Abstract
Circular RNAs (circRNAs), a subclass of noncoding RNAs, are reportedly involved in the progression of various diseases. However, the exact role of circRIMS1, also termed hsa_circ_0132246, in human bladder cancer remains unknown. By performing RNA sequencing comparing bladder cell lines and normal uroepithelial cells, circRIMS1 was selected as a research object. We further verified by qRT-PCR that circRIMS1 is upregulated in both bladder cancer tissue and cell lines. Proliferation, colony-formation, Transwell migration, invasion, apoptosis, western blotting, and in vivo experiments were utilized to clarify the roles of circRIMS1, microRNA (miR)-433-3p, and cell cycle and apoptosis regulator 1 (CCAR1). For mechanistic investigation, RNA pulldown, fluorescence in situ hybridization (FISH), and luciferase reporter assay confirmed the binding of circRIMS1 with miR-433-3p. Inhibition of circRIMS1 suppressed the proliferation, migration, and invasion of bladder cancer cells both in vitro and in vivo. Moreover, the circRIMS1/miR-433-3p/CCAR1 regulatory axis was confirmed to be responsible for the biological functions of circRIMS1. Taken together, our research demonstrated that circRIMS1 promotes tumor growth, migration, and invasion through the miR-433-3p/CCAR1 regulatory axis, representing a potential therapeutic target and biomarker in bladder cancer.
Keywords: circRIMS1, bladder cancer, miR-433-3p, CCAR1, progression
Graphical Abstract
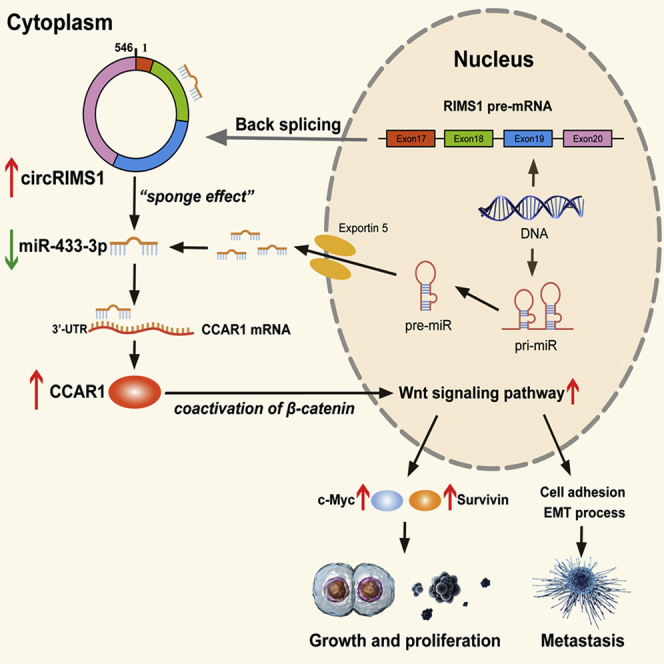
Jin and colleagues explored the expression and biological functions of circular RNA circRIMS1 in bladder cancer. Mechanically, a circRIMS1/miR-433-3p/CCAR1 regulatory axis was verified in vitro and in vivo. The study indicated that circRIMS1 might be a potential therapeutic target and biomarker in bladder cancer.
Introduction
Bladder cancer is the ninth most common cancer and ranks 13th in annual mortality among cancers worldwide. Depending on the depth of tumor infiltration, bladder cancer is classified into two types: nonmuscle invasive (75%) and muscle-invasive bladder cancer (25%).1,2 Due to distant metastasis, the 5-year overall survival rate of muscle-invasive bladder cancer patients is only 60% after surgery and chemotherapy.3 Thus, it is meaningful to elucidate the underlying molecular mechanisms to develop precise anticancer strategies for bladder cancer.
Circular RNA (circRNA) is a brand new subclass of endogenous noncoding RNAs that is characterized by a closed covalent loop structure with a specific joining site between the 3′ and 5′ ends formed by back-splicing.4, 5, 6 circRNAs exhibit high sequence conservation, abundant expression, and cell- or tissue-specific expression.7,8 Emerging findings have reported that circRNAs modulate various biological processes, whereby they act by sponging microRNAs (miRNAs), mediating RNA-protein interactions, or modulating expression of parental genes.4,9, 10, 11 In addition, numerous circRNAs may represent prognostic and diagnostic biomarkers in human diseases, especially cancer.11, 12, 13, 14 Nevertheless, further research is needed to determine the roles and functions of circRNAs in bladder cancer.
miRNAs regulate gene expression by binding directly to specific mRNA sequences.15 In recent decades, numerous studies have reported that miRNAs play vital roles in various biological behaviors and processes, making them potential molecular targets for cancer therapeutics.16,17 Some researchers have demonstrated aberrant expression of miRNAs in cancer, as well as their various influences on tumorigenesis, development, and progression in bladder cancer.18,19
Cell cycle and apoptosis regulator 1 (CCAR1/CARP-1) plays a vital role in apoptosis signaling through the retinoid CD437 and in chemotherapy in human breast cancer.20 CCAR1 is also necessary for human breast cell growth and gene expression, relying on an estrogen-dependent pathway.21 Moreover, CCAR1 enhances antitumor effects induced by the suppression of the epidermal growth factor receptor (EGFR).22 Recent studies reported that CCAR1 interacts with β-catenin to activate the Wnt signaling pathway, enhancing transcriptional activation of downstream genes in gastric and colon cancer cells,23,24 which indicated that CCAR1 was a coactivator of β-catenin and a key regulator in the Wnt pathway.
In the present study, we identified and named a circRNA derived from the RIMS1 gene, termed circRIMS1. circRIMS1 was found to be aberrantly upregulated in bladder cancer. We hypothesized that circRIMS1 might play a vital role in the progression of bladder cancer by sponging microRNA (miR)-433-3p to influence expression of CCAR1 and tumor biological behaviors. The underlying mechanisms and functions of circRIMS1 were explored, and circRIMS1 may represent a novel biomarker and a potential therapeutic target for clinical treatment in bladder cancer.
Results
circRIMS1 (hsa_circ_0132246) Is Upregulated in Bladder Cancer
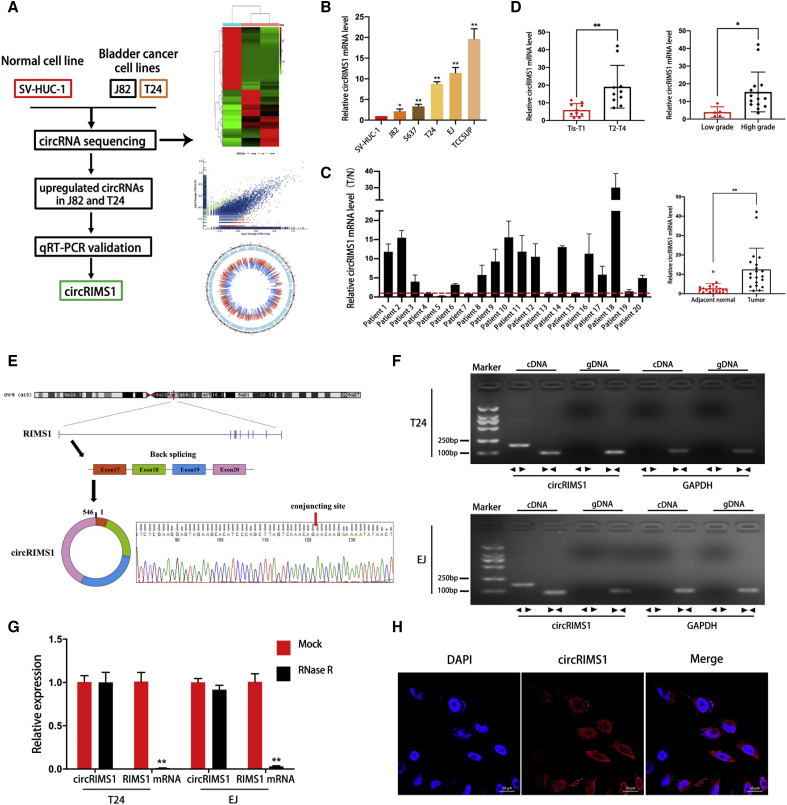
To investigate the roles of circRNAs in the progression and metastasis of bladder cancer, circRNA expression profiles between a normal urothelial cell line (SV-HUC-1) and two bladder cancer cell lines (J82 and T24) were examined by circRNA sequencing (Figure 1A; each sample was mixed with three replicates, GEO: GSE150142). Among significantly upregulated circRNAs in both J82 and T24 compared to SV-HUC-1 cells, circRIMS1 (hsa_circ_0132246) was selected as a candidate study object due to the repeatability of its upregulation in other bladder cancer cell lines. Expression levels of circRIMS1 were markedly higher in 5 bladder cancer cell lines compared to SV-HUC-1 cells (Figure 1B). To further investigate the correlation between circRIMS1 and bladder cancer, we performed qRT-PCR to identify differential expression of circRIMS1 in bladder cancer tissues and adjacent normal tissues (n = 20). As shown in Figure 1C, circRIMS1 was aberrantly expressed in tumor tissue. Meanwhile, we also found that circRIMS1 was associated with the pathological and histological characteristics in our cohort (Figure 1D; Table S3). Next, we performed Sanger sequencing to verify head-to-tail splicing with the existing sequence in circBase in the RT-PCR product of circRIMS1 (Figure 1E). Actually, both trans-splicing and genomic rearrangements lead to head-to-tail splicing.25 Therefore, we employed convergent primers of RIMS1 mRNA and divergent primers of circRIMS1 for amplification to rule out these possibilities. We performed nucleic acid electrophoresis detection with cDNA and genomic DNA (gDNA) acquired from T24 and EJ cells separately. Results in Figure 1F revealed that no amplified products were detected in the gDNA fraction, whereas circRIMS1 was detected in cDNA only. circRNAs are characteristically resistant to digestion by RNase R due to their unique and stable structure.25 In an experiment to confirm the stability of circRIMS1, the linear forms of RIMS1 were remarkably depleted by RNase R, but circRIMS1 could tolerate the digestion of RNase R (Figure 1G). Last, in the EJ cell line, RNA fluorescence in situ hybridization (FISH) results demonstrated that circRIMS1 was primarily localized in the cytoplasm (Figure 1H).
