Abstract
Ring canals in the female germline of Drosophila melanogaster are supported by a robust filamentous actin (F-actin) cytoskeleton, setting them apart from ring canals in other species and tissues. Previous work has identified components required for the expansion of the ring canal actin cytoskeleton, but has not identified the proteins responsible for F-actin recruitment or accumulation. Using a combination of CRISPR-Cas9 mediated mutagenesis and UAS-Gal4 overexpression, we show that HtsRC—a component specific to female germline ring canals—is both necessary and sufficient to drive F-actin accumulation. Absence of HtsRC in the germline resulted in ring canals lacking inner rim F-actin, while overexpression of HtsRC led to larger ring canals. HtsRC functions in combination with Filamin to recruit F-actin to ectopic actin structures in somatic follicle cells. Finally, we present findings that indicate that HtsRC expression and robust female germline ring canal expansion are important for high fecundity in fruit flies but dispensable for their fertility—a result that is consistent with our understanding of HtsRC as a newly evolved gene specific to female germline ring canals.
Keywords: ring canal, hu li tai shao, Drosophila oogenesis, actin cytoskeleton, intrinsically disordered protein
ACROSS the animal kingdom, gametogenesis occurs in a syncytium, or a group of interconnected cells (Haglund et al. 2011). Germline syncytia are formed through a modified cytokinesis pathway, wherein abscission does not occur and daughter cells remain connected by a cytoplasmic intercellular bridge (Haglund et al. 2011; Lu et al. 2017). In both male and female germlines, intercellular bridges retain some components of the contractile ring, but also recruit additional cytoskeletal proteins to stabilize or grow nascent ring canals (Robinson and Cooley 1996; Greenbaum et al. 2007; Haglund et al. 2011). However, the process by which intercellular bridges form and recruit components is poorly understood in most species.
Intercellular bridge formation and function are best characterized in the Drosophila ovary, where the bridges are called ring canals. In Drosophila, oogenesis occurs in egg chambers—the functional units of the ovary. Each egg chamber contains a syncytium of 16 germline cells: 15 nurse cells and one transcriptionally quiescent oocyte, formed through four rounds of mitosis via incomplete cytokinesis (Hinnant et al. 2020). Following the fourth mitotic division, ring canals increase in diameter, expanding from 0.5 to 1 µm to a final size of 10 µm or larger, a 20-fold increase (Tilney et al. 1996). Ring canals in egg chambers are necessary to support oocyte growth by allowing the flow of cytoplasm (Robinson et al. 1994) as well as the transfer of organelles, including mitochondria (Cox and Spradling 2003), from nurse cells to the oocyte. Compared to ring canals in Drosophila males and in females of other species, which range from 1 to 4 µm in diameter, female germline ring canals in Drosophila are the largest ring canals known to date (Haglund et al. 2011). Although these ring canals are among the best characterized, the evolutionary and mechanistic reasons for their large size remain a mystery.
The large size of Drosophila female germline ring canals depends on the recruitment and expansion of a robust ring canal F-actin cytoskeleton (Tilney et al. 1996). After completion of the mitotic divisions, maturing ring canals recruit noncontractile F-actin, HtsRC, a product of the hu li tai shao (hts) gene (Yue and Spradling 1992), and Filamin, encoded by the cheerio gene (Robinson et al. 1997; Sokol and Cooley 1999). F-actin recruitment requires both HtsRC and Filamin, as the absence of either protein results in F-actin-deficient ring canals. During the actin recruitment phase, both the number of actin filaments and the tube length of the ring canal increase without a large increase in the ring canal diameter (Tilney et al. 1996). Next, the expansion phase is dependent on the Arp2/3 complex, which acts on existing filaments to promote branching and an increase in the total length of actin filaments at the ring canal (Hudson and Cooley 2002; Zallen et al. 2002). It is not known how the branched actin networks formed by this mechanism are remodeled into mixed polarity bundles. It also remains unclear what other players or mechanisms may be involved in the expansion of ring canals. Work in the 1990s and early 2000s uncovered multiple kinases involved in ring canal expansion, suggesting that post-translational modification of ring canal components is required in this process (Dodson et al. 1998; Guarnieri et al. 1998; Jackson and Berg 2002). More recent work has also revealed a role for membrane tension sensing by Filamin in normal ring canal expansion (Huelsmann et al. 2016), and we have previously shown proteasome dependent HtsRC turnover of HtsRC is important for clearance of the ring canal lumen (Hudson et al. 2019).
Here, we investigate in detail the role of HtsRC in ring canal biogenesis. HtsRC is produced from one class of alternatively spliced mRNAs from the hts gene, referred to collectively as ovhts; the hts gene also produces other splice variants, including the conserved F-actin- and Spectrin-binding protein, Adducin (Figure 1A) (Ding et al. 1993; Whittaker et al. 1999; Petrella et al. 2007). Mutations in the hts locus were initially isolated from a screen for female sterile mutants; hts mutant egg chambers have too few germline cells and ring canals lacking F-actin (Yue and Spradling 1992). The aberrant egg chamber architecture has been attributed to the lack of Adducin, which is required to assemble fusomes needed during early development (Lin et al. 1994). An understanding of the contribution of HtsRC to oogenesis has lagged because of its complicated biogenesis and the lack of mutations that affect only the expression of HtsRC.
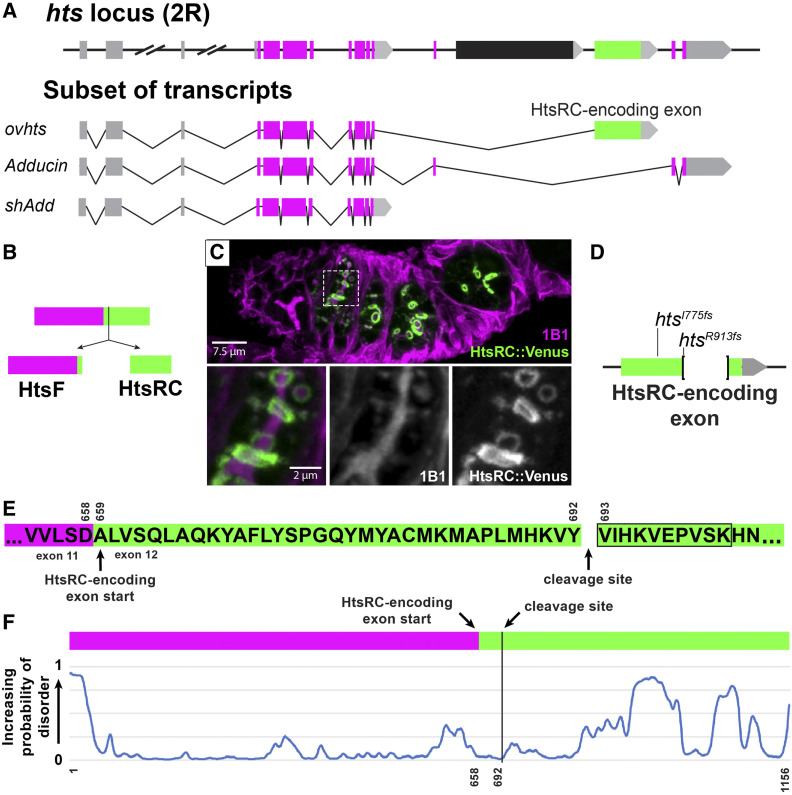
Figure 1.
HtsRC is produced from the HtsRC exon after cleavage of the Ovhts polyprotein. (A) Diagram of the hts locus, showing a subset of spliced exons (top) and the structure of three transcripts containing alternatively spliced terminal exons. Untranslated regions and exons are color coded as follows: 5′ UTRs (gray boxes), 3′ UTRs (gray arrows), conserved Adducin exons (magenta boxes), and the HtsRC exon (green box). The black exon corresponds to a testis-specific splice variant not shown. Three hts isoforms with unique terminal exon groups can be detected in the ovary: ovhts, Adducin, and shAdd. Distances are to scale, except where indicated by hash marks. (B) ovhts mRNA produces the Ovhts polyprotein, which is cleaved into HtsF and HtsRC. Exons are color coded as in (A). (C) A germarium expressing transgenic Ovhts::Venus polyprotein. Ovhts tagged at the C-terminus with Venus produces HtsRC::Venus and untagged HtsF following cleavage. HtsF is labeled with a monoclonal antibody (1B1), which also labels Add and shAdd in the fusome. Insets show 1B1 labeling and HtsRC::Venus at the time point where the fusome is dissolving and ring canals first recruit HtsRC. (D) Summary of mutations in the HtsRC exon induced by CRISPR-Cas9 mediated NHEJ. htsI775fs is a 2 bp deletion resulting in a frameshift and 62 novel amino acid residues followed by a stop codon. htsR913fs is a 559 bp deletion followed by a frameshift resulting in 10 novel residues and a stop codon. (E) Amino acid sequence of the Ovhts polyprotein, showing the start of the HtsRC exon and the cleavage site identified by mass spectrometry experiments at residue 693 (residue 35 of the HtsRC exon). Black outline indicates the identified semitryptic peptide. (F) Prediction of intrinsically disordered regions within the Ovhts polyprotein using the SPOT-Disorder-Single application (Hanson et al. 2018). Vertical black line indicates the polyprotein cleavage site described in (E).
The ovhts mRNA (Figure 1A) contains a terminal exon not present in adducin genes outside of flies (Figure 1A, green exon). In Drosophila, mRNA containing this exon has been detected only in the ovary, and antibody staining reveals that the protein product of this exon is germline specific. Previous work from our laboratory has shown that the Ovhts protein produced from this mRNA is a polyprotein that undergoes post-translational cleavage to produce HtsF, a truncated Adducin protein that localizes to fusomes (magenta, Figure 1, B and C), and HtsRC, which localizes to ring canals (green, Figure 1, B and C) (Petrella et al. 2007). In addition to ring canals, HtsRC is also found on cytoplasmic actin bundles in stage 10B egg chambers and on the oocyte cortex in later stages (Huelsmann et al. 2013; Pokrywka et al. 2014). The level of HtsRC at ring canals is under the control of a Cullin3 RING Ubiquitin ligase (CRL3) in which Kelch is the substrate adaptor that binds HtsRC, leading to polyubiquitylation of HtsRC and its destruction by the proteasome (Hudson et al. 2019). Mutations in the kelch gene lead to increased levels of ring canal HtsRC and F-actin, consistent with HtsRC being the target of CRL3Kelch.
In this work, we investigated how HtsRC controls F-actin levels at the ring canal by testing the impact of loss of function mutations and overexpression of HtsRC. We found that HtsRC is both necessary and sufficient for F-actin accumulation. HtsRC-specific mutants lacked F-actin at ring canals and ectopic expression of HtsRC in somatic follicle cells drove the formation of F-actin aggregates. The ability of HtsRC to drive F-actin accumulation ectopically depended on Filamin, but was independent of the Arp2/3 complex. The absence of HtsRC in germline cells resulted in small or collapsed ring canals, and mutant females were fertile but had dramatically reduced fecundity. These data suggest that large ring canals contribute to the high fecundity of Drosophila females.
Materials and Methods
Supplementary materials
Supplemental Figures PDF containing Supplemental Material, Figures S1–S6, Table S1 and Reagent Table.
Drosophila genetics
Fruit flies were maintained at room temperature on standard fly food medium, unless otherwise indicated. Females were fattened for 8–24 hr on wet yeast paste prior to dissection.
