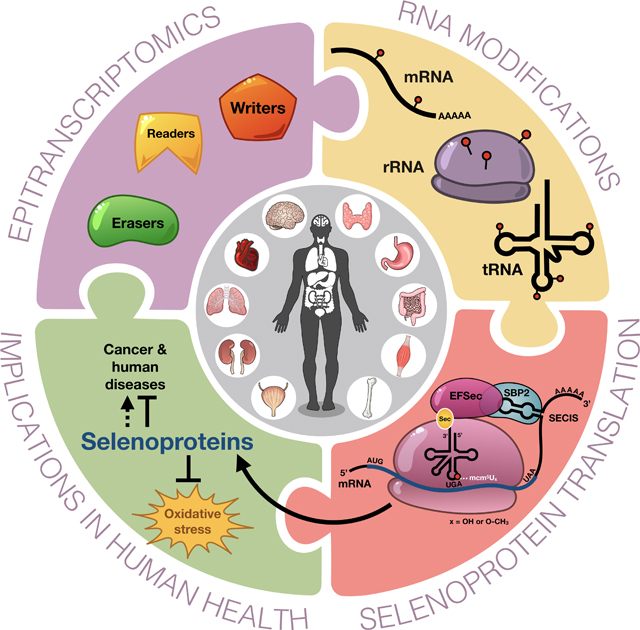
Abstract
Here we highlight the role of epitranscriptomic systems in post-transcriptional regulation, with a specific focus on RNA modifying writers required for the incorporation of the 21st amino acid selenocysteine during translation, and the pathologies linked to epitranscriptomic and selenoprotein defects. Epitranscriptomic marks in the form of enzyme-catalyzed modifications to RNA have been shown to be important signals regulating translation, with defects linked to altered development, intellectual impairment, and cancer. Modifications to rRNA, mRNA and tRNA can affect their structure and function, while the levels of these dynamic tRNA-specific epitranscriptomic marks are stress-regulated to control translation. The tRNA for selenocysteine contains five distinct epitranscriptomic marks and the ALKBH8 writer for the wobble uridine (U) has been shown to be vital for the translation of the glutathione peroxidase (GPX) and thioredoxin reductase (TRXR) family of selenoproteins. The reactive oxygen species (ROS) detoxifying selenocysteine containing proteins are a prime examples of how specialized translation can be regulated by specific tRNA modifications working in conjunction with distinct codon usage patterns, RNA binding proteins and specific 3’ untranslated region (UTR) signals. We highlight the important role of selenoproteins in detoxifying ROS and provide details on how epitranscriptomic marks and selenoproteins can play key roles in and maintaining mitochondrial function and preventing disease.
Graphical Abstract
1.1. Intro to RNA and Epitranscriptomics
Living systems use regulated gene expression for survival under stressful conditions. Gene expression involves transcription of the DNA message into RNA and translation of messenger RNA (mRNA) to proteins within the ribosome. During translation, transfer RNA (tRNA) carry amino acids to the ribosome where they are incorporated into a growing polypeptide via the direction of codon-anticodon interactions. Ribosomal RNA (rRNA), the predominant component of ribosomes, catalyzes peptide bond formation between two amino acids. While molecular mechanisms of stress responses at the epigenetic level function to modulate transcription and have been well studied and reviewed [1–3], epitranscriptomic regulation, using RNA modifications, to control the translation of stress response proteins has only recently been identified as an important determinant of protein expression[4–7]. In this review we highlight RNA modifications on different RNAs, detail tRNA modification systems and their role in translating selenoproteins. We also discuss recent epitranscriptomic findings on the control of stress induced protein expression by RNA modifications, and implications for mitochondrial function and selenoprotein based detoxification of reactive oxygen species (ROS), with defects linked to human disease.
RNA modifications are catalyzed by specific enzymes or RNA-protein complexes and may occur on the base and/or ribose sugar of the canonical adenosine (A), cytosine (C), guanosine (G) and uridine (U) nucleosides. The modifications can include simple changes such as deamination, methylation or acetylation, or hypermodifications such as queosine or N6-threonylcarbamoyladenosine (t6A) on tRNA [8]. The first mRNA modifications in the coding region were discovered in the 1970s [9] and were found to accelerate pre-mRNA processing and transport in HeLa cells [10]. In the 1980s, apolipoprotein B (apoB) mRNA was found to contain a tissue-specific cytidine-to-uridine base modification in the CAA codon at position 2153 which was not genomically encoded, resulting in a UAA stop codon and translational termination at this site [11,12]. The C to U change was present only in human and rabbit intestinal mRNA and resulted in a truncated version of the protein (termed APO-B48) while other tissues such as the liver did not have the edit and resulted in the full protein (APO-B100). It was the first example of a tissue-specific modification of a mRNA nucleotide resulting in two different proteins from the same primary transcript [12]. The enzyme APOBEC-1 (apolipoprotein B editing enzyme catalytic subunit 1) was found to catalyze the C to U editing of apoB, along with other factors [13–15], and APOBEC-1 was one of the early players which conceptually led to the modern field of epitranscriptomics.
Another early discovery of proteins that catalyze irreversible RNA modifications includes the ADAR (adenosine deaminase acting on RNA) family of enzymes, which deaminate adenosine on mRNA to inosine primarily in the 3’ UTR and intronic regions, suggesting a predominantly regulatory role of this edit (Fig. 1) [16]. While bioinformatics studies have identified more than 12,000 new A-to-I editing sites, only ~30 of those involve known protein-coding targets while the rest are located in noncoding regions such as introns and 5’ and 3’ UTRs [17,18]. The conversion of A to I is the most common type of RNA edit, and it serves critical functions in RNA processing, alternative splicing, protein function, development and immune response [19–22]. A related family of adenosine deaminases, the adenosine deaminases acting on tRNAs (ADATs), catalyze A to I at position 34 in 7–8 different cytosolic tRNAs (Fig. 2A) [25]. Inosine is also found in its methylated state (1-methylinosine) at position 37 and 57 [8]. The position 34 edit from A to I promotes pairing with codons ending in U, A and C [26].
Figure 1. Common epitranscriptomic marks on mRNA and rRNA.
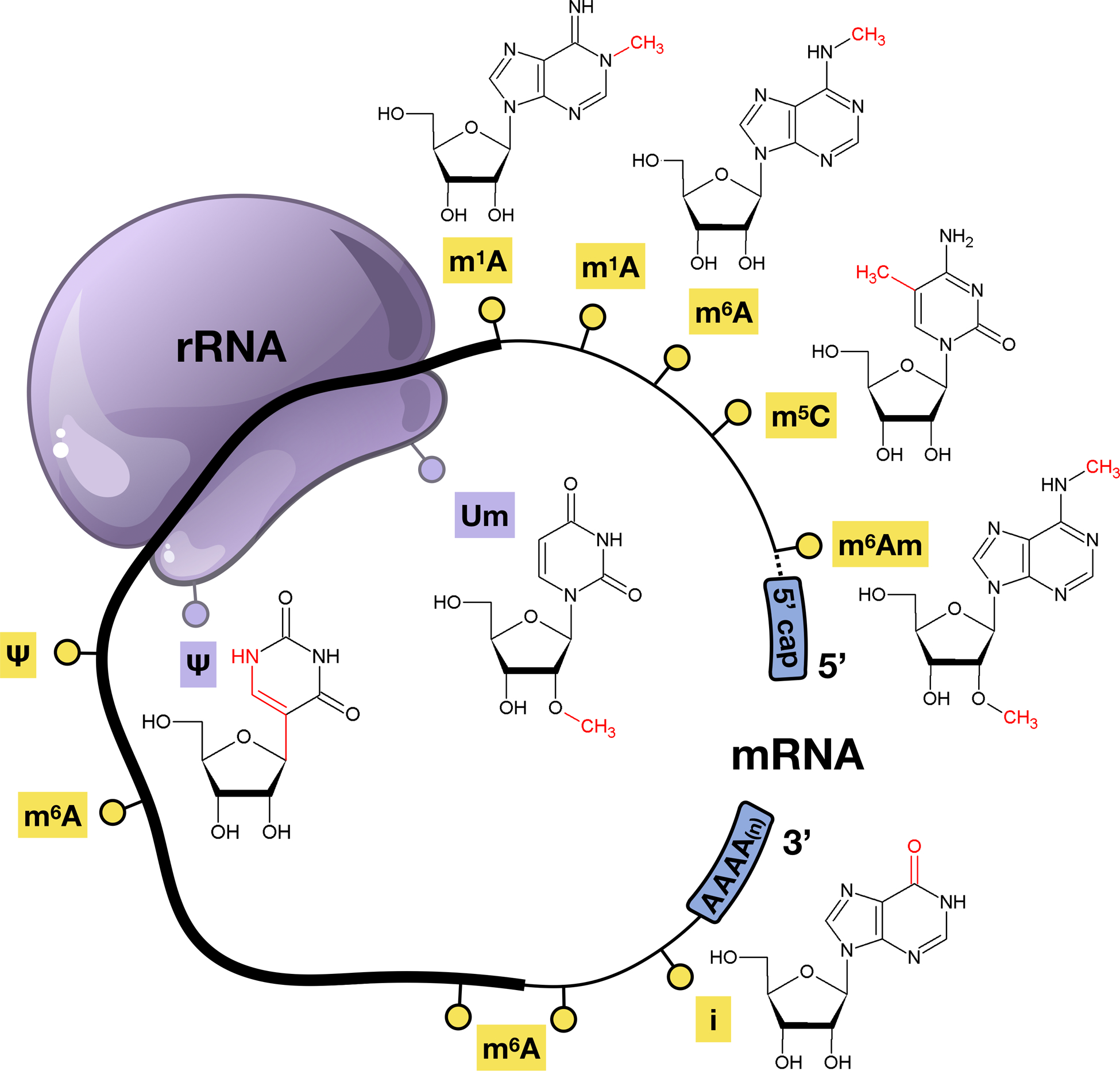
Six RNA modifications that are found on rRNA and mRNA, with the open reading frame (thick line) of mRNA and the 5’ and 3’ untranslated region (UTR)(thin line) highlighted to show where epitranscriptomic marks can be found.
Figure 2. Some epitranscriptomic marks found on tRNA.
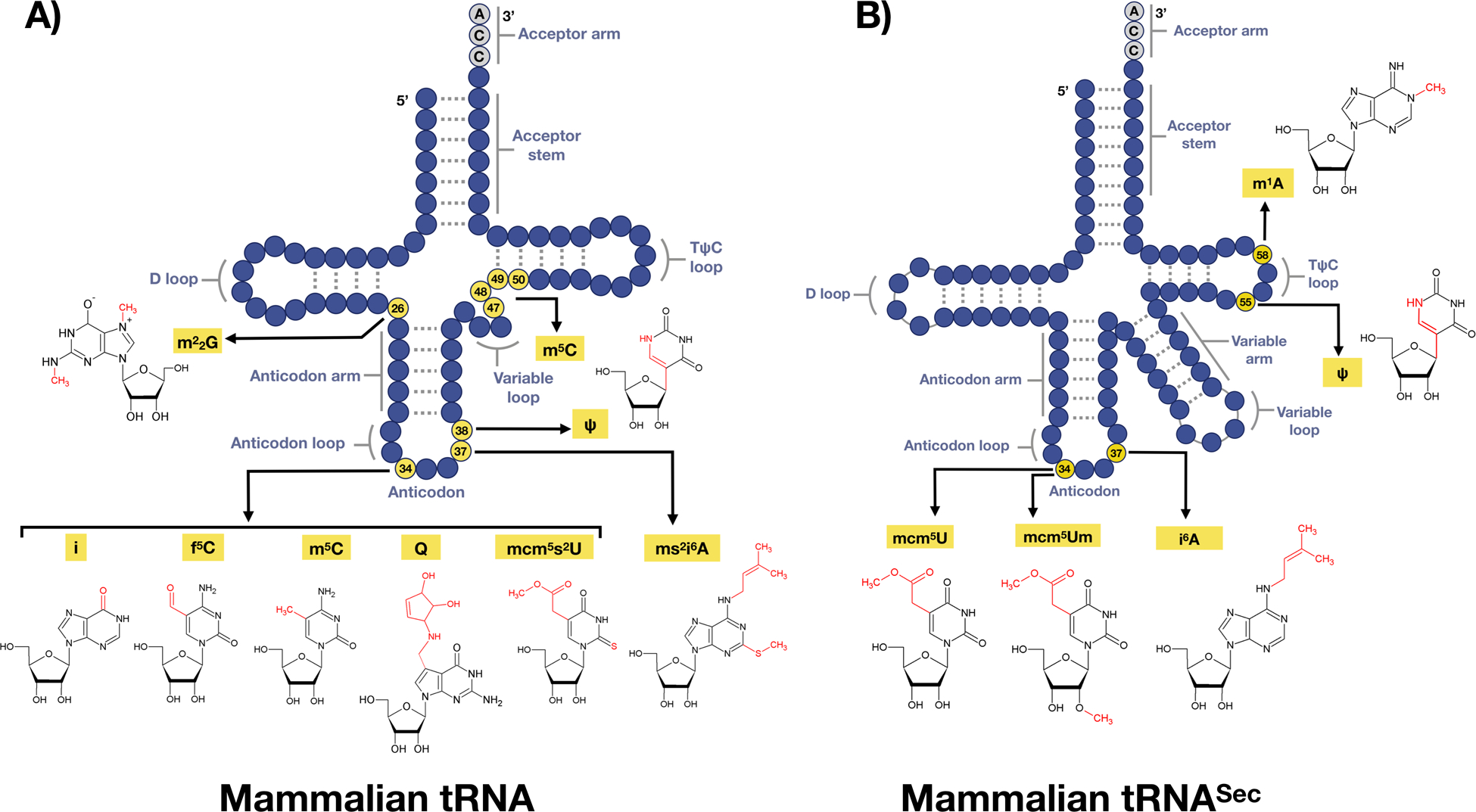
(A) There are on average 13 modifications on each tRNA species, with six epitranscriptomics marks highlighted. (B) tRNASec structure with 5 nucleoside modifications found on 4 positions.
Enzyme-catalyzed modifications can be found on almost every type of RNA and are able to fine-tune many aspects of gene expression. Over 140 post-transcriptional modifications across all types of RNA species have been identified to date (Fig. 1, Fig. 2), and these modifications directly influence RNA structure, function and have been linked to disease [8]. Various enzymes can add (“writers”), remove (“erasers”), and act upon (“readers”) RNA modifications, which can influence gene expression by altering charge, base-pairing potential, secondary structure, and protein-RNA interactions.
1.2. Epitranscriptomic marks on messenger RNA (mRNA) and ribosomal RNA (rRNA)
Epitranscriptomic marks on mRNA include I, N6-methyladenosine (m6A), N1-methyladenosine (m1A), 5-methylcytidine (m5C), and pseudouridine (Ψ), with m6A and Ψ being the most abundant modifications based on mass spectrometry [27] (Fig. 1). m6A, m5C, and m1A have been identified within the mRNA coding sequence: [28–31]. Much of our understanding of readers, writers and erasers comes from studies on the N6-methyladenosine (m6A) posttranscriptional modification on mRNA (Fig. 1), [32–39]. The m6A epitranscriptomic mark can be located in multiple areas of the transcript, with common enrichment occurring near the stop codon in the open reading frame (ORF) as well as in the 3’ UTR [28,40]. The m6A writer complex consists of the Methyltransferase Like 3 (METTL3) and 14 (METTL14) proteins, which together form a METTL3-METTL14 heterodimer with additional accessory proteins such as Wilms tumor 1(WT1)-associated protein (WTAP), all together forming the N6-methyltransferase complex [41–43]. Dysregulation of m6A writer enzyme expression can promote tumor progression in glioblastoma stem cells [44], embryonic lethality [45], and disruption of neurogenesis [46].
The m6A modification was recently found to be dynamically regulated and control meiosis and stem cell differentiation and proliferation [47], heat shock response [48], DNA damage response [49], and tumorigenesis [50]. Dynamic regulation of epitranscriptomic marks is an emerging theme and global increases or decreases in specific RNA modifications can occur by changes in RNA level or regulated activity of writers and erasers. In mammals, m6A and its 2’-O-ribose methylated variant m6Am, have been shown to be dynamically regulated in the mouse brain in response to acute stress as well as in writer and eraser deficient mouse models [33]. In humans dysregulated m6A has been identified in major depressive disorder patients [51]. Notably, m6A was found to be reversible by the action of fat mass and obesity-associated protein (FTO) and alkylation repair homolog 5 (ALKBH5), both exhibiting robust m6A demethylase activity [52,53]. The addition and removal of m6A plays a dynamic role in a number of biological processes including mRNA transport, splicing, association with RNA binding proteins, UV-induced DNA damage response, oncogenesis and drug response [48,49,53,54]. ALKBH5, is expressed in many tissues, but most abundantly in the testes where its demethylase activity promotes mouse spermatogenesis and fertility [53]. ALKBH5 has also been shown to participate in cancer pathogenesis [55]. Patient-derived gliobastoma stem-like cells (GSCs) show high ALKBH5 expression, with its m6A demethylation of transcripts for the transcription factor FOXM1 driving tumorgenesis. Suppressing ALKBH5 also suppresses FOXM1 translation and reduces GSC proliferation [55]. Furthermore, ALKBH5 demethylase activity is important for antiviral innate immunity. DEAD-box helicase member 46 (DDX46) is a regulator of the immune response after viral infection, and recruits ALKBH5 to the nucleus. As the m6A modification is normally required for export of viral mRNA from the nucleus and for protein translation, ALKBH5 traps m6A modified antiviral transcripts in the nucleus by removing this mark [56].
The Ψ modification is the isomerization of the uridine base and was first characterized in both rRNA and tRNA [57–59]. This modification is also the most abundant in a wide range of cellular RNAs and highly conserved through evolution. However, the identification of this modification in mRNA was not discovered until recently with PseudoU-seq technology. PseudoU-seq technology is the high-throughput, single nucleotide resolution transcriptome mapping that allows the exploitation of N-cyclohexyl-N’–(2-morpholinoethyl)-carbodiimide metho-p-toluenesulphonate (CMC) modification to determine the locations of Ψ [60]. The presence of an extra hydrogen bond donor on the non-Watson-Crick edge of pseudouridine affects structure to increase the thermodynamic stability of the Ψ-A base pairs by improving base pairing [61,62]. The Ψ modification is also known to affect RNA secondary structure and alter the stop codon read through [63,64]. In the case of mRNA, Ψ is catalyzed by the pseudouridine synthase (PUS) enzymes, specifically PUS1–4, PUS6, PUS7 and PUS9 [65].
Human ribosomes contain 80 ribosomal proteins and four rRNA chains, the 28S, 5S and 5.8S rRNAs in the 60S subunit, and 18S rRNA in the 40S subunit [66]. Among the human rRNA chemical modifications, approximately 95% are 2’-OH ribose methylations (2’-O-Me) and conversions of uridine (U) to pseudouridine (Ψ), (Fig. 2) while the remaining 5% of modifications are predicted to contain methylated bases and other modifications [67–69]. The exact biological effects of rRNA modifications are still under investigation, but the prevailing hypothesis is that they fine-tune translation, structure, and possibly ribosomal biogenesis [70]. There are 97 predicted Ψ in mammalian rRNA, including 28S, 18S, 5.8S and 5S rRNAs, and they predominately function in protein translation efficiency [71]. Pseudouridylation in rRNA is dependent on the Box H/ACA RNAs that are noncoding and fold into a hairpin-hinge-hairpin-tail secondary structure. There are few links between mammalian Ψ on rRNA and stress response and/or disease, with some links to cancer, mitochondrial myopathy and sideroblastic anemia [72,73]. Mutations in PUS proteins and box H/ACA RNAPs are correlated with a range of diseases such as sideroblastic anemia, pituitary adenoma and mitochondrial myopathies [72,74].
1.3. Epitranscriptomic marks on transfer RNA (tRNA)
tRNAs are best known as the adaptor molecules of the translation machinery, in which they deliver the appropriate amino acids to the ribosomes according to the interaction of their anticodons with the codons of messenger RNAs (mRNAs). They are also critically involved in other cellular processes. Charged tRNAs act as amino acid donors not only to peptide chains during translation, but also to membrane lipids, peptidoglycan precursors, and the amino-terminus of proteins [75]. tRNAs also participate in stress responses and function as stress sensors [76]. For example, during nutrient deprivation eukaryotic cells respond by inhibition of protein synthesis, which can be mediated by tRNAs acting as signaling molecules. Starvation leads to limited availability of extracellular amino acids, which leads to accumulation of uncharged tRNAs in the cytosol. Uncharged tRNA can act as a signaling molecule by directly binding to the protein kinase GCN2 [77,78]. The now-activated GCN2 kinase phosphorylates eukaryotic initiation factor 2 (eIF2), which regulates translation in amino-acid starved cells and promotes synthesis of transcription factor GCN4, which activates amino acid synthesis genes. Interestingly, in budding yeast the GCN4-regulated transcripts exhibit a codon bias that can be linked to regulation by modified wobble uridines in tRNA [79].
tRNAs are extensively post-transcriptionally modified by epitranscriptomic writer enzymes, with these RNA modifications having structural and functional roles, as well as playing downstream roles in distinct biological processes. tRNA ribonucleoside modifications vary greatly in their type, location, and abundance. Ribonucleoside modifications are located throughout tRNA molecules and can affect tRNA stability, folding, transport, processing, localization and function [80]. On average, ~17% of the 80–90 ribonucleosides in tRNA are modified, ranging from relatively small structural changes, such as methylation to m5C or m1A, to highly intricate chemical alterations that give rise to hypermodified bases such as queuosine (Q) (Fig. 2). To highlight the importance of tRNA based epitranscriptomic marks, in the hyperthermophilic bacterium Thermus thermophilius, depletion of m1A results in a thermosensitive phenotype implying a role of m1A in temperature adaptation. Bacterial, yeast, mouse and human cells deficient in tRNA modifications have been shown to be sensitive to DNA damaging agents and agents that promote increased ROS [5,81–86]. Global studies have demonstrated that the levels of epitranscriptomic marks in cytosolic tRNA are dynamically regulated during cellular stress [4–6]. For example, in response to oxidizing agents such as γ-radiation, hydrogen peroxide (H2O2), tert-Butyl hydroperoxide (TBHP) or peroxynitrite (ONOO−), there is a significant increase in m5C, ncm5U, and i6A levels in yeast tRNA while the same modifications were unaffected in response to alkylating agents. In contrast, Um, mcm5U, mcm5s2U, m2G, and m1A modification levels increase in response to alkylating agents ethylmethanesulfonate (EMS), methylmethanesulfonate (MMS), isopropyl methanesulfonate (IMS) and N-methyl-N’-nitro-N-mitrosoguanidine (MNNG), and are conversely unaffected by treatment with oxidizing agents. The patterns of tRNA modifications are highly predictive and are responsive to distinct toxicants or stressors, and serve to reprogram translation to more effectively translate codon-biased transcripts into critical survival proteins in order to overcome the stress[4–6]. Stress-induced changes in tRNA have been observed in bacteria [7], yeast [4,87–90], mouse embryonic fibroblasts [91], human colorectal cancer cells [92] and melanoma cells [93].
The anticodon loops of nearly all tRNAs, including cytosolic and mitochondrial tRNAs are almost always modified [8,94]. Modifications on position 34 and 37 are especially important and promote translational fidelity and optimized codon-anticodon interaction within the ribosome [95]. The U34 modifications 5-methoxycarbony-methyl-uridine (mcm5U) and 5-methoxycarbonyl-methyl-2-thiouridine (mcm5s2U) (Fig. 2), are catalyzed by elongator (ELP 1–6) enzymes, Alkylation repair homolog 8 (ALKBH8) methyltransferase, and the Ubiquitin-related modifier 1 (URM1) pathway [96]. The mcm5U modification can be found on tRNAs encoding lysine, arginine, glycine, glutamine, glutamic acid and selenocysteine[97–100], while the mcm5s2U is found on tRNAs for lysine, arginine glutamine and glutamic acid [99–101]. In budding yeast, the mcm5U and mcm5s2U modifications play key roles in preventing protein synthesis errors, which include amino acid misincorporation and −1 frameshifts, with writer deficiencies activating heat shock and unfolded protein response (UPR) pathways[102]. Wobble uridine modifications at position 34 (U34) are important for maintaining proteome integrity and loss of these modifications perturbs cellular signaling and causes proteotoxic stress [103,104]. Yeast studies have shown that tRNA modifications play important roles in reading frame maintenance such that tRNAs deficient in wobble U modifications enter the ribosome A-site with decreased efficiency [105,106]. Many genes encoding tRNA modification enzymes are critical for cell function during conditions of stress as well as general function, growth and development [107,108]. We have previously shown that Alkbh8-deficient MEFs have an impaired ability to catalyze mcm5Um modifications in response to H2O2 on tRNA for selenocysteine (tRNASec) and are sensitive to oxidizing agents [91]. The tRNA methyltransferase ALKBH8 has been directly linked to intellectual disability as a consequence of severe impairment in wobble uridine modifications [109]. Deficiencies in other tRNA-modifying enzymes are also linked to severe and complex human diseases and wide tissue vulnerability [107]. Kirino et al., 2004 linked mutations in mitochondrial methyltransferases on the tRNALeu(UUR) gene which caused mitochondrial myopathy, lactic acidosis, encephalopathy, and a specific group of mitochondrial encephalomyopathic diseases that cause stroke-like episodes (MELAS) [110]. A polyadenylation factor I subunit 1 (CLP1) is a multifunctional kinase that modifies tRNA, mRNA, and siRNA and is important for RNA maturation. As one example, a mutation in CLP1 destabilizes the tRNA endonuclease complex and results in impaired pre-tRNA cleavage as well as impaired ligation in the final step of tRNA maturation. This is a direct consequence of decreased CLP1-catalyzed phosphorylation required during tRNA maturation. The downstream result is cerebellar neurodegeneration due to the depletion of mature tRNAs and accumulation of unspliced pre-tRNAs [111]. CDK5 regulatory associated protein 1-like 1 (CDKAL 1) has been widely studied and linked to type 2 diabetes. CDKAL 1 is a mammalian methylthiotransferase that biosynthesizes 2-methylthio-N6-threonylcarbmoyladenosine (ms2t6A) in tRNALys(UUU) allowing accurate translation of AAA and AAG codons. Mice that were deficient in CDKAL 1, showed a decrease in insulin secretion, pancreatic islet hypertrophy and impaired blood glucose control. Cdkal1−/− mice were also hypersensitive to a high fat diet which induced ER stress. This study unveiled a suggested molecular pathogenesis of type 2 diabetes in patients who carried Cdkal 1 risk alleles[112].
