Summary:
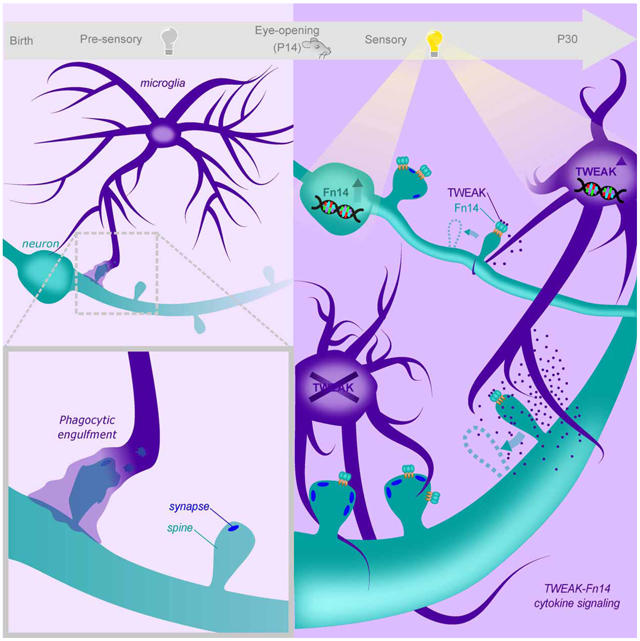
Sensory experience remodels neural circuits in the early postnatal brain through mechanisms that remain to be elucidated. Applying a new method of ultrastructural analysis to the retinogeniculate circuit, we find that visual experience alters the number and structure of synapses between the retina and the thalamus. These changes require the vision-dependent transcription of the receptor Fn14 in thalamic relay neurons and the induction of its ligand TWEAK in microglia. Fn14 functions to increase the number of bulbous spine-associated synapses at retinogeniculate connections likely contributing to the strengthening of the circuit that occurs in response to visual experience. However, at retinogeniculate connections nearby TWEAK-expressing microglia, TWEAK signals via Fn14 to restrict the number of bulbous spines on relay neurons, leading to the elimination of a subset of connections. Thus, TWEAK and Fn14 represent an intercellular signaling axis through which microglia shape retinogeniculate connectivity in response to sensory experience.
Keywords: Synapse, synaptic refinement, microglia, pruning, sensory experience, dendritic spine, development
Graphical Abstract
eToc summary:
Sensory experience induces Fn14 expression in relay neurons and TWEAK expression in microglia to drive the refinement of retinogeniculate connectivity. Microglial TWEAK signals through neuronal Fn14 to eliminate a subset of synapses proximal to TWEAK-expressing microglia while Fn14 acts alone at other synapses to strengthen connectivity.
Introduction:
The connectivity of the mature brain is established through a convergence of intrinsic biological factors (Nature) and environmental cues (Nurture). While early stages of neural circuit assembly are governed by genetic programs in utero, nascent circuits are extensively refined in response to sensory experience during postnatal brain development (Katz and Shatz, 1996; Wiesel and Hubei, 1963). This dynamic process of sensory-dependent (SD) refinement tunes the connectivity of a given neuron by determining which of its immature synaptic connections are strengthened and maintained and which connections are eliminated. Impairments in synaptic refinement contribute to neurodevelopmental disorders such as autism and schizophrenia, and the aberrant re-activation of refinement in the mature brain may contribute to neurodegeneration (Feinberg, 1982; Hammond et al., 2019). While these observations underscore the importance of refinement for brain function and human health, the cellular and molecular mechanisms through which sensory experience refines developing circuits remain incompletely understood.
Because many cellular processes occur simultaneously in the early postnatal brain, it has been difficult to study synaptic refinement in isolation. This challenge has been addressed in part by studies of the retinogeniculate pathway of the mouse, a visual circuit that undergoes a robust phase of refinement across the first postnatal month (Hooks and Chen, 2020). In this circuit, retinal ganglion cell (RGC) axons synapse onto the dendrites of excitatory relay neurons in the dorsal lateral geniculate nucleus (dLGN) of the thalamus, which relay visual information to the cortex (Fig. 1A). These connections between RGCs and relay neurons are complex, with each presynaptic RGC and its postsynaptic relay neuron partner sharing multiple synapses. Retinogeniculate refinement has been shown by electrophysiology to entail the strengthening of a few immature RGC inputs onto a given relay neuron and the concurrent elimination of inputs that fail to strengthen (Chen and Regehr, 2000). These physiological changes occur at the same time that a structural remodeling of retinal axons that synapse onto their targets in the dLGN is observed (Hong et al., 2014). While these changes in connectivity are initially driven by sensory-independent brain activity between birth and postnatal day 20 (P20), sensory experience drives further refinement during a critical period between P20 and P30 (Hooks and Chen, 2006, 2008).
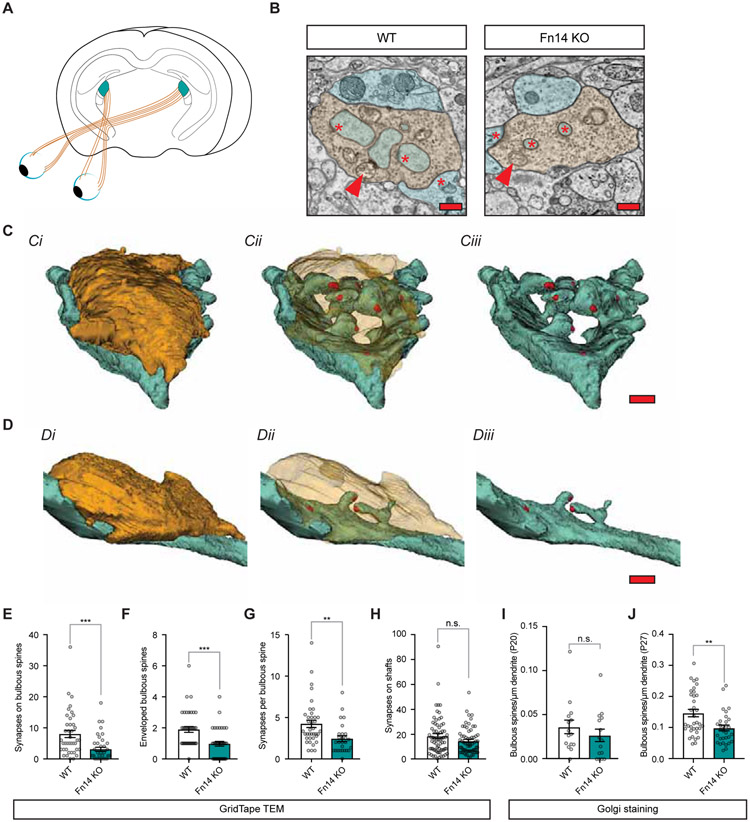
Figure 1. Sensory experience increases the number of spine-associated synapses through Fn14.
(A) Schematic of the retinogeniculate circuit. Axons of retinal ganglion cells (RGCs; orange) synapse onto relay neurons of the dorsal lateral geniculate nucleus (dLGN) of the thalamus (teal).
(B) Single panel cross-sections of retinogeniculate connections in Fn14 knockout (KO) and wildtype (WT) mice identified by GridTape. Synapses, red asterisks; dendrites, blue; spines, teal; boutons, orange; arrows, mitochondria. Scale bar, 500 nm.
(C) Three-dimensional reconstruction of a RGC bouton (orange) converging upon relay neuron dendrites (blue) and spines (teal) in the WT mouse. Synapses in red. Ci, Opague bouton; Cii, Transparent bouton; Ciii, No bouton. Scale bar, 500 nm.
(D) Reconstruction of a retinogeniculate connection in the Fn14 KO mouse, color conventions and figure organization as described in (C). Scale bar, 500 nm.
(E) The number of bulbous-spine-associated synapses is decreased in the Fn14 KO mouse compared to WT.
(F) The number of bulbous spines enveloped by RGC boutons is decreased in the Fn14 KO mouse compared to WT.
(G) The number of synapses per bulbous spine is decreased in the Fn14 KO mouse compared to WT.
(H) The number of synapses on dendritic shafts is unaffected by loss of Fn14.
(I) The density of bulbous spines as measured by Golgi staining is unaffected by loss of Fn14 at P20.
(J) The density of bulbous spines as measured by Golgi staining is significantly decreased in constitutive Fn14 KO mice at P27.
Student’s t-test. **p < 0.01; ***p < 0.001. Means plotted with individual data points, +/− S.E.M. N values in STAR methods.
Due to a lack of molecular analyses of the late postnatal dLGN, there is currently an incomplete understanding of the mechanisms that regulate the SD refinement of the retinogeniculate circuit. By contrast, several studies have shown that molecules commonly associated with the innate immune system, including MHC class I proteins, coordinate phases of retinogeniculate refinement that occur prior to the onset of experience (Corriveau et al., 1998; Datwani et al., 2009). More recently, it was discovered that microglia, the resident immune cells of the brain, sculpt neural circuits through the phagocytic engulfment of presynaptic inputs during this early sensory-independent phase of development (Paolicelli et al., 2011; Schafer et al., 2012; Stevens et al., 2007; Tremblay et al., 2010). Yet, it has been unclear whether the functions of immune-related molecules in brain development are restricted to early phases of refinement or are also engaged to coordinate SD refinement between P20 and P30.
To address this gap in knowledge, we previously characterized SD gene expression in the dLGN using single-cell transcriptomics, identifying the cell-surface cytokine receptor Fn14 (Fibroblast Growth Factor-Inducible protein, 14 kDa) as the most highly induced molecule in relay neurons in response to visual stimulation. Fn14, a member of the Tumor Necrosis Factor (TNF) Receptor Superfamily, binds the TNF family cytokine TWEAK (TNF-associated Weak Inducer of Apoptosis)(Wiley and Winkles, 2003), which we show here is induced by visual experience selectively in microglia. Electrophysiological analysis in an Fn14 knockout (KO) mouse demonstrated that, in the absence of Fn14, the connections between RGCs and relay neurons fail to strengthen, and that these weak connections between RGCs and relay neurons are not properly eliminated. Notably, this dual requirement of Fn14 for RGC input strengthening and elimination is restricted to the SD phase of refinement, as earlier phases of circuit development proceed normally in the absence of Fn14 (Cheadle et al., 2018).
In the current study, we combine a new method of automated serial transmission electron microscopy (TEM) (https://doi.org/10.1101/2020.01.10.902478; Graham et al, 2019) and Golgi staining to profile structural features of synapses at the peak of SD refinement in the dLGN. We find that Fn14 expressed by relay neurons promotes an increase in the number of bulbous-shaped relay neuron spines that form synapses with RGC axons. However, when microglia expressing TWEAK are near thalamic relay neuron dendritic spines, the Fn14- dependent increase in spine number is suppressed. This effect of TWEAK appears not to involve microglial engulfment of synapses, but instead requires experience-dependent signaling within dLGN relay neurons. Thus, we have identified a mechanism through which experience engages microglia to shape synaptic connectivity during refinement of the retinogeniculate circuit.
Results:
Analysis of retinogeniculate synapses at nanometer resolution
In WT mice, visual function is established in part through the strengthening of retinogeniculate connections between P20 and P27 in response to visual experience. One feature that is thought to inform the strength of a retinogeniculate connection is the number of synapses shared by a RGC and a given relay neuron in the dLGN (Hamos et al., 1987). Electrophysiological analysis revealed that when Fn14 function is disrupted synaptic strengthening fails to occur, and it has been suggested that this failure may arise from a decrease in the number of synapses linking RGCs to relay neurons (Cheadle et al., 2018). To determine whether Fn14 strengthens connectivity by increasing the number of synapses formed by RGC inputs, we carried out an ultrastructural analysis of individual retinogeniculate connections in the dLGNs of an Fn14 KO mouse and a WT littermate at P27, a timepoint at which sensory experience has been shown by electrophysiology to actively strengthen synapses via an Fn14-dependent mechanism (Cheadle et al., 2018). We employed a new semi-automated serial transmission EM (TEM) method called GridTape to visualize and quantify retinogeniculate synapses at nanometer resolution. This method leverages semi-automated section imaging and alignment, allowing us to reconstruct synapses in large volumes of dLGN tissue from a WT and an Fn14 KO mouse in parallel while blinded to condition (Fig. S1).
Using serial TEM of ultrathin 40 nm sections, we imaged ~6 million μm3 regions from the dLGNs of an Fn14 KO mouse and a WT mouse and identified the following features: (1) presynaptic RGC boutons, axonal compartments containing neurotransmitter-filled vesicles that converge upon dendrites of the dLGN. These boutons are distinguished from other inputs based upon their large size and the presence of pale mitochondria (Fig. 1B); (2) dendrites, postsynaptic branches of relay neurons that contain the molecular machinery for processing incoming neurotransmission from retinal boutons; (3) dendritic spines, actin-rich protrusions that contain neurotransmitter receptors and receive synaptic input from RGCs; and (4) synapses, specific loci at which docked neurotransmitter vesicles in the presynaptic bouton release glutamate onto postsynaptic specializations of the relay neuron dendrite or dendritic spine.
In the WT dLGN, we found that 60% of RGC boutons made contact with dendritic segments of relay neurons containing spines, while 40% of RGCs converged upon smooth segments of dendrite. RGC boutons are relatively large and the majority not only contact spines but envelope the spines, forming multiple synapses onto both the dendritic shaft and the spines themselves (Fig. 1B,C). The convergence of RGC inputs onto spines is unique compared to other sensory-processing brain regions such as the cortex, in which presynaptic inputs are smaller and do not typically envelop spines (Morgan et al., 2016; Rafols and Valverde, 1973). Here, we show that these uniquely complex interactions between RGC inputs and spines make up a significant proportion of retinogeniculate connections.
Since spines have not yet been systematically characterized in the mouse dLGN, we quantified the morphologies of spines within RGC boutons. While live imaging of spines in the cortex has shown spines to exist along a structural continuum (Berry and Nedivi, 2017; Holtmaat and Svoboda, 2009), analyses of spines by techniques such as Golgi staining and EM have focused on the distinct characteristics of spine morphology as viewed in fixed tissue. These characteristics include spine length and spine head diameter (Parajuli et al., 2017; Peters and Kaiserman-Abramof, 1970; Sorra and Harris, 2000). In this study we measured these objective structural parameters as well as the numbers of spines corresponding to structurally defined subclasses: “bulbous spines” (similar to ‘mushroom’ spines in other systems) which we defined as having a relatively large spine head that is at least twice as wide as the spine neck; “thin spines” defined by a uniform diameter (head width to neck width ratio less than 2) of less than 0.5 μm; and spines of uniform diameter greater than 0.5 μm which we called “broad non-bulbous spines” (these correspond in part to ‘stubby spines’ in other systems)(Fig. S2A). Results of hierarchical clustering of the distribution of spines based upon head-to-neck width ratio were consistent with these morphological categories representing distinct classes (Fig. S2B,C).
Applying these classifications to our WT dataset, we found that the majority of spines that were enveloped by retinal inputs, about 60%, have a bulbous morphology. Although retinal inputs represent only about 10% of total synaptic inputs to the dLGN, synaptic contacts deriving from other brain regions – most prominently visual cortex – were observed to converge upon bulbous spines less frequently, suggesting that bulbous spines may be specialized for mediating incoming sensory information from the retina. Interestingly, we found that the different classes of spines contained varying numbers of synapses: bulbous spines contained on average ~4 synapses per spine, non-bulbous spines ~2 synapses per spine, and thin spines ~1 synapse per spine, suggesting that the different spine types make distinct contributions to synaptic strength Based upon this observation, bulbous spines, which contain the most synapses, are likely to be mediators of strong retinogeniculate connections.
