Abstract
Aging is the progressive decline or loss of function at the cellular, tissue, and organismal levels that ultimately leads to death. A number of external and internal factors, including diet, exercise, metabolic dysfunction, genome instability, and epigenetic imbalance, affect the lifespan of an organism. These aging factors regulate transcriptome changes related to the aging process through chromatin remodeling. Many epigenetic regulators, such as histone modification, histone variants, and ATP-dependent chromatin remodeling factors, play roles in chromatin reorganization. The key to understanding the role of gene regulatory networks in aging lies in characterizing the epigenetic regulators responsible for reorganizing and potentiating particular chromatin structures. This review covers epigenetic studies on aging, discusses the impact of epigenetic modifications on gene expression, and provides future directions in this area.
Keywords: aging, histone level, histone modification, histone variant, chromatin remodeling, epigenetics
1. Introduction
An individual organism undergoes a series of developmental stages, including birth, growth, maturity, aging, and death. Aging is the gradual and continuous decline or loss of function at the cellular, tissue, and organismal levels with the passage of time. Aging is considered to begin in early adulthood, but is regarded as an integral part of life since other stages also affect the aging process [1]. Aging itself is regarded as a risk factor for numerous chronic diseases, such as neurodegenerative diseases, cardiovascular diseases, metabolic diseases, and osteoporosis. Due to an increase in average life expectancy and a decline in fertility, the global population is currently aging rapidly. Understanding the mechanisms of aging, longevity, and susceptibility to age-related disease is necessary to slow down the rate of aging and increase healthy lifespan [2]. Large numbers of studies have been performed to unravel why and how aging occurs; these studies have suggested that aging is regulated by complex cellular and molecular mechanisms at different developmental stages [1,3]. Numerous factors affecting the aging process and longevity have been reported. Genetic differences and genetic damage are widely known to play an important role in aging, but non-genetic factors such as stress, dietary manipulation, sexual stimuli, circadian rhythms, and exercise have also been implicated [4,5,6,7,8,9]. In particular, social insects such as ants and honey bees have emerged as models to show the influence of environmental stimuli on lifespan. Individuals with a common genome present caste-specific differences in morphology, behavior, and longevity that are determined by environmental inputs early in the developmental process. Recently, environmental cues have been shown to stimulate epigenetic changes, such as DNA methylation and histone modifications, which play critical roles in regulating caste-based differences in longevity and behavior [10,11].
Chromatin is a nucleosome polymer composed of DNA and histone protein. It is a flexible and dynamic structure that exists either as heterochromatin or euchromatin. The former is compact and transcriptionally inactive, while the latter is a decondensed type of chromatin and is transcriptionally active [12,13]. Since the accessibility of transcription regulatory machinery to chromatin is affected by its state, chromatin remodeling is required to expose or hide regions of DNA for transcription [14]. The chromatin structure is remodeled via several mechanisms, including histone modification, histone tail cleavage, exchange of histone variants, and ATP-dependent chromatin remodeling [15,16]. The state of chromatin can be altered by environmental stimuli, which subsequently affects the expression of genes associated with aging and longevity [4,8]. In this review, we discuss recent research focusing on the roles of histone protein levels, the exchange of histone variants, histone modifications, and chromatin remodeling in aging. We also review recent genome-wide profiling of chromatin accessibility patterns and histone modification in young and aged models. Links between chromatin architecture and aging may provide strategies to counter or reverse the aging process or age-related diseases.
2. Histone Changes
2.1. Histone Levels and Nucleosome Occupancy
Global loss of canonical histones is regarded as a common feature of aging in a range of organisms from yeast to humans (Table 1). In yeast, a decline in canonical histone occurs during replicative aging as a consequence of reduced histone protein synthesis, and ectopic expression of histones H3 and H4 strongly promotes replicative lifespan extension [17]. Micrococcal nuclease digestion followed by sequencing revealed ≈50% nucleosome loss across the whole genome and a fuzzy redistribution of the remaining nucleosomes during replicative aging, increasing the transcription of many genes and resulting in genomic instability [18]. In senescent human fibroblasts, a reduction in histone biosynthesis or lysosomal-mediated processing caused the depletion of histones [19,20]. One report notes that the transcription of histones decreases in quiescent muscle stem cells during chronological aging [21]. Interestingly, Yu et al. demonstrated that reducing the dosage to 85% of wild-type H3 and H4 by deleting the minor H3-H4-coding gene pair HHT1-HHF1 profoundly extends replicative lifespan, whereas reducing the dosage to 15% by deleting HHT2-HHF2 shortens the lifespan of yeast. The authors also showed that moderate reductions in H3-H4 activate a distinct stress response and block TOR signaling, thus promoting longevity [22]. However, it remains unclear whether moderate chromatin architectural defects promote longevity in other organisms. Recently, Chen et al. reanalyzed data on H3 chromatin immunoprecipitation and high-throughput sequencing (ChIP-seq) datasets derived from mouse tissues at three different stages of the mouse lifespan. They found a number of age-related changes in H3 occupancy, including increased and decreased H3 occupancy, but no dramatic changes in H3 expression levels [23]. However, the replacement of canonical histone H3 with histone H3 variants, such as H3.3, may occur during aging in mice since the H3 antibody used in these studies recognizes both canonical and variant H3 proteins (see the section of histone variants). Taken together, the results of these studies show that it is likely that histone loss or nucleosome loss occurs in a cell type-specific or context-specific manner during aging.
Table 1.
Modulation of histones and chromatin remodelers during aging.
| Name | Description | Epigenetic Changes Linked to Aging | Effect on Longevity/Health Span | Model | Reference |
|---|---|---|---|---|---|
| Histones | |||||
| H3, H4, H2A, H2B | Canonical histones | Canonical histone loss | + | Yeast | [17,18] |
| H3, H4 | Canonical histones | Canonical histone loss | + | Human senescent cell | [19,20] |
| H2A.Z and H3.3 | Histone variants | Accumulation of H2A.Z and H3.3 | − | Human senescent cell | [24] |
| MacroH2A | Histone variant | Increased protein level of macroH2A | − | Senescent cell, tissues of mice and primates | [25] |
| H3.3 | Histone variant | Accumulation of H3.3 | − | Mouse neuron and glial cells | [26] |
| H3.3 | Histone variant | Accumulation of H3.3 | − | Various mouse somatic tissues | [27] |
| H2A.Z | Histone variant | Accumulation of H2A.Z | − | Mouse hippocampus | [28] |
| H2A.J | Histone variant | Accumulation of H2A.J | − | Senescent human fibroblasts, mouse hair follicle stem and interfollicular epidermal cells, human epidermis | [29] |
| Chromatin Remodelers | |||||
| SWI/SNF | ATP-dependent chromatin remodeler | SWI/SNF is required for DAF-16/FOXO-mediated dauer formation, stress resistance and longevity | + | Caenorhabditis elegans | [30] |
| Isw2 and Itch | ATP-dependent chromatin remodeler | − | Yeast | [31] | |
| LET-418 | Mi2 homolog (NurD complex) | − | C. elegans | [32] | |
+: positive, −: negative.
2.2. Histone Variants
Histone variants are isoforms of histones, which have different primary sequences and specialized functional properties compared to canonical histones. Histone variants are expressed throughout the cell cycle and incorporated into chromatin in a replication-independent manner, although canonical histones are synthesized and preferentially incorporated into nucleosomes in a replication-dependent manner. The exchange of canonical histones with histone variants modulates the properties of the nucleosome, thereby affecting DNA replication and transcription [33]. A series of studies implicates histone variants in aging (Table 1). It was found that the level of macro H2A (mH2A) increased in human fibroblasts during replicative senescence. An upregulated level of mH2A was also observed in the liver and lung tissue of aged mice and in the muscle tissue of aged baboons [25]. In human fibroblasts, upregulation of H2A.Z and H3.3 as well as downregulation of H2A.1 and H3.1 was found during aging [24].
Using mass spectrometry, age-related H3.3 accumulation was observed in mouse neurons and postmortem human brains [26]. H3.3 accounts for only a small portion of the total H3 pool in embryonic neuronal chromatin, but in aged mice, H3.3 becomes dominant in the total H3 pool of neuronal chromatin, which in turn regulates cell type-specific gene expression. In mouse somatic tissues such as the liver, kidney, brain, and heart, histone variant H3.3 replaces canonical H3.1 and H3.2 with age, resulting in prominent changes in global levels of H3 methyl modifications [27]. In addition, H2A.Z has been reported to accumulate in the mouse hippocampus with age [28]. Another histone variant, H2A.J, accumulates in senescent human fibroblasts, upregulating the expression of inflammatory genes. It was also observed that the accumulation of H2A.J occurs in mouse hair follicle stem cells, mouse interfollicular epidermal cells, and human epidermal cells with age [29]. Thus, the replacement of canonical histones with histone variants is likely to be a common feature in aging. Additionally, histone chaperones affect nucleosome composition through deposition of histones on DNA or eviction of histones from nucleosomes. Although HIRA, a histone chaperone, is reported to be required for the dynamics of the chromatin landscape, including H3.3 deposition and H4K16ac retention, in senescent cells [34], very little is known about the functional importance of histone chaperones for histone variants during aging. Thus, it may be interesting to explore the role of histone chaperones in aging.
