Abstract
Pulsatilla (Ranunculaceae) consists of about 40 species, and many of them have horticultural and/or medicinal value. However, it is difficult to recognize and identify wild Pulsatilla species. Universal molecular markers have been used to identify these species, but insufficient phylogenetic signal was available. Here, we compared the complete chloroplast genomes of seven Pulsatilla species. The chloroplast genomes of Pulsatilla were very similar and their length ranges from 161,501 to 162,669 bp. Eight highly variable regions and potential sources of molecular markers such as simple sequence repeats, large repeat sequences, and single nucleotide polymorphisms were identified, which are valuable for studies of infra- and inter-specific genetic diversity. The SNP number differentiating any two Pulsatilla chloroplast genomes ranged from 112 to 1214, and provided sufficient data for species delimitation. Phylogenetic trees based on different data sets were consistent with one another, with the IR, SSC regions and the barcode combination rbcL + matK + trnH-psbA produced slightly different results. Phylogenetic relationships within Pulsatilla were certainly resolved using the complete cp genome sequences. Overall, this study provides plentiful chloroplast genomic resources, which will be helpful to identify members of this taxonomically challenging group in further investigation.
Subject terms: Plant sciences, Systems biology
Introduction
DNA barcoding is an effective tool to identify many plant species rapidly and accurately1–4. However, there is no single universal barcode that can be successfully used to identify all plants to the species level5. Consequently, two alternative strategies have been proposed to distinguish among plant species: the first one is the use of complete chloroplast genomes6, named ‘super-barcoding’, and the second one is an approach that involves searching for mutational hotspots7, or using comparative plastid analyses to find loci with suitable species-level divergence8,9. Analyses of entire chloroplast genome sequences provide an effective way to develop both of these strategies.
In most angiosperms, the chloroplast genomes are inherited maternally and have a consistent structure, including two inverted repeats (IR), one large (LSC) and one small (SSC) single copy region. The chloroplast genome always contains 110–130 genes that exhibit a range of levels of polymorphism4,5. Thus, chloroplast genome sequence data are extremely valuable for studies of plant population genetics, phylogeny reconstruction, species identification, and genome evolution2,4–6.
The Ranunculaceae is a large family, which includes approximately 59 genera and 2500 species. Many plants of Ranunculaceae are pharmaceutically important10. The genus Pulsatilla Adans. consists of about 40 species which are distributed in temperate subarctic and mountainous areas of the Northern Hemisphere10. There are always long, soft hairs covering plants of Pulsatilla species. Most of the flowers of Pulsatilla are large and showy, and therefore the genus has horticultural importance11,12. The flowers are solitary and bisexual. In one flower, there are always six tepals, numerous stamens and carpels, with the outermost stamens resembling degenerated petals, excluding P. kostyczewii11,13–15.
In China, there are eleven species of Pulsatilla. Some species of Pulsatilla have been used in traditional Chinese medicine for many years, such as for “detoxification” or “blood-cooling”, because Pulsatilla species contain numerous secondary metabolites, including phytosterols, triterpenoid saponins and anthocyanins16. At the same time, all members of Pulsatilla produced the lactone protoanemonin17–19.
In Europe, some species of Pulsatilla are rare, endangered and endemic. Those taxa are protected due to their small populations and disappearing localities, and those species have been placed on the Red Lists of Endangered Species20.
Taxonomically, Pulsatilla is an especially complex and challenging group. In all treatments published before, three subgenera have been recognized: subgenus Kostyczewianae (only one species), subgenus Preonanthus, and the largest subgenus Pulsatilla. However, the intragenic morphological variability of Pulsatilla was especially complicated12. The recognition and identification of wild Pulsatilla species is particularly difficult based on traditional approaches.
Molecular markers are significant to explore the phylogenetic relationships of the genus Pulsatilla. Phylogenetic relationships between Pulsatilla and closely related genera have been dedicated during the past years21–25. Previous studies have attempted to identify these species among Pulsatilla with universal molecular markers, but the species resolution was relatively low15.
In this study, we present seven complete cp genomes from two subgenera of Pulsatilla obtained through next-generation sequencing (NGS) and genomic comparative analyses with four previously published cp genome sequences of Pulsatilla from NCBI, with Anemoclema glaucifolium as the outgroup. We identify microsatellites (SSRs), larger repeat sequences, and highly variable regions, with the aim of developing DNA barcodes and testing the feasibility of phylogenetic analyses of Pulsatilla using the chloroplast genome.
Results and discussion
Chloroplast genome features
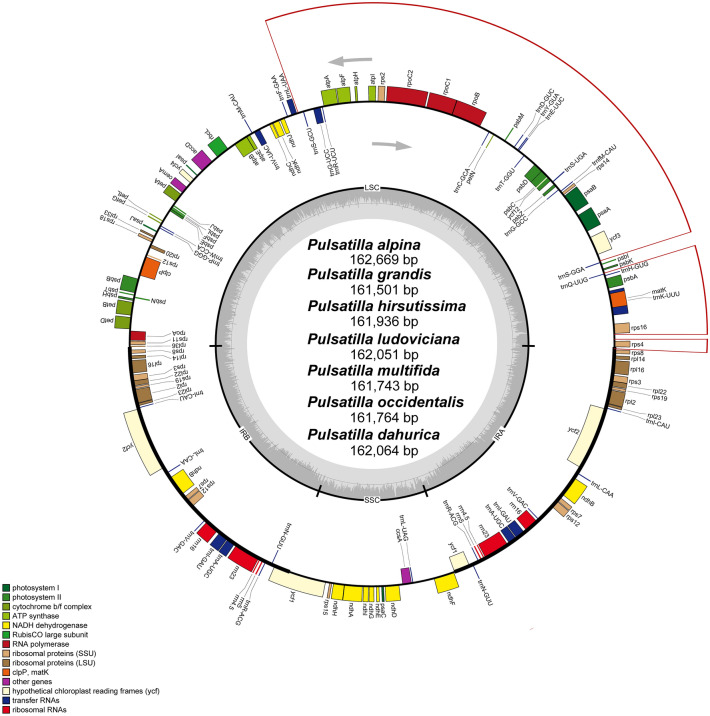
We have obtained 1.95 Gb of average NGS clean data for each species, with minimum and maximum values of 1.14 Gb (P. dahurica), and 3.56 Gb (Pulsatilla alpina), respectively. The read number for each species ranged from 6,468,944 (P. dahurica) to 15,816,765 (P. alpina). The average length of the reads was 150 bp on the Illumina Sequencing System. The seven new Pulsatilla cp genomes ranged from 161,501 bp (P. grandis) to 162,669 bp (P. alpina) in length and 151.5 × to 503.4 × coverage. These seven novel Pulsatilla cp genome sequences were submitted to GenBank (Tables 1, 2). Their quadripartite structure is similar to the majority of cp genomes of land plants, which are composed of a pair of IRs (31,184–31,416 bp), separated by the LSC (81,615–82,149 bp) and SSC (of 17,431–17,908 bp) regions26,27 (Fig. 1; Table 2). Previous studies of other angiosperm groups have found that chloroplast genomes are conserved28 or highly polymorphic29,30. These genomes which we reported are highly conserved in gene order, gene content and intron number, which is in accordance with the results from many other taxa26,27. However, in some taxa, e.g. Amorphophallus of Araceae, some genes were lost31, and in others, e.g. Pelargonium32, the structure and gene order diverges from what is reported here and in most other angiosperms. The cp genomes of P. alpina, P. grandis, P. hirsutissima, P. ludoviciana, P. multifida and P. occidentalis had the same GC content of 37.6%, while P. dahurica had a subtle difference (37.5%) compared with the others.