Fn14 increases retinal bouton convergence onto bulbous spines
Analysis of retinogeniculate synapses across the dLGN of a P27 Fn14 KO mouse revealed that 30% of RGC inputs converged upon spines in the absence of Fn14, in contrast to 60% in WT mice (Fig. 1B-D). Classification of spines by morphological subtype in the Fn14 KO dLGN revealed that this decrease in spine-convergent inputs is selective for RGC inputs converging upon bulbous spines (Fig. 1E) Not only was the number of retinal input-contacted bulbous spines ~50% lower in the Fn14 KO mouse (Fig. 1F), but the bulbous spines that were associated with retinal inputs contained ~40% fewer synapses on average (Fig. 1G). It is not clear whether the decrease in bulbous spines overall and the decrease in synapses per spine represent distinct parallel mechanisms or are two steps of a single process. In either case, the decrease in bulbous spine-associated synapses contributed to a substantial 30% decrease in the total number of synapses connecting the retina to the dLGN in the absence of Fn14. In contrast, the numbers of synapses associated with dendritic shafts, non-bulbous spines, and thin spines in the Fn14 KO dLGN were equivalent to those in the WT (Fig. 1H and Fig. S2D-I), suggesting that the regulation of synapses by Fn14 is selective to bulbous spines. Given that the reported deficits in synaptic strengthening in the Fn14 KO correlate with a significant decrease in bulbous-spine-associated synapses, we reasoned that the formation or maintenance of synapse-containing bulbous spines by Fn14 is likely to underlie SD synaptic strengthening in the dLGN.
Developmental spine changes are coordinated by experience and postsynaptic Fn14
We next employed morphological spine analysis via Golgi staining as a surrogate read-out of the changes in synaptic strength that occur during postnatal dLGN development. We performed Golgi staining on the dLGNs of constitutive Fn14 KO and WT mice at P20 and P27, the time points flanking the SD phase of retinogeniculate refinement, to determine whether we can detect the decrease in bulbous spines that we had observed by TEM using this more tractable method. We found that spines are normal in the absence of Fn14 at P20 (Fig. 1I) but the number of bulbous spines is significantly decreased in the Fn14 KO dLGN compared to WT at P27 (Fig. 1J), consistent with our TEM results (Fig. 1F) and our prior study (Cheadle et al., 2018).
Because the analyses described thus far have been performed in a constitutive KO mouse lacking Fn14 in all cell types, it remained unclear whether Fn14 regulates synaptic refinement through a presynaptic mechanism, a postsynaptic mechanism, or both. To address this gap in knowledge, we developed a Fn14fl/fl conditional KO mouse in which exons 2 – 4 of the Fn14 locus are flanked by LoxP sites and compared spine numbers in Fn14fl/fl Cre-negative control mice with spine numbers in Fn14fl/fl mice crossed to either a relay-neuron-specific VGLUT2-Cre driver or a retina-specific Chx10-Cre driver (Rowan and Cepko, 2004). We validated region-specific loss of Fn14 in Cre-positive mice by protein and mRNA analysis (Fig. S3A,B). We found that Fn14 expression in relay neurons is required to form or maintain bulbous spines, consistent with Fn14 functioning to regulate synapse number and strength via a postsynaptic mechanism that is consistent with its SD transcription in relay neurons (Fig. 2A-D). In contrast, genetic ablation of Fn14 in RGCs had no significant effect on bulbous spine number in the dLGN (Fig. 2E). However, we found that spine length is significantly decreased by ablation of Fn14 in either pre- or postsynaptic neurons, highlighting that the functions of Fn14 at synapses are likely to be multi-faceted (Fig. 2F,G).
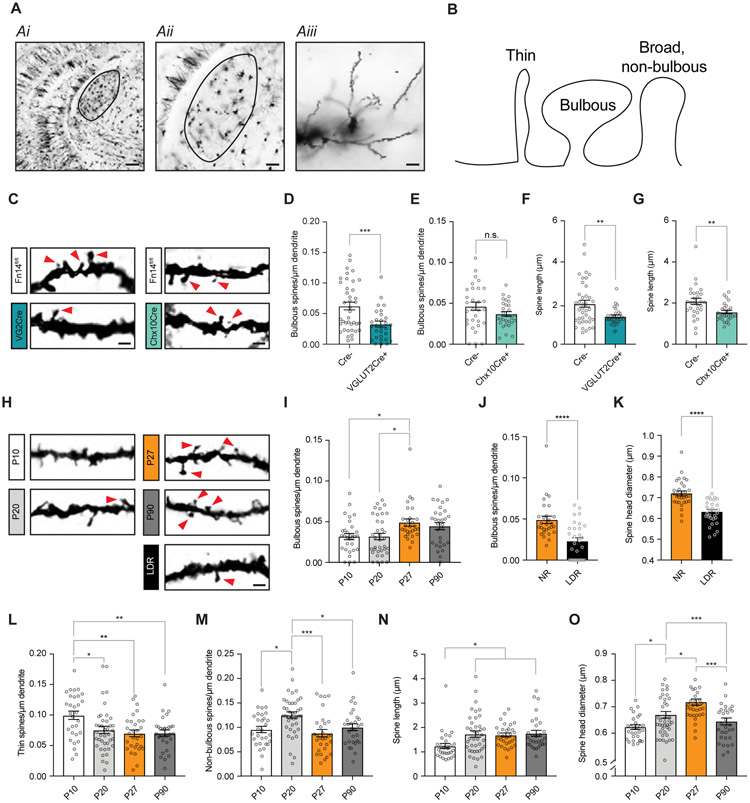
Figure 2. Developmental changes in spines require experience and postsynaptic Fn14.
(A) Low- and high-magnification images of a Golgi-stained brain section centered on the dLGN (outlined). Scale bars, 200 (Ai), 100 (Aii), and 10 (Aiii) μm.
(B) Schematic of spine types defined in the study.
(C) Example images of Golgi-stained spines in Fn14fl/fl; VGLUT2-Cre mice, Fn14fl/fl; Chx10-Cre mice, and Cre-negative littermates at P27. As in other panels, contrast increased to better display spine morphology. Arrows, bulbous spines. Scale bar, 2 μm.
(D) Bulbous spine density is decreased by genetic ablation of Fn14 in postsynaptic relay neurons.
(E) Bulbous spine density is unaffected by genetic ablation of Fn14 in presynaptic RGCs.
(F) Spine length is decreased by genetic ablation of Fn14 in relay neurons.
(G) Spine length is decreased by genetic ablation of Fn14 in RGCs.
(H) Examples of Golgi-stained dendrites and spines analyzed across postnatal development and in animals subjected to late-dark-rearing (LDR). Arrows, bulbous spines. Scale bar, 2 μm.
(I) Bulbous spine density increases between P20 and P27 in wildtype (WT) mice.
(J) Bulbous spine density is significantly decreased in LDR mice compared to normally reared (NR) mice.
(K) Spine head diameter across all spine types is decreased by LDR.
(L) Thin spine density decreases between P10 and P20 in WT mice.
(M) Non-bulbous spine density increases between P10 and P20 then decreases between P20 and P27 in WT mice.
(N) Spine length across all spine types increases during development.
(O) Spine head diameter increases between P10 and P27 in WT mice.
Statistical analysis for (D) – (G), (J), and (K), Student’s t-test. (I) and (L) - (O), One-way ANOVA with Tukey’s post hoc comparison. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. Means plotted with individual data points, +/− S.E.M.
An analysis of spine morphology across postnatal development in WT mice revealed that bulbous spine number increases between P20 and P27, highlighting that visual experience likely engages Fn14 during this period to regulate the number of bulbous spines in the course of development (Fig. 2H,I). If so, then we would expect depriving mice of experience during this phase, a manipulation that results in a significant decrease in Fn14 expression, to result in a decrease in the number of bulbous spines, phenocopying the Fn14 KO mouse. We find that, compared to normally reared mice (NR), mice reared in complete darkness between P20 and P27 (late-dark-reared, LDR) have 50% fewer bulbous spines with the average diameter of all spines decreasing by 12% (Fig. 2J,K). While changes in the overall structure of spines as well as in the numbers of thin and non-bulbous spines also occurred during postnatal development (Fig. 2L-O), the numbers of thin and non-bulbous spines and spine length overall were unaffected by LDR (Fig. S2J-L). Altogether, our data suggest that the Fn14-dependent addition of bulbous-spine-associated synapses to retinogeniculate connections is one mechanism by which experience strengthens connectivity during the vision-sensitive phase of retinogeniculate circuit maturation.
Sensory experience induces TWEAK expression in microglia
We next asked if the cytokine ligand of Fn14, TWEAK, collaborates with Fn14 to promote neural circuit development. We first assessed the pattern of TWEAK expression by interrogating a single-cell RNA-sequencing dataset from the dLGN (Kalish et al., 2018). While TWEAK is lowly expressed in multiple cell types at P5 and P10, TWEAK expression is significantly upregulated in microglia between P10 and P16, time points flanking the onset of visual experience at eye-opening (Fig. 3A,B). TWEAK expression continues to increase in microglia into the period of SD refinement that begins by P20.
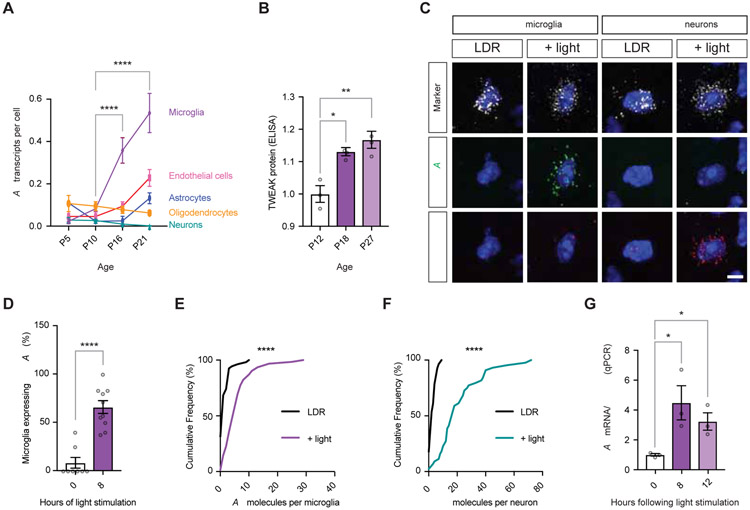
Figure 3. Experience drives the expression of TWEAK in microglia and Fn14 in relay neurons.
(A) Single-cell RNA-sequencing analysis of TWEAK expression across postnatal development in the dLGN. Y-axis, TWEAK mRNA transcripts per cell. X-axis, age.
(B) TWEAK protein levels as measured by ELISA increase across postnatal development. Levels normalized to P12.
(C) Confocal images of mRNA expression in the dLGNs of mice subjected to late-dark-rearing (LDR; unstimulated controls) or mice subjected to LDR then acutely re-exposed to light for eight hours (+ light). Sections were probed for markers of either microglia (Cx3cr1) or relay neurons (VGLUT2)(white), TWEAK (green), and Fn14 (red). Scale bar, 5 μm.
(D) Percentage of microglia expressing at least 3 TWEAK mRNA molecules is significantly increased in light-stimulated mice.
(E) Cumulative frequency distribution plot displaying increased TWEAK expression per microglia following light re-exposure.
(F) Cumulative frequency distribution plot displaying increased Fn14 expression per relay neuron following light re-exposure.
(G) qPCR quantification of TWEAK expression in microglia isolated from the visual cortices of mice following LDR and light re-exposure for eight or twelve hours. TWEAK expression normalized to Cx3cr1 expression to account for variability in enrichment efficiency.
Statistical analysis (A), (B), and (G): One-way ANOVA with Tukey’s post hoc comparison. (D), Student’s t-test. (E) and (F), Kolmogorov-Smirnov distribution test. *p < 0.05; **p < 0.01; ****p < 0.0001. Means plotted with individual data points, +/− S.E.M.
The temporal overlap between the onset of vision and the upregulation of TWEAK in microglia led us to hypothesize that, in addition to its ability to induce gene expression in relay neurons, experience might also promote transcriptional changes in dLGN microglia. To test this idea mice were subjected to LDR and then re-exposed to light for eight hours, a treatment that leads to Fn14 upregulation in relay neurons. Multiplexed single-molecule fluorescence in situ hybridization (smFISH) was then used to probe dLGN sections for TWEAK, Fn14, and cell-type-specific markers of microglia (Cx3cr1) or relay neurons (VGLUT2). We found that about 70% of microglia express TWEAK following visual stimulation while 8% of dLGN microglia express TWEAK in unstimulated mice (Fig. 3C,D). Analysis of the number of TWEAK mRNA molecules per microglia and the number of Fn14 molecules per relay neuron revealed strong light-driven increases in both TWEAK and Fn14 expression within individual cells of non-overlapping classes (Fig. 3E,F). A light-dependent increase in TWEAK expression was also observed by qPCR following the isolation of microglia from the visual cortices of stimulated and unstimulated mice (Fig. 3G). These patterns of stimulus-dependent TWEAK mRNA expression are largely recapitulated at the level of the TWEAK protein as measured by ELISA (Fig. S4A,B). While we also detected low levels of TWEAK in astrocytes and endothelial cells (Fig. 3A and Fig. S4C), TWEAK expression is notably higher in microglia, and only microglia induce TWEAK expression in response to visual experience (Fig. S4D-F).
TWEAK is dispensable for the phagocytic engulfment of synapses by microglia
We next considered what the function of TWEAK might be during retinogeniculate circuit refinement. Phagocytic engulfment of synapses is the best characterized mechanism by which microglia shape developing brain circuits (Cowan and Petri, 2018; Neniskyte and Gross, 2017). We therefore hypothesized that TWEAK expressed by microglia might mediate the elimination of retinogeniculate synapses by binding to Fn14 expressed by relay neuron synapses leading to their engulfment. This possibility is consistent with our previous finding that in addition to the failure to strengthen RGC inputs to relay neurons in Fn14 KO mice, we also observed that in these mice there is a failure to eliminate retinogeniculate connections that don’t strengthen in response to visual experience.
When we assessed the degree of synaptic engulfment by microglia in the dLGNs of WT mice we found that pre- but not postsynaptic elements are engulfed by microglia at the height of SD refinement (P27; Fig. S5). When we compared pre-synaptic engulfment by microglia in WT and TWEAK KO mice (Dohi et al., 2009) we found that TWEAK is not required for the phagocytic engulfment of synapses by microglia at both an early time point that precedes visual experience (P7) and at the height of SD refinement (P27) (Fig. 4A,B). Moreover, eye-specific segregation of ipsi- and contralateral retinal inputs, a developmental process that relies upon synaptic engulfment by microglia (Schafer et al., 2012; Stevens et al., 2007), proceeds normally in TWEAK KO mice (Fig. 4C,D). In addition, other aspects of microglial health and function, such as the number of microglia and their morphology, are also unaffected by the disruption of TWEAK function (Fig. 4E-G). Together, these data suggest a possible function for SD TWEAK activation in microglia that does not involve the engulfment of synapses.
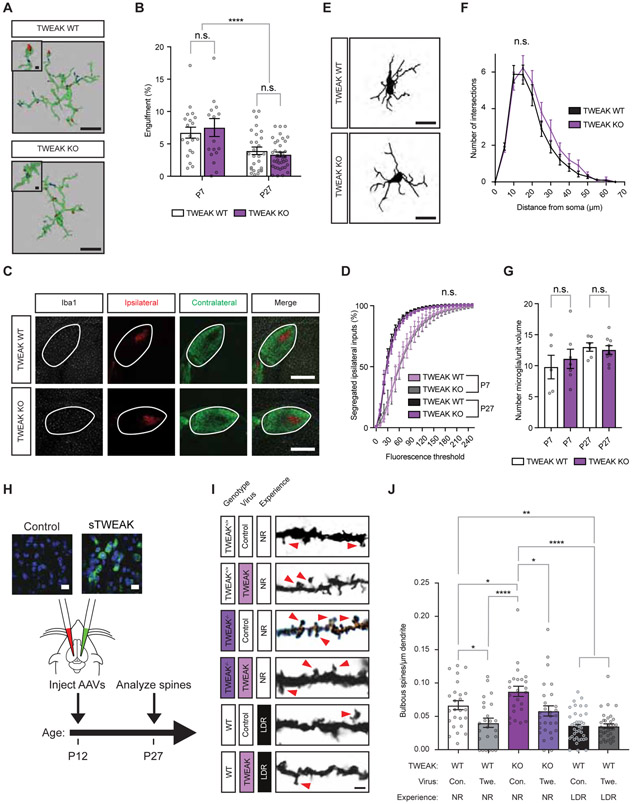
Figure 4. TWEAK promotes experience-dependent spine loss through a non-phagocytic mechanism.
