Abstract
Attention deficit hyperactivity disorder (ADHD) is a neurodevelopmental disorder of childhood with a strong genetic component. Despite the success of mapping ADHD risk loci, little work has been done to experimentally verify the contribution of these loci to ADHD phenotypes. Meta-analysis of four genome-wide association studies in ADHD suggested CHMP7 as a predisposing gene for ADHD. A DNA variant (rs2294123) mapped to CHMP7 has been shown (via bioinformatic analysis) to have a high likelihood for functionality and correlate with reduced transcript levels. We used CRISPR-Cas9 genome editing to generate a chmp7 zebrafish model for ADHD. chmp7+/− fish showed comparable reductions in mRNA levels to individuals homozygous for the CHMP7 ADHD risk allele. These fish displayed significant hyperactivity over a 24-h period at 6 days post-fertilisation compared to chmp7+/+, but this effect did not persist into juvenile and adulthood stages. In addition, chmp7+/− fish had significantly smaller total brain volumes than chmp7+/+ fish. Finally, the hyperactivity at 6 days post-fertilisation was significantly reduced through the application of methylphenidate, a mainstay pharmacological treatment for ADHD. Overall, this study highlights an important role for CHMP7 in the neurodevelopment of ADHD, and demonstrates the utility of zebrafish for modelling the functional effects of genes conferring risk to ADHD.
Subject terms: ADHD, Medical genetics
Introduction
Attention deficit hyperactivity disorder (ADHD) is a highly prevalent neurodevelopmental disorder that affects ~5% of school age children1 and persists into adulthood in 30–60% of cases2,3. Reductions in brain volume are also often associated with the disorder4.
It is estimated that genetic factors account for ~80% of ADHD’s aetiology5,6. Meta-analyses of genome wide association studies (GWAS) identified several DNA variants potentially involved in ADHD, including rs2294123, which maps to charged multivesicular body protein 7 (CHMP77). In addition, recent large scale GWAS8 demonstrates that rs2294123 is in significant LD with several SNPs showing nominal association with ADHD (Supplementary Fig. 1, Supplementary Table 1). However, functional validation of the contribution of these associated genes to the development of ADHD is lacking. Most variants detected via candidate gene and GWAS approaches map to non-coding regions of the genome9. Given the wide range of roles that non-coding regions play in gene expression, separating neutral ADHD-associated variants from causative/pathogenic ones remains a major challenge.
To tackle this, Tong et al.9 developed an in silico pipeline to prioritise a curated set of non-coding single nucleotide polymorphisms (SNPs) linked to ADHD for functional analysis. They identified an SNP (rs2294123, G→T) mapped to the 5ˈUTR of CHMP7, which was significantly associated with ADHD. Homozygosity for the risk allele (T) was significantly associated with lower neurocognitive function in ADHD individuals, higher ADHD symptom traits, and a 33% reduction in CHMP7 transcript levels compared to homozygous G individuals9. These findings suggest that a reduction in CHMP7 expression contributes to ADHD phenotypes and warrants further investigation.
CHMP7 plays an important role in endosomal sorting10, nuclear envelope formation11, and has recently been implicated in spinal and bulbar muscular atrophy12. It interacts with a member of the endosomal sorting complex required for transport (ESCRT) family, ESCRT-III10. Several CHMP family members are part of ESCRT-III, have been implicated in neurodevelopmental processes13, and are known to contribute to the development of neuropsychiatric conditions14–16. This supports the need for further examination of the role of CHMP7 in ADHD.
To examine the functional relevance of CHMP7 to ADHD, we generated a chmp7 zebrafish mutant line using CRISPR-Cas9 genome editing, and hypothesised that chmp7 heterozygous (chmp7+/−) animals would mimic the reduction in transcripts correlated with the ADHD-associated SNP (rs22941239). We demonstrate that chmp7+/− fish are more active than wildtype (chmp7+/+) and have decreased total brain volumes, similar to that reported in ADHD cases. Furthermore, we also show that hyperactivity of chmp7+/− fish can be significantly reduced through the application of methylphenidate, which is commonly used for the treatment of ADHD. Overall, we have provided experimental validation for CHMP7 as a risk factor for ADHD.
Materials and methods
Ethics
Fish were maintained in the Monash University Fish Core facility under breeding colony license MARP/2015/004/BC. Creation of transgenic lines was approved by the School of Biological Sciences Animal Ethics Committee (BSCI/2015/07). Experiments were carried out on embryos of wildtype (Tübingen, TU) background.
