Abstract
The death of cardiomyocytes either through apoptosis or necroptosis is the pathological feature of cardiac ischemia-reperfusion (I/R) injury. Phosphoglycerate mutase 5 (PGAM5), a mitochondrially-localized serine/threonine-protein phosphatase, functions as a novel inducer of necroptosis. However, intense debate exists regarding the effect of PGAM5 on I/R-related cardiomyocyte death. Using cardiac-specific PGAM5 knockout (PGAM5CKO) mice, we comprehensively investigated the precise contribution and molecular mechanism of PGAM5 in cardiomyocyte death. Our data showed that both PGAM5 transcription and expression were upregulated in reperfused myocardium. Genetic ablation of PGAM5 suppressed I/R-mediated necroptosis but failed to prevent apoptosis activation, a result that went along with improved heart function and decreased inflammation response. Regardless of PGAM5 status, mitophagy-related cell death was not apparent following I/R. Under physiological conditions, PGAM5 overexpression in primary cardiomyocytes was sufficient to induce cardiomyocyte necroptosis rather than apoptosis. At the sub-cellular levels, PGAM5 deficiency increased mitochondrial DNA copy number and transcript levels, normalized mitochondrial respiration, repressed mitochondrial ROS production, and prevented abnormal mPTP opening upon I/R. Molecular investigation demonstrated that PGAM5 deletion interrupted I/R-mediated DrpS637 dephosphorylation but failed to abolish I/R-induce Drp1S616 phosphorylation, resulting in partial inhibition of mitochondrial fission. In addition, declining Mfn2 and OPA1 levels were restored in PGAM5CKO cardiomyocytes following I/R. Nevertheless, PGAM5 depletion did not rescue suppressed mitophagy upon I/R injury. In conclusion, our results provide an insight into the specific role and working mechanism of PGAM5 in driving cardiomyocyte necroptosis through imposing mitochondrial quality control in cardiac I/R injury.
Keywords: PGAM5, Cardiac I/R injury, Death, Necroptosis, Mitochondrial quality control, Mitochondrial fission, Mitophagy
1. Introduction
Massive cardiomyocyte death occurs during myocardial infarction resulting from acute ischemic attack [1]. Although emergency coronary recanalization restores blood supply to ischemic tissue, clinical and laboratory data often reveal sustained cardiac injury due to cardiomyocyte death within the first few minutes of reperfusion, a phenomenon known as ischemia-reperfusion (I/R) injury. The occurrence of I/R injury extends infarct area, which is a critical determinant of ventricular function and cardiovascular mortality [2]. Therefore, substantial efforts are being devoted to clarifying the pathogenic mechanisms of I/R injury to develop therapeutic approaches to minimize its consequences [3]. It is generally accepted that excessive death of cardiomyocyte is the fundamental pathological feature of myocardial I/R injury [4]. While the involvement of apoptosis in cardiomyocyte death is widely recognized, the occurrence of necroptosis, a distinct form of regulated cell death, has been highlighted by our previous studies [5,6] and other investigations [7,8]. Importantly, pharmacological experiments and/or genetically-engineered mouse models of cardiac I/R injury have observed that ~30% of death is executed through apoptosis, whereas necroptosis accounts for ~50% of cardiomyocyte death during I/R episodes [7].
The fundamental mediators of necroptosis in cardiomyocytes include receptor-interacting protein kinase-3 (RIPK3), phosphoglycerate mutase 5 (PGAM5), and mixed lineage kinase domain-like pseudokinase (MLKL) [9,10]. RIPK3 acts as a sensor of environmental and cellular stress to initiate and transmit necroptotic signal to MLKL and PGAM5 [10]. MLKL, the principal RIPK3 substrate, acts as the terminal necroptosis executioner, triggering membrane rupture after phosphorylation at Ser358 [11,12]. PGAM5 is a serine/threonine-protein phosphatase localized in the outer mitochondria membrane. It was originally identified as a pivotal necroptotic regulator through serving as a platform for the assembly and activation of RIPK3-MLKL protein complexes [13]. However, the regulatory actions of PGAM5 on necroptosis are debated and may vary according to cellular context and specific pathology. During TNF-α-mediated inflammation response, PGAM5 expression is upregulated and promotes dynamin related protein-1 (Drp1) activation through Drp1S637 dephosphorylation [14], an obligatory step for necroptosis induction. These observations were recently complemented by a study showing that cardiac PGAM5 expression in rats is upregulated by I/R injury and consequently induces Drp1S616 phosphorylation and necroptosis activation [15]. By contrast, increased PGAM5 was shown to alleviate cardiomyocyte necroptosis by activating mitophagy in a mouse model of myocardial I/R injury [16]. Surprisingly, another study of mouse myocardial I/R injury reported a previously unforeseen role of PGAM5 in participation of apoptosis induction through the degradation of Bcl-xL [17]. In the present study, the cardiac-specific PGAM5 knockout mice were generated and used to clarify the regulatory roles and molecular mechanisms of PGAM5 in necroptosis and/or apoptosis (in)activation in the context of cardiac I/R injury.
A progressive disruption of mitochondrial function and structure is not only an adaptive response to reperfusion but also a critical process predisposing cardiomyocytes to apoptosis or necroptosis [18,19]. Under conditions of I/R injury, the mitochondrial quality control system is activated and principally devoted to restore mitochondrial homeostasis through the modification of mitochondrial redox reaction, fission, fusion, mitophagy, and bioenergetics. Superfluous ROS are produced at mitochondria correlating with cardiomyocyte oxidative stress at the stage of reperfusion [20]. In addition, mitochondrial fragmentation, caused by excessive mitochondrial fission, is not just a morphological adaptive response but also a contributor to mitochondrial membrane damage and consequent cell death [21,22]. Although mitochondrial fusion serves to counteract abnormal mitochondrial constriction and cleavage, its activity seems to be downregulated during the reperfusion phase [23]. As a mitochondria degradation system, mitophagy represents a double-edged sword for cardiomyocytes, as it can promote cell survival or trigger cell death depending on the physiopathogenic context [24]. Interestingly, cumulative evidence is emerging to support PGAM5 having diverse impacts on mitochondrial quality control. It is important to understand whether and how PGAM5 alters mitochondrial quality control participating in cardiac I/R injury. In the present study, we generated cardiac-specific PGAM5 knockout mice to attempt to clarify the molecular mechanisms underlying the regulatory roles of PGAM5 in mitochondrial bioenergetics, oxidative stress, fission, fusion, and mitophagy in cardiomyocytes enduring I/R injury.
2. Materials and methods
2.1. Experimental animals
All animal procedures were performed in accordance with the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health (NIH Publication No. 85-23, revised 1996) and approved by the Chinese PLA General Hospital and University of Wyoming Institutional Animal Use and Care Committees (Laramie, WY, USA). PGAM5f/f mice were generated as previously described [6] and bred to α-MHC (alpha myosin heavy chain)Cre transgenic mice to generate cardiac-specific PGAM5 knockout (PGAM5CKO, PGAM5f/f, α-MHCCre+) mice. All genetically modified mice were crossed on a C57BL/6 N background for at least 10 generations. Male mice (8 weeks old) were used for in vivo experiments.
2.2. I/R surgery and infarct size assessment
I/R injury was conducted as previously described [25]. In brief, mice were anesthetized with 1%–3% isoflurane through inhalation (Baxter, USA). The left anterior descending (LAD) coronary artery was temporarily ligated for 45 min, followed by 0–24 h reperfusion. Sham operation was performed by opening the chest and exposing the pericardium of anesthetized mice, leaving the LAD intact. For each experimental condition, a minimum of 6 mice were examined. After reperfusion, 2% Evans blue/saline were injected into mice to demarcate the ischemic area at risk (AAR). Then, the hearts were excised and cut into five 1-mm slices, which were incubated with 1% 2,3,5-triphenyltetrazolium chloride (TTC) for 10 min at 37 °C to demonstrate infarct size (IS). The percentage of IS and AAR of individual myocardial sections was multiplied by the corresponding section weight. To inhibit apoptosis or necroptosis, mice were intraperitoneally injected with Z-VAD-FMK (Selleck Chemicals, USA; No. S7023, 20 mg/kg body weight) or Necrostatin-1 (Selleck Chemicals; No. S8037, 5 mg/kg) 12 h before I/R injury. To activate or inhibit mitophagy, mice were intraperitoneally injected with a mitophagy activator (Urolithin A, Sigma, USA, No. SML1791, 10 mg/kg) or antagonist (MF-094, Sigma, No. SML2501, 20 mg/kg) 12 h before I/R injury.
2.3. Echocardiography and electron microscopy
Echocardiography was performed in mice after I/R surgery using a Vevo 2100 system (FUJIFILM VisualSonics, Canada) as our previously described [25]. For electron microscopy, heart tissue fragments were excised from the left ventricle and washed with 0.1 M sodium cacodylate, post-fixed in 1% OsO4 for 1 h, dehydrated in graded acetone, and embedded in Epon-Araldite resin. Thin (80 nm) and semi-thick (280 nm) sections were placed on formvar-coated slot-grids and post stained with 2% aqueous uranyl acetate and Reynold's lead citrate. Colloidal gold particles (15 nm) were added to both surfaces of the semi-thick sections to serve as fiducial markers for tilt-series alignment. Preparations were imaged at the EMBL Heidelberg Electron Microscopy Core Facility using a Tecnai F30 electron microscope operating at 300 kV and a Phillips CM120 BioTwin electron microscope operating at 120 kV. Images were captured on a 4K Eagle camera and a SIS 1K KeenView device.
2.4. Cell culture and sI/R injury in vitro
Primary cardiomyocytes were isolated from wild-type (WT), PGAM5f/f, and PGAM5CKO mice and cultured for 24–48 h in DMEM (Gibco, ThermoFisher, USA) supplemented with 1% l-glutamine, 20% fetal bovine serum (FBS), and 1% penicillin-streptomycin. Simulated I/R (sI/R) injury was induced in vitro through 45-min hypoxia and 4-h reoxygenation according to our previous protocols [24,25]. To inhibit apoptosis or necroptosis, cell-permeable Z-VAD-FMK (20 μM) and Necrostatin-1 (50 μM) were added to cultured cardiomyocytes 3 h before sI/R injury. To activate or inhibit mitophagy, cardiomyocytes were treated with Urolithin A (20 μM) or MF-094 (5 mM) for 6 h before sI/R injury.
2.5. Cardiomyocyte contraction measurements
After reperfusion, ventricular cardiomyocytes were isolated from PGAM5f/f and PGAM5CKO mice via Langendorff perfusion [24] and allowed to settle in a pacing chamber for 5 min prior to the initiation of laminar flow from an in-line solution heater set to 37 °C. Cell contraction was measured by bright-field microscopy with external field pacing at 1 Hz using the IonWizard acquisition system (IonOptix Corp., USA). Contraction parameters were automatically quantified and averaged over at least 10 contractions for each cell. Cells from 2 to 3 mice were isolated for each condition.
2.6. Immunoprecipitation analysis
Co-immunoprecipitation (Co-IP) was conducted using a magnetic IP kit (Thermo Fisher Scientific, USA). In short, protein lysates from cardiomyocytes (1000 μg total protein) were incubated with 10 μg of anti-Drp1 monoclonal antibody overnight at 4 °C [26]. Immune complexes were bound to protein A/G magnetic beads and collected with a magnetic stand. Proteins co-immunoprecipitated with Drp1 were eluted and subjected to gel electrophoresis and immunoblotting using the antibodies described in Supplemental Table 1. Normal IgG was used as a negative IP control [27].