Table 1.
Voucher information and GenBank accession numbers for Pulsatilla and outgroups.
| Taxon | Location | Date | Herbarium | Accession | SRA accession |
|---|---|---|---|---|---|
| P. grandis* | Europe MoaiJuni | 1904 | US | MN025344 | SRR12822481 |
| P. multifida* | U.S.S.R. | 1957 | US | MN025347 | SRR12822474 |
| P. alpina* | America Graubunden | 1936 | US | MN025343 | SRR12822486 |
| P. occidentalis* | America Siskiyon Calfornia | 1943 | US | MN025348 | SRR12822484 |
| P. ludoviciana* | America Albany county | 1898 | US | MN025346 | SRR12822477 |
| P. hirsutissima* | America | – | US | MN025345 | SRR12822480 |
| P. dahurica* | China | 2014 | WUK | MN025349 | SRR12822482 |
| P. patens | NA | NA | NA | KR_297058 | NA |
| P. pratensis | NA | NA | NA | KR_297060 | NA |
| P. vernalis | NA | NA | NA | KR_297062 | NA |
| P. chinensis | NA | NA | NA | MG_001341 | NA |
| Anemoclema glaucifolium | NA | NA | NA | MH_205609 | NA |
Species with asterisks were collected by this study, whereas others were obtained from Genbank.
NA not applicable.
Table 2.
Summary of complete chloroplast genomes of Pulsatilla.
| Species | Number of reads | Average depth of coverage (×) | Size (bp) | Length (bp) | Coding (bp) | Non-coding (bp) | GC% | ||
|---|---|---|---|---|---|---|---|---|---|
| LSC | SSC | IR | |||||||
| P. grandis | 13,012,108 | 194.8 | 161,501 | 81,672 | 17,431 | 31,199 | 78,377 | 78,377 | 37.6 |
| P. multifida | 11,405,519 | 151.5 | 161,743 | 81,653 | 17,648 | 31,221 | 81,800 | 81,800 | 37.6 |
| P. alpina | 15,816,765 | 367.1 | 162,669 | 82,149 | 17,688 | 31,416 | 81,246 | 81,246 | 37.6 |
| P. occidentalis | 12,554,200 | 276.9 | 161,764 | 81,615 | 17,755 | 31,197 | 79,089 | 79,089 | 37.6 |
| P. ludoviciana | 9,251,830 | 257.5 | 162,051 | 81,860 | 17,771 | 31,210 | 79,280 | 79,280 | 37.6 |
| P. hirsutissima | 13,337,339 | 503.4 | 161,936 | 81,866 | 17,702 | 31,184 | 80,206 | 80,206 | 37.6 |
| P. dahurica | 6,468,944 | 410.1 | 162,064 | 81,688 | 17,908 | 31,234 | 79,114 | 79,114 | 37.5 |
Figure 1.
Gene map of the Pulsatilla chloroplast genome. Dashed area in the inner circle indicates the GC content of the chloroplast genome. LSC, SSC and IR mean large single copy, small single copy and inverted repeat, respectively. Genes belonging to different functional groups are color-coded as indicated by icons on the lower left corner. The red line on the outside of the gene is three inversions.
Chloroplast genome comparison
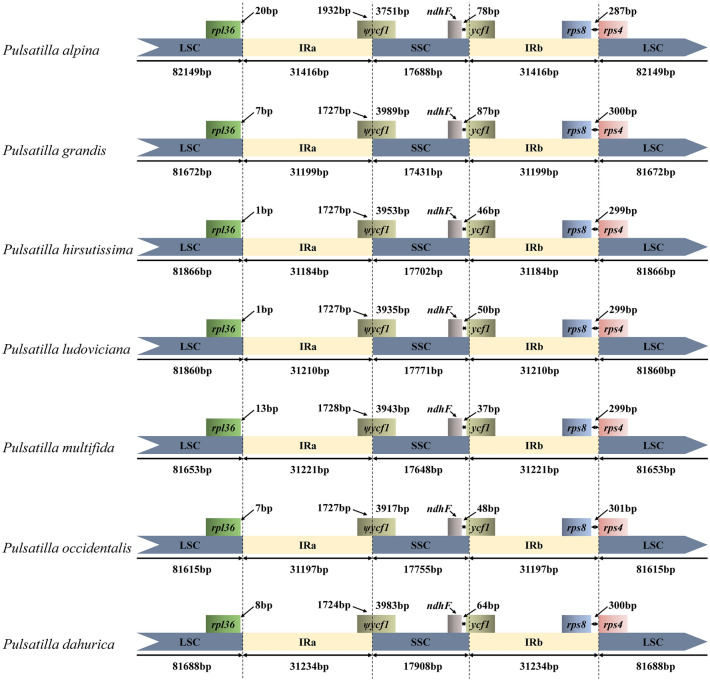
In most angiosperms, the IR regions of cp genomes of angiosperms are highly conserved, but the expansion and contraction of IR region boundaries are ever present33,34. At the same time, several lineages of land plant chloroplast genomes show great structural rearrangement, even loss of IR regions or some gene families35. The expansion and contraction in IRs are significant evolutionary events, because they can change gene content and chloroplast genome size30,36. Expansion of the IRs has been reported in Araceae36,37. Sometimes, the size of LSC increases and that of SSC decreases, becoming only 7000 bp in Pothos38. At the same time, a linear chloroplast genome was also reported in some groups, e.g. maize35,39. Expansion and contraction of the IR regions can also lead to duplication of certain genes or conversion of duplicate genes to single copy, respectively30,36. Changes in the size of the IRs can also cause rearrangement of the genes in the SSC as recently observed in Zantedeschia36.
The Pulsatilla chloroplast genomes were compared to previously published data and showed typical Anemoneae (Ranunculaceae) genome structure37,39. As reported for Anemoclema, Anemone, Clematis and Hepatica, the IR regions of genus Pulsatilla are roughly 4.4 kb longer than those of other genera of the family Ranunculaceae, such as Aconitum, Coptis, Thalictrum, Megaleranthis, Ranunculus, and Trollius37,39. The gene orders located within the IR-SSC and IR-LSC boundaries are similar among tribe Anemoneae but different from those of other genera of Ranunculaceae (Fig. 2, Fig. S1).
