Abstract
The present study was conducted to clarify the cooperative significance of epigenomic and genomic abnormalities during gastric carcinogenesis. Using 21 samples of normal control gastric mucosa (C), 109 samples of non-cancerous gastric mucosa (N) and 105 samples of cancerous tissue (T) from 109 patients with primary gastric adenocarcinomas, genome-wide DNA methylation analysis was performed using Infinium assay. Among these samples, 66 paired N and corresponding T samples were subjected to whole-exome and single nucleotide polymorphism array analyses. As had been shown in our previous study, 109 patients were clustered clinicopathologically into least aggressive Cluster A (n = 20), most aggressive Cluster B1 (n = 20) and Cluster B2 (n = 69). Most DNA methylation alterations in each cluster had already occurred even in N samples compared with C samples, and DNA methylation alterations at the precancerous N stage were inherited by the established cancers themselves. Recurrent single nucleotide variants and insertions/deletions resulting in functional disruption of the proteins encoded by the ABCA10, BNC2, CDH1, CTNNB1, SMAD4 and VAV2 genes were specific to Cluster B1, whereas those of the APC, EGFR, ERBB2, ERBB3, MLH1 and MUC6 genes were specific to Cluster A. MetaCore pathway analysis revealed that the epigenomically affected TWIST1 gene and genomically affected CDH1, CTNNB1, MMP9, TLN2, ROCK1 and SMAD4 genes were accumulated in signaling pathways related to cell adhesion, cytoskeleton remodeling and epithelial–mesenchymal transition in Cluster B1. These data indicate that epigenomic alterations at the precancerous stage are important in gastric carcinogenesis and that epigenomic and genomic alterations cooperatively underlie the aggressiveness of gastric adenocarcinomas.
Genome-wide analyses have revealed that the epigenomically affected TWIST1 gene and genomically affected CDH1, CTNNB1, MMP9, TLN2, ROCK1, SMAD4 and VAV2 genes cooperatively accumulate in signaling pathways related to cell adhesion, cytoskeleton remodeling and epithelial–mesenchymal transition in aggressive gastric adenocarcinomas.
Introduction
Gastric adenocarcinoma is a highly heterogeneous disease in terms of histological features and clinical phenotype (1). Despite improvements in surgical techniques and chemotherapy, patients with aggressive gastric carcinomas still have a poor clinical outcome (2). Therefore, there is a need to clarify the molecular backgrounds responsible for the clinicopathological diversity of these cancers. We and other groups have revealed that changes in DNA methylation, a major epigenomic event in human cancers, participate even in the very early and precancerous stages during multistage carcinogenesis (3–7). It is well known that the features of gastric carcinoma conform to the concept of field cancerization: in precancerous gastric mucosa, aberrant DNA methylation is reportedly induced by Helicobacter pylori (8) and Epstein–Barr virus (9) infection. Since DNA methylation profiles at the precancerous stage are inherited by the established carcinomas themselves (10–12), thus determining tumor aggressiveness and patient outcome, it is not surprising that the clinicopathological diversity of gastric carcinoma reflects genome-wide DNA methylation profiles. Indeed, our previous study demonstrated that epigenomic clustering of patients with gastric adenocarcinomas based on the genome-wide DNA methylation profile of the non-cancerous gastric mucosa (N) was significantly correlated with the clinicopathological aggressiveness of tumors that had developed from the N of individual patients (13). However, we focused primarily on DNA methylation profiles in N samples already at the precancerous stage based on the field cancerization concept, and those profiles at both the precancerous stage and in established cancer were not compared with normal control gastric mucosa (C) of patients without gastric carcinomas or carcinogenetic factors such as H. pylori or Epstein–Barr virus infection. Therefore, the impacts of epigenomic abnormalities on the clinicopathological diversity of gastric carcinomas themselves have yet to be fully elucidated, and further comparison with C is still needed.
On the other hand, genomic events can, of course, affect clinicopathological features (14). High-throughput sequencing and probe-based microarray technologies have been generally used for detection of alterations in numerous potential tumor-related genes (15–17), and somatic mutations of the TP53, CTNNB1, PIK3CA, RHOA and ARID1A genes and amplification of the ERBB2 gene have been considered important in gastric adenocarcinomas (18). However, large-scale analyses of both genomic and epigenomic events, such as studies conducted by The Cancer Genome Atlas (TCGA), have not fully focused on the cooperative impact of epigenomic and genomic alterations (18).
In the present study, to clarify the molecular backgrounds of clinicopathological diversity based on epigenomic clustering of gastric adenocarcinoma, we performed whole-exome sequencing and single nucleotide polymorphism microarray analysis of gastric adenocarcinomas belonging to each of the clusters defined by genome-wide DNA methylation analysis and clarified the heterogeneity of their clinicopathological features.
Materials and methods
Patient and tissue samples
Twenty-one C samples showing no remarkable histological features were obtained from 21 autopsied patients who were negative for H. pylori or Epstein–Barr virus infection at the Department of Pathology, Keio University School of Medicine. We then obtained 109 N and 105 cancerous tissue (T) samples from 109 patients [79 male, 30 female, median age 66 (range, 26–91) years] with primary gastric adenocarcinomas who had undergone total or partial gastrectomy at the National Cancer Center Hospital, Tokyo, Japan. Histological types were determined based on the World Health Organization classification (19). All tumors were classified according to the pathological Tumor–Node–Metastasis classification (20). None of the patients had received any preoperative treatment. Recurrence was diagnosed by clinicians on the basis of physical examination and imaging modalities such as computed tomography, magnetic resonance imaging, scintigraphy or positron emission tomography, and sometimes confirmed pathologically by biopsy. The clinicopathological parameters of the 109 patients are summarized in Supplementary Table S1, available at Carcinogenesis Online.
Tissue specimens were frozen immediately after autopsy and surgery and preserved in liquid nitrogen tanks at the Department of Pathology, Keio University School of Medicine, and the National Cancer Center Biobank, Tokyo, Japan, respectively, in accordance with the Japanese Society of Pathology Guidelines for the Handling of Pathological Tissue Samples for Genomic Research (21). Written informed consent for the use of these materials was obtained from all of the bereaved families and patients included in this study. The study was approved by the Ethics Committees of Keio University School of Medicine and the National Cancer Center, Tokyo, Japan, and performed in accordance with the Declaration of Helsinki.
Epigenomic clustering of gastric adenocarcinomas
In our previous study, the 109 patients with gastric adenocarcinomas were divided into Clusters A (n = 20), B1 (n = 20) and B2 (n = 69) based on their genome-wide DNA methylation profiles (Supplementary Table S1) (13). Briefly, genomic DNA extracted from fresh frozen tissue samples was subjected to bisulfite conversion using an EZ DNA methylation-Gold™ kit (Zymo Research, Irvine, CA), and then DNA methylation status at 27 578 CpG loci was examined using the Infinium HumanMethylation27 Bead Array (Illumina, San Diego, CA), all in accordance with the manufacturers’ instructions. The specifically hybridized DNA was detected using a Bead Scan reader (Illumina) and the data were assembled using Genome Studio methylation software (Illumina). At each CpG site, the ratio of the fluorescent signal was measured using a methylated probe relative to the sum of the methylated and unmethylated probes, i.e. the so-called β-value, which ranges from 0.00 to 1.00. The Infinium data were deposited in the Integrative Disease Omics Database (iDOx DB, https://gemdbj.ncc.go.jp/omics/). Then unsupervised hierarchical clustering (Euclidean distance, Ward’s method) was performed using the DNA methylation levels in N samples (βN) of the 3861 probes showing significant differences in DNA methylation levels between the 109 N samples and 105 T samples (13).
