Summary
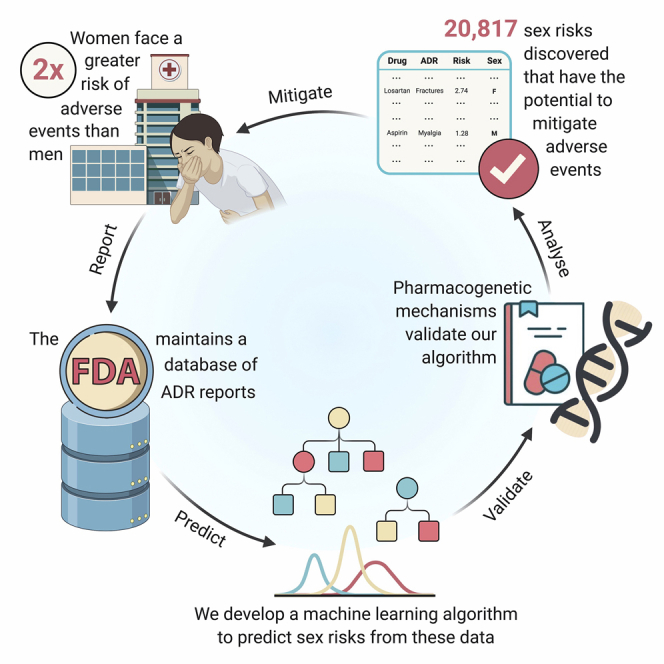
Adverse drug reactions are the fourth leading cause of death in the US. Although women take longer to metabolize medications and experience twice the risk of developing adverse reactions compared with men, these sex differences are not comprehensively understood. Real-world clinical data provide an opportunity to estimate safety effects in otherwise understudied populations, i.e., women. These data, however, are subject to confounding biases and correlated covariates. We present AwareDX, a pharmacovigilance algorithm that leverages advances in machine learning to predict sex risks. Our algorithm mitigates these biases and quantifies the differential risk of a drug causing an adverse event in either men or women. AwareDX demonstrates high precision during validation against clinical literature and pharmacogenetic mechanisms. We present a resource of 20,817 adverse drug effects posing sex-specific risks. AwareDX, and this resource, present an opportunity to minimize adverse events by tailoring drug prescription and dosage to sex.
Keywords: machine learning, data science, adverse drug reactions, sex, gender, women, bias, understudied populations, pharmacovigilance, pharmacogenetics
Graphical Abstract
Highlights
-
•
We develop a machine learning method, AwareDX, to predict sex risks of adverse events
-
•
AwareDX has high precision during validation against known pharmacogenetic mechanisms
-
•
We present a resource of 20,817 adverse drug effects posing sex-specific risks
The Bigger Picture
We present the first, and to our knowledge only, approach for predicting sex differences in drug response corroborated by pharmacogenomic data. Our algorithm AwareDX identifies drugs associated with increased rates of adverse events to either sex. AwareDX uses machine learning to dampen correlated covariates and mitigate confounding biases of sex. This approach has the potential to generalize to understudied populations in many data science domains. We introduce a resource of sex-specific adverse drug effects for use in drug discovery, repositioning, and pharmacogenetic studies and for further analysis through electronic health records and clinical trials. Ultimately, such analyses could potentially raise awareness of sex differences during clinical decision making.
Adverse events are unwanted effects of drugs that lead to injury and disease. Women face twice the risk of developing adverse events compared with men due to metabolic differences. Unfortunately, these sex differences are not comprehensively understood. Here, we leverage a public database of adverse event reports to systematically estimate safety effects in women. We develop a machine learning algorithm, AwareDX: Analysing Women At Risk for Experiencing Drug toXicity, that predicts sex-specific risks of adverse drug effects.
Introduction
Adverse drug reactions (ADRs) are unwanted effects of drugs that lead to injury and disease. In 2016, the cost of drug-related morbidity and mortality was estimated at $528.4 billion.1 ADRs are the fourth leading cause of death in the United States—ahead of pulmonary disease and diabetes.2 Although half of all ADRs are preventable,3 many population-specific ADRs remain unidentified. This is largely because clinical trials have historically been conducted in homogeneous patient populations (e.g., white males). Until 1993, the US Food and Drug Administration (FDA) designated women as a special “subgroup” of patients during clinical trials.4,5 A decade after this designation was lifted, women remained severely underrepresented in clinical trials.6
Women have a 2-fold greater risk of developing ADRs than men.7 This increased risk cannot be explained by the use of hormonal contraceptives.8 Rather, differences in pharmacokinetics and pharmacodynamics induce increased drug bioavailability and greater sensitivity to medication in women.9,10 For example, it is well known that being female is a risk factor for developing torsades de pointes from cardiovascular drugs.11,12 21 years after it was approved as an insomnia drug, the recommended dosage of Zolpidem (Ambien) was halved for women due to their decreased metabolic clearance.13 While a few sex risks such as these have been clinically established, a comprehensive understanding remains lacking.
The FDA maintains post-marketing drug surveillance data in its Adverse Event Reporting System (FAERS). This resource presents an opportunity to systematically quantify sex-specific risks of drugs. However, FAERS is subject to many biases because of differential prescription and the sex-specific nature of some diseases and drugs. When sex risks are calculated using simple disproportionality analysis, causative covariates can create selection bias that leads to “synthetic” associations being identified. A drug can show a synthetic association with an ADR that is more appropriately attributed to the sex-exclusive nature of the underlying disease. For example, chemotherapy drugs for breast cancer and prostrate cancer are commonly associated with sex-specific adverse drug effects (ADEs).14 Medications that have sex-exclusive prescription can also confound drug-effect associations. Ethinyl estradiol, commonly found in oral contraceptives, has many sex-specific effects simply due to differential consumption in women. These issues extend to other confounding factors, such as age and co-medication. Elderly age and concomitant drug use are known risk factors for ADRs.15 Because women tend to live longer than men and take more medications simultaneously, this can induce additional synthetic associations.
Although systematic work in this domain remains very limited, one seminal study used disproportionality analysis on FAERS to identify drugs with higher odds of having an adverse event for either sex.16 While the study corrected for differential reporting bias, it did not account for other covariates, such as sex-specificity of diseases, differential prescription, and demographic associations with sex. This analysis was limited because it only corrected for reporting bias after finding significant associations and potentially missed findings that remained hidden due to biases in the underlying data. Moreover, the study did not include an independent validation that connected its findings to pharmacogenetic mechanisms.
Here, we present an algorithm, AwareDX: Analysing Women At Risk for Experiencing Drug toXicity. AwareDX mitigates sex biases in the data by using a machine learning adaptation of propensity score matching. This approach has been used to effectively identify ADRs caused by drug-drug interactions.17 A random forest (RF) model predicts the likelihood of being “female” given confounding factors. Using this likelihood, drug-exposed females are matched to drug-exposed males to create balanced cohorts for downstream disproportionality analysis. We show that by building these cohorts, AwareDX mitigates 79% of underlying sex biases. Using this data-driven approach, we successfully flagged significant sex-specific ADEs, many of which were previously unknown. For independent validation of our algorithm, we not only used clinical literature but also explored the metabolic and genetic basis of sex differences in drug response. Given a set of genes with differential expression across sex18 and information about how their variants affect drug response,19 we hypothesized that genes with sex-differential expression should lead to sex-differential ADRs similarly to variants of those gene. We show that AwareDX recovers these expected sex risks with high precision.
Results
Adverse Event Reports Contain Many Confounding Biases and Covariates of Sex
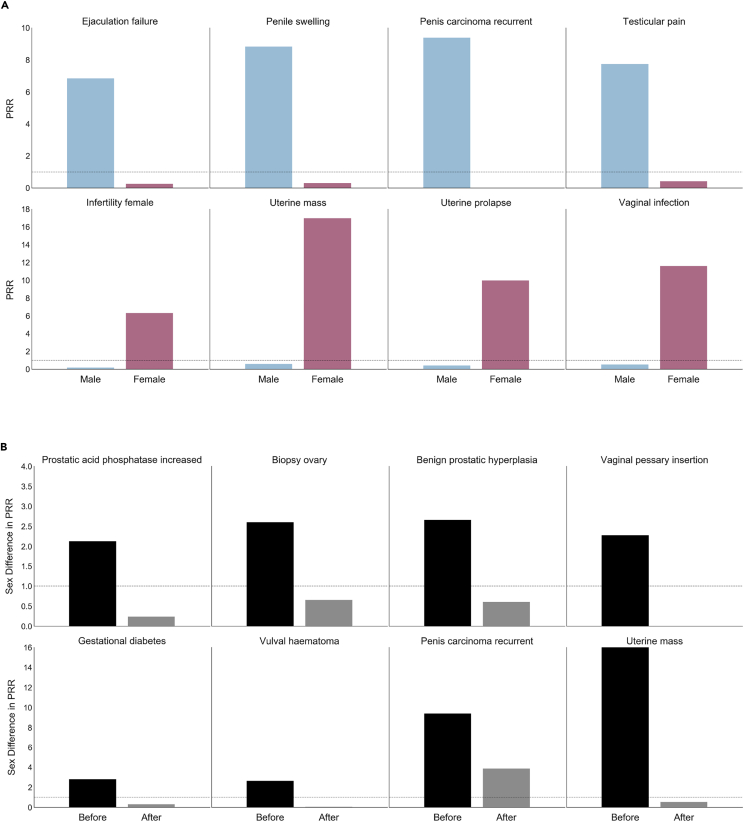
Of the 8.8 million patients in FAERS, 61.9% were female. Sex-differential drug exposure was evident from the biased reporting of sex for certain medications (Table 1). We characterized sex-specific bias between drugs and adverse events by calculating proportional reporting ratios (PRR). For sex-exclusive ADRs, we calculated the PRR averaged over all drugs for each sex. The sex bias for an ADR was quantified as the absolute difference in mean PRR across sex. Figure 2A visualizes adverse events with the largest sex biases. We established age (two-sample t test; t = 2.31, P = 0.0239) as a covariate of sex. No significant association was found between concomitant drug exposures and sex (Mann-Whitney U test; statistic = 5,388.0, P = 0.0800).