2.7. Immunofluorescence and TUNEL
The samples (cardiomyocytes or reperfused heart tissues) were fixed in ice-cold fixative (4% paraformaldehyde and 0.5% glutaraldehyde in PBS) for 18 min in the dark at room temperature, followed by permeabilization for 4 min with 0.2% Triton X-100 in PBS [28]. The samples were subsequently incubated with primary antibodies at 4 °C in a humidified chamber, washed in PBS, and incubated with secondary antibodies (1:500 dilution in blocking buffer) for 1 h at room temperature or overnight at 4 °C in a humidified chamber. After three washes in PBS, coverslips were mounted on slides with a Vectashield mounting medium containing DAPI (Vector Laboratories, USA; No. H-1200) [29]. Images were obtained using a Zeiss fluorescence microscope. The antibodies used for immunofluorescence are listed in the Supplemental Table 1. For TUNEL assays, tissue slides and cells were reacted with terminal deoxynucleotidyl transferase (TdT) enzyme and fluorescently-labeled 2′-Deoxyuridine 5′-Triphosphate (dUTP) at 37 °C for 1 h. The nuclei were stained with DAPI. The number of TUNEL-positive cells was determined from fluorescence microscopy images [30].
2.8. LDH cytotoxicity assay, MTT assay, and caspase-3 activity assay
Cell death was determined through LDH release assay. An LDH Cytotoxicity Assay Kit (Beyotime, China; No: C0016) was used to measure the levels of LDH released by necroptotic cardiomyocytes according to the manufacturer's manual [31]. The MTT assay was used to analyze cardiomyocyte viability. Briefly, cardiomyocytes (103/well) were seeded into 96-well plates and were subjected to sI/R injury 24 h later. Afterwards, the MTT assay was performed and cell viability examined by measuring wells' absorbance at 490 nm on a microplate reader (Synergy™ 2; BioTek Instruments, Inc., USA) [32]. Caspase-3 activity was determined through an ELISA kit (Beyotime, China, Cat. No:C1115) according to the manufacturer's manual [33].
2.9. Real time PCR
Heart tissue and cultured cardiomyocytes were homogenized in RLT buffer, and RNA was extracted using an RNeasy mini kit (Qiagen, USA; No. 74104). The extracted RNA was reverse transcribed into cDNA using a high-capacity cDNA reverse transcription kit (Applied Biosystems, Lithuania; No. 4368814). Quantitative real-time PCR was performed using a Power SYBR Green PCR Master Mix (Applied Biosystems, UK; No. 4368708) on an Applied Biosystems ABI Prism machine [34]. Amplification conditions consisted of one cycle at 50 °C for 2 min and one cycle at 95 °C for 10 min, followed by 40 cycles at 95 °C for 15 s and at 60 °C for 1 min. Fold-change gene expression was determined using cycle threshold (Ct) values and normalized to GAPDH expression [35]. The primers are listed in the Supplemental Table 2.
2.10. Immunoblot analysis
Cells and cardiac tissues were homogenized using 1 x Cell Lysis Buffer (Cell Signaling Technology, USA) supplemented with protease inhibitor cocktail and phosphatase inhibitors (Thermo Fisher Scientific, USA) and centrifuged at 4 °C for 10 min at 10,000×g [36]. The resulting supernatant was mixed with a 5x protein loading buffer. The protein lysates were boiled at 60 °C for 5 min, and the protein contents were determined using a BCA assay. Aliquots of 15–30 μg protein per sample were fractionated on 10% SDS-PAGE gels, transferred onto PVDF membranes, and blocked in blocking buffer (PBS with 5% BSA, 0.1% Tween 20). Membranes were incubated with primary antibodies overnight at 4 °C, extensively washed, and incubated with HRP-conjugated anti-mouse or anti-rabbit whole IgG secondary antibodies (1:5000, Thermo Fisher Scientific) for 1 h at room temperature. Western Bright ECL HRP substrate (Advansta, USA) was then applied, and protein bands were detected using a ChemiDoc MP system (BioRad Laboratories, USA) [37]. Protein band intensity quantification analysis was performed with ImageLab software version 5.1. The antibodies that were used for western blots are listed in the Supplemental Table 1.
2.11. Mitochondrial ROS and membrane potential measurements
Mitochondrial ROS production was determined using the MitoSOX red mitochondrial superoxide indicator (Thermo Fisher Scientific; No. M36008) according to the manufacturer's instructions [38,39]. In brief, cardiomyocytes were washed with warm PBS, loaded with 2 μmol/L MitoSOX red at 37 °C for 30 min, and washed with warm PBS again. Then, fluorescence (excitation 485 nm, emission 535 nm) was measured in a microplate reader. Representative photomicrographs of MitoSOX stained cells were taken using a Zeiss Axio Imager M1 fluorescence microscope (Carl Zeiss, Germany). Mitochondrial membrane potential was detected through a Mitochondrial Membrane Potential JC-1 Kit (Beyotime, China, No. C2006) [40,41]. In brief, the cardiomyocytes were washed with warm PBS and then loaded with 2.5 mg/ml JC-1 for 30 min at 37 °C. After DAPI staining, the fluorescent images were analyzed under a Zeiss Axio Imager M1 fluorescence microscope (Carl Zeiss, Oberkochen, Germany) [42,43]. Mitochondrial potential is expressed as relative aggregate-to-monomer (red/green) fluorescence intensity ratio.
ATP detection, mPTP opening rate measurement, and mitophagy assay.
Total ATP production was measured through an ATP Assay Kit (Abcam, UK; No. ab83355) [44,45]. In brief, cardiomyocytes were harvested, washed with PBS, and then resuspended in 100 μL of ATP assay buffer. Cells were homogenized and then centrifuged (4 °C at 13,000 g) to remove any insoluble material. The supernatants were then collected and incubated with the ATP probe. Absorbance was detected at 580 nm using a microplate reader (Epoch 2; BioTek Instruments, Inc.). The results are presented as the ratio between the test and control values [46].
The mPTP opening rate was determined by loading cardiomyocytes with 5 μmol/L Calcein-AM (Molecular Probes, USA) and 2–5 mmol/L cobalt chloride. The results are presented as fluorescence intensity relative to control [5]. The fluorescent, pH-sensitive mitophagy reporter Keima (pMT-mKeima-Red, MBL Medical & Biological Laboratories, USA; No. AM-V-251) has a bimodal excitation spectra defined by a 440 nm peak at neutral pH and a 568 nm peak under acidic pH conditions. The Kemia-Red plasmid was transfected into cardiomyocytes according to the manufacturer's instructions. Mitophagy was quantified as the 550 nm/440 nm excitation ratio [47,48].
2.12. Gene silencing and overexpression
Adenoviral (Ad) vectors containing the PGAM5 gene (Ad-PGAM5) and control adenovirus carrying the β-galactosidase (β-gal) reporter gene (Ad-β-gal) were constructed as previously described [6]. Briefly, Myc-tagged WT PGAM5 cDNA was sub-cloned into a pcDNA3 vector, followed by adenovirus generation. Viral particles were quantified using an Adeno-X-Rapid Titer Kit (Clontech, USA). A multiplicity of infection (MOI) of 100 was used to transduce Ad-PGAM5 into cardiomyocytes [49]. The transduction efficiency was verified by both Western blot and direct observation using fluorescence microscopy. shRNAs against CK2 and PGAM5 were purchased from GenePharma (Shanghai, China). Cell transfection was performed using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer's protocol.
2.13. Statistical analysis
Data are presented as the mean ± SEM of at least three independent experiments. After confirming by the Kolmogorov-Smirnov test followed by Q-Q plots analysis that all variables were normally distributed, statistical differences were determined by Student's t-test for comparison between two groups and ANOVA followed by Bonferroni's multiple comparison test for comparison among three or more groups. P < 0.05 was considered significant. Statistical calculations were carried out using GraphPad Prism 6.0 or SPSS version 17.0.
3. Results
3.1. PGAM5 promotes I/R-mediated cardiomyocytes necroptosis
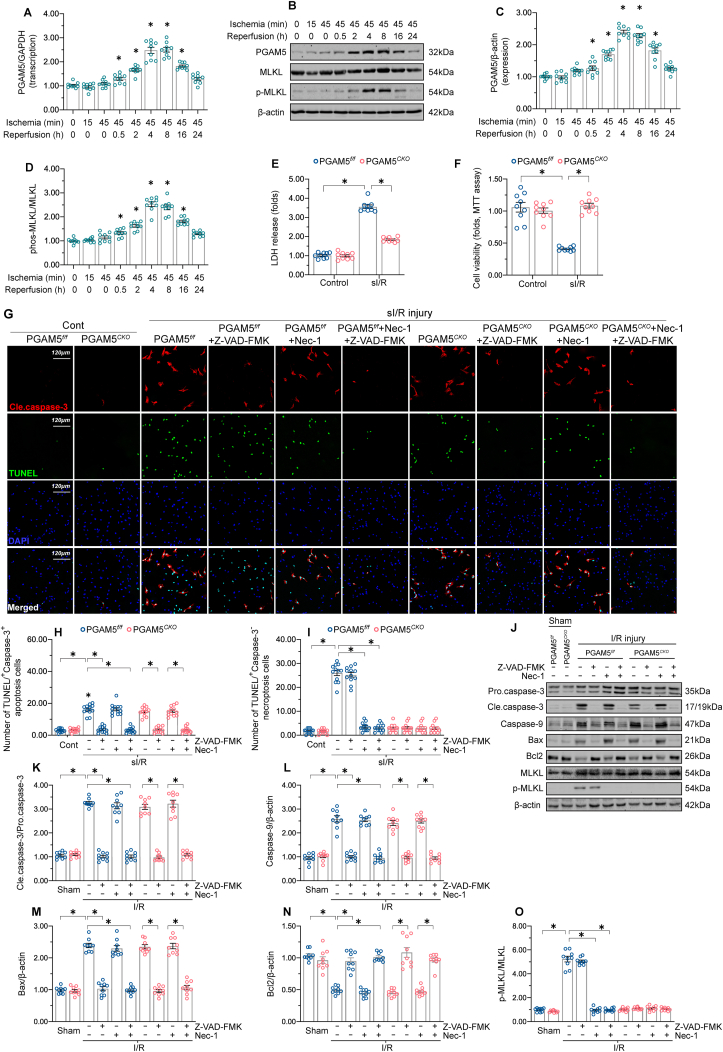
To characterize the effect of I/R on cardiomyocyte PGAM5 levels, we first isolated mouse primary cardiomyocytes and subjected them to simulated I/R (sI/R). Relative to the baseline, PGAM5 transcription (Supplemental Fig. 1A) and protein expression (Supplemental Fig. 1B–D) increased following sI/R injury. This effect was paralleled by an elevation in phospho-MLKL expression (Supplemental Fig. 1B–D). In vivo, cardiac PGAM5 mRNA (Fig. 1A) and protein levels (Fig. 1B–D) started to increase following I/R injury in a time-dependent manner, plateaued at 4-h reperfusion, and decreased in subsequent hours, returning to the baseline at 16-h reperfusion. Similar results were also observed in the expression of phospho-MLKL (Fig. 1B–D). Thus, 45-min ischemia and 4-h reperfusion were selected to explore the role of PGAM5 in I/R-mediated cardiac damage.
Fig. 1.
