Phenotypic convergence between taxa can be caused by divergent genetic evolution (different genetic pathways), parallel genetic evolution (convergent mutations), or collateral evolution (shared ancestry). Heliconius butterflies have bright mimetic color patterns shared between multiple species, making an excellent .....
Keywords: adaptation, cis-regulation, collateral evolution, genetic architecture
Abstract
Convergent evolution can occur through different genetic mechanisms in different species. It is now clear that convergence at the genetic level is also widespread, and can be caused by either (i) parallel genetic evolution, where independently evolved convergent mutations arise in different populations or species, or (ii) collateral evolution in which shared ancestry results from either ancestral polymorphism or introgression among taxa. The adaptive radiation of Heliconius butterflies shows color pattern variation within species, as well as mimetic convergence between species. Using comparisons from across multiple hybrid zones, we use signals of shared ancestry to identify and refine multiple putative regulatory elements in Heliconius melpomene and its comimics, Heliconius elevatus and Heliconius besckei, around three known major color patterning genes: optix, WntA, and cortex. While we find that convergence between H. melpomene and H. elevatus is caused by a complex history of collateral evolution via introgression in the Amazon, convergence between these species in the Guianas appears to have evolved independently. Thus, we find adaptive convergent genetic evolution to be a key driver of regulatory changes that lead to rapid phenotypic changes. Furthermore, we uncover evidence of parallel genetic evolution at some loci around optix and WntA in H. melpomene and its distant comimic Heliconius erato. Ultimately, we show that all three of convergence, conservation, and novelty underlie the modular architecture of Heliconius color pattern mimicry.
CONVERGENT evolution is a natural experiment in repeated evolution of similar traits, and offers unique insights into the evolutionary process (Blount et al. 2018). It is widespread across the tree of life, critical to the composition of ecosystems (Sage et al. 2012) (e.g., the repeated colonization of land/water/air by different taxonomic groups), and underpins the ability of organisms to exploit novel environments (e.g., the repeated evolution of drug/insecticide/drought resistance (Farhat et al. 2013). The genetic changes causing convergence can be categorized as (i) divergent genetic mechanisms, (ii) parallel genetic evolution, or (iii) collateral evolution (Stern 2013). With divergent genetic mechanisms, different loci cause the same phenotype in different lineages. In parallel genetic evolution, different alleles at the same locus cause trait convergence (this includes cases where the same mutation has arisen multiple times) (Tishkoff et al. 2007), whereas in collateral evolution, convergence results from the sharing of alleles that are identical by descent, either because the alleles were present in an ancestral population (Jones et al. 2012), or from the introgression of alleles from one species/taxon to another (Huerta-Sánchez et al. 2014).
Cis-regulatory evolution has been implicated in several examples of convergent evolution in vertebrates (Booker et al. 2016; Partha et al. 2017; Tollis et al. 2018; Feigin et al. 2019), suggesting that trait evolution proceeding via cis-regulatory changes to conserved regulatory pathways may be recurrent and predictable. Cis-regulatory evolution is a powerful mechanism that can result in rapid developmental and physiological changes (Wittkopp and Kalay 2012). This is because multiple enhancers at the same gene can each control the gene’s expression in different cell types or developmental times. In these cases, the modular architecture can isolate the effects of a mutation to a single trait (Feigin et al. 2019), circumventing the pleiotropic effects that might constrain adaptive evolution in the protein-coding sequence. Notable examples in which modular enhancers drive convergent evolution include the gain of melanic wing spots in Drosophila elegans and Drosophila tristis through enhancers of the gene yellow (Prud’homme et al. 2006), coat coloration phenotypes via regulation of the gene Agouti in Peromyscus mice (Steiner et al. 2007; Linnen et al. 2009, 2013), loss of Drosophila larval trichomes through mutations to regulatory regions of ovo/svb (Frankel et al. 2012), and pelvic reduction in sticklebacks due to enhancer deletions at the gene pitx1 (Chan et al. 2010). A modular cis-regulatory architecture has also been proposed as a flexible toolkit controlling wing color patterning in Heliconius butterflies (Wallbank et al. 2016; Van Belleghem et al. 2017).
Müllerian mimicry is ubiquitous among neotropical Heliconius butterflies, with multiple species evolving convergent, bright, aposematic wing color patterns. At the same time, in several species such as Heliconius erato and Heliconius melpomene, phenotypic divergence within species is also present in the form of geographic color pattern races (Figure 1), with color pattern loci easily identifiable in population genomic studies across hybrid zones as clear islands of divergence in the genome (Baxter et al. 2010; Counterman et al. 2010; Nadeau et al. 2013, 2014). Owing to the presence of repeated color pattern phenotypes, Heliconius butterflies are an excellent system for studying the genetic basis of convergent evolution (Baxter et al. 2008; Merrill et al. 2015). The small number of mimicry genes controlling the majority of color pattern elements, have been identified across Heliconius species using a combination of QTL mapping, genome-wide association studies across color pattern hybrid zones, and gene expression studies. Across multiple species, parallel genetic evolution at the genes optix, cortex, and WntA is known to control red-orange pattern elements (Baxter et al. 2008; Reed et al. 2011; Martin et al. 2014; Huber et al. 2015; Lewis et al. 2019), white and yellow pattern elements (Nadeau et al. 2016), and melanic patterning (Martin et al. 2012; Gallant et al. 2014; Mazo-Vargas et al. 2017; Moest et al. 2019; Morris et al. 2019; Van Belleghem et al. 2020), respectively.
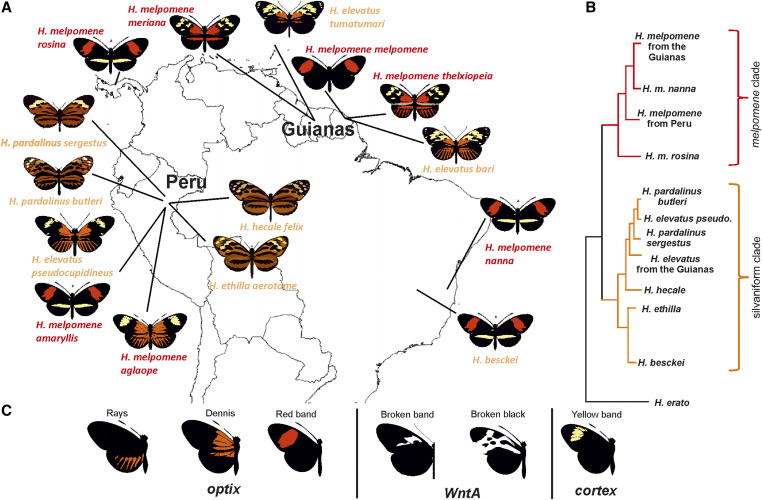
Figure 1.
(A) Map showing geographic distribution of color pattern races of the silvaniform clade species (orange; H. pardalinus, H. elevatus, H. besckei, H. ethilla, and H. hecale) and H. melpomene (red) used in the analyses. The postman color pattern is found in H. melpomene amaryllis/nanna/rosina/melpomene and H. besckei (H. m. melponene lacks the yellow hindwing bar). The dennis-rayed pattern is found in H. melpomene aglaope/thelxiopeia/meriana and H. elevatus pseudocupidineus/bari/tumatumari (H. m. meriana and H. e. tumatumari lack hindwing rays). (B) Cladogram showing the relationships between taxa, based on the species topology inferred here. Note the paraphyly of the species H. elevatus and H. pardalinus (Dasmahapatra et al. 2012). The species H. erato, used here as an outgroup, mimics the appearance of the races of H. melpomene with which it co-occurs. (C) Phenotypes investigated in this study and known to be controlled by the three major wing patterning genes, optix, cortex, and WntA.
Multiple putative regulatory elements have now been identified in H. erato around all three major wing patterning genes (optix, cortex, and WntA) using comparisons between phenotypically distinct races across multiple hybrid zones (Van Belleghem et al. 2017). In the H. melpomene clade the picture is less complete. ABBA-BABA comparisons and changes in phylogenetic topologies have shown that mimetic resemblance between some races of several H. melpomene-silvaniform clade species (H. melpomene, H. elevatus, H. timareta, and H. besckei; Figure 1B) are the result of collateral evolution, via the introgression of color pattern alleles at optix and cortex among the species (Dasmahapatra et al. 2012; Pardo-Diaz et al. 2012; Zhang et al. 2016). Association mapping across a number of H. melpomene and H. timareta taxa in conjunction with recombination breakpoint analysis (which included H. elevatus), was used to define both a 25 kb and an 11 kb regulatory element at optix associated with the presence and absence of the red hindwing rays and the forewing dennis phenotypes, respectively (Wallbank et al. 2016). However, these genomic regions are still relatively large, and no regulatory element for red band has yet been found. Similarly, around cortex it has also been shown that introgression between races of H. melpomene and H. cydno has likely allowed these species to share variation in the hindwing yellow bar phenotype (Enciso-Romero et al. 2017). However, no yellow band regulatory element has been identified at cortex. Furthermore, patterns of introgression and any regulatory elements around WntA are thus far unknown in the H. melpomene-silvaniform clade.
In this study, we investigate the contributions of three genetic modes of evolution, divergent genetic mechanisms, parallel genetic evolution, and collateral evolution to explain the remarkable convergent wing color pattern phenotypes found in Heliconius butterflies. In particular, we use phylogenetic analysis across multiple hybrid zone comparisons to delimit narrow regions of the genome associated with color pattern elements due to shared ancestry (either from introgression or ancestral polymorphism). We do this by identifying genomic regions that show genotype-by-phenotype associations and particular phylogenetic histories consistent with controlling specific wing color pattern phenotypes. This allows us for the first time to look at the mechanism of convergence between Guianese H. melpomene and H. elevatus. We propose that these narrow regions are putative modular regulatory elements, with each controlling a specific wing pattern phenotype. We identify these around all three of the major wing patterning genes; optix, cortex, and WntA, in H. melpomene and its silvaniform mimics and determine the ancestral origins of each element. Finally, we investigate the homology and conservation of these regulatory elements between H. melpomene and H. erato, as well as across other Lepidoptera species.