(A) Surface renderings of microglia (green) containing phagocytosed retinal boutons (blue and red) labeled by ocular injection of fluorophore-conjugated tracers in mice at P27. Microglial reconstructions based upon a combination of Iba-1 and P2ry12 marker immunostaining. Scale bars, 10 μm (inset, 1 μm).
(B) Quantification of the volume of individual microglia occupied by retinal inputs, plotted as the percentage of a given microglial cell occupied.
(C) Confocal images of fluorescently labeled RGC boutons in contralateral (green) and ipsilateral (red) dLGN. Microglia immunostained for Iba-1 shown in white. Scale bar, 400 μm.
(D) Quantification of the overlap between ipsi- and contralateral inputs measured by co-localized signal in Imaris.
(E) Example tracings of microglia analyzed by Sholl morphological analysis. Scale bar, 10 μm.
(F) Sholl analysis of morphological complexity indicating the number of microglial projections intersecting with a series of concentric circles radiating outward from the soma.
(G) Quantification of the number of microglia per dLGN volume based upon Iba-1 staining.
(H) Schematic of the viral overexpression experiments probing the role of TWEAK in spine development. Inset, fluorescence in situ hybridization probing for TWEAK mRNA (green). Scale bar, 12 μm.
(I) Example images of dendritic spines in the dLGNs of TWEAK KO and WT mice of different genotypes following viral infection. Arrows, bulbous spines. Scale bar, 2 μm.
(J) Quantification of bulbous spine densities in TWEAK KO and WT mice following viral infection, with or without experience. Con., control virus. Twe., TWEAK virus. NR, normally reared. LDR, late-dark-reared.
Statistical analysis: One-way ANOVA with Tukey’s post hoc comparison. *p < 0.05; **p < 0.01; ****p < 0.0001. Means plotted with individual data points, +/− S.E.M.
Microglial TWEAK-dependent regulation of bulbous spines requires light and postsynaptic Fn14
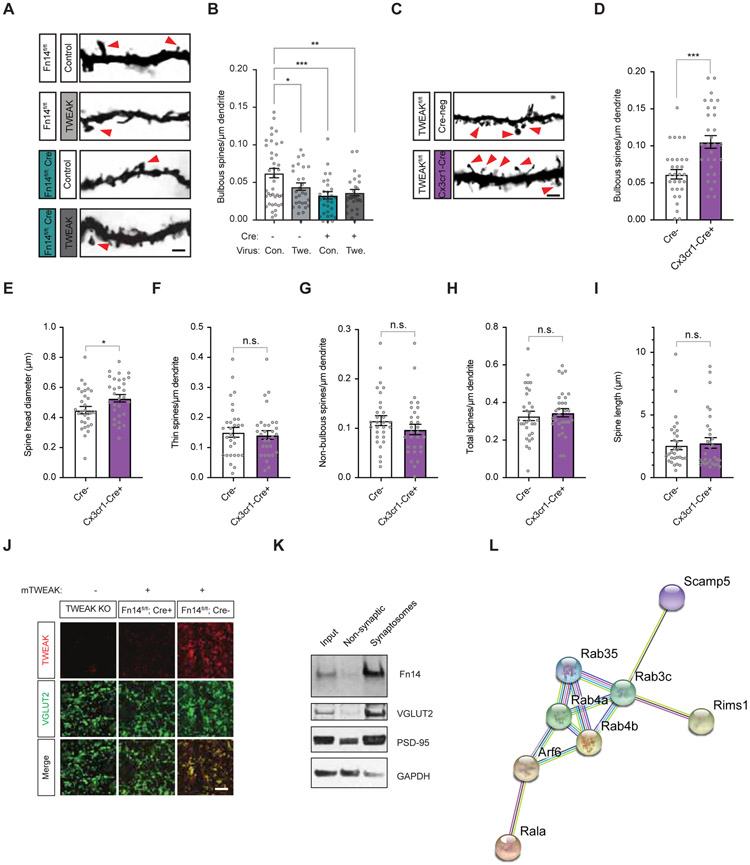
Having established spine analysis as a proxy for functional circuit changes downstream of experience and Fn14, we next assessed the effect of disrupting TWEAK function on the number and morphology of spines. Given that Fn14 promotes an increase in the number of bulbous spines in the dLGN, we hypothesized that, if TWEAK binding activates Fn14 function, then genetic ablation of TWEAK might lead to a decrease in the number of bulbous spines. Alternatively, if TWEAK binding to Fn14 inhibits Fn14 function then the disruption of TWEAK function might lead to an increase in the number of bulbous spines. We found that, in the absence of TWEAK, relay neurons display a significant increase in the number of bulbous spines. This suggests that TWEAK binding to Fn14 antagonizes the ability of Fn14 to enhance bulbous spine number, or alternatively might initiate the active disassembly of bulbous spines (Fig. 4H-J).
To examine further if TWEAK antagonizes the effect of Fn14 on bulbous spine number, we over-expressed TWEAK in the dLGN by bilaterally injecting adeno-associated viruses (AAVs) expressing either soluble TWEAK or mCherry (control) into the right and left dLGNs, respectively, of TWEAK KO and WT mice at P12 (Fig. 4H). We confirmed that at the area of injection glial activation is minimal in most of the dLGN sections analyzed at 15 days after injection (Fig. S6). The AAVs that we injected predominantly led to soluble TWEAK expression in relay neurons of the dLGN (Fig. 4H). Therefore, while this experiment does not allow us to assess specifically the role of microglial TWEAK in spine development, it nevertheless allowed us to determine if localized over-expression of soluble TWEAK is sufficient to affect dLGN synapses in vivo. We then assessed synaptic morphology in TWEAK-overexpressing mice by Golgi staining at P27. In a WT background, we found that overexpression of TWEAK was sufficient to decrease the number of bulbous spines by 25%. By contrast, mice in which TWEAK function was disrupted had 30% more bulbous spines than their WT littermates, and this increase in bulbous spines in the KO mice was reversed by re-expression of TWEAK (Fig. 4I,J).
Given that the Fn14-dependent enhancement of synaptic strengthening and bulbous spine number are both dependent upon visual experience (Cheadle et al., 2018), we next asked if the antagonistic effect of TWEAK on bulbous spine number also depends upon sensory input. To test this possibility, we overexpressed soluble TWEAK in the dLGNs of WT mice at P12, subjected the mice to LDR between P20 and P27, and analyzed spines. While we again found that LDR of WT mice leads to a significant decrease in the number of bulbous spines compared to WT mice housed under standard conditions (Fig. 2J), TWEAK overexpression had no further effect on bulbous spine number in LDR mice indicating that TWEAK signaling requires experience to decrease bulbous spine number (Fig. 4I,J).
To test directly if Fn14 expression in relay neurons is required for the TWEAK-dependent decrease in bulbous spines, we virally overexpressed soluble TWEAK or mCherry in Fn14fl/fl; VGLUT2-Cre mice or Cre-negative littermates and assessed spines. As shown in Fig. 2D, removal of Fn14 from relay neurons by crossing conditional KO mice to the VGLUT2-Cre line decreases bulbous spines by about 50%. We found that overexpressed TWEAK significantly decreased bulbous spine number in Cre-negative mice in which relay neurons express normal levels of Fn14 but did not affect spine number in Fn14fl/fl; VGLUT2-Cre mice where Fn14 expression is selectively ablated in relay neurons (Fig. 5A,B). This finding, taken together with the observation that TWEAK overexpression in the dLGNs of visually deprived mice has no effect on bulbous spine number (Fig. 4J), indicates that the TWEAK-dependent decrease in spines requires sensory-evoked Fn14 expression in relay neurons. Since we have shown that bulbous spines contain approximately four synapses per spine and interact with ~40% of incoming retinal terminals, these TWEAK-driven changes in bulbous spine number are likely to have a powerful functional impact on retinogeniculate connectivity.
Figure 5. Microglial TWEAK signals through postsynaptic Fn14 to decrease bulbous spine number.
(A) Example images of spines in the dLGNs of Fn14fl/fl Cre-negative or VGLUT2-Cre-positive mice following viral infection. Scale bar, 2 μm.
(B) Quantification of bulbous spine densities following TWEAK or mCherry expression in the dLGNs of Cre-negative (−) and VGLUT2-Cre-positive (+) mice. Comparison between mCherry-infected conditions also plotted in Fig. 2D. Con., control virus. Twe., TWEAK virus.
(C) Example images of spines in the dLGNs of TWEAKfl/fl Cre-negative or Cx3cr1-Cre-positive mice. Scale bar, 2 μm.
(D) Bulbous spine density is increased by genetic ablation of TWEAK in microglia.
(E) Spine head diameter is increased by genetic ablation of TWEAK in microglia.
(F) Thin spine density is unaffected by genetic ablation of TWEAK in microglia.
(G) Non-bulbous spine density is unaffected by genetic ablation of TWEAK in microglia.
(H) Total spine density is unaffected by genetic ablation of TWEAK in microglia.
(I) Spine length is unaffected by genetic ablation of TWEAK in microglia.
(J) Confocal images of dLGNs from a TWEAK KO mouse, a Fn14fl/fl Cre-negative mouse, and a Fn14fl/fl; VGLUT2-Cre+ mouse following bath application of recombinant mouse TWEAK and subsequent immunostaining for TWEAK (red) and VGLUT2 (green). Scale bar, 10 μm.
(K) Western blot of whole mouse forebrain fractionated to enrich for synaptosomes. Blots probed for Fn14, the retinal presynaptic marker VGLUT2, the postsynaptic marker PSD-95, and GAPDH, a non-synaptic control.
(L) Functional protein association network determined by STRING analysis illustrating known and predicted interactions between proteins identified as potential Fn14 interactors by mass spectrometry.
Statistical analysis: (B), One-way ANOVA with Tukey’s post hoc comparison; (D) – (I), Student’s t-test. *p < 0.05; **p < 0.01; ***p < 0.001. Means plotted with individual data points, +/− S.E.M.
The SD regulation of spine number by TWEAK-Fn14 signaling may be a mechanism by which microglia drive synapse loss. However, the experiments presented thus-far using TWEAK KO mice in which TWEAK function is disrupted in all cells do not explicitly show that microglia are the relevant expressers of TWEAK in the context of synaptic development, especially given the observation that in addition to microglia endothelial cells and astrocytes express low levels of TWEAK (Fig. S4C). Therefore, we next asked whether microglia-expressed TWEAK mediates the decrease in bulbous spine number. Towards this end we generated a TWEAK floxed conditional KO mouse and crossed it to the Cx3cr1-Cre microglial driver (Yona et al., 2013)(validation in Fig. S3C-G). Analysis of dendritic spines in these mice at P27 revealed a 70% increase in the number of bulbous spines compared to WT mice (Fig. 5C,D). We also noted a small but significant increase in spine head diameter upon microglial ablation of TWEAK consistent with bulbous spines having relatively large head diameters (Fig. 5E). Other spine parameters were unaffected by loss of microglial TWEAK (Fig. 5F-I). Together, these data identify a role for microglial TWEAK-to-neuronal Fn14 signaling in restricting the number of bulbous spines in response to sensory experience via a mechanism that does not involve synaptic engulfment.
TWEAK and Fn14 are likely to signal locally at retinogeniculate synapses
To further investigate the possibility that TWEAK and Fn14 signal locally at a subset of synaptic connections to regulate bulbous spine number, we developed a method for assessing the subcellular localization of TWEAK. Acute dLGN slices from a Fn14fl/fl; VGLUT2-Cre or an Fn14fl/fl; Cre-negative littermate were bathed with recombinant mouse TWEAK, and then washed and immunostained for VGLUT2, a retinogeniculate synapse marker, and TWEAK using an antibody capable of detecting TWEAK when overexpressed (Chicheportiche et al., 1997). Strikingly, we found that, in mice expressing neuronal Fn14, recombinant TWEAK was highly localized to retinogeniculate synapses while very little recombinant TWEAK was detected at synapses in the dLGN of mice lacking Fn14 in relay neurons, suggesting that recombinant TWEAK binds to synaptically localized Fn14 (Fig. 5J). Consistent with an Fn14-dependent localization of TWEAK at retinogeniculate synapses, synaptosomal fractionation of the mouse brain revealed an enrichment of Fn14 at synapses (Fig. 5K). In addition, an unbiased analysis of candidate Fn14-binding partners by co-immunoprecipitation and mass spectrometry identified a cohort of 24 likely Fn14 interactors that STRING analysis suggests make up a functional interaction network (Fig. 5L). Most of these candidates are known to localize to synapses and play roles in synaptic function, particularly membrane trafficking, neurotransmitter receptor recycling, and cytoskeletal dynamics (Table S1). Altogether, these data suggest that TWEAK is recruited to retinogeniculate synapses by Fn14, where TWEAK signals locally through Fn14 to restrict the number of bulbous spines.
Synapses near TWEAK-expressing microglia are preferentially lost
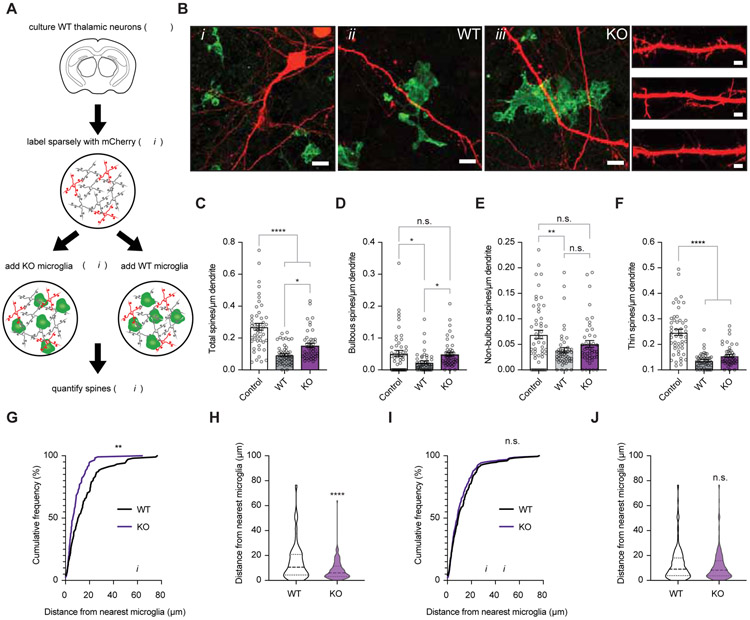
What factors determine which synapses are removed through TWEAK-Fn14 signaling and which synapses are strengthened by Fn14 in the absence of TWEAK? Since TWEAK is a soluble factor, we reasoned that synapses proximal to microglia that express TWEAK may be preferentially targeted for TWEAK-dependent removal. To test this possibility, we developed an in vitro co-culture system in which neurons are dissociated from the embryonic WT mouse thalamus and co-cultured with microglia derived from TWEAK KO or WT littermate mice. These mice had previously been crossed to the Cx3cr1-GFP line thereby allowing us to visualize microglia in both the TWEAK KO and WT co-cultures (Fig. 6A,B and Fig. S7)(Jung et al., 2000). We found that the addition of microglia to mCherry-labeled thalamic neurons leads to a significant decrease in the total number of spines that are present on neurons regardless of whether the microglia expressed TWEAK or not (Fig. 6C). However, when we specifically looked at bulbous spines, we found that this class of spines was unaffected by the addition of TWEAK KO microglia despite a 54% decrease in bulbous spines observed upon the addition of WT microglia (Fig. 6D-F). This TWEAK-dependent decrease in bulbous spines depended upon the proximity of the microglia to spines, as we observed a significantly greater average distance between bulbous spines and the nearest TWEAK-expressing microglial cell compared to the distance between bulbous spines and TWEAK-non-expressing microglia (Fig. 6G,H). By contrast, the distance between TWEAK-non-expressing microglia and thin spines was the same as for TWEAK-expressing microglia (Fig. 6I,J). These data suggest that microglial release of TWEAK provides a local signal that restricts the number of bulbous spines on nearby thalamic relay neurons.