Figure 1.
circRNA Expression Profiles and Validation of circRIMS1 in Bladder Cancer Tissues and Cells
(A) A flow diagram of circRNA sequencing in SV-HUC-1, J82, and T24 cell lines. (B) Levels of circRIMS1 in SV-HUC-1 cells and bladder cancer cells (J82, 5637, T24, EJ, and TCCSUP) were determined by qRT-PCR. circRIMS1 was upregulated in bladder cancer cell lines. (C) Analysis of circRIMS1 expression fold change in tumor tissues and adjacent normal tissues of bladder cancer patients (n = 20). (D) Expression of circRIMS1 in our bladder cancer patients during different pathological stages and histological grades. (E) Schematic illustration indicating the circularization of exons 17, 18, 19, and 20 of RIMS1, forming circRIMS1. RT-PCR was performed to verify the existence of circRIMS1. Sanger sequencing further proved the head-to-tail splice site in circRIMS1, and the specific junction is indicated by the red arrow. (F) circRIMS1 was detected in T24 and EJ cell lines by RT-PCR. Divergent primers could only amplify circRIMS1 from cDNA. GAPDH was used as a negative control. (G) circRIMS1 and linear RIMS1 mRNA levels in T24 and EJ cells were determined by qRT-PCR, with or without RNase R. (H) RNA FISH for circRIMS1 was detected in EJ cells. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI; blue), and circRIMS1 appeared red. Results indicated that circRIMS1 was primarily localized in the cytoplasm. Scale bars, 20 μm. Data are presented as the mean ± SD of three independent experiments. ∗p < 0.05, ∗∗p < 0.01 versus control group.
circRIMS1 Enhances the Proliferation, Migration, and Invasion of Bladder Cancer Cells
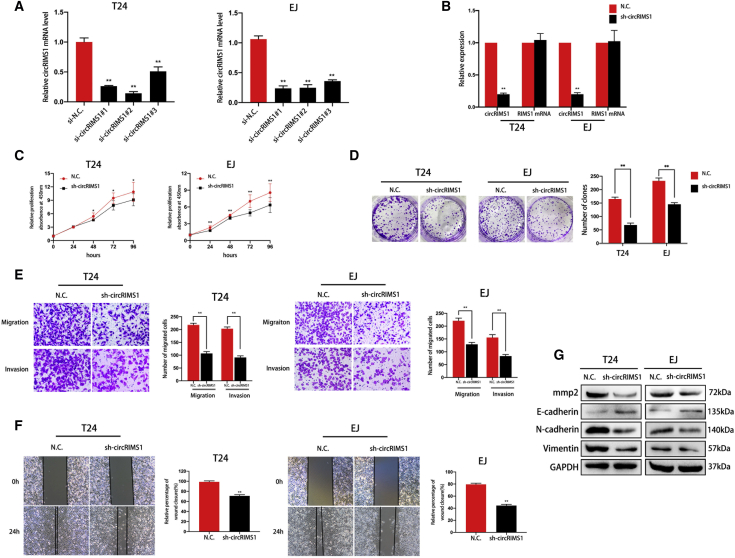
To verify the biological functions of circRIMS1 in bladder cancer cells, we first determined the knockdown efficiency of small interfering RNAs (siRNAs) and used si-circRIMS1#2 in the following studies (Figure 2A). circRIMS1-specific small hairpin RNAs (shRNAs) were generated, according to the sequence of si-circRIMS1#2, to stably knock down the expression of circRIMS1. Results showed that circRIMS1 expression was significantly reduced by sh-circRIMS1, but expression of the linear RIMS1 mRNA was unchanged (Figure 2B). A subsequent Cell Counting Kit 8 (CCK-8) assay indicated that circRIMS1 played a key role in cell proliferation (Figure 2C). In addition, colony-formation assays showed that the colony-forming abilities of circRIMS1-knockdown cells were reduced compared to control cells (Figure 2D). Transwell migration and invasion assays further confirmed that circRIMS1 knockdown markedly suppressed bladder cancer cell migration and invasion (Figure 2E). The subsequent wound-healing assay showed a similar trend in T24 and EJ cells (Figure 2F). Epithelial-to-mesenchymal transition (EMT), a vital process for both cell migration and cancer metastasis, is aberrantly activated in various cancers.26 Western blot results indicated that inhibition of circRIMS1 enhanced E-cadherin expression but suppressed the expressions of N-cadherin and vimentin in both cell lines, suggesting inhibition of the EMT process (Figure 2G). Meanwhile, matrix metalloproteases (MMPs) are necessary for migration and invasion of tumor cells.27 The expression of MMP2 was also decreased in response to circRIMS1 inhibition (Figure 2G). In summary, our results revealed that circRIMS1 is a key regulator in bladder cancer progression and metastasis in vitro.
Figure 2.
circRIMS1 Promotes Proliferation, Migration, and Invasion of Bladder Cancer Cells
(A) Efficiency of siRNAs on circRIMS1 in T24 and EJ detected by qRT-PCR. (B) Expression levels of circRIMS1 and RIMS1 mRNA in T24 and EJ cells transfected with sh-circRIMS1 or negative control vector were determined by qRT-PCR. (C) CCK-8 assay was performed to determine cell viability in sh-circRIMS1 cells. (D) Cell proliferation was evaluated by a colony-formation assay in circRIMS1-knockdown T24 and EJ cells. (E and F) Transwell migration and invasion assays (E) and wound-healing assay (F) indicated the biological effects of circRIMS1 knockdown in T24 and EJ cells. (G) Expression levels of MMP2 and EMT markers (E-cadherin, N-cadherin, and vimentin) detected at the protein level in circRIMS1-knockdown T24 and EJ cells. ∗p < 0.05, ∗∗p < 0.01 versus control group.
circRIMS1 Serves as a Sponge for miR-433-3p in Bladder Cancer
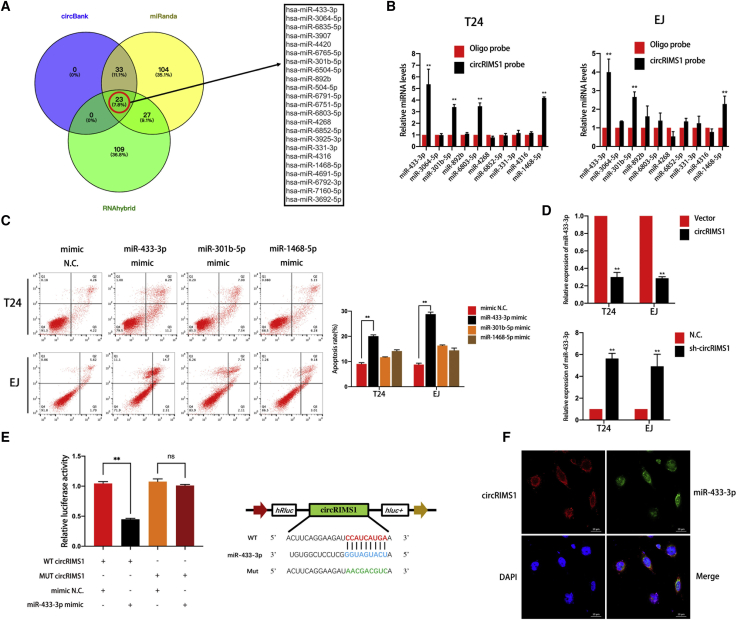
A growing number of studies have confirmed that circRNAs can act as ceRNAs (competing endogenous RNAs) to weaken the biological effects of targeted miRNAs.28,29 Given that circRIMS1 is predominantly localized in the cytoplasm and is remarkably stable, we hypothesized that circRIMS1 regulates the biological behaviors of bladder cancer cells by sponging miRNAs. Therefore, we predicted potential miRNAs that bind to circRIMS1 using miRanda, circBank, and RNAhybrid. Twenty-three miRNAs were obtained from the overlap among these databases. Next, we searched these miRNAs’ cancer-related literatures and identified 10 miRNAs that were reportedly involved in cancer progression and development in at least one study (Figure 3A). To verify the binding potential of the 10 selected miRNAs, we also performed a pulldown assay with a biotinylated circRIMS1 probe. Expression of target miRNAs, which were pulled down from T24 and EJ cells, was analyzed by qRT-PCR. According to Figure 3B, we selected three miRNAs with significantly increased fold changes for circRIMS1 capture. Then, we determined the biological function of these candidate miRNAs via apoptotic assessment. Figure 3C shows that miR-433-3p mimics significantly increased the apoptotic rate in T24 and EJ cells after 48 h, whereas other miRNAs had no obvious effects. In addition, we measured miR-433-3p expression levels in circRIMS1-overexpressing and circRIMS1-knockdown cells, and the results were consistent with our conclusions above (Figure 3D). We subsequently inserted wild-type and mutated circRIMS1 into a luciferase reporter vector. With transfection of the miR-433-3p mimics, the luciferase reporter assay demonstrated that luciferase activity of the mutated reporter was remarkably higher than the wild-type sequence in HEK293 cells (Figure 3E), indicating that miR-433-3p directly binds to circRIMS1. Furthermore, RNA FISH revealed that circRIMS1 and miR-433-3p both localized to the cytoplasm of bladder cancer cells (Figure 3F), suggesting that miR-433-3p can interact with circRIMS1.
