Here, Hu et al. show that AP-2 transcription factors have diverged to take on bidirectional control of sleep in mammals. This is the first instance where a sleep gene is shown to have diversified in evolution .....
Keywords: TFAP2, sleep, behavior, EEG, Mus musculus
Abstract
Sleep is a conserved behavioral state. Invertebrates typically show quiet sleep, whereas in mammals, sleep consists of periods of nonrapid-eye-movement sleep (NREMS) and REM sleep (REMS). We previously found that the transcription factor AP-2 promotes sleep in Caenorhabditis elegans and Drosophila. In mammals, several paralogous AP-2 transcription factors exist. Sleep-controlling genes are often conserved. However, little is known about how sleep genes evolved from controlling simpler types of sleep to govern complex mammalian sleep. Here, we studied the roles of Tfap2a and Tfap2b in sleep control in mice. Consistent with our results from C. elegans and Drosophila, the AP-2 transcription factors Tfap2a and Tfap2b also control sleep in mice. Surprisingly, however, the two AP-2 paralogs play contrary roles in sleep control. Tfap2a reduction of function causes stronger delta and theta power in both baseline and homeostasis analysis, thus indicating increased sleep quality, but did not affect sleep quantity. By contrast, Tfap2b reduction of function decreased NREM sleep time specifically during the dark phase, reduced NREMS and REMS power, and caused a weaker response to sleep deprivation. Consistent with the observed signatures of decreased sleep quality, stress resistance and memory were impaired in Tfap2b mutant animals. Also, the circadian period was slightly shortened. Taken together, AP-2 transcription factors control sleep behavior also in mice, but the role of the AP-2 genes functionally diversified to allow for a bidirectional control of sleep quality. Divergence of AP-2 transcription factors might perhaps have supported the evolution of more complex types of sleep.
SLEEP is a fundamental state that is defined by behavioral criteria that include the absence of voluntary movement, an increased arousal threshold, relaxed body posture, reversibility, and homeostatic regulation (Campbell and Tobler 1984) . By these criteria, sleep has been identified not only in mammals, but also in other vertebrates as well as in invertebrates (Campbell and Tobler 1984; Tobler 1995; Joiner 2016; Bringmann 2018; Keene and Duboue 2018). Sleep in invertebrates is characterized mostly as quiet sleep, with a reduction of neuronal and behavioral activity (Raizen and Zimmerman 2011; Miyazaki et al. 2017). In the more complex brains of mammals, two major stages of sleep have been defined. Rapid eye movement sleep (REMS) is characterized by a relatively active brain and muscle paralysis and is also called active sleep. Non-REM sleep (NREMS) is a type of quiet sleep characterized by a strong reduction of brain and muscle activity (Campbell and Tobler 1984). This suggests that sleep appeared first in evolution as a type of quiet sleep and then diversified into two types of sleep, which are manifested as NREMS and REMS in mammals (Miyazaki et al. 2017).
The molecular biology of sleep has been studied in all major genetic model animals such as mice, zebrafish, fruit flies, and nematodes (Joiner 2016; Miyazaki et al. 2017). Genetic analysis indicates that many genes play evolutionarily conserved roles in sleep control (Roberts and Hudson 2009). Thus, genes can be studied across model organisms to solve underlying molecular mechanisms of sleep regulation. For example, a gain-of-function mutation of salt-inducible kinase 3 (SIK-3) called sleepy increased NREM sleep in mice (Funato et al. 2016). Whereas a loss-of-function mutation of SIK-3 is lethal in mice, deletion of the Caenorhabditis elegans SIK-3 homolog KIN-29 is not lethal, but reduces sleep (Funato et al. 2016). It was shown that SIK-3 impacts sleep in C. elegans by controlling energy metabolism (Grubbs et al. 2019). We previously showed that knockout of the AP-2 transcription factor APTF-1 results in sleep loss in C. elegans (Turek et al. 2013). The AP-2 family of transcription factors is evolutionarily conserved in both invertebrates and vertebrates (Bringmann 2018). The basic and helix-span-helix (HSH) domains are necessary for DNA binding and dimerization functions, and are highly conserved among all TFAP2 orthologs and paralogues. The N-terminal portion of the protein contains the transactivation domain, which has an amino acid sequence that is poorly conserved among the AP-2 proteins (Williams and Tjian 1991).
AP-2 transcription factors are best known to control ontogenetic processes such as the development of face, limbs, and organs (Moser et al. 1997b; Werling and Schorle 2002; Zhao et al. 2003; Lin et al. 2018). We showed that, in C. elegans, APTF-1 is required for the functioning of the sleep-inducing RIS (Ring Interneuron S) neuron. Without APTF-1, crucial sleep-inducing neuropeptides are not expressed in RIS, and, thus, sleep is virtually abolished (Turek et al. 2013). Consistent with a conserved role in sleep control, neuronal knockdown of the sole AP-2 homolog in adult Drosophila almost completely abolishes night sleep but without affecting day sleep (Kucherenko et al. 2016).
In mammals, the AP-2 family consists of five paralogs, AP-2α–AP-2ε, encoded by genes Tfap2a–Tfap2e, respectively (Eckert et al. 2005). Here, we focus on Tfap2a and b, which are expressed prominently in neural crest cells starting around embryonic day 8 (E8) during early development of the central nervous system and are still detectable in adult brains (Chazaud et al. 1996; Moser et al. 1997b; Zhao et al. 2003). In humans, mutations affecting the basic domain in AP-2 can lead to the loss of function of these transcription factors. Heterozygous mutation of Tfap2a causes branchio-oculo-facial syndrome (BOFS; OMIM#113620) by a mechanism that mostly involves the loss of function of transcription factor activity (Li et al. 2013). BOFS is associated with multiple craniofacial abnormalities as well as eye, hearing, and skin defects. To our knowledge, no sleep abnormalities have been reported for BOFS individuals. Loss of Tfap2a causes severe developmental problems of the heart, brain, and skeletal systems leading to lethality (Schorle et al. 1996; Zhang et al. 1996; Brewer et al. 2004). Tfap2a+/− mice are viable and fertile. Heterozygous deletion is associated with mild developmental defects in craniofacial and brain development, providing a mouse model to study BOFS (Schorle et al. 1996; Zhang et al. 1996; Kohlbecker et al. 2002; Green et al. 2015). To our knowledge, Tfap2a+/− mice have not yet been tested for sleep abnormalities.
Heterozygous mutation of Tfap2b causes Char syndrome (CHAR, OMIM#169100) by either dominant negative or haploinsufficiency mechanisms (Satoda et al. 2000; Zhao et al. 2001; Mani et al. 2005). CHAR is characterized by developmental defects that include facial dysmorphism, abnormalities of the fifth finger, and failure of ductus arteriosus closure (patent ductus arteriosus, PDA) (Satoda et al. 1999). In two families with heterozygous loss of Tfap2b function, CHAR individuals showed self-reported sleep abnormalities. In the first family, sleepwalking was reported, whereas in the second family individuals reported shortened nocturnal sleep. However, the nature of these sleep changes remains unclear as these phenotypes were not confirmed using polysomnography (Mani et al. 2005). Homozygous deletion of Tfap2b in mice causes early lethality. By contrast, heterozygous deletion in mice causes PDA and fifth finger digit abnormalities but no obvious facial anomalies, providing a model of CHAR (Moser et al. 1997a; Satoda et al. 1999). However, sleep has not yet been studied in Tfap2b+/− mice.
AP-2 transcription factors have diverged in mammals to play nonredundant roles in development. In invertebrates, AP-2 plays a key role in sleep induction. Hence, AP-2 transcription factors provide a unique chance to study how sleep genes evolved from controlling simpler types of sleep in invertebrates to more complex types of sleep in mammals. Multiple hypotheses are conceivable for how AP-2 transcription factors may have evolved to control sleep. Three hypotheses may seem most plausible. (1) A sleep-promoting role might be present in one of the AP-2 paralogs, but such a role is not found in other AP-2 paralogs. (2) Multiple AP-2 paralogs might play a redundant role in promoting sleep. (3) Different AP-2 paralogs might serve specialized subfunctions in promoting sleep.
In this study, we studied sleep in Tfap2a+/− and Tfap2b+/− mice. Surprisingly, we found that Tfap2a and Tfap2b play opposing roles in sleep control. Tfap2a+/− causes an increase in sleep quality, and is also associated with hyperactivity during a stress test. By contrast, Tfap2b+/− reduces sleep time and quality and is associated with altered circadian rhythms, mildly depressive-like symptoms, and a learning defect. Thus, AP-2 transcription factors appear to have diverged to allow bidirectional control of sleep.
Materials and Methods
Animals
Neofloxed-Tfap2a conditional knockout mice were obtained from Trever Williams (Brewer et al. 2004) and bred with CMV-Cre mice to delete exons 5–6 of Tfap2a. Mice that are homozygous for this allele were perinatal lethal in our colony probably due to neural tube closure defects and cleft secondary palate (Zhang et al. 1996). Thus, heterozygous mice (Tfap2a+/−) were used in this study and their wild-type littermates (Tfap2a+/+) were used as controls.
Tfap2b knockout mice were provided by Markus Moser [Max Planck Institute (MPI) for Biochemistry] with PGK-neo cassette inserted into exon four of the Tfap2b gene. Tfap2b−/− mice die shortly after birth due to polycystic kidney disease (Moser et al. 1997a). Thus, heterozygous mice (Tfap2b+/−) were used in this study and their wild-type littermates (Tfap2b+/+) were used as controls.
Adult (2–6 M) males were used in this study, except for the running-wheel test (see section Wheel-running activity and circadian analysis). Mice were kept at the animal facility of the MPI of Biophysical Chemistry in accordance to Lower-Saxony animal welfare laws. All animal experiments were carried out in compliance with the German Law on Animal Welfare and were approved by the Office for Consumer Protection and Food Safety of the State of Lower Saxony. All animal experiments were approved by the Animal Experiments Ethical Committee of the Max Planck Institute for Biophysical Chemistry (Göttingen, Germany) and Laves, and were carried out in accordance with European Union (EU) Directive 2010/63/EU on the protection of animals used for scientific reasons. Mice were entrained in a 12:12 hr light-dark cycle from 6:00 to 18:00. For the entire behavioral testing, adult male mice were singly housed with ad libitum access to water and food pellets, in controlled constant temperature and humidity. The animals were individually housed at least 1 week prior to the experiment.
Genotyping
Ear biopsies of mice were collected and genomic DNA was extracted through incubation in PBND lysis buffer (PCR buffer with nonionic detergents: 50 mM KCl, 10 mM Tris HCl pH 8.3, 2.5 M MgCl2, 0.1 mg/ml Gelatin, 1 mg/ml proteinase K, 0.45% NP-40, 0.45% Tween) for at least 6 hr at 55°, followed by 45 min at 85° to deactivate the Proteinase K. Genotyping primers and conditions are listed in Supplemental material, Table S1.
Surgeries
Mice were anesthetized using isoflurane (1–2% in 100% O2) during the surgery. Miniature screw electrodes (2.16 mm diameter, Bilaney Consultants GmbH) with wire (5 mm) attached were implanted. Two electrodes were placed over the right and left frontal cortex [anteroposterior (AP) +1.5 mm from bregma; mediolateral (ML) 1.7 mm). One electrode was placed over the right parietal (AP +1.5 mm from lambda, ML, 1.7 mm) cortex. Two electrodes were placed bilaterally over the cerebellum (AP -1.5 mm from lambda, ML, 1.7 mm) as reference (left) and ground signal (right). One subcutaneous electrode (12 mm, Bilaney Consultants GmbH) was placed in the nuchal muscle for the electromyogram (EMG) recording. All attached wires were assembled in a plastic pedestal (MS363, PlasticsOne, Bilaney Consultants GmbH), which was fixed to the skull with dental cement. The mice were housed individually and left to recover for at least 8 days before they were attached to the recording cable. Mice were given a 2-day acclimation period to adjust to the cable before recording.
Electroencephalogram recording setups and schedule
The recording room was kept under 12 hr light/12 hr dark cycles and room temperature. Light was delivered from ZT0 (6 am ) to ZT12 every day. All electrodes were gathered into a light weight and flexible cable and connected to the recording system (Multi Channel Systems MCS GmbH). Electroencephalogram (EEG) and Electromyography (EMG) signals were collected continuously at a sampling rate of 2000 Hz. To examine the sleep–wake behavior under the baseline conditions, the EEG/EMG recordings were performed for two consecutive days, beginning at ZT0. By the end of the baseline recording, all mice were sleep-deprived (Bernard et al. 2015) by gentle handling and novel object interaction protocols for six consecutive hours started from ZT0 to ZT6 (Colavito et al. 2013). Any direct contact of the experimenter with the animals was avoided. At the end of the sleep deprivation (SD) period, the animals were left to move and sleep freely with free access to food and water, and recording was continued for the next 48 hr.
