0.060 | ||
| Representation | 1999 | 22 | 1.048 | 0.020 | ||
| natural language processing | 2003 | 86 | 5.059 | 0.200 | ||
| Ontology | 2003 | 72 | 4.235 | 0.110 | ||
| Text | 2004 | 61 | 3.813 | 0.150 | ||
| information extraction | 2004 | 22 | 1.375 | 0.090 | ||
| Network | 2004 | 18 | 1.125 | 0.050 | ||
| Classification | 2008 | 23 | 1.917 | 0.090 | ||
| P3 | Extraction | 2011 | 21 | 2.333 | 0.060 | |
| machine learning | 2011 | 20 | 2.222 | 0.100 | ||
| Expanded | P1 | System | 1992 | 1196 | 42.714 | 0.040 |
| Information | 1992 | 801 | 28.607 | 0.060 | ||
| Database | 1994 | 667 | 25.654 | 0.060 | ||
| information retrieval | 1994 | 362 | 13.923 | 0.030 | ||
| Terminology | 1994 | 348 | 13.385 | 0.040 | ||
| Ontology | 1995 | 1084 | 43.360 | 0.080 | ||
| Knowledge | 1995 | 556 | 22.240 | 0.050 | ||
| natural language processing | 1995 | 550 | 22.000 | 0.020 | ||
| Classification | 1995 | 409 | 16.360 | 0.080 | ||
| Care | 1995 | 345 | 13.800 | 0.050 | ||
| Umls | 1995 | 327 | 13.080 | 0.040 | ||
| Disease | 1996 | 293 | 12.208 | 0.040 | ||
| Model | 1997 | 443 | 19.261 | 0.050 | ||
| P2 | Text | 2001 | 489 | 25.737 | 0.020 | |
| Tool | 2001 | 329 | 17.316 | 0.020 | ||
| Extraction | 2002 | 292 | 16.222 | 0.020 | ||
| text mining | 2003 | 312 | 18.353 | 0.010 | ||
| Gene | 2003 | 296 | 17.412 | 0.010 | ||
| Identification | 2004 | 291 | 18.188 | 0.020 | ||
| electronic health record | 2006 | 350 | 25.000 | 0.040 |
In Core, 12 keywords were identified from CP2 (core phase 2) while CP3 had the lowest number of new keywords owing to the accumulation over time. This observation also suggests that the core literature is in a mature stage and recent themes have not received significant attention yet. Keywords “umls” and “system” are the leading ones based on all the metrics. Keyword “information” also shows the second highest count (87) and betweenness (0.240), indicating that “information” creation as a general purpose of UMLS has had the largest influence on the transfer of knowledge in Core. The exclusive concepts in this phase are “medline” and “vocabulary.” We suggest that the keywords from CP1 represent an interdisciplinary endeavor for the development of UMLS resources. CP2 can be divided into 2 groups: 1) CP2-1 between 1998 and 1999 and 2) CP2-2 between 2003 and 2008. In CP2-1, “database” and “information retrieval” indicate the scientific efforts for retrieval, integration, and aggregation of information; “knowledge” and “representation” are also shown to be dominant themes. CP2-2 starts with “natural language processing” (NLP) that has the third highest count (86), second highest density (5.059), and third highest betweenness (0.200). Applications such as clinical “text” mining and “information extraction,” together with the semantic “network” follow the “NLP” in this phase. Finally, recent applications such as “machine learning” represent CP3.
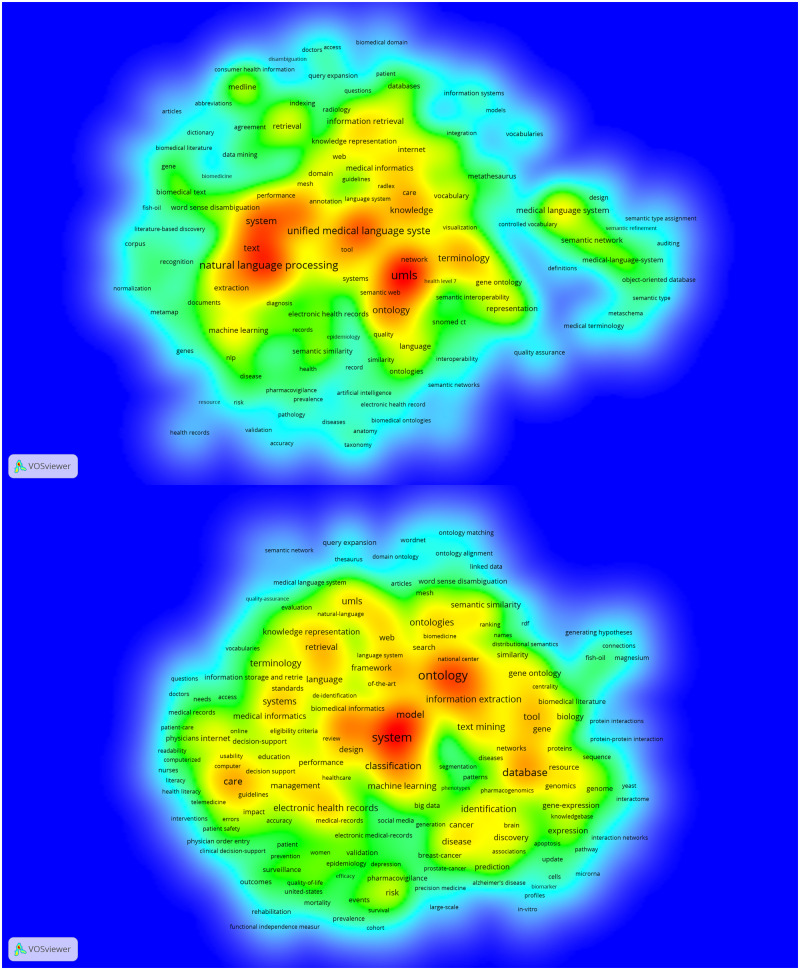
Figure 3 upper shows the keyword co-occurrence network in Core, which consists of 173 nodes and 945 links. We used density visualization in VOSviewer. In the figure, the closer a pair of keywords positions to the hotter zone, the more frequently the pair co-occurs in the literature. As depicted in the figure, 3 hot zones of co-words were identified around the leading keywords discussed above: 1) “system,” “natural language processing,” “text,” and “extraction” on the left, 2) “unified medical language system” in the middle, and 3) “umls,” “ontology,” “network,” and “semantic web” on the right. Keyword “machine learning” co-occurs closely to the left zone and other keywords regarding text analytics, such as “documents,” “word sense disambiguation,” and “recognition,” move toward the perimeter. The