Recent successes in malaria control have been seriously threatened by the emergence of Plasmodium falciparum parasite resistance to the frontline artemisinin drugs in Southeast Asia. P. falciparum artemisinin resistance is associated with mutations in the parasite K13 protein, which associates with a delay in the time required to clear the parasites upon drug treatment. Gene editing technologies have been used to validate the role of several candidate K13 mutations in mediating P. falciparum artemisinin resistance in vitro under laboratory conditions. Nonetheless, the causal role of these mutations under in vivo conditions has been a matter of debate. Here, we have used CRISPR/Cas9 gene editing to introduce K13 mutations associated with artemisinin resistance into the related rodent-infecting parasite, Plasmodium berghei. Phenotyping of these P. berghei K13 mutant parasites provides evidence of their role in mediating artemisinin resistance in vivo, which supports in vitro artemisinin resistance observations. However, we were unable to introduce some of the P. falciparum K13 mutations (C580Y and I543T) into the corresponding amino acid residues, while other introduced mutations (M476I and R539T equivalents) carried pronounced fitness costs. Our study provides evidence of a clear causal role of K13 mutations in modulating susceptibility to artemisinins in vitro and in vivo using the well-characterized P. berghei model. We also show that inhibition of the P. berghei proteasome offsets parasite resistance to artemisinins in these mutant lines.
KEYWORDS: malaria, Plasmodium berghei, Plasmodium falciparum, artemisinin resistance, K13, gene editing, ring-stage survival assays, parasite clearance times, proteasome, synergy
ABSTRACT
The recent emergence of Plasmodium falciparum parasite resistance to the first line antimalarial drug artemisinin is of particular concern. Artemisinin resistance is primarily driven by mutations in the P. falciparum K13 protein, which enhance survival of early ring-stage parasites treated with the artemisinin active metabolite dihydroartemisinin in vitro and associate with delayed parasite clearance in vivo. However, association of K13 mutations with in vivo artemisinin resistance has been problematic due to the absence of a tractable model. Herein, we have employed CRISPR/Cas9 genome editing to engineer selected orthologous P. falciparum K13 mutations into the K13 gene of an artemisinin-sensitive Plasmodium berghei rodent model of malaria. Introduction of the orthologous P. falciparum K13 F446I, M476I, Y493H, and R539T mutations into P. berghei K13 yielded gene-edited parasites with reduced susceptibility to dihydroartemisinin in the standard 24-h in vitro assay and increased survival in an adapted in vitro ring-stage survival assay. Mutant P. berghei K13 parasites also displayed delayed clearance in vivo upon treatment with artesunate and achieved faster recrudescence upon treatment with artemisinin. Orthologous C580Y and I543T mutations could not be introduced into P. berghei, while the equivalents of the M476I and R539T mutations resulted in significant growth defects. Furthermore, a Plasmodium-selective proteasome inhibitor strongly synergized dihydroartemisinin action in these P. berghei K13 mutant lines, providing further evidence that the proteasome can be targeted to overcome artemisinin resistance. Taken together, our findings provide clear experimental evidence for the involvement of K13 polymorphisms in mediating susceptibility to artemisinins in vitro and, most importantly, under in vivo conditions.
INTRODUCTION
Artemisinin (ART)-based combination therapies (ACTs) have been at the forefront of globally coordinated efforts to drive down the burden of malaria. A pharmacodynamic hallmark of ARTs and their derivatives is that they are highly active and fast acting against blood stages of malaria parasites. These drugs can achieve up to 10,000-fold parasite reductions in the first replication cycle upon drug exposure (1). Such is the effectiveness of ARTs that recently reported reductions in malaria morbidity and mortality are, indeed, partly attributed to ACTs (2). The use of ARTs in combination therapies originated from early clinical trials, which showed that despite achieving faster parasite clearance, ART monotherapies resulted in recrudescence rates of up to 40% (3). ACTs deliver a pharmacological cure by taking advantage of ARTs to rapidly clear the parasite biomass in the early days of treatment while relying on the partner drug to eliminate residual parasites (4). So far, ACTs remain highly effective in Sub-Saharan Africa, the region that harbors the highest disease burden, with efficacy rates of >98% (2). Nevertheless, ACTs have been threatened by the emergence of Plasmodium falciparum resistance to ARTs in Southeast Asia, and resistance has the potential to spread to other regions of malaria endemicity, as has been a historical trend with earlier first-line antimalarial drugs (2, 5–7). Recently, locally derived K13 variants that are able to mediate ART resistance in vitro have been identified in P. falciparum parasites in French Guiana and in Rwanda (8, 9), further illustrating the emergent threat to ART efficacy. Moreover, an aggressive expansion of a parasite lineage carrying the genetic determinants of resistance to both ART derivatives and the ACT partner drug piperaquine has been reported across Southeast Asia, resulting in a dramatic loss of clinical efficacy (10–13).
Clinically, P. falciparum resistance to ARTs manifests as reduced in vivo parasite clearance upon treatment with ACTs or ART monotherapies (2, 14, 15). These clearance rates are based on the Worldwide Antimalarial Resistance Network (WWARN) parasite clearance estimator (16), which quantifies relative resistance by estimating parasitemia lag phases and clearance half-lives upon treatment with artesunate (AS) or ACTs. This involves in vivo quantification of viable parasitemia (in patients) upon treatment with AS (2 to 4 mg/kg body weight/day) or ACTs at specified time intervals and subsequent calculation of parasite densities as a function of time (16). The parasite clearance estimator has been used to generate substantial baseline data that classify ART resistance as parasite clearance half-lives of >5.5 h and ART sensitivity as parasite clearance half-lives of <3 h (17, 18). However, interpretation of clearance half-lives can be confounded by differences in initial parasite biomass, the efficacy of the partner drug, and the level of host immunity (17, 19). Moreover, this in vivo phenotype does not correlate with decreased susceptibility to dihydroartemisinin (DHA) in standard growth inhibition assays where P. falciparum parasites (which have a ∼48-h intraerythrocytic developmental cycle) are exposed to the drug for a total of 72 h (15, 20, 21). The ring-stage survival assay (RSA), where highly synchronized early-ring-stage parasites (0 to 3 h postinvasion) are exposed for a short period of time (3 to 6 h) to DHA (at the pharmacologically relevant concentration of 700 nM), provides an improved correlate for the in vivo delayed parasite clearance phenotype and has been the principal in vitro assay for determining P. falciparum resistance to ARTs (22, 23). At the genetic level, polymorphisms in the P. falciparum K13 propeller domain have been strongly associated with ACT treatment failure (21, 24) and also correlate with delayed parasite clearance in vivo and increased parasite survival in vitro in RSAs (25–27). Reverse genetic approaches have been successfully used to show that the P. falciparum K13 mutations M476I, R539T, I543T, Y493H, and C580Y can confer DHA resistance in vitro, as defined by >1% survival in RSAs (28, 29). However, the parasite genetic background as well as underlying polymorphisms in drug resistance determinants such as pfcrt (P. falciparum chloroquine resistance transporter) and pfmdr2 (P. falciparum multidrug resistance protein-2) may play a role either by modulating different levels of susceptibility to DHA or by providing a suitable biological landscape upon which these K13 mutations are more likely to arise (25, 28).
ART resistance as typified by the “delayed clearance phenotype” is, however, still classified as “partial resistance,” primarily because most patients with parasites harboring the phenotype effectively clear the infection when an effective partner drug is used or duration of monotherapy is extended (4). ART partial resistance is, therefore, confirmed or suspected when patients carry parasites with certain K13 mutations, display a parasite clearance half-life of >5.5 h, or are microscopically smear positive on day three after initiation of treatment (2, 4). The full extent to which these parameters predict subsequent ACT treatment failure or define ART resistance remains an area of continuing debate (30–35). The definition of ART resistance in these contexts would thus benefit from experimentally accessible in vivo models that would help interrogate ART parasite susceptibility parameters, including clearance half-lives, recrudescence rates, and treatment failures. Such models would allow for a genetic dissection of the role of K13 mutations in mediating resistance in vivo in the absence of confounding factors such as secondary genetic factors and/or host factors (25, 28). Currently, the K13 C580Y polymorphism is the most prevalent and dominant ART-resistant mutation in Southeast Asia (14, 36). A recent genetic cross of the K13 C580Y ART-resistant line with an Aotus monkey-infecting P. falciparum strain provided evidence, in this nonhuman primate model, that parasites carrying the C580Y mutation can display increased survival in in vitro RSAs with no accompanying in vivo ART resistance (37).
Moreover, P. falciparum drug resistance mutations are known to often associate with significant fitness costs that limit the prevalence and eventual propagation of resistance-conferring alleles in natural infections. For example, mutations in the P. falciparum chloroquine (CQ) resistance transporter (pfcrt) that modulate resistance to CQ massively expanded when CQ was in use in the 1970s but eventually were outcompeted and replaced with parasites carrying wild-type alleles in African high-transmission settings following withdrawal of CQ use (38, 39). Similarly, P. falciparum K13 mutations have been shown to carry in vitro fitness costs; however, the degree to which a given mutation is detrimental for growth seems to depend on the parasite genetic background (40). Relative to other K13 mutations, P. falciparum R539T and I543T mutant parasites that are associated with the highest RSA survival rates (23, 28) and most significant delays in parasite clearance (41) also carried the most pronounced fitness costs (40). Intriguingly, the most prevalent K13 mutation in Southeast Asia, C580Y, was fitness neutral in vitro when gene edited into recent Cambodian clinical isolates, whereas it displayed a significant growth defect when introduced into ART-susceptible parasites isolated before ARTs were widely deployed (40, 42). Recently, it was demonstrated that P. falciparum K13 localizes to the parasite cytostomes and other intracellular vesicles and plays a role in parasite hemoglobin endocytosis and trafficking to the lysosome-like digestive vacuole (43–45). K13 mutations are thought to lead to a partial loss of protein function, which subsequently impairs hemoglobin endocytic uptake, thereby lessening ART activation and conferring ART resistance (43). This has pointed toward a K13-mediated hemoglobin-centric mechanism of ART resistance, which could possibly be shared with other drugs such as CQ that act by binding to heme moieties in the digestive vacuole, following cytostome-mediated hemoglobin endocytosis (44, 46–48). Of note, mutant K13-mediated ART resistance phenotypes are associated with upregulated cellular stress responses, which can be targeted by selective inhibition of the parasite 26S proteasome (49, 50).
Here, we report the in vitro and in vivo phenotypes of orthologous P. falciparum K13 mutations that were gene edited into an in vivo rodent model of malaria, Plasmodium berghei. We profiled the fitness of these P. berghei K13 mutant parasites relative to their isogenic wild-type counterparts as well as their sensitivity to combinations of DHA and proteasome inhibitors. Our data provide evidence that K13 mutations are causal for reduced susceptibility to ARTs in an in vivo model and link these mutations to in vitro and ex vivo phenotypes. Our findings also demonstrate that inhibition of the Plasmodium proteasome is an effective strategy to restore ART action in resistant parasites that survive treatment with ART alone.
RESULTS
CRISPR/Cas9-mediated introduction of P. berghei orthologous K13 mutations and in vivo mutant enrichment by AS.
