Abstract
The legacy endocrine disrupting chemical and aryl hydrocarbon receptor agonist, 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD), is produced as a byproduct of industrial processes and causes adverse health effects ranging from skin irritation to cancer. TCDD endpoints are also observed in subsequent, unexposed generations; however, the mechanisms of these multi- and transgenerational effects are unknown. We hypothesized an epigenetic mechanism, specifically DNA methylation for the transgenerational, male-mediated reproductive effects of developmental TCDD exposure. Using whole genome bisulfite sequencing, we evaluated DNA methylation changes in three generations of zebrafish, the first of which was exposed to TCDD during sexual development at 50 ppt for 1 h at both 3- and 7-week post-fertilization. We discovered that TCDD induces multi- and transgenerational methylomic changes in testicular tissue from zebrafish with decreased reproductive capacity, but most significantly in the indirectly exposed F1 generation. In comparing differentially methylated genes to concurrent transcriptomic changes, we identified several genes and pathways through which transgenerational effects of low level TCDD exposure are likely inherited. These include significant differential methylation of genes involved in reproduction, endocrine function, xenobiotic metabolism, and epigenetic processing. Notably, a number of histone modification genes were both differentially methylated and expressed in all generations, and many differentially methylated genes overlapped between multiple generations. Collectively, our results suggest that DNA methylation is a promising mechanism to explain male-mediated transgenerational reproductive effects of TCDD exposure in zebrafish, and these effects are likely inherited through integration of multiple epigenetic pathways.
Keywords: zebrafish; transgenerational; epigenetics; methylation; 2,3,7,8-tetrachlorodibenzo-p-dioxin; reproduction
Introduction
As our understanding of the effects of environmental contaminants expands, we now know that certain chemicals cause adverse health effects for both the directly exposed generation and subsequent generations that were either indirectly exposed through the germline or not exposed at all. Transgenerational effects that are passed from exposed to unexposed generations are thought to be propagated epigenetically by mechanisms such as DNA methylation, histone modifications, and/or small non-coding RNA (1). The list of contaminants known to produce transgenerational effects will continue to grow but currently includes chemicals such as phthalates (2–4), lead (5, 6), polychlorinated biphenyls (7, 8), and dioxins, specifically 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) (9–11).
TCDD is a persistent environmental toxicant formed as a byproduct of industrial processes including incineration, metal production, and paper and pulp bleaching (12). TCDD bioaccumulates (13), thus the main method of modern-day human exposure is ingesting animal products such as meat, fish, and dairy (12). Toxic effects of TCDD are generally propagated through binding and activation of the aryl hydrocarbon receptor (AhR) leading to acute health effects including headaches, nausea, and skin irritation (14). Additional long-term health impacts can include reproductive effects with decreased fertility, defects in spermatogenesis, and endometriosis (10, 15); liver effects including fibrosis, disrupted transcriptomics, and tumorigenesis (16–18); and several types of cancer including breast, stomach, colon, ovary, and testis (19). As the most potent AhR agonist, the effects of TCDD are well characterized; therefore, it can be used as a model chemical to better understand the mechanisms of other AhR agonists and/or endocrine disrupting chemicals. Trans- or multigenerational effects following TCDD exposure have been reported in rodents (2, 9), fish (10, 11), and humans (20–22) at concentrations as low as 50 ppt. Exposure levels of TCDD in humans are generally very small, estimated at an average 0.5 pg/kg (parts per quadrillion) body weight per day in the USA (23), so it is essential to determine the effects of such low level exposures, especially on sensitive populations such as developing organisms. Transgenerational mechanisms are thought to be propagated through epigenetic effects, and several studies have examined various epigenetic effects of TCDD (24); however, no study has yet evaluated the specific mechanisms through which epigenetic effects translate to transgenerational outcomes. Understanding the mechanism(s) underlying these transgenerational outcomes is necessary to identify prevention and treatment options for both exposed and unexposed generations.
Zebrafish (Danio rerio) are an ideal model organism to study transgenerational effects of chemical exposure. Since their eggs are externally fertilized, fewer generations are needed to study transgenerational effects in zebrafish than in rodent models (25). Females release hundreds of eggs per spawning event, thus many fish can be exposed simultaneously during early development allowing for medium- to high-throughput screening of environmental toxicants. Importantly, zebrafish are an NIH-approved human model organism with a fully sequenced genome allowing for translational genomic, transcriptomic, and epigenetic analysis.
Previous work in our lab revealed significant differential methylation of specific genes in the testes of zebrafish exposed to TCDD during sexual differentiation and maturation (26). Testes were the target organ of this previous study because of earlier work in which we discovered transgenerational reproductive effects, primarily male-mediated, in TCDD-exposed fish (11, 25). Adult zebrafish in the F0 generation and two subsequent unexposed generations (F1 and F2) displayed TCDD-induced fertility defects, as defined by fewer eggs released and a decreased percentage of eggs fertilized, as well as defects in spermatogenesis. In addition, transcriptomic analysis revealed that differentially expressed genes were involved in similar fertility-related pathways across all three generations, though the specific genes involved in these pathways varied.
In this study, we aim to integrate findings from these previous experiments by investigating DNA methylation as a mechanism of transgenerational inheritance of reproductive effects following TCDD exposure. We chose to investigate DNA methylation because it is the most well-studied, thus relatively well understood, epigenetic mechanism. In addition, other studies have demonstrated the presence of DNA methylation in response to TCDD exposure (9, 27–29), but no one has yet examined the transgenerational whole genome DNA methylation response to TCDD exposure. Here, we examine DNA methylation status of the testes of F1 and F2 offspring of the TCDD-exposed F0 generation. Using whole genome bisulfite sequencing, we found changes in the DNA methylation profile of all three generations in the form of differentially methylated sites (DMSs), regions (DMRs), and genes (DMGs), each of which reveal different types of information. DMSs represent a change in methylation at a single base while DMRs represent a change in the methylation status of a region of DNA, defined in this study as 1 kb. DMRs are often associated with diseases including inflammatory bowel syndrome, post-traumatic stress disorder, and colon cancer, and are generally more replicable in multiple individuals with the same disease status than DMSs (30–33). Nevertheless, DMSs provide useful information as well and should not be overlooked. Several of the DNA methylation changes found in this study overlap with differentially expressed genes and among generations.
Methods
Animal Husbandry
Zebrafish (AB strain) were kept at 28°C in buffered reverse osmosis water (60 mg/l Instant Ocean Salts; Aquarium Systems, Mentor, OH, USA) with a standard light/dark cycle of 14/10 h and fed Aquatox Fish Diet flakes (Zeigler, PA, USA) twice per day, supplemented with brine shrimp. Fish were raised in beakers with daily water changes of 40–60% at a density of five fish per 400 ml beaker between 3- and 6-week post-fertilization (wpf), and five fish per 800 ml beaker between 6- and 9-wpf. Adult fish were raised on a recirculating system at a maximum density of five fish per liter until euthanization at 1-year post-fertilization. Fish were euthanized with tricaine methanesulfonate (1.67 mg/ml). Animal use protocols were approved by the Institutional Animal Care and Use Committees at Wayne State University and the University of Wisconsin–Madison, according to the National Institutes of Health Guide to the Care and Use of Laboratory Animals (Protocol No. M00489).
TCDD Exposure
Exposures were performed as previously described (34). TCDD (>99% purity; Chemsyn) was used as a 0.4 ng/μl stock solution in dimethyl sulfoxide (DMSO). Zebrafish were exposed at 3-wpf and again at 7-wpf to waterborne TCDD (50 pg/ml) or vehicle (0.1% DMSO) for 1 h in glass beakers with gentle rocking. The number of fish per volume of dosing solution was 1 fish/ml at 3-wpf and 1 fish/2 ml at 7-wpf. All results are derived from three independent TCDD exposure experiments performed in successive blocks. These exposed fish are referred to as F0 fish. At maturity, F0 males and females were spawned to yield the F1 generation, which were kept as separate blocks, each derived from F0 fish from a single experiment. These were in turn crossed at adulthood to produce the F2 generation fish, again descended from discrete exposure experiments. The fish tissues used for methylation analysis originated from the same blocks as those used in the previously published microarray and histology studies (34, 35).
DNA Isolation
Testes were extracted from F1 and F2 generation zebrafish descended from TCDD- and DMSO- (vehicle) treated zebrafish and flash frozen in liquid nitrogen. Samples were kept at −80°C until DNA isolation. DNA isolation was performed on 4 DMSO control and 4 TCDD-exposed samples using the BioRobot EZ1 workstation following the provided protocol from Qiagen (Hilden, Germany). Concentration and quality of DNA was measured using Qubit (Invitrogen, Carlsbad, CA, USA) and DropSense 96 (Trinean, Gentbrugge, Belgium), respectively (Supplementary Table S1). DNA was collected from different testicular tissue than used for RNA collection and microarray analysis.
Microarray
Microarray data were previously analyzed (11, 35). Microarray data for the F0 generation (GSE77335), and the F1 and F2 generations (GSE111446) are available on the NCBI GEO database. Microarray data were analyzed with one-way between subject ANOVA using the Transcriptome Analysis Console (TAC, Affymetrix). Genes uploaded into Ingenuity Pathway Analysis (IPA; Qiagen Bioinformatics; Redwood City, CA, USA) for pathway analysis were defined as significantly altered with a P-value ≤0.05 and absolute fold change ≥1.5. Gene expression was validated by qPCR of nine genes of interest. The validation and IPA analysis data was previously reported (11). Although all microarray data were previously uploaded, fold changes for a subset of genes not reported in the previous paper are discussed here for the first time.
