Abstract
Fumonisin toxins are produced by Fusarium fungal pathogens. Fumonisins are structural analogs of sphingosine and potent inhibitors of ceramide synthases (CerSs); they disrupt sphingolipid metabolism and cause disease in plants and animals. Over the past three decades, researchers have used fumonisin B1 (FB1), the most common fumonisin, as a probe to investigate sphingolipid metabolism in yeast and animals. Although the physiological effects of FB1 in plants have yet to be investigated in detail, forward and reverse genetic approaches have revealed many genes involved in these processes. In this review, we discuss the intricate network of signaling pathways affected by FB1, including changes in sphingolipid metabolism and the effects of these changes, with a focus on our current understanding of the multiple effects of FB1 on plant cell death and plant growth. We analyze the major findings that highlight the connections between sphingolipid metabolism and FB1-induced signaling, and we point out where additional research is needed to fill the gaps in our understanding of FB1-induced signaling pathways in plants.
Keywords: fumonisin B1, sphingolipid, long chain bases, ceramides, cell death, plant growth
Introduction
Fumonisins are produced by several species of Fusarium molds such as Fusarium verticillioides and Fusarium moniliforme, which infect many cereal crops such as maize (Zea mays), wheat (Triticum aestivum), and barley (Hordeum vulgare) (Rheeder et al., 2002). Fumonisins are required for the development of foliar disease symptoms in infected maize seedlings (Glenn et al., 2008). In the field, fumonisins are synthesized in planta by fungi and can be taken up by roots and disseminated inside seedlings, possibly affecting the fungus–host interaction prior to the first contact between the pathogen and seedling (Arias et al., 2016). Contaminated corn and corn-based products are frequently ingested in food or feed, causing diseases such as equine leukoencephalomalacia, porcine pulmonary edema, and possibly kidney and liver cancer (as revealed in rats in the laboratory) (Riley and Merrill, 2019).
In 1988, fumonisin B1 (FB1) and FB2 were isolated from F. verticillioides (F. moniliforme) MRC 826 cultures as novel mycotoxins with cancer-promoting activity in rat livers (Gelderblom et al., 1988). FB1 and FB2 are diesters of propane-1,2,3-tricarboxylic acid with similar long-chain aminopolyol backbones. These compounds are structurally similar to the sphingoid bases sphinganine and sphingosine, with tricarboxylic acid groups added at the C14 and C15 positions (Supplementary Figure S1). However, the chain lengths of their backbones differ, with sphingosine mostly containing 18 carbons and FB1 containing 20 carbons (Supplementary Figure S1). Three years after the chemical structures of fumonisins were elucidated, these carcinogenic mycotoxins were found to disrupt the pathway for de novo sphingolipid biosynthesis (Wang et al., 1991). Accumulating evidence has confirmed that the wide spectrum of animal and plant diseases caused by fumonisins are due to changes in the levels of multiple bioactive lipids and related biological processes. Although the mode of action of these toxins has been studied in detail at the cellular level in animals, their role in plant diseases has not been thoroughly elucidated.
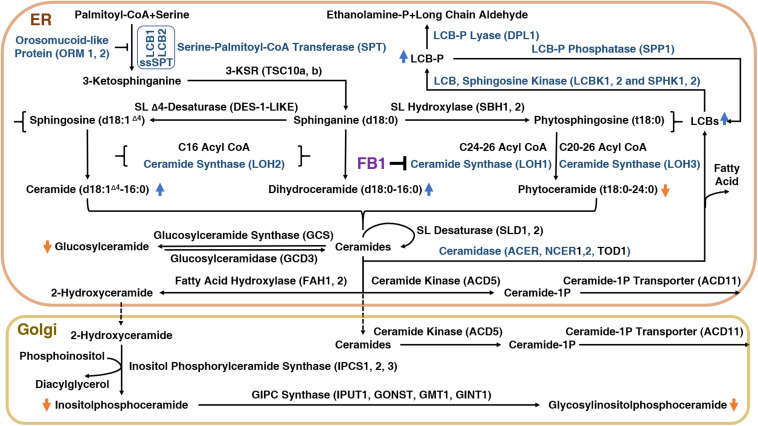
To explore how FB1 affects plants, it is important to understand the plant sphingolipid metabolism pathway (Figure 1). There are five major classes of plant sphingolipids: long-chain bases (LCBs), ceramides (Cers), hydroxyceramides (hCers), glucosylceramides (GluCers), and more complex glycosylated sphingolipids known as glycosyl inositol phosphoryl ceramides (GIPCs). The first four groups are produced in the endoplasmic reticulum (ER), whereas GIPC biosynthesis is initiated in the ER and completed in the Golgi apparatus (Figure 1). The serine palmitoyl-CoA transferase (SPT) protein complex catalyzes de novo biosynthesis of LCBs resulting from the condensation of serine and palmitoyl-CoA moieties (Kimberlin et al., 2013). The SPT product is subsequently reduced to form sphinganine, the precursor of the eight other LCBs found in plants.
FIGURE 1.
The sphingolipid biosynthetic pathway in plants and the inhibition of this pathway by FB1.
Ceramide synthases (CerSs) are encoded by a multigenic family named after the yeast protein Lag One Homolog (LOH) and are responsible for the formation of the amide bond that links long-chain (<C20) fatty acids (LCFs) or very-long-chain (≥C20) fatty acids (VLCFAs) to LCB, leading to Cer formation (Markham et al., 2011). Cers can then be phosphorylated to form ceramide 1-phosphate (ceramide-1P) (Liang et al., 2003), or α-hydroxylated at the fatty acid moiety to form hCers (Konig et al., 2012). Cers can also be used as backbones for the production of GluCer and GIPC by the addition of a glucose molecule or an inositolphosphoryl group, followed by one glycosylation step for GluCer or several glycosylations for GIPC (Markham et al., 2013).
Sphingolipid compounds have a multitude of functions. In addition to functioning as structural components of the plasma membrane and other endomembrane systems, sphingolipids serve as bioactive molecules and second messengers for an array of cellular signaling activities in plant cells, such development, biotic and abiotic stress responses, and programmed cell death (PCD). However, compared with animals, much less is known about sphingolipid metabolism and signaling in plants. For instance, the receptors, targets, and mediators involved in these processes are almost totally unclear (Ali et al., 2018), limiting our understanding of the FB1 signal transduction pathway in plants.
Schematic representation of the sphingolipid biosynthetic pathway in plants. 3-KSR, 3-ketosphinganine reductase; ACD11, accelerated cell death 11, Cer-1P transporter; ACD5, accelerated cell death 5, ceramide kinase; ACER, alkaline ceramidase; CoA, coenzyme A; d18:1Δ4, sphingosine; d18:0, sphinganine; DPL1, dihydrosphingosine phosphate lyase; FAH1/2, fatty acid hydroxylase; GCS, glucosylceramide synthase; GINT1, glucosamine inositolphosphorylceramide transferase 1; GMT1, GIPC mannosyl-transferase 1; GONST1, Golgi localized nucleotide sugar transporter 1; IPC, inositol phosphorylceramide; IPUT1, inositol phosphorylceramide glucuronosyltransferase 1; LCB, long-chain-base; LCB1/2, subunit of serine palmitoyltransferase 1/2; LCBK1/2, long-chain base kinase 1/2; LCB-P, long-chain-base phosphate; LOH1/2/3, LAG1 Homolog 1/2/3, ceramide synthase; NCER, neutral ceramidase; ORM1/2, orosomucoid-like protein 1/2; SBH1/2, sphingoid base hydroxylase 1/2; DPL1, dihydrosphingosine phosphate lyase; SL, sphingolipid; SPHK1/2, sphingosine kinase 1/2; SPP1, sphingoid phosphate phosphatase; SPT, serine palmitoyl transferase; ssSPT, small subunit of serine palmitoyl transferase; t18:0, phytosphingosine; TOD1, turgor regulation defect 1.
FB1 Disrupts Sphingolipid Homeostasis in Plants
Inhibition of Ceramide Synthase Activity
In mammals, FB1 inhibits the activity of the six known CerSs, which differ in terms of tissue distribution and fatty acyl-CoA specificity (Cingolani et al., 2016). Tomato (Solanum lycopersicum) Alternaria stem canker resistance-1 (Asc-1) was the first CerS identified in plants (Brandwagt et al., 2000; Spassieva et al., 2002). A tBLASTn search of the complete Arabidopsis thaliana genome using the sequence of the CerS gene longevity assurance gene 1 (LAG1) from yeast and Asc-1 as queries identified three Asc-1/LAG1 Homologs, LOH1–3 (Markham et al., 2011). Phylogenetic analysis revealed that LOH1 and LOH3 are closely related and belong to a cluster with most other plant LAG1 homologs, but LOH2 appears to be evolutionarily distinct from LOH1 and LOH3 (Markham et al., 2011; Ternes et al., 2011).
The varied chemical compositions of Cers in Arabidopsis are determined by the specificity of three different isoforms of CerS for a range of LCBs and acyl-CoA substrates. LOH1 and LOH3 prefer very-long chain acyl-CoA and trihydroxy LCB as substrates, but LOH2 prefers palmitoyl-CoA and dihydroxy LCB as substrates (Markham et al., 2011; Ternes et al., 2011; Luttgeharm et al., 2015, 2016). The differences between the two groups of CerSs are reflected by the phylogenetic separation of the LOHs, which preceded the separation of mosses and vascular plants (Ternes et al., 2011). Consistent with their redundant biochemical functions, LOH1 and LOH3 share high sequence similarity and a more recent evolutionary origin (Ternes et al., 2011). Nevertheless, when the physiological functions of these CerSs were examined by mutant analysis, only loh1 plants showed spontaneous cell death, perhaps due to the higher expression level of LOH1 and the stronger contribution of LOH1 to Cer biosynthesis compared with LOH3 (Ternes et al., 2011). In vitro ceramide synthase assays showed that LOH1 was the most sensitive to inhibition by FB1 (Luttgeharm et al., 2016). Furthermore, overexpressing LOH1 had no effect on FB1 resistance in Arabidopsis, whereas overexpressing LOH2 or LOH3 resulted in increased plant resistance to FB1 treatment, confirming the notion that LOH1 confers FB1 sensitivity to plants (Luttgeharm et al., 2015). In line with these observations, LOH2- and LOH3-overexpressing plants had reduced levels of free LCBs and LCB-Ps in response to FB1 treatment (Luttgeharm et al., 2015).
It remains unknown why FB1 preferentially inhibits LOH1 rather than LOH3, which share similar properties, but have slightly different substrate binding pockets. Supporting the hypothesis that the difference in substrate binding pockets affects their inhibition by FB1, LOH1 prefers FB1-like saturated trihydroxy LCBs, but LOH3 prefers unsaturated trihydroxy LCB (Luttgeharm et al., 2016). It is unclear if LOH1 and LOH3 in other plant species show parallel differences in substrate preference. Notably, in tomato, the presence of Asc-1 relieves blocked sphingolipid synthesis caused by AAL toxin (a tomato-specific toxin produced by the fungus Alternaria alternata f.sp. lycopersici) (Supplementary Figure S1), which would otherwise lead to PCD (Spassieva et al., 2002), pointing to the presence of at least one more ceramide synthesis enzyme besides Asc-1. Notably, maize (Z. mays) likely contains at least two ceramide synthase isoforms with different substrate specificities, which may result in susceptibility/resistance to F. verticillioides (Arias et al., 2016).
FB1 Treatment Increases LCB and C16 Sphingolipid Levels and Reduces VLC Sphingolipid Levels
When applied to plants, FB1 triggers the accumulation of LCBs and LCB-Ps (Shi et al., 2007; Tsegaye et al., 2007; Saucedo-García et al., 2011b; Yanagawa et al., 2017; Figure 1). Indeed, 1 h of FB1 treatment altered the levels of LCBs in Arabidopsis (Shi et al., 2007). Interestingly, like treatment with FB1, infection with Pseudomonas syringae pv. tomato DC3000 also provoked a rapid increase in t18:0 (phytosphinganine) levels, implying that LCBs function in pathogen resistance (Peer et al., 2010).
Fumonisin B1 suppresses the activity of all six known mammalian CerSs and therefore, LC and VLC sphingolipid levels decreased in response to FB1 treatment. Moreover, FB1 treatment resulted in the specific inhibition of C16:0 Cer biosynthesis and cured adipose triglyceride lipase macrophages from mitochondrial dysfunction and PCD (Aflaki et al., 2012), suggesting that FB1 counters C16 ceramide-mediated cell death. However, the situation is quite different in plants, since FB1 significantly increases C16 ceramide levels in plants. FB1 treatment or depletion of LOH1 and LOH3 resulted in a reduction in VLCF Cer levels (Markham et al., 2011). The levels of Cers and hCer with very long fatty acid chains decreased slightly after 24 h of FB1 treatment (Markham et al., 2011). Significant increases in the levels of 16:0-containing Cer and GluCer were observed in Arabidopsis loh1 null mutants (Ternes et al., 2011). Furthermore, a stronger depletion of VLC sphingolipids and enhanced accumulation of 16:0 sphingolipids were detected in FB1-treated loh1-1 loh3-1 double mutants (Markham et al., 2011).