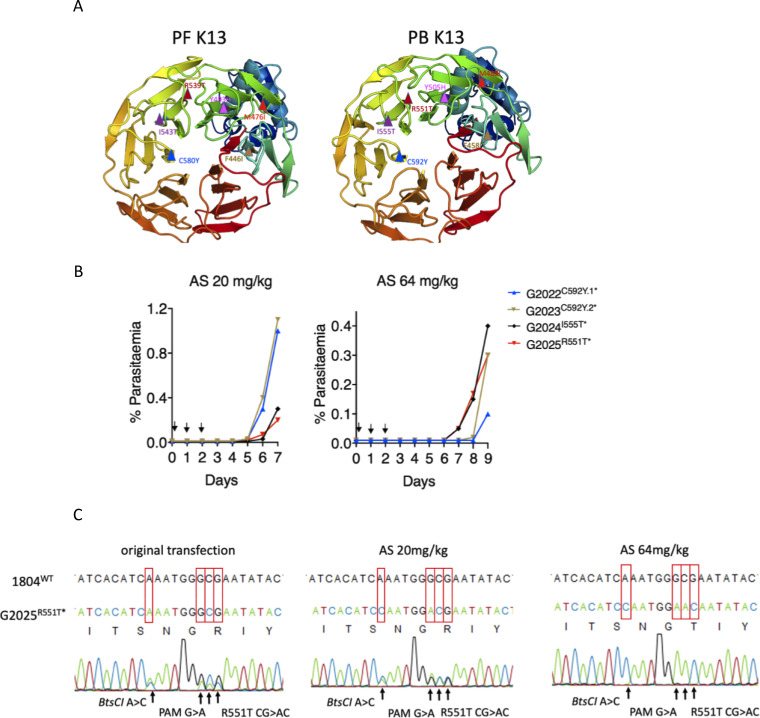
To generate P. berghei mutant parasites carrying orthologous P. falciparum K13 mutations, we attempted to introduce P. berghei equivalents of five P. falciparum K13 mutations (M476I, Y493H, R539T, I543T, and C580Y) that by reverse genetics were previously shown to confer enhanced P. falciparum survival in in vitro RSAs (28). We also introduced the equivalent of the F446I mutation that is predominant in Southern China along the Myanmar border (14). These mutations are all validated determinants of reduced P. falciparum susceptibility to ARTs (4). Structural homology modeling revealed that P. berghei and P. falciparum K13 (PBANKA_1356700 and PF3D7_1343700, respectively) are highly conserved (∼84% sequence identity overall) at the C-terminal propeller domain, especially where resistance-conferring mutations localize (Fig. 1A). P. berghei K13 carries 12 extra amino acids, resulting in 738 amino acids for P. berghei compared to 726 for P. falciparum. However, modeling suggests that the extra amino acids in P. berghei do not change the overall propeller structure of K13 or the amino acid identity at the orthologous positions of the mutations examined in this study (Fig. 1A; see also Fig. S1A and B in the supplemental material). Using a CRISPR/Cas9 system (Fig. S2A) (46), we designed Cas9 plasmids carrying single guide RNAs (sgRNAs) to target the P. berghei K13 locus with corresponding homology repair templates. The repair templates carried the mutations of interest as well as silent mutations that inactivated the protospacer adjacent motif (PAM) and introduced restriction sites for restriction fragment length polymorphism (RFLP) analyses (see Table S1). Electroporation of the plasmids pG1004 (C592Y), pG1005 (I555T), and pG1006 (R551T) into the K13 wild-type P. berghei 1804cl1 line yielded edited parasites (G2022C592Y.1*, G2023C592Y.2*, G2024I555T* and G2025R551T*) with calculated 13.4%, 18.5%, 7.7%, and 30.0% efficiencies, respectively, by RFLP analysis (see Fig. S2B; Table S1). Intriguingly, bulk DNA sequencing of these transformed parasites revealed that only the G2025R551T* line carried sequence traces for the R551T amino acid substitution and accompanying silent mutations (Fig. S3A), while the rest had traces only of the silent mutations (Fig. S3B and C). Our prior studies with refractory mutations have also revealed the parasite’s ability to restrict CRISPR/Cas9-mediated double-stranded break repair to the region immediately proximal to the cut site, thereby capturing the silent mutations without extending to nearby deleterious single nucleotide polymorphisms (SNPs) (46). We suspect this is a consequence of very short resection events (51). These data suggested that the C592Y and I555T mutations either result in extremely slow growing parasites or are entirely lethal in P. berghei. We attempted to clone the G2025R551T* line by limiting dilution, but this could not be achieved, possibly due to the low mutant population (30.0%) combined with a potentially low growth rate of the mutants compared to that of wild-type parasites.
FIG 1.
Introduction of orthologous K13 nucleotide substitutions in P. berghei. (A) Three-dimensional homology model of P. falciparum (PF3D7_1343700) and P. berghei (PBANKA_1356700) K13 for amino acid residues 350 to 726 and 362 to 738, respectively. P. falciparum K13 mutation sites (F446I, M476I, Y493H, R539T, I543T, and C592Y) are indicated in the structure on the left, and P. berghei orthologous mutation sites are modeled on the right. Models were created in SWISS-MODEL using PDB template 4zgc.1.A. Structures were visualized and annotated using PyMOL 2.3. (B) Parasitemia growth curves monitoring recrudescence of the G2022, G2023, G2024, and G2025 lines upon artesunate (AS) challenge. Mice were infected with 2 × 107 parasites by i.p. injection on day 0. Treatment with AS was commenced ∼3 h postinfection by i.p. injection and was continued for three consecutive days as indicated by arrows. Parasitemia was monitored microscopically until recrudescence was observed. Mice were bled when the parasitemia was less than 1.5% to minimize competition from wild-type parasites in case mutants carried growth defects. (C) Sanger sequencing of bulk DNA from the G2025 R551T line showing selective enrichment of this mutation upon AS treatment at 20 or 64 mg/kg. Enrichment of this mutation was also observed in the RFLP analysis (see also Fig. S2B in the supplemental material).
Schematic and amino acid alignment of P. falciparum and P. berghei K13. (A) P. falciparum K13 protein showing amino acid positions and the protein domains. Positions of K13 mutations that have been investigated in this study are indicated. Equivalent amino acid positions for P. berghei are indicated in parallel at the bottom (in blue). (B) Protein alignment of P. falciparum and P. berghei K13 showing conservation at the mutation sites. Alignments were carried out using Clustal Omega protein alignment tool. Conserved residues are indicated by “*.” Download FIG S1, TIF file, 2.9 MB (2.9MB, tif) .
Copyright © 2020 Simwela et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
CRISPR/Cas9 editing strategy and RFLP analysis of P. berghei K13 mutant lines. (A) Schematic of CRISPR/Cas9 strategy used to introduce K13 mutations into P. berghei asexual blood-stage parasites. For this, 20-bp sgRNA targeting regions within 0 to 30 bp of the mutation site (see Tables S1 and S2) were designed to contain Esp3I digestion overhangs and were cloned into the Cas9 plasmid ABR099, as previously described (46). Donor templates were generated by overlapping extension PCR as described in Materials and Methods and were subsequently cloned into the ABR099 plasmids carrying the appropriate sgRNA (Tables S1 and S2) at the HincII linker site. The donor templates carried the mutation of interest as well as silent mutations to introduce a restriction site for RFLP analysis and to inactivate the PAM site recognition sequence (as illustrated in the schematic). Details of plasmids, sgRNA, and lines generated are in Table S1. (B) RFLP analysis of transfected parasite populations before and after challenge with artesunate (AS) at 20 or 64 mg/kg for the G2022 (C592Y.1), G2023 (C592Y.2), G2024 (I555T), and G2025 (R551T) lines. RFLP analysis was carried out using the indicated restriction enzymes on PCR fragments amplified from genomic DNA of transfected parasites. PCRs used primers that bound on either side of the donor DNA (Table S1 and S2). (C) RFLP analysis of parasite bulk populations for the G1957 (F458I), G1979 (Y505H), and G1989 (M488I) transfected lines. RFLP analysis was carried out using the indicated restriction enzymes. In both RFLP analyses, “*” indicates that these lines were uncloned. Download FIG S2, TIF file, 2.8 MB (2.7MB, tif) .
Copyright © 2020 Simwela et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DNA sequence and further RFLP analysis of P. berghei K13 mutant lines. (A) Sequencing analysis of the G2025 line showing the presence of traces for both the silent mutations and the R551T substation in the original transfection. DNA sequencing showing the absence of the C592Y and I555T nucleotide substitutions and the presence of minor traces of the silent mutations in the G2023 (B) and G2024 (C) transfected lines; “*” on the transfectant parasite lines indicates that the line was not cloned. (D) RFLP analysis of the G2042 (C592Y, sgRNA 2, TAT codon), G2043 (C592Y, sgRNA 2, TAC codon), and G2044 (C592Y, sgRNA 1, TAC codon) transfected lines (Table S1), showing further unsuccessful attempts to introduce the C592Y in P. berghei. RFLP analysis of the G2045 control line (C592C, sgRNA 1, silent mutations control) where editing was readily achieved is shown for comparison. (E) Sequencing analysis of the G2045 line showing successful editing with high efficiency to introduce silent mutations without the C592Y substitution. DNA sequence analysis showing high efficiency editing to introduce silent mutations and mutations of interest in the G1957 (F458I) (F), G1989 (M488I) (G), and G1979 (Y505H) (H) lines. (I) RFLP analysis with indicated restriction enzymes for the cloned parasite lines G1957 (F458I), G1979 (Y505H), G1989 (M488I), and G2025 (R551T, AS 64 mg/kg). Download FIG S3, TIF file, 2.8 MB (2.8MB, tif) .
Copyright © 2020 Simwela et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Plasmids, generated lines, transfection efficiencies, and outcome genotypes. RFLP analysis of the bulk transfected parasites was carried out on PCR fragments amplified using diagnostic PCR primers exterior of the donor template: GU5300 plus GU5301 (1,111 bp) for K13 and GU5186 plus GU4895 (946 bp) for UBP-1. Download Table S1, XLSX file, 0.1 MB (11KB, xlsx) .
Copyright © 2020 Simwela et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
In earlier efforts to introduce UBP-1 mutations in P. berghei, we found that preemptive drug pressure to which the engineered mutation is anticipated to confer a protective advantage can selectively enrich for the mutant in a mixed, transfected parasite population, even when the mutant population is <1% in the mixture (46). Using this approach, we subjected a larger inoculum (2 × 107) of the G2022C592Y.1*, G2023C592Y.2*, G2024I555T*, and G2025R551T* lines to AS at 20 or 64 mg/kg to see if any enrichment in the recrudescent parasite populations could be achieved (Fig. 1B). Indeed, AS at both 20 and 64 mg/kg specifically enriched the R551T mutant population in the G2025R551T* line from 30.0% in the initial transfection to 49.7% at AS 20 mg/kg and >99% at 64 mg/kg (Fig. 1C; Fig. S2B and Table S1). In contrast, apart from a minor enrichment that was observed for the G2024I555T* line, no useful enrichments in both the G2022C592Y.1* and G2023C592Y.2* lines were observed by RFLP at either concentration of AS (Fig. S2B; Table S1). Furthermore, no I555T or C592Y amino acid substitution traces could be seen after population-level DNA sequencing of these lines. These data further supported the relative nonviability of P. berghei parasites bearing K13 C592Y and I555T mutations. In agreement with the above-described observations, further attempts to introduce the C592Y mutation using a different sgRNA and/or different codons for the tyrosine residue in the donor template (TAT or TAC) were also unsuccessful. We did, however, observe >90% editing efficiency when introducing only silent mutations that maintained the C592C wild-type genotype in the donor template (Fig. S3 and E; Table S1). This, plus other unsuccessful attempts to generate the I555T mutant, further implies that these two K13 mutations are not viable in P. berghei. Meanwhile, transfection of the P. berghei 1804cl1 line with pG983 (F458I), pG984 (Y505H), and pG1008 (M488I) successfully introduced these mutations in P. berghei K13, yielding the G1957F458I*, G1979Y505H*, and G1989M488I* lines with >93% efficiencies, as confirmed by RFLP analysis (Fig. S2C; Table S1) as well as population-level DNA sequencing (Fig. S3F, G, and H). These three lines (G1957F458I*, G1979Y505H*, and G1989M488I*) and the G2025R551T* AS 64 mg/kg-challenged line were all cloned by limiting dilution. Mutations were further confirmed by RFLP analysis (Fig. S3I) and sequencing. The V2721F UBP-1 mutant line, which we previously found to mediate reduced susceptibility to ARTs in P. berghei (46), was also generated in the 1804cl1 background and cloned (Table S1).
P. berghei K13 mutants display reduced susceptibility to DHA in 24-h assays and increased survival in P. berghei-adapted RSAs.
