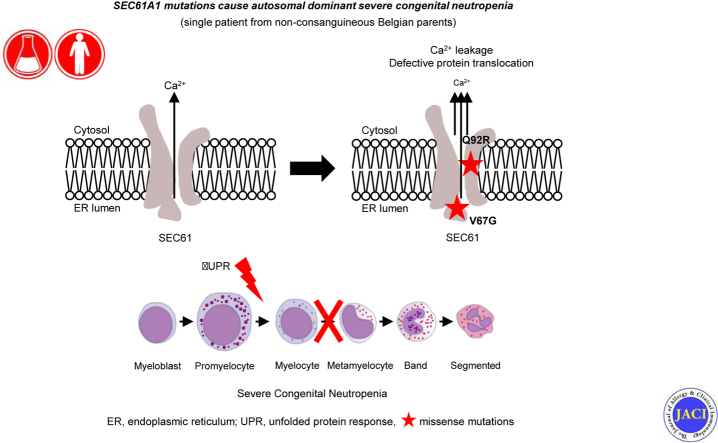
Abstract
Background
The molecular cause of severe congenital neutropenia (SCN) is unknown in 30% to 50% of patients. SEC61A1 encodes the α-subunit of the Sec61 complex, which governs endoplasmic reticulum protein transport and passive calcium leakage. Recently, mutations in SEC61A1 were reported to be pathogenic in common variable immunodeficiency and glomerulocystic kidney disease.
Objective
Our aim was to expand the spectrum of SEC61A1-mediated disease to include autosomal dominant SCN.
Methods
Whole exome sequencing findings were validated, and reported mutations were compared by Western blotting, Ca2+ flux assays, differentiation of transduced HL-60 cells, in vitro differentiation of primary CD34 cells, quantitative PCR for unfolded protein response (UPR) genes, and single-cell RNA sequencing on whole bone marrow.
Results
We identified a novel de novo missense mutation in SEC61A1 (c.A275G;p.Q92R) in a patient with SCN who was born to nonconsanguineous Belgian parents. The mutation results in diminished protein expression, disturbed protein translocation, and an increase in calcium leakage from the endoplasmic reticulum. In vitro differentiation of CD34+ cells recapitulated the patient’s clinical arrest in granulopoiesis. The impact of Q92R-Sec61α1 on neutrophil maturation was validated by using HL-60 cells, in which transduction reduced differentiation into CD11b+CD16+ cells. A potential mechanism for this defect is the uncontrolled initiation of the unfolded protein stress response, with single-cell analysis of primary bone marrow revealing perturbed UPR in myeloid precursors and in vitro differentiation of primary CD34+ cells revealing upregulation of CCAAT/enhancer-binding protein homologous protein and immunoglobulin heavy chain binding protein UPR-response genes.
Conclusion
Specific mutations in SEC61A1 cause SCN through dysregulation of the UPR.
Key words: Severe congenital neutropenia, SEC61A1, endoplasmic reticulum stress, unfolded protein response, whole exome sequencing
Abbreviations used: ADTKD, Autosomal dominant tubulointerstitial kidney disease; BiP, Immunoglobulin heavy chain binding protein; CHOP, CCAAT/enhancer-binding protein homologous protein; CVID, Common variable immune deficiency; DMSO, Dimethyl sulfoxide; ER, Endoplasmic reticulum; G-CSF, Granulocyte colony-stimulating factor; NE, Neutrophil elastase; qPCR, Quantitative polymerase chain reaction; SCN, Severe congenital neutropenia; TM, Transmembrane helix; UPR, Unfolded protein response; WT, Wild-type
Graphical abstract
Severe congenital neutropenias (SCNs) comprise a genetically heterogeneous group of inborn errors of immunity that are characterized by a differentiation arrest of granulopoiesis at the promyelocyte stage. From early childhood, the low absolute number of circulating mature neutrophils predisposes patients with SCN to life-threatening and recurrent infections, including otitis, skin infections, deep abscesses, gingivitis, and septicemia. However, syndromic forms of SCN with extrahematopoietic manifestations may also occur. Furthermore, patients are at increased risk for the development of myelodysplastic syndromes or acute myeloid leukaemia.1,2 Autosomal dominant mutations in ELANE, encoding for neutrophil elastase (NE) and autosomal recessive mutations in HAX1, are the most common cause of SCN,3 with mutations in 22 additional genes accounting for the remainder of the characterized cases. Despite genetic advances, around 30% to 50% of patients with a clinical diagnosis of SCN are cared for without an identified pathogenic mutation.1,4,5 The known monogenic defects disrupt distinct cellular pathways, demonstrating the value of gene discovery to provide insights into pathophysiologic mechanisms and potential targeted therapy.1 Upregulation of unfolded protein response (UPR) in patients with mutant ELANE,6,7 enhanced endoplasmic reticulum (ER) stress (G6PC3 and JAGN1),8,9 disturbed endosome trafficking (VPS13B and VPS45),10,11 and defective ribosome biogenesis (SBDS, DNAJC21, and EFL1) have all been implicated as potential mechanisms.12, 13, 14, 15 Cellular stress is a recurring theme in SCN, indicating that the neutrophil differentiation process is highly sensitive to deregulated cellular stress.
SEC61A1 encodes for the α-subunit of the heterotrimeric Sec61 complex, the principal component of the human translocon. This structure is responsible for cotranslational or posttranslational signal peptide–dependent transport of newly synthesized proteins into the ER and for the integration of nascent proteins into the ER membrane.16,17 In addition, the Sec61 channel also passively leaks calcium, contributing to the calcium balance of the cell. The term Sec61-channelopathies encompasses a family of inherited or acquired diseases derived either from direct detrimental effects on Sec61 subunits or from indirect influence on Sec61 channel gating.16 The intricate network involved in fulfilling Sec61 function implicates a broad spectrum of associated disease, including autosomal dominant polycystic liver disease (SEC6318 and SEC61B19) and diabetes (DNAJC320). Recently, autosomal dominant mutations were reported in tubulointerstitial kidney disease (ADTKD) with congenital anemia and in primary antibody deficiency with variable penetrance characterized by an intrinsic deficiency in plasma cell differentiation.21,22
Here, we identify a new SEC61A1 mutation with autosomal dominant SCN and report on the mechanisms by which the mutation disturbs neutrophil differentiation and maturation. Biochemical characterization reveals both a quantitative defect due to protein instability and functional impairment with dysregulated calcium homeostasis. Through in vitro modeling we propose a mechanistic pathway of mutation-induced upregulation of the UPR leading to cellular arrest of myeloid precursors.
Methods
Written informed consent was obtained from all participants, and the ethics committee of University Hospitals Leuven approved the study.
Genetic analysis
Whole exome sequencing and filtering were performed as previously described.23 For details, please refer to the Supplemental Data provided in this article’s Online Repository at www.jacionline.org.
Variant effect prediction on protein structure
Mutations were analyzed by using the available cryo-EM structure (Protein Data Bank identifier 2wwb). All structural visualization was done by using YASARA Structure (version 18.4.2424). Images were generated by using Ray-traced screenshots.
Western blotting
Fresh PBMCs and primary fibroblasts were solubilized in lysis buffer containing 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1 mM egtazic acid, 1% Triton-X, phosphatase inhibitor (PhosSTOP [Roche, Basel, Switzerland]), and protease inhibitor (Pierce Protease Inhibitor [ThermoFisher Scientific, Waltham, Mass]). A quantity of 50 μg of lysate, which was determined by using a Bradford Protein Assay (Bio-Rad, Hercules, Calif), was separated on a 4% to 12% bis-Tris acrylamide gel with 3-(N-morpholino)propanesulfonic acid buffer (NuPAGE Precast Gel System [Thermo Fisher Scientific]) before blotting on polyvinylidene fluoride membrane (GE Healthcare). After blocking, the membranes were incubated with specific primary antibodies: rabbit anti-Sec61α1 (Abcam, Cambridge, United Kingdom, Ab183046), rabbit anti-Actin (Sigma-Aldrich, St Louis, Mo, A2103), mouse anti–glyceraldehyde-3-phosphate dehydrogenase (Thermo Fisher Scientific, MA5-15738), and mouse anti-Vinculin (Sigma, V9264). Proteins were revealed by using ECL Prime (GE Healthcare, Chicago, Ill) or Western Lightning Plus-ECL (Perkin Elmer, Waltham, Mass) with the G:box Chemi-XRQ and quantified by using ImageJ software.
RNA isolation and quantification
Total RNA was isolated by using TRIzol reagent (Ambion, Thermo Fisher Scientific), except for CD34+ cells, in which case the ReliaPrep RNA Cell Miniprep System (Promega, Madison, Wis) was used. Complementary DNA was synthesized by using the GoScriptTM Reverse Transcription System (Promega). Quantitative PCR was performed on a StepOnePlus real-time PCR system (ABI) with Fast SYBR Green Master Mix (Applied Biosystems, Foster City, Calif) supplemented with gene-specific primers (see Table E1 in this article’s Online Repository at www.jacionline.org). Experiments were performed in duplicate and repeated thrice. For DDIT3 and HSPA5, TaqMan Gene Expression Assays were used (ThermoFisher Scientific) and expression was normalized to 18S.
Confocal microscopy
HL-60 cells were differentiated by using 1% dimethyl sulfoxide (DMSO) and allowed to attach to coverslips coated with poly-l-lysine (0.1%) (Sigma) by a 20-minute incubation in PBS at room temperature. After fixation with 4% paraformaldehyde (or with methanol for anti–protein disulphide isomerase), cells were permeabilized with 0.1% Triton X-100 in PBS and incubated in blocking buffer (PBS + 2% BSA + 10% donkey serum + 0.1% Triton X-100). The specific primary antibodies used include rabbit anti-GM130 (G7295, Sigma) for Golgi detection, rabbit anti-protein disulphide isomerase (3501S, Cell Signaling Technology, Danvers, Mass) for ER detection, and 4',6-diamino-2-phenylindole (D1306; Molecular Probes, Eugene, Ore). The secondary antibody used was donkey anti-rabbit Alexa Fluor 555 (A31572; Molecular Probes). Images were acquired with the Nikon A1R confocal unit mounted on an Eclipse Ti inverted microscope.
Calcium flux
After labeling with Zombie Aqua (Biolegend, San Diego, Calif, catalog No. 4231), cells were loaded with 1 μM FuraRed AM (Thermo Fisher Scientific, catalog No. F3020) in HBSS and incubated in calcium-free buffer (150 mM NaCl, 6 mM KCl, 10 mM HEPES, and 1.2 mM MgCl2 [pH 7.4]). Following acquisition of background signals, 1 μM thapsigargin was added and acquisition was immediately resumed for 10 minutes by using BD FACSCanto II. FuraRed was recorded as the ratio of emission at 450/50 nm following excitation at 405 nm and emission at 670 nm following excitation at 488 nm. The collected data (FACSDiva software, BD Biosciences, San Jose, Calif) were analyzed as mean fluorescence ratio by using the Kinetics platform provided in FlowJo version 10.6.0 software (Tree Star Inc, Ashland, Ore).
In vitro protein transport
Protein translocation experiments were performed as previously described.25,26 For a detailed description, please refer to the Supplemental Data.
Differentiation of CD34+ cells from peripheral blood
CD34+ cells were isolated from PBMCs by using the CD34 MicroBead Kit UltraPure (Miltenyi) and were differentiated by using SCF, FLT3L, TPO, IL-3, and G-CSF. For a detailed description, please refer to the Supplemental Data.
Flow cytometry
Thawed PBMCs were stained in Brilliant Stain Buffer (BD Biosciences) for live-dead and surface markers by using anti-human antibodies. Data were collected on BD Symphony (BD Biosciences) and analyzed by using FlowJo for Mac version 10.5 (Tree Star Inc). Primary neutrophils were isolated by using the EasySep Direct Human Neutrophil Isolation Kit (StemCell Technologies, Vancouver, British Columbia, Canada) and incubated with Fc block (Miltenyi Biotec, Bergisch Gladbach, Germany) before staining with mAbs. Flow cytometric analysis was performed with a FACS BD LSR Fortessa X20 (BD Biosciences). Analysis was performed with FlowJo software (LLC). For details on the antibodies used, refer to the Supplemental Data.
