Abstract
The mitotic kinesin superfamily protein KIF14 is essential for cytokinesis and chromosome segregation and increased KIF14 expression is related to a variety of human cancers. In this study, we investigate KIF14 expression in association with clinical variables and the role of KIF14 during tumorigenesis. We found that KIF14 is overexpressed in most primary hepatocellular carcinoma (HCC) tissues compared with the adjacent normal liver tissues and KIF14 overexpression is associated with tumor grade (P = 0.002), stage (P = 0.013) and poor survival (P < 0.001). Downregulation of KIF14 decreased the capacity of proliferation both in vitro and in vivo. Furthermore, suppression of KIF14 not only decreases cancer cell migration but also induces apoptosis of cells with inactivation of the phosphatidylinositol 3‐kinase‐Akt signaling pathway. Therefore, our current study indicates that KIF14 promotes HCC carcinogenesis and may serve as a potential therapeutic target for human HCC.
Hepatocellular carcinoma (HCC), one of the most frequent visceral neoplasms worldwide, is a common fatal malignant tumor that is characterized by a high incidence recurrence, development of resistance to chemotherapy, and low sensitivity to radiation therapy.1 Improved surgical techniques, new chemotherapeutic agents, altered modes of chemotherapy delivery and emerging molecular targets have improved life expectancy; however, the incomplete understanding of its carcinogenic mechanisms leads to difficulties in selecting targeted treatment and contributes to a low survival rate for patients with HCC.
Microtubule‐associated motor proteins have important roles in numerous cellular processes including intracellular transportation and cell division,2 with more than 45 members having been identified in mammalian cells.3 There is an indication that the abnormal expression and function of kinesins plays a key role in the development or progression of many kinds of human cancers. Better understanding of kinesin protein functions may translate into development of molecular‐targeted therapy for various human cancers.4, 5 Human KIF14 was first identified as a 6586 base‐pair kinesin family 10 cDNA clone.6 Like all kinesins, KIF14 is a microtubule‐dependent molecular motor, containing a kinesin motor domain and a forkhead‐associated domain.7 KIF14 also contains a C‐terminal citron kinase binding region and an N‐terminal protein regulating cytokinesis binding region.8, 9
KIF14 has been predicted to be a possible oncogene in the 1q region, and was found to overexpress in four breast cancer cell lines compared with the normal breast tissues.10 Furthermore, the KIF14 mRNA was also more over expressed in retinoblastoma than in retina and showed that patients with an older age at diagnosis express higher levels of KIF14 mRNA, probably leading to chromosomal/genetic instability during tumorigenesis.11 KIF14 is regulated during the cell cycle, and is correlated with mitotic progression. Furthermore, KIF14, along with the microtubule‐bundling protein PRC1 and citron kinase, with which it interacts, plays an important role in the cytokinesis, and depletion of KIF14 in HeLa cells results in the generation of binucleate cells, polyploidy and apoptosis.9 In addition, the transient KIF14 knockdown by siRNA in H1299 and HeLa cells significantly decreased the capacity of proliferation and colony formation, suggesting that KIF14 may have an important oncogenic role in cancer cells.12 Furthermore, KIF14 is also overexpressed in breast cancer and medulloblastoma cell lines, and some lung primary tumors, compared to appropriate normal tissues.10, 13 In addition, KIF14 mRNA expression is a prognostic marker in breast, lung and ovarian cancer and potential therapeutic target for ovarian cancer.7, 12, 14
In the present study, we show that 23 of 30 primary HCC tissues display 1.5‐ to 46‐fold increased KIF14 mRNA expression over matched normal samples. The KIF14 mRNA expression is associated with tumor grade and stage. Additionally, the patients with increased KIF14 levels show decreased disease free survival (DFS). The suppression of KIF14 inhibits proliferation and tumor formation both in vivo and in vitro. Furthermore, the downregulated expression KIF14 inhibits the migration and induces apoptosis via inactivation of Akt kinase in HCC cells.
Materials and Methods
Cell lines and patients
Human HCC cell line HepG2 and SMMC‐7721 were routinely maintained in DMEM (Gibco, Paisley, UK) medium supplemented with 10% FBS (Gibco) and 1% penicillin/streptomycin in a humidified atmosphere of 5% CO2 at 37°C.