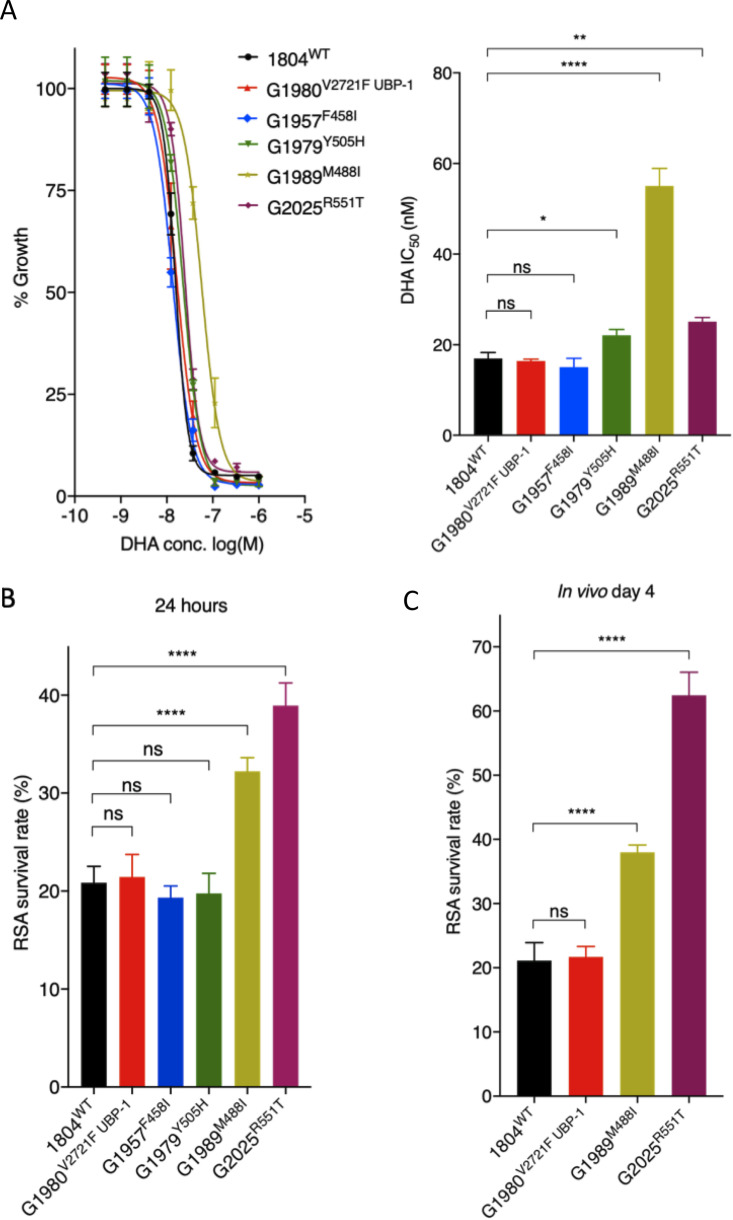
Unlike that for P. falciparum, P. berghei can only be maintained in one blood-stage cycle in vitro, which restricts drug susceptibility assays to one 24 h developmental cycle. Drug susceptibility readouts are therefore based on single-generation flow cytometry quantification of schizont maturation (46, 52, 53). Using this approach, we aimed to characterize the DHA dose-response profiles of the P. berghei K13 mutants compared to those of wild-type parasites or to a previously reported UBP-1 mutant with reduced ART susceptibility (46). Interestingly, in contrast to the equivalent P. falciparum K13 mutants, P. berghei M488I, R551T, and Y505H K13 mutant parasites displayed reduced susceptibility to DHA in standard growth inhibition assays with 3.3-, 1.4-, and 1.2-fold 50% inhibitory concentration (IC50) increases, respectively, compared to that of isogenic K13 wild-type parasites (Fig. 2A). The P. berghei F458I K13 mutant displayed equal sensitivity to DHA as the wild-type and the UBP-1 V2721F mutant (Fig. 2A), in agreement with our previous observations (46). These data suggest that, despite being limited to a single-cycle 24 h exposure, the P. berghei standard assay can distinguish even modestly ART-resistant parasites from sensitive ones. We next investigated the DHA susceptibility of early ring-stage P. berghei K13 mutant parasites by adapting the P. falciparum RSA (22). The P. falciparum RSA relies on exposure of early ring-stage parasites (0 to 3 h postinvasion) to 700 nM DHA for 4 to 6 h, followed by assessment of viability in the 2nd life cycle. This protocol allows drug-exposed parasites to reinvade fresh red blood cells. With this approach, current RSA parameters define in vitro ART resistance as survival of ≥1% and ART sensitivity as <1% survival (22). Using a similar approach, we exposed ∼1.5-h postinvasion K13 mutant P. berghei ring-stage parasites to DHA at 700 nM for 3 h (to accommodate for the shorter life cycle in P. berghei). Viability was assessed 24 h later by flow cytometry-based quantification of schizont maturation and mCherry expression. Interestingly, we observed that a significant fraction of P. berghei wild-type parasites survived exposure to DHA at 700 nM, with percentage survival rates of ∼20.9% (Fig. 2B). This is in agreement with our previous observations that P. berghei is less susceptible to ARTs than P. falciparum (46, 54). Both the UBP-1 mutant and F458I or Y505H K13 mutant parasites had the same survival rates as the wild-type line, whereas the M488I and R551T mutants exhibited significantly higher survival rates (32.3% or 39.0%, respectively, P < 0.001) (Fig. 2B). This is consistent with previous reports that, in P. falciparum, the R539T and I543T mutations are associated with the highest rates of RSA survival (28). However, we noted inconsistencies between drug susceptibility data of the mutants in the two in vitro tests (standard 24-h assay and adapted P. berghei RSA). This might result from the inability to maintain P. berghei in long-term culture and extend the analysis. We therefore developed a modified in vivo RSA, where we injected wild-type, UBP-1 V2721F, M488I, and R551T parasites back into mice 24 h after dimethyl sulfoxide (DMSO) or DHA exposure in the RSA as described above and then assessed viability by quantifying in vivo parasitemia on day 4. Remarkably, percentage survival in the R551T mutant parasites significantly increased from ∼39.0% (24 h readout) to ∼62.5%, while M488I mutant parasite survival increased from ∼32.3% (24 h readout) to ∼38.0% (Fig. 2C). In contrast, the percentage survival of the wild-type and UBP-1 mutant lines did not significantly change in the extended assay, despite the minor growth defect in the UBP-1 mutant, demonstrating that the P. berghei in vitro RSA and standard growth inhibition assays with 24-h readouts may be less robust in quantifying resistance phenotypes, especially if mutant parasites are less fit (Fig. 2C).
FIG 2.
In vitro and ex vivo susceptibility of P. berghei K13 mutants to DHA. (A) DHA dose-response curves and IC50 values for P. berghei K13 mutant lines compared to those of the wild-type 1804WT and the UBP-1 G1980V2721F mutant lines. (B) Survival of P. berghei K13 mutant lines in the P. berghei RSA. Results show the percentages of synchronized early ring-stage parasites (1.5-h postinvasion) that survived a 3 h exposure to 700 nM DHA relative to DMSO-treated parasites. Survival was quantified 24 h posttreatment by flow cytometry analysis based on Hoechst 33258 DNA staining and mCherry expression. (C) In vivo RSA survival for two K13 mutant lines (G1989M488I and G2025R551T) compared to that of the wild-type (1804WT) and UBP-1 mutant (G1980V2721F) controls. After in vitro exposure to DHA or DMSO as described above, parasites were i.v. injected back into mice as described in Materials and Methods. Parasitemia was quantified by flow cytometry analysis of mCherry expression on day 4 after i.v. injection, from which percentage survival rates were calculated. Error bars show standard deviations calculated from three biological repeats. Statistical significance (compared to the 1804WT line) was calculated using one-way analysis of variance (ANOVA) alongside the Dunnett’s multiple-comparison test. ns, not significant; *, P < 0.05; **, P < 0.01; ****, P < 0.0001.
P. berghei K13 mutants mimic the delayed parasite clearance phenotype in vivo upon AS treatment and achieve faster recrudescence than wild-type parasites at high ART doses.
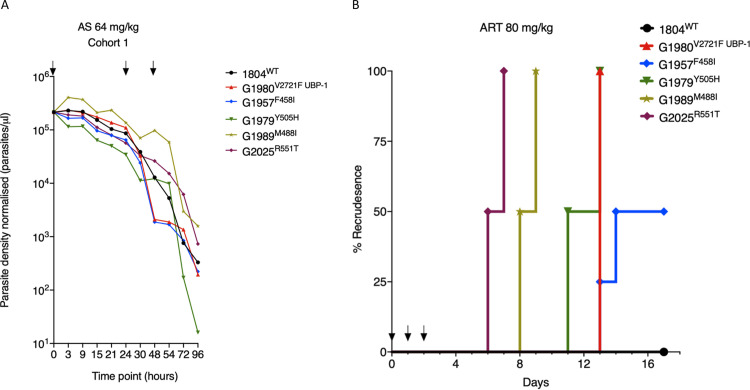
We next investigated the in vivo parasite clearance rates of P. berghei K13 mutant parasites in infected mice treated with AS. Mice were infected with a fixed inoculum of K13 and UBP-1 mutant parasites (105) in four cohorts, and parasitemias were allowed to rise to ∼10%. This was followed by dosing with AS at 64 mg/kg body weight, which is slightly higher than the equivalent of the maximal human clinical dose of 4 mg/kg (mouse equivalent = 49.2 mg/kg) to accommodate for the reduced ART susceptibility observed in P. berghei parasites. Parasitemias were quantified by flow cytometry (based on mCherry positivity) and microscopic analysis every 3 h for the first 24 h and at least once after the second and third doses at 24 and 48 h, respectively. Plotting parasite density in P. berghei K13 and UBP-1 mutant parasites against time revealed that in the first 24 h of sampling, parasite clearance kinetics did not sufficiently discriminate K13 or UBP-1 mutant parasites from the wild type. However, as the majority of dying parasites were being cleared by the host and mice received further doses, extended analysis revealed that P. berghei M488I and R551T mutant parasites consistently and significantly persisted compared to wild-type, F458I, Y505H, and UBP-1 mutant parasites (Fig. 3A; see also Fig. S4). Starting AS treatment at a high initial parasitemia (∼10%) also ensured that a good proportion of parasites would be within the early ring-stage window and, therefore, would be expected to preferentially survive the first AS dose. Surviving rings were easily distinguished as viable trophozoites at either 18-, 21-, or 24-h time points by microscopic examination of blood smears, which enabled comparisons between parasite lines. We therefore carried out concurrent collection and analysis of thin blood smears at all time points examined for flow analysis (Fig. 3A; Fig. S4). Results demonstrated that enhanced survival after the first AS dose was evident for all four P. berghei K13 mutant parasites as well as the UBP-1 mutant compared to wild-type parasites (see Fig. S5). Microscopy provided a more sensitive discrimination than flow cytometry-based estimation of clearance kinetics that was unable to distinguish mutant from wild-type parasites in the first 24 h. False positives could be due to the retention of mCherry positivity by dying parasites. For instance, we observed that a significant proportion of wild-type parasites remained mCherry positive and were counted as viable by flow cytometry (Fig. 3A; Fig. S4), whereas, microscopically, they were pyknotic forms (Fig. S5A and G). Remarkably, the M488I and R551T mutants remained smear positive after two consecutive AS doses (Fig. S5E, F, K, and L), whereas the wild-type, F458I, Y505H, and UBP-1 mutant parasites were cleared (microscopically smear negative) after 48 h. These data suggest that the M488I and R551T mutants meet the classical definition of ART resistance, as defined by the WHO based on day 3 (second generation) microscopy positivity, when accounting for the duration of the P. berghei life cycle and the dosing intervals (4). One of the four mice in the M488I treatment group remained smear positive after three consecutive AS doses (Fig. S5E). These data provide evidence that P. berghei K13 mutants modulate in vivo susceptibility to ARTs, resulting in a persister/delayed clearance phenotype under controlled conditions of initial parasite biomass and host immune status. Of note, we consistently used naive mice of same age, sex, breed, and genetic background.
FIG 3.
