Abstract
Cell–cell interactions orchestrate organismal development, homeostasis and single-cell functions. When cells do not properly interact or improperly decode molecular messages, disease ensues. Thus, the identification and quantification of intercellular signalling pathways has become a common analysis performed across diverse disciplines. The expansion of protein–protein interaction databases and recent advances in RNA sequencing technologies have enabled routine analyses of intercellular signalling from gene expression measurements of bulk and single-cell data sets. In particular, ligand–receptor pairs can be used to infer intercellular communication from the coordinated expression of their cognate genes. In this Review, we highlight discoveries enabled by analyses of cell–cell interactions from transcriptomic data and review the methods and tools used in this context.
Subject terms: RNA sequencing, Systems biology, Computational biology and bioinformatics
Cell–cell interactions and communication can be inferred from RNA sequencing data of, for example, ligand–receptor pairs. The authors review insights gained and the methods and tools used in studies of cell–cell interactions based on transcriptomic data.
Introduction
Multicellular life relies on the coordination of cellular activities, which depend on cell–cell interactions (CCIs) across an organism’s diverse cell types and tissues1–3. Thus, studies on cellular functions increasingly require consideration of the community context of each cell4. CCIs leverage diverse molecules, including ions, metabolites, integrins, receptors, junction proteins, structural proteins, ligands and secreted proteins of the extracellular matrix. Some molecules support structural CCIs (for example, cell adhesion molecules), whereas ligands such as hormones, growth factors, chemokines, cytokines and neurotransmitters mediate cell–cell communication (CCC) (Fig. 1a). The signalling events behind CCC are often mediated by interactions of various types of protein, encompassing ligand–receptor, receptor–receptor and extracellular matrix–receptor interactions. Receiver cells trigger downstream signalling through cognate receptors, generally culminating in altered transcription factor activity and gene expression. These cells with altered expression further interact with their microenvironment. To understand the role of each cell within its local community, one must identify the protein messages passed between cells; measuring expressed messenger molecules and their associated pathways is fundamental to understanding the directionality, magnitude and biological relevance of CCC.
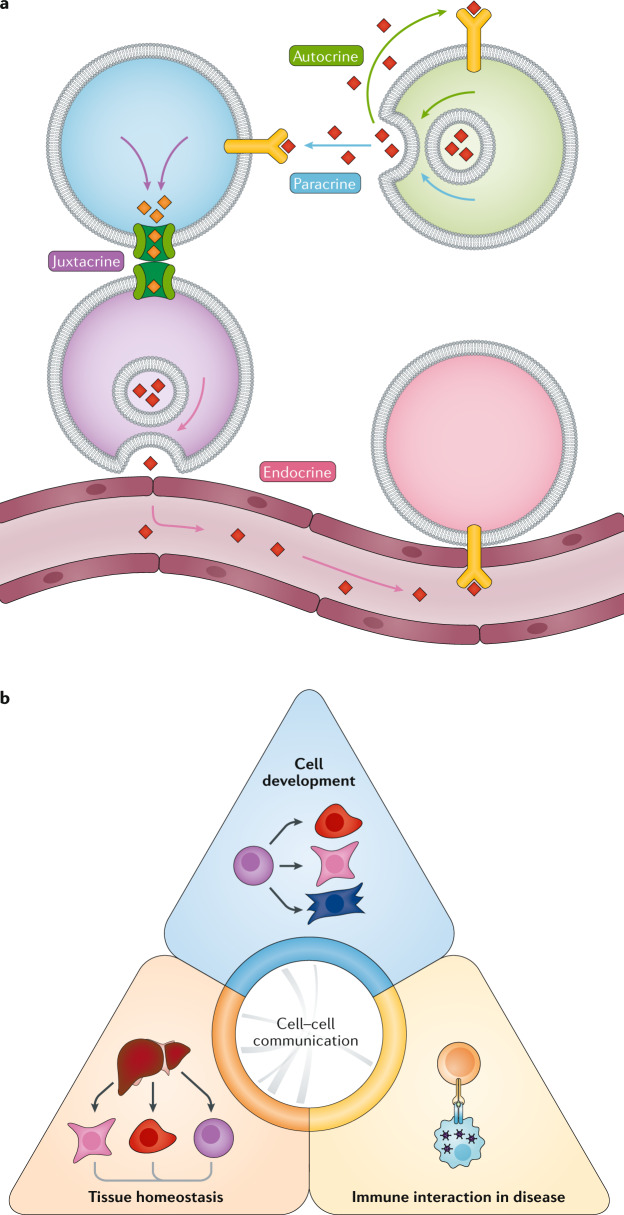
Fig. 1. Types and applications of cell–cell interactions and communication.
a | ‘Autocrine signalling’ refers to intracellular communication whereby cells secrete ligands that are used to induce a cellular response through cognate receptors for those molecules expressed on the same cell. Paracrine cell–cell communication does not require cell–cell contact, rather depending on the diffusion of signalling molecules from one cell to another after secretion. Juxtacrine, that is, contact-dependent, cell–cell communication relies on gap junctions or other structures such as membrane nanotubes to pass signalling molecules directly between cells, without secretion into the extracellular space. Endocrine cell–cell communication represents intercellular communication whereby signalling molecules are secreted and travel long distances through extracellular fluids such as the blood plasma; typical mediators of this communication are hormones. b | Overview of the main applications of cell–cell interaction methods: cell development, tissue and organ homeostasis, and immune interactions in disease (for more details on each study type, see Supplementary Table 1).
Direct measurement of proteins mediating CCC requires specialized biochemical assays and extensive domain knowledge; moreover, these proteins cannot always be studied in the native microenvironment. Traditional assays of the underlying protein–protein interactions (PPIs) include yeast two-hybrid screening, co-immunoprecipitation, proximity labelling proteomics, fluorescence resonance energy transfer imaging and X-ray crystallography5,6. These techniques have identified many interactions between proteins that are secreted or displayed extracellularly to mediate intercellular communication. Proteomics and transcriptomics can further reinforce such studies as evidence of expression supports the presence of PPIs. This approach has been applied to, for instance, the analysis of communication between 144 human primary cell types, which provided insights into pairs of cells that are more likely to interact and the specific pathways they use to communicate7. While proteomic technologies are preferable for these analyses owing to the direct measurement of protein abundances, RNA sequencing (RNA-seq) data sets are more numerous, easier to access and straightforward to analyse. They can also be generated from bulk samples8, microdissected specimens9 or single-cell suspensions10 and enable studies of CCC at different resolutions, whereas proteomics at single-cell resolution is a technology still under development11. Single-cell RNA-seq has benefits over bulk analysis, chiefly in quantifying expression in rare cell types and in identifying the cell type of origin of proteins mediating CCIs12,13. Results from transcriptomics must be cautiously considered and validated to avoid misleading hypotheses; however, the ubiquity and ease of analysis have enabled many recent studies to infer CCC from gene expression, generating testable hypotheses across diverse disciplines. In particular, the coordinated gene expression of ligands and receptors can be used to infer intercellular communication.
Here, we start by providing an overview of the range of fields that RNA-based CCI analyses have been applied to, illustrating the types of insight that can be gleaned. We then discuss the computational strategies adopted in those studies, detailing the PPI databases and mathematical models commonly used to decipher CCC. Additionally, we introduce the computational tools that facilitate these analyses, describing their main features as well as their strengths and weaknesses. Finally, we review approaches to validate CCI-derived results and discuss remaining challenges and future directions in the field.
Insights from RNA-based CCI analyses
The study of intercellular interactions has greatly accelerated as transcriptomics, in particular bulk and single-cell RNA-seq, has become commonplace. These approaches use transcriptional profiling to decipher CCCs at any stage of development and in any multicellular community. Many studies focus on signals mediating cellular differentiation, interactions of cell types within tissues and organs, and immune responses (Fig. 1b; Table 1; see Supplementary Table 1 for more details). Here, we review these studies and illustrate the types of insights gained from analysing CCC.
Table 1.
Illustrative studies and their strategies for deciphering cell–cell interactions and communication
| Sample or organ | Key input | Scoring | CS value | CCC score | Validation | Study focus | Ref |
|---|---|---|---|---|---|---|---|
| Cell development | |||||||
| Haematopoietic cells (human) | Microarray; LRIs | Expression thresholding | Binary | No score | Functional validation | Role of CCC between differentiated haematopoietic cells and HSCs in fate decisions | 15 |
| Brain (mouse embryonic cortex) | Microarray; LRIs | Expression thresholding | Binary | No score | Functional validation | Role of microenvironment in self-renewal versus differentiation decision of neural precursor cells during neurogenesis | 16 |
| Liver and iPS cells (human) | scRNA-seq; LRIs | Expression thresholding | Binary | Normalized sum of CS | Functional validation | 3D liver bud organoid from iPS cells to characterize CCC shaping hepatogenesis | 24 |
| Placenta (human) | scRNA-seq; LRIs | Expression thresholding | Binary | No score | Functional validation | CCI in the fetus–placenta interface before and after decidualization | 29 |
| Brain (mouse) | Bulk RNA-seq; LRIs | Expression thresholding | Binary | Sum of CS | Colocalization | Ligand–receptor pathways active during neural development; CCC between neural, vascular and microglial cells | 22 |
| iPS cells (mouse) | scRNA-seq; LRIs | Expression product | Continuous | Sum of CS | Functional validation | CCC at the beginning of differentiation | 19 |
| Bone marrow (mouse) | scRNA-seq; LRIs | RNA-Magnet | Continuous | No score | Colocalization | CCC and interactions between bone marrow cells | 53 |
| Tissue interactions | |||||||
| Multiple lineages (human) | scRNA-seq; LRIs | Expression thresholding | Binary | Sum of CS | None | CCC between multiple cell lineages | 7 |
| Lungs (human) | popRNA-seq; LRIs | Expression thresholding | Binary | Sum of CS | Colocalization functional validation | Signals sent by mesenchymal cells in lungs that are key for self-renewal of epithelial progenitors after tissue injury | 40 |
| Heart (mouse) | scRNA-seq; LRIs | Expression thresholding | Binary | Sum of CS | Functional validation | Transcriptional profiles of non-myocyte cells in heart and their CCC | 33 |
| Lungs (mouse) | scRNA-seq; LRIs | Expression correlation (Spearman) | Continuous | No score | Expression; colocalization functional validation | CCC between and within immune and non-immune cells during development | 41 |
| Immune system and structural cells (mouse) | Low-input RNA-seq; LRIs | Differential combinations | Binary | Odds ratios | Functional validation | Role of structural cells in immune responses | 58 |
| Heart (mouse) | scRNA-seq; LRIs | Differential combinations | Binary | Sum of CS | Expression; colocalization functional validation | CCC of cardiomyocytes and non-cardiomyocytes in human heart in health and under failure | 34 |
| Placenta (human) | scRNA-seq; LRIs | CellPhoneDB | Continuous | No score | Colocalization | Key ligand–receptor pairs based on subunit architecture; CCC at maternal–fetal interface | 30 |
| Tumour microenvironment | |||||||
| Melanoma (human) | scRNA-seq; LRIs | Expression thresholding | Binary | Sum of CS | None | CCI network of isolated cells | 70 |
| HNSCC (human) | scRNA-seq; LRIs | Expression thresholding | Binary | No score | Colocalization | CCC in patients with HNSCC generated by HPV or environmental carcinogens (HPV negative) | 61 |
| Five cancer types (mouse) | scRNA-seq; LRIs | Expression product | Continuous | No score | None | CCC within a tumoural microenvironment | 60 |
| Nine cancer types (human) | Microarray; LRIs | Expression correlation (Pearson) | Continuous | No score | None | Correlation between autocrine signalling pathways and mRNA levels of ligands and receptors | 69 |
| Lungs (human) | scRNA-seq; LRIs | Differential combination; expression thresholding | Binary | No score | Functional validation | Tumour–stroma CCC in lung cancer; introduced CCCExplorer | 67 |
| Ovary (human) | Microarray; LRIs; downstream target genes | Differential combination; expression thresholding | Binary | No score | Expression; functional validation | CCC between stromal and ovarian cancer cells | 68 |
| Head and neck and immune system (human) | scRNA-seq; LRIs; downstream target genes | NicheNet | Continuous | No score | None | Prediction of ligand–target links between interacting cells; tested on HNSCC data set | 54 |
CCC, cell–cell communication; CCI, cell–cell interaction; CS, communication score; HNSCC, head and neck squamous cell carcinoma; HPV, human papillomavirus; HSC, haematopoietic stem cell; iPS cell, induced pluripotent stem cell; LRI, ligand–receptor interaction; popRNA-seq, population RNA sequencing; RNA-seq; RNA sequencing; scRNA-seq, single-cell RNA sequencing. For additional studies and details, see Supplementary Table 1.
