Abstract
Objective:
The aim of the study was to perform mRNA-miRNA regulatory network analyses to identify a miRNA panel for molecular subtype identification and stratification of high-risk PDAC patients.
Summary and Background data:
Recent transcriptional profiling effort in pancreatic ductal adenocarcinoma (PDAC) has led to the identification of molecular subtypes that associate with poor survival; however, their clinical significance for risk-stratification in PDAC patients has been challenging.
Methods:
By performing a systematic analysis in TCGA and ICGC cohorts, we discovered a panel of miRNAs that associated with squamous and other poor molecular subtypes in PDAC. Subsequently, we used logistic regression analysis to develop models for risk-stratification and Cox’s proportional hazard analysis to determine survival prediction probability of this signature in multiple cohorts of 433 PDAC patients, including a tissue cohort (n=199) and a preoperative serum cohort (n=51).
Results:
We identified a panel of 9-miRNAs that were significantly upregulated (miR-205–5p and −934) or downregulated (miR-192–5p, 194–5p, 194–3p, 215–5p, 375–3p, 552–3p and 1251–5p) in PDAC molecular subtypes with poor survival (squamous,AUC=0.90; basal,AUC=0.89; and quasi-mesenchymal,AUC=0.83). The validation of this miRNA panel in a tissue clinical cohort was a significant predictor of OS (HR=2.48, p<0.0001), and this predictive accuracy improved further in a risk-nomogram which included key clinicopathological factors. Finally, we were able to successfully translate this miRNA predictive signature into a liquid-biopsy based assay in pre-operative serum specimens from PDAC patients (HR: 2.85, p=0.02).
Conclusion:
We report a novel miRNA risk-stratification signature that can be used as a non-invasive assay for the identification of high-risk patients and potential disease-monitoring in PDAC patients.
Keywords: Pancreatic ductal adenocarcinoma, overall survival, molecular subtypes, miRNA, prognosis, risk-stratification
MINI ABSTRACT
In this study, by performing a mRNA-miRNA regulatory network analyses, we identified a novel miRNA risk-stratification signature that can be used as a non-invasive assay for the identification of high-risk patients and potential disease-monitoring in patients with pancreatic ductal adenocarcinoma.
INTRODUCTION
Pancreatic ductal adenocarcinoma (PDAC) is currently the fourth leading cause of cancer-related deaths, but is projected to become the second most common cancer in the Western countries in the next decade 1. The prognosis for PDAC patients is quite poor, with 5-year survival rates less than 9%. This dismal prognosis is in part due to disease diagnosis at an advanced stage, when the patients are unable to undergo curative resection of their tumor 2. Furthermore, even those patients who undergo curative surgery generally experience high frequency of tumor recurrence; and neoadjuvant treatment or chemoradiotherapy has only moderately improved survival outcomes in PDAC patients 3. Moreover, unlike other cancers, conventional clinicopathological features are often inadequate at providing an accurate measure of prognosis in PDAC patients. One possible explanation for this variable prognosis in PDAC is the inherent tumor heterogeneity, as well as unusually higher stromal content 4. Several driver mutations have been identified in PDAC, including KRAS, p16, TP53, CDKN2A, and SMAD4 genes 5–7; and transcriptomic data has repeatedly confirmed that majority of patients have diverse genomic alterations 8, making it harder to precisely target this disease. This highlights the need to gain additional insights into the molecular characteristics of PDAC patients for tailoring personalized treatment strategies and improving disease outcomes in this lethal malignancy.
With recent advances in high-throughput molecular profiling technologies, most human cancers have now been comprehensively profiled, which has led to the recognition of previously unrecognized molecular subtypes 9–11. Most recently, the International Cancer Genome Consortium (ICGC) conducted an integrated genomic analysis of 456 PDAC patients to reveal four molecular subtypes in PDAC, which correlated with distinct histopathological characteristics: i) squamous, ii) pancreatic progenitor, iii) immunogenic, and iv) aberrantly differentiated endocrine exocrine (ADEX) 12. More importantly, this molecular classification discovered that PDAC patients with the squamous subtype exhibited significantly poor prognosis. Subsequently, three follow-up studies have identified similar molecular subtypes of their own, which particularly associate with poor outcomes in PDAC patients 13–15. Even though molecular subtypes are biologically intriguing, their translation into the clinic has been challenging. This has primarily because of a few key reasons including: each subtype comprises of several hundred genes, which makes it difficult to perform transcriptomic profiling in each patient, especially in RNA derived from paraffin-embedded tissues. In addition, each molecular subtyping effort proposed their own classification criteria for defining subtype(s) with poor prognosis, which leads to lack of a consensus gene panel for interrogation in clinical settings.
In this study, we attempted to address some of these concerns by performing a systematic and comprehensive biomarker discovery effort, which allowed us to develop a robust biomarker panel that represents all previously described molecular subtypes with poor outcomes in PDAC patients. We based our effort on the hypothesis that since a single microRNA (miRNA) can regulate expression of hundreds of target genes, perhaps a panel of miRNAs that control the expression of candidate genes within each PDAC subtype might offer a more robust and clinically meaningful signature. This approach gains further credence from the fact that previous studies have convincingly demonstrated that miRNA expression is frequently dysregulated in various malignancies 16, 17, including PDAC 18, 19; and emerging evidence indicates that miRNAs might serve as important biomarkers in PDAC patients 20, 21. Furthermore, considering that miRNA expression is generally stable in a variety of clinical specimens makes them attractive surrogates in situations where analysis of gene expression is challenging. In addition, due the ease of their detection in bodily fluids, miRNAs biomarkers have the potential for development into liquid biopsy-based assays for an easier translation into the clinic.
Herein, we undertook a comprehensive biomarker discovery effort by analyzing multiple high-throughput sequencing datasets to identify poor-prognostic subtype-associated miRNAs in PDAC. Following an extensive bioinformatic analysis and biomarker validation, we established a 9-miRNA panel that exhibited excellent prognostic potential in predicting survival in PDAC patients. More importantly, we successfully translated these tissue-based biomarkers into blood-based liquid biopsy assays, highlighting their exciting translational potential as non-invasive prognostic and potential disease-monitoring biomarkers in PDAC patients, considering that majority of these patients have either an unresectable or a borderline resectable disease.
