The main outcome is the molecular description of the fine-tuned binding of the cdM2-1/RNA complex and the provision of evidence that the domain alone has unfolding activity for long RNAs. This binding mode is essential in the understanding of the function in the full-length protein. Human respiratory syncytial virus (hRSV), an orthopneumovirus, stands out for the unique role of its M2-1 protein as a transcriptional antitermination factor able to increase RNA polymerase processivity.
KEYWORDS: hRSV M2-1 core domain, RNA binding protein, fine-tuned binding, RNA unfolding activity, NMR, molecular docking, molecular dynamics, nuclear magnetic resonance
ABSTRACT
The human respiratory syncytial virus (hRSV) M2-1 protein functions as a processivity and antitermination factor of the viral polymerase complex. Here, the first evidence that the hRSV M2-1 core domain (cdM2-1) alone has an unfolding activity for long RNAs is presented and the biophysical and dynamic characterization of the cdM2-1/RNA complex is provided. The main contact region of cdM2-1 with RNA was the α1-α2-α5-α6 helix bundle, which suffered local conformational changes and promoted the RNA unfolding activity. This activity may be triggered by base-pairing recognition. RNA molecules wrap around the whole cdM2-1, protruding their termini over the domain. The α2-α3 and α3-α4 loops of cdM2-1 were marked by an increase in picosecond internal motions upon RNA binding, even though they are not directly involved in the interaction. The results revealed that the cdM2-1/RNA complex originates from a fine-tuned binding, contributing to the unraveling interaction aspects necessary for M2-1 activity.
IMPORTANCE The main outcome is the molecular description of the fine-tuned binding of the cdM2-1/RNA complex and the provision of evidence that the domain alone has unfolding activity for long RNAs. This binding mode is essential in the understanding of the function in the full-length protein. Human respiratory syncytial virus (hRSV), an orthopneumovirus, stands out for the unique role of its M2-1 protein as a transcriptional antitermination factor able to increase RNA polymerase processivity.
INTRODUCTION
Human respiratory syncytial virus (hRSV) is the major causative agent of lower tract respiratory diseases, such as pneumonia and bronchiolitis, in children worldwide. This virus is also responsible for respiratory morbidity in the elderly and people with compromised immunity and cardiorespiratory disease. The main group at risk of hRSV-related morbidity is children with congenital immunodeficiency, bronchopulmonary dysplasia, heart disease, hypertension, prematurity, and low birth weight (1–3).
Of the 11 proteins encoded by hRSV, 2 are expressed by the M2 gene, the so-called M2-1 and M2-2 genes (1). The proteins encoded by the M2 gene are involved in the assembly of the active form of the ribonucleoprotein (RNP) complex, with M2-1 acting in transcription as a processivity and antitermination factor and M2-2 working as a molecular switch between transcription and RNA replication (4).
M2-1 prevents the RNA-dependent RNA polymerase (RdRp) complex from dissociating prematurely at intragenic sites and also allows it to read through gene and termination signals, thus increasing the transcription efficiency of genes near the 5′ end (5, 6). Blondot and collaborators hypothesized three possible explanations for how M2-1 acts as a factor that prevents the premature termination of transcription and that is thus important for mRNA transcription (7). (i) The M2-1 protein could bind to the nascent mRNA transcript to favor transcription elongation, preventing it either from rehybridizing to the template or from forming secondary structures, which would destabilize the transcription complex. (ii) The polymerase processivity-enhancing effect of M2-1 could be due to an increase in the affinity of the polymerase for the genomic RNA template in a sequence-nonspecific manner. (iii) M2-1 could recognize gene end sequences either on the nascent mRNA or on the RNA template, preventing the release of the polymerase complex from its template and favoring transcription reinitiation at the downstream gene start sequences (7).
The crystal structure of the hRSV M2-1 protein determined by Tanner and collaborators shows a tetrameric arrangement, with the monomers presenting three structurally distinct regions: the zinc finger domain, which is composed of N-terminal residues that bind to a zinc atom and that aid in the transcription process; the oligomerization helix, which is responsible for tetramer formation; and the core domain, which acts directly on the interaction with phosphoprotein P and RNA (8). The core domain of M2-1 (cdM2-1) is folded into six α-helices, which are structurally arranged in an α1-α2-α5-α6 helix bundle and an α3-α4 hairpin (7, 8). Blondot et al. determined by nuclear magnetic resonance (NMR) studies that the α2, α5, and α6 helices of cdM2-1 play a key role in the binding to nucleic acids and also demonstrated the preference of the core domain for purine-rich RNAs, especially adenine (7).
The crystal structure of the human metapneumovirus (hMPV) M2-1 protein, which is homologous to hRSV M2-1, presented a distinct conformation for one of the tetramer core domains, which was termed the open conformation (9). In this conformation, one of the tetramer core domains is located far from the rest of the protein due to a rotation in the flexible linker (residues with electronic density absent in the crystal structures) between the oligomerization and core domains. From molecular dynamics (MD) simulations and small-angle X-ray scattering (SAXS) experiments, a dynamic open-closed conformation equilibrium was characterized for hMPV M2-1, and it was determined that the closed conformation is structurally similar to that of the hRSV M2-1 protein. It was also reported that the M2-1 closed conformation is prevalent in the presence of RNA, which is stabilized by the simultaneous binding of the nucleic acid molecule to the zinc finger region and to the core domain of the protein (9).
Recently, Molina and coauthors observed that the RNA of 20 nucleotides (20-mer RNA) binds with positive cooperativity to two core domains in the hRSV M2-1 tetramer, allowing RNAs to bridge two adjacent RNA binding sites. No cooperativity was reported for the binding of the core domain to 20-mer RNA and of the M2-1 tetramer to 10-mer RNA (10). They showed by circular dichroism (CD) that 20-mer RNAs are unfolded upon the formation of their complex with the M2-1 tetramer but that, in the presence of only the core domain, significant structural changes are not detectable. The CD approach indicated that the secondary structure content of M2-1 shows no major changes upon the formation of the complex (10). This finding corroborates the first of the above-mentioned hypotheses proposed by Blondot and colleagues for the mechanisms by which M2-1 prevents the polymerase from terminating transcription (7). Molina and collaborators (10) also reported by fluorescence experiments that the process of association of nucleic acids with the M2-1 tetramer is composed of two steps, with the first being a fast phase related to conformational changes of the RNA molecules and the second being a slow phase related to subtle rearrangements of the M2-1/RNA complex that occur through an induced-fit mechanism. Based on the induced-fit mechanism, subtle conformational rearrangements in the complex take place in the protein moiety. Therefore, in the case of M2-1, as CD experiments indicated that no major secondary structure changes occur upon binding, loop or hinge motions probably reposition the RNA binding domains (core domains), as shown for the hMPV M2-1 protein, where the transition from the open to the closed conformation is observed for cdM2-1 upon its association with RNA (9, 10).
Taken together, the aforementioned studies characterize the M2-1 tetramer as a flexible platform for RNA recognition, with the core domain being the major binding site responsible for the first molecular contact with the nucleic acid molecules. Despite the well-described biological activity of M2-1 (11, 12) and structures of M2-1 (8, 9) and recent clarifications on the possible biochemical mechanism accounting for its antitermination activity (10), molecular details about the biophysical-chemical and dynamic features of the interaction between the core domain of M2-1 and RNA still need to be introduced. In this report, an integrated approach with experimental and computational techniques was employed to obtain information at the molecular level on the binding of hRSV cdM2-1 to long RNAs (RNAs of ∼80 bases). The fluorescence spectroscopy experiments under different conditions of temperature (T), ionic strength, and pH revealed a key role of the electrostatic interactions in the formation of the complex and the van der Waals interactions and hydrogen bonds in the bimolecular stabilization, while differential scanning calorimetry (DSC) measurements indicated a slight increase in the thermal stability of cdM2-1 upon RNA binding. Nuclear magnetic resonance (NMR) highlighted the structural features of cdM2-1 and its complex with RNA. Chemical shift perturbation (CSP) analysis identified the α1-α2-α5-α6 helix bundle in the core domain to be the main region interacting with RNA and, along with thermal susceptibility measurements of the amide hydrogen (1HN) chemical shifts, pointed out that this bundle helix may undergo local structural changes that extend to the α3-α4 hairpin. Solvent paramagnetic relaxation enhancement (sPRE) measurements showed that cdM2-1 is permanently protected from solvent exposure when complexed with the nucleic acid, while the changes in the intensity of imino proton resonances suggest a protein-induced unfolding of partial formations of RNA base pairing. 15N relaxation data revealed that cdM2-1 dynamics undergo significant changes after the interaction of cdM2-1 with RNA, presenting a decrease in the overall tumbling motion and, locally, an increase in the flexibility of the α2-α3 and α3-α4 loops of the protein. The structural models of the cdM2-1/RNA complex generated from docking calculations and evaluated by molecular dynamics (MD) simulations corroborated the experimental results and highlighted that Lys150 and Arg151 are pivotal residues for the stabilization of the complex. The results presented herein contribute to an understanding of the molecular behavior of the core domain of hRSV M2-1 in solution and upon binding to its biological partner, RNA.
RESULTS
cdM2-1/RNA interaction investigated by fluorescence spectroscopy.
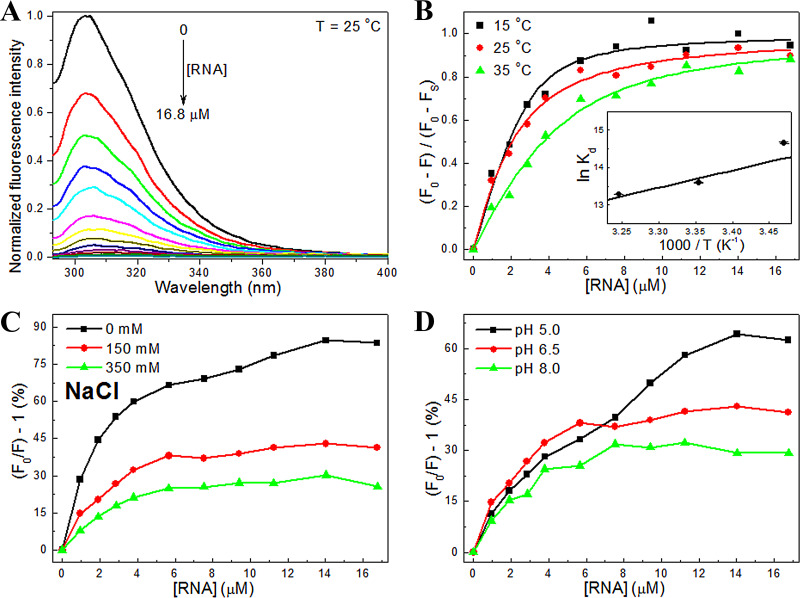
The binding of the hRSV cdM2-1 to RNA was investigated using fluorescence-quenching experiments at different temperatures (15, 25, and 35°C), salt concentrations (0, 150, and 350 mM NaCl), and pHs (5, 6.5, and 8). Figure 1A shows that the fluorescence intensity of cdM2-1 is quenched with increments of RNA concentrations, suggesting that the microenvironment of the fluorophores of the domain is affected by the presence of the nucleic acid. The changes in the fluorescence-quenching signal [, where F0 is the fluorescence intensity in the absence of the ligand (RNA), F is the fluorescence in the presence of RNA, and Fsat is the intensity of the bound protein saturated with RNA] as a function of the RNA concentration were analyzed, using equation 1 (see Materials and Methods), at temperatures of 15, 25, and 35°C (Fig. 1B). The binding isotherm curves provided the values of the dissociation constant (Kd) and the stoichiometry coefficient (nSC) for the cdM2-1/RNA complex, which are shown in Table 1. The values of Kd determined for the cdM2-1/RNA interaction were in the nanomolar-to-micromolar range, and the binding affinity was revealed to decrease with an increase in the temperature from 15 to 35°C. The average value of nSC (∼2.25) at the investigated temperatures indicates that at least two cdM2-1 molecules bind to one RNA molecule.
FIG 1.
Analysis of the fluorescence-quenching data for the cdM2-1/RNA interaction under different conditions of temperature, ionic strength, and pH. (A) Emission spectra of cdM2-1 in the absence and the presence of RNA concentration increments (pH 6.5, T = 25°C, excitation λ = 288 nm). The cdM2-1 concentration was 5.5 μM, and the RNA concentration was 0 to 16.8 μM. The fluorescence-quenching spectra at 15 and 35°C showed a profile similar to that seen at 25°C (data not shown). (B) Analysis of the fluorescence data for the cdM2-1/RNA interaction. The changes in the fluorescence-quenching signal of cdM2-1 as a function of the RNA concentration in 50 mM phosphate buffer (pH 6.5) containing 150 mM NaCl and 1.0 mM DTT at 15, 25, and 35°C are shown. (Inset) van’t Hoff plot used to determine the enthalpy change value in the formation of the cdM2-1/RNA complex. (C, D) Fluorescence-quenching percentage rates as a function of RNA concentrations under conditions of different salt concentrations (0, 150, and 350 mM NaCl) and different pHs (5, 6.5, and 8) at a temperature of 25°C.
TABLE 1.