Since C samples and N and T samples were obtained from different cohorts, that is, patients who underwent autopsy at Keio University and surgical resection at the National Cancer Center, respectively, we examined DNA methylation levels on 12 556 representative probes not showing any correlation with the steps of carcinogenesis or aging. The DNA methylation levels on these probes showed excellent concordance in both N and T samples relative to C samples (correlation coefficient r = 0.994 or more; P < 2.2E-16), indicating that differences in the processing procedures did not affect the DNA methylation profiles (Supplementary Figure S1 is available at Carcinogenesis Online).
Whole-exome sequencing
Whole-exome analysis of genomic DNA was performed for the 66 paired samples of N and the corresponding T from patients who had been included in our previous study (13) and for whom genomic DNA was available even after methylome analysis (Supplementary Table S1 is available at Carcinogenesis Online). About 3 µg aliquots of genomic DNA were fragmented by a Covaris-S2 instrument (Covaris, Woburn, MA) to provide DNA fragments with a base pair peak at 150–200 bp. The DNA fragments were end-repaired and ligated with paired-end adaptors (NEBNext DNA sample prep, New England Biolabs, Ipswich, MA) and then amplified by four cycles of PCR. About 500 ng aliquots of the adaptor-ligated libraries were hybridized with biotinylated oligo RNA bait, SureSelect Human All Exon 50 Mb (Agilent Technologies, Santa Clara, CA). After 10 cycles of PCR reamplification, the whole-exome DNA library was sequenced on an Illumina HiSeq 2000 (Illumina, San Diego, CA) using 75-bp paired-end reads in accordance with the manufacturer’s standard protocols.
After completion of the entire run, image analyses, error estimation and base calling were performed using the Illumina Pipeline (version 1.3.4) to generate primary data. The reads were aligned against the reference human genome from UCSC human genome 19 (hg19) using the Burrows-Wheeler Aligner Multi-Vision software package (22). The Genome Analysis Toolkit (Broad Institute, Cambridge, MA) was used for local alignment (23). Single nucleotide variants (SNVs) and insertions/deletions (indels) were identified using Genome Analysis Toolkit, and SNVs were also identified using MuTect software (Broad Institute) (24). Exome data were deposited in the Integrative Disease Omics Database (iDOx DB, https://gemdbj.ncc.go.jp/omics/). Effects of amino acid substitutions on protein function due to SNVs have been estimated using the Sorting Intolerant from Tolerent (SIFT; https://sift.bii.a-star.edu.sg/) (25) and polymorphism phenotyping (PolyPhen)-2 (http://genetics.bwh.harvard.edu/pph2/) (26), and those due to indels have been estimated using SIFT (27).
Single nucleotide polymorphism microarray analysis
About 200 ng aliquots of DNA were genotyped using the HumanOmniExpressExome-8 BeadChip (Illumina) in accordance with the manufacturer’s protocol. The data were assembled using Genome Studio software (Illumina). Copy number data were obtained using Allele-Specific Copy Number Analysis of Tumors (version 2.5.2; https://www.crick.ac.uk/research/labs/peter-van-loo/software) (28) and the Global Parameter Hidden Markov Model (version 1.3; http://bioinformatics.ustc.edu.cn/gphmm/) (29) software. Genes corresponding to those aberrated segments were identified by BEDTools (version 2.29.0; https://bedtools.readthedocs.io/en/latest/#) (30).
Pathway analysis
MetaCore™ software (version 19.3; Thomson Reuters, NY) is a pathway analysis tool based on a proprietary manually curated database of human protein–protein, protein–DNA and protein compound interactions. MetaCore pathway analysis by GeneGo was performed using genes showing epigenomic and genomic alterations in T samples belonging to each cluster. Such genes were considered significantly enriched in pathways for which the false discovery rate (FDR) was less than 0.05.
Statistical analysis
Differences in average DNA methylation levels between clusters or numbers of genes showing epigenomic and genomic aberrations were examined using Welch’s T-test and P values of less than 0.05 after Bonferroni correction were considered significant. Correlations between epigenomic clustering and clinicopathological parameters were evaluated by Fisher’s exact test. All statistical analyses were performed using the programming language R and SPSS 23 (IBM, Armonk, NY).
Results
DNA methylation profiles of gastric adenocarcinomas in each epigenomic cluster
In our previous study using tissue samples from the same 109 patients, gastric adenocarcinomas belonging to Cluster B1 frequently showed an undifferentiated histology, deeper invasion and a higher pathological tumor–node–metastasis stage in comparison with carcinomas belonging to Clusters A and B2 (13). When we focused on the clinicopathologically most aggressive Cluster B1 and the least aggressive Cluster A, the recurrence-free and overall survival rates for patients in Cluster B1 were significantly lower than those of patients in Cluster A (13). Such epigenomic clustering was established on the basis of DNA methylation data from the Infinium assay of N samples in our previous study.
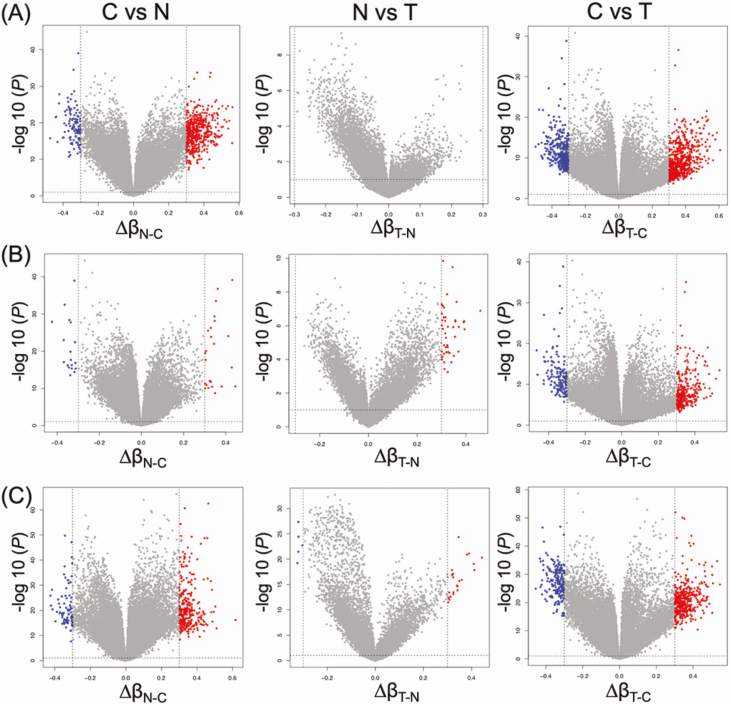
In the present study, we further characterized the DNA methylation profiles of such N samples by comparison with C samples. The volcano plots in Figure 1 clearly showed that DNA methylation alterations occurred in precancerous N samples compared with C samples: 608, 42 and 369 probes showed significant differences in DNA methylation levels in N samples compared with C samples in Clusters A, B1 and B2, respectively (P < 0.05 for Welch’s T test after Bonferroni correction and ∆β N-C≥0.3 or ∆β N-C ≤ −0.3). Supplementary Table S2, available at Carcinogenesis Online, summarizes 568, 45 and 378 genes for which 608, 42 and 369 probes were designed, respectively. Three hundred and fifty, 1 and 157 genes showed DNA methylation alterations at precancerous N stages in Cluster A, B1 and B2-specific manner, respectively: the only Cluster B1-specific gene was PKP2, the desmosomal cell adhesion molecule that is a direct transcriptional target of the Wnt/β-catenin pathway (31).
