Abstract
Classification of benign and malignant in lung nodules using chest CT images is a key step in the diagnosis of early-stage lung cancer, as well as an effective way to improve the patients’ survival rate. However, due to the diversity of lung nodules and the visual similarity of lung nodules to their surrounding tissues, it is difficult to construct a robust classification model with conventional deep learning–based diagnostic methods. To address this problem, we propose a multi-model ensemble learning architecture based on 3D convolutional neural network (MMEL-3DCNN). This approach incorporates three key ideas: (1) Constructed multi-model network architecture can be well adapted to the heterogeneity of lung nodules. (2) The input that concatenated of the intensity image corresponding to the nodule mask, the original image, and the enhanced image corresponding to which can help training model to extract advanced feature with more discriminative capacity. (3) Select the corresponding model to different nodule size dynamically for prediction, which can improve the generalization ability of the model effectively. In addition, ensemble learning is applied in this paper to further improve the robustness of the nodule classification model. The proposed method has been experimentally verified on the public dataset, LIDC-IDRI. The experimental results show that the proposed MMEL-3DCNN architecture can obtain satisfactory classification results.
Keywords: Benign and malignant classification, Computer-aided diagnosis, Image enhancement, Multi-model ensemble architecture, 3D CNN
Introduction
According to global cancer statistics in 2018, lung cancer, prostate cancer, breast cancer, and colorectal cancer are the most common causes of cancer death in humans, and they account for 45% of all cancer deaths, 25% of which are caused by lung cancer (5-year survival rate is 18%) [1]. However, if lung cancer can be diagnosed early, the patient’s 5-year survival rate can be tripled [2]. Early lung cancer diagnosis using lung-based computed tomography (CT) images is an important strategy to improve patient survival [3]. In particular, the use of deep learning–based methods in CT images to classify the benign and malignant lung nodules is a valuable task. Because it can help doctors to judge the benign and malignant of early lung nodules, to reduce the risk of misdiagnosis and missed diagnosis, which is one of the essential tasks in the early detection of lung cancer [4, 5]. This work has important clinical significance, and its typical application is to provide relevant evidence for the follow-up treatment plan for lung cancer diagnosis. For example, if the result of the classification is benign, CT monitoring can be performed to continue observation; if the result of the classification is malignant, in vivo detection or even surgical resection is required [6].
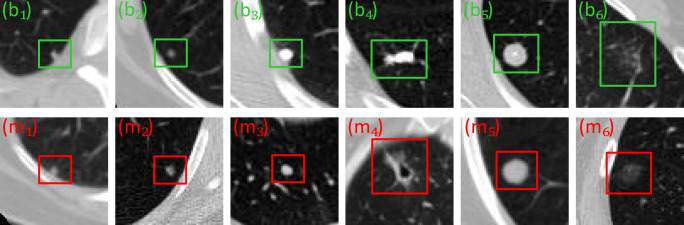
In recent years, the benign and malignant classification methods of lung nodules have been developed [7–16]. However, due to the heterogeneity of lung nodules on CT images, it is difficult to obtain a satisfactory classification result. For example, for the juxtapleural nodules (Fig. 1 (b1, m1)), calcific nodules (Fig. 1 (b3–4)), ground-glass opacity nodules (Fig. 1 (b6, m6), and cavitary nodules (Fig. 1 (m4)), they reflect the heterogeneity of lung nodules in terms of size, grayscale, shape, and texture. Besides, due to the high degree of similarity between benign nodules and malignant nodules, it is also a challenge to develop a robust classification model. For example, for some isolated nodules, although they are very similar, they do not belong to the same category (smaller scales are shown in Fig. 1 (b2, m2), and larger scales are shown in Fig. 1 (b5, M5))). Meanwhile, there is a similar situation for the juxtapleural nodule (Fig. 1 (b1, m1)) and the ground-glass opacity nodule (Fig. 1 (b6, m6)), that is, their appearance is very similar, but they belong to different categories.
Fig. 1.
Example image of benign and malignant nodules in CT image. Note that the green-labeled subgraph (b1–6) and the red-labeled subgraph (m1–6) represent six benign nodules and six malignant nodules, respectively
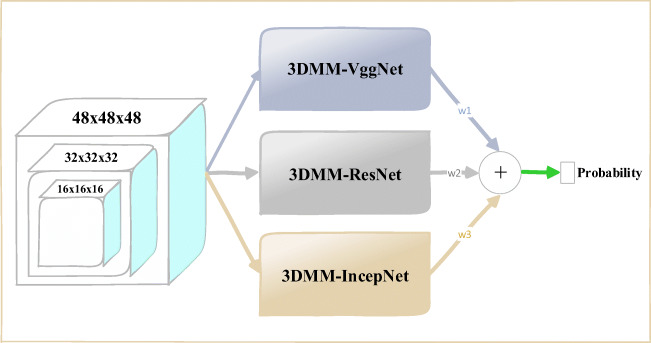
To solve the difficult problem of benign and malignant classification of lung nodules, we propose a multi-model ensemble learning architecture based on 3D convolutional neural network (MMEL-3DCNN). The architecture consists of three different types of network structures, namely a 3D multi-model network structure based on VggNet [17], ResNet [18], and InceptionNet [19]. In addition, to better adapt to different scales of lung nodules, we divide each network architecture into three sub-network structures of different input sizes. In general, MMEL-3DCNN can perform benign and malignant judgments on various types of lung nodules and achieve better classification results. Our technical contributions in this work are mainly in the following four aspects.
To take full advantage of the 3D spatial information of lung nodules, we designed three different structures of 3D CNN based on VggNet, ResNet, and InceptionNet (Fig. 2).
To make the model had better adapt to lung nodules of various sizes, we have designed three sub-networks with different input sizes under a specific architecture (“Network Architecture” section).
For purpose of more efficiently extracting advanced features with stronger discriminative ability, we concatenate the original image and its corresponding enhanced image, and then send it to the network for training (“Input Improvement” section).
To further improve the generalization ability of the model, we dynamically select the corresponding network model for prediction according to the size of the nodule, and then use their mean as the final prediction result (“Output Improvement” section).
Fig. 2.
The proposed 3D CNN-based multi-model ensemble learning framework: 3DMM-VggNet, 3DMM-IncepNet, and 3DMM-ResNet represent 3D multi-model VggNet, ResNet, and IncepNet (InceptionNet), respectively. The final prediction result is a weighted average of the output probabilities of the three modules, where w1, w2, and w3 are all one-third
Related Works
The classification task of lung nodules generally consists of three stages, namely segmentation, feature extraction, and classification. In recent years, many solutions have been proposed for the benign and malignant classification of lung nodules. These methods can generally be divided into classification methods based on traditional machine learning and classification methods based on deep convolutional neural networks.
In the classification method based on traditional machine learning, in order to better distinguish between benign nodules and malignant nodules, solutions based on support vector machines, random forests, clustering, and self-encoders have been widely used [20–26]. For example, Ashis et al. first used semi-automatic techniques to segment the nodules, then extracted the shape and texture features of the nodules, and then selected the features, and finally sent the relevant features to the support vector machine for classification [27]. Similarly, Sasidhar et al. first used the active contour model to segment the lungs, then extracted the texture features using the gray level co-occurrence matrix, and also extracted the Haralick texture features, and finally used the support vector machine to classify the nodules [28]. In addition, to ensure the validity of feature extraction, Hu et al. extracted the intensity, shape, and texture features of the nodules based on the nodule mask marked by four radiologists, and finally the improved random forest classifier was used to classify the benign and malignant nodules [29]. Different from the above method, to avoid the problem of data labeling (labeling of data in radiology field is relatively expensive), Wei et al. proposed a new unsupervised spectral clustering algorithm to distinguish benign and malignant nodules [30].
In the classification method based on deep convolutional neural networks, people use CNN to automatically extract the relevant features of lung nodules, replacing traditional hand-designed features (such as shape and texture features), and the classification model of the lung nodules is trained end-to-end in a supervised learning style. Such methods can be broadly divided into two categories, namely, a deep learning method based on 2D CNN and 3D CNN.
