Abstract
Approximately 30% of Major Depressive Disorder (MDD) patients develop treatment-resistant depression (TRD). Among the different causes that make TRD so challenging in both clinical and research contexts, major roles are played by the inadequate understanding of MDD pathophysiology and the limitations of current pharmacological treatments. Nevertheless, the field of psychiatry is facing exciting times. Combined with recent advances in genome editing techniques, human induced pluripotent stem cell (hiPSC) technology is offering novel and unique opportunities in both disease modelling and drug discovery. This technology has allowed innovative disease-relevant patient-specific in vitro models to be set up for many psychiatric disorders. Such models hold great potential in enhancing our understanding of MDD pathophysiology and overcoming many of the well-known practical limitations inherent to animal and post-mortem models.
Moreover, the field is approaching the advent of (es)ketamine, a glutamate N-methyl-d-aspartate (NMDA) receptor antagonist, claimed as one of the first and exemplary agents with rapid (in hours) antidepressant effects, even in TRD patients. Although ketamine seems poised to transform the treatment of depression, its exact mechanisms of action are still unclear but greatly demanded, as the resulting knowledge may provide a model to understand the mechanisms behind rapid-acting antidepressants, which may lead to the discovery of novel compounds for the treatment of depression. After reviewing insights into ketamine’s mechanisms of action (derived from preclinical animal studies) and depicting the current state of the art of hiPSC technology below, we will consider the implementation of an hiPSC technology-based TRD model for the study of ketamine’s fast acting antidepressant mechanisms of action.
Keywords: drug response, glutamate N-methyl-d-aspartate, in vitro model, induced pluripotent stem cells, ketamine, major depressive disorder, neurons, treatment-resistant depression
Introduction
Major depressive disorder (MDD), affecting roughly 300 million people (i.e. 4.5% of the global population), is a life-threatening mental illness, a leading cause of morbidity worldwide and a major medical and economic burden for society.1,2 Unsatisfactory response rates to currently approved antidepressant drugs, which alleviate symptoms in about half of treated patients, contribute to the enormous public health burden of MDD.3 Perhaps most critically, approximately 30% of MDD patients develop treatment-resistant depression (TRD).4 TRD is commonly defined as a difficult-to-treat condition characterised by failure to achieve a reduction in baseline symptomatology of at least 50%, as measured by widespread rating scales such as the Montgomery–Åsberg Depression Rating Scale, after at least two antidepressant treatment trials of adequate dosage and duration.5 In this condition, related sequelae include poor quality of life, loss of productivity, increased suicide risk and worse prognosis.6
Among the different causes that make TRD so challenging in both clinical and research contexts, major roles are played by the inadequate understanding of MDD pathophysiology and the limitations of current pharmacological treatments. The inadequate understanding of MDD pathophysiology reflects the more general lack of suitable scientific models for neuropsychiatric disorders. The development and use of animal models in psychiatry is challenging, due to the complex nature and heterogeneity of MDD. In this regard, according to the International Union of Basic and Clinical Pharmacology, currently available animal models have limited power and effectiveness in mimicking the whole MDD complexity and have not been subject to sequential application of different treatments – a key requisite to define TRD in clinical practice.7
Despite the overly simplistic role of the monoaminergic hypothesis of MDD, its current pharmacological treatment still relies on drugs that mainly target monoamine neurotransmitter systems and show delayed efficacy with lag times of several weeks to months before producing clinical improvement (if MDD patients show a response).
Nevertheless, the field of psychiatry is facing exciting times. Combined with recent advances in genome editing techniques, human induced pluripotent stem cell (hiPSC) technology is offering novel and unique opportunities in both disease modelling and drug discovery. This technology has allowed innovative disease-relevant patient-specific in vitro models to be set up for many psychiatric disorders.8 Such models hold great potential in enhancing our understanding of MDD pathophysiology and overcoming many of the well-known practical limitations inherent to animal and post-mortem models. Moreover, the field is facing the advent of (es)ketamine (the s-enantiomer of the racemic mixture is reported to be better tolerated),9 a glutamate N-methyl-d-aspartate (NMDA) receptor antagonist, claimed as one of the first and exemplary agents with rapid (in hours) antidepressant effects, even in TRD patients. Multiple controlled trials have demonstrated that 50–70% of TRD patients show clinical response after a single 40-min ketamine intravenous infusion.10 According to a recent trial,11 the optimal dose of intravenous ketamine must be administered in a specific subanaesthetic range comprised between 0.5 mg/kg and 1 mg/kg, with no clear or consistent evidence for clinically meaningful efficacy of lower doses. Although ketamine seems poised to transform the treatment of depression, its exact mechanisms of action are still unclear but their knowledge greatly demanded, as the resulting insight may provide a model to understand the mechanisms behind rapid-acting antidepressants, which may lead to the discovery of novel compounds for the treatment of depression.
After reviewing insights into ketamine’s mechanisms of action (derived from preclinical animal studies) and depicting the current state of the art of hiPSC technology below, we will consider the implementation of an hiPSC technology-based TRD model for the study of ketamine’s fast acting antidepressant mechanisms of action.
