Abstract
Background
Sarcomas are rare heterogeneous tumours, derived from primitive mesenchymal stem cells, with more than 100 distinct subtypes. Radioresistance remains a major clinical challenge for sarcomas, demanding urgent for effective biomarkers of radiosensitivity.
Methods
The radiosensitive gene Kinesin family member 18B (KIF18B) was mined through bioinformatics with integrating of 15 Gene Expression Omnibus (GEO) datasets and The Cancer Genome Atlas (TCGA) database. We used radiotherapy-sh-KIF18B combination to observe the anti-tumour effect in sarcoma cells and subcutaneous or orthotopic xenograft models. The KIF18B-sensitive drug T0901317 (T09) was further mined to act as radiosensitizer using the Genomics of Drug Sensitivity in Cancer (GDSC) database.
Findings
KIF18B mRNA was significantly up-regulated in most of the subtypes of bone and soft tissue sarcoma. Multivariate Cox regression analysis showed that KIF18B high expression was an independent risk factor for prognosis in sarcoma patients with radiotherapy. Silencing KIF18B or using T09 significantly improved the radiosensitivity of sarcoma cells, delayed tumour growth in subcutaneous and orthotopic xenograft model, and elongated mice survival time. Furthermore, we predicted that T09 might bind to the structural region of KIF18B to exert radiosensitization.
Interpretation
These results indicated that sarcomas with low expression of KIF18B may benefit from radiotherapy. Moreover, the radiosensitivity of sarcomas with overexpressed KIF18B could be effectively improved by silencing KIF18B or using T09, which may provide promising strategies for radiotherapy treatment of sarcoma.
Fundings
A full list of funding can be found in the Funding Sources section.
Keywords: KIF18B, Sarcoma, Radiosensitivity, Bioinformatics analysis
Research in context.
Evidence before this study
A total of 15 Gene Expression Omnibus (GEO) datasets of sarcomas including different subtypes were obtained in October, 2018. The differentially expressed genes (DEGs) between sarcomatous and non-sarcomatous samples were analysed using limma package. RobustRankAggreg (RRA) package was used to integrate genes to avoid problems related to low sample size and inconsistent results in different technological platforms. Gene expression and clinical information data were extracted from the TCGA dataset of sarcoma on September 15, 2018. Genomics of Drug Sensitivity in Cancer (GDSC) database provides an accurate data resource for relocation of existing drugs.
Added value of this study
The specific sarcoma-radiosensitivity related gene, KIF18B, was mined through bioinformatics analysis. It was found that KIF18B overexpression is closely related to radioresistance. The study confirmed that silencing KIF18B or using a KIF18B-sensitive drug (T0901317) could reduce the survival rate of clone formation, promote apoptosis and DNA damage, and effectively improve radiosensitivity on high KIF18B expressing sarcoma cells.
Implications of all the available evidence
This study indicated the promising prospect of KIF18B as a predictive biomarker and a novel target for radiosensitivity in sarcoma patients. Combining radiotherapy and KIF18B-based therapy could exert radiosensitization, which plays a possible role in reducing radiation exposure dose during radiotherapy. We will further explore KIF18B-targeted drugs to improve radiosensitivity of sarcoma.
Alt-text: Unlabelled box
1. Introduction
Sarcomas are rare and heterogeneous tumours with more than 100 distinct subtypes. They are derived from mesenchymal cells, and develop in soft tissues or skeleton in 80% and 20% of the cases, respectively [1]. At present, surgery and radiotherapy are main treatments for sarcomas at present [2]. Although these tumours are sometimes mistakenly called "radio-resistant", radiotherapy can alleviate metastatic patients’ symptoms such as metastasis pain and haemoptysis. Image-guided radiotherapy is more advantageous than surgery in patients with oligometastatic disease, especially in sarcoma [3]. Therefore, radiotherapy is a critical part of the treatment of sarcomas.
Sarcoma is one of the few clinical tumour models for neoadjuvant radiotherapy, which makes it a good model to investigate radiosensitive markers [4]. Moreover, the low sensitivity to radiotherapy of sarcomas makes the search for these markers imperative [5]. Identification of radiosensitivity-related markers could rely on high-throughput microarray platforms. However, the study of individual microarrays often provides insufficient samples, thereby neglecting high abundance molecules [6]. Here we integrated multiple microarray datasets to compensate for the low sample sizes and to provide more convincing results.
Kinesin family member 18B (KIF18B) is closely associated with the pairing and separation of chromosomes during mitosis, and controllings microtubule length, and centring the mitotic spindle at metaphase [7], [8], [9]. Furthermore, KIF18B expression correlates with tumorigenicity and poor prognosis of some tumours [10,11]. For instance, Gao [12] and Yang [13] showed that KIF18B promoted tumour progression by activating β-catenin. A bioinformatics study confirmed that overexpressed KIF18B could work as a potential prognostic biomarker [14]. However, the biological impact of KIF18B expression has not been verified in sarcoma.
We used bioinformatics tools, including the Gene Expression Omnibus (GEO) database [15] and the Cancer Genome Atlas (TCGA) database [16], to mine sarcoma gene profiles to screen radiosensitivity-related genes KIF18B was chosen for further analysis. We investigated the radiosensitivity of KIF18B-silenced sarcoma cells, showing that KIF18B was a biomarker for radioresistance. The KIF18B-sensitive drug T0901317 (T09) was mined from the Genomics of Drug Sensitivity in Cancer (GDSC) database. The combination beteen T09 and radiotherapy provides new insights into the treatment for sarcomas.
2. Materials and methods
2.1. Gene microarray datasets of GEO database and clinical characteristics of TCGA
All relevant data are available from the public database. The data collection related to osteosarcoma and soft tissue sarcoma were obtained from GEO database on October 15, 2018, in which fifteen datasets (GSE42352, GSE14359, GSE59568, GSE52390, GSE51049, GSE62747, GSE21122, GSE764, GSE68295, GSE6476, GSE42977, GSE28511, GSE62544, GSE2719, and GSE90592) were included in the study based on the following criteria: (1) The study of osteosarcoma or soft tissue sarcoma samples. (2) The study containing non-sarcomatous tissues or cell lines. The gene expression profile data characteristics from GEO datasets were shown in Table 1.
Table 1.
The gene expression profile data characteristics
| Reference | Tissue | GEO | Platform | Normal | Tumor | Upregulated Genes | Downregulated Genes |
|---|---|---|---|---|---|---|---|
| Kuijjer ML et al. 2012 | Osteosarcoma | GSE42352 | GPL10295 | 3 | 84 | 304 | 382 |
| Guenther R et al. 2010 | Osteosarcoma | GSE14359 | GPL96 | 2 | 10 | 460 | 452 |
| Iura K, Oda Y et al. 2015 | Liposarcoma | GSE59568 | GPL13915 | 3 | 6 | 603 | 682 |
| Wolf T, Mechtersheimer G et al. 2013 | Liposarcoma | GSE52390 | GPL6947 | 2 | 29 | 61 | 239 |
| Patil N, Heil O et al. 2013 | Liposarcoma | GSE51049 | GPL6884 | 3 | 3 | 11 | 3 |
| Künstlinger H et al. 2014 | Liposarcoma | GSE62747 | GPL4133 | 1 | 7 | 994 | 863 |
| Taylor BS et al. 2010 | Liposarcoma | GSE21122 | GPL96 | 9 | 89 | 246 | 449 |
| Quade BJ et al. 2004 | Uterinesarcoma | GSE764 | GPL80 | 4 | 13 | 49 | 59 |
| Miyata T et al. 2017 | Uterinesarcoma | GSE68295 | GPL6480 | 3 | 3 | 153 | 128 |
| Levine DA et al. 2015 | Uterinesarcoma | GSE64763 | GPL571 | 29 | 25 | 307 | 520 |
| De Rienzo A et al. 2013 | Pleuralmesothelioma | GSE42977 | GPL6790 | 7 | 8 | 711 | 970 |
| Sarver A et al. 2013 | Rhabdomyosarcoma | GSE28511 | GPL6947 | 3 | 18 | 1109 | 1246 |
| Guo X et al. 2015 | Leiomyosarcoma | GSE62544 | GPL11154 | 6 | 17 | 867 | 849 |
| Yoon SS et al. 2005 | Fibrosarcoma | GSE2719 | GPL96 | 1 | 7 | 3 | 17 |
| Laginestra MA | Fibrosarcoma | GSE90592 | GPL14951 | 3 | 26 | 932 | 1245 |
The data of gene expression and clinical information (Table 2) were extracted from TCGA dataset of sarcoma on September 15, 2018.
Table 2.
The clinical information of TCGA sarcoma dataset
| Categories | Frequency | Percent (%) | |
|---|---|---|---|
| Sex | Male | 118 | 45.6 |
| Female | 141 | 54.4 | |
| Age (years) | ≤60 | 128 | 49.4 |
| >60 | 131 | 50.6 | |
| Event | Alive | 161 | 62.2 |
| Dead | 98 | 37.8 | |
| Local disease recurrence | No | 143 | 83.1 |
| Yes | 29 | 16.9 | |
| Tumor necrosis percent | ≤10 | 48 | 50 |
| >10 | 48 | 50 | |
| Metastatic diagnosis (%) | No | 120 | 68.2 |
| Yes | 56 | 31.8 | |
| Radiation therapy | Yes | 73 | 34.3 |
| No | 140 | 65.7 |
2.2. Screening of radiosensitivity-related genes
The GEO database stores functional genomics data sets based on public arrays, allowing users to query and download experiments and gene expression profiles. The limma R package was used to filter differentially expressed genes (DEGs) in each GEO dataset (p-value adjustment<0.05 and |logFC|>1). Using RobustRankAggreg (RRA) R software package, we integrated DEGs from 15 datasets to significantly increase the sample size and to avoid generating less reliable results.
The TCGA dataset of sarcoma comprises 263 sarcomatous samples and two non-sarcomatous samples, including leiomyosarcomas (105), undifferentiated pleomorphic sarcomas (51), dedifferentiated liposarcomas (59), myxofibrosarcomas (25), synovial sarcomas (10), malignant peripheral nerve sheath tumours (9), desmoid tumours (2) and unclassified samples (2). The data were standardized using EdgeR (R package) to analyse DEGs between sarcomatous tissues and non-sarcomatous tissues based on TCGA database. We used a cut-off criteria of |log2FC≥1|, P<0.05 to identify DEGs. The clustering heatmap was generated by R package gplots 3.3.3.
The hub genes were obtained from up/down-regulated DEGs between integrated GEO datasets and TCGA database using VENN diagram.
The expression of hub genes was divided into the high expression group and the low expression group based on the median. Univariate and multivariate analysis of hub genes were performed using Cox regression in the radiotherapy patient group and the non-radiotherapy patient group (P<0.05). Survival (R package) was performed for Kaplan-Meier survival analysis.
2.3. Cell lines and culture
Gene expression data of sarcoma cell lines were obtained from the Cancer Cell Line Encyclopedia (CCLE) database (portals.broadinstitute.org/ccle) [17]. All relevant data are available from the public database.
MNNG/HOS Cl #5 and HT-1080 were cultured in Eagle's minimum basic Medium (#30-2003) with 10% FBS, RD and A-673 were cultured in Dulbecco's Modified Eagle's Medium (#30-2002) containing 10% FBS. These cell lines were maintained at 37 °C with 5% CO2. Above cells were obtained from Beina Bio (Beijing, China).