MARCM:
We conducted experiments in follicle cell clones using the MARCM system (Lee and Luo 1999) to generate GFP positive clones also expressing HtsRC protein. MARCM stocks were generated by crossing tubulin-Gal80 FRT40A (second chromosome) and UAS-mCD8::GFP, tubulin-Gal4 (third chromosome) into a stock containing hsFLP1 (X chromosome) to generate a balanced stock. Separately, an isogenized control FRT40A chromosome and the Arp2/3 complex alleles (AprC1Q29sd FRT40A and ArpC4SH1036 FRT40A) on the second chromosome were crossed with UASh-ovhts::V5::APEX (htsN4UTR) on the third chromosome. The resulting stocks were crossed to the MARCM stock. Heat shocks were conducted on pupae for 1 hr at 38°C, then flies were allowed to develop for 6 days before dissection. Following heat shock, clones homozygous for Arp2/3 complex subunits or the control FRT40A chromosome were identified by the expression of membrane localized CD8::GFP and the HtsRC::APEX transgene, both driven by tubulin-Gal4. The removal of any component of the Arp2/3 complex has previously been shown to inactivate the entire complex, so mutant clones were considered null for Arp2/3 complex activity (Hudson and Cooley 2002).
Molecular biology
Construction of UASp-ovhts::GFP (K10UTR) transgene:
To make UASp-Ovhts::GFP, the coding sequence of Ovhts::GFP was PCR-amplified from UASH-Ovhts::GFP (contains htsN4 3′UTR; Petrella et al. 2007) using primers to add Gateway recombination sequences to the 5′ and 3′ ends. PCR product encoding Ovhts::GFP was recombined into pDONR201, and a sequence-verified entry clone was recombined with pPW-attB—a UASp Gateway destination vector modified to include a phiC31 attB recombination site (gift from Michael Buszczak, UT Southwestern). The resulting pUASp-Ovhts::GFP plasmid was transformed into a strain containing the attP2 landing site on chromosome III (68A4) at Rainbow Transgenic Flies, Inc.
Construction of UASH-ovhts::V5::APEX (htsN4UTR) transgene: to make UASH-ovhts::V5::APEX, the coding sequence of APEX1 was PCR-amplified from pcDNA3-mito-APEX. The resulting PCR product contained a 5′ V5-tag linker sequence in addition to 5′ XhoI and 3′ NotI cut sites. pCOH-ovhts::GFP (Petrella et al. 2007) was digested with XhoI and NotI to excise GFP and the V5::APEX1 fragment was ligated into the plasmid inframe and in place of GFP. Transgenic flies were generated via P-element-mediated insertion at GenetiVision.
Analysis of MS/MS data for semitryptic peptides
Mass spectrometry data from Hudson et al. (2019) and Mannix et al. (2019) were searched for semitryptic peptides at MS Bioworks using Mascot (Matrix Science); searches were conducted against the UniProt Drosophila melanogaster database with common laboratory contaminants added as well as custom protein sequences for HtsRC that included known SNPs. The results were parsed into Scaffold (Proteome Software) for further analysis.
Ovary preparation and immunofluorescence
Ovaries were dissected in IMADS (Singleton and Woodruff 1994) or 1× PBS and fixed with 4% PFA (Electron Microscopy Sciences) in 1× PBS plus 0.3% Triton-X100. For samples with actin cable imaging, we used a modified protocol based on Starble and Pokrywka 2018 with an additional 1 hr BSA incubation step prior to primary antibody incubation. Primary antibody incubations were conducted for either 2 hr at room temperature or overnight at 4° and incubations with fluorescent secondaries plus TRITC-phalloidin were done for 2 hr at room temperature. Antibody dilutions are indicated in the reagent table. Samples were mounted in ProLong Gold Antifade Mountant (Thermo Fisher Scientific) and allowed to cure 24–48 hr before imaging.
Microscopy
Image acquisition:
Samples were imaged on one of three microscope setups. (1) A Leica SP8 scanning confocal system using a 40× Plan Apo 1.30 NA objective or 63× Plan APO 1.40 NA objective in combination with optical zoom. This set up was used to collect images presented throughout the paper. (2) A Zeiss Axio Observer 7 inverted microscope with a CrEST X-Light V2 spinning disc system, Photometrics Prime BSi sCMOS camera, and a 40× C-Apo 1.2 NA water-immersion objective. This set up was used to collect images for the quantifications shown in Figure 7 and Figure S4. (3) Lower resolution egg images for Figure 4, B and C were taken with a Leica MZFLIII stereo microscope (10× objective) fitted with a Leica MC120HD camera.
Figure 7.
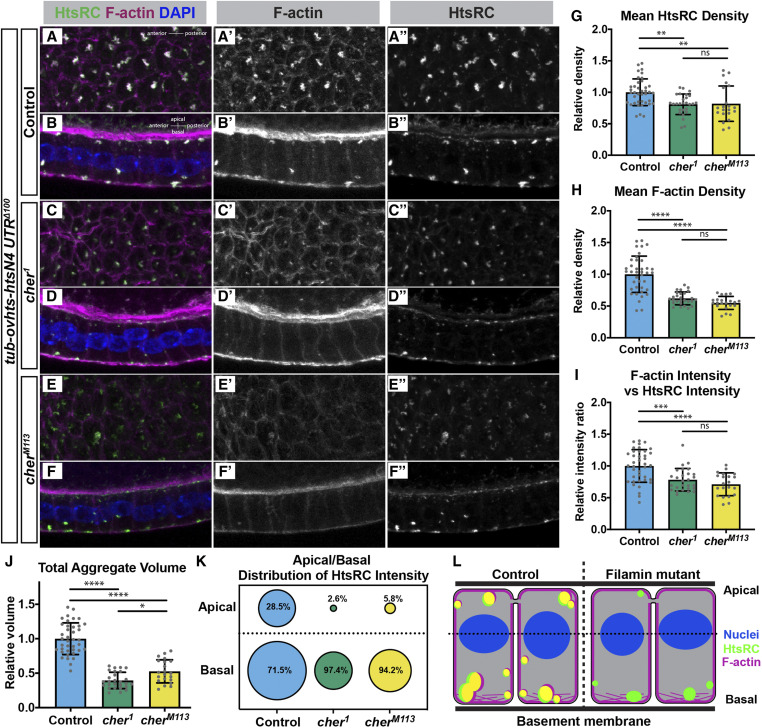
Filamin is important for F-actin recruitment to HtsRC aggregates. Ectopic HtsRC protein was driven from the tubulin-ovhts-N4Δ100 transgene in somatic follicle cells in either a wild-type background (A and B) or two independent cheerio mutant backgrounds (C–F). (A, C, and E) Z-projections from representative egg chambers showing the follicle cells from the basal surface. (B, D, and F) Z-projections of follicle cells showing the apical/basal distribution of aggregates with the apical side facing the top. Image pairs come from the same egg chamber. (G and H) Density calculated as HtsRC (G) or F-actin (H) intensity per μm3, averaged across all aggregates within a uniform region. All mean values were divided by the mean of the controls to obtain a relative density. (I) F-actin intensity per unit HtsRC intensity, relative to the mean of the controls. (J) Total volume occupied by aggregates normalized to the mean total aggregate volume for the controls. (G–J) All results were significant by both ANOVA and pairwise t-tests with P values adjusted for multiple comparisons, with pairwise t-tests displayed on graphs. Significance thresholds: *P < 0.05; **P < 0.01; ***P < 0.001, ****P < 0.0001. (K) Distribution of total HtsRC intensity within aggregates relative to the center of the nuclei (dotted line in L). Percentages indicate the distribution of HtsRC as a fraction of total HtsRC intensity within that genotype. The areas of the circles are scaled to indicate relative intensity between genotypes and between regions. (L) Model of HtsRC/F-actin aggregate formation in control (left) compared to cheerio mutants (right). HtsRC (green) and F-actin (magenta) colocalize (yellow) in aggregates at both the apical and basal ends of the cells in controls, but HtsRC does not recruit F-actin robustly in mutants and primarily forms aggregates at the basal end.
Figure 4.
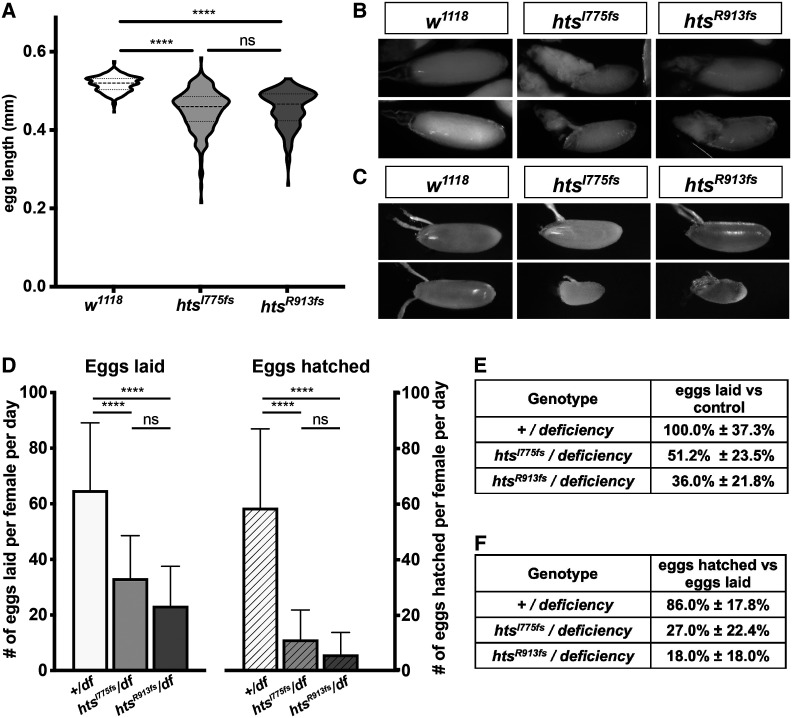
Loss of HtsRC protein impacts egg length and fecundity. (A) Violin plots showing distribution of egg length for two unique htsRC-specific mutants or w1118. n > 200 per genotype. Significance was determined using Tukey’s multiple comparison test. (B) Representative images from late stage egg chambers showing residual nurse cell cytoplasm. (C) Representative images showing the range of sizes among laid eggs. (D) Graph indicating number of eggs laid per day per female (solid bars) compared with the number of eggs hatched per female (patterned bars) for flies carrying either a wild-type allele or one of two independent htsRC-specific mutations over the Df(2R)BSC26 deficiency. Error bars indicate SD. n > 11 per genotype. Significance was determined using Tukey’s multiple comparison test. (E) Egg laying rates with SD as a percentage of egg laying in the wild-type control. (F) Egg hatching as a percentage of the number of eggs laid for each genotype, ± SD. Significance thresholds: ****P < 0.0001; ns, not significant.
Microscopy data analysis
Image processing:
All images shown in figures were processed and analyzed with ImageJ/FIJI. All processing and treatments in ImageJ/FIJI were matched within each data set.
Egg chamber staging:
Egg chamber stage was determined using both identifiable stage-specific features, such as shape, follicle cell behavior and oocyte size (Cummings and King 1969), and egg chamber area calculated for an ellipse based on the dimensions of the germ cells in a central z-slice, excluding the follicle cells (Jia et al. 2016). Where the two methods disagreed, visible features described in Cummings and King (1969) were given precedence.
Measurement of ring canal size:
Ring canals were measured on z-projections using ImageJ/FIJI. The freehand line tool was used to plot the profile of a line bisecting the widest point of the ring canal, then the diameter was measured as the distance from the outside of one peak to the outside of the other at ∼50% of the maximum fluorescence intensity for each peak. Ring canals were considered collapsed if no lumen was visible by eye or on a line scan of the ring canal, and diameter was measured at the half maximum for the single peak. Violin plots were generated in Jupyter notebook using the matplotlib and seaborn visualization packages to display the distribution of ring canal diameter measurements across genotypes.
