Abstract
Background
The emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and its disease COVID-19 has strongly encouraged the search for antiviral compounds. Most of the evaluated drugs against SARS-CoV-2 derive from drug repurposing of Food and Drug Administration-approved molecules. These drugs have as target three major processes: (1) early stages of virus-cell interaction, (2) viral proteases, and (3) the viral RNA-dependent RNA polymerase.
Summary
This review focused on the basic principles of virology and pharmacology to understand the importance of early stages of virus-cell interaction as therapeutic targets and other main processes vital for SARS-CoV-2 replication. Furthermore, we focused on describing the main targets associated with SARS-CoV-2 antiviral therapy and the rationale of drug combinations for efficiently suppressing viral replication.
Key Messages
We hypothesized that blocking of both entry mechanisms could allow a more effective antiviral effect compared to the partial results obtained with chloroquine or its derivatives alone. This approach, already used to achieve an antiviral effect higher than that offered by every single drug administered separately, has been successfully applied in several viral infections such as HIV and HCV. This review will contribute to expanding the perception of the possible therapeutic targets in SARS-CoV-2 infection and highlight the benefits of using combination therapies.
Keywords: Coronavirus, Virus-host interactions, SARS-CoV-2, Treatment, Combination, Protease, Entry
Introduction
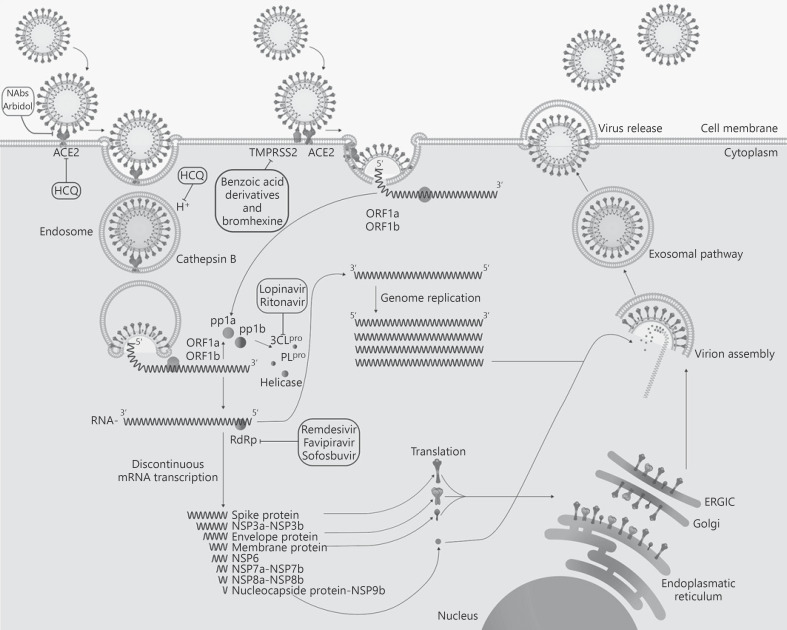
In the last trimester of 2019 a new coronavirus, named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and responsible for coronavirus disease 2019 (COVID-19), emerged in Wuhan, China. There is no yet an approved therapy to treat the infection by SARS-CoV-2 and its associated disease, COVID-19, and the therapeutic approaches used have been changing from the beginning of the outbreak [1, 2, 3]. The standard care of patients with COVID-19 is mainly focused on symptomatic and respiratory support according to the diagnosis and treatment of pneumonia and coagulopathies caused by COVID-19 [4]. Several research groups have been working to determine the possible targets to control the viral infection [5, 6, 7]. Three major targets have been identified and can be summarized in the following: early stages of virus-cell interaction, viral proteases, and the viral RNA-dependent RNA polymerase (RdRp) [7, 8, 9]. The replication cycle of SARS-CoV-2 and the main targets for the antiviral therapy described in this review are shown in Figure 1.
Fig. 1.
