Abstract
Purpose of the review
Significant numbers of patients worldwide are affected by various rare diseases, but the effective treatment options to these individuals are limited. Rare diseases remain underfunded compared to more common diseases, leading to significant delays in research progress and ultimately, to finding an effective cure. Here, we review the use of genome-editing tools to understand the pathogenesis of rare diseases and develop additional therapeutic approaches with a high degree of precision.
Recent findings
Several genome-editing approaches, including CRISPR/Cas9, TALEN and ZFN, have been used to generate animal models of rare diseases, understand the disease pathogenesis, correct pathogenic mutations in patient-derived somatic cells and iPSCs, and develop new therapies for rare diseases. The CRISPR/Cas9 system stands out as the most extensively used method for genome editing due to its relative simplicity and superior efficiency compared to TALEN and ZFN. CRISPR/Cas9 is emerging as a feasible gene-editing option to treat rare monogenic and other genetically defined human diseases.
Summary
Less than 5% of ~7000 known rare diseases have FDA-approved therapies, providing a compelling need for additional research and clinical trials to identify efficient treatment options for patients with rare diseases. Development of efficient genome-editing tools capable to correct or replace dysfunctional genes will lead to novel therapeutic approaches in these diseases.
Keywords: Genome editing, rare diseases, gene therapy, CRISPR/Cas9
Introduction
A rare disease (also known as orphan disease) affects a relatively small percentage of patients in the general population. There are approximately 7000 different types of rare diseases worldwide, which altogether account for significant numbers of patients and substantial healthcare cost [1–4]. There is no universal definition for rare diseases. For example, a rare disease in the USA is defined as a condition that affects fewer than 200,000 people, whereas the European Union uses a frequency of <1 patient per 2000 people as a definition for rare diseases [3]. Although individual occurrences of these diseases are rare, they cumulatively affect a significant proportion of the human population, accounting for approximately 20–30 million patients in the US [5]. There are several factors that often complicate the development of new therapeutic approaches for patients with rare diseases. For example, rarity of their occurrences hampers the advances in scientific research and delays medical referrals to disease experts who can correctly diagnose the disorder through specialized biochemical and genetic testing. Rare diseases often have limited clinical information and inadequate numbers of patients to calculate statistical significance and design clinical trials. Lack of biomarkers to evaluate the disease progression contributes to delays in treatments. Many rare diseases have early developmental or neonatal onsets resulting in poor prognosis without immediate therapeutic intervention.
Rare diseases are diverse and affect multiple organ systems. For example, Alveolar Capillary Dysplasia with Misalignment of Pulmonary Veins (ACDMPV) causes fatal lung disease in newborns and infants but is also associated with developmental abnormalities in cardiovascular, gastrointestinal and urogenital systems [6]. Recently, more than 70 pathogenic mutations in the Forkhead Box F1 (FOXF1) gene were identified in ACDMPV patients [6, 7]. Approximately 1700 mutations in the Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) gene are associated with Cystic fibrosis (CF), a severe, progressive disorder affecting the lung, pancreas, liver, kidney and other organs [8]. Niemann–Pick disease type C1 is a neurovisceral lipid storage disorder associated with over 200 mutations in the NPC1 gene [9]. Many rare diseases require organ transplantation, which is dependent on the availability of compatible donors and is associated with high healthcare costs [10–12]. Known mutational status in rare diseases provides a unique opportunity to use gene-editing approaches capable of achieving specific genomic modification within endogenous cells and tissues to repair the mutated gene itself.
Genome editing approaches have been used for several rare diseases with varied levels of success, aiming to correct or replace dysfunctional (mutated) genes [4, 13]. Specific nucleases, such as Transcription Activator-like Effector Nucleases (TALEN), meganucleases, Zinc Finger Nucleases (ZFN) and the Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) / CRISPR associated protein 9 (Cas9) complex, allow the addition, replacement and removal of DNA fragments to correct mutated gene in genome. CRISPR/Cas9 system is the most commonly used method among genome editing approaches due to its simplicity, accuracy, and relatively low cost [14–17]. CRISPR/Cas9-mediated genome editing is useful to repair non-functional genes as well as to correct pathogenic mutations encoding mutant proteins that are harmful or toxic. In the following four sections we summarized recent applications of CRISPR/Cas9 technology to rare diseases and discuss improvements required for its clinical translation.
Diversity of methods for genome editing
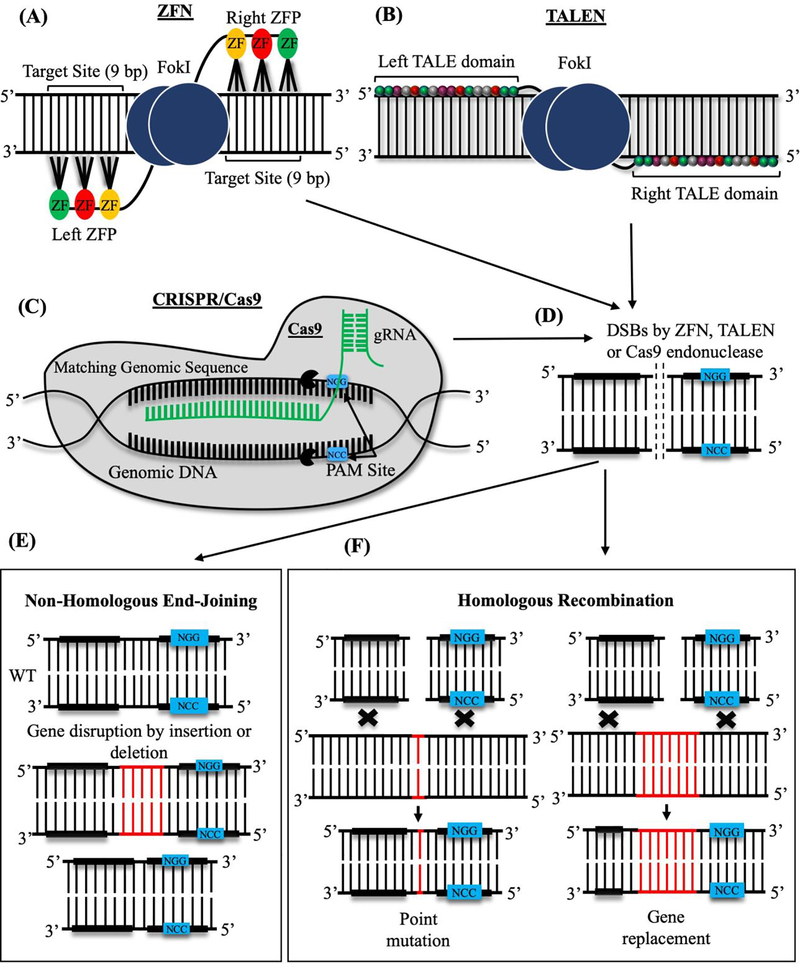
Genome editing methods use bioengineered nuclease enzymes to modify the organism’s genome in which DNA is inserted, deleted, or replaced in a precise and predetermined manner. Nuclease enzymes, also known as molecular scissors, cut genomic DNA to create site-specific double-strand breaks (DSBs) which later can be repaired through either Non-Homologous End Joining (NHEJ) or Homologous Recombination (HR), resulting in precise changes in genome (Figure 1). The main genome editing techniques include ZFN, TALEN and CRISPR/Cas9, and each method has its advantages and disadvantages. Both ZFN and TALEN use artificial fusion proteins consisting of various DNA-binding domains linked to the nuclease domain of FokI to make DNA breaks. ZFN uses a combination of zinc finger DNA binding domains to recognize specific DNA sequences, however, the 3-base pair requirement for DNA-binding for each zinc finger and the low efficiency of the method are the main challenges for ZFN. The TALEN system works similarly to ZFN but relies on the TAL effector DNA-binding domain and the FokI nuclease to cut DNA. Compared to ZFN, the TAL effector DNA-binding domain uses 33–35 amino acid repeats as recognition sites, therefore increasing the specificity of genome editing. TALEN is easier to engineer than ZFN, the latter of which is technically challenging and time-consuming to design. Disadvantages of TALEN are related to low editing efficiency in heavily methylated DNA regions and significant difficulties with gene delivery due to large sizes of TALEN constructs. Overall, bioengineering and testing of TALEN and ZFN constructs for the correction of individual mutations are often time-consuming and expensive [14–16].