2.4. Human and animal tissue specimens
In this study, tissue microarray was obtained from Fanpu Biotechnology Co., Ltd (chip number: SFT961, China), containing 36 cases of human bone and soft tissue sarcoma (17 cases of sarcoma, 11 cases of fibrosarcoma, four cases of malignant stromal tumour, two cases of liposarcoma, and two cases of osteosarcoma) and 12 cases of human benign mesenchymal tissues (five cases of leiomyoma, two cases of lipoma, two cases of benign stromal tumour, one case of fibroma, and two cases of other benign tumour). Paraffin sections were also purchased from Fanpu Biotechnology Co., Ltd, containing four cases of osteosarcoma, six cases of normal bone. The average age of all patients was 43.45±17.4 years old (range from 8 to 80 years old); the original clinical data recorded the patients’ age, gender, histotype.
Moreover, paraffin sections of tumour tissues of mice were obtained from subcutaneous xenograft models and orthotopic xenograft models.
2.5. Immunohistochemistry (IHC)
The sections were deparaffinized with xylene and hydrated in graded ethanol and distilled water. The sections were subjected to heat-induced epitope retrieval utilizing 0.01 M citrate buffer (pH 6.0) for 8 min at 80 kPa high pressure. 3% hydrogen peroxide was used to block endogenous peroxidase activity. The sections were incubated with the primary antibody (primary-rabbit Anti-KIF18B antibody, 1: 50, ab121798, abcam; primary-rabbit Anti-Ki67 polyclonal antibody, 1: 200, BS90769, Bioworld) at 4 °C overnight. Tumour sections were then incubated with Goat anti-rabbit IgG (1: 50, ZDR-5306; Zhongshan Gold Bridge Biotechnology, China), and the signal was detected using the DAB Kit (ZLI-9018; Zhongshan Gold Bridge Biotechnology, China).
2.6. Evaluation of Immunohistochemistry
All stained specimens were evaluated blindly by two experienced researchers (ZJ Y. and XL L.). KIF18B is mainly localized in the cytoplasmic and nuclear. Ki67 protein was located mainly in the nuclear. The percentage of positively stained cells (0 to 100%) and the score of immunohistochemical staining intensity (no stain, 0; weak positive, 1; medium intensity, 2;and strong positive, 3) were multiplied as immunoreactive score (IRS), ranged from 0 to 300%.
2.7. ShRNA and transfection
The four human shRNA sequences were provided by GenePharma (Shanghai, China). Cells were cultured with 2 mL medium containing 2 μg shRNA, P3000 Reagent, Lipofectamine 3000 Reagent (Invitrogen, USA) for 6 h at 37 °C, then the medium with vectors were replaced with complete medium. The cells were collected after 48 h.
To conduct the rescue experiments, 2000 ng of shKIF18B-resistant pcDNA3.1-KIF18B expression vectors (GenePharma, China) were transfected into sh-KIF18B MNNG/HOS cells. Transfection was performed as described above.
2.8. Real-Time quantitative RT-PCR
Total RNA was extracted from cells with TRIzol (R1100, Solarbio, China). The purity and concentration of RNA were calculated with optical density (OD) at 260 and 280 nm. The synthesis of cDNA was performed with FastKing RT Kit (KR116, Tiangen, China) by PCR Thermal Cycler Dice (TP600, TaKaRa, Japan). Real-time PCR analyses were performed by SuperReal PreMix Plus (FP205, Tiangen, China) in the Applied Biosystems 7300 Real Time PCR System (Applied Biosystems, Foster City, USA). The expression level of KIF18B was standardized to β-actin (B661102, Sangon, China). The relative gene expression was calculated using the 2-ΔΔCT method. The primer sequences used to amplify KIF18B were 5′-GCTGCAAGTAGTGGTACGGG-3′ (forward) and 5′-CCTCAGGGTTAAACACCAGCA-3′ (reverse), and the β-actin sequences were 5′-GCCGCTATGCCCTCTA-3′ (forward) and 5′-CAATTCATGCTTGATCCCT-3′ (reverse). Experiments were conducted independently in at least triplicate.
2.9. Radiation
Radiation was conducted using a SIEMENS linear accelerator (SIEMENS Medical Systems, Germany). The doses were 0, 2, 4, 6 and 8 Gy with the rate of 2 Gy/min.
2.10. Colony survival experiment
48 h after transfection, cells were seeded on 6-well plates. DMSO (dimethyl sulfoxide) or T0901317 (3 uM, A2249, APExBIO, USA) was added on corresponding groups for another 48h, respectively. Then the cells were irradiated with 0, 2, 4, 6, or 8 Gy every three days for a total of two weeks. 70% ethanol was applied to fixed cells, and crystal violet was applied to stain cells. The number of colonies, defined as >50 cells/colony, was counted. Plating efficiency (PE) was calculated as follows: PE=number of cloning/number of vaccination, and this was compared with the negative control group. Survival rate (SF) was calculated as follows: SF=number of cloning/number of vaccination × PE.
2.11. Measurement of apoptosis
The apoptosis of tumour cells was measured with Annexin-V/FITC kit (Beyotime, Shanghai, China) using MACSQuantTM Flow Cytometer (Miltenyi Biotec, Germany). The cells were irradiated at the dose of 0 or 8 Gy. After irradiation for 24 h, cells were collected and FITC-coupled annexin V was added for 15 min, then propidiumiodide (PI) was supplemented for 5 min. The experiments were performed in triplicate.
2.12. Immunofluorescence
The different transfection-treated cells were exposed to 0 or 8 Gy. After 24 h, cells were fixed for 15 min, permeabilized with 0.1% Triton X-100, and blocked in 5% bovine serum albumin. The anti-γ-H2AX antibody (1: 200, ab11268, Absci, USA) was added at 4 °C overnight. The secondary antibody (goat anti-rabbit fluorescent, 1: 200, E031220; EarthOx) then was added for 1 h in darkness. After soaking in DAPI (C0065, Solarbio, China), cells were observed under a confocal microscope (C2 plus, Nikon, Japan). Multiple fields of view over 200 cells were selected randomly for the counting.
2.13. Data mining of GDSC databases
We confirmed that the reproduction and publication of the GDSC [18] data complied with the organization. We searched for compounds with significant sensitivity of high expressed KIF18B. Scatter plots were generated by R package gplots 3.3.3.
2.14. CCK8 assay
48 h after transfection, cells were seeded on 96-well plates. T09 with the final concentration of 0.04, 0.2, 1, 5, 25 or 125 μM was added for another 48 h. Then the cell viability was measured with Cell Counting Kit-8 (CCK8 APExBIO, USA) assay. The OD values were measured at 450 nm to calculate the cell survival rate. Repeated three times for every test.
2.15. Docking T09 onto KIF18B
The homology modeling of KIF18B protein (accession code: Q86Y91, from NCBI database) based on the template sequence, was constructed using Molecular Operating Environment (MOE) 2018 package (Chemical Computing Group, Montreal, QC, Canada).The whole modeling process was described comprehensively in our previous study [19]. The energy of homology model was minimized with MOE. The feasibility of homology modeling was analysed with Ramachandran plots. The KIF18B homology model was preprocessed with QuickPrep and docked with T09 based on the instruction of MOE General Dock. The interaction and the key amino acid residues were displayed.
2.16. Western blotting (WB)
Whole cell proteins were extracted with the whole Protein Extraction Kit (BB-3101, BestBio, China) and were quantified with the BCA protein assays. Then the proteins were separated by 12% SDS-polyacrylamide gel electrophoresis (PAGE) and were electrotransferred onto 0.45 um polyvinylidene difluoride (PVDF) membranes. After blocked with 5% (w/v) non-fat milk for 2 h, the membranes were incubated at 4 °C overnight with the primary antibody (Primary-rabbit Anti-KIF18B antibody, 1: 2000, ab168812, abcam; β-Actin (13E5) Rabbit mAb, 1: 1000, #4970, CST), followed by incubation with goat anti-rabbit HRP-conjugated secondary antibody (1: 10000, E030120, EarthOx, USA) at room temperate for 1 h. The strip imprint was visualized with an ECL detection system (Pierce, Thermo Fisher Scientific, USA) and densitometry was performed using ImageJ (1.8.0). The experiments were performed in triplicate.
2.17. KIF18B stable transfection
MNNG/HOS Cl #5 cells were transfected with sh-KIF18B 3 plasmid (h) and sh-NC plasmid (h), respectively. The medium containing puromycin antibiotic (P8230, Solarbio, China) was replaced with fresh medium after transfection for 72 h. The method used to select stable cell strains was described in the study [20,21]. Western blot analysis was used to confirm the stable expression of plasmids in MNNG/HOS Cl #5 cells.
2.18. In vivo mouse studies
Animal experiments were approved by the Ethics Committee of China Medical University, and were carried out in accordance with guidelines prescribed by the Institutional Animal Care and Use Committee of China Medical University.
2.18.1. Subcutaneous xenograft model
Female nude mice (6 weeks old, 401, Vital River, China) were randomly divided into the following groups: 1) sh-NC MNNG/HOS cells, 2) sh-NC MNNG/HOS cells+T09, 3) sh-KIF18B MNNG/HOS cells, and 4) sh-KIF18B MNNG/HOS cells+T09. 2 × 106 cells per site with the different treatments were injected subcutaneously into the right sub axillary of the mice. When the tumour volume reached about 50 mm3, mice from different groups were divided randomly into the two subgroups: one sub-group of mice was irradiated (2 Gy × 4 days, total body irradiation), and the other sub-group of mice was non-irradiated (n=6/subgroup). T09 (50 mg/kg) was injected into the tumours 48 h before irradiation in corresponding groups. Tumour volume (T) was estimated every three days, and calculated by V = (L × S2)/2 formula (L and S represent long and short axis of tumours, respectively). Weight of mice was monitored every three days throughout the study period. In order to balance the time of testing across treatment groups, all mice were submitted to euthanasia when the volume of the xenograft was big enough or started to show obvious tumour necrosis. Tumours and main organs were isolated to perform relevant examinations. The survival time of another batch of mice was monitored (n=6/group).
2.18.2. Orthotopic xenograft model
Groups were divided in the same way as described above. Six-week-old nude mice were inoculated cell suspensions by intra-bone marrow administration (2 × 106 cells/mouse). Prior to the inoculation, mice were anesthetized with 2% isoflurane. The left hind limb was bent 90° to allow piercing in tibial plateau with a 25 G needle for the inoculation of 10 μL of cell suspension. Time/dose schedule of radiotherapy was the same as the subcutaneous xenograft model. Tumour volumes were estimated every three days, and were calculated by the following standard formula: length × width2/2. The mice were euthanized when any tumour reached 1.5 cm in diameter (n = 4/subgroup). Primary tumours were removed en bloc along with the surrounding involved bone, including the articulation between the femur and the tibia. The femur was transected with scissors at the proximal border of the tumour; the tibia was transected at the distal border and fixed with 4% paraformaldehyde. Bone was decalcified with decalcifying solution (G1105, G1107, Servicebio, China) before being embedded in paraffin. The blood samples from retrobulbar vein were centrifuged at 1000 g for 10 min to harvest the required serums. The levels of alanine aminotransferase (ALT), aspartate aminotransferase (AST), blood creatinine (CRE), and uric acid (UA) were measured using a chemistry analyser (BS-120, Mindray, China). Tissues (liver, spleen, kidney, bone marrow, intestine) harvested from mice in 0 Gy+sh-NC+T09 group and 0 Gy+sh-NC group were processed for haematoxylin-eosin (H&E) staining, and the stained slides were analysed by experienced researchers (ZJ Y. and XL L.)
2.19. TUNEL assay
One step Terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) apoptosis assay kit was used to analyse the cell apoptosis in tissue sections according to the manufacturer's instructions. Three visual fields were randomly picked.