EEG data analysis
A MatLab-based, custom-written auto-score system was first trained with EEG/EMG data by a human scorer. The EEG/EMG data were then analyzed by the auto-score system (Gao et al. 2016), followed by visual inspection by the same human scorer. In brief, a training set was selected for every 24 hr of data based on a random REMS epoch and preceding 90 epochs (15 min) as well as the following 90 epochs. This process was repeated until a total of 720 epochs (2 hr) of training was selected. The remaining 7920 epochs (22 hr) were subjected to short-time Fourier transformation and auto-scored with a multiple classifier system at a 5% rejection threshold using MatLab. Training and rejected epochs were scored using Sirenia Sleep Pro (Pinnacle Technologies, Lawrence, KS). Manual scoring of three vigilance states were performed for each 10-s epoch as either wake, NREMS and REMS. Wake was scored based on the presence of low amplitude, fast EEG, and high amplitude, variable EMG. NREMS was characterized by high-amplitude delta (0.5–4 Hz) EEG but low frequency EMG activity. REMS was characterized by low-amplitude rhythmic theta waves (6–10 Hz) with EMG atonia. The scorer was blind to the genotype within all the scoring process. The power for each 0.1-Hz bin (between 0.5 and 25 Hz) within the 10-s segment was calculated. For 48 hr of baseline recording, average values were calculated and plotted on a 24 hr-scale. Time spent per hour or total hours in wake/NREMS/REMS stages was calculated over ZT or during the 24 hr light/dark phase for each genotype. To evaluate the effect of SD, changes in time or bout length of NREMS/REMS during recovery sleep were calculated. In power spectral analysis, the EEG time series were decomposed into a voltage by frequency spectral graph, with power calculated as the square of the EEG magnitude, and magnitude being the integral average of the amplitude of the EEG signal (Kent et al. 2018). During baseline recording, derived power data were further grouped and analyzed based on frequency and vigilance states. Here, we presented the raw power data based on the vigilance state in combination with either the distribution of power in 0.1-Hz windows or frequency classes. After SD, the NREMS/REMS/wake powers during recovery sleep were expressed as percentage to the mean of the same ZT from basal recording. Mean values of power spectrum data were calculated for each genotype and the values were submitted to Wilcoxon signed rank tests to make comparison between the genotypes. All power data were expressed as averaged values from measurement of frontal and temporal lobes. For delta and theta power analysis following SD, Z06-12, Z12-18, Z18-24 time zones following the end of SD were used.
Behavioral testing
All behavioral tests were conducted between 08:30 am and 06:00 pm during the light phase. The order of testing was as follows: elevated plus maze (EPM), rotarod, Morris water maze (MWM), sucrose preference test (SPT), forced swim test (FST), tail suspension test (TST), and fear conditioning (FC).
Elevated plus maze
The EPM consisted of four arms, each 30 cm long and 9.7 cm wide, elevated 50 cm off the ground. Two arms were enclosed by walls 25 cm high and the other two arms were exposed. Mice were placed on the central platform. The behavior of each subject was tracked by an overhead camera and a computer equipped with VideoMot (TSE Systems GmbH, Germany), and recorded for 5 min. VideoMot was used to calculate the time spent in the open or closed arm. Time spent in open arm or central platform was used to evaluate exposure aversion.
Rotarod test
Motor functions and coordination were examined on the rotarod machine with automatic falling sensors (RotaRod Advanced, TSE Systems GmbH, Germany). For the habituation, mice were trained with the rotating speed of 10 rpm twice a day for two consecutive days. In each trial, mice were placed back to the rod immediately after falling off. After training, mice were tested under continuous acceleration from 5 rpm to 40 rpm with two sessions per day for two consecutive days. The latency to fall was recorded with a computer equipped with RotaRod software. Each measurement lasted 180 s with at least 6-hr intervals.
MWM test
A circular pool of 1.0 m in diameter was used for the MWM. The water (21°), made opaque by addition of nontoxic tempera paint, was 20 cm deep, and the wall of the pool extended 15 cm above the surface of the water. A square hidden platform (13 × 13 cm) was located 1 cm below the water surface approximately in the middle of one of the pool quadrants. Distal visual cues surrounding the pool included four colored labels of different shapes fixed around the edge of the water tank, as well as a door and a wall of cabinets. The test consisted of three phases: visible platform task, hidden platform task and probe test for a total of 11 consecutive days. To exclude mice with visual or motivational impairments and habituate them to the testing conditions, we performed visible platform task during the first 2 days. In this phase, mice were trained to swim to a visible platform placed 1 cm above water surface and in the middle of the water tank. Next, mice were tested with a hidden platform task where a fixed platform was hidden 1.0 cm below the water surface. Mice that failed to locate the platform within 90 sec were guided to it, and all mice were allowed to rest on the platform for at least 15 sec before being returned to their cage. On probe test day, the platform was removed, and mice were allowed to swim in the pool for up to 90 sec. The time spent in each quadrant was measured. On each of the 11 days, mice were given four trials per day starting from each of the four cardinal directions (N, S, E, W) in a pseudorandom order that changed every day. In each trial, mice were gently held close to the water surface facing the wall and then placed in the pool. The swim patterns were monitored by the video-tracking system VideoMot (TSE Systems GmbH). The escape latency, swim speed, path length, and trajectory of swimming were recorded for each mouse. Mice that were swimming slower than 60% of the average speed of wild type, Tfap2a, and Tfap2b mutants during each phase were classified as floaters. Two floaters were removed from the Tfap2a+/−, eight from Tfap2b+/+, six from Tfap2b+/−.
Sucrose preference test
The whole experiment was carried out in the home cage of the mice in the breeding area. Two identical bottles were used for each cage and placed in left and right sides of the cage. Mice were allowed to habituate to the bottles with standard drinking water for 48 hr. In the second 24 hr of the habituation period, weights of the bottles were recorded. Then, mice were given 48 hr of free choice between two bottles of either 2% sucrose or standard drinking water. At the end of the period the bottles were weighed again and the consumption was calculated. No previous food or water deprivation was applied before the test. The percentage of sucrose preference was calculated using the following formula: Sucrose preference = V(sucrose solution)/[V(sucrose solution)+V(water)] × 100%.
Forced swim test
The test apparatus consisted of an inescapable transparent cylinder (25 cm height × 15 cm diameter) containing 20 cm of water (23°). Dividers (35 cm height × 22 cm width) were used between cylinders to prevent mice from seeing each other during the test. Mice were individually placed into the cylinders, and the immobility was recorded over a 6-min test period. Immobility was analyzed by an observer according to the following criteria. Each mouse was judged to be immobile when it ceased struggling and remained floating motionlessly in the water, making only those movements necessary to keep its head above water.
Tail suspension test
The TST (Gibney et al. 2013) was performed as described previously (Can et al. 2012). Each mouse was suspended 30 cm above the floor by the tail with a 16 cm long piece of tape. Dividers (35 cm height × 15 cm width) were hanged between tapes to prevent mice from seeing each other during the test. The behavior was recorded for 6 min. Immobility was analyzed by an observer.
Contextual FC test
The contextual FC test was performed as described previously (Fischer et al. 2004; Sananbenesi et al. 2007). In brief, FC was carried out with a computerized fear conditioning system (TSE Systems GmbH) using a computer, equipped with Freeze Scan software (Clever Systems), connected to a control unit containing a shock and a noise generator. Animals were allowed to explore the training cage for 3 min followed by a mild electric shock (2 sec, 0.5 mA). Context-dependent freezing, defined as the absence of movements other than those required for breathing, was assessed for the following 2 days with a 24-hr interval without the electric shock. Freezing behavior and average movement were recorded for each mouse.
Wheel-running activity and circadian analysis
The setup included six controls and six mutant mice (three males and three females for each genotype of Tfap2a and Tfap2b) at the age of 2–5 months. Mice were placed in single cages with running wheels connected to a computer running ClockLab (Actimetrics) data collection software. Genotypes and sexes were evenly distributed over two boxes. Animals were not disturbed during the entire experiment. Wheel counts were checked every day to assess well-being of the animal. The experiment consisted of five phases: training phase, entrainment phase, phase advance (jetlag), light pulse at ZT14, and free-running phase. During the training phase, animals were in a 12 hr light: 12 hr dark (LD) cycle for >7 days with a light phase light intensity of 300 lux to habituate to the running-wheel. Next, daily activities were recorded and calculated as an average of 10 days. After the entrainment period in LD, mice were subjected to an abrupt shift in the light schedule by advancing the “lights off” time by 6 hr (jetlag paradigm). The number of days needed to completely re-entrain to the shifted LD cycle was compared between the genotypes. After mice were completely re-entrained to the new LD cycle, a light pulse was delivered at ZT14 for 30 min at 300 lux and mice released into constant darkness (DD), and phase shifts were calculated for activity onsets on the day after the light pulse. Mice were retained in DD for another 2 weeks to assess free-running period lengths by χ2 periodogram analysis. One female in Tfap2a+/+ was identified as an outlier using ROUT (Q = 1%) method and was removed from further analyses.
RNA-sequencing
RNA isolation and sequencing were carried out by Bernd Timmermann and Stefan Börno at the sequencing facility of the Max Planck Institute for Molecular Genetics, Berlin, according to their protocol: RNA was isolated from 20 to 80 mg of mouse brain tissue from B6N, Tfap2a+/− and Tfap2b+/− (stored in RNAlater) following the Qiagen RNeasy protocol. First, the tissue samples were homogenized with the TissueLyser (Qiagen) at 25 Hz for 2 × 2 min in Qiazol lysis buffer; 140 µl chloroform was added, and, after 15 min centrifugation at 12,000 × g, the aqueous phase containing the RNA was extracted. Ethanol (1.5 volumes) was added and the samples were washed with Qiagen’s RWT buffer on a Qiagen RNeasy spin column. RNA was treated with 10 µl of DNase I on column for 15 min followed by a wash with RWT. After further washes with RPE buffer, the purified RNA was eluted with 50 µl water, yielding between 5 and 22 µg RNA.
After quality control using Agilent’s Bioanalyzer, sequencing libraries were prepared from 500 ng of total RNA per sample following Roche’s stranded “KAPA RNA HyperPrep” library preparation protocol for single indexed Illumina libraries: First, the polyA-RNA fraction was enriched using oligo-dT-probed paramagnetic beads. Enriched RNA was heat-fragmented and subjected to first-strand synthesis using random priming. The second strand was synthesized incorporating dUTP instead of dTTP to preserve strand information. After A-tailing, Illumina sequencing compatible adapters were ligated. Following bead-based clean-up steps, the libraries were amplified using 11 cycles of PCR. Library quality and size was checked with qBit, Agilent Bioanalyzer, and quantitative PCR (qPCR). Sequencing was carried out on an Illumina HiSeq 4000 system in PE75bp mode, yielding between 27 and 37 million fragments per sample.
Data analysis of RNA-sequencing
Following base calling, adaptor clipping was performed using cutadapt 2.4 (Martin 2011). Data were mapped against the GRCm38.p6 genome using STAR v 2.6.1d (Dobin et al. 2013) and differentially expressed genes were analyzed using EdgeR.
Statistics
Statistical analysis was performed using GraphPad Prism 8.3.0 and IBM SPSS (Version 22). All data were subjected to a Shapiro-Wilk normality test for Gaussian distribution and Levene’s test for equality of variances. For the dataset that showed a Gaussian distribution (P > 0.05 in normality test), we performed parametric tests such as two-tailed paired/unpaired t-test and ANOVA followed by Sidak’s multiple comparisons. For the dataset that failed to show a Gaussian distribution, we performed nonparametric tests, such as a Mann-Whitney test and Wilcoxon signed-rank test. Significance levels in the figures are represented as *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. Error bars in the graphs represent mean ± SEM.