Methods based on 2D CNN have been widely proposed [31–36]. For example, Liu et al. proposed a multi-view convolutional neural network (MV-CNN) method to classify the benign and malignant lung nodules. Unlike traditional CNN methods, the MV-CNN input contains multiple views of the lung nodules [37]. In contrast to the above method, to reflect the spatial information of the lung nodules onto the 2D image block, Lee et al. propose a method of weighted averaging the multi-layer CT images to generate 2D image blocks for CNN training [38]. Besides, Sun et al. proposed a multi-channel ROI-based deep learning method. The input used to train the model contains original data, nodule mask, and gradient image of prominent texture features [39]. The most typical of these is a new end-to-end deep learning architecture (called the dense convolutional binary-tree networks) proposed by Liu et al. Also, to introducing central cropping operations into ResNet, the architecture separates the isolated transition layer of ResNet and merges it with dense blocks to compress the model [40].
At present, although there are not many methods for the classification of benign and malignant lung nodules using 3D CNN, some scholars have conducted related research in recent years [41–44]. For example, in order to make full use of the 3D spatial information of lung nodules, Kang et al. proposed a 3D multi-view convolutional neural network, combining the chain structure and the directed acyclic graph structure to explore the benign and malignant classification of lung nodules [45]. Besides, there is a hierarchical learning framework (multi-scale convolutional neural network) proposed by Shen et al., which captures the heterogeneity of nodules by extracting discriminative features from the stacked layers [46]. It is worth noting that Shen et al. proposed another deep learning architecture for the benign and malignant classification of lung nodules, namely a multi-cropping convolutional neural network (MC-CNN). MC-CNN is different from the nodule classification method, which relies on nodule segmentation and feature extraction. The main innovation is to propose a multi-cropping pooling strategy that can adapt well to nodule heterogeneity and directly to the original nodule data is modeled [47].
The MMEL-3DCNN architecture proposed in this paper has the following four differences from the existing methods: (1) changing the multi-branch network structure to a multi-model network architecture, (2) dynamically selecting an appropriate model for prediction based on the size of the nodule, (3) feeding the intensity stretched image together with the original image data into the network for training, (4) improving the robustness of benign and malignant classification of nodules by the ensemble learning style.
Methods
The proposed theoretical method is described in detail below. This section is divided into four subsections: the “Network Architecture” section outlines the proposed network architecture, the “Input Improvement” section describes our improvements in input, and the “Output Improvement” section describes our improvements in output, and the last section gives our training procedure.
Network Architecture
Our proposed 3D CNN-based integrated learning framework consists of three different network structures, namely VggNet-based, ResNet-based, and InceptionNet-based 3D multi-model network architecture. For a central voxel point for a given lung nodule, we extracted three different scales of 3D data blocks containing the lung nodules as inputs to the three multi-model network architectures. Moreover, by combining the output probabilities of the three network architectures to obtain the final prediction results, Figure 2 shows the overall architecture of the proposed benign and malignant classification of lung nodules.
3D Multi-model VggNet
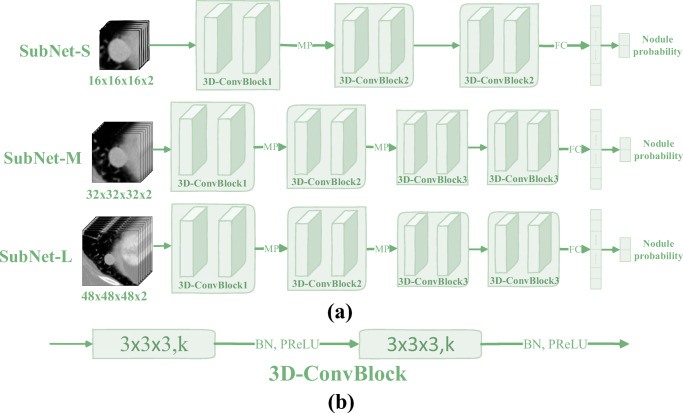
Figure 3 shows the network architecture diagram of the 3D multi-model VggNet (3DMM-VggNet) proposed in this paper. Table 1 lists the corresponding network parameters. The architecture combines three sub-networks of different structures, and the input corresponding to each sub-network is different. To improve the generalization performance of the model, we used the batch normalization operation [48], and after each convolution operation, the nonlinear parameter rectification linear unit (PReLU) was used as the activation function [49].
Fig. 3.
(a) The proposed 3D multi-model VggNet (3DMM-VggNet), where the symbols “MP” and “FC” represent the max pooling operation and the fully connected operation, respectively. (b) The structural diagram of 3D convolution block (3D-ConvBlock) in which the symbols “k,” “BN,” and “PReLU” represent the number of channels, the batch normalization operation, and the nonlinear activation function, respectively. In addition, unlike the traditional 3D input, the fourth dimension of the proposed input data has two channels, one is the original image and the other is its corresponding enhanced image
Table 1.
Network parameters of the 3DMM-VggNet. Building blocks are shown in brackets with the numbers of blocks stacked. Downsampling is done using the max pooling. The stride size of the convolution operation is one. The symbol “*” indicates that there is no such operation
| Layer name | SubNet-S | SubNet-M | SubNet-L |
|---|---|---|---|
| Input size | 16 × 16 × 16 × 2 | 32 × 32 × 32 × 2 | 48 × 48 × 48 × 2 |
| 3D-ConvBlock1 | [3 × 3 × 3, 64] × 2 | [3 × 3 × 3, 64] × 2 | [3 × 3 × 3, 64] × 2 |
| 3D-ConvBlock2 | [3 × 3 × 3, 64] × 2 | [3 × 3 × 3, 128] × 2 | [3 × 3 × 3, 128] × 2 |
| 3D-ConvBlock3 | * | [3 × 3 × 3, 128] × 2 | [3 × 3 × 3, 128] × 2 |
| FC | 150 | 200 | 250 |
| Output size | 8 × 8 × 8 | 8 × 8 × 8 | 12 × 12 × 12 |
| Nodule probability | 2-d FC, softmax | ||
3D Multi-model ResNet
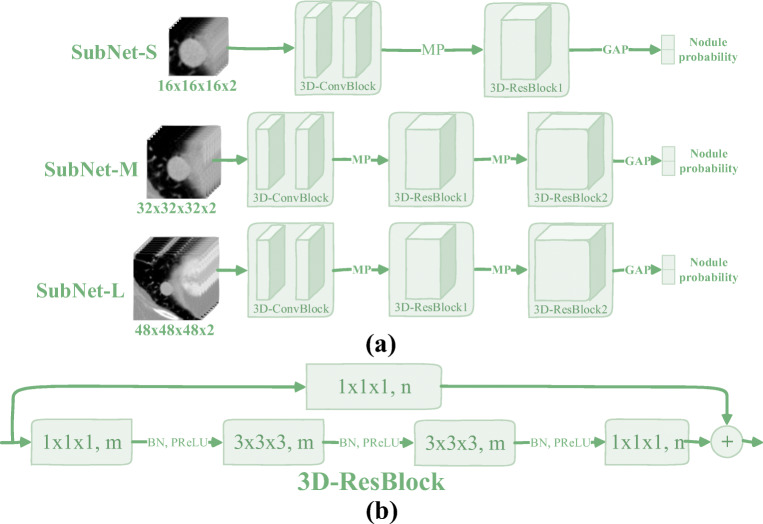
As shown in Fig. 4, the architectural diagram of the proposed 3D multi-model residual network (3DMM-ResNet) is shown, and Table 2 lists its corresponding network parameters. Similarly, the network fuses three sub-networks of different structures, and the input corresponding to each sub-network is different. In addition, batch normalization operations and nonlinear activation functions PReLU are also used to improve the robustness of the model.
Fig. 4.
(a) The proposed 3D multi-model ResNet (3DMM-ResNet), where the symbols “MP” and “GAP” represent the max pooling operation and the global average pooling operation, respectively. (b) The structural diagram of 3D residual block (3D-ResBlock) in which the symbols “m, n,” “BN,” and “PReLU” represent the number of channels, the batch normalization operation, and the nonlinear activation function, respectively. Besides, the input data and the 3D-ConvBlock are the same as described in Fig. 3
Table 2.