Ketamine’s mechanisms of action
Subanaesthetic doses of ketamine have shown promise for the rapid treatment of TRD patients.12–15 Over the last decade, a series of placebo-controlled studies confirmed the ability of intravenous ketamine (0.5 mg/kg infusion) to provide significant amelioration of symptoms in TRD patients within hours, although symptoms typically return just days after discontinuation of the acute intervention.14,15 As pointed out in a recent review,14 nine meta-analyses of acute-phase randomised short-term trials of ketamine for depression reported statistically significant advantages of ketamine over placebo or active control conditions across a variety of measures of depressive symptoms. These results stimulated basic and translational neuropsychiatric research. Nevertheless, the exact mechanism of action of ketamine is not yet clear.16 Preclinical animal studies suggest that the molecular mechanisms underlying ketamine’s effect are likely to be more complicated than NMDA-receptor blockade. Rapid responses in patients with TRD suggest fast changes in synaptic function and plasticity, processes thought to be deeply impaired in MDD and also closely correlated with the severity of depressive symptoms in humans.17
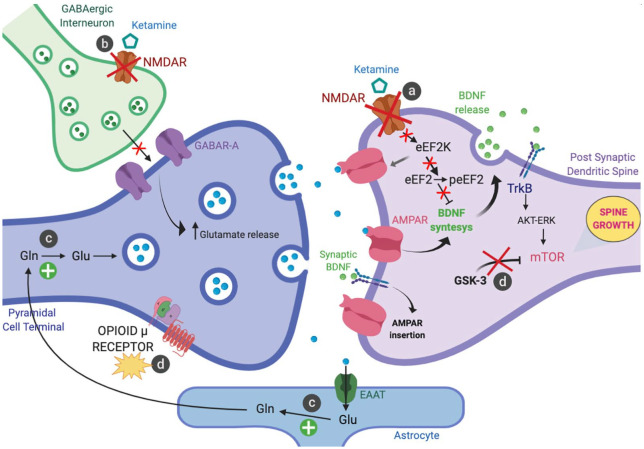
The latest emerging mechanistic hypotheses related to the antidepressant effects of ketamine are usually developed within a generic and prototypical glutamatergic synapse in the prefrontal cortex (PFC) (Figure 1).18
Figure 1.
Emerging mechanistic hypotheses related to the antidepressant effects of ketamine.
(a) Antagonism at postsynaptic NMDA receptors; (b) Blockade of NMDA receptors on GABAergic interneurons; (c) Increase of glutamate-glutamine cycling; and (d) Other hypothesised mechanism: opioid system activation and inhibition of GSK-3.
On this level, the specific effects of ketamine can be distinguished as follows:
– Antagonism at postsynaptic GluN2B-containing NMDA receptors prevents the eEF2K mediated phosphorylation and inhibition of eEF2 (which, when phosphorylated, usually reduces BDNF dendritic mRNA translation) leading to an increase of synaptic BDNF levels and shuttling of AMPA glutamate receptors to the synapse, thus enhancing synaptic efficacy (Figure 1a).19,20
– Preferential blockade of NMDA receptors on a subpopulation of GABAergic interneurons induces disinhibition of pyramidal cells leading to a glutamate surge and enhanced activation of AMPA receptors (Figure 1b).21,22 AMPA receptor activation promotes a signalling cascade that raises BDNF levels.23 Local release of BDNF is thought to stimulate TrkB receptors stimulating mTORC1 pathway via the activation of Akt and ERK signalling,23,24 leading to an increase of synaptic number and function in the PFC.25,26–29 In turn, this step activates the local protein synthesis necessary for increasing dendritic spine formation and restoring synaptic connectivity.26–29 These events share the temporal profile with antidepressant effects of ketamine.
– Ketamine and other NMDA antagonists also target metabolic pathways in neurons and glial cells by increasing glutamate-glutamine cycling and, consequently, extracellular glutamate levels in PFC, both in animal models and human patients (Figure 1c).30,31
– In animal models, ketamine has recently been shown to inhibit GSK-3 by its phosphorylation,32 linking this effect, again, to rapid activation of the mTOR signalling pathway and the related cascade of events, including increased dendritic spine density/diameter and synaptic strength in the medial PFC layer 5 pyramidal neurons with antidepressant behavioural responses persisting for up to 1 week (Figure 1d).33 In addition to the NMDA receptor antagonism and the related cellular events discussed above, as recent ground-breaking research has pointed out,34 ketamine’s acute antidepressant effect requires opioid system activation. Results from this randomised double-blind crossover trial demonstrate that concurrent administration of Naltrexone, a selective opioid receptor antagonist (mainly on µ-opioid receptors), with ketamine treatment in TRD-patients is able to attenuate, or block, the acute antidepressant effect.