Quantification of HtsRC aggregates:
Aggregates were characterized and quantified two ways: (1) using ImageJ/FIJI (Figure 6); (2) using Imaris 9.3.1 and Python code (Figure 7 and Figure S4).
Figure 6.
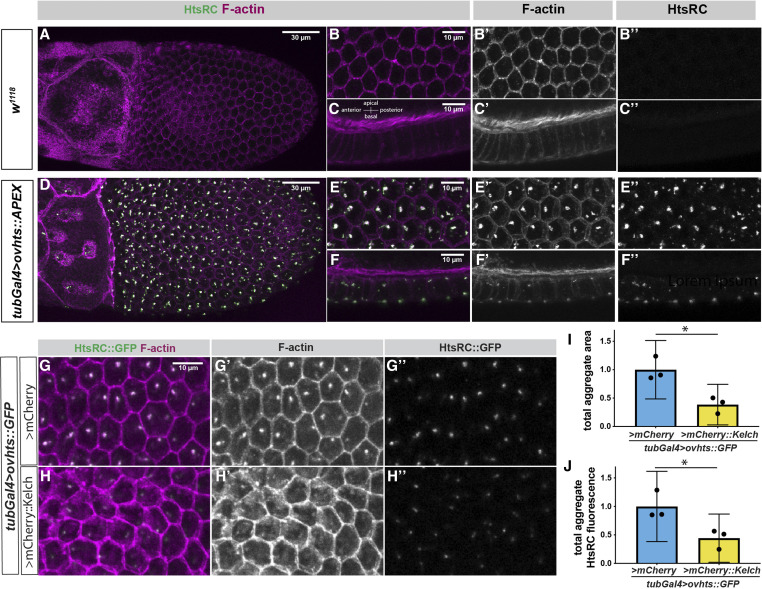
Ectopic HtsRC expression drives the formation of F-actin aggregates. (A–F) Stage 10B egg chambers focusing on the somatic follicle cell layer surrounding the oocyte either from the basal surface (A, B, D, and E) or from the side with the basal side at the bottom and the apical side and germline at the top (C and F). (A–C) Wild-type (w1118) control egg chamber labeled with Phalloidin and HtsRC antibody. All images shown were taken from the same egg chamber. (D–F) Ectopic expression of UASh-ovhts::APEX::V5 driven with the tubulin-Gal4 (tubGal4) driver. All images are taken from the same egg chamber. (G–H) tubulin-Gal4 driving UASh-ovhts::GFP along with either UASp-mCherry::kelch or a UASp-mCherry control. (I) Total area occupied by foci in a Z-projection the follicle cells, measured by HtsRC::GFP fluorescence. (J) Total HtsRC::GFP fluorescence contained within aggregates in control or UASp-mCherry::kelch overexpression. (I and J) Student’s t-tests were used to determine significance. Significance thresholds: *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; ns, not significant.
Z-projections of the follicle cell layer of stage 10B egg chambers were processed in ImageJ/FIJI. Positive F-actin foci were counted and measured in ImageJ/FIJI using the Auto-thresholding and Particle Analysis tools, with the minimum area of a foci set to 1 μm2. Egg chamber area was calculated in FIJI using the Freehand and Area/Measure tools.
Three-dimensional (3D) image files were processed in Imaris 9.3.1 as follows: 150 pixel (24.375 µm) × 150 pixel regions of follicle cells were duplicated from images of stage 10A egg chambers using ImageJ; care was taken to choose regions with minimal curvature. Although regions had consistent x and y dimensions, the z-depth varied depending on the thickness of the follicle cell layer in individual samples. For the experiments in Figure 7, three regions were chosen from approximately the same locations in each egg chamber. For the experiments with MARCM clones shown in Figure S4, regions were taken from within clones such that all the cells processed were mutant. The number of samples for this data set was limited by the size of individual clones and one to three regions were used per egg chamber. These images were imported into Imaris 9.3.1 and the Surfaces tool was used on the z-slices containing the somatic follicle cell layer and a small portion of the oocyte cortex. Aggregates (3D surfaces) were identified in the HtsRC (HtsRC antibody + Alexa 633) channel using background subtraction (local contrast) algorithm with automatic thresholding to distinguish objects (Surface Grain Size, 0.200 µm; Diameter of Largest Sphere, 0.200 µm). Nuclei were also identified by propagating the similar settings to the DAPI channel (Surface Grain Size, 0.325 µm; Diameter of Largest Sphere, 1.22 µm). Features of individual surfaces were exported including volume, center of mass, and sum of fluorescence intensity (the total fluorescence in an individual aggregate) for all channels. Custom Python scripts run in Jupyter notebook were used to import data files and calculate HtsRC density (summed fluorescence intensity divided by volume), F-actin density, and the ratio of F-actin to HtsRC. These calculations were done on individual aggregates and averaged to generate a summary result for the 150 × 150-pixel region. Values for individual aggregates were imported into Excel and used to calculate mean, standard deviation, and N for each region, and results for individual regions were graphed using Prism 3. The relative Z-position of each aggregate was calculated as follows: the center of fluorescent intensity for DAPI signal was calculated for each 150 × 150 pixel region and used as the origin for that region. Z-positions closer than the origin to the coverslip were considered negative (basal side was always facing coverslip). Values for individual aggregates were sorted using a Python script, where individual aggregates were identified as apical, if they were positive relative to the nuclei, or basal, if they were negative relative to the nuclei.
Fertility/fecundity tests
Fertility tests were performed in custom cages made from 14 ml polypropylene bacterial tubes attached in a honeycomb structure, which allowed tests to be performed on the same plates simultaneously for all genotypes. Cages were placed over grape juice or apple juice agar plates with charcoal added to increase visibility of eggs. One virgin and one w1118 male were mated in a single tube for 24 hr, then eggs were counted immediately (eggs laid) and 24 hr later (eggs remaining unhatched). Flies were initially left on plates for 24 hr to mature, then egg laying was measured for 1- to 2- and 2- to 3-day-old flies. Data presented is an aggregate of both days.
Measurement of mature egg length
Three virgins of each genotype and an equal number of w1118 males were placed in mesh cages over grape juice agar plates with charcoal added to increase visibility of eggs. Plates were changed every 12 hr. Immediately after the 12-hr laying period, eggs on laying plates were imaged on a Leica MZFLIII stereo microscope (10× objective) fitted with a Leica MC120HD camera. Egg length was measured in ImageJ/FIJI.
Protein structure and disorder prediction
Prediction of intrinsically disordered regions within the Ovhts polyprotein were done using Spot-Disorder-Single (Hanson et al. 2018). The full-length Ovhts polyprotein was used as the input. Disorder prediction was also run using the MetaDisorder (Kozlowski and Bujnicki 2012), which combines the results of multiple servers. Prediction of HtsRC structure and motifs was conducted using Phyre2 (Kelley et al. 2015).
Statistical tests
Statistical tests were conducted either in Prism 8 or in Microsoft Excel. Where appropriate, an F-test was conducted on paired samples to determine if samples had equal variance, then the appropriate t-tests were used to determine statistical significance of the results. Individual tests used are indicated in figure legends. Where three samples were compared, an ANOVA was used and the P-values displayed on graphs represent the results pairwise t-tests with P-values adjusted for multiple comparisons using built in statistical tests in Prism 8. Standard significance thresholds were used on graphs and are as follows: *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Identification of HtsRC orthologs in other species
Ovhts or just the HtsRC exon (D. melanogaster) was blasted against Drosophila as well as all available transcripts using NCBI blastn, blastx and blastp searches. Musca domestica sequence was identified by blasting D. melanogaster HtsRC against the other insect genomes using Vectorbase. Once identified, we repeated our search in Vectorbase using the M. domestica HtsRC sequence and identified the Glossina palpalis HtsRC ortholog, which appears to be mis-annotated as an independent transcript but is likely part of an ovhts transcript. Once obtained, 28 HtsRC sequences were aligned using ClustalW and ClustalO algorithms in the Jalview software package. For this comparison, sequences matching the HtsRC exon were generated by removing all sequences corresponding to the HtsF/Adducin exons of ovhts and only these trimmed sequences were aligned.
Data availability
Drosophila stocks and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Further information and requests for resources and reagents should be directed to and will be fulfilled by the corresponding author (L.C.). Supplemental material is available at figshare: https://doi.org/10.25386/genetics.12902582.
Results
Ovhts polyprotein is cleaved within the HtsRC exon-encoded peptide
We identified HtsRC in two independent mass spectrometry approaches designed to identify ring canal-associated proteins (Hudson et al. 2019; Mannix et al. 2019). Knowing that HtsRC is cleaved from the Ovhts polyprotein (Petrella et al. 2007), we searched our MS/MS data for peptides resulting from semitryptic cleavage, reasoning that an N-terminal peptide beginning at the Ovhts cleavage site could be identified in this way. A search of MS/MS data from proteins that copurify with the Kelch KREP domain (Hudson et al. 2019) identified three peptide spectra with the sequence VIHKVEPVSK (peptide false discovery rate threshold: 0.1%; Figure 1E, black box), consistent with cleavage prior to V693 in Ovhts. The same peptide was identified in a search for semitryptic peptides in a dataset from HtsRC::APEX proximity labeling experiments (Mannix et al. 2019). We therefore conclude that the Ovhts is cleaved following Y692. This result is consistent with our prior conclusion that HtsRC is encoded entirely within this exon. Furthermore, transgenes encoding either the entire ovhts cDNA (Hudson et al. 2019) or just the HtsRC-encoding exon (data not shown) rescued htsRC-specific mutants, confirming that HtsRC is produced from this exon. The following experiments were conducted with ovhts-based transgenes.
HtsRC is required for the accumulation of F-actin to RCs, but not to other actin-rich structures
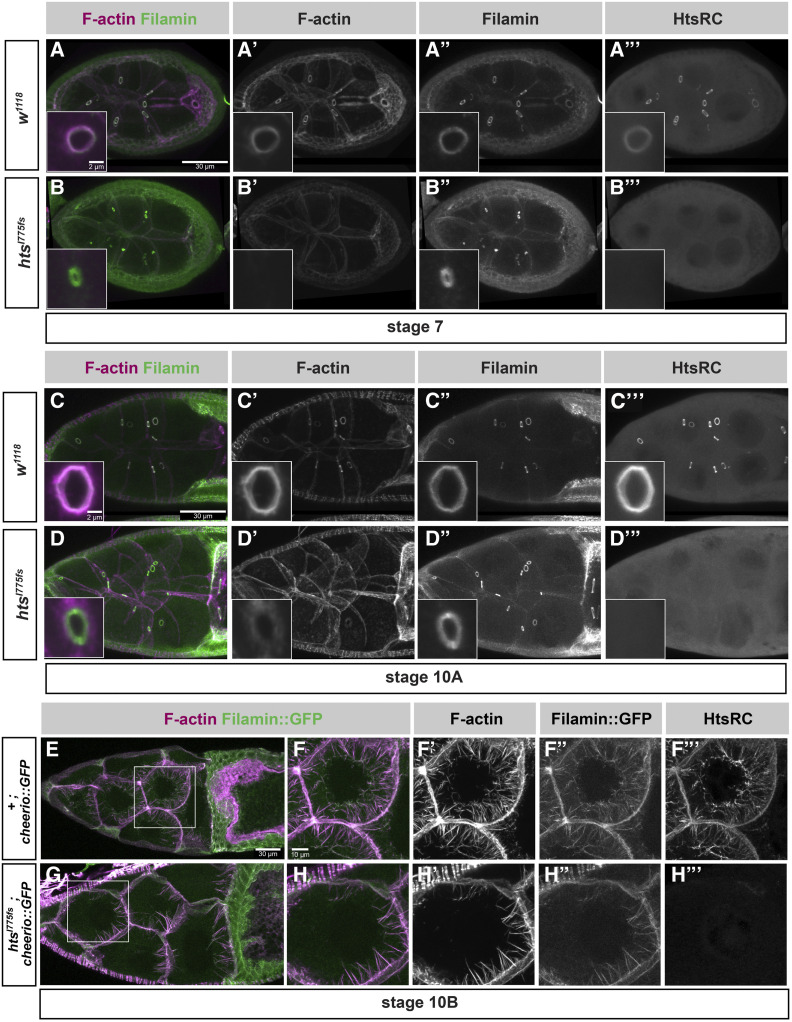
Functional characterization of HtsRC during oocyte development was complicated by the important role HtsF and other hts isoforms (Figure 1A) play in mitotic divisions and in oocyte specification (Yue and Spradling 1992). Previously isolated mutations in hts affected exons common to all hts isoforms (Figure 1A, magenta exons), and resulted in loss of the fusome in females, egg chamber arrest, and female sterility (Lin et al. 1994; Wilson 1999; Petrella et al. 2007). To better characterize the functions specific to HtsRC, we used CRISPR-Cas9 gene editing to generate several independent frameshift mutations in the HtsRC-encoding exon (Hudson et al. 2019) (Figure 1D). We have designated mutations in the HtsRC-encoding exon “htsRC-specific mutations” to differentiate them from previously isolated hts alleles. Flies homozygous for any of our htsRC-specific mutations lack detectable HtsRC protein (Figure 2, B” and D”), but have intact fusomes, indicating that Ovhts is processed and cleaved normally, and only HtsRC is impacted by the mutations (Hudson et al. 2019).