Quantitative PCR
To assess the mRNA reduction of AP-2α and β in Tfap2a and b mice, total RNA was extracted from the cortex using an RNA extraction kit (Qiagen GmbH). Total RNA was reverse transcribed into cDNA using the high capacity cDNA RT kit (Applied Biosystems) according to the manufacturer’s instructions. Subsequently, the mRNA expression levels of AP-2α and β were quantified by qPCR using the Fast SYBR Green Master Mix (Applied Biosystems) using specific primers for each gene (Table S2). cDNA amplification was performed following a PCR program of 40 cycles, with denaturation at 94° for 1 min and annealing at 62° for 30 sec, followed by elongation at 72° for 1 min using an ABI 7500 qPCR cycler. mRNA expression was analyzed using the 2−ΔΔCq method where the control was normalized to 1, and the treated samples were compared with their control. Primers and conditions are listed in Table S2.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. All data are available at Dryad: https://doi.org/10.5061/dryad.rv15dv45r. Raw RNA-seq data are available at GEO: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE155629. EEG-autoscore code is available at GitHub: https://zenodo.org/badge/latestdoi/240526669.
Results
Increased sleep pressure in Tfap2a+/− mice
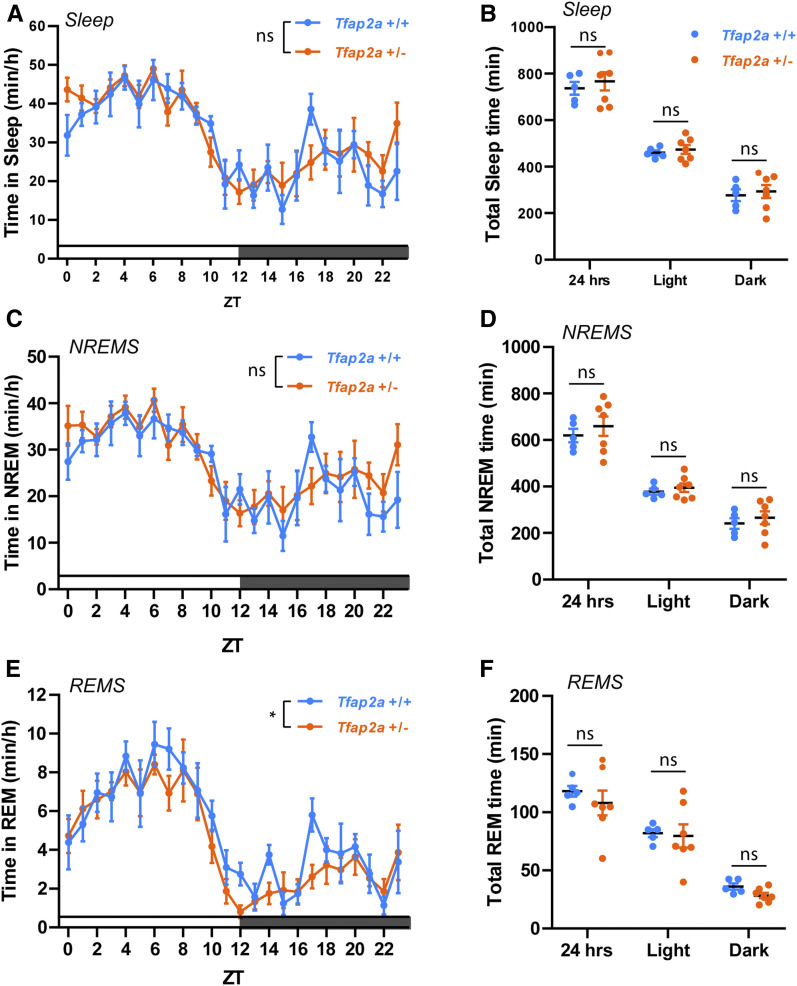
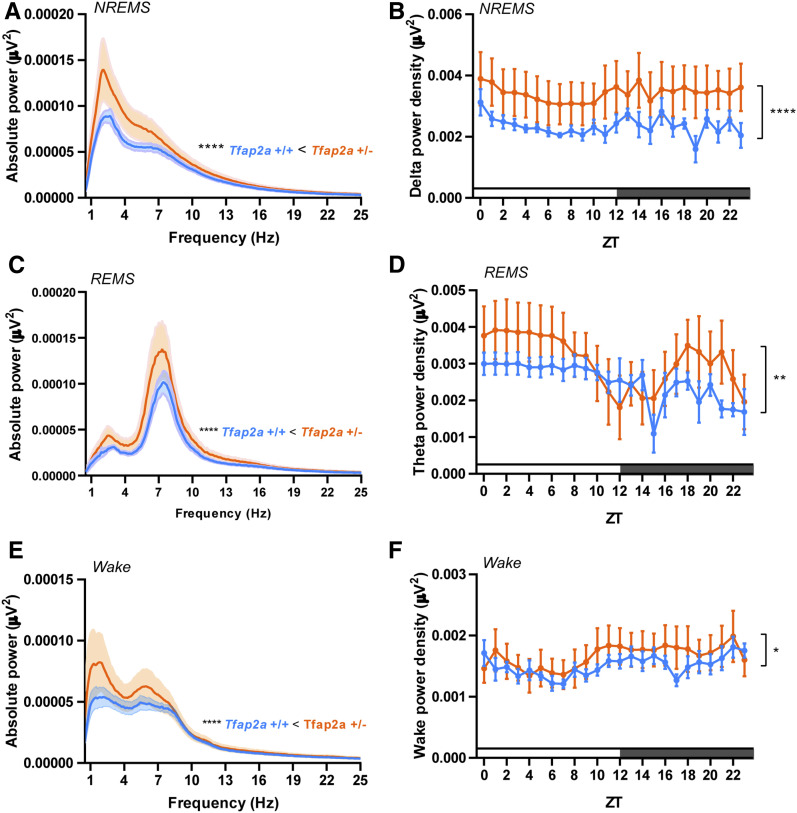
We first analyzed sleep in Tfap2a mice. To quantify sleep and wake, we used EEG and EMG recordings (Mang and Franken 2012). As homozygous knockouts of Tfap2a are not viable (Zhang et al. 1996), we studied Tfap2a heterozygous animals. qPCR showed that these mutants had a reduced Tfap2a mRNA amount by about half (Supplemental Material, Figure S1). We first analyzed the amount of NREMS and REMS from the electrophysiological recordings. Total sleep time was not significantly affected (Figure 1, A and B). We observed a trend toward increased NREMS (Figure 1, C and D), which did not, however, reach statistical significance. REMS time was decreased slightly (Figure 1, E and F). Sleep bout analysis did not show any significant changes in sleep architecture (Figure S2). We next investigated sleep quality using power spectrum analysis. Tfap2a+/− mice exhibited significantly increased delta power in NREMS (Figure 2, A and B) and theta power in REMS (Figure 2, C and D). Additionally, EEG spectral analysis of Tfap2a+/− mice during wakefulness showed increased low-frequency power (1–7 Hz) (Figure 2, E and F). Increased delta power in NREMS and increased theta power in REMS suggests that sleep intensity in Tfap2a+/− mice was increased.
Figure 1.
Sleep amount is not significantly altered in Tfap2a+/− mice. (A) Total sleep quantification over Zeitgeber times (ZT), two-way ANOVA followed by Sidak’s multiple comparisons test, the main effect of genotype, F (1,240) = 0.941, P = 0.333. (B) Total sleep time quantification, P = 0.576; light, P = 0.574; dark, P = 0.687. (C) NREMS quantification over ZT, two-way ANOVA followed by Sidak’s multiple comparisons test, the main effect of genotype, F (1,10) = 0.526, P = 0.485. (D) NREMS time quantification, P = 0.446; light, P = 0.521; dark, P = 0.543. (E) REMS quantification over ZT, two-way ANOVA followed by Sidak’s multiple comparisons test, the main effect of genotype, F (1,239) = 0.492, *P = 0.0353. (F) REMS time quantification, P = 0.466; light, P = 0.852; dark, P = 0.053. n = 5 for Tfap2a+/+, n = 7 for Tfap2a+/−. Two-tailed unpaired t-tests were used for (B, D, and F). Averaged data are shown as the mean ± SEM.
Figure 2.
EEG delta and theta power are increased in Tfap2a+/− mice. (A) EEG power spectra in NREMS. (B) NREMS delta power (1–4 Hz), two-way ANOVA followed by Sidak’s multiple comparisons test, the main effect of genotype, F (1,240) = 29.58, ****P < 0.0001. (C) EEG power spectra in REMS. (D) REMS theta power (6–10 Hz): two-way ANOVA followed by Sidak’s multiple comparisons test, the main effect of genotype, F (1,240) = 7.558, **P = 0.0064. (E) EEG power spectra during wakefulness. (F) Power analysis during wake (0.5–4 Hz), two-way ANOVA followed by Sidak’s multiple comparisons test, the main effect of genotype, F (1,240) = 4.028, *P = 0.0459. All data are shown as the mean ± SEM n = 5 for Tfap2a+/+, n = 7 for Tfap2a+/−. P values for 1–25 Hz were calculated using Wilcoxon two-sided signed-rank test, ****P < 0.0001. All data are shown as the mean ± SEM.
Sleep loss and reduced sleep quality in Tfap2b+/− mice
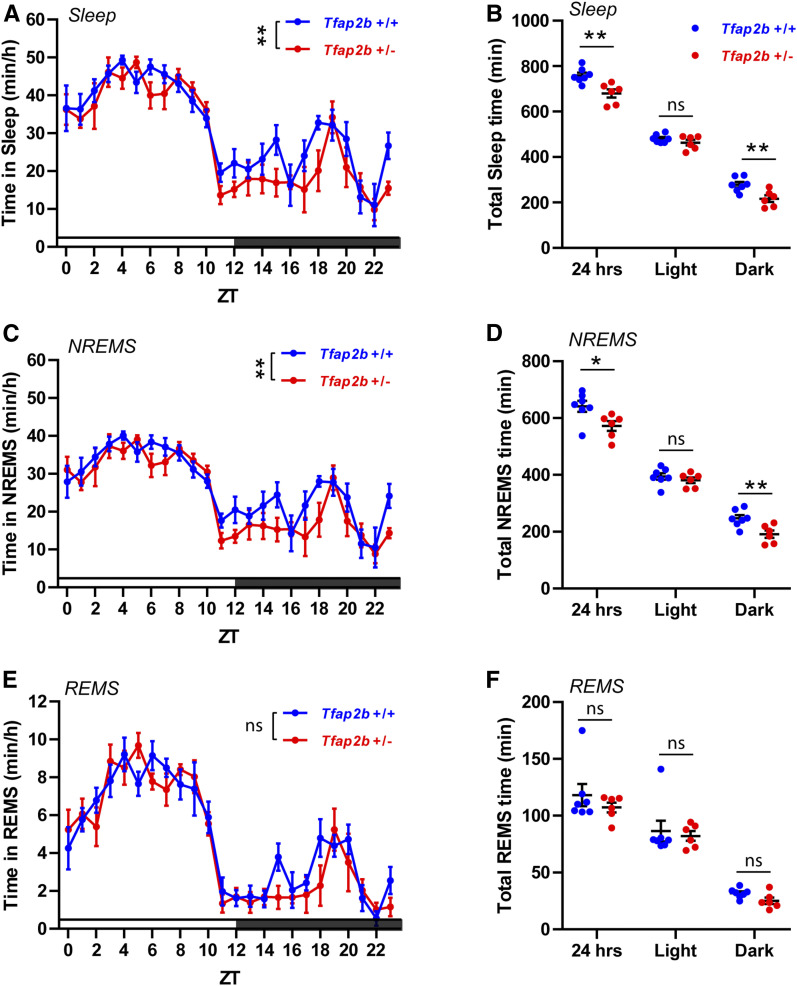
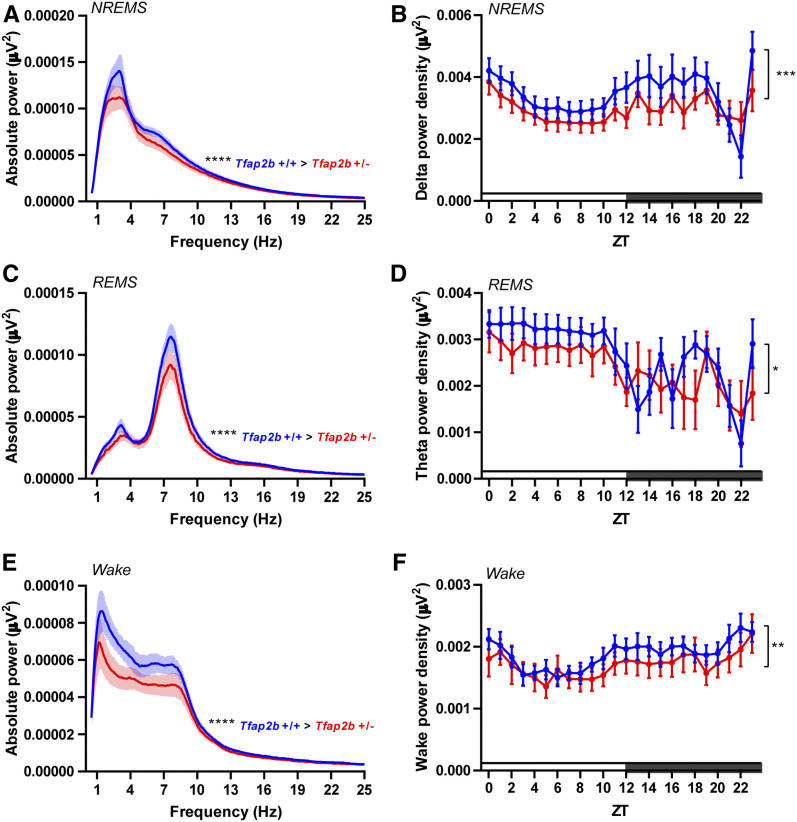
We next determined sleep amount and quality of Tfap2b+/− mice by EEG/EMG recordings. Heterozygous deletion of Tfap2b led to a reduction of mRNA by about half (Figure S1). Total sleep time was significantly reduced, an effect that was caused by a specific reduction of sleep during the dark phase (Figure 3, A and B). Analysis of NREMS showed that the reduction of total sleep was due mainly to a reduction in NREMS during the dark phase (Figure 3, C and D). By contrast, no difference could be detected for REMS (Figure 3, E and F). We next analyzed the distribution of sleep bouts in Tfap2b+/− mice. Longer sleep bouts were reduced, an effect that was particularly pronounced in the dark phase (Figure S3, A–D). NREMS bouts, particularly long ones, were significantly reduced, again most strongly in the dark phase (Figure S3, E–H). The distribution of REMS bouts remained unchanged (Figure S3, I–L). Power analysis showed that Tfap2b+/− mice had significantly decreased delta power in NREMS (Figure 4, A and B), as well as reduced theta power in REMS (Figure 4, C and D). Delta power was already reduced during wakefulness (Figure 4, E and F). In summary, these findings suggest that, in contrast to Tfap2a+/− mutants, Tfap2b+/− mice have reduced total sleep, which primarily is a consequence of reduced or shortened NREMS bouts in the dark phase. Tfap2b+/− mice have less NREMS/REMS power, suggesting a reduction not only of sleep amount, but also of sleep quality.