Table 1.
Drug Exposure Counts with Highest Disproportionatlity in Sex from FDA's Adverse Event Reporting System
| Drug | Male | Female |
|---|---|---|
| avanafil | 458 | 2 |
| udenafil | 458 | 2 |
| degarelix | 1,500 | 6 |
| radium dichloride | 2,403 | 16 |
| abiraterone | 15,210 | 96 |
| enzalutamide | 35,015 | 242 |
| drospirenone | 3 | 2,000 |
| norethisterone | 7 | 2,243 |
| ospemifene | 7 | 2,250 |
| medroxyprogesterone and estrogen | 17 | 21,776 |
| drospirenone and ethinylestradiol | 52 | 40,536 |
| conjugated estrogens | 176 | 57,524 |
Figure 2.
AwareDX Mitigates Sex Biases
To quantify sex biases in FAERS, we selected adverse events that were sex-exclusive and calculated their PRR for each sex averaged over all drugs. In (A), sex biases in the original data are evident from the large disparity in PRR between males and females. AwareDX corrects sex biases by building balanced cohorts by sampling from propensity scores. (B) Shows sex biases (PRR disparity) in original data (“Before” column) and after applying our algorithm (“After” column). Sex disparity across adverse events was significantly reduced after applying AwareDX. Thus, AwareDX effectively mitigated the effect of confounding biases and covariates of sex. PRR, proportional reporting ratio; FAERS, FDA Adverse Events Reporting System.
Our Machine Learning Method Mitigates Sex Biases
Our algorithm mitigates biases in FAERS by using covariate information to build sex-balanced cohorts for drug-exposed subpopulations (visualized in Figure 1). We used traditional machine learning models to predict propensity scores (i.e., the likelihood of being female) for each patient given information about drug exposure, co-medications, and age. All models had similar performance ( accuracy) during cross-validation (see Table 2). We selected random forest (RF) because its ability to predict out-of-bag scores allowed us to conserve 100% of reports for downstream disproportionality analysis. The RF model had an out-of-bag score of 0.63 and area under receiver operating characteristic curve (ROC-AUC) of 0.64. See Figure S2 for feature importances and ROC curve.
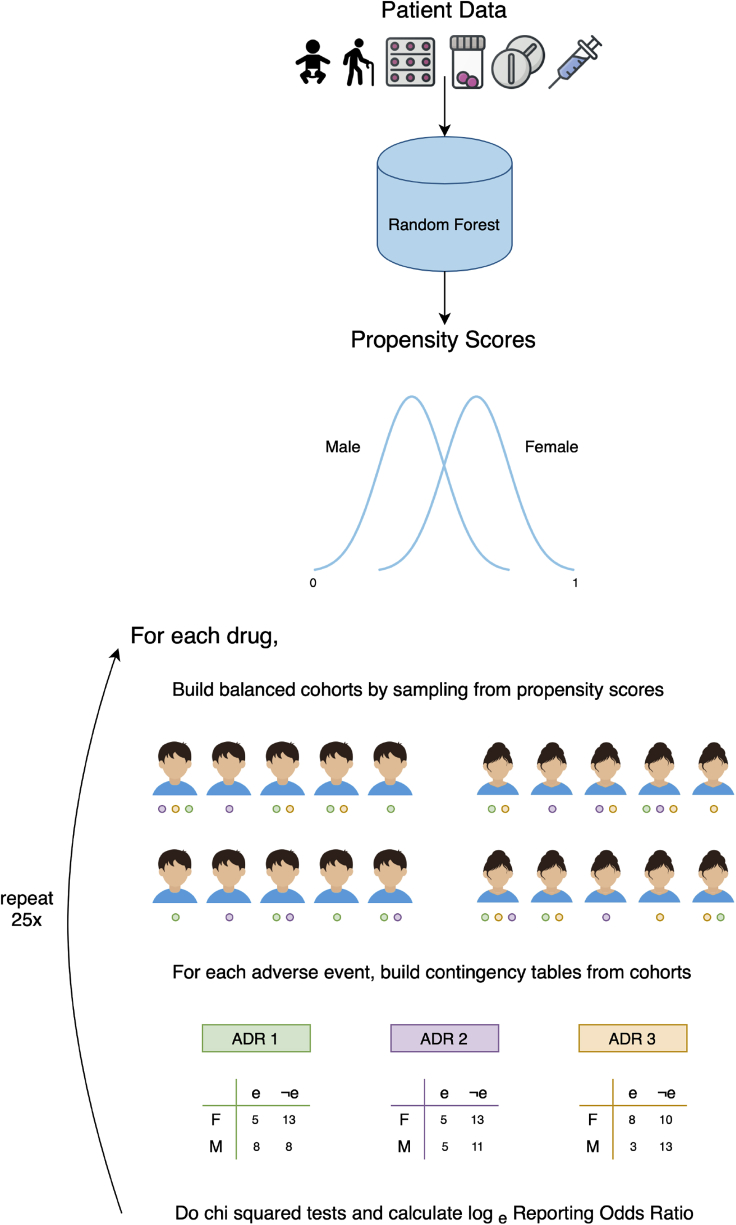
Figure 1.
How Does AwareDX Determine Sex Risks?
AwareDX evaluates sex risks in three steps: predicting propensity scores, building cohorts that mitigate bias, and doing disproportionality analysis. Starting at the top, for each patient, drug exposure, age, and co-medication features are curated from adverse event reports. A random forest model uses each patient's features to predict their propensity score, i.e., their likelihood of being female. These propensity scores are used to mitigate biases when evaluating sex risks. To analyze the sex risk of a drug and adverse event, the following steps are repeated for 25 iterations. The drug-exposed subpopulation is selected. Then propensity scores are used to build sex-balanced cohorts of that subpopulation. The patients within the cohorts have reported various adverse events, represented here by green, purple, and orange circles. For the adverse event of interest, a contingency matrix is constructed. A chi-square test supplies the P value for that iteration and the loge reporting odds ratio quantifies the sex risk. After 25 iterations, if all P values are significant, then the mean loge reporting odds ratio CI are used to quantify the sex risk. ADR, adverse drug event; F, female; M, male; e, has adverse event; e, does not have adverse event.
Table 2.
Performance of Machine Learning Models at Predicting Propensity Scores
| Model | Specification | Accuracy |
|---|---|---|
| Support vector machine | Radial basis function | 0.62 ± 0.0038 |
| Polynomial (p = 3) | 0.62 ± 0.0038 | |
| Linear | 0.65 ± 0.0058 | |
| Random forest | Gini impurity | 0.63 ± 0.0037 |
| Entropy | 0.63 ± 0.0039 | |
| Logistic regression | L1 | 0.65 ± 0.0064 |
| L2 | 0.65 ± 0.0063 | |
| ElasticNet (ratio = 0.5) | 0.65 ± 0.0063 |
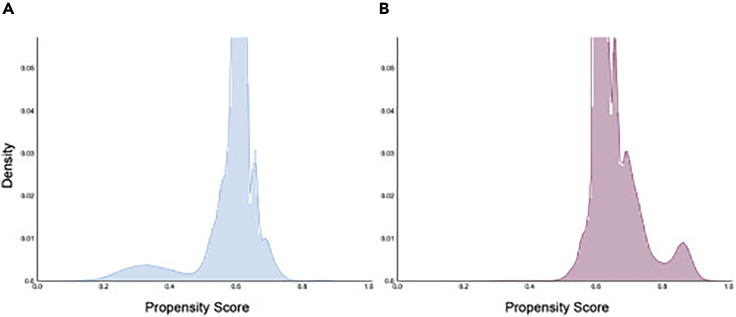
To characterize the effectiveness of our approach in migitating sex biases, we examined the PRR disparity before and after applying AwareDX. We calculated the mean PRR for sex-specific ADRs on cohorts built using our adaptation of propensity score matching. Overall, 79.2% of PRRs were removed or dampened after applying AwareDX. Figure 2B reinforces this finding by showing that PRR disparity is significantly reduced after applying AwareDX. Furthermore, we qualitatively explored the distribution of propensity scores predicted by the RF model for each sex (Figure 3). While the propensity scores overlap for a large majority of patients, many subsets of patients are clearly separated.
Figure 3.
Distribution of Propensity Scores
For each patient, a random forest model was used to predict a propensity score, i.e., the likelihood of being female. Both histograms visualize propensity score (x axis) against density or normalized frequency (y axis). (A) The distribution of propensity scores where the true sex label was male and (B) the distribution of propensity scores where the true sex label was female. There is clear separation for some subsets of patients, with the propensity scores for females tending toward 1 and those for males tending toward 0.