Figure 3.
circRIMS1 Sponges miR-433-3p in Bladder Cancer Cells
(A) Overlapping target miRNAs of circRIMS1 in miRanda, circBank, and RNAhybrid. (B) RNA pull-down and subsequent qRT-PCR detection of 10 miRNA candidates indicated that multiple miRNAs could be captured by the circRIMS1 probe in T24 and EJ lysates. miR-433-3p, miR-301-5p, and miR-1468-5p were pulled down by the circRIMS1 probe in both cell lines. (C) Apoptosis was assessed in cells transfected with miRNA mimics or mimic negative control by flow cytometry. (D) Expression levels of miR-433-3p were evaluated in sh-circRIMS1 and circRIMS1-overexpressing T24 and EJ cells. (E) Luciferase reporter assay was employed to determine the luciferase activity of the circRIMS1 wild-type/mutant reporter in HEK293 cells, with or without miR-433-3p mimics. Wild-type and mutant sequences. (F) RNA FISH indicated that miR-433-3p and circRIMS1 colocalized in EJ cells. Nuclei are stained with DAPI (blue), circRIMS1 is shown in red, and miR-433-3p is shown in green. Scale bars, 20 μm. ∗p < 0.05, ∗∗p < 0.01 versus control group.
miR-433-3p Is Downregulated in Bladder Cancer Cell Lines and Tissues and Acts as a Tumor Suppressor
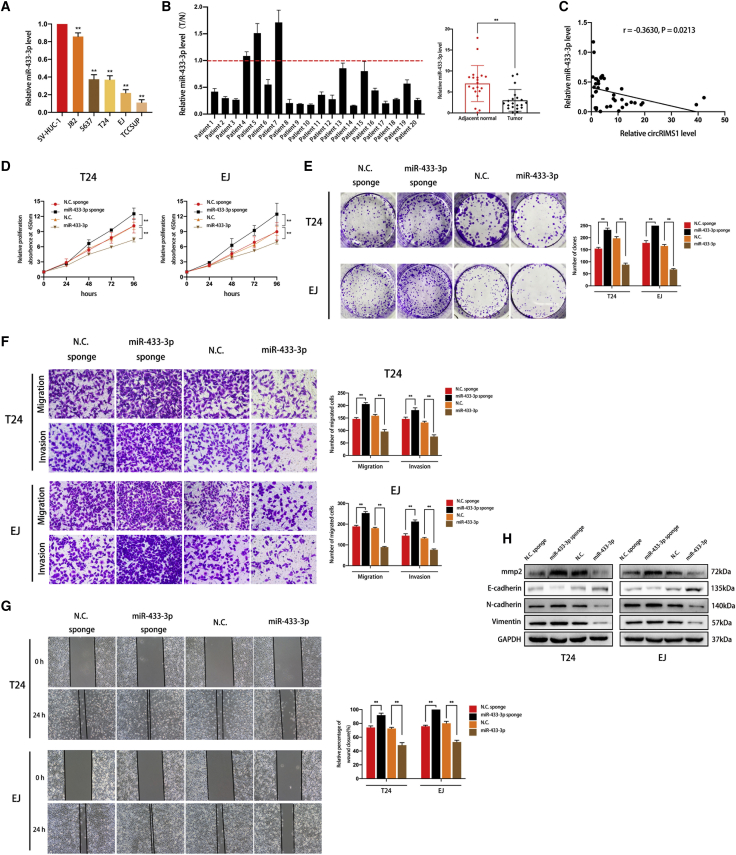
Our previous results showed that circRIMS1 was a tumor promoter and that miR-433-3p could interact with circRIMS1 in bladder cancer cells, indicating the vital roles of miR-433-3p in bladder cancer. Therefore, we next assessed miR-433-3p levels in bladder cancer cell lines and tissues. As shown in Figure 4A, in qRT-PCR experiments, miR-433-3p exhibited lower expression levels in bladder cancer cells than in SV-HUC-1 cells. We also confirmed lower expression levels of miR-433-3p in human bladder cancer tissues compared to adjacent normal bladder tissues (Figure 4B). Moreover, we found that levels of miR-433-3p were negatively correlated with levels of circRIMS1by performing Pearson correlation analysis (Figure 4C).
Figure 4.
miR-433-3p Is Vital for Cell Proliferation, Migration, and Invasion of Bladder Cancer Cells
(A) miR-433-3p levels were detected in bladder cancer cells (J82, 5637, T24, EJ, and TCCSUP) and SV-HUC-1 by qRT-PCR. (B) qRT-PCR indicated that miR-433-3p exhibited lower levels in bladder cancer tissues than in normal adjacent tissues (n = 20). (C) Pearson correlation analysis showing that expression levels of miR-433-3p are negatively correlated with circRIMS1 in 20 pairs of clinical specimens. Cells were transfected with miR-433-3p sponge and miR-433-3p and their negative controls, respectively. (D) Cell viability was measured by a CCK-8 assay to evaluate the biological effects of miR-433-3p. (E) Colony-formation assay indicated the vital role of miR-433-3p in cell proliferation. (F) Transwell migration and invasion assays indicated the role of miR-433-3p on cell migration and invasion. (G) Wound-healing assay showed that miR-433-3p influenced the cell migration ability. (H) Expression levels of MMP2, E-cadherin, N-cadherin, and vimentin in T24 and EJ cells transfected with pre-miR-433 or miR-433-3p sponge. ∗p < 0.05, ∗∗p < 0.01 versus control group.
To clarify whether miR-433-3p modulated biological behaviors in bladder cancer, miR-433-3p-overexpressing T24 and EJ cells were constructed via pre-miR-433 and cells with a miR-433-3p sponge. First, we performed CCK-8 and colony-formation assays to evaluate the influence of miR-433-3p on bladder cancer cell proliferation. As shown in Figures 4D and 4E, knockdown of miR-433-3p promoted cell growth and proliferation, whereas increased miR-433-3p exhibited the opposite effect. We also determined migratory and invasive abilities by wound-healing and Transwell assays. Our results indicated that miR-433-3p remarkably inhibited cell migration and invasion (Figures 4F and 4G). Detection of EMT markers and MMPs by western blotting demonstrated that miR-433-3p inhibited the EMT process and MMP2 expression in bladder cancer cells (Figure 4H). Overall, these results demonstrate that overexpression of miR-433-3p suppresses proliferation, migration, and invasion of T24 and EJ cells.
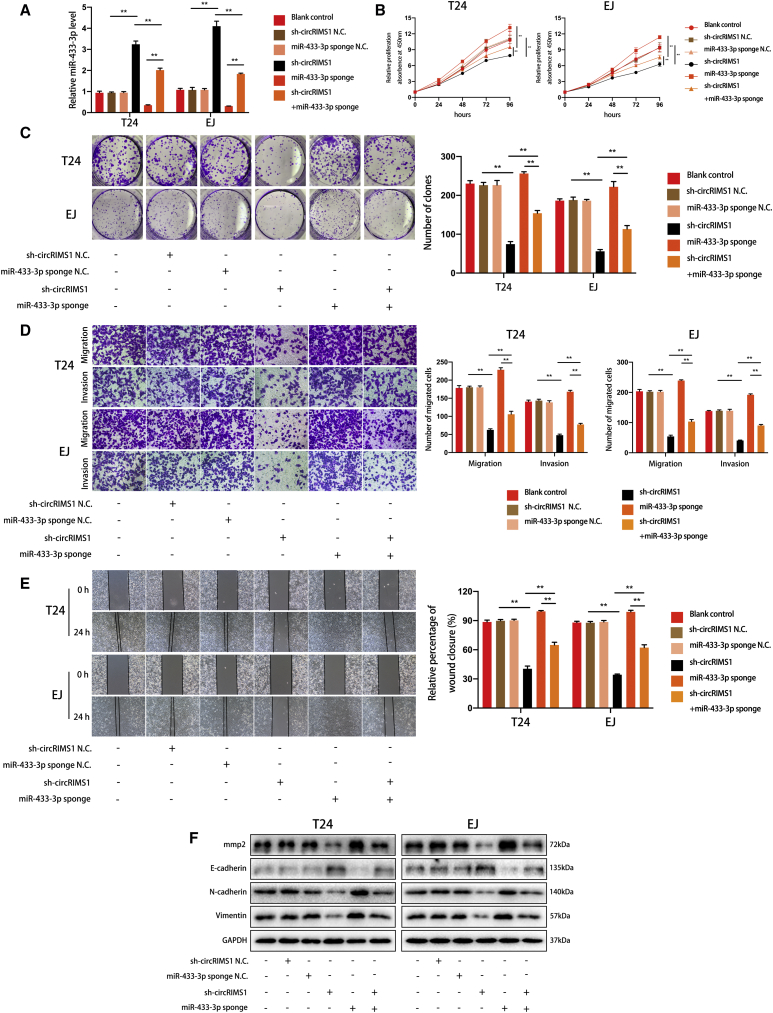
Knockdown of miR-433-3p Reverses the Antitumor Effects Induced by Inhibition of circRIMS1 in Bladder Cancer Cells
To confirm the circRIMS1/miR-433-3p regulatory axis, we transfected sh-circRIMS1, a miR-433-3p sponge, and negative control vectors into T24 and EJ cells. We first determined initial levels of miR-433-3p in the cells (Figure 5A). As predicted, results of CCK-8 and colony-formation assays indicated that miR-433-3p inhibition partially attenuated the suppression of cell viability and proliferation induced by knockdown of circRIMS1. In addition, inhibition of circRIMS1 also reversed miR-433-3p sponge-induced enhancement of cell growth and proliferation (Figures 5B and 5C). Moreover, Transwell migration, invasion assays, and wound-healing assays showed that circRIMS1-induced inhibition of migration and invasion was reversed in response to the miR-433-3p sponge, and miR-433-3p sponge-induced biological effects were attenuated by sh-circRIMS1 (Figures 5D and 5E). Accordingly, western blot confirmed the above results, demonstrating altered expression levels of MMP2 and EMT markers (Figure 5F). These findings indicated that circRIMS1 promotes bladder cancer progression by sponging miR-433-3p, which eliminates the antitumor effects induced by miR-433-3p.
Figure 5.
Inhibition of miR-433-3p Alleviates the Antitumor Effects of sh-circRIMS1
(A) Expression levels of miR-433-3p were verified in blank control, sh-circRIMS1 negative control, miR-433-3p sponge negative control, circRIMS1-knockdown, miR-433-3p sponge, or sh-circRIMS1 with miR-433-3p sponge bladder cancer cells. (B) CCK-8 assay was used to detect cell viability in different cells. (C) Colony-forming ability was determined by the colony-formation assay. (D and E) Transwell migration and invasion assays (D) and wound-healing assays (E) showed the effects of sh-circRIMS1, miR-433-3p sponge, or sh-circRIMS1 with miR-433-3p sponge on cell migration and invasion. (F) Expression levels of MMP2 and EMT-related genes were detected in cells with various treatments. ∗p < 0.05, ∗∗p < 0.01 versus control group.