Single-cell sequencing
Single-cell RNA sequencing libraries were generated by using a commercial droplet platform (Chromium Controller system [10x Genomics, Pleasanton, Calif]). Briefly, the EasySep RBC Depletion Reagent (StemCell Technologies) was used to deplete erythrocytes from whole bone marrow samples collected in heparinized tubes. The cell count and viability of the cells were assessed by using a LUNA dual fluorescence cell counter (Logos Biosystems, South Korea), and as per the 10x Genomics recommendations (Single-Cell 3' Reagent Kits Version 2 User Guide; CG00052 Rev B), we aimed at a target recovery of 6,000 cells per sample. After the cell count and quality control, the samples were immediately loaded individually onto the Chromium Controller. Single-cell RNA sequencing libraries were prepared by using the manufacturer recommendations, and at the different check points the library quality was assessed by using Qubit (ThermoFisher) and Bioanalyzer (Agilent, Santa Clara, Calif). With a sequencing coverage target of 50,000 reads per cell, single-cell libraries were sequenced on the Illumina HiSeq4000 platform by using paired-end sequencing workflow with 10x version 3 read parameters (28-8-0-91 cycles). For single-cell sequencing analysis, please refer to the Supplemental Data provided.
Annexin V and propidium iodide staining
PBMCs were treated for 24 hours with DMSO (vehicle control), thapsigargin (1 μM), brefeldin A (4 μg/mL), or tunicamycin (10 μg/mL). After the PBMCs had been stained with Zombie Aqua, cells were stained with allophycocyanin-annexin V (Invitrogen, Thermo Fisher Scientific) and propidium iodide (00-6990) according to the manufacturer’s instructions, followed by acquisition using the BD FACSCanto II system.
Lentiviral transduction
Human SEC61A1 (NM_013336.4) was cloned into the pWPXL backbone by using an In-Fusion HD Cloning Kit (Takara Bio USA, Mountainview, Calif). HEK293T cells were transfected with lentiviral vectors and second-generation packaging plasmids by using X-tremeGEne HP Transfection Reagent (Sigma-Aldrich). Confluent HL60 cells were resuspended in filtered undiluted virus-containing medium with 5 μg/mL of polybrene for 16 hours. Transduced HL-60 cells were sorted with BD FACSAria II gating on live GFP+ cells.
Differentiation of HL60 cells
HL-60 cells were plated in T75 flasks and treated with 1% DMSO for 6 days with a refreshment of media on day 4. Differentiated cells were stained with Zombie Aqua followed by surface staining with the antibodies anti-CD11b allophycocyanin-eFluor 780 (ICRF44, eBioscience, San Diego, Calif) and anti-CD16 PerCP (3G8, BioLegend). Cells were acquired by using the BD FACSCanto II system. For cytospin preparations, single-cell suspensions were spun on a glass slide and stained with hematoxylin and eosin. Cells were visualized on a Leica DM 2000 microscope (Leica Microsystems, Wetzlar, Germany) for manual counting by 3 independent researchers.
Statistics
Statistical analyses were performed by using a (un)paired t test or as indicated.
Data sharing statement
For original data please contact Stephanie Humblet-Baron (E-mail address: stephanie.humbletbaron@kuleuven.be). The 10X Genomics RNA sequencing data on bone marrow samples are available at the Gene Expression Omnibus under accession number GSE137496.
Results
Identification of a de novo mutation in SEC61A1 in a patient with SCN
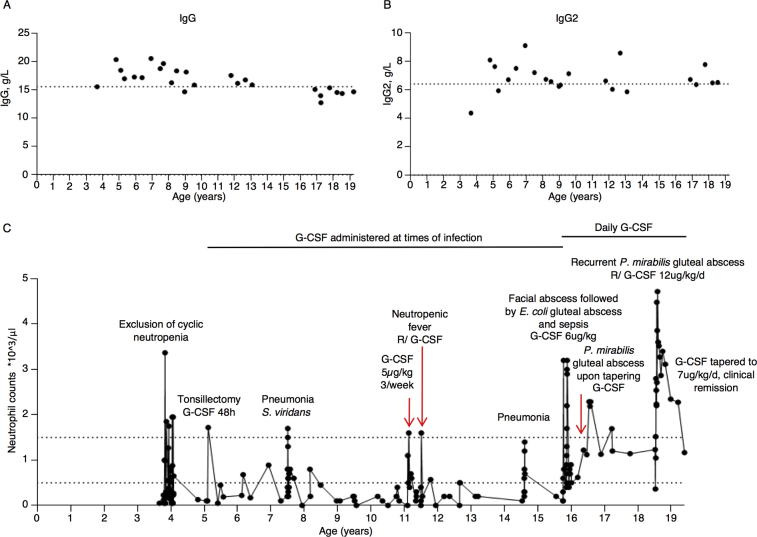
The index patient is a 19-year-old female who was born at term as the only child to nonconsanguineous parents of European-Belgian descent. Her birth weight was 2750 g (–1.8 SD) (Fig 1, A). At the age of 4 months she presented with a retroauricular Staphylococcus aureus abscess and subsequently suffered from recurrent otitis, bronchitis, sinusitis, and tonsillitis that necessitated a tonsillectomy at age 5 years. At the age of 1 year, she was treated for varicella that was complicated by bacterial superinfection treated with oral antibiotics. Aside from her infectious susceptibility, she presented with oral aphthosis, problematic wound healing, gingivitis, and an episode of limited verrucae plantaris. She was diagnosed with SCN when her neutrophil levels were analyzed during hospitalization for enteritis at the age of 2years. At 7 years of age, she developed pneumonia with Streptococcus viridans complicated by empyema. Aside from prophylactic antibiotics, she began receiving subcutaneous granulocyte colony-stimulating factor (G-CSF) at times of infection from the age of 7 years. Following an Escherichia coli abscess in her facial region at the age of 15 years and septicemia secondary to a gluteal E coli abscess a year later, she began receiving daily G-CSF. When G-CSF therapy was tapered to every 2 days, she developed a refractory perianal abscess with Proteus mirabilis that cleared only after surgical drainage, intravenous antibiotic treatment, and long-term administration of G-CSF at a rate of 12 μg/kg per day. Her neutrophil counts were repeatedly below 0.5 ×109/L but responsive to G-CSF treatment (see Fig E1 in this article’s Online Repository at www.jacionline.org). Before any initiation of G-CSF treatment at age 3 years, her complete blood count showed a total leukocyte level of 4.79 × 109/L with a neutrophil level of 0.048 × 109/L, lymphocyte level of 2.108 × 109/L, monocyte level of 1.916 × 109/L, eosinophil level of 0.287 × 109/L, and basophil level of 0.096 × x109/L. The result of testing for anti-granulocyte antibodies was negative, and further laboratory work-up revealed hypergammaglobulinemia before the initiation of daily G-CSF (in particular, IgG2 and IgA [see Fig E1]), mild thrombocytopenia (a nadir of 104 [reference range 150-450 × 109/L]), and discrete intermittent anemia (a nadir of 10.2 [reference range 12.0-16.0g/dL]). Under daily G-CSF treatment her total leukocyte numbers increased to 9.19 × 109/L (reference range 4.0-10.0), with a myelocyte level of 0.1 × 109/L (1% [reference percentage ≤0%]), neutrophil level of 3.4 × 109/L (37% [reference range 38%-77%]), lymphocyte level of 2.2 × 109/L (24% [reference range 20%-50%]), basophil level of 0.2 × 109/L (2% [reference percentage ≤1%]), relative monocytosis with a monocyte level of 2.4 × 109/L (26% [reference range 2.0%-10.0%]), and relative eosinophilia with an eosinophil level of 1.0 × 109/L (10% [reference range ≤6%]). Bone marrow analysis before G-CSF treatment showed a maturation arrest in granulopoiesis at the level of the promyelocytes and myelocytes (Fig 1, B).
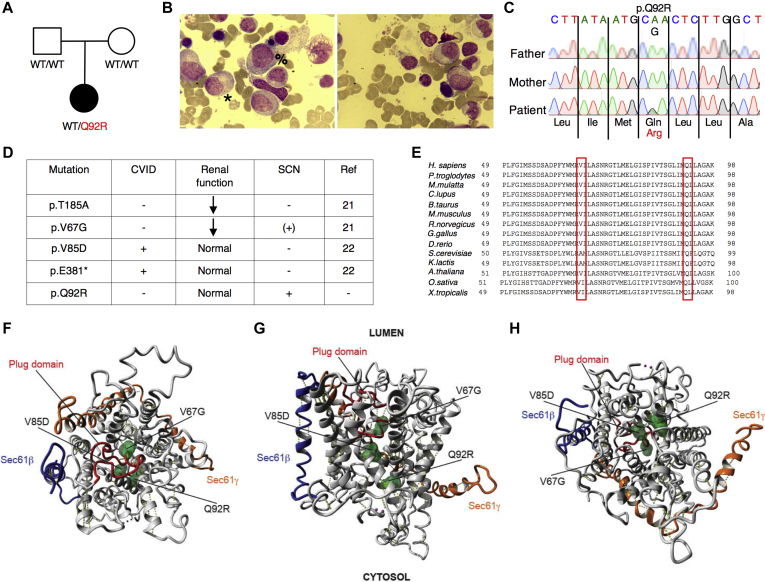
Fig 1.
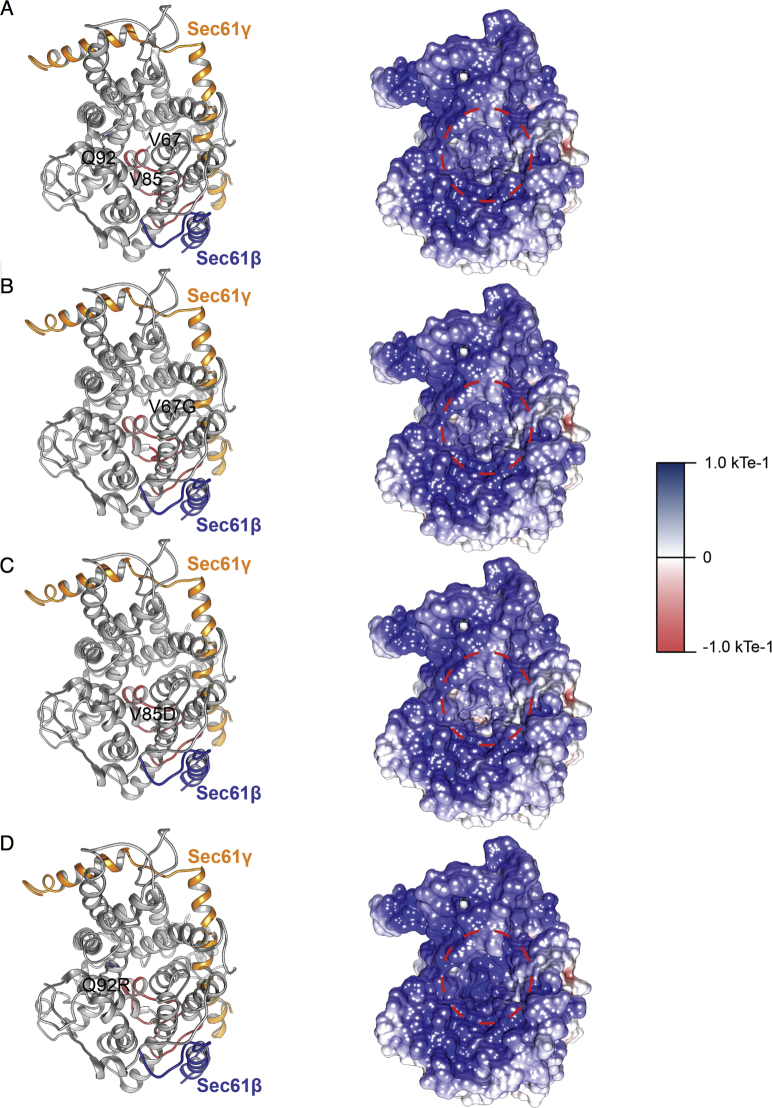
Autosomal dominant mutations in SEC61A1 cause SCN. A, Pedigree of a family with segregation of the p.Q92R mutant allele. The index patient is denoted by a black symbol. B, May-Grünwald Giemsa staining of representative bone marrow smears from the index patient. Promyelocytes are marked by the symbol %, and myelocytes are marked by the symbol ∗. Magnification, ×500. C, Sequencing profiles demonstrating the heterozygous SEC61A1 c.A275G:p.Gln92Arg mutation in the kindred. D, Table of all reported mutations in SEC61A1 with associated phenotypes. E, Multiple alignment to display conservation exported by using HomoloGene (National Center for Biotechnology Information). F-H, Illustration of mutated residues in green obtained using available cryo-EM structure (Protein Data Bank identifier 2wwb), with lumenal view in (F) and cytosolic view in (H). The plug domain is visible in red, the Sec61β protein is visible in blue, and the Sec61γ subunit is visible in orange.
