SUMMARY
In the developing human neocortex, progenitor cells generate diverse cell types prenatally. Progenitor cells and newborn neurons respond to signaling cues, including neurotransmitters. While single-cell RNA sequencing has revealed cellular diversity, physiological heterogeneity has yet to be mapped onto these developing and diverse cell types. By combining measurements of intracellular Ca2+ elevations in response to neurotransmitter receptor agonists and RNA sequencing of the same single cells, we show that Ca2+ responses are cell-type-specific and change dynamically with lineage progression. Physiological response properties predict molecular cell identity and additionally reveal diversity not captured by single-cell transcriptomics. We find that the serotonin receptor HTR2A selectively activates radial glia cells in the developing human, but not mouse, neocortex, and inhibiting HTR2A receptors in human radial glia disrupts the radial glial scaffold. We show highly specific neurotransmitter signaling during neurogenesis in the developing human neocortex and highlight evolutionarily divergent mechanisms of physiological signaling.
Graphical Abstract
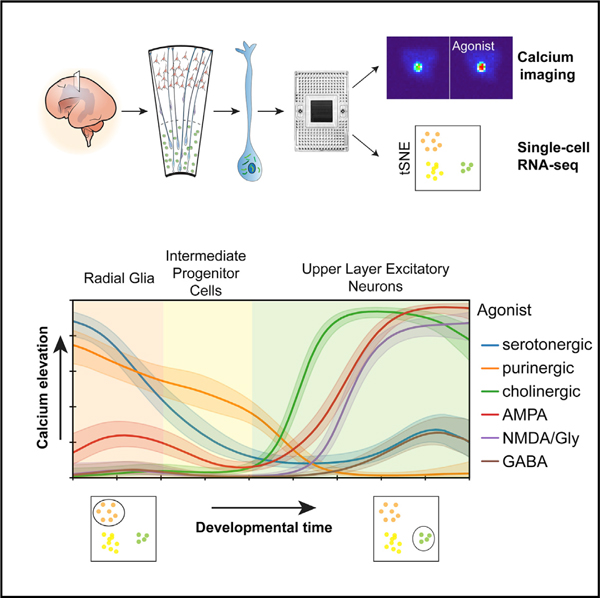
In Brief
Mayer et al. develop a microfluidics-based approach that links calcium imaging and single-cell transcriptomics to study cellular responses to neurotransmitters in the developing human neocortex. They reveal dynamically changing response profiles as progenitor cells differentiate to diverse types of neurons.
INTRODUCTION
The neocortex has expanded in mammalian evolution through an increase in the number of progenitor cells and the time period over which they produce neurons and glia (Azevedo et al., 2009). Human radial glia cells, the neural stem cells of the developing cortex, have been subdivided into several subtypes characterized by distinct morphologies, dynamic behavior, and transcriptomic profiles (Lui et al., 2011; Pollen et al., 2015; Thomsen et al., 2016). Ventricular radial glia (vRG) are embedded in an adhesion belt along the ventricle and maintain ventricular contact. Outer radial glia (oRG) (Fietz et al., 2010; Hansen et al., 2010) in the outer subventricular zone (OSVZ) arise from vRGs through a process resembling epithelial-mesenchymal transition. oRGs are very rare in mice (Shitamukai et al., 2011; Wang et al., 2011) and are thought to have significantly contributed to the evolutionary expansion of the neocortex in primates and carnivores (Lui et al., 2011). Intermediate progenitor cells (IPCs) are multipolar transit amplifying cells generated by radial glia that directly produce most of the excitatory neurons in the neocortex (Hansen et al., 2010; Kowalczyk et al., 2009; Noctor et al., 2004). Newborn excitatory neurons migrate along radial glial fibers from the germinal zones (GZ) to the cortical plate (CP), eventually forming six layers (Rakic, 2000), while inhibitory interneurons originate predominantly from the ganglionic eminences and migrate tangentially into the neocortex (Anderson et al., 2001; Hansen et al., 2013). Neurotransmitters can induce Ca2+ transients and influence progenitor cell proliferation, neuronal migration, and differentiation in rodents (Le Magueresse and Monyer, 2013; LoTurco et al., 1995; Weissman et al., 2004). Progenitor cells, maturing excitatory neurons and interneurons, endothelial cells, and the cerebrospinal fluid (CSF) are all potential sources of neurotransmitters (Bonnin et al., 2011; Le Magueresse and Monyer, 2013; Lehtinen et al., 2011; Li et al., 2018; Manent et al., 2005; Reillo et al., 2017; Weissman et al., 2004). Additionally, axons from extracortical regions such as the diencephalon and brain stem innervate the cortex and affect cortical development (Bonetti and Surace, 2010; Chen and Kriegstein, 2015; Duque et al., 2016; Reillo et al., 2017). Protracted development in humans may suggest a greater role of neurotransmitter signaling in regulating cellular processes. Differences in activity-dependent gene expression as well as differences in gene expression in serotonergic and cholinergic systems between different mammalian species indicate that neurotransmitter signaling has likely impacted cortical evolution (Ataman et al., 2016; Sousa et al., 2017).
The diverse cell types in the developing neocortex likely respond differently to neurotransmitter signals. In the developing human neocortex, single-cell RNA sequencing (scRNA-seq) has revealed molecular signatures of distinct cell types (Nowakowski et al., 2017; Pollen et al., 2015; Thomsen et al., 2016). Neuronal identity, however, is further determined by properties beyond gene expression, including morphology, connectivity, and physiology. Therefore, large-scale analyses of neuronal cell diversity will require multimodal characterization of cells that overlay physiological properties and transcriptomic signatures at the single-cell level. This has proven challenging due to technical difficulties (Regev et al., 2017; Zeng and Sanes, 2017). However, combining scRNA-seq with patch-clamp recording has enabled the integration of intrinsic physiological and molecular properties in relatively few cells (Cadwell et al., 2016; Chen et al., 2016; Fuzik et al., 2016).
We asked whether molecularly distinct cell types in the developing human neocortex also display unique physiological properties. To link neurotransmitter signaling to molecular profiles, we developed a multimodal single-cell approach that allows agonist dosing, response monitoring, and subsequent transcriptomic analysis in the same single cells. Using intracellular calcium elevations as a proxy for physiological responsiveness to a series of neurotransmitter receptor agonists, we found that not only do physiological response properties correlate with molecular identity, but they also capture additional aspects of diversity. Interestingly, inhibition of a serotonergic receptor expressed in human, but not mouse, radial glia disrupts the radial glial scaffold. Together, these findings highlight the cell-type specificity and functional importance of early neurotransmitter-mediated signaling events in the developing human neocortex and provide an avenue for analyzing early active signaling networks.
RESULTS
Neurotransmitter Receptor Expression Changes with Differentiation
To identify cell-type-specific neurotransmitter receptor systems in the developing human neocortex, we first analyzed the expression of neurotransmitter receptor genes in a scRNA-seq dataset (Table S1) (Nowakowski et al., 2017). While not all receptors are likely to be detectable with scRNA-seq, we did find receptor gene transcripts enriched in specific cell populations of the excitatory lineage using pseudoage analysis (Figure 1A). Two N-methyl-D-aspartic acid (NMDA) receptor subunits had opposing expression patterns: GRIN2A was highly expressed in progenitors, while GRIN2B expression was only detected in neurons (Figure 1B), consistent with findings from rodent neural stem cells (Muth-Köhne et al., 2010a). The transition from GRIN2A to GRIN2B expression yields channels with faster kinetics and less sensitivity and is important for developmental regulation of synaptic plasticity and the maturation of neuronal circuits (Flint et al., 1997; Muth-Köhne et al., 2010b; Paoletti et al., 2013). Similarly, we detected a developmental switch in gamma-amino butyric acid (GABA) A receptor subunit expression. GABRB1 and GABRG1 subunits were expressed in progenitor cells, a subunit combination yielding higher affinity, non-desensitizing GABA responses (Ma and Barker, 1995; Owens et al., 1999), while characteristic synaptic subunits GABRB3 and GABRG2 were expressed in neurons (Figure 1C) (Luscher et al., 2011). The purinergic P2RY1 receptor was highly expressed in progenitor cells, including radial glia and IPCs, and decreased sharply in neurons (Figure 1D), consistent with reports in rodents (Liu et al., 2008; Weissman et al., 2004). α-Amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor subunit GRIA1-3 expression increased as cells differentiated (Figure 1E), in line with increased chromatin accessibility in the promoter regions of GRIA1–4 in the CP compared to the GZ (de la Torre-Ubieta et al., 2018). We find that neurotransmitter receptor subunits of the same receptor type (e.g., different NMDA receptor subunits) have different and sometimes opposing expression trajectories in the human neocortex that correlate with the development of distinct physiological properties.
Figure 1. Neurotransmitter Receptor Expression during Neurogenesis in the Developing Human Neocortex.
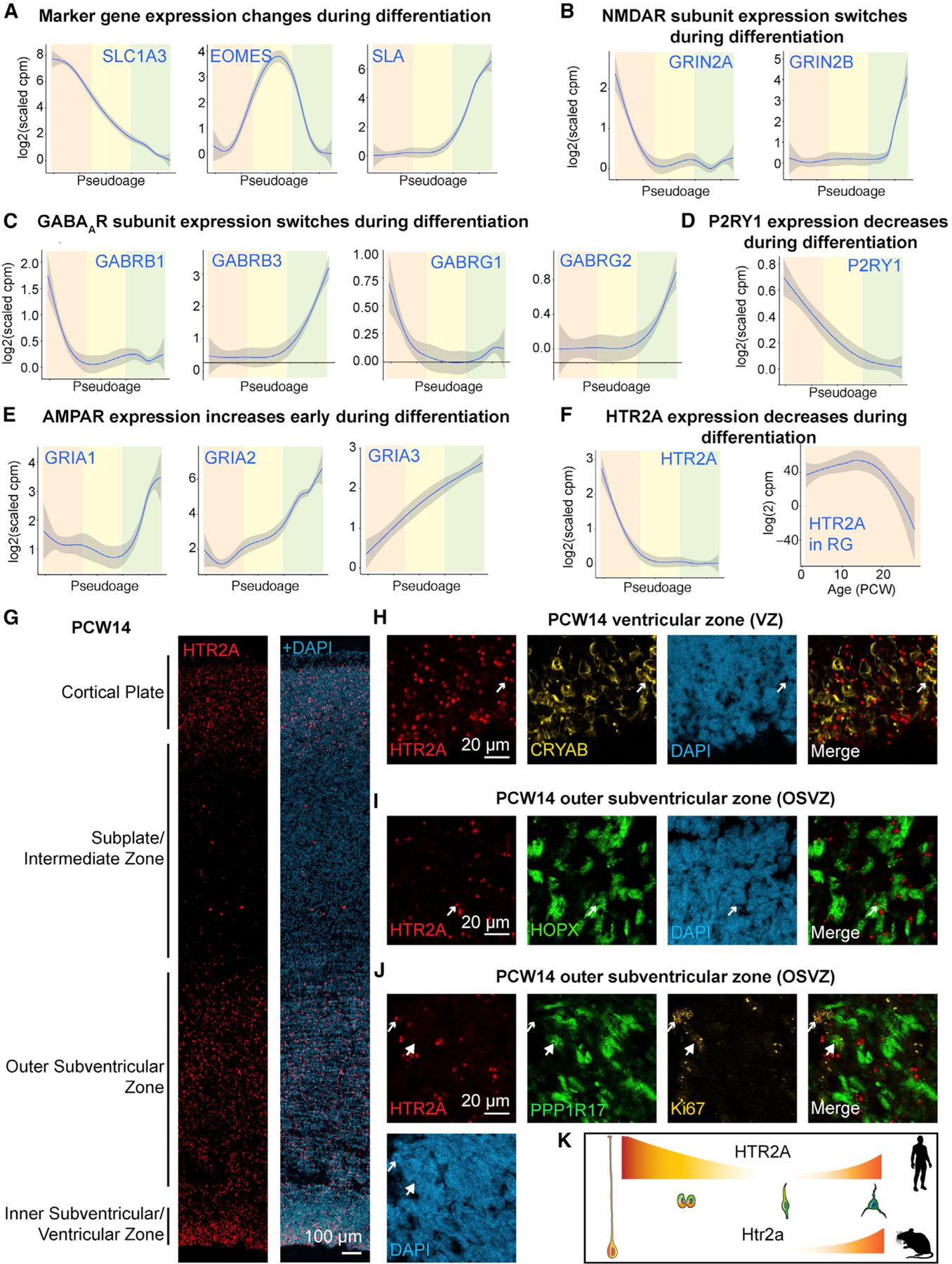
(A–F) Pseudoage analysis of the excitatory neuronal lineage (A) Marker gene expression of radial glia cells (SLC1A3, orange shading), intermediate progenitor cells (EOMES, yellow shading), and excitatory neurons (SLA, green shading). Expression trajectories of NMDA receptor (NMDAR) subunits (B), and GABAA receptor subunits (C), the purinergic receptor P2RY1 (D), AMPA receptor (AMPAR) subunits (E), and the serotonergic receptor HTR2A (F, left). Correlation of HTR2A expression values with age (PCW) in radial glia cells (F, right).
(G–J) Enrichment of serotonergic receptor HTR2A in specific progenitor cell types in the developing human neocortex using single molecule in situ hybridization (smFISH). (G) smFISH for HTR2A was combined with immunohistochemistry against CRYAB (H), HOPX (I), and PPP1R17 and Ki67 (J). Arrows indicate colocalization, large arrowheads lack of colocalization.
(K) Schematic showing HTR2A pseudoage expression trajectory in human and mouse.
The Serotonergic Receptor HTR2A Is Enriched in Outer Radial Glial Cells
We found that HTR2A, a receptor not previously reported in progenitor cells (Vitalis and Verney, 2017), was highly expressed in first and second trimester human radial glia (Figure 1F). The expression of HTR2A correlated with genes important for progenitor cell proliferation including CTNNA1 and SMARCC2 (Table S1) (Tuoc et al., 2013; Lien et al., 2006). Because vRG and oRG cells reside in distinct progenitor regions, we aimed to identify expression of HTR2A in specific laminae by single-molecule fluorescent in situ hybridization (smFISH). Starting at postconceptional week 8 (PCW8), HTR2A was strongly expressed in the ventricular and subventricular zones (Figures 1G, S1A, and S1C). Additionally, HTR2A was expressed in the CP at midgestation (Figures 1G and S1C). By performing immunohistochemistry after smFISH, we found that HTR2A colocalized with the vRG marker CRYAB (Figure 1H) and was enriched in cells positive for the oRG marker HOPX (Figure 1I and S1D). HTR2A also colocalized with the mitotic marker Ki67 (Figure 1J) but only rarely with the IPC marker PPP1R17 (Figure 1J). The Htr2a receptor is not expressed during neurogenesis in developing mouse neocortex (Figure S1E), indicating the potential evolutionary importance of this receptor in human radial glia. In contrast, P2RY1, which is expressed in progenitors in rodents (Liu et al., 2008; Weissman et al., 2004), was also expressed in the human GZ (Figures S1A–S1C), but not enriched in oRGs (Figure S1D). These findings indicate that, in addition to displaying unique molecular signatures (Pollen et al., 2015; Thomsen et al., 2016), human oRG cells may also be responsive to serotonergic stimulation.