LCBs Primarily Contribute to the Cytotoxicity of FB1 in Plants
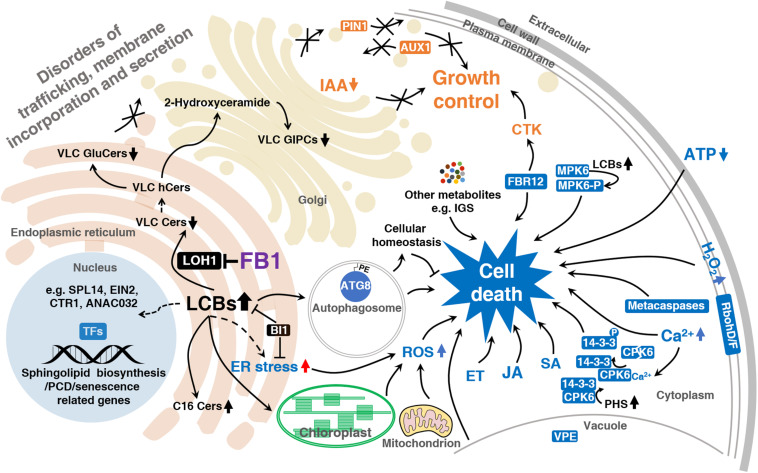
Long-chain bases serve as building blocks of sphingolipids and act as second messengers. These compounds are primarily responsible for FB1-induced cell death (Figure 2). The regulation of LCB homeostasis is as essential in plants as it is in animal models. Although the exact molecular mechanism underlying how LCBs control cell fate in plants is far from clear, it is assumed that LCB accumulation would kill plant cells, as it does in animals. The observation that FB1 treatment leads to LCB accumulation and cell death could be tentatively explained by the disequilibrium of the tightly regulated intracellular balance between free LCBs and Cers.
FIGURE 2.
Model summarizing how FB1 impairs and kills plant cells.
Early insight into the relationship between FB1 and the sphingolipid pathway in plants came from genetic studies focused on identifying Arabidopsis FB1-resistant mutants. FB1-resistant 11 was characterized as a deletion mutant in the gene encoding a long-chain base1 (LCB1) subunit of SPT. SPT mediates FB1-initiated PCD by catalyzing the first rate-limiting step of de novo sphingolipid biosynthesis (Shi et al., 2007; Saucedo-García et al., 2011b; Kimberlin et al., 2013, 2016; Li et al., 2016; Shao et al., 2019; Table 1). For example, increasing SPT activity by overexpressing small subunit of SPT (ssSPTa) resulted in the accumulation of LCBs and reduced tolerance to FB1, whereas ssSPTa suppression lines displayed lower levels of LCBs but enhanced tolerance to FB1 (Kimberlin et al., 2013). Notably, among all SPTs, only fumonisin b1 resistant 41 (FBR41) represses FB1-elicited cell death when overexpressed, consistent with its negative effect on SPT activity.
TABLE 1.
Sphingolipid metabolism genes involved in plant responses to FB1.
| Gene name | Function | Species | Mutant/transgenic plants | FB1 response | System | References |
| LCB1/FBR11 | Subunit of serine palmitoyltransferase | At | fbr11-1 | Tolerant | Leaves, seedlings | Shi et al., 2007 |
| LCB2a | Subunit of serine palmitoyltransferase | At | lcb2a-1, -2, -3 | Tolerant | Seedlings | Saucedo-García et al., 2011a,b |
| LCB2b/ | Subunit of serine | At | dominant mutant fbr41 | Tolerant | Leaves, seedlings | Shao et al., 2019 |
| FBR41 | palmitoyltransferase | At | FBR41 OE | Tolerant | Leaves | Shao et al., 2019 |
| ssSPTa | Small subunit of serine | At | ssSPTa RNAi | Tolerant | Seedlings | Kimberlin et al., 2013 |
| palmitoyltransferase | At | ssSPTa-OE | Sensitive | Seedlings | Kimberlin et al., 2013 | |
| ORM1 | Orosomucoid-like protein | At | ORM1-OE | Tolerant | Seedlings | Kimberlin et al., 2016; Li et al., 2016 |
| ORM2 | Orosomucoid-like protein | At | ORM2-OE | Tolerant | Seedlings | Kimberlin et al., 2016; Li et al., 2016 |
| ORM1/ | Orosomucoid-like protein | At | orm1 amiR-ORM2 | Sensitive | Seedlings | Li et al., 2016 |
| ORM2 | ORM RNAi | |||||
| LCBK1 | Long-chain base kinase | At | LCBK1-KD | Sensitive | Leaves, seedlings | Yanagawa et al., 2017 |
| At | LCBK1-OE | Tolerant | Leaves, seedlings | Yanagawa et al., 2017 | ||
| SPHK1 | Sphingosine kinase 1 | At | SPHK1-KD, sphk1-1 | Tolerant | Leaves | Qin et al., 2017 |
| At | SPHK1-KD | Sensitive | Leaves | Glenz et al., 2019 | ||
| At | SPHK1 OE | Sensitive | Leaves | Qin et al., 2017; Glenz et al., 2019 | ||
| SPHK2 | Sphingosine kinase 2 | At | sphk2-1 | Tolerant | Leaves | Qin et al., 2017 |
| At | SPHK2 OE | Sensitive | Leaves | Qin et al., 2017 | ||
| SPP1 | Sphingoid phosphate | At | Spp1 | Tolerant | Leaves | Yanagawa et al., 2017 |
| phosphatase | At | SPPASE RNAi (SPHK1-OE) | Sensitive | Leaves | Qin et al., 2017 | |
| At | SPP1-OE | Sensitive | Leaves | Glenz et al., 2019 | ||
| DPL1 | Dihydrosphingosine-1-phosphate lyase | At | dpl 1-1, -2, DPL1 RNAi (SPHK1-OE) | Sensitive | Leaves | Tsegaye et al., 2007; Nakagawa et al., 2012; Qin et al., 2017; Yanagawa et al., 2017; Glenz et al., 2019 |
| LOH2 | Ceramide synthase | At | loh2-1, -2 | Sensitive | Leaves | Mase et al., 2013 |
| At | LOH2-OE | Tolerant | Seedlings | Luttgeharm et al., 2015 | ||
| LOH3 | Ceramide synthase | At | LOH3-OE | Tolerant | Seedlings | Luttgeharm et al., 2015 |
| ACER | Alkaline ceramidase | At | amiR-ACER-1, acer-1 | Sensitive | Seedlings, Protoplasts | Wu et al., 2015a |
| At | ACER-OX-1 | Tolerant | Seedlings, Protoplasts | Wu et al., 2015a | ||
| NCER | Neutral ceramidase | At | ncer2 | Sensitive | Seedlings | Zienkiewicz et al., 2019 |
| BI1 | Protein that interacts with sphingolipid-modifying enzymes | At | atbi1-l, atbi1-2 | Sensitive | Leaves | Watanabe and Lam, 2006 |
| PAS1 | PASTICCINO1, required for VLCF synthesis | At | Pas1 | Sensitive | Roots | Markham et al., 2011 |
KD, knockdown; OE or OX, overexpressing line. At, Arabidopsis thaliana.
Orosomucoid proteins (ORMs) negatively regulate SPT activity to maintain sphingolipid levels in humans and yeast, and are required for sphingolipid homeostasis in plants, as well as FB1 resistance (Kimberlin et al., 2016; Li et al., 2016). The tolerance of Arabidopsis ORM overexpression plants to FB1 was accompanied by reduced levels of C16 Cers, LCBs, and their phosphorylated counterparts (Kimberlin et al., 2016). By contrast, AtORM RNA interference lines were more sensitive to FB1 treatment and contained more C16 Cers, LCBs, and LCB-Ps than the wild type (Kimberlin et al., 2016).
In addition to SPT, many enzymes related to LCB metabolism control plant responses to FB1. Overexpressing AtLCBK1 (A. thaliana sphingoid LCB kinase) increased plant resistance to FB1 treatment, whereas AtLCBK1 knockdown plants showed increased sensitivity to FB1 treatment (Yanagawa et al., 2017). Moreover, analysis of transgenic plants with altered levels of proteins including long-chain base kinase 1 (LCBK1), sphingoid phosphate phosphatase (SPP1), and dihydrosphingosine phosphate lyase (DPL1), which are involved in maintaining LCB/LCB-P homeostasis, revealed a positive correlation between the levels of free LCBs and the degree of FB1-induced cell death (Yanagawa et al., 2017). FB1-induced PCD is primarily due to the accumulation of free LCBs. Directly feeding plants with the free sphingoid bases dihydrosphingosine, phytosphingosine, and sphingosine efficiently induced cell death (Shi et al., 2007). Overexpressing LCBK1 in Arabidopsis led to hyposensitivity to FB1, whereas knockdown of LCBK1 conferred hypersensitivity to FB1 (Yanagawa et al., 2017).
Fumonisin B1-elicited cell death is mainly ascribed to the accumulation of dihydroxy-LCBs, but not trihydroxy-LCBs (Saucedo-García et al., 2011b). For instance, the sbh1-1 mutant, which is deficient in one of the two LCB hydroxylase genes (SBH1), contains lower levels of trihydroxy-LCBs and higher levels of total LCBs than wild-type plants (Chen et al., 2008). However, these plants appeared similar to wild-type plants when exposed to FB1 (Saucedo-García et al., 2011b).
Phosphorylated LCB derivatives appear to block cell death induced by the corresponding LCB in a dose-dependent manner, implying that the maintenance of homeostasis between a free sphingoid base and its phosphorylated derivative determines cell fate (Shi et al., 2007). The application of S1P or inhibitors of sphingosine kinase reduced apoptotic-like PCD and promoted cell survival in Arabidopsis cell suspension cultures (Alden et al., 2011). However, the antagonistic effects of phosphorylated LCBs after simultaneous treatment with non-phosphorylated LCBs were due to the reduced uptake of non-phosphorylated LCBs into the tissue (Glenz et al., 2019). Increasing the in vivo levels of phosphorylated LCBs did not reduce FB1-induced cell death (Glenz et al., 2019). Analysis of FB1-treated Arabidopsis lines with perturbed levels of phosphorylated LCBs highlighted a positive correlation between non-phosphorylated LCB levels and FB1-elicited cell death, rather than a negative correlation between phosphorylated LCB levels and this process (Glenz et al., 2019).
FB1 as a Tool to Investigate Sphingolipid Metabolism
Deciphering the role of a putative LCB/Cer metabolic enzyme in FB1 resistance is an excellent first step in confirming its function (Table 1). Ceramidases such as ACER maintain the homeostasis between LCB and Cer. ACER1-silenced plants and acer1 mutant plants were sensitive to FB1, and ACER1-overexpressing plants were relatively resistant to FB1 (Wu et al., 2015a). Disrupting neutral ceramidase 2 (NCER2), which likely functions as a ceramide synthase, increased sensitivity to FB1 in Arabidopsis (Zienkiewicz et al., 2019). Bax Inhibitor-1 (BI-1), an evolutionarily conserved cell death suppressor in mammals and plants (Kawai-Yamada et al., 2001; Mosblech et al., 2009; Christensen and Kolomiets, 2011), attenuates cell death progression triggered by FB1 in plants (Watanabe and Lam, 2006). Knockout mutants of AtBI1 (atbi1-1 and atbi1-2) exhibited increased sensitivity to FB1, whereas these phenotypes were rescued by the overexpression of AtBI1 (Watanabe and Lam, 2006). Similar to its function in mammals (Chae et al., 2004; Bailly-Maitre et al., 2006), BI-1 helps protect plant cells from ER stress-induced apoptosis (Gao et al., 2007, 2009). The ER membrane-localized protein BI-1 modifies sphingolipids by interacting with FATTY ACID HYDROXYLASE 1 (FAH1) and FAH2 via the electron donor cytochrome b5 (Cb5) in plant cells. BI-1 contributes to sphingolipid biosynthesis during cold stress in Arabidopsis by interacting with ceramide-modifying enzymes (Nagano et al., 2014). Moreover, BI-1 regulates VLCF synthesis by forming a complex with VLCF-synthesizing enzymes (Nagano et al., 2019).
FB1 Induces Cell Death in Plants
Fumonisin B1 induces cell death in a process involving multiple signaling pathways (Table 2 and Figure 2).
TABLE 2.
