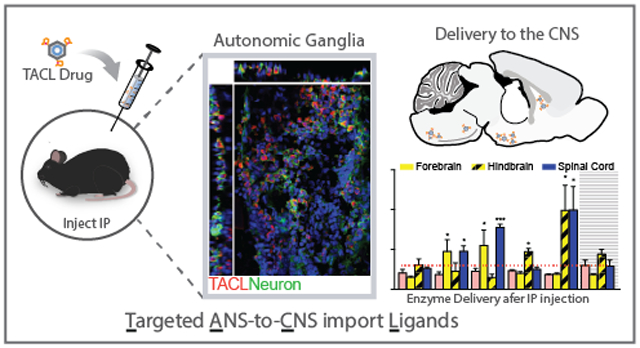
Abstract
While biologic drugs such as proteins, peptides, or nucleic acids have shown promise in the treatment of neurodegenerative diseases, the blood-brain barrier (BBB) severely limits drug delivery to the central nervous system (CNS) after systemic administration. Consequently, drug delivery challenges preclude biological drug candidates from the clinical armamentarium. In order to target drug delivery and uptake into to the CNS, we used an in vivo phage display screen to identify peptides able to target drug-uptake by the vast array of neurons of the autonomic nervous system (ANS). Using next-generation sequencing, we identified 21 candidate Targeted ANS-to-CNS uptake Ligands (TACL) that enriched bacteriophage accumulation and delivered protein-cargo into the CNS after intraperitoneal (IP) administration. The series of TACL peptides were synthesized and tested for their ability to deliver a model enzyme (NeutrAvidin-horseradish peroxidase fusion) to the brain and spinal cord. Three TACL-peptides facilitated significant active enzyme delivery into the CNS, with limited accumulation in off-target organs. Peptide structure and serum stability is increased when internal cysteine residues are cyclized by perfluoroarylation with decafluorobiphenyl, which increased delivery to the CNS further. TACL-peptide was demonstrated to localize in parasympathetic ganglia neurons in addition to neuronal structures in the hindbrain and spinal cord. By targeting uptake into ANS neurons, we demonstrate the potential for TACL-peptides to bypass the blood-brain barrier and deliver therapeutics into the brain and spinal cord.
Keywords: peptide, targeting ligands, drug delivery, brain, spinal cord, phage display
Graphical Abstract
The inability of biologic drugs to cross the BBB has severely limited the therapeutic options for neurodegenerative diseases of the CNS, such as Alzheimer’s Disease (AD), Parkinson’s Disease (PD), and amyotrophic lateral sclerosis (ALS).1–5 In an effort to circumvent the BBB, research efforts have used attempted to utilize sequential metabolism, chemical disruption, and focused ultrasound to transiently disrupt the BBB for drug delivery.6–12 Similar efforts have exploited receptor-mediated uptake and transcytosis13,14 across the brain endothelium to treat brain-afflicting diseases like AD.15–19 Still, delivery challenges restrict and preclude many promising biological drug candidates from being translated into the clinic.20 Therefore, additional technologies to target tissue uptake and promote biologic drug delivery to the CNS, without compromising the integrity of the BBB, would significantly enhance the development of drugs to cure or treat neurodegenerative diseases.13,21
For decades, viruses (e.g. pseudorabies virus; PRV, and herpes simplex viruses) have been known to enter the CNS by retrograde axonal transport after infection via peripheral lesions. Pseudorabies virus (PRV) has been widely used to map synaptically linked neural circuits and has demonstrated the ability to traffic within the sympathetic and autonomic nervous system.22–24 Consequently, viruses have been engineered for remote gene transfer in animal models of disease,25–28 and virus chimeras have been used in gene therapy experiments to treat spinal muscular atrophy, chronic pain and amyotrophic lateral sclerosis.29–35 In addition, a growing effort has begun to explore virus serotypes that show improved delivery into the CNS,36–39 and to discover targeting ligands that promote uptake and delivery via peripheral neurons.40
Given the diversity of viruses that demonstrate robust delivery into the CNS after remote administration, we expanded on our previous effort to elucidate CNS targeting ligands and performed an in vivo phage display screen to discover peptide-ligands to enrich therapeutic delivery into the CNS.41–44 In attempt to exploit a CNS delivery mechanism analogous to PRV, an in vivo phage display screen was designed to identify peptide ligands that mediate M13 bacteriophage delivery into the CNS after intraperitoneal (IP) administration. Importantly, the screen strategy was adopted to elucidate peptide ligands that target drug-uptake by the vast array of ANS neurons within the peritoneum to enhance delivery into the CNS, versus other peripheral routes (e.g. intramuscular injection). Furthermore, delivery via the ANS could facilitate efficient drug delivery into the brain or spinal cord without the toxic side-effects observed with excessive blood volume dosing. Moreover, the ANS is comprised of sympathetic and parasympathetic neurons, IP delivery could target a branch of the ANS to enhance delivery to the spinal cord and/or specific brain regions. Here, we report the identification of peptide targeting ligands that transit the ANS and enrich delivery into the CNS. After initial discovery of peptide sequences by phage display, synthesized peptide ligands were engineered to further enhance CNS delivery and peptide stability. We show accumulation of CNS targeting ligands and model protein cargo within the parasympathetic ganglia and along ANS neuronal processes. Thus, we demonstrate the potential for Targeted ANS-to-CNS uptake Ligand (TACL) peptides to utilize the vast network of autonomic neurons for biologics delivery into the CNS after IP administration.
Results and Discussion
Next-generation sequencing elucidates targeting peptides with frequency distributions exhibiting CNS selectivity.
A functional bacteriophage peptide library selection strategy was developed to identify recombinant bacteriophage that exploit autonomic neurons for delivery to the brain and spinal cord. A mixture of linear (Ph.D.12) and cyclic (C7C) bacteriophage peptide libraries were administered by IP injection and bacteriophage were recovered from the brain, spinal cord and heart either 8 or 24 hours post-injection. This selection paradigm was performed for three reiterative selection rounds (Supplementary Figure S1). The tissue harvest time was selected based on our previous experience with retrograde transport peptides40 as well as previous research that reports circulating bacteriophage are cleared from the blood-supply within 24 hours.45,46 However, since phage clearance was not monitored in the current study, animals were perfused with ≥10-blood volumes of saline to exsanguinate and fully dilute the blood supply. Additionally, heart tissue was collected to monitor any non-specific bacteriophage carry-over from the blood supply. Next, to identify and quantify phage in each lysate, single-stranded phage DNA was purified and PCR amplified with tissue-specific barcodes for next-generation sequencing (NGS) analysis (Figure 1A).47 After elimination of DNA sequences known to show preferential amplification, C7C bacteriophage with cyclic peptides comprised just 2% of the total sequences. Comparing the biodistribution of Ph.D.12 and C7C phage between the three collected tissue types, highest titers were found in the spinal cord (43% and 51%, respectively). Brain tissue showed 23% and 33% respective accumulation in contrast to heart tissue (34% and 15%, respectively), which was monitored as an off-target control (Figure 1B). The observed accumulation of phage in the CNS compared to the heart, and the increased distribution to spinal cord compared to brain, suggest that by the third round of screening significant fractions of bacteriophage were trafficking through the ANS to the spinal cord with secondary accumulation in the brain.
Figure 1.