2.3. ATP-Dependent Chromatin Remodeling
The chromatin landscape can be altered via nucleosome assembly, nucleosome repositioning, and exchange of histone variants by ATP-dependent chromatin remodelers, which play an important role in chromatin accessibility [35]. Chromatin remodelers are divided into four major subfamilies: imitation switch (ISWI), chromodomain helicase DNA-binding (CHD), switch/sucrose non-fermentable (SWI/SNF), and INO80 [36]. Several chromatin remodeling complexes have been reported to be associated with age-associated chromatin remodeling and regulation of lifespan (Table 1). It was previously reported that deletion of ISW2 causes nucleosome positioning changes at 1187 gene loci across the genome in yeast [37]. Interestingly, Dang and coauthors revealed that deletion of ISW2, which encodes the catalytic component of the ISW2 complex, promotes longevity through induction of genotoxic stress response, which partially mimics calorie restriction effects. These results suggest that ISW2 negatively modulates longevity through stress response pathways in yeast [31]. In Caenorhabditis elegans, the distinct chromatin remodeler SWI/SNF is linked to expansion of lifespan. SWI/SNF as a cofactor colocalizes with DAF-16/FOXO at target promoters to activate transcription. Inactivation of the SWI/SNF by RNAi reduced lifespan and stress resistance mediated by DAF-16/FOXO [30]. Another study showed that Mi2 (CHD-3/CHD-4) plays a negative regulator role in longevity. For example, mutations in LET-418, an Mi2 homolog in C. elegans, extend lifespan and enhance stress resistance in a DAF-16/FOXO-dependent manner [32]. Thus, ATP-dependent chromatin remodeling factors function as a positive and negative regulator of longevity by controlling stress resistance.
2.4. Histone Methylation
Histone methylation contributes to either transcriptional activation or transcriptional repression, depending on the histone, the residue modified, and the level of methylation. Generally, methylation of H3K4, H3K36, and H3K79 is linked to active transcription, whereas that of H3K9, H3K27, and H4K20 is associated with repressive transcription [38]. This modification can be added by histone methyltransferases and removed by histone demethylases [39]. A large number of studies have reported that histone methylations modified by histone methyltransferases or histone demethylases change during aging (Table 2).
Table 2.
Histone methylation-modulating proteins and aging.
| Name | Description | Change in Histone Modifications Linked to Aging | Effect on Longevity/Health Span | Model | Reference |
|---|---|---|---|---|---|
| Methyltransferases | |||||
| SET-2 | Histone lysine methyltransferase (ASH-2 complex subunit) | H3K4me3 ↑ | − | C. elegans | [38] |
| ASH-2 | ASH-2 complex subunit | ||||
| WDR-5 | ASH-2 complex subunit | ||||
| SUV39H1 | Histone methyltransferase | H3K9me3 ↓ | + | Human and mouse HSCs | [40] |
| EZH2 | Methyltransferase | H3K27me3 ↓ | + | Human senescent cell | [41] |
| E(Z) | Methyltransferase (PRC2 complex subunit) | H3K27me3 ↑ | − | Drosophila | [42,43] |
| ESC | PRC2 complex subunit | ||||
| EZH1, CBXs | Polycomb complex | H3K27me3 ↑ | − | Killifish brain | [44] |
| Set-2 | Histone lysine methyltransferase | H3K36me3 ↓ | + | Yeast | [45] |
| MET-1 | Histone lysine methyltransferase | H3K36me3 ↓ | + | C. elegans | [46] |
| SET-18 | H3K36 di-methyltransferase | H3K36me2 ↑ | − | C. elegans | [47] |
| Demethylases | |||||
| T08D10.2 | H3K4me/me2 demethylase, LSD-1 ortholog | − | C. elegans | [48] | |
| RBR-2 | H3K4me3 demethylase | H3K4me3 ↑ | + | C. elegans | [49] |
| Lid | Demethylase, RBR-2 ortholog | H3K4me3 ↑ | + | Drosophila | [50] |
| KDM4A | Histone demethylase | H3K9me3 ↑ | + | Male Drosophila | [51] |
| UTX-1 | H3K27me3 histone demethylase | H3K27me3 ↓ | − | C. elegans | [52,53] |
| RPH1 | H3K36me2/3 demethylase | H3K36me3 ↓ | − | Yeast | [45] |
| Others | |||||
| HP1 beta | Methylated histone reader, heterochromatin marker | − | Senescent cell, tissues of mice and primates | [25] | |
| Lamin B1 | A component of nuclear lamina | H3K27me3 ↓ | + | Human senescent cell | [54] |
↑: increase, ↓: decrease, +: positive, −: negative.
Heterochromatin remains condensed and transcriptionally silent. Heterochromatin is known to be associated with H3K9me3, H4K20me3, and H3K64me3 in addition to DNA methylation [55,56,57]. The heterochromatin loss model of aging proposes that heterochromatin domains established early in embryogenesis are decreased during the aging process, contributing to the global loss of heterochromatin-induced gene silencing and leading to aberrant gene expression patterns [58]. Global heterochromatin loss with aging has been reported in humans and several model organisms [59,60,61,62,63]. Models of premature aging diseases, such as Hutchinson–Gilford progeria syndrome (HGPS) and Werner syndrome, exhibit heterochromatin loss with a decrease of H3K9me3, HP1, or SUV39H, a H3K9me3 histone methyltransferase [62,64]. For example, HGPS is caused by germline mutations in the LMNA gene, leading to the expression of progerin, a truncated mutant form of lamin A. Cultured cells from HGPS patients show abnormal nuclear morphology, including enlarged nuclei and heterochromatin loss with decreased H3K9me3 and reduced HP1 expression [62,65,66]. Although heterochromatin loss is observed during senescence, localized heterochromatin referred to as senescence-associated heterochromatin foci (SAHFs) has also been reported in senescent cells [67,68,69,70]. Recently, this contradictory role of heterochromatin in cellular aging and SAHF formation was further investigated through Hi-C analysis [71]. It was observed that HGPS cells show a loss of local internal structure in heterochromatin but do not exhibit the spatial clustering of heterochromatin. In contrast, in senescent cells, a loss of internal structure in heterochromatic regions is accompanied by spatial clustering of the heterochromatic regions. These findings suggest that the senescence-specific spatial clustering of heterochromatic regions may be required for SAHF formation. Taken together, these studies indicate that higher order chromatin structures are altered during aging.
2.4.1. H3K4me3
H3K4me3, an active histone mark, is most abundant around transcription start sites [38]. There are several reports of the use of targeted RNAi to show that modifying H3K4me3 enzymes can regulate the lifespan of flies and worms (Table 2). Greer et al. reported that deficiencies in ASH-2, WDR-5, and SET-2, components of the ASH-2 Trithorax complex, cause a global decrease in H3K4me3 levels and extend lifespan in fertile worms. Meanwhile, knockdown of RBR-2, an H3K4me3 demethylase, increases H3K4me3 level and shortens lifespan, counteracting the effect of the ASH-2 methyltransferase complex. Overexpressing RBR-2 in the germ line extends lifespan [49]. Consistent with this, a deficiency in Lid, the ortholog of RBR-2, decreases lifespan and increases H3K4me3 levels in male flies, although it has no effect on the lifespan of female flies [50]. However, a deficiency in Trithorax, a component of another type of H3K4me3 methyltransferase complex in Drosophila, has no significant effect on the lifespan of male flies [43]. It seems, therefore, that regulators of H3K4me3 have different effects on lifespan depending on the H3K4me3 complex or organism.
2.4.2. H3K9me3
H3K9me3, a repressive mark, is known to be associated with heterochromatin. There are some reports that implicate H3K9me3-modifying enzymes in aging. In Drosophila, deletion of Kdm4A, an H3K9me3 demethylase, was observed to affect fly lifespan. The disruption of Kdm4A shortens the male lifespan, suggesting that KDM4A plays a role in longevity [51]. Another report demonstrated that expression of the histone methyltransferase SUV39H1 in both human and mouse hematopoietic stem cells (HSCs) decreased concomitant with age, leading to a global reduction in H3K9me3 and perturbed heterochromatin function [40]. The authors further found that expression of the microRNA miR-125b, a known regulator of HSC function, increased with age in human HSCs. Since miR-125b directly targets SUV39H1, overexpression of miR-125b caused changes in heterochromatin structure and loss of B cell potential, indicating that an age-dependent decrease in SUV39H1 is closely associated with the destruction of heterochromatin structure and B lymphocyte formation in HSCs. These studies suggest that global changes in H3K9me3 are accompanied by heterochromatin misregulation, affecting lifespan depending on organism.
2.4.3. H3K27me3
The trimethylated histone H3 at lysine 27 (H3K27me3) is likely to be modified dynamically during aging. H3K27me3 denotes transcriptional silencing, which is produced by polycomb repressive complex-2 (PRC2) and functionally maintained by PRC1 [72]. PRC2 contains the PcG proteins E(Z), SU(Z)12, ESC (or ESCL), and PCL. E(Z), the catalytic subunit of PRC2, along with the other PRC2 subunits, catalyzes H3K27 trimethylation [73]. In Drosophila, mutation of E(Z), the Drosophila homolog of EZH1/2, and its H3-targeting partner protein ESC, increased longevity and reduced H3K27me3 levels [43]. A further study demonstrated that a reduction in H3K27me3 due to PRC deficiency promotes glycolysis and healthy lifespan [42]. Consistent with this, upregulation of the transcript levels of EZH1 and CBX7/8 and an increase in H3K27me3 were observed in the killifish brain during aging [44]. Additionally, the global level of H3K27me3 was increased in quiescent mouse muscle stem cells during chronological aging [21]. These results suggest that a decrease H3K27me3 extends lifespan in some animals.