Slice Electrophysiology Shows Agonist-Induced Currents in Specific Cell Types
We next tested whether receptor mRNAs expressed by neural progenitor cells and immature neurons are translated into functional receptors. Based on transcriptomic (Figures 1 and S1) and physiological studies (Flint et al., 1999; Haydar et al., 2000; Liu et al., 2008; LoTurco et al., 1995; Ma et al., 2000; Weissman et al., 2004), we compiled a panel of neurotransmitter receptor agonists active during brain development that included two specific glutamate receptor agonists, NMDA and AMPA, as well as GABA, acetylcholine (ACh), and purinergic P2RY1 receptor agonist methylthioadenosine diphosphate (MeSADP). Additionally, we included TCB-2, a specific agonist for HTR2A, whose activation induces membrane currents and intracellular Ca2+ elevations in pyramidal neurons and astrocytes (Aghajanian and Marek, 1999; Hagberg et al., 1998). We used patch-clamp recordings in second trimester acute cortical slices to measure currents induced by these neurotransmitter receptor agonists. Cell types were identified based on their anatomical location, electrophysiological properties, morphology, and marker gene expression (Figures 2A, 2B, and S2A–S2C). We found a diversity of cellular responses to agonist application. Consistent with Mg2+ block of the NMDA receptor at hyperpolarized membrane potentials, NMDA only induced currents at positive membrane potentials in neurons (Figure 2C). However, NMDA induced currents at resting membrane potentials in radial glia (Figure 2C), consistent with Mg2+-insensitive NMDA currents recorded in immature neurons, such as subplate (SP) neurons in the developing rodent brain (Hanganu et al., 2002), as well as glial cells (Káradóttir et al., 2005; Lalo et al., 2006). AMPA and GABA induced currents in all cell types analyzed (Figure 2C). TCB-2 induced currents in vRGs and oRGs, while MeSADP induced hyperpolarization only in vRGs (Figure 2C). Receptor specificity was confirmed by antagonists that selectively blocked agonist-induced currents (Figures S2C–S2I). GABA-induced currents in vRGs were larger than in oRGs, which we hypothesized to be due to gap junction coupling among vRGs (Bittman et al., 1997). Indeed, application of gap junction blockers significantly reduced the GABA current (Figure S2I). Together, these recordings show that neurotransmitter receptors are functionally diverse in a cell-type-specific pattern in the developing human cortex.
Figure 2. Patch-Clamp Recordings in Cortical Slices Show In Situ Receptor Functionality.
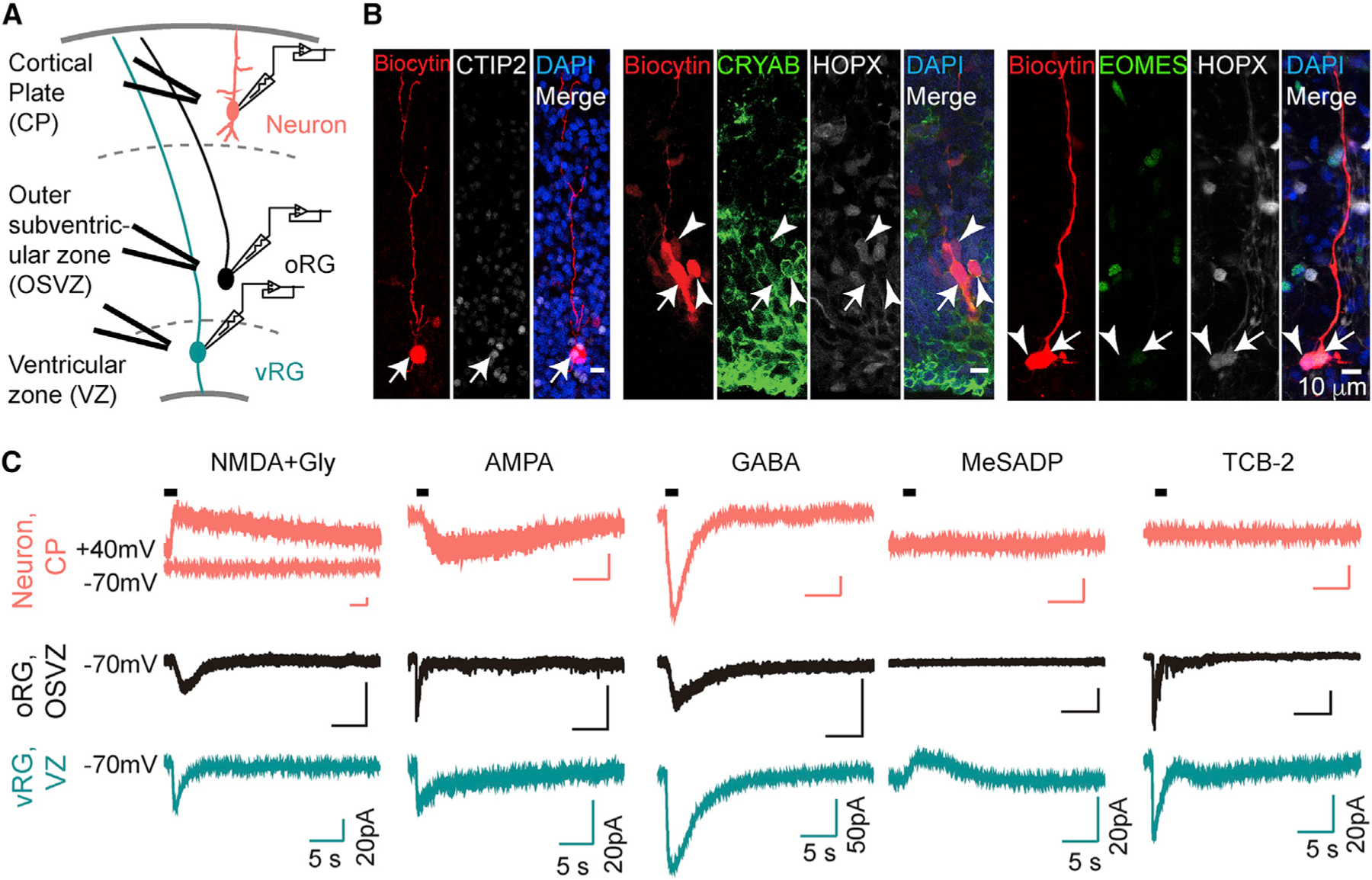
(A) Schematic diagram shows whole-cell patch clamp recordings with local application of agonists in slices.
(B) Cells were loaded with biocytin after recording and immunostained with cell-type-specific markers (arrows). Note that the vRG is coupled to both a vRG and an oRG cell (arrowheads, middle), while the oRG is coupled to another oRG cell (arrowheads, right).
(C) Example traces of currents in response to agonists. Neurons (red) were held at −60 mV except for NMDA + glycine (held at +40 mV). All oRGs (black) and vRGs (green) were recorded at a holding potential of −70 mV.
Ca2+ Imaging Confirms the Diversity of Neurotransmitter Responses in the Developing Human Neocortex and Helps Map Temporal Trajectories
To study response patterns to multiple agonists in a broad range of cell types, we microdissected the GZ or CP and SP of midgestation cortical tissue (PCW14–22), loaded dissociated cells with a Ca2+ indicator, and recorded fluorescence changes in response to sequential agonist applications (Figures 3A–3C). We generated dose-response curves for each agonist to determine supersaturating agonist concentrations that reliably induce responses with little desensitization (Figures S3A–S3C). The order of agonist application did not affect the probability of response to specific agonists (Figure S3D). The majority of cells from both CP and SP and GZ responded to at least one of the six agonists tested (91.5% ± 1.6% in GZ, 99.8% ± 0.08% in CP and SP). We found significantly more responses to NMDA, AMPA, ACh, and GABA in cells originating from the CP and SP than GZ (Figures 3B–3D). Consistent with receptor expression observed by smFISH (Figures 1 and S1), GZ cells responded preferentially to MeSADP (Figures 3B–3D), while cells from both the GZ and CP and SP responded to TCB-2 (Figures 3B–3D). Responses were dramatically different during the first and early second trimester (PCW8–12) compared to PCW14–22, with more than 90% of cells responding to TCB-2 and few cells responding to other agonists (Figure 3E). Interestingly, at an equivalent stage in mouse cortical development, cells did not respond to TCB-2 (Figures S3E and S3F), consistent with in situ RNA expression analysis. Responses to MeSADP were absent at PCW8–12 (Figure 3E), mirroring the age-dependent effect of purinergic regulation of progenitor proliferation in the rat (Weissman et al., 2004). Responses to agonists did not change through midgestation (PCW14–22) (Figure 3F), with the exception of GABA responses in the CP and SP, which decreased with age (Figure 3G). Hierarchical clustering of the response patterns of cells from PCW8–22 revealed 11 physiologically distinct response types (Figure 3H). Combinatorial responses at PCW8–12 were largely distinct from those at later stages, and at PCW14–22, clusters mostly differed by anatomical source (Figure 3H). Thus, physiological response patterns diversify during development and may reflect a range of cell-type-specific response patterns as cells differentiate.
Figure 3. Ca2+ Imaging Reveals Lamina-Specific Responses to Neurotransmitter Receptor Agonists.
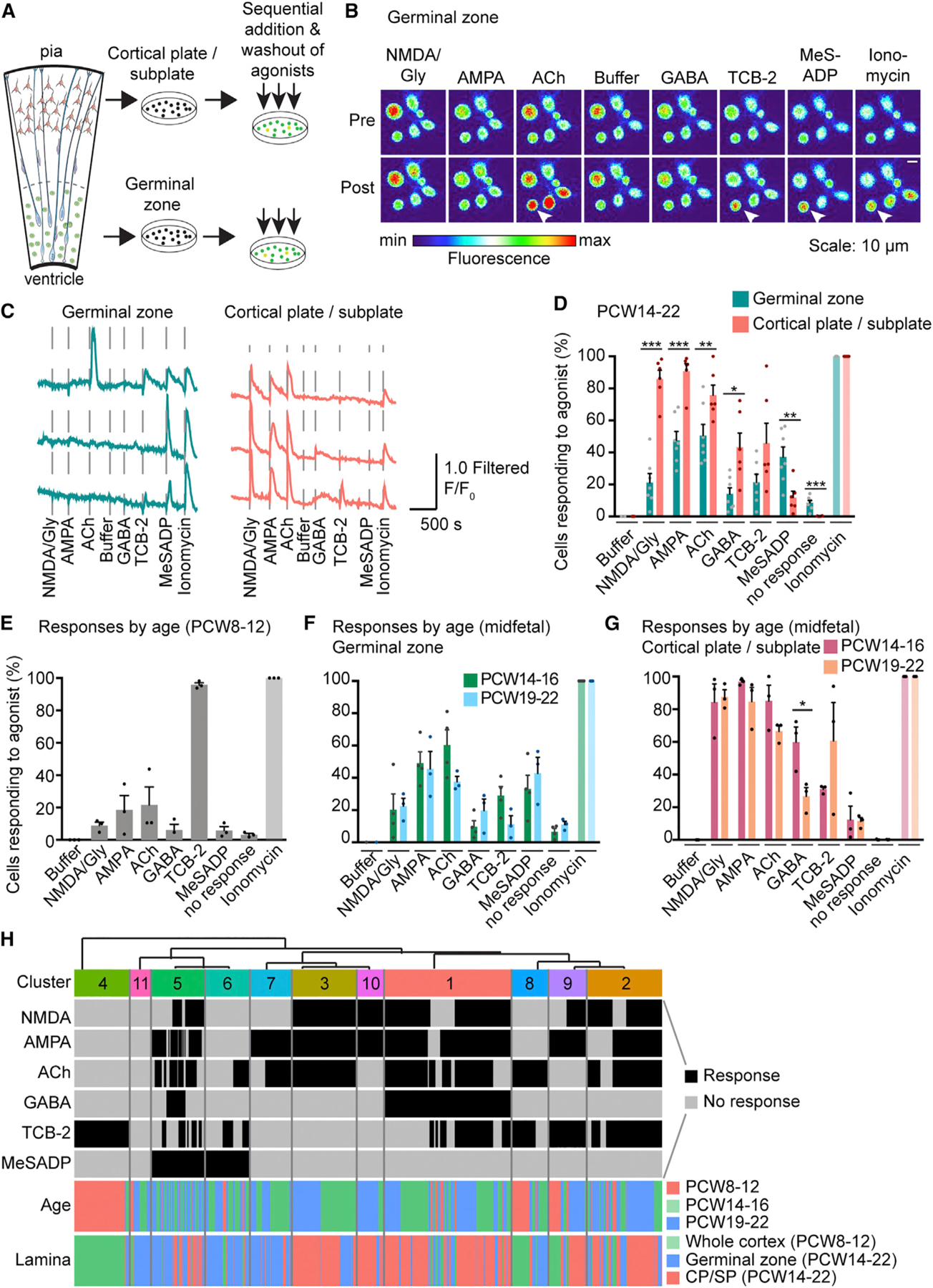
(A) Schematic of the experimental workflow for Ca2+ imaging of dissociated cells.
(B) Images of [Ca2+]i (pseudocolored) pre and post agonist application in cells dissociated from the GZ. Arrowheads indicate Ca2+ elevations of one representative cell in response to specific agonists.
(C) Traces of [Ca2+]i changes in response to agonist dosing.
(D) Summarized results of the percentage of cells showing Ca2+ elevation upon agonist stimulation in GZ and CP and SP, respectively (PCW14–22, binary quantification). N = 10, n = 2,613 cells.
(E–G) Summarized results of Ca2+ responses to the panel of neurotransmitter receptor agonists at PCW8–12 (N = 3, n = 608 cells) (E), and in the GZ (F) and in the CP and SP (G) by age range in midfetal development. No response indicates cells that only responded to ionomycin. A significant difference in GABA responses between PCW14–16 and PCW19–22 in CP and SP was found using multiple t tests (Holm-Sidak method). Data are represented as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, dots indicate biological replicates.
(H) Single-cell Ca2+ responses to different agonists at different stages of development were co-clustered to reveal 11 distinct physiological clusters (n = 3,221 cells). GZ, germinal zone. SP, subplate.
Single-Cell Approach Links Physiological Response Patterns to Molecularly Defined Cell Types
To map physiological response patterns onto cellular diversity, we developed an approach using the microfluidic Polaris device (Ramalingam et al., 2016) to couple the recording of Ca2+ responses to transcriptomic analysis of the same single cells (Figure 4A). Cells dissociated from the GZ or CP and SP of mid-gestational cortical tissue were loaded with a Ca2+ dye, and single cells were captured on microfluidic chips. Cells were successively dosed with six agonists at 10-min intervals, while intracellular Ca2+ elevations were simultaneously monitored. Cells were lysed within 1 h of final agonist application (Figure 4A). The Ca2+ responses observed in dissociated cells on the rig were recapitulated in single cells captured on the microfluidic chips (Figures 3, 4B, 4C, and S4A). Physiologically characterized single cells were processed for scRNA-seq and sequenced at approximately four million reads/cell (Table S3). Unbiased clustering of 476 cells by gene expression from 14 samples ranging from PCW14 to PCW22 revealed ten transcriptomic clusters (Figures 4D and S4B; Table S4). Cells clustered mostly according to their anatomical source, which was not affected by order of agonist application (Figure S4C). Importantly, cells that were not dosed co-clustered with dosed cells, indicating that major transcriptome changes were not induced by agonist exposure during this short period (Figure S4C). We assigned transcriptomic clusters to known cell types based on marker gene expression (Figure S4B; Table S4). One cluster expressed high levels of mitotic genes, as well as radial glia and IPC marker genes, and was thus classified “Dividing Progenitors.” We next mapped Ca2+ responses onto molecularly defined clusters (Figure 4E). Progenitor cells responded predominantly to TCB-2 and MeSADP, while excitatory neurons and interneurons responded predominantly to glutamatergic and GABAergic agonists (Figure 4E). Interestingly, ACh and TCB-2 induced responses preferentially in upper layer excitatory neurons (Figure 4E). AMPA induced responses in medial ganglionic eminence (MGE)-derived interneurons and excitatory neurons, while NMDA responses predominated in excitatory neurons (Figure 4E). Caudal ganglionic eminence (CGE)-derived interneurons displayed very few responses to any of the six agonists (Figure 4E). In order to obtain a finer classification of cells, we co-clustered cells captured on Polaris microfluidic chips with 3,528 neocortical cells captured on C1 microfluidic chips (Nowakowski et al., 2017). Co-clustering revealed 24 transcriptomic clusters, most containing cells captured on both systems (Figure 4F; Table S4). We found that while radial glia responded to TCB-2 with a frequency of 42%–58%, 100% of dividing oRGs responded to TCB-2 (Figure 4G). Differential gene expression analysis followed by Gene Ontology (GO) analysis between dividing oRGs that were dosed with TCB-2 on the Polaris device and those that were not dosed with TCB-2 and captured on the C1 device revealed no significant differences, confirming that the enrichment of TCB-2 responsive cells in dividing oRGs was not due to induction of cell-cycle genes by TCB-2. Because no significant enrichment of biological processes was associated with dosed compared to non-dosed dividing oRGs (Table S4), we concluded that mitotic oRGs respond preferentially to TCB-2. All progenitors responded to MeSADP stimulation (51%–83%), but fewer than 20% of IPCs responded to TCB-2 stimulation (Figure 4G). In summary, our multimodal approach synthesizes two distinct measurements of cellular identity—physiological responses and transcriptomic profiles at the single-cell level—that together provide evidence for cell-type- and cell-state-specific neurotransmitter signaling.