2.20. Statistical analysis
The correlation between KIF18B expression and clinicopathological parameters of sarcoma patients from TCGA was evaluated by Pearson Chi-square test or Fisher's Exact Test. Univariate and multivariate Cox proportional hazards regression models were used for assessing the association between potential confounding variables and prognosis. Survival analysis was conducted using the Kaplan-Meier method, and differences between cohorts were assessed using the log-rank test. Wilcoxon rank-sum test was used to analyse the differences between groups with non-normally distributed variables. The independent-samples t test was used to compare the continuous variable among the different groups. All statistical analyses were carried out using SPSS statistical software package (ver.16.0, SPSS). P<0.05 was considered to statistically significant.
3. Results
3.1. Identification of differential genes in sarcoma
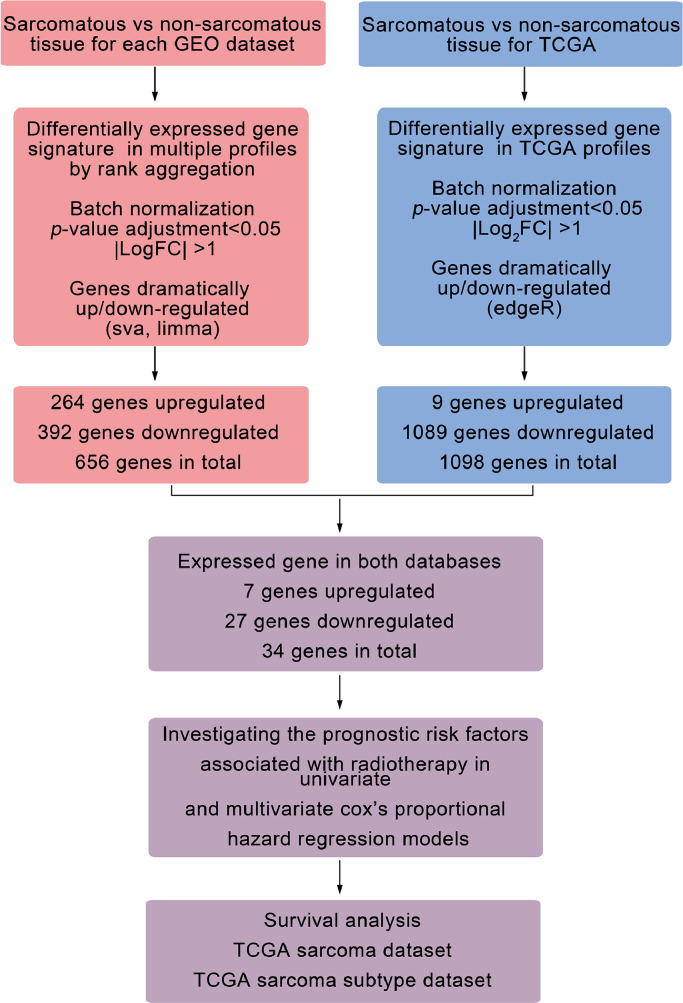
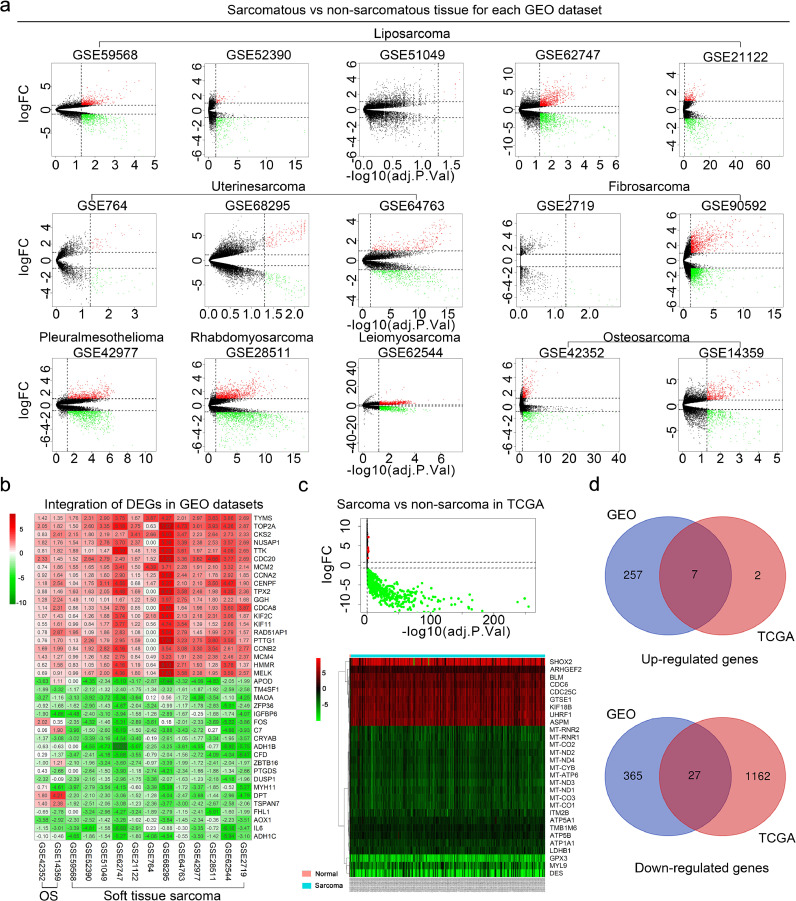
A bioinformatics analysis workflow was shown in Fig. 1. DEGs between sarcomatous and non-sarcomatous gene profiles were obtained from each GEO dataset (Fig. 2a, padj<0.05 and [logFC]>1). The DEGs from each dataset were integrated and analysed using the RobustRankAggreg. The integrated DEGs, including 392 down-regulated genes and 264 up-regulated genes, were identified for further screening (Fig. 2b, Table S1).
Fig. 1.
Flowchart for bioinformatics analysis from GEO and TCGA databases. GEO, gene expression omnibus; NCBI, national center for biotechnology information; TCGA, the cancer genome atlas; logFC, logarithmic foldness; log2FC, log2 counts-per-million.
Fig. 2.
Convergence of DEGs across sarcoma and non-sarcoma. a, The volcanic plot of gene profiles of each GEO dataset (limma R package, padj<0.05 and [logFC]>1). b, The heat maps showed DEGs integrated from the GEO datasets. We showed the top 20 genes with up-regulated and down-regulated expressions. The numbers in each rectangle represented standardized levels of gene expression. OS, osteosarcoma. c, The volcano plot and heatmap analysis were performed by R software with edgeR package (padj<0.05 and [log2FC]>1). Data were downloaded from the TCGA database. The volcano plot showed nine up-regulated DEGs and 1089 down-regulated. The heatmap showed all up-regulated DEGs and top 20 downed-regulated DEGs, respectively. d, The intersection of DEGs between TCGA database and GEO datasets. GEO, gene expression omnibus; TCGA, the cancer genome atlas. (a-c) Up-regulated genes, red; down-regulated genes, green.
Using the TCGA database, 1098 DEGs, 1089 of them down-regulated and nine up-regulated, were found between 263 sarcomatous tissues and two non-sarcomatous (Fig. 2c, Table S2, padj<0.05 and [log2FC]> 1).
The Venn analysis demonstrated 34 intersections of DEGs from TCGA and GEO databases (Fig. 2d), including 27 down-regulated genes and 7 up-regulated genes (Table 3).
Table 3.
Differentially expressed genes in sarcoma
| Total | Genes | |
|---|---|---|
| Up-regulated Genes | 7 | BLM, ASPM, KIF18B, UHRF1, CDC25C, GTSE1, CDC6 |
| Down-regulated Genes | 27 | HSPB8, LIMS2, ALDH2, GPD1, SORBS2, BHMT2, DES, C1orf115, PCK1, GABARAPL1, ECHDC3, CLIC5, ALDH1L1, COL4A4, AZGP1, ACSL1, TST, FAM189A2, AQP1, CCDC69, MYL9, SCNN1A, GPX3, CYB5A, SERPINA5, MAL2, AQP7 |
3.2. KIF18B high expression is a specific risk factor for radiotherapeutic sarcoma patients
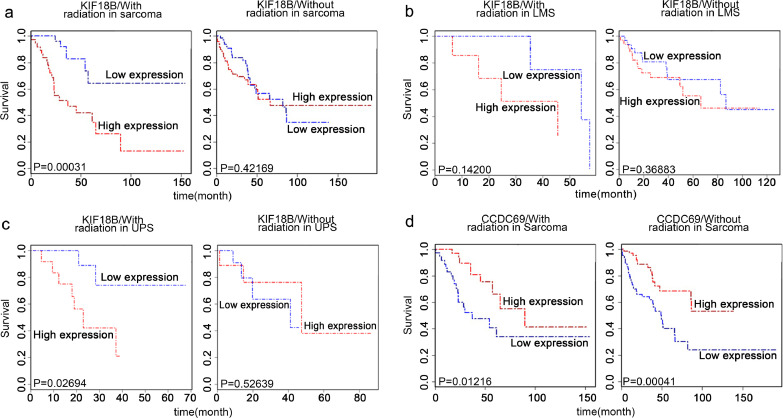
We downloaded the clinical characteristics of 263 soft tissue sarcoma patients from TCGA database (Table S3), including 73 patients that had undergone radiotherapy, 140 patients that had not, and 50 patients unknown, to mine genes related to the radiotherapy outcome. We performed univariate Cox regression analysis (Table S4) in the radiotherapy and the non-radiotherapy groups to assess the association among DEGs and the survival of patients. In the radiotherapy group, recurrence, metastasis, ASPM, GTSE1, KIF18B, UHRF1, DES, CLIC5, and CCDC69 were significantly correlated with overall survival (OS). Further multivariate Cox regression analysis revealed that high expression of KIF18B (P=0.039) and CCDC69 (P=0.038) were independent risk factors of sarcoma with radiotherapy (Table 4). In the group of patients without radiotherapy, univariate Cox regression analysis showed that CCDC69 was also related to OS (Table S4). Besides, we found that the OS of patients that had undergone radiotherapy treatment with high KIF18B expression was significantly worse than that of those with low KIF18B expression; however, no significant differences were observed in patients without radiotherapy (Fig. 3a). The similar phenomenon was also found in two subgroups of sarcomas (Fig. 3b, c; LMS, leiomyosarcoma; UPS, undifferentiated pleomorphic sarcoma). However, the expression of CCDC69 was related to OS of patients regardless of whether radiotherapy was used (Fig. 3d). Therefore, KIF18B was further analysed as a radiosensitivity-related gene. We hypothesised that KIF18B-based therapy might specifically affect the radiotherapy effect.
Table 4.
Univariate and multivariate Cox regression analysis of of clinicopathological data correlated with OS in radiation-treated sarcoma.
| Factors | n | Univariate |
Multivariate (n=60) |
||
|---|---|---|---|---|---|
| HR (95% CI)a | Pa | HR (95% CI)b | Pb | ||
| Sex (Male/Female) | 73 | 1.218 (0.568-2.611) | 0.613 | •• | •• |
| Age,years (≤62/>62) | 73 | 1.547 (0.722-3.316) | 0.262 | 1.819 (0.667-4.963) | 0.242 |
| Local disease recurrence (No/Yes) | 60 | 2.964 (1.192-7.368) | 0.019 | 3.139 (0.996-9.891) | 0.051 |
| Tumor depth (Superficial/Deep) | 63 | 2.947 (0.395-21.961) | 0.292 | •• | •• |
| Tumor necrosis percent (≤15/>15) | 38 | 1.405 (0.503-3.928) | 0.517 | •• | •• |
| Race (Other/White) | 70 | 0.921 (0.216-3.927) | 0.912 | •• | •• |
| New tumor event after initial Treatment | |||||
| (No/YES) | 46 | 111.064 (0.333-37090.821) | 0.112 | •• | •• |
| Metastatic diagnosis (No/YES) | 63 | 3.090 (1.330-7.180) | 0.009 | 1.397 (0.437-4.465) | 0.573 |
| ASPM (low/high) | 73 | 3.004 (1.323-6.820) | 0.009 | 1.030 (0.180-5.893) | 0.973 |
| GTSE1 (low/high) | 73 | 2.213 (1.006-4.868) | 0.048 | 0.619 (0.097-3.972) | 0.613 |
| KIF18B (low/high) | 73 | 4.179 (1.761-9.920) | 0.001 | 7.773 (1.113-54.315) | 0.039 |
| UHRF1 (low/high) | 73 | 2.228 (1.014-4.897) | 0.046 | 0.863 (0.159-4.692) | 0.864 |
| DES (low/high) | 73 | 3.458 (1.458-8.198) | 0.005 | 2.721 (0.832-8.904) | 0.098 |
| CLIC5 (low/high) | 73 | 0.372 (0.169-0.822) | 0.014 | 0.471 (0.159-1.401) | 0.176 |
| CCDC69 (low/high) | 73 | 0.372 (0.167-0.831) | 0.016 | 0.285 (0.087-0.932) | 0.038 |
P value, HR and 95% CI were assessed using univariate Cox regression analysis.
