Abstract
Background:
Regulatory roles of long noncoding RNAs (lncRNAs) during viral infection has become more evident in last decade, but are yet to be explored for SARS-CoV-2.
Materials & methods:
We analyzed RNA-seq dataset of SARS-CoV-2 infected lung epithelial cells to identify differentially expressed genes.
Results:
Our analyses uncover 21 differentially expressed lncRNAs broadly involved in cell survival and regulation of gene expression. These lncRNAs can directly interact with six differentially expressed protein-coding genes, and ten host genes that interact with SARS-CoV-2 proteins. Also, they can block the suppressive effect of nine microRNAs induced in viral infections.
Conclusion:
Our investigation determines that deregulated lncRNAs in SARS-CoV-2 infection are involved in viral proliferation, cellular survival, and immune response, ultimately determining disease outcome.
Keywords: : COVID-19, lncRNA, long noncoding RNA, miRNA, SARS-CoV-2, viral pathogenesis
By September 2020, the confirmed cases of COVID-19 have crossed 27 million [1], making it one of the largest pandemics in modern times. Since the first reported case at Wuhan, China in December 2019 [2], this viral disease has been responsible for nearly 900,000 deaths worldwide [1], and the number is steadily rising. The causative agent behind the disease is a novel beta-coronavirus [3], which has been named SARS coronavirus 2 (abbreviated as SARS-CoV-2) due to its similarity to the earlier SARS coronavirus first detected in 2002 [4]. The rapid spread of the virus, the ever-increasing death toll and absence of a sufficient treatment strategy has affected societies and economies all over the globe.
The molecular mechanism of SARS-CoV-2 is complex and interrelated with host mechanisms, as is common with most pathogenic viruses [5]. It is possible that infected lung epithelial cells trigger innate immune pathways, leading to immune effector cells releasing high levels of chemokines and pro-inflammatory cytokines; the resultant unconfined or uncontrolled systemic inflammatory response leads to fatality [6,7]. Recent studies also indicate the virus may cause viral sepsis [8] and infection can lead to deregulation in blood coagulation [9,10]. However, no one conjecture has been able to formulate a clear and concise explanation of how the virus spreads so effectively and affects so profoundly. With emergence of more and more data, new dimensions arise as possible modes of pathogenesis and progression of infection.
Noncoding RNAs (ncRNAs), RNAs that do not code for any proteins, are key players in the regulation of gene expression and influence the interplay involved in host defense mechanisms [11]. Regulatory ncRNAs, such as microRNAs (miRNAs) and long noncoding RNAs (lncRNAs) act as important regulators of the cellular antiviral response. Consequently, viruses have been found to utilize cellular ncRNA to evade immune response and exploit cellular machinery to their advantage [12].
LncRNAs are a type of noncoding RNA having a size of more than 200 nts that can function as primary or spliced transcripts [13]. LncRNAs have become increasingly crucial in explaining cellular processes and understanding molecular progression of diseases. They may regulate gene expression through epigenetic modification of chromatin structure, transcriptional control, regulation of gene transcription via direct binding or transcription factor recruitment and post-transcriptional processing through protein-RNA interaction [14–16]. Additionally, lncRNAs may function as competing endogenous RNAs (ceRNA) [17] to function as regulators of microRNA targeting of genes involved in important pathways.
Previous studies have found lncRNAs to be involved in viral infection and subsequent host response [18–20]. A wide selection of lncRNAs get aberrantly modulated in many viral infections like- HSV, influenza, HIV etc [21]. Upon viral infections, dysregulation of cellular lncRNAs occur which in turn abnormally regulates several host processes resulting in the progression of the viral infection [20]. Apart from the general host routes, abnormalities in the expression of lncRNAs mainly affect host’s different antiviral innate immune responses, particularly interferon signaling and IFN-stimulated genes (ISGs) [20,21]. Even in SARS-CoV infection, Peng et al. showed that differential expression of lncRNAs could aberrantly regulate several host responses along with the innate immune signaling [22], which suggests a similar deregulation pattern of lncRNAs could also occur in SARS-CoV-2 infection.
Some attempts have been made to decipher the roles of ncRNAs in SARS-CoV-2 infection, For example, Bartoszewski et al. have proposed that SARS-CoV-2 could modulate host miRNA levels by acting as miRNA sponges [23]. Khan et al. [24] and Chow et al. [25] both predicted host miRNAs that target the virus. But the focus is yet to turn on to deregulated host lncRNAs and their interaction with SARS-CoV-2.
Blanco-Melo et al. [26] investigated the host response to SARS-CoV-2 by infecting primary human lung epithelium (NHBE) cells and A549 alveolar cell lines with the virus and performing RNA-seq analysis to identify differentially expressed (DE) genes. But no such study yet concluded the possible outcomes of the deregulated lncRNAs in SARS-CoV-2. In our present study we have identified lncRNAs that are differentially expressed in SARS-CoV-2 infected cell’s transcriptome compared with uninfected cells, and then correlated the putative effects of the deregulated lncRNAs in the tug-of-war between SARS-CoV-2 and the host. We have also investigated the possible aftermaths of lncRNA deregulation in COVID-19 disease pathobiology.
Materials & methods
Identification of differentially expressed lncRNAs
Raw FastQ reads of RNA-Seq performed by Illumina were extracted from GEO database (GEO accession GSE147507) [27]. The experimental data included independently assessed biological triplicates of primary human lung epithelium (NHBE) cells, which were either mock treated or infected with SARS-CoV-2 and cultured for 24 h. The reads were mapped with TopHat (TopHat v2.1.1 with Bowtie v2.4.1) [28]. Using the latest version of human reference genome GRCh38, as downloaded from UCSC database [29], short reads were uniquely aligned, allowing at best two mismatches. Sequences that were exact matches to multiple regions with equal quality were discarded to avoid bias [30]. The reads not mapped to the genome were utilized by mapping against the transcriptome. The latest version of Ensembl gene model [31] (version 99, as extracted from UCSC) was utilized for this process. After completion of mapping, we used SubRead package featureCount v2.21 [32] to calculate the absolute read abundance, leading to read count (r.c.) for each of the Ensembl genes. For the subsequent differential expression (DE) analysis, we used DESeq2 v1.26.0 with R v3.6.2 (2019-07-05) [33] that employs a model based on negative binomial distribution. To avoid false positives, transcripts with at least ten reads annotated in at least one of the samples used in this study were considered.
PPI network construction
From RNA-seq data analysis, 638 DE protein-coding genes were identified. These were combined with the recently reported 332 SARS-CoV-2 protein interactors [34] retrieved from BioGRID [35] to build a protein–protein interaction (PPI) network. Edge information for this network were extracted from the STRING [36] database, where only interactions with high confidence (score >0.700) were selected and retrieved. A network file was prepared in simple interaction format to be visualized using Cytoscape v3.7.2 [37].
Retrieval of RNA–RNA interactions & lncRNA functions
We retrieved previously established RNA–RNA interactions between the DE lncRNAs and other RNAs from NPInter v4.0 [38]. Among the interacting RNAs, we searched for DE protein-coding genes and viral protein-interactor genes. Functions of the DE lncRNAs were retrieved from LncBook [39]. The viral proteins that bind to lncRNA–interacting host proteins were also identified.
Identification of virally-induced miRNAs
miRwayDB [40] provided information about human miRNAs that were previously identified as involved in viral-mediated diseases. Additionally, information about host miRNAs involved in viral infection as identified by Girardi et al. [41] and miRNAs involved in viral acute respiratory infections as described by Leon-Icaza et al. [42] were retrieved. These miRNAs were manually curated for their involvement in viral pathogenesis and possible connection to SARS-CoV-2.
Extraction of microRNA targets
Gene targets for the curated set of miRNAs were retrieved from experimentally validated miRNA–gene interactions from miRTarBase [43] and filtered for the DE protein-coding genes. DIANA-LncBase v3 [44] was used to retrieve miRNAs that target the DE lncRNAs, considering only high-confidence interactions. Combining the lists led to identification of the miRNAs that target both DE protein-coding genes and lncRNAs. The set of upregulated target genes for each miRNA were analyzed using NetworkAnalyst 3.0 [45] tools for functional over-representation and network enrichment. miRNAs that mostly targeted upregulated genes were finally selected.
Construction of biological networks
Networks depicting the interactions between DE lncRNAs, DE genes, viral proteins and curated miRNAs were built using Cytoscape v3.7.2 [37]. Gradient coloring scheme was used to depict the change in expression of DE genes.
Results
lncRNAs differentially modulated in SARS-CoV-2 infection
To investigate the probable deregulation of lncRNAs in SARS-CoV-2 infection, first we have analyzed the 24 h postinfection transcriptome data of SARS-CoV-2 infected NHBE cells. Analyzing the RNA-seq data led to identification of 687 DE genes (Supplementary file 1). Intriguingly among those DE genes, we discovered 21 lncRNA genes, nine of them upregulated while 12 were downregulated (Figure 1).
Figure 1. . Differentially expressed long noncoding RNAs in SARS-CoV-2 infected primary human lung epithelium (NHBE) cells.
Analyzing the RNA-seq data, nine lncRNAs were found to be upregulated, while twelve were downregulated. Gene expressions are presented in log2 fold change (compared with uninfected control cells) value color coded heatmap. Color toward red indicates more upregulation and color toward green indicates further downregulation, while yellow color indicates absence of differential expression.
lncRNAs interact with several differentially modulated proteins in SARS-CoV-2 infected cells
We now sought to elucidate the putative effects of these deregulated lncRNAs in SARS-CoV-2 infection. In order to achieve that, we built a network of the deregulated lncRNAs along with their interacting protein coding target genes. Among the differentially expressed protein-coding genes, six were found to interact with the DE lncRNAs (Table 1). Among them, there are direct RNA–RNA interactions for four genes and RNA–protein interactions for the rest (Figure 2). These lncRNA interacting protein coding genes have potential roles during the viral infections (Table 1).