Figure 1: Site-specific genome editing methods that use endonuclease activity.
(A) The Zinc-Finger Nuclease complex (ZFN) consists of the FokI endonuclease domain (blue) and several Zinc finger (ZF) DNA-binding domains (yellow, red and green). Each ZF domain recognizes a 3-base pair (bp) DNA sequence. (B) Transcription Activator-Like Effector Nuclease (TALEN) is shown as a fusion protein consisting of FokI endonuclease domain and TALE DNA-binding domains. Sixteen colored subunits in each TALE domain recognize a single base pair of DNA sequences. (C) CRISPR/Cas9 system is comprised of Clustered Regulatory Interspaced Short Palindromic Repeats (CRISPR) and CRISPR associated (Cas) protein 9. Guide RNA (gRNA, green) directs Cas9 endonuclease (grey) to specific DNA sites, which are located close to short Protospacer Adjacent Motifs (PAM, blue). Cas9 cleaves DNA upstream of PAM. (D) FokI and Cas9 endonucleases cause double-strand DNA breaks (DSBs). DSBs are repaired through either NHEJ or HR mechanisms. (E) NHEJ repair is based on nucleotide deletions or insertions into cleaved DNA. (F) Repair through HR mechanism occurs in the presence of the donor DNA template, which contains homology regions on both sides of the template. Exogenous DNA is specifically inserted into the target site to modify the gene.
CRISPR/Cas9, a two-component genome editing system [18, 19], consists of a guide RNA and the CRISPR associated protein 9 (Cas9), which function as nuclease (Figure 1). Guide RNA of the CRISPR system can be designed to bind specific DNA sequences in genome. Compared to ZFN and TALEN, the CRISPR/Cas9 system is a simple, cost-effective, and highly efficient method. CRISPR/Cas9, like TALEN, was originally discovered in bacteria, where it functions as a part of internal defense mechanisms to protect the bacterial host from invading viruses [18, 20]. The main characteristic feature of CRISPR/Cas9 is that it uses RNA sequences instead of DNA-binding proteins to recognize specific DNA motifs (Figure 1). Guide RNAs can be synthesized to target any specific DNA sequence in the genome, and specialized CRISPR/Cas9 algorithms have been developed to predict the possibility of off-target effects. CRISPR/Cas9 system has completely revolutionized the genome-editing field, opening multiple possibilities for development of new therapies for rare diseases.
Generation of animal models for rare diseases using CRISPR/Cas9 genome editing
CRISPR/ Cas9 has been extensively used to generate animal models for rare human diseases [21] (Table 1). Niemann-Pick disease type C1 (NPC1) is a rare lysosomal storage disease caused by inactivating mutations in the NPC1 gene. This disease is characterized by abnormal accumulation of cholesterol and glycolipids in late endosomes and lysosomes. Both mouse and feline models of NPC1 have been generated and used in preclinical studies to model the disease progression in humans [22–23]. However, the use of these models for high throughput screens to identify novel therapeutic agents is not economically feasible due to large time and labor costs to maintain and screen these animals. Recently, Tseng and coworkers [22] have generated two Zebrafish Npc1-null mutants using CRISPR/Cas9-mediated gene targeting. The Npc1 mutants exhibited the early hepatic and the late neurological abnormality, both phenotypes being similar to human NPC1 [22]. These Zebrafish models of NPC1 can be used for rapid in vivo screening of potential therapeutic agents for this lysosomal storage disease. In another study, a mouse model of Desbuquois dysplasia (DBQD) type 1, a rare skeletal dysplasia characterized by short stature, round face and skeletal abnormalities, has been generated by CRISPR/Cas9 through disruption of the Cant1 gene encoding Calcium-Activated Nucleotidase 1 enzyme, which preferentially hydrolyzes UDP to UMP and phosphate [24]. Newly generated Cant1−/− mice exhibited multiple skeletal defects, recapitulating clinical features seen in patients with DBQD type 1 [24]. Recently, Dreano and coworkers [25] reported the characterization of two rat models of cystic fibrosis (CF) using CRISPR/Cas9 genome editing. Rats homozygous for the F508del mutation in the transmembrane conductance regulator (Cftr) gene were compared with Cftr−/− rats. F508del and Cftr−/− rats exhibited abnormalities that are typical to CF [25]. Authors concluded that F508del mutant rats represent a novel CF model, which can be useful for development of CF therapeutics.