SARS-CoV-2 replication. The viral cycle begins with the interaction between the viral spike and the cellular receptor. Several membrane proteins have been proposed as possible receptors for SARS-CoV-2; however, ACE2 is likely to be the most important one [10]. After the viral spike has interacted with the receptor, the virus gains entry into the host cytosol by two mechanisms: (1) by late endocytosis, releasing the viral RNA after the fusion with the lysosome (this entry is blocked by HCQ), or (2) by early endocytosis by fusion of the viral and host membrane without the participation of the lysosome. In these early stages, the priming processing by cellular proteases is the key for the exposure of the viral fusion motive. TMPRSS2 could act in both early and late endosome entry processes and could be inhibited by benzoic acid derivatives such as nafamostat, camostat, and bromhexine [18, 21]. Furthermore, other proteases such as cathepsin B could mediate the entry in the late lysosomal pathway, while only TMPRSS2 has been related to the early entry endosome [11, 13, 15, 18]. After fusion has occurred, the viral RNA is released into the cytoplasm and open reading frame 1 (ORF1) is translated to produce the RdRp. Subgenomic mRNAs are produced by discontinuous transcription, a process characteristic of this RdRp, which favors recombination. Compounds such as remdesivir, favipiravir, and sofosbuvir block this enzyme [39, 40, 41, 45]. The subgenomic mRNAs are then translated into protein. The genome has eight ORFs. The gene segments that encode nonstructural polyproteins are processed first and translated into ORF1a and ORF1b producing pp1a and pp1ab proteins, respectively. Protein pp1a and pp1ab are cleaved by the viral proteases (3CLpro and PLpro). The main protease is also the target of protease inhibitors such as lopinavir/ritonavir [7]. The structural proteins − spike, envelope, and membrane proteins − enter into the endoplasmic reticulum/Golgi complexes. Then, the nucleoprotein combines with the (+) strand genomic RNA (nucleoprotein complex) and merges with the other structural proteins in the endoplasmic reticulum-Golgi apparatus compartment [64]. Finally, the virion is excreted to the extracellular region through the exosomal pathway [64]. HCQ, hydroxychloroquine; RdRp, RNA-dependent RNA polymerase; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; TMPRSS2, transmembrane serine protease 2; ORF, open reading frame.
Early Infection Stages: Blocking Viral Entry
After interaction with its receptor ACE2, SARS-CoV-2 enters the cell through the fusion of its viral membrane with the cellular one (Fig. 1) [10]. The viral fusion protein is exposed by priming with a proteolytic enzyme. Three different types of proteases have been proposed for this cleavage: (1) Priming by the cellular transmembrane serine protease 2 (TMPRSS2) and also TMPRSS4 in enterocytes. This priming allows viral entry by the early endosomal pathway, although the exact mechanism of entry is unknown [8, 11, 12]. (2) Priming by cathepsin B in the late endosomes [13]. (3) Activation during virus egress from the cell by furin [14]. Furin-dependent pre-cleavage of the spike has been shown to be necessary for activation through the TMPRSS2 pathway [15].
In the early stages of infection, different therapeutic agents have been used, including hydroxychloroquine (HCQ) [16, 17], benzoic acid derivatives such as nafamostat and camostat [18], and neutralizing antibodies [19]. The antiviral effect of HCQ is related to an elevation of the pH in endosomes/lysosomes, which is essential for the fusion between the viral and vesicle membranes. In addition, HCQ could inhibit SARS-CoV cellular entry by changing the glycosylation pattern of ACE2, the key receptor for SARS-CoV-2 entry [16, 17]. Furthermore, HCQ exerts immunomodulatory effects through attenuation of cytokine production [20]. The effects of HCQ on cellular targets are considered nonspecific antiviral effects.
Another early target evaluated against SARS-CoV-2 is a cellular protease related to the priming of the spike protein (S), which exposes the fusion motive and allows the release of viral RNA into the cytosol. The proteolytic processing of the viral spike protein is a key step during the infection process of SARS-CoV-2. This reaction is driven by TMPRSS2 and could be inhibited by camostat, nafamostat, and bromhexine [18, 21]. Interestingly, some reports showed that SARS-CoV-2 could gain entry into the host cell without using the late endocytic pathway. The processing of TMPRSS2 in the extracellular lumen could likely be sufficient to activate the fusion between the viral and host membrane [22, 23]. This alternative entry pathway would explain the low success rates in the use of HCQ/chloroquine alone in COVID-19 therapy. In addition, chloroquine does not seem to prevent SARS-CoV-2 entry into human lung cells, suggesting that the TMPRSS2-primed pathway predominates in the lung [24].
Arbidol (umifenovir) is an antiviral agent that targets S/ACE2 interaction, inhibiting membrane fusion of the viral envelope [2]. This compound could be considered as an early-stage inhibitor of SARS-CoV-2. Another therapy for COVID-19 is the use of convalescent plasma or hyperimmune immunoglobulins (NAbs) [25]. Antibodies from recovered patients or neutralizing monoclonal antibodies may neutralize the free virus and induce the immune clearance of infected cells. On March 24, 2020 the Food and Drug Administration (FDA) released an emergency approval for the use of COVID-19 convalescent plasma. The development of neutralizing monoclonal antibodies against SARS-CoV-2 infection is ongoing [19, 26].