Dissociation constant, stoichiometry coefficient, enthalpy change, Gibbs free energy change, and entropy change of the cdM2-1/RNA interaction, determined using fluorescence-quenching experiments at 15, 25, and 35°Ca
| Temp (°C) | Kd (10−6 M) | nSC | ΔH (kJ · mol−1) | ΔG (kJ · mol−1) | ΔS (J · mol−1 · K−1) |
|---|---|---|---|---|---|
| 15 | 0.43 ± 0.18 | 2.0 ± 0.4 | −45 ± 10 | −35.1 ± 0.7 | −34 ± 35 |
| 25 | 1.24 ± 0.18 | 3.3 ± 1.1 | −33.7 ± 0.4 | −37 ± 34 | |
| 35 | 1.70 ± 0.28 | 1.4 ± 0.2 | −34.0 ± 0.4 | −36 ± 32 |
All experiments were performed in 50 mM phosphate buffer (pH 6.5) containing 150 mM NaCl and 1.0 mM DTT. Kd, dissociation constant; nSC, stoichiometry coefficient; ΔH, enthalpy change; ΔG, Gibbs free energy change; ΔS, entropy change.
The van’t Hoff plot (equation 2) [ln(Kd) and 1/T; Fig. 1B] was linear over the investigated temperature range. The enthalpy change () was −45 ± 10 kJ · mol−1 (Table 1), denoting an enthalpically favorable exothermic binding reaction. The negative values of the changes in the Gibbs free energy () and entropy () indicate that the interaction is spontaneous and entropically unfavorable. The enthalpic term provides the major contribution to , suggesting that the binding process is enthalpically driven. The ΔH value of <0 and the ΔS value of <0 indicate that van der Waals interactions and hydrogen bonds are important noncovalent interactions responsible for the stabilization of the cdM2-1/RNA complex (13).
Figure 1C and D show the effect of ionic strength and pH at 25°C. With an increase in the salt concentration, the fluorescence-quenching percentage rates [(), in percent] decreased in each step of the titration of RNA with cdM2-1 (Fig. 1B), indicating that the rise in ionic strength caused a reduction in the quenching efficiency, with a tendency to increase the RNA binding constant (1.13 ± 0.11 and 1.70 ± 0.19 μM for nSC equal to 2.25 at 0 and 350 mM NaCl, respectively). The decrease in quenching efficiency is probably related to an electrostatic shielding of the protein and/or nucleic acid. The decrease in pH from 8 to 5 induced an increase in () (in percent) that suggests an increase in the quenching efficiency of cdM2-1 by RNA, which may be explained by the strengthening of the electrostatic interactions involved in the formation of the cdM2-1/RNA complex. Protonation of His147 and His168 at pH 5 may play a key role. Despite the increase in () (in percent), there was a decrease in the cdM2-1/RNA affinity (Kd = 0.90 ± 0.14 and 4.04 ± 0.41 μM for nSC equal to 2.25 at pH 8 and 5, respectively) due to the reduction in the pH, which reinforces the role of electrostatic interactions in the formation of the complex, while hydrogen bonds and van der Waals interactions play a key role in the binding stabilization. Circular dichroism (CD) experiments showed that there are no significant changes in the secondary structure elicited by the changes in pH (data not shown). The variations in the fluorescence-quenching percentage rates under different conditions of pH did not arise from secondary structure changes.
Thermal stability of cdM2-1 and the cdM2-1/RNA complex determined by DSC.
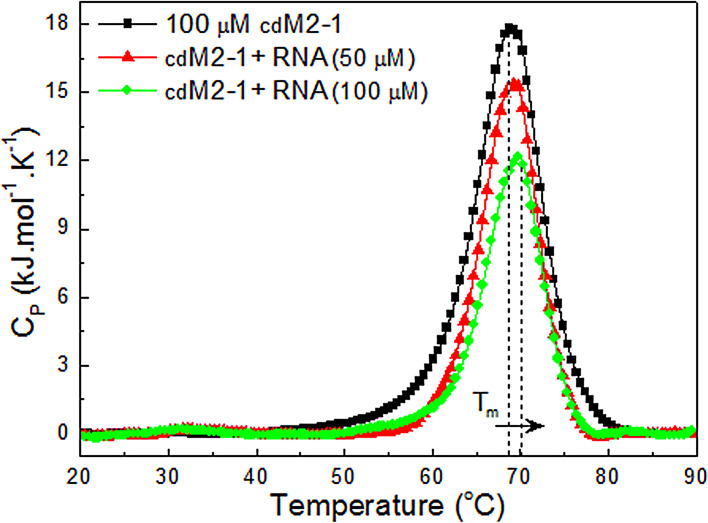
Thermal denaturation analysis by differential scanning calorimetry (DSC) showed that cdM2-1 has an endothermic transition with a melting temperature () of 68.7 ± 0.4°C and a calorimetry enthalpy change () of 174 ± 18 kJ · mol−1 (Fig. 2). Interestingly, Teixeira and collaborators showed a value of ∼55°C for the full-length hRSV M2-1, which is at least 13°C lower than that of its core domain (14). The van’t Hoff enthalpy change () calculated from the integration of the cdM2-1 thermogram revealed a value of 341 ± 30 kJ · mol−1, which is different from . This result indicated that the temperature-induced unfolding of cdM2-1 did not follow a two-state model. The thermal denaturation of the protein is irreversible, showing no transition in the cooling process. The and values for the thermal unfolding of the cdM2-1/RNA complex were 69.4 ± 0.1°C and 136 ± 10 kJ · mol−1, respectively, for a protein/RNA molar ratio of 1:0.5 and 69.8 ± 0.1°C and 103 ± 8 kJ · mol−1, respectively, for a molar ratio of 1:1 (Fig. 2), indicating that the binding of cdM2-1 to RNA promoted a slight change in the thermal stability of the protein.
FIG 2.
Thermal stability of free and RNA-bound cdM2-1. DSC thermograms (showing specific heat capacity [Cp] as a function of the temperature) of the protein and its complex with RNA collected in 50 mM phosphate buffer (pH 6.5) containing 150 mM NaCl and 1.0 mM DTT using a scan rate of 1.0°C/min are shown. The dotted lines denote the melting temperature (Tm) for cdM2-1 in the absence and the presence of RNA (100 μM). The concentration of the protein was 100 μM, and that of nucleic acid was 0, 50, and 100 μM.
Mapping cdM2-1 interaction with RNA.
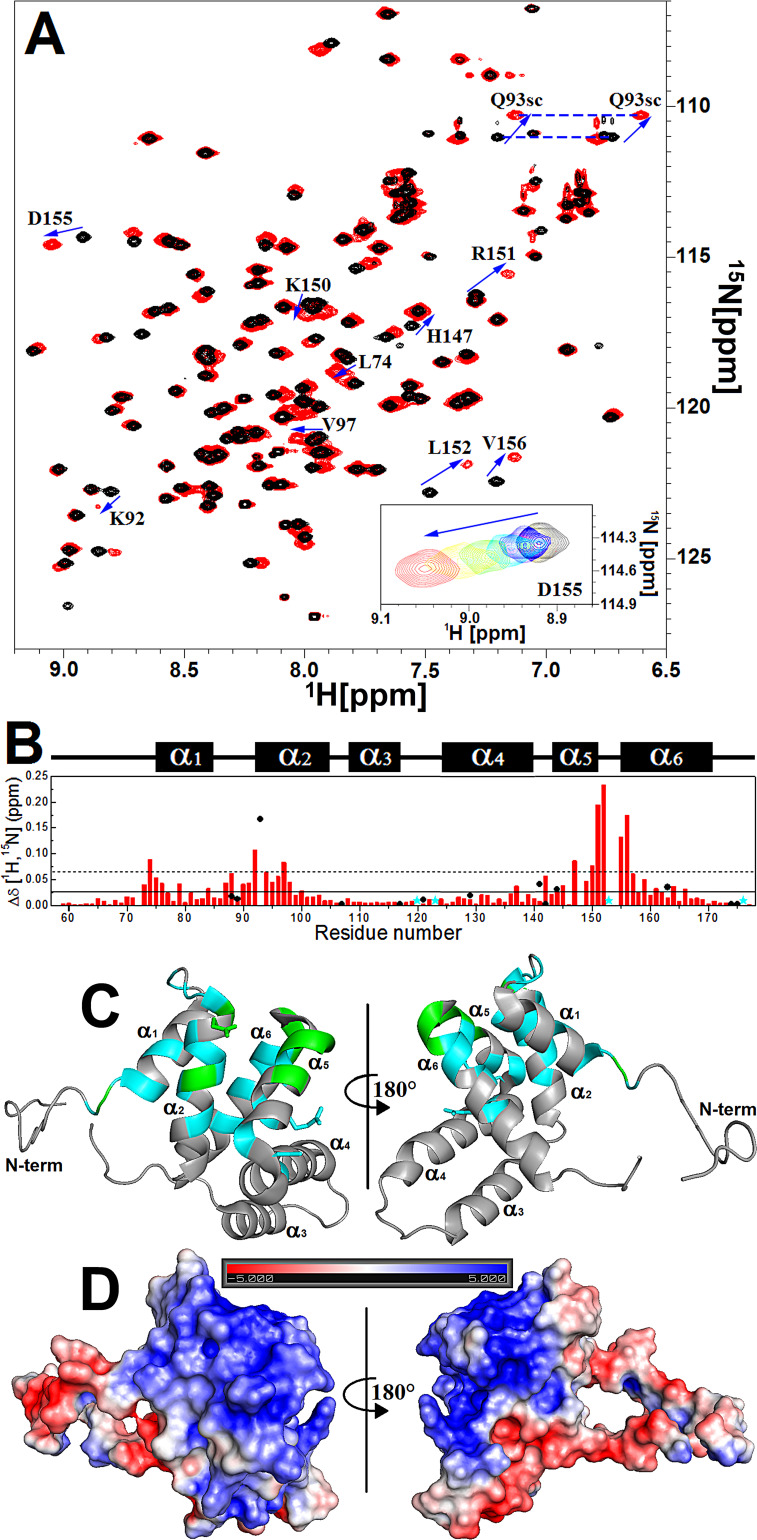
Analysis of the chemical shift perturbation (CSP) by NMR spectroscopy was also used to characterize cdM2-1/RNA binding (Fig. 3A). The CSP experiments presented a fast chemical exchange process between the free and bound forms on the NMR chemical shift timescale (Fig. 3A, inset). The amino acid residues associated with resonances that showed a chemical shift perturbation [Δδ(1H,15N)] value higher than the average value of Δδ (Δδave) plus the standard deviation (SD) were as follows (Fig. 3B and C): at the N terminus, Leu74; in the α2 helix, Lys92, the Gln93 side chain, and Val97; in the α5 helix, His147, Lys150, and Arg151; in the α5-α6 loop, Leu152; and in the α6 helix, Asp155 and Val156. These residues of cdM2-1 promoted the binding to RNA either by a direct interaction or by conformational rearrangements remote from the interaction interface. The resonances of residues Gly62, Ala63, Lys92, Gln93, Leu152, and Ala154 were broadened or even disappeared under binding to nucleic acid. The chemical shift changes of the amide group of [U-15N]cdM2-1 under binding to RNA revealed that the resonances of regions in the α1-α2-α5-α6 helix bundle presented the most significant Δδ(1H,15N) values, as seen by Blondot et al. (7). These regions denote a positive electrostatic potential (Fig. 3D) that is favorable for the interaction with a negative charge of the phosphate groups from the nucleic acid. In particular, Lys92, Lys150, and Arg151, which showed large chemical shift changes (Fig. 3B), are residues conserved between hRSV M2-1 and hMPV M2-1. Recently, Gao et al. characterized these charged residues to be essential for the RNA interaction from the crystal structure of hRSV M2-1 complexed with 7-mer RNA (15). It is worth mentioning that Lys150 and Arg151 were identified from mutagenesis assays to be key residues for transcription enhancement in vivo by hRSV M2-1 (7).
FIG 3.
cdM2-1/RNA binding investigated by NMR spectroscopy. (A) Two-dimensional 1H-15N HSQC spectra of the free (black) and RNA-bound (red) [U-15N]cdM2-1 collected by using an NMR spectrometer of 14.1 T (1H frequency, 600 MHz) at a temperature of 25°C. The arrows indicate the residues that presented a chemical shift perturbation (Δδ) upon RNA binding higher than the average chemical shift perturbation (Δδave) plus the SD. (Inset) Titration effect on the behavior of a fast exchange regime for Asp155 upon RNA binding. The spectra were recorded at a protein concentration of 100 μM (black) and at an RNA concentration of 15 μM (blue), 30 μM (cyan), 55 μM (green), 80 μM (yellow), and 115 μM (red). The dashed blue line links the two resonances of the side chain of Gln93 (Q93sc). (B) Chemical shift perturbation for the formation of the [U-15N]dgM2-1/RNA complex, where Δδ = (ΔδHN2 + ΔδN2/25). The solid line denotes Δδave, and the dashed line indicates Δδave + SD. The black circles are the Δδ values for the side chains of the Asn and Gln residues. The cyan stars indicate the proline residues (Pro120, Pro123, Pro153, and Pro176). The secondary structures along the sequence are indicated at the top. (C) Identification on the cdM2-1 structure of residues that present a significant Δδ upon RNA binding. Values of Δδ higher than Δδave are in cyan (Ala73, Gly75, Val76, Val79, Ile84, Ile87, Asn88, Ile90, Thr91, Ser94, Ala95, Cys96, Ala98, Ser100, Asn142, Thr145, Leu149, Leu157, Lys158, Lys159, Ile161, and Thr164), and values higher than Δδave plus SD are in green (for Leu74, Lys92, Gln93, Val97, His147, Lys150, Arg151, Leu152, Asp155, and Val156). N-term, N terminus. (D) Electrostatic potential surface of cdM2-1 calculated with APBS software, using the charge values and protonation states (pH 6.5, 150 mM NaCl, 25°C) determined by the PDB2PQR web server along with the PROPKA program. The bar denotes the electrostatic potential range from −5 kT (red) to +5 kT (blue).
sPRE for free and RNA-bound cdM2-1.