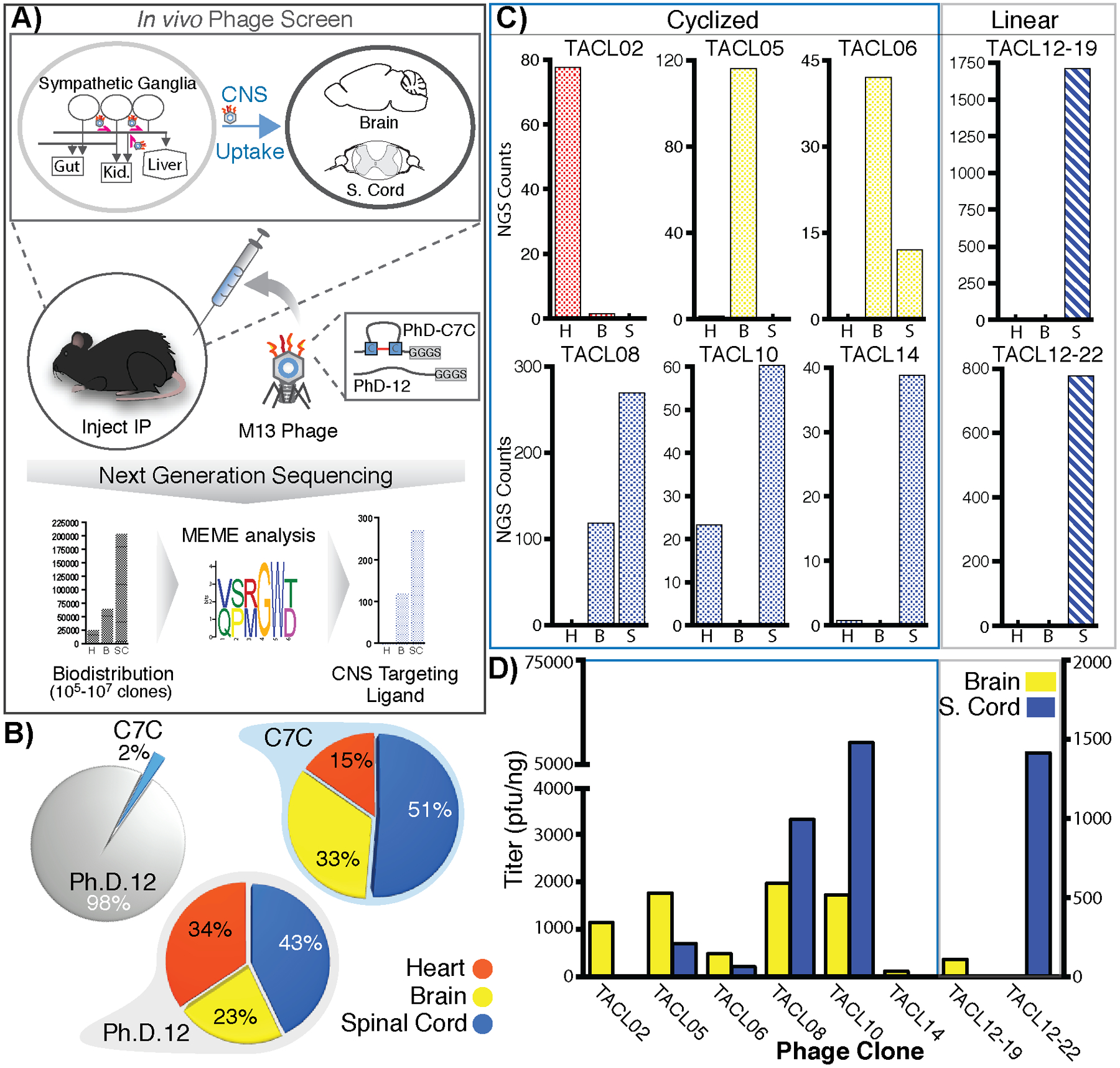
Elucidation of targeted ANS-to-CNS uptake ligands (TACL) that enrich bacteriophage accumulation in the CNS after uptake into the autonomic nervous system. After three rounds of an in vivo screen, bacteriophage isolated from heart, brain, and spinal cord were analyzed by NGS to deduce tissue enrichment of a consensus domain (A). While an initial 50:50 mix of circular (C7C) and linear (Ph.D.12) phage was injected, only 2% of phage-sequences in the final CNS lysate were derived from a C7C background, yet both phage-populations yielded 84% and 66% CNS accumulation versus heart tissue (15% and 34%, respectively) (B). Based on NGS data, phage candidates for TACL peptides sequences were identified with ≥10-fold targeted enrichment in either heart (H; red), brain (B; yellow), spinal cord (S; blue) (C). Five bacteriophage clones showed enrichment in brain and spinal cord tissue after IP injection (D).
Since NGS is able to identify a comprehensive list of phage sequences present within each tissue (~1.0×10e6 total yield), selection criterion was established to qualify phage candidates based on tissue distribution and enrichment (as a function of NGS read counts) for subsequent characterization. From the >900,000 linear and >30,000 cyclic phage recovered, we set the selection criteria to filter the NGS data and isolate phage-clones with ≥500 (linear peptides) or ≥30 (cyclic peptides) read counts within CNS tissues. At the second step of selection, the 71 linear-peptide and 17 cyclic-peptide candidate bacteriophages were pared down further to prioritize sequences with ≥10-fold enrichment in brain or spinal cord compared to heart, with additional weight applied to prioritize the selection of sequences with zero reads within heart lysate. For subsequent comparisons and targeting validation, phage clones with elevated off-target enrichment (in the heart) were selected. In total, we settle on a list of 6 linear peptide and 15 cyclic peptide-displaying bacteriophage to produce high titer clonal populations to administer by IP injection and evaluate tissue biodistribution. The NGS distribution of phage-reads in the brain, spinal cord and heart of all 21 candidates is shown in Supplementary Figure S2. Selected clones with high NGS representation in the CNS (Figure 1C) were produced at high-titer concentrates (107-109 cfu/ml) for IP injection, and phage accumulation in the brain and spinal cord was determined 24hours after injection. The biodistribution of 5 high-titer clones showed consistent CNS enrichment (Figure 1D: TACL05, TACL06, TACL08, TACL10, and TACL12–22).
TACL peptides traffic protein cargo into the central nervous system.
We next sought to determine whether TACL peptides could be used to deliver exogenous proteins into the CNS after IP injection. To accomplish this, TACL peptide sequences from select phage shown in Figure 1C were synthesized by microwave assisted solid-phase peptide synthesis (SPPS), purified and characterized by MALDI-ToF mass spectrometry (Table 1). Each peptide was synthesized with biotin at the amino- or carboxy-terminal end; however, the location of biotin conjugation did not affect delivery (data not shown). Thus, in all subsequent studies, peptides synthesized with amino-terminal biotin were used. To mimic the multimeric display of recombinant peptides on the bacteriophage pIII coat-protein, NeutrAvidin (NA) was complexed with biotin-TACL peptides prior to IP injection. After full-body perfusion with saline, tissues were harvested 24-hours post-injection and homogenized in reaction lysis buffer and TACL-mediated protein delivery was quantified in each tissue lysate as a function of NeutrAvidin horse-radish peroxidase (NA-HRP) activity. As shown, both TACL05, TACL08 increased NA-HRP delivery 2- and 2.3-fold to the spinal cord versus control C7con-peptide (p < 0.01; Figure 2A), while TACL06 and TACL10 trended toward significant CNS accumulation. After normalization against corresponding tissues from saline-treated animals (naive tissues) to account for the endogenous peroxidase activity of each organ, TACL peptides (TACL05, TACL06, TACL08, and TACL10) did not show significant NA-HRP delivery to off-target organs (Figure 2B), aside from elevated peroxidase activity observed in liver. Surprisingly, peptides derived from clones with elevated off-target (heart; e.g. TACL02) accumulation did not recapitulate prior heart delivery. As shown, TACL10 (SACHQSQSRMCGGGS) showed a similar delivery profile and nearly identical peptide sequence to our previously described peptide for Targeted Axonal Import (TAxI: SACQSQSQMRCGGGS: >92% homology to TAC10; 13/14 amino acids) that localizes to the CNS after intramuscular administration.40 Thus, the TAxI sequence, which is a likely consensus sequence for a CNS targeting ligand via ANS or somatic nervous system (SNS), was used in subsequent peptide optimization studies.
Table 1.
TACL peptide sequences used for NeutrAvidin delivery to the CNS. Phage clones enrichment (by NGS) in the Heart, Brain, and Spinal Cord are listed.
| Peptide | Phage | Sequence | Weight (g/mol) | |||
|---|---|---|---|---|---|---|
| S.Cord | ||||||
| TACL02 | Circular | SACNTTLNGLCGGG | 1266.4 | 80X | 0 | 0 |
| TACL05 | Circular | SACPSHLTKMCGGG | 1347.6 | 2X | 120X | 0 |
| TACL06 | Circular | SACSQKNFTHCGGG | 1395.5 | 0 | 40X | 10X |
| TACL08 | Circular | SACPLSKGKLCGGG | 1276.5 | 0 | 60X | 90X |
| TACL10 | Circular | SACHQSQSRMCGGG | 1407.6 | 20X | 0 | 60X |
| TACL14 | Circular | SACTMSTTQVCGGG | 1301.5 | 0 | 0 | 40X |
| TACL12–19 | Linear | GFPSVRDLSPLRGGGS | 1600.8 | 0 | 0 | 1700X |
| TAC12–19scr | Linear | SGVPLFDLRPSRGGGS | 1600.8 | N/A | N/A | N/A |
| TACL12–22 | Linear | WATLDLGPQPYSGGGS | 1604.7 | 0 | 0 | 780X |
| TAxI† | Circular | SACQSQSQMRCGGG | 1398.6 | N/A | N/A | N/A |
| C7con | Control | SACGAGGGAGCGGG | 980.04 | N/A | N/A | N/A |
, peptide sequenced isolated from previously publish phage display screen.
Figure 2.