Network parameters of the 3DMM-ResNet. Building blocks are shown in brackets with the numbers of blocks stacked. Downsampling is performed using Max Pooling before the first layer of 3D-ResBlock1 and 3D-ResBlock2. The stride size of the convolution operation is one. The symbol “*” indicates that there is no such operation
| Layer name | SubNet-S | SubNet-M | SubNet-L |
|---|---|---|---|
| Input size | 16 × 16 × 16 × 2 | 32 × 32 × 32 × 2 | 48 × 48 × 48 × 2 |
| 3D-ConvBlock | [3 × 3 × 3, 96] × 2 | [3 × 3 × 3, 64] × 2 | [3 × 3 × 3, 32] × 2 |
| 3D-ResBlock1 | |||
| 3D-ResBlock2 | * | ||
| Output size | 8 × 8 × 8 | 8 × 8 × 8 | 12 × 12 × 12 |
| Nodule probability | Global average pooling, 2-d fc, softmax | ||
3D Multi-model IncepNet
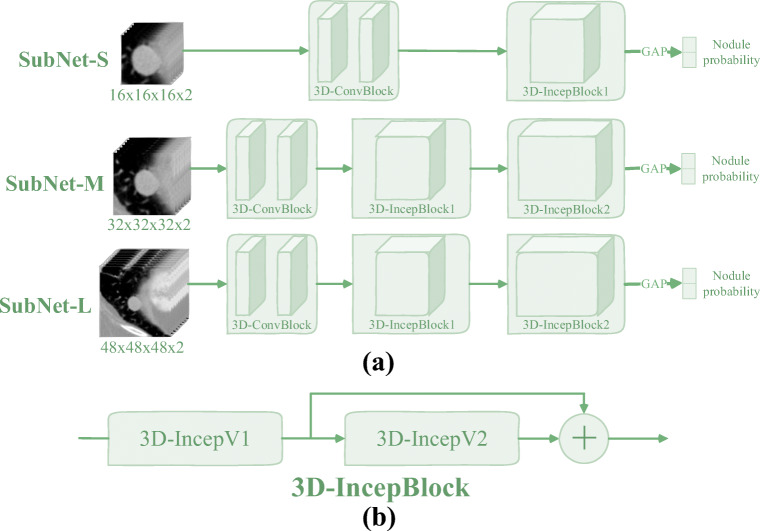
Figure 5 shows the architecture of the proposed 3D multi-model inception network (3DMM-IncepNet). Table 3 lists the corresponding network parameters. The overall architecture of the network is similar to 3DMM-ResNet and includes three sub-networks of different structures. Similarly, these three sub-networks also use batch normalization operations and PReLU activation functions.
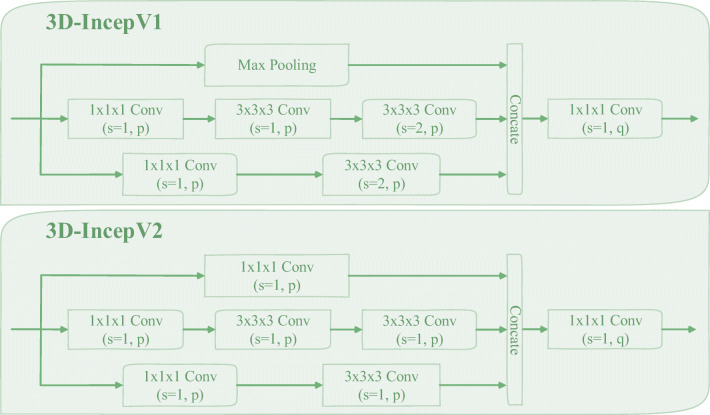
Fig. 6.
The structural diagram of “3D-IncepV1” and “3D-IncepV2”, in which the symbols “p, q” and “s” indicate the number of channels and the stride of the convolution operation, respectively, and “Concate” indicates the concatenate operation
Fig. 5.
(a) The proposed 3D multi-model InceptionNet (3DMM-IncepNet), where the symbol “GAP” represent the global average pooling operation. (b) The structural diagram of 3D Inception block (3D-IncepBlock), in which the detailed structures represented by “3D-IncepV1” and “3D-IncepV2” are shown in Fig. 6. In addition, the input data and the 3D-ConvBlock are the same as described in Fig. 3
Table 3.
Network parameters of the 3DMM-IncepNet. Building blocks are shown in brackets with the number of blocks stacked. The symbol “*” indicates there is no such operation
| Layer name | SubNet-S | SubNet-M | SubNet-L |
|---|---|---|---|
| Input size | 16 × 16 × 16 × 2 | 32 × 32 × 32 × 2 | 48 × 48 × 48 × 2 |
| 3D-ConvBlock | [3 × 3 × 3, 96] × 2 | [3 × 3 × 3, 64] × 2 | [3 × 3 × 3, 32] × 2 |
| 3D-IncepBlock1 | p = 32, q = 128 | p = 24, q = 96 | p = 16, q = 64 |
| 3D-IncepBlock2 | * | p = 32, q = 128 | p = 24, q = 96 |
| Output size | 8 × 8 × 8 | 8 × 8 × 8 | 12 × 12 × 12 |
| Nodule probability | Global average pooling, 2-d fc, softmax | ||
Input Improvement
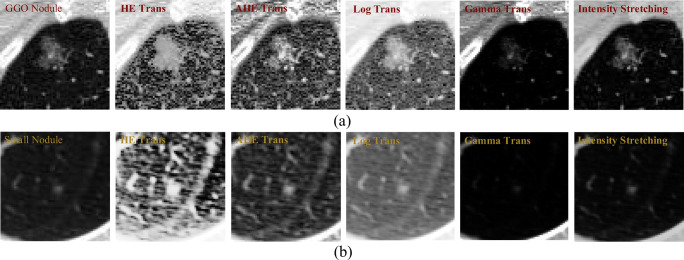
Due to the wide variety of lung nodules, some of which have low contrast with their surrounding environment, such nodule networks often fail to adequately extract their features, making it difficult to identify such nodules. The most typical are ground-glass opacity nodules (as shown in Fig. 7(a)) and darker small nodules (as shown in Fig. 7(b)). To solve this problem, we perform image enhancement on the data sent to the network, thereby suppressing features that are not of interest in the image, and expanding the difference between the lung nodules and the background.
Fig. 7.
The visual comparison of five image enhancement methods, where (a) shows a ground-glass opacity (GGO) nodule and (b) shows a small nodule with a lower contrast. Besides, “HE” represents histogram equalization, “AHE” represents adaptive histogram equalization, and “Tran” is an abbreviation for “transformation”
Specifically, in order for the proposed network architecture to better handle low-contrast nodules, we set the fourth dimension (number of channels) of the input data to two; one is the original image and the other is its corresponding enhanced image. Image enhancement methods can be generally divided into two categories, one is to adjust the distribution of the intensity histogram to achieve image enhancement and the other is to directly transform the intensity according to the formula to improve the contrast of the image. This paper attempts five image enhancement methods, namely histogram equalization (HE) [50], adaptive histogram equalization (AHE) [51], gamma transformation (Gamma Trans) [52], logarithmic transformation (Log Trans) [53], and intensity stretch transformation. The intensity stretching method differs from the Log Trans and Gamma Trans methods in that the intensity stretching can improve the dynamic range of the image and convert the original low-contrast image into a high contrast image.
| 1 |
The calculation formula of the intensity stretch transformation used in this paper is shown in Eq. (1). Wherein, the symbol “ni” represents the input image after normalization, the symbol “ce” represents the transformed output image, and the parameter “k” represents the slope of the control function. In the experiment, the value of k is 2. In addition, it should be noted that the values of ni and ce are all [0, 1], τ is a very small number (preventing the zero-dividing error), and the value in the experiment is e−8.
As can be seen from Fig. 7, the image enhancement method based on the histogram adjustment, although the nodules are largely highlighted, the surrounding tissues (blood vessels, lung walls, interlobular fissures, etc.) are also enhanced to some extent. In particular, the HE transformation method in Fig. 7. Similarly, the Log Trans–based image enhancement method also significantly enhanced the tissue surrounding the lung nodules, such as the blood vessels in Fig. 7(a) and the interlobular fissures in Fig. 7(b). Furthermore, as can be seen from Fig. 7(a), the image enhancement method based on the Gamma Trans can well suppress the tissue around the nodule. However, for small nodules with lower intensity, it does not show a good enhancement effect (see the image corresponding to Gamma Trans in Fig. 7(b)). The intensity stretching method we used not only highlights the nodules to a certain extent but also further inhibits the tissue around the nodules. The quantitative indicators corresponding to the above five image enhancement methods are discussed in the “Ablation Study” section.
Finally, to make the model better focus on the location, shape, and intensity information of the nodule, we concatenate the intensity image corresponding to the nodule mask image with the original image and the enhanced image to form a three-channel 3D data patch for the input of the model. In other words, sending the mask image as input to the model allows the model to be better extracted to representative features, similar to the idea of an attention mechanism. In the “Ablation Study” section, we will demonstrate its effectiveness from an experimental perspective.
