Abstract
Background
Chlorophyll is the most important factor enabling plants to absorb, transfer and transform light energy and plays an important role in yield formation. Brassica napus is one of the most important oil crops. Breeding Brassica napus for high light efficiency by improving photosynthetic efficiency has considerable social and economic value. In Brassica napus, there have been studies of the initial location of chlorophyll in seed embryos and pericarps, but there are few reports on the fine mapping of chlorophyll QTLs. We constructed near-isogenic lines (NIL), fine-mapped a chlorophyll locus, and evaluated the effect of this dominant locus on agronomic traits.
Results
The cqSPDA2 locus was mapped to an interval of 21.87–22.91 Mb on the chromosome A02 of Brassica napus using doubled haploid (DH) lines. To fine-map cqSPDA2, we built NIL and designed Indel primers covering the mapping interval. The 469 individuals in the BC3F2 population were analyzed using these indel primers. Among these indel primers, 15 could narrow the mapping interval to 188 kb between Indel3 and Indel15. Next, 16 indel primers and 19 SSR primers were designed within the new narrower mapping interval, and 5 of the primer-amplified fragments were found to be polymorphic and tightly linked to the cqSPDA2 locus in the BC4F2 population. The mapping interval was narrowed to 152 kb on A02 between SSR2 and Indel15. By gene expression analysis, we found three annotated genes in the mapping interval, including BnaA02g30260D, BnaA02g30290D and BnaA02g30310D, which may be responsible for chlorophyll synthesis.
Conclusions
The locus cqSPDA2, a dominant QTL for chlorophyll content in Brassica napus, was fine-mapped to a 21.89–22.04 Mb interval on A02. Three annotated genes (BnaA02g30260D, BnaA02g30290D and BnaA02g30310D) that may be responsible for chlorophyll synthesis were found.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12870-020-02710-y.
Keywords: Brassica napus, Chlorophyll content, Near-isogenic line, Fine mapping, qRT-PCR
Background
The material basis of crop yield formation is derived from photosynthesis, and high yield based on photosynthesis has long been a hot topic in crop breeding [1]. Chlorophyll is the most important factor enabling plants to absorb, transfer and transform light energy and plays an important role in the growth and development of plants [2]. Maintaining a high level of chlorophyll content in leaves is an important factor in increasing photosynthetic activity [3]. Within a certain range, there is a positive correlation between chlorophyll content and photosynthetic rate, which directly determines the yield [4, 5]. Therefore, chlorophyll content plays an important role in yield formation [6, 7]. Robust seedling development in Brassica napus also leads to higher yield stability and has a high importance for plant breeders. Chlorophyll content is a quantitative characteristic that is primarily controlled by nuclear genes and has high heritability [8, 9]. Previous studies have suggested that at least 27 genes are involved in 15 steps of chlorophyll synthesis [10], and biosynthetic defects are considered to be one of the main reasons for low chlorophyll content. Other reasons for chlorophyll deficiency include deficient signal transduction, restrained heme feedback, harmful photooxidation and so on. Overall, the molecular mechanisms of chlorophyll synthesis are very complex [11]. In recent years, researchers have analyzed QTLs for chlorophyll content in the seedling leaves of different populations of multiple crops from different perspectives, making considerable progress and establishing a foundation for future research attempting to elucidate the molecular genetic mechanisms that determine of chlorophyll content [12–18].
The completion of the whole genome sequencing of Brassica napus indicates that research on the Brassica napus genome has entered a new era. In recent years, with the rapid development of molecular marker technology, it has became possible to construct a high-density molecular marker genetic map of Brassica napus. Therefore, the study of chlorophyll QTLs that will allow efficient breeding of Brassica napus for greater light efficiency will provide important information for improving the yield potential and direction of high-yield Brassica napus breeding in the future. Breeding Brassica napus for high light efficiency by improving photosynthetic efficiency is socially and economically important. At present, studies of the initial location of chlorophyll content in the seed embryos [19] and pericarps [20] of winter Brassica napus have been conducted, QTLs have been identified under drought and salt stress, and candidate genes related to salt tolerance have even been predicted [21–23]. However, there are few reports of the fine mapping of chlorophyll content QTLs in Brassica napus [11].
In a previous study, we discovered a dominant major QTL named cqSPDA2 located in 21.87–22.91 Mb on A02 using DH lines derived from a cross between Zhongshuang11 (ZS11, a semi-winter variety) and QU (a spring variety), which explained 15.72% variance of phenotype and was detected in six environments stably (under review). In this study, a near- isogenic line (NIL) population for cqSPDA2 was constructed to further narrow the interval of cqSPDA2. This study established a foundation for the cloning of chlorophyll genes controlling photosynthetic function and provided a theoretical basis for improving germplasm resources and selecting new high-yield varieties with molecular markers.
Results
Phenotypic and genetic analysis
The first fully developed leaves counting from the tops of the 2061 individuals in the BC4F2 population at the six-leaf stage were measured by SPAD (SPAD 502, Japan). The distribution of chlorophyll content in the BC4F2 population was bimodal, with the lowest point of the column chart as the dividing line (SPAD = 43). SPAD≥43 was regarded as the high chlorophyll content (n = 1514), and SPAD<43 was regarded as the low chlorophyll content (n = 547). A chi-square test showed that the segregation pattern of the chlorophyll content trait was in keeping with the expected Mendelian segregation ratio of 3:1 (χ2 = 2.53) (high-chlorophyll content vs. low-chlorophyll content) (Fig. 1). Among the random selection of 198 individuals in the BC6F1 population, the markers SSR2 and Indel100 were used to validate the effect of cqSPDA2. The result of a chi-square test was in keeping with a 1:1 (χ2 = 1.46; χ2 = 1.82) (AA:Aa) Mendelian ratio (Additional file 1: Table S1).
Fig. 1.