Generation and genotyping of the chmp7 mutant line
To create a chmp7 mutant zebrafish line, a guide RNA targeting chmp7 (ENSDARG00000041362) exon 2 was generated according to Gagnon et al.17. One-cell stage embryos were injected with a mixture containing: 150 ng/µL guide RNA, 5 µg/µL Cas9 protein (PNA Bio, Newbury Park, CA, USA), 20 µM STOP cassette, 0.25 µL Phenol Red, 0.25 µL Cascade Blue (Molecular Probes, Waltham, MA, USA), and ultra-pure H2O to a final volume of 2.5 µL. Embryos were selected for Cascade Blue, indicating successful injection, at 24 hpf and raised to adulthood. F0 founders were identified by outcrossing to TU fish, collecting DNA from 15 to 20 offspring, and the pooled DNA used as a template for PCR amplification of the region surrounding the target site. Polyacrylamide gel electrophoresis was used to visualise heterodimers. Founders were outcrossed to wildtype fish, and F1 individuals screened for the presence of mutations using PCR and gel electrophoresis. The mutation was determined using Sanger sequencing. Experiments utilised F3 and subsequent generations. Guide RNAs and primer sequences are presented in Supplementary Table 2. Genotyping was performed using allele-specific KASP fluorescence assays (LGC Biosearch Technologies, Teddington, Middlesex, UK).
Whole-mount in situ hybridisation
To generate a probe to examine chmp7 expression, a chmp7 fragment was amplified from genomic DNA using primers (Supplementary Table 2) and cloned into pGEM-T Easy (Promega, Madison, WI, USA). Sequence orientation was determined via PCR using combinations of chmp7 and M13 primers (Supplementary Table 2). Probe templates were amplified from the plasmid using the chmp7 in situ template reverse and pGEM-T Easy M13 forward primers, and digoxygenin riboprobes generated using T7 RNA polymerase as previously described18. Whole-mount in situ hybridisation was performed as per Ruparelia et al.19.
Reverse transcription-PCR
RT-PCR was utilised to determine when chmp7 is expressed. cDNA synthesis was performed on RNA extracted from wildtype embryos at the 8- and 16-somite stages, 1, 1.5, 2, 3, 4, and 5 dpf. Total RNA was isolated using TRIzol® reagent as described by the manufacturer (Sigma, St. Louis, MO, USA) and treated with DNAse (Promega) to remove possible DNA contamination. One µg of total RNA was reverse transcribed using the Superscript III first-strand synthesis kit (Invitrogen, Waltham, MA, USA). RT-PCR primers are presented in Supplementary Table 2. PCR cycles were: initial denaturing cycle at 96 °C for 2 min, 30 cycles of 96 °C, 57 °C, and 72 °C for 30 s each; followed by a final extension cycle at 72 °C for 5 min. Twenty-five µL of PCR product was run on a 1% Tris-acetate-EDTA agarose gel for visualisation.
Quantitative RT-PCR
qRT-PCR was used to compare chmp7 mRNA levels between genotypes. RNA was extracted from chmp7+/+, chmp7+/−, and chmp7−/− embryos at 6 dpf, and pooled with a constant number of fish (20–25) per genotype within each biological replicate. cDNA was prepared as above. qRT-PCR was performed using a Lightcycler 480 (Roche, Basel, Switzerland) and SYBR Green master mix (Roche). An average of actin, beta 1 (actb1), 18s ribosomal RNA (18SrRNA), and eukaryotic translation elongation factor 1 alpha 1 (eef1α1) expression values was used as a reference. Primers are presented in Supplementary Table 2. Three technical replicates were conducted for each biological replicate. Generalised mixed linear modelling was used to examine differences in mRNA levels. Genotype was examined as a fixed effect, with biological replicate as a random effect. F tests were performed using Satterthwaite estimated degrees of freedom.
24-h locomotion assay: 6 dpf
Zebrafish activity levels were examined using locomotion assays at 6 dpf to examine activity at a larval stage that demonstrated autonomous movement, while still utilising a high throughput behavioural assay. Embryos were collected between 09:00 and 10:00 and raised in Petri dishes in a 14-h day (09:00-23:00, 300 lux ± 20 lux) and 10-h night (23:00-09:00, full darkness) cycle until 6 dpf. Larvae were fed 0.5 ml concentrated paramecium between 09:00 and 10:00 on day 5 and 6, and medium was changed daily between 14:00 and 16:00. Between 14:00 and 16:00 on day 6, larvae were transferred to 24-well plates containing 1.5 ml of E3 embryo medium (5 mm NaCl, 0.17 mm KCl, 0.33 mm CaCl, 0.33 mm MgSO4 in water) per well to acclimatise. Between 22:30 and 22:50 on day 6, plates were transferred to Zebraboxes (Viewpoint, Lyon, France). At 22:50, Zebraboxes were closed to allow fish to habituate to the darkness for 10 min, and video recording began at 23:00. The experiment ran for 24 h and 30 min in full darkness, after which embryos were sacrificed and genotyped.
24-h locomotion assay: drug treatment at 6 dpf
To examine the effect of methylphenidate, locomotion assays were performed as described above. However, at 22:00 on day 6, 150 µL of dH2O (vehicle control) or 100 µM of Threo-methylphenidate hydrochloride (Tocris Bioscience, Bristol, United Kingdom) was added to wells containing 1.35 ml of E3 medium and the fish, to yield a final volume of 1.5 ml per well and 10 µM methylphenidate, as described by Lange et al.20. Raised lids were used to prevent evaporation, any wells showing condensation were removed from analysis. For each experiment, methylphenidate and vehicle were randomised across the plate, and the investigator blinded to treatment by a third party. Blinding was removed after initial mixed model tests.