Genes in various signaling pathways involved in plant responses to FB1.
| Gene name | Function | Species | Mutant/transgenic plants | FB1 response | System | References |
| MPK6 | SA, ET | At | mpk6 | Tolerant | Seedlings | Saucedo-García et al., 2011b |
| ACD2 | SA | At | acd2-2 | Sensitive | Protoplasts | Asai et al., 2000 |
| CPR1 | SA | At | cpr1-1 | Sensitive | Protoplasts | Asai et al., 2000 |
| CPR6 | SA | At | cpr6-l | Sensitive | Protoplasts | Asai et al., 2000 |
| PAD4 | SA | At | pad4-l | Tolerant | Protoplasts | Asai et al., 2000 |
| ETR1 | ET | At | etr1-1 | Tolerant | Protoplasts | Asai et al., 2000 |
| At | etr1-1 | Tolerant | Leaves | Mase et al., 2013 | ||
| At | etr1-1 | Sensitive | Leaves | Plett et al., 2009 | ||
| At | etr1-1 | Sensitive | Seedlings | Wu et al., 2015b | ||
| EIN2 | ET | At | ein2 | Sensitive | Leaves | Wu et al., 2015b |
| EIN3 | ET | At | ein3 | Tolerant | Leaves | Mase et al., 2013 |
| EIN4 | ET | At | ein4-1 | Tolerant | Leaves | Plett et al., 2009 |
| ETO1 | ET | At | eto1-1 | Tolerant | Seedlings | Wu et al., 2015b |
| CTR1 | ET | At | ctr1-1 | Tolerant | Seedlings | Wu et al., 2015b |
| ERF4/MACD1 | ET | Nu | TRV:NuERF4 | Tolerant | Leaves | Mase et al., 2013 |
| Nu | MACD1 OE | Sensitive | Leaves | Mase et al., 2013 | ||
| ERF102 | ET | At | erf 102, | Tolerant | Leaves | Mase et al., 2013 |
| erf 102 (loh2) | Tolerant | Leaves | Mase et al., 2013 | |||
| ERF103 | ET | At | erf 103 (erf 102) | Tolerant | Leaves | Mase et al., 2013 |
| JAR1 | JA | At | jar1-1 | Tolerant | Protoplasts | Asai et al., 2000 |
| RGLG3 | JA | At | rglg3 | Tolerant | Leaves | Zhang et al., 2015 |
| At | RGLG3OE | Sensitive | Leaves | Zhang et al., 2015 | ||
| RGLG4 | JA | At | rglg4 | Tolerant | Leaves | Zhang et al., 2015 |
| At | RGLG4OE | Sensitive | Leaves | Zhang et al., 2015 | ||
| RGLG3/4 | JA | At | rglg3 rglg4 | Tolerant | Leaves | Zhang et al., 2015 |
| COI1 | JA | At | coi1-2 | Tolerant | Leaves | Zhang et al., 2015 |
| MYC2 | JA | At | myc2-2 | Sensitive | Leaves | Zhang et al., 2015 |
| CPK3 | Ca2+ | At | cpk3-1, cpk3-2 | Tolerant | Leaves | Lachaud et al., 2013 |
| VPE | Vacuole | At | VPE-null mutant | Tolerant | Leaves | Kuroyanagi et al., 2005 |
| ATG5 | Autophagy | At | atg5 | Sensitive | Leaves | Lenz et al., 2011 |
| ATG10 | Autophagy | At | atg10 | Sensitive | Leaves | Lenz et al., 2011 |
| ATG18 | Autophagy | At | atg18 | Sensitive | Leaves | Coll et al., 2014 |
| Atg18-1 | Sensitive | Leaves | Lenz et al., 2011 | |||
| ATG18 and MC1 | Autophagy and Metacaspases | atg18 mc1 | Sensitive | Leaves | Coll et al., 2014 | |
| MC1 | Metacaspases | At | Mc1 | Sensitive | Leaves | Coll et al., 2014 |
| MC4/MCP2d | Metacaspases | At | mcp2d-1, mcp2d-3 | Tolerant | Seedlings | Watanabe and Lam, 2011 |
| At | AtMCP2d OE | Sensitive | Leaves | Watanabe and Lam, 2011 | ||
| KTI1 | Kunitz Trypsin Inhibitor | At | kti1 RNAi | Sensitive | Leaves | Li et al., 2008 |
| At | KTI1 OE | Tolerant | Leaves | Li et al., 2008 | ||
| RD21 | Papain-like cysteine protease | At | rd21-1, -4 | Sensitive | Root | Ormancey et al., 2019 |
| LSD1 | Zinc finger protein | Os | OsLSD1 | Tolerant | Tobacco Seedlings | Wang et al., 2005 |
| SPL14/FBR6 | Transcription | At | fbr6 | Tolerant | Seedlings | Stone et al., 2005 |
| ANAC032 | Transcription, senescence | At | ANAC032 SRDX lines | Tolerant | Seedlings | Mahmood et al., 2016 |
| CYP79B2/CYP79B3 | Glucosinolates | At | cyp79B2 cyp79B3 | Sensitive | Leaves | Zhao et al., 2015 |
| TGG1 TGG2 | Glucosinolates | At | tgg1 tgg2 | Sensitive | Leaves | Zhao et al., 2015 |
| UGP1 | UDP-glucose pyrophosphorylase | At | SALK_020808 SALK_100183 | Tolerant | Leaves | Chivasa et al., 2013 |
| ATP SYNTHASE β- SUBUNIT | ATP synthase | At | SALK_024990 | Tolerant | Leaves | Chivasa et al., 2011 |
| SALK_135351 SALK_005252 | ||||||
| CYCLASE1 | Cyclase-family protein, SA, ATP | At | cyclase1-1, -2 | Sensitive | Leaves | Smith et al., 2015 |
| RING1 | RING motif protein | At | amiR-R1159 #1-16, #39-1 | Tolerant | Seedlings | Lin et al., 2008 |
| At | RING1-3HA #2-26, #4-1 | Sensitive | Seedlings | Lin et al., 2008 | ||
| DAL1 | Ring finger protein | dal1-1, -2 | Sensitive | Leaves | Basnayake et al., 2011 | |
| DAL2 | Ring finger protein | dal2-1, -2 | Sensitive | Leaves | Basnayake et al., 2011 | |
| BI1 | ER stress | At | atbi1-1/atbi1-2 | Sensitive | Leaves | Watanabe and Lam, 2006; Nagano et al., 2009 |
| RabG3b | GTPase, ATG | At | constitutively active RabG3b (RabG3bCA) and RabG3b OE sad2-2, -5 | Sensitive | Leaves | Kwon et al., 2009; Kwon et al., 2013 |
| SAD2 | Importin beta-like protein | At | Sensitive | Seedlings | Zheng et al., 2019 | |
| ILP | Inhibitor of apoptosis-like protein, ring finger protein | At | ILP | Tolerant | Seedlings | Kim et al., 2011 |
| ATR1 | Unknown | At | Atr1 | Tolerant | Seedlings | Gechev et al., 2008 |
KD, knockdown; OE or OX, overexpressing line. At, Arabidopsis thaliana.
Hormone Signaling
The link between ethylene and plant responses to FB1 is well known. Initially, it was reported that ethylene-dependent signaling pathways are required for FB1-induced cell death in Arabidopsis protoplasts (Asai et al., 2000). Arabidopsis ethylene receptors play distinct roles in FB1-induced cell death (Plett et al., 2009). Intriguingly, different reports describe different phenotypes of ethylene response 1-1 (etr1-1) mutants, possibly due to different growth conditions such as photoperiod (Asai et al., 2000; Plett et al., 2009; Mase et al., 2013; Wu et al., 2015b). By investigating the responses of various ethylene mutants to FB1 and the corresponding sphingolipid profiles of the plants, as well as the effects of 1-aminocyclopropane-1-carboxylic acid on FB1-induced cell death and sphingolipid metabolism, our group demonstrated that ethylene signaling inhibits sphingolipid synthesis, thereby playing a negative role in FB1-induced cell death (Wu et al., 2015b). Despite the finding that ethylene signaling modulates sphingolipid metabolism, the possibility that ethylene signaling acts downstream of sphingolipids (such LCBs and Cers) cannot be excluded. Notably, C24:1-Cer interacts with constitutive triple response 1 (CTR1) and inhibits its kinase activity, thereby modulating CTR1-mediated ethylene signaling (Xie et al., 2015), supporting this hypothesis.
The role of the plant hormone jasmonate (JA) in plant resistance to fungi is well known (Pieterse et al., 2009). The role of JA in mediating FB1-induced PCD was first described by Asai et al. (2000). The authors found that Arabidopsis protoplasts isolated from jasmonate resistant 1-1 (jar1-1) plants, which are insensitive to JA, exhibited much lower susceptibility to cell death induced by FB1 than FB1-treated wild-type protoplasts (Asai et al., 2000). Similarly, the deletion of oxophytodienoate reductase 3 (OPR3), which is involved in JA biosynthesis, enhanced the resistance of protoplasts to FB1 upon a dark/light shift (Danon et al., 2004). However, these findings may need to be further explored, since OPR3 is only partially required for JA biosynthesis and data obtained with the opr3 mutants are often unclear (Chini et al., 2018). FB1 hijacks the JA pathway to initiate PCD. FB1 induces JA-responsive defense genes but represses growth-related and JA biosynthesis-related genes, thereby reducing JA contents in plants (Zhang et al., 2015). Furthermore, the ubiquitin ligases ring domain ligase 3 (RGLG3) and RGLG4 coordinately and positively regulate FB1-triggered PCD by modulating the JA signaling pathway in a coronatine insensitive 1 (COI1)- and MYC2-dependent manner in Arabidopsis (Zhang et al., 2015).
In addition to ethylene and JA, plant susceptibility to FB1 appears to be associated with salicylic acid (SA) signaling, whereas FB1-induced PCD does not require the SA signal transmitter non-expresser of PR genes 1 (Asai et al., 2000). Indeed, SA promotes FB1-induced cell death (Smith et al., 2015). Moreover, FB1-induced SA likely suppresses the JA pathway to facilitate cell death, since exogenously applied SA inhibited JA signaling additively with FB1 treatment (Zhang et al., 2015).
Taken together, these findings indicate that FB1-elicited cell death requires ethylene, JA, and SA. Nevertheless, the molecular mechanisms underlying the roles of these hormone-signaling pathways in FB1-induced cell death remain to be investigated.
MPK6, Ca2+, Reactive Oxygen Species, and ATP Signaling
Mitogen-activated protein kinases (MAPKs) were activated in FB1-treated monkey kidney samples, but the functional role of MAPKs in FB1-induced toxicity has not been addressed in this system (Yin et al., 2016). In Arabidopsis, MAP kinase 6 (MPK6) was described as a novel transducer in the pathway leading to LCB-induced PCD (Saucedo-García et al., 2011b). In addition to mediating PCD downstream of LCBs, MPK6 can be activated by phytosphingosine−1−phosphate (PHS−P), a response rapidly and transiently evoked by chilling (Dutilleul et al., 2012). Nevertheless, how MPK6 triggers FB1-induced cell death downstream of PCD signaling remains unknown.
Calcium signaling is also required for FB1-induced cell death. Upon exposure to FB1, intracellular Ca2+ levels increase via FB1-induced phytosphingosine (PHS) (Lachaud et al., 2013). Meanwhile, PHS activates calcium-dependent kinase 3 (CPK3), which phosphorylates its associated partners, the 14-3-3 proteins, thus leading to the disruption of the 14-3-3-CPK3 complex and CPK3 degradation, ultimately triggering PCD in plants (Lachaud et al., 2013).
Reactive oxygen species (ROS) have key roles in PCD; in fact, cell death is considered to be triggered by ROS (Mittler, 2017) and ROS production precedes LCB-induced PCD in many cases (Shi et al., 2007; Saucedo-García et al., 2011a). The cytosolic increase in Ca2+ levels and the accumulation of ROS are critical for inducing PCD. The disruption of Arabidopsis super sensitive to aba and drought 2 (SAD2) (an importin beta-like gene) enhanced H2O2 accumulation and cell death in the sad2-5 mutant under FB1 treatment (Zheng et al., 2019). The FB1 sensitivity of sad2-5 is partially dependent on Ca2+ signaling (Zheng et al., 2019). SAD2-mediated Ca2+ and ROS signaling appear to function downstream of LCB, since the sad2-5 fbr11-1 double mutant exhibited the same FB1-insensitive phenotype as fbr11-1 (Zheng et al., 2019). In fact, many signaling pathways involved in regulating FB1-induced cell death are associated with ROS. For instance, the breakdown products of indole glucosinolate (IGS), which function in innate immune responses (Clay et al., 2009), attenuate FB1-induced cell death through their ROS-scavenging activity and function independently of indole-3-acetic acid signaling (Zhao et al., 2015).