Interactions drive cellular differentiation and organ development
Cellular differentiation depends on temporally and locally precise cell communication, so inspecting intercellular communication has increased our understanding of stem cell fates and revealed ligand–receptor interactions that initiate self-renewal and differentiation14–20. For example, a CCC network of haematopoietic cells built using ligand–receptor pairs showed that fate decisions are regulated through precise coordination of an antagonistic feedback circuit involving megakaryocyte-derived stimulatory factors and monocyte-derived inhibitory factors14. Another analysis of CCC networks interrogating how differentiated cells influence haematopoietic stem cell fate revealed that ligand production is cell type specific, whereby some cells can produce signals with the same function, whereas receptors are less specific15. Given the promiscuity of receptors, physical compartmentalization of cells is key to limit ligand signalling and confer specificity to stem cell fates.
Tissue and organ development also depends on signals that progenitor cells send and receive21. The analysis of brain CCC showed crosstalk involved in neurogenesis and identified novel mediators16,22, such as apolipoprotein E (APOE), a protein associated with Alzheimer disease. CCC analysis also elucidated how erythroblasts interact with macrophages during haematopoiesis in the fetal liver23 and was applied to liver organoid development to investigate how multilineage communication shapes the differentiation of hepatic cells24. Following similar principles, other studies have used CCC analyses to understand differentiation during epidermal regeneration25–27, development of the interfollicular epidermis28 and formation of the maternal–fetal interface in early pregnancy29,30.
Cell-type communication in tissue and organ homeostasis
Expression profiling of different cell types in adult tissues and organs has shown how intercellular communication contributes to organ function. To date, this approach has been applied to brain31,32, heart33,34, kidney35–37, liver38,39, lungs40–43, placenta29,30,44, retina45 and visual cortex46. Remarkably, this approach revealed new roles of cells within a tissue41 and helped explain how ageing32,47, diseases34–36,48–51, infections52 and injuries31,40,43 shape multicellular organization. Intercellular crosstalk has been investigated in healthy and diseased liver50, kidneys during homeostasis and rejected kidney transplants35,36, heart during failure and recovery conditions34, and healthy and asthmatic lungs51. RNA-based strategies for studying CCIs and CCC have helped elucidate, for instance, how cells communicate during ageing of the mouse brain. This revealed that CST3 and CXCL12 are mediators that differentiate intercellular interactions in young and old brains and may modulate ageing-related processes32. Thus, within-tissue CCC studies continue to deepen our understanding, for basic and therapeutic purposes, of how cellular communities work.
During the study of CCC, considering spatial context clarifies relationships between cells across tissues and organs. Several studies have incorporated imaging to spatially map cells and integrated transcriptomics to measure CCC. For example, researchers interrogated the communication and response of lung cells to tissue injury40,43; spatial information aided the identification of a new population of lung endothelial cells (Car4 high) and elucidated how these cells communicate with neighbouring alveolar type 1 cells through VEGF signalling given both their proximity and their expression of the cognate genes43. Spatial maps have also helped investigate the communication of T cells during their development in the human thymus13. As this process is spatially coordinated, knowing the cellular localizations was crucial to understand, for example, interactions between XCR1+ dendritic cells and T cells with high expression of XCL1, which are important to recruit dendritic cells into the medulla of the thymus. Yet another study inferred the 3D organization of bone marrow — instead of using tissue imaging to map the cells — ultimately identifying signalling between immune and non-immune cells53. These examples show how cell localization can help elucidate interactions between spatially proximal regions.
Immune interactions in disease
The immune system receives signals from multiple tissues, but only specific signals allow it to coordinate healthy immune responses. For instance, CCL2- and CX3CL1-mediated communication coordinate the recruitment and positioning of immune cells, as determined from single-cell transcriptomes37,39. Specifically, these CCCs were associated with the recruitment of monocytes that later became liver-resident macrophages39 and the positioning of mononuclear phagocytes in kidney37, which are crucial processes to combat ascending uropathogenic Escherichia coli infection. CCC is also involved in the response to viral infections52,54–56. Studies of respiratory diseases investigated the crosstalk between lung and T cells in Sendai virus-infected mice55 or CCC associated with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection52,56, where interactions between immune and epithelial cells correlated with COVID-19 severity. CCC-based studies have also provided more general insights. For example, they helped build a social network of immune cells by identifying communication pathways between immune cells57. Furthermore, they revealed the roles of structural cells in immune responses by elucidating how fibroblasts, endothelial cells and epithelial cells are primed for organ-specific immune gene activation through upregulation of ligands and receptors, including β2M, CD74, CXCL10, VCAM1 and TNFRSF1A58.
Tumours and their surrounding microenvironments are complex communities of cells that modify local immune cell functions. Studying CCC within these communities can reveal how cells communicate in these ecosystems54,59–64 and help guide the development of effective cancer immunotherapies just as the inhibition of CCC through PD1 and PDL1 has revolutionized the field65,66. CCC analyses have also elucidated crosstalk between tumour and stroma67,68 and communication pathways used by tumours69–71. Several studies have developed statistical models to connect inferred CCC mechanisms to cancer phenotypes. One case observed clear correlations between the activity of specific ligand–receptor interactions and the degree of regulatory T cell infiltration and tumour growth60. In another study, active ligand–receptor pairs were associated with invasiveness and proliferation of malignant cells under a partial epithelial–mesenchymal transition programme in patients with head and neck squamous cell carcinoma59. Moreover, the expression levels of key mediators of CCC were used as inputs for training a decision tree to predict prognosis for patients with glioma71; this model classified patients into high-risk and low-risk groups, defined on the basis of the difference in patient survival time. Thus, studying CCC within the tumour microenvironment provides opportunities to identify druggable pathways and develop new cancer therapeutics68.
Deciphering CCC
The aforementioned studies provide a glimpse of the insights attainable when studying CCC. While the methods and tools that these studies used have in common that they infer CCC from gene expression (Fig. 2a), a diverse range of strategies can be applied (Fig. 2b; Table 1). For simplicity, we refer to them as methods for studying CCC, but these strategies can decipher any type of gene product-based CCIs, including proteins that participate in structural interactions between cells. Furthermore, although we focus on mammalian CCIs, the approaches apply to any prokaryotic or eukaryotic cells with a characterized interactome (for example, Drosophila melanogaster72,73 and Caenorhabditis elegans74,75).
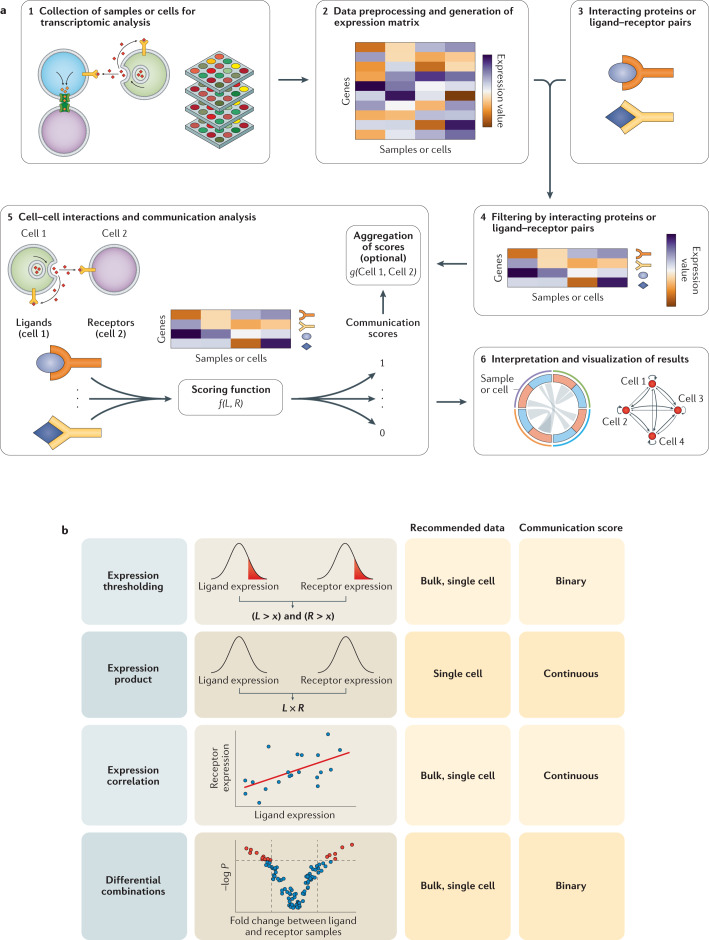
Fig. 2. Analysis workflow for inferring cell–cell interactions and communication from gene expression.
a | Samples or cells are analysed by transcriptomics to measure the expression of genes (step 1). Then the data generated are preprocessed to build a gene expression matrix, which contains the transcript levels of each gene across different samples or cells (step 2). A list of interacting proteins that are involved in intercellular communication is generated or obtained from other sources (step 3), often including interactions between secreted and membrane-bound proteins (commonly ligands and receptors, respectively). Only the genes associated with the interacting proteins are held in the gene expression matrix (step 4). Their expression levels are used as inputs to compute a communication score for each ligand–receptor pair using a scoring function (function f(L, R), where L and R are the expression values of the ligand and the receptor, respectively). These communication scores may be aggregated to compute an overall state of interaction between the respective samples or cells using an aggregation function (function g(Cell 1, Cell 2), where Cell 1 and Cell 2 are all communication scores of those cells or corresponding samples) (step 5). Finally, communication and aggregated scores can be represented by, for instance, Circos plots and network visualizations to facilitate the interpretation of the results (step 6). b | Main scoring functions of communication pathways based on the expression of their components. Recommended data to use with these functions and the type of their resulting communication score are indicated.
Building from PPIs
Inferring CCC from transcriptomics relies on gene co-expression, whereby one gene in a given pair comes from one interacting cell and the other gene comes from the second interacting cell. Several studies focused on intercellular signalling using co-expression of all genes or specific cell markers31,76, the similarity between expression profiles77 or the properties of regulatory networks78. However, most studies rely on literature-curated lists of interacting proteins, which facilitates the biological interpretation of results (Fig. 2a). Although several studies have used interactions between any class of cell-surface protein and secreted protein32,53, the predominant class of interactions used for studying CCC are known ligands and their cognate receptors.
By focusing on ligands and receptors, an early study investigated autocrine signalling loops in cancer69. The authors analysed the correlation between ligand expression and cognate receptor expression. They first created a literature-curated list of ligand–receptor pairs (Database of Ligand-Receptor Partners), which was subsequently integrated with microarray-based expression data to analyse autocrine signalling in cancer cells. At the time, the Database of Ligand-Receptor Partners consisted of 451 interactions in humans; since then, many other databases have catalogued substantially more interacting ligand–receptor pairs, enabling more comprehensive testing of communication processes between cells.
Many studies have enumerated known ligand–receptor pairs using different approaches (Box 1). To facilitate further use and comparison, we collated publicly available lists into a single ligand–receptor pair repository. One extensively used database7 contains ~2,400 human ligand–receptor pairs, obtained from multiple databases and literature curation. Similar approaches allowed researchers to increase the number of known ligand–receptor pairs and to build databases for other organisms (Supplementary Table 1). However, integrating multiple sources of data is challenging and requires reconciliation of the different ways ligand–receptor pair confidence was assessed or how orthologues were determined. Furthermore, few predicted PPIs in databases have been validated, raising the concern of false positives.
While early efforts increased the number of reported ligand–receptor pairs, many lacked information about protein complexes involving multiple subunits. This scenario greatly increases false-positive CCC predictions79. Some proteins, such as transforming growth factor-β receptors80 and cytokine receptors81, require multisubunit assembly for function. A lack of expression of any subunit blocks ligand–receptor interactions and the resulting communication82. Thus, more recent computational tools such as CellPhoneDB30,83, CellChat79 and ICELLNET84 include multimeric proteins and interactions between complexes of both ligands and receptors. Accounting for subunit co-expression better represents functional ligand–receptor interactions. For example, CellChat79 includes ~2,000 ligand–receptor interactions, ~48% of which represent interactions of heterodimers, a substantial increase from the ~900 ligand–receptor pairs in CellPhoneDB.
Additional efforts have incorporated information beyond ligand–receptor pairs to reveal other aspects of CCC, such as the interchange of metabolites or activation of intracellular signalling. For example, metabolite secretion data have been included in communication studies by considering the expression of the producing enzymes85. In this regard, the production of specific metabolites can be inferred from transcriptomic data86,87; however, metabolite concentrations cannot be directly predicted, so concentration-dependent interactions cannot be assessed. Among other efforts, information about downstream signalling gene products and gene regulatory networks can be included25,54 by weighing the potential of using a communication pathway given the intracellular effect on a receiver cell. For this, genes in a downstream pathway to each receptor are obtained from signalling and regulatory networks; then their expression provides additional inputs for scoring the respective ligand–receptor pairs. However, more extensive information on ligand–receptor pairs is required to include downstream genes. A limitation of this information is that rules of gene regulation are not considered, leading to potential false positives and false negatives.