middle zone plays a bridging role between the left and right clusters. Above this cluster, the keywords in CP2-1, such as “information retrieval,” “knowledge,” and “(knowledge) representation,” form a cooler zone, co-occurring with “care,” “medical informatics,” and “radiology”. It indicates that “tool”-ing the UMLS resources in broader clinical contexts is also important. Finally, “terminology” is near the right zone, and “gene ontology” is closely located to “terminology.” The co-occurrence of “language” and “health level 7” with this cluster suggests that the extension of UMLS resources is an emerging topic, and the existence of “semantic interoperability” triangulates our interpretation about the semantic network being of interest.
Figure 3.
Keyword co-occurrence networks: Core (upper; node = 173; edges = 945; density = 0.063) and Expanded (lower; node = 483; edges = 5337; density = 0.046).
Compared to the Core, the values of betweenness centrality in Expanded are relatively lower across all the keywords. A total of 12 and 8 keywords are identified in EP1 (expanded phase 1) and EP2, respectively, and none in EP3, which suggests that, similar to Core, the latest concepts in the expanded literature have not received significant attention yet, compared to the maturity of those in the earlier phases. The keywords that appeared in CP2-1 such as “database,” “information retrieval,” “terminology,” and “knowledge” and in CP2-2, such as “ontology,” “natural language processing,” and “classification,” are the leading concepts in EP1. In EP1, “care” and “disease” are the exclusive keywords, and in EP2, “text” and “extraction” appear earlier than in CP2-2 and CP-3. Moreover, “text mining,” “gene,” “identification,” and “electronic health record” are newly discovered concepts in Expanded. These findings suggest that systems development and “tool”-ing the biomedical ontologies have already been the greatest concerns in the expanded contexts of the UMLS research. The results also confirm that applications and extensions are among the most important themes.
Figure 3 lower visualizes the keyword co-occurrence network in Expanded. It consists of 483 vertices with 5337 edges having lower density (0.046) than Core. The co-word network is spread out more (see Figure 3 upper for comparison), which indicates the existence of a variety of subtopics. Two hot zones of keywords are identified: 1) “system,” “classification,” and “model” on the left with “machine learning” on the perimeter and 2) “ontology” and information “extraction” on the right with “text mining” on the perimeter. The other keywords listed in Expanded tend to form their own clusters: 1) “database” with “networks,” “resource,” “genomics,” and “pharmacogenomics,” 2) “care” with “guideline,” “computer,” and “usability,” 3) “disease” and “identification” with “cancer,” “discovery,” and “brain,” 4) “tool” and “gene,” and 5) “electronic health record” with “management” and “medical records.” Findings suggest that scientific landscapes have accessed much broader contexts, such as resource usability, knowledge discovery, ontology extension, health records management, as well as derivative databases.
Bursting keywords
We investigated the bursting activities in keywords occurrence to add time-aware interpretations. The burstiness of a keyword is calculated by the weighted sum of its frequency during 1 or multiple times windows. If the probability of these occurrences is higher than a data-dependent global threshold, that keyword is said to have a burst(s). Table 5 is sorted in ascending order of Begin. The exclusive keywords are colored in gray. The burst charts start from 1992 because that was the first year a burst appeared.
Table 5.