Figure 6. Microglial TWEAK selectively eliminates proximal bulbous spines in vitro.
(A) Schematic of the in vitro co-culture experiment. Neurons from embryonic thalami (thal.) were sparsely transfected to express mCherry then seeded with microglia isolated from TWEAK knockout (KO); Cx3cr1-GFP or wildtype (WT); Cx3cr1-GFP mice. Spines were analyzed 24 hours later.
(B) Confocal images of neurons (red) co-cultured with TWEAK KO or WT microglia (green). Bi, mCherry-filled neuron contacted by WT GFP+ microglia. Scale bar, 15 μm. Bii, example of a WT microglia contacting the dendrite of a neuron. Scale bar, 5 μm. Biii, example of a TWEAK KO microglia contacting the dendrite of a neuron. Scale bar, 5 μm. Dendrites with spines from each condition, Scale bar, 2 μm.
(C) Total spine density in cultures without microglia (control) or with TWEAK WT or TWEAK KO microglia. Neurons co-cultured with TWEAK KO microglia maintained significantly more spines than those co-cultured with WT microglia.
(D) Quantification of bulbous spine density reveals that bulbous spines are protected when microglial TWEAK is ablated.
(E) Quantification of non-bulbous spine density in co-cultures.
(F) Quantification of thin spine density in co-cultures.
(G) Cumulative frequency distribution plot reflecting the proximity of bulbous spines to the nearest microglia with or without TWEAK expression. Bulbous spines were maintained closer to microglia when microglia lacked TWEAK.
(H) Violin plot reflecting median (dashed line) and quartile values (dotted lines) of the distance between bulbous spines and microglia with or without TWEAK.
(I) Cumulative frequency distribution plot reflecting the proximity of thin spines to the nearest microglia in the co-cultures with WT microglia (black) or TWEAK KO microglia (purple).
(J) Violin plot reflecting median (dashed line) and quartile values (dotted lines) of the distance between thin spines and microglia with or without TWEAK.
Means plotted with individual data points +/− S.E.M. Statistical analysis, One-way ANOVA with Tukey’s post hoc comparison. Statistical analysis (G) and (I), Kolmogorov-Smirnov distribution comparison. (H) and (J), Student’s T-test. *p < 0.05; **p < 0.01; ****p < 0.0001.
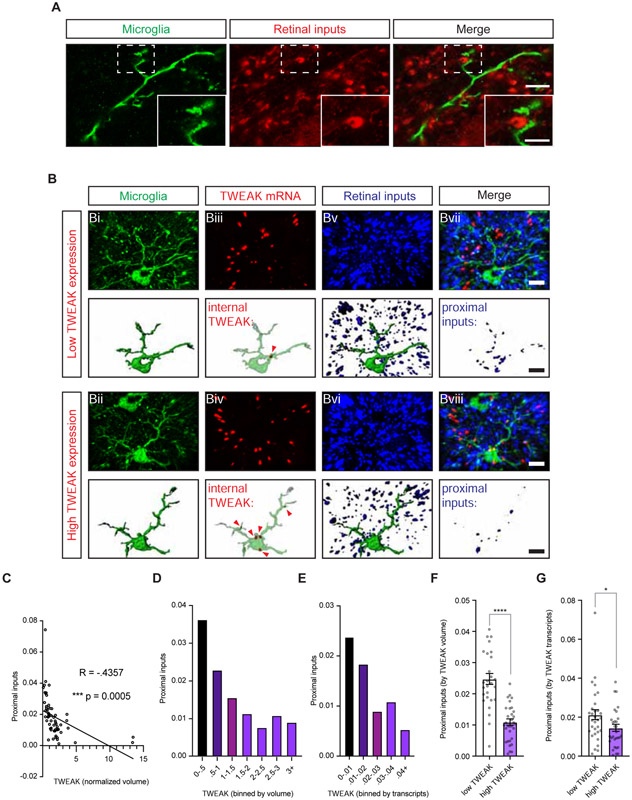
To complement our in vitro co-culture experiments we next combined FISH with protein immunostaining to analyze the relationship between the amount of TWEAK mRNA expressed by a microglial cell and the proximity of retinal inputs to that same cell in the dLGNs of visually stimulated mice. This approach takes advantage of the observation that microglia are heterogeneous in their expression of TWEAK such that some express high levels while some express low levels of TWEAK, even in the context of visual stimulation (Fig. 3D). Stimulated emission depletion (STED) microscopy indicated that microglia are commonly found within 50 – 200 nm of retinogeniculate synapses (Fig. 7A), therefore we designed our strategy to measure the number of retinal inputs within ~200 nm of a given microglia.
Figure 7. Synapse number is negatively correlated with the level of TWEAK expression in nearby microglia.
(A) Stimulated emission depletion (STED) microscopy images of a microglial cell process visualized by immunostaining for both Iba-1 and P2ry12 (green) within nanometer distance of VGLUT2-immunostained retinogeniculate synapses (red). Scale bar, 4 μm; inset, 2 μm.
(B) Confocal images and volumetric reconstructions of microglia (Iba-1 and P2ry12, Bi and Bii), TWEAK mRNA (Biii and Biv, red), and retinogeniculate synapses (VGLUT2, Bv and Bvi, blue). Bvii and Bviii, channels merged. Arrows mark TWEAK mRNA transcripts within microglia. Top rows, a microglia expressing only 1 mRNA of TWEAK. Bottom rows, a microglia expressing 5 mRNAs of TWEAK. Scale bar, 5 μm.
(C) Scatter plot of TWEAK expression (x-axis) and proximal retinogeniculate synapses (y-axis).
(D) Bar graph displaying the number of proximal retinogeniculate synapses (y-axis) corresponding with increasing expression of TWEAK mRNA volume binned as shown.
(E) Bar graph displaying the number of proximal retinogeniculate synapses (y-axis) corresponding with increasing number of TWEAK mRNA transcripts binned as shown.
(F) Comparison of proximal synapses in microglia expressing less than the median level of TWEAK (low TWEAK) vs. those expressing greater than the median level of TWEAK (high TWEAK) assigned based upon microglial volume occupied by mRNA signal.
(G) Same as (F) but mRNA plotted as transcripts per microglia. Values normalized to microglial volume.
(C) Pearson’s correlation coefficient R = −.4357. ***p < 0.001. (F) and (G), Student’s t-test. *p < 0.05; ***p < 0.001. (F) and (G), means plotted with individual data points, +/− S.E.M. (D) and (E), average proximal input value across microglia per bin.
To determine whether sensory-induced microglial TWEAK regulates the number of nearby retinogeniculate synapses, we analyzed dLGN tissue from mice that were dark-reared then acutely re-exposed to light to induce TWEAK expression in microglia. Remarkably, this analysis revealed a significant negative correlation between the level of TWEAK expressed by a microglial cell and the number of retinal synapses proximal to the microglial cell (Fig. 7B,C; Pearson’s coefficient R = - 0.4357; p < 0.001). This correlation was also apparent when we plotted proximal inputs (y-axis) versus microglial expression of TWEAK in bins (x-axis) (Fig. 7D,E), or when we divided microglia into low-expressers and high-expressers based upon whether their TWEAK expression level fell below or above the median (Fig. 7F,G). These data support the conclusion that in response to visual stimulation a subset of microglia express TWEAK which in turn binds Fn14 on the surface of nearby thalamic neurons to locally suppress the number of retinogeniculate synapses.
Discussion:
Proposed model of TWEAK-Fn14 function in synaptic refinement
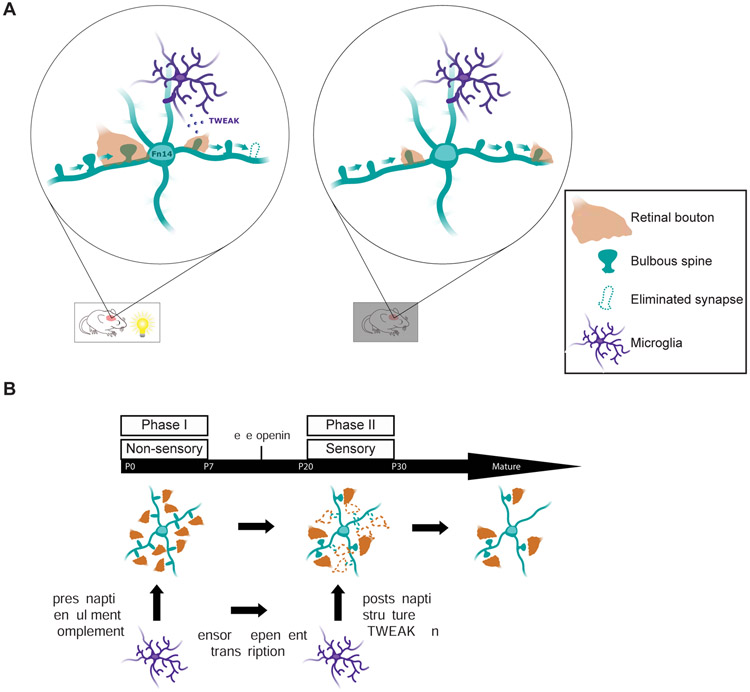
Based upon our findings, we propose a mechanism by which Fn14 differentially strengthens some synapses and eliminates others (Fig. 8A). In this model, during the visual experience-dependent period of retinogeniculate circuit maturation the induction of Fn14 promotes the formation and/or maintenance of bulbous spines on thalamic relay neurons thereby increasing the number of RGC-relay neuron synapses and ultimately contributing to synapse strengthening within this neural circuit. However, when a retinogeniculate synapse is proximal to a TWEAK-expressing microglia, microglial TWEAK binds neuronal Fn14 to inhibit its ability to promote spine formation/maintenance leading to a spatially localized suppression of bulbous spine number. Thus, retinogeniculate synapses that are near TWEAK-expressing microglia remain weak and are ultimately eliminated specifically during the vision-sensitive phase of retinogeniculate circuit maturation. Taken together, these findings provide a potential explanation for how the protein product of an experience-induced gene such as Fn14 can effect the strengthening of some synapses and the elimination of others within the same neuron. They also define a role for microglia in synapse development that goes beyond phagocytic engulfment and is uniquely shaped by sensory experience (Fig. 8B).
Figure 8. Model of TWEAK/Fn14-dependent synapse regulation during experience-dependent refinement.
(A) Schematic of retinal inputs (orange) converging onto the dendrites of a relay neuron (teal). Alone, Fn14 increases bulbous spines to strengthen and maintain synapses, while TWEAK binding at other synapses leads to their ultimate disassembly. In the absence of experience, neither TWEAK nor Fn14 is expressed so neither of these processes occur and synapses remain in a weakened state but are not properly removed.
(B) We propose that the sensory-dependent period of postsynaptic regulation by microglia identified in this study constitutes a later phase of microglia-driven circuit-sculpting that follows earlier phases of phagocytic pruning and is driven by distinct molecular mechanisms.
SD synapse elimination is mechanistically distinct from phagocytic engulfment
Since the discovery that dLGN relay neurons induce the expression of MHC class I molecules in response to spontaneous activity, it has been appreciated that immune signaling molecules are expressed in the brain and play various roles in neural development (Corriveau et al., 1998). For example, microglia have been shown to engage the classical complement cascade to engulf less active synapses during the first week of postnatal life, consistent with the idea that immune signaling pathways function in the brain at least in part in response to changes in neural activity (Gunner et al., 2019; Schafer et al., 2012). Notably, we found that the complement protein C1qa which is required for activity-dependent engulfment at P5 does not regulate spines at P27 (Fig. S8), indicating that this early phagocytosis-based process of microglial synapse removal is mechanistically distinct from the SD regulation of synaptic structure that we describe. It makes sense that, in the context of development, the removal of the presynaptic inputs would precede the disassembly of the postsynaptic site since performing these functions in reverse would potentially lead to excessive glutamate spillover resulting in excitotoxicity. Our findings raise the intriguing question of whether the same microglial cell that engulfs presynaptic terminals might later remodel the postsynaptic specialization, or whether different populations of microglia carry out these distinct processes. That TWEAK is induced in a subset of microglia suggests that different populations of microglia may be tuned to either engulf or remodel synapses selectively. In any case, our results suggest that microglial roles in retinogeniculate synapse development extend beyond the phagocytic pruning of synapses that occurs prior to eye-opening (Fig. 8B).
Roles for microglia in regulating synaptic structure outside of the dLGN
Our findings regarding the function of microglia in the dLGN are complemented by recent studies of microglia in other brain regions. For example, in the hippocampus, microglia regulate spine structure without engulfing spines (Weinhard et al., 2018). Additionally, live imaging studies of microglial dynamics in the cortex have shown that microglia survey and contact synapses in response to sensory experience, and that these microglia/synapse interactions influence the number, morphology, and physiology of spines (Akiyoshi et al., 2018; Tremblay et al., 2010). In a related study, Parkhurst et al. showed that depleting microglia from the mouse brain limits the spine turnover that usually occurs with motor learning (Parkhurst et al., 2013). Most of these studies used live imaging of spines to study spine turnover and dynamics in the brains of living mice, an approach that will be useful for determining the mechanisms by which the TWEAK-Fn14 pathway regulates spines in vivo.
Implications for human disease
Brain disorders ranging from autism to Alzheimer’s disease are characterized by changes in the number and structure of spines (Faludi and Mimics, 2011; Glantz and Lewis, 2000; Hammond et al., 2019). These disorders are increasingly thought to involve the misregulation of microglial function (Hong et al., 2016; Velmeshev et al., 2019). Our finding that microglia via TWEAK-Fn14 signaling shape the number and structure of spines during key periods of sensory experience-dependent circuit maturation raises the possibility that the mis-regulation of this feature of microglial signaling may contribute to these disorders. A further examination of microglial roles in postsynaptic remodeling may therefore provide much-needed insight into how synapses become altered in pathological states that involve the mis-regulation of microglial function.
STAR Methods:
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Michael E. Greenberg (meg@hms.harvard.edu).
Materials Availability
All unique resources generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement. B6.Tnfrsf12atm1(KO)Biogen (Fn14 KO; Jakubowski et al, 2005), B6.Tnfrsf12a(fl/fl)Biogen mice, and B6.Tnfsf12tm1(KO)Biogen (Dohi et al., 2009) are subject to restrictions imposed in an MTA by Biogen (Cambridge, MA).