On the contrary, reduction of H3K27me3 is associated with aging in other animal models. UTX/KDM6A has been identified as a demethylase for H3K27me3 [74,75]. By removing the transcriptionally repressive H3K27me3 mark, UTX can antagonize transcriptional repression via PRCs. A decrease in H3K27me3 and an increase in UTX-1 concomitant with aging were observed in C. elegans and humans [52,53,76]. RNA interference (RNAi) in the utx-1 gene extends lifespan in C. elegans in a daf-16-dependent manner. Furthermore, it was observed that H3K27me3 on the daf-2 gene, which encodes the insulin-like receptor DAF-2, is profoundly reduced in aged worms and that utx-1 RNAi significantly increased the H3K27me3 level on the daf-2 gene. These results suggest that UTX-1 plays an important role in lifespan in C. elegans via modulation of the insulin/IGF-1 signaling pathway. Additionally, Bracken et al. demonstrated that EZH2 is downregulated in stressed and senescent human lung cells, leading to the loss of H3K27me3 [41]. Although the results of studies on several models, including worms, flies, fish, and senescent cells, showed that the link between H3K27me3 levels and aging is complex, it is possible that depending on the specific loci and cells, different H3K27me3 regulators may influence lifespan. Thus, aging may be associated with both an increase and decrease in H3K27me3.
2.4.4. H3K36me3
H3K36me3 is associated with transcriptional elongation and is located in gene bodies rather than promoters [77]. Recently, it was reported that H3K36 methylation plays an important role in transcriptional precision, affecting longevity. Mutation of H3 at the K36 residue and deletion of Set2, an H3K36 methyltransferase, shortened lifespan; deletion of the H3K36 demethylase, Rph1, increased H3K36me3 levels and extended lifespan in wild-type yeast but not H3K36 mutant yeast. These results indicate that methylation at H3K36 is required for lifespan extension. In addition, loss of sustained histone H3K36 methylation is associated both with increased cryptic transcription in a subset of genes in old cells and with decreased lifespan [45]. Consistent with this, Pu et al. revealed that knockdown of Met1 methyltransferase induces a decline in global H3K36me3 levels, resulting in an increase in mRNA expression change with age and shortened lifespan in C. elegans [46]. While there is no significant change in H3K36me3 distribution during aging in C. elegans somatic cells, H3K36me3 marking negatively correlates with changes in gene expression during aging in C. elegans as well as Drosophila [46]. These results suggest a conserved role for H3K36me3 marks in maintaining transcriptional consistency and longevity.
While decreased trimethylation of H3K36 in worms and yeast is known to be associated with reduced lifespan, the role of dimethylation of H3K36me2 in aging is yet to be clarified. Recently, C. elegans SET-18 was identified as a histone H3K36 dimethyltransferase. The deletion of SET-18 increased lifespan and oxidative stress resistance depending on daf-16 activity in the insulin/IGF pathway. Muscle-specific expression of SET-18 increased in aged worms, resulting in elevation of global H3K36me2 and inhibition of daf-16a expression and, consequently, decreased longevity. These results suggest that H3K36me2 and H3K36me3 modification have distinct functions in regulating aging [47].
2.5. Histone Acetylation
An acetyl group can be added to the ε-amino group of a histone lysine, resulting in neutralization of the positive charge of the lysine and weakening of the interaction between the histone and DNA. The resultant decondensed chromatin structure leads to transcription activation [78,79]. Generally, histone acetyltransferases (HATs) are regarded as coactivators in transcription as they catalyze lysine acetylation and loosen chromatin structure, whereas histone deacetylases (HDACs) are considered to function as corepressors. HATs and HDACs are known to play a critical role in longevity (Table 3). For example, in Drosophila, an age-related decrease in H3K5 acetylation level is suppressed by dietary restriction but accelerated by RNAi of the histone acetyltransferase CBP-1 [80,81]. Recently, an interesting report suggested that CBP plays a role in modulating longevity in the pea aphid (Acyrthosiphon pisum), a laboratory insect model; RNAi of A. pisum p300/CBP reduces both lifespan and number of offspring, suggesting that the manipulation of CBP contributes to accelerated aging [82]. On the other hand, it was observed that deletion of the histone acetyltransferase gene GCN5 reduces the extension of replicative lifespan in yeast [83]. In summary, a series of histone acetyltransferases or deacetylases are involved in the process of aging.
Table 3.
Histone acetylation-modulating proteins and aging.
| Name | Description | Change in Histone Modifications Linked to Aging | Effect on Longevity/Health Span | Model | Reference |
|---|---|---|---|---|---|
| Acetyltransferases | |||||
| Gcn5 | Histone acetyltransferase | + | Yeast | [83] | |
| CBP | Histone acetyltransferase | H4K5ac ↓ | + | C. elegans | [81] |
| CBP | Histone acetyltransferase | + | Acyrthosiphon pisum | [82] | |
| Chameau | MYST domain acetyltransferase, human HBO1 homolog | H4K12ac ↑ | − | Drosophila | [84] |
| Sas2 | Histone acetyltransferase | H4K16ac ↑ | − | Yeast | [85] |
| Asf1 | Histone chaperone | H3K56ac ↓ | + | Yeast | [17] |
| RTT109 | Histone acetyltransferase | H3K56ac ↓ | |||
| Deacetylases | |||||
| Rpd3 | Histone deacetylase, HDAC1 ortholog | − | Yeast; Drosophila | [86,87] | |
| Sir2 | NAD+-dependent deacetylase | H4K16ac ↑ | + | Yeast | [85,88] |
| Sir-2.1 | Sir2 ortholog | + | C. elegans | [89] | |
| dSir2 | Sir2 ortholog | + | Drosophila | [90] | |
| SIRT1 | NAD+-dependent deacetylase | + | Mice | [91] | |
| SIRT1 | NAD+-dependent deacetylase | + | Human and mouse fibroblast senescent cells | [92] | |
| SIRT6 | NAD+-dependent deacetylase | H3K9ac ↑ | + | Mice | [93,94] |
| SIRT6 | NAD+-dependent deacetylase | + | Mice | [95] | |
| SIRT6 | NAD+-dependent deacetylase | + | Rat and human nucleus pulposus senescent cells | [96] | |
| Others | |||||
| ZMPSTE24 | Lamin A processing protease | Acetylation of H4 and H2B ↑ | + | Premature aging mouse model | [97] |
| HIRA | Histone chaperone | H4K16ac retention | − | Human senescent cells | [34] |
↑: increase, ↓: decrease, +: positive, -: negative.
2.5.1. H3K9ac
In mice, a deficiency of SIRT6, a member of the sirtuin (SIRT) family of NAD+-dependent deacetylases, causes a decrease in lifespan and a premature aging-like phenotype [94]. SIRT6 is reported to function as an NAD+-dependent H3K9 deacetylase that modulates telomeric chromatin [98]. Notably, Kawahara et al. showed that SIRT6 deacetylates H3K9ac and blocks hyperactive NF-κB signaling, which may modulate lifespan of mice [93]. Consistent with the above mentioned studies, SIRT6 overexpression increased longevity in transgenic mice [95] and inhibited the senescence of rat and human nucleus pulposus cells in a model of intervertebral disc degeneration [96]. These results suggest that SIRT6 may contribute to longevity via H3K9ac deacetylation.
2.5.2. H3K56ac
The acetylation of lysine 56 on histone H3 (H3K56ac) promotes de novo nucleosome assembly, genomic stability, transcription, and formation of heterochromatin/euchromatin boundaries [99,100,101]. In yeast, the level of H3K56 acetylation decreased with age, while the level of H4K16 acetylation increased [85]. Consistent with this, Feser et al. showed that deletion of Rtt109, a major acetyltransferase for H3K56, reduces lifespan in yeast [17]. Unexpectedly, they also found that lifespan is somewhat reduced by deletion of the H3K56-associated deacetylases Hst3 and Hst4. These contradictory results suggest that a delicate balance of acetylated H3 K56Ac may be required to promote longevity.
2.5.3. H4K12ac
A recent report showed that during early aging in Drosophila, elevated acetyl-CoA levels cause changes in histone acetylation. Specifically, a reduction in the levels of the histone lysine acetyltransferase Chameau (HBO1, KAT7) decreases H4K12 acetylation and extends the lifespan of Drosophila males [84].
2.5.4. H4K16ac
The sirtuin family, a group of NAD+-dependent deacetylases, has been strongly linked to increased lifespan [102,103]. Sir2, a member of the sirtuin family, was first identified in the budding yeast Saccharomyces cerevisiae and was found to remove H4K16ac and H3K56ac [104]. During replicative yeast aging, sir2 mutants exhibit a shortened lifespan [88]. Furthermore, an age-associated decrease in Sir2 protein is coupled with an increase in H4K16 acetylation, resulting in compromised transcriptional silencing in subtelomeric regions [85]. In C. elegans, increased dosage of the SIR2 ortholog sir-2.1 extends mean lifespan by up to 50% [89]. This extension requires the FOXO transcription factor DAF-16. Rogina et al. reported that an increase in Drosophila Sir2 (dSir2) extends lifespan [90]. Sas2 is the major H4K16 acetyltransferase that establishes boundaries between telomeres and euchromatin [105,106]. Deletion of Sas2, a histone acetylase, extends lifespan [85]. The antagonizing activities of Sir2 and Sas2 establish an H4K16ac gradient in close proximity to telomeres, maintaining a silencing boundary. In mammals, there are seven sirtuins (SIRT1–SIRT7) that exhibit different enzymatic activities and subcellular compartmentation [107]. While many reports have shown that SIRT1 and SIRT6 are involved in longevity (Table 3), it is a mystery whether H4K16ac is a main target of those SIRT proteins in mammalian aging.