Whole exome sequencing was performed after negative genetic screening of SBDS, GATA2, and CXCR4 through Sanger sequencing and negative genetic screening of known genes for SCN (ELANE, G6PC3, GFI1, HAX1, LAMTOR2, SLC37A4, USB1, VPS13B, SBDS, VPS45, WAS, and JAGN1) and inborn errors of immunity through custom-designed gene panels. Through filtering for rare de novo or recessive variants, we identified a novel heterozygous mutation in SEC61A1 (NM_013336), c.A275G:p.Gln92Arg, that was not registered in any public database and had functional scores predicted as damaging by Polymorphism Phenotyping, Sorting Intolerant from Tolerant, and Combined Annotation-Dependent Depletion (CADD 24.500; mutation significance cut-off 6.099) analyses (Fig 1, C). To date, mutations in SEC61A1 have been reported in 2 kindreds with ADTKD (c.T200G:p.Val67Gly and c.A553G:p.Thr185Ala)21 and 2 kindreds with common variable immune deficiency (CVID) with incomplete penetrance (c.T254A:p.Val85Asp and c.G1325T:p.Glu381∗)22 (Fig 1, D and see Table E2 in this article’s Online Repository at www.jacionline.org). Although neutropenia was observed in the p.V67G kindred in addition to the well-described late-onset ADTKD and congenital anemia, it was not further investigated.21 In light of our patient, the cosegregation of SCN in the p.V67G kindred may also be attributable to mutant SEC61A1. Conversely, we investigated the possibility of subclinical renal dysfunction in our patient. Ultrasound imaging revealed small bilateral kidneys (right kidney, 9.6 cm [–3.2 SD]; left kidney 9.8 cm [–2.5 SD]) that were morphologically normal. Her urine sediment analysis results and kidney function remained normal. However, her uric acid levels were slightly elevated over the past year (6.6 mg/dL [reference range 2.6-6.0 mg/dL]). Otherwise, no evidence for extramedullary involvement was found. In view of our current findings, we conclude that mutations in SEC61A1 can give rise to a clinical spectrum of disease including SCN.
Biochemical impact of SEC61A1 mutations
First, we investigated the structural consequences of the resulting mutations. Both of the neutropenia-associated mutations affect the well-conserved amino acid residues Gln92 (Q92) and Val67 (V67) (Fig 1, E). The heterotrimeric Sec61 translocon complex comprises Sec61β, Sec61γ, and Sec61α1, with the latter containing 10 transmembrane helices (TMs) that form the central channel of Sec61. In the closed conformation, a so-called plug domain between TM1 and TM2 interacts with the center of transmembrane helix bundle, thereby blocking and stabilizing the pore.21 It is believed that signal peptides of nascent proteins intercalate between TM2 and TM7, with displacement of TM2 and subsequent opening of the “lateral gate” of the Sec61 complex comprising these 2 TMs. Utilizing the available cryo-EM structure (Protein Data Bank identifier 2wwb),27 we determined that both Q92 and V85 are located in TM2 (Fig 1, F-H). In the closed conformation, Q92 points into the core to form a hydrogen bond with a neighboring residue, whereas in the open conformation, it forms a hydrogen bond with the translocating peptide. V85 is located closer to the core of the helix bundle. Both Q92R and V85D introduce a buried charge in the hydrophobic core of the TM bundle without the presence of a counter charge and are therefore predicted to severely destabilize the helix bundle thermodynamically. V67 is situated in the middle of an α-helical part of the plug domain, and mutating this residue to a glycine is likely destabilizing (Fig 1, F-H). Glycine mutations notoriously destabilize α-helices thermodynamically, especially when occurring at internal positions.28 Thus, this mutation potentially affects the protein translocation regulatory function of the plug domain. Additionally, in the closed conformation, the wild-type (WT) side chain of V67 forms hydrophobic interactions with TM5 and TM10 that are completely removed by the truncation to glycine, further decreasing the interaction energy between the plug domain and the 2 transmembrane helices. Mapping the electrostatic potential on the molecular surface of Sec61 revealed that the WT is positively charged, maintaining a constant distribution of the potential around the internal surface of the channel (see Fig E2, A in this article’s Online Repository at www.jacionline.org). Although V67G does not disturb the electrostatic potential (see Fig E2, B), V85D increases the negative charge at the bottom of the channel (see Fig E2, C), in contrast to the Q92R, which instead raises the positive charge on the inside of the channel (see Fig E2, D). These opposing alterations may directly disturb the conformational changes of the Sec61 complex or affect its ability to interact with or translocate newly synthesized polypeptide chains. Using these structural predictions, we noted that pathogenic mutations cluster in the pore-forming region.
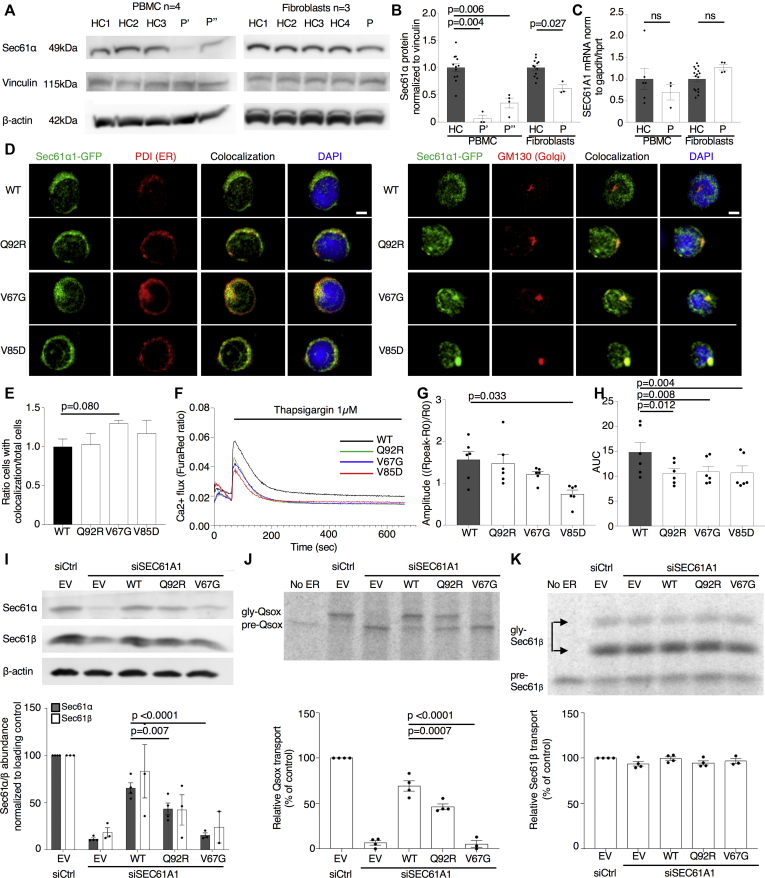
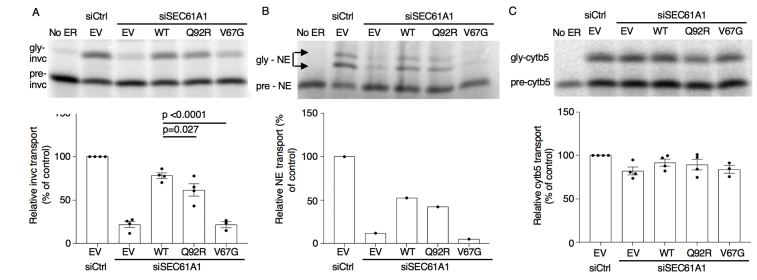
In support of the predicted impact of the Q92R mutation on protein stability, we demonstrated reduced Sec61α1 protein expression in primary PBMCs and primary fibroblasts from our index patient (Fig 2, A and B), whereas the mRNA levels of SEC61A1 remained unaffected (Fig 2, C). This parallels work on the V67G mutation, associated with diminished expression of Sec61α1.21,22 Colocalization studies through immunofluorescence imaging of transduced HL-60 cells revealed that the WT and all 3 mutant proteins were located in the ER; however, unlike the WT, the V67G and (to a lesser extent) V85D mutations demonstrated signs of Golgi accumulation (Fig 2, D and E). As proposed by Bolar et al, this may be consistent with mislocalization of mutant proteins to the ER-Golgi intermediate compartment to undergo ER-associated protein degradation.21 Notably, the reduced protein expression correlated with reduced Sec61-dependent protein translocation across the ER, including the NE among other substrates tested (Fig 2, I-K and see Fig E3 in this article’s Online Repository at www.jacionline.org).
Fig 2.
Mutations in SEC61A1 affect protein stability and disrupt ER/cytosol calcium homeostasis. A, Sec61α1 expression in healthy controls (HCs) and the patient (with P' indicating the patient's status as of April 2019 and P'' indicating the patients status as of June 2018). B, Quantification of (A) normalized to the average of the HCs. C,SEC61A1 mRNA expression in PBMCs (n = 3) and primary fibroblasts from 2 different passages (n = 4), normalized to the average of the HCs. Unpaired t test. D, HL-60 cells transduced with SEC61A1–green fluorescent protein (GFP) stained with anti–protein disulphide isomerase (PDI) (left) and anti-GM130 (right) (n = 2). E, Ratio of cells with colocalization of Sec61α1 and GM130 over the total number of cells from (D), normalized to WT. F, Calcium flux measurements in transduced HL-60 cells. Mean of 3 independent experiments with duplicates. G, Amplitude of calcium efflux following thapsigargin treatment. R represents the ratio of the FuraRed fluorescence intensity at 450/50 nm and 670 nm. Paired t test. H, Area under the curve between t = 60 and 660 seconds (duration of treatment). I,SEC61A1 expression in HeLa cells transfected with small interfering RNA and plasmid as indicated (empty vector [EV]). J and K, Phosphorimaging autoradiogram and quantification of Sec61-dependent cotranslational transport efficiency of Quiescin-sulfhydryl oxidase (K) and Sec61-independent posttranslational transport efficiency of Sec61β (J), normalized to the control (Ctrl). Gly, glycosylated. Ordinary 1-way ANOVA with Sidak correction. Data are represented as means ± SEMs. DAPI, 4',6-Diamino-2-phenylindole.
To ascertain the function of mutant SEC61A1, we assessed calcium leakage from the ER into the cytosol. HL-60 cells transduced with WT or mutant forms of SEC61A1 were loaded with a Ca2+ indicator dye (FuraRed) and treated with thapsigargin, a known inhibitor of the sacroplasmic/endoplasmic reticulum (Fig 2, F and see Fig E4 in this article’s Online Repository at www.jacionline.org). We observed a significant decrease in the amplitude of the Ca2+ flux for the V85D mutant and significantly lower integrated cytosolic Ca2+ levels in all 3 mutants following treatment (Fig 2, G and H). We can postulate that these mutations result in increased Ca2+ leakiness causing neutralization of the ER/cytosol Ca2+ gradient, and therefore, the calcium flux elicited by thapsigargin is reduced. Together, our results indicate that SEC61A1 mutations give rise to disease through a combined quantitative and functional protein defect.
