Abstract
The human body is host to several distinct microbial communities. Disruption of these communities increases susceptibility to a wide range of diseases, including respiratory tract infections. While commensal bacteria in the gut contribute to this effect, recent studies point to a role for commensals occupying the upper respiratory tract through direct pathogen killing and by modifying nasal and lung immune homeostasis. Clinical trials exploring ‘probiotic’ respiratory tract commensals are an exciting development in this area. Upper respiratory tract microbiome sequencing has revealed that destabilization of this community precedes infection, indicating that microbiome profiling of individuals has predictive value. Further investigation of respiratory tract commensal–host interactions will be critical to translate bacterial-mediated protection toward new therapeutic approaches for respiratory tract disease.
Introduction
The upper respiratory tract (URT) contains a well-documented bacterial community, or microbiome, residing in the nasal cavity and nasopharynx. Opportunistic bacterial pathogens, referred to here as opportunistic pathogens, are transient members of this community that cause illness upon invasion of other host tissues. Respiratory tract infections caused by opportunistic pathogens constitute a major burden of disease. Among these, pneumonia is the number one cause of death worldwide in children under five years old and the leading infectious cause of death in the elderly [1–3]. Otitis media, or ear infection, is the most frequent diagnosis for antibiotic prescription in young children [4]. The four predominant opportunistic pathogens of the URT are Streptococcus pneumoniae, Haemophilus influenzae, Moraxella catarrhalis and Staphylococcus aureus. While these bacteria often occupy the URT asymptomatically, colonization is a prerequisite for invasive disease [5–7]. Cooperative and competitive interactions between these bacterial pathogens influence susceptibility to infection [8–11]. In addition, co-infection with viral pathogens including influenza A, influenza B, or respiratory syncytial virus (RSV) pre-dispose bacterial invasion and are associated with more severe disease [12–14]. However, it is less clear how the remaining members of the URT microbial community modify pathogen acquisition.
Many bacteria inhabiting the gut and URT are never or only rarely associated with disease, referred to as commensals. URT commensals predominantly reside in the nasal cavity and nasopharynx, but also reach the lung through aspiration [15–17]. The URT microbiome is composed of several ‘core’ genera present in the majority of healthy individuals, the most abundant of which include Staphylococcus, Streptococcus, Corynebacterium, Prevotella, Veillonella, Propionibacterium, and Fusobacterium in adults with Moraxella also prominent in young children [17,18,19••,20]. It is clear that the microbiome as a whole contributes to resistance against diverse (fungal, viral and bacterial) lung infections, which are frequently more severe in antibiotic treated and germ-free animals [21–26]. However, the depletion (or absence) of bacteria from both the gut and URT obscures the contributions of microbiota from each site to immune homeostasis. Antibiotic treatment also differentially enriches genera in the URT as well as the gut [23], which may influence susceptibility to infection depending on which bacteria remain. In people, the increased abundance of Haemophilus, Streptococcus, and Staphylococcus following antibiotic therapy is likely in part due the rise in antibiotic resistance among opportunistic pathogens including H. influenzae, S. pneumoniae and S. aureus [20,27,28]. This review focuses on new developments in our understanding of how commensal bacteria regulate susceptibility to respiratory tract infection, with an emphasis on the role of the URT microbiota.
Shared mechanisms for the regulation of lung immunity by commensal bacteria
URT commensal bacteria occupy the same niche as opportunistic pathogens, making it difficult to distinguish between direct competition and indirect modulation of the immune response. In contrast, bacteria colonizing the gut must engage circulating immune factors to influence lung immunity. Some of the signaling pathways activated by gut commensals are also induced by bacteria in the URT. This is particularly evident for innate immune receptors, which recognize conserved bacterial ligands. For example, Toll-like receptor (TLR)4 recognition of bacterial lipopolysaccharide (LPS) improves protection against influenza A virus following either intranasal or rectal exposure in mice [23]. More recently, this was also shown for the innate immune Nod-like receptor (Nod)2[29••].Reconstitution of antibiotic treated mice with a compilation of potent Nod2-stimulators from either the URT (intranasal reconstitution) or gut (oral reconstitution) rescues protection against S. pneumoniae and Klebsiella pneumoniae infections [29••]. Nod2 activation primes alveolar macrophages in the lung to produce reactive oxygen species (ROS), contributing to pathogen clearance [29••]. Similarly, the respiratory tract commensal Staphylococcus sciuri promotes the adjuvanticity of cholera toxin in a Nod2-dependent manner [30]. In this case, stimulation of lung CD11c+ cells, the majority of which are alveolar macrophages, by intranasal cholera toxin requires activation of Nod2 by commensal bacteria including S. sciuri [30]. Alveolar macrophage ROS is also induced by oral treatment with the gut commensal Bifidobacterium longum, boosting protection against lethal K. pneumoniae lung infection in mice [31]. These studies demonstrate that recognition of either gut or URT commensals by innate immune receptors activates alveolar macrophages in the lung, improving resistance to lung infection.
Commensal bacteria also influence lung immune homeostasis by regulating production of the cytokines IL-17A and IL-22 from several immune cell types. This has been shown for gut segmented filamentous bacteria (SFB), which improve resistance against both S. aureus and the fungus Aspergillus fumigatus in mice by activating lung IL-17A+ T helper (Th)17 cells and IL-22 production [25,32]. SFB are also protective against lethal S. pneumoniae infection in Rag1−/− mice, which lack mature T and B cells, indicating a role for other cell types [33]. Aside from Th17 cells, gut commensal bacteria regulate lung mucosal-associated invariant T (MAIT) cell production of IL-17A and innate lymphoid cell type 3 (ILC3) production of IL-22, increasing resistance in murine models of Mycobacterium tuberculosis and S. pneumoniae infections respectively [24,34]. The bacterial ligands from SFB and other gut commensals that are critical for regulating these responses, and whether these ligands are shared by URT commensals, have not been established. However, the observation that Nod2 signaling induced by URT bacteria correlates with IL-17A-dependent GM-CSF production in the lung supports a role for bacteria from both the lung and gut in modulating lung IL-17A [29••]. Together, these studies indicate that commensal URT and gut bacteria activate shared innate immune pathways that regulate resistance to lung infection.