From February 2005 to December 2010, 102 patients underwent surgery for HCC at the first hospital of Shijiazhuang city. Specimens from 102 patients with HCC and 30 cases of adjacent histologic normal tissues were immediately frozen in liquid nitrogen and stored at −80°C until use. Follow‐up data were obtained by reviewing the hospital records, and direct communication with the patients after hepatic resection for all 102 patients. The follow‐up period was defined from the date of surgical excision of the tumor to the date of death or last follow‐up. The DFS was defined as the length of time after hepatectomy for HCC during which a patient survives with no sign of HCC. This study was approved by the Institutional Review Board of the first hospital of Shijiazhuang city and written consent was obtained from all participants.
Plasmid construction, generation of stable cell lines
Small interfering RNA (siRNA) expressing vectors were constructed by introducing synthetic double‐stranded oligonucleotides (KIF14: 5′‐TTCCCGATCTCATTCAGTTTT‐TTCAAGAG‐AAAACTGAATGAGATCGGGAA or non‐silencing: 5′‐ TTCTCCGAACGTTTCTATCTT‐TTCAAGAG‐AAGATAGAAACGTTCGGAGAA‐3′) into pGCsi‐U6/Neo/GFP vector (Genechem, Shanghai, China). All constructs were fully sequenced.
Following the manufacturer's instructions, four independent HepG2 and SMMC‐7721 stable transfectants were screened for depleted KIF14 after 21 days selection in 800 μg/mL G418 (Calbiochem, La Jolla, CA, USA). Transfectants were routinely cultured under selection.
Tissue RNA isolation and RT‐qPCR
Total RNA was extracted with TRIZOL reagent according to the manufacturer's instructions. Five micrograms of total RNA was used to perform reverse transcription by using SuperScript II and oligo dT following the manufacturer's recommendations (Invitrogen, Carlsbad, CA, USA). The RT‐qPCR analysis was performed using the Fast SYBR Green MasterMix System (Invitrogen) according to the manufacturer's instructions. Primers for KIF14 and GAPDH were as follows: KIF14, 5′‐TGGTGAAATGGCCTGTACAAGT‐3′ (forward), 5′‐GGCAACCAGTTAACCCTTTGAG‐3′ (reverse); GAPDH, 5′‐GAAATCCCATCACCATCTTCCAGG‐3′ (forward), 5′‐ GAGCCCCAGCCTTCTCCATG‐3′ (reverse). The PCR conditions were as follows: 95°C for 20 s; 95°C for 3 min; 60°C for 30 s; for 40 cycles by using ABI 7500 (Applied Biosystems, Foster City, CA, USA). The relative quantification was given by the CT values, determined by triplicate reactions for all of the samples for KIF14 and GAPDH. The triplicate CT values of detectable gene were averaged, and the CT value of GAPDH was subtracted to obtain ΔCT. The relative mRNA expression level of KFI14 was determined as.16
Evaluation copy number by real‐time quantitative PCR
DNA was extracted from the paraffin‐embedded tissues with the QIAamp DNA mini kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. KIF14 copy number was performed by Applied Biosystems using TaqMan Gene Copy Number Assays. One set of primers for ASXL2 (Applied Biosystems) was used as the reference gene as previously described by Reis–Filho et al.15 The ratio of KIF14 to ASXL2 copy number obtained in each sample as the KIF14 copy number for each specimen.
Western blot analysis
A quantity of 30 μg of lysates per sample was separated by SDS‐PAGE using 10% polyacrylamide gels and transferred to PVDF membrane, which was subsequently incubated with rabbit polyclonal antibody to KIF14 (1:2000; Abcam, Cambridge, MA, USA), anti‐Akt (1:1000, Cell Signaling, Beverly, MA, USA), anti‐phospho‐Akt (Ser473, 1:1000; Cell Signaling) for 4°C overnight, and corresponding were immunodetected by incubation with HRP‐linked anti rabbit secondary antibody (1:3000; Sigma, St Louis, MO, USA) using an ECL detection kit (Pierce Biotechnology, Rockford, IL, USA). Rabbit polyclonal antibody to β‐actin (1:5000; Sigma) was used as gel loading control.
