Abstract
RNA interference (RNAi) is mediated by an ∼21-nt double-stranded small interfering RNA (siRNA) and shows great promise in delineating gene functions and in developing therapeutics for human diseases. However, effective gene silencing usually requires the delivery of multiple siRNAs for a given gene, which is often technically challenging and time-consuming. In this study, by exploiting the type IIS restriction endonuclease-based synthetic biology methodology, we developed the fast assembly of multiplex siRNAs (FAMSi) system. In our proof-of-concept experiments, we demonstrated that multiple fragments containing three, four, or five siRNA sites targeting common Smad4 and/or BMPR-specific Smad1, Smad5, and Smad8 required for BMP9 signaling could be assembled efficiently. The constructed multiplex siRNAs effectively knocked down the expression of Smad4 and/or Smad1, Smad5, and Smad8 in mesenchymal stem cells (MSCs), and they inhibited all aspects of BMP9-induced osteogenic differentiation in bone marrow MSCs (BMSCs), including decreased expression of osteogenic regulators/markers, reduced osteogenic marker alkaline phosphatase (ALP) activity, and diminished in vitro matrix mineralization and in vivo ectopic bone formation. Collectively, we demonstrate that the engineered FAMSi system provides a fast-track platform for assembling multiplexed siRNAs in a single vector, and thus it may be a valuable tool to study gene functions or to develop novel siRNA-based therapeutics.
Keywords: RNA interference, RNAi, double-stranded small interfering RNA, siRNA, BMP9/Smad signaling, mesenchymal stem cells, osteoblastic differentiation, multiplex siRNA expression
Graphical Abstract

RNA interference shows great promise in delineating gene functions and in developing therapeutics for human diseases. He et al. exploited the type IIS restriction endonuclease-based synthetic biology and developed the fast assembly of multiplex siRNAs (FAMSi) system, which streamlines RNAi-based gene function studies and the development of siRNA-based therapeutics.
Introduction
RNA interference (RNAi) was initially discovered in 1998 as a self-protection mechanism against invasion by RNA viruses in C. elegans. RNAi has since been found in a broad range of eukaryotes.1, 2, 3, 4, 5 It was subsequently demonstrated that 21- and 22-nt double-stranded RNAs could induce RNAi-mediated gene silencing without eliciting interferon responses in mammalian cells.6,7 Mechanistically, it is now well recognized that the RNAi posttranscriptional silencing process is carried out by the small interfering RNAs (siRNAs) homologous to a target gene through the action of the RNA-induced silencing complex (RISC).2,3,5 Such siRNAs have become ubiquitous and powerful tools for biomedical research both in vitro and in vivo, as they offer opportunities for facile inhibition of any gene simply based on its nucleotide sequence.4,5 Moreover, RNAi technology provides unique avenues for developing innovative therapeutics, which has been highlighted by the recent approval of the first RNAi therapeutic drug, Onpattro (patisiran), to treat polyneuropathy of hereditary transthyretin-mediated (hATTR) amyloidosis.3,5,8, 9, 10, 11
In practice, RNAi is usually accomplished either by transfecting chemically synthesized siRNAs into cells, or by expressing short hairpin RNAs (shRNAs) or siRNAs in cells.10, 11, 12, 13, 14, 15 Unfortunately, effective RNAi silencing frequently requires multiple siRNAs for a given gene, which is time-consuming to build multiple siRNA expression constructs. To simplify this technical process, we previously developed the pSOS system, in which a single siRNA is produced from the converging U6 and H1 promoters.16 To improve the construction efficiency, we explored the use of the Gibson DNA assembly technique and developed a one-step system (i.e., pSOK) to express multiple siRNA sites in a single vector.17 While the pSOK system has been successfully used for numerous siRNA-based studies,18, 19, 20, 21, 22, 23, 24, 25 the Gibson Assembly technique is highly empirical and finicky, rendering it not user-friendly for the researchers with limited experience in molecular cloning. To overcome such practical and technical challenges, we have recently developed a two-step BstXI-based shotgun cloning (BSG) system, consisting of multiple entry vectors with siRNA expression units (SiEUs) flanked with distinct BstXI sites, and a retroviral destination vector for shotgun SiEU assembly.26 While functional, the BSG system requires two-stage cloning and more entry vectors if more siRNAs are desired. Thus, a more simplistic and user-friendly system is warranted for the rapid assembly of multiplexed siRNAs in mammalian cells.
For the past decade, synthetic biology has revolutionized genome biology and offers unique approaches for creating new biological systems or redesigning existing ones for new purposes through the use of different techniques to assemble genetic fragments.27 One such technique is to use the type IIS restriction enzyme-dependent DNA assembly methods for rapid construction of large genetic fragments through ordering multi-fragment assembly dictated by the ligation of Watson-Crick base-paired overhangs.28 The type IIS restriction enzymes, such as BsaI and BbsI, recognize asymmetric DNA sequences and cleave outside of their recognition sequences. Thus, the fragments and recipient vectors are designed to place the type IIS recognition sites distal to the cleavage sites, so that type IIS enzymes can remove the recognition sequences after assembly. The type IIS enzyme-based seamless assembly system offers at least three advantages. First, the overhang sequence created is not dictated by the restriction enzymes, and yet no footprint sequence is retained. Second, the overhangs contain fragment-specific sequences, allowing orderly assembly of multiple fragments in one reaction. Third, the restriction sites can be eliminated from the final assembled products.
In this study, we exploited the type IIS restriction endonuclease-based synthetic biology methodology and developed a fast assembly of multiplex siRNAs (FAMSi) system to silence gene expression in mammalian cells. Through a series of proof-of-concept experiments, we demonstrated that multiple fragments containing three, four, or five siRNAs targeting the common Smad4, and/or BMP receptor-specific Smad1, Smad5, and Smad8 required for BMP9 osteogenic signaling could be efficiently assembled. The constructed multiplex siRNAs effectively knocked down the expression of Smad4 and/or Smad1, Smad5, and Smad8 in mesenchymal stem cells (MSCs), and inhibited all aspects of BMP9-induced osteogenic differentiation in bone marrow mesenchymal stem cells (BMSCs) in vitro and in vivo. Thus, we demonstrate that the engineered FAMSi system provides a one-step fast-track platform to assemble multiplexed siRNAs in a single vector, and thus may be a valuable tool to interrogate gene function and/or to develop novel and efficacious siRNA-based therapeutics.
Results
The FAMSi System Facilitates the Expression and Delivery of Multiplexed siRNAs
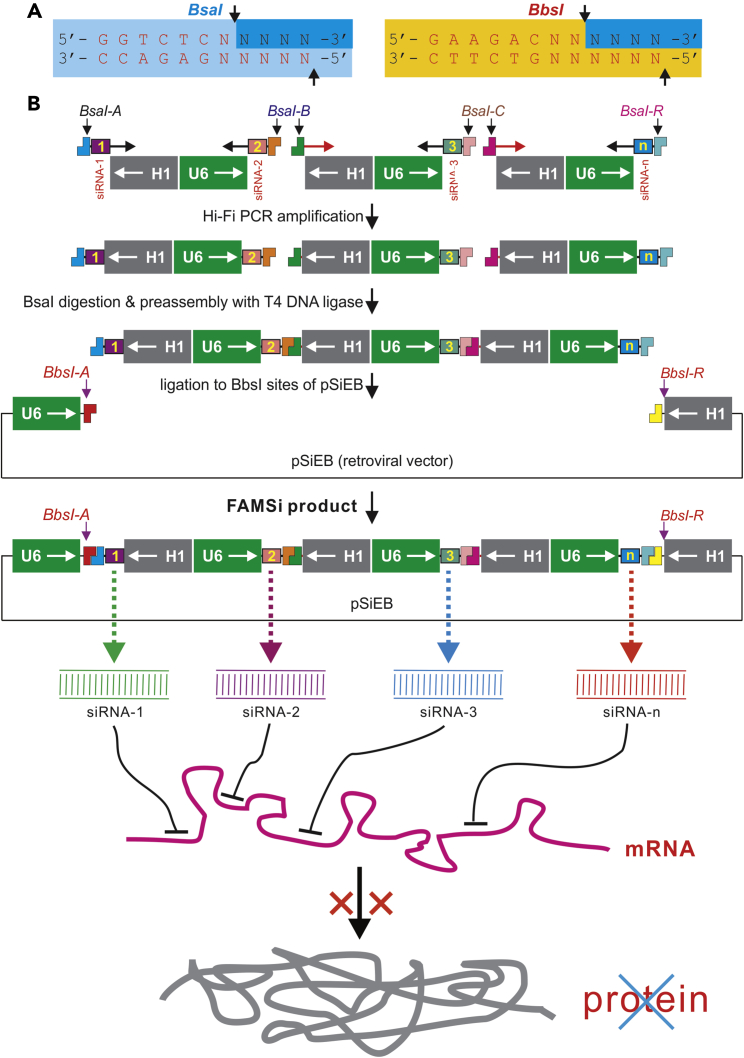
We have demonstrated that retroviral and adenoviral vector-mediated expression of siRNAs driven by convergent U6 and H1 promoters effectively silences target gene expression in mammalian cells in vitro and in vivo.16,17,19,26,29 To develop the FAMSi system that enables rapid construction of multiple siRNA expression cassettes through an ordered multi-fragment assembly determined by ligating Watson-Crick base-paired overhangs, we took advantage of two commonly used type IIS restriction enzymes, BsaI and BbsI, both of which cut downstream random sequences and leave a 4-nt 5′ end protruding overhang (Figure 1A). The choice of these two enzymes should broaden the utility of this system, as either or both should be unique in most vectors or potential siRNA target sites.
Figure 1.