Figure 2.
Comparisons of LSC, SSC, and IR region borders among the seven Pulsatilla chloroplast genomes.
We compared the IR/SC boundary regions of Pulsatilla, and the junction positions are very similar and conserved within genus Pulsatilla. In the four boundary regions (LSC, IRa, SSC, IRb) of seven Pulsatilla cp genomes, the LSC/IRa and IRb/LSC border was in the intergenic region, and the adjacent genes is rps36, rps8 and rps4, respectively. The genes ycf1 andψycf1 have crossed the SSC/IRb and IRa/SSC boundary, respectively, which was also found in Monsteroideae (Araceae)28. The pseudogene ycf1 has been found in other groups30,36. The IR regions were highly conserved, with nucleotide diversity values in those regions less than 2%.
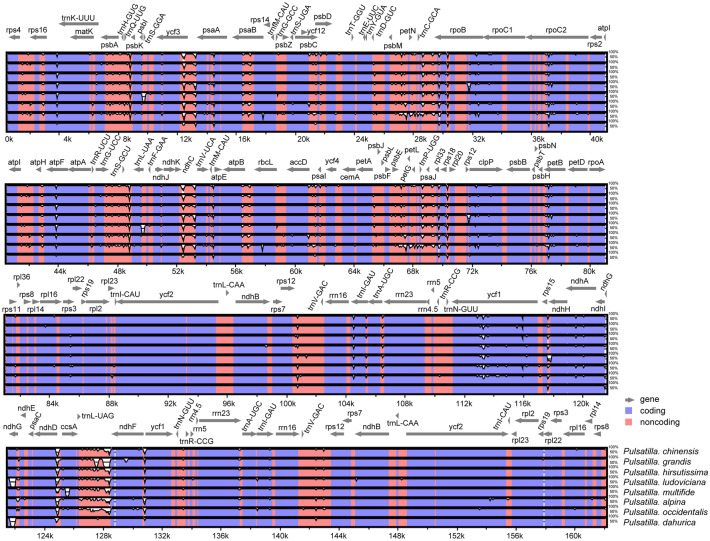
Sequence divergence
Multiple alignments of plastid genomes were performed to investigate levels of genome divergence. Based on MAFFT analysis, there are three inversions in LSC of Pulsatilla, same as the tribe Anemoneae (Fig. 1, Fig. S1)37. The mVISTA analysis has revealed high sequence similarity across the coding region and there exists more variability in non-coding regions. Sequence identity among the seven species was 96.68–98.66%. The number of nucleotide substitutions and sequence distance (Pi) were the highest (1214, 0.0063) between P. alpine and P. dahurica, with the lowest (112, 0.0005) between P. vernalis and P. patens (Fig. 3; Table 3).
Figure 3.
Visualized alignment of the eight Pulsatilla chloroplast genomes. The mVISTA-based identity plots show the sequence identity among the seven chloroplast genomes, with P. chinensis serving as a reference. Blue represents coding regions, and pink represents non-coding regions.
Table 3.
Numbers of nucleotide substitutions and sequence distance (Pi) in eleven complete cp genomes.
| P. chinensis | P. dahurica | P. patens | P. vernalis | P. hirsutissima | P. ludoviciana | P. pratensis | P. grandis | P. multifide | P. alpina | P. occidentalis | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| P. chinensis | 0.0028 | 0.0027 | 0.0029 | 0.0030 | 0.0035 | 0.0024 | 0.0034 | 0.0028 | 0.0058 | 0.0064 | |
| P. dahurica | 563 | 0.0033 | 0.0034 | 0.0034 | 0.0038 | 0.0029 | 0.0039 | 0.0033 | 0.0063 | 0.0068 | |
| P. patens | 558 | 642 | 0.0005 | 0.0011 | 0.0015 | 0.0019 | 0.0028 | 0.0010 | 0.0046 | 0.0052 | |
| P. vernalis | 572 | 638 | 112 | 0.0011 | 0.0015 | 0.0019 | 0.0028 | 0.0010 | 0.0046 | 0.0051 | |
| P. hirsutissima | 652 | 676 | 205 | 229 | 0.0009 | 0.0020 | 0.0029 | 0.0008 | 0.0048 | 0.0053 | |
| P. ludoviciana | 685 | 735 | 290 | 303 | 200 | 0.0024 | 0.0034 | 0.0013 | 0.0052 | 0.0058 | |
| P. pratensis | 470 | 516 | 378 | 383 | 392 | 547 | 0.0020 | 0.0019 | 0.0050 | 0.0055 | |
| P. grandis | 662 | 750 | 586 | 577 | 604 | 701 | 414 | 0.0029 | 0.0059 | 0.0064 | |
| P. multifide | 650 | 735 | 215 | 233 | 198 | 284 | 463 | 527 | 0.0046 | 0.0052 | |
| P. alpina | 1212 | 1214 | 915 | 903 | 916 | 1007 | 964 | 1128 | 908 | 0.0045 | |
| P. occidentalis | 1207 | 1293 | 979 | 968 | 1006 | 1081 | 1046 | 1219 | 930 | 869 |
The uppertriangle shows the number of nucleotide substitutions and the lower triangle indicates the number of sequence distance in complete cp genomes.
Identification of highly variable regions
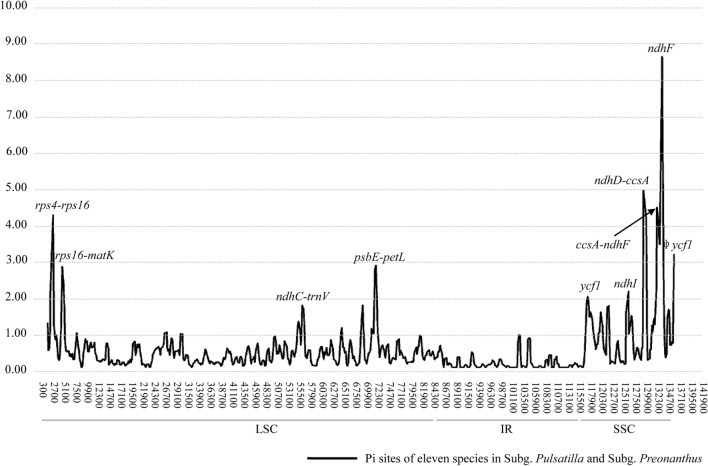
Chloroplast genome markers, especially several universal chloroplast regions, have been widely used in plant systematics and identification at multiple taxonomic levels. Highly suitable polymorphic chloroplast loci have been identified and designed as unique markers in different groups28,40. However, relationships within the genus Pulsatilla have not been well resolved because of the low polymorphism of these universal markers15. In order to facilitate identification of closely related species of Pulsatilla, we sought to identify highly variable regions of the chloroplast genome, as previously described9,27,41–44. As a result, we identified nine divergent hotspot regions, including six intergenic spacer regions (rps4-rps16, rps16-matK, ndhC-trnV, psbE-petL, ndhD-ccsA, ccsA-ndhF) and four protein-coding regions (ycf1, ndhF, ndhI) (Fig. 4; Table 4). Most commonly employed loci, e.g. trnL-trnF, trnH-psbA were not selected in our finding. The nine highly variable regions included 684 variable sites, including 181 indels. However, these indels are not suitable for the phylogenetic inference because Maximum likelihood model used only substitutions not indels28. Their nucleotide diversity values ranged from 0.00802 to 0.02212. The region of ccsA-ndhF showed the highest variability, the next most variable regions were rps4-rps16, ndhC-trnV, and psbE-petL. The diversity level of two protein-coding regions (ycf1, ndhF) was the lowest.