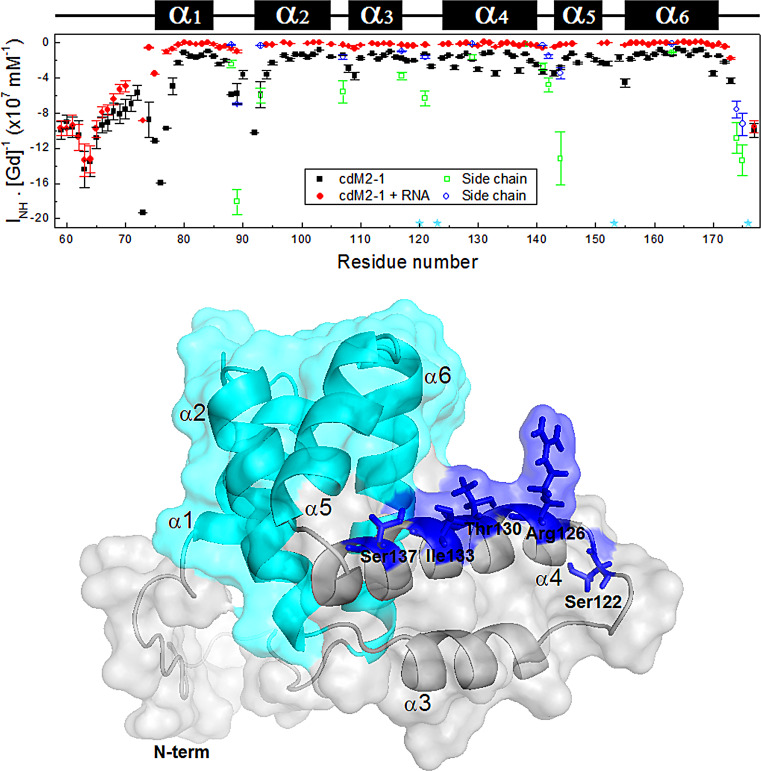
Solvent exposure measures of each cdM2-1 backbone amide group and its complex with RNA were obtained from solvent paramagnetic relaxation enhancement (sPRE) experiments (Fig. 4). The more negative that the sPRE effect (the intensity of each 1HN/15N cross-peak of the residues [INH] as a function of the Gd-diethylenetriamine pentaacetic acid-bismathylamide [Gd-DTPA] concentration and the slope of the adjusted straight line [INH · [Gd]−1]) is (Fig. 4, top), the more exposed the residue to the solvent is. For the domain in the absence of RNA, it was possible to observe that the greatest sPRE effects occurred in the N and C termini of cdM2-1 and that they mainly occurred in the N-terminal residues of the α1 helix, characterizing these regions as being significantly exposed to the solvent. Significant sPRE effects were also observed in virtually all C-terminal residues of the protein loop regions, highlighting the α1-α2 loop (Fig. 4, top). In general, the helical secondary structure regions had the lowest sPRE effects, indicating that the amide groups of the residues in these regions are protected from solvent exposure. An exception to this is the α4 helix, which exhibited an i + 4 pattern of solvent-exposed residues (Ser122, Arg126, Thr130, Ile133, and Ser137) corresponding to a nearly α-helix turn (Fig. 4, bottom). These data revealed the presence of a solvent-accessible cleft between the α1-α2-α5-α6 helix bundle and the α3-α4 hairpin (Fig. 4, bottom).
FIG 4.
Solvent paramagnetic relaxation enhancement for free and RNA-bound cdM2-1. (Top) Solvent exposure (INH · [Gd]−1) of each backbone NH amide of the protein (black squares) and its complex with RNA (red circles) measured as a function of the residue number. The cdM2-1 concentration was 350 μM, and the RNA concentration was 115 μM, the temperature was 25°C, and the NMR spectrometer was 14.1 T (1H frequency, 600 MHz). The green squares and blue circles denote the solvent exposure for the side chains of the Asn and Gln residues, respectively. The cyan stars indicate the proline residues (Pro120, Pro123, Pro153, and Pro176). The secondary structures along the sequence are indicated at the top. (Bottom) Structural representation of hRSV cdM2-1, shown as a cartoon with a transparent surface. The residues Ser122, Arg126, Thr130, Ile133, and Ser137 in the α4 helix (blue sticks) exhibit an i + 4 pattern of solvent exposure corresponding to a nearly α-helix turn. The cyan color denotes the α1-α2-α5-α6 helix bundle.
For the cdM2-1/RNA complex, a significant reduction of the sPRE effect can be seen throughout all the residues of the domain, except the N and C termini (Fig. 4, top). This result indicates that the helical secondary structure regions, along with their loops, are permanently protected from solvent exposure when cdM2-1 is complexed with long RNAs, while the termini of the domain remain exposed to the solvent, as observed for free cdM2-1. Thus, the N and C termini of the core domain of hRSV M2-1 might be considered intrinsically negative internal controls for the experiments performed.
Thermal susceptibility measurements of the amide hydrogen chemical shifts.
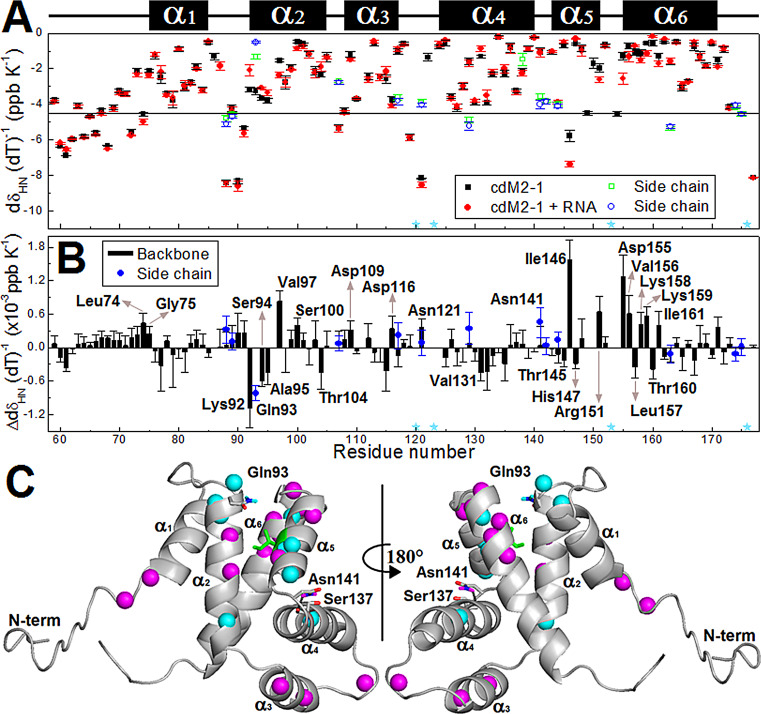
The thermal susceptibility of the amide hydrogen (1HN) chemical shifts () of cdM2-1 was measured in the absence and the presence of RNA. is correlated to the strength of the hydrogen bonds formed by 1HN, with values less than −5.0 ppb · K−1 being interpreted as weak hydrogen bonds (more expandable), which are usually found in unstructured and loop regions, and values greater than −5.0 ppb · K−1 being interpreted as strong hydrogen bonds, which are typical of secondary structure elements (Fig. 5A) (see Materials and Methods for details about the interpretation of values). In general, cdM2-1 residues in secondary structure elements presented values greater than −5.0 ppb · K−1, except for Ile146 in the α5 helix (green sticks in Fig. 5C). Ile146 showed an unusual value (less than −5.0 ppb · K−1) (Fig. 5A) for a residue in the secondary structure, which stands out as a possible point of a break in the hydrogen bond network of the α5 helix. On the other hand, most values of lower than −5.0 ppb · K−1 were reported for residues in the N and C termini and loops between helices of the core domain. However, Glu59, Ala63, Asp67, Thr69, Glu70, Glu71, and Ala73 in the N terminus of cdM2-1 showed values that were greater than −5.0 ppb · K−1, indicating that the amide group of these residues might form less expandable hydrogen bonds, as is typically observed for 1HN residues in secondary structures (16). These residues are not structured in the N terminus of cdM2-1 (7), and, interestingly, they are in α-helix secondary structures in full-length hRSV M2-1. These data revealed a helical propensity of this region for cdM2-1.
FIG 5.
Thermal susceptibility data for the amide hydrogen (1HN) chemical shifts of the free and RNA-bound cdM2-1. (A) values of the protein (black squares) and its complex with RNA (red circles) as a function of the residue number. The temperature dependence measurements were obtained from 2D 1H-15H HSQC spectra at 15, 20, 25, 30, and 35°C. The cdM2-1 concentration was 350 μM, the RNA concentration was 115 μM, and the NMR spectrometer was 14.1 T (1H frequency, 600 MHz). The green squares and blue circles denote the values of for the side chains of the Asn and Gln residues, respectively. The cyan stars indicate the proline residues (Pro120, Pro123, Pro153, and Pro176). The secondary structures along the sequence are indicated at the top. (B) Difference between the values of for free and RNA-bound cdM2-1 as a function of the residue number (). plus SD values far from zero were considered significant changes, and the correspondent residue names are denoted. (C) Significant changes of values between the free and bound states of the cdM2-1 are indicated in the protein structure. The values of (increase in ) and (decrease in ) for the 1HN chemical shifts are denoted as cyan and magenta spheres, respectively. The side chain of Gln93 and Asn141 are displayed as sticks, with amide hydrogens colored according to . The side chain of Asn141 forms a hydrogen bond (black dashed line) with the backbone of Ser137. Ile146 is shown as green sticks because it presents a value of greater than −5.0 ppb · K−1.
The binding of cdM2-1 to long RNAs promoted significant changes in the values for residues all over the domain (Fig. 5A). Changes in the values of () significantly different from zero were taken into consideration (Fig. 5B). Most of these residues map the protein/RNA interaction interface delimited by regions in the N terminus, α1, α2, α5, and α6 (Fig. 5C), which corroborates the results of the CSP experiments (Fig. 3B). Leu74, Gly75, Val97, Ser100, Asp109, Asp116, the Asn141 side chain, Ile146, Arg151, Asp155, Val156, Lys158, Lys159, and Ile161 showed an increase in values (), which indicates that the hydrogen bonds formed by the 1HN of these residues became more expandable and possibly weaker, while the hydrogen bonds established by 1HN of Lys92, the Gln93 side chain, Ser94, Ala95, Thr104, Val131, Thr145, His147, Leu157, and Thr160 turned into less expandable, possibly stronger hydrogen bonds (). In particular, the decrease for the Gln93 side chain could be explained by a direct hydrogen bond with RNA since it is exposed to the solvent in free cdM2-1 (Fig. 4, top) (7), while the increase for the Asn141 side chain could be due to a weakening of its hydrogen bond formed with Ser137 in the α4-α5 loop (7), which possibly comes from a conformational rearrangement of the α3-α4 hairpin. In the cdM2-1/RNA binding interface, there are two clusters of residues, with their 1HN residues forming hydrogen bonds that became less and more expandable, with the first cluster being composed of the Lys92, Gln93 side chain, Ser94, and Ala95 residues in the N terminus of the α2 helix and the second one being formed by Asp155, Val156, Lys158, Lys159, and Ile161 in the N terminus of the α6 helix. Altogether, these results indicate that the α1-α2-α5-α6 helix bundle suffers local structural changes upon RNA binding and that the α3-α4 hairpin shows punctual conformational rearrangements remote from the binding region.
Evidence of cdM2-1-induced structural changes in RNA.
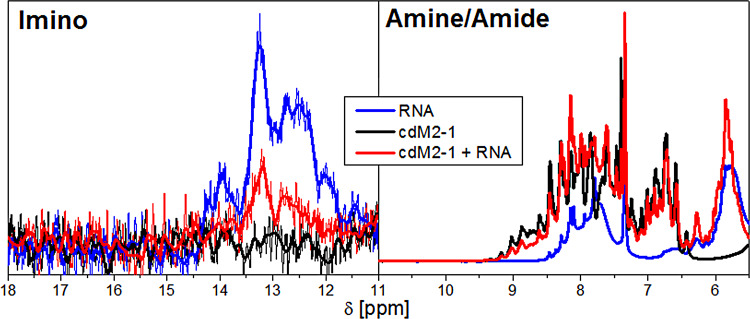
The one-dimensional (1D) 1H NMR spectra of protein/nucleic acid complexes reveal that most of the resonance signals from the nucleic acid and protein overlap, as can be observed from the amine/amide proton region (6 to 10 ppm) in Fig. 6. The only spectral region that is not disturbed by the protein resonances is the imino proton region from 11 to 15 ppm (17). The resonances of imino protons are directly involved in base pairing, which is remarkable evidence of the secondary structure organization of the nucleic acid. Figure 6 shows the imino proton signals of the protein-free and bound forms of the RNA, as well as the spectrum of free cdM2-1 as a negative control, since proteins have no observable resonances at this spectral region. The 1D 1H spectrum of the protein-free RNA presented resonance signals of imino protons in the 11- to 15-ppm range (Fig. 6, blue line), which indicates a structural arrangement level due to partial formations of RNA base pairing. On the other hand, the 1H spectrum of the cdM2-1-bound RNA showed a significant decrease in the intensities of the imino proton signals (Fig. 6, red line), which suggests a protein-induced unfolding of the RNA in its secondary structure elements. No significant line broadening was observed in the amine/amide region (6 to 10 ppm in Fig. 6) of the 1D 1H spectra, which shows that the decrease in the intensities of the imino proton signals is not due to the larger size of the cdM2-1/RNA complex.