Table 1. . Differentially expressed genes, their encoded protein’s functions and associated interacting long non-coding RNAs.
| Gene name | Protein function | lncRNA interactor | Interaction Type | Ref. |
|---|---|---|---|---|
| ADAR | ADAR works as a regulator in RIG-I/MDA5 mediated induction of IFN-α/β pathways and controls innate immune response against dsRNA in the cell. It exerts an antiviral effect on HCV, but works in favor of VSV, MV, HDV and type 1 HIV-1. |
KCNQ1OT1 GAS6-AS1 MEG3 INHBA-AS1 |
Protein–RNA | [46–54] |
| EDN1 | Endothelin-1 is an endothelium-derived vasoconstrictor peptide that belongs to the endothelin/sarafotoxin family. The peptide works as a potent vasoconstrictor and its cognate receptors are therapeutic targets in the treatment of pulmonary arterial hypertension. It is involved in downstream GPCR-controlled signaling. | UGDH-AS1 | RNA–RNA | [55,56] |
| KYNU | KYNU gene encodes Kynureninase enzyme, which catalyzes the cleavage of L-kynurenine and L-3-hydroxykynurenine into anthranilic acid and 3-hydroxyanthranilic acid, respectively. Among its functional pathways are NAD metabolism and tryptophan utilization. | FAM230J | RNA–RNA | [57,58] |
| MALL | MALL gene encodes an element of the machinery for raft-mediated trafficking in endothelial cells. The encoded protein, a member of the MAL proteolipid family, predominantly localizes in glycolipid- and cholesterol-enriched membrane (GEM) rafts. It interacts with caveolin-1 and is involved in cholesterol homeostasis. | AC006058.1 | RNA–RNA | [59,60] |
| TLR2 | TLR2 encodes the toll-like receptor 2 protein, a fundamental protein in the pathways involved with pathogen recognition and activation of innate immunity. A cell-surface protein that can form heterodimers with other TLR family members, it can recognize conserved molecules derived from microorganisms known as PAMPs. Activation of TLRs by PAMPs switches on signaling pathways that modulate the host’s inflammatory response. TLR2 acts via MYD88 and TRAF6, leading to NF-κB activation, cytokine secretion and inflammatory response. This gene may also promote apoptosis in response to bacterial lipoproteins. | KCNQ1OT1 | RNA–RNA | [61–65] |
| YWHAG | YWHAG encodes the 14-3-3 protein gamma, an adapter protein implicated in the regulation of a large spectrum of both general and specialized signaling pathways. It is induced by growth factors in human vascular smooth muscle cells, and is also highly expressed in skeletal and heart muscles, suggesting an important role for this protein in muscle tissue. Among its interactors are RAF1 and PKC, which link it to various signal transduction pathways. The protein is involved in apoptosis and PI3K-Akt signaling pathway. YWHAG was identified as a direct target of miR-181b-3p, a metastasis activator which downregulated YWHAG to promote Snail-induced EMT in breast cancer cells. MiR-182 was found to suppress esophageal squamous cell carcinoma cell proliferation and metastasis via regulating YWHAG |
KCNQ1OT1 SNHG8 GAS6-AS1 |
Protein–RNA | [66–73] |
EMT: Epithelial-mesenchimal transformation; HDV: Hepatitis delta virus; MV: Measles virus; GPCR: G protein-coupled receptors; PAMP: Pathogen-associated molecular pattern; TLR: Toll like receptor; VSV: Vesicular stomatitis virus.
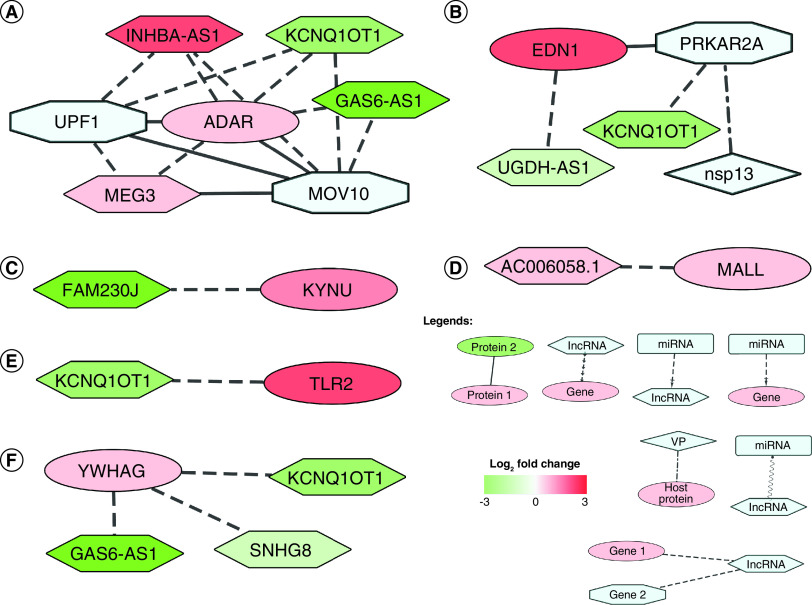
Figure 2. . Differentially expressed long noncoding RNAs interact with differentially expressed protein-coding genes.
(A) Upregulated ADAR interacts with downregulated KCNQ1OT1 and GAS6-AS1, and upregulated MEG3 and INHBA-AS1; it also interacts with UPF1 and MOV10. (B) Upregulated EDN1 interacts with downregulated UGDH-AS1 and Nsp13 interactor PRKAR2A. (C) KYNU mRNA interacts with downregulated FAM230J. (D) Upregulated MALL interacts with upregulated AC006058.1. (E) Upregulated TLR2 interacts with downregulated KCNQ1OT1. (F) YWHAG protein interacts with downregulated KCNQ1OT1, SNHG8 and GAS6-AS1. Node shape legends: ellipse: DE gene; hexagon: lncRNA; diamond: VP; octagon: viral protein interactor; rectangle: microRNA; types of edges are- dot-dash: VP-host protein interaction; dash: lncRNA–gene interaction; dash-arrow: miRNA–gene interaction; solid: PPI; sine wave: ceRNA; separate arrows: alternate target. Log2 fold change color scale is as depicted in Figure 1.
ceRNA: Competing endogenous RNA; DE: Differentially expressed; lncRNA: Long noncoding RNA; PPI: Protein–protein interaction; VP: Viral protein.
Upregulated ADAR interacts directly with downregulated KCNQ1OT1 and GAS6-AS1, and upregulated MEG3 and INHBA-AS1 (Figure 2A). ADAR protein also interacts with UPF1 and MOV10 – two proteins that are also interactors of other DE lncRNAs. EDN1 is found to be upregulated in SARS-CoV-2 infection, which interacts directly with downregulated UGDH-AS1 (Figure 2B). EDN1 protein can also interact with PRKAR2A, a protein that interacts with KCNQ1OT1 and viral protein Nsp13. KYNU mRNA interacts directly with FAM230J, which was downregulated (Figure 2C) [74]. MALL, a gene found upregulated in SARS-CoV-2 infection, interacts directly with AC006058.1, which was also upregulated (Figure 2D). Upregulated TLR2 interacts directly with downregulated KCNQ1OT1 (Figure 2E). YWHAG protein interacts directly with KCNQ1OT1, SNHG8, GAS6-AS1, all downregulated (Figure 2F).
Protein interactors of SARS-CoV-2 viral proteins also interacted with DE lncRNAs
Gordon et al. investigated possible protein–protein interactions (PPI) between SARS-CoV-2 proteins and host proteins and identified 332 high-confidence interactions [34]. We retrieved these interactions from BioGRID [35], which are provided in supplementary file 2. We wanted to link whether these interacting proteins can also interact with the DE lncRNAs in SARS-CoV-2 infection. Subsequently, we constructed a network with the DE lncRNAs and their interacting host proteins, which also bind with viral proteins. Among these 332 proteins, we found ten such proteins that also interact with the DE lncRNAs (Table 2). Among them, seven proteins interacted at the protein–RNA level, whereas the rest were RNA–RNA interactions. SARS-CoV-2 proteins M (Membrane), N (Nucleocapsid), Nsp8, Nsp12, Nsp13, and ORF8 were found to interact with these host proteins (Figure 3).
Table 2. . SARS-CoV-2 interactor host proteins and associated interacting long non-coding RNAs.
