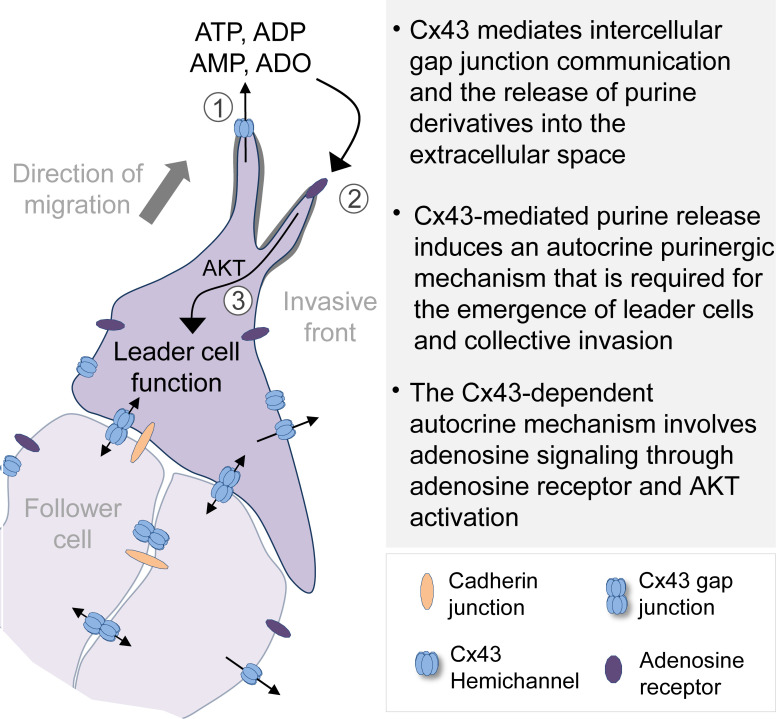
Khalil et al. show that breast cancer cells release purines into the extracellular space via connexin-43 hemichannels. Extracellular purines activate adenosine receptors and the AKT pathway to induce and maintain leader cell function and collective cancer cell invasion.
Abstract
Progression of epithelial cancers predominantly proceeds by collective invasion of cell groups with coordinated cell–cell junctions and multicellular cytoskeletal activity. Collectively invading breast cancer cells express the gap junction protein connexin-43 (Cx43), yet whether Cx43 regulates collective invasion remains unclear. We here show that Cx43 mediates gap-junctional coupling between collectively invading breast cancer cells and, via hemichannels, adenosine nucleotide/nucleoside release into the extracellular space. Using molecular interference and rescue strategies, we identify that Cx43 hemichannel function, but not intercellular communication, induces leader cell activity and collective migration through the engagement of the adenosine receptor 1 (ADORA1) and AKT signaling. Accordingly, pharmacological inhibition of ADORA1 or AKT signaling caused leader cell collapse and halted collective invasion. ADORA1 inhibition further reduced local invasion of orthotopic mammary tumors in vivo, and joint up-regulation of Cx43 and ADORA1 in breast cancer patients correlated with decreased relapse-free survival. This identifies autocrine purinergic signaling, through Cx43 hemichannels, as a critical pathway in leader cell function and collective invasion.
Graphical Abstract
Introduction
Collective invasion of multicellular groups with intact cell–cell junctions is an important mechanism to local invasion and metastasis in epithelial cancers (Gaggioli et al., 2007; Friedl and Gilmour, 2009; Cheung and Ewald, 2016). Collective invasion is initiated and maintained by leader cells that extend actin-rich protrusions, engage with the ECM to exert traction and ECM remodeling, and maintain cell–cell coupling with follower cells (Cheung et al., 2013; Westcott et al., 2015). Mechanical connection and cell-to-cell signaling between moving cells are provided by adherens junctions, through E-, N-, and other cadherins (Theveneau and Mayor, 2012). Cadherins coordinate the actomyosin cytoskeleton through catenins and vinculin (Hartsock and Nelson, 2008; Han and de Rooij, 2016) and secure supracellular force balance, front–rear and apicobasal polarity, and multicellular branching (Tambe et al., 2011).
In nonneoplastic collective processes during morphogenesis and regeneration, moving cell sheets are further connected by gap junctions (Ashton et al., 1999; Huang et al., 1998; Kotini et al., 2018; Marins et al., 2009). Gap junctions consist of connexins (Cxs) oligomerized into hemichannels that engage across cell membranes (Goodenough and Paul, 2009). The resulting transmembrane connections mediate gap-junctional transfer of ions and small molecules (<1 kD), including Ca2+, phosphoinosites, and nucleotides (Goodenough and Paul, 2009). Cxs mediate multicellular contractility of cardiomyocytes and the myoepithelial layer of mammary ducts (Kumai et al., 2000; Mroue et al., 2015) and, by unknown mechanisms, collective movement of neural and endothelial cells during morphogenesis (Ashton et al., 1999; Huang et al., 1998; Marins et al., 2009). The mechanisms by which Cxs support coordinated cytoskeletal contractility and multicellular dynamics vary. Direct cell-to-cell signaling occurs through gap-junctional intercellular transfer of second messengers, including IP3 and cAMP, energy equivalents (ATP and glucose), and Ca2+ wave propagation (Boitano et al., 1992; Goldberg et al., 2004; Howe, 2004). Cxs further contribute to gene expression of cadherins (Kotini et al., 2018) and/or the release of chemotactic factors to induce cell polarity and migration (Barletta et al., 2012; Haynes et al., 2006; Kaczmarek et al., 2005). In morphogenesis, Cxs are indispensable for coordinated tissue growth (Sinyuk et al., 2018). By regulating cell differentiation, Cxs further counteract neoplastic transformation (Bazzoun et al., 2019; Fostok et al., 2019; Saunders et al., 2001; Zhang et al., 2003), and forced expression of Cxs in transformed cells reduces tumor cell growth and inhibits invasion by reverting the epithelial-to-mesenchymal transition (Kazan et al., 2019; McLachlan et al., 2006). On the other hand, Cxs remain expressed in several solid tumors, and their expression increases in metastases (Bos et al., 2009; Elzarrad et al., 2008; Kanczuga-Koda et al., 2006; Stoletov et al., 2013). Interference with Cx43 expression or channel function inhibits cancer cell migration in vitro (Ogawa et al., 2012) and reduces metastatic seeding by reducing binding of circulating tumor cells to vascular endothelial cells (el-Sabban and Pauli, 1991; Elzarrad et al., 2008; Stoletov et al., 2013). Because Cxs exert a range of functions, and these may vary between tumor types and experimental conditions, an integrating concept on how Cxs either suppress or enhance neoplastic progression is lacking (Aasen et al., 2019; Naus and Laird, 2010).
Whereas a role of Cxs in collective processes in morphogenesis has been established, the contribution of gap-junctional communication in collective tumor cell invasion remains unclear (Friedl and Gilmour, 2009). We here revisited Cx43 expression and function in breast cancer models of collective invasion. Using pharmacological inhibition, molecular interference, and rescue strategies, we show that Cx43 mediates gap-junctional intercellular communication (GJIC) between collectively invading cells and, via hemichannels, the release of purine derivatives into the extracellular space. Cx43 hemichannel-mediated release of adenosine (ADO) nucleotides initiate an autocrine loop inducing leader cell function and collective invasion, but not single-cell motility, through ADO receptor 1 (ADORA1) and AKT/protein kinase B signaling.
Results
Breast cancer cells express Cx43 during collective invasion
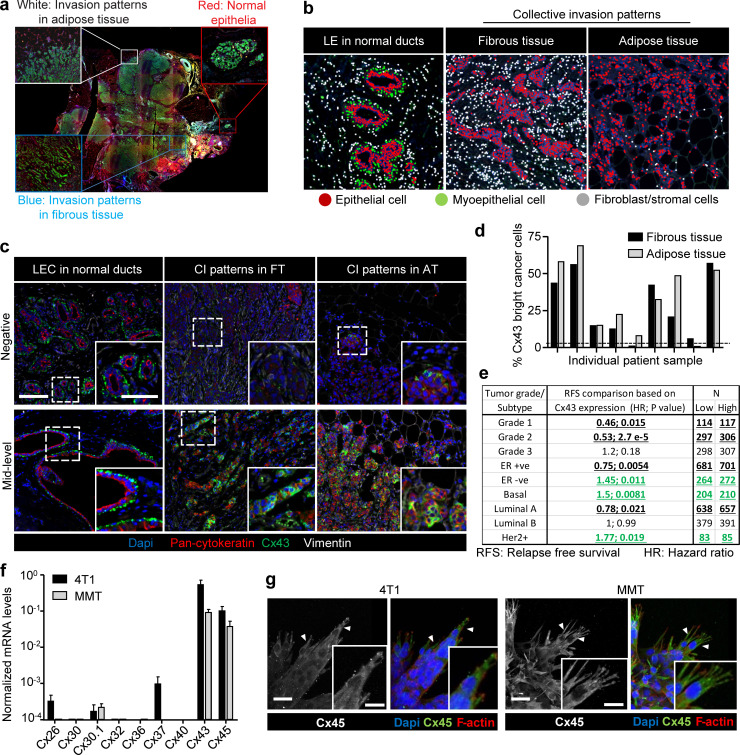
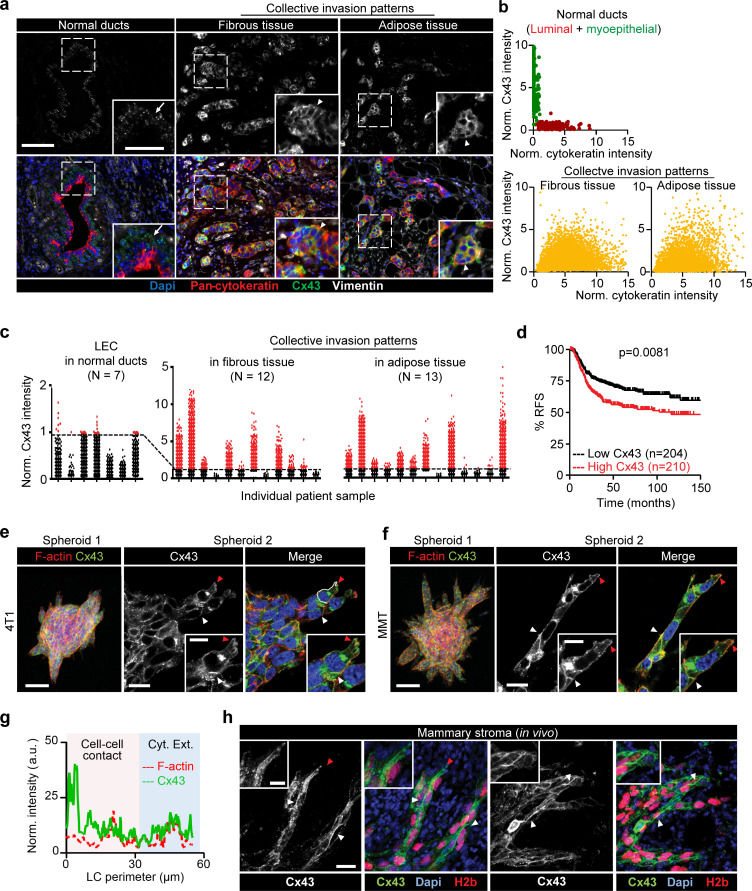
To address whether Cxs are expressed in invasive ductal breast carcinoma infiltrating the fibrous tumor stroma and adipose tissue, Cx43 expression was detected in patient samples by multispectral imaging (Fig. S1, a and b; and Mascaux et al., 2019). Nonneoplastic ducts consisted of luminal epithelial cells with negative to low Cx43 expression and Cx43-positive myoepithelial cells (Fig. 1, a and b; and Fig. S1 c). In tumors, however, multicellular epithelial strands or roundish clusters with cytokeratin expression located in the fibrous or adipose tissue showed increased Cx43 expression in 9 out of 12 samples, and expression was increased compared with normal luminal epithelium (Fig. 1 a, arrowheads; Fig. 1 c; and Fig. S1, c and d). The fraction of Cx43bright cells, as quantified by single-cell in situ cytometry in Cx43-positive samples (Fig. 1 c, red label), varied from 5% to 75% (Fig. S1 d). This indicates that invasive ductal carcinomas contain subregions of high Cx43 expression in the invasion zones. Using meta-analysis of the Györffy cohort of breast cancer patients (Györffy et al., 2010), high Cx43 gene expression was associated with decreased relapse-free survival (RFS) in patients with estrogen receptor (ER)–negative subtypes, including the basal subtype (Fig. 1 d), and Her2-positive tumors (Fig. S1 e). No inverse association was detected when patients were classified based on tumor grade or on the ER-positive subtype (Fig. S1 e).
Figure S1.
Expression of Cx43 in breast cancer cells during collective cancer cell invasion. (a) Original colored fluorescence overview of breast cancer lesions with zoomed insets on normal ducts and tumor invasive margins. (b) Trainable cell phenotyping function of the Inform software to identify different cell types within a multicellular tissue based on spectral fluorescence fingerprinting. Images display the results of the per-cell phenotyping analysis in normal ducts (luminal epithelial cells [LE]) and collective invasion (CI) patterns in fibrous and in adipose tissue. In each region, different cell types are marked by colored dots, as indicated. For all regions devoid of bilayered ducts, cells of epithelial origin, based on pan-cytokeratin, were considered as invasive breast cancer cells. (c) Unmixed composite images of Cx43 intensity in epithelial cancer cells within the tumor margins of representative samples with negative (upper panel) and mid-level (lower panel) Cx43 expression. (d) Heterogeneous expression of Cx43 within cancer cells of the collective invasion patterns in different tumor regions and samples. Values represent the percentage of Cx43 positive cancer cells. (e) Summary of Kaplan–Meier survival analysis correlating RFS with Cx43 expression in breast cancer patients categorized based on grade or molecular subtype. Bold and underlined values indicate positive (black) or inverse (green) association with Cx43 expression. P values, log-rank test. (f) Quantitative PCR for nine connexin subtypes relevant for the mammary gland (Cx26, Cx30, Cx30.1, Cx32, Cx36, Cx37, Cx40, Cx43, and Cx45). Connexin mRNA expression in 4T1 (black) and MMT (gray) cells from 3D collagen invasion cultures. Values represent mean normalized mRNA levels and SEM from three independent experiments. (g) Cx45 localization in collectively invading strands in 3D collagen. Confocal images represent maximum intensity projections from 3D confocal stacks. Arrowheads, cytoplasmic protrusions. Scale bars: 100 µm (c), 50 µm (c, inset), 20 µm (g).
Figure 1.
Increased expression of Cx43 in breast cancer cells during collective cancer cell invasion in vivo and in vitro. (a) Multispectral microscopy of breast cancer tissue sections stained for Cx43 (green), pan-cytokeratin (red; epithelial cells), vimentin (white; stromal cells, cancer cells after epithelial-to-mesenchymal transition). The three panels represent example slices for normal ducts and the invasion zones in the collagen-rich fibrous or adipose tissue. Detection of Cx43 in myoepithelial cells surrounding the luminal epithelium and epithelial cancer cells along cell–cell junctions. Arrows and arrowheads depict Cx43 at cell–cell contacts with puncta or linear patterns, respectively. (b) Colocalization analysis of Cx43 versus pan-cytokeratin expression and classified myoepithelial cells, luminal epithelial cells, and invading cancer cells in fibrous or adipose tissue. Cx43 and pan-cytokeratin levels were acquired using supervised automated tissue segmentation software (Inform). (c) Cx43 levels in luminal epithelium cells (LEC) and cancer cells within the fibrous or adipose tissue from 13 clinical samples (see Table 1 for sample details). Dotted line represents the threshold level for Cx43 negativity, determined by receiving operator characteristic (ROC) analysis with intensity values obtained from the luminal epithelial cells as control. (d) Kaplan–Meier survival plot predicting RFS for high versus low Cx43 expression in basal-type breast cancer patients (Györffy et al., 2010). P values, log-rank test. (e and f) Distribution of Cx43 along cell–cell junctions (white arrowheads) and at the polar extensions (red arrowheads) of leader cells during collective invasion of 4T1 (e) and MMT (f) spheroids in 3D collagen. (g) Intensity distribution of Cx43 and F-actin along the circumference of a leader cell (LC; e, white dashed border). Values show the pixel intensity with background subtraction. Confocal images are displayed as maximum-intensity projections from a 3D confocal stack. (h) Cx43 expression and distribution during collective invasion of 4T1 cells in the stroma of the mouse mammary gland, 4 d after implantation and monitored by confocal microscopy. Scale bars: 100 µm (a, e, and f, overview), 50 µm (a, inset), 20 µm (e and f, spheroid 2; h), 10 µm (e, f, and h, inset). Cyt. Ext., cytoplasmic extension. Norm, normal.