Data and Code Availability
The published article includes all datasets generated or analyzed during the study.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All animal experiments were performed in compliance with protocols approved by the Institutional Animal Care and Use Committee (IACUC) at Harvard Medical School. The following mouse lines were used in the study: C57BI/6J (the Jackson Laboratory, JAX:000664); B6.129P2(Cg)-Cx3cr1tm1Litt/J (Cx3cr1-GFP; the Jackson Laboratory, JAX: 005582); B6.Tnfrsf12atm1(KO)Biogen (Fn14 KO)(Jakubowski et al., 2005); B6.Tnfsf12tm1(KO)Biogen (TWEAK KO)(Dohi et al., 2009); a novel Fn14fl/fl mouse line (B6.Tnfrsf12a(fl/fl)Blogen; targeting strategy and validation shown in Fig. S3); a novel TWEAKfl/fl mouse line (B6.Tnfsf12(fl/fl)Gree/J; targeting strategy and validation shown in Fig. S3); Slc17a6tm2(cre)Lowl/J (VGLUT2-Cre; the Jackson Laboratory, JAX:028863); Tg(Prkcd-glc-1/CFP,-Cre) mice backcrossed to a C57BI6/J background (Haubensak et al., 2010) then crossed to B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J (Ai14; the Jackson Laboratory, JAX:07914); B6J.B6N(Cg)-Cx3cr1tm1.1(cre)Jung/J (Cx3cr1-Cre; the Jackson Laboratory, JAX: 025524), the Tg(Chx10-EGFP/cre,-ALPP)2Clc/J line (Chx10-Cre; the Jackson Laboratory, JAX:005105)(Rowan and Cepko, 2004), and B6.C1qa(KO) (lab of Beth Stevens). B6.Tnfrsf12atm1(KO)Biogen, B6.Tnfsf12tm1(KO)Biogen, and B6.Tnfrsf12a(fl/fl)Biogen mice were provided by co-authors Dr. Linda Burkly and Katelin Ennis at Biogen. Most analyses were performed on mice at P27, though the developmental spine analysis also included P10, P20, and P90 ages, and some cohorts of mice were injected with AAVs at P12 to allow for spine analysis at P27. Thalamic neurons were cultured from pups at E15.5, and microglia were acutely isolated from mice between P15 and P20. Both males and females were included in all analyses. We did not note an effect of sex on any of the parameters measured in the study.
In most studies, mice were housed under standard conditions according to a 12-hour light/dark cycle (normally reared, NR). For the late-dark-rearing (LDR) paradigm, pups were bred in-house and housed with moms under standard light/dark conditions until P20, at which time they were weaned and moved into the dark, light-proof chamber of a ventilated cabinet. At P27, mice were moved into a separate, well-lit compartment of the cabinet for re-exposure to light for eight or twelve hours. Conversely, dark-reared, unstimulated control animals were euthanized by isoflurane and the brain removed in the dark by an investigator using night-vision goggles.
METHOD DETAILS
Generation of Fn14fl/fl, Fn14fl/fl; VGLUT2-Cre, and Fn14fl/fl; Chx10-Cre mice
Fn14fl/fl mice were generated by Taconic for Biogen. The strategy involved the insertion of two LoxP sites flanking the 2nd – 4th exons. To remove Fn14 from thalamic relay neurons, Fn14fl/fl mice were bred in-house with VGLUT2-Cre mice (JAX:028863). To remove Fn14 from retinal ganglion cells, mice were bred in-house with Chx10-Cre mice (JAX:005105). Note that, although VGLUT2 expression has been reported in the retina, FISH revealed remaining Fn14 expression in the retina but not the dLGNs of Fn14fl/fl; VGLUT2-Cre mice. We therefore considered this mouse to be a dLGN-specific knockout.
Generation of TWEAKfl/fl and TWEAKfl/fl; Cx3cr1-Cre mice
TWEAKfl/fl mice were designed to excise the 3rd coding exon by floxing this region with LoxP sites in the introns directly before and after exon three. This strategy was predicted to introduce a STOP codon and lead to nonsense-mediated decay of the message. LoxP sites were introduced by CRISPR technology at the Genome Modification Facility in Cambridge, Massachusetts. Microglia-specific loss of TWEAK in TWEAKfl/fl mice bred to the microglial Cre driver Cx3cr1-Cre was validated by RNAscope. In addition, significant decreases (but not complete ablation) of TWEAK at the whole-tissue level was demonstrated by qPCR.
GridTape serial transmission electron microscopy
Tissue preparation
An Fn14 KO mouse and a WT littermate at P27 were trans-cardially perfused with 2% paraformaldehyde and 2.5% glutaraldehyde in 0.1 M Cacodylate buffer with 0.04% CaCl2. Coronal sections of the brain containing the dLGN (300 μm thickness) were cut on a Vibratome (VT1000S, Leica) and stained with 1% osmium tetroxide and 1.5% potassium ferrocyanide followed by 1% uranyl acetate, then lead aspartate. Sections were dehydrated with a graded ethanol series and embedded in epoxy resin (TAAB 812 Epon, Cane MCO).
Sectioning and collection
For both the WT and Fn14 KO samples, 250 ultrathin sections (~45 nm) of the dLGN tissue were cut using an ultramicrotome (UC7, Leica) with a diamond knife (Diatome). Sections were collected onto GridTape using a modified automated tape-collecting ultramicrotome (ATUM) equipped with an optical interrupter (GP1A57HRJ00F, Sharp) and a hall-effect sensor (A1301EUA-T, Allegro Microsystems) and magnet. The tape’s movement speed was varied based on signals from the interrupter and hall-effect sensor to consistently place sections of the dLGN over GridTape’s slots (Graham, Hildebrand et al. 2019).
Post-section staining and TEM imaging
Following sectioning and collection, sample contrast was enhanced via post-section staining with Reynolds lead citrate (Ultrostain II, Leica). Electron microscopy imaging of the collected thin sections from both samples was performed at 4.3 nm pixel size on a custom JEOL 1200EX transmission electron microscope modified to hold and automatically image reels of GridTape (Graham, Hildebrand et al. 2019). After acquisition, images were stitched and aligned with AlignTK (http://mmbios.org/installation) into three-dimensional (3D) volumes.
Quantification of bouton properties
Aligned volumes were uploaded to CATMAID (Saalfeld et al., 2009; Schneider-Mizell et al., 2016) for manual annotation of retinal boutons, their synapses, and the type of the postsynaptic structure (i.e. shaft, bulbous spine, thin spine, or broad non-bulbous spine). The dLGNs were divided into 4 quadrants to ensure synapses were analyzed across all regions for both Fn14 KO and WT sections. The proportions of retinal inputs converging onto spines versus dendritic shafts was determined by randomly sampling 250 boutons per quadrant (1,000 total) for each condition. Retinal boutons were identified based upon morphological properties (Colonnier and Guillery, 1964; Guillery and Colonnier, 1970). Specifically, retinal boutons were identified by their mitochondria, which are uniquely large and pale, in combination with the relatively large size of the boutons themselves. 3D reconstructions of selected boutons (as seen in Fig. 1C,D) were performed in ITK-SNAP (www.itksnap.org) (Yushkevich et al., 2006) and Gaussian image smoothing was applied according to the following parameters: standard deviation, 0.90; approximation error max, 0.10. All analyses were performed with the researcher blinded to condition.
Golgi-staininq and dendritic spine analysis
Golgi staining was performed with the FD Rapid GolgiStain kit (FC Neurotechnologies, Inc) according to the manufacturer’s protocol. Following sample processing, dendritic segments were traced in x, y, and z planes using a Zeiss Axioskop microscope (63X objective) and Neurolucida (Microbrightfield Bioscience). To categorize spines by morphology, we defined bulbous spines as those whose head diameter is at least 2X that of its neck, spines with a relatively uniform width (head to neck width ratio less than 2) of less than 0.5 μm as thin spines, and spines with a relatively uniform width of greater than 0.5 μm as broad non-bulbous spines. In a majority of cases, head-to-neck ratio cutoffs were clear by eye after some analysis on training datasets by the investigator. In cases where this was not clear, the criteria were determined by measurement. We find by hierarchical clustering that these morphological subtypes may represent distinct biological categories (Fig. S2B,C). In addition to these categorizations, we also measured the spine head diameter and the length of spines overall regardless of classification. Although spines are thought to exist along a structural continuum, complementing our measurements of spine structure overall with an analysis of spine subtypes is useful in allowing our analysis to be compared with analyses in other brain regions, many of which describe spine morphology according to subclasses similar to those used in this study (Peters and Kaiserman-Abramof, 1970; Sorra and Harris, 2000). Example images of Golgi-stained spines were obtained on an Olympus BX63 fluorescence microscope with a 100X objective.
Protein detection by ELISA
The dLGNs and primary visual cortices of normally reared, dark-reared, and visually stimulated mice were microdissected in ice-cold PBS following sectioning on a vibratome (VT1000S, Leica) and flash-frozen in liquid nitrogen. On the day of the assay, the tissues were thawed and resuspended in 500 μL of ice-cold RIPA buffer (Life Technologies) including complete protease inhibitor cocktail tablet (Roche) and phosphatase cocktails two and three (Sigma). Homogenization was performed by douncing 25X in a 2 mL douncer and the protein concentration of each sample was determined by BCA assay (Pierce). The samples were then diluted to achieve a concentration of 100 μg/μL for each sample. TWEAK protein expression was measured by ELISA (R&D Systems, DY1237) according to the manufacturer’s instructions, with 30 μg of sample loaded per well in triplicate.
Single-molecule fluorescence in situ hybridization (RNAscope)
The brains of euthanized mice were harvested and immediately embedded in OCT (Optimum Cutting Temperature) on dry ice then stored at −80° C. Twenty μm sections were cut on a Leica CM 1950 Cryostat, collected on Superfrost Plus slides, and stored at −80° C until the day of the experiment. smFISH was performed using RNAscope (Advanced Cell Diagnostics) according to the manufacturer’s protocol for fresh-frozen sections and as described (Cheadle et al., 2018). Sections were probed for the microglial markers Cx3cr1 (314221-C2), C1qa (441221-C3), and P2ry12 (317601-C2). In addition, the excitatory relay neuron marker VGLUT2 (319171-C2), the interneuron marker Gad1 (400951-C2), the endothelial cell marker Cldn5 (491611-C3), the astrocyte markers Aldh1l1 (405891-C2) and Aldoc (429531-C3), the oligodendrocyte marker Olig1 (480651-C3), TWEAK (552051), and Fn14 (505311-C3) were also used.
Blinded multi-channel images were imported into ImageJ. Following background subtraction, cell types were identified based on robust expression of at least eight marker gene mRNAs (Cx3cr1 or P2ry12 for microglia, Cldn5 for endothelial cells, Aldh1l1 or Aldoc for astrocytes, VGLUT2 for relay neurons, Gad1 for interneurons, and Olig1 for oligodendrocytes), and the DAPI-stained nuclei of these cells were traced. Next, the channel representing TWEAK or Fn14 signal was opened, and the number of TWEAK mRNAs per microglial cell body or Fn14 per relay neuron was quantified.
Combined FISH and Immunofluorescence (FISH-IF)
We combined RNAscope FISH and immunofluorescence in order to visualize and measure the relationship between microglial TWEAK expression and retinogeniculate synapse number. Mice were perfused in 4% paraformaldehyde and post-fixed overnight at 4° C. Brains were cryoprotected by incubation in 15% then 30% sucrose. Sections of 20 μm were made and stored at −80° C until use.
Tissue was processed with the RNAscope Multiplex Fluorescent V2 Assay (Advanced Cell Diagnostics, #323100) according to the manufacturer’s instructions. Several modifications to the manufacturer’s protocol were made to optimize co-visualization of mRNA and protein signal. First, the pretreatment fixation in 4% PFA was performed for 20 minutes. Next, the hydrogen peroxide incubation step was 10 minutes. For the target retrieval step, the ACD target retrieval reagent was brought to a boil and allowed to cool to 90° C on the bench top, at which point sections were added and incubated for 15 minutes. Sections were incubated with Protease III for 20 minutes.
After performing all TWEAK mRNA probe detection steps, sections were blocked for 30 minutes at room temperature in PBS adjusted to 0.3% Triton X-100 and 5% normal goat serum. Sections were incubated at 4° C overnight with primary antibodies: rabbit anti-Iba1 (Wako, 019-19741; 1:500), rabbit anti-P2ry12 (Sigma, HPA013796; 1:500), and guinea pig anti-VGLUT2 (AB2251, Millipore; 1:500) in PBS adjusted to 0.1% Triton X-100 and 5% normal goat serum. Sections were washed in RNAscope wash buffer 2 times for 2 minutes per wash. Secondary Alexa Fluors (Invitrogen; 1:500) were used to detect primary antibodies via incubation with sections for 2 hours at RT. Sections were then washed and coverslipped in Fluoromount G + DAPI (ThermoFisher).
Quantification of FISH-IF experiments
Sections were imaged in three dimensions on a laser-scanning confocal microscope (Zeiss FV1000) with a 60X objective. After pre-processing to enhance contrast and subtract background in ImageJ, Z-stacks of individual microglia containing all three channels were imported into Imaris (Bitplane). Using the “Surface” function, volumetric reconstructions were generated of (1) the microglia; (2) TWEAK mRNA signal; (3) retinogeniculate synapses (VGLUT2 signal); (4) “masked” TWEAK mRNA puncta within the boundaries of the microglial volume; and (5) “masked” retinogeniculate synapses co-localizing with the microglial volume. Since the resolution of confocal images is roughly 200 nm, we denoted colocalized synapses as those most proximal to the microglia in question. The amount of TWEAK within the microglial cell was measured based upon (1) the percentage of the microglial volume occupied by TWEAK signal, and (2) the number of discrete TWEAK puncta detected per microglial volume. Because in several cases individual TWEAK mRNA were so close together that they appeared as one continuous unit, we believe the volumetric measurement is most accurate. In our final quantifications, we not only normalized the amount of TWEAK mRNA detected to the volume of the microglia analyzed, we also normalized the amount of colocalizing retinogeniculate inputs to the microglial volume as well. Furthermore, because retinal synapse density varies across the dLGN, we normalized all inputs associated with the microglia from the same section to the overall average input density across the image.
Synaptic engulfment and eye-specific segregation assays
Retinal inputs to the dLGN were labeled through ocular injections of choleratoxin-B anterograde tracers conjugated to Alexa Fluor dyes (Invitrogen). Conversely, Prkcd-Cre/LSL-TdTomato animals in which relay neurons are labeled with TdTomato were analyzed. After 48 hours, mice were euthanized and perfused with 4% paraformaldehyde (PFA) then the brains were sectioned and immunostained for microglia using rabbit anti-Iba1 (Wako, 019-19741) and rabbit anti-P2ry12 (Sigma, HPA013796) in combination. Sections were imaged in three dimensions on a laser-scanning confocal microscope (Zeiss FV1000) with a 60X objective. Z-stacks containing all three channels were imported into Imaris (Bitplane) and the volumes of individual microglia were reconstructed based upon Iba-1 and Psry12 staining. The volume of each microglial cell occupied by phagocytosed retinal inputs was quantified as described (Schafer et al., 2012).
For eye-specific segregation analysis, z-stacks were imaged at 10X to allow visualization of the ipsi- and contralateral input regions across a coronal section of the dLGN. Images were imported into Metamorph (Molecular Devices). The fluorescence intensity of the labeled ipsilateral retinal inputs was increased to include all signal above background. Next, the intensity of the contralateral input signal was gradually increased from 0 to 250, and the overlap between the signals at each contralateral threshold was measured.
Biochemical isolation of microglia-enriched fractions
Microglia were purified from the visual cortices of dark-reared, unstimulated mice and mice re-exposed to light for eight or twelve hours as previously described (Cardona et al., 2006). After anesthetization with isoflurane, mice were perfused with ice cold 1X HBSS (Gibco) and their brains dissected and placed in RPMI buffer (Sigma). After discarding the cerebellum, the brains were minced with a razor blade and transferred to a 2 mL dounce homogenizer containing 1.5 mL of RPMI buffer. The brains of eight mice per condition were pooled at this point and dounced 30X with the loose pestle then 25X with the tight pestle. Homogenates were transferred to a 15 mL falcon tube containing 5.5 mLs of RPMI (total volume, 7 mL). Standard isotonic percoll (SIP; 4.5 mL percoll with 0.5 mL 10X HBSS) was then added to the homogenate and mixed by pipetting. The homogenate was then overlaid onto 3 mLs of 70% SIP in a 15 mL falcon tube and centrifuged at 500xg for 30 minutes, 18° C. After removal of the lipid layer at the top, the microglial fraction was recovered from the 30%/70% SIP interface (about 1 mL) and transferred to a falcon tube containing 10 mL HBSS. Following centrifugation at 900xg for 7 minutes at 18° C, the supernatant was discarded and pelleted microglia were resuspended in Trizol (Sigma), flash-frozen in liquid nitrogen, and stored at −80° C.