| Gene name | Protein function | SARS interactor protein | lncRNA interactor | Interaction type | Ref. |
|---|---|---|---|---|---|
| AKAP8L | AKAP8L (A-kinase anchor protein 8-like) protein probably plays a role in CTE-mediated gene expression by association with DHX9 by increasing nuclear unspliced mRNA export. In EBV infected cells, it may target PRKACA to nuclear sites containing EBNA-LP (an EBV protein) to modulate transcription from specific promoters. In synergy with DHX9, it can activate the CTE-mediated gene expression of type D retroviruses. In case of HIV-1 infection, it is involved in the DHX9-promoted annealing of host tRNA (Lys3) to viral genomic RNA as a primer in reverse transcription. | M |
KCNQ1OT1 GAS6-AS1 SNHG8 AC048341.2 AL109615.3 |
Protein–RNA | [75–78] |
| EXOSC5 | EXOSC5 is a noncatalytic component of the RNA exosome complex, which has 3′ to 5′ exoribonuclease activity and participates in a multitude of cellular RNA processing and degradation events. | Nsp8 |
KCNQ1OT1 SNHG8 |
Protein–RNA | [79] |
| GDF15 | GDF15 is a member of the GDNF family which binds to GDNF family receptor α-like (GFRAL) protein, a transmembrane receptor exclusively expressed in the hind brain. The protein is expressed in a broad range of cell types, acts as a pleiotropic cytokine and is involved in the stress response program of cells after cellular injury. Increased protein levels are associated with disease states such as tissue hypoxia, inflammation, acute injury and oxidative stress. | ORF8 | MEG3 | RNA–RNA | [80–83] |
| HECTD1 | HECTD1 is an E3 ubiquitin-protein ligase which accepts ubiquitin from an E2 ubiquitin-conjugating enzyme in the form of a thioester and then directly transfers the ubiquitin to targeted substrates and mediates ‘Lys-63’-linked polyubiquitination of HSP90AA1, which leads to its intracellular localization and reduced secretion. The protein is involved in class I MHC mediated antigen processing and presentation and innate immune system. | Nsp8 | KCNQ1OT1 | RNA–RNA | [84] |
| LARP4B | LARP4B encodes a La-module containing factor that can bind AU-rich RNA sequence directly and promotes mRNA accumulation and translation. It was deemed a candidate tumor suppressor gene in glioma, as it was consistently decreased in human glioma stem cells and cell lines compared with normal neural stem cells. LARP4B overexpression strongly inhibited cell proliferation by inducing mitotic arrest and apoptosis and CDKN1A and BAX were upregulated. | Nsp12 |
UGDH-AS1 SNHG8 |
Protein–RNA | [85,86] |
| LARP7 | LARP7 works as a negative transcriptional regulator of polymerase II genes, acting by means of the 7SK RNP system. This snRNP complex inhibits a cyclin-dependent kinase, positive transcription elongation factor b, which is required for paused RNA polymerase II at a promoter to begin transcription elongation. | Nsp8 |
KCNQ1OT1 SNHG8 |
Protein–RNA | [87,88] |
| MIPOL1 | MIPOL1 encodes a coiled-coil domain-containing protein, which may function as a tumor suppressor. | Nsp13 | KCNQ1OT1 | RNA–RNA | [89] |
| UPF1 | UPF1 protein, an RNA-dependent helicase and ATPase, is required for NMD of mRNAs containing premature stop codons. It is recruited to mRNAs upon translation termination by release factors to stalled ribosomes together with the SMG1C protein kinase complex to form the transient SURF (SMG1-UPF1-eRF1-eRF3) complex. | N |
AC048341.2 LINC00937 GAS6-AS1 KCNQ1OT1 UGDH-AS1 SNHG8 AC087473.1 AL109615.3 INHBA-AS1 MEG3 LINC02015 MIR3142HG AC006058.1 C1RL-AS1 |
Protein–RNA | [90–92] |
| MOV10 | MOV10 is a 5′ to 3′ RNA helicase contributing to UPF1 mRNA target degradation by translocation along 3′ UTRs and is involved in miRNA-mediated gene silencing by the RISC. It plays a role for both miRNA-mediated translational repression and miRNA-mediated cleavage of complementary mRNAs by RISC. UPF1-MOV10 is involved in antiviral activity through both NMD pathway and IFN induction. | N |
LINC00504 AC048341.2 LINC00937 GAS6-AS1 KCNQ1OT1 UGDH-AS1 SNHG8 AC087473.1 AL109615.3 INHBA-AS1 MEG3 LINC02015 |
Protein–RNA | [93–98] |
| PRKAR2A | PRKAR2A is the cAMP-dependent protein kinase type II-alpha regulatory subunit, which works as the regulatory subunit of the cAMP-dependent protein kinases involved in cAMP signaling in cells. | Nsp13 | KCNQ1OT1 | RNA–RNA | [89] |
CTE: Constitutive transport element; GDNF: Glial cell-derived neurotropic factor; NMD: Nonsense-mediated decay; RISC: RNA-induced silencing complex.
Figure 3. . Differentially expressed long noncoding RNAs interact with SARS-CoV-2 protein interacting genes.
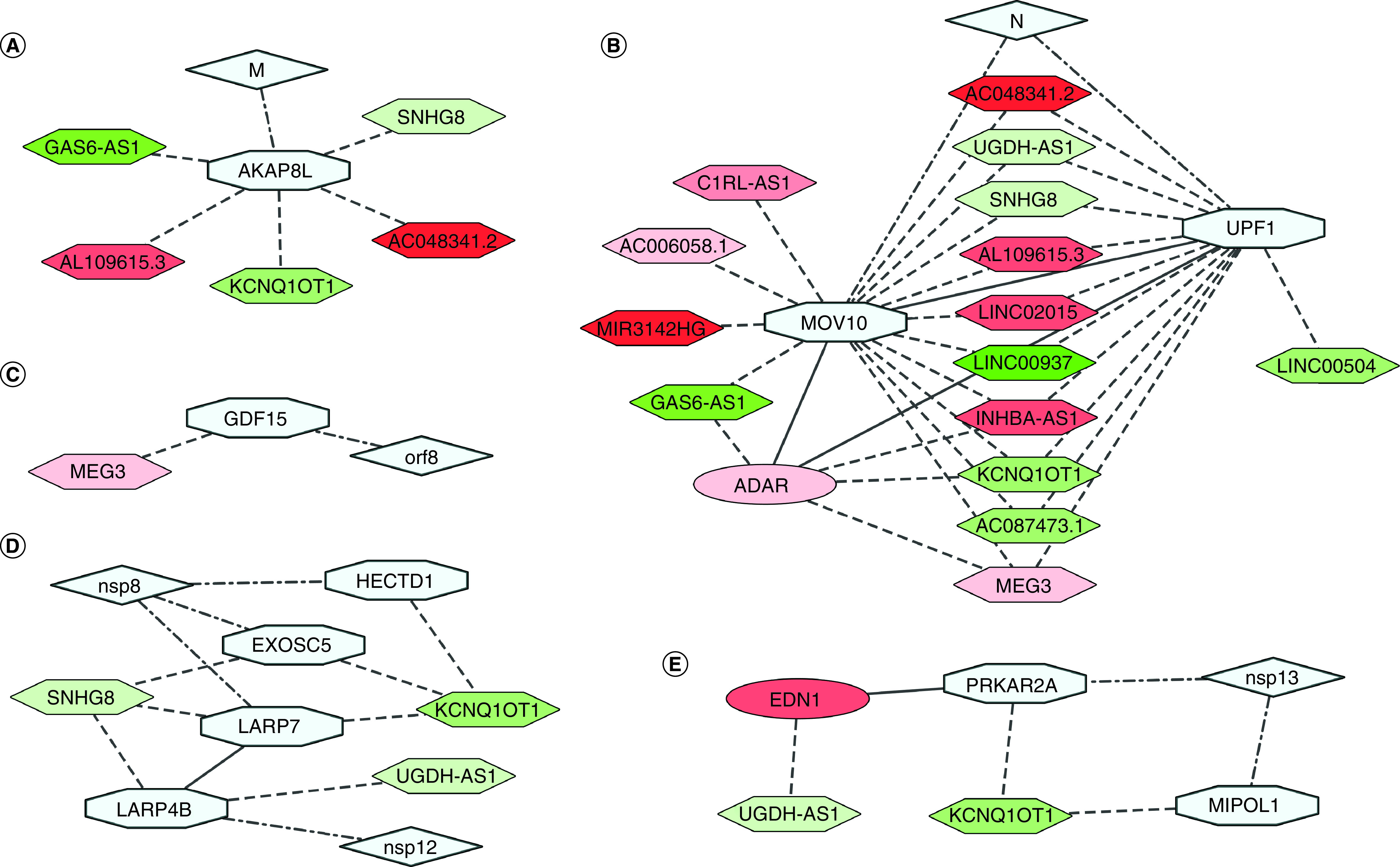
(A) M protein interactor AKAP8L interacts with downregulated KCNQ1OT1, GAS6-AS1, SNHG8, AC048341.2 and upregulated AL109615.3. (B) Among N protein interactors, UPF1 and MOV10 proteins interacts directly with multiple DE lncRNAs, and each other. Both also interact with ADAR protein. (C) MEG3 can stimulate expression of GDF15, an interactor of viral protein orf8. (D) Nsp8 interactors EXOSC5 and LARP7 proteins interact with downregulated KCNQ1OT1 and SNHG8; HECTD1 mRNA interacts with KCNQ1OT1. Nsp12 interactor LARP4B protein interacts with downregulated UGDH-AS1 and SNHG8, and also with LARP7. (E) Nsp13 interactors MIPOL1 and PRKAR2A interact with downregulated KCNQ1OT1; PRKAR2A also interacts with UGDH-AS1 interactor EDN1. Color codes, node and edge notations are similar as Figure 2.
DE: Differentially expressed; lncRNAs: Long noncoding RNAs.
AKAP8L protein, an M protein interactor, interacts directly with downregulated KCNQ1OT1, GAS6-AS1, SNHG8, AC048341.2 and upregulated AL109615.3 (Figure 3A). Among N protein interactors, UPF1 and MOV10 proteins interact directly with DE lncRNAs, and also interact with each other (Figure 3B). UPF1 protein interacts with downregulated AC048341.2, LINC00937, GAS6-AS1, KCNQ1OT1, UGDH-AS1, SNHG8 and AC087473.1, and upregulated AL109615.3, INHBA-AS1, MEG3, LINC02015, MIR3142HG, AC006058.1 and C1RL-AS1. MOV10 protein interacts with LINC00504, AC048341.2, LINC00937, GAS6-AS1, KCNQ1OT1, UGDH-AS1, SNHG8 and AC087473.1, which were downregulated, and AL109615.3, INHBA-AS1, MEG3 and LINC02015, which were upregulated. Both UPF1 and MOV10 proteins also interact with ADAR protein, an interactor of KCNQ1OT1, GAS6-AS1, MEG3 and INHBA-AS1 (Figure 3B). MEG3 can stimulate expression of GDF15 by enhancing p53 binding to the GDF15 gene promoter [80], while GDF15 protein can be targeted by viral orf8 (Figure 3C).
Among Nsp8 interactors, EXOSC5 and LARP7 proteins interact with KCNQ1OT1 and SNHG8, both downregulated; another interactor HECTD1 mRNA interacts directly with KCNQ1OT1 (Figure 3D). Among Nsp12 protein interactors, LARP4B protein interacts with UGDH-AS1 and SNHG8, both downregulated, and with LARP7 (Figure 3D). Among Nsp13 interactors, MIPOL1 and PRKAR2A both have RNA–RNA interactions with downregulated KCNQ1OT1. PRKAR2A also interacts with EDN1, which interacts with UGDH-AS1 (Figure 3E).