P value, HR and 95% CI were assessed using multivariate Cox regression analysis.
HR, hazard ratio; 95% CI, 95% confidence interval. OS, overall survival. ••, no data to display.
Fig. 3.
High expression of KIF18B is a specific risk factor for sarcoma patients with radiotherapy. a, The effect of mRNA expression of KIF18B on survival time of sarcoma patients (including the two main subtypes, LMS and UPS, and other subtypes) with or without radiotherapy. b, The effect of KIF18B mRNA expression on survival time of LMS patients with or without radiotherapy. c, The effect of KIF18B mRNA expression on survival time of UPS patients with or without radiotherapy. d, The effect of CCDC69 mRNA expression on survival time of sarcoma patients with or without radiotherapy. LMS, leiomyosarcoma. UPS, undifferentiated pleomorphic sarcoma. (a-d) Survival (R package) was performed for Kaplan-Meier survival analysis.
3.3. Silencing KIF18B enhances the radiosensitivity in sarcoma cells
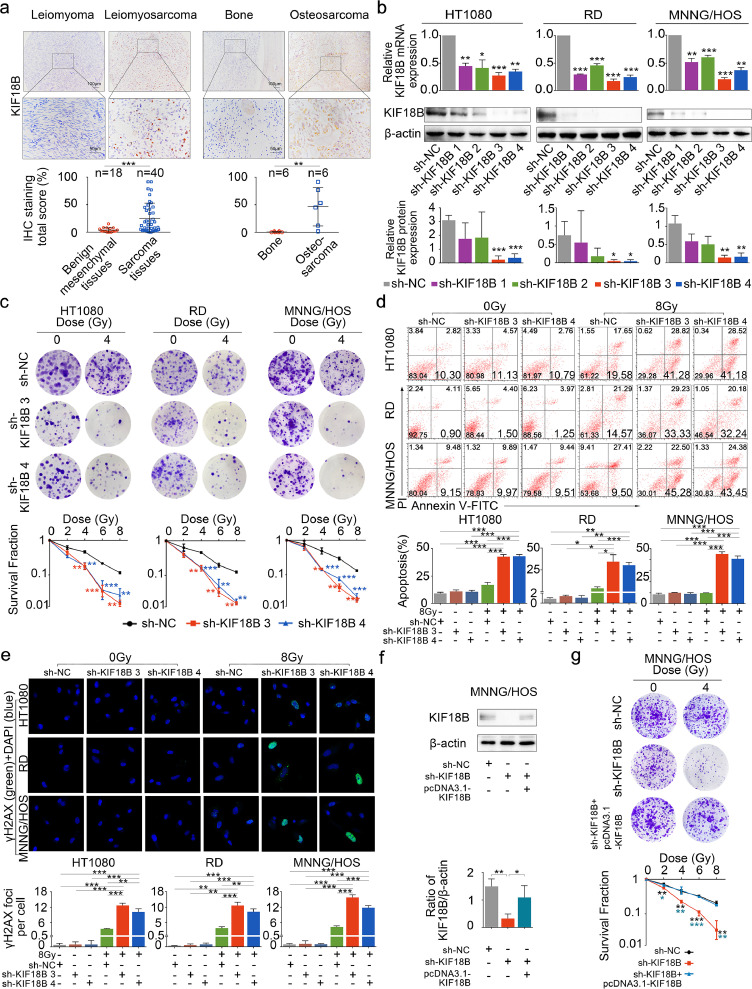
After validation with TCGA and GEO data, the expression of KIF18B also was examined by immunohistochemistry. Results suggested that, compared with benign mesenchymal tissues, KIF18B was significantly overexpressed in bone and soft tissue sarcomas, such as leiomyosarcoma, osteosarcoma, etc (Fig. 4a). We chose high KIF18B expression cell lines, HT1080, RD, MNNG/HOS Cl #5, among bone and soft tissue sarcoma cell lines for the following experiments. To verify the relationship between KIF18B expression and radiosensitivity, the expression of KIF18B was disturbed significantly with sh-KIF18B 3 and sh-KIF18B 4 (Fig. 4b).
Fig. 4.
Inhibition of KIF18B enhances the radiotherapeutic effect on sarcoma cells. a, Representative micrographs presenting immunohistochemical staining of KIF18B in human sarcoma tissues and benign mesenchymal tissues. Human sarcoma tissue samples were collected from 34 cases of soft tissue sarcoma and six cases of osteosarcoma. The six cases of osteosarcoma were shown independently. b, The transfection efficiency of shRNA demonstrated that sh-KIF18B 3 and sh-KIF18B 4 could down-regulate mRNA and protein expression of KIF18B in HT1080, RD and MNNG/HOS cells. c, 48 h after transfection with sh-NC or sh-KIF18B 3, sh-KIF18B 4, the clone formation assay was performed on the cells irradiated with different doses, ranging from 0 Gy, 2 Gy, 4 Gy, 6 Gy, and 8 Gy, every three days for a total of two weeks. Representative images and the surviving fraction (SF) were shown. d, The early apoptosis increased in sh-KIF18B 3, sh-KIF18B 4 group compared with sh-NC group at 8 Gy by flow cytometer analysis. There was no significant change in each group at 0 Gy. e, The expression of γH2AX was detected by immunofluorescence; γH2AX was stained with secondary antibody (green) and its nuclear counter stain was DAPI (blue). f, Relative protein expression of KIF18B was rescued with overexpression of shKIF18B-resistant KIF18B cDNA. g, The clone formation assay was performed in sh-KIF18B cells with re-expression of shKIF18B-resistant KIF18B cDNA. Cells were irradiated with different doses, ranging from 0 Gy, 2 Gy, 4 Gy, 6 Gy, and 8 Gy every three days for a total of two weeks. (a)P values were obtained with Wilcoxon signed ranks test; Error bars denote medians and interquartile ranges; ⁎⁎P<0.01, ⁎⁎⁎P<0.001. (b-g)P values were obtained from independent-samples t test; Error bars denote SD (n=3). (b, c) Compared to sh-NC group, *P<0.05, ⁎⁎P<0.01, ⁎⁎⁎P<0.001. (d-g) *P<0.05, ⁎⁎P<0.01, ⁎⁎⁎P<0.001.
We performed the clonal formation assay to analyse the cell survival fraction (SF). The results showed that the SF in the sh-KIF18B group significantly decreased compared with the sh-NC group at the dose of 4 Gy, 6 Gy and 8 Gy (Fig. 4c, Fig. S1). Although KIF18B knockdown also caused a substantial growth inhibition on sarcoma cells without radiation (Fig. S2), the calculation of the SF excluded the possibility that lower colony numbers in the sh-KIF18B+8 Gy group than in the sh-NC+8 Gy group was just due to KIF18B knockdown. Similarly, the levels of early apoptosis and DNA damage marker (γH2AX) significantly increased in the sh-KIF18B+8 Gy group compared with the sh-NC+8 Gy group, but no significant differences were found between the shKIF18B+0Gy group and the shNC+0Gy group (Fig. 4d, e). Furthermore, sh-KIF18B MNNG/HOS cells were transfected with shKIF18B-resistant pcDNA3.1-KIF18B expression vectors to rescue expression of KIF18B (Fig. 4f). We found a significant recovery of resistance to radiation in the sh-KIF18B+pcDNA3.1-KIF18B group compared with the sh-KIF18B group (Fig. 4g). Besides, we found that overexpression of KIF18B in A-673 cell line (low expression of KIF18B among sarcoma cell lines in CCLE database) induced radiotherapy resistance (Fig. S3a-c). These results indicated that silencing KIF18B enhanced the radiosensitivity of sarcoma cells and that KIF18B played a critical role as a potential target in radiotherapy for sarcoma.
3.4. Silencing KIF18B improves the efficacy of radiotherapy for sarcoma in vivo
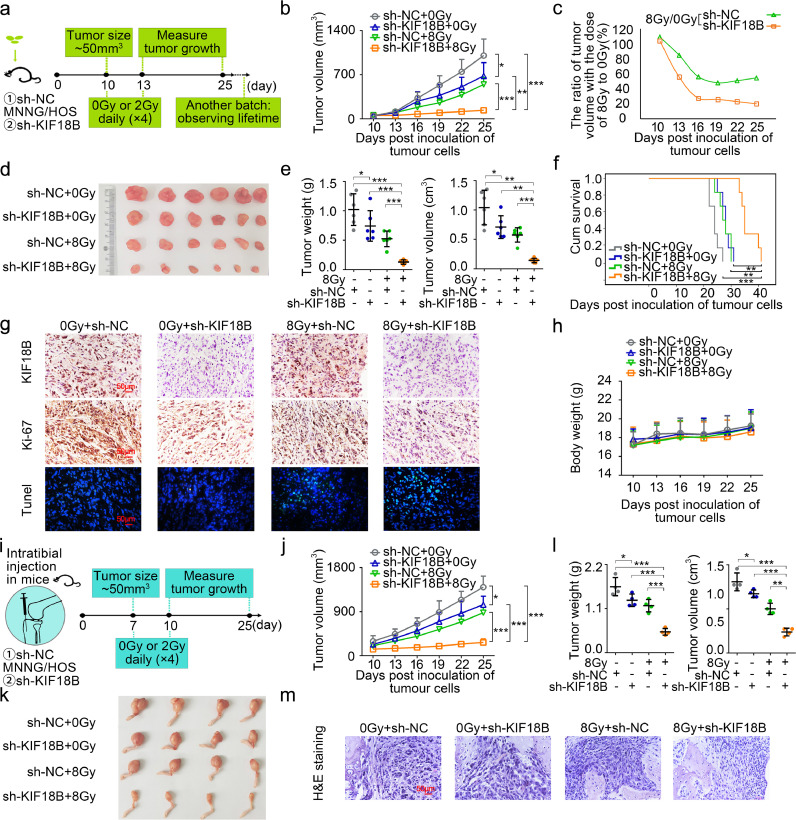
To establish the subcutaneous xenograft model, sh-NC and sh-KIF18B transfected MNNG/HOS cells were injected subcutaneously into mice, respectively (Fig. 5a). Mice were radiated with a dose of 0 (without radiation) or 8 Gy (2 Gy × 4 days). Although radiotherapy or sh-KIF18B alone could moderately delay the tumour growth, radiotherapy combined with sh-KIF18B significantly inhibited tumour growth (Fig. 5b, d, e). By calculating the tumour volume ratio of 8 Gy to 0 Gy, we found that the transplanted tumours in mice of sh-KIF18B group were more sensitive to radiotherapy compared to those in the sh-NC group (Fig. 5c). Kaplan-Meier survival analysis revealed that mice in the sh-KIF18B group with radiotherapy had significant prolongation of survival time compared with the other three groups (Fig. 5f). Immunohistochemistry (IHC) analysis confirmed that expression of KIF18B was decreased in the sh-KIF18B group (Fig. 5g). The Ki-67 expression was significantly inhibited in the sh-KIF18B group treated with radiotherapy, and moderately decreased in the radiotherapy or sh-KIF18B groups alone compared with sh-NC group (Fig. 5g). The TUNEL staining assay found that radiotherapy combined with sh-KIF18B induced the highest levels of apoptosis of all treatments in vivo (Fig. 5g). No significant change in weight was observed in any of the groups during this trial. (Fig. 5h).