24-h locomotion assay: 6- & 12-weeks post-fertilisation
Locomotion assays were used to examine activity levels in larval and adult fish. Fish were genotyped at 3 dpf by extracting DNA from fin clippings using 50 mM NaOH and 1 M Tris-HCl (pH 7.5), and then sorted by genotype. Fish were raised under a day-night cycle of 12 h day and 12 h night (08:00-20:00, and 20:00-08:00 respectively). Between 12:00 and 14:00 on day 41 and day 83, fish were transferred to individual tanks to acclimatise. Between 19:00 and 19:50 on day 41 and 83, tanks were transferred to Zebracubes (Viewpoint, 9 tanks per system). At 19:50, Zebracubes were closed to allow fish to habituate to the darkness for 10 min, and tracking began at 20:00. Positions of genotypes were randomised, and the investigator blinded to genotype. Video tracking ran for 24 h and 30 min in full darkness, after which fish were returned to their tanks.
Video analysis
Videos were analysed using Ethovision (Noldus, version 14, Wageningen, Netherlands). Thresholds were: Moving, 1 mm/sec; Stopping, 0.75 mm/sec; Detection threshold, Dynamic Subtraction, Darker, 9. Voxel smoothing was used to remove errors in 6 dpf analyses, with movements < 0.04 mm and > 12 mm per frame excluded.
Locomotion assay statistical analysis
Data were processed using Microsoft Excel 2013 (Microsoft, Redmond, WA, USA) and statistical analyses performed using SPSS Statistics 26 (IBM, Armonk, NY, USA). Data were ordered chronologically into 10-min bins. Time points at the end of videos less than 300 s long were excluded. For each fish, activity data were summed by hour. A normalised value for each fish per hour was determined by comparing activity, per fish per hour, to the average activity value of all fish per hour, from the respective replicate. Genotypes were then assigned to individual fish, and fish with ambiguous genotypes removed from analysis. Data points from the 30 min past the initial 24 h were excluded. Data were visualised using a line graph in GraphPad Prism Version 8 (GraphPad Software, San Diego, CA, USA).
To examine differences in activity, a mixed linear model was used. For the 6, 42, and 84 dpf locomotion assays, main effects of time and genotype, and an interaction effect of time by genotype were used. Repeated measures of time (h) were modelled using a first-order autoregressive variance structure for the 6 dpf assay, and a diagonal variance structure was used for the 42 dpf and 84 dpf assays. Random effects of Zebrabox tracking system, as well as individual animals grouped by genotype, were used. A natural log transformation was applied to the normalised data to meet assumptions of normality which were checked by inspection of the residuals. F tests were performed using a maximal likelihood model, with Satterthwaite estimated degrees of freedom. For the drug treatment assays main effects of time, treatment, and genotype, and an interaction effect of time by genotype by treatment were used. All pairwise comparisons for time points were two-tailed, performed using Bonferroni adjustment for multiple comparisons.
Confocal microscopy live imaging
chmp7+/− fish were crossed to a green fluorescent protein (GFP)-tagged HuC reporter (HuC:eGFP21) transgenic (Tg) line and raised to adulthood. Tg(HuC:eGFP);chmp7+/− fish were crossed to chmp7+/− fish, and embryos raised in E3 containing 200 µM N-Phenylthiourea (PTU, Sigma) from 6 h to suppress melanocyte formation, changing medium every 48 h. Embryos were sorted for fluorescence at 2 dpf. At 3 dpf, fish were anaesthetised using 0.0016% Tricaine methanesulfonate (Sigma) in E3, their tails clipped, DNA extracted, and fish sorted by genotype. In order to examine brain volume at a larval stage that was in line with the locomotion assays, at 6 dpf, embryos were again anesthetised and set in 1% low melting agarose in E3 containing tricaine in 0.8 mm fluorinated ethylene propylene (FEP) tubing (Bola, Grünsfeld, Germany). Genotypes were randomised, and the investigator blinded to genotype. Images were taken using a Thorlabs confocal microscope (Newton, NJ, USA), with an Olympus 20x water dipping 1.0 NA objective (Tokyo, Japan), pinhole 25 µm, 2.005 µm/pixel, step size = 1 µm, averaging = 16 frames.
Brain image registration and analysis
Image registration of live confocal stacks was achieved using Advanced Normalization Tools (ANTs) registration software (3.0.0.0), running on Monash University’s MASSIVE computing cluster. Registered images were analysed using cobraZ brain volume analysis software as described by Gupta et al.22. In addition to total volume, brain regions homologous to human regions known to have volume differences in ADHD individuals4 (telencephalon (pallium, subpallium, anterior commissure), thalamus, and ventral thalamus) were examined using generalised mixed linear modelling. Genotype was examined as a fixed effect, with biological replicate as a random effect. F tests were performed with Satterthwaite estimated degrees of freedom. Bonferroni corrections for multiple comparisons were applied to each test.