Targeting Viral Enzymes
Other promising therapeutic targets against SARS-CoV-2 infection are viral proteases (Fig. 1) due to their low homology with host protease and the structural knowledge available for SARS-CoV [7, 27]. SARS-CoV-2 has two viral proteases related to the proteolytic processing of the viral polyprotein into mature and functional proteins. These enzymes are the main protease (3CLpro) and the papain-like protease (PLpro), both belonging to the family of cysteine proteases. They exhibit a high structural and functional homology with SARS-CoV proteases [27, 28, 29]. The main protease is responsible for the proteolysis of pre-proteins associated with the viral replication machinery such as the RdRp, the helicase, and the exoribonuclease, among others [29]. Several groups have shown by using in silico and in vitro approaches that this protease may be blocked by protease inhibitors effective in HIV therapy [7, 27, 29, 30]. Furthermore, lopinavir/ritonavir, an FDA-approved oral drug combination for treating HIV, exhibited in vitro activity against other coronaviruses. However, in the case of SARS-CoV-2 infection, the timing of the administration of these drugs during the early peak of the viral replication phase (at initial 7–10 days) appears to be critical. Delayed initiation of therapy with lopinavir/ritonavir did not result in positive clinical outcomes mainly due to the high viral load [31]. Another important concern with lopinavir/ritonavir therapy against SARS-CoV-2 is related to the administration route. This drug combination is administered to patients with COVID-19 by the oral pathway, applicable for drugs with good gastrointestinal absorption and low volume of distribution (the volume of distribution for lopinavir/ritonavir is around 17 L) [32, 33]. However, these compounds bind tightly to plasma proteins (in the plasma, >98% of the drug is in bound form), which decreases the ability to reach tissues such as the lung [32]. This important pharmacological issue related to the pharmacokinetic parameters of lopinavir/ritonavir needs to be addressed in order to improve the therapeutic outcome of treatment with this drug combination. The second protease related to the SARS-CoV-2 replication cycle, PLpro, is responsible for the cleavages of the N-terminus of the polyprotein to release Nsp1, Nsp2, and Nsp3, nonstructural proteins associated with viral replication [34, 35, 36]. In addition, PLpro also possesses de-ubiquitination and de-ISGylation activities that could antagonize the host's innate immunity [37].
The SARS-CoV-2 genome replication is carried out by an RdRp and could be one of the best targets in the SARS-CoV-2 replication (Fig. 1) [38]. This enzymatic activity has no parallel process in the eukaryotic cells. Thus, attacking this target might reduce side effects that might occur by affecting any similar protein in the host. The sequences of RdRp in SARS-CoV and SARS-CoV-2 have a high homology and encode structurally similar proteins. Remdesivir is a nucleotide analog inhibitor of RdRp and has shown a broad spectrum of antiviral activity against several RNA viruses [38, 39]. Remdesivir similarly to sofosbuvir, a direct-acting antiviral used in HCV therapy, is a chain-terminating nucleotide analog. These drugs produce their therapeutic effect by directly interacting with the RdRp and incorporating the active form of the inhibitor into the growing RNA strand, preventing the replication to continue [40, 41]. Several clinical trials have been conducted evaluating the activity of remdesivir and other direct-acting antivirals used in HCV against SARS-CoV-2 [42]. Only remdesivir reached the FDA emergency approval status for its use in patients with severe COVID-19. However, the clinical improvements by using remdesivir alone in SARS-CoV-2 patients are modest. Favipiravir, another antiviral agent with broad activity against other RNA viruses by inhibiting the RdRp, halting viral replication, was evaluated against SARS-CoV-2, showing effects in vitro and in vivo [43, 44, 45].
Combined Therapy
Drug combinations that have been widely used in antiviral therapy have become the leading choice for treating HIV and HCV, among other viral pathologies [46, 47, 48, 49, 50]. The use of multiple drugs with different mechanisms of action increases the efficacy of the therapeutic effect. It also allows decreasing the dose of a single drug, thus preventing host toxicity [51, 52, 53, 54]. Moreover, one important advantage of combination therapies besides enhancing the selective effect over the target is that this approach minimizes or slows down the development of drug resistance, which is the main issue in antiviral therapy of RNA viruses. Experimental conditions for drug combinations in vitro can be easily defined. Information for most individual drugs used against SARS-CoV-2 is available, thus conducting in vitro assays for the effect of their combinations could take a very short time (approximately <2 weeks) by using methods such as the combination index-isobologram method of Chou [55]. However, there are some issues associated with the evaluation of drug combinations in clinical trials related to dose, timing, route of administration, efficacy, and toxicity, among others, that make the extrapolation of in vitro data difficult. These factors are associated with, but not limited to, variables in the patient population such as sex, age, race, and disease stage. Furthermore, drug combination trials need to fulfill all the ethical concerns to treat patients with placebo or suboptimal therapeutic doses as required for a drug combination study design according to the guidelines of regulatory agencies, such as the FDA.