It less clear whether URT commensal bacteria modulate Th1/Th2 immunity in the lung similar to bacteria from the gut. For example, short-chain fatty acids (SCFAs) produced by gut commensals are protective against Th2 associated allergic lung inflammation, without influencing accumulation of regulatory T cells (Tregs), in mice [35,36]. Gut bacteria including Lactobacillus rhamnosus also improve Th1 responses in the lungs of mice infected with influenza A virus, S. pneumoniae, and M. tuberculosis [37–39]. While the URT harbors anaerobic SCFA-producers including Prevotella, the concentrations of SCFAs found in the URT are extremely low [40], suggesting a limited role. Commensal bacteria found in the skin and nose can differentially induce Th1 cytokines in the lung [41], but the impact of these responses on pathogen challenge remains largely unexplored. Instead, URT commensal bacteria have been shown to improve resistance to infection by mechanisms unique to the environment of the respiratory tract, as discussed below.
Distinct mechanisms for the regulation of pathogen acquisition and invasion by URT commensal bacteria
URT commensal bacteria regulate the mucosal barrier, the initial site of pathogen exposure. For example, the URT commensal Staphylococcus epidermidis induces nasal epithelial production of interferon (IFN)-λ, which increases resistance to influenza A virus infection in mice [42••]. IFN-λ is observed in the nasal secretions of people colonized with S. epidermidis [42••], though it is unclear whether these levels are sufficient for protection in humans. Resistance to influenza A virus infection is also associated with modulation of type I IFN signaling in the lung epithelium by gut bacteria [43]. However, there is no evidence to date that gut commensals regulate the nasal mucosa similar to URT bacteria. Another way that S. epidermidis regulates the nasal epithelium is by the stimulation of antimicrobial peptide (AMP) production [44••]. This has direct consequences for pathogen resistance, as S. epidermidis blocks S. aureus and M. catarrhalis acquisition in an AMP-dependent manner in mice [44••]. In addition to stimulating mucosal immune factors, S. epidermidis produces bacteriocins that restrict the growth of S. aureus and M. catarrhalis [45,46] and protect against S. aureus colonization in mice [47]. Similarly, commensals closely related to H. influenzae produce bacteriocins and modify nasal pro-inflammatory cytokine production [48,49•], indicating that other URT commensals contribute to pathogen resistance through similar mechanisms. Collectively, these findings illustrate that URT commensal bacteria reduce susceptibility to pathogen acquisition by regulating the nasal mucosa.
URT commensals also influence the adaptive immune response, which has the potential for long-term consequences through the generation of immune memory. This has been shown most clearly for commensal Streptococcus species. Streptococcus mitis induces cross-reactive protection against S. pneumoniae in mice characterized by systemic antibody production and IL-17+ Th17 cells in the lung [50••]. Memory T helper cells, including Th17 cells, with cross-reactivity against S. mitis and S. pneumoniae have also been identified in humans [51]. Several commensal Streptococcus species express capsule, the predominant Streptococcus antigen, with genetic and antigenic similarities to that found in S. pneumoniae [52•,53,54•]. However, capsule-specific memory was not prevalent in a pool of cross-reactive memory T cells identified in humans [51], indicating importance for other antigens. Commensal Streptococcus species may thus contribute to baseline resistance against S. pneumoniae by promoting cross-reactive immunity. Taken together, URT commensal bacteria improve protection against respiratory tract pathogen colonization and infection through both direct competition and indirect immune modulation (Figure 1).
Figure 1.
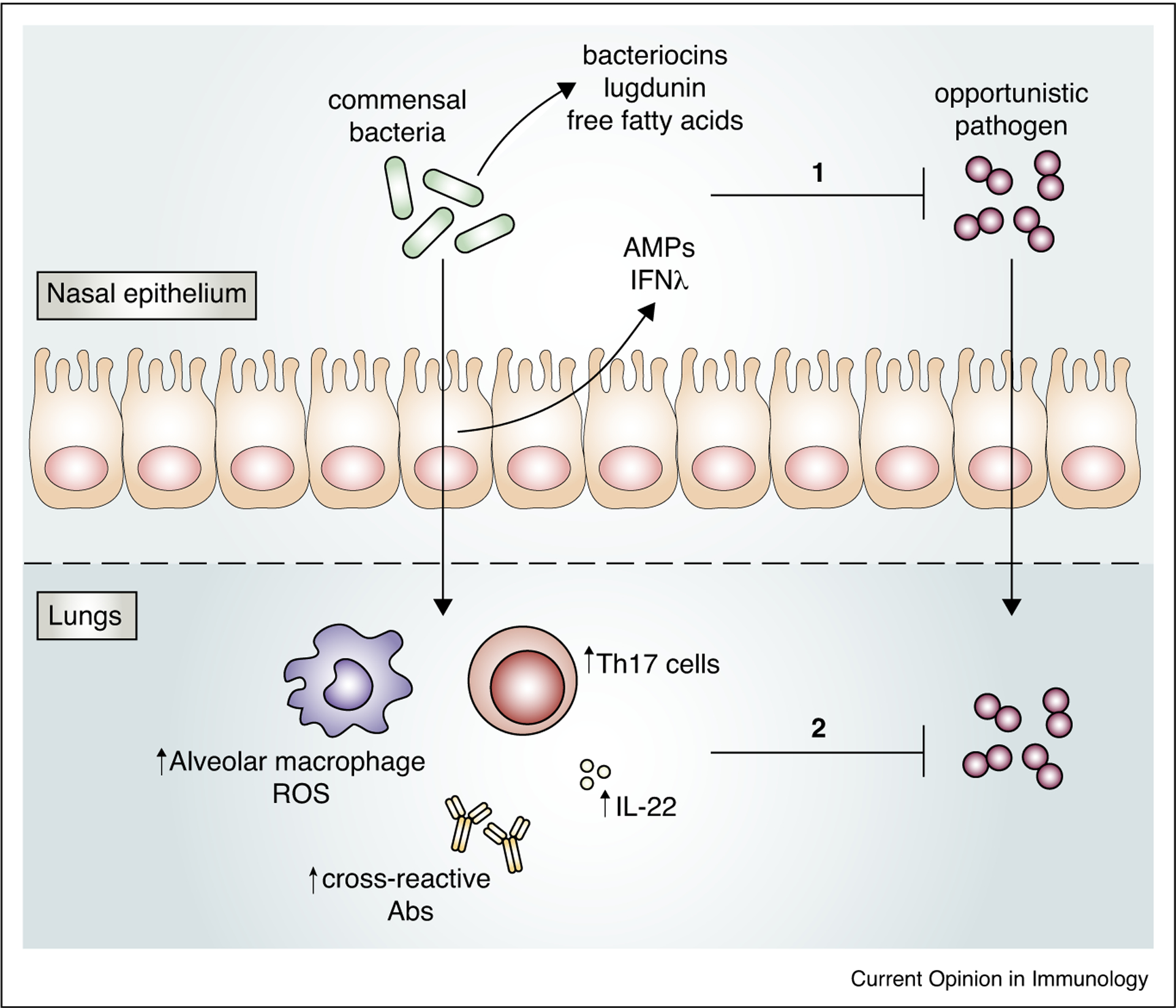
URT commensal bacteria protect against respiratory tract infection. Colonization of the nasal epithelium with opportunistic pathogens is restricted by URT commensal bacteria through direct competition, for example by production of bacteriocins, lugdunin, and free fatty acids that restrict pathogen growth, and indirectly through modulation of the mucosal barrier, resulting in secretion of factors including antimicrobial peptides (AMPs) and IFN-l, which contribute to pathogen clearance (1). URT commensal bacteria also reduce pathogen invasion by promoting innate and adaptive lung immune responses including alveolar macrophage production of reactive oxygen species (ROS), IL-22, Th17 cells, and cross-reactive antibodies (2).