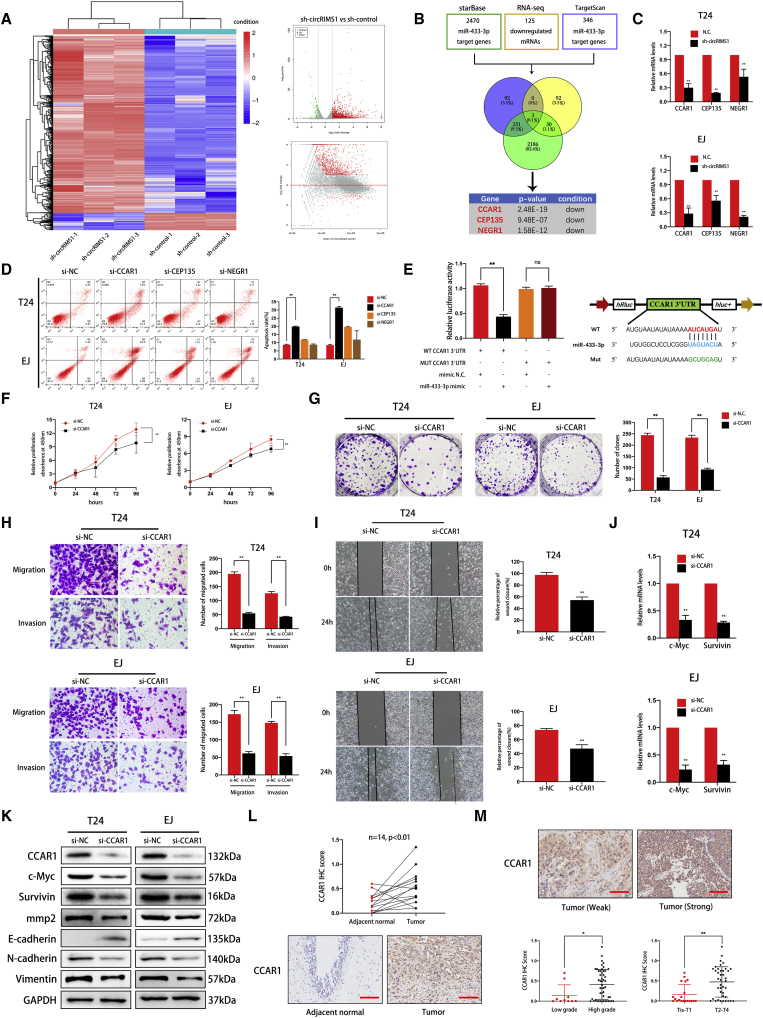
CCAR1 Is Targeted by miR-433-3p and Acts as an Oncogene in Bladder Cancer
To elucidate which genes are regulated by the circRIMS1/miR-433-3p axis in bladder cancer, we performed RNA sequencing and identified 125 downregulated genes in the transcriptome between control T24 cells and T24 cells with stable inhibition of circRIMS1 (Figure 6A; GEO: GSE150143). In addition, StarBase analysis revealed 2,470 potential target genes, whereas TargetScan predicted 346 genes potentially targeted by miR-433-3p. We overlapped these results, and three genes (CCAR1, CEP135, and NEGR1) were selected as research objects (Figure 6B). We further confirmed the mRNA levels of these genes in sh-circRIMS1 and control T24 or EJ cells. The results indicated that CCAR1, CEP135, and NEGR1 were simultaneously decreased in response to circRIMS1 inhibition in both cell lines (Figure 6C). We then designed siRNAs for these three targeted genes and detected the apoptotic rate by cytometry after transfection. The effects of siRNAs were confirmed by western blotting (Figures S1A–S1C). As shown in Figure 6D, depletion of CCAR1 induced the most remarkable apoptosis levels, whereas knockdown of CEP135 and NEGR1 did not trigger apoptosis. Moreover, CCAR1 was negatively correlated with levels of miR-433-3p, and inhibition of miR-433-3p reversed the CCAR1 depletion phenotype induced by sh-circRIMS1 (Figures S2A and S2B). Next, we performed luciferase reporter assays to confirm the interaction between miR-433-3p and CCAR1. Results showed that miR-433-3p significantly reduced the activity of the wild-type CCAR1 3′ UTR luciferase reporter compared to the miR-433-3p mimic negative control in HEK293 cells. In contrast, the mutated luciferase reporter showed no significant changes (Figure 6E). Based on these results, we then evaluated the biological functions of CCAR1 in T24 and EJ cells using si-CCAR1. Knockdown of CCAR1 inhibited cell proliferation and colony formation (Figures 6F and 6G). Moreover, as determined by Transwell assays and wound-healing assay, migration and invasion in CCAR1-depleted cells were also impaired (Figures 6H and 6I). In addition, western blot analysis further confirmed the critical role of CCAR1 in migration and invasion of bladder cancer (Figure 6K). We next verified the correlation between CCAR1 and clinical features in another cohort. Figure 6L demonstrates that protein levels of CCAR1 were strikingly higher in bladder cancer than in adjacent normal tissue (n = 14). Furthermore, CCAR1 was associated with the pathological and histological characteristics of bladder cancer (n = 60; Figure 6M; Table S5). These series of experiments verified that CCAR1 is directly targeted by miR-433-3p and acts as a tumor promoter.
Figure 6.
CCAR1 Is Directly Targeted by miR-433-3p and Serves as a Tumor Promoter in Bladder Cancer
(A) Significantly differentially expressed mRNAs are presented as a clustered heatmap in T24 cells (negative control versus sh-circRIMS1). Red dots represent upregulated genes, and green dots indicate downregulated genes in the volcano plot. (B) Overlap of downregulated mRNAs identified by RNA sequencing and target genes of miR-433-3p predicted from TargetScan and StarBase databases. (C) Expression levels of CCAR1, CEP135, and NEGR1 in control or circRIMS1-knockdown cells were further verified. (D) The biological effect of CCAR1 was determined by siRNA transfections and Annexin V-FITC/PI staining. (E) Luciferase reporter assay confirmed the interaction between miR-433-3p and the CCAR1 3′ UTR. miR-433-3p mimics (or negative control mimic) were cotransfected with wild-type or mutated CCAR1 3′ UTR luciferase reporter vector into HEK293 cells. (F and G) Cell viability and colony-forming ability in control or CCAR1-deficient cells evaluated by CCK-8 assay (F) and colony-formation assay (G), respectively. (H and I) To confirm the role of CCAR1 in cell migration and invasion abilities, Transwell migration and invasion assays (H) and wound-healing assay (I) were used to detect cell migration ability. (J) mRNA expression levels of two Wnt signaling target genes (c-Myc and survivin) were downregulated by CCAR1 depletion, as determined by qRT-PCR. (K) Expression of CCAR1, c-Myc, survivin, MMP2, and EMT markers at the protein level by western blotting in cells with various treatments. (L and M) Immunohistochemistry (IHC) detection of CCAR1 in tissue chip (n = 60). Representative images are shown, and IHC score was calculated from immunostaining intensity and positive staining rate. CCAR1 was abundantly expressed in bladder cancer tissue (L) and was associated with pathological stage and histological grade (M). Scale bars, 100 μm. ∗p < 0.05, ∗∗p < 0.01 versus control group.
CCAR1 Regulates the Expression of c-Myc and Survivin to Influence Tumor Phenotype
Previous studies have reported that CCAR1 interacts with β-catenin to enhance the expression of downstream Wnt signaling target genes to modulate the growth of gastric and colon cancer cells.23,24 Therefore, we evaluated expression levels of c-Myc and survivin, which are closely associated with cell proliferation and survival and are regulated by the Wnt signaling pathway, using qRT-PCR and western blotting in CCAR1-knockdown cells. As shown in Figures 6J and 6K, reduction in CCAR1 had a remarkable impact on both c-Myc and survivin at both the mRNA and protein levels. Therefore, our results demonstrated that CCAR1, which was targeted by miR-433-3p, regulates the expression of c-Myc and survivin to medicate survival and proliferation in bladder cancer cells. However, the exact regulatory mechanism among CCAR1, c-Myc, and survivin requires further study.
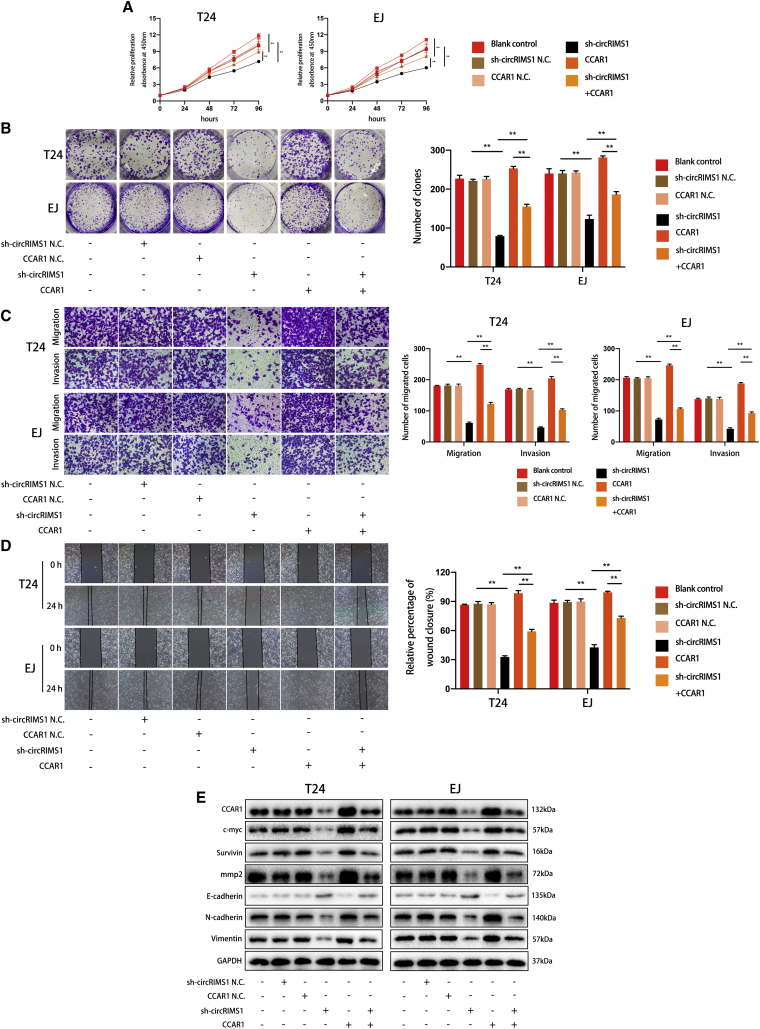
circRIMS1 Regulates Bladder Cancer Progression by Restoring the Expression of CCAR1
To further validate the circRIMS1/miR-433-3p/CCAR1 regulatory axis, we overexpressed CCAR1 in circRIMS1-knockdown T24 and EJ cells and observed the subsequent effects. CCK-8 assay and colony-formation assay showed that restoring CCAR1 expression partially reversed the inhibition of cell viability and proliferation induced by circRIMS1 knockdown, whereas knockdown of circRIMS1 attenuated CCAR1-induced biological effects (Figures 7A and 7B). Additionally, consistent results were found in Transwell migration and invasion assays and wound-healing assays in blank control, negative control, cricRIMS1-knockdown, CCAR1-overexpressing, and sh-circRIMS1 with CCAR1 overexpression cells (Figures 7C and 7D). As expected, changes in MMP2 and EMT markers also indicated the key role of CCAR1 in cell adhesion and EMT in bladder cancer cells. Interestingly, c-Myc and survivin were also upregulated in response to overexpression of CCAR1. Meanwhile, alterations of the above proteins induced by CCAR1 were attenuated by sh-circRIMS1 (Figure 7E). In summary, our results indicated that circRIMS1 regulates the biological functions of bladder cancer cells through modulating levels of CCAR1.