Figure 4. Mapping Ca2+ Responses onto Cell Types at the Single-Cell Level.
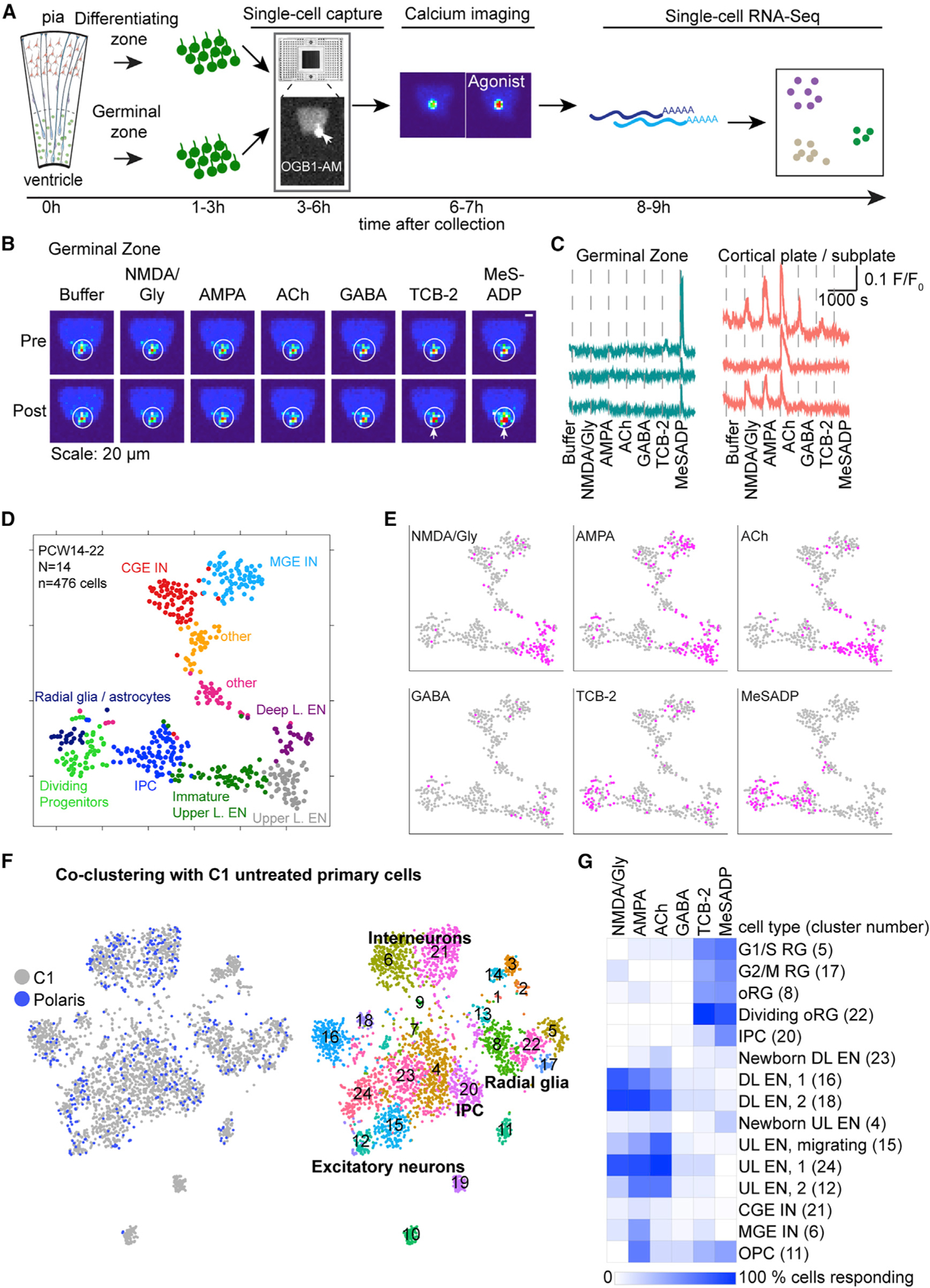
(A) Schematic of experimental workflow using the Polaris microfluidic system.
(B) Representative images of [Ca2+]i changes (pseudocolored) pre and post agonist application. Arrows indicate cell responding to agonist with Ca2+ elevation.
(C) Representative traces of Ca2+ changes in response to agonists.
(D) Unbiased clustering of cells into 10 molecular clusters based on their transcriptomes represented in t-distributed stochastic neighbor embedding (tSNE) space.
(E) Feature plots show responses to agonists (pink).
(F) Feature plots showing co-clustering of cells captured on Polaris or C1 microfluidic chips revealing 24 transcriptomic clusters.
(G) Heatmap shows the percentage of cells in each C1-Polaris transcriptomic cluster (row, cluster number indicated in brackets) responding to agonists (columns). Only clusters with more than 5 Polaris cells were included. RG, radial glia; L, layer; CGE, caudal ganglionic eminence; MGE, medial ganglionic eminence; UL, upper layer; DL, deep layer; OPC, oligodendrocyte progenitor cell; EN, excitatory neuron; IN, interneuron.
HTR2A Receptor Activation Controls Radial Fiber Morphology
Given our finding of HTR2A receptor responses in dividing oRGs (Figure 4G), we asked whether HTR2A stimulation promotes progenitor proliferation. Thymidine analog incorporation assays in organotypic slice cultures revealed that stimulation of HTR2A by TCB-2 or inhibition with the specific antagonist (EMD 281014) did not affect proliferation (Figures 5A and 5B). Thus, although TCB-2 responses are enriched in cycling cells, activation of this specific serotonin receptor does not appear to play a role in cell-cycle progression. Htr2a stimulation affects dendritic growth in mouse neurons (Ohtani et al., 2014; Roppongi et al., 2013). We therefore tested whether HTR2A stimulation in radial glia could affect their morphology. We applied the antagonist EMD281014 to cortical organotypic slice cultures for 72 h and observed a disruption of the radial glial scaffold (Figures 5C–5G). Additionally, individual fibers labeled with GFP through viral infection showed aberrant morphology in the presence of EMD281014 (Figure 5C). Co-staining with the oRG marker HOPX revealed that oRG cells in the OSVZ were labeled with the CMV-GFP virus (Figure 5E). Quantification of fiber length in the OSVZ showed a significant reduction in fiber length in the presence of EMD281014 (Figures 5F and 5G; Videos S1 and S2). Taken together, these experiments reveal that serotonergic stimulation has an effect on radial fiber morphology of human oRG cells.
Figure 5. Signaling through HTR2A Regulates Radial Glia Morphology.
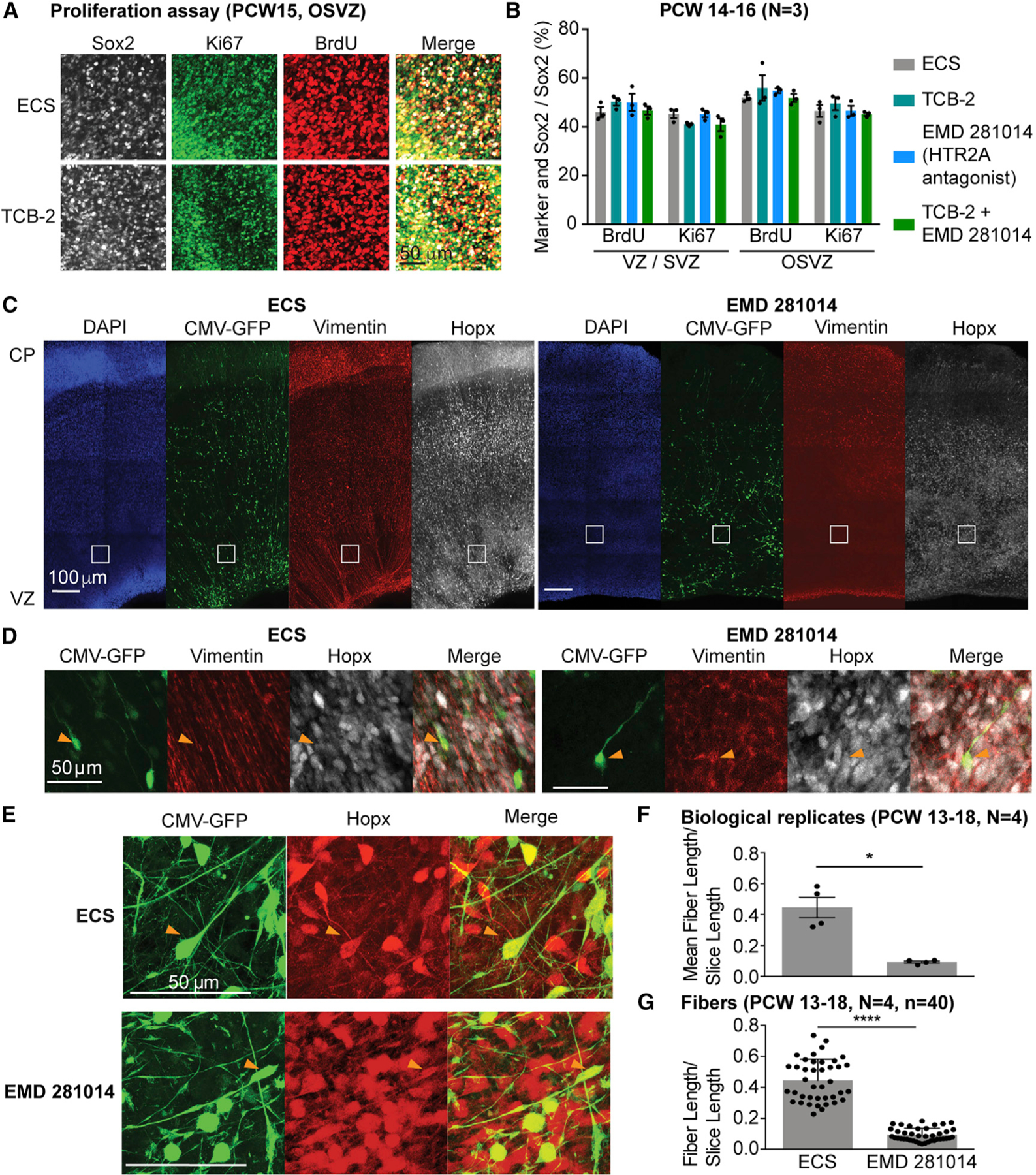
(A) BrdU incorporation and Ki67-positive proliferating cells are not changed after HTR2A receptor activation or inhibition in cortical slices.
(B) Quantification of proliferation assay shown in (A).
(C) Treatment of cortical slices with HTR2A antagonist EMD 281014 leads to loss of radial fiber organization as visualized by vimentin staining and GFP-staining of cells infected with a CMV-GFP adenovirus.
(D) Higher magnification of inserts shown in (C) highlighting the lack of radial vimentin fiber organization after antagonist treatment.
(E) CMV-GFP-positive fibers in OSVZ are oRGs based on co-staining with Hopx.
(F and G) Quantification of fiber length reveals a significant reduction in average fiber length normalized by slice length (F) and fiber length normalized by slice length (G) in the EMD 281014-treated cortical slices compared to the control using the Wilcoxon rank-sum test. Data are represented as mean ± SEM. *p < 0.05, ****p < 0.0001.
Physiological Response Trajectories in Upper Layer Neuron Differentiation
Based on the cell-type-specific responses, we hypothesized that physiological and molecular properties might change in concert as cells differentiate and mature. Using transcriptomic data, we analyzed physiological responses in the upper layer excitatory neuron lineage (Figure 6A). As cells became postmitotic, a dramatic shift in responsiveness occurred: while progenitors responded to TCB-2 and MeSADP, newborn neurons responded to ACh. As neurons matured, they began to respond to AMPA, concomitant with upregulation of neuronal genes such as SYT4, a synaptic protein (Figure 6A). Subsequently, cells became responsive to NMDA, which coincided with expression of GRIN2B, an NMDA receptor subunit, and a subset of more mature upper layer neurons responded to TCB-2 and GABA (Figure 6A). We also analyzed whether the detection of receptor mRNA coincided with responses to specific agonists in the same cell. We found a significant correlation of receptor detection and agonist response in specific cases, such as GRIA1, GRIA2, and GRIA4 expression correlating with AMPA responses, GRIN2B expression coinciding with NMDA responses, and GABA responses coinciding with GABBR2 and GABRG2 expression (Figure S5A; Table S5). Despite a sequencing depth of four million reads, we did not detect mRNA expression for all receptors in cells displaying responses to specific agonists, highlighting the need to assay rare transcript expression with more sensitive methods such as smFISH (Figure 1) (Moffitt et al., 2018; Torre et al., 2018; Zhu et al., 2018) or gene product function (Figure 4).
Figure 6. Physiological Response Patterns Predict Cell Identity.
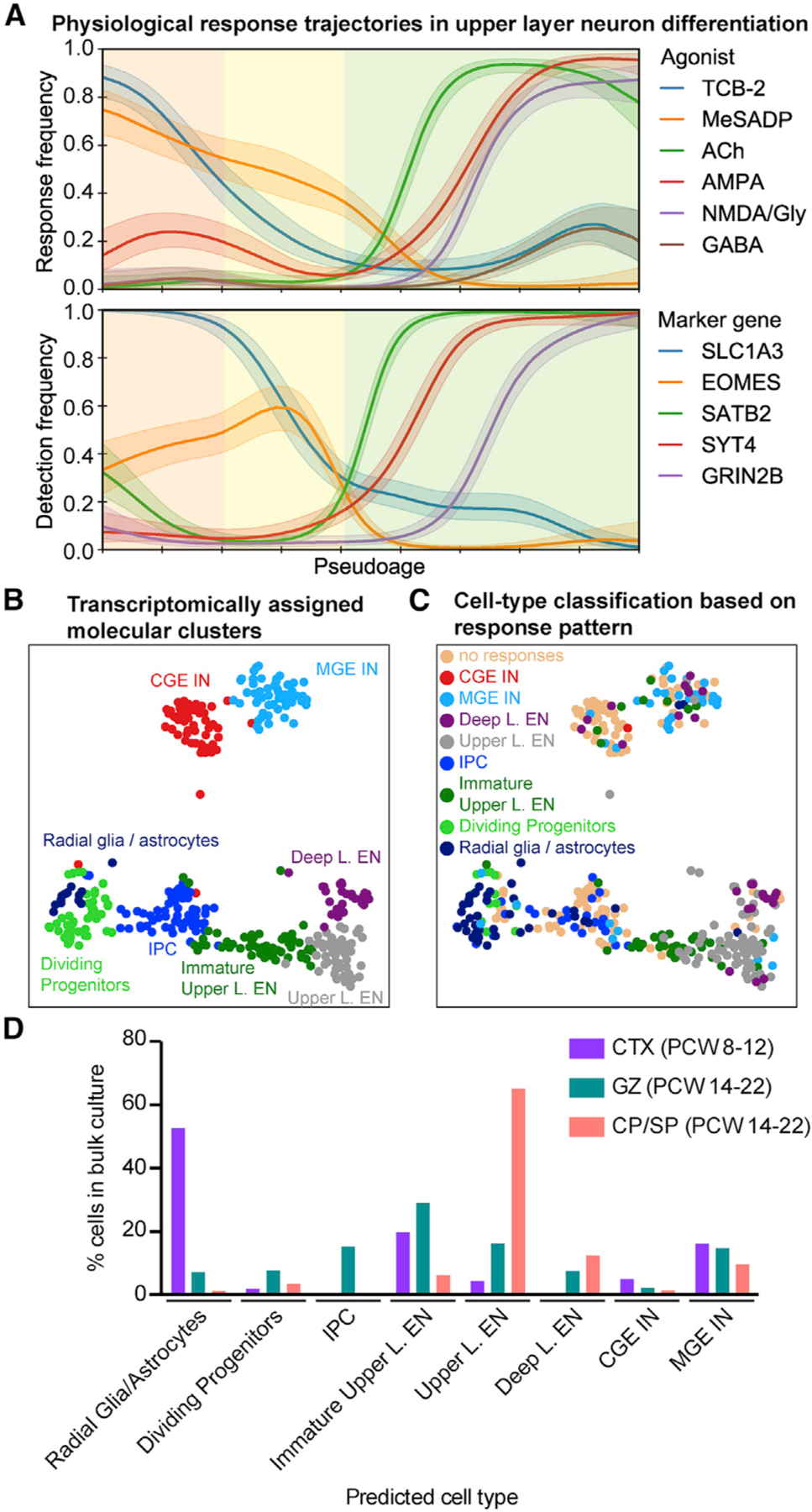
(A) The frequency of responses to agonist application varies as a function of pseudoage (top). See detection frequencies of marker genes (bottom). Major cell types are highlighted by shading: radial glia cells, orange; intermediate progenitor cells, yellow; excitatory neurons, green.