Treatment with URT commensals protects against infection in humans
The development of probiotics is centered on the concept that commensal bacteria with beneficial properties can be used to improve resistance to disease. While originally developed for intestinal diseases, probiotic gut bacteria also increase resistance to influenza A virus infection in mice [55–57], indicating the potential utility of probiotics for respiratory tract infections. More recently, clinical trials have explored the probiotic potential of URT commensal bacteria. In one study, serial treatment with nasal sprays containing commensal Streptococcus salivarius and Streptococcus oralis bacteria reduced acute otitis media recurrence and severity in children [58•,59]. Similarly, an oral spray containing S. salivarius and S. oralis reduced episodes of pharyngotonsillitis infection and need for antibiotic treatment in children infected with group A beta-hemolytic Streptococcus (GABHS) [60•]. Successful therapy with commensal Streptococcus bacteria relies on repeated inoculation over a period of several weeks to months, and lower doses are not effective [61], indicating the need for further development of this approach. The protective effect of these commensals may depend on direct pathogen competition in addition to the generation of cross-reactive immunity discussed above. Streptococcus commensals can disrupt pre-formed biofilms from several pathogens [62] and produce bacteriocins that kill S. pneumoniae [63], though these observations have not been confirmed in vivo. S. salivarius also restricts S. pneumoniae binding to a human epithelial cell line independent of bacteriocin production [64], further demonstrating that multiple mechanisms contribute to the inhibitory effect of these commensals on pathogen infection.
Other genera in the respiratory tract have similarly antagonistic commensal-pathogen interactions in humans. The nasal commensal Staphylococcus lugdunensis produces lugdunin, an antibiotic that is bactericidal for S. aureus and S. pneumoniae [65•]. While the therapeutic potential of lugdunin has not been evaluated in clinical trials, carriage of S. lugdunensis correlates with reduced S. aureus in people [65•]. In another example of competition between closely related bacteria, nasal inoculation of the commensal Neisseria lactamica reduces carriage of Neisseria meningitidis in young adults [66]. Similar to evidence for cross-reactive Streptococcus immunity, cross-reactive opsonophagocytic antibodies generated in people colonized with N. lactamica may contribute to this effect [67]. These studies identify URT commensal bacteria with probiotic potential for protection against infection with closely related opportunistic pathogens in humans.
URT microbiome composition predicts disease risk
URT microbiome sequencing has revealed that this community undergoes substantial changes during early development and in the context of disease. The identification of respiratory tract commensals that are predominant in healthy, but not infected, people has emerged as a common theme (Figure 2). For example, Corynebacterium propinquum, Dolosigranulum pigrum and Moraxella bacteria in the nasopharynx negatively correlate with lower respiratory tract infection (lower RTI) in young children [68•].
Figure 2.
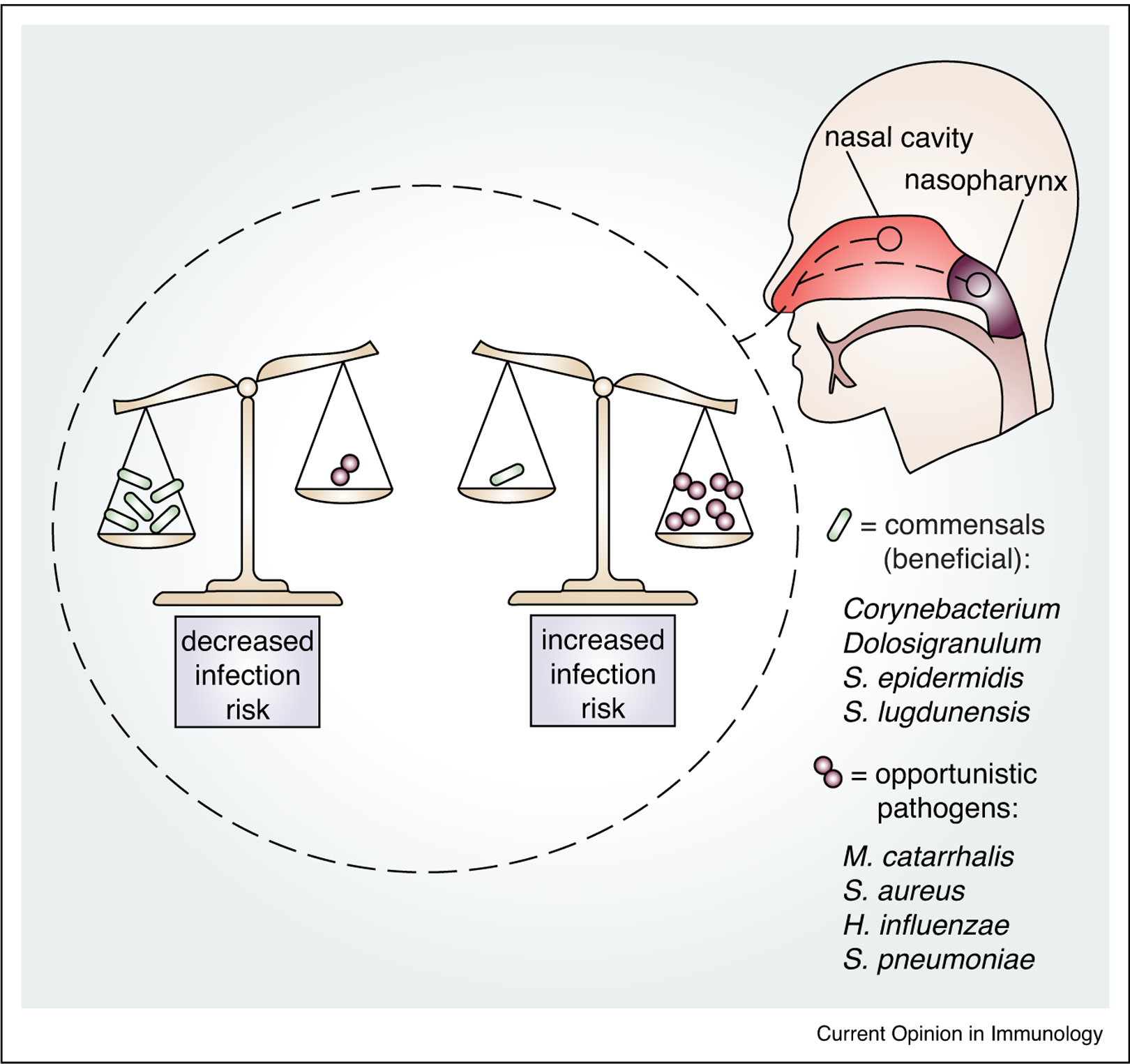
The composition of the URT microbiome of the nasal cavity and nasopharynx influences susceptibility to respiratory tract infection. Scales represent how the balance of colonization with different types of bacteria reflects an individual’s risk of respiratory tract infection. In this example, the scale on the left depicts a higher burden of potentially beneficial commensal bacteria, which is associated with health, while scale on the right depicts a higher burden of opportunistic pathogens, associated with increased risk of respiratory tract infection. URT commensal bacteria that negatively correlate with disease include Corynebacterium, Dolosigranulum, S. epidermidis and S. lugdunensis. URT opportunistic pathogens that increase infection risk include M. catarrhalis, S. aureus, H. influenzae and S. pneumoniae.