B-cell maturation defect with increased plasmablast number
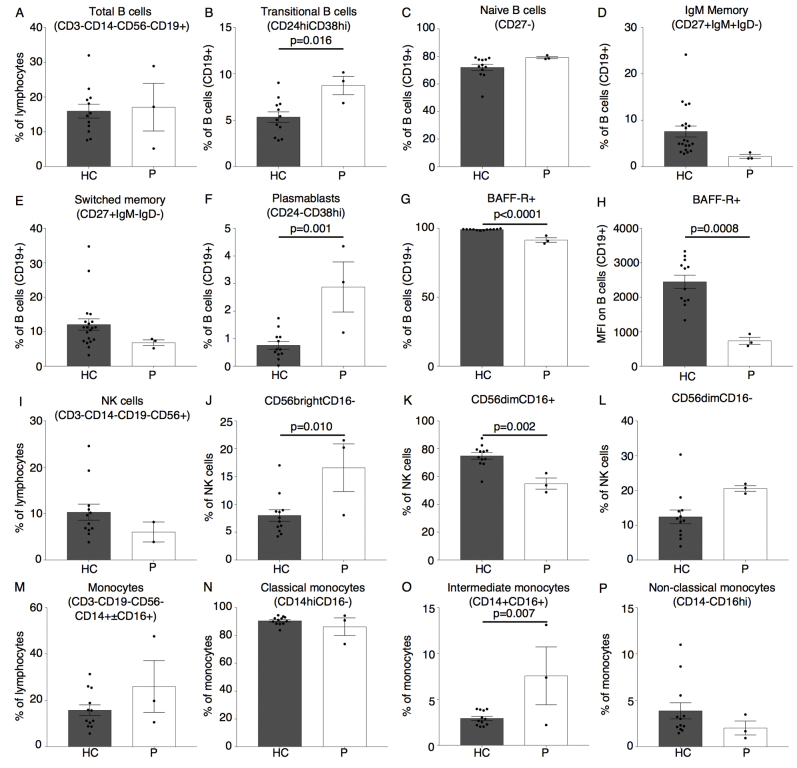
In light of the recent report on CVID in a kindred harboring the p.V85D mutation, we performed a thorough analysis of our patient’s immunophenotype.22 Although the index patient's total B-cell numbers were normal, she exhibited a maturation arrest of B-cell precursors at the transitional B-cell stage (see Fig E5, A-E in this article’s Online Repository at www.jacionline.org). In contrast to patients with CVID and SEC61A1 mutations, the index patient had plasmablast numbers that were significantly elevated, corresponding to the persistent hypergammaglobulinemia (see Figs E1 and E5, F). B-cell maturation antigen signaling is crucial for B-cell differentiation from the transitional B-cell stage. Both the percentage and mean fluorescence index of B-cell maturation antigen receptor on CD19+ B cells was significantly reduced in our patient, likely downregulated in response to the decreased consumption of BAFF because of the observed B-cell maturation defect (see Fig E5, G and H).29 Furthermore, we perceived a marked increase in CD56bright natural killer cells, with a parallel reduction in CD56dimCD16+ natural killer cells (see Fig E5, I-L). Considering the proposed linear differentiation model of natural killer cells, this would indicate a maturation defect.30 The combination of low birth weight with SCN and a natural killer cell maturation defect with a history of warts was reminiscent of GINS1 deficiency.31 Therefore, rare variants affecting the origin of replication complex (of which GINS1 is a member) were specifically sought, but none was identified. Finally, monocyte percentages were increased in line with G-CSF treatment, with a notable increase in intermediate CD14+CD16+ monocytes (see Fig E5, M-P). Together, these results indicate that although the primary clinical presentation of the index patient was SCN, mutation in SEC61A1 also affected other leukocyte compartments.
Defective myeloid differentiation and neutrophil maturation
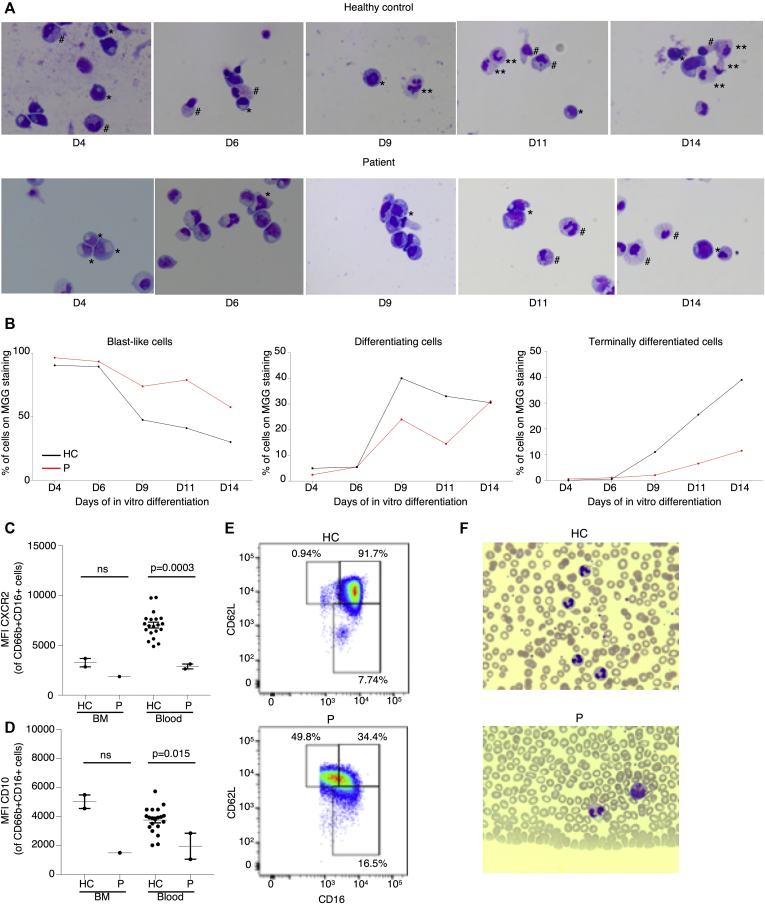
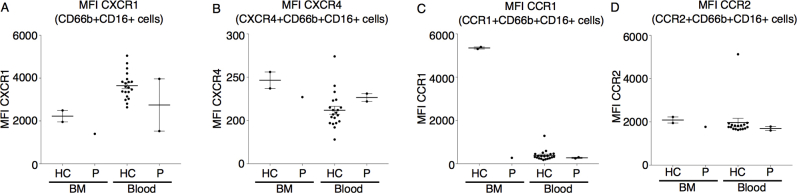
To determine whether myeloid precursors from our patient had an intrinsic differentiation defect, we isolated peripheral blood CD34+ cells for in vitro differentiation. Monitoring by Giemsa staining showed productive differentiation into mature neutrophils with granulation in the healthy control, whereas the majority of the patient cells remained in a blast-like state, with few cells reaching the final stage of maturation (Fig 3, A and B). In contrast, CD34+ cells from a healthy control generated mature cells with clear segmentation of the nucleus and normal granulation from day 9 onward. Considering that the patient's neutrophil levels were responsive to G-CSF treatment, we performed in-depth immune phenotyping to determine the maturity of her primary circulating neutrophils. Primary neutrophils from the index patient had lower expression of the IL-8 receptors CXCR1 and CXCR2, where the latter is typically upregulated during granulopoiesis to promote release from the bone marrow (Fig 3, C and see Fig E6 in this article’s Online Repository at www.jacionline.org). Both expression of CD10, a surface marker used to distinguish mature from immature neutrophils, and the percentage of CD16highCD62Lhigh cells were shown to be reduced, indicating an immature phenotype (Fig 3, D and E). Also, cytologic analysis of patient's peripheral neutrophils showed hyposegmentation of the nucleus despite G-CSF treatment (Fig 3, F). Collectively, these findings indicated that although the patient was clinically G-CSF–responsive, the circulating neutrophils remained immature with hyposegmentation of the nucleus.
Fig 3.
Defective in vitro granulocytic differentiation and maturation in patient primary cells. A, May-Grünwald Giemsa (MGG) staining of representative cytospins from the index patient (P) and an unrelated healthy control (HC) monitoring in vitro differentiation of CD34+ cells on day 4 (D4) to day 14 (D14) as indicated. Blast-like cells are marked by the symbol ∗, differentiating cells are marked by the symbol #, and terminally differentiated cells are marked by the symbol ∗∗. Magnification, ×100. (B) Graphic representation of the different stages of differentiation quantified by expert cytologic evaluation of cytospins in (A). (C) Mean fluorescence index (MFI) of CXCR2 gated on CD66b+CD16+ neutrophils isolated from whole blood or bone marrow (BM) from the HC and the patient. Unpaired t test. (D) MFI of CD10 on CD66b+CD16+ neutrophils isolated from whole blood or BM from the HC and the P. Unpaired t test. (E) Flow cytometry plot showing CD62L and CD16 expression gated on live CD66b+CD16med-high cells. (F) Representative MGG staining of peripheral blood smears from the HC and the patient. Magnification, ×50.