Adding information can be laborious, requiring careful curation of mediator molecules and development of algorithms that incorporate these data. Furthermore, inference methods based on PPI databases are sensitive to the quality of the underlying databases. Nevertheless, PPIs, especially ligand–receptor pairs, have been central to decipher CCC in all strategies.
Box 1 Computational strategies for building lists of interacting proteins.
To reduce the dependency on laborious experiments, lists of interacting proteins can be compiled computationally using the resources mentioned in the table below. These strategies include a heuristic search of gene products as candidates for interaction and manual curation of interacting protein pairs by using available databases or previously curated lists of interacting partners from the literature.
Several high-quality lists of ligand–receptor pairs exist for model organisms7,14–16,22,29,30,32,33,38,53–55,67,84. However, studying cell–cell communication in organisms with fewer protein–protein interaction (PPI) data requires additional effort, especially in imputing ligand–receptor pairs through heuristic approaches and subsequent manual curation to increase the confidence in the list. For instance, some studies use orthologues of human ligand–receptor pair proteins from databases as indicated in the table below to adapt those pairs to the organism of interest, thus delineating core pathways of communication16,33,60. When reference PPI networks or orthologue data are not available, other heuristic procedures to impute protein interactions could be used. A preliminary selection of candidate genes can be done according to their functional annotations. Some genes may lack annotation, so their potential interactions can be imputed with strategies such as protein co-evolution162, modelling of protein structures163 and other in silico techniques for inferring PPIs5. Additionally, machine learning164,165 and data mining166 can also be used, facilitating the search of candidate ligand–receptor pairs.
| Resource | URL | Ref. |
|---|---|---|
| Functional annotations | ||
| Gene Ontology | http://geneontology.org/ | 167 |
| UniProt | https://www.uniprot.org/ | 168 |
| KEGG | https://www.genome.jp/kegg/ | 169 |
| Orthologues | ||
| OrthoDB | https://www.orthodb.org/ | 170 |
| gProfiler | https://biit.cs.ut.ee/gprofiler/orth | 171 |
| Databases of protein–protein interactions | ||
| The Human Protein Atlas | https://www.proteinatlas.org/ | 172 |
| STRING | https://string-db.org/ | 173 |
| BioGRID | https://thebiogrid.org/ | 174 |
| PICKLE | http://www.pickle.gr | 175 |
| APID | http://apid.dep.usal.es | 176 |
| IntAct | https://www.ebi.ac.uk/intact | 177 |
| Pathway Commons | http://www.pathwaycommons.org/ | 178 |
| Ligand–receptor interactions | ||
| IUPHAR/BPS Guide to Pharmacology | https://www.guidetopharmacology.org/ | 179 |
| Resources for cell–cell interactions from the Bader laboratory | http://baderlab.org/CellCellInteractions | 32 |
| Compendium of ligand–receptor pairs in the literature | https://github.com/LewisLabUCSD/Ligand-Receptor-Pairs | 10.1038/s41576-020-00292-x |
BPS, British Pharmacological Society; IUPHAR, International Union of Basic and Clinical Pharmacology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Which communication pathways are cells using?
To elucidate which biological processes are used by interacting cells to communicate, a score is usually estimated for each pair of interacting proteins (Box 1) using the expression of their cognate genes as input to a scoring function (Fig. 2a). Importantly, the main assumption of these methods is that (1) gene expression reflects protein abundance and (2) protein abundance is sufficient to infer the PPI strength, ignoring essential factors for their binding, such as post-translational modifications or multisubunit complex assembly. Here, we focus on ligand–receptor pairs as PPIs (Fig. 2a, step 3); however, these strategies can be extended to any intercellular PPIs.
Communication scores can be binary or continuous (Fig. 2b), each providing different insights into the signalling pathways that cells use. Binary scores are simpler, whereas continuous scores enable more precise quantification of intercellular signalling. For both types of communication scores, we identified two core scoring functions in the literature. The ‘expression thresholding’ and ‘differential combinations’ methods are defined for the binary category, whereas the ‘expression product’ and ‘expression correlation’ approaches quantify continuous scores.
In binary scoring, expression thresholding is widely used because of its easy implementation and interpretation (Table 1). By thresholding expression values of both interacting partners in each ligand–receptor pair, we can measure all communication processes used between cells. If both genes are expressed above a threshold, the ligand–receptor pair is considered ‘active’; otherwise it is ‘inactive’ (assigning ones and zeros, respectively; Fig. 3). Different thresholds can be used for the ligand and receptor44,67. By contrast, the differential combinations method uses any approach to quantify differential gene expression88,89 to identify ‘active’ interacting partners. Thus, this strategy measures communication pathways in a sample- or cell type-specific manner. Regarding input data sets, both binary functions are suitable for either bulk or single-cell data (Fig. 2b); however, bulk-derived samples may miss certain interacting ligand–receptor pairs, as averaging gene expression across all cells can mask signals from low-abundance cells. These binary methods both assume that higher expression is needed for interaction and require the choosing of a gene expression threshold (either significance or gene expression magnitude), which can lead to false positives and/or false negatives7. General thresholds may fail as (1) some proteins have different bioactivities in a concentration-dependent manner90 and (2) mRNA–protein level correlations vary across genes82. Hence, gene-specific thresholds should be developed, as a general threshold may not properly represent the presence and activity of proteins.
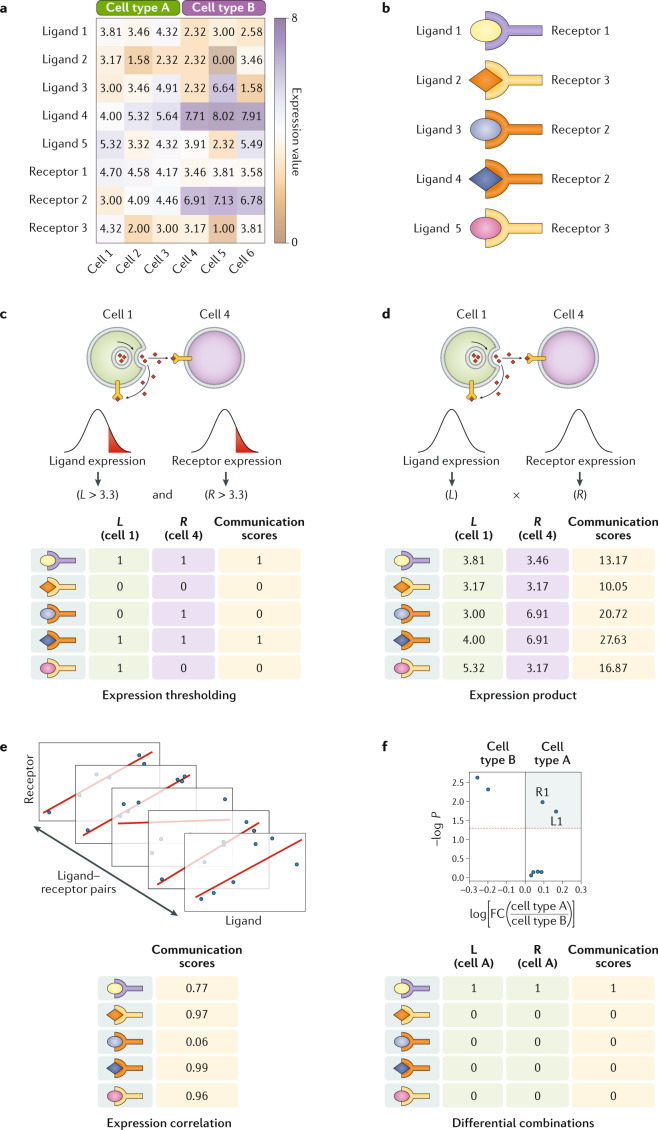
Fig. 3. Toy examples of using core functions to compute communication scores.
Two primary inputs are used for quantifying communication scores: a preprocessed gene expression matrix (part a) and a list of interacting proteins to supervise the analysis (for example, ligand–receptor pairs) (part b). Then a communication score (CS) can be computed for every ligand–receptor pair in a given pair of cells. Here, we show how to perform these calculations for four core functions (parts c–f). These are applied to elucidate paracrine (parts c,d) and autocrine (parts e,f) communication. To assess cell–cell communication, a CS can be computed for each ligand–receptor pair by accounting for the presence of both partners if their expression is greater than a given threshold, which for demonstrative purposes was set arbitrarily to a value of 3.3 (part c), or by multiplying their expression values (part d). Similarly, the CS for each ligand–receptor pair can be the correlation score obtained from their expression across all cell types for autocrine communication (part e). To reveal non-autocrine interactions, the correlation can be computed across pairs of different cells. Particular signatures of each cell type can be extracted through analysing differentially expressed ligands and receptors. Using the cell type-specific differentially expressed genes, we can assign a binary CS and study the ligand–receptors used for autocrine communication (part f). In this example, autocrine communication is evaluated for cell type A by using its differentially expressed genes with respect to cell type B (cell type A-specific genes are located in the coloured quadrant). Analogously to the correlation score, for non-autocrine communication we would need to consider differentially expressed genes in each of the cell types or samples. For a given pair of cells, we can say that a communication pathway is active when the ligand is differentially expressed in one cell and its cognate receptor is differentially expressed in the other. FC, fold change.
Modelling interactions as non-binary processes provides biologically relevant information91 and helps infer active communication pathways. The expression product method computes a continuous value by multiplying the expression of both interacting proteins, and it has successfully found differences in ligand–receptor pair use between cells55,60. For example, communication pathways that were previously described to enhance the expansion of regulatory T cells were also identified to have communication scores linearly correlated with the infiltration of those cells in human metastatic melanoma60, which would not have been possible with binary scores. However, this approach may become problematic if interacting proteins have vastly different transcript levels wherein one protein dominates the interaction signal. Additional data preprocessing, such as cell-type normalization of gene expression using housekeeping genes60 or accounting for correlation between transcripts and proteins7, may mitigate this effect. Although this method can be applied to bulk data, microdissected or single-cell data sets are preferred as they capture the full heterogeneity of expression across cells19,48,49,55,60,62, resulting in clearer differences in expression92.
The second continuous scoring function is the expression correlation method. This score is the correlation between expression values of the interacting proteins across many samples. Thus, the score represents the general behaviour of each ligand–receptor pair in the evaluated groups rather than the individual importance for each pair of samples or cells. Whereas the previous methods can use any gene expression data type, the expression correlation method requires data sets containing many measurements of two populations to compute correlation for each ligand–receptor pair. A potential obstacle for this method is the sparsity of single-cell data sets, which can increase or decrease correlation coefficients in undesirable ways93,94, leading to correlation values that measure sparsity, rather than biology. In addition, the formation of subgroups of data points (representing pairs of cells) due to low expression variance of the ligand or the receptor in an interaction is known to bias correlation and can make this score inappropriate95.
In addition to these core scoring functions, more advanced approaches can generate interaction scores, such as RNA-Magnet53, a method using fuzzy logic to identify candidate ligands and receptors active in cell communication. As mRNAs encoding surface receptors frequently show low abundance, this strategy better accounts for the variance in receptor expression. Other advanced approaches have been wrapped into tools (see the section entitled A growing toolbox to facilitate CCC analysis). Each strategy can decipher CCC but relies on different assumptions that may influence the results. Therefore, knowing the limitations and following best practices in data set preprocessing93,96,97 are crucial to minimize false discoveries.
Distinguishing which pair of cells are more likely to interact
Measurement of individual communication scores facilitates the study of CCC, exposing the roles of specific signalling mechanisms; however, it does not reveal the entire interaction state between cells. Thus, it may be desirable to use an aggregate score to define the interactions between pairs of cells84.
From the few studies that aggregated communication scores into an overall score (Fig. 2a, step 5), the most common approach quantifies the number of active ligand–receptor pairs between cells (that is, the sum of binary communication scores). This score suggests which cells interact more strongly and enables the building of CCC networks to perform graph-based analyses. However, different cell pairs could have similar counts but completely different circuits of communication7, causing inaccuracies. Newer methods attempted to deal with those inaccuracies by weighting CCIs and CCC on the basis of additional data. Hence, these approaches enable the comparison of interactions weighted by their importance, among those that are considered active. One approach computes a probability of intracellular communication using a given ligand–receptor pair between a sender and a receiver cell and then aggregates this into a global CCC score25. Another study proposed a statistical model to evaluate expression variation of individual genes given a cell’s CCIs in its neighbourhood98. This method yields coefficients interpretable as an overall CCI score. Here, there is no aggregation of communication scores as the coefficient is computed from the covariance of different factors, such as the distance between cells, the cellular composition of the neighbourhood and transcriptomes including all genes instead of only ligands and receptors. Although these approximations seem more biologically relevant than just counting the expressed ligand–receptor pairs, they require more detailed information that might not be available in all experimental settings.
