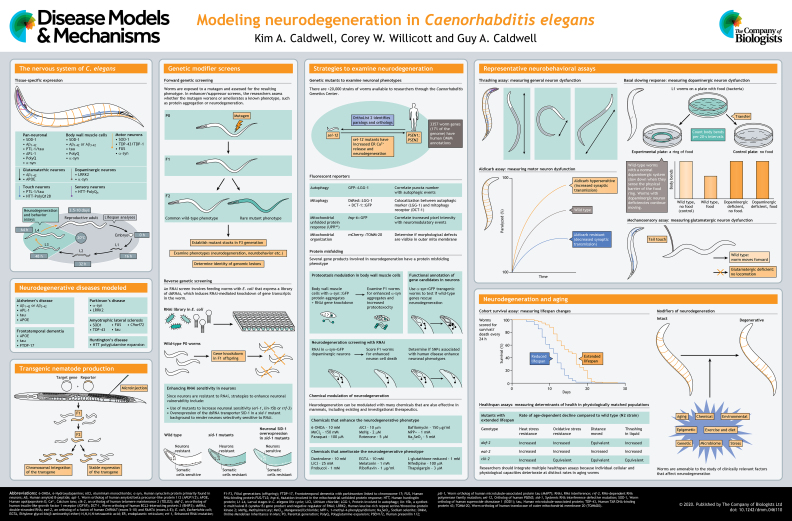
ABSTRACT
The global burden of neurodegenerative diseases underscores the urgent need for innovative strategies to define new drug targets and disease-modifying factors. The nematode Caenorhabditis elegans has served as the experimental subject for multiple transformative discoveries that have redefined our understanding of biology for ∼60 years. More recently, the considerable attributes of C. elegans have been applied to neurodegenerative diseases, including amyotrophic lateral sclerosis, Alzheimer's disease, Parkinson's disease and Huntington's disease. Transgenic nematodes with genes encoding normal and disease variants of proteins at the single- or multi-copy level under neuronal-specific promoters limits expression to select neuronal subtypes. The anatomical transparency of C. elegans affords the use of co-expressed fluorescent proteins to follow the progression of neurodegeneration as the animals age. Significantly, a completely defined connectome facilitates detailed understanding of the impact of neurodegeneration on organismal health and offers a unique capacity to accurately link cell death with behavioral dysfunction or phenotypic variation in vivo. Moreover, chemical treatments, as well as forward and reverse genetic screening, hasten the identification of modifiers that alter neurodegeneration. When combined, these chemical-genetic analyses establish critical threshold states to enhance or reduce cellular stress for dissecting associated pathways. Furthermore, C. elegans can rapidly reveal whether lifespan or healthspan factor into neurodegenerative processes. Here, we outline the methodologies employed to investigate neurodegeneration in C. elegans and highlight numerous studies that exemplify its utility as a pre-clinical intermediary to expedite and inform mammalian translational research.
KEY WORDS: Aging, Behavior, Genetics, Modeling, Phenotype, Proteostasis
Summary: While unsurpassed as an experimental system for fundamental biology, Caenorhabditis elegans remains undervalued for its translational potential. Here, we highlight significant outcomes from, and resources available for, C. elegans-based research into neurodegenerative disorders.
Introduction
Neurodegenerative diseases represent a significant and growing burden globally and the identification of factors that reduce their impact necessitates considerable research effort. Undoubtedly, rodent models have expanded our understanding of neurodegenerative disorders over the past few decades. However, a range of phenotypic variations can manifest in humans; therefore, it seems prudent to use diverse model organisms to hasten research into causes and cures for neurodegenerative diseases. In this At a Glance article, and the accompanying poster, we survey Caenorhabditis elegans as a model for neurodegenerative disease research. Many consider this nematode roundworm to be the most completely understood animal on the planet. Therefore, given the sense of urgency to accelerate discovery, the assays, molecular tools and genetic strategies developed through years of collaborative and collective effort in the worm community should be seriously considered within the arsenal for eradicating neurodegenerative diseases from the human population.
Despite the lack of evolutionary complexity, C. elegans shares many conserved molecular pathways and cellular mechanisms with mammals, thus allowing for comparative studies. OrthoList 2 is a searchable compendium of all orthologs and paralogs between C. elegans and humans and includes 7943 genes, or ∼41% of the C. elegans protein-coding genome (Shaye and Greenwald, 2011; Kim et al., 2018a). Moreover, 3357 worm genes possess human orthologs with Online Mendelian Inheritance in Man (OMIM) annotations; this is 17% of the worm genome (Kim et al., 2018a; http://ortholist.shaye-lab.org/). This conservation provides great potential for modeling human genetic disease. For example, many C. elegans neuronal genes are expressed in comparable mammalian cell types (Hobert, 2013).
While mammalian nervous systems consist of billions of neurons, the adult C. elegans hermaphrodite has ∼300 neurons throughout its body, thereby reducing the complexity and increasing the accuracy of neuronal analyses (Cook et al., 2019). The major functional components of mammalian synaptic transmission, such as neurotransmitters, receptors, transporters and ion channels, are conserved in C. elegans. Based on differences in function, morphology and connectivity, the neurons of this animal have been sorted into separate classes: mechano-, chemo- and thermo-sensory neurons, interneurons, motor neurons and neuromuscular junction components, among others. Importantly, all neuron types can be further subcategorized by neurotransmitter biosynthesis and activity; neuronal signaling molecules include glutamate (Li et al., 1997), GABA (McIntire et al., 1993), acetylcholine (Alfonso et al., 1994), dopamine (Sulston et al., 1975), serotonin and octopamine (Horvitz et al., 1982), as well as neuropeptides (Li and Kim, 2008). Stereotypical behavioral phenotypes are associated with distinct neuronal subclasses (see poster, ‘Representative neurobehavioral assays’). The C. elegans neuronal circuitry has also been fully mapped since the 1980s (White et al., 1986), and researchers recently updated the compendium of the neuronal connectomes of both the male and hermaphrodite using state-of-the-art technologies (Cook et al., 2019). Moreover, the constituent gene expression patterns, as discerned by the collective efforts of researchers in the CeNGEN Project, facilitate enhanced understanding of neuronal signaling at a level of detail simply not yet possible in other species (Hammarlund et al., 2018). Furthermore, because C. elegans is anatomically transparent, neurons are easily visualized by expressing fluorescent proteins in live animals to study neuronal properties throughout development and over time.
Genetic models of neurodegeneration
C. elegans is informative for basic studies of neuronal function. Perhaps more impressively, it can be effectively harnessed to rapidly and cost effectively identify gene targets and drug candidates that modify neurodegenerative processes. In contrast, rodent neurodegeneration studies are time consuming and economically challenging. Despite the abundance of cellular and animal models to study neurodegenerative disease, we contend that C. elegans has been underutilized with respect to its predictive capacity for translational biology. In this At a Glance article, we describe common forms of neurodegeneration modeled in C. elegans (see poster, ‘Neurodegenerative diseases modeled’), including Alzheimer's disease (AD), amyotrophic lateral sclerosis (ALS), frontotemporal dementia (FTD), Huntington's disease (HD) and Parkinson's disease (PD). We discuss the genetic and environmental modulators that share common pathways leading to cellular malfunction and eventually neurodegeneration within the context of these C. elegans human disease models. There are additional models of these neurodegenerative diseases that are not covered within the article, and we apologize for this omission due to space constraints. Furthermore, we have largely excluded discussions of any models in which neurodegeneration was not an endpoint phenotype evaluated.
AD
Two defining characteristics of AD are the presence of insoluble amyloid β-peptide (Aβ) plaques and of tau-associated neurofibrillary tangles in the brain. In mammals, Aβ is accumulated into extracellular plaques, followed by endocytic uptake of these neurotoxic oligomers. This process induces tau phosphorylation and aggregation into neurofibrillary tangles. Aβ accumulation is an outcome of sequential enzymatic processing of the human amyloid precursor protein (APP) by membrane-bound secretase enzymes mediated by the subcomponent PS1 and PS2 presenilin proteins. Although the product of the worm sel-12 gene provided some of the first evidence for presenilins as functional subunits of human γ-secretase (Levitan and Greenwald, 1995), the C. elegans genome lacks both a clear β-secretase ortholog and an ortholog of APP that would be processed to yield Aβ peptides. The worm APL-1 protein is an ortholog of the human amyloid beta precursor-like proteins 1 and 2 (APLP1 and APLP2), and has 71% sequence similarity to the intracellular domain of APP, but completely lacks an Aβ domain. Thus, AD modeling in C. elegans has primarily focused on the transgenic expression of mature human Aβ and tau (Griffin et al., 2017). Nevertheless, single-copy insertion and expression of human APP results in neurodegeneration and neurobehavioral dysfunction in C. elegans (Yi et al., 2017). Likewise, although apl-1 is essential for viability, overexpression of apl-1 yields pleiotropic phenotypes that include neuronal deficits (Hornsten et al., 2007; Ewald et al., 2012). Owing to spatial constraints, the AD section of this article will focus on Aβ modeling, while the ALS section will describe tau expression in C. elegans. However, we direct the reader to studies in which transgenic nematodes expressing human tau have been generated and include the fine work of several groups with interests in AD modeling (Kraemer et al., 2006; Ayyadevara et al., 2016; Pir et al., 2016; Wang et al., 2018; Fang et al., 2019).
Modeling AD in C. elegans has largely been based on the ‘amyloid cascade hypothesis’ (Hardy and Selkoe, 2002). Here, the Aβ1-42 peptide is thought to be the most toxic species generated by the cleavage of APP. The original and most commonly used C. elegans AD model was a result of pioneering research by Chris Link (1995). In this model, and related variants, the Aβ peptide is tagged with a secretion signaling sequence that is expressed from the unc-54 promoter in the body wall muscles, where it causes paralysis, thereby allowing for quantitative analysis of modulators. Aβ1-42-mediated neurodegeneration has been studied primarily in mechanosensory and in glutamatergic neurons, which are commonly susceptible to hyperexcitability and are clinically associated with the early stages of degeneration in AD (Palop and Mucke, 2009, 2016). Wild-type C. elegans have five glutamatergic neurons in their tails. Models of AD that overexpress GFP-fused human Aβ1-42 specifically in these neurons under the eat-4 glutamate transporter promoter demonstrate progressive, age-dependent neurodegeneration (Treusch et al., 2011). Researchers utilized a yeast model expressing Aβ1-42 targeted to the secretory pathway to identify genetic modifiers of Aβ toxicity. This genome-wide screen revealed 23 suppressors and 17 enhancers of toxicity, with 12 of these having human homologs. These modifiers were then tested in C. elegans to determine their neurotoxicity. Phosphatidylinositol-binding clathrin-assembly protein (PICALM) protected against Aβ neurotoxicity, which was further verified in primary rat cortical neurons and corroborated the human genetic data implicating this gene in AD (Treusch et al., 2011). In a separate study, a high-throughput compound screen in yeast and C. elegans models of AD identified a class of metal-binding compounds that were neuroprotective through the degradation of Aβ (Matlack et al., 2014). Further investigation showed that these small molecules were related to clioquinol, which was previously found to reduce Aβ toxicity in mouse models (Cherny et al., 2001). Using the same transgenic C. elegans Aβ1-42 model, Lu et al. (2014) showed that Wnt/β-catenin signaling attenuated glutamatergic neuron loss through regulation of spr-4, a homolog of the stress regulatory transcription factor REST.
The ε2 isoform of apolipoprotein E (APOE) has been associated with reduced risk for developing AD, whereas the APOEε4 allele increases risk, with APOEε3, the most frequent isoform, being neutral (Spinney, 2014). Our group developed a suite of transgenic C. elegans models expressing human APOE alleles, with and without Aβ1-42 (Griffin et al., 2019). Co-expressing APOEε2 with Aβ protected glutamatergic neurons from degeneration and restored normal mechanosensory behavior. In contrast, the APOEε3 worms displayed an intermediate phenotype, and expression of APOEε4 did not protect from Aβ neurotoxicity. In the absence of Aβ, the APOE alleles did not alter glutamatergic neuron function. These allele-specific neuroprotective effects could be modulated in response to endoplasmic reticulum (ER)-associated calcium changes, both pharmacologically and via RNA interference (RNAi) knockdown. Moreover, lifespan was reduced in C. elegans expressing Aβ and APOEε4; it was rescued by APOEε2 and APOEε3 alleles (Griffin et al., 2019). Thus, transgenic C. elegans lines recapitulate the established clinical profile of APOE polymorphism-associated impact on neurodegeneration, which can be used to discern new mechanistic insights for AD.
ALS and FTD
ALS is a neurodegenerative disorder that affects the upper and lower motor neurons, causing progressive muscle weakness. More recently, FTD, a dementia syndrome that can present with changes in behavior or language dysfunction, has become recognized in many ALS patients (Caga et al., 2019; Zucchi et al., 2019). Most ALS is sporadic, with only ∼10% of cases displaying a familial inheritance pattern. A causative mutation has been identified in ∼80% of these kindreds; notably, some of these same mutations also lead to sporadic ALS and many are autosomal dominant (Al-Chalabi et al., 2017). C. elegans has been used to model the four most common ALS-causing mutations in C9orf72-SMCR8 complex subunit (C9orf72), superoxide dismutase (SOD1), TAR DNA-binding protein (TARDBP) and RNA-binding protein FUS/TLS (FUS), as well as a mutation associated with FTD [microtubule-associated protein tau (MAPT)].
Mutations in C9orf72, where a hexanucleotide repeat (GGGGCC) within the first intron of C9orf72 undergoes expansion, are responsible for 10-15% of familial ALS cases (van Damme et al., 2017). The mechanism associated with this expansion remains under investigation. One hypothesis is that the translation of these repeats is AUG independent. This example of repeat-associated non-AUG (RAN) translation (Cleary and Ranum, 2017) can result in five separate dipeptide-containing proteins from the GGGGCC repeat. C. elegans researchers created transgenic models of four possible dipeptides that could potentially lead to toxic cellular consequences. Subsequent characterization revealed that arginine-containing dipeptides exhibited potent motor neuron and muscle cell toxicity (Rudich et al., 2017). A distinct effort involved C. elegans overexpressing nine and 29 GGGGCC repeats under the broadly active hsp-16 promoter. The phenotypes were more severe in animals expressing 29 repeats versus nine repeats and included paralysis followed by lethality (Wang et al., 2018). A forward genetic screen of the 29-repeat transgenic worms identified uncharacterized genes, still under analysis, that reversed this severe phenotype. In addition to overexpressing the hexanucleotide repeat, the C. elegans genome has an ortholog of C9orf72 termed alfa-1; a loss-of-function alfa-1 mutant that exhibits motor neuron degeneration and a motility defect (Therrien et al., 2013). Non-neuronally, these worms have an endocytosis defect that can be partially rescued by introducing wild-type human C9orf72 (Corrionero and Horvitz, 2018). C. elegans alfa-1 has also been reported to be involved in nutrient sensing and metabolic control (Ji et al., 2020).