Upregulated targets of virally-induced microRNAs might be protected by competing lncRNAs
ceRNA hypothesis illustrates that lncRNAs and other RNA molecules harboring miRNA response elements can suppress the expression and biological function of each other by competing for miRNAs that can bind to the complementary regions, thus regulating miRNA-mediated gene silencing of the target genes [17,99]. To reveal if such networks exists in SARS-CoV-2 infected cells, a list of miRNAs induced in viral infection was curated from miRwayDB database [40], Girardi et al. [41], and Leon-Icaza et al. [42]. Among them, few were found to target genes upregulated in the cells, which were apparently not targeted as DE lncRNAs were targeted simultaneously (Figure 4). The names and functions of all identified target genes of the selected miRNAs that were differentially expressed in the infected cells are provided in supplementary file 3.
Figure 4. . Potential virally induced miRNAs and their targets.
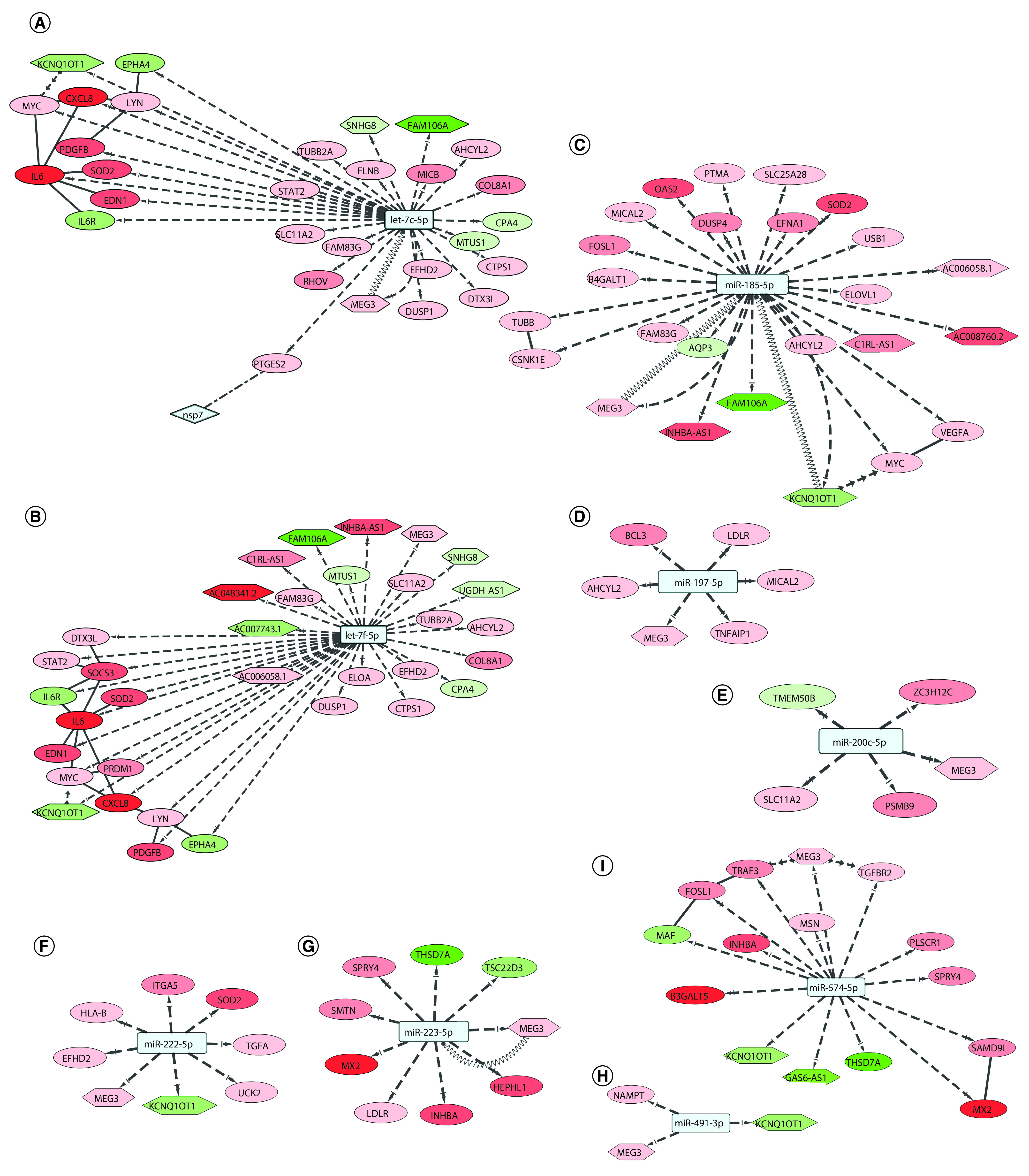
(A) miRNA let-7c can target 21 upregulated genes and four differentially expressed long non-coding RNAs; MEG3 can act as a competing endogenous RNA. (B) miRNA let-7f can target 20 upregulated genes and nine DE lncRNAs; MEG3 can act as a ceRNA. (C) miR-185-5p can target 17 upregulated genes and seven DE lncRNAs; MEG3 and KCNQ1OT1 can act as ceRNAs. (D) miR-197-5p can target five upregulated genes and lncRNA MEG3. (E) miR-200c-5p can target three upregulated genes and lncRNA MEG3. (F) miR-222-5p can target six upregulated genes and lncRNAs KCNQ1OT1 and MEG3. (G) miR-223-5p can target six upregulated genes and lncRNA MEG3, which can act as a ceRNA. (H) miR-491-3p can target upregulated NAMPT and lncRNA MEG3. (I) miR-574-5p can target ten upregulated genes and lncRNAs MEG3, KCNQ1OT1 and GAS6-AS1. Color codes, node and edge notations are similar as Figure 2.
ceRNA: Competing endogenous RNA; DE: Differentially expressed; lncRNAs: Long noncoding RNAs.
In SARS-CoV-2 infection, miRNA let-7c might target 21 upregulated genes and can also target MEG3, KCNQ1OT1, SNHG8 and FAM106A lncRNAs (Figure 4A). The 21 upregulated targets of let-7c-5p are significantly enriched for KEGG pathways like – Kaposi’s sarcoma-associated herpesvirus infection, and JAK-STAT signaling pathway (data not shown). Among the target genes, PTGES2 protein was found to interact with SARS-CoV-2 Nsp7 protein (Figure 4).
miRNA let-7f can target 20 of the upregulated genes in SARS-CoV-2 infection. It can also target lncRNAs AC006058.1, AC007743.1, AC048341.2, C1RL-AS1, FAM106A, INHBA-AS1, KCNQ1OT1, MEG3 and SNHG8 (Figure 4B). The 20 upregulated targets of the let-7f-5p are involved in pathways like Kaposi’s sarcoma-associated herpesvirus infection-related, JAK-STAT signaling, and hepatitis B related signaling (data not shown).
miR-185-5p targets 17 upregulated genes, but also targets lncRNAs AC006058.1, AC008760.2, C1RL-AS1, FAM106A, INHBA-AS1, KCNQ1OT1 and MEG3 (Figure 4C). Among the 17 upregulated targets of the miR-185-5p miRNA, most significantly enriched KEGG pathways were MAPK signaling pathway, and Wnt signaling pathway (data not shown).
miR-197-5p, miR-200c-5p, miR-222-5p and miR-223-5p target five, three, six and six upregulated genes, respectively (Figure 4D–G). They can all target lncRNA MEG3 while miR-222-5p can additionally target KCNQ1OT1. The six upregulated targets of the miR-222-5p are involved in phagosome, and PI3K-Akt signaling pathway (data not shown).
miR-491-3p targets upregulated NAMPT gene, but also targets MEG3 lncRNA (Figure 4H). miR-574-5p targets ten upregulated genes, but also targets lncRNAs MEG3, KCNQ1OT1 and GAS6-AS1 (Figure 4I). The ten upregulated targets of the miR-574-5p are found significantly enriched in biological process/pathways like TGF-β signaling pathway, IL-17 signaling pathway and apoptotic process (data not shown).
Involvement of lncRNA may modulate important cellular pathways
Cell survival can be beneficial for continued viral replication and growth, but apoptosis is also necessary for further spread. Pathways related to cell survival are targeted by virally-induced microRNAs. IL-6/JAK/STAT3 signaling acts in favor of cell survival [100]. Upregulation of IL-6 during certain viral infections may promote virus survival and/or exacerbation of clinical disease [101]. Among let-7c-5p targets and let-7f-5p targets, four upregulated genes, IL6, MYC, PDGFB and STAT2 belong to JAK-STAT signaling pathway. KCNQ1OT1 can act as a sponge for MYC gene, a target of miR-185-5p along with these miRNAs (Figure 4C [102]). Also, both let-7c and let-7f target IL-6, and absence of this interaction may lead to cell transformation progressing from initial inflammation, implying cell survival [103].
Wnt signaling pathway is known to regulate apoptosis through a variety of mechanisms [104] and it can respond to viral infections through modulation of β-catenin stabilization [105]. MYC, CSNK1E and FOSL1 belong to Wnt signaling pathway and are targeted by miR-185-5p. FOSL1 is also targeted by miR-574-5p (Figure 4I).
Activation of PI3K-Akt signaling is a known viral strategy to delay apoptosis and prolong viral replication, as seen in acute and persistent infection [106]. Three upregulated genes, EFNA1, MYC and VEGFA, targeted by miR-185-5p, and two upregulated genes targeted by miR-222-5p, ITGA5 and TGFA, belong to PI3K-Akt signaling pathway (Figure 4). Also, miR-185 was found to inhibit cell proliferation and induce apoptosis by targeting VEGFA directly in von Hippel-Lindau-inactivated clear cell renal cell carcinoma [107].
NF-κB signaling pathway is involved in upregulating various pro-inflammatory genes that encode cytokines and chemokines [108]. It is a major regulator of antiviral response, and NF-κB activation pathways are manipulated by viruses to avoid cellular mechanisms that eliminate the infection [109,110]. Two upregulated gene targets of let-7c-5p and let-7f-5p, CXCL8 and LYN, and two upregulated targets of miR-574-5p, TRAF3 and TGFBR2 is involved in NF-κB signaling pathway (Figure 4). MEG3 can act as a sponge to promote upregulation of TRAF3 [111] and TGFBR2 [112]. BCL3, a target of miR-197-5p, is involved in regulation of cell proliferation and participates in NF-κB signaling pathway (Figure 4D).
