We read with great interest the article by Henry et al. [1] showed that elevated lactate dehydrogenase (LDH) values were associated with 6-fold increased odds of severe COVID-19 disease. Lactate dehydrogenase increases in the early stage of myocardial infarction as well as in states of hemolysis. It is most active in the liver, striated muscles, heart, kidneys, lungs, brain, and red blood cells (erythrocytes). In the case of cell damage, lactate dehydrogenase is released from inside them, its concentration and activity in the blood increase. High serum LDH activity is a negative prognostic factor in such patients. LDH is a marker of various inflammatory states, e.g., infections, malignancies, MI, sepsis, or cardio-pulmonary compromise. Denese et al. showed that lactate dehydrogenase is a potential marker of vascular permeability in immune-mediated lung injury [2]. Early data Henry et al. reported in COVID-19 patients have suggested significant differences in LDH levels between patients and without the severe disease [3].
A systematic review and meta-analysis were performed to verify the usefulness of using lactate dehydrogenase as a predictor of a patient's severity with COVID-19.
Two authors (M.P. and L.S.) searched electronic resources (Medline, EMBASE, and the Cochrane Central register from databases inception to 9 November 2020). A review of the bibliographies of the relevant articles was also performed. The retrieved articles were screened for relevance on title and abstract, followed by two independent investigators (L.S. and J.S.). The key search words were: „lactate dehydrogenase” OR „LDH” AND „COVID-19” OR „SARS-CoV-2″.
All statistical analyses were performed with Review Manager Software 5.4 (The Cochrane Collaboration, Oxford, Copenhagen, Denmark). All results are presented with their 95% confidence interval (CI). When the continuous outcome was reported in a study as median, range, and interquartile range, we estimated means and standard deviations using the formula described by Hozo et al. [4]. The random-effects model was used for I2 > 50%. P < 0.05 was taken to show statistical significance. Statistical testing was two-tailed.
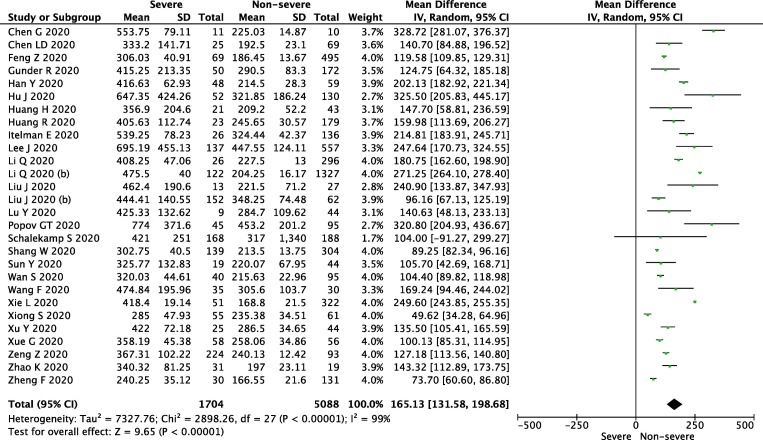
Twenty-eight studies reported LDH levels in severe vs. non-sever groups. The level of LDH in the individual groups varied (MD = 154.49; 95% CI: 121.24, 191.73; P < 0.001, I2 = 99%; Fig. 1 ). A statistically significant higher level of LDH was also observed in terms of ICU vs. Non-ICU (MD = 272.98; 95% CI: 195.46, 350.51; p < 0.001; I2 = 99%), patients and in nonsurvival patients vs. survival patients (MD = 259.21; 95% CI: 166.91, 351.51; p < 0.001, I2 = 100%). Supplementary Digital Content, SDC). The full list of publications included in this meta-analysis is presented in SDC.
Fig. 1.
Forest plot of lactate dehydrogenase level in sever vs. non-sever group. The center of each square represents the weighted mean difference for individual trials, and the corresponding horizontal line stands for a 95% confidence interval. The diamonds represent pooled results.
In conclusion, the current meta-analysis confirmed that lactate dehydrogenase level can be used as a COVID-19 severity marker and is a predictor of survival.
Declaration of Competing Interest
Authors don't declare any conflict of interest.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ajem.2020.11.025.
Appendix A. Supplementary data
References
- 1.Henry B.M., Aggarwal G., Wong J., et al. Lactate dehydrogenase levels predict coronavirus disease 2019 (COVID-19) severity and mortality: a pooled analysis. Am J Emerg Med. 2020 Sep;38(9):1722–1726. doi: 10.1016/j.ajem.2020.05.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Danese E., Montagnana M. An historical approach to the diagnostic biomarkers of acute coronary syndrome. Annals of translational medicine. 2016;4(10):194–196. doi: 10.21037/atm.2016.05.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Henry B., De Olivera M.H.S., S. B, M. P, G L Hematologic, biochemical and immune marker abnormalities associated with severe illness and mortality in corona virus disease 2019 (COVID 19): a meta-analysis. Clin Chem Lab Med. 2020;58(7):1021–1028. doi: 10.1515/cclm-2020-0369. [DOI] [PubMed] [Google Scholar]
- 4.Hozo S.P., Djulbegovic B., Hozo I. Estimating the mean and variance from the median, range, and the size of a sample. BMC Med Res Methodol. 2005;5 doi: 10.1186/1471-2288-5-13. 13, indexed in Pubmed: 15840177. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.