Abstract
Background
Mobile genetic elements are found in genomes throughout the microbial world, mediating genome plasticity and important prokaryotic phenotypes. Even the cell wall-less mycoplasmas, which are known to harbour a minimal set of genes, seem to accumulate mobile genetic elements. In Mycoplasma hominis, a facultative pathogen of the human urogenital tract and an inherently very heterogeneous species, four different MGE-classes had been detected until now: insertion sequence ISMhom-1, prophage MHoV-1, a tetracycline resistance mediating transposon, and ICEHo, a species-specific variant of a mycoplasma integrative and conjugative element encoding a T4SS secretion system (termed MICE).
Results
To characterize the prevalence of these MGEs, genomes of 23 M. hominis isolates were assembled using whole genome sequencing and bioinformatically analysed for the presence of mobile genetic elements. In addition to the previously described MGEs, a new ICEHo variant was found, which we designate ICEHo-II. Of 15 ICEHo-II genes, five are common MICE genes; eight are unique to ICEHo-II; and two represent a duplication of a gene also present in ICEHo-I. In 150 M. hominis isolates and based on a screening PCR, prevalence of ICEHo-I was 40.7%; of ICEHo-II, 28.7%; and of both elements, 15.3%. Activity of ICEHo-I and -II was demonstrated by detection of circularized extrachromosomal forms of the elements through PCR and subsequent Sanger sequencing.
Conclusions
Nanopore sequencing enabled the identification of mobile genetic elements and of ICEHo-II, a novel MICE element of M. hominis, whose phenotypic impact and potential impact on pathogenicity can now be elucidated.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13100-020-00225-9.
Keywords: Mobile genetic element, Mycoplasma, M. hominis, Nanopore sequencing
Background
Mycoplasma hominis is a facultative pathogen of the human urogenital tract and associated with bacterial vaginosis, pelvic inflammatory disease, septic arthritis, preterm birth or even neonatal meningitis [1–3]. The factors accounting for the pathogenic potential of this heterogeneous species with the second smallest genome described so far are not fully understood. Several studies were conducted to characterize host-pathogen interactions in vitro [4–6] and in vivo [7–9], including microarray-based characterization of host [10] and pathogen [11] transcriptome changes in M. hominis infection. With increasing numbers of completely resolved M. hominis genomes (20 at the time of writing; https://www.ncbi.nlm.nih.gov/genome/), however, it became increasingly clear that mobile genetic elements, such as of MhoV-1 [12], ISMhom-1 [13], the tet(M)-carrying transposon [14], and the recently detected ICEHo element [15], significantly contribute to genomic plasticity of M. hominis [16].
The present study was conducted to elucidate the presence and prevalence of mobile genetic elements in selected clinical strains of M. hominis. To ensure the correct resolution and localization of MGE-associated genomic repeats, a Nanopore-based long-read sequencing approach was combined with an Illumina-based assembly polishing strategy.
Results
Generation of high-quality assemblies of 11 M. hominis strains
A hybrid approach combining short- and long-read sequencing data (Table 1) was used to generate high-quality assemblies of 11 isolates of M. hominis. Briefly, the Oxford Nanopore and Pacific Biosciences technologies were used to generate ≥500X of long-read sequencing data for each isolate genome; these data were assembled using Canu [17] or HGAP [18] and polished using ≥100X of short-read Illumina sequencing data for each sample. All assemblies were manually inspected for quality. A full description of the sequencing and assembly process is given in the Methods section. Genome lengths of all 11 isolates were larger than that of type strain PG21 (665 kbp [19]), ranging from 673 kbp (SS25) to 780 kbp (FBG); the number of annotated genes, predicted by Prokka [20], ranged from 580 (SS25) to 680 genes (FBG). Two additional publicly available genome sequences were also incorporated into the analysis (TO0613 and PL5).
Table 1.
Whole-genome sequencing of 23 M. hominis strains
| Strain | Short-read sequencing | Long-read sequencing | ||||||
|---|---|---|---|---|---|---|---|---|
| Protocol | Generated data (Mb) | Est. coverage (X) | Technology | Kit | Generated data (Mb) | Est. coverage (X) | Median read length/kb | |
| FBG | 2 × 300 | 1612 | 2067 | Nanopore | SQK-RAD003 | 3542 | 4541 | 2.5 |
| 8958 | 2 × 300 | 157 | 231 | Nanopore | SQK-RAD003 | 657 | 966 | 4.5 |
| 2539 | 2 × 300 | 2344 | 3119 | Nanopore | SQK-LSK108 | 384 | 511 | 12.6 |
| A136 | 2 × 300 | 240 | 344 | Nanopore | EXP-NBD103 + SQK-LSK108 | 707 | 1016 | 13.6 |
| SP2565 | 2 × 300 | 205 | 287 | Nanopore | 1092 | 1533 | 10.5 | |
| 475 | 2 × 300 | 184 | 257 | Nanopore | 638 | 889 | 5.3 | |
| SS10 | 2 × 300 | 1648 | 2353 | Nanopore | 756 | 1080 | 5.7 | |
| SS25 | 2 × 300 | 1865 | 2772 | Nanopore | 732 | 1088 | 8.3 | |
| VO31120 | 2 × 250 | 89 | 130 | Nanopore | 728 | 1069 | 6.3 | |
| SP10291 | 2 × 250 | 80 | 106 | Nanopore | 562 | 749 | 4.4 | |
| SP3615 | 2 × 250 | 99 | 138 | PacBio | SMRTbell Template Prep Kit 1.0 + Sequel Binding and Internal Control Kit 2.1 | 7447 | 10,401 | 2.9 |
| 727 J | Nanopore | EXP-NBD103 + SQK-LSK108 | 17 | 21 | 5.2 | |||
| 942 J | Nanopore | 38 | 47 | 44.1 | ||||
| 2740 | Nanopore | 15 | 16 | 4.1 | ||||
| 7388VA | Nanopore | 39 | 47 | 41.2 | ||||
| 7447VA | Nanopore | 59 | 66 | 1.7 | ||||
| 10936VA | Nanopore | 53 | 66 | 3.0 | ||||
| 12256 U | Nanopore | 35 | 42 | 14.5 | ||||
| 14352VA | Nanopore | 37 | 47 | 32.3 | ||||
| 16753 | Nanopore | 7 | 9 | 4.4 | ||||
| 18847 | Nanopore | 38 | 49 | 45.7 | ||||
| 19791 | Nanopore | 44 | 52 | 4.6 | ||||
| 21127VA | Nanopore | 38 | 44 | 42.4 | ||||
Detection of mobile genetic elements (MGE) in selected M. hominis strains
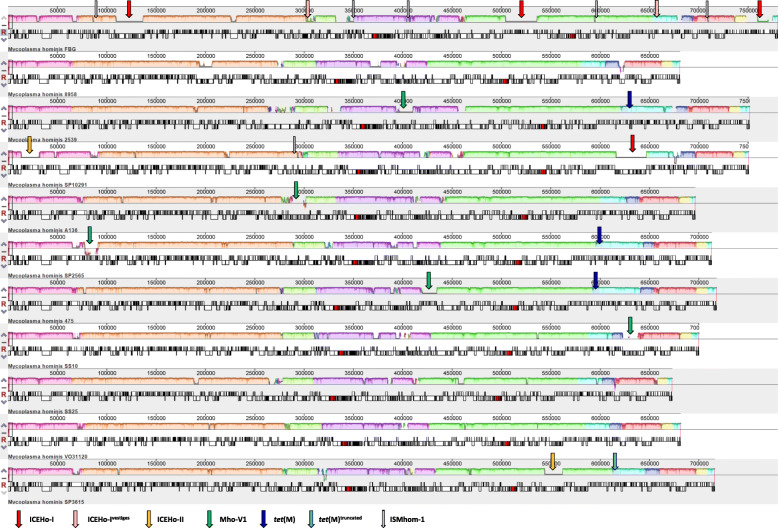
The online software tool Mauve [21] was used for genome alignments (Fig. 1) illustrating homologous regions by colour. Thus, larger isolate-specific regions of gene gain were evident by blocks of zero similarity (e.g. uncoloured sections) and classified as putative mobile genetic elements (MGE).
Fig. 1.
Mauve alignment of M. hominis genomes. In Mauve progressive alignment of genomes of M. hominis strains FBG, 8958, 2539, SP10291, A136, SP2565, 475, SS10, SS25, VO31120 and SP3615 the FBG genome served as a reference. Regions with the same colour represent locally collinear blocks without rearrangement of the homologous backbone sequences. Open reading frames of both strands are depicted below with rRNA genes in red. Local positions of MGE are marked above genomes by vertical arrows in specific colouring: ICEHo-I in red, ICEHo-I vestiges in pink, ICEHo-II in yellow, MHoV-1 in green, tet(M) in dark blue, truncated tet(M) in light blue and ISMhom-1 in light purple
Four different classes of MGE were characterized in the M. hominis genomes: i) insertion sequence ISMhom-1, first described in 2008 [13], ii) prophage MHoV-1 [12], iii) a tetracycline resistance mediating transposon [14], and iv) ICEHo-I and -II, two M. hominis-specific variants of MICE, a mycoplasma integrative and conjugative element [22], of which ICEHo-I corresponds to ICEHo recently published [15]. All MGE insertion sites are shown in Table 2 and visualized in Fig. 1. We detected between 0 (isolates 8958, SS25, and VO31120) and 8 MGEs (isolate FBG) per genome; of note, the three isolates in which no MGEs were detected had the smallest genome sizes.
Table 2.