Table 1:
Animal models of rare diseases generated using CRISPR/Cas9 genome editing approach
| Disease | Species | Type of mutation | Reference | |
|---|---|---|---|---|
| 1. | Niemann-Pick disease type C1 (NPC1) | Zebrafish | Null mutant of Npc1 gene | 22 |
| 2. | Desbuquois dysplasia (DD) type 1 | Mouse | Knockout of Cant1 gene | 24 |
| 3. | Cystic fibrosis | Rat | Knockout/ deletion of Cftr gene | 25 |
| 4. | Alveolar Capillary Dysplasia with Misalignment of Pulmonary Veins (ACDMPV) | Mouse | Knock-in mutation in Foxf1 gene | 7 |
| 5. | Primary hyperoxaluria type I (PH1) | Mouse | Gene correction in Agxt1 gene | 26 |
| 6. | X-linked retinitis pigmentosa (XLRP) | Mouse | Transgenic expression of Rpgr gene | 27 |
| 7. | Bronchopulmonary dysplasia (BPD) | Mouse | Expression of exogenes Foxf1 and Foxm1 from nanoparticle vector | 47 |
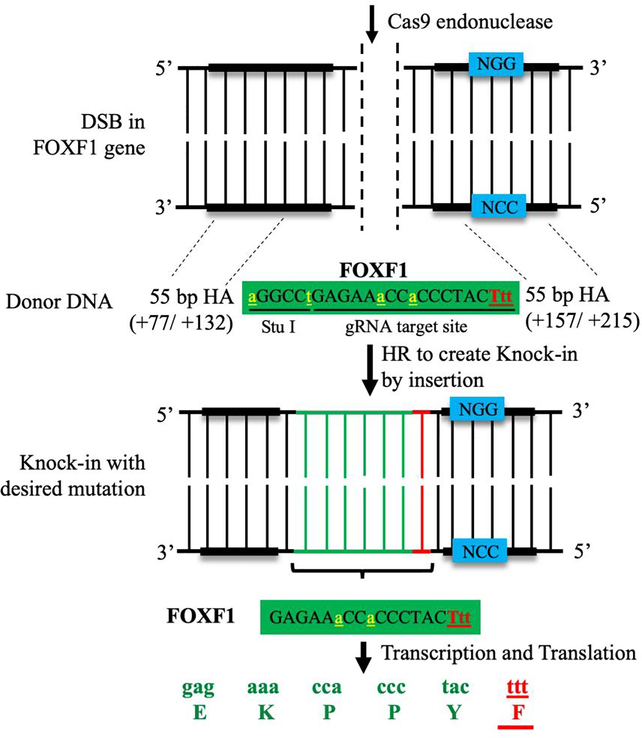
The generation of new mouse models by CRISPR/Cas9 has been recently reported for Hyperoxaluria Type 1 (PH1) [26] and X-linked Retinitis Pigmentosa (XLRP) [27]. Our laboratory has created a clinically relevant mouse model of Alveolar Capillary Dysplasia with Misalignment of Pulmonary Veins (ACDMPV) by introducing the S52F FOXF1 mutation (found in ACDMPV patients) [7] into the mouse Foxf1 gene locus using CRISPR/Cas9 technology (Figure 2). ACDMPV is a severe congenital disorder characterized by lung hypoplasia, a paucity of alveolar capillaries and gallbladder agenesis in newborns and infants [6]. All phenotypes were recapitulated in genetically engineered Foxf1WT/S52F knock-in mice [7] and previously described mice heterozygous for the Foxf1 null allele (Foxf1+/−) [28, 29]. Similar to ACDMPV patients, Foxf1WT/S52F and Foxf1+/− mice exhibited reduced endothelial cell proliferation, decreased lung angiogenesis, and increased mortality after birth [7, 30]. FOXF1, a transcription factor from the Forkhead Box (FOX) family, is expressed in mesenchyme-derived cells such as endothelial cells, fibroblasts, pericytes, and visceral smooth muscle cells [31–34]. Published studies demonstrated that FOXF1 is critical for embryonic development [35–38], carcinogenesis [39, 40], organ regeneration [35, 41, 42], and lung repair after various injuries [43–46]. The Foxf1WT/S52F and Foxf1+/− mutant mice were recently used to develop two potential therapeutic approaches for ACDMPV patients. These approaches are based on increasing perinatal lung angiogenesis and endothelial proliferation through either nanoparticle delivery of proangiogenic genes directly into endothelial cells [7, 47, 48] or cell transplantation with pulmonary endothelial progenitor cells [49].
Figure 2: Generation of Foxf1WT/S52F knock-in mouse model of ACDMPV using CRISPR/Cas9 genome editing.
The schematic diagram shows the generation of Foxf1WT/S52F knock-in mice by homology-directed repair method. Single amino acid substitution of serine-52 (S52) to phenylalanine (F) is performed using a donor DNA template containing 55 nucleotide base pairs (bp) homology arms (HA) on either side of the template. HAs allow the donor DNA template to recombine into the host genome through homologous recombination (HR), inserting the S52F Foxf1 mutation. The mutated amino acid and triplet nucleotide codon (F/Ttt) are labeled in red, whereas endogenous nucleotides are labeled in green. Silent mutations (yellow) are inserted into the donor DNA template to create a new Stu I restriction site needed for identification of the S52F Foxf1 mutant allele.
Using CRISPR/Cas9 to correct pathogenic mutations in patient-derived primary cells and iPSCs
CRISPR/Cas9 genome editing has been extensively used to correct pathogenic mutations in primary cells and induced pluripotent stem cells (iPSCs) generated from patients with rare diseases [21] (Table 2). Recently, an intronic variant of Structural Maintenance of Chromosomes Flexible Hinge Domain Containing 1 gene (SMCHD1) was repaired by CRISPR/Cas9 method in primary and immortalized myoblasts obtained from patients with Facioscapulohumeral dystrophy type 2 (FSHD2), a rare autosomal dominant muscular disorder [50]. This genome modification restored expression of SMCHD1 and inhibited transcription of the DUX4 gene encoding highly cytotoxic myogenic protein, which is increased in Facioscapulohumeral dystrophy type 1 (FSHD1). These studies suggest that CRISPR/Cas9-mediated targeting of SMCHD1 can be explored in the development of new therapeutics for patients with FSHD 1 and FSHD2. CRISPR/Cas9 has been also used for disruption of the NPC1 gene in human cell lines, modeling cellular defects in cholesterol storage that occur in patients with Niemann-Pick disease [51]. Multiple groups have performed CRISPR/Cas9-mediated disruption of GLA gene (which encodes alpha-galactosidase A) in either immortalized patient-derived cells or human embryonic stem cells (ESCs) to model cellular abnormalities that occur in Fabry disease (FD), a rare lysosomal storage disorder. Specifically, Lenders and co-workers [52] have used GLA-deficient cells to examine a possibility of chaperone therapy to correct FD defects, whereas GLA-deficient human ESCs were used to model autophagic dysfunction and exosome secretion in FD [53, 54]. CRISPR/Cas9-mediated disruption of GLA in human iPSCs led to the identification of additional molecular mechanisms critical for FD pathogenesis [55].
Table 2:
Use of CRISPR/Cas9 to correct pathogenic mutations in patient-derived primary cells and iPSCs.
| Disease | Species/ Cell type | Type of mutation | Reference | |
|---|---|---|---|---|
| 1. | Facioscapulohumeral dystrophy | Human / myoblasts | Pseudogene SMCHD1 deletion | 50 |
| 2. | Niemann-Pick Type C | Human/ HeLa cells | NPC1 gene disruption | 51 |
| 3. | Fabry disease | Human - HEK293T and primary urinary cells | Knockout of GLA gene | 52 |
| 4. | Fabry disease | Human Embryonic Stem Cell | Knockout of GLA gene | 53 |
| 5. | Fabry disease | Patient-derived fibroblasts and HEK-293T cells | Knockout of GLA gene | 54 |
| 6. | Fabry disease | Human-induced pluripotent stem cells/ Fibroblast | Knockout of GLA gene | 55 |
| 7. | Sickle cell disease | Human/ hematopoietic stem and progenitor cells from peripheral blood or bone marrow | HBB gene correction | 56 |
| 8. | Sickle cell disease | Human cell lines | HBB gene correction | 57 |
| 9. | Sickle cell disease | Induced pluripotent stem cells (iPSCs) | HBB gene correction | 58 |
| 10. | Sickle cell disease | Patient-derived hematopoietic stem/progenitor cells | Insertion/ deletion in HBG gene | 59 |
| 11. | Sickle cell disease | Human hematopoietic stem and progenitor cells | HBG1/HBG2 gene disruption | 60 |
| 12. | Chronic granulomatous disease | Human hematopoietic cells and induced pluripotent stem cells (iPSCs) | CYBB and NCF1 gene correction | 61, 62 |
| 13. | Junctional epidermolysis bullosa | Mouse/ Keratinocytes | LAMB3 gene correction | 63 |
| 14. | Cockayne syndrome | Human induced pluripotent stem cells (iPSCs) from fibroblasts | ERCC6 gene correction | 64 |
| 15. | Limb-Girdle Muscular Dystrophy 2G (LGMD2G) and Hermansky-Pudlak Syndrome Type 1 (HPS1) | LGMD2G iPSC-derived myotubes and HPS1 patient-derived B-lymphoblastoid cell line | TCAP gene correction | 65 |
Several research groups have reported the use of CRISPR/Cas9 genome editing to target β-globin gene (HBB) in Sickle Cell Disease (SCD) and β-thalassemia [4]. SCD is a monogenic disorder caused by a point mutation in the human β-globin gene. Using a short single-stranded oligonucleotide template, the sickle mutation in β-globin was repaired in hematopoietic stem and progenitor cells obtained from peripheral blood and bone marrow of patients with SCD [56]. Compared to β-globin mutants, erythrocytes derived from gene-edited cells showed fewer sickle cells after transplantation into non-obese diabetic mice in vivo [56]. Efficient targeting of the human β-globin gene to repair SCD point mutations by CRISPR/Cas9 genome editing was also performed by Kalkan et al [57]. Erythroid cells generated from biallelic-corrected iPSCs from SCD patients showed normal expression of β-globin [58]. Interestingly, CRISPR/Cas9-mediated genome editing of the γ-globin repressor binding site in patient-derived hematopoietic stem and progenitor cells restored fetal hemoglobin synthesis and corrected the SCD phenotype [59]. Activation of fetal hemoglobin in erythroid cell lineage was also achieved using CRISPR/Cas9-mediated de-repression of γ-globin genes HBG1 and HBG2 [60]. These studies provide novel and promising therapeutic approaches for hemoglobinopathies.