MAPK pathway positively regulates virus replication in diverse group of viruses [113]. Three upregulated gene targets of let-7c-5p, MYC, PDGFB and DUSP1, belong to MAPK signaling pathway (Figure 4A). LDLR, involved with cholesterol homeostasis, is targeted by miR-197-5p and miR-223-5p (Figure 4).
Discussion
Long noncoding RNAs are regulators and modulators of complex cellular pathways and viral infection is no exception [19,20]. Dysregulation of lncRNA expression and interaction, either directly with viral RNA, or indirectly with host RNA, affect the progression of viral infection [12]. LncRNAs can work as a sponge for miRNA, bind protein as a competitive inhibitor, inhibit PPI, influence post-translational modification, or affect the activity of target protein. They can modulate viral life cycle, regulate innate immune response or assist adaptive immunity [18,21]. In our exploration of the deregulated lncRNA in SARS-CoV-2 infected NHBE cells, all of these functionalities come to fore. In our study, we find lncRNA-interacting DE protein-coding genes and SARS-CoV-2 protein interactors to be involved in various pathways. Both RNA–RNA and RNA–protein interactions were present, indicating a complex interplay between the components involved. Additionally, these lncRNAs may perform as sponges to protect upregulated genes that would otherwise be targeted by virally-induced miRNAs. Our findings shed light on the possible role of DE lncRNAs in specific processes involved with viral infection, proliferation and cellular response.
SARS-CoV-2 Nsp7-Nsp8-Nsp12 proteins interact to form a multi-subunit RNA-synthesis complex, where Nsp12 works as the RNA-dependent RNA polymerase. EXOSC5, interactor of Nsp8 protein, is a noncatalytic component of the RNA exosome complex. RNA exosome complex is involved in 3′ processing of various stable RNA species and is crucial for RNA quality control in the nucleus [114], thus Nsp8 binding may be fundamental to diminishing the capacity of exosome to act against viral mRNAs. Antiviral drug remdesivir has been predicted to target and disassemble this complex [115,116]. Interaction of EXOSC5 protein with KCNQ1OT1 and SNHG8 can modulate this interaction. The putative RNA–RNA interaction between upregulated AC006058.1 and MALL mRNA, a gene involved in cholesterol homeostasis and membrane trafficking, can exert an influence in maturation of SARS-CoV-2, an enveloped virus. FAM230J, which has no reported function, is downregulated in the infected cells. Its absence may activate the upregulated KYNU, which is involved in metabolite biosynthesis pathways, through lack of RNA–RNA interaction.
Host RNA-binding proteins that interact with SARS-CoV-2 proteins are involved in regulating viral transcription and mRNA stability. This is evident through the interaction of SARS-CoV-2 nucleocapsid (N) protein with UPF1 and MOV10 proteins. In case for murine hepatitis virus, a model coronavirus, the N protein carries out this interaction to inhibit nonsense-mediated decay (NMD) of viral mRNAs containing multiple stop codons, thus favoring viral mRNA transcription. NMD pathways recognized cytoplasmic viral mRNAs as a substrate for degradation, but viral replication induced the inhibition of the NMD pathway through N protein [117]. In human T-lymphotropic virus type 1-infected cells, viral protein tax bound to components of NMD pathways, including UPF1, to inhibit the process [118]. SARS-CoV-2 Membrane (M) protein interacts with AKAP8L, which assists in viral infection progression through favoring transcription. Both LARP7 (nsp8 interactor) and LARP4B (nsp12 interactor) are RNA-binding proteins involved in RNA transcription regulation. Interaction of these RNA-binding proteins with the DE lncRNAs may be crucial for viral mRNA transcription and stability against cellular defense.
ADAR can be regarded as the ‘Editor-in-Chief’ of innate immunity against viral infection [119]. MEG3 was found to act as a biomarker and regulate cell functions by targeting ADAR in colorectal cancer. The cells overexpressing MEG3 exhibited increased ADAR expression, and downregulation of MEG3 was found in colorectal cancer tissues, cell lines and serum [120]. In the infected cells, MEG3 upregulation may lead to ADAR overexpression, which could have been favorable to the virus, as evidenced in influenza A [121]. INHBA-AS1 upregulation in virus-infected cells may also contribute to cell survival through interaction with ADAR, as evidenced in gastric cancer [122] and oral squamous cell carcinoma [123]. As the role of ADAR as a controlling element in cellular response to viral infection is paramount, its regulation by MEG3 and interactions with lncRNAs in SARS-CoV-2 infected cells may influence progression of the disease.
Cell survival can be both beneficial to virus and be inhibitory. Viruses need the cell to survive for a certain period to undergo replication, but they also need apoptosis to exit the cell and infect others. Thus, cell survival and apoptosis becomes a key indicator of pathogenesis. Among lncRNAs, downregulated SNHG8 has possible link to apoptosis and cell death [124], as does KCNQ1OT1 [125–128]. Unlike both, downregulation of GAS6-AS1 may be linked with cell proliferation and survival [129,130]. All three interact with YWHAG, which is involved in signal transduction. IL-6/JAK/STAT3 signaling, Wnt signaling pathway, and PI3K-Akt signaling, involved in cell survival, are also protected by probable ceRNA function by lncRNA.
Cellular response to viral infection can make or break the progression of viral life cycle. Innate immune response is inevitable, but the complex interactions underlying its activation and effect may have been the target of lncRNAs. HECTD1 is an E3 ubiquitin-protein ligase that interacts with Nsp8. As it is involved in class I MHC mediated antigen processing and presentation and innate immune system, the binding may lead to modulation of that response. In case of HECTD1 mRNA, absence of RNA–RNA interaction with downregulated KCNQ1OT1 lncRNA can facilitate the response. PTGES2 gene was upregulated and also found to interact with SARS-CoV-2 Nsp7 protein. As it is involved in innate immune system and signaling pathways, this binding may exert an indirect influence. TLR2, as part of the innate immune response, has been connected to antiviral action against multiple viral infections [131–134]. KCNQ1OT1, the lncRNA having putative RNA–RNA interaction with TLR2, was found to attenuate sepsis-induced myocardial injury via regulating miR-192-5p/XIAP axis. Downregulation of the lncRNA advanced cardiac injury by allowing miR-192-5p to target XIAP [135]. XIAP functions in the inhibition of apoptosis, whereas TLR2 is known to promote apoptosis. The downregulation of KCNQ1OT1 in infected cells thus will promote apoptosis, in accordance with its interaction with overexpressed TLR2 mRNA. Virally-induced microRNAs can also be involved in targeting pathways related with inflammation and host defense. Response by NF-κB signaling pathway against the viral infection is probably protected by lncRNA against inhibition by these miRNAs.
DE lncRNAs also had probable involvement in regulating cellular processes, including signaling. The downregulation of UGDH-AS1 can be related to EDN1 overexpression, which can be crucial in vasoconstriction, leading to severe symptoms, as seen in SARS-CoV-2 infections [136]. SARS-CoV-2 Nsp13 protein works as a viral helicase and interacts with MIPOL1 and PRKAR2A. Function of MIPOL1 is unclear, whereas PRKAR2A is involved in cAMP-mediated signaling, which is the probable target for modulation by Nsp13. The RNA–RNA interactions of downregulated KCNQ1OT1 with the mRNAs of these two genes may have implications on viral infection progression. Additionally, MAPK pathway is probably regulated through lncRNA involvement.
Growing evidence indicates that the ceRNA regulation mechanism plays a role in disease progression and drug efficiency [137–140]. We checked if any of the DE lncRNAs in the infected cells were exerting such effects to ensure a particular miRNA does not downregulate a gene. In turn, the upregulated genes play a crucial role in viral replication, disease progression and immune response. Although some of these miRNAs also target a few downregulated genes, their decrease in expression may be controlled by separate mechanisms. There were previous instances where some of these miRNAs were blocked by a specific lncRNA that was deregulated in the infected cells. For instance, KCNQ1OT1 was found to regulate Rab14 expression, a target of miR-185-5p, in oral squamous cell carcinoma cells as a ceRNA [141]. MEG3 aggravated palmitate-induced insulin resistance by regulating miR-185-5p/Egr2 axis as a ceRNA in insulin-resistant hepatocytes [142]. Additionally, miRNA let-7c-5p was found to negatively regulate NLRC5 expression in ethanol-induced hepatic injury, where MEG3 functioned as a ceRNA against the miRNA [143]. MEG3 acted as an endogenous sponge to suppress the function of miR-223 and to increase NLRP3 expression and enhance endothelial cell pyroptosis in ox-LDL-treated human aortic endothelial cells [144]. KCNQ1OT1 plays the role of a ceRNA to regulate MYC, an upregulated target of let-7c-5p, let-7f-5p and miR-185-5p [102]. We can deduce that these lncRNAs can be involved in a similar role in infected cells.
Conclusion
Our findings explore the molecular footprint of SARS-CoV-2 infection from the lens of lncRNAs. The integrated analysis has linked multiple actors in the complex interplay of molecules in exacerbating the infection and identified possible drug targets. We trace the involvement of lncRNAs with cellular behavior in this situation and illuminate their role in cell survival, viral replication and immune defense. The lethality and swift transmission of this virus is entwined with the deregulated cellular environment, and lncRNA regulation is crucial for understanding its parameters. Our results could provide insights for scheming some novel RNA therapeutics during the lacking of effective cure option.
Future perspective
SARS-CoV-2 has wreaked havoc across the world within a short period. This virus may persist in the population and assume a seasonal nature. The dysregulation in cellular level is ultimately responsible for the disease, and lncRNAs are crucial pieces of the puzzle. The lncRNAs can control cellular response and determine disease outcome. Our elucidation of these molecular connections can deepen our understanding of the virus and the nature of its infection. Effective treatment methods of the future will certainly rely on this knowledge.