Several reports have shown that H4K16 acetylation is somewhat decreased in an age-dependent manner in mammals. A deficiency of Zmpste24 induces accumulation of prelamin A, which interferes with the retention of MOF, a histone acetyltransferase of H4K16, inducing hypoacetylation of H4K16 and cellular senescence [97,108]. Furthermore, supplementation with a histone deacetylase inhibitor, sodium butyrate, extends the lifespan of Zmpste24-deficient mice [108]. Recent studies have demonstrated that aging in HSCs is correlated with a reduction in H4K16ac and loss of polar distribution of H4K16ac [109,110]. Therefore, the role of H4K16ac in regulating lifespan needs to be clarified.
2.5.5. H4 N-Terminal Acetylation
Calorie restriction decreases the level of Nat4, which belongs to the N-terminal acetyltransferases family and shows high substrate selectivity for histone H4 and H2A. In yeast, Nat4 deletion and the loss of H4 N-terminal acetylation extend lifespan as part of a calorie restriction–mediated pathway [111]. These findings reveal that histone N-acH4 functions as a modulator of lifespan linked to calorie restriction to promote longevity.
2.6. Histone Phosphorylation
A recent report showed links between nutrition sensing pathways and chromatin regulation in aging. It was found that in yeast under nutritional stress, the level of histone H3 threonine 11 phosphorylation (H3pT11) is specifically increased at stress-responsive genes, regulating the transcription of genes involved in metabolic transition. Sch9 and Cka1, a catalytic subunit of CK2, are required for the phosphorylation of H3T11 under such stress [112]. These results suggest that H3pT11 functions as a marker of nutritional stress and aging.
In Drosophila, histone H3S28A mutants (mimicking H3S28 phosphorylation) are associated with increased longevity and improved resistance to starvation and paraquat-induced oxidative stress [113] Additionally, whole-exome deep gene sequencing has revealed that H3S28A mutants exhibit a differential expression pattern of longevity-promoting and mitochondrial biogenesis and respiration genes. These findings present a potential role for H3S28 phosphorylation in controlling longevity and stress resistance.
2.7. Histone Ubiquitination
Histone H2B monoubiquitination is required for the trimethylation of both H3K4 and H3K79 by COMPASS and Dot1 methyltransferases, respectively [114]. Interestingly, the accumulation of H2B monoubiquitination is observed at the telomere-proximal regions of replicative aged yeast, accompanied by an increase in H3K4me3, H3K79me3, and H4K16ac. Rhie et al. demonstrated that H2BK123 mutants have short lifespans in yeast; deletion of the components of the H2B ubiquitinase complex Rad6/Bre1 and the deubiquitinase Ubp10 decreases lifespan [115]. Furthermore, deletion of some components of the SAGA/SLIK histone deubiquitinase complex, including Sgf73, Ubp8, and Sgf11, extends lifespan in yeast [116]. Collectively, an increase of H2B ubiquitination appears to be linked to replicative aging. However, the role of H2B ubiquitination in aging is yet to be investigated.
2.8. Genome-Wide Profiles of Chromatin Accessibility and Histone Modifications during Aging
Comparative chromatin analysis of young cells and old cells reveals changes in their gene expression patterns, an increase in variation in gene expression, and, further, elevated cell-to-cell variability. Recent technological breakthroughs, such as ChIP-seq, ATAC-seq, and RNA-seq, have allowed researchers to investigate genome-wide chromatin dynamics using a small number of cells or even single cells [117].
2.8.1. Chromatin Immunoprecipitation followed by Sequencing (ChIP-seq) and RNA-seq
A recent study revealed that depletion of lamin B1 triggers disruption of nuclear lamin interaction and causes large-scale changes in the chromatin landscape during senescence [54]. The authors found that relative levels of H3K4me3 and H3K27me3 normalized to H3 do not differ between proliferating and senescent cells. Interestingly, H3K4me3 and H3K27me3 marks were redistributed during senescence. The formation of K4me3-enriched domains and K27me3-enriched domains occurs within lamin-associated domains. H3K27me3-depleted regions often contain enhancers near key senescence genes, and the H3K27me3 loss is associated with the upregulation of senescence gene expression. In addition, the distribution of H4K20me3 in senescent cells was determined using ChIP-seq and immunofluorescence; H4K20me3 was enriched at SAHFs, and this was linked with the presence of H3K9me3 and the depletion of H4K16ac and DNA methylation [118].
Genome-wide ChIP-seq and RNA-seq analyses have revealed that downregulation of lamin B1 during senescence triggers global and local chromatin changes that impact gene expression and aging. Other ChIP-seq data shows a unique pattern of H3K4me3 changes during aging in C. elegans [119]. It was shown that around 30% of H3K4me3-enriched regions exhibit significant and reproducible changes with age. These dynamic H3K4me3 regions are largely assigned to protein coding genes and are enriched in areas that span the gene body, where they lead to changes in gene expression. Sun et al. performed the first extensive study integrating epigenomic information on multiple histone modifications and DNA methylation in young and old HSCs. ChIP-seq with H3K4me3 and H3K27me3 has shown that old HSCs exhibit a 6.3% increase in the number of H3K4me3 peaks and broader H3K4me3 peaks across genes related to HSC identity and self-renewal. H3K27me3 peak counts in aged HSCs are similar to those in young HSCs, but the peaks show increased length of coverage. The authors also examined bivalent domains with H3K4me3 and H3K27me3; 335 bivalent domains disappeared in aged HSCs, while 1245 bivalent domains appeared [120]. These age-associated changes in histone modifications may contribute to functional changes in aged HSCs. Most recently, Benayoun et al. generated chromatin maps and transcriptomes from four tissues and one cell type from young, middle-aged, and old mice. The results showed general rules and patterns in age-related chromatin and transcriptional changes with age [121]. Machine-learning analysis with potential predictors revealed that age-related epigenomic changes can predict transcriptional changes and that immune responses are upregulated during aging across tissues and species. These comprehensive findings are likely to aid in the identification of strategies to control aging and age-related diseases.
2.8.2. Assay for Transposase-Accessible Chromatin Using Sequencing and RNA-seq
Moskowitz et al. published genome-wide maps of chromatin accessibility in purified CD8+ T cells from human young and old donors [122]. Integrated assay for transposase-accessible chromatin using sequencing (ATAC-seq) and transcriptome analysis have shown that naïve and central memory cells from older individuals genetically shift toward more differentiated patterns of chromatin openness. In addition, aged naïve cells displayed a loss in chromatin accessibility at gene promoters, largely associated with a decrease in nuclear respiratory factor 1 (NRF1) binding, resulting in lower metabolic activity in aged T cells. Through combined analysis of chromatin accessibility and the transcriptome, Ucar et al. showed the epigenomic signature of aging in peripheral blood mononuclear cells (PBMCs) with chromatin closing at promoters and enhancers and chromatin opening at quiescent and repressed sites. The authors also defined the chromatin accessibility signature of aging-related reduced immune responses in PBMCs. The chromatin accessibility profiles of purified cells show that this aging-related signature in PBMCs stems from CD8+ T cells, which account for 10%–15% of PBMCs [123]. Most recently, Koohy and coauthors performed RNA-seq, ChIP-seq, ATAT-seq, and Hi-C to examine how gene expression, chromatin states, and genome organization are altered in developing B cells during mice aging. The authors observed that aging affected the expression of a limited number of genes and identified transcriptional downregulation of insulin-like growth factor signaling pathway as a hallmark of aging in the aged B cell precursors. These changes are accompanied by changes in microRNAs, histone modifications, and higher-order chromatin structure. It is also found that active gene expression is more strongly linked to H3K4me3 and H3K27ac than to chromatin accessibility, suggesting the combined analysis of the epigenome and transcriptome provides more detail and genome-wide information [124].
2.8.3. Single-Cell Approach
Single-cell RNA sequencing and single-nucleus RNA sequencing have emerged as a powerful tool to reveal the cellular complexity and heterogeneity during aging [125,126,127]. However, epigenetic changes at single-cell resolution during aging remain largely unknown. Recently, Cheung et al. developed a highly multiplexed mass cytometry analysis to profile the global levels of a number of chromatin modifications in human immune cells at the single-cell level [117]. Comparative single-cell ChIP analysis of younger and older adults indicated increased variation between individuals and elevated cell-to-cell variability in chromatin marks as signatures of aging. Through analysis of twin pairs, the authors identified divergent chromatin modification profiles in older monozygotic twin pairs compared to younger pairs, suggesting that non-heritable factors contribute considerably to increased variation in chromatin marks with age.