Alternative strategies to quantify CCC include Euclidian-like or Jaccard-like distance metrics that represent the number of active ligand–receptor pairs in relation to all ligand–receptors in interacting cells. Furthermore, spatial transcriptomics technologies99–104 may help train machine learning models to predict a physical distance-dependent potential of CCIs105. Expression of ligands and receptors can serve as inputs and distances between cells as outputs.
Although there are many approaches, it remains unclear which metric best captures the underlying biology. Moreover, any strategy relying on gene expression will remain limited to signals captured in the ligand–receptor list, which may miss ways that cells can communicate. Thus, there remains a need for strategies that determine an aggregate CCC potential and functional–spatial relationships of cells.
A growing toolbox to facilitate CCC analysis
In addition to the core scoring functions (Fig. 2b), many tools use advanced statistical methods to identify intercellular communication (Table 2). Existing computational tools can be grouped into one of four categories on the basis of the mathematical models used for identifying ligand–receptor interactions, classified here as (1) differential combination-based, (2) network-based, (3) expression permutation-based and (4) array-based tools.
Table 2.
Existing tools for measuring cell–cell communication
| Tool | Method overview | Output | Visualization | Available in | URL | Refs |
|---|---|---|---|---|---|---|
| Differential combinations | ||||||
| CellTalker | Uses differentially expressed ligands and receptors in each cluster to identify unique interactions between clusters | Upregulated and downregulated interactions between all clusters | Circos plot of differential interactions between clusters | R | https://github.com/arc85/celltalker | 61 |
| iTALK | Enumerates differentially expressed ligand and receptor values to identify LRIs between different clusters | Upregulated and downregulated interactions between all clusters | CCI networks, Circos plots and boxplots | R | https://github.com/Coolgenome/iTALK | 106 |
| PyMINEr | Uses differentially expressed ligand and receptor pairs to identify altered signalling pathways. Detects both activation and inhibition | Upregulated and downregulated interactions between all clusters | Network visualization and Circos plots | Python and standalone application | https://www.sciencescott.com/pyminer | 107 |
| Expression permutation | ||||||
| CellChat | Modelling of LRI is generalized from the Hill equation, including expression of both agonists and antagonists. Significance is computed by permutation | Likelihood of CCC between all clusters for all interactions | Alluvial and Circos plots of communication pathways, dot plots of interactions between clusters | R and Web interface | https://github.com/sqjin/CellChat | 79 |
| CellPhoneDB | Randomly permutes cluster labels to generate a null distribution of LRI scores using protein complex subunit architecture to identify significant interactions | Upregulated and downregulated interactions between all clusters | Heatmap of significant interaction counts, dot plot of LRIs and cluster combinations | Python and Web interface | https://github.com/Teichlab/cellphonedb | 30,83 |
| Giotto | Randomly permutes cluster labels to generate a null distribution of LRI scores using spatial information to identify significant interactions | Upregulated and downregulated interactions between all clusters | Heatmap of significant interaction counts, dot plot of LRIs and cluster combinations | R | https://github.com/RubD/Giotto | 111 |
| ICELLNET | Sums the product of all LRI scores between two clusters to compute an overall CCI score | Intergroup communication scores, P values for these scores and LRI scores | Stacked bar plot of LRIs, network visualization of interacting groups and pathway-level analysis | R | https://github.com/soumelis-lab/ICELLNET | 84 |
| SingleCellSignalR | Uses a regularized ligand–receptor expression product to measure extent of CCC | Interaction scores for each LRI between all clusters in the dataset | Circos plot, tables and graph visualizations of interactions between clusters | R | https://github.com/SCA-IRCM | 112 |
| Graph or network | ||||||
| CCCExplorer | A graph of all signalling pathways is built, then a Fisher-like statistic is computed using ligand, receptor and downstream TF expression to identify significant interactions | Graph visualizations of all interactions | Interactive directed graphs | Standalone application | https://github.com/methodistsmab/CCCExplorer | 67 |
| NicheNet | A network of ligand–receptor pathway interactions is used to measure the predictive power of the ligand for its downstream pathway targets as an interaction score, based on a personalized PageRank algorithm | Ligand interaction scores and expressing cell types for provided target pathway | Circos plot of interactions between cells or clusters | R | https://github.com/saeyslab/nichenetr | 54 |
| SoptSC | Integrates downstream signalling measurements into an LRI scoring function | Upregulated and downregulated interactions between all clusters | Circos plot of interactions between cells | MATLAB and R | 25 | |
| SpaOTsc | An optimal transport model is used to infer CCC from ligand, receptor and downstream component expression | Likelihood of CCC between all clusters for all LRIs | Not implemented | Python | https://github.com/zcang/SpaOTsc | 108 |
| Tensor based | ||||||
| scTensor | Tucker decomposition on a tensor of order three to identify key LRIs present in certain cell types | HTML file with summaries of clustering, decomposition and interaction components | Many options for interaction, expression and pattern visualization | R | https://github.com/rikenbit/scTensor | 113 |
CCC, cell–cell communication; CCI, cell–cell interaction; LRI, ligand–receptor interaction; TF, transcription factor.
In differential combination-based tools, significantly differentially expressed genes between cell clusters in single-cell RNA-seq data are identified, and these lists are analysed for ligand–receptor pairs. Two such tools, iTALK106 and CellTalker61, use slightly different downstream analysis methods to curate the final list of significantly interacting ligand–receptor pairs. Similarly, PyMINEr107 annotates interacting cell types but labels interactions as ‘activating’ or ‘inhibitory’ according to public interaction databases. Another tool, CCCExplorer67, uses differential gene expression and PPI database-guided network analysis of downstream targets and transcription factors to determine activated or deactivated signalling events between two groups of samples or cells. These methods are powerful for discerning ligand–receptor interactions in the background of the rest of the data set but are blind to interactions that are common between all groups.
Network methods are used by several tools and exploit properties of connections between genes. For example, CCCExplorer uses them to identify concerted movements in the expression levels of genes involved in ligand–receptor signalling67. The tools SoptSC25 and NicheNet54 use known interactions between ligands, receptors and downstream targets to build a network of ligand–receptor relationships. The former computes a probability, while the latter uses a personalized PageRank algorithm, in both cases to evaluate the effect of ligand–receptor co-expression on downstream signalling genes in the receiver cell and, thus, obtain a continuous score for ranking ligands and receptors based on this effect. Most recently, a method called ‘SpaOTsc’108 formulates intercellular communication as an optimal transport problem109 using RNA-seq and spatial information. All these tools use not only the expression levels of ligands and receptors to compute interaction scores but also expression levels of downstream signalling targets, which is intended to be a strength of these techniques. However, they do not account for the rules of gene regulation, so a limitation of these methods is that they are blind to signalling crosstalk, where the signals triggered by one receptor could interfere with the signals triggered by another receptor. Pitfall scenarios could be when an intracellular pathway is highly scored because of the expression of its downstream genes, but its activity may be strongly diminished because of inactivation due to another activated pathway that regulates it with post-translational modifications rather than expression control, as may happen in some cytokine signalling pathways110, leading to false positives or false negatives not seen with other strategies.
Expression permutation-based tools are the most widely used among those listed in this Review. These methods compute a communication score for each ligand–receptor pair and evaluate its significance either through cluster label permutation, non-parametric tests to assess differences with the null model, or through empirical methods. To increase confidence, CellPhoneDB30,83, CellChat79 and ICELLNET84 address one of the limitations common to most CCC methods: not considering multisubunit protein complexes. Lists with multimeric proteins are used to assess whether all subunits are simultaneously expressed to identify likely functional ligand–receptor interactions. CellChat also allows multisubunit complexes and incorporates positive and negative effectors into its Hill function-inspired framework. Also, other important features are present in different tools in this category: Giotto111 includes spatial expression information, accounting for the potential of cells to interact given their physical proximity, whereas ICELLNET is the only tool that returns a global CCC score aggregated from all ligand–receptor interactions using percentile normalization and the expression product core function. SingleCellSignalR112 uses a regularized expression product to compute ligand–receptor interaction scores and is the only tool reviewed that provides explicit cut-off values for this score to achieve appropriate false discovery rates based on empirical results.
Tensor-based tools are the most mathematically sophisticated group of tools. Although network computations can be formulated as matrix operations, scTensor113 explicitly models ligand–receptor interactions using a tensor. A tensor of rank three is generated from the data, wherein two dimensions are for ligand and receptor expression by each cell type or cluster within the single-cell RNA-seq data set, respectively, and the third dimension represents all ligand–receptor interactions. Then non-negative Tucker decomposition is performed to decompose this tensor114, resulting in three matrices with coefficients representing the relationship between interacting cells and their respective ligands and receptors. Thus, this tool captures communication pathways in a context where all pairs of cells are simultaneously considered and extracts the relationships between different CCIs and further produces lower error rates than are obtained with independent hypothesis tests. However, interpreting the scores from a tensor decomposition may not be as straightforward as interpreting the scores from other groups of tools.
The tools described also include powerful visualization features that facilitate the interpretation of results. Several of the more common visualization methods are outlined in Fig. 4 and display data by directly plotting ligand–receptor co-expression patterns and communication scores (Fig. 4a–c) and provide higher-level intuition concerning overall CCI levels and the directionality of these effects between cell types (Fig. 4d–f). Thus, several tools not only quantify CCIs and CCC but also facilitate their analysis and interpretation.
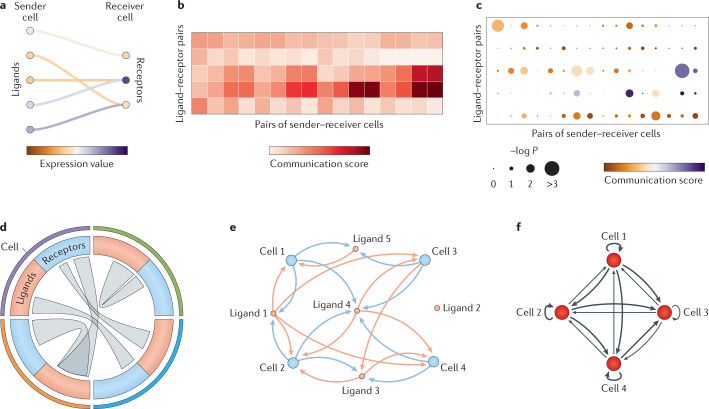
Fig. 4. Common visualization techniques for cell–cell interactions and communication.
a | A Sankey diagram for connecting key ligands from a sender cell to cognate receptors in the receiver cell. Node colour (ligand or receptor) indicates the expression level. b | Heatmap to represent the communication scores for each ligand–receptor interaction in each cell pair. c | Dot plot to show the communication score (colour of dots) and at the same time its significance (size), often obtained from a statistical model or permutation analysis. d | Circos plot or chord diagram to show key communication pathways used by different cell types to communicate. The links start from a ligand (red) and end in a receptor (blue), which are grouped for each cell type (coloured outer arcs). e | Bipartite network where nodes can be either cells or ligands. Edges can be directed only from a cell to a ligand it produces or from a ligand to a cell that expresses its cognate receptor. f | Cell–cell interaction network to represent the potential of cells to interact. Nodes correspond to cells and edges correspond to their interactions. These are directed from a sender cell to a receiver cell, and their thicknesses are proportional to the respective global cell–cell communication scores (for example, number of active ligand–receptor pairs).
Assessing predicted CCC mechanisms
CCC inference should be considered a data-driven process for generating hypotheses, which can lead to different results depending on the strategy adopted (Fig. 3). Thus, validating inferred mechanisms is crucial to confirm associations with phenotypes and behaviours of cell communities. In this section, we discuss current approaches used for this purpose, emphasizing both computational methods to minimize false discoveries and experiments used to validate results, and illustrate hallmark studies that successfully implemented this important step.
Computational minimization of false discoveries
Robust inferences are essential to minimize false positives and false negatives and help reduce the number of validations to perform, which is especially useful when experiments are expensive. From the total pool of inferred ligand–receptor interactions between cells, statistical modelling can assess whether they may result from the null hypothesis and help discard artefactual or non-specific CCC inferences.
Permutation-based analyses can help discard results arising from random noise by prioritizing cell type-enriched ligand–receptor pairs. The cluster labels, representing the cell type, are permuted for each single cell, and the mean gene expression within each permuted cluster is computed, followed by a recalculation of the communication scores. With all communication scores generated for each ligand–receptor pair in a given pair of clusters, a null distribution is built and a P value of the measured communication score can be computed. A full list of tools that use this method is included in the discussion of permutation-based tools (see the section entitled A growing toolbox to facilitate CCC analysis and Table 2). As a representative example, the CellPhoneDB tool was applied on single-cell RNA-seq of human first-trimester placentae to understand the regulation of the immune response and how it prevents harmful maternal responses30.