Whole Genome Bisulfite Sequencing
Bisulfite conversion of methylated cytosines of DNA from F1 and F2 zebrafish testes was performed using the EZ DNA Methylation Kit (Zymo, Irvine, CA, USA). The resulting bisulfite-converted ssDNA was converted to Illumina libraries with the TruSeq DNA Methylation Kit (Illumina, San Diego, CA, USA). DNA libraries were sequenced with 100-bp paired-end reads on an Illumina HiSeq 2500 run in high output mode. Reads from six F1 and six F2 fish testes (three controls and two descendants of TCDD-treated fish for the F1 generation, and two controls and four descendants of TCDD-treated fish for the F2 generation after removing samples that did not pass our quality control cutoffs post-amplification) were aligned (bismark 0.18.1; bowtie2 2.2.3) to the zebrafish genome (dR10) and DMSs were determined (methylKit 1.6.3) between conditions (36–38). Methylated sites (percent methylation change >5%; P-value ≤0.05) were annotated to the nearest gene. Five percent methylation change was used as a cutoff based on previous (39–41). These cutoff values for significant methylation change are consistent with our previous publication examining changes in DNA methylation in the F0 generation (26) and other DNA methylation studies (40, 42). Raw data and processed files were uploaded to the NCBI GEO database (record GSE149919). Differentially expressed genes from the microarray data were overlapped with annotations for DMSs. Individual replicates from each condition were kept separate and not pooled.
Ingenuity Pathway Analysis
Genes associated with differential methylation (methylation change >5%, P-value ≤0.05) were converted to homologous human genes and uploaded into IPA software and analyzed using RefSeq ID as the identifier. T Pathway analysis was performed on the available F1 (1094 molecules for DMSs and 3057 molecules for DMRs) , and F2 (21 molecules for DMSs and 17 molecules for DMRs) for enriched disease and biologic functions.
Results
DMSs and Regions
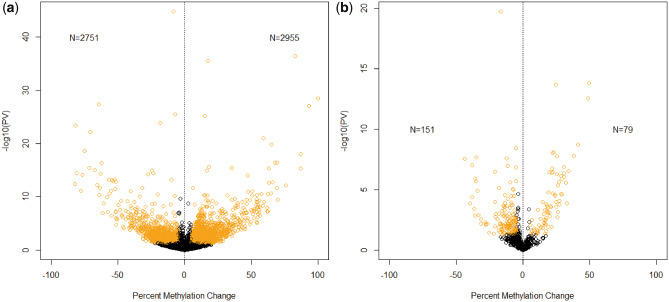
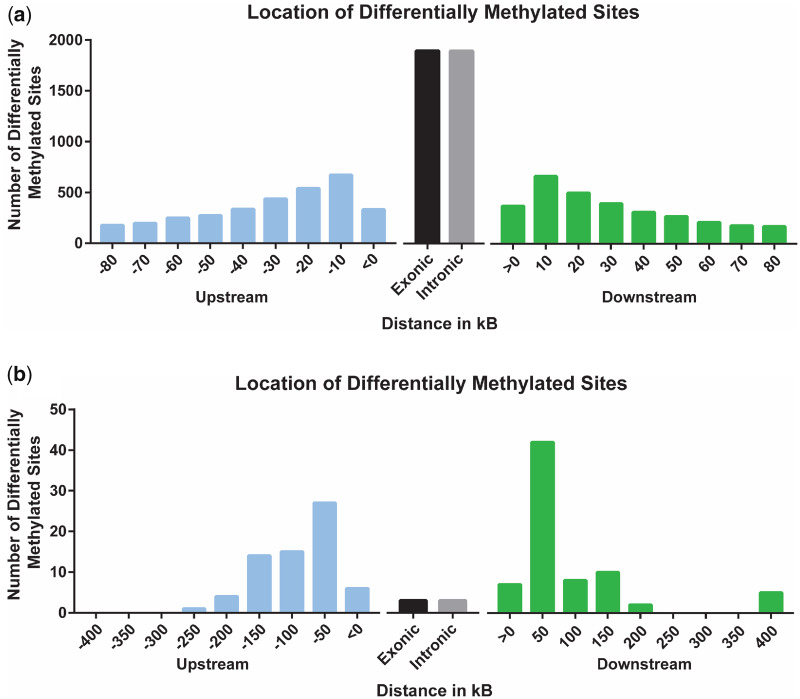
A total of 5706 sites (2955 hypermethylated, 2751 hypomethylated; P-value <0.05) were significantly differentially methylated in the testicular tissue from the F1 generation (Fig. 1A). For the F2 generation, 230 DMSs were identified with 79 hypermethylated and 151 hypomethylated (Fig. 1B). We previously reported 397 DMSs in the F0 generation with 113 sites hypermethylated and 284 hypomethylated (26). No changes in global methylation levels were discovered in either generation, similar to the F0 generation findings (26). In the F1 generation, most DMSs are within the gene body, relatively equally divided between introns and exons (Fig. 2A). This was not the case for both the F0 and F2 generations in which the majority of DMSs are in the intergenic region (Fig. 2B) (26).
Figure 1:
Volcano plot of significantly DMSs between F1 generation (A) and F2 generation (B) descendants of TCDD-treated and -untreated zebrafish testes depicting percent methylation change and P-value of methylated sites (circles). Significantly changed CpG sites (percent methylation change 5%; P-value ≤0.05) are in yellow
Figure 2:
Distribution of significant (percent methylation change ≥5%; P-value ≤0.05) F1 generation (A) and F2 generation (B) DMSs relative to nearest gene. x-Axis represents location of DMS relative to nearest gene and y-axis represents number of sites at each location
The F1 generation had 11 583 DMRs and the F2 generation had 130; 6730 of the DMRs in the F1 generation were hypomethylated and 4854 were hypermethylated. Like the F1 DMSs, an even distribution of intragenic DMRs was observed between exons and introns; however, there were overall more intergenic than intragenic F1 DMRs. In the F2 generation, 94 of the DMRs were hypomethylated and the remaining 36 were hypermethylated. Most of the DMRs in the F2 generation were located outside of the gene body.
Differentially Methylated Genes
In total, there were 132 DMGs in the F0, 5353 genes in the F1, and 53 in the F2 generation (Fig. 3). Fewer DMGs were found in the F0 and F2 generations compared to the F1. A large proportion of these genes (93% for F0 and 96% for F2), however, were also differentially methylated in at least one other generation, and 34 genes were differentially methylated in all three generations (Supplementary Table S3). Only 2.5% of DMGs in the F1 generation overlapped with a DMG in another generation. Of the 5353 DMGs in the F1 generation, 383 were associated with only DMSs, 3382 were associated with only DMRs, and the remaining 1242 were associated with both DMSs and DMRs. In the F2 generation, 16 were associated with just DMSs, 13 with just DMRs, and 21 with both DMSs and DMRs. Many of the DMGs in both the F1 and F2 generations are involved in pathways such as reproduction, endocrine system, xenobiotic metabolism, and epigenetics, as shown in Tables 1–3. Though there were many more DMGs in the F1 generation, more IPA pathways were related to male reproduction in the F2 generation than the F1 (Tables 4 and 5).
Figure 3:
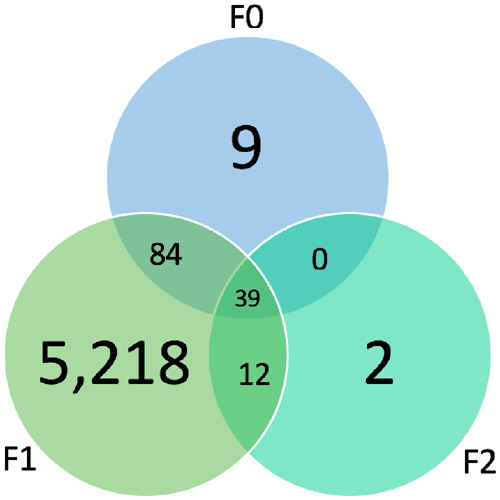
Venn diagram of significantly differential methylated genes and overlap in F0, F1, and F2 generations (P-value <0.05)
Table 1:
Differentially methylated F1 genes involved in reproduction
| Gene symbol | Gene name | DMRs | DMSs |
|---|---|---|---|
| Reproduction | |||
| General | |||
| adcyap1r1aa,b | Adenylate cyclase activating polypeptide 1a (pituitary) receptor type I | 4 ↑ | 1 ↓ |
| cftra,b | CF transmembrane conductance regulator | 2 ↑ | 1 ↑ |
| csf1raa | Colony stimulating factor 1 receptor, a | 2 ↑, 1 ↓ | 1 ↓ |
| ptenba | Phosphatase and tensin homolog b | 2 ↑ | 0 |
| gata4a | GATA-binding protein 4 | 6 ↓ | 0 |
| fshra,b | Follicle stimulating hormone receptor | 2 ↑, 1 ↓ | 1 ↑, 2 ↓ |
| inhbaaa | Inhibin subunit beta Aa | 3 ↓ | 0 |
| inhbaba,b | Inhibin subunit beta Ab | 4 ↑, 2 ↓ | 1 ↑ |
| inhbba | Inhibin subunit beta Bb | 2 ↑ | 0 |
| Sexual differentiation | |||
| zfpm2aa,b | Zinc finger protein, FOG family member 2a | 6 ↓ | 2 ↓ |
| zfpm2ba | Zinc finger protein, FOG family member 2b | 3 ↑ | 0 |
| sox9aa,b | SRY-box transcription factor 9a | 1 ↑, 2 ↓ | 1 ↓ |
| sox9ba | SRY-box transcription factor 9b | 1 ↑ | 0 |
| wnt4ba | Wingless-type MMTV integration site family, member 4b | 2 ↓ | 0 |
| nr0b1a,b | Nuclear receptor subfamily 0, group B, member 1 | 1 ↓ | 1 ↓ |
| Spermatogenesis | |||
| dhx36a,b | DEAH (Asp-Glu-Ala-His) box polypeptide 36 | 2 ↑, 6 ↓ | 4 ↑, 8 ↓ |
| m1apa,b | Meiosis 1-associated protein | 1 ↑, 2 ↓ | 1 ↑, 1 ↓ |
| fmr1a,b | FMRP translational regulator 1 | 10 ↑, 22 ↓ | 12 ↑, 15 ↓ |
| brip1a | BRCA1 interacting protein C-terminal helicase 1 | 6 ↓ | 0 |
| cadm1aa,b | Cell adhesion molecule 1a | 9 ↑, 1 ↓ | 1 ↑ |
| cadm1ba | Cell adhesion molecule 1b | 2 ↑ | 0 |
| dazla | Deleted in azoospermia-like | 2 ↓ | 0 |
| dzip1a | DAZ interacting zinc finger protein 1 | 2 ↑ | 0 |
| cx43a | Connexin 43 | 1 ↑, 1 ↓ | 0 |
| hook1a | Hook microtubule-tethering protein 1 | 2 ↑, 1 ↓ | 0 |
| hsf2a | Heat shock transcription factor 2 | 1 ↑, 1 ↓ | 0 |
| pdgfaba | Platelet-derived growth factor alpha polypeptide b | 3 ↑ | 0 |
| piwil2a | Piwi-like RNA-mediated gene silencing 2 | 2 ↓ | 0 |
| sbf1a | SET-binding factor 1 | 2 ↑ | 0 |
| dnaja1a | DnaJ heat shock protein family (Hsp40) member A1 | 1 ↑, 2 ↓ | 0 |
| elavl1a | ELAV like RNA-binding protein 1 | 3 ↓ | 0 |
| jam3ba | Junctional adhesion molecule 3b | 1 ↑, 3 ↓ | 0 |
| pacrga | PARK2 co-regulated | 4 ↓ | 0 |
| prlrba | Prolactin receptor b | 2 ↓ | 0 |
| gpx4ba,b | Glutathione peroxidase 4b | 1 ↓ | 1 ↓ |
| psme4ba,b | Proteasome activator subunit 4b | 1 ↑, 1 ↓ | 1 ↑ |
| cnr1a,b | Cannabinoid receptor 1 | 1 ↓ | 1 ↓ |
| rfx2a,b | Regulator factor X, 2 | 2 ↓ | 2 ↓ |
| brdta | Bromodomain, testis-specific | 4 ↑ | 0 |
| Sertoli cell junction | |||
| cldn11ba | Claudin 11b | 4 ↑ | 0 |
| cldn7ba | Claudin 7b | 2 ↓ | 0 |
| mtmr2a | Myotubularin-related protein 2 | 2 ↓ | 0 |
| myo7aaa | Myosin 7aa | 2 ↑ | 0 |
| tjp2aa | Tight junction protein 2a | 2 ↑, 2 ↓ | 0 |
| tjp2ba,b | Tight junction protein 2b | 1 ↓ | 1 ↓ |
| itgb1aa | Integrin, beta 1a | 4 ↓ | 0 |
| limk1aa | LIM domain kinase 1a | 2 ↑ | 0 |
| tgfb2a | Transforming growth factor, beta 2 | 2 ↓ | 0 |
| klf6aa | Kruppel-like factor 6a | 5 ↑ | 0 |
Numbers of DMSs and regions are shown with direction of methylation change.