Top 20 most bursting keywords (sorted in descending order of Begin)
| Phase | Keywords | Burst | Begin | End | 1992–2019 |
|---|---|---|---|---|---|
| CP1 | Vocabulary | 5.766 | 1994 | 2003 |
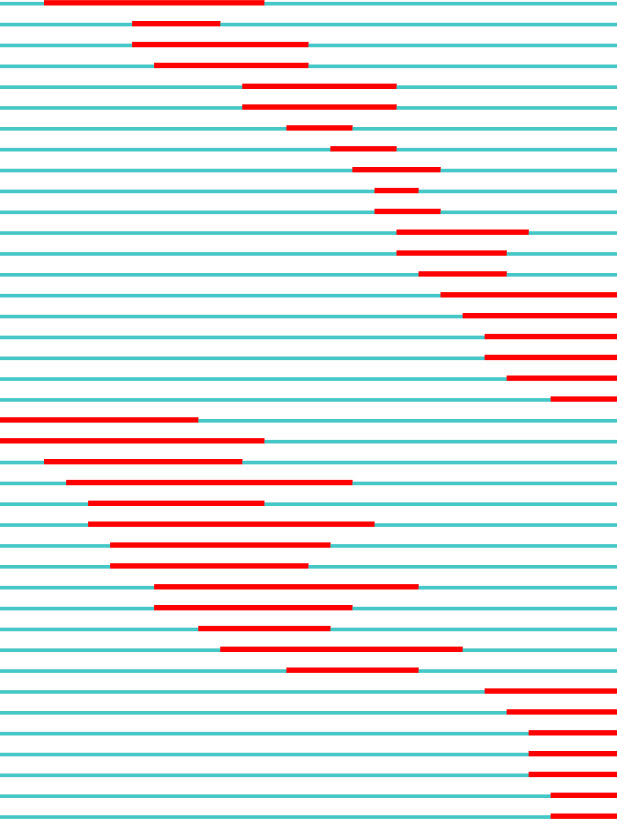
|
| CP2 | Internet | 6.144 | 1998 | 2001 | |
| Language | 6.341 | 1998 | 2005 | ||
| Representation | 8.582 | 1999 | 2005 | ||
| semantic network | 4.432 | 2003 | 2009 | ||
| Database | 6.936 | 2003 | 2009 | ||
| information retrieval | 5.461 | 2005 | 2007 | ||
| Informatics | 3.593 | 2007 | 2009 | ||
| Classification | 3.760 | 2008 | 2011 | ||
| CP3 | medical language system | 3.845 | 2009 | 2010 | |
| Gene | 4.348 | 2009 | 2011 | ||
| word sense disambiguation | 4.590 | 2010 | 2015 | ||
| Term | 3.556 | 2010 | 2014 | ||
| Identification | 3.986 | 2011 | 2014 | ||
| electronic health record | 4.150 | 2012 | 2019 | ||
| snomed ct | 3.806 | 2013 | 2019 | ||
| machine learning | 3.689 | 2014 | 2019 | ||
| information extraction | 5.874 | 2014 | 2019 | ||
| Extraction | 5.011 | 2015 | 2019 | ||
| word embedding | 4.105 | 2017 | 2019 | ||
| EP1 | System | 29,573 | 1992 | 2000 | |
| Computer | 15.947 | 1992 | 2003 | ||
| Language | 40.535 | 1994 | 2002 | ||
| knowledge representation | 26.810 | 1995 | 2007 | ||
| Representation | 37.132 | 1996 | 2003 | ||
| information system | 20.357 | 1996 | 2008 | ||
| Internet | 33.931 | 1997 | 2006 | ||
| world wide web | 21.627 | 1997 | 2005 | ||
| EP2 | medical language system | 19.617 | 1999 | 2010 | |
| health care | 14.730 | 1999 | 2007 | ||
| medical informatics | 17.403 | 2001 | 2006 | ||
| Bioinformatics | 20.724 | 2002 | 2012 | ||
| Biology | 16.252 | 2005 | 2010 | ||
| EP3 | electronic health record | 29.462 | 2014 | 2019 | |
| Pharmacovigilance | 17.768 | 2015 | 2019 | ||
| big data | 24.361 | 2016 | 2019 | ||
| Risk | 17.189 | 2016 | 2019 | ||
| Prevalence | 16.220 | 2016 | 2019 | ||
| machine learning | 35.858 | 2017 | 2019 | ||
| word embedding | 19.075 | 2017 | 2019 |
Table 5 presents the top 20 bursting keywords in Core and Expanded. Compared to the 8 exclusive keywords identified between Core and Expanded in Table 4, 13 exclusive keywords appear between Core and Expanded in Table 5, which suggests that the concepts receiving increasing attention are more diverse. Unlike in Table 4, burst detection identified 19 keywords from CP2 (8) and CP3 (11). In CP1, “vocabulary” is the only keyword with the longest burst between 1994 and 2003. In CP2-1, “representation” is the keyword with the strongest burst; “database” and “information retrieval” were previously identified in CP2-1 (see Table 4) and also burst in CP2-2. Keyword “informatics” as a novel way of knowledge discovery receives increasing attention. Keyword “word sense disambiguation” is a relatively recent concept receiving burst between 2010 and 2015, and it co-occurred with the “natural language processing” cluster in Figure 3 upper. Keyword “electronic health record” (EHR), which was an exclusive keyword in Expanded (see Table 4), has received the longest burst in CP3. With the existence of successive keywords such as “machine learning,” “information extraction,” and “word embedding,” we suggest that knowledge discovery from EHR is the most recent and emerging application of UMLS resources. Keyword “snomed ct” has been receiving recent burst, given that it is an established medical ontology.