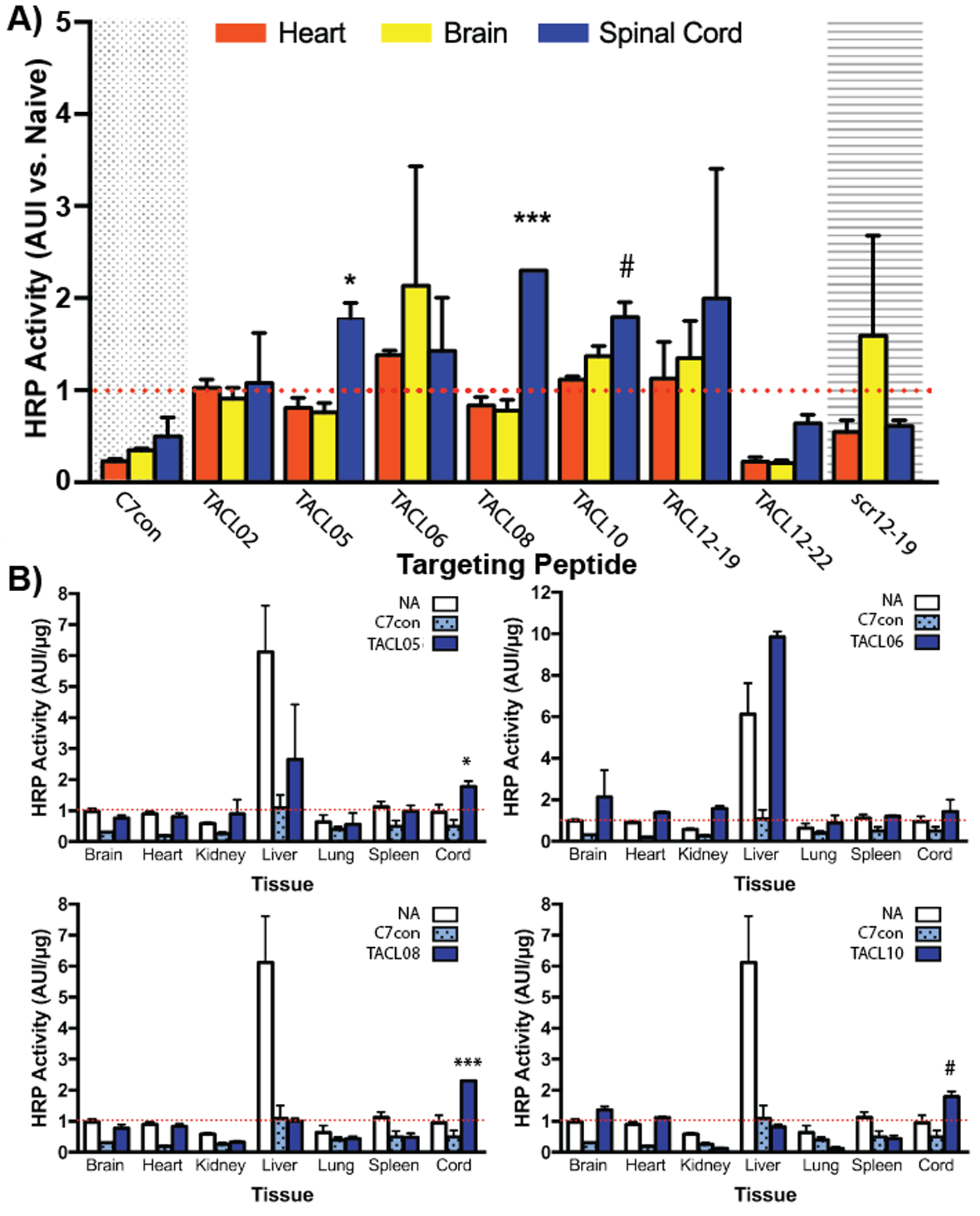
TACL peptides deliver model protein drug cargo into the CNS after IP injection. Biotinylated TACL peptides designed and synthesized by standard SPPS protocols, in concordance with each respective recombinant phage insert, were complexed with NeutrAvidin-HRP to quantify drug-delivery into the heart (red), brain (yellow), or spinal cord (blue) 24 hours after IP injection (A). Accumulation in all vital organs (B) was quantified by HRP activity and normalized against protein concentration for each NA-HRP (clear bars), C7con-NA (dotted bars), and TACL-NA complexes (solid blue bars) that showed CNS delivery (***, p<0.0001; *, p<0.01; #, p=0.052. n=3)
Peptide stapling via cysteine thiols stabilizes peptide structure and improves serum stability.
Despite the predominant recovery of Ph.D.12 (linear peptide library) phage from our in vivo screen, few synthesized linear peptides (if any) recapitulated the CNS accumulation observed with bacteriophage display (Figure 2A). While puzzling, the disparity in phage versus peptide distribution (i.e. Table 1; TACL 12–19) could be a consequence of the instability and short half-life of synthetic peptides in vivo. Peptides are known to have increased stability and peptidase resistance when immobilized versus free in solution.48–51 Furthermore, previous research has demonstrated that peptide stability is increased by cyclization.52,53 While linear peptides displayed from the Ph.D.12 library were dominantly selected by in vivo screening, the isolated cyclic peptide sequences from the C7C library are likely a better starting point for applications as in vivo targeting ligands due to the increase structural stability of the cyclized peptides.
We hypothesized that peptide stability and resistance to peptidase degradation could increase the efficacy of CNS targeting ligands. The C7C library displays peptides cyclized by disulfides between cysteines flanking a variable 7-amino acid sequence. However, disulfide cyclization can be reversed by thiol-disulfide exchange.54,55 We therefore evaluated alternate peptide stapling methods that utilize cysteine perfluoroarylation to produce constrained structures56 and improve peptide activity.57 Previously, our lab has demonstrated that cysteine perfluoroarylation with decafluorobiphenyl (DFBP) significantly affects peptide resistance to degradation and target binding affinity when compared against peptides stabilized by disulfides, amides, or triazoles formed by click cyclization strategies.58 Therefore, TAxI peptide was cyclized by DFBP stapling of flanking cysteines and the structure of the cyclized peptide was characterized by circular dichroism (CD) measurements. While none of the CD spectra show conversion to an alpha-helix or beta-sheet conformation,59 a step-wise progression toward a lower percent random-coil conformation was observed from linear to disulfide bridge, and DFBP-stapled peptides (Figure 3). DFBP-stapled peptides showed less random coil structure than peptides with cysteine-cysteine disulfides despite consistent ligation between Cys-4 and Cys-8 and suggests that the biphenyl (BP) moiety reduces random-coil anisotropy and imparts improved structural stability than disulfide cyclization.60 Thus, DFBP reacted peptides displayed more structural stability than disulfide-cyclized peptides.
Figure 3.
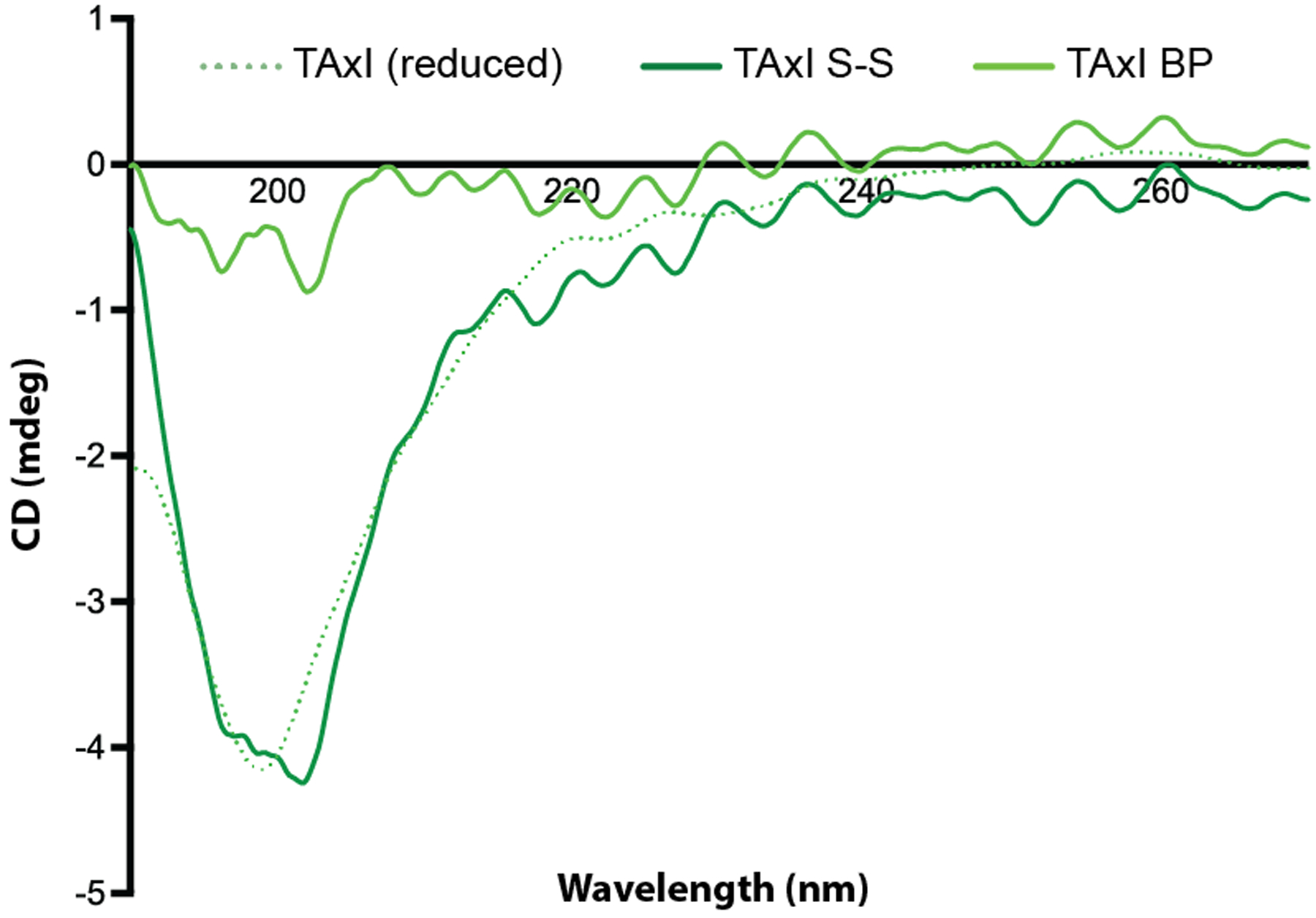
Perfluoroarylation of TAxI-peptide reduces random-coil structure. Circular dichroism measurements of linear TAxI peptide (reduced; dotted line), circular disulfide-bridge TAxI peptide (S-S; dark-green line), and TAxI peptide cyclized by cysteine perfluoroarylation (BP; light-green line).