(B) Transcriptomically defined cell types, excluding clusters “other,” which are outliers.
(C) Predicted cell-type assignments of a classifier trained on the physiological response patterns in a multimodal dataset, relative to their transcriptomically defined cell types (B).
(D) Predicted cell type based on responses to agonists of cells analyzed in bulk imaging experiments (Figures 3E and 3F). Percentage of cells relative to the total number of cells responding to at least one agonist is shown. CTX, neocortex.
Physiological Response Patterns Predict Cell Identities
We next leveraged our high-dimensional dataset to integrate combinatorial responses to the six agonists with transcriptomic profiling. Using a machine-learning approach, we trained a naive Bayes classifier to recognize response patterns associated with transcriptomically distinct cell types in our multimodal dataset (Figure 4). We found that response patterns alone were highly predictive of cell identity (Figures 6B, 6C, and S5B; Table S5). We then validated the classifier on an independent dataset using the cells recorded in dissociated culture (Figure 3) and found that cell-type assignments matched their region of origin (Figure 6D). While cell-type assignments based on transcriptomes and physiological response patterns were mostly correlated, some differences in classification were found (Figures 6 and S5B). For example, several cells identified as immature upper layer excitatory neurons based on scRNA-seq displayed functional responses of mature upper layer excitatory neurons, illustrating how physiological profiling can help refine transcriptomic clustering of cells into more phenotypically relevant subtypes (Regev et al., 2017).
Physiological Response Patterns Reveal Heterogeneity within Molecularly Defined Cell Types
We next sought to study combinatorial responses to the six agonists in an unbiased manner using an unsupervised clustering approach. In order to achieve higher confidence in the physiological response patterns, we combined physiological response patterns in 2,613 cells recorded in culture (Figure 3D) and 347 cells recorded on microfluidic chips (Figure 4). Hierarchical clustering revealed 12 distinct physiological response types (P) (Figure 7A). The same physiological profiles that we observed in culture were recapitulated on microfluidic chips (Figure 7A), allowing us to use the combined mRNA and response profiling as a “Rosetta Stone” to translate between physiological profiling and gene expression datasets. Differential gene expression analysis between physiological response types revealed cell-type-specific gene expression profiles for P-10 (progenitors), P-5 and P-7 (radial glia), and P-2 (upper layer excitatory neurons) (Table S6), in agreement with the correlation of physiological response profiles and molecular clusters (Figure 6B). We next mapped response profiles onto transcriptomic cell clusters (Figure 4D) and found a strong correlation (p < 0.05, χ2 test; Figures 7B and S6C). Within molecular clusters, cells often fell into several different physiological types (Figure 7B). We hypothesized that functional heterogeneity may relate to cell state or maturation not resolved by unbiased clustering of cells according to their transcriptomes. To understand which molecular features underlie this physiological heterogeneity, we analyzed genes differentially expressed between the two major physiological types (i.e., combination of responses to the six different agonists) in a molecularly defined cell type (Table S6). We found that while cell-type-defining genes were highly expressed in all cells of a certain molecular cluster, other genes were enriched in cells with a specific response type (Figure 7C; Table S6). Importantly, very few immediate early genes, which are transcribed within 1 h following a stimulus (Ataman et al., 2016), were found to be differentially expressed between different physiological types, indicating that transcriptomic differences are unlikely to be induced by agonists but rather reflect cell-state specificity. We performed GO analysis of differentially expressed genes to gain insight into processes that differ between physiologically distinct cells. Genes differentially expressed between the two main response types, P-1 and P-2, in upper layer excitatory neurons were enriched for GO terms related to differentiation, posttranscriptional regulation of gene expression, regulation of RNA, and protein metabolic processes (Figure 7D). Among the differentially expressed genes, we observed regulators of translation initiation (EIF3L, EIF4E, EIF4H) and genes involved in neurotransmitter receptor trafficking (ARHGEF9) (Table S6). In “Dividing Progenitors,” genes differentially expressed between P-7 and P-5 were enriched for terms related to cell cycle (Figure 7E). Many genes enriched in P-7 were known regulators of the cell cycle, such as cyclins (CCNB1, CCNB2, CCNE2) (Table S6) corroborating TCB-2 responses are cell-cycle dependent (Figure 4G). Genes differentially expressed between P-6 and P-10, the two main response types in IPCs, were related to posttranscriptional regulation of gene expression (Figure 7F), possibly reflecting transcriptional priming and translational repression in progenitor cells (Zahr et al., 2018). Additionally, genes related to neurogenesis and differentiation were differentially expressed in IPCs with distinct responses (Figure 7F). Thus, lineage progression and posttranscriptional mechanisms of gene expression underlie physiological responsivity.
Figure 7. Physiological Heterogeneity Exists within Transcriptomically Defined Cell Types.
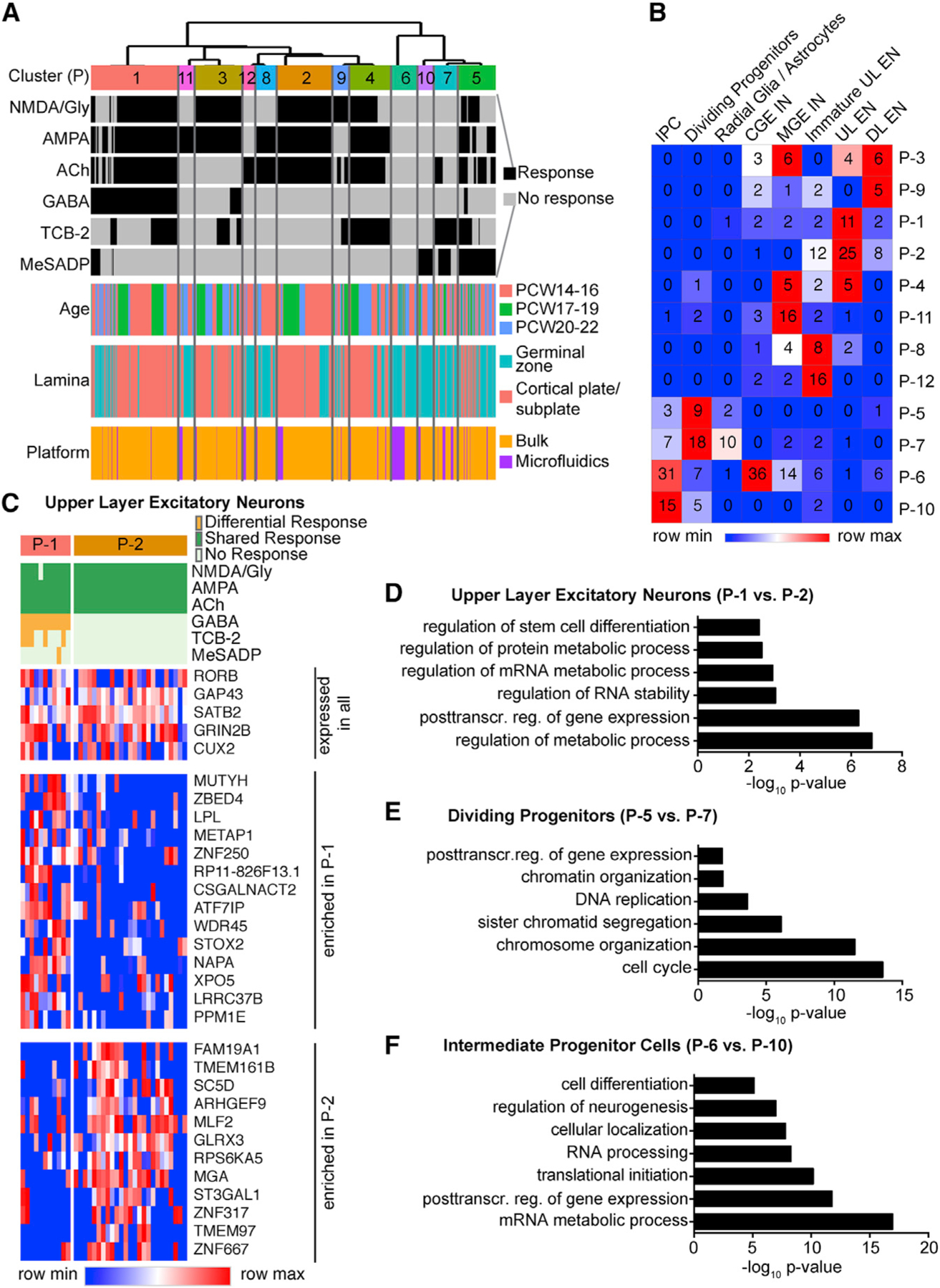
(A) Single-cell Ca2+ responses to different agonists analyzed on microfluidic chips and on physiological rig were co-clustered to reveal physiological types (P).
(B) Heatmap showing the relative responses to each neurotransmitter (columns) in the different physiological types (P, rows). Numbers in heatmap indicate number of cells in each cluster analyzed on microfluidic chips at the single-cell level.
(C) Heatmap highlighting representative genes that are expressed in all mature upper layer excitatory neurons or enriched in physiological cluster P-1 or P-2.
(D–F) Differential gene expression between distinct physiological types within one cell type. Top Gene Ontology terms enriched in differentially expressed genes between physiological types P-1 and P-2 in mature upper layer excitatory neurons (D), between physiological types P-5 and P-7 in dividing progenitors (E), and between physiological types P-6 and P-10 in intermediate progenitor cells (F).
DISCUSSION
Our multimodal single-cell approach reveals a diverse cellular repertoire of neurotransmitter responses in second trimester human neocortex that can be resolved into cell-type-specific response patterns that change as cells differentiate and mature. By linking physiological and molecular features in single cells, we found that cell-intrinsic physiological properties are predictive of molecular identity derived from unbiased clustering of scRNA-seq data, but we also identified physiological heterogeneity within transcriptomically defined cell types. These findings reveal a significant degree of physiological response specialization at stages when synapses are just starting to form, highlighting the potential for neurotransmitter-mediated signaling to influence very early stages of human cortical development.
Classical Neurotransmitters Induce Cell-type-Specific Responses in Maturing Neurons
At the early stages studied here, when synapses are mostly restricted to the SP and marginal zone (Molliver et al., 1973; Moore et al., 2011; Ohtaka-Maruyama et al., 2018; Sarnat et al., 2010), neurotransmitters may be released through non-canonical mechanisms (Manent et al., 2005), such as transporters (Unichenko et al., 2015) or connexin hemichannels (Moore et al., 2014). Additionally, neurotransmitters may be present in the CSF (Bonnin et al., 2011; Lehtinen et al., 2011; Sundström et al., 1993; Toda et al., 2013) and released from endothelial cells (Li et al., 2018). There are thus multiple endogenous neurotransmitter sources during stages of neurogenesis and differentiation.
We find that newborn excitatory neurons have a response profile that is very distinct from that of progenitor cells. Newborn human neurons respond to ACh (Figure 6A), which promotes neuronal maturation and neurite development in mouse (Bruel-Jungerman et al., 2011). Subsequently, neurons become responsive to glutamatergic agonists (Figures 6A and S3F). Excitatory neurons respond to both NMDA and AMPA, while interneurons respond most strongly to AMPA stimulation (Figure 4) (Yuzwa et al., 2016). Radial migration of excitatory neurons can be influenced by NMDA-receptor-mediated signaling, whereas tangential migration of interneurons is predominantly controlled by non-NMDA receptors (Bortone and Polleux, 2009; Luhmann et al., 2015; Manent et al., 2005; Ohtaka-Maruyama et al., 2018). Interestingly, the GRIN2B NMDA receptor subtype is important for migration of neurons to upper cortical layers (Jiang et al., 2015), and we find that responsiveness to NMDA in upper layer excitatory neurons correlates with expression of this receptor (Figures 6 and S5; Table S5). MGE-derived interneurons display more responses to AMPA than CGE-derived interneurons, which may be related to migration that occurs earlier in MGE-derived interneurons than CGE-derived interneurons (Anderson et al., 2001).
Interestingly, GABA responses in mouse progenitor cells were very robust, consistent with previous reports (LoTurco et al., 1995), but were much less abundant in human progenitors (Figures 3 and S3E). In human, most GABA Ca2+ transients were recorded in a subset of excitatory neurons and are likely to impact their migration and maturation (Cancedda et al., 2007; Chen and Kriegstein, 2015; Luhmann et al., 2015). The developmental switch of chloride transporters NKCC1 and KCC2 determines intracellular chloride concentration and dictates whether GABAA receptor activation is depolarizing (Ben-Ari, 2014). In mouse, the switch occurs at P7 (Garaschuk et al., 2000), while in human, expression of KCC2 is first evident in the SP at PCW18 and becomes abundant in the CP and SP at PCW25 (Sedmak et al., 2016). This could explain the decrease in GABA responses in the CP at PCW19–22 compared to PCW14–16 (Figure 3G). Alternatively, changes in the proportion of cell types present in the CP during later stages of neurogenesis could change the percentage of GABA-responsive cells. Drugs used during pregnancy, for example, to treat epilepsy, often target GABAergic signaling (Morrow et al., 2006; Rogawski and Löscher, 2004) and may thus affect endogenous GABAergic signaling in the fetal brain.