As explained above, HCQ could produce a non-direct-acting specific antiviral effect against SARS-CoV-2 by several mechanisms. However, the results obtained in the clinical trial by using HCQ alone were less satisfactory than expected [56]. Although HCQ affects the early stages of the viral infection, i.e., the endosome-mediated entry, some reports showed that SARS-CoV-2 could enter the host cells bypassing the pathways blocked by HCQ [15]. Thus, we hypothesize that this could be avoided by combining HCQ and camostat or nafamostat [18] or bromhexine [21, 57]. These TMPRSS2 inhibitors could block the other mechanism associated with SARS-CoV-2 entry. In addition to interfering with the different proteases involved in the activation of the fusion peptide of S, a combined scheme might include an inhibitor of this domain [8]. Furthermore, we support the fact that other TMPRSS2 inhibitors such as bromhexine, a drug used as mucolytic, could be an important pharmacological tool to design prophylactic therapies. Bromhexine has a well-known drug safety profile and it is marketed as an over-the-counter medication. Thus, the design of clinical trials and extrapolation into effective therapy should be less complicated in comparison with drugs as camostat or nafamostat. Furthermore, some reports showed that bromhexine could overcome the main pharmacokinetic problem in SARS-CoV-2 therapy, reaching the pulmonary and bronchial epithelial cells in concentrations higher than those found in the plasma [58].
The above combination targets viral entry into the cell. Similarly, neutralizing antibodies and fusion inhibitors could be combined in a triple combination format. Most of the targets associated with the early stages of the viral infection are host targets that significantly decrease the probability of developing resistance in comparison with viral targets [51].
The early study published by Cao et al. [59] showed that lopinavir/ritonavir could produce a modest positive effect in comparison to the standard care in patients with SARS-CoV-2. However, several researchers have criticized the structural design of the study of Cao et al. [59], suggesting that the antiviral effect of lopinavir/ritonavir against SARS-CoV-2 could be higher; thus, more studies are needed [60, 61, 62]. Nevertheless, the side effects associated with these protease inhibitors are important. Thus, antiviral therapy based on combining these protease inhibitors with another antiviral compound such as remdesivir could decrease the effective dose required to produce the antiviral effect and then decreasing the possibility of side effects. The possibility of the occurrence of viral resistance with this strategy should be kept in mind.
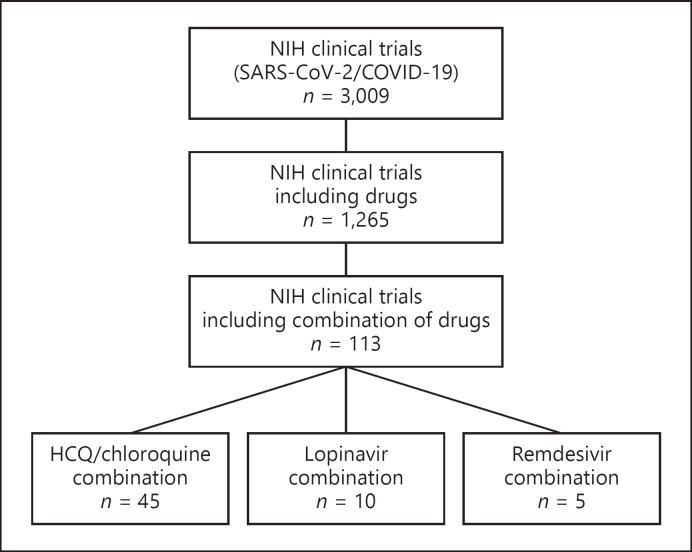
Of the 1,265 clinical trials based on drugs to treat SARS-CoV-2 registered in the National Institutes of Health Library, only 113 (<10%) are based on drug combinations. One-third of them include HCQ under combination therapy (Fig. 2). Table 1 summarizes the ongoing clinical trials combining drugs targeting early viral replication events or viral enzymes.
Fig. 2.
Clinical trials using a combination of drugs targeting early replication steps or viral enzymes. An exhaustive revision of the ongoing clinical trials related to SARS-CoV-2 was performed using the database of the National Institutes of Health (NIH) of the United States. The data were accessed from the NIH webserver ClinicalTrials.gov. The search terms were “COVID” or “SARS-CoV-2,” resulting in 3,009 trials. Then, the search parameter drug was applied as a further term, retrieving 1,616 clinical trials. In each search, the trials included those not yet recruiting, recruiting, active, or completed. A visual inspection of each of the 1,616 trials was conducted and only 1,265 included drugs as the main intervention. Moreover, the drug combination trials were also reviewed, and only 113 included a combination of pharmacological therapy for the treatment of COVID-19 patients. The database search was conducted on August 14, 2020. COVID-19, coronavirus disease 2019; HCQ, hydroxychloroquine; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Table 1.