Figure 1.
Volcano plots of probes showing DNA hypermethylation (P < 0.05 after Bonferroni correction and ∆β ≥ 0.3; red) and DNA hypomethylation (P < 0.05 after Bonferroni correction and ∆β ≤ −0.3; blue) based on Infinium assay in Clusters A (n = 20) (A), B1 (n = 20) (B) and B2 (n = 69) (C). Probes shown by red or blue are listed in Supplementary Tables S2–S4, available at Carcinogenesis Online.
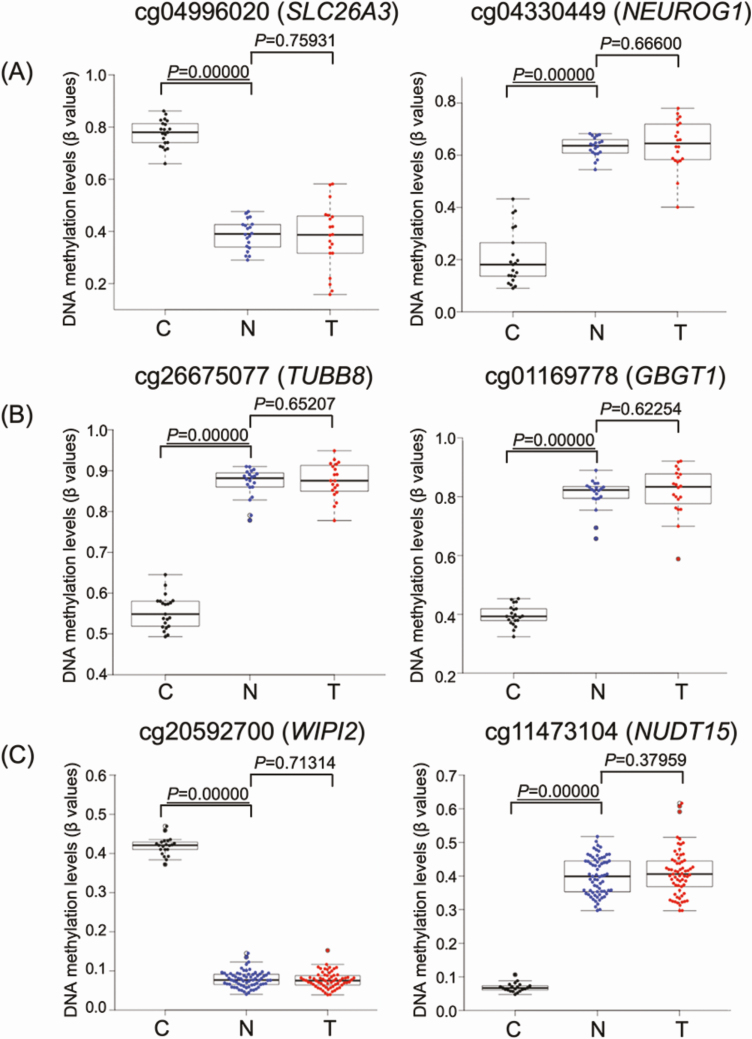
Next, we focused on DNA methylation alterations during the establishment of T from N. Although—based simply on Welch’s T test (P < 0.05) with Bonferroni correction (gray dots above the dashed line at P = 0.05 in Figure 1)—many probes showed differences between T and N, as had been shown in our previous study (13), in the present study we adapted the definition of ‘significant differences’ to include the absolute value of the difference (∆β T-N ≥ 0.3 or ∆β T-N ≤ −0.3) in order to focus on more obvious differences. Then, differing from the early steps of carcinogenesis from C to N, the volcano plots in Figure 1 clearly showed that DNA methylation alterations during the N to T transition were very limited. Only 0, 23 and 29 probes, designed for 0, 21 and 28 genes, showed significant differences in DNA methylation levels between N and T samples in Clusters A, B1 and B2, respectively (P < 0.05 for Welch’s T test after Bonferroni correction and ∆β T-N ≥ 0.3 or ∆β T-N ≤ −0.3; Supplementary Table S3 is available at Carcinogenesis Online), indicating that most of the DNA methylation alterations characterizing epigenomic clustering has already occurred at the precancerous N stages and that DNA methylation alterations added during the transition from N to T may have had less significance for multistage carcinogenesis. As shown in Figure 2, DNA methylation levels of the representative genes in each cluster suggested that DNA methylation alterations had actually occurred at the precancerous N stages and that such alterations had been inherited by the T samples themselves.
Figure 2.
DNA methylation levels of representative genes in normal control gastric mucosa (C; black), non-cancerous gastric mucosa (N; blue) and cancerous tissue (T; red) samples in Clusters A (n = 20) (A), B1 (n = 20) (B) and B2 (n = 69) (C). Infinium probe ID and gene names are shown at the top of each panel. DNA methylation alterations actually occurred at the precancerous N stages compared with C samples and such alterations were inherited by the T samples themselves. P values less than 0.05 have been underlined (Welch’s T-test).
To obtain an overall picture of DNA methylation alterations in T samples relative to C samples, we again identified 874, 382 and 581 probes showing significant differences in DNA methylation levels between T and C samples (P < 0.05 for Welch’s T-test after Bonferroni correction and ∆β T-C ≥ 0.3 or ∆β T-C ≤ −0.3) in Clusters A, B1 and B2, respectively. Supplementary Table S4, available at Carcinogenesis Online, summarizes the 783, 367 and 550 genes, for which 874, 382 and 581 probes were designed, respectively. Although not necessarily specific to Cluster B1, in addition to tumor-suppressor genes such as DCC, RUNX3 and VHL, and the oncogenic genes FZD10 and WT1, genes encoding cell adhesion-related molecules such as CDH13, CDH18, CTNNA3, PCDHA7, PCDHAC1 and PKP2, cytoskeletal proteins such as ACTA1, ACTB, KRT1, TLN2, TUBB8, VIM, kinases phosphorylating cytoskeletal proteins such as MYLK and ROCK1, and an epithelial–mesenchymal transition (EMT)-related transcription factor, TWIST1, showed DNA methylation alterations in T samples relative to C samples in Cluster B1.