Output Improvement
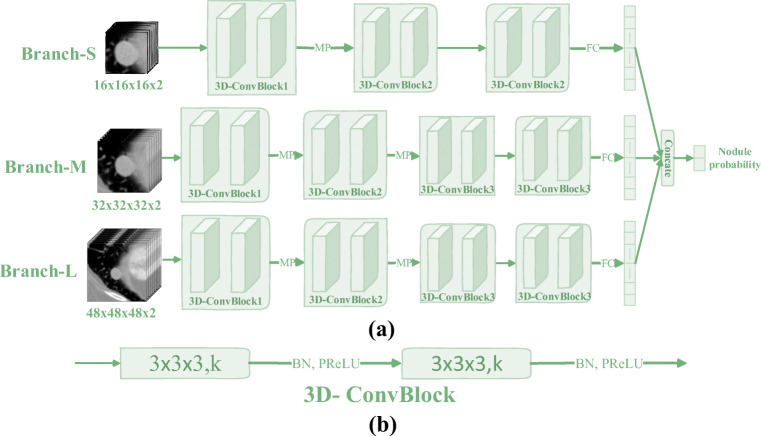
At the beginning of the network architecture design, to adapt to different sizes of lung nodules, we have designed three multi-branch network architectures based on VggNet, ResNet, and InceptionNet. As shown in Fig. 8, the multi-branch network architecture based on VggNet is shown. The other two multi-branch network architectures based on ResNet and InceptionNet are transformed in a similar way to VggNet, which combines them into a multi-branch network through concatenate operations at the end of the three sub-networks. In addition, the quantitative indicators of the multi-branch network architecture are described in the ablation experiments in the “Ablation Study” section.
Fig. 8.
Multi-branch network architecture based on VggNet (3DMB-VggNet). The corresponding network parameters are the same as those corresponding to Fig. 3 (Table 1). The only difference is that there is one more concatenate operation at the end of the multi-branch network architecture
The following are two reasons why we use the multi-model network architecture to replace the multi-branch network architecture.
One is that the training time of the network model can be greatly reduced (if the computer used for training must have at least three or more GPUs). After we split a multi-branch network architecture into three sub-networks, the complexity of the network is greatly reduced. Since the three sub-networks are independent, the training can be performed in parallel, that is, the time for the completion of the training of the three sub-networks depends only on the training time required for the sub-network that trains the slowest. It is assumed that the slowest training sub-network is the sub-network SubNet-L, but the training time of the sub-network SubNet-L is largely less than its corresponding multi-branch network architecture. Because it is far less than the corresponding multi-branch network architecture in terms of the size of the input data and the network parameters that need to be trained. In addition, if the experimental hardware environment has enough GPUs and large enough video memory and memory, then the nine sub-networks can be trained at the same time, further reducing the training time of the entire classification network. The time we spend training the network will be explained in the description of the “Experimental Environment” section.
The second is that you can use the size information provided by LIDC to describe the size of the lung nodules. That is, an appropriate network model can be dynamically selected for prediction according to the size of the nodule. For example, we train three network models M1, M2, and M3 using 16 × 16 × 16, 32 × 32 × 32, and 48 × 48 × 48 sizes, respectively. If the size of a given candidate nodule is less than 16 mm, then we can use M1 and M2 for prediction and then take their average as the final benign and malignant probability. This is because, for lung nodules less than 16 mm in diameter, models trained using larger scale 48 × 48 × 48 image blocks often bring a lot of redundant information or even noise to the network, which makes it not well adapted to such nodules. Similarly, if the size of the lung nodules is greater than 16 mm, we will use M2 and M3 for prediction, and then take the average of the two prediction results as the final output. The reason for not selecting the model M1 to predict a nodule larger than 16 mm is the 16 × 16 × 16 image block corresponding to a large nodule larger than 16 mm contains almost no context information,, and the model trained using it does not extract the high-level semantic features of the image well. Besides, the quantitative indicators corresponding to the measure of selecting the appropriate model for prediction based on the size of the nodule will be described in the “Ablation Study” section.
Training Procedure
The ratio of positive and negative samples we analyzed from the lung image database consortium and image database resource initiative (LIDC-IDRI) dataset is approximately 1:2. To alleviate the problem of the imbalance in the number of positive samples (malignant nodules) and negative samples (benign nodules), we oversampled the positive samples in the training set so that the proportion of positive and negative samples approached 1:1. To further improve the generalization ability of the model, we expand the positive and negative samples in the training set by the traditional data expansion method (flip in three directions of X-axis, Y-axis, and Z-axis, and 90° rotation of XOY plane), and finally expand the size of the training set to 16 times.
Specifically, during the training process, we use the Adam optimizer to update the model parameters [54]. To prevent over-fitting, we use the early stop training strategy [55] (continue training if the performance of the model is not improved), where the epoch of continuing training is 30 and the total training algebra is 100. Besides, the batch size and learning rate of the nine sub-networks corresponding to 3DMM-VggNet, 3DMM-ResNet, and 3DMM-IncepNet are listed in Table 4.
Table 4.
The batch size and learning rate of nine sub-networks corresponding to 3DMM-VggNet, 3DMM-ResNet, and 3DMM-IncepNet
| Network Name | 3DMM-VggNet | 3DMM-ResNet | 3DMM-IncepNet | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Input Size | 16 | 32 | 48 | 16 | 32 | 48 | 16 | 32 | 48 |
| Batch Size | 32 | 24 | 16 | 32 | 32 | 24 | 32 | 32 | 24 |
| Learning Rate | 3 × e−5 | 3 × e−5 | 1 × e−5 | 3 × e−4 | 3 × e−4 | 1 × e−4 | 3 × e−4 | 3 × e−4 | 1 × e−5 |
Since the training and validation of these nine sub-network architectures are independent of each other and the training objectives are the probabilities of maximizing the correct class, we achieve this by minimizing the cross entropy loss of each training sample. For a given input sample with positive and negative labels, assuming y is their true label, and then the cross entropy loss function is defined as shown in Eq. (2).
| 2 |
Where y′ represents the predicted probability of the model and N represents the number of samples.
Data and Experiment
This chapter is divided into three subsections, the “Data” section describes the data used, the “Evaluation Criteria” section describes the evaluation criteria, and the “Experimental Environment” section describes the experimental environment.
Data
This paper uses a public dataset from the lung image database consortium and image database resource initiative (LIDC-IDRI). Among them, the slice interval ranges from 0.45 to 5.0 mm, and the axial plane resolution ranges from 0.46mm × 0.46 mm to 0.98 mm × 0.98 mm. In addition, the dataset was scored by four experienced radiologists for each nodule with a malignancy ranging from 1 to 5. The greater the value, the higher the degree of malignancy. To classify the benign and malignant lung nodules, we consider the average malignancy less than 3 as benign nodules (LMN), the average malignancy greater than 3 as malignant nodules (HMN), and the average malignancy equal to 3 as uncertain nodules (not involved in training and evaluation) [47, 56, 57].
Overall, the number of nodules with an average degree of malignancy of 1, 2, 3, 4, and 5 was 288, 575, 1182, 343, and 62, after excluding the nodule samples with ID blur and nodule size from 3 to 30 mm. Finally, in order to better evaluate the robustness of the algorithm, we adopt the 5-fold cross validation, in which the ratio of each fold, training set, validation set, and testing set, is 3:1:1, and the specific division of the experimental data is shown in Table 5.
Table 5.
The distribution of data for the training set, validation set, and testing set in each fold cross validation
| Malignancy level | Training set (764) | Validation set (252) | Testing set (252) |
|---|---|---|---|
| 1 | 174 | 57 | 57 |
| 2 | 345 | 115 | 115 |
| 3 | * | * | * |
| 4 | 207 | 68 | 68 |
| 5 | 38 | 12 | 12 |
Evaluation Criteria
In addition to using the AUC score to assess the performance of benign and malignant classification of lung nodules, three more common indicators were used, sensitivity (SEN), specificity (SPC), and accuracy (ACC). Their definitions are shown in Eqs. (3)–(5), respectively.
| 3 |
| 4 |
| 5 |
where TP represents the number of samples that will predict the positive sample correctly; TN represents the number of samples that will predict the negative sample correctly; FP represents the number of samples that predict the negative sample as the positive sample; FN represents the number of samples that predict the positive sample as the negative sample.
Experimental Environment
We use a dynamic, object-oriented scripting language (Python) for related experiments, the corresponding version number is 3.6, and the integrated development environment is PyCharm. Besides, we use the high-level neural network API (Keras) to build the designed network architecture. To speed up the training of the network model, all of our experiments were performed on a server with an Intel(R) Xeon(R) processor and 125GB of memory, and the server had ten GPUs. However, there are actually three GPUs that can be applied for (GPU model NVIDIA GTX-1080Ti GPU, video memory size 11GB).