Fumonisin B1 triggers the rapid depletion of extracellular ATP in Arabidopsis, which initiates cell death; this process was rescued by exogenous ATP treatment (Chivasa et al., 2005). ADP, AMP, and inorganic phosphate had no effect on this process, indicating that intact ATP is required for the rescue of FB1-induced cell death. In line with this notion, Arabidopsis knockout mutants lacking the gene encoding the mitochondrial ATP synthase β-subunit are resistant to FB1 (Chivasa et al., 2011). ATP and SA antagonistically regulate the cell death process (Smith et al., 2015). Further study based on this hypothesis revealed that CYCLASE1, an extracellular matrix proteins whose response to SA is suppressed by ATP, is a negative regulator of FB1-induced cell death (Smith et al., 2015).
Autophagy, Disruption of the Vacuolar Membrane, and ER Stress
Autophagy, a mechanism used to degrade unwanted constituents in eukaryotic cells, is essential for maintaining cell homeostasis and nutrient recycling. We recently demonstrated that FB1 and LCBs induce autophagy in Arabidopsis (Zheng et al., 2018). The loss of Arabidopsis ACER, an ACER that hydrolyzes Cer to LCB, inhibited autophagy, and its overexpression promoted autophagy under various abiotic stress conditions (Zheng et al., 2018).
In animals, the induction of autophagy has either cytoprotective or cytotoxic effects in certain types of mycotoxin-mediated cytotoxicity, but how the role (pro-survival or pro-death) of autophagy in regulating cell death is determined remains elusive (Yin et al., 2018). In plants, autophagy has also been assigned pro-death and pro-survival functions in controlling cell death (Zeng et al., 2019). Treatment with FB1 caused spreading lesions to form in several autophagy-related (atg) mutants (Lenz et al., 2011; Coll et al., 2014), implying that autophagy constitutes a pro-survival mechanism that contains unrestricted cell death in the presence of FB1. Consistent with this finding, autophagy participates in controlling plant lipid metabolism and catabolism (Have et al., 2019). Cer and GIPC levels are significantly altered in atg5 mutants in a SA-independent manner (Have et al., 2019). This observation suggests that autophagy modifies the composition of endomembrane lipids, especially plasma membrane lipids, thereby altering the outcome of FB1 treatment. Nevertheless, overexpressing or constitutively active Rab GTPase RabG3b, which contributes to cell death during the hypersensitive response (HR) in Arabidopsis by activating autophagy, led to unrestricted hypersensitive PCD in response to FB1 (Kwon et al., 2009, 2013), confirming another role of autophagy in FB1-associated cell death. In summary, autophagy functions in life and death of the cell through multiple pathways, but the underlying molecular mechanisms remain to be revealed.
The disruption of vacuolar membranes is one of the early events in FB1-induced cell death, followed by lesion formation (Kuroyanagi et al., 2005). The deletion of all four vacuolar processing enzyme (VPE) genes or treatment with VPE-specific inhibitors significantly suppressed FB1-induced cell death in Arabidopsis (Kuroyanagi et al., 2005). VPE exhibits caspase-1 activity and is required for the collapse of the tonoplast during FB1 treatment (Kuroyanagi et al., 2005). Treatment with caspase-1 inhibitors prevented the formation of FB1-induced lesions, indicating that caspase activity is required during FB1-induced cell death (Kuroyanagi et al., 2005). Notably, a deficiency of the retromer complex leads to defects in late endocytic/lytic compartments and impairs autophagy-associated vacuolar processes (Munch et al., 2015). vacuolar protein sorting 35, part of the retromer complex, functions in endosomal protein sorting and vacuolar trafficking and is involved in acd11-associated cell death (Munch et al., 2015). It would be interesting to investigate whether this process is required for FB1 resistance.
In animals, ER stress is responsible for autophagy and autophagic cell death induced by FB1 (Yin et al., 2016). Although we lack direct evidence in plants, the accumulation of LCB appears to increase ER stress-related responses. We frequently observed irregular ER and vacuolization in orm1 amiR-ORM2 cells, with many tiny membrane sacs located close to the plasma membrane and cell wall (Li et al., 2016). In line with this observation, the expression of ER stress marker genes significantly increased in orm1 amiR-ORM2 plants compared to the wild type (Li et al., 2016). Apart from mediating sphingolipid metabolism, AtBI1 plays a pivotal role as a highly conserved survival factor during ER stress that acts in parallel with the unfolded protein response pathway. Compared to wild-type plants, atbi1-1 and atbi1-2 exhibit hypersensitivity to tunicamycin, an inducer of ER stress, whereas overexpressing AtBI1 markedly reduced the sensitivity of Arabidopsis seedlings to tunicamycin (Watanabe and Lam, 2008). Whether the role of BI1 in the ER stress response is associated with its sphingolipid-modifying function remains to be investigated.
Other Participants in FB1-Induced PCD
Plant proteases, such as serine proteases and cysteine proteases, have been implicated in PCD (Rotari et al., 2005). The serine protease kunitz trypsin inhibitor 1 (KTI1) acts as a functional protease in Arabidopsis when produced in bacteria and in planta. RNAi silencing of KTI1 resulted in enhanced lesion development in plants after FB1 treatment (Li et al., 2008). Conversely, overexpressing AtKTI1 reduced lesion development in plants after FB1 treatment (Li et al., 2008). The cysteine protease metacaspase 2D (MCP2d)/MC4 also participates in FB1-induced PCD. The AtMCP2d mutants mcp2d-1 and mcp2d-3 show reduced sensitivity to FB1, whereas AtMCP2d overexpression lines are more sensitive to FB1 and display accelerated progression of cell death (Li et al., 2008). AtMCP2d exclusively localizes to the cytosol (Li et al., 2008). AtMCP2d is engaged in both oxidative stress-induced cell death and pathogen-induced cell death (Li et al., 2008). The type I metacaspase AtMC1 likely plays a positive role in the HR, methyl viologen-induced cell death, and FB1-induced cell death (Coll et al., 2014). Finally, the Arabidopsis papain-like cysteine protease responsive to desiccation 21 is a negative regulator of FB1-induced cell death (Ormancey et al., 2019). Together, these findings support the notion that the cell death triggered by FB1 requires general cell death factors, such as various proteases.
Membrane microdomains have also been implicated in PCD. RING1, a RING finger domain protein with E3 ligase activity, is localized to the lipid rafts of plasma membranes (Lin et al., 2008). Knockdown of RING1 led to FB1 hyposensitivity, whereas overexpressing RING1 conferred hypersensitivity to this treatment (Lin et al., 2008). RING1 acts as a signal from lipid rafts in the plasma membrane that trigger the FB1-induced plant PCD pathway (Lin et al., 2008). Transient expression of Capsicum annuum RING1 induced cell death in pepper leaves (Lee et al., 2011). It would be interesting to further investigate how lipid rafts in the plasma membrane respond to FB1. Suppressor of PPI1 locus 1 (SP1) regulates chloroplast biosynthesis, and both SP1 and SP1-like 1 (SPL1) regulate peroxisome biosynthesis in Arabidopsis (Ling et al., 2012, 2019; Pan et al., 2016, 2018). These proteins negatively regulate FB1-induced PCD as well (Basnayake et al., 2011). The involvement of these two regulators of organelle biogenesis in FB1-induced cell death points to a possible link between organelles and FB1 resistance.
Various proteins involved in FB1-induced cell death, such FBR6 and FBR12, also regulate plant growth and development. Misexpression of AtSPL14, encoding a plant-specific SQUAMOSA promoter (SBP)-domain putative transcription factor, conferred FB1 resistance in fbr6 plants (Stone et al., 2005). In addition to participating in FB1-associated cell death, AtSPL14 modifies normal plant development, as the fbr6 mutant displays altered plant architecture in the absence of FB1, most notably elongated petioles and enhanced leaf margin serration (Stone et al., 2005). These findings point to a possible association between insensitivity to FB1-induced cell death and altered plant development. Similar observations were made for Arabidopsis fbr12, a mutant deficient in eukaryotic translation initiation factor 5A. FBR12 positively regulates PCD caused by infection with virulent P. syringae pv. tomato DC3000 (Pst DC3000), and transgenic plants constitutively overexpressing FBR12 exhibited phenotypes consistent with precocious cell death (Hopkins et al., 2008).
FBR12 mediates processes beyond cell death, such as cell division and cell growth (Feng et al., 2007). The fbr12 mutant is extremely dwarf, with substantially reduced sizes and numbers of all adult organs, and shows abnormal floral organ development and defective sporogenesis, leading to the abortion of both female and male germline cells during reproductive development (Feng et al., 2007). FBR12 also functions in cytokinin-mediated specification of the root protoxylem, as a mutation in FBR12 led to defective protoxylem development and reduced sensitivity to cytokinin (CTK) (Ren et al., 2013). These findings point to a connection between PCD and inhibited development induced by FB1.
The solid arrows indicate established links and the dashed arrows indicate putative links. Reduced VLC sphingolipid levels lead to disordered trafficking, membrane incorporation, and secretion. LCBs produced in response to FB1 to trigger cell death signaling, such as ET, JA, SA, ROS, Ca2+, ATP signaling, and so on. LCBs regulate the activities of cell death-associated proteins such as MPK6, CPK6, and 14-3-3 proteins. LCBs also induce autophagosome formation, which maintains cellular homeostasis, protecting the cell from unnecessary death. ER stress appears to be involved in this process. LCBs induce the expression of sphingolipid biosynthesis, PCD, and senescence-related genes through transcription factors such as EIN2, CTR1, SPL14, and ANAC032. 14-3-3, 14-3-3 family protein; ANAC032, NAC domain containing protein 32; ATG8, autophagy 8; AUX1, auxin resistant 1; BI1, bax inhibitor 1; Cers, ceramides; CPK6, calcium dependent protein kinase 6; CTK, cytokinin; CTR1, constitutive triple response 1; EIN2, ethylene insensitive 2; ET, ethylene, ER stress, endoplasmic reticulum stress; FB1, fumonisin B1; FBR12, fumonisin B1 resistant 12; GluCers, glucosylceramides; GIPCs, glycosyl-inositolphosphoryl-ceramides; hCers, hydroxyceramides; IAA, indole-3-acetic acid; IGS, indole glucosinolate; JA, jasmonate; LCBs, long-chain bases; LOH1, LAG1 Homolog 1; MPK6, MAP kinase 6; MPK6-P, MPK6 phosphorylated form; PE, phosphatidylethanolamine; PHS, phytosphingosine; PIN1, PIN-FORMED 1; RbohD/F, respiratory burst oxidase homolog; ROS, reactive oxygen species; SA, salicylic acid; SPL14, squamosa promoter binding protein-like 14; TFs, transcription factors; VLC, very-long-chain; VPE, vacuolar processing enzyme.
FB1 Impairs Plant Growth and Affects Plant Development
Sphingolipids, particularly those containing VLCFAs, are essential for protein sorting and secretion in eukaryotes. The hydrophobicity, membrane-leaflet interdigitation, and transition of a membrane from a fluid to a gel phase, which are required for microdomain formation, are highly correlated with the presence of VLC sphingolipids (Markham et al., 2011). Several reports speculated that sphingolipids may represent the main reservoir of VLCFAs in leaves; since phosphatidylserine and phosphatidylethanolamine harbor VLCFAs as well in their backbone and no quantitative comparison with these lipid classes exist, this needs to be analyzed in more detail in the future (Pata et al., 2010; Maatta et al., 2012; Cacas et al., 2016; De Bigault Du Granrut and Cacas, 2016). Plants overexpressing LOH1 and LOH3 showed increased growth and cell division, with enhanced production of Cers with VLCFAs and trihydroxy LCBs (Luttgeharm et al., 2015). Interestingly, unlike LOH2, overexpressing LOH1/3 had only a minor effect on the sphingolipid profiles of plants.
Fumonisin B1-mediated reductions in VLCFA levels remodel the membrane structure and influence numerous processes that require the foundation maintained by VLC sphingolipids (Figure 2). Upon FB1 application, tobacco (Nicotiana tabacum) BY2 cells showed severe effects on cell growth and cell shape and a delay in cell division (Aubert et al., 2011). These changes were accompanied by the formation of ER-derived tubular aggregates, as well as inhibited ER-to-Golgi cargo transport (Aubert et al., 2011). A change in the polar localization of the auxin transporter PIN-FORMED 1 (PIN1) was also observed, but there was little effect on endocytic processes. FB1 targets molecules distinct from those targeted by Brefeldin A, an ER-to-Golgi transport inhibitor (Ritzenthaler et al., 2002; Langhans and Robinson, 2007), as revealed by electron microscopy. These findings reflect the importance of sphingolipids in cell growth and the establishment of cell polarity in plant cells, especially their contribution to the functional organization of the ER or its differentiation into distinct compartments (Aubert et al., 2011).