Schematic Representation of the FAMSi System
(A) The recognition sequences and cutting features of the type IIS restriction endonucleases BbsI and BsaI used in the FAMSi system. The recognition sites are shown, where N represents any nucleotide, and the cleavage sites on both strands are indicated with arrows. (B) One-step assembly of the multiplexed U6-H1-driven siRNA expression cassettes into the pSiEB retroviral destination vector. A series of unique downstream NNNN sites (BsaI-A, BsaI-B, …, BsaI-R) for BsaI site is designed, while the pSiEB contains the U6- and H1-flanked, unique BbsI-A and BbsI-R sites, which share complementary overhangs to BsaI-A and BsaI-R, respectively. The siRNA sequences are incorporated into the BsaI site-containing PCR primers that amplify the back-to-back H1-U6 promoters engineered in the template vectors (pH1U6-T1 and pH1U6-T2). After parallel high-fidelity PCR amplification, multiple siRNA fragments are digested with BsaI, followed by preassembly and ligation into the BbsI-digested pSiEB vector. The correctly assembled multiplex siRNA expression vectors are identified after bacterial transformation and then functionally validated. The vector maps and sequences for pH1U6-T1, pH1U6-T2, and pSiEB are available in Figures S1, S2, and S3, respectively.
The FAMSi system consists of three vectors, that is, two template vectors, pH1U6-T1 and pH1U6-T2, and the destination vector pSiEB. The pH1U6-T1 and pH1U6-T2 vectors contain the H1 and U6 promoters in a back-to-back configuration and are used as templates for Hi-Fi PCR amplification with siRNA sequence-containing primers (Figures S1 and S2). Alternatively, the retroviral vector pSiEB contains a built-in U6-H1 promoter unit flanking the unique BbsI-A and BbsI-R sites, which share unique but complementary overhangs to BsaI-A and BsaI-R, respectively (Figure S3).
The one-step assembly of multiplexed U6-H1-driven siRNA cassettes into pSiEB starts with simultaneous Hi-Fi PCR amplification of back-to-back H1-U6 fragments with primers containing siRNA sequences and unique BsaI sites. The PCR-amplified fragments are then digested with BsaI and subjected to a quick preassembly with T4 DNA ligase. The desired preassembled fragments are gel-purified and subcloned into the BbsI-digested pSiEB vector, followed by transformation of competent bacterial cells and colony PCR identification (Figure 1B).
Three siRNA Sites Targeting Mouse Smad4 Can Be Efficiently Assembled into pSiEB Using the FAMSi System
To carry out the proof-of-principle experiments on the technical and functional feasibility of the FAMSi system, we chose to construct a series of siRNA vectors that target the BMP9-dependent Smad signaling pathway, as we and others have demonstrated that Smad4 and BMP receptor-specific Smad1/Smad5/Smad8 play an important role in BMP9-induced osteoblastic differentiation of MSCs.30, 31, 32, 33, 34, 35, 36, 37, 38
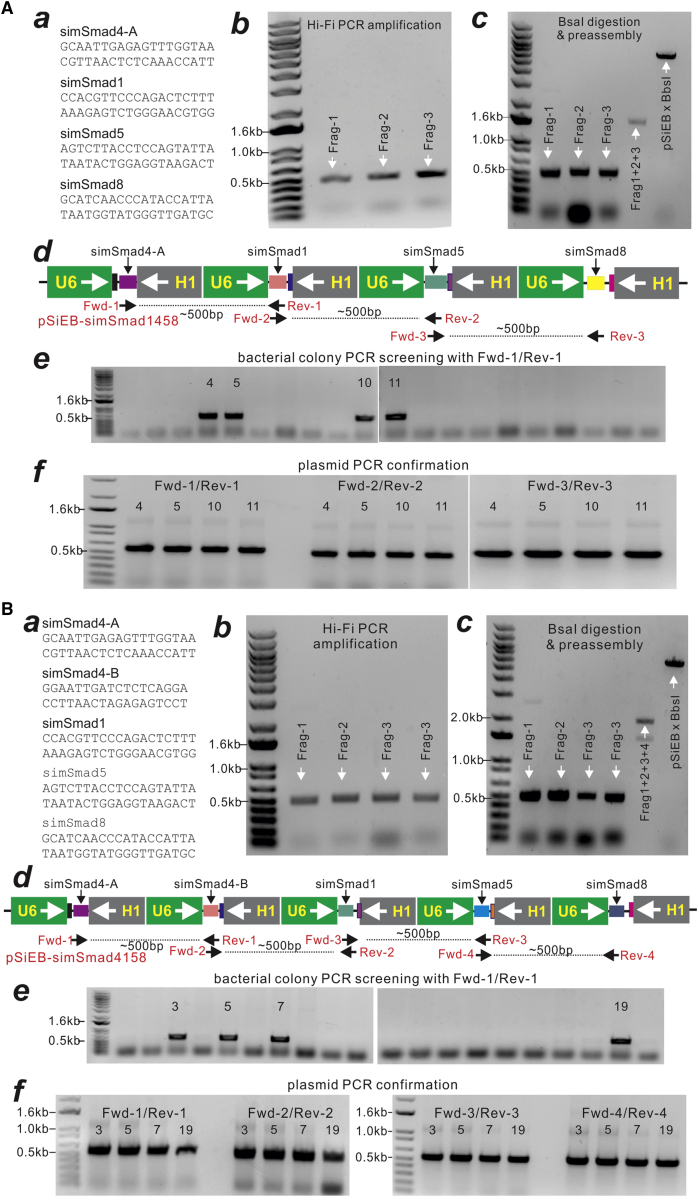
To simplify the validation process, we started with the construction of a vector that expresses three siRNAs targeting mouse Smad4 or simSmad4 (Figure 2A, a). We demonstrated that the two siRNA-containing fragments (∼0.5 kb each) were readily obtained through PCR amplification (Figure 2A, b). After BsaI digestion, these two fragments were effectively preassembled into a larger ∼1-kb fragment and subcloned into BbsI-digested pSiEB (Figure 2A, c). For bacterial colony PCR screening, primers were designed based on the ordered siRNA cassettes using primer pairs to amplify the sequences between neighboring siRNA sequences (Figure 2B, a). Among 20 randomly picked bacterial colonies, 15 showed positive PCR screening results (Figure 2B, b), which was further confirmed by two pairs of PCR primers at the plasmid DNA level (Figure 2B, c). All the positive clones were also confirmed by restriction enzyme digestions and DNA sequencing reactions (data not shown). These constructs were pooled and designated as pSiEB-simSmad4 for further functional assays. Therefore, the above results demonstrate that the FAMSi system offers a highly efficient approach to the construction of three siRNA sites for any RNAi studies.
Figure 2.
One-Step Assembly of Three siRNAs Targeting Mouse Smad4 (pSiEB-simSmad4)
(A) The three siRNA sequences targeting mouse Smad4 (i.e., simSmad4) (a) are designed to PCR amplify and generate two fragments (b), which are further cut with BsaI, preassembled, and then cloned into the BbsI-cut pSiEB vector (c). (B) (a) The assembled siRNA fragments, the locations of PCR primers, and expected product sizes for colony PCR screening are depicted. (b) Fifteen of the randomly picked 20 clones were positive by PCR screening with the indicated primer pair. (c and d) The plasmid DNA purified from the same 15 clones was further confirmed by PCR using the indicated primer pairs. All PCR products were resolved on 1% agarose gels and visualized by ethidium bromide. Representative results are shown.
Four and Five siRNA Sites Targeting Mouse Smad1/Smad4/Smad5/Smad8 Can Be Assembled into pSiEB in One Step Using the FAMSi System
An ideal versatile siRNA expression system should allow one to express more than three siRNAs in one vector in order to silence the expression of multiple genes of interest. Thus, we further tested how efficiently multiple siRNA sites could be effectively assembled into pSiEB with the FAMSi system. We first tested the feasibility of assembling four siRNAs that target mouse Smad1/Smad4/Smad5/Smad8, with one siRNA site for each Smad gene (Figure 3A, a). We found that the three fragments containing the four simSmad sites were effectively PCR amplified (∼0.5 kb each) (Figure 3A, b). Furthermore, the three BsaI-digested fragments were readily preassembled into one large fragment of ∼1.5 kb (Figure 3A, c), followed by subcloning into BbsI-digested pSiEB, yielding pSiEB-simSmad1458 (Figure 3A, d). From the bacterial colony screening, 4 of the 20 randomly picked colonies were PCR positive (Figure 3A, e), and the four positive clones were confirmed by three pairs of PCR primers (Figure 3A, f), which were further confirmed by restriction digestions and DNA sequencing (data not shown). These confirmed clones were pooled and designated as pSiEB-simSmad1458.
Figure 3.
One-Step Assembly of Four and Five siRNAs Targeting Mouse Smad1/4/5/8 (pSiEB-simSmad1458 and pSiEB-simSmad4158)
(A) Assembly of four siRNAs targeting mouse Smad1/4/5/8 (pSiEB-simSmad1458). The four siRNA sequences targeting mouse Smad1/4/5/8 (i.e., simSmad1458) (a) are designed to PCR amplify and generate the three fragments (b), which are further cut with BsaI, preassembled, and then subcloned into the BbsI-cut pSiEB vector (c). (d) The assembled siRNA fragments, the locations of PCR primers, and expected product sizes for colony PCR screening are depicted. (e) Four of the randomly picked 20 clones were positive by PCR screening with the indicated primer pair. (f) The plasmid DNA purified from the same four clones was further confirmed by PCR using three pairs of primers. (B) Assembly of five siRNAs targeting mouse Smad4/1/5/8 (pSiEB-simSmad4158). (a–c) The five siRNA sequences targeting mouse Smad1/4/5/8 (i.e., simSmad4158) (a) are designed to PCR amplify and generate the four fragments (b), which are further cut with BsaI, preassembled, and then subcloned into BbsI-cut pSiEB vector (c). (d) The assembled siRNA fragments, the locations of PCR primers, and the expected product sizes for colony PCR screening are depicted. (e) Four of the randomly picked 20 clones were positive by PCR screening with the indicated primer pair. (f) The plasmid DNA purified from the same four clones was further confirmed by PCR using four pairs of primers. All PCR products were resolved on 1% agarose gels and visualized by ethidium bromide. Representative results are shown.