Figure 2.
htsRC-specific mutants lose F-actin at ring canals but not at cytoplasmic actin bundles. Maximum intensity projections of wild-type (w1118) and htsRC-specific mutant (htsI775 fs) egg chambers at (A and B) stage 7, (C and D) stage 10A, and (E–H) stage 10B. (A–D) Egg chambers are stained with TRITC-Phalloidin and labeled with Filamin and HtsRC antibodies. The left column shows a merge of F-actin (magenta) and Filamin (green) channels, where white indicates overlap between channels. Insets contain a representative nurse cell ring canal from the same genotype and stage, although not the same egg chamber. Bar, 30 µm, inset Bar, 2 µm. (E–H) Egg chambers from wild-type and htsRC-specific mutant flies expressing Filamin::GFP. Size differences between these egg chambers represent the normal variation among stage 10B egg chambers. (E and G) Egg chambers are labeled with TRITC-Phalloidin (magenta). Bar, 30 µm. (F and H) Nurse cells marked by white boxes in (E) and (G) are magnified to show labeling of the cytoplasmic actin bundles. Bar, 10 µm.
F-actin is present in several subcellular locations within the germline cells of egg chambers: ring canals; cell cortexes, including enrichment at microvilli-like structures surrounding ring canals (Tilney et al. 1996; Loyer et al. 2015); and robust cytoplasmic bundles that form specifically during stage 10B (Huelsmann et al. 2013; Spracklen et al. 2014). Electron microscopy of ring canals revealed they have an inner rim of highly crosslinked mixed-polarity bundles of F-actin facing the lumen and an electron dense outer rim at the plasma membrane (Cooley and Theurkauf 1994; Tilney et al. 1996). Initial analysis of hts mutants by electron microscopy revealed that the F-actin rich inner rim was absent; ring canal components that remained in hts mutants detected by immunofluorescence were therefore defined as components of the outer rim (Yue and Spradling 1992; Cooley and Theurkauf 1994; Sokol and Cooley 1999). These outer rim proteins include phosphotyrosine-containing protein(s), a Mucin-like glycoprotein, and Filamin (Sokol and Cooley 1999). Filamin is also a component of the inner rim based on its colocalization with HtsRC and F-actin in kelch mutants, in which inner rim components accumulate in the ring canal lumen (Tilney et al. 1996; Sokol and Cooley 1999).
In htsRC-specific mutants, the ring canal inner rim F-actin was completely absent in all stages of egg chamber development (Figure 2, A–D), while cortical F-actin surrounding ring canals remained intact (Figure 2, D and D’). Filamin was present at the plasma membrane in ring canals lacking F-actin (Figure 2, B–B” and D–D”) and did not colocalize with cortical F-actin in mutant ring canals (compare insets in Figure 2, C and D).
Intriguingly, the impact of htsRC-specific mutations on F-actin was less severe in the four ring canals connecting the nurse cells directly to the oocyte. In stages 7–10, oocyte ring canals contained some F-actin, while nurse cell ring canals did not (Figure S1). The oocyte has more cortical F-actin than the nurse cells, including at the interface between the oocyte and the nurse cells where the ring canals are located (Gates et al. 2009). The absence of HtsRC did not cause any apparent change in cortical F-actin in either cell type. Although HtsRC is required to establish an F-actin rich rim at nurse cell ring canals, the accumulation of F-actin in oocyte ring canals in the context of the robust oocyte cortical F-actin may use a modified mechanism.
HtsRC is also present on cytoplasmic F-actin bundles that form in stage 10B nurse cells (Huelsmann et al. 2013); however, it was not known whether HtsRC is required for bundle formation. The F-actin bundles are monopolar, with barbed, elongating ends at the plasma membrane and pointed ends that reach the nuclear envelope as the bundles lengthen. The bundles contain at least three F-actin crosslinking proteins: Fascin, encoded by the singed gene (Cant et al. 1994; Cant and Cooley 1996); Villin, encoded by the quail gene (Mahajan-Miklos and Cooley 1994); and Filamin, encoded by the cheerio gene (Sokol and Cooley 1999). Singed and Quail localize along the length of the cytoplasmic bundles and are important for normal bundle formation. HtsRC is also detected along the length of the cytoplasmic actin bundles, although it is most highly concentrated at the pointed ends where they meet the nuclear envelope (Huelsmann et al. 2013). Flies with strong mutations in singed or quail are sterile due to a failure to form F-actin bundles, resulting in ring canals plugged by the large nurse cell nuclei during nurse cell dumping in stage 10B and 11 egg chambers (Cant et al. 1994; Mahajan-Miklos and Cooley 1994). In contrast, stage 10B egg chambers in htsRC-specific mutants produced cytoplasmic F-actin bundles containing Filamin::GFP (Figure 2, E–H) and nurse cell nuclei did not interfere with ring canals. This result suggests that HtsRC is not essential for cytoplasmic F-actin bundle formation late in oogenesis.
We also tested whether HtsRC localization at cytoplasmic F-actin bundles depends on Filamin. In three independent cheerio mutants, HtsRC protein localized to the bundles as in the wild type (Figure S2) indicating that HtsRC can localize to bundles independently of Filamin. This is different from ring canals where HtsRC localization is dependent on Filamin (Robinson et al. 1997).
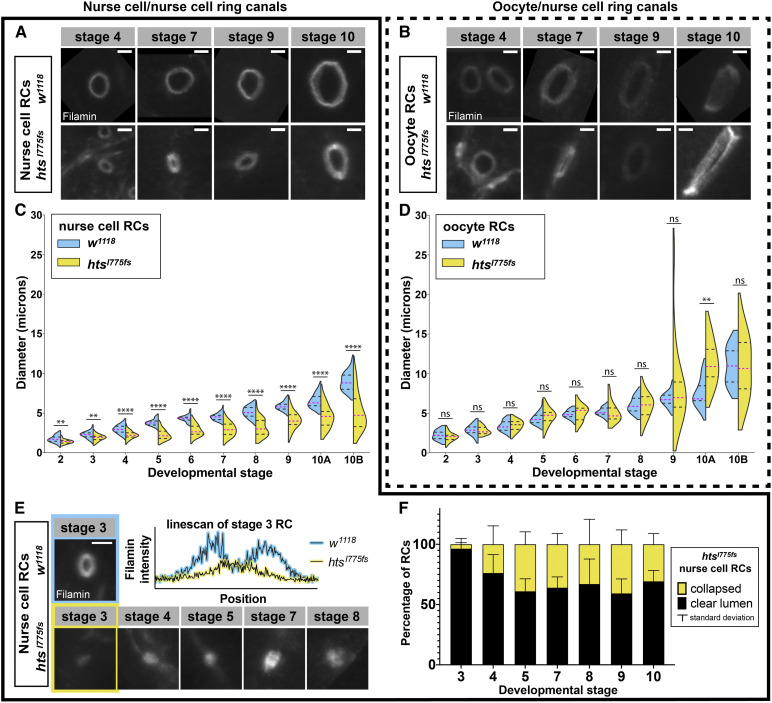
Ring canals lacking F-actin are small and unstable
Ring canal size is known to depend on the recruitment and elongation of F-actin. Previous work characterizing the role of the Arp2/3 complex in ring canal expansion showed that ring canal diameter decreased by 30–50% in ArpC1Q25st and Arp3EP(3)3640 mutant clones, where truncation of the ArpC1 subunit or absence of the Arp3 subunit rendered the Arp2/3 complex inactive (Hudson and Cooley 2002). Ring canals in ArpC1Q25st and Arp3EP(3)3640 mutant cells still contained F-actin but had a significant reduction in size beginning at stage 5, the point at which rapid ring canal expansion begins (Tilney et al. 1996; Hudson and Cooley 2002). As our htsRC-specific mutants failed to recruit F-actin at any stage, we expected to see at least as severe an effect on ring canal size.
Filamin persists on ring canal membranes in HtsRC mutants, allowing us to measure ring canal diameter. In htsRC-specific mutants, ring canal diameter was smaller compared to wild type, but only for ring canals connecting two nurse cells (Figure 3, A–D). The difference in nurse cell ring canal diameter was easily observable starting at stages 4 and 5, but was statistically significant even at stage 2 (Figure 3C). At stage 2, the htsRC-specific mutant ovaries had nurse cell ring canal diameters that were 18% smaller than in wild-type ovaries on average (1.4 vs. 1.7 µm); after stage 5, the diameter of these ring canals was reduced by 30–40% compared to wild type. By stage 10B, the average nurse cell ring canal in htsRC-specific mutants was 5.2 µm in diameter, compared to 9.0 µm in wild-type controls. Unlike nurse cell to nurse cell ring canals, the four ring canals connecting nurse cells to the oocyte did not show a significant change in diameter (Figure 3D). The difference we measured in stage 10A (Figure 3D) may have been due to the wide range of ring canal sizes during stage 10. The difference between nurse cell and oocyte ring canal phenotypes was consistent with the results reported for ArpC1Q25sd (Hudson and Cooley 2002), and may be related to the residual F-actin we saw specifically at oocyte ring canals (Figure S1). Although there was not a significant decrease in size for oocyte ring canals, they did exhibit a statistically significant change in variance, reflecting the presence of both extremely large and extremely small ring canals in that data set (Figure 3D). This increase in variance suggests that the normal mechanisms regulating oocyte ring canal expansion were disrupted in htsRC-specific mutants. Our results for htsRC mutants, combined with previous results for the Arp2/3 complex, strongly suggest that the ring canal F-actin cytoskeleton is required for ring canal expansion. HtsRC functions to recruit F-actin and Arp2/3 promotes F-actin nucleation and an overall increase in total filament length.
Figure 3.