Dysregulated UPR in myeloid progenitors and primary cells harboring the SEC61A1 mutation
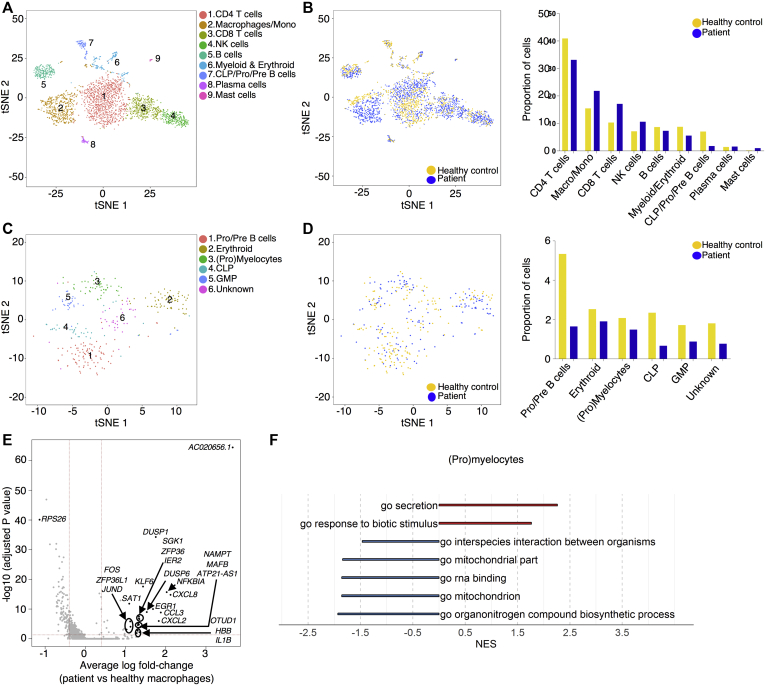
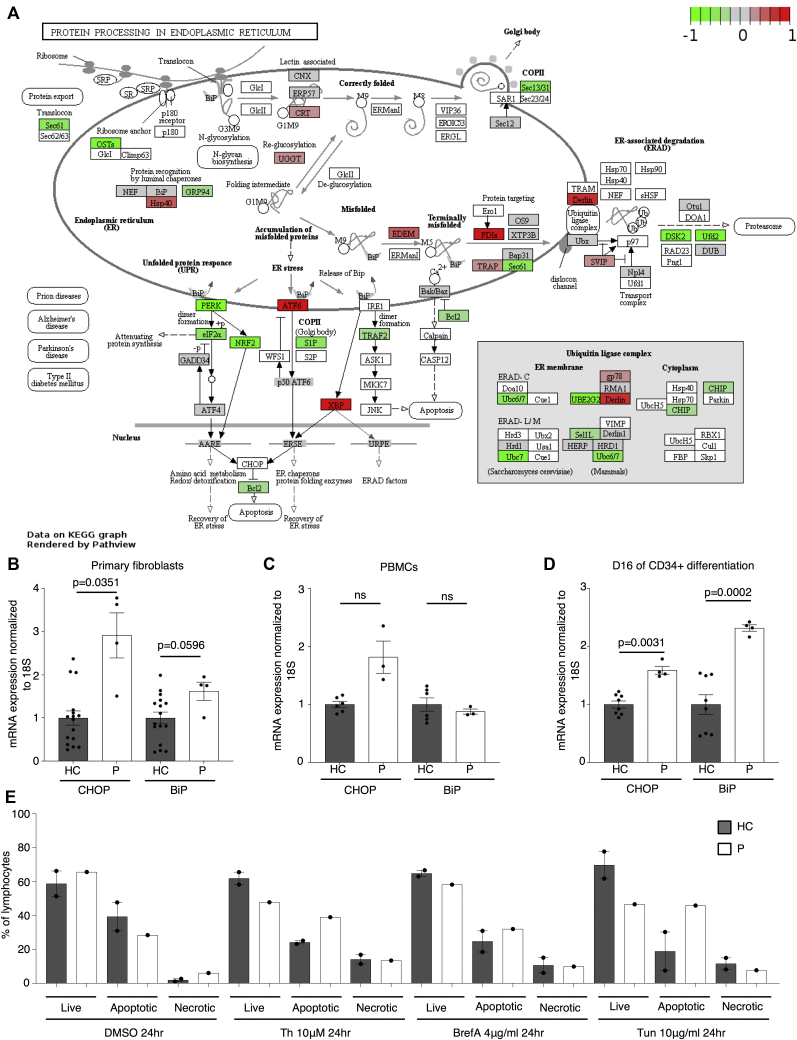
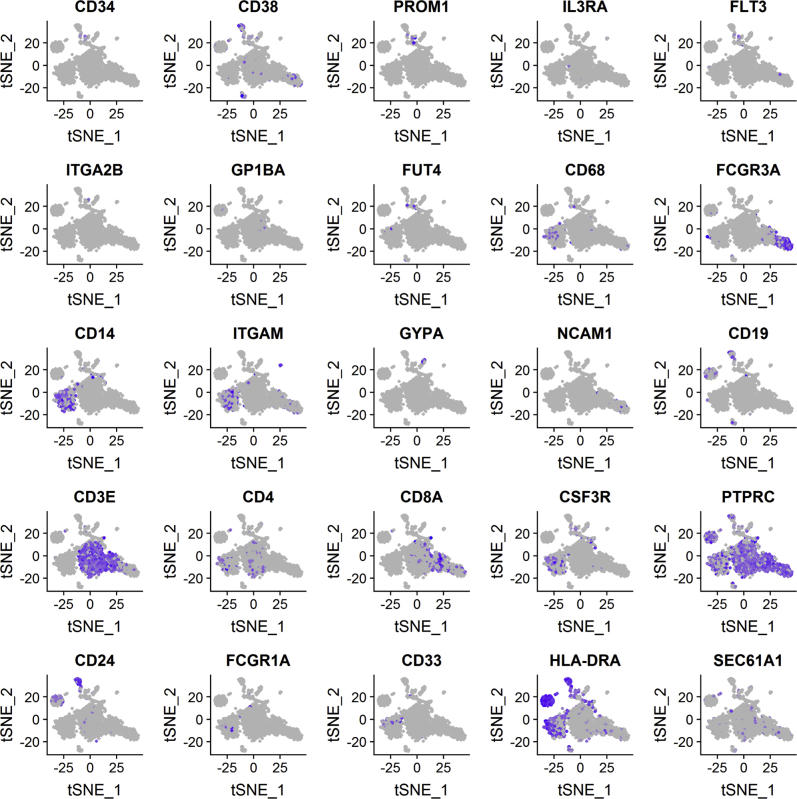
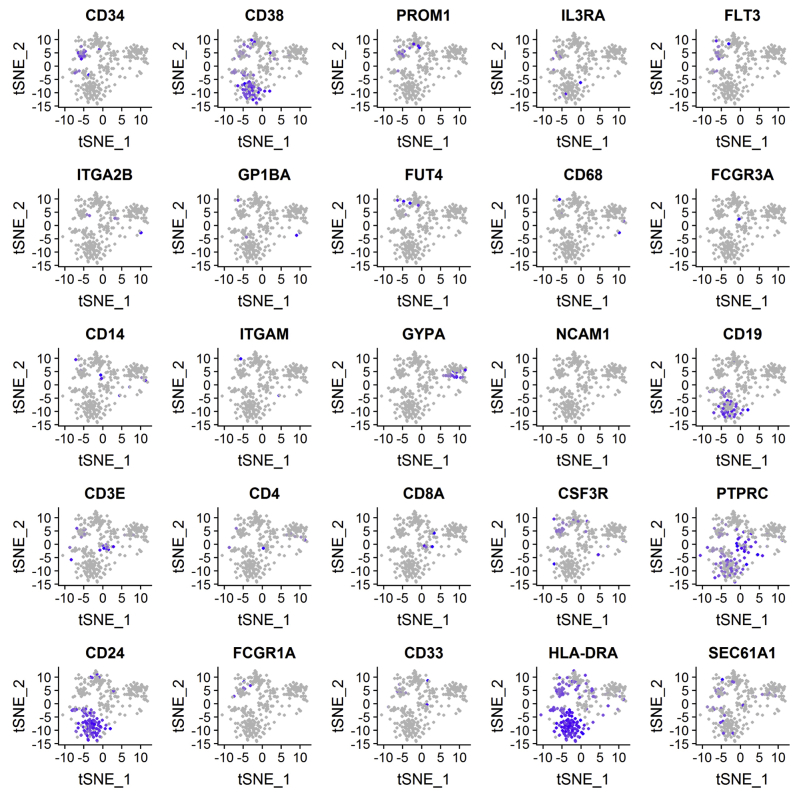
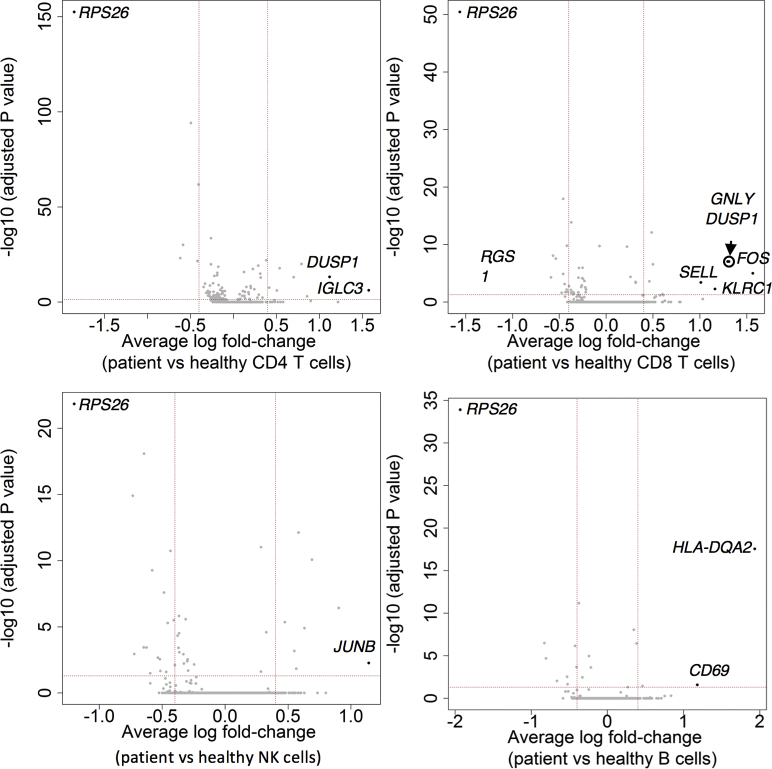
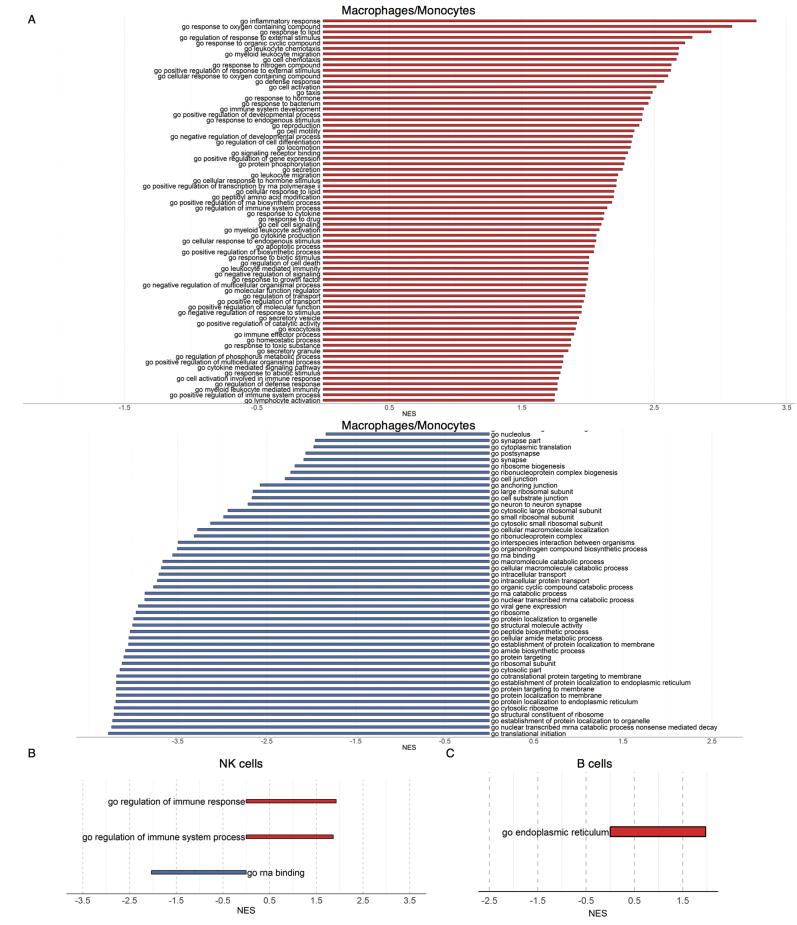
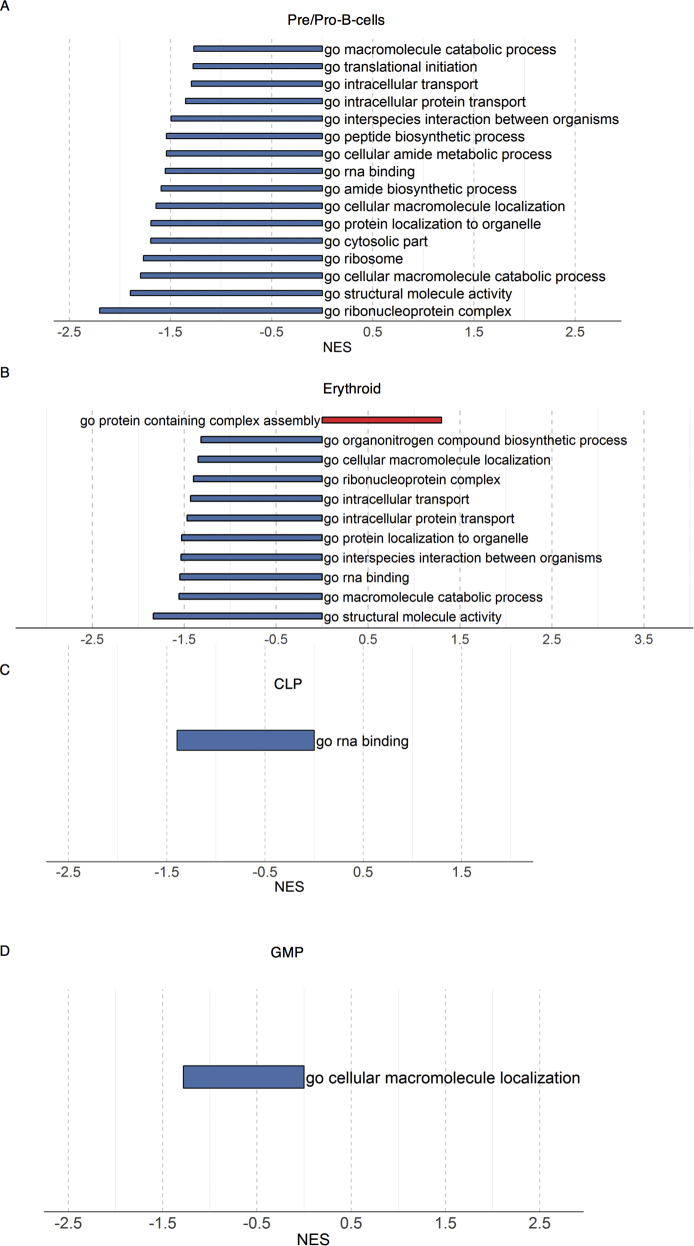
To evaluate the effects of Sec61α1 dysfunction on hematopoiesis, we performed a single-cell sequencing experiment on red blood cell–depleted bone marrow from the index patient and a healthy sex-matched control. The patient did not receive G-CSF the day before the bone marrow sampling, and according to morphologic expert assessment, she exhibited a myeloid maturation arrest at the (pro)myelocyte stage (blast percentage 0.3% [reference range 0.3%-2.9%], promyelocyte percentage 11.7% [reference range 1.7%-8.4%], myelocyte percentage 16.3% [reference range 3.3%-12.0%], metamyelocyte percentage 1.7% [reference range 3.1%-10.7%], band neutrophil percentage 0.3% [reference range 5.4%-16.2%], segmented neutrophil percentage 2.3% [reference range 15.1%-35%], eosinophil precursor percentage 25% [reference range 1.6%-6.3%], lymphocyte percentage 15.7% [reference range 6.9%-24.5%], (pro)monocyte percentage 4.3% [reference range 0.4%-5.2%], and erythroblast percentage 22.3% [reference range 16.1%-39.5%]). With use of a t-distributed stochastic neighbor embedding approach, 9 distinct populations were annotated on the basis of expression of key lineage markers (Fig 4, A and see Figs E7 and E8 and Tables E3 and E4 in this article’s Online Repository at www.jacionline.org), with a shift in the frequency of macrophage/monocyte and CD4 T-cell populations observed in the patient (Fig 4, B). Reclustering of progenitor cells into 6 distinct populations (Fig 4, C) revealed that the index patient had a decreased proportion of progenitor subsets (in particular, pre-B cells and/or pro-B cells, common lymphoid progenitors, and granulocyte/myeloid progenitors) (Fig 4, D). Although systematic analysis was undertaken for all cell clusters, the macrophage/monocyte cluster demonstrated the most differentially expressed genes (Fig 4, E and see Figs E9-E11 and Tables E5 and E6 in this article’s Online Repository at www.jacionline.org). Significantly upregulated genes in the macrophage/monocyte grouping are known to stimulate monocyte/macrophage differentiation (ZFP36L1, MAFB, and EGR1) and promote neutrophil recruitment and activation (CXCL2 and CXCL8) and “emergency granulopoiesis” (NAMPT). However, these alterations do not elucidate the etiology of the granulocytic maturation arrest and rather may be secondary to the presence of peripheral neutropenia or G-CSF treatment.32, 33, 34, 35 Gene set enrichment analysis in the macrophage/monocyte cluster reflected similar perturbations, whereas downregulated pathways were more directly related to reduced Sec61α1 expression (in particular, processes related to cotranslational protein localization or targeting to the ER or membrane] see Fig E10, A and B]). Notably, the fold-change increase in XBP1 (1.6X) and TMBIM4 (1.4X) and the observation that 10 of the top 35 ranked genes differentially expressed in the (pro)myelocyte cluster encode mitochondrial proteins indicate that upregulation of the UPR and mitochondrial dysfunction may contribute to disease pathogenesis (see Table E6 in this article’s Online Repository at www.jacionline.org).36 Both autosomal recessive HAX1 and AK2 deficiency present with SCN related to disturbed mitochondrial membrane potential, implicating mitochondrial dysfunction in the pathogenesis of SCN.37, 38, 39, 40 This was also reflected in the gene set enrichment analysis of (pro)myelocytes, where the gene sets “mitochondrion” and “mitochondrial part” were downregulated compared with in the control (Fig 4, F). The disturbed Ca2+ homeostasis associated with mutant Sec61α1 can thus potentially impede the ability of mitochondria to restore ATP levels in the ER, thereby inducing apoptosis. In support of enhanced UPR, ER stress-related genes in the (pro)myelocyte cluster, which were plotted using Pathview, demonstrated reduced Sec61 expression and selective upregulation of the ATF6 pathway, with increased XBP1 expression and reduced expression of the anti-apoptotic BCL2 downstream (Fig 5, A).
Fig 4.