The reduction in VLC sphingolipid levels leads to auxin-dependent inhibition of lateral root emergence, which is correlated with the selective aggregation of the plasma membrane auxin carriers auxin resistant 1 (AUX1) and PIN1 in the cytosol (Markham et al., 2011). Defective targeting of polar auxin carriers is characterized by the specific aggregation of Rab-A2a- and Rab-A1e-labeled early endosomes along the secretory pathway (Markham et al., 2011). The formation of these aggregates is associated with the accumulation of membrane structures and vesicle fragmentation in the cytosol (Markham et al., 2011), indicating that sphingolipids with very long acyl chains define a trafficking pathway with specific endomembrane compartments and polar auxin transport protein cargoes (Markham et al., 2011). Finally, VLC sphingolipids, particularly VLC glycosphingolipids, are essential structural determinants of vesicle dynamics and membrane fusion during cytokinesis (Molino et al., 2014). Inhibiting VLC sphingolipids biosynthesis by FB1 led to the formation of defective cell plates with persistent vesicular structures and large gaps (Molino et al., 2014).
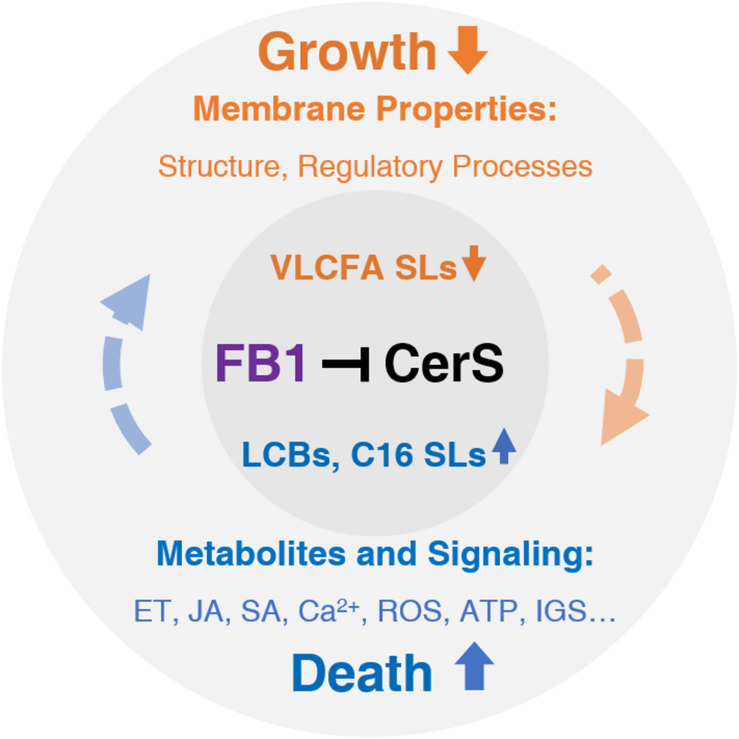
Route 1 (Orange): FB1-induced decreases in VLCF levels strongly affect membrane properties, which are responsible for normal plant growth. Route 2 (Blue): FB1-induced increases in LCB and C16 Cer levels primarily affect hormone, Ca2+, ROS, and ATP signaling, which ultimately contribute to cell death. However, this process is sufficiently complex such that interplay with other factors in each route and even across routes determines the final outcomes. Black, targeted by FB1 (purple), orange, inhibited or reduced by FB1; blue, promoted or elevated by FB1. CerS, ceramide synthase; ET, ethylene, FB1, fumonisin B1; IGS, indole glucosinolate; JA, jasmonate; LCBs, long-chain bases; ROS, reactive oxygen species; SA, salicylic acid; SLs, sphingolipids; VLCFA, very-long-chain fatty acid.
Maize–F. verticillioides Interaction
Fumonisin-producing pathogens are the major causal agents of fusarium ear rot, one of the most important diseases affecting maize production worldwide (Picot et al., 2010). FB1 is the predominant fumonisin in maize kernels (Bartók et al., 2006). The fungal genes required for fumonisin biosynthesis are organized in a cluster designated the fumonisin (FUM) gene cluster, which contains 22 genes with a length of 42 kb (Proctor et al., 1999,2003). This cluster may be regulated by multiple environmental factors including pH, water availability or nutrient sources, and various fungal genes to favor or restrain fumonisin biosynthesis (Picot et al., 2010). Among the 22 genes, 15 are co-regulated, including the key gene FUM1, which encodes a polyketide synthase (PKS) (Proctor et al., 1999).
In the complex interaction between F. verticillioides and maize, investigation of the role of fumonisin production on disease development has yielded controversial results. Only strains of F. verticillioides that produce fumonisins cause foliar disease symptoms on seedlings of the sweet maize hybrid “Silver Queen” (Williams et al., 2006; Lanubile et al., 2012a,b). However, the FUM1 deletion mutants, which do not produce fumonisin, still cause ear rot, implying that production of fumonisins is not required for ear rot (Jardine and Leslie, 1999; Desjardins et al., 2002). Lanubile et al. (2013) found that PKS is a relevant gene, essential not only for the fumonisin biosynthetic pathway, but also for pathogen colonization (Lanubile et al., 2013).
Sarocladium zeae, a fungal endophyte of maize, can co-inhabit a seed with F. verticillioides due to its capability to produce pyrrocidines A and B, which inhibit the growth of F. verticillioides (Gao et al., 2020). FvZBD1 (FVEG_00314) encodes a genetic repressor of fumonisin production, is directly adjacent to the FUM cluster, and was induced in response to pyrrocidines. This suggests that other microbes may manipulate the fumonisin biosynthetic program of F. verticillioides to control F. verticillioides growth in the host (Gao et al., 2020). Interestingly, a high dose of fumonisin induced necrosis and wilting in maize seedlings, but a low dose activated detoxification processes, indicating that maize has recovery mechanisms (Arias et al., 2012).
In maize, batteries of defense genes involved in pathogen-associated molecular pattern-triggered immunity and effector-triggered immunity, as well as multiple signaling, such as MAPK, Ca2+, ROS, hormones (including SA, auxin, abscisic acid, ethylene, JA) signaling are activated in response to F. verticillioides infection (Lanubile et al., 2017). A complex network of metabolic pathways is required for resistance to F. verticillioides (Lanubile et al., 2017). Lipoxygenase (LOX) genes encode non-heme iron-containing dioxygenases that catalyze the oxygenation of polyunsaturated fatty acids (Vick and Zimmerman, 1983), which are processed into an estimated 400 metabolites including JA (Mosblech et al., 2009). In plants, LOXs are involved in host susceptibility to fungal invasion and mycotoxin production (Kock et al., 2003; Christensen and Kolomiets, 2011; Maschietto et al., 2015). Depending on which carbon on the fatty acid chain is oxygenated, LOXs are classified into two main functional groups, 9-LOXs and 13-LOXs. It seems that a specific plant 9-LOX isoform is required for fungal pathogenesis, including disease development and production of spores and mycotoxins (Gao et al., 2007, 2009; Christensen et al., 2014). In addition, many factors contribute to resistance to F. verticillioides and fumonisin contamination, including the biochemical composition of the endosperm, and fatty acid composition, as well as many other metabolites, such phenylpropanoid pathway-related metabolites (flavonoids, phenolic compounds, and phytoalexins), flavones, and 2-amino-3H-phenoxazin-3-one (Lanubile et al., 2017).
Concluding Remarks and Outlook
Inhibition of ceramide synthesis by FB1 disturbs the homeostasis of sphingolipid metabolism, resulting in changes in structural and signaling sphingolipids, including elevated LCB and LCF Cer levels and decreased formation of Cers and complex VLC sphingolipids, accounting for the broad spectrum of plant disease symptoms. LCBs, as well as LCF Cers, contribute to cell death by functioning as signals, while Cers and complex VLC sphingolipids are critical components of the plant plasma membrane and endomembrane system that are closely associated with the fluidity and biophysical order of the membrane. Thus, the perturbations caused by FB1 not only induce cell death signals, but they also damage the structural components of the cell and impair plant growth. Small molecules and metabolites such as certain hormones (ethylene, JA, SA), ROS, NO, ATP, and Ca2+ likely affect FB1-triggered PCD, highlighting the strong association between sphingolipids and cell death. Nevertheless, the molecular details of the underlying mechanism are largely unclear.
In addition to being used as a ceramide inhibitor or a fungal toxicant, FB1 represents a highly useful tool for revealing details about sphingolipid-controlled cell death and growth inhibition. These details would benefit agriculture by helping us overcome the subtle strategy of FB1-producing pathogens and others foes sharing similar mechanisms in plant–pathogen interactions.
Based on the complicated signaling network induced by FB1, we propose a model to classify the global effects of this compound into basic two routes (Figure 3). However, it should be noted that most of these events interact with each other in a complex manner. For example, FB1 manipulates the JA signaling pathway through SA signaling (Zhang et al., 2015). In general, the activation of cell death signaling represses plant growth, which in turn sometimes leads to cell death.
FIGURE 3.
The effects of FB1 treatment.
Sphingolipids remain an underexplored, large group of metabolites (Ali et al., 2018). The study of FB1, the best-characterized inhibitor of sphingolipid biosynthesis, provides a fascinating illustration of the remarkably sophisticated sphingolipid-signaling network that is established in plants and animals. However, our understanding of the mechanisms underlying FB1 toxicity in plants is still in its infancy in terms of identifying the direct target(s) of FB1 and key elements in the responses to FB1.
Recent studies using molecular genetics and biochemical approaches have enhanced our understanding of this issue in plants and provided a deeper understanding of the roles of sphingolipids in plant life and death. Despite these important findings, our understanding of the importance of sphingolipids for the regulation of plant cell death and growth, and consequently crop yields remains limited and sphingolipids represent an exciting area for further study. Many efforts have been made to functionally characterize individual genes downstream of FB1. However, little is known about the FB1 signal transduction network. Therefore, apart from screening more participants involved in the FB1 response, studies should be designed to elaborate how the known participants operate simultaneously in a coordinated signaling network under FB1 treatment. In particular, it is crucial to investigate the connection between the regulators of FB1 resistance and sphingolipid metabolism. Specifically, sphingolipid profile analysis of FB1-resistant or -sensitive mutants with and without FB1 treatment should be performed to determine whether these proteins engage in sphingolipid metabolism. Furthermore, our lack of knowledge about the molecular targets of individual sphingolipid components, as well as their biological significance to plants, is still a major impediment to understanding FB1 signaling. Thus, there is a need for interactome association analysis between proteins downstream of FB1 and LCB, as well as sphingolipid enzymes, to untangle the interplay between these signals and sphingolipid metabolites or metabolism pathways. Recent studies have demonstrated the great potential of using the photoactivatable and clickable analog of sphingosine (pacSph) and azide-tagged sphingolipids to identify sphingolipid targets (Haberkant et al., 2016; Schulte-Zweckel et al., 2018). In addition to high-throughput screening of sphingolipid-interacting proteins, it will be important to gain deep insight into the roles of essential components of the HR, particularly enhanced disease susceptibility 1, activated disease resistance 1, and non-race-specific disease resistance 1, in FB1-triggered cell death, given that these HR signaling pathways are active in response to FB1.
Beside Arabidopsis, the FB1 signaling network should be elaborated in crop plants. Relatively little is known about the physiological effects of fumonisins from fungal pathogens in the natural environment on the development of plant diseases. The introduction and manipulation of parts of the FB1 signaling pathway in crop plants will have broad and overarching impacts on agriculture to help overcome the effects of FB1-producing pathogens. In view of the broad host range of FB1-producing pathogens, the eradication of plant diseases (as well as animal diseases) caused by FB1 appears to be elusive. However, better insight into the global effects of FB1 on plants will undoubtedly contribute to the development of a rational, effective approach to reducing the hazards of FB1 contamination.