Presence and genomic position of mobile genetic elements
| Strain | Genome / bp |
ISMhom-1a / bp x – bp y |
MHoV-1b /bp x – bp y |
tet(M)c / bp x – bp y |
ICEHo-I d / bp x – bp y |
ICEHo-II e / bp x – bp y |
|---|---|---|---|---|---|---|
| FBG | 780,024 |
85,753–87,012 349,147–350,406 404,580–403,321 593,967–595,226 708,740–709,999 |
– | – |
136,576–109,789 508,629–535,415 747,727–774,523 307,652 – 309385f (675938–675,828) g |
– |
| 8958 | 680,851 | – | – | – | – | – |
| 2539 | 751,326 | – | 407,989–392,742 | 619,357–644,618 | – | – |
| SP10291 | 750,518 | 293,110–294,369 | – | – | 646,629–616,362 | 13,153–31,485 |
| A136 | 696,338 | – | 301,276–286,023 | – | – | – |
| SP2565 | 712,781 | – | 91,014–75,747 | 593,982–619,248 | – | – |
| 475 | 717,789 | – | 419,145–434,419 | 590,550–615,528 | – | – |
| SS10 | 700,146 | 637,906–622,632 | – | – | – | |
| SS25 | 672,843 | – | – | – | – | – |
| VO31120 | 681,374 | – | – | – | – | – |
| SP3615 | 715,990 | – | – | 613,432–620,439 h | – | 543,628–561,978 |
| TO0613i | 766,228 | – | 694,774–710,019 | – |
49,522–18,905 310,644–280,027 |
– |
| PL5 |
767,767 (JRXA01) j |
– | – | 74,216–99,476(_000009.1) j |
[89,704–90,264] – [1950–3140] (_000010.1) j (_000001.1) [1814–1029] – [18,377–17,187] (_000005.1) (_000004.1) |
– |
MGE elements used in BLAST analysis: a ISMhom-1 (acc.-no. dq973625); b MHoV-1 prophage region from repB to exiS (acc.- no. CP009652; bp 596,991-bp 581,744); c tet(M) of SPROTT (acc.- no. CP011538; bp 573,817–599,077); d ICEHo-I region of FBG (CDS1 to CDS22, bp 508,629 – bp 535,415); e ICEHo-II region of SP3615 (CDS1 to CDS22, bp 543,628 – bp 561,978); fICEHo-I vestige (corresponding FBG ICEHo-I-2; bp 508,629 – bp 510,361); g ICEHo-I vestige (corresponding to FBG ICEHo-I-2 untranslated region 535,715–535,827); h truncated tet(M) transposon corresponding to bp 577,017 - bp 584,024 of SPROTT; accession numbers of i TO0613 genome (acc.-no CP033021.1) and j Pl5 contigs (JRXA01_000001.1 to JRXA01_000010.1)
ISMhom-1
ISMhom-1 (1.26 kb) was found in two isolates; isolate SP10291 contained one copy, and isolate FBG carried five copies. ISMhom-1 was highly conserved in sequence, carrying an open reading frame similar to transposase gene tnpA of the IS30 family [13], which was flanked by a nontranslated region (108 bp on the 5′ end and 140 bp 3′) with terminal inverted repeats of 27 bp. Generation of inverted repeats by IS elements was first described for an IS30-type insertion element of M. fermentans [23]. ISMhom-1 insertion positions included un-translated regions (FBG ISMhom-1_1, ISMhom-1_2, ISMhom-1_3, and SP10291) and the 3′ ends of the annotated genes BHBFJMJE_00532 and BHBFJMJE_00625 (FBG ISMhom-1_4 and ISMhom-1_5, respectively). The concomitant generation of insertion site-specific inverted repeats resulted in integrity of both ORFs (Table 3).
Table 3.
Integration sites of mobile genetic elements (MGE)
| MGE | Gene / contiga | PG21-homologueb | Gene product | Gene length (nt) |
Insertion site in gene (nt) | DR / IR c |
|---|---|---|---|---|---|---|
| FBG_ISMhom-1_1 | dBHBFJMJE_00070rc d | IGR e (dMHO_0640) | – | – | – | AAAATA |
| FBG_ISMhom-1_2 | dBHBFJMJE_00328 d | IGR e (dMHO_2560) | – | – | – | ATATTT |
| FBG_ISMhom-1_3 | dBHBFJMJE_00386 d |
IGR e (dMHO_3070) |
– | – | – | TAAATTATTTTGGGT |
| FBG_ISMhom-1_4 | BHBFJMJE_00532rc | MHO_4140rc | put. HAD hydrolase | 807 | 807 | AAATAAf |
| FBG_ISMhom-1_5 | BHBFJMJE_00625rc | MHO_5020rc | put. Sporulation transcription regulator WhiA | 828 | 828 | AAATAGGCf |
| SP10291_ISMhom-1 | dHPAMDCMO_00271 d | IGR e (dMHO_2210) | – | – | – | TGTGCTTTTT |
| 2539_MHoV1 | KLHMLDFE_00358 | MHO_3090 | hypothetical protein | 1134 | 808 | ATTTTTAT / ATTTTTTT |
| A136_MHoV1 | dMNIFKBBE_00264 d | IGR e (dMHO_2260rc) | – | – | – | TTTTTTTT / CTTTTTTT |
| SP2565_MHoV1 | dHHAOGLDO_00056 d | IGR e (dMHO_0530) | – | – | – | ATTTTTATA / ATTTTTCTA |
| 475_MHoV1 | dMOHCKDGE_00391 d | IGR e (dMHO_3470) | – | – | – | TTTTTT / CTTTTT |
| SS10_MHoV1 | MFOAEKDO_00548 | MHO_4930 | hypothetical protein | 903 | 867 |
CTTTTT (866–867) g |
| TO0613_MHoV1 | dKN71_002970 d | IGR e (dMHO_4930) | – | – | – | ATTTTTA |
| FBG_ICEHo-I-1rc | BHBFJMJE_00092 | MHO_0820 | hypothetical protein | 651 | 394 |
AAATAATA (387–394) g |
| FBG_ICEHo-I-2 | BHBFJMJE_00459 | MHO_3720 | P75 precursor | 1950 | 1950 + 1h | CAAATAAA/AATCTTT i (1323–1950) g |
| FBG_ICEHo-I-3 | BHBFJMJE_00651 | MHO_5300 | conserved hypothetical protein | 432 | 432 + 2h |
AAATAAAAf (427–432) g |
| FBG_ICEHo-I-4 vestige | BHBFJMJE_00287 | MHO_2250 | hypothetical protein | 663 | 44 | |
| FBG_ICEHo-I-5 vestigerc | dBHBFJMJE_00595 d |
IGR e (dMHO_4730) |
– | – | – | |
| SP10291_ICEHo-I | dHDENHCDK_00935 d | IGR e (dMHO_4550) | – | – | – | AAATTTTT |
| TO0613_ICEHo-I-1rc | dKN71_000220 d |
IGR e (dMHO_0170rc) |
– | – | – | TTTAAAAT |
| TO0613_ICEHo-I-2rc | KN71_001280 | MHO_2080 | conserved hypothetical lipoprotein | 1734 | 1407 |
TGGAAAT (1401–1407) g |
| PL5_ICEHo-I-1 |
JRX01A000001.1 |
MHO_0120 | type-III restriction enzyme | 2453 | 853 |
TTTTTAAA (846–853) g |
| PL5_ICEHo-I-2 | MHO_1960 | hypothetical protein | 369 | 4 |
AAATAAAG (4–12) g |
|
| SP3615_ICEHo-II | dKGPEAEHF_00485rc d |
IGR e (ddpnA) |
methylase DpnIIB | – | – | TAATATTA |
| SP10291_ICEHo-II | HPAMDCMO_00015 | MHO_0130 |
site-specific DNA methyl-transferase (DEAD/DEAH box helicase family protein) |
1200 | 1152 |
AAATCTTT (1145–1152) g |
aContigs only shown for strain PL5, due to assembly fragmentation; printed in italics; b MHO genes of PG21 according to acc.-no: FP236530.1; c DR = direct repeat; IR = inverted repeat; d dgene_X = insertion site downstream of gene X; rc = gene on reverse complementary strand; e IGR (= intergenic region) downstream of MHO-homologous gene X (dMHO_X); f downstream DR identical to 3′ end of the gene, protein encoding regions in bold; g DR region in the affected gene (nt x to nt y); h + 1 /+ 2 = direct repeat terminates 1 nt / 2 nt downstream of gene x resulting in downstream DR containing 3′ end of the gene; i no DR, but duplicated region of p75 (nt 1323–1950)
Prophage MHoV-1
Prophage MHoV-1 was detected in six isolates (five de novo assembled genomes and one publicly available genome, TO0613). Presence and sequence of genes (from repB to exiS, i.e. spanning the complete MHoV-1 element as defined by [12], and terminated by indirect (IR; AAAGTCCC) repeats of the phage) were highly conserved across the de novo assembled genomes (Table 3). The respective prophage region in TO0613 was structurally consistent with the de novo assembled sequences; several annotated TO0613 genes, however, were disrupted, suggesting a potential assembly problem in the published MiSeq-based assembly of TO0613. No systematic patterns of MHoV-1 integration positions were observed (Table 3). In four cases (strains A136, SP2565, 475 and TO0613), MHoV-1 integrated into intergenic regions; in two cases (strains 2539 and SS10), into open reading frames encoding hypothetical genes of unknown function, leading to premature disruption of the predicted hypothetical genes.
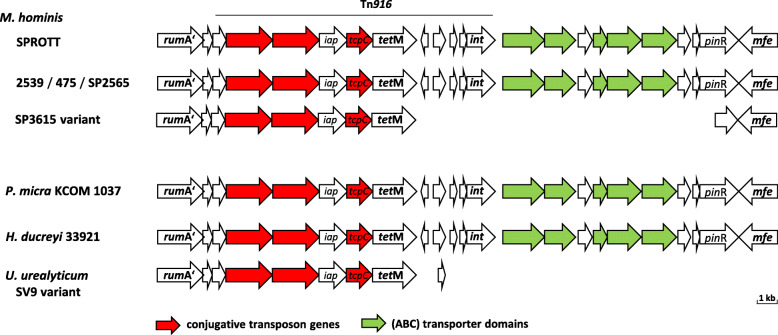
tet(M)-harbouring transposon
A tet(M)-harbouring transposon of 25 kb length, mediating tetracycline resistance, was detected in four M. hominis strains (2539, 475, SP2565, and PL5). The transposon was highly conserved in gene organisation (see Fig. 2) and sequence (> 94% nucleotide identity), and comprised a 13.3 kb region homologous to transposon Tn916 [14]. Insertion sites of the tet(M)-harbouring transposon were highly conserved, targeting the 3′ end of the rumA gene and leading to RumA C-terminal extension, consistent with findings in strain SPROTT [14], in which a homologous full-length transposon is also present (Fig. 2). Truncated versions of the element were found in strain SP3615 (encompassing conjugative transposon genes but missing integrase gene int), as well as in Ureaplasma urealyticum, serovar 9 (Fig. 2). The functional relevance of these truncations remains unclear. Further BLAST analyses identified a homologous transposon in Parvimonas micra (> 87% nucleotide identity), a pathogen which is commonly found in the oral cavity or gastrointestinal tract [24], and two homologous regions in Haemophilus ducreyi strain 33,921 (acc.-no. CP011228.1), covering the entire transposon with > 87% nucleotide identity.
Fig. 2.