Cell proliferation, cell migration and apoptosis assays
Both MTT and colony formation assays were used to observe and compare cell proliferation ability. Briefly, 1 × 103 cells in 100 μL culture medium were plated into a well of 96‐well plates. After culturing cells for an appropriate time, 10 μL of 5 mg/mL MTT was added into each well and continued to culture for 4 h. Then, the cell culture medium was replaced by 100 μL of DMSO. Thirty minutes after dimethyl sulfoxide addition, the plates were placed on a microplate autoreader (Bio‐Rad, Hercules, CA, USA). Optical density was read at 570 nm wavelength and cell growth curves were determined according to the optical density value. For colony formation assay, cell suspensions were seeded into six‐well plates, 300 cells per well. Cells were incubated for 2 weeks then fixed in methyl hydrate for 10 min. Colonies were then stained and counted using an optical microscope.
The ability of HCC cells and transfectants to invade was examined through Matrigel‐coated transwell inserts (8 μm pore size, BD Biosciences, San Jose, CA, USA). In brief, 1 × 105 cells were cultured in the upper chamber with serum‐free medium. The lower chamber contained complete medium (10% FBS). After incubation at 5% at 37°C, 5% CO2 for 24 h, cells adherent to the top surface of the membrane were removed with a cotton applicator, whereas cells that migrated to the bottom surface were fixed with 70% methanol and stained with crystal violet. The migrated cells on the bottom surface of the membrane were photographed and counted on an inverted microscope.
Apoptosis assays were performed as described.17 In brief, cells suspended in complete medium were seeded in polyhydroxyethylmethacrylate (Sigma) coated 35 mm wells and cultured for 20 h. After incubation, cells were harvested, stained with Annexin V‐FITC apoptosis detection kit (BD Biosciences) and then analyzed by flow cytometry.
Cell cycle analysis
Cell cycle phase distribution was assayed by flow cytometry.18 MultiCycle software program from Phoenix Flow Systems (San Diego, CA, USA) was used to deconvolute the cellular DNA histograms and quantify the percentage of cells in the G1, S, and G2/M phases.
Xenografts assays in nude mice
Cells stably expressing pGCsi‐U6/Neo/GFP‐non silencing (designated as control RNAi), cells stably expressing pGCsi‐U6/Neo/GFP‐KIF14 (designated as KIF14 RNAi) or parental cells (1 × 106) were inoculated into the right subaxillary region of athymic nude mice (n = 5, per group). Tumor growth was recorded once a week with a caliper‐like instrument. Tumor volume was calculated according to the formula volume = (width2 × length)/2. Seven weeks after inoculation, mice were killed, and the final volume and weight of tumor tissues were determined.
Statistical analysis
Survival analysis was carried out according to the methods of Kaplan and Meier. The relationship between KIF14 and various clinicopathological variables was analyzed by the chi‐squared test. All calculations were performed with the spss for Windows statistical software package (SPSS Inc., Chicago, IL, USA). Results of in vitro and in vivo experiments were depicted as mean ± SD. Student's two‐sided t‐test was used to compare values of test and control samples. The level of significance was set to P < 0.05.
Results
KIF14 is highly expressed in HCC
To determine the expression level of KIF14 in primary HCC, we performed RT‐qPCR to detect the KIF14 mRNA expression in primary HCC tissues and the paired normal liver. Our results indicated that KIF14 mRNA expression was upregulated in 23 of 30 primary HCC tissues compared with the adjacent normal tissues (1.5‐ to 46‐fold) (Fig. 1a).
Figure 1.
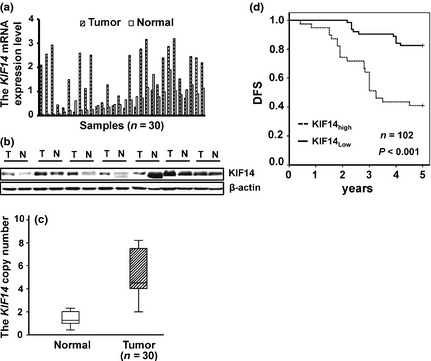
KIF14 is upregulated in human hepatocellular carcinoma (HCC) and predicts poor outcome. (a) Expression of KIF14 mRNA in HCC tissues and their adjacent normal tissues by RT‐qPCR. (b) Expression of KIF14 protein in HCC tissues and their adjacent normal tissues by western blot. (c) Expression of KIF14 copy number in HCC tissues and their adjacent normal tissues by qPCR. (d) The Kaplan–Meier plot of 5‐year disease free survival (DFS)curves stratified by KIF14 mRNA expression in 102 HCC patients.