Discovery of SOD1 mutations in 1993 allowed for the initial modeling of ALS (Rosen et al., 1993), even though SOD1 mutations are now understood to be responsible for only ∼2% of familial ALS (van Damme et al., 2017). As a cytosolic enzyme, SOD1 catalyzes the detoxification of superoxide. There are more than 100 mutant alleles of SOD1 associated with disease, and most mutations cause a toxic gain of function in motor neurons (Cleveland and Rothstein, 2001). Some of these mutations, such as G85R and G93A, misfold and then eventually aggregate in motor neurons, as well as in vitro (Ghosh et al., 2020). Other mutations, such as A4V, cause ER stress, but the underlying mechanism for this stress remains unclear (Perri et al., 2020). Dysfunctional proteostasis correlates with ER stress in a mouse model of G93A SOD1 (Kikuchi et al., 2006); moreover, motor neuron subclasses vulnerable to degeneration also exhibit increased ER stress upon G93A or G85A SOD1 expression (Saxena et al., 2009). Pan-neuronal expression of human G85R SOD1 in C. elegans leads to locomotor defects, development of aggregates and axonal abnormalities, such as reduced numbers and diameter of axons and fewer organelles within the remaining axons (Wang et al., 2009). Thompson et al. (2014) demonstrated that co-expression of torsinA, a highly-expressed neuronal chaperone in humans, can attenuate the ER stress caused by human G85R SOD1, as well as restore normal locomotion to transgenic C. elegans. Overexpression of a different human SOD1 mutation, G93A, specifically in motor neurons led to age-dependent paralysis as a consequence of axonal guidance defects (Li et al., 2013). Notably, these same two mutations (G85R and G93R) were subsequently modeled as single-copy insertions, along with three other common SOD-1 mutations (A4V, H71Y, L84) in C. elegans. The impact on both glutamatergic and cholinergic neuron degeneration was examined for all five of these mutations (Baskoylu et al., 2018). G93R displayed phenotypes consistent with a toxic gain-of-function phenotype in cholinergic neurons, while G85R caused glutamatergic neurodegeneration following exposure to the neurotoxin paraquat (Baskoylu et al., 2018).
Although the associated pathogenic mechanisms have yet to be understood, TAR DNA-binding protein-43 (TDP-43) (Arai et al., 2006) and FUS (Kwiatkowski et al., 2009) have been identified in proteinaceous cytoplasmic inclusions in motor neurons of ALS patients. ALS-causing mutations within the TDP-43-encoding gene, TARDBP, are associated with 0.9% of patients, whereas 0.7% of patients harbor mutations in FUS (van Damme et al., 2017). TDP-43 and FUS, which are DNA/RNA-binding proteins, regulate transcription, splicing, RNA transport and stress granule formation (Aksoy et al., 2020), and are predominantly nuclear under wild-type conditions. Notably, when mutated, both proteins display cytoplasmic mislocalization, along with protein aggregates, as a hallmark of ALS (Liu et al., 2017).
Multiple research groups have examined the phenotypic consequences of TBP-43 expression within C. elegans neurons. For example, pan-neuronal expression of human wild-type TDP-43 under control of the snb-1 promoter caused slowed and uncoordinated movement, as well as defasciculation of the motor neurons of the transgenic worms (Ash et al., 2010). In a separate effort, using the same pan-neuronal promoter, motility defects, aggregation and neurodegeneration specifically within the GABAergic neurons, resulted from the expression of mutant variants of TDP-43 (Liachko et al., 2013). These TDP-43 models have led to mechanistic insights, including the observation that calcineurin, a phosphatase, removes the pathological C-terminal phosphate from TDP-43 (Liachko et al., 2016). A phenotypic drug screen discovered that α-methyl-α-phenylsuccinimide (MPS) reversed the locomotion defects associated with TDP-43 mutant worms. MPS is the active metabolite of methsuximide, a currently used anti-epileptic drug, suggesting the utility of repurposing this compound in treating TDP-43 proteinopathies (Wong et al., 2018).
The C. elegans genome encodes an ortholog of TDP-43, called TDP-1, that is primarily expressed in the nuclei of neurons and body wall muscle cells (Vaccaro et al., 2012a; Zhang et al., 2012). TDP-1 contains most motifs found in human TDP-43, including two RNA-binding motifs, and nuclear import and export signals. However, the glycine-rich domain of human TDP-43 is missing. Nonetheless, there is still functional conservation between these two proteins; specifically, the toxicity phenotypes associated with the C. elegans tdp-1 mutant can be rescued by expressing wild-type human TDP-43 (Zhang et al., 2012). Deletion of endogenous tdp-1 could rescue the neurodegeneration associated with overexpression of mutant human TDP-43 in C. elegans GABAergic neurons (Vaccaro et al., 2012a).
FUS variants have also been examined in transgenic C. elegans. Expressing a FUS variant prone to aggregation in GABAergic neurons via the unc-47 promoter resulted in neurodegeneration, synaptic dysfunction, paralysis and aggregation, whereas expression of wild-type FUS did not cause these phenotypes (Vaccaro et al., 2012b). In a separate study, when FUS mutants were expressed pan-neuronally under control of the rgef-1 promoter, multiple mutations associated with aggregation resulted in motor dysfunction, while variants that do not aggregate did not cause observable phenotypes (Murakami et al., 2012). The C. elegans genome has an ortholog of FUS named fust-1. The fust-1 gene product is well conserved and has been reported to interact with the ortholog of AGO2 in C. elegans, ALG-1, a core microRNA-induced silencing complex component. Additionally, FUST-1 is required to achieve maximum microRNA (miRNA)-mediated gene silencing (Zhang et al., 2018).
FTD with parkinsonism linked to chromosome 17 (FTDP-17) is characterized by the accumulation of the microtubule-associated protein tau. Symptoms include neurodegeneration and cognitive decline. However, the underlying mechanisms remain unclear. Researchers have developed transgenic C. elegans models expressing either the wild-type human tau or FTDP-17-causing variants (P301L or V337M) under control of a pan-neuronal promoter (Kraemer et al., 2003). Expression of the mutant alleles resulted in cholinergic signaling defects and GABAergic neurodegenerative phenotypes. Additionally, these animals were uncoordinated, displayed egg-laying defects and had a reduced lifespan. Western blotting demonstrated the accumulation of phosphorylated insoluble tau in these animals (Kraemer et al., 2003). Another study showed that expression of either normal human tau or pseudohyperphosphorylated tau within the GABAergic neurons caused uncoordinated movement without neurodegeneration (Brandt et al., 2009). Notably, pseudohyperphosphorylated tau-expressing animals did exhibit a developmental abnormality with visible gaps in the motor neurons.
The tau mutation A152T is a rare risk factor for FTD and AD. Worms with pan-neuronal expression of human tau A152T displayed GABAergic neuron degeneration (Pir et al., 2016). A subsequent study found that pan-neuronal tau A152T expression effectuated the loss of glutamatergic neurons significantly more than GABAergic neurons and attributed this loss to differential vulnerability to excitotoxicity (Choudhary et al., 2018). Results from a genome-wide RNAi screen (Kraemer et al., 2006) and a follow-up study using a C. elegans model overexpressing human tau pan-neuronally discovered that the unfolded protein response (UPR) of the ER (UPRER) is a potential modulator of tau proteostasis (Waldherr et al., 2019). Furthermore, the expression of the constitutively active X-box binding protein 1 (XBP-1), the driving transcription factor of the UPRER, improved tau clearance in the cytoplasm and enhanced neuron survival (Waldherr et al., 2019).
HD
An autosomal-dominant disorder characterized by the progressive and age-associated decline in motor control and cognition, typically beginning in mid-life, HD is a profoundly tragic neurodegenerative disease because of its strong familial association and highly predictive mortality, owing to the absence of any disease-modifying therapy. In humans, HD is characterized by an increase in polyglutamine repeats (polyQs) in the N-terminus of the huntingtin protein (HTT). Repeat length correlates with the age of onset and severity, with 35 repeats being the threshold of disease development (Orr and Zoghbi, 2007). In C. elegans, HD has proven archetypical for modeling the protein misfolding pathology and altered proteostasis shared among neurodegenerative diseases.
Despite the absence of a C. elegans ortholog of HTT, several groups have generated transgenic animals expressing polyQ tracts and human HTT fragments with varying polyQ-repeat lengths in different neuronal subtypes to study HD pathology (Satyal et al., 2000; Parker et al., 2001; Morley et al., 2002; Nollen et al., 2004). One of these is limited to the ASH sensory neuron, achieved by placing the transgene under the control of the osm-10 promoter. Degeneration correlates with polyQ length and is readily detected by the failure of this ciliated neuron to uptake a fluorescent dye (Faber et al., 1999). Expression of HTT with normal and expanded polyQ tracts specifically in the touch receptor neurons, under control of the mec-3 promoter, caused mechanosensory neuron dysfunction and degeneration of axons in a polyQ-length-dependent manner (Parker et al., 2001). These models have been used to investigate the mechanisms underlying HD and the therapeutic potential of various compounds.
Because mouse models of neurodegenerative diseases showed that loss of autophagy enhanced degeneration, researchers tested the effect of neuron-specific autophagy gene loss to evaluate the role of protein turnover in an HD C. elegans model. Inactivation of these genes in the presence of polyQ expansions increased the accumulation of aggregates and their toxicity, resulting in augmented neurodegeneration (Jia et al., 2007). Similarly, researchers observed that glucose was neuroprotective in the 128-repeat polyQ HD model. This protection was DAF-16 dependent and glucose operated by reducing misfolded protein levels (Tauffenberger et al., 2012). As a final example, the protective activity of rutin, previously shown to be therapeutic in rat models of HD, was demonstrated through a reduction in polyQ-induced neuron death and elevated activation of DAF-16 (Cordeiro et al., 2020).
PD
Following AD in prevalence, PD is the second most common neurodegenerative disease, afflicting ∼10 million people worldwide. Unlike HD, PD is primarily idiopathic, with only ∼10% showing familial inheritance (Hernandez et al., 2019). Although early-onset PD accounts for a small amount of cases, the vast majority of individuals display symptomality late in life, typically after age 65, with increased age being the predominantly accepted risk factor. The primary clinical hallmarks of PD include postmortem evidence of Lewy bodies and loss of dopaminergic neurons in the substantia nigra region of the brain. Owing to its central role in PD pathogenesis, α-synuclein (α-syn), which is encoded by the PARK1/SNCA locus in humans and is associated with autosomal-dominant PD (Meade et al., 2019), has been modeled extensively in C. elegans. α-syn is also a major component of Lewy bodies and multiplication of the PARK1/SNCA locus has been linked to enhanced aggregation and dopaminergic neuron death (Mezey et al., 1998; Singleton et al., 2003). C. elegans has orthologs of many PARK genes, with the notable exception of PARK1/SNCA. This conveniently allows C. elegans researchers to overexpress α-syn without interference from endogenous α-syn or a dominant-negative effect, as the worm essentially serves as ‘an α-syn null genetic background’. Researchers have developed numerous C. elegans models of PD and, in most, α-syn and GFP are selectively expressed from either a dopaminergic or pan-neuronal promoter. The dopamine transporter (dat-1) promoter delivers higher expression levels and concomitant dopaminergic neurodegeneration from overexpression of either wild-type or familial mutant forms of α-syn (Lakso et al., 2003; Cao et al., 2005; Karpinar et al., 2009). Worm models of α-syn-induced neurodegeneration have been widely used in combination with genetic and drug screening in a discovery pipeline comprised of more than one model system to successfully demonstrate the evolutionary conservation of function for modifiers and pathways.
Cooper et al. (2006) demonstrated that overexpression of human α-syn in yeast blocks ER-to-Golgi vesicular trafficking; this was the first report of cellular malfunction due to α-syn. Moreover, this study employed multiple model systems to provide additional support with results from Drosophila, C. elegans and primary rat neuronal cultures confirming that increased expression of a key vesicular trafficking pathway protein, Rab1, could rescue dopaminergic neuron loss induced by α-syn (Cooper et al., 2006). A separate screen of 190,000 compounds for reversal of α-syn-induced yeast toxicity identified N-aryl benzimidazole (NAB). Notably, NAB activity was conserved in neurons, as it protected C. elegans dopaminergic neurons and α-syn-expressing rat primary neurons from α-syn-induced neurodegeneration (Tardiff et al., 2013), as it did in dopaminergic neurons differentiated from patient-derived induced pluripotent stem cells (Chung et al., 2013). Another large-scale candidate gene screen conducted in C. elegans expressing α-syn::GFP in a daf-2 receptor-mutant background, a pivotal regulator of aging and lifespan, identified the glycolytic enzyme GPI-1/GPI1 as a neuromodulatory factor. Concurrently, GPI-1/GPI1 was shown to protect against α-syn proteotoxicity in Drosophila and mouse primary neuronal cultures (Knight et al., 2014). These collaborative pipeline-type studies emphasize the importance of basic cell biology in neurodegeneration. Furthermore, they exemplify the power of using multiple model systems to identify evolutionarily conserved pathways and new targets as functional effectors of neurodegeneration.
One of the enduring mysteries of PD is the selective vulnerability of dopaminergic neurons to degeneration. It has been hypothesized that dopamine itself may be a key contributor to pathogenesis, as it is more readily oxidized than other neurotransmitters. In vitro experiments with purified α-syn polypeptides demonstrated that addition of dopamine enhances and stabilizes the oligomeric state of α-syn, whereas deletion of five amino acid residues (125YEMPS129) in the C-terminal region of α-syn abolished this effect (Herrera et al., 2008). Using transgenic C. elegans expressing the aggregation-prone A53T α-syn variant, researchers investigated the interaction between α-syn and dopamine with and without the dopamine interaction site (Mor et al., 2017). The animals were crossed into an established strain that overproduces dopamine by the overexpression of tyrosine hydroxylase, the product of cat-2 (Cao et al., 2005), to reveal that excess dopamine exacerbates neurodegeneration. These data corroborated results showing that enhanced dopamine levels in mice expressing human A53T α-syn induced nigrostriatal degeneration (Mor et al., 2017). Importantly, worms expressing A53T α-syn lacking the 125YEMPS129 sequence exhibited baseline levels of neurodegeneration. This differential effect depended on the presence of cat-2 overexpression (Mor et al., 2017). The use of C. elegans in this study provided mechanistic clarity to help resolve a long-standing question of PD neuropathology with clinical implications.