Quantitative PCR
Microglia-enriched fractions from visual cortex flash-frozen in Trizol were thawed and purified using the RNeasy Micro kit with on-column DNAse digestion (Qiagen). Reverse transcription was performed using the Superscript III first-strand synthesis kit (Invitrogen). Real-time quantitative PCR was performed using SYBR green mix (Life Technologies). Levels of endogenous TWEAK were detected with the following primers: GCTGGGCAACGCTGTCT (F) and GCGGTCCTCTGCTGTCA (R), and normalized to the expression of the microglial marker Cx3cr1: GAGCATCACTGACATCTACCTCC (F) and AGAAGGCAGTCGTGAGCTTGCA (R). Validation of loss of TWEAK in TWEAKfl/fl; Cx3cr1-Cre was performed by the same qPCR approach on whole-brain tissue with TWEAK levels normalized to GAPDH and P2ry12 expression using the following primers: GAPDH forward (GGGTGTGAACCACGAGAAATA), GAPDH reverse (CTGTGGTCATGAGCCCTTC), P2ry12 forward (CATTGACCGCTACCTGAAGACC), and P2ry12 reverse (GCCTCCTGTTGGTGAGAATCATG).
Microglial number and morphology analysis
Microglia were visualized by immunostaining for Iba1 and P2ry12 and z-stacks of sections were taken on a laser-scanning confocal microscope (Zeiss FV1000). Images were imported into ImageJ, background subtracted, and the numbers of Iba1-positive cells were measured per unit dLGN area. For Sholl analysis, z-stacks of individual microglia were opened in Neurolucida (Microbrightfield) and the cell bodies and primary protrusions were traced in 3D using a 40X objective. The tracings were then imported into Neurolucida Explorer and a Sholl analysis was performed to measure the morphological complexity of each microglial cell by quantifying the number of intersections between microglial processes and a series of concentric circles radiating from the soma outward (Sholl, 1953).
Bilateral AAV injections into the dLGN
AAV9-CASI-sTWEAK and AAV9-CASI-mCherry were designed, produced, and characterized by co-authors Dr. Linda Burkly and Katelin Ennis at Biogen. We further confirmed the efficacy of the TWEAK-expressing virus using RNAscope as described above (see inset of Fig. 4H). For injections, P12 mice were anesthetized with isoflurane and stereotaxically injected with 1 μl of AAV-sTWEAK (right dLGN) or AAV-mCherry (left dLGN) at the following stereotaxic coordinates: −2.0y, +/− 2.1x, 2.95z (mm). Animals were administered Flunixin twice daily for three days following the injections. Most mice appeared normal and comfortable within one day following injection. Injected mice were usually housed according to standard light/dark conditions until P27 at which point the animals were euthanized and the brains subjected to Golgi-staining. In some cases, mice were reared according to standard conditions until P20 and then late-dark-reared (LDR) until P27.
TWEAK localization experiment
TWEAK KO mice, Fn14fl/fl mice, and Fn14fl/fl; VGLUT2-Cre mice at P27 were perfused with ice cold oxygenated artificial cerebrospinal fluid (ACSF) containing (in mM): 125 NaCl, 26 NaHCO3, 1.25 NaH2PO4, 2.5 KCl, 1.0 MgCl2, 2.0 CaCl2, and 25 glucose (Sigma), adjusted to 310-312 mOsm with water. Coronal sections of dLGN at 150 μm were sliced on a vibratome (VT1000S, Leica) and kept on ice in ACSF. Sections were incubated with either ACSF or ACSF containing 200 ng/mL of recombinant mouse TWEAK (Biogen) and rotated at 4° C for 2 hours. ACSF was aspirated and 500 μL of 4% PFA was added to fix slices for 20 minutes at room temperature. Sections were then washed in 1X PBS (Gibco) 3x for 10 minutes per wash and blocked in PBS adjusted to 1.25% Triton X-100 (United States Biological) and 10% fetal bovine serum (Gibco; IHC buffer) for 1 hour at room temperature. Sections were then stained in IHC buffer overnight at 4° C with guinea pig anti-VGLUT2 (AB2251, Millipore) and goat anti-TWEAK (AF1237, R&D Systems) at 1:1000. The next day, sections were washed 3 times in PBS then incubated in secondary Alexafluor Dyes (Molecular Probes) in IHC buffer at 1:500 for 1 hour. Sections were washed in PBS and mounted onto slides in Fluoromount G plus DAPI (SouthernBiotech). Imaging parameters given below.
Immunofluorescence
Fifty μm sections of 4% PFA-fixed brains were made on a vibratome and blocked in PBS adjusted to 1.25% Triton X-100 and 10% fetal bovine serum for one hour at room temperature. Primary antibody incubations were performed at 4° C overnight in blocking buffer. The next day, sections were washed three times in PBS then probed with secondary Alexafluor dyes (Molecular Probes or Thermo Fisher) for one hour at room temperature. Sections were again washed three times in PBS and mounted onto Superfrost Plus slides (Fisher) using Fluoromount G plus DAPI (SouthernBiotech). Primary antibodies used include guinea pig anti-VGLUT2 (AB2251, Millipore), rabbit anti-Iba-1 (Wako, 019-19741), rabbit anti-P2ry12 (Sigma, HPA013796), and rabbit anti-GFAP (Abeam, ab4674).
Fluorescence imaging
Imaging of immunofluorescence and FISH was performed on an Olympus FluoView 1000 laser scanning confocal microscope equipped with 405 nm, 440 nm, 488 nm, 515 nm, 559 nm, and 635 nm excitation lasers, and 10x air 0.4NA, 20x air 0.75NA, 40x oil 1.3NA, 60x oil 1.42NA, and 100x oil 1.4NA objectives. STED imaging was performed on a Leica Sp8 STED/FLIM system using a 100X oil objective and a white light laser providing continuous excitation tuning from 470 nm to 670 nm.
Synaptosomal fractionation
Synaptosomal fractionation was performed using Syn-PER synaptic protein extraction reagent (ThermoFisher, 87793) according to the manufacturer’s instructions. Briefly, Complete protease inhibitor tablet (Roche) and phosphatase inhibitor cocktails 2 and 3 (Sigma) were added to 10 ml_ of ice cold Syn-PER reagent. The forebrains of 4 WT adult mice were minced and added to 2 mL of Syn-PER reagent in a 2 mL dounce homogenizer and homogenized ~10 times. The sample was then centrifuged at 1200xg for 10 minutes at 4° C. The supernatant was then centrifuged again at 15,000xg for 20 minutes at 4° C. The supernatant (cytosolic fraction) was then transferred to a separate tube and the synaptosomal pellet was resuspended in 200 pL of Syn-PER reagent. The protein concentrations of homogenate, cytosolic non-synaptic fraction, and synaptosomal fraction were determined by BCA protein assay (Pierce) and 30 μg of each were combined with protein sample buffer and run on a western blot as described below.
Immunoprecipitation
Validation of Fn14fl/fl mice was performed by immunoprecipitating Fn14 from the whole forebrain using a rabbit anti-Fn14 antibody (Cell Signaling Technologies, 4403s) at 1:50 as described (Cheadle et al., 2018). Briefly, the brains of WT, Fn14fl/fl mice, and Fn14fl/fl; VGLUT2-Cre mice were homogenized in buffer containing 10 mM HEPES-KOH pH 7.5, 25 mM KAc, 320 mM sucrose, 1% triton-X100, and 250 mM NaCl, along with a complete protease inhibitor cocktail tablet (Roche) and phosphatase cocktails two and three (Sigma). Following homogenization in a 2 mL douncer, homogenates were rotated at 4° C for 10 minutes and then centrifuged at 14,000 RPM for 15 minutes at 4° C. Supernatants were cleared by rotating for 1 hour with 50 μl of protein A dynabeads (Invitrogen). Beads were collected on a magnet and supernatants transferred to a new eppendorf tube. Rabbit anti-Fn14 (4403s) was added to each sample at 1:50 and samples were rotated for 2 hours at 4° C. Samples were then rotated with 50 μL of protein A dynabeads at 4° C which were then captured on a magnet. The supernatant was discarded and the beads were washed in 1 mL of wash buffer (homogenization buffer above but without sucrose) 4 times for 10 minutes/wash at 4° C. Proteins were then eluted from beads in 100 μl 1X Nupage LDS 4X Sample Buffer (Life Technologies) with 10% 2-mercaptoethanol by boiling at 95° C for one minute.
For western blotting, samples were run on 12% Bis-tris gels (Life Technologies) and transferred to 0.2 μm pore nitrocellulose (Bio-RAD). Blocking and antibody incubations were performed in TBS-T with 5% dry milk. Primary antibodies used were rabbit anti-Fn14 (4403s), guinea pig anti-VGLUT2 (AB2251, Millipore), mouse anti-PSD-95 (in-house), and rabbit anti-GAPDH (G9545, Sigma). Secondary antibodies included anti-rabbit, anti-guinea pig, and anti-mouse conjugated to HRP. Blots were developed with Western Lightning ECL Pro (Perkin-Elmer). In some cases, western blots were quantified using the Li-Cor Odyssey system and fluorescent secondaries (Li-Cor biosciences).
Mass spectrometry
For unbiased discovery of Fn14 interactors, Fn14 was immunoprecipitated from the brains of an adult WT mouse and an Fn14 KO littermate as described above. The immunoprecipitate was run on a bis-tris gel which was then silver stained using the SilverQuest kit from Invitrogen according to manufacturer’s instructions. The protein bands were excised and submitted to the Taplin Biological Mass Spectrometry Facility where the samples were analyzed by LC-MS/MS. Briefly, gel pieces were trypsinized and dehydrated, then rehydrated in 50 mM ammonium bicarbonate solution containing 12.5 ng/μl modified sequencing-grade trypsin (Promega, Madison, WI) at 4° C. After 45 min, the excess trypsin solution was removed and replaced with 50 mM ammonium bicarbonate solution to just cover the gel pieces. Samples were then incubated at 37° C overnight and extracted by removing the ammonium bicarbonate solution, followed by one wash with a solution containing 50% acetonitrile and 1% formic acid. The extracts were then dried in a speed-vac for 1 hour.
Samples were reconstituted in 5 - 10 μl of HPLC solvent A (2.5% acetonitrile, 0.1% formic acid). Each sample was loaded onto a Famos auto sampler (LC Packings, San Francisco, CA). Peptides were eluted with increasing concentrations of solvent B (97.5% acetonitrile, 0.1% formic acid). Eluted peptides were subjected to electrospray ionization and then entered into an LTQ Orbitrap Velos Pro ion-trap mass spectrometer (Thermo Fisher Scientific, Waltham, MA). Peptides were detected, isolated, and fragmented to produce a tandem mass spectrum of specific fragment ions for each peptide. Peptide sequences (and hence protein identity) were determined by matching protein databases with the acquired fragmentation pattern by the software program, Sequest (Thermo Fisher Scientific, Waltham, MA).
We performed 3 individual bioreplicates of the experiment. All peptide hits listed in the manuscript were identified in at least 2 of 3 bioreplicates and were never detected in IgG controls from the same wild-type mouse brains or in separate Fn14 co-IPs from the brains of Fn14 KO mice; n = 3 experiments from 3 mice per genotype. STRING analysis was performed to assess functional interactions between molecules identified as possible Fn14 interactors (STRING; https://string-db.org/)(Szklarczyk et al., 2019).
In vitro thalamic neuron-microglia co-culture system
The night before dissections, glass coverslips were coated in Poly-D-Lysine (Sigma) and Laminin (Life Technologies) diluted in sterile, autoclaved water, and incubated overnight in a sterile cell culture incubator set to 37° C, 5% CO2. The next morning, the coverslips were washed with sterile water 2 times and once with Neurobasal plain (Gibco).
Prior to dissection, Dissociation Media (DM) was prepared as a 10X stock. 100 mM MgCl2, 100 mM Hepes, and 10 mM kynurenic acid were added to HBSS w/o Mg++ or Ca++ (Gibco). The solution was pH-ed to 7.2. 10X stocks were stored up to 6 months at −20° C, and 1X dilutions in HBSS were stored up to 1 month at 4° C.
In a tissue culture hood, 0.06 grams of ovomucoid trypsin inhibitor (Worthington Biochemical) and 0.06 grams of BSA (Sigma) were added to 6 mL of 1X DM in a falcon tube, which was labeled “Heavy”. The Heavy solution was warmed in a water bath at 37° C until both the inhibitor and BSA were dissolved (about 10 minutes). Heavy was pH-ed with sterile, filtered 1N NaOH from a P20 pipetteman tip to about 7.2 (~20 μL of 1N NaOH accomplished this). Next, 1 mL of this solution was added to 9 mL of 1X DM to make a solution labeled “Light”.
Pregnant female mice at E15.5 gestation day were euthanized by isoflurane and fetuses surgically removed from the amniotic sac. The fetuses were decapitated and the brains removed. The meninges were removed using fine forceps and the brains were bisected down the longitudinal fissure. Subcortical thalamic tissue was removed from each hemisphere and placed in 1X DM on ice until all fetuses had been processed. Two to three litters were typically pooled for each experiment.
Papain solution was prepared by combining 5 mL of 1X DM, a few grains of L-Cysteine (Sigma), and 100 μL of papain (10108014001, Sigma) and warming the solution in a water bath until dissolved. Next, the solution was filtered through a .02 μm filter into a separate falcon tube containing 2.5 mg of DNAse (Sigma). The 1X DM in which the thalami were pooled was aspirated from the tube, and 3 mL of the papain/DNAse solution was added to the tissue. The falcon tube was placed in the 37° C incubator for 10 minutes and stirred about every 2.5 minutes. The papain solution was aspirated and the tissue washed in 3 mL of Light solution 3 times, then 3 mL of Heavy solution 1 time. Next, the tissue was washed 1 time in 3 mL of Neurobasal complete [Neurobasal (Gibco) adjusted to 1% Penicillin/Streptomycin, 1% Glutamax, and 10 mL B27 Supplement (1X)].
Tissue was dissociated by adding 2 mL of Neurobasal complete and pipetting 1 time with a P1000. Tissue chunks were allowed to settle then 1 mL of the supernatant was added to a clean falcon tube through a 50 μm cell strainer. One mL of fresh Neurobasal complete was added back to the tube with the tissue and this trituration/filtering step was repeated 10 to 15 times. Finally, the remaining solution was strained through the cell strainer and 3 mL of Neurobasal complete was added to the strainer to wash it. Cells were counted on a hemocytometer and plated at a concentration of 100,000 cells/well in 1 mL of Neurobasal complete. After incubating for about 1 hour, the media was changed to fresh pre-warmed Neurobasal complete. Media was supplemented about every 3 days by removing 300 μL of the media from each well and adding back 400 μL fresh Neurobasal. At 5 DIV, neurons were sparsely transfected with 1 μg of CAG-mCherry using Lipofectamine 2000 (ThermoFisher) according to the manufacturer’s instructions.