FIG 6.
Evidence of the structural changes of RNA induced by cdM2-1. The 1D 1H NMR spectra of the imino (11 to 15 ppm) and amine/amide (6 to 10 ppm) proton regions of the protein-free (blue line) and bound (red line) states of RNA are shown. The black line presents the 1D 1H spectrum of free cdM2-1 as a negative control, especially for the imino region. The cdM2-1 concentration was 350 μM, the RNA concentration was 115 μM, the temperature was 25°C, and the NMR spectrometer was 14.1 T (1H frequency, 600 MHz). In the imino proton region, the thick lines denote smoothed profiles that assist with the visualization of the spectra.
Backbone dynamics of hRSV cdM2-1 and its complex with RNA.
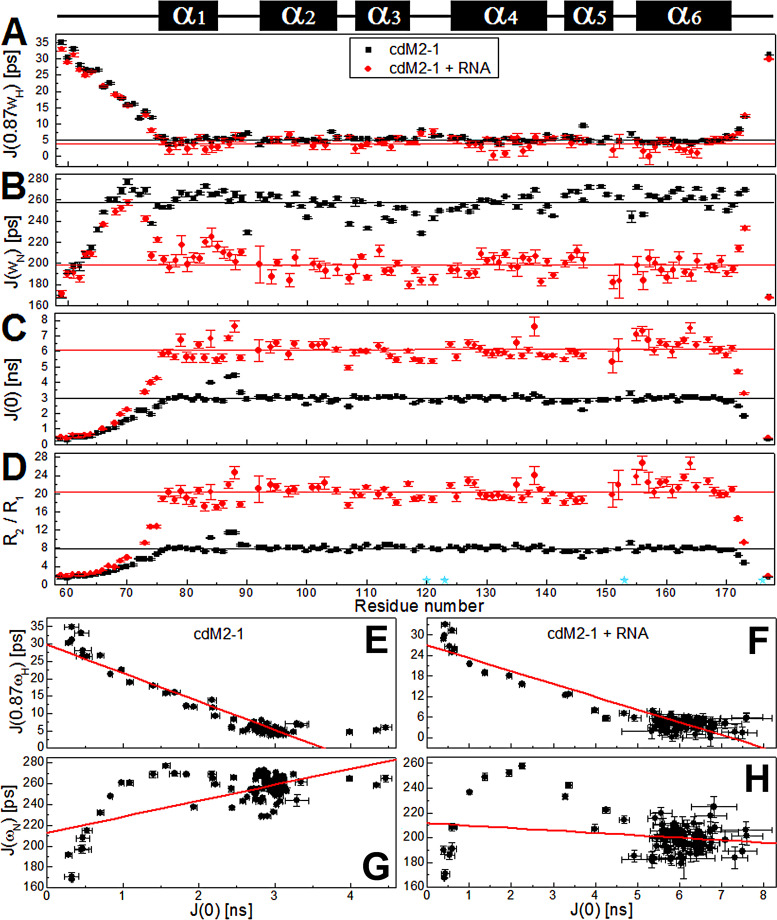
The 15N relaxation data for the protein backbone by NMR on picosecond-to-nanosecond timescales revealed the molecular dynamics of the protein in the absence and the presence of RNA. The results of the spin nuclear relaxation (15N R1 [longitudinal relaxation rate], R2 [transverse relaxation rate], and heteronuclear 1H-15N nuclear Overhauser effect [hetNOE]) experiments with the cdM2-1 and cdM2-1/RNA complex are shown in Table S1 in the supplemental material. In the presence of RNA, there was a significant change in the values of 15N R1, R2, and hetNOE, highlighting the increase in the average value of 15N R2 from 11.32 ± 1.10 to 22.24 ± 2.07 s−1 upon RNA binding (Table S2), which confirmed the formation of the cdM2-1/RNA complex. The N and C termini presented low hetNOE values in the absence and the presence of RNA, meaning that they are flexible in the free and bound states.
The reduced spectral density mapping, calculated from the relaxation parameters, provides insights into the internal motions of a protein with no a priori assumptions about its diffusion model. The spectral density (J) functions (see Materials and Methods), (spectral density at the nitrogen frequency), and (spectral density at the zero frequency) are sensitive to overall and intramolecular motions on picosecond-to-nanosecond timescales. and report fast motions in picoseconds, while probes motions in nanoseconds, with possible contributions from the chemical or conformational exchange term. The values of , , and for free and RNA-bound cdM2-1 were calculated from equations 3 to 5 and are presented in Fig. 7A to C. As with the relaxation parameters, the spectral density values of cdM2-1 showed outstanding changes in the presence of RNA, except for those for the N and C termini. The highest and lowest values of and , respectively, for residues in the N and C termini indicate that these regions undergo fast internal motion on a picosecond timescale and are highly flexible even after binding to long RNAs. The plots of and (Fig. 7E and F) indicated that these two spectral densities were significantly correlated with correlation coefficients of −0.934 and −0.931 for cdM2-1 in the absence and the presence of RNA, respectively. On the other hand, the and plots (Fig. 7G and H) showed smaller correlation coefficients, with values of 0.642 and −0.227 for the RNA-free and bound forms, respectively. It is noteworthy that the and plot presented a negative correlation for the bimolecular complex and a positive correlation for free cdM2-1. The average values of , , and when the terminal residues for the free core domain were excluded were 5.2 ± 1.0 ps, 258 ± 10 ps, and 3.0 ± 0.3 ns, respectively, whereas the corresponding values for the RNA-bound cdM2-1 were 4.1 ± 1.6 ps, 198 ± 10 ps, and 6.1 ± 0.5 ns, respectively (Table S2).
FIG 7.
15N nuclear spin relaxation data for the free and RNA-bound cdM2-1 backbone amide by NMR on a picosecond-to-nanosecond timescale. (A to C) Reduced spectral density functions (A), (B), and (C) of cdM2-1 and its complex with RNA, which were calculated from the 15N R1, R2, and hetNOE relaxation data. (D) R2/R1 ratio calculated from relaxation rates R1 and R2. The black and red lines denote the average values of , , and for free and RNA-bound cdM2-1, respectively. To determine the average values, those corresponding to the N- and C-terminal residues were excluded. The cyan stars indicate the proline residues (Pro120, Pro123, Pro153, and Pro176). The secondary structures along the sequence are indicated at the top. (E to H) Plots of the correlations of and and of and for free cdM2-1 (E and G) and RNA-bound cdM2-1 (F and H). The cdM2-1 concentration was 350 μM, the RNA concentration was 115 μM, the temperature was 25°C, and the NMR spectrometer was 14.1 T (1H frequency, 600 MHz). The red lines denote the linear fitting adjustment.
R2/R1 ratios are proportional to the protein rotational correlation time (), and values above the average may be involved in conformational exchange. The R2/R1 ratios calculated for the residues of cdM2-1 in the absence and the presence of nucleic acid demonstrated complex formation, as the average R2/R1 value obtained when the N- and C-terminal residues were excluded increased from 8.0 ± 0.8 to 20.4 ± 2.2 ns upon the addition of RNA (Fig. 7D). For free cdM2-1, the R2/R1 values for Ile84, Ile87, and Asn88 in the α1 helix and the α1-α2 loop were above the average, suggesting that these residues are in conformational exchange (Fig. 7D). After the formation of the cdM2-1/RNA complex, the residues Ile87 and Asn88 in the α1-α2 loop still presented R2/R1 values higher than the average, while other residues, such as Asp155, Val156, Asn163, and Thr164, in the α6 helix, had an increase in ratio values above the average, possibly due to chemical exchange (binding equilibrium). Interestingly, Asn138 in the α4 helix had R2/R1 and values above the average upon RNA binding, which reinforces the idea of α3-α4 hairpin conformational rearrangements remote from the interaction interface.
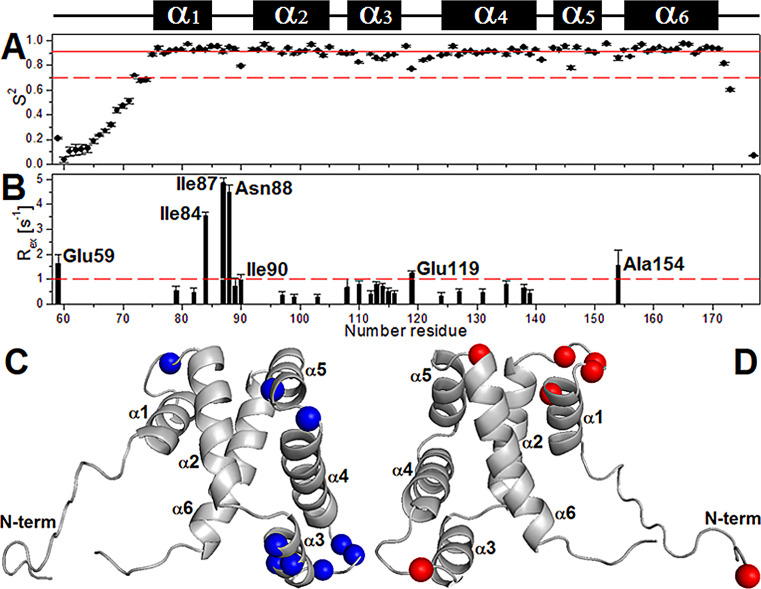
The extended Lipari-Szabo model-free formalism was used to analyze the relaxation parameters to obtain a hydrodynamic description of the protein informing the global diffusion anisotropy and the local motions of cdM2-1 (Table S3). A rotational correlation time () for the protein tumbling of 8.53 ± 0.07 ns at 25°C, compatible with that for the monomeric protein, was calculated using a fully anisotropic model for molecular rotational diffusion (see the details in Material and Methods). For comparison with experimental data, an estimate of the value was calculated using the HYDRONMR program (18) and the 20 NMR structures of cdM2-1 (Protein Data Bank [PDB] accession number 2L9J) (7), excluding amino acid residues from the flexible termini, and the calculated was 8.1 ± 0.3 ns at 25°C. Values of the order parameter (S2) of <0.7 for the N- and C-terminal residues indicated significant thermal fluctuations and a high degree of internal mobility for these protein regions (Fig. 8). Residues located in the α1-α2 loop (Ile90), the α3 helix (Asp110, Lys113, Leu114), the α3-α4 loop (Glu119, Asn121, Ser122), the α4-α5 loop (Asn141), and the α5 helix (Ile146) showed significantly lower S2 values than the helical-core region average (0.91 ± 0.04, excluding the terminal regions), which suggests a certain level of flexibility (Fig. 8A and C). The below-average S2 value, along with a value of less than −5.0 ppb · K−1, for Ile146 in the α5 helix reinforces the assumption that this residue is a point of break of the cdM2-1 secondary structure. Conformational exchange contributions with conformational exchange rate (Rex) values of >1.0 Hz were observed for residues Glu59, Ile84, Ile87, Asn88, Ile90, Glu119, and Ala154 of cdM2-1 (Fig. 8B and D). The model-free analysis of Lipari-Szabo for the RNA-bound cdM2-1 did not provide a satisfactory result.
FIG 8.
The extended model-free formalism of Lipari-Szabo applied to analyze the nuclear spin relaxation data. (A and B) Order parameter (S2) (A) and conformational exchange rate (Rex) (B), calculated with TENSOR2 software from the 15N nuclear spin relaxation data for the backbone of cdM2-1 using the Lipari-Szabo model-free formalism. The continuous and dashed red lines in panel A denote the average value of S2 (0.91 ± 0.04), excluding terminal residues, and an S2 value of 0.7, respectively. The dashed red line in panel B depicts the cutoff value of Rex equal to 1.0 Hz. The cdM2-1 concentration was 350 μM, the RNA concentration was 115 μM, the temperature was 25°C, and the NMR spectrometer field was 14.1 T (1H frequency, 600 MHz). (C and D) Cartoon representations of cdM2-1. Blue and red show residues with significant values of S2 (<0.91 ± 0.04; Ile90, Asp110, Lys113, Leu114, Glu119, Asn121, Ser122, and Asn141) (C) and Rex (>1.0 Hz; Glu59, Ile84, Ile87, Asn88, Ile90, Glu119, and Ala154) (D), respectively.
Computational approach of the cdM2-1/RNA interaction.