Figure 4.
Sliding window analysis of the whole chloroplast genomes of Pulsatilla taxa.
Table 4.
Sequence characteristics of eight high variable regionsamong eleven complete cp genomes of Pulsatilla.
| Region | Aligned length | Variable sites | Indels | Nucleotide diversity (Pi) | ||
|---|---|---|---|---|---|---|
| No | % | No | Length range | |||
| rps4-rps16 | 986 | 22 | 2.23 | 11 | 1–44 | 0.01497 |
| rps16-matK | 1985 | 62 | 3.12 | 23 | 1–79 | 0.00998 |
| ndhC-trnV | 1484 | 61 | 4.11 | 18 | 1–34 | 0.01502 |
| psbE-petL | 1469 | 70 | 4.77 | 24 | 1–139 | 0.01140 |
| ndhD-ccsA | 732 | 39 | 5.33 | 10 | 1–87 | 0.04368 |
| ccsA-ndhF | 2757 | 196 | 7.11 | 69 | 1–43 | 0.02212 |
| ycf1 | 5807 | 148 | 2.55 | 18 | 1–24 | 0.00802 |
| ndhF | 2345 | 86 | 3.67 | 8 | 1–18 | 0.00813 |
Among the nine divergent hotspot regions, the ndhI is difficult to align. There are large numbers of indels in ndhI and the intergenic spacer between ndhI and ndhG, these regions were not considered suitable for the phylogenetic inference of the Pulsatilla. Thus, we selected eight regions, four (rps4-rps16, rps16-matK, ndhC-trnV, psbE-petL) in the LSC and four (ndhD-ccsA, ccsA-ndhF, ycf1, ndhF) in the SSC, with relatively high variability as potential molecular markers for the study of species identification and phylogeny in Pulsatilla. Five hotpots were found in chloroplast genome of Veroniceae (Plantaginaceae), and two universal marker, trnH-psbA and matK were identified, respectively45,46. Ten highly variable regions were selected as potential molecular markers for Fritillaria, including ycf144,47, which was also selected in this study. Sequences of these variable regions founded in this study could be regarded as potential molecular markers for species identification and evolutionary studies and have been shown to be valuable for studies in other groups (e.g., Fritillaria)44.
SSRs and large repeat sequences
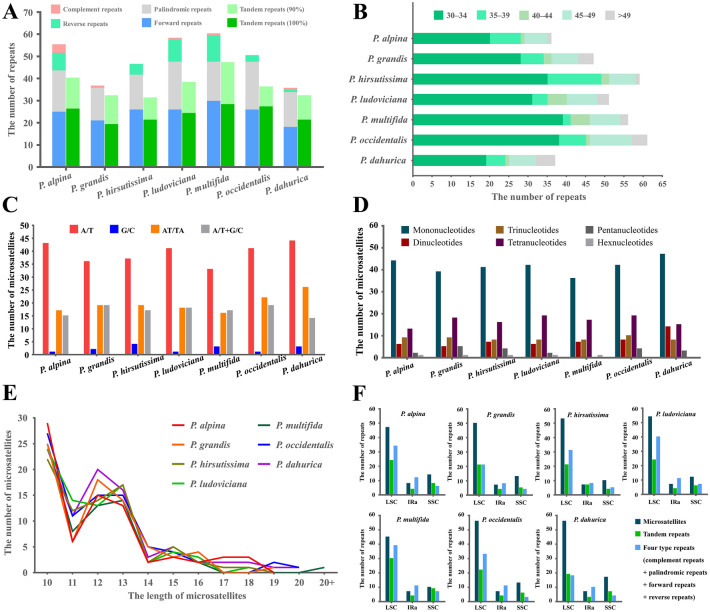
Oligonucleotide repeats play an important role for generating indels, inversion and substitutions29. Repeat sequences in the chloroplast genome could provide valuable information for understanding not only the sequence divergence but the evolutionary history of the plants48–50. We have detected five types of large repeats (forward, reverse, palindromic, complement and tandem repeats) in the seven Pulsatilla cp genomes. Among them, the most common repeat types are forward and palindromic repeats, followed by reverse repeats, and only little complement repeats were found in Pulsatilla cp genomes (Fig. 5A). Most of the repeats were short, ranging from 30–49 bp (Fig. 5B).
Figure 5.
Analyses of repeated sequences in seven newly sequenced chloroplast genomes. (A) Number of five repeat types; (B) frequency of four repeats by length; (C) frequency of microsatellites by base composition; (D) frequency of microsatellites by types; (E) frequency of microsatellites by length; (F) number of all repeats by location.
We also identified multiple microsatellite repeats, also known as simple sequence repeats (SSR) or short tandem repeats (STR)49. Due to their codominant inheritance and high variability, SSRs are robust and effective markers for species identification and population genetic analyses49–53. Most of the mononucleotide repeats were composed of A/T. The other microsatellites types were also dominated by AT/TA, with very little G/C (Fig. 5C). In this study, plentiful microsatellite loci were found through the comparative analysis of Pulsatilla cp genome sequences. In total, we detected six types of microsatellite (mononucleotide, dinucleotide, trinucleotide, tetranucleotide, pentanucleotide and hexanucleotide repeats) based on the comparison of seven Pulsatilla cp genomes (Fig. 5D). Each Pulsatilla cp genome had 69–87 microsatellites. The lengths of repeat motifs of these microsatellites ranged from 10 to 21 bp (Fig. 5E). Among the four structural regions in the cp genomes, most of the repeats and microsatellites were distributed in LSC, followed by SSC, and fewest in IRa/IRb (Fig. 5F), which were also reported in other studies in angiosperms29,30. These SSRs and repeat sequences are uncorrelated with genome size and phylogenetic position of the species36, but will provide important information for further studies of phylogenetic reconstruction and infra- and inter-specific genetic diversity54,55.