To further investigate if KIF14 protein levels were also upregulated in primary HCC tissues, we examined KIF14 expression in eight matched samples of human HCC (T) and adjacent histologic normal tissues (N) using western blot with KIF14. As shown in Figure 1(b), KIF14 protein levels were upregulated in six of eight malignant tumor samples. Kim et al. reported that copy number gains on 1q induce overexpression of KIF14, subsequently influencing hepatocarcinogenesis. To test whether KIF14 expression was elevated in DNA‐dependent manner, we detected the KIF14 copy number by qPCR. The result showed that the DNA copy number of KIF14 was also upregulated in HCC tissues than the adjacent normal tissues (Fig. 1c). Together, these results indicate that KIF14 is overexpressed in most human HCC.
Relationship between KIF14 mRNA expression and clinicopathological factors
Because of the distribution of KIF14 mRNA expression in all primary HCC tissues did not accord with normal distribution, ROC curve was made based on the relative level of KIF14 mRNA and DFS status of HCC patients. Of 1.5 × 10−3 defined as cut‐off value to group all the patients into a low KIF14 expression group (KIF14low) and a high KIF14 expression group (KIF14high).
To evaluate whether KIF14 mRNA expression was associated with clinicopathological factors of patients with HCC, we correlated RT‐qPCRs KIF14 results with age, size, grade, stage, portal vein tumor thrombus, and HBV infection (Table 1). As a result, we found that high levels of KIF14 were correlated with tumor grade (P = 0.002) and stage (P = 0.013). However, no significant differences were found between the mRNA levels of KIF14 and age, tumor size, portal vein tumor thrombus, and HBV infection. Furthermore, the Kaplan–Meier plot of 5‐year DFS curves stratified by KIF14 mRNA expression was shown in Figure 1(d). A significant relationship was found between KIF14 mRNA expression and 5‐year DFS (P < 0.001). These results suggest that high expression level of KIF14 significantly related to poor outcome in patients with HCC.
Table 1.
Relationship between clinicopathological factors and KIF14 expression in 102 hepatocellular carcinoma (HCC) patients
| Clinicopathological factors | KIF14 high | KIF14 low | P |
|---|---|---|---|
| Age | |||
| < 55 | 28 | 38 | 0.289 |
| ≥ 55 | 11 | 25 | |
| Size (cm) | |||
| ≤ 2 | 14 | 34 | 0.076 |
| > 2 | 25 | 29 | |
| Stage | |||
| I–II | 22 | 50 | 0.013* |
| III | 17 | 13 | |
| Grade | |||
| I–II | 22 | 53 | 0.002* |
| III | 17 | 10 | |
| Portal vein tumor thrombus | |||
| Negative | 24 | 34 | 0.851 |
| Positive | 7 | 11 | |
| Missing | 8 | 18 | |
| HBV infection | |||
| Negative | 16 | 26 | 0.921 |
| Positive | 18 | 28 | |
| Missing | 5 | 9 | |
Suppression of KIF14 expression in HCC cells inhibits cell proliferation and migration in vitro
For an in‐depth understanding of the biological functions of KIF14 and its role in carcinogenesis, we ablated expression of KIF14 in HCC cell line, HepG2 and SMMC‐7721, using mammalian vector‐based RNA interference. A pair of oligonucleotides targeting 1701–1721 bp in the coding region of the KIF14 gene was synthesized and inserted into the pGCsi‐U6/Neo/GFP siRNA expression vector. Hepatocellular carcinoma cells were transfected with KIF14 siRNA expression vector or control. After G418 selection, stable transfectants were obtained. We selected two KIF14 RNAi HepG2 clones (RNAi‐15 and RNAi‐22) and two KIF14 RNAi SMMC‐7721 clones (RNAi‐3 and RNAi‐11) as well as their control shRNA clones for further analyses. RT‐qPCR and western blot analysis indicated that expression levels of KIF14 mRNA and protein were downregulated in KIF14 shRNA transfected cells when compared with those in control cells or parental cells (Fig. 2a,b). Cell cycle profile examined by fluorescence activated cell sorting analysis indicated more G2/M cells in KIF14 RNAi cells compared with control shRNA cells (Fig. 2c,d). Given the fact that KIF14 was stably silenced in these cells, we proposed that these cells may have prolonged G2/M phase so that cell proliferation might be inhibited.
Figure 2.