Single-cell transcriptomics data from patient and control bone marrow demonstrating reduced myeloid progenitors and downregulated mitochondrial proteins. A, tSNE projection of 3046 red blood cell–depleted bone marrow cells. Following alignment, each cell was grouped into clusters denoted by number and color. Single joint clustering identified 9 immune subsets annotated on the basis of expression of key lineage markers. B, tSNE projection of 3046 cells from (A) split between the patient and the healthy control. Proportion of the total number of cells from each sample belonging to each immune population. C, tSNE projection of 318 cells from clusters 6 and 7 in (A) after reclustering. In all, 6 immune populations were identified and are denoted by number and color. D, tSNE projection of 318 cells from (C) split between the patient and the control. Proportion of the total number of cells from each sample belonging to each immune population. E, Volcano plot showing differentially expressed genes in patient versus healthy macrophages/monocytes. Vertical lines mark the fold changes 0.4 and –0.4 and horizontal line marks the adjusted P value of .05. F, Gene set enrichment analysis in the (pro)myelocyte population as annotated in (C). CLP, Common lymphoid progenitor; GMP, granulocyte-monocyte progenitor; NK, natural killer; tSNE, t-Distributed Stochastic Neighbor Embedding.
Fig 5.
Primary cells harboring SEC61A1 mutation exhibit increased ER stress and susceptibility to apoptosis. A, Pathview plot using the average log fold changes from the 158 available ER stress genes in the (pro)myelocyte cluster. B-D, mRNA expression of DDIT3 (CHOP) and HSPA5 (BiP) in primary fibroblasts from 2 different passages (n = 4) (B), PBMCs (n = 3) (C), and primary cells remaining after 16 days of in vitro granulocytic differentiation (n = 3) (D) in healthy controls (HCs) and the patient (P) normalized to 18S and normalized to the average of the HCs within each repeat experiment. Paired t test. E Live (annexin V–negative [A–]–propidium iodide–negative [PI–]), apoptotic (Ap) (A+PI+/–), and necrotic (A–PI+) cells as a percentage of lymphocytes from the HCs and P following treatment with vehicle control DMSO, 10 μg/mL of tunicamycin (Tun), 4μg/mL of brefeldin A (BrefA), or 10 μM thapsigargin (Th) for 24 hours.
To further explore ER stress as a potential pathogenic mechanism, we sought to study the UPR in the patient's cells. In the index patient, primary fibroblasts demonstrated upregulation of both DDIT3, encoding CCAAT/enhancer-binding protein homologous protein (CHOP), and HSPA5, encoding immunoglobulin heavy chain binding protein (BiP), with primary PBMCs showing a similar fold increase for CHOP (Fig 5, B and C). Likewise, the patient's cells upregulated both CHOP and BiP following 16 days of in vitro neutrophilic differentiation, indicating that SEC61A1-mediated disease may act through initiation of ER stress (Fig 5, D). To determine whether these changes sensitize patient cells to apoptosis, we treated primary PBMCs from the index patient with ER stress–inducing agents. Healthy control cells were resistant to cell death under these conditions, whereas the patient's PBMCs consistently showed a reduction of live cells with an increased percentage of apoptotic cells compared with in the vehicle control (Fig 5, E). Together, these results suggest that the patient's cells were more prone to apoptosis on triggering of the UPR.
Mutant SEC61A1 is directly responsible for ER stress and a genotype-specific arrest in neutrophilic differentiation
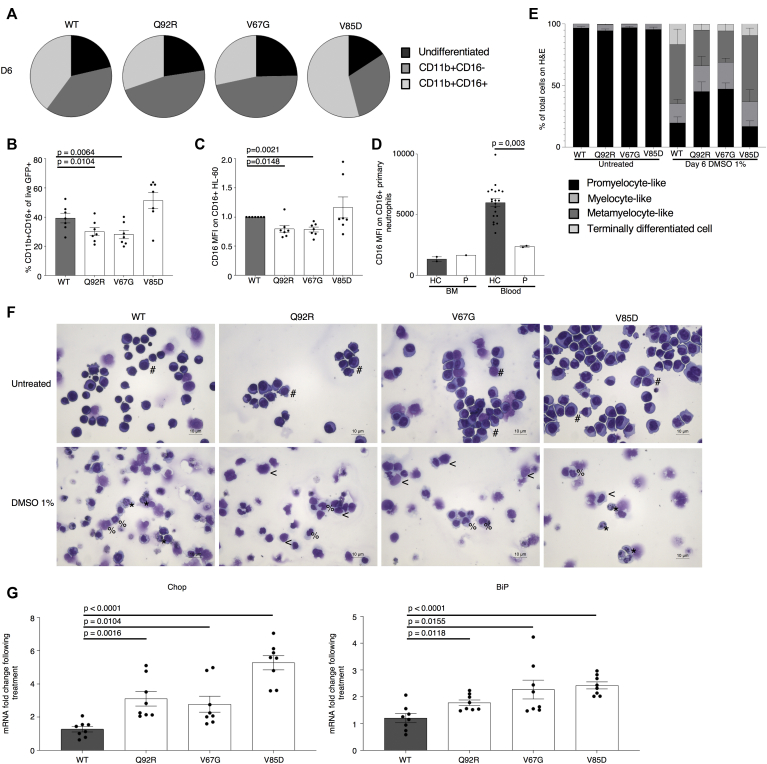
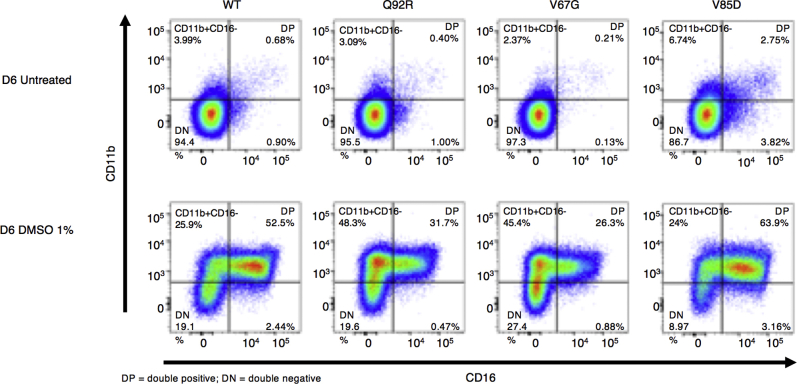
Having established both an intrinsic deficiency in granulocytic differentiation and increased ER stress in patient cells, we explored the potential experimental induction of these cellular phenotypes by using an appropriate model. The chosen HL-60 cell line was generated from a patient with acute promyelocytic leukemia and offers an in vitro model to study human myeloid cell differentiation. On treatment with chemical stimuli such as DMSO, cells terminally differentiate to granulocyte-like cells. HL-60 cells stably transduced with WT SEC61A1 and 3 mutants (ie, Q92R-SEC61A1, V67G-SEC61A1, and V85D-SEC61A1) were treated for 6 days with 1% DMSO. Both Q92R-SEC61A1– and V67G-SEC61A1–producing HL-60 cells generated fewer CD11b+CD16+ cells on differentiation compared with WT-SEC61A1 and V85D-SEC61A1 (Fig 6, A and B and see Fig E12 in this article’s Online Repository at www.jacionline.org). In addition, expression of CD16 on terminally differentiated cells was significantly diminished only for the neutropenia-associated mutations Q92R-SEC61A1 and V67G-SEC61A1, corresponding to the reduced CD16 expression observed on primary patient neutrophils (Fig 6, C and D). Hematoxylin and eosin staining confirmed that the surface markers chosen to distinguish terminally differentiated cells corresponded to neutrophil-like morphologic changes (Fig 6, E and F). Finally, to assess whether the observed upregulation of the UPR in primary cells could also be recapitulated by using the same model, we treated the stably transduced HL-60 cell line for 24 hours with 1 μM thapsigargin. This treatment failed to significantly upregulate CHOP or BiP in WT transduced cells but clearly showed increased ER stress in all 3 mutant cell lines (Fig 6, G), verifying that increased susceptibility to ER stress is a direct result of defective Sec61α1. Together, these data validate the role of SEC61A1 mutations in SCN.