Nurse cell ring canals are smaller and less stable in HtsRC mutants. Representative ring canals and quantification of ring canal diameter for ring canals connecting pairs of nurse cells (A, C, and E) or connecting nurse cells to the oocyte (B and D). (A and B) Representative ring canals labeled with Filamin antibody from w1118 (top) and htsI775 fs (bottom) ovaries. (C and D) Violin plots showing ring canal diameter across stages 2–10B of oogenesis in w1118 (blue) vs. htsI775 (yellow). Two or more egg chambers were sampled at each stage; Table S1 contains additional information about the means and sample numbers for the data displayed in this figure panel. Significance was determined by Welch’s t-test. Significance thresholds: **P < 0.01; ****P < 0.0001; ns, not significant. (E) Representative examples of collapsed nurse cell ring canals in htsRC-specific mutants (bottom) compared with a wild-type w1118 ring canal (top) with a clear lumen. Graph shows pixel intensity vs. position along a 3 μm horizontal linescan through the center of the stage three ring canals highlighted. (F) Number of collapsed nurse cell ring canals (no visible lumen; yellow) compared to nurse cell ring canals with a clear open lumen (black). Each egg chamber contains 11 nurse cell ring canals. n = 3 egg chambers per stage. Bars, 2 µm.
In addition to the effects on ring canal diameter, a subset of ring canals in htsRC-specific mutants had no visible lumen and appeared collapsed into foci that formed a single peak on a line scan of pixel intensity (Figure 3E). Collapsed ring canals were present starting at stage 3–4; some stage 3 egg chambers did not contain a single collapsed ring canal, while every stage 4 or older egg chamber contained at least one. This suggests that ring canals formed but grew slowly and sometimes collapsed, rather than failing to expand at all. No collapsed oocyte ring canals were observed, consistent with the more stable nature of oocyte ring canals.
Defective cytoplasm transfer causes fecundity defects in htsRC-specific mutants
Previously studied mutations in hts, as well as mutations in ring canal components encoded by kelch and cheerio, resulted in completely sterile female flies. In kelch and cheerio ovaries, egg chambers containing undersized oocytes progressed to stage 11 when the final rapid transfer of nurse cell cytoplasm to the oocyte was hampered by mutant ring canals, resulting in “dumpless” egg chambers containing residual nurse cells that produced small, infertile eggs (Xue and Cooley 1993; Robinson et al. 1994). The phenotype of hts null mutant egg chambers was more severe, with arrested development and degeneration of early egg chambers (Yue and Spradling 1992).
Surprisingly, htsRC-specific mutants were fertile; however, egg chamber development was not normal and hatching of eggs laid by mutant females was markedly reduced. Approximately one-quarter of oocytes were the same size and shape as in wild-type ovaries, while the remaining three-quarters of egg chambers showed a variably penetrant dumpless phenotype that we quantified by measuring egg length for two independent htsRC-specific mutants (Figure 4A). Eggs laid by htsRC mutant females ranged in size from full-sized (>0.5 mm) to less than half the length of wild-type eggs (<0.25 mm; Figure 4, A–C). Small eggs could be attributed to a failure in cytoplasmic transfer—ovaries contained dumpless egg chambers with small oocytes that had growing dorsal appendages (Figure 4B). Both egg laying and hatching rates were significantly decreased in htsRC-specific mutants (4D–F). Although most eggs in the control hatched (86%), eggs laid by two independent htsRC-specific mutants hatched <30% of the time and small eggs (produced from dumpless egg chambers) never hatched (Figure 4F). In order to avoid possible interference from secondary site mutations, the experiments shown in Figure 4, D–F were conducted using both controls and htsRC-specific mutants crossed to a deficiency that deletes the hts gene. However, a significant decrease in fecundity was also observed for htsI775fs and htsR913fs homozygotes, leading to the conclusion that HtsRC is required for normal levels of fecundity.
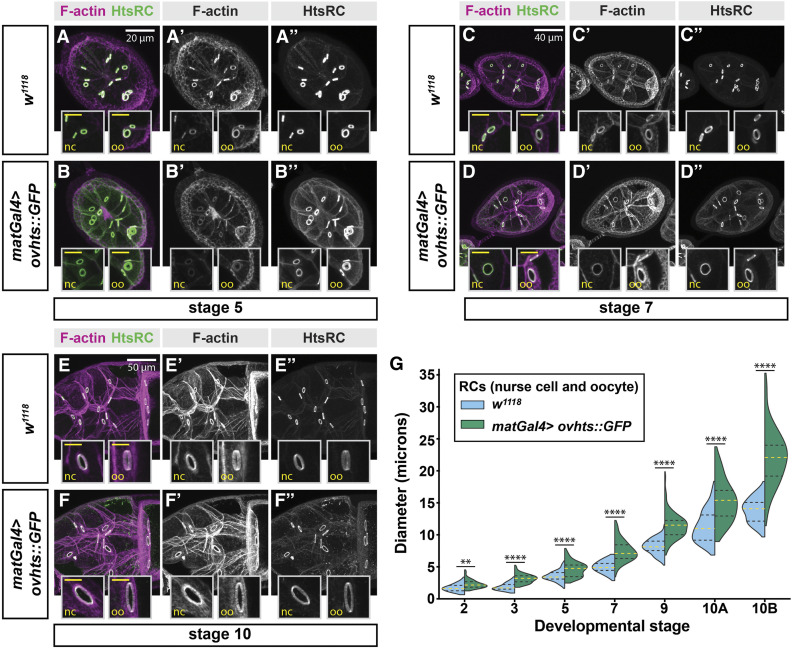
Overexpression of HtsRC in the germline drives increased RC diameter
As loss of HtsRC resulted in smaller ring canals, we sought to further investigate the role of HtsRC in the regulation of ring canal expansion by examining the effects of HtsRC overexpression. To achieve overexpression, matGAL4 was used to drive UAS-ovhts::GFP in stage 2 and older egg chambers. Ovhts polyprotein produced by this transgene undergoes complete cleavage to produce functional HtsRC protein that is detectable with HtsRC antibody or by GFP fluorescence (Petrella et al. 2007). In control egg chambers (w1118), ring canal expansion occurred gradually until reaching peak size at stage 10B (Figure 5, A, C, and E). In contrast, overexpression of HtsRC resulted in larger ring canals at all stages of oogenesis examined (Figure 5, B, D, and F). The increase in ring canal diameter was observed in both nurse cell and oocyte ring canals (insets, Figure 5, A–F). The differences in ring canal sizes were statistically significant throughout oogenesis, and became more apparent in late-stage egg chambers (Figure 5G).
Figure 5.
Overexpression of ovhts results in larger ring canals. (A–F) Fluorescence micrographs of F-actin staining and HtsRC antibody labeling in egg chambers throughout oogenesis show that maternal alpha tubulin-GAL4 (matGal4)-driven UASH-ovhts::GFP leads to the formation of large ring canals, compared to w1118 control. Insets contain higher-resolution micrographs of nurse cell and oocyte ring canals from the same egg chamber. Inset scale bars are 10 μm. (G) Quantification of ring canal diameter throughout development, visualized by violin plots. Refer to Table S1 for information about the means and sample numbers for the data in this figure panel. Yellow dotted lines indicate median ring canal diameter, and black dotted lines represent the upper and lower quartiles. Welch’s t-test was performed between w1118 control and matGal4>ovhts::GFP ring canal diameters at each stage; Significance thresholds: **P < 0.01; ****P < 0.0001.
In addition to larger ring canal diameter, we also observed changes in ring canal thickness in the presence of increased HtsRC protein levels. In stage 4 and 5 egg chambers with HtsRC::GFP overexpression, the F-actin ring was thicker, similar to the phenotype we observed at stage 6 in kelch null mutations or when the substrate binding domain of Kelch was overexpressed (Figure 5, A’’ and B’’) (Hudson and Cooley 2010). This phenotype was especially strong in ring canals connecting the oocyte to the nurse cells (Figure 5, A’’ and B’’). Conversely, overexpression of HtsRC::GFP at stage 10 caused the thickness of the F-actin and the tube length of the ring canal to decrease, resulting in ring canal rims that appeared thinner (Figure 5, E and F). This decrease in ring canal rim thickness was accompanied by a patchier, more irregular, appearance of the F-actin. Together, these results indicate that HtsRC is important for driving ring canal expansion, likely by regulating F-actin filaments. However, it was unclear from these results if the role of HtsRC in driving ring canal expansion is through the recruitment of additional actin filaments, or through the modulation of existing filaments at the ring canal.
HtsRC ectopically expressed in somatic follicle cells drives the formation of F-actin aggregates containing known ring canal proteins
Genetic analysis provided strong evidence that HtsRC functions in F-actin regulation. Efforts to investigate the biochemical activity of HtsRC using standard biochemical assays of F-actin polymerization and bundling were unproductive. Numerous attempts at expression and purification of HtsRC were unsuccessful, most likely because of the intrinsically disordered nature of the HtsRC protein. Seeking an alternative approach, we developed an ectopic expression system in which to further test HtsRC function.
We expressed HtsRC ectopically in the somatic epithelial cells of egg chambers, which are polarized with the apical side in contact with germline cells (Figure 6, A–C) (Horne-Badovinac and Bilder 2005). The cortex of follicle cells is F-actin-rich (Figure 6, B’ and C’). Follicle cells develop in syncytia of variable cell number in which cells remain connected by ring canals about 1 µm in diameter located near the apical end (McLean and Cooley 2013). HtsRC is not normally expressed in these cells (Figure 6, B” and C”) and their ring canals do not contain Filamin or Kelch.
When ovhts cDNA transgenes were expressed in the somatic follicle cells using the tubulin-Gal4 driver, prominent aggregates containing HtsRC and F-actin were visible (Figure 6, D–F). Aggregates were first visible at stages 8 and 9 of egg chamber development, and persisted through the formation of the eggshell at stage 14. The aggregates always formed near the cortex of these cells, with most of them near basal or apical surfaces (Figure 6F). Smaller aggregates were also present near the lateral cortex. Each cell contained at least one basal aggregate, but the number and location of the other aggregates were variable. We tested the ability of several HtsRC-expressing transgenes to cause the formation of aggregates, including transgene constructs containing different untranslated regions and tags, and either full length ovhts cDNA (Figure S3) or just the HtsRC-encoding exon (data not shown). The location and frequency of aggregate formation remained consistent, suggesting their formation was driven by HtsRC, and not by aggregation of GFP or APEX protein tags (Figure S3).
HtsRC did not colocalize with follicle cell ring canals as we initially predicted; however, HtsRC/F-actin aggregates did contain several components found in germline ring canals, including Kelch, Filamin, and Arp3, suggesting they could be used as an alternative system to study functional interplay between HtsRC and other ring canal proteins. We have shown that Kelch regulates HtsRC levels by targeting it for destruction by the proteasome in the germline (Hudson et al. 2019). To test whether HtsRC was similarly regulated by Kelch in follicle cells, we coexpressed Ovhts::GFP with either mCherry as a control or mCherry::Kelch (Figure 6, G and H). In the presence of additional Kelch, the number of aggregates and overall aggregate area were reduced (Figure 6I). HtsRC levels were also reduced (Figure 6J). These data suggest that HtsRC in ectopic aggregates undergoes Kelch-dependent turnover, as it does in germline ring canals.
To further test the impact of Kelch on HtsRC, we coexpressed an overactive form of Kelch lacking its N-terminal regulatory sequence, Kelch∆NTR (Figure S3). In this background, HtsRC/F-actin aggregates were almost entirely absent, consistent with our previous results in germline cells (Hudson et al. 2019). These data indicate that ectopically expressed HtsRC can be regulated in a manner similar to its regulation in the female germline.
The ability of HtsRC to cause the assembly of F-actin structures in germline ring canals and somatic follicle cells likely involves other proteins. The formin Diaphanous has been implicated in ring canal expansion but not specifically in regulating ring canal actin accumulation (Thestrup et al. 2020), and RNAi knockdown of each of the six formins did not reveal any defects in actin accumulation (data not shown). Enabled/VASP is known to be important for the formation of nurse cell cytoplasmic actin bundles, but it is dispensable for the formation of ring canals (Gates et al. 2009). In light of this information, we focused on investigating the role of the Arp2/3 complex and Filamin in actin aggregate formation.