Author Contributions
H-YZ and NY designed the manuscript and wrote the manuscript. C-YL and NY contributed to the reagents and materials. All authors have discussed and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by the National Natural Science Foundation of China (31771357) and the Natural Science Foundation of Guangdong Province (2019B1515120088 and 2017A030311005).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.600458/full#supplementary-material
References
- Aflaki E., Doddapattar P., Radovic B., Povoden S., Kolb D., Vujic N., et al. (2012). C16 ceramide is crucial for triacylglycerol-induced apoptosis in macrophages. Cell Death Dis. 3:e280. 10.1038/cddis.2012.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alden K. P., Dhondt-Cordelier S., McDonald K. L., Reape T. J., Ng C. K., McCabe P. F., et al. (2011). Sphingolipid long chain base phosphates can regulate apoptotic-like programmed cell death in plants. Biochem. Biophys. Res. Commun. 410 574–580. 10.1016/j.bbrc.2011.06.028 [DOI] [PubMed] [Google Scholar]
- Ali U., Li H., Wang X., Guo L. (2018). Emerging roles of sphingolipid signaling in plant response to biotic and abiotic stresses. Mol. Plant 11 1328–1343. 10.1016/j.molp.2018.10.001 [DOI] [PubMed] [Google Scholar]
- Arias S. L., Mary V. S., Otaiza S. N., Wunderlin D. A., Rubinstein H. R., Theumer M. G. (2016). Toxin distribution and sphingoid base imbalances in Fusarium verticillioides-infected and fumonisin B1-watered maize seedlings. Phytochemistry 125 54–64. 10.1016/j.phytochem.2016.02.006 [DOI] [PubMed] [Google Scholar]
- Arias S. L., Theumer M. G., Mary V. S., Rubinstein H. R. (2012). Fumonisins: probable role as effectors in the complex interaction of susceptible and resistant maize hybrids and Fusarium verticillioides. J. Agr. Food Chem. 60 5667–5675. 10.1021/jf3016333 [DOI] [PubMed] [Google Scholar]
- Asai T., Stone J. M., Heard J. E., Kovtun Y., Yorgey P., Sheen J., et al. (2000). Fumonisin B1–induced cell death in Arabidopsis protoplasts requires jasmonate-, ethylene-, and salicylate-dependent signaling pathways. Plant Cell 12 1823–1835. 10.2307/3871195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubert A., Marion J., Boulogne C., Bourge M., Abreu S., Bellec Y., et al. (2011). Sphingolipids involvement in plant endomembrane differentiation: the BY2 case. Plant J. 65 958–971. 10.1111/j.1365-313X.2011.04481.x [DOI] [PubMed] [Google Scholar]
- Bailly-Maitre B., Fondevila C., Kaldas F., Droin N., Luciano F., Ricci J. E., et al. (2006). Cytoprotective gene bi-1 is required for intrinsic protection from endoplasmic reticulum stress and ischemia-reperfusion injury. Proc. Natl. Acad. Sci. U.S.A. 103 2809–2814. 10.1073/pnas.0506854103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartók T., Szécsi Á, Szekeres A., Mesterházy Á, Bartók M. (2006). Detection of new fumonisin mycotoxins and fumonisin-like compounds by reversed-phase high-performance liquid chromatography/electrospray ionization ion trap mass spectrometry. Rapid Commun. Mass. Sp. 20 2447–2462. 10.1002/rcm.2607 [DOI] [PubMed] [Google Scholar]
- Basnayake B. M., Li D., Zhang H., Li G., Virk N., Song F. (2011). Arabidopsis DAL1 and DAL2, two RING finger proteins homologous to Drosophila DIAP1, are involved in regulation of programmed cell death. Plant Cell Rep. 30 37–48. 10.1007/s00299-010-0941-6 [DOI] [PubMed] [Google Scholar]
- Brandwagt B. F., Mesbah L. A., Takken F. L. W., Laurent P. L., Kneppers T. J. A., Hille J., et al. (2000). A longevity assurance gene homolog of tomato mediates resistance to Alternaria alternata f. sp. lycopersici toxins and fumonisin B1. Proc. Natl. Acad. Sci. U.S.A. 97:4961. 10.1073/pnas.97.9.4961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacas J. L., Buré C., Grosjean K., Gerbeau-Pissot P., Lherminier J., Rombouts Y., et al. (2016). Revisiting plant plasma membrane lipids in tobacco: a focus on sphingolipids. Plant Physiol. 170 367–384. 10.1104/pp.15.00564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae H.-J., Ke N., Kim H.-R., Chen S., Godzik A., Dickman M., et al. (2003). Evolutionarily conserved cytoprotection provided by Bax Inhibitor-1 homologs from animals, plants, and yeast. Gene 323 101–113. 10.1016/j.gene.2003.09.011 [DOI] [PubMed] [Google Scholar]
- Chae H.-J., Kim H.-R., Xu C., Bailly-Maitre B., Krajewska M., Krajewski S., et al. (2004). BI-1 regulates an apoptosis pathway linked to endoplasmic reticulum stress. Mol. Cell 15 355–366. 10.1016/j.molcel.2004.06.038 [DOI] [PubMed] [Google Scholar]
- Chen M., Markham J. E., Dietrich C. R., Jaworski J. G., Cahoon E. B. (2008). Sphingolipid long-chain base hydroxylation is important for growth and regulation of sphingolipid content and composition in Arabidopsis. Plant Cell 20 1862–1878. 10.1105/tpc.107.057851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A., Monte I., Zamarreño A. M., Hamberg M., Lassueur S., Reymond P., et al. (2018). An OPR3-independent pathway uses 4,5-didehydrojasmonate for jasmonate synthesis. Nat. Chem. Biol. 14 171–178. 10.1038/nchembio.2540 [DOI] [PubMed] [Google Scholar]
- Chivasa S., Ndimba B. K., Simon W. J., Lindsey K., Slabas A. R. (2005). Extracellular ATP functions as an endogenous external metabolite regulating plant cell viability. Plant Cell 17 3019–3034. 10.1105/tpc.105.036806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chivasa S., Tome D. F., Hamilton J. M., Slabas A. R. (2011). Proteomic analysis of extracellular ATP-regulated proteins identifies ATP synthase beta-subunit as a novel plant cell death regulator. Mol. Cell Proteomics 10:M110.003905. 10.1074/mcp.M110.003905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chivasa S., Tomé D. F. A., Slabas A. R. (2013). UDP-glucose pyrophosphorylase is a novel plant cell death regulator. J. Plant Res. 12 1743–1753. 10.1021/pr3010887 [DOI] [PubMed] [Google Scholar]
- Christensen S. A., Kolomiets M. V. (2011). The lipid language of plant-fungal interactions. Fungal Genet. Biol. 48 4–14. 10.1016/j.fgb.2010.05.005 [DOI] [PubMed] [Google Scholar]
- Christensen S. A., Nemchenko A., Park Y.-S., Borrego E., Huang P.-C., Schmelz E. A., et al. (2014). The novel monocot-specific 9-lipoxygenase ZmLOX12 is required to mount an effective jasmonate-mediated defense against Fusarium verticillioides in Maize. Mol. Plant Microbe Interact. 27 1263–1276. 10.1094/MPMI-06-13-0184-R [DOI] [PubMed] [Google Scholar]
- Cingolani F., Futerman A. H., Casas J. (2016). Ceramide synthases in biomedical research. Chem. Phys. Lipids 197 25–32. 10.1016/j.chemphyslip.2015.07.026 [DOI] [PubMed] [Google Scholar]
- Clay N. K., Adio A. M., Denoux C., Jander G., Ausubel F. M. (2009). Glucosinolate metabolites required for an Arabidopsis innate immune response. Science 323 95–101. 10.1126/science.1164627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll N. S., Smidler A., Puigvert M., Popa C., Valls M., Dangl J. L. (2014). The plant metacaspase AtMC1 in pathogen-triggered programmed cell death and aging: functional linkage with autophagy. Cell Death Differ. 21 1399–1408. 10.1038/cdd.2014.50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danon A., Miersch O., Felix G., Camp R. G. L., Apel K. (2004). Concurrent activation of cell death-regulating signaling pathways by singlet oxygen in Arabidopsis thaliana. Plant J. 41 68–80. 10.1111/j.1365-313X.2004.02276.x [DOI] [PubMed] [Google Scholar]
- De Bigault Du Granrut A., Cacas J. L. (2016). How very-long-chain fatty acids could signal stressful conditions in plants? Front Plant Sci. 7:1490. 10.3389/fpls.2016.01490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjardins A. E., Munkvold G., Plattner R., Proctor R. (2002). FUM1 —A gene required for fumonisin biosynthesis but not for maize ear rot and ear infection by Gibberella moniliformis in field tests. Mol. Plant Microbe Interact. 15 1157–1164. 10.1094/MPMI.2002.15.11.1157 [DOI] [PubMed] [Google Scholar]
- Dutilleul C., Benhassaine-Kesri G., Demandre C., Reze N., Launay A., Pelletier S., et al. (2012). Phytosphingosine-phosphate is a signal for AtMPK6 activation and Arabidopsis response to chilling. New Phytol. 194 181–191. 10.1111/j.1469-8137.2011.04017.x [DOI] [PubMed] [Google Scholar]
- Feng H., Chen Q., Feng J., Zhang J., Yang X., Zuo J. (2007). Functional characterization of the Arabidopsis eukaryotic translation initiation factor 5A-2 that plays a crucial role in plant growth and development by regulating cell division, cell growth, and cell death. Plant Physiol. 144 1531–1545. 10.1104/pp.107.098079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M., Glenn A. E., Gu X., Mitchell T. R., Satterlee T., Duke M. V., et al. (2020). Pyrrocidine, a molecular off switch for fumonisin biosynthesis. PLoS Pathog. 16:e1008595. 10.1371/journal.ppat.1008595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Brodhagen M., Isakeit T., Brown S. H., Gobel C., Betran J., et al. (2009). Inactivation of the lipoxygenase ZmLOX3 increases susceptibility of Maize to Aspergillus spp. Mol. Plant Microbe Interact. 22 222–231. 10.1094/MPMI-22-2-0222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Shim W. B., Göbel C., Kunze S., Feussner I., Meeley R., et al. (2007). Disruption of a maize 9-lipoxygenase results in increased resistance to fungal pathogens and reduced levels of contamination with mycotoxin fumonisin. Mol. Plant Microbe Interact. 20 922–933. [DOI] [PubMed] [Google Scholar]
- Gechev T. S., Ferwerda M. A., Mehterov N., Laloi C., Qureshi M. K., Hille J. (2008). Arabidopsis AAL-toxin-resistant mutant atr1 shows enhanced tolerance to programmed cell death induced by reactive oxygen species. Biochem. Biophys. Res. Commun. 375 639–644. 10.1016/j.bbrc.2008.08.056 [DOI] [PubMed] [Google Scholar]
- Gelderblom W. C., Jaskiewicz K., Marasas W. F., Thiel P. G., Horak R. M., Vleggaar R., et al. (1988). Fumonisins–novel mycotoxins with cancer-promoting activity produced by Fusarium moniliforme. Appl. Environ. Microb. 54 1806–1811. 10.1002/bit.260320218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glenn A., Zitomer N., Zimeri A., Williams L., Riley R. (2008). Transformation-mediated complementation of a FUM gene cluster deletion in Fusarium verticillioides restores both fumonisin production and pathogenicity on maize seedlings. Mol. Plant Microbe Interact. 21 87–97. [DOI] [PubMed] [Google Scholar]
- Glenz R., Schmalhaus D., Krischke M., Mueller M. J., Waller F. (2019). Elevated levels of phosphorylated sphingobases do not antagonize sphingobase- or fumonisin B1-induced plant cell death. Plant Cell Physiol. 60 1109–1119. 10.1093/pcp/pcz033 [DOI] [PubMed] [Google Scholar]
- Haberkant P., Stein F., Hoglinger D., Gerl M. J., Brugger B., Van Veldhoven P. P., et al. (2016). Bifunctional sphingosine for cell-based analysis of protein-sphingolipid interactions. ACS Chem. Biol. 11 222–230. 10.1021/acschembio.5b00810 [DOI] [PubMed] [Google Scholar]
- Have M., Luo J., Tellier F., Balliau T., Cueff G., Chardon F., et al. (2019). Proteomic and lipidomic analyses of the Arabidopsis atg5 autophagy mutant reveal major changes in endoplasmic reticulum and peroxisome metabolisms and in lipid composition. New Phytol. 223 1461–1477. 10.1111/nph.15913 [DOI] [PubMed] [Google Scholar]