Materials and Methods
Sample collection and sequencing
We used whole genome sequences of 53 individuals representing six species (H. melpomene, H. elevatus, H. besckei, H. pardalinus, H. ethilla, and H. hecale) and 15 races (Supplemental Materials, Table S1) from two hybrid zones in Peru and the Guianas, as well as two taxa from the Eastern Amazon in Brazil (H. m. nanna and H. besckei) (Figure 1). This includes data from newly sequenced samples of two H. elevatus tumatumari, two H. pardalinus butleri, and one H. elevatus bari. For these new samples, RNA-free genomic DNA was extracted from thoracic tissue using a Qiagen DNeasy Blood and Tissue Kit. Libraries were prepared using Illumina TruSeq DNA PCR-Free Library Preparation Kits with an insert size of ∼350 bp. Libraries were 100 or 125 bp pair-end sequenced to 30–40× coverage on an Illumina HiSeq 2500 instrument at the FAS Center for Systems Biology, Harvard (ENA accession number PRJEB37067).
Variant calling
We aligned sequences to the H. melpomene reference genome v2 (Davey et al. 2016; LepBase http://ensembl.lepbase.org) using BWA MEM (Li and Durbin 2009). We then sorted BAM files using Samtools (Li et al. 2009) and marked duplicate reads using PicardTools (http://broadinstitute.github.io/picard/). We then called genotypes in gVCF format with GATK’s HaplotypeCaller with the parameters -baq CALCULATE_AS_NECESSARY, -hets 0.02 and-emitRefConfidence GVCF, -gt_mode DISCOVERY and–dontUseSoftClippedBases. Subsequently we combined GVCFs before genotyping them, using CombineGVCFs and GenotypeGVCFs respectively (Van der Auwera et al. 2013). Genotypes were then marked as missing (N) with Bcftools v1.3.1 if minimum read depth was <5 or GQ <20, while sites with a minor allele frequency lower than 2/53 across all samples were removed. Python scripts (available at https://github.com/simonhmartin) were used to parse variant call formats (VCFs) to prepare files for use in phylogenetic weighting analyses. SNPs with >10% missing data across taxa were removed.
Identification of candidate regulatory modules based on shared ancestry
Three major loci (cortex, optix, and WntA) control the main color pattern differences between the postman and dennis-rayed races of H. melpomene we examine here (Figure 1; Baxter et al. 2010). Previous studies have shown that mimicry between H. melpomene races and closely related silvaniform species such as H. elevatus and H. besckei is a consequence of shared alleles (Dasmahapatra et al. 2012; Zhang et al. 2016). Therefore, the signal of shared ancestry between comimics can be used to identify narrow genomic intervals that may control color pattern elements (Wallbank et al. 2016; Van Belleghem et al. 2017). We look for these patterns of shared ancestry around cortex, optix, and WntA. Where previous studies have delimited intervals (Wallbank et al. 2016), we constrain our search to these regions.
In order to identify regions that show shared ancestry among different comimetic species at these loci, we employed a descriptive phylogenetic weighting method called Topology Weighting by Iterative Sampling of Subtrees (Twisst; Martin and Van Belleghem 2017). This method provides a quantitative summary of a tree by weighting different subtree topologies according to their occurrence within the tree. Each of our Twisst comparisons used six taxa. We first used RAxML v8.2.4 (Stamatakis 2014) with model -GTRCAT to build maximum-likelihood trees for 100 SNP sliding windows (slide every 25 SNPs) across the entirety of chromosomes 10, 15, and 18 (which contain the major color pattern loci WntA, cortex, and optix, respectively). Trees were built only for windows where all samples had ≥30 SNPs. We used a dynamic threshold as implemented by Twisst to estimate weightings in all analyses, such that trees were sampled until the 95% binomial confidence interval around each weighting was <5%.
For each six taxon Twisst comparison there are 105 potential tree topologies, with just five topologies for each phenotype clearly indicative of shared ancestry related to phenotype (see Figures S2–S6). These trees show monophyly of the nonsister taxa with convergent pattern elements, while the other four taxa group as expected based on the species tree. A topology weighting of 0 indicates that none of the trees for that genomic window were of these five topologies, while a topology weighting of +1 indicates that all of the trees were among these five topologies. We used these six taxon Twisst comparisons to identify putative regions controlling wing pattern elements across three geographic contexts: Peruvian hybrid zone, Guianese hybrid zone, and H. besckei-H. melpomene taxa. These geographically distant comparisons allow semi-independent inferences about genetic elements controlling six wing pattern phenotypes; hindwing red/orange “rays,” forewing proximal red/orange “dennis” patch, and forewing red “band” controlled by optix expression, broken black and broken band forewing variation controlled by WntA expression, and the yellow forewing band controlled by cortex expression (Figure 1C).
Taxon selection for Twisst comparisons
For each Twisst comparison, the six taxa comprised three H. melpomene races and three silvaniform species (see Figure 2, Figure 3, Figure 4, Figure 5, Figure 6, and Table S7 for taxa used in each comparison). In each comparison, two of the H. melpomene taxa are from either side of a hybrid zone across which at least one phenotype of interest differs (e.g., rays vs. no-rays). Across these hybrid zones, gene flow occurs freely along the genome except at regions controlling the phenotypic differences (Nadeau et al. 2012; Martin et al. 2013). The third H. melpomene taxon came from a geographically distant area, and while being genetically distinct from both, always shared the phenotype of one of the two hybrid zone H. melpomene (e.g., no-rays). A silvaniform species (H. elevatus or H. besckei) was included in each comparison that shares the phenotype of interest (e.g., rays) with one of the hybrid zone H. melpomene. The other two silvaniform species included H. ethilla, H. hecale, or H. pardalinus.
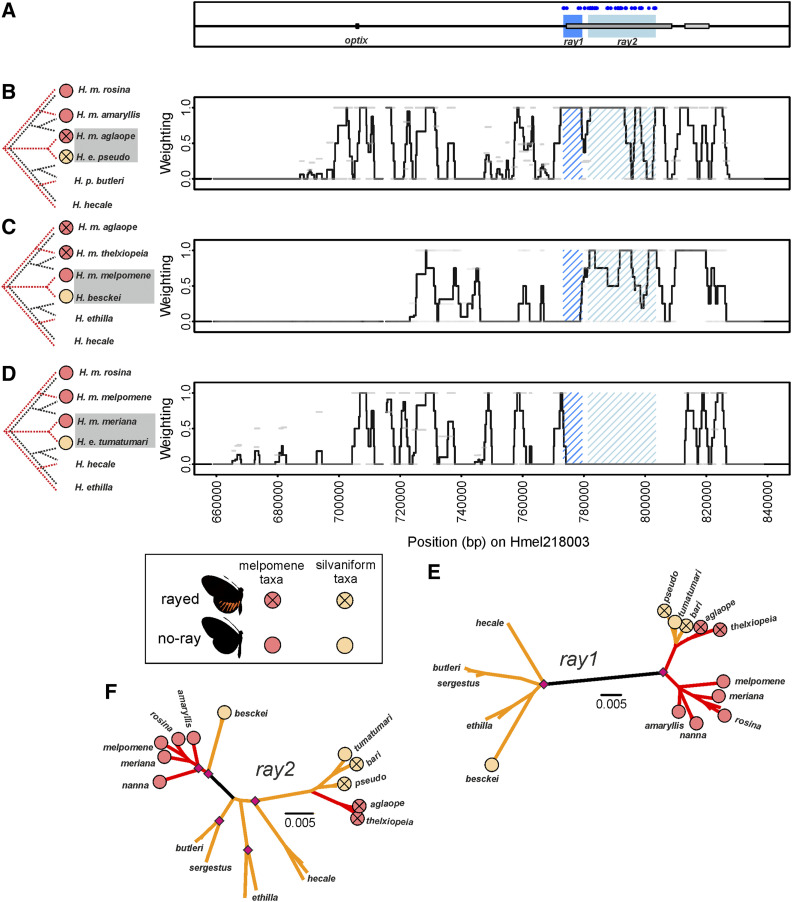
Figure 2.
Twisst comparisons across the optix region of scaffold Hmel218003. (A) Location of optix and ray elements. Dark blue (ray1) and light blue (ray2) shading shows putative functional elements. Gray boxes show ray and dennis regulatory elements respectively as previously delimited in Wallbank et al. (2016). Diagnostic fixed SNPs between phenotypes shown with blue dots in (A). Twisst comparison (100 SNP windows sliding by 25 SNPs) using (B) Peruvian hybrid zone taxa, (C) H. besckei and Guianese hybrid zone H. melpomene, and (D) Guianese hybrid zone taxa. Black trees to left show species topology, while red trees shows groupings (taxa shaded in gray) that indicate shared ancestry between the heterospecific mimetic taxa. Weighting (black line) is the mean from all four overlapping windows for that region, light gray bars show weighting for each 100 SNP window. A weighting of +1 means 100% of trees at that genomic interval show shared ancestry between the heterospecific mimetic taxa. Mimetic phenotypes for taxa are shown by circles; red circles for H. melpomene clade and orange circles for silvaniform taxa. (E and F) Maximum likelihood phylogenies of the ray elements with red branches joining H. melpomene taxa and orange branches joining silvaniform taxa. Node bootstrap support; pink diamonds ≥ 95%, green diamonds 75–94%. Black branch (illustrative only) separates the silvaniform and H. melpomene clades (excluding those taxa where introgression appears to have occurred).
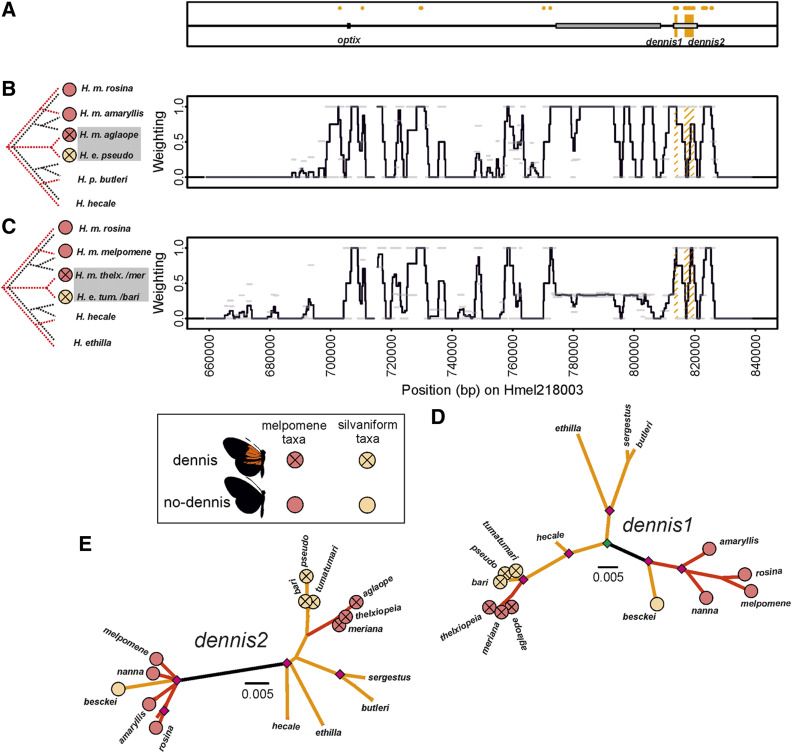
Figure 3.