We then tested the feasibility and efficiency of assembling five siRNA sites using the FAMSi system by constructing five siRNAs that target mouse Smad1/Smad4/Smad5/Smad8 (with two siRNA sites for Smad4 and one siRNA site each for the other three Smads) (Figure 3B, a). We found that the four fragments containing the five simSmad sites were effectively PCR amplified (∼0.5 kb each) (Figure 3B, b). The four BsaI-digested fragments were readily preassembled into a larger fragment of ∼2.0 kb (Figure 3B, c), followed by subcloning into BbsI-digested pSiEB, yielding pSiEB-simSmad4158 (Figure 3B, d). It is noteworthy that a partially preassembled product of ∼1.5 kb was also observed (Figure 3B, c), suggesting that the preassembly of more than four (including four) fragments may require a longer ligation time to ensure the full assembly. From the bacterial colony screening, 4 of the 20 randomly picked colonies were PCR positive (Figure 3B, e), and the four positive clones were confirmed by the four pairs of PCR primers (Figure 3B, f), which were further confirmed by restriction digestions and DNA sequencing (data not shown). These confirmed clones were pooled and designed as pSiEB-simSmad4158.
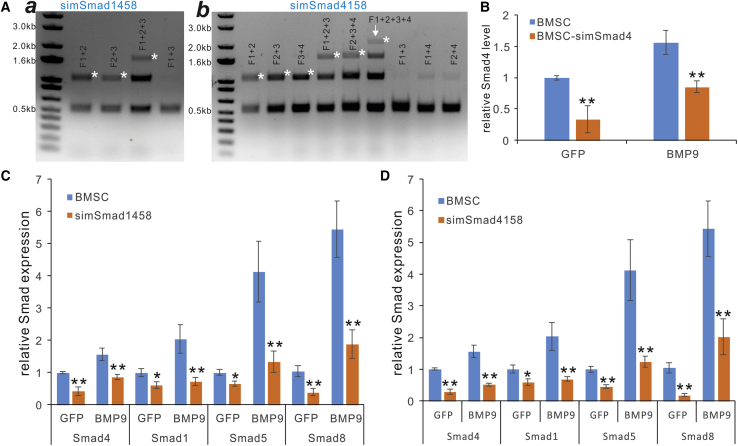
We also analyzed the assembly specificity of the PCR-amplified fragments. For the four-siRNA three-fragment assembly, we found that Frag1+2, Frag2+3, and Frag1+2+3 yielded the expected assembled product of ∼1.5 kb, whereas no assembled product was found in the Frag1+3 assembly reaction (Figure 4A, a). Similarly, for the assembly of five-siRNA four fragments, we detected the presence of expected products assembled from the neighboring siRNA fragments, whereas no or very few products were observed in random assembly reactions (Figure 4A, b). It is noteworthy that intermediate products were detected in these assembly reactions, suggesting that longer ligation time may be required to ensure the full assembly (Figure 4A). Nonetheless, the specificity analysis indicated that the tested four and five siRNA-containing fragments with unique BsaI sites were assembled in the predesigned orderly fashion with high specificity. Taken together, the above results demonstrate that the one-step assembly of multiple siRNA sites using the FAMSi system is technically feasible with high efficiency.
Figure 4.
Seamless and Orderly Assembly of Multiplex siRNA Fragments and Functional Characterization of the Multiplex siRNA Expression Vectors
(A) (a and b) The four and five siRNA fragments containing unique BsaI sites were assembled in the predesigned orderly fashion with high specificity. The desired PCR products and preassembled products are indicated with asterisks. Note that fragments that were out of order yielded no or very weak “assembled” products. (B) The simSmad4 stably expressed in BMSCs (BMSC-simSmad4) effectively knocked down Smad4 expression at the basal level (GFP) or stimulated by BMP9 as assessed by touchdown quantitative real-time PCR. (C) The simSmad1458 stably expressed in BMSCs simultaneously knocked down Smad1, Smad4, Smad5, and Smad8 expression at the basal level (GFP) or stimulated by BMP9 as assessed by touchdown quantitative real-time PCR. (D) The simSmad4158 stably expressed in BMSCs simultaneously knocked down Smad1, Smad4, Smad5, and Smad8 expression at the basal level (GFP) or stimulated by BMP9 as assessed by touchdown quantitative real-time PCR. ∗p < 0.05, ∗∗p < 0.01, compared with that of the siControl counterpart (i.e., BMSCs).
The FAMSi-Mediated Expression of simSmad Effectively Silences Smad Expression and Inhibits BMP9-Induced Osteoblastic Differentiation of MSCs
We analyzed the silencing efficiency for the above multiplex siRNAs by establishing retrovirally transduced stable lines in the mouse MSC line BMSCs, resultant BMSC-simSmad4, BMSC-simSmad1458, and BMSC-simSmad4158, and the siControl line BMSCs. In BMSC-simSmad4 cells, touchdown quantitative real-time PCR analysis revealed that Smad4 expression significantly decreased to approximately 33% and 54% of that of BMSCs when infected with Ad-GFP and Ad-BMP9, respectively (Figure 4B). The silencing efficiency of the Smads was further confirmed by western blotting analysis with Smad4 and Smad1/5/8 antibodies (Figure S4).
In the BMSC-simSmad1458 cells infected with Ad-GFP (i.e., silencing efficiency of endogenous Smads), we found that the expression levels for Smad4, Smad1, Smad5, and Smad8 decreased to approximately 42%, 60%, 65%, and 37% of that of the Ad-GFP-infected BMSCs. Accordingly, in the BMSC-simSmad1458 cells infected with Ad-BMP9, the expression levels of the four Smad genes significantly decreased, compared with that of the Ad-BMP9-infected BMSCs (Figure 4C). In particular, simSmad1458 effectively blunted the BMP9-induced expression of BMPR-specific Smad1, Smad5, and Smad8 (Figure 4C). Similar results were obtained in Ad-GFP- and Ad-BMP9-infected BMSC-simSmad4158 cells when compared with the infected BMSCs (Figure 4D). Collectively, these results strongly demonstrated that FAMSi-mediated expression of multiplexed siRNAs that either target a single gene or multiple genes can accomplish profound silencing effects on target expression.
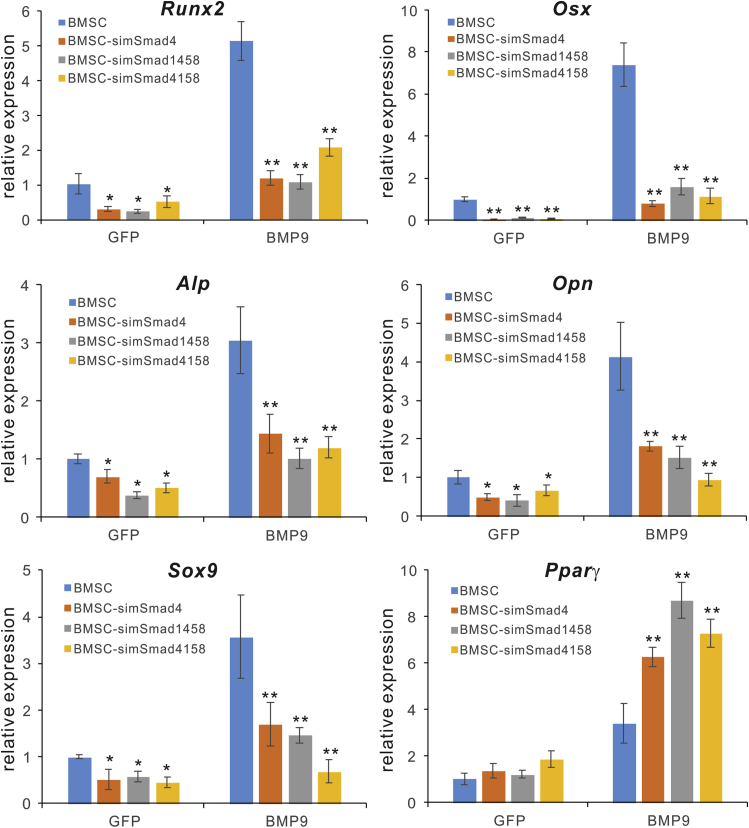
We further tested whether silencing Smad genes would impact BMP9-induced osteogenic signaling in MSCs. The three BMSC-simSmad lines, along with the control BMSCs, were infected with Ad-GFP or Ad-BMP9. The basal expression (i.e., GFP groups) of osteogenic regulators Runx2 and osterix (Osx), as well as osteogenic markers Alp and osteopontin (Opn), was noticeably reduced in targeted cells, compared with that of the control BMSCs (Figure 5). More importantly, BMP9-induced expression of Runx2, Osx, Alp, and Opn, which are known to be upregulated by BMP9,38, 39, 40, 41 was significantly inhibited in the three BMSC-simSmad lines, compared with that of the control BMSC line (Figure 5). Furthermore, BMP9-induced Sox9 expression was also reduced in the three BMSC-simSmad lines, compared with that of the control BMSC line (Figure 5). Interestingly, silencing Smads in BMSCs significantly enhanced BMP9-induced expression of Pparγ (Figure 5), suggesting that blockade of BMP9-induced osteogenic differentiation may be beneficial to adipogenic differentiation of MSCs.
Figure 5.
The Smad-Targeting siRNAs Effectively Inhibit BMP9-Stimulated Osteogenic Markers in BMSCs
The BMSCs stably expressing simSmad4, simSmad1458, simSmad4158 or siControl cells (i.e., BMSCs) were infected with AdGFP or AdBMP9 for 48 h. Total RNA was isolated and subjected to touchdown quantitative real-time PCR using primers specific for mouse Rnux2, Osx, Alp, Opn, Sox9, and Pparγ (Gapdh as the reference gene). ∗p < 0.05, ∗∗p < 0.01, compared with that of the siControl counterpart (BMSCs).