Clinical trials registered in the United States National Library of Medicine that included drug combinations against SARS-CoV-2 targeting early steps of viral replication or viral enzymes
| Drug combination | Clinical trial number (NIH) | Country | Registration date |
|---|---|---|---|
| Camostat mesilate, HCQ | NCT04338906 | Germany | April 8, 2020 |
| NCT04355052 | Israel | April 21, 2020 | |
| Baricitinib, HCQ, lopinavir/ritonavir, remdesivir | NCT04373044 | US | May 4, 2020 |
| HCQ, azithromycin, ivermectin, camostat mesilate | NCT04374019 | US | May 5, 2020 |
| Favipiravir, HCQ | NCT04411433 | Turkey | June 2, 2020 |
| Favipiravir, lopinavir | NCT04499677 | UK | August 5, 2020 |
The studies shown were retrieved from the website ClinicalTrials.gov (see also Fig. 2). SARS-CoV-2/COVID-19 as condition or disease and the drug names and their combinations were used as keywords for the search. Drugs related to blocking early stages of viral replication or viral enzymes are shown in bold. COVID-19, coronavirus disease 2019; NIH, National Institutes of Health; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Conclusions
Altogether, the virology and pharmacological principles discussed in this review emphasize the importance of early stages of the infection as therapeutic targets and the advantage of simultaneously blocking alternative processes critical for viral replication. The development of effective therapies against this new viral pathogen has proven to be a challenge. Useful in vitro and in vivo models are needed to address the effectiveness of drugs or their combination to be used as preclinical data, followed by randomized clinical trials. Prior studies with SARS-CoV might help to develop and validate models for this new infection. More studies will help to fully understand the possible advantages of drug combinations against SARS-CoV-2 infection. Based on the experience obtained with the treatment of other viral infections, the combination of two or more antiviral drugs could produce a better effect than the one reached with a single drug. These antiviral effects could be additive or synergistic, depending on the degree of inhibition of viral load. Furthermore, besides the improvement in the antiviral effect achieved, an eventual reduction of the dosage used could represent fewer side effects associated with the therapy. Thus, enhancing the antiviral effect and reducing side effects could represent a main change in SARS-CoV-2 therapy. There are at present many concerns because of the toxicity of chloroquine or its derivative [24]. Drug combination might help to reduce the dose or the time of application of this mixed therapy. Nasal administration is also a promising alternative that might overcome both toxicity effects and effective concentration challenges [63].
Conflict of Interest Statement
The authors have no conflicts of interest to declare.
Funding Sources
None.
Author Contributions
F.H. Pujol, J.T. Ortega, and H.R. Rangel visualized and designed the manuscript. F.H. Pujol and J.T. Ortega wrote the manuscript. J.L. Zambrano performed imaging and art creation. F. Liprandi and B. Jastrzebska were responsible for data curation and revision. All authors read and contributed to the final version of the manuscript.
References
- 1.Li H, Liu SM, Yu XH, Tang SL, Tang CK. Coronavirus disease 2019 (COVID-19): current status and future perspectives. Int J Antimicrob Agents. 2020 May;55((5)):105951. doi: 10.1016/j.ijantimicag.2020.105951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sanders JM, Monogue ML, Jodlowski TZ, Cutrell JB. Pharmacologic Treatments for Coronavirus Disease 2019 (COVID-19): A Review. JAMA. 2020 Apr;••• doi: 10.1001/jama.2020.6019. [DOI] [PubMed] [Google Scholar]
- 3.Tobaiqy M, Qashqary M, Al-Dahery S, Mujallad A, Hershan AA, Kamal MA, et al. Therapeutic management of patients with COVID-19: a systematic review. Infection Prevention in Practice. 2020 Apr;100061((3)):100061. doi: 10.1016/j.infpip.2020.100061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peng F, Tu L, Yan Y, Hu P, Wang R, Hu Q, Cao F, Jiang T, Sun J, Xu G, Chang C. Management and Treatment of COVID-19: The Chinese Experience. Can J Cardiol. 2020 Apr 17; doi: 10.1016/j.cjca.2020.04.010. S0828-282X(20)30395-0. doi: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Asai A, Konno M, Ozaki M, Otsuka C, Vecchione A, Arai T, et al. COVID-19 Drug Discovery Using Intensive Approaches. Int J Mol Sci. 2020 Apr;21((8)):2839. doi: 10.3390/ijms21082839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jin Z, Du X, Xu Y, Deng Y, Liu M, Zhao Y, et al. (2020). Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020 Jun;582((7811)):289–93. doi: 10.1038/s41586-020-2223-y. Advance online publication. [DOI] [PubMed] [Google Scholar]