In this analysis the species identity for Moraxella was not defined, making the role of the opportunistic pathogen M. catarrhalis unclear. Corynebacterium and Dolosigranulum are also largely absent in the nasopharynx of children with otitis media and other upper RTIs compared with healthy individuals [69•,70–72]. While it is tempting to speculate that these respiratory tract commensals are protective against pathogen acquisition and/or invasion, causation has not been established for most of these relationships. However, accumulating evidence suggests that Corynebacterium species directly compete with URT pathogens. Corynebacterium accolens produces free fatty acids that inhibit S. pneumoniae growth [73•], and cell-free medium from Corynebacterium striatum reduces S. aureus adhesion to epithelial cells [74•]. These findings are supported by the observation that live, but not inactivated, Corynebacterium pseudodiptheriticum reduces RSV and S. pneumoniae lung burdens in an infant rat co-infection model [75••]. In summary, microbiome sequencing has identified URT commensal bacteria with the potential to contribute to pathogen resistance.
In the absence of mechanistic studies in people, longitudinal analysis of URT microbiome profiles in the same individuals over time supports the concept that the composition of this community influences susceptibility to infection. One such study paired microbiome analysis and RTI incidence in infants throughout their first year of life [19••]. Infants with increased RTIs had shorter periods of colonization with Corynebacterium and Dolosigranulum species and more rapid dominance of Moraxella [19••]. This is consistent with Corynebacterium and Dolosigranulum as predominantly associated with the absence of disease, while the impact of Moraxella may be species or time dependent. Moraxella nonliquefaciens is also increased in children with acute sinusitis [76], indicating that species beyond M. catarrhalis are associated with disease. Importantly, changes in the URT microbiome precede the first RTIs in children [19••,20,77]. The predictive power of URT microbiome profiling is consistent with a direct relationship between the composition of this community and susceptibility to pathogen infection. In contrast to the consensus for RTIs considered as a whole, a separate cohort of respiratory tract commensal bacteria negatively correlate with tuberculosis (TB) [78•], indicating that the relationships between the URT microbiome and disease risk are pathogen specific.
Opportunistic pathogens destabilize the URT microbiome
While commensal URT bacteria contribute protective benefits, opportunistic pathogens have the opposite effect. For example, asymptomatic colonization with Streptococcus earlier in life correlates with a younger age of first lower RTI [20]. Pathogen colonization disturbs the microbial community of the URT even in the absence of disease presentation. This has been shown in children, as colonization with Haemophilus and Streptococcus is associated with reduced microbiome stability compared with Corynebacterium, Dolosigranulum and Moraxella [77]. The concept of microbiome disruption by opportunistic pathogens has been directly tested in a human challenge study, where establishment of S. pneumoniae carriage in healthy adults increased URT microbiome diversity [79]. These findings suggest a nuanced relationship between opportunistic pathogens and disease, where asymptomatic changes in the URT microbiome influence subsequent pathogen invasion. Whether these changes modulate invasion of colonizing pathogens or newly acquired opportunistic bacteria remains an important area for further study.
Conclusions and future directions
Despite substantial interest in the influence of the microbiome on human disease, the importance of commensal bacteria from sites beyond the gut remains poorly characterized. Given the dynamic relationships in the URT between commensal and pathogenic species within the same genera, more in-depth microbiome analysis may reveal species-specific relationships that have been previously overlooked. Models of respiratory tract infection that incorporate the manipulation of URT commensal species and pathogen co-infection will improve our ability to predict and manipulate these relationships. Collectively, the work highlighted here demonstrates that URT commensals influence susceptibility to infection through direct and indirect mechanisms, some of which are unique to the environment of the nasal mucosa. Further investigation of how URT commensal bacteria alter pathogen acquisition and immune homeostasis is critical for the development of new therapeutic approaches, the most promising of which include probiotics from and for the respiratory tract.
Acknowledgements
This work was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of HealthK22AI143922.