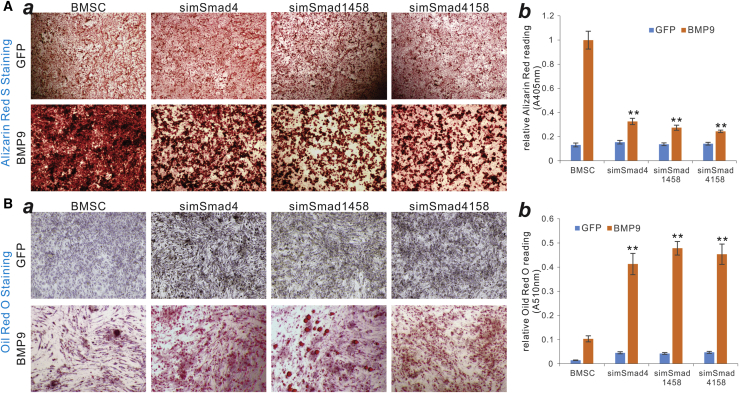
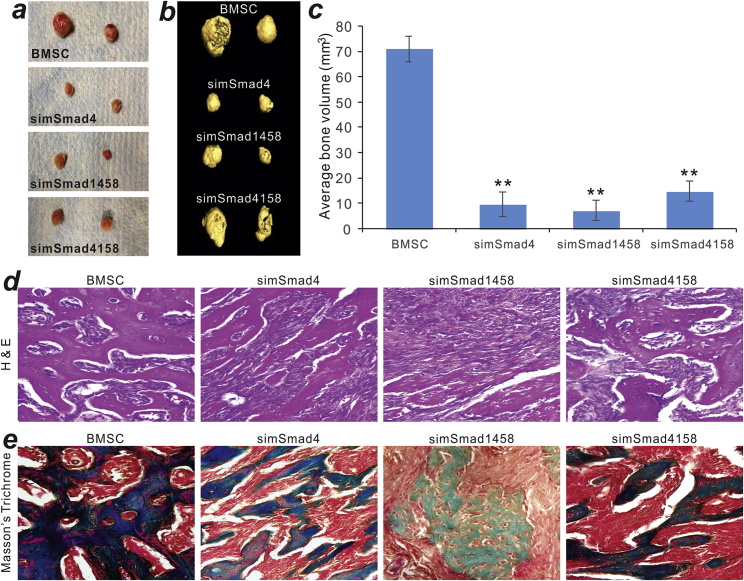
Next, we assessed the effect of silencing Smads on BMP9-induced early osteogenic marker ALP activity. Histochemical staining assays revealed that, compared with the control BMSCs, BMP9-induced ALP activity staining was inhibited in the three BSMC-simSmad lines at days 3 and 7 after Ad-BMP9 infection (Figure 6A, a and b). The quantitative assays further demonstrated that BMP9-induced ALP activity was significantly inhibited in the three BSMC-simSmad lines at days 3, 5, and 7 after Ad-BMP9 infection, compared with that in the control BMSCs (Figure 6B). Furthermore, silencing Smads in MSCs significantly diminished the BMP9-induced in vitro matrix mineralization as revealed by alizarin red S staining (Figure 7A, a and b). On the contrary, knockdown of Smad expression in MSCs enhanced BMP9-induced adipogenic differentiation as indicated by oil red O staining (Figure 7B, a and b), suggesting that BMP9 may induce adipogenic differentiation at least in part through a Smad-independent mechanism. Lastly, we analyzed the effect of silencing Smads on BMP9-induced ectopic bone formation capability and found that silencing Smad4 and/or Smad1/5/8 significantly diminished the sizes and volumetric measures of the ectopic bone masses in athymic nude mice (Figures 8A–8C). Histological analysis further indicated that, compared to that of the control BMSC group, silencing Smad expression effectively decreased BMP9-induced trabecular bone formation (Figure 8D). Trichrome staining also demonstrated that more immature bone or unmineralized bone matrix was detected in the three simSmad lines (Figure 8E).
Figure 6.
The Smad-Targeting siRNAs Effectively Inhibit BMP9-Induced Early Osteogenic Marker Alkaline Phosphatase (ALP) Activity in BMSCs
The BMSCs stably expressing simSmad4, simSmad1458, simSmad4158 or siControl cells (i.e., BMSCs) were infected with AdGFP or AdBMP9. (A and B) ALP activity was histochemically stained (A) and quantitatively assessed (B) at day 3 (a), day 5, and day 7(b). ALP activity was quantitatively determined by using a modified Great Escape SEAP chemiluminescence assay. Representative results are shown. ∗∗p < 0.01, compared with that of the siControl counterpart (BMSCs).
Figure 7.
The Smad-Targeting siRNAs Effectively Inhibit BMP9-Induced Bone Matrix Mineralization while Enhancing BMP9-Induced Adipogenic Differentiation in BMSCs
The BMSCs stably expressing simSmad4, simSmad1458, simSmad4158, or siControl cells (BMSCs) were infected with AdGFP or AdBMP9. (A) At 10 days after infection, the cells were fixed and subjected to alizarin red S staining and oil red O staining (a, day 3; b, day 7). Representative images are shown. (B) Both alizarin red S stain and oil red O stain were dissolved and quantitatively measured. ∗∗p < 0.01, compared with the BMSC-siControl.
Figure 8.
Silencing Mouse Smads Effectively Inhibits BMP9-Induced Osteogenic Differentiation of BMSCs
The BMSCs stably expressing simSmad4, simSmad1458, simSmad4158, or siControl cells (BMSCs) were infected with AdGFP or AdBMP9 for 36 h and collected for subcutaneous injection into the flanks of athymic nude mice (n = 5 per group). At 4 weeks after implantation, animals were sacrificed, and bony masses at the injection sites were retrieved. (A) Representative gross images are shown. No detectable masses were retrieved from the GFP group. (B) The retrieved masses were subjected to μCT imaging, and representative results of the 3D reconstruction of scanning are shown. (C) The average bone volumes of the retrieved bony masses were quantitatively analyzed by using Amira software. ∗∗p < 0.01, simSmad groups versus the BMSC group. (D and E) The retrieved samples were further decalcified and subjected to hematoxylin and eosin staining (D) and Masson’s trichrome staining (E). Representative images are shown.
Nonetheless, it is noteworthy that the decrease in BMP9-induced ALP activity, in vitro matrix mineralization, and ectopic bone formation in MSCs were not statistically different among the three simSmad lines, suggesting that the FAMSi-mediated expression of three siRNAs targeting Smad4 expression alone (i.e., BMSC-simSmad4) may exert similar biological effects on BMP9-induced osteogenic signaling in MSCs, compared to the two lines expressing siRNAs targeting Smad1, Smad5, and Smad8, in addition to Smad4 (i.e., BMSC-simSmad1458 and BMSC-simSmad41258).
Discussion
By exploiting the type IIS restriction endonuclease-based synthetic biology approach, we have developed the simple, efficient, and user-friendly FAMSi technology for the rapid assembly of multiplex siRNA expression cassettes. In our proof-of-principle experiments, we demonstrated that fragments containing three, four, or five siRNA sites targeting the BMP9/Smad signaling pathway could be efficiently assembled, and their gene expression silencing efficacy was fully validated by a series of in vitro and in vivo biological functional assays in MSCs. Therefore, the engineered FAMSi system may be a valuable tool to explore siRNA biology.
During the process of developing and optimizing the FAMSi system, we learned several important technical considerations. First, while it is theoretically feasible to bypass the preassembly step of all desirable fragments, we found practically that a direct shotgun approach by pooling all digested fragments and destination vector yielded 5- to 10-fold lower cloning efficiency. Second, we performed gel purifications of PCR-amplified and BsaI-digested fragments and achieved high assembly efficiency, while the unpurified fragments decreased the assembly efficiency by approximately 6-fold. Third, it is important to use the neighboring siRNA sequences as PCR primer pairs to perform bacterial colony PCR screening due to the repeated U6 and H1 promoter sequences. Otherwise, tandem or laddered products lead to inconclusive screening results. Lastly, while we only designed up to five siRNA cassettes in this proof-of-concept study, it is conceivable that the FAMSi system could accommodate more than five siRNA sites, as long as unique sequences of the 5′ end overhang of BsaI are used and validated. Therefore, the FAMSi system is highly versatile and flexible in this regard. Furthermore, it is noteworthy that the current FAMSi system can be easily reconfigured and expressed in different viral vector systems, such as adenoviral vectors, adenovirus-associated viral vectors, lentiviral vectors, and HSV vectors, to name a few.
Numerous efforts have been devoted to the development of effective construction and/or delivery systems for multiple siRNAs. In one of our early efforts, we developed the pSOS vector system, in which a single siRNA is produced by convergent U6 and H1 promoters.16 In order to accommodate the expression of multiple siRNA cassettes in a single vector, we later developed a one-step pSOK system by exploiting the use of the Gibson DNA assembly technology.17 However, the Gibson DNA assembly methodology is finicky and empirical. More recently, we developed the BSG restriction enzyme-based two-stage system to express three siRNA sites.26 While we and others have successfully used these vector systems to accomplish siRNA-mediated gene silencing,18,19,22, 23, 24, 25,42 these above systems are not user-friendly and require prospective users to possess significant experience in molecular cloning. In this study, we developed a simple and more user-friendly FAMSi system to accomplish the expression of multiple siRNAs in mammalian cells. It is noteworthy that the FAMSi system can be easily adapted to express multiple single guide RNAs (sgRNAs) for CRISPR-Cas9-based genome-editing investigation.43, 44, 45, 46
Several other strategies for single vector-based multiple siRNA and/or sgRNA expression have also been reported. Similar to our pSOK system,17 the string assembly guide RNA (gRNA) cloning (STAgR) technique involves a single-step multiplexing of gRNAs by using the N20 targeting sequences as necessary homologies for Gibson assembly.47 The adaptable system for assembly of multiplexed plasmids (ASAP) methodology allows the assembly of multiple gRNA cassettes via the “PCR-on-ligation” process.48 However, it is technically challenging to modify these systems for siRNA expression, as siRNAs necessitate the expression of both strands, which is not required for sgRNA expression.
In summary, we exploited the type IIS restriction endonuclease-based synthetic biology approach and developed the efficient and user-friendly FAMSi technology for the rapid assembly of multiplex siRNA expression cassettes. In a series of proof-of-concept experiments, we demonstrated that multiple fragments containing three, four, or five siRNA sites targeting the common Smad4 and/or Smad1, Smad5, and Smad8 required for the BMP9/Smad signaling pathway could be efficiently assembled. The constructed multiplex siRNAs were shown to effectively knock down the expression of Smad4 and/or Smad1, Smad5, and Smad8 in MSCs, and to inhibit all aspects of BMP9-induced osteogenic differentiation in BMSCs, including the decreased expression of osteogenic regulators and markers, a reduced ALP activity, and diminished in vitro matrix mineralization and in vivo ectopic bone formation. Collectively, our findings demonstrate that the engineered FAMSi system provides a one-step fast-track technology to express multiplexed siRNAs in a single vector, and thus it should be a valuable tool to investigate gene functions and/or to develop innovative siRNA-based therapeutics.