Genome editing methods have been used to study Chronic Granulomatous Disease (CGD), a rare immunodeficient disorder caused by defective NAPDH oxidase enzyme and mutations in either of five genes: CYBA, CYBB, NCF1, NCF2, or NCF4. Patient-specific point mutations in the Cytochrome b-245 heavy chain gene (CYBB) have been repaired in hematopoietic cells using non-integrating lentiviral vectors carrying RNA-guided Cas9 endonucleases [61]. In another study, iPSCs with patient-specific c.75_76delGT (ΔGT) mutation in the NCF1 gene were generated by CRISPR/Cas9 and later used to develop a gene-correction approach. Functional analysis of iPSC-derived cells revealed a restoration of NADPH oxidase activity and increased anti-bacterial properties in corrected phagocytes [62]. Recently, Benati et al. [63] reported the use of CRISPR/Cas9-mediated in situ correction of the LAMB3 gene in keratinocytes derived from a patient with Junctional Epidermolysis Bullosa (JEB), a rare disorder characterized by fragile skin and formation of blisters. CRISPR/Cas9 genome editing of the ERCC6 (CSB) gene was successfully used to correct iPSCs from a patient with Cockayne syndrome (CS), a rare autosomal recessive disorder characterized by short stature, progeria, severe photosensitivity and learning disabilities [64]. Likewise, Iyer et al. [65] employed CRISPR/Cas9 for precise correction of pathogenic mutations in cell lines derived from patients with Limb-Girdle Muscular Dystrophy 2G and Hermansky-Pudlak Syndrome Type 1. These published studies emphasize the importance of precise genome editing tools to repair pathogenic mutations in patient-derived primary cells and iPSCs.
The use of CRISPR/Cas9 to correct pathogenic mutations in vivo
CRISPR/Cas9 has been used to correct or replace dysfunctional genes in rare monogenic and other diseases [4, 13] (Table 3). In the case of recessive diseases, allogeneic cell transplantation can compensate for the mutated gene [14, 16]. This approach was explored in animal models and patients with muscular dystrophy [66–69]. Furthermore, a specifically designed viral vector can deliver normal exogenous gene (exogene) into cells or tissues to compensate for the mutated gene. Lentiviruses and adeno-associated viruses (AAV) are the most commonly used vectors for the delivery of exogenes. In dominantly inherited diseases, in which the correction of pathogenic gene variants has to be done precisely, genome editing approaches have become methods of choice.
Table 3:
Correction of pathogenic mutations in vivo using CRISPR/ Cas9
| Disease | Species | Type of mutation | Reference | |
|---|---|---|---|---|
| 1. | Duchenne muscular dystrophy (DMD) | Mouse | Dmd gene correction | 68, 69, 76, 77 |
| 2. | Tay-Sachs or Sandhoff disease | Mouse | HEXA and HEXB gene correction | 70 |
| 3. | Sandhoff disease | iPS cells/ Organoids | HEXB gene correction | 71 |
| 4. | Tay-Sachs and Sandhoff Diseases | Mouse | HEXA and HEXB gene correction | 72 |
| 5. | Tyrosinemia type I and mucopolysaccharidosis type I | Mouse | Allelic exchange in Fah and Idua genes | 73 |
| 6. | Mucopolysaccharidosis type II (MPS II) | Mouse | Ids gene correction | 74 |
| 7. | Duchenne muscular dystrophy (DMD) | Mouse | Permanent exon skipping in Dmd gene | 75 |
| 8. | Interstitial lung disease | Mouse | Inactivation of Sftpc gene | 78 |
Tay-Sachs and Sandhoff diseases are rare lipid storage disorders associated with inactivating mutations in HEXA and HEXB genes and are characterized by progressive loss of neurons in the brain and spinal cord. Several groups used CRISPR/Cas9 to generate HEXA- or HEXB-deficient immortalized cell lines as well as to perform gene corrections in patient-derived cells [70, 71]. Recently, Qu and co-workers [72] developed the CRISPR/Cas9-mediated in vivo strategy, which is based on the expression of the exogenous lysosomal enzyme in hepatocytes of neonatal Sandhoff mice. This approach decreased the disease severity in Sandhoff mice, demonstrating the future potential for the treatment of patients with Tay-Sachs and Sandhoff diseases through in vivo genome editing [72]. AAV vector delivery of CRISPR/Cas9 components repaired compound heterozygous recessive mutations and rescued the disease phenotype in mouse models of Hereditary Tyrosinemia Type I and Mucopolysaccharidosis Type I [73], whereas delivery of AAV vector carrying the iduronate-2-sulfatase gene into cerebrospinal fluid significantly attenuated brain disease in a mouse model of Mucopolysaccharidosis Type II [74]. In vivo genome editing partially restored expression of dystrophin and improved muscle function in a mouse model of Duchenne Muscular Dystrophy (DMD) [75, 76]. CRISPR/Cas9-mediated correction of DMD in mice was also performed using a self-complementary AAV delivery system [77]. In a landmark study for Interstitial Lung Disease, the delivery of CRISPR/Cas9 genome-editing components directly into the amniotic fluid led to in utero inactivation of the pathogenic SftpcI73T mutant allele, improving survival and decreasing lung fibrosis in SftpcI73T transgenic mice [78]. This study raises the possibility that in utero gene correction can be beneficial for monogenic lung diseases such as Interstitial Lung Disease, Cystic Fibrosis, and Alpha-1 Antitrypsin Deficiency.