Position 37 on tRNA can carry several types of modifications, which vary among species. In eukaryotes, they include N6-isopentyladenosine (i6A) (Fig. 2), N6-threonylcarbamoyladenosine (t6A) and wybutosine (yW). tRNA isopentenyltransferase 1 (TRIT1) catalyzes the addition of i6A at position 37 on both cytosolic and mitochondrial tRNAs in humans (Fig. 3). Re-expression of functional TRIT1 in A549 lung cancer cells significantly reduced colony formation, implicating TRIT1 as a potential lung tumor suppressor [113]. i6A is further hypermodified to 2-methylthio-N6-isopentenyl-A37 (ms2i6A) in four mitochondrial DNA-encoded tRNAs, mt-tRNATrp, mt-tRNATyr, mt-tRNAPhe, and mt-tRNASer(UCN) (Fig. 3) by a nuclear-encoded, mitochondrial enzyme, Cdk regulatory subunit-associated protein 1 (CDK5RAP1) [114]. Absence of ms2i6A due to knock down of Cdk5rap1 decreases translation of mitochondrial respiratory chain subunits, complex I, III and IV, compromising proper respiratory complex assembly and decreasing oxidative phosphorylation (OXPHOS) [114]. Impairment of proper complex I and III translation in Cdk5rap1-null mice increases ROS leakage, and consequently, these mice are susceptible to oxidative stress and they exhibit accelerated myopathy and cardiac dysfunction under stressed conditions [114,115]. There are only 2 cytoplasmic tRNAs that carry i6A, cy-tRNASer(UCN) and tRNASec in mammalian cells and they are not hypermodified further [116]. The mt- tRNA t6A modification is also sensitive to CO2 as incubation of human embryonic kidney cells (HEK293T) in sodium bicarbonate-free tissue culture media significantly decreased t6A modifications which are rescued by sodium bicarbonate addition. The authors speculate that hypoxic conditions in solid tumors could also affect mt-tRNA t6A levels as mitochondrial CO2 is primarily provided by the TCA cycle whose activity is directly reliant on OXPHOS efficiency. Lin et al. noted that mitochondrial tRNA isolated from solid tumor xenografts decreased levels of t6A37 on tRNASer [117] suggesting that solid tumors may also display a CO2 dependency.
Figure 3. Readers, writers and erasers for the five major tRNASec modifications.
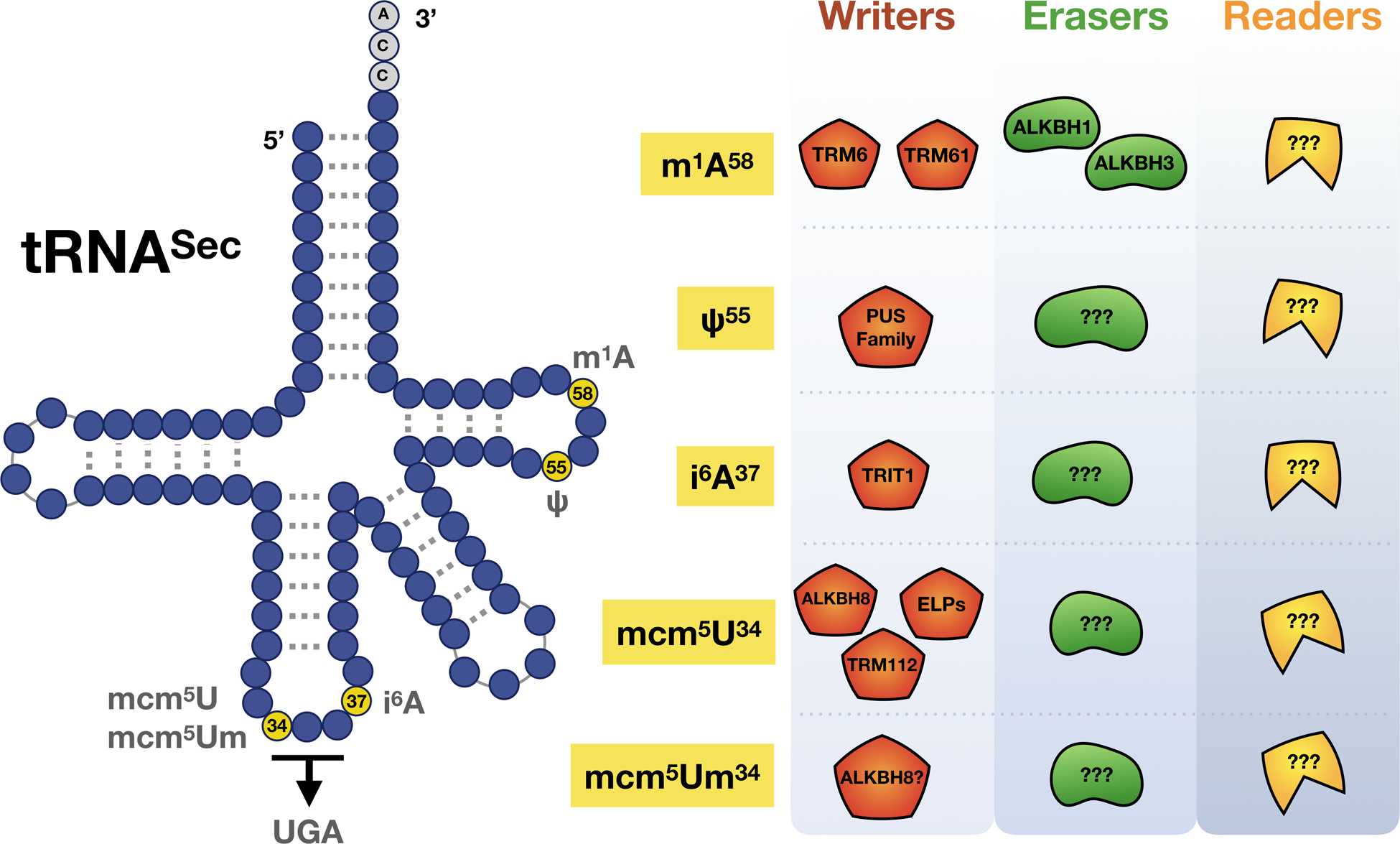
Known proteins and codons linked to the mcm5U, mcm5Um, i6A, Ψ and m1A modifications that play roles in regulating selenoprotein synthesis.
1.3. Epitranscriptomic Marks on Mitochondrial tRNA and their links to metabolism
Mitochondria translate some very specific transcripts into proteins that are essential for energy production and organelle function. Altered tRNA modification levels that disrupt mitochondrial function have been directly linked to the mitochondrial diseases myoclonus epilepsy associated with ragged-red fibers (MERRF) and mitochondrial encephalopathy, lactic acidosis, and stroke-like episodes (MELAS) [110,118,119]. These diseases are caused by a mutation in mitochondrial tRNA genes, with the change in sequence preventing the formation of the tRNA modifications s2 and 5-taurinomethyluridine (tm5U), both disturbing codon–anticodon interactions to disrupt protein synthesis [119,120]. Defects or deficiencies in methyltransferase NSUM3 or Fe(II) and α-ketoglutarate-dependent dioxygenase ALKBH1, which are responsible for f5C modification (Fig. 2) in mitochondria, can impair respiratory chain activity, resulting development of mitochondrial disease symptoms such as developmental disability, microcephaly, and OXPHOS deficiency in skeletal muscle [121–123]. As mitochondrial translation is used to synthesize key enzymes involved in metabolism, the resulting mistranslation defects can alter translation of mitochondrial proteins involved in the electron transport chain and ATP synthesis, and it can also dysregulate ROS levels [124]. The resulting defect in energy production is linked to the muscle weakness and neurological dysfunction associated with MERRF and MELAS, and provides a mechanistic link between tRNA modifications and energy metabolism [6].
Mitochondria are a major source of reactive oxygen species (ROS) production, which are generated during OXPHOS primarily as a result of electron leak and the 1-e− reduction of molecular oxygen (O2) to superoxide (O2•-) [125]. OXPHOS utilizes 5 protein complexes, ubiquinone oxidoreductase (complex I), succinate dehydrogenase (complex II), ubiquinol-cytochrome c oxidoreductase (complex III), cytochrome c oxidase (Complex IV) and ATP synthase (complex V), to provide chemical energy for cell survival [126]. Most of the subunits of complex I, III, IV and V are synthesized on cytosolic ribosomes, followed by transportation and assembly into mitochondrial membrane, and 13 subunits of these complexes are synthesized in mitoribosome and rapidly inserted into the mitochondrial inner membrane [127]. During OXPHOS, ATP synthesis is achieved by generating proton motive force through series of electron transfer processes, in which electron donors, NADH and succinate, are oxidized by complex I and II respectively, followed by electrons being transferred to complex IV through complex III and the electrons are reduced to molecular oxygen [127]. The concentration of potential electron donors and the production rate of ATP can influence the flux of O2•- [128] that is generated from complexes I and II in the mitochondrial matrix, and in both matrix and the intermembrane space by complex III [129,130]. O2•- generated from complex III can travel into the cytosol for signaling purposes [131] or enzymatically dismuted to H2O2 by superoxide dismutase proteins, SOD1 and SOD2 [132,133]. Although mitochondrial ROS (mtROS) are vital for maintaining cellular homeostasis via redox signaling[134–137], overproduction of mtROS can induce oxidative DNA damage and a wide range of pathologies [138]. As H2O2 and other ROS inducing agents have been shown to lead to epitranscriptomic reprogramming in cytosolic tRNA [139,140], there may be changes in RNA modification levels in mitochondrial tRNAs when metabolism, ATP levels and ROS levels change. Many ROS detoxifying proteins are selenoproteins which play important roles in limiting mtROS by consuming H2O2 and protecting mitochondria from aberrant redox activity [141,142]. Thioredoxin reductase 2 (TRXR2), a member of the mitochondrial thioredoxin system, controls the level of mt- H2O2 emission by maintaining thioredoxin 2 (TRX2) in a reduced state [143]. Increased steady-state H2O2 level, followed by cell death, decreased basal mitochondrial oxygen consumption rates, and decreased maximal and reserve respiratory capacity were observed in the cells treated with auranofin, a TRXR2 inhibitor [144]. In addition, TRXR2 deficient embryonic fibroblasts are highly sensitive to ROS when glutathione synthesis is inhibited. Further, Trxr2-null embryos are smaller in size, display increased apoptosis in the liver and show decreased cardiomyocyte proliferation [145].
1.4. Epitranscriptomic marks and systems linked to transfer RNA (tRNA) for selenocysteine
tRNASec is required for selenocysteine (Sec) biosynthesis and the production of ROS detoxifying selenoproteins. The production of selenoproteins requires a form of specialized translation called stop codon recoding, which utilizes specialized trans and cis-acting factors and is further detailed in section 1.5. Here we describe the distinct set of epitranscriptomic marks found on tRNASec that help regulate stop-codon recoding and selenoprotein synthesis, as tRNASec is the only known tRNA that governs the expression of an entire group of proteins. While canonical tRNAs contain on average 75 nucleotides, tRNASec is the longest tRNA species with 90 nucleotides in mammals and other eukaryotes (Fig. 2B and 3). This is in part due to its long variable arm and possession of the longest aminoacyl acceptor stem of any other tRNA [146,147]. tRNASec is under-modified relative to other tRNAs in humans, which contain 13 modifications on average but can have as many as 15–17, while tRNASec possesses only 5 possible epitranscriptomic marks [98,148]. tRNASec has 2 major isoacceptors: one containing mcm5U at wobble position 34, and the other mcm5Um at the same position, differing from the prior by a single methyl group addition. The 2’-O-ribosyl moiety found on mcm5Um is added to its mcm5U precursor in an ALKBH8 and selenium-dependent manner [82,100,149–151] (Fig. 2).
Multiple steps and enzymes are involved in the creation of both mcm5U and mcm5Um on tRNASec (Fig. 3). First, Uridine (U) is carboxymethylated to methyluridine (cm5U) by the ELP protein complex [152,153]. The tRNA methyltransferase ALKBH8, along with a small accessory protein TRM112, is required for the final step in the formation of mcm5U [100,154], with this precursor required for the 2’-O-ribose methylation to form mcm5Um (Fig. 4) [91]. Both tRNASec isoforms also have i6A, ψ55, and m1A modifications [150]. The i6A at position 37 is immediately 3’ of the anticodon and is catalyzed by TRIT1 in mammals. Knockdown of Trit1 results in decreased i6A and decreased selenoprotein expression in HepG2 cells [155]. The Ψ modification is catalyzed by PUS4 in eukaryotes [156–158]. The m1A mark is present at position 58 on tRNASec with this position being linked to structural stability and correct folding of the tRNA. [159]. In eukaryotes, the m1A writer is a complex consisting of catalytic protein TRM61A and RNA binding protein TRM6 [160]. The recent discovery of the m1A eraser protein ALKBH1 in mammals highlights the importance of this dynamic modification in regulating translation as ALKBH1-catalyzed demethylation results in decreased translation and usage of the unmodified tRNAs in protein synthesis [161]. The oxidative demethylase ALKBH3 is also able to remove m1A, as well as m3C, on RNA including tRNA [162,163]. ALKBH3 plays a protective function against alkylation damage on RNA via direct reversal of m1A and m3C to their original nucleosides. At the same time, it promotes angiogenin-catalyzed tRNA cleavage, an event occurring at greater frequency on unmodified tRNA, with the resulting tRNA-derived small RNAs having oncogenic functions by promoting ribosome assembly and preventing apoptosis [162,163].
Figure 4. The ALKBH8 epitranscriptomic writer regulates specialized translation by stop codon recoding.
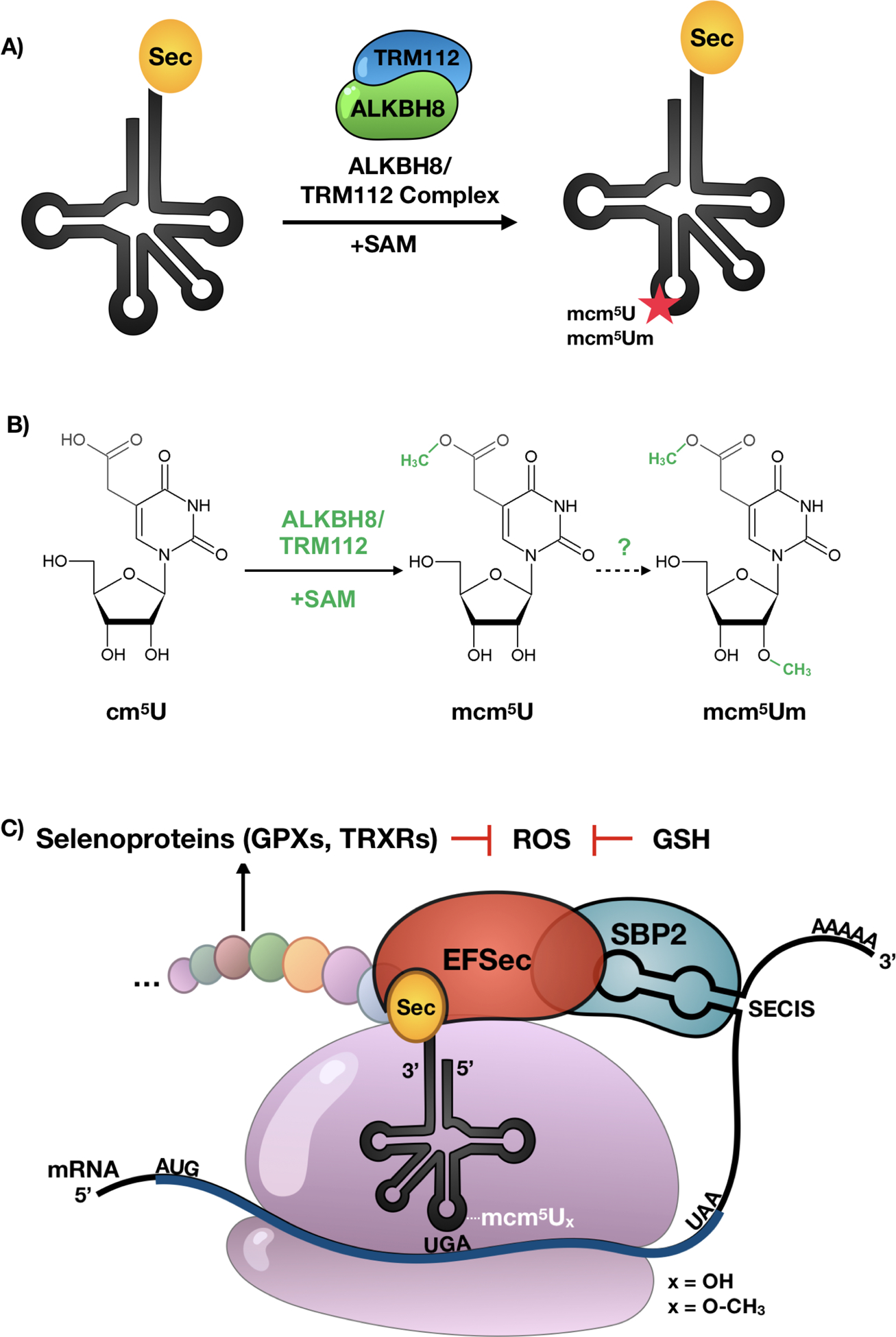
(A-B) ALKBH8 requires a small accessory protein TRM112 along with the SAM to modify tRNASec from cm5U to mcm5U, a key step required for the formation of mcm5Um. (C) Epitranscriptomic marks work with cis and trans factors to regulate UGA stop codon recoding. Wobble uridine modifications at position 34 of tRNASec along the elongation factor EFSec, a selenocysteine insertion sequence (SECIS) in the 3’ UTR and an RNA binding protein SBP2 are required for selenoprotein synthesis. Many selenoproteins use glutathione (GSH) to regulate ROS levels.
The two tRNASec isoforms are correlated with expression levels of 2 classes of selenoproteins. In general, the mcm5U isoform is thought to serve the synthesis of housekeeping selenoproteins and, while the mcm5Um isoform is involved in translation of stress response selenoproteins and is more sensitive to selenium status [150,164,165]. These findings were elucidated using transgenic mice encoding an A→G mutation in the tRNASec gene at position 37, which results in decreased expression of selenoproteins [155]. Overexpression of the G37 tRNASec mutant led to changes in the distribution of the mcm5U and mcm5Um modifications and dysregulated specific stress responsive selenoproteins. These observations led to the realization that many of the selenoproteins responsive to selenium status are involved in stress-related functions, while those unaffected serve housekeeping functions [165,166] Epitranscriptomic marks on tRNASec are dependent not only on their writers for modification, but also on each other. For example, the m1A mod at position 37 is required for synthesis of ψ at position 55, a modification that governs tertiary structure. Likewise, mutation at any of the sites containing the i6A, m1A or ψ, or indeed any mutations that influence tRNASec secondary structure severely decreases or completely inhibit the 2’-O ribose methylation at position 34, a modification that also results in the conformational change of the tRNA [149,150].
1.5. Epitranscriptomic marks work with cis and trans factors to help regulate selenoprotein synthesis
Selenoproteins rely on a unique method of translation based on stop codon recoding because there is no dedicated codon for Sec incorporation. Sec insertion into proteins relies on cis-acting factors on the mRNA and trans-acting factors involved in bringing Sec-charged and properly modified tRNASec to the ribosome, where it recognizes and decodes the UGA stop codon (Fig. 4C). UGA recoding uses a 3’ UTR stem-loop structure termed the selenocysteine insertion sequence (SECIS) [167–169] that can help recruit machinery for specialized translation. It also uses a Sec-specific elongation factor (eEFSec), a GTP binding protein that was found to recognize charged tRNASec in vitro and in vivo [170]. Selenoproteins have expanded our understanding of the universal genetic code, which is comprised of 61 codons encoding for 20 amino acids, and 3 stop codons signaling the end of translation. The UGA stop codon has the added function of encoding for the 21st amino acid Sec, via the process of stop codon recoding [91,171–174]. Translational recoding of the UGA codon in eukaryotes is regulated by selenium availability and other core factors such as tRNASec modifications, Type I & II selenocysteine insertion sequence (SECIS) elements, SECIS Binding proteins (SBP2), elongation factors (eFSec), as well as enzyme-catalyzed epitranscriptomic marks on tRNASec [91,166,175]. Disruption of selenoprotein synthesis can occur due to selenium deficiency, mutations or changes in mRNA elements including UGA codon and SECIS elements, changes in RNA binding proteins or elongation factors involved in Sec translation, tRNASec mutations and writer deficiencies [82,168,169,175–179], with the latter two leading to decreased epitranscriptomic marks.
1.6. Selenoprotein function
25 and 24 selenoproteins have been discovered in humans and rodents, respectively. Selenoproteins can be classified into 6 functional groups: those with peroxidase/reductase activity, redox signaling, hormone metabolism, protein folding, selenium transport and Sec synthesis. The majority of selenoproteins confer protection against oxidative stress and damage, described below, but some are also involved in homeostatic processes such as calcium signaling, biosynthesis of phospholipids, and maintaining and transporting selenium within the body [180,181] (Table 1). Selenoproteins V, W and GPX4 function as essential proteins for spermatogenesis and overall vitality. The synthesis and transportation of selenium is aided by selenophosphate synthase, selenoprotein P (SEPP), and selenoprotein 15 (SEL15). The deiodinase family (DIO1–3) of selenoproteins that regulate thyroid hormone level and thyroid function[182]. Some selenoproteins have not been assigned a function. These include, Selenoprotein O, V and M.
Table 1.