Organisation of tet(M) transposons. An identical organisation of genes was found in tet(M) transposons of M. hominis strains SPROTT (acc.- no. CP011538.1; nt 573,817-599,077), 2539, 475, SP2565, Parvimonas micra strain KCOM 1037 (CP031971.1; nt 249,283-277,971)) and H. ducreyi strain 33,921 (CP011228.1; nt 678,636–662,601 and nt 1,365,672-1,378,984). The region comprising the truncated Tn916 unit (13.3 kb) is marked by a solid line. Truncated variants with loss of the int gene were found in M. hominis strain SP3615 (nt 609,362-623,230) and U. urealyticum serovar 9 str. ATCC 33175 (AAYQ02000002.1; nt 59,407-45,316))
Mycoplasma integrative and conjugative elements
Two different MICE variants, ICEHo-I and ICEHo-II, were detected in the M. hominis genomes sequenced in this study. The first of these, ICEHo-I, was previously characterized by Meygret el al [15]. and named ICEHo.
Integrative and conjugative element ICEHo-I
ICEHo-I was detected in four isolate genomes, with copy numbers varying between 1 (SP10291), 2 (strains TO0613 and PL5), and 3 (strain FBG); the genomic locations and features of the integration sites of ICEHo-I are summarized in Tables 2 and 3.
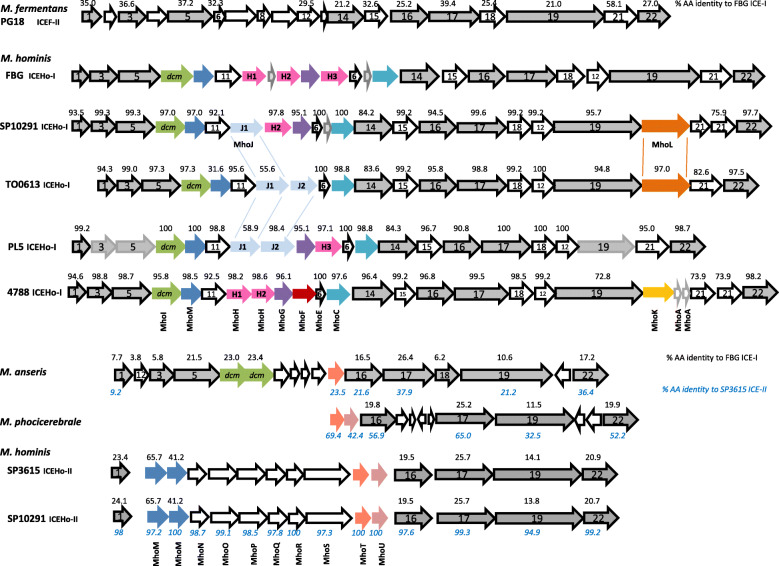
ICEHo-I carried a set of 13 MICE core genes as defined in an analysis of MICE of M. fermentans M64 and M. agalactiae 5632 [22]. MICE core genes exhibited a high degree of conservation across the assembled M. hominis genomes; inter-strain homologies of the MICE core proteins ranged from 76 to 100% with respect to strain FBG (Fig. 3). Inter-species homologies, by contrast, were lower; for example, protein homologies with respect to MICEF-II of M. fermentans [22] ranged from 21% (CDS19) to 58% (CDS21). Of note, the set of MICE core genes present in ICEHo-I included CDS6. In the original description of ICEHo-I [15], a highly homologous gene (EVJ69_RS02240 in strain 4788; 100% amino acid identity) had been classified as a non-core MICE gene [15]; identification with CDS6, however, was justified by 32.3% amino acid identity and 53.2% amino acid similarity to ICEF-ORF6 of M. fermentans (Additional file 1).
Fig. 3.
Structural organisation of ICEHo elements. Protein sequences of annotated MICE genes were obtained from NCBI for M. fermentans PG18 ICEF-II (MBIO_0551–0567 and MBIO_0555–5 (inserted by hand; nt 616,241 to 616,429 of AP009608.1)), M. hominis strains FBG ICEHo-I (BHBFJME_00460–00482), SP10291 ICEHo-I (HPAMDCMO_00562–00540) and ICEHo-II (HPAMDCMO_00016–00030), SP3615 ICEHo-II (KGPEAHF_00500–00483), TO0613 ICEHo-I (KN71_RS02000–01900), PL5 ICEHo-I (V136_RS01075/_03225–03305/VE10_RS00015), 4788_ICEHo-I (EVJ69_RS02290–02175), M. anseris ICEHo-I/II (DP065_01405–01485) and M. phocicerebrale ICEHo-II (DMC14_02545–02600) and analysed by multiple protein sequence alignments. Sequence identities of homologous proteins, estimated by ClustalW, are shown above the encoding ORFs in relation to FBG ICEHo-I, and below in relation to SP3615 ICEHo-II. Percent amino acid identities of MhoJ1 and MhoJ2 of ICEHo-I were calculated with respect to MhoJ1 of SP10291 ICEHo-I and MhoJ2 of TO0613 and respectively marked by lines. Incomplete recovery of CDS3, CDS5 and CDS19 of strain PL5, which did not enable calculation of homologies, is indicated by transparent framing
An analysis of MICE non-core (i.e. cargo) genes in ICEHo-I showed that the genes dcm, MhoM, MhoE, and MhoC were always present at a single copy, and their relative position was conserved (Fig. 3). MhoH, MhoG, MhoF, and MhoJ were consistently located between CDS11 and MhoE, and their copy number was variable (ranging from 0 to 1 for MhoG and MhoF; from 0 to 2, for MhoJ; and from 0 and 3, for MhoH). MhoA, MhoK, and MhoL were located between CDS19 and CDS22 and varied in copy number between 0 and 1 (MhoL and MhoK) and 0 and 2 (MhoA). In a phylogenetic analysis of MhoH, MhoJ, and MhoF, MhoF of strain 4788 clustered with MhoH (Additional file 2), demonstrating that MhoH and MhoF are closely related.
ICEHo-I untranslated regions (210 bp upstream of CDS1 and 413 bp downstream of CDS22) were highly conserved and terminated by an inverted repeat. ICEHo-I integration into host genomes resulted in the generation of direct repeats (Table 3). In two instances, integration was associated with a premature stop of translation, affecting a hypothetical protein (strain FBG; at nucleotide 394/651 of the MHO-0820-homologous BHBFJMJE_00092) and a lipoprotein (strain TO0613; at nucleotide 1407/1734 of the MHO-2080 homologue). In strain PL5, analysis of insertion sites was limited by incomplete genome resolution, but BLAST analysis suggested an insertion into the MHO-0120- and MHO-1960-homologous genes, putatively encoding a type III restriction enzyme and a hypothetical protein, respectively. In strain FBG, integration of ICEHo-I-2 was associated with a large duplication within the P75 precursor gene resulting in an upstream intact P75-precursor gene (nucleotide 1–1950) and a downstream remnant (nucleotide 1323–1950).
The three complete copies of ICEHo-I in strain FBG exhibited a high degree of conservation and differed by only four nucleotides, associated with a single amino acid exchange in CDS14 of ICEHo-I_3 (Asn485Ile). In addition to three complete copies of ICEHo-I, strain FBG also harboured two ICEHo-I vestiges (FBG ICEHo-I-4 and -5; see Fig. 1 and Table 3).
Integrative and conjugative element ICEHo-II
The detection of two additional regions of zero similarity with respect to the other M. hominis genomes in strains SP3615 and SP10291 (highlighted in Fig. 1) led to the discovery of another mycoplasma integrative and conjugative element, referred to as ICEHo-II. ICEHo-II was conserved in length (~18kbp) and sequence of 15 open reading frames (Fig. 3; 94.9–100% AA identity).
Protein homology analyses classified five of the open reading frames as MICE-core genes CDS-1, − 16, − 17, − 19, and − 22, with homologies of the encoded proteins to the respective ICEHo-I proteins of FBG-ICEHo-I ranging from 14.1% (CDS19) to 25.7% (CDS17). Of the ICEHo-II cargo gene encoded proteins (MhoM to MhoU), only protein MhoM, duplicated in ICEHo-II, was also found in ICEHo-I with 30–50% AA identity. A phylogenetic analysis showed that MhoM generally clustered distinctly from CDS11 into ICEHo-I- or –II-specific branches; except for ICEHo-I MhoM protein of TO0613, which was phylogenetically positioned between ICEHo-II MhoM and CDS11 (Additional file 3).
ICEHo-II untranslated regions (207 bp upstream of CDS1 and 210 bp downstream of CDS 22) were terminated by inverted repeats (IRL: GGCCGTGTAAAAAATATAAGGAAT and IRR: ATTCCTTTAATAATAAACACGACC). In strain SP10291, ICEHo-II insertion led to a premature stop in the MHO-0130-homologous gene, putatively encoding a site-specific DNA methyltransferase belonging to the DEAD/DEAH box helicase family (see Table 3). In strain SP3615, ICEHo-II was reversely inserted between the MHO-4180- and MHO-4190-homologous genes, and three additional genes (KGPEAEHF_0485 to KGPEAEHF_0483) were detected between MHO_4180 and ICEHo-II. A Phyre2 analysis of these genes showed homologies to two methyltransferases (KGPEAEHF_0485 and KGPEAEHF_0486) and a S. pneumoniae endonuclease encoded in the DpnII gene cassette (KGPEAEHF_0483).
A BLAST analysis identified ICEHo-II-homologous regions in other mycoplasma species, M. phocicerebrale [25] and M. anseris [26] (Fig. 3). In the seal pathogen M. phocicerebrale a truncated ICEHo-II region was detected, extending from gene MhoT to CDS22. In the duck and goose pathogen M. anseris a hybrid ICEHo element was found, carrying the ICEHo-I- homologous MICE genes CDS3, − 5, − 12, − 18, and dcm, and the ICEHo-II homologous MICE genes CDS1, − 16, − 17, − 19, − 22, and MhoT, suggesting a common ancestor or a product of recombination of both ICEHo elements.
Prevalence of ICEHo-I and ICEHo-II elements
Using a Real time PCR (qPCR) screening approach targeting ICEHo-I and -II-specific small gene fragments, 150 isolates from the M. hominis strain collection of our institute were tested for the presence of ICEHo elements (see Methods). For ICEHo-I, 57.3% of the M. hominis strains (86/150) were rated as unambiguously ICEHo-I-negative, and 28% (42/150) were classified as unambiguously ICEHo-I-positive. Of the remaining 22 isolates with ambiguous ICEHo-I-specific probe detection results, 19 isolates were rated as ICEHo-I positive, yielding an overall ICEHo-I detection rate of 40.7% (61/150). For ICEHo-II, 28.7% of the M. hominis strains (43/150) were tested ICEHo-II-positive; including 15.3% strains (23/150) also positive for ICEHo-I.