In contrast to Core, the bursting keywords in Expanded are more evenly distributed across all the phases. In the earliest phase, knowledge representation represented by “knowledge representation” and “representation” is 1 of the bursting concepts. Previously, it appeared to be a prevailing theme in CP2-1 (see Table 4). Bursting attention to “world wide web” together with “internet” suggests that public accessibility is an important consideration in developing medical ontologies as a new form of knowledge representation in biomedicine. In EP2, like “informatics” in Core, “medical informatics” and “bioinformatics” received considerable attention as promising approaches of knowledge creation that use medical ontologies. In EP3, like in Core, “big data” along with “electronic health record,” “machine learning,” and “word embedding” represents the recent interest in advanced text analytics. Keywords “pharmacovigilance” and “risk” also indicate the current and future direction of the UMLS research in drug safety.
Document co-citation networks
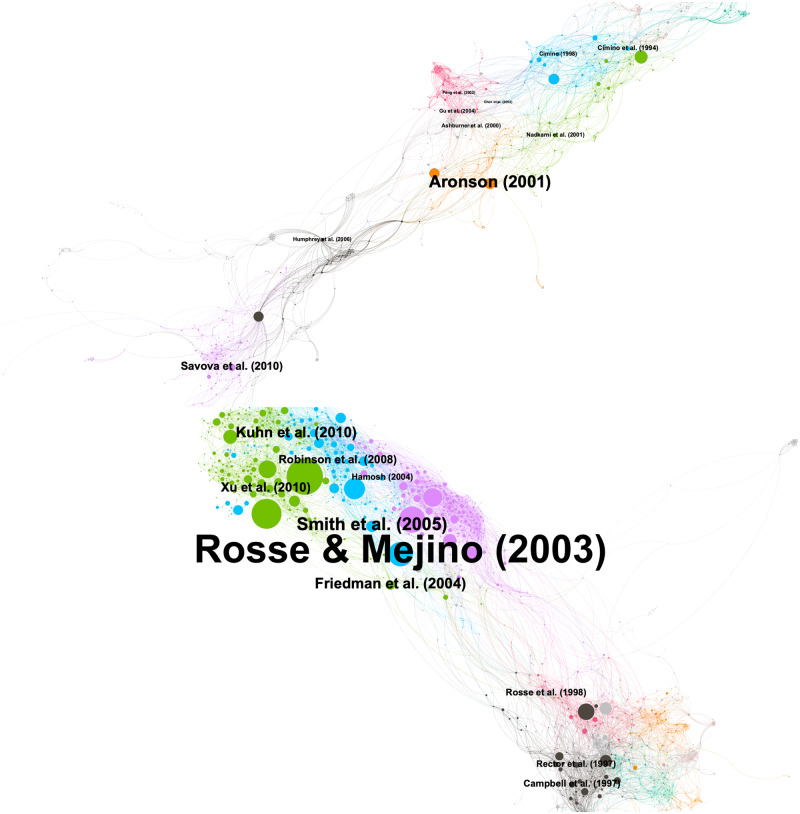
We used CiteSpace to create document co-citation networks in Core and Expanded. The visualizations (see Figure 4 upper and lower) were created by Gephi for enhanced legibility. In each graph, a vertex is a cited reference where the size is proportional to the cited count. Vertices are linked when they are co-cited in the literature. Therefore, neighboring nodes are assumed to be intellectually close to each other. Clusters were identified by community detection; the cluster membership is represented by the colors of nodes and edges. Table 6 describes 10 select articles in each data set. The references were also annotated in black in Figure 4 upper and lower. The size of a label is proportional to a corresponding node’s cited frequency. In the remainder of the paper, we call these select articles “landmark articles” as determined by the following inclusion/exclusion criteria:
Figure 4.
Document co-citation networks: Core (upper; node = 816; edges = 3311; density = 0.010; modularity = 0.773; clusters = 84) and Expanded (lower; node = 1833; edges = 9600; density = 0.006; modularity = 0.798; clusters = 156).
Table 6.
Ten landmark articles (sorted in ascending order of publication year and descending order of citation frequency)
| No. | Publication (year) | Region | Cluster | Citation frequency | Between | PageRank |
|---|---|---|---|---|---|---|
| C0 | Cimino et al (1994)18 | UR | 64 | 23 | 0.097 | 0.004 |
| C1 | Cimino (1998)19 | 64 | 20 | 0.030 | 0.003 | |
| C2 | Ashburner et al (2000)20 | 43 | 19 | 0.032 | 0.004 | |
| C3 | Aronson (2001)21 | 34 | 57 | 0.062 | 0.006 | |
| C4 | Nadkarni et al (2001)22 | 14 | 20 | 0.026 | 0.004 | |
| C5 | Peng et al (2002)23 | 47 | 14 | 0.036 | 0.003 | |
| C6 | Chen et al (2002)24 | 43 | 12 | 0.025 | 0.003 | |
| C7 | Gu et al (2004)25 | 47 | 18 | 0.034 | 0.004 | |
| C8 | Humphrey et al (2006)26 | LL | 77 | 19 | 0.082 | 0.006 |
| C9 | Savova et al (2010)27 | 33 | 38 | 0.039 | 0.006 | |
| E0 | Campbell et al (1997)28 | LR | 78 | 69 | 0.014 | 0.002 |
| E1 | Rector et al (1997)29 | 78 | 65 | 0.013 | 0.002 | |
| E2 | Rosse et al (1998)30 | 6 | 67 | 0.051 | 0.002 | |
| E3 | Rosse and Mejino (2003)31 | UL | 32 | 281 | 0.057 | 0.002 |
| E4 | Hamosh (2004)32 | 32 | 63 | 0.025 | 0.002 | |
| E5 | Smith et al (2005)33 | 32 | 128 | 0.013 | 0.002 | |
| E6 | Robinson et al (2008)34 | 32 | 83 | 0.014 | 0.002 | |
| E7 | Friedman et al (2004)35 | 0 | 107 | 0.072 | 0.002 | |
| E8 | Kuhn et al (2010)36 | 0 | 106 | 0.013 | 0.002 | |
| E9 | Xu et al (2010)37 | 0 | 93 | 0.033 | 0.002 |
Using the complete list of references, we derived 3 subtables based on cited frequency, betweenness, and PageRank, each containing the top 100 results;
Articles were selected if they appeared in all 3 derived tables;
Documents that are not original research articles were omitted;
(Expanded only) Articles that have already appeared in Core were excluded;
The top 10 references were selected in each data set and re-arranged in chronological order/cluster membership for better interpretability.