Mutations in leucine-rich repeat kinase 2 (LRRK2) are associated with autosomal dominant PD. Based on biochemical experiments, mutations are linked to modifications in the GTPase and kinase activities of LRRK2 that are likely the basis of the neuronal toxicity in these PD patients. Transgenic expression of wild-type or disease-causing mutant LRRK2 under pan-neuronal or dopaminergic-specific promoters results in neuronal loss in C. elegans (Saha et al., 2009; Liu et al., 2011; Johnson et al., 2015). Using the dopaminergic LRRK2 model, Liu et al. (2011) confirmed the effects of select inhibitors of LRRK2 kinase activity identified from chemical library screening. Additionally, the reduced glutaredoxin (Grx1; also known as GLRX) levels observed in postmortem PD patient midbrain samples was subsequently validated in a C. elegans LRRK2 model. Specifically, loss of worm Grx1, combined with LRRK2 overexpression, resulted in significantly greater levels of neurodegeneration (Johnson et al., 2015). While its kinase activity offers an attractive target for drug intervention, LRRK2 is a complex multidomain protein comprised of over 2500 amino acids, and largely remains a functional enigma underlying PD pathology. Interestingly, phosphoproteomic analysis has identified Rab GTPases as biological substrates for LRRK2-dependent phosphorylation (Steger et al., 2016). It is notable that among the Rab proteins modified by LRRK2 (Rab1, Rab3, Rab8, Rab10 and others), several were identified as modulators of α-syn-dependent neurodegeneration using C. elegans well over a decade ago (Cooper et al., 2006; Gitler et al., 2008). More recently, a convergence of α-syn and LRRK2 dysfunction around intracellular trafficking has coincided with evidence highlighting the significance of endolysosomal vesicular fusion and lysosome function in PD-associated neurodegeneration (Winckler et al., 2018). In this context, there is increased interest in factors affecting lipid homeostasis as a potential avenue of therapeutic target development (Fanning et al., 2020). C. elegans has already been used to demonstrate the neuroprotective effect of stearoyl-CoA desaturase inhibition (Fanning et al., 2019), an effect that translates to human-derived neurons cultured in the presence of neurotoxic α-syn (Vincent et al., 2018). Likewise, phosphatidylethanolamine (PE) deficiency enhances neurodegeneration in transgenic α-syn animals and ethanolamine supplementation reverses this effect (Wang et al., 2014). These data correlate with reports of an age-dependent decline in PE found in PD patients.
The role of the lysosome in misfolded protein degradation (autophagy) and damaged mitochondrial clearance (mitophagy) has emerged as a substantial factor in PD. Again, over a decade ago, C. elegans RNAi screening to identify modifiers of α-syn misfolding and neurodegeneration revealed in vivo effects of proteins that function in autophagy or mitophagy (ATG7, ULK1, PINK1, PRKN), endolysosomal trafficking (VPS41) and lysosome function (cathepsin D, ATP13A2/PARK9) on dopaminergic neurodegeneration (Hamamichi et al., 2008; Gitler et al., 2009; Qiao et al., 2008). Most significantly, Sidransky et al. (2009) defined an association between PD and mutations in the human glucocerebrosidase gene, GBA1 (also known as GBA), a causal factor in the rare lysosomal storage disorder Gaucher's disease. Whereas homozygous mutations in GBA1 cause Gaucher's disease, heterozygous carriers are of significantly higher risk of PD. Thus, a deficit in glucocerebrosidase leads to accumulation of glycolipids. Using transgenic C. elegans overexpressing human α-syn in the dopaminergic neurons, Mazzulli et al. (2011) reported that neuron-specific RNAi knockdown of gba-1 enhanced dopaminergic neurodegeneration. This supported an initial hypothesis that α-syn misfolding and GBA-1 loss of function combine in a pathogenic loop of exacerbated neurotoxicity. As of writing, GBA1 mutations comprise the most common class of genetic cases of PD.
These examples are only a small sampling of the numerous contributions C. elegans models have made to neurodegenerative disease research. Through the collective efforts of worm researchers worldwide, functional characterization of previously uncharacterized or poorly understood disease-associated gene products has provided mechanistic insights to illuminate the translational path forward. As discussed below, the toolbox of resources available in the C. elegans research community continues to expand, further establishing this model system as a means to enable and accelerate discovery in this area of urgent global need.
Tools and approaches for the functional characterization of neurodegenerative mechanisms
Transgenic nematodes
Researchers incorporate exogenous DNA into C. elegans for numerous purposes, such as to rescue mutant alleles, analyze gene function, or to examine gene expression (see poster, ‘Transgenic nematode production’). In all cases, the gene of interest is engineered to be controlled by an endogenous C. elegans promoter. This promoter permits expression in all or in selected tissues. Additionally, for strain construction, a fluorescent or phenotypic marker (GFP, mCherry, rol-6) is also used to distinguish transgenic progeny. For many years, these expression constructs were microinjected, via the germ line, into C. elegans, where they become linearized and naturally form concatemers that result in overexpression in the progeny of successfully injected animals (Mello et al., 1991; Berkowitz et al., 2008). A caveat to this method is that it is not possible to control the amount of transgene incorporated into separate transgenic lines. To correct for this, researchers typically analyze multiple lines of transgenic animals and determine a consensus phenotype for transgenic overexpression. More recently, worm researchers have turned to CRISPR/Cas9-guided genome engineering approaches for the generation of transgenic animals. This method can achieve single-copy insertion of transgenes, thus circumventing the issues of overexpression described above. Additionally, if one were to express a human disease mutation in a C. elegans strain that was concomitantly null mutant for the same gene, this would result in a ‘humanized’ worm. As an example, Zhu et al. (2020) sought to functionally validate epilepsy-related mutations and generated a strain in which a null mutation was introduced into the C. elegans ortholog of STXBP1, unc-18, via CRISPR/Cas9, which resulted in paralysis. They found that the human ortholog functionally replaced the worm gene and that the epilepsy-associated variants caused seizure-like activity. Although CRISPR/Cas9 and other means by which single-copy insertion or editing can be achieved are often desirable approaches, in the case of neurodegenerative disease modeling, multicopy transgene expression can actually be preferable. For example, dose-dependent neurotoxicity of α-syn and Aβ are established characteristics of human pathology that can be recapitulated in transgenic C. elegans harboring tandem multicopy arrays. Lower copy number arrays or less robust promoters can result in different levels of neurodegeneration, which is useful for threshold-based genetic screening of suppressors and enhancers, or a complete lack of degenerative phenotype in vivo (Harrington et al., 2010; Therrien and Parker, 2014; Wang et al., 2016).
Forward genetic screening
In the early 1960s, Nobel laureate Sydney Brenner began a nascent interest in developing a model to understand an organism's nervous system in its entirety. He chose C. elegans to uncover cellular details of the mechanistic foundation of the nervous system. This nematode was a wise choice because of its ease of genetic manipulation. While males exist among populations as a result of chromosomal non-disjunction events, C. elegans is predominantly a hermaphrodite in nature. Thus, for example, if a hermaphrodite has one mutation and is crossed with a male that has a different mutation, the next generation hermaphrodite will inherit both mutations as a heterozygote. Achieving homozygosity is simply a matter of ‘selfing’ this individual hermaphrodite, which will produce up to 300 offspring.
Brenner began understanding the nervous system in C. elegans by identifying synaptic transmission genes via a forward genetic screen (see poster, ‘Genetic modifier screens’). He treated worms with ethyl methanesulfonate (EMS) and screened for uncoordinated (or ‘Unc’) worms, identifying 77 Unc genes (Brenner, 1974). Mapping these genes was arduous, and cloning mutations had no guarantee of success. Now, with advances in genome sequencing techniques, there has been a renaissance in the approach. Countless forward genetic screens have been performed, based on individual phenotypes or as modifier screens. As examples, an EMS screen of 80,000 haploid C. elegans genomes expressing GFP in the dopaminergic neurons followed by fluorescence-activated sorting of individual worms with defective dopaminergic neuron differentiation, identified 22 alleles organized into six complementation groups. It was estimated that the screen reached ∼78% saturation based on the allele recovery rate (Doitsidou et al., 2008). In a separate effort, following the development of C. elegans models of tauopathy by pan-neuronally expressing wild-type or FTD-associated mutant tau (tau-FTDP-17) (Kraemer et al., 2003), forward genetic screens identified two new molecular factors, SUT-1 and SUT-2, that participate in the activation of tau (Kraemer and Schellenberg, 2007; Guthrie et al., 2009). SUT2 is now an established susceptibility factor for tau pathology in the mammalian brain (Wheeler et al., 2019). Worms harboring deletions and point mutations that have been isolated from genetic screens and carefully annotated are deposited in the Caenorhabditis Genetics Center, with over 20,000 strains available to the research community. These include mutations in many genes associated with neurodegenerative diseases.
Reverse genetic screening
Neurodegeneration-associated phenotypes have also served as the basis of reverse genetic screens (see poster, ‘Genetic modifier screens’). The discovery and application of RNAi has revolutionized phenotypic analysis in C. elegans. Unlike in most animals, RNAi is typically performed by feeding parental C. elegans with bacteria that produce double-stranded RNA (dsRNA) against the target of interest (Timmons et al., 2001). The progeny exhibit altered phenotypes that are scored. RNAi feeding of individually cloned dsRNA of nearly every predicted open-reading frame has been used in high-throughput screening of embryonic and post-embryonic phenotypes in C. elegans (Kamath et al., 2001, 2003). As an example, an RNAi screen covering 85% of the genome was performed in transgenic worms that express normal and FTDP-17 mutant human tau display uncoordinated phenotypes (Unc). Notably, 75 genes enhanced the Unc phenotype. Among these, the 46 candidates with human homologs were preferentially examined further and could be broadly classified as genes involved in dysregulated cell signaling (Kraemer et al., 2006).
RNAi in neurons
Historically, experimental and anecdotal reports suggested that RNAi was less effective in neurons (Asikainen et al., 2005), although mutant worm strains with enhanced sensitivity to RNAi, such as rrf-3, lin-15b or eri-1, bypass this limitation (Simmer et al., 2002; Kennedy et al., 2004). However, when performing RNAi in these strains, the silencing is systemic and occurs in all tissues of the animal. An alternative method for neuronal RNAi involves selective expression of sid-1, which encodes the ATP-independent dsRNA transporter (see poster, ‘Genetic modifier screens’). This is accomplished by generating sid mutant animals expressing sid-1 under the control of a neuron-specific promoter that restricts RNAi to neurons. Calixto et al. (2010) first used this approach to demonstrate successful selective pan-neuronal knockdown of targets. Similar strains have been developed for dopaminergic or glutamatergic neuron-specific RNAi (Harrington et al., 2012; Griffin et al., 2019). Using a neuronal-specific RNAi strain (versus eri-1 or rff-3) is an important strategy for studying the effects of depleting essential gene targets that would be lethal for C. elegans if knocked down systemically.
Comprehensive libraries of individual RNAi ‘feeding vectors’ are available from multiple sources, enabling large-scale screening efforts in neuronal contexts (Kamath et al., 2003; Rual et al., 2004). For example, a genome-wide RNAi screen for neurodegenerative phenotypes utilized a transgenic C. elegans strain in which human mutant TDP-43 was expressed pan-neuronally and caused motor neuron dysfunction. The authors identified 46 genes (out of 16,767 screened) that, when knocked down, partially suppressed the TDP-43-induced motor phenotype (Liachko et al., 2019). They then focused their efforts on the 24 candidates with human homologs. Knocking down one of these candidates, glucuronic acid epimerase (GLCE), in human cultures protected against the phosphorylated TDP-43 accumulation caused by oxidative stress. Additionally, GLCE expression was significantly decreased in the brains of patients with frontotemporal lobar degeneration with TDP-43 inclusions (FTD-TDP) relative to normal controls (Liachko et al., 2019). This exemplifies the value of C. elegans as a means to accelerate the translational path through defining previously undiscovered genetic modifiers, susceptibility factors, or potential therapeutic targets with direct disease relevance.
Strategies to examine neurodegeneration
The pathological loss of neurons in neurodegenerative diseases is recapitulated in C. elegans models, where conditions that promote either neuron survival or loss can be assessed in live animals that express fluorescent proteins in the neuronal class of interest (see poster, ‘Strategies to examine neurodegeneration’). Neurodegeneration is induced by expressing pathogenic proteins within specific the neuronal class(es), followed by scoring subtle attributes of neuronal health, including cell body rounding (a sign of apoptosis or necrosis), the disappearance of axons and/or axonal blebbing, broken neurites and/or retreat of dendritic terminals. Given the limited numbers of neurons comprising specific classes in C. elegans, researchers can score neurodegeneration with unparalleled accuracy in hundreds of animals per condition to provide robust and rigorous analyses.
Genetic crosses to examine neuronal phenotypes
To tackle familial forms of neurodegenerative disease, C. elegans investigators often take a genetic approach by examining mutant variants of endogenous proteins. As previously discussed, a form of familial AD results from mutations in PSEN1 or PSEN2. When mutated, the C. elegans presenilin ortholog, sel-12, causes ER Ca2+ dysregulation similar to that found in mammals (Sarasija and Norman, 2015). Notably, further research with the sel-12 mutant revealed mitochondrial morphological and metabolic phenotypes that fostered neurodegeneration (Sarasija et al., 2018). Another sel-12 mutant phenotype, an egg-laying defect, can be rescued by mutating the worm REST ortholog, spr-4 (Lakowski et al., 2003). These spr-4 mutations, originally isolated from a forward genetic screen for egg-laying defects, have since revealed insights into pathways regulating neurodegeneration. For example, mutations in SPR-4/REST enhance Aβ-induced glutamatergic neurodegeneration in transgenic C. elegans; this result correlated with those from companion studies in mammals (Lu et al., 2014).
Notably, mutations in C. elegans pink-1 and pdr-1, which are homologous to the recessive familial PD proteins PINK1 and PRKN, do not display neurodegenerative phenotypes even though they are null and strong loss-of-function mutations, respectively. However, these genetic lesions do cause dopamine-dependent behavioral deficiencies (Cooper et al., 2017) that could potentially be used in screens for gene-by-gene or gene-by-environment interactions. As examples, exposing pdr-1 mutant worms to either a secondary stressor such as α-syn, a proteasome inhibitor (i.e. MG-132) or the neurotoxic dopamine analog 6-hydroxydopamine (6-OHDA) enhances neurodegeneration (Martinez et al., 2015; Hartman et al., 2019). Conversely, pink-1 mutant worms in which α-syn is expressed in the dopaminergic neurons have significantly enhanced neurodegeneration, but no sensitivity to the proteasome inhibitor MG-132, and exhibit reduced neurodegeneration upon 6-OHDA exposure (Martinez et al., 2015; Hartman et al., 2019). These studies highlight the ability to dissect mechanisms in a genetically tractable organism as an informative prelude to more expensive and time-consuming validation studies in mammalian models.
Modeling protein misfolding
Many researchers also want to establish whether neurodegeneration is associated with alterations in protein handling. However, the small size of nematode neurons render visual inspection of protein misfolding a challenge and impractical for screening or serial analyses. To circumvent this limitation, models of pathogenic proteins conjugated to fluorescent molecules have been expressed in the large body wall muscle cells in C. elegans. This enables researchers to assess changes due to alterations in protein misfolding, apparent aggregate density or count following RNAi or small-molecule exposures, in distinct genetic backgrounds and over the course of aging. Although these are not neurodegenerative readouts, the ease with which rapid visual screening can be conducted in muscles has afforded researchers a means toward preliminary identification of putative functional modifiers of pathogenic proteins that can be subsequently investigated in neurons and/or mammalian cells. As outlined in the following sections, this strategy has identified multiple gene and drug modifiers of neurodegeneration that have successfully predicted outcomes in mammalian models.