The 25 human selenoproteins, their identifying information and current known functions. Selenoprotein gene nomenclature was recently updated as in [240]. We have therefore included the selenoprotein name in bold, and new and all previous abbreviations in parentheses. The abbreviation used in this manuscript is listed first.
| Selenoprotein | Number of Selenocysteine residues | Subcellular Localization | Molecular Wt (kDa) | Function | References |
|---|---|---|---|---|---|
| Selenoprotein K (SELK, SELENOK) | 1 | Endoplasmic reticulum, Plasma membrane | 10.64 | Required for T-cell proliferation and calcium flux in immune cells. Known to protect cardiomyocytes from oxidative stress. | [180,241–243] |
| Selenoprotein S (SELS, SEPS1, VIMP, SELENOS) | 1 | Endoplasmic reticulum | 21.16 | Protects cells from oxidative damage and ER stress-induced apoptosis. Removes misfolded proteins from ER lumen for degradation. | [180,197,202,223,241,244–247] |
| Selenoprotein O (SELO, SELENOO) | 1 | Mitochondrion | 73.49 | Cys-X-X-Sec motif suggests redox function; however, importance remains unknown. | [180,181,241] |
| Selenoprotein I (SELI, EPT1, SELENOI) | 1 | Transmembrane | 45.22 | Biosynthesis of phospholipids. Formation and maintenance of vesicular membranes. | [180,241,248] |
| Methionine sulfoxide reductase B1 (MSRB1, SELR, SELX, SEPX1) | 1 | Cytoskeleton, Nucleus | 12.76 | Methionine sulfoxide reductase. Known to be protective against neurodegeneration, lens cell viability, and oxidative damage during aging. | [180,241,249–252] |
| Selenoprotein H (SELH, SELENOH, C11orf31) | 1 | Nuclear | 13.45 | Gene expression regulator for glutathione synthesis, provides antioxidant defense. Involved in transcription. | [180,194,195,241,253] |
| Selenoprotein M (SELM, SEPM, SELENOM) | 1 | Golgi apparatus, Endoplasmic reticulum | 16.23 | Thiol-disulfide oxioreductase in endoplasmic reticulum. Role in protein folding. | [180,241,254,255] |
| Selenoprotein N (SEPN1, SELN, SEPN) | 1 | Endoplasmic reticulum | 65.81 | Regulates ryanodine receptor (RyR)-mediated calcium mobilization required for normal muscle development and differentiation. Regulator of redox-related calcium homeostasis and protection against oxidative stress. | [180,241,256–260] |
| Selenoprotein P (SEPP1, SELP, SELENOP) | 10 | Secreted, Cytoplasmic | 43.17 | Selenium transporter throughout organism, especially in brain and testes. Regulates glutathione peroxidase activity, heparin binding and heavy metal ion complexation. | [180,236,237,241,261–268] |
| Selenoprotein T (SELT, SELENOT) | 1 | Endoplasmic reticulum | 22.32 | Role in embryonic development and calcium mobilization. | [180,241,253,269] |
| Selenoprotein V (SELV, SELENOV) | 1 | Unknown | 36.8 | Expressed in testes. More functional evidence needed. | [180,241,253] |
| Selenoprotein W (SELW, SEPW1, SELENOW) | 1 | Cytoplasmic | 94.48 | Interacts with glutathione and plays an antioxidant role in cells. Protects developing myoblasts from oxidative stress and functions in muscle growth and differentiation. | [180,241,253,270–274] |
| Selenophosphate Synthetase 2 (SPS2, SEPHS2) | 1 | Cytosol | 47.31 | Participates in selenocysteine biosynthesis by providing selenium donor compound, selenophoasphate. Involved in synthesis of selenoproteins, including itself. | [180,241,275–277] |
| Selenoprotein 15 (SEP15, SELENOF) | 1 | Endoplasmic reticulum | 15 | Thiol-disulfide oxioreductase in endoplasmic reticulum. Maintains selenoproteins in the ER, role in protein folding. | [180,241,254,255,278] |
| Glutathione Peroxidase 1 (GPX1, GSHPX1, Cytosolic Glutathione Peroxidase) | 1 | Cytoplasmic | 22 | Plays an overall recovery in cells after oxidative stress. Catalyzes the reduction of organic hydroperoxidases and H2O2. | [180,209,241,279–282] |
| Glutathione Peroxidase 2 (GPX2, GSHPX-GI, Gastrointestinal glutathione peroxidase) | 1 | Cytoplasmic | 21.95 | Protects intestinal epithelium from oxidative stress and defense when exposure to induced oxidative stress by ingested prooxidants or gut microbiota. | [182,283–285] |
| Glutathione Peroxidase 3 (GPX3, Plasma gluthathione peroxidase) | 1 | Secreted | 25.55 | Main source in plasma is from the kidney. Serves as a local source of extracellular antioxidant capacity to protect against oxidative damage to extracellular matrix. Regulates bioavailability of nitric oxide (NO) produced from platelets and vascular cells. | [180,241,286,287] |
| Glutathione Peroxidase 4 (GPX4, Phosholipid hydroperoxide gluthathione peroxidase) | 1 | Mitochondrion, Cytoplasmic | 22.18 | Facilitates reverses oxidation of lipid peroxides and metabolism of lipids such as arachidonic acid and linoleic acid. Protective role in cardiovascular disease by decreasing lipid peroxidation. Dual roles as an enzyme and structural protein, transformed in later stages of spermatogenesis into structural protein that becomes a constituent of mitochondrial sheath of spermatozoa. | [180,211,241,288–292] |
| Glutathione Peroxidase 6 (GPX6, Olfactory gluthathione peroxidase) | 1 | Secreted | 24.97 | Expression restricted in developing embryo and olfactory epithelium in adults. Expected to have antioxidant role, more definitive evidence for role is needed. | [180,241,293] |
| Thioredoxin reductase I (TRXR1, TXNRD1, TR1) | 1 | Cytoplasmic, nuclear | 70.91 | Gene deletion is embryonic lethal. May possess glutaredoxin activity. Critical electron donor system in DNA replication during S-phase growth. | [180,241,294–299] |
| Thioredoxin reductase II (TRXR2, TXNRD2, TR3, Mitochondrial thioredoxin reductase) | 1 | Mitochondrion | 56.51 | Deletion is embryonic lethal. Regulation of mitochondrial redox homeostasis. Role in cardiomyocyte viability. | [180,191,193,241,300,301] |
| Thioredoxin-glutathione reductase (TRXR3, TXNRD3, TR2, TGR, Thioredoxin reductase 3) | 1 | Endoplasmic reticulum, Nucleus | 70.68 | Promotes disulfide bond formation in various sperm proteins. | [180,241,302] |
| Iodothyronine deiodinase I (DIO1, D1) | 1 | Endoplasmic reticulum | 28.92 | Important for systemic active thyroid hormone levels. Providing source of plasma T3 by deiodination of T4 in peripheral tissues such as liver and kidney. | [180,224,241,303,304] |
| Iodothyronine deiodinase II (DIO2, D2) | 1 | Membrane | 30.55 | Important for local active thyroid hormone levels. Essential for providing the brain with appropriate levels of T3 during critical development period. | [180,241,303,305–309] |
| Iodothyronine deiodinase III (DIO3, D3) | 1 | Endosome membrane, Plasma membrane | 33.95 | Inactivates thyroid hormone. | [180,241,303,304] |
1.7. Selenoproteins as essential participants in stress responses
Selenoproteins are key regulators of stress responses, metabolism, and immunity. A majority of selenoproteins play a role in maintaining cellular redox homeostasis and viability, serving as antioxidant enzymes to mitigate damage caused by reactive oxygen species (ROS). Examples of these selenoproteins include selenoprotein K, S, H, N, GPX1–4, TRXR1–3. Many selenoproteins play vital roles in embryonic vitality and development. More than half of the human selenoproteins have a thioredoxin-like fold and are involved in thiol-dependent reactions and play roles in redox homeostasis. Redox signaling is involved in regulating many of the characteristics of cancer cells, which often exhibit dysregulated redox homeostasis and continuous and elevated production of ROS. Selenoprotein links to cancer are extensive and can be found as part of a number of excellent reviews and broad population studies [183–186]. GPX1 remains one of the best-characterized selenoproteins, and given its broad function as an antioxidant is a prime candidate in cancer risk, discussed in section 1.8. GPX1 is one of five Sec-containing GPXs capable of reducing H2O2 and lipid hydroperoxides using glutathione as the electron donor, protecting cells against oxidative damage via the reaction:
Glutathione disulfide (GSSG) is returned to its reduced and active state by glutathione reductase (GR) using NADPH as the electron donor. GPX1 is expressed in almost all cell types and is critical to maintaining proper redox balance within the body. This is especially true under stress as GPX1 deficient mice are more susceptible to ROS-inducing agents such as H2O2 and lung inflammation and damage due to influenza infection and cigarette smoke [187–189].
TRXRs are Sec-containing proteins critical for a variety of cellular functions, including all signaling pathways in which thioredoxin is involved as a reducing agent. Thioredoxins can reduce oxidized protein cysteine residues to form reduced disulfide bonds, and are maintained in their reduced and active state by TRXRs using NADPH as an electron donor. This system is critical for signaling pathways involved in the protection from oxidants [190]. TRXRs are also required for development as complete knockout of both TRXR1 and TRXR2 in mice result in severe growth abnormality and embryonic death at day 8.5 and 13, respectively [191–193]. Like GPXs, TRXRs play roles in the etiology of many cancers, further discussed in section 1.8.
Selenoprotein H (SELH), a dual-function selenoprotein acting as both a transcription factor for antioxidants and phase II metabolism enzymes, and also exhibiting oxidoreductase activity [194]. SELH was originally described as a thioredoxin reductase-like protein with significant glutathione peroxidase-like activity [195]. Overexpression of SELH protects neuronal HT22 cells against UVB-induced injury and death by inhibiting apoptotic cell death pathways and promoting mitochondrial biogenesis and function, and suppression of superoxide production. Another selenoprotein involved in the response to stress is Selenoprotein S (SELS, also called VIMP). SELS is a widely expressed transmembrane protein located in the endoplasmic reticulum (ER) important for managing ER stress in mammals via elimination of misfolded proteins [196–198]. SELS is also involved in regulating many important processes, such as oxidative stress [199], inflammation [200–202], adipogenesis [203], and apoptosis [198]. It has been shown that a polymorphism in the promoter region of the SelS gene (−105G→A) results in increased inflammatory markers IL-6, IL-1β, and TNF-α [202].
1.8. Selenoproteins and their link to human diseases
Studies have shown a link between dysregulation in selenoprotein biosynthesis and an increased risk of disorders affecting many major organ systems in the body. Mutations in the promoter region of genes for selenoproteins, the SECIS element, single-nucleotide polymorphisms, or deletions in the coding frame have also been linked to the etiology of many cancers (Table 2). GPX1 is decreased in some tumors [204], while overexpression of GPX1 has been found to reduce tumor growth [205,206]. Associations between GPX1 and cancer risk have been found to include prostate [207], thyroid [208], head/neck cancer [209] and breast cancer [183,210]. Two Gpx1 polymorphisms in European populations are associated with advanced-stage prostate cancer risk: rs17650792 and rs1800668 [207]. GPX4 is unique among the protein family in that it can reduce phospholipid hydroperoxides [211]. Human population studies have shown that Gpx4 polymorphism is associated with increased risk for colon [212] and prostate cancer [213]. TRXR1 and TRXR2 have been linked to cancer etiology of the prostate, colon, and rectum [214–216]. Colorectal cancer has also been associated with polymorphisms in the selenoprotein SEP15 [217]. An in vitro study using mouse colon carcinoma CT26 cells suggests tumor-promoting effects of TRXR1, as well as SEP15, as cells with targeted knockdown of either selenoproteins exhibited reduced growth. Surprisingly, knockdown of both reversed the tumor-suppressive property suggesting that the two selenoproteins participate in interfering regulatory pathways in colon cancer cells [218].
Table 2.
Human population studies or mammalian models for specific selenoproteins and their link to human diseases.
| Disease Category | Selenoprotein | Cause | Disorder | Link or contributing factors | References |
|---|---|---|---|---|---|
| Immune | Selenoprotein K | Deletion in all tissues | Immune disorder | Deficient Ca2+ flux in immune cells. | [310] |
| Cancer (Gastric) | Selenoprotein S | Polymorphism | Gastric cancer | Chinese Hunan Han and Japanese populations. | [220,311] |
| Intestinal | Selenoprotein S | Polymorphism | Inflammatory bowel disease | SelS mRNA expression is upregulated by proinflammatory cytokines in Intestinal epithelial cells. However, the SelS –105G>A polymorphism is not associated with inflammatory bowel disease susceptibility. | [221] |
| Endocrine | Selenoprotein S | Polymorphism | Hashimoto’s thyroiditis | Polymorphism in the promoter region. | [222] |
| Cardiac | Selenoprotein S | Polymorphism | Coronary heart disease and ischemic stroke | Genetic variant rs8025174 increased coronary heart disease risk by 2.95 fold. Genetic variant rs7178239 increased risk of ischemic stroke by 3.35 fold, both in females in Finland. |
[223] |
| Cancer (Colorectal) | Selenoprotein S | Polymorphism | Colorectal cancer | SNP in SelS gene (rs34713741) associated with 2.25 increase in odds of colorectal cancer in females in a Korean population. | [217] |
| - | Selenoprotein O | - | Unknown | - | - |
| Neurological | Selenoprotein I | Exon skipping | Neurological disorders, inhibited neurodevelopment, inadequate myelination in the brain. | From case study of one patient, and in vitro in SELI deficient HeLa cells. | [312] |
| Neurological | Methionine sulfoxide reductase B1 | Deficiency | Alzheimer’s | MSRB1 possibly plays a protective role against Alzheimer’s disease (AD). A human fetal brain cDNA screen showed that MSRB1 interacts with Clusterin (CRU). This was shown to be a synergistic interaction, with CRU also interacting with the β-amyloid peptide, a pathological protein in AD. | [313,314] |
| Whole-body | Methionine sulfoxide reductase B1 | Knockout | Growth delays & increased oxidative stress | MSRB1 deficient mice have growth delays and increased oxidative stress, a phenotype exacerbated by dietary methionine deficiency. | [315,316] |
| Cancer (Colorectal) | Selenoprotein H | Overexpression | Colorectal cancer | In vivo & in vitro link between SELH overexpression and colorectal cancer. | [219] |
| Neurological | Selenoprotein M | Decreased expression | Alzheimer’s disease | SELM may prevent β-Amyloid aggregation by resisting oxidative stress generated during β-Amyloid oligomer formation. In vivo link, SELM may play a protective role in the pathology of patients with Alzheimer’s. | [317,318] |
| Muscular | Selenoprotein N | Nonsense mutation | Rigid spine muscular dystrophy | Nonsense mutation in selenocysteine codon (TGA → TAA), preventing Sec insertion and resulting in short proteins. Missense and frameshift mutations in the ORF, and mutation in SECIS element (3’ UTR) all lead to rigid spine muscular dystrophy. | [178,257,258] |
| Muscular | Selenoprotein N | Knockout | Impaired muscle repair | SepN1−/− mouse model shows depletion of satellite cells required for muscle repair after injury resulting in impaired and imperfect repair. Muscle biopsies of patients with SepN1 mutation shows decreased satellite cells. | [319] |
| Muscular | Selenoprotein N | Mutation in SRE element | SEPN1-related myopathy | A single missense mutation (c.1397G>A) in the SepN1 SRE significantly reduces Sec insertion, destabilizes the mRNA and leads to decreased SEPN1 protein levels. | [320] |
| Intestinal | Selenoprotein P | Polymorphism | Colorectal cancer | Polymorphism in promoter region (rs2972994) | [212] |
| Intestinal | Selenoprotein P | Deficiency | Crohn’s disease, inflammatory bowel disease | Decrease in serum SEPP1 levels | [321] |
| Metabolic | Selenoprotein P | Increase in secreted protein from liver | Insulin resistance, hyperglycemia & (inferred) type 2 diabetes | In vitro, in vivo, and population studies | [322] |
| Metabolic | Selenoprotein P | Increase in circulating Selenoprotein P | Hyperglycemia | Japanese population. Elevation of circulating SEPP1 positively correlated with onset of hyperglycemia. | [323] |
| Musculoskeletal | Selenoprotein P | Polymorphism | Kashin-Beck disease | Du et al show that SNP in Sepp1 −105G>A gene associated with Kashin-Beck disease (KBD), with the AA genotype conferring significant risk. Sun et al show that KBD patients have significantly decreased serum SEPP1 levels compared to controls. | [324,325] |
| Renal | Selenoprotein P | Deficiency | Renal Cell Carcinoma | Decrease in SEPP1 and Se levels | [326] |
| Neurological | Selenoprotein P | Knockdown | Alzheimer’s Disease | In vitro study in Neuronal N2A cells knows that RNAi knockdown of SEPP1 causes amyloid-beta induced toxicity and cell death. | [327] |
| Neurological | Selenoprotein P | Decreased expression | Parkinson’s Disease | SEPP1 expression reduced in neurons of substantia nigra of Parkinson’s patients (post-mortem analysis of brain) | [328] |
| Neurological | Selenoprotein P | Deletion | Impaired neurological function | Sepp1−/− mouse models show impaired function of parbalbumin (PV)-interneurons and changes in fear learning and sensorimotor gating. | [233] |
| Multiple | Selenoprotein P | Knockout | Reproductive disorder, neurological disorder | Required for Se homeostasis and metabolism. Diet dependent movement abnormality and weight loss when Sepp1−/− mice fed a low Se diet. | [236,237] |
| Neurological | Selenoprotein T | Under- and overexpression | Parkinson’s disease | In vitro, in vivo, and patient studies. Complete deletion in mice results in embryonic lethality. | [329] |
| Neurological | Selenoprotein T | Conditional Knockout in nerve cells | Neurodevelopmental abnormalities, hyperactive behavior | SelT−/− young mice had reduced volume of hippocampus, cerebellum and cerebral cortex, and although compensated in adulthood the mice had hyperactive behavior. | [330] |
| Endocrine | Selenoprotein T | Conditional knockout of SelT in pancreatic β cells | Impaired glucose tolerance, decreased insulin production and secretion | In mouse pancreatic β cells. | [331] |
| - | Selenoprotein V | - | Unknown | - | - |
| Hypertension |
Selenoprotein W | Polymorphism | Pre-Eclampsia | SelW polymorphism (rs3786777) did not affect risk factor for pre-eclampsia (PE) in pregnant women, but PE patients had significantly lower circulating SELW protein levels. | [332] |
| Cancer (Colorectal) | Selenoprotein 15 | Polymorphism | Colorectal Cancer | In a Korean population, polymorphism in rs5845 and rs5859 in Sep15 significantly correlated with increased rectal cancer in males. | [217] |
| Eye | Selenoprotein 15 | Knockout | Cataract Development | Mice were viable and fertile, but had elevated oxidative stress in livers and cataract formation early in life. | [333] |
| Cancer (Colon) | Selenoprotein 15 | Targeted downregulationof Sep15 | Colon Cancer | In vitro mouse colon carcinoma study using mouse colon carcinoma CT26 cells. Downregulation of Sep15 suppresses cancer cell growth, suggesting a tumor-promoting property of SEP15. | [218] |
| Bone | Glutathione peroxidase 1 | Polymorphism | Kashin-Beck disease | Tibetian population, Chinese Han population | [334,335] |
| Cancer (Thyroid) | Glutathione peroxidase 1 | Oxidative stress | Thyroid cancer | Data from cancer patients. | [208] |
| Cancer (Bladder) | Glutathione peroxidase 1 | Polymorphism | Bladder cancer | A polymorphism in Gpx1 resulting in an amino acid substitution (Pro198Leu) played a protective role against bladder cancer recurrence. WT patients had marginally shorter survival (p = 0.066), which was reduced to (p = 0.036) in whites. | [336] |
| Cancer (Head/Neck)/ Neurological | Glutathione peroxidase 1 | Gpx1 expression level | Head/Neck cancer | A negative correlation between tumor stage and GPX1 expression was found in pre-treatment biopsies of head/neck cancer patients. | [209] |
| Neurological | Glutathione peroxidase 1 | IncreasedGpx1 expression and activity | Huntington’s Disease | Increased GPX1 expression and activity in Huntington Disease patients | [337] |
| Cancer (Breast) | Glutathione peroxidase 1 | Loss of hetero-zygosity | Breast Cancer | Leucine vs. proline at GPX1 position 198 assessed, with leucine-containing allele more frequently associated with breast cancer samples. | [210] |
| Cancer (Prostate) | Glutathione peroxidase 1 | Polymorphism | Prostate Cancer | Polymorphism in rs17650792 and rs1800668 associated with increased advanced colorectal cancer risk. | [207] |
| Gastrointestinal | Glutathione peroxidase 2 | Double knockout of Gpx1 & Gpx2 | Colitis | Combined disruption of Gpx1 and Gpx2 in mice. | [281] |
| Cancer (Rectal) | Glutathione peroxidase 3 | Polymorphism | Rectal Cancer | Genetic variablility in Gpx3 alters risk of rectal cancer, but not colon cancer (study identified 3 different polymorphisms, and in this case resulted in reduced risk of cancer). | [338] |
| Cancer (Colorectal) | Glutathione peroxidase 4 | Polymorphism | Colorectal Cancer | rs713041 polymorphism in Gpx4 increases risk for colorectal cancer. | [212] |
| Cancer (Prostate) | Glutathione peroxidase 4 | Polymorphism | Prostate Cancer | Gpx4 polymorphism (rs2074452) linked to increase in prostate cancer risk and prostate-cancer specific mortality. | [213] |
| Neurological | Glutathione peroxidase 4 | Decreased expression | Parkinson’s Disease | Reduced GPX4 expression in neurons of substantia nigra of Parkinson’s disease patients (post-mortem analysis of brain). | [339] |
| Neurological | Glutathione peroxidase 4 | Conditional knockout in neurons | Neurodegeneration in the hippocampus, seizures, | Conditional knockout in mouse neurons. | [231] |
| Embryonic | Glutathione peroxidase 4 | Knockout | Embryonic lethality day 7.5 | Mouse embryos display lack of normal structural compartmentalization. | [230] |
| Bone | Glutathione peroxidase 4 | Decreased mRNA levels | Kashin-Beck Disease | Decreased Gpx4 mRNA results in increased risk for Kashin-Beck Disease. | [340] |
| Bone/Neurological | Glutathione peroxidase 4 | Exon skipping | Sedaghatian-type spondylometaphyseal dysplasia (SSMD) | 2 unrelated human cases with mutation in Gpx4 gene resulting in exon skipping, part of exon 4 in one case and exon 5 in another case. Severe bone defects and neurological abnormalities noted. | [232] |
| Renal | Glutathione peroxidase 4 | Knockout | Renal failure | Gpx−/− mice have increased ferroptosis-mediated cell death and lipid oxidation induced renal failure | [341] |
| Neurological | Glutathione peroxidase 6 | Increased expression level |
Huntington’s Disease | GPX6 increased in striatum. | [337] |
| Cancer (Colon) | Thioredoxin reductase I | Targeted downregulationof TrxR1 | Colon Cancer | In vitro mouse colon carcinoma study using mouse colon carcinoma CT26 cells. Downregulation of TrxR1 suppresses cancer cell growth, suggesting a tumor-promoting property of TRXR1. | [218] |
| Embryonic | Thioredoxin reductase I | Knockout | Embryonic lethality day 8.5 | Severe growth abnormalities, primitive streak mesoderm does not form (i.e. failure to gastrulate). | [191,192] |
| Cancer (Prostate) | Thioredoxin reductase I | Polymorphism | Prostate Cancer | TrxR1 SNPs rs11610799 and rs7138318 associated with increased risk for prostate cancer. | [215] |
| Cancer (Prostate) | Thioredoxin reductase II | Polymorphism | Prostate Cancer | TrxR2 SNPs rs3788317 and rs599245 associated with increased risk for prostate cancer. | [215] |
| Cardiac | Thioredoxin reductase II | Knockout | Cardiac failure and dilated cardiomyopathy | Complete knockout of TrxR2 in mice results in embryonic lethality at day 13. | [193] |
| Thyroid | Iodothyronine deiodinase I | Targeted disruption | Thyroid hormone disorder | Changes in thyroid hormone metabolism and excretion. | [224] |
| Neurological | Iodothyronine deiodinase II | Deletion of major coding exon of DIO2 | Auditory defects | Hearing loss and failed cochlear development in Dio2−/− mice. | [225] |
| Muscle | Iodothyronine deiodinase III | Ablated SECIS region of Dio3 gene | Muscle disorder | Defects in muscle regeneration. | [342] |
| Thyroid | Iodothyronine deiodinase III | Knockout | Hypothyroidism | Dio3−/− mice have hypothyroidism and defects in formation and function of thyroid axis. | [343] |
| Neurological | Iodothyronine deiodinase III | Deletion | Auditory defects | Failed cochlear development. | [226] |
| Neurological/ metabolic | Iodothyronine deiodinase III | Deletion | Disruption of circadian rhythm | Dio3−/− mice display leptin resistance, disrupted circadian rhythm and elevated energy expenditure & disrupted energy balance, and increased fat loss in response to T3 treatment. | [228] |
| Neurological, Metabolic | SECIS binding protein 2 (SBP2) | Nonsense Mutation, nonfunctional SBP2 protein | Abnormal thyroid hormone metabolism, delayed bone maturation, congenial myopathy, impaired mental development and motor coordination | Case study of 1 patient with mutation in SBP2 resulting in absence of all functional domains of the protein and severe developmental, neurological and metabolic phenotype. | [344] |
| Embryonic | SECIS binding protein 2 (SBP2) | Knockout | Embryonic lethal day 7.5 | Mouse SBP2 knockout model | [345] |
| Cancer | Alkylation repair homolog 8 | Overexpression | Bladder cancer | ALKBH8 is overexpressed in bladder cancer and promotes cancer progression.Knockdown causes death of cancer cells via downregulation of survivin. | [346,347] |
| Neurological | Alkylation repair homolog 8 | Truncating Mutation | Intellectual Disability | Study of two consanguineous families, truncating mutations in ALKBH8 lead to complete lack of ALKBH8-dependent tRNA modifications and intellectual disabilities. | [348] |
| Embryonic | Trsp | Knockout | Embryonic lethal day 6.5 | Mice with a targeted deletion of trsp gene died on day 6.5. | [176] |
| Neurological | Trsp | Conditional knockout in neurons | Neurodegeneration | Mouse model with inactivated trsp in neurons resulted complete loss of expressions. Loss of balance, growth delays, seizures and neurodegeneration evident. | [229] |
| Metabolic/Endocrine | Selenocysteine tRNA | i6A mutant | Diabetes | Reduced synthesis of selenoproteins due to an overexpression of i6A-mutant tRNASec. | [349] |
| Mitochondrial | TRIT1 (i6A writer) | TRIT1 mutation | Combined mitochondrial oxidative phosphorylation deficiency | Defective i6A tRNA modifications | [350] |
| Reproductive/Neurological | ALKBH1 (m1A eraser) | Deletion | Sex ratio distortion and unilateral eye defects | One female mouse born for every 3–4 males (sex ratio distortion). Eye defects and craniofocal malformations also observed. | [351] |
| Neurological | ALKBH1 (m1A eraser) | Deletion | Embryonic lethal, histone H2A methylation defects | Most mice die at embryonic stage due to defects in embryonic stem cell differentiation. Also evidence of defects in histone H2A methylation. | [352] |
| Cancer (lung) | ALKBH3 (m1A eraser) | Overexpression | Non-small-cell lung cancer | Knockdown of ALKBH3 reduced both number and diameter of tumors in the peritoneum after mice were inoculated with the A549 human lung carcinoma cell line. | [353] |
| Cancer (prostate) | ALKBH3 (m1A eraser) | Overexpression | Human Prostate Carcinoma | ALKBH3 (alt name prostate cancer antigen 1 (PCA-1)) highly expressed in human prostate cancer. | [354,355] |
SelH transcripts and protein are overexpressed in some cancer cells such as LCC1 (lung cancer) and LNCaP (human prostate carcinoma)[195]. Overexpression of SELH has also been shown in human and mouse-derived colorectal cancer tissue [219]. Mutations specifically in the promoter region of SELS have been linked to gastric cancer [220], inflammatory bowel disease [221] and hashimoto’s thyroiditis [222]. SELS population studies also suggest disease risks due to genetic variants; polymorphism rs8025174 has been linked to increased risk of coronary heart disease, and rs7178239 has been linked to more than a 3-fold increase in risk of ischemic stroke in females [223]. Deletion of selenoproteins DIO2 and DIO3 has been linked to defects in the auditory and visual systems. In general, deiodinases catalyze the elimination of iodide from thyroid hormones. Although DIO1 deletion was not associated with any neurological phenotype [224], deletion of DIO2 resulted in hearing loss and malformations in the cochlea in mice [225]. Dio3−/− mice also display hearing loss, but unlike the Dio2−/− mice they had accelerated cochlear differentiation [226]. In addition, Dio3−/− mice had defects in retinal photoreceptor development and a loss of about 80% of cone photoreceptors affecting light and color vision [227]. Lastly, Dio3−/− mice were shown to have disrupted energy balance, leptin resistance, and a disrupted circadian rhythm [228].