As shown in Figure 4 upper, the cited references in Core resulted in a sparse graph (density = 0.010), consisting of 816 nodes and 3311 links among them, and 84 communities were detected with relatively high modularity (0.773). These metrics suggest the existence of a variety of subtopics while the visual prominence confirms 2 main clusters of communities (upper right, UR, and lower left, LL). In Table 6, visual cluster membership is denoted as Region using either UR or LL. The table shows that 7 articles belong to the earlier phases, namely CP1 (C0) and CP2-1 (C1–C6).
Figure 4 lower depicts the document co-citation network in Expanded. The graph became sparser (density = 0.006) with 1833 vertices and 9600 edges. We found 156 communities with higher modularity (0.798). Again, we identified 2 distinctively larger regions of communities (upper left and lower right). The visual cluster membership is denoted as either UL or LR. Table 6 shows that 7 references come from the later phases, namely EP2-2 (E3–E7) and EP3 (E8, E9).
Content analysis of landmark literature
We conducted a content analysis for an in-depth understanding of the landmark literature. These papers were assigned to 1 of the following categories: 1) Extension, denoted as “Ext”; 2) Tooling denoted as “Tool”; and 3) Application, denoted as “App.” The main objectives of the papers in Ext include extensions to UMLS resources and development of new ontologies. The tooling research is about a concerted effort for evaluating and enhancing usability, interoperability, and integrity of controlled vocabularies in biomedicine. The App papers use UMLS and other ontology resources in informatics research and applications. Table 7 summarizes the objectives, methods, and findings of the papers.
Table 7.
Content analysis matrix (category code: Ext[ension]; Tool[ing]; App[lication])
| No. | Category | Objectives | Methods/criteria | Findings/products |
|---|---|---|---|---|
| C0 | Ext | Developing a controlled medical terminology | Knowledge-based semantic networking | Medical Entities Dictionary (MED) |
| C1 | Tool | Examining Metathesaurus’ semantic inconsistencies | String-matching with semantics and synonyms | Ambiguity, redundancy, and incorrect tree structure |
| C2 | Ext | Building a vocabulary for genes and gene products | Describing and annotating biological elements | Gene Ontology (GO) |
| C3 | Tool | Creating a concept-mapping tool to Metathesaurus | Natural language processing and text similarity | MetaMap, an algorithm mapping text to concepts |
| C4 | App | Using Metathesaurus for concept matching/indexing | Training and testing a concept-finding algorithm | 82.6%/76.3% true positive rates for training/testing |
| C5 | Tool | Identifying semantic types’ redundant classifications | Inspection of classification overlaps in type pedigrees | 12 657 redundant semantic type classifications |
| C6 | Tool | Creating simpler views of the Semantic Network of UMLS | Expert-based portioning with semantic considerations | 28 cohesive collections of semantic types |
| C7 | Tool | Detecting concept errors and inconsistencies in UMLS | Expert reviews on pure intersections in metaschema | Miscategorizations from 657 meta-type intersections |
| C8 | App | Disambiguating word sense for mapping text to concepts | Journal Descriptor Indexing based on MEDLINE citations | 78.7% precision with the highest-scoring JDI version |
| C9 | App | Implementing a clinical knowledge extraction system | Natural language processing and named-entity recognition | cTAKES, a text mining app with an F-score of 0.924 |
| E0 | Tool | Assessing medical ontologies: READ, UMLS, and SNOMED | Integrity, taxonomy, clarity, mapping, and definitions | Strengths and weaknesses scored by an expert panel |
| E1 | Ext | Proposing a language for modeling ontology concepts | Clarification of requirements for clinical concepts | GALEN representation and integration language (GRAIL) |
| E2 | Ext | Developing an anatomical module for physical entities | Definition and validation of anatomical entities’ attributes | An extendible ontology with structure, space, & substance |
| E3 | Ext | Proposing FMA as ontology reference to human anatomy | Disciplined modeling for restructuring vocabularies | Taxonomy, structural and transformation abstractions |
| E4 | Ext | Devising a knowledge base of genes and genetic disorders | Expert curation and editorial effort at Johns Hopkins | Online Mendelian Inheritance in Man (OMIM) |
| E5 | Ext | Devising a relation ontology for biomedical coding errors | Extensive, manual reviews of experts in life sciences | Relation Ontology with consistency and unambiguity |
| E6 | Ext | Developing a new ontology for human phenotypes | Text parsing and extensive, manual curation of terms | HPO, representing over 8000 human phenotypic anomalies |
| E7 | App | Proposing an NLP-based method for code-mapping | Adaptation of an existing NLP system, MedLEE | 84% recall and 89% precision in extracting UMLS codes |
| E8 | Ext | Developing a public resource for mapping side effects | Text mining using public labels and UMLS COSTART | SIDER, connecting 888 drugs to 1450 adverse reactions |
| E9 | App | Devising an information extraction app for medication | Sentence boundary detection; semantic tagging; parsing | MedEx, a text mining system run against clinical narratives |
Abbreviations: FMA, foundational model of anatomy; HPO, human phenotype ontology; NLP, natural language processing; UMLS, unified medical language system.