Gene is associated with a DMR.
Gene is associated with a DMS.
Table 4:
F1 IPA pathways related to male reproduction
| F1 male reproduction pathways | ||
|---|---|---|
| Disease/function | P-value | Number of genes |
| Genital tumor | 7.75E−17 | 589 |
| Genital tract cancer | 2.46E−16 | 578 |
| Tumorigenesis of reproductive tract | 4.25E−12 | 420 |
| Development of genital tumor | 6.63E−11 | 407 |
| Male genital neoplasm | 7.6E−11 | 360 |
| Malignant neoplasm of male genital organ | 2.44E−10 | 355 |
| Development of reproductive system | 5.27E−07 | 89 |
| Morphology of reproductive system | 1.44E−05 | 77 |
| Fertility | 1.41E−06 | 49 |
| Proliferation of Sertoli cells | 9.55E−06 | 6 |
| Production of sperm | 1.70E−05 | 6 |
Table 5:
F2 IPA pathways related to male reproduction
| F2 male reproductive pathways | ||
|---|---|---|
| Disease/function | P-value | Number of genes |
| Morphology of genital organ | 0.00043 | 5 |
| Abnormal morphology of internal genitalia | 0.000883 | 4 |
| Morphology of testis | 0.000951 | 4 |
| Development of reproductive system | 0.0143 | 4 |
| Mass of testis | 0.0017 | 3 |
| Abnormal morphology of testis | 0.00371 | 3 |
| Development of genital organ | 0.0324 | 3 |
| Morphology of vas deferens | 0.00177 | 2 |
| Concentration of testosterone | 0.00332 | 2 |
| Disorder of epididymis | 0.0036 | 2 |
| Cell death of male germ cells | 0.00687 | 2 |
| Development of internal genitalia | 0.034 | 2 |
| Lack of epididymis | 0.00453 | 1 |
| Elongation of spermatids | 0.00544 | 1 |
| Lack of vas deferens | 0.00634 | 1 |
| Abnormal morphology of enlarged testis | 0.00994 | 1 |
| Proliferation of spermatogonia | 0.0153 | 1 |
| Morphogenesis of male genital organ | 0.0171 | 1 |
| Cell death of spermatocytes | 0.0278 | 1 |
| Abnormal morphology of Sertoli cells | 0.034 | 1 |
| Atrophy of testis | 0.0392 | 1 |
| Sterility | 0.0392 | 1 |
| Mass of seminal vesicle | 0.0225 | 1 |
Differentially Methylated and Differentially Expressed Genes
In the F1 generation, 159 genes were both differentially methylated and expressed (Table 6 and Supplementary Table S2) while in the F2 generation, there were five (pbx3b, si: dkey-266f7.5, snai3, tmem132e, and capn7; Table 7), four of which (all except tmem132e) were differentially methylated in all three generations (Supplementary Table S3). We previously found that the F0 generation had seven DMGs that were also differentially expressed (26), one of which (rab11bb) was differentially methylated in the F1 and F2 generations as well (Supplementary Table S3). Of the 159 genes in the F1 generation, 29 have DMSs or regions located in the exon, 21 in the intron, 90 in a location outside of the gene body, and 19 had methylated sites or regions in multiple locations. In addition, 34% of the F1 genes have the same direction of gene expression and DNA methylation change (e.g. up-regulated and hypermethylated or down-regulated and hypomethylated), 30% have opposite directions of gene expression and DNA methylation change, and the remaining 27% have multiple sites of differential methylation that are both hyper- and hypomethylated.
Table 6:
Genes both differentially methylated and differentially expressed in the F1 generation with roles in reproduction
| Gene | Methylation |
Gene expression |
Function | ||
|---|---|---|---|---|---|
| Percent change | P-value | Fold change | P-value | ||
| otud6ba,b | −24.2 | 9.38E−03 | 1.23 | 2.43E−02 | Deubiquitinating enzyme; variants in this gene can cause Intellectual Disability Syndrome Associated with Seizures and Dysmorphic Features including cryptorchidism Santiago-Sim et al. (51) |
| −12.0 | 1.37E−06 | ||||
| −6.64 | 2.90E−03 | ||||
| dirc2a,b | −11.4 | 1.96E−02 | 1.23 | 4.32E−02 | Membrane-bound electrogenic metabolite transporter; increased expression in prostate cancer tissue [Nesslinger et al. (85)] |
| −6.26 | 2.63E−03 | ||||
| plxna4a,b | −14.7 | 4.10E−05 | −1.2 | 2.58E−02 | Involved in axon guidance, axonal defasciculation, and morphogenesis of a branching structure; also found to be differentially methylated with BPA exposure in zebrafish [Olsvik et al. (86)], lead exposure in human embryonic stem cells [Senut et al. (87)], and folic acid exposure in mouse [Barua et al. (88)] |
| 62.0 | 1.06E−08 | ||||
| −9.25 | 6.45E−03 | ||||
| 22.8 | 8.52E−06 | ||||
| −10.4 | 4.71E−03 | ||||
| −10.4 | 4.71E−03 | ||||
| −6.19 | 2.86E−02 | ||||
| −5.69 | 4.62E−02 | ||||
| 58.4 | 2.68E−15 | ||||
| 58.4 | 2.68E−15 | ||||
| rhouba,b | 9.52 | 2.44E−03 | 1.31 | 4.38E−02 | Upstream regulator of the actin cytoskeleton through PAK1 and can initiate cell cycle reentry of quiescent cells; endometriosis (GWAS) |
| −10.3 | 6.46E−03 | ||||
| sall1aa,b | −22.9 | 3.40E−04 | 1.8 | 3.25E−02 | Positive regulation of fibroblast growth factor receptor signaling pathway, cryptorchidism, abnormal testis morphology (GWAS) |
| 11.7 | 3.44E−03 | ||||
| 11.7 | 3.44E−03 | ||||
| −9.76 | 2.21E−02 | ||||
| −9.76 | 2.21E−02 | ||||
| −6.40 | 1.24E−02 | ||||
| −6.32 | 7.73E−04 | ||||
| −5.81 | 1.85E−03 | ||||
| 6.61 | 3.69E−03 | ||||
| xpab | 7.90 | 2.20E−02 | −1.82 | 4.35E−02 | Nucleotide excision repair protein; decreased expression associated with defects in spermatogenesis Nakane et al. (53) and Singh et al. (54) |
| arrb1b | 13.6 | 1.71E−03 | 1.25 | 1.39E−02 | |
| 6.98 | 2.20E−02 | ||||
| acox1a,b | 17.9 | 2.40E−03 | 10.7 | 7.12E−03 | Fatty acid beta oxidation; necessary for fertility in mice Fan et al. (55) |
| 18.2 | 2.29E−02 | ||||
| 9.06 | 1.45E−03 | ||||
| −10.1 | 7.30E−05 | ||||
| −24.6 | 1.20E−03 | ||||
| 6.1 | 1.50E−02 | ||||
| −10.5 | 4.87E−06 | ||||
| −7.87 | 3.40E−03 | ||||
| ptenaa | 5.37 | 1.12E−02 | 1.17 | 3.19E−02 | Involved in gonadal development |
| 5.37 | 1.12E−02 | ||||
All DMSs and/or DMRs are listed.