PGAM5 upregulation promotes cardiomyocyte necroptosis after I/R injury. (A) Evaluation of cardiac PGAM5 expression through qPCR in mice subjected to I/R surgery. (B) Western blot detection of cardiac PGAM5 and phospho-MLKL expression following I/R injury. (C–D) Quantitative analysis of PGAM5 and phospho-MLKL levels in heart samples processed for western blotting. (E) Evaluation of cardiomyocyte necrosis through the LDH release assay. (F) MTT assay was used to evaluate cell death. (G–I) Representative images of TUNEL and caspase-3 double immunofuorescence in PGAM5f/f and PGAM5CKO cardiomyocytes. The apoptosis inhibitor Z-VAD-FMK and the necroptosis blocker Necrostatin-1 were applied to isolated cardiomyocytes before sI/R injury. (J–O) Western blot detection of apoptosis- and necroptosis-related proteins in heart extracts from PGAM5f/f and PGAM5CKO mice subjected to I/R injury. Before I/R injury, the mice were injected with the apoptosis inhibitor Z-VAD-FMK or the necroptosis blocker Necrostatin-1. Experiments were repeated three times with similar results. Data are shown as the means ± SEM, n = 6 mice or 3 independent cell isolations per group. *p < 0.05.
What is the biologic function within cells of increased PGAM5 in cardiac I/R injury? Previous research showed an important regulatory role for PGAM5 in cell death mediated by apoptosis or necroptosis [13,50]. To test whether PGAM5 participates in apoptosis and/or necroptosis activation in the reperfused heart, the cardiac-specific PGAM5 deletion mice (PGAM5CKO) were generated, and primary cardiomyocytes were isolated from PGAM5CKO mice and their corresponding PGAM5f/f mice. Cell death index, as assessed through LDH release assay (Fig. 1E) and MTT assay (Fig. 1F), was elevated in PGAM5f/f cardiomyocytes and markedly lower in PGAM5CKO cardiomyocytes after sI/R injury. To access whether PGAM5 regulates cardiomyocyte apoptosis and/or necroptosis induced by I/R injury, cardiomyocytes were treated with the apoptosis inhibitor Z-VAD-FMK and the necroptosis blocker Necrostatin-1 (Nec-1). Then, TUNEL and caspase-3 double-immunofluorescence assay was applied to observe apoptosis and necroptosis. After sI/R injury, ~18% PGAM5f/f cardiomyocytes were TUNEL+/Caspase-3+ (apoptotic marker), while ~27% PGAM5f/f cardiomyocytes were TUNEL+/Caspase-3- (necroptosis index) (Fig. 1G–I). This suggests that both apoptosis and necroptosis are activated by reperfusion damage. In sI/R-treated PGAM5CKO cardiomyocytes, only ~4% of cells were TUNEL+/Caspase-3-, whereas the ratio of TUNEL+/Caspase-3+ apoptotic cells was unchanged (~18%) (Fig. 1G–I), suggesting that necroptosis rather than apoptosis was interrupted by PGAM5 deletion. Interestingly, supplementation with Z-VAD-FMK further reduced the ratio of PGAM5CKO TUNEL+/Caspase-3+ cardiomyocytes to ~3%; this reduction was akin to that observed in PGAM5f/f cardiomyocytes co-administered with Z-VAD-FMK and Nec-1 (Fig. 1G–I). These data highlight that the inhibition of apoptosis further represses the sI/R-induced death of PGAM5CKO cardiomyocytes, which suggests that PGAM5 controls necroptosis rather than apoptosis upon cardiac I/R injury. To substantiate these findings, we analyzed the cardiac expression of apoptosis- and necroptosis-related proteins after I/R in vivo. Relative to the sham-operated control mice, the levels of pro-apoptotic proteins (caspase-3, caspase-9, Bax/Bcl2) and pro-necroptotic markers (phospho-MLKL) were markedly increased in I/R-treated PGAM5f/f hearts (Fig. 1J-O). By comparison, the expression of phospho-MLKL, but not of pro-apoptotic proteins, was lower in the hearts of PGAM5CKO mice following I/R injury (Fig. 1J-O).
To evaluate whether PGAM5 upregulation is sufficient to induce cardiomyocyte necroptosis under non-stress conditions, adenovirus-mediated PGAM5 overexpression (Ad-PGAM5) was established in cardiomyocytes isolated from wild-type (WT) mice (Supplemental Fig. 2A and B). Compared to control (Ad-β-gal-transduced) cells, higher LDH release was detected in Ad-PGAM5-transduced cardiomyocytes (Supplemental Fig. 2C). Besides, increased PGAM5 expression was correlated with a rise in the ratio of TUNEL+/Caspase-3- necroptotic cardiomyocytes (Supplemental Fig. 2D–F), underscoring the fact that cardiomyocytes with elevated PGAM5 expression have a pronounced susceptibility to necroptosis in a physiological context. Overall, necroptosis rather than apoptosis is induced by PGAM5 in cardiac I/R injury.
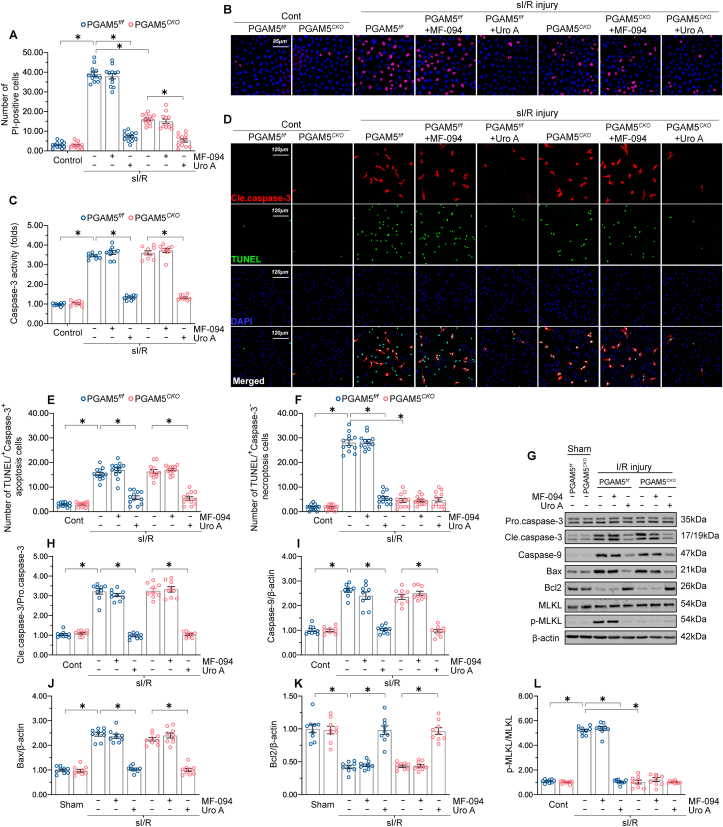
PGAM5 does not trigger mitophagy-related cardiomyocyte death in the reperfused heart.
In addition to apoptosis or necroptosis, PGAM5 has also been shown to positively regulate mitophagic cell death (or mitophagy-related cell death) [51], a newly identified cell death program reported to be a key event in the pathogenesis of cardiovascular disorders [52]. Although mitochondrial autophagy was found to be suppressed at the stage of reperfusion [53,54], it is unclear whether PGAM5 expression influences mitophagic cell death during cardiac I/R injury. To address this, PI staining was conducted to evaluate necroptosis-related cell membrane rupture in primary mouse cardiomyocytes treated with a mitophagy activator (Urolithin A) or a mitophagy antagonist (MF-094) under sI/R injury. First, compared to sI/R-treated cardiomyocytes, administration of MF-094 showed no impact on the ratio of PI+ necroptotic cells in either PGAM5f/f or PGAM5CKO cardiomyocytes (Fig. 2A–B), supporting that cardiomyocytes die independently of mitophagy and that PGAM5 has no association with mitophagy-related cell death. By comparison, following a sI/R challenge, significantly fewer PGAM5f/f and PGAM5CKO cardiomyocytes were PI+ after Urolithin A exposure (Fig. 2A–B). These data might indicate that mitophagy activation provides a pro-survival signal in the reperfused heart. In addition to necroptotic marker PI staining, caspase-3 activity, an apoptotic parameter, was increased by sI/R injury but reduced to near-normal levels by prior exposure to Urolithin A (Fig. 2C). By contrast, MF-094 supplementation had no influence on sI/R-mediated caspase-3 activation in either PGAM5f/f or PGAM5CKO cardiomyocytes (Fig. 2C). With the help of TUNEL and Caspase-3 double immunofluorescence assay, we also observed that MF-094 treatment had no influence on sI/R-induced cardiomyocytes apoptosis (TUNEL+/Caspase-3+) or necroptosis (TUNEL+/Caspase-3-) (Fig. 2D–F). In contrast, supplementation of Urolithin A was associated with the repression of both necroptosis and apoptosis in sI/R-treated cardiomyocytes (Fig. 2D–F).
Fig. 2.
Mitophagy-related cell death is absent in the reperfused heart. (A–B) Representative PI staining images from PGAM5f/f and PGAM5CKO cardiomyocytes treated with either Urolithin A or MF-094 and exposed to sI/R injury. (C) Caspase-3 activity assay results. (D–F) Representative images of TUNEL and caspase-3 double immunofuorescence in PGAM5f/f and PGAM5CKO cardiomyocytes treated with Urolithin A or MF-094 prior to I/R surgery. (G–L) Western blot detection of apoptosis- and necroptosis-related markers in cardiac samples from PGAM5f/f and PGAM5CKO mice treated with Urolithin A or MF-094 prior to I/R surgery. Experiments were repeated three times with similar results. Data are shown as the means ± SEM, n = 6 mice or 3 independent cell isolations per group. *p < 0.05.
In line with the findings in vitro, administration of MF-094 prior to I/R injury in vivo did not affect the expression of pro-apoptotic proteins or necroptosis-related factors in either PGAM5f/f or PGAM5CKO hearts (Fig. 2G-L). However, pre-treatment of Urolithin A to activate mitophagy significantly reduced the levels of pro-apoptotic proteins and necroptosis-related factors in PGAM5f/f and PGAM5CKO hearts (Fig. 2G-L). These data indicate that I/R-mediated PGAM5 upregulation does not promote mitophagy-related cell death in cardiomyocytes, and otherwise suggest that mitophagy has a pro-survival effect in the reperfused heart.
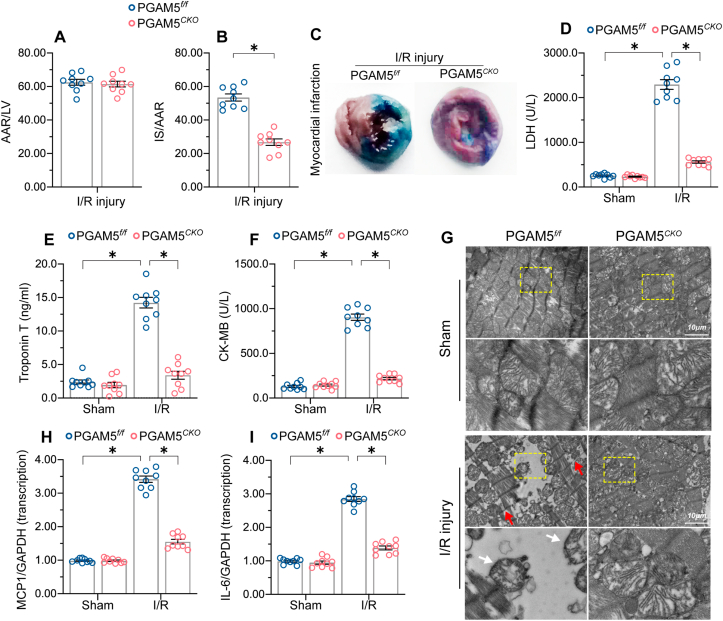
3.2. Cardiac-specific PGAM5 deletion reduces myocardial infarction area and attenuates cardiac inflammation
Compared to PGAM5f/f mice, PGAM5CKO mice exhibited decreased myocardial infarction area after I/R injury (Fig. 3A–C). Serum concentrations of myocardial damage indicators such as LDH, troponin T, and CK-MB were not significantly different between untreated PGAM5f/f and PGAM5CKO mice, but increased significantly, compared to the latter, in PGAM5f/f mice following I/R (Fig. 3D–F). Consistently, histopathological evidence obtained through electron microscopy showed mitochondrial swelling, cardiomyocyte destruction, and Z-line disappearance in heart tissue from PGAM5f/f mice (Fig. 3G). By comparison, improved myocardial structure and mitochondrial morphology following I/R injury was observed in PGAM5CKO mice (Fig. 3G).