Closing/ Conclusions
In summary, CRISPR/Cas9 genome editing technology has shown great potential for developing clinically relevant animal models that will be useful for testing new therapeutics for human rare diseases. While the correction of pathogenic mutations in cultured cells and patient-derived iPSCs are routinely performed in laboratory settings, CRISPR/Cas9 genome editing is emerging as a feasible option to target somatic cells in rare monogenic and other genetically defined human diseases. Recent progress in CRISPR/Cas9 technology has also led to clinical trials in the area of gene therapy. Several commercially available products that are based on CRISPR/Cas9 are on different stages of clinical testing to gain FDA approval [4, 79]. The global gene editing market is expected to expand at a high rate to accommodate the clinical needs of many rare diseases lacking efficient treatment options. While this technology accelerates the development of novel therapeutics for rare diseases, many important questions about the use of CRISPR/Cas9 genome editing in patients, such as efficacy, safety, and off-target effects, remain to be addressed. Furthermore, potential clinical applications of genome editing involve ethical and regulatory hurdles among other challenges [80–83]. Gene therapy applications mostly focus on targeting somatic cells, such as blood and bone marrow cells, so the genome changes cannot be inherited and passed to the next generations. Potential therapies targeting germ cells are considered to be controversial due to unknown long-term consequences that the germline gene therapy could cause.
Acknowledgments
We thank Anna Kohrs and to Erika Smith (Cincinnati Children’s Hospital Medical Center) for help with manuscript preparation and Gregory Kalin (Yale University) for critical comments. This work was supported by NIH Grants HL84151 (to V.V.K.), HL141174 (to V.V.K.), HL149631 (to V.V.K.) and HL132849 (to T.V.K.).
Footnotes
Compliance with Ethics Guidelines
Conflict of Interest
Arun Pradhan, Tanya V. Kalin and Vladimir V. Kalinichenko declare that they have no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of a an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
References
Papers of particular interest, published recently, have been highlighted as:
* Of importance
** Of major importance
- 1.Sun W, Zheng W, Simeonov A. Drug discovery and development for rare genetic disorders. Am J Med Genet A 2017. September;173(9):2307–2322. Doi: 10.1002/ajmg.a.38326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Szajner P, Yusufzai T. Introducing Rare Diseases. Rare Dis 2013; 1: e24735 Doi: 10.4161/rdis.24735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moliner AM, Waligora J. The European Union policy in the field of rare diseases. Adv Exp Med Biol 2017; 1031:561–587. Doi: 10.1007/978-3-319-67144-4_30. [DOI] [PubMed] [Google Scholar]
- 4.Papasavva P, Kleanthous M, Lederer CW. Rare Opportunities: CRISPR/Cas-Based Therapy Development for Rare Genetic Diseases. Mol Diagn Ther 2019. April;23(2):201–222. Doi: 10.1007/s40291-019-00392-3.** Comprehensive review about CRISPR/Cas9 technology in rare diseases.
- 5.Schieppati A, Henter JI, Daina E, Aperia A. Why rare diseases are an important medical and social issue. Lancet 2008. June 14;371(9629):2039–41. Doi: 10.1016/S0140-6736(08)60872-7. [DOI] [PubMed] [Google Scholar]
- 6.Dharmadhikari AV, Szafranski P, Kalinichenko VV, Stankiewicz P. Genomic and Epigenetic Complexity of the FOXF1 Locus in 16q24.1: Implications for Development and Disease. Curr Genomics 2015. April;16(2):107–16. Doi: 10.2174/1389202916666150122223252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pradhan A, Dunn A, Ustiyan V, Bolte C, Wang G, et al. The S52F FOXF1 Mutation Inhibits STAT3 Signaling and Causes Alveolar Capillary Dysplasia. Am J Respir Crit Care Med 2019. October 15;200(8):1045–1056. Doi: 10.1164/rccm.201810-1897OC.** Generation of mouse model of ACDMPV using CRISPR/ Cas9 approach, and improvement of pulmonary vasculature by nanoparticle-mediated gene delivery.
- 8.Cuthbert AW. New horizons in the treatment of cystic fibrosis. Br J Pharmacol 2011. May;163(1):173–83. Doi: 10.1111/j.1476-5381.2010.01137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Runz H, Dolle D, Schlitter AM, Zschocke J. NPC-db, a Niemann-Pick type C disease gene variation database. Hum Mutat 2008. March;29(3):345–50. Doi: 10.1002/humu.20636. [DOI] [PubMed] [Google Scholar]
- 10.Chabannon C, Kuball J, Bondanza A, Dazzi F, Pedrazzoli P, Toubert A, et al. Hematopoietic stem cell transplantation in its 60s: A platform for cellular therapies. Sci Transl Med 2018. April 11;10(436). Doi: 10.1126/scitranslmed.aap9630. [DOI] [PubMed] [Google Scholar]
- 11.Bernstein DL, Lobritto S, Iuga A, Remotti H, Schiano T, Fiel MI, Balwani M. Lysosomal acid lipase deficiency allograft recurrence and liver failure- clinical outcomes of 18 liver transplantation patients. Mol Genet Metab 2018. May;124(1):11–19. Doi: 10.1016/j.ymgme.2018.03.010. Epub 2018 Mar 27. [DOI] [PubMed] [Google Scholar]
- 12.Choi KA, Choi Y, Hong S. Stem cell transplantation for Huntington’s diseases. Methods 2018. January 15; 133:104–112. Doi: 10.1016/j.ymeth.2017.08.017. Epub 2017 Sep 1. [DOI] [PubMed] [Google Scholar]
- 13.Santos R, Amaral O. Advances in Sphingolipidoses: CRISPR-Cas9 Editing as an Option for Modelling and Therapy. Int J Mol Sci 2019. November 24;20(23). pii: E5897 Doi: 10.3390/ijms20235897.** Comprehensive review about CRISPR/ Cas9 technology in generation of animal models of inherited genetic diseases caused by accumulation of glycosphingolipids.
- 14.Cox DB, Platt RJ, Zhang F. Therapeutic genome editing: prospects and challenges. Nat Med 2015. February;21(2):121–31. Doi: 10.1038/nm.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doudna JA, Gersbach CA. Genome editing: the end of the beginning. Genome Biol 2015. December 23; 16:292 Doi: 10.1186/s13059-015-0860-5.* Comprehensive review about genome editing approaches.
- 16.Kanchiswamy CN, Maffei M, Malnoy M, Velasco R, Kim JS. Fine-Tuning Next-Generation Genome Editing Tools. Trends Biotechnol 2016. July;34(7):562–574. Doi: 10.1016/j.tibtech.2016.03.007. Epub 2016 May 8.* Comprehensive review about recent advances of genome editing tools.
- 17.Greely HT. Human Germline Genome Editing: An Assessment. CRISPR J 2019. October;2(5):253–265. Doi: 10.1089/crispr.2019.0038. [DOI] [PubMed] [Google Scholar]
- 18.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 2014. November 28;346(6213):1258096 Doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 19.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell 2014. June 5;157(6):1262–78. Doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barrangou R, Dudley EG. CRISPR-based typing and next-generation tracking technologies. Annu Rev Food Sci Technol 2016; 7:395–411. Doi: 10.1146/annurev-food-022814-015729. Epub 2016 Jan 11. [DOI] [PubMed] [Google Scholar]
- 21.Kang Y, Chu C, Wang F, Niu Y. CRISPR/Cas9-mediated genome editing in nonhuman primates. Dis Model Mech 2019. October 16;12(10). pii: dmm039982 Doi: 10.1242/dmm.039982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tseng WC, Loeb HE, Pei W, Tsai-Morris CH, Xu L, Cluzeau CV, et al. Modeling Niemann-Pick disease type C1 in zebrafish: a robust platform for in vivo screening of candidate therapeutic compounds. Dis Model Mech 2018. August 15;11(9). pii: dmm034165 Doi: 10.1242/dmm.034165.** Generation of zebrafish model of NPC1 for screening of therapeutic compounds.