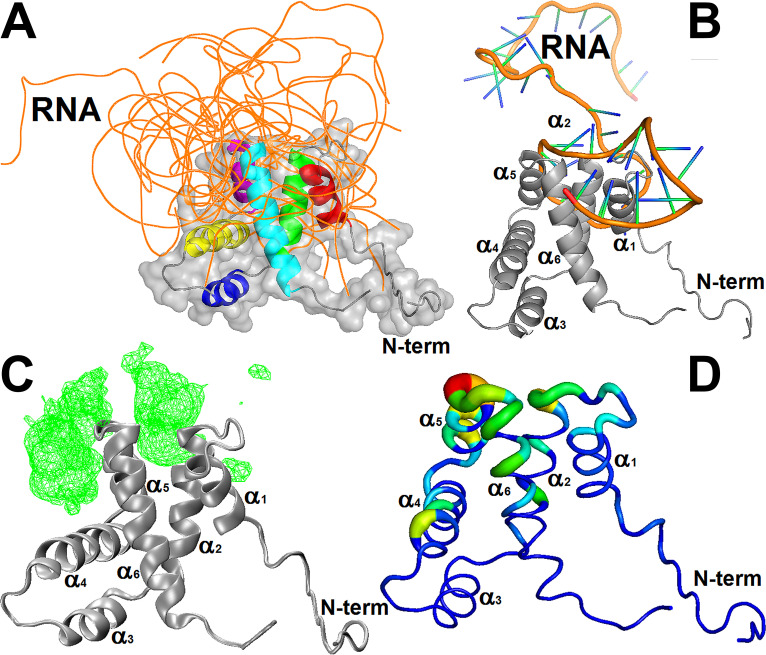
The structural models of the cdM2-1/RNA complex were generated using the 3dRPC web server (19). The structural restraints of the complex were defined from CSP analysis, defining residues to be involved in the binding interface when Δδ was greater than Δδave. Figure 9A shows 10 structural models of the cdM2-1/RNA interaction determined by the use of independent docking calculations, which correspond to the lowest-energy structures from 10 complex models predicted by the 3dRPC-Score function (19). The 10 RNA molecules crowded around the α1-α2 and α5-α6 loops, protruding like an umbrella over the α1, α2, α5, and α6 helices and the solvent-accessible cleft formed between the α1-α2-α5-α6 helix bundle and the α3-α4 hairpin. Such behavior can be evidenced by analysis of the mass density map of the protein-bound RNA (Fig. 9C) and by the change in the absolute accessible surface area (ΔASA) of the RNA-bound cdM2-1 (Fig. 9D). The docking calculations also showed that cdM2-1 preferably binds close to the secondary structure regions of the RNA molecules, where the nucleic acids presented a conformational organization level due to base pairing. This fact is exemplified by the cdM2-1/RNA complex model in Fig. 9B, where the α1-α2-α5-α6 helix bundle of the protein lies between two stem-loop secondary structures of the nucleic acid. The evaluation of the noncovalent interactions carried out with the PLIP server (20) revealed that residues Lys150 and Arg151 are involved in salt bridges and hydrogen bonds in 9 and 8 of 10 cdM2-1/RNA complex models, respectively (Tables S4 to S13). In general, Lys150 formed two salt bridges with phosphate groups of adjacent nucleotides, while Arg151 established hydrogen bonds with nitrogenous bases and ribose groups of the nucleic acids. These two residues also took place in π-cation interactions with the heterocyclic rings of nitrogenous bases in at least 3 of 10 structural models of the cdM2-1/RNA complex. Recently, the participation of Lys150 in salt bridges was also reported by Gao et al. for the crystal structure of the hRSV M2-1/7-mer RNA complex (15).
FIG 9.
Analysis of the molecular dockings of the structural models of the cdM2-1/RNA complex. (A) Structural models of the cdM2-1/RNA complex calculated by the 3dRPC web server. The backbone of the protein (different colors) and the 10 nucleic acids (orange) are presented as a cartoon and ribbons, respectively. The protein is also shown as a surface representation, and its helices are highlighted using different colors (red, α1; green, α2; yellow, α3; blue, α4; purple α5; cyan, α6). (B) Structural model of the cdM2-1/RNA complex, which exemplified the preferential binding of the protein (gray cartoon) close to secondary structural regions of the RNA molecules (shown as a cartoon; orange, backbone; green/blue, nucleosides). (C) Mass density map (green mesh with an isovalue of 0.5) of 10 RNA molecules bound to the protein (gray cartoon) generated by the VMD program using the VolMap tool. (D) Absolute accessible surface area change (ΔASA) of cdM2-1 (as a colored worm) bound to the nucleic acids. The thickness and colors represent the degree of ΔASA, with the strongly and weakly accessible residues shown in blue (thin) and red (thick), respectively. The structural representations were made with the PyMOL and VMD programs.
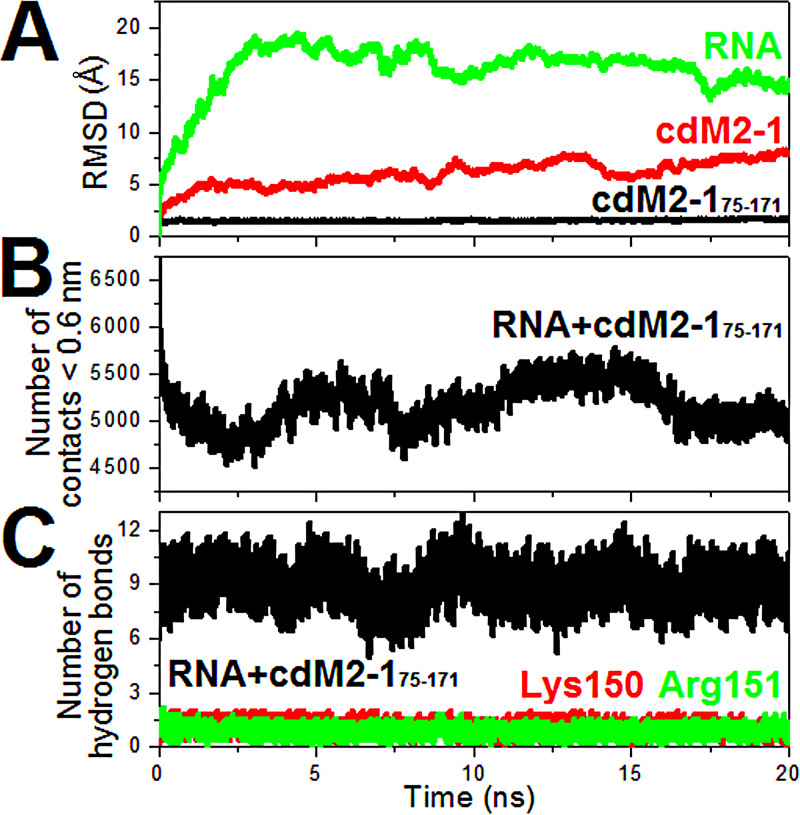
The MD calculations were used to check the structural stability of the cdM2-1/RNA complex models generated from the 3dRPC server. The number of hydrogen bonds, the number of contacts, and the root mean square deviation (RMSD) analyzed from the 20-ns MD simulations for the complex models are presented as average values for five independent calculations in Fig. 10. RMSD values for nonhydrogen atoms of RNA and backbone atoms of the helical-core region of the domain (residues 75 to 171 of cdM2-1 [cdM2-175–171]) reached stable levels after 2.5 and 1.0 ns, respectively, whereas significant differences for the entire cdM2-1 were observed due to the flexible terminal region contributions (Fig. 10A). The number of contacts between atoms of the cdM2-175–171 and RNA molecules for distances of <0.6 nm was almost stable, being approximately 5,200 (Fig. 10B), indicating that the protein and nucleic acids remained interacting throughout the simulation time. Figure 10C shows that the number of hydrogen bonds formed between cdM2-175–171 and nucleic acids were very stable, as were all the MD simulations, with an average value of about 9, with Lys150 and Arg151 being responsible for at least one hydrogen bond each. Therefore, the molecular dynamics calculations proved the stability of the structural models of the cdM2-1/RNA complex over 20-ns simulations.
FIG 10.
Analysis of the molecular dynamics of the structural models of the cdM2-1/RNA complex. (A) RMSD values for the nonhydrogen atoms of RNAs (green line), for the backbone atoms of the helical-core region of the protein (cdM2-175–171; black line), and for the backbone atoms of the entire cdM2-1 (red line). (B) Number of contacts between the atoms of cdM2-175–171 and RNAs for distances of <0.6 nm. (C) Number of hydrogen bonds formed between the RNA molecules and cdM2-175–171 (black line), the Lys150 residue (red line), and the Arg151 residue (green line).
DISCUSSION
This study has presented the first evidence that cdM2-1 alone has an unfolding activity for long RNAs and therefore has an active role in the function of the full-length protein as a processivity and antitermination factor of the RdRp complex. The intensity of the imino proton resonances is a direct measure of the base pairing that stabilizes RNA secondary structures. The reduction in these intensities was explained by the destabilizing activity of cdM2-1 (Fig. 6), promoting an unfolding of RNA secondary structures. This structural finding is different from that reported by Molina et al., in which they observed no significant structural change from that for the 20-mer RNA band at 270 nm in the presence of cdM2-1 via CD measurements (10). The evidence that cdM2-1 alone induces secondary structure unfolding in long nucleic acid molecules may be a consequence of the molecular recognition of RNA base pairing by this domain. Such logical thinking sums with the outcome reported by Blondot et al., in which cdM2-1 presented a higher affinity for the double-stranded RNA (12-mer) than for its corresponding single-stranded ones, highlighting the key role of base pairing of the nucleic acid for the cdM2-1/RNA interaction (7).
The molecular docking calculations revealed that the structural models of the cdM2-1/RNA complex are supported by the findings of the CSP and sPRE analyses (Fig. 3 and 4), which suggest that even though the nucleic acids specifically bind to the distal portions of the α1, α2, α5, and α6 helices, the RNA molecules may surround the entire helical-core region of the protein (Fig. 9A, C, and D). The theoretical structural models of the complexes also corroborate evidence of the RNA unfolding induced by cdM2-1 verified by NMR (Fig. 6), since the models indicate that the domain preferably binds to the RNA secondary structure regions (Fig. 9B). Therefore, the hypothesis is that the unfolding activity of long RNAs would be triggered by molecular recognition of base pairing.
The cdM2-1/RNA binding presents Kds in the nanomolar-to-micromolar range (Table 1), which were similar to values reported by Blondot et al. (7) and Molina et al. (10). On the other hand, the full-length M2-1 has Kds of 1/10s of nanomolar that were at least 10-fold lower than those recorded for cdM2-1 (8, 10, 21). This indicates that although the core domain is responsible for the first molecular contact with the nucleic acid molecules, there is another region on M2-1 that provides an increase in the affinity with RNA. These regions may be the zinc finger domains, which provide a type of binding specificity for adenine bases at specific positions in the nucleotide sequence (9, 15), and/or the neighboring core domains in the tetrameric arrangement, promoting a positive cooperative binding to two RNA molecules of 13-mer or longer per tetramer of M2-1 (10). Interestingly, Gao et al. recently revealed from crystal structures and molecular dynamics analysis of the hRSV M2-1/7-mer RNA complex that the interactions of RNA with two separate domains of M2-1, the zinc finger domain and the core domain, can be independent of each other (15).
The temperature, ionic strength, and pH dependence on the cdM2-1/RNA binding affinity suggests that electrostatic interactions drive the formation of the encounter complex, while van der Waals interactions and hydrogen bonds are responsible for complex stabilization ( and ) (13). An analysis of noncovalent interactions of the cdM2-1/RNA docking models shows the occurrence of hydrogen bonds, salt bridges, and π-cation interactions (see Tables S3 to S12 in the supplemental material), which is in line with the interpretation of the thermodynamic parameters. These results are important to understand the M2-1/RNA interaction, since they indicate that this process is fine-tuned by the molecular environment. In the cell, M2-1 is found in cytoplasmic inclusion bodies, in which all the components of the viral polymerase concentrate and where RNA synthesis occurs (22).
DSC data revealed that cdM2-1 is more thermally stable than the full-length M2-1. This behavior is not observed when the domains intimately interact as tethered domains in the multidomain proteins (23). This indicates that the thermal stability of cdM2-1 is affected due to contacts in the full-length protein, which may promote a type of negative interdomain coupling interaction, as defined by Bhaskara and Srinivasan (23). The results also pointed out a slight increase in thermal stability upon RNA binding, indicating a preferential binding to the native state of the core domain (24).
The α1-α2-α5-α6 helix bundle is the main region involved in the cdM2-1/RNA interaction, as determined by CSP. This helix bundle delimits a positive electrostatic potential surface on cdM2-1 (Fig. 3D) that contributes to the formation of the encounter complex, considering the ionic strength and pH dependence on the affinity of binding to RNA (Fig. 1C and D). The thermal susceptibility measurements of 1HN chemical shifts suggest that the interaction with RNA promoted local structural rearrangements of the helix bundle, with residues in the N termini of the α2 helix and the α6 helix showing stronger (less expandable) and weaker (more expandable) hydrogen bonds, respectively (Fig. 5C). Except for the N and C termini of cdM2-1, the sPRE experiments pointed out that the helical-core region is permanently protected from solvent exposure when complexed with the long RNA (Fig. 4, top). This result, along with the analysis of CSP and the thermal susceptibility of 1HN chemical shifts, suggests that an RNA region anchors mainly in α1-α2 and α5-α6 loops, while other noninteracting portions of its length protrude over the α1-α2-α5-α6 helix bundle, wrapping the helical region of cdM2-1.