Footnotes
Conflict of interest statement
Nothing declared.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Global Burden of Disease Pediatrics Collaboration, Kyu HH, Pinho C, Wagner JA, Brown JC, Bertozzi-Villa A, Charlson FJ, Coffeng LE, Dandona L, Erskine HE et al. : Global and national burden of diseases and injuries among children and adolescents between 1990 and 2013. JAMA Pediatr 2016, 170:267–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Broulette J, Yu H, Pyenson B, Iwasaki K, Sato R: The incidence rate and economic burden of community-acquired pneumonia in a working-age population. Am Health Drug Benefits 2013, 6:494–503. [PMC free article] [PubMed] [Google Scholar]
- 3.Centers for Disease Control, Prevention: Deaths: final data for 2017. Natl Vital Stat Rep 2019, 68:1–80. [PubMed] [Google Scholar]
- 4.Vaz LE, Kleinman KP, Raebel MA, Nordin JD, Lakoma MD, Dutta-Linn MM, Finkelstein JA: Recent trends in outpatient antibiotic use in children. Pediatrics 2014, 133:533–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weiser JN, Ferreira DM, Paton JC: Streptococcus pneumoniae: transmission, colonization and invasion. Nat Rev Microbiol 2018, 16:355–367 10.1038/s41579-018-0001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prat C, Lacoma A: Bacteria in the respiratory tract—how to treat? Or do not treat?. Int J Infect Dis 2016, 51:113–122. [DOI] [PubMed] [Google Scholar]
- 7.Sillanpää S, Kramna L, Oikarinen S, Sipilä M, Rautiainen M, Aittoniemi J, Laranne J, Hyöty H, Cinek O: Next-generation sequencing combined with specific PCR assays to determine the bacterial 16S rRNA gene profiles of middle ear fluid collected from children with acute otitis media. mSphere 2017, 2 1417–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Regev-Yochay G, Trzcinski K, Thompson CM, Malley R, Lipsitch M: Interference between Streptococcus pneumoniae and Staphylococcus aureus: in vitro hydrogen peroxide-mediated killing by Streptococcus pneumoniae. J Bacteriol 2006, 188:4996–5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lysenko ES, Ratner AJ, Nelson AL, Weiser JN: The role of innate immune responses in the outcome of interspecies competition for colonization of mucosal surfaces. PLoS Pathog 2005, 1:e1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tan TT, Mörgelin M, Forsgren A, Riesbeck K: Haemophilus influenzae survival during complement-mediated attacks is promoted by Moraxella catarrhalis outer membrane vesicles. J Infect Dis 2007, 195:1661–1670. [DOI] [PubMed] [Google Scholar]
- 11.Briaud P, Camus L, Bastien S, Doléans-Jordheim A, Vandenesch F, Moreau K: Coexistence with Pseudomonas aeruginosa alters Staphylococcus aureus transcriptome, antibiotic resistance and internalization into epithelial cells. Sci Rep 2019, 9:16564 10.1038/s41598-019-52975-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brealey JC, Chappell KJ, Galbraith S, Fantino E, Gaydon J, Tozer S, Young PR, Holt PG, Sly PD: Streptococcus pneumoniae colonization of the nasopharynx is associated with increased severity during respiratory syncytial virus infection in young children. Respirology 2017, 23:220–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Metersky ML, Masterton RG, Lode H, File TM Jr, Babinchak T: Epidemiology, microbiology, and treatment considerations for bacterial pneumonia complicating influenza. Int J Infect Dis 2012, 16:e321–e331. [DOI] [PubMed] [Google Scholar]
- 14.Bakaletz LO: Viral–bacterial co-infections in the respiratory tract. Curr Opin Microbiol 2017, 35:30–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huxley EJ, Viroslav J, Gray WR, Pierce AK: Pharyngeal aspiration in normal adults and patients with depressed consciousness. Am J Med 1978, 64:564–568. [DOI] [PubMed] [Google Scholar]
- 16.Marsh RL, Kaestli M, Chang AB, Binks MJ, Pope CE, Hoffman LR, Smith-Vaughan HC: The microbiota in bronchoalveolar lavage from young children with chronic lung disease includes taxa present in both the oropharynx and nasopharynx. Microbiome 2016, 4:37 10.1186/s40168-016-0182-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bassis CM, Erb-Downward JR, Dickson RP, Freeman CM, Schmidt TM, Young VB, Beck JM, Curtis JL, Huffnagle GB: Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. mBio 2015, 6 e00037–15–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Segal LN, Alekseyenko AV, Clemente JC, Kulkarni R, Wu B, Chen H, Berger KI, Goldring RM, Rom WN, Blaser MJ et al. : Enrichment of lung microbiome with supraglottic taxa is associated with increased pulmonary inflammation. Microbiome 2013, 1 337–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.••.Bosch AATM, de Steenhuijsen Piters WAA, van Houten MA, Chu MLJN, Biesbroek G, Kool J, Pernet P, de Groot P-KCM, Eijkemans MJC, Keijser BJF et al. : Maturation of the infant respiratory microbiota, environmental drivers, and health consequences. A prospective cohort study. Ame J Respir Crit Care Med 2017, 196:1582–1590. [DOI] [PubMed] [Google Scholar]; In this study authors tracked URT microbial profiles in infants from birth through the first year of life. They found that children with more frequent RTIs had altered URT microbiome maturation, including reduced microbiome stability and less Corynebacterium and Dolosigranulum colonization.
- 20.Teo SM, Mok D, Pham K, Kusel M, Serralha M, Troy N, Holt BJ, Hales BJ, Walker ML, Hollams E et al. : The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe 2015, 17:704–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schuijt TJ, Lankelma JM, Scicluna BP, de Sousa e Melo F, Roelofs JJTH, de Boer JD, Hoogendijk AJ, de Beer R, de Vos A, Belzer C et al. : The gut microbiota plays a protective role in the host defence against pneumococcal pneumonia. Gut 2016, 65:575–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abt MC, Osborne LC, Monticelli LA, Doering TA, Alenghat T, Sonnenberg GF, Paley MA, Antenus M, Williams KL, Erikson J et al. : Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity 2012, 37:158–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ichinohe T, Pang IK, Kumamoto Y, Peaper DR, Ho JH, Murray TS, Iwasaki A: Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc Natl Acad Sci U S A 2011, 108:5354–5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dumas A, Corral D, Colom A, Levillain F, Peixoto A, Hudrisier D, Poquet Y, Neyrolles O: The host microbiota contributes to early protection against lung colonization by Mycobacterium tuberculosis. Front Immunol 2018, 9:e183–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McAleer JP, Nguyen NLH, Chen K, Kumar P, Ricks DM, Binnie M, Armentrout RA, Pociask DA, Hein A, Yu A et al. : Pulmonary Th17 antifungal immunity is regulated by the gut microbiome. J Immunol 2016, 197:97–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fagundes CT, Amaral FA, Vieira AT, Soares AC, Pinho V, Nicoli JR, Vieira LQ, Teixeira MM, Souza DG: Transient TLR activation restores inflammatory response and ability to control pulmonary bacterial infection in germfree mice. J Immunol 2012, 188:1411–1420. [DOI] [PubMed] [Google Scholar]
- 27.Wang L-M, Qiao X-L, Ai L, Zhai J-J, Wang X-X: Isolation of antimicrobial resistant bacteria in upper respiratory tract infections of patients. 3 Biotech 2016, 6:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feazel LM, Robertson CE, Ramakrishnan VR, Frank DN: Microbiome complexity and Staphylococcus aureus in chronic rhinosinusitis. Laryngoscope 2012, 122:467–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.••.Brown RL, Sequeira RP, Clarke TB: The microbiota protects against respiratory infection via GM-CSF signaling. Nat Commun 2017, 8:1512. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study identified a common immune signaling pathway activated by commensals from distinct host sites. Nod2 stimulation by URT or gut bacteria promotes IL-17A production in the lung, resulting in GM-CSF-dependent activation of alveolar macrophages and increased protection against S. pneumoniae and K. pneumoniae lung infection in mice.