In terms of the distribution of categories, Core has 2 extensions, 5 toolings, and 3 applications; Expanded has 7 extensions, 1 tooling, and 2 applications. Although this distribution should not be generalized as the thematic categorization of the entire data sets, it confirms that the landmark papers in Core have put large efforts toward 1) auditing and enhancing UMLS resources and 2) developing novel approaches for knowledge creation in biomedicine. These findings can also be mapped to the visual region formation in Figure 4 upper. Two extensions (C0 and C2), 5 toolings (C1, C3, C5, C6, and C7), and 1 application (C4) formed the UR region. Despite the categorical differences, 1 consistent theme identified in these papers is to make UMLS and its related resources more accessible and error-free. Lastly, C8 and C9 belong to the LL region where advanced text analytics with the clinical text is a major theme. In Expanded, landmark papers have brought a wide range of UMLS extensions and new terminologies for specific applications. As illustrated in Figure 4 lower, 1 tooling (E0) and 2 extensions (E1 and E2) formed the LR region. The UL region consisted of 5 extensions (E3, E4, E5, E6, and E8) and 2 applications (E7 and E9). Except for the applications, the landmark articles in Expanded made a concerted effort for extensible ontologies with profound biomedical concepts.
DISCUSSION
RQ1: What are the intellectual driving forces of UMLS resources and projects?
The dual-map overlays and subject category assignments revealed the following. First, the core UMLS research is interdisciplinary. The studies published in medicine and biology heavily cited each other and other technical papers (systems, computing, and computer). The subject domains, such as medical informatics and computer and healthcare sciences, are the driving forces of the emergence of UMLS resources and projects. Second, the diffusion of knowledge represented in the Expanded data set was led by more multidisciplinary efforts such as biological and medical sciences and mathematics. Although they are not yet fully interdisciplinary, scientific communities are characterized by medical informatics, computer and healthcare sciences, biochemical technologies, and genetics, and further by software engineering and pharmacology and pharmaceutics. In conclusion, our analysis confirms that scientific profile and knowledge diffusion of the UMLS research are boundary-spanning to applications in a variety of contexts.
RQ2: What thematic patterns characterize the domain’s historical footprint?
The investigation of keyword co-occurrence revealed the early-phase research themes regarding UMLS resources, such as 1) development and 2) knowledge representation and information creation, as general objectives. These topics were followed by efforts for tooling and applications in broader contexts, such as semantic networking, information extraction, and text mining. The bursting keywords confirmed that informatics as a novel approach for knowledge discovery received significant attention. In the expanded context, thematic patterns, identified in the later phases of Core, were of greatest interest during the earlier phase. Moreover, applications, such as knowledge discovery in health records, were among the important themes. The bursting keywords in Expanded confirmed the medical informatics and bioinformatics as methodological driving forces of such endeavors. The following topics are also confirmed as prevalent concepts in the expanded data set: resource usability, ontology extension, health records management, and derivative databases. The document co-citation networks revealed that most core landmark papers belong to the earlier phases. The canonical endeavors include 1) auditing and enhancing UMLS resources and tools and 2) developing novel approaches for knowledge creation. The landmarks in Expanded focused more on the extensions of medical ontologies and advanced analytics. With the high concentration of keywords in the earlier phases, we argue that these topics have reached intellectual maturity. We also observed that scientific communities of UMLS research and its broader contexts have coevolved, exerting influence on each other.
RQ3: What are the emerging tools and applications, and current challenges?
Machine learning and information extraction against unstructured records were identified as the emerging application areas. Word sense ambiguation in clinical texts appeared to be the most challenging task. To this end, NLP and advanced algorithms, such as word embedding, have been recently considered. While showing similar trends to Core, there has been new initiatives arising in Expanded such as big data and drug safety surveillance. The existence of fewer concepts in the recent phases of Core and Expanded suggests that these topics have not had sufficient attention yet, while being the domain-leading concerns. We also identified the following potential challenges in the domain from our analyses. First, thematic trends suggest that recent studies heavily focus on computational techniques. This is not surprising given 1) the velocity and variety of data generated in biomedicine and 2) the remarkable advancement of NLP techniques with deep learning. The use of big data along with advanced machine learning techniques is, without a doubt, a promising approach for the discovery of less biased, more generalizable knowledge at scale. However, we observed limited coverage of the other methods. The potential challenge for the domain is to accommodate a variety of research topics as such diversity has advanced this field to date. Moreover, the heavy focus on applications in a few domains such as pharmaceutical sciences could explain the fact that there are a limited number of themes identified recently. This may be an indication that current research does not use UMLS resources and tools to its fullest extent of coverage and capacity. We contend that the domain should embrace more diverse applications to further advance the UMLS research and increasingly achieve quality care.