The 15N backbone dynamics data revealed that the N and C termini of cdM2-1 are equally flexible in the free and RNA-bound states, corroborating the results of analyses of sPRE, CSP, and , which showed that the terminal residues do not take part in the interaction with the nucleic acid and remain solvent exposed. The free cdM2-1 helical core has well-defined structures with few localized points of thermal flexibility in the loops (Fig. 8), which may serve as hinges that confer plasticity to the domain. Residues in the α1-α2 loop of free cdM2-1 undergo dynamics on a microsecond-to-millisecond timescale (conformational exchange) that are important for the molecular recognition of RNA since this region participates directly in the interaction interface. There are significant changes in the dynamics of the RNA-bound cdM2-1, showing R2/R1 ratios that characterize a decrease in the overall tumbling motion compared with that of the free domain and thus confirm the complexation. The values of and the R2/R1 ratio reveal that residues in the α6 helix displayed dynamics on a microsecond-to-millisecond timescale upon RNA binding, while the residues in the α1-α2 loop remained in conformational exchange. It is worth pointing out that the helix α1-α2 loop and α6-helix cdM2-1 are important for the binding to nucleic acids, as determined herein by CSP analysis and thermal susceptibility measurements and as also reported by Blondot et al. (7).
The best way to describe the dynamics of the cdM2-1/RNA complex is through reduced spectral density mapping. Here, this analysis enriched the dynamic information, indicating that the α2-α3 and α3-α4 loops of the protein became more flexible after binding to RNA, since these undergo dynamics on a picosecond timescale (fast internal motions). The mean values of the α2-α3 and α3-α4 loops decreased significantly with respect to the helical-core region average for the RNA-bound cdM2-1, while the mean values for these loops were slightly higher than the overall average (Table S2). The inverse correlation between and (Fig. 7E and F) has been widely reported (25–27), and it can be exemplified for Asn107 in the α2-α3 loop and Glu119 in the α3-α4 loop (Fig. 7A and C). The mean values of the α2-α3 and α3-α4 loops were significantly lower than the helical-core region average for the free domain, and after the interaction with RNA, these values approached the overall average (Table S2). Asp110, Lys113, and Lys114 in the α3 helix showed behavior similar to that shown by these loops (Fig. 7B). The direct and inverse correlations observed from the and plot for the protein in the absence and the presence of RNA (Fig. 7G and H), respectively, suggest that fast internal motions took place in the α2-α3 and α3-α4 loops and in the α3 helix before complexation, and after that, these fast dynamics increased. This result corroborates that from the analysis of and .
In conclusion, the present work describes that cdM2-1 alone has an unfolding activity for long RNAs, which is probably triggered by molecular recognition of the base pairing. The α1-α2-α5-α6 helix bundle is characterized as the major RNA binding region of cdM2-1 which undergoes local conformational changes due to the interaction. Although the α3-α4 hairpin is not directly involved in the binding to RNA, it presents conformational rearrangements remote from the interaction interface and also an increase in the picosecond motions of its α2-α3 and α3-α4 loops. cdM2-1 as a whole is surrounded by long RNAs, and the most likely hypothesis is that the helix bundle preferentially binds to the base-pairing region, while its other regions protrude over the helical-core region of the domain. Therefore, it is revealed that the cdM2-1/RNA complex originates from a fine-tuned binding which likely contributes to the interaction aspects required for the processivity and antitermination activity of M2-1.
MATERIALS AND METHODS
Sample preparation.
The core domain (residues 58 to 177) of hRSV M2-1 (cdM2-1) was expressed in Escherichia coli BL21(DE3) RIL cells with a pD441-NHT vector (ATUM, USA) in M9 minimal medium containing 15NH4Cl as the sole nitrogen source for the production of isotopically labeled protein, as described previously (28). The construct of the vector includes an N-terminal hexahistidine affinity tag (His6 tag) and a tobacco etch virus (TEV) cleavage site. After expression, the cell suspension was centrifuged and the pellet was resuspended in buffer A (50 mM Tris-HCl, pH 8.0, 500 mM NaCl, 1.0 mM β-mercaptoethanol, 5% [vol/vol] glycerol). The cells were lysed by sonication, and next, the crude extract was centrifuged. The supernatant was loaded on a Ni-nitrilotriacetic acid column for affinity chromatography, in which the column was preequilibrated with buffer A. The column was washed extensively with buffer A containing 10, 20, and 40 mM imidazole, and the protein was eluted with a 60 to 500 mM imidazole gradient. The eluted protein fractions were digested to remove the His6 tag using TEV protease at 20°C for 14 h. This step was performed via dialysis using a solution containing 20 mM Tris-HCl (pH 8.0), 1.0 mM dithiothreitol (DTT), and 0.5 mM EDTA. Next, the protein without the His6 tag was injected into a Superdex 75 10/300 GL (GE Healthcare) size exclusion column with buffer B (50 mM NaH2PO4/Na2HPO4, pH 6.5, 150 mM NaCl, 1.0 mM DTT), which was used for the fluorescence-quenching, CD, DSC, and NMR experiments. In the fluorescence-quenching and CD experiments, the protein sample was dialyzed against a citrate/phosphate and monobasic/dibasic phosphate buffer to reach pH 5 and 8, respectively. For NMR measurements, the protein sample contained 0.1% (wt/vol) NaN3 and 7% (vol/vol) D2O in phosphate buffer solution (buffer B). The RNA from Saccharomyces cerevisiae Torula yeast type VI (∼80 bases) was purchased from Sigma-Aldrich. The stock solution of RNA was prepared in the same buffer solution used for the protein, taking into account the pH control because of the solubilization of the nucleic acid. The concentrations of cdM2-1 and RNA were determined by determination of the absorbance at 280 and 260 nm, using extinction coefficients of 5,960 M−1 · cm−1 (by use of the ProtParam tool on the Expasy server) (29) and 8,050 M−1 · cm−1 · base−1 (DNA and RNA molecular weights and conversions; Thermo Fisher Scientific), respectively.
Fluorescence-quenching measurements.
The fluorescence-quenching experiments were performed using a fluorescent spectrometer (Lumina; Thermo Fisher Scientific, USA) equipped with a Peltier system for temperature control and a 10-mm-optical-path quartz cuvette. The excitation wavelength at 288 nm was set to promote the fluorescence emission of the protein. The emission spectra were reported in the 300- to 450-nm range with an increment of 1.0 nm, which was corrected for background fluorescence (from the buffer) and inner filter effects (30). Both excitation and emission bandwidths were set at 5 nm. Each point in the emission spectrum is the average of 10 accumulations. The titrations were performed by adding small aliquots of the RNA stock solution (715 μM) to 2 ml of cdM2-1 at a constant concentration (5.5 μM). The titration experiments were reported at different temperatures (15, 25, and 35°C) for determining the binding thermodynamic profile of the cdM2-1/RNA complex, as well as under different conditions of salt concentration (0, 150, and 350 mM NaCl) and pH (5, 6.5, and 8). The measurements were performed in duplicate.
The fluorescence-quenching data for the cdM2-1/RNA interaction were analyzed using the following equation (31):
| (1) |
where is the fluorescence intensity in the absence of the ligand (RNA), is the fluorescence in the presence of RNA, Fsat is the intensity of the bound protein saturated with RNA, is the dissociation constant, nSC is the stoichiometry coefficient of the cdM2-1/RNA complex and corresponds to the number of protein molecules bound to one nucleic acid molecule, is the total concentration of the protein, and is the total concentration of the ligand. The and nSC values were determined from the process of fitting of the data to the experimental data by nonlinear least-squares optimization using Levenberg-Marquardt interactions.
Thermodynamic profile analysis.
The driving forces responsible for the binding between biomolecules may include electrostatic interactions, hydrogen bonds, van der Waals interactions, and hydrophobic contacts (13). To elucidate the interactions involved in the cdM2-1/RNA complex, the thermodynamic parameters were calculated from the van’t Hoff equation:
| (2) |
where is the enthalpy change, is the universal gas constant, and is the dissociation constant at the corresponding temperature (T; 15, 25, and 35°C). The value was obtained from the slope of the van’t Hoff plot, with the respective values of the Gibbs free energy changes () being determined from the equation and the entropy change () being determined from the equation .
Circular dichroism measurements.
The circular dichroism (CD) experiments were carried out using a Jasco 710 spectropolarimeter (Jasco, USA) equipped with a quartz cell with an optical path length of 0.5 mm. The far-UV CD spectra of 5.5 μM cdM2-1 were recorded at 25°C and pHs 5, 6.5, and 8. The spectra were averaged over 20 scans in the 260- to 200-nm range with a resolution of 0.2 nm at a scan speed of 50 nm · min−1 and a spectral bandwidth of 1.0 nm. The CD signal, taken as millidegrees (θ), was corrected for the background contribution of the buffer and next expressed in terms of the mean residues of ellipticity ([Θ]), in degrees · square centimeter · decimoles−1, using the equation , where is the molar concentration of the protein, is the optical path length (in centimeters), and is the number of amino acid residues and where θ is in millidegrees.
Differential scanning calorimetry.
Differential scanning calorimetry (DSC) experiments were performed using an N-DSC III calorimeter (TA Instruments, USA) over a temperature range of 10 to 90°C at a heating and cooling scan rate of 1.0°C/min. Both calorimetry cells were loaded with the buffer solution, equilibrated at 10°C for 10 min, and scanned repeatedly as described above until the baseline was reproducible. Next, the sample cell was loaded with 100 μM cdM2-1 plus 0, 50, and 100 μM RNA and scanned. The thermograms of RNA in buffer solution were recorded as a control for the baseline. The baseline corrections were obtained by subtracting the buffer (or RNA) scan from the corresponding protein scan. Measurements were performed in duplicate. The calorimetry enthalpy change () of the unfolding process of cdM2-1 was calculated from the area under the thermogram curve. The van’t Hoff enthalpy change () of the thermal denaturation process was obtained from equation 2, replacing the dissociation constant by the unfolding constant (). The values of were calculated by , where is the fraction of unfolding protein determined from an integral process of the denaturation thermogram.
Chemical shift perturbation analysis.
The chemical shift perturbation (CSP) method was used to map the amino acid residues of cdM2-1 involved in the binding to RNA. The two-dimensional (2D) 1H-15N heteronuclear single quantum coherence (HSQC) experiments were carried out with increasing amounts of RNA ranging from 0 to 115 μM, which was added to the [U-15N]cdM2-1 solution at constant concentration of 350 μM. The 15N HSQC spectra were recorded at 25°C on an NMR Bruker Avance III spectrometer of 14.1 T operating at a 1H frequency of 600 MHz (Bruker BioSpin GmbH, Germany) and equipped with a cryogenically cooled Z-gradient probe. The data matrix of the spectra consisted of 1,024* × 128* data points (were n* refers to the number of complex points) with acquisition times of 106.5 ms (the time for the amide hydrogen [tHN]) and 87.6 ms (the time for the nitrogen [tN]). A total of 16 scans per complex tN increment were collected with a recycle delay of 1.25. The resonance assignment of the core domain of hRSV M2-1 was obtained from accession number 17451 in the BMRB repository (www.bmrb.wisc.edu) (28).
The 1H-15N HSQC spectra were processed using the NMRPipe system (32) and analyzed using CcpNMR analysis (33). The chemical shift perturbation (δ) of the amino acid residues of the protein recorded in each titration step of RNA against the [U-15N]cdM2-1 solution was normalized using (34), where and denote the chemical shift difference of 1H and 15N, respectively, recorded in the absence and the presence of RNA. The mean of the amino acid residues as well as its standard deviation of CSP was used as the cutoff value to identify the residues of the protein involved in the cdM2-1/RNA interaction.
Amide hydrogen chemical shift temperature coefficient.
The amide hydrogen (1HN) chemical shift temperature coefficient of cdM2-1 was determined by recording a series of 2D 1H-15H HSQC spectra at 15, 20, 25, 30, and 35°C in the absence and the presence of 115 μM RNA, using a Bruker Avance III NMR spectrometer of 14.1 T operating at a 1H frequency of 600 MHz (Bruker BioSpin GmbH, Germany). All spectra were referenced to the water signal for each temperature, next processed using NMRPipe (32), and analyzed using CcpNMR analysis (33). The chemical shift values () of all residues at different temperatures were plotted as a function of temperature. The resulting slope () of every curve was plotted for each residue. For evaluation of the values, it was used as a straightforward interpretation compilation, as published by Morando et al. (35). In principle, the 1HN chemical shift temperature coefficient reports on the thermal susceptibility of the HN-C′ amide hydrogen bonds, since is correlated to the length of the hydrogen bond () and hydrogen bond J coupling () (16). On the one hand, residues with a value of less than −5.0 ppb · K−1 form more expandable hydrogen bonds, which may be interpreted as weaker, since they present smaller hydrogen bond J coupling (). For this reason, an amide group in the protein structure with a value of less than −5.0 ppb · K−1 may be considered a weak point of a secondary structure or, when this amide is in a loop or an intrinsically disordered region (IDR), may be more exposed to a hydrogen bond with water. On the other hand, residues with a value of greater than −5.0 ppb · K−1 make less expandable hydrogen bonds, which may be interpreted to be stronger, since they present larger hydrogen bond J coupling (). In this sense, an amide group with a value of greater than −5.0 ppb · K−1 is involved in secondary structures of hydrogen bonds or, when this amide is in a loop or IDR, may tend to form intramolecular hydrogen bonds and, consequently, to increase order (16, 35).