Twisst comparisons across the optix region of scaffold Hmel218003. (A) Location of optix and dennis elements. Gray boxes show ray and dennis regulatory elements respectively as previously delimited in Wallbank et al. (2016). Orange shading shows putative functional elements, dennis1 and dennis2. Diagnostic fixed SNPS between phenotypes shown with orange dots in (A). Twisst comparison (100 SNP windows sliding by 25 SNPs) using (B) Peruvian hybrid zone taxa; (C) Guianese hybrid zone taxa. Mimetic phenotypes for taxa are shown by circles; red circles for H. melpomene clade and orange circles for silvaniform taxa. (D and E) Maximum likelihood phylogenies of dennis elements; see Figure 2 legend for more detailed explanations. Note position of black branch in dennis2 phylogeny, which has been drawn on the longest branch from where H. besckei connects to phylogeny.
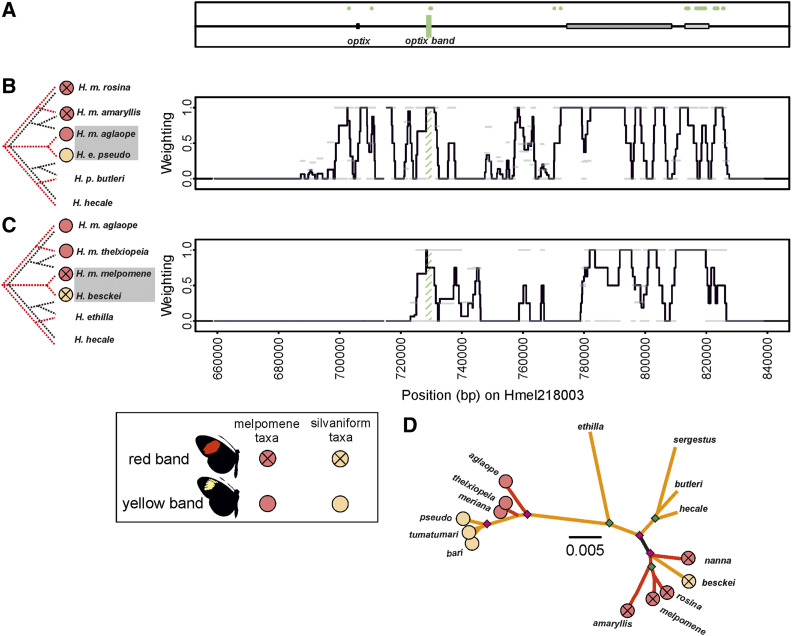
Figure 4.
Twisst comparisons across the optix region of scaffold Hmel218003. (A) Shows location of optix and putative optix band element. Gray boxes show ray and dennis regulatory elements respectively as previously delimited in Wallbank et al. (2016). Green shading shows putative functional elements. Diagnostic fixed SNPS between phenotypes shown with green dots in (A). Twisst comparison (100 SNP windows sliding by 25 SNPs) using (B) Peruvian hybrid zone taxa, and (C) H. besckei and Guianese hybrid zone taxa. Mimetic phenotypes for taxa are shown by circles; red circles for H. melpomene clade and orange circles for silvaniform taxa. (D) Maximum likelihood phylogeny of putative optix band element; see Figure 2 legend for more detailed explanations.
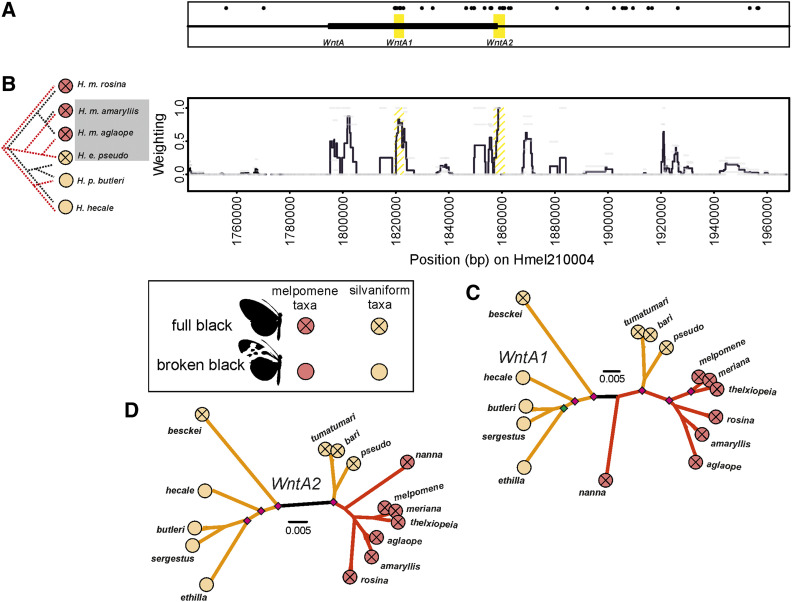
Figure 5.
Paired phylogenetic Twisst comparison across the WntA region of scaffold Hmel210004. (A) Shows location of WntA, and putative functional elements. Black bar shows the WntA gene, yellow shading shows putative regulatory elements. Diagnostic fixed SNPS between phenotypes shown with black dots in (A). (B) Twisst comparison (100 SNP windows sliding by 25 SNPs) with Peruvian hybrid zone taxa. Mimetic phenotypes for taxa are shown by circles; red circles for H. melpomene clade and orange circles for silvaniform taxa. (C and D) Maximum likelihood phylogenies of putative elements; see Figure 2 legend for more detailed explanations. Note positions of black branches on phylogenies which have been drawn on the longest branches from where H. besckei connects to phylogeny.
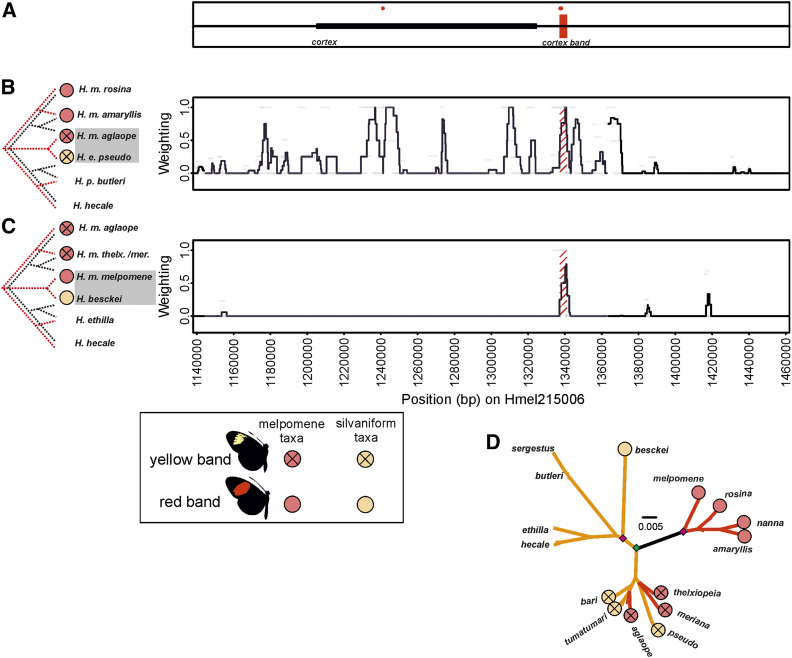
Figure 6.
Paired phylogenetic Twisst comparisons across the cortex region of scaffold Hmel215006. (A) Shows location of cortex, and putative cortex band element. Black bar shows Cortex gene, red shading shows putative regulatory element. Diagnostic fixed SNPS between phenotypes shown with red dots in (A). Twisst comparison (100 SNP windows sliding by 25 SNPs) using (B) Peruvian hybrid zone taxa and (C) H. besckei and Guianese hybrid zone taxa. Mimetic phenotypes for taxa are shown by circles; red circles for H. melpomene clade and orange circles for silvaniform taxa. (D) Maximum likelihood phylogeny of the putative cortex band element; see Figure 2 legend for more detailed explanations.
For comparisons with Guianese taxa, H. ethilla and H. hecale were used alongside Guianese H. elevatus taxa. However, for the Peruvian hybrid zone comparisons, H. ethilla and H. pardalinus butleri were used alongside H. e. pseudocupidineus. H. pardalinus butleri and H. e. pseudocupidineus are very closely related to each other across most of the genome (FST ∼0 across 95% of the genome) (Kryvokhyzha 2014), and so including these taxa together with races of H. melpomene provides the sensitivity to differentiate shared variation that is associated with the phenotype (introgression or ancestral polymorphism) from shared variation that is unrelated to the phenotype (ancestry). If the mimetic silvaniform and H. melpomene taxa cluster together without the nonmimetic H. melpomene and silvaniform taxa, this is suggestive of shared variation associated with phenotype. Such a pattern should only occur at narrow color pattern regions due to our careful choice of taxa.
For comparisons with H. besckei, there is no nonmimetic H. melpomene race forming a hybrid zone with H. m. nanna available. Using H. m. nanna in Twisst analyses with other H. melpomene taxa (H. m. thelxiopeia and H. m. aglaope) resulted in a noisy analysis due to divergence between the H. melpomene taxa. Therefore, we instead compared H. besckei to H. m. melpomene from the Guianese hybrid zone (alongside H. m. thelxiopeia and H. m. aglaope).