Phylogenetic analyses
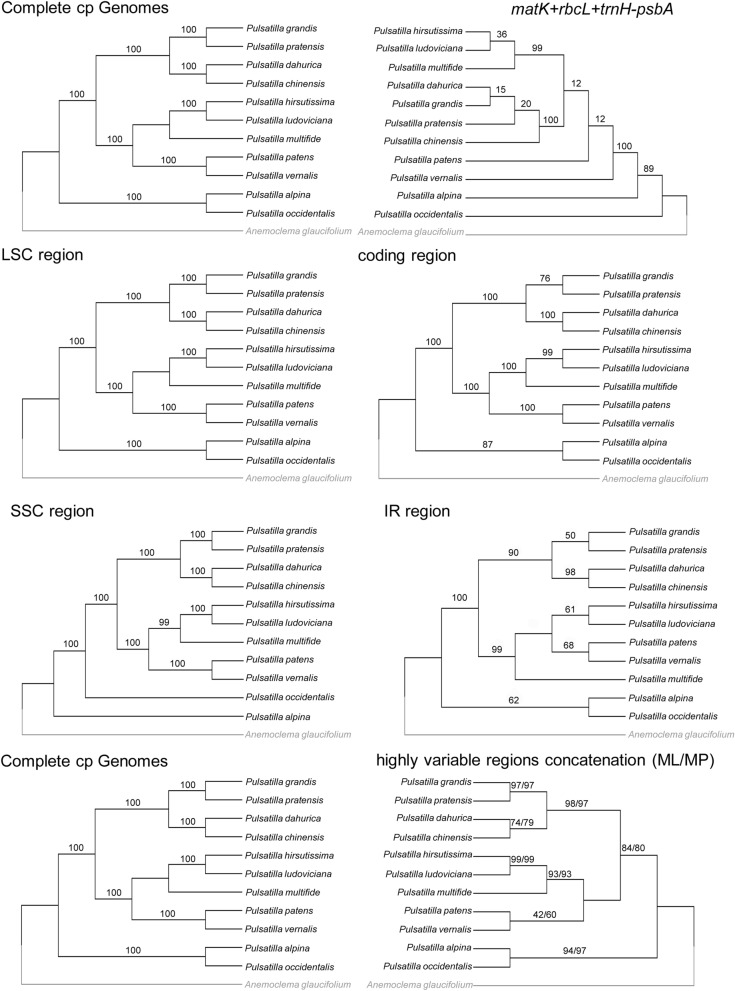
Chloroplast genomes have been widely used and have made significant contributions to phylogeny reconstruction at different taxonomic levels in plants7,9,24,35. To better clarify the evolutionary relationships within Pulsatilla, we used each data set to construct phylogenetic trees using the ML analytical methods. We also construct phylogenetic trees with those eight highly variable regions using the ML, MP analytical methods. All tree topology structures were identical. Therefore, here we presented the phylogenetic studies using the ML tree with the support values from the MP analyses recorded at the corresponding nodes (Fig. 6). The phylogenetic tree based on all data sets (except the IR and SSC regions) from the complete plastid genome sequences yielded the same topology. The phylogenetic tree based on chloroplast genome differed from that of the DNA barcode combination rbcL + matK + trnH-psbA, but with higher support values. The phylogenetic trees based on data from complete plastid genome sequences showed that the species of Pulsatilla formed a monophyletic group which in turn includes two strongly supported (bootstrap = 100) clades. One clade comprised P. alpina and P. occidentalis, members of subg. Preonanthus. The other comprised two subclades: (1) members of P. hirsutissima, P. ludoviciana, P. multifidi, P. patens and P. vernalis, and (2) species of P. chinensis, P. dahurica, P. grandis and P. pratensis. All the species of the two subclades are members of the subg. Pulsatilla. These results were congruent with our former results based on universal markers15.
Figure 6.
Phylogenetic relationships of the eleven Pulsatilla species inferred from maximum likelihood (ML). Including whole chloroplast genome, rbcL + matK + trnH-psbA, LSC region, coding region, SSC region, IR region, and the concatenation of the eight highly variable regions mentioned in Table 4 (Numbers above nodes are support values with ML bootstrap values on the left, and MP bootstrap values on the right).
In phylogenetic analyses, compared to the combination of barcodes, the full chloroplast genome sequence data formed distinct clades with high bootstrap support, improving the inadequate resolution of barcodes combination. The LSC regions and coding regions have the same topology structures with robust support. However, sequencing of genomic DNA is still expensive. It is necessary to utilize variation within chloroplast regions for rapid species-specific assay5,9,31,33,42. Here we found that phylogenetic inference based on highly variable regions yielded a tree with the same topology as the one recovered based on complete chloroplast genome sequences, demonstrating the high utility of hotspots of variability for species identification and phylogenetic analysis. More samples and laboratory works are needed in the future to increase the number of these variable regions available for study.
Conclusions
In this study, we generated complete chloroplast genomes of seven species of Pulsatilla and compared them to four previously published cp genome sequences of Pulsatilla. Chloroplast genomes of Pulsatilla share many features with those of other angiosperms. Informative differences between cp genomes of Pulsatilla were most evident in inversions in the large single copy region and expansion of the inverted repeat region. We identified multiple potentially valuable genetic markers, including large repeat sequences, numerous SSRs, and eight highly variable regions. Genetic markers have provided a reference for the improvement of plants fingerprints and the identification of similar Pulsatilla. In addition, it will be better to construct the phylogenetic relationships of Pulsatilla species using these highly variable regions and genetic markers in the future. Totally, this study provides a basis for future studies of horticultural cultivation, conservation, population genetics, phyletic evolution, development of DNA barcodes, and diverse research in Pulsatilla.
Materials and methods
Plant material, DNA extraction and sequencing
Fresh leaves of P. dahurica were collected from Jilin province of China and dried with silica gel. Dry leaves of other six Pulsatilla species were taken from herbarium specimens. We extracted total genomic DNA with the DNeasy Plant mini kits (QIAGEN, Guangzhou, China). The genomic DNA was sequenced using the Illumina Miseq platform (Illumina, San Diego, CA, USA).
Chloroplast genome assembly and annotation
Whole chloroplast genome sequencing was done for the seven species of Pulsatilla. For each species, high-quality Illumina sequencing reads were assembled into scaffolds with de novo sequence assembly software Spades, SOAPdenovo and CLC Genomics Workbench v.6.5 (CLC Bio), respectively56. We checked the contigs against the reference genome of P. chinensis (MG001341), using BLAST (https://blast.ncbi.nlm.nih.gov/) and oriented aligned contigs according to the reference genome. We mapped all the raw reads back to assembled sequences to check the assembly and then constructed the complete cp genomes using Geneious v.9.057. We submitted all the newly sequence data in raw format (fastq) and obtained SRA accesions (Table 1).
Annotations of cp genome sequences were performed using Plastid Genome Annotator (https://github.com/quxiaojian/PGA)58 and adjusted in Geneious v.9.0. To verify the accuracy of the annotations, we also used GeSeq59 to annotate each chloroplast genome in this study. We checked every boundary of tRNAs using tRNAscan-SE v.2.060. The circular genome maps were generated in OGDRAW (https://ogdraw.mpimp-golm.mpg.de/)61.