According to our experiments, the time required for the convergence of the three network models 3DMM-VggNet, 3DMM-ResNet, and 3DMM-IncepNet is shown in Table 6. Correspondingly, the time required for their original three multi-branch network architectures to converge is shown in Table 7. Comparing the experimental results of Tables 6 and 7, it can be seen that the improvement from the original multi-branch network architecture to the multi-model network architecture can greatly shorten the training time of the network, which also confirms the theoretical analysis described in the “Output Improvement” section of this paper.
Table 6.
Time required for the three 3D CNN-based multi-model network architectures to converge. Among them, the unit is minutes (m)
| Network name | 3DMM-VggNet | 3DMM-ResNet | 3DMM-IncepNet | ||||||
| Input size | 16 | 32 | 48 | 16 | 32 | 48 | 16 | 32 | 48 |
| Train time | 7 m | 16 m | 36 m | 6 m | 20 m | 31 m | 5 m | 12 m | 36 m |
Table 7.
Time required for the original three 3D CNN-based multi-branch network architectures to converge
| Network name | 3DMB-VggNet | 3DMB-ResNet | 3DMB-IncepNet |
| Input size | 16 & 32 & 48 | 16 & 32 & 48 | 16 & 32 & 48 |
| Train time | 79 m | 52 m | 50 m |
Results and Discussion
This chapter is divided into four subsections, of which the “Overall Performance” section analyzes the overall performance of the proposed method, the “Ablation Study” section describes the ablation experiment of the proposed architecture, the “Experimental Comparison” section introduces our comparative experiments and experimental results, and the last section explain why we chose VggNet, ResNet, and IncepNet for integration.
Overall Performance
For purpose of more intuitively observing the overall performance of the proposed method, we plot the ROC curves corresponding to the three network architectures (3DMM-VggNet, 3DMM-IncepNet, and 3DMM-ResNet), and the corresponding ROC curves for their ensemble (as shown in Fig. 9). It can be seen from Fig. 9 that the sensitivity of the three separate network architectures can reach more than 90% with the false positive rate of 0.3, which indicates that the 3D CNN-based network architecture proposed in this paper can extract features with strong nodule discrimination ability from CT images. Besides, we can clearly see that the ensemble thinking is effective for the classification of benign and malignant lung nodules.
Fig. 9.
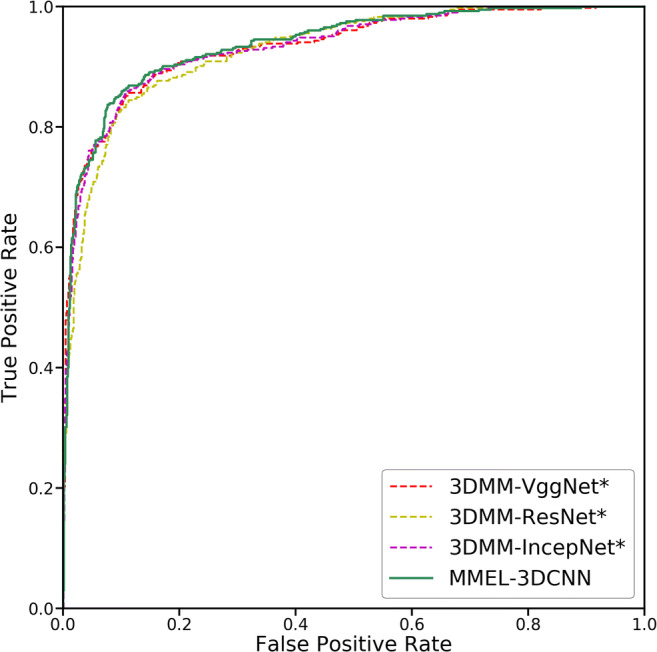
The ROC (receiver operating characteristic) curves for the three networks and their integration. Among them, 3DMM-VggNet* indicates the fusion result of its corresponding three sub-networks. Similarly, 3DMM-ResNet* and 3DMM-IncepNet* are also a fusion of their corresponding three sub-networks
Table 8 lists the four quantitative metrics used to evaluate the performance of the algorithm, namely the sensitivity, specificity, accuracy, and AUC score for the three network architectures and their ensemble. As can be seen from Table 8, the three AUC scores corresponding to the three network architectures are higher than 90%, and after they are integrated, the four evaluation indicators are improved. Especially for 3DMM-IncepNet, in addition to the specificity, the other three indicators have increased by more than two percentage points. The results of these experiments show that the ensemble learning approach can complement each other’s defects between different network architectures, resulting in an overall performance improvement.
Table 8.
The quantitative indicator used to evaluate the performance of the algorithm. Also, the meanings of 3DMM-VggNet*, 3DMM-ResNet*, and 3DMM-IncepNet* are the same as explained in Fig. 9
| Method | SEN | SPC | ACC | AUC |
|---|---|---|---|---|
| 3DMM-VggNet* | 0.815 | 0.918 | 0.885 | 0.938 |
| 3DMM-ResNet* | 0.815 | 0.922 | 0.888 | 0.935 |
| 3DMM-IncepNet* | 0.798 | 0.927 | 0.886 | 0.934 |
| MMEL-3DCNN | 0.837 | 0.939 | 0.906 | 0.939 |
Ablation Study
To validate the effectiveness of the components in the MMEL-3DCNN architecture, we designed an ablation experiment based on 2DMB-VggNet (the same as the overall architecture of 3DMB-VggNet, the difference is that it uses a two-dimensional convolutional neural network). The relevant results of this experiment are shown in Table 9.
Table 9.
Note that “MB” indicates the original multi-branch structure, “MM” indicates the proposed multi-model structure, “DP” indicates that the appropriate model is selected according to the diameter for dynamic prediction; “IE” indicates image enhancement, “MI” indicates mask image, “TSA” indicates testing set augmentation, and “ParaA” means the amount of parameters that need to be trained
| Method | ParaA | SEN | SPC | ACC | AUC |
|---|---|---|---|---|---|
| 2DMB-VggNet | 0.7 × 107 | 0.731 | 0.898 | 0.845 | 0.895 |
| 3DMB-VggNet | 7.8 × 107 | 0.756 | 0.899 | 0.853 | 0.908 |
| 3DMM-VggNet | 7.8 × 107 | 0.788 | 0.905 | 0.867 | 0.925 |
| 3DMM-VggNet_DP | 7.8 × 107 | 0.810 | 0.897 | 0.869 | 0.924 |
| 3DMM-VggNet_DP_IE | 7.8 × 107 | 0.802 | 0.917 | 0.880 | 0.923 |
| 3DMM-VggNet_DP_IE_MI | 7.8 × 107 | 0.807 | 0.920 | 0.884 | 0.923 |
| 3DMM-VggNet_DP_IE_MI_TSA | 7.8 × 107 | 0.815 | 0.918 | 0.885 | 0.938 |
| 3DMM-ResNet_DP_IE_MI_TSA | 3.7 × 107 | 0.815 | 0.922 | 0.888 | 0.935 |
| 3DMM-IncepNet_DP_IE_MI_TSA | 2.8 × 107 | 0.798 | 0.927 | 0.886 | 0.934 |
| MMEL-3DCNN | * | 0.837 | 0.939 | 0.906 | 0.939 |
Effect of the Dimensional Space
In the experiment shown in Table 9, 2DMM-VggNet is based on the 2D convolutional neural network, the same classification model as 3DMB-VggNet (shown in Fig. 8). 3DMB-VggNet is a classification model based on 3D convolutional neural network (see the “Output Improvement” section for details). The experimental results show that the recall rate is increased by two percentage points compared with 2DMB-VggNet, and the other three indicators are also improved. By comparing the results of these two experiments, we can see that although the model parameters of the algorithm have increased, the performance has been greatly improved, which also verifies the effectiveness of using more spatial information.
Effect of the Multi-model
Two reasons why we have changed the multi-branch network architecture to the multi-model network architecture have been described in detail in the “Output Improvement” section, and its strength in training time is also confirmed in the “Experimental Environment” section. Here, by comparing the 3rd and 4th rows in Table 9, we can clearly see that the sensitivity and AUC have been greatly improved, and the specificity and accuracy have also been improved to some extent. This validates the effectiveness of the multi-model network architecture proposed in this paper from an experimental perspective.