Given the observed changes in random-coil properties with perfluoroarylation, we hypothesized that peptides with alternative cyclization chemistries would show greater serum stability.61 As predicted, the linear biotin-TAxI peptide showed early degradation-products within 2 hours (Figure 4A: 1625 Da = start; 656 Da QSQMR product) and complete loss of intact biotin-TAxI after 4 hours. In comparison, the presence of a disulfide linkage in the cyclized TAxI peptide, prolonged biotin-TAxI serum stability. Early degradation products at 8 hours show biotin-TAxI disulfide (S-S) undergoes both cleavage between the cysteine-linkages (Figure 4B: 1623 Da= start; 1452, 1509, 1531 Da products), as well as a n-terminal degradation after reduction of the disulfide bridge (1623 Da), which will linearize with changes in the redox environment. Still, intact biotin-TAxI S-S peptide persisted in mouse serum for 48 hours. DFPB-stapled biotin-TAxI (b-TAxI BP) serum stability was markedly improved and showed few endolytic degradation products until 24 hours (Figure 4C: 1918 Da = start; 652 Da QMRCGGG product). It should be noted that MALDI-ToF measurements are not quantitative and attempts to quantify the extent of degradation are highly variable after acetonitrile precipitation and extraction; thus, we could not quantitatively compare the improved serum stability (see Supplementary Table S1 for a complete fragment list and sequence identification). Nonetheless, by identifying the degradation products, we demonstrate that non-reversible cysteine linkages greatly improve peptide molecular and serum stability. These results highlight the potential to utilize peptide cyclization as a strategy to stabilize linear peptide-candidates shown to enrich phage CNS accumulation (i.e. TALC12–22), which warrant further investigation.
Figure 4.
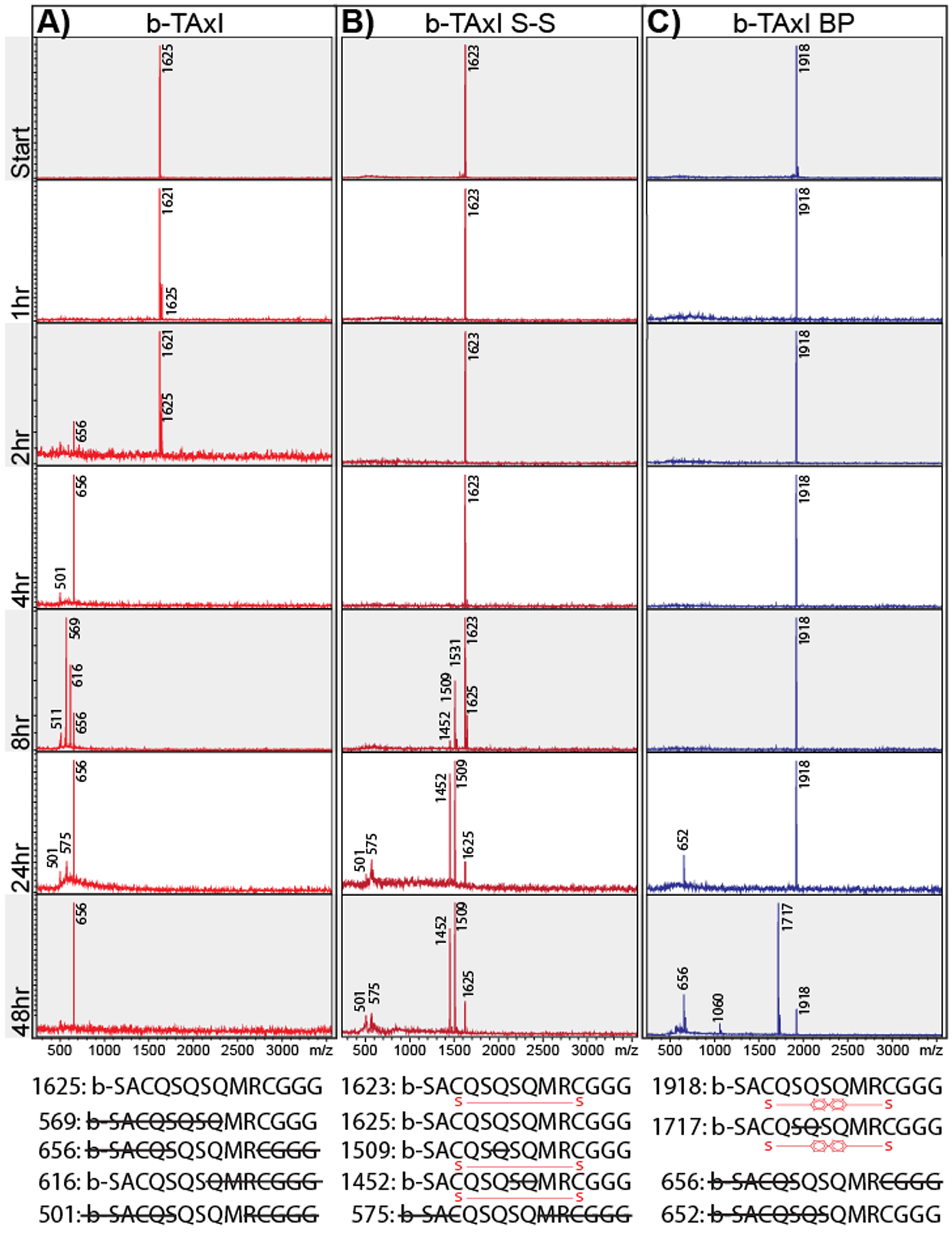
Non-reversible peptide stapling increases peptide stability and protease resistance. MALDI-ToF spectra of biotin-TAxI (A), biotin-TAxI S-S (B), and biotin-TAxI BP (C) peptides incubated in normal mouse serum for 1hr, 2hr, 4hr, 8hr, 24h, and 48 hr. Primary peaks of each peptide-fragment and molecular weight is shown. Bottom: predicted amino acid sequence of degradation products, based on molecular weight.
Stapled TACL- and TAxI-peptide enhances delivery and shows labeling in ANS neurons.
The ability to deliver proteins into the spinal cord from distal administration sites provides tremendous potential for the development of CNS therapeutics.62 After demonstrating the molecular structure and stability benefits imparted by DFBP-cyclization, we next examined whether perfluoroarylation impacted protein-delivery into the CNS. As with previous experiments, 24 hours after IP injection of peptide-neutravidin-HRP constructs (10 μg per mouse), CNS tissue was harvested after full-body saline perfusion to quantify protein delivery (HRP activity) within heart, brain and spinal cord. With the recent elucidation of divergent vagal nerve circuits,63 we further dissected brain tissue and separated the forebrain from the hindbrain (i.e. cerebellum and brainstem). Surprisingly, TACL08- and TAxI-peptide both showed robust delivery of perfluoroarylated versus disulfide cyclized peptide into the spinal cord 3.1- and 4.0-fold enrichment, respectively (***, p< 0.001; *, p<0.05, n=3). Yet TAxI- and TACL-peptide showed enhanced delivery to different brain regions (Figure 5A). TAxI(BP) enhanced NA-delivery 4.0-fold (2X vs. TaxI S-S) to the hindbrain with minimal delivery to the forebrain, while TACL08 increased NA-delivery to the forebrain when compared against controls, seemingly with no added benefit by DFBP-cyclization (*, p< 0.05; #, p=0.053; n=3). Therefore, TAxI- and TACL-peptides showed differential fore- and hindbrain accumulation, which could have been masked in previous studies as a consequence of signal dilution in the whole brain lysate (Figure 2A). Moreover, when evaluated against a NA-HRP dilutions series, the levels of HRP activity measured in the CNS (after a single IP injection) correlate to a 1×10−6 enzyme dilution. Thus, animals injected with a micromolar drug dose IP (e.g. NA: 0.5mg/kg) show picomolar levels of drug activity within the CNS, which has been proposed to be sufficient for the delivery of growth factors to impact prognosis and disease progression.64–67
Figure 5.