To verify the accuracy of the qPCR screen, additional Nanopore long-read sequencing data were generated on 12 isolates (Table 1), draft de novo assembly was carried out, and ICEHo-I and -II positions in the de novo assemblies were determined. ICEHo detection results and genomic locations in the draft de novo assembled genomes are summarized in Additional file 4. The determined ICEHo-I and -II copy number counts agreed with the screening-based results for each evaluated isolate, confirming the accuracy of the qPCR screen. Variability in ICEHo-I structure, as already observed in the set of genomes assembled to high quality, was also found in the newly sequenced isolates; by contrast, the structure of ICEHo-II was found to be highly conserved (Additional file 4).
MGE co-occurrence analysis
MGE copy numbers were tabulated across the assembled genomes (Table 4) and statistical tests were carried out to assess the evidence for non-random co-occurrence of different MGEs, using the Chi-Square test to detect associations at the level of presence and absence and Spearman’s rank correlation test to detect associations at the level of MGE multiplicity. No statistically significant association at p = 0.05 was found between the presence or multiplicity of ICEHo-II in a given strain and presence of any other MGE; the lowest p-values were achieved for MhoV-1 being present more often in the absence of ICEHo-I (p = 0.066) and ISMhom-1 only occurring when ICEHo-I was present (p = 0.096).
Table 4.
MGE copy number in assembled M. hominis genomes
| Isolate | ISMhom1a | MhoV1b | tet(M) transposonc | ICEHo-Id | ICEHo-IIe |
|---|---|---|---|---|---|
| FBG | 5 | 0 | 0 | 3 | 0 |
| 8958 | 0 | 0 | 0 | 0 | 0 |
| 2539 | 0 | 1 | 1 | 0 | 0 |
| SP10291 | 1 | 0 | 0 | 1 | 1 |
| A136 | 0 | 1 | 0 | 0 | 0 |
| SP2565 | 0 | 1 | 1 | 0 | 0 |
| 475 | 0 | 1 | 1 | 0 | 0 |
| SS10 | 0 | 1 | 0 | 0 | 0 |
| SS25 | 0 | 0 | 0 | 0 | 0 |
| VO31120 | 0 | 0 | 0 | 0 | 0 |
| SP3615 | 0 | 0 | 1 | 0 | 1 |
| TO0613 | 0 | 1 | 0 | 2 | 0 |
| PL5 | 0 | 0 | 1 | 2 | 0 |
| 18847 | 0 | 0 | 0 | 1 | 0 |
| 21127 | 0 | 0 | 1 | 3 | 0 |
| 7388 | 0 | 1 | 1 | 0 | 0 |
| 727 J | 0 | 0 | 0 | 0 | 1 |
| 7447VA | 0 | 1 | 0 | 0 | 1 |
| 2740 | 0 | 1 | 0 | 0 | 2 |
| 16753 | 0 | 1 | 1 | 0 | 2 |
| 942 J | 0 | 1 | 0 | 1 | 1 |
| 10936 | 0 | 0 | 1 | 1 | 1 |
| 12256 | 0 | 0 | 0 | 1 | 2 |
| 14352 | 0 | 0 | 0 | 1 | 1 |
| 19791 | 0 | 1 | 1 | 1 | 2 |
MGEs were detected using pairwise sequence alignments between the assembly and MGE query sequences a ISMhom-1, query sequence acc.-no. dq973625, detection threshold: > 80% identity; b MHoV-1 prophage, query sequences CP009652:581744–584,733 (covering repB) and CP009652:596991–598,991 (covering exiS), detection threshold: > 80% identity for both query sequences; c tet(M), query sequence CP011538: 573817–599,077 (truncated tet(M) transposon, query sequence CP011538:577017–584,024) (derived from SPROTT), detection threshold: > 80% identity;; d ICEHo-I, query sequences: 14 MICE core genes of FBG, detection threshold: > 80% identity for > 80% of query sequences in a genomic region ≤35 kb; e ICEHo-II, query sequences: 5 MICE core genes of SP3615, detection threshold: > 80% identity for > 80% of query sequences in a genomic region ≤20 kb
Episomal occurrence of ICEHo elements
Nanopore reads of the 23 sequenced M. hominis strains were mapped to circularized ICEHo-I (strains FBG and SP10291) and ICEHo-II (SP3615 and SP10291). Reads overlapping the IRR-IRL junction site were only detected in strains FBG (ICEHo-I) and 19791 (ICEHo-II). To detect the presence of episomal ICEHo-I and ICEHo-II with increased sensitivity, a Real time PCR assay, designed to exclusively amplify episomal circularized ICEHo (cICEHo), was employed (see Methods). Application of this cICEHo screening assay to 80 ICEHo-positive isolates from our collection showed that more than two thirds (49/60) of the ICEHo-I- and more than half (27/43) of the ICEHo-II-carrying strains harbour episomal circularized versions of ICEHo-I and -II, respectively (see Additional file 5).
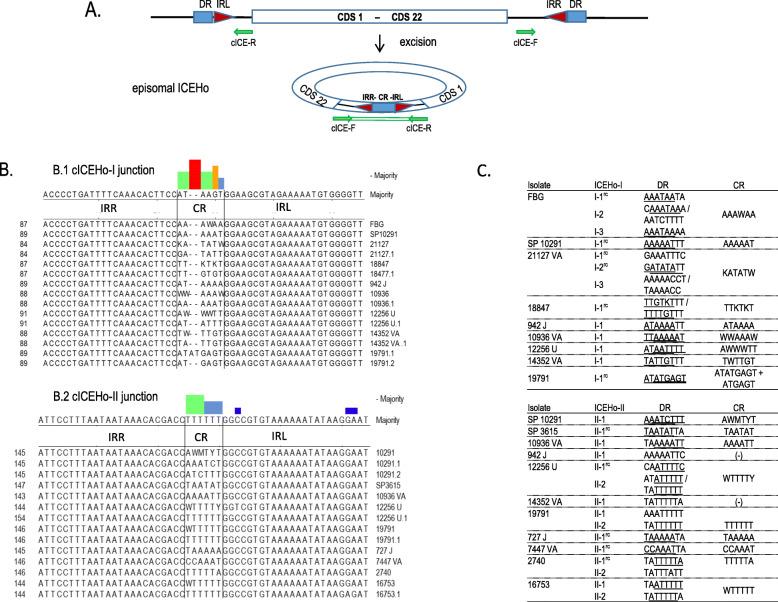
In all whole-genome-sequenced samples, the coupling region (CR) of the episomal ICEs was characterized with Sanger sequencing. In all cases except for cICEHo-I of strain 19791, the detected cICEHO-I and cICEHo-II CR sequences had a length of 6 nucleotides (Fig. 4). The CR of cICEHo-I in strain 19791 consisted of a mixture of six- and eight-nucleotide sequences (ATGAGT and ATATGAGT), with the longer version dominating (see Methods). CR sequences were characterized by a dominance of weak nucleotides (W = A or T) and generally corresponded to the genomic sequences of the IRR−/IRL-flanking direct repeats. The CR of circularized ICEHo-I was typically composed of nucleotides 1–6 of the DR (n = 11), less often of nucleotides 3–8 (n = 4) or 1–8 (n = 1). The CR of circularized ICHo-II, by contrast, was typically composed of nucleotides 3–8 of the DR (n = 7), less often of nucleotides 1–6 (n = 4) or 2–7 (n = 1).
Fig. 4.
Chromosomal and episomal ICEHo-I and –II. a Schematic representation of the excision of a chromosomal integrated ICEHo element to build the circularized episomal cICE-I or -II. ICEHo-flanking left- and right-positioned inverted repeats (IRL and IRR) are represented by red triangles, and the direct repeats (DRs) are represented by blue squares. After excision of ICEHo, direct repeats are fused to the coupling region (CR) in the episomal cICEHo. For the detection of episomal cICEHo-I and -II, specific primer pairs (represented as green arrows) were used that do not lead to an amplification when targeting the chromosomal ICEHo-I and –II in outwards-facing position, and which result in 0.2 (cICE-I) or 0.3 kb (cICE-II) PCR products when targeting the circularized ICEHo (see Methods). b Multiple sequence alignments of IRR-IRL junction sequences with the central CR of cICE-I (B.1) and cICE-II (B.2), determined by Sanger sequencing of cICE PCR products. Ambiguity characters in the coupling region (M = A or C; K = G or T; W = A or T; Y = C or T) indicate the presence of minor sequence variants as detected by Mixed Sequence Reader [27]. The respective sequences are listed as strain.1 for the major sequence and strain.2 for the minor sequence. Coloured bar charts above the consensus sequence, created with the Lasergene software package (DNAStar, Madison, WI), indicate the presence of non-consensus characters (disagreements) in the corresponding column, ranging from red (high frequency of non-consensus characters or gaps) to blue (low frequency of non-consensus characters). c List of direct repeats (DR) of the chromosomal ICEHo-I and cICEHo-II copies (I-1 to I-3; II-1 to II-2) detected in the nanopore-sequenced M. hominis strains in comparison to the episomal coupling regions (CR). Identical (or comparable) sequences are underlined; if two different regions of one DR are underlined, both regions used for recombination explain the mismatch in CR. IUAPC ambiguity characters in the genomic DR sequences were introduced during the removal of circular contig overlaps (see Methods). I-xrc = the respective ICEHo-element is inserted in a reverse-complementary fashion with respect to the genome assembly, requiring evaluation of the reverse complementary DR
The detection of major and minor CR sequence variants may reflect (i) simultaneous usage of different DR subregions from the same ICEHo element (see underlined sequence regions of DR in Fig. 4.C), (ii) simultaneous observation of multiple ICEHo-I/−II elements with different DR sequences (ICEHo-II of strain 12256 U), (iii) circularisation- or recombination-associated mutagenesis in the circularized ICEHo product, (iv) sequencing error. Of note, minor CR sequence variants were also observed in isolates in which only one ICEHo copy was present (e.g. ICEHo-II of SP10291), and we observed mismatches between CR and the underlying genomic DR sequences in both high quality and draft de novo assembled genomes (e.g. AAAAAA in ICEHo-I of FBG, TTTTGTT in ICEHo-I of 14352VA, and TTTTTT ICEHo-II of 16753). Of note, joint analysis of CR and DR sequences enabled the mapping of circularized ICEHo copies to their respective genomic origins in strains FBG (for ICEHo-I_1 and _3), 21127 (ICEHo-I_2), 19791 (ICEHo-II_2), and 2740 (ICEHo-II_1). In strain 16753, both copies of ICEHo-II were found in circularized form.