Whole-exome analysis
Clinicopathological parameters for 66 patients (Supplementary Table S1 is available at Carcinogenesis Online), from whom additional DNA samples were available for genomic analysis even after the Infinium assay, are summarized in Supplementary Table S5, available at Carcinogenesis Online. As described in our previous article detailing the clinicopathological features of all 109 patients (13), even in the latter 66 patients, gastric adenocarcinomas belonging to the most aggressive Cluster B1 (n = 12) showed an undifferentiated histology significantly more frequently than those belonging to the less aggressive Clusters A (n = 12) and B2 (n = 42; Supplementary Table S5 is available at Carcinogenesis Online).
Exome analysis detected 45 375 synonymous or non-synonymous SNVs and 6837 indels in the 66 T samples. The incidence of the top 50 genes most frequently showing genetic aberrations (both SNVs and indels) in gastric carcinomas deposited in the TCGA database was compared with that in our 66 T samples (Supplementary Figure S2 is available at Carcinogenesis Online). Only 10 genes showed a significantly higher incidence of SNVs and indels in our cohort than in the TCGA datasets, and no significant differences in incidence were observed for the remaining 40 genes included in Supplementary Figure S2A, available at Carcinogenesis Online. Although the incidence of ARID1A gene mutations tended to be low in our cohort compared with the TCGA dataset (Supplementary Figure S2A is available at Carcinogenesis Online), the difference did not reach a statistically significant level.
When we focused on genomic aberrations in each epigenomic cluster, the average number of mutated genes (both SNVs and indels per case) in Cluster B1 (356 and 19) tended to be lower than in Clusters A (446 and 26) and B2 (379 and 23), respectively, although the differences did not reach statistical significance (Supplementary Table S6 is available at Carcinogenesis Online). When the SIFT score was less than 0.05 (damaging) and the Polyphen-2 score was more than 0.85 (probably damaging) in SNVs and the SIFT score was ‘damaging’ in indels, we considered that the protein function of the mutated gene was disrupted. Supplementary Table S7, available at Carcinogenesis Online, summarizes the 515, 345 and 112 recurrently mutated genes that showed SNVs or indels in more than 15% of cancers belonging to each cluster and for which protein function was disrupted due to at least one genetic aberration (SNVs or indels) in Clusters A, B1 and B2, respectively. With respect to genes for which disruptions have been reported in a previous study by TCGA (18), Supplementary Table S7, available at Carcinogenesis Online, shows that mutations of the ABCA10, BNC2, CDH1, CTNNB1 and SMAD4 genes potentially resulting in protein function disruption were observed only in Cluster B1, whereas those of the APC, EGFR, ERBB2, ERBB3, MLH1 and MUC6 genes were observed only in Cluster A.
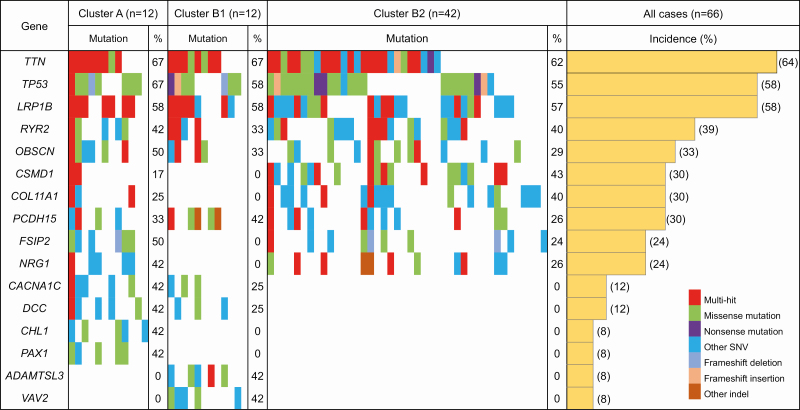
Figure 3 shows a waterfall plot (oncoplot) of the distribution of SNVs and indels in the top 16 genes for which mutations would potentially result in protein function disruption (accounting for more than 40% of the incidence of mutations in each cluster in Supplementary Table S7, available at Carcinogenesis Online) in Clusters A, B1 and B2. This highlights the fact that mutations of the ADMTSL3 gene, which reportedly promotes cancer cell proliferation (32), and the VAV2 gene, which reportedly promotes EMT (33), occur exclusively in Cluster B1, and not in Clusters A and B2.
Figure 3.
Waterfall plot (oncoplot) of the distribution of SNVs (i.e. missense mutation, nonsense mutation and other SNV) and insertions/deletions (indels; i.e. frameshift deletion, frameshift insertion and other indel) of the top 16 genes for which mutations would potentially result in protein function disruption (accounting for more than 40% of the incidence of mutations in each cluster in Supplementary Table S7, available at Carcinogenesis Online) in each tumor (vertical row) belonging to Clusters A (n = 12), B1 (n = 12) and B2 (n = 42). Samples showing more than one type of mutation in the same gene are indicated as the ‘multi-hit’ type. In addition, the incidence (%) of gene mutations in each cluster and in all samples (n = 66) is shown.
Copy number aberration analysis
Aberrated chromosomal segments showing gain (copy number of 6 or more) and loss (copy number of 0) across 22 autosomes in the 66 T samples are summarized in Supplementary Table S8, available at Carcinogenesis Online. The incidence of the top 50 genes most frequently showing copy number aberrations (both gain and loss) in gastric carcinomas deposited in the TCGA database was compared with that in our 66 T samples (Supplementary Figure S2B is available at Carcinogenesis Online). In our cohort, only 9 genes showed significantly higher or lower incidences of gains and losses than in the TCGA datasets, and no significant differences in these incidences were observed for the remaining 41 genes included in Supplementary Figure S2B, available at Carcinogenesis Online. Supplementary Table S9, available at Carcinogenesis Online, summarizes the 214, 180 and 18 genes located on the aberrated chromosomal segments recurrently showing gains or losses in more than 15% of cancers belonging to each of Clusters A, B1 and B2, respectively.