- 7.Ortega JT, Serrano ML, Pujol FH, Rangel HR. Unrevealing sequence and structural features of novel coronavirus using in silico approaches: the main protease as molecular target. EXCLI J. 2020 Mar;19:400–9. doi: 10.17179/excli2020-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang T, Bidon M, Jaimes JA, Whittaker GR, Daniel S. Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antiviral Res. 2020 Apr;178:104792. doi: 10.1016/j.antiviral.2020.104792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang N, Shen HM. Targeting the Endocytic Pathway and Autophagy Process as a Novel Therapeutic Strategy in COVID-19. Int J Biol Sci. 2020 Mar;16((10)):1724–31. doi: 10.7150/ijbs.45498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ortega JT, Serrano ML, Pujol FH, Rangel HR. Role of changes in SARS-CoV-2 spike protein in the interaction with the human ACE2 receptor: an in silico analysis. EXCLI J. 2020 Mar;19:410–7. doi: 10.17179/excli2020-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsuyama S, Nao N, Shirato K, Kawase M, Saito S, Takayama I, et al. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc Natl Acad Sci USA. 2020 Mar;117((13)):7001–3. doi: 10.1073/pnas.2002589117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zang R, Gomez Castro MF, McCune BT, Zeng Q, Rothlauf PW, Sonnek NM, et al. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci Immunol. 2020 May;5((47)) doi: 10.1126/sciimmunol.abc3582. eabc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ou X, Liu Y, Lei X, Li P, Mi D, Ren L, et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. 2020 Mar;11((1)):1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coutard B, Valle C, de Lamballerie X, Canard B, Seidah NG, Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res. 2020 Apr;176:104742. doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoffmann M, Kleine-Weber H, Pöhlmann S. A Multibasic Cleavage Site in the Spike Protein of SARS-CoV-2 Is Essential for Infection of Human Lung Cells. Mol Cell. 2020 May;78((4)):779–784.e5. doi: 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Devaux CA, Rolain JM, Colson P, Raoult D. New insights on the antiviral effects of chloroquine against coronavirus: what to expect for COVID-19? Int J Antimicrob Agents. 2020 May;55((5)):105938. doi: 10.1016/j.ijantimicag.2020.105938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu J, Cao R, Xu M, Wang X, Zhang H, Hu H, et al. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discov. 2020 Mar;6((1)):16. doi: 10.1038/s41421-020-0156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020 Apr;181((2)):271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang C, Li W, Drabek D, Okba NM, van Haperen R, Osterhaus AD, et al. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat Commun. 2020 May;11((1)):2251. doi: 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perricone C, Triggianese P, Bartoloni E, Cafaro G, Bonifacio AF, Bursi R, et al. The anti-viral facet of anti-rheumatic drugs: lessons from COVID-19. J Autoimmun. 2020 Jul;111:102468. doi: 10.1016/j.jaut.2020.102468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ansarin K, Tolouian R, Ardalan M, Taghizadieh A, Varshochi M, Teimouri S, et al. Ardalan4 M, Taghizadieh A, Varshochi M, Teimouri S, Vaezi T, Valizadeh H, Saleh P, Safiri S, Chapman K. Effect of bromhexine on clinical outcomes and mortality in COVID-19 patients: A randomized clinical trial. Bioimpacts. 2020;10((4)):209–15. doi: 10.34172/bi.2020.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glowacka I, Bertram S, Müller MA, Allen P, Soilleux E, Pfefferle S, et al. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J Virol. 2011 May;85((9)):4122–34. doi: 10.1128/JVI.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kupferschmidt K, Cohen J. Race to find COVID-19 treatments accelerates. Science. 2020 Mar;367((6485)):1412–3. doi: 10.1126/science.367.6485.1412. [DOI] [PubMed] [Google Scholar]
- 24.Hoffmann M, Mösbauer K, Hofmann-Winkler H, Kaul A, Kleine-Weber H, Krüger N, et al. Chloroquine does not inhibit infection of human lung cells with SARS-CoV-2. Nature. 2020 Jul;585((7826)):588–90. doi: 10.1038/s41586-020-2575-3. [DOI] [PubMed] [Google Scholar]