Discussion
Mobile genetic elements play an important role in mediating prokaryotic genome plasticity, often contributing to important phenotypes such as virulence and antibiotic resistance. MGEs can exert their effect by expanding the gene set of the host, or via the disruption of existing genes in the case of integration events. In the present study, we detected and characterized four types of MGEs in clinical isolates of M. hominis: ISMhom-1, an insertion sequence; prophage MHoV-1; a tet(M)-carrying transposon; and ICEHo-I and -II, two M. hominis-specific integrative and conjugative elements.
MGE insertion patterns
In our study, ISMhom-1 was found exclusively in noncoding chromosomal regions; in other studies, IS element insertions are also reported in MICEs [28] or MICE vestiges [29]. The other types of MGEs detected here were also found to be inserted in coding regions; 2 of 7 detected MhoV-1 insertions led to the interruption of a gene; 6 of 10 ICEHo-I insertion events; and 1 of 2 ICEHo-II insertion events. In more than half of the 11 M. hominis genomes assembled to high quality, a chromosomal gene was found to be disrupted by insertion of an MGE. No statistically significant effects were identified during the MGE co-occurrence analysis; the conjecture that ICEHo-I-free isolates may be less susceptible to the entry of other mobile elements [15] was not replicated here.
ICEHo-I and -II gene content
Horizontal transfer of ICEs from one host to the other is mediated by type IV secretion systems (T4SS), typically comprising the surface-localized pilus, the integral membrane core channel, a protein complex at the cytoplasmic site of the membrane, and ATPases at the cytoplasmic site of the channel [30]. In addition, mobilization and integration of ICEs typically require the presence of a relaxase or integrase enzyme. Genes that participate in the mobilization or conjugation process are referred to as core genes, whereas cargo genes often encode ICE-associated phenotypes of interest, such as resistance [14], metabolic traits [31], or virulence [32]. Characterization of ICEs in mycoplasmas (MICE) has enabled the definition of a MICE core gene set [22], including a mycoplasma-minimized T4SS [22]. ICEHo-II contains a smaller set of MICE core genes than ICEHo-I, but the impact of this on the transfer potential of ICEHo-II remains to be studied. Table 5 shows results of a bioinformatics analysis of putative gene functions, and an integrated view of putative MICE gene functions incorporating results from the literature is shown in Additional file 6. Low levels of homology present significant challenges for the in silico characterization of MICE genes; follow-up experimental studies will be necessary to better characterize the functions of ICEHo-I and -II genes.
Table 5.
Sequence-based characteristics of ICEHo-II encoded proteins
| MICE-/ICEHo- | SP3615_ ICEHo-II | SP10291_ ICEHo-II | Homologues proteins | Species | Accession number | TMHMM v.2.0 | SignalP v. 5.0 | Motif | Reference of motif | DEPP S - T - Yc |
Protein length | Query cover | E-value | Identity |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CDS | KGPEAEHFa | HPAMDCMOa | BLASTp /[PSI-BLASTb] | (x/y) | (AA) | (%) | (%) | |||||||
| MICE_ CDS1 | _00500 | _00016 | ICEF-IA CDS1 | M. fermentans | AY168953.1 | – | – | rpoC2 | CHL00117 | 2/21–0/10–0/12 | 252 | 88.89 | 4.22E-03 | 36.60 |
| MhoM | _00499 | _00017 | hypothetical protein | M. hominis 4235 | WP_158532045.1 | – | – | – | – | 6/7–1/2–1/6 | 108 | 90.00 | 1.00E-58 | 87.76 |
| MhoM | _00498 | _00018 | hypothetical protein | M. hominis 4235 | WP_158532045.1 | – | – | – | – | 1/6–1/4–2/5 | 97 | 89.00 | 1.00E-21 | 48.31 |
| MhoN | _00497 | _00019 | hypothetical protein | M. hominis PL5 | WP_044329953.1 | – | – | – | – | 3/13–1/1–0/9 | 157 | 21.00 | 3.30E-01 | 61.76 |
| MhoO | _00496 | _00020 | hypothetical protein | M. primatum | WP_029513615.1 | – | – | – | – | 0/8–0/8–0/20 | 217 | 64.98 | 2.00E-14 | 37.59 |
| MhoP | _00495 | _00021 | variable lipoprotein | M. arginini | WP_129694644.1 | 1 | II | Lipoprot. 7 | pfam01540 | 0/14–0/13–1 / 14 | 272 | 45.96 | 3.86E-09 | 33.60 |
| MhoQ | _00494 | _00022 | hypothetical protein | I. bacterium | OIO22386.1 | – | – | AcrB | COG0841 | 0/4–0/3–0/2 | 93 | 59.00 | 7.40E-02 | 38.18 |
| MhoR | _00493 | _00023 | response regulator b | C. estertheticum | WP_152751962.1 | – | – | – | – | 1/10–0/2–0/2 | 102 | 87.00 | 9.90E-01 | 29.59 |
| MhoS | _00492 | _00024 | hypothetical lipoprotein | M. phocicerebrale | WP_116171775.1 | – | II | – | – | 5/27–1/21–2/22 | 415 | 99.00 | 1.00E-109 | 45.93 |
| BspA leu-rich repeat surface protein | M. bovis | WP_141790766.1 | – | – | 100.00 | 5.00E-101 | 13.17 | |||||||
| MhoT | _00491 | _00025 | hypothetical protein | M. phocicerebrale | WP_116171528.1 | 2 | – | – | – | 0/7–0/9–0/1 | 98 | 94.00 | 1.00E-19 | 72.04 |
| MhoU | _00490 | _00026 | hypothetical protein | M. phocicerebrale | WP_116171527.1 | 2 | – | – | – | 0/3–0/5–0/5 | 85 | 97.00 | 1.00E-14 | 42.86 |
| MICE_ CDS16 | _00489 | _00027 | hypothetical protein | M. phocicerebrale | WP_116171526.1 | 7 | – | smc_procA | TIGR02169 | 6/36–2/18–2/20 | 536 | 98.00 | 0.0 | 56.96 |
| ICEF_-A CDS16 b | M. fermentans | AAN85226.1 | 67.35 | 1.00E-34 | 14.20 | |||||||||
| MICE_ CDS17 | _00488 | _00028 | hypothetical protein | M. phocicerebrale | WP_116171521.1 | 3 / 2 | – | TraC_F-type | TIGR02746 | 0/58–5/49–1/35 | 885 | 99.00 | 0.0 | 65.34 |
| TraE/TraCb | Mycoplasma spp. | WP_013526664.1 | 99.00 | 0.0 | 37.02 | |||||||||
| MICE_ CDS19 | _00487 | _00029 | DNA polymerase III | M. phocicerebrale | WP_116171520.1 | 2 | I | polC | PRK00448 | 15/112–7/113–5/99 | 1821 | 99.00 | 0.0 | 44.38 |
| ICE-M CDS19b | M. mycoides | CBW53951.1 | 82.00 | 2.00E-123 | 26.29 | |||||||||
| MICE_ CDS22 | _00486 | _00030 | hypothetical protein | M. phocicerebrale | WP_116171517.1 | – | – | UPF0236 | pfam06782 | 0/21–0/12–0/24 | 387 | 100.00 | 8.00E-125 | 52.20 |
| ISLre2 transposase b | H. hemicellulosilytica | WP_125474456.1 | 99.00 | 9.00E-161 | 19.11 |
- = no detection; a number of the respective ICEHo_II ORFs in SP3615_(KGPEAEHF_00500 to _00468) and SP10291 (HPAMDCMO_00016 to _00030) according to Additional file 7; b homologous proteins identified by PSI-BLAST; c DEPP predicted phosphorylation sites (x) compared to total numbers of possible phosphorylation sites (y) of S serine, T threonine, Y tyrosine
Circularization is likely indicative of ICEHo-I and -II transfer potential
For ICEs, excision and circularization represent key steps in the mobilization process [33]. We used a specifically developed PCR assay to demonstrate the presence of ICEHo-I and -II in their episomal circularized forms across many isolates in our screened cohort. The detection of circularized ICEHo-I and -II demonstrates the first step in the potential horizontal transfer of these elements and indicates that ICEHo-I and -II likely retain their mobile potential. Interestingly, we also detected the presence of minor sequence variants in the coupling region of circularized ICEHo-I and -II elements that could not readily be explained based on the respective genomic DR regions. Follow-up studies to confirm the existence of these minor CR sequence variants and to characterize their potential functional are an important direction for future work.
Conclusions
Nanopore sequencing enabled the characterization of mobile genetic elements and the identification of ICEHo-II, a novel MICE element of M. hominis. Our characterization provides a starting point for the elucidation of the function of the ICEHo-I and -II cargo genes and their phenotypic impact, in particular with respect to a potential impact on the pathogenicity of this genetically heterogeneous human facultative pathogen.
Methods
M. hominis strains
M. hominis strains were isolated from human specimens. Strains FBG, 8958 and 2539 were part of a collection of clinical M. hominis strains, created at the Institute of Pathology of the Johannes Gutenberg University Mainz, Germany, and transferred in 1988 to the Institute of Med. Microbiology and Hospital Hygiene at the Medical Faculty of the Heinrich-Heine-University of Duesseldorf; strains 475 and A136 derived from the Institute of Microbiology, University of Veterinary Medicine Vienna, Austria; strains SS10 and SS25 from the Institute for Specific Prophylaxis and Tropical Medicine, Centre for Pathophysiology, Immunology and Infectiology, Medical University of Vienna, Austria; and strains SP3615, VO31120, SP10291 and SP2656 were part of the strain collection of our institute, collected within the last 10 years. FBG, 8958 and 2539 were isolated from women; only for isolate 8958, the donor’s age (64) and strain location (vaginal) are known. Strain 475 was isolated from vaginal specimen; A136 and SP3615 were isolated from placenta after preterm birth; VO31120 was isolated from pleura of a patient with pneumonia; SP10291 was isolated from brain material after cerebral infarction; SP2565 was derived from blood culture of a patient in NHL remission [34]; SS10 and SS25 were isolated from in vitro cultured T. vaginalis (as endosymbionts). Protozoa were isolated from women affected by acute trichomoniasis respectively in 1996 and 1999, at the Department of Biomedical Sciences, University of Sassari, Italy [35]. All other M. hominis strains were taken from the strain collection of our institute in Duesseldorf, lacking information about associated diseases.
M. hominis culturing and genomic DNA preparations
M. hominis strains were cultivated in arginine-medium as described in detail previously [36]. Genomic DNA of the strains was isolated by the use of the QIAamp Blood and Tissue kit (Hilden, Germany) following the tissue protocol with minor modifications as published [37]. Concentration of genomic DNA was measured by Invitrogen Qubit 4 Fluorometer Qubit and its quality verified spectrophotometrically by NanoDrop 1000 Spectrophotometer and on a Fragment Analyzer System (Agilent, Santa Clara, CA USA) with method DNF-464-33 for high sensitivity large fragment 50 kb analysis.