Identifying SNPs associated with phenotypes
We also looked for SNPs on our three focal chromosomes that were “diagnostically fixed” between taxa in different phenotype groups (Table S8). We excluded H. e. tumatumari from the no-ray taxa grouping as it appears to have independently evolved its no-ray phenotype. We used the same SNPs as in the Twisst analysis, but with up to 20% of genotype calls across taxa allowed to be missing for each SNP. For optix phenotypes, genotype calls were required to be present in all H. melpomene meriana and H. besckei samples, as these taxa are essential in differentiating diagnostically fixed SNPs for rays—no-rays from those for dennis/no-band—no-dennis/band. At WntA we looked for SNPs “diagnostically fixed” between H. pardalinus in one group (with the silvaniform WntA phenotype) and all H. melpomene, H. elevatus, and H. besckei in the other group (with a nonsilvaniform WntA phenotype). In this WntA analysis, genotypes had to be present in all H. butleri and H. besckei samples. As one of these groups is only made up of a single taxon (H. pardalinus), the results of the WntA analysis were noisier.
Phylogenetic reconstruction at putative functional elements
At each of the putative functional elements we identified, we constructed unrooted trees using all 53 samples to determine the ancestral origin of each element. We included both variable and invariant sites within the boundaries of each element as delineated in our previous analyses using Twisst and fixed differences. Bcftools was used to remove poor quality genotype calls, and mark genotypes as missing if minimum read depth was <5 or GQ < 20. In addition, we also constructed a phylogenetic tree to determine the overall species relationships using all sites from across chromosomes 1 and 2, which we expect to show the species phylogeny as they do not contain any of the main color loci. We used RAxML to build all trees, with the GTRCAT model and 100 maximum likelihood trees to find the best tree, followed by 1000 bootstrap pseudoreplicates.
Conservation and homology
Functional elements controlling gene expression may be conserved across taxa. In order to identify whether there was sequence conservation across Lepidoptera and in particular with H. erato (a distant relative and mimic of H. melpomene) at the putative functional elements we have identified, we retrieved the genome sequences of nine additional species from Lepbase (http://ensembl.lepbase.org); H. erato demophoon v1, Junonia coenia v1.0, Bicyclus anynana v1.2, Danaus plexippus v3, Papilio machoan v1.0, Papilio polytes v1.0, Pieris napi v1.1, Amyelois transitella v1, and Bombyx mori GCA000151625.1. We then identified scaffolds corresponding to the WntA, optix, and cortex loci with BLAST (Altschul et al. 1990). Fine-scale sequence conservation between H. melpomene and H. erato was visualized using the Artemis Comparison Tool (Carver et al. 2005). We then calculated pairwise conservation between H. melpomene and each of the other species using mVISTA (Frazer et al. 2004), an mLAGAN (Brudno et al. 2003) alignment and a conservation cut-off of 70% sequence identity.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. All original raw sequence data files are available via the ENA (accession number PRJEB37067). Supplementary tables and figures referred to in the text are available via figshare: https://doi.org/10.25386/genetics.12911180.
Results
To identify putative regulatory elements in the H. melpomene-silvaniform clade, we use Twisst analyses across multiple hybrid zones to find signals of shared ancestry relating to wing pattern phenotypes and looked for diagnostically fixed SNPs between taxon groupings with these different phenotypes. We look for these patterns around loci known to control particular patterns, and where existing data have previously delimited an interval, we looked within that region. We first refine the rays and dennis loci, finding two putative loci for each of these phenotypes. Furthermore, we identify a 1.5 kb locus that we putatively delimit as the optix band region, as well as two loci at WntA that are associated with the full black discal forewing phenotype that H. elevatus shares with these H. melpomene races rather than the broken black of its sister species H. pardalinus. In contrast, our comparison using taxa from the Guianese hybrid zone shows no regions of shared derived ancestry between the broken band and no-rays forms of H. elevatus and H. melpomene, suggesting both these phenotypes evolved independently in mimetic H. melpomene and H. elevatus in the Guianas. We also identify a putative locus near the cortex gene associated with the yellow band phenotype. Finally, we use phylogenetic analysis to infer the ancestral origins of these putative regulatory elements across the H. melpomene-silvaniform clade, uncovering a complex history of introgression with different evolutionary origins for the various regulatory elements.
Broader patterns of shared ancestry
On chromosome 18, our analyses identified only one additional ∼4 kb peak of shared ancestry (weighting of +1) between comimics that was consistent across hybrid zones and that was not located in the vicinity of optix. This was positioned ∼30 Mb from optix; at ∼3,014,000 bp on chromosome 18; and ∼227,000 bp on Hmel218005 (Figures S9–S11) within the gene HMEL034250g1. As a tblastx search of this gene in the Drosophila melanogaster genome does not return any good hits we are unable to speculate about its function. No peaks of shared ancestry with a weighting of +1 were found outside of the vicinity of WntA on chromosome 10 (Figure S12), and no large peaks showing consistent shared ancestry between comimics across comparisons (Figures S13–S15) were found on chromosome 15 (which contains the cortex gene). Thus, we demonstrate the near absence of regions of consistent shared ancestry between comimics outside the proximity of WntA, optix, and cortex on chromosomes 10, 15, and 18, respectively.
The ray loci
Inferred recombination breakpoints around shared haplotypes have previously been used to delimit a 37 kb ray region on chromosome 18, further narrowed to 25 kb using SNPs perfectly associated with the hindwing rays (Wallbank et al. 2016). Our Twisst comparisons allow us to define two separable loci within this region with different evolutionary histories, a ∼6 kb region (Hmel218003 773301–779400) we call ray1 and another ∼22 kb region (Hmel218003 781427–803436) we call ray2 (Figure 2). Across both regions, we see a pattern of shared ancestry between rayed H. elevatus and H. melpomene mimics from the Peruvian hybrid zone (Figure 2B). However, in our H. besckei comparison, we see shared ancestry only across ray2 (Figure 2C). Our phylogeny at ray2 (Figure 2F) also shows that both no-ray and rayed Guianese and Peruvian H. elevatus show shared ancestry with rayed H. melpomene taxa.
Our Twisst comparison using the Guianese taxa without rays does not show shared ancestry between no-ray H. elevatus and H. melpomene from the Guianas at either ray1 or ray2 (Figure 2D). This suggests that the Guianese no-ray phenotypes in H. elevatus and in H. melpomene have evolved independently and are not the result of shared ancestry, with the no-ray phenotype in H. e. tumatumari evolving after introgression of ray alleles. Other peaks of shared ancestry are also found in this comparison, some of these relate to dennis (Figure 3, B and C; peaks at 813370–814253 bp; 816686–819634 bp) and band (Figure 4, B and C; peak at 728308–729971) phenotypes found in both H. m. meriana and H. e. tumatumari. It is unclear what might cause the additional peaks; however, as these are not consistent across phenotypes/comparisons they do not contribute to the locations of putative functional elements.
Due to independent evolution of the no-ray phenotype, H. e. tumatumari was excluded from the “diagnostically fixed” SNP analysis as its inclusion in the no-ray group would wrongly remove all “diagnostically fixed” SNPs (Table S8). We find that ray1 contains 5 “diagnostically fixed” SNPs while ray2 contains 67. Our phylogenies inferred using all taxa (Figure 2, E and F) show that all rayed and no-ray H. elevatus and all rayed H. melpomene group together at both loci, but that the no-ray H. besckei groups with no-ray H. melpomene only at ray2. These phylogenies also suggest the alleles in rayed taxa at these loci derive from different clades. The ray1 rayed allele appears to originate from the H. melpomene clade (Figure 2E) as rayed individuals at ray1 are clustered within the melpomene samples. In contrast, the ray2 rayed allele appears to originate from the silvaniform clade, as rayed individuals at ray2 are clustered within the silvaniform samples (Figure 2F).
The dennis loci
Previously an ∼11 kb dennis region had been defined (Wallbank et al. 2016). We narrow this region by including H. e. tumatumari and H. besckei in separate Twisst comparisons and comparing these to Guianese H. melpomene taxa. As H. e. tumatumari and H. besckei are separate species and allopatric to each other, we used separate comparisons so as not to introduce noise. We define our loci using “diagnostically fixed” SNPs between our two phenotype groups, and signals of shared ancestry that are both consistent across Twisst comparisons and within the previously delimited ∼11 kb dennis region from Wallbank et al. (2016). On this basis, we delimit ∼1 kb (Hmel218003 813370–814253) and ∼3 kb (Hmel218003 816686–819634) regions that we term dennis1 and dennis2, respectively (Figure 3). We split these as a consequence of a dip in shared ancestry consistent across all our comparisons. Both regions show shared ancestry between H. elevatus and H. melpomene mimics with the dennis phenotype across both the Peruvian (H. e. pseudocupidineus and H. m. aglaope) and Guianese hybrid zones (H. e. tumatumari/bari and H. m. meriana/thelxiope) (Figure 3, B and C). Our diagnostic fixed SNP analysis found that the dennis1 region contained 20 “diagnostically fixed” SNPs with the correct pattern for dennis (or optix band as all red band taxa lack the dennis phenotype and so cannot be separated), while dennis2 contains 18 such SNPs. There were only 40 other “diagnostically fixed SNPs” across the whole of Chromosome 18, with nine of these coming from our putative optix band region as expected. However, some of these other SNPs are also clustered, 15 SNPs clustered just upstream (Hmel218003 822665–825712) but outside the dennis region defined by Wallbank et al. (2016) and therefore outside of our defined dennis region (Figure 3D), and eight SNPs were clustered at the peak at ∼227,000 bp on Hmel218005. Phylogenies allowed us to define two separate phenotypic groups: one containing all no-dennis H. melpomene and H. besckei and the other containing mimetic dennis H. melpomene and H. elevatus. This is consistent with Wallbank et al. (2016). As the dennis H. melpomene races are nested within the silvaniform clade, these phylogenies suggest that the dennis phenotype originated in the silvaniform clade (Figure 3, D and E) and introgressed into H. melpomene, while the no-dennis alleles appear to have introgressed from H. melpomene into H. besckei.