- 23.Kim YK, Yu JH, Min SH, Park SW. Generation of a GLA knock-out human-induced pluripotent stem cell line, KSBCi002-A-1, using CRISPR/Cas9. Stem Cell Res 2020. January; 42:101676 Doi: 10.1016/j.scr.2019.101676. Epub 2019 Dec 4. [DOI] [PubMed] [Google Scholar]
- 24.Kodama K, Takahashi H, Oiji N, Nakano K, Okamura T, Niimi K, et al. CANT1 deficiency in a mouse model of Desbuquois dysplasia impairs glycosaminoglycan synthesis and chondrocyte differentiation in growth plate cartilage. FEBS Open Bio 2020. April 11 Doi: 10.1002/2211-5463.12859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dreano E, Bacchetta M, Simonin J, Galmiche L, Usal C, Slimani L, et al. Characterization of two rat models of cystic fibrosis—KO and F508del CFTR—Generated by Crispr-Cas9. Animal Model Exp Med 2019;2:297–311. Doi: 10.1002/ame2.12091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zabaleta N, Barberia M, Martin-Higueras C, Zapata-Linares N, Betancor I, Rodriguez S, et al. CRISPR/Cas9-mediated glycolate oxidase disruption is an efficacious and safe treatment for primary hyperoxaluria type I. Nat Commun 2018. December 21;9(1):5454 Doi: 10.1038/s41467-018-07827-1.* The paper demonstrates that CRISPR/Cas9-mediated substrate reduction therapy in Agxt1−/− mice is a promising therapeutic option for primary hyperoxaluria type I.
- 27.Schlegel J, Hoffmann J, Röll D, Müller B, Günther S, Zhang W, et al. Toward genome editing in X-linked RP-development of a mouse model with specific treatment relevant features. Transl Res 2019. January; 203:57–72. Doi: 10.1016/j.trsl.2018.08.006. Epub 2018 Aug 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sen P, Dharmadhikari AV, Majewski T, Mohammad MA, Kalin TV, Zabielska J, et al. Comparative analyses of lung transcriptomes in patients with alveolar capillary dysplasia with misalignment of pulmonary veins and in foxf1 heterozygous knockout mice. PLoS One 2014. April 10; 9(4):e94390 Doi: 10.1371/journal.pone.0094390. eCollection 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bolte C, Whitsett JA, Kalin TV, Kalinichenko VV. Transcription Factors Regulating Embryonic Development of Pulmonary Vasculature. Adv Anat Embryol Cell Biol 2018;228:1–20. Doi: 10.1007/978-3-319-68483-3_1. [DOI] [PubMed] [Google Scholar]
- 30.Whitsett JA, Kalin TV, Xu Y, Kalinichenko VV. Building and Regenerating the Lung Cell by Cell. Physiol Rev 2019. January 1;99(1):513–554. Doi: 10.1152/physrev.00001.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim IM, Zhou Y, Ramakrishna S, Hughes DE, Solway J, Costa RH, Kalinichenko VV. Functional characterization of evolutionarily conserved DNA regions in forkhead box f1 gene locus. J Biol Chem 2005. November 11;280(45):37908–16. Epub 2005 Sep 6. Doi: 10.1074/jbc.M506531200 [DOI] [PubMed] [Google Scholar]
- 32.Kalinichenko VV, Gusarova GA, Shin B, Costa RH. The forkhead box F1 transcription factor is expressed in brain and head mesenchyme during mouse embryonic development. Gene Expr Patterns 2003. May;3(2):153–8. Doi: 10.1016/s1567-133x(03)00010-3 [DOI] [PubMed] [Google Scholar]
- 33.Hoggatt AM, Kim JR, Ustiyan V, Ren X, Kalin TV, Kalinichenko VV, Herring BP. The transcription factor Foxf1 binds to serum response factor and myocardin to regulate gene transcription in visceral smooth muscle cells. J Biol Chem 2013. October 4;288(40):28477–87. Doi: 10.1074/jbc.M113.478974. Epub 2013 Aug 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bolte C, Ren X, Tomley T, Ustiyan V, Pradhan A, Hoggatt A, et al. Forkhead box F2 regulation of platelet-derived growth factor and myocardin/serum response factor signaling is essential for intestinal development. J Biol Chem 2015. March 20;290(12):7563–75. Doi: 10.1074/jbc.M114.609487. Epub 2015 Jan 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoffmann AD, Yang XH, Burnicka-Turek O, Bosman JD, Ren X, Steimle JD, et al. Foxf genes integrate tbx5 and hedgehog pathways in the second heart field for cardiac septation. PLoS Genet 2014. October 30;10(10): e1004604 Doi: 10.1371/journal.pgen.1004604. eCollection 2014 Oct. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu J, Liu H, Lan Y, Aronow BJ, Kalinichenko VV, Jiang R. A Shh-Foxf-Fgf18-Shh Molecular Circuit Regulating Palate Development . PLoS Genet 2016. January 8;12(1):e1005769 Doi: 10.1371/journal.pgen.1005769. eCollection 2016 Jan. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dharmadhikari AV, Sun JJ, Gogolewski K, Carofino BL, Ustiyan V, Hill M, et al. Lethal Lung Hypoplasia and Vascular Defects in Mice with Conditional Foxf1 Overexpression. Biol Open 2016. November 15;5(11):1595–1606. Doi: 10.1242/bio.019208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ustiyan V, Bolte C, Zhang Y, Han L, Xu Y, Yutzey KE, et al. FOXF1 Transcription Factor Promotes Lung Morphogenesis by Inducing Cellular Proliferation in Fetal Lung Mesenchyme. Dev Biol 2018. November 1;443(1):50–63. Doi: 10.1016/j.ydbio.2018.08.011. Epub 2018 Aug 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Milewski D, Pradhan A, Wang X, Cai Y, Le T, Turpin B, et al. FoxF1 and FoxF2 Transcription Factors Synergistically Promote Rhabdomyosarcoma Carcinogenesis by Repressing Transcription of p21Cip1 CDK Inhibitor. Oncogene 2017. February 9;36(6):850–862. Doi: 10.1038/onc.2016.254. Epub 2016 Jul 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pradhan A, Ustiyan V, Zhang Y, Kalin TV, Kalinichenko VV. Forkhead Transcription Factor FoxF1 Interacts With Fanconi Anemia Protein Complexes to Promote DNA Damage Response. Oncotarget 2016. January 12; 7(2):1912–26. Doi: 10.18632/oncotarget.6422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Flood HM, Bolte C, Dasgupta N, Sharma A, Zhang Y, Gandhi CR, et al. The Forkhead box F1 transcription factor inhibits collagen deposition and accumulation of myofibroblasts during liver fibrosis. Biol Open 2019. February 11;8(2). pii: bio039800 Doi: 10.1242/bio.039800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bolte C, Kalin TV, Kalinichenko VV. Molecular, cellular, and bioengineering approaches to stimulate lung regeneration after injury. Semin Cell Dev Biol 2020. April; 100:101–108. Doi: 10.1016/j.semcdb.2019.10.006. Epub 2019 Oct 25. [DOI] [PubMed] [Google Scholar]
- 43.Kalinichenko VV, Zhou Y, Shin B, Stolz DB, Watkins SC, Whitsett JA, Costa RH. Wildtype levels of the mouse Forkhead Box f1 gene are essential for lung repair. Am J Physiol Lung Cell Mol Physiol 2002. June;282(6):L1253–65. Doi: 10.1152/ajplung.00463.2001. [DOI] [PubMed] [Google Scholar]
- 44.Kalin TV, Meliton L, Meliton AY, Zhu X, Whitsett JA, Kalinichenko VV. Pulmonary mastocytosis and enhanced lung inflammation in mice heterozygous null for the Foxf1 gene. Am J Respir Cell Mol Biol 2008. October;39(4):390–9. Doi: 10.1165/rcmb.2008-0044OC. Epub 2008 Apr 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cai Y, Bolte C, Le T, Goda C, Xu Y, Kalin TV, Kalinichenko VV. FOXF1 maintains endothelial barrier function and prevents edema after lung injury. Sci Signal 2016. April 19;9(424):ra40 Doi: 10.1126/scisignal.aad1899. [DOI] [PubMed] [Google Scholar]
- 46.Black M, Milewski D, Le T, Ren X, Xu Y, Kalinichenko VV, Kalin TV. FOXF1 Inhibits Pulmonary Fibrosis by Preventing CDH2-CDH11 Cadherin Switch in Myofibroblasts. Cell Rep 2018. April 10;23(2):442–458. Doi: 10.1016/j.celrep.2018.03.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bolte C, Ustiyan V, Ren X, Dunn AW, Pradhan A, Wang G. et al. Nanoparticle Delivery of Proangiogenic Transcription Factors into the Neonatal Circulation Inhibits Alveolar Simplification Caused by Hyperoxia. Am J Respir Crit Care Med 2020. April 2 Doi: 10.1164/rccm.201906-1232OC.** The paper provides evidence that nanoparticle-mediated delivery of proangiogenic transcription factors stimulates lung angiogenesis and alveolarization during recovery from neonatal hyperoxic injury.