In vivo clearance and recrudescence rate of P. berghei K13 mutants following treatment with AS or ART. (A) Parasite clearance curves in mice infected with P. berghei K13 mutant lines following treatment with AS. Six mice (in each of four cohorts) were infected with 105 parasites of each of the four K13 mutants, the UBP-1 mutant, and wild-type control on day 0. On day 5, at a parasitemia of ∼10%, mice were dosed with AS at 64 mg/kg body weight. Day 5 was the designated 0 h time point for the dosing regimen. Parasite density per microliter of blood was quantified based on absolute counts of mCherry-positive parasites at staggered time points for each of the two cohorts, with 5 time points in the first 24 h (corresponding to at least 3 h interval coverage between the two cohorts) and at least once daily thereafter. Mice were dosed three times at 0, 24, and 48 h as indicated by arrows. Concurrent thin blood smears were prepared at each time point for microscopic analysis (Fig. S4). (B) Kaplan-Meier plots of recrudescence in wild-type and UBP-1 mutant controls compared to that of K13 mutants. A modified Peters’ 4-day suppressive test was used to monitor susceptibility of the K13 mutants to 80 mg/kg ART, a dose that effectively suppresses wild-type parasites for up to 18 days. Groups of three (UBP-1 mutant, 1804WT) or four mice (K13 mutants) were infected with 1 × 106 parasites on day 0. ART treatment was initiated ∼3 h later and continued every 24 h for three consecutive days (treatment days shown by arrows). Parasitemias were monitored by microscopic analysis of Giemsa-stained blood smears up to day 18 (Table S3). Recrudescence rates were plotted as the proportion of mice in the treatment groups that became smear positive on every individual day for the 18 days of follow-up.
Clearance kinetics of P. berghei K13 mutants upon AS treatment. Parasite clearance curves in mice with established parasitemias of K13 mutant lines following treatment with 64 mg/kg artesunate (AS) at the indicated times (see arrows) for the three cohorts. See Materials and Methods and Fig. 3A. Download FIG S4, TIF file, 2.7 MB (3.3MB, tif) .
Copyright © 2020 Simwela et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Microscopic analysis of P. berghei K13 mutants upon AS treatment. Microscopic analysis of Giemsa-stained thin blood smears showing preferential survival of UBP-1 (B and H) and K13 mutant parasites G1957F458I (C and I), G1979Y505H (D and J), G1989M488I (E and K), and G2025R551T (F and L) compared to wild-type parasites (A and G) upon treatment with AS. Cohorts 1 and 2 are shown in panels A to F, and cohorts 3 and 4 are shown in panels G to L. Smears were taken at time points corresponding to those shown in the clearance plots in Fig. 3A and in Fig. S4. Second and third dose treatment days are indicated by black arrows. Red arrows indicate viable parasites. Viability was deemed significant if at least 4 viable parasites were observed upon observation of at least 10 microscopic fields. Download FIG S5, TIF file, 2.8 MB (2.7MB, tif) .
Copyright © 2020 Simwela et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Another in vivo marker of reduced ART susceptibility in P. falciparum is the rate of recrudescence upon AS treatment, which acts as a possible indicator of AS treatment failure. However, at pharmacologically safe doses in humans (2 to 4 mg/kg), ART monotherapy treatment leads to >40% recrudescence rates (1, 3), making it difficult to use this approach to separate clinically ART-sensitive from ART-resistant parasites. P. berghei K13 mutants, therefore, provide the opportunity to test for recrudescence rates using controlled parasite inocula as well as AS or ART dose ascendency. We treated groups of mice initially infected with 106 K13 mutant, ART-resistant UBP-1 mutant, or wild-type parasites with a daily ART dose of 80 mg/kg for three consecutive days. This ART dose sufficiently suppresses the P. berghei wild type at equivalent parasite inocula for up to 18 days of follow-up (46). All UBP-1 mutant infections recrudesced 11 days after the last ART dose, whereas no recrudescence (0%) was observed for the wild type (Fig. 3B; see also Table S3). These data are consistent with our previous observations (46). However, R551T mutant parasite infections achieved even faster recrudescence, namely, 50% on day 4 after the last dosing and 100% a day later, indicating a higher level of in vivo resistance for this K13 mutation compared to that of the UBP-1 mutant. M488I mutant parasites had a similar recrudescence profile beginning on day 6. The Y505H and F458I mutant lines both achieved recrudescence at approximately the same time as the UBP-1 mutant; however, the latter achieved only 50% recrudescence across the 18-day follow-up period (Fig. 3B; Table S3). These data further confirm that P. berghei K13 mutants modulate in vivo susceptibility to ARTs and, crucially, that recrudescence rates strongly correlate with our in vitro DHA RSA profiles (Fig. 2) as well as with in vivo clearance kinetics in established infections (Fig. 3A; Fig. S4 and S5).
P. berghei K13 mutants are associated with an in vivo fitness cost but are preferentially selected for in the presence of AS or CQ.
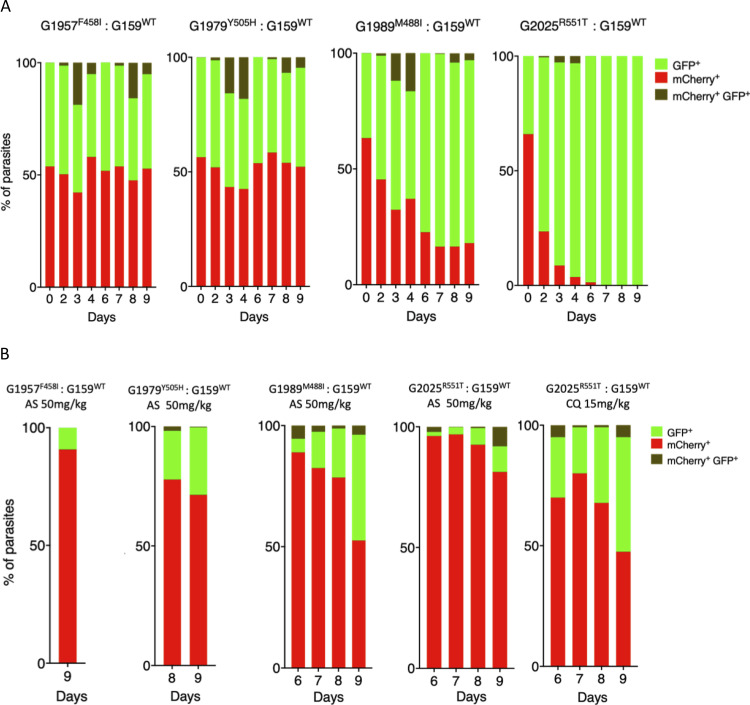
To assess the fitness of our P. berghei K13 mutants, we performed direct head-to-head competitions with wild-type parasites under in vivo growth conditions. P. berghei K13 or UBP-1 mutant lines or the parental 1804WT (mCherry positive) line were mixed at a 1:1 ratio with the G159WT (green fluorescent protein [GFP] positive) line and injected into mice. Changes in the proportion of GFP- or mCherry-positive parasites in the competition mixture were then quantified by flow cytometry over 9 days. These assays revealed that the F458I and Y505H mutant parasites were fitness neutral relative to the G159WT line, whereas the M488I and R551T mutants carried significant fitness costs (Fig. 4A). Both the M488I and R551T mutations were associated with high levels of reduced susceptibility to DHA in vitro (Fig. 2), delayed clearance kinetics (Fig. 3A; Fig. S4), and faster recrudescence following ART treatment in vivo (Fig. 3B; Table S3). Comparatively, the R551T mutant parasites had a more severe growth defect than the M488I mutants and were completely outcompeted by the GFP-positive wild-type line by day 7 (Fig. 4A). This is consistent with previous observations of high in vitro fitness costs for the equivalent P. falciparum R539T mutation (40). In comparison to the G159WT line, the parental wild-type line (1804WT) was fitness neutral, whereas the UBP-1 V2721F mutant carried a minor growth defect as previously observed (46) (see Fig. S6A and B). We also examined the proportions of GFP-positive versus mCherry-positive parasites over time in P. berghei K13 mutant and wild-type parasites upon treatment with AS. Mutant parasites were mixed at 1:1 ratios with the G159WT line and injected into mice that were treated with AS at 50 mg/kg beginning 3 h after infection for three consecutive days. Monitoring of recrudescence up to day 9 revealed that, upon AS treatment, the M488I and R551T mixtures recrudesced slightly faster than the wild-type mixture and were highly enriched for the mutant population (>90%) at the time of recrudescence (Fig. 4B). The F458I and Y505H mutant mixtures recrudesced slightly later (Fig. 4B), as did the UBP-1 V2721F mutant (Fig. S6B), and were all significantly enriched for the mutants. In contrast, the proportions of GFP-positive versus mCherry-positive parasites in the parent 1804WT and G159WT competition mixture after AS treatment did not change at the time of recrudescence (Fig. S6A). These data show that mutant P. berghei K13 parasites are preferentially selected for upon AS treatment, despite some carrying growth defects that rendered them at a complete competitive disadvantage in the absence of drug.
FIG 4.
Relative fitness of P. berghei K13 mutants in presence or absence of AS or CQ. Growth competition assays with K13 mutant lines that constitutively express mCherry compared to the wild-type G159WT line that constitutively expresses GFP in the presence or absence of drug pressure. The G159WT line was mixed with a given mutant line at a 1:1 ratio in three groups of mice on day 0. The first group was left untreated, the second group received a dose of AS at 50 mg/kg starting from 3 h after i.p. injection for three consecutive doses, while the third group consisting of the 1804 WT, G1980V2721F, and K13 mutant G2025R551T lines received CQ at 15 mg/kg at similar dosing times as AS. Percentages of mCherry- or GFP-positive parasites were determined by flow cytometry as described in Materials and Methods. (A) Percentage population changes as measured by flow cytometry of the G1957F458I, G1979Y505H, G1989M488I, and G2025R551T mutant lines relative to that of the G159WT wild-type line. (B) Proportion representation of the G159WT line in mixtures with G1957F458I, G1979Y505H, G1989M488I, and G2025R551T lines on the days of recrudescence upon treatment with AS or CQ as indicated.
Growth competition of the parent 1804WT and UBP-1 V2721F mutant line compared to the G159WT in the presence or absence of AS or CQ. Growth competition assays with the wild-type parent 1804WT or the UBP-1 G1980V2721F line compared to wild-type GFP-expressing G159WT line in the presence or absence of artesunate (AS) or chloroquine (CQ) drug pressure. Parasites were mixed at a 1:1 ratio, injected into mice, and left treated or untreated with AS at 50 mg/kg or CQ at 15 mg/kg as described in Materials and Methods and Fig. 4. (A) Percentage population changes of the 1804WT and wild-type G159WT in the absence of drug and on the day of recrudescence on day 9 for AS or CQ. (B) Proportion representations of the G159WT line in mixture with G1980V2721F, in the absence of drug or upon AS or CQ treatment, on the days of recrudescence. Download FIG S6, TIF file, 2.7 MB (2.7MB, tif) .
Copyright © 2020 Simwela et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
With the supposed role of P. falciparum K13 in mediating parasite hemoglobin endocytosis (43–45), we also speculated that P. berghei K13 mutant parasites with strong ART resistance phenotypes might be able to modulate susceptibility to CQ (to some degree) through a similar dysregulation of the endocytic machinery. Using the in vivo competition assay under drug pressure as with AS as described above, the parental 1804WT line, the UBP-1 V2721F line, and the K13 R551T mutant line were each mixed at 1:1 ratios with the G159WT line and treated with CQ at 15 mg/kg. At the time of recrudescence, the proportion of 1804WT parasites (mCherry positive) did not significantly change compared to the proportion of GFP-positive G159WT parasites (Fig. S6A). In comparison, the UBP-1 V2721F mutant was enriched to ∼70% (Fig. S6B), which mirrors our previous observations that this mutation can indeed be selectively enriched by CQ (46). Interestingly, upon CQ treatment, the combination of R551T mutant parasites and the G159WT line achieved recrudescence at almost the same rate as that under AS pressure, with mutant parasites enriched to ∼72% (Fig. 4B). These data suggest that K13 mutations can also contribute to low-level protection to CQ (43, 44).
A Plasmodium-selective proteasome inhibitor is potent against P. berghei wild-type and K13 mutant parasites and synergizes DHA action.