The optix band locus
Although our taxa do not allow us to tell shared ancestry due to band phenotypes from shared ancestry due to dennis phenotypes, we postulate that an optix band element would most likely be outside of the dennis (or ray) regions (Wallbank et al. 2016). This is based on (i) the existence of H. timareta races that lack red bands and dennis phenotypes (while other taxa have dennis phenotypes with rays but lack red bands) (Giraldo et al. 2008), and (ii) specimens of H. melpomene from the Guianese hybrid zone that have both dennis and red band phenotypes together (rather than having only one of these as seen in the main Guianese H. melpomene taxa) (J.M. personal observation). We hypothesize that any putative optix band element should also show shared ancestry across both Twisst comparisons. This is because the red band silvaniform H. besckei should show shared ancestry with red band races of H. melpomene, while H. elevatus races that have a yellow band should show shared ancestry with H. melpomene races that also have this phenotype. Using shared ancestry and diagnostically fixed SNPs we identify a single 1.5-kb region (Hmel218003 728308–729971), close to the 5′ end of optix that we putatively delimit as the previously unidentified optix band region (Figure 4, A and B). This region spans the only consistent peak of shared ancestry (outside of the dennis and ray regions) across all hybrid zones and includes nine “diagnostically fixed” SNPs between mimetic red and yellow band taxa. Furthermore, the wider region around this 1.5 kb also contains windows that inconsistently show a signal of shared ancestry between mimetic red band taxa and mimetic yellow band taxa (shown as gray bars with weighting of +1 in Figure 4, B and C). However, because the red band is always found in the absence of dennis in our taxa (and vice-versa), it should be noted that we cannot rule out that this region is in fact involved in the control of the dennis phenotype, or that the dennis regions are not involved in the control of the red band phenotype. The phylogeny at this locus shows that H. besckei is nested within the clade containing the red band H. melpomene races. While H. m. aglaope, H. m. meriana and H. m. thelxiopeia which all lack the red forewing band group with the other silvaniform species (Figure 4D), suggesting an ancestral silvaniform clade origin for the allele causing loss of the red band.
Two WntA loci
Around WntA we identify two separate loci that we term WntA1 (Hmel210004 1819577–1822844) and WntA2 (Hmel210004 1857021–1860913), which show a signal of shared ancestry between H. melpomene aglaope/amaryllis and H. elevatus pseudocupidineus, but not between H. melpomene aglaope/amaryllis and H. pardalinus butleri (Figure 5B). This pattern is consistent with what we would expect in regions involved in controlling the full black discal forewing phenotype that H. elevatus shares with these H. melpomene races (and which replaces the broken black patterning found in H. pardalinus and other silvaniforms in this part of the wing). These two loci were the only peaks also supported by the results from our “diagnostically fixed” SNP analysis between H. pardalinus butleri and all taxa with a H. melpomene type phenotype (all H. melpomene and H. elevatus races and our H. besckei). In total WntA1 and WntA2 contained, respectively, 12 and 6 of the 252 “diagnostically fixed” SNPs on chromosome 10. Thus, WntA1 and WntA2 together contain 7% of these “diagnostically fixed” SNPs in just 0.04% of the length of the chromosome.
The phylogenies at WntA1 and WntA2 clearly show H. elevatus races grouping within (WntA1) or close to (WntA2) the H. melpomene clade. In both phylogenies, H. besckei was found outside the rest of the silvaniforms (excluding H. elevatus), rather than grouping with H. ethilla as it does in the species tree (Figure S16). This suggests that there may be some shared ancestry in these regions between H. besckei and the other H. melpomene races. Both phylogenies suggest that the full black discal forewing phenotype which replaces broken black in H. e. pseudocupidineus originates from the H. melpomene clade. In contrast to our results with Peruvian taxa, our comparison using taxa from the Guianese hybrid zone (Figure S17) shows no regions of shared derived ancestry on chromosome 10 between the broken band forms of H. elevatus (H. e. tumatumari and H. e. bari) and H. melpomene in the Guianas (H. m. meriana and H. m. thelxiopeia), relative to H. m. melpomene, which lacks the broken band phenotype. Given previous research has shown that this phenotype is controlled by WntA in H. melpomene (Morris et al. 2019), our results here suggests that just like the no-rays phenotype, the broken band phenotype in the Guianas also evolved independently in mimetic H. melpomene and H. elevatus.
The cortex band locus
Only the yellow forewing band phenotype (and not the yellow hindwing bar) is found in both our Peruvian and Guianese hybrid zone taxa. We identified a ∼3.5 kb region near cortex (Hmel215006 1337470–1340886) that shows shared derived ancestry between yellow band H. e. pseudocupidineus and H. m. aglaope from the Peruvian hybrid zone (Figure 6B) and between red band H. besckei and H. m. melpomene (Figure 6C). This region also contained two of the three “diagnostically fixed” SNPs on chromosome 15, between red band H. melpomene and H. besckei and our yellow band H. melpomene and H. elevatus. These results suggest that this region may be involved in controlling the yellow band phenotype. We term this locus “cortex band.” Phylogenetic reconstruction at cortex band, shows that this region is also shared between all yellow band H. elevatus and H. melpomene taxa (Figure 6D), and that the putative yellow band allele appears to have originated in the silvaniform clade before then introgressing into the H. melpomene taxa with yellow bands. However, H. besckei was not found on the branch with red band H. melpomene. Therefore, the signal of shared ancestry seen between H. besckei and red band H. melpomene in the Twisst comparison may not be due to shared ancestry between these two taxa in an allele that results in a lack of yellow band. Instead it may be an artifact of the shared ancestry between yellow band H. melpomene races and H. elevatus, which would make the H. melpomene haplotype in these races more closely related to H. hecale and H. ethilla haplotype than that in H. besckei. However, given that we only detect this phylogenetic signal at this region, and the location of our diagnostic fixed differences, this single peak is still our best candidate for a “cortex band” element, even if we cannot rule out other regions that we miss due to complex pleiotropic interactions that do not give such an expected phylogenetic signal of shared ancestry.
Modular conservation across Heliconius and the Lepidoptera
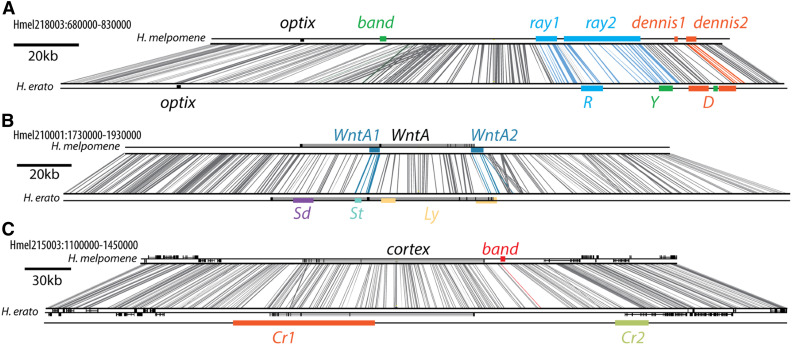
Fine-scale sequence conservation between H. melpomene and H. erato at and around optix, WntA, and cortex is shown in Figure 7. As expected, we find that these two genomes are largely colinear, with large sections of homology across the genomes. Ray1 contains a substantial amount of conserved sequence between the H. melpomene and H. erato, as well as a 2000 bp region (Hmel218003:778000–780000; Figure S18) deeply conserved across the Lepidoptera, indicating the presence of a conserved functional element within ray1 of both species (See Table S19 for scaffold locations in other Lepidoptera). Our analysis also finds narrow regions of homology (Table S20) between the ray2 locus in H. melpomene and the R locus controlling the ray phenotype in H. erato (Van Belleghem et al. 2017). The H. melpomene dennis1 element has very low sequence conservation with H. erato and no conserved sequence with other Lepidoptera. However, the H. melpomene dennis2 element contains extensive sequence conservation with H. erato (as well as a short region of conservation with other nymphalids; Hmel218003: 819500–82000), and we also find a narrow region of homology between dennis2 (Table S20) and the D locus controlling the dennis phenotype in H. erato. The H. melpomene optix band element identified here is located within 50 kb of the gene optix, but at a substantial distance from the Y element that controls the corresponding phenotype in the H. erato clade (Van Belleghem et al. 2017) in a region of low conservation with H. erato. The physical distance between the elements in the two species indicates that the evolution of the red band occurred by evolution of regulatory changes at unrelated loci. At the WntA locus, the WntA1 and WntA2 elements we identify contain homologous sequence with the H. erato St (a likely promoter of the WntA gene) and Ly elements respectively (Table S20). The WntA1 element contains two peaks of conservation with multiple other lepidopteran species (Figure S18), corresponding to the two 3′ coding exons of the WntA gene. At cortex, the H. melpomene cortex band element was located between the two large yellow bar-linked regions identified in H. erato (Van Belleghem et al. 2017). Overall we find that several of the putative regulatory elements we have identified as controlling patterning in H. melpomene and the silvaniform clade (ray2, dennis2, WntA1, and WntA2) appear to be at least partly homologous with regulatory elements proposed to control similar phenotypes in H. erato, a species from which they diverged ∼12 MYA (Kozak et al. 2015).
Figure 7.
ACT-BLAST alignment of three color pattern regions between H. melpomene and its comimic H. erato. In each panel, the H. melpomene scaffold (top) is compared to the H. erato scaffold (bottom) with each BLAST hit connected by a line. H. melpomene is annotated with elements identified herein, and H. erato is annotated with elements controlling convergent phenotypes as identified in Van Belleghem et al. (2017). In (A) one of the H. melpomene elements associated with the ray pattern element (blue; ray2 in H. melpomene; R in H. erato) contains homologous sequence with the ray pattern element in H. erato, as indicated by the connecting colored lines. As does one of the elements associated with dennis pattern in H. melpomene and the H. erato dennis associated element (orange; dennis2 in H. melpomene; D in H. erato) (Table S20). The elements associated with band (green; band in H. melpomene; Y in H. erato) are in distant positions, but H. erato Y is situated in conserved sequence which is included in the H. melpomene ray2 element. In (B) the H. melpomene WntA1 and WntA2 elements both contain homologous sequence to the H. erato elements St and Ly (Table S20). In (C) band is shown to not be homologous to either of the previously identified elements (Cr1 and Cr2) in H. erato. H. erato elements are colored based on scheme used in Van Belleghem et al. (2017).
Discussion
In this study, we have investigated the contributions of three genetic modes of evolution (divergent genetic mechanisms, parallel genetic evolution, and collateral evolution) to explain convergent wing color pattern phenotypes found in Heliconius butterflies (H. melpomene, H. elevatus, H. besckei, and H. erato). Using phylogenetic analyses, we have identified strong and narrow signals of shared ancestry related to wing pattern phenotypes in H. melpomene, H. elevatus, and H. besckei around all three main color pattern genes (optix, WntA, and cortex). This is indicative of collateral evolution of putative regulatory elements among these closely related species. In contrast, signals of consistent shared ancestry among these taxa were low outside of these regions. However, we also find that convergent phenotypes between H. melpomene and H. elevatus in the Guianas appear to have arisen independently and so are not a result of collateral evolution. We also show that four out of the seven putative regulatory elements around optix and WntA in H. melpomene also show some homology to regulatory elements controlling similar phenotypes in its distant comimic H. erato. Thus, convergent phenotypes between these more distantly related species appear to result from a combination of convergent parallel evolution and divergent genetic mechanisms. Overall, our results show that all three genetic modes of evolution underlie convergent phenotypes among mimetic Heliconius species, but that these likely operate at different evolutionary timescales.