Evolutionary Changes in Responses to Neuromodulatory Neurotransmitters in the Excitatory Neuronal Lineage
Serotonergic fibers innervate the developing neocortex starting in the first trimester, prior to arrival of thalamocortical afferents (Berger et al., 1993; Kostović and Judaš, 2010; Verney et al., 2002), and are a potential source of serotonin. Additionally, serotonin may be present in the CSF (Bonnin et al., 2011). We found that expression of HTR2A is enriched in human radial glia and that activation was most prominent in dividing oRGs (Figure 4). Our calcium imaging experiments in mouse (Figure S3E), together with gene expression data from public databases, indicate that expression of HTR2A in progenitors may be specific to primates (Bakken et al., 2016; Thompson et al., 2014). Previous studies in rodents have demonstrated a role for serotonin regulating neurogenesis during development and in adult (Tong et al., 2014; Vitalis et al., 2007). However, we do not find evidence for a role of HTR2A stimulation in proliferation (Figures 5A and 5B). Instead, tonic inhibition of the HTR2A receptor affects radial fiber morphology (Figures 5C–5G). Htr2a is expressed in deep-layer subcortical projection neurons in the mouse (Figure S1E) (Molyneaux et al., 2015), where it regulates cell survival in addition to dendritic outgrowth (Dooley et al., 1997; Ohtani et al., 2014; Roppongi et al., 2013). In contrast, a subset of upper layer excitatory neurons that are more abundant in primates than other mammals (Marín-Padilla, 1992) also responds to the HTR2A-specific agonist TCB-2 (Figure 4). Aberrant serotonergic signaling has been linked to several neurodevelopmental disorders, including Down syndrome and autism (Sztainberg and Zoghbi, 2016), and HTR2A expression is specifically disturbed during neurogenic stages in Down syndrome (Olmos-Serrano et al., 2016). Together with the potential of maternal antidepressants to affect fetal serotonergic signaling, our findings may help explain how aberrant serotonin signaling during fetal development can lead to congenital disorders (Casper et al., 2003; Gingrich et al., 2017).
Multimodal Single-Cell Analysis Reveals Functional Heterogeneity of Molecularly Defined Cell Types
The importance of multidimensional analysis of molecular and physiological features for determining cellular taxonomy has been widely recognized but hindered by technological challenges (Regev et al., 2017; Zeng and Sanes, 2017). The approach we developed here to achieve multimodal analysis of single cells is highly versatile because it relies on dynamic imaging of a ubiquitous signal transducer in response to diverse stimuli. Patch-seq has been applied to cells in intact tissue slices (Cadwell et al., 2016; Fuzik et al., 2016). In contrast, our approach can be broadly applied to diverse cell and tissue types using biosensors, allowing the analysis of cell-intrinsic physiological properties. In the future, multimodal surveys at the single-cell level that include physiological features will be critical for evaluating the functional relevance of transcriptomic cell classification and to achieving a meaningful definition of the “ground truth” of a cell type. This may also be helpful for evaluating the fidelity of cell types generated in vitro (Bredenoord et al., 2017; Di Lullo and Kriegstein, 2017).
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Arnold R. Kriegstein (Arnold.Kriegstein@ucsf.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human tissue samples
The first trimester material was provided by the Joint MRC / Wellcome Trust (grant # 099175/Z/12/Z) Human Developmental Biology Resource (http://www.hdbr.org). De-identified second trimester tissue samples were collected with previous patient consent in strict observance of the legal and institutional ethical regulations. Protocols were approved by the Human Gamete, Embryo, and Stem Cell Research Committee (institutional review board) at the University of California, San Francisco
Mouse cells
All animal experiments were approved by the UCSF Institutional Animal Care and Use Committee. Cerebral cortex was dissected from E14 and E18 C57/BL6 or Swiss Webster mouse embryos in ice cold HBSS. Cortices from one litter were pooled together. Tissue was dissociated at room temperature for 5 min in a papain solution (Worthington). Tissue was subsequently bathed in a trypsin inhibitor solution for 1 min. Cortices were gently triturated in NPC media, which was composed of DMEM (ThermoFisher Scientific) supplemented with N2 (ThermoFisher Scientific), B27 (ThermoFisher Scientific), Glutamax (ThermoFisher Scientific) and 1% penicillin-streptomycin (ThermoFisher Scientific), to achieve a single cell suspension. Cells were cultured in NPC media on polyornithine (Sigma) coated glass coverslips at 1.5 million cells per well of a 24 well plate. Coverslips were prepared the following day for calcium imaging.
METHOD DETAILS
Single-cell receptor expression analysis
Trajectory plots from published C1 datasets were constructed by correlating cells to excitatory differentiation and ordering cells along this axis. The lines depicting the correlation are shown using loess smooth regression from R package ggplot2. (Figures 1A–1F). Correlation of HTR2A expression values to age (pcw) in any radial glia cells as defined by the cluster annotations was plotted (Figure 1F). Networks annotated in Nowakowski et al. (2017) were correlated to HTR2A expression. Networks that correlated to HTR2A expression > 0.4 were used to generate a correlated gene list by including all genes within those networks (Table S1).
Single molecule in situ hybridization
Tissue blocks were drop-fixed in 4% PFA in PBS overnight at 4°C. Subsequently, tissue was cryoprotected in 30% sucrose in PBS, and embedded in a 1:1 mixture of 30% sucrose and optimal cutting temperature compound (TissueTek). 16 µm cryosections were cut on Superfrost Plus slides (VWR) using Leica CM3050S cryostat. Slides were stored at −80°C until use.
Single molecule in situ hybridization was performed according to the RNAscope manual (2.5 manual assay red chromogen). Sequences of target probes, preamplifier, amplifier, and label probe are proprietary and commercially available (Advanced Cell Diagnostics (ACD), Hayward, CA). Typically, the probes contain 20 ZZ probe pairs (approx. 50 bp/pair) covering 1000bp. Here, we used probes against human HTR2A and P2YR1 as single-plex probe. Slides were dried at 60°C for 1 h and fixed in 4% PFA for 2 h. After several washes in PBS, slides were treated with ACD Hydrogen peroxide for 10 min and then washed in water 2x before treatment in 1x target retrieval buffer (ACD) for 5 min (at 95–100°C). After washing in water and then 100% alcohol, the slides were left to dry overnight. Protease treatment was performed for 15 min at 40°C in the RNAscope oven. Hybridization of probes and amplification was performed according to the manufacturer’s instructions. In short, tissue sections were incubated in desired probe (2–3 drops/section) for 2 h at 40°C. The slides were washed twice times in 1x wash buffer (ACD) for 2 min each. Amplification and detection steps were performed using the RNAscope 2.5 HD Red Detection Kit reagents (ACD, 320497) for single-plex probes. Sections were incubated with Amp1 for 30 min at 40°C and then washed two times in wash buffer for 2 min each. Amp2 was incubated on the sections for 15 min at 40°C, followed by two washes in wash buffer. Sections were incubated in Amp3 for 30 min at 40°C and washed two times in wash buffer for 2 min each, followed by incubation of Amp4 for 15 min at 40°C. Slides were washed two times in wash buffer for 2 min each. Slides were incubated with Amp5 for 30 min at RT using the HybEZ humidity control tray and slide rack to maintain humidity. The slides were washed two times in 1x wash buffer for 2 min each and incubated in Amp6 for 15 min at RT before washing two times in wash buffer for 2 min each. Signal was detected by diluting Fast RED-B in Fast RED-A solution (1:60 ratio) and incubating sections in this solution for 10 min. Slides were washed in water 2 times to stop the reaction.
Subsequently, slides were either stained with DAPI and mounted in Aqua Mount (Lerner) or subjected to immunohistochemistry for cell type markers using the Perkin-Elmer Tyramide Signal Amplification technology according to manufacturer’s instructions. Sections were blocked using TNB for 45 min before applying primary antibodies in TNB overnight at 4°C. Slides were washed in PBS with 0.05% Triton X-100 (PBST), secondary antibodies were applied for 2 h in TNB together with DAPI. Slides were washed in PBST and incubated in Streptavidin-HRP in TNB (1:200) for 30 min. Slides were washed in PBST and developed in a 1:50 dilution of Fluorescent Tyramide in AMP for 5 min. Slides were washed in PBST and mounted in Aqua Mount (Lerner). Images were taken using 40x objectives on Leica confocal microscopes SP5 and SP8.
Primary antibodies used after RNAscope: mouse anti Cryab (1:100, ab13496, abcam, detected with Alexa 647), rabbit anti Hop (1:200, sc30216, Santa Cruz Biotechnology, detected with fluorescent tyramide), rabbit anti PPP1R17 (1:100, HPA 047819, Sigma, detected with fluorescent tyramide), mouse anti Ki67 (1:200, M7240, Dako, detected with Alexa647). For immunohistochemistry, goat anti Sox2 (sc17320, Santa Cruz Biotechnology, detected with Alexa647) was used according to a previous protocol (Pollen et al., 2015).
Unbiased quantification of fluorescent immunohistochemistry and RNAscope FISH was performed using a custom semi-automatic macro written for the FIJI distribution of ImageJ (Schindelin et al., 2012). Nuclei and stained cell regions were identified using multiple noise-cancelling filters and an intensity threshold using the Li method (Li and Tam, 1998), segregated using a watershed algorithm, and automatically quantified using the Analyze Particles function. Small regions of high intensity were regarded as noise and were automatically eliminated using the Shape Filter plugin (Wagner and Lipinski, 2013). Periodic manual quality checks were programmed into the macro to ensure proper thresholding and to manually eliminate blood vessel regions falsely identified as cells. Quantification of colocalization was performed by running previously processed and segmented regions through the Morphological Reconstruction plugin from the MorphoLibJ library (Legland et al., 2016). For example, to identify Hopx+ nuclei, the processed DAPI channel was used as the mask image and the processed Hopx channel was used as the marker, yielding an output of all nuclei that stained positive for Hopx. Subsequently, this Hopx+ nuclei channel was used as a mask and the processed RNAscope channel used as a marker to identify Hopx+ nuclei containing RNAscope target mRNA transcripts. All Morphological Reconstruction outputs were quantified automatically using the Analyze Particles function.
Electrophysiological recordings
Chemicals were from Sigma unless otherwise noted. Modified tissue transfer solutions were used to improve cell survival as described previously (Chen and Kriegstein, 2015). Cortical slabs were embedded in 4% low melting point agarose (Fisher Scientific) and placed in ice-cold oxygenated (5% CO2/95% O2) artificial CSF (aCSF) solution (pH 7.3 – 7.4, 290 – 300 mOsm) for sectioning containing (in mM): 110 Choline Cl, 2.5 KCl, 0.5 CaCl2, 7 MgCl2, 1.3 NaH2PO4, 25 NaHCO3, 10 glucose. 400 µm slice sections were obtained by vibratome (Leica VT1200S) and maintained in an incubation chamber in recovery and recording aCSF containing (in mM): 125 NaCl, 2.5 KCl, 2 CaCl2, 1.3 MgCl2, 1.3 NaH2PO4, 25 NaHCO3, 10 glucose, at room temperature.
During experiments, individual slices were transferred to a submerged recording chamber and continuously perfused with the recovery and recording aCSF (4.0 mL/min) at 32°C. Slices were visualized under a microscope (Olympus BX51WI) using infrared video microscopy and differential interference contrast optics. Different types of cells were first identified by their location, shape of cell bodies, and current-voltage (I-V) curves, subsequently confirmed by immunostaining with cell type specific markers post recordings. Recordings were made with an Axon 700B patch-clamp amplifier and 1320A interface (Axon Instruments). Signals were filtered at 2kHz using amplifier circuitry, sampled at 10 kHz, and analyzed using Clampex 10.2 (Axon Instruments). Patch electrodes were made from borosilicate glass capillaries (BF 150-86-10, Sutter Instruments) and pulled on a P-97 Micropipette Puller (Sutter Instruments) for a resistance in the range of 4–6 MΩ. The pipettes were tip- and back-filled with internal solutions containing (in mM): 125 K-gluconate, 10 KCl, 10 HEPES, 4 MgCl2,4 Na2ATP, 0.4 Na4GTP, 0.5 EGTA, 0.2% biocytin for whole-cell recordings. Drugs loaded in micropipettes (tip diameter 50 µm) were delivered locally to the recorded cells by pressure pulses using microinjector (Picospritzer III, 20 – 50 ms duration, 15 s interval). All antagonists were bath applied. Small depolarizing current pulses (50 ms duration, 50 ms interval, 200–500 pA) were injected into the cell to facilitate biocytin labeling. Each slice was fixed with 4% PFA immediately after recordings overnight at 4°C for subsequent immunohistochemistry experiments. Primary antibodies: rat anti Ctip2 (1:500, Abcam ab18465), mouse anti EOMES (1:200, Affymetrix 14-4877-82). Biocytin was detected by Streptavidin Dylight 549 (1:500, Vector Labs SA-5549).
Human neocortex dissociation
The germinal and the CP and SP were microdissected in artificial cerebrospinal fluid containing 125 mM NaCl, 2.5 mM KCl, 1 mM MgCl2, 1 mM CaCl2, 1.25 mM NaH2PO4 using a microsurgical blade. To obtain a single-cell suspension, tissue samples were dissociated in a pre-warmed working solution of Papain and 2000 units/mL of DNase freshly diluted in Earl’s Balanced Salt Solution (Worthington Biochem. Corp.) or using a trypsin-based neural dissociation kit (Miltenyi, 130-093-231). The samples were incubated at 37°C for 20–30 min, gently triturated and resuspended in either extracellular solution (ECS) with Mg2+ (145 mM NaCl, 3 mM KCl, 3 mM CaCl2, 2 mM MgCl2, 10 mM HEPES, 10 mM D-glucose, pH 7.4, 290–300 mOsm) and 10% FBS (Sigma) for Polaris experiments, or cell culture media (DMEM, 1% N2, 1% B27, and 1x penicillin, streptomycin, glutamine) with 10% FBS, for overnight culture as described previously (Pollen et al., 2015). Cells were centrifuged for 5 min at 300 x g and washed in the resuspension media without FBS. The suspension was passed through a 40 µm strainer cap (BD Falcon) to yield a uniform single-cell suspension. Viability was assessed using Trypan blue staining and automatic quantification using the cell counter Countess (Invitrogen). Viability was normally 90%–98%.
Mouse cell type identification
Dissociated mouse embryonic cortical cells were fixed in 4% PFA for 10 min after calcium imaging. Cells were washed 3 times in PBS and were blocked for 40 min in donkey blocking buffer containing 0.2% gelatin, 10% donkey serum (Sigma D9663), and 0.1% Triton X-100 in PBS. Primary antibody incubation was performed in donkey blocking buffer at 4°C overnight with the following antibodies: Rabbit polyclonal IgG Beta-III Tubulin (Covance PRB-435P) and Goat polyclonal Sox-2 (Santa Cruz Biotechnology sc-17320). The following day, cells were washed 3 times in PBS and were incubated for 1 h at room temperature with secondary antibody solutions composed of fluorophore conjugated Alexa Fluor dyes (ThermoFisher Scientific) diluted at 1:1000 in donkey blocking buffer. Cells were subsequently stained with the Hoechst nuclear dye (Life Technologies 1:10,000) for 10 min and then washed 3 times in PBS. Coverslips were mounted onto slides and imaged on a Leica epifluorescence microscope. Immunocytochemistry (ICC) images then were used to match a cell’s protein expression to its calcium responses. A python script was employed to find the same field of cells that were imaged for their calcium responses in the ICC images. To do this, the first frame of a calcium imaging experiment was taken and manually thresholded using ImageJ to remove background fluorescence. Features in this calcium imaging frame were then matched to the nuclear marker stain using a python script that employed packages in the Open Source Computer Vision Library (OpenCV). Specifically, features were detected with the scale invariant feature transform (SIFT) package (Lowe, 2004) and subsequently matched with The Fast Library for Approximate Nearest Neighbors (FLANN) (Muja and Lowe, 2009). Together, these packages identified and matched similar features from the calcium imaging frame with the ICC nuclear marker image regardless of size and scale. Once the matched coordinates in the calcium recording and ICC image were found, the images were manually overlaid and counted to associate protein expression with calcium response.