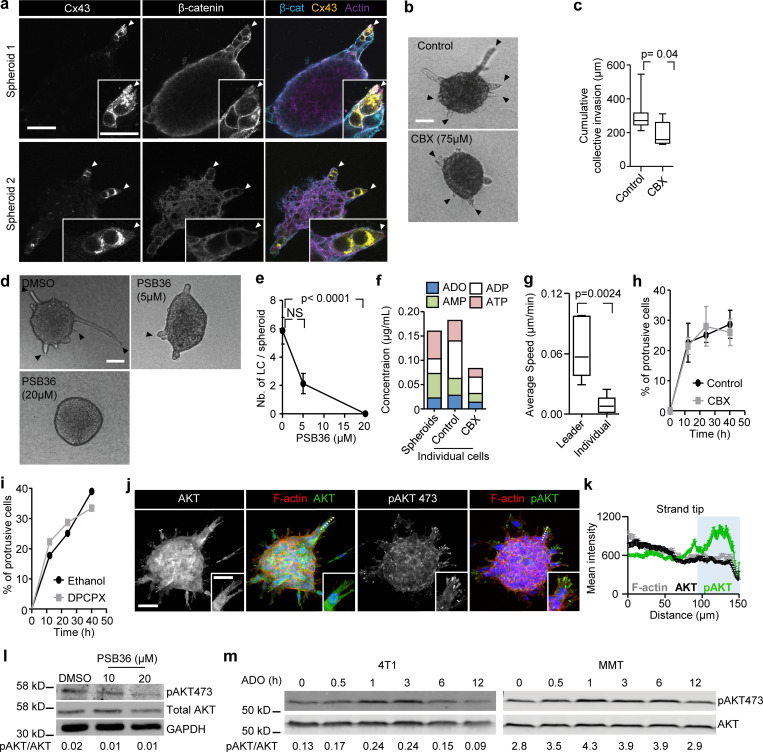
To address whether Cx43 supports invasion, the ER-negative metastatic breast cancer cells 4T1, which express E-cadherin, and mouse mammary tumor (MMT) cells, which lack E-cadherin but expresses N-cadherin (Ilina et al., 2018), were embedded as multicellular spheroids in 3D collagen lattices and monitored for invasion over 24–48 h (Videos 1 and 2). 4T1 and MMT cells invaded as collective strands and expressed high levels of Cx43 and Cx45 mRNA and low levels of other Cxs after isolation from collagen matrix culture (Fig. S1 f). During collective invasion, Cx43 was localized along cell–cell junctions (Fig. 1, e and f, white arrowheads) and in nonjunctional domains, along cell protrusions (Fig. 1, e and f, red arrowheads; and Fig. 1 g). Junctional and nonjunctional Cx43 localization was further observed during invasion into the mammary fat pad in vivo (Fig. 1 h). In contrast to Cx43, Cx45 was absent along cell–cell junctions but localized in the cytosol (Fig. S1 g). Thus, Cx43 is expressed at the surface of collectively invading breast cancer cells in vitro and in vivo.
Video 1.
Bright-field time-lapse microscopy of 4T1 spheroids showing protrusive tips during initiation of collective invasion in 3D collagen I. Field size: 529 × 301 µm. The playback speed is indicated in the time stamp of the video.
Video 2.
Time-lapse imaging by confocal microscopy of MMT spheroids (H2b-GFP and cytosolic dsRed) during initiation and elongation of collective invasion in 3D collagen I. Field size: 1,463 × 1,463 µm. The playback speed is indicated in the time stamp of the video.
Cx43 mediates GJIC during collective invasion
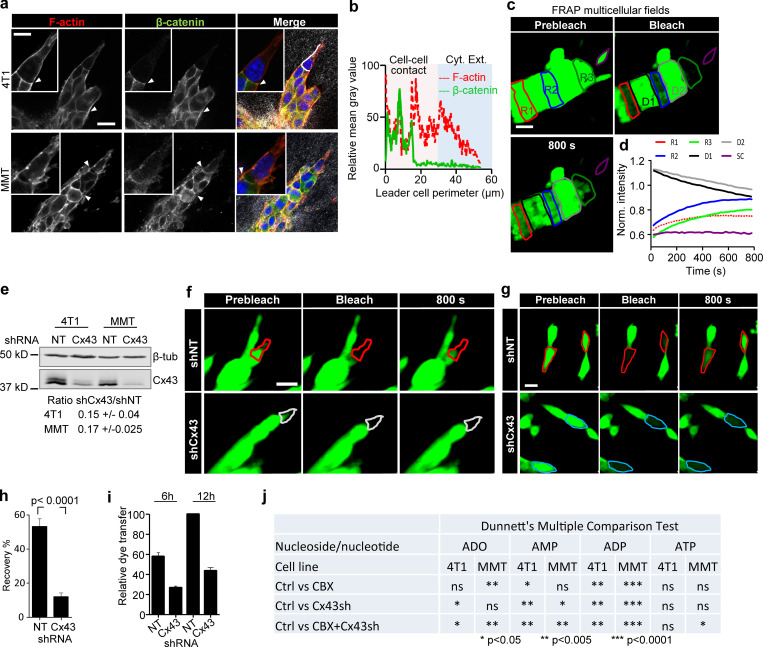
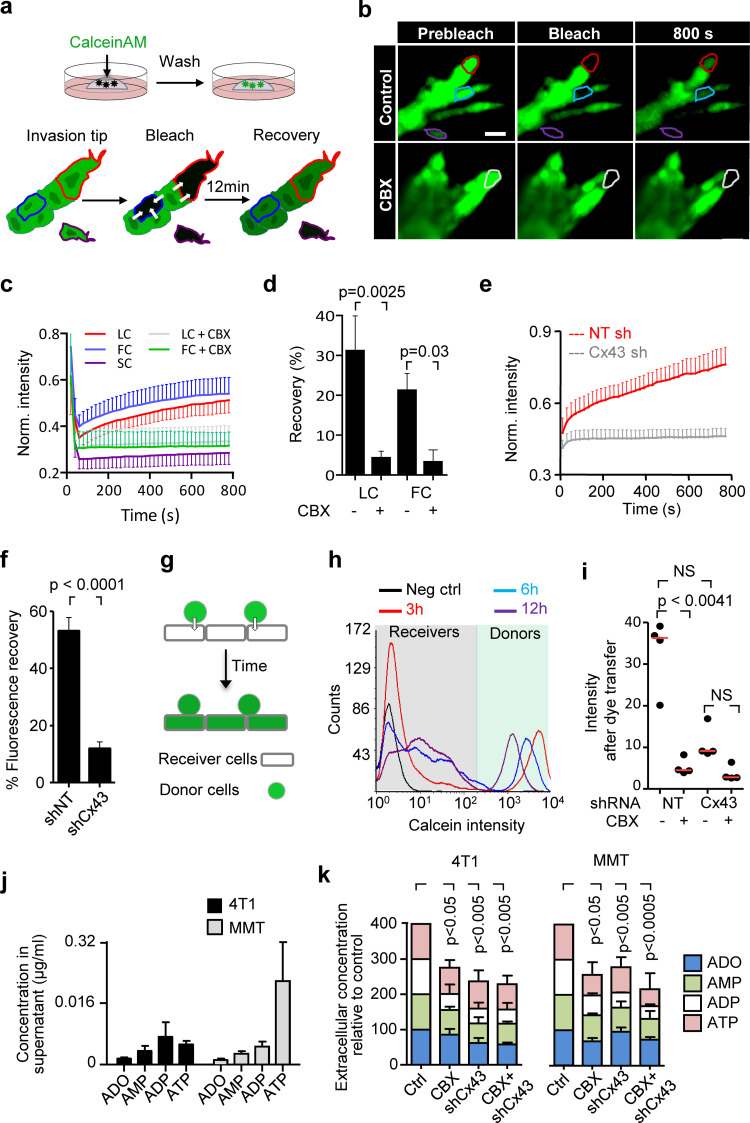
Collective invasion in 3D spheroid culture was initiated by single or few leader cells, which extended actin-rich protrusions to the front and maintained cadherin-based junctions with following cells (Fig. S2, a and b). To examine whether gap junctions are active during collective invasion, intercellular dye transfer was measured in 3D spheroid invasion cultures using gap FRAP (Video 3 and Fig. 2 a). Within minutes after photobleaching, leader and follower cells recovered the calcein signal by 20–30% (Fig. 2, b and c), with unbleached neighboring cells acting as donors (Video 3; and Fig. S2, c and d). As internal control, detached single cells lacked dye transfer after photobleaching (Fig. 2, b and c; Fig. S2, c and d, purple cell outline; Video 3). Fluorescence recovery was reduced by 70–85% after inhibition of Cx channel function by carbenoxolone (CBX; Fig. 2, b–d; and Video 3). Fluorescence recovery was further impaired when Cx43 expression was stably down-regulated (by >80%) using shRNA (Fig. 2, e and f; and Fig. S2, e–h).
Figure S2.
Collectively invading cancer cells maintain Cx43-mediated GJIC and extracellular nucleotide release. (a) β-Catenin localization along cell–cell junctions between both leader cells and follower cells in collective strands of 4T1 and MMT cells invading 3D fibrillar collagen. Arrowheads, cell–cell junctions. (b) Intensity distribution of β-catenin and F-actin along the circumference of a leader cell (white dashed border). Values show the pixel intensity with background subtraction. (c) Multicellular 3D gap FRAP. Single confocal slices of calcein-labeled invasive 4T1 strands before and after photobleaching. Contours indicate regions of interest for bleaching, including follower regions (red: R1; blue: R2), an invasive front (green: R3), and a single detached cell (SC; purple), in addition to the unbleached neighboring regions D1 and D2. (d) Normalized mean calcein fluorescence intensity of the selected regions. (e) Western blot for Cx43 and β-tubulin as loading control. Whole-cell lysates extracted from 4T1 and MMT cells with stable expression of shNT or shCx43; ±SD. (f and g) Fluorescence recovery of calcein-labeled MMT leader cells expressing shNT or shCx43 during invasion in 3D collagen (f) and physically connected cells in 2D culture (g). (h) Values represent the means and SEM of at least 19 bleached cells from 2D culture per condition from three independent experiments. P values, two-tailed unpaired Mann–Whitney test. (i) Relative dye transfer in parachute assay of 4T1 with nontargeting shRNA (NT) and Cx43 shRNA (Cx43) after 6 or 12 h of cell–cell (donor–receiver) encounter. Values represent the means and SEM from two independent experiments. (j) Summary of the effect of Cx43 down-regulation and/or CBX treatment on the reduction of individual nucleotide and nucleosides concentration detected in the media of 4T1 and MMT collagen cultures by HPLC (related to Fig. 2 k). Concentrations from CBX-treated and Cx43 shRNA cultures were compared with control cells (Ctrl, untreated cells with nontargeting shRNA). P values, ANOVA with Dunnett’s multiple comparison test (ns, not significant). Scale bars: 20 µm (a, c, f, and g), 10 µm (a, inset).
Video 3.
Time-lapse confocal imaging of calcein-labeled 4T1 spheroids before and after photobleaching of cells and multicellular regions within invasive strands in the absence (control media) and presence of the gap junction inhibitor (CBX) in 3D collagen I. Field size: 213 × 122 µm. The playback speed is indicated in the time stamp of the video. Related to Figs. 2 b and S2 c.
Figure 2.
Cx43 mediates GJIC and extracellular release of nucleotides/nucleosides during collective invasion. (a) Workflow of time-resolved 3D gap FRAP, including calcein labeling of multicellular spheroids in 3D collagen cultures and 3D gap-FRAP procedure of leader and follower cells within a 3D invasion strand by asymmetric region of interest selection and photobleaching, followed by recording the change in fluorescence intensity over time. (b) Single confocal slices of calcein-labeled invasive 4T1 strands before and after photobleaching. Dashed contours represent the bleached leader cell (LC; red), FC (blue), and SC (purple) in control media and LC in the presence of CBX (white). (c) Normalized calcein fluorescence intensity in bleached LCs, FCs, and SCs in the presence or absence of CBX. Values represent normalized mean fluorescence intensities with SEM; 11–16 cells per treatment condition from four independent experiments. (d) Effect of CBX on percentage fluorescence recovery after photobleaching of LCs and FCs. Values are represented as the means and SEM of three independent experiments. P values, two-tailed unpaired Mann–Whitney test. (e) Average fluorescence recovery in cells stably expressing control vector (shNT) or Cx43 shRNA (shCx43) during invasion in 3D collagen. Values represent the normalized mean intensities and SEM of four leader cells for shNT and pooled two leader cells and two follower cells for shCx43 condition. (f) Inhibition of fluorescence recovery of MMT cells after Cx43 down-regulation (cell groups in Petri dish culture). Normalized mean intensity and SEM of 19–22 cells from three independent experiments. P value, two-tailed unpaired Mann–Whitney test. (g and h) Parachute assay to measure de novo junction formation (g) followed by dye transfer into nonlabeled cells (h) and example histograms obtained by flow cytometry showing the time-dependent alterations of calcein label in donor and recipient cells. (i) Intensity of calcein in 4T1 receiver cells after 12 h of incubation with 4T1 donor cells in control conditions and during inhibition with CBX or after Cx43 down-regulation. Median (red line) intensities from four independent experiments. P values, Kruskal-Wallis test with Dunn’s multiple comparison test. (j and k) HPLC analysis of purines released from 4T1 and MMT spheroids into the supernatant after 24 h of invasion in 3D collagen. Values represent average concentrations of each purine (j) and the relative change of the total purine concentrations after treatment with CBX and/or Cx43 down-regulation in 4T1 and MMT cells (k). Mean values and SEM from four (j) or SD from three (k) independent experiments. P values, ANOVA Dunnett’s multiple comparison test. Scale bar: 20 µm (b). Neg ctrl, negative control; Norm., normalized; FC, follower cell; LC, leader cell; SC, single detached cell.
To test whether all cells maintain GJIC or only specific cell subsets, dye transfer between freshly interacting breast cancer cells was measured (parachute assay; Abbaci et al., 2008; Fig. 2 g). Within a 12-h observation period, the majority of control 4T1 cells (90–95%) received dye from the calcein-loaded monolayer (Fig. 2 h). The extent of dye transfer was strongly reduced by 67–84% by CBX treatment or Cx43 down-regulation (Figs. 2 i and S2 i). These data indicate that collectively invading breast cancer cells interact by GJIC through Cx43.
Cx43 hemichannels release purine nucleotides
Besides gap junctions, Cxs can form transmembrane hemichannels (Ye et al., 2003), which release small molecules from the cytosol into the extracellular space, including glutamate, prostaglandins, and the purine nucleotides ATP and ADP (Baroja-Mazo et al., 2013; Eltzschig et al., 2006; Retamal et al., 2007). Extracellular ATP and ADP and their degradation product, ADO, are important energy equivalents. In addition, extracellular nucleotides/nucleosides are effective extracellular signaling molecules, which promote the directional migration in endothelial cells (Kaczmarek et al., 2005), microglia (Haynes et al., 2006), and neutrophils (Barletta et al., 2012), as well as in individually moving cancer cells (transwell assay; Zhou et al., 2015). In addition to its localization at cell–cell contacts between invading cells, Cx43-positive foci were present in protrusions of leader cells (Fig. 1, e, f, and h, red arrowheads; and Fig. 1 g). To address whether breast cancer cells release purine nucleotides during invasion, supernatants from 3D collagen cultures were analyzed for ADO, AMP, ADP, and ATP by HPLC. Whereas cell-free media contained no purine metabolites, 3D spheroid invasion cultures after 24 h contained high levels of extracellular ATP (0.1–0.25 µg/ml) and moderate levels of ADO, AMP, and ADP (10–50 ng/ml; Fig. 2 j).
Inhibition of Cx channel-function by CBX and Cx43 knockdown both reduced the extracellular levels of ADO (by 26–42%), AMP (by 40–41%), ADP (by 60–64%), and ATP (by 28–52%; Figs. 2 k and S2 j). This indicates that breast cancer cells release purine nucleosides and nucleotides through Cx43 hemichannels in 3D invasion culture.