Figure 3.
Sleep during the dark phase is reduced in Tfap2b+/− mice. (A) Total sleep time quantification over ZT, two-way ANOVA followed by Sidak’s multiple comparisons test, the main effect of genotype, F (1,264) = 10.01, **P = 0.002. (B) Total sleep time quantification, **P = 0.004; light, P = 0.222; dark, **P = 0.007. (C) NREMS quantification over ZT, two-way ANOVA followed by Sidak’s multiple comparisons test, the main effect of genotype, F (1,264) = 10.26, **P = 0.002. (D) NREMS total time quantification, *P = 0.024; light, P = 0.411; dark, **P = 0.008. (E) REMS quantification over ZT, two-way ANOVA followed by Sidak’s multiple comparisons test, the main effect of genotype, F (1,263) = 1.492, P = 0.223. (F) REMS total time quantification, P = 0.668; light, P = 1.000; dark, P = 0.055. n = 7 for Tfap2b+/+, n = 6 for Tfap2b+/−. Two-tailed unpaired t-tests were used for (B, D, and F). All data are shown as the mean ± SEM.
Figure 4.
EEG delta and theta power are decreased in Tfap2b+/− mice. (A) EEG power spectra in NREMS. (B) NREMS delta power (1–4 Hz), two-way ANOVA followed by Sidak’s multiple comparisons test, the main effect of genotype, F (1,264) = 13.68, ***P = 0.0003. (C) EEG power spectra in REMS. (D) REMS theta power (6–10 Hz), two-way ANOVA followed by Sidak’s multiple comparisons test, the main effect of genotype, F (1,264) = 5.525, *P = 0.0195. (E) EEG power spectra during wakefulness. (F) Wake power analysis (0.5–4 Hz), two-way ANOVA followed by Sidak’s multiple comparisons test, the main effect of genotype, F (1,264) = 9.126, **P = 0.0028. All data are shown as the mean ± SEM, n = 7 for Tfap2b+/+, n = 6 for Tfap2b+/−. P values for 1–25 Hz were calculated using Wilcoxon two-sided signed-rank test, ****P < 0.0001. All data are shown as the mean ± SEM.
Opposing effects of Tfap2a+/− and Tfap2b+/− on homeostatic responses to SD
Baseline sleep characterization showed that reductions in the function of Tfap2a and Tfap2b have opposing roles in sleep regulation. To test whether these different roles of the two transcription factors extend to sleep homeostasis, we tested the response of Tfap2a+/− and Tfap2b+/− mice to SD. Using gentle handling (Colavito et al. 2013), we kept mice awake for 6 hr starting from light onset and measured and quantified the increase in sleep amount and quality by EEG.
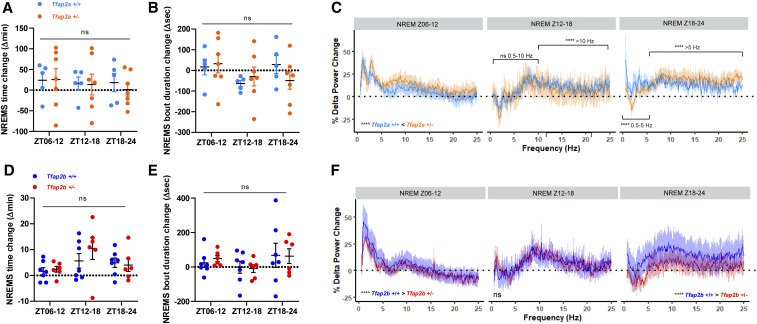
Sleep time and delta power increased after SD in all conditions tested (Figures S4 and S5), but the magnitude of delta power responses differed in the different mutants. Tfap2a+/− mutant and control animals did not show statistically significant differences in sleep time and bout length (Figure 5, A and B). However, mutant animals exhibited higher rebound delta power compared to control animals within the first 6 hr after SD (Z06–12) (Figure 5C). The delta power rebound diminished during the next (Z12–18) and following 6 hr periods (Z18–24) in both control and mutant animals (Figure 5C). Tfap2b+/− animals also showed no difference in NREMS time (Figure 5D) or bout duration (Figure 5E) after SD compared to control animals, but lower rebound delta power during Z06–12 (Figure 5F). During Z12–18, no rebound was observed in both control and mutant animals, but the power change was smaller in mutants during Z18–24 (Figure 5F).
Figure 5.
NREMS delta power following sleep deprivation is increased more strongly in Tfap2a+/− mice but less strongly in Tfap2b+/− mice. (A) NREMS time change in Tfap2a+/−, two-way ANOVA tests followed by Sidak’s pairwise comparison, the main effect of genotype: F(1, 30) = 0.103, P = 0.750; the main effect of time, F(2, 30) = 0.791, P = 0.791. (B) Average NREMS bout duration change in Tfap2a+/−, two-way ANOVA tests followed by Sidak’s pairwise comparison, the main effect of genotype: F(1, 30) = 0.072, P = 0.790; the main effect of time, F(2, 30) = 1.407, P = 0.261. (C) NREMS delta power changes (0.5–25 Hz) in Tfap2a+/− mice. Z06-12: 0.5–25 Hz, ****P < 0.0001. Z12-18: 0.5–10 Hz, P = 0.334; 10–25 Hz, ****P < 0.0001. Z18–24: 0.5–5 Hz, P = 0.334; 5–25 Hz, ****P < 0.0001. (D) NREMS time change in Tfap2b+/−, two-way ANOVA tests followed by Sidak’s pairwise comparison, the main effect of genotype: F(1, 33) = 0.595, P = 0.446; the main effect of time, F(2, 33) = 3.008, P = 0.063. (E) Average NREMS bout duration change in Tfap2b+/−, two-way ANOVA tests followed by Sidak’s pairwise comparison, the main effect of genotype: F(1, 33) = 0.014, P = 0.906; the main effect of time, F(2, 33) = 1.476, P = 0.243. (F) NREMS delta power changes (0.5–25 Hz) in Tfap2b+/− mice. Z06–12: 0.5–25Hz, ****P < 0.0001. Z12–18: 0.5–25 Hz, P = 0.715. Z18–24: 0.5–25 Hz, ****P < 0.0001. n = 5 for Tfap2a+/+, n = 7 for Tfap2a+/−, n = 7 for Tfap2b+/+, n = 6 for Tfap2b+/−. Wilcoxon two-sided signed-rank tests were used for power changes in (C and F). BSL, baseline sleep; R, recovery sleep. All data are shown as the mean ± SEM.
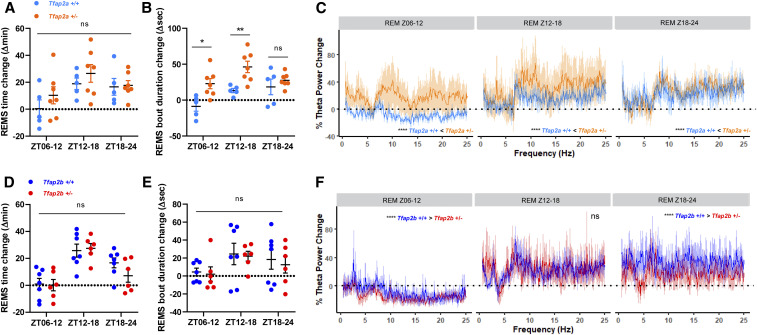
We next quantified REMS time and theta power following SD. A delayed REMS rebound was observed in both Tfap2a and Tfap2b mutants for REMS time as well as for theta power (Figure S6 and S7). REMS bout duration increased more strongly in Tfap2a+/− compared to wild-type littermates (P < 0.01) (Figure 6, A and B). Tfap2a+/− also showed a higher theta power rebound compared to their littermates (Figure 6C). In contrast, REMS time and REMS bout duration increased less in Tfap2b+/− after SD compared to wild-type controls during ZT18-24, although these differences were not significant (Figure 6, D and E). Supporting this trend, the theta power rebound after SD in Tfap2b+/− mice was significantly weaker compared to wild-type control animals during Z06-12 and Z18-24 (Figure 6F). Thus, Tfap2a+/− mice respond more strongly to SD, whereas Tfap2b+/− animals exhibit a weaker response to SD compared to wild-type controls.
Figure 6.
REMS theta power following sleep deprivation is increased more strongly in Tfap2a+/− mice but less strongly in Tfap2b+/− mice compared with wild-type controls. (A) REMS time change in Tfap2a+/−, two-way ANOVA tests followed by Sidak’s pairwise comparison, the main effect of genotype: F(1, 30) = 1.698, P = 0.202; the main effect of time, F(2, 30) = 4.585, P = 0.018. (B) Average REMS bout duration change in Tfap2a+/−, two-way ANOVA tests followed by Sidak’s pairwise comparison, the main effect of genotype: F(1, 30) = 17.95, P = 0.010; the main effect of time, F(2, 30) = 5.365, P = 0.0002. ZT06–12: *P = 0.011. Z12–18: **P = 0.0079. Z18–24: P = 0.749. (C) Rebound differences of REMS in theta power (0.5–25 Hz) in Tfap2a+/−. Z06–12: 0.5–25 Hz, ****P < 0.0001. Z12–18: 0.5–25 Hz, ****P < 0.0001. Z18–24: 0.5–25 Hz, ****P < 0.0001. (D) REMS time change in Tfap2b+/−, two-way ANOVA tests followed by Sidak’s pairwise comparison, the main effect of genotype: F(1, 33) = 0.673, P = 0.418; the main effect of time, F(2, 33) = 16.86, P < 0.0001. (E) Average REMS bout duration in Tfap2b+/−, two-way ANOVA tests followed by Sidak’s pairwise comparison, the main effect of genotype: F(1, 33) = 0.234, P = 0.632; the main effect of time, F(2, 33) = 2.521, P = 0.096. (F) Rebound differences of REMS in theta power (0.5–25 Hz) in Tfap2b+/−. Z06–12: 0.5–25 Hz, ****P < 0.0001. Z12–18: 0.5–25 Hz, P = 0.119. Z18–24: 0.5–25 Hz, ****P < 0.0001. n = 5 for Tfap2a+/+, n = 7 for Tfap2a+/−, n = 7 for Tfap2b+/+, n = 6 for Tfap2b+/−. Wilcoxon two-sided signed-rank tests were used for power changes in (C and F). BSL, baseline sleep; R, recovery sleep. All data are shown as the mean ± SEM.