As many neurodegenerative diseases have concomitant defects in protein handling, C. elegans researchers have developed models of pathogenic protein misfolding (i.e. Aβ1-42, polyQn, tau and α-syn) in the nematode body wall muscle cells (see poster, ‘Strategies to examine neurodegeneration’). Misfolded protein density and/or count of aggregated pathogenic proteins can be assayed in the α-syn models (Hamamichi et al., 2008; van Ham et al., 2008). Following hypothesis-based bioinformatics approaches to systematically define candidate targets potentially associated with PD, RNAi was used to identify genetic modifiers of α-syn::GFP misfolding and accumulation in screens of hundreds of candidate targets, initially in body wall muscle cells (Hamamichi et al., 2008; Knight et al., 2014). Upon a winnowing of candidates, the significantly shorter lists of hits were then examined for knockdown or overexpression in dopaminergic neurons (Hamamichi et al., 2008; Knight et al., 2014). Using this tiered strategy in screens for α-syn-related phenotypes, neuroprotective candidates identified in C. elegans have been repeatedly validated in mammals and/or in human genetic studies. For example, overexpression of the lysosomal trafficking protein VPS41 (VPS-41 in C. elegans) protects cells against several PD-related neurotoxins, including 6-OHDA and rotenone (Ruan et al., 2010). Moreover, overexpression of VPS41 decreases the levels of α-syn protein levels in human neuroglioma cells (Harrington et al., 2012). Considering the pathological overlap between PD and AD, VPS-41 was examined for therapeutic potential using C. elegans Aβ paralysis and neurodegeneration models. Notably, VPS-41 protected in both diseases, but with notable functional distinctions between the modes of neuroprotection between PD and AD. Specifically, in the α-syn model, neuroprotection is mediated via RAB-7 and AP-3, while in the Aβ model, it occurred through an ARF-like GTPase gene product, ARL-8 (Griffin et al., 2018). Given the high conservation of endocytic components among cells and species, the ability to dissect such mechanistic distinctions in neuroprotective pathways using C. elegans warrants further consideration when prioritizing targets for in vivo modeling.
In the Aβ models of AD, Aβ peptide accumulation in muscle cells induces paralysis, which can be readily quantified. Two different muscle expression models are widely used in the field; one constitutively expresses Aβ3-42 and is utilized for studies of toxicity and metabolism (Link, 1995), while the other provides inducible expression of Aβ1-42 (Link et al., 2003). RNAi screens can identify the targets of drugs previously shown to inhibit Aβ aggregation. Studies of the antioxidant resveratrol in C. elegans have demonstrated that it is neuroprotective and prevents the formation of Aβ aggregates (Regitz et al., 2016). Following verification of this protection in C. elegans constitutively expressing Aβ3-42 in the body wall muscles, RNAi was performed on proteostasis-related targets. Notably, UBL-5, part of the mitochondrial UPR (UPRmt), was identified as a critical component of resveratrol-mediated aggregate prevention and XBP-1, a key regulator in the UPRER, was also necessary (Regitz et al., 2016).
The polyQn HD body wall muscle expression models can provide both quantitative and behavioral information. Aggregate accumulation positively correlated with adverse effects on motility, which worsened with increasing polyQ lengths (Q29 versus Q35 versus Q82) (Satyal et al., 2000). These models have been used in concert with either α-syn or Aβ models in comparative RNAi screens to parse gene candidates into subcategories for those that have a common basis in proteostasis malfunction versus those that drive a more pathogenic-protein specific misfolding event (Nollen et al., 2004; van Ham et al., 2008; Knight et al., 2014). For example, a genome-wide RNAi screen performed on animals expressing polyQ expansions identified genes involved in RNA processing and in the synthesis, folding, trafficking and degradation of proteins as important in polyQ aggregate formation (Nollen et al., 2004). A subsequent screen using animals expressing misfolded α-syn identified ER/Golgi vesicle-trafficking and quality control genes (van Ham et al., 2008).
Chemical modulation of neurodegeneration
A variety of chemical compounds can alter the extent of neurodegeneration in C. elegans (see poster, ‘Strategies to examine neurodegeneration’). Neurodegeneration enhancers such as 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) and 6-OHDA induce PD-like phenotypes in C. elegans, rodents and other model organisms (Betarbet et al., 2002; Braungart et al., 2004; Cao et al., 2005). Conversely, chemical modulators can also attenuate neurodegeneration through distinct mechanisms. Bafilomycin, for example, reduces neurodegeneration by inhibiting autophagic vesicle maturation (Pivtoraiko et al., 2010), dantrolene reduces neurodegeneration through inhibition of intracellular calcium release in the ER (Xu et al., 2001), and probucol attenuates neurodegeneration via its antioxidant properties (Ray et al., 2014). Diverse modulators, applied alone or in tandem with transgenes, coupled with medium- or high-throughput screening, facilitate the discovery of drugs to combat neurodegenerative diseases.
In addition to the aforementioned examples, a transgenic C. elegans ALS model expressing mutant TARDBP was an initial platform for screening chemical compounds; the hits, neuroleptics, were then validated in a zebrafish model, and the most potent molecules were subsequently examined in mice and in a small clinical trial (Patten et al., 2017). This screen is among the first in the neurodegenerative disease class to realize the true translational potential for target engagement in humans. In another example, after screening >14,000 chemical candidates that reduce aggregation and fibril formation in a human neuroblastoma cell line (SH-SY5Y), a hit compound also reduced α-syn-induced neurodegeneration in C. elegans (Pujols et al., 2018). The predictive nature of these screening results, based on genomic and cellular homology across species, demonstrates the utility of this nematode as part of a comprehensive discovery scheme.
Monitoring neuronal cell health
The transparent nature of C. elegans is useful for gauging cellular health via fluorescent reporters (see poster, ‘Strategies to examine neurodegeneration’). As both autophagy and mitochondrial maintenance or impairment have emerged as pivotal to disease and aging (Chan, 2006; Malpass, 2013), several such reporters inform cellular health within the context of neurodegenerative diseases.
The autophagy marker LGG-1 [the worm homolog of LC3 (also known as MAP1LC3A)] is expressed in the muscles, intestine, pharynx, vulva, hypodermis, somatic gonad and nervous system of C. elegans, and allows detection of autophagosomes using the fluorescent reporter GFP::LGG-1 (Palmisano and Melendez, 2016). This marker shows diffuse intracellular expression under baseline conditions, with increased expression becoming evident in conditions that induce autophagy, such as starvation. Monitoring the localization and/or intensity of GFP::LGG-1 can query gene targets that functionally affect autophagy. Another transgenic line to evaluate mitophagy consists of the worm ortholog of human mitophagic regulator BNIP3, DCT-1, tagged with GFP along with dsRed::LGG-1 (Palikaras et al., 2015). DCT-1::GFP localizes to the outer mitochondrial membrane and dsRed::LGG-1 labels autophagosomes. Mitophagy-inducing stimuli can be visualized by the colocalization of these markers. Likewise, mitochondrial fragmentation can be visualized by expressing TOMM-20::mCherry in C. elegans (Jiang et al., 2015). The localization of the mitochondrial import receptor TOMM-20 to the outer mitochondrial membrane allows researchers to study conditions that interrupt the fusion/fission balance.
Assays can quantify mitochondrial stress response through the activation of the UPRmt. This mitochondrial quality control program is activated during periods of stress to promote survival and recovery of mitochondrial function. One response of this pathway is the induction of the chaperone HSP-6. By tracking the fluorescence intensity of an hsp-6::GFP transcriptional reporter, researchers can measure the activation of UPRmt (Haynes et al., 2007). This readout has proven key to deciphering the impact of the UPRmt in neurodegenerative disease. Notably, in transgenic C. elegans PD models, α-syn overexpression appears to co-opt this pathway from a normally protective response to transient stressors into a potential contributor to proteostatic collapse and neurodegeneration when chronically activated (Martinez et al., 2017).
C. elegans research ushered in the revolution in live imaging with the Nobel prize-winning report of GFP by Marty Chalfie and colleagues (Chalfie et al., 1994) and the toolbox of in vivo reporters of bioactivity for worm research has always been substantial, as the examples discussed here highlight. With the advent of CRISPR/Cas9 editing, novel optogenetic reporters, increasingly diverse indicators and high-resolution microscopy methods, C. elegans is poised to remain at the forefront of technologies that can evaluate neuronal function and health in disease modeling.
Evaluation of neurobehavior
Researchers have developed several common assays with relevance to neurodegenerative disorders that we describe here (see poster, ‘Representative neurobehavioral assays’). For many, automated image software programs assist with accuracy. It should be noted that this section is not a comprehensive survey, as we are unable to describe all the variations and related assays in this space. For a complete review and applications of worm behavioral assays and detailed protocols go to www.wormbook.org.
General neuronal dysfunction assay
The thrashing assay is a basic assay for generalized neuronal dysfunction. Worms placed in liquid rapidly initiate a response termed ‘thrashing’; a thrash is defined as a directional change in mid-body bending. During the thrashing assay, the frequency of lateral swimming is measured over a short time (30-60 s) for each animal. This assay can be used as a functional readout of neuronal health of transgenic and drug-treated worms. Kraemer et al. (2003) used thrashing assays to discern behavioral distinctions between normal and mutant forms of human tau linked to FTDP-17 expressed pan-neuronally in C. elegans. The FTDP-17 variants exhibited decreased thrashing independent of tau expression levels. Similarly, another C. elegans model in which neurodegeneration was induced via GABAergic neuron expression of arginine-rich dipeptide repeat proteins designed to functionally mimic the effects induced by such repeats in the human C9orf72 protein, a prevalent cause of ALS and FTD. The altered motility in these animals was associated with dipeptide-repeat expression and correlated with morphological changes in neuron structure indicative of neurodegeneration (Rudich et al., 2017).
Motor neuron dysfunction
One convenient tool for the identification of genes that code for synaptic transmission regulators is the quantification of sensitivity to aldicarb, an acetylcholinesterase inhibitor. Beginning with the seminal research of Rand and Russell (1984), aldicarb has been used to define many conserved components of cholinergic neurotransmission. Aldicarb causes paralysis in wild-type C. elegans, while animals with mutations in acetylcholine neuromuscular signaling at the synaptic cleft are resistant. In contrast, some animals might be more sensitive to aldicarb, indicating increased acetylcholine release. Worms are exposed to aldicarb on a Petri dish, and populations are assessed for paralysis every 10-30 min for a total of 150 min. A study of the neurotoxic selectivity of perfluorooctane sulfonate, a chemical widely used in industry, showed that exposure causes dopaminergic neuron deficits and, through the use of aldicarb, found that this was independent of acetylcholine function (Sammi et al., 2019). In another study, an aldicarb-based assay showed that DNJ-14, a worm homolog of the human cysteine-string protein (DNAJC5), a co-chaperone necessary for synaptic maintenance, was not required for neuromuscular transmission in aging (Mulcahy et al., 2019). Conversely, in an ALS/FTD model, Vaccaro et al. (2012a,b) showed that transgenic worms expressing either mutant or wild-type human TDP-43 and FUS differed in their response to aldicarb, with the mutants exhibiting hypersensitivity to paralysis. In this example and others, aldicarb assays provide a means to correlate behavioral deficits with neurodegeneration.
Dopaminergic neuron dysfunction
Wild-type C. elegans display a dopamine-mediated slowing of locomotion upon entry into a bacterial lawn, owing to a mechanosensory process that detects textural change (Sawin et al., 2000). This dopamine-regulated behavior is termed the basal slowing response (BSR) and is measured by counting the number of body bends in 20-s intervals. Rates are compared between worms placed on plates with food (Escherichia coli) versus rates of the same worms on plates without food. This method is a phenotypic readout to assess functional changes in dopaminergic neurons. Experiments typically compare the change in body bends over time to positive control worms that do not slow in the presence of food, such as cat-2 mutants, which lack tyrosine hydroxylase, the rate-limiting enzyme for dopamine synthesis. The BSR assay can elucidate pathways leading to the dysfunction and death of dopaminergic neurons when exposed to toxic substances. As examples, lead (Pb2+) exposure increases dopaminergic neurotoxicity in wild-type C. elegans, but protein kinase (pkc) mutant animals are resistant, an effect that can be quantified via the BSR assay (Akinyemi et al., 2019). Similarly, BSR assays have revealed that Akt signaling provides resistance to manganese-induced dopaminergic neurotoxicity (Peres et al., 2019). This assay is also informative when analyzing the contributions of candidate genes to dopaminergic function. For example, the BSR assay was used to reveal that depletion of the C. elegans RAC1 GTPase CED-10, in the presence of α-syn overexpression in dopaminergic neurons, contributes to dopaminergic dysfunction (Kim et al., 2018b). Importantly, BSR defects precede neurodegeneration in C. elegans PD models (Martinez et al., 2017); the influence of temporal factors mediating the aging process can thus be evaluated for their progressive impact on dopaminergic neurons over time (Gaeta et al., 2019).
Glutamatergic neuron dysfunction
A wild-type hermaphrodite C. elegans has six sensory neurons spanning its length that detect light touch as administered by stroking the animal with an eyebrow hair glued to a toothpick. This is a mechanosensory response where, normally, animals will exhibit either forward or backward reversal in locomotion when stroked toward the posterior or anterior, respectively (Goodman and Sengupta, 2019). Touch neuron degeneration results in a mutant mechanosensory (Mec) locomotive phenotype with defective forward or backward, or both, movement indicative of glutamatergic dysfunction. As previously described, this assay was used as a readout to examine phenotypic rescue of Aβ-induced degeneration of glutamatergic neurons in the presence of different isoforms of human APOE (Griffin et al., 2019). As another example, animals treated with quinolinic acid exhibited neurodegeneration due to glutamatergic neurotransmission defects (da Silveira et al., 2018). Similarly, in single-copy knock-in SOD1 models of ALS, loss of sod-1 function produced defects in light touch response indicative of a disruption in glutamate signaling (Baskoylu et al., 2018).
Correlation between neurodegeneration and aging
Assays that assess whether a treatment or condition affects lifespan and/or healthspan are well established in the field (see poster, ‘Neurodegeneration and aging’). Lifespan consists of healthspan and gerospan. Long-lived animals experience a frailty period termed gerospan, whereas healthspan is defined as the period when an animal has greater than 50% maximal functional capacity (Bansal et al., 2015). Considering that aging is the main risk factor for AD, PD and other neurodegenerative diseases, lifespan and healthspan benchmarks of animal health are appropriate companion assays. Aging-associated molecular signatures, such as epigenetic changes, telomere shortening, proteostasis inhibition, mitochondrial dysfunction, altered lipid metabolism and nutrient sensing dysregulation, among others, can be assessed (Lopez-Otin et al., 2013). Notably, neurodegeneration is associated with many of these cellular processes, and the intersection of aging and neurodegeneration can often be efficiently examined using C. elegans models (Knight et al., 2014).