3. Conclusions and Future Perspectives
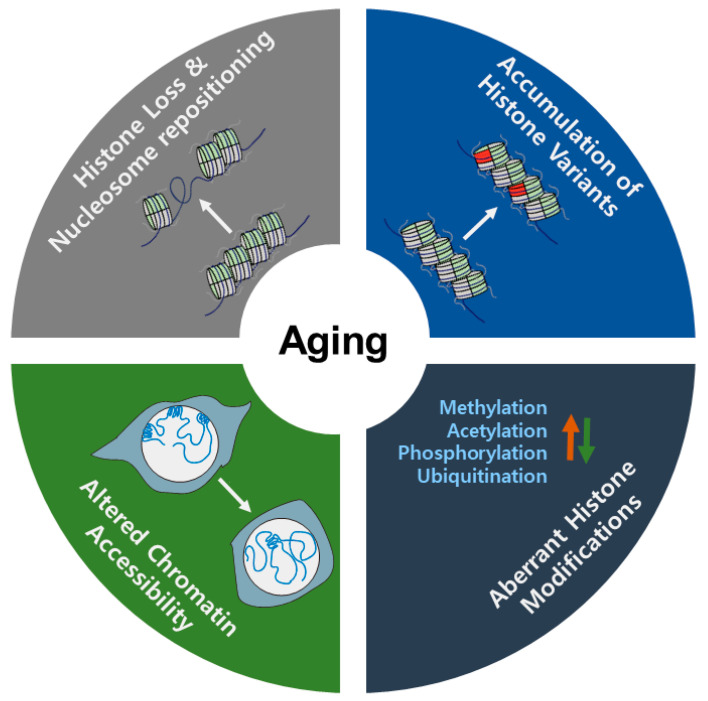
Over time, many researchers have focused on the mechanisms underlying aging. The aging process is regulated by various internal (e.g., genomic instability, epigenetic changes, and metabolic dysfunction) and external (e.g., nutrients and exercise) factors. Epigenetic regulation is interconnected with other factors. During the aging process, epigenetic changes, including variation in histone levels, exchange of histone variants, histone modifications, and nucleosome occupancy, are dynamically changed, modulating chromatin states (see Figure 1).
Figure 1.
Changes in chromatin states linked to aging.
In this review, we provided comprehensive information on the roles of epigenetic modifications in aging. In general, histone levels are decreased and aberrant nucleosome occupancies occur during aging. The accumulation of histone variants is another common feature in the aging process. In contrast, histone modifications such as acetylation, methylation, phosphorylation, and ubiquitylation are more complex. These modifications are differentially implicated in aging; some active (e.g., H3K9ac and H4K16ac) or repressive marks (e.g., H3K9me3 and H3K27me3) may be globally increased or decreased, respectively, during the aging process. In contrast, the active mark H3K36me3 is generally decreased in aging. In other cases, gene-specific changes in histone modifications (e.g., H3K27me3) modulate the expression of key genes related to longevity. Despite increased knowledge on these topics, many questions remain unanswered regarding the mechanism of the effects of epigenetic marks on longevity. Although genetic studies using loss of function have revealed the biological significance of histone modifications in aging, we need to extend our understanding of exactly how these modifications regulate the aging process. Several recent technological advances provide promising clues that may help us to tackle these challenges. For example, ChIP-seq and ATAC-seq combined with transcriptome analysis from the tissue to the single cell level, provide novel findings. As discussed in this review, age-related epigenetic marks are somewhat different between invertebrates and vertebrates. The epigenetic mechanisms of aging are more complicated in vertebrates due to their specialized systems. The development of new experimental models for aging research will provide key insights into the longevity and epigenetic pathways that underlie aging. Indeed, a better understanding of the epigenetic mechanisms of aging and longevity will help us to develop innovative therapeutics that will improve lifespan and counter aging.
Acknowledgments
All sources of funding of the study should be disclosed. Please clearly indicate grants that you have received in support of your research work. Clearly state if you received funds for covering the costs to publish in open access.
Author Contributions
Writing—original draft preparation, S.-J.Y. and K.K.; Writing—review and editing, S.-J.Y. and K.K.; Visualization, S.-J.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by a grant from the National Research Foundation of Korea (2020R1A6A1A06046235 and 2020R1A2C1008179 to K.K.; 2019R1I1A1A01061125 to S.-J.Y. to S.-J.Y.).
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kirkwood T.B. Understanding the odd science of aging. Cell. 2005;120:437–447. doi: 10.1016/j.cell.2005.01.027. [DOI] [PubMed] [Google Scholar]
- 2.Chang A.Y., Skirbekk V.F., Tyrovolas S., Kassebaum N.J., Dieleman J.L. Measuring population ageing: An analysis of the Global Burden of Disease Study 2017. Lancet Public Health. 2019;4:e159–e167. doi: 10.1016/S2468-2667(19)30019-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lopez-Otin C., Blasco M.A., Partridge L., Serrano M., Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benayoun B.A., Pollina E.A., Brunet A. Epigenetic regulation of ageing: Linking environmental inputs to genomic stability. Nat. Rev. Mol. Cell Biol. 2015;16:593–610. doi: 10.1038/nrm4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Booth L.N., Brunet A. The aging epigenome. Mol Cell. 2016;62:728–744. doi: 10.1016/j.molcel.2016.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fraga M.F. Genetic and epigenetic regulation of aging. Curr. Opin. Immunol. 2009;21:446–453. doi: 10.1016/j.coi.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 7.Molina-Serrano D., Kyriakou D., Kirmizis A. Histone modifications as an intersection between diet and longevity. Front. Genet. 2019;10:192. doi: 10.3389/fgene.2019.00192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sen P., Shah P.P., Nativio R., Berger S.L. Epigenetic mechanisms of longevity and aging. Cell. 2016;166:822–839. doi: 10.1016/j.cell.2016.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song S., Johnson F.B. Epigenetic mechanisms impacting aging: A focus on histone levels and telomeres. Genes (Basel) 2018;9:201. doi: 10.3390/genes9040201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glastad K.M., Graham R.J., Ju L., Roessler J., Brady C.M., Berger S.L. Epigenetic regulator CoREST controls social behavior in Ants. Mol. Cell. 2020;77:338–351 e336. doi: 10.1016/j.molcel.2019.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simola D.F., Ye C., Mutti N.S., Dolezal K., Bonasio R., Liebig J., Reinberg D., Berger S.L. A chromatin link to caste identity in the carpenter ant Camponotus floridanus. Genome Res. 2013;23:486–496. doi: 10.1101/gr.148361.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kornberg R.D., Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/S0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 13.Wolffe A.P., Kurumizaka H. The nucleosome: A powerful regulator of transcription. Prog. Nucleic. Acid. Res. Mol. Biol. 1998;61:379–422. doi: 10.1016/s0079-6603(08)60832-6. [DOI] [PubMed] [Google Scholar]
- 14.Demeret C., Vassetzky Y., Mechali M. Chromatin remodelling and DNA replication: From nucleosomes to loop domains. Oncogene. 2001;20:3086–3093. doi: 10.1038/sj.onc.1204333. [DOI] [PubMed] [Google Scholar]
- 15.Kornberg R.D., Lorch Y. Chromatin-modifying and-remodeling complexes. Curr. Opin. Genet. Dev. 1999;9:148–151. doi: 10.1016/S0959-437X(99)80022-7. [DOI] [PubMed] [Google Scholar]
- 16.Yi S.J., Kim K. Histone tail cleavage as a novel epigenetic regulatory mechanism for gene expression. BMB Rep. 2018;51:211–218. doi: 10.5483/BMBRep.2018.51.5.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feser J., Truong D., Das C., Carson J.J., Kieft J., Harkness T., Tyler J.K. Elevated histone expression promotes life span extension. Mol. Cell. 2010;39:724–735. doi: 10.1016/j.molcel.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu Z., Chen K., Xia Z., Chavez M., Pal S., Seol J.H., Chen C.C., Li W., Tyler J.K. Nucleosome loss leads to global transcriptional up-regulation and genomic instability during yeast aging. Genes Dev. 2014;28:396–408. doi: 10.1101/gad.233221.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ivanov A., Pawlikowski J., Manoharan I., van Tuyn J., Nelson D.M., Rai T.S., Shah P.P., Hewitt G., Korolchuk V.I., Passos J.F., et al. Lysosome-mediated processing of chromatin in senescence. J. Cell Biol. 2013;202:129–143. doi: 10.1083/jcb.201212110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Sullivan R.J., Kubicek S., Schreiber S.L., Karlseder J. Reduced histone biosynthesis and chromatin changes arising from a damage signal at telomeres. Nat. Struct. Mol. Biol. 2010;17:1218–1225. doi: 10.1038/nsmb.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu L., Cheung T.H., Charville G.W., Hurgo B.M., Leavitt T., Shih J., Brunet A., Rando T.A. Chromatin modifications as determinants of muscle stem cell quiescence and chronological aging. Cell Rep. 2013;4:189–204. doi: 10.1016/j.celrep.2013.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu R., Sun L., Sun Y., Han X., Qin L., Dang W. Cellular response to moderate chromatin architectural defects promotes longevity. Sci. Adv. 2019;5:eaav1165. doi: 10.1126/sciadv.aav1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Y., Bravo J.I., Son J.M., Lee C., Benayoun B.A. Remodeling of the H3 nucleosomal landscape during mouse aging. Transl. Med. Aging. 2020;4:22–31. doi: 10.1016/j.tma.2019.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rogakou E.P., Sekeri-Pataryas K.E. Histone variants of H2A and H3 families are regulated during in vitro aging in the same manner as during differentiation. Exp. Gerontol. 1999;34:741–754. doi: 10.1016/S0531-5565(99)00046-7. [DOI] [PubMed] [Google Scholar]