As underlined above, most of our insights into the mechanisms associated with MDD endophenotypes and ketamine’s antidepressant efficacy derive from animal models of depression. Limitations and translational issues of this animal modelling of an exquisitely human mental illness such as MDD have been debated for years.35
hiPSC technology: the state of the art
Until a few decades ago, irreversibility of the developmental path that leads from pluripotent cells to progressively mature cells was considered a dogma of biology, implying that a differentiated cell could no longer regress to a primordial state of pluripotency (i.e. the capacity to generate each of the discrete somatic cells types present in the body). However, in 2006, Shinya Yamanaka reported a sensational discovery that subverted this vision and led to the definition of a ‘cellular reprogramming’ process by which functionally specialised cells, such as skin cells, can be converted into pluripotent cells through the transient expression of a cocktail of four genes (called Oct-3/4, Sox2, c-Myc, and Klf4).36,37 These cells, artificially created in the laboratory setting, are pluripotent and have been named ‘induced pluripotent stem cells’ (iPSCs). They offer the pluripotency of embryonic stem cells without the intrinsic ethical implications associated with the sacrifice of embryos, with the advantage of being potentially obtained from any healthy or sick individual. Since 2006, reprogramming technology has been increasingly improved, both in terms of efficiency and quality of the cells obtained. Today, there are thousands of iPSC lines derived from different donor cells (skin cells, blood cells, even from ‘waste’ cells in urine) and numerous individuals.38 Initially, reprogramming was delivered into cells using modified viral particles that, however, had the defect of integrating into the genome of the recipient cells, with the risk of unpredictably altering the functioning of potentially risky genes. Due to this technical constraint, the first iPSCs obtained showed tumour-like features that precluded the potential use of their cellular derivatives in the clinical setting. Furthermore, efficiency was very limited, with less than one cell in 100,000 actually reprogrammed.36,37 However, in recent years, these limitations have been progressively overcome and footprint-free (i.e. without genome alterations caused by reprogramming) as well as clinical grade (i.e. suitable for applications in the clinical setting) iPSCs can be efficiently produced, thanks to viral particles that do not integrate in the genome or other non viral-based delivery methods.39 Although the molecular mechanisms responsible for the reprogramming process are not yet fully known, the discovery of iPSCs has revolutionised the work of many scientists and profoundly changed the concepts of cell identity and functioning, opening up new and exciting implications in the field of epigenetics and tremendous opportunities in the field of biomedical research.39
In recent years, the reprogramming technique has provided researchers with an excellent platform to generate iPSCs from which to produce and study specialised sick cells with the same genetic material as patients.40 A fundamental step for the development of effective therapies for a given pathology is to have ‘disease models’ that allow disease-specific cellular dysfunctions to be precisely understood. The most reliable approach to do so is the study of live tissue-derived cells of patients affected with the disease. However, this is not always feasible, especially for tissues that cannot be easily accessed, such as cerebral neurons. The reprogramming technique now allows scientists to overcome this limitation by accessing a large number of putatively ‘sick’ neurons through the differentiation of neurons on which to perform assays and dissect mechanisms from patient-specific iPSCs.41–43 These neurons have the same genetic background as the cells of the originating patients. Besides their use to study mechanisms underlying the onset and progression of specific diseases, iPSC-based disease models also play an important role in drug discovery processes.44 To date, large numbers of iPSC lines have been obtained from cells of normal individuals or patients with specific diseases, both monogenic and multifactorial, such as muscular dystrophies, Parkinson’s disease, Huntington’s disease, Down’s syndrome, juvenile diabetes mellitus, heart disease and many others.45
Much of the public interest in iPSCs stems from their immense potential in the field of regenerative medicine.46 This concerns the possibility of converting iPSCs into several cell types exploitable for cell-based therapies aimed at the regeneration of tissues or organs with poor intrinsic reparative capacity (such as the brain, pancreas or heart), damaged by disease or trauma. Among other things, patient-specific iPSCs have the potential advantage of being exploitable for autologous transplants, thereby excluding rejection problems.45 Thus, theoretically, clinical reprogramming technology would allow the development of customised cellular therapies based on the transplant of specialised cells of interest obtained from patient-specific iPSCs. Furthermore, recent developments in the field of genomic editing would allow combined approaches of personalised gene and cell replacement therapy.47 For example, in the case of a patient suffering from an inherited disease, the genetic defect could be corrected in the laboratory in his own iPSCs, which would then be used to produce the healthy specialised cells of interest for transplant. Over the past 10 years, all of these approaches have been successfully tested in the treatment of sickle cell disease, Parkinson’s disease, heart injuries and other pathological conditions, opening way to the first clinical trials in macular degeneration and other studies currently on the way for the treatment of Parkinson’s disease and heart failure.47 There are high expectations from these studies, which are fundamental to assess clinical benefits, identify potential critical issues, optimise these therapeutic approaches in humans and draw regenerative medicine a step closer to reality.