Subsampling analysis has been applied to evaluate the robustness of the CCC results. Random subsamples of the original data set are used to rerun the CCC analysis. Using the subsampled results, one can compute true-positive and false-positive rates with respect to the original data set results79 and quantify how variable the inferences are, given subtle changes in the composition of cell clusters. CellChat is one tool that has applied this method to compare its performance with that of other tools and measure robustness when identifying the role of a specific population of mouse myeloid cells, termed ‘MYL-A’, in wound healing through transforming growth factor-β signalling79.
Enrichment analyses have also helped reduce false discoveries. A recent study applied this strategy to study the role of structural cells (that is, fibroblasts, endothelial cells and epithelial cells) in priming immune responses and how they interact with immune cells58. It identified ligand–receptor pairs that were enriched between interacting cell types using a Fisher’s exact test over all possible pairs of differentially expressed genes. Supported by other experiments, these interactions revealed the crucial role of structural cells in priming immune response in an organ-specific fashion58.
Data generated with other technologies, such as proteomics, have shaped CCC methods and their potential performance. Specifically, they have helped tune the parameters of methods before making predictions on the gene expression data set of interest, addressing issues originating from imperfect mRNA–protein correlations. For example, Browaeys et al.54 used several gene and protein expression data sets to optimize NicheNet. Similarly, another study112 used gene and protein expression data sets to evaluate false-positive rates and tune the parameters of CCC measurement methods. Thus, integrating other types of data is also important to benchmark and optimize tools.
As discussed here, computational strategies can help reduce false discoveries and facilitate the selection of important results for further research or experimental validation. Although we highlighted cases for which these methods helped drive new hypotheses, many other strategies can be used, such as Wilcoxon’s test60, random generation of data sets68, cross-validation105 and Welch’s test48.
Experimental validation
Many studies use experimental methods to validate CCC mechanisms inferred computationally (Table 1; Supplementary Table 1). These cover three levels of experimental tests: (1) confirmation of the expression of proteins thought to be involved in CCC (for example, through proteomics, enzyme-linked immunosorbent assay, western blot or immunohistochemistry); (2) visualization of interactions between gene products expressed in neighbouring cells (for example, through microscopy coupled with immunostaining, single-molecule fluorescence in situ hybridization or measurement of co-occurrence through flow cytometry); and (3) assessment of the functional role of CCC mediators by performing in vivo or in vitro experiments using activators or inhibitors of the interacting molecules or genetic manipulation of cells to observe effects consistent with the predicted communication.
Although providing only tangential information, rather than validation, studies can confirm the presence of proteins to test the potential of signals contributing to CCC events. Immunohistochemistry or western blots have measured the cell type-specific presence of involved signalling complex members. A recent analysis of human liver development used immunohistochemistry to verify VCAM1 production in macrophages and ITGA4 production in erythroid cells of the human fetal liver to suggest a possible interaction between these two cell types and their relevance to lineage differentiation23. However, these approaches only support and do not validate a predicted interaction.
Immunohistochemistry and other tagging techniques, such as single-molecule fluorescence in situ hybridization, are frequently used to gain further information about cellular localization for predicted CCIs. This set-up verifies the spatial colocalization of CCC mediators and therefore supports the potential occurrence of a CCC event. For example, in addition to immunostaining to confirm protein expression, statistically significant co-occurrence of IL-33 and its receptor was validated on adjacent AT-2 cells and basophils, respectively41, whereas another study used imaging flow cytometry to show interaction between macrophages and liver progenitor cells23. In addition, immunofluorescence was used to colocalize the expression of four ligand–receptor interactions (APOE–LRP1, APOE–LDLR, VTN–KDR and LAMA4–ITGB1) to adjacent cells in mouse brain on embryonic day 14.5 (ref.22), supporting the relevance of these interactions to the developing brain. Thus, these approaches confirm spatial colocalization of predicted interactions.
Functional assessment is the most informative validation technique. This approach evaluates whether the phenotypes observed in the interacting cells are the result of specific CCC events. For example, the interaction between VEGFA and KDR (also known as VEGFR2) in liver bud development was confirmed by dosing KDR inhibitors in vitro into micro-liver buds, significantly reducing hepatoblast abundance and impairing differentiation24. Elsewhere, 33 candidate ligands affecting haematopoietic differentiation were dosed in vitro in haematopoietic stem cells, and 27 significantly affected the differentiation rate and trajectory of these cells15. Similarly, organ structural cells thought to contribute to the immune response were exposed to candidate cytokines, which induced expression changes similar to those seen upon lymphocytic choriomeningitis virus infection, as determined with a mouse model of lymphocytic choriomeningitis virus infection as a reference58.
Challenges and future directions
As the methods measuring CCIs improve and the associated results are experimentally validated, new research opportunities will arise that may improve the reliability of inferred CCIs. Furthermore, novel insights will emerge through the study of interactions between cells from different species and the engineering of phenotypes by manipulating communication events.
Multi-omics integration
Although mRNA and protein levels are qualitatively correlated, transcriptomics may not represent a fully accurate view of intercellular communication, as transcript and protein abundances can be uncoupled by post-transcriptional and post-translational processes82,115,116. For example, the inferred presence of ligand–receptor pairs from transcriptomics may not coincide with their actual presence in proteomic data7,82. Borrowing information from other omics technologies can improve confidence in results16,117, especially for emerging technologies such as Nativeomics118, which detects intact ligand–receptor assemblies using mass spectrometry, and INs-seq119, which couples single-cell RNA-seq with intracellular protein measurements to simultaneously profile transcription factors, signalling activity and metabolism. Moreover, emerging technologies such as single-cell proteomics11 will become important inputs for CCC methods and complement single-cell RNA-seq to improve CCI predictions.
Omics integration can extend beyond gene expression. Mammalian cells are covered by a thick glycocalyx, and most hormones and receptors are glycoproteins. Hence, glycomic data can add contextual information to CCI analyses. Glycosylation has an impact on protein interactions, especially in ligand–receptor binding as many ligands are glycosylated120, and glycosylation can change receptor affinity121–123. This phenomenon may rewire CCIs and affect, for example, development or T cell activation124. Moreover, glycans are involved in interspecies crosstalk, contributing to the specificity of communication125–127. Therefore, integration of additional omics technologies, such as proteomics and glycomics, will help identify additional CCIs that are missed with use of RNA-based methods.
Adding a spatial dimension
Ligand mRNA expression and ligand abundance are not the only factors required for communication: the ligand must also be localized in the correct cellular compartment, which RNA-based methods are blind to, and interacting cells are usually close to each other, which routine single-cell RNA-seq experiments cannot measure. To improve reliability of computed CCIs, it is crucial to account for spatial positions of mediators and interacting cells. Single-cell analyses have considered cell location in mouse bone marrow53,128, demonstrating that cell proximity is key to the study of intercellular communication. The study of spatial proximity in interacting cells is an emerging approach129. One technology for profiling these physical interactions, PIC-seq, uses cell sorting to acquire and transcriptionally profile physically interacting cells (PICs) through massively parallel single-cell RNA-seq130. The study authors presented an algorithm for deconvolving the data to capture signals from intercellular physical interactions. One advantage of this method is that it captures more details in the crosstalk of PICs than single-cell RNA-seq. Although this technology is promising, it is currently limited to studying PICs from only two populations of cells. Nevertheless, similarly to other PIC-based methods128,131,132, PIC-seq can be readily integrated into the pipelines described here and leverage the analysis of CCIs by including the intrinsic spatial information that PICs encode. As more approaches emerge for spatial-based transcriptomics99–104 and proteomics133,134, future studies and algorithms should include this information. Accounting for the physical distance between cells will lead to the generation of new scoring functions that may better capture the potential of cells to communicate and interact53,98,108,111,135. As an example, ligand-specific diffusion constants can be considered to reflect how effectively gene products can mediate long-distance communication136,137.
Adding a temporal dimension
Time is another important factor for studying CCC. It can help elucidate how communication evolves and detect important changes in dynamic processes, such as cellular differentiation and the immune response. One can use samples taken at different time points or infer pseudotimes from the RNA-seq data set as happens when one is studying cellular differentiation through lineage tracing138,139. Then CCC analyses on each (pseudo) time point can identify active ligand–receptor pairs and test the overall interaction potential between cells25. However, time-dependent behaviours might be uncoupled from mRNA levels, hindering the detection of changes through transcriptomics-based CCC predictions. mRNA expression can be temporarily disconnected from the activity of the products owing to long half-lives or storage of the products140. It can also take longer to reach the appropriate abundance of a ligand or a receptor than the production of the cognate mRNA82. For example, endothelial cells store cell adhesion molecules, such as P-selectins, in granules, which are quickly mobilized to the cell surface to start the recruitment of immune cells, instead of expressing those proteins de novo141. Thus, time-dependent analyses can improve CCC discoveries, but other data types should be integrated to distinguish whether phenotypes stem from CCC or other dynamics of gene expression.
Shedding light on interspecies interactions
Comprehensive lists of ligand–receptor pairs or interacting surface-secreted proteins are crucial for algorithms that study intraspecies CCIs. However, study of interspecies interactions — for example, humans and pathogens142–145 — requires lists of molecular interactions between species. An opportunity awaits to define interspecies PPI lists with omics methods. Considering the avalanche of data generated in the microbiome field146–149 and recent approaches for modelling microbial communities150, lists of interacting proteins or cross-species ligand–receptor pairs to enable CCI analyses would evidently yield novel discoveries. For example, a study mapped the interaction between inclusion membrane proteins secreted by Chlamydia trachomatis and cognate human proteins145, providing insights into the host machinery this pathogen uses to establish the intracellular niche needed for infection. Thus, even small-scale lists of interacting partners between the host and a single microorganism, instead of the whole microbiota, will open great opportunities to use CCC methods to understand infections and diseases on the basis of host–pathogen interactions151,152. Although this endeavour will require validation of putative interactions, as done for human–virus protein interactions153–156, results from such studies will have a considerable impact on biomedicine and microbiome fields.
Predicting and manipulating phenotypes
Models for predicting phenotypes were previously trained with the active communication pathways underlying intercellular interactions71. As more models are developed and their predictive power is increased, they will enable the identification of drug targets and manipulation of phenotypes through cell engineering. Particularly, removing or adding communication pathways with genome editing and cell engineering technologies157 will modify cellular phenotypes to control how pairs of cells interact. This approach will have a great impact on many fields, such as developmental biology158, wherein CCIs are fundamental for cell differentiation into specific lineages. Biomedicine and biotherapeutics will benefit further from controlling CCIs, particularly in modifying disease courses, as is currently done with immune checkpoint inhibitors159,160. As a proof of concept, a tool that induces gene activation when specific cells interact or are directly in contact was recently built by combining a synthetic Notch receptor and the CRISPR–Cas9 system161. This biological device enabled customization of cell behaviours as observed through the expression of reporter genes. As reporters may be replaced by activators or inhibitors of other communication pathways, this tool holds great potential for therapeutic uses. Thus, manipulation of interaction pathways to control CCIs is feasible, with predictive models helping to decide which modifications to perform.
Conclusions
Incredible advances are now emerging to infer CCIs and CCC from gene expression. The diverse strategies applied have elucidated fundamental roles of cells within their communities and how they shape cellular functions, with great potential for future applications, especially in biomedicine and biotherapeutics. Each approach for inferring CCIs and CCC has its own assumptions and limitations to consider; when one is using such strategies, it is important to be aware of these strengths and weaknesses and to choose appropriate parameters for analyses. Methodological and technological challenges remain, but many opportunities exist to increase our understanding of intercellular interactions.
Supplementary information
Acknowledgements
E.A. is supported by the Chilean Agencia Nacional de Investigación y Desarrollo through its scholarship programme DOCTORADO BECAS CHILE/2018 - 72190270 and by the Fulbright Commission. A.O. is supported by the US NLM (T15LM011271). O.H. is supported by the US NIH (U01CA196406). The authors also thank A. Perez-Lopez, C. Zuñiga, J. Tibocha-Bonilla, L. Zaramela, M. Kumar and P. Tamayo for providing meaningful feedback and discussion. This work was also supported by the US NIGMS (R35 GM119850; N.E.L.).
Glossary
- Cell–cell interactions
(CCIs). Physical interactions between two or more cells, which can be mediated by proteins, ligands, sugars or other biomolecules.
- Receptors
Proteins that bind to other biomolecules to receive or amplify a signal. They are most commonly membrane-bound but can also be found in the cytoplasm.
- Ligands
Biomolecules that bind to receptors and change the activity, conformation or other biological properties of the receptor, triggering a signalling event.
- Extracellular matrix
Three-dimensional organization of biomolecules located in the extracellular space. It provides structural and functional support to neighbouring cells.
- Cell–cell communication
(CCC). Subset of cell–cell interactions involving biochemical signals that are sent between or within cells and generate an intracellular effect.
- Protein–protein interactions
(PPIs). Physical interaction between two proteins, often involved in structural systems, signal transduction or metabolic processes.
- Network
A set of nodes with defined pairwise attributes. For example, a protein–protein interaction network would consist of proteins as nodes with attributes linking nodes that are known to interact with each other.