Materials and Methods
Cell Culture and Chemicals
The mouse MSC line imBMSCs (referred to as BMSCs hereafter) is conditionally immortalized mouse BMSCs as previously described.49 The 293pTP and RAPA cells were previously characterized HEK293 derivative cells.50,51 The above cells were cultured in DMEM containing 10% fetal bovine serum (FBS, Gemini Bio-Products, West Sacramento, CA, USA), supplemented with 100 U/mL penicillin and 100 μg/mL streptomycin at 37°C in 5% CO2 as described.19,26,41,52, 53, 54 Restriction endonucleases, NEBuilder HiFi DNA assembly master mix, Phusion high-fidelity DNA polymerase, and the electrocompetent DH10B cells were purchased from New England Biolabs (NEB, Ipswich, MA, USA). Blasticidin S was ordered from InvivoGen (San Diego, CA, USA). Unless indicated otherwise, other chemicals/reagents were purchased from Thermo Fisher Scientific (Waltham, MA, USA) or Sigma-Aldrich (St. Louis, MO, USA).
Development of the FAMSi Vectors
The FAMSi system consists of three vectors, two template vectors (pH1U6-T1 and pH1U6-T2) and a destination vector (pSiEB). Both template vectors were constructed on the base of our homemade pMOLuc and pMOK vectors17,55,56 by incorporating the back-to-back human H1 and U6 promoters. The pH1U6-T1 and pH1U6-T2 vectors have a slight sequence difference at the 3′ end of the H1 promoter region. The pH1U6-T1 vector is used for the amplification of the first fragment, while the pH1U6-T2 vector is used for amplifying all other fragments. The full-length sequences and vector maps for pH1U6-T1 and pH1U6-T2 are shown in Figures S1 and S2.
The destination vector pSiEB was constructed through a series of subcloning experiments on the base of our previously reported retroviral siRNA expression systems, such as the pSEB61 vector, which has been used to express single siRNA driven by convergent U6 and H1 promoters.16,17,26 Briefly, the BbsI site in the blasticidin S coding region was first silently mutated through the Gibson assembly approach by following the manufacturer’s instructions. Then, a long oligonucleotide cassette containing both BbsI-A and BbsI-R sites was inserted between the convergent U6 and H1 promoters through Gibson assembly. The full-length sequence and vector map for pSiEB are shown in Figure S3. All cloning junctions and inserted oligonucleotide fragments were DNA sequencing verified. Details about the vector construction are available upon request.
General Guidelines for Hi-Fi PCR Amplification and Assembly of Multiplex siRNA Fragments Using the FAMSi System
The siRNA targeting sequences were designed by using either BLOCK-iT RNAi designer (Invitrogen, Carlsbad, CA, USA) or siDESIGN (Dharmacon/Horizon Discovery). The siRNA sites were incorporated into the PCR primers that were used to amplify the back-to-back H1-U6 fragments from pH1U6-T1 and pH1U6-T2 using the Phusion Hi-Fi PCR system. The amplified fragments were cut with BsaI and preassembled with T4 DNA ligase for 30 min at room temperature, followed by the gel purification of the desired preassembled products. Lastly, the purified preassembled products were subcloned into the BbsI-digested pSiEB, followed by electroporation transformation into DH10B cells for bacterial culture. Colony PCR was performed to identify potential positive recombinants for further analyses.
Establishment of BMSC Stable Lines Expressing siRNAs Targeting Smads
The retroviral vector-mediated transduction and establishment of stable cell lines were carried out as previously described.19,22,53,54,57, 58, 59, 60 Briefly, the retroviral transfer vectors pSiEB-simSmad4, pSiEB-simSmad1458, and pSiEB-simSmad4158 were co-transfected with pCL-Ampho into subconfluent 293 Phoenix Ampho (293PA) cells to package retroviruses. The retrovirus-containing supernatants were harvested at 36–72 h after transfection and pooled to infect subconfluent BMSCs. At 48 h post-infection, the cells were subjected to blasticidin S selection (5 μg/mL). The resulting stable lines were designated as BMSC-simSmad4, BMSC-simSmad1458, and BMSC-simSmad4158. The previously constructed pSEB-siControl26 was used to establish a stable control line, designated as BMSCs. All stable lines were kept in LN2 tanks for future use.
Construction and Amplification of Recombinant Adenoviral Vectors Ad-BMP9 and Ad-GFP
Recombinant adenoviruses were created by using the AdEasy technology as reported.38,41,61, 62, 63 Briefly, the coding region of human BMP9 was PCR amplified and subcloned into an adenoviral shuttle vector to generate the recombinant adenoviral vector pAd-BMP9, which was then used to produce and/or amplify the adenovirus Ad-BMP9 in 293pTP or RAPA cells.50,51 Ad-BMP9 also co-expressed GFP as reported.20,21,64,65 The Ad-GFP was used as a mock virus control.66, 67, 68 Polybrene (4–8 μg/mL) was added to all adenoviral infections to enhance viral transduction efficiency as reported.69
RNA Isolation and Touchdown Quantitative Real-Time PCR
Total RNA was purified by using the NucleoZOL RNA isolation kit (Takara Bio USA, Mountain View, CA, USA) and subjected to reverse transcriptase (RT) reactions using hexamer and Moloney murine leukemia virus (M-MuLV) RT (NEB). The resultant RT cDNA products were properly diluted and used as touchdown quantitative real-time PCR templates. Touchdown quantitative real-time PCR primers were designed with the Primer3 Plus program.70 Touchdown quantitative real-time PCR reactions were carried out as described.21,71, 72, 73, 74. Briefly, the SYBR Green (Bimake, Houston, TX, USA) qPCR reactions were carried out by using the following cycling program: 95°C for 3 min for 1 cycle; 95°C for 20 s, 66°C for 10 s per cycle, and then −3°C per cycle for four cycles; followed by 95°C for 10 s, 55°C for 15 s, and 70°C for 1 s for 40 cycles. Gapdh was used as a reference gene. The gene expression calculations were performed using the 2−ΔΔCt method.37,56,57 Touchdown quantitative real-time PCR primer sequences are listed in Table S1.
ALP Assays
ALP activities were assessed both qualitatively and quantitatively by using histochemical staining and a modified Great EscAPe secreted alkaline phosphatase (SEAP) chemiluminescence assay (Takara Bio USA), respectively, as described.75, 76, 77 Each assay condition was done in triplicate.
Matrix Mineralization Assay (Alizarin Red S Staining)
The cells were infected with Ad-BMP9 or Ad-GFP and cultured with 10% FBS DMEM containing ascorbic acid (50 mg/mL) and β-glycerophosphate (10 mM) for the indicated time periods. Alizarin red S staining was carried out to assess mineralized matrix nodules as described.20,64,78, 79, 80, 81 The staining of calcium mineral deposits was documented under a bright field microscope. For quantitative assays, the alizarin red stain was dissolved in 10% acetic acid, and the absorbance was measured photometrically at 405 nm. Each assay was done in triplicate.
Oil Red O Staining Assay
Subconfluent BMSCs were infected with indicated adenoviruses for 7 or 14 days. Oil red O staining was carried out as described.22,49,82,83 Briefly, the cells were fixed with 10% formalin at room temperature for 10 min and washed with PBS. After residual PBS being aspirated, the cells were stained with oil red O solution, placed in a 37°C incubator for 1 h, and washed with tap water. The stained lipid droplets were documented under a bright field microscope. For quantitative assays, the oil red O stain was eluted in 2-propanol and the absorbance was measured photometrically at 510 nm. Each assay was done in triplicate.
Subcutaneous Stem Cell Implantation and Ectopic Bone Formation
The use and care of athymic nude mice were approved by the Institutional Animal Care and Use Committee. All experimental procedures were carried out according to the approved guidelines. The subcutaneous stem cell implantation procedure was carried out as previously described.24,25,66,84 Briefly, subconfluent BMSCs were co-infected with Ad-BMP9 or Ad-GFP for 36 h and then collected, resuspended in PBS (∼50 μL per injection), and subcutaneously injected into the flanks of nude mice (Envigo/Harlan Laboratories; n = 5/group, female, 5 weeks old; 2 × 106 cells per injection). At the indicated endpoints, nude mice were sacrificed and the masses were retrieved for micro-computed tomography (μCT) imaging and histologic evaluation as described.22,24,59,85
μCT Analysis
The retrieved masses were fixed in 10% formalin and imaged using the X-CUBE (Molecubes, Gent, Belgium) at the Integrated Small Animal Imaging Research Resource (iSAIRR) of The University of Chicago. Spiral high-resolution CT acquisitions were performed with an X-ray source of 50 kVp and 200 μA. Volumetric CT images were reconstructed in a 350 × 350 × 840 format with voxel dimensions of 200 μm3. All image data were analyzed by Amira 5.3 (Visage Imaging), and 3D volumetric data and bone density were calculated as described.24,52,76,86
Hematoxylin and Eosin (H&E) and Masson’s Trichrome Staining
After μCT imaging, the fixed samples were decalcified, paraffin-embedded, and subjected to serial sections and H&E staining and Masson’s trichrome staining as described.59,75,85,87
Statistical Analysis
All quantitative experiments were carried out in triplicate and/or repeated in three independent batches. Data are expressed as mean ± standard deviation (SD). Statistical significance was analyzed by using the one-way analysis of variance. p < 0.05 was considered statistically significant.