Towards a hiPSC-based TRD model for the study of ketamine’s fast-acting antidepressant mechanisms of action
In recent years, several iPSCs lines have been derived from fibroblasts and peripheral blood mononuclear cells of patients affected by different neuropsychiatric disorders including schizophrenia, autism spectrum disorders, MDD and bipolar spectrum disorders.41,44–45,48,49
Cellular reprogramming via hiPSC technology offers a unique opportunity to generate neural cells directly from subsets of psychiatric patients with specific clinical characteristics,36 enabling the study of cellular and molecular aspects of neurotransmission and treatment response on a previously inaccessible level.8 Indeed, psychiatric disorders such as schizophrenia, bipolar disorder, and autism spectrum disorders have already been studied successfully using patient-derived neurons.50–54
MDD, perhaps due to the current diagnostic definition leading to clinical and genetic heterogeneity,55 is more challenging to study as a biologically distinct phenomenon. The weight of environmental risk factors such as childhood abuse or socioeconomic adversity also contributes to this complexity and a recent re-examination of polymorphisms, polymorphism-by-environment interactions and gene-level effects failed to confirm the relevance of the 18 most studied candidate genes to depression phenotypes.56 However, psychiatric disorders are known to share common genetic risk variants that were recently confirmed by assessing their relationship to 17 phenotypes from 1,191,588 individuals pooled from large-scale genome-wide association studies.57
In order to identify MDD subtype-associated cellular and molecular phenotypes, the only two available hiPSC-based studies on MDD are from the same research group and focused on TRD, stratifying patients on the basis of SSRI-response, a clear biological phenotype.58,59 In the first study,58 Vadodaria and colleagues generated iPSCs from 3 SSRI-remitters (R) and 3 SSRI-non-remitter (NR) MDD patients and studied serotonergic neurotransmission in patient’s forebrain neurons in vitro. They observed that NR patient-derived neurons surprisingly displayed serotonin-induced hyperactivity downstream of upregulated excitatory serotonergic receptors (5-HT2A and 5-HT7) in contrast to what was seen in healthy and R patient-derived neurons. Pharmacological blockade of these receptors by Lurasidone (a high affinity 5-HT2A and 5-HT7 antagonist recently approved for bipolar depression) partially rescued 5-HT-induced hyperactivity in NR patient-derived neurons. In the second study,59 NR patient-derived serotonergic neurons exhibited altered neurite growth and morphology downstream of lowered expression of key protocadherin alpha genes as compared with healthy controls and R. Furthermore, knockdown of protocadherin alpha genes directly regulated iPSC-derived neurite length and morphology, suggesting intrinsic differences in serotonergic neuron morphology and resulting circuitry of TRD patients.
Although promising, these findings from one research team should be considered preliminary. Stem cell technology is still considered a low throughput method that can only be applied to few samples. Both studies employed neuronal cell lines from the same six female patients. Given the broad diversity of clinical MDD phenotypes, the neuronal populations examined probably explain a minimal portion of the complex neuronal network involved in the pathogenesis of the disorder and in treatment response.
Only one research group has recently conducted three hiPSC-based technology studies on ketamine’s antidepressant mechanisms of action, proposing a dopaminergic neuron model derived from one healthy person. In the first study,60 authors confirmed the leading hypothesis that ketamine is also able to enhance structural plasticity via AMPA receptor-driven BDNF and mTOR signalling in their hiPSC-derived dopaminergic neurons.24 In the second study,61 they demonstrated that a ketamine metabolite (2R,6R-hydroxynorketamine) also produced similar effects in the same model. In the last study,62 ketamine was shown to increase the expression of AMPA receptor -GluR1 and -GluR2 subunits, suggesting their involvement in driving structural plasticity in human dopaminergic neurons depending on ketamine transient exposure.
To unravel the ketamine’s fast acting antidepressant mechanisms of action, an optimisation of the disease-model is necessary by adopting a new and completely translational approach. First, it is important to focus on precise clinical stratification and characterisation of patients’ resistance profile based on their treatment history. Because only one every six intervention studies enrol patients that meet these criteria, information on this population can be considered sparse.63 True resistance to the effect of specific drugs might only play a minor part,64 so deep phenotyping must include multiple strategies to address the persistence of depression in individual subjects when small-sample protocols are implemented. Within stratified samples of TRD patients, exceptional ketamine responders (eK-Rs) and ketamine non-responders (K-NRs) must be identified. Second, once hiPSCs have been obtained from these selected cohorts, the cell subtypes to derive should be planned carefully. Pioneer hiPSC-studies in the neuropsychiatric field adopted a mainly single cell modelling approach. As shown in Figure 1, ketamine’s efficacy seems to involve a system consisting of different cell types and mechanisms, finely inter-dependent and inter-regulated. A modern disease-modelling should take this complexity into account and switch from single type cell model perspective to that of co-cultured multiple cell types.
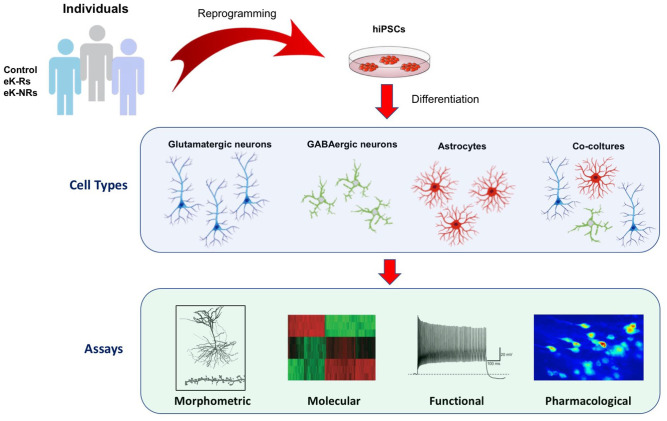
Our and other groups have been focussing on optimising single type differentiation protocols and co-culturing techniques.65,66 The technology can now be considered mature enough to reassemble the patient derived-glutamatergic synapse depicted in Figure 1 along with all forebrain neurons (glutamatergic cells, GABAergic interneurons) and glial cells (astrocytes) in vitro, in a culture plate. These populations can be experimentally interrogated at multiple levels to highlight possible phenotypic alterations specifically segregating with eK-Rs and K-NRs experimental groups (Figure 2).