Genome comparisons
We aligned the cp genomes of Pulsatilla using multiple alignment of MAFFT v762 and manually edited in Geneious v.9.0. The contraction and expansion of inverted repeat regions were also examined among the seven species (excluded P. chinensis, MG001341) of the genus Pulsatilla using Irscope63. Then, we performed multiple alignments of the eight genomes of Pulsatilla in the mVISTA program64 under Shuffle-LAGAN mode, with default parameters for other options, using the annotation genome of P. chinensis as a reference, with the aim of comparing and visualized the similarities and differences among different plastid genomes.
To analyse chloroplast genome organisation and gene arrangement, we perform the analyses of collinear blocks with Mauve v 2.3.165 plugin in Geneious v.9.0, including only one copy of the IR, assuming collinear genomes for the full alignment. Detailed gene inversions were identified by comparing the gene order of Pulsatilla samples and Anemoclema to Berberis.
To observe the plastid genome divergence and determine parsimony informative sites, we conducted sliding window analysis after alignment to determine the nucleotide diversity (Pi) of the cp genome using DnaSP v5, with 200 bp of step size and 600 bp window length66. We defined hotspots as those regions with a higher value of Pi. We computed the variable sites across the complete cp genomes and the sequence characteristics of hotspots by DnaSP v5.0.
Repeated sequences identification
We identified repeat sequences, including palindromic, reverse and forward repeats, using the online software REPuter (https://bibiserv.cebitec.uni-bielefeld.de/reputer), with the following settings: Hamming distance of 3 and minimum repeat size of 30 bp67. We used the online program Tandem Repeats Finder (https://tandem.bu.edu/trf/trf.html) to find the tandem repeat sequences, in which the similarity percentage of two repeat copies was at least 90% and the minimal repeat size was 10 bp. The alignment parameters for match, mismatch, and indels were set at 2, 7, and 7, respectively. We identified microsatellites (SSRs) by MISA68 with thresholds of 10, 5, 4, 3, 3, and 3 for mono-, di-, tri-, tetra-, penta-, and hexa-nucleotide, respectively.
Phylogenetic analyses
For the purpose of reconstructing the phylogenetic relationships, four published complete cp genome sequences from the genus Pulsatilla and Anemoclema were also included in our analyses. The monotypic genus Anemoclema (MH205609) was selected as the outgroup. Because molecular evolutionary rates among the different cp genome regions are diverse, analyses of phylogenetic relationships were performed based on the following seven datasets: (a) the complete cp genome sequences; (b) coding genes (CDS); (c) one inverted repeat (IR) region (IRb); (d) the large single copy region (LSC); (e) the small single copy region (SSC), (f) the consensus sequences of eight highly variable regions; and (g) the DNA barcodes combination (rbcL + matK + trnH-psbA). We applied Maximum Likelihood (ML) analysis for each of the seven datasets to construct tree-sets. Maximum Parsimony (MP) analyses were also applied for the consensus sequences of eight highly variable regions and the DNA barcodes combination.
We conducted ML analyses with RAxMLHPC2 v.8.0.969 on the Cyberinfrastructure for Phylogenetic Research (CIPRES) Science Gateway v.3.370. Then the analysis of 1000 rapid bootstrap replicates (-x) was followed by a search for the best-scoring ML tree in one program (-f a). The best-fit model for nucleotide and amino acid sequences were evaluated using jModelTest 271. We applied the GTR + G model to nucleotide data for both bootstrapping and best-tree searching phases, with other parameters as the default settings. We performed the maximum Parsimony (MP) analysis on PAUP* v.4.0b1072. All the characters were treated as unordered and equally weighted. The heuristic search specified 1000 random sequence addition replicates with TBR branch swapping, saving only 10 trees per replicate. We obtained the strict consensus tree from all the most-parsimonious trees (MPTs) detected during the search. We calculated bootstrap percentages (BP) from 10,000 rapid bootstrap replicates, each comprising 10 random sequence addition replicates and saving only one tree per replicate.
Supplementary information
Acknowledgements
We sincerely thank Prof. Zhong-hu Li, Dr. Jian-jun Jin, De-long Guan and Molecular Biology Experiment Center, Germplasm Bank of Wild Species in Southwest China for help with data analysis. This project was supported by the National Nature Science Foundation of China (No. 31770200, 31300158, 31770203, 31660048) and the Fundamental Research for the Central Universities (No. 2452020179).
Author contributions
L.Zhao designed the research. Q.J.L., L.Zhang, X.H.Z, Z.Y.C., and L.Zhao. collected the samples, Q.J.L., N.S., L.Zhang, R.C.T., and J.R.W. conceived the experiments, Q.J.L., N.S., L.Z., R.C.T. and J.R.W. did computational analysis and deposited sequences. Q.J.L., N.S., L.Zhang, X.H.Z., Z.Y.C., L.Zhao and D.P. wrote the manuscript. All authors have read and approved the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Qiu-jie Li and Na Su.
Supplementary information
is available for this paper at 10.1038/s41598-020-76699-7.
References
- 1.Hebert PDN, Cywinska A, Ball SL, Waard JR. Biological identifications through DNA barcodes. Proc. R. Soc. Lond. B Biol. Sci. 2003;270:313–321. doi: 10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kress WJ, Wurdack KJ, Zimmer EA, Weigt LA, Janzen DH. Use of DNA barcodes to identify flowering plants. Proc. Natl. Acad. Sci. U.S.A. 2005;102:8369–8374. doi: 10.1073/pnas.0503123102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kress WJ, et al. Plant DNA barcodes and a community phylogeny of a tropical forest dynamics plot in Panama. Proc. Natl. Acad. Sci. U.S.A. 2009;106:18621–18626. doi: 10.1073/pnas.0909820106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.CBOL Plant Working Group A DNA barcode for land plants. Proc. Natl. Acad. Sci. U.S.A. 2009;106:12794–12797. doi: 10.1073/pnas.0905845106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li XW, et al. Plant DNA barcoding: from gene to genome. Biol. Rev. Camb. Philos. Soc. 2015;90:157–166. doi: 10.1111/brv.12104. [DOI] [PubMed] [Google Scholar]
- 6.Hollingsworth PM, Graham SW, Little DP. Choosing and using a plant DNA barcode. PLoS ONE. 2011;6:e19254. doi: 10.1371/journal.pone.0019254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ahmed I, et al. Identification of chloroplast genome loci suitable for highresolution phylogeographic studies of Colocasiaesculenta (L.) Schott (Araceae) and closely related taxa. Mol. Ecol. Resour. 2013;13:929–937. doi: 10.1111/1755-0998.12128. [DOI] [PubMed] [Google Scholar]
- 8.Kuang DY, et al. Complete chloroplast genome sequence of Magnolia kwangsiensis (Magnoliaceae): implication for DNA barcoding and population genetics. Genome. 2011;54:663–673. doi: 10.1139/g11-026. [DOI] [PubMed] [Google Scholar]
- 9.Dong W, Liu J, Yu J, Wang L, Zhou S. Highly variable chloroplast markers for evaluating plant phylogeny at low taxonomic levels and for DNA barcoding. PLoS ONE. 2012;7:e35071. doi: 10.1371/journal.pone.0035071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tamura, M. Ranunculaceae. in Die natürlichen pflanzenfamilien, Aufl. II. 2nd ed. 17a IV (ed Hiepko, P.) (Duncker und Humblot, Berlin, 1995).