Figure 7.
circRIMS1 Regulates Bladder Cancer Progression through Restoring the Expression of CCAR1
Cells under different conditions (blank control, sh-circRIMS1 negative control, CCAR1 overexpression negative control, circRIMS1 knockdown, CCAR1 overexpressing, or sh-circRIMS1 with CCAR1 overexpression) were used in the following assays. (A) Cell viability was evaluated to determine the effects on proliferation rate in T24 and EJ cells. (B) Colony-forming ability was detected by colony-formation assay. (C and D) Transwell migration and invasion assays (C) and wound-healing assays (D) indicated that CCAR1 overexpression promoted migration and invasion of circRIMS1-knockdown cells and inhibition of circRIMS1 attenuated CCAR1-induced biological effects. (E) Effects of sh-circRIMS1, CCAR1 overexpression, or sh-circRIMS1 with CCAR1 overexpression on expression levels of MMP2 and EMT-related genes were detected at the protein level. ∗p < 0.05, ∗∗p < 0.01 versus control group.
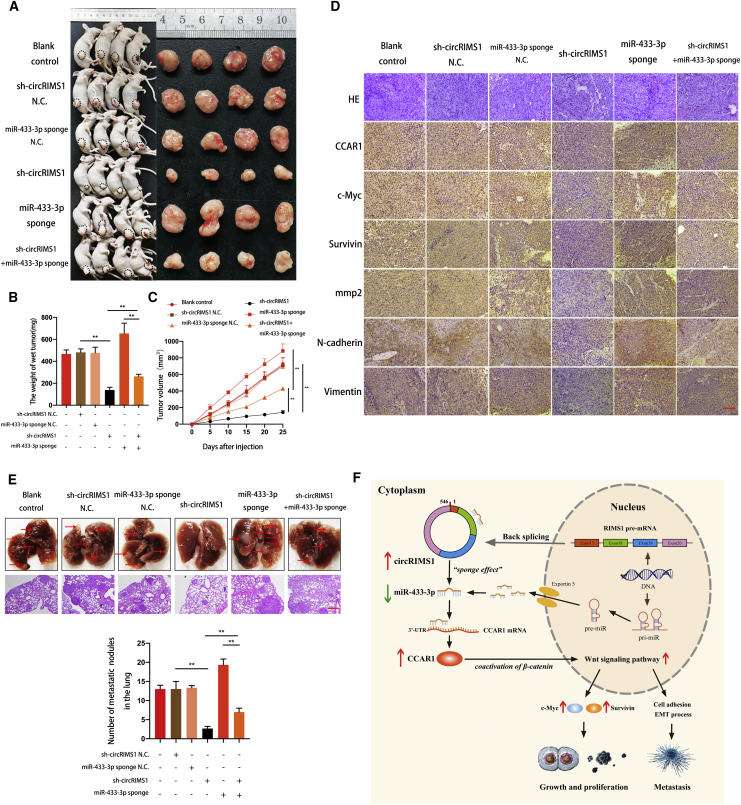
circRIMS1 Is Vital for Bladder Cancer Growth and Metastasis In Vivo
To further confirm a circRIMS1/miR-433-3p/CCAR1 axis in vivo, T24 cells under different conditions (blank control, sh-circRIMS1 negative control, miR-433-3p sponge negative control, circRIMS1 knockdown, miR-433-3p sponge, and sh-circRIMS1 with miR-433-3p sponge) were subcutaneously injected into nude mice. As shown in Figures 8A–8C, inhibition of circRIMS1 led to a remarkable reduction in tumor weight and growth rate. Interestingly, miR-433-3p knockdown remarkably rescued the suppression of tumor growth caused by circRIMS1 silencing. Meanwhile, inhibition of circRIMS1 reversed the enhancement of tumor growth induced by miR-433-3p sponge. Immunohistochemistry (IHC) indicated that the mean immunopositive area for CCAR1, c-Myc, and survivin was decreased due to the depletion of circRIMS1, and the miR-433-3p sponge reversed this change (Figure 8D).
Figure 8.
circRIMS1 Enhances Tumor Growth and Metastasis of Bladder Cancer Cells In Vivo
(A) An equal number of different T24 cells (blank control, sh-circRIMS1 negative control, miR-433-3p sponge negative control, circRIMS1 knockdown, miR-433-3p sponge, and sh-circRIMS1 with miR-433-3p sponge) were used to establish subcutaneous xenograft tumors. Tumors were harvested and imaged (n = 4 each group). (B) Tumor weight was determined when mice were sacrificed. (C) Tumor volumes were measured and calculated every 5 days after the cells were injected into mice. (D) H&E staining and IHC detection of CCAR1, c-Myc, survivin, MMP2, and EMT markers in tumors. Scale bar, 200 μm. (E) T24 cells (blank control, sh-circRIMS1 negative control, miR-433-3p sponge negative control, circRIMS1 knockdown, miR-433-3p sponge, and sh-circRIMS1 with miR-433-3p sponge) were injected into mice via the tail veins (n = 3 for each group). Lung metastatic colonies were counted and recorded. Scale bar, 100 μm. (F) Schematic diagram illustrating the circRIMS1 regulatory axis in bladder cancer cells. circRIMS1 promotes bladder cancer progression by acting as a ceRNA to sponge miR-433-3p and restore CCAR1 expression, which serves as a coactivator of β-catenin to enhance the Wnt pathway. ∗p < 0.05, ∗∗p < 0.01 versus control group.
To investigate the effects of circRIMS1 on tumor metastasis in an animal model, T24 cells were injected into the tail vein of BALB/c nude mice. As shown in Figure 8E, mice treated with sh-circRIMS1 contained fewer metastatic colonies in the lung than those treated with blank control or negative control vectors. miR-433-3p sponge enhanced the formation of lung metastatic colonies. Moreover, sh-circRIMS1-induced suppression was reversed by miR-433-3p sponge, and miR-433-3p sponge-induced enhancement of tumor metastasis was attenuated by inhibition of circRIMS1. IHC analysis confirmed expression of MMP2 and EMT markers, indicating the interaction between circRIMS1 and miR-433-3p in cell adhesion and the EMT process (Figure 8D). To illustrate our study more clearly, we created a schematic of the circRIMS1/miR-433-3p/CCAR1 regulatory axis (Figure 8F). Altogether, our results demonstrated that inhibition of circRIMS1 dramatically inhibits bladder cancer cell growth and metastasis primarily through the circRIMS1/miR-433-3p/CCAR1 axis in vivo.
Discussion
During the past decade, a paradigm shift has occurred, and circRNAs are no longer regarded as simply the junk products of pre-mRNA splicing.30 Dysregulation of circRNAs has been confirmed in numerous cancer types, such as hepatocellular carcinoma,31 oral cancer,12 esophageal squamous cell carcinoma,32 and bladder cancer.11,33 However, there are still numerous enigmas about the molecular mechanisms of circRNAs that remain to be investigated in bladder cancer. In this study, we identified a circRNA, termed circRIMS1, which exhibits aberrantly higher expression in bladder cancer cells and tissue, and explored its underlying regulatory mechanism.
circRNAs are derived from exons of existing genes via back-splicing.6 Here, we verified that circRIMS1 consisted of exons 17, 18, 19, and 20 of the RIMS1 gene and formed the circular structure by joining the 3′ and 5′ sites. Previous studies have verified the stability of circRNAs.25 Our subsequent results showed that circRIMS1 could tolerate RNase R treatment with little degradation. In addition, we also confirmed that circRIMS1 was abundantly expressed in bladder both cancer cell lines and clinical specimens. Our correlation analysis indicated that circRIMS1 might be related to bladder cancer development and progression. However, due to the sample size of our cohort (n = 20), further verification was necessary and meaningful. Meanwhile, our in vitro and in vivo assays confirmed the important roles of circRIMS1 on bladder cancer carcinogenesis.