Fig 6.
Mutant SEC61A1 is directly responsible for ER stress and a genotype-specific arrest in neutrophilic differentiation. A, Mean percentage of undifferentiated, CD11b+CD16–, and CD11b+CD16+ HL-60 cells stably transduced as indicated on day 6 (D6) of differentiation by using 1% DMSO (n = 7). B, Percentage of CD11b+CD16+ HL-60 cells from (A) gated on live, GFP+ cells on day 6 of differentiation (n = 7). C, Mean fluorescence index (MFI) of CD16 on CD16+ HL-60 cells gated on live GFP+ cells on day 6 of differentiation (n = 7). Paired t test. D, MFI of CD16 on CD16+ neutrophils isolated from whole blood or bone marrow (BM) from the HC and P normalized to WT MFI within each independent repeat. E, Quantification of hematoxylin and eosin (H&E) staining of untreated and 1% DMSO-treated stably transduced HL-60 cells for 6 days from representative repeat in (A). F, Representative H&E staining from differentiation in (A-C). The symbol # indicates promyelocyte-like, the symbol < means myelocyte-like, the symbol % means metamyelocyte-like, and the symbol ∗ means terminally differentiated cell. G, mRNA expression of DDIT3 (CHOP [left]) and HSPA5 (BiP [right]) normalized to 18S in transduced HL-60 cells left untreated or treated with 1 μM thapsigargin for 24 hours. Each fold change represents treated over average of untreated (n = 8). Unpaired t test.
Discussion
Here we have reported specific mutations in SEC61A1 as a novel genetic cause of autosomal dominant SCN. To date, including in our study, 5 kindreds harboring pathogenic mutations in SEC61A1 have been reported, with diverse clinical phenotypes including late-onset ADTKD (2 of 5),21 primary antibody deficiency (2 of 5), and SCN (2 of 5).21 Although SCN was previous observed in 1 family with ADTKD, causality with the SEC61A1 mutation was not investigated.21 Although our index patient first presented with a nonsyndromic form of SCN, caution is warranted as ultrasound imaging guided by her genetic diagnosis revealed small bilateral kidneys. Considering that clinically evident renal disease in patients with mutations in SEC61A1 occurred later in life, we propose that patients with mutations in SEC61A1 be serially monitored for renal dysfunction, regardless of the initial clinical phenotype. The recent report on CVID prompted in-depth immunophenotyping of the B-cell compartment, revealing impaired B-cell maturation. However, in contrast to what has been observed patients with SEC61A1 mutation and CVID, we noted an elevated plasmablast number and persistent hypergammaglobulinemia, indicating that although the Q92R mutation affects other leukocyte subsets, the B-cell phenotype remains distinct from that reported in patients with CVID and mutant SEC61A1. The origin of clinical diversity in patients with SEC61A1 mutation is currently unclear. With our present patient set, a particular phenotype cannot be predicted on the basis of location or nature of the mutation. Haploinsufficiency has been formally demonstrated in primary cells from patients harboring the p.E381∗ nonsense or p.Q92R missense mutation. Likewise, in vitro silencing of SEC61A1 in relevant cell lines induces the terminal UPR. Structural predictions and transient in vitro expression of other reported missense mutations indicate that these other mutations also have a detrimental impact on protein stability, further implicating haploinsufficiency as a disease mechanism. Potential dominant effects on protein function are more challenging to ascertain, and combined with variation in the remaining level of protein, they potentially explain the emerging clinical diversity. Our data support genotype-specific effects contributing to the clinical presentation, with only the neutropenia-associated mutations and not the CVID-associated mutation showing defects in our HL-60 model of neutrophilic maturation. This suggests that the clinical heterogeneity may be driven by dominant separation-of-function mutations in SEC61A1 rather than by secondary polymorphisms or externalities.
SEC61A1-related disease was shown to result from both a quantitative and qualitative defect, including protein instability, disturbed protein translocation, and dysregulated calcium homeostasis, which may also affect protein folding. Considering the function of the Sec61 complex, it is reasonable to hypothesize that such disruption will lead to insurmountable ER stress with initiation of the terminal UPR. Analogous to what has been observed in patients with ELANE mutation and SCN, the observed granulocytic differentiation arrest is cell intrinsic, and successful recapitulation of the maturation defect in human cell lines revealed a mechanistic pathway of mutation-induced upregulation of BiP and proapoptotic CHOP, supporting a model of increased UPR.41,42 Notably, single-cell RNA sequencing of myeloid progenitors further supported increased ER stress and implicated secondary mitochondrial dysfunction as a potential pathogenic mechanism in SCN, analogous to HAX1 and AK2 deficiency.41 It is becoming increasingly evident that ER-mitochondrial exchange of Ca2+ and other metabolites is involved in the regulation of mitochondrial bioenergetics and apoptotic signaling, sensitizing mitochondria to the effects of ER stress.39,43 The early adaptive ER stress response is aimed at increasing Ca2+ flux into mitochondria to promote mitochondrial respiratory chain activity with increased ATP synthesis.43 However, chronic ER stress reduces mitochondrial respiration and cellular ATP production, with depletion of ER Ca2+ stores and ultimately apoptosis, with the Sec61 complex involved in the maintenance of the ER ATP supply.16 Furthermore, it was shown that when Sec61 ER import function is limiting, some precursors of secretory proteins may be redirected for aberrant mitochondrial targeting to the translocase of the inner mitochondrial membrane, dissipating the membrane potential.44
Our data provide further support for a link between ER stress and SCN. Although the UPR is a feature of all cell types, the reoccurring theme of SCN in patients with mutated UPR genes indicates that the differentiating myeloid progenitor is highly susceptible to ER stress, a feature shared with plasma cells, pancreatic β-cells, and the developing kidney. Defects in this shared pathway thus manifest in specific pathologies. In the context of the CVID phenotype, previous work validated upregulation of the UPR in multiple myeloma cell lines stably expressing V85D.22 Plasma cells undergo expansion of their ER to accommodate the massive production of immunoglobulins, with constitutive activation of the UPR key to their survival.45 Defects in the UPR can therefore lead to impaired plasma cell differentiation and survival. Likewise, the compensatory insulin upregulation in diabetic pancreatic β-cells is thought to drive the UPR, resulting in β-cell decline.46 Clustered regularly interspaced short palindromic repeat–mediated knockout of the SEC61A1 ortholog sec61al2 in zebrafish established involvement of SEC61A1 in the normal development of the pronephros.21 However, with missense mutations across clinically distinct phenotypes demonstrating upregulation of the UPR in our study, it has become apparent that ER stress alone may not reconcile the emerging clinical diversity. Protein translocation of Sec61α1-dependent substrates may be differentially affected by the different mutations driving cell type–specific defects. Autosomal dominant REN mutations affect the signal sequence, thereby impairing ER translocation and processing of nascent preprorenin. This results in reduced renin levels in juxtaglomerular cells and exposure to chronic ER stress with detrimental effects on cell survival that ultimately lead to nephron loss.47,48 As previously speculated, SEC61A1 mutations in ADTKD may also specifically impair the translocation of preprorenin, thereby resulting in a clinical phenotype reminiscent of mutations in REN itself.22 With regard to SCN, transport of NE, which is encoded by the most commonly mutated gene in SCN (ELANE), is also shown to be Sec61α1 dependent (see Fig E3, B). The mechanism for the divergent phenotypes remains to be fully elucidated, and it may be worthwhile to specifically study the transport of such substrates to establish a functional genotype-phenotype correlation. Our current work demonstrates there are cell type–specific clinical phenotypes related to SEC61A1 mutations, expanding the clinical spectrum to include SCN.
Key messages.
-
•
Mutations in SEC61A1 cause autosomal dominant SCN.
-
•
Defects in Sec61α1 drive the UPR in primary cells.
Acknowledgments
We thank all the institutions that provided funding for this work. We also thank Dr Margaux Gerbaux for providing clinical information.
Footnotes
This work was supported by the VIB Grand Challenges program, the Research Foundation Flanders (Fonds voor Wetenschappelijk Onderzoek [FWO]) (grants G0A3218N and G080818N), the KU Leuven (C1 grant C16/17/010), the Biotechnology and Biological Sciences Research Council through Institute Strategic Program Grant funding BBS/E/B/000C0427 and BBS/E/B/000C0428, and the Biotechnology and Biological Sciences Research Council Core Capability Grant to the Babraham Institute. This project has received funding from the European Union's Horizon 2020 research and innovation programme under grant agreement No 779295. The Nikon A1R Eclipse Ti Confocal microscope was funded by a Hercules type 1AKUL/09/037 grant. The VIB Flow Core provided research support in this study. E.V.N. is funded by an SB grant (1S22716N). The following authors are funded by postdoctoral grants from the FWO: S.H.-B. (grant 1272517N), EP (grant 12S8918N), and VL (grant 12H3319N). I.M. is funded by the Jeffrey Modell Foundation and by the CSL Chair in Primary Immunodeficiencies. C.W. and I.M. are members of the European Reference Network for Rare Immunodeficiency, Autoinflammatory and Autoimmune Diseases (Project ID No. 739543). S.L. and R.Z. were supported by the DFG via IRTG1830.
Disclosure of potential conflict of interest: I. Meyts is funded by the CSL Chair in Primary Immunodeficiencies. The rest of the authors declare that they have no relevant conflicts of interest.
Contributor Information
Isabelle Meyts, Email: isabelle.meyts@uzleuven.be.
Adrian Liston, Email: adrian.liston@babraham.ac.uk.
Supplementary data
Fig E1.
Fig E2.
Fig E3.
Fig E4.
Fig E5.
Fig E6.
Fig E7.
Fig E8.
Fig E9.
Fig E10.
Fig E11.
Fig E12.