Summary points.
21 long noncoding RNAs (lncRNAs) are dysregulated in SARS-CoV-2 infected lung epithelial cells.
These lncRNAs are involved in cell survival and regulation of gene expression.
Six differentially expressed genes interact with these lncRNAs.
Ten proteins that interact with SARS-CoV-2 viral proteins also interact with these lncRNAs.
These protein-coding genes function in cellular signaling, metabolism, immune response and RNA homeostasis.
The lncRNAs can act as competing endogenous RNAs for virally induced microRNAs.
Effect of these microRNAs center on host defense and cell survival.
The interactions affect cellular processes underlying disease progression.
Footnotes
Author contributions
ABMMK Islam conceived the project. ABMMK Islam and RR Turjya designed the workflow. RR Turjya, MAAK Khan and ABMMK Islam performed the analyses. RR Turjya, MAAK Khan and ABMMK Islam wrote the manuscript. All authors read and approved the final manuscript.
Financial & competing interests disclosure
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
No writing assistance was utilized in the production of this manuscript.
References
- 1.Dong E, Du H, Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Inf Dis. 20(5), 533–534 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Du Toit A. Outbreak of a novel coronavirus. Nat. Rev. Microbiol. 18(3), 123 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu F, Zhao S, Yu B. et al. A new coronavirus associated with human respiratory disease in China. Nature 579(7798), 265–269 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhong NS, Zheng BJ, Li YM. et al. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People's Republic of China, in February, 2003. Lancet 362(9393), 1353–1358 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rothan HA, Byrareddy SN. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J. Autoimmun. 109, 102433 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kumar S, Nyodu R, Maurya VK, Saxena SK. Host immune response and immunobiology of human SARS-CoV-2 infection. Coronavirus Disease 2019 (COVID-19) 43–53 (2020) (Epub ahead of print). [Google Scholar]
- 7.Huang C, Wang Y, Li X. et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395(10223), 497–506 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li H, Liu L, Zhang D. et al. SARS-CoV-2 and viral sepsis: observations and hypotheses. Lancet 395(10235), 1517–1520 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou F, Yu T, Du R. et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet 395(10229), 1054–1062 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang N, Li D, Wang X, Sun Z. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J. Thromb. Haemost. 18(4), 844–847 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharma N, Singh SK. Implications of non-coding RNAs in viral infections. Rev. Med. Virol. 26(5), 356–368 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Damas ND, Fossat N, Scheel TKH. Functional interplay between RNA viruses and non-coding RNA in mammals. Noncoding RNA 5(1), 1 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma L, Bajic VB, Zhang Z. On the classification of long non-coding RNAs. RNA Biol. 10(6), 925–933 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ernst C, Morton CC. Identification and function of long non-coding RNA. Front. Cell. Neurosci. 7(168), 168 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol. Cell. 43(6), 904–914 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dhanoa JK, Sethi RS, Verma R, Arora JS, Mukhopadhyay CS. Long non-coding RNA: its evolutionary relics and biological implications in mammals: a review. J. Anim. Sci. Technol. 60, 25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 146(3), 353–358 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang P. The opening of pandora's box: an emerging role of long noncoding rna in viral infections. Front. Immunol. 9(3138), 3138 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu W, Ding C. Roles of LncRNAs in viral infections. Front. Cell Infect. Microbiol. 7(205), 205 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yi K, Zhang Y, Wang Y. et al. Long noncoding RNA and its role in virus infection and pathogenesis. Front. Biosci. (Landmark Ed) 24, 777–789 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Qiu L, Wang T, Tang Q, Li G, Wu P, Chen K. Long non-coding rnas: regulators of viral infection and the interferon antiviral response. Front. Microbiol. 9, 1621 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peng X, Gralinski L, Armour CD. et al. Unique signatures of long noncoding RNA expression in response to virus infection and altered innate immune signaling. mBio 1(5), e00206–00210 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bartoszewski R, Dabrowski M, Jakiela B. et al. SARS-CoV-2 may regulate cellular responses through depletion of specific host miRNAs. Am. J. Physiol. Lung Cell. Mol. Physiol. 319(3), L444–L455 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khan Ma-a-K, Sany MRU, Islam MS, Islam ABMMK. Epigenetic regulator mirna pattern differences among SARS-CoV, SARS-CoV-2, and SARS-CoV-2 world-wide isolates delineated the mystery behind the epic pathogenicity and distinct clinical characteristics of pandemic COVID-19. Front. Genet. 11(765), (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chow JT-S, Salmena L. Prediction and Analysis of SARS-CoV-2-targeting MicroRNA in human lung epithelium. Genes 11(9), 1002 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blanco-Melo D, Nilsson-Payant BE, Liu WC. et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell 181(5), 1036–1045 e1039 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barrett T, Wilhite SE, Ledoux P. et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 41 D991–995 (2013) (Database issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25(9), 1105–1111 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lander ES, Linton LM, Birren B. et al. Initial sequencing and analysis of the human genome. Nature 409 (6822), 860–921 (2001). [DOI] [PubMed] [Google Scholar]
- 30.Hansen KD, Brenner SE, Dudoit S. Biases in Illumina transcriptome sequencing caused by random hexamer priming. Nucleic Acids Res. 38(12), e131 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hubbard TJ, Aken BL, Beal K. et al. Ensembl 2007. Nucleic Acids Res. 35, D610–617 (2007) (Database issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liao Y, Smyth GK, Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 41(10), e108 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 11(10), R106 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gordon DE, Jang GM, Bouhaddou M. et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 583(7816), 459–468 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 34 D535–539 (2006) (Database issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Szklarczyk D, Gable AL, Lyon D. et al. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47(D1), D607–D613 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shannon P, Markiel A, Ozier O. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13(11), 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Teng X, Chen X, Xue H. et al. NPInter v4.0: an integrated database of ncRNA interactions. Nucleic Acids Res. 48(D1), D160–D165 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma L, Cao J, Liu L. et al. LncBook: a curated knowledgebase of human long non-coding RNAs. Nucleic Acids Res. 47 (D1), D128–D134 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Das SS, Saha P, Chakravorty N. miRwayDB: a database for experimentally validated microRNA-pathway associations in pathophysiological conditions. Database (Oxford). 2018, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Girardi E, Lopez P, Pfeffer S. On the importance of Host MicroRNAs during viral infection. Front. Genet. 9, 439 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leon-Icaza SA, Zeng M, Rosas-Taraco AG. microRNAs in viral acute respiratory infections: immune regulation, biomarkers, therapy, and vaccines. ExRNA 1(1), 1 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang HY, Lin YC, Li J. et al. miRTarBase 2020: updates to the experimentally validated microRNA-target interaction database. Nucleic Acids Res. 48(D1), D148–D154 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karagkouni D, Paraskevopoulou MD, Tastsoglou S. et al. DIANA-LncBase v3: indexing experimentally supported miRNA targets on non-coding transcripts. Nucleic Acids Res. 48(D1), D101–D110 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou G, Soufan O, Ewald J, Hancock REW, Basu N, Xia J. NetworkAnalyst 3.0: a visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res. 47(W1), W234–W241 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Samuel CE. Adenosine deaminase acting on RNA (ADAR1), a suppressor of double-stranded RNA-triggered innate immune responses. J. Biol. Chem. 294(5), 1710–1720 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liddicoat BJ, Piskol R, Chalk AM. et al. RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science 349(6252), 1115–1120 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taylor DR, Puig M, Darnell ME, Mihalik K, Feinstone SM. New antiviral pathway that mediates hepatitis C virus replicon interferon sensitivity through ADAR1. J. Virol. 79(10), 6291–6298 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li Z, Wolff KC, Samuel CE. RNA adenosine deaminase ADAR1 deficiency leads to increased activation of protein kinase PKR and reduced vesicular stomatitis virus growth following interferon treatment. Virology 396(2), 316–322 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Toth AM, Li Z, Cattaneo R, Samuel CE. RNA-specific adenosine deaminase ADAR1 suppresses measles virus-induced apoptosis and activation of protein kinase PKR. J. Biol. Chem. 284(43), 29350–29356 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li Z, Okonski KM, Samuel CE. Adenosine deaminase acting on RNA 1 (ADAR1) suppresses the induction of interferon by measles virus. J. Virol. 86(7), 3787–3794 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hartwig D, Schutte C, Warnecke J. et al. The large form of ADAR 1 is responsible for enhanced hepatitis delta virus RNA editing in interferon-alpha-stimulated host cells. J. Viral Hepat. 13(3), 150–157 (2006). [DOI] [PubMed] [Google Scholar]