MATERIALS AND METHODS
Patients and datasets
During the biomarker discovery phase, datasets from the International Cancer Genome Consortium (ICGC, n=96) and the Cancer Genome Atlas (TCGA, n=183), were analyzed for the identification of candidate miRNAs specifically associated with the ‘squamous subtype’, and those which predicted poor overall survival in PDAC patients. The robustness and prognostic performance of candidate biomarkers was validated in two independent patient cohorts, comprised of tissue and serum specimens. A written informed consent was obtained from all patients, and the Institutional Review Boards of all participating institutions approved the study.
In the validation phase, we examined 199 retrospectively collected formalin-fixed paraffin-embedded (FFPE) primary PDAC tissue and 51 preoperative serum specimens. The tissue specimens were collected from PDAC patients enrolled at the Kumamoto University Hospital, Japan from 2009–2014, and Nagoya University Hospital, Japan from 2002–2010. The serum specimens were collected from PDAC patients enrolled from 2010–2014, at the Medical College of Wisconsin, USA. Patient demographics and clinicopathological characteristics are summarized in Table 1.
Table 1:
Clinicopathological characteristics of patients in the clinical cohorts
| Tissue cohort | Serum cohort | ||
|---|---|---|---|
|
| |||
| Variable | number | number | |
|
| |||
| Gender | male | 116 | 30 |
| female | 83 | 21 | |
| Age at operation | median (range) | 67 (35 – 90) | 67 (45 – 87) |
| CA19-9 value (U/ml) | median | 133.8 | 79.5 |
| Neoadjuvant chemotherapy | No | 183 | 8 |
| Yes | 16 | 35 | |
| Missing | 0 | 8 | |
| Tumor location | pancreatic head | 146 | 37 |
| pancreatic body or tail | 53 | 14 | |
| Tumor size (mm) | median (range) | 30 (4 – 65) | 30 (9 – 110) |
| Histological type | well or moderate | 169 | 45 |
| poorly | 30 | 6 | |
| T factor | T1 or T2 | 10 | 9 |
| T3 or T4 | 189 | 42 | |
| Lymph node metastasis | negative | 56 | 27 |
| positive | 143 | 24 | |
| Distant metastasis | negative | 186 | 41 |
| positive | 13 | 10 | |
| UICC Stage | IA or IB or IIA | 55 | 22 |
| IIB or III | 131 | 19 | |
| IV | 13 | 10 | |
| Lymphatic invasion | negative | 34 | 27 |
| positive | 165 | 24 | |
| Venous invasion | negative | 63 | NA |
| positive | 136 | NA | |
Identification of squamous subtype-associated miRNA biomarkers
For these analyses, we acquired publicly available RNA sequencing data from the ICGC, as well as RNA and miRNA sequencing data from the TCGA HiSeq dataset. Within the TCGA cohort of 183 PDAC patients, we successfully analyzed expression profiles of 1046 miRNAs and 20,520 mRNAs. Since transcriptomic subtyping information was only available for the patients within the ICGC dataset, we used these data to predict PDAC subtypes in the TCGA cohort using ‘Prediction Analysis for Microarrays R’ tool (PAMR) 22. This allowed us to identify miRNAs specific for the squamous subtype using TCGA subtype labels. Detailed methodology for these analyses and a study design flow chart is provided in the Supplementary Figure 1.
RNA isolation and qRT-PCR assays
RNA enriched for small non-coding RNAs was extracted from the FFPE tissue and serum specimens using the miRNeasy FFPE Kit or the miRNeasy serum kit (Qiagen, Valencia, CA). The expression of miRNAs was quantified by TaqMan real-time qRT-PCR assays (Applied Biosystems, Foster City, CA) using QuantStudio™ 7 Flex Real-Time PCR System (Applied Biosystems). The assay IDs for qRT-PCR of miR-192–5p, miR-194–5p, miR-194–3p, miR-205–5p, miR-215–5p, miR-375–3p, miR-552–3p, miR-934, and miR-1251–5p were as follows (Assay No: 000491, 000493, 002379, 000509, 000518, 000564, 001520, 002177, and 002820, Catalog No: 4427975, Thermo Fisher Scientific, Waltham, MA). U6 and RNU44 (Assay No: 001973, and 001094, Cat no: 4427975, Thermo Fisher Scientific) were used as endogenous controls for data normalization. Expression of each miRNA was calculated using 2-ΔCT method.
Statistical analysis
The overall survival (OS) was determined from the date the patient underwent surgery until the date of death resulting from any cause (i.e. cancer-unrelated deaths were not censored), or last known follow-up for patients that were still alive. The Cox’s proportional hazards model was used to estimate hazard ratios (HRs) for death, and to predict other factors influencing OS. Receiver operating characteristic (ROC) curves were established to determine the cut-off thresholds for analyzing OS. Youden’s index was used to determine the optimal cutoff values for individual miRNAs, as well as the combined miRNA panel risk scores for dichotomizing patients into low and high expression groups. For time-to-event analyses, survival estimates were calculated using the Kaplan-Meier analysis and the groups were compared using the log-rank test. Forced-entry regression was used to include these variables in all multivariate analysis to analyze whether each of the predictors affected the outcome after adjustment for known confounders. To determine the accuracy of our 9-miRNA panel in determining the squamous and other PDAC subtypes with poor outcomes published previously, we used a logistic regression model by including all 9-miRNAs. The logistic regression model derived from the 9-miRNA panel was used to plot the AUCs in all three cohorts. All p-values were 2-sided, and values <0.05 were considered statistically significant. All statistical analyses were performed using the Medcalc statistical software Ver.16.2.0 (Medcalc Software bvba, Ostend, Belgium), JMP software 10.0.2 (SAS Institute, Cary, NC), and GraphPad Prism Ver. 7.0 (GraphPad Software, San Diego, CA).