- 30.Kim D, Kim Y-G, Seo S-U, Kim D-J, Kamada N, Prescott D, Chamaillard M, Philpott DJ, Rosenstiel P, Inohara N et al. : Nod2-mediated recognition of the microbiota is critical for mucosal adjuvant activity of cholera toxin. Nat Med 2016, 22:524–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vieira AT, Rocha VM, Tavares L, Garcia CC, Teixeira MM, Oliveira SC, Cassali GD, Gamba C, Martins FS, Nicoli JR: Control of Klebsiella pneumoniae pulmonary infection and immunomodulation by oral treatment with the commensal probiotic Bifidobacterium longum 51A. Microbes Infect 2016, 18:180–189. [DOI] [PubMed] [Google Scholar]
- 32.Gauguet S, D’Ortona S, Ahnger-Pier K, Duan B, Surana NK, Lu R, Cywes-Bentley C, Gadjeva M, Shan Q, Priebe GP et al. : Intestinal microbiota of mice influences resistance to Staphylococcus aureus pneumonia. Infect Immun 2015, 83:4003–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Felix KM, Jaimez IA, Nguyen T-VV, Ma H, Raslan WA, Klinger CN, Doyle KP, Wu H-JJ: Gut microbiota contributes to resistance against pneumococcal pneumonia in immunodeficient Rag−/− mice. Front Cell Infect Microbiol 2018, 8 371–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gray J, Oehrle K, Worthen G, Alenghat T, Whitsett J, Deshmukh H: Intestinal commensal bacteria mediate lung mucosal immunity and promote resistance of newborn mice to infection. Sci Transl Med 2017, 9:eaaf9412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C, Blanchard C, Junt T, Nicod LP, Harris NL et al. : Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med 2014, 20:159–166. [DOI] [PubMed] [Google Scholar]
- 36.Cait A, Hughes MR, Antignano F, Cait J, Dimitriu PA, Maas KR, Reynolds LA, Hacker L, Mohr J, Finlay BB et al. : Microbiome-driven allergic lung inflammation is ameliorated by short-chain fatty acids. Mucosal Immunol 2017, 11:785–795. [DOI] [PubMed] [Google Scholar]
- 37.Tonetti FR, Islam MA, Vizoso-Pinto MG, Takahashi H, Kitazawa H, Villena J: Nasal priming with immunobiotic lactobacilli improves the adaptive immune response against influenza virus. Int Immunopharmacol 2020, 78:106115. [DOI] [PubMed] [Google Scholar]
- 38.Barbieri N, Herrera M, Salva S, Villena J, Alvarez S: Lactobacillus Rhamnosus CRL1505 nasal administration improves recovery of T-cell mediated immunity against pneumococcal infection in malnourished mice. Beneficial Microbes 2017, 8:393–405. [DOI] [PubMed] [Google Scholar]
- 39.Khan N, Vidyarthi A, Nadeem S, Negi S, Nair G, Agrewala JN: Alteration in the gut microbiota provokes susceptibility to tuberculosis. Front Immunol 2016, 7:3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Das B, Dobrowolski C, Shahir A-M, Feng Z, Yu X, Sha J, Bissada NF, Weinberg A, Karn J, Ye F: Short chain fatty acids potently induce latent HIV-1 in T-cells by activating P-TEFb and multiple histone modifications. Virology 2015, 474:65–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCaskill JG, Chason KD, Hua X, Neuringer IP, Ghio AJ, Funkhouser WK, Tilley SL: Pulmonary immune responses to Propionibacterium acnes in C57BL/6 and BALB/c mice. Am J Respir Cell Mol Biol 2006, 35:347–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.••.Kim HJ, Jo A, Jeon YJ, An S, Lee K-M, Yoon SS, Choi JY: Nasal commensal Staphylococcus epidermidis enhances interferon-λ- dependent immunity against influenza virus. Microbiome 2019, 7:80 10.1186/s40168-019-0691-9. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study demonstrated that the respiratory tract commensal S. epidermidis promotes IFN-λ production by the nasal epithelium, which correlates with increased resistance to influenza A virus infection in mice. A positive association between S. epidermidis colonization and nasal IFN-λ in human nasal secretions is also shown.
- 43.Bradley KC, Finsterbusch K, Schnepf D, Crotta S, Llorian M, Davidson S, Fuchs SY, Staeheli P, Wack A: Microbiota-driven tonic interferon signals in lung stromal cells protect from influenza virus infection. Cell Rep 2019, 28:245–256.e4. [DOI] [PubMed] [Google Scholar]
- 44.••.Liu Q, Liu Q, Meng H, Lv H, Liu Y, Liu J, Wang H, He L, Qin J, Wang Y et al. : Staphylococcus epidermidis contributes to healthy maturation of the nasal microbiome by stimulating antimicrobial peptide production. Cell Host Microbe 2019, 27:68–78.e5 10.1016/j.chom.2019.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study showed that the stimulation of AMP production during nasal colonization with the respiratory tract commensal S. epidermidis restricts S. aureus and M. catarrhalis colonization following nasal challenge in mice.
- 45.Janek D, Zipperer A, Kulik A, Krismer B, Peschel A: High frequency and diversity of antimicrobial activities produced by nasal Staphylococcus strains against bacterial competitors. PLoS Pathog 2016, 12:e1005812–e1005820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Iwase T, Uehara Y, Shinji H, Tajima A, Seo H, Takada K, Agata T, Mizunoe Y: Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature 2010, 465:346–349 10.1038/nature09074. [DOI] [PubMed] [Google Scholar]
- 47.Park B, Iwase T, Liu GY: Intranasal application of S. epidermidis prevents colonization by methicillin-resistant Staphylococcus aureus in mice. PLoS One 2011, 6:e25880–e25885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Latham RD, Gell DA, Fairbairn RL, Lyons AB, Shukla SD, Cho KY, Jones DA, Harkness NM, Tristram SG: An isolate of Haemophilus haemolyticus produces a bacteriocin-like substance that inhibits the growth of nontypeable Haemophilus influenzae. Int J Antimicrob Agents 2017, 49:503–506. [DOI] [PubMed] [Google Scholar]
- 49.•.Granland CM, Scott NM, Lauzon-Joset J-F, Langereis JD, de Gier C, Sutherland K, Clark SL, Pickering JL, Thornton RB, Richmond PC et al. : Nasal delivery of a commensal Pasteurellaceae species inhibits nontypeable Haemophilus influenzae colonisation and delays onset of otitis media in mice. Infect Immun 2020, 88:e00685–19 10.1128/IAI.00685-19. [DOI] [PMC free article] [PubMed] [Google Scholar]; In this study the authors found that mice inoculated with Muribacter muris, a commensal in the same family as H. influenzae that is naturally present in mice, reduces H. influenzae colonization and invasion of the middle ear using an influenza A virus co-infection model. M. muris inoculation modulated the expression of pro-inflammatory cytokines in the URT.