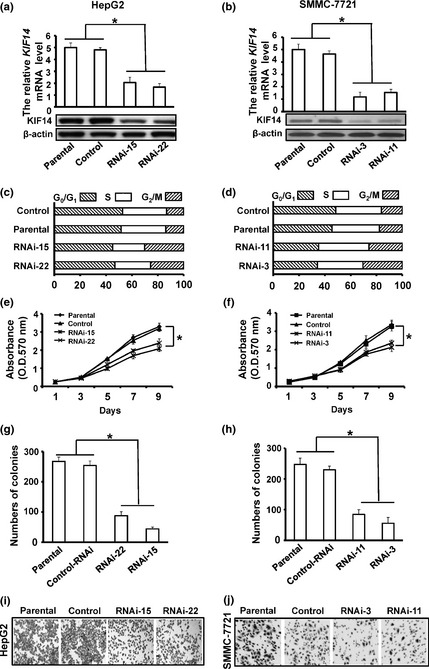
Suppression of KIF14 inhibits hepatocellular carcinoma (HCC)cells proliferation and migration in vitro. (a, b) HepG2 and SMMC‐7721 cells stably expressing KIF14 siRNA (RNAi‐15 and RNAi‐22; RNAi‐3 and RNAi‐11) or non‐sense siRNA (control) were lysed and cells were analyzed for KIF14 by RT‐qPCR and western blot, respectively. (c, d) Cell cycle distribution of KIF14 siRNA and controls serum starved for 12 h and then re‐stimulated with 10% FBS‐containing medium for the indicated time. (e, f) Cells were cultured in 96‐well plates and analyzed by MTT assay. Cell proliferation curves were shown in 9 days. Three independent experiments were conducted. (g, h) Cells were cultured in 6‐well plates and analyzed by colony formation assay. After 2 weeks, cells were stained and counted. Three independent experiments were conducted. (i, j) Cell migration motility was examined in 24‐well plates with Transwell chambers. After addition of 24 h incubation, cells adherent to the upper surface of the filter were removed using a cotton applicator. Cells migrated into the lower chambers were fixed with 70% methanol, stained with crystal violet. *P < 0.05.
Cell proliferation analyses showed that suppression of KIF14 in HepG2 and SMMC‐7721 significantly inhibited cell growth. KIF14 RNAi cells grew much slower than parental or control cells in MTT assay (Fig. 2e,f). Colony formation assay also revealed that KIF14 RNAi cells formed fewer colonies than parental or control cells (Fig. 2g,h). To test whether suppression of KIF14 affect the cancer cell migration, we performed cell migration assay using polycarbonate membrane transwell chambers in KIF14 RNAi HepG2 cells (RNAi‐15 and RNAi‐22) and KIF14 RNAi SMMC‐7721 cells (RNAi‐3 and RNAi‐11) as well as their controls. As shown in Figure 2(i,j), KIF14 RNAi cells displayed much lower transwell migratory rates than control cells. Thus, these results indicate that suppression of KIF14 expression not only affects cell division but also cell migration thereby preventing tumorigenesis and tumor development.
Suppression of KIF14 expression in HCC cells inhibits tumor formation in vivo
We next performed xenograft assays in nude mice to determine if long‐term suppression of KIF14 in HepG2 cells was able to affect tumor growth in vivo. KIF14 RNAi, control RNAi or parental cells (1 × 106) were injected into the right subaxillary region of nude mice (n = 5 per group). Tumor formation and volume of the tumor in each mouse were examined, measured and recorded for 7 weeks and tumor growth curves were determined. As shown in Figure 3(a), tumor formation and tumor growth of KIF14 RNAi cells were significantly reduced in nude mice when compared with those of control RNAi or parental cells. Seven weeks after inoculation, mice were killed and tumor xenografts were obtained. Consistently, weight measurement showed that tumor xenografts from control or parental cells were much heavier than those from KIF14 RNAi cells (Fig. 3b).
Figure 3.
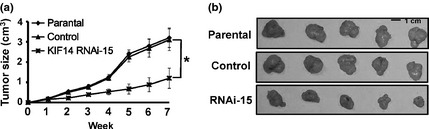
Downregulation of KIF14 in HepG2 cells decreases tumor formation in vivo. (a, b) 1 × 106 cells were injected into the right subaxillary of athymic nude mice and tumor growth was recorded with a caliper‐like instrument (n = 5, per group). Tumor volumes were calculated according to the formula volume 5 (width 2 × length)/2 (a). After 7 weeks, all mice were killed and final tumor tissues were isolated and photographed. Representative tumor tissues were shown (b). *P < 0.05.