Figure 2.
The proposed hiPSC-based model for studying the molecular basis of ketamine response. Patients can be easily characterised and stratified according to clinical characteristics and treatment response profiles. PBMC isolated from drawn blood samples can be reprogrammed to hiPSC and then differentiated to disease-specific cell type according to the hypothesised disease model. Different assays can be identified and used to study and test the model.
hiPSC, human induced pluripotent stem cell; PBMC, peripheral blood mononuclear cells.
Mature neuronal populations can be investigated at morphological level (area of the somata, level of branching, interconnectivity, length of neurites of differentiated neurons, etc.), antigenic level (expression of mature markers for specific neuronal subtypes, etc.), functional level (establishment of functional neuronal networks capable of spontaneous activity and stimulus-evoked responses by means of high-density multi-electrode array-based approaches) and molecular level. As noted before, most of the core neurobiological insights about ketamine’s mechanisms of action come from preclinical animal studies, which underscored the activation of AMPA receptors, BDNF-TrkB, eEF2K/eEF2 and mTOR signalling pathways.16,18,60,61 Yet recent findings suggest that ketamine’s acute antidepressant effect requires opioid system activation and, therefore, may not be directly ascribed to NMDA antagonism.34 The proposed model offers multiple options to dissect different ketamine molecular pathways and mechanisms of action. Certainly, it is possible to use traditional sets of specific inhibitors (AMPA receptor antagonists, BDNF-TrkB or TrkB/MEK signalling inhibitors, etc.) in combination with ketamine treatment to determine if specific pathway inhibitors are able to block the dendritic and synaptic changes induced by ketamine, identified in previous steps. Moreover, the same goal can be pursued by performing a transcriptomic profiling and accurate bioinformatics analysis of hiPSC-derived patient-specific samples to delineate ketamine related alterations in networks of transcriptional activity and identify the co-expression of genes responsible for the response/non-response mechanisms.67
If applied to the proposed hiPSC-based model, all these techniques could offer unique contributions to the unveiling of molecular basis of ketamine response, and they might help to develop new and personalised treatments, more individually tailored and less hazardous.
Footnotes
Conflict of interest statement: The authors declare that there is no conflict of interest.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work has been supported by a grant (n° 2019-3396 to C.S., A.D. and L.C.) from the Italian Cariplo Foundation, which had not any involvement in manuscript preparation, or decision to submit the article for publication.
ORCID iD: Matteo Marcatili  https://orcid.org/0000-0002-1436-1961
https://orcid.org/0000-0002-1436-1961
Contributor Information
Matteo Marcatili, Psychiatric Department, San Gerardo Hospital, ASST Monza, Monza, Italy.
Carlo Sala, National Research Council Neuroscience Institute, Milan, Italy; Department of Medical Biotechnology and Translational Medicine, Università degli Studi di Milano, Milan, Italy; NeuroMI Milan Center for Neuroscience, University of Milano-Bicocca, Italy.
Antonios Dakanalis, Department of Medicine and Surgery, University of Milano Bicocca, Monza, Italy.
Fabrizia Colmegna, Psychiatric Department, San Gerardo Hospital, ASST Monza, Monza, Italy.
Armando D’Agostino, Department of Health Sciences, Università degli Studi di Milano, Milan, Italy.
Orsola Gambini, Department of Health Sciences, Università degli Studi di Milano, Milan, Italy; CRC “Aldo Ravelli” for Neurotechnology and Experimental Brain Therapeutics, University of Milan, Milan, Italy.
Bernardo Dell’Osso, Psychiatry Unit, Department of Biomedical and Clinical Sciences “Luigi Sacco”, University of Milan, Milan, Italy; CRC “Aldo Ravelli” for Neurotechnology and Experimental Brain Therapeutics, University of Milan, Milan, Italy; Department of Psychiatry and Behavioural Sciences, Bipolar Disorders Clinic, Stanford University, CA, USA.
Beatrice Benatti, Psychiatry Unit, Department of Biomedical and Clinical Sciences “Luigi Sacco”, University of Milan, Milan, Italy; CRC “Aldo Ravelli” for Neurotechnology and Experimental Brain Therapeutics, University of Milan, Milan, Italy.
Luciano Conti, Laboratory of Stem Cell Biology, Department of Cellular, Computational and Integrative Biology (CIBIO), Università degli Studi di Trento, Trento, Italy.
Massimo Clerici, Psychiatric Department, San Gerardo Hospital, ASST Monza, Monza, Italy; Department of Medicine and Surgery, University of Milano Bicocca, Monza, Italy.