Pathway analysis
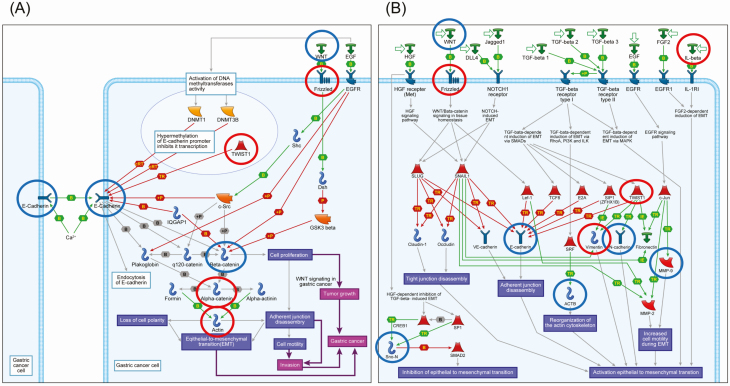
MetaCore pathway analysis was performed using all of the genes showing DNA methylation alterations between C and T samples (P < 0.05 for Welch’s T-test after Bonferroni correction and ∆β N-C ≥ 0.3 or ∆β N-C ≤ −0.3; Supplementary Table S4, available at Carcinogenesis Online), those showing genetic aberrations (SNVs and/or indels) in more than 15% of cases and affecting protein function (Supplementary Table S7 is available at Carcinogenesis Online) or copy number alterations (gains and/or losses) detected in more than 15% of cases (Supplementary Table S9 is available at Carcinogenesis Online). The numbers of genes included in Supplementary Tables S4, S7 and S9, available at Carcinogenesis Online, and used for MetaCore pathway analysis in Clusters A, B1 and B2 are summarized in Supplementary Table S10, available at Carcinogenesis Online. For Clusters A, B1 and B2, such genes were significantly accumulated in 40, 116 and 55 pathways (FDR < 0.05), respectively. After elimination of pathways participating solely in organs or tissues other than the stomach or diseases other than cancer, 9, 39 and 8 pathways in Clusters A, B1 and B2 are summarized in Supplementary Table S11, available at Carcinogenesis Online. Among them, GeneGo pathways relating to cell adhesion, cytoskeletal remodeling, EMT and WNT/β-catenin signaling, in which epigenetically and genetically affected genes in gastric carcinomas belonging to Clusters A, B1 and B2 were significantly accumulated, are summarized in Table 1. Representative maps of the pathways that showed modest significance and were included in Table 1 and Supplementary Table S11, available at Carcinogenesis Online, such as ‘E-cadherin signaling and its regulation in gastric cancer (FDR = 0.004 and P < 0.001 for Cluster B1)’ and ‘Development_Regulation of EMT (FDR = 0.002 and P < 0.001 for Cluster B1)’, are illustrated schematically in Figure 4 using MetaCore software. Epigenetic alterations of ACTA1, CTNNA3, FZD10 and TWIST1 and genetic alterations of CDH1, CTNNB1 and WNT16 appear to cooperatively disrupt the ‘E-cadherin signaling and its regulation in gastric cancer’ pathway. Epigenetic alterations of FZD10, IL1B, VIM and TWIST1 and genetic alterations of ACTB, CDH1, CDH2, MMP9 and WNT16 appear to cooperatively participate in the ‘Development_Regulation of EMT’ pathway.
Table 1.
GeneGo pathways related to cell adhesion, cytoskeleton remodeling, epithelial–mesenchymal transition (EMT) and WNT/β-catenin signaling, in which epigenetically and genetically affected genes in gastric carcinomas belonging to Clusters A, B1 and B2 were significantly accumulated (FDR < 0.05)
| Category | Cluster A | Cluster B1 | Cluster B2 | |||
|---|---|---|---|---|---|---|
| Pathways | Involved gens | Pathways | Involved gens | Pathways | Involved gens | |
| Cell adhesion | E-cadherin signaling and its regulation in gastric cancer | ACTN2, TWIST1, WNT2, ACTA1, EGFR, CTNNA1, FZD2, ZEB2 | Cell adhesion_Histamine H1 receptor signaling in the interruption of cell barrier integrity | TLN2, CDH1, CTNNB1, PRKCD, ROCK1, ACTB, CTNNA3, MYLK, ITPR3, GNA15 | ||
| Cell adhesion_Role of CDK5 in cell adhesion | TLN2, CDH1, CTNNB1, CTNNA3, ERBB2 | |||||
| E-cadherin signaling and its regulation in gastric cancer | CDH1, CTNNB1, WNT16, ACTA1, CTNNA3, FZD10, TWIST1 | |||||
| Cell adhesion_Cadherin-mediated cell adhesion | CDH2, CDH1, CTNNB1, ACTB, ACTA1, CTNNA3 | |||||
| Cell adhesion_ECM remodeling | CXCR1, NID1, ACTB, FLOT2, MMP9, IGF2, ERBB4, COL4A1 | |||||
| Cell adhesion_Endothelial cell contacts by non-junctional mechanisms | CDH2, CTNNB1, ACTB, CTNNA3, COL4A1 | |||||
| Cell adhesion_Role of tetraspanins in the integrin- mediated cell adhesion | TLN2, CD53, ROCK1, ACTB, ACTA1, FLNA | |||||
| Cell adhesion_Endothelial cell contacts by junctional mechanisms | CDH2, CTNNB1, VIM, ACTB, CTNNA3 | |||||
| Cell adhesion_Integrin-mediated cell adhesion and migration | TLN2, MYLK, ROCK1, ACTB, COL4A1 | |||||
| Cytoskeleton remodeling | Cytoskeleton remodeling_Keratin filaments | ACTN2, TWIST1, WNT2, ACTA1, EGFR, CTNNA1, FZD2, ZEB2 | Cytoskeleton remodeling_Keratin filaments | PLEC, TUBB8, PKP2, EPPK1, VIM, ACTB, KRT1, KRT13 | Cytoskeleton remodeling_ Thyroliberin in cytoskeleton remodeling | TRHR, PRKCB, PRKAR1B, ADCY4, PLCB1, TRH |
| Cytoskeleton remodeling_Role of PKA in cytoskeleton reorganization | ROCK1, PRKAR1B, ADCY4, ACTB, MYLK, ITPR3, ADD2 | |||||
| Cytoskeleton remodeling_ Regulation of actin cytoskeleton organization by the kinase effectors of Rho GTPases | TLN2, PIP5K1A, ROCK1, ACTB, FLNA, SPTBN5, MYLK | |||||
| Cytoskeleton remodeling_ Thyroliberin in cytoskeleton remodeling | TRHR , CACNA1C, PRKAR1B, ADCY4, TRH, ITPR3 | |||||
| EMT | Development_Regulation of epithelial-to-mesenchymal transition (EMT) | IL1B, CDH2, CDH1, VIM, WNT16, MMP9, FZD10, ACTB, SKIL, TWIST1 | Development_ Regulation of epithelial-to- mesenchymal transition (EMT) | IL1B, CDH2 , VIM, WNT2, FGF2, FZD10, TWIST1, TNF | ||
| Development_TGF-β-dependent induction of EMT via RhoA, PI3K and ILK | CDH1, CTNNB1, VIM, ACTA1, ROCK1, ACTB | |||||
| Development_TGF-β-dependent induction of EMT via SMADs | CDH2, CDH1, VIM, SMAD4, TWIST1 | |||||
| WNT/β- catenin signaling | WNT signaling in gastric cancer | SKP2, SFRP1, SFRP2, APC, WNT8A, FZD2, WNT2, TNF | Development_Negative feedback regulation of WNT/β-catenin signaling | CTNNB1, PRKCD, RNF43, WNT16, HUWE1, TRRAP, FZD10 | ||
| Development_ Negative regulation of WNT/β-catenin signaling in the nucleus | FHIT, KDM2B, RNF43, PAX7, TLE4, RUNX3, VHL, WNT2, APC, SOX17, CTNNA1, FZD2, NOS3, YWHAZ | Development_Negative regulation of WNT/β-catenin signaling in the nucleus | RUNX3, FHIT, VHL, CTNNB1, PRKCD, RNF43, PAX7, WNT16, GLI3, TRRAP, FZD10 | |||
| Development_Negative regulation of WNT/β-catenin signaling in the cytoplasm | DAB2, CDH1, TP53, VHL, CTNNB1, WNT16, HUWE1, FZD10 | |||||
All statistically significant GeneGo pathways revealed by MetaCore software using epigenetically and genetically affected genes in gastric carcinomas belonging to Clusters A, B1 and B2 are shown in Supplementary Table S11, available at Carcinogenesis Online.