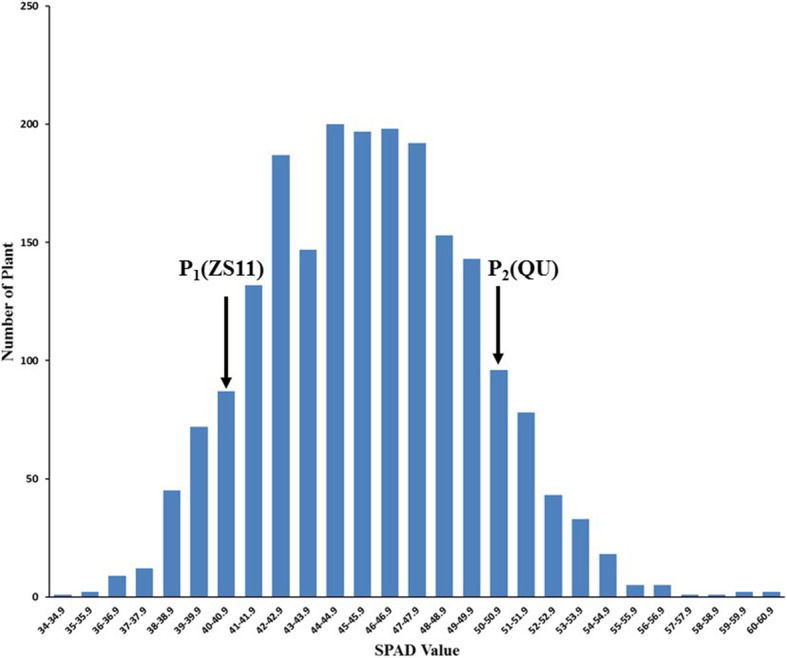
Phenotypic frequency distribution of chlorophyll SPAD in the BC4F2 population. The lowest point of the column chart represents the dividing value (SPAD = 43). Leaves were considered to have high chlorophyll content(n = 1514) with a SPAD value≥43 and a low chlorophyll content(n = 547) with a SPAD value < 43. A chi-square test showed that the segregation pattern of the chlorophyll content trait was in keeping with the expected Mendelian segregation ratio of 3:1 (χ2 = 2.53)
Fine mapping of cqSPDA2
To fine-map the cqSPDA2 locus and identify the candidate genes, 87 primer pairs of indel markers were designed to uniformly cover the preliminary mapping interval (A02 21.87–22.91 Mb). As a result, 28 polymorphic markers were detected in the two parental lines and selected DH lines. Twenty-three of these markers were found to cosegregated in the DH lines. The 469 individuals in the BC3F2 population were then analyzed using these indel primers. The linkage map constructed using the indel data and corresponding chlorophyll content phenotypes showed that 15 of the indel primers were tightly linked to the cqSPDA2 locus (Additional file 2: Table S2). The cqSPDA2 locus was delimited to an interval of 5.2 cM between Indel3 and Indel15 (Additional file 3: Fig. S1). The fragments amplified by the primers Indel3, Indel6, Indel15 and Indel17 near the QTL were recovered. TA cloning was performed with the PMD18-T vector, and the physical location of the region was found to be within the 188 kb range of 21.88–22.07 Mb (Additional file 4: Fig. S2).
Next, 16 indel and 19 SSR primers were designed within the new, narrow mapping interval, and the fragments amplified from 5 primers were polymorphic and tightly linked to the cqSPDA2 locus (Additional file 5: Table S3). These new primers helped to narrow the interval for the 250 individuals in the BC4F2 population. As a result, the cqSPDA2 locus was mapped to a 152 kb interval between SSR2 and Indel15 (Fig. 2). BSNP88 and BSNP90 were developed kompetitive allele-specific PCR (KASP) markers based on SNP analysis in theis interval (Additional file 6: Table S4). SSR2 is a codominant marker closely linked to cqSPDA2. Twenty BC4F2 plants with low-, medium- and high-chlorophyll phenotypes were selected, and their SSR2 genotypes were determined. The results showed that the three groups of different phenotypes can be divided into three genotypes: AA (dominant homozygous, high- chlorophyll phenotype), Aa (heterozygote, medium-chlorophyll phenotype) and aa (recessive homozygous, low-chlorophyll phenotype). These results suggested that SSR2 was closely linked to cqSPDA2 and could be effectively used in marker-assisted selection (MAS) (Fig. 3).
Fig. 2.
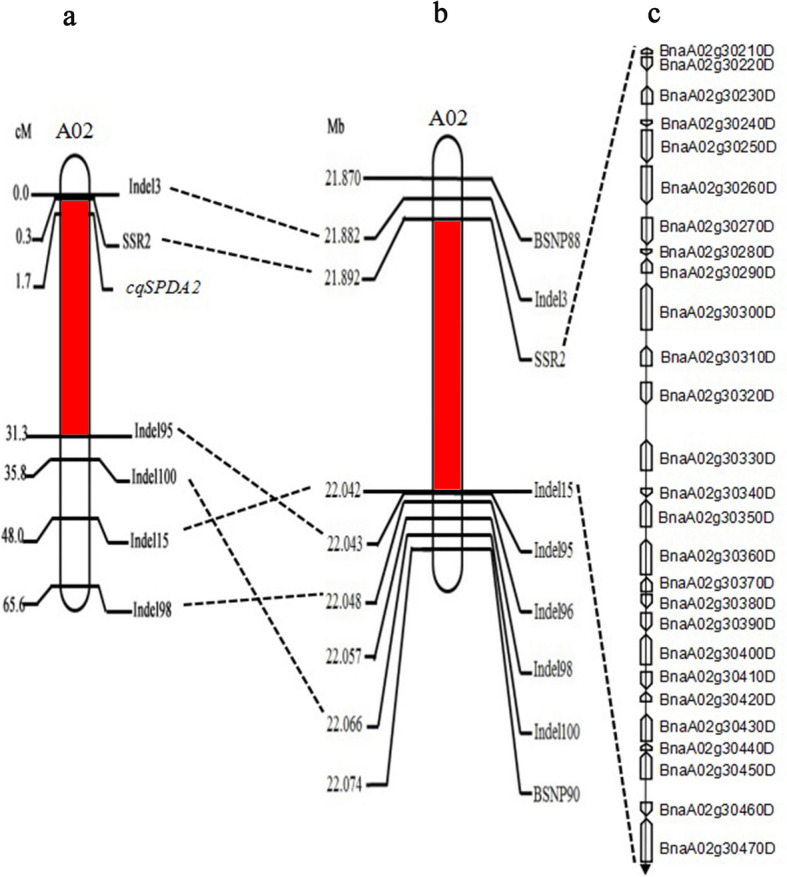
Genetic and physical maps of the cqSPDA2 gene locus and candidate gene analysis. a Genetic linkage map of the cqSPDA2 region on chromosome A02. The numbers between the markers indicate genetic distance in centimorgans. b Fine mapping of the cqSPDA2 locus in the BC4F2 population. The cqSPDA2 was narrowed down to a 152-kb interval between the markers SSR2 and Indel15. The numbers between the markers indicate physical distance. c Genetic and physical maps of the candidate genes in the targeted interval and the annotated genes in the Brassica napus genome annotation database (http://www.genoscope.cns.fr/brassicanapus/)
Fig. 3.