Cx43 is required for leader cell function
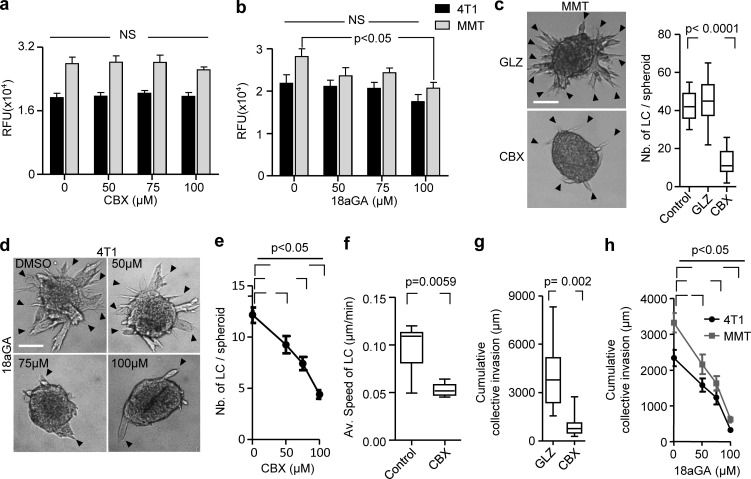
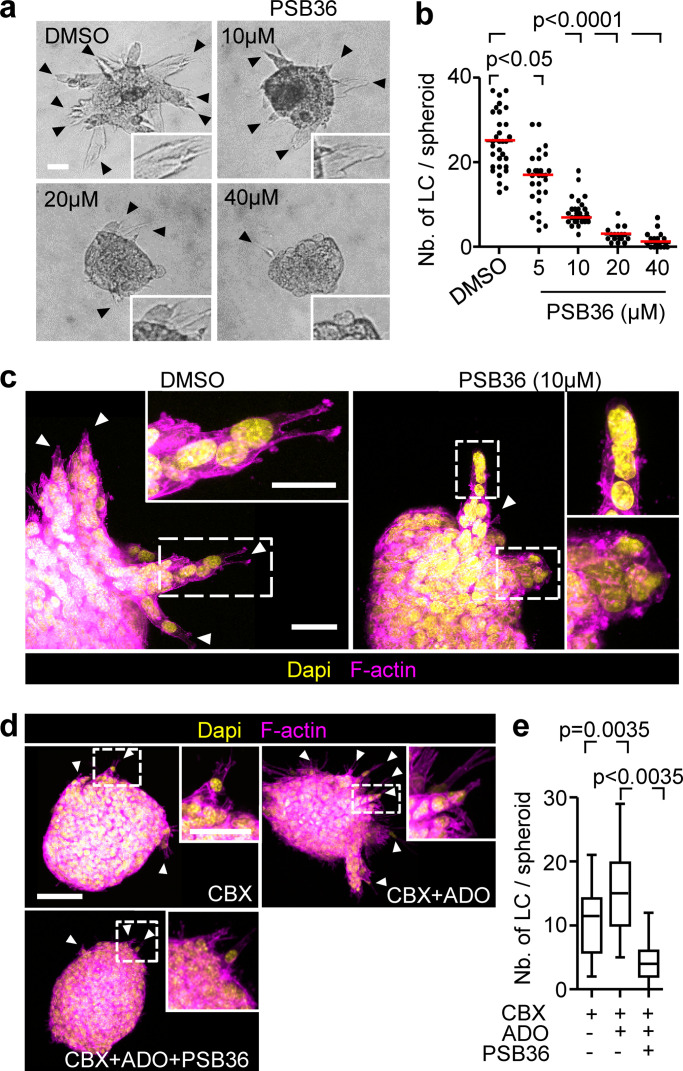
To determine whether Cx43 function, including GJIC and hemichannel activity, is required for collective cell invasion, 3D spheroid cultures were treated with the pharmacological channel blockers CBX or 18 α-glycyrrhetinic acid (18aGA) at doses that did not compromise growth (Fig. S3, a and b). Cultures were treated either before (prevention) or after collective invasion has started (intervention). Both compounds dose-dependently reduced strand initiation and impaired the emergence of leader cells compared with the inactive homologue glycyrrhizic acid ammonium salt (GLZ) or vehicle control (Fig. 3 a, arrowheads, Fig. 3, b and c; Fig. S3, c and d, arrowheads; and Fig. 3 e). Consequently, the invasion speed and cumulative length of collective invasion strands after 24 h were reduced by up to 10-fold (Fig. 3 b; and Fig. S3, f–h).
Figure S3.
Cx43 dependence of leader cell function. (a and b) Resazurin cytotoxicity assay for 4T1 and MMT cells in 3D collagen for 24 h in the presence of vehicle control and escalating doses of CBX (a) and 18aGA (b). Values represent the mean relative fluorescence unit (RFU) with SEM of two or three independent experiments. P values, one-way ANOVA with Bonferroni multiple comparison test. (c) Bright-field images after 24 h of MMT cell invasion in 3D collagen in the presence of CBX or an inactive structurally homologous control compound (GLZ). Data represent the medians (black line), 25th/75th percentiles (box) and maximum/minimum (whiskers) of 8–11 MMT spheroids per condition pooled from three independent experiments. P value, two-tailed unpaired Mann–Whitney test. (d) Efficacy of collective invasion in 4T1 spheroids in the presence of escalating doses of 18aGA. Bright-field images after 24 h of culture. Arrowheads (c and d), tips of invasion strands.(e) 4T1 leader cell (LC) initiation in the presence of escalating doses of CBX. Data represent mean values and SEM from 23 to 40 spheroids per condition pooled from at least three independent experiments. P value, ANOVA with Bonferroni multiple comparison test. (f) Speed of MMT leader cells during collective invasion in 3D collagen. Values represent median (black line), 25th/75th percentiles (box) and maximum/minimum (whiskers) of seven or eight leader cells from one experiment. P value, two-tailed unpaired Mann–Whitney test. (g) Cumulative collective invasion of MMT cells. Data represent the medians (black line), 25th/75th percentiles (box) and maximum/minimum (whiskers) of 8–11 MMT spheroids per condition pooled from three independent experiments. P value, two-tailed unpaired Mann–Whitney test. (h) Cumulative collective invasion of 4T1 and MMT spheroids in the presence of escalating doses of 18aGA. Data represent mean values and SEM from 12 spheroids per condition pooled from three independent experiments. P value, ANOVA with Bonferroni multiple comparison test. Scale bars: 100 µm (c and d).
Figure 3.
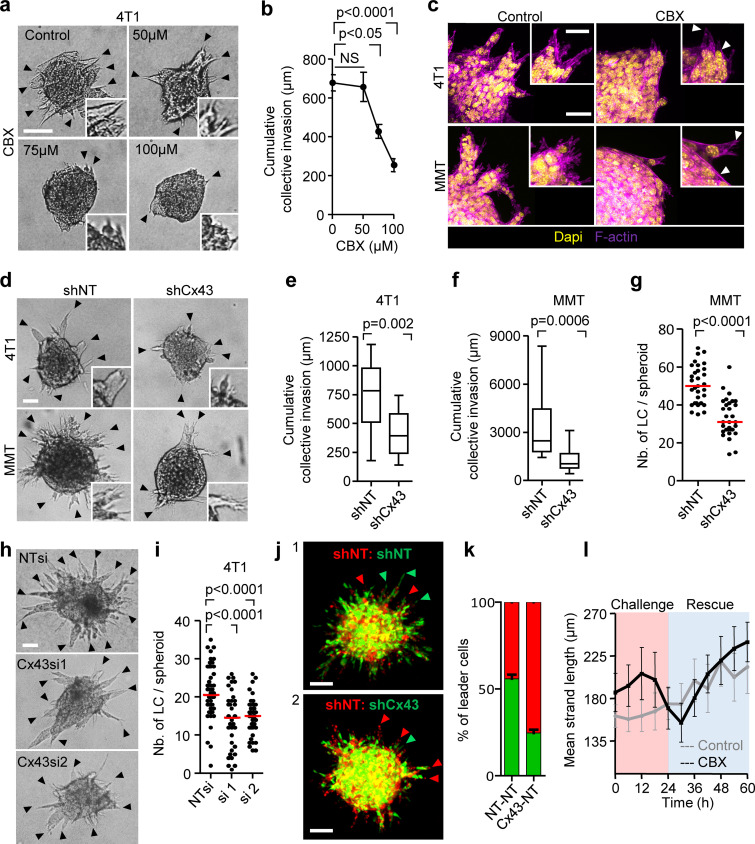
Cx43 dependence of leader cell function. (a) Bright-field images of 4T1 spheroids after 24 h of invasion in 3D collagen in the presence of increasing concentration of CBX. Arrowheads, tips of invasion strands. (b) Mean cumulative length of collective invasion strands in response to CBX. Mean values and SEM from three independent experiments each comprising seven spheroids/condition. P values, Kruskal–Wallis test with Dunn’s multiple comparison test. (c) Maximum intensity projection from a confocal 3D stack, showing F-actin and DAPI in control and CBX-treated spheroids (100 µM). (d–g) Cx43-dependent collective invasion. Bright-field images (d), cumulative collective invasion (e and f), and leader cell initiation (g) of spheroids composed of 4T1 or MMT cells stably expressing Cx43 or nontargeting shRNA (NT). Values represent the medians (black line), 25th/75th percentiles (box) and maximum/minimum (whiskers) from three or four independent experiments with three to eight spheroids each. P values, two-tailed unpaired Mann–Whitney test. (h) Bright-field images of 4T1 spheroids in response to transient Cx43 down-regulation using two different Cx43 RNAi probes. (i) Number of leader cells (LCs) per spheroid. P values, Kruskal–Wallis test with Dunn’s multiple comparison test. (j) Maximum intensity projection from a confocal 3D stack showing mosaic spheroids composed of fluorescence-coded MMT cells stably expressing NT or Cx43 shRNA as indicated (1:1 ratio); arrowheads depict red or green leader cells. (k) Frequency of green or red leader cells. Bars represent the mean values and SEM pooled from one (j1) and three (j2) independent experiments. (l) Effect of CBX on the kinetics of established invasion strands. Mean length per invasion strand after addition of CBX at 0 h (intervention) and washout at 24 h (rescue). Data show the mean values and SEM from six to eight invasion strands from one spheroid per condition. Scale bars: 100 µm (a), 50 µm (c, d, and h), 25 µm (c, inset). LC, leader cell.
Likewise, stable down-regulation of Cx43 by shRNA compromised both the number and length of collectively invading strands (Fig. 3, d–g). To rule out adaptation or off-target effects of stable Cx43 down-regulation, transient Cx43 down-regulation using individual RNAi probes was used, leading to reduced invasive strand initiation and elongation (Fig. 3, h and i). Neither pharmacological nor RNAi with GJIC altered cell–cell cohesion or shape. Invasion strands, emerging despite interference, retained intact morphology and cortical F-actin organization (Fig. 3 c, arrowheads; and Fig. 3 d).
The decreased initiation and prolongation of invasion strands after interference with Cx43 suggested that the activity of leader cells was compromised. Leader cells are critical in initiating and maintaining collective invasion (Cheung et al., 2013; Zhang et al., 2019). To test whether leader cell function depends on Cx43, fluorescent mosaic spheroids of cells expressing nontargeting or Cx43 shRNA at equal ratio were allowed to invade. Cells with down-regulated Cx43 were 50% less likely to acquire leader cell position (Fig. 3, j and k). CBX treatment reduced the speed of leader cells as assessed by cell tracking (Fig. S3 f and Video 4). Besides decreasing leader cell initiation, intermittent inhibition of GJIC by CBX in spheroids with established collective invasion affected leader cell function. CBX caused near-instantaneously collapsing protrusive tips, followed by speed reduction, and, ultimately, strand retraction (Fig. 3 l and Video 5). Leader cell functions were restored after CBX washout (Video 5), indicating that Cx channels were required for the initiation and maintenance of collective invasion.
Video 4.
Time-lapse confocal imaging of collective invasion of MMT cells (H2b-GFP) in 3D collagen I in the presence and absence of CBX. Field size: 540 × 550 µm. The playback speed is indicated in the time stamp of the video. Related to Fig. S3 f.
Video 5.
Intervention with collective invasion by the gap junction channel blocker (CBX). Time-lapse imaging by bright-field microscopy of established invasive 4T1 strands in 3D collagen I after challenge with connexin inhibitor CBX and followed by drug washout. Field size: 1,174 × 881 µm. The playback speed is indicated in the time stamp of the video. Related to Fig. 3 l.
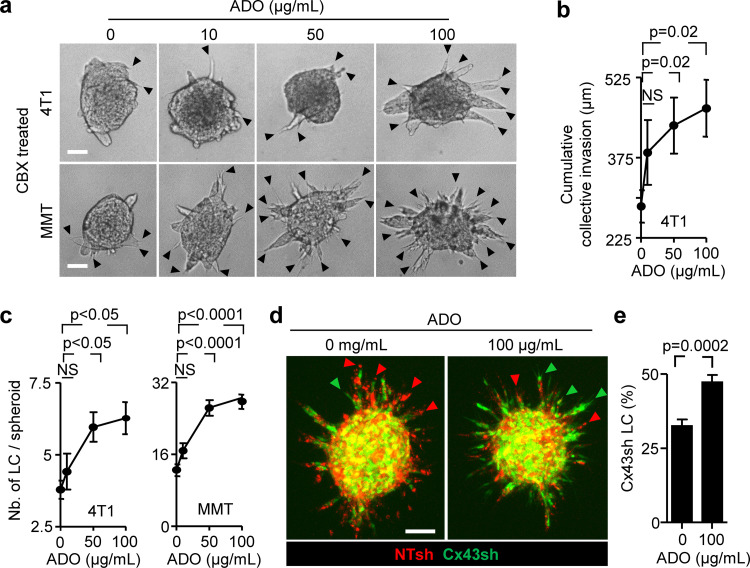
We next aimed to understand which of the two channel functions of Cx43, GJIC or hemichannel activity, is required for collective invasion. Therefore, we bypassed hemichannel function by exogenous addition of ADO in Cx-inhibited cultures and tested whether invasion was restored. Exogenous ADO was added to 4T1 and MMT spheroid cultures in the presence of CBX or after Cx43 down-regulation. Increasing concentrations of ADO partially (35–50%) restored the frequency of leader cells compared with untreated cells (Fig. 4, a–c; and Fig. S4 a). ADO substitution further restored defective leader cell function caused by down-regulation of Cx43 in mosaic spheroids (Fig. 4 d, green cells; and Fig. 4 e). These data suggest that nucleotide/nucleoside release via hemichannels is essential, while GJIC is dispensable for collective invasion. The restored leader cell function by exogenous ADO thus indicates an autocrine Cx43-hemichannel-dependent purinergic signaling loop to secure collective migration.
Figure 4.
Cx43 hemichannel function in leader cell induction by purine nucleotide release. (a–c) Rescue of invasion in 4T1 and MMT spheroids in 3D collagen in the presence of CBX and escalating doses of ADO; for comparison with untreated baseline levels refer to Figs. 3 b and S3 (c and e). Representative bright-field images (a), cumulative collective invasion (b), and number of leader cells (LC) per spheroid (c). Mean values and SEM from four independent experiments, each comprising 5–11 spheroids/condition. P values, Kruskal–Wallis test with Dunn’s multiple comparison test. (d and e) Rescue of leader cell deficiency after Cx43 down-regulation by exogenous ADO. (d) Maximum intensity projection from a 3D confocal stack showing mosaic spheroids with MMT cells stably expressing NT or Cx43 shRNA (1:1 ratio) in the absence or presence of ADO and (e) frequency of LC with green color expressing Cx43 shRNA. Bars represent the means and SEM from three independent experiments, each comprising three to six spheroids. P values, two-tailed unpaired Mann–Whitney test. Scale bars: 50 µm (a), 100 µm (d).
Figure S4.
Cx43-mediated nucleotide release in LC initiation. (a) Leader cell (LC) initiation in MMT spheroids expressing shCx43 after 24 h of invasion in collagen in the presence and absence of exogenous ADO. Values display median (black line), 25th/75th percentiles (boxes) and maximum/minimum values (whiskers) of 30–35 spheroids per condition pooled from three independent experiments. P value, two-tailed unpaired Mann–Whitney test. (b) Purinergic receptor mRNA expression in 4T1 (black) and MMT (gray) cells during collective invasion in 3D collagen assessed by quantitative PCR. Values represent the mean normalized mRNA levels and SEM from three independent experiments. (c) MMT leader cell initiation in the presence of vehicle and escalating doses of PSB36. Values represent mean values and SEM of at least 20 spheroids per condition pooled from three independent experiments. P values, Kruskal–Wallis test with Dunn’s multiple comparison test. (d) Leader cell initiation in response to DCPX treatment 12–15 h of invasion of 4T1cells in collagen. Values display medians (black lines) 25th/75th percentiles (boxes) and maximum/minimum values (whiskers), 23–27 spheroids from three independent experiments. P values, two-tailed unpaired Mann–Whitney test. (e) Mitotic figures in 4T1 spheroid after 24 h of invasion in collagen. Values represent average number and SEM of mitotic figures per spheroids, three spheroids (≥1,000 cell/spheroid) per condition. P value, two-tailed unpaired Mann–Whitney test. (f) Leader cell initiation of 4T1 and MMT spheroids after 12–15 h of invasion in collagen in the presence of ADORA2b inhibitor PSB1115. Values display medians (black lines) 25th/75th percentiles (boxes) and maximum/minimum values (whiskers), 4–12 spheroids from one experiment. P values, Kruskal–Wallis test with Dunn’s multiple comparison test. (g) Expression and distribution of ADORA1 in 4T1 spheroids treated with vehicle control or CBX for 24 h. Images represent single confocal slices. (h) ADORA1 expression of the outer cellular layer. Data represent the background-corrected mean fluorescence and SD of four (control) or six (CBX) spheroids from one experiment. P value, two-tailed unpaired Mann–Whitney test. Scale bars: 50 µm (g), 25 µm (g, inset).