- 25.Shen C, Wang Z, Zhao F, Yang Y, Li J, Yuan J, et al. Treatment of 5 Critically Ill Patients With COVID-19 With Convalescent Plasma. JAMA. 2020 Mar;323((16)):1582–9. doi: 10.1001/jama.2020.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dhama K, Sharun K, Tiwari R, Dadar M, Malik YS, Singh KP, et al. COVID-19, an emerging coronavirus infection: advances and prospects in designing and developing vaccines, immunotherapeutics, and therapeutics. Hum Vaccin Immunother. 2020 Mar;16((6)):1–7. doi: 10.1080/21645515.2020.1735227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu C, Liu Y, Yang Y, Zhang P, Zhong W, Wang Y, et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm Sin B. 2020 Feb;10((5)):766–88. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xue X, Yu H, Yang H, Xue F, Wu Z, Shen W, et al. Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design. J Virol. 2008 Mar;82((5)):2515–27. doi: 10.1128/JVI.02114-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang L, Lin D, Sun X, Curth U, Drosten C, Sauerhering L, et al. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020 Apr;368((6489)):409–12. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choy KT, Wong AY, Kaewpreedee P, Sia SF, Chen D, Hui KP, et al. Remdesivir, lopinavir, emetine, and homoharringtonine inhibit SARS-CoV-2 replication in vitro. Antiviral Res. 2020 Apr;178:104786. doi: 10.1016/j.antiviral.2020.104786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yao TT, Qian JD, Zhu WY, Wang Y, Wang GQ. A systematic review of lopinavir therapy for SARS coronavirus and MERS coronavirus-A possible reference for coronavirus disease-19 treatment option. J Med Virol. 2020 Feb;92((6)):556–63. doi: 10.1002/jmv.25729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, Grant JR, et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018 Jan;46(D1):D1074–82. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marzolini C, Stader F, Stoeckle M, Franzeck F, Egli A, Bassetti S, Hollinger A, Osthoff M, Weisser M, Gebhard CE, Baettig V, Geenen J, Khanna N, Tschudin-Sutter S, Mueller D, Hirsch HH, Battegay M, Sendi P. Effect of Systemic Inflammatory Response to SARS-CoV-2 on Lopinavir and Hydroxychloroquine Plasma Concentrations. Antimicrob Agents Chemother. 2020 Jul 8; doi: 10.1128/AAC.01177-20. AAC.01177-20. doi: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arya R, Das A, Prashar V, Kumar M. Potential inhibitors against papain-like protease of novel coronavirus (SARS-CoV2) from FDA approved drugs. ChemRxiv. 2020 Preprint. [Google Scholar]
- 35.Barretto N, Jukneliene D, Ratia K, Chen Z, Mesecar AD, Baker SC. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J Virol. 2005 Dec;79((24)):15189–98. doi: 10.1128/JVI.79.24.15189-15198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shin D, Mukherjee R, Grewe D, Bojkova D, Baek K, Bhattacharya A, et al. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature. 2020 doi: 10.1038/s41586-020-2601-5. doi: 10.1038/s41586-020-2601-5 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Niemeyer D, Mösbauer K, Klein EM, Sieberg A, Mettelman RC, Mielech AM, et al. The papain-like protease determines a virulence trait that varies among members of the SARS-coronavirus species. PLoS Pathog. 2018 Sep;14((9)):e1007296. doi: 10.1371/journal.ppat.1007296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yin W, Mao C, Luan X, Shen DD, Shen Q, Su H, Wang X, Zhou F, Zhao W, Gao M, Chang S, Xie YC, Tian G, Jiang HW, Tao SC, Shen J, Jiang Y, Jiang H, Xu Y, Zhang S, Zhang Y, Xu HE. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020 May 1; doi: 10.1126/science.abc1560. eabc1560. doi: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tchesnokov EP, Feng JY, Porter DP, Götte M. Mechanism of Inhibition of Ebola Virus RNA-Dependent RNA Polymerase by Remdesivir. Viruses. 2019 Apr;11((4)):326. doi: 10.3390/v11040326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gordon CJ, Tchesnokov EP, Woolner E, Perry JK, Feng JY, Porter DP, et al. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J Biol Chem. 2020 May;295((20)):6785–97. doi: 10.1074/jbc.RA120.013679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sadeghi A, Ali Asgari A, Norouzi A, Kheiri Z, Anushirvani A, Montazeri M, Hosamirudsai H, Afhami S, Akbarpour E, Aliannejad R, Radmard AR, Davarpanah AH, Levi J, Wentzel H, Qavi A, Garratt A, Simmons B, Hill A, Merat S. Sofosbuvir and daclatasvir compared with standard of care in the treatment of patients admitted to hospital with moderate or severe coronavirus infection (COVID-19): a randomized controlled trial. J Antimicrob Chemother. 