Fig. 5.
Sarcomas progression is inhibited in vivo by silencing KIF18B combined with radiotherapy. a, The experimental process of establishing subcutaneous xenograft model (n=6). b, The tumour growth volume curve of mice. c, The ratio of tumour volume with the dose of 8 Gy to 0 Gy. d, The subcutaneous xenograft tumour was removed surgically at day 25 post-tumour implantation. e, The weight and volume of xenograft tumour. f, Kaplan–Meier survival plot showed survival of mice. g, The KIF18B and Ki-67 expression were detected in tumour tissues by IHC assay. Fluorescence staining was performed on TUNEL (green) and chromatin was stained with DAPI (blue). Scale bars, 50 μm. h, Body weight changes in mice. i, The experimental process of orthotopic xenograft model (n=4). j, Tibial xenograft tumour volumes (mm3) were measured every three days. k, The orthotopic xenograft tumours were removed surgically at day 25 post-tumour implantation. l, The weight and volume of orthotopic xenografts tumours. m, Representative photographs of H&E-stained sections of tibial xenograft tumour at day 25 after inoculation. (b, e, j, l)P values were obtained from independent-samples t test; Error bars denote SD. *P<0.05, ⁎⁎P<0.01, ⁎⁎⁎P<0.001. (f)P values were obtained with log-rank test. ⁎⁎P<0.01, ⁎⁎⁎P<0.001.
To better mimic the clinical scenario, we used an orthotopic xenograft model to further explore the sh-KIF18B mediated radiosensitivity in sarcoma (Fig. 5i). Consistent with the results of the subcutaneous xenograft model, the orthotopic tumour volumes were decreased significantly in sh-KIF18B group treated with radiotherapy compared to that in all other groups (Fig. 5j-l). Histologic staining showed that orthotopic tumours broke through the bone cortex and extended into the surrounding soft tissue in the control groups, but not in the sh-KIF18B group treated with radiotherapy (Fig. 5m). In conclusion, these results suggested that silencing KIF18B was beneficial for the efficacy of radiotherapy for sarcoma in vivo.
3.5. The improved radiosensitivity of sarcoma by T09 is associated with KIF18B high expression
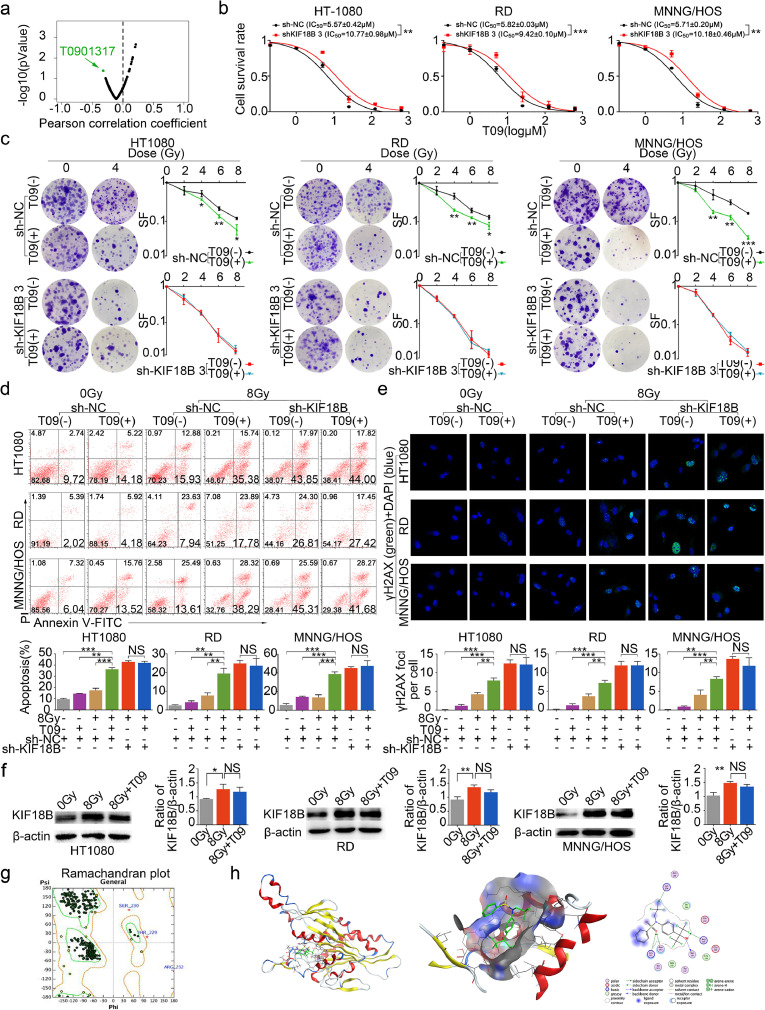
We aimed to mine drugs related to KIF18B to enhance the radiosensitivity of sarcoma with KIF18B high expression. The GDSC database contains data from ∼75,000 experiments, revealing statistically significant drug-gene correlations [22]. We selected drugs whose half-maximal inhibitory concentration (IC50) values were significantly associated with high KIF18B expression. Based on IC50 of drugs in cell lines with different KIF18B expression from the GDSC database, we found that T0901317 [T09, N-(2,2,2-trifluoroethyl)-N-[4-[2,2,2-trifluoro-1-hydroxy-1-(trifluoromethyl)ethyl]phenyl]-benzenesulfonamide], an agonist of liver X receptor (LXR), was a drug that was sensitive to high KIF18B-expressing cells (P=0.042, Fig. 6a). Without radiation, the cell counting kit-8 (CCK8) assay showed statistically significant increases of IC50 for T09 in shKIF18B sarcoma cells compared to that in the sh-NC group (Fig. 6b), indicating that high KIF18B-expressing sarcoma cells were more sensitive to T09. We further evaluated the radiosensitization of T09-treated high KIF18B-expressing sarcoma cells. Using sh-NC sarcoma cells, we found that the SF significantly decreased (Fig. 6c), and early apoptosis and DNA damage significantly increased (Fig. 6d, e) in T09 group treated with radiotherapy compared with the radiotherapy group alone, the T09 group alone, or the untreated group. These results suggested that T09, as a sensitive drug for KIF18B, could effectively increase the radiosensitivity of high KIF18B-expressing sarcoma cells.
Fig. 6.
T09 increases the radiosensitivity of cells with KIF18B high expression. a, The green-labeled compound in volcano plot was sensitive to cells with KIF18B high expression based on GDSC database (P<0.05). b, 48 h after transfection with sh-NC or sh-KIF18B 3, the sarcoma cells were treated with different concentrations of T09 for another 48 h, and then cell survival rate was analysed using CCK8 assay. c, The clone formation assay was performed with a specified dose of X-radiation. Representative images and the surviving fraction were shown. d, Treated cells were stained with Annexin V-FITC/PI to detect the early apoptosis, and then analysed using flow cytometry. e, Treated cells were stained for secondary antibody (green) and DAPI (blue), then analysed by confocal microscopy. f, WB assay confirmed that KIF18B expression increased in sarcoma cells at 8 Gy compared with 0 Gy. T09 could not inhibit KIF18B expression in sarcoma cells at 8 Gy. g, The crystal structure of KIF18B was constructed using the homology model by MOE. The ramachandran plot was used to evaluate the homology modeling of KIF18B. h, Docking T09 onto KIF18B. KIF18B homology model with minimizing energy combined with T09. SF, survival fraction. NS, not significant. (b-f)P values were obtained from independent-samples t test; Error bars denote SD (n=3). (c) Compared to sh-NC group, *P<0.05, ⁎⁎P<0.01, ⁎⁎⁎P<0.001. (b, d-f)⁎⁎P<0.01, ⁎⁎⁎P<0.001.
To investigate whether the increased radiosensitivity by T09 was related to KIF18B, the sh-KIF18B sarcoma cells treated with or without T09 were radiated at a dose of 8 Gy. There were no significant differences in SF (Fig. 6c), cell apoptosis (Fig. 6d) and DNA damage (Fig. 6e) between the T09 group and the drug-free group after silencing of KIF18B. These results suggested that T09 could not improve the radiosensitivity of sarcoma cells after silencing of KIF18B. Based on these studies, we speculated that the improvement of radiosensitivity by T09 might be related to KIF18B.
In order to explore how T09 influenced KIF18B to increase radiosensitivity, we performed a WB assay and found that T09 could not inhibit the expression of KIF18B in sarcoma cells with radiation (Fig. 6f). We further speculated that T09 might interfere with KIF18B through direct binding to it. The molecular operating environment (MOE) docking results indicated that T09 could interact with KIF18B stably(S=-6.941) and the analysis of the binding sites showed that KIF18B-protein 267Ser, 269Arg, 304Lys, 349Asn, 352Lys might bind T09 (Fig. 6g, h). Therefore, T09 might interact with the above amino acid residues of KIF18B to exert inhibiting effects.
In addition, trametinib was selected as a high KIF18B-resistant drug (PCC=0.21, P=0.002) based on the GDSC database (Fig. S4a). Although the IC50 value of trametinib was positively correlated with KIF18B expression (Fig. S4b), the clone formation assay showed trametinib did not affect efficacy of radiotherapy on sarcoma cells (Fig. S4c).
3.6. T0901317 enhances radiosensitivity of sarcoma cells in vivo
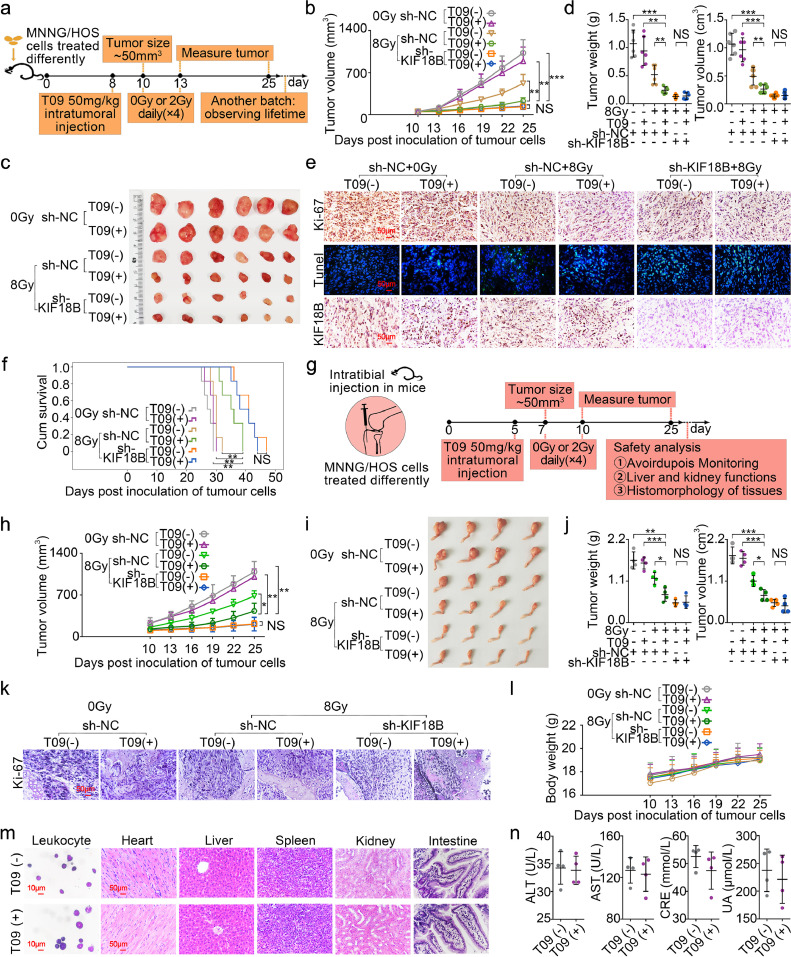
In order to investigate the inhibitory effect of T09 on the growth of sarcoma cells in vivo, we used a subcutaneous xenograft model (Fig. 7a). Using the sh-NC sarcoma cells, the tumour volumes in the T09+8Gy group was significantly smaller than the radiotherapy group alone, the T09 group alone, or the untreated group (Fig. 7b-d). Similarly, Ki67 expression was significantly lower in the T09 group with radiotherapy (Fig. 7e), and the TUNEL assay showed that T09 group treated with radiotherapy had higher ratio of cell apoptosis (Fig. 7e). Additionally, the combination of T09 and radiotherapy significantly extended the survival time of the sarcoma-bearing mice (Fig. 7f). The intratibial inoculation of MNNG/HOS cell suspensions in mice was also performed to determine the contribution of T09 to radiosensitivity in the orthotopic xenograft model. Tibial xenograft tumour growth was suppressed significantly in the T09 group treated with radiotherapy (Fig. 7g-k). In summary, T09 could significantly enhance the radiosensitivity of sarcoma cells in vivo.