Ca2+ imaging
Coverslips (Pretreated German coverslips, Neuvitro, GG-12-Pre) were coated with 10 µg/mL Fibronectin (Sigma, F1141) or Matrigel (354234, Corning) in Dulbecco’s phosphate buffered saline (DPBS) overnight at 37°C. Cells were cultured in cell culture media (see above) overnight at 37°C (5% CO2, 8% O2). Cells were loaded with 5 µM Oregon Green 488 BAPTA-1, AM (Invitrogen) in ECS with Mg2+ for 30 min at 37°C. Cells were washed in ECS with Mg2+. Cells were continuously perfused with ECS without Mg2+ (150 mM NaCl, 3 mM KCl, 3 mM CaCl2, 10 mM HEPES, 10 mM D-glucose, pH 7.4, 290–300 mOsm) while imaging. Images were collected using an epifluorescence microscope (Olympus BX51WI) and a Xenon arc light source excitation light with GFP excitation bandpass filter. Light pulses were delivered through a filter wheel and Smartshutter (Lambda XL, Sutter). A short exposure (10–20 ms) and low sampling frequency (1 Hz) were used to avoid photobleaching. Fluorescence signals were collected using Metafluor software. Agonists were applied through a pipette (50 µL per agonist diluted in ECS without Mg2+). The interval between agonist applications was 5–10 min. At the end of each recording, ionomycin was applied as a positive control. See table below for concentrations of agonists used.
Ca2+ imaging data analysis
For analysis of Ca2+ transients, cells were identified as regions of interest using a custom ImageJ macro (Schindelin et al., 2012). First, the image stack was summed in the Z axis. The phansalkar threshold method (Phansalkar et al., 2011) was then applied to increase contrast, using a radius of 15 and standard parameters. ImageJ’s particle analyzer function was subsequently used to identify all possible cells. The identified regions of interest were used to determine the integrated fluorescence intensity in the original image stack. A custom R-based script was then used for plotting normalized traces and assisting with identifying responses to agonists. Gaps in the data (data collection artifacts) were filled with an average of the cell’s fluorescence over the whole recording. The data collected on the physiological rig was filtered using a Chebyshev type II filter (‘signal’ package) (signal Developers, 2013). Changes in fluorescence were calculated and normalized against the baseline by F/F0, where F0 is the average fluorescence across the whole recording. Peaks at least 5% above the baseline were detected (‘pracma’ package) (Borchers, 2018), in the period between drug injection and up to 150 frames after the application. Additionally, manual curation assessed detected peaks individually and adjusted as required, generating a binary matrix of responses to each agonist in each cell. Only cells responding to ionomycin were included. Population responses were analyzed using Prism (unpaired t tests).
Polaris single-cell imaging and RNA-Seq
The Polaris system has been described in detail recently (Ramalingam et al., 2016). Polaris Integrated Fluidic Circuit (IFC) chips were operated according to the manual (PN 101–0075 A1). Priming followed the adherent protocol with 25 µg/mL Fibronectin. Dissociated cells were stained with 5 µM OGB-1 AM (Thermo Fisher) in ECS with Mg2+ (see above) for 30 min at 37°C. Stained cells were captured on microfluidic chips. A custom python script enabled dosing of captured cells with agonists diluted in ECS without Mg2+ followed (duration: approx. 2.25 min) by washing in ECS without Mg2+. Agonists were added in 10 min intervals. Concurrently, the same custom script allowed images to be recorded at a frequency of 0.4 Hz. After dosing, cells were immediately processed for lysis, reverse transcription and PCR on the Polaris IFC (Fluidigm) using the SMARTer Ultra Low RNA Kit (Clontech). The time between the application of the last agonist and the start of the lysis step was approximately 30–60 min. Single-cell sequencing libraries were prepared using the Nextera XT DNA Sample Preparation Kit (Illumina). Libraries were sequenced using the Illumina HiSeq flow cell. Fastq files from individual cells were processed using an automated in-house pipeline on a high-performance computing cluster. Sequencing reads were trimmed off library adapters using TrimGalore! and aligned to the GRCh38 genome reference using HISAT2 (Kim et al., 2015). Aligned reads were summarized to calculate gene counts using featureCounts (Liao et al., 2014) and the ENSEMBL GRCh38.88 gene reference. We detected 3118 ± 47 genes per cell and 3,939,629 ± 105,874 reads per cell.
Analysis of scRNA-seq data through clustering
Counts per million (CPM) were calculated by dividing counts for each gene and cell by total number of counts in the given cell and multiplying by a million. Matrix of CPM values was log-transformed and filtered to remove any cells expressing less than 1000 genes and genes expressed in less than 2 cells. Then, principal component analysis was performed on the transformed matrix. To identify the number of principle components to retain, scree plot analysis was performed to detect the point on the curve when variation associated with additional components reached a plateau. Scores of significant principle components were projected onto two-dimensional t-SNE space (van der Maaten and Hinton, 2008). To determine the number of cell clusters, Gap Statistic (Tibshirani et al., 2002) was used with partitioning around medoids (PAM) clustering over 1000 bootstraps. PAM method was then used to cluster the data into the number of clusters identified (Tibshirani et al., 2002). To identified gene signatures of each cluster, we used MAST (Finak et al., 2015) and zero-inflated regression (zlm) method to compare cells in each given cluster to all the other cells and identify genes upregulated in each cluster (FDR < 0.05; fold change ≥ 2). To analyze differential gene expression for cells belonging to the same cluster but displaying differential responses, we utilized RankProd method (Hong et al., 2006). Gene Ontology enrichment analysis of differentially expressed genes was performed using Panther Statistical Overrepresentation tests (comparing to all genes expressed in a specific cluster, including correction for multiple testing) (Mi et al., 2013). Gene Ontology Terms were condensed using REVIGO (SimRel algorithm) (Supek et al., 2011). Integration of Polaris and published C1 datasets (Nowakowski et al., 2017; Pollen et al., 2015) was performed using canonical correlation analysis in the space of 20 dimensions (Butler et al., 2018). Subsequent tSNE maps were generated in the reduced dimensionality CCA space. Cluster markers were generated using the zero-inflated log ratio test distribution and interpreted to correspond to published cell types (Nowakowski et al., 2017). Differential gene expression between Polaris and C1-captured single cells was performed using the log-ratio test (LRT) with a bimodal distribution well suited to zero-inflated data such as single-cell transcriptomics. Gene ontology analysis was performed using Panther and the list of genes expressed all oRGs as a background.
Heatmaps were constructed using Morpheus. Clusters with downregulation of housekeeping genes (“Other”) were defined as outliers and excluded from further analysis. Responses to agonists were compared between pairs of transcriptomic clusters using a two-sided Fisher’s exact test. To control for the false discovery rate across the multiple comparisons we use the Benjamini-Hochberg procedure.
Physiological and transcriptomic pseudoage
For the pseudotemporal analysis we first project the single cell transcriptomes of the cells belonging to clusters along the lineage on the first two principal components. In this dimensionality-reduced space we then fit a principal curve through the points (Hastie and Stuetzle, 1989) (using the R ‘princurve’ package), which we initialize with the mean coordinates of the first and last cluster in the lineage. All cells are then projected onto this principal curve. A cells pseudoage is defined as the arclength along the curve up to their projected position.
We analyze the dependence of the agonist response frequency on pseudoage using nonparametric logistic regression implemented in PyGAM (https://zenodo.org/record/1213536). In short, the frequencies are modeled as being dependent on the pseudoage via a logit link function of an arbitrary smooth function. This smooth function is assumed to be composed of 20 third-order penalized B-splines with a second derivative penalty of weight 0.6. To allow an interpretation of the pseudoage in terms of different cell types we analyze in the same way how the frequency of detection of marker genes in the transcriptome varies with pseudoage.
Classification of physiological responses
A naive Bayes classifier was trained on the multimodal single-cell data of all cells responding to at least one agonist. This type of classifier has been shown to learn efficiently from relatively small amounts of data (Ng and Jordan, 2002). Additionally, it is the natural classifier under a plausible generative model of response distributions in which each cell type can be described by a set of independent response probabilities to the different agonists. Pseudocounts of 0.5 were used in the training. No attempt was made to learn a prior distribution of cell type frequencies as we may expect these to differ across conditions. Accuracy of the classifier as assessed by ten-fold cross-validation was found to be 53% ± 3% (mean ± SE).
Clustering of physiological response data
The response data were clustered in R using binary clustering metrics and Ward hierarchical clustering. To determine the number of significant clusters, Dynamic Tree Cut (Langfelder et al., 2008) method was used. The analysis of the contingency tables uses Pearson residuals, which are defined as the difference between the observed counts and the expected counts under an independent model normalized by the square root of the expected counts.
Differential gene expression analysis
MAST was utilized to compare single-cell gene expression profiles in each physiological cluster identified when contrasted with the rest of the cells. Genes with FDR < 0.05 and abs(log2(FC)) > 0.5 were considered differentially expressed.
Cortical organotypic slice culture
This protocol was adapted from Lui et al. (2014).
Human cortical tissue was embedded in 3% low melting point agarose in artificial cerebrospinal fluid (ACSF; 125 mM NaCl, 2.5 mM KCl, 1 mM MgCl2, 2 mM CaCl2, 1.25 mM NaH2PO4, 25 mM NaHCO3, 25 mM d-(+)-glucose, bubbled with 95% O2/5% CO2), and sliced at 300 µm in ice-chilled ACSF in the vibratome at the plane most closely approximating a coronal section. Slices were transferred to and suspended on Millicell-CM slice culture inserts (Millipore) over culture medium (66% BME, 25% Hanks, 5% FBS, 1% N-2, 1% penicillin, streptomycin, and glutamine (all Invitrogen) and 0.66% d-(+)-glucose (Sigma)) in 6-well plates and incubated at 37°C, 8% O2, 5% CO2 for 2 days.
For morphology assays, slices were incubated with Ad-CMV-GFP virus (1:2000, Vector Biolabs, 1060) for 30 min prior to positioning on slice culture inserts. After an overnight recovery, slices were incubated with either HTR2A agonist (100 µM TCB-2) or antagonist (100 µM EMD 281014 hydrochloride) dissolved in ECS (150 mM NaCl, 10 mM HEPES, 3 mM KCl, 2 mM CaCl2, 2 mM MgCl2, 5.5 mM glucose, pH 7.3), or ECS only as the negative control. 72 h after agonist application, slices were fixed in 4% PFA. The tissue was subsequently subjected to heat-induced 10 mM citrate antigen retrieval for 20 min. Slices were permeabilized and blocked overnight in 0.5% Triton X-100, 10% donkey serum, and 0.2% gelatin in PBS. Primary antibody incubation was performed in the blocking solution for 2 days at 4°C. The following primary antibodies were used: chicken anti GFP 1:1000 (Aves, GFP-1020), rabbit anti HopX 1:200 (Santa Cruz, sc30216), and mouse anti Vimentin 1:500 (Sigma, V5255). Secondary antibody and DAPI incubation was performed in blocking solution overnight at 4°C. The following secondary antibodies were used: Alexa Fluor 488 donkey anti chicken (Jackson, 703-545-155), Alexa Fluor 546 donkey anti mouse (Life Technologies, A10036), Alexa Fluor 647 donkey anti rabbit (Life Technologies, A31573). After washing slices in PBS, slides were mounted using Aqua Polymount. Images were taken on a Leica SP5 confocal with a 40X objective. Maximum intensity projections were performed using FIJI, and images were adjusted for brightness and contrast. For fiber length quantification, confocal image stacks were taken as tile scans using a 40X objective. Outer subventricular zone was delineated from rest of slice, and segmented into five equidistant areas. Two Hopx+ fibers were chosen from each area and measured using the “filament” function in Imaris imaging software (Bitplane). Ten total fibers were measured and recorded in the fiber quantification of each slice to determine the mean fiber length of Hopx+ cells in the outer subventricular zone.
For BrdU incorporation assays, BrdU (Sigma, 15 µg/mL) was added to the culture medium at the beginning of all culture periods. In these experiments BrdU was present for the entire two-day culture period either HTR2A agonist (100 µM TCB-2) or antagonist (100 µM EMD 281014 hydrochloride) or HTR2A agonist + antagonist dissolved in ECS, or ECS only as the negative control. Experiments lasted 1 or 2 days and slices were fixed overnight with 4% PFA afterward. The fixed cortical slices were stained for SOX2 and Ki67 before staining for BrdU was performed. In the first step, the tissue was subjected to heat-induced 10mM citrate antigen retrieval for 20 min. Slices were permeabilized / blocked overnight in PBS plus 0.5% Triton X-100, 10% serum, and 0.2% gelatin. Primary and secondary incubations were each performed in the blocking solution for 2–3 days at 4°C. The following primary antibodies were used: goat anti Sox2 (1:250, Santa Cruz, sc17320), mouse anti Ki67 (1:200, Dako, M7240). The following secondary antibodies were used: Alexa Fluor 488 donkey anti mouse (Life Technologies, A-21202), Alexa Fluor 546 donkey anti goat (Life Technologies, A-11056). The slices were then re-fixed in 4% PFA for 1 hr. To stain for BrdU, slices were treated with 2N HCl for 40 min at 37°C, neutralized with boric acid, and primary (rat anti BrdU, 1:500, Abcam, ab6326) and secondary (Alexa Fluor 647 donkey anti rat, Jackson, 712-605-153) antibodies were used as previously described. The slices were washed in PBS, and mounted using Aqua Polymount. Images were taken on a Leica SP5 confocal, and adjusted for brightness and contrast using FIJI.
For cell counting, single-plane confocal images were collected using a 20x objective. Fluorescence signal from nuclear immunoreactivity was first counted individually using the ‘spots’ function in Imaris imaging software (Bitplane). A threshold for intensity mean was set to find fluorescent spots of ~6-μm in diameter for each channel as a representation of the number of cells. The preliminary sets of positive cells were then manually edited to correct software error. Double and triple co-localized spots were then calculated by the ImarisXT (Bitplane) module, and further edited to correct software errors. The numbers of total and co-localized spots were recorded and subtracted from each other to determine the number of single-, double- and triple-labeled cells.
Agonists
| Agonist | Product number | Concentration in pipette for Ca imaging | Final concentration in Polaris | Concentration in pipette for slice recordings |
|---|---|---|---|---|
| NMDA / Gly | Sigma M3262 / G6761 | 100 mM / 1 mM | 1 mM / 10 µM | 100 mM / 1 mM |
| AMPA | Tocris 0169 | 10 mM | 100 µM | 10 mM |
| Acetylcholine | Sigma A6625 | 10 mM | 100 µM | 10 mM |
| GABA | Sigma A2129 | 100 mM | 1 mM | 100 mM |
| TCB-2 | Tocris 2592 | 10 mM | 1 mM | 10 mM |
| MeSADP | Tocris 1624 | 10 mM | 100 µM | 10 mM |
| Ionomycin | Sigma I3909 | 10 µM | - | - |
Antagonists
| Antagonist | Product number | Bath Concentration |
|---|---|---|
| DNQX | Sigma D0540 | 20 µM |
| EMD281014 | Tocris 4470 | 200 µM |
| Suramin | Tocris 1472 | 100 µM |
| D-APV | Tocris 0106 | 100 µM |
| Carbenoxolone | Tocris 3096 | 100 µM |
| BMI | Sigma B7561 | 20 µM |
QUANTIFICATION AND STATISTICAL ANALYSIS
Specific statistical tests are described in the relevant sections. Throughout the manuscript, N refers to the number of biological replicates, n refers to the number of cells, error bars show SEM (related to N).