Results
chmp7 is expressed throughout early development
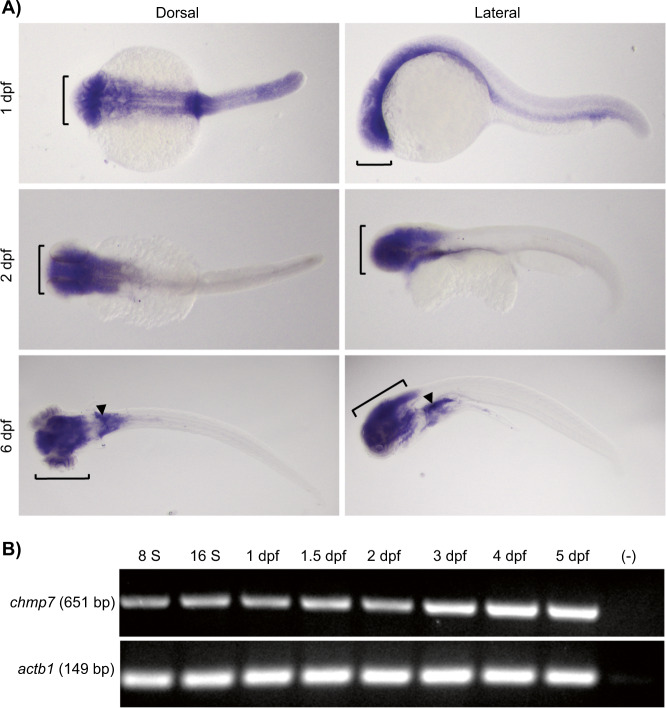
Zebrafish possess orthologues of all members of the human CHMP family genes (Supplementary Fig. 2, Supplementary Table 3.). More importantly, the zebrafish Chmp7 protein has a sequence identity of 51% and similarity of 70% to that of human. To identify where and when chmp7 is expressed in zebrafish, in situ hybridisations and RT-PCR were performed on wildtype embryos. We observed that chmp7 was expressed ubiquitously in the embryo at 1 day post-fertilisation (dpf), with higher expression levels in the brain, becoming more restricted to the head by 2 dpf. It remained visible only in the head and kidney at 6 dpf (Fig. 1A). RT-PCR showed that chmp7 was expressed at the 8-somite stage through to at least 5 dpf (Fig. 1B).
Fig. 1. Characterisation of chmp7 expression.
a Whole-mount in situ hybridisation on zebrafish at 1, 2, and 6 dpf, using DIG-labelled RNA probes specific to zebrafish chmp7. [: head, ▼: kidney. b RT-PCR for chmp7 during early zebrafish development. actb1 was amplified as a positive control. – is the no template negative control.
chmp7 heterozygotes have reduced mRNA levels
After confirming that chmp7 expression was detectable during early zebrafish development, CRISPR-Cas9 genome editing was used to mutate the gene, resulting in a 7 bp deletion in exon 2 (Supplementary Fig. 3A). This results in the addition of 20 amino acids, and a stop codon following the 142nd amino acid (Supplementary Fig. 3B). This is predicted to remove the sucrose non-fermenting protein 7 (Snf7) domain, the main catalytic domain for CHMP7 responsible for its interaction with CHMP4B and thus, ESCRT-III10.
This mutation is predicted to trigger nonsense mediated decay, and a loss of protein, rather than the production of a truncated isoform. To determine if chmp7+/− fish have reduced chmp7 mRNA, thereby mimicking the reduction observed in individuals homozygous for the ADHD risk allele (T) of the ADHD-associated SNP, quantitative real-time PCR (qRT-PCR) on cDNA from chmp7+/+, chmp7+/−, and chmp7−/− 6 dpf fish was performed. Mixed linear modelling demonstrated a significant reduction in mRNA levels (F = 71.41 (2, 4), p = 0.001, Fig. 2), and chmp7+/− fish had 53% total chmp7 mRNA compared to chmp7+/+ (t = −5.13 (4), p = 0.007), supporting the use of chmp7+/− fish as a model of the reduction in CHMP7 mRNA levels correlated with the rs2294123 homozygous ADHD risk allele (T).
Fig. 2. chmp7 qRT-PCR on chmp7+/+, chmp7+/−, and chmp7−/− embryos.