Gene is associated with a DMR.
Gene is associated with a DMS.
Table 7:
All genes both differentially methylated and differentially expressed in the F2 generation
| Gene | Methylation |
Gene expression |
Function | ||
|---|---|---|---|---|---|
| Percent change | P-value | Fold change | P-value | ||
| si: dkey-266f7.5a | −11.3 | 2.09E−02 | 2.11 | 8.10E−03 | Unknown |
| snai3a,b | −7.69 | 4.76E−02 | −1.45 | 4.83E−02 | Roles in mesodermal formation during embryogenesis; involved in spermatogenesis |
| tmem132ea | −17.2 | 2.28E−02 | −1.32 | 4.78E−02 | Required for normal inner ear hair cell function and hearing |
| pbx3bb | −18.1 | 2.50E−06 | 1.45 | 4.37E−02 | Transcriptional activator |
| −14.9 | 2.04E−06 | ||||
| −14.6 | 4.92E−03 | ||||
| 6.76 | 2.71E−02 | ||||
| capn7b | −25.3 | 1.68E−05 | −1.34 | 1.99E−02 | Member of the calpain family of proteins |
| −25.3 | 1.68E−05 | ||||
All DMSs and/or DMRs are listed.
Gene is associated with a DMS.
Gene is associated with a DMR.
Several of the F1 genes that are both differentially methylated and differentially expressed have reproductive effects (Table 6), and one gene in the F2 generation (snai3) is involved in spermatogenesis (43).
Differentially Expressed Genes Involved in Epigenetic Regulation
Transcriptomic analysis identified 25 differentially expressed genes in the F1 generation and 19 genes in the F2 generation with roles in epigenetic regulation (Tables 8 and 9). Of these genes, 56% were down-regulated in the F1 generation and 74% down-regulated in the F2 generation. The majority of these epigenetic genes were involved in either methyl or acetyl group histone modifications.
Table 8:
Differentially expressed F1 and F2 genes involved in epigenetic processes
| Epigenetic role | F1 genes | F2 genes |
|---|---|---|
| Histone methyltransferase/demethylases and related genes | kdm6bb, nlk2, prdm6, smyd1a | kmt2d, kmt5aa, men1, setd5, smyd1b, smyd2b |
| Other methylation-related genes | loxl2a, mat2b, mthfd1l, mtrr, zbtb38 | mbd2, mbd5 |
| Histone acetyltransferase/deacetylases and related genes | atf3, hoxa10b, jade2, mark3a, sall1a, supt7l, vrk1 | brms1la, hdac10, jdp2b, sf3b3 |
| Other epigenetic genes | anp32e, gfi1b, ino80c, insm1b, l3mbtl2, smarca4a, smarcc1b, ube2a, usp49 | actr5, arid1ab, auts2a, phc2b, phf21ab, ruvbl1, wdr61 |
Table 9:
Fold changes and P-values of differentially expressed F1 and F2 genes involved in epigenetic processes
| Gene symbol | Gene name | Fold change | P-value |
|---|---|---|---|
| F1 genes | |||
| anp32e | Acidic (leucine-rich) nuclear phosphoprotein 32 family, member E | −1.21 | 4.31E−02 |
| atf3 | Activating transcription factor 3 | 1.80 | 2.45E−03 |
| gfi1b | Growth factor-independent 1B transcription repressor | 1.81 | 4.81E−02 |
| hoxa10b | Homeobox A10b | −1.33 | 2.34E−02 |
| ino80c | INO80 complex subunit C | −1.26 | 3.39E−02 |
| insm1b | Insulinoma-associated 1b | 1.22 | 5.57E−03 |
| jade2 | Jade family PHD finger 2 | 1.29 | 8.97E−03 |
| kdm6bb | Lysine (K)-specific demethylase 6B, b | 1.61 | 3.99E−02 |
| l3mbtl2 | L3MBTL histone methyl-lysine-binding protein 2 | −1.37 | 2.83E−02 |
| loxl2a | Lysyl oxidase-like 2a | 2.26 | 1.10E−02 |
| mark3a | MAP/microtubule affinity-regulating kinase 3a | −1.60 | 3.17E−02 |
| mat2b | Methionine adenosyltransferase II, beta | −1.53 | 3.16E−02 |
| mthfd1l | Methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like | −2.10 | 9.39E−03 |
| mtrr | 5-Methyltetrahydrofolate-homocysteine methyltransferase reductase | −1.42 | 1.93E−02 |
| nlk2 | Nemo-like kinase, type 2 | 1.24 | 3.17E−02 |
| prdm6 | PR domain containing 6 | 1.28 | 6.04E−03 |
| sall1a | Spalt-like transcription factor 1a | 1.80 | 3.25E−02 |
| smarca4a | SWI/SNF related, matrix associated, actin-dependent regulator of chromatin, subfamily a, member 4a | −1.39 | 2.99E−02 |
| smarcc1b | SWI/SNF related, matrix associated, actin-dependent regulator of chromatin, subfamily c, member 1b | −1.79 | 3.49E−02 |
| smyd1a | SET and MYND domain containing 1a | −3.60 | 6.41E−03 |
| supt7l | SPT7 like, STAGA complex subunit gamma | −1.39 | 3.75E−02 |
| ube2a | Ubiquitin-conjugating enzyme E2A (RAD6 homolog) | −1.47 | 2.78E−02 |
| usp49 | Ubiquitin-specific peptidase 49 | −1.49 | 4.90E−02 |
| vrk1 | VRK serine/threonine kinase 1 | 1.23 | 4.12E−02 |
| zbtb38 | Zinc finger and BTB domain containing 38 | 1.32 | 4.08E−02 |
| F2 genes | |||
| actr5 | Actin-related protein 5 | −1.45 | 2.29E−02 |
| arid1ab | AT rich interactive domain 1Ab (SWI-like) | −1.23 | 3.33E−02 |
| auts2a | Activator of transcription and developmental regulator AUTS2 a | −1.22 | 1.08E−02 |
| brms1la | BRMS1 like transcriptional repressor a | −2.57 | 2.57E−02 |
| hdac10 | Histone deacetylase 10 | 1.29 | 3.38E−02 |
| jdp2b | JUN dimerization protein 2b | −1.20 | 1.35E−02 |
| kmt2d | Lysine (K)-specific methyltransferase 2D | −1.30 | 2.02E−02 |
| kmt5aa | Lysine methyltransferase 5Aa | −1.21 | 2.90E−02 |
| mbd2 | Methyl-CpG-binding domain protein 2 | 1.26 | 2.65E−02 |
| mbd5 | Methyl-CpG-binding domain protein 5 | −1.33 | 2.79E−02 |
| men1 | Multiple endocrine neoplasia I | −1.20 | 2.87E−03 |
| phc2b | Polyhomeotic homolog 2b (Drosophila) | −1.37 | 1.81E−03 |
| phf21ab | PHD finger protein 21Ab | −1.27 | 3.41E−02 |
| ruvbl1 | RuvB-like AAA ATPase 1 | 1.29 | 8.25E−03 |
| setd5 | SET domain containing 5 | −1.32 | 2.09E−04 |
| sf3b3 | Splicing factor 3b, subunit 3 | 1.20 | 2.20E−02 |
| smyd1b | SET and MYND domain containing 1b | −1.53 | 1.39E−02 |
| smyd2b | SET and MYND domain containing 2b | −1.25 | 3.17E−02 |
| wdr61 | WD repeat domain 61 | 1.3 | 1.96E−03 |
Discussion
We discovered that TCDD exposure during sexual differentiation and maturation causes multi- and transgenerational DNA methylation changes in testicular tissue from zebrafish with decreased reproductive capacity. While all three generations had site-specific changes in DNA methylation, the F1 generation had substantially more DMSs, DMRs, and DMGs than either the F0 or F2 generations. In addition, of the DMSs in the F1 generation, most were located within the gene body, almost equally distributed between introns and exons, while in the F0 and F2 generations most DMSs were found in intergenic regions that consist of non-coding DNA. This could explain the small percentage of overlapping DMGs between the F1 and F0/F2 generations compared to the higher percentage of overlapping DMGs between the F0 and F2 generations and also suggests that TCDD has the largest effect on DNA methylation multigenerationally (i.e. indirectly on the F1 generation), as opposed to directly (F0) or transgenerationally (F2). In fact, the F1 generation had the greatest number of genes that were both differentially methylated and expressed. Although these findings do not correlate with phenotypes, as the most severe decreases in reproductive capacity were observed in the F0 generation (10, 35), or transcriptomic outcomes, which were most altered in the F2 generation (11), it is not unusual for patterns of epigenetic inheritance to be generationally distinct (9, 44, 45). These outcomes are likely due to the shifting balance between direct and heritable effects of exposure in each generation and align with a dynamic view of epigenetic inheritance, in which multiple epigenetic mechanisms interact with the transcriptome in a nonlinear fashion across generations (46).
Another potential explanation of the differences in methylation between generations could be different proportions of testicular cell types. We previously found a reduction in mature sperm cells (spermatozoa) and an increase in immature sperm cells (spermatogonia) in the F0 and F1 generations of TCDD-exposed zebrafish compared to control, but no change from control in the F2 generation (11, 35). DNA methylation status changes by cell type (47), so it is possible that these histological findings have some impact on the differential methylation status we find between experimental and control fish and between generations. However, this does not fully account for the increase in DMSs and DMRs in the F1 over the F0 generation (Fig. 3), because the histological outcomes between the F0 and F1 generations were similar (11, 35).
Several genes that were differentially methylated in all three generations are involved in reproductive processes, including mitochondrial regulator stoml2 (48), fmr1, implicated in the progression of spermatogenesis and RNA regulation (49, 50), and snai3, also involved in spermatogenesis as well as embryonic mesodermal formation (43). A subset of DMGs involved in reproduction also had differential gene expression, including: otud6 and sall1a, which are both associated with cryptorchidism (51, 52); xpa and arrb1, which are both nucleotide excision repair genes likely involved in spermatogenesis (53, 54); and acox1, involved in fatty acid beta oxidation and required for fertility in mice (55) in the F1 generation, as well as snai3 in the F2 generation (Tables 6 and 7). Interestingly, acox1 is differentially expressed in both the F1 and F2 generations, although only differentially methylated in the F1.