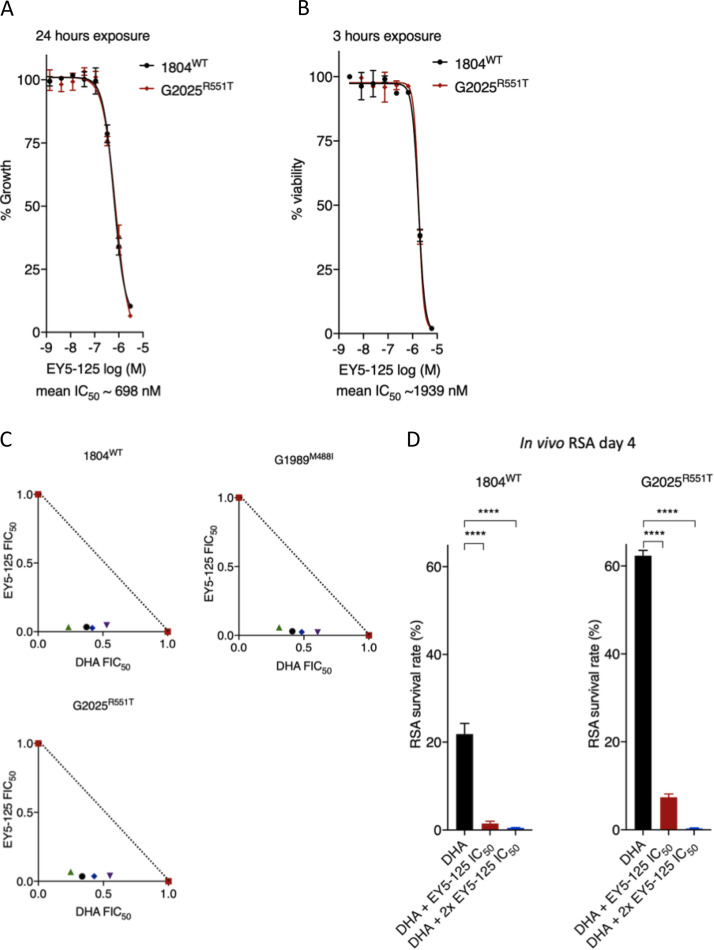
An enhanced cell stress response characterized by upregulation of genes in the unfolded protein response (UPR) is a typical signature of ART-resistant parasites (50). Resistant parasites (K13 mutants) also display enhanced activity of the ubiquitin proteasome system (UPS), a conserved eukaryotic pathway that acts downstream of the UPR by degrading unfolded proteins (49, 55). UPS inhibitors are available for cancer treatment and have been shown to synergize DHA activity in wild-type and K13 mutant P. falciparum both in vitro and in vivo, marking them as promising agents for overcoming ART resistance (49, 56). The Plasmodium-selective proteasome inhibitor EY5-125 is a potent antimalarial (standard IC50 against P. falciparum = 19 nM) that acts in synergy with ART against both ART-resistant and -sensitive P. falciparum strains in vitro (57). Here, we tested the efficacy of EY5-125 against P. berghei wild-type and K13 mutant parasites and examined its potential ability to synergize DHA action. P. berghei wild-type and the most ART-resistant K13 mutant (R551T) parasites were found to be equally sensitive to EY5-125 (Fig. 5A and B). Compared to that in P. falciparum (72-h IC50 of ∼19 nM and 1-h IC50 of ∼648 nM), EY5-125 is much less potent in P. berghei in both standard in vitro growth inhibition (IC50 = ∼700 nM) and 3-h assays (IC50 = ∼1,900 nM), respectively (Fig. 5A and B). These differences could be due to species-specific differences in drug sensitivity as we have observed with ARTs (46, 54) and many other drugs (58). However, combinations of DHA and EY5-125 in fixed-ratio isobologram analyses revealed a strong synergistic interaction against the P. berghei K13 wild-type and M488I and R551T mutant lines (Fig. 5C; see also Table S4). We also employed our in vivo RSA to examine whether a combination of DHA at 700 nM and EY5-125 at the equivalent 3-h IC50 (1.94 μM) or 2× IC50 (3.88 μM) could impact parasite survival rates. Indeed, at both the 3-h IC50 and 2× IC50 concentrations, EY5-125 strongly synergized with DHA (700 nM), as evidenced by significant abrogation of survival for both the wild-type and R551T mutant lines (Fig. 5D). These data demonstrate that proteasome inhibitors synergize DHA action in P. berghei K13 mutants equally as well as wild-type parasites both in vitro and in vivo and have the potential to be used to overcome ART resistance.
FIG 5.
Activity and DHA synergy of proteasome inhibitor in P. berghei K13 mutants. Dose-response curves and mean IC50 values for the Plasmodium-selective proteasome inhibitor EY5-125 for the wild-type 1804WT and K13 mutant G2025R551T lines in standard 24-h assays (A) or 3-h exposure assays conducted on early ring-stage parasites (B). Mean IC50 is a calculated average for the two lines independently screened in three biological repeats. (C) Isobologram plots representing the interaction between DHA and EY5-125 in the wild-type 1804WT, G1989M488I, and G2025R551T lines. Plots show mean FIC50 values (Table S4) for each drug calculated from three biological repeats. (D) Synergy of EY5-125 proteasome inhibitor with DHA in the in vivo RSA. Parasites were exposed to DMSO or DHA at 700 nM alone or in combination with EY5-125 at 3-h IC50 or 2× IC50 and then injected back into mice 24 h later as described in Materials and Methods. Parasitemias in mice infected with drug or DMSO-treated parasites were determined by flow analysis of mCherry expression on day 4 after i.v. injection and were used to calculate percent survivals relative to that of DMSO-treated parasites. Error bars are standard deviations from three biological repeats. Statistical significance was calculated using one-way ANOVA alongside the Dunnett’s multiple-comparison test. ****, P < 0.001.
DISCUSSION
In this study, we successfully employed CRISPR/Cas9 editing to introduce four of the six targeted orthologous P. falciparum K13 (F446I, M476I, Y493H, and R539T) mutations into the K13 gene of the rodent model of malaria P. berghei. Meanwhile, introduction of two mutations (C580Y and I543T) could not be achieved. As debate continues on the role of K13 in mediating in vivo susceptibility to ARTs (37), phenotyping of these P. berghei K13 (F458I, M488I, Y505H, and R551T) mutants provides experimental evidence for the ability of mutant K13 to confer in vivo resistance to ARTs in a naive parasite genome background. These mutants displayed reduced in vitro susceptibility to DHA and phenocopied P. falciparum delayed clearance phenotypes upon AS treatment. Moreover, these K13 mutants achieved faster recrudescence upon ART treatment under in vivo growth conditions. As in P. falciparum, certain P. berghei K13 mutations were found to cause significant growth defects, which highlights the structural and functional conservation of this protein across the two Plasmodium species and illustrates the fitness trade-offs that the acquisition of such mutations exerts on malaria parasite physiology.
ART resistance, principally associated with mutations in K13, is now almost endemic in Southeast Asia, with risks of global spread threatening the utility of ACTs that are at the forefront of malaria control programs (2). The P. falciparum C580Y K13 mutation is the most frequently observed (with >50% prevalence) and has reached fixation in most parts of Southeast Asia (25, 59). Why the P. falciparum C580Y mutation is so successful compared to other K13 mutations remains unclear. This mutation does not associate with high RSA survival rates compared to P. falciparum R539T or I543T mutant parasites, and treatment failure rates and parasite clearance rates are not more significant in C580Y-harboring parasites than those with other K13 mutations (27, 28, 60). Do fitness constraints, founder genetic landscapes, or species-specific differences between P. berghei and P. falciparum K13 explain our failed attempts to introduce the C592Y or I555T mutations in P. berghei? The structural homology model of the K13 propeller domain presented here demonstrates that this region is highly conserved between P. berghei and P. falciparum K13, with identical amino acids at the sites of mutations associated with ART resistance. Our unsuccessful attempts to introduce the P. berghei C592Y or P. berghei I555T mutations could therefore be more related to growth disadvantages or other deleterious effects. For example, in P. falciparum, the equivalent I543T and R539T mutations carry the most pronounced fitness costs (40), which could partly explain our inability to introduce the P. berghei I555T mutation in P. berghei. Moreover, P. berghei K13 mutations were introduced into PBANKA parasites with no history of ART exposure. These parasites might therefore be more sensitive to fitness impacts conferred by the P. berghei I555T or P. berghei C592Y substitution, as introduction of the equivalent P. falciparum C580Y in parasites isolated before ART was clinically introduced carried significant growth defects, as opposed to more recent Cambodian isolates where it was fitness neutral (40). A less prevalent K13 allele, P. falciparum R561H, that associates with significant delays in parasite clearance and peaked in prevalence in 2012 in Southeast Asia but has since declined (60) also easily outcompeted the P. falciparum C580Y mutation in head-to-head competitions (42). These data suggest that acquisition and propagation of certain P. falciparum K13 alleles, notably the C580Y substitution, might require appropriate founder genome architectures to compensate for the deleterious phenotypes. In these situations, K13 mutations (P. falciparum C580Y, for example), would arise in a necessary compensatory background that mitigates the deleterious growth effects leading to an initial soft sweep. In the case of ACTs, these compensatory backgrounds may also serve as general templates upon which partner drug resistance mutations might arise. This seems to be the case with the recent aggressive expansion of parasite colineages carrying the P. falciparum C580Y mutation and piperaquine resistance determinants (10, 11).
Despite the obstacles to introducing the P. berghei C592Y and I555T mutations, introduction of the P. falciparum R539T equivalent was achieved in P. berghei (R551T) despite low editing efficiency in the initial transfection. We were, however, able to enrich for this mutation with AS selection applied in vivo, yielding almost clonal levels of the P. berghei R551T mutant. Similar to the P. falciparum R539T mutant, clonal P. berghei R551T mutant parasites carried the strongest DHA resistance phenotypes in vitro as well as the clearest AS or ART resistance profiles in vivo. The P. falciparum R539T and P. falciparum I543T mutations occur at relatively low frequencies in Southeast Asia, with the prevalence of both mutations ranging between 0.3% and 3.5% (36, 41, 59). This could be due to the pronounced fitness cost of these mutations (40) limiting their expansion, which we also observed with the P. berghei R551T mutant parasites. The combination of a naive genomic background and species-specific differences can also be invoked to explain some phenotypic differences (growth rate and level of ART resistance) seen between mutant lines of P. falciparum and P. berghei K13, as observed in this study. For example, P. falciparum Y493H mutants clearly associate with increased RSA survival (23, 28) and delayed parasite clearance phenotypes (23, 41, 61), unlike the P. berghei counterpart (Y505H) that displayed low-level resistance to ARTs in vitro (in the standard assay but not in the adapted RSA) and in vivo. This could be due to additional underlying genetic factors in P. falciparum isolates providing an additive effect to the observed phenotypes, which would be absent in P. berghei. Nevertheless, the other P. berghei K13 mutations tested here appear to directly reflect the impact of the equivalent mutations in P. falciparum. Both P. berghei F458I (this study) and P. falciparum F446I K13 mutants are fitness neutral (62) and do not enhance RSA survival in vitro (62, 63) yet carry ART-protective phenotypes in vivo (64–66). Furthermore, P. berghei M488I K13 mutants display a significant growth defect that has not yet been characterized in the P. falciparum equivalent (M476I) and might explain its relative scarcity in Southeast Asia (67, 68).
Enhanced proteostasis is a characteristic signature of P. falciparum K13 ART-resistant parasites, which is typified by upregulation of genes in the UPR as well as enhanced activity of the UPS (49, 50, 55). Inhibition of the UPS by 26S proteasome inhibitors synergizes DHA action both in vitro and in vivo, which has offered a potential avenue to overcome ART resistance (49). Despite UPS inhibitors (which are clinically available for treatment of certain cancers) displaying activity in malaria parasites and synergizing DHA action, their translation into animal studies has been limited by host toxicity (69, 70). Recent structure-based design of Plasmodium-selective proteasome inhibitors has yielded vinyl sulfone inhibitors with a wider therapeutic window and improved host toxicity profiles (56, 57). These inhibitors not only display activity in diverse P. falciparum backgrounds, including those harboring K13 mutations, but also strongly synergize with DHA (71). Even though P. berghei proteasome structures have not been solved, functional and life cycle conservation between this parasite and P. falciparum is pronounced. Using EY5-125, an inhibitor selective for the P. falciparum proteasome (57), we demonstrate similar activity and synergy with DHA in P. berghei wild-type and K13 ART-resistant mutants. Importantly, we demonstrate these properties in vivo, which significantly strengthens the potential of these compounds in overcoming ART resistance in infected hosts.