Whole genome sequencing and assembly of M. hominis strains
Generation of short-read sequencing data
Short-read Illumina sequencing was carried out for 11 isolates. Sequencing libraries were prepared according to the manufacturer’s instructions and sequenced on the MiSeq platform with 2 × 300 bp or 2 × 250 bp paired-end sequencing protocols (Table 1).
Nanopore sequencing and assembly
22 M. hominis strains were sequenced on a MinION MK1B device. Sequencing libraries from quality-controlled genomic DNA were prepared according to the manufacturer’s instructions, employing the rapid (2 samples), ligation-based (1 sample), and barcoded ligation-based (19 samples) protocols (Table 1). Basecalling and demultiplexing were carried out with MinKNOW (basecalling only) and Albacore (basecalling and demultiplexing).
Canu [17] 1.6 (with parameters -genomeSize = 1 m -nanopore-raw) was used for the assembly of the generated long-read data, yielding one large contig for each sample but one. Two smaller contigs in the assemblies of samples SS25 and SP2565 had only spurious read support as reported by Canu and were removed from the assembly. To generate “high quality” assemblies for the samples for which short-read data were available, the assemblies of the first 10 samples (Table 1) were polished with Nanopolish [38] 0.8.4, circular overlaps at the ends of contigs were removed, and orientation to the PG21 type strain genome was carried out. Two rounds of Pilon [39] 1.22 were used for further polishing based on short reads for each of the “high-quality” assemblies. Finally, short-read data were aligned against the Pilon-polished assemblies; GATK [40] 3.7 (with parameters -T HaplotypeCaller -ploidy 1) was used to call variants; and reference alleles were substituted with variant alleles whenever the reference allele frequency, measured via samtools mpileup -q0 -Q10 [41], was ≤10%. All short-read alignments were generated with bwa mem [42] 0.7.15-r1140. The genome structure of the generated assemblies was examined with nucmer [43] and the effectiveness of the polishing strategy was assessed by visually screening for potential base errors in IGV [44]. For the remaining 12 samples for which no short-read data were available, Nanopore-only based assemblies (referred to as “draft assemblies”) as produced by Canu were only used to characterize the MGEs contain within them. Draft genomes were oriented to the PG21 type strain and the circular contig overlap region was substituted with a consensus sequence of the two underlying overlaps, computed with SeqMan Version 6.0 (DNASTAR. Madison, WI). Of note, ambiguities in the computed consensus were represented using IUPAC ambiguity characters.
To further improve sequence quality for a triplicate repeat (later identified as ICEHo-I) identified by our inspection strategy in the genome of sample FBG, we applied a modification of the GATK-based polishing strategy described above. First, all short reads aligned to any of the three copies of the repeat in the genome of sample FBG were extracted. Second, for the three assembled repeat sequences independently, the complete set of extracted reads was aligned against the individual instance of the repeat and variants were called with GATK (using -ploidy 3). Finally, reference alleles were substituted with variant alleles at homozygous variant positions with reference allele frequency ≤ 10%. A manuscript describing a generalization of our approach and presenting a stand-alone software implementation is currently under preparation.
PacBio sequencing and assembly
Library preparation for long-read sequencing of M. hominis isolate SP3615 on the Sequel system was carried out with the SMRTbell Template Prep Kit 1.0 and the Sequel Binding and Internal Control Kit 2.1, using the “Greater than 10kb Template Protocol” and 10 h movie time. Assembly was carried out with HGAP4 (SMRT Link Version 5.1.0.26412) [18] and polished with Arrow. Orientation and removal of circular overlaps were carried out as described above,. Visual inspection was used to confirm the quality of the generated assembly.
qPCR
Oligonucleotides used in qPCRs were designed using Probefinder (Roche Applied Science) (https://qpcr.probefinder.com). Primers are listed in Table 6.
Table 6.
Primers used
| Gene | qPCR primer | Sequence (5′-3′) | Amplicon length (nt) | PCR protocol |
|---|---|---|---|---|
| ICEHo-I_dcm | 463_F | CACGGATCTCCTTGTCAAGAT | ||
| 463_R | TGTTTCCCACAATAAACTACTTCG | 91 | 1 | |
| ICEHo-I_CDS5 | 462_F | AGAAGATTTGTCAAAAACTCCTAAAGA | ||
| 462_R | ACCACTTTGTGCTTCGGCTAA | 64 | 1 | |
| ICEHo-I_CDS14 | 474_F | CCAAATCCTTCAAACCCAACT | ||
| 474_R | TCTGGTTTAACTTCAGGGGTTG | 62 | 1 | |
| ICEHo-I_CDS16 | 476_F | GCAATTGCTTTTGTTGGAAGT | ||
| 476_R | CTGATCTTGCTCCAGACATAGC | 73 | 1 | |
| ICEHo-I_CDS17.1 | 477_F1 | GATTTTGTGCCGTCATCGTA | ||
| 477_R1 | TTTAAAATGGCAGGATTATCAGG | 67 | 1 | |
| ICEHo-I_CDS17.2 | 477_F2 | RAAAATATTTGCAAGAACATAACATTA | ||
| 477_R2 | ATTTTCTAACCGTTTTTGTCATTT | 165 | 1 | |
| ICEHo-II_CDS17 | 17-II_F | CCCAATAAATCCGATAGCATTA | ||
| 17-II_R | GTTCCCAACACTAACATTCCTC | 84 | 1 | |
| circular ICEHo-I | cICE_I-F | GCGGGCGCGTAGAGCAT | ||
| cICE_I-R | TATTTGGAATTAACCCCACATTTT | 185 | 2 | |
| circular ICEHo-II | cICE_II-F | CAAAATTCAGATTAATTACTAATAAACAAA | ||
| cICE_II-R | AGAGCATGAGCAAGAAAAAAAGTA | 290 | 2 | |
| hitA | hitA_F | TTGAGGCACAGCAATAGC | ||
| hitA_R | AAGGCTTAGGTAAGGAATTGATTAG | 81 | 1 | |
| gap | gap_F | GCAGGCTCAATATTTGACTCACT | ||
| gap_R | GATGATTCATTGTCGTATCATGC | 95 | 1 |
qPCR assays were carried out in a total volume of 25 μl consisting of 1 × MesaGreen MasterMix, 5 mM MgCl2, Amperase, 300 nM of each primer and 2.5 μl of genomic DNA or cDNA solution, which was derived from 20 ng RNA. Thermal cycling conditions were as follows: 1 cycle at 50 °C for 10 min, 1 cycle at 95 °C for 5 min followed by 45 cycles of 95 °C for 15 s and 60 °C for 1 min (protocol 1) or 1 cycle at 95 °C for 5 min followed by 35 cycles of 95 °C for 15 s, 30 s 55 °C and 60 °C for 45 s (protocol 2). The product was than heated from 65 °C to 95 °C with an increment of 0.5 °C/15 s and the plate read for melt curve analysis to check the identity of the amplicon. Each sample was analysed in duplicate. Cycling, fluorescent data collection and analysis were carried out on a CFX-Cycler of BioRad Laboratories (Munich, Germany) according to the manufacturer’s instructions.
ICEHo qPCR screening assay
Real time PCR (qPCR) was used to screen for the presence of ICEHo-I and -II. For ICEHo-I, qPCR was used to determine the presence of MICE core genes CDS5, − 14, − 16, − 17, and of the ICEHo-I specific dcm gene. For ICEHo-II, qPCR was used to determine the presence of a conserved region of the ICEHo-II CDS17 gene. Utilized primers are listed in Table 6. Ct values were interpreted relative to the chromosomal M. hominis-specific hitA gene [45, 46] (see Additional file 5), with ∆Ct values (defined as Ct (ICEHo gene X) – Ct (hitA)) ≥ 10 classified as negative, and ∆Ct-values < 10 classified as positive. The utilized ∆Ct value threshold of 10 was determined based on strains FBG (ICEHo-I), SP13615 (ICEHo-II), and SP10291 (ICEHo-I and –II) as positive controls, and ICEHo-free strains PG21, 8958, 2539, SP2565, SS10, and VO31120 as negative controls. For ICEHo-I, isolates in which all PCRs were positive were classified as unambiguously positive; isolates in which at least two PCRs were positive were classified as positive; and isolates in which 0 or 1 PCRs were positive were classified as negative. With the chosen threshold values and decision algorithm, assembly- and qPCR-based results were in perfect agreement for the sequenced strains (Additional file 5).
qPCR screening for episomal circularized ICEHo (cICEHo)
Real time PCR (qPCR) was used to screen for the presence of ICEHo-I and -II in their episomal circularized forms, utilizing outwards-facing primer pairs (cICE_I-F/_I-R and cICE_II-F/_II-R; see Table 6). For ICEHo-I, these primers targeted the conserved untranslated ICEHo-I regions 266 bp downstream of CDS22 (cICE_I-F) and 175 bp upstream of CDS1 (cICE_I-R), leading to PCR products of 0.2 kb in case of episomal circularisation. For ICEHo-II, they targeted the conserved untranslated ICEHo-II regions just downstream of CDS22 (cICE_II-F) and 152 bp upstream of CDS1, leading to cICE-II PCR products of 0.3 kb in case of episomal circularisation (see Fig. 4.A). In the whole-genome-sequenced samples, all cICE amplification products were sequenced with Sanger sequencing, confirming cICE detection results through the detection of a valid IRR-IRL junction and coupling region (CR) in all but two cases with high qPCR Ct values (33 and 31; Additional file 5). At higher cycle counts (> 30), SybrGreen-based qPCRs are known to be prone to false-positives due to the generation of primer dimers or mispriming to imperfect binding sites. For the wider cohort of samples that were only screened with qPCR, all cICE-PCRs with Ct values > 30 were thus classified as negative, unless Sanger sequencing of the PCR product proved the presence of a CR region in the amplification product (Additional file 5). Major and minor CR sequence variants were detected by applying the algorithm Mixed Sequence Reader [27] to the Sanger chromatogram data.