Unexpectedly, we found minimal overlap between differential methylation and expression in each generation, despite the association of many DMGs with pathways related to TCDD exposure endpoints including reproduction, endocrine system function, xenobiotic metabolism, and epigenetics (Tables 1–3). One possible explanation for the mismatch between transcriptomic and methylomic outcomes is that methylation patterns, in general, are established during early development and remain consistent throughout the majority of adulthood, whereas the transcriptome is dynamic across the lifespan. It is possible that altered methylation patterns established early in life are dysregulating development-specific transcriptomic networks, which are no longer evident in adulthood when the methylome and transcriptome were evaluated. Several DMGs in both the F1 and F2 generations with roles in reproduction (discussed in detail below) have shown distinct patterns of expression specifically during gonadal development (43, 56, 57). Future work will evaluate acute gene expression changes in zebrafish gonads during reproductive development in response to TCDD exposure.
The influence of other epigenetic mechanisms on gene expression may offer another potential explanation for the discrepancy between differentially methylated and expressed genes. In addition to DNA methylation, other mechanisms including histone modifications and non-coding RNAs, are often involved in regulation of gene expression (58–60). For instance, histone modifying proteins can interact with DNA methyltransferases (61) and conversely, the status of DNA methylation can affect the status of chromatin structure and histone modifications (62). In this study, the F1 generation had at least 28 DMGs involved in epigenetic pathways (Table 2), including the histone acetyltransferase mrgbp (63), which was differentially methylated in all three generations and was the only epigenetic-related DMG found in the F2 generation. These epigenetic-related DMGs have a known role in histone modifications with the exception of 3 DMGs in the F1 generation, specifically dnmt3aa, dnmt3ab, and dnmt3ba, which are de novo DNA methyltransferases (64). Similarly, we uncovered a large subset of differentially expressed genes involved in histone modification in both the F1 and F2 generations, including: histone demethylase, kdm6bb; histone methyltransferases, prdm6 and kmt2d; histone deacetylases, sall1a and hdac10 (65–69). Our data suggest that DNA methylation is likely one of multiple epigenetic mechanisms regulating transgenerational reproductive transgenerational changes in responses to TCDD exposure in zebrafish.
Table 2:
Differentially methylated F1 genes involved in the endocrine system, drug metabolism, histone modifications, or DNA methylation
| Gene symbol | Gene name | DMRs | DMSs |
|---|---|---|---|
| Endocrine | |||
| esr1a | Estrogen receptor 1 | 1 ↓ | 1 ↓ |
| esr2ab | Estrogen receptor2a | 2 ↓ | 0 |
| esr2bb | Estrogen receptor 2b | 5 ↓ | 0 |
| hsd17b10a | Hydroxysteroid (17-beta) dehydrogenase 10 | 1 ↑, 3 ↓ | 1 ↑, 1 ↓ |
| mdm2a | MDM2 proto-oncogene | 4 ↓ | 2 ↓ |
| nr5a2b | Nuclear receptor subfamily 5, group A, member 2 | 4 ↑, 5 ↓ | 0 |
| ncoa4a | Nuclear receptor coactivator 4 | 3 ↑, 1 ↓ | 1 ↑ |
| Xenobiotic metabolism | |||
| ahr1ab | Aryl hydrocarbon receptor 1a | 2 ↑ | 0 |
| ahr1ba | Aryl hydrocarbon receptor 1b | 1 ↑, 2 ↓ | 2 ↓ |
| ahr2a | Aryl hydrocarbon receptor 2 | 1 ↑, 6 ↓ | 3 ↑, 2 ↓ |
| Histone modification | |||
| setd7a | SET domain containing 7, histone lysine methyltransferase | 1 ↑ | 1 ↑ |
| jak2ab | Janus kinase 2a | 3 ↑ | 0 |
| jak2b | Janus kinase 2b | 0 | 1 ↑ |
| rbl1a | Retinoblastoma-like 1 | 1 ↑, 2 ↓ | 2 ↑ |
| rcor1a | REST corepressor 1 | 3 ↓ | 1 ↓ |
| taf6l | TAF6-like RNA polymerase II | 0 | 2 ↑ |
| mrgbpa | MRG/MORF4L-binding protein | 4 ↑, 6 ↓ | 2 ↑ |
| brpf1b | Bromodomain and PHD finger containing, 1 | 2 ↑, 3 ↓ | 0 |
| carm1b | Coactivator-associated arginine methyltransferase 1 | 2 ↓ | 0 |
| ehmt2b | Euchromatic histone-lysine N-methyltransferase 2 | 2 ↓ | 0 |
| hmga2b | High mobility group AT-hook 2 | 1 ↓ | 0 |
| kdm5bbb | Lysine (K)-specific demethylase 5Bb | 2 ↓ | 0 |
| ncor1b | Nuclear receptor corepressor 1 | 2 ↑ | 0 |
| ncor2b | Nuclear receptor corepressor 2 | 2 ↑, 2 ↓ | 0 |
| pak2bb | p21 protein (Cdc42/Rac)-activated kinase 2b | 2 ↓ | 0 |
| phc2aa | Polyhomeotic homolog 2a | 2 ↓ | 1 ↑, 1 ↓ |
| prkcbbb | Protein kinase C beta b | 2 ↓ | 0 |
| raraab | Retinoic acid receptor, alpha a | 2 ↑ | 0 |
| rarabb | Retinoic acid receptor, alpha b | 1 ↑, 3 ↓ | 0 |
| psme4ba | Proteasome activator subunit 4b | 1 ↑, 1 ↓ | 1 ↑ |
| cnr1a | Cannabinoid receptor 1 | 1 ↓ | 1 ↓ |
| rfx2a | Regulatory factor X, 2 | 2 ↓ | 2 ↓ |
| alkbh1 | AlkB homolog 1, histone H2A dioxygenase | 0 | 2 ↑, 2 ↓ |
| DNA methylation | |||
| dnmt3aab | DNA (cytosine-5-)-methyltransferase 3 alpha a | 1 ↑ | 0 |
| dnmt3abb | DNA (cytosine-5-)-methyltransferase 3 alpha b | 2 ↓ | 0 |
| dnmt3ba | DNA (cytosine-5-)-methyltransferase 3 beta a | 1 ↓ | 1 ↑ |
| ppm1dba | Protein phosphatase, Mg2+/Mn2+ dependent, 1Db | 2 ↑ | 1 ↑ |
Numbers of DMSs and regions are shown with direction of methylation change.
Gene is associated with a DMS.
Gene is associated with a DMR.
Several findings were not surprising based on previous TCDD exposure studies. For instance, transgenerational reproductive effects of TCDD exposure are mainly male-mediated (10, 11). Similarly, exposure to endocrine-disrupting compounds in rodents led to male germline-associated transgenerational outcomes, including reproductive effects and DNA methylation changes (70–74). We observed a large percentage of DMGs overlapping among all three generations, suggesting these sites were inherited transgenerationally and supporting a mechanism of paternal inheritance for DNA methylation. In fact, Potok et al. (75) showed that DNA methylation of the early embryo is nearly identical to that of the parental sperm. Furthermore, many of the DMGs in the F1 and F2 generations are involved in reproductive processes, including spermatogenesis, Sertoli cell junctions, and gonadal development (Tables 1 and 3). Collectively, our findings support the hypothesis that defects in spermatogenesis are a mechanism for decreased male fertility due to altered sperm development in the F0–F2 generation following TCDD exposure in the F0 generation (11).
Table 3:
Differentially methylated F2 genes involved in reproduction, the endocrine system, and epigenetic processes
| Gene symbol | Gene name | DMRs | DMSs |
|---|---|---|---|
| Reproduction | |||
| wnt9ba | Wingless-type MMTV integration site family, member 9B | 0 | 2 ↑ |
| elavl1a | ELAV like RNA-binding protein 1a | 0 | 1 ↓ |
| mycbpa | MYC-binding protein | 0 | 2 ↑ |
| rab11bba | RAB11B, member RAS oncogene family, b | 0 | 1 ↑ |
| fmr1a,b | FMRP translational regulator 1 | 1 ↑, 11 ↓ | 1 ↑, 2 ↓ |
| stoml2a,b | Stomatin (EPB72)-like 2 | 3 ↑, 7 ↓ | 2 ↑, 1 ↓ |
| nf2bb | Neurofibromin 2b | 2 ↓ | 0 |
| otx2b | Orthodenticle homeobox 2 | 1 ↓ | 0 |
| Endocrine | |||
| nr1h3a | Nuclear receptor subfamily 1, group H, member 3 | 0 | 2 ↓ |
| nr5a2a,b | Nuclear receptor subfamily 5, group A, member 2 | 1 ↓ | 2 ↑, 2 ↓ |
| Epigenetic | |||
| mrgbpa,b | MRG/MORF4L-binding protein | 6 ↓ | 2 ↑, 4 ↓ |
Numbers of DMSs and regions are shown with direction of methylation change.
Gene is associated with a DMS.
Gene is associated with a DMR.
Our previous research also showed F0–F2 zebrafish with female secondary sex characteristics and appearance, but testicular tissue upon dissection (10). In the F1 generation, genes related to sexual differentiation were also differentially methylated (Table 1). One of these genes, nr0b1, plays an important role in sexual differentiation in multiple organisms including humans (76, 77), rodents (78), and fish (79). Chen et al. (79) found that homozygous nr0b1 knockout zebrafish underwent a female to male sex reversal. Two isoforms of Sox9, a regulator of sexual differentiation and maturation in many species (80, 81), were also differentially methylated in the F1 generation. Zebrafish have two isoforms of sox9: sox9a, which is expressed in testes, and sox9b, which is expressed in ovaries. All juvenile zebrafish develop a premature ovary early in development that either matures to become a functional ovary or undergoes apoptosis and develops into testes. This process is thought to be mediated by the sox9 axis, specifically an increase in sox9a is found in gonads transitioning from ovaries to testes (57). Our findings suggest that dysregulation of many genes, not a single pathway, contributes to TCDD’s sexual differentiation effects and support the hypothesis that sox9a and/or sox9b cause dysregulation of gonad development followed by uncoupling from other sexual-development functions (11).