A Resource of Sex-Specific ADEs
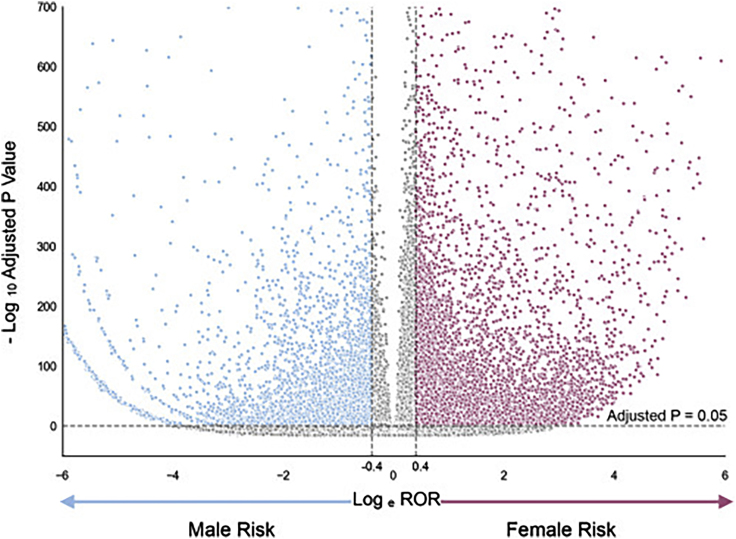
We present a resource of 20,817 ADEs that AwareDX predicted as posing sex-specific risks (Table S1). This resource comprises of 792 unique drugs and 297 unique ADRs. The sex risks are summarized using a volcano plot (Figure 4) and histogram (Figure S1). 62.7% of significant hits posed increased risks to women. Each drug was associated with 26 sex risks of ADRs on average. For the 10 most prescribed drugs in the US,20 this average increased to 66 ADRs per drug. For the 10 drugs with the highest adverse event reports in FAERS, there were 87 sex-specific ADR risk associated with each drug. We identify the most prominent sex risks in Table 3, which shows the top 5 drug-ADR pairs that pose the highest risk to each sex. The “highest risk” ADEs were defined as those with the largest absolute logarithm of the reporting odds ratio (logROR) where the associated ADRs had a severity score21 greater than 0.5. Table 4 shows the most prominent risks to each sex from the 10 most prescribed drugs. A complete database of all 287,605 sex risks evaluated by AwareDX is included in the Supplemental Information (Table S2).
Figure 4.
Volcano Plot of Significant Sex Risks
The volcano plot visualizes the magnitude of each sex risks (loge ROR; x axis) against their significance ( adjusted P value; y axis). Each point represents a drug-adverse event pair. Blue points indicate male risks. Pink points indicate female risks. Gray points indicate sex risks that did not pass the significance threshold ( after applying a Bonferroni correction) or had a low magnitude (ROR < 1.5). The significance threshold is denoted by the horizontal dotted line and the magnitude thresholds are denoted by the vertical dotted lines. See also Figure S1.
Abbreviations: ROR, reporting odds ratio.
Table 3.
Top Sex Risks Predicted by AwareDX
| Drug | Description | Indication | ADR | Sex | logROR | 95% CI |
|---|---|---|---|---|---|---|
| anakinra | interleukin-1 receptor antagonist | rheumatoid arthritis |
diverticular disorders | M | 4.52 | (4.51, 4.54) |
| chlordiazepoxide | benzodiazepine | anxiety disorders | central nervous system infections and inflammations | M | 3.86 | (3.82, 3.91) |
| desoximetasone | topical anti-inflammatory glucocorticoid | skin irritation, allergic reactions | miscellaneous and site unspecified neoplasms | M | 3.76 | (3.73, 3.78) |
| sirolimus | immunosuppressant | transplant and heart stent | non-hodgkin's B cell lymphomas | M | 3.09 | (3.04, 3.13) |
| misoprostol | prostaglandin analog | stomach ulcers | metabolism disorders | M | 2.93 | (2.91, 2.96) |
| anakinra | interleukin-1 receptor antagonist | rheumatoid arthritis |
vascular inflammations | F | 3.25 | (3.03, 3.48) |
| rizatriptan | triptan drug | migraine headaches | pulmonary vascular disorders | F | 3.14 | (2.92, 3.36) |
| abatacept | soluble fusion protein | rheumatoid arthritis |
parathyroid gland disorders | F | 2.72 | (2.54, 2.91) |
| phentermine | atypical amphetamine | weight loss | central nervous system vascular disorders | F | 2.57 | (2.41, 2.74) |
| anakinra | interleukin-1 receptor antagonist | rheumatoid arthritis |
musculoskeletal and connective tissue deformities | F | 2.54 | (2.37, 2.71) |
ADR, adverse drug reaction; CI, confidence interval; F, female; logROR, loge reporting odds ratio; M, male.
Table 4.
Top Sex Risks Posed by the 10 Most Prescribed Drugs in the US
| Drug | Indication | ADR | Sex | logROR | 95% CI |
|---|---|---|---|---|---|
| atorvastatin | dyslipidemia | glucose metabolism disorders (including diabetes mellitus) | F | 1.33 | (1.33, 1.34) |
| vascular therapeutic procedures | M | 0.59 | (0.61, 0.57) | ||
| levothyroxine sodium | hypothyroidism | synovial and bursal disorders | F | 0.94 | (0.91, 0.97) |
| congenital cardiac disorders | M | 2.15 | (2.17, 2.13) | ||
| lisinopril | hypertension and myocardial infarction | anterior eye structural change, deposit, and degeneration | F | 1.06 | (1.04, 1.07) |
| suicidal and self-injurious behaviors NEC | M | 0.39 | (0.40, 0.38) | ||
| gabapentin | seizures | aural disorders NEC | F | 0.58 | (0.56, 0.60) |
| hemolyses and related conditions | M | 0.72 | (0.74, 0.70) | ||
| amlodipine | hypertension, coronary artery disease, angina | anterior eye structural change, deposit, and degeneration | F | 0.74 | (0.72, 0.76) |
| hepatobiliary neoplasms: malignant and unspecified | M | 0.91 | (0.94, 0.88) | ||
| amoxicillin | bacterial infections | allergic conditions | F | 0.46 | (0.45, 0.47) |
| fatal outcomes | M | 0.67 | (0.69, 0.65) | ||
| omeprazole | gastric acid-related disorders | connective tissue disorders (excluding congenital) | F | 0.99 | (0.96, 1.02) |
| hepatobiliary neoplasms: malignant and unspecified | M | 0.89 | (0.92, 0.87) | ||
| metformin | type 2 diabetes mellitus | anterior eye structural change, deposit, and degeneration | F | 0.66 | (0.65, 0.68) |
| hepatobiliary neoplasms: malignant and unspecified | M | 0.90 | (0.92, 0.87) | ||
| losartan | hypertension | fractures | F | 0.77 | (0.74, 0.79) |
| fatal outcomes | M | 0.62 | (0.64, 0.61) | ||
| paracetamol | moderate pain and fever | connective tissue disorders (excluding congenital) | F | 0.73 | (0.71, 0.76) |
| urethral disorders (excluding calculi) | M | 1.19 | (1.23, 1.16) |
ADR, adverse drug reaction; CI, confidence interval; F, female; logROR, loge reporting odds ratio; M, male; NEC, not elsewhere classified.
Distribution of Sex Risks by Disease Indications and Drug Mechanisms
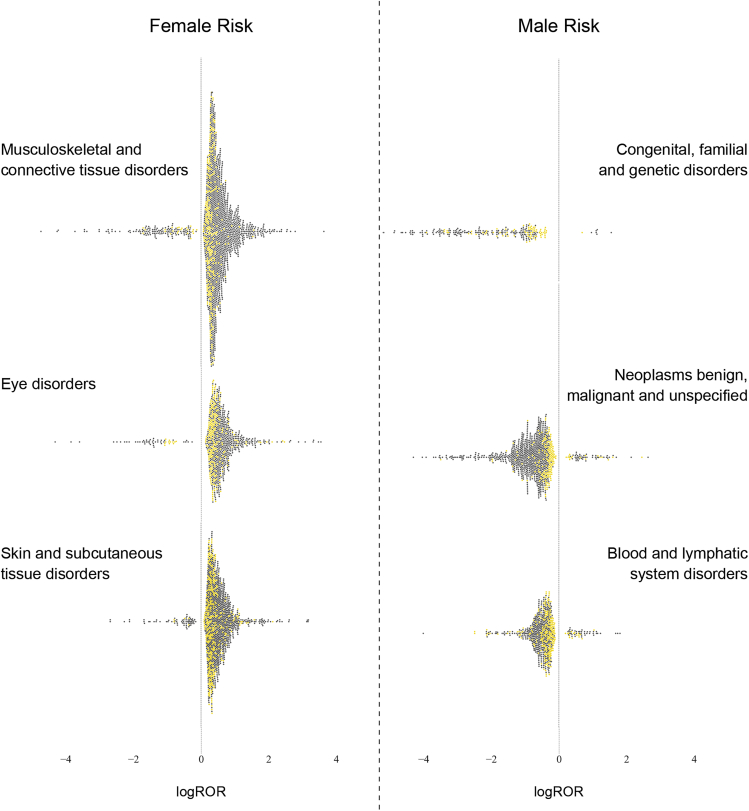
We characterized the variation of sex risks within drug and adverse event classes. To explore the variation of adverse events within disease indication groupings, we compared the distribution of drug-event pairs by sex in each system organ class (SOC). Figure 5 shows the top SOCs with the most disproportionate distribution in each sex. Women are at greatest risk for experiencing disorders of musculoskeletal and connective tissue; of skin and subcutaneous tissue; and of the eye. On the other hand, men are vulnerable to congenital and genetic disorders; benign and malignant neoplasms; and blood and lymphatic system disorders. Figure 5 demonstrates a nuanced understanding that goes beyond simply assigning the risk of an SOC to either sex. Across ADR classes, the yellow dots (indicating SOC risks) are much closer to zero than the dark gray dots (indicating high level group term [HLGT] risks). Sex risks have a much smaller effect size (logROR) at the SOC level than the HLGT level.
Figure 5.
Variation in Sex Risks of Adverse Events Grouped by Etiology
For each sex, the three SOCs that posed the most disproportionate risk of ADRs to that sex are visualized here. The SOCs posing greatest risk to women (left) are separated from those posing risks to men (right) by a dashed line. Each individual swarm plot visualizes the distribution of sex risks within the labeled SOC. Positive sex risks are associated with women and negative risks are associated with men. Each point represents a significant sex risk from one drug-ADR pair. Yellow dots indicate that the ADR was at the SOC level. Gray dots indicate that the ADR was at the HLGT level.
SOC, system organ class; HLGT, high level group term; logROR, loge reporting odds ratio.