Paramagnetic relaxation enhancement experiments.
To monitor the solvent accessibility of residues of cdM2-1 (350 μM) in the absence and the presence of 115 μM RNA, the gadolinium-based paramagnetic relaxation agent Gd-diethylenetriamine pentaacetic acid-bismathylamide (Gd-DTPA; GE Life Science, United Kingdom) was used. The 1HN/15N amide cross-peak intensities of the protein were determined by recording a series of 2D 1H-15H HSQC spectra at different concentration of Gd-DTPA (0, 1.0, 2.0, 3.0, and 4.0 mM) at 25°C using a Bruker Avance III NMR spectrometer of 14.1 T operating at a 1H frequency of 600 MHz (Bruker BioSpin GmbH, Germany). All spectra were processed using an NMRPipe system (32) and analyzed using CcpNMR analysis (33). The intensity of each 1HN/15N cross-peak of the residues (INH) was plotted as a function of the Gd-DTPA concentration, and the slope of the adjusted straight line (INH · [Gd]−1) was plotted for each residue. To calculate an accurate slope INH · [Gd]−1, the points where the intensities were close to zero (a lack of linearity) were not taken into account.
Imino proton resonances of RNA.
Unidimensional 1H excitation sculpting spectra were collected at 25°C on Bruker Avance III NMR spectrometer of 14.1 T operating at a 1H frequency of 600 MHz (Bruker BioSpin GmbH, Germany) using a 180° water-selective pulse of 2 ms for solvent suppression. The free induction decays (FIDs) were recorded with 32,768 data points using a spectral width of 21,000 Hz (35 ppm), a relaxation delay of 1.25 s, and an acquisition time of 0.78 s. The experiment was performed for the free protein at 350 μM and the free nucleic acid at 115 μM, and after that it was performed for the cdM2-1/RNA complex by maintaining the same concentrations used for the free protein and the free nucleic acid. The spectral region from 10 to 18 ppm was used for analyzing the structural changes in the RNA induced by cdM2-1, probing the imino protons of the nucleic acid involved in base pair formation (17).
NMR relaxation experiments.
The backbone dynamics of [U-15N]cdM2-1 at 350 μM and the core domain complexed with 115 μM RNA were investigated from 15N nuclear spin relaxation experiments (36) at 25°C by using a Bruker Avance III NMR spectrometer of 14.1 T operating at a 1H frequency of 600 MHz (Bruker BioSpin GmbH, Germany). The experiments were collected using 14 delay times of inversion recovery of 54, 104, 204, 304, 404 (twice), 604, 804, 904 (twice), 1,204, 1,504 (twice), and 1,804 ms. The (longitudinal relaxation rate at the rotating frame) experiments were performed as described by Korzhnev et al. (37) with 14 spin-lock periods of 10, 20, 30 (twice), 40, 50 (twice), 60, 70, 80, 90 (twice), 100, and 110 ms and 15N spin-lock field strengths of 2.0 kHz. A recycle delay of 3.0 s was used for the and experiments. The values of and were determined from nonlinear least-square fittings of the intensities using two-parameter monoexponential equations. values were determined from the measured and rates (38). The reported errors for the rates were calculated from the estimated uncertainties for the relaxation delay duplicates. 1H-15N steady-state heteronuclear nuclear Overhauser effects (hetNOE) were measured from pairs of interleaved spectra recorded with (nuclear Overhauser effect [NOE]) and without (control) HN proton saturation during a recycle delay of 12 s. The 1H-15N hetNOE values were calculated from the resonance intensity ratios obtained from the NOE and control spectra, with uncertainties being estimated from the background noise of the spectra. The data were processed using NMRPipe (32) and analyzed using CcpNMR analysis (33).
Reduced spectral density mapping approach.
The reduced spectral density mapping (rSDM) is a simplified approach for analysis of nuclear spin relaxation data (R1, R2, and hetNOE) developed by Farrow et al. (39), Ishima and Nagayama (40), and Lefèvre et al. (41). rSDM maps the spectral density for determining the accurate values of the spectral density (J) function at three frequencies: at the zero frequency, at the nitrogen frequency, and at an effective proton frequency. The approach exploits the assumption that at higher frequencies, the spectral density terms contributing to the relaxation processes are equal in magnitude, i.e., , and is replaced by a single equivalent term , which corresponds to . This assumption is based on the premise that at frequencies higher than those for and , spectral densities and show minimal variation. The spectral density functions at three frequencies, as derived through reduced spectral density mapping, are given by the following equations:
| (3) |
| (4) |
| (5) |
where the cross-relaxation rate is defined by , , and and where is the permeability of the vacuum; is the reduced Planck constant; and are the gyromagnetic ratios of 15N and 1H, respectively; rNH is the bond length; is the Larmor frequency of the 15N nucleus; and is the 15N chemical shift anisotropy (in parts per million).
Model free analysis.
The relaxation parameters were fitted according to the extended model-free formalism of Lipari-Szabo for obtaining intramolecular dynamics (42). The TENSOR2 program (43) was employed to define a motional model for hRSV cdM2-1 using an anisotropic tensor with a fully asymmetry diffusion model (DX = 1.81 × 107 ± 0.03 × 107 s−1, DY = 1.95 × 107 ± 0.02 × 107 s−1, DZ = 2.11 × 107 ± 0.03 × 107 s−1, where Dx, Dy, and Dz are axial diffusion tensors). The calculations of the rotational correlation time () were performed from the relaxation data for the amino acid residues with values of R2/R1 ratios within 1 standard deviation relative to the calculated average and with values of {1H}-15N hetNOE greater than 0.65. The anisotropic diffusion tensor was also used for calculating the internal molecular dynamics parameters, such as the order parameter () and the conformational exchange rate (Rex), which reflect movements of the NH bond in picosecond-to-nanosecond and microsecond-to-millisecond timescales, respectively.
Charge, protonation state, and electrostatic potential calculation.
The electrostatic potential calculations were performed by APBS software (44), using charge values and protonation states obtained from the PDB2PQR server (version 2.1.1) (45), along with the PROPKA program (version 3.0) (46). The physical-chemical parameters used for the calculations were 150 mM NaCl, pH 6.5, and 25°C. The electrostatic potential surface of the core domain of hRSV M2-1 was displayed using the PyMOL program (47).
Molecular modeling and molecular docking.
The primary sequences of 10 RNAs were generated from the Random Sequence Generator (RSG) web server, setting the length of the sequence equal to 40 bases and the AU content at 80%. All RNA sequences generated by the RSG server are presented in Table S14 in the supplemental material. These random RNA sequences were submitted to the Direct Coupling Analysis (DCA) server (http://biophy.hust.edu.cn/home.html), which is a statistical inference framework used to infer direct coevolutionary couplings among nucleotide pairs in multiple-sequence alignments and which aims at disentangling direct from indirect correlations. The information provided by the DCA server was subsequently used in the 2dRNA web server (http://biophy.hust.edu.cn/home.html), which is a secondary structure prediction method for RNA based on DCA data. The parameters of the 2dRNA server were set at the default. Next, the predicted secondary structure information was used in the 3dRNA web server (version 2.0) (19), which is an automatic molecular modeling method for building the three-dimensional structure of RNA. The assembly calculations performed by the 3dRNA server were followed by an optimization step using all the parameters at the default setting.
The molecular docking calculations for predicting the molecular model of the complex formed between the random RNA sequences (Table S14) and cdM2-1 were performed by using the 3dRPC web server (version 2.0) (48). The three-dimensional structure of the hRSV M2-1 core domain was downloaded from the Protein Data Bank for the structure with accession number 2L9J (7), and three-dimensional (3D) structures of 10 RNAs were obtained from the 3dRNA web server (version 2.0) (19). A total of 10 complex models were predicted using the 3dRPC-Score function. The amino acid residues of cdM2-1 involved in the RNA binding interface, which was determined from the CSP method by NMR experiments, were specified as a constraint set in the advanced setting of the 3dRPC server. Following docking, the lowest-energy structural models of the 10 cdM2-1/RNA complexes were analyzed by VMD software (49), using the VolMap tool for evaluating the mass density map of collocated RNA (1.0 Å of resolution, 1.5 Å × radius of atom size, 10 frames combined using the average, and isosurfaces with an isovalue of 0.5) and by the PLIP web server (20) for characterizing the protein/nucleic acid noncovalent interactions, such as the hydrogen bonds, π-cation interactions, π-stacking, and salt bridges. The structural conformation of the constructed models was displayed using PyMOL (47) and VMD (49) software.
Molecular dynamics simulation.
The molecular dynamics (MD) simulations were carried out with the GROMACS program (version 5.0.7) (50). The molecular systems were modeled by using the AMBER99SB-IDLN protein and AMBER94 nucleic acid force field (51) and the TIP3P water model (52). The three-dimensional structures of 5 randomly selected cdM2-1/RNA complexes calculated from molecular docking (3dRPC server) were used in the MD simulations. These structures were placed in the center of 89- to 135-Å cubic boxes filled with TIP3P water molecules and Na+/Cl– ions ([NaCl] = 150 mM). The protonation states of charged residues were set considering a pH 6.5 from the PROPKA results (46). All simulations were performed in the NPT ensemble using periodic boundary conditions and keeping the system at 298 K (Nose-Hoover thermostat, = 2.0 ps) and 1.0 bar (Parrinello-Rahman barostat, = 2.0 ps, and compressibility = 4.5 × 10−5 bar−1) ( is the time constant for temperature coupling, and is the time constant for pressure coupling). Lennard-Jones and Coulomb potentials were applied using a cutoff distance of 12 Å. The long-range electrostatic interactions were calculated using the particle mesh Ewald (PME) algorithm. The covalent bonds involving hydrogen atoms were constrained to their equilibrium distance. A conjugate gradient minimization algorithm was utilized to relax the superposition of atoms generated in the box construction process. The energy minimizations were performed with the steepest descent integrator and conjugate gradient algorithm, using 500 kJ · mol−1 · nm−1 as the maximum force criterion. One hundred thousand steps of molecular dynamics were performed for each NVT and NPT equilibration, applying force constants of 1,000 kJ · mol−1 · nm−2 to all heavy atoms of the cdM2-1/RNA complexes. Lastly, MD simulations of 20 ns were accomplished for data acquisition. Following the simulations, the trajectories were concatenated and analyzed according to the number of hydrogen bonds (cutoff distance = 3.5 Å and maximum angle = 30°), number of contacts (<0.6 nm), and the root mean square deviation (RMSD) of nonhydrogen atoms for the nucleic acid and backbone atoms for cdM2-1 (the entire protein and the helical-core region). These parameters (hydrogen bonds, number of contacts, RMSD) from 5 MD simulations were presented as averages.
Accessible surface area calculations.
The accessible surface areas (ASA) of free cdM2-1 and its docked complex with RNAs were calculated using the NACCESS program (53). The structural models of the cdM2-1/RNA complexes were obtained from the molecular docking calculations (3dRPC server). Changes in the absolute ASA for residue were calculated using , where and are the absolute accessible surface area for free and RNA-bound cdM2-1 residues, respectively. The values of were averaged, and next, the average value was normalized to a range of from 0 and 1.0 unit for coloring the cdM2-1 structure.
Supplementary Material
ACKNOWLEDGMENTS
I.P.C. gratefully acknowledges the financial support provided by a postdoctoral fellowship from FAPERJ (fellowship 202.279/2018) and the PROPe UNESP. G.C.G., V.B.M., and F.P.S. thank FAPESP for scholarships (scholarships 2019/08739-1 and 2018/08900-4). The study was funded by the Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ), Brazil (grants 202.279/2018, 210.361/2015, 239.229/2018, and 204.432/2014), the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Brazil (grant 309564/2017-4), and the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP), Brazil (grants 2019/08739-1, 2018/08900-4, and 2010/18169-3, 2009/53989-4). The Calix cluster is funded by FAPESP grant 2010/18169-3, the NMR Laboratory of the Multiuser Center for Biomolecular Innovation is funded by FAPESP grant 2009/53989-4, and NMRbox: National Center for Biomolecular NMR Data Processing and Analysis, a Biomedical Technology Research Resource (BTRR), is supported by NIH grant P41GM111135 (NIGMS).
We thank Marcelo de Freitas Lima for access to the Fluorescent Spectrometer Lumina, located in the Bio-organic Environmental Laboratory of the Department of Chemistry and Environmental Sciences, and Alexandre Suma de Araújo for access to the Calix cluster for performing the molecular dynamics simulations. We also recognize GridUNESP for the availability of the software package for molecular dynamics simulations. We thank the NMR Laboratory of the Multiuser Center for Biomolecular Innovation in IBILCE/UNESP for access. We thank NMRbox: National Center for Biomolecular NMR Data Processing and Analysis, a Biomedical Technology Research Resource (BTRR).