Modularity of mimicry facilitates pattern switching
Recent studies in a variety of organisms have demonstrated the importance of combinatorial evolution, where ancient alleles are reused in novel combinations to generate new phenotypes and adaptive combinations (Marques et al. 2019). This can lead to adaptive convergent changes more rapidly than evolution via divergent genetic mechanisms or parallel genetic evolution. This can be seen in cichlids, where regulatory changes at the gene agouti-related peptide 2 are associated with the convergent stripe evolution across species (Kratochwil et al. 2018), and in sticklebacks, where recurrent deletions of the same pitx1 enhancer in different populations have led to reduced or lost pelvic structures (Chan et al. 2010). Another excellent example are the diverse wing patterns of Heliconius butterflies. These butterflies appear to have a flexible toolkit of cis-regulatory enhancers (Wallbank et al. 2016; Van Belleghem et al. 2017) through which gene expression changes can rapidly alter phenotypes and drive adaptive evolution (Wray 2007), with a single mutation at an enhancer potentially enough to have major phenotypic effects (Chan et al. 2010; Frankel et al. 2012). Such genetic architecture combined with introgression can facilitate adaptive evolution through the swapping of these enhancers among lineages of Heliconius (Wallbank et al. 2016; Moest et al. 2019; Lewis and Van Belleghem 2020). For example, the evidence suggests that the ancestral sources of the ray and dennis elements were different, with the rays phenotype originating in the H. melpomene clade and the dennis phenotype originating in the silvaniform clade, before being brought together as the dennis-rayed phenotype in both H. melpomene and H. elevatus (Wallbank et al. 2016).
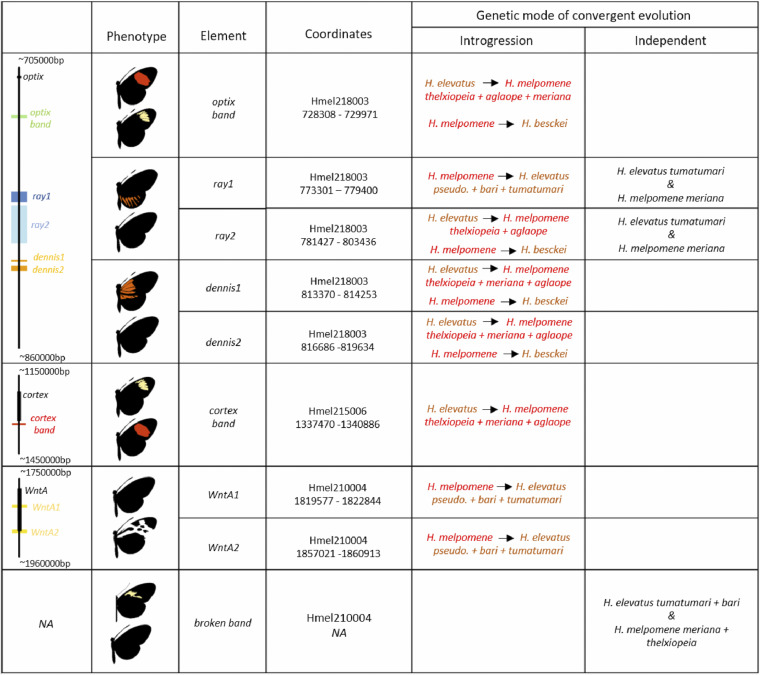
Our analysis, which narrows the dennis and ray elements and splits them into two loci each, is consistent with this finding of multiple origins, even finding separate origins for each ray locus. We also identify a putative optix band locus near optix that suggests the red band is ancestral to the H. melpomene clade, while its absence is ancestral to the silvaniform clade. The two putative loci near WntA that we propose control the full black melanic discal forewing phenotype of H. elevatus (which replaces the broken black pattern in other silvaniforms) also appear to be ancestral to the H. melpomene clade. Finally, our putative cortex band loci suggests that the yellow band phenotype was ancestral to the silvaniforms. By expanding on the range of taxa and loci considered in previous studies, our results paint a picture of multiple loci originating in different clades and then being brought together via introgression to derive the Amazonian Heliconius races that we see today. For example, around optix, it appears that the ray2 rays allele, the dennis1 and dennis2 dennis alleles, and the optix band no-band allele originated in the silvaniform clade, and, through gene flow with H. elevatus, introgressed into H. melpomene. In contrast, the ray1 ray allele introgressed from H. melpomene into H. elevatus, while the optix band red band allele, dennis1 and (possibly) dennis2 no-dennis alleles, and the ray1 no-ray allele introgressed from H. melpomene into H. besckei (summarized in Figure 8). These loci may all have introgressed separately, alternatively for example, the two dennis loci may have introgressed together, perhaps even along with ray2 and optix band as a single haplotype, and then subsequently been broken up by recombination. Reconstructing the exact evolutionary history with the order and timing of introgression events at these narrow regions may prove to be difficult.
Figure 8.
A summary of the locations of putative cis-regulatory elements reported here, and the genetic mode of evolution causing mimetic convergence between H. melpomene races and silvaniform species at each element. In the introgression column: red to orange names indicate introgression from H. melpomene (H. melpomene clade origin) into silvaniform taxa, while orange to red names indicate introgression from silvaniform taxa (silvaniform clade origin) into H. melpomene. There is no identified element for broken band due to a lack of shared ancestry and its independent evolution in each species. The schematic in the left most column shows the locations of each element (colored labels) with respect to neighboring genes (black labels).
Although modularity is just one hypothetical mode by which these putative regulatory elements may impact on wing color patterns, our results are currently the most compelling evidence for modularity in the cis-regulatory architecture of Heliconius wing pattern variation. Our work therefore adds to a growing body of evidence for a profound role of modularity and introgression in the evolution of mimicry in Heliconius butterflies. Furthermore, our results, along with examples such as pesticide resistance in mice and Helicoverpa moths (Song et al. 2011; Valencia-Montoya et al. 2020), beak adaptations in Darwin’s finches (Grant and Grant 2010; Lamichhaney et al. 2015), vectorial capacity in mosquitoes (Fontaine et al. 2015), and human evolution (Abi-Rached et al. 2011; Huerta-Sánchez et al. 2014; Sankararaman et al. 2016) highlight the potential for introgression to act as an adaptive facilitator.
Independent evolution driving mimetic convergence in the Guianas
Our analyses have allowed us to putatively identify regulatory elements shared between H. melpomene and silvaniform taxa, with evidence suggesting that this has occurred via introgression (Dasmahapatra et al. 2012; Pardo-Diaz et al. 2012; Wallbank et al. 2016; Enciso-Romero et al. 2017). However, although introgression is one way in which major cis-regulatory changes to expression can occur, changes can also occur via de novo evolution of cis-regulatory enhancers (Wray 2007). This can occur via sequence duplications (Eichenlaub and Ettwiller 2011), transposable elements that lead to the translocation of regulatory elements from one gene to another (Daborn 2002; Domené et al. 2013), or the co-option of existing regulatory sequences to derive novel expression patterns (Rebeiz et al. 2011). While our analyses cannot identify the elements that are not shared across taxa, we are able to show that the convergent no-ray phenotypes in mimetic H. melpomene meriana and H. elevatus tumatumari appears to have resulted from independent evolution within each species because there is no signal of shared ancestry at the ray1 or ray2 regions between these taxa (Figure 2, E and F). Given our evidence that introgression of phenotypes between H. melpomene and silvaniform taxa appears to have occurred in Peru, and that H. elevatus tumatumari appears to have the rayed allele found in other rayed races of H. melpomene and H. elevatus, we hypothesize that H. elevatus tumatumari has secondarily and independently evolved a no-ray phenotype. In contrast, the lack of rays in H. melpomene meriana appears to be due to having the no-ray allele found in other H. melpomene races (Figure 2, E and F). This lack of rays may either be the ancestral state in H. melpomene, with H. melpomene meriana then later gaining the dennis and yellow band phenotypes through introgression, or alternatively, a result of the ray allele of H. melpomene meriana being replaced by a no-ray allele through recombination with H. melpomene melpomene. In addition, our analyses also show that the forewing broken band phenotype found both in H. melpomene meriana/thelxiopeia and H. elevatus tumatumari/bari in the Guianas must also have independent origins as we see no signal of shared ancestry between these taxa around WntA.
Our results indicate that mimicry via introgression between H. elevatus and H. melpomene has therefore not occurred consistently across their ranges. A possible scenario is that introgression first occurred in Peru allowing species to switch or perhaps create new mimicry rings, with these newly introgressed alleles and then simply persisting in the Guianas, where independent convergent evolution has refined local mimicry leading to H. elevatus losing the rays and melanic WntA phenotypes. This means that both introgression and the independent convergent evolution of novel cis-regulatory elements, has been important in driving mimicry between these two species.
Convergence and sequence conservation
Our results further support the notion that cis-regulatory modularity is common across mimicry genes in Heliconius. Having refined and identified putative cis-regulatory elements, we investigated whether these intervals showed sequence conservation between H. erato, H. melpomene and other Lepidoptera, using sequence conservation as a proxy for cis-regulatory function. While the band element regulates the expression of optix in both H. erato and H. melpomene, this was achieved by divergent genetic mechanisms. On the other hand, we have found evidence of parallel evolution in the modification to the 5′ noncoding region of WntA in both H. melpomene and H. erato; these modifications occurred in evidently homologous regulatory elements, despite their independent evolution.