Together, these results indicate that suppression of KIF14 expression in human HCC cells inhibits tumor growth in vivo.
Suppression of KIF14 promotes apoptosis via inactivation of Akt kinase
Finally, we sought to explore the ultimate consequence of long‐term suppression of KIF14 in human HCC cells. Since suppression of KIF14 expression inhibited cancer cell growth, we monitored cell survival of RNAi‐15 and RNAi‐22 as well as control RNAi and parental cells after detachment and replanting in dishes coated with polyhydroxyethylmethacrylate. Cell apoptosis assay revealed that, after 20 h, KIF14 RNAi cells displayed significantly increased apoptosis compared with control siRNA‐treated or parental cells (Fig. 4a). Thus, these results indicate that suppression of KIF14 in HepG2 cells decrease growth and induce cell apoptosis.
Figure 4.
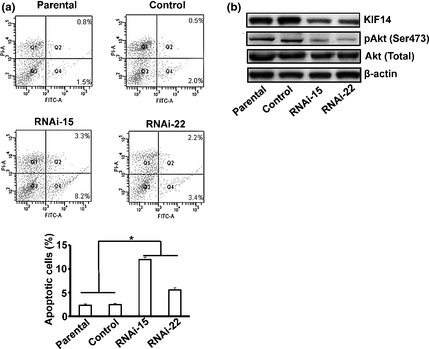
Downregulation of KIF14 in HepG2 Cells promotes cell apoptosis. (a) HepG2 cells stably expressing KIF14 siRNA (RNAi‐15 and RNAi‐22) or nonsense siRNA were suspended in the polyhydroxyethylmethacrylate‐coated wells and cultured in a humidified incubator. After 18 h of growth in suspension, cells were harvested and measured for apoptosis using Annexin V‐FITC apoptosis detection kit followed by flow cytometry analysis. The percentage of apoptotic cells from three independent assays is shown in lower panel. (b) Cells were lysed and analyzed by western blot. *P < 0.05.
To further explore the mechanisms by which KIF14 depletion induces apoptosis of HepG2 cells, we examined the potential effects of KIF14 depletion on signaling pathways associated with apoptosis by western blot. As shown in Figure 4(b), suppression of KIF14 expression resulted in downregulation of Akt Ser473 phosphorylation. These results indicate that suppression of KFI14 disrupts cell survival via suppression of the phosphatidylinositol 3‐kinase‐Akt signaling pathway in HepG2 cells.
Discussion
In the present study, we investigated the involvement of the kinesin motor protein, KIF14, in human HCC. Our results indicate that KIF14 is overexpressed in human HCC and such overexpression is correlated significantly with tumors in advanced grade, metastasis and decreased DFS rate, suggesting that KIF14 plays an important role in human HCC carcinogenesis. Consistent with these findings, we show that ablation of KIF14 expression in human HCC cells inhibits cell proliferation in vitro and tumor formation in vivo. Furthermore, the suppression of KIF14 decreases the cell migration and promotes apoptosis via inactivation of Akt kinase.
KIF14 is essential for cytokinesis and chromosome segregation.8 Several reports have been showed that the increased KIF14 expression is related to a variety of different cancers tumorigenesis, including HCC.7, 11, 12, 13, 14, 19, 20, 21 In this study, although we observed that KIF14 is upregulation in most primary HCC tissues compared to the paired normal tissues, KIF14 was overexpressed in 5 of 30 non‐tumor tissues. These conflicting data suggested that KIF14 may have a different role in HCC carcinogenesis. In fact, previous studies indicate that increased KIF14 expression seems to be associated with more advanced, aggressive tumors and its expression increases pronouncedly with grade, fraction of tumor‐positive nodes removed and percent invasive cells in patients with breast cancer.14 Because of the limited number of samples, Kim et al.19 did not observe any significant association between KIF14 expression levels and invasiveness, tumor grade, size and survival However, we observed an association between expression of KIF14 level and grade, stage or survival in HCC. Moreover, KIF14 has been reported to be an independent prognostic marker and potential therapeutic target in different cancers.7, 12, 21 Further large‐scale studies will be required to validate this possibility in HCC.