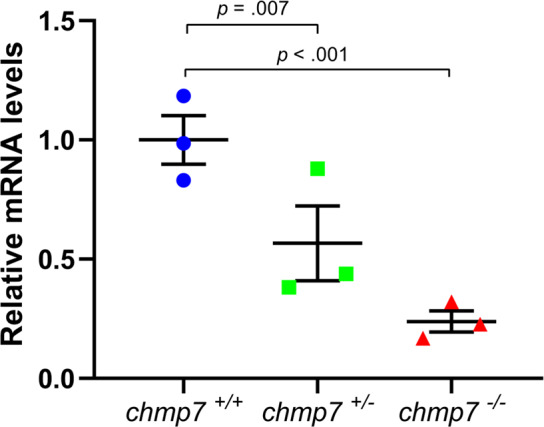
Mixed linear modelling demonstrated a significant difference between genotypes (p = 0.001). Data are from three biological replicates, and is normalised to chmp7+/+ values. Centre lines = mean, error bars = ± standard error of the mean (SEM).
chmp7 heterozygous larvae display a hyperactivity phenotype
Given that chmp7+/− fish possess similar reductions in chmp7 mRNA levels to individuals homozygous for the CHMP7 ADHD risk allele, we examined if reduced chmp7 mRNA levels lead to a hyperactivity phenotype at embryonic (6 dpf), juvenile (42 dpf), and adult (84 dpf) stages.
The activity of chmp7+/+ (n = 153) and chmp7+/− (n = 131) zebrafish at 6 dpf was tracked over a period of 24 h, starting at 6 days, 14 h post-fertilisation (hpf) (Fig. 3). Mixed linear modelling analysis demonstrated a significant main effect of genotype (F = 4.70 (1, 287.06), p = 0.031). There was no significant interaction effect of genotype and time (F = 0.77 (23, 3543.20), p = 0.78). This demonstrates that chmp7+/− fish are consistently more active than chmp7+/+ fish over the 24-h period.
Fig. 3. Activity analysis of chmp7+/+ (n = 153) and chmp7+/− (n = 131) zebrafish 6 dpf embryos over a 24-h period.
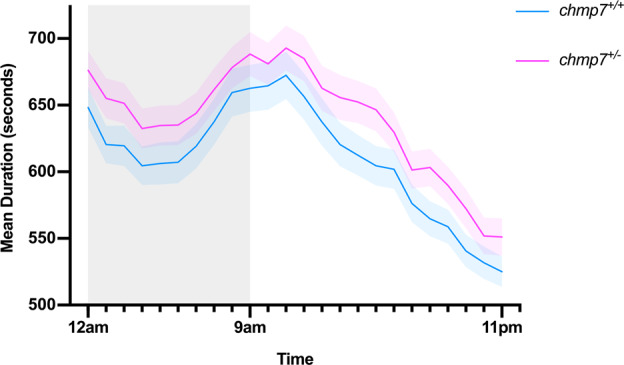
Average time spent moving in seconds each hour per genotype is displayed on the Y-axis. Data are from three biological replicates. Error bars = ± SEM.
To determine if the hyperactivity phenotype persists into juvenile and adult stages, activity of chmp7+/+ and chmp7+/− fish was tracked over a period of 24 h, starting at 41 days 11 hpf for juveniles, and 83 days 11 hpf for adults. There were no significant differences between genotypes over the entire experimental period for either juveniles (chmp7+/+, n = 41, chmp7+/−, n = 50, F = 0.29 (1, 61.52), p = 0.59, Supplementary Fig. 4A) or adults (chmp7+/+, n = 30, chmp7+/−, n = 36, F = 0.008 (1, 60.55), p = 0.93, Supplementary Fig. 4B), suggesting the hyperactivity phenotype does not persist past the larval stage.
chmp7 heterozygotes have significantly smaller brain volumes
Given the consistently reported reduced brain volumes for individuals with ADHD, the heads of chmp7+/+ (n = 12) and chmp7+/− (n = 12) zebrafish on a pan-neuronal fluorescent Tg(HuC:eGFP) background, were imaged live using confocal microscopy at 6 dpf (Fig. 4A). We observed a 9.2% total brain volume reduction in chmp7+/− fish compared to chmp7+/+ fish (F = 11.49 (1, 20), p = 0.018, one-tailed, Fig. 4B). However, we did not observe significant differences for specific selected brain regions (Supplementary Table 4).
Fig. 4. Total brain volume is decreased in chmp7 heterozygotes at 6 dpf.
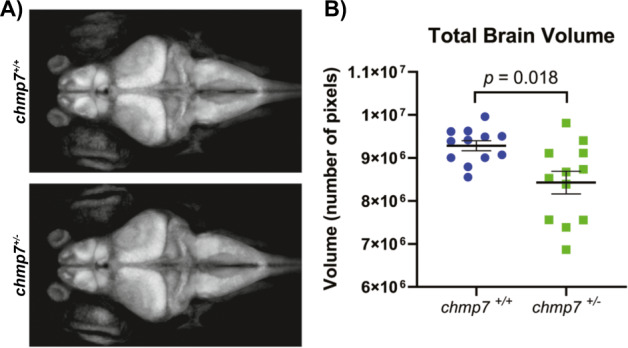
a Maximum intensity projections of whole brains from Tg(HuC:eGFP);chmp7+/+ (n = 12) and Tg(HuC:eGFP);chmp7+/− (n = 12) 6 dpf fish. b chmp7+/− fish had significantly reduced total brain volumes when compared to chmp7+/+ fish. Data are from three biological replicates. Centre lines = mean, error bars = ± SEM.