The broad region around the ray and the dennis elements contains a high density of deeply conserved sequences, but appears to be a hotspot for the modification by selection within Heliconius. Likewise, repeated modifications in the 5′ promoter region of WntA could indicate a role for this region as a hotspot for modification by selection. Possible explanations for the repeated reuse of the same noncoding regions in regulatory evolution focus on aspects of the structure and function of ancestral cis-regulatory elements. Aspects such as low pleiotropy, and the greater variety of mutations that can cause functional changes, makes these regions predisposed toward gaining new regulatory functions (Stern 2013). Potentiating mutations could arise stochastically and neutrally in nonfunctional sequence, or they could occur in pre-existing regulatory elements, which already have the structure and function necessary to act as a regulator (Blount et al. 2008). Modification of a site with ancestrally shared potentiating mutations would require fewer mutational steps as opposed to de novo generation of a regulatory element, increasing the probability that the novel function will evolve at that site; i.e., pre-adaptation, as was observed in the evolution of citrate metabolism in populations of Escherichia coli (Blount et al. 2008). If potentiating mutations were present in the ancestor, then we would expect to find regulatory mutations to occur close together in convergent species, and indeed, we observed this in the dennis2, ray2, WntA1, and WntA2 elements. There are still only a few well-studied examples of independent convergence in regulatory sequence (Booker et al. 2016; Partha et al. 2017; Kratochwil et al. 2018; Tollis et al. 2018; Feigin et al. 2019) as not many cis-regulatory mutations that pertain to convergent phenotypes have yet been identified. It is therefore yet to be seen whether there is a general trend of convergence across taxa at this level of granularity, but, as more examples are characterized, whether or not such a trend persists will be revealed.
Acknowledgments
We thank Ewout Eriks (Neotropical Butterfly Park, Suriname) for help collecting wild specimens, and organizing collection and export permits as well as access to his facilities. We also thank Neil Rosser for specimen collection, Mathieu Joron for supplying specimen MJ09-4014, and James Mallet for comments and thoughts on the manuscript. We would like to thank the governments of Peru and Suriname for permission to collect butterflies. Surinamese samples were collected and exported on permit (No. 10865) from the Suriname Forest Service and Nature Conservation Division. Peruvian samples were collected with permits from SERFOR and the Peruvian Ministry of Agriculture (288-2009-AG-DGFFS-DGEFFS and 0148-2011-AG-DGFFS-DGEFFS). H. elevatus bari MJ09-4014 was collected in French Guiana as a nonprotected species in a nonprotected area in a territory of France, for which no permit was needed at the time. The work by K.K.D. and J.M. was funded by the Natural Environment Research Council (NERC; grant: NE/K012886/1; https://nerc.ukri.org/). J.M. was also supported by a NERC studentship (https://nerc.ukri.org/) and a Heredity fieldwork grant from the Genetics Society (http://www.genetics.org.uk/). Work by J.J.H., S.H.M., S.M.V.B., and C.D.J. was funded by the European Research Council (grant: Speciation Genetics 339873) (https://erc.europa.eu/). J.J.H. was also supported by the Wellcome Trust (https://wellcome.ac.uk/) and Cambridge Philosophical Society (https://www.cambridgephilosophicalsociety.org/), and S.H.M. was supported by a St Johns College (Cambridge) Fellowship (https://johnian.joh.cam.ac.uk/giving/fellowships). C.S. was funded by Fondos Concursables Big - grant IV-FGD005/ IV-FGI006 Universidad del Rosario.
Footnotes
Supplemental material available at figshare: https://doi.org/10.25386/genetics.12911180.
Communicating editor: A. Sweigart
Literature Cited
- Abi-Rached L., Jobin M. J., Kulkarni S., McWhinnie A., Dalva K. et al. , 2011. The shaping of modern human immune systems by multiregional admixture with archaic humans. Science 334: 89–94. 10.1126/science.1209202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., and Lipman D. J., 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Baxter S. W., Papa R., Chamberlain N., Humphray S. J., Joron M. et al. , 2008. Convergent evolution in the genetic basis of Müllerian mimicry in Heliconius butterflies. Genetics 180: 1567–1577. 10.1534/genetics.107.082982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter S. W., Nadeau N. J., Maroja L. S., Wilkinson P., Counterman B. A. et al. , 2010. Genomic hotspots for adaptation: the population genetics of Müllerian mimicry in the Heliconius melpomene clade. PLoS Genet. 6: e1000794 10.1371/journal.pgen.1000794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blount Z. D., Borland C. Z., and Lenski R. E., 2008. Historical contingency and the evolution of a key innovation in an experimental population of Escherichia coli. Proc. Natl. Acad. Sci. USA 105: 7899–7906. 10.1073/pnas.0803151105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blount Z. D., Lenski R. E., and Losos J. B., 2018. Contingency and determinism in evolution: Replaying life’s tape. Science. 362: eaam5979 10.1126/science.aam5979 [DOI] [PubMed] [Google Scholar]
- Booker B. M., Friedrich T., Mason M. K., VanderMeer J. E., Zhao J. et al. , 2016. Bat accelerated regions identify a bat forelimb specific enhancer in the HoxD locus. PLoS Genet. 12: e1005738 10.1371/journal.pgen.1005738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brudno M., Do C. B., Cooper G. M., Kim M. F., Davydov E. et al. , 2003. LAGAN and Multi-LAGAN: efficient tools for large-scale multiple alignment of genomic DNA. Genome Res. 13: 721–731. 10.1101/gr.926603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver T. J., Rutherford K. M., Berriman M., Rajandream M. A., Barrell B. G. et al. , 2005. ACT: the Artemis comparison tool. Bioinformatics 21: 3422–3423. 10.1093/bioinformatics/bti553 [DOI] [PubMed] [Google Scholar]
- Chan Y. F., Marks M. E., Jones F. C., Villarreal G., Shapiro M. D., et al. , 2010. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science 327: 302–305. 10.1126/science.1182213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Counterman B. A., Araujo-Perez F., Hines H. M., Baxter S. W., Morrison C. M. et al. , 2010. Genomic hotspots for adaptation: the population genetics of Müllerian mimicry in Heliconius erato. PLoS Genet. 6: e1000796 10.1371/journal.pgen.1000796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daborn P. J., 2002. A single P450 allele associated with insecticide resistance in Drosophila. Science 297: 2253–2256. 10.1126/science.1074170 [DOI] [PubMed] [Google Scholar]
- Dasmahapatra K. K., Walters J. R., Briscoe A. D., Davey J. W., Whibley A. et al. , 2012. Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature 487: 2–6. 10.1038/nature11041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey J. W., Chouteau M., Barker S. L., Maroja L., Baxter S. W., et al. , 2016. Major improvements to the Heliconius melpomene genome assembly used to confirm 10 chromosome fusion events in 6 million years of butterfly evolution. G3 (Bethesda) 6: 695–708. 10.1534/g3.115.023655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domené S., Bumaschny V. F., de Souza F. S. J., Franchini L. F., Nasif S. et al. , 2013. Enhancer turnover and conserved regulatory function in vertebrate evolution. Philos. Trans. R. Soc. B Biol. Sci. 368: 20130027 10.1098/rstb.2013.0027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichenlaub M. P., and Ettwiller L., 2011. De novo genesis of enhancers in vertebrates. PLoS Biol. 9: e1001188 10.1371/journal.pbio.1001188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enciso-Romero J., Pardo-Díaz C., Martin S. H., Arias C. F., Linares M. et al. , 2017. Evolution of novel mimicry rings facilitated by adaptive introgression in tropical butterflies. Mol. Ecol. 26: 5160–5172. 10.1111/mec.14277 [DOI] [PubMed] [Google Scholar]
- Farhat M. R., Shapiro B. J., Kieser K. J., Sultana R., Jacobson K. R. et al. , 2013. Genomic analysis identifies targets of convergent positive selection in drug-resistant Mycobacterium tuberculosis. Nat. Genet. 45: 1183–1189. 10.1038/ng.2747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feigin C. Y., Newton A. H., and Pask A. J., 2019. Widespread cis-regulatory convergence between the extinct Tasmanian tiger and gray wolf. Genome Res. 29: 1648–1658. 10.1101/gr.244251.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine M. C., Pease J. B., Steele A., Waterhouse R. M., Neafsey D. E. et al. , 2015. Extensive introgression in a malaria vector species complex revealed by phylogenomics. Science 347: 1258524 10.1126/science.1258524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel N., Wang S., and Stern D. L., 2012. Conserved regulatory architecture underlies parallel genetic changes and convergent phenotypic evolution. Proc. Natl. Acad. Sci. USA 109: 20975–20979. 10.1073/pnas.1207715109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer K. A., Pachter L., Poliakov A., Rubin E. M., and Dubchak I., 2004. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 32: W273–W279. 10.1093/nar/gkh458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallant J. R., Imhoff V. E., Martin A., Savage W. K., Chamberlain N. L. et al. , 2014. Ancient homology underlies adaptive mimetic diversity across butterflies. Nat. Commun. 5: 4817 10.1038/ncomms5817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldo N., Salazar C., Jiggins C. D., Bermingham E., and Linares M., 2008. Two sisters in the same dress: Heliconius cryptic species. BMC Evol. Biol. 8: 324 10.1186/1471-2148-8-324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant P. R., and Grant B. R., 2010. Conspecific vs. heterospecific gene exchange between populations of Darwin’s finches. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365: 1065–1076. 10.1098/rstb.2009.0283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber B., Whibley A., Poul Y. L., Navarro N., Martin A. et al. , 2015. Conservatism and novelty in the genetic architecture of adaptation in Heliconius butterflies. Heredity 114: 515–524. 10.1038/hdy.2015.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Sánchez E., Jin X., Asan Z. Bianba B. M. Peter et al. , 2014. Altitude adaptation in Tibetans caused by introgression of Denisovan-like DNA. Nature 512: 194–197. 10.1038/nature13408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones F. C., Grabherr M. G., Chan Y. F., Russell P., Mauceli E. et al. , 2012. The genomic basis of adaptive evolution in threespine sticklebacks. Nature 484: 55–61. 10.1038/nature10944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak K. M., Wahlberg N., Neild A. F. E., Dasmahapatra K. K., Mallet J. et al. , 2015. Multilocus species trees show the recent adaptive radiation of the mimetic Heliconius butterflies. Syst. Biol. 64: 505–524. 10.1093/sysbio/syv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratochwil C. F., Gerwin J., Woltering J. M., Meyer A., Hulsey C. D. et al. , 2018. Agouti-related peptide 2 facilitates convergent evolution of stripe patterns across cichlid fish radiations. Science 362: 457–460. 10.1126/science.aao6809 [DOI] [PubMed] [Google Scholar]