CONCLUSION
Celebrating the monumental 30-year anniversary of UMLS resources, we explored the historical footprint and landmark milestones of the UMLS research in a bibliometric fashion. The triangulated findings from domain-level citations, subject categories, keyword co-occurrence and bursts, document co-citation networks, and manuscript survey characterized our investigation. Our multilevel analyses identified thematic patterns, emerging technologies, and current challenges as well as the domain’s epistemological characteristics. Methodologically, our review demonstrated high research validity by synthesizing quantitative and qualitative approaches in deriving richer interpretations and implications. We expect that the detailed scientific profile of the UMLS research evaluated in this study will help scientists and communities better align their work in progress and future studies with the identified thematic trends and challenges.
The present study has several limitations, some of which direct our future studies. First, we discovered data loss while conducting the PMID search on the WoS. Two-step data collection was employed because our study leveraged the scientometrics tool kits that use bibliographic records retrieved from the WoS. To complement the data gap between PubMed and WoS, we adopted citation indexing to expand our analysis toward the much broader context of the UMLS research. In addition, we aimed to avoid any overgeneralization of findings per subsection, triangulating consistently identified themes across all the result sections. In the future, we plan to develop 1) an Extract-Transform-Load pipeline incorporating multisource records and 2) an interoperable tool kit leveraging across a variety of bibliographic databases. Second, the collected data sets may underrepresent some of the document types, especially conference papers, due to the WoS’s indexing policy.6,38 Furthermore, the authors could only collect records from the core collection of the bibliographic database because of the affiliated institutions’ subscription status. Therefore, some relevant literature could have been omitted. In the future, we plan to use complementary data sources such as Scopus to enhance the variety as well as inclusiveness of the records. This will let us not only generate a more complete, detailed scientific profile of UMLS research but also better direct future editorship and readership in the research community. Next, we conducted network reduction by selecting only the top 10% most frequently occurring entities per year. Although this threshold is in part intuitive, it can be further strengthened by using refined selection criteria such as h-index or g-index. Finally, we plan to apply the present work’s analytical procedure to other academic domains to discover more generalized understandings of the creation and diffusion of knowledge.
FUNDING
This work was supported by the Ministry of Education of the Republic of Korea and the National Research Foundation of Korea grant number NRF-2019S1A5A8033338.
AUTHOR CONTRIBUTIONS
All authors significantly contributed to the present work. All authors give approval for the final version to be published and agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
CONFLICT OF INTEREST STATEMENT
None declared.
REFERENCES
- 1. Humphreys BL, Lindberg DAB, Schoolman HM, Barnett GO. The Unified Medical Language System: an informatics research collaboration. J Am Med Inform Assoc 1998; 5 (1): 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chen C, Dubin R, Kim MC. Orphan drugs and rare diseases: a scientometric review (2000–2014). Expert Opin Orphan Drugs 2014; 2 (7): 709–24. [Google Scholar]
- 3. Chen C, Dubin R, Kim MC. Emerging trends and new developments in regenerative medicine: a scientometric update (2000–2014). Expert Opin Biol Ther 2014; 14 (9): 1295–317. [DOI] [PubMed] [Google Scholar]
- 4. Kim MC, Jeong YK, Song M. Investigating the integrated landscape of the intellectual topology of bioinformatics. Scientometrics 2014; 101 (1): 309–35. [Google Scholar]
- 5. Kim MC, Zhu Y, Chen C. How are they different? A quantitative domain comparison of information visualization and data visualization (2000–2014). Scientometrics 2016; 107 (1): 123–65. [Google Scholar]
- 6. Zhu Y, Kim MC, Chen C. An investigation of the intellectual structure of opinion mining research. Inf Res 2017; 22 (1): paper 739. [Google Scholar]
- 7. Deerwester S, Dumais ST, Furnas GW, Landauer TK, Harshman R. Indexing by latent semantic analysis. J Am Soc Inf Sci 1990; 41 (6): 391–407. [Google Scholar]
- 8. Garfield E. Citation Indexing: Its Theory and Applications in Science Technology, and Humanities. New York: Wiley; 1979. [Google Scholar]
- 9. Chen C. CiteSpace II: detecting and visualizing emerging trends and transient patterns in scientific literature. J Am Soc Inf Sci Technol 2006; 57 (3): 359–77. [Google Scholar]
- 10. Chen C, Ibekwe-SanJuan F, Hou J. The structure and dynamics of co-citation clusters: a multiple-perspective co-citation analysis. J Am Soc Inf Sci Technol 2010; 61 (7): 1386–409. [Google Scholar]