Annotation and bioinformatic analysis of M. hominis genomes
Prokka [20] was used to annotate the assembled genomes. PHAST (PHAge Search Tool) (http://phast.wishartlab.com/) was used to identify and annotate prophage sequences [47]. BLAST Microbes (https://BLAST.ncbi.nlm.nih.gov/BLAST.cgi) was used for detection of homologous genes and plasmids. Multiple sequence alignments were calculated by using Genious Pro (vers. 5.5.8) and MegAlign version 6.0 of the Lasergene software package (DNAStar, Madison, WI). Genome alignments illustrating gene gain, loss and rearrangement were done with Mauve [21]. The Phyre2 web portal was used for protein modelling, prediction and analysis [48]; RADAR for detection and alignment of repeats in protein sequences ( [49] https://www.ebi.ac.uk/Tools/services/web_radar/toolform.ebi); MEME for discovering novel, ungapped motifs (recurring, fixed-length patterns) ( [50] http://meme-suite.org/tools/meme); Disorder Enhanced Phosphorylation Predictor (DEPP) (http://www.pondr.com/cgi-bin/depp.cgi).
Statistical programs used
Statistical tests were performed in Stata 14 (StataCorp, TX). Associations of presence of different MGEs were assessed by Chi-square test, associations of abundance by Spearman’s rank correlation.
Supplementary Information
Additional file 1. Homology of CDS6. Deduced protein sequences of CDS6 derived from M. hominis strains FBG (BHBFJMJE_00471), SP10291 (HDENHCDK_00553), TO0613 (WP_036439043.1), PL5 (WP_036439043.1), and 4788 (WP_036439043.1) and of M. fermentans strains M64 (ADV34390.1 (I) and ADV34456.1(II)) and PG18 (ICEF-IA (AAN85216.1 (IA)) and ICEF-II-A (AAN85259.1 (II-A)). Two chromosomal proteins of M. hominis PG21 (MHO_0070 (CAX37141.1) and MHO_1280 (CAX37262.1)) were used as unrelated ICE-outliners in ClustalW-based Multiple Sequence Alignment. A. Phylogenetic tree; B. Percent amino acid identities and divergences; and C. Multiple Sequence Alignment of CDS6 encoded proteins. Identical amino acids are marked in green, isofunctional amino acids marked in yellow.
Additional file 2. Clustering of MhoH and MhoJ. MhoH and MhoJ proteins of FBG, SP10291, PL5 and TO0613 were clustered in multiple sequence alignment using Clustal W and divided in five subgroups (MhoH1 to MhoH3 and MhoJ1 and MhoJ2) according to their phylogenetic relationship (A.). All MhoH and MhoJ proteins carried the TAL-effector motif in the C-terminal part (B.). Percent amino acid identities and divergences are shown in C.).
Additional file 3. Phylogeny of MhoM and CDS11. MhoM encoded proteins of ICEHo-I and -II elements of strains FBG, SP10291, PL5, 4788 and TO0613 were clustered with the respective CDS11 genes in multiple sequence alignment using Clustal W. A.) Phylogenetic tree of MhoM and CDS11 encoded proteins. B.) Percent identities and divergences of MhoM and CDS11 proteins.
Additional file 4. ICEHo locations in draft genomes. Positions and gene presence patterns of ICEHo-I and -II in draft de novo assemblies of 12 M. hominis strains sequenced only with Nanopore. ICEHo positions in the draft assemblies were determined by aligning the sequences of ICEHo-I of strain FBG and of ICEHo-II of strain SP3615 to the draft assemblies. The additional columns show the homology (nucleotide identity %) between the genes present in the draft assembly ICEHo elements and the genes present in ICEHo-I of FBG and the genes present in ICEHo-II of SP3615 (gene order and names correspond to Fig. 3).
Additional file 5. qPCR data of ICEHo and cICE. Ct and ∆Ct values for ICEHo-I, −II and cICE detection, as well as ICEHo-I and -II status based on the assembled genomes, and confirmatory detection of cICE by Sanger sequencing.
Additional file 6. ICEHo-I and ICEHo-II putative gene functions. ICEHo-I and -II putative gene functions, based on bioinformatics analyses (see main text) and literature review.
Additional file 7. M. hominis genomes in GB.
Acknowledgements
We thank two members of the Institute of Medical Microbiology and Hospital Hygiene, Dana Belick for excellent technical assistance, and Malte Kohns Vasconcelos for statistical support. Computational support and infrastructure were provided by the “Centre for Information and Media Technology” (ZIM) at the University of Duesseldorf (Germany).
Abbreviations
- CDS
CoDing sequence
- CR
Coupling Region
- DEPP
Disorder enhanced phosphorylation predictor
- DR
Direct repeat
- ICE
Integrative and conjugative element
- ICEHo
Integrative and conjugative element of M. hominis
- MICE
Mycoplasmal integrative and conjugative element
- MGE
Mobile genetic element
- MTase
Methyltransferase
- PHAST
PHAge search tool
- qPCR
Real time quantitative PCR
- R-M system
Restriction-modification system
- SRP
Signal recognition particle
- T4SS
Type IV secretion system
- TAL
Transcription activator-like
Authors’ contributions
SH and DH performed the PCR-based screening of M. hominis strains for ICEHo prevalence; UF, PLF and JS isolated and characterised some of the pathogenic clinical strains of M. hominis. KK, SS, LP and ATD performed NGS sequencing and bioinformatics analyses; BH, ATD and KP defined the study design; BH and AD were major contributors in writing the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the Jürgen Manchot Foundation and departmental funds from the Institute of Medical Microbiology and Hospital Hygiene, Heinrich-Heine-University of Duesseldorf, Germany.
Availability of data and materials
Genome sequences of strains LBD-4 (acc.-no. CP009652.1), PG21 (acc.-no. FP236530.1), SPROTT (acc.-no. CP011538.1), TO0613 (acc.-no: CP033021.1) and contigs of strain PL5 (acc.-nos: JRXA01000001.1 - JRXA010000016.1) were downloaded from NCBI (https://www.ncbi.nlm.nih.gov/nuccore/).
All raw sequencing data and high-quality assemblies are made available under BioProject PRJNA429440; all draft assemblies are available at OSF (DOI: 10.17605/OSF.IO/CZRBT). Generated genome sequences in FASTA format are available at NCBI. Generated genome sequences in GenBank format and annotated using this publication’s annotation terminology are provided as an Additional file 7.
Ethics approval and consent to participate
The study was approved by the Ethical Committee of the Medical Faculty of the Heinrich-Heine University (Study-No.: 018–98-RetroDEuA). Consent to participate is not applicable due to the retrospective analysis of M. hominis strains.
Consent for publication
Not applicable.
Competing interests
Not applicable.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Taylor-Robinson D, Furr PM. Genital mycoplasma infections. Wien Klin Wochenschr. 1997;109:578–583. [PubMed] [Google Scholar]
- 2.Xiang L, Lu B. Infection due to mycoplasma hominis after left hip replacement: case report and literature review. BMC Infect Dis. 2019;19:50. doi: 10.1186/s12879-019-3686-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Waites KB, Katz B, Schelonka RL. Mycoplasmas and ureaplasmas as neonatal pathogens. Clin Microbiol Rev. 2005;18:757–789. doi: 10.1128/CMR.18.4.757-789.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goret J, Beven L, Faustin B, Contin-Bordes C, Le Roy C, Claverol S, Renaudin H, Bebear C, Pereyre S. Interaction of mycoplasma hominis PG21 with human dendritic cells: Interleukin-23-inducing Mycoplasmal lipoproteins and Inflammasome activation of the cell. J Bacteriol. 2017;199:e00213–17. [DOI] [PMC free article] [PubMed]
- 5.Goret J, Le Roy C, Touati A, Mesureur J, Renaudin H, Claverol S, Bebear C, Beven L, Pereyre S. Surface lipoproteome of mycoplasma hominis PG21 and differential expression after contact with human dendritic cells. Future Microbiol. 2016;11:179–194. doi: 10.2217/fmb.15.130. [DOI] [PubMed] [Google Scholar]
- 6.Stalberg C, Noda N, Polettini J, Jacobsson B, Menon R. Anti-inflammatory Elafin in human fetal membranes. J Perinat Med. 2017;45:237–244. doi: 10.1515/jpm-2016-0139. [DOI] [PubMed] [Google Scholar]
- 7.Rappelli P, Carta F, Delogu G, Addis MF, Dessi D, Cappuccinelli P, Fiori PL. Mycoplasma hominis and Trichomonas vaginalis symbiosis: multiplicity of infection and transmissibility of M. hominis to human cells. Arch Microbiol. 2001;175:70–74. doi: 10.1007/s002030000240. [DOI] [PubMed] [Google Scholar]
- 8.Cacciotto C, Dessi D, Cubeddu T, Cocco AR, Pisano A, Tore G, Fiori PL, Rapelli P, Pittau M, Alberti A. MHO_0730 is a surface-exposed calcium-dependent nuclease of mycoplasma hominis promoting neutrophils extracellular traps formation and escape. J Infect Dis. 2019. 10.1093/infdis/jiz406. [DOI] [PubMed]
- 9.Pekmezovic M, Mogavero S, Naglik JR, Hube B. Host-pathogen interactions during female genital tract infections. Trends Microbiol. 2019. 10.1016/j.tim.2019.07.006. [DOI] [PubMed]
- 10.Hopfe M, Deenen R, Degrandi D, Kohrer K, Henrich B. Host cell responses to persistent mycoplasmas--different stages in infection of HeLa cells with mycoplasma hominis. Plos One. 2013;8:e54219. doi: 10.1371/journal.pone.0054219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henrich B, Kretzmer F, Deenen R, Kohrer K. Validation of a novel mho microarray for a comprehensive characterisation of the mycoplasma hominis action in HeLa cell infection. Plos One. 2017;12:e0181383. doi: 10.1371/journal.pone.0181383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calcutt MJ, Foecking MF. Analysis of the complete mycoplasma hominis LBD-4 genome sequence reveals strain-variable Prophage insertion and distinctive repeat-containing surface protein arrangements. Genome Announc. 2015;3:e01582–14. [DOI] [PMC free article] [PubMed]
- 13.Degrange S, Renaudin H, Charron A, Bebear C, Bebear CM. Tetracycline resistance in Ureaplasma spp. and mycoplasma hominis: prevalence in Bordeaux, France, from 1999 to 2002 and description of two tet(M)-positive isolates of M. hominis susceptible to tetracyclines. Antimicrob Agents Chemother. 2008;52:742–744. doi: 10.1128/AAC.00960-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Calcutt MJ, Foecking MF. An excision-competent and exogenous mosaic transposon harbors the tetM gene in multiple mycoplasma hominis lineages. Antimicrob Agents Chemother. 2015;59:6665–6666. doi: 10.1128/AAC.01382-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meygret A, Peuchant O, Dordet-Frisoni E, Sirand-Pugnet P, Citti C, Bebear C, Beven L, Pereyre S. High prevalence of integrative and conjugative elements encoding transcription activator-like effector repeats in mycoplasma hominis. Front Microbiol. 2019;10:2385. doi: 10.3389/fmicb.2019.02385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Citti C, Baranowski E, Dordet-Frisoni E, Faucher M, Nouvel LX. Genomic Islands in mycoplasmas. Genes (Basel). 2020;11:836. [DOI] [PMC free article] [PubMed]