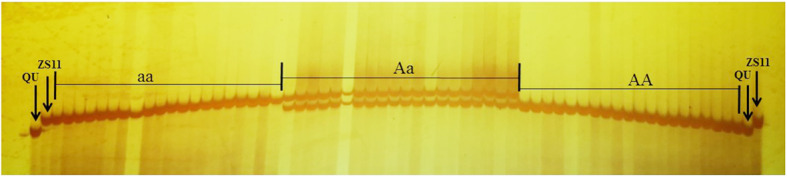
Codominant marker and closely linked to cqSPDA2. The aa group (low-chlorophyll phenotype): the mean of SPAD = 40.0 ± 0.27; the Aa group (medium-chlorophyll phenotype): the mean of SPAD = 44.8 ± 1.44; the AA group (high chlorophyll phenotype): the mean of SPAD = 53.3 ± 0.80
Quantitative RT-PCR of genes in the mapping interval
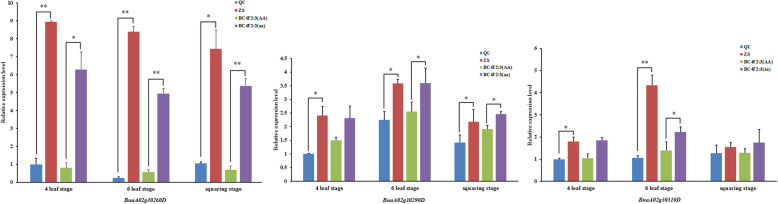
According to the Brassica napus genome annotation database (http://www.genoscope.cns.fr/ brassicanapus/), twenty-seven genes were identified in the targeted mapping interval of 21.89–22.04 Mb on A02 (Additional file 7: Table S5). Quantitative real-time PCR (qRT-PCR) was used to identify the expression levels of the genes in the targeted mapping interval between the leaves of different genotypes. The melting and amplification curves of twenty-seven genes were analyzed, and the results showed that 24 primer pairs could be used to analyze gene expression (Additional file 8: Table S6). Thus, twenty-four genes in the mapping interval and the housekeeping gene Actin7 were quantified by qRT-PCR (Additional file 9: Table S7). The results showed that the expression levels of the three genes (BnaA02g30260D, BnaA02g30290D and BnaA02g30310D) were all higher in ZS11 and BC4F2:3(aa) than in QU and BC4F2:3(AA) at three stages. Student’s t-test was used to compare QU with ZS11 and BC4F2:3 (AA) with BC4F2:3 (aa). BnaA02g30290D and BnaA02g30310D showed significant differences in expression between BC4F2:3(AA) and BC4F2:3(aa) at the 6-leaf stage (p < 0.05), and the difference in BnaA02g30260D was highly significant (p < 0.01) (Fig. 4). There was no consistent expression difference in the other genes tested among QU, BC4F2:3(AA), ZS11 and BC4F2:3(aa) at the three stages. Therefore, BnaA02g30260D, BnaA02g30290D and BnaA02g30310D were likely candidate genes for cqSPDA2.
Fig. 4.
Differential expression of 3 genes in the mapping interval in the parents and NILs. The relative expression levels were calculated by the 2−△△Ct method based on the QU samples, and three replicates were performed for each sample. The housekeeping gene Actin7 was used as the internal control. Values shown are means ± SD (n = 3). Student’s t-test was used to compare QU with ZS11 and BC4F2:3 (AA) with BC4F2:3 (aa). * denotes significance at the probability level of 0.05. ** denotes significance at the probability level of 0.01
Agronomic traits analysis
To investigate the effect of cqSPDA2 on agronomic traits, 50 plants with the AA genotype (high-chlorophyll content) and 50 plants with the aa genotype (low-chlorophyll content) were selected from the BC4F2 population by molecular marker and SPAD analysis. We investigated plant height, silique length, number of seeds per silique, number of siliques per plant, 1000-grain weight, and individual plant yield. The results showed that plant height, number of seeds per silique, number of siliques per plant and individual plant yield were highly significantly different between the AA-genotype plants and the aa-genotype plants (P < 0.01). There was a difference in 1000-grain weight but no difference in silique length between groups (P < 0.05) (Table 1).
Table 1.