Author Contributions
J.F., F.H., H.G., and T.-C.H. conceived and designed the study. F.H., N.N., Z.Z., D.W., A.J.L., B.L., A.F.L., and K.Q. performed the experiments and collected data. Y.F., E.W., X. Wang, X. Wu, H.L., J.Z., M.Z., Y.M., M.P., W.W., Y.Z., C.N., H.W., L.H., D.S., Q.L., X.Z., and K.F. participated in molecular cloning experiments, provided essential experimental materials, assisted in histological preparations, and provided qPCR data analysis and interpretations. T.-C.H., J.F., F.H., H.G., R.R.R., J.M.W., M.J.L., K.H., J.S., and M.E.D. drafted and revised the manuscript. All authors reviewed and approved the manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
The authors want to thank the technical support provided by the Integrated Small Animal Imaging Research Resource (iSAIRR) Faculty at The University of Chicago. The reported work was supported in part by research grants from the National Institutes of Health (CA226303 to T.-C.H.) and the Scoliosis Research Society (to T.-C.H. and MJL). Z.Z. was supported by the China Postdoctoral Science Foundation (2019M663446) and the Postdoctoral Program of the Natural Science Foundation of Chongqing, China (cstc2019jcyj-bsh0006). W.W. was supported by the Medical Scientist Training Program of the National Institutes of Health (T32 GM007281). This project was also supported in part by The University of Chicago Cancer Center Support Grant (P30CA014599) and the National Center for Advancing Translational Sciences of the National Institutes of Health through grant no. UL1 TR000430. T.-C.H. was supported by the Mabel Green Myers Research Endowment Fund and The University of Chicago Orthopaedics Alumni Fund. Funding sources were not involved in the study design; in the collection, analysis and/or interpretation of data; in the writing of the report; or in the decision to submit the paper for publication.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.10.007.
Contributor Information
Tong-Chuan He, Email: tche@uchicago.edu.
Jiaming Fan, Email: fanjiaming1988@cqmu.edu.cn.
Supplemental Information
References
- 1.Fire A., Xu S., Montgomery M.K., Kostas S.A., Driver S.E., Mello C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Hammond S.M., Bernstein E., Beach D., Hannon G.J. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 3.Bobbin M.L., Rossi J.J. RNA interference (RNAi)-based therapeutics: delivering on the promise? Annu. Rev. Pharmacol. Toxicol. 2016;56:103–122. doi: 10.1146/annurev-pharmtox-010715-103633. [DOI] [PubMed] [Google Scholar]
- 4.Castel S.E., Martienssen R.A. RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat. Rev. Genet. 2013;14:100–112. doi: 10.1038/nrg3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Setten R.L., Rossi J.J., Han S.P. The current state and future directions of RNAi-based therapeutics. Nat. Rev. Drug Discov. 2019;18:421–446. doi: 10.1038/s41573-019-0017-4. [DOI] [PubMed] [Google Scholar]
- 6.Elbashir S.M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 7.Caplen N.J., Parrish S., Imani F., Fire A., Morgan R.A. Specific inhibition of gene expression by small double-stranded RNAs in invertebrate and vertebrate systems. Proc. Natl. Acad. Sci. USA. 2001;98:9742–9747. doi: 10.1073/pnas.171251798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weng Y., Xiao H., Zhang J., Liang X.J., Huang Y. RNAi therapeutic and its innovative biotechnological evolution. Biotechnol. Adv. 2019;37:801–825. doi: 10.1016/j.biotechadv.2019.04.012. [DOI] [PubMed] [Google Scholar]
- 9.Whitehead K.A., Langer R., Anderson D.G. Knocking down barriers: advances in siRNA delivery. Nat. Rev. Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Czech M.P., Aouadi M., Tesz G.J. RNAi-based therapeutic strategies for metabolic disease. Nat. Rev. Endocrinol. 2011;7:473–484. doi: 10.1038/nrendo.2011.57. [DOI] [PubMed] [Google Scholar]
- 11.Pecot C.V., Calin G.A., Coleman R.L., Lopez-Berestein G., Sood A.K. RNA interference in the clinic: challenges and future directions. Nat. Rev. Cancer. 2011;11:59–67. doi: 10.1038/nrc2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fellmann C., Lowe S.W. Stable RNA interference rules for silencing. Nat. Cell Biol. 2014;16:10–18. doi: 10.1038/ncb2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iorns E., Lord C.J., Turner N., Ashworth A. Utilizing RNA interference to enhance cancer drug discovery. Nat. Rev. Drug Discov. 2007;6:556–568. doi: 10.1038/nrd2355. [DOI] [PubMed] [Google Scholar]
- 14.Kim D.H., Rossi J.J. Strategies for silencing human disease using RNA interference. Nat. Rev. Genet. 2007;8:173–184. doi: 10.1038/nrg2006. [DOI] [PubMed] [Google Scholar]
- 15.de Fougerolles A., Vornlocher H.P., Maraganore J., Lieberman J. Interfering with disease: a progress report on siRNA-based therapeutics. Nat. Rev. Drug Discov. 2007;6:443–453. doi: 10.1038/nrd2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo Q., Kang Q., Song W.X., Luu H.H., Luo X., An N., Luo J., Deng Z.L., Jiang W., Yin H. Selection and validation of optimal siRNA target sites for RNAi-mediated gene silencing. Gene. 2007;395:160–169. doi: 10.1016/j.gene.2007.02.030. [DOI] [PubMed] [Google Scholar]
- 17.Deng F., Chen X., Liao Z., Yan Z., Wang Z., Deng Y., Zhang Q., Zhang Z., Ye J., Qiao M. A simplified and versatile system for the simultaneous expression of multiple siRNAs in mammalian cells using Gibson DNA assembly. PLoS ONE. 2014;9:e113064. doi: 10.1371/journal.pone.0113064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang X., Wang F., Zhao C., Yang S., Cheng Q., Tang Y., Zhang F., Zhang Y., Luo W., Wang C. Dentinogenesis and tooth-alveolar bone complex defects in BMP9/GDF2 knockout mice. Stem Cells Dev. 2019;28:683–694. doi: 10.1089/scd.2018.0230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan S., Zhang R., Wu K., Cui J., Huang S., Ji X., An L., Yuan C., Gong C., Zhang L. Characterization of the essential role of bone morphogenetic protein 9 (BMP9) in osteogenic differentiation of mesenchymal stem cells (MSCs) through RNA interference. Genes Dis. 2018;5:172–184. doi: 10.1016/j.gendis.2018.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liao J., Wei Q., Zou Y., Fan J., Song D., Cui J., Zhang W., Zhu Y., Ma C., Hu X. Notch signaling augments BMP9-induced bone formation by promoting the osteogenesis-angiogenesis coupling process in mesenchymal stem cells (MSCs) Cell. Physiol. Biochem. 2017;41:1905–1923. doi: 10.1159/000471945. [DOI] [PubMed] [Google Scholar]
- 21.Liao J., Yu X., Hu X., Fan J., Wang J., Zhang Z., Zhao C., Zeng Z., Shu Y., Zhang R. lncRNA H19 mediates BMP9-induced osteogenic differentiation of mesenchymal stem cells (MSCs) through Notch signaling. Oncotarget. 2017;8:53581–53601. doi: 10.18632/oncotarget.18655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang J., Liao J., Zhang F., Song D., Lu M., Liu J., Wei Q., Tang S., Liu H., Fan J. NEL-like molecule-1 (Nell1) is regulated by bone morphogenetic protein 9 (BMP9) and potentiates BMP9-induced osteogenic differentiation at the expense of adipogenesis in mesenchymal stem cells. Cell. Physiol. Biochem. 2017;41:484–500. doi: 10.1159/000456885. [DOI] [PubMed] [Google Scholar]
- 23.Deng Y., Wang Z., Zhang F., Qiao M., Yan Z., Wei Q., Wang J., Liu H., Fan J., Zou Y. A blockade of IGF signaling sensitizes human ovarian cancer cells to the anthelmintic niclosamide-induced anti-proliferative and anticancer activities. Cell. Physiol. Biochem. 2016;39:871–888. doi: 10.1159/000447797. [DOI] [PubMed] [Google Scholar]
- 24.Zhang H., Wang J., Deng F., Huang E., Yan Z., Wang Z., Deng Y., Zhang Q., Zhang Z., Ye J. Canonical Wnt signaling acts synergistically on BMP9-induced osteo/odontoblastic differentiation of stem cells of dental apical papilla (SCAPs) Biomaterials. 2015;39:145–154. doi: 10.1016/j.biomaterials.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y., Wagner E.R., Yan Z., Wang Z., Luther G., Jiang W., Ye J., Wei Q., Wang J., Zhao L. The calcium-binding protein S100A6 accelerates human osteosarcoma growth by promoting cell proliferation and inhibiting osteogenic differentiation. Cell. Physiol. Biochem. 2015;37:2375–2392. doi: 10.1159/000438591. [DOI] [PubMed] [Google Scholar]
- 26.Wang X., Yuan C., Huang B., Fan J., Feng Y., Li A.J., Zhang B., Lei Y., Ye Z., Zhao L. Developing a versatile shotgun cloning strategy for single-vector-based multiplex expression of short interfering RNAs (siRNAs) in mammalian cells. ACS Synth. Biol. 2019;8:2092–2105. doi: 10.1021/acssynbio.9b00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.El Karoui M., Hoyos-Flight M., Fletcher L. Future trends in synthetic biology—a report. Front. Bioeng. Biotechnol. 2019;7:175. doi: 10.3389/fbioe.2019.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Potapov V., Ong J.L., Kucera R.B., Langhorst B.W., Bilotti K., Pryor J.M., Cantor E.J., Canton B., Knight T.F., Evans T.C., Jr., Lohman G.J.S. comprehensive profiling of four base overhang ligation fidelity by T4 DNA ligase and application to DNA assembly. ACS Synth. Biol. 2018;7:2665–2674. doi: 10.1021/acssynbio.8b00333. [DOI] [PubMed] [Google Scholar]