The brain expresses almost all selenoproteins and mutations or deficiencies in selenoproteins are linked to a number of neurological dysfunctions. A conditional inactivation of the mouse tRNASec gene trsp in neurons resulted in total loss of selenoprotein expression in neurons and severely decreased expression in the cerebrum phenotypically resulting in loss of balance, growth retardation, seizures and neurodegeneration in mutant mice [229]. Complete inactivation of Gpx4 is embryonic lethal in mice [230,231], but mice with a conditional knockout of Gpx4 in neurons were found to have massive degeneration [229]. Mutations in Gpx4 in two separate human cases resulted in the embryonic lethal bone defect Sedaghatian-type spondylometaphyseal dysplasia (SSMD), a defining feature of which is also central nervous system abnormality [232]. Deletion of Selenoprotein P (Sepp1) in mice has been associated with neurological dysfunction including prevalence of seizures and decrease in activity, grooming behavior and motor coordination and delays in fear learning [233–235]. Sepp1 deletion results in drastically reduced selenium delivery to the brain [236,237]. These neurological phenotypes increased in male relative to female mice suggesting that male mice are more dependent on SEPP1 and Se for normal brain function [235].
1.8. Concluding thoughts, epitranscriptomic marks could be linked to many diseases
In this review we have highlighted examples of enzyme-catalyzed epitranscriptomic marks on RNA, and described how epitranscriptomic systems can function as regulatory elements during translation. Defects in RNA epitranscriptomic marks and their associated epitranscriptomic readers/writers/erasers have been linked to a number of human diseases, including developmental defects, neurological disorders, and cancer. The epitranscriptomic writer ALKBH8 is essential for promoting translation of selenoproteins, which are critical for development, homeostatic functions and stress responses. To date, studies have elucidated the function many human selenoproteins, outlined in Table 1, which includes redox signaling, oxidoreductase activity, selenium transport and storage, Sec synthesis, protein folding, and hormone metabolism. While it is clear that selenoproteins play critical roles in development and proper function of cells, organs, and organ systems, the human disease associations are complex. We highlight the importance of selenoproteins in detoxifying ROS, of which more than half of selenoproteins play important roles. Human and animal studies demonstrate the importance of properly regulated selenoprotein synthesis in preventing diseases of nearly every major organ system in the body; often involving biochemical pathways in the oxidative stress response (Table 2). The study of broad selenoprotein deficiency, such as in the case of disrupted UGA stop codon translation as a result of epitranscriptomic deficiencies, is also helpful in identifying the global role of selenoproteins in health and development.
The term “epitranscriptomics” was coined in 2012 [238,239], and a growing body of literature has identified more than 140 post-transcriptional modifications present on every type of RNA. Many studies discussed herein support the idea that RNA modifications function as an added level of translational control in stress responses. Deficiencies in writers and erasers have been linked to disrupted function and disease, with strong links between ALKBH1–3 and TRIT1 to ROS, mitochondrial function and cancer. Mitochondria are major sources of ROS production and rely on epitranscriptomic marks for regulated translation of ROS-management proteins, with mitochondrial thioredoxin reductase (TRXR2) being a prime example. Disruption of various RNA marks in mitochondria can also lead to disrupted OXPHOS and/or ATP synthesis often culminating in dysregulated ROS levels and resulting in devastating disorders such as developmental and neurological disability, cancer, and a host of other free radical diseases. We have endeavored to shed light on the significance of epitranscriptomic systems in regulating translation and mitochondrial function and their links to disease states, especially as they pertain to the specialized translation and function of selenoproteins.
Figure 5. Diseases linked to selenoproteins and dysregulated epitranscriptomic marks on tRNASec.
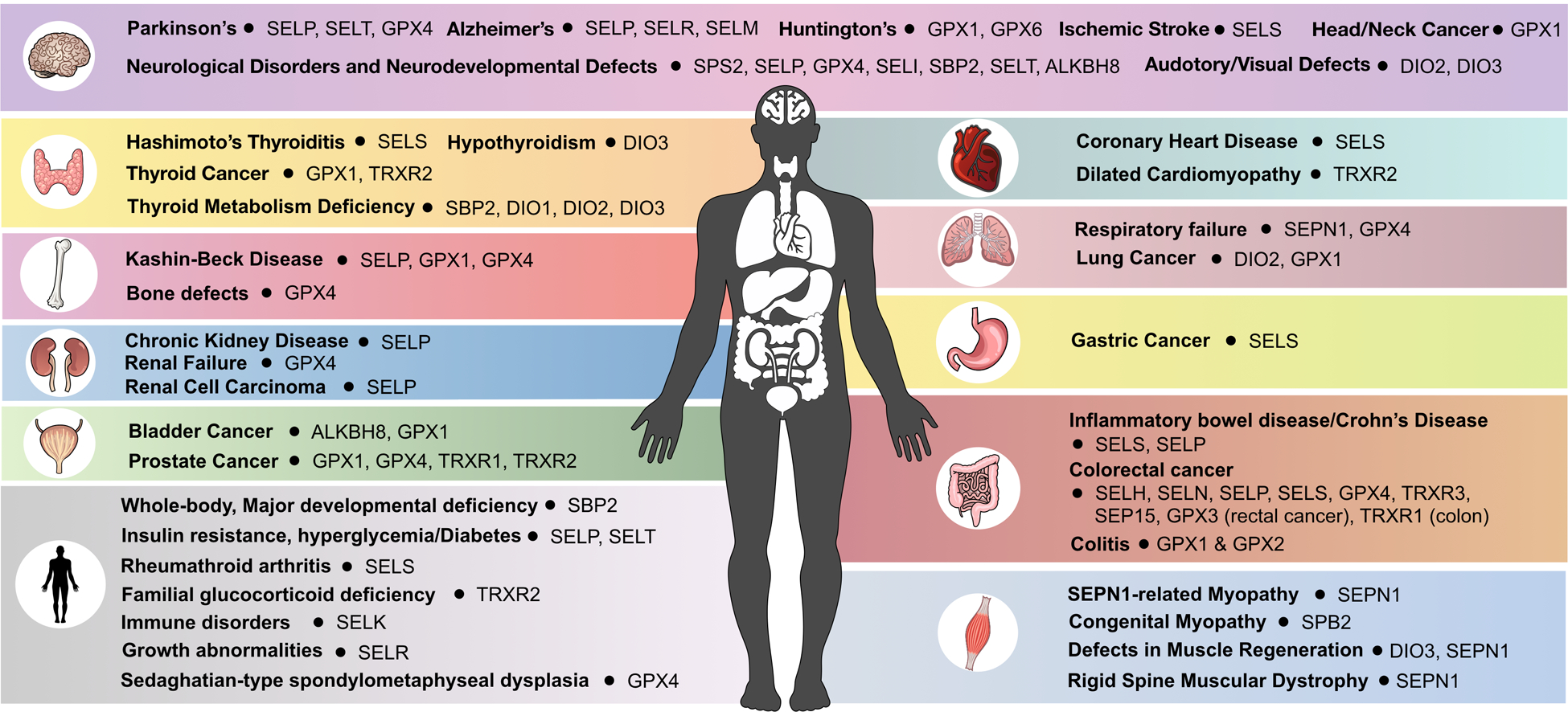
The 25 human selenoproteins and tRNASec writers and erasers were linked to diseases based on studies in mammalian systems (mammalian cells or animal models) and human diseases.
1.9. References
- [1].Bohacek J, Mansuy IM, Epigenetic inheritance of disease and disease risk, Neuropsychopharmacology. 38 (2013) 220–236. doi: 10.1038/npp.2012.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Johnstone SE, Baylin SB, Stress and the epigenetic landscape: A link to the pathobiology of human diseases?, Nat. Rev. Genet 11 (2010) 806–812. doi: 10.1038/nrg2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Lian CG, Xu Y, Ceol C, Wu F, Larson A, Dresser K, Xu W, Tan L, Hu Y, Zhan Q, Lee CW, Hu D, Lian BQ, Kleffel S, Yang Y, Neiswender J, Khorasani AJ, Fang R, Lezcano C, Duncan LM, Scolyer RA, Thompson JF, Kakavand H, Houvras Y, Zon LI, Mihm MC, Kaiser UB, Schatton T, Woda BA, Murphy GF, Shi YG, Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of Melanoma, Cell. 150 (2012) 1135–1146. doi: 10.1016/j.cell.2012.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Chan CTY, Deng W, Li F, Demott MS, Babu IR, Begley TJ, Dedon PC, Highly Predictive Reprogramming of tRNA Modifications Is Linked to Selective Expression of Codon-Biased Genes, Chem. Res. Toxicol 28 (2015) 978–988. doi: 10.1021/acs.chemrestox.5b00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Chan CTY, Dyavaiah M, DeMott MS, Taghizadeh K, Dedon PC, Begley TJ, A Quantitative Systems Approach Reveals Dynamic Control of tRNA Modifications during Cellular Stress, PLoS Genet. 6 (2010) e1001247. doi: 10.1371/journal.pgen.1001247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Dewe JM, Fuller BL, Lentini JM, Kellner SM, Fu D, TRMT1-catalyzed tRNA modifications are required for redox homeostasis to ensure proper cellular proliferation and oxidative stress survival, Mol. Cell. Biol 37 (2017) MCB.00214–17. doi: 10.1128/MCB.00214-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Chionh YH, McBee M, Babu IR, Hia F, Lin W, Zhao W, Cao J, Dziergowska A, Malkiewicz A, Begley TJ, Alonso S, Dedon PC, tRNA-mediated codon-biased translation in mycobacterial hypoxic persistence, Nat. Commun 7 (2016) 1–12. doi: 10.1038/ncomms13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Boccaletto P, MacHnicka MA, Purta E, Pitkowski P, Baginski B, Wirecki TK, De Crécy-Lagard V, Ross R, Limbach PA, Kotter A, Helm M, Bujnicki JM, MODOMICS: A database of RNA modification pathways. 2017 update, Nucleic Acids Res. 46 (2018) D303–D307. doi: 10.1093/nar/gkx1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Dubin DT, Taylor RH, The methylation state of poly A-containing-messenger RNA from cultured hamster cells, Nucleic Acids Res. 2 (1975) 1653–1668. doi: 10.1093/nar/2.10.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Camper SA, Albers RJ, Coward JK, Rottman FM, Effect of undermethylation on mRNA cytoplasmic appearance and half-life., Mol. Cell. Biol 4 (1984) 538–43. http://www.ncbi.nlm.nih.gov/pubmed/6201720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Chen S, Habib G, Yang C, Gu Z, Lee B, Weng S, Silberman, Cai S, Deslypere J, Rosseneu M, Et A, Apolipoprotein B-48 is the product of a messenger RNA with an organ-specific in-frame stop codon, Science (80-.) 238 (1987) 363–366. doi: 10.1126/science.3659919. [DOI] [PubMed] [Google Scholar]
- [12].Powell LM, Wallis SC, Pease RJ, Edwards YH, Knott TJ, Scott J, A novel form of tissue-specific RNA processing produces apolipoprotein-B48 in intestine, Cell. 50 (1987) 831–840. doi: 10.1016/0092-8674(87)90510-1. [DOI] [PubMed] [Google Scholar]
- [13].Teng B, Burant C, Davidson N, Molecular cloning of an apolipoprotein B messenger RNA editing protein, Science (80-.) 260 (1993) 1816–1819. doi: 10.1126/science.8511591. [DOI] [PubMed] [Google Scholar]
- [14].Davidson NO, Innerarity TL, Scott J, Smith H, Driscoll DM, Teng B, Chan L, Proposed nomenclature for the catalytic subunit of the mammalian apolipoprotein B mRNA editing enzyme: APOBEC-1., RNA. 1 (1995) 3 http://www.ncbi.nlm.nih.gov/pubmed/7489485. [PMC free article] [PubMed] [Google Scholar]
- [15].Smith HC, Bennett RP, Kizilyer A, McDougall WM, Prohaska KM, Functions and regulation of the APOBEC family of proteins, Semin. Cell Dev. Biol 23 (2012) 258–268. doi: 10.1016/j.semcdb.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Bass BL, Weintraub H, A developmentally regulated activity that unwinds RNA duplexes, Cell. 48 (1987) 607–613. doi: 10.1016/0092-8674(87)90239-X. [DOI] [PubMed] [Google Scholar]
- [17].Levanon EY, Eisenberg E, Yelin R, Nemzer S, Hallegger M, Shemesh R, Fligelman ZY, Shoshan A, Pollock SR, Sztybel D, Olshansky M, Rechavi G, Jantsch MF, Systematic identification of abundant A-to-I editing sites in the human transcriptome., Nat. Biotechnol 22 (2004) 1001–5. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- [18].Maas S, Rich A, Nishikura K, A-to-I RNA editing: recent news and residual mysteries., J. Biol. Chem 278 (2003) 1391–4. doi: 10.1074/jbc.R200025200. [DOI] [PubMed] [Google Scholar]
- [19].Higuchi M, Maas S, Single FN, Hartner J, Rozov A, Burnashev N, Feldmeyer D, Sprengel R, Seeburg PH, Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2., Nature. 406 (2000) 78–81. doi: 10.1038/35017558. [DOI] [PubMed] [Google Scholar]
- [20].Rueter SM, Dawson TR, Emeson RB, Regulation of alternative splicing by RNA editing., Nature. 399 (1999) 75–80. doi: 10.1038/19992. [DOI] [PubMed] [Google Scholar]
- [21].Scadden ADJ, O’Connell MA, Cleavage of dsRNAs hyper-edited by ADARs occurs at preferred editing sites., Nucleic Acids Res. 33 (2005) 5954–64. doi: 10.1093/nar/gki909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Scadden AD, Smith CW, Specific cleavage of hyper-edited dsRNAs., EMBO J. 20 (2001) 4243–52. doi: 10.1093/emboj/20.15.4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Chen CX, Cho DS, Wang Q, Lai F, Carter KC, Nishikura K, A third member of the RNA-specific adenosine deaminase gene family, ADAR3, contains both single- and double-stranded RNA binding domains., RNA. 6 (2000) 755–67. doi: 10.1017/s1355838200000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Mladenova D, Barry G, Konen LM, Pineda SS, Guennewig B, Avesson L, Zinn R, Schonrock N, Bitar M, Jonkhout N, Crumlish L, Kaczorowski DC, Gong A, Pinese M, Franco GR, Walkley CR, Vissel B, Mattick JS, Adar3 is involved in learning and memory in mice, Front. Neurosci 12 (2018) 1–17. doi: 10.3389/fnins.2018.00243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Maas S, Gerber AP, Rich A, Identification and characterization of a human tRNA-specific adenosine deaminase related to the ADAR family of pre-mRNA editing enzymes, Proc. Natl. Acad. Sci 96 (1999) 8895–8900. doi: 10.1073/pnas.96.16.8895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].V Murphy F, Ramakrishnan V, Structure of a purine-purine wobble base pair in the decoding center of the ribosome, Nat. Struct. Mol. Biol 11 (2004) 1251–1252. doi: 10.1038/nsmb866. [DOI] [PubMed] [Google Scholar]
- [27].Dominissini D, Rechavi G, Loud and Clear Epitranscriptomic m1A Signals: Now in Single-Base Resolution, Mol. Cell 68 (2017) 825–826. doi: 10.1016/j.molcel.2017.11.029. [DOI] [PubMed] [Google Scholar]
- [28].Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, Sorek R, Rechavi G, Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq, Nature. 485 (2012) 201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- [29].Squires JE, Patel HR, Nousch M, Sibbritt T, Humphreys DT, Parker BJ, Suter CM, Preiss T, Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA, Nucleic Acids Res. 40 (2012) 5023–5033. doi: 10.1093/nar/gks144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Pullirsch D, Jantsch MF, Proteome diversification by adenosine to inosine RNA-editing, RNA Biol. 7 (2010) 205–212. doi: 10.4161/rna.7.2.11286. [DOI] [PubMed] [Google Scholar]
- [31].Li X, Xiong X, Zhang M, Wang K, Chen Y, Zhou J, Mao Y, Lv J, Yi D, Chen X-W, Wang C, Qian S-B, Yi C, Base-resolution mapping reveals distinct m1A methylome in nuclear- and mitochondrial-encoded transcripts, 68 (2017) 993–1005. doi: 10.1002/stem.1868.Human. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Niu Y, Zhao X, Wu YS, Li MM, Wang XJ, Yang YG, N6-methyl-adenosine (m6A) in RNA: An Old Modification with A Novel Epigenetic Function, Genomics, Proteomics Bioinforma 11 (2013) 8–17. doi: 10.1016/j.gpb.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Maity A, Das B, N6-methyladenosine modification in mRNA: Machinery, function and implications for health and diseases, FEBS J. 283 (2016) 1607–1630. doi: 10.1111/febs.13614. [DOI] [PubMed] [Google Scholar]
- [34].Liu N, Zhou KI, Parisien M, Dai Q, Diatchenko L, Pan T, N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein, Nucleic Acids Res. 45 (2017) 6051–6063. doi: 10.1093/nar/gkx141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Yue Y, Liu J, He C, RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation, Genes Dev. 29 (2015) 1343–1355. doi: 10.1101/gad.262766.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Cao G, Li H-B, Yin Z, Flavell RA, Recent advances in dynamic m 6 A RNA modification, Open Biol. 6 (2016) 160003. doi: 10.1098/rsob.160003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Wu B, Li L, Huang Y, Ma J, Min J, Readers, writers and erasers of N6-methylated adenosine modification, Curr. Opin. Struct. Biol 47 (2017) 67–76. doi: 10.1016/j.sbi.2017.05.011. [DOI] [PubMed] [Google Scholar]
- [38].Roignant JY, Soller M, m6A in mRNA: An Ancient Mechanism for Fine-Tuning Gene Expression, Trends Genet. 33 (2017) 380–390. doi: 10.1016/j.tig.2017.04.003. [DOI] [PubMed] [Google Scholar]
- [39].Meyer KD, Jaffrey SR, The dynamic epitranscriptome: N6-methyladenosine and gene expression control, Nat. Rev. Mol. Cell Biol 15 (2014) 313–326. doi: 10.1038/nrm3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Meyer KD, Jaffrey SR, The dynamic epitranscriptome: N6-methyladenosine and gene expression control, 15 (2014) 313–326. doi: 10.1038/nrm3785.The. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Wang X, Feng J, Xue Y, Guan Z, Zhang D, Liu Z, Gong Z, Wang Q, Huang J, Tang C, Zou T, Yin P, Structural basis of N6-adenosine methylation by the METTL3-METTL14 complex, Nature. 534 (2016) 575–578. doi: 10.1038/nature18298. [DOI] [PubMed] [Google Scholar]
- [42].Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, Jia G, Yu M, Lu Z, Deng X, Dai Q, Chen W, He C, A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation., Nat. Chem. Biol 10 (2014) 93–95. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, Adhikari S, Shi Y, Lv Y, Chen YS, Zhao X, Li A, Yang Y, Dahal U, Lou XM, Liu X, Huang J, Yuan WP, Zhu XF, Cheng T, Zhao YL, Wang X, Danielsen JMR, Liu F, Yang YG, Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase, Cell Res. 24 (2014) 177–189. doi: 10.1038/cr.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Cui Q, Shi H, Ye P, Li L, Qu Q, Sun G, Sun G, Lu Z, Huang Y, Yang CG, Riggs AD, He C, Shi Y, m6A RNA Methylation Regulates the Self-Renewal and Tumorigenesis of Glioblastoma Stem Cells, Cell Rep. 18 (2017) 2622–2634. doi: 10.1016/j.celrep.2017.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Geula S, m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation, Science (80-.) 347 (2015) 1002. doi: 10.1126/science.1261417. [DOI] [PubMed] [Google Scholar]
- [46].Yoon KJ, Ringeling FR, Vissers C, Jacob F, Pokrass M, Jimenez-Cyrus D, Su Y, Kim NS, Zhu Y, Zheng L, Kim S, Wang X, Doré LC, Jin P, Regot S, Zhuang X, Canzar S, He C, li Ming G, Song H, Temporal Control of Mammalian Cortical Neurogenesis by m6A Methylation, Cell. 171 (2017) 877–889.e17. doi: 10.1016/j.cell.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Klungland A, Dahl JA, Greggains G, Fedorcsak P, Filipczyk A, Reversible RNA modifications in meiosis and pluripotency, Nat. Methods 14 (2016) 18–22. doi: 10.1038/nmeth.4111. [DOI] [PubMed] [Google Scholar]
- [48].Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR, Qian S, Dynamic m6A mRNA methylation directs translational control of heat shock response, Nature. 526 (2015) 591–594. doi: 10.1038/nature15377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Xiang Y, Laurent B, Hsu CH, Nachtergaele S, Lu Z, Sheng W, Xu C, Chen H, Ouyang J, Wang S, Ling D, Hsu PH, Zou L, Jambhekar A, He C, Shi Y, RNA m6 A methylation regulates the ultraviolet-induced DNA damage response, Nature. 543 (2017) 573–576. doi: 10.1038/nature21671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Cui Q, Shi H, Ye P, Li L, Qu Q, Sun G, Sun G, Lu Z, Huang Y, Yang C, Riggs AD, He C, Shi Y, m 6 A RNA Methylation Regulates the Self-Renewal and Tumorigenesis of Glioblastoma Stem Cells, Cell Rep. 18 (2017) 2622–2634. doi: 10.1016/j.celrep.2017.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Engel M, Eggert C, Kaplick PM, Eder M, Röh S, Tietze L, Namendorf C, Arloth J, Weber P, Rex-Haffner M, Geula S, Jakovcevski M, Hanna JH, Leshkowitz D, Uhr M, Wotjak CT, V Schmidt M, Deussing JM, Binder EB, Chen A, The Role of m6A/m-RNA Methylation in Stress Response Regulation., Neuron. 99 (2018) 389–403.e9. doi: 10.1016/j.neuron.2018.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang YG, He C, N6-Methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO, Nat. Chem. Biol 8 (2012) 1008. doi: 10.1038/nchembio1212-1008a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, Vågbø CB, Shi Y, Wang WL, Song SH, Lu Z, Bosmans RPG, Dai Q, Hao YJ, Yang X, Zhao WM, Tong WM, Wang XJ, Bogdan F, Furu K, Fu Y, Jia G, Zhao X, Liu J, Krokan HE, Klungland A, Yang YG, He C, ALKBH5 Is a Mammalian RNA Demethylase that Impacts RNA Metabolism and Mouse Fertility, Mol. Cell 49 (2013) 18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Li Z, Weng H, Su R, Weng X, Zuo Z, Li C, Huang H, Nachtergaele S, Dong L, Hu C, Qin X, Tang L, Wang Y, Hong G, Huang H, Wang X, Chen P, Gurbuxani S, Arnovitz S, Li Y, Li S, Strong J, Neilly MB, Larson RA, Jiang X, Zhang P, Jin J, He C, Chen J, FTO Plays an Oncogenic Role in Acute Myeloid Leukemia as a N 6 -Methyladenosine RNA Demethylase, Cancer Cell. 31 (2017) 127–141. doi: 10.1016/j.ccell.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Zhang S, The m6A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-Like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program, 31 (2017) 591–606. doi: 10.1016/j.ccell.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Zheng Q, Hou J, Zhou Y, Li Z, Cao X, The RNA helicase DDX46 inhibits innate immunity by entrapping m 6 A-demethylated antiviral transcripts in the nucleus, Nat. Immunol 18 (2017) 1094–1103. doi: 10.1038/ni.3830. [DOI] [PubMed] [Google Scholar]
- [57].DAVIS FF, ALLEN FW, Ribonucleic acids from yeast which contain a fifth nucleotide., J. Biol. Chem 227 (1957) 907–15. http://www.ncbi.nlm.nih.gov/pubmed/13463012. [PubMed] [Google Scholar]
- [58].Cohn WE, Pseudouridine, a carbon-carbon linked ribonucleoside in ribonucleic acids: isolation, structure, and chemical characteristics., J. Biol. Chem 235 (1960) 1488–98. http://www.ncbi.nlm.nih.gov/pubmed/13811056. [PubMed] [Google Scholar]
- [59].Yu C-T, Allen FW, Studies of an isomer of uridine isolated from ribonucleic acids, Biochim. Biophys. Acta 32 (1959) 393–406. doi: 10.1016/0006-3002(59)90612-2. [DOI] [PubMed] [Google Scholar]
- [60].Carlile TM, Rojas-Duran MF, Zinshteyn B, Shin H, Bartoli KM, Gilbert WV, Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells, Nature. 515 (2014) 143–146. doi: 10.1038/nature13802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Arnez JG, Steitz TA, Crystal Structure of Unmodified tRNAGln Complexed with Glutaminyl-tRNA Synthetase and ATP Suggests a Possible Role for Pseudo-Uridines in Stabilization of RNA Structure, Biochemistry. 33 (1994) 7560–7567. doi: 10.1021/bi00190a008. [DOI] [PubMed] [Google Scholar]
- [62].Charette M, Gray MW, Critical Review Pseudouridine in RNA: What, Where, How, and Why, IUBMB Life. 49 (2000) 341–351. [DOI] [PubMed] [Google Scholar]
- [63].Karijolich J, Yi C, Yu YT, Transcriptome-wide dynamics of RNA pseudouridylation, Nat. Rev. Mol. Cell Biol 16 (2015) 581–585. doi: 10.1038/nrm4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Fernández IS, Ng CL, Kelley AC, Wu G, Yu YT, Ramakrishnan V, Unusual base pairing during the decoding of a stop codon by the ribosome, Nature. 500 (2013) 107–110. doi: 10.1038/nature12302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Carlile TM, Rojas-duran MF, Zinshteyn B, Shin H, Kristen M, V Gilbert W, Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells, Nature. 515 (2014) 143–146. doi: 10.1038/nature13802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Rodnina MV, Fischer N, Maracci C, Stark H, Ribosome dynamics during decoding, Philos. Trans. R. Soc. B Biol. Sci 372 (2017). doi: 10.1098/rstb.2016.0182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Sharma S, Lafontaine DLJ, “View From A Bridge”: A New Perspective on Eukaryotic rRNA Base Modification, Trends Biochem. Sci 40 (2015) 560–575. doi: 10.1016/j.tibs.2015.07.008. [DOI] [PubMed] [Google Scholar]