References
- 1. World Health Organization. Global health estimates 2018: disease burden by cause, sex, by country and region, 2000-2016. Geneva: World Health Organization, 2018. [Google Scholar]
- 2. Vos T, Flaxman AD, Naghavi M, et al. Years lived with disability (YLDs) for 1160 sequelae of 289 diseases and injuries 1990-2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 2012; 380: 2163–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Undurraga J, Baldessarini RJ. Randomized, placebo-controlled trials of antidepressants for acute major depression: thirty-year meta-analytic review. Neuropsychopharmacology. Epub ahead of print 14 December 2011. DOI: 10.1038/npp.2011.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rush AJ, Trivedi MH, Wisniewski SR, et al. Acute and longer-term outcomes in depressed outpatients requiring one or several treatment steps: a STAR*D report. Am J Psychiatry 2006; 163: 1905–1917. [DOI] [PubMed] [Google Scholar]
- 5. Malhi GS, Byrow Y. Is treatment-resistant depression a useful concept? Evid Based Ment Health. Epub ahead of print 14 January 2016. DOI: 10.1136/eb-2015-102299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mrazek DA, Hornberger JC, Altar CA, et al. A review of the clinical, economic, and societal burden of treatment-resistant depression: 1996-2013. Psychiatr Serv 2014; 65: 977–987. [DOI] [PubMed] [Google Scholar]
- 7. Drago F, Calabrese F, Leggio GM, et al. International union of basic and clinical pharmacology CIV: the neurobiology of treatment-resistant depression: from antidepressant classifications to novel pharmacological targets. Pharmacol Rev 2018; 70: 475–504. [DOI] [PubMed] [Google Scholar]
- 8. Soliman MA, Aboharb F, Zeltner N, et al. Pluripotent stem cells in neuropsychiatric disorders. Mol Psychiatry 2017; 22: 1241–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Correia-Melo FS, Leal GC, Carvalho MS, et al. Comparative study of esketamine and racemic ketamine in treatment-resistant depression: protocol for a non-inferiority clinical trial. Medicine (Baltimore) 2018; 97: e12414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Newport DJ, Carpenter LL, McDonald WM, et al. Ketamine and other NMDA antagonists: early clinical trials and possible mechanisms in depression. Am J Psychiatry 2015; 172: 950–966. [DOI] [PubMed] [Google Scholar]
- 11. Fava M, Freeman MP, Flynn M, et al. Double-blind, placebo-controlled, dose-ranging trial of intravenous ketamine as adjunctive therapy in treatment-resistant depression (TRD). Mol Psychiatry. Epub ahead of print 3 October 2018. DOI: 10.1038/s41380-018-0256-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Berman RM, Cappiello A, Anand A, et al. Antidepressant effects of ketamine in depressed patients. Biol Psychiatry 2000; 47: 351–354. [DOI] [PubMed] [Google Scholar]
- 13. Zarate CA, Jr, Singh JB, Carlson PJ, et al. A randomized trial of an N-methyl-d-aspartate antagonist in treatment-resistant major depression. Arch Gen Psychiatry 2006; 63: 856–864. [DOI] [PubMed] [Google Scholar]
- 14. Bobo WV, Voort JLV, Croarkin PE, et al. Ketamine for treatment-resistant unipolar and bipolar major depression: critical review and implications for clinical practice. Depress Anxiety. Epub ahead of print 6 April 2016. DOI: 10.1002/da.22505. [DOI] [PubMed] [Google Scholar]
- 15. Loo CK, Gálvez V, O’Keefe E, et al. Placebo-controlled pilot trial testing dose titration and intravenous, intramuscular and subcutaneous routes for ketamine in depression. Acta Psychiatr Scand. Epub ahead of print 30 March 2016. DOI: 10.1111/acps.12572. [DOI] [PubMed] [Google Scholar]
- 16. Wohleb ES, Gerhard D, Thomas A, et al. Molecular and cellular mechanisms of rapid-acting antidepressants ketamine and scopolamine. Curr Neuropharmacol 2016; 15: 11–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Holmes SE, Scheinost D, Finnema SJ, et al. Lower synaptic density is associated with depression severity and network alterations. Nat Commun 2019; 10: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Krystal JH, Abdallah CG, Sanacora G, et al. Ketamine: a paradigm shift for depression research and treatment. Neuron 2019; 101: 774–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Monteggia LM, Gideons E, Kavalali ET. The role of eukaryotic elongation factor 2 kinase in rapid antidepressant action of ketamine. Biol Psychiatry 2013; 73: 1199–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Heise C, Gardoni F, Culotta L, et al. Elongation factor-2 phosphorylation in dendrites and the regulation of dendritic mRNA translation in neurons. Front Cell Neurosci 2014; 8: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Moghaddam B, Javitt D. From revolution to evolution: the glutamate hypothesis of schizophrenia and its implication for treatment. Neuropsychopharmacology 2011; 37: 4–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Moghaddam B, Adams B, Verma A, et al. Activation of glutamatergic neurotransmission by ketamine: a novel step in the pathway from NMDA receptor blockade to dopaminergic and cognitive disruptions associated with the prefrontal cortex. J Neurosci 1997; 