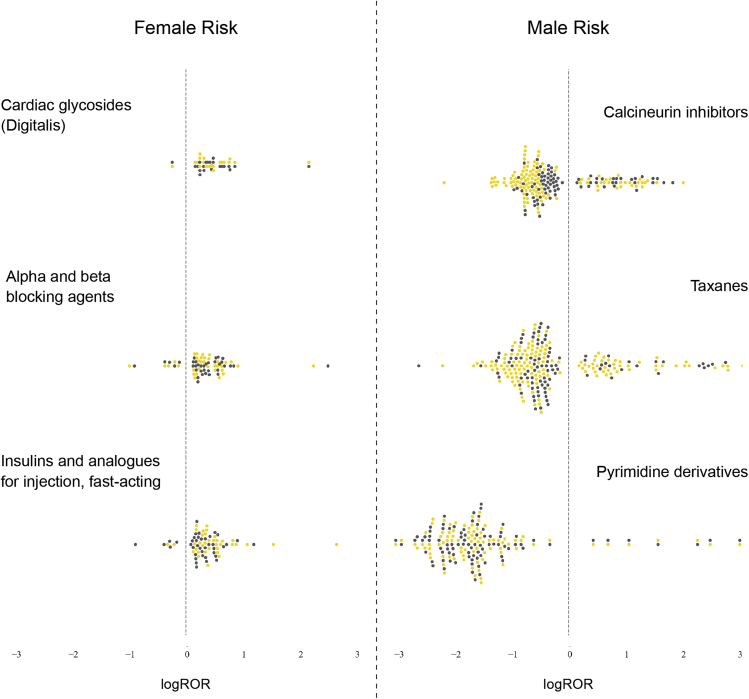
Similarly, we analyzed the distribution of drugs when grouped by mechanism at the ATC 4 level. Figure 6 shows the top drug classes with the most disproportionate risk distribution across sex. Men are at greatest risk from taxanes, pyrimidine derivatives, and calcineurin inhibitors. Cardiac glycosides (digitalis); alpha and beta blocking agents; fast-acting insulins and analogs for injection pose the largest risk of ADRs to women. Across drug classes, there is an even mix of yellow dots (indicating ATC 4 risks) and dark gray dots (indicating ATC 5 risks). This signals that effect sizes (logROR) are relatively similar for individual drugs and those grouped by mechanism.
Figure 6.
Variation in Sex Risks of Drugs Grouped by Mechanism
For each sex, the ATC 4 drug classes that posed the most disproportionate risk to that sex are visualized here. The drug classes posing greatest risk to women (left) are separated from those posing risks to men (right) by a dashed line. Each individual swarm plot visualizes the distribution of sex risks within the labeled ATC 4 drug class. Positive sex risks are associated with women and negative risks are associated with men. Each point represents a significant sex risk from one drug-ADR pair. Yellow dots indicate that the ADR was at the ATC 4 level. Gray dots indicate that the ADR was at the ATC 5 level.
ADR, adverse drug reaction; ATC 4, drugs grouped by mechanism; ATC 5, individual drugs; logROR, loge reporting odds ratio.
Clinical and Pharmacogenetic Validation Show that AwareDX has High Precision
To validate our algorithm, we used ADEs known to have sex differences according to clinical literature and pharmacogenetic mechanisms. We curated 22 expected sex differences in drug-ADR pairs from clinical literature. Of these, 3 drugs had insufficient patient data to test reliably. Of the remaining 19 associations, AwareDX recovered 1 male and 8 female sex risks correctly and the rest were predicted as no risk (see the Confusion matrix in Table 5). Our method had perfect precision but low recall (see Performance in Table 5). Critically, AwareDX did not classify expected male risks as female or vice versa since incorrect predictions were limited to predicting “No Risk.” The exact odds and confidence intervals (CI) predicted for each drug-ADR pair are listed in Table S4.
Table 5.
AwareDX Performance during Literature Validation
| Actual | Predicted |
||
|---|---|---|---|
| Female | Male | No risk | |
| Female | 8 | 0 | 8 |
| Male | 0 | 1 | 2 |
| No risk | 0 | 0 | 0 |
| Precision | Recall | F1 Score | |
|---|---|---|---|
| Female | 1.00 | 0.50 | 0.67 |
| Male | 1.00 | 0.33 | 0.50 |
| No risk | – | – | – |
| Weighted average | 1.00 | 0.47 | 0.64 |
Top, confusion matrix. Bottom, performance metrics. These results do not include cases that AwareDX was not tested against due to insufficient data or unavailable drugs.
For further validation, we explored genes with sex-differential expression and pharmacogenetic variants of these genes. We tested whether AwareDX could recover sex risks expected according to pharmacogenetic mechanisms. We curated 28 expected sex differences in drug-ADR pairs using reported sex-differential expression in hepatic metabolism and transport genes18 and knowledge from PharmGKB19 on their pharmacogenetic effects. Of these, 2 drugs were not found in FAERS and 2 drugs had insufficient patient data to test reliably. Our algorithm correctly recovered 9 of the remaining 24 risks and found no significant associations with the other 15. To ensure that our method was not predicting excessive false positives, we tested AwareDX against negative examples (i.e., drugs known not to have sex differences). Because a proper negative control set did not exist, we generated a set of pseudo-negative examples from variants without statistically significant associations in PharmGKB. Of the 14 pseudo-negative examples, 1 drug was not found and the remaining were correctly predicted as having no risk. The Confusion matrix in Table 6 combines results for positive and pseudo-negative examples. As with literature validation, AwareDX had 100% precision and low recall for both sexes. After including the no risk category, the weighted average precision was 81% and recall was 59% (see Performance in Table 6). As with literature validation, no false positives were predicted for the male and female categories. To the best of our knowledge, our algorithm is the first and only pharmacogenetically validated method for predicting sex risks. Predicted odds and CI for all expected sex risks are available from the Supplemental Information.
Table 6.
AwareDX Performance in Pharmacogenetic Validation
| Actual | Predicted |
||
|---|---|---|---|
| Female | Male | No risk | |
| Female | 5 | 0 | 9 |
| Male | 0 | 4 | 6 |
| No risk | 0 | 0 | 13 |
| Precision | Recall | F1 score | |
|---|---|---|---|
| Female | 1.00 | 0.36 | 0.53 |
| Male | 1.00 | 0.40 | 0.57 |
| No risk | 0.46 | 1.00 | 0.63 |
| Weighted average | 0.81 | 0.59 | 0.58 |
Top, confusion matrix. Bottom, performance metrics. These results do not include cases that AwareDX was not tested against due to insufficient data or unavailable drugs.
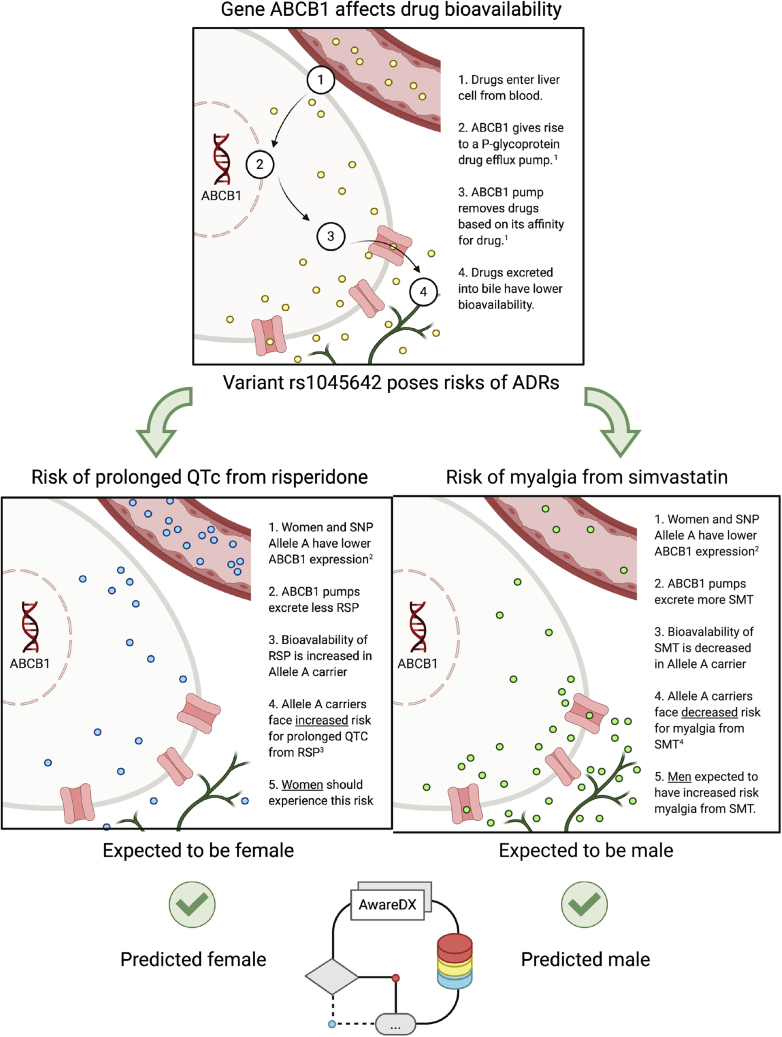
AwareDX Recovers Known Pleiotropic Effects of Gene ABCB1 for Adverse Events across Sexes
During validation against pharmacogenetic mechanisms, our algorithm confirmed that a single gene can pose disparate risks for men and women. ABCB1 was expected to pose a risk to men from simvastatin and another to women from risperidone (Figure 7). AwareDX correctly recovered both risks, thereby showing that sex-differential gene expression can have complex downstream effects on pharmacokinetics and pharmacodynamics (Figure 7).
Figure 7.