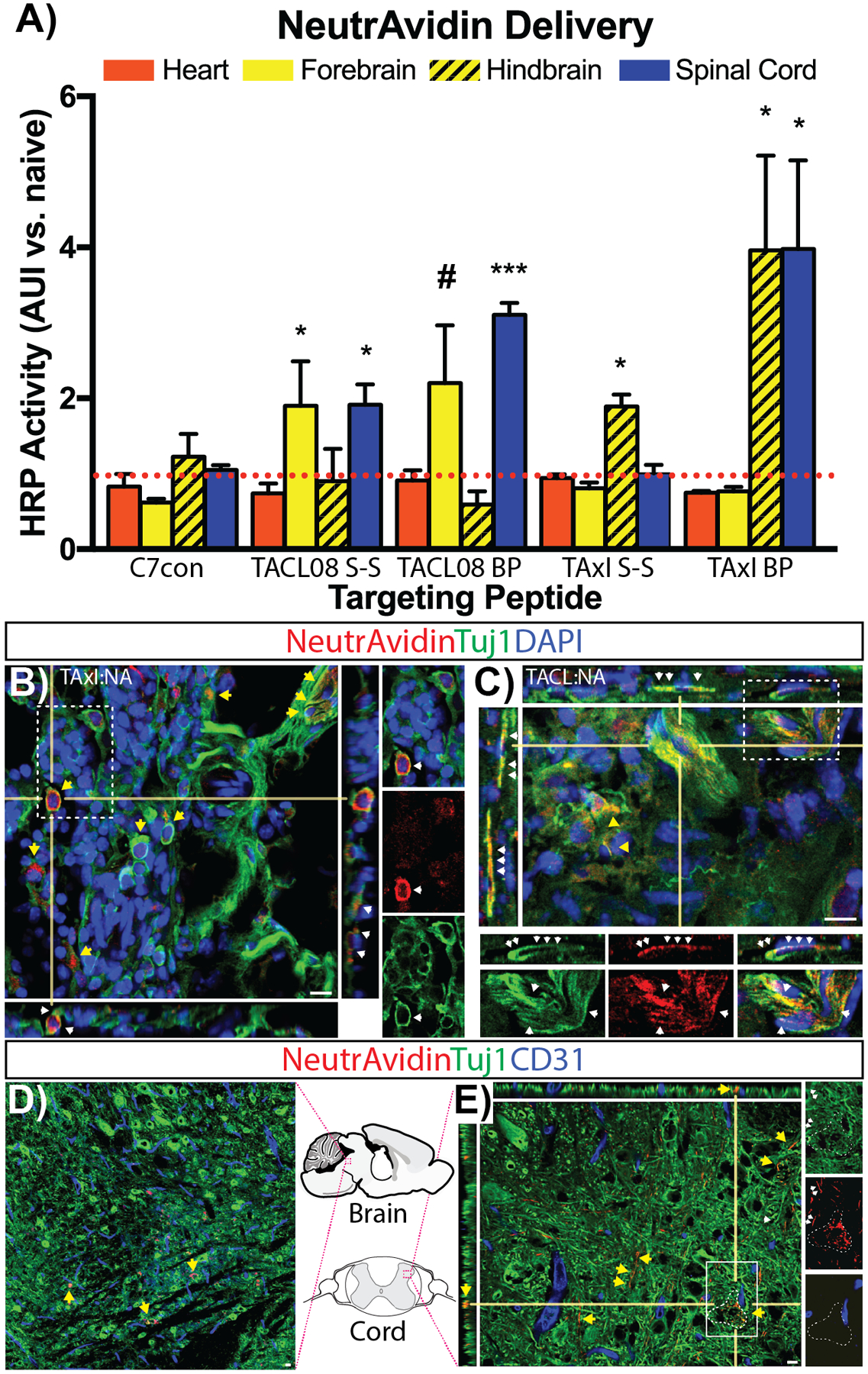
Cysteine cyclization by perfluoroarylation enhances TACL- and TAxI-drug delivery into the CNS. Biotin-TACL08 and Biotin-TAxI peptide cyclized by disulfide bridge formation (S-S; cysteine oxidation) or DFBP-cyclization (BP) were complexed with NA-HRP for IP injection. After 24 hours, HRP activity was quantified in heart (red), fore- and hindbrain (yellow), and spinal cord (blue) and normalized against endogenous peroxidase in naive tissues (A). ***, p<0.001; *, p<0.05; #, p=0.053 versus C7con tissues. n=3. Parasympathetic ganglia harvested 4–6 hours post-injection, processed and stained by immunofluorescence to visualize NeutrAvidin (red) and neuron-specific class III beta-tubulin (Tuj1, green) colocalization in TAxI:NA (B) and TACL:NA (C) injected animals (DAPI, blue). High-resolution confocal micrographs show TAxI:NA (B, inset) and TACL:NA (C, inset) colocalization in neuronal cell bodies (yellow arrows) and along Tuj1+ neuronal processes (traced by white arrows). CNS tissue stained with CD31 (blue; vascular endothelium) in hindbrain (D, outlined region) and spinal cord (E, outlined region) reveal NA (red) on Tuj1+-neurons (yellow arrows) and processes (white arrows, inset). White dotted-box highlights region of zoomed insets from (B), (C) and (E). Bar, 10 μm.
In order to verify trafficking of TAxI- and TACL-peptides within the ANS, parasympathetic ganglia were isolated from the peritoneal cavity 4–6 hrs after IP injection of peptide-NeutrAvidin complexes and stained by immunofluorescence. As shown by confocal microscopy, both TAxI- and TACL-NA decorated neuronal cell bodies throughout the ganglia. Moreover, high-resolution confocal micrographs showed prominent TAxI- and TACL-NA labeling along ganglia-efferent Tuj1+-processes (Figure 5B and C). Together, these data demonstrate that IP injection of TAxI- and TACL-loaded NeutrAvidin (NA) results in ANS uptake via ganglia in the peritoneal cavity. Next, to confirm neuronal uptake in the brain and spinal cord tissue harvested 24-hours post-injection were stained for NA delivery. In both brain and spinal tissue, NA localization within cell bodies was sparse yet observable within the hindbrain (Figure 5D). Similarly, NA immunoreactivity could be observed along neuronal Tuj1-labeled processes and an occasional cell body within spinal cord grey matter (Figure 5E, highlighted inset). As expected (based on HRP levels and dilution effects), NA immunostaining in the CNS was sparse. While, NA was observed along Tuj1+ neuronal process, we did not observe NA colocalization on vascular endothelium (i.e. CD31).
Since ANS pathways converge in the brainstem and hindbrain,68 the observed difference in forebrain versus hindbrain accumulation was an unexpected result. With the recent publication of a polysynaptic vagal-to-striatum circuit,63 the observed difference in TACL08- and TAxI-peptide could suggest alternate selective routes of transmission through the ANS (i.e. sympathetic vs. parasympathetic neurons). While these results do not rule out alternate routes for CNS entry, the data support further development and testing of TACL- and TAxI-peptides for CNS drug delivery.
Conclusion
In this study, in vivo bacteriophage peptide library screening was combined with next generation sequencing to identify peptides that facilitate drug delivery to the CNS after intraperitoneal administration. By targeting the expansive network of autonomic neurons to enhance drug uptake, we demonstrate the ability of targeting ligands to utilize the ANS to deliver therapeutically viable dosages into the CNS in a minimally invasive manner.64 We have identified TACL-peptides that enhance delivery of recombinant bacteriophage and a model protein-drug into the brain and spinal cord. Our results reveal that by utilizing perfluoroarylation cysteine stapling, an evolved synthesis significantly increased structural stability and endopeptidase resistance to further enhance delivery. Thus, TACL peptides demonstrate the ability to deliver protein drugs to sites of interest within the CNS, potentially avoiding toxicity observed with systemic or direct tissue injection.71,72
Interestingly, the TACL08 and TAxI peptide each showed a distinct distribution within the CNS. As demonstrated (Figure 5A), TACL08 enhanced protein delivery into the spinal cord and forebrain, while TAxI enhanced protein uptake in the spinal cord and hindbrain. In contrast to initial NeutrAvidin delivery (Figure 2), brain tissue was subdivided to quantify HRP activity in the fore- and hindbrain. Although speculative, the differences in forebrain and hindbrain accumulation suggests a possible preference for parasympathetic versus sympathetic neurons. While TAxI and TACL08 each show uptake to the spinal cord via parasympathetic ganglia (e.g. celiac ganglia: Figure 5B), differential trafficking via the vagal nerve could affect polysynaptic transmission into the striatum (e.g. TACL08) versus the brainstem and hindbrain (e.g. TAxI).63
While the mechanism of uptake remains unknown, the potential to target distinct branches of the ANS to modulate uptake by the sympathetic ganglia to enhance targeted drug delivery to the spinal cord versus brain. A protein-database search revealed TACL08 shares homology with receptors (GABA-receptors and a vomeronasal receptor) and intracellular proteins that would facilitate uptake and transmission in inhibitory parasympathetic neurons (Supplementary Table S2). Preferential uptake and protein delivery to distinct sympathetic or parasympathetic neurons suggests that ANS import-ligands could be further tailored to specific neuropathies or reward centers in the brain. Thus, TACL08 and TAxI peptide could be used for select or broad targeting of drugs to treat neurodegenerative diseases (e.g. Alzheimer’s Disease) or reward and addiction disorders (e.g. obesity and/or drug addiction).69
Methods
Animals
As required, all animal work was review and approved by the University of Washington institutional animal care and use committee (IACUC). Mice (C57bl/6 strain) were purchased from Jackson Laboratory.