The abnormal expression and critical roles of circRIMS1 in bladder cancer indicated that this circRNA might represent an ideal target for clinical application in bladder cancer. Indeed, circRNAs can be detected in body fluids. A growing body of evidence has shown that circRNAs exist in circulating system and plasma.34, 35, 36 Of note, some researchers verified the dysregulation of circRNAs in urine of bladder cancer patients.37, 38, 39 Therefore, with the consideration of the characteristics and accessibility of circRNAs, they are suitable for clinical diagnosis and prognostic prediction in human cancers. Chen and his colleagues40 found that hsa_circ_0000190 has better sensitivity and specificity for a gastric cancer diagnosis compared to carcinoembryonic antigen (CEA) and CA19-9 (classic biomarkers of gastric cancer). Moreover, a recent study reported that expression levels of ciRS-7 were correlated with hepatic microvascular invasion and α-fetoprotein (AFP) levels.41 circRNAs also enhance the efficiency of Kirsten rat sarcoma 2 viral oncogene homolog (KRAS) diagnosis in colorectal cancer.42 Based on these valuable studies and our results, our future research will focus on the detection of circRIMS1 in body fluids, such as plasma and urine, or tumor tissue acquired from a greater number of bladder cancer patients to establish a diagnostic system based on the levels of circRIMS1 and other existing biomarkers in bladder cancer, such as circRNAs, miRNAs, cancer-related genes, and biochemical index, which might promote the precise and early diagnosis of bladder cancer. Meanwhile, correlations between circRIMS1 and clinical or pathological stages and histological grades will also be investigated. circRNAs have been reported to participate in the initiation and progression of sepsis,43 schizophrenia, depression,44 inflammatory bowel disease,45 autoimmune diseases,46 and especially human cancers.47, 48, 49, 50, 51, 52 Therefore, it is reasonable to speculate that they are also potential therapeutic targets. Since our results showed that circRIMS1 was remarkably overexpressed in bladder cancer, siRNA targeting its back-splice junction might represent a novel therapeutic method. Although there are not any preclinical trials concerning circRNAs in clinical treatment for cancer, numerous studies and attempts to utilize siRNAs as therapeutics have been reported.53, 54, 55
circRNAs contain abundant miRNA target-binding sites and could weaken the downregulation of target genes mediated by miRNA,4 widely known as the “miRNA sponge” effect that has been reported in emerging studies.11,12,14,31,32 In the present study, circRIMS1 was found to be localized in the cytoplasm, and a subsequent targeted miRNA prediction indicated that circRIMS1 contained numerous miRNA-binding sites, suggesting that circRIMS1 may exert its biological effects as a miRNA sponge. For further verification, we performed RNA pulldown and luciferase assays to elucidate this interaction in detail and confirmed the existence of the circRIMS1/miR-433-3p axis. Next, we found that miR-433-3p served as a tumor suppressor in bladder cancer. Furthermore, RNA sequencing, bioinformatics prediction, and luciferase assays revealed that miR-433-3p is a negative regulator of CCAR1 and that circRIMS1 exerts its biological functions primarily via CCAR1.
CCAR1 is a key modulator of apoptosis signaling and chemotherapy in human breast cancer.20 Moreover, CCAR1 is also involved in biological behaviors via an estrogen-dependent pathway in human breast cancer cells.21 In addition, a previous study found that CCAR1 was vital for cell growth and invasion in gastric cancer, serving as a coactivator of β-catenin and enhancing transcriptional activation of Wnt signaling pathway target genes.24 In this research, our experiments confirmed that CCAR1 was a tumor promoter. Interestingly, we identified a regulatory effect of CCAR1 on c-Myc and survivin, which are both Wnt signaling target genes associated with cell proliferation and growth. Furthermore, we verified enhanced expression of CCAR1 in bladder cancer tissues compared to adjacent normal tissues. Furthermore, CCAR1 was positively correlated with pathological stage and histological grade. Because the Wnt pathway is closely related to cell adhesion and EMT,56, 57, 58 it is reasonable to speculate that CCAR1 may represent a key regulator in the progression of bladder cancer by activating the Wnt pathway. However, the exact downstream genes of CCAR1 and Wnt signaling and the underlying molecular mechanism responsible for this biological regulatory process still need further investigation. Overall, by upregulating CCAR1 and sponging miR-433-3p, circRIMS1 promotes the progression of bladder cancer and is a promising target for clinical application in bladder cancer in the near future.
Materials and Methods
Ethical Approval
All animal experiments were approved by the Ethics Committee of The First Affiliated Hospital, Zhejiang University School of Medicine. All procedures were performed according to the NIH Laboratory Guidelines for animals.
Clinical Samples
Bladder cancer tissue samples and matched adjacent normal bladder tissues were acquired from the surgical specimens, and this research was approved by the Ethics Committee of First Affiliated Hospital of Zhejiang University School of Medicine. Written, informed consent was signed by patients before the study began. The World Health Organization (WHO) Consensus Classification and tumor node metastasis (TNM) staging system were used to classify the pathological and clinical stage of all the specimens. Detailed patient information is shown in Tables S2 and S4.
Cell Lines and Culture
HEK293 cells; a human immortalized uroepithelium cell line (SV-HUC-1); and human bladder cancer cell lines J82, 5637, T24, EJ, and TCCSUP were obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA). Dulbecco’s modified Eagle’s medium and RPMI-1640 medium with 10% fetal bovine serum (FBS; Gibco, Grand Island, NY, USA), penicillin, and streptomycin were used to culture HEK293 cells and the other cell lines, respectively. All cells were cultured at 37°C with 5% CO2.
RNA Extraction, Reverse Transcription, and Quantitative Real-Time PCR Analysis
Cells were collected, and total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). For the RNase R degradation assay, total RNA (2 mg) was incubated with or without 3 U/mg RNase R (Epicenter, WI, USA) for 15 min at 37°C. To determine levels of circRNA and mRNA, total RNA (1 μg) was used for reverse transcription by employing the PrimeScript RT Reagent Kit (Takara Bio, PR China). TB Green Premix Ex Taq II (Takara Bio, PR China) was used for subsequent detection on a QUANT5 PCR system (Applied Biosystems by Life Technologies). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an endogenous control. For miRNA analyses, the All-in-One miRNA qRT-PCR Detection Kit (GeneCopoeia, USA) was used, and human U6 was used as a control. Results were analyzed using the 2−ΔΔCt method. All primers are listed in Table S1.
siRNA, miRNA Mimics, Plasmid, and Lentivirus Construction and Transfection
RiboBio (Guangzhou, PR China) synthesized siRNA and miRNA mimics for the study. Lipofectamine RNAiMAX (Invitrogen) was used for transfections. The sequences used are listed in Table S1. For the construction of circRIMS1 overexpression plasmids, cDNA containing circRIMS1 was constructed and cloned into the pcD-ciR vector by GeneSeed (Guangzhou, PR China). The pcD-ciR vector contains a front circular frame and a back circular frame and was transfected into cells using Lipofectamine 3000 (Invitrogen). Lentiviruses were obtained from GeneChem (Shanghai, PR China). Puromycin was used for selection for at least 6 days after infection.
FISH
Cy3-circRIMS1 and fluorescein isothiocyanate (FITC)-miR-433-3p probes were synthesized and obtained from RiboBio (Guangzhou, PR China). Hybridization assays were performed using a FISH Detection Kit (RiboBio, PR China). All images were captured by a confocal microscope (FV1000; Olympus, Tokyo, Japan). The sequences of the probes are listed in Table S1.
Nucleic Acid Electrophoresis
PCR products were verified using 2% agarose gel electrophoresis with Tris base, acetic acid, and EDTA (TAE) running buffer (120 V, 30 min). NormalRun 250 bp-II DNA ladder (Generay, Shanghai, PR China) was used as the DNA marker. Bands were observed by UV irradiation. Primer sequences are provided in Table S1.
Target Prediction of circRIMS1 and miR-433-3p
We predicted potential miRNA-binding sites of circRIMS1 using the bioinformatics databases miRanda (http://www.miranda.org/), RNAhybrid (https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid/), and circBank (http://www.circbank.cn). We used StarBase59 and TargetScan (http://www.targetscan.org/vert_72/) to predict target genes of miR-433-3p.
Pulldown Assay with Biotinylated circRIMS1 Probe
Bladder cancer cells (1 × 107) were obtained, lysed, and sonicated as indicated. To generate the probe-coated beads, C-1 magnetic beads (Life Technologies) were coincubated with the circRIMS1 probe for 2.5 h at 25°C. Then, the circRIMS1 probe or oligo probe was coincubated with cell lysates at 4°C overnight. RNA was eluted and extracted by wash buffer and used for qRT-PCR. The circRIMS1 probe with biotinylation was synthesized and purchased from RiboBio (Guangzhou, PR China). The sequences of the biotinylated circRIMS1 probe are listed in Table S1.
Luciferase Reporter Assay
First, a luciferase reporter vector (pGL3-Firefly_Luciferase-Renilla_Luciferase) with CCAR1 3′ UTR or circRIMS1 was built, and the mutant vectors were constructed by GeneChem (Shanghai, PR China). A luciferase vector and miR-433-3p mimic or mimic negative control were cotransfected into cells and incubated for 24 h. A dual-luciferase reporter assay detection kit (Promega, WI, USA) was used to detect firefly and Renilla luciferase activities, which were measured on a Fluoroskan Ascent device (Thermo Fisher Scientific, USA).
Colony-Formation Assay
Cells were counted and seeded into 12-well plates (500 cells per well). After 8 days of incubation, cell colonies were fixed in 4% paraformaldehyde and stained with crystal violet. Only cell colonies containing 50 cells or more were counted and recorded.
Migration and Invasion Assays
For the migration assay, 200 μL serum-free medium containing 3 × 104 cells was added into the upper chambers of Transwells (Costar, NY, USA) for 24 h. Similarly, for the invasion assay, cells were added to Matrigel (Becton Dickinson [BD], MA, USA)-precoated Transwell upper chambers and incubated for 48 h. A total of 600 μL RPMI-1640 medium with 10% FBS was added to the bottom chambers as an attractant. Migrated or invaded cells were fixed in 4% paraformaldehyde and then stained with crystal violet. After removing the remaining cells on the upper surface of the membrane, the stained cells were visualized and imaged by microscopy (100×) in five randomly chosen fields.
Wound-Healing Assay
Cells were seeded into 12-well plates with Culture-Inserts (ibidi, Germany), according to the manufacturer’s instructions. The next day, Culture-Inserts were removed, and cells were incubated without serum for another 24 h. Images were recorded at 0 and 24 h on a microscope (Nikon, Tokyo, Japan), and the migration rate of the cells was measured using ImageJ software.
Apoptosis Analysis
Cells were collected and stained with Annexin V-FITC and propidium iodide (PI) in binding buffer, according to the instructions of the Apoptosis Detection Kit (BD Biosciences, San Diego, CA, USA). After 30 min of incubation, the apoptotic rate was determined by flow cytometry (Becton Dickinson, USA) and was analyzed using FlowJo software.