- 17.Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017;27:722–736. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE, Turner SW, Korlach J. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods. 2013;10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 19.Pereyre S, Sirand-Pugnet P, Beven L, Charron A, Renaudin H, Barre A, Avenaud P, Jacob D, Couloux A, Barbe V, de Daruvar A, Blanchard A, Bebear C. Life on arginine for mycoplasma hominis: clues from its minimal genome and comparison with other human urogenital mycoplasmas. Plos Genet. 2009;5:e1000677. doi: 10.1371/journal.pgen.1000677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 21.Darling AE, Mau B, Perna NT. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. Plos One. 2010;5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baranowski E, Dordet-Frisoni E, Sagne E, Hygonenq MC, Pretre G, Claverol S, Fernandez L, Nouvel LX, Citti C. The integrative conjugative element (ICE) of mycoplasma agalactiae: key elements involved in horizontal dissemination and influence of Coresident ICEs. MBio. 2018;9:e00873–18. [DOI] [PMC free article] [PubMed]
- 23.Calcutt MJ, Lavrrar JL, Wise KS. IS1630 of mycoplasma fermentans, a novel IS30-type insertion element that targets and duplicates inverted repeats of variable length and sequence during insertion. J Bacteriol. 1999;181:7597–7607. doi: 10.1128/JB.181.24.7597-7607.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murdoch DA. Gram-positive anaerobic cocci. Clin Microbiol Rev. 1998;11:81–120. doi: 10.1128/CMR.11.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ayling RD, Bashiruddin S, Davison NJ, Foster G, Dagleish MP, Nicholas RA. The occurrence of mycoplasma phocicerebrale, mycoplasma phocidae, and mycoplasma phocirhinis in grey and common seals (Halichoerus grypus and Phoca vitulina) in the United Kingdom. J Wildl Dis. 2011;47:471–475. doi: 10.7589/0090-3558-47.2.471. [DOI] [PubMed] [Google Scholar]
- 26.Grozner D, Forro B, Sulyok KM, Marton S, Kreizinger Z, Banyai K, Gyuranecz M. Complete genome sequences of mycoplasma anatis, M. anseris, and M. cloacale type strains. Microbiol Resour Announc. 2018;7:e00939–18. [DOI] [PMC free article] [PubMed]
- 27.Chang C-T, Tsai C-N, Tang CY, Chen C-H, Lian J-H, Hu C-Y, Tsai C-L, Chao A, Lai C-H, Wang T-H, Lee Y-S. Mixed sequence reader: a program for analyzing DNA sequences with Heterozygous Base calling. Sci World J. 2012;2012:365104. doi: 10.1100/2012/365104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tardy F, Mick V, Dordet-Frisoni E, Marenda MS, Sirand-Pugnet P, Blanchard A, Citti C. Integrative conjugative elements are widespread in field isolates of mycoplasma species pathogenic for ruminants. Appl Environ Microbiol. 2015;81:1634–1643. doi: 10.1128/AEM.03723-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Citti C, Dordet-Frisoni E, Nouvel LX, Kuo CH, Baranowski E. Horizontal gene transfers in mycoplasmas (Mollicutes) Curr Issues Mol Biol. 2018;29:3–22. doi: 10.21775/cimb.029.003. [DOI] [PubMed] [Google Scholar]
- 30.Cabezon E, Ripoll-Rozada J, Pena A, de la Cruz F, Arechaga I. Towards an integrated model of bacterial conjugation. FEMS Microbiol Rev. 2015;39:81–95. doi: 10.1111/1574-6976.12085. [DOI] [PubMed] [Google Scholar]
- 31.Johnson CM, Grossman AD. Integrative and conjugative elements (ICEs): what they do and how they work. Annu Rev Genet. 2015;49:577–601. doi: 10.1146/annurev-genet-112414-055018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leanza AG, Matteo MJ, Crespo O, Antelo P, Olmos J, Catalano M. Genetic characterisation of helicobacter pylori isolates from an Argentinean adult population based on cag pathogenicity island right-end motifs, lspA-glmM polymorphism and iceA and vacA genotypes. Clin Microbiol Infect. 2004;10:811–819. doi: 10.1111/j.1198-743X.2004.00940.x. [DOI] [PubMed] [Google Scholar]
- 33.Szuplewska M, Czarnecki J, Bartosik D. Autonomous and non-autonomous Tn3-family transposons and their role in the evolution of mobile genetic elements. Mob Genet Elements. 2014;4:1–4. doi: 10.1080/2159256X.2014.998537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.MacKenzie CR, Nischik N, Kram R, Krauspe R, Jager M, Henrich B. Fatal outcome of a disseminated dual infection with drug-resistant mycoplasma hominis and Ureaplasma parvum originating from a septic arthritis in an immunocompromised patient. Int J Infect Dis. 2010;14(Suppl 3):e307–e309. doi: 10.1016/j.ijid.2010.02.2253. [DOI] [PubMed] [Google Scholar]
- 35.Fichorova R, Fraga J, Rappelli P, Fiori PL. Trichomonas vaginalis infection in symbiosis with Trichomonasvirus and mycoplasma. Res Microbiol. 2017;168:882–891. doi: 10.1016/j.resmic.2017.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Henrich B, Feldmann RC, Hadding U. Cytoadhesins of mycoplasma hominis. Infect Immun. 1993;61:2945–2951. doi: 10.1128/IAI.61.7.2945-2951.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Henrich B, Lang K, Kitzerow A, MacKenzie C, Hadding U. Truncation as a novel form of variation of the p50 gene in mycoplasma hominis. Microbiology. 1998;144(Pt 11):2979–2985. doi: 10.1099/00221287-144-11-2979. [DOI] [PubMed] [Google Scholar]
- 38.Loman NJ, Quick J, Simpson JT. A complete bacterial genome assembled de novo using only nanopore sequencing data. Nat Methods. 2015;12:733–735. doi: 10.1038/nmeth.3444. [DOI] [PubMed] [Google Scholar]
- 39.Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, Earl AM. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. Plos One. 2014;9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing S The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. ArXiv e-prints. arXiv. 2013;1303.3997.
- 43.Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hopfe M, Hegemann JH, Henrich B. HinT proteins and their putative interaction partners in Mollicutes and Chlamydiaceae. BMC Microbiol. 2005;5:27. doi: 10.1186/1471-2180-5-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. PHAST: a fast phage search tool. Nucleic Acids Res. 2011;39:W347–W352. doi: 10.1093/nar/gkr485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Madeira F, Ym P, Lee J, Buso N, Gur T, Madhusoodanan N, Basutkar P, Tivey ARN, Potter SC, Finn RD, Lopez R. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019;47:W636–W641. doi: 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Homology of CDS6. Deduced protein sequences of CDS6 derived from M. hominis strains FBG (BHBFJMJE_00471), SP10291 (HDENHCDK_00553), TO0613 (WP_036439043.1), PL5 (WP_036439043.1), and 4788 (WP_036439043.1) and of M. fermentans strains M64 (ADV34390.1 (I) and ADV34456.1(II)) and PG18 (ICEF-IA (AAN85216.1 (IA)) and ICEF-II-A (AAN85259.1 (II-A)). Two chromosomal proteins of M. hominis PG21 (MHO_0070 (CAX37141.1) and MHO_1280 (CAX37262.1)) were used as unrelated ICE-outliners in ClustalW-based Multiple Sequence Alignment. A. Phylogenetic tree; B. Percent amino acid identities and divergences; and C. Multiple Sequence Alignment of CDS6 encoded proteins. Identical amino acids are marked in green, isofunctional amino acids marked in yellow.
Additional file 2. Clustering of MhoH and MhoJ. MhoH and MhoJ proteins of FBG, SP10291, PL5 and TO0613 were clustered in multiple sequence alignment using Clustal W and divided in five subgroups (MhoH1 to MhoH3 and MhoJ1 and MhoJ2) according to their phylogenetic relationship (A.). All MhoH and MhoJ proteins carried the TAL-effector motif in the C-terminal part (B.). Percent amino acid identities and divergences are shown in C.).
Additional file 3. Phylogeny of MhoM and CDS11. MhoM encoded proteins of ICEHo-I and -II elements of strains FBG, SP10291, PL5, 4788 and TO0613 were clustered with the respective CDS11 genes in multiple sequence alignment using Clustal W. A.) Phylogenetic tree of MhoM and CDS11 encoded proteins. B.) Percent identities and divergences of MhoM and CDS11 proteins.
Additional file 4. ICEHo locations in draft genomes. Positions and gene presence patterns of ICEHo-I and -II in draft de novo assemblies of 12 M. hominis strains sequenced only with Nanopore. ICEHo positions in the draft assemblies were determined by aligning the sequences of ICEHo-I of strain FBG and of ICEHo-II of strain SP3615 to the draft assemblies. The additional columns show the homology (nucleotide identity %) between the genes present in the draft assembly ICEHo elements and the genes present in ICEHo-I of FBG and the genes present in ICEHo-II of SP3615 (gene order and names correspond to Fig. 3).
Additional file 5. qPCR data of ICEHo and cICE. Ct and ∆Ct values for ICEHo-I, −II and cICE detection, as well as ICEHo-I and -II status based on the assembled genomes, and confirmatory detection of cICE by Sanger sequencing.
Additional file 6. ICEHo-I and ICEHo-II putative gene functions. ICEHo-I and -II putative gene functions, based on bioinformatics analyses (see main text) and literature review.
Additional file 7. M. hominis genomes in GB.
Data Availability Statement
Genome sequences of strains LBD-4 (acc.-no. CP009652.1), PG21 (acc.-no. FP236530.1), SPROTT (acc.-no. CP011538.1), TO0613 (acc.-no: CP033021.1) and contigs of strain PL5 (acc.-nos: JRXA01000001.1 - JRXA010000016.1) were downloaded from NCBI (https://www.ncbi.nlm.nih.gov/nuccore/).
All raw sequencing data and high-quality assemblies are made available under BioProject PRJNA429440; all draft assemblies are available at OSF (DOI: 10.17605/OSF.IO/CZRBT). Generated genome sequences in FASTA format are available at NCBI. Generated genome sequences in GenBank format and annotated using this publication’s annotation terminology are provided as an Additional file 7.