Autocrine purinergic receptor signaling maintains collective cell invasion
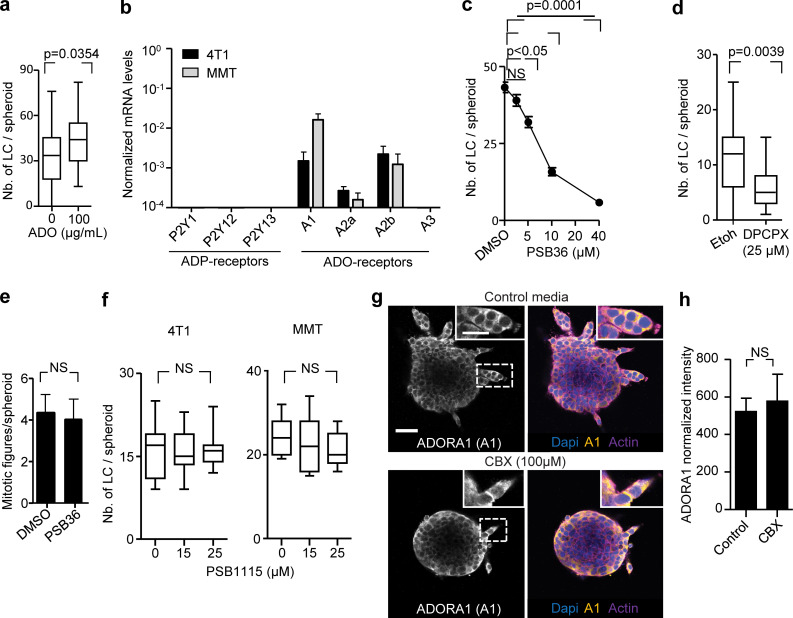
The signaling function of extracellular ADO depends on its binding to P1 purinergic receptors (Barletta et al., 2012). P1 receptors belong to the G-protein–coupled family of receptors, with the members ADORA1, ADORA2a, ADORA2b, and ADORA3 (Antonioli et al., 2013). 4T1 and MMT cells isolated from 3D invasion culture expressed ADORA1 and ADORA2b mRNA but no other ADO or ADP receptor (Fig. S4 b). We thus tested whether ADORA1 or ADORA2b mediate collective invasion using pharmacological interference. PSB36 and 8-cyclopentyl-1,3-dipropylxanthine (DPCPX), both selective ADORA1 antagonists with established activity in vitro and in vivo (Abo-Salem et al., 2004; Töpfer et al., 2008), caused dose-dependent inhibition of collective invasion in 4T1 and MMT cells (Fig. 5, a–c; Fig. S4 c; and Fig. S4 d). PSB36 did not affect the mitotic frequency of the cells (Fig. S4 e). Unlike ADORA1, interference with ADORA2b using the antagonist PSB1115 showed no effect on the emergence of leader cells of 4T1 and MMT cells (Fig. S4 f).
Figure 5.
Autocrine purinergic receptor signaling maintains leader cell functions. (a) Bright-field images of 4T1 spheroids cultured in 3D collagen for 24 h in the presence of escalating concentrations of PSB36. (b) Median numbers of leader cells (LC) per spheroid (red line) from three independent experiments. P values, Kruskal–Wallis test with Dunn’s multiple comparison test. (c) Maximum intensity projection from a 3D confocal stack of 4T1 spheroid in the presence or absence of PSB36. Arrowheads, leading extensions of individual LCs. (d) Maximum intensity projections of 3D MMT spheroids with different treatments and (e) resulting number of leader cells per spheroid. Data represent the medians (black line), 25th/75th percentiles (boxes) and maximum/minimum values (whiskers) from two independent experiments each comprising 10 to 16 spheroids/condition. P values, ANOVA with Bonferroni multiple comparison test. Scale bars: 50 µm (a and d), 20 µm (c), 25 µm (d, inset), 10 µm (c, inset).
PSB36 abolished the effects of exogenous ADO on rescuing collective invasion (Fig. 5, d and e) and prevented leader cells to develop pointed anterior protrusions (Video 6). As a consequence of leader cell collapse, invasion strands retracted, and this was reversible after washout of the inhibitor (Video 6). Thus, leader cell function in collective invasion critically depends on ADO signaling and ADORA1.
Video 6.
Intervention with collective invasion by ADORA1 inhibitor (PSB36). Time-lapse imaging by bright-field microscopy of collective invasion of 4T1 cells in 3D collagen I before and after the addition of PSB36. Field size: 1,174 × 881 µm. The playback speed is indicated in the time stamp of the video.
ADORA1 localization during collective invasion in vitro was predominantly cytosolic, and its expression levels were not affected by CBX treatment (Fig. S4, g and h). The localization of ADORA1 is in line with previously reported cytosolic distribution of ADORA1 and other chemokine receptors (G-protein–coupled receptors) such as in nerve terminals and gastric cancer tissues (Iwasa et al., 2009; Rebola et al., 2003).
To address whether Cx43/ADORA1 regulates collective movement of nontumorigenic cells, we embedded spheroids of the normal mammary cell line NMuMG (Nemir et al., 2000) in 3D collagen. Normal Mus muculus mammary gland (NMuMG) cells are deficient in both junctional expression of Cxs (including Cx43) and GJIC (Sheng et al., 2007). Consistent with defective GJIC, Cx43 was enriched in the cytosols of NMuMG leader cells guiding collective invasion strands (Fig. S5 a, arrowheads). Treatment with the channel blocker CBX or ADORA1 inhibitor PSB36 reduced invasion of NMuMG (Fig. S5, b–e), suggesting that Cx43 hemichannels and extracellular ADO signaling rather than GJIC support collective movement. Thus, the autocrine nucleotide loop is not restricted to cancer cells but may represent a general collective steering mechanism.
Figure S5.
Hemichannel-ADORA1 loop in other collective but not single-cell invasion models and downstream AKT activation. (a) Distribution of Cx43, β-catenin, and F-actin in NMuMG spheroids during collective invasion in 3D collagen after 68 h. Images represent maximum intensity projections from a 3D confocal stacks. Arrowheads, focal Cx43 hemichannel localization in leader cells. (b) Bright-field images of NMuMG spheroids embedded in 3D collagen for 68 h in the presence or absence of CBX. Arrowheads, protrusive leader cells. (c) Cumulative median length of invasion per spheroid (black line), 25th/75th percentiles (box) and maximum/minimum (whiskers) from 42 to 75 stands from six (control) and eight (CBX) spheroids (one experiment). P values, two-tailed unpaired Mann–Whitney test. (d) Bright-field images of NMuMG spheroids in 3D collagen after 48 h culture in the presence of PSB36 (5 or 20 µM). (e) Mean number of leader cells per spheroid and SEM from seven to nine spheroids per condition (one experiment). P values, Kruskal–Wallis test with Dunn’s multiple comparison test. (f) HPLC analysis of purines released from individualized 4T1 cells into the supernatant after 24 h of invasion in 3D collagen. Values represent concentrations of each purine from one experiment. (g) Speed of 4T1 leader vs individual cells in 3D collagen. Values represent the medians (black line), 25th/75th percentiles (box) and maximum/minimum (whiskers) of six leader cells (one experiment) and 19 individual cells tracked over 24 h (two independent experiments). P value, two-tailed unpaired Mann–Whitney test. (h and i) Percentage of individualized 4T1 cells with elongated morphology after 12, 24, and 40 h in collagen and treatment with CBX (h) and PSB36 (i). Data show the means and SEM from 17–33 cells per condition from two (h) or one (i) experiments. DPCPX, 8-cyclopentyl-1,3-dipropylxanthine. (j) Distribution of AKT, phospho-AKT473, and F-actin in MMT spheroid invasion culture. 3D confocal maximum intensity projections after 24 h of culture. Dashed line indicates the region of analysis in k. (k) Distribution of AKT and phospho-AKT473 and F-actin along extending protrusions of leader cells. Data represent the mean intensities and SEM of F-actin, AKT, and phospho-AKT473 from at least 12 leader cells from one experiment. (l) Phospho-AKT473 levels in response to 24 h of PSB36 treatment during 4T1 spheroid culture in 3D collagen. (m) Phospho-AKT473 levels in of 4T1 and MMT 2D cultures after ADO treatment for 0.5, 1, 3, 6, and 12 h. Numbers in (l, m) represent the ratio of phospho-AKT473 over total AKT mean intensities from one experiment. Scale bars: 50 µm (a, b, d, and j), 25 µm (a and j, insert).
We next tested whether single-cell invasion is also regulated by Cx hemichannels and ADORA1. 4T1 cells were individualized and embedded in 3D collagen and their movement followed by cell tracking over a time course of 40 h. Although individual cells maintained the release of nucleotides and nucleosides into the culture supernatant (Fig. S5 f), only 20–30% of individualized 4T1 cells developed elongated morphology, and single cells only migrated on the spot with an average speed of 0.009 µm/min, which is at least six times below the average speed of collectively invading 4T1 cells (0.06 µm/min; Fig. S5 g). Treatment with the pan-Cx inhibitor CBX or the ADORA1 inhibitor PSB36 did not affect the number of protrusive cells (Fig. S5, h and i). Thus, the hemichannel–ADORA loop does not support cell protrusion of individual breast cancer cells.
AKT phosphorylation in Cx/ADORA-mediated invasion
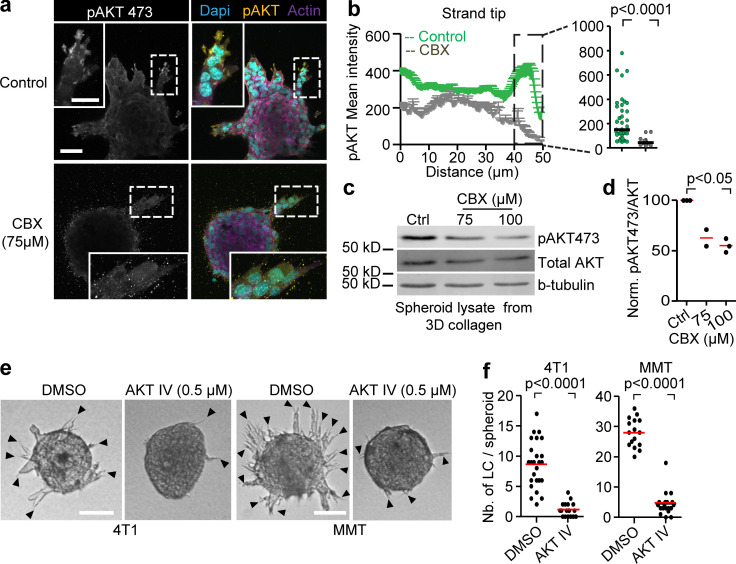
In 2D sheet migration of nontransformed epithelial cells, leader cell function depends on phosphoinositide 3-kinase and AKT signaling (Yamaguchi et al., 2015). Since AKT activation is a major event downstream of ADO signaling via ADORA1 (Park et al., 2010; Umapathy et al., 2013), we investigated the involvement of AKT in Cx/ADORA1-mediated invasion. In 3D invasion culture, the active form of AKT (AKT-pSer473) was abundant near the protrusion tips of leaders, but not in follower cells (Fig. 6 a; and Fig. S5, j and k). The focal AKT-Ser473 intensity in leader cells was reduced after addition of CBX (Fig. 6, a–d). Likewise, PSB36 reduced AKT-pSer473 levels (Fig. S5 l), whereas AKT-pSer473 levels increased within 1 h of ADO addition (Fig. S5 m), in line with previous reports showing time-dependent regulation of AKT phosphorylation in response to purinergic signaling (Jacques-Silva et al., 2004). To determine whether AKT activation is required for invasion, we treated 4T1 and MMT 3D invasion cultures with the AKT inhibitor IV. The emergence of leader cells and strand formation in both 4T1 and MMT spheroids were reduced by >84%, compared with the DMSO controls (Fig. 6, e and f). These data suggest that the hemichannel/ADO loop regulates collective invasion by activating AKT in leader cells.
Figure 6.
AKT regulation and requirement for leader cell function and invasion. (a) Maximum intensity projection of phospho-AKT473 and F-actin staining in MMT spheroids treated with vehicle control or CBX. Maximum intensity projection of 3D confocal stack. (b) Distribution of phospho-AKT473 along protrusions of leader cells in both conditions with dot plot comparing phospho-AKT473 levels in the last 10 µm of leader cell protrusions in vehicle- and CBX-treated condition. Line graphs and dot plot represent intensities of phospho-AKT473 from 11 (control) or 21 (CBX) leader cells from three or four spheroids per respective condition. P values, two-tailed unpaired Mann–Whitney test. (c) Western blot for phospho-AKT473, AKT and β-tubulin as loading control. Whole-cell lysates extracted from MMT spheroids after 24 h of invasion into 3D collagen in the presence of 0, 75, and 100 µM of CBX. (d) Values represent normalized intensities of phospho-AKT473/total AKT with median values (red line) from two (75 µM) or three independent experiments (0 and 100 µM). P value, Kruskal–Wallis test with Dunn’s multiple comparison test. (e) Bright-field images of 4T1 and MMT spheroids embedded in 3D collagen for 15 h in the presence of DMSO or AKT inhibitor IV. (f) Number of leader cells (LC) per spheroid; red line represents the median value from 18 to 25 spheroids (4T1) and 16–19 spheroids (MMT) pooled from two independent experiments. P values, two-tailed unpaired Mann–Whitney test. Scale bars: 50 µm (a), 25 µm (a, inset), 100 µm (e).
ADORA1 in breast cancer invasion and progression in vivo
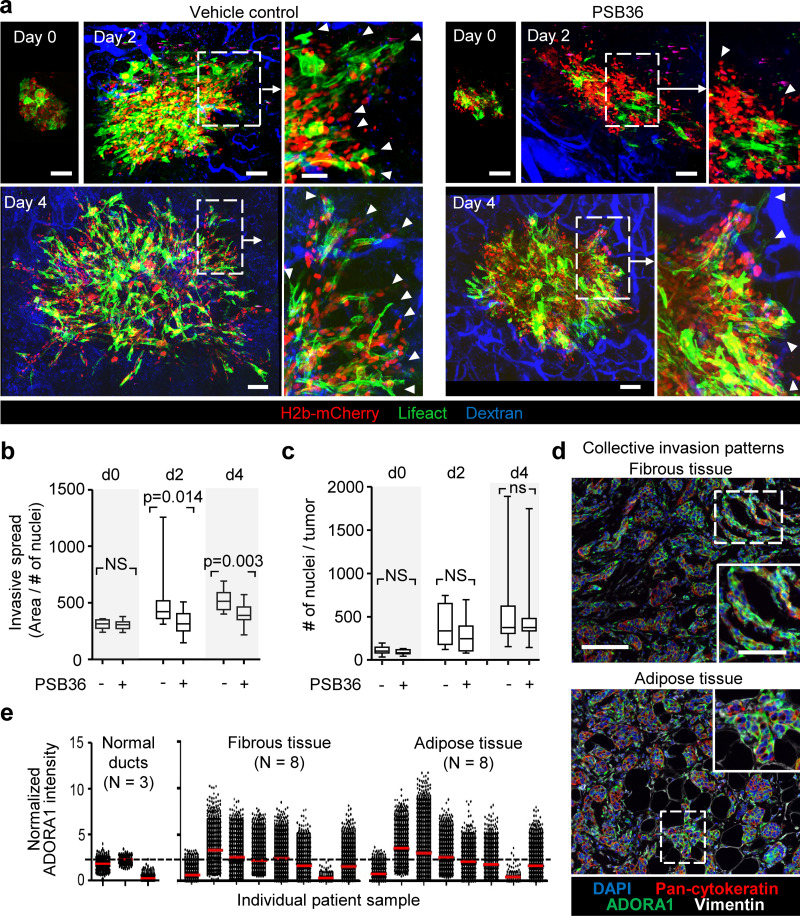
To address whether ADORA1 signaling supports collective breast cancer cell invasion in vivo, multicellular tumoroids were implanted into the mammary fat pad and the effect of systemic administration of PSB36 on progressing invasion was monitored by intravital multiphoton microscopy through a body window for 4 d (Ilina et al., 2018). Untreated tumors invaded into the mammary fat pad via collective strands with elongated leading tip cells (Fig. 7 a, arrowheads), as described previously (Ilina et al., 2018). PSB36 inhibited the extent of invasion with fewer strands with tips and reduced strand length (Fig. 7 a, arrowheads; and Fig. 7 b). PSB36 did not affect tumor growth, as assessed by the number of nuclei per spheroid (Fig. 7 c), and this resulted in more compact tumors with increased packing density of cells. These data establish ADORA1 in promoting collective breast cancer cell invasion in vivo.
Figure 7.