RESULTS
Identification of a miRNA network that associated with molecular subtypes associated with poor prognosis in PDAC
We first analyzed ICGC and TCGA datasets using PAMR and LIMMA-VOOM tools to identify miRNAs that were specifically deregulated in the squamous PDAC subtype. Firstly, for subtype prediction in the TCGA cohort, we computed the gene-wise variance for ICGC RNA-seq data and determined top 3,000 highest discriminant genes. Next, we identified a total of 2,049 overlapping genes between the TCGA and ICGC RNA-seq data. Next, for the training set analyses, we applied PAMR tool on the ICGC data containing these 2,049 common genes and 96 labeled samples with known subtype information and obtained a cut-off threshold (=2.0768) through cross-validation of samples where error-rate was minimum (=0.052; Figure 1A). As a test set, we then analyzed the TCGA dataset containing these 2,049 common genes and 183 unlabeled samples with no subtype information and predicted poor molecular subtypes of this cohort using the same cut-off threshold (=2.0768) obtained in the test set.
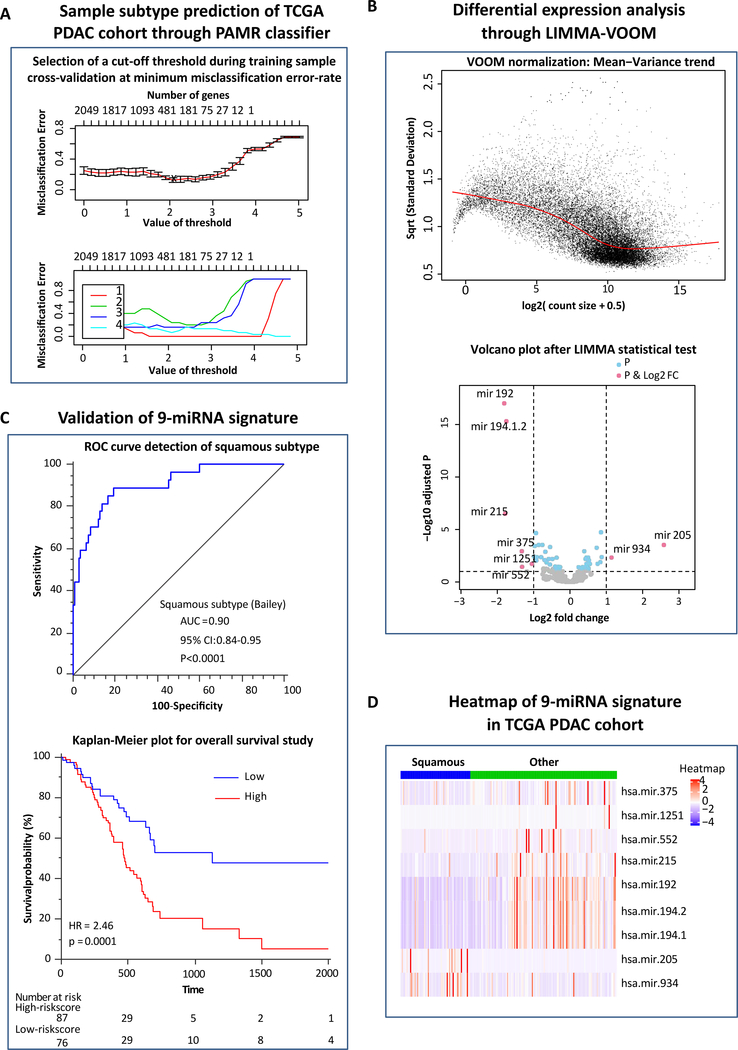
Figure 1: Identification of a panel poor molecular PDAC-subtype associated miRNAs and their potential for predicting survival outcomes in PDAC patients.
(A) Selection of threshold through cross-validation of underlying PDAC cases with a minimum misclassification error-rate used for sample subtype prediction in the TCGA RNA-seq data through PAMR classifier. (B) Voom normalization plot prior to Limma fit (empirical Bayes test using linear model) in which the read counts were converted into logCPMs (log2 counts per million) while considering the mean-variance relationship in the profile. (C) Volcano plot showing p-value and log2 fold change filtering to identify differentially expressed miRNAs (up-regulated miRNAs and down-regulated miRNAs). (D) Heatmap of the 9-squamous subtype associated miRNAs in the TCGA PDAC cohort. (E) Receiver operating characteristic (ROC) curve of the combined 9-miRNA signature for the detection of squamous subtype in Bailey classification. (F) Kaplan-Meier plot of the 9-miRNA signature for overall survival (OS) in the TCGA discovery cohort.
During these analyses, 12 cases segregated into ADEX subtype, 19 into Immunogenic, 59 into Squamous and remaining 93 into Pancreatic Progenitor subtype. We then conducted LIMMA-VOOM (Figure 1B) statistical analysis using pairwise class-label information and the p-value correction and identified a panel of 9 differentially expressed miRNAs (DEMIRs) associated with a poor molecular PDAC subtypes (Figure 1C and D). This included two miRNAs (hsa-mir-205 and hsa-mir-934) that were up-regulated (URMIRs), while the remaining seven (hsa-mir-192, hsa-mir-215, hsa-mir-375, hsa-mir-1251, hsa-mir-552, hsa-mir-194.1 and hsa-mir-194.2) were down-regulated (DRMIRs). The AUC values of all individual miRNAs for identifying squamous subtype from Bailey, basal from Moffitt and quasi-mesenchymal from Collisson classifications are illustrated in Supplementary Table 1. Our combined 9-miRNA panel yielded an AUC of 0.90, 0.89 and 0.83 for the identification of squamous (Figure 1E), basal and quasi-mesenchymal subtypes respectively (Supplementary Figures 2A and 2B). Taken together, our miRNA-mRNA network-based approach allowed us to identify a clinically translatable miRNA classifier that robustly detected poor molecular PDAC subtypes.
The 9-miRNA panel significantly allowed risk-stratification and predicting survival outcomes in PDAC patients
To evaluate the prognostic significance of the 9-miRNA panel identified in the discovery phase, we first interrogated the relationship between their expression levels and overall survival (OS) in PDAC patients in the TCGA cohort. The patients with high expression of miR-205–5p and miR-934 exhibited worse survival vs. the low-expression group (p=0.001 and p=0.005, respectively). Likewise, low expression of miR-192–5p, miR-194–5p, miR-194–3p and miR-375–3p, significantly associated with poor OS (p=0.009, 0.04, 0.04 and 0.003, respectively) in PDAC patients. On similar lines, low-expression of miR-215–5p and miR-1251–5p also depicted a similar trend for unfavorable survival (p=0.06 and p=0.05; Supplementary Figure 3).