- 50.••.Shekhar S, Khan R, Schenck K, Petersen FC: Intranasal immunization with the commensal Streptococcus mitis confers protective immunity against pneumococcal lung infection. Appl Environ Microbiol 2019, 85 893–13. [DOI] [PMC free article] [PubMed] [Google Scholar]; In this study the authors found that immunization with the URT commensal S. mitis is protective against S. pneumoniae lung infection in mice. Systemic cross-reactive IgG and IgA antibodies as well as increased lung IL-17A and Th17 cells were observed in S. mitis immunized mice.
- 51.Engen SA, Valen Rukke H, Becattini S, Jarrossay D, Blix IJ, Petersen FC, Sallusto F, Schenck K: The oral commensal Streptococcus mitis shows a mixed memory Th cell signature that is similar to and cross-reactive with Streptococcus pneumoniae. PLoS One 2014, 9:e104306–e104309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.•.Skov Sørensen UB, Yao K, Yang Y, Tettelin H, Kilian M: Capsular polysaccharide expression in commensal Streptococcus species: genetic and antigenic similarities to Streptococcus pneumoniae. mBio 2016, 7 762–17. [DOI] [PMC free article] [PubMed] [Google Scholar]; In this study the genetic loci for expression of capsular polysaccharide were observed in a large number of commensal Streptococcus species, and cross-reactivity with capsule expressed by S. pneumoniae was identified.
- 53.Pimenta F, Gertz RE Jr, Park SH, Kim E, Moura I, Milucky J, Rouphael N, Farley MM, Harrison LH, Bennett NM et al. : Streptococcus infantis, Streptococcus mitis, and Streptococcus oralis strains with highly similar cps5 loci and antigenic relatedness to serotype 5 pneumococci. Front Microbiol 2019, 9:3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.•.Lessa FC, Milucky J, Rouphael NG, Bennett NM, Talbot HK, Harrison LH, Farley MM, Walston J, Pimenta F, Gertz RE et al. : Streptococcus mitis expressing pneumococcal serotype 1 capsule. Sci Rep 2018, 8:17959. [DOI] [PMC free article] [PubMed] [Google Scholar]; In this study authors identified commensal S. mitis bacteria that express capsule with antigenic and immunogenic similarity to serotype 1 capsule expressed by S. pneumoniae. It should be noted that serotype 1 is targeted by the currently licensed PCV-13 vaccine.
- 55.Kechaou N, Chain F, Gratadoux J-J, Blugeon S, Bertho N, Chevalier C, Le Goffic R, Courau S, Molimard P, Chatel JM et al. : Identification of one novel candidate probiotic Lactobacillus plantarum strain active against influenza virus infection in mice by a large-scale screening. Appl Environ Microbiol 2013, 79:1491–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nakayama Y, Moriya T, Sakai F, Ikeda N, Shiozaki T, Hosoya T, Nakagawa H, Miyazaki T: Oral administration of Lactobacillus gasseri SBT2055 is effective for preventing influenza in mice. Sci Rep 2014, 4:523–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kawase M, He F, Kubota A, Yoda K, Miyazawa K, Hiramatsu M: Heat-killed Lactobacillus gasseri TMC0356 protects mice against influenza virus infection by stimulating gut and respiratory immune responses. FEMS Immunol Med Microbiol 2011, 64:280–288. [DOI] [PubMed] [Google Scholar]
- 58.•.La Mantia I, Varricchio A, Ciprandi G: Bacteriotherapy with Streptococcus salivarius 24SMB and Streptococcus oralis 89a nasal spray for preventing recurrent acute otitis media in children: a real-life clinical experience. IJGM 2017, 10:171–175. [DOI] [PMC free article] [PubMed] [Google Scholar]; In this study the authors found that children treated with a nasal spray containing the URT commensals S. salivarius and S. oralis had reduced otitis media recurrence after an initial episode of acute otitis media, compared to the control group.
- 59.Marchisio P, Santagati M, Scillato M, Baggi E, Fattizzo M, Rosazza C, Stefani S, Esposito S, Principi N: Streptococcus salivarius 24SMB administered by nasal spray for the prevention of acute otitis media in otitis-prone children. Eur J Clin Microbiol Infect Dis 2015, 34:2377–2383. [DOI] [PubMed] [Google Scholar]
- 60.•.Andaloro C: Bacteriotherapy with Streptococcus salivarius 24SMB and Streptococcus oralis 89a oral spray for children with recurrent streptococcal pharyngotonsillitis: a randomized placebo-controlled clinical study. Eur Arch Oto Rhino Laryngol 2019, 276:879–887. [DOI] [PubMed] [Google Scholar]; In this study authors treated children with confirmed group A beta-hemolytic Streptococcus (GABHS) with an oral spray containing the URT commensals S. salivarius and S. oralis. Children treated with bacterial sprays had less frequent and shorter episodes of GABHS pharyngotonsillitis and less antibiotic treatment compared to the control group.
- 61.Tano K, Kakansson EG, Holm SE, Hellstrom S: A nasal spray with alpha-haemolytic streptococci as long term prophylaxis against recurrent otitis media. Int J Pediatr Otorhinolaryngol 2001, 62:17–23. [DOI] [PubMed] [Google Scholar]
- 62.Bidossi A, De Grandi R, Toscano M, Bottagisio M, De Vecchi E, Gelardi M, Drago L: Probiotics Streptococcus salivarius 24SMB and Streptococcus oralis 89a interfere with biofilm formation of pathogens of the upper respiratory tract. BMC Infect Dis 2018, 18:653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Santagati M, Scillato M, Patanè F, Aiello C, Stefani S: Bacteriocin-producing oral streptococci and inhibition of respiratory pathogens. FEMS Immunol Med Microbiol 2012, 65:23–31. [DOI] [PubMed] [Google Scholar]
- 64.Manning J, Dunne EM, Wescombe PA, Hale JDF, Mulholland EK, Tagg JR, Robins-Browne RM, Satzke C: Investigation of Streptococcus salivarius-mediated inhibition of pneumococcal adherence to pharyngeal epithelial cells. BMC Microbiol 2016, 16:225 10.1186/s12866-016-0843-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.•.Zipperer A, Konnerth MC, Laux C, Berscheid A, Janek D, Weidenmaier C, Burian M, Schilling NA, Slavetinsky C, Marschal M et al. : Human commensals producing a novel antibiotic impair pathogen colonization. Nature 2016, 535:511–516. [DOI] [PubMed] [Google Scholar]; In this study authors describe a new antibiotic called lugdunin produced by the commensal S. lugdunensis that is bactericidal for S. aureus in vitro and reduces S. aureus growth in a rat nasal colonization model. S. lugdunensis is also negatively associated with S. aureus colonization in humans.