Agronomic trait comparisons of AA-genotype plants and aa-genotype plants in the BC4F2 population
| Trait | AA genotype plant | aa genotype plant |
|---|---|---|
| Plant height (cm) | 192.016 ± 11.70 | 182.534 ± 14.23b |
| Silique length (cm) | 94.234 ± 10.91 | 92.672 ± 4.35 |
| Seeds per silique | 280.06 ± 28.98 | 263.88 ± 19.86b |
| 1000-grain weight(g) | 3.6388 ± 0.47 | 3.8256 ± 0.42a |
| Yield per plant(g) | 21.58 ± 10.39 | 13.3122 ± 8.08b |
| Siliques per plant | 274.84 ± 111.19 | 162.08 ± 83.81b |
Fifty AA-genotype plants with high chlorophyll content and 50 aa- genotype plants with low chlorophyll content were selected by molecular markers and SPAD. Student’s t-test was used to compare the AA- and aa-genotype plants. aindicates significance at the 0.05 probability level; bindicates significance at the 0.01 probability level. Data are shown as mean ± SD (n = 50 for each sample)
Discussion
Significance and QTL analysis of chlorophyll content
The leaf is the main photosynthetic organ in plants, and its chlorophyll content is an important agronomic trait for crop yield. Ninety to 95 % of plant dry matter is produced by photosynthesis, and crop yield is primarily derived from the photosynthetic products of leaves [24]. Chlorophyll is an important pigment involved in photosynthesis in chloroplasts, which can absorb and transform light energy, and an important index used to evaluate leaf photosynthetic capacity [25]. Increasing crop yield by increasing chlorophyll content is one of the important objectives of high-light-efficiency breeding [26]. The emergence of high-throughput sequencing technology provides a new method for the development of molecular markers. Molecular marker technology promotes rapid genetic map construction, and provides convenient data for fine mapping. Chlorophyll content is a quantitative characteristic that is primarily controlled by nuclear genes and has high heritability. At present, research on chlorophyll QTLs has been performed in different populations of various crops, such as rice [12, 14, 16, 17], wheat [13, 15], soybean [27, 28], and cabbage [18], and especially rice [17]. Previous studies have examined the location of chlorophyll in the embryo [19] and pericarp [20] of winter Brassica napus., In addition, QTLs were detected under drought and salt stress conditions, and a candidate gene related to salt tolerance was predicted [21–23]. Wang et al. [11] detected a QTL on chromosome C08 in Brassica napus using chlorophyll deficient mutants by measuring the absolute chlorophyll content in leaves.
Fine mapping using NILs and MAS
The development of NILs is a productive strategy for fine mapping and evaluating genetic effects during chlorophyll QTL studies [29]. In this study, a NIL population for the cqSPDA2 regions was constructed using ZS11 as the recurrent parent with flanking markers. The flanking markers’ Indel1, 3, 15, 87 were used to carry out foreground selection for construction the population of BC3F2 and BC4F2 populations, and the mapping interval was reduced to 152 kb (Fig. 2). This strategy for mapping the target genes is reasonable, inexpensive, and highly efficient. Based on BC4F2 phenotype and BC6F1 genotype analysis, the segregation of chlorophyll content was consistent with chi-square test (Fig. 1 and Additional file 1: Table S1). These results indicated that cqSPDA2 is the dominant QTL controlling chlorophyll content in Brassica napus. The NIL analysis also showed positive correlations between cqSPDA2 and agronomic traits such as yield, plant height, seeds per silique and siliques per plant (Table 1).
Molecular marker SSR2 with close linkage was developed through fine mapping (Fig. 3). Indel15 was also next to cqSPDA2, these markers could accelerate the breeding process for molecular marker assisted selection in the future.
Candidate genes prediction
According to qRT-PCR analysis, BnaA02g30260D, BnaA02g30290D and BnaA02g30310D were suitable candidate genes for cqSPDA2 among the twenty-four genes annotated in the Brassica napus genome in the mapping interval (Additional file 9: Table S7). BnaA02g30260D, which is part of a disease-resistance protein family, has transmembrane receptor activity, nucleoside-triphosphatase activity, nucleotide binding funtion, and ATP binding function and is involved in signal transduction, defense response, apoptosis, and innate immune response according to its annotations. Further study is needed to determine whether BnaA02g30260D affects chlorophyll synthesis. BnaA02g30290D is FK506- and rapamycin-binding protein 15 kD-2 (FKBP15–2) and has peptidyl-prolyl cis-trans isomerase activity related to protein folding. Luan et al. [30] found that AtFKBP15–1 and AtFKBP15–2 had the highest homology to FKBP13 and encoded functional homologs of FKBP13. AtFKBP13 was reported to be associated with Rieske protein both before and after the import of proteins into the chloroplast stroma, and AtFKBP13 can play a role in the downregulation of Rieske protein accumulation. Rieske is a subunit of the cytochrome b6f complex, which is one of the four complexes of the photosynthetic electron transport chain [31]. It was also reported that when ScFKBP12 was transferred into Arabidopsis, chloroplast formation and the expression of genes related to chloroplast formation were inhibited [32]. In this study, the expression levels of BnaA02g30290D (AtFKBP15–2) in the NIL (aa) and ZS11 plants were all higher than those in the NIL (AA) and QU at the three stages; if this gene inhibited the formation of chlorophyll, this finding would be consistent with the abovementioned results. BnaA02g30310D is homologous to GCH-1 in Arabidopsis thaliana. GCH-1 is the first enzyme in tetrahydrobiopterin (BH4) biosynthesis [33]. BH4 is an essential coenzyme for all three kinds of nitric oxide synthase (NOS) [34]. AtNOA1 (AtNOS1) is located in Arabidopsis chloroplasts, and OsNOA1 (OsNOS1) is also located in rice chloroplasts [35–37]. Yang et al. [37] found that the chlorophyll content decreased with increasing OsNOA1 at a low temperature (22 °C). He [38] suggested that OsNOA1 directly regulates chloroplast-encoded proteins by affecting the function of the chloroplast ribosome and then transmits a signal to the nucleus through the chloroplast retrograde signaling pathway mediated by Mg protoporphyrin IX, further affecting the expression of chloroplast proteins encoded by the nuclear genes. The location of this study, Qinghai Province, is on the Qinghai-Tibet Plateau, where the average temperature during crop growth is low. In this location, the expression level of BnaA02g30310D (GCH-1) in NIL (aa) and ZS11 was high at the three stages observed, especially at the six-leaf stage, which is consistent with the results obtained by He [37, 38].
Additional experiments, such as transgenic complementation test, CRISPR/Cas9, VIGS and RNAi, are warranted to investigate whether BnaA02g30260D, BnaA02g30290D and BnaA02g30310D are the genes underlying cqSPDA2. Analysis of the regulatory network controlling chlorophyll synthesis will facilitate molecular breeding of Brassica napus for high yield.