In mammals, the lengthy time requirements to evaluate age-associated changes can be considered a hindrance to higher-throughput analyses. In contrast, the average ∼20-day lifespan of the C. elegans wild-type strain Bristol N2 accelerates the discovery of fundamental modulators of neurodegenerative processes within the context of lifespan. A common misconception is that the short survival of C. elegans precludes its utility in translating lifespan modifiers in this animal to mammals. A variety of mutants and chemical modifiers of aging have been identified that directly affect highly conserved metabolic pathways and mechanisms shared among metazoans. The most notable example is the daf-2/insulin-like signaling pathway (Martins et al., 2016), whereby C. elegans research has led to the discovery of critical modulators of cellular and organismal health (i.e. mTOR, Nrf2, FOXO/DAF-16). Among the biological modulators that can be readily examined in C. elegans are genetic and epigenetic modifiers, chemical modifiers, microbiome exposures and putative environmental toxicants, in addition to exercise and diet.
Lifespan is assessed by the cohort survival assay (Lionaki and Tavernarakis, 2013), in which at least 120 synchronized L4-stage worms are grown at 20°C and scored for survival/death every 24 h by gently tapping with a platinum wire at both head and tail. A log-rank (Mantel–Cox) test is used to compare survival between strains. Investigation of the role of nicotinamide-N-methyltransferase (NNMT) found that expression of the C. elegans homolog, ANMT-1, in dopaminergic neurons increased neuron survival and overall lifespan through the regulation of autophagy (Schmeisser and Parker, 2018). A second study showed that the transcription factor SPR-4/REST regulates aging by reducing neural activity and maintaining neural homeostasis (Zullo et al., 2019). Reproducibility of lifespan data is contingent on rigorous experimental design and repetition. To evaluate variability in quantifying lifespan, Lucanic et al. (2017) reported results from a coalition of multiple laboratories termed the Caenorhabditis Intervention Testing Program that assessed longevity for 22 strains, inclusive of three Caenorhabditis species. Multiple replicates were collected from three independent laboratories to reveal that diversity in replicate data within a single laboratory for a given strain was more common than variation in lifespan determined across different laboratories. This study highlights the importance of tightly controlled experiments and internal consistency in the replication of lifespan data.
Healthspan
Like humans, C. elegans exhibits a decline in physical ability with age and loss of ability to recover from stress, which manifest as reduced body movement and increased sensitivity to heat and oxidative stress, respectively (Bansal et al. 2015). When searching for genetic modifiers, increased healthspan should be prioritized because long-lived mutants also have increased gerospan (Bansal et al., 2015), which is counterintuitive to enhancing the quality of life. Multiple assays can be performed to assess healthspan. For a comprehensive review, see Bansal et al. (2015). For example, an automated digital video microscopy system examines the average distance C. elegans travel on solid agar (Hahm et al., 2015). In a second assay, healthspan can be examined in liquid media. Using this method, researchers learned that C. elegans models of neurodegenerative disease had better outcomes following swim training exercise, which improved their neuronal healthspan (Laranjeiro et al., 2019). A third healthspan assay consists of oxidative stress analysis. Worms are transferred to plates containing paraquat, and stress-resistant survival is recorded every 5 days until death (Bansal et al., 2015). Using this assay, it was discovered that a gene product associated with cholesterol metabolism that was neuroprotective in a C. elegans PD model maintained healthspan but did not extend lifespan (Zhang et al., 2017).
Addressing lifespan and healthspan, along with other cellular signatures such as mitochondrial and/or ER function and stress responses, as well as alterations in proteostasis and transcriptional or macromolecule profiling (RNA/DNA/miRNA/lipids), provides the C. elegans researcher with an arsenal of complementary approaches with which to investigate neurodegenerative diseases. As noted below, the ‘big data’ increasingly emerging from human genomic analyses provides a greater impetus for the application of a more rapid and cost-effective system whereby the significance of genetic variation can be discerned for its impact not only on disease, but also on quality of life.
Concluding remarks
Among the largely untapped potential avenues of C. elegans models of neurodegeneration is the functional annotation of genomic variation among humans to discern factors that confer either susceptibility or resilience. The ever-expanding databases of human sequence information have resulted in an informational overload of variants of uncertain significance (Cooper, 2015). Characterizing these and other changes via functional analyses with the bioassays outlined in this article could greatly advance our understanding of therapeutic options. While it may superficially appear illogical to use an invertebrate such as C. elegans for human personalized medicine, there are successful examples of highly specialized drug discovery tailored to the genomic composition of cancer patients (Bangi et al., 2019). C. elegans is also being used in the drug discovery efforts for rare glycosylation disorders, N-glycanase 1 (NGLY1) deficiency and phosphomannomutase 2 (PMM2) deficiency (Iyer et al., 2019a,b).
In closing, this article prompted us to reflect on a Primer that we had the privilege of co-authoring for the inaugural issue of Disease Models & Mechanisms. At the inception of this journal as a platform to communicate the increasingly important role of model systems in advancing disease research, we titled our article ‘Traversing a wormhole to combat Parkinson's disease’ (Caldwell and Caldwell, 2008). The choice of this celestial analogy now appears prescient in capturing the exceptional contributions of C. elegans models in rapidly advancing discoveries from the experimental space through to the other side of clinical investigation. We contend that the expedience, accuracy, molecular tools and mechanistic capacity of C. elegans present an attractive, bioethical alternative to more costly, time-consuming and sometimes redundant in vitro or mammalian models. With an increasingly strong track record of translational outcomes that continue to emerge from C. elegans research, this microscopic nematode is positioned to help fill the remaining black holes in our understanding of neurodegeneration. Indeed, the gravity of the societal burden from these diseases is worldwide, and weighs on tens of millions of individuals each day. A sense of urgency and innovative strategies that coalesce into model systems research are essential to accelerate discovery and progress. In the words of an admired and fearless explorer known to go where no one (or no worm) has gone before, “Warp speed ahead. Engage!” (Jean-Luc Picard).
Acknowledgements
We are grateful to Dan Shaye for providing us OrthoList 2 data and to Steve Cook for his insights into C. elegans neuroanatomy described in the Introduction section of this article. To our colleagues whose C. elegans models and/or assays have not been covered, we apologize for the space limitations that necessitated these omissions. We further acknowledge the collective contributions of the C. elegans research community in developing shared resources that have generously made the advancements outlined in this article possible.
Footnotes
Funding
K.A.C. was funded by National Institutes of Health (NIH) grant R15NS106460; G.A.C. was funded by NIH grant R15NS104857.
At a glance
A high-resolution version of this poster is available for downloading in the online version of the article at http://dmm.biologists.org/content/13/10/dmm046110/F1.poster.jpg.
References
- Alfonso A., Grundahl K., McManus J. R. and Rand J. B. (1994). Cloning and characterization of the choline acetyltransferase structural gene (cha-1) from C. elegans. J. Neurosci. 14, 2290-2300. 10.1523/JNEUROSCI.14-04-02290.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Chalabi A., van den Berg L. H. and Veldink J. (2017). Gene discovery in amyotrophic lateral sclerosis: implications for clinical management. Nat. Rev. Neurol. 13, 96-104. 10.1038/nrneurol.2016.182 [DOI] [PubMed] [Google Scholar]
- Akinyemi A. J., Miah M. R., Ijomone O. M., Tsatsakis A., Soares F. A. A., Tinkov A. A., Skalny A. V., Venkataramani V. and Aschner M. (2019). Lead (Pb) exposure induces dopaminergic neurotoxicity in Caenorhabditis elegans: Involvement of the dopamine transporter. Toxicol Rep. 6, 833-840. 10.1016/j.toxrep.2019.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aksoy Y. A., Deng W., Stoddart J., Chung R., Guillemin G., Cole N. J., Neely G. G. and Hesselson D. (2020). “STRESSED OUT”: The role of FUS and TDP-43 in amyotrophic lateral sclerosis. Int. J. Biochem. Cell Biol. 126, 105821 10.1016/j.biocel.2020.105821 [DOI] [PubMed] [Google Scholar]
- Arai T., Hasegawa M., Akiyama H., Ikeda K., Nonaka T., Mori H., Mann D., Tsuchiya K., Yoshida M., Hashizume Y. et al. (2006). TDP-43 is component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem. Biophys. Res. Commun. 351, 602-611. 10.1016/j.bbrc.2006.10.093 [DOI] [PubMed] [Google Scholar]
- Ash P. E. A., Zhang Y.-J., Roberts C. M., Saldi T., Hutter H., Buratti E., Petrucelli L. and Link C. D. (2010). Neurotoxic effects of TDP-43 overexpression in C. elegans. Hum. Mol. Genet. 19, 3206-3218. 10.1093/hmg/ddq230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asikainen S., Vartiainen S., Lakso M., Nass R. and Wong G. (2005). Selective sensitivity of Caenorhabditis elegans neurons to RNA interference. Neuroreport 16, 1995-1999. 10.1097/00001756-200512190-00005 [DOI] [PubMed] [Google Scholar]
- Ayyadevara S., Balasubramaniam M., Parcon P. A., Barger S. W., Griffin W. S., Alla R., Tackett A. J., Mackintosh S. G., Petricoin E., Zhou W. et al. (2016). Proteins that mediate protein aggregation and cytotoxicity distinguish Alzheimer's hippocampus from normal controls. Aging Cell 15, 924-939. 10.1111/acel.12501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bangi E., Ang C., Smibert P., Uzilov A. V., Teague A. G., Antipin Y., Chen R., Hecht C., Gruszczynski N., Yon W. J. et al. (2019). A personalized platform identifies trametinib plus zoledronate for a patient with KRAS-mutant metastatic colorectal cancer. Sci. Adv. 5, eaav6528 10.1126/sciadv.aav6528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal A., Zhu L. J., Yen K. and Tissenbaum H. A. (2015). Uncoupling lifespan and healthspan in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 112, E277-E286. 10.1073/pnas.1412192112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskoylu S. N., Yersak J., O'Hern P., Grosser S., Simon J., Kim S., Schuch K., Dimitriadi M., Yanagi K. S., Lins J. et al. (2018). Single copy/knock-in models of ALS SOD1 in C. elegans suggest loss and gain of function have different contributions to cholinergic and glutamatergic neurodegeneration. PLoS Genet. 14, e1007682 10.1371/journal.pgen.1007682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkowitz L. A., Knight A. L., Caldwell G. A. and Caldwell K. A. (2008). Generation of stable transgenic C. elegans using microinjection. J. Vis. Exp. 18, 833 10.3791/833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betarbet R., Sherer T. B. and Greenamyre J. T. (2002). Animal models of Parkinson's disease. BioEssays 24, 308-318. 10.1002/bies.10067 [DOI] [PubMed] [Google Scholar]
- Brandt R., Gergou A., Wacker I., Fath T. and Hutter H. (2009). A Caenorhabditis elegans model of tau hyperphosphorylation: induction of developmental defects by transgenic overexpression of Alzheimer's disease-like modified tau. Neurobiol. Aging 30, 22-33. 10.1016/j.neurobiolaging.2007.05.011 [DOI] [PubMed] [Google Scholar]
- Braungart E., Gerlach M., Riederer P., Baumeister R. and Hoener M. C. (2004). Caenorhabditis elegans MPP+ model of Parkinson's disease for high-throughput drug screenings. Neurodegener. Dis. 1, 175-183. 10.1159/000080983 [DOI] [PubMed] [Google Scholar]
- Brenner S. (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caga J., Hsieh S., Lillo P., Dudley K. and Mioshi E. (2019). The impact of cognitive and behavioral symptoms of ALS patients and their caregivers. Front. Neurol. 10, 192 10.3389/fneur.2019.00192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell G. A. and Caldwell K. A. (2008). Traversing a wormhole to combat Parkinson's disease. Dis. Model Mech. 1, 32-36. 10.1242/dmm.000257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calixto A., Chelur D., Topalidou I., Chen X. and Chalfie M. (2010). Enhanced neuronal RNAi in C. elegans using SID-1. Nat. Meth. 7, 554-559. 10.1038/nmeth.1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S., Gelwix C. C., Caldwell K. A. and Caldwell G. A. (2005). Torsin-mediated neuroprotection from cellular stresses to dopaminergic neurons of C. elegans. J. Neurosci. 25, 3801-3812. 10.1523/JNEUROSCI.5157-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie M., Tu Y., Euskirchen G., Ward W. W. and Prasher D. C. (1994). Green fluorescent protein as a marker for gene expression. Science 263, 802-805. 10.1126/science.8303295 [DOI] [PubMed] [Google Scholar]
- Chan D. C. (2006). Mitochondria: dynamic organelles in disease, aging, and development. Cell 125, 1241-1252. 10.1016/j.cell.2006.06.010 [DOI] [PubMed] [Google Scholar]
- Cherny R. A., Atwood C. S., Xilinas M. E., Gray D. N., Jones W. D., McLean C. A., Barnham K. J., Volitakis I., Fraser F. W., Kim Y. S. et al. (2001). Treatment with a copper-zinc chelator markedly and rapidly inhibits β-amyloid accumulation in Alzheimer's Disease transgenic mice. Neuron 30, 665-676. 10.1016/S0896-6273(01)00317-8 [DOI] [PubMed] [Google Scholar]
- Choudhary B., Mandelkow E., Mandelkow E.-M. and Pir G. J. (2018). Glutamatergic nervous system degeneration in a C. elegans TauA152T tauopathy model involves pathways of excitotoxicity and Ca2+ dysregulation. Neurobiol. Dis. 117, 189-202. 10.1016/j.nbd.2018.06.005 [DOI] [PubMed] [Google Scholar]