- 53.Doria M, Neri F, Gallo A, Farace MG, Michienzi A. Editing of HIV-1 RNA by the double-stranded RNA deaminase ADAR1 stimulates viral infection. Nucleic Acids Res. 37(17), 5848–5858 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Doria M, Tomaselli S, Neri F. et al. ADAR2 editing enzyme is a novel human immunodeficiency virus-1 proviral factor. J. Gen. Virol. 92(Pt 5), 1228–1232 (2011). [DOI] [PubMed] [Google Scholar]
- 55.Rufanova VA, Alexanian A, Wakatsuki T, Lerner A, Sorokin A. Pyk2 mediates endothelin-1 signaling via p130Cas/BCAR3 cascade and regulates human glomerular mesangial cell adhesion and spreading. J. Cell. Physiol. 219(1), 45–56 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chester AH, Yacoub MH. The role of endothelin-1 in pulmonary arterial hypertension. Glob. Cardiol. Sci. Pract. 2014(2), 62–78 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Walsh HA, Botting NP. Purification and biochemical characterization of some of the properties of recombinant human kynureninase. Eur. J. Biochem. 269(8), 2069–2074 (2002). [DOI] [PubMed] [Google Scholar]
- 58.Toma S, Nakamura M, Tone S. et al. Cloning and recombinant expression of rat and human kynureninase. FEBS Lett. 408(1), 5–10 (1997). [DOI] [PubMed] [Google Scholar]
- 59.De Marco MC, Kremer L, Albar JP. et al. BENE, a novel raft-associated protein of the MAL proteolipid family, interacts with caveolin-1 in human endothelial-like ECV304 cells. J. Biochem. 276(25), 23009–23017 (2001). [DOI] [PubMed] [Google Scholar]
- 60.Saunier S, Calado J, Heilig R. et al. A novel gene that encodes a protein with a putative src homology 3 domain is a candidate gene for familial juvenile nephronophthisis. Hum. Mol. Genet. 6(13), 2317–2323 (1997). [DOI] [PubMed] [Google Scholar]
- 61.Oliveira-Nascimento L, Massari P, Wetzler LM. The role of TLR2 in infection and immunity. Front. Immunol. 3, 79 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lancioni CL, Li Q, Thomas JJ. et al. Mycobacterium tuberculosis lipoproteins directly regulate human memory CD4(+) T cell activation via Toll-like receptors 1 and 2. Infect. Immun. 79(2), 663–673 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Drage MG, Pecora ND, Hise AG. et al. TLR2 and its co-receptors determine responses of macrophages and dendritic cells to lipoproteins of Mycobacterium tuberculosis. Cell. Immunol. 258(1), 29–37 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brightbill HD, Libraty DH, Krutzik SR. et al. Host defense mechanisms triggered by microbial lipoproteins through toll-like receptors. Science 285(5428), 732–736 (1999). [DOI] [PubMed] [Google Scholar]
- 65.Aliprantis AO, Yang RB, Mark MR. et al. Cell activation and apoptosis by bacterial lipoproteins through toll-like receptor-2. Science 285(5428), 736–739 (1999). [DOI] [PubMed] [Google Scholar]
- 66.Strochlic L, Cartaud A, Mejat A. et al. 14-3-3 gamma associates with muscle specific kinase and regulates synaptic gene transcription at vertebrate neuromuscular synapse. Proc. Natl Acad. Sci. USA 101(52), 18189–18194 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Howlett KF, Sakamoto K, Garnham A, Cameron-Smith D, Hargreaves M. Resistance exercise and insulin regulate AS160 and interaction with 14-3-3 in human skeletal muscle. Diabetes 56(6), 1608–1614 (2007). [DOI] [PubMed] [Google Scholar]
- 68.Autieri MV, Carbone CJ. 14-3-3Gamma interacts with and is phosphorylated by multiple protein kinase C isoforms in PDGF-stimulated human vascular smooth muscle cells. DNA Cell Biol. 18(7), 555–564 (1999). [DOI] [PubMed] [Google Scholar]
- 69.Yoshida K, Yamaguchi T, Natsume T, Kufe D, Miki Y. JNK phosphorylation of 14-3-3 proteins regulates nuclear targeting of c-Abl in the apoptotic response to DNA damage. Nat. Cell Biol. 7(3), 278–285 (2005). [DOI] [PubMed] [Google Scholar]
- 70.Subramanian RR, Masters SC, Zhang H, Fu H. Functional conservation of 14-3-3 isoforms in inhibiting bad-induced apoptosis. Exp. Cell Res. 271(1), 142–151 (2001). [DOI] [PubMed] [Google Scholar]
- 71.Kakinuma N, Roy BC, Zhu Y, Wang Y, Kiyama R. Kank regulates RhoA-dependent formation of actin stress fibers and cell migration via 14-3-3 in PI3K-Akt signaling. J. Cell Biol. 181(3), 537–549 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yoo JO, Kwak SY, An HJ, Bae IH, Park MJ, Han YH. miR-181b-3p promotes epithelial-mesenchymal transition in breast cancer cells through Snail stabilization by directly targeting YWHAG. Biochim. Biophys. Acta 1863 (7 Pt A), 1601–1611 (2016). [DOI] [PubMed] [Google Scholar]
- 73.Bai Q, Yu J, Li Y, Ma J, Gou Y. MicroRNA-182 promoted esophageal squamous cell carcinoma cell growth and metastasis via targeting YWHAG. J. BUON 23(5), 1439–1447 (2018). [PubMed] [Google Scholar]
- 74.Lu Z, Zhang QC, Lee B. et al. RNA duplex map in living cells reveals higher-order transcriptome structure. Cell 165(5), 1267–1279 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Westberg C, Yang JP, Tang H, Reddy TR, Wong-Staal F. A novel shuttle protein binds to RNA helicase A and activates the retroviral constitutive transport element. J. Biol. Chem. 275(28), 21396–21401 (2000). [DOI] [PubMed] [Google Scholar]
- 76.Yang JP, Tang H, Reddy TR, Wong-Staal F. Mapping the functional domains of HAP95, a protein that binds RNA helicase A and activates the constitutive transport element of type D retroviruses. J. Biol. Chem. 276(33), 30694–30700 (2001). [DOI] [PubMed] [Google Scholar]
- 77.Han I, Xue Y, Harada S, Orstavik S, Skalhegg B, Kieff E. Protein kinase A associates with HA95 and affects transcriptional coactivation by Epstein-Barr virus nuclear proteins. Mol. Cell. Biol. 22(7), 2136–2146 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xing L, Zhao X, Guo F, Kleiman L. The role of A-kinase anchoring protein 95-like protein in annealing of tRNALys3 to HIV-1 RNA. Retrovirology 11, 58 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mukherjee D, Gao M, O'connor JP. et al. The mammalian exosome mediates the efficient degradation of mRNAs that contain AU-rich elements. EMBO J. 21(1–2), 165–174 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tsai VW, Macia L, Johnen H. et al. TGF-b superfamily cytokine MIC-1/GDF15 is a physiological appetite and body weight regulator. PloS ONE 8(2), e55174 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Baek SJ, Eling T. Growth differentiation factor 15 (GDF15): a survival protein with therapeutic potential in metabolic diseases. Pharmacol. Ther. 198, 46–58 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Adela R, Banerjee SK. GDF-15 as a target and biomarker for diabetes and cardiovascular diseases: a translational prospective. J. Diabetes Res. 2015, 490842 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mullican SE, Lin-Schmidt X, Chin CN. et al. GFRAL is the receptor for GDF15 and the ligand promotes weight loss in mice and nonhuman primates. Nat. Med. 23(10), 1150–1157 (2017). [DOI] [PubMed] [Google Scholar]
- 84.Sarkar AA, Zohn IE. Hectd1 regulates intracellular localization and secretion of Hsp90 to control cellular behavior of the cranial mesenchyme. Journal Cell Biol. 196(6), 789–800 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kuspert M, Murakawa Y, Schaffler K. et al. LARP4B is an AU-rich sequence associated factor that promotes mRNA accumulation and translation. RNA 21(7), 1294–1305 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Koso H, Yi H, Sheridan P. et al. Identification of RNA-binding protein larp4b as a tumor suppressor in glioma. Cancer Res. 76(8), 2254–2264 (2016). [DOI] [PubMed] [Google Scholar]
- 87.Markert A, Grimm M, Martinez J. et al. The La-related protein LARP7 is a component of the 7SK ribonucleoprotein and affects transcription of cellular and viral polymerase II genes. EMBO Rep. 9(6), 569–575 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.He N, Jahchan NS, Hong E. et al. A La-related protein modulates 7SK snRNP integrity to suppress P-TEFb-dependent transcriptional elongation and tumorigenesis. Mol. Cell 29(5), 588–599 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Leong MML, Cheung AKL, Kwok TCT, Lung ML. Functional characterization of a candidate tumor suppressor gene, Mirror Image Polydactyly 1, in nasopharyngeal carcinoma. Int. J. Cancer 146(10), 2891–2900 (2020). [DOI] [PubMed] [Google Scholar]
- 90.Lykke-Andersen J, Shu MD, Steitz JA. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell 103(7), 1121–1131 (2000). [DOI] [PubMed] [Google Scholar]
- 91.Chakrabarti S, Jayachandran U, Bonneau F. et al. Molecular mechanisms for the RNA-dependent ATPase activity of Upf1 and its regulation by Upf2. Mol. Cell 41(6), 693–703 (2011). [DOI] [PubMed] [Google Scholar]
- 92.Kashima I, Yamashita A, Izumi N. et al. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 20(3), 355–367 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gregersen LH, Schueler M, Munschauer M. et al. MOV10 Is a 5′ to 3′ RNA helicase contributing to UPF1 mRNA target degradation by translocation along 3′ UTRs. Mol. Cell 54(4), 573–585 (2014). [DOI] [PubMed] [Google Scholar]
- 94.Meister G, Landthaler M, Peters L. et al. Identification of novel argonaute-associated proteins. Curr. Biol. 15(23), 2149–2155 (2005). [DOI] [PubMed] [Google Scholar]
- 95.Liu C, Zhang X, Huang F. et al. APOBEC3G inhibits microRNA-mediated repression of translation by interfering with the interaction between Argonaute-2 and MOV10. J. Biol. Chem. 287(35), 29373–29383 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chendrimada TP, Finn KJ, Ji X. et al. MicroRNA silencing through RISC recruitment of eIF6. Nature 447(7146), 823–828 (2007). [DOI] [PubMed] [Google Scholar]