Next, we performed a multivariate cox-regression analysis by combining all 9-miRNAs together and used Youden’s index to dichotomize all patients into high- and low-risk groups. As expected, patients with miRNA high-risk scores exhibited a significantly reduced 5-year OS (5.1%) vs. the miRNA low-risk group (47.6%), with a corresponding HR of 2.46 (95% CI: 1.60–3.79, p=0.0001; Figure 1F). The waterfall plot shown in Supplementary Figure 2C, also demonstrates the robust prognostic performance of our 9-miRNA signature. Collectively, these results illustrate the clinical significance of our miRNA signature in risk-stratification for predicting survival in PDAC patients.
The 9-miRNA signature successfully validated in an independent PDAC patient cohort
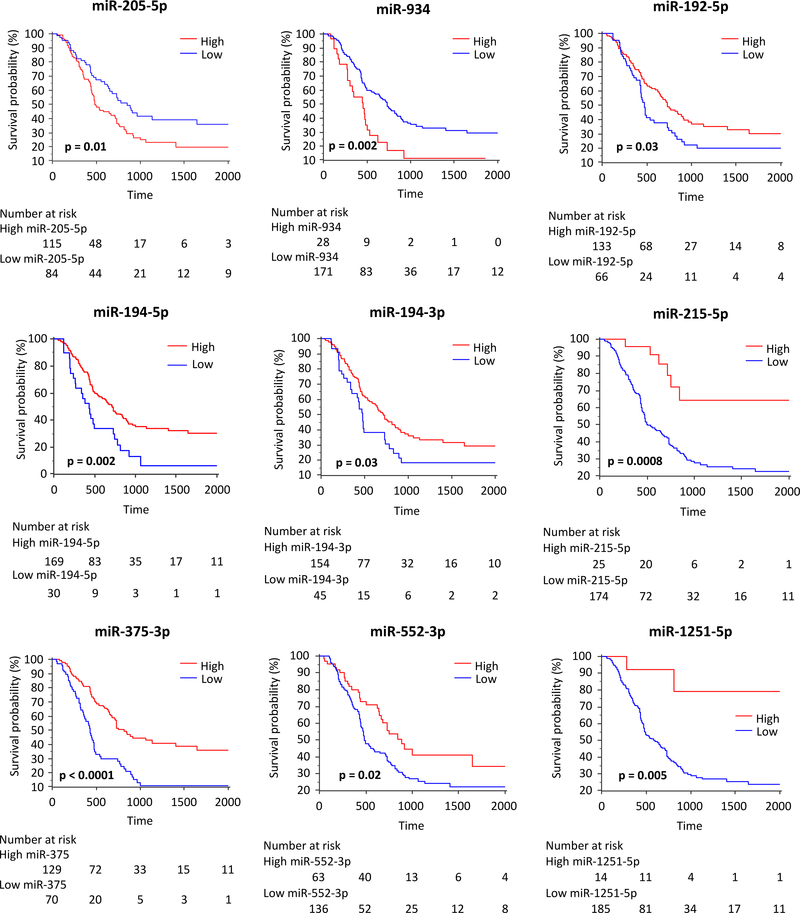
To determine the translational potential of our miRNA signature in identifying high-risk PDAC patients, we next examined its performance in an independent patient cohort. The association between various clinicopathological factors and the relative expression level of individual miRNAs is shown in Supplementary Table 2. Using Kaplan Meier analysis, we examined whether the expression of 9-miRNAs panel is predictive of survival outcomes in PDAC patients. In agreement with the TCGA discovery cohort data, among the group of miRNAs overexpressed in the poor molecular subtype, patients with high expression of miR-205–5p and miR-934 significantly associated with poor OS compared to the low expression group (p=0.01 and 0.002, respectively). Similarly, for all seven downregulated miRNAs (miR-192–5p, miR-194–5p, miR-194–3p, miR-215–5p, miR-375–3p, miR-552–3p and miR-1251–5p), the low expression group significantly correlated with poor survival (p=0.03, 0.002, 0.03, 0.0008, <0.0001, 0.02, and 0.005, respectively, Figure 2).
Figure 2: Expression of individual miRNAs and their association with OS in the independent clinical tissue validation cohort.
OS analyses of the expression of individual squamous subtype associated miRNAs in the independent clinical tissue validation cohort. Each miRNA is dichotomized using Youden’s index and the p-values are derived from log-rank test.
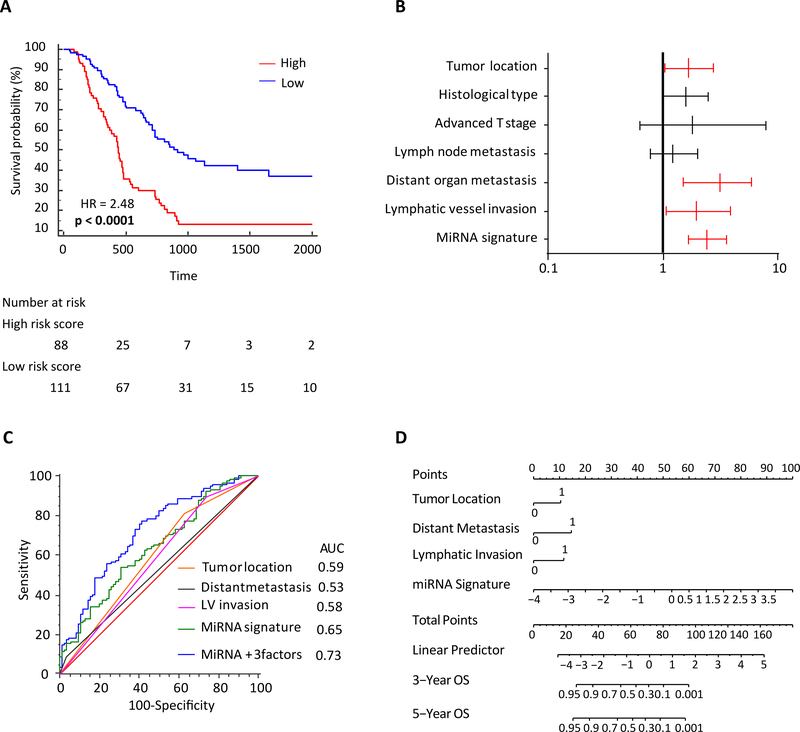
Reassuringly, patients with high-risk scores derived from the combined miRNA panel exhibited significantly poorer 5-year survival vs. the ones with the low risk scores (HR: 2.48 [95% CI-1.67–3.66]; p<0.0001; Figure 3A). Collectively, these findings from the clinical cohort are in line with our data from the discovery cohort, and yet again highlight that our miRNA panel is robust for risk stratification and predicts survival outcomes in PDAC patients.