- [68].Czerwoniec A, Dunin-Horkawicz S, Purta E, Kaminska KH, Kasprzak JM, Bujnicki JM, Grosjean H, Rother K, MODOMICS: A database of RNA modification pathways. 2008 update, Nucleic Acids Res. 37 (2009) 118–121. doi: 10.1007/s00270-014-0858-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Cantara WA, Crain PF, Rozenski J, McCloskey JA, Harris KA, Zhang X, Vendeix FAP, Fabris D, Agris PF, The RNA modification database, RNAMDB: 2011 update, Nucleic Acids Res. 39 (2011) 195–201. doi: 10.1093/nar/gkq1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Wang X, He C, Dynamic RNA modifications in posttranscriptional regulation, Mol. Cell 56 (2014) 5–12. doi: 10.1016/j.molcel.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Schattner P, Barberan-Soler S, Lowe TM, A computational screen for mammalian pseudouridylation guide H/ACA RNAs, Rna. 12 (2006) 15–25. doi: 10.1261/rna.2210406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Bykhovskaya Y, Casas K, Mengesha E, Inbal A, Fischel-Ghodsian N, Missense Mutation in Pseudouridine Synthase 1 (PUS1) Causes Mitochondrial Myopathy and Sideroblastic Anemia (MLASA), Am. J. Hum. Genet 74 (2004) 1303–1308. doi: 10.1086/421530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Zeharia A, Mitochondrial Myopathy, Sideroblastic Anemia, and Lactic Acidosis: An Autosomal Recessive Syndrome in Persian Jews Caused by a Mutation in the PUS1 Gene, J Child Neurol. 20 (2005) 449–452. [DOI] [PubMed] [Google Scholar]
- [74].Bellodi C, Krasnykh O, Haynes N, Theodoropoulou M, Peng G, Montanaro L, Ruggero D, Loss of function of the tumor suppressor DKC1 perturbs p27 translation control and contributes to pituitary tumorigenesis, Cancer Res. 70 (2010) 6026–6035. doi: 10.1158/0008-5472.CAN-09-4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Banerjee R, Chen S, Dare K, Gilreath M, Praetorius-Ibba M, Raina M, Reynolds NM, Rogers T, Roy H, Yadavalli SS, Ibba M, tRNAs: cellular barcodes for amino acids., FEBS Lett. 584 (2010) 387–95. doi: 10.1016/j.febslet.2009.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Zhong J, Xiao C, Gu W, Du G, Sun X, He Q-Y, Zhang G, Transfer RNAs Mediate the Rapid Adaptation of Escherichia coli to Oxidative Stress, PLOS Genet. 11 (2015) e1005302. doi: 10.1371/journal.pgen.1005302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Wek SA, Zhu S, Wek RC, The histidyl-tRNA synthetase-related sequence in the eIF-2 alpha protein kinase GCN2 interacts with tRNA and is required for activation in response to starvation for different amino acids., Mol. Cell. Biol 15 (1995) 4497–506. http://www.ncbi.nlm.nih.gov/pubmed/7623840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Dong J, Qiu H, Garcia-Barrio M, Anderson J, Hinnebusch AG, Uncharged tRNA activates GCN2 by displacing the protein kinase moiety from a bipartite tRNA-binding domain., Mol. Cell 6 (2000) 269–79. http://www.ncbi.nlm.nih.gov/pubmed/10983975. [DOI] [PubMed] [Google Scholar]
- [79].Doyle F, Leonardi A, Endres L, Tenenbaum SA, Dedon PC, Begley TJ, Gene- and genome-based analysis of significant codon patterns in yeast, rat and mice genomes with the CUT Codon UTilization tool., Methods. 107 (2016) 98–109. doi: 10.1016/j.ymeth.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Jackman JE, Alfonzo JD, Transfer RNA modifications: nature’s combinatorial chemistry playground., Wiley Interdiscip. Rev. RNA 4 (2013) 35–48. doi: 10.1002/wrna.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Begley U, Dyavaiah M, Patil A, Rooney JP, DiRenzo D, Young CM, Conklin DS, Zitomer RS, Begley TJ, Trm9-Catalyzed tRNA Modifications Link Translation to the DNA Damage Response, Mol. Cell 28 (2007) 860–870. doi: 10.1016/j.molcel.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Endres L, Begley U, Clark R, Gu C, Dziergowska A, Małkiewicz A, Melendez JA, Dedon PC, Begley TJ, Alkbh8 Regulates Selenocysteine-Protein Expression to Protect against Reactive Oxygen Species Damage., PLoS One. 10 (2015) e0131335. doi: 10.1371/journal.pone.0131335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Gu C, Begley TJ, Dedon PC, TRNA modifications regulate translation during cellular stress, FEBS Lett. 588 (2014) 4287–4296. doi: 10.1016/j.febslet.2014.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Patil A, Dyavaiah M, Joseph F, Rooney JP, Chan CTY, Dedon PC, Begley TJ, Increased tRNA modification and gene-specific codon usage regulate cell cycle progression during the DNA damage response, Cell Cycle. 11 (2012) 3656–3665. doi: 10.4161/cc.21919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Chan CTY, Pang YLJ, Deng W, Babu IR, Dyavaiah M, Begley TJ, Dedon PC, Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins, Nat. Commun 3 (2012) 937. doi: 10.1038/ncomms1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Dedon PC, Begley TJ, A system of RNA modifications and biased codon use controls cellular stress response at the level of translation, Chem. Res. Toxicol 27 (2014) 330–337. doi: 10.1021/tx400438d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Begley TJ, Dedon PC, Chan CTY, Dyavaiah M, Pang YLJ, Deng W, Babu IR, Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins, Nat. Commun 3 (2012). doi: 10.1038/ncomms1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Bauer F, Hermand D, A coordinated codon-dependent regulation of translation by Elongator, Cell Cycle. 11 (2012) 4524–4529. doi: 10.4161/cc.22689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Bauer F, Matsuyama A, Candiracci J, Dieu M, Scheliga J, Wolf DA, Yoshida M, Hermand D, Translational Control of Cell Division by Elongator, Cell Rep. 1 (2012) 424–433. doi: 10.1016/j.celrep.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Deng W, Babu IR, Su D, Yin S, Begley TJ, Dedon PC, Trm9-Catalyzed tRNA Modifications Regulate Global Protein Expression by Codon-Biased Translation, PLoS Genet. 11 (2015) 1–24. doi: 10.1371/journal.pgen.1005706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Endres L, Begley U, Clark R, Gu C, Dziergowska A, Małkiewicz A, Melendez JA, Dedon PC, Begley TJ, Alkbh8 regulates selenocysteine-protein expression to protect against reactive oxygen species damage, PLoS One. 10 (2015) 1–24. doi: 10.1371/journal.pone.0131335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Begley U, Sosa MS, Avivar-Valderas A, Patil A, Endres L, Estrada Y, Chan CTY, Su D, Dedon PC, Aguirre-Ghiso JA, Begley T, A human tRNA methyltransferase 9-like protein prevents tumour growth by regulating LIN9 and HIF1-α, EMBO Mol. Med 5 (2013) 366–383. doi: 10.1002/emmm.201201161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Rapino F, Delaunay S, Rambow F, Zhou Z, Tharun L, De Tullio P, Sin O, Shostak K, Schmitz S, Piepers J, Ghesquière B, Karim L, Charloteaux B, Jamart D, Florin A, Lambert C, Rorive A, Jerusalem G, Leucci E, Dewaele M, Vooijs M, Leidel SA, Georges M, Voz M, Peers B, Büttner R, Marine J-C, Chariot A, Close P, Codon-specific translation reprogramming promotes resistance to targeted therapy, Nature. 558 (2018) 605–609. doi: 10.1038/s41586-018-0243-7. [DOI] [PubMed] [Google Scholar]
- [94].Cantara WA, Crain PF, Rozenski J, McCloskey JA, Harris KA, Zhang X, Vendeix FAP, Fabris D, Agris PF, The RNA Modification Database, RNAMDB: 2011 update., Nucleic Acids Res. 39 (2011) D195–201. doi: 10.1093/nar/gkq1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Suzuki T, Nagao A, Suzuki T, Human Mitochondrial tRNAs: Biogenesis, Function, Structural Aspects, and Diseases, Annu. Rev. Genet 45 (2011) 299–329. doi: 10.1146/annurev-genet-110410-132531. [DOI] [PubMed] [Google Scholar]
- [96].Rapino F, Delaunay S, Zhou Z, Chariot A, Close P, tRNA Modification: Is Cancer Having a Wobble?, Trends in Cancer. 3 (2017) 249–252. doi: 10.1016/j.trecan.2017.02.004. [DOI] [PubMed] [Google Scholar]
- [97].Fu D, Begley U, Samson LD, Brophy JAN, Paules RS, Atmore KA, Dedon PC, Begley TJ, Chan CTY, Human AlkB Homolog ABH8 Is a tRNA Methyltransferase Required for Wobble Uridine Modification and DNA Damage Survival, Mol. Cell. Biol 30 (2010) 2449–2459. doi: 10.1128/mcb.01604-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Hatfield DL, Gladyshev VN, How selenium has altered our understanding of the genetic code, Mol. Cell. Biol 22 (2002) 3565–3576. doi: 10.1128/MCB.22.11.3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Sprinzl M, Vassilenko KS, Compilation of tRNA sequences and sequences of tRNA genes, Nucleic Acids Res 33 (2005) D139–D140. doi: 10.1093/nar/gki012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Songe-Moller L, van den Born E, Leihne V, Vagbo CB, Kristoffersen T, Krokan HE, Kirpekar F, Falnes PO, Klungland A, Mammalian ALKBH8 Possesses tRNA Methyltransferase Activity Required for the Biogenesis of Multiple Wobble Uridine Modifications Implicated in Translational Decoding, Mol. Cell. Biol 30 (2010) 1814–1827. doi: 10.1128/MCB.01602-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Lentini JM, Ramos J, Fu D, Monitoring the 5-methoxycarbonylmethyl-2-thiouridine (mcm5s2U) modification in eukaryotic tRNAs via the γ-toxin endonuclease, Rna. 24 (2018) 749–758. doi: 10.1261/rna.065581.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Patil A, Chan CTY, Dyavaiah M, Rooney JP, Dedon PC, Begley TJ, Translational infidelity-induced protein stress results from a deficiency in Trm9-catalyzed tRNA modifications., RNA Biol. 9 (2012) 990–1001. doi: 10.4161/rna.20531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Zinshteyn B, Gilbert WV, Loss of a Conserved tRNA Anticodon Modification Perturbs Cellular Signaling, PLoS Genet. 9 (2013) e1003675. doi: 10.1371/journal.pgen.1003675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Nedialkova DD, Leidel SA, Optimization of Codon Translation Rates via tRNA Modifications Maintains Proteome Integrity, Cell. 161 (2015) 1606. doi: 10.1016/J.CELL.2015.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Paredes JA, Carreto L, Simões J, Bezerra AR, Gomes AC, Santamaria R, Kapushesky M, Moura GR, Santos MAS, Low level genome mistranslations deregulate the transcriptome and translatome and generate proteotoxic stress in yeast, BMC Biol. 10 (2012). doi: 10.1186/1741-7007-10-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Tükenmez H, Xu H, Esberg A, Byström AS, The role of wobble uridine modifications in +1 translational frameshifting in eukaryotes, Nucleic Acids Res. 43 (2015) 9489–9499. doi: 10.1093/nar/gkv832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Kirchner S, Ignatova Z, Emerging roles of tRNA in adaptive translation, signalling dynamics and disease, Nat. Rev. Genet 16 (2015) 98–112. doi: 10.1038/nrg3861. [DOI] [PubMed] [Google Scholar]
- [108].Lyko F, Tuorto F, Genome recoding by tRNA modifications, Open Biol. (2016) 0–2. doi: 10.1098/rsob.160287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Monies D, Vågbø CB, Al-Owain M, Alhomaidi S, Alkuraya FS, Recessive Truncating Mutations in ALKBH8 Cause Intellectual Disability and Severe Impairment of Wobble Uridine Modification, Am. J. Hum. Genet 104 (2019) 1202–1209. doi: 10.1016/j.ajhg.2019.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Kirino Y, Yasukawa T, Ohta S, Akira S, Ishihara K, Watanabe K, Suzuki T, Codon-specific translational defect caused by a wobble modification deficiency in mutant tRNA from a human mitochondrial disease., Proc. Natl. Acad. Sci. U. S. A 101 (2004) 15070–5. doi: 10.1073/pnas.0405173101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Schaffer AE, Eggens VRC, Caglayan AO, Reuter MS, Scott E, Coufal NG, Silhavy JL, Xue Y, Kayserili H, Yasuno K, Rosti RO, Abdellateef M, Caglar C, Kasher PR, Cazemier JL, Weterman MA, Cantagrel V, Cai N, Zweier C, Altunoglu U, Satkin NB, Aktar F, Tuysuz B, Yalcinkaya C, Caksen H, Bilguvar K, Fu XD, Trotta CR, Gabriel S, Reis A, Gunel M, Baas F, Gleeson JG, CLP1 founder mutation links tRNA splicing and maturation to cerebellar development and neurodegeneration, Cell. 157 (2014) 651–663. doi: 10.1016/j.cell.2014.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Wei F, Suzuki T, Tomizawa K, Wei F, Suzuki T, Watanabe S, Kimura S, Kaitsuka T, Deficit of tRNA Lys modification by Cdkal1 causes the development of type 2 diabetes in mice Find the latest version : Deficit of tRNA Lys modification by Cdkal1 causes the development of type 2 diabetes in mice, J Clin Invest. 121 (2011) 3598–3608. doi: 10.1172/JCI58056DS1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Spinola M, Galvan A, Pignatiello C, Conti B, Pastorino U, Nicander B, Paroni R, Dragani TA, Identification and functional characterization of the candidate tumor suppressor gene TRIT1 in human lung cancer, Oncogene. 24 (2005) 5502–5509. doi: 10.1038/sj.onc.1208687. [DOI] [PubMed] [Google Scholar]
- [114].Wei FY, Zhou B, Suzuki T, Miyata K, Ujihara Y, Horiguchi H, Takahashi N, Xie P, Michiue H, Fujimura A, Kaitsuka T, Matsui H, Koga Y, Mohri S, Suzuki T, Oike Y, Tomizawa K, Cdk5rap1-mediated 2-methylthio modification of mitochondrial tRNAs governs protein translation and contributes to myopathy in mice and humans, Cell Metab. 21 (2015) 428–442. doi: 10.1016/j.cmet.2015.01.019. [DOI] [PubMed] [Google Scholar]
- [115].Fakruddin M, Wei FY, Emura S, Matsuda S, Yasukawa T, Kang D, Tomizawa K, Cdk5rap1-mediated 2-methylthio-N 6-isopentenyladenosine modification is absent from nuclear-derived RNA species, Nucleic Acids Res. 45 (2017) 11954–11961. doi: 10.1093/nar/gkx819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Schweizer U, Bohleber S, Fradejas-Villar N, The modified base isopentenyladenosine and its derivatives in tRNA, (2017). doi: 10.1080/15476286.2017.1294309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Lin H, Miyauchi K, Harada T, Okita R, Takeshita E, Komaki H, Fujioka K, Yagasaki H, Goto YI, Yanaka K, Nakagawa S, Sakaguchi Y, Suzuki T, CO 2 -sensitive tRNA modification associated with human mitochondrial disease, Nat. Commun 9 (2018). doi: 10.1038/s41467-018-04250-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Shoffner JM, Lott MT, Lezza AM, Seibel P, Ballinger SW, Wallace DC, Myoclonic epilepsy and ragged-red fiber disease (MERRF) is associated with a mitochondrial DNA tRNA(Lys) mutation., Cell. 61 (1990) 931–7. http://www.ncbi.nlm.nih.gov/pubmed/2112427. [DOI] [PubMed] [Google Scholar]
- [119].Yasukawa T, Suzuki T, Ishii N, Ohta S, Watanabe K, Wobble modification defect in tRNA disturbs codon-anticodon interaction in a mitochondrial disease., EMBO J. 20 (2001) 4794–802. doi: 10.1093/emboj/20.17.4794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Yasukawa T, Suzuki T, Ueda T, Ohta S, Watanabe K, Modification defect at anticodon wobble nucleotide of mitochondrial tRNAs(Leu)(UUR) with pathogenic mutations of mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes., J. Biol. Chem 275 (2000) 4251–7. http://www.ncbi.nlm.nih.gov/pubmed/10660592. [DOI] [PubMed] [Google Scholar]
- [121].Nakano S, Suzuki T, Kawarada L, Iwata H, Asano K, Suzuki T, NSUN3 methylase initiates 5-formylcytidine biogenesis in human mitochondrial tRNAMet, Nat. Chem. Biol 12 (2016) 546–551. doi: 10.1038/nchembio.2099. [DOI] [PubMed] [Google Scholar]
- [122].Haag S, Sloan KE, Ranjan N, Warda AS, Kretschmer J, Blessing C, Hübner B, Seikowski J, Dennerlein S, Rehling P, V Rodnina M, Höbartner C, Bohnsack MT, NSUN3 and ABH1 modify the wobble position of mt-tRNA Met to expand codon recognition in mitochondrial translation, (n.d.) doi: 10.15252/embj.201694885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Van Haute L, Dietmann S, Kremer L, Hussain S, Pearce SF, Powell CA, Rorbach J, Lantaff R, Blanco S, Sauer S, Kotzaeridou U, Hoffmann GF, Memari Y, Kolb-Kokocinski A, Durbin R, Mayr JA, Frye M, Prokisch H, Minczuk M, Deficient methylation and formylation of mt-tRNA Met wobble cytosine in a patient carrying mutations in NSUN3, Nat. Commun 7 (2016). doi: 10.1038/ncomms12039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Boczonadi V, Horvath R, Mitochondria: impaired mitochondrial translation in human disease., Int. J. Biochem. Cell Biol 48 (2014) 77–84. doi: 10.1016/j.biocel.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Chen Q, Vazquez EJ, Moghaddas S, Hoppel CL, Lesnefsky EJ, Production of Reactive Oxygen Species by Mitochondria, J. Biol. Chem 278 (2003) 36027–36031. doi: 10.1074/jbc.m304854200. [DOI] [PubMed] [Google Scholar]
- [126].Sousa JS, D’Imprima E, Vonck J, Mitochondrial respiratory chain complexes, in: Subcell. Biochem, Springer, Singapore, 2018: pp. 167–227. doi: 10.1007/978-981-10-7757-9_7. [DOI] [PubMed] [Google Scholar]
- [127].Neupert W, A perspective on transport of proteins into mitochondria: A myriad of open questions, J. Mol. Biol 427 (2015) 1135–1158. doi: 10.1016/j.jmb.2015.02.001. [DOI] [PubMed] [Google Scholar]
- [128].Murphy MP, How mitochondria produce reactive oxygen species, Biochem. J 417 (2009) 1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Turrens JF, Mitochondrial formation of reactive oxygen species, J. Physiol 552 (2003) 335–344. doi: 10.1113/jphysiol.2003.049478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Muller FL, Liu Y, Van Remmen H, Complex III releases superoxide to both sides of the inner mitochondrial membrane, J. Biol. Chem 279 (2004) 49064–49073. doi: 10.1074/jbc.M407715200. [DOI] [PubMed] [Google Scholar]
- [131].Orr AL, Vargas L, Turk CN, Baaten JE, Matzen JT, Dardov VJ, Attle SJ, Li J, Quackenbush DC, Goncalves RLS, V Perevoshchikova I, Petrassi HM, Meeusen SL, Ainscow EK, Brand MD, Suppressors of superoxide production from mitochondrial complex III, Nat. Chem. Biol 11 (2015) 834–836. doi: 10.1038/nchembio.1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Han D, Antunes F, Canali R, Rettori D, Cadenas E, Voltage-dependent anion channels control the release of the superoxide anion from mitochondria to cytosol, J. Biol. Chem 278 (2003) 5557–5563. doi: 10.1074/jbc.M210269200. [DOI] [PubMed] [Google Scholar]
- [133].Fridovich I, Superoxide anion radical (O2-.), superoxide dismutases, and related matters., J. Biol. Chem 272 (1997) 18515–7. doi: 10.1074/JBC.272.30.18515. [DOI] [PubMed] [Google Scholar]
- [134].Sena LA, Chandel NS, Physiological roles of mitochondrial reactive oxygen species, (2012). doi: 10.1016/j.molcel.2012.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Hamanaka RB, Chandel NS, Mitochondrial reactive oxygen species regulate cellular signaling and dictate biological outcomes Production of ROS by the Mitochondrial Electron Transport Chain, Trends Biochem Sci. 35 (2010) 505–513. doi: 10.1016/j.tibs.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Chandel NS, Cell Metabolism Essay Evolution of Mitochondria as Signaling Organelles, Cell Metab. 22 (2015) 204–206. doi: 10.1016/j.cmet.2015.05.013. [DOI] [PubMed] [Google Scholar]
- [137].Holmström KM, Finkel T, Cellular mechanisms and physiological consequences of redox-dependent signalling, Nat. Rev. Mol. Cell Biol 15 (2014) 411–421. doi: 10.1038/nrm3801. [DOI] [PubMed] [Google Scholar]
- [138].Georgieva E, Ivanova D, Zhelev Z, Mitochondrial Dysfunction and Redox Imbalance as a Diagnostic Marker of “Free Radical Diseases,” Anticancer Res. 37 (2017) 5373–5381. doi: 10.21873/anticanres.11963. [DOI] [PubMed] [Google Scholar]
- [139].Endres L, Dedon PC, Begley TJ, Codon-biased translation can be regulated by wobble-base tRNA modification systems during cellular stress responses, RNA Biol. 12 (2015) 603–614. doi: 10.1080/15476286.2015.1031947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Huber S, Leonardi A, Dedon P, Begley T, The Versatile Roles of the tRNA Epitranscriptome during Cellular Responses to Toxic Exposures and Environmental Stress, Toxics. 7 (2019) 17. doi: 10.3390/toxics7010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Yoshioka J, Thioredoxin Reductase 2 (Txnrd2) Regulates Mitochondrial Integrity in the Progression of Age-Related Heart Failure, J. Am. Heart Assoc 4 (2015). doi: 10.1161/JAHA.115.002278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Handy DE, Lubos E, Yang Y, Galbraith JD, Kelly N, Zhang Y, Leopold JA, Loscalzo J, Glutathione Peroxidase-1 Regulates Mitochondrial Function to Modulate Redox-dependent Cellular Responses, J. Biol. Chem 284 (2009) 11913–11921. doi: 10.1074/jbc.M900392200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Stanley BA, Sivakumaran V, Shi S, McDonald I, Lloyd D, Watson WH, Aon MA, Paolocci N, Thioredoxin reductase-2 is essential for keeping low levels of H 2O 2 emission from isolated heart mitochondria, J. Biol. Chem 286 (2011) 33669–33677. doi: 10.1074/jbc.M111.284612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Lopert P, Day BJ, Patel M, Thioredoxin Reductase Deficiency Potentiates Oxidative Stress, Mitochondrial Dysfunction and Cell Death in Dopaminergic Cells, PLoS One. 7 (2012) e50683. doi: 10.1371/journal.pone.0050683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Conrad M, Jakupoglu C, Moreno SG, Lippl S, Banjac A, Schneider M, Beck H, Hatzopoulos AK, Just U, Sinowatz F, Schmahl W, Chien KR, Wurst W, Bornkamm GW, Brielmeier M, Essential Role for Mitochondrial Thioredoxin Reductase in Hematopoiesis, Heart Development, and Heart Function, Mol. Cell. Biol 24 (2004) 9414–9423. doi: 10.1128/mcb.24.21.9414-9423.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Hubert N, Sturchler C, Westhof E, Carbon P, Krol A, The 9/4 secondary structure of eukaryotic selenocysteine tRNA: More pieces of evidence, Rna. 4 (1998) 1029–1033. doi: 10.1017/S1355838298980888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Sturchler-Pierrat C, Hubert N, Totsuka T, Mizutani T, Carbon P, Krol A, Selenocysteylation in eukaryotes necessitates the uniquely long aminoacyl acceptor stem of selenocysteine tRNASec, J. Biol. Chem 270 (1995) 18570–18574. doi: 10.1074/jbc.270.31.18570. [DOI] [PubMed] [Google Scholar]