- 97.Balistreri G, Bognanni C, Muhlemann O. Virus escape and manipulation of cellular nonsense-mediated mRNA decay. Viruses 9(1), 24 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cuevas RA, Ghosh A, Wallerath C, Hornung V, Coyne CB, Sarkar SN. MOV10 provides antiviral activity against RNA viruses by enhancing RIG-I-MAVS-independent IFN induction. J. Immunol. 196(9), 3877–3886 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature 505(7483), 344–352 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Johnson DE, O'keefe RA, Grandis JR. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat. Rev. Clin. Oncol. 15(4), 234–248 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Velazquez-Salinas L, Verdugo-Rodriguez A, Rodriguez LL, Borca MV. The role of interleukin 6 during viral infections. Front. Microbiol. 10, 1057 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cheng P, Lu P, Guan J. et al. LncRNA KCNQ1OT1 controls cell proliferation, differentiation and apoptosis by sponging miR-326 to regulate c-Myc expression in acute myeloid leukemia. Neoplasma 67(2), 238–248 (2020). [DOI] [PubMed] [Google Scholar]
- 103.Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell 139(4), 693–706 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pecina-Slaus N. Wnt signal transduction pathway and apoptosis: a review. Cancer Cell Int. 10, 22 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ljungberg JK, Kling JC, Tran TT, Blumenthal A. Functions of the WNT signaling network in shaping host responses to infection. Front. Immunol. 10(2521), 2521 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ji WT, Liu HJ. PI3K-Akt signaling and viral infection. Rec. Pat. Biotechnol. 2(3), 218–226 (2008). [DOI] [PubMed] [Google Scholar]
- 107.Ma X, Shen D, Li H. et al. MicroRNA-185 inhibits cell proliferation and induces cell apoptosis by targeting VEGFA directly in von Hippel-Lindau-inactivated clear cell renal cell carcinoma. Urol. Oncol. 33(4), 169 e161–111 (2015). [DOI] [PubMed] [Google Scholar]
- 108.Liu T, Zhang L, Joo D, Sun SC. NF-kappaB signaling in inflammation. Signal Transduct. Target. Ther. 2(1), 17023 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Freaney JE, Kim R, Mandhana R, Horvath CM. Extensive cooperation of immune master regulators IRF3 and NFκB in RNA Pol II recruitment and pause release in human innate antiviral transcription. Cell Rep. 4(5), 959–973 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rajsbaum R, Garcia-Sastre A. Viral evasion mechanisms of early antiviral responses involving regulation of ubiquitin pathways. Trends Microbiol. 21(8), 421–429 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lu WX. Long non-coding RNA MEG3 represses cholangiocarcinoma by regulating miR-361-5p/TRAF3 axis. Eur. Rev. Med. Pharmacol. Sci. 23(17), 7356–7368 (2019). [DOI] [PubMed] [Google Scholar]
- 112.Chen K, Zhu H, Zheng MQ, Dong QR. LncRNA MEG3 inhibits the degradation of the extracellular matrix of chondrocytes in osteoarthritis via targeting miR-93/TGFBR2 axis. Cartilage (2019) (Epub ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kumar R, Khandelwal N, Thachamvally R. et al. Role of MAPK/MNK1 signaling in virus replication. Virus Res. 253, 48–61 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kilchert C, Wittmann S, Vasiljeva L. The regulation and functions of the nuclear RNA exosome complex. Nat. Rev. Mol. Cell Biol. 17(4), 227–239 (2016). [DOI] [PubMed] [Google Scholar]
- 115.Olotu F, Omolabi K, Soliman M. Piece of the Puzzle: remdesivir disassemble the multimeric SARS-CoV-2 RNA-dependent RNA polymerase non-structural proteins (RdRp-NSPs) complex. Research Square (2020) (Epub ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yin W, Mao C, Luan X. et al. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science 368(6498), 1499–1504 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wada M, Lokugamage KG, Nakagawa K, Narayanan K, Makino S. Interplay between coronavirus, a cytoplasmic RNA virus, and nonsense-mediated mRNA decay pathway. Proc. Natl Acad. Sci. USA 115(43), E10157–E10166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Mocquet V, Neusiedler J, Rende F. et al. The human T-lymphotropic virus type 1 tax protein inhibits nonsense-mediated mRNA decay by interacting with INT6/EIF3E and UPF1. J. Virol. 86(14), 7530–7543 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lamers MM, Van Den Hoogen BG, Haagmans BL. ADAR1: ‘editor-in-chief” of cytoplasmic innate immunity. Front. Immunol. 10(1763), 1763 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wang W, Xie Y, Chen F. et al. LncRNA MEG3 acts a biomarker and regulates cell functions by targeting ADAR1 in colorectal cancer. World J. Gastroenterol. 25(29), 3972–3984 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.De Chassey B, Aublin-Gex A, Ruggieri A. et al. The interactomes of influenza virus NS1 and NS2 proteins identify new host factors and provide insights for ADAR1 playing a supportive role in virus replication. PLoS Pathog. 9(7), e1003440 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ke D, Li H, Zhang Y. et al. The combination of circulating long noncoding RNAs AK001058, INHBA-AS1, MIR4435-2HG, and CEBPA-AS1 fragments in plasma serve as diagnostic markers for gastric cancer. Oncotarget 8(13), 21516–21525 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ma WQ, Chen J, Fang W. et al. LncRNA INHBA-AS1 promotes cell growth, migration, and invasion of oral squamous cell carcinoma by sponging miR-143-3p. Eur. Rev. Med. Pharmacol. Sci. 24(4), 1821–1828 (2020). [DOI] [PubMed] [Google Scholar]
- 124.Liu J, An P, Xue Y. et al. Mechanism of Snhg8/miR-384/Hoxa13/FAM3A axis regulating neuronal apoptosis in ischemic mice model. Cell Death Dis. 10(6), 441 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Li C, Miao R, Zhang J, Qu K, Liu C. Long non-coding RNA KCNQ1OT1 mediates the growth of hepatocellular carcinoma by functioning as a competing endogenous RNA of miR-504. Int. J. Oncol. 52(5), 1603–1612 (2018). [DOI] [PubMed] [Google Scholar]
- 126.Feng W, Wang C, Liang C. et al. The dysregulated expression of KCNQ1OT1 and its interaction with downstream factors miR-145/CCNE2 in breast cancer cells. Cell Physiol. Biochem. 49(2), 432–446 (2018). [DOI] [PubMed] [Google Scholar]
- 127.Sun H, Li Y, Kong H, Dai S, Qian H. Dysregulation of KCNQ1OT1 promotes cholangiocarcinoma progression via miR-140-5p/SOX4 axis. Arch. Biochem. Biophys. 658, 7–15 (2018). [DOI] [PubMed] [Google Scholar]
- 128.Dong Z, Yang P, Qiu X. et al. KCNQ1OT1 facilitates progression of non-small-cell lung carcinoma via modulating miRNA-27b-3p/HSP90AA1 axis. J. Cell. Physiol. 234(7), 11304–11314 (2019). [DOI] [PubMed] [Google Scholar]
- 129.Han L, Kong R, Yin DD. et al. Low expression of long noncoding RNA GAS6-AS1 predicts a poor prognosis in patients with NSCLC. Med. Oncol. 30(4), 694 (2013). [DOI] [PubMed] [Google Scholar]
- 130.Li X, Zhang R, Liu Z, Li C, Xu H. Low expression of long noncoding RNA GAS6-AS1 as a novel biomarker of poor prognosis for breast cancer. Int. J. Clin. Exp. Med. 9, 15820–15827 (2016). [Google Scholar]
- 131.Ge Y, Mansell A, Ussher JE. et al. Rotavirus NSP4 triggers secretion of proinflammatory cytokines from macrophages via Toll-like receptor 2. J. Virol. 87(20), 11160–11167 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kurt-Jones EA, Popova L, Kwinn L. et al. Pattern recognition receptors TLR4 and CD14 mediate response to respiratory syncytial virus. Nat. Immunol. 1(5), 398–401 (2000). [DOI] [PubMed] [Google Scholar]
- 133.Mogensen TH, Paludan SR. Reading the viral signature by toll-like receptors and other pattern recognition receptors. J. Mol. Med. (Berl). 83(3), 180–192 (2005). [DOI] [PubMed] [Google Scholar]
- 134.Bieback K, Lien E, Klagge IM. et al. Hemagglutinin protein of wild-type measles virus activates toll-like receptor 2 signaling. J. Virol. 76(17), 8729–8736 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Sun F, Yuan W, Wu H. et al. LncRNA KCNQ1OT1 attenuates sepsis-induced myocardial injury via regulating miR-192-5p/XIAP axis. Exp. Biol. Med. (Maywood) 245(7), 620–630 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Gattinoni L, Coppola S, Cressoni M, Busana M, Rossi S, Chiumello D. COVID-19 does not lead to a ‘typical’ acute respiratory distress syndrome. Am. J. Respir. Crit. Care Med. 201(10), 1299–1300 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Qi X, Zhang DH, Wu N, Xiao JH, Wang X, Ma W. ceRNA in cancer: possible functions and clinical implications. J. Med. Gen. 52(10), 710–718 (2015). [DOI] [PubMed] [Google Scholar]
- 138.Su Q, Lv X. Revealing new landscape of cardiovascular disease through circular RNA–miRNA–mRNA axis. Genomics 112(2), 1680–1685 (2020). [DOI] [PubMed] [Google Scholar]
- 139.Lang Y, Zhang J, Yuan Z. Construction and dissection of the ceRNAceRNA network reveals critical modules in depression. Mol. Med. Rep. 19(5), 3411–3420 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zhang Y, Zhang H, An M. et al. Crosstalk in competing endogenous RNA networks reveals new circular RNAs involved in the pathogenesis of early HIV infection. J. Transl. Med. 16(1), 332 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bao Q, Liao X, Li R, Ding N. KCNQ1OT1 promotes migration and inhibits apoptosis by modulating miR-185-5p/Rab14 axis in oral squamous cell carcinoma. Dev. Growth Differ. 61(9), 466–474 (2019). [DOI] [PubMed] [Google Scholar]
- 142.Chen DL, Shen DY, Han CK, Tian Y. LncRNA MEG3 aggravates palmitate-induced insulin resistance by regulating miR-185-5p/Egr2 axis in hepatic cells. Eur. Rev. Med. Pharmacol. Sci. 23(12), 5456–5467 (2019). [DOI] [PubMed] [Google Scholar]
- 143.Wang Q, Li M, Shen Z. et al. The long non-coding RNA MEG3/miR-let-7c-5p axis regulates ethanol-induced hepatic steatosis and apoptosis by targeting NLRC5. Front. Pharmacol. 9(302), 302 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zhang Y, Liu X, Bai X. et al. Melatonin prevents endothelial cell pyroptosis via regulation of long noncoding RNA MEG3/miR-223/NLRP3 axis. J. Pineal Res. 64(2), e12449 (2018). [DOI] [PubMed] [Google Scholar]