- Hopkins M. T., Lampi Y., Wang T. W., Liu Z., Thompson J. E. (2008). Eukaryotic translation initiation factor 5A is involved in pathogen-induced cell death and development of disease symptoms in Arabidopsis. Plant Physiol. 148 479–489. 10.1104/pp.108.118869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardine D., Leslie J. (1999). Aggressiveness to mature maize plants of Fusarium strains differing in ability to produce fumonisin. Plant Dis. 83 690–693. 10.1094/PDIS.1999.83.7.690 [DOI] [PubMed] [Google Scholar]
- Kawai-Yamada M., Jin L., Yoshinaga K., Hirata A., Uchimiya H. (2001). Mammalian Bax-induced plant cell death can be down-regulated by overexpression of Arabidopsis Bax Inhibitor-1 (AtBI-1). Proc. Natl. Acad. Sci. U.S.A. 98 12295–12300. 10.1073/pnas.211423998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W. Y., Lee S. Y., Jung Y. J., Chae H. B., Nawkar G. M., Shin M. R., et al. (2011). Inhibitor of apoptosis (IAP)-like protein lacks a Baculovirus IAP Repeat (BIR) domain and attenuates cell death in plant and animal systems. J. Biol. Chem. 286 42670–42678. 10.1074/jbc.M111.262204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimberlin A. N., Han G., Luttgeharm K. D., Chen M., Cahoon R. E., Stone J. M., et al. (2016). ORM expression alters sphingolipid homeostasis and differentially affects ceramide synthase activity. Plant Physiol. 172 889–900. 10.1104/pp.16.00965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimberlin A. N., Majumder S., Han G., Chen M., Cahoon R. E., Stone J. M., et al. (2013). Arabidopsis 56-amino acid serine palmitoyltransferase-interacting proteins stimulate sphingolipid synthesis, are essential, and affect mycotoxin sensitivity. Plant Cell 25 4627–4639. 10.2307/23598494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kock J. L. F., Strauss C. J., Pohl C. H., Nigam S. (2003). The distribution of 3-hydroxy oxylipins in fungi. Prostag. Oth. Lipid M 71 85–96. 10.1016/S1098-8823(03)00046-7 [DOI] [PubMed] [Google Scholar]
- Konig S., Feussner K., Schwarz M., Kaever A., Iven T., Landesfeind M., et al. (2012). Arabidopsis mutants of sphingolipid fatty acid alpha-hydroxylases accumulate ceramides and salicylates. New Phytol. 196 1086–1097. 10.1111/j.1469-8137.2012.04351.x [DOI] [PubMed] [Google Scholar]
- Kuroyanagi M., Yamada K., Hatsugai N., Kondo M., Nishimura M., Hara-Nishimura I. (2005). Vacuolar processing enzyme is essential for mycotoxin-induced cell death in Arabidopsis thaliana. J. Biol. Chem. 280 32914–32920. 10.1074/jbc.M504476200 [DOI] [PubMed] [Google Scholar]
- Kwon S. I., Cho H. J., Bae K., Jung J. H., Jin H. C., Park O. K. (2009). Role of an Arabidopsis Rab GTPase RabG3b in pathogen response and leaf senescence. J. Plant Biol. 52 79–87. 10.1007/s12374-009-9011-4 [DOI] [Google Scholar]
- Kwon S. I., Cho H. J., Kim S. R., Park O. K. (2013). The Rab GTPase RabG3b positively regulates autophagy and immunity-associated hypersensitive cell death in Arabidopsis. Plant Physiol. 161 1722–1736. 10.1104/pp.112.208108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachaud C., Prigent E., Thuleau P., Grat S., Da Silva D., Briere C., et al. (2013). 14-3-3-regulated Ca(2+)-dependent protein kinase CPK3 is required for sphingolipid-induced cell death in Arabidopsis. Cell Death Differ. 20 209–217. 10.1038/cdd.2012.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langhans M., Robinson D. G. (2007). 1-Butanol targets the Golgi apparatus in tobacco BY-2 cells, but in a different way to Brefeldin A. J. Exp. Bot. 58 3439–3447. 10.1093/jxb/erm194 [DOI] [PubMed] [Google Scholar]
- Lanubile A., Bernardi J., Battilani P., Logrieco A., Marocco A. (2012a). Resistant and susceptible maize genotypes activate different transcriptional responses against Fusarium verticillioides. Physiol. Mol. Plant Pathol. 77 52–59. 10.1016/j.pmpp.2011.12.002 [DOI] [Google Scholar]
- Lanubile A., Bernardi J., Marocco A., Logrieco A., Paciolla C. (2012b). Differential activation of defense genes and enzymes in maize genotypes with contrasting levels of resistance to Fusarium verticillioides. Environ. Exp. Bot. 78 39–46. 10.1016/j.envexpbot.2011.12.006 [DOI] [Google Scholar]
- Lanubile A., Logrieco A., Battilani P., Proctor R., Marocco A. (2013). Transcriptional changes in developing maize kernels in response to fumonisin-producing and nonproducing strains of Fusarium verticillioides. Plant Sci. 210 183–192. 10.1016/j.plantsci.2013.05.020 [DOI] [PubMed] [Google Scholar]
- Lanubile A., Maschietto V., Borrelli V. M., Stagnati L., Logrieco A. F., Marocco A. (2017). Molecular basis of resistance to Fusarium ear rot in Maize. Front Plant Sci. 8:1774. 10.3389/fpls.2017.01774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D. H., Choi H. W., Hwang B. K. (2011). The pepper E3 ubiquitin ligase RING1 gene, CaRING1, is required for cell death and the salicylic acid-dependent defense response. Plant Physiol. 156 2011–2025. 10.1104/pp.111.177568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz H. D., Haller E., Melzer E., Kober K., Wurster K., Stahl M., et al. (2011). Autophagy differentially controls plant basal immunity to biotrophic and necrotrophic pathogens. Plant J. 66 818–830. 10.1111/j.1365-313X.2011.04546.x [DOI] [PubMed] [Google Scholar]
- Li J., Brader G., Palva E. T. (2008). Kunitz trypsin inhibitor: an antagonist of cell death triggered by phytopathogens and fumonisin B1 in Arabidopsis. Mol. Plant 1 482–495. 10.1093/mp/ssn013 [DOI] [PubMed] [Google Scholar]
- Li J., Yin J., Rong C., Li K. E., Wu J. X., Huang L. Q., et al. (2016). Orosomucoid proteins interact with the small subunit of serine palmitoyltransferase and contribute to sphingolipid homeostasis and stress responses in Arabidopsis. Plant Cell 28 3038–3051. 10.1105/tpc.16.00574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H., Yao N., Song J. T., Luo S., Lu H., Greenberg J. T. (2003). Ceramides modulate programmed cell death in plants. Genes Dev. 17 2636–2641. 10.1101/gad.1140503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S. S., Martin R., Mongrand S., Vandenabeele S., Chen K. C., Jang I. C., et al. (2008). RING1 E3 ligase localizes to plasma membrane lipid rafts to trigger FB1-induced programmed cell death in Arabidopsis. Plant J. 56 550–561. 10.1111/j.1365-313X.2008.03625.x [DOI] [PubMed] [Google Scholar]
- Ling Q., Broad W., Trösch R., Töpel M., Demiral Sert T., Lymperopoulos P., et al. (2019). Ubiquitin-dependent chloroplast-associated protein degradation in plants. Science 363:eaav4467. 10.1126/science.aav4467 [DOI] [PubMed] [Google Scholar]
- Ling Q., Huang W., Baldwin A., Jarvis P. (2012). Chloroplast biogenesis is regulated by direct action of the ubiquitin-proteasome system. Science 338 655–659. 10.1126/science.1225053 [DOI] [PubMed] [Google Scholar]
- Luttgeharm K. D., Cahoon E. B., Markham J. E. (2016). Substrate specificity, kinetic properties and inhibition by fumonisin B1 of ceramide synthase isoforms from Arabidopsis. Biochem. J. 473 593–603. 10.1042/BJ20150824 [DOI] [PubMed] [Google Scholar]
- Luttgeharm K. D., Chen M., Mehra A., Cahoon R. E., Markham J. E., Cahoon E. B. (2015). Overexpression of Arabidopsis ceramide synthases differentially affects growth, sphingolipid metabolism, programmed cell death, and mycotoxin resistance. Plant Physiol. 169 1108–1117. 10.1104/pp.15.00987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maatta S., Scheu B., Roth M. R., Tamura P., Li M., Williams T. D., et al. (2012). Levels of Arabidopsis thaliana leaf phosphatidic acids, phosphatidylserines, and most trienoate-containing polar lipid molecular species increase during the dark period of the diurnal cycle. Front. Plant Sci. 3:49. 10.3389/fpls.2012.00049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmood K., El-Kereamy A., Kim S. H., Nambara E., Rothstein S. J. (2016). ANAC032 positively regulates age-dependent and stress-induced senescence in Arabidopsis thaliana. Plant Cell Physiol. 57 2029–2046. 10.1093/pcp/pcw120 [DOI] [PubMed] [Google Scholar]
- Markham J. E., Lynch D. V., Napier J. A., Dunn T. M., Cahoon E. B. (2013). Plant sphingolipids: function follows form. Curr. Opin. Plant Biol. 16 350–357. 10.1016/j.pbi.2013.02.009 [DOI] [PubMed] [Google Scholar]
- Markham J. E., Molino D., Gissot L., Bellec Y., Hématy K., Marion J., et al. (2011). Sphingolipids containing very-long-chain fatty acids define a secretory pathway for specific polar plasma membrane protein targeting in Arabidopsis. Plant Cell 23 2362–2378. 10.1105/tpc.110.080473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maschietto V., Marocco A., Malachova A., Lanubile A. (2015). Resistance to Fusarium verticillioides and fumonisin accumulation in maize inbred lines involves an earlier and enhanced expression of lipoxygenase (LOX) genes. J. Plant Physiol. 188 9–18. 10.1016/j.jplph.2015.09.003 [DOI] [PubMed] [Google Scholar]
- Mase K., Ishihama N., Mori H., Takahashi H., Kaminaka H., Kodama M., et al. (2013). Ethylene-responsive AP2/ERF transcription factor MACD1 participates in phytotoxin-triggered programmed cell death. Mol. Plant Microbe Interact. 26 868–879. 10.1094/MPMI-10-12-0253-R [DOI] [PubMed] [Google Scholar]
- Mittler R. (2017). ROS are good. Trends Plant Sci. 22 11–19. 10.1016/j.tplants.2016.08.002 [DOI] [PubMed] [Google Scholar]
- Molino D., Van der Giessen E., Gissot L., Hematy K., Marion J., Barthelemy J., et al. (2014). Inhibition of very long acyl chain sphingolipid synthesis modifies membrane dynamics during plant cytokinesis. Biochim. Biophys. Acta 1842 1422–1430. 10.1016/j.bbalip.2014.06.014 [DOI] [PubMed] [Google Scholar]
- Mosblech A., Feussner I., Heilmann I. (2009). Oxylipins: structurally diverse metabolites from fatty acid oxidation. Plant Physiol. Bioch. 47 511–517. 10.1016/j.plaphy.2008.12.011 [DOI] [PubMed] [Google Scholar]
- Munch D., Teh O. K., Malinovsky F. G., Liu Q., Vetukuri R. R., El Kasmi F., et al. (2015). Retromer contributes to immunity-associated cell death in Arabidopsis. Plant Cell 27 463–479. 10.1105/tpc.114.132043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano M., Ihara-Ohori Y., Imai H., Inada N., Fujimoto M., Tsutsumi N., et al. (2009). Functional association of cell death suppressor, Arabidopsis Bax inhibitor-1, with fatty acid 2-hydroxylation through cytochrome b (5). Plant J. 58 122–134. 10.1111/j.1365-313X.2008.03765.x [DOI] [PubMed] [Google Scholar]
- Nagano M., Ishikawa T., Ogawa Y., Iwabuchi M., Nakasone A., Shimamoto K., et al. (2014). Arabidopsis Bax inhibitor-1 promotes sphingolipid synthesis during cold stress by interacting with ceramide-modifying enzymes. Planta 240 77–89. 10.1007/s00425-014-2065-7 [DOI] [PubMed] [Google Scholar]
- Nagano M., Kakuta C., Fukao Y., Fujiwara M., Uchimiya H., Kawai-Yamada M. (2019). Arabidopsis Bax inhibitor-1 interacts with enzymes related to very-long-chain fatty acid synthesis. J. Plant Res. 132 131–143. 10.1007/s10265-018-01081-8 [DOI] [PubMed] [Google Scholar]
- Nakagawa N., Kato M., Takahashi Y., Shimazaki K., Tamura K., Tokuji Y., et al. (2012). Degradation of long-chain base 1-phosphate (LCBP) in Arabidopsis: functional characterization of LCBP phosphatase involved in the dehydration stress response. J. Plant Res. 125 439–449. 10.1007/s10265-011-0451-9 [DOI] [PubMed] [Google Scholar]
- Ormancey M., Thuleau P., van der Hoorn R. A. L., Grat S., Testard A., Kamal K. Y., et al. (2019). Sphingolipid-induced cell death in Arabidopsis is negatively regulated by the papain-like cysteine protease RD21. Plant Sci. 280 12–17. 10.1016/j.plantsci.2018.10.028 [DOI] [PubMed] [Google Scholar]
- Pan R., Satkovich J., Chen C., Hu J. (2018). The E3 ubiquitin ligase SP1-like 1 plays a positive role in peroxisome biogenesis in Arabidopsis. Plant J. 94 836–846. 10.1111/tpj.13900 [DOI] [PubMed] [Google Scholar]