Fig. 7.
Sarcoma radiosensitivity enhancement in vivo by T09 is associated with inhibition of KIF18B. The mice were randomly into the following groups: 1) sh-NC MNNG/HOS cells, 2) sh-NC MNNG/HOS cells+T09, 3) sh-KIF18B MNNG/HOS cells, 4) sh-KIF18B MNNG/HOS cells+T09. Nude mice in the radiotherapy group were treated with 8 Gy (2 Gy × 4 days). a, The establishment of experimental model of subcutaneous xenograft tumour (n=6). b, The tumour growth volume curve of each group of mice. c, The subcutaneous xenograft tumours at day 25 post-tumour implantation. d, The weight and volume of subcutaneous xenograft tumours. e, IHC assay of Ki-67 and KIF18B in tumour tissues. Immunofluorescence staining was performed on TUNEL (green) and chromatin was stained with DAPI (blue). Scale bars, 50 μm. f, Kaplan–Meier survival plot showed survival of mice. g, The experimental process of orthotopic xenograft model (n=4). h, Tibial xenograft volumes (mm3) were measured every three days. i, The orthotopic xenograft tumours were removed surgically at day 25 post-tumour implantation. j, The weight and volume of orthotopic xenografts tumours. k, Representative photographs of H&E-stained sections of tibial xenograft tumour at day 25 after inoculation. Scale bars, 50 μm. l, Body weight changes in mice. m, Histopathological studies in various treatment groups. Bone marrow leukocyte (wright-giemsa staining, scale bar, 10 μm); heart, liver, spleen, kidney, intestine (H&E staining, scale bar, 50 μm). n, The detection results of serum biochemical parameters of mice in each treatment group. ALT, alanine aminotransferase; AST, aspartate aminotransferase; CRE: creatinine; UA: uric acid. NS, not significant. (b, d, h, j)P values were obtained from independent-samples t test; Error bars denote SD. *P< 0.05, ⁎⁎P< 0.01, ⁎⁎⁎P< 0.001. (f)P values were obtained with log-rank test. *P< 0.05, ⁎⁎P<0.01. (b-d) The data in 8Gy sh-NC (T09-) group presented was the same as the data in Fig. 5. (h-j) The data presented was the same as the data in Fig. 5 in 0Gy sh-NC (T09-) group, 8Gy sh-NC (T09-) group, and 8Gy sh-KIF18B (T09-) group.
Using the sh-KIF18B sarcoma cells, we found that there were no significant differences in tumour volume and survival time between the T09 group and the drug-free group with radiation (Fig. 7b-d, h-j). IHC and TUNEL assays showed that there were no significant differences in the expression of Ki-67 (Fig. 7e) and the proportion of apoptosis (Fig. 7e). The results suggested that T09 cannot exert additional radiosensitization in vivo after silencing of KIF18B. Moreover, the IHC assay showed that T09 cannot inhibit the KIF18B expression of sarcoma in sh-NC sarcoma cells (Fig. 7e). This was consistent with the previous results, indicating that T09 increased the radiosensitivity of sarcoma cells might through interfering with KIF18B rather than inhibiting KIF18B expression.
Finally, we evaluated the safety of T09 treatment in vivo. There were no significant differences in the weight of mice among disparate groups (Fig. 7l). The T09 (+) and T09 (-) groups without radiation showed no difference of serum biochemical parameters (Fig. 7n). Microscopic evaluations of stained bone marrow smear did not manifest any morphological abnormalities of bone marrow cells in the above two groups (Fig. 7m). The vital organs of the mice were assessed by H&E staining, there was no observable organ/tissue damage including liver, kidney, heart, spleen, and intestine (Fig. 7m). The results showed that treatment of T09 would not impair the health of mice.
4. Discussion
Clinical studies have demonstrated that resistance to radiotherapy is a critical factor for restricting the therapy of sarcoma and contributes to its low survival rate and poor prognosis. The low incidence and the complexly heterogenic sarcoma subtypes hindered the development of effective treatments. In this study, we identified DEGs through bioinformatics analysis with integrating of TCGA and GEO datasets. The found DEGs could be used as targets in different sarcoma subtypes. We conducted GSEA and Gene Ontology (GO) analysis of the DEGs as well, and found that the occurrence of sarcoma could be induced by the dysregulation of cell cycle and mitosis (Fig. S5). We further screened out the radiotherapy resistance-related gene KIF18B, which is also a mitosis related gene. We confirmed that inhibition of KIF18B could enhance the radiosensitivity of sarcoma cells. Our results indicated that KIF18B plays a critical role as a potential sarcoma-specific target in radiation therapy. Furthermore, we selected T0901317, a drug sensitive to KIF18B, to provide a potential treatment for radiosensitizing sarcoma tumours.
Kinesin superfamily proteins (KIFs) that participate in cell mitosis were first identified by Vale et al. [23] Their dysregulation might be closely related to the occurrence of tumours such as liver cancer, cervical cancer, and breast cancer [10,11,24]. This study found that KIF18B was significantly up-regulated in different sarcoma subtypes, which correlated with poor prognosis (Table S5). Furthermore, members of the KIF family could also respond to cellular DNA repair [25], and inhibiting DNA repair has become an attractive treatment to enhance the cytotoxic effects of ionizing radiation [26]. We found that KIF18B was an independent risk factor that affected specifically the prognosis of sarcoma patients with radiotherapy. Besides, KIF18B expression in sarcoma cells was significantly increased when exposed to X-ray irradiation (Fig. 6f), which might facilitate the radiotherapy resistance of sarcoma. Moreover, this study confirmed that inhibition of KIF18B could enhance the efficacy of radiotherapy. Repair of DNA lesions and the inhibition of apoptosis represent two main mechanisms of tumour radioresistance [27]. H2AX could rapidly phosphorylate and locate at the DNA lesions site upon DNA damage and could be used as a sensitive marker for DNA lesions. Our data also suggested that up-regulation of H2AX phosphorylation and increase of apoptosis played a vital role in KIF18B knockdown-mediated enhancement of radiosensitivity in sarcoma cells. Tumour cells undergo clonogenic death via multiple mechanisms in addition to apoptosis, such as mitotic catastrophe. Cells in the G2/M phase have been found to have the highest sensitivity to the radiation [28,29]. Microtubule stabilizing agents can lead to tumour cell cycle arrest in the G2/M-phase [30], and hence, increase radiosensitivity of cells [31]. Similarly, microtubule destabilizers such as vincristine, which inhibits microtubule polymerization to cause cell cycle arrest at the M phase [32], might potentially exert radiosensitization theoretically, even though this has not been reported yet. KIF18B is important for microtubule polymerization, and its expression level is maximal at late G2/M-phase [9], suggesting KIF18B knockdown might exert similar effects like vincristine to regulate cell cycle and to increase radiosensitivity of cells. In our study, we found that KIF18B knockdown alone (without radiation) exerted an antiproliferative action on sarcoma cells, but not induced apoptosis of cells. The phenomon might also be due to cell-cycle arrest, which was similar with the results by Hua Wang and CorachánA [33,34]. We cannot rule out the possibility that more than one mechanisms contribute to KIF18B knockdown-mediated radiosensitization, and we will explore the mechanisms in-depth in future research.
Additionally, we explored the association between DEGs and the survival of patients undergoing chemotherapy based on TCGA database. The results showed that KIF18B expression was not related to prognosis of chemotherapy patients (Fig. S6), indicating that KIF18B knockdown might not improve chemosensitivity of sarcoma. A possible explanation for this might be that chemosensitivity and radiosensitivity are affected through different DNA damage response (DDR) paths. For example, ionizing radiation (IR) directly induces double-strand breaks (DSB), while many chemotherapeutic agents indirectly induce DSBs via interfering DNA topoisomerases [35,36]. According to the GSEA analysis, KIF18B functional enrichment was significantly related to nucleotide excision repair (Fig. S7), which was directly associated with DDR. Therefore, KIF18B knockdown might specifically affect radiosensitivity, instead of chemosensitivity. Our data provided compelling evidence that KIF18B could be a viable target from improving radiation therapy, and therefore, exploring new drugs for inhibiting KIF18B could be useful.
We mined the high KIF18B expression (radioresistance)-sensitive drug T09, based on the IC50 of different drugs in cell lines with different KIF18B expression levels from GDSC. Liver X receptor agonist T09 was reported to exert synergies in the treatment of lung, colon, and prostate cancer [37], [38], [39]. Our study proved that T09 could sensitize high-KIF18B expressing sarcoma cells to radiotherapy, whereas it did not exert additive radionsensitization in KIF18B-knockdown sarcoma cells, indicating that the increased radiosensitivity by T09 might be related to KIF18B. Previous studies reported the correlation between radiosensitivity and LXR signaling. For example, LXR activation was indirectly related to radioresistance on the head and neck squamous cell carcinomas cell line SCC-61 [40] and radiosensitization linked to LXR inactivation or deficiency in irradiated macrophages [41]. These results are not consistent with our study. We demonstrated that LXR agonist T09 could effectively increase the radiosensitivity of KIF18B high-expressing sarcoma cells. These results suggested that the radiosensitization effect of T09 could not correlate with LXR activation. Another possibility is that the difference might be caused by heterogenous KIF18B expression in cells or the different cell of origin in cancer or sarcoma. There is no published study reporting any correlation between LXR signaling and KIF18B, or even KIF family. Conversely, if T09 increased radiosensitivity through other pathways rather than KIF18B, T09 and sh-KIF18B would exert an additive action, the radiotherapy of the T09+sh-KIF18B group should be more effective than that of the sh-KIF18B group. However, the radiosensitivity of the T09+sh-KIF18B group was similar to that of the sh-KIF18B group, indicating that T09 could not exert additive radiosensitization in sarcoma cells with sh-KIF18B (Fig. S8). Therefore, we speculated that T09 improved the curative effect of radiotherapy via affecting KIF18B. According to our analysis, T09 could affect KIF18B in either of the following ways: direct regulation of KIF18B expression or function; indirect regulation of upstream signal of KIF18B to affect KIF18B expression. Our study found that the expression of KIF18B in T09 group was not suppressed compared with the drug-free group at 8 Gy (Fig. 6f), indicating that T09 could not influence the expression of KIF18B directly or influence it indirectly via regulation of upstream signals. Therefore, we speculated that T09 might affect the function of KIF18B. Drugs might affect the function of proteins or interfere with their activity through direct binding to protein functional domains. However, no specific binding domain of KIF18B has yet been described as there are few studies on them. In this study, the crystal structure of KIF18B was simulated by MOE, and the results indicated that T09 might bind KIF18B through 267Ser, 269Arg, 304Lys, 349Asn, and 352Lys to interfere KIF18B.