17: 2921–2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jourdi H, Hsu YT, Zhou M, et al. Positive AMPA receptor modulation rapidly stimulates BDNF release and increases dendritic mRNA translation. J Neurosci 2009; 29: 8688–8697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Takei N, Inamura N, Kawamura M, et al. Brain-derived neurotrophic factor induces mammalian target of rapamycin-dependent local activation of translation machinery and protein synthesis in neuronal dendrites. J Neurosci 2004; 24: 9760–9769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Duncan GE, Moy SS, Knapp DJ, et al. Metabolic mapping of the rat brain after subanesthetic doses of ketamine: potential relevance to schizophrenia. Brain Res 1998; 787: 181–190. [DOI] [PubMed] [Google Scholar]
- 26. Li N, Lee B, Liu R-J, et al. MTOR-dependent synapse formation underlies the rapid antidepressant effects of NMDA antagonists. Science 2010; 329: 959–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Miller OH, Yang L, Wang C-C, et al. GluN2B-containing NMDA receptors regulate depression-like behavior and are critical for the rapid antidepressant actions of ketamine. Elife 2014; 3: e03581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Paul RK, Singh NS, Khadeer M, et al. (R,S)-Ketamine metabolites (R,S)-norketamine and (2S,6S)-hydroxynorketamine increase the mammalian target of rapamycin function. Anesthesiology 2014; 121: 149–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhou W, Wang N, Yang C, et al. Ketamine-induced antidepressant effects are associated with AMPA receptors-mediated upregulation of mTOR and BDNF in rat hippocampus and prefrontal cortex. Eur Psychiatry. Epub ahead of print 8 December 2014. DOI: 10.1016/j.eurpsy.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 30. Chowdhury GMI, Zhang J, Thomas M, et al. Transiently increased glutamate cycling in rat PFC is associated with rapid onset of antidepressant-like effects. Mol Psychiatry 2017; 22: 120–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Abdallah CG, De Feyter HM, Averill LA, et al. The effects of ketamine on prefrontal glutamate neurotransmission in healthy and depressed subjects. Neuropsychopharmacology 2018; 43: 2154–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Beurel E, Song L, Jope RS. Inhibition of glycogen synthase kinase-3 is necessary for the rapid antidepressant effect of ketamine in mice. Mol Psychiatry. Epub ahead of print 19 April 2011. DOI: 10.1038/mp.2011.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu R-J, Fuchikami M, Dwyer JM, et al. GSK-3 inhibition potentiates the synaptogenic and antidepressant-like effects of subthreshold doses of ketamine. Neuropsychopharmacology. Epub ahead of print 17 May 2013. DOI: 10.1038/npp.2013.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Williams NR, Heifets BD, Blasey C, et al. Attenuation of antidepressant effects of ketamine by opioid receptor antagonism. Am J Psychiatry 2018; 175: 1205–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nestler EJ, Hyman SE. Animal models of neuropsychiatric disorders. Nat Neurosci. Epub ahead of print 27 September 2010. DOI: 10.1038/nn.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006; 126: 663–676. [DOI] [PubMed] [Google Scholar]
- 37. Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007; 131: 861–872. [DOI] [PubMed] [Google Scholar]
- 38. Takahashi K, Yamanaka S. A decade of transcription factor-mediated reprogramming to pluripotency. Nat Rev Mol Cell Biol. Epub ahead of print 17 February 2016. DOI: 10.1038/nrm.2016.8. [DOI] [PubMed] [Google Scholar]
- 39. Li M, Izpisua Belmonte JC. Deconstructing the pluripotency gene regulatory network. Nat Cell Biol. Epub ahead of print 28 March 2018. DOI: 10.1038/s41556-018-0067-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hamazaki T, El Rouby N, Fredette NC, et al. Concise review: induced pluripotent stem cell research in the era of precision medicine. Stem Cells. Epub ahead of print 5 February 2017. DOI: 10.1002/stem.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Corti S, Faravelli I, Cardano M, et al. Human pluripotent stem cells as tools for neurodegenerative and neurodevelopmental disease modeling and drug discovery. Expert Opin Drug Discov 2015; 10: 615–629. [DOI] [PubMed] [Google Scholar]
- 42. Ardhanareeswaran K, Mariani J, Coppola G, et al. Human induced pluripotent stem cells for modelling neurodevelopmental disorders. Nat Rev Neurol. Epub ahead of print 18 April 2017. DOI: 10.1038/nrneurol.2017.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang M, Zhang L, Gage FH. Modeling neuropsychiatric disorders using human induced pluripotent stem cells. Protein Cell. Epub ahead of print 27 May 2019. DOI: 10.1007/s13238-019-0638-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rowe RG, Daley GQ. Induced pluripotent stem cells in disease modelling and drug discovery. Nat Rev Genet 2019; 20: 377–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Huang C-Y, Liu C-L, Ting C-Y, et al. Human iPSC banking: barriers and opportunities. J Biomed Sci 2019; 26: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Garreta E, Sanchez S, Lajara J, et al. Roadblocks in the path of iPSC to the clinic. Curr Transplant Rep. Epub ahead of print 8 February 2018. DOI: 10.1007/s40472-018-0177-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ortuño-Costela MDC, Cerrada V, García-López M, et al. The challenge of bringing iPSCs to the patient. Int J Mol Sci 2019; 20: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Marcatili M, Marsoner F, Karnavas T, et al. Establishment of an induced pluripotent stem cell (iPSC) line from a patient with clozapine-responder schizophrenia. Stem Cell Res 2016; 17: 630–633. [DOI] [PubMed] [Google Scholar]