Methylphenidate significantly reduces hyperactivity in chmp7 heterozygotes
To determine if methylphenidate could ameliorate the hyperactivity seen in chmp7+/− fish, chmp7+/+ + dH2O (n = 179), chmp7+/− + dH2O (n = 160), chmp7+/+ + 10 µM methylphenidate (n = 171), chmp7+/− + 10 µM methylphenidate (n = 166) zebrafish were tracked over a period of 24 h starting at 6 days, 14 hpf. chmp7+/− + dH2O fish demonstrated hyperactivity compared to chmp7+/+ + dH2O fish during the 10-h night period (Fig. 5). However, this effect was diminished in the chmp7+/− + methylphenidate fish. Mixed linear modelling demonstrated a significant interaction between genotype, drug treatment, and time (F = 1.60 (69, 8038.37), p = 0.001). Given the significant interaction of genotype and treatment over time, we investigated the differences between groups across time.
Fig. 5. Activity analysis of chmp7+/+ and chmp7+/− zebrafish 6 dpf embryos over a 24-h period, both treated with either 10 µM methylphenidate (MpH) or dH2O.
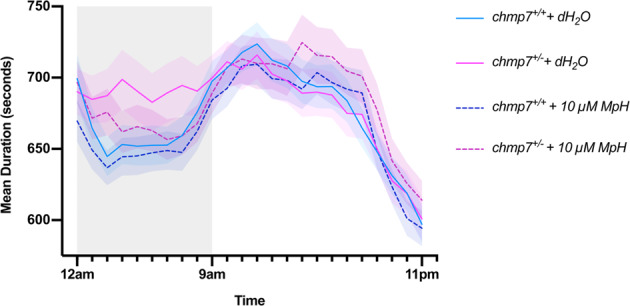
Average time spent moving in seconds each hour per genotype is displayed on the Y axis. Data are from six biological replicates. Error bars = ± SEM.
chmp7+/− + dH2O fish demonstrated significant hyperactivity compared to chmp7+/+ + dH2O fish across the majority of the night period (hour 3, p = 0.002; hour 4, p = 0.007; hour 5, p = 0.013; hour 6, p = 0.020; hour 7, p = 0.025; hour 8, p = 0.014). Application of methylphenidate gradually reduced the activity of chmp7+/− + methylphenidate fish until it was significantly less than chmp7+/− + dH2O fish (hour 8, p = 0.045). In addition, chmp7+/− + methylphenidate fish were not significantly different from chmp7+/+ + dH2O fish for the majority of the night period. This demonstrates that the application of methylphenidate was sufficient to significantly reduce the hyperactivity seen in chmp7+/− fish to levels comparable to that of wildtype.
Discussion
Zebrafish are emerging as a promising model for neuropsychiatric disorders23. We demonstrate here for the first time the utility and versatility of zebrafish models to validate ADHD genetic associations through analysis of swimming activity and brain volume as phenotypes of ADHD. The hyperactivity observed in 6 dpf chmp7+/− fish did not persist into adulthood, demonstrating the use of zebrafish for testing the temporal progression of ADHD phenotypes. Zebrafish can also be used to examine changes in brain volume commonly observed in ADHD individuals4, providing anatomical evidence for ADHD-associations. Finally, the response to methylphenidate suggests that dysregulation of dopamine (or noradrenaline) signalling could be contributing to the observed hyperactivity. In addition, given the effectiveness of methylphenidate varies between individuals with ADHD24, the use of zebrafish for testing ADHD-associated gene models for their response to drugs may be beneficial for understanding drug response variability.
The molecular mechanism behind the hyperactivity of chmp7+/− fish requires further investigation. Given the positive response of chmp7+/− fish to methylphenidate treatment, this is suggestive of abnormalities in either dopamine or noradrenaline signalling. While it is possible that other neurotransmitter pathways could be contributing to the phenotype we see in chmp7+/− fish, abnormal dopaminergic signalling, in particular, has been repeatedly associated with ADHD25. As such, investigating the role of CHMP7 in dopamine signalling requires further investigation. Additionally, defects in neuronal pruning are seen in knockdown26, loss of function27, and dominant negative mutations28, of ESCRT-III proteins. Given CHMP7’s known interactions with ESCRT-III proteins, it is possible that neuronal pruning is also disrupted when CHMP7 mRNA levels are reduced. The hyperactivity phenotype of chmp7+/− fish could therefore be due to delayed maturation of neural networks important for impulse control, or motor control/coordination, due to a lack of efficient neural pruning. This would be consistent with the neurodevelopmental delay seen in ADHD individuals (reviewed in Dark et al.29).