- 66.Deasy AM, Guccione E, Dale AP, Andrews N, Evans CM, Bennett JS, Bratcher HB, Maiden MCJ, Gorringe AR, Read RC: Nasal inoculation of the commensal Neisseria lactamica inhibits carriage of Neisseria meningitidis by young adults: a controlled human infection study. Clin Infect Dis 2015, 60:1512–1520. [DOI] [PubMed] [Google Scholar]
- 67.Evans CM, Pratt CB, Matheson M, Vaughan TE, Findlow J, Borrow R, Gorringe AR, Read RC: Nasopharyngeal colonization by Neisseria lactamica and induction of protective immunity against Neisseria meningitidis. Clin Infect Dis 2011, 52:70–77. [DOI] [PubMed] [Google Scholar]
- 68.•.Man WH, van Houten MA, Mérelle ME, Vlieger AM, Chu MLJN, Jansen NJG, Sanders EAM, Bogaert D: Bacterial and viral respiratory tract microbiota and host characteristics in children with lower respiratory tract infections: a matched case-control study. Lancet Respir 2019, 7:417–426 10.1016/S2213-2600(18)30449-1. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study demonstrates an association between the composition of the URT microbiome and the presence and severity of lower RTIs in children. Bacteria including Corynebacterium, Dolosigranulum and Moraxella negatively correlate with lower RTIs.
- 69.•.Lappan R, Imbrogno K, Sikazwe C, Anderson D, Mok D, Coates H, Vijayasekaran S, Bumbak P, Blyth CC, Jamieson SE et al. : A microbiome case-control study of recurrent acute otitis media identified potentially protective bacterial genera. BMC Microbiol 2018, 18:13 10.1186/s12866-018-1154-3. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study identified bacteria including Corynebacterium and Dolosigranulum as more abundant in the nasopharynx of healthy children compared to those with recurrent acute otitis media.
- 70.Pettigrew MM, Laufer AS, Gent JF, Kong Y, Fennie KP, Metlay JP: Upper respiratory tract microbial communities, acute otitis media pathogens, and antibiotic use in healthy and sick children. Appl Environ Microbiol 2012, 78:6262–6270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Laufer AS, Metlay JP, Gent JF, Fennie KP, Kong Y, Pettigrew MM: Microbial communities of the upper respiratory tract and otitis media in children. mBio 2011, 2:e34–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kelly MS, Surette MG, Smieja M, Pernica JM, Rossi L, Luinstra K, Steenhoff AP, Feemster KA, Goldfarb DM, Arscott-Mills T et al. : The nasopharyngeal microbiota of children with respiratory infections in Botswana. Pediatr Infect Dis J 2017, 36:e211–e218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.•.Bomar L, Brugger SD, Yost BH, Davies SS, Lemon KP: Corynebacterium accolens releases antipneumococcal free fatty acids from human nostril and skin surface triacylglycerols. mBio 2016, 7 144–13. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study identifies a mechanism for direct competition between commensal Corynebacterium and S. pneumoniae. The authors found that C. accolens hydrolysis of fatty acids releases free fatty acid products that inhibit S. pneumoniae growth in vitro.
- 74.•.Ramsey MM, Freire MO, Gabrilska RA, Rumbaugh KP, Lemon KP: Staphylococcus aureus shifts toward commensalism in response to Corynebacterium species. Front Microbiol 2016, 7 279–15. [DOI] [PMC free article] [PubMed] [Google Scholar]; In this study the authors observed that cell-free medium from Corynebacterium striatum blocks S. aureus adhesion to epithelial cells. C. striatum co-culture also reduces virulence gene expression in S. aureus.
- 75.••.Kanmani P, Clua P, Vizoso-Pinto MG, Rodriguez C, Alvarez S, Melnikov V, Takahashi H, Kitazawa H, Villena J: Respiratory commensal bacteria Corynebacterium pseudodiphtheriticum improves resistance of infant mice to respiratory syncytial virus and Streptococcus pneumoniae superinfection. Front Microbiol 2017, 8:1613. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study demonstrates an inhibitory effect for Corynebacterium pseudodiptheriticum on RSV and S. pneumoniae co-infection in the lung. Pre-treatment with live, but not heat-killed, C. pseudodiptheriticum reduced viral and bacterial lung burdens and inflammation in an infant rat co-infection model.
- 76.Santee CA, Nagalingam NA, Faruqi AA, DeMuri GP, Gern JE, Wald ER, Lynch SV: Nasopharyngeal microbiota composition of children is related to the frequency of upper respiratory infection and acute sinusitis. Microbiome 2016, 4:34 10.1186/s40168-016-0179-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Biesbroek G, Tsivtsivadze E, Sanders EAM, Montijn R, Veenhoven RH, Keijser BJF, Bogaert D: Early respiratory microbiota composition determines bacterial succession patterns and respiratory health in children. Am J Respir Crit Care Med 2014, 190:1283–1292. [DOI] [PubMed] [Google Scholar]
- 78.•.Hong B-Y, Paulson JN, Stine OC, Weinstock GM, Cervantes JL: Meta-analysis of the lung microbiota in pulmonary tuberculosis. Tuberculosis 2018, 109:102–108. [DOI] [PubMed] [Google Scholar]; This study identified bacteria including Propionibacterium acnes, Haemophilus parahaemolyticus and Tumebacillus ginsengisoli that negatively correlate with Mycobacterium tuberculosis infection.
- 79.Cremers AJ, Zomer AL, Gritzfeld JF, Ferwerda G, van Hijum SA, Ferreira DM, Shak JR, Klugman KP, Boekhorst J, Timmerman HM et al. : The adult nasopharyngeal microbiome as a determinant of pneumococcal acquisition. Microbiome 2014, 2 44–10. [DOI] [PMC free article] [PubMed] [Google Scholar]