Figure 3: Predictive potential of the 9-miRNA panel in an independent clinical validation cohort.
(A) Kaplan-Meier analysis of the 9-miRNA signature for OS. (B) Forest plot depicting hazard ratios and 95% confidence intervals of the univariate significant clinicopathological variables as well as miRNA signature. Parameters colored in red are significant risk factors in multivariate analysis. (C) ROC for OS of the combined 9-miRNA signature and key clinicopathological parameters. (D) Nomogram derived from the combination of 9-miRNA signature and key clinicopathological parameters.
Establishment of a nomogram for easier clinical translation and risk stratification in PDAC patients
We performed univariate and multivariate Cox regression analyses to determine whether our miRNA panel might serve as an independent predictor of patient prognosis, in the presence of other clinicopathological variables. Using Youden’s index, we dichotomized patients into low and high risk-groups. The univariate analysis revealed that pancreatic tumor location (pancreatic head; p<0.0001), poor differentiation (p=0.0005), advanced T-Stage (>T3; p=0.003), lymph node metastasis (p=0.0003), distant metastasis (p=0.002), lymphatic vessel invasion (p=0.0003), and the miRNA signature (p<0.0001) were all associated with poor OS in PDAC patients (Table 2). Subsequent multivariate analysis which included only the significant variables from the univariate model identified that tumor location within pancreatic head (p=0.03), distant metastasis (p=0.004), lymphatic vessel invasion (p=0.03), and the miRNA signature (p<0.0001) emerged as independent risk factors for OS in PDAC patients (Table 2 and Figure 3B).
Table 2:
Multivariate cox proportional hazard model for overall survival in clinical cohorts
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
|
| ||||||
| Variable | HR | 95%CI | P-value | HR | 95%CI | P-value |
|
| ||||||
| Tissue cohort (n=199) | ||||||
| Age ≧ 68 | 0.91 | 0.63–1.32 | 0.63 | |||
| Male | 1.09 | 0.76–1.58 | 0.64 | |||
| Pancreatic head cancer | 2.37 | 1.53–3.81 | <0.0001 | 1.66 | 1.04–2.74 | 0.03 |
| Tumor size > 30mm | 1.4 | 0.95–2.04 | 0.08 | |||
| Poorly differentiated histology | 2.37 | 1.48–3.64 | 0.0005 | 1.59 | 0.98–2.49 | 0.06 |
| T-Stage ≧3 | 4 | 1.50–16.26 | 0.003 | 1.82 | 0.63–7.71 | 0.29 |
| Lymph node metastasis positive | 2.17 | 1.41–3.45 | 0.0003 | 1.22 | 0.77–2.01 | 0.41 |
| Distant organ metastasis positive | 3.28 | 1.59–6.03 | 0.002 | 3.12 | 1.49–5.89 | 0.004 |
| Lymphatic vessel invasion positive | 2.58 | 1.51–4.84 | 0.0003 | 1.94 | 1.07–3.81 | 0.03 |
| Venous invasion positive | 1.09 | 0.75–1.61 | 0.65 | |||
| MiRNA combined signature high risk | 2.52 | 1.74–3.66 | <0.0001 | 2.43 | 1.65–3.59 | <0.0001 |
|
| ||||||
| Serum cohort (n=51) | ||||||
| Age ≧ 68 | 1.31 | 0.67–2.53 | 0.43 | |||
| Male | 1.11 | 0.56–2.26 | 0.77 | |||
| Pancreatic head cancer | 0.6 | 0.31–1.22 | 0.15 | |||
| Tumor size > 30mm | 1.06 | 0.55–2.09 | 0.85 | |||
| Poorly differentiated histology | 0.49 | 0.14–1.27 | 0.15 | |||
| T-Stage ≧3 | 1.74 | 0.73–5.10 | 0.22 | |||
| Lymph node metastasis positive | 2.87 | 1.45–5.75 | 0.003 | 2.39 | 0.83–6.93 | 0.11 |
| Distant organ metastasis positive | 9.44 | 3.89–22.16 | <0.0001 | 7.19 | 2.51–21.76 | 0.0003 |
| Lymphatic vessel invasion positive | 3.27 | 1.61–6.91 | 0.001 | 1.27 | 0.44–3.48 | 0.65 |
| Venous invasion positive | NA | NA | NA | |||
| MiRNA combined signature high risk | 2.88 | 1.22–8.45 | 0.01 | 4.46 | 1.73–14.16 | 0.001 |
HR, Hazard Ratio; CI, Confidence Interval; NA, Not available. Bold indicates a statistically significant.
We next asked whether inclusion of key clinicopathological factors that were found to be significantly associated with OS in multivariate analysis (tumor location, distant metastasis, and lymphatic vessel invasion), might further improve the robustness of our 9-miRNA panel. In support of our hypothesis, we discovered that a combination of the miRNA panel with these clinicopathological factors significantly improved its robustness in predicting patient survival in this clinical cohort (AUC improved from 0.65 to 0.73, Figure 3C).
Finally, for an easier translation of our findings into the clinic, we established a risk nomogram, which included data from the miRNA panel as well as significant clinicopathological variables (Figure 3D). Higher total score based on the sum of the assigned points for each of the factors within the nomogram were associated with a worse 3-year and 5-year OS. For instance, a patient with a combined total of at least 80 points exhibited a less than 10% probability of 3-year and 5-year OS rates. Collectively, our data suggest that inclusion of clinicopathological factors into the miRNA panel further enhanced its predictive accuracy for the identification of high-risk PDAC patients with poor survival outcomes.