In conclusion, our work provides robust experimental evidence that K13 mutations modulate in vitro and in vivo susceptibility to ARTs in the P. berghei rodent model of malaria. The cause and effect link between P. falciparum K13 mutations and reduced ART susceptibility is strong (23, 28). However, the reason for ART clinical failure has remained obscure because, in some cases, delayed parasite clearance phenotypes have been reported in parasites carrying wild-type K13 alleles (35, 72). This lack of clarity is further compounded by a reduced correlation between K13 mutations and parasite clearance half-lives or the frequencies of recrudescence in certain cases of ART monotherapy (35). As we demonstrate in this study, some of these observations may be attributable to fitness defects in mutant parasites that could confound the interpretation of recrudescence rates. These fitness differences might be especially relevant at the relatively low ART doses used in humans, which are already known to permit higher rates of recrudescence (3). Although a recent genetic cross between a P. falciparum K13 C580Y mutant parasite and an Aotus-infecting K13 wild-type parasite demonstrated a lack of association of this mutation with in vivo ART resistance metrics (recrudescence and clearance half-life) (37), we propose that this could be due to (i) the AS doses used being insufficiently high to clearly separate the lineages, (ii) the small sample sizes used, and (iii) the inherent limitation of using heterogeneous Aotus monkeys with various individual histories of parasite exposure and spleen status (spleen intact or splenectomized). Our in vitro and in vivo phenotypes for the P. falciparum F446I, M476I, Y493H, and R539T K13 mutation equivalents in P. berghei support their direct involvement in mediating resistance to ARTs. Our data also provide a robust immune-replete rodent host model to test for synergistic antimalarial combinations that can restore ART efficacy and overcome resistance.
MATERIALS AND METHODS
CRISPR/Cas9 generation of P. berghei K13 mutant lines.
The Cas9 plasmid ABR099 was used to target mutations of interest into the P. berghei K13 locus (PlasmoDB gene identifier [ID] PBANKA_1356700) (46). To obtain P. berghei equivalents of P. falciparum ART-resistant K13 mutations (PlasmoDB gene ID PF3D7_1343700), the amino acid sequences of the two proteins were retrieved and aligned using Clustal Omega (73). To structurally align the equivalent mutations in P. berghei K13, three-dimensional homology models of P. berghei and P. falciparum K13 were constructed using SWISS-MODEL (PDB template 4zgc.1.A) for amino acid residues 362 to 738 for P. berghei and 350 to 726 for P. falciparum. Models were visualized using pyMol 2.3. sgRNAs designed to target a region within 0 to 30 bp of the mutation of interest within the P. berghei K13 locus were initially cloned into the ABR099 plasmid (see Fig. S2A in the supplemental material). Donor DNA repair templates were designed to carry the mutation of interest in addition to silent mutations that introduced restriction sites for RFLP and that inactivated the PAMs. These templates were generated by overlap extension PCR (74) and were subsequently cloned into ABR099 plasmids carrying corresponding sgRNAs at the linker sites (Fig. S2A). Generated plasmids and all corresponding sgRNAs are listed in Table S1 in the supplemental material.
Parasite lines and animal infections.
This study employed two P. berghei ANKA-derived parasite lines, 1804cl1 and G159. The 1804cl1 (75) and G159 (Katie Hughes, unpublished) lines express mCherry and GFP, respectively, under the control of the strong constitutive hsp70 promoter. Infections were carried out in female Theiler’s Original mice (Envigo), 6 to 8 weeks old, weighing 25 to 30 g. Infections were established either by intraperitoneal (i.p.) injections of ∼200 μl of cryopreserved parasite stocks or by intravenous (i.v.) injections of purified schizonts or mixed-stage parasites diluted in phosphate-buffered saline (PBS). Parasitemias in infected mice were monitored by microscopic examination of methanol-fixed thin blood smears stained with Giemsa (Sigma) or flow cytometry-based analysis of infected blood stained with Hoechst 33342 (Invitrogen). Blood from infected mice was collected by cardiac puncture under terminal anesthesia. All animal work was performed in compliance with UK home office licensing (project reference P6CA91811) and ethical approval from the University of Glasgow animal welfare and ethical review body.
Transfections.
Primary transfections were carried out in the 1804cl1 line. Approximately 10 μg of episomal plasmid DNA from the vectors described above (Table S1) was transfected by electroporation of Nycodenz-purified schizonts using the Amaxa Nucleofector Device II program U-o33, as previously described (76). Parasites were then immediately i.v. injected into mice. Positive selection of transfected parasites was commenced 24 h later by adding pyrimethamine (0.07 mg/ml; Sigma) to their drinking water.
Genotyping of transformed parasites.
Parasite pellets were prepared from infected mouse blood that was lysed by resuspension in 1× E-lysis buffer (Thermo). Genomic DNA was extracted from the pellets using the Qiagen DNeasy blood and tissue kit according to the manufacturer’s instructions. Initial analysis of the transfected or cloned parasite lines was performed using a dual PCR-RFLP approach. PCR using primers exterior to the donor templates (Table S1 and S2) was used to amplify fragments from the genomic DNA of the mutant lines, followed by restriction digests with the artificially introduced RFLP restriction enzymes. Relative transformation efficiencies were estimated by densitometric quantification of wild-type and mutant RFLP fragments by ImageJ2 (77). Mutations and initial RFLP analyses were further confirmed by Sanger DNA sequencing.
List of primers used in the study and their descriptions. Download Table S2, XLSX file, 0.1 MB (11.9KB, xlsx) .
Copyright © 2020 Simwela et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Recrudescence of P. berghei K13 and UBP-1 mutants compared to that of the wild type in infected mice treated with ART at 80 mg/kg as described for Fig. 3B. Groups of three or four mice (M0 to M4) were infected with ∼106 parasites on day 0 and treated from 3 h with ART at 80 mg/kg as indicated by arrows. A recrudescent event was recorded as “−” for negative smears or “+” with associated parasitemia (% infected erythrocytes). Refers to Fig. 3B. Download Table S3, XLSX file, 0.1 MB (15KB, xlsx) .
Copyright © 2020 Simwela et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ΣFIC50 values for DHA and EY5-125 drug combination ratios in the 1804WT, G1989M488I, and G2025R551T lines. ΣFIC50 values are derived from FIC50 data as plotted in Fig. 5C. Download Table S4, XLSX file, 0.1 MB (9.9KB, xlsx) .
Copyright © 2020 Simwela et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Antimalarial agents.
DHA (Selleckchem) at 10 mM was diluted to a working concentration in schizont culture medium. The Plasmodium-selective proteasome inhibitor EY5-125, also known as compound 28 (57), was used to test for proteasome inhibitor synergy with DHA in K13 mutant and wild-type parasites. For in vivo drug treatment, AS (Sigma) was dissolved in 5% sodium bicarbonate prepared in 0.9% sodium chloride. CQ diphosphate (Sigma) was dissolved in 1× PBS. ART (Sigma) was prepared at 50 mg/ml in a 1:1 mixture of DMSO and Tween 80 (Sigma) and diluted 10-fold in sterile distilled water immediately before administration. All drugs were prepared fresh before in vivo administration, and drug delivery was carried out by i.p. injection.
Twenty-four-hour P. berghei in vitro culture and drug susceptibility assays.
In vitro culture and drug susceptibility assays were carried out beginning with synchronized ring-stage parasites over 24-h schizont maturation cycles, as P. berghei can only be maintained for one intraerythrocytic developmental cycle in vitro. Parasites were cultured and exposed to drugs as previously described (46), after which schizont maturation was analyzed by flow cytometry. Infected cells were stained with the DNA dye Hoechst 33258. Schizont maturation was used as a surrogate marker of growth inhibition and was quantified based on Hoechst 33258 fluorescence intensity or mCherry expression. To determine growth inhibition and calculate half-maximal inhibitory concentrations (IC50s), the percentage of schizonts in no-drug controls was set to 100% growth, and subsequent growth percentages in the presence of drugs were calculated accordingly. Dose-response curves were plotted in GraphPad Prism.
Adapted P. berghei ring-stage survival assays.
The P. falciparum RSA was adapted for P. berghei to further assess the in vitro phenotypes of K13-mutant parasites based on a previously published protocol (22). Schizonts were obtained from in vitro cultured parasites as previously described (76) and injected i.v. into naive mice to obtain synchronous in vivo infections containing >90% rings at parasitemias of 0.5% to 1.5%. Approximately 1.5 h postinjection, blood was collected from the infected mice, adjusted to 0.5% hematocrit, and exposed to 700 nM DHA or 0.1% DMSO (Thermo Fisher Scientific) in 96-well plates or 10-ml culture flasks. The plates and flasks were incubated with drug under standard culture conditions for 3 h, after which, the drug was washed off at least three times. Parasites were then returned to standard culture conditions in new plates and flasks with fresh schizont medium for in vitro maturation. After 24 h of incubation, parasite survival was assessed by flow cytometry analysis of Hoechst 33258-stained infected cells. Viability was assessed by gating on the Hoechst 33258 DNA stain and live mCherry expression. DHA-treated samples were compared to DMSO-treated controls processed in parallel. Percent survival was calculated using the following formula: survival (%) = (viability [%] [DHA − treated])/(viability [%] [mock DMSO − treated]).
To improve the robustness of the viability readouts beyond the 24-h flow cytometry counts, an in vivo expansion of the 3 h DHA- or DMSO-exposed parasites was used for selected mutants and the wild-type control. After 24 h of recovery, 2 ml of DHA- or DMSO-treated parasites was pelleted and resuspended in a 1-ml volume, from which, 200 μl was injected i.v. into mice. In vivo parasitemias were quantified on day 4 postinjection, from which percentage survivals based on in vivo parasitemia (absolute counts of mCherry positive parasites) were calculated using the following slightly modified formula: survival (%) = (parasitemia [DHA − treated])/(parasitemia [mock DMSO − treated]).
In vitro isobologram drug combinations.
DHA and EY5-125 drug interaction analyses in fixed ratios were carried out using a modified fixed-ratio interaction assay as previously described (78). DHA and EY5-125 combinations were prepared in molar concentration combination ratios of 5:0, 4:1, 3:2, 2:3, 1:4, and 0:5 and were dispensed into 96-well plates. This was followed by a 3-fold serial dilution with precalculated estimates to ensure that the test wells containing the 3-h IC50s of the two drugs were located near the middle of the plate. The drug combinations were then incubated with synchronized ∼1.5-h-old ring-stage wild-type or K13 mutant parasites for 3 h, after which, the drugs were washed off at least 3 times. Percent viability was quantified 24 h later by flow cytometry analysis of Hoechst 33258-stained infected cells and mCherry expression. Dose-response curves were calculated for each drug alone or in combination, from which fractional inhibitory concentrations (FIC50) were obtained and summed to obtain the ∑FIC50 using the following formula: ΣFIC50 = (IC50 of DHA in combination/IC50 of drug DHA alone) + (IC50 of EY5-125 in combination/IC50 of EY5-125 alone).
An ΣFIC50 of >1 was used to denote antagonism, ΣFIC50 <1 synergism, and ΣFIC50 = 1 additivity. FIC50 values for the drug combinations were plotted to obtain isobolograms for the drug combination ratios.
In vivo drug assays.
(i) Parasite clearance. Parasite clearance upon treatment with AS was used to evaluate potential delayed clearance phenotypes in K13 mutant parasites. These studies were based on a modified Rane’s curative test in established mice infections as previously described (79). Donor mice were infected with mutant lines and the wild-type control. Once a parasitemia of ∼2% was reached, blood was obtained from the donor mice and diluted in 1× PBS. Approximately 105 parasites were inoculated in 4 cohorts of mice (4 mice per line) by i.p. injections on day 0, and parasitemias were allowed to rise to ∼10%, typically on day 5. On day 5, at time zero, 2 μl of blood was collected and diluted 200-fold in 1× PBS. Thin blood smears were also collected at this time. All four cohorts were then dosed with AS at 64 mg/kg at 0, 24, and 48 h. Blood sampling was performed for flow cytometry analysis, and thin blood smears were prepared five times during the first 24 h for each cohort and at least daily thereafter in a staggered manner that allowed for a 3 h life cycle coverage in the first 24 h for at least two cohorts. Parasite density at each time point was determined by absolute cell counts and mCherry expression in 0.1 μl of whole blood diluted in PBS analyzed on a MACSQuant Analyzer 10. Thin blood smears of parasite morphologies were analyzed by microscopy. Significant viability counts in microscopy smears were based on microscopic confirmation of at least four viable parasites in a minimum of 10 fields. Clearance kinetics of normalized parasite densities versus time were plotted in GraphPad prism.