- 29.Sharff K.A., Song W.X., Luo X., Tang N., Luo J., Chen J., Bi Y., He B.C., Huang J., Li X. Hey1 basic helix-loop-helix protein plays an important role in mediating BMP9-induced osteogenic differentiation of mesenchymal progenitor cells. J. Biol. Chem. 2009;284:649–659. doi: 10.1074/jbc.M806389200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luu H.H., Song W.X., Luo X., Manning D., Luo J., Deng Z.L., Sharff K.A., Montag A.G., Haydon R.C., He T.C. Distinct roles of bone morphogenetic proteins in osteogenic differentiation of mesenchymal stem cells. J. Orthop. Res. 2007;25:665–677. doi: 10.1002/jor.20359. [DOI] [PubMed] [Google Scholar]
- 31.Wang R.N., Green J., Wang Z., Deng Y., Qiao M., Peabody M., Zhang Q., Ye J., Yan Z., Denduluri S. Bone morphogenetic protein (BMP) signaling in development and human diseases. Genes Dis. 2014;1:87–105. doi: 10.1016/j.gendis.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang F., Song J., Zhang H., Huang E., Song D., Tollemar V., Wang J., Wang J., Mohammed M., Wei Q. Wnt and BMP signaling crosstalk in regulating dental stem cells: implications in dental tissue engineering. Genes Dis. 2016;3:263–276. doi: 10.1016/j.gendis.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pakvasa M., Haravu P., Boachie-Mensah M., Jones A., Coalson E., Liao J., Zeng Z., Wu D., Qin K., Wu X. Notch signaling: its essential roles in bone and craniofacial development. Genes Dis. 2020 doi: 10.1016/j.gendis.2020.04.006. Published online April 11, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luther G., Wagner E.R., Zhu G., Kang Q., Luo Q., Lamplot J., Bi Y., Luo X., Luo J., Teven C. BMP-9 induced osteogenic differentiation of mesenchymal stem cells: molecular mechanism and therapeutic potential. Curr. Gene Ther. 2011;11:229–240. doi: 10.2174/156652311795684777. [DOI] [PubMed] [Google Scholar]
- 35.Deng Z.L., Sharff K.A., Tang N., Song W.X., Luo J., Luo X., Chen J., Bennett E., Reid R., Manning D. Regulation of osteogenic differentiation during skeletal development. Front. Biosci. 2008;13:2001–2021. doi: 10.2741/2819. [DOI] [PubMed] [Google Scholar]
- 36.Lamplot J.D., Qin J., Nan G., Wang J., Liu X., Yin L., Tomal J., Li R., Shui W., Zhang H. BMP9 signaling in stem cell differentiation and osteogenesis. Am. J. Stem Cells. 2013;2:1–21. [PMC free article] [PubMed] [Google Scholar]
- 37.Liu W., Deng Z., Zeng Z., Fan J., Feng Y., Wang X., Cao D., Zhang B., Yang L., Liu B. Highly expressed BMP9/GDF2 in postnatal mouse liver and lungs may account for its pleiotropic effects on stem cell differentiation, angiogenesis, tumor growth and metabolism. Genes Dis. 2019;7:235–244. doi: 10.1016/j.gendis.2019.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mostafa S., Pakvasa M., Coalson E., Zhu A., Alverdy A., Castillo H., Fan J., Li A., Feng Y., Wu D. The wonders of BMP9: from mesenchymal stem cell differentiation, angiogenesis, neurogenesis, tumorigenesis, and metabolism to regenerative medicine. Genes Dis. 2019;6:201–223. doi: 10.1016/j.gendis.2019.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peng Y., Kang Q., Cheng H., Li X., Sun M.H., Jiang W., Luu H.H., Park J.Y., Haydon R.C., He T.C. Transcriptional characterization of bone morphogenetic proteins (BMPs)-mediated osteogenic signaling. J. Cell. Biochem. 2003;90:1149–1165. doi: 10.1002/jcb.10744. [DOI] [PubMed] [Google Scholar]
- 40.Peng Y., Kang Q., Luo Q., Jiang W., Si W., Liu B.A., Luu H.H., Park J.K., Li X., Luo J. Inhibitor of DNA binding/differentiation helix-loop-helix proteins mediate bone morphogenetic protein-induced osteoblast differentiation of mesenchymal stem cells. J. Biol. Chem. 2004;279:32941–32949. doi: 10.1074/jbc.M403344200. [DOI] [PubMed] [Google Scholar]
- 41.Zhang L., Luo Q., Shu Y., Zeng Z., Huang B., Feng Y., Zhang B., Wang X., Lei Y., Ye Z. Transcriptomic landscape regulated by the 14 types of bone morphogenetic proteins (BMPs) in lineage commitment and differentiation of mesenchymal stem cells (MSCs) Genes Dis. 2019;6:258–275. doi: 10.1016/j.gendis.2019.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Si W., Kang Q., Luu H.H., Park J.K., Luo Q., Song W.X., Jiang W., Luo X., Li X., Yin H. CCN1/Cyr61 is regulated by the canonical Wnt signal and plays an important role in Wnt3A-induced osteoblast differentiation of mesenchymal stem cells. Mol. Cell. Biol. 2006;26:2955–2964. doi: 10.1128/MCB.26.8.2955-2964.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dominguez A.A., Lim W.A., Qi L.S. Beyond editing: repurposing CRISPR-Cas9 for precision genome regulation and interrogation. Nat. Rev. Mol. Cell Biol. 2016;17:5–15. doi: 10.1038/nrm.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Doudna J.A., Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 46.Wang H., La Russa M., Qi L.S. CRISPR/Cas9 in genome editing and beyond. Annu. Rev. Biochem. 2016;85:227–264. doi: 10.1146/annurev-biochem-060815-014607. [DOI] [PubMed] [Google Scholar]
- 47.Breunig C.T., Durovic T., Neuner A.M., Baumann V., Wiesbeck M.F., Köferle A., Götz M., Ninkovic J., Stricker S.H. One step generation of customizable gRNA vectors for multiplex CRISPR approaches through string assembly gRNA cloning (STAgR) PLoS ONE. 2018;13:e0196015. doi: 10.1371/journal.pone.0196015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zuckermann M., Hlevnjak M., Yazdanparast H., Zapatka M., Jones D.T.W., Lichter P., Gronych J. A novel cloning strategy for one-step assembly of multiplex CRISPR vectors. Sci. Rep. 2018;8:17499. doi: 10.1038/s41598-018-35727-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hu X., Li L., Yu X., Zhang R., Yan S., Zeng Z., Shu Y., Zhao C., Wu X., Lei J. CRISPR/Cas9-mediated reversibly immortalized mouse bone marrow stromal stem cells (BMSCs) retain multipotent features of mesenchymal stem cells (MSCs) Oncotarget. 2017;8:111847–111865. doi: 10.18632/oncotarget.22915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu N., Zhang H., Deng F., Li R., Zhang W., Chen X., Wen S., Wang N., Zhang J., Yin L. Overexpression of Ad5 precursor terminal protein accelerates recombinant adenovirus packaging and amplification in HEK-293 packaging cells. Gene Ther. 2014;21:629–637. doi: 10.1038/gt.2014.40. [DOI] [PubMed] [Google Scholar]
- 51.Wei Q., Fan J., Liao J., Zou Y., Song D., Liu J., Cui J., Liu F., Ma C., Hu X. Engineering the rapid adenovirus production and amplification (RAPA) cell line to expedite the generation of recombinant adenoviruses. Cell. Physiol. Biochem. 2017;41:2383–2398. doi: 10.1159/000475909. [DOI] [PubMed] [Google Scholar]
- 52.Cui J., Zhang W., Huang E., Wang J., Liao J., Li R., Yu X., Zhao C., Zeng Z., Shu Y. BMP9-induced osteoblastic differentiation requires functional Notch signaling in mesenchymal stem cells. Lab. Invest. 2019;99:58–71. doi: 10.1038/s41374-018-0087-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shu Y., Wu K., Zeng Z., Huang S., Ji X., Yuan C., Zhang L., Liu W., Huang B., Feng Y. A simplified system to express circularized inhibitors of miRNA for stable and potent suppression of miRNA functions. Mol. Ther. Nucleic Acids. 2018;13:556–567. doi: 10.1016/j.omtn.2018.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu X., Chen L., Wu K., Yan S., Zhang R., Zhao C., Zeng Z., Shu Y., Huang S., Lei J. Establishment and functional characterization of the reversibly immortalized mouse glomerular podocytes (imPODs) Genes Dis. 2018;5:137–149. doi: 10.1016/j.gendis.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Feng T., Li Z., Jiang W., Breyer B., Zhou L., Cheng H., Haydon R.C., Ishikawa A., Joudeh M.A., He T.C. Increased efficiency of cloning large DNA fragments using a lower copy number plasmid. Biotechniques. 2002;32:992–, 994, 996 passim. doi: 10.2144/02325bm03. [DOI] [PubMed] [Google Scholar]
- 56.Zeng Z., Huang B., Huang S., Zhang R., Yan S., Yu X., Shu Y., Zhao C., Lei J., Zhang W. The development of a sensitive fluorescent protein-based transcript reporter for high throughput screening of negative modulators of lncRNAs. Genes Dis. 2018;5:62–74. doi: 10.1016/j.gendis.2018.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liao J., Wei Q., Fan J., Zou Y., Song D., Liu J., Liu F., Ma C., Hu X., Li L. Characterization of retroviral infectivity and superinfection resistance during retrovirus-mediated transduction of mammalian cells. Gene Ther. 2017;24:333–341. doi: 10.1038/gt.2017.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang E., Bi Y., Jiang W., Luo X., Yang K., Gao J.L., Gao Y., Luo Q., Shi Q., Kim S.H. Conditionally immortalized mouse embryonic fibroblasts retain proliferative activity without compromising multipotent differentiation potential. PLoS ONE. 2012;7:e32428. doi: 10.1371/journal.pone.0032428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang J., Zhang H., Zhang W., Huang E., Wang N., Wu N., Wen S., Chen X., Liao Z., Deng F. Bone morphogenetic protein-9 effectively induces osteo/odontoblastic differentiation of the reversibly immortalized stem cells of dental apical papilla. Stem Cells Dev. 2014;23:1405–1416. doi: 10.1089/scd.2013.0580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang J., Weng Y., Liu X., Wang J., Zhang W., Kim S.H., Zhang H., Li R., Kong Y., Chen X. Endoplasmic reticulum (ER) stress inducible factor cysteine-rich with EGF-like domains 2 (Creld2) is an important mediator of BMP9-regulated osteogenic differentiation of mesenchymal stem cells. PLoS ONE. 2013;8:e73086. doi: 10.1371/journal.pone.0073086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Luo J., Deng Z.L., Luo X., Tang N., Song W.X., Chen J., Sharff K.A., Luu H.H., Haydon R.C., Kinzler K.W. A protocol for rapid generation of recombinant adenoviruses using the AdEasy system. Nat. Protoc. 2007;2:1236–1247. doi: 10.1038/nprot.2007.135. [DOI] [PubMed] [Google Scholar]