The miRNA signature demonstrated excellent prognostic performance in pre-operative serum
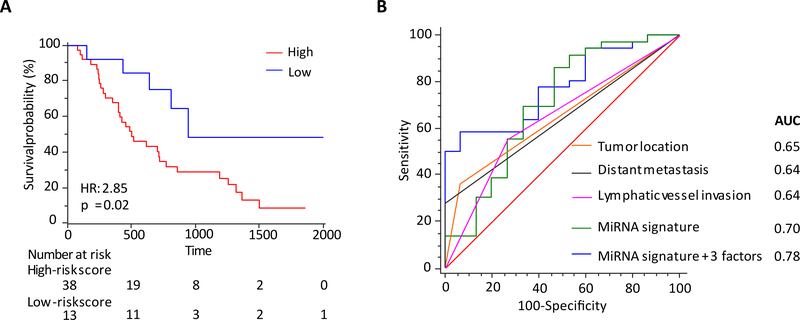
Availability of non-invasive prognostic biomarkers in PDAC patients could be of significant clinical impact, as these could improve clinical decision-making for which subsets of patients might be candidate for neoadjuvant treatments (chemotherapy or chemo-radiotherapy). Accordingly, we evaluated the feasibility of translating our tissue-based biomarkers into a liquid-biopsy assay by analyzing their performance in an independent cohort of patients where pre-operative serum specimens were available (n=51). The correlation between clinicopathological factors and the relative expression of each miRNA is shown in Supplementary Table 3. We observed that 6 of the 9 miRNAs demonstrated expression profiles in serum specimens that were consistent with the tissue-data; hence, we interrogated their prognostic potential in determining 5-year overall survival in PDAC patients (Supplementary Figure 4). Intriguingly, PDAC patients with high-risk scores for these miRNAs in serum exhibited significantly shorter 5-year OS rates vs. the patients with low-risk scores (8.6% vs. 48.4%; HR=2.85 [95% CI-1.41–5.76]; p=0.02, Figure 4A); highlighting that our miRNA signature has the potential to serve as a non-invasive survival prediction tool in PDAC patients.
Figure 4: Predictive value of 6-miRNA signature in an independent clinical serum validation cohort.
(A) Kaplan-Meier analysis of the 6-miRNA signature for OS. (B) ROC for OS of the combined 6-miRNA signature and key clinicopathological parameters.
Furthermore, in the serum cohort, univariate analysis revealed that lymph node metastasis (p=0.003), distant metastasis (p<0.0001), lymphatic vessel invasion (p=0.001) and the miRNA signature (p=0.01) were significant predictors of poor survival in PDAC (Table 2). Likewise, in multivariate analysis, distant metastasis (p=0.0003) and the miRNA signature (p=0.001) emerged as independent risk factors for OS (Table 2). As was the case in the tissue cohort, the serum cohort also exhibited a significant improvement in prognostic accuracy when the miRNA signature was combined together with significant clinicopathological factors (AUC increased from 0.70 to 0.78; Figure 4B). Collectively, these data demonstrate that our miRNA signature, both in tissue and serum, offers an attractive potential for predicting patient survival in PDAC patients.
DISCUSSION
Recently, multiple transcriptomic-based efforts have led to the identification of distinct molecular subtypes in PDAC, including the most recent classification from Bailey and colleagues 12, which categorized all PDACs into four distinct subtypes; with the squamous subtype exhibiting the worst prognosis. Given the challenge of measuring expression of hundreds of genes in clinical settings, we exploited miRNA-mRNA regulatory network, to identify a panel of miRNA biomarkers that represent gene expression changes associated with poor molecular subtypes in PDAC. Given the ubiquitous and cancer-specific expression of miRNAs in clinical specimens, these are emerging as important biomarkers candidates with a promise for easier clinical translation – both in tissues, as well as from non-invasive bodily fluids. Previously, we discovered that miR-200 family is an important driver for mesenchymal subtype with poor prognosis in colorectal cancer 23. While miRNA-mRNA regulatory network is a well-known concept that can provide additional insights into the biology of PDAC, subtype-specific miRNAs and their prognostic significance remains unexplored. Accordingly, in this by undertaking a systematic biomarker discovery effort through analyses of multiple high-throughput datasets, we identified a panel of 9 miRNAs that strongly associated with all three major molecular subtypes associated with poor survival outcomes in PDAC patients (Basal by Moffitt et al 15, Quasi-mesenchymal by Collison et al 14 and Squamous by Bailey et al 12). Subsequently we evaluated their prognostic potential in tissue and pre-operative serum specimens from two independent clinical cohorts. In the end, for an easier clinical translation, we established a risk nomogram that included the miRNA panel and other significant clinicopathological features.
Currently, FOLFIRINOX (oxaliplatin, irinotecan, fluorouracil, and leucovorin) or gemcitabine plus abraxane (albumin-bound paclitaxel) is the standard of care in the first-line metastatic PDAC patients 2, 24. Despite these newer regimens have somewhat improved the survival rates, the overall survival in PDAC remains quite poor compared to other solid cancers. Therefore, it is important to develop risk-stratification biomarkers that can help predict prognosis and identify high-risk PDAC patients. Several clinicopathological factors such as tumor size, histologic grade, vascular invasion, perineural invasion, lymph node metastases, and distant metastases have been recognized as prognostic factors, but identifying clinically translatable molecular markers is an important step towards implementation of precision medicine in pancreatic cancer 25. In this regard, the miRNA classifier we identified has an exciting potential, and the fact that we can quantify the expression these miRNAs non-invasively in serum is even more encouraging.