We first tested whether the Arp2/3 complex is needed for HtsRC-induced F-actin in follicle cells. In germline clones homozygous for ArpC1Q25st or Arp3EP(3)3640, null mutations in components of the Arp2/3 complex, HtsRC and F-actin were still present on ring canals (Hudson and Cooley 2002). These data suggested that HtsRC may act upstream, or independently, of the Arp2/3 complex to recruit or regulate F-actin in the context of a ring canal. Similarly, large mitotic clones of ArpC1Q29sd or ArpC4SH1036 mutant follicle cells expressing the HtsRC::APEX transgene contained F-actin aggregates with the same size and localization pattern as neighboring heterozygous cells (Figure S4, A–C). To confirm this observation, we quantified the HtsRC and F-actin densities within the clones (Figure S4, D–E) as well as the total aggregate volume (Figure S4F) and total HtsRC intensity (Figure S4G) within uniform regions of epithelium. Aggregates within mutant clones did not have a significant decrease in F-actin density compared to control clones (Figure S4E), suggesting F-actin recruitment was not impacted by the absence of functional Arp2/3 complex. These results suggest that HtsRC generates F-actin rich structures independently of the Arp2/3 complex in both germline and follicle cells.
Filamin is important for HtsRC-mediated F-actin aggregate formation
Filamin is required for recruitment of HtsRC to ring canals (Robinson et al. 1997) and we recently identified Filamin as a nearest neighbor and likely interaction partner for HtsRC (Mannix et al. 2019). However, it is not possible to test the function of HtsRC independently of Filamin at the ring canal, as ring canals do not form or recruit HtsRC in cheerio mutants (Robinson et al. 1997). By leveraging the HtsRC/F-actin aggregates in somatic follicle cells, we were able to separate HtsRC-dependent recruitment of F-actin from ring canal formation.
To test whether HtsRC F-actin aggregation activity in follicle cells depends on Filamin, we examined the formation of F-actin aggregates in cheerio mutant somatic follicle cells overexpressing HtsRC from the tubulin-ovhts-htsN4∆100 transgene, which was previously shown to produce aggregates (Pokrywka et al. 2014) (Figure S3). In control follicle cells, HtsRC-positive F-actin aggregates were readily observed at the basal surface (Figure 7, A–A”) as well as at the apical and lateral membranes (Figure 7, B–B”). In comparison, F-actin aggregates were largely disrupted in the absence of Filamin, as shown with two independent cheerio alleles (Figure 7, C–F”). To quantify the phenotype, we measured the intensity of HtsRC antibody labeling and phalloidin staining within each aggregate in a defined region as well as aggregate volume and derived a density value; the average density of all aggregates was calculated for each region (see Materials and Methods). We observed a decrease in the density of both HtsRC (Figure 7G) and F-actin (Figure 7H) in the absence of Filamin. The loss of F-actin was more severe than the loss of HtsRC as seen in representative images (Figure 7, A’–F’) and quantification (Figure 7H). Furthermore, there was less F-actin per unit of HtsRC in the aggregates (Figure 7I), suggesting the ability of HtsRC to recruit F-actin was diminished in the absence of Filamin.
The total volume of aggregates was also markedly reduced in cheerio mutant follicle cells (Figure 7J), with the greatest effect on the apical population of aggregates, which were nearly absent (Figure 7K). We also plotted location of the aggregates as a function of relative z-position to show the distribution in control vs. cheerio mutants (Figure S5). A bimodal distribution of aggregates can be seen in controls, but this distribution pattern is lost in cheerio mutants, with the remaining aggregates located toward basal surface (summarized in Figure 7L). Like the oocyte cortex in the germline, the basal cortex of the follicle cells has a greater enrichment of F-actin than the other surfaces in this cell type (Gutzeit 1990), which may contribute to the persistence of basal aggregates in the absence of Filamin.
Discussion
In this work, we have determined that HtsRC is essential for the presence of F-actin at ring canals. When HtsRC-dependent accumulation of F-actin was disrupted, ring canals did not fully expand. Although some ring canals in htsRC-specific mutants were unstable and collapsed, the lumens of most ring canals remained clear. Unlike most other mutations impacting ring canals, the loss of HtsRC did not render females sterile. Instead, cytoplasm transfer to oocytes was compromised and fecundity was reduced. We also found evidence that HtsRC is a potent driver of F-actin recruitment when expressed ectopically, and that Filamin function is linked to the ability of HtsRC to accumulate F-actin. In the germline cells of the egg chamber, Filamin is required for HtsRC localization to ring canals (Robinson et al. 1997). In somatic follicle cells, the lack of Filamin impaired the ability of HtsRC to accumulate F-actin in ectopic structures. We conclude that HtsRC is a potent actin regulator and the primary driver of ring canal F-actin accumulation in egg chamber germline cells, and that the recruitment of F-actin is important, but not essential, for ring canal stability and fertility.
HtsRC functions with Filamin to promote ring canal F-actin accumulation
The localization pattern of HtsRC points to a role for HtsRC in stabilizing F-actin rather than nucleating actin polymerization. HtsRC colocalizes ubiquitously with the mixed-polarity actin bundles at ring canals, and is also enriched near pointed ends of filaments in two contexts: the cytoplasmic bundles in nurse cells (Figure 2) (Huelsmann et al. 2013; Spracklen et al. 2014) and ectopic F-actin filaments formed at the oocyte cortex during later stages of oogenesis (Pokrywka et al. 2014). This localization pattern is similar to what would be expected of Cofilin, which interacts preferentially with filaments containing ADP-actin at pointed ends (Carlier et al. 1997) and at high concentrations, coats F-actin filaments and stabilizes them (Andrianantoandro and Pollard 2006; Ngo et al. 2015); HtsRC could function similarly. The actin nucleation factors Arp2/3 and Spire are retained at the ends of the filaments following nucleation (Mullins et al. 1998; Quinlan et al. 2005) while HtsRC coats the sides of filaments. Furthermore, HtsRC is not present at the barbed ends of nurse cell actin bundles (at the plasma membrane), which also argues against a role for HtsRC as a nucleation or elongation factor. In addition, the F-actin aggregates formed by ectopic expression of HtsRC in follicle cells formed only at the F-actin-rich cortex, most prominently the basal surfaces, suggesting it requires existing filaments to initiate aggregate formation.
We propose that HtsRC functions as a scaffold for the ring canal F-actin cytoskeleton. Fluorescence recovery after photobleaching (FRAP) experiments showed that GFP-tagged F-actin recovers rapidly with a t1/2 of 65 sec (Kelso et al. 2002) while GFP-tagged HtsRC does not recover in 30 min (Lisa Petrella and Lynn Cooley, unpublished data), suggesting HtsRC could be part of a stable matrix for collecting actin filaments. We also know that the levels of HtsRC are regulated by ubiquitin-mediated proteasome turnover orchestrated by CRL3Kelch (Hudson et al. 2019). Since most of the HtsRC at the ring canal is stable to turnover, CRL3Kelch is likely acting on a subset of HtsRC in order to keep the lumen clear of F-actin as the ring canal grows. CRL3Kelch also controls HtsRC expressed in follicle cells (Figure 6), again indicating that HtsRC’s potent F-actin accumulation activity is restricted by CRL3Kelch-mediated turnover. Overall, our results point to recruitment or stabilization of existing filaments as the function of HtsRC.
We have uncovered functional and physical links between HtsRC and Filamin. Proximity labeling experiments support the conclusion that HtsRC and Filamin are direct binding partners (Mannix et al. 2019) and Filamin is required for HtsRC localization to ring canals. The simplest model is that Filamin directly recruits HtsRC, and HtsRC recruits F-actin. Filamin’s numerous documented interactions (Nakamura et al. 2011; Razinia et al. 2012) include membrane-associated proteins and F-actin, making it an attractive candidate for a role in anchoring the ring canal cytoskeleton to the ring canal plasma membrane. The behavior of HtsRC when expressed ectopically in follicle cells suggests that the presence of Filamin may also enhance HtsRC activity in promoting F-actin structures. In follicle cells, HtsRC-dependent actin structures are less robust without Filamin, with smaller aggregates containing less HtsRC and less F-actin per unit of HtsRC. Filamin may remodel ring canal F-actin recruited by HtsRC into condensed and ordered bundles. Alternatively, HtsRC itself may depend on an interaction with Filamin to stimulate its activity, possibly by changing the structure of HtsRC protein to a more active conformation or by sequestering HtsRC away from CRL3Kelch ubiquitin ligase activity.
We were unable to conduct standard biochemical assays to directly test how HtsRC interacts with actin owing to the insoluble nature of purified HtsRC. Computational methods for predicting protein structure did not identify any likely actin-binding motifs, but instead indicated intrinsic disorder (Figure 1F). Even so, it is still possible that HtsRC contains an F-actin-binding motif. Historically, protein–protein interactions were thought to be modulated through large, globular structured domains. Recent insights from the study of peptide motifs and post-translational modifications reveal that short motifs, often <10 amino acids in length, actually confer the majority of specificity to protein interactions (Davey et al. 2012). These motifs are frequently within regions of disorder and contain only a handful of conserved, essential residues, allowing for a great deal of sequence variability, and, therefore, evolutionary plasticity (Tompa et al. 2014). The well-known actin binding motif, the WH2 domain, follows this trend. WH2 domains are composed of 17–20 amino acids located within regions of disorder that fold into helical structures upon interaction with actin monomers (Panchal et al. 2003; Carlier et al. 2011; Dominguez 2016). Although we do not have evidence to suggest that HtsRC contains a WH2 domain or any other previously characterized actin binding motif, it is possible that one or more short peptide motifs are present within the disordered regions and underlie HtsRC function.
Intrinsically disordered proteins and protein motifs can become structured through interaction with a ligand, as is the case for myelin basic protein (MBP) (Majava et al. 2010) and the actin-monomer binding protein Tarp (Tolchard et al. 2018). One avenue for future research is to determine if HtsRC becomes ordered in the presence of Filamin.
Ring canal expansion and stability
This study adds to a body of work documenting the importance of cytoskeletal regulation for ring canal expansion and stability. We found that ring canal diameter is sensitive to the level of HtsRC—in its absence, ring canals are too small and when over-expressed, ring canals are wider than normal. Ring canal expansion also involves unidentified phospho-tyrosine species; flies mutant for either Src64B or Btk29A, nonreceptor tyrosine kinases known to localize to ring canals, have defects in ring canal expansion despite normal recruitment and localization of Filamin, HtsRC, F-actin, and Kelch (Dodson et al. 1998; Guarnieri et al. 1998; Roulier et al. 1998). In addition to showing a 30% decrease in ring canal diameter, which is comparable to the impact of both htsRC and Arp2/3 complex mutations (Hudson and Cooley 2002), Src64B mutants also exhibited frequent ring canal collapse (Dodson et al. 1998; O’Reilly et al. 2006).
The opposite phenotype, ring canal overexpansion, has been reported for mutations affecting other kinases. Mutations in Akap200, encoding an anchoring protein for PKA regulatory subunits associated with ring canals (Jackson and Berg 2002) result in increased ring canal expansion and thinner HtsRC rims. In contrast, overexpression of Akap200 resulted in smaller, thicker ring canals, and both conditions caused ring canal collapse. Intriguingly, Akap200 and Src64B may function in the same pathway, as a single mutant Akap200 allele rescues ring canal size defects Src64B mutants (Jackson and Berg 2002). Knockdown of the Ste20 family kinase Misshapen also promotes increases in ring canal diameter and frequent ring canal collapse (Kline et al. 2018) and Misshapen overexpression resulted in decreased ring canal diameter, similar to the Akap200 overexpression phenotype (Jackson and Berg 2002; Kline et al. 2018).