- Communication pathways
Molecular components used for an intercellular communication event, usually corresponding to a ligand–receptor pair.
- Interactome
Network of biomolecule interactions within and between cells.
- False positives
In classification, a false positive occurs when a negative example is assigned a positive label. For the purpose of this Review, this means a non-interacting ligand–receptor pair is labelled as interacting.
- False negatives
A false negative occurs when a positive example is assigned a negative label. For the purpose of this Review, this means a true interacting ligand–receptor pair is labelled as not interacting.
- Post-translational modifications
Covalent modification of amino acid residues on a protein, commonly altering function, structure or localization. Phosphorylation, acetylation and glycosylation are among the most common.
- Signalling pathways
The network of biomolecules that serve to transmit signals and induce cellular responses. Post-translational modification of proteins is the most common way signals are propagated.
- Fuzzy logic
Extension of standard Boolean logic to define truth values of variables, encompassing real values between 0 and 1 both inclusive, instead of binary values.
- Permutation
Random reassignment of sample labels, frequently used to compute null models in biological systems.
- Differentially expressed genes
Genes identified as more highly (or lowly) expressed in one condition versus the other after comparison of their expression values between two conditions.
- PageRank algorithm
Algorithm that takes as input a network and quantifies the importance of each node on the basis of centrality and connectedness to other central nodes.
- Null model
Statistical model under which there is no interaction or difference between the groups being tested.
- Multisubunit protein complexes
Quaternary structures of proteins involving the non-covalent interaction of two or more proteins to generate a functional unit.
- Tensor
Higher N-dimensional generalizations of matrices and vectors. Vectors are tensors of rank 1, matrices are tensors of rank 2.
- Tucker decomposition
Decomposing of a tensor of rank N as a product of a set of N matrices and one core tensor. Used to summarize data, similarly to principal component analysis.
Author contributions
E.A. and A.O. researched the literature. All authors contributed to discussions of the content and wrote, reviewed and/or edited the manuscript before submission.
Competing interests
The authors declare no competing interests.
Footnotes
Peer review information
Nature Reviews Genetics thanks Q. Nie; V. Soumelis; and R. Vento-Tormo, who co-reviewed with A. Arutyunyan, for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Ligand–receptor pair repository: https://github.com/LewisLabUCSD/Ligand-Receptor-Pairs
These authors contributed equally: Erick Armingol, Adam Officer.
Contributor Information
Olivier Harismendy, Email: oharismendy@ucsd.edu.
Nathan E. Lewis, Email: nlewisres@ucsd.edu
Supplementary information
Supplementary information is available for this paper at 10.1038/s41576-020-00292-x.
References
- 1.Zhou X, et al. Circuit design features of a stable two-cell system. Cell. 2018;172:744–757.e17. doi: 10.1016/j.cell.2018.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rouault H, Hakim V. Different cell fates from cell-cell interactions: core architectures of two-cell bistable networks. Biophys. J. 2012;102:417–426. doi: 10.1016/j.bpj.2011.11.4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonnans C, Chou J, Werb Z. Remodelling the extracellular matrix in development and disease. Nat. Rev. Mol. Cell Biol. 2014;15:786–801. doi: 10.1038/nrm3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bich L, Pradeu T, Moreau J-F. Understanding multicellularity: the functional organization of the intercellular space. Front. Physiol. 2019;10:1170. doi: 10.3389/fphys.2019.01170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rao VS, Srinivasa Rao V, Srinivas K, Sujini GN, Sunand Kumar GN. Protein-protein interaction detection: methods and analysis. Int. J. Proteom. 2014;2014:1–12. doi: 10.1155/2014/147648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou Y, et al. Evaluation of single-cell cytokine secretion and cell-cell interactions with a hierarchical loading microwell chip. Cell Rep. 2020;31:107574. doi: 10.1016/j.celrep.2020.107574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramilowski JA, et al. A draft network of ligand-receptor-mediated multicellular signalling in human. Nat. Commun. 2015;6:7866. doi: 10.1038/ncomms8866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 9.Fend F, et al. Immuno-LCM: laser capture microdissection of immunostained frozen sections for mRNA analysis. Am. J. Pathol. 1999;154:61–66. doi: 10.1016/S0002-9440(10)65251-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang F, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods. 2009;6:377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 11.Marx V. A dream of single-cell proteomics. Nat. Methods. 2019;16:809–812. doi: 10.1038/s41592-019-0540-6. [DOI] [PubMed] [Google Scholar]
- 12.Grün D, et al. Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature. 2015;525:251–255. doi: 10.1038/nature14966. [DOI] [PubMed] [Google Scholar]
- 13.Park J-E, et al. A cell atlas of human thymic development defines T cell repertoire formation. Science. 2020;367:eaay3224. doi: 10.1126/science.aay3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kirouac DC, et al. Dynamic interaction networks in a hierarchically organized tissue. Mol. Syst. Biol. 2010;6:417. doi: 10.1038/msb.2010.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qiao W, et al. Intercellular network structure and regulatory motifs in the human hematopoietic system. Mol. Syst. Biol. 2014;10:741. doi: 10.15252/msb.20145141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yuzwa SA, et al. Proneurogenic ligands defined by modeling developing cortex growth factor communication networks. Neuron. 2016;91:988–1004. doi: 10.1016/j.neuron.2016.07.037. [DOI] [PubMed] [Google Scholar]
- 17.Paik DT, et al. Large-scale single-cell RNA-seq reveals molecular signatures of heterogeneous populations of human induced pluripotent stem cell-derived endothelial cells. Circ. Res. 2018;123:443–450. doi: 10.1161/CIRCRESAHA.118.312913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li G, et al. Single cell expression analysis reveals anatomical and cell cycle-dependent transcriptional shifts during heart development. Development. 2019;146:dev173476. doi: 10.1242/dev.173476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schiebinger G, et al. Optimal-transport analysis of single-cell gene expression identifies developmental trajectories in reprogramming. Cell. 2019;176:928–943.e22. doi: 10.1016/j.cell.2019.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xue Y, et al. A 3D atlas of hematopoietic stem and progenitor cell expansion by multi-dimensional RNA-seq analysis. Cell Rep. 2019;27:1567–1578.e5. doi: 10.1016/j.celrep.2019.04.030. [DOI] [PubMed] [Google Scholar]
- 21.Basson MA. Signaling in cell differentiation and morphogenesis. Cold Spring Harb. Perspect. Biol. 2012;4:a008151. doi: 10.1101/cshperspect.a008151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sheikh BN, et al. Systematic identification of cell-cell communication networks in the developing brain. iScience. 2019;21:273–287. doi: 10.1016/j.isci.2019.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Popescu D-M, et al. Decoding human fetal liver haematopoiesis. Nature. 2019;574:365–371. doi: 10.1038/s41586-019-1652-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Camp JG, et al. Multilineage communication regulates human liver bud development from pluripotency. Nature. 2017;546:533–538. doi: 10.1038/nature22796. [DOI] [PubMed] [Google Scholar]
- 25.Wang S, Karikomi M, MacLean AL, Nie Q. Cell lineage and communication network inference via optimization for single-cell transcriptomics. Nucleic Acids Res. 2019;47:e66. doi: 10.1093/nar/gkz204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joost S, et al. Single-cell transcriptomics reveals that differentiation and spatial signatures shape epidermal and hair follicle heterogeneity. Cell Syst. 2016;3:221–237.e9. doi: 10.1016/j.cels.2016.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Joost S, et al. Single-cell transcriptomics of traced epidermal and hair follicle stem cells reveals rapid adaptations during wound healing. Cell Rep. 2018;25:585–597.e7. doi: 10.1016/j.celrep.2018.09.059. [DOI] [PubMed] [Google Scholar]
- 28.Wang S, et al. Single cell transcriptomics of human epidermis reveals basal stem cell transition states. Preprint at. bioRxiv. 2019 doi: 10.1101/784579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pavličev M, et al. Single-cell transcriptomics of the human placenta: inferring the cell communication network of the maternal-fetal interface. Genome Res. 2017;27:349–361. doi: 10.1101/gr.207597.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vento-Tormo R, et al. Single-cell reconstruction of the early maternal-fetal interface in humans. Nature. 2018;563:347–353. doi: 10.1038/s41586-018-0698-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arneson D, et al. Single cell molecular alterations reveal target cells and pathways of concussive brain injury. Nat. Commun. 2018;9:3894. doi: 10.1038/s41467-018-06222-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ximerakis M, et al. Single-cell transcriptomic profiling of the aging mouse brain. Nat. Neurosci. 2019;22:1696–1708. doi: 10.1038/s41593-019-0491-3. [DOI] [PubMed] [Google Scholar]
- 33.Skelly DA, et al. Single-cell transcriptional profiling reveals cellular diversity and intercommunication in the mouse heart. Cell Rep. 2018;22:600–610. doi: 10.1016/j.celrep.2017.12.072. [DOI] [PubMed] [Google Scholar]
- 34.Wang L, et al. Single-cell reconstruction of the adult human heart during heart failure and recovery reveals the cellular landscape underlying cardiac function. Nat. Cell Biol. 2020;22:108–119. doi: 10.1038/s41556-019-0446-7. [DOI] [PubMed] [Google Scholar]
- 35.Wu H, et al. Single-cell transcriptomics of a human kidney allograft biopsy specimen defines a diverse inflammatory response. J. Am. Soc. Nephrol. 2018;29:2069–2080. doi: 10.1681/ASN.2018020125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu H, Kirita Y, Donnelly EL, Humphreys BD. Advantages of single-nucleus over single-cell RNA sequencing of adult kidney: rare cell types and novel cell states revealed in fibrosis. J. Am. Soc. Nephrol. 2019;30:23–32. doi: 10.1681/ASN.2018090912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stewart BJ, et al. Spatiotemporal immune zonation of the human kidney. Science. 2019;365:1461–1466. doi: 10.1126/science.aat5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ding C, et al. A cell-type-resolved liver proteome. Mol. Cell. Proteom. 2016;15:3190–3202. doi: 10.1074/mcp.M116.060145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bonnardel J, et al. Stellate cells, hepatocytes, and endothelial cells imprint the Kupffer cell identity on monocytes colonizing the liver macrophage niche. Immunity. 2019;51:638–654.e9. doi: 10.1016/j.immuni.2019.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zepp JA, et al. Distinct mesenchymal lineages and niches promote epithelial self-renewal and myofibrogenesis in the lung. Cell. 2017;170:1134–1148.e10. doi: 10.1016/j.cell.2017.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cohen M, et al. Lung single-cell signaling interaction map reveals basophil role in macrophage imprinting. Cell. 2018;175:1031–1044.e18. doi: 10.1016/j.cell.2018.09.009. [DOI] [PubMed] [Google Scholar]
- 42.Raredon MSB, et al. Single-cell connectomic analysis of adult mammalian lungs. Sci. Adv. 2019;5:eaaw3851. doi: 10.1126/sciadv.aaw3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Niethamer TK, et al. Defining the role of pulmonary endothelial cell heterogeneity in the response to acute lung injury. eLife. 2020;9:e53072. doi: 10.7554/eLife.53072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suryawanshi H, Morozov P, Straus A, Sahasrabudhe N. A single-cell survey of the human first-trimester placenta and decidua. Sci. Adv. 2018 doi: 10.1126/sciadv.aau4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hu Y, et al. Dissecting the transcriptome landscape of the human fetal neural retina and retinal pigment epithelium by single-cell RNA-seq analysis. PLoS Biol. 2019;17:e3000365. doi: 10.1371/journal.pbio.3000365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hrvatin S, et al. Single-cell analysis of experience-dependent transcriptomic states in the mouse visual cortex. Nat. Neurosci. 2018;21:120–129. doi: 10.1038/s41593-017-0029-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Solé-Boldo L, et al. Single-cell transcriptomes of the human skin reveal age-related loss of fibroblast priming. Commun. Biol. 2020;3:188. doi: 10.1038/s42003-020-0922-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fernandez DM, et al. Single-cell immune landscape of human atherosclerotic plaques. Nat. Med. 2019;25:1576–1588. doi: 10.1038/s41591-019-0590-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martin JC, et al. Single-cell analysis of Crohn’s disease lesions identifies a pathogenic cellular module associated with resistance to anti-TNF therapy. Cell. 2019;178:1493–1508.e20. doi: 10.1016/j.cell.2019.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiong X, et al. Landscape of intercellular crosstalk in healthy and NASH liver revealed by single-cell secretome gene analysis. Mol. Cell. 2019;75:644–660.e5. doi: 10.1016/j.molcel.2019.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vieira Braga FA, et al. A cellular census of human lungs identifies novel cell states in health and in asthma. Nat. Med. 2019;25:1153–1163. doi: 10.1038/s41591-019-0468-5. [DOI] [PubMed] [Google Scholar]