Divergent behavioral changes in Tfap2a+/− and Tfap2b+/− mice
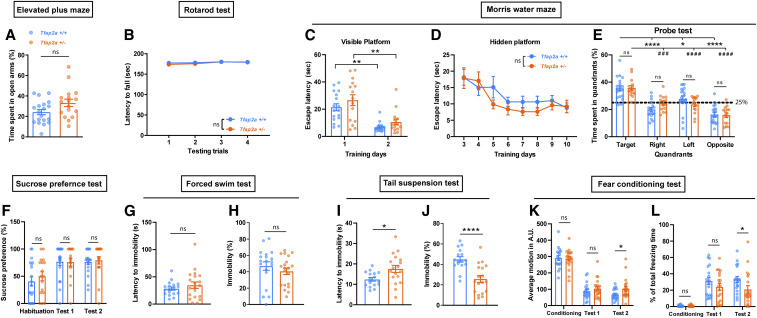
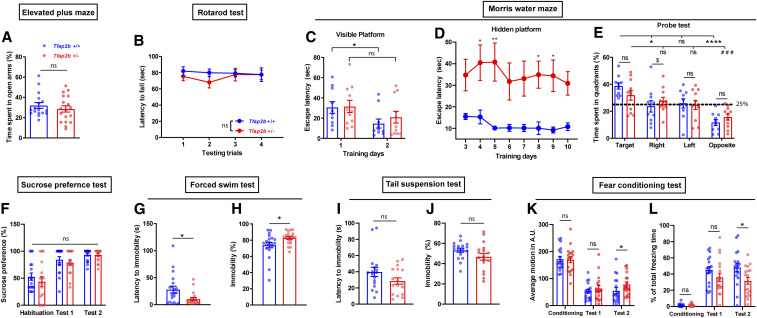
Sleep loss is correlated with emotional instability, anxiety (Verbitsky 2017), depression (Matsuda et al. 2017), and cognitive defects (Bezdicek et al. 2018). We hence tested how reduction of function of Tfap2a and Tfap2b affects anxiety, depression, as well as learning and memory (Figures 7 and 8). To assess anxiety-associated behavior, we performed the EPM test (Walf and Frye 2007). Explorative behavior assayed in the EPM was similar across groups (Figure S8, A–D). The time spent in open arms and entries into the open arms were not changed relative to the corresponding littermates. Thus, we could not detect altered anxiety-associated exploration behavior in Tfap2a+/− or Tfap2b+/− (Figures 7A and 8A). Accelerating rotarod tests (Deacon 2013) showed that there was no difference in latency to fall during four testing trials in both mutants compared to their respective littermates (Figures 7B and 8B), indicating that motor coordination and balance in both mutants are not affected. Interestingly, Tfap2b+/− showed minor motor learning deficiency during the training trials, but this was not observed in Tfap2a+/− (Figure S8, E and F).
Figure 7.
Behavioral phenotyping of Tfap2a+/− mice reveals signs of mild hyperactivity. (A) Elevate plus maze, two-tailed unpaired t-test, P = 0.136. (B) Rotarod test, two-way ANOVA test, the main effect of genotype: F(1, 148) = 0.927, P = 0.337; the main effect of time, F(3, 148) = 2.512, P = 0.061. (C) Morris water maze (MWM), time spent searching for visible platform during two consecutive training days: to compare mutants and their controls, unpaired Student t-test was used for day 1, P = 0.274; Mann-Whitney U-test for day 2, P = 0.096. Wilcoxon two-sided signed-rank tests for two related samples were used for day 1 and day 2 within each genotype: Tfap2a+/+, **P = 0.001; Tfap2a+/−, **P = 0.004. (D) MWM, time spent searching for hidden platform during eight consecutive training days from day 3 to day 10. Two-way ANOVA tests followed by Sidak’s pairwise comparison, the main effect of genotype: F(1, 256) = 1.049, P = 0.307; the main effect of time, F(7, 256) = 8.652, P < 0.0001. (E) MWM, time spent in each quadrant during probe test: one-way ANOVA followed by Sidak’s pairwise comparison, F = 21.125, P < 0.0001. Tfap2a+/+ vs. Tfap2a+/−: target, P = 1.000; right, P = 0.819; left, P = 0.918; opposite, P = 1.000. Target vs. right/left/opposite quadrant in Tfap2a+/+: ****P < 0.0001, *P = 0.024, ****P < 0.0001. Target vs. right/left/opposite in Tfap2a+/−: ###P = 0.001, ####P < 0.0001, ####P < 0.0001. (F) Sucrose preference test: habituation, P = 0.379; test 1, P = 0.781; test 2, P = 0.415, Mann Whitney test. (G) Forced swim test (FST), latency to immobility, P = 0.830, Mann Whitney test. (H) FST, time spent immobile, P = 0.147, Mann Whitney test. (I) Tail suspension test (TST), latency to immobility, *P = 0.039, Mann Whitney test. (J) TST, time spent immobile, ****P < 0.0001, Mann Whitney test. (K) Fear conditioning test, average motion, training, P = 0.296; test 1, P = 0.428; test 2, *P = 0.028, Mann Whitney test. (L) Total freezing time, training, P = 0.771; test 1, P = 0.258; test 2, *P = 0.018, Mann Whitney test. Data are shown as the mean ± SEM n ≥ 16 for Tfap2a+/+, n ≥ 15 for Tfap2a+/−.
Figure 8.
Behavioral phenotyping of Tfap2b+/− mice reveals signs of mild depressive-like symptoms. (A) Elevated plus maze, P = 0.463, Mann Whitney test. (B) Rotarod test, two-way ANOVA test, the main effect of genotype: F(1, 136) = 1.313, P = 0.254; the main effect of time, F(3, 136) = 0.249, P = 0.862. (C) Morris water maze (MWM), time spent searching for visible platform during two consecutive training days: to compare mutants and their controls, unpaired Student t-test was used for day 1, P = 0.908; Mann-Whitney U-test for day 2, P = 0.557. Wilcoxon two-sided signed-rank tests for two related samples were used for day 1 and day 2 within each genotype: Tfap2b+/+, *P = 0.028; Tfap2b+/−, P = 0.091. (D) MWM, time spent searching for hidden platform during eight consecutive training days, Two-way ANOVA tests, the main effect of genotype: F(1, 148) = 63.40, P < 0.0001; the main effect of time, F(7, 148) = 0.377, P = 0.915; Sidak’s pairwise comparison between genotype,*P < 0.05,**P < 0.01. (E) MWM, time spent in each quadrant during probe test, one-way ANOVA followed by Sidak’s pairwise comparison, F = 7.381, P < 0.0001. Tfap2b+/+ vs. Tfap2b+/−: target, P = 1.000; right, $P = 0.034; left, P = 0.141; opposite, P = 1.000. Target vs. right/left/opposite quadrant in Tfap2b+/+: *P = 0.034, P = 0.141, ****P < 0.0001. Target vs. right/left/opposite in Tfap2b+/−: P = 1.000, P = 0.950, ###P = 0.009. (F) Sucrose preference test, habituation, P = 0.465; test 1, P = 0.372; test 2, P = 0.961, Mann Whitney test. (G) Forced swim test (FST), latency to immobility, *P = 0.039, Mann-Whitney test. (H) FST, time spent immobile, *P = 0.020, Mann-Whitney test. (I) Tail suspension test, latency to immobility, P = 0.231, Mann-Whitney test. (J) Tail suspension test, time spent immobile, P = 0.087, Mann-Whitney test. (K) Fear conditioning test, average motion, training, P = 0.815; test 1, P = 0.673; test 2, *P = 0.047, Mann Whitney test. (L) Fear conditioning test, total freezing time, training, P = 0.696; test 1, P = 0.152; test 2, *P = 0.027. Mann Whitney test. All data are shown as the mean ± SEM n ≥ 10 for Tfap2b+/+, n ≥ 11 for Tfap2b+/−.
To assess spatial learning and memory, we performed the MWM test (Radyushkin et al. 2009). Tfap2a+/− mice as well as controls required similar times to reach the visible platform during the 2 days of training, and the latency was significantly shortened on the second training day (Figure 7C). Tfap2b+/− and control mice did not differ significantly when exposed to the visual water maze paradigm (Figure 8C). The mice were next subjected to an 8-day hidden platform training test. Over time, the latency to locate the hidden platform decreased, and the learning pattern in Tfap2a+/− mice was comparable to wild-type litter mates (Figure 7D). By contrast, Tfap2b+/− mice had severe problems finding the hidden platform (Figure 8D). Finally, the platform was removed in the probe test, and the percentage of time spent in the target quadrant where the platform was previously located was examined in comparison to other quadrants. Tfap2a+/− mice and their littermates spent most of the time in the target quadrant, suggesting that they learnt and remembered the previous location of the platform (Figure 7E). By contrast, Tfap2b+/− mice did not show the preference for the target quadrant as clearly as the wild type (Figure 8E). Together, these data suggest that spatial learning and memory might be impaired in Tfap2b+/− but not in Tfap2a+/− mice.Depression-associated behavior was assessed by SPT (Alkhlaif et al. 2017), FST (Yankelevitch-Yahav et al. 2015) and TST (Can et al. 2012). In Tfap2a+/− and Tfap2b+/− mice, no difference was observed in sucrose preference across all days of the experiment (Figures 7F and 8F). This result suggests that both Tfap2a+/− and Tfap2b+/− mice are able to experience pleasure from the reward (sucrose water). In the FST, there was no significant difference in latency to immobility and time spent immobile in Tfap2a+/− mice, but Tfap2b+/− mice showed a reduced immobility latency and spent more time immobile (Figures 7, G–H and 8, G–H). During the TST, increased immobilization latency and less immobility time were observed in Tfap2a+/− mutants but not in Tfap2b+/− mice (Figures 7, I and J and 8, I and J). Taken together, Tfap2a+/− mice showed signs of hyperactive behavior in the TST. By contrast, Tfap2b+/− mice showed longer periods of immobility in the FST.
We next performed the contextual FC test (Fischer et al. 2004). In the conditioning session, average motion and freezing did not differ significantly between mutants and controls in both Tfap2a+/− and Tfap2b+/− mice. On the first test day after conditioning, both groups of mice showed signs of fear memory as evidenced by increased freezing time. On the second test day, both Tfap2a+/− and Tfap2b+/− mice exhibited higher motions and less freezing time compared to the respective controls, indicating impaired reconsolidation (Figures 7, K and L and 8, K and L).
In summary, motor function and balance in both Tfap2a+/− and Tfap2b+/− mice are comparable to wild-type controls. Emotion-associated behavior, assayed by the response to positive sensation (sucrose water) or mild environmental stress (e.g., EPM), in both Tfap2a+/− and Tfap2b+/− mice also appears to be comparable to wild-type controls. In MWM tests, Tfap2b+/− mice showed severe impairment in finding the platform during training. However, the difference between Tfap2b+/− and controls was less severe in the probe test, where Tfap2b+/− still showed preference of the target quadrant, even though this preference did not reach statistical significance to the neighboring left quadrant. This suggests that, perhaps, spatial memory in Tfap2b+/− is less accurate. In addition, the lack of platform arrival might have stimulated expanded search behavior also in other quadrants. Interestingly, Tfap2a+/− mice appear to be less stressed when exposed to the TST, whereas Tfap2b+/− mice tend to react negatively in this test. Tfap2b+/− mice have impaired spatial as well as contextual fear memory. Tfap2a+/− mutants have reduced freezing time during FC, but this might be due to the hyperactivity rather than impaired fear memory. These results suggest that the different AP-2 mutants display at least partially divergent behavioral characteristics.