Finally, TCDD is a strong agonist for AhR, and most of the effects of TCDD exposure are mediated through this receptor (82). Therefore, it was unsurprising that three AhR genes, ahr1a, ahr1b, and ahr2, were differentially methylated in the F1 generation (Table 2). Of the three, Ahr2 had the most DMSs and regions. Though humans have only one AhR gene, in zebrafish, TCDD binds both ahr1b and ahr2 (not ahr1a), but only ahr2 is necessary for a toxic response to TCDD exposure, including skeletal and fin abnormalities and yolk sac and cardiac edema (83). In the F0 and F2 generation, none of the three AhR genes were differentially methylated; however, ahr2 expression was up-regulated in both the F0 and F2 generations, but not in the F1. The F0 up-regulation could be explained by a positive feedback mechanism in which AhR activation leads to increased AhR gene expression, which has been found in innate lymphoid cells (84). In addition, it is conceivable that differential methylation of the AhR genes in the F1 prevented up-regulation, and could be protective mechanism following indirect exposure.
In conclusion, developmental TCDD exposure led to changes in DNA methylation of zebrafish testes in the F0, F1, and F2 generations. Significantly more F1 genes were differentially methylated with a much smaller number of changes in the F0 and F2 generations. Few of these DMGs correlated with differential gene expression, which could potentially be explained by the variation of gene expression changes throughout the lifetime of an organism and/or the interplay of several epigenetic processes controlling gene expression (i.e. not just DNA methylation). In support of this, a number of histone modification genes were both differentially methylated and expressed in all generations, but particularly in the F1. Many DMGs overlapped between multiple generations, providing further evidence of male-mediated inheritance of DNA methylation patterns. In addition, several DMGs in all generations were involved in reproductive pathways, like those found in response to TCDD exposure (decreased fertility, defects in spermatogenesis, and skewed sex ratios). Collectively, our results suggest that DNA methylation is one mechanism of male-mediated transgenerational reproductive effects of TCDD exposure in zebrafish, but these effects are likely inherited through integration of multiple epigenetic pathways.
Supplementary data
Supplementary data are available at EnvEpig online.
Supplementary Material
Acknowledgments
We would like to acknowledge the Wayne State University Genome Sciences Core for providing WGBS services and the use of Ingenuity Pathway Analysis software, and the University of Wisconsin Biotechnology Gene Expression Center for providing Affymetrix GeneChip services. We are grateful to Kim Bauman, Emily Crofts, Bridget Baker, and the other members of the WATER Laboratory for the time and effort they have dedicated to fish husbandry, laboratory maintenance, and their general advice and support on this manuscript.
Data availability
Microarray data for the F0 generation (GSE77335), and the F1 and F2 generations (GSE111446) are available on the NCBI GEO database. Whole genome bisulfite sequencing (F0, F1, and F2 generations) raw data and processed files were uploaded to the NCBI GEO database (record GSE149919).
Funding
This work was supported by the National Institute of Environmental Health Sciences (R01 ES030722 to T.R.B., F31 ES030278 to D.N.M.), the National Center for Advancement of Translational Sciences (K01 OD01462 to T.R.B.), and the WSU Center for Urban Responses to Environmental Stressors (P30 ES020957 to D.N.M., T.R.B.). Additional funding was provided by the National Science Foundation (1735038 to C.A.), and by the NIH Center (P30 CA022453 to K.G.) to the Karmanos Cancer Institute at Wayne State University and the Perinatology Research Branch of the National Institutes of Child Health and Development at Wayne State University.
Conflict of interest statement. None declared.
References
- 1. Xavier MJ, Roman SD, Aitken RJ, Nixon B.. Transgenerational inheritance: how impacts to the epigenetic and genetic information of parents affect offspring health. Hum Reprod Update 2019;25:519–41. [DOI] [PubMed] [Google Scholar]
- 2. Zhou C, Gao L, Flaws JA.. Exposure to an environmentally relevant phthalate mixture causes transgenerational effects on female reproduction in mice. Endocrinology 2017;158:1739–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brehm E, Rattan S, Gao L, Flaws JA.. Prenatal exposure to di(2-ethylhexyl) phthalate causes long-term transgenerational effects on female reproduction in mice. Endocrinology 2018;159:795–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rattan S, Brehm E, Gao L, Flaws JA.. Di (2-ethylhexyl) phthalate exposure during prenatal development causes adverse transgenerational effects on female fertility in mice. Soc Toxicol 2018;163:420–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Meyer DN, Crofts EJ, Akemann C, Gurdziel K, Farr R, Baker BB, Weber D, Baker TR.. Developmental exposure to Pb2+ induces transgenerational changes to zebrafish brain transcriptome. Chemosphere 2020;244:125527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sobolewski M, Abston K, Conrad K, Marvin E, Harvey K, Susiarjo M, Cory-Slechta DA.. Lineage- and sex-dependent behavioral and biochemical transgenerational consequences of developmental exposure to lead, prenatal stress, and combined lead and prenatal stress in mice. Environ Health Perspect 2020;128:027001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Aluru N, Karchner SI, Krick KS, Zhu W, Liu J.. Role of DNA methylation in altered gene expression patterns in adult zebrafish (Danio rerio) exposed to 3,3′,4,4′,5-pentachlorobiphenyl (PCB 126). Environ Epigenet 2018;4:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mennigen JA, Thompson LM, Bell M, Tellez Santos M, Gore AC.. Transgenerational effects of polychlorinated biphenyls: 1. Development and physiology across 3 generations of rats. Environ Health 2018;17:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Manikkam M, Tracey R, Guerrero-Bosagna C, Skinner MK.. Dioxin (TCDD) induces epigenetic transgenerational inheritance of adult onset disease and sperm epimutations. PLoS One 2012;7:e46249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Baker TR, Peterson RE, Heideman W.. Using zebrafish as a model system for studying the transgenerational effects of dioxin. Toxicol Sci 2014;138:403–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Meyer DN, Baker BB, Baker TR.. Ancestral TCDD exposure induces multigenerational histologic and transcriptomic alterations in gonads of male zebrafish. Soc Toxicol 2018;16:603–612. 10.1093/toxsci/kfy115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.National Toxicology Program. Report on Carcinogens: 2,3,7,8-Tetrachlorodibenzo-p-dioxin, 2016. https://ntp.niehs.nih.gov/whatwestudy/assessments/cancer/roc/index.html? utm_source=direct&utm_medium=prod&utm_campaign=ntpgolinks&utm_term=roc#toc1 (20 February 2020, date last accessed).
- 13. Isensee AR. Bioaccumulation of 2,3,7,8-tetrachlorodibenzo-para-dioxin. Ecol Bull 1978;27:255–62. [Google Scholar]
- 14. Signorini S, Gerthoux PM, Dassi C, Cazzaniga M, Brambilla P, Vincoli N, Mocarelli P.. Environmental exposure to dioxin: the Seveso experience. Andrologia 2000;32:263–70. [DOI] [PubMed] [Google Scholar]
- 15. Bruner-tran KL, Gnecco J, Ding T, Glore DR, Pensabene V, Osteen KG.. Exposure to the environmental endocrine disruptor TCDD and human reproductive dysfunction: translating lessons from murine models. Reprod Toxicol 2017;68:59–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Harrill JA, Parks BB, Wauthier E, Rowlands JC, Reid LM, Thomas RS.. Lineage-dependent effects of aryl hydrocarbon receptor agonists contribute to liver tumorigenesis. Hepatology 2015;61:548–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lee J, Prokopec SD, Watson JD, Sun RX, Pohjanvirta R, Boutros PC.. Male and female mice show significant differences in hepatic transcriptomic response to 2,3,7,8-tetrachlorodibenzo-p-dioxin. BMC Genomics 2015;16:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lamb CL, Cholico GN, Pu X, Hagler GD, Cornell KA, Mitchell KA.. 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) increases necroinflammation and hepatic stellate cell activation but does not exacerbate experimental liver fibrosis in mice. Toxicol Appl Pharmacol 2016;311:42–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xu J, Ye Y, Huang F, Chen H, Wu H, Huang J, Hu J, Xia D, Wu Y.. Association between dioxin and cancer incidence and mortality: a meta-analysis. Sci Rep 2016;6:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mocarelli P, Gerthoux PM, Ferrari E, Patterson DG, Kieszak SM, Brambilla P, Vincoli N, Signorini S, Tramacere P, Carreri V, Sampson EJ, Turner WE, Needham LL.. Paternal concentrations of dioxin and sex ratio of offspring. Lancet 2000;355:1858–63. [DOI] [PubMed] [Google Scholar]
- 21. Ryan JJ, Amirova Z, Carrier G.. Sex ratios of children of Russian pesticide producers exposed to dioxin. Child Health Articles 2002;110:699–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. ‘t Mannetje A, Eng A, Walls C, Dryson E, Kogevinas M, Brooks C, McLean D, Cheng S, Smith AH, Pearce N.. Sex ratio of the offspring of New Zealand phenoxy herbicide producers exposed to 2,3,7,8-tetrachlorodibenzo-p-dioxin. Occup Environ Med 2017;74:24–9. [DOI] [PubMed] [Google Scholar]