- [148].Pan T, Modifications and functional genomics of human transfer RNA, Cell Res. 28 (2018) 395–404. doi: 10.1038/s41422-018-0013-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Kim LK, Matsufuji T, Matsufuji S, Carlson BA, Kim SS, Hatfield DL, Lee BJ, Methylation of the ribosyl moiety at position 34 of selenocysteine tRNA[Ser]Sec is governed by both primary and tertiary structure., RNA. 6 (2000) 1306–15. http://www.ncbi.nlm.nih.gov/pubmed/10999607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Diamond AM, Choi In Shon, Crain PF, Hashizume T, Pomerantz SC, Cruz R, Steer CJ, Hill KE, Burk RF, McCloskey JA, Hatfield DL, Dietary selenium affects methylation of the wobble nucleoside in the anticodon of selenocysteine tRNA([Ser]Sec), J. Biol. Chem 268 (1993) 14215–14223. [PubMed] [Google Scholar]
- [151].Hatfield D, Lee BJ, Hampton L, Diamond AM, Selenium induces changes in the selenocysteine tRNA[Ser]Secpopulation in mammalian cells, Nucleic Acids Res. 19 (1991) 939–943. doi: 10.1093/nar/19.4.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Karlsborn T, Tükenmez H, Mahmud AKMF, Xu F, Xu H, Byström AS, Elongator, a conserved complex required for wobble uridine modifications in Eukaryotes, RNA Biol. 11 (2014) 1519–1528. doi: 10.4161/15476286.2014.992276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Huang B, Johansson MJO, Byström AS, An early step in wobble uridine tRNA modification requires the Elongator complex., RNA. 11 (2005) 424–36. doi: 10.1261/rna.7247705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Van Den Born E, Vågbø CB, Songe-Møller L, Leihne V, Lien GF, Leszczynska G, Malkiewicz A, Krokan HE, Kirpekar F, Klungland A, Falnes P, ALKBH8-mediated formation of a novel diastereomeric pair of wobble nucleosides in mammalian tRNA, Nat. Commun 2 (2011) 172–177. doi: 10.1038/ncomms1173. [DOI] [PubMed] [Google Scholar]
- [155].Fradejas N, Carlson BA, Rijntjes E, Becker N-P, Tobe R, Schweizer U, Mammalian Trit1 is a tRNA [Ser]Sec -isopentenyl transferase required for full selenoprotein expression, Biochem. J 450 (2013) 427–432. doi: 10.1042/BJ20121713. [DOI] [PubMed] [Google Scholar]
- [156].Roovers M, Hale C, Tricot C, Terns MP, Terns RM, Grosjean H, Droogmans L, Formation of the conserved pseudouridine at position 55 in archaeal tRNA, Nucleic Acids Res. 34 (2006) 4293–4301. doi: 10.1093/nar/gkl530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Becker HF, Motorin Y, Planta RJ, Grosjean H, The yeast gene YNL292w encodes a pseudouridine synthase (Pus4) catalyzing the formation of psi55 in both mitochondrial and cytoplasmic tRNAs., Nucleic Acids Res. 25 (1997) 4493–9. doi: 10.1093/nar/25.22.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Gurha P, Gupta R, Archaeal Pus10 proteins can produce both pseudouridine 54 and 55 in tRNA, Rna. 14 (2008) 2521–2527. doi: 10.1261/rna.1276508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Oerum S, Dégut C, Barraud P, Tisné C, m1A post-transcriptional modification in tRNAs, Biomolecules. 7 (2017) 1–15. doi: 10.3390/biom7010020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Wang M, Zhu Y, Wang C, Fan X, Jiang X, Ebrahimi M, Qiao Z, Niu L, Teng M, Li X, Crystal structure of the two-subunit tRNA m1 A58 methyltransferase TRM6-TRM61 from Saccharomyces cerevisiae, Sci. Rep 6 (2016) 1–10. doi: 10.1038/srep32562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Liu F, Clark W, Luo G, Wang X, Fu Y, Wei J, Wang X, Hao Z, Dai Q, Zheng G, Ma H, Han D, Evans M, Klungland A, Pan T, He C, ALKBH1-Mediated tRNA Demethylation Regulates Translation, Cell. 167 (2016) 816–828.e16. doi: 10.1016/j.cell.2016.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Aas PA, Otterlei M, Falnes PØ, Vågbø CB, Skorpen F, Akbari M, Sundheim O, Bjørås M, Slupphaug G, Seeberg E, Krokan HE, Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA, Nature. 421 (2003) 859–863. doi: 10.1038/nature01363. [DOI] [PubMed] [Google Scholar]
- [163].Chen Z, Qi M, Shen B, Luo G, Wu Y, Li J, Lu Z, Zheng Z, Dai Q, Wang H, Transfer RNA demethylase ALKBH3 promotes cancer progression via induction of tRNA-derived small RNAs, Nucleic Acids Res. 47 (2019) 2533–2545. doi: 10.1093/nar/gky1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Carlson BA, Yoo MH, Tsuji PA, Gladyshev VN, Hatfield DL, Mouse models targeting selenocysteine tRNA expression for elucidating the role of selenoproteins in health and development, Molecules. 14 (2009) 3509–3527. doi: 10.3390/molecules14093509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Carlson BA, Moustafa ME, Sengupta A, Schweizer U, Shrimali R, Rao M, Zhong N, Wang S, Feigenbaum L, Byeong JL, Gladyshev VN, Hatfield DL, Selective restoration of the selenoprotein population in a mouse hepatocyte selenoproteinless background with different mutant selenocysteine tRNAs lacking Um34, J. Biol. Chem 282 (2007) 32591–32602. doi: 10.1074/jbc.M707036200. [DOI] [PubMed] [Google Scholar]
- [166].Hatfield DL, Carlson BA, Xu XM, Mix H, Gladyshev VN, Selenocysteine Incorporation Machinery and the Role of Selenoproteins in Development and Health, Prog. Nucleic Acid Res. Mol. Biol 81 (2006) 97–142. doi: 10.1016/S0079-6603(06)81003-2. [DOI] [PubMed] [Google Scholar]
- [167].Berry MJ, Banu L, Chen YY, Mandel SJ, Kieffer JD, Harney JW, Larsen PR, Recognition of UGA as a selenocysteine codon in type I deiodinase requires sequences in the 3’ untranslated region., Nature. 353 (1991) 273–6. doi: 10.1038/353273a0. [DOI] [PubMed] [Google Scholar]
- [168].Lesoon A, Mehta A, Singh R, Chisolm GM, Driscoll DM, An RNA-binding protein recognizes a mammalian selenocysteine insertion sequence element required for cotranslational incorporation of selenocysteine., Mol. Cell. Biol 17 (1997) 1977–1985. doi: 10.1128/MCB.17.4.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Copeland PR, Fletcher JE, Carlson BA, Hatfield DL, Driscoll DM, A novel RNA binding protein, SBP2, is required for the translation of mammalian selenoprotein mRNAs, EMBO J. 19 (2000) 306–314. doi: 10.1093/emboj/19.2.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Fagegaltier D, Hubert N, Yamada K, Mizutani T, Carbon P, Krol A, Characterization of mSelB, a novel mammalian elongation factor for selenoprotein translation., EMBO J. 19 (2000) 4796–805. doi: 10.1093/emboj/19.17.4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Copeland PR, Regulation of gene expression by stop codon recoding: selenocysteine., Gene. 312 (2003) 17–25. doi: 10.1016/j.neuroimage.2013.08.045.The. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Gu C, Begley TJ, Dedon PC, TRNA modifications regulate translation during cellular stress, FEBS Lett. 588 (2014) 4287–4296. doi: 10.1016/j.febslet.2014.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Low SC, Berry MJ, Knowing when not to stop: selenocysteine incorporation in eukaryotes, Trends Biochem. Sci 21 (1996) 203–208. doi: 10.1016/0968-0004(96)10025-6. [DOI] [PubMed] [Google Scholar]
- [174].Songe-Moller L, van den Born E, Leihne V, Vagbo CB, Kristoffersen T, Krokan HE, Kirpekar F, Falnes PO, Klungland A, Mammalian ALKBH8 Possesses tRNA Methyltransferase Activity Required for the Biogenesis of Multiple Wobble Uridine Modifications Implicated in Translational Decoding, Mol. Cell. Biol 30 (2010) 1814–1827. doi: 10.1128/MCB.01602-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Howard MT, Carlson BA, Anderson CB, Hatfield DL, Translational redefinition of UGA codons is regulated by selenium availability, J. Biol. Chem 288 (2013) 19401–19413. doi: 10.1074/jbc.M113.481051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Bösl MR, Takaku K, Oshima M, Nishimura S, Taketo MM, Early embryonic lethality caused by targeted disruption of the mouse selenocysteine tRNA gene (Trsp)., Proc. Natl. Acad. Sci. U. S. A 94 (1997) 5531–4. doi: 10.1073/pnas.94.11.5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Kumaraswamy E, Carlson BA, Morgan F, Miyoshi K, Robinson GW, Su D, Wang S, Southon E, Tessarollo L, Lee BJ, Gladyshev VN, Hennighausen L, Hatfield DL, Selective removal of the selenocysteine tRNA [Ser]Sec gene (Trsp) in mouse mammary epithelium., Mol. Cell. Biol 23 (2003) 1477–88. doi: 10.1128/MCB.23.5.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Allamand V, Richard P, Lescure A, Ledeuil C, Desjardin D, Petit N, Gartioux C, Ferreiro A, Krol A, Pellegrini N, Urtizberea JA, Guicheney P, A single homozygous point mutation in a 3′ untranslated region motif of selenoprotein N mRNA causes SEPN1-related myopathy, EMBO Rep. 7 (2006) 450–454. doi: 10.1038/sj.embor.7400648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Aubee JI, Olu M, Thompson KM, The i6A37 tRNA modification is essential for proper decoding of UUX-Leucine codons during rpoS and iraP translation, RNA. 22 (2016) 729–742. doi: 10.1261/rna.053165.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Reeves MA, Hoffmann PR, The human selenoproteome: Recent insights into functions and regulation, Cell. Mol. Life Sci 66 (2009) 2457–2478. doi: 10.1007/s00018-009-0032-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Kryukov GV, Castellano S, Novoselov SV, Lobanov AV, Zehtab O, Guigo R, Gladyshev VN, Characterization of mammalian selenoproteomes., Science (80-.) 300 (2003) 1439–1443. [DOI] [PubMed] [Google Scholar]
- [182].Reeves MA, Hoffmann PR, The human selenoproteome: Recent insights into functions and regulation, Cell. Mol. Life Sci 66 (2009) 2457–2478. doi: 10.1007/s00018-009-0032-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Méplan C, Selenium and chronic diseases: A nutritional genomics perspective, Nutrients. 7 (2015) 3621–3651. doi: 10.3390/nu7053621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Short SP, Williams CS, Selenoproteins in Tumorigenesis and Cancer Progression, in: 2017: pp. 49–83. doi: 10.1016/bs.acr.2017.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Fedirko V, Jenab M, Méplan C, Jones JS, Zhu W, Schomburg L, Siddiq A, Hybsier S, Overvad K, Tjønneland A, Omichessan H, Perduca V, Boutron-Ruault M-C, Kühn T, Katzke V, Aleksandrova K, Trichopoulou A, Karakatsani A, Kotanidou A, Tumino R, Panico S, Masala G, Agnoli C, Naccarati A, Bueno-de-Mesquita B, Vermeulen RCH, Weiderpass E, Skeie G, Nøst TH, Lujan-Barroso L, Quirós JR, Huerta JM, Rodríguez-Barranco M, Barricarte A, Gylling B, Harlid S, Bradbury KE, Wareham N, Khaw K-T, Gunter M, Murphy N, Freisling H, Tsilidis K, Aune D, Riboli E, Hesketh JE, Hughes DJ, Association of Selenoprotein and Selenium Pathway Genotypes with Risk of Colorectal Cancer and Interaction with Selenium Status, Nutrients. 11 (2019) 935. doi: 10.3390/nu11040935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Peters KM, Carlson BA, Gladyshev VN, Tsuji PA, Selenoproteins in colon cancer, Free Radic. Biol. Med 127 (2018) 14–25. doi: 10.1016/j.freeradbiomed.2018.05.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Haan D, Bladier JB,C, Griffiths P, Kelner M, O’Shea RD, Cheung NS, Bronson RT, Silvestro MJ, Wild S, Zheng SS, Beart PM, Hertzog PJ, Kola I, Mice with a Homozygous Null Mutation for the Most Abundant Glutathione Peroxidase, Gpx1, Show Increased Susceptibility to the Oxidative Stress-Inducing Agents Paraquat and Hydrogen Peroxide, J. Biol. Chem 273 (1998) 22528–22536. doi: 10.1074/jbc.273.35.22528. [DOI] [PubMed] [Google Scholar]
- [188].Duong C, Seow HJ, Bozinovski S, Crack PJ, Anderson GP, Vlahos R, Glutathione peroxidase-1 protects against cigarette smoke-induced lung inflammation in mice, Am. J. Physiol. Cell. Mol. Physiol 299 (2010) L425–L433. doi: 10.1152/ajplung.00038.2010. [DOI] [PubMed] [Google Scholar]
- [189].Yatmaz S, Seow HJ, Gualano RC, Wong ZX, Stambas J, Selemidis S, Crack PJ, Bozinovski S, Anderson GP, Vlahos R, Glutathione Peroxidase-1 Reduces Influenza A Virus–Induced Lung Inflammation, Am. J. Respir. Cell Mol. Biol 48 (2013) 17–26. doi: 10.1165/rcmb.2011-0345OC. [DOI] [PubMed] [Google Scholar]
- [190].Saccoccia F, Angelucci F, Boumis G, Carotti D, Desiato G, Miele A, Bellelli A, Thioredoxin Reductase and its Inhibitors, Curr. Protein Pept. Sci 15 (2014) 621–646. doi: 10.2174/1389203715666140530091910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Jakupoglu C, Przemeck GKH, Schneider M, Moreno SG, Mayr N, Hatzopoulos AK, de Angelis MH, Wurst W, Bornkamm GW, Brielmeier M, Conrad M, Cytoplasmic Thioredoxin Reductase Is Essential for Embryogenesis but Dispensable for Cardiac Development, Mol. Cell. Biol 25 (2005) 1980–1988. doi: 10.1128/MCB.25.5.1980-1988.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Bondareva AA, Capecchi MR, Iverson SV, Li Y, Lopez NI, Lucas O, Merrill GF, Prigge JR, Siders AM, Wakamiya M, Wallin SL, Schmidt EE, Effects of thioredoxin reductase-1 deletion on embryogenesis and transcriptome, Free Radic. Biol. Med 43 (2007) 911–923. doi: 10.1016/j.freeradbiomed.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Conrad M, Jakupoglu C, Moreno G, Lippl S, Banjac A, Schneider M, Beck H, Hatzopoulos AK, Just U, Sinowatz F, Schmahl W, Chien KR, Wurst W, Bornkamm GW, Brielmeier M, Essential Role for Mitochondrial Thioredoxin Reductase in Hematopoiesis, Heart Development, and Heart Function, Mol. Cell. Biol 24 (2004) 9414–9423. doi: 10.1128/MCB.24.21.9414-9423.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Panee J, Stoytcheva ZR, Liu W, Berry MJ, Selenoprotein H is a redox-sensing high mobility group family DNA-binding protein that up-regulates genes involved in glutathione synthesis and phase II detoxification, J. Biol. Chem 282 (2007) 23759–23765. doi: 10.1074/jbc.M702267200. [DOI] [PubMed] [Google Scholar]
- [195].Novoselov SV, Kryukov GV, Xu XM, Carlson BA, Hatfield DL, Gladyshev VN, Selenoprotein H is a nucleolar thioredoxin-like protein with a unique expression pattern, J. Biol. Chem 282 (2007) 11960–11968. doi: 10.1074/jbc.M701605200. [DOI] [PubMed] [Google Scholar]
- [196].Turanov AA, Shchedrina VA, Everley RA, Lobanov AV, Yim SH, Marino SM, Gygi SP, Hatfield DL, Gladyshev VN, Selenoprotein S is involved in maintenance and transport of multiprotein complexes, Biochem. J 462 (2014) 555–565. doi: 10.1042/BJ20140076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Ye Y, Shibata Y, Yun C, Ron D, Rapoport TA, A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol, Nature. 429 (2004) 841–847. doi: 10.1038/nature02656. [DOI] [PubMed] [Google Scholar]
- [198].Du S, Liu H, Huang K, Influence of SelS gene silence on beta-Mercaptoethanol-mediated endoplasmic reticulum stress and cell apoptosis in HepG2 cells., Biochim. Biophys. Acta 1800 (2010) 511–7. doi: 10.1016/j.bbagen.2010.01.005. [DOI] [PubMed] [Google Scholar]
- [199].Liu J, Li F, Rozovsky S, The Intrinsically Disordered Membrane Protein Selenoprotein S Is a Reductase in Vitro, Biochemistry. 52 (2013) 3051–3061. doi: 10.1021/bi4001358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Wright CR, Allsopp GL, Addinsall AB, McRae NL, Andrikopoulos S, Stupka N, A Reduction in Selenoprotein S Amplifies the Inflammatory Profile of Fast-Twitch Skeletal Muscle in the mdx Dystrophic Mouse, Mediators Inflamm. 2017 (2017) 1–12. doi: 10.1155/2017/7043429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Gao Y, Hannan NRF, Wanyonyi S, Konstantopolous N, Pagnon J, Feng HC, Jowett JBM, Kim K-H, Walder K, Collier GR, Activation of the selenoprotein SEPS1 gene expression by pro-inflammatory cytokines in HepG2 cells., Cytokine. 33 (2006) 246–51. doi: 10.1016/j.cyto.2006.02.005. [DOI] [PubMed] [Google Scholar]
- [202].Curran JE, Jowett JBM, Elliott KS, Gao Y, Gluschenko K, Wang J, Azim DMA, Cai G, Mahaney MC, Comuzzie AG, Dyer TD, Walder KR, Zimmet P, MacCluer JW, Collier GR, Kissebah AH, Blangero J, Genetic variation in selenoprotein S influences inflammatory response, Nat. Genet 37 (2005) 1234–1241. doi: 10.1038/ng1655. [DOI] [PubMed] [Google Scholar]
- [203].Kim CY, Kim K, Dexamethasone-induced selenoprotein S degradation is required for adipogenesis, J. Lipid Res 54 (2013) 2069–2082. doi: 10.1194/jlr.M034603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Lubos E, Loscalzo J, Handy DE, Glutathione Peroxidase-1 in Health and Disease: From Molecular Mechanisms to Therapeutic Opportunities, Antioxid. Redox Signal 15 (2011) 1957–1997. doi: 10.1089/ars.2010.3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [205].Liu J, Hinkhouse MM, Sun W, Weydert CJ, Ritchie JM, Oberley LW, Cullen JJ, Redox regulation of pancreatic cancer cell growth: role of glutathione peroxidase in the suppression of the malignant phenotype., Hum. Gene Ther 15 (2004) 239–50. doi: 10.1089/104303404322886093. [DOI] [PubMed] [Google Scholar]
- [206].Nasr MA, Fedele MJ, Esser K, Diamond AM, GPx-1 modulates Akt and P70S6K phosphorylation and Gadd45 levels in MCF-7 cells., Free Radic. Biol. Med 37 (2004) 187–95. doi: 10.1016/j.freeradbiomed.2004.04.038. [DOI] [PubMed] [Google Scholar]
- [207].Geybels MS, van den Brandt PA, Schouten LJ, van Schooten FJ, van Breda SG, Rayman MP, Green FR, Verhage BAJ, Selenoprotein Gene Variants, Toenail Selenium Levels, and Risk for Advanced Prostate Cancer, JNCI J. Natl. Cancer Inst 106 (2014). doi: 10.1093/jnci/dju003. [DOI] [PubMed] [Google Scholar]
- [208].Metere A, Frezzotti F, Graves CE, Vergine M, De Luca A, Pietraforte D, Giacomelli L, A possible role for selenoprotein glutathione peroxidase (GPx1) and thioredoxin reductases (TrxR1) in thyroid cancer: Our experience in thyroid surgery, Cancer Cell Int. 18 (2018) 1–8. doi: 10.1186/s12935-018-0504-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [209].Dequanter D, Dok R, Koolen L, Vander Poorten V, Nuyts S, Prognostic Significance of Glutathione Peroxidase Levels (GPx1) in Head and Neck Cancers, Front. Oncol 7 (2017) 1–5. doi: 10.3389/fonc.2017.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [210].Hu YJ, Diamond AM, Role of Glutathione Peroxidase 1 in Breast Cancer : Loss of Heterozygosity and Allelic Differences in the Response to Selenium 1, Cancer Res. 63 (2003) 3347–3351. [PubMed] [Google Scholar]
- [211].Conrad M, Moreno SG, Sinowatz F, Ursini F, Kolle S, Roveri A, Brielmeier M, Wurst W, Maiorino M, Bornkamm GW, The Nuclear Form of Phospholipid Hydroperoxide Glutathione Peroxidase Is a Protein Thiol Peroxidase Contributing to Sperm Chromatin Stability, Mol. Cell. Biol 25 (2005) 7637–7644. doi: 10.1128/mcb.25.17.7637-7644.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212].Méplan C, Hughes DJ, Pardini B, Naccarati A, Soucek P, Vodickova L, Hlavatá I, Vrána D, Vodicka P, Hesketh JE, Genetic variants in selenoprotein genes increase risk of colorectal cancer, Carcinogenesis. 31 (2010) 1074–1079. doi: 10.1093/carcin/bgq076. [DOI] [PubMed] [Google Scholar]
- [213].Geybels MS, Hutter CM, Kwon EM, Ostrander EA, Fu R, Feng Z, Stanford JL, Peters U, Variation in selenoenzyme genes and prostate cancer risk and survival, Prostate. 73 (2013) 734–742. doi: 10.1002/pros.22617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [214].Slattery ML, Lundgreen A, Welbourn B, Corcoran C, Wolff RK, Genetic variation in selenoprotein genes, lifestyle, and risk of colon and rectal cancer, PLoS One. 7 (2012). doi: 10.1371/journal.pone.0037312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [215].Gerstenberger JP, Bauer SR, Van Blarigan EL, Sosa E, Song X, Witte JS, Carroll PR, Chan JM, Selenoprotein and antioxidant genes and the risk of high-grade prostate cancer and prostate cancer recurrence, Prostate. 75 (2015) 60–69. doi: 10.1002/pros.22892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [216].Méplan C, Rohrmann S, Steinbrecher A, Schomburg L, Jansen E, Linseisen J, Hesketh J, Polymorphisms in Thioredoxin Reductase and Selenoprotein K Genes and Selenium Status Modulate Risk of Prostate Cancer, PLoS One. 7 (2012) 1–10. doi: 10.1371/journal.pone.0048709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [217].Sutherland A, Kim DH, Relton C, Ahn YO, Hesketh J, Polymorphisms in the selenoprotein S and 15-kDa selenoprotein genes are associated with altered susceptibility to colorectal cancer, Genes Nutr. 5 (2010) 215–223. doi: 10.1007/s12263-010-0176-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [218].Tsuji PA, Carlson BA, Yoo MH, Naranjo-Suarez S, Xu XM, He Y, Asaki E, Seifried HE, Reinhold WC, Davis CD, Gladyshev VN, Hatfield DL, The 15kDa selenoprotein and thioredoxin reductase 1 promote colon cancer by different pathways, PLoS One. 10 (2015) 1–18. doi: 10.1371/journal.pone.0124487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [219].Bertz M, Kühn K, Koeberle SC, Müller MF, Hoelzer D, Thies K, Deubel S, Thierbach R, Kipp AP, Selenoprotein H controls cell cycle progression and proliferation of human colorectal cancer cells, Free Radic. Biol. Med 127 (2018) 98–107. doi: 10.1016/j.freeradbiomed.2018.01.010. [DOI] [PubMed] [Google Scholar]