Effect of the Dynamic Prediction
Based on the multi-model classification network proposed in this paper, we can dynamically select the appropriate model to predict the benign and malignant according to the size of the nodule, and then use the average of their prediction results as the final prediction result. The corresponding theoretical analysis can be found in the “Output Improvement” section. Here, by comparing the experimental data of the 4th and 5th rows in Table 9, we can see that the sensitivity is increased by 2.2 percentage points when the specificity, correct rate, and AUC remain almost unchanged. This also validates the effectiveness of dynamic prediction using the size information of nodules from an experimental perspective.
Effect of the Image Enhancement
We introduced five methods for image enhancement of input data in the “Input Improvement” section. By comparing the results of the 5th and 6th rows in Table 9, we can see that the addition of the “IE” (Intensity Stretching) component caused the sensitivity to drop by 0.8%. However, the specificity and accuracy rate increased by 2.0% and 1.1%, respectively. Overall, image enhancement of the data is advantageous.
The “Input Improvement” section only visually demonstrates that the grayscale stretching method is superior to the other four image enhancement methods, while Table 10 shows their corresponding quantitative indicators. As can be seen from Table 10, the intensity stretching method we use can be better applied to the classification of benign and malignant lung nodules.
Table 10.
The quantitative indicators for the five image enhancement methods
| Method | SEN | SPC | ACC | AUC |
|---|---|---|---|---|
| HE Trans | 0.741 | 0.931 | 0.870 | 0.927 |
| AHE Trans | 0.753 | 0.948 | 0.876 | 0.921 |
| Log Trans | 0.716 | 0.936 | 0.866 | 0.927 |
| Gamma Trans | 0.704 | 0.960 | 0.878 | 0.911 |
| Intensity stretching | 0.802 | 0.917 | 0.880 | 0.923 |
Effect of the Mask Image
By comparing the experimental results of the 6th and 7th rows in Table 9, it can be seen that under the condition that the AUC remains unchanged, the other three indicators have a certain degree of improvement. This verifies from an experimental point of view that it is effective to add an intensity image corresponding to the nodule mask to the input of the model.
Effect of the Testing Set Augmentation
Testing set enhancement is a common strategy for improving the generalization ability of models. Specifically, when predicting the sample S in the testing set, we not only predict itself but also predict the expansion samples (the expansion method is the same as the training set) corresponding to it, and then use the mean of these predicted values corresponding to the sample S as its final prediction result. By comparing the experimental results of the 7th and 8th rows in Table 9, we can see that although the specificity decreased by 0.2%, the sensitivity and AUC increased by 0.8% and 1.5%, respectively. In general, the testing set augmentation method has played a role.
Effect of the Ensemble Learning
The results of the last four lines of Table 9 demonstrate the effectiveness of the ensemble learning method. Among them, 3DMM-ResNet_DP_IE_MI_TSA and 3DMM-IncepNet_DP_IE_MI_TSA, except for the network architecture, are different from 3DMB-VggNet_DP_IE_MI_TSA, and the other conditions are the same. MMEL-3DCNN is the integration of the above three. By comparing the experimental data of the last four lines, it can be seen that the experimental results corresponding to MMEL-3DCNN have a small increase in AUC, but the other three indicators have improved significantly. In particular, the specificity and accuracy of MMEL-3DCNN increased by 3.9% and 2.0%, respectively, relative to 3DMM-IncepNet_DP_IE_MI_TSA.
Experimental Comparison
To verify the superiority of the proposed method, we compared it with the benign and malignant classification of lung nodules published in recent years. Table 11 shows the quantitative results in the comparison of traditional machine learning–based classification methods and CNN-based classification methods.
Table 11.
The comparison on the quantitative results of various lung nodule malignancy suspiciousness classification methods. Please note that “TML” indicates the traditional machine learning
| Type | Method | SEN | SPC | ACC | AUC |
|---|---|---|---|---|---|
| TML | Lee et al. [20] | 0.870 | 0.840 | 0.810 | 0.889 |
| Kaya et al. [23] | 0.559 | 0.947 | 0.825 | * | |
| Dhara et al. [27] | * | * | * | 0.946 | |
| Wei et al. [30] | * | * | 0.854 | * | |
| CNN | Liu et al. [37] | 0.664 | 0.853 | 0.779 | 0.852 |
| Sun et al. [39] | 0.692 | 0.880 | 0.806 | 0.867 | |
| Causey et al. [36] | * | * | 0.750 | 0.860 | |
| Lee et al. [38] | 0.716 | 0.894 | 0.838 | 0.889 | |
| Liu et al. [40] | * | * | 0.882 | 0.934 | |
| Shen et al. [47] | 0.770 | 0.930 | 0.871 | 0.930 | |
| Ours | 0.837 | 0.939 | 0.906 | 0.939 |
As can be seen from Table 11, although the AUC of the SVM-based lung nodule classification method proposed by Dhara et al. is the highest, the method requires manual intervention and only partial data of the LIDC-IDRI dataset is evaluated [27]. Lee et al. [20] combined genetic algorithm and random subspace method to classify benign and malignant pulmonary nodules; Kaya et al. [23] proposed a classification method based on weighting rules to predict the malignancy of lung nodules, and they all achieved good results. In addition, both Liu et al. [37] and Sun et al. [39] proposed the method based on 2D CNN, which only utilizes the characteristic information of the central layer of the nodule, and its sensitivity is less than 70%, and the effect is not ideal. The method proposed by Lee et al. [38], by sending multiple slices of different views and different angles together into the network for training, enables the network to learn more spatial information. This type of method has some improvement over the single view and single angle method. To make fuller use of the spatial information of nodules, Liu et al. [40] and Shen et al. [47] proposed a 3D CNN-based classification of benign and malignant lung nodules, and they obtained better results. The method proposed in this paper is also based on the method of 3D CNN construction. Through comparative experiments, it can be seen from the experimental results in Table 11 that this method is superior to other existing lung and nodule classification methods.
Conclusion
In the study of benign and malignant judgment of lung nodules, to adapt to the heterogeneity of lung nodules, we designed a multi-model ensemble network framework based on 3D CNN. Meanwhile, in order to better identify lung nodules with low contrast to surrounding tissues, we performed image enhancement on the input data to improve the contrast of such samples. Furthermore, we have improved the multi-branch network architecture into a multi-model network architecture, which can greatly shorten the training time of the model, and dynamically select the appropriate model for prediction based on the size of the nodule. Besides, we draw on the idea of “expert consultation” to build multiple independent networks to simulate different expert behaviors, and then use ensemble learning to fuse the prediction results of multiple models to reflect the joint decision of experts. This complements each other’s defects, making the system more stable. Finally, through the comparison of ablation research and experiment, we verified the overall performance of the proposed network architecture and the effectiveness of each component. According to the experimental results shown in Tables 9, 10, and 11, it can be seen that for the benign and malignant classification of lung nodules, the method proposed in this paper can obtain ideal experimental results.
Acknowledgments
The authors acknowledge the National Cancer Institute and the Foundation for the National Institutes of Health and their critical role in the creation of the free publicly available LIDC-IDRI Database used in this study.
Funding Information
The National Key R&D Program of China (Grant No. 2017YFC0112804) supported this work.
Compliance with Ethical Standards
Conflict of Interest
The authors declare that they have no conflicts of interest.