- Chung C. Y., Khurana V., Auluck P. K., Tardiff D. F., Mazzulli J. R., Soldner F., Baru V., Lou Y., Freyzon Y., Cho S. et al. (2013). Identification and rescue of α-synuclein toxicity in Parkinson patient-derived neurons. Science 342, 983-987. 10.1126/science.1245296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary J. D. and Ranum L. P. W. (2017). New developments in RAN translation: insights from multiple diseases. Genet Develop 44, 125-134. 10.1016/j.gde.2017.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland D. W. and Rothstein J. D. (2001). From charcot to lou gehrig: decipherin selective motor neuron death in ALS. Nat. Rev. Neurosci. 2, 806-819. 10.1038/35097565 [DOI] [PubMed] [Google Scholar]
- Cook S. J., Jarrell T. A., Brittin C. A., Wang Y., Bloniarz A. E., Yakovlev M. A., Nguyen K. C. Q., Tang L. T.-H., Bayer E. A., Duerr J. S. et al. (2019). Whole-animal connectomes of both Caenorhabditis elegans sexes. Nature 571, 63-71. 10.1038/s41586-019-1352-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper G. M. (2015). Parlez-vous VUS? Genome Res. 25, 1423-1426. 10.1101/gr.190116.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper A. A., Gitler A. D., Cashikar A., Haynes C. M., Hill K. J., Bhullar B., Liu K., Xu K., Strathearn K. E., Liu F. et al. (2006). Alpha-synuclein blocks ER-Golgi traffic and Rab1 rescues neuron loss in Parkinson's models. Science 313, 324-328. 10.1126/science.1129462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper J. F., Machiela E., Dues D. J., Spielbauer K. K., Senchuk M. M. and Van Raamsdonk J. M. (2017). Activation of the mitochondrial unfolded protein response promotes longevity and dopamine neuron survival in Parkinson's disease models. Sci. Rep. 7, 16441 10.1038/s41598-017-16637-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordeiro L. M., Machado M. L., da Silva A. F., Baptista F. B. O., da Silveira T. L., Soares F. A. A. and Arantes L. P. (2020). Rutin protects Huntington's disease through the insulin/IGF1 (IIS) signaling pathway and autophagy activity: study in Caenorhabditis elegans model. Food Chem. Toxicol. 141, 111323 10.1016/j.fct.2020.111323 [DOI] [PubMed] [Google Scholar]
- Corrionero A. and Horvitz H. R. (2018). A C9orf72 ALS/FTD ortholog acts in endolysosomal degradation and lysosomal homeostasis. Curr. Biol. 28, 1522-1535.e5. 10.1016/j.cub.2018.03.063 [DOI] [PubMed] [Google Scholar]
- da Silveira T. L., Zamberlan D. C., Arantes L. P., Machado M. L., da Silva T. C., de Freitas Câmara D., Santamaria A., Aschner M. and Soares F. A. A. (2018). Quinolinic acid and glutamatergic neurodegeneration in Caenorhabditis elegans. Neurotoxicology 67, 94-101. 10.1016/j.neuro.2018.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doitsidou M., Flames N., Lee A. C., Boyanov A. and Hobert O. (2008). Automated screening for mutants affecting dopaminergic-neuron specification in C. elegans. Nat. Methods 5, 869-872. 10.1038/nmeth.1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald C. Y., Cheng R., Tolen L., Shah V., Gillani A., Nasrin A. and Li C. (2012). Pan-neuronal expression of APL-1, and APP-related protein, disrupts olfactory, gustatory, and touch plasticity in Caenorhabditis elegans. J. Neurosci 32, 10156-10169. 10.1523/JNEUROSCI.0495-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber P. W., Alter J. R., MacDonald M. E. and Hart A. C. (1999). Polyglutamine-mediated dysfunction and apoptotic death of a Caenorhabditis elegans sensory neuron. Proc. Natl. Acad. Sci. USA 96, 179-184. 10.1073/pnas.96.1.179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang E. F., Hou Y., Palikaras K., Adriaanse B. A., Kerr J. S., Yang B., Lautrup S., Hasan-Olive M. M., Caponio D., Dan X. et al. (2019). Mitophagy inhibits amyloid-β and tau pathology and reverses cognitive deficits in models of Alzheimer's disease. Nat. Neurosci. 22, 401-412. 10.1038/s41593-018-0332-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning S., Haque A., Imberdis T., Baru V., Barrasa M. I., Nuber S., Termine D., Ramalingam N., Ho G., Noble T. et al. (2019). Lipidomic analysis of α-synuclein neurotoxicity identifies Stearoyl CoA desaturase as a target for Parkinson treatment. Mol. Cell 73, 1001-1014. 10.1016/j.molcel.2018.11.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning S., Selkoe D. and Dettmer U. (2020). Vesicle trafficking and lipid metabolism in synucleinopathy. Acta Neuropathol. 10.1007/s00401-020-02177-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaeta A. L., Caldwell K. A. and Caldwell G. A. (2019). Found in translation: the utility of C. elegans Alpha-synuclein models of Parkinson's disease. Brain Sci. 2, 73 10.3390/brainsci9040073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh D. K., Shrikondawar A. N. and Ranjan A. (2020). Local structural unfolding at the edge-strands of beta sheets is the molecular basis for instability and aggregation of G85R and G93A mutants of superoxide dismutase 1. J. Biomol. Struct. Dyn. 38, 647-659. 10.1080/07391102.2019.1584125 [DOI] [PubMed] [Google Scholar]
- Gitler A. D., Bevis B. J., Shorter J., Strathearn K. E., Hamamichi S., Su L. J., Caldwell K. A., Caldwell G. A., Rochet J.-C., McCaffery J. M. et al. (2008). The Parkinson's disease protein alpha-synuclein disrupts cellular Rab homeostasis. Proc. Natl. Acad. Sci. USA 105, 145-150. 10.1073/pnas.0710685105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitler A. D., Chesi A., Geddie M. L., Strathearn K. E., Hamamichi S., Hill K. J., Caldwell K. A., Caldwell G. A., Cooper A. A., Rochet J. C. et al. (2009). Alpha-synuclein is part of a diverse and highly conserved interaction network that includes PARK9 and manganese toxicity. Nat. Genet. 41, 308-315. 10.1038/ng.300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman M. B. and Sengupta P. (2019). How Caenorhabditis elegans senses mechanical stress, temperature, and other physical stimuli. Genetics 212, 25-51. 10.1534/genetics.118.300241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin E. F., Caldwell K. A. and Caldwell G. A. (2017). Genetic and pharmacological discovery for Alzheimer's Disease using Caenorhabditis elegans. ACS Chem. Neurosci. 8, 2596-2606. 10.1021/acschemneuro.7b00361 [DOI] [PubMed] [Google Scholar]
- Griffin E. F., Yan X., Caldwell K. A. and Caldwell G. A. (2018). Distinct functional roles of Vps41-mediated neuroprotection in Alzheimer's and Parkinson's disease models of neurodegeneration. Hum. Mol. Genet. 27, 4176-4193. 10.1093/hmg/ddy308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin E. F., Scopel S. E., Stephen C. A., Holzhauer A. C., Vaji M. A., Tuckey R. A., Berkowitz L. A., Caldwell K. A. and Caldwell G. A. (2019). ApoE-associated modulation of neuroprotection from Ab-mediated neurodegeneration in transgenic Caenorhabditis elegans. Dis. Models Mech. 12, dmm037218 10.1242/dmm.037218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C. R., Schellenberg G. D. and Kraemer B. C. (2009). SUT-2 potentiates tau-induced neurotoxicity in Caenorhabditis elegans. Hum. Mol. Genet. 18, 1825-1838. 10.1093/hmg/ddp099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahm J.-H., Kim S., DiLoreto R., Shi C., Lee S.-J. V., Murphy C. T. and Nam H. G. (2015). C. elegans maximum velocity correlates with healthspan and is maintained in worms with an insulin receptor mutation. Nat. Commun. 6, 8919 10.1038/ncomms9919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamamichi S., Rivas R. N., Knight A. L., Cao S., Caldwell K. A. and Caldwell G. A. (2008). Hypothesis-based RNAi screening identifies neuroprotective genes in a Parkinson's disease model. Proc. Natl. Acad. Sci. USA 105, 728-733. 10.1073/pnas.0711018105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarlund M., Hobert O., Miller D. M. III and Sestan N. (2018). The CeNGEN project: the complete gene expression map of an entire nervous system. Neuron 99, 430-433. 10.1016/j.neuron.2018.07.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy J. and Selkoe D. J. (2002). The amyloid hypothesis of Alzheimer's disease: progress and problems on the road to therapeutics. Science 297, 353-356. 10.1126/science.1072994 [DOI] [PubMed] [Google Scholar]
- Harrington A. J., Hamamichi S., Caldwell G. A. and Caldwell K. A. (2010). C. elegans as a model organism to investigate molecular pathways involved with Parkinson's disease. Devel Dyn. 239, 1282-1295. 10.1002/dvdy.22184 [DOI] [PubMed] [Google Scholar]
- Harrington A. J., Yacoubian T. A., Slone S. R., Caldwell K. A. and Caldwell G. A. (2012). Functional analysis of VPS41-mediated neuroprotection in Caenorhabdiditis elegans and mammalian models of Parkinson's disease. J. Neurosci. 32, 2142-2153. 10.1523/JNEUROSCI.2606-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman J. H., Gonzalez-Hunt C., Hall S. M., Ryde I. T., Caldwell K. A., Caldwell G. A. and Meyer J. N. (2019). Genetic defects in mitochondrial dynamics in Caenorhabditis elegans impact ultraviolet C radiation- and 6-hydroxydopamine-induced neurodegeneration. Int. J. Mol. Sci. 20, 3202 10.3390/ijms20133202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes C. M., Petrova K., Benedetti C., Yang Y. and Ron D. (2007). ClpP mediates activation of a mitochondrial unfolded protein response in C. elegans. Dev. Cell 13, 467-480. 10.1016/j.devcel.2007.07.016 [DOI] [PubMed] [Google Scholar]
- Hernandez D. G., Reed X. and Singleton A. B. (2019). Genetics in Parkinson disease: Mendelian versus non-Mendelian inheritance. J. Neurochem. 139, 59-74. 10.1111/jnc.13593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera F. E., Chesi A., Paleologou K. E., Schmid A., Munoz A., Vendruscolo M., Gustincich S., Lashuel H. A. and Carloni P. (2008). Inhibition of alpha-synuclein fibrillization by dopamine is mediated by interactions with five C-terminal residues and with E83 in the NAC region. PloS One 3, e3394 10.1371/journal.pone.0003394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O. (2013). The neuronal genome of Caenorhabditis elegans. In WormBook (ed. The C. elegans Research Community ). WormBook, pp. 1-106. http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornsten A., Lieberthal J., Fadia S., Malins R., Ha L., Xu X., Daigle I., Markowitz M., O'Connor G., Plasterk R. et al. (2007). APL-1, a Caenorhabditis elegans protein related to the human beta-amyloid precursor protein, is essential for viability. Proc. Natl. Acad. Sci. USA 104, 1971-1976. 10.1073/pnas.0603997104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvitz H. R., Chalfie M., Trent C., Sulston J. E. and Evans P. D. (1982). Serotonin and octopamine in the nematode Caenorhabditis elegans. Science 216, 1012-1014. 10.1126/science.6805073 [DOI] [PubMed] [Google Scholar]
- Iyer S., Mast J. D., Tsang H., Rodriguez T. P., DiPrimio N., Prangley M., Sam F. S., Parton Z. and Perlstein E. O. (2019a). Drug screens of NGLY1 deficiency in worm and fly models reveal catecholamine, NRF2 and anti-inflammatory-pathway activation as potential clinical approaches. Dis. Models Mech. 12, dmm040576 10.1242/dmm.040576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer S., Sam F. S., DiPrimio N., Preston G., Verheijen J., Murthy K., Parton Z., Tsang H., Lao J., Morava E. et al. (2019b). Repurposing the aldose reductase inhibitor and diabetic neuropathy drug epalrestat for the congenital disorder of glycosylation PMM2-CDG. Dis. Models Mech. 12, dmm040584 10.1242/dmm.040584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y. J., Ugolino J., Zhang T., Lu J., Kim D. and Wang J. (2020). C9orf72/ALFA-1 controls TFEB/HLH-30-dependent metabolism through dynamic regulation of Rag GTPases. PLoS Genet. 16, e1008738 10.1371/journal.pgen.1008738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia K., Hart A. C. and Levine B. (2007). Autophagy genes protect against disease caused by polyglutamine expansion proteins in Caenorhabditis elegans. Autophagy 3, 21-25. 10.4161/auto.3528 [DOI] [PubMed] [Google Scholar]
- Jiang H.-C., Hsu J.-M., Yen C.-P., Chao C.-C., Chen R.-H. and Pan C.-L. (2015). Neural activity and CaMKII protect mitochondria from fragmentation in aging Caenorhabditis elegans neurons. Proc. Natl. Acad. Sci. USA 112, 8768-8773. 10.1073/pnas.1501831112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson W. M., Yao C., Siedlak S. L., Wang W., Zhu X., Caldwell G. A., Wilson-Delfosse A. L., Mieyal J. J. and Chen S. G. (2015). Glutaredoxin deficiency exacerbates neurodegeneration in C. elegans models of Parkinson's disease. Hum. Mol. Genet. 24, 1322-1335. 10.1093/hmg/ddu542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath R. S., Martinez-Campos M., Zipperlen P., Fraser A. G. and Ahringer J. (2001). Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2, RESEARCH0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath R. S., Fraser A. G., Dong Y., Poulin G., Durbin R., Gotta M., Kanapin A., Le Bot N., Moreno S., Sohrmann M. et al. (2003). Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421, 231-237. 10.1038/nature01278 [DOI] [PubMed] [Google Scholar]
- Karpinar D. P., Balija M. B., Kügler S., Opazo F., Rezaei-Ghaleh N., Wender N., Kim H. Y., Taschenberger G., Falkenburger B. H., Heise H. et al. (2009). Pre-fibrillar alpha-synuclein variants with impaired beta-structure increase neurotoxicity in Parkinson's disease models. EMBO J. 28, 3256-3268. 10.1038/emboj.2009.257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy S., Wang D. and Ruvkun G. (2004). A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature 427, 645-649. 10.1038/nature02302 [DOI] [PubMed] [Google Scholar]
- Kikuchi H., Almer G., Yamashita S., Guégan C., Nagai M., Xu Z., Sosunov A. A., McKhann G. M. II and Przedborski S. (2006). Spinal cord endoplasmic reticulum stress associated with a microsomal accumulation of mutant superoxide dismutase-1 in an ALS model. Proc. Natl. Acad. Sci. USA 103, 6025-6030. 10.1073/pnas.0509227103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W., Underwood R. S., Greenwald I. and Shaye D. D. (2018a). OrthoList 2: a new comparative genomic analysis of human and Caenorhabditis elegans Genes. Genetics 210, 445-461. 10.1534/genetics.118.301307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Calatayud C., Guha S., Fernández-Carasa I., Berkowitz L., Carballo-Carbajal I., Ezquerra M., Fernández-Santiago R., Kapahi P., Raya Á. et al. (2018b). The small GTPase RAC1/CED-10 is essential in maintaining dopaminergic neuron function and survival against α-synuclein-induced toxicity. Mol. Neurobiol. 55, 7533-7552. 10.1007/s12035-018-0881-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight A. L., Yan X., Hamamichi S., Ajjuri R. R., Mazzulli J. R., Zhang M. W., Daigle J. G., Zhang S., Borom A. R., Roberts L. R. et al. (2014). The glycolytic enzyme, GPI, is a functionally conserved modifier of dopaminergic neurodegeneration in Parkinson's models. Cell Metab. 20, 145-157. 10.1016/j.cmet.2014.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer B. C. and Schellenberg G. D. (2007). SUT-1 enables tau-induced neurotoxicity in C. elegans. Hum. Mol. Genet. 16, 1959-1971. 10.1093/hmg/ddm143 [DOI] [PubMed] [Google Scholar]