Figure 4.
Schematically illustrated representative pathway maps, ‘E-cadherin signaling and its regulation in gastric cancer (FDR = 0.004 and P < 0.001)’ (A) and ‘Development_Regulation of EMT (FDR = 0.002 and P < 0.001)’ (B), in which epigenomically and genomically affected genes were accumulated in Cluster B1, by MetaCore software. Original pathway maps generated by MetaCore software have been simplified by eliminating items that were not correlated with genes affected epigenetically and/or genetically in our Cluster B1. Genes showing DNA methylation alterations and those showing genetic aberrations (SNVs, insertions/deletions and/or copy number aberrations) are indicated in red and blue, respectively.
Discussion
Hierarchical clustering based on genome-wide DNA methylation profiles has allowed patients with cancers of various organs to be stratified into subclasses associated with distinct clinicopathological features (34), probably because DNA methylation alterations arise even from the precancerous stages and are then stably preserved on DNA double strands by covalent bonds due to maintenance methylation mechanisms attributable to DNA methyltransferase DNMT1 (35). For example, in relation to treatment strategy, our previous studies have shown that epigenomic clustering of renal cell carcinomas of the kidney (10) and young-onset endometrial cancers of the uterine corpus (36) are closely associated with amenability to Aurora kinase inhibitors and fertility preservation therapy, respectively. In terms of prognostic impact, such clustering of lung adenocarcinomas was shown to be associated with history of smoking and chronic obstructive pulmonary disease and poor prognosis (11).
With respect to gastric adenocarcinomas, our previous study using epigenomic clustering based on genome-wide DNA methylation profiling of N samples at the precancerous stage has identified distinct Clusters A and B1, each showing favorable and poor outcome, respectively (13). In that study, however, we did not perform methylome analysis of normal control gastric mucosae from patients without gastric carcinomas, and no comparison of N samples with C samples was done (13). Therefore, it has not fully clarified whether DNA methylation alterations were present at the precancerous N stage compared with C samples. In the present study, volcano plots (Figure 1) and scatter plots (Figure 2) revealed that DNA methylation alterations had actually occurred in N samples compared with C samples. In addition, DNA methylation profiles were not simply altered in N samples but also targeted molecules including transcription factors and kinases potentially involved in tumor-related signaling pathways. On the other hand, DNA methylation alterations during transition from the precancerous stage to established cancer (N to T transition stage) were very limited (Figures 1 and 2), indicating that DNA methylation profiles during multistage carcinogenesis are already established at the precancerous stage and inherited by the cancers themselves.
The number of genes showing DNA methylation alterations in the most aggressive Cluster B1 was lower than that in the less aggressive Clusters A and B2 (Supplementary Tables S2–S4 are available at Carcinogenesis Online). This finding is consistent with our previous data for non-small cell lung cancers (11): lung cancers belonging to the most aggressive epigenetic cluster showed DNA methylation alterations in N samples relative to C samples on a smaller number of probes compared with the less aggressive cluster. Patients belonging to the most aggressive cluster appeared to show more rapid progression than those in the less aggressive cluster, whose lung cancers developed slowly through the long-term cumulative effects of cigarette smoking and subsequent chronic obstructive lung disease. By analogy with lung cancers, we assume that when DNA methylation alterations occur and affect important tumor-related genes during the field cancerization process, such precancerous tissue may progress rapidly to the N to T transition step before DNA methylation alterations have accumulated on many genes, thus generating more aggressive cancers. This may explain why the number of genes showing DNA methylation alterations in Cluster B1 was lower than that in Clusters A and B2.
In fact, the affected genes in Cluster B1 were effectively concentrated in the GO protein class of transcription factors, which can disturb the expression profiles of downstream genes (data not shown). In addition to the fact that many genes showed differences of DNA methylation levels between the most aggressive Cluster B1 and the least aggressive Cluster A even at the precancerous stage (13), only one specifically affected gene in Cluster B1, PKP2, encoding the desmosomal protein plakophilin-2, is a direct transcriptional target of the Wnt/β-catenin pathway (31). Plakophilin-2 has been shown to promote tumor development by binding to the epidermal growth factor receptor and enhancing its activity (37). By analogy with the reported invasiveness and metastatic ability of cancers showing overexpression of plakophilin-2 (38), DNA hypomethylation of the PKP2 gene in Cluster B1 may participate in the aggressiveness of cancers belonging to this cluster through its overexpression. These data indicated that the clinicopathological diversity among the clusters may be at least partly attributable to the differences in the DNA methylation profiles evident at the precancerous stage.
On the other hand, not only epigenomic but also genomic events of course determine the carcinogenetic pathway. Although we have not performed exome sequencing in N samples, some recent papers have reported non-synonymous mutations that are commonly dysregulated in gastric adenocarcinomas, such as the MUC5AC, KRAS, BRAF and EZH2 genes, in microdissected specimens of precancerous lesions, such as spasmolytic polypeptide-expressing metaplasia and pyloric gland adenoma, based on exome sequencing (39,40). Moreover, microsatellite instability has reportedly been observed in non-cancerous gastric mucosa without remarkable histological features obtained from gastric cancer patients (40), suggesting the participation of genomic events in the precancerous stages during field cancerization in the stomach. Therefore, genetic alterations should be further examined even in N samples.
In T samples, we performed exome-sequencing and copy number analysis for each cluster. This revealed that the incidence of SNVs and indels and the copy number alteration profiles of all the cancers examined in our cohort were almost consistent with data deposited in the TCGA database (Supplementary Figure S2 is available at Carcinogenesis Online), indicating the reliability of our analyses. Even though the incidence of SNVs and indels in the ARID1A gene, the major player in gastric carcinogenesis, tended to be lower than that in the TCGA database, such a tendency has been observed in previous Asian cohorts (41,42) and our data are consistent with such previous reports. With respect to differences among the epigenomic clusters, although the number of mutated genes showing SNVs and indels in the most aggressive Cluster B1 was low (Supplementary Table S6 is available at Carcinogenesis Online), the number of genes showing copy number alterations was not so different between Clusters B1 and A (Supplementary Table S9 is available at Carcinogenesis Online), suggesting that Cluster B1 may represent copy number alterations reflecting chromosomal instability rather than gene mutations. It is feasible that chromosomal instability plays a significant role in the epigenomically defined Cluster B1, since genome-wide DNA methylation alterations generally result in chromosomal instability through aberrant chromatin configurations (43).
When focusing on differences in the mutation profiles of genes well known to participate in gastric adenocarcinomas (18), it was found that among epigenomic clusters, recurrent mutations (SNVs and/or indels) associated with protein function disruption (Supplementary Table S7 is available at Carcinogenesis Online) of the ABCA10, BNC2, CDH1, CTNNB1 and SMAD4 genes were specific to Cluster B1, whereas those of the APC, EGFR, ERBB2, ERBB3, MLH1 and MUC6, genes were specific to Cluster A. Although our clustering was based on the DNA methylation profiles at the precancerous stage, probably through epigenomic-genomic interaction, distinct genetic profiles may be established in cancers belonging to each cluster.