`1AwareDX Recovers Known Pleiotropic Effects of Gene ABCB1 for Adverse Events across Sexes
Top: gene ABCB1 codes for a hepatic drug efflux pump that removes active drugs from circulation. The capillary delivers some drug (yellow dots) to a hepatic cell. In the liver, gene ABCB1 is expressed and mobilizes a drug efflux pump (pink) on the apical membrane. This ABCB1 pump excretes the drug into bile ducts (green). Middle: ABCB1 has an SNP rs1045642 (A > G). Allele A of this variant poses two varying risks of adverse events: one from risperidone and the other from simvastatin. Both allele A and women18 are associated with lower expression of ABCB1. Using this information, we identify which sex is likely to experience these risks. Middle left: rs1045642 allele A is linked to increased risk of prolonged QTc from risperidone. Because both allele A and women have decreased expression of ABCB1, there is increased bioavailability of risperidone (blue dots) in women. This puts women at risk of prolonged QTc from risperidone. Middle right: rs1045642 allele A is linked to decreased risk of myalgia from simvastatin. Because allele A and women have similar expression of ABCB1, there is decreased bioavailability of simvastatin (green dots) in women. Women are at decreased risk, or, men are at increased risk of myalgia from simvastatin. Bottom: one drug-ADR risk is expected to be female while the other is expected to be male due to known effects of ABCB1. In the center, the AwareDX algorithm is visualized as a flowchart. AwareDX predicts both these risks correctly. ABCB1, ATP binding cassette subfamily B member 1; SNP, single-nucleotide polymorphism; RSP, risperidone; QTc, corrected QT interval; SMT, simvastatin. (1) https://www.pharmgkb.org/vip/PA166170352; (2) https://www.pharmgkb.org/variantAnnotation/1444687158; (3) https://www.pharmgkb.org/variantAnnotation/1296599320; (4) https://www.pharmgkb.org/variantAnnotation/982046440.
Sex Risks Identify Genes that Could Be Potentially Very Important Pharmacogenes
After independently validating AwareDX, we leveraged its findings to identify genes that could have variants with important, and possibly undiscovered, pharmacokinetic and pharmacodynamic effects. For each gene, we compared the counts of drugs associated in DrugBank to the count of significant sex risks identified by AwareDX (Table 7). Applying linear regression to both log counts resulted in line with a coefficient of 0.941 and intercept of −0.0373 (y = 0.941x − 0.0373). Each gene's level of importance as a pharmacogene was quantified as the residual from this line. The top 5 genes were CYP2A13, FMO1, ALDH5A1, CYP27A1, and FMO3 (see Figure S4).
Table 7.
Genes that Could Potentially Be Very Important Pharmacogenes, with Associated Drugs and Number of Adverse Drug Events Predicted to Have Sex Risks
| Gene | Drug | ADEs |
|---|---|---|
| ALDH5A1 | sodium oxybate | 7 |
| CYP27A1 | colecalciferol | 29 |
| CYP27A1 | ergocalciferol | 6 |
| FMO3 | clozapine | 1 |
| FMO3 | olanzapine | 1 |
| FMO3 | dasatinib | 1 |
| FMO1 | lorcaserin | 1 |
Discussion
AwareDX Identifies and Mitigates Confounding Biases in Underlying Data
The effectiveness of AwareDX in correcting sex biases is evident in (1) the ability of the RF model to clearly separate patients by sex and (2) the improvement in the sex disparity of PRRs for sex-specific ADRs. We used an RF model to predict propensity scores (i.e., the likelihood of being female) from drug exposure, co-medication, and age features. Analyzing the feature importances (Figure S2A) showed that the top features were logically linked to knowledge of sex. For example, Niraparib, the fourth most important feature, is used to treat ovarian cancer. This suggests that propensity scores were generated by using information from confounding factors, as expected.
The propensity scores generated by the RF model demonstrate the high predictive ability of the curated features for some patients. The distribution of propensity scores (Figure 3) shows clear separation for some subsets of patients, with the propensity scores for women tending toward 1 and those for men tending toward 0. The model clearly singled out patients associated with sex biases using covariate features. Furthermore, we used the sex disparity in the PRR of MedDRA's gender-specific ADRs to quantify sex bias in FAERS. Applying AwareDX reduced or removed 79.2% of these PRR differences. Thus, our machine learning approach was able to identify patients associated with covariates and effectively dampen these biases.
Sex Risks Are Highly Precise and Consistent with Clinical Literature
During both literature and pharmacogenetic validation (Tables 5 and 6), AwareDX had 100% precision for predicting male and female sex risks. After including the No Risk category during pharmacogenetic validation, the weighted average precision was 81%. Critically, there were zero false positives in the male and female categories. Furthermore, we identified the disease indications and drug mechanisms that most disproportionately affect the sexes (Figures 5 and 6). Many of these results have been reported previously in clinical settings. The discovery that women are at risk of musculoskeletal and connective tissue disorders is well known.22 Zopf and colleagues22 found that female patients experienced a significantly higher incidence of musculoskeletal system ADRs than their male counterparts. AwareDX predicted that women are prone to ADRs from alpha and beta blocking agents and from cardiac glycosides. Because of pharmacodynamic differences, females are known to have greater sensitivity and enhanced bioavailability of beta blockers.10 Digoxin, the only cardiac glycoside that is frequently used in clinical settings, notoriously causes increased mortality in women at high serum concentration.10,23 Thus, the sex risks identified are precise and broadly in accordance with clinical literature. AwareDX is the first, and to our knowledge only, approach for predicting sex risks that is not only in agreement with clinical literature but has also been validated against pharmacogenetic mechanisms.
Sex Risks Increase with Increasing Specificity of ADRs but Are Stable across Drug Mechanism Groupings
We clustered sex risks by adverse events and drugs to explore patterns of incidence across sex risks. We studied the variation in effect size (mean logROR) across groupings of adverse events by etiology and of drugs by mechanism. For ADRs, effect sizes were generally larger at the HLGT level than at the SOC level (Figure 5). The pharmacogenetic analysis, conducted at the preferred term (PT) level, also found significant sex risks. This suggested that neither sex was at great risk for experiencing all events within the umbrella SOC term. Rather, sex risks increased with increasing specificity of the ADR. Effect sizes were relatively similar for drugs grouped by mechanism and for individual drugs (Figure 6). Because drugs with the same mechanism of action tend to have analogous metabolic pathways, they could pose similar sex risks. This pattern may not hold on drug groupings by therapy or pharmacology, such as ATC 3 and above.
Anakinra Poses Severe, Disparate, and Previously Unknown Risks to Both Sexes
Anakinra is an interleukin-1 receptor antagonist that is most commonly used to treat rheumatoid arthritis. Between 5% and 10% of patients on Anakinra experience severe side effects, such as neutropenia and acute infections. Anakinra was associated with three of the 10 most prominent sex risks reported (Table 3). AwareDX predicted that Anakinra puts men at risk of a common disease of the colon: diverticular disorders (logROR = 4.52; 95% CI = 4.51–4.54). Women exposed to Anakinra are vulnerable to vascular inflammations (logROR = 3.25; 95% CI = 3.03–3.48) and musculoskeletal and connective tissue deformities (logROR = 2.54; 95% CI = 2.37–2.71). To the best of our knowledge, neither diverticular disorders nor vascular inflammations have been associated with Anakinra in the past. The highly disparate nature of the predictions suggests that AwareDX is capable of predicting risks that are not skewed toward either sex.
ABCB1 Provides a Pharmacogenetic Basis for Disparate Sex Risks
The sex-differential expression of a single gene can lead to complex ADEs in both sexes. Existing knowledge maintains that a single gene, ABCB1, can pose disparate risks for men and women. ABCB1 is a very important pharmacogene that codes for a hepatic efflux pump.24 It has been shown to have a 1.13-fold higher expression in males.18 Based on information about its variants, we expected ABCB1 to pose (1) an increased risk to males for myalgia from exposure to simvastatin and (2) an increased risk to females for prolonged QTc from exposure to risperidone (Figure 7). AwareDX correctly predicted both these results, thereby demonstrating that it can recover disparate sex risks that are grounded in pharmacogenetic mechanisms. This validation of our algorithm lends further support to putative sex risks that may be disparate, such as with Anakinra.
Predictions of Pharmacogenes Are Supported by Known Metabolic Pathways
We leveraged our results to flag genes that could be very important pharmacogenes (see Table 7). All flagged genes had zero to few clinical annotations on PharmGKB. We explored whether clinical literature could establish drug-gene interactions for the two most compelling genes: CYP27A1 because it was associated with the highest number of drug-ADR pairs and FMO3 because it was associated with the most number of drugs.
CYP27A1 was most strongly associated with cholecalciferol (vitamin D3) and then with ergocalciferol (vitamin D2). CYP27A1 had no clinical or variant annotations on PharmGKB. The pharmacogenetic associations of CYP27A1 and vitamin D are stated as “unknown” on DrugBank, and to the extent of our knowledge, have not been discovered elsewhere. One function of CYP27A1 is to metabolize vitamin D; specifically having vitamin D3 25-hydroxylase activity.25, 26, 27 A study showed that vitamin D supplementation could impair organ function even in hypovitaminosis D.28 In our findings, all (29 of 29) sex risks from cholecalciferol were posed to women and the majority (4 of 6) of sex risks from ergocalciferol were posed toward men. Specifically, women might be at risk of coagulopathies and bleeding diathesis (logROR = 1.61; 95% CI = 1.53–1.69) and gastrointestinal conditions (logROR = 1.73; 95% CI = 1.63–1.82) from cholecalciferol. Men exposed to ergocalciferol could be vulnerable to cytogenetic investigations (logROR = 0.93; 95% CI = 0.90–0.95). This may suggest that women respond better to vitamin D2 and men respond better to vitamin D3.