The free-running period of the circadian rhythm is shortened in Tfap2b+/− mice
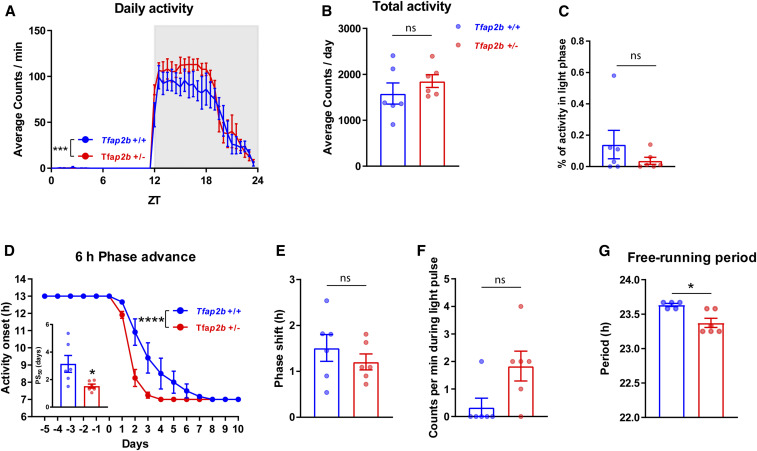
Sleep is strongly regulated by the circadian system. We thus explored circadian rest-activity regulation in Tfap2a+/− and Tfap2b+/− mice. We measured wheel-running activity under baseline conditions (12 hr light: 12 hr dark - LD), during re-entrainment to a 6 hr phase advance of the LD cycle, in response to a nocturnal light pulse, and under constant dark (DD) conditions (Zheng et al. 2001) (Figures 9 and 10). Under LD conditions, Tfap2a+/− and Tfap2b+/− mice did not show significant differences in their rest-activity patterns compared to controls (Figures 9, A–C and 10, A–C). We next measured the response of Tfap2a+/− and Tfap2b+/− mice to a shift in the LD cycle by advancing the dark phase by 6 hr (jetlag paradigm). Re-entrainment to the new cycle was normal in Tfap2a+/− mice compared with wild-type controls (Figure 9D), but it was significantly faster in Tfap2b+/− mice (Figure 10D).
Figure 9.
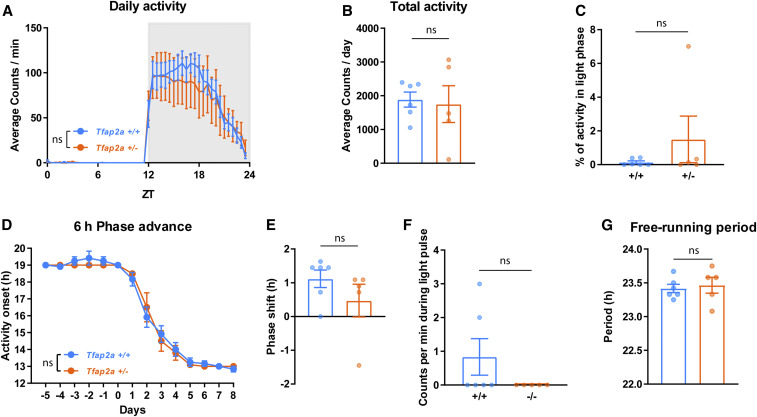
Circadian rhythms are comparable to wild-type controls in Tfap2a+/− mice. (A) Daily activity, two-way ANOVA tests followed by Sidak’s pairwise comparison, the main effect of genotype: F(1, 432) = 1.133, P = 0.288; the main effect of time, F(47, 432) = 21.64, P < 0.0001. (B) Total activity, P = 0.931, Mann-Whitney test. (C) Activity in light phase, P = 0.784, Mann-Whitney test. (D) Six hours phase advance, Two-way ANOVA tests followed by Sidak’s pairwise comparison, the main effect of genotype: F(1, 126) = 0.169, P = 0.682; the main effect of time, F(13, 126) = 156.8, P < 0.0001. (E) Phase shift, P = 0.197, Mann-Whitney test. (F) Locomotor activity during light pulse, P = 0.455, Mann-Whitney test. (G) Free-running period during constant darkness, P = 0.626, Mann-Whitney test. All data are shown as the mean ± SEM n = 6 for Tfap2a+/+, n = 5 for Tfap2a+/−.
Figure 10.
Circadian period is shortened and jetlag re-entrainment is accelerated in Tfap2b+/− mice. (A) Daily activity, two-way ANOVA test followed by Sidak’s pairwise comparison, the main effect of genotype: F(1, 480) = 13.12, ***P = 0.0003; the main effect of time, F(47, 480) = 65.66, P < 0.0001. (B) Total activity, P = 0.310, Mann-Whitney test. (C) Activity in light phase, P = 0.558, Mann-Whitney test. (D) Six hours phase advance two-way ANOVA test followed by Sidak’s pairwise comparison, the main effect of genotype, F(1, 160) = 24.82, ****P < 0.0001; the main effect of time, F(15, 160) = 161.1, P < 0.0001; 50% phase shift (PS50): two-tailed unpaired t-test, *P = 0.0273. (E) Phase shift, P = 0.452, Mann-Whitney test. (F) Locomotor activity during light pulse, P = 0.558, Mann-Whitney test. (G) Free-running period during constant darkness, *P = 0.022, Mann-Whitney test. All data are shown as the mean ± SEM n = 6 for Tfap2b+/+, n = 6 for Tfap2b+/−.
To measure the capacity of light to phase reset the circadian clock system of Tfap2a+/− and Tfap2b+/− mice, a 30-min light pulse was delivered at ZT14 (2 hr after “lights off”) and mice were released into DD, which, in wild-type mice, normally induces a phase delay in their rest-activity rhythm (Schwartz and Zimmerman 1990). Light induced similar amounts of phase delays in both Tfap2a+/− and Tfap2b+/− mice, and the acute reduction of activity during the 30-min light pulse (a.k.a. negative light masking) was not significantly different between mutants and controls (Figures 9, E and F and 10, E and F). There was no difference in DD free-running periods in Tfap2a+/− mice, but a shorter period length was observed in Tfap2b+/− mice (Figures 9G and 10G). In summary, circadian rhythms in Tfap2a+/− mice were comparable to wild-type controls. In Tfap2b+/− mice, a shorter intrinsic clock period may have facilitated the faster entrainment in the jetlag paradigm. Speculatively, the observed reduction in sleep amount in these mice might also facilitate such clock acceleration.
Divergent differential gene expression in Tfap2a+/− and Tfap2b+/− mice
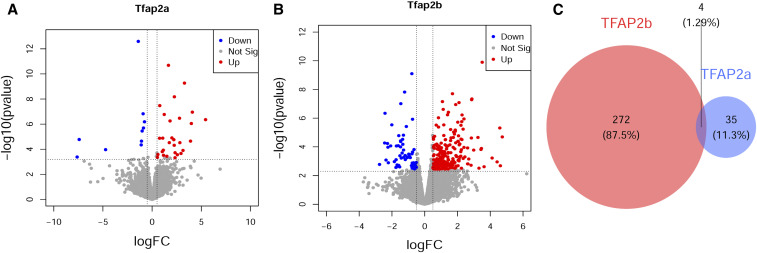
The divergent effects of Tfap2a+/− and Tfap2b+/− on behavior might stem from different gene expression caused by reduction of the amount of the respective transcription factor. We thus measured gene expression changes in the brain in Tfap2a+/−, Tfap2b+/−, and wild type controls. For this, we took brain samples from these mutants and controls, determined their transcriptomes using RNA sequencing, and extracted differentially expressed genes. We found 276 genes to be significantly differentially expressed in Tfap2b+/− compared with wild types, whereas only 39 genes were found to be differentially expressed in Tfap2a+/−. Thus, the more severe behavioral and sleep phenotypes in Tfap2b+/− correlate with increased differences in gene expression (Figure 11). Only one gene was downregulated and three genes were upregulated in both Tfap2a+/− and Tfap2b+/− (Supplemental excel workbook), while all other genes were unique to either of the mutants. This broad divergence in gene expression is consistent with the divergence in sleep and behavior. The differentially expressed genes could potentially explain the sleep and behavioral phenotypes observed in Tfap2a+/− and Tfap2b+/−. We hence searched the literature for these genes to find phenotypes associated with sleep and the behavioral tests that we have performed. Only one gene that is differentially expressed in Tfap2a+/− has been previously associated with sleep and none of the genes has been described to have phenotypes in the behavioral tests. By contrast, four genes that are differentially expressed in Tfap2b+/− have been associated with sleep phenotypes, and eight genes have been associated with phenotypes in the behavioral tests that we have performed (see details in Table 1). Future studies will be required to establish causality and to solve mechanisms by which Tfap2a+/− and Tfap2b+/− affect behavior.
Figure 11.
Divergent gene expression changes in Tfap2a+/− and Tfap2b+/−. (A) Volcano plot of genes that are differentially expressed in Tfap2a+/− mice. (B) Volcano plot of genes that are differentially expressed in Tfap2b+/− mice. (C) Venn diagram showing distinct and overlapping genes that are differentially expressed [false discovery rate (FDR) < 0.20] in Tfap2a+/− and Tfap2b+/−. The overlapping P value was calculated using Fischer’s Exact tests, P = 0.005. n = 3 for B6N, n = 3 for Tfap2a+/−, n = 3 for Tfap2b+/−. Genes that are differentially expressed at FDR < 0.20 and logFC > 0.5 are highlighted and labeled in the volcano plot.
Table 1. Association of Tfap2b and Tfap2a-controlled genes with sleep and behavioral phenotypes.
| Assay | Tfap2a+/− | Tfap2b+/− | ||
|---|---|---|---|---|
| Phenotype | DE genes | Phenotype | DE genes | |
| EEG/EMG recording | Normal sleep amount but increased sleep power | Slc5a5a | Shortened sleep amount and impaired sleep quality | Fosa, Fosba, Slc5a5a Slc18a2b |
| EPM | Normal anxiety-associated behavior | — | Normal anxiety-associated behavior | — |
| Rotarod | Normal motor function | — | Normal motor function | — |
| MWM | Normal spatial learning and memory | — | Impaired spatial learning and memory | Arca, Fosa, Fosba, Vgfa |
| SPT, FST, TST | Hyperactive in TST, but normal in SPT or TST | — | Depression-like behavior only in FST, but normal in SPT or TST | Fosba, Dusp1a |
| Slc6a4b | ||||
| Contextual fear conditioning | Shortened freezing behavior | — | Shortened freezing behavior | Arca, Vgfa |
| Slc6a4b | ||||
| Wheel-running | Normal circadian activity | — | Shortened circadian period and accelerated re-entrainment | Vgfa |
Literature analysis of genes that are differentially expressed in Tfap2a+/− and TFAP2b+/−. All interpretations are purely speculative. Fos and Fosb from the Fos family of transcription factors as well as Slc5a5 and Slc18a2 from the solute carrier (SLC) group of membrane transport proteins were found to be differentially regulated in Tfap2b+/− mice, which might contribute to the shortened sleep (Figure 3). Slc5a5, which encodes a sodium/iodide symporter (NIS), was down regulated in both Tfap2a+/− and Tfap2b+/− mice. The NIS plays a fundamental role in the first step in thyroid hormone biosynthesis (Dohán et al. 2003). Multiple phenotypes relating to neurological features, skeleton, vision, and metabolism have been discovered in Slc5a5 knockouts. Among those phenotypes, abnormal sleep behavior, such as shorter sleep bout duration during the dark phase, has been found in male Slc5a5−/− mice (Dickinson et al. 2016). Fos (c-fos) is a nuclear proto-oncogene, whose expression is used as an indirect marker of neuronal activity. Fosb expression is induced often in the same cells as Fos, but at a later time (Gass et al. 1992; Peters et al. 1994). In Tfap2b+/− mice, Fos and Fosb were downregulated. Abnormal sleep has been found in Fos or Fosb-deficient mice such that Fos-null mice have less NREMS and normal REM sleep, but Fosb-deficient mice have less REMS but unchanged NREMS (Shiromani et al. 2000). Dopamine is transported into synaptic vesicles by the vesicular monoamine transporter (VMAT2), which is encoded by Slc18a2. VMAT2-deficient mice are used as a model of Parkinson’s Disease (PD) that exhibits shorter latency to sleep and lower circadian activity except for a major phenotype of the motor dysfunction (Taylor et al. 2009). In Tfap2b+/− animals, Slc18a2 was upregulated, and overexpression of this gene was reported to have neuroprotective effects, such as antidepressive and anxiolytic activity and increased ambulation during the dark phase (Lohr et al. 2014). The downregulation of Arc, Fos, Fosb, and Vgf might be associated with the impaired spatial learning and memory that we found in Tfap2b+/− animals (Figure 8, C–E). The deletion of either Arc, Fos, Fosb, or Vgf in mice result in impaired spatial learning (Paylor et al. 1994; Plath et al. 2006; Bozdagi et al. 2008; Ohnishi et al. 2011). Increased immobility was observed in Tfap2b+/− mice (Figure 8H), which is consistent with downregulation of Fosb and Duspt1 as well as upregulation of Slc6a4. Fosb null mice have increased immobility in FST compared with their controls (Ohnishi et al. 2011). On the contrary, overexpression of Dusp1 causes depressive behaviors and mice lacking Dusp-1 are resilient to stress (Duric et al. 2010). Slc6a4 encodes the serotonin transporter (SERT) in mice. It is reported that mice that are more susceptible to stress have increased expression of SERT and exhibit longer immobility in FST (Couch et al. 2013). In addition, SERT overexpression (5-HTTOE) mice have reduced freezing time in the cued fear conditioning test (McHugh et al. 2015). In Tfap2b+/− mice, we found a shortened freezing time in the contextual fear conditioning test (Figure 8L). The downregulation of Arc and Vgf might also contribute to this fear-related phenotype, as Arc and Vgf null mice exhibit shortened freezing time in the contextual fear conditioning test (Plath et al. 2006; Bozdagi et al. 2008). Moreover, Vgf−/− mice have a slightly shortened circadian period length (Hahm et al. 1999), which might help explain the similar phenotype we observed in Tfap2b+/− mice (Figure 10).