Conclusions
In this study, we constructed NILs and narrowed the interval of cqSPDA2 to 152 kb on A02 between SSR2 and Indel15. According to the Brassica napus genome annotation database, there were twenty-seven genes in this mapping interval. BnaA02g30260D, BnaA02g30290D and BnaA02g30310D were identified as suitable candidate genes for cqSPDA2 according to a qRT-PCR analysis, and thus, these genes may be responsible for chlorophyll synthesis. In addition, the dominant locus cqSPDA2 has positive effects on agronomic traits.
Methods
Plant materials
The leaves of ZS11 have low chlorophyll content, and the leaves of QU have high chlorophyll content (Fig. 5). To obtain a relatively simple genetic background and to fine-map cqSPDA2, we constructed an NIL population. An F1 line with the QU genotype in the cqSPDA2 region was selected and backcrossed to ZS11 for three generations. BC3F1 individuals were selfed to generate a BC3F2 mapping population backcrossed to ZS11. The flanking markers Indel1 and Indel87 were used to construct the NIL population with foreground selection (Additional file 2: Table S2).
Fig. 5.
Leaf color phenotypes of the parents QU and ZS11. QU: deep green leaves, high- chlorophyll content, SPAD value = 50.4; ZS11: light green leaves, low-chlorophyll content, SPAD value = 40.6
The BC4F1 individuals with a QU genetic background in the cqSPDA2 region as selected by the flanking markers Indel3 and Indel15 (Additional file 3: Fig. S1) were selfed to generate a BC4F2 population, which was used for fine mapping of the cqSPDA2 locus. The detailed process of population development is illustrated in Additional file 10: Fig. S3. BC4F2:3 individuals with the AA genotype (homozygous for cqSPDA2) and aa genotype (without cqSPDA2) were detected by the flanking markers Indel3 and Indel15 and subjected to qRT-PCR analysis. In addition, each population was grown in the experimental plots with a spacing of 30 cm (between rows) × 15 cm (within rows). The BC3F2 and BC4F2 populations were grown at the same density in fields in Yuanmou, Yunnan (altitude 898 m, 101°52′N, 25°42′E) and Xining, Qinghai (altitude 2225 m, 101°49′N, 36°34′E), respectively. The BC4F2:3 and BC6F1 populations were grown in a greenhouse at the Academy of Agricultural and Forestry Sciences, Qinghai University (Xining, Qinghai, China). Standard crop management practices were followed.
Phenotypic trait and data analysis
The testing targets included every plant of the population that was lacking diseases and insect pests. According to the previous method of chlorophyll determination, we measured three positions in the distal third of the first fully developed leaf from the top of each plant at the six-leaf stage by SPAD (SPAD 502, Japan) during the 9:00–11:00 am interval [39]. Each measurement was repeated three times per leaf, and the veins were avoided. Statistical analysis was performed with Excel. Chi-square tests were performed on the segregation data to determine the genetic regulation of the chlorophyll content.
DNA extraction and development of molecular markers development
Total DNA was extracted from fresh leaves using the CTAB method [40]. PCR was performed in a 20-μL reaction solution containing 2 μL DNA, 2 μL 2 mM dNTPs, 2 μL 10× PCR buffer, 1 μL Taq, 1 μL of 2 μM forward and reverse primers and 12 μL of ddH2O. The PCR program was carried out according to Yang’s method with minor modifications [41]. The PCR products were separated on 6% nondenatured polyacrylamide gels and detected by silver staining [42]. Indel (insertion/deletion) markers were developed from the resequencing data of the parents according to the B. napus ‘Darmor-bzh’ reference genome sequence in the primary mapping interval. SSR markers were developed based on the B. napus ‘Darmor-bzh’ reference genome sequence corresponding to the interval. The sequences of the SSR markers were designed using SSR Hunter 1.3 and Primer Premier 5.0 [43, 44].
Mapping of the cqSPDA2 locus
The BC3F2 and BC4F2 family populations were used to fine map the cqSPDA2 locus using indel and SSR markers. First, we designed 87 indel markers in the primer mapping interval (21.87–22.91 Mb on chromosome A02) to map the cqSPDA2 locus. A linkage map for the cqSPDA2 locus was constructed using JoinMap 4.0 [45]. The mapping interval for the cqSPDA2 locus was gradually reduced using the mapping results of the BC3F2 population. Finally, additional indel and SSR markers within the new narrow mapping interval were designed to fine-map the cqSPDA2 locus based on the BC4F2 population with WinQTLCart 2.5. The physical location was obtained by blasting the Brassica napus genome database using the indel and SSR sequences. The physical linkage map was produced with MapDraw 2.1 [46].
TA cloning
The specific markers closely linked to cqSPDA2 were sequenced by NIL population scanning. Specific fragments were collected according to Yi et al. [47]. The product was ligated into the PMD18-T vector (Takara), and the transformed clone was detected with M13 primers. Six positive clones were randomly selected and sequenced by Sangon Biotech (Shanghai) Co., Ltd. [48].
Genes in the mapping interval
All genes within the targeted mapping interval on A02 were identified using annotations from the Brassica napus genomes (http://www.genoscope.cns.fr/brassicanapus/) and annotated according to the BRAD annotations (http://brassicadb.org/brad/blastPage.php). The homologous sequences were aligned using BLASTN (http://blast.ncbi.nlm.nih.gov/).
RNA extraction and qRT-PCR analysis
BC4F2:3 (91 AA-genotype plants homozygous for cqSPDA2 and 104 aa-genotype plants without cqSPDA2) were grown in a greenhouse in September 2019. Total RNA was isolated from the first fully developed leaves counted from the top of each plant (at the 4-leaf stage, 6-leaf stage and squaring stage) of the BC4F2:3 population and the parental lines using TRNzol-A+ Total RNA Reagent (Takara, Dalian, China) according to the manufacturer’s protocol. RNA integrity was monitored using 1% agarose gel electrophoresis. cDNA was obtained via reverse transcription of total RNA using the PrimeScript RT Reagent Kit (Takara, Dalian, China) and following the manufacturer’s instructions.