Fig. 3.
Cardiac-specific deletion of PGAM5 reduces myocardial infarction area and attenuates the inflammatory response. (A–C). Assessment of area at risk (AAR) and infarct size (IS) using Evans blue and TTC staining, respectively, following 45 min ischemia/6 h reperfusion. Percentual IS and AAR values were multiplied by the corresponding section weight. IS/AAR are presented as percentages. (D–F) ELISA analysis of serum markers of cardiac damage. (G) Representative electron microscopy images of reperfused heart samples. White arrowheads indicate mitochondrial swelling and red arrowheads indicate Z-line disappearance. (H–I) Transcriptional analysis of pro-inflammatory gene expression in cardiomyocytes exposed to sI/R. Experiments were repeated three times with similar results. Data are shown as the means ± SEM, n = 3 independent cell isolations per group. *p < 0.05. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
We next applied qPCR to analyze the expression of pro-inflammatory factors in reperfused heart tissues. The results showed that I/R induced significant upregulation of IL-6 and MCP1 expression in cardiac samples from PGAM5f/f mice, whereas this effect was attenuated in PGAM5CKO mice (Fig. 3H–I), possibly due to a reduction in cardiomyocyte necroptosis after genetic ablation of PGAM5. Under non-stress conditions, the transcription of IL-6 and MCP1 was drastically upregulated in cardiomyocytes transfected with Ad-PGAM5 compared to those transfected with the control Ad-β-gal (Supplemental Fig. 3A and B).
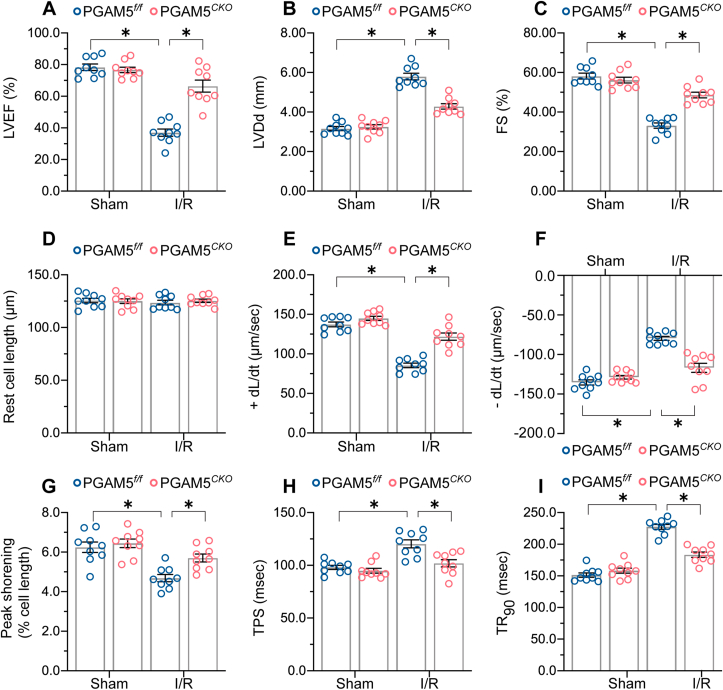
3.3. Genetic ablation of PGAM5 sustains myocardial function upon I/R injury
To investigate whether PGAM5 deletion affects cardiac function, we performed echocardiography in PGAM5f/f and PGAM5CKO mice. PGAM5 deletion had no impact on baseline myocardial performance but attenuated I/R-mediated cardiac depression (Fig. 4A–C). Then, we analyzed the mechanical properties of single cardiomyocytes acutely isolated from I/R-treated mice. Compared to cells from sham-operated mice, resting cardiomyocyte length was not influenced by I/R (Fig. 4D–I). However, the cardiomyocytes from I/R-treated PGAM5f/f mice presented obviously decreased PS and ±dL/dt as well as prolonged TPS and TR90, whereas these alterations were normalized in the cardiomyocytes isolated from PGAM5CKO mice (Fig. 4D–I). In turn, under non-stress conditions, decreased PS and ±dL/dt and extended TPS and TR90 were detected after the transfection of Ad-PGAM5 into primary cardiomyocytes isolated from WT mice (Supplementary Fig. 4A–F). These data suggest that PGAM5 expression compromises myocardial function through impairing cardiomyocyte contractile/relaxation properties.
Fig. 4.
Genetic ablation of PGAM5 sustains myocardial function after I/R injury. (A–C) Echocardiographic measurements performed after I/R injury. n = 6 mice per group. (D–I) Evaluation of mechanical properties of single, acutely isolated cardiomyocytes from I/R-treated mice. +dL/dt: maximal velocity of shortening; −dL/dt: maximal velocity of relengthening; TPS: time-to-peak shortening; TR90: time-to-90% relengthening. n = 70–80 cells from 2 mice per group. Experiments were repeated three times with similar results. Data are shown as the means ± SEM. *p < 0.05.
3.4. PGAM5 deletion improves cardiomyocyte mitochondrial function
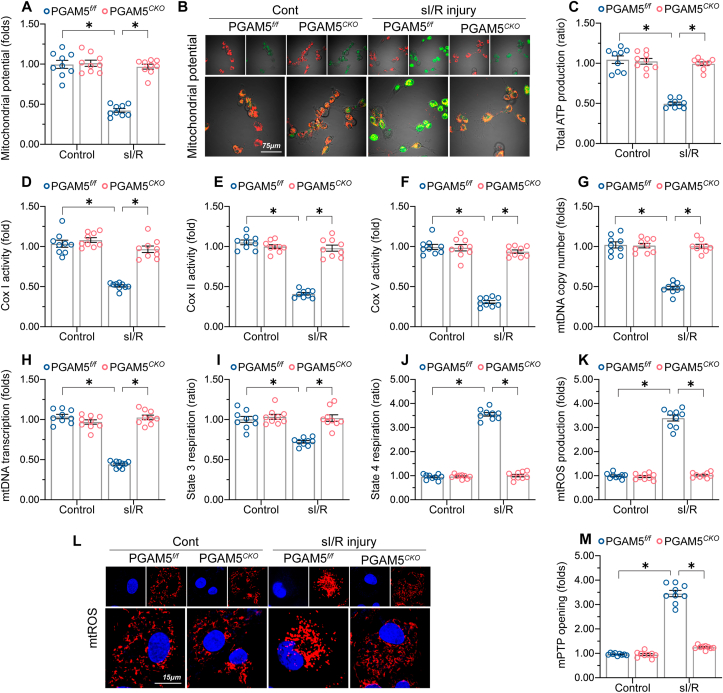
Mitochondrial quality control has emerged as a critical cardioprotective target in the setting of I/R-mediated myocardial damage [20,55,56]. Interestingly, the mechanisms responsible for mitochondrial quality control have been shown to be finely tuned by PGAM5 through pleiotropic mechanisms involving mitochondrial oxidative stress [57], mitochondrial dynamics [14], and mitophagy [[58], [59], [60]]. These findings prompted us to investigate the interdependency of PGAM5 and mitochondrial function in I/R-mediated cardiomyocyte damage. Mitochondrial membrane potential, an index of mitochondrial bioenergetics, was dissipated by sI/R in PGAM5f/f but not in PGAM5CKO cardiomyocytes (Fig. 5A–B). Consistently, cellular ATP production was significantly repressed by sI/R injury but enhanced instead after PGAM5 depletion (Fig. 5C). Interestingly, we observed that the activities of mitochondrial electron transport chain complexes (ETCx), including Cox-I, Cox-II, and Cox-V, were drastically downregulated following sI/R injury in PGAM5f/f but not in PGAM5CKO cardiomyocytes (Fig. 5D–F). Several ETCx components are encoded by mitochondrial DNA (mtDNA). In our study, both mtDNA copy number (Fig. 5G) and mtDNA-specific transcript expression (Fig. 5H) were decreased in sI/R-treated PGAM5f/f cardiomyocytes, an effect paralleled by a decrease in state-3/4 respiration (Fig. 5I–J). In turn, sI/R-treated PGAM5CKO cardiomyocytes showed partial but significant preservation of mtDNA copy number and transcript levels (Fig. 5G–H) as well as improved state-3/4 respiration (Fig. 5I–J). Moreover, due to blunted mitochondrial bioenergetics, mitochondrial ROS (mtROS), a byproduct of mitochondrial respiration, was significantly increased by sI/R injury in PGAM5f/f cardiomyocytes but was maintained at normal levels after the deletion of PGAM5 (Fig. 5K-L). In addition, following sI/R, the opening of the mitochondrial permeability transition pore (mPTP), an event predictive of mitochondria-regulated necroptosis [7,61], was apparently augmented in PGAM5f/f cardiomyocytes but unaffected in PGAM5CKO cardiomyocytes (Fig. 5M). On the other hand, under non-stress conditions, cardiomyocytes transfected with Ad-PGAM5 exhibited lower mitochondrial potential (Supplemental Fig. 5A and B), increased mtROS generation (Supplemental Fig. 5C and D), and augmented mPTP opening rate (Supplemental Fig. 5E). Therefore, these data support the functional importance of PGAM5 in managing cardiomyocytes mitochondrial homeostasis.
Fig. 5.