- 11.Wang, W. C. et al. Flora of China Vol. 6, 133–438 (Science Press, Beijing; Missouri Botanical Garden Press, St. Louis, 2001).
- 12.Grey-Wilson C. Pasque-flowers. The Genus Pulsatilla. Kenning Hall: . The Charlotte Louise Press; 2014. [Google Scholar]
- 13.Ren Y, Gu TQ, Chang HL. Floral development of Dichocarpum, Thalictrum, and Aquilegia (Thalictroideae, Ranunculaceae) Plant Syst. Evol. 2011;292:203–213. doi: 10.1007/s00606-010-0399-6. [DOI] [Google Scholar]
- 14.Ren Y, Chang HL, Endress PK. Floral development in Anemoneae (Ranunculaceae) Bot. J. Linn. Soc. 2015;162:77–100. doi: 10.1111/j.1095-8339.2009.01017.x. [DOI] [Google Scholar]
- 15.Li QJ, et al. Efficient identification of Pulsatilla (Ranunculaceae) using DNA barcodes and micro-morphological characters. Front. Plant Sci. 2019;10:1196. doi: 10.3389/fpls.2019.01196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.China Pharmacopoeia Committee . Pharmacopoeia of the People’s Republic of China. Beijing: Chemical Industry Press; 2015. [Google Scholar]
- 17.Xu Q, et al. Antitumor activity of Pulsatilla chinensis (Bunge) Regel saponins in human liver tumor 7402 cells in vitro and in vivo. Phytomedicine. 2012;19:293–300. doi: 10.1016/j.phymed.2011.08.066. [DOI] [PubMed] [Google Scholar]
- 18.Wang XW, Fan FG, Cao Q. Modified Pulsatilla decoction attenuates oxazolone-induced colitis in mice through suppression of inflammation and epithelial barrier disruption. Mol. Med. Rep. 2016;14:1173–1179. doi: 10.3892/mmr.2016.5358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suh SY, An WG. Systems pharmacological approach of Pulsatillae radix on treating Crohn's disease. Evid. Based Complement. Altern. Med. 2017;2017:1–21. doi: 10.1155/2017/4198035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szczecińska M, Sawicki J. Genomic resources of three Pulsatilla species reveal evolutionary hotspots, species-specific sites and variable plastid structure in the family Ranunculaceae. Int. J. Mol. Sci. 2015;16:22258–22279. doi: 10.3390/ijms160922258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoot SB, Reznicek AA, Palmer JD. Phylogenetic relationship in Anemone (Ranunculaceae) based on morphology and chloroplast DNA. Syst. Bot. 1994;19:169–200. doi: 10.2307/2419720. [DOI] [Google Scholar]
- 22.Hoot SB. Phylogenetic relationships in Anemone (Ranunculaceae) based on DNA restriction site variation and morphology. In: Jensen U, Kadereit JW, editors. Systematics and Evolution of the Ranunculiflorae. Vienn: Springer; 1995. pp. 295–300. [Google Scholar]
- 23.Szczecińska M, Gabor S, Katarzyna W, Jakub S, Dusan G. Genetic diversity and population structure of the rare and endangered plant species Pulsatillapatens (L.) Mill in east central Europe. PLoS ONE. 2016;11:e0151730. doi: 10.1371/journal.pone.0151730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang N, et al. Phylogenetic reassessment of tribe Anemoneae (Ranunculaceae): non-monophyly of Anemone s.l. revealed by plastid datasets. PLoS ONE. 2017;12:e0174792. doi: 10.1371/journal.pone.0174792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sramkó G, Laczkó L, Volkova PA, Bateman RM, Mlinarec J. Evolutionary history of the Pasque-flowers (Pulsatilla, Ranunculaceae): molecular phylogenetics, systematics and rDNA evolution. Mol. Phylogenet. Evol. 2019;135:45–61. doi: 10.1016/j.ympev.2019.02.015. [DOI] [PubMed] [Google Scholar]
- 26.Yu XQ, Yang D, Guo C, Gao LM. Plant phylogenomics based on genome-partitioning strategies: progress and prospects. Plant Divers. 2018;40:158–164. doi: 10.1016/j.pld.2018.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang Y, Yukawa T, Bateman RM, Jiang H, Peng H. Phylogeny and classification of the East Asian Amitostigma alliance (Orchidaceae: Orchideae) based on six DNA markers. BMC Evol. Biol. 2015;15:96. doi: 10.1186/s12862-015-0376-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henriquez CL, et al. Molecular evolution of chloroplast genomes in Monsteroideae (Araceae) Planta. 2020;251:72. doi: 10.1007/s00425-020-03365-7. [DOI] [PubMed] [Google Scholar]
- 29.Abdullah, et al. Correlations among oligonucleotide repeats, nucleotide substitutions, and insertion–deletion mutations in chloroplast genomes of plant family Malvaceae. J. Syst. Evol.10.1111/jse.12585 (2020).