- [220].Mao H, Cui R, Wang X, Association analysis of selenoprotein S polymorphisms in Chinese Han with susceptibility to gastric cancer, Int. J. Clin. Exp. Med 8 (2015) 10993–10999. [PMC free article] [PubMed] [Google Scholar]
- [221].Seiderer J, Dambacher J, Kühnlein B, Pfennig S, Konrad A, Török HP, Haller D, Göke B, Ochsenkühn T, Lohse P, Brand S, The role of the selenoprotein S (SELS) gene −105G>A promoter polymorphism in inflammatory bowel disease and regulation of SELS gene expression in intestinal inflammation, Tissue Antigens. 70 (2007) 238–246. doi: 10.1016/j.trpro.2016.11.026. [DOI] [PubMed] [Google Scholar]
- [222].Santos LR, Durães C, Mendes A, Prazeres H, Alvelos MI, Moreira CS, Canedo P, Esteves C, Neves C, Carvalho D, Sobrinho-Simões M, Soares P, A polymorphism in the promoter region of the selenoprotein S gene (SEPS1) contributes to hashimoto’s thyroiditis susceptibility, J. Clin. Endocrinol. Metab 99 (2014). doi: 10.1210/jc.2013-3539. [DOI] [PubMed] [Google Scholar]
- [223].Alanne M, Kristiansson K, Auro K, Silander K, Kuulasmaa K, Peltonen L, Salomaa V, Perola M, Variation in the selenoprotein S gene locus is associated with coronary heart disease and ischemic stroke in two independent Finnish cohorts, Hum. Genet 122 (2007) 355–365. doi: 10.1007/s00439-007-0402-7. [DOI] [PubMed] [Google Scholar]
- [224].Schneider MJ, Fiering SN, Thai B, Wu S, St Germain E, Parlow AF, St Germain DL, Galton VA, Targeted disruption of the type 1 selenodeiodinase gene (Dio1) results in marked changes in thyroid hormone economy in mice., Endocrinology. 147 (2006) 580–9. doi: 10.1210/en.2005-0739. [DOI] [PubMed] [Google Scholar]
- [225].Ng L, Goodyear RJ, Woods CA, Schneider MJ, Diamond E, Richardson GP, Kelley MW, Germain DL St., Galton VA, Forrest D, Hearing loss and retarded cochlear development in mice lacking type 2 iodothyronine deiodinase, Proc. Natl. Acad. Sci 101 (2004) 3474–3479. doi: 10.1073/pnas.0307402101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [226].Ng L, Hernandez A, He W, Ren T, Srinivas M, Michelle M, Galton VA, St Germain DL, Forrest D, A protective role for type 3 deiodinase, a thyroid hormone-inactivating enzyme, in cochlear development and auditory function, Endocrinology. 150 (2009) 1952–1960. doi: 10.1210/en.2008-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [227].Ng L, Lyubarsky A, Nikonov SS, Ma M, Srinivas M, Kefas B, Germain DL St., Hernandez A, Pugh EN, Forrest D, Type 3 Deiodinase, a Thyroid-Hormone-Inactivating Enzyme, Controls Survival and Maturation of Cone Photoreceptors, J. Neurosci 30 (2010) 3347–3357. doi: 10.1523/JNEUROSCI.5267-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [228].Wu Z, Martinez ME, Germain DL St., Hernandez A, Type 3 Deiodinase Role on Central Thyroid Hormone Action Affects the Leptin-Melanocortin System and Circadian Activity, Endocrinology. (2016) en.2016–1680. doi: 10.1210/en.2016-1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [229].Wirth EK, Conrad M, Winterer J, Wozny C, Carlson BA, Roth S, Schmitz D, Bornkamm GW, Coppola V, Tessarollo L, Schomburg L, Köhrle J, Hatfield DL, Schweizer U, Neuronal selenoprotein expression is required for interneuron development and prevents seizures and neurodegeneration., FASEB J. 24 (2010) 844–52. doi: 10.1096/fj.09-143974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [230].Yant LJ, Ran Q, Rao L, Van Remmen H, Shibatani T, Belter JG, Motta L, Richardson A, Prolla TA, The selenoprotein GPX4 is essential for mouse development and protects from radiation and oxidative damage insults, Free Radic. Biol. Med 34 (2003) 496–502. doi: 10.1016/S0891-5849(02)01360-6. [DOI] [PubMed] [Google Scholar]
- [231].Seiler A, Schneider M, Förster H, Roth S, Wirth EK, Culmsee C, Plesnila N, Kremmer E, Rådmark O, Wurst W, Bornkamm GW, Schweizer U, Conrad M, Glutathione Peroxidase 4 Senses and Translates Oxidative Stress into 12/15-Lipoxygenase Dependent- and AIF-Mediated Cell Death, Cell Metab. 8 (2008) 237–248. doi: 10.1016/j.cmet.2008.07.005. [DOI] [PubMed] [Google Scholar]
- [232].Smith AC, Mears AJ, Bunker R, Ahmed A, MacKenzie M, Schwartzentruber JA, Beaulieu CL, Ferretti E, Majewski J, Bulman DE, Celik FC, Boycott KM, Graham GE, Mutations in the enzyme glutathione peroxidase 4 cause Sedaghatian-type spondylometaphyseal dysplasia, J. Med. Genet 51 (2014) 470–474. doi: 10.1136/jmedgenet-2013-102218. [DOI] [PubMed] [Google Scholar]
- [233].Pitts MW, Raman AV, Hashimoto AC, Todorovic C, Nichols RA, Berry MJ, Deletion of selenoprotein P results in impaired function of parvalbumin interneurons and alterations in fear learning and sensorimotor gating, Neuroscience. 208 (2012) 58–68. doi: 10.1016/j.neuroscience.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [234].Schweizer U, Michaelis M, Kohrle J, Schomburg L, Efficient selenium transfer from mother to offspring in selenoprotein-P-deficient mice enables dose-dependent rescue of phenotypes associated with selenium deficiency, Biochem. J 378 (2004) 21–26. doi: 10.1042/bj20031795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [235].V Raman A, Pitts MW, Seyedali A, Hashimoto AC, Seale LA, Bellinger FP, Berry MJ, Absence of selenoprotein P but not selenocysteine lyase results in severe neurological dysfunction, Genes, Brain Behav. 11 (2012) 601–613. doi: 10.1111/j.1601-183X.2012.00794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [236].Schomburg L, Schweizer U, Holtmann B, Flohe L, Semdtner M, Kohrle J, Gene disruption discloses role of selenoprotein P in selenium delivery to target tissues, Biochem. J 370 (2003) 397–402. doi: 10.1042/bj20021853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [237].Hill KE, Zhou J, McMahan WJ, Motley AK, Atkins JF, Gesteland RF, Burk RF, Deletion of selenoprotein P alters distribution of selenium in the mouse, J. Biol. Chem 278 (2003) 13640–13646. doi: 10.1074/jbc.M300755200. [DOI] [PubMed] [Google Scholar]
- [238].Meyer K, Jaffrey S, Saletore Y, Korlach J, Mason CE, Vilfan ID, The birth of the Epitranscriptome: deciphering the function of RNA modifications, Genome Biol. 13 (2012) 175. doi: 10.1186/gb-2012-13-10-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [239].Meyer Kate D., Saletore Y, Zumbo P, Elemento C.E.M.S.R.J. Olivier, Comprehensive Analysis of mRNA Methylation Reveals Enrichment in 3’ UTRs and Near Stop Codons, Cell. 149 (2012) 1635–1646. doi: 10.1016/j.cell.2012.05.003.Comprehensive. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [240].Regina BF, Gladyshev VN, Arnér ES, Berry MJ, Bruford EA, Burk RF, Carlson BA, Castellano S, Chavatte L, Conrad M, Copeland PR, Diamond AM, Driscoll DM, Ferreiro A, Flohé L, Green FR, Guigó R, Handy DE, Hatfield DL, Hesketh J, Hoffmann PR, Holmgren A, Hondal RJ, Howard MT, Huang K, Kim HY, Kim IY, Köhrle J, Krol A, Kryukov GV, Lee BJ, Lee BC, Lei XG, Liu Q, Lescure A, Lobanov AV, Loscalzo J, Maiorino M, Mariotti M, Prabhu KS, Rayman MP, Rozovsky S, Salinas G, Schmidt EE, Schomburg L, Schweizer U, Simonović M, Sunde RA, Tsuji PA, Tweedie S, Ursini FF, Whanger PD, Zhang Y, Selenoprotein gene nomenclature, J. Biol. Chem 291 (2016) 24036–24040. doi: 10.1074/jbc.M116.756155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [241].Gladyshev VN, Selenoproteins and selenoproteomes, in: Selenium, Springer US, Boston, MA, 2009: pp. 99–110. doi: 10.1007/0-387-33827-6_9. [DOI] [Google Scholar]
- [242].Lu C, Qiu F, Zhou H, Peng Y, Hao W, Xu J, Yuan J, Wang S, Qiang B, Xu C, Peng X, Identification and characterization of selenoprotein K: an antioxidant in cardiomyocytes., FEBS Lett. 580 (2006) 5189–97. doi: 10.1016/j.febslet.2006.08.065. [DOI] [PubMed] [Google Scholar]
- [243].Shchedrina VA, Everley RA, Zhang Y, Gygi SP, Hatfield DL, Gladyshev VN, Selenoprotein K Binds Multiprotein Complexes and Is Involved in the Regulation of Endoplasmic Reticulum Homeostasis, J. Biol. Chem 286 (2011) 42937–42948. doi: 10.1074/jbc.M111.310920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [244].Gao Y, Feng HC, Walder K, Bolton K, Sunderland T, Bishara N, Quick M, Kantham L, Collier GR, Regulation of the selenoprotein SelS by glucose deprivation and endoplasmic reticulum stress - SelS is a novel glucose-regulated protein., FEBS Lett. 563 (2004) 185–90. doi: 10.1016/S0014-5793(04)00296-0. [DOI] [PubMed] [Google Scholar]
- [245].Kim K-H, Gao Y, Walder K, Collier GR, Skelton J, Kissebah AH, SEPS1 protects RAW264.7 cells from pharmacological ER stress agent-induced apoptosis, Biochem. Biophys. Res. Commun 354 (2007) 127–132. doi: 10.1016/j.bbrc.2006.12.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [246].Moses EK, Johnson MP, Tømmerdal L, Forsmo S, Curran JE, Abraham LJ, Charlesworth JC, Brennecke SP, Blangero J, Austgulen R, Genetic association of preeclampsia to the inflammatory response gene SEPS1., Am. J. Obstet. Gynecol 198 (2008) 336.e1–5. doi: 10.1016/j.ajog.2007.09.024. [DOI] [PubMed] [Google Scholar]
- [247].Martínez A, Santiago JL, Varadé J, Márquez A, Lamas JR, Mendoza JL, de la Calle H, Díaz-Rubio M, de la Concha EG, Fernández-Gutiérrez B, Urcelay E, Polymorphisms in the selenoprotein S gene: Lack of association with autoimmune inflammatory diseases, BMC Genomics. 9 (2008) 1–6. doi: 10.1186/1471-2164-9-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [248].Horibata Y, Hirabayashi Y, Identification and characterization of human ethanolaminephosphotransferase1., J. Lipid Res 48 (2007) 503–8. doi: 10.1194/jlr.C600019-JLR200. [DOI] [PubMed] [Google Scholar]
- [249].Kim H-Y, Gladyshev VN, Methionine sulfoxide reductases: selenoprotein forms and roles in antioxidant protein repair in mammals, Biochem. J 407 (2007) 321–329. doi: 10.1042/BJ20070929. [DOI] [PubMed] [Google Scholar]
- [250].Kim H-Y, Fomenko DE, Yoon Y-E, Gladyshev VN, Catalytic Advantages Provided by Selenocysteine in Methionine- S -Sulfoxide Reductases, Biochemistry. 45 (2006) 13697–13704. doi: 10.1021/bi0611614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [251].Hansel A, Heinemann SH, Hoshi T, Heterogeneity and function of mammalian MSRs: enzymes for repair, protection and regulation., Biochim. Biophys. Acta 1703 (2005) 239–47. doi: 10.1016/j.bbapap.2004.09.010. [DOI] [PubMed] [Google Scholar]
- [252].Lee T-H, Kim H-Y, An anaerobic bacterial MsrB model reveals catalytic mechanisms, advantages, and disadvantages provided by selenocysteine and cysteine in reduction of methionine-R-sulfoxide, Arch. Biochem. Biophys 478 (2008) 175–180. doi: 10.1016/j.abb.2008.07.028. [DOI] [PubMed] [Google Scholar]
- [253].Dikiy A, Novoselov SV, Fomenko DE, Sengupta A, Carlson BA, Cerny RL, Ginalski K, Grishin NV, Hatfield DL, Gladyshev VN, SelT, SelW, SelH, and Rdx12: Genomics and Molecular Insights into the Functions of Selenoproteins of a Novel Thioredoxin-like Family, Biochemistry. 46 (2007) 6871–6882. doi: 10.1021/bi602462q. [DOI] [PubMed] [Google Scholar]
- [254].Ferguson AD, Labunskyy VM, Fomenko DE, Araç D, Chelliah Y, Amezcua CA, Rizo J, Gladyshev VN, Deisenhofer J, NMR Structures of the Selenoproteins Sep15 and SelM Reveal Redox Activity of a New Thioredoxin-like Family, J. Biol. Chem 281 (2006) 3536–3543. doi: 10.1074/jbc.M511386200. [DOI] [PubMed] [Google Scholar]
- [255].Labunskyy VM, Ferguson AD, Fomenko DE, Chelliah Y, Hatfield DL, Gladyshev VN, A Novel Cysteine-rich Domain of Sep15 Mediates the Interaction with UDP-glucose:Glycoprotein Glucosyltransferase, J. Biol. Chem 280 (2005) 37839–37845. doi: 10.1074/jbc.M508685200. [DOI] [PubMed] [Google Scholar]
- [256].Petit N, Selenoprotein N: an endoplasmic reticulum glycoprotein with an early developmental expression pattern, Hum. Mol. Genet 12 (2003) 1045–1053. doi: 10.1093/hmg/ddg115. [DOI] [PubMed] [Google Scholar]
- [257].Moghadaszadeh B, Petit N, Jaillard C, Brockington M, Roy SQ, Merlini L, Romero N, Estournet B, Desguerre I, Chaigne D, Muntoni F, Topaloglu H, Guicheney P, Mutations in SEPN1 cause congenital muscular dystrophy with spinal rigidity and restrictive respiratory syndrome, Nat. Genet 29 (2001) 17–18. doi: 10.1038/ng713. [DOI] [PubMed] [Google Scholar]
- [258].Ferreiro A, Quijano-Roy S, Pichereau C, Moghadaszadeh B, Goemans N, Bönnemann C, Jungbluth H, Straub V, Villanova M, Leroy J-P, Romero NB, Martin J-J, Muntoni F, Voit T, Estournet B, Richard P, Fardeau M, Guicheney P, Mutations of the Selenoprotein N Gene, Which Is Implicated in Rigid Spine Muscular Dystrophy, Cause the Classical Phenotype of Multiminicore Disease: Reassessing the Nosology of Early-Onset Myopathies, Am. J. Hum. Genet 71 (2002) 739–749. doi: 10.1086/342719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [259].Clarke NF, Kidson W, Quijano-Roy S, Estournet B, Ferreiro A, Guicheney P, Manson JI, Kornberg AJ, Shield LK, North KN, SEPN1 : Associated with congenital fiber-type disproportion and insulin resistance, Ann. Neurol 59 (2006) 546–552. doi: 10.1002/ana.20761. [DOI] [PubMed] [Google Scholar]
- [260].Jurynec MJ, Xia R, Mackrill JJ, Gunther D, Crawford T, Flanigan KM, Abramson JJ, Howard MT, Grunwald DJ, Selenoprotein N is required for ryanodine receptor calcium release channel activity in human and zebrafish muscle, Proc. Natl. Acad. Sci 105 (2008) 12485–12490. doi: 10.1073/pnas.0806015105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [261].Arteel GE, Mostert V, Oubrahim H, Briviba K, Abel J, Sies H, Protection by selenoprotein P in human plasma against peroxynitrite-mediated oxidation and nitration., Biol. Chem 379 (n.d.) 1201–5. http://www.ncbi.nlm.nih.gov/pubmed/9792455. [PubMed] [Google Scholar]
- [262].Hoffmann PR, Hoge SC, Li P-A, Hoffmann FW, Hashimoto AC, Berry MJ, The selenoproteome exhibits widely varying, tissue-specific dependence on selenoprotein P for selenium supply, Nucleic Acids Res. 35 (2007) 3963–3973. doi: 10.1093/nar/gkm355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [263].Saito Y, Takahashi K, Characterization of selenoprotein P as a selenium supply protein, Eur. J. Biochem 269 (2002) 5746–5751. doi: 10.1046/j.1432-1033.2002.03298.x. [DOI] [PubMed] [Google Scholar]
- [264].SCHWEIZER U, STRECKFUß F, PELT P, CARLSON BA, HATFIELD DL, KÖHRLE J, SCHOMBURG L, Hepatically derived selenoprotein P is a key factor for kidney but not for brain selenium supply, Biochem. J 386 (2005) 221–226. doi: 10.1042/BJ20041973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [265].Valentine WM, Abel TW, Hill KE, Austin LM, Burk RF, Neurodegeneration in Mice Resulting From Loss of Functional Selenoprotein P or Its Receptor Apolipoprotein E Receptor 2, J. Neuropathol. Exp. Neurol 67 (2008) 68–77. doi: 10.1097/NEN.0b013e318160f347. [DOI] [PubMed] [Google Scholar]
- [266].Olson GE, Winfrey VP, NagDas SK, Hill KE, Burk RF, Apolipoprotein E Receptor-2 (ApoER2) Mediates Selenium Uptake from Selenoprotein P by the Mouse Testis, J. Biol. Chem 282 (2007) 12290–12297. doi: 10.1074/jbc.M611403200. [DOI] [PubMed] [Google Scholar]
- [267].Olson GE, Winfrey VP, Hill KE, Burk RF, Megalin Mediates Selenoprotein P Uptake by Kidney Proximal Tubule Epithelial Cells, J. Biol. Chem 283 (2008) 6854–6860. doi: 10.1074/jbc.M709945200. [DOI] [PubMed] [Google Scholar]
- [268].Saito Y, Hayashi T, Tanaka A, Watanabe Y, Suzuki M, Saito E, Takahashi K, Selenoprotein P in Human Plasma as an Extracellular Phospholipid Hydroperoxide Glutathione Peroxidase, J. Biol. Chem 274 (1999) 2866–2871. doi: 10.1074/jbc.274.5.2866. [DOI] [PubMed] [Google Scholar]
- [269].Grumolato L, Ghzili H, Montero-Hadjadje M, Gasman S, Lesage J, Tanguy Y, Galas L, Ait-Ali D, Leprince J, Guérineau NC, Elkahloun AG, Fournier A, Vieau D, Vaudry H, Anouar Y, Selenoprotein T is a PACAP-regulated gene involved in intracellular Ca 2+ mobilization and neuroendocrine secretion, FASEB J. 22 (2008) 1756–1768. doi: 10.1096/fj.06-075820. [DOI] [PubMed] [Google Scholar]
- [270].Gu QP, Sun Y, Ream LW, Whanger PD, Selenoprotein W accumulates primarily in primate skeletal muscle, heart, brain and tongue., Mol. Cell. Biochem 204 (2000) 49–56. http://www.ncbi.nlm.nih.gov/pubmed/10718624. [DOI] [PubMed] [Google Scholar]
- [271].Yeh JY, Beilstein MA, Andrews JS, Whanger PD, Tissue distribution and influence of selenium status on levels of selenoprotein W., FASEB J. 9 (1995) 392–6. http://www.ncbi.nlm.nih.gov/pubmed/7896009. [DOI] [PubMed] [Google Scholar]
- [272].Beilstein MA, Vendeland SC, Barofsky E, Jensen ON, Whanger PD, Selenoprotein W of rat muscle binds glutathione and an unknown small molecular weight moiety., J. Inorg. Biochem 61 (1996) 117–24. http://www.ncbi.nlm.nih.gov/pubmed/8576706. [DOI] [PubMed] [Google Scholar]
- [273].Loflin J, Lopez N, Whanger PD, Kioussi C, Selenoprotein W during development and oxidative stress, J. Inorg. Biochem 100 (2006) 1679–1684. doi: 10.1016/j.jinorgbio.2006.05.018. [DOI] [PubMed] [Google Scholar]
- [274].Aachmann FL, Fomenko DE, Soragni A, Gladyshev VN, Dikiy A, Solution Structure of Selenoprotein W and NMR Analysis of Its Interaction with 14-3-3 Proteins, J. Biol. Chem 282 (2007) 37036–37044. doi: 10.1074/jbc.M705410200. [DOI] [PubMed] [Google Scholar]
- [275].Kim IY, Stadtman TC, Selenophosphate synthetase: detection in extracts of rat tissues by immunoblot assay and partial purification of the enzyme from the archaean Methanococcus vannielii., Proc. Natl. Acad. Sci 92 (1995) 7710–7713. doi: 10.1073/pnas.92.17.7710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [276].Low SC, Harney JW, Berry MJ, Cloning and Functional Characterization of Human Selenophosphate Synthetase, an Essential Component of Selenoprotein Synthesis, J. Biol. Chem 270 (1995) 21659–21664. doi: 10.1074/jbc.270.37.21659. [DOI] [PubMed] [Google Scholar]
- [277].Xu X-M, Carlson BA, Irons R, Mix H, Zhong N, Gladyshev VN, Hatfield DL, Selenophosphate synthetase 2 is essential for selenoprotein biosynthesis, Biochem. J 404 (2007) 115–120. doi: 10.1042/BJ20070165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [278].Korotkov KV, Kumaraswamy E, Zhou Y, Hatfield DL, Gladyshev VN, Association between the 15-kDa Selenoprotein and UDP-glucose:Glycoprotein Glucosyltransferase in the Endoplasmic Reticulum of Mammalian Cells, J. Biol. Chem 276 (2001) 15330–15336. doi: 10.1074/jbc.M009861200. [DOI] [PubMed] [Google Scholar]
- [279].Cheng W, Fu YX, Porres JM, Ross DA, Lei XG, Selenium-dependent cellular glutathione peroxidase protects mice against a pro-oxidant-induced oxidation of NADPH, NADH, lipids, and protein., FASEB J. 13 (1999) 1467–75. http://www.ncbi.nlm.nih.gov/pubmed/10428770. [DOI] [PubMed] [Google Scholar]
- [280].Reszka E, Selenoproteins in bladder cancer, Clin. Chim. Acta 413 (2012) 847–854. doi: 10.1016/j.cca.2012.01.041. [DOI] [PubMed] [Google Scholar]
- [281].Esworthy RS, Aranda R, Martín MG, Doroshow JH, Binder SW, Chu F-F, Mice with combined disruption of Gpx1 and Gpx2 genes have colitis, Am. J. Physiol. Liver Physiol 281 (2001) G848–G855. doi: 10.1152/ajpgi.2001.281.3.G848. [DOI] [PubMed] [Google Scholar]
- [282].Rudenko EE, V Gerashchenko G, V Lapska Y, Bogatyrova OO, Vozianov SO, Zgonnyk YM, Kashuba VI, Genetic and epigenetic changes of GPX1 and GPX3 in human clear-cell renal cell carcinoma, Biopolym. Cell 29 (2013) 395–401. doi: 10.7124/bc.00082F. [DOI] [Google Scholar]
- [283].Chu FF, Doroshow JH, Esworthy RS, Expression, characterization, and tissue distribution of a new cellular selenium-dependent glutathione peroxidase, GSHPx-GI, J. Biol. Chem 268 (1993) 2571–2576. [PubMed] [Google Scholar]
- [284].Labunskyy VM, Hatfield DL, Gladyshev VN, Selenoproteins: molecular pathways and physiological roles., Physiol. Rev 94 (2014) 739–77. doi: 10.1152/physrev.00039.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [285].Esworthy RS, Swiderek KM, Ho YS, Chu FF, Selenium-dependent glutathione peroxidase-GI is a major glutathione peroxidase activity in the mucosal epithelium of rodent intestine., Biochim. Biophys. Acta 1381 (1998) 213–26. http://www.ncbi.nlm.nih.gov/pubmed/9685647. [DOI] [PubMed] [Google Scholar]
- [286].Yoshimura S, Watanabe K, Suemizu H, Onozawa T, Mizoguchi J, Tsuda K, Hatta H, Moriuchi T, Tissue Specific Expression of the Plasma Glutathione Peroxidase Gene in Rat Kidney1, J. Biochem 109 (1991) 918–923. doi: 10.1093/oxfordjournals.jbchem.a123480. [DOI] [PubMed] [Google Scholar]
- [287].Schmutzler C, Mentrup B, Schomburg L, Hoang-Vu C, Herzog V, Köhrle J, Selenoproteins of the thyroid gland: expression, localization and possible function of glutathione peroxidase 3, Biol. Chem 388 (2007). doi: 10.1515/BC.2007.122. [DOI] [PubMed] [Google Scholar]