The sensitivity of the ring canal expansion process to the levels of several kinases suggests an exquisite balance between opposing activities must be maintained for wild-type ring canal growth. One possible mechanism is that kinases like PKA (regulated by Akap200), Misshapen, Src64, and Tec29 impact ring canal expansion by regulating the phosphorylation and activity of HtsRC or other cytoskeletal proteins in ring canals. HtsRC contains one or more phosphorylated residues (L. Petrella and L. Cooley, unpublished results) that may be important for regulating HtsRC activity at ring canals.
Ring canal expansion may also require coordination between the ring canal cytoskeleton and the cortical actin network at the nurse cell and oocyte plasma membranes. Ring canal Filamin could play a role in sensing the tension of the membrane surrounding ring canals. When the mechanosensory region of Filamin is deleted or locked in one conformation, ring canal expansion was impaired (Huelsmann et al. 2016). Interestingly, ring canal expansion appears to be affected by the different cortical cytoskeletal environments of nurse cells and oocytes. Ring canals at the oocyte cortex, which has more cortical F-actin, have a weaker phenotype in htsRC-specific mutants than ring canals connecting nurse cells. Decreased penetrance of ring canal phenotypes at the oocyte cortex have also been reported for mutations affecting ring canal growth (Hudson and Cooley 2002) and ring canal adhesion (Loyer et al. 2015). Thus, the oocyte cortical cytoskeleton appears to be more robust and can stabilize compromised ring canals in mutants.
HtsRC evolution and fecundity
Unlike other ring canal components including Kelch, Filamin, and the Arp2/3 complex, HtsRC is not essential for fertility, is specifically expressed in the female germline and is an evolutionary newcomer to the Drosophila proteome. We have not found evidence of a HtsRC ortholog outside of flies, although it is present in a recognizable form in the genomes of 26 species of Drosophila, as well as in house fly (M. domestica) and in tsetse fly (G. palpalis) genomes (Figure S6). Comparison of HtsRC protein from these 28 species reveals regions of conservation interspersed with short, less conserved, segments; the less conserved regions tended to be longer and more divergent in the two non-Drosophila species. Conserved sequence regions did not show any apparent correlation with regions of either order or disorder suggesting disordered regions are not less likely to be conserved. The exon encoding HtsRC may have been acquired in a fly lineage as recently as 60 MYA, after the divergence of Dipteran flies.
Some insects have large ring canals while others do not (Haglund et al. 2011). Ovarian ring canals range from 2 μm or less in aphids (Pyka-Fosciak and Szklarzewicz 2008) and up to 4 μm in bees (Ramamurty and Engels 1977; Patrício and Cruz-Landim 2006) to 10 μm in D. melanogaster and the butterfly Pieris napi (Mazurkiewicz-Kania et al. 2019), both of which are supported by an actin-rich cytoskeleton. The ring canals in tsetse fly egg chambers also reach 10 μm in diameter and have a prominent cytoskeleton visible in electron micrographs (Huebner et al. 1975), bringing up the intriguing possibility that the presence of a HtsRC ortholog in the genome (Figure S6) is responsible for the development of large ring canals.
Genes specific to reproduction are more likely to be rapidly evolving than the rest of the genome, especially those with testis-specific functions (Haerty et al. 2007). Although HtsRC is expressed only in the female germline, there is a testis-specific natural antisense transcript (CR43430) corresponding to the htsRC exon that may be blocking the expression of HtsRC in developing sperm. The hts locus additionally contains a poorly understood testis-specific splice variant, which also appears to be a recent evolutionary event in flies and is detectable in house flies and tsetse flies. We do not understand the function of either the noncoding RNA or the testis specific hts variant, but the presence of these transcripts hints at the complex evolutionary history of the gene and its regulation in both the male and female germline. Although the germline is permissive for the evolution of new genes or new gene functions, recent CRISPR-mediated mutagenesis of a subset of recently evolved genes determined that very few new genes are essential for fertility, and none of those tested were essential for viability (Kondo et al. 2017).
Given that HtsRC is a newly evolved gene functioning in the germline, it is not surprising to find that it is not essential for either viability or fertility. Yet, we have demonstrated an impact on fecundity—the presence of HtsRC confers an increase in the number of viable offspring a female can produce. This evidence supports the idea that the presence and expansion of a robust F-actin cytoskeleton may enhance the ability of ring canals to promote increased fecundity, and may have been under positive selection.
Acknowledgments
We thank Nancy Pokrywka, David Ish-Horowicz, Mary Baylies, and Tian Xu for stocks used in this study. Stocks obtained from the Bloomington Drosophila Stock Center [National Institutes of Health (NIH) P40OD018537] were also used. We thank Mike Buszczak for sharing plasmids. We thank Tian Xu and Kaelyn Sumigray for providing access to the Leica SP8 confocal microscope. We thank Alexander Scherer and Anthony Koleske for their help with attempts to purify HtsRC for actin biochemistry. J.A.G. and K.M.M. were funded in part by a Gruber Science Fellowship from Yale University and by NIH training grants [T32 GM007223 for K.M.M., T32 GM007499 for J.A.G.]. This work was supported by the NIH R01 GM043301 grant to L.C.
Footnotes
Supplemental material available at figshare: https://doi.org/10.25386/genetics.12902582.
Communicating editor: M. Wolfner
Literature Cited
- Andrianantoandro E., and Pollard T. D., 2006. Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol. Cell 24: 13–23. 10.1016/j.molcel.2006.08.006 [DOI] [PubMed] [Google Scholar]
- Cant K., and Cooley L., 1996. Single amino acid mutations in Drosophila fascin disrupt actin bundling function in vivo. Genetics 143: 249–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cant K., Knowles B. A., Mooseker M. S., and Cooley L., 1994. Drosophila singed, a fascin homolog, is required for actin bundle formation during oogenesis and bristle extension. J. Cell Biol. 125: 369–380. 10.1083/jcb.125.2.369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier M. -F., Husson C., Renault L., and Didry D., 2011. Control of actin assembly by the WH2 domains and their multifunctional tandem repeats in Spire and Cordon-Bleu. Int. Rev. Cell Mol. Biol. 290: 55–85. 10.1016/B978-0-12-386037-8.00005-3 [DOI] [PubMed] [Google Scholar]
- Carlier M.-F., and Pantaloni D., 1997. Control of actin dynamics in cell motility. J. Mol. Bio. 269: 459–467. 10.1006/jmbi.1997.1062 [DOI] [PubMed] [Google Scholar]
- Cooley L., and Theurkauf W. E., 1994. Cytoskeletal functions during Drosophila oogenesis. Science 266: 590–596. 10.1126/science.7939713 [DOI] [PubMed] [Google Scholar]
- Cox R. T., and Spradling A. C., 2003. A Balbiani body and the fusome mediate mitochondrial inheritance during Drosophila oogenesis. Development 130: 1579–1590. 10.1242/dev.00365 [DOI] [PubMed] [Google Scholar]
- Cummings M. R., and King R. C., 1969. The cytology of the vitellogenic stages of oogenesis in Drosophila melanogaster. I. General staging characteristics. J. Morphol. 128: 427–441. 10.1002/jmor.1051280404 [DOI] [Google Scholar]
- Davey N. E., Van Roey K., Weatheritt R. J., Toedt G., Uyar B. et al. , 2012. Attributes of short linear motifs. Mol. Biosyst. 8: 268–281. 10.1039/C1MB05231D [DOI] [PubMed] [Google Scholar]
- Ding D., Parkhurst S. M., and Lipshitz H. D., 1993. Different genetic requirements for anterior RNA localization revealed by the distribution of Adducin-like transcripts during Drosophila oogenesis. Proc. Natl. Acad. Sci. USA 90: 2512–2516. 10.1073/pnas.90.6.2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodson G. S., Guarnieri D. J., and Simon M. A., 1998. Src64 is required for ovarian ring canal morphogenesis during Drosophila oogenesis. Development 125: 2883–2892. [DOI] [PubMed] [Google Scholar]
- Dominguez R., 2016. The WH2 domain and actin nucleation: necessary but insufficient. Trends Biochem. Sci. 41: 478–490. 10.1016/j.tibs.2016.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gates J., Nowotarski S. H., Yin H., Mahaffey J. P., Bridges T. et al. , 2009. Enabled and Capping protein play important roles in shaping cell behavior during Drosophila oogenesis. Dev. Biol. 333: 90–107. 10.1016/j.ydbio.2009.06.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum M. P., Ma L., and Matzuk M. M., 2007. Conversion of midbodies into germ cell intercellular bridges. Dev. Biol. 305: 389–396. 10.1016/j.ydbio.2007.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarnieri D. J., Dodson G. S., and Simon M. A., 1998. SRC64 regulates the localization of a Tec-family kinase required for Drosophila ring canal growth. Mol. Cell 1: 831–840. 10.1016/S1097-2765(00)80082-9 [DOI] [PubMed] [Google Scholar]
- Gutzeit H. O., 1990. The microfilament pattern in the somatic follicle cells of mid-vitellogenic ovarian follicles of Drosophila. Eur. J. Cell Biol. 53: 349–356. [PubMed] [Google Scholar]
- Haerty W., Jagadeeshan S., Kulathinal R. J., Wong A., Ravi Ram K. et al. , 2007. Evolution in the fast lane: rapidly evolving sex-related genes in Drosophila. Genetics 177: 1321–1335. 10.1534/genetics.107.078865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haglund K., Nezis I. P., and Stenmark H., 2011. Structure and functions of stable intercellular bridges formed by incomplete cytokinesis during development. Commun. Integr. Biol. 4: 1–9. 10.4161/cib.13550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson J., Paliwal K., Litfin T., Yang Y., and Zhou Y., 2018. Accurate prediction of protein contact maps by coupling residual two-dimensional bidirectional long short-term memory with convolutional neural networks. Bioinformatics 34: 4039–4045. 10.1093/bioinformatics/bty1006 [DOI] [PubMed] [Google Scholar]
- Hinnant T. D., Merkle J. A., and Ables E. T., 2020. Coordinating proliferation, polarity, and cell fate in the Drosophila female germline. Front. Cell Dev. Biol. 8: 19 10.3389/fcell.2020.00019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horne-Badovinac S., and Bilder D., 2005. Mass transit: epithelial morphogenesis in the Drosophila egg chamber. Dev. Dyn. 232: 559–574. 10.1002/dvdy.20286 [DOI] [PubMed] [Google Scholar]
- Hudson A. M., and Cooley L., 2002. A subset of dynamic actin rearrangements in Drosophila requires the Arp2/3 complex. J. Cell Biol. 156: 677–687. 10.1083/jcb.200109065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson A. M., and Cooley L., 2010. Drosophila Kelch functions with Cullin-3 to organize the ring canal actin cytoskeleton. J. Cell Biol. 188: 29–37. 10.1083/jcb.200909017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson A. M., Mannix K. M., Gerdes J. A., Kottemann M. C., and Cooley L., 2019. Targeted substrate degradation by Kelch controls the actin cytoskeleton during ring canal expansion. Development 146: dev169219 10.1242/dev.169219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huebner E., Tobe S. S., and Davey K. G., 1975. Structural and functional dynamics of oogenesis in Glossina austeni: general features, previtellogenesis and nurse cells. Tissue Cell 7: 297–317. 10.1016/0040-8166(75)90007-5 [DOI] [PubMed] [Google Scholar]