DATA AND SOFTWARE AVAILABILITY
Next Generation Sequencing data will be available on dbGaP: phs000989.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Chicken anti GFP | Aves | GFP-1020; RRID: AB_10000240 |
| Goat anti Sox2 | Santa Cruz | sc17320; RRID: AB_2286684 |
| Mouse anti Ki67 | Dako | M7240; RRID: AB_2142367 |
| Mouse anti Vimentin | Sigma | V5255; RRID: AB_477625 |
| Rabbit anti HopX | Santa Cruz | sc30216; RRID: AB_2120833 |
| Alexa Fluor 546 donkey anti mouse | Life Technologies | A10036; RRID: AB_2534012 |
| Alexa Fluor 488 donkey anti chicken | Jackson | 703-545-155; RRID: AB_2340375 |
| Alexa Fluor 488 donkey anti mouse | Life Technologies | A-21202; RRID: AB_141607 |
| Alexa Fluor 546 donkey anti goat | Life Technologies | A-11056; RRID: AB_142628 |
| Alexa Fluor 647 donkey anti rabbit | Life Technologies | A31573; RRID: AB_2536183 |
| Rabbit polyclonal IgG Beta-III Tubulin | Covance | PRB-435P; RRID: AB_291637 |
| Rat anti Ctip2 | Abcam | ab18465; RRID: AB_2064130 |
| Mouse anti EOMES | Affymetrix | 14-4877-82; RRID: AB_2572882 |
| Streptavidin Dylight 549 | Vector Labs | SA-5549; RRID: AB_2336408 |
| Mouse anti Cryab | Abcam | ab13496; RRID: AB_300400 |
| Rabbit anti PPP1R17 | Sigma | HPA 047819; RRID: AB_2680166 |
| Chemicals | ||
| NMDA / Gly | Sigma | M3262 / G6761 |
| AMPA | Tocris | 169 |
| Acetylcholine | Sigma | A6625 |
| GABA | Sigma | A2129 |
| TCB-2 | Tocris | 2592 |
| MeSADP | Tocris | 1624 |
| Ionomycin | Sigma | I3909 |
| DNQX | Sigma | D0540 |
| EMD281014 | Tocris | 4470 |
| Suramin | Tocris | 1472 |
| D-APV | Tocris | 106 |
| Carbenoxolone | Tocris | 3096 |
| BMI | Sigma | B7561 |
| Oregon Green 488 BAPTA-1, AM | Invitrogen | O6807 |
| Critical Commercial Assays | ||
| RNAscope 2.5 HD Assay - RED | Advanced Cell Diagnostics | 322350 |
| Polaris Single-Cell Dosing mRNA Seq IFC | Fluidigm | PN 101–0082 |
| Deposited Data | ||
| RNA sequencing data | This paper | dbGaP: phs000989 |
| Software and Algorithms | ||
| ImarisXT | Bitplane | http://www.bitplane.com/ |
| GraphPad Prism 5 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| Fiji (ImageJ) | NIH | https://fiji.sc/ |
| ggplot2 (R package) | https://cran.r-project.org/web/packages/ggplot2/index.html | |
| Fast Library for Approximate Nearest | Muja and Lowe, 2009 | https://www.cs.ubc.ca/research/flann/ |
| Neighbors (Python package) | ||
| signal (R package) | signal Developers, 2013 | https://cran.r-project.org/web/packages/signal/index.html |
| pracma (R package) | Borchers, 2018 | https://cran.r-project.org/web/packages/pracma/index.html |
| MAST (R package) | Finak et al., 2015 | https://www.bioconductor.org/packages/release/bioc/html/MAST.html |
| princurve (R package) | https://cran.r-project.org/web/packages/princurve/index.html | |
| cluster (R package) | https://cran.r-project.org/web/packages/cluster/index.html | |
| TrimGalore (Python package) | Kim et al., 2015 | http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ |
| Rsubread (R package) | Liao et al., 2014 | http://bioconductor.org/packages/release/bioc/html/Rsubread.html |
| MorphoLibJ (ImageJ plugin) | Legland et al., 2016 | https://imagej.net/MorphoLibJ |
| Shape Filter (ImageJ plugin) | Wagner and Lipinski, 2013 | https://imagej.net/Shape_Filter |
| Other | ||
| Pretreated German coverslips | Neuvitro | GG-12-Pre |
| Fibronectin | Sigma | F1141 |
| Matrigel | Corning | 354234 |
Highlights.
Multimodal analysis differentiates cells beyond transcriptomic classification
Single-cell analysis links stimulus-induced calcium elevations to transcriptomes
Cell-type-specific responses to neurotransmitters are associated with maturation
Serotonergic signaling in human radial glia promotes radial fiber formation
ACKNOWLEDGMENTS
Data presented here will be uploaded on dbGaP (phs000989). We would like to thank Adam Fries, William Walantus, Yulia Oganian, Aaron Diaz, Jerry Wang, and Carmen Sandoval Espinosa for technical help and Joseph J. Loturco and Kirsten Obernier for fruitful discussions. We are grateful for funding from the NIH (1U01MH105989–01, 1U01MH114825–01, and R35NS097305 to A.R.K.), EMBO (ALTF 393–2015 to S.M.), DFG (MA 7374/1–1; fellowship to S.M.), NIGMS (MSTP fellowship to U.C.E.), University of California, San Francisco (UCSF) Program for Breakthrough Biomedical Research, Sandler Foundation (to G.P.), and NSFC (81870898, 91732301, and LR18H090002 to J.C.).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes six figures, six tables, and two videos and can be found with this article online at https://doi.org/10.1016/j.neuron.2019.01.027.
DECLARATION OF INTERESTS
A.A.P. is a scientific advisor to System1 Biosciences. A.R.K. is a co-founder, board member, and scientific advisor of Neurona Therapeutics. A.A.-B. is co-founder and scientific advisor of Neurona Therapeutics.
REFERENCES
- Aghajanian GK, and Marek GJ (1999). Serotonin, via 5-HT2A receptors, increases EPSCs in layer V pyramidal cells of prefrontal cortex by an asynchronous mode of glutamate release. Brain Res 825, 161–171. [DOI] [PubMed] [Google Scholar]
- Anderson SA, Marín O, Horn C, Jennings K, and Rubenstein JL (2001). Distinct cortical migrations from the medial and lateral ganglionic eminences. Development 128, 353–363. [DOI] [PubMed] [Google Scholar]
- Ataman B, Boulting GL, Harmin DA, Yang MG, Baker-Salisbury M, Yap E-L, Malik AN, Mei K, Rubin AA, Spiegel I, et al. (2016). Evolution of Osteocrin as an activity-regulated factor in the primate brain. Nature 539, 242–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo FAC, Carvalho LRB, Grinberg LT, Farfel JM, Ferretti REL, Leite REP, Jacob Filho W, Lent R, and Herculano-Houzel S (2009). Equal numbers of neuronal and nonneuronal cells make the human brain an isometrically scaled-up primate brain. J. Comp. Neurol 513, 532–541. [DOI] [PubMed] [Google Scholar]
- Bakken TE, Miller JA, Ding S-L, Sunkin SM, Smith KA, Ng L, Szafer A, Dalley RA, Royall JJ, Lemon T, et al. (2016). A comprehensive transcriptional map of primate brain development. Nature 535, 367–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Ari Y (2014). The GABA excitatory/inhibitory developmental sequence: a personal journey. Neuroscience 279, 187–219. [DOI] [PubMed] [Google Scholar]
- Berger B, Alvarez C, and Goldman-rakic PS (1993). Neurochemical development of the hippocampal region in the fetal Rhesus monkey. I. Early appearance of peptides, innervation in the entorhinal cortex during the first half of gestation (E47 to E90). Hippocampus 3, 279–305. [DOI] [PubMed] [Google Scholar]
- Bittman K, Owens DF, Kriegstein AR, and LoTurco JJ (1997). Cell coupling and uncoupling in the ventricular zone of developing neocortex. J. Neurosci 17, 7037–7044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonetti C, and Surace EM (2010). Mouse embryonic retina delivers information controlling cortical neurogenesis. PLoS ONE 5, e15211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnin A, Goeden N, Chen K, Wilson ML, King J, Shih JC, Blakely RD, Deneris ES, and Levitt P (2011). A transient placental source of serotonin for the fetal forebrain. Nature 472, 347–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchers HW (2018). CRAN - Package pracma
- Bortone D, and Polleux F (2009). KCC2 expression promotes the termination of cortical interneuron migration in a voltage-sensitive calcium-dependent manner. Neuron 62, 53–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredenoord AL, Clevers H, and Knoblich JA (2017). Human tissues in a dish: the research and ethical implications of organoid technology. Science 355, eaaf9414. [DOI] [PubMed] [Google Scholar]
- Bruel-Jungerman E, Lucassen PJ, and Francis F (2011). Cholinergic influences on cortical development and adult neurogenesis. Behav. Brain Res 221, 379–388. [DOI] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell CR, Palasantza A, Jiang X, Berens P, Deng Q, Yilmaz M, Reimer J, Shen S, Bethge M, Tolias KF, et al. (2016). Electrophysiological, transcriptomic and morphologic profiling of single neurons using Patch-seq. Nat. Biotechnol 34, 199–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancedda L, Fiumelli H, Chen K, and Poo MM (2007). Excitatory GABA action is essential for morphological maturation of cortical neurons in vivo. J. Neurosci 27, 5224–5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casper RC, Fleisher BE, Lee-Ancajas JC, Gilles A, Gaylor E, DeBattista A, and Hoyme HE (2003). Follow-up of children of depressed mothers exposed or not exposed to antidepressant drugs during pregnancy. J. Pediatr 142, 402–408. [DOI] [PubMed] [Google Scholar]
- Chen J, and Kriegstein AR (2015). A GABAergic projection from the zona incerta to cortex promotes cortical neuron development. Science 350, 554–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Zhang K, Zhou L, Gao X, Wang J, Yao Y, He F, Luo Y, Yu Y, Li S, et al. (2016). Coupled electrophysiological recording and single cell transcriptome analyses revealed molecular mechanisms underlying neuronal maturation. Protein Cell 7, 175–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Torre-Ubieta L, Stein JL, Won H, Opland CK, Liang D, Lu D, and Geschwind DH (2018). The dynamic landscape of open chromatin during human cortical neurogenesis. Cell 172, 289–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Lullo E, and Kriegstein AR (2017). The use of brain organoids to investigate neural development and disease. Nat. Rev. Neurosci 18, 573–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooley AE, Pappas IS, and Parnavelas JG (1997). Serotonin promotes the survival of cortical glutamatergic neurons in vitro. Exp. Neurol 148, 205–214. [DOI] [PubMed] [Google Scholar]
- Duque A, Krsnik Z, Kostović I, and Rakic P (2016). Secondary expansion of the transient subplate zone in the developing cerebrum of human and nonhuman primates. Proc. Natl. Acad. Sci. USA 113, 9892–9897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fietz SA, Kelava I, Vogt J, Wilsch-Brӓuninger M, Stenzel D, Fish JL, Corbeil D, Riehn A, Distler W, Nitsch R, and Huttner WB (2010). OSVZ progenitors of human and ferret neocortex are epithelial-like and expand by integrin signaling. Nat. Neurosci 13, 690–699. [DOI] [PubMed] [Google Scholar]
- Finak G, McDavid A, Yajima M, Deng J, Gersuk V, Shalek AK, Slichter CK, Miller HW, McElrath MJ, Prlic M, et al. (2015). MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol 16, 278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint AC, Maisch US, Weishaupt JH, Kriegstein AR, and Monyer H (1997). NR2A subunit expression shortens NMDA receptor synaptic currents in developing neocortex. J. Neurosci 17, 2469–2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint AC, Dammerman RS, and Kriegstein AR (1999). Endogenous activation of metabotropic glutamate receptors in neocortical development causes neuronal calcium oscillations. Proc. Natl. Acad. Sci. USA 96, 12144–12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuzik J, Zeisel A, Máté Z, Calvigioni D, Yanagawa Y, Szabó G, Linnarsson S, and Harkany T(2016). Integration of electrophysiological recordings with single-cell RNA-seq data identifies neuronal subtypes. Nat. Biotechnol 34, 175–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garaschuk O, Linn J, Eilers J, and Konnerth A (2000). Large-scale oscillatory calcium waves in the immature cortex. Nat. Neurosci 3, 452–459. [DOI] [PubMed] [Google Scholar]
- Gingrich JA, Malm H, Ansorge MS, Brown A, Sourander A, Suri D, Teixeira CM, Caffrey Cagliostro MK, Mahadevia D, and Weissman MM (2017). New insights into how serotonin selective reuptake inhibitors shape the developing brain. Birth Defects Res 109, 924–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagberg GB, Blomstrand F, Nilsson M, Tamir H, and Hansson E (1998). Stimulation of 5-HT2A receptors on astrocytes in primary culture opens voltage-independent Ca2+ channels. Neurochem. Int 32, 153–162. [DOI] [PubMed] [Google Scholar]
- Hanganu IL, Kilb W, and Luhmann HJ (2002). Functional synaptic projections onto subplate neurons in neonatal rat somatosensory cortex. J. Neurosci 22, 7165–7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen DV, Lui JH, Parker PRL, and Kriegstein AR (2010). Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature 464, 554–561. [DOI] [PubMed] [Google Scholar]
- Hansen DV, Lui JH, Flandin P, Yoshikawa K, Rubenstein JL, Alvarez-Buylla A, and Kriegstein AR (2013). Non-epithelial stem cells and cortical interneuron production in the human ganglionic eminences. Nat. Neurosci 16, 1576–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie T, and Stuetzle W (1989). Principal curves. J. Am. Stat. Assoc 84, 502–516. [Google Scholar]
- Haydar TF, Wang F, Schwartz ML, and Rakic P (2000). Differential modulation of proliferation in the neocortical ventricular and subventricular zones. J. Neurosci 20, 5764–5774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong F, Breitling R, McEntee CW, Wittner BS, Nemhauser JL, and Chory J (2006). RankProd: a bioconductor package for detecting differentially expressed genes in meta-analysis. Bioinformatics 22, 2825–2827. [DOI] [PubMed] [Google Scholar]
- Jiang H, Jiang W, Zou J, Wang B, Yu M, Pan Y, Lin Y, Mao Y, and Wang Y (2015). The GluN2B subunit of N-methy-D-asparate receptor regulates the radial migration of cortical neurons in vivo. Brain Res 1610, 20–32. [DOI] [PubMed] [Google Scholar]
- Káradóttir R, Cavelier P, Bergersen LH, and Attwell D (2005). NMDA receptors are expressed in oligodendrocytes and activated in ischaemia. Nature 438, 1162–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, and Salzberg SL (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostović I, and Judaš M (2010). The development of the subplate and thalamocortical connections in the human foetal brain. Acta Paediatr 99, 1119–1127. [DOI] [PubMed] [Google Scholar]
- Kowalczyk T, Pontious A, Englund C, Daza R, Bedogni F, Hodge RD, Attardo A, Bell C, Huttner WB, and Hevner RF (2009). Intermediate neuronal progenitors (basal progenitors) produce pyramidal–projection neurons for all layers of cerebral cortex. Cereb. Cortex 19, 2439–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalo U, Pankratov Y, Kirchhoff F, North RA, and Verkhratsky A (2006). NMDA receptors mediate neuron-to-glia signaling in mouse cortical astrocytes. J. Neurosci 26, 2673–2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Zhang B, and Horvath S (2008). Defining clusters from a hierarchical cluster tree: the Dynamic Tree Cut package for R. Bioinformatics 24, 719–720. [DOI] [PubMed] [Google Scholar]
- Le Magueresse C, and Monyer H (2013). GABAergic interneurons shape the functional maturation of the cortex. Neuron 77, 388–405. [DOI] [PubMed] [Google Scholar]
- Legland D, Arganda-Carreras I, and Andrey P (2016). MorphoLibJ: integrated library and plugins for mathematical morphology with ImageJ. Bioinformatics 32, 3532–3534. [DOI] [PubMed] [Google Scholar]
- Lehtinen MK, Zappaterra MW, Chen X, Yang YJ, Hill AD, Lun M, Maynard T, Gonzalez D, Kim S, Ye P, et al. (2011). The cerebrospinal fluid provides a proliferative niche for neural progenitor cells. Neuron 69, 893–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CH, and Tam PKS (1998). An iterative algorithm for minimum cross entropy thresholding. Pattern Recognit. Lett 19, 771–776. [Google Scholar]