- [288].Kenet G, Freedman J, Shenkman B, Regina E, Brok-Simoni F, Holzman F, Vavva F, Brand N, Michelson A, Trolliet M, Loscalzo J, Inbal A, Plasma glutathione peroxidase deficiency and platelet insensitivity to nitric oxide in children with familial stroke., Arterioscler. Thromb. Vasc. Biol 19 (1999) 2017–23. http://www.ncbi.nlm.nih.gov/pubmed/10446087. [DOI] [PubMed] [Google Scholar]
- [289].Conrad M, Schneider M, Seiler A, Bornkamm GW, Physiological role of phospholipid hydroperoxide glutathione peroxidase in mammals, Biol. Chem 388 (2007). doi: 10.1515/BC.2007.130. [DOI] [PubMed] [Google Scholar]
- [290].Ursini F, Heim S, Kiess M, Maiorino M, Roveri A, Wissing J, Flohé L, Dual function of the selenoprotein PHGPx during sperm maturation., Science. 285 (1999) 1393–6. http://www.ncbi.nlm.nih.gov/pubmed/10464096. [DOI] [PubMed] [Google Scholar]
- [291].Flohé L, Selenium in mammalian spermiogenesis, Biol. Chem 388 (2007). doi: 10.1515/BC.2007.112. [DOI] [PubMed] [Google Scholar]
- [292].Foresta C, Flohé L, Garolla A, Roveri A, Ursini F, Maiorino M, Male Fertility Is Linked to the Selenoprotein Phospholipid Hydroperoxide Glutathione Peroxidase, Biol. Reprod 67 (2002) 967–971. doi: 10.1095/biolreprod.102.003822. [DOI] [PubMed] [Google Scholar]
- [293].GV K, S C, SV N, AV L, O Z, R G, VN G, Characterization of mammalian selenoproteomes., Science (80-.) 300 (2003) 1439–1443. http://www.wormbase.org/db/misc/paper?name=WBPaper00013278. [DOI] [PubMed] [Google Scholar]
- [294].Kalantari P, Narayan V, Natarajan SK, Muralidhar K, Gandhi UH, Vunta H, Henderson AJ, Prabhu KS, Thioredoxin Reductase-1 Negatively Regulates HIV-1 Transactivating Protein Tat-dependent Transcription in Human Macrophages, J. Biol. Chem 283 (2008) 33183–33190. doi: 10.1074/jbc.M807403200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [295].Powis G, Kirkpatrick DL, Thioredoxin signaling as a target for cancer therapy, Curr. Opin. Pharmacol 7 (2007) 392–397. doi: 10.1016/j.coph.2007.04.003. [DOI] [PubMed] [Google Scholar]
- [296].Yoo MH, Xu XM, Carlson BA, Gladyshev VN, Hatfield DL, Thioredoxin reductase 1 deficiency reverses tumor phenotype and tumorigenicity of lung carcinoma cells, J. Biol. Chem 281 (2006) 13005–13008. doi: 10.1074/jbc.C600012200. [DOI] [PubMed] [Google Scholar]
- [297].Yoo M-H, Xu X-M, Carlson BA, Patterson AD, Gladyshev VN, Hatfield DL, Targeting Thioredoxin Reductase 1 Reduction in Cancer Cells Inhibits Self-Sufficient Growth and DNA Replication, PLoS One. 2 (2007) e1112. doi: 10.1371/journal.pone.0001112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [298].Avval FZ, Holmgren A, Molecular Mechanisms of Thioredoxin and Glutaredoxin as Hydrogen Donors for Mammalian S Phase Ribonucleotide Reductase, J. Biol. Chem 284 (2009) 8233–8240. doi: 10.1074/jbc.M809338200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [299].Cheng Q, Sandalova T, Lindqvist Y, Arnér ESJ, Crystal Structure and Catalysis of the Selenoprotein Thioredoxin Reductase 1, J. Biol. Chem 284 (2009) 3998–4008. doi: 10.1074/jbc.M807068200. [DOI] [PubMed] [Google Scholar]
- [300].Crosley LK, Méplan C, Nicol F, Rundlöf AK, Arnér ESJ, Hesketh JE, Arthur JR, Differential regulation of expression of cytosolic and mitochondrial thioredoxin reductase in rat liver and kidney, Arch. Biochem. Biophys 459 (2007) 178–188. doi: 10.1016/j.abb.2006.12.029. [DOI] [PubMed] [Google Scholar]
- [301].Turanov AA, Su D, Gladyshev VN, Characterization of Alternative Cytosolic Forms and Cellular Targets of Mouse Mitochondrial Thioredoxin Reductase, J. Biol. Chem 281 (2006) 22953–22963. doi: 10.1074/jbc.M604326200. [DOI] [PubMed] [Google Scholar]
- [302].Holmgren A, Lyckeborg C, Enzymatic reduction of alloxan by thioredoxin and NADPH-thioredoxin reductase., Proc. Natl. Acad. Sci 77 (1980) 5149–5152. doi: 10.1073/pnas.77.9.5149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [303].Bianco AC, Salvatore D, Gereben B, Berry MJ, Larsen PR, Biochemistry, Cellular and Molecular Biology, and Physiological Roles of the Iodothyronine Selenodeiodinases, Endocr. Rev 23 (2002) 38–89. doi: 10.1210/edrv.23.1.0455. [DOI] [PubMed] [Google Scholar]
- [304].Hernandez A, Germain D St., Thyroid hormone deiodinases: physiology and clinical disorders, Curr. Opin. Pediatr 15 (2003) 416–420. [DOI] [PubMed] [Google Scholar]
- [305].Gereben B, Goncalves C, Harney JW, Larsen PR, Bianco AC, Selective Proteolysis of Human Type 2 Deiodinase: A Novel Ubiquitin-Proteasomal Mediated Mechanism for Regulation of Hormone Activation, Mol. Endocrinol 14 (2000) 1697–1708. doi: 10.1210/mend.14.11.0558. [DOI] [PubMed] [Google Scholar]
- [306].Steinsapir J, Harney J, Larsen PR, Type 2 iodothyronine deiodinase in rat pituitary tumor cells is inactivated in proteasomes., J. Clin. Invest 102 (1998) 1895–1899. doi: 10.1172/JCI4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [307].Steinsapir J, Bianco AC, Buettner C, Harney J, Larsen PR, Substrate-Induced Down-Regulation of Human Type 2 Deiodinase (hD2) Is Mediated through Proteasomal Degradation and Requires Interaction with the Enzyme’s Active Center 1, Endocrinology. 141 (2000) 1127–1135. doi: 10.1210/endo.141.3.7355. [DOI] [PubMed] [Google Scholar]
- [308].Schneider MJ, Fiering SN, Pallud SE, Parlow AF, Germain DL St., Galton VA, Targeted Disruption of the Type 2 Selenodeiodinase Gene (DIO 2) Results in a Phenotype of Pituitary Resistance to T 4, Mol. Endocrinol 15 (2001) 2137–2148. doi: 10.1210/mend.15.12.0740. [DOI] [PubMed] [Google Scholar]
- [309].Galton VA, Wood ET, Germain EA St., Withrow C-A, Aldrich G, Germain GM St., Clark AS, Germain DL St., Thyroid Hormone Homeostasis and Action in the Type 2 Deiodinase-Deficient Rodent Brain during Development, Endocrinology. 148 (2007) 3080–3088. doi: 10.1210/en.2006-1727. [DOI] [PubMed] [Google Scholar]
- [310].Verma S, Hoffmann FW, Kumar M, Huang Z, Roe K, Nguyen-Wu E, Hashimoto AS, Hoffmann PR, Selenoprotein K Knockout Mice Exhibit Deficient Calcium Flux in Immune Cells and Impaired Immune Responses, J. Immunol 186 (2011) 2127–2137. doi: 10.4049/jimmunol.1002878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [311].Shibata T, Arisawa T, Tahara T, Ohkubo M, Yoshioka D, Maruyama N, Fujita H, Kamiya Y, Nakamura M, Nagasaka M, Iwata M, Takahama K, Watanabe M, Hirata I, Selenoprotein S (SEPS1) gene −105G>A promoter polymorphism influences the susceptibility to gastric cancer in the Japanese population, BMC Gastroenterol. 9 (2009) 1–7. doi: 10.1186/1471-230X-9-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [312].Horibata Y, Elpeleg O, Eran A, Hirabayashi Y, Savitzki D, Tal G, Mandel H, Sugimoto H, EPT1 (selenoprotein I) is critical for the neural development and maintenance of plasmalogen in humans, J. Lipid Res 59 (2018) 1015–1026. doi: 10.1194/jlr.P081620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [313].Chen P, Wang C, Ma X, Zhang Y, Liu Q, Qiu S, Liu Q, Tian J, Ni J, Direct Interaction of Selenoprotein R with Clusterin and Its Possible Role in Alzheimer’s Disease, PLoS One. 8 (2013) 1–12. doi: 10.1371/journal.pone.0066384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [314].Moskovitz J, Maiti P, Lopes DHJ, Oien DB, Attar A, Liu T, Mittal S, Hayes J, Bitan G, Induction of Methionine-Sulfoxide Reductases Protects Neurons from Amyloid β-Protein Insults in Vitro and in Vivo, Biochemistry. 50 (2011) 10687–10697. doi: 10.1021/bi201426b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [315].Fomenko DE, Novoselov SV, Natarajan SK, Lee BC, Koc A, Carlson BA, Lee TH, Kim HY, Hatfield DL, Gladyshev VN, MsrB1 (methionine-R-sulfoxide reductase 1) knock-out mice: Roles of MsrB1 in redox regulation and identification of a novel selenoprotein form, J. Biol. Chem 284 (2009) 5986–5993. doi: 10.1074/jbc.M805770200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [316].Zhao H, Kim G, Levine RL, Methionine sulfoxide reductase contributes to meeting dietary methionine requirements, Arch. Biochem. Biophys 522 (2012) 37–43. doi: 10.1016/j.abb.2012.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [317].Chen P, Wang RR, Ma XJ, Liu Q, Ni JZ, Different forms of selenoprotein M differentially affect Aβ aggregation and ROS generation, Int. J. Mol. Sci 14 (2013) 4385–4399. doi: 10.3390/ijms14034385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [318].Hwang DY, Cho JS, Oh JH, Shim SB, Jee SW, Lee SH, Seo SJ, Lee SK, Lee SH, Kim YK, Differentially expressed genes in transgenic mice carrying human mutant presenilin-2 (N141I): Correlation of selenoprotein M with Alzheimer’s disease, Neurochem. Res 30 (2005) 1009–1019. doi: 10.1007/s11064-005-6787-6. [DOI] [PubMed] [Google Scholar]
- [319].Castets P, Bertrand AT, Beuvin M, Ferry A, Le Grand F, Castets M, Chazot G, Rederstorff M, Krol A, Lescure A, Romero NB, Guicheney P, Allamand V, Satellite cell loss and impaired muscle regeneration in selenoprotein N deficiency, Hum. Mol. Genet 20 (2011) 694–704. doi: 10.1093/hmg/ddq515. [DOI] [PubMed] [Google Scholar]
- [320].Maiti B, Arbogast S, Allamand V, Moyle MW, Anderson CB, Richard P, Guicheney P, Ferreiro A, Flanigan KM, Howard MT, A mutation in the SEPN1 selenocysteine redefinition element (SRE) reduces selenocysteine incorporation and leads to SEPN1 -related myopathy, Hum. Mutat 30 (2009) 411–416. doi: 10.1002/humu.20879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [321].Andoh A, Hirashima M, Maeda H, Hata K, Inatomi O, Tsujikawa T, Sasaki M, Takahashi K, Fujiyama Y, Serum selenoprotein-P levels in patients with inflammatory bowel disease, Nutrition. 21 (2005) 574–579. doi: 10.1016/j.nut.2004.08.025. [DOI] [PubMed] [Google Scholar]
- [322].Misu H, Takamura T, Takayama H, Hayashi H, Matsuzawa-Nagata N, Kurita S, Ishikura K, Ando H, Takeshita Y, Ota T, Sakurai M, Yamashita T, Mizukoshi E, Yamashita T, Honda M, Miyamoto KI, Kubota T, Kubota N, Kadowaki T, Kim HJ, Lee IK, Minokoshi Y, Saito Y, Takahashi K, Yamada Y, Takakura N, Kaneko S, A liver-derived secretory protein, selenoprotein P, causes insulin resistance, Cell Metab. 12 (2010) 483–495. doi: 10.1016/j.cmet.2010.09.015. [DOI] [PubMed] [Google Scholar]
- [323].Oo SM, Misu H, Saito Y, Tanaka M, Kato S, Kita Y, Takayama H, Takeshita Y, Kanamori T, Nagano T, Nakagen M, Urabe T, Matsuyama N, Kaneko S, Takamura T, Serum selenoprotein P, but not selenium, predicts future hyperglycemia in a general Japanese population, Sci. Rep 8 (2018) 1–10. doi: 10.1038/s41598-018-35067-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [324].Du XA, Wang HM, Dai XX, Kou Y, Wu RP, Chen Q, Cao JL, Mo XY, Xiong YM, Role of selenoprotein S (SEPS1) −105G>A polymorphisms and PI3K/Akt signaling pathway in Kashin-Beck disease, Osteoarthr. Cartil 23 (2015) 210–216. doi: 10.1016/j.joca.2014.11.017. [DOI] [PubMed] [Google Scholar]
- [325].Sun W, Wang X, Zou X, Song R, Du X, Hu J, Xiong Y, Selenoprotein P gene r25191g/a polymorphism and quantification of selenoprotein P mRNA level in patients with Kashin-Beck disease, Br. J. Nutr 104 (2010) 1283–1287. doi: 10.1017/S0007114510002199. [DOI] [PubMed] [Google Scholar]
- [326].Meyer HA, Endermann T, Stephan C, Stoedter M, Behrends T, Wolff I, Jung K, Schomburg L, Selenoprotein P Status Correlates to Cancer-Specific Mortality in Renal Cancer Patients, PLoS One. 7 (2012) 1–7. doi: 10.1371/journal.pone.0046644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [327].Takemoto AS, Berry MJ, Bellinger FP, Role of selenoprotein P in Alzheimer’s disease, Ethn. Dis 20 (2010) 1–7. [PMC free article] [PubMed] [Google Scholar]
- [328].Bellinger FP, Raman AV, Rueli RH, Bellinger MT, Dewing AS, Seale LA, Andres MA, Uyehara-Lock JH, White LR, Ross GW, Berry MJ, Changes in Selenoprotein P in Substantia Nigra and Putamen in Parkinson’s Disease, J Park. Dis 2 (2012) 115–126. doi: 10.3233/JPD-2012-11052.Changes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [329].Boukhzar L, Hamieh A, Cartier D, Tanguy Y, Alsharif I, Castex M, Arabo A, El Hajji S, Bonnet J-J, Errami M, Falluel-Morel A, Chagraoui A, Lihrmann I, Anouar Y, Selenoprotein T Exerts an Essential Oxidoreductase Activity That Protects Dopaminergic Neurons in Mouse Models of Parkinson’s Disease, Antioxid. Redox Signal 24 (2016) 557–574. doi: 10.1089/ars.2015.6478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [330].Castex MT, Arabo A, Bénard M, Roy V, Le Joncour V, Prévost G, Bonnet JJ, Anouar Y, Falluel-Morel A, Selenoprotein T Deficiency Leads to Neurodevelopmental Abnormalities and Hyperactive Behavior in Mice, Mol. Neurobiol 53 (2016) 5818–5832. doi: 10.1007/s12035-015-9505-7. [DOI] [PubMed] [Google Scholar]
- [331].Prevost G, Arabo A, Jian L, Quelennec E, Cartier D, Hassan S, Falluel-Morel A, Tanguy Y, Gargani S, Lihrmann I, Kerr-Conte J, Lefebvre H, Pattou F, Anouar Y, The PACAP-regulated gene selenoprotein T is abundantly expressed in mouse and human β-cells and its targeted inactivation impairs glucose tolerance, Endocrinology. 154 (2013) 3796–3806. doi: 10.1210/en.2013-1167. [DOI] [PubMed] [Google Scholar]
- [332].Cinemre FBS, Cinemre H, Erdoğan E, Dilaveroglu N, Tüten A, Kaya B, Yılmaz N, Gulyasar T, Yıldız M, Bahtiyar N, Kızıler AR, Aydemir B, Association of selenoprotein W1 (rs3786777) polymorphism, maternal plasma selenoprotein W (SelW), and selenium levels in patients with pre-eclampsia, Trace Elem. Electrolytes (2018). doi: 10.5414/TEX01542. [DOI] [Google Scholar]
- [333].Kasaikina MV, Fomenko DE, Labunskyy VM, Lachke SA, Qiu W, Moncaster JA, Zhang J, Wojnarowicz MW, Natarajan SK, Malinouski M, Schweizer U, Tsuji PA, Carlson BA, Maas RL, Lou MF, Goldstein LE, Hatfield DL, Gladyshev VN, Roles of the 15-kDa Selenoprotein (Sep15) in Redox Homeostasis and Cataract Development Revealed by the Analysis of Sep 15 Knockout Mice, J. Biol. Chem 286 (2011) 33203–33212. doi: 10.1074/jbc.M111.259218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [334].Huang L, Shi Y, Lu F, Zheng H, Liu X, Gong B, Yang J, Lin Y, Cheng J, Ma S, Lin H, Yang Z, Association Study of Polymorphisms in Selenoprotein Genes and Kashin-Beck Disease and Serum Selenium/Iodine Concentration in a Tibetan Population, PLoS One. 8 (2013) 1–8. doi: 10.1371/journal.pone.0071411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [335].Xiong YM, Mo XY, Zou XZ, Song RX, Sun WY, Lu W, Chen Q, Yu YX, Zang WJ, Association study between polymorphisms in selenoprotein genes and susceptibility to Kashin-Beck disease, Osteoarthr. Cartil 18 (2010) 817–824. doi: 10.1016/j.joca.2010.02.004. [DOI] [PubMed] [Google Scholar]
- [336].Zhao H, Liang D, Grossman HB, Wu X, Glutathione peroxidase 1 gene polymorphism and risk of recurrence in patients with superficial bladder cancer, Urology. 66 (2005) 769–774. doi: 10.1016/j.urology.2005.04.033. [DOI] [PubMed] [Google Scholar]
- [337].Sorolla MA, Reverter-Branchat G, Tamarit J, Ferrer I, Ros J, Cabiscol E, Proteomic and oxidative stress analysis in human brain samples of Huntington disease, Free Radic. Biol. Med 45 (2008) 667–678. doi: 10.1016/j.freeradbiomed.2008.05.014. [DOI] [PubMed] [Google Scholar]
- [338].Haug U, Poole EM, Xiao L, Curtin K, Duggan D, Hsu L, Makar KW, Peters U, Kulmacz RJ, Potter JD, Koepl L, Caan BJ, Slattery ML, Ulrich CM, Glutathione peroxidase tagSNPs: Associations with rectal cancer but not with colon cancer, Genes Chromosom. Cancer 51 (2012) 598–605. doi: 10.1002/gcc.21946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [339].Bellinger FP, Bellinger MT, Seale LA, Takemoto AS, Raman AV, Miki T, Manning-Boǧ AB, Berry MJ, White LR, Ross GW, Glutathione peroxidase 4 is associated with neuromelanin in substantia nigra and dystrophic axons in putamen of Parkinson’s brain, Mol. Neurodegener 6 (2011). doi: 10.1186/1750-1326-6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [340].Du XH, Dai XX, Song RX, Zou XZ, Sun WY, Mo XY, Bai GL, Xiong YM, SNP and mRNA expression for glutathione peroxidase 4 in Kashin-Beck disease, Br. J. Nutr 107 (2012) 164–169. doi: 10.1017/S0007114511002704. [DOI] [PubMed] [Google Scholar]
- [341].Friedmann Angeli JP, Schneider M, Proneth B, Tyurina YY, Tyurin VA, Hammond VJ, Herbach N, Aichler M, Walch A, Eggenhofer E, Basavarajappa D, Rådmark O, Kobayashi S, Seibt T, Beck H, Neff F, Esposito I, Wanke R, Förster H, Yefremova O, Heinrichmeyer M, Bornkamm GW, Geissler EK, Thomas SB, Stockwell BR, O’Donnell VB, Kagan VE, Schick JA, Conrad M, Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice, Nat. Cell Biol 16 (2014) 1180–1191. doi: 10.1038/ncb3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [342].Dentice M, Ambrosio R, Damiano V, Sibilio A, Luongo C, Guardiola O, Yennek S, Zordan P, Minchiotti G, Colao A, Marsili A, Brunelli S, Del Vecchio L, Larsen PR, Tajbakhsh S, Salvatore D, Intracellular inactivation of thyroid hormone is a survival mechanism for muscle stem cell proliferation and lineage progression, Cell Metab. 20 (2014) 1038–1048. doi: 10.1016/j.cmet.2014.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [343].Hernandez A, Martinez ME, Fiering S, Galton VA, Germain D St., Type 3 deiodinase is critical for the maturation and function of the thyroid axis, J. Clin. Invest 116 (2006) 476–484. doi: 10.1172/JCI26240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [344].Azevedo MF, Barra GB, Naves LA, Velasco LFR, Castro PGG, De Castro LCG, Amato AA, Miniard A, Driscoll D, Schomburg L, Neves FDAR, Selenoprotein-related disease in a young girl caused by nonsense mutations in the SBP2 gene, J. Clin. Endocrinol. Metab 95 (2010) 4066–4071. doi: 10.1210/jc.2009-2611. [DOI] [PubMed] [Google Scholar]
- [345].Seeher S, Atassi T, Mahdi Y, Carlson BA, Braun D, Wirth EK, Klein MO, Reix N, Miniard AC, Schomburg L, Hatfield DL, Driscoll DM, Schweizer U, Secisbp2 is essential for embryonic development and enhances selenoprotein expression., Antioxid. Redox Signal 21 (2014) 835–49. doi: 10.1089/ars.2013.5358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [346].Ohshio I, Kawakami R, Tsukada Y, Nakajima K, Kitae K, Shimanoe T, Saigo Y, Hase H, Ueda Y, Jingushi K, Tsujikawa K, ALKBH8 promotes bladder cancer growth and progression through regulating the expression of survivin, Biochem. Biophys. Res. Commun 477 (2016) 413–418. doi: 10.1016/j.bbrc.2016.06.084. [DOI] [PubMed] [Google Scholar]
- [347].Shimada K, Nakamura M, Anai S, De Velasco M, Tanaka M, Tsujikawa K, Ouji Y, Konishi N, A novel human AIkB homologue, ALKBH8, contributes to human bladder cancer progression, Cancer Res. 69 (2009) 3157–3164. doi: 10.1158/0008-5472.CAN-08-3530. [DOI] [PubMed] [Google Scholar]
- [348].Monies D, Vågbø CB, Al-Owain M, Alhomaidi S, Alkuraya FS, Recessive Truncating Mutations in ALKBH8 Cause Intellectual Disability and Severe Impairment of Wobble Uridine Modification, Am. J. Hum. Genet (2019). doi: 10.1016/j.ajhg.2019.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [349].Labunskyy VM, Lee BC, Handy DE, Loscalzo J, Hatfield DL, Gladyshev VN, Both Maximal Expression of Selenoproteins and Selenoprotein Deficiency Can Promote Development of Type 2 Diabetes-Like Phenotype in Mice, Antioxid. Redox Signal 14 (2011) 2327–2336. doi: 10.1089/ars.2010.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [350].Yarham JW, Lamichhane TN, Pyle A, Mattijssen S, Baruffini E, Bruni F, Donnini C, Vassilev A, He L, Blakely EL, Griffin H, Santibanez-Koref M, Bindoff LA, Ferrero I, Chinnery PF, McFarland R, Maraia RJ, Taylor RW, Defective i6A37 Modification of Mitochondrial and Cytosolic tRNAs Results from Pathogenic Mutations in TRIT1 and Its Substrate tRNA, PLoS Genet. 10 (2014). doi: 10.1371/journal.pgen.1004424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [351].Nordstrand LM, Svärd J, Larsen E, Nilsen A, Ougland R, Furu K, Lien GF, Rognes T, Namekawa SH, Lee JT, Klungland A, Mice lacking Alkbh1 display sex-ratio distortion and unilateral eye defects, PLoS One. 5 (2010) 1–12. doi: 10.1371/journal.pone.0013827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [352].Ougland R, Lando D, Jonson I, Dahl JA, Moen MN, Nordstrand LM, Rognes T, Lee JT, Klungland A, Kouzarides T, Larsen E, ALKBH1 is a histone H2A dioxygenase involved in neural differentiation, Stem Cells. 30 (2012) 2672–2682. doi: 10.1002/stem.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [353].Tasaki M, Shimada K, Kimura H, Tsujikawa K, Konishi N, ALKBH3, a human AlkB homologue, contributes to cell survival in human non-small-cell lung cancer, Br. J. Cancer 104 (2011) 700–706. doi: 10.1038/sj.bjc.6606012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [354].Konishi N, Nakamura M, Ishida E, Shimada K, Mitsui E, Yoshikawa R, Yamamoto H, Tsujikawa K, High expression of a new marker PCA-1 in human prostate carcinoma, Clin. Cancer Res 11 (2005) 5090–5097. doi: 10.1158/1078-0432.CCR-05-0195. [DOI] [PubMed] [Google Scholar]
- [355].Yamato I, Sho M, Shimada K, Hotta K, Ueda Y, Yasuda S, Shigi N, Konishi N, Tsujikawa K, Nakajima Y, PCA-1/ALKBH3 contributes to pancreatic cancer by supporting apoptotic resistance and angiogenesis, Cancer Res. 72 (2012) 4829–4839. doi: 10.1158/0008-5472.CAN-12-0328. [DOI] [PubMed] [Google Scholar]