- Kryvokhyzha, D., 2014 Whole genome resequencing of Heliconius butterflies revolutionizes our view of the level of admixture between species. Master’s Thesis, Evol. Biol. Uppsala Univ. Harvard Univ. 10.1038/ng.953 10.1038/ng.953 [DOI] [Google Scholar]
- Lamichhaney S., Berglund J., Almén M. S., Maqbool K., Grabherr M. et al. , 2015. Evolution of Darwin’s finches and their beaks revealed by genome sequencing. Nature 518: 371–375. 10.1038/nature14181 [DOI] [PubMed] [Google Scholar]
- Lewis J. J., and Van Belleghem S. M., 2020. Mechanisms of change: unraveling the roles of modularity and pleiotropy in diversification. Front. Ecol. Evol. 8: 261. [Google Scholar]
- Lewis J. J., Geltman R. C., Pollak P. C., Rondem K. E., Van Belleghem S. M. et al. , 2019. Parallel evolution of ancient, pleiotropic enhancers underlies butterfly wing pattern mimicry. Proc. Natl. Acad. Sci. USA 116: 24174–24183. 10.1073/pnas.1907068116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27: 2987–2993. 10.1093/bioinformatics/btr509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., and Durbin R., 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J. et al. , 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnen C. R., Kingsley E. P., Jensen J. D., and Hoekstra H. E., 2009. On the origin and spread of an adaptive allele in deer mice. Science 325: 1095–1098. 10.1126/science.1175826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnen C. R., Larson J. G., Poh Y.-P., Peterson B. K., Barrett R. D. H. et al. , 2013. Adaptive evolution of multiple traits through multiple mutations at a single gene. Science 339: 1312–1316. 10.1126/science.1233213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques D. A., Meier J. I., and Seehausen O., 2019. A combinatorial view on speciation and adaptive radiation. Trends Ecol. Evol. 34: 531–544. 10.1016/j.tree.2019.02.008 [DOI] [PubMed] [Google Scholar]
- Martin A., Papa R., Nadeau N. J., Hill R. I., Counterman B. A. et al. , 2012. Diversification of complex butterfly wing patterns by repeated regulatory evolution of a Wnt ligand. Proc. Natl. Acad. Sci. USA 109: 12632–12637. 10.1073/pnas.1204800109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A., McCulloch K. J., Patel N. H., Briscoe A. D., Gilbert L. E. et al. , 2014. Multiple recent co-options of Optix associated with novel traits in adaptive butterfly wing radiations. Evodevo 5: 7 10.1186/2041-9139-5-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S. H., and Van Belleghem S. M., 2017. Exploring evolutionary relationships across the genome using topology weighting. Genetics 206: 429–438. 10.1534/genetics.116.194720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S. H., Dasmahapatra K. K., Nadeau N. J., Salazar C., Walters J. R. et al. , 2013. Genome-wide evidence for speciation with gene flow in Heliconius butterflies. Genome Res. 23: 1817–1828. 10.1101/gr.159426.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazo-Vargas A., Concha C., Livraghi L., Massardo D., Wallbank R. W. R. et al. , 2017. Macroevolutionary shifts of WntA function potentiate butterfly wing-pattern diversity. Proc. Natl. Acad. Sci. USA 114: 10701–10706. 10.1073/pnas.1708149114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill R. M., Dasmahapatra K. K., Davey J. W., Dell’Aglio D. D., Hanly J. J. et al. , 2015. The diversification of Heliconius butterflies: what have we learned in 150 years? J. Evol. Biol. 28: 1417–1438. 10.1111/jeb.12672 [DOI] [PubMed] [Google Scholar]
- Moest M., Van Belleghem S. M., James J. E., Salazar C., Martin S. H., et al. , 2020. Selective sweeps on novel and introgressed variation shape mimicry loci in a butterfly adaptive radiation PLOS Biol. 18: e3000597. 10.1371/journal.pbio.3000597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris J., Navarro N., Rastas P., Rawlins L. D., Sammy J. et al. , 2019. The genetic architecture of adaptation: convergence and pleiotropy in Heliconius wing pattern evolution. Heredity 123: 138–152. 10.1038/s41437-018-0180-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau N. J., Whibley A., Jones R. T., Davey J. W., Dasmahapatra K. K. et al. , 2012. Genomic islands of divergence in hybridizing Heliconius butterflies identified by large-scale targeted sequencing. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367: 343–353. 10.1098/rstb.2011.0198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau N. J., Martin S. H., Kozak K. M., Salazar C., Dasmahapatra K. K., et al. , 2013. Genome-wide patterns of divergence and gene flow across a butterfly radiation. Mol. Ecol. 22: 814–826. 10.1111/j.1365-294X.2012.05730.x [DOI] [PubMed] [Google Scholar]
- Nadeau N. J., Ruiz M., Salazar P., Counterman B., Medina J. A. et al. , 2014. Population genomics of parallel hybrid zones in the mimetic butterflies, H. melpomene and H. erato. Genome Res. 24: 1316–1333. 10.1101/gr.169292.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau N. J., Pardo-Diaz C., Whibley A., Supple M. A., Saenko S. V. et al. , 2016. The gene cortex controls mimicry and crypsis in butterflies and moths. Nature 534: 106–110. 10.1038/nature17961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo-Diaz C., Salazar C., Baxter S. W., Merot C., Figueiredo-Ready W. et al. , 2012. Adaptive introgression across species boundaries in Heliconius butterflies. PLoS Genet. 8: e1002752 10.1371/journal.pgen.1002752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partha R., Chauhan B. K., Ferreira Z., Robinson J. D., Lathrop K. et al. , 2017. Subterranean mammals show convergent regression in ocular genes and enhancers, along with adaptation to tunneling. eLife 6: e25884 10.7554/eLife.25884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prud’homme B., Gompel N., Rokas A., Kassner V. A., Williams T. M. et al. , 2006. Repeated morphological evolution through cis-regulatory changes in a pleiotropic gene. Nature 440: 1050–1053. 10.1038/nature04597 [DOI] [PubMed] [Google Scholar]
- Rebeiz M., Jikomes N., Kassner V. A., and Carroll S. B., 2011. Evolutionary origin of a novel gene expression pattern through co-option of the latent activities of existing regulatory sequences. Proc. Natl. Acad. Sci. USA 108: 10036–10043. 10.1073/pnas.1105937108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R. D., Papa R., Martin A., Hines H. M., Counterman B. A. et al. , 2011. Optix drives the repeated convergent evolution of butterfly wing pattern mimicry. Science 333: 1137–1141. 10.1126/science.1208227 [DOI] [PubMed] [Google Scholar]
- Sage R. F., Sage T. L., and Kocacinar F., 2012. Photorespiration and the evolution of C4 photosynthesis. Annu. Rev. Plant Biol. 63: 19–47. 10.1146/annurev-arplant-042811-105511 [DOI] [PubMed] [Google Scholar]
- Sankararaman S., Mallick S., Patterson N., and Reich D., 2016. The combined landscape of Denisovan and Neanderthal ancestry in present-day humans. Curr. Biol. 26: 1241–1247. 10.1016/j.cub.2016.03.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y., Endepols S., Klemann N., Richter D., Matuschka F. R. et al. , 2011. Adaptive introgression of anticoagulant rodent poison resistance by hybridization between Old World mice. Curr. Biol. 21: 1296–1301. 10.1016/j.cub.2011.06.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A., 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner C. C., Weber J. N., and Hoekstra H. E., 2007. Adaptive variation in beach mice produced by two interacting pigmentation genes. PLoS Biol. 5: e219 [corrigenda: PLoS Biol. 6: e36 (2008)]. 10.1371/journal.pbio.0050219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern D. L., 2013. The genetic causes of convergent evolution. Nat. Rev. Genet. 14: 751–764. 10.1038/nrg3483 [DOI] [PubMed] [Google Scholar]
- Tishkoff S., Reed F. A., Ranciaro A., Voight B. F., Babbitt C. C. et al. , 2007. Convergent adaptation of human lactase persistence in Africa and Europe. Nat. Genet. 39: 31–40. 10.1038/ng1946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollis M., Hutchins E. D., Stapley J., Rupp S. M., Eckalbar W. L. et al. , 2018. Comparative genomics reveals accelerated evolution in conserved pathways during the diversification of anole lizards. Genome Biol. Evol. 10: 489–506. 10.1093/gbe/evy013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia-Montoya W. A., Elfekih S., North H. L., Meier J. I., Warren I. A. et al. , 2020. Adaptive introgression across semipermeable species boundaries between local Helicoverpa zea and invasive Helicoverpa armigera moths. Mol. Biol. Evol. 37: 2568–2583. 10.1093/molbev/msaa108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Belleghem S. M., Rastas P., Papanicolaou A., Martin S. H., Hanly J. J. et al. , 2017. Complex modular architecture around a simple toolkit of wing pattern genes. Nat. Evol. Ecol. 1: 52 10.1038/s41559-016-0052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Belleghem S. M., Alicea Roman P. A., Carbia Gutierrez H., Counterman B. A., and Papa R., 2020. Perfect mimicry between Heliconius butterflies is constrained by genetics and development. Proceedings. Biol. Sci. B 287: 20201267 10.1098/rspb.2020.1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Auwera G. A, Carneiro M. O., Hartl C., Poplin R., Levy-Moonshine A., et al. , 2013. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinformatics 43: 11.10.1–11.10.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallbank R. W. R., Baxter S. W., Pardo-Diaz C., Hanly J. J., Martin S. H. et al. , 2016. Evolutionary novelty in a butterfly wing pattern through enhancer shuffling. PLoS Biol. 14: e1002353 10.1371/journal.pbio.1002353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp P. J., and Kalay G., 2012. Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat. Rev. Genet. 13: 59–69. 10.1038/nrg3095 [DOI] [PubMed] [Google Scholar]
- Wray G., 2007. The evolutionary significance of cis-regulatory mutations. Nat. Rev. Genet. 8: 206–216. 10.1038/nrg2063 [DOI] [PubMed] [Google Scholar]
- Zhang W., Dasmahapatra K. K., Mallet J., Moreira G. R. P., and Kronforst M. R., 2016. Genome-wide introgression among distantly related Heliconius butterfly species. Genome Biol. 17: 25 10.1186/s13059-016-0889-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. All original raw sequence data files are available via the ENA (accession number PRJEB37067). Supplementary tables and figures referred to in the text are available via figshare: https://doi.org/10.25386/genetics.12911180.