- Li S, Kumar TP, Joshee S, Kirschstein T, Subburaju S, Khalili JS, Kloepper J, Du C, Elkhal A, Szabó G, et al. (2018). Endothelial cell-derived GABA signaling modulates neuronal migration and postnatal behavior. Cell Res 28, 221–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, and Shi W (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. [DOI] [PubMed] [Google Scholar]
- Lien WH, Klezovitch O, Fernandez TE, Delrow J, and Vasioukhin V (2006). alphaE-catenin controls cerebral cortical size by regulating the hedge-hog signaling pathway. Science 311, 1609–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Hashimoto-Torii K, Torii M, Haydar TF, and Rakic P (2008). The role of ATP signaling in the migration of intermediate neuronal progenitors to the neocortical subventricular zone. Proc. Natl. Acad. Sci. USA 105, 11802–11807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LoTurco JJ, Owens DF, Heath MJS, Davis MBE, and Kriegstein AR (1995). GABA and glutamate depolarize cortical progenitor cells and inhibit DNA synthesis. Neuron 15, 1287–1298. [DOI] [PubMed] [Google Scholar]
- Lowe DG (2004). Distinctive image features from scale-invariant keypoints. Int. J. Comput. Vis 60, 91–110. [Google Scholar]
- Luhmann HJ, Fukuda A, and Kilb W (2015). Control of cortical neuronal migration by glutamate and GABA. Front. Cell. Neurosci 9, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui JH, Hansen DV, and Kriegstein AR (2011). Development and evolution of the human neocortex. Cell 146, 18–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui JH, Nowakowski TJ, Pollen AA, Javaherian A, Kriegstein AR, and Oldham MC (2014). Radial glia require PDGFD-PDGFRβ signalling in human but not mouse neocortex. Nature 515, 264–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luscher B, Fuchs T, and Kilpatrick CL (2011). GABAA receptor trafficking-mediated plasticity of inhibitory synapses. Neuron 70, 385–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, and Barker JL (1995). Complementary expressions of transcripts encoding GAD67 and GABAA receptor a4, bl, and gl subunits in the proliferative zone of the embryonic rat central nervous system. J. Neurosci 15, 2547–2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Maric D, Li BS, Hu Q, Andreadis JD, Grant GM, Liu QY, Shaffer KM, Chang YH, Zhang L, et al. (2000). Acetylcholine stimulates cortical precursor cell proliferation in vitro via muscarinic receptor activation and MAP kinase phosphorylation. Eur. J. Neurosci 12, 1227–1240. [DOI] [PubMed] [Google Scholar]
- Manent J-B, Demarque M, Jorquera I, Pellegrino C, Ben-Ari Y, Aniksztejn L, and Represa A (2005). A noncanonical release of GABA and glutamate modulates neuronal migration. J. Neurosci 25, 4755–4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín-Padilla M (1992). Ontogenesis of the pyramidal cell of the mammalian neocortex and developmental cytoarchitectonics: a unifying theory. J. Comp. Neurol 321, 223–240. [DOI] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Casagrande JT, and Thomas PD (2013). Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc 8, 1551–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffitt JR, Bambah-Mukku D, Eichhorn SW, Vaughn E, Shekhar K, Perez JD, Rubinstein ND, Hao J, Regev A, Dulac C, et al. (2018). Molecular, spatial and functional single-cell profiling of the hypothalamic preoptic region. Science 362, eaau5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molliver ME, Kostović I, and van der Loos H (1973). The development of synapses in cerebral cortex of the human fetus. Brain Res 50, 403–407. [DOI] [PubMed] [Google Scholar]
- Molyneaux BJ, Goff LA, Brettler AC, Chen HH, Hrvatin S, Rinn JL, and Arlotta P (2015). DeCoN: genome-wide analysis of in vivo transcriptional dynamics during pyramidal neuron fate selection in neocortex. Neuron 85, 275–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore AR, Zhou W-L, Jakovcevski I, Zecevic N, and Antic SD (2011). Spontaneous electrical activity in the human fetal cortex in vitro. J. Neurosci 31, 2391–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore AR, Zhou W-L, Sirois CL, Belinsky GS, Zecevic N, and Antic SD (2014). Connexin hemichannels contribute to spontaneous electrical activity in the human fetal cortex. Proc. Natl. Acad. Sci. USA 111, E3919–E3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow J, Russell A, Guthrie E, Parsons L, Robertson I, Waddell R, Irwin B, McGivern RC, Morrison PJ, and Craig J (2006). Malformation risks of antiepileptic drugs in pregnancy: a prospective study from the UK Epilepsy and Pregnancy Register. J. Neurol. Neurosurg. Psychiatry 77, 193–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muja M, and Lowe DG (2009). Fast approximate nearest neighbors with automatic algorithm configuration. VISAPP 2009 - Proceedings of the 4th International Conference on Computer Vision Theory and Applications 1, 331–340. [Google Scholar]
- Muth-Köhne E, Terhag J, Pahl S, Werner M, Joshi I, and Hollmann M (2010a). Functional excitatory GABAA receptors precede ionotropic glutamate receptors in radial glia-like neural stem cells. Mol. Cell. Neurosci 43, 209–221. [DOI] [PubMed] [Google Scholar]
- Muth-Köne E, Pachernegg S, Karus M, Faissner A, and Hollmann M (2010b). Expression of NMDA receptors and Ca2+-impermeable AMPA receptors requires neuronal differentiation and allows discrimination between two different types of neural stem cells. Cell. Physiol. Biochem 26, 935–946. [DOI] [PubMed] [Google Scholar]
- Ng AY, and Jordan MI (2002). On discriminative vs. generative classifiers: A comparison of logistic regression and naive Bayes. Adv. Neural Inf. Process. Syst 2, 841–848. [Google Scholar]
- Noctor SC, Martínez-Cerdeño V, Ivic L, and Kriegstein AR (2004). Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat. Neurosci 7, 136–144. [DOI] [PubMed] [Google Scholar]
- Nowakowski TJ, Bhaduri A, Pollen AA, Alvarado B, Mostajo-Radji MA, Di Lullo E, Haeussler M, Sandoval-Espinosa C, Liu SJ, Velmeshev D, et al. (2017). Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science 358, 1318–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtaka-Maruyama C, Okamoto M, Endo K, Oshima M, Kaneko N, Yura K, Okado H, Miyata T, and Maeda N (2018). Synaptic transmission from subplate neurons controls radial migration of neocortical neurons. Science 360, 313–317. [DOI] [PubMed] [Google Scholar]
- Ohtani A, Kozono N, Senzaki K, and Shiga T (2014). Serotonin 2A receptor regulates microtubule assembly and induces dynamics of dendritic growth cones in rat cortical neurons in vitro. Neurosci. Res. 81–82, 11–20. [DOI] [PubMed] [Google Scholar]
- Olmos-Serrano JL, Kang HJ, Tyler WA, Silbereis JC, Cheng F, Zhu Y, Pletikos M, Jankovic-Rapan L, Cramer NP, Galdzicki Z, et al. (2016). Down syndrome developmental brain transcriptome reveals defective oligodendrocyte differentiation and myelination. Neuron 89, 1208–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens DF, Liu X, Kriegstein AR, Porcher C, Hatchett C, Longbottom RE, Mcainch K, Talvinder S, Moss SJ, Thomson AM, et al. (1999). Changing properties of GABA A receptor-mediated signaling during early neocortical development. J. Neurophysiol. 82, 570–583. [DOI] [PubMed] [Google Scholar]
- Paoletti P, Bellone C, and Zhou Q (2013). NMDA receptor subunit diversity: impact on receptor properties, synaptic plasticity and disease. Nat. Rev. Neurosci 14, 383–400. [DOI] [PubMed] [Google Scholar]
- Phansalkar N, More S, Sabale A, and Joshi M (2011). Adaptive local thresholding for detection of nuclei in diversity stained cytology images. 2011 International Conference on Communications and Signal Processing, 218–220. [Google Scholar]
- Pollen AA, Nowakowski TJ, Chen J, Retallack H, Sandoval-Espinosa C, Nicholas CR, Shuga J, Liu SJ, Oldham MC, Diaz A, et al. (2015). Molecular identity of human outer radial glia during cortical development. Cell 163, 55–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P (2000). Radial unit hypothesis of neocortical expansion. Novartis Found. Symp 228, 30–42, discussion 42–52. [DOI] [PubMed] [Google Scholar]
- Ramalingam N, Fowler B, Szpankowski L, Leyrat AA, Hukari K, Maung MT, Yorza W, Norris M, Cesar C, Shuga J, et al. (2016). Fluidic logic used in a systems approach to enable integrated single-cell functional analysis. Front. Bioeng. Biotechnol 4, 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regev A, Teichmann SA, Lander ES, Amit I, Benoist C, Yosef N, Birney E, Bodenmiller B, Campbell P, Carninci P, et al. (2017). The human cell atlas. Elife 6, e27041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reillo I, de Juan Romero C, Cárdenas A, Clascá F, Martίnez-Martinez MÁ, and Borrell V (2017). A complex code of extrinsic influences on cortical progenitor cells of higher mammals. Cereb. Cortex 27, 4586–4606. [DOI] [PubMed] [Google Scholar]
- Rogawski MA, and Löscher W (2004). The neurobiology of antiepileptic drugs. Nat. Rev. Neurosci. 5, 553–564. [DOI] [PubMed] [Google Scholar]
- Roppongi RT, Kojima N, Hanamura K, Yamazaki H, and Shirao T (2013). Selective reduction of drebrin and actin in dendritic spines of hippocampal neurons by activation of 5-HT(2A) receptors. Neurosci. Lett 547, 76–81. [DOI] [PubMed] [Google Scholar]
- Sarnat HB, Flores-Sarnat L, and Trevenen CL (2010). Synaptophysin immunoreactivity in the human hippocampus and neocortex from 6 to 41 weeks of gestation. J. Neuropathol. Exp. Neurol 69, 234–245. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedmak G, Jovanov-Milošević N, Puskarjov M, Ulamec M, Krušlin B, Kaila K, and Judaš M (2016). Developmental expression patterns of KCC2 and functionally associated molecules in the human brain. Cereb. Cortex 26, 4574–4589. [DOI] [PubMed] [Google Scholar]
- Shitamukai A, Konno D, and Matsuzaki F (2011). Oblique radial glial divisions in the developing mouse neocortex induce self-renewing progenitors outside the germinal zone that resemble primate outer subventricular zone progenitors. J. Neurosci 31, 3683–3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- signal Developers (2013). signal: Signal processing http://r-forge.r-project.org/projects/signal/
- Sousa AMM, Zhu Y, Raghanti MA, Kitchen RR, Onorati M, Tebbenkamp ATN, Stutz B, Meyer KA, Li M, Kawasawa YI, et al. (2017). Molecular and cellular reorganization of neural circuits in the human lineage. Science 358, 1027–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundström E, Kölare S, Souverbie F, Samuelsson EB, Pschera H, Lunell NO, and Seiger A (1993). Neurochemical differentiation of human bulbospinal monoaminergic neurons during the first trimester. Brain Res. Dev. Brain Res 75, 1–12. [DOI] [PubMed] [Google Scholar]
- Supek F, Bošnjak M, Škunca N, and Šmuc T (2011). REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 6, e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sztainberg Y, and Zoghbi HY (2016). Lessons learned from studying syndromic autism spectrum disorders. Nat. Neurosci 19, 1408–1417. [DOI] [PubMed] [Google Scholar]
- Thompson CL, Ng L, Menon V, Martinez S, Lee CK, Glattfelder K, Sunkin SM, Henry A, Lau C, Dang C, et al. (2014). A high-resolution spatiotemporal atlas of gene expression of the developing mouse brain. Neuron 83, 309–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen ER, Mich JK, Yao Z, Hodge RD, Doyle AM, Jang S, Shehata SI, Nelson AM, Shapovalova NV, Levi BP, and Ramanathan S (2016). Fixed single-cell transcriptomic characterization of human radial glial diversity. Nat. Methods 13, 87–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibshirani R, Hastie T, Narasimhan B, and Chu G (2002). Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc. Natl. Acad. Sci. USA 99, 6567–6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T, Homma D, Tokuoka H, Hayakawa I, Sugimoto Y, Ichinose H, and Kawasaki H (2013). Birth regulates the initiation of sensory map formation through serotonin signaling. Dev. Cell 27, 32–46. [DOI] [PubMed] [Google Scholar]
- Tong CK, Chen J, Cebrián-Silla A, Mirzadeh Z, Obernier K, Guinto CD, Tecott LH, García-Verdugo JM, Kriegstein A, and Alvarez-Buylla A (2014). Axonal control of the adult neural stem cell niche. Cell Stem Cell 14, 500–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torre E, Dueck H, Shaffer S, Gospocic J, Gupte R, Bonasio R, Kim J, Murray J, and Raj A (2018). Rare cell detection by single-cell RNA sequencing as guided by single-molecule RNA FISH. Cell Syst 6, 171–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuoc TC, Boretius S, Sansom SN, Pitulescu ME, Frahm J, Livesey FJ, and Stoykova A (2013). Chromatin regulation by BAF170 controls cerebral cortical size and thickness. Dev. Cell 25, 256–269. [DOI] [PubMed] [Google Scholar]
- Unichenko P, Kirischuk S, and Luhmann HJ (2015). GABA transporters control GABAergic neurotransmission in the mouse subplate. Neuroscience 304, 217–227. [DOI] [PubMed] [Google Scholar]
- van der Maaten L, and Hinton G (2008). Visualizing high data using t-sne. J. Mach. Learn. Res 9, 2579–2605. [Google Scholar]
- Verney C, Lebrand C, and Gaspar P (2002). Changing distribution of monoaminergic markers in the developing human cerebral cortex with special emphasis on the serotonin transporter. Anat. Rec 267, 87–93. [DOI] [PubMed] [Google Scholar]
- Vitalis T, and Verney C (2017). Sculpting cerebral cortex with serotonin in rodent and primate. In Serotonin - A Chemical Messenger Between All Types of Living Cells (InTech)
- Vitalis T, Cases O, Passemard S, Callebert J, and Parnavelas JG (2007). Embryonic depletion of serotonin affects cortical development. Eur. J. Neurosci 26, 331–344. [DOI] [PubMed] [Google Scholar]
- Wagner T, and Lipinski H-G (2013). IJBlob: an ImageJ library for connected component analysis and shape analysis. J. Open Res. Softw 1, e6. [Google Scholar]
- Wang X, Tsai J-W, LaMonica B, and Kriegstein AR (2011). A new subtype of progenitor cell in the mouse embryonic neocortex. Nat. Neurosci 14, 555–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman TA, Riquelme PA, Ivic L, Flint AC, and Kriegstein AR (2004). Calcium waves propagate through radial glial cells and modulate proliferation in the developing neocortex. Neuron 43, 647–661. [DOI] [PubMed] [Google Scholar]
- Yuzwa SA, Yang G, Borrett MJ, Clarke G, Cancino GI, Zahr SK, Zandstra PW, Kaplan DR, and Miller FD (2016). Proneurogenic ligands defined by modeling developing cortex growth factor communication networks. Neuron 91, 988–1004. [DOI] [PubMed] [Google Scholar]
- Zahr SK, Yang G, Kazan H, Borrett MJ, Yuzwa SA, Voronova A, Kaplan DR, and Miller FD (2018). A translational repression complex in developing mammalian neural stem cells that regulates neuronal specification. Neuron 97, 520–537. [DOI] [PubMed] [Google Scholar]
- Zeng H, and Sanes JR (2017). Neuronal cell-type classification: challenges, opportunities and the path forward. Nat. Rev. Neurosci 18, 530–546. [DOI] [PubMed] [Google Scholar]
- Zhu Q, Shah S, Dries R, Cai L, and Yuan G-C (2018). Identification of spatially associated subpopulations by combining scRNAseq and sequential fluorescence in situ hybridization data. Nat. Biotechnol 36, 1183–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Next Generation Sequencing data will be available on dbGaP: phs000989.