- 52.Qi F, Qian S, Zhang S, Zhang Z. Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses. Biochem. Biophys. Res. Commun. 2020 doi: 10.1016/j.bbrc.2020.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baccin C, et al. Combined single-cell and spatial transcriptomics reveal the molecular, cellular and spatial bone marrow niche organization. Nat. Cell Biol. 2020;22:38–48. doi: 10.1038/s41556-019-0439-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Browaeys R, Saelens W, Saeys Y. NicheNet: modeling intercellular communication by linking ligands to target genes. Nat. Methods. 2019 doi: 10.1038/s41592-019-0667-5. [DOI] [PubMed] [Google Scholar]
- 55.Cain MP, Hernandez BJ, Chen J. Quantitative single-cell interactomes in normal and virus-infected mouse lungs. Preprint at. bioRxiv. 2020 doi: 10.1101/2020.02.05.936054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chua RL, et al. COVID-19 severity correlates with airway epithelium–immune cell interactions identified by single-cell analysis. Nat. Biotechnol. 2020 doi: 10.1038/s41587-020-0602-4. [DOI] [PubMed] [Google Scholar]
- 57.Rieckmann JC, et al. Social network architecture of human immune cells unveiled by quantitative proteomics. Nat. Immunol. 2017;18:583–593. doi: 10.1038/ni.3693. [DOI] [PubMed] [Google Scholar]
- 58.Krausgruber T, et al. Structural cells are key regulators of organ-specific immune responses. Nature. 2020 doi: 10.1038/s41586-020-2424-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Puram SV, et al. Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell. 2017;171:1611–1624.e24. doi: 10.1016/j.cell.2017.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kumar MP, et al. Analysis of single-cell RNA-seq identifies cell-cell communication associated with tumor characteristics. Cell Rep. 2018;25:1458–1468.e4. doi: 10.1016/j.celrep.2018.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cillo AR, et al. Immune landscape of viral- and carcinogen-driven head and neck cancer. Immunity. 2019 doi: 10.1016/j.immuni.2019.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang Q, et al. Landscape and dynamics of single immune cells in hepatocellular carcinoma. Cell. 2019;179:829–845.e20. doi: 10.1016/j.cell.2019.10.003. [DOI] [PubMed] [Google Scholar]
- 63.Song Q, et al. Dissecting intratumoral myeloid cell plasticity by single cell RNA-seq. Cancer Med. 2019;68:7. doi: 10.1002/cam4.2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang M, et al. Single cell transcriptomic architecture and intercellular crosstalk of human intrahepatic cholangiocarcinoma. J. Hepatol. 2020 doi: 10.1016/j.jhep.2020.05.039. [DOI] [PubMed] [Google Scholar]
- 65.Finotello F, Rieder D, Hackl H, Trajanoski Z. Next-generation computational tools for interrogating cancer immunity. Nat. Rev. Genet. 2019;20:724–746. doi: 10.1038/s41576-019-0166-7. [DOI] [PubMed] [Google Scholar]
- 66.Zitvogel L, Kroemer G. Targeting PD-1/PD-L1 interactions for cancer immunotherapy. Oncoimmunology. 2012;1:1223–1225. doi: 10.4161/onci.21335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Choi H, et al. Transcriptome analysis of individual stromal cell populations identifies stroma-tumor crosstalk in mouse lung cancer model. Cell Rep. 2015;10:1187–1201. doi: 10.1016/j.celrep.2015.01.040. [DOI] [PubMed] [Google Scholar]
- 68.Yeung T-L, et al. Systematic identification of druggable epithelial–stromal crosstalk signaling networks in ovarian cancer. J. Natl. Cancer Inst. 2019;111:272–282. doi: 10.1093/jnci/djy097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Graeber TG, Eisenberg D. Bioinformatic identification of potential autocrine signaling loops in cancers from gene expression profiles. Nat. Genet. 2001;29:295–300. doi: 10.1038/ng755. [DOI] [PubMed] [Google Scholar]
- 70.Zhou JX, Taramelli R, Pedrini E, Knijnenburg T, Huang S. Extracting intercellular signaling network of cancer tissues using ligand-receptor expression patterns from whole-tumor and single-cell transcriptomes. Sci. Rep. 2017;7:8815. doi: 10.1038/s41598-017-09307-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yuan D, Tao Y, Chen G, Shi T. Systematic expression analysis of ligand-receptor pairs reveals important cell-to-cell interactions inside glioma. Cell Commun. Signal. 2019;17:48. doi: 10.1186/s12964-019-0363-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Murali T, et al. DroID 2011: a comprehensive, integrated resource for protein, transcription factor, RNA and gene interactions for Drosophila. Nucleic Acids Res. 2011;39:D736–D743. doi: 10.1093/nar/gkq1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thurmond J, et al. FlyBase 2.0: the next generation. Nucleic Acids Res. 2019;47:D759–D765. doi: 10.1093/nar/gky1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Girard LR, et al. WormBook: the online review of Caenorhabditis elegans biology. Nucleic Acids Res. 2007;35:D472–D475. doi: 10.1093/nar/gkl894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Harris TW, et al. WormBase: a comprehensive resource for nematode research. Nucleic Acids Res. 2010;38:D463–D467. doi: 10.1093/nar/gkp952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Oh E-Y, et al. Extensive rewiring of epithelial-stromal co-expression networks in breast cancer. Genome Biol. 2015;16:128. doi: 10.1186/s13059-015-0675-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Han X, et al. Mapping the mouse cell atlas by microwell-seq. Cell. 2018;172:1091–1107.e17. doi: 10.1016/j.cell.2018.02.001. [DOI] [PubMed] [Google Scholar]
- 78.Krämer A, Green J, Pollard J, Jr, Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jin S, et al. Inference and analysis of cell-cell communication using CellChat. Preprint at. bioRxiv. 2020 doi: 10.1101/2020.07.21.214387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wrana JL, et al. TGFβ signals through a heteromeric protein kinase receptor complex. Cell. 1992;71:1003–1014. doi: 10.1016/0092-8674(92)90395-s. [DOI] [PubMed] [Google Scholar]
- 81.Sato N, Miyajima A. Multimeric cytokine receptors: common versus specific functions. Curr. Opin. Cell Biol. 1994;6:174–179. doi: 10.1016/0955-0674(94)90133-3. [DOI] [PubMed] [Google Scholar]
- 82.Buccitelli C, Selbach M. mRNAs, proteins and the emerging principles of gene expression control. Nat. Rev. Genet. 2020 doi: 10.1038/s41576-020-0258-4. [DOI] [PubMed] [Google Scholar]
- 83.Efremova M, Vento-Tormo M, Teichmann SA, Vento-Tormo R. CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat. Protoc. 2020 doi: 10.1038/s41596-020-0292-x. [DOI] [PubMed] [Google Scholar]
- 84.Noël F, et al. ICELLNET: a transcriptome-based framework to dissect intercellular communication. Preprint at. bioRxiv. 2020 doi: 10.1101/2020.03.05.976878. [DOI] [Google Scholar]
- 85.Komurov K. Modeling community-wide molecular networks of multicellular systems. Bioinformatics. 2012;28:694–700. doi: 10.1093/bioinformatics/btr718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Richelle A, Joshi C, Lewis NE. Assessing key decisions for transcriptomic data integration in biochemical networks. PLoS Comput. Biol. 2019;15:e1007185. doi: 10.1371/journal.pcbi.1007185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Richelle A, et al. What does your cell really do? Model-based assessment of mammalian cells metabolic functionalities using omics data. Preprint at. bioRxiv. 2020 doi: 10.1101/2020.04.26.057943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Costa-Silva J, Domingues D, Lopes FM. RNA-Seq differential expression analysis: an extended review and a software tool. PLoS ONE. 2017;12:e0190152. doi: 10.1371/journal.pone.0190152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang T, Li B, Nelson CE, Nabavi S. Comparative analysis of differential gene expression analysis tools for single-cell RNA sequencing data. BMC Bioinformatics. 2019;20:40. doi: 10.1186/s12859-019-2599-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Maedler K, et al. Low concentration of interleukin-1beta induces FLICE-inhibitory protein-mediated beta-cell proliferation in human pancreatic islets. Diabetes. 2006;55:2713–2722. doi: 10.2337/db05-1430. [DOI] [PubMed] [Google Scholar]
- 91.Middendorf TR, Aldrich RW. The structure of binding curves and practical identifiability of equilibrium ligand-binding parameters. J. Gen. Physiol. 2017;149:121–147. doi: 10.1085/jgp.201611703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Goldman SL, et al. The impact of heterogeneity on single-cell sequencing. Front. Genet. 2019;10:8. doi: 10.3389/fgene.2019.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.AlJanahi AA, Danielsen M, Dunbar CE. An introduction to the analysis of single-cell RNA-sequencing data. Mol. Ther. Methods Clin. Dev. 2018;10:189–196. doi: 10.1016/j.omtm.2018.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lähnemann D, et al. Eleven grand challenges in single-cell data science. Genome Biol. 2020;21:31. doi: 10.1186/s13059-020-1926-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Aggarwal R, Ranganathan P. Common pitfalls in statistical analysis: the use of correlation techniques. Perspect. Clin. Res. 2016;7:187–190. doi: 10.4103/2229-3485.192046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Conesa A, et al. A survey of best practices for RNA-seq data analysis. Genome Biol. 2016;17:13. doi: 10.1186/s13059-016-0881-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Luecken MD, Theis FJ. Current best practices in single-cell RNA-seq analysis: a tutorial. Mol. Syst. Biol. 2019;15:e8746. doi: 10.15252/msb.20188746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Arnol D, Schapiro D, Bodenmiller B, Saez-Rodriguez J, Stegle O. Modeling cell-cell interactions from spatial molecular data with spatial variance component analysis. Cell Rep. 2019;29:202–211. doi: 10.1016/j.celrep.2019.08.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Navarro JF, Sjöstrand J, Salmén F, Lundeberg J, Ståhl PL. ST Pipeline: an automated pipeline for spatial mapping of unique transcripts. Bioinformatics. 2017;33:2591–2593. doi: 10.1093/bioinformatics/btx211. [DOI] [PubMed] [Google Scholar]
- 101.Rodriques SG, et al. Slide-seq: a scalable technology for measuring genome-wide expression at high spatial resolution. Science. 2019;363:1463–1467. doi: 10.1126/science.aaw1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Vickovic S, et al. High-definition spatial transcriptomics for in situ tissue profiling. Nat. Methods. 2019;16:987–990. doi: 10.1038/s41592-019-0548-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Moncada R, et al. Integrating microarray-based spatial transcriptomics and single-cell RNA-seq reveals tissue architecture in pancreatic ductal adenocarcinomas. Nat. Biotechnol. 2020 doi: 10.1038/s41587-019-0392-8. [DOI] [PubMed] [Google Scholar]
- 104.Ståhl PL, et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science. 2016;353:78–82. doi: 10.1126/science.aaf2403. [DOI] [PubMed] [Google Scholar]
- 105.Li D, Ding J, Bar-Joseph Z. Identifying signaling genes in spatial single cell expression data. Preprint at. bioRxiv. 2020 doi: 10.1101/2020.07.27.221465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang Y, et al. iTALK: an R package to characterize and illustrate intercellular communication. Preprint at. bioRxiv. 2019 doi: 10.1101/507871. [DOI] [Google Scholar]
- 107.Tyler SR, et al. PyMINEr finds gene and autocrine-paracrine networks from human islet scRNA-Seq. Cell Rep. 2019;26:1951–1964.e8. doi: 10.1016/j.celrep.2019.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cang Z, Nie Q. Inferring spatial and signaling relationships between cells from single cell transcriptomic data. Nat. Commun. 2020;11:2084. doi: 10.1038/s41467-020-15968-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Titouan, V., Courty, N., Tavenard, R., Laetitia, C. & Flamary, R. in Proceedings of the 36th International Conference on Machine Learning vol. 97 (eds Chaudhuri, K. & Salakhutdinov, R.) 6275–6284 (PMLR, 2019).
- 110.Yasukawa H, Sasaki A, Yoshimura A. Negative regulation of cytokine signaling pathways. Annu. Rev. Immunol. 2000;18:143–164. doi: 10.1146/annurev.immunol.18.1.143. [DOI] [PubMed] [Google Scholar]
- 111.Dries R, et al. Giotto, a pipeline for integrative analysis and visualization of single-cell spatial transcriptomic data. Preprint at. bioRxiv. 2019 doi: 10.1101/701680. [DOI] [Google Scholar]
- 112.Cabello-Aguilar S, et al. SingleCellSignalR: inference of intercellular networks from single-cell transcriptomics. Nucleic Acids Res. 2020;48:e55. doi: 10.1093/nar/gkaa183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tsuyuzaki K, Ishii M, Nikaido I. Uncovering hypergraphs of cell-cell interaction from single cell RNA-sequencing data. Preprint at. bioRxiv. 2019 doi: 10.1101/566182. [DOI] [Google Scholar]
- 114.Kim, Y. & Choi, S. in 2007 IEEE Conference on Computer Vision and Pattern Recognition 1–8 (IEEE, 2007).