We performed a qRT-PCR analysis to identify the genes in the mapping interval. Real-time PCR was conducted using LightCycler 480 II 96-Well PCR Plates (Roche, Rotkreuz, Switzerland). The utilized reaction system contained 10 μL of 2 × SG Fast qPCR Master Mix (B639271, BBI), 2 μL cDNA, and 10 μM gene-specific primers in a final volume of 20 μL. The thermal cycling conditions used were 95 °C for 3 min, followed by 45 cycles at 95 °C for 5 s and 60 °C for 30 s followed by a final extension stage. The relative expression levels of all the genes in the mapping interval were calculated by the 2−△△Ct method based on the QU samples and three replicates were performed for each sample [49]. The housekeeping gene Actin7 was used as the internal control to calculate the relative expression levels of each gene. Three biological replicates were performed in this experiment and t-test was used for statistical analysis.
Phenotyping for agronomic traits
To evaluate the agronomic efficiency of cqSPDA2, 100 individuals (50 AA- and 50 aa- genotype plants) from the BC4F2 population were sampled using the markers Indel3 and Indel15, and their chlorophyll content was characterized. The agronomic traits investigated were as follows: plant height (cm), total siliques per plant, silique length (cm), seeds per silique, 1000-grain weight (g), and yield per plant (g). The mean values, standard deviations and significance analyses of all the agronomic traits were compared between the AA- and aa-genotype plants by Minitab16 and Excel2010.
Supplementary Information
Additional file 1: Table S1. Genotyping of cqSPDA2 in BC6F1 population.
Additional file 2: Table S2. Primer sequences designed in this study.
Additional file 3: Fig. S1. Genetic linkage map of cqSPDA2.
Additional file 4: Fig. S2. The local genetic linkage map and physical map of cqSPDA2 on chromosome A02 in BC3F2 population.
Additional file 5: Table S3. Primer sequences designed in this study.
Additional file 6: Table S4. Primer sequences of KASP designed in this study.
Additional file 7: Table S5. Genes in the mapping interval on the chromosome A02 and their orthologs in Arabidopsis.
Additional file 8: Table S6. Primers designed for qRT-PCR of genes in the mapping interval.
Additional file 9: Table S7. Gene expression in the mapping interval on A02.
Additional file 10: Fig. S3. The scheme of NIL development for fine mapping.
Acknowledgements
Not applicable.
Abbreviations
- QTL
Quantitative trait locus
- NIL
Near-isogenic line
- DH
Doubled haploid
- qRT-PCR
Quantitative real-time PCR
- SNP
Single nucleotide polymorphism
- Indel
Insertion-deletion
- SSR
Simple sequence repeat
- KASP
kompetitive allele-specific PCR
- SPAD
Soil and plant analyzer development
- FKBP
FK506- and rapamycin-binding protein
- GCH-1
GTP cyclohydrolase І
- BH4
Tetrahydroboipterin
- NOS/NOA
nitric oxide synthase
Authors’ contributions
YJX, LHD and DDZ conceived and designed the research; YJX and LHD performed the experiments; YJX, LHD and DDZ discussed the results and strategies; YJX analyzed the data and wrote the manuscript. ZZ, XL and LKX corrected the manuscript. All authors read and approved the manuscript.
Funding
This research was supported by the National Key Research and Development Plan (2016YFD0101304), the National Natural Science Foundation of China (31460354), the National System of Technology of the Rapeseed Industry (CARS-12), the Laboratory of Spring Rape Genetic Improvement of Qinghai Province (2017-ZJ-Y09; 2020-ZJ-Y10), and the Qinghai Academy of Agriculture and Forestry Sciences Research Project (2018-NKY-011).
Availability of data and materials
All data used during the study are included in this published article and its additional files.
Ethics approval and consent to participate
This study does not contain any research requiring ethical consent or approval.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing financial interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jingxiu Ye and Haidong Liu contributed equally to this work.
References
- 1.Raines CA. Increasing photosynthetic carbon assimilation in C3 plants to improve crop yield: current and future strategies. Plant Physiol. 2011;155:36–42. doi: 10.1104/pp.110.168559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eggink L, Park H, Hoober JK. The role of chlorophyll b in photosynthesis: hypothesis. BMC Plant Biol. 2001;1:2. doi: 10.1186/1471-2229-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo P, Baum M, Varshney RK, Graner A, Grando S, Ceccarelli S. QTLs for chlorophyll and chlorophyll fluorescence parameters in barley under post-flowering drought. Euphytica. 2008;163:203–214. doi: 10.1007/s10681-007-9629-6. [DOI] [Google Scholar]