- Huelsmann S., Ylänne J., and Brown N. H., 2013. Filopodia-like actin cables position nuclei in association with perinuclear actin in Drosophila nurse cells. Dev. Cell 26: 604–615. 10.1016/j.devcel.2013.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsmann S., Rintanen N., Sethi R., Brown N. H., and Ylänne J., 2016. Evidence for the mechanosensor function of filamin in tissue development. Sci. Rep. 6: 32798 10.1038/srep32798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S. M., and Berg C. A., 2002. An A-kinase anchoring protein is required for protein kinase A regulatory subunit localization and morphology of actin structures during oogenesis in Drosophila. Development 129: 4423–4433. [DOI] [PubMed] [Google Scholar]
- Jia D., Xu Q., Xie Q., Mio W., and Deng W. M., 2016. Automatic stage identification of Drosophila egg chamber based on DAPI images. Sci. Rep. 6: 18850 10.1038/srep18850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley L. A., Mezulis S., Yates C. M., Wass M. N., and Sternberg M. J., 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10: 845–858. 10.1038/nprot.2015.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelso R. J., Hudson A. M., and Cooley L., 2002. Drosophila Kelch regulates actin organization via Src64-dependent tyrosine phosphorylation. J. Cell Biol. 156: 703–713. 10.1083/jcb.200110063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline A., Curry T., and Lewellyn L., 2018. The Misshapen kinase regulates the size and stability of the germline ring canals in the Drosophila egg chamber. Dev. Biol. 440: 99–112. 10.1016/j.ydbio.2018.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo S., Vedanayagam J., Mohammed J., Eizadshenass S., Kan L. et al. , 2017. New genes often acquire male-specific functions but rarely become essential in Drosophila. Genes Dev. 31: 1841–1846. 10.1101/gad.303131.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlowski L. P., and Bujnicki J. M., 2012. MetaDisorder: a meta-server for the prediction of intrinsic disorder in proteins. BMC Bioinformatics 13: 111 10.1186/1471-2105-13-111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T., and Luo L., 1999. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 22: 451–461. 10.1016/S0896-6273(00)80701-1 [DOI] [PubMed] [Google Scholar]
- Lin H., Yue L., and Spradling A. C., 1994. The Drosophila fusome, a germline-specific organelle, contains membrane skeletal proteins and functions in cyst formation. Development 120: 947–956. [DOI] [PubMed] [Google Scholar]
- Loyer N., Kolotuev I., Pinot M., and Le Borgne R., 2015. Drosophila E-cadherin is required for the maintenance of ring canals anchoring to mechanically withstand tissue growth. Proc. Natl. Acad. Sci. USA 112: 12717–12722 [corrigenda: Proc. Natl. Acad. Sci. USA 115: E9260 (2018)]. 10.1073/pnas.1504455112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K., Jensen L., Lei L., and Yamashita Y. M., 2017. Stay connected: a germ cell strategy. Trends Genet. 33: 971–978. 10.1016/j.tig.2017.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan-Miklos S., and Cooley L., 1994. The villin-like protein encoded by the Drosophila quail gene is required for actin bundle assembly during oogenesis. Cell 78: 291–301. 10.1016/0092-8674(94)90298-4 [DOI] [PubMed] [Google Scholar]
- Majava V., Wang C., Myllykoski M., Kangas S. M., Kang S. U. et al. , 2010. Structural analysis of the complex between calmodulin and full-length myelin basic protein, an intrinsically disordered molecule. Amino Acids 39: 59–71. 10.1007/s00726-009-0364-2 [DOI] [PubMed] [Google Scholar]
- Mannix K. M., Starble R. M., Kaufman R. S., and Cooley L., 2019. Proximity labeling reveals novel interactomes in live Drosophila tissue. Development 146: dev17644 10.1242/dev.176644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazurkiewicz-Kania M., Simiczyjew B., and Jędrzejowska I., 2019. Differentiation of follicular epithelium in polytrophic ovaries of Pieris napi (Lepidoptera: pieridae)-how far to Drosophila model. Protoplasma 256: 1433–1447. 10.1007/s00709-019-01391-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins R. D., Heuser J. A., and Pollard T. D., 1998. The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc. Natl. Acad. Sci. USA 95: 6181–6186. 10.1073/pnas.95.11.6181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura F., Stossel T. P., and Hartwig J. H., 2011. The filamins: organizers of cell structure and function. Cell Adhes. Migr. 5: 160–169. 10.4161/cam.5.2.14401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo K. X., Kodera N., Katayama E., Ando T., and Uyeda T. Q., 2015. Cofilin-induced unidirectional cooperative conformational changes in actin filaments revealed by high-speed atomic force microscopy. eLife 4: e04806 10.7554/eLife.04806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Reilly A. M., Ballew A. C., Miyazawa B., Stocker H., Hafen E. et al. , 2006. Csk differentially regulates Src64 during distinct morphological events in Drosophila germ cells. Development 133: 2627–2638. 10.1242/dev.02423 [DOI] [PubMed] [Google Scholar]
- Panchal S. C., Kaiser D. A., Torres E., Pollard T. D., and Rosen M. K., 2003. A conserved amphipathic helix in WASP/Scar proteins is essential for activation of Arp2/3 complex. Nat. Struct. Biol. 10: 591–598. 10.1038/nsb952 [DOI] [PubMed] [Google Scholar]
- Patrício K., and Cruz-Landim C., 2006. Ultrastructural aspects of the intercellular bridges between female bee germ cells. Braz. J. Biol. 66: 309–315. 10.1590/S1519-69842006000200013 [DOI] [PubMed] [Google Scholar]
- Petrella L. N., Smith-Leiker T., and Cooley L., 2007. The Ovhts polyprotein is cleaved to produce fusome and ring canal proteins required for Drosophila oogenesis. Development 134: 703–712. 10.1242/dev.02766 [DOI] [PubMed] [Google Scholar]
- Pokrywka N. J., Zhang H., and Raley-Susman K., 2014. Distinct roles for hu li tai shao and swallow in cytoskeletal organization during Drosophila oogenesis. Dev. Dyn. 243: 906–916. 10.1002/dvdy.24132 [DOI] [PubMed] [Google Scholar]
- Pyka-Fosciak G., and Szklarzewicz T., 2008. Germ cell cluster formation and ovariole structure in viviparous and oviparous generations of the aphid Stomaphis quercus. Int. J. Dev. Biol. 52: 259–265. 10.1387/ijdb.072338gp [DOI] [PubMed] [Google Scholar]
- Quinlan M. E., Heuser J. E., Kerkhoff E., and Mullins R. D., 2005. Drosophila Spire is an actin nucleation factor. Nature 433: 382–388. 10.1038/nature03241 [DOI] [PubMed] [Google Scholar]
- Ramamurty P. S., and Engels W., 1977. Occurrence of intercellular bridges between follicle epithelial cells in the ovary of Apis mellifica queens. J. Cell Sci. 24: 195–202. [DOI] [PubMed] [Google Scholar]
- Razinia Z., Mäkelä T., Ylänne J., and Calderwood D. A., 2012. Filamins in mechanosensing and signaling. Annu. Rev. Biophys. 41: 227–246. 10.1146/annurev-biophys-050511-102252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson D. N., Cant K., and Cooley L., 1994. Morphogenesis of Drosophila ovarian ring canals. Development 120: 2015–2025. [DOI] [PubMed] [Google Scholar]
- Robinson D. N., and Cooley L., 1996. Stable intercellular bridges in development: the cytoskeleton lining the tunnel. Trends Cell Biol. 6: 474–479. 10.1016/0962-8924(96)84945-2 [DOI] [PubMed] [Google Scholar]
- Robinson D. N., Smith-Leiker T. A., Sokol N. S., Hudson A. M., and Cooley L., 1997. Formation of the Drosophila ovarian ring canal inner rim depends on cheerio. Genetics 145: 1063–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roulier E. M., Panzer S., and Beckendorf S. K., 1998. The Tec29 tyrosine kinase is required during Drosophila embryogenesis and interacts with Src64 in ring canal development. Mol. Cell 1: 819–829. 10.1016/S1097-2765(00)80081-7 [DOI] [PubMed] [Google Scholar]
- Singleton K., and Woodruff R. I, 1994. The osmolarity of adult Drosophila hemolymph and its effect on oocyte-nurse cell electrical polarity. Dev. Bio. 161: 154–167. 10.1006/dbio.1994.1017 [DOI] [PubMed] [Google Scholar]
- Sokol N. S., and Cooley L., 1999. Drosophila filamin encoded by the cheerio locus is a component of ovarian ring canals. Curr. Biol. 9: 1221–1230. 10.1016/S0960-9822(99)80502-8 [DOI] [PubMed] [Google Scholar]
- Spracklen A. J., Kelpsch D. J., Chen X., Spracklen C. N., and Tootle T. L., 2014. Prostaglandins temporally regulate cytoplasmic actin bundle formation during Drosophila oogenesis. Mol. Biol. Cell 25: 397–411. 10.1091/mbc.e13-07-0366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starble R., and Pokrywka N. J., 2018. The retromer subunit Vps26 mediates Notch signaling during Drosophila oogenesis. Mech. Dev. 149: 1–8. 10.1016/j.mod.2017.10.001 [DOI] [PubMed] [Google Scholar]
- Thestrup J., Tipold M., Kindred A., Stark K., Curry T. et al. , 2020. The Arp2/3 complex and the formin, Diaphanous, are both required to regulate the size of germline ring canals in the developing egg chamber. Dev. Biol. 461: 75–85. 10.1016/j.ydbio.2020.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilney L. G., Tilney M. S., and Guild G. M., 1996. Formation of actin filament bundles in the ring canals of developing Drosophila follicles. J. Cell Biol. 133: 61–74. 10.1083/jcb.133.1.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolchard J., Walpole S. J., Miles A. J., Maytum R., Eaglen L. A. et al. , 2018. The intrinsically disordered Tarp protein from chlamydia binds actin with a partially preformed helix. Sci. Rep. 8: 1960 10.1038/s41598-018-20290-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompa P., Davey N. E., Gibson T. J., and Babu M. M., 2014. A million peptide motifs for the molecular biologist. Mol. Cell 55: 161–169. 10.1016/j.molcel.2014.05.032 [DOI] [PubMed] [Google Scholar]
- Whittaker K. L., Ding D., Fisher W. W., and Lipshitz H. D., 1999. Different 3′ untranslated regions target alternatively processed hu-li tai shao (hts) transcripts to distinct cytoplasmic locations during Drosophila oogenesis. J. Cell Sci. 112: 3385–3398. [DOI] [PubMed] [Google Scholar]
- Wilson P. G., 1999. BimC motor protein KLP61F cycles between mitotic spindles and fusomes in Drosophila germ cells. Curr. Biol. 9: 923–926. 10.1016/S0960-9822(99)80400-X [DOI] [PubMed] [Google Scholar]
- Xue F., and Cooley L., 1993. Kelch encodes a component of intercellular bridges in Drosophila egg chambers. Cell 72: 681–693. 10.1016/0092-8674(93)90397-9 [DOI] [PubMed] [Google Scholar]
- Yue L., and Spradling A. C., 1992. hu-li tai shao, a gene required for ring canal formation during Drosophila oogenesis, encodes a homolog of adducin. Genes Dev. 6: 2443–2454. 10.1101/gad.6.12b.2443 [DOI] [PubMed] [Google Scholar]
- Zallen J. A., Cohen Y., Hudson A. M., Cooley L., Wieschaus E. et al. , 2002. SCAR is a primary regulator of Arp2/3-dependent morphological events in Drosophila. J. Cell Biol. 156: 689–701. 10.1083/jcb.200109057 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Drosophila stocks and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Further information and requests for resources and reagents should be directed to and will be fulfilled by the corresponding author (L.C.). Supplemental material is available at figshare: https://doi.org/10.25386/genetics.12902582.