2020 Aug 19; doi: 10.1093/jac/dkaa334. dkaa334. doi: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elfiky AA. Anti-HCV, nucleotide inhibitors, repurposing against COVID-19. Life Sci. 2020 May;248:117477. doi: 10.1016/j.lfs.2020.117477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Şimşek Yavuz S, Ünal S. Antiviral treatment of COVID-19. Turk J Med Sci. 2020 Apr;50(SI-1):611–9. doi: 10.3906/sag-2004-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang M, Cao R, Zhang L, Yang X, Liu J, Xu M, et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30((3)):269–71. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cai Q, Yang M, Liu D, Chen J, Shu D, Xia J, et al. Experimental Treatment with Favipiravir for COVID-19: An Open-Label Control Study. Engineering (Beijing) 2020 Mar;••• doi: 10.1016/j.eng.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cihlar T, Fordyce M. Current status and prospects of HIV treatment. Curr Opin Virol. 2016 Jun;18:50–6. doi: 10.1016/j.coviro.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 47.Spengler U. Direct antiviral agents (DAAs) - A new age in the treatment of hepatitis C virus infection. Pharmacol Ther. 2018 Mar;183:118–26. doi: 10.1016/j.pharmthera.2017.10.009. [DOI] [PubMed] [Google Scholar]
- 48.Fanning GC, Zoulim F, Hou J, Bertoletti A. Therapeutic strategies for hepatitis B virus infection: towards a cure. Nat Rev Drug Discov. 2019 Nov;18((11)):827–44. doi: 10.1038/s41573-019-0037-0. [DOI] [PubMed] [Google Scholar]
- 49.Britt WJ, Prichard MN. New therapies for human cytomegalovirus infections. Antiviral Res. 2018 Nov;159:153–74. doi: 10.1016/j.antiviral.2018.09.003. [DOI] [PubMed] [Google Scholar]
- 50.Jaspe RC, Ortega J, Zambrano JL, Pujol FH. Presente y futuro de la terapia contra la hepatitis C. Invest Clin. 2016 Mar;57((1)):93–107. [PubMed] [Google Scholar]
- 51.Ortega JT, Rangel HR, Pujol FH. Cellular targets as an alternative mechanism in antiviral therapy. Interciencia. 2013 Jan;38:836–42. [Google Scholar]
- 52.De Clercq E. Anti-HIV drugs. Verh K Acad Geneeskd Belg. 2007;69((2)):81–104. [PubMed] [Google Scholar]
- 53.Shanks RH, Rizzieri DA, Flowers JL, Colvin OM, Adams DJ. Preclinical evaluation of gemcitabine combination regimens for application in acute myeloid leukemia. Clin Cancer Res. 2005 Jun;11((11)):4225–33. doi: 10.1158/1078-0432.CCR-04-2106. [DOI] [PubMed] [Google Scholar]
- 54.Zhang G, Lu H, Lu Y, Jiang S, Chen YH. Neutralization of HIV-1 primary isolate by ELDKWA-specific murine monoclonal antibodies. Immunobiology. 2005;210((9)):639–45. doi: 10.1016/j.imbio.2005.05.025. [DOI] [PubMed] [Google Scholar]
- 55.Chou TC. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol Rev. 2006 Sep;58((3)):621–81. doi: 10.1124/pr.58.3.10. [DOI] [PubMed] [Google Scholar]
- 56.Hashem AM, Alghamdi BS, Algaissi AA, Alshehri FS, Bukhari A, Alfaleh MA, et al. Therapeutic use of chloroquine and hydroxychloroquine in COVID-19 and other viral infections: A narrative review. Travel Med Infect Dis. 2020 May;101735:101735. doi: 10.1016/j.tmaid.2020.101735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Depfenhart M, Lemperle G, Meyer M, Rautenbach M, Bertossi D, de Villiers DA. SARS-CoV-2 Prophylactic and Treatment; A Counter Argument Against The Sole Use of Chloroquine. Am J Biomed Sci Res. 2020 Apr;8((4)):248–51. [Google Scholar]
- 58.Maggio R, Corsini GU. Repurposing the mucolytic cough suppressant and TMPRSS2 protease inhibitor bromhexine for the prevention and management of SARS-CoV-2 infection. Pharmacol Res. 2020 Apr;157:104837. doi: 10.1016/j.phrs.2020.104837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cao B, Wang Y, Wen D, Liu W, Wang J, Fan G, et al. A Trial of Lopinavir-Ritonavir in Adults Hospitalized with Severe Covid-19. N Engl J Med. 2020 May;382((19)):1787–99. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Carmona-Bayonas A, Jimenez-Fonseca P, Castañón E. A Trial of Lopinavir-Ritonavir in Covid-19. N Engl J Med. 2020 May;382((21)):e68. doi: 10.1056/NEJMc2008043. [DOI] [PubMed] [Google Scholar]
- 61.Corrao S, Natoli G, Cacopardo B. A Trial of Lopinavir-Ritonavir in Covid-19. N Engl J Med. 2020 May;382((21)):e68. doi: 10.1056/NEJMc2008043. [DOI] [PubMed] [Google Scholar]
- 62.Havlichek D., Jr A Trial of Lopinavir-Ritonavir in Covid-19. N Engl J Med. 2020 May;382((21)):e68. doi: 10.1056/NEJMc2008043. [DOI] [PubMed] [Google Scholar]
- 63.Higgins TS, Wu AW, Illing EA, Sokoloski KJ, Weaver BA, Anthony BP, et al. Intranasal Antiviral Drug Delivery and Coronavirus Disease 2019 (COVID-19): A State of the Art Review. Otolaryngol Head Neck Surg. 2020 Jul;194599820933170((4)):682–94. doi: 10.1177/0194599820933170. [DOI] [PubMed] [Google Scholar]
- 64.Kumar S, Nyodu R, Maurya VK, Saxena SK. Morphology, Genome Organization, Replication, and Pathogenesis of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Coronavirus Disease 2019 (COVID-19) 2020 Apr 30;23 doi: . 0. [Google Scholar]