- 25.Kreiling J.A., Tamamori-Adachi M., Sexton A.N., Jeyapalan J.C., Munoz-Najar U., Peterson A.L., Manivannan J., Rogers E.S., Pchelintsev N.A., Adams P.D., et al. Age-associated increase in heterochromatic marks in murine and primate tissues. Aging Cell. 2011;10:292–304. doi: 10.1111/j.1474-9726.2010.00666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maze I., Wenderski W., Noh K.M., Bagot R.C., Tzavaras N., Purushothaman I., Elsasser S.J., Guo Y., Ionete C., Hurd Y.L., et al. Critical role of histone turnover in neuronal transcription and plasticity. Neuron. 2015;87:77–94. doi: 10.1016/j.neuron.2015.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tvardovskiy A., Schwammle V., Kempf S.J., Rogowska-Wrzesinska A., Jensen O.N. Accumulation of histone variant H3.3 with age is associated with profound changes in the histone methylation landscape. Nucleic Acids Res. 2017;45:9272–9289. doi: 10.1093/nar/gkx696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stefanelli G., Azam A.B., Walters B.J., Brimble M.A., Gettens C.P., Bouchard-Cannon P., Cheng H.M., Davidoff A.M., Narkaj K., Day J.J., et al. Learning and age-related changes in genome-wide H2A.Z binding in the mouse hippocampus. Cell Rep. 2018;22:1124–1131. doi: 10.1016/j.celrep.2018.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Contrepois K., Coudereau C., Benayoun B.A., Schuler N., Roux P.F., Bischof O., Courbeyrette R., Carvalho C., Thuret J.Y., Ma Z., et al. Histone variant H2A.J accumulates in senescent cells and promotes inflammatory gene expression. Nat. Commun. 2017;8:14995. doi: 10.1038/ncomms14995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Riedel C.G., Dowen R.H., Lourenco G.F., Kirienko N.V., Heimbucher T., West J.A., Bowman S.K., Kingston R.E., Dillin A., Asara J.M., et al. DAF-16 employs the chromatin remodeller SWI/SNF to promote stress resistance and longevity. Nat. Cell Biol. 2013;15:491–501. doi: 10.1038/ncb2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dang W., Sutphin G.L., Dorsey J.A., Otte G.L., Cao K., Perry R.M., Wanat J.J., Saviolaki D., Murakami C.J., Tsuchiyama S., et al. Inactivation of yeast Isw2 chromatin remodeling enzyme mimics longevity effect of calorie restriction via induction of genotoxic stress response. Cell Metab. 2014;19:952–966. doi: 10.1016/j.cmet.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Vaux V., Pfefferli C., Passannante M., Belhaj K., von Essen A., Sprecher S.G., Muller F., Wicky C. The Caenorhabditis elegans LET-418/Mi2 plays a conserved role in lifespan regulation. Aging Cell. 2013;12:1012–1020. doi: 10.1111/acel.12129. [DOI] [PubMed] [Google Scholar]
- 33.Henikoff S., Smith M.M. Histone variants and epigenetics. Cold Spring Harb. Perspect. Biol. 2015;7:a019364. doi: 10.1101/cshperspect.a019364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rai T.S., Cole J.J., Nelson D.M., Dikovskaya D., Faller W.J., Vizioli M.G., Hewitt R.N., Anannya O., McBryan T., Manoharan I., et al. HIRA orchestrates a dynamic chromatin landscape in senescence and is required for suppression of neoplasia. Genes Dev. 2014;28:2712–2725. doi: 10.1101/gad.247528.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Becker P.B., Workman J.L. Nucleosome remodeling and epigenetics. Cold Spring Harb. Perspect. Biol. 2013;5 doi: 10.1101/cshperspect.a017905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clapier C.R., Iwasa J., Cairns B.R., Peterson C.L. Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat. Rev. Mol. Cell Biol. 2017;18:407–422. doi: 10.1038/nrm.2017.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whitehouse I., Rando O.J., Delrow J., Tsukiyama T. Chromatin remodelling at promoters suppresses antisense transcription. Nature. 2007;450:1031–1035. doi: 10.1038/nature06391. [DOI] [PubMed] [Google Scholar]
- 38.Greer E.L., Shi Y. Histone methylation: A dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 2012;13:343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Strahl B.D., Allis C.D. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 40.Djeghloul D., Kuranda K., Kuzniak I., Barbieri D., Naguibneva I., Choisy C., Bories J.C., Dosquet C., Pla M., Vanneaux V., et al. Age-associated decrease of the histone methyltransferase SUV39H1 in HSC perturbs heterochromatin and B Lymphoid differentiation. Stem Cell Rep. 2016;6:970–984. doi: 10.1016/j.stemcr.2016.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bracken A.P., Kleine-Kohlbrecher D., Dietrich N., Pasini D., Gargiulo G., Beekman C., Theilgaard-Monch K., Minucci S., Porse B.T., Marine J.C., et al. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev. 2007;21:525–530. doi: 10.1101/gad.415507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma Z., Wang H., Cai Y., Wang H., Niu K., Wu X., Ma H., Yang Y., Tong W., Liu F., et al. Epigenetic drift of H3K27me3 in aging links glycolysis to healthy longevity in Drosophila. Elife. 2018;7 doi: 10.7554/eLife.35368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Siebold A.P., Banerjee R., Tie F., Kiss D.L., Moskowitz J., Harte P.J. Polycomb repressive complex 2 and trithorax modulate drosophila longevity and stress resistance. Proc. Natl. Acad. Sci. USA. 2010;107:169–174. doi: 10.1073/pnas.0907739107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baumgart M., Groth M., Priebe S., Savino A., Testa G., Dix A., Ripa R., Spallotta F., Gaetano C., Ori M., et al. RNA-seq of the aging brain in the short-lived fish N. furzeri-conserved pathways and novel genes associated with neurogenesis. Aging Cell. 2014;13:965–974. doi: 10.1111/acel.12257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sen P., Dang W., Donahue G., Dai J., Dorsey J., Cao X., Liu W., Cao K., Perry R., Lee J.Y., et al. H3K36 methylation promotes longevity by enhancing transcriptional fidelity. Genes Dev. 2015;29:1362–1376. doi: 10.1101/gad.263707.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pu M., Ni Z., Wang M., Wang X., Wood J.G., Helfand S.L., Yu H., Lee S.S. Trimethylation of Lys36 on H3 restricts gene expression change during aging and impacts life span. Genes Dev. 2015;29:718–731. doi: 10.1101/gad.254144.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Su L., Li H., Huang C., Zhao T., Zhang Y., Ba X., Li Z., Zhang Y., Huang B., Lu J., et al. Muscle-specific histone H3K36 Dimethyltransferase SET-18 Shortens lifespan of Caenorhabditis elegans by repressing daf-16a expression. Cell Rep. 2018;22:2716–2729. doi: 10.1016/j.celrep.2018.02.029. [DOI] [PubMed] [Google Scholar]
- 48.McColl G., Killilea D.W., Hubbard A.E., Vantipalli M.C., Melov S., Lithgow G.J. Pharmacogenetic analysis of lithium-induced delayed aging in Caenorhabditis elegans. J. Biol. Chem. 2008;283:350–357. doi: 10.1074/jbc.M705028200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Greer E.L., Maures T.J., Hauswirth A.G., Green E.M., Leeman D.S., Maro G.S., Han S., Banko M.R., Gozani O., Brunet A. Members of the H3K4 trimethylation complex regulate lifespan in a germline-dependent manner in C. elegans. Nature. 2010;466:383–387. doi: 10.1038/nature09195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li L., Greer C., Eisenman R.N., Secombe J. Essential functions of the histone demethylase lid. PLoS Genet. 2010;6:e1001221. doi: 10.1371/journal.pgen.1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lorbeck M.T., Singh N., Zervos A., Dhatta M., Lapchenko M., Yang C., Elefant F. The histone demethylase Dmel\Kdm4A controls genes required for life span and male-specific sex determination in Drosophila. Gene. 2010;450:8–17. doi: 10.1016/j.gene.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jin C., Li J., Green C.D., Yu X., Tang X., Han D., Xian B., Wang D., Huang X., Cao X., et al. Histone demethylase UTX-1 regulates C. elegans life span by targeting the insulin/IGF-1 signaling pathway. Cell Metab. 2011;14:161–172. doi: 10.1016/j.cmet.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 53.Maures T.J., Greer E.L., Hauswirth A.G., Brunet A. The H3K27 demethylase UTX-1 regulates C. elegans lifespan in a germline-independent, insulin-dependent manner. Aging Cell. 2011;10:980–990. doi: 10.1111/j.1474-9726.2011.00738.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shah P.P., Donahue G., Otte G.L., Capell B.C., Nelson D.M., Cao K., Aggarwala V., Cruickshanks H.A., Rai T.S., McBryan T., et al. Lamin B1 depletion in senescent cells triggers large-scale changes in gene expression and the chromatin landscape. Genes Dev. 2013;27:1787–1799. doi: 10.1101/gad.223834.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Daujat S., Weiss T., Mohn F., Lange U.C., Ziegler-Birling C., Zeissler U., Lappe M., Schubeler D., Torres-Padilla M.E., Schneider R. H3K64 trimethylation marks heterochromatin and is dynamically remodeled during developmental reprogramming. Nat. Struct. Mol. Biol. 2009;16:777–781. doi: 10.1038/nsmb.1629. [DOI] [PubMed] [Google Scholar]
- 56.Kourmouli N., Jeppesen P., Mahadevhaiah S., Burgoyne P., Wu R., Gilbert D.M., Bongiorni S., Prantera G., Fanti L., Pimpinelli S., et al. Heterochromatin and tri-methylated lysine 20 of histone H4 in animals. J. Cell Sci. 2004;117:2491–2501. doi: 10.1242/jcs.01238. [DOI] [PubMed] [Google Scholar]