The assays presented in Figs. 3 and 5 identified significant hyperactivity in chmp7+/− fish compared to chmp7+/+. However, for the chmp7+/− + dH2O control in the methylphenidate assay which essentially replicated the previous activity experiment, the hyperactivity was restricted to the night period. This could indicate that a reduction in chmp7 mRNA has stronger, more consistent impacts on sleep patterns rather than waking cognition. This is consistent with sleep impairments often seen in ADHD, as well as inter- and intra-subject variability in circadian rhythms30. In addition, hyperactivity during night periods was observed in pan-neuronal knockdown of the dopamine transporter, or latrophilin, in Drosophila, which was suggested to be characteristic of dysregulation of dopamine signalling31. This highlights that dopamine imbalance could be contributing to the hyperactivity phenotype observed in chmp7+/− fish. This is consistent with the amelioration of the phenotype by methylphenidate application (Fig. 5).
This study is the first of its kind to functionally examine CHMP7 as a risk gene for ADHD using an animal model. It is also the first functional validation using zebrafish of an ADHD-associated gene identified through GWAS. We demonstrate here that chmp7+/− fish are an appropriate model for the CHMP7 ADHD-associated alleles, and that the decrease in chmp7 mRNA levels is correlated with a hyperactivity phenotype and reduced brain volume. Furthermore, this hyperactivity is alleviated by treatment with methylphenidate, contributing to our understanding of specific ADHD risk factors to variability in drug responsiveness. More generally, we advocate for the use of zebrafish to model the large number of candidate genes for ADHD that have recently emerged from large-scale GWAS8.
Supplementary information
Acknowledgements
The authors would like to thank members of the Bryson-Richardson and Bellgrove laboratories for their support. We would also like to extend thanks to A/Prof Sean Cain and Angus Burns for their advice with the locomotion assay, Dr Harold Burgess for advice on brain registration, Dr Keyne Monro for statistical advice, and to FishCore for housing the chmp7 strain. This research was supported by an Australian Government Research Training Program (RTP) Scholarship to C.D. M.A.B is supported by a Senior Research Fellowship (SRF B) from the Australian National Health and Medical Research Council.
Data availability
Raw data and statistical analysis files are available at https://bridges.monash.edu/projects/Functional_validation_of_CHMP7_as_an_ADHD_risk_gene/83780.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41398-020-01077-w).
References
- 1.Polanczyk G, de Lima MS, Horta BL, Biederman J, Rohde LA. The worldwide prevalence of ADHD: a systematic review and metaregression analysis. Am. J. Psychiatry. 2007;164:942–948. doi: 10.1176/ajp.2007.164.6.942. [DOI] [PubMed] [Google Scholar]
- 2.Faraone SV, Biederman J, Mick E. The age-dependent decline of attention deficit hyperactivity disorder: a meta-analysis of follow-up studies. Psychol. Med. 2006;36:159–165. doi: 10.1017/S003329170500471X. [DOI] [PubMed] [Google Scholar]
- 3.Faraone SV, Biederman J. What Is the prevalence of adult ADHD? Results of a population screen of 966 adults. J. Atten. Disord. 2005;9:384–391. doi: 10.1177/1087054705281478. [DOI] [PubMed] [Google Scholar]
- 4.Hoogman M, et al. Subcortical brain volume differences in participants with attention deficit hyperactivity disorder in children and adults: a cross-sectional mega-analysis. Lancet Psychiatry. 2017;4:310–319. doi: 10.1016/S2215-0366(17)30049-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hawi Z, et al. The molecular genetic architecture of attention deficit hyperactivity disorder. Mol. Psychiatry. 2015;20:289–297. doi: 10.1038/mp.2014.183. [DOI] [PubMed] [Google Scholar]
- 6.Levy F, Hay DA, McStephen M, Wood C, Waldman I. Attention-deficit hyperactivity disorder: a category or a continuum? Genetic analysis of a large-scale twin study. J. Am. Acad. Child Adolesc. Psychiatry. 1997;36:737–744. doi: 10.1097/00004583-199706000-00009. [DOI] [PubMed] [Google Scholar]
- 7.Neale BM, et al. Meta-analysis of genome-wide association studies of attention-deficit/hyperactivity disorder. J. Am. Acad. Child Adolesc. Psychiatry. 2010;49:884–897. doi: 10.1016/j.jaac.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Demontis D, et al. Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat. Genet. 2019;51:63–75. doi: 10.1038/s41588-018-0269-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tong, J. H. S. et al. Separating the wheat from the chaff: systematic identification of functionally relevant noncoding variants in ADHD. Mol Psychiatry. 10.1038/mp.2016.2 (2016). [DOI] [PubMed]