Western Blot Analysis and Antibodies
Briefly, total protein was extracted using radioimmunoprecipitation assay (RIPA) buffer (C1053; Applygen, Beijing, PR China). After measurement of protein concentrations, equal amounts of proteins (30 μg) were separated by SDS-PAGE and were transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, USA) and incubated with 5% skim milk for 2 h at room temperature. After washing the membrane with Tris-buffered saline Tween 20 (TBST) three times, primary antibodies were coincubated with the membrane at 4°C overnight. The next day, the membrane was incubated with secondary antibody (1:5,000) for 2 h at room temperature. Bands were detected by a Bio-Rad CD Touch Detection System. The antibodies used in this study were anti-GAPDH (catalog #5174), anti-E-cadherin (catalog #3195), anti-N-cadherin (catalog #13116), and anti-vimentin (catalog #5741) were all obtained from Cell Signaling Technology. Anti-c-myc (ab32072), anti-survivin (ab76424), anti-CEP135 (ab75005), and anti-NEGR1 (ab237798) were purchased from Abcam (Cambridge, UK); anti-CCAR1 (PA5-52473) was purchased from Invitrogen (Carlsbad, CA, USA); and anti-MMP2 (catalog number: 10373-2-AP) was obtained from Proteintech (Chicago, IL, USA).
Other In Vitro Experiments
Other in vitro experiments, such as CCK-8 assay, H&E staining, IHC, and confocal microscopy, have been previously described by us.60
In Vivo Studies
To establish the orthotopic xenograft tumor models, 4-week-old female BALB/c nude mice were obtained and randomly divided into six groups (n = 4 for each group). An equal amount of T24 cells (5 × 106) was subcutaneously injected into the nude mice. 25 days later, tumors were harvested from mice. Tumor volume (mm3) = ab2/2. For tail-vein metastasis models, 6-week-old female BALB/c nude mice were randomly divided and were injected with different T24 cells via the tail vein (5 × 106 cells per mouse). After 40 days, mice were sacrificed, and the numbers of lung metastatic colonies were examined and recorded.
Statistical Analysis
Statistical analyses were performed using SPSS 20 (Abbott Laboratories, Chicago, IL, USA). All data are presented as the mean ± standard deviation (SD). Student’s t test and chi-square tests were used to analyze differences between the two groups. A paired t test was used to analyze expression levels of circRIMS1, miR-433-3p, and CCAR1 in cancer tissues and matched adjacent normal tissues. Pearson’s analysis was used to clarify the relationship between circRIMS1 and miR-433-3p. A p value <0.05 indicated meaningful results in this study.
Author Contributions
X.J. and F.W. conceived and designed the study. F.W., M.F., Y.C., X.Z., and N.H. performed the experiments. F.W., M.F., H.W., Y.Z., J.L., S.H., and S.T. conducted the statistical analyses. F.W. wrote the paper, and X.J. and Z.H. revised the paper. F.W., M.F., Y.C., X.Z., and Y.Y. revised the manuscript and conducted the in vitro and in vivo experiments.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
We thank Dr. Mengjing Fan from the Department of Pathology, Sir Run Shaw Hospital, Zhejiang University School of Medicine. She patiently instructed us in performing our in vivo studies. We also thank Guangzhou GeneSeed for designing the divergent primers and constructing the overexpression plasmids for circRIMS1. This study was approved by the Ethics Committee of The First Affiliated Hospital l, School of Medicine, Zhejiang University, and the Animal Research Ethics Committee of Zhejiang University. This work was supported by the National Natural Science Foundation of China, NSFC grant numbers 81370799 and 81900595. The Natural Science Foundation of Zhejiang Province also supported this work under grant number LY18H160013.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.10.003.
Supplemental Information
References
- 1.Kamat A.M., Hahn N.M., Efstathiou J.A., Lerner S.P., Malmström P.U., Choi W., Guo C.C., Lotan Y., Kassouf W. Bladder cancer. Lancet. 2016;388:2796–2810. doi: 10.1016/S0140-6736(16)30512-8. [DOI] [PubMed] [Google Scholar]
- 2.Sanli O., Dobruch J., Knowles M.A., Burger M., Alemozaffar M., Nielsen M.E., Lotan Y. Bladder cancer. Nat. Rev. Dis. Primers. 2017;3:17022. doi: 10.1038/nrdp.2017.22. [DOI] [PubMed] [Google Scholar]
- 3.Soloway M.S. Bladder cancer: Lack of progress in bladder cancer--what are the obstacles? Nat. Rev. Urol. 2013;10:5–6. doi: 10.1038/nrurol.2012.219. [DOI] [PubMed] [Google Scholar]
- 4.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 5.Jeck W.R., Sorrentino J.A., Wang K., Slevin M.K., Burd C.E., Liu J., Marzluff W.F., Sharpless N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 7.Salzman J., Chen R.E., Olsen M.N., Wang P.L., Brown P.O. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9:e1003777. doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rybak-Wolf A., Stottmeister C., Glažar P., Jens M., Pino N., Giusti S., Hanan M., Behm M., Bartok O., Ashwal-Fluss R. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol. Cell. 2015;58:870–885. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 9.Du W.W., Yang W., Liu E., Yang Z., Dhaliwal P., Yang B.B. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846–2858. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang Q., Du W.W., Wu N., Yang W., Awan F.M., Fang L., Ma J., Li X., Zeng Y., Yang Z. A circular RNA promotes tumorigenesis by inducing c-myc nuclear translocation. Cell Death Differ. 2017;24:1609–1620. doi: 10.1038/cdd.2017.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu Q., Liu T., Feng H., Yang R., Zhao X., Chen W., Jiang B., Qin H., Guo X., Liu M. Circular RNA circSLC8A1 acts as a sponge of miR-130b/miR-494 in suppressing bladder cancer progression via regulating PTEN. Mol. Cancer. 2019;18:111. doi: 10.1186/s12943-019-1040-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen L., Zhang S., Wu J., Cui J., Zhong L., Zeng L., Ge S. circRNA_100290 plays a role in oral cancer by functioning as a sponge of the miR-29 family. Oncogene. 2017;36:4551–4561. doi: 10.1038/onc.2017.89. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Hsiao K.Y., Lin Y.C., Gupta S.K., Chang N., Yen L., Sun H.S., Tsai S.J. Noncoding Effects of Circular RNA CCDC66 Promote Colon Cancer Growth and Metastasis. Cancer Res. 2017;77:2339–2350. doi: 10.1158/0008-5472.CAN-16-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Y., Zheng F., Xiao X., Xie F., Tao D., Huang C., Liu D., Wang M., Wang L., Zeng F., Jiang G. CircHIPK3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. EMBO Rep. 2017;18:1646–1659. doi: 10.15252/embr.201643581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carthew R.W., Sontheimer E.J. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kloosterman W.P., Plasterk R.H. The diverse functions of microRNAs in animal development and disease. Dev. Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 17.Trang P., Weidhaas J.B., Slack F.J. MicroRNAs as potential cancer therapeutics. Oncogene. 2008;27(Suppl 2):S52–S57. doi: 10.1038/onc.2009.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoshino H., Seki N., Itesako T., Chiyomaru T., Nakagawa M., Enokida H. Aberrant expression of microRNAs in bladder cancer. Nat. Rev. Urol. 2013;10:396–404. doi: 10.1038/nrurol.2013.113. [DOI] [PubMed] [Google Scholar]
- 19.Catto J.W., Alcaraz A., Bjartell A.S., De Vere White R., Evans C.P., Fussel S., Hamdy F.C., Kallioniemi O., Mengual L., Schlomm T., Visakorpi T. MicroRNA in prostate, bladder, and kidney cancer: a systematic review. Eur. Urol. 2011;59:671–681. doi: 10.1016/j.eururo.2011.01.044. [DOI] [PubMed] [Google Scholar]
- 20.Rishi A.K., Zhang L., Boyanapalli M., Wali A., Mohammad R.M., Yu Y., Fontana J.A., Hatfield J.S., Dawson M.I., Majumdar A.P., Reichert U. Identification and characterization of a cell cycle and apoptosis regulatory protein-1 as a novel mediator of apoptosis signaling by retinoid CD437. J. Biol. Chem. 2003;278:33422–33435. doi: 10.1074/jbc.M303173200. [DOI] [PubMed] [Google Scholar]
- 21.Kim J.H., Yang C.K., Heo K., Roeder R.G., An W., Stallcup M.R. CCAR1, a key regulator of mediator complex recruitment to nuclear receptor transcription complexes. Mol. Cell. 2008;31:510–519. doi: 10.1016/j.molcel.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rishi A.K., Zhang L., Yu Y., Jiang Y., Nautiyal J., Wali A., Fontana J.A., Levi E., Majumdar A.P. Cell cycle- and apoptosis-regulatory protein-1 is involved in apoptosis signaling by epidermal growth factor receptor. J. Biol. Chem. 2006;281:13188–13198. doi: 10.1074/jbc.M512279200. [DOI] [PubMed] [Google Scholar]