- 23. Lorber M, Huwe J, Rawn D.. An update to estimates of intake of dioxin-like compounds for the general population of the United States. Organohalogen Compd 2010;72:316–9. [Google Scholar]
- 24. Patrizi B, Siciliani de Cumis M.. TCDD toxicity mediated by epigenetic mechanisms. IJMS 2018;19:4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Baker TR, King-Heiden TC, Peterson RE, Heideman W.. Dioxin induction of transgenerational inheritance of disease in zebrafish. Mol Cell Endocrinol 2014;398:36–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Akemann C, Meyer DN, Gurdziel K, Baker TR.. Developmental dioxin exposure alters the methylome of adult male zebrafish gonads. Front Genet 2019;9:719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shen ES, Whitlock JP.. The potential role of DNA methylation in the response to 2,3,7,8-tetrachlorodibenzo-p-dioxin. J Biol Chem 1989;264:17754–8. [PubMed] [Google Scholar]
- 28. Aluru N, Kuo E, Helfrich LW, Karchner SI, Linney EA, Pais JE, Franks DG.. Developmental exposure to 2,3,7,8-tetrachlorodibenzo-p-dioxin alters DNA methyltransferase (DNMT) expression in zebrafish (Danio rerio). Toxicol Appl Pharmacol 2015;284:142–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ma J, Chen X, Liu Y, Xie Q, Sun Y, Chen J, Leng L, Yan H, Zhao B, Tang N.. Ancestral TCDD exposure promotes epigenetic transgenerational inheritance of imprinted gene Igf2: methylation status and DNMTs. Toxicol Appl Pharmacol 2015;289:193–202. [DOI] [PubMed] [Google Scholar]
- 30. Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, Ji H, Potash JB, Sabunciyan S, Feinberg AP.. Genome-wide methylation analysis of human colon cancer reveals similar hypo-and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet 2009;41:178–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ventham NT, Kennedy NA, Adams AT, Kalla R, Heath S, O’Leary KR, Drummond H, Wilson DC, Gut IG, Nimmo ER, Satsangi J.. Integrative epigenome-wide analysis demonstrates that DNA methylation may mediate genetic risk in inflammatory bowel disease. Nat Commun 2016;7:1–14.; doi:10.1038/ncomms13507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rutten BPF, Vermetten E, Vinkers CH, Ursini G, Daskalakis NP, Pishva E, De Nijs L, Houtepen LC, Eijssen L, Jaffe AE, Kenis G, Viechtbauer W, Van Den Hove D, Schraut KG, Lesch KP, Kleinman JE, Hyde TM, Weinberger DR, Schalkwyk L, Lunnon K, Mill J, Cohen H, Yehuda R, Baker DG, Maihofer AX, Nievergelt CM, Geuze E, Boks MPM.. Longitudinal analyses of the DNA methylome in deployed military servicemen identify susceptibility loci for post-traumatic stress disorder. Mol Psychiatry 2018;23:1145–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gomez L, Odom GJ, Young JI, Martin ER, Liu L, Chen X, Griswold AJ, Gao Z, Zhang L, Wang L.. coMethDMR: accurate identification of co-methylated and differentially methylated regions in epigenome-wide association studies with continuous phenotypes. Nucleic Acids Res 2019;47:e98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Baker TR, Peterson RE, Heideman W.. Early dioxin exposure causes toxic effects in adult zebrafish. Toxicol Sci 2013;135:241–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Baker BB, Yee JS, Meyer DN, Yang D, Baker TR.. Histological and transcriptomic changes in male zebrafish testes due to early life exposure to low level 2,3,7,8-tetrachlorodibenzo-p-dioxin. Zebrafish 2016;13:413–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Langmead B, Trapnell C, Pop M, Salzberg SL.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 2009;10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Krueger F, Andrews SR.. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 2011;27:1571–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Akalin A, Kormaksson M, Li S, Garrett-Bakelman FE, Figueroa ME, Melnick A, Mason CE.. MethylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol 2012;13:R87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yang AS, Doshi KD, Choi S-W, Mason JB, Mannari RK, Gharybian V, Luna R, Rashid A, Shen L, Estecio MRH, Kantarjian HM, Garcia-Manero G, Issa J-PJ.. DNA methylation changes after 5-Aza-2′-deoxycytidine therapy in patients with leukemia. Cancer Res 2006;66:5495–503. [DOI] [PubMed] [Google Scholar]
- 40. Byun H-M, Motta V, Panni T, Bertazzi P, Apostoli P, Hou L, Baccarelli AA.. Evolutionary age of repetitive element subfamilies and sensitivity of DNA methylation to airborne pollutants. Part Fibre Toxicol 2013;10:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dayeh T, Volkov P, Salö S, Hall E, Nilsson E, Olsson AH, Kirkpatrick CL, Wollheim CB, Eliasson L, Rönn T, Bacos K, Ling C.. Genome-wide DNA methylation analysis of human pancreatic islets from type 2 diabetic and non-diabetic donors identifies candidate genes that influence insulin secretion. PLoS Genet 2014;10:e1004160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jiang L, Zhang J, Wang J-J, Wang L, Zhang L, Li G, Yang X, Ma X, Sun X, Cai J, Zhang J, Huang X, Yu M, Wang X, Liu F, Wu C-I, He C, Zhang B, Ci W, Liu J.. Sperm, but not oocyte, DNA methylome is inherited by zebrafish early embryos. Cell 2013;153:773–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Micati DJ, Hime GR, McLaughlin EA, Abud HE, Loveland KL.. Differential expression profiles of conserved Snail transcription factors in the mouse testis. Andrology 2018;6:362–73. [DOI] [PubMed] [Google Scholar]
- 44. Ben Maamar M, Sadler-Riggleman I, Beck D, McBirney M, Nilsson E, Klukovich R, Xie Y, Tang C, Yan W, Skinner MK.. Alterations in sperm DNA methylation, non-coding RNA expression, and histone retention mediate vinclozolin-induced epigenetic transgenerational inheritance of disease. Environ Epigenet 2018;4:dvy010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Siklenka K, Erkek S, Godmann M, Lambrot R, McGraw S, Lafleur C, Cohen T, Xia J, Suderman M, Hallett M, Trasler J, Peters AHFM, Kimmins S.. Disruption of histone methylation in developing sperm impairs offspring health transgenerationally. Science 2015;350:aab2006. [DOI] [PubMed] [Google Scholar]
- 46. Burggren WW. Dynamics of epigenetic phenomena: intergenerational and intragenerational phenotype ‘washout’. J Exp Biol 2015;218:80–7. [DOI] [PubMed] [Google Scholar]
- 47. Zhang B, Zhou Y, Lin N, Lowdon RF, Hong C, Nagarajan RP, Cheng JB, Li D, Stevens M, Lee HJ, Xing X, Zhou J, Sundaram V, Elliott G, Gu J, Shi T, Gascard P, Sigaroudinia M, Tlsty TD, Kadlecek T, Weiss A, O'Geen H, Farnham PJ, Maire CL, Ligon KL, Madden PAF, Tam A, Moore R, Hirst M, Marra MA, Zhang B, Costello JF, Wang T.. Functional DNA methylation differences between tissues, cell types, and across individuals discovered using the M&M algorithm. Genome Res 2013;23:1522–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Christie DA, Lemke CD, Elias IM, Chau LA, Kirchhof MG, Li B, Ball EH, Dunn SD, Hatch GM, Madrenas J.. Stomatin-like protein 2 binds cardiolipin and regulates mitochondrial biogenesis and function. Mol Cell Biol 2011;31:3845–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tian H, Cao Y-X, Zhang X-S, Liao W-P, Yi Y-H, Lian J, Liu L, Huang H-L, Liu W-J, Yin M-M, Liang M, Shan G, Sun F.. The targeting and functions of miRNA-383 are mediated by FMRP during spermatogenesis. Cell Death Dis 2013;4:e617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Alpatov R, Lesch BJ, Nakamoto-Kinoshita M, Blanco A, Chen S, Stützer A, Armache KJ, Simon MD, Xu C, Ali M, Murn J, Prisic S, Kutateladze TG, Vakoc CR, Min J, Kingston RE, Fischle W, Warren ST, Page DC, Shi Y.. A chromatin-dependent role of the fragile X mental retardation protein FMRP in the DNA damage response. Cell 2014;157:869–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Santiago-Sim T, Burrage LC, Ebstein F, Tokita MJ, Miller M, Bi W, Braxton AA, Rosenfeld JA, Shahrour M, Lehmann A, Cogné B, Küry S, Besnard T, Isidor B, Bézieau S, Hazart I, Nagakura H, Immken LL, Littlejohn RO, Roeder E, Kara B, Hardies K, Weckhuysen S, May P, Lemke JR, Elpeleg O, Abu-Libdeh B, James KN, Silhavy JL, Issa MY, Zaki MS, Gleeson JG, Seavitt JR, Dickinson ME, Ljungberg MC, Wells S, Johnson SJ, Teboul L, Eng CM, Yang Y, Kloetzel P-M, Heaney JD, Walkiewicz MA, Afawi Z, Balling R, Barisic N, Baulac S, Craiu D, De Jonghe P, Guerrero-Lopez R, Guerrini R, Helbig I, Hjalgrim H, Jähn J, Klein KM, Leguern E, Lerche H, Marini C, Muhle H, Rosenow F, Serratosa J, Sterbová K, Suls A, Moller RS, Striano P, Weber Y, Zara F.. Biallelic variants in OTUD6B cause an intellectual disability syndrome associated with seizures and dysmorphic features. Am J Hum Genet 2017;100:676–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Botzenhart EM, Green A, Ilyina H, König R, Lowry RB, Lo IFM, Shohat M, Burke L, McGaughran J, Chafai R, Pierquin G, Michaelis RC, Whiteford ML, Simola KOJ, Rösler B, Kohlhase J.. SALL1 mutation analysis in Townes-Brocks syndrome: twelve novel mutations and expansion of the phenotype. Hum Mutat 2005;26:282. [DOI] [PubMed] [Google Scholar]