We analysed the efficacy and preliminary safety of radiotherapy combined with T09 or KIF18B knockdown in vivo. The inhibition of KIF18B induced a robust and long-lasting radiation response, eliminating tumour growth and extending survival time in a subcutaneous xenograft model. T09 was also been shown to increase the radiosensitivity of sarcoma in vivo. Furthermore, post-treatment measurements of these tumour tissues, including proliferation and apoptosis, were consistent with the results in vitro. Implanting the tumour at the orthotopic site, or site corresponding to the original tumour in the patient, makes it behave more similar to the original tumour in patients. Besides, intratibial injection offers the ability to study orthotopic tumour growth in bone microenvironment [42]. Therefore, an orthotopic model is especially appropriate concerning radiotherapy. The results of this orthotopic tumour model showed that KIF18B-based therapy could increase the sarcoma radiosensitivity, which is consistent with the subcutaneous xenograft model. Some studies used the patient-derived orthotopic xenograft (PDOX) model that showed more sarcoma-like behaviours and deeper invasion [43,44], which give us a hint of potential research directions in the future. In addition, we found that the combination of radiotherapy with KIF18B knockdown could inhibit tumour growth, but it could not induce tumour stasis. Tumour-bearing mice would reach the terminal criteria from 36 to 47 days. This might because the time point of radiotherapy mainly focused on early stage (from Day 10 to Day 13), and the remaining tumour cells might continue to proliferate. Meanwhile, resistance of xenograft to the combination of T09 and radiotherapy could not be ruled out. Nevertheless, the survival time was significant elongated in sh-KIF18B+8 Gy group, further research will be carried out.
We preliminarily investigated the safety of KIF18B-based therapy. No significant apoptosis was found in sarcoma cells treated with sh-KIF18B at 0Gy, and the results of the bioinformatics analysis and immunohistochemistry showed that KIF18B has almost no expression in most normal tissues (Fig. S9). This suggested that decrease of KIF18B might not cause apoptosis and other side effects, and hence KIF18B might be a useful therapeutic target for radiosensitization in sarcoma. Moreover, there was no detectable weight loss and no abnormal changes of biochemical indexes (ALT, AST, CRE, UA) and organ pathology in both T09 (+) and T09 (-) groups, indicating that T09 had nearly no toxicity in these mouse models. Additionally, KIF18B-based therapy radiosensitization might play a role in reducing radiation exposure dose during radiotherapy. Our results provided hope for the clinical application, and we will further explore the value of T09 in the radiosensitization of sarcoma.
In summary, based on bioinformatics methods integrating GEO microarray data and RNAseq from TCGA database, we identified that KIF18B was overexpressed in multiple sarcoma subtypes and was associated with poor prognosis of sarcoma patients. We confirmed that high KIF18B expression was related to sarcoma radiation resistance, which indicated that KIF18B could be used as a predictive biomarker to evaluate the radiotherapy efficacy and as a potential therapeutic target in sarcoma patients. Meanwhile, the knockdown of KIF18B or using T09 to target KIF18B could significantly enhance radiosensitivity of sarcoma cells, and could also improve tumour sensitivity to radiotherapy in subcutaneous and orthotopic xenograft models. The mechanism of radiosensitization of T09might be related to that T09 interfering with the functional region of KIF18B.
These results indicated that sarcomas with low expression of KIF18B may benefit from radiotherapy, and the radiosensitivity of sarcoma with high KIF18B expression could be effectively improved by silencing KIF18B or using T09, which may be a window for overcoming radioresistance and provide potential strategies for radiosensitization of sarcoma.
Funding sources
This work was supported by following foundations: (a). NSFC-Liaoning Joint Fund Key Program (No. U1608281) hosted by Minjie Wei who contributed to experiment design and guidance. (b). National Natural Science Foundation of China (No. 81601370) hosted by Zhaojin Yu who contributed to experiment design .
Declaration of Competing Interests
The authors declare no conflict of interest.
Author Contributions
Z.Y. and M.W. conceived this study. W.L. and B.Z. conceived and performed the experiments. X.W. analysed the data. X.L. developed statistical analysis. H.T. wrote the manuscript. J.Z. and M.W. supervised the research. W.L. and Z.Y. contributed equally in this paper. All authors participated in revising the manuscript and agreed to the final version.
Acknowledgements
The authors would like to acknowledge the Key Laboratory of Precision Diagnosis and Treatment of Gastrointestinal Tumors, Ministry of Education (China Medical University, Shenyang, China) for providing the space and equipment for conducting the experiments.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2020.103056.
Contributor Information
Jingdong Zhang, Email: jdzhang@cancerhosp-ln-cmu.com.
Minjie Wei, Email: mjwei@cmu.edu.cn.
Appendix. Supplementary materials
Fig. S1. 48 h after transfection with sh-NC or sh-KIF18B 3, sh-KIF18B 4, the clone formation assay was performed on the cells irradiated with different doses, ranging from 0 Gy, 2 Gy, 4 Gy, 6 Gy, and 8 Gy, every three days for a total of two weeks. P values were obtained from independent-samples t test; Error bars denote SD (n=3). Compared to sh-NC group, *P<0.05, ⁎⁎P<0.01, ⁎⁎⁎P<0.001.
Fig. S2. Cell proliferation assays in sarcoma cell lines transfected with sh-NC, sh-KIF18B 3, or sh-KIF18B 4. P values were obtained from independent-samples t test; Error bars denote SD (n=3). Compared to sh-NC group, *P<0.05, ⁎⁎P<0.01, ⁎⁎⁎P<0.001.
Fig. S3. Overexpression of KIF18B enables A-673 to be more resistant to radiotherapy. a, KIF18B expression levels of sarcoma cell lines. b, WB assay showed that pcDNA3.1-KIF18B could up-regulate KIF18B protein expression in A-673. c, The clone formation assay was performed in A-673 cells transfected NC or pcDNA3.1-KIF18B with different doses of radiation, ranging from 0 Gy, 2 Gy, 4 Gy, 6 Gy, and 8 Gy every three days for a total of two weeks (n=3). (b, c)P values were obtained from independent-samples t test; Error bars denote SD (n=3). Compared to NC group, *P<0.05, ⁎⁎P<0.01.
Fig. S4. Trametinib cannot enhance the radiotherapeutic effect on sarcoma cells. a, KIF18B mRNA expression was associated with drug sensitivity described in the GDSC project. PCC, pearson correlation coefficient. b, Cell survival rate was analysed using CCK8 assay in MNNG/HOS cells transfected with sh-NC or sh-KIF18B by treatment with different concentrations of trametinib for 48 h (n=3). c, The clone formation assay was performed in MNNG/HOS cells treated with or without trametinib at different doses every three days for a total of two weeks (n=3).
Fig. S5. GSEA and GO analysis of DEGs in sarcoma compared to non-sarcomatous tissue. a, GSEA, gene set enrichment analysis; NES, normalized enrichment score; FDR, false discovery rate. P < 0.01 and FDR < 0.05 were considered statistically significant. b, GO, Gene Ontology; BP, Biological process.
Fig. S6. KIF18B expression is irrelevant to survival time of chemotherapeutic patients in sarcoma, based on TCGA database. a, The effect of mRNA expression of KIF18B on survival time of sarcoma patients with or without doxorubicin. b, The effect of mRNA expression of KIF18B on survival time of sarcoma patients with or without gemcitabine. c, The effect of mRNA expression of KIF18B on survival time of sarcoma patients with or without paclitaxel. (a-c) Survival (R package) was performed for Kaplan-Meier survival analysis.
Fig. S7. GSEA results of KIF18B reference gene set of sarcoma patients. GSEA, gene set enrichment analysis; NES, normalized enrichment score; FDR, false discovery rate; FWER P, familywise-error rate p-value <0.05.
Fig. S8. T09 could not increase apoptosis and DNA repair of sh-KIF18B sarcoma cells compared with sh-NC.P values were obtained from independent-samples t test; Error bars denote SD (n=3). *P<0.05, ⁎⁎P<0.01, compared to sh-NC group.
Fig. S9. KIF18B expression in various tissues. a, KIF18B protein expression was detected in many tissues by immunohistochemistry assay. The samples, the same with the samples in Fig. 4a, were shown independently. b, The mRNA expression of KIF18B in many tissues, such as lung, breast, thyroid et al., were analysed based on TCGA.