- Kraemer B. C., Zhang B., Leverenz J. B., Thomas J. H., Trojanowski J. Q. and Schellenberg G. D. (2003). Neurodegeneration and defective neurotransmission in a Caenorhabditis elegans model of tauopathy. Proc. Natl. Acad. Sci. USA 100, 9980-9985. 10.1073/pnas.1533448100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer B. C., Burgess J. K., Chen J. H., Thomas J. H. and Schellenberg G. D. (2006). Molecular pathways that influence human tau-induced pathology in Caenorhabditis elegans. Hum. Mol. Genet. 15, 1483-1496. 10.1093/hmg/ddl067 [DOI] [PubMed] [Google Scholar]
- Kwiatkowski T. J., Bosco D. A., LeClerc A. L., Tamrazian E., Vanderburg C. R., Russ C., Davis A., Gilchrist J., Kasarskis E. J., Munsat T. et al. (2009). Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science 323, 1205-1208. 10.1126/science.1166066 [DOI] [PubMed] [Google Scholar]
- Lakowski B., Eimer S., Göbel C., Böttcher A., Wagler B. and Baumeister R. (2003). Two suppressors of sel-12 encode C2H2 zinc-finger proteins that regulate presenilin transcription in Caenorhabditis elegans. Development 130, 2117-2128. 10.1242/dev.00429 [DOI] [PubMed] [Google Scholar]
- Lakso M., Vartiainen S., Moilanen A. M., Sirviö J., Thomas J. H., Nass R., Blakely R. D. and Wong G. (2003). Dopaminergic neuronal loss and motor deficits in Caenorhabditis elegans overexpressing human alpha-synuclein. J. Neurochem. 86, 165-172. 10.1046/j.1471-4159.2003.01809.x [DOI] [PubMed] [Google Scholar]
- Laranjeiro R., Harinath G., Hewitt J. E., Hartman J. H., Royal M. A., Meyer J. N., Vanapalli S. A. and Driscoll M. (2019). Swim exercise in Caenorhabditis elegans extends neuromuscular and gut healthspan, enhances learning ability, and protects against neurodegeneration. Proc. Natl. Acad. Sci. USA 116, 23829-23839. 10.1073/pnas.1909210116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitan D. and Greenwald I. (1995). Facilitation of lin-12-mediated signalling by sel-12, a Caenorhabditis elegans S182 Alzheimer's disease gene. Nature 377, 351-354. 10.1038/377351a0 [DOI] [PubMed] [Google Scholar]
- Li C. and Kim K. (2008). Neuropeptides. WormBook: The Online Review of C. elegans Biology 1-36. 10.1895/wormbook.1.142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Huang K.-x and Le W.-d. (2013). Establishing a novel C. elegans model to investigate the role of autophagy in amyotrophic lateral sclerosis. Acta Pharmacol. Sin. 34, 644-650. 10.1038/aps.2012.190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Avery L., Denk W. and Hess G. P. (1997). Identification of chemical synapses in the pharynx of Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 94, 5912-5916. 10.1073/pnas.94.11.5912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liachko N. F., McMillan P. J., Guthrie C. R., Bird T. D., Leverenz J. B. and Kraemer B. C. (2013). CDC7 inhibition blocks pathological TDP-43 phosphorylation and neurodegeneration. Ann. Neurol. 74, 39-52. 10.1002/ana.23870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liachko N. F., Saxton A. D., McMillan P. J., Strovas T. J., Currey H. N., Taylor L. M., Wheeler J. M., Oblak A. L., Ghetti B., Montine T. J. et al. (2016). The phosphatase calcineurin regulates pathological TDP-43 phosphorylation. Acta Neuropath 132, 545-561. 10.1007/s00401-016-1600-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liachko N. F., Saxton A. D., McMillan P. J., Strovas T. J., Keene C. D., Bird T. D. and Kraemer B. C. (2019). Genome wide analysis reveals heparan sulfate epimerase modulates TDP-43 proteinopathy. PLoS Genet. 15, e1008526 10.1371/journal.pgen.1008526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link C. D. (1995). Expression of human beta-amyloid peptide in transgenic Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 92, 9368-9372. 10.1073/pnas.92.20.9368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link C. D., Taft A., Kapulkin V., Duke K., Kim S., Fei Q., Wood D. E. and Sahagan B. G. (2003). Gene expression analysis in a transgenic Caenorhabditis elegans Alzheimer's disease model. Neurobiol. Aging 24, 397-413. 10.1016/S0197-4580(02)00224-5 [DOI] [PubMed] [Google Scholar]
- Lionaki E. and Tavernarakis N. (2013). Assessing aging and senescent decline in Caenorhabditis elegans: cohort survival analysis. Meth. Molec. Biol. 965, 473-484. 10.1007/978-1-62703-239-1_31 [DOI] [PubMed] [Google Scholar]
- Liu Z., Hamamichi S., Lee B. D., Yang D., Ray A., Caldwell G. A., Caldwell K. A., Dawson T. M., Smith W. W. and Dawson V. L. (2011). Inhibitors of LRRK2 kinase attenuate neurodegeneration and Parkinson-like phenotypes in Caenorhabditis elegans and Drosophila Parkinson's disease models. Hum. Mol. Genet. 20, 3933-3942. 10.1093/hmg/ddr312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu E. Y., Cali C. P. and Lee E. B. (2017). RNA metabolism in neurodegenerative disease. Dis. Models Mech. 10, 509-518. 10.1242/dmm.028613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Otin C., Blasco M. A., Partridge L., Serrano M. and Kroemer G. (2013). The hallmarks of aging. Cell 153, 1194-1217. 10.1016/j.cell.2013.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T., Aron L., Zullo J., Pan Y., Kim H., Chen Y., Tun-Hsiang Y., Kim H.-M., Drake D., Liu X. S. et al. (2014). REST and stress resistance in ageing and Alzheimer's disease. Nature 507, 448-454. 10.1038/nature13163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucanic M., Plummer W. T., Chen E., Harke J., Foulger A. C., Onken B., Coleman-Hulbert A. L., Dumas K. J., Guo S., Johnson E. et al. (2017). Impact of genetic background and experimental reproducibility on identifying chemical compounds with robust longevity effects. Nat. Comm. 8, 14256 10.1038/ncomms14256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malpass K. (2013). Neurodegenerative disease: Defective mitochondrial dynamics in the hot seat – a therapeutic target common to many neurological disorders? Nat. Rev. Neurol. 9, 417 10.1038/nrneurol.2013.138 [DOI] [PubMed] [Google Scholar]
- Martinez B. A., Kim H., Ray A., Caldwell G. A. and Caldwell K. A. (2015). A bacterial metabolite induces glutathione-tractable proteostatic damage, proteasomal disturbances, and PINK1-dependent autophagy in C. elegans. Cell Death Dis. 6, e1908 10.1038/cddis.2015.270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez B. A., Petersen D. A., Gaeta A. L., Stanley S. P., Caldwell G. A. and Caldwell K. A. (2017). Dysregulation of the mitochondrial unfolded protein response induces non-apoptotic dopaminergic neurodegeneration in C. elegans models of Parkinson's disease. J. Neurosci. 37, 11085-11100. 10.1523/JNEUROSCI.1294-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins R., Lithgow G. J. and Link W. (2016). Long live FOXO: unraveling the role of FOXO proteins in aging and longevity. Aging Cell 15, 196-207. 10.1111/acel.12427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlack K. E. S., Tardiff D. F., Narayan P., Hamamichi S., Caldwell K. A., Caldwell G. A. and Lindquist S. (2014). Clioquinol promotes the degradation of metal-dependent amyloid-β(Aβ) oligomers to restore endocytosis and ameliorate Aβ toxicity. Proc. Natl. Acad. Sci. USA 111, 4013-4018. 10.1073/pnas.1402228111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzulli J. R., Xu Y.-H., Sun Y., Knight A. L., McLean P. J., Caldwell G. A., Sidransky E., Grabowski G. A. and Krainc D. (2011). Gaucher disease glucocerebrosidase and α-synuclein form a bidirectional pathogenic loop in Synucleinopathies. Cell 146, 37-52. 10.1016/j.cell.2011.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntire S. L., Jorgensen E. and Horvitz H. R. (1993). Genes required for GABA function in Caenorhabditis elegans. Nature 364, 334-337. 10.1038/364334a0 [DOI] [PubMed] [Google Scholar]
- Meade R. M., Fairlie D. P. and Mason J. M. (2019). Alpha-synuclein structure and Parkinson's disease – lessons and emerging principles. Mol. Neurodegen 14, 29 10.1186/s13024-019-0329-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello C. C., Kramer J. M., Stinchcomb D. and Ambros V. (1991). Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10, 3959-3970. 10.1002/j.1460-2075.1991.tb04966.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mezey E., Dehejia A. M., Harta G., Tresser N., Suchy S. F., Nussbaum R. L., Brownstein M. J. and Polymeropoulos M. H. (1998). Alpha-synuclein is present in Lewy bodies in sporadic Parkinson's disease. Mol. Psychiatr. 2, 493-499. 10.1038/sj.mp.4000446 [DOI] [PubMed] [Google Scholar]
- Mor D. E., Tsika E., Mazzulli J. R., Gould N. S., Kim H., Daniels M. J., Doshi S., Gupta P., Grossman J. L., Tan V. X. et al. (2017). a-synuclein oligomers and nigrostriatal degeneration. Nat. Neurosci. 20, 1560-1568. 10.1038/nn.4641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley J. F., Brignull H. R., Weyers J. J. and Morimoto R. I. (2002). The threshold for polyglutamine-expansion protein aggregation and cellular toxicity is dynamic and influenced by aging in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 99, 10417-10422. 10.1073/pnas.152161099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulcahy B., Ibbett P., Holden-Dye L. and O'Connor V. (2019). The C. elegans cysteine-string protein homologue, DNJ-14, is dispensable for neuromuscular junction maintenance across ageing. J. Exp. Biol. 222, jeb205450 10.1242/jeb.205450 [DOI] [PubMed] [Google Scholar]
- Murakami T., Yang S.-P., Xie L., Kawano T., Fu D., Mukai A., Bohm C., Chen F., Robertson J. and Suzuki H. (2012). ALS mutations in FUS cause neuronal dysfunction and death in Caenorhabditis elegans by a dominant gain-of-function mechanism. Hum. Mol. Genet. 21, 1-9. 10.1093/hmg/ddr417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nollen E. A., Garcia S. M., van Haaften G., Kim S., Chavez A., Morimoto R. I. and Plasterk R. H. (2004). Genome-wide RNA interference screen identifies previously undescribed regulators of polyglutamine aggregation. Proc. Natl. Acad. Sci. USA 101, 6403-6408. 10.1073/pnas.0307697101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr H. T. and Zoghbi H. Y. (2007). Trinucleotide repeat disorders. Annu. Rev. Neurosci. 30, 575-621. 10.1146/annurev.neuro.29.051605.113042 [DOI] [PubMed] [Google Scholar]
- Palop J. J. and Mucke L. (2009). Epilepsy and cognitive impairments in Alzheimer disease. Arch. Neurol. 66, 435-440. 10.1001/archneurol.2009.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palop J. J. and Mucke L. (2016). Network abnormalities and interneuron dysfunction in Alzheimer disease. Nat. Rev. Neurosci. 17, 777-792. 10.1038/nrn.2016.141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palikaras K., Lionaki E. and Tavernarakis N. (2015). Coordination of mitophagy and mitochondrial biogenesis during ageing in C. elegans. Nature 521, 525-528. 10.1038/nature14300 [DOI] [PubMed] [Google Scholar]
- Palmisano N. J. and Melendez A. (2016). Detection of autophagy in Caenorhabditis elegans using GFP::LGG-1 as an autophagy marker. Cold Spring Harb. Protoc. 1, pdb-prot086496 10.1101/pdb.prot086496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker J. A., Connolly J. B., Wellington C., Hayden M., Dausset J. and Neri C. (2001). Expanded polyglutamines in Caenorhabditis elegans cause axonal abnormalities and severe dysfunction of PLM mechanosensory neurons without cell death. Proc. Natl. Acad. Sci. USA 98, 13318-13323. 10.1073/pnas.231476398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patten S. A., Aggad D., Martinez J., Tremblay E., Petrillo J., Armstrong G. A., La Fontaine A., Maios C., Liao M., Ciura S. et al. (2017). Neuroleptics as therapeutic compounds stabilizing neuromuscular transmission in amyotrophic lateral sclerosis. JCI Insight 2, 97152 10.1172/jci.insight.97152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peres T. V., Arantes L., Miah M. R., Bornhorst J., Schwerdtle T., Bowman A. B., Leal R. B. and Aschner M. (2019). Role of Caenorhabditis elegans AKT1/2 and SGK-1 in manganese toxicity. Neurotox. Res. 34, 584-596. 10.1007/s12640-018-9915-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perri E. R., Parakh S., Vidal M., Mehta P., Ma Y., Walker A. K. and Atkin J. D. (2020). The cysteine (Cys) residues Cys-6 and Cys-111 in mutant superoxide dismutase 1 (SOD1) A4V are required for induction of endoplasmic reticulum stress in amyotrophic lateral sclerosis. J. Mol. Neurosci. 70, 1357-1368. 10.1007/s12031-020-01551-6 [DOI] [PubMed] [Google Scholar]
- Pir G. J., Choudhary B., Mandelkow E. and Mandelkow E. M. (2016). Tau mutant A152T, a risk factor for FTD/PSP, induces neuronal dysfunction and reduced lifespan independently of aggregation in a C. elegans tauopathy model. Mol. Neurodegener 11, 33 10.1186/s13024-016-0096-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pivtoraiko V. N., Harrington A. J., Mader B. J., Luker A. M., Caldwell G. A., Caldwell K. A., Roth K. A. and Shacka J. J. (2010). Low-dose bafilomycin attenuates neuronal cell death associated with autophagy-lysosome pathway dysfunction. J. Neurochem. 114, 1193-1204. 10.1111/j.1471-4159.2010.06838.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujols J., Peña-Díaz S., Lázaro D. F., Peccati F., Pinheiro F., González D., Carija A., Navarro S., Conde-Giménez M., García J. et al. (2018). Small molecule inhibits α-synuclein aggregation, disrupts amyloid fibrils, and prevents degeneration of dopaminergic neurons. Proc. Natl. Acad. Sci. USA 115, 10481-10486. 10.1073/pnas.1804198115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao L., Hamamichi S., Caldwell K. A., Caldwell G. A., Yacoubian T. A., Wilson S., Xie Z. L., Speake L. D., Parks R., Crabtree D. et al. (2008). Lysosomal enzyme cathepsin D protects against alpha-synuclein aggregation and toxicity. Mol Brain 1, 1-17. 10.1186/1756-6606-1-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand J. B. and Russell R. L. (1984). Choline acetyltransferase-deficient mutants of the nematode Caenorhabditis elegans. Genetics 106, 227-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A., Martinez B. A., Berkowitz L. A., Caldwell G. A. and Caldwell K. A. (2014). Mitochondrial dysfunction, oxidative stress, and neurodegeneration elicited by a bacterial metabolite in a C. elegans Parkinson's model. Cell Death Dis. 5, e984 10.1038/cddis.2013.513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regitz C., Fitzenberger E., Mahn F. L., Dußling L. M. and Wenzel U. (2016). Resveratrol reduces amyloid-beta (Aβ1-42)-induced paralysis through targeting proteostasis in an Alzheimer model of Caenorhabditis elegans. Eur. J. Nutr. 55, 741-747. 10.1007/s00394-015-0894-1 [DOI] [PubMed] [Google Scholar]