- 48.Dunn AW, Kalinichenko VV, Shi D. Highly Efficient In Vivo Targeting of the Pulmonary Endothelium Using Novel Modifications of Polyethylenimine: An Importance of Charge. Adv Healthc Mater 2018. December;7(23): e1800876 Doi: 10.1002/adhm.201800876. Epub 2018 Nov 6. [DOI] [PubMed] [Google Scholar]
- 49.Ren X, Ustiyan V, Guo M, Wang G, Bolte C, Zhang Y et al. Postnatal Alveologenesis Depends on FOXF1 Signaling in c-KIT+ Endothelial Progenitor Cells. Am J Respir Crit Care Med 2019. November 1;200(9):1164–1176. Doi: 10.1164/rccm.201812-2312OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goossens R, van den Boogaard ML, Lemmers RJLF, Balog J, van der Vliet PJ, Willemsen IM, et al. Intronic SMCHD1 variants in FSHD: testing the potential for CRISPR-Cas9 genome editing. J Med Genet 2019. December;56(12):828–837. Doi: 10.1136/jmedgenet-2019-106402. Epub 2019 Nov 1. [DOI] [PubMed] [Google Scholar]
- 51.Du X, Lukmantara I, Yang H. CRISPR/Cas9-Mediated Generation of Niemann-Pick C1 Knockout Cell Line. Methods Mol Biol 2017; 1583:73–83. Doi: 10.1007/978-1-4939-6875-6_7. [DOI] [PubMed] [Google Scholar]
- 52.Lenders M, Stappers F, Niemietz C, Schmitz B, Boutin M, Ballmaier PJ, et al. Mutation-specific Fabry disease patient-derived cell model to evaluate the amenability to chaperone therapy. J Med Genet 2019. August;56(8):548–556. Doi: 10.1136/jmedgenet-2019-106005. Epub 2019 Apr 22. [DOI] [PubMed] [Google Scholar]
- 53.Song HY, Chien CS, Yarmishyn AA, Chou SJ, Yang YP, Wang ML, et al. Generation of GLA-Knockout Human Embryonic Stem Cell Lines to Model Autophagic Dysfunction and Exosome Secretion in Fabry Disease-Associated Hypertrophic Cardiomyopathy. Cells 2019. April 8;8(4). pii: E327 Doi: 10.3390/cells8040327.** The paper provides evidence that GLA−/− human embryonic stem cells (generated using CRISPR/ Cas9) can be used as a promising tool to study hypertrophic cardiomyopathy and to develop new therapies for Fabry disease.
- 54.Song HY, Chiang HC, Tseng WL, Wu P, Chien CS, Leu HB, et al. Using CRISPR/Cas9-Mediated GLA Gene Knockout as an In Vitro Drug Screening Model for Fabry Disease. Int J Mol Sci 2016. December 13;17(12). pii: E2089 Doi: 10.3390/ijms17122089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim YK, Yu JH, Min SH, Park SW. Generation of a GLA knock-out human-induced pluripotent stem cell line, KSBCi002-A-1, using CRISPR/Cas9. Stem Cell Res 2020. January; 42:101676 Doi: 10.1016/j.scr.2019.101676. Epub 2019 Dec 4. [DOI] [PubMed] [Google Scholar]
- 56.Park SH, Lee CM, Dever DP, Davis TH, Camarena J, Srifa W, et al. Highly efficient editing of the β-globin gene in patient-derived hematopoietic stem and progenitor cells to treat sickle cell disease. Nucleic Acids Res 2019. September 5;47(15):7955–7972. Doi: 10.1093/nar/gkz475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kalkan BM, Kala EY, Yuce M, Karadag Alpaslan M, Kocabas F. Development of gene editing strategies for human β-globin (HBB) gene mutations. Gene 2020. April 15; 734:144398 Doi: 10.1016/j.gene.2020.144398. Epub 2020 Jan 24. [DOI] [PubMed] [Google Scholar]
- 58.Haro-Mora JJ, Uchida N, Demirci S, Wang Q, Zou J, Tisdale JF. Biallelic correction of sickle cell disease-derived iPSCs confirmed at the protein level through serum-free iPS-sac/erythroid differentiation. Stem Cells Transl Med 2020. February 7 Doi: 10.1002/sctm.19-0216. [DOI] [Google Scholar]
- 59.Weber L, Frati G, Felix T, Hardouin G, Casini A, Wollenschlaeger C, et al. Editing a γ-globin repressor binding site restores fetal hemoglobin synthesis and corrects the sickle cell disease phenotype. Sci Adv 2020. February 12;6(7): eaay9392 Doi: 10.1126/sciadv.aay9392. eCollection 2020 Feb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Métais JY, Doerfler PA, Mayuranathan T, Bauer DE, Fowler SC, Hsieh MM, et al. Genome editing of HBG1 and HBG2 to induce fetal hemoglobin. Blood Adv 2019. November 12;3(21):3379–3392. Doi: 10.1182/bloodadvances.2019000820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sürün D, Schwäble J, Tomasovic A, Ehling R, Stein S, Kurrle N, et al. High Efficiency Gene Correction in Hematopoietic Cells by Donor-Template-Free CRISPR/Cas9 Genome Editing. Mol Ther Nucleic Acids 2018. March 2; 10:1–8. Doi: 10.1016/j.omtn.2017.11.001. Epub 2017 Nov 10.** The paper demonstrated the possibility of CRISPR/Cas9-mediated repair of patient-specific point mutations in CYBB gene, whose inactivation causes chronic granulomatous disease (XCGD).