- 62.Wang N., Zhang H., Zhang B.Q., Liu W., Zhang Z., Qiao M., Zhang H., Deng F., Wu N., Chen X. Adenovirus-mediated efficient gene transfer into cultured three-dimensional organoids. PLoS ONE. 2014;9:e93608. doi: 10.1371/journal.pone.0093608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee C.S., Bishop E.S., Zhang R., Yu X., Farina E.M., Yan S., Zhao C., Zheng Z., Shu Y., Wu X. Adenovirus-mediated gene delivery: potential applications for gene and cell-based therapies in the new era of personalized medicine. Genes Dis. 2017;4:43–63. doi: 10.1016/j.gendis.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhao C., Qazvini N.T., Sadati M., Zeng Z., Huang S., De La Lastra A.L., Zhang L., Feng Y., Liu W., Huang B. A pH-triggered, self-assembled, and bioprintable hybrid hydrogel scaffold for mesenchymal stem cell based bone tissue engineering. ACS Appl. Mater. Interfaces. 2019;11:8749–8762. doi: 10.1021/acsami.8b19094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Z., Liu J., Zeng Z., Fan J., Huang S., Zhang L., Zhang B., Wang X., Feng Y., Ye Z. lncRNA Rmst acts as an important mediator of BMP9-induced osteogenic differentiation of mesenchymal stem cells (MSCs) by antagonizing Notch-targeting microRNAs. Aging (Albany NY) 2019;11:12476–12496. doi: 10.18632/aging.102583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Song D., Zhang F., Reid R.R., Ye J., Wei Q., Liao J., Zou Y., Fan J., Ma C., Hu X. BMP9 induces osteogenesis and adipogenesis in the immortalized human cranial suture progenitors from the patent sutures of craniosynostosis patients. J. Cell. Mol. Med. 2017;21:2782–2795. doi: 10.1111/jcmm.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang H., Cao Y., Shu L., Zhu Y., Peng Q., Ran L., Wu J., Luo Y., Zuo G., Luo J. Long non-coding RNA (lncRNA) H19 induces hepatic steatosis through activating MLXIPL and mTORC1 networks in hepatocytes. J. Cell. Mol. Med. 2020;24:1399–1412. doi: 10.1111/jcmm.14818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang B., Yang L., Zeng Z., Feng Y., Wang X., Wu X., Luo H., Zhang J., Zhang M., Pakvasa M. Leptin potentiates BMP9-induced osteogenic differentiation of mesenchymal stem cells through the activation of JAK/STAT signaling. Stem Cells Dev. 2020;29:498–510. doi: 10.1089/scd.2019.0292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhao C., Wu N., Deng F., Zhang H., Wang N., Zhang W., Chen X., Wen S., Zhang J., Yin L. Adenovirus-mediated gene transfer in mesenchymal stem cells can be significantly enhanced by the cationic polymer polybrene. PLoS ONE. 2014;9:e92908. doi: 10.1371/journal.pone.0092908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Untergasser A., Cutcutache I., Koressaar T., Ye J., Faircloth B.C., Remm M., Rozen S.G. Primer3—new capabilities and interfaces. Nucleic Acids Res. 2012;40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang Q., Wang J., Deng F., Yan Z., Xia Y., Wang Z., Ye J., Deng Y., Zhang Z., Qiao M. TqPCR: a touchdown qPCR assay with significantly improved detection sensitivity and amplification efficiency of SYBR Green qPCR. PLoS ONE. 2015;10:e0132666. doi: 10.1371/journal.pone.0132666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fan J., Feng Y., Zhang R., Zhang W., Shu Y., Zeng Z., Huang S., Zhang L., Huang B., Wu D. A simplified system for the effective expression and delivery of functional mature microRNAs in mammalian cells. Cancer Gene Ther. 2020;27:424–437. doi: 10.1038/s41417-019-0113-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fan J., Wei Q., Liao J., Zou Y., Song D., Xiong D., Ma C., Hu X., Qu X., Chen L. Noncanonical Wnt signaling plays an important role in modulating canonical Wnt-regulated stemness, proliferation and terminal differentiation of hepatic progenitors. Oncotarget. 2017;8:27105–27119. doi: 10.18632/oncotarget.15637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huang B., Huang L.F., Zhao L., Zeng Z., Wang X., Cao D., Yang L., Ye Z., Chen X., Liu B. Microvesicles (MIVs) secreted from adipose-derived stem cells (ADSCs) contain multiple microRNAs and promote the migration and invasion of endothelial cells. Genes Dis. 2019;7:225–234. doi: 10.1016/j.gendis.2019.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hu N., Jiang D., Huang E., Liu X., Li R., Liang X., Kim S.H., Chen X., Gao J.L., Zhang H. BMP9-regulated angiogenic signaling plays an important role in the osteogenic differentiation of mesenchymal progenitor cells. J. Cell Sci. 2013;126:532–541. doi: 10.1242/jcs.114231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Huang E., Zhu G., Jiang W., Yang K., Gao Y., Luo Q., Gao J.L., Kim S.H., Liu X., Li M. Growth hormone synergizes with BMP9 in osteogenic differentiation by activating the JAK/STAT/IGF1 pathway in murine multilineage cells. J. Bone Miner. Res. 2012;27:1566–1575. doi: 10.1002/jbmr.1622. [DOI] [PubMed] [Google Scholar]
- 77.Gao Y., Huang E., Zhang H., Wang J., Wu N., Chen X., Wang N., Wen S., Nan G., Deng F. Crosstalk between Wnt/β-catenin and estrogen receptor signaling synergistically promotes osteogenic differentiation of mesenchymal progenitor cells. PLoS ONE. 2013;8:e82436. doi: 10.1371/journal.pone.0082436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen L., Jiang W., Huang J., He B.C., Zuo G.W., Zhang W., Luo Q., Shi Q., Zhang B.Q., Wagner E.R. Insulin-like growth factor 2 (IGF-2) potentiates BMP-9-induced osteogenic differentiation and bone formation. J. Bone Miner. Res. 2010;25:2447–2459. doi: 10.1002/jbmr.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lu S., Wang J., Ye J., Zou Y., Zhu Y., Wei Q., Wang X., Tang S., Liu H., Fan J. Bone morphogenetic protein 9 (BMP9) induces effective bone formation from reversibly immortalized multipotent adipose-derived (iMAD) mesenchymal stem cells. Am. J. Transl. Res. 2016;8:3710–3730. [PMC free article] [PubMed] [Google Scholar]
- 80.Ye J., Wang J., Zhu Y., Wei Q., Wang X., Yang J., Tang S., Liu H., Fan J., Zhang F. A thermoresponsive polydiolcitrate-gelatin scaffold and delivery system mediates effective bone formation from BMP9-transduced mesenchymal stem cells. Biomed. Mater. 2016;11:025021. doi: 10.1088/1748-6041/11/2/025021. [DOI] [PubMed] [Google Scholar]
- 81.Zou Y., Qazvini N.T., Zane K., Sadati M., Wei Q., Liao J., Fan J., Song D., Liu J., Ma C. Gelatin-derived graphene-silicate hybrid materials are biocompatible and synergistically promote BMP9-induced osteogenic differentiation of mesenchymal stem cells. ACS Appl. Mater. Interfaces. 2017;9:15922–15932. doi: 10.1021/acsami.7b00272. [DOI] [PubMed] [Google Scholar]
- 82.Kang Q., Song W.X., Luo Q., Tang N., Luo J., Luo X., Chen J., Bi Y., He B.C., Park J.K. A comprehensive analysis of the dual roles of BMPs in regulating adipogenic and osteogenic differentiation of mesenchymal progenitor cells. Stem Cells Dev. 2009;18:545–559. doi: 10.1089/scd.2008.0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shu Y., Yang C., Ji X., Zhang L., Bi Y., Yang K., Gong M., Liu X., Guo Q., Su Y. Reversibly immortalized human umbilical cord-derived mesenchymal stem cells (UC-MSCs) are responsive to BMP9-induced osteogenic and adipogenic differentiation. J. Cell. Biochem. 2018;119:8872–8886. doi: 10.1002/jcb.27140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li R., Zhang W., Cui J., Shui W., Yin L., Wang Y., Zhang H., Wang N., Wu N., Nan G. Targeting BMP9-promoted human osteosarcoma growth by inactivation of notch signaling. Curr. Cancer Drug Targets. 2014;14:274–285. doi: 10.2174/1568009614666140305105805. [DOI] [PubMed] [Google Scholar]
- 85.Liu X., Qin J., Luo Q., Bi Y., Zhu G., Jiang W., Kim S.H., Li M., Su Y., Nan G. Cross-talk between EGF and BMP9 signalling pathways regulates the osteogenic differentiation of mesenchymal stem cells. J. Cell. Mol. Med. 2013;17:1160–1172. doi: 10.1111/jcmm.12097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhao C., Zeng Z., Qazvini N.T., Yu X., Zhang R., Yan S., Shu Y., Zhu Y., Duan C., Bishop E. Thermoresponsive citrate-based graphene oxide scaffold enhances bone regeneration from BMP9-stimulated adipose-derived mesenchymal stem cells. ACS Biomater. Sci. Eng. 2018;4:2943–2955. doi: 10.1021/acsbiomaterials.8b00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang X., Wu X., Zhang Z., Ma C., Wu T., Tang S., Zeng Z., Huang S., Gong C., Yuan C. Monensin inhibits cell proliferation and tumor growth of chemo-resistant pancreatic cancer cells by targeting the EGFR signaling pathway. Sci. Rep. 2018;8:17914. doi: 10.1038/s41598-018-36214-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.