PGAM5 impairs cardiomyocyte mitochondrial function. (A–B) Representative images of cardiomyocytes loaded with the mitochondrial membrane potential indicator JC-1. (C) Cellular ATP production measurements in cardiomyocytes isolated from PGAM5f/f and PGAM5CKO mice. (D–F) Analysis of ETCx activity (ELISA assay) in primary cardiomyocytes. (G–H) mtDNA copy number was assessed by complex IV segment. The transcript level of mtDNA was assessed by NADH dehydrogenase subunit 1 (ND1). (I–J) Mitochondrial respiration assay. State 3 respiration was initiated by adding ADP; state 4 was defined as the rate of oxygen consumption after ADP phosphorylation. (K–L) Representative images of sI/R-treated cardiomyocytes loaded with the mitochondrial ROS (mtROS) indicator MitoSox red. (M) Measurement of mPTP opening rate through analysis of arbitrary mPTP opening time. Experiments were repeated three times with similar results. Data are shown as the means ± SEM, n = 3 independent cell isolations per group. *p < 0.05. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.5. PGAM5 deletion normalizes mitochondrial dynamics by suppressing Drp1Ser637 dephosphorylation
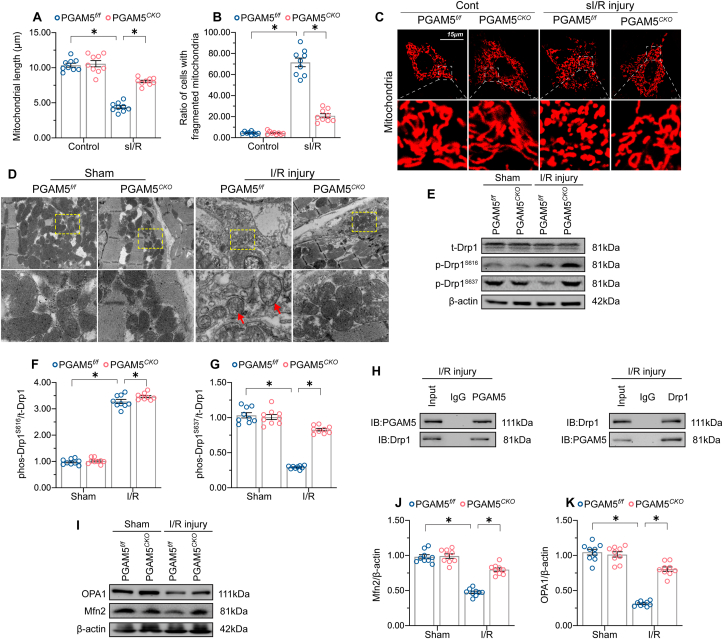
To complement the above observations on mitochondrial bioenergetics and oxidative stress, we monitored whether PGAM5 expression would also affect mitochondrial morphology in cardiomyocytes. As shown in Fig. 6A–C, sI/R injury resulted in mitochondrial fragmentation denoted by organelle cleavage into small spheres or short rods. This effect was counteracted by PGAM5 depletion, which clearly restricted mitochondrial fragmentation after sI/R (Fig. 6A–C). Under non-stress conditions, shorter mitochondria were observed in PGAM5-overexpressing cardiomyocytes compared to control cells (Supplemental Fig. 6A–C). Furthermore, electron microscopy (EM) confirmed the presence of mitochondrial swelling, disorganized cristae, and fragmented, shorter mitochondria in I/R-treated PGAM5f/f heart tissue. In contrast, these alterations were largely absent in heart samples from I/R-treated PGAM5CKO mice (Fig. 6D).
Fig. 6.
PGAM5 deletion normalizes mitochondrial dynamics by suppressing Drp1 dephosphorylation. (A–C) Representative images illustrating mitochondrial morphology in primary isolated cardiomyocytes. The average length of mitochondria and the number of cardiomyocytes with fragmented mitochondria were recorded. (D) Electron microscopy images depicting mitochondrial structure in primary cardiomyocytes. White arrowheads indicate mitochondrial swelling and disorganized mitochondrial cristae. (E–G) Western blot analysis of phospho-Drp1S616 and phospho-Drp1S637 expression in cardiomyocytes isolated from I/R-treated hearts. (H) Results of co-immunoprecipitation assays detecting endogenous Drp1 and PGAM5 interaction in reperfused heart tissues. (I–K) Western blot analysis of OPA1 and Mfn2 expression in cardiomyocytes isolated from I/R-treated hearts. Experiments were repeated three times with similar results. Data are shown as the means ± SEM, n = 6 mice or 3 independent cell isolations per group. *p < 0.05.
PGAM5 was initially discovered as a factor promoting mitochondrial fission through dephosphorylation of Drp1S637 [14]. Evidence suggests that either Drp1S616 phosphorylation or Drp1S637 dephosphorylation promotes Drp1 oligomerization and subsequently drives mitochondria constriction and cleavage [62]. In our study, myocardial Drp1S637 dephosphorylation was enhanced by I/R stimulus, whereas this conformational switch was attenuated by PGAM5 deletion (Fig. 6E–G). Under non-stress conditions, PGAM5 overexpression induced Drp1S637 dephosphorylation (Supplemental Fig. 6D–F). Unlike Drp1S637, PGAM5 deletion did not affect I/R-mediated Drp1S616 phosphorylation (Fig. 6E–G). Similar observations were also noted in cardiomyocytes transfected with Ad-PGAM5 under non-stress conditions (Supplemental Fig. 6D–F). Mechanistically, PGAM5 is a serine/threonine-protein phosphatase, and the crosslink between PGAM5 and Drp1 was predicted theoretically to induce Drp1 dephosphorylation [14]. Endogenous interaction between PGAM5 and Drp1 was confirmed in I/R-treated hearts through co-immunoprecipitation assays (Fig. 6H). Consistently, exogenous PGAM5 was pulled down by Drp1 in cardiomyocytes cultured under non-stress conditions (Supplemental Fig. 6G). These data indicate that PGAM5 acts as an upstream mediator of Drp1S637 dephosphorylation to promote I/R-mediated mitochondrial fission.
Pathological mitochondrial fragmentation may be a combined result of activated fission and suppressed fusion [63]. It remains unclear whether mitochondrial fusion is also influenced by PGAM5. To address this possibility, we compared the expression of mitochondrial fusion-related proteins in PGAM5f/f and PGAM5CKO mice following I/R injury. As shown in Fig. 6I–K, a slight decline in Mfn2 and OPA1 levels occurred in I/R-treated PGAM5f/f cardiomyocytes; however, these proteins maintained baseline expression levels in I/R-treated PGAM5CKO cardiomyocytes. Thus, these data provide preliminary evidence for a regulatory role of PGAM5 in sustaining mitochondrial fusion.
3.6. PGAM5 had no impact on impaired mitophagy in I/R-treated heart
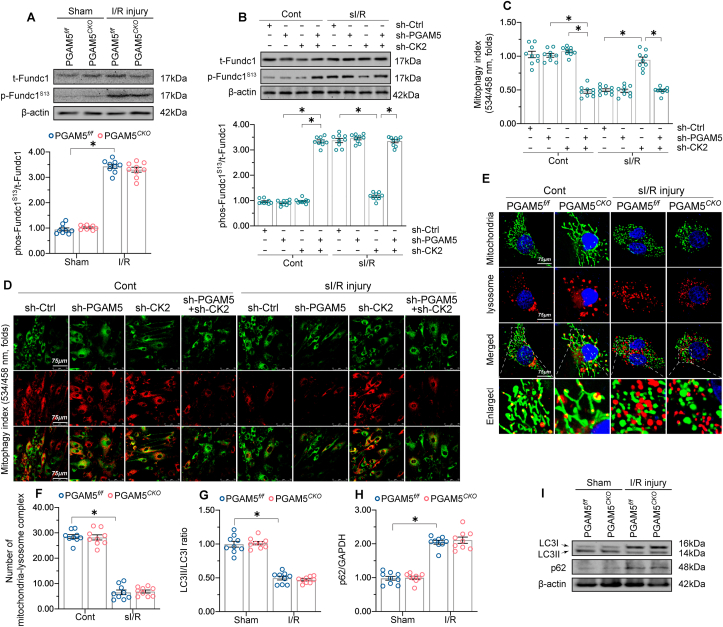
In addition to mitochondrial dynamic, mitophagy eliminates damaged mitochondria and protects the heart against various types of stresses. FUN14 domain-containing protein 1 (Fundc1)-induced mitophagy can be inactivated by casein kinase 2 (CK2)‐mediated phosphorylation of Fundc1S13, but it is then reactivated by PGAM5‐mediated Fundc1S13 dephosphorylation under hypoxic stimulation [60]. These observations prompted us to investigate the potential role of PGAM5 on mitophagy during cardiac I/R. Western blot analysis demonstrated that Fundc1S13 dephosphorylation (i.e., active status) was drastically attenuated by sI/R injury, but PGAM5 depletion did not rescue this effect (Fig. 7A). This suggests that mitophagy was impaired by reperfusion injury independently of PGAM5. Interestingly, CK2 knockdown increased Fundc1S13 dephosphorylation in sI/R-treated cardiomyocytes (Fig. 7B), suggesting that CK2 deletion activates Fundc1. In turn, PGAM5 depletion substantially suppressed the increase in Fundc1S13 dephosphorylation observed in CK2 knockdown cardiomyocytes after sI/R (Fig. 7B). These data indicate that Fundc1 dephosphorylation and activation are prevented by CK2, but enabled by PGAM5 under CK2-deficient conditions. This finding was further validated in cardiomyocytes cultured under non-stress conditions, in which PGAM5 overexpression failed to alter Fundc1S13 phosphorylation levels relative to baseline (Supplemental Fig. 7A and B). By comparison, CK2 deletion induced Fundc1S13 dephosphorylation, suggesting that baseline Fundc1 activity is repressed by CK2 (Supplemental Fig. 7C). Further, in CK2 knockdown cardiomyocytes, PGAM5 deficiency attenuated (Fig. 7B) while PGAM5 overexpression augmented (Supplemental Fig. 7C) Fundc1S13 dephosphorylation. These results suggest that Fundc1 activation is mainly and negatively governed by CK2, whereas PGAM5 functions to restore Fundc1 activation in the absence of CK2.
Fig. 7.
PGAM5 deletion does not alter impaired mitophagy in I/R-treated hearts. (A) proteins were isolated from reperfused herat tissues and then the expression of p-Fundc1S13 expression was detected through western blots. (B) Western blot analysis of p-Fundc1S13 expression in sI/R-treated, WT cardiomyocytes previously transfected with shRNAs against CK2 or PGAM5. (C–D) The mt-Kemia reporter assay was performed in sI/R-exposed cardiomyocytes to detect mitophagic activity in response to PGAM5 deletion. (E–F) Representative images of double-immunofluorescence for mitochondrial (Tom20) and lysosomal (Lamp1) markers. (G–I) Western blot analysis of LC3II/I and p62 expression in PGAM5f/f and PGAM5CKO mice subjected to I/R injury. Experiments were repeated three times with similar results. Data are shown as the means ± SEM, n = 6 mice or 3 independent cell isolations per group. *p < 0.05.
In light of the necessary role of Fundc1 as a mitophagy inducer, the pH-sensitive, mitochondria-targeted mt-Kemia probe was used to detect mitophagic activity in response to PGAM5 deletion under cardiac I/R injury. At the baseline, several acidic mitochondria (or lysosome-mitochondria) complexes were detected in isolated cardiomyocytes (Fig. 7C–D). After exposure to sI/R, the number of acidic complexes was reduced regardless of PGAM5 expression levels (Fig. 7C–D). Indeed, neither Ad-PGAM5 nor Ad-β-gal transfection affected basal mitophagy in cultured cardiomyocytes under non-stress conditions (Supplemental Fig. 7D and E). The formation of lysosome-mitochondria complexes was further validated through double-immunofluorescence for Lamp-1 (lysosome marker) and Tom20 (mitochondrial marker). In untreated cardiomyocytes, normal, filamentous mitochondria appeared partly engulfed by lysosomes, indicative of moderate mitophagy at baseline (Fig. 7E–F). Upon sI/R injury, PGAM5f/f cardiomyocytes exhibited fragmented mitochondria and very scarce lysosome-mitochondrial complexes, suggestive of mitophagy inhibition. However, PGAM5CKO cardiomyocytes contained elongated mitochondria and a preserved mitochondrial network, showing also little interaction with lysosomal vesicles (Fig. 7E–F). Meanwhile, we found in addition that the myocardial expression of the autophagy/mitophagy markers (LC3I/II and p62) was altered by I/R injury but not by PGAM5 deletion (Fig. 7G–I). Therefore, although PGAM5 was first revealed as a contributor to hypoxia-induced mitophagy through the dephosphorylation of Fundc1, our data indicates that PGAM5 levels do not influence post-I/R mitophagic activity in mouse cardiomyocytes.