FMO3 was associated with three different drugs in our results: clozapine, olanzapine, and dasatinib. Clozapine and olanzapine are anti-psychotics that have very similar chemical structures.29 Dasatinib is a tyrosine kinase inhibitor used to treat leukemia. On PharmGKB, FMO3 was only annotated as affecting the serum concentration of olanzapine and no risks for ADRs from olanzapine were noted. Clozapine and dasatinib were not associated with FMO3 on PharmGKB. FMO3 codes for a liver enzyme that oxidatively metabolizes olazapine,30 clozapine,31 and dasatinib.32 According to our results, all three drugs could be associated with ADR risks to women: clozapine may cause uterine, pelvic, and broad ligament disorders (logROR = 2.86; 95% CI = 2.62–3.10); olanzapine may cause genitourinary tract disorders (logROR = 0.82; 95% CI = 0.79–0.85); and dasatinib may cause skin appendage conditions (logROR = 0.92; 95% CI = 0.88–0.95).
Limitations
Literature and pharmacogenetic validation showed that AwareDX has a low recall, 47% and 59% weighted averages respectively. In part, the low recall could be due to the algorithm's minimum data requirement of 250 patients per sex to prevent oversampling of rare ADRs. The performance of our method with respect to detecting rare events exhaustively may be less reliable. Nevertheless, it is important to note that sex differences in drug-induced adverse events are a relatively rare phenomenon. In similar analyses we published in the past,33, 34, 35,17 where we used data-mining algorithms to identify and experimentally validate unexpected adverse effects, we found similarly low recall. It is not recall that is critical in these cases, but the enrichment of true positives against the predicted positives (i.e., precision).
Our method assigns P values to each iteration of stochastic sampling and risk calculation rather than drug-ADR pairs. When iterations are assimilated into the mean logROR with CIs, the P values are lost. We apply strict cut offs to predicted risks by ensuring that each iteration produces a significant P value before labeling the drug-event pair as having a sex risk. While our approach is extremely conservative, there remains potential to develop a method that can quantify the significance of each sex-specific ADE.
Evaluation of side effect prediction algorithms, in general, is not straightforward; no gold standard of known ADEs exists. In lieu of a standard, we evaluated our proposed methodology against known pharmacogenetic mechanisms and clinical literature. To compensate for the lack of a proper negative control set, we generated a set of pseudo-negative examples from statistically insignificant pharmacogenetic mechanisms. Although these methods of validation independently evaluated our algorithm, they could not evaluate the resource of sex risks generated by said algorithm. Sex risks in validation were reported at a lower MedDRA level compared with sex risks in our resource (HLGT/SOC). Mapping these preferred terms (PT) up to HLGTs or SOCs for validation would not be semantically consistent. Furthermore, it was not practical to conduct the initial study using PTs due to statistical and data reasons mentioned in the “Data Processing and Mapping” section of the Experimental Procedures. A publicly available resource of sex risks would greatly enhance the evaluation of the sex risks identified in our resource. Finally, our findings are limited by the data we use. Because FAERS consists of reports from the US, these results may not generalize to non-representative patient populations. It would be valuable to compare the results of this and other algorithms across adverse event reporting systems.
Future Directions
We present a new resource of sex-specific ADEs (Table S1). This resource could vastly advance the consideration of sex during drug discovery, repositioning, and pharmacogenetic studies. The sex risks proposed here could be examined retrospectively in electronic health records or prospectively through pre-clinical studies and clinical trials. Furthermore, unsupervised learning techniques could be applied to identify patterns in sex-specific ADEs. Ultimately, these analyses would contribute toward advancing knowledge of sex differences during drug prescription.
AwareDX is able to identify patient reports associated with covariates of sex and effectively dampen confounding biases. It can be applied to systematically quantify sex risks from any post-marketing surveillance dataset. The algorithm can also be restructured to correct biases in any discrete variables of interest. For instance, sex can be replaced with age to study pediatric ADEs. In essence, our method corrects for confounding biases found in an understudied population. This approach of using machine learning to build balanced cohorts for disproportionality analysis has the potential to generalize to many data science domains.
Conclusion
Adverse reactions are the fourth leading cause of death in the US and women have twice the risk of developing them as compared with men. Unfortunately, sex differences in drug response have neither been systematically studied nor clinically applied. Here, we present an algorithm that is the first, and to our knowledge only, validated approach for predicting sex risks. Our algorithm AwareDX identifies drugs posing increased risks of adverse events to either sex. We show that AwareDX mitigates confounding biases in data and recovers known clinical and pharmacogenetic sex risks with high precision. Our resource of sex-specific ADEs would be suitable for use in drug discovery, repositioning, and pharmacogenetic studies. We believe that AwareDX could vastly advance the incorporation of sex in considerations of drug safety and efficacy. Knowledge of sex differences during drug prescription has the potential to significantly reduce adverse events, making AwareDX a valuable tool for the advancement of precision medicine.
Experimental Procedures
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Nicholas Tatonetti (nick.tatonetti@columbia.edu).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
All source data used in the paper are publically available. We downloaded the following: (1) FAERS from openFDA; (2) Gender Specific Adverse Events List from MedDRA; (3) Severity Scores for MedDRA Adverse Events from Table S2 from Gottlieb and colleagues21; (4) Sex Differences in the gene expression from supplementary materials from Yang and Li18; and (5) a vocabulary linking MedDRA, SNOMED CT, and RxNorm from OHDSI Athena. The exact database used in this study is available on request. The published article includes all datasets generated during this study. See Tables S1 and S2 for sex risks generated by AwareDX and Tables S3 and S4 for expected sex risks curated from PharmGKB and literature. The code generated during this study are available on our GitHub repository tatonetti-lab/sex_risks.
Data Processing and Mapping
Using an API key with extended permissions, we extracted and processed all available drug-event json files through the second quarter of 2019 from FAERS. We developed an extraction, transfer, and load pipeline to get the json embedded data in a tabular format for conversion into an SQL database. The code to pull embeddings and to generate the database can be found on GitHub openFDA drug-event parsing and TABELIZER!, respectively. From the FAERS data, all patients with missing sex were excluded from this analysis. Patients over the age of 85 and under the age of 18 were excluded, which led to 5.25% of patients being excluded. Ultimately, the dataset consisted of 8,860,677 patients.
In FAERS, drugs and adverse events are identified using RxNorm and MedDRA, respectively. Although AwareDX can be applied to any vocabulary, we mapped drugs and ADRs in the following manner to ultimately allow for effective generalization of our results. We mapped drugs from RxNorm to ATC using Athena by OHDSI. A total of 1,458 drugs were identified at the ATC 5 level, and were mapped to 481 drug classes at the ATC 4 level.
AwareDX can be applied at any MedDRA level, including PTs, HLGTs, and SOCs. Although FAERS data are reported at the PT level, we leveraged the MedDRA heirarchy to reduce noise and errors by mapping PTs to HLGTs and SOCs. Analyzing SOCs and HLGTs required only 700,000 tests, as opposed to 35 million tests with PTs. With only 8.8 million adverse event reports, mapping to higher levels allowed us to preserve the power of our analysis. By aggregating up to SOC/HLGT, we leverage information theory to reduce the effect of noise at the PT level. Many PTs can be synonymous with other PTs and the choice of a term may be arbitrary and dependent on the specific reporter or the specific data abstractor at the FDA. For example, the following PTs all encoded a specification of heart failure but had noisy and imbalanced reporting: Cardiac failure (36,404 reports), Cardiac failure acute (2,559), Cardiac failure chronic (1,359), Cardiac failure congestive (53,487), Cardiac failure high output (75). All of these terms mapped to the HLGT “Heart failures” and the SOC “Cardiac disorders.” These higher-level terms served as more robust statistics that counteracted semantic and reporting variations in the FAERS dataset. Because of these merits, we mapped ADRs from 18,335 PTs to 335 MedDRA HLGTs and 27 SOCs. If a PT belonged to multiple groups, it was mapped separately to each higher-level group. Severity scores in Gottlieb and colleagues21 were reported for MedDRA PTs. At the HLGT and SOC level, we calculated severity scores as the mean severity score across PTs within that grouping.
Sex Biases
To understand the relationship between sex, drugs, and adverse events, we explored the distribution of ADRs that are known to differentially occur in either sex. We selected sex-exclusive ADRs from MedDRA's Gender Specific Adverse Events List. For each combination of drugs, sex, and sex-exclusive ADRs, we calculated a PRR. We quantified sex bias as the absolute difference in mean PRR across sex. Although some of these PRR differences could have been due to real differential risks between males and females, we assumed that the disparity could be completely attributed to sex biases.
We explored whether AwareDX is effective in mitigating these biases. To do so, we noted the PRR disparity in FAERS as the original or “before” sex biases and we applied AwareDX to generate the corrected or “after” sex biases. As explained in the following section, AwareDX builds balanced cohorts for any given drug-consuming subpopulation of FAERS. For each combination of drugs, sex, and sex-exclusive ADRs, we calculated the PRR using patient counts from these balanced cohorts. Sex biases were defined as the difference in mean PRRs. We said that AwareDX mitigated a particular sex bias if the PRR disparity after was reduced or removed compared with the PRR disparity before.
The AwareDX Algorithm
Our algorithm, AwareDX evaluated whether a drug-ADR pair had a significant sex risk in three key steps. First, AwareDX generated propensity scores for each patient. Next, these propensity scores were used to build sex-balanced cohorts for each drug-consuming subpopulation. This process helped to mitigate the sex biases found in FAERS. Finally, the algorithm applied disproportionality analysis on the balanced cohorts to quantify the sex risk of a given drug and adverse event. Figure 1 provides a visual overview of this process.
Propensity Score Matching with Machine Learning
We adapted existing propensity score-matching methods to mitigate the confounding effects on sex. Each patient's propensity score was defined as their predicted likelihood of being female given information about their demographics and drug exposures. A classification model was trained to predict propensity scores for each patient from curated features such as age, polypharmacy, and one-hot-encodings of drug exposures. Because age was missing for 38.5% of patients, the mean age (54.7) was imputed.