Pharmacological interference with ADORA1 inhibits collective invasion in vivo. (a) Invasion of 4T1 dual-color cells (H2B/mCherry; LifeAct/YFP) from microtumors implanted into the mouse mammary fat pad and imaged by multiphoton microscopy through a mammary window at days 0, 2, and 4 after implantation. Mice were treated with DMSO (vehicle) or 30 mg/kg PSB36 via intraperitoneal injection. Arrowhead, tip of collective invasion strand. (b and c) Analysis of invasion as the average area covered by the lesion normalized to the number of nuclei (b) and the tumor growth expressed as number of cells per lesion (c). Data in b and c represent the medians (black line), 25th/75th percentiles (boxes), and maximum/minimum values (whiskers) from at least nine implanted tumors from four independent experiments. P values, two-tailed unpaired Mann–Whitney test. (d) Detection of ADORA1 in fibrous or adipose tissue invasion zones of breast cancer samples using multispectral microscopy. (e) Relative ADORA1 levels in epithelial cancer cells quantified from eight independent lesions (see Table 1 for sample characteristics). Dotted line represents the threshold level for ADORA1 negativity, determined by ROC analysis with intensity values obtained from the luminal epithelial cells as control. Scale bars: 100 µm (a and d), 50 µm (a and d, inset).
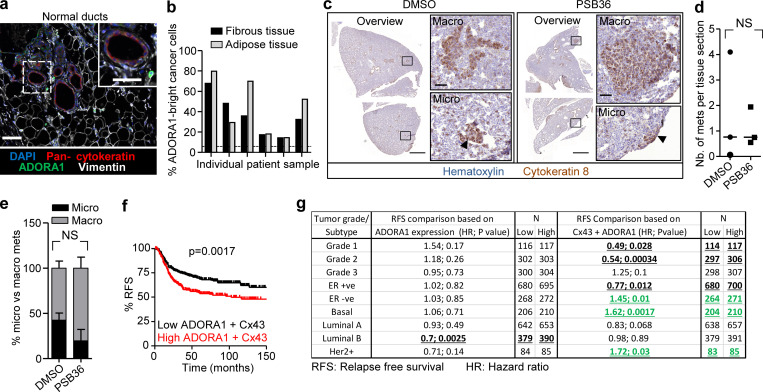
In tumor lesions in vivo, extracellular ADO levels are elevated to micromolar ranges (Blay et al., 1997; Kumar, 2013), and ADORA1 mRNA and protein expression were reported to be increased in whole-tissue lysates of primary tumors compared with normal tissue (Khoo et al., 1996; Mirza et al., 2005). To verify that ADORA1 was also present in the invasion zone of ductal breast cancer, clinical samples were stained for ADORA1, and the collective invasion patterns located in the fibrous and adipose tissues were analyzed by multispectral microscopy (Fig. 7 d). Multicellular strands and nests containing cytokeratin-positive epithelial cancer cells expressed ADORA1 (in six out of eight samples) with increased levels and heterogeneity compared with the luminal epithelial cells of normal ducts (Fig. 7, d and e; and Fig. S6 a). In ADORA1-positive samples, 14–68% of the cancer cells within the invasion patterns were ADORA1 bright (Fig. S6 b) with a predominant cytosolic distribution, consistent with the ADORA1 distribution detected in 3D invasion culture in vitro.
Figure S6.
Role of ADORA1 in preclinical spontaneous metastasis and prognostic relevance in clinical samples. (a) Detection of ADORA1 in normal ducts within clinical samples using multispectral microscopy. (b) Frequency of ADORA1-positive cancer cells with collective invasion patterns in breast lesions. Data represent the fibrous and adipose tissue from individual patients. (c) Overviews of lung sections isolated from DMSO and PSB36 (30 mg/kg) treated mice, stained for cytokeratin-8. Insets and arrowheads depict macro- and micrometastases. (d) Average number of micro- and macrometastases normalized per lung section. Data represent a total of 135–145 lung sections from three independent mice per group. (e) Relative frequency of micro- or macrometastases per treatment group: DMSO: 39 micrometastases, 59 macrometastases, 98 total; PSB36: 39 micrometastases, 91 macrometastases, 130 in total. Data show the mean percentages and SD from three mice. P value, two-tailed unpaired Mann–Whitney test. (f and g) Kaplan–Meier survival analysis predicting RFS in (f) basal-type breast cancer patients. (g) Correlation of ADORA1 or combined ADORA1 and Cx43 expression in clinical samples. Classification was based on tumor grade and molecular subtype (Györffy et al., 2010). Bold and underlined values show the positive (black) or inverse (green) association with ADORA1/Cx43 expression. (f and g) P values, log-rank test. Scale bars: 100 µm (a), 50 µm (a and c, inset), 1,000 µm (c, overview).
We finally tested whether inhibition of ADORA1 also reduces spontaneous metastasis to the lungs. 4T1 cells were implanted into the mammary fat pad, and the mice were treated daily with DMSO or PSB36. PSB36 therapy had no effect on the number or size of metastases to the lungs 30–36 d after tumor implantation (Fig. S6, c–e). Thus, although PSB36 inhibited local invasion in vivo, it had no effect on spontaneous metastasis in the 4T1 model. ADORA1 expression was inversely correlated with RFS in ER-negative patients with high Cx43 expression, but not ADORA1 alone (Fig. S6, f and g). This indicates that Cx43/ADORA1 expression correlates with poor outcome in ER-negative cancer, including the basal subtype.
Discussion
We here identify an autocrine mechanism of leader cell activation and maintenance through Cx43 hemichannels, which release nucleotides that induce AKT activation in leader cells and collective invasion of breast cancer cells via a signaling cascade involving ADORA1. ADORA1 drives local invasion and dissemination from implanted tumors in mice, and ER-negative breast cancer patients with high Cx43/ADORA1 expression suffer from worsened prognosis. Autocrine nucleotide signaling thus provides a cell-autonomous mechanism sustaining collective invasion in breast cancer and may deliver a rational basis for targeted intervention using hemichannel and ADORA inhibitors.
The data indicate a previously unappreciated role for nucleotide release via Cx43 hemichannels and autocrine ADO signaling in inducing leader cells that guide collective invasion of breast cancer cells. The concept of hemichannel-mediated nucleotide release in our study is supported by (1) focal Cx43 localization at cytoplasmic extensions of leader cells and lateral sites of follower cells, (2) dependence of extracellular nucleotide levels in 3D tumor spheroid culture on Cx43 channel function, (3) the decreased probability of cells with low Cx43 expression to reach leader cell position, (4) the rapid loss of leader cell activity after Cx channel inhibition or antagonization of ADORA1, and (5) reversion of invasion inhibition by CBX or Cx43 down-regulation by exogenous ADO addition. These data strongly suggest Cx43 hemichannel-mediated nucleotide release as major pathway to collective invasion.
The release of nucleotides by Cx hemichannels reveals a tumor-cell–autonomous, autocrine mechanism of induction and continuation of collective invasion. Hemichannel/purinergic signaling loops are known to regulate polarized protrusions of microglial cells during response to brain injury (Davalos et al., 2005) and single-cell migration of neutrophils during chemotaxis (Wang et al., 2017). In contrast to microglia and leukocytes, the autocrine nucleotide loop was not sufficient to induce polarization and migration of individualized breast cancer cells. Several parameters may account for this differential hemichannel function in epithelial contexts. Collective, but not individual-cell, polarity is reinforced by cell–cell junctions, including cadherins and Cxs, and receptor–ligand intercellular interactions, such as notch/delta and ephrin/Eph signaling (Friedl and Mayor, 2017). Intercellular junctions and interactions mediate mechanical and biochemical transductions between cells that enhance promigratory signals such as ECM contact and paracrine factors. As example, cadherin-dependent mechanocoupling promotes front–rear asymmetry by directing protrusion orientation and ECM binding toward the cell-free edge (Khalil and de Rooij, 2019; Mayor and Carmona-Fontaine, 2010; Plutoni et al., 2016). Thus, to induce leader cell activity and collective invasion, purinergic signaling may cooperate with the cell–cell junction machinery.
Our data further indicate that Cx43 hemichannel activity is essential but GJIC is dispensable for leader cell function and collective invasion. Whether, beyond the short-term effects detected here, GJIC contributes to mid- and long-term sustainability of collective invasion remains to be addressed. By allowing cytoplasmic continuity between invading cells, Cx43 intercellular- and hemichannels may act in series and contribute to ADO delivery and availability to the invasion front for release into the extracellular space.
In the extracellular space, ATP and ADP are degraded into ADO by ectonucleotidases, including CD39 and CD73 (de Andrade Mello et al., 2017), and ADO levels in the high-micromolar range are present in the tumor microenvironment (Blay et al., 1997); therefore ADO released in 3D invasion cultures and in vivo acts as both, metabolite and signaling effector of cancer progression (Antonioli et al., 2013). The ADO levels released by 4T1 and MMT cells during in vitro culture were one to five orders lower than ADO levels detected in the extracellular space of the tumor microenvironment (10–100 µM; Vaupel and Mayer, 2016; Vaupel and Multhoff, 2016). It is difficult to compare ADO levels present in the tumor tissue to the levels detected in spheroid invasion culture. Extracellular ADO in intact tissues is contributed by both tumor and stromal cells (de Lourdes Mora-García et al., 2016) over long periods of time. The resulting steady-state level of ADO reached in vivo may thus be higher compared with levels reached in invasion cultures over 24 h, with cancer cells as the sole source of ADO. Leader cell induction by auto- or paracrine nucleotides depends on downstream signaling function through ADORAs to induce polarized cell–substrate interaction, and this is shared with nucleotide activity inducing single-cell migration in other contexts. AKT signaling, which is induced by G-protein–coupled receptors (ADORA1, ADORA2a, ADORA2b, and ADORA3), mediates polarization of moving cells downstream of ADORA signaling (Chen et al., 2006; Gao et al., 2001; Othman et al., 2003; Stagg et al., 2010; Umapathy et al., 2013; Wen et al., 2011). Active AKT was increased in leader, but not follower, cells and required both Cx channel and ADORA1 function. In single-cell migration, individualized keratinocyte migration along electrical gradient depends on the Cx/G-protein–coupled receptor axis (Riding and Pullar, 2015). In brain-damage responses, astrocyte processes protrude in a Cx-ADORA–dependent manner to interact and close the microdefect (Davalos et al., 2005; Riding and Pullar, 2015). Although both ADORA1 and ADORA2b are expressed, only ADORA1 mediated ADO-dependent invasion of 4T1 and MMT cells. Possibly, the reported 70-fold higher affinity of ADO to ADORA1 (Fredholm et al., 2001) accounts for its dominance in leader cell activation. However, in difference to ADORA-stimulated single-cell migration, autocrine stimulation of tip cells favors their engagement toward ECM while remaining coupled to follower cells, thus mediating the maintenance and elongation of collective invasion strands.
While ADORA1 function was clearly involved in local tissue invasion, and collective invasion in breast carcinoma samples is positively associated with clinical progression to metastasis (Khalil et al., 2017), spontaneous metastasis to the lungs was not affected by pharmacological ADORA1 antagonization. The 4T1 in vivo model resulted in 100% penetrance of spontaneous micro- and macrometastases, and the used dosing of PSB36 inhibited invasion in mammary fat pads, suggesting that the drug is efficient in vivo. The lack of inhibition of metastases may indicate that leader cell function maintained by ADORA1 is of lower relevance in secondary organ colonization and metastatic outgrowth. A similar disconnect between local invasion and metastatic ability in distant organs was recently shown after deletion of E-cadherin, which caused increased local invasion in breast cancer models but strongly compromised metastatic ability due to a survival defect (Padmanaban et al., 2019). Interfering with ADORA1 pathways may thus be justified to limit locally invasive disease, which cannot be effectively treated by surgery (Bakst et al., 2019). Beyond enhancing invasion, hemichannels and ADORAs also contribute to tumor growth and immunomodulation (Schalper et al., 2014) and orchestrate an integrated program to enhance local cancer progression. Purinergic signaling thus establishes a proinvasive microenvironment through multipronged mechanisms, rendering pharmacological interference of nucleotide signaling through the Cx/ADORA axis in patient subsets as an attractive route to reprogram the tumor stroma and neoplastic invasion.
Materials and methods
Antibodies and reagents
The following antibodies were used: rabbit anti-human Cx43 (3512; CST), chicken anti-human vimentin (ab24525; Abcam), mouse anti-mouse β-catenin (clone 14/β-Catenin; BD Biosciences), rabbit anti-mouse AKT (9272; CST), mouse anti-human AKT (2920; CST); rabbit anti-human phospho-AKT Ser473 (4060; CST); mouse anti-human Pan Keratin (4545; CST); rabbit anti-rat ADORA1 (ab82477; Abcam); rabbit anti-human ADORA1 (ab124780; Abcam); and secondary polyclonal Alexa Fluor 488/647–conjugated goat anti-mouse, anti-rabbit, or anti-chicken antibody. For F-actin visualization, Alexa Fluor 568–conjugated phalloidin (Invitrogen) was used. ADO (A4036; Sigma-Aldrich) was dissolved in water at a concentration of 1 mg/ml and stored in aliquots at −20°C. The following inhibitors were used: gap junction channel inhibitors CBX disodium salt, 18aGA, and its inactive homologue, GLZ (Sigma-Aldrich; Kenny et al., 2002; Marins et al., 2009); the ADORA1 selective and competitive antagonists 1-butyl-3-(3-hydroxypropyl)-8-(3-noradamantyl) xanthine (PSB36) and 8-cyclopentyl-1,3-dipropylxanthine (Sigma-Aldrich); and the ADORA2b selective antagonist 4-(2,3,6,7-tetrahydro-2, 6-dioxo-1-propyl-1H-purin-8-yl)-benzenesulfonic acid (PSB1115; Tocris; Lin et al., 2010). CBX was dissolved in water (stock concentration: 163 mM); all other inhibitors and antagonists were dissolved in DMSO (stock concentration 100–200 mM). All drugs were aliquoted (5–20 µl) and stored at −20°C.
Cell lines and culture
The following metastatic breast cancer cell lines were used: 4T1 (CRL-2539 ATCC); MMT wild-type and MMT dual-color cells (H2b-GFP and cytosolic RFP; Tsuji et al., 2006; a generous gift from R. Hoffman, AntiCancer Inc., San Diego, CA); and NMuMG cells (a generous gift from M. Vooijs, Maastricht University Medical Center, Netherlands; Owens et al., 1974). The identity of 4T1, MMT and NMuMG cells was verified by short tandem repeat DNA profiling (IDEXX BioResearch). No mammalian interspecies contamination was detected. Cells were routinely tested for mycoplasma contamination (MycoAlert; Lonza). All cell lines were negative for mycoplasma contamination. Cells were maintained (37°C, 5% CO2, humidified atmosphere) in RPMI media (Invitrogen) supplemented with 10% fetal bovine serum (Sigma-Aldrich), penicillin (100 U/ml) and streptomycin (100 µg/ml; both PAA), and l-glutamine (2 mM; Invitrogen). For generation of 4T1 and MMT cells with stable Cx43 RNA down-regulation, mission lentiviral vector targeting mouse GJA1 (TRCN0000068473, sequence: 5′-CCGGCCCACCTTTGTGTCTTCCATACTCGAGTATGGAAGACACAAAGGTGGGTTTTTG-3′) and nontargeting control sequence (5′-CCGGCAACAAGATGAAGAGCACCAACTCGAGTTGGTGCTCTTCATCTTGTTGTTTTT-3′) was used (Sigma-Aldrich). For tumor implantation into the mammary fat pad in vivo, a stable 4T1/H2B-mCherry/LifeAct-GFP cells were used, as described previously (Ilina et al., 2018). Fluorescence-expressing cells did not deviate from the wild-type counterparts in parameters such as growth and invasion (data not shown).
Cancer spheroid and single-cell culture
Spheroids from 4T1, MMT, and NMuMG cells used in short-term in vitro migration assays were generated using the hanging drop method (Venhuizen et al., 2019). In brief, cells from subconfluent culture were detached with EDTA (1 mM) and trypsin (0.075%; Invitrogen), washed in PBS, suspended in medium/methylcellulose (2.4%; Sigma), and maintained as hanging droplets (25 µl), each containing 1,000 cells. Hanging drops were kept in 37°C, 5% CO2 humidified atmosphere for 24 h (Del Duca et al., 2004). Spheroids were harvested and placed into DMEM media without FCS and without penicillin/streptomycin in a six-well plate.
After cell aggregation, spheroids were washed in PBS and incorporated into 3D type I collagen lattices consisting of nonpepsinized rat-tail collagen (BD Biosciences) at a final concentration of 4 or 6 mg/ml for 4T1 or MMT cells (Ilina et al., 2018). In brief, spheroids were washed in complete medium, mixed with collagen solution, and prevented from sinking during collagen polymerization (37°C, 5% CO2) by rotation of the sample. Only spheroids invading into the 3D phase of the collagen matrix were analyzed, whereas spheroids moving as 2D sheets underneath or on top of the culture were excluded.