Among the panel of 9 miRNAs we identified, miR-375–3p is the most widely studied. In fact, miR-375–3p was first identified from murine pancreatic beta-cell line as a pancreatic islet-specific miRNA regulating insulin secretion 26, and its low expression was reported to associate with poor prognosis in PDAC 20 and other malignancies 27, 28. Furthermore, miR-375 targets PDK1 and suppresses proliferation through Akt signaling pathway 29 in PDAC. Upregulation of miR-205–5p is reported to be associated with poor prognosis in lung cancer 30. Although its overexpression is reported in PDAC tissues 18, 20, 31, ours is the first report for its prognostic potential. With regards to miR-192–5p, it is reported to play a tumor suppressor role in hepatocellular carcinoma via silencing oncogenic LncRNA HOTTIP, and its low expression has been shown to associate with poor prognosis in hepatocellular carcinoma patients 32. In addition, miR-194–5p was reported to play a major role in suppressing proliferation, migration or metastatic potential in lung, gastric, and breast cancer 33–35. Finally, as for miR-934, it is linked with increased cellular proliferation of head and neck squamous carcinoma 36. Intriguingly, the expression levels of miR-192–5p and miR-375 are also reported to be downregulated in epithelial mesenchymal transition (EMT) related subtype of colorectal cancer (consensus molecular subtype: CMS4 class subtype) 23. Recently, three molecular subtypes have been reported based on the interaction between tumor cells and the immune cells within the tumor microenvironment. Intriguingly, the patients belonging to the “immune escape” subtype showed the worst prognosis. Though we are unable to directly compare our miRNA classifier with the proposed immune subtypes, it is exciting to see that the poor “immune escape” subtype is correlated with squamous and quasi-mesenchymal subtypes as seen with our miRNA risk classifier. Overall, these biological roles for the panel of miRNAs we have identified in this report are of great interest, as all of them have already been shown to play bona fide roles in cancer and PDAC; hence providing further support in their role as important risk-assessment biomarkers in PDAC. The risk nomogram developed in this study can be used to accurately predict outcomes in patients with pancreatic cancer. Implementing this nomogram-based risk stratification in the clinic can help the clinicians in identifying patients belonging to high-risk groups and this can further help in refining treatment decisions.
Our present study has been quite comprehensive; however, the patient cohorts analyzed were retrospective in nature, and these findings must be validated in other retrospective and prospective cohorts prior to further consideration for their clinical translation. In addition, while our serum data is quire robust and encouraging, the patient cohort that was available to us was rather modest in size.
In conclusion, we for the first time report a novel panel of miRNA biomarkers, that can help identify poor molecular subtypes, allow identification of high-risk patients and predict prognosis in PDAC patients. Pending further validation in future prospective cohorts, these data highlight the potential clinical significance of these biomarkers in improved risk-stratification and improving survival outcomes in patients suffering from a lethal malignancy such as pancreatic cancer.
Supplementary Material
Supplementary Table 1: Accuracy of individual miRNAs in identifying poor molecular subtypes in TCGA cohort.
Supplementary Table 2: Correlation between clinicopathological factors and expression of individual miRNAs in tissue cohort.
Supplementary Table 3: Correlation between clinicopathological factors and expression of individual miRNAs in serum cohort
Supplementary Figure 1: Overall schematic flow-chart of the experimental design of this study.
Supplementary Figure 2: The efficacy of 9-miRNA signature for the identification of poor molecular subtypes in the TCGA cohort. Receiver operating characteristic (ROC) curve of the combined 9-miRNA signature for the detection of Basal subtype in Moffitt classification (A), and Quasimesenchymal subtype in Collisson classification (B). The waterfall plot representing risk-score of dead or alive patients based on the combined 9-miRNA signature (C).
Supplementary Figure 3: Expression of individual miRNAs and their association with OS in the TCGA discovery cohort. OS analyses of the expression of individual squamous subtype associated miRNAs in the TCGA discovery cohort. Each miRNA is dichotomized using Youden’s index and the p-values are derived from log-rank test.
Supplementary Figure 4: Expression of individual miRNAs and their association with OS in the serum validation cohort. OS analyses of the expression of individual squamous subtype associated miRNAs in the serum validation cohort. Each miRNA is dichotomized using Youden’s index and the p-values are derived from log-rank test.
Acknowledgements
We would like to acknowledge Jasjit Kaur Banwait for statistical support and acknowledge Lauren Patterson for her critical reading and valuable insights provided in improving the quality of this article.
Grant support: The present work was supported by the CA72851, CA187956, CA202797 and CA214254 grants from the National Cancer Institute, National Institute of Health; and institutional grants from the Sammons Cancer Center and Baylor Foundation, The Lee Hanley Foundation, as well as funds from the Baylor Scott & White Research Institute, Dallas, TX, USA
Footnotes
Conflict of interest: None of the authors have any conflicts to disclose.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018; 68(1):7–30. [DOI] [PubMed] [Google Scholar]
- 2.Conroy T, Desseigne F, Ychou M, et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N Engl J Med 2011; 364(19):1817–25. [DOI] [PubMed] [Google Scholar]
- 3.Neoptolemos JP, Palmer DH, Ghaneh P, et al. Comparison of adjuvant gemcitabine and capecitabine with gemcitabine monotherapy in patients with resected pancreatic cancer (ESPAC-4): a multicentre, open-label, randomised, phase 3 trial. Lancet 2017; 389(10073):1011–1024. [DOI] [PubMed] [Google Scholar]
- 4.Pandol S, Edderkaoui M, Gukovsky I, et al. Desmoplasia of pancreatic ductal adenocarcinoma. Clin Gastroenterol Hepatol 2009; 7(11 Suppl):S44–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hruban RH, Iacobuzio-Donahue C, Wilentz RE, et al. Molecular pathology of pancreatic cancer. Cancer J 2001; 7(4):251–8. [PubMed] [Google Scholar]