References
- 1.Florou V, Nascimento AG, Gulia A, de Lima Lopes G., Jr Global health perspective in sarcomas and other rare cancers. Am Soc Clin Oncol Educ Book. 2018;38:916–924. doi: 10.1200/EDBK_200589. [DOI] [PubMed] [Google Scholar]
- 2.Greto D, Loi M, Terziani F. A matched cohort study of radio-chemotherapy versus radiotherapy alone in soft tissue sarcoma patients. Radiol Med. 2018 doi: 10.1007/s11547-018-0939-7. [DOI] [PubMed] [Google Scholar]
- 3.Grilley-Olson JE, Webber NP, Demos DS, Christensen JD, Kirsch DG. Multidisciplinary management of oligometastatic soft tissue sarcoma. Am Soc Clin Oncol Educ Book. 2018;38:939–948. doi: 10.1200/EDBK_200573. [DOI] [PubMed] [Google Scholar]
- 4.Chan CH, Wong P. Molecular predictors of radiotherapy response in sarcoma. Curr Treat Options Oncol. 2016;17(1):2. doi: 10.1007/s11864-015-0381-1. [DOI] [PubMed] [Google Scholar]
- 5.Olsen CE, Sellevold S, Theodossiou T, Patzke S, Berg K. Impact of genotypic and phenotypic differences in sarcoma models on the outcome of photochemical internalization (PCI) of bleomycin. Photodiagnosis Photodyn Ther. 2017;20:35–47. doi: 10.1016/j.pdpdt.2017.08.010. [DOI] [PubMed] [Google Scholar]
- 6.Li Y, Gu J, Xu F, Zhu Q, Ge D, Lu C. Transcriptomic and functional network features of lung squamous cell carcinoma through integrative analysis of GEO and TCGA data. Sci Rep. 2018;8(1):15834. doi: 10.1038/s41598-018-34160-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharp DJ, Rogers GC, Scholey JM. Microtubule motors in mitosis. Nature. 2000;407(6800):41–47. doi: 10.1038/35024000. [DOI] [PubMed] [Google Scholar]
- 8.McHugh T, Gluszek AA, Welburn JPI. Microtubule end tethering of a processive kinesin-8 motor Kif18b is required for spindle positioning. J Cell Biol. 2018;217(7):2403–2416. doi: 10.1083/jcb.201705209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee YM, Kim E, Park M. Cell cycle-regulated expression and subcellular localization of a kinesin-8 member human KIF18B. Gene. 2010;466(1-2):16–25. doi: 10.1016/j.gene.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 10.Itzel T, Scholz P, Maass T. Translating bioinformatics in oncology: guilt-by-profiling analysis and identification of KIF18B and CDCA3 as novel driver genes in carcinogenesis. Bioinformatics. 2015;31(2):216–224. doi: 10.1093/bioinformatics/btu586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu YQ, Wang AP, Zhu BQ. KIF18B promotes tumor progression through activating the Wnt/beta-catenin pathway in cervical cancer. Oncotargets Ther. 2018;11:1707–1720. doi: 10.2147/OTT.S157440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao T, Yu L, Fang ZW. KIF18B promotes tumor progression in osteosarcoma by activating beta-catenin. Cancer Biol Med. 2020;17(2):371–386. doi: 10.20892/j.issn.2095-3941.2019.0452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang B, Wang SZ, Xie H. KIF18B promotes hepatocellular carcinoma progression through activating Wnt/beta-catenin-signaling pathway. J Cell Physiol. 2020 doi: 10.1002/jcp.29444. [DOI] [PubMed] [Google Scholar]
- 14.Li TF, Zeng HJ, Shan Z. Overexpression of kinesin superfamily members as prognostic biomarkers of breast cancer. Cancer Cell Int. 2020;20:123. doi: 10.1186/s12935-020-01191-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edgar R, Domrachev M, Lash AE. Gene Expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cancer Genome Atlas Research N, Weinstein JN, Collisson EA. The cancer genome atlas pan-cancer analysis project. Nat Genet. 2013;45(10):1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barretina J, Caponigro G, Stransky N. The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483(7391):603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang W, Soares J, Greninger P. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013;41(Database issue) doi: 10.1093/nar/gks1111. D955-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng A, Zhang L, Song X, Wang Y, Wei M, Jin F. Clinical implications of a novel prognostic factor AIFM3 in breast cancer patients. BMC Cancer. 2019;19(1):451. doi: 10.1186/s12885-019-5659-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salazar N, Munoz D, Hoy J, Lokeshwar BL. Use of shRNA for stable suppression of chemokine receptor expression and function in human cancer cell lines. Methods Mol Biol. 2014;1172:209–218. doi: 10.1007/978-1-4939-0928-5_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berlivet S, Houlard M, Gerard M. Loss-of-function studies in mouse embryonic stem cells using the pHYPER shRNA plasmid vector. Methods Mol Biol. 2010;650:85–100. doi: 10.1007/978-1-60761-769-3_7. [DOI] [PubMed] [Google Scholar]
- 22.Nguyen L, Dang CC, Ballester PJ. Systematic assessment of multi-gene predictors of pan-cancer cell line sensitivity to drugs exploiting gene expression data. F1000Res. 2016;5 doi: 10.12688/f1000research.10529.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J, Li S, Zhou S. Kinesin superfamily protein expression and its association with progression and prognosis in hepatocellular carcinoma. J Cancer Res Ther. 2017;13(4):651–659. doi: 10.4103/jcrt.JCRT_491_17. [DOI] [PubMed] [Google Scholar]
- 24.Lucanus AJ, Yip GW. Kinesin superfamily: roles in breast cancer, patient prognosis and therapeutics. Oncogene. 2018;37(7):833–838. doi: 10.1038/onc.2017.406. [DOI] [PubMed] [Google Scholar]
- 25.Guerrero-Preston R, Hadar T, Ostrow KL. Differential promoter methylation of kinesin family member 1a in plasma is associated with breast cancer and DNA repair capacity. Oncol Rep. 2014;32(2):505–512. doi: 10.3892/or.2014.3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morgan MA, Lawrence TS. Molecular pathways: overcoming radiation resistance by targeting DNA damage response pathways. Clin Cancer Res. 2015;21(13):2898–2904. doi: 10.1158/1078-0432.CCR-13-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dumont F, Le Roux A, Bischoff P. Radiation countermeasure agents: an update. Expert Opin Ther Pat. 2010;20(1):73–101. doi: 10.1517/13543770903490429. [DOI] [PubMed] [Google Scholar]
- 28.Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Ann Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 29.Cerciello F, Hofstetter B, Fatah SA. G(2)/M cell cycle checkpoint is functional in cervical cancer patients after initiation of external beam radiotherapy. Int J Radiat Oncol. 2005;62(5):1390–1398. doi: 10.1016/j.ijrobp.2004.12.086. [DOI] [PubMed] [Google Scholar]
- 30.Rohrer Bley C, Furmanova P, Orlowski K. Microtubule stabilising agents and ionising radiation: multiple exploitable mechanisms for combined treatment. Eur J Cancer. 2013;49(1):245–253. doi: 10.1016/j.ejca.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 31.Oehler C, von Bueren AO, Furmanova P. The microtubule stabilizer patupilone (epothilone B) is a potent radiosensitizer in medulloblastoma cells. Neuro Oncol. 2011;13(9):1000–1010. doi: 10.1093/neuonc/nor069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zollner SK, Selvanathan SP, Graham GT. Inhibition of the oncogenic fusion protein EWS-FLI1 causes G2-M cell cycle arrest and enhanced vincristine sensitivity in Ewing's sarcoma. Sci Signal. 2017;10(499) doi: 10.1126/scisignal.aam8429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang H, Yang GH, Bu H. Systematic analysis of the TGF-beta/Smad signalling pathway in the rhabdomyosarcoma cell line RD. Int J Exp Pathol. 2003;84(3):153–163. doi: 10.1046/j.1365-2613.2003.00347.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Corachan A, Ferrero H, Aguilar A. Inhibition of tumor cell proliferation in human uterine leiomyomas by vitamin D via Wnt/beta-catenin pathway. Fertil Steril. 2019;111(2):397–407. doi: 10.1016/j.fertnstert.2018.10.008. [DOI] [PubMed] [Google Scholar]
- 35.Salles B, Calsou P, Frit P, Muller C. The DNA repair complex DNA-PK, a pharmacological target in cancer chemotherapy and radiotherapy. Pathol Biol (Paris) 2006;54(4):185–193. doi: 10.1016/j.patbio.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 36.Goldstein M, Kastan MB. The DNA damage response: implications for tumor responses to radiation and chemotherapy. Ann Rev Med. 2015;66:129–143. doi: 10.1146/annurev-med-081313-121208. [DOI] [PubMed] [Google Scholar]
- 37.Ni J, Zhou LL, Ding L. Efatutazone and T0901317 exert synergistically therapeutic effects in acquired gefitinib-resistant lung adenocarcinoma cells. Cancer Med. 2018;7(5):1955–1966. doi: 10.1002/cam4.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Q, Ren M, Feng F, Chen K, Ju X. Treatment of colon cancer with liver X receptor agonists induces immunogenic cell death. Mol Carcinog. 2018;57(7):903–910. doi: 10.1002/mc.22811. [DOI] [PubMed] [Google Scholar]
- 39.Yang CM, Lu YL, Chen HY, Hu ML. Lycopene and the LXR alpha agonist T0901317 synergistically inhibit the proliferation of androgen-independent prostate cancer cells via the PPAR-gamma-LXR alpha-ABCA1 pathway. J Nutr Biochem. 2012;23(9):1155–1162. doi: 10.1016/j.jnutbio.2011.06.009. [DOI] [PubMed] [Google Scholar]
- 40.Chen X, Liu L, Mims J. Analysis of DNA methylation and gene expression in radiation-resistant head and neck tumors. Epigenetics. 2015;10(6):545–561. doi: 10.1080/15592294.2015.1048953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tabraue C, Lara PC, De Mirecki-Garrido M. LXR Signaling Regulates Macrophage Survival and Inflammation in Response to Ionizing Radiation. Int J Radiat Oncol Biol Phys. 2019;104(4):913–923. doi: 10.1016/j.ijrobp.2019.03.028. [DOI] [PubMed] [Google Scholar]
- 42.Maloney C, Edelman MC, Kallis MP, Soffer SZ, Symons M, Steinberg BM. Intratibial injection causes direct pulmonary seeding of osteosarcoma cells and is not a spontaneous model of metastasis: a mouse osteosarcoma model. Clin Orthop Relat Res. 2018;476(7):1514–1522. doi: 10.1007/s11999.0000000000000291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Igarashi K, Kawaguchi K, Murakami T. Patient-derived orthotopic xenograft models of sarcoma. Cancer Lett. 2020;469:332–339. doi: 10.1016/j.canlet.2019.10.028. [DOI] [PubMed] [Google Scholar]
- 44.Igarashi K, Kawaguchi K, Kiyuna T. Patient-derived orthotopic xenograft (PDOX) mouse model of adult rhabdomyosarcoma invades and recurs after resection in contrast to the subcutaneous ectopic model. Cell Cycle. 2017;16(1):91–94. doi: 10.1080/15384101.2016.1252885. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. 48 h after transfection with sh-NC or sh-KIF18B 3, sh-KIF18B 4, the clone formation assay was performed on the cells irradiated with different doses, ranging from 0 Gy, 2 Gy, 4 Gy, 6 Gy, and 8 Gy, every three days for a total of two weeks. P values were obtained from independent-samples t test; Error bars denote SD (n=3). Compared to sh-NC group, *P<0.05, ⁎⁎P<0.01, ⁎⁎⁎P<0.001.
Fig. S2. Cell proliferation assays in sarcoma cell lines transfected with sh-NC, sh-KIF18B 3, or sh-KIF18B 4. P values were obtained from independent-samples t test; Error bars denote SD (n=3). Compared to sh-NC group, *P<0.05, ⁎⁎P<0.01, ⁎⁎⁎P<0.001.
Fig. S3. Overexpression of KIF18B enables A-673 to be more resistant to radiotherapy. a, KIF18B expression levels of sarcoma cell lines. b, WB assay showed that pcDNA3.1-KIF18B could up-regulate KIF18B protein expression in A-673. c, The clone formation assay was performed in A-673 cells transfected NC or pcDNA3.1-KIF18B with different doses of radiation, ranging from 0 Gy, 2 Gy, 4 Gy, 6 Gy, and 8 Gy every three days for a total of two weeks (n=3). (b, c)P values were obtained from independent-samples t test; Error bars denote SD (n=3). Compared to NC group, *P<0.05, ⁎⁎P<0.01.
Fig. S4. Trametinib cannot enhance the radiotherapeutic effect on sarcoma cells. a, KIF18B mRNA expression was associated with drug sensitivity described in the GDSC project. PCC, pearson correlation coefficient. b, Cell survival rate was analysed using CCK8 assay in MNNG/HOS cells transfected with sh-NC or sh-KIF18B by treatment with different concentrations of trametinib for 48 h (n=3). c, The clone formation assay was performed in MNNG/HOS cells treated with or without trametinib at different doses every three days for a total of two weeks (n=3).
Fig. S5. GSEA and GO analysis of DEGs in sarcoma compared to non-sarcomatous tissue. a, GSEA, gene set enrichment analysis; NES, normalized enrichment score; FDR, false discovery rate. P < 0.01 and FDR < 0.05 were considered statistically significant. b, GO, Gene Ontology; BP, Biological process.
Fig. S6. KIF18B expression is irrelevant to survival time of chemotherapeutic patients in sarcoma, based on TCGA database. a, The effect of mRNA expression of KIF18B on survival time of sarcoma patients with or without doxorubicin. b, The effect of mRNA expression of KIF18B on survival time of sarcoma patients with or without gemcitabine. c, The effect of mRNA expression of KIF18B on survival time of sarcoma patients with or without paclitaxel. (a-c) Survival (R package) was performed for Kaplan-Meier survival analysis.
Fig. S7. GSEA results of KIF18B reference gene set of sarcoma patients. GSEA, gene set enrichment analysis; NES, normalized enrichment score; FDR, false discovery rate; FWER P, familywise-error rate p-value <0.05.
Fig. S8. T09 could not increase apoptosis and DNA repair of sh-KIF18B sarcoma cells compared with sh-NC.P values were obtained from independent-samples t test; Error bars denote SD (n=3). *P<0.05, ⁎⁎P<0.01, compared to sh-NC group.
Fig. S9. KIF18B expression in various tissues. a, KIF18B protein expression was detected in many tissues by immunohistochemistry assay. The samples, the same with the samples in Fig. 4a, were shown independently. b, The mRNA expression of KIF18B in many tissues, such as lung, breast, thyroid et al., were analysed based on TCGA.