Moreover, when both epigenomic and genomic alterations cooperatively accumulate in certain signaling pathways, such pathways may be completely disrupted and have a larger clinicopathological impact in each cluster than pathways that are impaired by either epigenomic or genomic alteration alone. Therefore, MetaCore pathway analysis was performed using all of the genes showing DNA methylation alterations between C and T samples (Supplementary Table S4 is available at Carcinogenesis Online), recurrent genetic aberrations (SNVs or indels) affecting protein function (Supplementary Table S7 is available at Carcinogenesis Online) and recurrent copy number alterations (gains or losses; Supplementary Table S9 is available at Carcinogenesis Online). Even though the number of genes showing DNA methylation alterations and genetic mutations in the most aggressive Cluster B1 was lower than that in the least aggressive Cluster A, the total number of cooperatively affected pathways in Cluster B1 was larger than that in Cluster A (Supplementary Table S11, available at Carcinogenesis Online, and Table 1), indicating that epigenomic and genomic alterations effectively cooperate during cancer development and progression, especially in Cluster B1.
In Table 1, with respect to the most aggressive Cluster B1, 9 and 4 pathways including the top pathway in Supplementary Table S11, available at Carcinogenesis Online, were significantly related to cell adhesion and cytoskeleton remodeling (which is closely related to cell adhesion; 13 pathways in total), whereas only 2 pathways related to cell adhesion and cytoskeleton remodeling were disrupted in the least aggressive Cluster A. Moreover, cell adhesion abnormality is one of the key components of EMT, and 3 EMT-related pathways were affected in Cluster B1. Even though Wnt/β-catenin-related pathways seem to be activated in Cluster A, mutations resulting in protein function aberration of the CTNNB1 gene itself were observed only in Cluster B1. Key players of cell adhesion abnormalities and EMT in Cluster B1 appear to be Cluster B1-specific mutations of the CDH1 (44), CTNNB1 (44), MMP9 (45), TLN2 (46), ROCK1 (47), SMAD4 (48) and VAV2 (33) genes. In addition, TWIST1 is a master regulator of morphogenesis repressing CDH1 expression, and is required for EMT initiation (49) and expression regulation of the TWIST1 (50) gene due to DNA methylation alterations has been reported. Based on these previous findings, it has been assumed that epigenomic alteration of the TWIST1 gene, along with additional epigenomic alterations shown in Figure 4, may effectively cooperates with Cluster B1-specific mutations. On the other hand, based on epigenomic and genomic analyses of Cluster B2, clear accumulation of abnormalities in any of the pathways related to cell adhesion or Wnt/β-catenin signaling has not been proven. To obtain a complete picture of cancers belonging to Cluster B2 and to further classify such cancers based on their molecular signatures, more integrated analyses, including the transcriptome and proteome, may be needed in addition to epigenomic and genomic analyses.
In conclusion, epigenomic alterations at the precancerous stage, which may generally precede genomic alteration and, at least partially, affect mutational profiles, are important in the development of gastric adenocarcinomas. Epigenomic and genomic alterations underlie the clinicopathological aggressiveness of gastric adenocarcinomas in a coordinated manner, especially by enhancing abnormalities of cell–cell adhesiveness and inducing EMT. Integrated omics analysis is a powerful tool for clarifying molecular events that are complementary to each other during multistage carcinogenesis and can reflect the diversity of clinicopathological characteristics.
Supplementary Material
Glossary
Abbreviations
- ASCAT
allele-specific copy number analysis of tumors
- C
normal control gastric mucosa
- EMT
epithelial–mesenchymal transition
- FDR
false discovery rate
- GO
Gene ontology
- GPHMM
Global Parameter Hidden Markov Model
- indel
insertion/deletion
- N
non-cancerous gastric mucosa
- PolyPhen
polymorphism phenotyping
- SIFT
Sorting intolerant from tolerant
- SNP
single nucleotide polymorphism
- SNV
single nucleotide variant
- T
cancerous tissue
- TCGA
The Cancer Genome Atlas
- TNM
Tumor–Node–Metastasis
Funding
This study was supported by the Funding for Research to Expedite Effective Drug Discovery by Government, Academia and Private Partnership (19ak0101043h0105) from the Japan Agency for Medical Research and Development (AMED), and also KAKENHI (19K07444) from the Japan Society for the Promotion of Science.
Conflict of Interest Statement: None declared.
References
- 1. Van CE et al. (2016) Gastric cancer. Lancet, 388, 2654–2664. [DOI] [PubMed] [Google Scholar]
- 2. Ferro A et al. (2014) Worldwide trends in gastric cancer mortality (1980–2011), with predictions to 2015, and incidence by subtype. Eur. J. Cancer, 50, 1330–1344. [DOI] [PubMed] [Google Scholar]
- 3. Jones PA et al. (2016) Targeting the cancer epigenome for therapy. Nat. Rev. Genet., 17, 630–641. [DOI] [PubMed] [Google Scholar]
- 4. Watari J et al. (2019) DNA methylation silencing of microRNA gene methylator in the precancerous background mucosa with and without gastric cancer: analysis of the effects of H. pylori eradication and long-term aspirin use. Sci. Rep., 9, 12559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tsumura K et al. (2019) Establishment of permutation for cancer risk estimation in the urothelium based on genome-wide DNA methylation analysis. Carcinogenesis, 40, 1308–1319. [DOI] [PubMed] [Google Scholar]
- 6. Kuramoto J et al. (2017) Genome-wide DNA methylation analysis during non-alcoholic steatohepatitis-related multistage hepatocarcinogenesis: comparison with hepatitis virus-related carcinogenesis. Carcinogenesis, 38, 261–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ohara K et al. (2017) Genes involved in development and differentiation are commonly methylated in cancers derived from multiple organs: a single-institutional methylome analysis using 1007 tissue specimens. Carcinogenesis, 38, 241–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mizuguchi A et al. (2018) Genetic features of multicentric/multifocal intramucosal gastric carcinoma. Int. J. Cancer, 143, 1923–1934. [DOI] [PubMed] [Google Scholar]
- 9. Kaneda A et al. (2012) Epstein-Barr virus infection as an epigenetic driver of tumorigenesis. Cancer Res., 72, 3445–3450. [DOI] [PubMed] [Google Scholar]
- 10. Arai E et al. (2015) Alterations of the spindle checkpoint pathway in clinicopathologically aggressive CpG island methylator phenotype clear cell renal cell carcinomas. Int. J. Cancer, 137, 2589–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sato T et al. (2014) Epigenetic clustering of lung adenocarcinomas based on DNA methylation profiles in adjacent lung tissue: its correlation with smoking history and chronic obstructive pulmonary disease. Int. J. Cancer, 135, 319–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kanai Y et al. (2014) Multilayer-omics analyses of human cancers: exploration of biomarkers and drug targets based on the activities of the International Human Epigenome Consortium. Front. Genet., 5, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yamanoi K et al. (2015) Epigenetic clustering of gastric carcinomas based on DNA methylation profiles at the precancerous stage: its correlation with tumor aggressiveness and patient outcome. Carcinogenesis, 36, 509–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Oue N et al. (2019) Molecular carcinogenesis of gastric cancer: Lauren classification, mucin phenotype expression, and cancer stem cells. Int. J. Clin. Oncol., 24, 771–778. [DOI] [PubMed] [Google Scholar]