Upregulated genes.
Downregulated genes. “—” indicates that no information was found.
Discussion
Divergent phenotypes in sleep, behavior, circadian rhythms, and gene expression in Tfap2a+/– and Tfap2b+/–
After gene duplication events, paralogs can take on different functions. In mice, both Tfap2a and b genes play an important role during the development of the neural crest (Mander et al. 2013) that gives rise to most of the peripheral nervous system (PNS) and to several non-neural cell types, including smooth muscle cells of the cardiovascular system, pigment cells in the skin, and craniofacial bones (Moser et al. 1997b). Despite their common expression pattern during early embryonic stages, deletion of Tfap2a or Tfap2b produces strikingly different phenotypes (Zhang et al. 1996). The loss of Tfap2a causes neuronal, craniofacial, skeletal, and body wall defects, whereas Tfap2b-deficient mice show brain development, ductus arteriosus, and renal impairments (Zhang et al. 1996; Moser et al. 1997a; Brewer et al. 2004).
Functional divergence in AP-2 is also seen for sleep phenotypes. Tfap2a+/− displays a rather increased sleep quality, whereas sleep length and quality in Tfap2b+/− are reduced. Homeostatic regulation exists in both Tfap2a+/− and Tfap2b+/−, but a stronger response was found in the former and a weaker one in the latter. Our work from C. elegans and Drosophila indicated that the ancient function of AP-2 appears to be to promote sleep (Turek et al. 2013; Kucherenko et al. 2016). This suggests that Tfap2b+/− may have kept the original sleep-promoting function, whereas Tfap2a+/− may have taken on a new, opposing function in sleep control. Thus, as sleep evolved in more complex brains, there might have emerged a need for negative control of sleep that favored the divergence of AP-2 transcription factors.
We show that this functional divergence extended to additional behaviors. Tfap2a+/− was more robust in the behavioral assays, at least in the TST, which indicated a mild hyperactivity. By contrast, the performance of Tfap2b+/− revealed mildly depressive-like symptoms. Consistent with diverging roles in development and behavior, our RNA-sequencing data of Tfap2a+/− and Tfap2b+/− mice revealed divergent patterns of gene expression. The divergent behavioral changes observed in Tfap2a+/− and Tfap2b+/− mice may, thus, speculatively result from the modulation of expression of Tfap2a and Tfap2b target genes. Consistent with this view, Tfap2a and Tfap2b have been shown to play distinct roles in the specification of GABAergic neurons (Zainolabidin et al. 2017). This finding is intriguing as sleep-active, sleep-inducing neurons typically are GABAergic. Alternatively, changes in depressive-like symptoms in the mutants may be the consequence of sleep quality alterations. As poor sleep quality impairs memory consolidation (Mander et al. 2013; Bezdicek et al. 2018), the memory deficits shown in Tfap2b+/− mice might be the result of a loss of sleep time or quality. Tfap2a+/− mice showed less freezing in FC despite a better sleep quality. However, since Tfap2a+/− mice exhibited hyperactive behavior in a stressful situation (TST), this effect might speculatively have blunted the freezing response in FC.
Different phenotypes were also found in circadian rhythm regulation for Tfap2a+/− and Tfap2b+/− mice. Circadian rhythmicity was unaffected in Tfap2a+/−, whereas Tfap2b+/− showed a shortened period of the internal clock system. The circadian period shortening in Tfap2b+/− most likely is too mild to be the cause of the sleep impairment. For example, mice lacking the clock gene Per1 with 1 hr shorter intrinsic period and preserved homeostatic responses do not exhibit overall sleep changes (Kopp et al. 2002).
Our results suggest that AP-2 transcription factors have diverged to take on divergent control of sleep and other behaviors. To our knowledge, this is the first instance where a sleep gene is shown to have diversified in evolution from a sleep-promoting role in invertebrates to serve bidirectional control of sleep in mammals.
Acknowledgments
We thank Ahmed Mansouri, Mayumi Kimura, Anja Ronnenberg, Ulrike Teichmann, and Sara Kimmina for advice on electroencephalogram (EEG), animal experimentation, and help with obtaining permits. We are grateful to Gregor Eichele for providing laboratory space and resources. We thank Markus Moser and Trever Williams for mouse strains. Sequencing was carried out by Bernd Timmermann and Stefan Börno, Max Planck Institute for Molecular Genetics, Berlin. This work was supported by the Max Planck Society (Max Planck Research Group “Sleep and Waking”), by a European Research Council Starting Grant (ID: 637860, SLEEPCONTROL), and by Deutsche Forschungsgemeinschaft (DFG) grants AS547/1-1 and OS353-10/1.
Footnotes
Supplemental material available at figshare: https://doi.org/10.6084/m9.figshare.12738545.v1.
Communicating editor: J. Schimenti
Literature Cited
- Alkhlaif Y., Bagdas D., Jackson A., Park A. J., and Damaj I. M., 2017. Assessment of nicotine withdrawal-induced changes in sucrose preference in mice. Pharmacol. Biochem. Behav. 161: 47–52. 10.1016/j.pbb.2017.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard K., Logsdon N. J., Ravi S., Xie N., Persons B. P. et al. , 2015. Metabolic reprogramming is required for myofibroblast contractility and differentiation. J. Biol. Chem. 290: 25427–25438. 10.1074/jbc.M115.646984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezdicek O., Nikolai T., Nepozitek J., Perinova P., Kemlink D. et al. , 2018. Prospective memory impairment in idiopathic REM sleep behavior disorder. Clin. Neuropsychol. 32: 1019–1037. 10.1080/13854046.2017.1394493 [DOI] [PubMed] [Google Scholar]
- Bozdagi O., Rich E., Tronel S., Sadahiro M., Patterson K. et al. , 2008. The neurotrophin-inducible gene Vgf regulates hippocampal function and behavior through a brain-derived neurotrophic factor-dependent mechanism. J. Neurosci. 28: 9857–9869. 10.1523/JNEUROSCI.3145-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer S., Feng W., Huang J., Sullivan S., and Williams T., 2004. Wnt1-Cre-mediated deletion of AP-2alpha causes multiple neural crest-related defects. Dev. Biol. 267: 135–152. 10.1016/j.ydbio.2003.10.039 [DOI] [PubMed] [Google Scholar]
- Bringmann H., 2018. Sleep-active neurons: conserved motors of sleep. Genetics 208: 1279–1289. 10.1534/genetics.117.300521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell S. S., and Tobler I., 1984. Animal sleep: a review of sleep duration across phylogeny. Neurosci. Biobehav. Rev. 8: 269–300. 10.1016/0149-7634(84)90054-X [DOI] [PubMed] [Google Scholar]
- Can A., Dao D. T., Terrillion C. E., Piantadosi S. C., Bhat S. et al. , 2012. The tail suspension test. J. Vis. Exp. 59: e 3769 10.3791/3769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chazaud C., Oulad-Abdelghani M., Bouillet P., Decimo D., Chambon P. et al. , 1996. AP-2.2, a novel gene related to AP-2, is expressed in the forebrain, limbs and face during mouse embryogenesis. Mech. Dev. 54: 83–94. 10.1016/0925-4773(95)00463-7 [DOI] [PubMed] [Google Scholar]
- Colavito V., Fabene P. F., Grassi-Zucconi G., Pifferi F., Lamberty Y. et al. , 2013. Experimental sleep deprivation as a tool to test memory deficits in rodents. Front. Syst. Neurosci. 7: 106 10.3389/fnsys.2013.00106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couch Y., Anthony D. C., Dolgov O., Revischin A., Festoff B. et al. , 2013. Microglial activation, increased TNF and SERT expression in the prefrontal cortex define stress-altered behaviour in mice susceptible to anhedonia. Brain Behav. Immun. 29: 136–146 [Corrigenda: Brain Behav. Immun. 36: 215 (2014)]. 10.1016/j.bbi.2012.12.017 [DOI] [PubMed] [Google Scholar]
- Deacon R. M., 2013. Measuring motor coordination in mice. J. Vis. Exp. 75: e 2609 10.3791/2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson M. E., Flenniken A. M., Ji X., Teboul L., Wong M. D. et al. , 2016. High-throughput discovery of novel developmental phenotypes. Nature 537: 508–514 [Corrigenda: Nature 551: 398 (2017)]. 10.1038/nature19356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C. et al. , 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohán O., De la Vieja A., Paroder V., Riedel C., Artani M. et al. , 2003. The sodium/iodide Symporter (NIS): characterization, regulation, and medical significance. Endocr. Rev. 24: 48–77. 10.1210/er.2001-0029 [DOI] [PubMed] [Google Scholar]
- Duric V., Banasr M., Licznerski P., Schmidt H. D., Stockmeier C. A. et al. , 2010. A negative regulator of MAP kinase causes depressive behavior. Nat. Med. 16: 1328–1332. 10.1038/nm.2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert D., Buhl S., Weber S., Jager R., and Schorle H., 2005. The AP-2 family of transcription factors. Genome Biol. 6: 246 10.1186/gb-2005-6-13-246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer A., Sananbenesi F., Schrick C., Spiess J., and Radulovic J., 2004. Distinct roles of hippocampal de novo protein synthesis and actin rearrangement in extinction of contextual fear. J. Neurosci. 24: 1962–1966. 10.1523/JNEUROSCI.5112-03.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funato H., Miyoshi C., Fujiyama T., Kanda T., Sato M. et al. , 2016. Forward-genetics analysis of sleep in randomly mutagenized mice. Nature 539: 378–383. 10.1038/nature20142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao V., Turek F., and Vitaterna M., 2016. Multiple classifier systems for automatic sleep scoring in mice. J. Neurosci. Methods 264: 33–39. 10.1016/j.jneumeth.2016.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gass P., Herdegen T., Bravo R., and Kiessling M., 1992. Induction of immediate early gene encoded proteins in the rat hippocampus after bicuculline-induced seizures: differential expression of KROX-24, FOS and JUN proteins. Neuroscience 48: 315–324. 10.1016/0306-4522(92)90493-L [DOI] [PubMed] [Google Scholar]
- Gibney P. A., Lu C., Caudy A. A., Hess D. C., and Botstein D., 2013. Yeast metabolic and signaling genes are required for heat-shock survival and have little overlap with the heat-induced genes. Proc. Natl. Acad. Sci. USA 110: E4393–E4402. 10.1073/pnas.1318100110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green R. M., Feng W., Phang T., Fish J. L., Li H. et al. , 2015. Tfap2a-dependent changes in mouse facial morphology result in clefting that can be ameliorated by a reduction in Fgf8 gene dosage. Dis. Model. Mech. 8: 31–43. 10.1242/dmm.017616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubbs J. J., Lopes L. L., van der Linden A. M., and Raizen D. M., 2019. Sik-Hdac4 signaling is required for the metabolic regulation of sleep. Sleep 42: A11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahm S., Mizuno T. M., Wu T. J., Wisor J. P., Priest C. A. et al. , 1999. Targeted deletion of the Vgf gene indicates that the encoded secretory peptide precursor plays a novel role in the regulation of energy balance. Neuron 23: 537–548. 10.1016/S0896-6273(00)80806-5 [DOI] [PubMed] [Google Scholar]