4. Discussion
This study employed cardiac-specific PGAM5 knockout mice to explore the role of PGAM5 in cardiac I/R injury. Our results illustrate that: (1) PGAM5 transcription and expression are upregulated at the stage of reperfusion; (2) I/R-mediated PGAM5 upregulation promotes necroptosis rather than apoptosis in cardiomyocytes; (3) cardiac-specific deletion of PGAM5 attenuates I/R-mediated necroptosis but not apoptosis; (4) PGAM deletion rescues deficits in mitochondrial homeostasis induced by I/R by increasing mtDNA copy number and transcript levels, improving respiratory capacity, and decreasing mtROS production; (5) I/R promotes mitochondrial fission through the induction of Drp1S616 phosphorylation and Drp1S637 dephosphorylation; PGAM5 deficiency interrupts Drp1S637 dephosphorylation but has no impact on I/R-mediated Drp1S616 phosphorylation, partially attenuating mitochondrial division; (6) mitochondrial fusion, which is inhibited by I/R injury, might be stimulated by PGAM5 deletion; (7) I/R suppresses mitophagy through the upregulation of CK2, whereas PGAM5 deletion fails to rescue this effect; (8) mitophagy-related cell death was not apparent in the reperfused myocardium; instead, mitophagy activation prevents necroptosis and apoptosis in cardiomyocytes exposed to sI/R injury. These data highlight that PGAM5 determines pathology in cardiac I/R injury through the induction of necroptosis rather than apoptosis. Mechanistically, PGAM5 blunts mitochondrial bioenergetics, triggers mitochondrial oxidative stress, partially induces mitochondrial dynamics disorder, and has no effect on mitophagy in the reperfused heart. Taken together, utilizing cardiac-specific conditional PGAM5 knockout assays and adenovirus-mediated gene overexpression experiments, we describe the precise role of PGAM5 in sensitizing cardiomyocytes to necroptosis through the perturbation of mitochondrial quality control.
PGAM5 is a mitochondrial outer-membrane serine/threonine-protein phosphatase first discovered as a Keap1 substrate in eukaryotic cells [64]. Oxidative stress inhibits Keap1-mediated ubiquitination and degradation of PGAM5, suggesting a link between PGAM5 and cellular adaptive responses [57]. In addition, although data from mammalian cells is lacking, the interaction between PGAM5 and apoptosis signal-regulated kinase 1 (ASK1), one of the MAPK kinases accounting for apoptosis induction through the activation of the MAPK-JNK and MAPK-p38 signaling pathways, has been reported in Drosophila and Caenorhabditis elegans [65,66]. Interestingly, a recent study reports that PGAM5 is able to dephosphorylate BCL-xL at Ser62, thus reversing Bcl-xL sequestration of BAX/BAK and conferring resistance to apoptosis [67]. Moreover, PGAM5 has been shown to function as a predominant mediator of TNFα-activated necroptosis and a driver of inflammatory processes through the induction of mitochondrial fission [14,68]. Such a scenario is supported by recent studies that specifically delineate the mechanisms whereby PGAM5 contributes to inflammation-related necroptosis in the liver [69,70] and during acute pancreatitis [71]. Intriguingly, studies addressing the role of PAGM5 in cardiac I/R injury have yielded conflicting results, as both necroptosis stimulation [15] and inhibition [16] have been described in I/R-exposed rat and mouse hearts, respectively, upon PGAM5 inhibition or knockout. Nevertheless, there were clear methodological differences between these studies; whereas in the rat study 1 h/3 h I/R injury was performed in vivo after systemic pharmacological PGAM5 inhibition [15], in the mouse study isolated hearts from PGAM5-KO and PGAM5-WT mice were exposed to a 25 min/90 min I/R protocol ex-vivo [16]. Conceivably, these seemingly contradictory results may be reconciled if PGAM actions were determined by the degree and duration of the ischemic challenge; under this hypothesis, PGAM activity would protect cells from hypoxic cell damage by activating mitophagy early after an ischemic insult, and mediate instead necroptosis once mitophagy becomes ineffective or is suppressed at the stage of reperfusion. Our data using PGAM5CKO mice provide strong evidence that necroptosis rather than apoptosis or mitophagy-related death is driven by PGAM5 in cardiac I/R injury. These interlinked mechanisms between PGAM5 and necroptosis could offer new targets to prevent cardiomyocyte death.
I/R-mediated cardiomyocyte death is associated with mitochondrial dysfunction. Research has shown that oxidative stress, abnormal mitochondrial division, defective mitochondrial fusion, and dysregulated mitophagy may act synergistically to trigger cardiomyocyte death through pleiotropic effects [72,73]. We found that I/R failed to induce mitochondrial fragmentation in PGAM5-depleted cardiomyocytes, an effect linked to blunted PGAM5-mediated Drp1 dephosphorylation. Drp1 possesses two critical phosphorylation sites, which in turn stimulate (Drp1S616) and inhibit (Drp1S637) mitochondrial cleavage upon phosphorylation. Indeed, PGAM5 had been predicted to induce Drp1 S637 dephosphorylation [14]. Interestingly, there is little phosphorylated Drp1S616 under physiological conditions, while Drp1S637 is phosphorylated instead to restrict mitochondrial fission. Upon I/R injury, Drp1S637 becomes dephosphorylated due to PGAM5 upregulation, while Drp1S616 is phosphorylated through PGAM5-independent mechanisms, ultimately facilitating mitochondrial fission. Since PGAM5 deletion interrupts I/R-mediated Drp1S637 dephosphorylation but has not impact on Drp1S616 phosphorylation, mitochondrial fission is partially inhibited. Notably, mitochondrial fission is reported to be an upstream mediator of either apoptosis or necroptosis in cardiomyocytes, although it remains as open questions whether PGAM5-dependent mitochondrial fission is required for necroptosis induction and whether PGAM5-independent mitochondrial cleavage is involved in apoptosis activation.
The reciprocal regulation of mitochondrial fusion and fission has attracted increasing attention in recent years [63]. The present work showed that, in addition to attenuating mitochondrial fission, mitochondrial fusion, which is repressed by I/R injury, was enhanced by PGAM5 knockout. Because many fusion-related proteins participate in the regulation of mitochondrial fission, this effect may be secondary to normalized fission in PGAM5-KO cells [74]. In contrast, we found no obvious association between PGAM5 and mitophagy under our experimental settings. Previous studies demonstrated that Fundc1, a mitophagy inducer, can be inactivated by CK2‐mediated phosphorylation at Ser13, but is then reactivated by PGAM5‐mediated dephosphorylation under hypoxic stimulation [60]. However, abundant phosphorylated Fundc1S13 was noted in the reperfused heart regardless of PGAM5 deletion. Interestingly, in CK2-depleted cardiomyocytes, PGAM5 expression is correlated with dephosphorylated Fundc1S13 levels. This suggests that CK2 plays a prominent role in managing cardiomyocyte mitophagy, whereas PGAM5 may play a compensatory role once CK2 action on Fundc1 is inhibited or lost. This finding is in accordance with our previous study indicating that reperfusion-mediated mitophagy depression is attributable to CK2 upregulation [24]. Along these lines, we excluded the existence of mitophagy-related death in the reperfused heart and confirmed through in vivo treatment with the mitophagy activator urolithin A that mitophagy activation protects cardiomyocytes from I/R-induced death.
In conclusion, our results indicate that PGAM5 upregulation facilitates necroptosis during myocardial I/R injury by impairing mitochondrial quality control mechanisms. These findings lay the foundation for future research addressing the connection between PGAM5, mitochondrial quality surveillance, and cardiomyocyte survival in the context of I/R and suggest the therapeutic potential of modulating cardiac PGAM5 levels in coronary reperfusion surgery patients.
Declaration of competing interest
The authors have declared that they have no conflicts of interest.
Acknowledgements
None.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2020.101777.
Contributor Information
Feng Tian, Email: tianfeng@126.com.
Hao Zhou, Email: zhouhao301@outlook.com, zhouhao@plagh.org.