We considered various machine learning models for prediction of propensity scores, including logistic regression, support vector machines, and RF. All models were fit over 25 iterations, where each iteration used 10,000 patient reports uniformly subsampled from FAERS. We expected the sampled reports for a given iteration to closely follow the distribution of total patient reports in FAERS. Each experiment's performance was consolidated using mean and 95% CI over iterations. All models performed similarly during 5-fold cross-validation (see Table 2). We selected the RF model because it is an ensemble model that allows the computation of out-of-bag scores. For any sample, the out-of-bag score is calculated as the average prediction of all the trees that have not seen the given sample during training. Importantly, out-of-bag scores produce similar estimates to cross-validation and hold-out validation.36 Using out-of-bag scores allowed us to conserve 100% of our data, as opposed to just the test subset, for downstream disproportionality analysis. This would not be possible with other machine learning methods and would unnecessarily reduce the power of AwareDX for rare ADEs. To reduce dimensionality and accelerate computation, we selected the top 10% of features using a chi-square scoring function. Using grid search, we identified the ideal hyperparameters as gini criterion, 100 trees, and a maximum depth of 9.
We showed that the addition of indication features and sophisticated imputation techniques does not improve model performance (see Figure S3). Although drug indications can be important confounding factors to ADE signals, model performance remained largely unchanged on adding up to 5,000 indication features. Indication features did not add substantial new information to the model because they were tightly tied to drug exposures and their data in FAERS was sparse (only 29.5% of indications had more than 25 patient reports). Because imputing age with the mean is a simple technique, we explored whether using a k-nearest neighbors regressor to predict age would boost model performance. For various values of k, we found that the RF model's out-of-bag score was remained constant. Because neither modification had a significant impact on model performance and since other ML methods achieved similar performance, we concluded that the RF model developed here was sufficiently robust.
Building Cohorts with Bootstrapping
After generating propensity scores for all patients, we evaluated sex risks for each drug. For each drug-consuming subpopulation, we built balanced cohorts by bootstrapping. Patients were assigned to 1 of 100 bins according to their propensity score. From each bin, we selected all the women and sampled an equal number of men for comparison. When a bin contained only male/female patients, no patients were selected from that bin. The resulting cohorts were sex balanced and had uniform contributions from the covariates.
For drugs with extremely limited data for either sex, bootstrapping led to over-representation and oversampling of the limited reports. To prevent such imbalanced prescription from leading to synthetic associations, drugs with fewer than 250 patients for each sex were excluded from analysis. To identify ADRs that exclusively occur in either sex, we used MedDRA's “Gender Specific Adverse Events List.” All reports associated with these ADRs were excluded to correct for biases in underlying diseases.
Evaluating Sex Risks with Disproportionality Analysis
To evaluate sex-specific risks of drug-event pairs, we conducted disproportionality analysis on the balanced cohorts. From the cohorts of drug-exposed patients, we constructed a contingency matrix as shown in Figure 1. We used a chi-square test to identify whether a significant sex risk was present, and calculated the logarithm of the logROR to quantify that sex risk.
Because the bootstrapping process was stochastic, the entire process from building cohorts to calculating the logROR was repeated 25 times. After applying Bonferroni correction to the P values, significant results were retained. For each drug-event pair, if all 25 association tests were significant, the sex risk of the pair was deemed to have a significant sex-specific risk. This risk was quantified as the mean logROR with 95% CI, where a positive value indicated female risk and negative value indicated male risk. For ease of interpretation, risks are reported with sex and absolute log odds throughout.
Validation
We used ADEs known to have sex differences to validate our algorithm. We explored clinical literature, drug product labels, and pharmacogenetic mechanisms to identify drug-ADR pairs with established sex-specific risks. To validate, we used our algorithm to predict each of the expected sex risks given the drug-ADR pair and tested whether the generated sex risks matched the expected sex risks. All expected sex risks were reported at the MedDRA PT level. We did not map these PTs to SOC/HLGT because sex risks expected at the PT level need not exist at higher levels. Because ADRs were specified as MedDRA PTs, the expected sex risks were not part of the tests initially run on FAERS. Thus, each expected sex risk was run ad-hoc and AwareDX was independently validated on an unseen set of expected ADEs.
Clinical Literature
We validated our algorithm by showing that it can recover sex risks from clinical literature. To identify studies that explored sex-specific ADEs, we searched PubMed and Google Scholar for the following terms: sex, gender, men, man, women, woman, males, male, females, female, adverse event, adverse drug reaction, drug, drug response, pharmacodynamics, and pharmacokinetics. From the search results, we curated a selection of 23 relevant articles and reviews. We excluded studies that only discussed the genetic basis for sex risks if they did not mention a sex-specific ADE. From the selected studies, we identified drug-ADR pairs that had been clinically shown to pose sex risks. We excluded agents that could not be mapped to ATC 4 or 5. For validation, we tested AwareDX against these drug-ADR pairs with clinically established sex risks.
We attempted to mine drug label annotations for sex risks. To search for relevant annotations, we used DailyMed, which is the National Library of Medicine's official provider of FDA's Structured Product Labels. When we did a string matching search for any of the following terms in the Adverse Reactions Section and the Contraindications Section, no results were returned. The terms included: males, male, females, female, men, man, women, woman, sex, and gender. Hence, we concluded that sex-specific ADEs are not found frequently enough on drug labels to support a validation analysis.
Pharmacogenetic Mechanisms
Pharmacogenetics has the potential to explain sex risks by identifying the genetic variants responsible for differences in drug metabolism between males and females. We validated AwareDX by recovering sex risks that were expected according to pharmacogenetic mechanisms. To identify expected sex risks, we explored 77 hepatic drug-metabolizing and transporter genes that have sex differences in expression.18 We assumed that increased expression of a gene leads to increased production of its corresponding protein. Based on this assumption, genes with sex-differential expression should affect the bioavailability of drugs and pose risks for ADRs similarly to their pharmacogenetic variants. We used PharmGKB19 to identify variants and their associated drug-event risks. Information about the expected sex differences, along with relevant supporting knowledge from Yang and Li18 and PharmGKB to explain the hypotheses, can be found in the Supplemental Information. These expected sex risks provided a set of pharmacogenetically grounded positive examples to test AwareDX against.
It was important to test AwareDX against negative examples (i.e., drugs known not to have sex differences) to ensure that the true positives did not drown in an overwhelming majority of false positives. Unfortunately, to the best of our knowledge, no resource exists for such negative examples. Nonetheless, to address the limitation faced without a proper negative control set, we generated a set of pseudo-negative examples. For genes with known sex differences in expression from Yang and Li,18 we used PharmGKB's variant annotations to identify variants without statistically significant associations with adverse drug outcomes as pseudo-negative examples. If a sex-varying gene's variant had no ADEs then we expected that AwareDX should be less likely to produce sex risks for that drug-ADR pair. We used statistical insignificance () as a substitute for the lack of a variant's ADEs. These expected sex risks provided a set of pseudo-negative examples to test AwareDX against.
Prediction of Pharmacogenes from Sex Risks
We leveraged the sex risks identified by AwareDX to flag genes that could have variants with important, and possibly undiscovered, pharmacokinetic and pharmacodynamic effects. For each gene, we compared the counts of drugs associated in DrugBank to the count of significant drug-event pairs identified by AwareDX. We expect that the number of adverse drug events associated with the gene may be proportional to the number of drugs for which that gene is a metabolizer. We confirmed this in a scatterplot shown on linear and log-log scales (Figure S4). The choice of log-log was motivated by the typical scale-free nature of biological networks,37 which this gene-drug network appeared to follow. We then applied linear regression to the logarithm of both counts and used the residuals as a ranking mechanism to identify top sex-varying pharmcogene candidates.
Acknowledgments
All data analysis conducted in this work is available upon request. We thank Nicholas Giangreco for helpful discussions and technical guidance. N.P.T. is supported by R35GM131905 from the US NIH NIGMS. The graphical abstract and Figure 7 were created using Biorender.
Author Contributions
Conceptualization, P.C. and N.T.; Methodology, P.C. and N.T.; Software, P.C. and N.T.; Validation, P.C. and N.T.; Formal Analysis, P.C. and N.T.; Investigation, P.C. and N.T.; Resources, P.C. and N.T.; Data Curation, P.C. and N.T.; Writing – Original Draft, P.C. and N.T.; Writing – Review & Editing, P.C. and N.T.; Visualization, P.C. and N.T; Supervision, N.T.; Funding Acquisition, N.T.
Declaration of Interests
The authors declare no competing interests.
Published: September 22, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.patter.2020.100108.
Supplemental Information
Tab 1: Positive Examples
Tab 2: Pseudo-negative Examples.