- 4.von Korff M, Grando S, Del Greco A, This D, Baum M, Ceccarelli S. Quantitative trait loci associated with adaptation to Mediterranean dryland conditions in barley. Theor Appl Genet. 2008;117:653–669. doi: 10.1007/s00122-008-0787-2. [DOI] [PubMed] [Google Scholar]
- 5.Mae T. Physiological nitrogen efficiency in rice: nitrogen utilization, photosynthesis, and yield potential. Plant Soil. 1997;196:201–210. doi: 10.1023/A:1004293706242. [DOI] [Google Scholar]
- 6.Teng S, Qian Q, Zeng D, Kunihiro Y, Fujimoto K, Huang D, et al. QTL analysis of leaf photosynthetic rate and related physiological traits in rice (Oryza sativa L.) Euphytica. 2004;135:1–7. doi: 10.1023/B:EUPH.0000009487.89270.e9. [DOI] [Google Scholar]
- 7.Zhang GH, Xu Q, Zhu XD, Qian Q, Xue HW. SHALLOT-LIKE1 is a KANADI transcription factor that modulates rice leaf rolling by regulating leaf abaxial cell development. Plant Cell. 2009;21:719–735. doi: 10.1105/tpc.108.061457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang ZH, Wu XS, Chang XP, Li RZ, Jing RL. Chlorophyll content and chlorophyll fluorescence kinetics parameters of flag leaf and their gray relational grade with yield in wheat. Acta Agronom Sin. 2010;36(2):217–227. doi: 10.3724/SP.J.1006.2010.00217. [DOI] [Google Scholar]
- 9.Shi DK, Yao TL, Liu NN, Deng M, Duan HY, Wang LL, Wan J, Gao JH, Xie HL, Tang JH, Zhang XH. Genome-wide association study of chlorophyll content in maize. Sci Agri Sin. 2019;52(11):1839–1857. [Google Scholar]
- 10.Beale SI. Green genes gleaned. Trends Plant Sci. 2005;10:309–312. doi: 10.1016/j.tplants.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 11.Wang YK, He YJ, Yang M, He JB, Xu P, Shao MQ, Chu P, Guan RZ. Fine mapping of a dominant gene conferring chlorophyll-deficiency in Brassica napus. Sci Rep. 2016;6:31419. doi: 10.1038/srep31419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang L, Dai L, Wang L, Leng Y, Yang Y, Xu J, et al. Genetic dissection for chlorophyll content of the top three leaves during grain filling in rice (Oryza sativa L.) J Plant Growth Regul. 2015;34:381–391. doi: 10.1007/s00344-014-9474-5. [DOI] [Google Scholar]
- 13.Graziani M, Maccaferri M, Royo C, Salvatorelli F, Tuberosa R. QTL dissection of yield components and morpho-physiological traits in a durum wheat elite population tested in contrasting thermo-pluviometric conditions. Crop Pasture Sci. 2014;65:80–95. doi: 10.1071/CP13349. [DOI] [Google Scholar]
- 14.Zhang GH, Li SY, Wang L, Ye WJ, Zeng DL, Rao YC, et al. LSCHL4 from Japonica cultivar, which is allelic to NAL1, increases yield of Indica super rice 93-11. Mol Plant. 2014;7:1350–1364. doi: 10.1093/mp/ssu055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar S, Sehgal SK, Kumar U, Prasad PVV, Joshi AK, Gill BS. Genomic characterization of drought tolerance-related traits in spring wheat. Euphytica. 2012;186:265–276. doi: 10.1007/s10681-012-0675-3. [DOI] [Google Scholar]
- 16.Takai T, Kondo M, Yano M, Yamamoto T. A quantitative trait locus for chlorophyll content and its association with leaf photosynthesis in rice. Rice. 2010;3:172–180. doi: 10.1007/s12284-010-9047-6. [DOI] [Google Scholar]
- 17.Ye W, Hu S, Wu L, Chang W, Cui Y, Xu J, et al. Fine mapping a major QTL qFCC7 L for chlorophyll content in rice (Oryza sativa L.) cv. PA64s. Plant Growth Regul. 2017;81:81–90. doi: 10.1007/s10725-016-0188-5. [DOI] [Google Scholar]
- 18.Ge Y, Wang T, Wang N, Wang Z, Liang C, Ramchiary N, et al. Genetic mapping and localization of quantitative trait loci for chlorophyll content in Chinese cabbage (Brassica rapa ssp. pekinensis) Sci Hortic. 2012;147:42–48. doi: 10.1016/j.scienta.2012.09.004. [DOI] [Google Scholar]
- 19.Huang JH, Xu XF, Qu C, Yan XY, Fu F, Chen L, et al. Mapping of QTLs for embryonic chlorophyll in Brassica napus L. J Plant Genet Resour. 2010;11:766–771. [Google Scholar]
- 20.Yan X, Li J, Jin M, Chen L, Wang J, Qu C, et al. QTL analysis of chlorophyll content in silique wall in Brassica napus L. Chin J Oil Crop Sci. 2009;31:269–273. [Google Scholar]
- 21.Ding J. Physiology of salt tolerance and QTL mapping of related genes at the seedling stage in Brassica napus L. Dissertation. Yangling: Northwest A& F University; 2015.
- 22.Lang LN, Xu AX, Ding J, Yan Z, Zhao N, Tian ZS, Liu YP, Wang Y, Liu X, Liang FH, et al. Quantitative trait locus mapping of salt tolerance and identification of salt-tolerant genes in Brassica napus L. Front Plant Sci. 2017;8:1000. doi: 10.3389/fpls.2017.01000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu JH. QTL analysis of drought tolerance traits at seedling stage in Brassica napus L. Dissertation. Chongqing: Southwest University; 2016.