For neuron-microglia co-cultures, microglia were acutely isolated from the brains of Cx3cr1-GFP; TWEAK KO or Cx3cr1-GFP; TWEAK WT mice between P15 and P20 as described in the “biochemical isolation of microglia-enriched fractions” section above, with the exception that, following the final centrifugation to pellet the microglia, the pellet was re-suspended in Neurobasal complete and the cells were counted on a hemocytometer. Cells were then plated atop thalamic neurons at 9 DIV at a concentration of 100,000 microglia/well. Following 24 hours, co-cultures were fixed in 4% PFA and 4% sucrose for 15 minutes at room temperature, washed in PBS, and mounted onto microscope slides in Fluoromount G + DAPI (SouthernBiotech). Imaging was performed on a Zeiss FV1000 confocal microscope as described above. Primary antibodies used include those listed above as well as mouse anti-PSD-95 (in-house), chicken anti-MAP2 (Lifespan Biosciences, LS-C61805), and rabbit anti-GFAP (Abcam, ab4674).
Statistical Analysis
N values for all experiments described in the main figures
Figure 1: TEM analysis and Golgi staining
Eighty total boutons (40 with spines, 40 without spines) per condition were reconstructed and quantified in (E) and (F). (G), n = 38 (WT) and n = 27 (KO) bulbous spines. (H), n = 68 boutons per condition. (I), n = 15 WT and 15 KO neurons analyzed across 3 mice per condition. (J), n = 35 WT and 31 KO neurons analyzed across 3 mice per condition.
Figure 2: Golgi analysis of spines
(D), n = 40 (Cre-) and 29 (Cre+) neurons analyzed from 3 mice per condition. (E), n = 30 neurons across 3 mice per condition. (F), averaged spine lengths from n = 39 (Cre-) and 30 (Cre+) neurons from 3 mice per condition. (G), Averaged spine lengths from n = 29 neurons each from 3 mice per condition. In panels (I) - (O), n = 30 (P10, P90, and P27 LDR), 36 (P20), and 28 (P27 NR) neurons across 3 mice per condition.
Figure 3: Molecular analysis of TWEAK expression
(B) and (G): n = 3 mice per condition; (D), n = 8 (0) and 10 (8) mice per condition. (E), n = 54 (0) and 62 (8) microglia from 3 mice per condition. (F), n = 28 (0) and 44 (8) relay neurons from 3 mice per condition.
Figure 4: Engulfment assays and spine analysis
(B), n = 20 (P7 WT), 17 (P7 KO), 28 (P27 WT), and 37 (P27 KO) microglia from 4 mice per condition. (D), n = 42 (WT) and 30 (KO) microglia from 4 mice per condition. (F), n = 5 (P7 WT), 4 (P7 KO), 6 (P27 WT), and 8 (P27 KO) mice per condition. (G), n = 5 (P7 WT), 7 (P7 KO), 6 (P27 WT), and 9 (P27 KO) mice per condition. (J), conditions left to right, n = 25, 24, 24, 29, 38, and 37 neurons from 3 mice per condition.
Figure 5. Spine analyses and TWEAK localization
(B), conditions left to right, n = 40, 29, 29, and 31 neurons from 3 mice per condition. (D) – (I), n = 30 neurons across 3 mice per condition.
Figure 6: Microglia-neuron co-cultures
For (C) – (F), n = 51 (control), 47 (WT), and 45 (KO) neurons from 3 independent rounds of co-cultures. For (G) and (H), n = 104 and 117 bulbous spines, WT and KO respectively, pooled across 3 independent rounds of co-cultures. For (I) and (J), N = 174 (WT) and 228 (KO) thin spines across three separate bioreplicates.
Figure 7: FISH-IF analysis of microglial TWEAK and proximal synapses.
(C) – (G), n = 60 total microglia, 28 low TWEAK expressers and 32 high TWEAK expressers.
Blinding
All experiments were performed with investigators blinded to the conditions at each stage. For GridTape analysis, Fn14 KO and WT littermates were identified by S.R. via genotyping, and the mouse numbers were given to L.C. L.C. perfusion-fixed brains, microdissected out the dLGNs, and provided both sections to the Electron Microscopy Core for further processing (see above). The core stained then provided the coded sections to J.M-S. and W-C.L., who sectioned and aligned the datasets. L.C. then performed quantifications and reconstructions. Following all data analysis, L.C. was unblinded to the conditions.
For spine analysis, L.C. performed Golgi staining of comparable samples in parallel and blinded S.R. to the sample identities of brains upon freezing. S.R. then sectioned, stained, and quantified spines prior to being unblinded by L.C. For microglial engulfment assay, Sholl assay, and eye-specific segregation, S.R. blinded L.C. to the samples and L.C. performed imaging in parallel. L.C. then further blinded the samples for analysis by S.R. All other analyses, including RNAscope and immunoprecipitation, were similarly blinded.
Statistical tests and software used
Data processing, figure generation, and statistical analyses were performed in Graphpad Prism 7 and 8. For the comparison of conditions including only 2 conditions, Student’s t-test (unpaired) was used. For the comparison of more than 2 conditions within the same graph, One-way ANOVAs were performed, with post hoc comparison of all conditions to one another by Tukey’s test. For comparison of microglial levels of TWEAK to other cell types, One-way ANOVA with Dunnett’s test was used. Cumulative frequency distributions were statistically analyzed using Kolmogorov-Smirnov test.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-Fn14 | Cell Signaling Technology | Cat # 4403S; RRID:AB_10693941 |
| Guinea Pig anti-VGLUT2 | Millipore | Cat # AB2251; RRID:AB_2665454 |
| Rabbit anti-Iba-1 | Wako | Cat # 019-19741; RRID:AB_839504 |
| Rabbit anti-P2ry12 | Sigma | Cat # HPA013796; RRID:AB_1854884 |
| Goat anti-TWEAK | R&D Systems | Cat # AF1237; RRID:AB_2206219 |
| Rabbit anti-GAPDH | Sigma-Aldrich | Cat # G9545; RRID:AB_796208 |
| Mouse anti-PSD95 | In-house | Cheadle et al, 2018 |
| Chicken anti-MAP2 | Lifespan Biosciences | Cat # LS-C61805; RRID:AB_1509808 |
| Rabbit anti-GFAP | Abcam | Cat # ab4674; RRID:AB_304558 |
| Goat anti-guinea pig AlexaFluor 647 | Molecular Probes | Cat # A-21450; RRID:AB_141882 |
| Goat anti-rabbit AlexaFluor 488 | Thermo Fisher | Cat # A-11001; RRID:AB_2534069 |
| Goat anti-mouse AlexaFluor 555 | Thermo Fisher | Cat # A-21428; RRID:AB_2535849 |
| Donkey anti-goat AlexaFluor 488 | Thermo Fisher | Cat # A-11055; RRID:AB_2534102 |
| Goat anti-guinea pig AlexaFluor 488 | Molecular Probes | Cat # A-11073; RRID:AB_2534117 |
| Goat anti-rabbit Complete IRdye 800 CW | Li-Cor biosciences | Cat # 827–08365; RRID:AB_10796098 |
| Bacterial and Virus Strains | ||
| N/A | ||
| Biological Samples | ||
| N/A | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| DAPI Fluoromount-G | Southern Biotech | Cat # 0100-20 |
| NuPAGE LDS Sample Buffer (4X) | Novex | Ref # NP0007 |
| Syn-PER synaptic protein extraction reagent | Thermo Fisher | Cat # 87793 |
| Paraformaldehyde, 16% | Electron Microscopy Sciences | Cat # 15710 |
| Protein A dynabeads | Life Technologies | Cat # 10002D |
| Triton X-100 | Sigma-Aldrich | Cat # X100 |
| Trizol | Life Technologies | Ref # 15596026 |
| Choleratoxin-B conjugated to 488 | Life Technologies | Cat # C34775 |
| Choleratoxin-B conjugated to 555 | Life Technologies | Cat # C34776 |
| Lipofectamine 2000 | Life Technologies | Cat # 11668500 |
| Critical Commercial Assays | ||
| FD Rapid Golgistain kit | FD NeuroTechnologies, Inc. | Cat #PK401A |
| PowerUp SYBR Green Master Mix | Life Technologies | Cat #A25743 |
| RNAscope Multiplexed Fluorescence Detection kit | ACDBio | Cat #320850 |
| RNeasy Micro Kit | Qiagen | Cat #74004 |
| RNAscope Multiplex Fluorescent Reagent Kit v2 | ACDBio | Cat #323100 |
| Mouse TWEAK DuoSet ELISA | R&D Systems | Cat #DY1237 |
| BCA protein assay | Thermo Fisher | Cat # 23225 |
| Silverquest Silver Staining kit | Thermo Fisher | Cat # LC6070 |
| Western Lightning ECL Pro | Perkin-Elmer | Cat # NEL121001EA |
| Deposited Data | ||
| N/A | ||
| Experimental Models: Cell Lines | ||
| N/A | ||
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6J | The Jackson Laboratory | 000664; RRID:IMSR_JAX:000664 |
| Mouse: B6.Tnfrsf12atm1(KO)Biogen (Fn14 KO) | Jakubowski et al, 2005 | N/A |
| Mouse: B6.Tnfsf12tm1(KO)Biogen | Dohi et al, 2009 | N/A |
| Mouse: B6.Tnfrsf12a(fl/fl)Biogen | This paper | N/A |
| Mouse: B6.Tnfsf12(fl/fl)Gree/J | This paper | N/A |
| Mouse: B6.129P2(Cg)-Cx3cr1tm1Litt/J | The Jackson Laboratory | 005582; RRID:IMSR_JAX:005582 |
| Mouse: Slc17a6tm2(cre)Lowl/J | The Jackson Laboratory | 028863; RRID:IMSR_JAX:016963 |
| Mouse: Tg(Prkcd-glc-1/CFP,-Cre) | Haubensak et al, 2010 | N/A |
| Mouse: B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J | The Jackson Laboratory | 007914; RRID:IMSR_JAX:007914 |
| Mouse: Tg(Chx10-EGFP/cre,-ALPP)2Clc/J | The Jackson Laboratory | 005105; RRID:IMSR_JAX:005105 |
| Mouse: B6J.B6N(Cg)-Cx3cr1tm1.1(cre)Jung/J | The Jackson Laboratory | 025524; RRID:IMSR_JAX:025524 |
| Mouse: B6.C1qa(KO) | Lab of Beth Stevens | N/A |
| Oligonucleotides | ||
| qPCR primer: Gapdh (Forward): GGGTGTGAACCACGAGAAATA | Origene | Cat #: MP205604 |
| qPCR primer: Gapdh (Reverse): CTGTGGTCATGAGCCCTTC | Origene | Cat #: MP205604 |
| qPCR primer: Tnfsf12 (TWEAK) (Forward): GCTGGGCAACGCTGTCT | Biogen | N/A |
| qPCR primer: Tnfsf12 (TWEAK) (Reverse): GCGGTCCTCTGCTGTCA | Biogen | N/A |
| qPCR: Cx3cr1 (Forward): GAGCATCACTGACATCTACCTCC | Origene | Cat #: MP202408 |
| qPCR: Cx3cr1 (Reverse): AGAAGGCAGTCGTGAGCTTGCA | Origene | Cat #: MP202408 |
| qPCR: P2ry12 (Forward): CATTGACCGCTACCTGAAGACC | Origene | Cat #: MP212229 |
| qPCR: P2ry12 (Reverse): GCCTCCTGTTGGTGAGAATCATG | Origene | Cat #: MP212229 |
| Recombinant DNA | ||
| AAV9-CASI-sTWEAK | Biogen | N/A |
| AAV9-CASI-mCherry | Biogen | N/A |
| pCAG-mCherry | In-house | N/A |
| Software and Algorithms | ||
| ImageJ | NIH | https://fiji.sc/ or https://imagej.nih.gov/ij/ |
| Prism | Graphpad | version 7.0b; RRID:SCR_002798 |
| Neurolucida | Microbrightfield | RRID:SCR_001775 |
| Imaris | Bitplane | ImarisColoc |
| Metamorph | Molecular Devices | Version 1.0 |
| Odyssey infrared imaging system | Li-Cor biosciences | Version 3.0 |
| CATMAID | Saalfeld et al., 2009 | https://catmaid.readthedocs.io/en/stable/ |
| iTK-SNAP | Yushkevich et al., 2006 | http://www.itksnap.org/pmwiki/pmwiki.php |
| AlignTK | N/A | http://mmbios.org/installation |
| STRING | Szklarczyk et al., 2019 | https://string-db.org/ |
| Other | ||
| FISH probe: C1qa, Channel 3 | ACDBio | Cat # 441221-C3 |
| FISH probe: Cx3cr1, Channel 2 | ACDBio | Cat # 314221-C2 |
| FISH probe: Olig1, Channel 3 | ACDBio | Cat # 480651-C3 |
| FISH probe: Aldh1l1, Channel 2 | ACDBio | Cat # 405891-C2 |
| FISH probe: Cldn5, Channel 3 | ACDBio | Cat # 491611-C3 |
| FISH probe: Tnfrsf12a (Fn14), Channel 3 | ACDBio | Cat # 505311-C3 |
| FISH probe: P2ry12, Channel 2 | ACDBio | Cat # 317601-C2 |
| FISH probe: Vglut2, Channel 2 | ACDBio | Cat # 319171-C2 |
| FISH probe: Tnfsf12 (TWEAK), Channel 1 | ACDBio | Cat # 552051 |
| FISH probe: Aldoc, Channel 3 | ACDBio | Cat # 429531-C3 |
Highlights:
Experience induces Fn14 expression in neurons and TWEAK expression in microglia
Fn14 increases the number of spine-associated synapses when not bound by TWEAK
Microglial TWEAK signals through neuronal Fn14 to decrease synapse number locally
Microglia-driven synapse loss occurs through a non-phagocytic mechanism
Acknowledgements:
We thank Drs. C. Chen, E. Griffith, S. Ashrafi, E. Duffy Lacy, B. Kalish, and E. Pollina for their thoughtful input on the manuscript. We thank Vance Soares for artistic work on the model. We thank Dr. Corey Harwell for Prkcd-Cre/Ai14 mouse tissue. We thank Dr. D.G.C. Hildebrand for help with sectioning and collection of dLGN tissue using GridTape. This work was supported by several core facilities at Harvard Medical School: Neurobiology Imaging Facility supported by NINDS P30 core center grant No. NS072030, the Vision Core supported by grant P30 EY12196, Taplin Mass Spectrometry Facility, Genome Modification Facility, and the EM core at HMS. This work was supported by K99 MH120051 (NIMH) and Goldenson Postdoctoral Fellowship (L.C.) and R01 NS028829 (NINDS) (M.E.G.), as well as NIH grants (R21NS085320, RF1MH114047) to W-C.A.L., the Bertarelli Program in Translational Neuroscience and Neuroengineering, Edward R. and Anne G. Lefler Center, and the Stanley and Theodora Feldberg Fund.