- 23.Ou C.Y., Kim J.H., Yang C.K., Stallcup M.R. Requirement of cell cycle and apoptosis regulator 1 for target gene activation by Wnt and beta-catenin and for anchorage-independent growth of human colon carcinoma cells. J. Biol. Chem. 2009;284:20629–20637. doi: 10.1074/jbc.M109.014332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang T.S., Wei K.L., Lu C.K., Chen Y.H., Cheng Y.T., Tung S.Y., Wu C.S., Chiang M.K. Inhibition of CCAR1, a Coactivator of β-Catenin, Suppresses the Proliferation and Migration of Gastric Cancer Cells. Int. J. Mol. Sci. 2017;18:E460. doi: 10.3390/ijms18020460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jeck W.R., Sharpless N.E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014;32:453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Santamaria P.G., Moreno-Bueno G., Portillo F., Cano A. EMT: Present and future in clinical oncology. Mol. Oncol. 2017;11:718–738. doi: 10.1002/1878-0261.12091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shay G., Lynch C.C., Fingleton B. Moving targets: Emerging roles for MMPs in cancer progression and metastasis. Matrix Biol. 2015;44-46:200–206. doi: 10.1016/j.matbio.2015.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu J., Li P., Song Y., Ge Y.X., Meng X.M., Huang C., Li J., Xu T. Progress and prospects of circular RNAs in Hepatocellular carcinoma: Novel insights into their function. J. Cell. Physiol. 2018;233:4408–4422. doi: 10.1002/jcp.26154. [DOI] [PubMed] [Google Scholar]
- 29.Qiu M., Xia W., Chen R., Wang S., Xu Y., Ma Z., Xu W., Zhang E., Wang J., Fang T. The Circular RNA circPRKCI Promotes Tumor Growth in Lung Adenocarcinoma. Cancer Res. 2018;78:2839–2851. doi: 10.1158/0008-5472.CAN-17-2808. [DOI] [PubMed] [Google Scholar]
- 30.Chen L.L. The biogenesis and emerging roles of circular RNAs. Nat. Rev. Mol. Cell Biol. 2016;17:205–211. doi: 10.1038/nrm.2015.32. [DOI] [PubMed] [Google Scholar]
- 31.Zheng Q., Bao C., Guo W., Li S., Chen J., Chen B., Luo Y., Lyu D., Li Y., Shi G. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xia W., Qiu M., Chen R., Wang S., Leng X., Wang J., Xu Y., Hu J., Dong G., Xu P.L., Yin R. Circular RNA has_circ_0067934 is upregulated in esophageal squamous cell carcinoma and promoted proliferation. Sci. Rep. 2016;6:35576. doi: 10.1038/srep35576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xie F., Li Y., Wang M., Huang C., Tao D., Zheng F., Zhang H., Zeng F., Xiao X., Jiang G. Circular RNA BCRC-3 suppresses bladder cancer proliferation through miR-182-5p/p27 axis. Mol. Cancer. 2018;17:144. doi: 10.1186/s12943-018-0892-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ouyang Q., Huang Q., Jiang Z., Zhao J., Shi G.P., Yang M. Using plasma circRNA_002453 as a novel biomarker in the diagnosis of lupus nephritis. Mol. Immunol. 2018;101:531–538. doi: 10.1016/j.molimm.2018.07.029. [DOI] [PubMed] [Google Scholar]
- 35.Tang W., Fu K., Sun H., Rong D., Wang H., Cao H. CircRNA microarray profiling identifies a novel circulating biomarker for detection of gastric cancer. Mol. Cancer. 2018;17:137. doi: 10.1186/s12943-018-0888-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Memczak S., Papavasileiou P., Peters O., Rajewsky N. Identification and Characterization of Circular RNAs As a New Class of Putative Biomarkers in Human Blood. PLoS ONE. 2015;10:e0141214. doi: 10.1371/journal.pone.0141214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen X., Chen R.X., Wei W.S., Li Y.H., Feng Z.H., Tan L., Chen J.W., Yuan G.J., Chen S.L., Guo S.J. PRMT5 Circular RNA Promotes Metastasis of Urothelial Carcinoma of the Bladder through Sponging miR-30c to Induce Epithelial-Mesenchymal Transition. Clin. Cancer Res. 2018;24:6319–6330. doi: 10.1158/1078-0432.CCR-18-1270. [DOI] [PubMed] [Google Scholar]
- 38.Goel A., Ward D.G., Gordon N.S., Abbotts B., Zeegers M.P., Cheng K.K., James N.D., Bryan R.T., Arnold R. Back-Splicing Transcript Isoforms (Circular RNAs) Affect Biologically Relevant Pathways and Offer an Additional Layer of Information to Stratify NMIBC Patients. Front. Oncol. 2020;10:812. doi: 10.3389/fonc.2020.00812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Song Z., Zhang Q., Zhu J., Yin G., Lin L., Liang C. Identification of urinary hsa_circ _0137439 as potential biomarker and tumor regulator of bladder cancer. Neoplasma. 2020;67:137–146. doi: 10.4149/neo_2018_181214N970. [DOI] [PubMed] [Google Scholar]
- 40.Chen S., Li T., Zhao Q., Xiao B., Guo J. Using circular RNA hsa_circ_0000190 as a new biomarker in the diagnosis of gastric cancer. Clin. Chim. Acta. 2017;466:167–171. doi: 10.1016/j.cca.2017.01.025. [DOI] [PubMed] [Google Scholar]
- 41.Qin M., Liu G., Huo X., Tao X., Sun X., Ge Z., Yang J., Fan J., Liu L., Qin W. Hsa_circ_0001649: A circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomark. 2016;16:161–169. doi: 10.3233/CBM-150552. [DOI] [PubMed] [Google Scholar]
- 42.Dou Y., Cha D.J., Franklin J.L., Higginbotham J.N., Jeppesen D.K., Weaver A.M., Prasad N., Levy S., Coffey R.J., Patton J.G., Zhang B. Circular RNAs are down-regulated in KRAS mutant colon cancer cells and can be transferred to exosomes. Sci. Rep. 2016;6:37982. doi: 10.1038/srep37982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beltrán-García J., Osca-Verdegal R., Nacher-Sendra E., Pallardó F.V., García-Giménez J.L. Circular RNAs in Sepsis: Biogenesis, Function, and Clinical Significance. Cells. 2020;9:E1544. doi: 10.3390/cells9061544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Z., Liu S., Li X., Zhao W., Li J., Xu Y. Circular RNA in Schizophrenia and Depression. Front. Psychiatry. 2020;11:392. doi: 10.3389/fpsyt.2020.00392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin L., Zhou G., Chen P., Wang Y., Han J., Chen M., He Y., Zhang S. Which long noncoding RNAs and circular RNAs contribute to inflammatory bowel disease? Cell Death Dis. 2020;11:456. doi: 10.1038/s41419-020-2657-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lodde V., Murgia G., Simula E.R., Steri M., Floris M., Idda M.L. Long Noncoding RNAs and Circular RNAs in Autoimmune Diseases. Biomolecules. 2020;10:E1044. doi: 10.3390/biom10071044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cristóbal I., Caramés C., Rubio J., Sanz-Alvarez M., Luque M., Madoz-Gúrpide J., Rojo F., García-Foncillas J. Functional and Clinical Impact of CircRNAs in Oral Cancer. Cancers (Basel) 2020;12:E1041. doi: 10.3390/cancers12041041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dai F., Dai L., Zheng X., Guo Y., Zhang Y., Niu M., Lu Y., Li H., Hou R., Zhang Y. Non-coding RNAs in drug resistance of head and neck cancers: A review. Biomed. Pharmacother. 2020;127:110231. doi: 10.1016/j.biopha.2020.110231. [DOI] [PubMed] [Google Scholar]
- 49.Dong P., Xu D., Xiong Y., Yue J., Ihira K., Konno Y., Watari H. The Expression, Functions and Mechanisms of Circular RNAs in Gynecological Cancers. Cancers (Basel) 2020;12:E1472. doi: 10.3390/cancers12061472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nie H., Wang Y., Liao Z., Zhou J., Ou C. The function and mechanism of circular RNAs in gastrointestinal tumours. Cell Prolif. 2020;53:e12815. doi: 10.1111/cpr.12815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ren S., Lin P., Wang J., Yu H., Lv T., Sun L., Du G. Circular RNAs: Promising Molecular Biomarkers of Human Aging-Related Diseases via Functioning as an miRNA Sponge. Mol. Ther. Methods Clin. Dev. 2020;18:215–229. doi: 10.1016/j.omtm.2020.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang X., Mei J., Wang H., Gu D., Ding J., Liu C. The emerging roles of circular RNAs in ovarian cancer. Cancer Cell Int. 2020;20:265. doi: 10.1186/s12935-020-01367-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hu B., Zhong L., Weng Y., Peng L., Huang Y., Zhao Y., Liang X.J. Therapeutic siRNA: state of the art. Signal Transduct. Target. Ther. 2020;5:101. doi: 10.1038/s41392-020-0207-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mainini F., Eccles M.R. Lipid and Polymer-Based Nanoparticle siRNA Delivery Systems for Cancer Therapy. Molecules. 2020;25:E2692. doi: 10.3390/molecules25112692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang M., Weng Y., Cao Z., Guo S., Hu B., Lu M., Guo W., Yang T., Li C., Yang X., Huang Y. ROS-Activatable siRNA-Engineered Polyplex for NIR-Triggered Synergistic Cancer Treatment. ACS Appl. Mater. Interfaces. 2020;12:32289–32300. doi: 10.1021/acsami.0c06614. [DOI] [PubMed] [Google Scholar]
- 56.Ghahhari N.M., Babashah S. Interplay between microRNAs and WNT/β-catenin signalling pathway regulates epithelial-mesenchymal transition in cancer. Eur. J. Cancer. 2015;51:1638–1649. doi: 10.1016/j.ejca.2015.04.021. [DOI] [PubMed] [Google Scholar]
- 57.Royer P.J., Henrio K., Pain M., Loy J., Roux A., Tissot A., Lacoste P., Pison C., Brouard S., Magnan A., COLT consortium TLR3 promotes MMP-9 production in primary human airway epithelial cells through Wnt/β-catenin signaling. Respir. Res. 2017;18:208. doi: 10.1186/s12931-017-0690-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang S., Liu Y., Li M.Y., Ng C.S.H., Yang S.L., Wang S., Zou C., Dong Y., Du J., Long X. FOXP3 promotes tumor growth and metastasis by activating Wnt/β-catenin signaling pathway and EMT in non-small cell lung cancer. Mol. Cancer. 2017;16:124. doi: 10.1186/s12943-017-0700-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang J.H., Li J.H., Shao P., Zhou H., Chen Y.Q., Qu L.H. starBase: a database for exploring microRNA-mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data. Nucleic Acids Res. 2011;39:D202–D209. doi: 10.1093/nar/gkq1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang F., Wu H., Fan M., Yu R., Zhang Y., Liu J., Zhou X., Cai Y., Huang S., Hu Z., Jin X. Sodium butyrate inhibits migration and induces AMPK-mTOR pathway-dependent autophagy and ROS-mediated apoptosis via the miR-139-5p/Bmi-1 axis in human bladder cancer cells. FASEB J. 2020;34:4266–4282. doi: 10.1096/fj.201902626R. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.