In vivo phage display selection of CNS-targeted phage
For library screening, Ph.D.−12 and C7C phage library (New England Biolabs, USA) were mixed, 1.5 × 1013 pfu/mL, and injected intraperitoneally (IP) into C57bl/6 mice (100 μl, n=2 per group). Prior to tissue harvest, mice were euthanized and perfused with 20 mL saline to ensure complete tissue exsanguination. Spinal cord, brain, and cardiac tissue was harvested and flash-frozen in DMEM (Invitrogen, Inc.) with 1% BSA (Fisher Scientific). After homogenization, tissue homogenates were centrifuged and resuspended in 1% Triton-X 100 (Sigma-Aldrich, Inc.) and passed through a 25-gauge needle to ensure cell lysis. Similar to previous studies, phage titer was quantified and spinal- and brain-tissue homogenate was amplified for subsequent screens.38
Phage DNA Isolation and Purification for Next-Generation Sequencing
DNA amplicons were prepared as previously described with some modifications.40,47 Single-stranded DNA (ssDNA) was isolated from amplified phage eluate after three rounds of in vivo phage display using QIAprep Spin M13 Kit (QIAGEN). ssDNA was amplified by PCR using primers containing Illumina-compatible sequencing adaptors and sample-specific barcodes.
Illumina Next-Generation Sequencing and Data Processing
Sequencing and data processing was performed as previously described with some modifications.47 Briefly, phage pools were sequenced on an Illumina MiSeq system. After completion, sequence reads were identified and tabulated. The relative abundance of specific recombinant phage was calculated as the fraction of total reads in the library with “PepRS” (https://bitbucket.org/stevesal/peprs).
Recombinant Phage Clones
Oligonucleotides encoding the peptides of interest were synthesized Operon (Eurofins) and annealed into M13KE dsDNA vector. After transduction into bacterial, phage plaques were used to isolate ssDNA and confirm sequences by DNA sequencing prior to production of high-titer pools for in vivo usage.
Phage Biodistribution Study
The selected phage clones (Figure S2) were tested for CNS-targeting in adult C57bl/6 mice (8 weeks old). Mice injected with (1 ×106, 107, 108, and 109 pfu) were exsanguinated by transcardiac saline perfusion (20 mls) prior to vital organs being harvested and snap-frozen on liquid nitrogen (n=3–4 per phage clone, one mouse per concentrate). Tissues homogenized in PBS+1% BSA (PBSA) were exposed to E. coli ER2738 cultures and tittered on IPTG/X-gal plates (according to standard protocols). To account for volume and tissue density variability, phage titers were normalized against tissue-lysate protein concentration as determined by a standard BCA assay (pfu/mg of tissue).
Peptide synthesis
Peptide synthesis was performed on a CEM microwave peptide synthesizer by standard FMOC solid phase peptide synthesis on a Rink Amide resin (Novabiochem). A tri-lysine block was appended at the n-terminus to increase peptide solubility, as needed.70 On-resin cyclization with decafluorobiphenyl (DFBP) or disulfide formation in solution (0.1M Sodium Carbonate) with each peptide proceeded efficiently based on HPLC traces during purification.58 Full peptide synthesis schematics are shown in Table1.
Serum Stability
TAxI peptide (30μg) was incubated in normal mouse serum (300 μl) at 37 °C. At 2hr, 4hr, 8hr, 24hr, and 48hr increments an aliquot of the serum (40 μl) was precipitated with in cold ACN (40 μl). The mixture was centrifuged at 15 000 rpm for 5 min. A 50:50 H2O−ACN solution was added (1:1 v/v, 80 μl) to each pellet and sonicated for 10 min (to further extract residual peptides). The mixture was then centrifuged, and the supernatant was collected and pooled with the first sample. The combined supernatants were dried on a Speedvac. The peptide pellet was resuspended in ACN/water and sonicated for analysis on MALDI-ToF MS. Fragmentation product molecular weights were used by a custom Java program (stability.jar) to analyze peptide serum stability, peptide-fragment sequences, and cleavage sites. The stability.jar program is hosted on GitHub and is available for download (https://github.com/juliomarcopineda/peptide-serum-stability/releases).
Circular Dichroism
TAxI peptide was prepared in H2O at 50 μM. CD measurement of the peptides was performed on a Jasco 720 circular dichroism spectrophotometer (Jasco Inc., Easton, MD) at 25 °C with an average from eight scans. The presented measurements are solvent blank-subtracted and zeroed to the 270nm measurement.
NeutrAvdidin Biodistribution analysis by ELISA
Prior to injection, NeutrAvidin-HRP (Thermo-fisher, Peirce) was complexed with each peptide (4:1 molar ratio) in phosphate buffered saline. Each peptide was synthesized with biotin at the amino terminus to facilitate complexation. After 30 minutes at room temperature, NA-complexes were isolated from unbound peptide by size-exclusion chromatography on a 10 kDa spin-column (Millipore). NA:peptide complexes were eluted and brought to volume in saline prior to use. NA:peptide concentration was confirmed by absorbance (A280) on a Nanodrop (ThermoFisher, Inc). After IP injection of NeutrAvidin:peptide complexes (10μg NA per injection), the organs of exsanguinated mice were isolated and snap-frozen with liquid nitrogen. Tissues were homogenized in Reporter Lysis Buffer (RLB, Promega) supplemented with Protease Inhibitors (Roche) and DNase (1U/ml; Worthington). After centrifugation (21,000 RFC, 10–15 minutes) to remove debris, the protein concentration of each tissue lysate was determined by BCA assay. The delivery of NA:peptide complexes was quantified by addition of ELISA substrate (R&D Systems) and HRP activity was quantified as absorbance units (AUI) at 450 nm (540 nm the reference) after 20 (heart, kidney, liver, and spleen) and 40 mins (brain, lung and spinal cord). NA-HRP distribution was normalized by protein content (as above). To express the level of NA delivery, in all HRP ELISA assays, experimental tissues were normalized against HRP activity in tissues from sham treated animals (saline injection: naïve tissue) to express values as a ratio above endogenous peroxidase activity (i.e. NA-HRP versus naive tissue AUI).
Immunohistochemistry
After treatment and transcardiac perfusion, parasympathetic ganglia embedded in OTC were sectioned with a Leica SM1850 cryostat. Primary antibodies previously shown to identify neurons (NeuN, Millipore clone A60, MAB377, diluted 1:200; Tuj1, Biolegends, diluted 1:1000) were used to demonstrate colocalization with NeutrAvidin (goat anti-Avidin; Life Technologies, 31850) animals. After rinses in 0.1% Tween-20 TBS, secondary antibodies (donkey α-mouse IgG, α-goat, conjugates at 1:500 dilution, Jackson Labs) were applied in blocking buffer for 2 hours at room temperature, or overnight at 4°C.
Microscopy
Confocal microscopy with a Nikon TS2000E equipped with a krypton/argon laser, a red diode laser, and an infinity-corrected 40X 1.4 NA lens was used to visualize NeutrAvidin distribution along neuronal processes. Imaris Software (Bitplane Software) managed digital images and reconstructed z-series stacks. Final image presentation was produced with Photoshop (Adobe Software).
Statistical analysis
A non-paired students t-test evaluated differences in pair-wise comparisons versus control phage distribution. NeutrAvidin tissue biodistribution was analyzed by ANOVA and a Dunnett’s multiple comparison post-hoc analysis was implemented when significant differences occurred. For all analyses, statistical significance was accepted at a p value ≤ 0.05.
Supplementary Material
ACKNOWLEDGMENT
We would like to acknowledge the contributions of Lynn and Mike Garvey to the Institute for Stem Cell and Regenerative Medicine (ISCRM) Imaging Core at the University of Washington. A special thank you to Dr. K. Woodrow (University of Washington, Department of Bioengineering) for continued use of her Tecan infinite 200Pro plate reader, and Dr. C. Giachelli (University of Washington, Department of Bioengineering) for access to a Leica C1850 cryostat.
Funding Sources
This work was supported by NIH R21 NS099654, NIH R01 NS064404 and NSF CBET 0448547.
Footnotes
SUPPORTING CONTENT
Figures highlighting the bacteriophage titers throughout each round of the in vivo screen, and a list of the 21 TACL-phage sequence qualifiers from NGS enrichment data can be found in supplementary figures. Similarly, supplementary tables containing list of stability.jar derived peptide fragments (for serum stability degradation product prediction) and the results of a BLASTp search for proteins with TACL08 sequences homology can be found in the Supporting Information available online.