Footnotes
Declaration of interests:
Dr. Linda C. Burkly and Katelin A. Ennis are employees and shareholders at Biogen.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- Akiyoshi R, Wake H, Kato D, Horiuchi H, Ono R, Ikegami A, Haruwaka K, Omori T, Tachibana Y, Moorhouse AJ, et al. (2018). Microglia Enhance Synapse Activity to Promote Local Network Synchronization. eNeuro 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assali A, Le Magueresse C, Bennis M, Nicol X, Gaspar P, and Rebsam A (2017). RIM1/2 in retinal ganglion cells are required for the refinement of ipsilateral axons and eye-specific segregation. Sci Rep 7, 3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry KP, and Nedivi E (2017). Spine Dynamics: Are They All the Same? Neuron 96, 43–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardona AE, Huang D, Sasse ME, and Ransohoff RM (2006). Isolation of murine microglial cells for RNA analysis or flow cytometry. Nat Protoc 1, 1947–1951. [DOI] [PubMed] [Google Scholar]
- Cheadle L, Tzeng CP, Kalish BT, Harmin DA, Rivera S, Ling E, Nagy MA, Hrvatin S, Hu L, Stroud H, et al. (2018). Visual Experience-Dependent Expression of Fn14 Is Required for Retinogeniculate Refinement. Neuron 99, 525–539 e510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, and Regehr WG (2000). Developmental remodeling of the retinogeniculate synapse. Neuron 28, 955–966. [DOI] [PubMed] [Google Scholar]
- Chicheportiche Y, Bourdon PR, Xu H, Hsu YM, Scott H, Hession C, Garcia I, and Browning JL (1997). TWEAK, a new secreted ligand in the tumor necrosis factor family that weakly induces apoptosis. J Biol Chem 272, 32401–32410. [DOI] [PubMed] [Google Scholar]
- Choi S, Ko J, Lee JR, Lee HW, Kim K, Chung HS, Kim H, and Kim E (2006). ARF6 and EFA6A regulate the development and maintenance of dendritic spines. J Neurosci 26, 4811–4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colonnier M, and Guillery RW (1964). Synaptic Organization in the Lateral Geniculate Nucleus of the Monkey. Z Zellforsch Mikrosk Anat 62, 333–355. [DOI] [PubMed] [Google Scholar]
- Corriveau RA, Huh GS, and Shatz CJ (1998). Regulation of class I MHC gene expression in the developing and mature CNS by neural activity. Neuron 21, 505–520. [DOI] [PubMed] [Google Scholar]
- Cowan M, and Petri WA Jr. (2018). Microglia: Immune Regulators of Neurodevelopment. Front Immunol 9, 2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datwani A, McConnell MJ, Kanold PO, Micheva KD, Busse B, Shamloo M, Smith SJ, and Shatz CJ (2009). Classical MHCl molecules regulate retinogeniculate refinement and limit ocular dominance plasticity. Neuron 64, 463–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Kaeser PS, Xu W, and Sudhof TC (2011). RIM proteins activate vesicle priming by reversing autoinhibitory homodimerization of Munc13. Neuron 69, 317–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohi T, Borodovsky A, Wu P, Shearstone JR, Kawashima R, Runkel L, Rajman L, Dong X, Scott ML, Michaelson JS, et al. (2009). TWEAK/Fn14 pathway: a nonredundant role in intestinal damage in mice through a TWEAK/intestinal epithelial cell axis. Gastroenterology 136, 912–923. [DOI] [PubMed] [Google Scholar]
- Faludi G, and Mirnics K (2011). Synaptic changes in the brain of subjects with schizophrenia. Int J Dev Neurosci 29, 305–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg I (1982). Schizophrenia: caused by a fault in programmed synaptic elimination during adolescence? J Psychiatr Res 17, 319–334. [DOI] [PubMed] [Google Scholar]
- Fischer von Mollard G, Stahl B, Li C, Sudhof TC, and Jahn R (1994). Rab proteins in regulated exocytosis. Trends Biochem Sci 19, 164–168. [DOI] [PubMed] [Google Scholar]
- Fukuda M (2003). Distinct Rab binding specificity of Rim1, Rim2, rabphilin, and Noc2. Identification of a critical determinant of Rab3A/Rab27A recognition by Rim2. J Biol Chem 278, 15373–15380. [DOI] [PubMed] [Google Scholar]
- Fukuda M (2004). Alternative splicing in the first alpha-helical region of the Rab-binding domain of Rim regulates Rab3A binding activity: is Rim a Rab3 effector protein during evolution? Genes Cells 9, 831–842. [DOI] [PubMed] [Google Scholar]
- Gerges NZ, Brown TC, Correia SS, and Esteban JA (2005). Analysis of Rab protein function in neurotransmitter receptor trafficking at hippocampal synapses. Methods Enzymol 403, 153–166. [DOI] [PubMed] [Google Scholar]
- Glantz LA, and Lewis DA (2000). Decreased dendritic spine density on prefrontal cortical pyramidal neurons in schizophrenia. Arch Gen Psychiatry 57, 65–73. [DOI] [PubMed] [Google Scholar]
- Graf ER, Daniels RW, Burgess RW, Schwarz TL, and DiAntonio A (2009). Rab3 dynamically controls protein composition at active zones. Neuron 64, 663–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromova KV, Muhia M, Rothammer N, Gee CE, Thies E, Schaefer I, Kress S, Kilimann MW, Shevchuk O, Oertner TG, et al. (2018). Neurobeachin and the Kinesin KIF21B Are Critical for Endocytic Recycling of NMDA Receptors and Regulate Social Behavior. Cell Rep 23, 2705–2717. [DOI] [PubMed] [Google Scholar]
- Guillery RW, and Colonnier M (1970). Synaptic patterns in the dorsal lateral geniculate nucleus of the monkey. Z Zellforsch Mikrosk Anat 103, 90–108. [DOI] [PubMed] [Google Scholar]
- Gunner G, Cheadle L, Johnson KM, Ayata P, Badimon A, Mondo E, Nagy MA, Liu L, Bemiller SM, Kim KW, et al. (2019). Sensory lesioning induces microglial synapse elimination via ADAM10 and fractalkine signaling. Nat Neurosci 22, 1075–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond TR, Marsh SE, and Stevens B (2019). Immune Signaling in Neurodegeneration. Immunity 50, 955–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamos JE, Van Horn SC, Raczkowski D, and Sherman SM (1987). Synaptic circuits involving an individual retinogeniculate axon in the cat. J Comp Neurol 259, 165–192. [DOI] [PubMed] [Google Scholar]
- Han K, Kim MH, Seeburg D, Seo J, Verpelli C, Han S, Chung HS, Ko J, Lee HW, Kim K, et al. (2009). Regulated RalBP1 binding to RalA and PSD-95 controls AMPA receptor endocytosis and LTD. PLoS Biol 7, e1000187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Kaeser PS, Sudhof TC, and Schneggenburger R (2011). RIM determines Ca(2)+ channel density and vesicle docking at the presynaptic active zone. Neuron 69, 304–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haubensak W, Kunwar PS, Cai H, Ciocchi S, Wall NR, Ponnusamy R, Biag J, Dong HW, Deisseroth K, Callaway EM, et al. (2010). Genetic dissection of an amygdala microcircuit that gates conditioned fear. Nature 468, 270–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtmaat A, and Svoboda K (2009). Experience-dependent structural synaptic plasticity in the mammalian brain. Nat Rev Neurosci 10, 647–658. [DOI] [PubMed] [Google Scholar]
- Hong S, Dissing-Olesen L, and Stevens B (2016). New insights on the role of microglia in synaptic pruning in health and disease. Curr Opin Neurobiol 36, 128–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong YK, Park S, Litvina EY, Morales J, Sanes JR, and Chen C (2014). Refinement of the retinogeniculate synapse by bouton clustering. Neuron 84, 332–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooks BM, and Chen C (2006). Distinct roles for spontaneous and visual activity in remodeling of the retinogeniculate synapse. Neuron 52, 281–291. [DOI] [PubMed] [Google Scholar]
- Hooks BM, and Chen C (2008). Vision triggers an experience-dependent sensitive period at the retinogeniculate synapse. J Neurosci 28, 4807–4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooks BM, and Chen C (2020). Circuitry Underlying Experience-Dependent Plasticity in the Mouse Visual System. Neuron 106, 21–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubowski A, Ambrose C, Parr M, Lincecum JM, Wang MZ, Zheng TS, Browning B, Michaelson JS, Baetscher M, Wang B, et al. (2005). TWEAK induces liver progenitor cell proliferation. J Clin Invest 115, 2330–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S, Aliberti J, Graemmel P, Sunshine MJ, Kreutzberg GW, Sher A, and Littman DR (2000). Analysis of fractalkine receptor CX(3)CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol Cell Biol 20, 4106–4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeser PS, Deng L, Fan M, and Sudhof TC (2012). RIM genes differentially contribute to organizing presynaptic release sites. Proc Natl Acad Sci U S A 109, 11830–11835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalish BT, Cheadle L, Hrvatin S, Nagy MA, Rivera S, Crow M, Gillis J, Kirchner R, and Greenberg ME (2018). Single-cell transcriptomics of the developing lateral geniculate nucleus reveals insights into circuit assembly and refinement. Proc Natl Acad Sci U S A 115, E1051–E1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz LC, and Shatz CJ (1996). Synaptic activity and the construction of cortical circuits. Science 274, 1133–1138. [DOI] [PubMed] [Google Scholar]
- Kim Y, Lee SE, Park J, Kim M, Lee B, Hwang D, and Chang S (2015). ADP-ribosylation factor 6 (ARF6) bidirectionally regulates dendritic spine formation depending on neuronal maturation and activity. J Biol Chem 290, 7323–7335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan JL, Berger DR, Wetzel AW, and Lichtman JW (2016). The Fuzzy Logic of Network Connectivity in Mouse Visual Thalamus. Cell 165, 192–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neniskyte U, and Gross CT (2017). Errant gardeners: glial-cell-dependent synaptic pruning and neurodevelopmental disorders. Nat Rev Neurosci 18, 658–670. [DOI] [PubMed] [Google Scholar]
- Paolicelli RC, Bolasco G, Pagani F, Maggi L, Scianni M, Panzanelli P, Giustetto M, Ferreira TA, Guiducci E, Dumas L, et al. (2011). Synaptic pruning by microglia is necessary for normal brain development. Science 333, 1456–1458. [DOI] [PubMed] [Google Scholar]
- Parajuli LK, Tanaka S, and Okabe S (2017). Insights into age-old questions of new dendritic spines: From form to function. Brain Res Bull 129, 3–11. [DOI] [PubMed] [Google Scholar]
- Park D, Lee U, Cho E, Zhao H, Kim JA, Lee BJ, Regan P, Ho WK, Cho K, and Chang S (2018). Impairment of Release Site Clearance within the Active Zone by Reduced SCAMP5 Expression Causes Short-Term Depression of Synaptic Release. Cell Rep 22, 3339–3350. [DOI] [PubMed] [Google Scholar]
- Parkhurst CN, Yang G, Ninan I, Savas JN, Yates JR 3rd, Lafaille JJ, Hempstead BL, Littman DR, and Gan WB (2013). Microglia promote learning-dependent synapse formation through brain-derived neurotrophic factor. Cell 155, 1596–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters A, and Kaiserman-Abramof IR (1970). The small pyramidal neuron of the rat cerebral cortex. The perikaryon, dendrites and spines. Am J Anat 127, 321–355. [DOI] [PubMed] [Google Scholar]
- Raemaekers T, Peric A, Baatsen P, Sannerud R, Declerck I, Baert V, Michiels C, and Annaert W (2012). ARF6-mediated endosomal transport of Telencephalin affects dendritic filopodia-to-spine maturation. EMBO J 31, 3252–3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafols JA, and Valverde F (1973). The structure of the dorsal lateral geniculate nucleus in the mouse. A Golgi and electron microscopic study. J Comp Neurol 150, 303–332. [DOI] [PubMed] [Google Scholar]
- Rowan S, and Cepko CL (2004). Genetic analysis of the homeodomain transcription factor ChxlO in the retina using a novel multifunctional BAC transgenic mouse reporter. Dev Biol 271, 388–402. [DOI] [PubMed] [Google Scholar]
- Saalfeld S, Cardona A, Hartenstein V, and Tomancak P (2009). CATMAID: collaborative annotation toolkit for massive amounts of image data. Bioinformatics 25, 1984–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer DP, Lehrman EK, Kautzman AG, Koyama R, Mardinly AR, Yamasaki R, Ransohoff RM, Greenberg ME, Barres BA, and Stevens B (2012). Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron 74, 691–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider-Mizell CM, Gerhard S, Longair M, Kazimiers T, Li F, Zwart MF, Champion A, Midgley FM, Fetter RD, Saalfeld S, et al. (2016). Quantitative neuroanatomy for connectomics in Drosophila. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan P, and Waites CL (2019). Coordination of synaptic vesicle trafficking and turnover by the Rab35 signaling network. Small GTPases 10, 54–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan P, Zhu M, Beskow A, Vollmer C, and Waites CL (2016). Activity-Dependent Degradation of Synaptic Vesicle Proteins Requires Rab35 and the ESCRT Pathway. J Neurosci 36, 8668–8686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sholl DA (1953). Dendritic organization in the neurons of the visual and motor cortices of the cat. J Anat 87, 387–406. [PMC free article] [PubMed] [Google Scholar]
- Sorra KE, and Harris KM (2000). Overview on the structure, composition, function, development, and plasticity of hippocampal dendritic spines. Hippocampus 10, 501–511. [DOI] [PubMed] [Google Scholar]
- Stahl B, von Mollard GF, Walch-Solimena C, and Jahn R (1994). GTP cleavage by the small GTP-binding protein Rab3A is associated with exocytosis of synaptic vesicles induced by alpha-latrotoxin. J Biol Chem 269, 24770–24776. [PubMed] [Google Scholar]
- Stevens B, Allen NJ, Vazquez LE, Howell GR, Christopherson KS, Nouri N, Micheva KD, Mehalow AK, Huberman AD, Stafford B, et al. (2007). The classical complement cascade mediates CNS synapse elimination. Cell 131, 1164–1178. [DOI] [PubMed] [Google Scholar]
- Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, et al. (2019). STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47, D607–D613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teodoro RO, Pekkurnaz G, Nasser A, Higashi-Kovtun ME, Balakireva M, McLachlan IG, Camonis J, and Schwarz TL (2013). Ral mediates activity-dependent growth of postsynaptic membranes via recruitment of the exocyst. EMBO J 32, 2039–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay ME, Lowery RL, and Majewska AK (2010). Microglial interactions with synapses are modulated by visual experience. PLoS Biol 8, e1000527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velmeshev D, Schirmer L, Jung D, Haeussler M, Perez Y, Mayer S, Bhaduri A, Goyal N, Rowitch DH, and Kriegstein AR (2019). Single-cell genomics identifies cell type-specific molecular changes in autism. Science 364, 685–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Lv X, Wu Y, Xu T, Jiao M, Yang R, Li X, Chen M, Yan Y, Chen C, et al. (2018). Postsynaptic RIM1 modulates synaptic function by facilitating membrane delivery of recycling NMDARs in hippocampal neurons. Nat Commun 9, 2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Sugita S, and Sudhof TC (2000). The RIM/NIM family of neuronal C2 domain proteins. Interactions with Rab3 and a new class of Src homology 3 domain proteins. J Biol Chem 275, 20033–20044. [DOI] [PubMed] [Google Scholar]
- Weinhard L, di Bartolomei G, Bolasco G, Machado P, Schieber NL, Neniskyte U, Exiga M, Vadisiute A, Raggioli A, Schertel A, et al. (2018). Microglia remodel synapses by presynaptic trogocytosis and spine head filopodia induction. Nat Commun 9, 1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesel TN, and Hubei DH (1963). Effects of Visual Deprivation on Morphology and Physiology of Cells in the Cats Lateral Geniculate Body. J Neurophysiol 26, 978–993. [DOI] [PubMed] [Google Scholar]
- Wiley SR, and Winkles JA (2003). TWEAK, a member of the TNF superfamily, is a multifunctional cytokine that binds the TweakR/Fn14 receptor. Cytokine Growth Factor Rev 14, 241–249. [DOI] [PubMed] [Google Scholar]
- Yona S, Kim KW, Wolf Y, Mildner A, Varol D, Breker M, Strauss-Ayali D, Viukov S, Guilliams M, Misharin A, et al. (2013). Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity 38, 79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, Gee JC, and Gerig G (2006). User- guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage 31, 1116–1128. [DOI] [PubMed] [Google Scholar]
- Zhao H, Kim Y, Park J, Park D, Lee SE, Chang I, and Chang S (2014). SCAMP5 plays a critical role in synaptic vesicle endocytosis during high neuronal activity. J Neurosci 34, 10085–10095. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The published article includes all datasets generated or analyzed during the study.