- 49. Marsoner F, Marcatili M, Karnavas T, et al. Generation and characterization of an induced pluripotent stem cell (iPSC) line from a patient with clozapine-resistant schizophrenia. Stem Cell Res 2016; 17: 1–4. [DOI] [PubMed] [Google Scholar]
- 50. Brennand KJ, Simone A, Jou J, et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature 2011; 473: 221–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mertens J, Wang Q-W, Kim Y, et al. Differential responses to lithium in hyperexcitable neurons from patients with bipolar disorder. Nature 2015; 527: 95–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Marchetto MC, Belinson H, Tian Y, et al. Altered proliferation and networks in neural cells derived from idiopathic autistic individuals. Mol Psychiatry. Epub ahead of print 5 July 2017. DOI: 10.1038/mp.2016.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yoon K-J, Nguyen HN, Ursini G, et al. Modeling a genetic risk for schizophrenia in iPSCs and mice reveals neural stem cell deficits associated with adherens junctions and polarity. Cell Stem Cell 2014; 15: 79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Madison JM, Zhou F, Nigam A, et al. Characterization of bipolar disorder patient-specific induced pluripotent stem cells from a family reveals neurodevelopmental and mRNA expression abnormalities. Mol Psychiatry. Epub ahead of print 3 March 2015. DOI: 10.1038/mp.2015.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wray NR, Ripke S, Mattheisen M, et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet. Epub ahead of print 26 April 2018. DOI: 10.1038/s41588-018-0090-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Border R, Johnson EC, Evans LM, et al. No support for historical candidate gene or candidate gene-by-interaction hypotheses for major depression across multiple large samples. Am J Psychiatry 2019; 176: 376–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Anttila V, Bulik-Sullivan B, Finucane HK, et al. Analysis of shared heritability in common disorders of the brain. Science 2018; 360: eaap8757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Vadodaria KC, Ji Y, Skime M, et al. Serotonin-induced hyperactivity in SSRI-resistant major depressive disorder patient-derived neurons. Mol Psychiatry 2019; 24: 795–807. [DOI] [PubMed] [Google Scholar]
- 59. Vadodaria KC, Ji Y, Skime M, et al. Altered serotonergic circuitry in SSRI-resistant major depressive disorder patient-derived neurons. Mol Psychiatry 2019; 24: 808–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Cavalleri L, Merlo Pich E, Millan MJ, et al. Ketamine enhances structural plasticity in mouse mesencephalic and human iPSC-derived dopaminergic neurons via AMPAR-driven BDNF and mTOR signaling. Mol Psychiatry 2018; 23: 812–823. [DOI] [PubMed] [Google Scholar]
- 61. Collo G, Cavalleri L, Chiamulera C, et al. (2R,6R)-Hydroxynorketamine promotes dendrite outgrowth in human inducible pluripotent stem cell-derived neurons through AMPA receptor with timing and exposure compatible with ketamine infusion pharmacokinetics in humans. Neuroreport 2018; 29: 1425–1430. [DOI] [PubMed] [Google Scholar]
- 62. Collo G, Cavalleri L, Chiamulera C, et al. Ketamine increases the expression of GluR1 and GluR2 α-amino-3-hydroxy-5-methy-4-isoxazole propionate receptor subunits in human dopaminergic neurons differentiated from induced pluripotent stem cells. Neuroreport 2019; 30: 207–212. [DOI] [PubMed] [Google Scholar]
- 63. Gaynes BN, Lux L, Gartlehner G, et al. Defining treatment-resistant depression. Depress Anxiety. Epub ahead of print 22 October 2020. DOI: 10.1002/da.22968. [DOI] [PubMed] [Google Scholar]
- 64. Anderson IM. We all know what we mean by treatment-resistant depression – don’t we? Br J Psychiatry 2018; 212: 259–261. [DOI] [PubMed] [Google Scholar]
- 65. McComish SF, Caldwell MA. Generation of defined neural populations from pluripotent stem cells. Philos Trans R Soc Lond B Biol Sci 2018; 373: 20170214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Tukker AM, Wijnolts FMJ, de Groot A, et al. Human iPSC-derived neuronal models for in vitro neurotoxicity assessment. Neurotoxicology. Epub ahead of print 19 June 2018. DOI: 10.1016/j.neuro.2018.06.007. [DOI] [PubMed] [Google Scholar]
- 67. Lin M, Lachman HM, Zheng D. Transcriptomics analysis of iPSC-derived neurons and modeling of neuropsychiatric disorders. Mol Cell Neurosci. Epub ahead of print 26 November 2016. DOI: 10.1016/j.mcn.2015.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]