REFERENCES
- (1).Bodor N; Buchwald P Barriers to Remember: Brain-Targeting Chemical Delivery Systems and Alzheimer’s Disease. Drug Discov. Today 2002, 7, 766–774. [DOI] [PubMed] [Google Scholar]
- (2).Barchet TM; Amiji MM Challenges and Opportunities in CNS Delivery of Therapeutics for Neurodegenerative Diseases. Expert Opin. Drug Delivery 2009, 6, 211–225. [DOI] [PubMed] [Google Scholar]
- (3).Elliott Donaghue I; Tam R; Sefton MV; Shoichet MS Cell and Biomolecule Delivery for Tissue Repair and Regeneration in the Central Nervous System. J. Controlled Release 2014, 190, 219–227. [DOI] [PubMed] [Google Scholar]
- (4).Dong X Current Strategies for Brain Drug Delivery. Theranostics 2018, 8, 1481–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Lochhead JJ; Thorne RG Intranasal Delivery of Biologics to the Central Nervous System. Adv. Drug Delivery Rev 2012, 64, 614–628. [DOI] [PubMed] [Google Scholar]
- (6).Bodor N; Prokai L; Wu WM; Farag H; Jonalagadda S; Kawamura M; Simpkins J A Strategy for Delivering Peptides Into the Central Nervous System by Sequential Metabolism. Science 1992, 257, 1698–1700. [DOI] [PubMed] [Google Scholar]
- (7).Choi JJ; Wang S; Tung Y-S; Morrison B; Konofagou EE Molecules of Various Pharmacologically-Relevant Sizes Can Cross the Ultrasound-Induced Blood-Brain Barrier Opening in Vivo. Ultrasound Med. Biol 2010, 36, 58–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Choi JJ; Pernot M; Small SA; Konofagou EE Noninvasive, Transcranial and Localized Opening of the Blood-Brain Barrier Using Focused Ultrasound in Mice. Ultrasound Med. Biol 2007, 33, 95–104. [DOI] [PubMed] [Google Scholar]
- (9).Burgess A; Hynynen K Noninvasive and Targeted Drug Delivery to the Brain Using Focused Ultrasound. ACS Chem. Neurosci 2013, 4, 519–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Aryal M; Arvanitis CD; Alexander PM; McDannold N Ultrasound-Mediated Blood–Brain Barrier Disruption for Targeted Drug Delivery in the Central Nervous System. Adv. Drug Delivery Rev 2014, 72, 94–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Mesiwala AH; Farrell L; Wenzel HJ; Silbergeld DL; Crum LA; Winn HR; Mourad PD High-Intensity Focused Ultrasound Selectively Disrupts the Blood-Brain Barrier in Vivo. Ultrasound Med. Biol 2002, 28, 389–400. [DOI] [PubMed] [Google Scholar]
- (12).Tan J-KY; Pham B; Zong Y; Perez C; Maris DO; Hemphill A; Miao CH; Matula TJ; Mourad PD; Wei H; Sellers DL; Horner PH; Pun SH Microbubbles and Ultrasound Increase Intraventricular Polyplex Gene Transfer to the Brain. J. Controlled Release 2016, 231, 86–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Pardridge WM Targeted Delivery of Protein and Gene Medicines Through the Blood-Brain Barrier. Clin. Pharmacol. Ther 2014, 97, 347–361. [DOI] [PubMed] [Google Scholar]
- (14).Pardridge WM Drug Transport Across the Blood-Brain Barrier. J. Cereb. Blood Flow Metab 2012, 32, 1959–1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Spencer B; Marr RA; Gindi R; Potkar R; Michael S; Adame A; Rockenstein E; Verma IM; Masliah E Peripheral Delivery of a CNS Targeted, Metalo-Protease Reduces Aβ Toxicity in a Mouse Model of Alzheimer’s Disease. PLoS ONE 2011, 6, e16575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Spencer BJ; Verma IM Targeted Delivery of Proteins Across the Blood-Brain Barrier. Proc. Natl. Acad. Sci. USA 2007, 104, 7594–7599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Zhou QH; Boado RJ; Hui EKW; Lu JZ; Pardridge WM Chronic Dosing of Mice with a Transferrin Receptor Monoclonal Antibody-Glial-Derived Neurotrophic Factor Fusion Protein. Drug Metab. Dispos 2011, 39, 1149–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Yu YJ; Zhang Y; Kenrick M; Hoyte K; Luk W; Lu Y; Atwal J; Elliott JM; Prabhu S; Watts RJ; Dennis MS Boosting Brain Uptake of a Therapeutic Antibody by Reducing Its Affinity for a Transcytosis Target. Sci. Transl. Med 2011, 3, 84ra44. [DOI] [PubMed] [Google Scholar]
- (19).Yu YJ; Atwal JK; Zhang Y; Tong RK; Wildsmith KR; Tan C; Bien-Ly N; Hersom M; Maloney JA; Meilandt WJ; Bumbaca D; Gadkar K; Hoyte K; Luk W; Lu Y; Ernst JA; Scearce-Levie K; Couch JA; Dennis MS; Watts RJ Therapeutic Bispecific Antibodies Cross the Blood-Brain Barrier in Nonhuman Primates. Sci. Transl. Med 2014, 6, 261ra154. [DOI] [PubMed] [Google Scholar]
- (20).Zlokovic BV The Blood-Brain Barrier in Health and Chronic Neurodegenerative Disorders. Neuron 2008, 57, 178–201. [DOI] [PubMed] [Google Scholar]
- (21).Mäger I; Meyer AH; Li J; Lenter M; Hildebrandt T; Leparc G; Wood MJA Targeting Blood-Brain-Barrier Transcytosis – Perspectives for Drug Delivery. Neuropharmacology 2017, 120, 4–7. [DOI] [PubMed] [Google Scholar]
- (22).Kerman IA; Enquist LW; Watson SJ; Yates BJ Brainstem Substrates of Sympatho-Motor Circuitry Identified Using Trans-Synaptic Tracing with Pseudorabies Virus Recombinants. J. Neurosci 2003, 23, 4657–4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Card JP; Rinaman L; Schwaber JS; Miselis RR; Whealy ME; Robbins AK; Enquist LW Neurotropic Properties of Pseudorabies Virus: Uptake and Transneuronal Passage in the Rat Central Nervous System. J. Neurosci 1990, 10, 1974–1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Standish A; Enquist LW; Schwaber JS Innervation of the Heart and Its Central Medullary Origin Defined by Viral Tracing. Science 1994, 263, 232–234. [DOI] [PubMed] [Google Scholar]
- (25).Goodpasture EW; Teague O Transmission of the Virus of Herpes Febrilis Along Nerves in Experimentally Infected Rabbits. J. Med. Res 1923, 44, 139–184.7. [PMC free article] [PubMed] [Google Scholar]
- (26).Kristensson K Morphological Studies of the Neural Spread of Herpes Simplex Virus to the Central Nervous System. Acta Neuropathol 1970, 16, 54–63. [DOI] [PubMed] [Google Scholar]
- (27).Stoeckel K; Thoenen H Retrograde Axonal Transport of Nerve Growth Factor: Specificity and Biological Importance. Brain Res 1975, 85, 337–341. [DOI] [PubMed] [Google Scholar]
- (28).Davidson BL; Breakefield XO Viral Vectors for Gene Delivery to the Nervous System. Nat. Rev. Neurosci 2003, 4, 353–364. [DOI] [PubMed] [Google Scholar]
- (29).Azzouz M; Ralph GS; Storkebaum E; Walmsley LE; Mitrophanous KA; Kingsman SM; Carmeliet P; Mazarakis ND VEGF Delivery with Retrogradely Transported Lentivector Prolongs Survival in a Mouse ALS Model. Nature 2004, 429, 413–417. [DOI] [PubMed] [Google Scholar]
- (30).Kaspar BK; Lladó J; Sherkat N; Rothstein JD; Gage FH Retrograde Viral Delivery of IGF-1 Prolongs Survival in a Mouse ALS Model. Science 2003, 301, 839–842. [DOI] [PubMed] [Google Scholar]
- (31).Glorioso JC; Fink DJ Herpes Vector-Mediated Gene Transfer in the Treatment of Chronic Pain. Mol. Ther 2009, 17, 13–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Foust KD; Wang X; McGovern VL; Braun L; Bevan AK; Haidet AM; Le TT; Morales PR; Rich MM; Burghes AHM; Kaspar BK Rescue of the Spinal Muscular Atrophy Phenotype in a Mouse Model by Early Postnatal Delivery of SMN. Nat. Biotechnol 2010, 28, 271–274. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- (33).Boyce VS; Park J; Gage FH; Mendell LM Differential Effects of Brain-Derived Neurotrophic Factor and Neurotrophin-3 on Hindlimb Function in Paraplegic Rats. Eur. J. Neurosci 2011, 35, 221–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Fortun J; Puzis R; Pearse DD; Gage FH; Bunge MB Muscle Injection of AAV-NT3 Promotes Anatomical Reorganization of CST Axons and Improves Behavioral Outcome Following SCI. J. Neurotrauma 2009, 26, 941–953. [DOI] [PubMed] [Google Scholar]
- (35).Snyder BR; Gray SJ; Quach ET; Huang JW; Leung CH; Samulski RJ; Boulis NM; Federici T Comparison of Adeno-Associated Viral Vector Serotypes for Spinal Cord and Motor Neuron Gene Delivery. Hum. Gene Ther 2011, 22, 1129–1135. [DOI] [PubMed] [Google Scholar]