For single-cell invasion, cells from subconfluent culture were detached with EDTA (2 mM), washed and embedded in 3D nonpepsinized rat-tail collagen (4 mg/ml; BD Biosciences) at a density of 20,000 cells/100 μl collagen.
Transient down-regulation of Cx43 by RNAi
4T1 and MMT cells were seeded in a 12-well plate at a density of 60 × 103/well, in the presence of siRNA duplex (Dharmacon, Thermo Fisher Scientific). ON-Target plus nontargeting siRNA (D-001810-10) and two Cx43-specific targeting sequences (J-051694-05 and J-051694-06) were used at a final concentration of 50 nM using Dharmafect transfection reagent 4 (Thermo Fisher Scientific). After 12 h, RNAi containing supernatant was removed and replaced by penicillin/streptomycin free media. After a recovery period of at least 6 h, cells were used for spheroid generation. For assessment of knockdown efficiency, whole-cell lysates were analyzed after 24, 48, and 72 h after removal of siRNA by Western blot.
Resazurin cytotoxicity assay
Mitochondrial activity was assessed by resazurin labeling (Czekanska, 2011). 4T1 and MMT spheroids embedded in 3D collagen were incubated with resazurin-containing media (100 µg/ml) for 4h (at 37°C, 5% CO2, humidified atmosphere). Gels were gently washed with PBS, and cell-bound fluorescence was measured (excitation/emission 560/590 nm) using an ELISA reader.
Real-time quantitative PCR
Collagen gels containing spheroids were dissolved with Trizol (250 µl/100 µl gel; Invitrogen) and homogenized before chloroform (Merck) phase separation (Farhat, 2012). After centrifugation (15 min, 12 × 103 g, 4°C), the upper phase was mixed with 70% vol/vol ethanol and transferred to an RNeasy MinElute spin column for RNA extraction according to the manufacturer’s instructions (Qiagen’s RNeasy kit). After RNA quality control, reverse transcription (SuperScript II Reverse transcription; Thermo Fisher Scientific) was performed. Primers for Cxs and purinergic receptors (Table S1) were designed using HomoloGene (http://www.ncbi.nlm.nih.gov) and validated in silico using Oligoanalyzer software and in vitro using positive controls (RNA isolated from mouse tissues including brain and heart). Quantitative PCR was performed using CFX96 Real-Time PCR Detection System with C1000 Thermocycler (Bio-Rad) and analyzed using the Bio-Rad CFX Manager software (version 2.0). RNA levels for the genes of interest were normalized to the pooled levels of three housekeeping genes (β-actin, GAPDH, and ΥWhaz).
Western blot analysis
Cells from 2D culture were lysed using SDS sample buffer (62.5 mM Tris-HCl, 2% SDS, 10% glycerol, 50 mM DTT, and bromophenol blue). Collagen gels containing spheroids were digested by highly purified type VII collagenase (C0773; Sigma-Aldrich), followed by whole-cell lyses using SDS buffer. Western blotting was performed by SDS-PAGE. Briefly, whole-cell lysates were loaded and separated on a 10% acrylamide gel with Tris-glycine running buffer followed by blotting to polyvinylidene difluoride membrane by wet transfer. Polyvinylidene difluoride membranes were blotted with monoclonal mouse anti-β-tubulin (E7; Department of Cell Biology, Radboud University Medical Center, Nijmegen, Netherlands) as loading control, anti-Cx43 (dilution 1:1,000), anti-AKT (1:1,000), and anti-phospho-AKT Ser473 (1:500) followed by fluorescence detection (Odyssey; LI-COR Biosciences) and densitometric analysis.
HPLC analysis for purine content in media
Supernatants from 3D single-cell or spheroid cultures (24 h) were used for purine analysis. Media contained 80°C heated FCS, which minimizes the degradation of nucleotides and nucleosides (Gendaszewska-Darmach et al., 2003). ADO, AMP, ADP, and ATP levels were determined by HPLC using a modified method from Bhatt et al. (2012). In brief, culture supernatant (1 ml) was mixed with chloroacetaldehyde (250 µl; 6x diluted in 1 M acetate buffer, pH 4.5; Sigma-Aldrich), followed by derivatization (60 min, 70°C, 500 rpm) using Thermomixer comfort (Eppendorf AG) and centrifugation (3 min, RT, 12,100 × g). For HPLC, supernatant (400 µl) was transferred to a HPLC vial and injected. Purines were separated by HPLC (1200 Series; Agilent Technologies) using a Polariz C18-A column (150 × 4.6 mm) with a gradient elution using eluent A (0.1 M K2HPO4, 10 mM tetrabutylammonium hydrogen sulfate, pH 6.5, and 2% MeOH) and eluent B (H2O/ACN/THF 50:49:1). Retention times were 6.5 (ADO), 7.7 (AMP), 11.8 (ADP), 15.6 (ATP), and 15.0 min (cAMP). Quantification was based on peak areas of the samples, and reference standards were measured with fluorescence detection (excitation, 280 nm; emission, 420 nm).
Bright-field microscopy
Single or collective invasion was detected by digital time-lapse bright-field microscopy, as described previously (Wolf et al., 2013). In brief, single cells or 3D spheroid cultures were maintained at 37°C and images were taken with 5-min intervals for up to 72 h. For pharmacological intervention, spheroids were allowed to migrate for 24 h, followed by addition of inhibitor or vehicle (solvent) for 24 h and washout using 1 ml of growth media and follow-up for additional 24 h. Single cells and leader cells were tracked using the manual tracking plugin from ImageJ (1.40v; National Institutes of Health). The number, length, and elongation speed of invading single cells or strands were quantified using ImageJ. For endpoint experiments, bright-field micrographs of spheroid cultures in 3D collagen after 15, 24, and 48 h of incubation were recorded using a CCD camera (Sentech), 10x/0.20 NA air objective (Leica) and frame grabber system (Vistek). The number of pointed and protrusive tips (leader cells) per spheroid was counted manually; length of invading strands were quantified using the image analysis software ImageJ.
Parachute assay and FACS analysis
Receiver 4T1 cells (150 × 103 cells/well) were incubated for 24 h (37°C, 5% CO2 humidified atmosphere) in 48-well plates to reach 95–100% confluency. Donor 4T1 cells were loaded with calcein-AM (1 µM; Life Technologies) for 30 min, washed, and incubated for a further 45 min (Abbaci et al., 2008). Calcein-labeled cells were detached with EDTA (1 mM) and trypsin (0.075%), centrifuged (251 g, 5 min), resuspended in media, and 75 × 103 cells/well were added over the receiver cell culture at a ratio of 1:2 (donor/receiver) and co-cultured (3, 6, or 12 h). For analysis of dye transfer, cells were detached with EDTA (1 mM) and trypsin (0.075%), assayed by flow cytometry (FACS Caliber), and analyzed using FCS Express software.
Immunofluorescence staining and confocal microscopy
3D spheroid samples were fixed with 4% paraformaldehyde at room temperature (RT) for 15 min. For detection of intracellular antigens, gels were washed with PBS, permeabilized using 5–10% normal goat serum, 0.3% Triton X-100 or 0.2% Saponin in PBS (1 h, 20°C), incubated with antibody in PBS containing 0.1% BSA, 0.3% Triton X-100, or 0.2% Saponin. Antibody dilutions were as follows: Cx43 (1:50), AKT (1:50), S473 pAKT (1:50), β-Catenin (1:200), ZO-1 (1:100), and vimentin (1:200). Primary antibody was incubated at 4°C with shaking overnight, followed by at least five washing steps of 15 min each with PBS. Samples were then incubated with secondary antibody (1:400) in PBS buffer for intracellular staining, together with DAPI (5 µg/ml) and phalloidin (1:200; 4 h, RT), washed at least five times (15 min each) and imaged by confocal microscopy (Olympus FV1000; long working distance objectives 20×/NA 0.50, 40×/NA 0.80, or 60×/NA 1.35). Subcellular localization and quantification of Cx43, β-catenin, AKT, and pAKT at cell–cell contacts and cytoplasmic extensions were determined by manually outlining F-actin–positive regions followed by background correction using ImageJ.
For time-lapse confocal microscopy, 3D spheroid cultures consisting of MMT dual-color cells or 4T1/H2B-mCherry within 3D collagen were imaged (37°C, 5% CO2, humidified atmosphere). Time-lapse confocal microscopy (20×/NA 0.5 air objective, LSM 510 system; Carl Zeiss) was performed with time intervals of 10–15 min for 24 h with usually four to seven z-scans comprising all of the strands per spheroid (∼100 µm in depth). The confocal slices were reconstructed as maximum intensity projection and image analysis, including leader cell tracking, was performed using ImageJ. The minimum and maximum pixel intensity values of different samples were equally adjusted per experiment using the color balance tool of ImageJ.
Gap FRAP
Cancer cells grown as monolayers on glass-bottom dishes or as multicellular spheroids in 3D collagen were labeled with calcein-AM (2–3 µM, 30 min) using a modified method described previously (Kuzma-Kuzniarska et al., 2014). Regions of interest were defined for single cells or small regions and bleaching was performed using 100% Argon laser output 1 mW with 200 iterations using a large pinhole (∼8 airy units) and 20-s interval at 37°C, 5% CO2 and humidified atmosphere. For 12 min of follow-up, calcein transfer between cells seeded on 2D coverglass or within collagen gels was monitored using confocal microscopy (LSM 510; 20×/NA 0.5 air; Carl Zeiss; 37°C, 5% CO2). Inhibition of GJIC was achieved using CBX (100 µM) at least 45 min before gap FRAP imaging. Time-resolved bleaching and postbleaching recovery was quantified using ImageJ as the mean fluorescence intensity. For normalizing fluorescence after photobleaching, cells in nonbleached control regions were coregistered and the signal used for background correction. Detached single cells were used as an internal negative control for GJIC-dependent fluorescence recovery.
Breast cancer tissue section immunofluorescence
For multispectral imaging of Cx43 and ADORA1, tissue sections with positive invasion margin were selected from 19 breast cancer patients (Table 1). The use of coded tumor tissue was approved by the institutional review board, according to national law (Nagelkerke et al., 2011). Formalin-fixed paraffin-embedded breast tissue sections (5 µm thickness) were deparaffinized, followed by antigen retrieval using Tris-EDTA buffer (15 min, 95–100°C) followed by a washing step with PBS. Sections were blocked with 5% normal goat serum in 0.2% Tween for 1 h at 20°C. Primary antibody was incubated at 4°C with shaking overnight using the following dilutions: Cx43 (1:100), vimentin (1:200), pan-cytokeratin (1:200), and ADORA1 (1:200). Samples were then washed four times (PBS, 5 min each), incubated with secondary Alexa Fluor 488/546/647–conjugated goat anti-mouse, anti-rabbit, and anti-chicken antibodies, together with DAPI (1 μg/ml), for 1 h at 20°C and again washed four times (PBS, 5 min each). Dried sections were embedded in Mowiol (Sigma-Aldrich) and scanned using the automated Vectra Intelligent Slide Analysis System (version 2.0.8; PerkinElmer).
Table 1. Histological and clinical classification of patient samples analyzed for Cx43 and/or ADORA1 expression.
| ID | Tumor type | Year | Age (yr) | Menopausal status | Surgical removal |
|---|---|---|---|---|---|
| 1 | ND | 1996 | 48.47 | Pre | Mastectomy |
| 2 | IDC | 1994 | 46.71 | Pre | Mastectomy |
| 3 | IDC | 1993 | 76.10 | Post | Mastectomy |
| 4 | IDC | 1992 | 39.85 | Pre | Mastectomy |
| 5 | ND | 1996 | 67.27 | Post | Lumpectomy |
| 6 | IDC | 1995 | 55.57 | Post | Lumpectomy |
| 7 | IDC | 1992 | 64.39 | Post | Lumpectomy |
| 8 | IDC | 1992 | 43.29 | Pre | Lumpectomy |
| 9 | IDC | 1993 | 47.90 | Pre | Lumpectomy |
| 10 | IDC | 1996 | 34.35 | Pre | Mastectomy |
| 11 | ND | 1995 | 30.50 | Pre | Lumpectomy |
| 12 | IDC | 1996 | 47.93 | Pre | Mastectomy |
| 13 | IDC | 1996 | 52.81 | Post | Lumpectomy |
| 14 | ILC | 1991 | 59.54 | Post | Lumpectomy |
| 15 | IDC | 1991 | 68.69 | Post | Lumpectomy |
| 16 | IDC | 1995 | 68.95 | Post | Lumpectomy |
| 17 | IDC | 1992 | 36.67 | Pre | Lumpectomy |
| 18 | IDC | 1993 | 76.51 | Post | Lumpectomy |
| 19 | IDC | 1995 | 80.56 | Post | Mastectomy |
Red shading indicates samples stained for Cx43, blue shading indicates samples stained for ADORA1, and green shading indicates samples stained for both Cx43 and ADORA1. IDC, invasive ductal carcinoma; ILC, invasive lobular carcinoma.
Multispectral imaging and quantitative digital analysis
Tissue slides were imaged using Vectra Intelligent Slide Analysis System (Version 2.0.8; PerkinElmer), as described (Mascaux et al., 2019). This imaging technique combines imaging with spectroscopy by collecting the spectral range from 400 to 720 nm in 10-nm steps in an automatic manner. To define the spectrum of each fluorophore as well as of tissue autofluorescence, stained single-fluorescence and unstained tissues were recorded with the Nuance Multispectral Imaging System (Version 3.0.2; PerkinElmer). A spectral library was built from the extracted spectra to enable the quantitative separation and spectral unmixing, thus removing cross-talk between fluorophores and interfering tissue autofluorescence. A selection of 10 representative original multispectral images was loaded into an advanced user-trainable morphological image analysis software (InForm Version 2.1; PerkinElmer) which utilizes machine learning for user-trained assignment of morphological and spectral intensity patterns to discriminate tumor cells, stromal regions, and normal ducts. The analysis software utilized the multimarker, multispectral data to extract and calculate single-cell parameters, including nuclear, cytoplasmic, and membrane intensity values used to classify each cell (e.g., as luminal, cancerous, or stroma cell; Fig. S1 b). After training, the algorithm was applied as batch analysis of multiple original multispectral images of different samples. Cx43, pan-cytokeratin, and vimentin expression levels were quantified in normal bilayered ducts, tumor cells located within the fibrous and adipose tissue, and their surrounding stroma. The expression levels of Cx43 were measured in each cell and the positivity threshold was determined by receiver operating characteristic (ROC) analysis whereby Cx43 and ADORA1 values of all the luminal epithelium cells available in different patients were used as negative control. With 99% specificity and 50% sensitivity for Cx43 and 93% specificity and 31% selectivity for ADORA1, the minimum threshold positive staining was calculated to be 0.875 and 2.500, respectively (Figs. 1 c and 7 e, dashed line).
In silico analysis of Cx43 and ADORA1 expression in breast cancers
We used the publicly available database (http://kmplot.com), which contains expression data on several genes, including Cx43 and ADORA1, in a cohort consisting of 3,951 breast cancer patients. The database classifies patients by trichotomizing the expression levels of the gene of interest, into high (T3) or low expression (T1), to extract correlations between gene expression level and patient outcome.
Mice
BALB/c female mice (6–8 wk old) were purchased from Charles River Laboratories. All animal procedures were approved by the Ethical Committee on Animal Experiments at Radboud University, Nijmegen (protocol number 140201) and performed according to guidelines of the Animal Welfare Committee of the Royal Netherlands Academy of Arts and Sciences, the Netherlands.
Implantation of tumor cell spheroids in the mammary fat pad
All surgical procedures were performed under isoflurane inhalation anesthesia (1–2% isoflurane/O2 mixture) and performed under conditions of microsurgery, as described (Ilina et al., 2018). In brief, 4T1 dual-color spheroids (500 cells/spheroid) were implanted adjacent to the third mammary fat pad into a central area of the mammary imaging window by nontraumatic microimplantation. After implantation, a sterile coverglass was placed into the window frame, and the integrity of both implanted tumoroid and stroma was verified by fluorescence microscopy.
Intravital microscopy and image analysis
Tumor growth and cell invasion were monitored for up to 4 d by intravital epifluorescence and multiphoton microscopy, as described (Ilina et al., 2018). In brief, mice were anesthetized with isoflurane, and the imaging window was stably fixed on a temperature-controlled stage (37°C) using a custom holder. Blood vessels were visualized by i.v. injection of 70-kD dextran labeled with Alexa Fluor 750. Invasion was detected using multiphoton microscopy (TrimScope II, LaVision BioTec; 20x objective, NA 0.95). Images were analyzed to quantify both, the number of nuclei and the surface/invasion area of implanted tumors at different time points using ImageJ.