Ethical Approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed Consent
Informed consent was obtained from all individual participants included in the study.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Bach PB, Mirkin JN, Oliver TK, Azzoli CG, Berry DA, Brawley OW, Byers T, Colditz GA, Gould MK, Jett JR, Sabichi AL, Smith-Bindman R, Wood DE, Qaseem A, Detterbeck FC. Benefits and harms of CT screening for lung cancer: a systematic review. JAMA. 2012;307:2418–2429. doi: 10.1001/jama.2012.5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atwater T, Cook CM, Massion PP. The Pursuit of Noninvasive Diagnosis of Lung Cancer. Semin Respir Crit Care Med. 2016;37:670–680. doi: 10.1055/s-0036-1592314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wahidi MM, Govert JA, Goudar RK, Gould MK, McCrory DC. Evidence for the Treatment of Patients With Pulmonary Nodules: When Is It Lung Cancer?: ACCP Evidence-Based Clinical Practice Guidelines (2nd Edition) Chest. 2007;132:94S–107S. doi: 10.1378/chest.07-1352. [DOI] [PubMed] [Google Scholar]
- 5.Gould MK, Ananth L, Barnett PG. A Clinical Model To Estimate the Pretest Probability of Lung Cancer in Patients With Solitary Pulmonary Nodules. Chest. 2007;131:383–388. doi: 10.1378/chest.06-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gould MK, Donington J, Lynch WR, Mazzone PJ, Midthun DE, Naidich DP, Wiener RS. Evaluation of Individuals With Pulmonary Nodules: When Is It Lung Cancer?: Diagnosis and Management of Lung Cancer, 3rd ed: American College of Chest Physicians Evidence-Based Clinical Practice Guidelines. Chest. 2013;143:e93S–e120S. doi: 10.1378/chest.12-2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mao K, Deng Z. Lung Nodule Image Classification Based on Local Difference Pattern and Combined Classifier. Comput Math Methods Med. 2016;2016:1091279. doi: 10.1155/2016/1091279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Froz BR, de Carvalho Filho AO, Silva AC, de Paiva AC, Acatauassú Nunes R, Gattass M. Lung nodule classification using artificial crawlers, directional texture and support vector machine. Expert Syst Appl. 2017;69:176–188. doi: 10.1016/j.eswa.2016.10.039. [DOI] [Google Scholar]
- 9.Wei G, Cao H, Ma H, Qi S, Qian W, Ma Z. Content-based image retrieval for Lung Nodule Classification Using Texture Features and Learned Distance Metric. J Med Syst. 2017;42:13. doi: 10.1007/s10916-017-0874-5. [DOI] [PubMed] [Google Scholar]
- 10.R. Dey, Z. Lu, Y. Hong, Diagnostic classification of lung nodules using 3D neural networks, in: 2018 IEEE 15th Int. Symp. Biomed. Imaging (ISBI 2018), 2018: pp. 774–778. 10.1109/ISBI.2018.8363687.
- 11.G. Polat, Y.S. Dogrusöz, U. Halici, Effect of input size on the classification of lung nodules using convolutional neural networks, in: 2018 26th Signal Process. Commun. Appl. Conf., 2018: pp. 1–4. 10.1109/SIU.2018.8404659.
- 12.Xie Y, Zhang J, Xia Y, Fulham M, Zhang Y. Fusing texture, shape and deep model-learned information at decision level for automated classification of lung nodules on chest CT. Inf Fusion. 2018;42:102–110. doi: 10.1016/j.inffus.2017.10.005. [DOI] [Google Scholar]
- 13.M. Sakamoto, H. Nakano, K. Zhao, T. Sekiyama, Lung nodule classification by the combination of fusion classifier and cascaded convolutional neural networks, in: 2018 IEEE 15th Int. Symp. Biomed. Imaging (ISBI 2018), 2018: pp. 822–825. 10.1109/ISBI.2018.8363698.
- 14.R.V.M. d. Nóbrega, S.A. Peixoto, S.P.P. d. Silva, P.P.R. Filho, Lung Nodule Classification via Deep Transfer Learning in CT Lung Images, in: 2018 IEEE 31st Int. Symp. Comput. Med. Syst., 2018: pp. 244–249. 10.1109/CBMS.2018.00050.
- 15.Zhao D, Zhu D, Lu J, Luo Y, Zhang G. Synthetic Medical Images Using F&BGAN for Improved Lung Nodules Classification by Multi-Scale VGG16. Symmetry (Basel) 2018;10:519. doi: 10.3390/sym10100519. [DOI] [Google Scholar]
- 16.J. Lyu, S.H. Ling, Using Multi-level Convolutional Neural Network for Classification of Lung Nodules on CT images, in: 2018 40th Annu. Int. Conf. IEEE Eng. Med. Biol. Soc., 2018: pp. 686–689. 10.1109/EMBC.2018.8512376. [DOI] [PubMed]
- 17.K. Simonyan, A. Zisserman, Very Deep Convolutional Networks for Large-Scale Image Recognition., CoRR. abs/1409.1 (2014). http://arxiv.org/abs/1409.1556.
- 18.K. He, X. Zhang, S. Ren, J. Sun, Deep Residual Learning for Image Recognition, in: 2016 IEEE Conf. Comput. Vis. Pattern Recognit., 2016: pp. 770–778. 10.1109/CVPR.2016.90.
- 19.C. Szegedy, S. Ioffe, V. Vanhoucke, A.A. Alemi, Inception-v4, Inception-ResNet and the Impact of Residual Connections on Learning. BT - Proceedings of the Thirty-First AAAI Conference on Artificial Intelligence, February 4-9, 2017, San Francisco, California, USA., (2017) 4278–4284. http://aaai.org/ocs/index.php/AAAI/AAAI17/paper/view/14806.
- 20.Lee MC, Boroczky L, Sungur-Stasik K, Cann AD, Borczuk AC, Kawut SM, Powell CA. Computer-aided diagnosis of pulmonary nodules using a two-step approach for feature selection and classifier ensemble construction. Artif Intell Med. 2010;50:43–53. doi: 10.1016/j.artmed.2010.04.011. [DOI] [PubMed] [Google Scholar]
- 21.Barros Netto SM, Silva AC, Acatauassú Nunes R, Gattass M. Analysis of directional patterns of lung nodules in computerized tomography using Getis statistics and their accumulated forms as malignancy and benignity indicators. Pattern Recognit Lett. 2012;33:1734–1740. doi: 10.1016/j.patrec.2012.05.010. [DOI] [Google Scholar]
- 22.Elizabeth DS, Nehemiah HK, Raj CSR, Kannan A. Computer-aided diagnosis of lung cancer based on analysis of the significant slice of chest computed tomography image. IET Image Process. 2012;6:697–705. doi: 10.1049/iet-ipr.2010.0521. [DOI] [Google Scholar]
- 23.Kaya A, Can AB. A weighted rule based method for predicting malignancy of pulmonary nodules by nodule characteristics. J. Biomed. Inform. 2015;56:69–79. doi: 10.1016/j.jbi.2015.05.011. [DOI] [PubMed] [Google Scholar]
- 24.Firmino M, Angelo G, Morais H, Dantas MR, Valentim R. Computer-aided detection (CADe) and diagnosis (CADx) system for lung cancer with likelihood of malignancy. Biomed Eng Online. 2016;15:2. doi: 10.1186/s12938-015-0120-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Sousa Costa RW, da Silva GLF, de Carvalho Filho AO, Silva AC, de Paiva AC, Gattass M. Classification of malignant and benign lung nodules using taxonomic diversity index and phylogenetic distance. Med. Biol. Eng. Comput. 2018;56:2125–2136. doi: 10.1007/s11517-018-1841-0. [DOI] [PubMed] [Google Scholar]
- 26.Rodrigues MB, Da Nóbrega RVM, Alves SSA, Filho PPR, Duarte JBF, Sangaiah AK, De Albuquerque VHC. Health of Things Algorithms for Malignancy Level Classification of Lung Nodules. IEEE Access. 2018;6:18592–18601. doi: 10.1109/ACCESS.2018.2817614. [DOI] [Google Scholar]
- 27.A.K. Dhara, S. Mukhopadhyay, A. Dutta, M. Garg, N. Khandelwal, P. Kumar, Classification of pulmonary nodules in lung CT images using shape and texture features, in: 2016: pp. 97852Y-9785–6. [DOI] [PMC free article] [PubMed]
- 28.B. Sasidhar, G. Geetha, B.I. Khodanpur, D.R. Ramesh Babu, Automatic Classification of Lung Nodules into Benign or Malignant Using SVM Classifier BT - Proceedings of the 5th International Conference on Frontiers in Intelligent Computing: Theory and Applications, in: S.C. Satapathy, V. Bhateja, S.K. Udgata, P.K. Pattnaik (Eds.), Springer Singapore, Singapore, 2017: pp. 551–559.
- 29.H. Hu, S. Nie, Classification of malignant-benign pulmonary nodules in lung CT images using an improved random forest (Use style: Paper title), in: 2017 13th Int. Conf. Nat. Comput. Fuzzy Syst. Knowl. Discov., 2017: pp. 2285–2290. 10.1109/FSKD.2017.8393127.