References
- 1.Skokowa J., Dale D.C., Touw I.P., Zeidler C., Welte K. Severe congenital neutropenias. Nat Rev Dis Prim. 2017;3:17032. doi: 10.1038/nrdp.2017.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Germeshausen M., Ballmaier M., Welte K. Incidence of CSF3R mutations in severe congenital neutropenia and relevance for leukemogenesis: results of a long-term survey. Blood. 2019;109:93–99. doi: 10.1182/blood-2006-02-004275. [DOI] [PubMed] [Google Scholar]
- 3.Makaryan V., Kelley M.L., Fletcher B., Bolyard A.A., Aprikyan A.A., Dale D.C. Elastase inhibitors as potential therapies for ELANE-associated neutropenia. J Leukoc Biol. 2017;102:1143–1151. doi: 10.1189/jlb.5A1016-445R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donadieu J., Beaupain B., Mahlaoui N., Bellanné-chantelot C. Epidemiology of congenital neutropenia. Hematol Oncol Clin North Am. 2013;27:1–17. doi: 10.1016/j.hoc.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 5.Xia J., Bolyard A.A., Rodger E., Stein S., Aprikyan A.A., Dale C. Prevalence of mutations in ELANE, GFI1, HAX1, SBDS, WAS, and G6PC3 in patients with severe congenital neutropenia. Br J Haematol. 2009;147:535–542. doi: 10.1111/j.1365-2141.2009.07888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grenda D.S., Murakami M., Ghatak J., Xia J., Boxer L.A., Dale D. Mutations of the ELA2 gene found in patients with severe congenital neutropenia induce the unfolded protein response and cellular apoptosis. Blood. 2007;110:4179–4187. doi: 10.1182/blood-2006-11-057299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dannenmann B., Zahabi A., Mir P., Oswald B., Bernhard R., Klimiankou M. Human iPSC-based model of severe congenital neutropenia reveals elevated UPR and DNA damage in CD34+ cells preceding leukemic transformation. Exp Hematol. 2019;71:51–60. doi: 10.1016/j.exphem.2018.12.006. [DOI] [PubMed] [Google Scholar]
- 8.Boztug K., Järvinen P.M., Salzer E., Racek T., Mönch S., Garncarz W. JAGN1 deficiency causes aberrant myeloid cell homeostasis and congenital neutropenia. Nat Genet. 2014;46:1021–1027. doi: 10.1038/ng.3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boztug K., Appaswamy G., Ashikov A., Schäffer A.A., Salzer U., Diestelhorst J. A syndrome with congenital neutropenia and mutations in G6PC3. N Engl J Med. 2009;360:32–43. doi: 10.1056/NEJMoa0805051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vilboux T., Lev A., Malicdan M.C.V., Simon A.J., Sc B., Järvinen P. A congenital neutrophil defect syndrome associated with mutations in VPS45. N Engl J Med. 2013;369:54–65. doi: 10.1056/NEJMoa1301296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kolehmainen J., Black G.C.M., Saarinen A., Chandler K., Clayton-Smith J., Traskelin A. Cohen syndrome is caused by mutations in a novel gene, COH1, encoding a transmembrane protein with a presumed role in vesicle-mediated sorting and intracellular protein transport. Am J Hum Genet. 2003;72:1359–1369. doi: 10.1086/375454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bellann C., Schmaltz-panneau B., Marty C., Fenneteau O., Callebaut I. Mutations in the SRP54 gene cause severe congenital neutropenia as well as Shwachman-Diamond-like. syndrome. 2019;132:1318–1331. doi: 10.1182/blood-2017-12-820308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finch A.J., Hilcenko C., Basse N., Drynan L.F., Simpson P., Goyenechea B. Uncoupling of GTP hydrolysis from eIF6 release on the ribosome causes Shwachman-Diamond syndrome. Genes Dev. 2011;25:917–929. doi: 10.1101/gad.623011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tummala H., Walne A.J., Williams M., Bockett N., Collopy L., Cardoso S. DNAJC21 mutations link a cancer-prone bone marrow failure syndrome to corruption in 60S ribosome subunit maturation. Am J Hum Genet. 2016;99:115–124. doi: 10.1016/j.ajhg.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stepensky P., Kim K.H., Abuzaitoun O., Bautista-santos A., Simanovsky N., Siliqi D. Mutations in EFL1, an SBDS partner, are associated with infantile pancytopenia, exocrine pancreatic insufficiency and skeletal anomalies in a Shwachman-Diamond like syndrome. J Med Genet. 2017;54:558–566. doi: 10.1136/jmedgenet-2016-104366. [DOI] [PubMed] [Google Scholar]
- 16.Lang S., Pfeffer S., Lee P., Cavalié A., Helms V., Forster F. An update on Sec61 channel functions, mechanisms, and related diseases. Front Physiol. 2017;8:887. doi: 10.3389/fphys.2017.00887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Linxweiler M., Schick B., Zimmermann R. Let’s talk about Secs: Sec61, Sec62 and Sec63 in signal transduction, oncology and personalized medicine. Signal Transduct Target Ther. 2017;2:17002. doi: 10.1038/sigtrans.2017.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davila S., Furu L., Gharavi A.G., Tian X., Onoe T., Qian Q. Mutations in SEC63 cause autosomal dominant polycystic liver disease. Nat Genet. 2004;36:575–577. doi: 10.1038/ng1357. [DOI] [PubMed] [Google Scholar]
- 19.Besse W., Dong K., Choi J., Punia S., Fedeles S.V., Choi M. Isolated polycystic liver disease genes define effectors of polycystin-1 function. J Clin Invest. 2017;127:3558. doi: 10.1172/JCI96729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Synofzik M., Haack T.B., Kopajtich R., Gorza M., Rapaport D., Greiner M. Absence of BiP Co-chaperone DNAJC3 causes diabetes mellitus and multisystemic neurodegeneration. Am J Hum Genet. 2014;95:689–697. doi: 10.1016/j.ajhg.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bolar N.A., Golzio C., Živná M., Hayot G., Van Hemelrijk C., Schepers D. Heterozygous loss-of-function SEC61A1 mutations cause autosomal-dominant tubulo-interstitial and glomerulocystic kidney disease with anemia. Am J Hum Genet. 2016;99:174–187. doi: 10.1016/j.ajhg.2016.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schubert D., Klein M.C., Hassdenteufel S., Caballero-Oteyza A., Yang L., Proietti M. Plasma cell deficiency in human subjects with heterozygous mutations in Sec61 translocon alpha 1 subunit (SEC61A1) J Allergy Clin Immunol. 2018;141:1427–1438. doi: 10.1016/j.jaci.2017.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Nieuwenhove E., Garcia-Perez J.E., Helsen C., Rodriguez P.D., van Schouwenburg P.A., Dooley J. A kindred with mutant IKAROS and autoimmunity. J Allergy Clin Immunol. 2018;142:699–702.e12. doi: 10.1016/j.jaci.2018.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krieger E., Vriend G. YASARA View - molecular graphics for all devices - from smartphones to workstations. Bioinformatics. 2014;30:2981–2982. doi: 10.1093/bioinformatics/btu426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dudek J., Lang S., Schorr S., Linxweiler J., Greiner M., Zimmermann R. Analysis of protein translocation into the endoplasmic reticulum of human cells. Methods Mol Biol. 2013;1033:285–299. doi: 10.1007/978-1-62703-487-6_18. [DOI] [PubMed] [Google Scholar]
- 26.Lang S., Benedix J., Fedeles S.V., Schorr S., Schirra C., Schauble N. Different effects of Sec61α , Sec62 and Sec63 depletion on transport of polypeptides into the endoplasmic reticulum of mammalian cells. J Cell Sci. 2012;125(pt 8):1958–1969. doi: 10.1242/jcs.096727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Becker T., Bhushan S., Jarasch A., Armache J., Funes S., Jossinet F. Structure of monomeric yeast and mammalian sec61 complexes interacting with the translating ribosome. Science. 2009;326:1369–1373. doi: 10.1126/science.1178535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Serrano L., Neira J., Sancho J., Fersht A.R. Effect of alanine versus glycine in α-helices on protein stability. Nature. 1992;4:453–455. doi: 10.1038/356453a0. [DOI] [PubMed] [Google Scholar]
- 29.Thompson J.S., Bixler S.A., Qian F., Vora K., Scott M.L., Cachero T.G. BAFF-R, a newly identified TNF receptor that specifically interacts with BAFF. Science. 2001;293:2108–2111. doi: 10.1126/science.1061965. [DOI] [PubMed] [Google Scholar]
- 30.Michel T., Poli A., Cuapio A., Briquemont B., Iserentant G., Ollert M. Human CD56 bright NK cells: an update. J Immunol. 2019;196:2923–2931. doi: 10.4049/jimmunol.1502570. [DOI] [PubMed] [Google Scholar]
- 31.Cottineau J., Kottemann M.C., Lach F.P., Kang Y., Vély F., Deenick E.K. Inherited GINS1 deficiency underlies growth retardation along with neutropenia and NK cell deficiency. J Clin Invest. 2017;127:1991–2006. doi: 10.1172/JCI90727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tillmanns S., Otto C., Jaffray E., Du Roure C., Bakri Y., Vanhille L. SUMO modification regulates MafB-driven macrophage differentiation by enabling Myb-dependent transcriptional repression. Mol Cell Biol. 2007;27:5554–5564. doi: 10.1128/MCB.01811-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krishnaraju B.K., Hoffman B., Liebermann D.A. The zinc finger transcription factor Egr-1 activates macrophage differentiation in M1 myeloblastic leukemia cells. Blood. 1998;92:1957–1966. [PubMed] [Google Scholar]
- 34.Rajarathnam K., Schnoor M., Richardson R.M., Rajagopal S. How do chemokines navigate neutrophils to the target site: dissecting the structural mechanisms and signaling pathways. Cell Signal. 2019;54:69–80. doi: 10.1016/j.cellsig.2018.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Travelli C., Colombo G., Mola S., Genazzani A.A., Porta C. NAMPT: a pleiotropic modulator of monocytes and macrophages. Pharmacol Res. 2018;135:25–36. doi: 10.1016/j.phrs.2018.06.022. [DOI] [PubMed] [Google Scholar]
- 36.Rojas-Rivera D., Hetz C. TMBIM protein family: ancestral regulators of cell death. Oncogene. 2015;34:269–280. doi: 10.1038/onc.2014.6. [DOI] [PubMed] [Google Scholar]
- 37.Morishima T., Watanabe K., Niwa A., Hirai H., Saida S., Tanaka T. Genetic correction of HAX1 in induced pluripotent stem cells from a patient with severe congenital neutropenia improves defective granulopoiesis. Haematologica. 2014;99:19–27. doi: 10.3324/haematol.2013.083873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hauck F., Klein C. Pathogenic mechanisms and clinical implications of congenital neutropenia syndromes. Curr Opin Allergy Clin Immunol. 2013;13:596–606. doi: 10.1097/ACI.0000000000000014. [DOI] [PubMed] [Google Scholar]
- 39.Klein C. Genetic defects in severe congenital neutropenia: emerging insights into life and death of human neutrophil granulocytes. Annu Rev Immunol. 2011;29:399–413. doi: 10.1146/annurev-immunol-030409-101259. [DOI] [PubMed] [Google Scholar]
- 40.Six E., Lagresle-Peyrou C., Susini S., De Chappedelaine C., Sigrist N., Sadek H. AK2 deficiency compromises the mitochondrial energy metabolism required for differentiation of human neutrophil and lymphoid lineages. Cell Death Dis. 2015;6 doi: 10.1038/cddis.2015.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nayak R.C., Trump L.R., Aronow B.J., Myers K., Mehta P., Kalfa T. Pathogenesis of ELANE-mutant severe neutropenia revealed by induced pluripotent stem cells. J Clin Invest. 2015;125:3103–3116. doi: 10.1172/JCI80924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu H., Tian M., Ding C., Yu S. The C/EBP homologous protein (CHOP) transcription factor functions in endoplasmic reticulum stress-induced apoptosis and microbial infection. Front Immunol. 2019;9:3083. doi: 10.3389/fimmu.2018.03083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rainbolt T.K., Saunders J.M., Wiseman R.L. Stress-responsive regulation of mitochondria through the ER unfolded protein response. Trends Endocrinol Metab. 2014;25:528–537. doi: 10.1016/j.tem.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 44.Pfeiffer N.V., Dirndorfer D., Lang S., Resenberger U.K., Restelli L.M., Hemion C. Structural features within the nascent chain regulate alternative targeting of secretory proteins to mitochondria. EMBO. 2013;32:1036–1051. doi: 10.1038/emboj.2013.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cenci S., Sitia R. Managing and exploiting stress in the antibody factory. FEBS Lett. 2007;581:3652–3657. doi: 10.1016/j.febslet.2007.04.031. [DOI] [PubMed] [Google Scholar]
- 46.Liston A., Todd J.A., Lagou V. Beta-cell fragility as a common underlying risk factor in type 1 and type 2 diabetes. Trends Mol Med. 2017;23:181–194. doi: 10.1016/j.molmed.2016.12.005. [DOI] [PubMed] [Google Scholar]
- 47.Zivna M., Hůlková H., Matignon M., Hodanová K., Vylet’al P., Kalbácová M. Dominant renin gene mutations associated with early-onset hyperuricemia, anemia, and chronic kidney failure. Am J Hum Genet. 2009;85:204–213. doi: 10.1016/j.ajhg.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schaeffer C., Izzi C., Vettori A., Pasqualetto E., Cittaro D., Lazarevic D. Autosomal dominant tubulointerstitial kidney disease with adult onset due to a novel renin mutation mapping in the mature protein. Sci Rep. 2019;9:11601. doi: 10.1038/s41598-019-48014-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.