Spontaneous metastasis analysis
The development of spontaneous lung metastasis was obtained from orthotopically implanted mammary tumors (Ilina et al., 2018). In brief, 4T1 cells (105 in 50 µl) were injected into the fourth mammary fat pad of BALB/c 8-wk-old female mice (Charles River Laboratories). DMSO (0.2%) and PSB36 (30 mg/kg) dissolved in PBS (Bilkei-Gorzo et al., 2008) were injected intraperitoneally once per day. Tumor size was recorded weekly by a caliper, and mice were euthanized 30–36 d after cancer cell injection or at the humane endpoint (tumor volume of 2 cm3). Lungs were harvested, fixed in buffered formalin, embedded in paraffin, and sectioned (5 µm thickness). Metastatic foci were detected by immunohistochemistry of cytokeratin-8 and quantified by operator-based blinded scoring (Ilina et al., 2018). In brief, the number of neoplastic events was counted from sequential sections and represented as the number of metastatic foci per slice. Metastasis was classified as micrometastasis (3–15 cytokeratin-8–positive cells) and macrometastasis (>15 tumor cells in nodular topology).
Statistical analysis
Graph Prism (version 5.0) was used for statistical analysis. The two-tailed unpaired Mann–Whitney (two groups) or Kruskal–Wallis test (more than two groups) with Dunns’ correction were used for all invasion analysis in vitro (number of leader cells/spheroid, cumulative invasion distance, and average speed). As an exception, data in Fig. S3 (e and h) were analyzed using ANOVA with Bonferroni correction after confirming normal distribution of the data (Kolmogorov–Smirnov normality test). The two-tailed unpaired Mann–Whitney test was further used for comparing invasion and tumor growth between DMSO and PSB36 treated mice in vivo. Data from Resazurin assay and HPLC analysis were tested for normal distribution using Kolmogorov–Smirnov test followed by ANOVA analysis with Bonferroni correction. Kaplan–Meier survival curves were compared using log-rank test.
Online supplemental material
Fig. S1 shows the expression of Cx43 in collective invasion patterns within breast lesions and Cx expression profile of 4T1 and MMT cells in 3D collagen (related to Fig. 1). Fig. S2 shows that collectively invading cancer cells maintain Cx43-mediated GJIC and extracellular nucleotide release (related to Fig. 2). Fig. S3 shows the dependence of leader cell function on Cx43 expression and channel function (related to Fig. 3). Fig. S4 shows purinergic receptor mRNA expression and the effect of ADORA inhibition on invasion and proliferation (related to Figs. 4 and 5). Fig. S5 shows the requirement of the Cx hemichannel-ADORA1 loop in other collective, but not single-cell, invasion models and downstream AKT activation (related to Fig. 6). Fig. S6 shows ADORA1 in normal human mammary epithelium, its role in preclinical spontaneous metastasis, and its prognostic relevance in clinical samples (related to Fig. 7). Video 1 shows 4T1 protrusive tips during initiation of collective invasion in 3D collagen I by bright-field time-lapse microscopy. Video 2 shows the initiation and elongation of collective invasion of MMT cells (H2b-GFP and cytosolic dsRed) in 3D collagen I by time-lapse confocal microscopy. Video 3 shows calcein intercellular dye transfer between collectively invading strands in the presence and absence of CBX. Video 4 shows MMT leader cells driving invasion (H2b-GFP) in 3D collagen I in the presence and absence of CBX. Video 5 and Video 6 show changes in the dynamics of established strands after intervention with CBX and PSB36 treatment, respectively. Table S1 shows primer sequences.
Supplementary Material
lists primer sequences.
Acknowledgments
We gratefully acknowledge Huib Croes, Gert-Jan Bakker, and Dagmar Verweij for technical support and Michael Dustin and Pavlo G. Gritsenko for helpful discussions.
This work was supported by a German Excellence Initiative grant to the Graduate School of Life Sciences, University of Würzburg (to A.A. Khalil), the Netherlands Science Organization (NWO-VICI 918.11.626), the European Research Council (617430-DEEPINSIGHT), and the Cancer Genomics Center, Netherlands (to P. Friedl).
The authors declare no competing financial interests.
Author contributions: A.A. Khalil and P. Friedl designed the experiments. A.A. Khalil, O. Ilina, A. Vasaturo, J.-H. Venhuizen, M. Vullings, V. Venhuizen, and A. Bilos performed the experiments. A.A. Khalil, O. Ilina, A. Vasaturo, J.-H. Venhuizen, M. Vullings, V. Venhuizen, A. Bilos, and P. Friedl analyzed the data. C.G. Figdor provided the platform for multispectral imaging. P.N. Span provided and interpreted clinical samples. A.A. Khalil and P. Friedl wrote the manuscript. All authors read the manuscript.
References
- Aasen T., Leithe E., Graham S.V., Kameritsch P., Mayán M.D., Mesnil M., Pogoda K., and Tabernero A.. 2019. Connexins in cancer: bridging the gap to the clinic. Oncogene. 38:4429–4451. 10.1038/s41388-019-0741-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbaci M., Barberi-Heyob M., Blondel W., Guillemin F., and Didelon J.. 2008. Advantages and limitations of commonly used methods to assay the molecular permeability of gap junctional intercellular communication. Biotechniques. 45:33–52–62. 10.2144/000112810 [DOI] [PubMed] [Google Scholar]
- Abo-Salem O.M., Hayallah A.M., Bilkei-Gorzo A., Filipek B., Zimmer A., and Müller C.E.. 2004. Antinociceptive effects of novel A2B adenosine receptor antagonists. J. Pharmacol. Exp. Ther. 308:358–366. 10.1124/jpet.103.056036 [DOI] [PubMed] [Google Scholar]
- Antonioli L., Blandizzi C., Pacher P., and Haskó G.. 2013. Immunity, inflammation and cancer: a leading role for adenosine. Nat. Rev. Cancer. 13:842–857. 10.1038/nrc3613 [DOI] [PubMed] [Google Scholar]
- Ashton A.W., Yokota R., John G., Zhao S., Suadicani S.O., Spray D.C., and Ware J.A.. 1999. Inhibition of endothelial cell migration, intercellular communication, and vascular tube formation by thromboxane A(2). J. Biol. Chem. 274:35562–35570. 10.1074/jbc.274.50.35562 [DOI] [PubMed] [Google Scholar]
- Bakst R.L., Glastonbury C.M., Parvathaneni U., Katabi N., Hu K.S., and Yom S.S.. 2019. Perineural Invasion and Perineural Tumor Spread in Head and Neck Cancer. Int. J. Radiat. Oncol. Biol. Phys. 103:1109–1124. 10.1016/j.ijrobp.2018.12.009 [DOI] [PubMed] [Google Scholar]
- Barletta K.E., Ley K., and Mehrad B.. 2012. Regulation of neutrophil function by adenosine. Arterioscler. Thromb. Vasc. Biol. 32:856–864. 10.1161/ATVBAHA.111.226845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroja-Mazo A., Barberà-Cremades M., and Pelegrín P.. 2013. The participation of plasma membrane hemichannels to purinergic signaling. Biochim. Biophys. Acta. 1828:79–93. 10.1016/j.bbamem.2012.01.002 [DOI] [PubMed] [Google Scholar]
- Bazzoun D., Adissu H.A., Wang L., Urazaev A., Tenvooren I., Fostok S.F., Chittiboyina S., Sturgis J., Hodges K., Chandramouly G., et al. 2019. Connexin 43 maintains tissue polarity and regulates mitotic spindle orientation in the breast epithelium. J. Cell Sci. 132 jcs223313 10.1242/jcs.223313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt D.P., Chen X., Geiger J.D., and Rosenberger T.A.. 2012. A sensitive HPLC-based method to quantify adenine nucleotides in primary astrocyte cell cultures. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 889-890:110–115. 10.1016/j.jchromb.2012.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilkei-Gorzo A., Abo-Salem O.M., Hayallah A.M., Michel K., Müller C.E., and Zimmer A.. 2008. Adenosine receptor subtype-selective antagonists in inflammation and hyperalgesia. Naunyn Schmiedebergs Arch. Pharmacol. 377:65–76. 10.1007/s00210-007-0252-9 [DOI] [PubMed] [Google Scholar]
- Blay J., White T.D., and Hoskin D.W.. 1997. The extracellular fluid of solid carcinomas contains immunosuppressive concentrations of adenosine. Cancer Res. 57:2602–2605. [PubMed] [Google Scholar]
- Boitano S., Dirksen E.R., and Sanderson M.J.. 1992. Intercellular propagation of calcium waves mediated by inositol trisphosphate. Science. 258:292–295. 10.1126/science.1411526 [DOI] [PubMed] [Google Scholar]
- Bos P.D., Zhang X.H., Nadal C., Shu W., Gomis R.R., Nguyen D.X., Minn A.J., van de Vijver M.J., Gerald W.L., Foekens J.A., et al. 2009. Genes that mediate breast cancer metastasis to the brain. Nature. 459:1005–1009. 10.1038/nature08021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Corriden R., Inoue Y., Yip L., Hashiguchi N., Zinkernagel A., Nizet V., Insel P.A., and Junger W.G.. 2006. ATP release guides neutrophil chemotaxis via P2Y2 and A3 receptors. Science. 314:1792–1795. 10.1126/science.1132559 [DOI] [PubMed] [Google Scholar]
- Cheung K.J., and Ewald A.J.. 2016. A collective route to metastasis: Seeding by tumor cell clusters. Science. 352:167–169. 10.1126/science.aaf6546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung K.J., Gabrielson E., Werb Z., and Ewald A.J.. 2013. Collective invasion in breast cancer requires a conserved basal epithelial program. Cell. 155:1639–1651. 10.1016/j.cell.2013.11.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czekanska E.M. 2011. Assessment of cell proliferation with resazurin-based fluorescent dye. Methods Mol. Biol. 740:27–32. 10.1007/978-1-61779-108-6_5 [DOI] [PubMed] [Google Scholar]
- Davalos D., Grutzendler J., Yang G., Kim J.V., Zuo Y., Jung S., Littman D.R., Dustin M.L., and Gan W.B.. 2005. ATP mediates rapid microglial response to local brain injury in vivo. Nat. Neurosci. 8:752–758. 10.1038/nn1472 [DOI] [PubMed] [Google Scholar]
- de Andrade Mello P., Coutinho-Silva R., and Savio L.E.B.. 2017. Multifaceted Effects of Extracellular Adenosine Triphosphate and Adenosine in the Tumor-Host Interaction and Therapeutic Perspectives. Front. Immunol. 8:1526 10.3389/fimmu.2017.01526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lourdes Mora-García M., García-Rocha R., Morales-Ramírez O., Montesinos J.J., Weiss-Steider B., Hernández-Montes J., Ávila-Ibarra L.R., Don-López C.A., Velasco-Velázquez M.A., Gutiérrez-Serrano V., et al. 2016. Mesenchymal stromal cells derived from cervical cancer produce high amounts of adenosine to suppress cytotoxic T lymphocyte functions. J. Transl. Med. 14:302 10.1186/s12967-016-1057-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Duca D., Werbowetski T., and Del Maestro R.F.. 2004. Spheroid preparation from hanging drops: characterization of a model of brain tumor invasion. J. Neurooncol. 67:295–303. 10.1023/B:NEON.0000024220.07063.70 [DOI] [PubMed] [Google Scholar]
- el-Sabban M.E., and Pauli B.U.. 1991. Cytoplasmic dye transfer between metastatic tumor cells and vascular endothelium. J. Cell Biol. 115:1375–1382. 10.1083/jcb.115.5.1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eltzschig H.K., Eckle T., Mager A., Küper N., Karcher C., Weissmüller T., Boengler K., Schulz R., Robson S.C., and Colgan S.P.. 2006. ATP release from activated neutrophils occurs via connexin 43 and modulates adenosine-dependent endothelial cell function. Circ. Res. 99:1100–1108. 10.1161/01.RES.0000250174.31269.70 [DOI] [PubMed] [Google Scholar]
- Elzarrad M.K., Haroon A., Willecke K., Dobrowolski R., Gillespie M.N., and Al-Mehdi A.B.. 2008. Connexin-43 upregulation in micrometastases and tumor vasculature and its role in tumor cell attachment to pulmonary endothelium. BMC Med. 6:20 10.1186/1741-7015-6-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhat Y. 2012. RNA Extraction from Collagen Gels Using Qiagen’s RNeasy Kit. The Protocol Place. [Google Scholar]
- Fostok S., El-Sibai M., Bazzoun D., Lelièvre S., and Talhouk R.. 2019. Connexin 43 Loss Triggers Cell Cycle Entry and Invasion in Non-Neoplastic Breast Epithelium: A Role for Noncanonical Wnt Signaling. Cancers (Basel). 11:339 10.3390/cancers11030339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredholm B.B., Irenius E., Kull B., and Schulte G.. 2001. Comparison of the potency of adenosine as an agonist at human adenosine receptors expressed in Chinese hamster ovary cells. Biochem. Pharmacol. 61:443–448. 10.1016/S0006-2952(00)00570-0 [DOI] [PubMed] [Google Scholar]
- Friedl P., and Gilmour D.. 2009. Collective cell migration in morphogenesis, regeneration and cancer. Nat. Rev. Mol. Cell Biol. 10:445–457. 10.1038/nrm2720 [DOI] [PubMed] [Google Scholar]
- Friedl P., and Mayor R.. 2017. Tuning Collective Cell Migration by Cell-Cell Junction Regulation. Cold Spring Harb. Perspect. Biol. 9 a029199 10.1101/cshperspect.a029199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaggioli C., Hooper S., Hidalgo-Carcedo C., Grosse R., Marshall J.F., Harrington K., and Sahai E.. 2007. Fibroblast-led collective invasion of carcinoma cells with differing roles for RhoGTPases in leading and following cells. Nat. Cell Biol. 9:1392–1400. 10.1038/ncb1658 [DOI] [PubMed] [Google Scholar]
- Gao Z., Li B.S., Day Y.J., and Linden J.. 2001. A3 adenosine receptor activation triggers phosphorylation of protein kinase B and protects rat basophilic leukemia 2H3 mast cells from apoptosis. Mol. Pharmacol. 59:76–82. 10.1124/mol.59.1.76 [DOI] [PubMed] [Google Scholar]
- Gendaszewska-Darmach E., Maszewska M., Zakłos M., and Koziołkiewicz M.. 2003. Degradation of extracellular nucleotides and their analogs in HeLa and HUVEC cell cultures. Acta Biochim. Pol. 50:973–984. 10.18388/abp.2003_3627 [DOI] [PubMed] [Google Scholar]
- Goldberg G.S., Valiunas V., and Brink P.R.. 2004. Selective permeability of gap junction channels. Biochim. Biophys. Acta. 1662:96–101. 10.1016/j.bbamem.2003.11.022 [DOI] [PubMed] [Google Scholar]
- Goodenough D.A., and Paul D.L.. 2009. Gap junctions. Cold Spring Harb. Perspect. Biol. 1 a002576 10.1101/cshperspect.a002576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Györffy B., Lanczky A., Eklund A.C., Denkert C., Budczies J., Li Q., and Szallasi Z.. 2010. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res. Treat. 123:725–731. 10.1007/s10549-009-0674-9 [DOI] [PubMed] [Google Scholar]
- Han Mitchell K.L., and de Rooij Johan. 2016. Converging and Unique Mechanisms of Mechanotransduction at Adhesion Sites. Trends in Cell Biology. 10.1016/j.tcb.2016.03.005 [DOI] [PubMed] [Google Scholar]
- Hartsock A., and Nelson W.J.. 2008. Adherens and tight junctions: structure, function and connections to the actin cytoskeleton. Biochim. Biophys. Acta. 1778:660–669. 10.1016/j.bbamem.2007.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes S.E., Hollopeter G., Yang G., Kurpius D., Dailey M.E., Gan W.B., and Julius D.. 2006. The P2Y12 receptor regulates microglial activation by extracellular nucleotides. Nat. Neurosci. 9:1512–1519. 10.1038/nn1805 [DOI] [PubMed] [Google Scholar]
- Howe A.K. 2004. Regulation of actin-based cell migration by cAMP/PKA. Biochim. Biophys. Acta. 1692:159–174. 10.1016/j.bbamcr.2004.03.005 [DOI] [PubMed] [Google Scholar]
- Huang G.Y., Cooper E.S., Waldo K., Kirby M.L., Gilula N.B., and Lo C.W.. 1998. Gap junction-mediated cell-cell communication modulates mouse neural crest migration. J. Cell Biol. 143:1725–1734. 10.1083/jcb.143.6.1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilina O., Campanello L., Gritsenko P.G., Vullings M., Wang C., Bult P., Losert W., and Friedl P.. 2018. Intravital microscopy of collective invasion plasticity in breast cancer. Dis. Model. Mech. 11 dmm034330 10.1242/dmm.034330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasa S., Yanagawa T., Fan J., and Katoh R.. 2009. Expression of CXCR4 and its ligand SDF-1 in intestinal-type gastric cancer is associated with lymph node and liver metastasis. Anticancer Res. 29:4751–4758. [PubMed] [Google Scholar]