- 30.Abdullah, et al. Chloroplast genome of Hibiscus rosa-sinensis (Malvaceae): comparative analyses and identification of mutational hotspots. Genomics112, 581–591 (2020). [DOI] [PubMed]
- 31.Liu E, et al. Comparative analysis of complete chloroplast genome sequences of four major Amorphophallus species. Sci. Rep. 2019;9:809. doi: 10.1038/s41598-018-37456-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chumley, T. W. et al. The complete chloroplast genome sequence of Pelargonium × hortorum: organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Mol. Biol. Evol.23, 2175–2190. [DOI] [PubMed]
- 33.Ogihara Y, et al. Structural features of a wheat plastome as revealed by complete sequencing of chloroplast DNA. Mol. Genet. Genomics. 2002;266:740–746. doi: 10.1007/s00438-001-0606-9. [DOI] [PubMed] [Google Scholar]
- 34.Raman G, Park S. The complete chloroplast genome sequence of Ampelopsis: gene organization, comparative analysis, and phylogenetic relationships to other angiosperms. Front. Plant Sci. 2016;7:341. doi: 10.3389/fpls.2016.00341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Daniell H, Lin CS, Yu M, Chang WJ. Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol. 2016;17:134. doi: 10.1186/s13059-016-1004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Henriquez CL, et al. Evolutionary dynamics in chloroplast genomes of subfamily Aroideae (Araceae) Genomics. 2020;112:2349–2360. doi: 10.1016/j.ygeno.2020.01.006. [DOI] [PubMed] [Google Scholar]
- 37.Wang W, Messing J. High-throughput sequencing of three Lemnoideae (duckweeds) chloroplast genomes from total DNA. PLoS ONE. 2011;6:e24670. doi: 10.1371/journal.pone.0024670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adbulch, et al. Complete chloroplast genomes of Anthurium huixtlense and Pothos scandens (Pothoideae, Araceae): unique inverted repeat expansion and contraction affect rate of evolution. J. Mol. Evol. 2020;88:562–574. doi: 10.1007/s00239-020-09958-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oldenburg DJ, Bendich AJ. The linear plastid chromosomes of maize: terminal sequences, structures, and implications for DNA replication. Curr. Genet. 2016;62:431–442. doi: 10.1007/s00294-015-0548-0. [DOI] [PubMed] [Google Scholar]
- 40.Sun JH, et al. Evolutionary and phylogenetic aspects of the chloroplast genome of Chaenomeles species. Sci. Rep. 2020;10:11466. doi: 10.1038/s41598-020-67943-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu HJ, et al. Comparative analysis of complete chloroplast genomes of Anemoclema, Anemone, Pulsatilla, and Hepatica revealing structural variations among genera in tribe Anemoneae (Ranunculaceae) Front. Plant Sci. 2018;9:1097. doi: 10.3389/fpls.2018.01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meng J, et al. Comparative analysis of the complete chloroplast genomes of four Aconitum medicinal species. Molecules. 2018;23:1015. doi: 10.3390/molecules23051015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Du YP, et al. Complete chloroplast genome sequences of Lilium: insights into evolutionary dynamics and phylogenetic analyses. Sci. Rep. 2017;7:5751. doi: 10.1038/s41598-017-06210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bi Y, Zhang MF, Xue J, Dong R, Du YP, Zhang XH. Chloroplast genomic resources for phylogeny and DNA barcoding: a case study on Fritillaria. Sci. Rep. 2018;8:1184. doi: 10.1038/s41598-018-19591-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Choi KS, Chung MG, Park S. The complete chloroplast genome sequences of three Veroniceae species (Plantaginaceae): comparative analysis and highly divergent regions. Front. Plant Sci. 2016;7:1–8. doi: 10.3389/fpls.2016.00355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Menezes APA, et al. Chloroplast genomes of Byrsonima species (Malpighiaceae): Comparative analysis and screening of high divergence sequences. Sci. Rep. 2018;8:1–12. doi: 10.1038/s41598-017-17765-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Y, Zhang Z, Yang J, Lv G. Complete chloroplast genome of seven Fritillaria species, variable DNA markers identification and phylogenetic relationships within the genus. PLoS ONE. 2018;13:e0194613. doi: 10.1371/journal.pone.0194613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grassi F, Labra M, Scienza A, Imazio S. Chloroplast SSR markers to assess DNA diversity in wild and cultivated grapevines. Vitis. 2002;41:157–158. [Google Scholar]
- 49.Chen C, Zhou P, Choi YA, Huang S, Gmitter FG., Jr Mining and characterizing microsatellites from citrus ESTs. Theor. Appl. Genet. 2006;112:1248–1257. doi: 10.1007/s00122-006-0226-1. [DOI] [PubMed] [Google Scholar]
- 50.Powell W, Morgante M, McDevitt R, Vendramin GG, Rafalski JA. Polymorphic simple sequence repeat regions in chloroplast genomes: applications to the population genetics of pines. Proc. Natl. Acad. Sci. U.S.A. 1995;92:7759–7763. doi: 10.1073/pnas.92.17.7759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kaundun SS, Matsumoto S. Heterologous nuclear and chloroplast microsatellite amplification and variation in tea, Camellia sinensis. Genome. 2002;45:1041–1048. doi: 10.1139/g02-070. [DOI] [PubMed] [Google Scholar]
- 52.Doorduin L, et al. The complete chloroplast genome of 17 individuals of pest species Jacobaea vulgaris: SNPs, microsatellites and barcoding markers for population and phylogenetic studies. DNA Res. 2011;18:93–105. doi: 10.1093/dnares/dsr002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jiao Y, et al. Development of simple sequence repeat (SSR) markers from a genome survey of Chinese bayberry (Myrica rubra) BMC Genomics. 2012;13:201. doi: 10.1186/1471-2164-13-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.He S, Wang Y, Volis S, Li D, Yi T. Genetic diversity and population structure: implications for conservation of wild soybean (Glycinesoja Sieb. et Zucc) based on nuclear and chloroplast microsatellite variation. Int. J. Mol. Sci. 2012;13:12608–12628. doi: 10.3390/ijms131012608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang N, et al. An analysis of Echinacea chloroplast genomes: implications for future botanical identification. Sci. Rep. 2017;7:216. doi: 10.1038/s41598-017-00321-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li HT, et al. Origin of angiosperms and the puzzle of the Jurassic gap. Nat. Plants. 2019;5:461–470. doi: 10.1038/s41477-019-0421-0. [DOI] [PubMed] [Google Scholar]
- 57.Kearse M, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Qu XJ, Moore MJ, Li DZ, Yi TS. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 2019;15:12. doi: 10.1186/s13007-019-0435-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tillich M, et al. GeSeq-versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017;45:W6–W11. doi: 10.1093/nar/gkx391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lowe TM, Chen PP. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016;44:W54–W57. doi: 10.1093/nar/gkw413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lohse M, Drechsel O, Bock R. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr. Genet. 2007;52:267–274. doi: 10.1007/s00294-007-0161-y. [DOI] [PubMed] [Google Scholar]
- 62.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Amiryousefi A, Hyvönen J, Poczai P. IRscope: an online program to visualize the junction sites of chloroplast genomes. Bioinformatics. 2018;34:3030–3031. doi: 10.1093/bioinformatics/bty220. [DOI] [PubMed] [Google Scholar]
- 64.Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids. Res. 2004;32:W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Darling AC, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 67.Kurtz S, et al. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids. Res. 2001;29:4633–4642. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thiel T, Michalek W, Varshney R, Graner A. Exploiting EST databases for the development and characterization of gene-derived SSR markers in barley (Hordeumvulgare L.) Theor. Appl. Genet. 2003;106:411–422. doi: 10.1007/s00122-002-1031-0. [DOI] [PubMed] [Google Scholar]
- 69.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Miller, M. A., Pfeiffer, W. & Schwartz, T. Creating the CIPRES science gateway for inference of large phylogenetic trees. New Orleans, LA. Proceedings of the Gateway Computing Environments Workshop (GCE), pp. 1–8 (2010).
- 71.Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Swofford, D. L. PAUP*: phylogenetic analysis using Parsimony (*and other methods), version 4.0b10 (Sinauer Associates, Sunderland, 2003).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.