- 24.Liu HM, Zhou XY, Liu JF, Qiu YB, Fan FF, Xu QG. Analysis of combining ability of photosynthetic characteristics in Indica hybrid rice. J Plant Genet Resour. 2014;15:699–705. [Google Scholar]
- 25.Liu J, Wang JY, Yao XY, Zhang Y, Li JQ, Wang XX, Xu ZJ, Chen WF. Characterization and fine mapping of thermo-sensitive chlorophyll deficit mutant1 in rice (Oryza sativa L.) Breeding Sci. 2015;65:161–169. doi: 10.1270/jsbbs.65.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi DK, Yao TM, Liu NN, Deng M, Duan HY, Wang LL, et al. Genome-wide association study of chlorophyll content in maize. Sci Agric Sin. 2019;52:1839–1857. [Google Scholar]
- 27.Li W, Pan XC, Yu HX, Qi HD, Mao XR, Huang SY, et al. QTL mapping for chlorophyll content and candidate gene predictionin soybean. Genom Appl Biol. 2016;35:1793–1799. [Google Scholar]
- 28.Liang HZ, Yu YL, Yang HQ, Dong W, Xu LJ, Niu YG, et al. Epistatic and QTL×environment interaction effects of QTLs for leaf traits and leaf chlorophyll content in soybean. Acta Agron Sin. 2015;41:889–899. doi: 10.3724/SP.J.1006.2015.00889. [DOI] [Google Scholar]
- 29.Liu Y, Anning Zhang AN, Wang FM, Kong DY, Li MS, Bi JG, Zhang FY, Wang JH, Luo XX, Pan ZQ, Yu XQ, Liu GL, Luo LJ. Fine mapping a quantitative trait locus, qSER-7, that controls stigma exsertion rate in rice (Oryza sativa L.) Rice. 2019;12:46. doi: 10.1186/s12284-019-0304-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luan S, Kudla J, Gruissem W, Schreiber SL. Molecular characterization of a FKBP-type immunophilin from higher plants. Proc Natl Acad Sci U S A. 1996;93:6964–6969. doi: 10.1073/pnas.93.14.6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gupta R, Mould RM, He Z, Luan S. A chloroplast FKBP interacts with and affects the accumulation of Rieske subunit of cytochrome bf complex. Proc Natl Acad Sci U S A. 2002;99:15806–15811. doi: 10.1073/pnas.222550399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deng K, Yu L, Zheng X, Zhang K, Wang W, Dong P, et al. Target of rapamycin is a key player for auxin signaling transduction in Arabidopsis. Front Plant Sci. 2016;7:291. doi: 10.3389/fpls.2016.00291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Werner-Felmayer G, Golderer G, Werner ER. Tetrahydrobiopterin biosynthesis, utilization and pharmacological effects. Curr Drug Metab. 2002;3:159–173. doi: 10.2174/1389200024605073. [DOI] [PubMed] [Google Scholar]
- 34.Kotsonis P, Fröhlich LG, Shutenko ZV, Horejsi R, Pfleiderer W, Schmidt HH. Allosteric regulation of neuronal nitric oxide synthase by tetrahydrobiopterin and suppression of auto-damaging superoxide. Biochem J. 2000;346(Pt 3):767–776. doi: 10.1042/bj3460767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Flores-Pérez U, Sauret-Güeto S, Gas E, Jarvis P, Rodríguez-Concepción M. A mutant impaired in the production of plastome-encoded proteins uncovers a mechanism for the homeostasis of isoprenoid biosynthetic enzymes in Arabidopsis plastids. Plant Cell. 2008;20:1303–1315. doi: 10.1105/tpc.108.058768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu H, Lau E, Lam MP, Chu H, Li S, Huang G, et al. OsNOA1/RIF1 is a functional homolog of AtNOA1/RIF1: implication for a highly conserved plant cGTPase essential for chloroplast function. New Phytol. 2010;187:83–105. doi: 10.1111/j.1469-8137.2010.03264.x. [DOI] [PubMed] [Google Scholar]
- 37.Yang Q, He H, Li H, Tian H, Zhang J, Zhai L, et al. NOA1 functions in a temperature-dependent manner to regulate chlorophyll biosynthesis and Rubisco formation in rice. PLoS One. 2011;6:e20015. doi: 10.1371/journal.pone.0020015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He H, Yang QS, Shen BR, Zhang S, Peng XX. OsNOA1 functions in a threshold-dependent manner to regulate chloroplast proteins in rice at lower temperatures. BMC Plant Biol. 2018;18(1):44. doi: 10.1186/s12870-018-1258-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ling QH, Huang WH, Jarvis P. Use of a SPAD-502 meter to measure leaf chlorophyll concentration in Arabidopsis thaliana. Photosynth Res. 2011;107(2):209–214. doi: 10.1007/s11120-010-9606-0. [DOI] [PubMed] [Google Scholar]
- 40.Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW. Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc Natl Acad Sci U S A. 1984;81:8014–8018. doi: 10.1073/pnas.81.24.8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang M, Huang C, Wang M, Fan H, Wan S, Wang Y, et al. Fine mapping of an up-curling leaf locus (BnUC1) in Brassica napus. BMC Plant Biol. 2019;19:324. doi: 10.1186/s12870-019-1938-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Creste S, Tulmann AN, Figueira A. Detection of single sequence repeat polymorphisms in denaturing polyacrilamide sequencing gels by silver staining. Plant Mol Biol Report. 2012;19:299–306. doi: 10.1007/BF02772828. [DOI] [Google Scholar]
- 43.Li Q, Wan JM. SSRHUNTER: development of a local searching software for SSR sites. Hereditas. 2005;27(5):808–810. [PubMed] [Google Scholar]
- 44.Lalitha S. Primer Premier 5[J] Biotech Software Internet Rep. 2000;1(6):270–272. doi: 10.1089/152791600459894. [DOI] [Google Scholar]
- 45.Ooijen JWV. JoinMap 4, software for the calculation of genetic linkage maps in experimental populations. Wageningen: Kyazma B.V; 2006. [Google Scholar]
- 46.Liu RH, Meng JL. MapDraw: a microsoft excel macro for drawing genetic linkage maps based on given genetic linkage data. Hereditas. 2003;25(3):317–321. [PubMed] [Google Scholar]
- 47.Yi B, Chen Y, Lei S, Tu J, Fu T. Fine mapping of the recessive genic male-sterile gene (Bnms1) in Brassica napus L. Theor Appl Genet. 2006;113:643–650. doi: 10.1007/s00122-006-0328-9. [DOI] [PubMed] [Google Scholar]
- 48.Yen TY, Li KP, Ou SC, Shien JH, Lu HM, Chang PC. Construction of an infectious plasmid clone of Muscovy duck parvovirus by TA cloning and creation of a partially attenuated strain. Avian Pathol. 2015;44:124–128. doi: 10.1080/03079457.2015.1008399. [DOI] [PubMed] [Google Scholar]
- 49.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−△△C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Genotyping of cqSPDA2 in BC6F1 population.
Additional file 2: Table S2. Primer sequences designed in this study.
Additional file 3: Fig. S1. Genetic linkage map of cqSPDA2.
Additional file 4: Fig. S2. The local genetic linkage map and physical map of cqSPDA2 on chromosome A02 in BC3F2 population.
Additional file 5: Table S3. Primer sequences designed in this study.
Additional file 6: Table S4. Primer sequences of KASP designed in this study.
Additional file 7: Table S5. Genes in the mapping interval on the chromosome A02 and their orthologs in Arabidopsis.
Additional file 8: Table S6. Primers designed for qRT-PCR of genes in the mapping interval.
Additional file 9: Table S7. Gene expression in the mapping interval on A02.
Additional file 10: Fig. S3. The scheme of NIL development for fine mapping.
Data Availability Statement
All data used during the study are included in this published article and its additional files.