- 57.Schotta G., Lachner M., Sarma K., Ebert A., Sengupta R., Reuter G., Reinberg D., Jenuwein T. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 2004;18:1251–1262. doi: 10.1101/gad.300704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Villeponteau B. The heterochromatin loss model of aging. Exp. Gerontol. 1997;32:383–394. doi: 10.1016/S0531-5565(96)00155-6. [DOI] [PubMed] [Google Scholar]
- 59.Haithcock E., Dayani Y., Neufeld E., Zahand A.J., Feinstein N., Mattout A., Gruenbaum Y., Liu J. Age-related changes of nuclear architecture in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 2005;102:16690–16695. doi: 10.1073/pnas.0506955102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Larson K., Yan S.J., Tsurumi A., Liu J., Zhou J., Gaur K., Guo D., Eickbush T.H., Li W.X. Heterochromatin formation promotes longevity and represses ribosomal RNA synthesis. PLoS Genet. 2012;8:e1002473. doi: 10.1371/journal.pgen.1002473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee J.H., Kim E.W., Croteau D.L., Bohr V.A. Heterochromatin: An epigenetic point of view in aging. Exp. Mol. Med. 2020 doi: 10.1038/s12276-020-00497-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shumaker D.K., Dechat T., Kohlmaier A., Adam S.A., Bozovsky M.R., Erdos M.R., Eriksson M., Goldman A.E., Khuon S., Collins F.S., et al. Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging. Proc. Natl. Acad. Sci. USA. 2006;103:8703–8708. doi: 10.1073/pnas.0602569103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tsurumi A., Li W.X. Global heterochromatin loss: A unifying theory of aging? Epigenetics. 2012;7:680–688. doi: 10.4161/epi.20540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang W., Li J., Suzuki K., Qu J., Wang P., Zhou J., Liu X., Ren R., Xu X., Ocampo A., et al. Aging stem cells. A Werner syndrome stem cell model unveils heterochromatin alterations as a driver of human aging. Science. 2015;348:1160–1163. doi: 10.1126/science.aaa1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Machida S., Takizawa Y., Ishimaru M., Sugita Y., Sekine S., Nakayama J.I., Wolf M., Kurumizaka H. Structural basis of Heterochromatin formation by human HP1. Mol. Cell. 2018;69:385–397 e388. doi: 10.1016/j.molcel.2017.12.011. [DOI] [PubMed] [Google Scholar]
- 66.Scaffidi P., Misteli T. Lamin A-dependent nuclear defects in human aging. Science. 2006;312:1059–1063. doi: 10.1126/science.1127168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Adams P.D. Remodeling of chromatin structure in senescent cells and its potential impact on tumor suppression and aging. Gene. 2007;397:84–93. doi: 10.1016/j.gene.2007.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Collado M., Blasco M.A., Serrano M. Cellular senescence in cancer and aging. Cell. 2007;130:223–233. doi: 10.1016/j.cell.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 69.Hanahan D., Weinberg R.A. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 70.Herbig U., Ferreira M., Condel L., Carey D., Sedivy J.M. Cellular senescence in aging primates. Science. 2006;311:1257. doi: 10.1126/science.1122446. [DOI] [PubMed] [Google Scholar]
- 71.Chandra T., Ewels P.A., Schoenfelder S., Furlan-Magaril M., Wingett S.W., Kirschner K., Thuret J.Y., Andrews S., Fraser P., Reik W. Global reorganization of the nuclear landscape in senescent cells. Cell Rep. 2015;10:471–483. doi: 10.1016/j.celrep.2014.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cao R., Wang L., Wang H., Xia L., Erdjument-Bromage H., Tempst P., Jones R.S., Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 73.Schwartz Y.B., Pirrotta V. Polycomb silencing mechanisms and the management of genomic programmes. Nat. Rev. Genet. 2007;8:9–22. doi: 10.1038/nrg1981. [DOI] [PubMed] [Google Scholar]
- 74.Agger K., Cloos P.A., Christensen J., Pasini D., Rose S., Rappsilber J., Issaeva I., Canaani E., Salcini A.E., Helin K. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 75.Hong S., Cho Y.W., Yu L.R., Yu H., Veenstra T.D., Ge K. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc. Natl. Acad. Sci. USA. 2007;104:18439–18444. doi: 10.1073/pnas.0707292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ni Z., Ebata A., Alipanahiramandi E., Lee S.S. Two SET domain containing genes link epigenetic changes and aging in Caenorhabditis elegans. Aging Cell. 2012;11:315–325. doi: 10.1111/j.1474-9726.2011.00785.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bannister A.J., Schneider R., Myers F.A., Thorne A.W., Crane-Robinson C., Kouzarides T. Spatial distribution of di- and tri-methyl lysine 36 of histone H3 at active genes. J. Biol. Chem. 2005;280:17732–17736. doi: 10.1074/jbc.M500796200. [DOI] [PubMed] [Google Scholar]
- 78.Bannister A.J., Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shahbazian M.D., Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- 80.Cai H., Dhondt I., Vandemeulebroucke L., Vlaeminck C., Rasulova M., Braeckman B.P. CBP-1 acts in GABAergic neurons to double life span in axenically cultured Caenorhabditis elegans. J. Gerontol. A Biol. Sci. Med. Sci. 2019;74:1198–1205. doi: 10.1093/gerona/glx206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang M., Poplawski M., Yen K., Cheng H., Bloss E., Zhu X., Patel H., Mobbs C.V. Role of CBP and SATB-1 in aging, dietary restriction, and insulin-like signaling. PLoS Biol. 2009;7:e1000245. doi: 10.1371/journal.pbio.1000245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kirfel P., Vilcinskas A., Skaljac M. Lysine Acetyltransferase p300/CBP plays an important role in reproduction, Embryogenesis and longevity of the Pea Aphid Acyrthosiphon pisum. Insects. 2020;11:265. doi: 10.3390/insects11050265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kim S., Ohkuni K., Couplan E., Jazwinski S.M. The histone acetyltransferase GCN5 modulates the retrograde response and genome stability determining yeast longevity. Biogerontology. 2004;5:305–316. doi: 10.1007/s10522-004-2568-x. [DOI] [PubMed] [Google Scholar]
- 84.Peleg S., Feller C., Forne I., Schiller E., Sevin D.C., Schauer T., Regnard C., Straub T., Prestel M., Klima C., et al. Life span extension by targeting a link between metabolism and histone acetylation in Drosophila. EMBO Rep. 2016;17:455–469. doi: 10.15252/embr.201541132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dang W., Steffen K.K., Perry R., Dorsey J.A., Johnson F.B., Shilatifard A., Kaeberlein M., Kennedy B.K., Berger S.L. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature. 2009;459:802–807. doi: 10.1038/nature08085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kim S., Benguria A., Lai C.Y., Jazwinski S.M. Modulation of life-span by histone deacetylase genes in Saccharomyces cerevisiae. Mol. Biol. Cell. 1999;10:3125–3136. doi: 10.1091/mbc.10.10.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rogina B., Helfand S.L., Frankel S. Longevity regulation by Drosophila Rpd3 deacetylase and caloric restriction. Science. 2002;298:1745. doi: 10.1126/science.1078986. [DOI] [PubMed] [Google Scholar]
- 88.Kaeberlein M., McVey M., Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tissenbaum H.A., Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- 90.Rogina B., Helfand S.L. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc. Natl. Acad. Sci. USA. 2004;101:15998–16003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Satoh A., Brace C.S., Rensing N., Cliften P., Wozniak D.F., Herzog E.D., Yamada K.A., Imai S. Sirt1 extends life span and delays aging in mice through the regulation of Nk2 homeobox 1 in the DMH and LH. Cell Metab. 2013;18:416–430. doi: 10.1016/j.cmet.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sasaki T., Maier B., Bartke A., Scrable H. Progressive loss of SIRT1 with cell cycle withdrawal. Aging Cell. 2006;5:413–422. doi: 10.1111/j.1474-9726.2006.00235.x. [DOI] [PubMed] [Google Scholar]
- 93.Kawahara T.L., Michishita E., Adler A.S., Damian M., Berber E., Lin M., McCord R.A., Ongaigui K.C., Boxer L.D., Chang H.Y., et al. SIRT6 links histone H3 lysine 9 deacetylation to NF-kappaB-dependent gene expression and organismal life span. Cell. 2009;136:62–74. doi: 10.1016/j.cell.2008.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mostoslavsky R., Chua K.F., Lombard D.B., Pang W.W., Fischer M.R., Gellon L., Liu P., Mostoslavsky G., Franco S., Murphy M.M., et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 95.Kanfi Y., Naiman S., Amir G., Peshti V., Zinman G., Nahum L., Bar-Joseph Z., Cohen H.Y. The sirtuin SIRT6 regulates lifespan in male mice. Nature. 2012;483:218–221. doi: 10.1038/nature10815. [DOI] [PubMed] [Google Scholar]
- 96.Chen J., Xie J.J., Jin M.Y., Gu Y.T., Wu C.C., Guo W.J., Yan Y.Z., Zhang Z.J., Wang J.L., Zhang X.L., et al. Sirt6 overexpression suppresses senescence and apoptosis of nucleus pulposus cells by inducing autophagy in a model of intervertebral disc degeneration. Cell Death Dis. 2018;9:56. doi: 10.1038/s41419-017-0085-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Osorio F.G., Varela I., Lara E., Puente X.S., Espada J., Santoro R., Freije J.M., Fraga M.F., Lopez-Otin C. Nuclear envelope alterations generate an aging-like epigenetic pattern in mice deficient in Zmpste24 metalloprotease. Aging Cell. 2010;9:947–957. doi: 10.1111/j.1474-9726.2010.00621.x. [DOI] [PubMed] [Google Scholar]