We declare that no conflict of interest exists.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Collins PL, Karron RA. 2013. Respiratory syncytial virus and metapneumovirus, p 1086–1123. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Hall CB, Weinberg GA, Iwane MK, Blumkin AK, Edwards KM, Staat MA, Auinger P, Griffin MR, Poehling KA, Erdman D, Grijalva CG, Zhu Y, Szilagyi P. 2009. The burden of respiratory syncytial virus infection in young children. N Engl J Med 360:588–598. doi: 10.1056/NEJMoa0804877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Falsey AR, Hennessey PA, Formica MA, Cox C, Walsh EE. 2005. Respiratory syncytial virus infection in elderly and high-risk adults. N Engl J Med 352:1749–1759. doi: 10.1056/NEJMoa043951. [DOI] [PubMed] [Google Scholar]
- 4.Bermingham A, Collins PL. 1999. The M2-2 protein of human respiratory syncytial virus is a regulatory factor involved in the balance between RNA replication and transcription. Proc Natl Acad Sci U S A 96:11259–11264. doi: 10.1073/pnas.96.20.11259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collins PL, Hill MG, Cristina J, Grosfeld H. 1996. Transcription elongation factor of respiratory syncytial virus, a nonsegmented negative-strand RNA virus. Proc Natl Acad Sci U S A 93:81–85. doi: 10.1073/pnas.93.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hardy RW, Wertz GW. 1998. The product of the respiratory syncytial virus M2 gene ORF1 enhances readthrough of intergenic junctions during viral transcription. J Virol 72:520–526. doi: 10.1128/JVI.72.1.520-526.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blondot M-L, Dubosclard V, Fix J, Lassoued S, Aumont-Nicaise M, Bontems F, Eléouët J-F, Sizun C. 2012. Structure and functional analysis of the RNA- and viral phosphoprotein-binding domain of respiratory syncytial virus M2-1 protein. PLoS Pathog 8:e1002734. doi: 10.1371/journal.ppat.1002734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanner SJ, Ariza A, Richard CA, Kyle HF, Dods RL, Blondot ML, Wu W, Trincão J, Trinh CH, Hiscox JA, Carroll MW, Silman NJ, Eléouët JF, Edwards TA, Barr JN. 2014. Crystal structure of the essential transcription antiterminator M2-1 protein of human respiratory syncytial virus and implications of its phosphorylation. Proc Natl Acad Sci U S A 111:1580–1585. doi: 10.1073/pnas.1317262111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leyrat C, Renner M, Harlos K, Huiskonen JT, Grimes JM. 2014. Drastic changes in conformational dynamics of the antiterminator M2-1 regulate transcription efficiency in Pneumovirinae. Elife 3:e02674. doi: 10.7554/eLife.02674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Molina IG, Esperante SA, Marino-Buslje C, Chemes LB, de Prat-Gay G. 2018. Cooperative RNA recognition by a viral transcription antiterminator. J Mol Biol 430:777–792. doi: 10.1016/j.jmb.2018.01.018. [DOI] [PubMed] [Google Scholar]
- 11.Fearns R, Collins PL. 1999. Role of the M2-1 transcription antitermination protein of respiratory syncytial virus in sequential transcription. J Virol 73:5852–5864. doi: 10.1128/JVI.73.7.5852-5864.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hardy RW, Wertz GW. 2000. The Cys3-His1 motif of the respiratory syncytial virus M2-1 protein is essential for protein function. J Virol 74:5880–5885. doi: 10.1128/JVI.74.13.5880-5885.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ross PD, Subramanian S. 1981. Thermodynamics of protein association reactions: forces contributing to stability. Biochemistry 20:3096–3102. doi: 10.1021/bi00514a017. [DOI] [PubMed] [Google Scholar]
- 14.Teixeira TSP, Caruso ÍP, Lopes BRP, Regasini LO, de Toledo KA, Fossey MA, de Souza FP. 2017. Biophysical characterization of the interaction between M2-1 protein of hRSV and quercetin. Int J Biol Macromol 95:63–71. doi: 10.1016/j.ijbiomac.2016.11.033. [DOI] [PubMed] [Google Scholar]
- 15.Gao Y, Cao D, Pawnikar S, John KP, Ahn HM, Hill S, Ha JM, Parikh P, Ogilvie C, Swain A, Yang A, Bell A, Salazar A, Miao Y, Liang B. 2020. Structure of the human respiratory syncytial virus M2-1 protein in complex with a short positive-sense gene-end RNA. Structure 28:979–990.e4. doi: 10.1016/j.str.2020.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hong J, Jing Q, Yao L. 2013. The protein amide 1HN chemical shift temperature coefficient reflects thermal expansion of the N-H···O=C hydrogen bond. J Biomol NMR 55:71–78. doi: 10.1007/s10858-012-9689-3. [DOI] [PubMed] [Google Scholar]
- 17.Kyogoku Y, Kojima C, Lee SJ, Tochio H, Suzuki N, Matsuo H, Shirakawa M. 1995. Induced structural changes in protein-DNA complexes. Methods Enzymol 261:524–541. doi: 10.1016/s0076-6879(95)61023-5. [DOI] [PubMed] [Google Scholar]
- 18.De La Torre JG, Huertas ML, Carrasco B. 2000. HYDRONMR: prediction of NMR relaxation of globular proteins from atomic-level structures and hydrodynamic calculations. J Magn Reson 147:138–146. doi: 10.1006/jmre.2000.2170. [DOI] [PubMed] [Google Scholar]
- 19.Jian W, Mao K, Zhao Y, Chen Z, Jianjin X, Yi Z, Xiao Y. 2017. Optimization of RNA 3D structure prediction using evolutionary restraints of nucleotide-nucleotide interactions from direct coupling analysis. Nucleic Acids Res 45:6299–6309. doi: 10.1093/nar/gkx386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salentin S, Schreiber S, Haupt VJ, Adasme MF, Schroeder M. 2015. PLIP: fully automated protein-ligand interaction profiler. Nucleic Acids Res 43:W443–W447. doi: 10.1093/nar/gkv315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cuesta I, Geng X, Asenjo A, Villanueva N. 2000. Structural phosphoprotein M2-1 of the human respiratory syncytial virus is an RNA binding protein. J Virol 74:9858–9867. doi: 10.1128/JVI.74.21.9858-9867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Richard CA, Rincheval V, Lassoued S, Fix J, Cardone C, Esneau C, Nekhai S, Galloux M, Rameix-Welti MA, Sizun C, Eléouët JF. 2018. RSV hijacks cellular protein phosphatase 1 to regulate M2-1 phosphorylation and viral transcription. PLoS Pathog 14:e1006920. doi: 10.1371/journal.ppat.1006920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhaskara RM, Srinivasan N. 2011. Stability of domain structures in multi-domain proteins. Sci Rep 1:40. doi: 10.1038/srep00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cooper A, Nutley MA, Wadood A. 2000. Differential scanning microcalorimetry, p 287–318. In Harding SE, Chowdhry BZ (ed), Protein-ligand interactions: hydrodynamics and calorimetry. Oxford University Press, Oxford, United Kingdom. [Google Scholar]
- 25.Dayie KT, Wagner G, Lefèvre J-F. 1996. Theory and practice of nuclear spin relaxation in proteins. Annu Rev Phys Chem 47:243–282. doi: 10.1146/annurev.physchem.47.1.243. [DOI] [PubMed] [Google Scholar]
- 26.Bracken C, Carr PA, Cavanagh J, Palmer AG. 1999. Temperature dependence of intramolecular dynamics of the basic leucine zipper of GCN4: implications for the entropy of association with DNA. J Mol Biol 285:2133–2146. doi: 10.1006/jmbi.1998.2429. [DOI] [PubMed] [Google Scholar]
- 27.Berlow RB, Martinez-Yamout MA, Dyson HJ, Wright PE. 2019. Role of backbone dynamics in modulating the interactions of disordered ligands with the TAZ1 domain of the CREB-binding protein. Biochemistry 58:1354–1362. doi: 10.1021/acs.biochem.8b01290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dubosclard V, Blondot ML, Eléouët JF, Bontems F, Sizun C. 2011. 1H, 13C, and 15N resonance assignment of the central domain of hRSV transcription antitermination factor M2-1. Biomol NMR Assign 5:237–239. doi: 10.1007/s12104-011-9308-3. [DOI] [PubMed] [Google Scholar]
- 29.Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. 2005. Protein identification and analysis tools on the ExPASy server, p 571–607. In Walker MJ. (ed), The proteomics protocols handbook. Humana Press, Totowa, NJ. [Google Scholar]
- 30.Lakowicz JR. 2006. Instrumentation for fluorescence spectroscopy, p 27–61. In Principles of fluorescence spectroscopy. Springer US, New York, NY. [Google Scholar]
- 31.Beckett D. 2011. Measurement and analysis of equilibrium binding titrations: a beginner’s guide. Methods Enzymol 488:1–16. doi: 10.1016/B978-0-12-381268-1.00001-X. [DOI] [PubMed] [Google Scholar]
- 32.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. 1995. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 33.Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, Laue ED. 2005. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins Struct Funct Genet 59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 34.Grzesiek S, Stahl SJ, Wingfield PT, Bax A. 1996. The CD4 determinant for downregulation by HIV-1 Nef directly binds to Nef. Mapping of the Nef binding surface by NMR. Biochemistry 35:10256–10261. doi: 10.1021/bi9611164. [DOI] [PubMed] [Google Scholar]
- 35.Morando MA, Barbosa GM, Cruz-Oliveira C, Da Poian AT, Almeida FCL. 2019. Dynamics of Zika virus capsid protein in solution: the properties and exposure of the hydrophobic cleft are controlled by the α-helix 1 sequence. Biochemistry 58:2488–2498. doi: 10.1021/acs.biochem.9b00194. [DOI] [PubMed] [Google Scholar]
- 36.Farrow NA, Muhandiram R, Pascal SM, Kay LE, Singer AU, Forman-Kay JD, Kay CM, Gish G, Pawson T, Shoelson SE. 1994. Backbone dynamics of a free and a phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry 33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 37.Korzhnev DM, Skrynnikov NR, Millet O, Torchia DA, Kay LE. 2002. An NMR experiment for the accurate measurement of heteronuclear spin-lock relaxation rates. J Am Chem Soc 124:10743–10753. doi: 10.1021/ja0204776. [DOI] [PubMed] [Google Scholar]
- 38.Tjandra N, Wingfield P, Stahl S, Bax A. 1996. Anisotropic rotational diffusion of perdeuterated HIV protease from 15N NMR relaxation measurements at two magnetic fields. J Biomol NMR 8:273–284. doi: 10.1007/BF00410326. [DOI] [PubMed] [Google Scholar]
- 39.Farrow NA, Zhang O, Szabo A, Torchia DA, Kay LE. 1995. Spectral density function mapping using 15N relaxation data exclusively. J Biomol NMR 6:153–162. doi: 10.1007/BF00211779. [DOI] [PubMed] [Google Scholar]
- 40.Ishima R, Nagayama K. 1995. Protein backbone dynamics revealed by quasi spectral density function analysis of amide N-15 nuclei. Biochemistry 34:3162–3171. doi: 10.1021/bi00010a005. [DOI] [PubMed] [Google Scholar]
- 41.Lefèvre JF, Dayie KT, Peng JW, Wagner G. 1996. Internal mobility in the partially folded DNA binding and dimerization domains of GAL4: NMR analysis of the N-H spectral density functions. Biochemistry 35:2674–2686. doi: 10.1021/bi9526802. [DOI] [PubMed] [Google Scholar]
- 42.Lipari G, Szabo A. 1982. Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 2. Analysis of experimental results. J Am Chem Soc 104:4559–4570. doi: 10.1021/ja00381a010. [DOI] [Google Scholar]
- 43.Dosset P, Hus JC, Blackledge M, Marion D. 2000. Efficient analysis of macromolecular rotational diffusion from heteronuclear relaxation data. J Biomol NMR 16:23–28. doi: 10.1023/A:1008305808620. [DOI] [PubMed] [Google Scholar]
- 44.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. 2001. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A 98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dolinsky TJ, Czodrowski P, Li H, Nielsen JE, Jensen JH, Klebe G, Baker NA. 2007. PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res 35:W522–W525. doi: 10.1093/nar/gkm276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Olsson MHM, Søndergaard CR, Rostkowski M, Jensen JH. 2011. PROPKA3: consistent treatment of internal and surface residues in empirical pKa predictions. J Chem Theory Comput 7:525–537. doi: 10.1021/ct100578z. [DOI] [PubMed] [Google Scholar]
- 47.Delano WL. 2002. The PyMOL molecular graphics system. DeLano Scientific, San Carlos, CA. [Google Scholar]
- 48.Li H, Huang Y, Xiao Y. 2017. A pair-conformation-dependent scoring function for evaluating 3D RNA-protein complex structures. PLoS One 12:e0174662. doi: 10.1371/journal.pone.0174662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Humphrey W, Dalke A, Schulten K. 1996. VMD: visual molecular dynamics. J Mol Graph 14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 50.Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, Lindah E. 2015. Gromacs: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2:19–25. doi: 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- 51.Lindorff-Larsen K, Piana S, Palmo K, Maragakis P, Klepeis JL, Dror RO, Shaw DE. 2010. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins 78:1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. 1983. Comparison of simple potential functions for simulating liquid water. J Chem Phys 79:926–935. doi: 10.1063/1.445869. [DOI] [Google Scholar]
- 53.Hubbard SJ, Thornton JM. 1993. NACCESS: computer program. Department of Biochemistry and Molecular Biology, University College London, London, United Kingdom. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.