(ii) Recrudescence. A modified Peters’ 4-day suppressive test was used to assess in vivo response profiles and recrudescence rates of wild-type and mutant lines as previously described (46, 80). Infections were initiated by i.p. inoculation of 106 parasites diluted from donor mice and were followed by three daily consecutive drug doses of ART at 80 mg/kg, with the first initiated ∼3 h postinoculation. Parasitemia was monitored by microscopic analysis of methanol-fixed Giemsa-stained smears up to day 18 or until recrudescence was observed.
In vivo growth competition assays in presence or absence of drug treatment.
Mutant lines in the 1804cl1 mCherry background line were mixed with the G159 GFP line at 1:1 ratios and injected i.p. (total parasite inocula of 106) into 3 groups of mice. The groups were either left untreated or treated with AS at 50 mg/kg for 3 consecutive days starting 3 h postinfection or CQ at 15 mg/kg. Parasitemias and fractions of mutant versus wild-type parasites were determined by flow cytometry-based quantification of mCherry- or GFP-positive parasite populations.
Reagent availability.
Parasite lines and plasmids are available upon request from A. Waters.
ACKNOWLEDGMENTS
We thank Diane Vaughan and the University of Glasgow flow cytometry facility for assistance. We also thank Euna Yoo (Stanford University and NCI-Frederick) for providing the EY5-125 proteasome inhibitor.
This work was supported in part by grants from the Wellcome Trust to A.P.W. (083811/Z/07/Z, 107046/Z/15/Z, and 104111/Z/14/Z). Partial funding for this work was provided by the NIH (R01 AI109023 to D.A.F. and R33 AI127581 to M.B. and D.A.F.), the Department of Defense (W81XWH-19-1-0086 to D.A.F.), and the Columbia University—University of Glasgow Research Exchange Program. N.V.S. is a Commonwealth Doctoral Scholar (MWCS-2017-789), funded by the UK government. B.H.S. gratefully acknowledges earlier support from the Columbia University Graduate Training Program in Microbiology and Immunology (T32 AI106711; Program Director, D. A. Fidock).
Footnotes
Citation Simwela NV, Stokes BH, Aghabi D, Bogyo M, Fidock DA, Waters AP. 2020. Plasmodium berghei K13 mutations mediate in vivo artemisinin resistance that is reversed by proteasome inhibition. mBio 11:e02312-20. https://doi.org/10.1128/mBio.02312-20.
REFERENCES
- 1.White NJ. 2008. Qinghaosu (artemisinin): the price of success. Science 320:330–334. doi: 10.1126/science.1155165. [DOI] [PubMed] [Google Scholar]
- 2.WHO. 2019. World malaria report. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 3.Li GQ, Arnold K, Guo XB, Jian HX, Fu LC. 1984. Randomised comparative study of mefloquine, qinghaosu, and pyrimethamine-sulfadoxine in patients with falciparum malaria. Lancet 2:1360–1361. doi: 10.1016/s0140-6736(84)92057-9. [DOI] [PubMed] [Google Scholar]
- 4.WHO. 2018. Artemisinin resistance and artemisinin-based combination therapy efficacy: status report. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 5.Gregson A, Plowe CV. 2005. Mechanisms of resistance of malaria parasites to antifolates. Pharmacol Rev 57:117–145. doi: 10.1124/pr.57.1.4. [DOI] [PubMed] [Google Scholar]
- 6.Wongsrichanalai C, Pickard AL, Wernsdorfer WH, Meshnick SR. 2002. Epidemiology of drug-resistant malaria. Lancet Infect Dis 2:209–218. doi: 10.1016/S1473-3099(02)00239-6. [DOI] [PubMed] [Google Scholar]
- 7.Blasco B, Leroy D, Fidock DA. 2017. Antimalarial drug resistance: linking Plasmodium falciparum parasite biology to the clinic. Nat Med 23:917–928. doi: 10.1038/nm.4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mathieu LC, Cox H, Early AM, Mok S, Lazrek Y, Paquet J-C, Ade M-P, Lucchi NW, Grant Q, Udhayakumar V, Alexandre JS, Demar M, Ringwald P, Neafsey DE, Fidock DA, Musset L. 2020. Local emergence in Amazonia of Plasmodium falciparum K13 C580Y mutants associated with in vitro artemisinin resistance. eLife 9:e51015. doi: 10.7554/eLife.51015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Uwimana A, Legrand E, Stokes BH, Ndikumana JM, Warsame M, Umulisa N, Ngamije D, Munyaneza T, Mazarati JB, Munguti K, Campagne P, Criscuolo A, Ariey F, Murindahabi M, Ringwald P, Fidock DA, Mbituyumuremyi A, Menard D. 2020. Emergence and clonal expansion of in vitro artemisinin-resistant Plasmodium falciparum Kelch13 R561H mutant parasites in Rwanda. Nat Med 26:1602–1608. doi: 10.1038/s41591-020-1005-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamilton WL, Amato R, van der Pluijm RW, Jacob CG, Quang HH, Thuy-Nhien NT, Hien TT, Hongvanthong B, Chindavongsa K, Mayxay M, Huy R, Leang R, Huch C, Dysoley L, Amaratunga C, Suon S, Fairhurst RM, Tripura R, Peto TJ, Sovann Y, Jittamala P, Hanboonkunupakarn B, Pukrittayakamee S, Chau NH, Imwong M, Dhorda M, Vongpromek R, Chan XHS, Maude RJ, Pearson RD, Nguyen T, Rockett K, Drury E, Goncalves S, White NJ, Day NP, Kwiatkowski DP, Dondorp AM, Miotto O. 2019. Evolution and expansion of multidrug-resistant malaria in Southeast Asia: a genomic epidemiology study. Lancet Infect Dis 19:943–951. doi: 10.1016/S1473-3099(19)30392-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van der Pluijm RW, Imwong M, Chau NH, Hoa NT, Thuy-Nhien NT, Thanh NV, Jittamala P, Hanboonkunupakarn B, Chutasmit K, Saelow C, Runjarern R, Kaewmok W, Tripura R, Peto TJ, Yok S, Suon S, Sreng S, Mao S, Oun S, Yen S, Amaratunga C, Lek D, Huy R, Dhorda M, Chotivanich K, Ashley EA, Mukaka M, Waithira N, Cheah PY, Maude RJ, Amato R, Pearson RD, Gonçalves S, Jacob CG, Hamilton WL, Fairhurst RM, Tarning J, Winterberg M, Kwiatkowski DP, Pukrittayakamee S, Hien TT, Day NP, Miotto O, White NJ, Dondorp AM. 2019. Determinants of dihydroartemisinin-piperaquine treatment failure in Plasmodium falciparum malaria in Cambodia, Thailand, and Vietnam: a prospective clinical, pharmacological, and genetic study. Lancet Infect Dis 19:952–961. doi: 10.1016/S1473-3099(19)30391-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Imwong M, Dhorda M, Myo Tun K, Thu AM, Phyo AP, Proux S, Suwannasin K, Kunasol C, Srisutham S, Duanguppama J, Vongpromek R, Promnarate C, Saejeng A, Khantikul N, Sugaram R, Thanapongpichat S, Sawangjaroen N, Sutawong K, Han KT, Htut Y, Linn K, Win AA, Hlaing TM, van der Pluijm RW, Mayxay M, Pongvongsa T, Phommasone K, Tripura R, Peto TJ, von Seidlein L, Nguon C, Lek D, Chan XHS, Rekol H, Leang R, Huch C, Kwiatkowski DP, Miotto O, Ashley EA, Kyaw MP, Pukrittayakamee S, Day NPJ, Dondorp AM, Smithuis FM, Nosten FH, White NJ. 14 July 2020. Molecular epidemiology of resistance to antimalarial drugs in the greater mekong subregion: an observational study. Lancet Infect Dis doi: 10.1016/S1473-3099(20)30228-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amato R, Pearson RD, Almagro-Garcia J, Amaratunga C, Lim P, Suon S, Sreng S, Drury E, Stalker J, Miotto O, Fairhurst RM, Kwiatkowski DP. 2018. Origins of the current outbreak of multidrug-resistant malaria in Southeast Asia: a retrospective genetic study. Lancet Infect Dis 18:337–345. doi: 10.1016/S1473-3099(18)30068-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ashley EA, Dhorda M, Fairhurst RM, Amaratunga C, Lim P, Suon S, Sreng S, Anderson JM, Mao S, Sam B, Sopha C, Chuor CM, Nguon C, Sovannaroth S, Pukrittayakamee S, Jittamala P, Chotivanich K, Chutasmit K, Suchatsoonthorn C, Runcharoen R, Hien TT, Thuy-Nhien NT, Thanh NV, Phu NH, Htut Y, Han K-T, Aye KH, Mokuolu OA, Olaosebikan RR, Folaranmi OO, Mayxay M, Khanthavong M, Hongvanthong B, Newton PN, Onyamboko MA, Fanello CI, Tshefu AK, Mishra N, Valecha N, Phyo AP, Nosten F, Yi P, Tripura R, Borrmann S, Bashraheil M, Peshu J, Faiz MA, Ghose A, Hossain MA, Samad R, Tracking Resistance to Artemisinin Collaboration (TRAC), et al. 2014. Spread of artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med 371:411–423. doi: 10.1056/NEJMoa1314981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dondorp AM, Nosten F, Yi P, Das D, Phyo AP, Tarning J, Lwin KM, Ariey F, Hanpithakpong W, Lee SJ, Ringwald P, Silamut K, Imwong M, Chotivanich K, Lim P, Herdman T, An SS, Yeung S, Singhasivanon P, Day NPJ, Lindegardh N, Socheat D, White NJ. 2009. Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med 361:455–467. doi: 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flegg JA, Guerin PJ, White NJ, Stepniewska K. 2011. Standardizing the measurement of parasite clearance in falciparum malaria: the parasite clearance estimator. Malar J 10:339. doi: 10.1186/1475-2875-10-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.WWARN Parasite Clearance Study Group, Abdulla S, Ashley EA, Bassat Q, Bethell D, Björkman A, Borrmann S, D'Alessandro U, Dahal P, Day NP, Diakite M, Djimde AA, Dondorp AM, Duong S, Edstein MD, Fairhurst RM, Faiz MA, Falade C, Flegg JA, Fogg C, Gonzalez R, Greenwood B, Guérin PJ, Guthmann J-P, Hamed K, Hien TT, Htut Y, Juma E, Lim P, Mårtensson A, Mayxay M, Mokuolu OA, Moreira C, Newton P, Noedl H, Nosten F, Ogutu BR, Onyamboko MA, Owusu-Agyei S, Phyo AP, Premji Z, Price RN, Pukrittayakamee S, Ramharter M, Sagara I, Se Y, Suon S, Stepniewska K, Ward SA, White NJ, et al. 2015. Baseline data of parasite clearance in patients with falciparum malaria treated with an artemisinin derivative: an individual patient data meta-analysis. Malar J 14:359–359. doi: 10.1186/s12936-015-0874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.WWARN K13 Genotype-Phenotype Study Group. 