- Jacques-Silva M.C., Rodnight R., Lenz G., Liao Z., Kong Q., Tran M., Kang Y., Gonzalez F.A., Weisman G.A., and Neary J.T.. 2004. P2X7 receptors stimulate AKT phosphorylation in astrocytes. Br. J. Pharmacol. 141:1106–1117. 10.1038/sj.bjp.0705685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczmarek E., Erb L., Koziak K., Jarzyna R., Wink M.R., Guckelberger O., Blusztajn J.K., Trinkaus-Randall V., Weisman G.A., and Robson S.C.. 2005. Modulation of endothelial cell migration by extracellular nucleotides: involvement of focal adhesion kinase and phosphatidylinositol 3-kinase-mediated pathways. Thromb. Haemost. 93:735–742. 10.1160/TH04-09-0576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanczuga-Koda L., Sulkowski S., Lenczewski A., Koda M., Wincewicz A., Baltaziak M., and Sulkowska M.. 2006. Increased expression of connexins 26 and 43 in lymph node metastases of breast cancer. J. Clin. Pathol. 59:429–433. 10.1136/jcp.2005.029272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan J.M., El-Saghir J., Saliba J., Shaito A., Jalaleddine N., El-Hajjar L., Al-Ghadban S., Yehia L., Zibara K., and El-Sabban M.. 2019. Cx43 Expression Correlates with Breast Cancer Metastasis in MDA-MB-231 Cells In Vitro, In a Mouse Xenograft Model and in Human Breast Cancer Tissues. Cancers (Basel). 11:460 10.3390/cancers11040460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny L.C., Baker P.N., Kendall D.A., Randall M.D., and Dunn W.R.. 2002. The role of gap junctions in mediating endothelium-dependent responses to bradykinin in myometrial small arteries isolated from pregnant women. Br. J. Pharmacol. 136:1085–1088. 10.1038/sj.bjp.0704817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil A.A., and de Rooij J.. 2019. Cadherin mechanotransduction in leader-follower cell specification during collective migration. Exp. Cell Res. 376:86–91. 10.1016/j.yexcr.2019.01.006 [DOI] [PubMed] [Google Scholar]
- Khalil A.A., Ilina O., Gritsenko P.G., Bult P., Span P.N., and Friedl P.. 2017. Collective invasion in ductal and lobular breast cancer associates with distant metastasis. Clin. Exp. Metastasis. 34:421–429. 10.1007/s10585-017-9858-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoo H.E., Ho C.L., Chhatwal V.J., Chan S.T., Ngoi S.S., and Moochhala S.M.. 1996. Differential expression of adenosine A1 receptors in colorectal cancer and related mucosa. Cancer Lett. 106:17–21. 10.1016/0304-3835(96)04289-9 [DOI] [PubMed] [Google Scholar]
- Kotini M., Barriga E.H., Leslie J., Gentzel M., Rauschenberger V., Schambony A., and Mayor R.. 2018. Gap junction protein Connexin-43 is a direct transcriptional regulator of N-cadherin in vivo. Nat. Commun. 9:3846 10.1038/s41467-018-06368-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumai M., Nishii K., Nakamura K., Takeda N., Suzuki M., and Shibata Y.. 2000. Loss of connexin45 causes a cushion defect in early cardiogenesis. Development. 127:3501–3512. [DOI] [PubMed] [Google Scholar]
- Kumar V. 2013. Adenosine as an endogenous immunoregulator in cancer pathogenesis: where to go? Purinergic Signal. 9:145–165. 10.1007/s11302-012-9349-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzma-Kuzniarska M., Yapp C., Pearson-Jones T.W., Jones A.K., and Hulley P.A.. 2014. Functional assessment of gap junctions in monolayer and three-dimensional cultures of human tendon cells using fluorescence recovery after photobleaching. J. Biomed. Opt. 19:15001 10.1117/1.JBO.19.1.015001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z., Yin P., Reierstad S., O’Halloran M., Coon V.J., Pearson E.K., Mutlu G.M., and Bulun S.E.. 2010. Adenosine A1 receptor, a target and regulator of estrogen receptoralpha action, mediates the proliferative effects of estradiol in breast cancer. Oncogene. 29:1114–1122. 10.1038/onc.2009.409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marins M., Xavier A.L., Viana N.B., Fortes F.S., Fróes M.M., and Menezes J.R.. 2009. Gap junctions are involved in cell migration in the early postnatal subventricular zone. Dev. Neurobiol. 69:715–730. 10.1002/dneu.20737 [DOI] [PubMed] [Google Scholar]
- Mascaux C., Angelova M., Vasaturo A., Beane J., Hijazi K., Anthoine G., Buttard B., Rothe F., Willard-Gallo K., Haller A., et al. 2019. Immune evasion before tumour invasion in early lung squamous carcinogenesis. Nature. 571:570–575. 10.1038/s41586-019-1330-0 [DOI] [PubMed] [Google Scholar]
- Mayor R., and Carmona-Fontaine C.. 2010. Keeping in touch with contact inhibition of locomotion. Trends Cell Biol. 20:319–328. 10.1016/j.tcb.2010.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLachlan E., Shao Q., Wang H.L., Langlois S., and Laird D.W.. 2006. Connexins act as tumor suppressors in three-dimensional mammary cell organoids by regulating differentiation and angiogenesis. Cancer Res. 66:9886–9894. 10.1158/0008-5472.CAN-05-4302 [DOI] [PubMed] [Google Scholar]
- Mirza A., Basso A., Black S., Malkowski M., Kwee L., Pachter J.A., Lachowicz J.E., Wang Y., and Liu S.. 2005. RNA interference targeting of A1 receptor-overexpressing breast carcinoma cells leads to diminished rates of cell proliferation and induction of apoptosis. Cancer Biol. Ther. 4:1355–1360. 10.4161/cbt.4.12.2196 [DOI] [PubMed] [Google Scholar]
- Mroue R., Inman J., Mott J., Budunova I., and Bissell M.J.. 2015. Asymmetric expression of connexins between luminal epithelial- and myoepithelial- cells is essential for contractile function of the mammary gland. Dev. Biol. 399:15–26. 10.1016/j.ydbio.2014.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagelkerke A., van Kuijk S.J., Sweep F.C., Nagtegaal I.D., Hoogerbrugge N., Martens J.W., Timmermans M.A., van Laarhoven H.W., Bussink J., and Span P.N.. 2011. Constitutive expression of γ-H2AX has prognostic relevance in triple negative breast cancer. Radiother. Oncol. 101:39–45. 10.1016/j.radonc.2011.07.009 [DOI] [PubMed] [Google Scholar]
- Naus C.C., and Laird D.W.. 2010. Implications and challenges of connexin connections to cancer. Nat. Rev. Cancer. 10:435–441. 10.1038/nrc2841 [DOI] [PubMed] [Google Scholar]
- Nemir M., Bhattacharyya D., Li X., Singh K., Mukherjee A.B., and Mukherjee B.B.. 2000. Targeted inhibition of osteopontin expression in the mammary gland causes abnormal morphogenesis and lactation deficiency. J. Biol. Chem. 275:969–976. 10.1074/jbc.275.2.969 [DOI] [PubMed] [Google Scholar]
- Ogawa K., Pitchakarn P., Suzuki S., Chewonarin T., Tang M., Takahashi S., Naiki-Ito A., Sato S., Takahashi S., Asamoto M., et al. 2012. Silencing of connexin 43 suppresses invasion, migration and lung metastasis of rat hepatocellular carcinoma cells. Cancer Sci. 103:860–867. 10.1111/j.1349-7006.2012.02228.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Othman T., Yan H., and Rivkees S.A.. 2003. Oligodendrocytes express functional A1 adenosine receptors that stimulate cellular migration. Glia. 44:166–172. 10.1002/glia.10281 [DOI] [PubMed] [Google Scholar]
- Owens, Smith, Hackett, et al. 1974. Epithelial cell cultures from normal glandular tissue of mice. J Natl Cancer Inst. 10.1093/jnci/53.1.261. [DOI] [PubMed] [Google Scholar]
- Padmanaban V., Krol I., Suhail Y., Szczerba B.M., Aceto N., Bader J.S., and Ewald A.J.. 2019. E-cadherin is required for metastasis in multiple models of breast cancer. Nature. 573:439–444. 10.1038/s41586-019-1526-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.W., Chen S.W., Kim M., Brown K.M., D’Agati V.D., and Lee H.T.. 2010. Protection against acute kidney injury via A(1) adenosine receptor-mediated Akt activation reduces liver injury after liver ischemia and reperfusion in mice. J. Pharmacol. Exp. Ther. 333:736–747. 10.1124/jpet.110.166884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plutoni C., Bazellieres E., Le Borgne-Rochet M., Comunale F., Brugues A., Séveno M., Planchon D., Thuault S., Morin N., Bodin S., et al. 2016. P-cadherin promotes collective cell migration via a Cdc42-mediated increase in mechanical forces. J. Cell Biol. 212:199–217. 10.1083/jcb.201505105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebola N., Pinheiro P.C., Oliveira C.R., Malva J.O., and Cunha R.A.. 2003. Subcellular localization of adenosine A(1) receptors in nerve terminals and synapses of the rat hippocampus. Brain Res. 987:49–58. 10.1016/S0006-8993(03)03247-5 [DOI] [PubMed] [Google Scholar]
- Retamal M.A., Froger N., Palacios-Prado N., Ezan P., Sáez P.J., Sáez J.C., and Giaume C.. 2007. Cx43 hemichannels and gap junction channels in astrocytes are regulated oppositely by proinflammatory cytokines released from activated microglia. J. Neurosci. 27:13781–13792. 10.1523/JNEUROSCI.2042-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riding A., and Pullar C.E.. 2015. ATP release and P2Y Receptor Signalling are Essential for Keratinocyte Galvanotaxis. J. Cell. Physiol. 231:181–191. [DOI] [PubMed] [Google Scholar]
- Saunders M.M., Seraj M.J., Li Z., Zhou Z., Winter C.R., Welch D.R., and Donahue H.J.. 2001. Breast cancer metastatic potential correlates with a breakdown in homospecific and heterospecific gap junctional intercellular communication. Cancer Res. 61:1765–1767. [PubMed] [Google Scholar]
- Schalper K.A., Carvajal-Hausdorf D., and Oyarzo M.P.. 2014. Possible role of hemichannels in cancer. Front. Physiol. 5:237 10.3389/fphys.2014.00237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng W., Dong H., Lee D.Y., Lu W.Y., and Yang B.B.. 2007. Versican modulates gap junction intercellular communication. J. Cell. Physiol. 211:213–219. 10.1002/jcp.20921 [DOI] [PubMed] [Google Scholar]
- Sinyuk M., Mulkearns-Hubert E.E., Reizes O., and Lathia J.. 2018. Cancer Connectors: Connexins, Gap Junctions, and Communication. Front. Oncol. 8:646 10.3389/fonc.2018.00646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stagg J., Divisekera U., McLaughlin N., Sharkey J., Pommey S., Denoyer D., Dwyer K.M., and Smyth M.J.. 2010. Anti-CD73 antibody therapy inhibits breast tumor growth and metastasis. Proc. Natl. Acad. Sci. USA. 107:1547–1552. 10.1073/pnas.0908801107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoletov K., Strnadel J., Zardouzian E., Momiyama M., Park F.D., Kelber J.A., Pizzo D.P., Hoffman R., VandenBerg S.R., and Klemke R.L.. 2013. Role of connexins in metastatic breast cancer and melanoma brain colonization. J. Cell Sci. 126:904–913. 10.1242/jcs.112748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tambe D.T., Hardin C.C., Angelini T.E., Rajendran K., Park C.Y., Serra-Picamal X., Zhou E.H., Zaman M.H., Butler J.P., Weitz D.A., et al. 2011. Collective cell guidance by cooperative intercellular forces. Nat. Mater. 10:469–475. 10.1038/nmat3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theveneau E., and Mayor R.. 2012. Cadherins in collective cell migration of mesenchymal cells. Curr. Opin. Cell Biol. 24:677–684. 10.1016/j.ceb.2012.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Töpfer M., Burbiel C.E., Müller C.E., Knittel J., and Verspohl E.J.. 2008. Modulation of insulin release by adenosine A1 receptor agonists and antagonists in INS-1 cells: the possible contribution of 86Rb+ efflux and 45Ca2+ uptake. Cell Biochem. Funct. 26:833–843. 10.1002/cbf.1514 [DOI] [PubMed] [Google Scholar]
- Tsuji K., Yamauchi K., Yang M., Jiang P., Bouvet M., Endo H., Kanai Y., Yamashita K., Moossa A.R., and Hoffman R.M.. 2006. Dual-color imaging of nuclear-cytoplasmic dynamics, viability, and proliferation of cancer cells in the portal vein area. Cancer Res. 66:303–306. 10.1158/0008-5472.CAN-05-2958 [DOI] [PubMed] [Google Scholar]
- Umapathy S.N., Kaczmarek E., Fatteh N., Burns N., Lucas R., Stenmark K.R., Verin A.D., and Gerasimovskaya E.V.. 2013. Adenosine A1 receptors promote vasa vasorum endothelial cell barrier integrity via Gi and Akt-dependent actin cytoskeleton remodeling. PLoS One. 8 e59733 10.1371/journal.pone.0059733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaupel P., and Mayer A.. 2016. Hypoxia-Driven Adenosine Accumulation: A Crucial Microenvironmental Factor Promoting Tumor Progression. Adv. Exp. Med. Biol. 876:177–183. 10.1007/978-1-4939-3023-4_22 [DOI] [PubMed] [Google Scholar]
- Vaupel P., and Multhoff G.. 2016. Adenosine can thwart antitumor immune responses elicited by radiotherapy: Therapeutic strategies alleviating protumor ADO activities. Strahlenther. Onkol. 192:279–287. [DOI] [PubMed] [Google Scholar]
- Venhuizen J.H., Sommer S., Span P.N., Friedl P., and Zegers M.M.. 2019. Differential expression of p120-catenin 1 and 3 isoforms in epithelial tissues. Sci. Rep. 9:90 10.1038/s41598-018-36889-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Qin W., Xu X., Xiong Y., Zhang Y., Zhang H., and Sun B.. 2017. Endotoxin-induced autocrine ATP signaling inhibits neutrophil chemotaxis through enhancing myosin light chain phosphorylation. Proc. Natl. Acad. Sci. USA. 114:4483–4488. 10.1073/pnas.1616752114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J., Grenz A., Zhang Y., Dai Y., Kellems R.E., Blackburn M.R., Eltzschig H.K., and Xia Y.. 2011. A2B adenosine receptor contributes to penile erection via PI3K/AKT signaling cascade-mediated eNOS activation. FASEB J. 25:2823–2830. 10.1096/fj.11-181057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westcott J.M., Prechtl A.M., Maine E.A., Dang T.T., Esparza M.A., Sun H., Zhou Y., Xie Y., and Pearson G.W.. 2015. An epigenetically distinct breast cancer cell subpopulation promotes collective invasion. J. Clin. Invest. 125:1927–1943. 10.1172/JCI77767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf K., Te Lindert M., Krause M., Alexander S., Te Riet J., Willis A.L., Hoffman R.M., Figdor C.G., Weiss S.J., and Friedl P.. 2013. Physical limits of cell migration: control by ECM space and nuclear deformation and tuning by proteolysis and traction force. J. Cell Biol. 201:1069–1084. 10.1083/jcb.201210152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi N., Mizutani T., Kawabata K., and Haga H.. 2015. Leader cells regulate collective cell migration via Rac activation in the downstream signaling of integrin β1 and PI3K. Sci. Rep. 5:7656 10.1038/srep07656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z.C., Wyeth M.S., Baltan-Tekkok S., and Ransom B.R.. 2003. Functional hemichannels in astrocytes: a novel mechanism of glutamate release. J. Neurosci. 23:3588–3596. 10.1523/JNEUROSCI.23-09-03588.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.W., Nakayama K., Nakayama K., and Morita I.. 2003. A novel route for connexin 43 to inhibit cell proliferation: negative regulation of S-phase kinase-associated protein (Skp 2). Cancer Res. 63:1623–1630. [PubMed] [Google Scholar]
- Zhang J., Goliwas K.F., Wang W., Taufalele P.V., Bordeleau F., and Reinhart-King C.A.. 2019. Energetic regulation of coordinated leader-follower dynamics during collective invasion of breast cancer cells. Proc. Natl. Acad. Sci. USA. 116:7867–7872. 10.1073/pnas.1809964116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J.Z., Riquelme M.A., Gao X., Ellies L.G., Sun L.Z., and Jiang J.X.. 2015. Differential impact of adenosine nucleotides released by osteocytes on breast cancer growth and bone metastasis. Oncogene. 34:1831–1842. 10.1038/onc.2014.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists primer sequences.