- Pan R., Satkovich J., Hu J. (2016). E3 ubiquitin ligase SP1 regulates peroxisome biogenesis in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 113 E7307–E7316. 10.1073/pnas.1613530113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pata M. O., Hannun Y. A., Ng C. K. (2010). Plant sphingolipids: decoding the enigma of the Sphinx. New Phytol. 185 611–630. 10.1111/j.1469-8137.2009.03123.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer M., Stegmann M., Mueller M. J., Waller F. (2010). Pseudomonas syringae infection triggers de novo synthesis of phytosphingosine from sphinganine in Arabidopsis thaliana. FEBS Lett. 584 4053–4056. 10.1016/j.febslet.2010.08.027 [DOI] [PubMed] [Google Scholar]
- Picot A., Barreau C., Pinson-Gadais L., Caron D., Lannou C., Richard-Forget F. (2010). Factors of the Fusarium verticillioides-maize environment modulating fumonisin production. Curr. Opin. Microbiol. 36 221–231. 10.3109/10408411003720209 [DOI] [PubMed] [Google Scholar]
- Pieterse C. M. J., Leon-Reyes A., Van der Ent S., Van Wees S. C. M. (2009). Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 5 308–316. 10.1038/nchembio.164 [DOI] [PubMed] [Google Scholar]
- Proctor R. H., Desjardins A. E., Plattner R. D., Hohn T. M. (1999). A polyketide synthase gene required for biosynthesis of fumonisin mycotoxins in Gibberella fujikuroi mating population A. Fungal Genet. Biol. 27 100–112. 10.1006/fgbi.1999.1141 [DOI] [PubMed] [Google Scholar]
- Proctor R. H., Brown D. W., Plattner R. D., Desjardins A. E. (2003). Co-expression of 15 contiguous genes delineates a fumonisin biosynthetic gene cluster in Gibberella moniliformis. Fungal Genet. Biol. 38 237–249. 10.1016/S1087-1845(02)00525-X [DOI] [PubMed] [Google Scholar]
- Plett J. M., Cvetkovska M., Makenson P., Xing T., Regan S. (2009). Arabidopsis ethylene receptors have different roles in fumonisin B1-induced cell death. Physiol. Mol. Plant Pathol. 74 18–26. 10.1016/j.pmpp.2009.08.004 [DOI] [Google Scholar]
- Qin X., Zhang R. X., Ge S., Zhou T., Liang Y. K. (2017). Sphingosine kinase AtSPHK1 functions in fumonisin B1-triggered cell death in Arabidopsis. Plant Physiol. Bioch. 119 70–80. 10.1016/j.plaphy.2017.08.008 [DOI] [PubMed] [Google Scholar]
- Ren B., Chen Q., Hong S., Zhao W., Feng J., Feng H., et al. (2013). The Arabidopsis eukaryotic translation initiation factor eIF5A-2 regulates root protoxylem development by modulating cytokinin signaling. Plant Cell 25 3841–3857. 10.1105/tpc.113.116236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rheeder J. P., Marasas W. F. O., Vismer H. F. (2002). Production of fumonisin analogs by Fusarium Species. Appl. Environ. Microb. 68 2101–2105. 10.1128/AEM.68.5.2101-2105-2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley R. T., Merrill A. H. (2019). Ceramide synthase inhibition by fumonisins: a perfect storm of perturbed sphingolipid metabolism, signaling, and disease. J. Lipid Res. 60 1183–1189. 10.1194/jlr.S093815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritzenthaler C., Nebenführ A., Movafeghi A., Stussi-Garaud C., Behnia L., Pimpl P., et al. (2002). Reevaluation of the effects of Brefeldin A on plant cells using Tobacco Bright Yellow 2 cells expressing Golgi-targeted green fluorescent protein and COPI Antisera. Plant Cell 14 237–261. 10.1105/tpc.010237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotari V. I., He R., Gallois P. (2005). Death by proteases in plants: whodunit. Physiol. Plantarum 123 376–385. 10.1111/j.1399-3054.2005.00465.x [DOI] [Google Scholar]
- Saucedo-García M., González-Solís A., Rodríguez-Mejía P., Olivera M. T., Vazquez Santana S., Cahoon E., et al. (2011a). Reactive oxygen species as transducers of sphinganine-mediated cell death pathway. Plant Signal Behav. 6 1616–1619. 10.4161/psb.6.10.16981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saucedo-García M., Guevara-García A., González-Solís A., Cruz-García F., Vázquez-Santana S., Markham J. E., et al. (2011b). MPK6, sphinganine and the LCB2a gene from serine palmitoyltransferase are required in the signaling pathway that mediates cell death induced by long chain bases in Arabidopsis. New Phytol. 191 943–957. 10.1111/j.1469-8137.2011.03727.x [DOI] [PubMed] [Google Scholar]
- Schulte-Zweckel J., Schneidewind T., Abad J. L., Brockmeyer A., Janning P., Triola G. (2018). Azide-tagged sphingolipids for the proteome-wide identification of C16-ceramide-binding proteins. Chem. Commun. 54 13742–13745. 10.1039/C8CC05691A [DOI] [PubMed] [Google Scholar]
- Shao Z., Zhao Y., Liu L., Chen S., Li C., Meng F., et al. (2019). Overexpression of FBR41 enhances resistance to sphinganine analog mycotoxin-induced cell death and Alternaria stem canker in tomato. Plant Biotechnol. J. 18 141–154. 10.1111/pbi.13182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L., Bielawski J., Mu J., Dong H., Teng C., Zhang J., et al. (2007). Involvement of sphingoid bases in mediating reactive oxygen intermediate production and programmed cell death in Arabidopsis. Cell Res. 17 1030–1040. 10.1038/cr.2007.100 [DOI] [PubMed] [Google Scholar]
- Smith S., Kroon J., Simon W., Slabas A., Chivasa S. (2015). A novel function for Arabidopsis CYCLASE1 in programmed cell death revealed by isobaric tags for relative and absolute quantitation (iTRAQ) analysis of extracellular matrix proteins. Mol. Cell Proteomics 14 1556–1568. 10.1074/mcp.M114.045054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spassieva S. D., Markham J. E., Hille J. (2002). The plant disease resistance gene Asc-1 prevents disruption of sphingolipid metabolism during AAL-toxin-induced programmed cell death. Plant J. 32 561–572. 10.1046/j.1365-313X.2002.01444.x [DOI] [PubMed] [Google Scholar]
- Stone J. M., Liang X., Nekl E. R., Stiers J. J. (2005). Arabidopsis AtSPL14, a plant-specific SBP-domain transcription factor, participates in plant development and sensitivity to fumonisin B1. Plant J. 41 744–754. 10.1111/j.1365-313X.2005.02334.x [DOI] [PubMed] [Google Scholar]
- Ternes P., Feussner K., Werner S., Lerche J., Iven T., Heilmann I., et al. (2011). Disruption of the ceramide synthase LOH1 causes spontaneous cell death in Arabidopsis thaliana. New Phytol. 192 841–854. 10.1111/j.1469-8137.2011.03852.x [DOI] [PubMed] [Google Scholar]
- Tsegaye Y., Richardson C. G., Bravo J. E., Mulcahy B. J., Lynch D. V., Markham J. E., et al. (2007). Arabidopsis mutants lacking long chain base phosphate lyase are fumonisin-sensitive and accumulate trihydroxy-18:1 long chain base phosphate. J. Biol. Chem. 282 28195–28206. 10.1074/jbc.M705074200 [DOI] [PubMed] [Google Scholar]
- Vick B. A., Zimmerman D. C. (1983). The biosynthesis of jasmonic acid: a physiological role for plant lipoxygenase. Biochem. Biophys. Res. Commun. 111 470–477. 10.1016/0006-291X(83)90330-3 [DOI] [PubMed] [Google Scholar]
- Wang E., Norred W. P., Bacon C. W., Riley R. T., Merrill A. H. (1991). Inhibition of sphingolipid biosynthesis by fumonisins. Implications for diseases associated with Fusarium moniliforme. J. Biol. Chem. 266 14486–14490. 10.1016/0014-5793(91)80056-9 [DOI] [PubMed] [Google Scholar]
- Wang L., Pei Z., Tian Y., He C. (2005). OsLSD1, a rice zinc finger protein, regulates programmed cell death and callus differentiation. Mol. Plant Microbe Interact. 18 375–384. 10.1094/MPMI-18-0375 [DOI] [PubMed] [Google Scholar]
- Watanabe N., Lam E. (2006). Arabidopsis Bax inhibitor-1 functions as an attenuator of biotic and abiotic types of cell death. Plant J. 45 884–894. 10.1111/j.1365-313X.2006.02654.x [DOI] [PubMed] [Google Scholar]
- Watanabe N., Lam E. (2008). BAX inhibitor-1 modulates endoplasmic reticulum stress-mediated programmed cell death in Arabidopsis. J. Biol. Chem. 283 3200–3210. 10.1074/jbc.M706659200 [DOI] [PubMed] [Google Scholar]
- Watanabe N., Lam E. (2011). Arabidopsis metacaspase 2d is a positive mediator of cell death induced during biotic and abiotic stresses. Plant J. 66 969–982. 10.1111/j.1365-313X.2011.04554.x [DOI] [PubMed] [Google Scholar]
- Williams L. D., Glenn A. E., Bacon C. W., Smith M. A., Riley R. T. (2006). Fumonisin production and bioavailability to maize seedlings grown from seeds inoculated with Fusarium verticillioides and grown in natural soils. J. Agr. Food Chem. 54 5694–5700. 10.1021/jf0610209 [DOI] [PubMed] [Google Scholar]
- Wu J. X., Li J., Liu Z., Yin J., Chang Z. Y., Rong C., et al. (2015a). The Arabidopsis ceramidase AtACER functions in disease resistance and salt tolerance. Plant J. 81 767–780. 10.1111/tpj.12769 [DOI] [PubMed] [Google Scholar]
- Wu J.-X., Wu J.-L., Yin J., Zheng P., Yao N. (2015b). Ethylene modulates sphingolipid synthesis in Arabidopsis. Front. Plant Sci. 6:1122. 10.3389/fpls.2015.01122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L. J., Chen Q. F., Chen M. X., Yu L. J., Huang L., Chen L., et al. (2015). Unsaturation of very-long-chain ceramides protects plant from hypoxia-induced damages by modulating ethylene signaling in Arabidopsis. PLoS Genet. 11:e1005143. 10.1371/journal.pgen.1005143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagawa D., Ishikawa T., Imai H. (2017). Synthesis and degradation of long-chain base phosphates affect fumonisin B1-induced cell death in Arabidopsis thaliana. J. Plant Res. 130 571–585. 10.1007/s10265-017-0923-7 [DOI] [PubMed] [Google Scholar]
- Yin S., Guo X., Li J., Fan L., Hu H. (2016). Fumonisin B1 induces autophagic cell death via activation of ERN1-MAPK8/9/10 pathway in monkey kidney MARC-145 cells. Arch. Toxicol. 90 985–996. 10.1007/s00204-015-1514-9 [DOI] [PubMed] [Google Scholar]
- Yin S., Liu X., Fan L., Hu H. (2018). Mechanisms of cell death induction by food-borne mycotoxins. Crit. Rev. Food Sci. 58 1406–1417. 10.1080/10408398.2016.1260526 [DOI] [PubMed] [Google Scholar]
- Zeng H. Y., Zheng P., Wang L. Y., Bao H. N., Sahu S. K., Yao N. (2019). Autophagy in plant immunity. Adv. Exp. Med. Biol. 1209 23–41. 10.1007/978-981-15-0606-2_3 [DOI] [PubMed] [Google Scholar]
- Zhang X., Wu Q., Cui S., Ren J., Qian W., Yang Y., et al. (2015). Hijacking of the jasmonate pathway by the mycotoxin fumonisin B1 (FB1) to initiate programmed cell death in Arabidopsis is modulated by RGLG3 and RGLG4. J. Exp. Bot. 66 2709–2721. 10.1093/jxb/erv068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Wang J., Liu Y., Miao H., Cai C., Shao Z., et al. (2015). Classic myrosinase-dependent degradation of indole glucosinolate attenuates fumonisin B1-induced programmed cell death in Arabidopsis. Plant J. 81 920–933. 10.1111/tpj.12778 [DOI] [PubMed] [Google Scholar]
- Zheng P., Wu J. X., Sahu S. K., Zeng H. Y., Huang L. Q., Liu Z., et al. (2018). Loss of alkaline ceramidase inhibits autophagy in Arabidopsis and plays an important role during environmental stress response. Plant Cell Environ. 41 837–849. 10.1111/pce.13148 [DOI] [PubMed] [Google Scholar]
- Zheng Y., Zhan Q., Shi T., Liu J., Zhao K., Gao Y. (2019). The nuclear transporter SAD2 plays a role in calcium- and H2O2 -mediated cell death in Arabidopsis. Plant J. 101 324–333. 10.1111/tpj.14544 [DOI] [PubMed] [Google Scholar]
- Zienkiewicz A., Gömann J., König S., Herrfurth C., Liu Y. T., Meldau D., et al. (2019). Disruption of Arabidopsis neutral ceramidases 1 and 2 results in specific sphingolipid imbalances triggering different phytohormone-dependent plant cell death programs. New Phytol. 228 170–188. 10.1111/nph.16336 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.