We next determined the potential role of KIF14 in tumor formation and progression. Our results indicate that suppression of KIF14 expression by RNAi in HCC cells inhibits cell proliferation in vitro and tumor formation in vivo. We show here that downregulation of KIF14 expression by siRNA moderately decreases proliferation but strongly decreases the ability of HepG2 and SMMC‐7721 cells to form colonies. These in vitro results suggest that downregulation of KIF14 is not only cytotoxic, but also affects the capability of independent growth of cancer cells. This makes KIF14 a potential target for cancer therapy. The work of Carleton et al.9 showed that knockdown of KIF14 by siRNA in HeLa cells induced binucleation and apoptosis. Although, in our work, stable depletion of HepG2 cells induced significant apoptosis, stable deletion of KIF14 by shRNA did not induce binucleation in HepG2 cells (data not shown). This discrepancy could be explained in that the type (stable versus transient) and level of KIF14 knockdown may produce different cellular phenotypes, which could point to unique functions for KIF14 in different cell lines. Furthermore, our stable cones may be sensitive to multinucleation and induce apoptosis before any such phenotype can be characterized. Furthermore, our findings indicate that suppression of KIF14 not only inhibits cell division, but also decreases cell migration.
Previous studies demonstrated that KIF14 interacts with citron kinase, localizing this Rho effector kinase to the central spindle8 and citron kinase was a prognostic marker for DFS and the KIF14 was negatively related to citron kinase expression in lung cancer.22 These findings suggest that the expressions of KIF14 and citron kinase may be coordinately controlled to deregulate the central spindle and final phase of cytokinesis in HCC cells. It will be required for evaluating if KIF14 expression is predictive for response to treatment with spindle poisons such as Vinca Alkaloids and taxanes in future studies.
Apoptosis is a process of programmed cell death. Although normal cells use apoptosis to prevent abnormal cell proliferation, cancer cells often overcome the process and migrate to new sites for their abnormal growth. Therefore, the increased migration ability and resistance of apoptosis take an advantage for metastasis. Our results indicated that suppression of KIF14 in HCC cells decreases the growth and induces apoptosis. Although the exact mechanism by which KIF14 is directly involved in the regulation of apoptosis remains to be clarified, our results show that the cell proliferation/survival of phosphatidylinositol 3‐kinase‐Akt signaling pathway might be inhibited in depleted KIF14 expression cells, which is similar to the role of KIF18A during breast carcinogenesis.23 Further study is required to determine how KIF14 activates the phosphatidylinositol 3‐kinase‐Akt signaling pathway.
Although the exact function of KIF14 remains unclear, its role in aligning chromosomes at metaphase3 suggests that it could play an important role in the formation of aneuploidy, and its overexpression may function to accelerate the cell cycle during cancer progression. Whatever its mode of action, its specific overexpression in HCC and association with grade and survival, and inhibition of cell proliferation in vitro and tumor formation in vivo by suppression of KIF14 expression in HCC cells demonstrate its importance as a putative oncogene, therapeutic target and prognostic indicator.
Disclosure Statement
The authors have no conflict of interest.