- 53. Nakane H, Hirota S, Brooks PJ, Nakabeppu Y, Nakatsu Y, Nishimune Y, Iino A, Tanaka K.. Impaired spermatogenesis and elevated spontaneous tumorigenesis in xeroderma pigmentosum group A gene (Xpa)-deficient mice. DNA Repair (Amst) 2008;7:1938–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Singh V, Jaiswal D, Singh K, Trivedi S, Agrawal NK, Gupta G, Rajender S, Singh K.. Azoospermic infertility is associated with altered expression of DNA repair genes. DNA Repair (Amst) 2019;75:39–47. [DOI] [PubMed] [Google Scholar]
- 55. Fan C-Y, Pan J, Chu R, Lee D, Kluckman KD, Usuda N, Singh I, Yeldandi AV, Rao MS, Maeda N, Reddy JK.. Targeted disruption of the peroxisomal fatty acyl-CoA oxidase gene: generation of a mouse model of pseudoneonatal adrenoleukodystrophy. Ann N Y Acad Sci 1996;804:530–541. [DOI] [PubMed] [Google Scholar]
- 56. Furusawa M, Ohnishi T, Taira T, Iguchi-Ariga SMM, Ariga H.. AMY-1, a c-Myc-binding protein, is localized in the mitochondria of sperm by association with S-AKAP84, an anchor protein of cAMP-dependent protein kinase. J Biol Chem 2001;276:36647–51. [DOI] [PubMed] [Google Scholar]
- 57. Sun D, Zhang Y, Wang C, Hua X, Zhang XA, Yan J.. Sox9-related signaling controls zebrafish juvenile ovary-testis transformation. Cell Death Dis 2013;4:e930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Day JJ, Sweatt JD.. Epigenetic mechanisms in cognition. Neuron 2011;70:813–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Dunn J, McCuaig R, Tu WJ, Hardy K, Rao S.. Multi-layered epigenetic mechanisms contribute to transcriptional memory in T lymphocytes. BMC Immunol 2015;16:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kelsey G, Stegle O, Reik W.. Single-cell epigenomics: recording the past and predicting the future. Science 2017;358:69–75. [DOI] [PubMed] [Google Scholar]
- 61. D’Alessio AC, Szyf M.. Epigenetic tête-à-tête: the bilateral relationship between chromatin modifications and DNA methylation. Biochem Cell Biol 2006;84:463–76. [DOI] [PubMed] [Google Scholar]
- 62. Cervoni N, Szyf M.. Demethylase activity is directed by histone acetylation. J Biol Chem 2001;276:40778–87. [DOI] [PubMed] [Google Scholar]
- 63. Krogan NJ, Baetz K, Keogh M-C, Datta N, Sawa C, Kwok TCY, Thompson NJ, Davey MG, Pootoolal J, Hughes TR, Emili A, Buratowski S, Hieter P, Greenblatt JF.. Regulation of chromosome stability by the histone H2A variant Htz1, the Swr1 chromatin remodeling complex, and the histone acetyltransferase NuA4. Proc Natl Acad Sci USA 2004;101:13513–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bheemanaik S, Reddy YVR, Rao DN.. Structure, function and mechanism of exocyclic DNA methyltransferases. Biochem J 2006;399:177–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Johnstone AL, Andrade NS, Barbier E, Khomtchouk BB, Rienas CA, Lowe K, Van Booven BJ, Domi E, Esanov R, Vilca S, Tapocik JD, Rodriguez K, Maryanski D, Keogh MC, Meinhardt MW, Sommer WH, Heilig M, Zeier Z, Wahlestedt C.. Dysregulation of the histone demethylase KDM6B in alcohol dependence is associated with epigenetic regulation of inflammatory signaling pathways. Addict Biol 2019;e12816. 10.1111/adb.12816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Li N, Subrahmanyan L, Smith E, Yu X, Zaidi S, Choi M, Mane S, Nelson-Williams C, Behjati M, Kazemi M, Hashemi M, Fathzadeh M, Narayanan A, Tian L, Montazeri F, Mani M, Begleiter ML, Coon BG, Lynch HT, Olson EN, Zhao H, Ruland J, Lifton RP, Mani A.. Mutations in the histone modifier PRDM6 are associated with isolated nonsyndromic patent ductus arteriosis. Am J Hum Genet 2016;99:1000–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lv S, Ji L, Chen B, Liu S, Lei C, Liu X, Qi X, Wang Y, Lai-Han Leung E, Wang H, Zhang L, Yu X, Liu Z, Wei Q, Lu L.. Histone methyltransferase KMT2D sustains prostate carcinogenesis and metastasis via epigenetically activating LIFR and KLF4. Oncogene 2018;37:1354–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lauberth SM, Rauchman M.. A conserved 12-amino acid motif in Sall1 recruits the nucleosome remodeling and deacetylase corepressor complex. J Biol Chem 2006;281:23922–31. [DOI] [PubMed] [Google Scholar]
- 69. Kao HY, Lee CH, Komarov A, Han CC, Evans RM.. Isolation and characterization of mammalian HDAC10, a novel histone deacetylase. J Biol Chem 2002;277:187–93. [DOI] [PubMed] [Google Scholar]
- 70. Anway MD, Rekow SS, Skinner MK.. Transgenerational epigenetic programming of the embryonic testis transcriptome. Genomics 2008;91:30–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Stouder C, Paoloni-Giacobino A.. Specific transgenerational imprinting effects of the endocrine disruptor methoxychlor on male gametes. Reproduction 2011;141:207–16. [DOI] [PubMed] [Google Scholar]
- 72. Kubsad D, Nilsson EE, King SE, Sadler-riggleman I, Beck D, Skinner MK.. Assessment of glyphosate induced epigenetic transgenerational inheritance of pathologies and sperm epimutations. Sci Rep 2019;9:6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Pandey A, Rudraiah M.. Analysis of endocrine disruption effect of Roundup® in adrenal gland of male rats. Toxicol Rep 2015;2:1075–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Smith CM, Vera MKM, Bhandari RK.. Developmental and epigenetic effects of Roundup and glyphosate exposure on Japanese medaka (Oryzias latipes). Aquat Toxicol 2019;210:215–26. [DOI] [PubMed] [Google Scholar]
- 75. Potok ME, Nix DA, Parnell TJ, Cairns BR.. Reprogramming the maternal zebrafish genome after fertilization to match the paternal methylation pattern. Cell 2013;153:759–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Goodfellow PN, Camerino G.. Dax-1, an “antitestis” gene. Cell Mol Life Sci 1999;55:857–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Bardoni B, Zanaria E, Guioli S, Floridia G, Worley KC, Tonini G, Ferrante E, Chiumello G, McCabe ERB, Fraccaro M, Zuffardi O, Camerino G.. A dosage sensitive locus at chromosome Xp21 is involved in male to female sex reversal. Nat Genet 1994;7:497–501. [DOI] [PubMed] [Google Scholar]
- 78. Park SY, Lee E-J, Emge D, Jahn CL, Jameson JL.. A phenotypic spectrum of sexual development in Dax1 (Nr0b1)-deficient mice: consequence of the C57BL/6J strain on sex determination. Biol Reprod 2008;79:1038–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Chen S, Zhang H, Wang F, Zhang W, Peng G.. Nr0b1 (DAX1) mutation in zebrafish causes female-to-male sex reversal through abnormal gonadal proliferation and differentiation. Mol Cell Endocrinol 2016;433:105–16. [DOI] [PubMed] [Google Scholar]
- 80. Foster JW, Dominguez-Steglich MA, Guioli S, Kwok C, Weller PA, Stevanović M, Weissenbach J, Mansour S, Young ID, Goodfellow PN, Brook JD, Schafer AJ.. Campomelic dysplasia and autosomal sex reversal caused by mutations in an SRY-related gene. Nature 1994;372:525–30. [DOI] [PubMed] [Google Scholar]
- 81. Wagner T, Wirth J, Meyer J, Zabel B, Held M, Zimmer J, Pasantes J, Bricarelli FD, Keutel J, Hustert E, Wolf U, Tommerup N, Schempp W, Scherer G.. Autosomal sex reversal and campomelic dysplasia are caused by mutations in and around the SRY-related gene SOX9. Cell 1994;79:1111–20. [DOI] [PubMed] [Google Scholar]
- 82. Mimura J, Yamashita K, Nakamura K, Morita M, Takagi TN, Nakao K, Ema M, Sogawa K, Yasuda M, Katsuki M, Fujii-Kuriyama Y.. Loss of teratogenic response to 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) in mice lacking the Ah (dioxin) receptor. Genes Cells 2003;2:645–54. [DOI] [PubMed] [Google Scholar]
- 83. Garcia GR, Bugel SM, Truong L, Spagnoli S, Tanguay RL.. AHR2 required for normal behavioral responses and proper development of the skeletal and reproductive systems in zebrafish. PLoS One 2018;13:e0193484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Li S, Bostick JW, Ye J, Qiu J, Zhang B, Urban JF, Avram D, Zhou L.. Aryl hydrocarbon receptor signaling cell intrinsically inhibits intestinal group 2 innate lymphoid cell function. Immunity 2018;49:915–28.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Nesslinger N J, Ng A, Tsang K-Y, Ferrara T, Schlom J, Gulley J L, Nelson B H. A viral vaccine encoding prostate-specific antigen induces antigen spreading to a common set of self-proteins in prostate cancer patients. Clinical Cancer Research 2010;16:4046–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Olsvik PA. Whatmore P. Penglase SJ, Skjærven KH. D’Auriac MA. Ellingsen S.. Associations between behavioral effects of bisphenol A and DNA methylation in zebrafish embryos. Front Genet 2019;10: 1 10.3389/fgene.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Senut M-C, Sen A, Cingolani P, Shaik A, Land S J, Ruden D M. Lead exposure disrupts global DNA methylation in human embryonic stem cells and alters their neuronal differentiation. Toxicol Sci 2014;139:142–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Barua S, Kuizon S, Brown WT, Junaid MA. DNA methylation profiling at single-baseresolution reveals gestational folic acid supplementation influences theepigenome of mouse offspring cerebellum. Front Neurosci 2016;10: [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Microarray data for the F0 generation (GSE77335), and the F1 and F2 generations (GSE111446) are available on the NCBI GEO database. Whole genome bisulfite sequencing (F0, F1, and F2 generations) raw data and processed files were uploaded to the NCBI GEO database (record GSE149919).