- Joiner W. J., 2016. Unraveling the evolutionary determinants of sleep. Curr. Biol. 26: R1073–R1087. 10.1016/j.cub.2016.08.068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene A. C., and Duboue E. R., 2018. The origins and evolution of sleep. J. Exp. Biol. 221: jeb159533. 10.1242/jeb.159533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent B. A., Strittmatter S. M., and Nygaard H. B., 2018. Sleep and EEG power spectral analysis in three transgenic mouse models of Alzheimer’s disease: APP/PS1, 3xTgAD, and Tg2576. J. Alzheimers Dis. 64: 1325–1336. 10.3233/JAD-180260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlbecker A., Lee A. E., and Schorle H., 2002. Exencephaly in a subset of animals heterozygous for AP-2alpha mutation. Teratology 65: 213–218. 10.1002/tera.10037 [DOI] [PubMed] [Google Scholar]
- Kopp C., Albrecht U., Zheng B., and Tobler I., 2002. Homeostatic sleep regulation is preserved in mPer1 and mPer2 mutant mice. Eur. J. Neurosci. 16: 1099–1106. 10.1046/j.1460-9568.2002.02156.x [DOI] [PubMed] [Google Scholar]
- Kucherenko M. M., Ilangovan V., Herzig B., Shcherbata H. R., and Bringmann H., 2016. TfAP-2 is required for night sleep in Drosophila. BMC Neurosci. 17: 72 10.1186/s12868-016-0306-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J. M., Taroc E. Z. M., Frias J. A., Prasad A., Catizone A. N. et al. , 2018. The transcription factor Tfap2e/AP-2ε plays a pivotal role in maintaining the identity of basal vomeronasal sensory neurons. Dev. Biol. 441: 67–82. 10.1016/j.ydbio.2018.06.007 [DOI] [PubMed] [Google Scholar]
- Li H., Sheridan R., and Williams T., 2013. Analysis of TFAP2A mutations in Branchio-Oculo-Facial Syndrome indicates functional complexity within the AP-2alpha DNA-binding domain. Hum. Mol. Genet. 22: 3195–3206. 10.1093/hmg/ddt173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr K. M., Bernstein A. I., Stout K. A., Dunn A. R., Lazo C. R. et al. , 2014. Increased vesicular monoamine transporter enhances dopamine release and opposes Parkinson disease-related neurodegeneration in vivo. Proc. Natl. Acad. Sci. USA 111: 9977–9982. 10.1073/pnas.1402134111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mander B. A., Rao V., Lu B., Saletin J. M., Lindquist J. R. et al. , 2013. Prefrontal atrophy, disrupted NREM slow waves and impaired hippocampal-dependent memory in aging. Nat. Neurosci. 16: 357–364. 10.1038/nn.3324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mang G. M., and Franken P., 2012. Sleep and EEG phenotyping in mice. Curr. Protoc. Mouse Biol. 2: 55–74. [DOI] [PubMed] [Google Scholar]
- Mani A., Radhakrishnan J., Farhi A., Carew K. S., Warnes C. A. et al. , 2005. Syndromic patent ductus arteriosus: evidence for haploinsufficient TFAP2B mutations and identification of a linked sleep disorder. Proc. Natl. Acad. Sci. USA 102: 2975–2979. 10.1073/pnas.0409852102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M., 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. 17: 3. [Google Scholar]
- Matsuda R., Kohno T., Kohsaka S., Fukuoka R., Maekawa Y. et al. , 2017. The prevalence of poor sleep quality and its association with depression and anxiety scores in patients admitted for cardiovascular disease: a cross-sectional designed study. Int. J. Cardiol. 228: 977–982. 10.1016/j.ijcard.2016.11.091 [DOI] [PubMed] [Google Scholar]
- McHugh S. B., Barkus C., Lima J., Glover L. R., Sharp T. et al. , 2015. SERT and uncertainty: serotonin transporter expression influences information processing biases for ambiguous aversive cues in mice. Genes Brain Behav. 14: 330–336. 10.1111/gbb.12215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki S., Liu C. Y., and Hayashi Y., 2017. Sleep in vertebrate and invertebrate animals, and insights into the function and evolution of sleep. Neurosci. Res. 118: 3–12. 10.1016/j.neures.2017.04.017 [DOI] [PubMed] [Google Scholar]
- Moser M., Pscherer A., Roth C., Becker J., Mucher G. et al. , 1997a Enhanced apoptotic cell death of renal epithelial cells in mice lacking transcription factor AP-2beta. Genes Dev. 11: 1938–1948. 10.1101/gad.11.15.1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser M., Ruschoff J., and Buettner R., 1997b Comparative analysis of AP-2 alpha and AP-2 beta gene expression during murine embryogenesis. Dev. Dyn. 208: 115–124. [DOI] [PubMed] [Google Scholar]
- Ohnishi Y. N., Ohnishi Y. H., Hokama M., Nomaru H., Yamazaki K. et al. , 2011. FosB is essential for the enhancement of stress tolerance and antagonizes locomotor sensitization by DeltaFosB. Biol. Psychiatry 70: 487–495. 10.1016/j.biopsych.2011.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paylor R., Johnson R. S., Papaioannou V., Spiegelman B. M., and Wehner J. M., 1994. Behavioral assessment of c-fos mutant mice. Brain Res. 651: 275–282. 10.1016/0006-8993(94)90707-2 [DOI] [PubMed] [Google Scholar]
- Peters R. V., Aronin N., and Schwartz W. J., 1994. Circadian regulation of Fos B is different from c-Fos in the rat suprachiasmatic nucleus. Brain Res. Mol. Brain Res. 27: 243–248. 10.1016/0169-328X(94)90006-X [DOI] [PubMed] [Google Scholar]
- Plath N., Ohana O., Dammermann B., Errington M. L., Schmitz D. et al. , 2006. Arc/Arg3.1 is essential for the consolidation of synaptic plasticity and memories. Neuron 52: 437–444. 10.1016/j.neuron.2006.08.024 [DOI] [PubMed] [Google Scholar]
- Radyushkin K., Hammerschmidt K., Boretius S., Varoqueaux F., El-Kordi A. et al. , 2009. Neuroligin-3-deficient mice: model of a monogenic heritable form of autism with an olfactory deficit. Genes Brain Behav. 8: 416–425. 10.1111/j.1601-183X.2009.00487.x [DOI] [PubMed] [Google Scholar]
- Raizen D. M., and Zimmerman J. E., 2011. Non-mammalian genetic model systems in sleep research. Sleep Med. Clin. 6: 131–139. 10.1016/j.jsmc.2011.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts G. G. 3rd, and Hudson A. P., 2009. Rsf1p is required for an efficient metabolic shift from fermentative to glycerol-based respiratory growth in S. cerevisiae. Yeast 26: 95–110. 10.1002/yea.1655 [DOI] [PubMed] [Google Scholar]
- Sananbenesi F., Fischer A., Wang X., Schrick C., Neve R. et al. , 2007. A hippocampal Cdk5 pathway regulates extinction of contextual fear. Nat. Neurosci. 10: 1012–1019. 10.1038/nn1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoda M., Pierpont M. E., Diaz G. A., Bornemeier R. A., and Gelb B. D., 1999. Char syndrome, an inherited disorder with patent ductus arteriosus, maps to chromosome 6p12-p21. Circulation 99: 3036–3042. 10.1161/01.CIR.99.23.3036 [DOI] [PubMed] [Google Scholar]
- Satoda M., Zhao F., Diaz G. A., Burn J., Goodship J. et al. , 2000. Mutations in TFAP2B cause Char syndrome, a familial form of patent ductus arteriosus. Nat. Genet. 25: 42–46. 10.1038/75578 [DOI] [PubMed] [Google Scholar]
- Schorle H., Meier P., Buchert M., Jaenisch R., and Mitchell P. J., 1996. Transcription factor AP-2 essential for cranial closure and craniofacial development. Nature 381: 235–238. 10.1038/381235a0 [DOI] [PubMed] [Google Scholar]
- Schwartz W. J., and Zimmerman P., 1990. Circadian timekeeping in BALB/c and C57BL/6 inbred mouse strains. J. Neurosci. 10: 3685–3694. 10.1523/JNEUROSCI.10-11-03685.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiromani P. J., Basheer R., Thakkar J., Wagner D., Greco M. A. et al. , 2000. Sleep and wakefulness in c-fos and fos B gene knockout mice. Brain Res. Mol. Brain Res. 80: 75–87. 10.1016/S0169-328X(00)00123-6 [DOI] [PubMed] [Google Scholar]
- Taylor T. N., Caudle W. M., Shepherd K. R., Noorian A., Jackson C. R. et al. , 2009. Nonmotor symptoms of Parkinson’s disease revealed in an animal model with reduced monoamine storage capacity. J. Neurosci. 29: 8103–8113. 10.1523/JNEUROSCI.1495-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobler I., 1995. Is sleep fundamentally different between mammalian species? Behav. Brain Res. 69: 35–41. 10.1016/0166-4328(95)00025-O [DOI] [PubMed] [Google Scholar]
- Turek M., Lewandrowski I., and Bringmann H., 2013. An AP2 transcription factor is required for a sleep-active neuron to induce sleep-like quiescence in C. elegans. Curr. Biol. 23: 2215–2223. 10.1016/j.cub.2013.09.028 [DOI] [PubMed] [Google Scholar]
- Verbitsky E. V., 2017. Anxiety and sleep in experiment and clinic. Zh. Nevrol. Psikhiatr. Im. S. S. Korsakova 117: 12–18. 10.17116/jnevro20171174212-18 [DOI] [PubMed] [Google Scholar]
- Walf A. A., and Frye C. A., 2007. The use of the elevated plus maze as an assay of anxiety-related behavior in rodents. Nat. Protoc. 2: 322–328. 10.1038/nprot.2007.44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werling U., and Schorle H., 2002. Transcription factor gene AP-2 gamma essential for early murine development. Mol. Cell. Biol. 22: 3149–3156. 10.1128/MCB.22.9.3149-3156.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams T., and Tjian R., 1991. Characterization of a dimerization motif in AP-2 and its function in heterologous DNA-binding proteins. Science 251: 1067–1071. 10.1126/science.1998122 [DOI] [PubMed] [Google Scholar]
- Yankelevitch-Yahav R., Franko M., Huly A., and Doron R., 2015. The forced swim test as a model of depressive-like behavior. J. Vis. Exp. 97: 52587 10.3791/52587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zainolabidin N., Kamath S. P., Thanawalla A. R., and Chen A. I., 2017. Distinct activities of Tfap2A and Tfap2B in the specification of GABAergic interneurons in the developing cerebellum. Front. Mol. Neurosci. 10: 281 10.3389/fnmol.2017.00281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Hagopian-Donaldson S., Serbedzija G., Elsemore J., Plehn-Dujowich D. et al. , 1996. Neural tube, skeletal and body wall defects in mice lacking transcription factor AP-2. Nature 381: 238–241. 10.1038/381238a0 [DOI] [PubMed] [Google Scholar]
- Zhao F., Weismann C. G., Satoda M., Pierpont M. E., Sweeney E. et al. , 2001. Novel TFAP2B mutations that cause Char syndrome provide a genotype-phenotype correlation. Am. J. Hum. Genet. 69: 695–703. 10.1086/323410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F., Lufkin T., and Gelb B. D., 2003. Expression of Tfap2d, the gene encoding the transcription factor Ap-2 delta, during mouse embryogenesis. Gene Expr. Patterns 3: 213–217. 10.1016/S1567-133X(02)00067-4 [DOI] [PubMed] [Google Scholar]
- Zheng B., Albrecht U., Kaasik K., Sage M., Lu W. et al. , 2001. Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell 105: 683–694. 10.1016/S0092-8674(01)00380-4 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. All data are available at Dryad: https://doi.org/10.5061/dryad.rv15dv45r. Raw RNA-seq data are available at GEO: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE155629. EEG-autoscore code is available at GitHub: https://zenodo.org/badge/latestdoi/240526669.