- 98.Michishita E., McCord R.A., Berber E., Kioi M., Padilla-Nash H., Damian M., Cheung P., Kusumoto R., Kawahara T.L., Barrett J.C., et al. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;452:492–496. doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Li Q., Zhou H., Wurtele H., Davies B., Horazdovsky B., Verreault A., Zhang Z. Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell. 2008;134:244–255. doi: 10.1016/j.cell.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lu P.Y., Kobor M.S. Maintenance of heterochromatin boundary and nucleosome composition at promoters by the Asf1 histone chaperone and SWR1-C chromatin remodeler in Saccharomyces cerevisiae. Genetics. 2014;197:133–145. doi: 10.1534/genetics.114.162909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yuan J., Pu M., Zhang Z., Lou Z. Histone H3-K56 acetylation is important for genomic stability in mammals. Cell Cycle. 2009;8:1747–1753. doi: 10.4161/cc.8.11.8620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lee S.H., Lee J.H., Lee H.Y., Min K.J. Sirtuin signaling in cellular senescence and aging. BMB Rep. 2019;52:24–34. doi: 10.5483/BMBRep.2019.52.1.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Longo V.D., Kennedy B.K. Sirtuins in aging and age-related disease. Cell. 2006;126:257–268. doi: 10.1016/j.cell.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 104.Moazed D. Enzymatic activities of Sir2 and chromatin silencing. Curr. Opin. Cell Biol. 2001;13:232–238. doi: 10.1016/S0955-0674(00)00202-7. [DOI] [PubMed] [Google Scholar]
- 105.Kimura A., Umehara T., Horikoshi M. Chromosomal gradient of histone acetylation established by Sas2p and Sir2p functions as a shield against gene silencing. Nat. Genet. 2002;32:370–377. doi: 10.1038/ng993. [DOI] [PubMed] [Google Scholar]
- 106.Suka N., Luo K., Grunstein M. Sir2p and Sas2p opposingly regulate acetylation of yeast histone H4 lysine16 and spreading of heterochromatin. Nat. Genet. 2002;32:378–383. doi: 10.1038/ng1017. [DOI] [PubMed] [Google Scholar]
- 107.Frye R.A. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem. Biophys. Res. Commun. 2000;273:793–798. doi: 10.1006/bbrc.2000.3000. [DOI] [PubMed] [Google Scholar]
- 108.Krishnan V., Chow M.Z., Wang Z., Zhang L., Liu B., Liu X., Zhou Z. Histone H4 lysine 16 hypoacetylation is associated with defective DNA repair and premature senescence in Zmpste24-deficient mice. Proc. Natl. Acad. Sci. USA. 2011;108:12325–12330. doi: 10.1073/pnas.1102789108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Florian M.C., Dorr K., Niebel A., Daria D., Schrezenmeier H., Rojewski M., Filippi M.D., Hasenberg A., Gunzer M., Scharffetter-Kochanek K., et al. Cdc42 activity regulates hematopoietic stem cell aging and rejuvenation. Cell Stem. Cell. 2012;10:520–530. doi: 10.1016/j.stem.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Grigoryan A., Guidi N., Senger K., Liehr T., Soller K., Marka G., Vollmer A., Markaki Y., Leonhardt H., Buske C., et al. LaminA/C regulates epigenetic and chromatin architecture changes upon aging of hematopoietic stem cells. Genome. Biol. 2018;19:189. doi: 10.1186/s13059-018-1557-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Molina-Serrano D., Schiza V., Demosthenous C., Stavrou E., Oppelt J., Kyriakou D., Liu W., Zisser G., Bergler H., Dang W., et al. Loss of Nat4 and its associated histone H4 N-terminal acetylation mediates calorie restriction-induced longevity. EMBO Rep. 2016;17:1829–1843. doi: 10.15252/embr.201642540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Oh S., Suganuma T., Gogol M.M., Workman J.L. Histone H3 threonine 11 phosphorylation by Sch9 and CK2 regulates chronological lifespan by controlling the nutritional stress response. Elife. 2018;7 doi: 10.7554/eLife.36157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Joos J.P., Saadatmand A.R., Schnabel C., Viktorinova I., Brand T., Kramer M., Nattel S., Dobrev D., Tomancak P., Backs J., et al. Ectopic expression of S28A-mutated Histone H3 modulates longevity, stress resistance and cardiac function in Drosophila. Sci. Rep. 2018;8:2940. doi: 10.1038/s41598-018-21372-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Nakanishi S., Lee J.S., Gardner K.E., Gardner J.M., Takahashi Y.H., Chandrasekharan M.B., Sun Z.W., Osley M.A., Strahl B.D., Jaspersen S.L., et al. Histone H2BK123 monoubiquitination is the critical determinant for H3K4 and H3K79 trimethylation by COMPASS and Dot1. J. Cell Biol. 2009;186:371–377. doi: 10.1083/jcb.200906005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rhie B.H., Song Y.H., Ryu H.Y., Ahn S.H. Cellular aging is associated with increased ubiquitylation of histone H2B in yeast telomeric heterochromatin. Biochem. Biophys. Res. Commun. 2013;439:570–575. doi: 10.1016/j.bbrc.2013.09.017. [DOI] [PubMed] [Google Scholar]
- 116.McCormick M.A., Mason A.G., Guyenet S.J., Dang W., Garza R.M., Ting M.K., Moller R.M., Berger S.L., Kaeberlein M., Pillus L., et al. The SAGA histone deubiquitinase module controls yeast replicative lifespan via Sir2 interaction. Cell Rep. 2014;8:477–486. doi: 10.1016/j.celrep.2014.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Cheung P., Vallania F., Warsinske H.C., Donato M., Schaffert S., Chang S.E., Dvorak M., Dekker C.L., Davis M.M., Utz P.J., et al. Single-cell chromatin modification profiling reveals increased epigenetic variations with aging. Cell. 2018;173:1385–1397 e1314. doi: 10.1016/j.cell.2018.03.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Nelson D.M., Jaber-Hijazi F., Cole J.J., Robertson N.A., Pawlikowski J.S., Norris K.T., Criscione S.W., Pchelintsev N.A., Piscitello D., Stong N., et al. Mapping H4K20me3 onto the chromatin landscape of senescent cells indicates a function in control of cell senescence and tumor suppression through preservation of genetic and epigenetic stability. Genome Biol. 2016;17:158. doi: 10.1186/s13059-016-1017-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Pu M., Wang M., Wang W., Velayudhan S.S., Lee S.S. Unique patterns of trimethylation of histone H3 lysine 4 are prone to changes during aging in Caenorhabditis elegans somatic cells. PLoS Genet. 2018;14:e1007466. doi: 10.1371/journal.pgen.1007466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sun D., Luo M., Jeong M., Rodriguez B., Xia Z., Hannah R., Wang H., Le T., Faull K.F., Chen R., et al. Epigenomic profiling of young and aged HSCs reveals concerted changes during aging that reinforce self-renewal. Cell Stem. Cell. 2014;14:673–688. doi: 10.1016/j.stem.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Benayoun B.A., Pollina E.A., Singh P.P., Mahmoudi S., Harel I., Casey K.M., Dulken B.W., Kundaje A., Brunet A. Remodeling of epigenome and transcriptome landscapes with aging in mice reveals widespread induction of inflammatory responses. Genome Res. 2019;29:697–709. doi: 10.1101/gr.240093.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Moskowitz D.M., Zhang D.W., Hu B., Le Saux S., Yanes R.E., Ye Z., Buenrostro J.D., Weyand C.M., Greenleaf W.J., Goronzy J.J. Epigenomics of human CD8 T cell differentiation and aging. Sci. Immunol. 2017;2 doi: 10.1126/sciimmunol.aag0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ucar D., Marquez E.J., Chung C.H., Marches R., Rossi R.J., Uyar A., Wu T.C., George J., Stitzel M.L., Palucka A.K., et al. The chromatin accessibility signature of human immune aging stems from CD8(+) T cells. J. Exp. Med. 2017;214:3123–3144. doi: 10.1084/jem.20170416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Koohy H., Bolland D.J., Matheson L.S., Schoenfelder S., Stellato C., Dimond A., Varnai C., Chovanec P., Chessa T., Denizot J., et al. Genome organization and chromatin analysis identify transcriptional downregulation of insulin-like growth factor signaling as a hallmark of aging in developing B cells. Genome Biol. 2018;19:126. doi: 10.1186/s13059-018-1489-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kimmel J.C., Penland L., Rubinstein N.D., Hendrickson D.G., Kelley D.R., Rosenthal A.Z. Murine single-cell RNA-seq reveals cell-identity- and tissue-specific trajectories of aging. Genome Res. 2019;29:2088–2103. doi: 10.1101/gr.253880.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ma S., Sun S., Geng L., Song M., Wang W., Ye Y., Ji Q., Zou Z., Wang S., He X., et al. Caloric restriction reprograms the single-cell transcriptional landscape of Rattus Norvegicus aging. Cell. 2020;180:984–1001 e1022. doi: 10.1016/j.cell.2020.02.008. [DOI] [PubMed] [Google Scholar]
- 127.Tabula Muris C. A single-cell transcriptomic atlas characterizes ageing tissues in the mouse. Nature. 2020;583:590–595. doi: 10.1038/s41586-020-2496-1. [DOI] [PMC free article] [PubMed] [Google Scholar]