- 6.Zavoral M, Minarikova P, Zavada F, et al. Molecular biology of pancreatic cancer. World J Gastroenterol 2011; 17(24):2897–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maitra A, Hruban RH. Pancreatic cancer. Annu Rev Pathol 2008; 3:157–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Waddell N, Pajic M, Patch AM, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 2015; 518(7540):495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012; 487(7407):330–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cancer Genome Atlas Research N. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014; 513(7517):202–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cancer Genome Atlas Research Network. Electronic address aadhe, Cancer Genome Atlas Research N. Integrated Genomic Characterization of Pancreatic Ductal Adenocarcinoma. Cancer Cell 2017; 32(2):185–203 e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bailey P, Chang DK, Nones K, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016; 531(7592):47–52. [DOI] [PubMed] [Google Scholar]
- 13.Hezel AF, Kimmelman AC, Stanger BZ, et al. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev 2006; 20(10):1218–49. [DOI] [PubMed] [Google Scholar]
- 14.Collisson EA, Sadanandam A, Olson P, et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med 2011; 17(4):500–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moffitt RA, Marayati R, Flate EL, et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat Genet 2015; 47(10):1168–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garzon R, Fabbri M, Cimmino A, et al. MicroRNA expression and function in cancer. Trends Mol Med 2006; 12(12):580–7. [DOI] [PubMed] [Google Scholar]
- 17.Di Leva G, Croce CM. miRNA profiling of cancer. Curr Opin Genet Dev 2013; 23(1):3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bloomston M, Frankel WL, Petrocca F, et al. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA 2007; 297(17):1901–8. [DOI] [PubMed] [Google Scholar]
- 19.Lee EJ, Gusev Y, Jiang J, et al. Expression profiling identifies microRNA signature in pancreatic cancer. Int J Cancer 2007; 120(5):1046–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma MZ, Kong X, Weng MZ, et al. Candidate microRNA biomarkers of pancreatic ductal adenocarcinoma: meta-analysis, experimental validation and clinical significance. J Exp Clin Cancer Res 2013; 32:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frampton AE, Krell J, Jamieson NB, et al. microRNAs with prognostic significance in pancreatic ductal adenocarcinoma: A meta-analysis. Eur J Cancer 2015; 51(11):1389–404. [DOI] [PubMed] [Google Scholar]
- 22.Tibshirani R, Hastie T, Narasimhan B, et al. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci U S A 2002; 99(10):6567–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fessler E, Jansen M, De Sousa EMF, et al. A multidimensional network approach reveals microRNAs as determinants of the mesenchymal colorectal cancer subtype. Oncogene 2016; 35(46):6026–6037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Von Hoff DD, Ervin T, Arena FP, et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N Engl J Med 2013; 369(18):1691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bilici A Prognostic factors related with survival in patients with pancreatic adenocarcinoma. World J Gastroenterol 2014; 20(31):10802–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poy MN, Eliasson L, Krutzfeldt J, et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature 2004; 432(7014):226–30. [DOI] [PubMed] [Google Scholar]
- 27.Kong KL, Kwong DL, Chan TH, et al. MicroRNA-375 inhibits tumour growth and metastasis in oesophageal squamous cell carcinoma through repressing insulin-like growth factor 1 receptor. Gut 2012; 61(1):33–42. [DOI] [PubMed] [Google Scholar]
- 28.Shao Y, Geng Y, Gu W, et al. Prognostic significance of microRNA-375 downregulation in solid tumors: a meta-analysis. Dis Markers 2014; 2014:626185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou J, Song S, He S, et al. MicroRNA-375 targets PDK1 in pancreatic carcinoma and suppresses cell growth through the Akt signaling pathway. Int J Mol Med 2014; 33(4):950–6. [DOI] [PubMed] [Google Scholar]
- 30.Duan B, Guo T, Sun H, et al. miR-205 as a biological marker in non-small cell lung cancer. Biomed Pharmacother 2017; 91:823–830. [DOI] [PubMed] [Google Scholar]
- 31.Schultz NA, Werner J, Willenbrock H, et al. MicroRNA expression profiles associated with pancreatic adenocarcinoma and ampullary adenocarcinoma. Mod Pathol 2012; 25(12):1609–22. [DOI] [PubMed] [Google Scholar]
- 32.Ge Y, Yan X, Jin Y, et al. MiRNA-192 [corrected] and miRNA-204 Directly Suppress lncRNA HOTTIP and Interrupt GLS1-Mediated Glutaminolysis in Hepatocellular Carcinoma. PLoS Genet 2015; 11(12):e1005726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu X, Liu T, Fang O, et al. miR-194 suppresses metastasis of non-small cell lung cancer through regulating expression of BMP1 and p27(kip1). Oncogene 2014; 33(12):1506–14. [DOI] [PubMed] [Google Scholar]
- 34.Bao J, Zou JH, Li CY, et al. miR-194 inhibits gastric cancer cell proliferation and tumorigenesis by targeting KDM5B. Eur Rev Med Pharmacol Sci 2016; 20(21):4487–4493. [PubMed] [Google Scholar]
- 35.Le XF, Almeida MI, Mao W, et al. Modulation of MicroRNA-194 and cell migration by HER2-targeting trastuzumab in breast cancer. PLoS One 2012; 7(7):e41170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saad MA, Kuo SZ, Rahimy E, et al. Alcohol-dysregulated miR-30a and miR-934 in head and neck squamous cell carcinoma. Mol Cancer 2015; 14:181. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1: Accuracy of individual miRNAs in identifying poor molecular subtypes in TCGA cohort.
Supplementary Table 2: Correlation between clinicopathological factors and expression of individual miRNAs in tissue cohort.
Supplementary Table 3: Correlation between clinicopathological factors and expression of individual miRNAs in serum cohort
Supplementary Figure 1: Overall schematic flow-chart of the experimental design of this study.
Supplementary Figure 2: The efficacy of 9-miRNA signature for the identification of poor molecular subtypes in the TCGA cohort. Receiver operating characteristic (ROC) curve of the combined 9-miRNA signature for the detection of Basal subtype in Moffitt classification (A), and Quasimesenchymal subtype in Collisson classification (B). The waterfall plot representing risk-score of dead or alive patients based on the combined 9-miRNA signature (C).
Supplementary Figure 3: Expression of individual miRNAs and their association with OS in the TCGA discovery cohort. OS analyses of the expression of individual squamous subtype associated miRNAs in the TCGA discovery cohort. Each miRNA is dichotomized using Youden’s index and the p-values are derived from log-rank test.
Supplementary Figure 4: Expression of individual miRNAs and their association with OS in the serum validation cohort. OS analyses of the expression of individual squamous subtype associated miRNAs in the serum validation cohort. Each miRNA is dichotomized using Youden’s index and the p-values are derived from log-rank test.