- 10.Horii M, et al. CHMP7, a novel ESCRT-III-related protein, associates with CHMP4b and functions in the endosomal sorting pathway. Biochem J. 2006;400:23–32. doi: 10.1042/BJ20060897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olmos Y, Perdrix-Rosell A, Carlton JG. Membrane binding by CHMP7 coordinates ESCRT-III-dependent nuclear envelope reformation. Curr. Biol. 2016;26:2635–2641. doi: 10.1016/j.cub.2016.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malik B, et al. Gene expression analysis reveals early dysregulation of disease pathways and links Chmp7 to pathogenesis of spinal and bulbar muscular atrophy. Sci. Rep. 2019;9:1–16. doi: 10.1038/s41598-018-37186-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sadoul R, et al. The role of ESCRT during development and functioning of the nervous system. Semin Cell Dev. Biol. 2018;74:40–49. doi: 10.1016/j.semcdb.2017.08.013. [DOI] [PubMed] [Google Scholar]
- 14.Chidambaram SB, et al. Dendritic spines: Revisiting the physiological role. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2019;92:161–193. doi: 10.1016/j.pnpbp.2019.01.005. [DOI] [PubMed] [Google Scholar]
- 15.Lau CG, Zukin RS. NMDA receptor trafficking in synaptic plasticity and neuropsychiatric disorders. Nat. Rev. Neurosci. 2007;8:413–426. doi: 10.1038/nrn2153. [DOI] [PubMed] [Google Scholar]
- 16.Mathews, P. M. & Levy, E. Exosome production is key to neuronal endosomal pathway integrity in neurodegenerative diseases. Front Neurosci. 10.3389/fnins.2019.01347 (2019). [DOI] [PMC free article] [PubMed]
- 17.Gagnon, J. A. et al. Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS ONE. 10.1371/journal.pone.0098186 (2014). [DOI] [PMC free article] [PubMed]
- 18.Broadbent J, Read EM. Wholemount in situ hybridization of Xenopus and zebrafish embryos. Methods Mol. Biol. Clifton NJ. 1999;127:57–67. doi: 10.1385/1-59259-678-9:57. [DOI] [PubMed] [Google Scholar]
- 19.Ruparelia AA, Zhao M, Currie PD, Bryson-Richardson RJ. Characterization and investigation of zebrafish models of filamin-related myofibrillar myopathy. Hum. Mol. Genet. 2012;21:4073–4083. doi: 10.1093/hmg/dds231. [DOI] [PubMed] [Google Scholar]
- 20.Lange M, et al. The ADHD-susceptibility gene lphn3.1 modulates dopaminergic neuron formation and locomotor activity during zebrafish development. Mol. Psychiatry. 2012;17:946–954. doi: 10.1038/mp.2012.29. [DOI] [PubMed] [Google Scholar]
- 21.Park H-C, et al. Analysis of upstream elements in the HuC promoter leads to the establishment of transgenic Zebrafish with fluorescent neurons. Dev. Biol. 2000;227:279–293. doi: 10.1006/dbio.2000.9898. [DOI] [PubMed] [Google Scholar]
- 22.Gupta T, et al. Morphometric analysis and neuroanatomical mapping of the zebrafish brain. Methods. 2018;150:49–62. doi: 10.1016/j.ymeth.2018.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fontana BD, Mezzomo NJ, Kalueff AV, Rosemberg DB. The developing utility of zebrafish models of neurological and neuropsychiatric disorders: a critical review. Exp. Neurol. 2018;299:157–171. doi: 10.1016/j.expneurol.2017.10.004. [DOI] [PubMed] [Google Scholar]
- 24.Polanczyk, G., Bigarella, M. P., Hutz, M. H. & Rohde, L. A. Pharmacogenetic approach for a better drug treatment in children. Curr Pharm Des. 10.2174/138161210791959872 (2010). [DOI] [PubMed]
- 25.Barr CL, Misener VL. Dopamine system genes and ADHD: a review of the evidence. Future Neurol. Lond. 2008;3:705–728. doi: 10.2217/14796708.3.6.705. [DOI] [Google Scholar]
- 26.Loncle N., Agromayor M., Martin-Serrano J. & Williams D. W. An ESCRT module is required for neuron pruning. Sci Rep. 10.1038/srep08461 (2015). [DOI] [PMC free article] [PubMed]
- 27.Sweeney NT, Brenman JE, Jan YN, Gao F-B. The coiled-coil protein shrub controls neuronal morphogenesis in Drosophila. Curr. Biol. CB. 2006;16:1006–1011. doi: 10.1016/j.cub.2006.03.067. [DOI] [PubMed] [Google Scholar]
- 28.Belly A, et al. CHMP2B mutants linked to frontotemporal dementia impair maturation of dendritic spines. J. Cell Sci. 2010;123:2943–2954. doi: 10.1242/jcs.068817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dark C, Homman-Ludiye J, Bryson-Richardson RJ. The role of ADHD associated genes in neurodevelopment. Dev. Biol. 2018;438:69–83. doi: 10.1016/j.ydbio.2018.03.023. [DOI] [PubMed] [Google Scholar]
- 30.Becker SP. ADHD and sleep: recent advances and future directions. Curr. Opin. Psychol. 2020;34:50–56. doi: 10.1016/j.copsyc.2019.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van der Voet M, Harich B, Franke B, Schenck A. ADHD-associated dopamine transporter, latrophilin and neurofibromin share a dopamine-related locomotor signature in Drosophila. Mol. Psychiatry. 2016;21:565–573. doi: 10.1038/mp.2015.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data and statistical analysis files are available at https://bridges.monash.edu/projects/Functional_validation_of_CHMP7_as_an_ADHD_risk_gene/83780.