- 62.Klatt D, Cheng E, Philipp F, Selich A, Dahlke J, Schmidt RE, et al. Targeted Repair of p47-CGD in iPSCs by CRISPR/Cas9: Functional Correction without Cleavage in the Highly Homologous Pseudogenes. Stem Cell Reports 2019. October 8;13(4):590–598. Doi: 10.1016/j.stemcr.2019.08.008. Epub 2019 Sep 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Benati D, Miselli F, Cocchiarella F, Patrizi C, Carretero M, Baldassarri S, et al. CRISPR/Cas9-Mediated in Situ Correction of LAMB3 Gene in Keratinocytes Derived from a Junctional Epidermolysis Bullosa Patient. Mol Ther 2018. November 7;26(11):2592–2603. Doi: 10.1016/j.ymthe.2018.07.024. Epub 2018 Aug 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang S, Min Z, Ji Q, Geng L, Su Y, Liu Z, et al. Rescue of premature aging defects in Cockayne syndrome stem cells by CRISPR/Cas9-mediated gene correction. Protein Cell 2020. January;11(1):1–22. Doi: 10.1007/s13238-019-0623-2. Epub 2019 Apr 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Iyer S, Suresh S, Guo D, Daman K, Chen JCJ, Liu P, et al. Precise therapeutic gene correction by a simple nuclease-induced double-stranded break. Nature 2019. April;568(7753):561–565. Doi: 10.1038/s41586-019-1076-8. Epub 2019 Apr 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ferrari G, Muntoni F, Tedesco FS. Generation of two genomic-integration-free DMD iPSC lines with mutations affecting all dystrophin isoforms and potentially amenable to exon-skipping. Stem Cell Res 2020. March; 43:101688 Doi: 10.1016/j.scr.2019.101688. Epub 2020 Feb 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Morisaka H, Yoshimi K, Okuzaki Y, Gee P, Kunihiro Y, Sonpho E, et al. CRISPR-Cas3 induces broad and unidirectional genome editing in human cells. Nat Commun 2019. December 6;10(1):5302 Doi: 10.1038/s41467-019-13226-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Amoasii L, Li H, Zhang Y, Min YL, Sanchez-Ortiz E, Shelton JM, et al. In vivo non-invasive monitoring of dystrophin correction in a new Duchenne muscular dystrophy reporter mouse. Nat Commun 2019. October 4;10(1):4537 Doi: 10.1038/s41467-019-12335-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Min YL, Li H, Rodriguez-Caycedo C, Mireault AA, Huang J, Shelton JM, et al. CRISPR-Cas9 corrects Duchenne muscular dystrophy exon 44 deletion mutations in mice and human cells. Sci Adv 2019. March 6;5(3): eaav4324 Doi: 10.1126/sciadv.aav4324. eCollection 2019 Mar. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tropak MB, Yonekawa S, Karumuthil-Melethil S, Thompson P, Wakarchuk W, Gray SJ, et al. Construction of a hybrid β-hexosaminidase subunit capable of forming stable homodimers that hydrolyze GM2 ganglioside in vivo. Mol Ther Methods Clin Dev 2016. March 2;3:15057 Doi: 10.1038/mtm.2015.57. eCollection 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Allende ML, Cook EK, Larman BC, Nugent A, Brady JM, Golebiowski D, et al. Cerebral organoids derived from Sandhoff disease-induced pluripotent stem cells exhibit impaired neurodifferentiation. J Lipid Res 2018. March;59(3):550–563. Doi: 10.1194/jlr.M081323. Epub 2018 Jan 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ou L, Przybilla MJ, Tăbăran AF, Overn P, O’Sullivan MG, Jiang X, et al. A novel gene editing system to treat both Tay-Sachs and Sandhoff diseases. Gene Ther 2020. January 2 Doi: 10.1038/s41434-019-0120-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang D, Li J, Song CQ, Tran K, Mou H, Wu PH, et al. Cas9-mediated allelic exchange repairs compound heterozygous recessive mutations in mice. Nat Biotechnol 2018. October;36(9):839–842. Doi: 10.1038/nbt.4219. Epub 2018 Aug 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hinderer C, Katz N, Louboutin JP, Bell P, Yu H, Nayal M, et al. Delivery of an Adeno-Associated Virus Vector into Cerebrospinal Fluid Attenuates Central Nervous System Disease in Mucopolysaccharidosis Type II Mice. Hum Gene Ther 2016. November;27(11):906–915. Epub 2016 Aug 10. Doi: 10.1089/hum.2016.101. [DOI] [PubMed] [Google Scholar]
- 75.Long C, Amoasii L, Mireault AA, McAnally JR, Li H, Sanchez-Ortiz E, et al. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science 2016. January 22;351(6271):400–3. Doi: 10.1126/science.aad5725. Epub 2015 Dec 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nelson CE, Hakim CH, Ousterout DG, Thakore PI, Moreb EA, Castellanos Rivera RM, et al. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science 2016. January 22;351(6271):403–7. Doi: 10.1126/science.aad5143. Epub 2015 Dec 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang Y, Li H, Min YL, Sanchez-Ortiz E, Huang J, Mireault AA, et al. Enhanced CRISPR-Cas9 correction of Duchenne muscular dystrophy in mice by a self-complementary AAV delivery system. Sci Adv 2020. February 19;6(8): eaay6812 Doi: 10.1126/sciadv.aay6812. eCollection 2020 Feb.** The paper demonstrates that a low dose of scAAV-delivered genome editing components is sufficient to restore dystrophin protein expression and improve the muscle function in a DMD mouse model.
- 78.Alapati D, Zacharias WJ, Hartman HA, Rossidis AC, Stratigis JD, Ahn NJ, et al. In utero gene editing for monogenic lung disease. Sci Transl Med 2019. April 17;11(488). pii: eaav8375 Doi: 10.1126/scitranslmed.aav8375.** In utero CRISPR-Cas9-mediated inactivation of the mutant SftpcI73T gene led to improved lung morphology and respiratory function.
- 79.Shahryari A, Saghaeian Jazi M, Mohammadi S, Razavi Nikoo H, Nazari Z, Hosseini ES, et al. Development and Clinical Translation of Approved Gene Therapy Products for Genetic Disorders. Front Genet 2019. September 25; 10:868 Doi: 10.3389/fgene.2019.00868. eCollection 2019.* Comprehensive review about gene therapies and cell-based gene therapy products that have been approved for clinical use.
- 80.Smith DM, Culme-Seymour EJ, Mason C. Evolving industry partnerships and investments in cell and gene therapies. Cell Stem Cell 2018. May 3;22(5):623–626. Doi: 10.1016/j.stem.2018.03.004. Epub 2018 Apr 12. [DOI] [PubMed] [Google Scholar]
- 81.Capps B Can a good tree bring forth evil fruit? The funding of medical research by industry. Br Med Bull 2016. June;118(1):5–15. Doi: 10.1093/bmb/ldw014. Epub 2016 May 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Capps B, Chadwick R, Joly Y, Mulvihill JJ, Lysaght T, Zwart H. Falling giants and the rise of gene editing: ethics, private interests and the public good. Hum Genomics 2017. August 29;11(1):20 Doi: 10.1186/s40246-017-0116-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Riva Luciana, Petrini Carlo. A few ethical issues in translational research for gene and cell therapy. J Transl Med 2019. November 28;17(1):395 Doi: 10.1186/s12967-019-02154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]