- Rosen D. R., Siddique T., Patterson D., Figlewicz D. A., Sapp P., Hentati A., Donaldson D., Goto J., O'Regan J. P. and Deng H. X. (1993). Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 362, 59-62. 10.1038/362059a0 [DOI] [PubMed] [Google Scholar]
- Rual J.-F., Ceron J., Koreth J., Hao T., Nicot A.-S., Hizozane-Kishikawa T., Vandenhaute J., Orkin S. H., Hill D. E., van den Heuvel S. et al. (2004). Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 14, 2162-2168. 10.1101/gr.2505604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Q., Harrington A. J., Caldwell K. A., Caldwell G. A. and Standaert D. G. (2010). VPS41, a protein involved in lysosomal trafficking, is protective in mammalian cellular models of Parkinson's disease. Neurobiol. Dis. 37, 330-338. 10.1016/j.nbd.2009.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudich P., Snoznik C., Watkins S. C., Monaghan J., Pandey U. B. and Lamitina S. T. (2017). Nuclear localized C9orf72-associated arginine-containing dipeptides exhibit age-dependent toxicity in C. elegan. Hum. Mol. Genet. 26, 4916-4928. 10.1093/hmg/ddx372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S., Guillily M. D., Ferree A., Lanceta J., Chan D., Ghosh J., Hsu C. H., Segal L., Raghavan K., Matsumoto K. et al. (2009). LRRK2 modulates vulnerability to mitochondrial dysfunction in Caenorhabditis elegans. J. Neurosci. 29, 9210-9218. 10.1523/JNEUROSCI.2281-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sammi S. R., Foguth R. M., Nieves C. S., De Perre C., Wipf P., McMurray C. T., Lee L. S. and Cannon J. R. (2019). Perfluorooctane sulfonate (PFOS) produces dopaminergic neuropathology in Caenorhabditis elegans. Toxicol. Sci. 172, 417-434. 10.1093/toxsci/kfz191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarasija S. and Norman K. R. (2015). A γ-secretase independent role for presenilin in calcium homeostasis impacts mitochondrial function and morphology in Caenorhabditis elegans. Genetics 201, 1453-1466. 10.1534/genetics.115.182808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarasija S., Laboy J. T., Ashkavand Z., Bonner J., Tang Y. and Norman K. R. (2018). Presenilin mutations deregulate mitochondrial Ca2+ homeostasis and metabolic activity causing neurodegeneration in Caenorhabditis elegans. Elife 7, e33052 10.7554/eLife.33052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satyal S. H., Schmidt E., Kitagawa K., Sondheimer N., Lindquist S., Kramer J. M. and Morimoto R. I. (2000). Polyglutamine aggregates alter protein folding homeostasis in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 97, 5750-5755. 10.1073/pnas.100107297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin E. R., Ranganathan R. and Horvitz H. R. (2000). C. elegans locomotory rate is modulated by the environment through a dopaminergic pathway and by experience through a serotonergic pathway. Neuron 26, 619-631. 10.1016/S0896-6273(00)81199-X [DOI] [PubMed] [Google Scholar]
- Saxena S., Cabuy E. and Caroni P. (2009). A role for motoneuron subtype-selective ER stress in disease manifestations of FALS mice. Nat. Neurosci. 12, 627-636. 10.1038/nn.2297 [DOI] [PubMed] [Google Scholar]
- Schmeisser K. and Parker J. A. (2018). Nicotinamide-N-methyltransferase controls behavior, neurodegeneration, and lifespan by regulating neuronal autophagy. PLoS Genet. 14, e1007561 10.1371/journal.pgen.1007561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaye D. D. and Greenwald I. (2011). OrthoList: a compendium of C. elegans genes with human orthologs. PLoS ONE 6, e20085 10.1371/journal.pone.0020085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidransky E., Nalls M. A., Aasly J. O., Aharon-Peretz J., Annesi G., Barbosa E. R., Bar-Shira A., Berg D., Bras J., Brice A. et al. (2009). Multicenter analysis of glucocerebrosidase mutations in Parkinson's disease. New Engl. J. Med. 361, 1651-1661. 10.1056/NEJMoa0901281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmer F., Tijsterman M., Parrish S., Koushika S. P., Nonet M. L., Fire A., Ahringer J. and Plasterk R. H. (2002). Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr. Biol. 12, 1317-1319. 10.1016/S0960-9822(02)01041-2 [DOI] [PubMed] [Google Scholar]
- Singleton A. B., Farrer M., Johnson J., Singleton A., Hague S., Kachergus J., Hulihan M., Peuralinna T., Dutra A., Nussbaum R. et al. (2003). Alpha-synuclein locus triplication causes Parkinson's disease. Science 302, 841 10.1126/science.1090278 [DOI] [PubMed] [Google Scholar]
- Spinney L. (2014). Alzheimer's disease: the forgetting gene. Nature 510, 26-28. 10.1038/510026a [DOI] [PubMed] [Google Scholar]
- Steger M., Tonelli F., Ito G., Davies P., Trost M., Vetter M., Wachter S., Lorentzen E., Duddy G., Wilson S. et al. (2016). Phosphoproteomics reveals that Parkinson's disease kinase LRRK2 regulates a subset of Rab GTPases. eLife 5, e12813 10.7554/eLife.12813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston J. E., Dew M. and Brenner S. (1975). Dopaminergic neurons in the nematode Caenorhabditis elegans. J. Comp. Neuro.l 163, 215-226. 10.1002/cne.901630207 [DOI] [PubMed] [Google Scholar]
- Tardiff D. F., Jui N. T., Khurana V., Tambe M. A., Thompson M. L., Chung C. Y., Kamadurai H. B., Kim H. T., Lancaster A. K., Caldwell K. A. et al. (2013). Yeast reveal a “druggable” Rsp5/Nedd4 network that ameliorates α-synuclein toxicity in neurons. Science 342, 979-983. 10.1126/science.1245321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauffenberger A., Vaccaro A., Aulas A., Velde C. V. and Parker J. A. (2012). Glucose delays age-dependent proteotoxicity. Aging Cell 11, 856-866. 10.1111/j.1474-9726.2012.00855.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therrien M. and Parker J. A. (2014). Worming forward: amyotrophic lateral sclerosis toxicity mechanisms and genetic interactions in Caenorhabditis elegans. Front. Genet. 5, 85 10.3389/fgene.2014.00085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therrien M., Rouleau G. A., Dion P. A. and Parker J. A. (2013). Deletion of C9ORF72 results in motor neuron degeneration and stress sensitivity in C. elegans. PLoS ONE 8, e83450 10.1371/journal.pone.0083450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson M. L., Chen P., Yan X., Kim H., Borom A. R., Roberts N. B., Caldwell K. A. and Caldwell G. A. (2014). TorsinA rescues ER-associated stress and locomotive defects in C. elegans models of ALS. Dis. Model. Mech. 7, 233-243. 10.1242/dmm.013615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L., Court D. L. and Fire A. (2001). Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263, 103-112. 10.1016/S0378-1119(00)00579-5 [DOI] [PubMed] [Google Scholar]
- Treusch S., Hamamichi S., Goodman J. L., Matlack K. E., Chung C. Y., Baru V., Shulman J. M., Parrado A., Bevis B. J., Valastyan J. S. et al. (2011). Functional links between Aβ toxicity, endocytic trafficking, and Alzheimer's disease risk factors in yeast. Science 334, 1241-1245. 10.1126/science.1213210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaccaro A., Tauffenberger A., Ash P. E. A., Carlomagno Y., Petrucelli L. and Parker J. A. (2012a). TDP-1/TDP-43 regulates stress signaling and age-dependent proteotoxicity in Caenorhabditis elegans. PLoS Genet. 8, e1002806 10.1371/journal.pgen.1002806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaccaro A., Tauffenberger A., Aggad D., Rouleau G., Drapeau P. and Parker J. A. (2012b). Mutant TDP-43 and FUS cause age-dependent paralysis and neurodegeneration in C. elegans. PLoS ONE 7, e31321 10.1371/journal.pone.0031321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Damme P., Robberecht W. and Van Den Bosch L. (2017). Modelling amyotrophic lateral sclerosis: progress and possibilities. Dis. Model. Mech. 10, 537-549. 10.1242/dmm.029058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ham T. J., Thijssen K. L., Breitling R., Hofstra R. M., Plasterk R. H. and Nollen E. A. (2008). C. elegans model identifies genetic modifiers of α-synuclein inclusion formation during aging. PLoS Genet. 4, e1000027 10.1371/journal.pgen.1000027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent B. M., Tardiff D. F., Piotrowski J. S., Aron R., Lucas M. C., Chung C. Y., Bacherman H., Chen Y., Pires M., Subramaniam R. et al. (2018). Inhibiting Stearoyl-CoA desaturase ameliorates α-synuclein cytotoxicity. Cell Rep. 25, 2742-2754. 10.1016/j.celrep.2018.11.028 [DOI] [PubMed] [Google Scholar]
- Waldherr S. M., Strovas T. J., Vadest T. A., Liachko N. F. and Kraemer B. C. (2019). Constitutive XBP-1s-mediated activation of the endoplasmic reticulum unfolded protein response protects against pathological tau. Nat. Commun. 10, 4443 10.1038/s41467-019-12070-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Farr G. W., Hall D. H., Li F., Furtak K., Dreier L. and Horwich A. L. (2009). An ALS-linked mutant SOD1 produces a locomotor defect associated with aggregation and synaptic dysfunction when expressed in neurons of Caenorhabditis elegans. PLoS Genet. 5, e1000350 10.1371/journal.pgen.1000350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Hao L., Saur T., Joyal K., Zhao Y., Zhai D., Li J., Pribadi M., Coppola G., Cohen B. M. et al. (2016). Forward genetic screen in Caenorhabditis elegans suggests F57A10.2 and acp-4 as suppressors of C9orf72 related phenotypes. Front. Mol. Neurosci. 9, 113 10.3389/fnmol.2016.00113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Saar V., Leung K. L., Chen L. and Wong G. (2018). Human amyloid β peptide and tau co-expression impairs behavior and causes specific gene expression changes in Caenorhabditis elegans. Neurobiol. Dis. 109, 88-101. 10.1016/j.nbd.2017.10.003 [DOI] [PubMed] [Google Scholar]
- Wang S., Zhang S., Liou L. C., Ren Q., Zhang Z., Caldwell G. A., Caldwell K. A., and Witt S. N. (2014). Phosphatidylethanolamine deficiency disrupts α-synuclein homeostasis in yeast and worm models of Parkinson disease. Proc. Natl. Acad. Sci. USA 111, E3976-E3985. 10.1073/pnas.1411694111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler J. M., McMillan P., Strovas T. J., Liachko N. F., Amlie-Wolf A., Kow R. L., Klein R. L., Szot P., Robinson L., Guthrie C. et al. (2019). Activity of the poly(A) binding protein MSUT2 determines susceptibility to pathological tau in the mammalian brain. Sci. Transl. Med. 11, eaao6545 10.1126/scitranslmed.aao6545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J. G., Southgate E., Thomson J. N. and Brenner S. (1986). The structure of the nervous system of the nematode Caenorhabditis elegans. Phil. Trans. R. Soc. Lond. B 314, 1-340. 10.1098/rstb.1986.0056 [DOI] [PubMed] [Google Scholar]
- Winckler B., Faundez V., Maday S., Cai Q., Guimas Almeida C. and Zhang H. (2018). The endolysosomal system and proteostasis: from development to degeneration. J. Neurosci. 38, 9364-9374. 10.1523/JNEUROSCI.1665-18.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S. Q., Pontifex M. G., Phelan M. M., Pidathala C., Kraemer B. C., Barclay J. W., Berry N. G., O'Neill P. M., Burgoyne R. D. and Morgan A. (2018). α-Methyl-α-phenylsuccinimide ameliorates neurodegeneration in a C. elegans model of TDP-43 proteinopathy. Neurobiol. Dis. 118, 40-54. 10.1016/j.nbd.2018.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K., Tavernarakis N. and Driscoll M. (2001). Necrotic cell death in C. elegans requires the function of calreticulin and regulators of Ca2+ release from the endoplasmic reticulum. Neuron 31, 957-971. 10.1016/S0896-6273(01)00432-9 [DOI] [PubMed] [Google Scholar]
- Yi B., Sahn J. J., Ardestani P. M., Evans A. K., Scott L. L., Chan J. Z., Iyer S., Crisp A., Zuniga G., Pierce J. T. et al. (2017). Small molecule modulator of sigma 2 receptor is neuroprotective and reduces cognitive deficits and neuroinflammation in experimental models of Alzheimer's disease. J. Neurochem. 140, 561-575. 10.1111/jnc.13917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Hwang H.-Y., Hao H., Talbot C. and Wang J. (2012). Caenorhabditis elegans RNA-processing protein TDP-1 regulates protein homeostasis and life span. J. Biol. Chem. 287, 8371-8382. 10.1074/jbc.M111.311977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Glukhova S. A., Caldwell K. A. and Caldwell G. A. (2017). NCEH-1 modulates cholesterol metabolism and protects against a-synuclein toxicity in a C. elegans model of Parkinson's disease. Hum. Mol. Genet. 26, 3823-3836. 10.1093/hmg/ddx269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Wu Y.-C., Mullane P., Ji Y. J., Liu H., He L., Arora A., Hwang H.-Y., Alessi A. F., Niaki A. G. et al. (2018). FUS regulates activity of MicroRNA-mediated gene silencing. Mol. Cell 69, 787-801. 10.1016/j.molcel.2018.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B., Mak J. C. H., Morris A. P., Marson A. G., Barclay J. W., Sills G. J. and Morgan A. (2020). Functional analysis of epilepsy-associated variants in STXBP1/Munc18-1 using humanized Caenorhabditis elegans. Epilepsia 61, 810-821. 10.1111/epi.16464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucchi E., Ticozzi N. and Mandrioli J. (2019). Psychiatric symptoms in Amyotrophic Lateral Sclerosis: Beyond a motor neuron disorder. Front. Neurosci. 13, 175 10.3389/fnins.2019.00175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zullo J. M., Drake D., Aron L., O'Hern P., Dhamme S. C., Davidsohn N., Mao C.-A., Klein W. H., Rotenberg A., Bennett D. A. et al. (2019). Regulation of lifespan by neural excitation and REST. Nature 574, 359-364. 10.1038/s41586-019-1647-8 [DOI] [PMC free article] [PubMed] [Google Scholar]