- 15. Zhang J et al. (2018) Genomic alterations in gastric cancers discovered via whole-exome sequencing. BMC Cancer, 18, 1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vogelaar IP et al. (2017) Unraveling genetic predisposition to familial or early onset gastric cancer using germline whole-exome sequencing. Eur. J. Hum. Genet., 25, 1246–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen K et al. (2015) Mutational landscape of gastric adenocarcinoma in Chinese: implications for prognosis and therapy. Proc. Natl. Acad. Sci. U.S.A., 112, 1107–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cancer Genome Atlas Research Network (2014) Comprehensive molecular characterization of gastric adenocarcinoma. Nature, 513, 202–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fukayama M et al. (2019) Tumours of the stomach. In The WHO Classification of Tumours Editorial Board (ed) WHO Classification of Tumours of Digestive System Tumours. 5th edn. Vol. 1 IARC Press, Lyon, France, pp. 59–104. [Google Scholar]
- 20. Wittekind CH et al. (2017) Stomach. In Brierley, J.D., Gospodarowicz, M.K. and Wittekind, C. (eds) TNM Classification of Malignant Tumours. 8th edn.Wiley-Blackwell Press, Hoboken, NJ, pp. 63–66. [Google Scholar]
- 21. Kanai Y et al. (2018) The Japanese Society of Pathology guidelines on the handling of pathological tissue samples for genomic research: standard operating procedures based on empirical analyses. Pathol. Int., 68, 63–90. [DOI] [PubMed] [Google Scholar]
- 22. Li H et al. (2010) Fast and accurate long-read alignment with burrows-wheeler transform. Bioinformatics, 26, 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. McKenna A et al. (2010) The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res., 20, 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cibulskis K et al. (2013) Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol., 31, 213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ng PC et al. (2003) SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res., 31, 3812–3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hicks S et al. (2011) Prediction of missense mutation functionality depends on both the algorithm and sequence alignment employed. Hum. Mutat., 32, 661–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sim NL et al. (2012) SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res., 40(Web Server issue), W452–W457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Van LP et al. (2010) Allele-specific copy number analysis of tumors. Proc. Natl. Acad. Sci. U.S.A., 107, 16910–16915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li A et al. (2011) GPHMM: an integrated hidden Markov model for identification of copy number alteration and loss of heterozygosity in complex tumor samples using whole genome SNP arrays. Nucleic Acids Res., 39, 4928–4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Quinlan AR et al. (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics, 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Niell N et al. (2018) The human PKP2/plakophilin-2 gene is induced by Wnt/β-catenin in normal and colon cancer-associated fibroblasts. Int. J. Cancer, 142, 792–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhou X et al. (2020) Genome-wide CRISPR knockout screens identify ADAMTSL3 and PTEN genes as suppressors of HCC proliferation and metastasis, respectively. J. Cancer Res. Clin. Oncol., 146, 1509–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li X et al. (2019) MicroRNA-331-3p inhibits epithelial-mesenchymal transition by targeting ErbB2 and VAV2 through the Rac1/PAK1/β-catenin axis in non-small-cell lung cancer. Cancer Sci., 110, 1883–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Arai E et al. (2012) Single-CpG-resolution methylome analysis identifies clinicopathologically aggressive CpG island methylator phenotype clear cell renal cell carcinomas. Carcinogenesis, 33, 1487–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jones PA et al. (2009) Rethinking how DNA methylation patterns are maintained. Nat. Rev. Genet., 10, 805–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Makabe T et al. (2019) Genome-wide DNA methylation profile of early-onset endometrial cancer: its correlation with genetic aberrations and comparison with late-onset endometrial cancer. Carcinogenesis, 40, 611–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Arimoto K et al. (2014) Plakophilin-2 promotes tumor development by enhancing ligand-dependent and -independent epidermal growth factor receptor dimerization and activation. Mol. Cell. Biol., 34, 3843–3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Takahashi H et al. (2012) Up-regulation of plakophilin-2 and Down-regulation of plakophilin-3 are correlated with invasiveness in bladder cancer. Urology, 79, 240.e1–240.e8. [DOI] [PubMed] [Google Scholar]
- 39. Srivastava S et al. (2020) An LCM-based genomic analysis of SPEM, gastric cancer and pyloric gland adenoma in an Asian cohort. Mod. Pathol. doi: 10.1038/s41379-020-0520-5. [DOI] [PubMed] [Google Scholar]
- 40. Chen J et al. (2020) Evidence for heightened genetic instability in precancerous spasmolytic polypeptide expressing gastric glands. J. Med. Genet., 57, 385–388. [DOI] [PubMed] [Google Scholar]
- 41. Wong SS et al. (2014) Genomic landscape and genetic heterogeneity in gastric adenocarcinoma revealed by whole-genome sequencing. Nat. Commun., 5, 5477. [DOI] [PubMed] [Google Scholar]
- 42. Kakiuchi M et al. (2014) Recurrent gain-of-function mutations of RHOA in diffuse-type gastric carcinoma. Nat. Genet., 46, 583–587. [DOI] [PubMed] [Google Scholar]
- 43. Gaudet F et al. (2003) Induction of tumors in mice by genomic hypomethylation. Science, 300, 489–492. [DOI] [PubMed] [Google Scholar]
- 44. Hirohashi S et al. (2003) Cell adhesion system and human cancer morphogenesis. Cancer Sci., 94, 575–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Moirangthem A et al. (2016) Simultaneous knockdown of uPA and MMP9 can reduce breast cancer progression by increasing cell-cell adhesion and modulating EMT genes. Sci. Rep., 6, 21903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Thapa N et al. (2017) PIPKIγ and talin couple phosphoinositide and adhesion signaling to control the epithelial to mesenchymal transition. Oncogene, 36, 899–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shi D et al. (2019) LncRNA AFAP1-AS1 promotes tumorigenesis and epithelial-mesenchymal transition of osteosarcoma through RhoC/ROCK1/p38MAPK/Twist1 signaling pathway. J. Exp. Clin. Cancer Res., 38, 375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tong X et al. (2020) MYOCD and SMAD3/SMAD4 form a positive feedback loop and drive TGF-β-induced epithelial-mesenchymal transition in non-small cell lung cancer. Oncogene, 39, 2890–2904. [DOI] [PubMed] [Google Scholar]
- 49. Yang J et al. (2004) Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell, 117, 927–939. [DOI] [PubMed] [Google Scholar]
- 50. Marofi F et al. (2019) Gene expression of TWIST1 and ZBTB16 is regulated by methylation modifications during the osteoblastic differentiation of mesenchymal stem cells. J. Cell. Physiol., 234, 6230–6243. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.