- 30.Wei G, Ma H, Qian W, Han F, Jiang H, Qi S, Qiu M. Lung nodule classification using local kernel regression models with out-of-sample extension. Biomed. Signal Process. Control. 2018;40:1–9. doi: 10.1016/j.bspc.2017.08.026. [DOI] [Google Scholar]
- 31.Bobadilla JCM, Pedrini H. In: Lung Nodule Classification Based on Deep Convolutional Neural Networks BT - Progress in Pattern Recognition, Image Analysis, Computer Vision, and Applications. Beltrán-Castañón C, Nyström I, Famili F, editors. Cham: Springer International Publishing; 2017. pp. 117–124. [Google Scholar]
- 32.Sun W, Zheng B, Qian W. Automatic feature learning using multichannel ROI based on deep structured algorithms for computerized lung cancer diagnosis. Comput Biol Med. 2017;89:530–539. doi: 10.1016/j.compbiomed.2017.04.006. [DOI] [PubMed] [Google Scholar]
- 33.Xie Y, Xia Y, Zhang J, Feng DD, Fulham M, Cai W. In: Transferable Multi-model Ensemble for Benign-Malignant Lung Nodule Classification on Chest CT BT - Medical Image Computing and Computer-Assisted Intervention − MICCAI 2017. Descoteaux M, Maier-Hein L, Franz A, Jannin P, Collins DL, Duchesne S, editors. Cham: Springer International Publishing; 2017. pp. 656–664. [Google Scholar]
- 34.Lückehe D, von Voigt G. Evolutionary image simplification for lung nodule classification with convolutional neural networks. Int. J. Comput. Assist. Radiol. Surg. 2018;13:1499–1513. doi: 10.1007/s11548-018-1794-7. [DOI] [PubMed] [Google Scholar]
- 35.Buty M, Xu Z, Gao M, Bagci U, Wu A, Mollura DJ. In: Characterization of Lung Nodule Malignancy Using Hybrid Shape and Appearance Features BT - Medical Image Computing and Computer-Assisted Intervention – MICCAI 2016. Ourselin S, Joskowicz L, Sabuncu MR, Unal G, Wells W, editors. Cham: Springer International Publishing; 2016. pp. 662–670. [Google Scholar]
- 36.Causey JL, Zhang J, Ma S, Jiang B, Qualls JA, Politte DG, Prior F, Zhang S, Huang X. Highly accurate model for prediction of lung nodule malignancy with CT scans. Sci Rep. 2018;8:9286. doi: 10.1038/s41598-018-27569-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu K, Kang G. Multiview convolutional neural networks for lung nodule classification. Int J Imaging Syst Technol. 2017;27:12–22. doi: 10.1002/ima.22206. [DOI] [Google Scholar]
- 38.H. Lee, H. Lee, M. Park, J. Kim, Contextual convolutional neural networks for lung nodule classification using Gaussian-weighted average image patches, in: Proc.SPIE, 2017. 10.1117/12.2253978.
- 39.W. Sun, B. Zheng, X. Huang, W. Qian, Balance the nodule shape and surroundings: a new multichannel image based convolutional neural network scheme on lung nodule diagnosis, in: Proc.SPIE, 2017. 10.1117/12.2251297.
- 40.Liu Y, Hao P, Zhang P, Xu X, Wu J, Chen W. Dense Convolutional Binary-Tree Networks for Lung Nodule Classification. IEEE Access. 2018;6:49080–49088. doi: 10.1109/ACCESS.2018.2865544. [DOI] [Google Scholar]
- 41.Yan X, Pang J, Qi H, Zhu Y, Bai C, Geng X, Liu M, Terzopoulos D, Ding X. In: Classification of Lung Nodule Malignancy Risk on Computed Tomography Images Using Convolutional Neural Network: A Comparison Between 2D and 3D Strategies BT - Computer Vision – ACCV 2016 Workshops. Chen C-S, Lu J, Ma K-K, editors. Cham: Springer International Publishing; 2017. pp. 91–101. [Google Scholar]
- 42.S. Shen, S.X. Han, D.R. Aberle, A.A.T. Bui, W. Hsu, An Interpretable Deep Hierarchical Semantic Convolutional Neural Network for Lung Nodule Malignancy Classification., CoRR. abs/1806.0 (2018). http://arxiv.org/abs/1806.00712. [DOI] [PMC free article] [PubMed]
- 43.X. Li, Y. Kao, W. Shen, X. Li, G. Xie, Lung nodule malignancy prediction using multi-task convolutional neural network, in: Proc.SPIE, 2017. 10.1117/12.2253836.
- 44.Ardila D, Kiraly AP, Bharadwaj S, Choi B, Reicher JJ, Peng L, Tse D, Etemadi M, Ye W, Corrado G, Naidich DP, Shetty S. End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nat. Med. 2019;25:954–961. doi: 10.1038/s41591-019-0447-x. [DOI] [PubMed] [Google Scholar]
- 45.Kang G, Liu K, Hou B, Zhang N. 3D multi-view convolutional neural networks for lung nodule classification. PLoS One. 2017;12:e0188290. doi: 10.1371/journal.pone.0188290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shen W, Zhou M, Yang F, Yang C, Tian J. Multi-scale Convolutional Neural Networks for Lung Nodule Classification. Inf Process Med Imaging. 2015;24:588–599. doi: 10.1007/978-3-319-19992-4_46. [DOI] [PubMed] [Google Scholar]
- 47.Shen W, Zhou M, Yang F, Yu D, Dong D, Yang C, Zang Y, Tian J. Multi-crop Convolutional Neural Networks for lung nodule malignancy suspiciousness classification. Pattern Recognit. 2017;61:663–673. doi: 10.1016/j.patcog.2016.05.029. [DOI] [Google Scholar]
- 48.Ioffe S, Szegedy C, Normalization B. Accelerating Deep Network Training by Reducing Internal Covariate Shift. BT - Proceedings of the 32nd International Conference on Machine Learning, ICML. Lille, France, 6-11 July. 2015;2015(2015):448–456. [Google Scholar]
- 49.K. He, X. Zhang, S. Ren, J. Sun, Delving Deep into Rectifiers: Surpassing Human-Level Performance on ImageNet Classification, in: 2015 IEEE Int. Conf. Comput. Vis., 2015: pp. 1026–1034. 10.1109/ICCV.2015.123.
- 50.H. Mzoughi, I. Njeh, M. Ben Slima, A. Ben Hamida, Histogram equalization-based techniques for contrast enhancement of MRI brain Glioma tumor images: Comparative study, in: 2018 4th Int. Conf. Adv. Technol. Signal Image Process., 2018: pp. 1–6. 10.1109/ATSIP.2018.8364471.
- 51.P. Amorim, T. Moraes, J. Silva, H. Pedrini, 3D Adaptive Histogram Equalization Method for Medical Volumes. BT - Proceedings of the 13th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2018) - Volume 4: VISAPP, Funchal, Madeira, Po, (2018) 363–370. 10.5220/0006615303630370.
- 52.S. Roy, P. Ghosh, S.K. Bandyopadhyay, Contour Extraction and Segmentation of Cerebral Hemorrhage from MRI of Brain by Gamma Transformation Approach BT - Proceedings of the 3rd International Conference on Frontiers of Intelligent Computing: Theory and Applications (FICTA) 2014, in: S.C. Satapathy, B.N. Biswal, S.K. Udgata, J.K. Mandal (Eds.), Springer International Publishing, Cham, 2015: pp. 383–394.
- 53.Zhan T, Gong M, Jiang X, Li S. Log-Based Transformation Feature Learning for Change Detection in Heterogeneous Images. IEEE Geosci Remote Sens Lett. 2018;15:1352–1356. doi: 10.1109/LGRS.2018.2843385. [DOI] [Google Scholar]
- 54.D.P. Kingma, J. Ba, Adam: A Method for Stochastic Optimization., CoRR. abs/1412.6 (2014). http://arxiv.org/abs/1412.6980.
- 55.R. Caruana, S. Lawrence, C.L. Giles, Overfitting in Neural Nets: Backpropagation, Conjugate Gradient, and Early Stopping. BT - Advances in Neural Information Processing Systems 13, Papers from Neural Information Processing Systems (NIPS) 2000, Denver, CO, USA, (2000) 402–408. http://papers.nips.cc/paper/1895-overfitting-in-neural-nets-backpropagation-conjugate-gradient-and-early-stopping.
- 56.Han F, Wang H, Zhang G, Han H, Song B, Li L, Moore W, Lu H, Zhao H, Liang Z. Texture Feature Analysis for Computer-Aided Diagnosis on Pulmonary Nodules. J. Digit. Imaging. 2015;28:99–115. doi: 10.1007/s10278-014-9718-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.F. Han, G. Zhang, H. Wang, B. Song, H. Lu, D. Zhao, H. Zhao, Z. Liang, A texture feature analysis for diagnosis of pulmonary nodules using LIDC-IDRI database, in: 2013 IEEE Int. Conf. Med. Imaging Phys. Eng., 2013: pp. 14–18. 10.1109/ICMIPE.2013.6864494.