- 11. Van Eck NJ, Waltman L. Software survey: VOSviewer, a computer program for bibliometric mapping. Scientometrics 2010; 84 (2): 523–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen C, Leydesdorff L. Patterns of connections and movements in dual-map overlays: A new method of publication portfolio analysis. J Assoc Inf Sci Technol 2014; 65 (2): 334–51. [Google Scholar]
- 13. Coleman TF, Moré JJ. Estimation of sparse Jacobian Matrices and Graph Coloring Blems. SIAM J Numer Anal 1983; 20 (1): 187–209. [Google Scholar]
- 14. Brandes U. A faster algorithm for betweenness centrality. J Math Sociol 2001; 25 (2): 163–77. [Google Scholar]
- 15. Brin S, Page L. The anatomy of a large-scale hypertextual Web search engine. Comput Netw ISDN Syst 1998; 30 (1-7): 107–17. [Google Scholar]
- 16. Kleinberg J. Bursty and hierarchical structure in streams In: proceedings of the Eighth ACM SIGKDD International Conference on Knowledge Discovery and Data mining-KDD ‘02 [Internet]. Edmonton, Alberta, Canada: ACM Press; 2002: 91 http://portal.acm.org/citation.cfm? doid=775047.775061 Accessed February 3, 2020 [Google Scholar]
- 17. Blondel VD, Guillaume J-L, Lambiotte R, Lefebvre E. Fast unfolding of communities in large networks. J Stat Mech Theory Exp 2008; 2008 (10): P10008. [Google Scholar]
- 18. Cimino JJ, Clayton PD, Hripcsak G, Johnson SB. Knowledge-based approaches to the maintenance of a large controlled medical terminology. J Am Med Inform Assoc 1994; 1 (1): 35–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cimino JJ. Auditing the unified medical language system with semantic methods. J Am Med Inform Assoc 1998; 5 (1): 41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. Nat Genet 2000. May; 25 (1): 25–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proc Annu AMIA Symp 2001; 2001: 17–21. [PMC free article] [PubMed] [Google Scholar]
- 22. Nadkarni P, Chen R, Brandt C. UMLS concept indexing for production databases: a feasibility study. J Am Med Inform Assoc 2001; 8 (1): 80–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Peng Y, Halper M, Perl Y, Geller J. Auditing the UMLS for redundant classifications. Proc Annu AMIA Symp; 2002. 2002: 612–6. [PMC free article] [PubMed] [Google Scholar]
- 24. Chen Z, Perl Y, Halper M, Geller J, Gu H. Partitioning the UMLS semantic network. IEEE Trans Inf Technol Biomed 2002; 6 (2): 102–8. [DOI] [PubMed] [Google Scholar]
- 25. Gu H, Perl Y, Elhanan G, Min H, Zhang L, Peng Y. Auditing concept categorizations in the UMLS. Artif Intell Med 2004; 31 (1): 29–44. [DOI] [PubMed] [Google Scholar]
- 26. Humphrey SM, Rogers WJ, Kilicoglu H, Demner-Fushman D, Rindflesch TC. Word sense disambiguation by selecting the best semantic type based on journal descriptor indexing: preliminary experiment. J Am Soc Inf Sci Technol 2006; 57 (1): 96–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Savova GK, Masanz JJ, Ogren PV, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation, and applications. J Am Med Inform Assoc 2010; 17 (5): 507–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Campbell JR, Carpenter P, Sneiderman C, Cohn S, Chute CG, Warren J. Phase II evaluation of clinical coding schemes: completeness, taxonomy, mapping, definitions, and clarity. J Am Med Inform Assoc 1997; 4 (3): 238–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rector AL, Bechhofer S, Goble CA, Horrocks I, Nowlan WA, Solomon WD. The GRAIL concept modelling language for medical terminology. Artif Intell Med 1997; 9 (2): 139–71. [DOI] [PubMed] [Google Scholar]
- 30. Rosse C, Mejino JL, Modayur BR, Jakobovits R, Hinshaw KP, Brinkley JF. Motivation and organizational principles for anatomical knowledge representation: the digital anatomist symbolic knowledge base. J Am Med Inform Assoc 1998; 5 (1): 17–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rosse C, Mejino J. A reference ontology for biomedical informatics: the foundational model of anatomy. J Biomed Inform 2003; 36 (6): 478–500. [DOI] [PubMed] [Google Scholar]
- 32. Hamosh A. Online Mendelian Inheritance in Man (OMIM), a knowledge base of human genes and genetic disorders. Nucleic Acids Res 2004; 33 (Database issue): D514–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Smith B, Ceusters W, Klagges B, et al. Relations in biomedical ontologies. Genome Biol 2005; 6 (5): R46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Robinson PN, Köhler S, Bauer S, Seelow D, Horn D, Mundlos S. The human phenotype ontology: a tool for annotating and analyzing human hereditary disease. Am J Hum Genet 2008; 83 (5): 610–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Friedman C, Shagina L, Lussier Y, Hripcsak G. Automated encoding of clinical documents based on natural language processing. J Am Med Inform Assoc 2004; 11 (5): 392–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kuhn M, Campillos M, Letunic I, Jensen LJ, Bork P. A side effect resource to capture phenotypic effects of drugs. Mol Syst Biol 2010; 6 (1): 343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xu H, Stenner SP, Doan S, Johnson KB, Waitman LR, Denny JC. MedEx: a medication information extraction system for clinical narratives. J Am Med Inform Assoc 2010; 17 (1): 19–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mongeon P, Paul-Hus A. The journal coverage of Web of Science and Scopus: a comparative analysis. Scientometrics 2016; 106 (1): 213–28. [Google Scholar]