- (36).Koerber JT; Klimczak R; Jang J-H; Dalkara D; Flannery JG; Schaffer DV Molecular Evolution of Adeno-Associated Virus for Enhanced Glial Gene Delivery. Mol. Ther 2009, 17, 2088–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Bourdenx M; Dutheil N; Bezard E; Dehay B Systemic Gene Delivery to the Central Nervous System Using Adeno-Associated Virus. Front. Mol. Neurosci 2014, 7, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Tervo DGR; Hwang B-Y; Viswanathan S; Gaj T; Lavzin M; Ritola KD; Lindo S; Michael S; Kuleshova E; Ojala D; Huang C; erfen CR; Schiller J; Dudman JT; Hantman AW; Looger LL; Schaffer DV; Karpova AY A Designer AAV Variant Permits Efficient Retrograde Access to Projection Neurons. Neuron 2016, 92, 372–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Chan KY; Jang MJ; Yoo BB; Greenbaum A; Ravi N; Wu W-L; Sánchez-Guardado L; Lois C; Mazmanian SK; Deverman BE; Gradinaru V Engineered AAVs for Efficient Noninvasive Gene Delivery to the Central and Peripheral Nervous Systems. Nat. Neurosci 2017, 20, 1172–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Sellers DL; Bergen JM; Johnson RN; Back H; Ravits JM; Horner PJ; Pun SH Targeted Axonal Import (TAxI) Peptide Delivers Functional Proteins Into Spinal Cord Motor Neurons After Peripheral Administration. Proc. Natl. Acad. Sci. USA 2016, 113, 2514–2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Urich E; Schmucki R; Ruderisch N; Kitas E; Certa U; Jacobsen H; Schweitzer C; Bergadano A; Ebeling M; Loetscher H; Freskgård P-O. Cargo Delivery Into the Brain by in Vivo Identified Transport Peptides. Sci. Rep 2015, 5, 14104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Li J; Zhang Q; Pang Z; Wang Y; Liu Q; Guo L; Jiang X Identification of Peptide Sequences That Target to the Brain Using in Vivo Phage Display. Amino Acids 2011. [DOI] [PubMed] [Google Scholar]
- (43).Frenkel D; Solomon B Filamentous Phage as Vector-Mediated Antibody Delivery to the Brain. Proc. Natl. Acad. Sci. U S A 2002, 99, 5675–5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Pasqualini R; Ruoslahti E Organ Targeting in Vivo Using Phage Display Peptide Libraries. Nature 1996, 380, 364–366. [DOI] [PubMed] [Google Scholar]
- (45).Zou J; Dickerson MT; Owen NK; Landon LA; Deutscher SL Biodistribution of Filamentous Phage Peptide Libraries in Mice. Mol. Biol. Rep 2004, 31, 121–129. [DOI] [PubMed] [Google Scholar]
- (46).Molenaar TJM; Michon I; de Haas SAM; van Berkel TJC; Kuiper J; Biessen EAL Uptake and Processing of Modified Bacteriophage M13 in Mice: Implications for Phage Display. Virology 2002, 293, 182–191. [DOI] [PubMed] [Google Scholar]
- (47).Liu GW; Livesay BR; Kacherovsky NA; Cieslewicz M; Lutz E; Waalkes A; Jensen MC; Salipante SJ; Pun SH Efficient Identification of Murine M2 Macrophage Peptide Targeting Ligands by Phage Display and Next-Generation Sequencing. Bioconjug. Chem 2015, 26, 1811–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Bonaccorso RL; Chepurny OG; Becker-Pauly C; Holz GG; Doyle RP Enhanced Peptide Stability Against Protease Digestion Induced by Intrinsic Factor Binding of a Vitamin B 12Conjugate of Exendin-4. Mol. Pharm 2015, 12, 3502–3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Angelini A; Morales-Sanfrutos J; Diderich P; Chen S; Heinis C Bicyclization and Tethering to Albumin Yields Long-Acting Peptide Antagonists. J. Med. Chem 2012, 55, 10187–10197. [DOI] [PubMed] [Google Scholar]
- (50).Trüssel S; Dumelin C; Frey K; Villa A; Buller F; Neri D New Strategy for the Extension of the Serum Half-Life of Antibody Fragments. Bioconjug. Chem 2009, 20, 2286–2292. [DOI] [PubMed] [Google Scholar]
- (51).Weinstock MT; Francis JN; Redman JS; Kay MS Protease-Resistant Peptide Design—Empowering Nature’s Fragile Warriors Against HIV. Peptide Science 2012, 98, 431–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Vlieghe P; Lisowski V; Martinez J; Khrestchatisky M Synthetic Therapeutic Peptides: Science and Market. Drug Discov. Today 2010, 15, 40–56. [DOI] [PubMed] [Google Scholar]
- (53).Zhang X-X; Eden HS; Chen X Peptides in Cancer Nanomedicine: Drug Carriers, Targeting Ligands and Protease Substrates. J. Controlled Release 2012, 159, 2–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Aubry S; Burlina F; Dupont E; Delaroche D; Joliot A; Lavielle S; Chassaing G; Sagan S Cell-Surface Thiols Affect Cell Entry of Disulfide-Conjugated Peptides. FASEB J 2009, 23, 2956–2967. [DOI] [PubMed] [Google Scholar]
- (55).Chandrasekhar S; Topp EM Thiol–Disulfide Exchange in Peptides Derived From Human Growth Hormone During Lyophilization and Storage in the Solid State. J. Pharm. Sci 2015, 104, 1291–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Spokoyny AM; Zou Y; Ling JJ; Yu H; Lin Y-S; Pentelute BL A Perfluoroaryl-Cysteine SNAr Chemistry Approach to Unprotected Peptide Stapling. J. Am. Chem. Soc 2013, 135, 5946–5949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Pelay-Gimeno M; Glas A; Koch O; Grossmann TN Structure-Based Design of Inhibitors of Protein-Protein Interactions: Mimicking Peptide Binding Epitopes. Angew. Chem. Int. Ed 2015, 54, 8896–8927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Ngambenjawong C; Pineda JMB; Pun SH Engineering an Affinity-Enhanced Peptide Through Optimization of Cyclization Chemistry. Bioconjug. Chem 2016, 27, 2854–2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Greenfield N; Fasman GD Computed Circular Dichroism Spectra for the Evaluation of Protein Conformation. Biochemistry 1969, 8, 4108–4116. [DOI] [PubMed] [Google Scholar]
- (60).Roberts RMG Conformational Analysis of Biphenyls using13C NMR Spectroscopy. Magn. Reson. Chem 1985, 23, 52–54. [Google Scholar]
- (61).Ngambenjawong C; Gustafson HH; Pineda JM; Kacherovsky NA; Cieslewicz M; Pun SH Serum Stability and Affinity Optimization of an M2 Macrophage-Targeting Peptide (M2pep). Theranostics 2016, 6, 1403–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Dietz G; Valbuena PC; Dietz B; Meuer K; Müller P Application of a Blood–Brain-Barrier-Penetrating Form of GDNF in a Mouse Model for Parkinson’s Disease. Brain Res 2006, 1, 61–66. [DOI] [PubMed] [Google Scholar]
- (63).Han W; Tellez LA; Perkins MH; Perez IO; Qu T; Ferreira J; Ferreira TL; Quinn D; Liu Z-W; Gao X-B; Kaelberer MM; Bohorquez DV; Shammah-Lagnado SJ; de Lartigue G; de Araujo IE A Neural Circuit for Gut-Induced Reward. Cell 2018, 175, 665–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Thorne RG; Frey WH Delivery of Neurotrophic Factors to the Central Nervous System. Clin. Pharmacokinet 2001, 40, 907–946. [DOI] [PubMed] [Google Scholar]
- (65).Brenneman DE; Gozes I A Femtomolar-Acting Neuroprotective Peptide. The J. Clin. Invest 1996, 97, 2299–2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Gozes I; Giladi E; Pinhasov A; Bardea A; Brenneman DE Activity-Dependent Neurotrophic Factor: Intranasal Administration of Femtomolar-Acting Peptides Improve Performance in a Water Maze. J. Pharmacol. Exp. Ther 2000, 293, 1091–1098. [PubMed] [Google Scholar]
- (67).Huwyler J; Wu D; Pardridge WM Brain Drug Delivery of Small Molecules Using Immunoliposomes. Proc. Natl. Acad. Sci 1996, 93, 14164–14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Powley TL Central Control of Autonomic Functions: Organization of the Autonomic Nervous System Fundamental Neuroscience; Academic Press, 2013; pp. 729–747. [Google Scholar]
- (69).Bonaz B; Sinniger V; Pellissier S Anti-Inflammatory Properties of the Vagus Nerve: Potential Therapeutic Implications of Vagus Nerve Stimulation. J. Physiol 2016, 594, 5781–5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Shi J; Johnson RN; Schellinger JG; Carlson PM; Pun SH Reducible HPMA-Co-Oligolysine Copolymers for Nucleic Acid Delivery. Int. J. Pharm 2012, 427, 113–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Allen SJ, Watson JJ, Shoemark DK, Barua NU, Patel NK. GDNF, NGF and BDNF as therapeutic options for neurodegeneration. Pharmacol. Ther 2013, 38, 155–75. [DOI] [PubMed] [Google Scholar]
- (72).Luz M, Mohr E, Fibiger HC. GDNF-induced cerebellar toxicity: A brief review. Neurotoxicology 2016, 52, 46–56. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