- 115.Liu Y, Beyer A, Aebersold R. On the dependency of cellular protein levels on mRNA abundance. Cell. 2016;165:535–550. doi: 10.1016/j.cell.2016.03.014. [DOI] [PubMed] [Google Scholar]
- 116.Wegler C, et al. Global variability analysis of mRNA and protein concentrations across and within human tissues. NAR Genom. Bioinform. 2019;2:lqz010. doi: 10.1093/nargab/lqz010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Grandclaudon M, et al. A quantitative multivariate model of human dendritic cell-T helper cell communication. Cell. 2019;179:432–447.e21. doi: 10.1016/j.cell.2019.09.012. [DOI] [PubMed] [Google Scholar]
- 118.Gault J, et al. Combining native and ‘omics’ mass spectrometry to identify endogenous ligands bound to membrane proteins. Nat. Methods. 2020;17:505–508. doi: 10.1038/s41592-020-0821-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Katzenelenbogen Y, et al. Coupled scRNA-seq and intracellular protein activity reveal an immunosuppressive role of TREM2 in cancer. Cell. 2020 doi: 10.1016/j.cell.2020.06.032. [DOI] [PubMed] [Google Scholar]
- 120.Madsen TD, et al. An atlas of O-linked glycosylation on peptide hormones reveals diverse biological roles. Nat. Commun. 2020;11:4033. doi: 10.1038/s41467-020-17473-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Arey, B. J. in Glycosylation (ed. Petrescu, S.) (InTech, 2012).
- 122.Lux, A. & Nimmerjahn, F. in Crossroads between Innate and Adaptive Immunity III 113–124 (Springer, 2011).
- 123.Boscher C, Dennis JW, Nabi IR. Glycosylation, galectins and cellular signaling. Curr. Opin. Cell Biol. 2011;23:383–392. doi: 10.1016/j.ceb.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 124.De Bousser E, Meuris L, Callewaert N, Festjens N. Human T cell glycosylation and implications on immune therapy for cancer. Hum. Vaccin. Immunother. 2020 doi: 10.1080/21645515.2020.1730658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Beltrao P, Bork P, Krogan NJ, van Noort V. Evolution and functional cross-talk of protein post-translational modifications. Mol. Syst. Biol. 2013;9:714. doi: 10.1002/msb.201304521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Tytgat HLP, de Vos WM. Sugar coating the envelope: glycoconjugates for microbe-host crosstalk. Trends Microbiol. 2016;24:853–861. doi: 10.1016/j.tim.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 127.Gagneux, P., Aebi, M. & Varki, A. in Essentials of Glycobiology (eds Varki, A. et al.) (Cold Spring Harbor Laboratory Press, 2017)
- 128.Boisset J-C, et al. Mapping the physical network of cellular interactions. Nat. Methods. 2018;15:547–553. doi: 10.1038/s41592-018-0009-z. [DOI] [PubMed] [Google Scholar]
- 129.Shao X, Lu X, Liao J, Chen H, Fan X. New avenues for systematically inferring cell-cell communication: through single-cell transcriptomics data. Protein Cell. 2020 doi: 10.1007/s13238-020-00727-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Giladi A, et al. Dissecting cellular crosstalk by sequencing physically interacting cells. Nat. Biotechnol. 2020 doi: 10.1038/s41587-020-0442-2. [DOI] [PubMed] [Google Scholar]
- 131.Halpern KB, et al. Paired-cell sequencing enables spatial gene expression mapping of liver endothelial cells. Nat. Biotechnol. 2018;36:962–970. doi: 10.1038/nbt.4231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Szczerba BM, et al. Neutrophils escort circulating tumour cells to enable cell cycle progression. Nature. 2019;566:553–557. doi: 10.1038/s41586-019-0915-y. [DOI] [PubMed] [Google Scholar]
- 133.Schapiro D, et al. histoCAT: analysis of cell phenotypes and interactions in multiplex image cytometry data. Nat. Methods. 2017;14:873–876. doi: 10.1038/nmeth.4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lundberg E, Borner GHH. Spatial proteomics: a powerful discovery tool for cell biology. Nat. Rev. Mol. Cell Biol. 2019;20:285–302. doi: 10.1038/s41580-018-0094-y. [DOI] [PubMed] [Google Scholar]
- 135.Ren X, et al. Reconstruction of cell spatial organization from single-cell RNA sequencing data based on ligand-receptor mediated self-assembly. Cell Res. 2020 doi: 10.1038/s41422-020-0353-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Francis K, Palsson BO. Effective intercellular communication distances are determined by the relative time constants for cyto/chemokine secretion and diffusion. Proc. Natl Acad. Sci. USA. 1997;94:12258–12262. doi: 10.1073/pnas.94.23.12258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Atakan, B. in Molecular Communications and Nanonetworks: From Nature to Practical Systems (ed. Atakan, B.) 105–143 (Springer, 2014).
- 138.Wagner DE, Klein AM. Lineage tracing meets single-cell omics: opportunities and challenges. Nat. Rev. Genet. 2020;21:410–427. doi: 10.1038/s41576-020-0223-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhang J, Nie Q, Zhou T. Revealing dynamic mechanisms of cell fate decisions from single-cell transcriptomic data. Front. Genet. 2019;10:1280. doi: 10.3389/fgene.2019.01280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kuchta K, et al. Predicting proteome dynamics using gene expression data. Sci. Rep. 2018;8:13866. doi: 10.1038/s41598-018-31752-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Krieglstein CF, Granger DN. Adhesion molecules and their role in vascular disease. Am. J. Hypertens. 2001;14:44S–54S. doi: 10.1016/s0895-7061(01)02069-6. [DOI] [PubMed] [Google Scholar]
- 142.Nourani E, Khunjush F, Durmuş S. Computational approaches for prediction of pathogen-host protein-protein interactions. Front. Microbiol. 2015;6:94. doi: 10.3389/fmicb.2015.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Durmuş S, Çakır T, Özgür A, Guthke R. A review on computational systems biology of pathogen-host interactions. Front. Microbiol. 2015;6:235. doi: 10.3389/fmicb.2015.00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Schulze S, Henkel SG, Driesch D, Guthke R, Linde J. Computational prediction of molecular pathogen-host interactions based on dual transcriptome data. Front. Microbiol. 2015;6:65. doi: 10.3389/fmicb.2015.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Mirrashidi KM, et al. Global mapping of the Inc-human interactome reveals that retromer restricts chlamydia infection. Cell Host Microbe. 2015;18:109–121. doi: 10.1016/j.chom.2015.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Thompson LR, et al. A communal catalogue reveals Earth’s multiscale microbial diversity. Nature. 2017;551:457–463. doi: 10.1038/nature24621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.McDonald, D. et al. American gut: an open platform for citizen science microbiome research. mSystems3, (2018). [DOI] [PMC free article] [PubMed]
- 148.Kyrpides NC, Eloe-Fadrosh EA, Ivanova NN. Microbiome data science: understanding our microbial planet. Trends Microbiol. 2016;24:425–427. doi: 10.1016/j.tim.2016.02.011. [DOI] [PubMed] [Google Scholar]
- 149.Gonzalez A, et al. Qiita: rapid, web-enabled microbiome meta-analysis. Nat. Methods. 2018;15:796–798. doi: 10.1038/s41592-018-0141-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zuñiga C, et al. Environmental stimuli drive a transition from cooperation to competition in synthetic phototrophic communities. Nat. Microbiol. 2019;4:2184–2191. doi: 10.1038/s41564-019-0567-6. [DOI] [PubMed] [Google Scholar]
- 151.Lapek JD, Jr, et al. Defining host responses during systemic bacterial infection through construction of a murine organ proteome atlas. Cell Syst. 2018;6:579–592.e4. doi: 10.1016/j.cels.2018.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Penaranda C, Hung DT. Single-cell RNA sequencing to understand host–pathogen interactions. ACS Infect. Dis. 2019;5:336–344. doi: 10.1021/acsinfecdis.8b00369. [DOI] [PubMed] [Google Scholar]
- 153.Jäger S, et al. Global landscape of HIV-human protein complexes. Nature. 2011;481:365–370. doi: 10.1038/nature10719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Batra J, et al. Protein interaction mapping identifies RBBP6 as a negative regulator of ebola virus replication. Cell. 2018;175:1917–1930.e13. doi: 10.1016/j.cell.2018.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Shah PS, et al. Comparative flavivirus-host protein interaction mapping reveals mechanisms of Dengue and Zika virus pathogenesis. Cell. 2018;175:1931–1945.e18. doi: 10.1016/j.cell.2018.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Gordon DE, et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020 doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Adli M. The CRISPR tool kit for genome editing and beyond. Nat. Commun. 2018;9:1911. doi: 10.1038/s41467-018-04252-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Hsu M-N, et al. CRISPR technologies for stem cell engineering and regenerative medicine. Biotechnol. Adv. 2019;37:107447. doi: 10.1016/j.biotechadv.2019.107447. [DOI] [PubMed] [Google Scholar]
- 159.Kwon ED, et al. Manipulation of T cell costimulatory and inhibitory signals for immunotherapy of prostate cancer. Proc. Natl Acad. Sci. USA. 1997;94:8099–8103. doi: 10.1073/pnas.94.15.8099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Xu W, Atkins MB, McDermott DF. Checkpoint inhibitor immunotherapy in kidney cancer. Nat. Rev. Urol. 2020;17:137–150. doi: 10.1038/s41585-020-0282-3. [DOI] [PubMed] [Google Scholar]
- 161.Huang H, et al. Cell-cell contact-induced gene editing/activation in mammalian cells using a synNotch-CRISPR/Cas9 system. Protein Cell. 2020;11:299–303. doi: 10.1007/s13238-020-00690-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.de Juan D, Pazos F, Valencia A. Emerging methods in protein co-evolution. Nat. Rev. Genet. 2013;14:249–261. doi: 10.1038/nrg3414. [DOI] [PubMed] [Google Scholar]
- 163.Kuhlman B, Bradley P. Advances in protein structure prediction and design. Nat. Rev. Mol. Cell Biol. 2019;20:681–697. doi: 10.1038/s41580-019-0163-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Chupakhin V, Marcou G, Baskin I, Varnek A, Rognan D. Predicting ligand binding modes from neural networks trained on protein-ligand interaction fingerprints. J. Chem. Inf. Model. 2013;53:763–772. doi: 10.1021/ci300200r. [DOI] [PubMed] [Google Scholar]
- 165.Fenoy E, Izarzugaza JMG, Jurtz V, Brunak S, Nielsen M. A generic deep convolutional neural network framework for prediction of receptor-ligand interactions-NetPhosPan: application to kinase phosphorylation prediction. Bioinformatics. 2019;35:1098–1107. doi: 10.1093/bioinformatics/bty715. [DOI] [PubMed] [Google Scholar]
- 166.Kveler K, et al. Immune-centric network of cytokines and cells in disease context identified by computational mining of PubMed. Nat. Biotechnol. 2018;36:651–659. doi: 10.1038/nbt.4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Ashburner M, et al. Gene ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.UniProt Consortium UniProt: a hub for protein information. Nucleic Acids Res. 2015;43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Waterhouse RM, Tegenfeldt F, Li J, Zdobnov EM, Kriventseva EV. OrthoDB: a hierarchical catalog of animal, fungal and bacterial orthologs. Nucleic Acids Res. 2013;41:D358–D365. doi: 10.1093/nar/gks1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Reimand J, Kull M, Peterson H, Hansen J, Vilo J. g:profiler — a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 2007;35:W193–W200. doi: 10.1093/nar/gkm226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Uhlén M, et al. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 173.Szklarczyk D, et al. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Stark C, et al. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Gioutlakis A, Klapa MI, Moschonas NK. PICKLE 2.0: a human protein-protein interaction meta-database employing data integration via genetic information ontology. PLoS ONE. 2017;12:e0186039. doi: 10.1371/journal.pone.0186039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Prieto C, De Las Rivas J. APID: agile protein interaction DataAnalyzer. Nucleic Acids Res. 2006;34:W298–W302. doi: 10.1093/nar/gkl128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Hermjakob H, et al. IntAct: an open source molecular interaction database. Nucleic Acids Res. 2004;32:D452–D455. doi: 10.1093/nar/gkh052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Cerami EG, et al. Pathway commons, a web resource for biological pathway data. Nucleic Acids Res. 2011;39:D685–D690. doi: 10.1093/nar/gkq1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Pawson AJ, et al. The IUPHAR/BPS Guide to PHARMACOLOGY: an expert-driven knowledgebase of drug targets and their ligands. Nucleic Acids Res. 2014;42:D1098–D1106. doi: 10.1093/nar/gkt1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.