Funding
This study was supported by grants from the National Natural Science Foundation of China (No. 81900252, 81870249, 82000537). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author contributions
HZ, YT, and ST were involved in the conception and design of the study, performance of experiments, data analysis and interpretation, and manuscript writing; HZ and DM were involved in development of the methodology; FT and HZ acquired the data; HZ and ST analyzed and interpreted data; FT and HZ obtained financial support, supervised the study and approved the final manuscript.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Murphy E., Steenbergen C. Mechanisms underlying acute protection from cardiac ischemia-reperfusion injury. Physiol. Rev. 2008;88(2):581–609. doi: 10.1152/physrev.00024.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davidson S.M., Ferdinandy P., Andreadou I., Botker H.E., Heusch G., Ibanez B., Ovize M., Schulz R., Yellon D.M., Hausenloy D.J., Garcia-Dorado D., Action C.C. Multitarget strategies to reduce myocardial ischemia/reperfusion injury: JACC review topic of the week. J. Am. Coll. Cardiol. 2019;73(1):89–99. doi: 10.1016/j.jacc.2018.09.086. [DOI] [PubMed] [Google Scholar]
- 3.Ibanez B., Heusch G., Ovize M., Van De Werf F. Evolving therapies for myocardial ischemia/reperfusion injury. J. Am. Coll. Cardiol. 2015;65(14):1454–1471. doi: 10.1016/j.jacc.2015.02.032. [DOI] [PubMed] [Google Scholar]
- 4.Hausenloy D.J., Yellon D.M. Myocardial ischemia-reperfusion injury: a neglected therapeutic target. J. Clin. Invest. 2013;123(1):92–100. doi: 10.1172/JCI62874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu P., Hu S., Jin Q., Li D., Tian F., Toan S., Li Y., Zhou H., Chen Y. Ripk3 promotes ER stress-induced necroptosis in cardiac IR injury: a mechanism involving calcium overload/XO/ROS/mPTP pathway. Redox. Biol. 2018;16:157–168. doi: 10.1016/j.redox.2018.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou H., Li D., Zhu P., Ma Q., Toan S., Wang J., Hu S., Chen Y., Zhang Y. Inhibitory effect of melatonin on necroptosis via repressing the Ripk3-PGAM5-CypD-mPTP pathway attenuates cardiac microvascular ischemia-reperfusion injury. J. Pineal Res. 2018;65(3) doi: 10.1111/jpi.12503. [DOI] [PubMed] [Google Scholar]
- 7.Zhang T., Zhang Y., Cui M., Jin L., Wang Y., Lv F., Liu Y., Zheng W., Shang H., Zhang J., Zhang M., Wu H., Guo J., Zhang X., Hu X., Cao C.M., Xiao R.P. CaMKII is a RIP3 substrate mediating ischemia- and oxidative stress-induced myocardial necroptosis. Nat. Med. 2016;22(2):175–182. doi: 10.1038/nm.4017. [DOI] [PubMed] [Google Scholar]
- 8.Oerlemans M.I., Koudstaal S., Chamuleau S.A., De Kleijn D.P., Doevendans P.A., Sluijter J.P. Targeting cell death in the reperfused heart: pharmacological approaches for cardioprotection. Int. J. Cardiol. 2013;165(3):410–422. doi: 10.1016/j.ijcard.2012.03.055. [DOI] [PubMed] [Google Scholar]
- 9.Frank D., Vince J.E. Pyroptosis versus necroptosis: similarities, differences, and crosstalk. Cell Death Differ. 2019;26(1):99–114. doi: 10.1038/s41418-018-0212-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim E.H., Wong S.W., Martinez J. Programmed Necrosis and Disease:We interrupt your regular programming to bring you necroinflammation. Cell Death Differ. 2019;26(1):25–40. doi: 10.1038/s41418-018-0179-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sarhan J., Liu B.C., Muendlein H.I., Weindel C.G., Smirnova I., Tang A.Y., Ilyukha V., Sorokin M., Buzdin A., Fitzgerald K.A., Poltorak A. Constitutive interferon signaling maintains critical threshold of MLKL expression to license necroptosis. Cell Death Differ. 2019;26(2):332–347. doi: 10.1038/s41418-018-0122-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davies K.A., Tanzer M.C., Griffin M.D.W., Mok Y.F., Young S.N., Qin R., Petrie E.J., Czabotar P.E., Silke J., Murphy J.M. The brace helices of MLKL mediate interdomain communication and oligomerisation to regulate cell death by necroptosis. Cell Death Differ. 2018;25(9):1567–1580. doi: 10.1038/s41418-018-0061-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou Z., Han V., Han J. New components of the necroptotic pathway. Protein Cell. 2012;3(11):811–817. doi: 10.1007/s13238-012-2083-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Z., Jiang H., Chen S., Du F., Wang X. The mitochondrial phosphatase PGAM5 functions at the convergence point of multiple necrotic death pathways. Cell. 2012;148(1–2):228–243. doi: 10.1016/j.cell.2011.11.030. [DOI] [PubMed] [Google Scholar]
- 15.She L., Tu H., Zhang Y.Z., Tang L.J., Li N.S., Ma Q.L., Liu B., Li Q., Luo X.J., Peng J. Inhibition of phosphoglycerate mutase 5 reduces necroptosis in rat hearts following ischemia/reperfusion through suppression of dynamin-related protein 1. Cardiovasc. Drugs Ther. 2019;33(1):13–23. doi: 10.1007/s10557-018-06848-8. [DOI] [PubMed] [Google Scholar]
- 16.Lu W., Sun J., Yoon J.S., Zhang Y., Zheng L., Murphy E., Mattson M.P., Lenardo M.J. Mitochondrial protein PGAM5 regulates mitophagic protection against cell necroptosis. PLoS One. 2016;11(1) doi: 10.1371/journal.pone.0147792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang C., Liu X., Yang F., Zhang W., Chen Z., Yan D., You Q., Wu X. Mitochondrial phosphatase PGAM5 regulates Keap1-mediated Bcl-xL degradation and controls cardiomyocyte apoptosis driven by myocardial ischemia/reperfusion injury. In Vitro Cell. Dev. Biol. Anim. 2017;53(3):248–257. doi: 10.1007/s11626-016-0105-2. [DOI] [PubMed] [Google Scholar]
- 18.Fan H., He Z., Huang H., Zhuang H., Liu H., Liu X., Yang S., He P., Yang H., Feng D. Mitochondrial quality control in cardiomyocytes: a critical role in the progression of cardiovascular diseases. Front. Physiol. 2020;11:252. doi: 10.3389/fphys.2020.00252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang J., Toan S., Zhou H. Mitochondrial quality control in cardiac microvascular ischemia-reperfusion injury: new insights into the mechanisms and therapeutic potentials. Pharmacol. Res. 2020;156 doi: 10.1016/j.phrs.2020.104771. [DOI] [PubMed] [Google Scholar]
- 20.Kuznetsov A.V., Javadov S., Margreiter R., Grimm M., Hagenbuchner J., Ausserlechner M.J. The role of mitochondria in the mechanisms of cardiac ischemia-reperfusion injury. Antioxidants. 2019;8(10) doi: 10.3390/antiox8100454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou H., Shi C., Hu S., Zhu H., Ren J., Chen Y. BI1 is associated with microvascular protection in cardiac ischemia reperfusion injury via repressing Syk-Nox2-Drp1-mitochondrial fission pathways. Angiogenesis. 2018;21(3):599–615. doi: 10.1007/s10456-018-9611-z. [DOI] [PubMed] [Google Scholar]
- 22.Zhou H., Wang J., Zhu P., Zhu H., Toan S., Hu S., Ren J., Chen Y. NR4A1 aggravates the cardiac microvascular ischemia reperfusion injury through suppressing FUNDC1-mediated mitophagy and promoting Mff-required mitochondrial fission by CK2alpha. Basic Res. Cardiol. 2018;113(4):23. doi: 10.1007/s00395-018-0682-1. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y., Wang Y., Xu J., Tian F., Hu S., Chen Y., Fu Z. Melatonin attenuates myocardial ischemia-reperfusion injury via improving mitochondrial fusion/mitophagy and activating the AMPK-OPA1 signaling pathways. J. Pineal Res. 2019;66(2) doi: 10.1111/jpi.12542. [DOI] [PubMed] [Google Scholar]
- 24.Zhou H., Zhu P., Wang J., Zhu H., Ren J., Chen Y. Pathogenesis of cardiac ischemia reperfusion injury is associated with CK2alpha-disturbed mitochondrial homeostasis via suppression of FUNDC1-related mitophagy. Cell Death Differ. 2018;25(6):1080–1093. doi: 10.1038/s41418-018-0086-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou H., Toan S., Zhu P., Wang J., Ren J., Zhang Y. DNA-PKcs promotes cardiac ischemia reperfusion injury through mitigating BI-1-governed mitochondrial homeostasis. Basic Res. Cardiol. 2020;115(2):11. doi: 10.1007/s00395-019-0773-7. [DOI] [PubMed] [Google Scholar]
- 26.Akbari P., Huijbers E.J.M., Themeli M., Griffioen A.W., Van Beijnum J.R. The tumor vasculature an attractive CAR T cell target in solid tumors. Angiogenesis. 2019;22(4):473–475. doi: 10.1007/s10456-019-09687-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Florido A., Saraiva N., Cerqueira S., Almeida N., Parsons M., Batinic-Haberle I., Miranda J.P., Costa J.G., Carrara G., Castro M., Oliveira N.G., Fernandes A.S. The manganese(III) porphyrin MnTnHex-2-PyP(5+) modulates intracellular ROS and breast cancer cell migration: impact on doxorubicin-treated cells. Redox. Biol. 2019;20:367–378. doi: 10.1016/j.redox.2018.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Derosa L., Bayar M.A., Albiges L., Le Teuff G., Escudier B. A new prognostic model for survival in second line for metastatic renal cell carcinoma: development and external validation. Angiogenesis. 2019;22(3):383–395. doi: 10.1007/s10456-019-09664-2. [DOI] [PubMed] [Google Scholar]
- 29.Galjart B., Nierop P.M.H., Van Der Stok E.P., Van Den Braak R., Höppener D.J., Daelemans S., Dirix L.Y., Verhoef C., Vermeulen P.B., Grünhagen D.J. Angiogenic desmoplastic histopathological growth pattern as a prognostic marker of good outcome in patients with colorectal liver metastases. Angiogenesis. 2019;22(2):355–368. doi: 10.1007/s10456-019-09661-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mapuskar K.A., Wen H., Holanda D.G., Rastogi P., Steinbach E., Han R., Coleman M.C., Attanasio M., Riley D.P., Spitz D.R., Allen B.G., Zepeda-Orozco D. Persistent increase in mitochondrial superoxide mediates cisplatin-induced chronic kidney disease. Redox. Biol. 2019;20:98–106. doi: 10.1016/j.redox.2018.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mo J., Enkhjargal B., Travis Z.D., Zhou K., Wu P., Zhang G., Zhu Q., Zhang T., Peng J., Xu W., Ocak U., Chen Y., Tang J., Zhang J., Zhang J.H. AVE 0991 attenuates oxidative stress and neuronal apoptosis via Mas/PKA/CREB/UCP-2 pathway after subarachnoid hemorrhage in rats. Redox. Biol. 2019;20:75–86. doi: 10.1016/j.redox.2018.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Narzt M.S., Nagelreiter I.M., Oskolkova O., Bochkov V.N., Latreille J., Fedorova M., Ni Z., Sialana F.J., Lubec G., Filzwieser M., Laggner M., Bilban M., Mildner M., Tschachler E., Grillari J., Gruber F. A novel role for NUPR1 in the keratinocyte stress response to UV oxidized phospholipids. Redox. Biol. 2019;20:467–482. doi: 10.1016/j.redox.2018.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hou P., Li H., Yong H., Chen F., Chu S., Zheng J., Bai J. PinX1 represses renal cancer angiogenesis via the mir-125a-3p/VEGF signaling pathway. Angiogenesis. 2019;22(4):507–519. doi: 10.1007/s10456-019-09675-z. [DOI] [PubMed] [Google Scholar]
- 34.Guo S., Lu J., Zhuo Y., Xiao M., Xue X., Zhong S., Shen X., Yin C., Li L., Chen Q., Zhu M., Chen B., Zhao M., Zheng L., Tao Y., Yin H. Endogenous cholesterol ester hydroperoxides modulate cholesterol levels and inhibit cholesterol uptake in hepatocytes and macrophages. Redox. Biol. 2019;21 doi: 10.1016/j.redox.2018.101069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bellot G.L., Dong X., Lahiri A., Sebastin S.J., Batinic-Haberle I., Pervaiz S., Puhaindran M.E. MnSOD is implicated in accelerated wound healing upon Negative Pressure Wound Therapy (NPWT): a case in point for MnSOD mimetics as adjuvants for wound management. Redox. Biol. 2019;20:307–320. doi: 10.1016/j.redox.2018.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang H.H., Wu Y.J., Tseng Y.M., Su C.H., Hsieh C.L., Yeh H.I. Mitochondrial fission protein 1 up-regulation ameliorates senescence-related endothelial dysfunction of human endothelial progenitor cells. Angiogenesis. 2019;22(4):569–582. doi: 10.1007/s10456-019-09680-2. [DOI] [PubMed] [Google Scholar]