References
- 1.Watanabe J.H., McInnis T., Hirsch J.D. Cost of prescription drug-related morbidity and mortality. Ann. Pharmacother. 2018 doi: 10.1177/1060028018765159. [DOI] [PubMed] [Google Scholar]
- 2.FDA . FDA; 2018. Preventable Adverse Drug Reactions: A Focus on Drug Interactions.https://www.fda.gov/drugs/drug-interactions-labeling/preventable-adverse-drug-reactions-focus-drug-interactions#ADRs:%20Prevalence%20and%20Incidence [Google Scholar]
- 3.Hakkarainen K.M., Hedna K., Petzold M., Hägg S. Percentage of patients with preventable adverse drug reactions and preventability of adverse drug reactions—a meta-analysis. PLoS One. 2012;7:e33236. doi: 10.1371/journal.pone.0033236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tannenbaum C., Day D. Age and sex in drug development and testing for adults. Pharmacol. Res. 2017;121:83–93. doi: 10.1016/j.phrs.2017.04.027. [DOI] [PubMed] [Google Scholar]
- 5.FDA FDA OWH established and begins advocating for the inclusion of women in clinical trials. 1994. https://www.fda.gov/files/science%20&%20research/published/1994- FDA- OWHestablished- and- begins- advocating- for- the- inclusion- of- women- in- clinical- trials. .pdf
- 6.Kwiatkowski K., Coe K., Bailar J.C., Swanson G.M. Inclusion of minorities and women in cancer clinical trials, a decade later: have we improved? Cancer. 2013;119:2956–2963. doi: 10.1002/cncr.28168. [DOI] [PubMed] [Google Scholar]
- 7.Tharpe N. Adverse drug reactions in women’s health care. J. Midwifery Women’s Health. 2011;56:205–213. doi: 10.1111/j.1542-2011.2010.00050.x. [DOI] [PubMed] [Google Scholar]
- 8.Watson S., Caster O., Rochon P.A., Ruijter H.d. Reported adverse drug reactions in women and men: aggregated evidence from globally collected individual case reports during half a century. EClinicalMedicine. 2019;17 doi: 10.1016/j.eclinm.2019.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soldin O.P., Chung S.H., Mattison D.R. Sex differences in drug disposition. J. Biomed. Biotechnol. 2011;2011:1–14. doi: 10.1155/2011/187103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whitley H.P., Lindsey W. Sex-based differences in drug activity. Am. Fam. Phys. 2009;80:1254–1258. [PubMed] [Google Scholar]
- 11.Drici M.-D., Clement N. Is gender a risk factor for adverse drug reactions? The example of drug-induced long QT syndrome. Drug Saf. 2001;24:575–585. doi: 10.2165/00002018-200124080-00002. [DOI] [PubMed] [Google Scholar]
- 12.Makkar R., Fromm B., Steinman R., Meissner M., Lehmann M. Female gender as a risk factor for torsades de pointes associated with cardiovascular drugs. JAMA. 1993;270:8. doi: 10.1001/jama.270.21.2590. [DOI] [PubMed] [Google Scholar]
- 13.FDA . Edluar, and Zolpimist; 2013. Risk of Next-Morning Impairment after Use of Insomnia Drugs; FDA Requires Lower Recommended Doses for Certain Drugs Containing Zolpidem. [Google Scholar]
- 14.Schmetzer O., Flörcken A. Sex differences in the drug therapy for oncologic diseases. Handbook Exp. Pharmacol. 2012;214:411–442. doi: 10.1007/978-3-642-30726-3_19. [DOI] [PubMed] [Google Scholar]
- 15.Zhou L., Rupa A.P. Categorization and association analysis of risk factors for adverse drug events. Eur. J. Clin. Pharmacol. 2018;74:389–404. doi: 10.1007/s00228-017-2373-5. [DOI] [PubMed] [Google Scholar]
- 16.Yu Y., Chen J., Li D., Wang L., Wang W., Liu H. Systematic analysis of adverse event reports for sex differences in adverse drug events. Sci. Rep. 2016;6 doi: 10.1038/srep24955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tatonetti N.P., Ye P.P., Daneshjou R., Altman R.B. Data-driven prediction of drug effects and interactions. Sci. Transl. Med. 2012;4:125ra31. doi: 10.1126/scitranslmed.3003377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang L., Li Y. Sex differences in the expression of drug-metabolizing and transporter genes in human liver. J. Drug Metab. Toxicol. 2012;3 doi: 10.4172/2157-7609.1000119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thorn C.F., Klein T.E., Altman R.B. PharmGKB: the pharmacogenomics knowledge base. Methods Mol. Biol. 2013;1015:311–320. doi: 10.1007/978-1-62703-435-7_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paavola A. 10 most prescribed drugs in the US in Q1. 2019. https://www.beckershospitalreview.com/pharmacy/10-most-prescribed-drugs-in-the-u-s-in-q1.html
- 21.Gottlieb A., Hoehndorf R., Dumontier M., Altman R.B. Ranking adverse drug reactions with crowdsourcing. J. Med. Int. Res. 2015;17:e80. doi: 10.2196/jmir.3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zopf Y., Rabe C., Neubert A., Janson C., Brune K., Hahn E., Dormann H. Gender-based differences in drug prescription: relation to adverse drug reactions. Pharmacology. 2009;84:333–339. doi: 10.1159/000248311. [DOI] [PubMed] [Google Scholar]
- 23.Adams K.F., Patterson J.H., Gattis W.A., O’Connor C.M., Lee C.R., Schwartz T.A., Gheorghiade M. Relationship of serum digoxin concentration to mortality and morbidity in women in the digitalis investigation group trial: a retrospective analysis. J. Am. Coll. Cardiol. 2005;46:497–504. doi: 10.1016/j.jacc.2005.02.091. [DOI] [PubMed] [Google Scholar]
- 24.Hodges L.M., Markova S.M., Chinn L.W., Gow J.M., Kroetz D.L., Klein T.E., Altman R.B. Very important pharmacogene summary: ABCB1 (MDR1, P-glycoprotein) Pharmacogenet. Genom. 2011;21:152–161. doi: 10.1097/FPC.0b013e3283385a1c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Araya Z., Hosseinpour F., Bodin K., Wikvall K. Metabolism of 25-hydroxyvitamin D3 by microsomal and mitochondrial vitamin D3 25-hydroxylases (CYP2D25 and CYP27A1): a novel reaction by CYP27A1. Biochim. Biophys. Acta. 2003;1632:40–47. doi: 10.1016/s1388-1981(03)00062-3. [DOI] [PubMed] [Google Scholar]
- 26.Guo Y.D., Strugnell S., Back D.W., Jones G. Transfected human liver cytochrome P-450 hydroxylates vitamin D analogs at different side-chain positions. Proc. Natl. Acad. Sci. U S A. 1993;90:8668–8672. doi: 10.1073/pnas.90.18.8668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu J.G., Ochalek J.T., Kaufmann M., Jones G., DeLuca H.F. CYP2R1 is a major, but not exclusive, contributor to 25-hydroxyvitamin D production in vivo. Proc. Natl. Acad. Sci. U S A. 2013;110:15 650–715 655. doi: 10.1073/pnas.1315006110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Razzaque M.S. Can adverse effects of excessive vitamin D supplementation occur without developing hypervitaminosis D? J. Steroid Biochem. Mol. Biol. 2018;180:81–86. doi: 10.1016/j.jsbmb.2017.07.006. [DOI] [PubMed] [Google Scholar]
- 29.Schatzberg A.F., Nemeroff C.B. American Psychiatric Association Publishing; 2017. The American Psychiatric Association Publishing Textbook of Psychopharmacology. OCLC: 971615789, ISBN: 978-1-61537-122-8 978-1-61537-162-4. [Google Scholar]
- 30.Söderberg M., Haslemo T., Molden E., Dahl M.-L. Influence of FMO1 and 3 polymorphisms on serum olanzapine and its N-oxide metabolite in psychiatric patients. Pharmacogenom. J. 2013;13:7. doi: 10.1038/tpj.2012.47. [DOI] [PubMed] [Google Scholar]
- 31.Sachse C. Flavin monooxygenase 3 (FMO3) polymorphism in a white population: allele frequencies, mutation linkage, and functional effects on clozapine and caffeine metabolism. Clin. Pharmacol. Ther. 1999;66:431–438. doi: 10.1053/cp.1999.v66.a102203. [DOI] [PubMed] [Google Scholar]
- 32.Wang L., Christopher L.J., Cui D., Li W., Iyer R., Humphreys W.G., Zhang D. Identification of the human enzymes involved in the oxidative metabolism of dasatinib: an effective approach for determining metabolite formation kinetics. Drug Metab. Dispos. 2008;36:1828–1839. doi: 10.1124/dmd.107.020255. [DOI] [PubMed] [Google Scholar]
- 33.Lorberbaum T., Nasir M., Keiser M.J., Vilar S., Hripcsak G., Tatonetti N.P. Systems pharmacology augments drug safety surveillance. Clin. Pharmacol. Ther. 2015;97:151–158. doi: 10.1002/cpt.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lorberbaum T., Sampson K.J., Chang J.B., Iyer V., Woosley R.L., Kass R.S., Tatonetti N.P. Coupling data mining and laboratory experiments to discover drug interactions causing QT prolongation. J. Am. Coll. Cardiol. 2016;68:1756–1764. doi: 10.1016/j.jacc.2016.07.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tatonetti N.P., Denny J.C., Murphy S.N., Fernald G.H., Krishnan G., Castro V., Yue P., Tsao P.S., Tsau P.S., Kohane I. Detecting drug interactions from adverse-event reports: interaction between paroxetine and pravastatin increases blood glucose levels. Clin. Pharmacol. Ther. 2011;90:133–142. doi: 10.1038/clpt.2011.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hastie T., Tibshirani R., Friedman J. Second edition. Springer; 2009. The Elements of Statistical Learning: Data Mining, Inference and Prediction. [Google Scholar]
- 37.Jeong H., Tombor B., Albert R., Oltvai Z.N., Barabási A.-L. The large-scale organization of metabolic networks. Nature. 2000;407:651–654. doi: 10.1038/35036627. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tab 1: Positive Examples
Tab 2: Pseudo-negative Examples.
Data Availability Statement
All source data used in the paper are publically available. We downloaded the following: (1) FAERS from openFDA; (2) Gender Specific Adverse Events List from MedDRA; (3) Severity Scores for MedDRA Adverse Events from Table S2 from Gottlieb and colleagues21; (4) Sex Differences in the gene expression from supplementary materials from Yang and Li18; and (5) a vocabulary linking MedDRA, SNOMED CT, and RxNorm from OHDSI Athena. The exact database used in this study is available on request. The published article includes all datasets generated during this study. See Tables S1 and S2 for sex risks generated by AwareDX and Tables S3 and S4 for expected sex risks curated from PharmGKB and literature. The code generated during this study are available on our GitHub repository tatonetti-lab/sex_risks.