(Cancer Sci 2013; 104: 552–557)
References
- 1. Thorgeirsson SS, Grisham JW. Molecular pathogenesis of human hepatocellular carcinoma. Nat Genet 2002; 31: 339–46. [DOI] [PubMed] [Google Scholar]
- 2. Miki H, Okada Y, Hirokawa N. Analysis of the kinesin superfamily: insights into structure and function. Trends Cell Biol 2005; 15: 467–76. [DOI] [PubMed] [Google Scholar]
- 3. Zhu C, Zhao J, Bibikova M et al Functional analysis of human microtubule‐based motor proteins, the kinesins and dyneins, in mitosis/cytokinesis using RNA interference. Mol Biol Cell 2005; 16: 3187–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Huszar D, Theoclitou ME, Skolnik J, Herbst R. Kinesin motor proteins as targets for cancer therapy. Cancer Metastasis Rev 2009; 28: 197–208. [DOI] [PubMed] [Google Scholar]
- 5. Yu Y, Feng YM. The role of kinesin family proteins in tumorigenesis and progression: potential biomarkers and molecular targets for cancer therapy. Cancer 2010; 116: 5150–60. [DOI] [PubMed] [Google Scholar]
- 6. Nomura N, Nagase T, Miyajima N et al Prediction of the coding sequences of unidentified human genes. II. The coding sequences of 40 new genes (KIAA0041‐KIAA0080) deduced by analysis of cDNA clones from human cell line KG‐1 (supplement). DNA Res 1994; 1: 251–62. [DOI] [PubMed] [Google Scholar]
- 7. Theriault BL, Pajovic S, Bernardini MQ, Shaw PA, Gallie BL. Kinesin family member 14: an independent prognostic marker and potential therapeutic target for ovarian cancer. Int J Cancer 2012; 130: 1844–54. [DOI] [PubMed] [Google Scholar]
- 8. Gruneberg U, Neef R, Li X et al KIF14 and citron kinase act together to promote efficient cytokinesis. J Cell Biol 2006; 172: 363–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Carleton M, Mao M, Biery M et al RNA interference‐mediated silencing of mitotic kinesin KIF14 disrupts cell cycle progression and induces cytokinesis failure. Mol Cell Biol 2006; 26: 3853–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Corson TW, Huang A, Tsao MS, Gallie BL. KIF14 is a candidate oncogene in the 1q minimal region of genomic gain in multiple cancers. Oncogene 2005; 24: 4741–53. [DOI] [PubMed] [Google Scholar]
- 11. Madhavan J, Coral K, Mallikarjuna K et al High expression of KIF14 in retinoblastoma: association with older age at diagnosis. Invest Ophthalmol Vis Sci 2007; 48: 4901–6. [DOI] [PubMed] [Google Scholar]
- 12. Corson TW, Zhu CQ, Lau SK, Shepherd FA, Tsao MS, Gallie BL. KIF14 messenger RNA expression is independently prognostic for outcome in lung cancer. Clin Cancer Res 2007; 13: 3229–34. [DOI] [PubMed] [Google Scholar]
- 13. Madhavan J, Mitra M, Mallikarjuna K et al KIF14 and E2F3 mRNA expression in human retinoblastoma and its phenotype association. Mol Vis 2009; 15: 235–40. [PMC free article] [PubMed] [Google Scholar]
- 14. Corson TW, Gallie BL. KIF14 mRNA expression is a predictor of grade and outcome in breast cancer. Int J Cancer 2006; 119: 1088–94. [DOI] [PubMed] [Google Scholar]
- 15. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real‐time quantitative PCR and the 2(‐Delta Delta C(T)) Method. Methods 2001; 25: 402–8. [DOI] [PubMed] [Google Scholar]
- 16. Reis‐Filho JS, Drury S, Lambros MB et al ESR1 gene amplification in breast cancer: a common phenomenon? Nat Genet 2008; 40: 809–10; author reply 810‐812. [DOI] [PubMed] [Google Scholar]
- 17. Reginato MJ, Mills KR, Paulus JK et al Integrins and EGFR coordinately regulate the pro‐apoptotic protein Bim to prevent anoikis. Nat Cell Biol 2003; 5: 733–40. [DOI] [PubMed] [Google Scholar]
- 18. Darzynkiewicz Z, Bedner E, Smolewski P. Flow cytometry in analysis of cell cycle and apoptosis. Semin Hematol 2001; 38: 179–93. [DOI] [PubMed] [Google Scholar]
- 19. Kim TM, Yim SH, Shin SH et al Clinical implication of recurrent copy number alterations in hepatocellular carcinoma and putative oncogenes in recurrent gains on 1q. Int J Cancer 2008; 123: 2808–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huang Q, Lin B, Liu H et al RNA‐Seq analyses generate comprehensive transcriptomic landscape and reveal complex transcript patterns in hepatocellular carcinoma. PLoS ONE 2011; 6: e26168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang Q, Wang L, Li D et al Kinesin family member 14 is a candidate prognostic marker for outcome of glioma patients. Cancer Epidemiol 2013; 37: 79–84. [DOI] [PubMed] [Google Scholar]
- 22. Blackhall FH, Wigle DA, Jurisica I et al Validating the prognostic value of marker genes derived from a non‐small cell lung cancer microarray study. Lung Cancer 2004; 46: 197–204. [DOI] [PubMed] [Google Scholar]
- 23. Zhang C, Zhu C, Chen H et al Kif18A is involved in human breast carcinogenesis. Carcinogenesis 2010; 31: 1676–84. [DOI] [PubMed] [Google Scholar]