- 37.Feng W., Gong H., Wang Y., Zhu G., Xue T., Wang Y., Cui G. circIFT80 functions as a ceRNA of miR-1236-3p to promote colorectal cancer progression. Mol. Ther. Nucleic Acids. 2019;18:375–387. doi: 10.1016/j.omtn.2019.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Zhou H., Hu S., Jin Q., Shi C., Zhang Y., Zhu P., Ma Q., Tian F., Chen Y. Mff-dependent mitochondrial fission contributes to the pathogenesis of cardiac microvasculature ischemia/reperfusion injury via induction of mROS-mediated cardiolipin oxidation and HK2/VDAC1 disassociation-involved mPTP opening. J. Am. Heart Assoc. 2017;6(3) doi: 10.1161/JAHA.116.005328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kowaltowski A.J. Strategies to detect mitochondrial oxidants. Redox. Biol. 2019;21 doi: 10.1016/j.redox.2018.101065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou H., Zhu P., Guo J., Hu N., Wang S., Li D., Hu S., Ren J., Cao F., Chen Y. Ripk3 induces mitochondrial apoptosis via inhibition of FUNDC1 mitophagy in cardiac IR injury. Redox. Biol. 2017;13:498–507. doi: 10.1016/j.redox.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim Y.R., Baek J.I., Kim S.H., Kim M.A., Lee B., Ryu N., Kim K.H., Choi D.G., Kim H.M., Murphy M.P., Macpherson G., Choo Y.S., Bok J., Lee K.Y., Park J.W., Kim U.K. Therapeutic potential of the mitochondria-targeted antioxidant MitoQ in mitochondrial-ROS induced sensorineural hearing loss caused by Idh2 deficiency. Redox. Biol. 2019;20:544–555. doi: 10.1016/j.redox.2018.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guo Y., Nong Y., Tukaye D.N., Rokosh G., Du J., Zhu X., Book M., Tomlin A., Li Q., Bolli R. Inducible cardiac-specific overexpression of cyclooxygenase-2 (COX-2) confers resistance to ischemia/reperfusion injury. Basic Res. Cardiol. 2019;114(5):32. doi: 10.1007/s00395-019-0741-2. [DOI] [PubMed] [Google Scholar]
- 43.Fuhrmann D.C., Wittig I., Brune B. TMEM126B deficiency reduces mitochondrial SDH oxidation by LPS, attenuating HIF-1alpha stabilization and IL-1beta expression. Redox. Biol. 2019;20:204–216. doi: 10.1016/j.redox.2018.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou H., Zhu P., Wang J., Toan S., Ren J. DNA-PKcs promotes alcohol-related liver disease by activating Drp1-related mitochondrial fission and repressing FUNDC1-required mitophagy. Signal Transduct. Target Ther. 2019;4(1):56. doi: 10.1038/s41392-019-0094-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang J., Zhu P., Li R., Ren J., Zhang Y., Zhou H. Bax inhibitor 1 preserves mitochondrial homeostasis in acute kidney injury through promoting mitochondrial retention of PHB2. Theranostics. 2020;10(1):384–397. doi: 10.7150/thno.40098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Imber M., Pietrzyk-Brzezinska A.J., Antelmann H. Redox regulation by reversible protein S-thiolation in Gram-positive bacteria. Redox. Biol. 2019;20:130–145. doi: 10.1016/j.redox.2018.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang J., Zhu P., Li R., Ren J., Zhou H. Fundc1-dependent mitophagy is obligatory to ischemic preconditioning-conferred renoprotection in ischemic AKI via suppression of Drp1-mediated mitochondrial fission. Redox. Biol. 2020;30 doi: 10.1016/j.redox.2019.101415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kelly S.C., Patel N.N., Eccardt A.M., Fisher J.S. Glucose-dependent trans-plasma membrane electron transport and p70(S6k) phosphorylation in skeletal muscle cells. Redox. Biol. 2019;27 doi: 10.1016/j.redox.2018.101075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Motterlini R., Nikam A., Manin S., Ollivier A., Wilson J.L., Djouadi S., Muchova L., Martens T., Rivard M., Foresti R. HYCO-3, a dual CO-releaser/Nrf2 activator, reduces tissue inflammation in mice challenged with lipopolysaccharide. Redox. Biol. 2019;20:334–348. doi: 10.1016/j.redox.2018.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu D., Dasgupta A., Chen K.H., Neuber-Hess M., Patel J., Hurst T.E., Mewburn J.D., Lima P.D.A., Alizadeh E., Martin A., Wells M., Snieckus V., Archer S.L. Identification of novel dynamin-related protein 1 (Drp1) GTPase inhibitors: therapeutic potential of Drpitor1 and Drpitor1a in cancer and cardiac ischemia-reperfusion injury. Faseb. J. 2020;34(1):1447–1464. doi: 10.1096/fj.201901467R. [DOI] [PubMed] [Google Scholar]
- 51.Lenhausen A.M., Wilkinson A.S., Lewis E.M., Dailey K.M., Scott A.J., Khan S., Wilkinson J.C. Apoptosis inducing factor binding protein PGAM5 triggers mitophagic cell death that is inhibited by the ubiquitin ligase activity of X-linked inhibitor of apoptosis. Biochemistry. 2016;55(23):3285–3302. doi: 10.1021/acs.biochem.6b00306. [DOI] [PubMed] [Google Scholar]
- 52.Denton D., Kumar S. Autophagy-dependent cell death. Cell Death Differ. 2019;26(4):605–616. doi: 10.1038/s41418-018-0252-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang M., Linn B.S., Zhang Y., Ren J. Mitophagy and mitochondrial integrity in cardiac ischemia-reperfusion injury. Biochim. Biophys. Acta (BBA) - Mol. Basis Dis. 2019;1865(9):2293–2302. doi: 10.1016/j.bbadis.2019.05.007. [DOI] [PubMed] [Google Scholar]
- 54.Ma X., Liu H., Foyil S.R., Godar R.J., Weinheimer C.J., Diwan A. Autophagy is impaired in cardiac ischemia-reperfusion injury. Autophagy. 2012;8(9):1394–1396. doi: 10.4161/auto.21036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Honda H.M., Korge P., Weiss J.N. Mitochondria and ischemia/reperfusion injury. Ann. N. Y. Acad. Sci. 2005;1047:248–258. doi: 10.1196/annals.1341.022. [DOI] [PubMed] [Google Scholar]
- 56.Lee B.W.L., Ghode P., Ong D.S.T. Redox regulation of cell state and fate. Redox. Biol. 2019;25 doi: 10.1016/j.redox.2018.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lo S.C., Hannink M. PGAM5 tethers a ternary complex containing Keap1 and Nrf2 to mitochondria. Exp. Cell Res. 2008;314(8):1789–1803. doi: 10.1016/j.yexcr.2008.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Imai Y., Kanao T., Sawada T., Kobayashi Y., Moriwaki Y., Ishida Y., Takeda K., Ichijo H., Lu B., Takahashi R. The loss of PGAM5 suppresses the mitochondrial degeneration caused by inactivation of PINK1 in Drosophila. PLoS Genet. 2010;6(12) doi: 10.1371/journal.pgen.1001229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu H., Xue D., Chen G., Han Z., Huang L., Zhu C., Wang X., Jin H., Wang J., Zhu Y., Liu L., Chen Q. The BCL2L1 and PGAM5 axis defines hypoxia-induced receptor-mediated mitophagy. Autophagy. 2014;10(10):1712–1725. doi: 10.4161/auto.29568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen G., Han Z., Feng D., Chen Y., Chen L., Wu H., Huang L., Zhou C., Cai X., Fu C., Duan L., Wang X., Liu L., Liu X., Shen Y., Zhu Y., Chen Q. A regulatory signaling loop comprising the PGAM5 phosphatase and CK2 controls receptor-mediated mitophagy. Mol. Cell. 2014;54(3):362–377. doi: 10.1016/j.molcel.2014.02.034. [DOI] [PubMed] [Google Scholar]
- 61.Lu L.Q., Tian J., Luo X.J., Peng J. Targeting the pathways of regulated necrosis: a potential strategy for alleviation of cardio-cerebrovascular injury. Cell. Mol. Life Sci. 2020;06:625–626. doi: 10.1007/s00018-020-03587-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Feng S.T., Wang Z.Z., Yuan Y.H., Wang X.L., Sun H.M., Chen N.H., Zhang Y. Dynamin-related protein 1: a protein critical for mitochondrial fission, mitophagy, and neuronal death in Parkinson's disease. Pharmacol. Res. 2020;151 doi: 10.1016/j.phrs.2019.104553. [DOI] [PubMed] [Google Scholar]
- 63.Sabouny R., Shutt T.E. Reciprocal regulation of mitochondrial fission and fusion. Trends Biochem. Sci. 2020;45(7):564–577. doi: 10.1016/j.tibs.2020.03.009. [DOI] [PubMed] [Google Scholar]
- 64.Lo S.C., Hannink M. PGAM5, a Bcl-XL-interacting protein, is a novel substrate for the redox-regulated Keap1-dependent ubiquitin ligase complex. J. Biol. Chem. 2006;281(49):37893–37903. doi: 10.1074/jbc.M606539200. [DOI] [PubMed] [Google Scholar]
- 65.Takeda K., Komuro Y., Hayakawa T., Oguchi H., Ishida Y., Murakami S., Noguchi T., Kinoshita H., Sekine Y., Iemura S., Natsume T., Ichijo H. Mitochondrial phosphoglycerate mutase 5 uses alternate catalytic activity as a protein serine/threonine phosphatase to activate ASK1. Proc. Natl. Acad. Sci. U. S. A. 2009;106(30):12301–12305. doi: 10.1073/pnas.0901823106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stepkowski T.M., Kruszewski M.K. Molecular cross-talk between the NRF2/KEAP1 signaling pathway, autophagy, and apoptosis. Free Radic. Biol. Med. 2011;50(9):1186–1195. doi: 10.1016/j.freeradbiomed.2011.01.033. [DOI] [PubMed] [Google Scholar]
- 67.Ma K., Zhang Z., Chang R., Cheng H., Mu C., Zhao T., Chen L., Zhang C., Luo Q., Lin J., Zhu Y., Chen Q. Dynamic PGAM5 multimers dephosphorylate BCL-xL or FUNDC1 to regulate mitochondrial and cellular fate. Cell Death Differ. 2020;27(3):1036–1051. doi: 10.1038/s41418-019-0396-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kang T.B., Yang S.H., Toth B., Kovalenko A., Wallach D. Caspase-8 blocks kinase RIPK3-mediated activation of the NLRP3 inflammasome. Immunity. 2013;38(1):27–40. doi: 10.1016/j.immuni.2012.09.015. [DOI] [PubMed] [Google Scholar]
- 69.Brenner C., Galluzzi L., Kepp O., Kroemer G. Decoding cell death signals in liver inflammation. J. Hepatol. 2013;59(3):583–594. doi: 10.1016/j.jhep.2013.03.033. [DOI] [PubMed] [Google Scholar]
- 70.Takemoto K., Hatano E., Iwaisako K., Takeiri M., Noma N., Ohmae S., Toriguchi K., Tanabe K., Tanaka H., Seo S., Taura K., Machida K., Takeda N., Saji S., Uemoto S., Asagiri M. Necrostatin-1 protects against reactive oxygen species (ROS)-induced hepatotoxicity in acetaminophen-induced acute liver failure. FEBS Open Bio. 2014;4:777–787. doi: 10.1016/j.fob.2014.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mukherjee R., Mareninova O.A., Odinokova I.V., Huang W., Murphy J., Chvanov M., Javed M.A., Wen L., Booth D.M., Cane M.C., Awais M., Gavillet B., Pruss R.M., Schaller S., Molkentin J.D., Tepikin A.V., Petersen O.H., Pandol S.J., Gukovsky I., Criddle D.N., Gukovskaya A.S., Sutton R., Unit N.P.B.R. Mechanism of mitochondrial permeability transition pore induction and damage in the pancreas: inhibition prevents acute pancreatitis by protecting production of ATP. Gut. 2016;65(8):1333–1346. doi: 10.1136/gutjnl-2014-308553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ramachandra C.J.A., Hernandez-Resendiz S., Crespo-Avilan G.E., Lin Y.H., Hausenloy D.J. Mitochondria in acute myocardial infarction and cardioprotection. EBioMedicine. 2020;57 doi: 10.1016/j.ebiom.2020.102884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Poznyak A.V., Ivanova E.A., Sobenin I.A., Yet S.F., Orekhov A.N. The role of mitochondria in cardiovascular diseases. Biology. 2020;9(6) doi: 10.3390/biology9060137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dorn G.W. Evolving concepts of mitochondrial dynamics. Annu. Rev. Physiol. 2019;81:1–17. doi: 10.1146/annurev-physiol-020518-114358. 2nd. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article.