2019. Association of mutations in the Plasmodium falciparum Kelch13 gene (Pf3D7_1343700) with parasite clearance rates after artemisinin-based treatments-a WWARN individual patient data meta-analysis. BMC Med 17:1. doi: 10.1186/s12916-018-1207-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ataide R, Ashley EA, Powell R, Chan JA, Malloy MJ, O'Flaherty K, Takashima E, Langer C, Tsuboi T, Dondorp AM, Day NP, Dhorda M, Fairhurst RM, Lim P, Amaratunga C, Pukrittayakamee S, Hien TT, Htut Y, Mayxay M, Faiz MA, Beeson JG, Nosten F, Simpson JA, White NJ, Fowkes FJ. 2017. Host immunity to Plasmodium falciparum and the assessment of emerging artemisinin resistance in a multinational cohort. Proc Natl Acad Sci U S A 114:3515–3520. doi: 10.1073/pnas.1615875114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Amaratunga C, Sreng S, Suon S, Phelps ES, Stepniewska K, Lim P, Zhou C, Mao S, Anderson JM, Lindegardh N, Jiang H, Song J, Su X-z, White NJ, Dondorp AM, Anderson TJC, Fay MP, Mu J, Duong S, Fairhurst RM. 2012. Artemisinin-resistant Plasmodium falciparum in Pursat Province, Western Cambodia: a parasite clearance rate study. Lancet Infect Dis 12:851–858. doi: 10.1016/S1473-3099(12)70181-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Phyo AP, Nkhoma S, Stepniewska K, Ashley EA, Nair S, McGready R, Ler Moo C, Al-Saai S, Dondorp AM, Lwin KM, Singhasivanon P, Day NPJ, White NJ, Anderson TJC, Nosten F. 2012. Emergence of artemisinin-resistant malaria on the Western border of Thailand: a longitudinal study. Lancet 379:1960–1966. doi: 10.1016/S0140-6736(12)60484-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Witkowski B, Amaratunga C, Khim N, Sreng S, Chim P, Kim S, Lim P, Mao S, Sopha C, Sam B, Anderson JM, Duong S, Chuor CM, Taylor WR, Suon S, Mercereau-Puijalon O, Fairhurst RM, Menard D. 2013. Novel phenotypic assays for the detection of artemisinin-resistant Plasmodium falciparum malaria in Cambodia: in-vitro and ex-vivo drug-response studies. Lancet Infect Dis 13:1043–1049. doi: 10.1016/S1473-3099(13)70252-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ariey F, Witkowski B, Amaratunga C, Beghain J, Langlois A-C, Khim N, Kim S, Duru V, Bouchier C, Ma L, Lim P, Leang R, Duong S, Sreng S, Suon S, Chuor CM, Bout DM, Ménard S, Rogers WO, Genton B, Fandeur T, Miotto O, Ringwald P, Le Bras J, Berry A, Barale J-C, Fairhurst RM, Benoit-Vical F, Mercereau-Puijalon O, Ménard D. 2014. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature 505:50–55. doi: 10.1038/nature12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spring MD, Lin JT, Manning JE, Vanachayangkul P, Somethy S, Bun R, Se Y, Chann S, Ittiverakul M, Sia-Ngam P, Kuntawunginn W, Arsanok M, Buathong N, Chaorattanakawee S, Gosi P, Ta-Aksorn W, Chanarat N, Sundrakes S, Kong N, Heng TK, Nou S, Teja-Isavadharm P, Pichyangkul S, Phann ST, Balasubramanian S, Juliano JJ, Meshnick SR, Chour CM, Prom S, Lanteri CA, Lon C, Saunders DL. 2015. Dihydroartemisinin-piperaquine failure associated with a triple mutant including Kelch13 C580Y in Cambodia: an observational cohort study. Lancet Infect Dis 15:683–691. doi: 10.1016/S1473-3099(15)70049-6. [DOI] [PubMed] [Google Scholar]
- 25.Miotto O, Amato R, Ashley EA, MacInnis B, Almagro-Garcia J, Amaratunga C, Lim P, Mead D, Oyola SO, Dhorda M, Imwong M, Woodrow C, Manske M, Stalker J, Drury E, Campino S, Amenga-Etego L, Thanh TN, Tran HT, Ringwald P, Bethell D, Nosten F, Phyo AP, Pukrittayakamee S, Chotivanich K, Chuor CM, Nguon C, Suon S, Sreng S, Newton PN, Mayxay M, Khanthavong M, Hongvanthong B, Htut Y, Han KT, Kyaw MP, Faiz MA, Fanello CI, Onyamboko M, Mokuolu OA, Jacob CG, Takala-Harrison S, Plowe CV, Day NP, Dondorp AM, Spencer CC, McVean G, Fairhurst RM, White NJ, Kwiatkowski DP. 2015. Genetic architecture of artemisinin-resistant Plasmodium falciparum. Nat Genet 47:226–234. doi: 10.1038/ng.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mbengue A, Bhattacharjee S, Pandharkar T, Liu H, Estiu G, Stahelin RV, Rizk SS, Njimoh DL, Ryan Y, Chotivanich K, Nguon C, Ghorbal M, Lopez-Rubio JJ, Pfrender M, Emrich S, Mohandas N, Dondorp AM, Wiest O, Haldar K. 2015. A molecular mechanism of artemisinin resistance in Plasmodium falciparum malaria. Nature 520:683–687. doi: 10.1038/nature14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Phyo AP, Ashley EA, Anderson TJC, Bozdech Z, Carrara VI, Sriprawat K, Nair S, White MM, Dziekan J, Ling C, Proux S, Konghahong K, Jeeyapant A, Woodrow CJ, Imwong M, McGready R, Lwin KM, Day NPJ, White NJ, Nosten F. 2016. Declining efficacy of artemisinin combination therapy against P falciparum malaria on the Thai-Myanmar border (2003-2013): the role of parasite genetic factors. Clin Infect Dis 63:784–791. doi: 10.1093/cid/ciw388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Straimer J, Gnadig NF, Witkowski B, Amaratunga C, Duru V, Ramadani AP, Dacheux M, Khim N, Zhang L, Lam S, Gregory PD, Urnov FD, Mercereau-Puijalon O, Benoit-Vical F, Fairhurst RM, Menard D, Fidock DA. 2015. K13-propeller mutations confer artemisinin resistance in Plasmodium falciparum clinical isolates. Science 347:428–431. doi: 10.1126/science.1260867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ghorbal M, Gorman M, Macpherson CR, Martins RM, Scherf A, Lopez-Rubio J-J. 2014. Genome editing in the human malaria parasite Plasmodium falciparum using the CRISPR-Cas9 system. Nat Biotechnol 32:819–821. doi: 10.1038/nbt.2925. [DOI] [PubMed] [Google Scholar]
- 30.Krishna S, Kremsner PG. 2013. Antidogmatic approaches to artemisinin resistance: reappraisal as treatment failure with artemisinin combination therapy. Trends Parasitol 29:313–317. doi: 10.1016/j.pt.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 31.Ferreira PE, Culleton R, Gil JP, Meshnick SR. 2013. Artemisinin resistance in Plasmodium falciparum: what is it really? Trends Parasitol 29:318–320. doi: 10.1016/j.pt.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 32.Hastings IM, Kay K, Hodel EM. 2016. The importance of scientific debate in the identification, containment, and control of artemisinin resistance. Clin Infect Dis 63:1527–1528. doi: 10.1093/cid/ciw581. [DOI] [PubMed] [Google Scholar]
- 33.Bethell D, Se Y, Lon C, Tyner S, Saunders D, Sriwichai S, Darapiseth S, Teja-Isavadharm P, Khemawoot P, Schaecher K, Ruttvisutinunt W, Lin J, Kuntawungin W, Gosi P, Timmermans A, Smith B, Socheat D, Fukuda MM. 2011. Artesunate dose escalation for the treatment of uncomplicated malaria in a region of reported artemisinin resistance: a randomized clinical trial. PLoS One 6:e19283. doi: 10.1371/journal.pone.0019283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saunders D, Khemawoot P, Vanachayangkul P, Siripokasupkul R, Bethell D, Tyner S, Se Y, Rutvisuttinunt W, Sriwichai S, Chanthap L, Lin J, Timmermans A, Socheat D, Ringwald P, Noedl H, Smith B, Fukuda M, Teja-Isavadharm P. 2012. Pharmacokinetics and pharmacodynamics of oral artesunate monotherapy in patients with uncomplicated Plasmodium falciparum malaria in western Cambodia. Antimicrob Agents Chemother 56:5484–5493. doi: 10.1128/AAC.00044-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kheang ST, Sovannaroth S, Ek S, Chy S, Chhun P, Mao S, Nguon S, Lek DS, Menard D, Kak N. 2017. Prevalence of K13 mutation and day-3 positive parasitaemia in artemisinin-resistant malaria endemic area of Cambodia: a cross-sectional study. Malar J 16:372. doi: 10.1186/s12936-017-2024-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.MalariaGEN Plasmodium falciparum Community Project. 2016. Genomic epidemiology of artemisinin resistant malaria. Elife 5:e08714. doi: 10.7554/eLife.08714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sa JM, Kaslow SR, Krause MA, Melendez-Muniz VA, Salzman RE, Kite WA, Zhang M, Moraes Barros RR, Mu J, Han PK, Mershon JP, Figan CE, Caleon RL, Rahman RS, Gibson TJ, Amaratunga C, Nishiguchi EP, Breglio KF, Engels TM, Velmurugan S, Ricklefs S, Straimer J, Gnadig NF, Deng B, Liu A, Diouf A, Miura K, Tullo GS, Eastman RT, Chakravarty S, James ER, Udenze K, Li S, Sturdevant DE, Gwadz RW, Porcella SF, Long CA, Fidock DA, Thomas ML, Fay MP, Sim BKL, Hoffman SL, Adams JH, Fairhurst RM, Su XZ, Wellems TE. 2018. Artemisinin resistance phenotypes and K13 inheritance in a Plasmodium falciparum cross and Aotus model. Proc Natl Acad Sci U S A 115:12513–12518. doi: 10.1073/pnas.1813386115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gabryszewski SJ, Modchang C, Musset L, Chookajorn T, Fidock DA. 2016. Combinatorial genetic modeling of pfcrt-mediated drug resistance evolution in Plasmodium falciparum. Mol Biol Evol 33:1554–1570. doi: 10.1093/molbev/msw037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laufer MK, Takala-Harrison S, Dzinjalamala FK, Stine OC, Taylor TE, Plowe CV. 2010. Return of chloroquine-susceptible falciparum malaria in Malawi was a reexpansion of diverse susceptible parasites. J Infect Dis 202:801–808. doi: 10.1086/655659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Straimer J, Gnädig NF, Stokes BH, Ehrenberger M, Crane AA, Fidock DA. 2017. Plasmodium falciparum K13 mutations differentially impact ozonide susceptibility and parasite fitness in vitro. mBio 8:e00172-17. doi: 10.1128/mBio.00172-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takala-Harrison S, Jacob CG, Arze C, Cummings MP, Silva JC, Dondorp AM, Fukuda MM, Hien TT, Mayxay M, Noedl H, Nosten F, Kyaw MP, Nhien NTT, Imwong M, Bethell D, Se Y, Lon C, Tyner SD, Saunders DL, Ariey F, Mercereau-Puijalon O, Menard D, Newton PN, Khanthavong M, Hongvanthong B, Starzengruber P, Fuehrer H-P, Swoboda P, Khan WA, Phyo AP, Nyunt MM, Nyunt MH, Brown TS, Adams M, Pepin CS, Bailey J, Tan JC, Ferdig MT, Clark TG, Miotto O, MacInnis B, Kwiatkowski DP, White NJ, Ringwald P, Plowe CV. 2015. Independent emergence of artemisinin resistance mutations among Plasmodium falciparum in Southeast Asia. J Infect Dis 211:670–679. doi: 10.1093/infdis/jiu491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nair S, Li X, Arya GA, McDew-White M, Ferrari M, Nosten F, Anderson TJC. 2018. Fitness costs and the rapid spread of Kelch13-C580Y substitutions conferring artemisinin resistance. Antimicrob Agents Chemother 62:e00605-18. doi: 10.1128/AAC.00605-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang T, Yeoh LM, Tutor MV, Dixon MW, McMillan PJ, Xie SC, Bridgford JL, Gillett DL, Duffy MF, Ralph SA, McConville MJ, Tilley L, Cobbold SA. 2019. Decreased K13 abundance reduces hemoglobin catabolism and proteotoxic stress, underpinning artemisinin resistance. Cell Rep 29:2917.e5–2928.e5. doi: 10.1016/j.celrep.2019.10.095. [DOI] [PubMed] [Google Scholar]
- 44.Birnbaum J, Scharf S, Schmidt S, Jonscher E, Hoeijmakers WAM, Flemming S, Toenhake CG, Schmitt M, Sabitzki R, Bergmann B, Fröhlke U, Mesén-Ramírez P, Blancke Soares A, Herrmann H, Bártfai R, Spielmann T. 2020. A Kelch13-defined endocytosis pathway mediates artemisinin resistance in malaria parasites. Science 367:51–59. doi: 10.1126/science.aax4735. [DOI] [PubMed] [Google Scholar]
- 45.Gnädig NF, Stokes BH, Edwards RL, Kalantarov GF, Heimsch KC, Kuderjavy M, Crane A, Lee MCS, Straimer J, Becker K, Trakht IN, Odom John AR, Mok S, Fidock DA. 2020. Insights into the intracellular localization, protein associations and artemisinin resistance properties of Plasmodium falciparum K13. PLoS Pathog 16:e1008482. doi: 10.1371/journal.ppat.1008482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Simwela NV, Hughes KR, Roberts AB, Rennie MT, Barrett MP, Waters AP. 2020. Experimentally engineered mutations in a ubiquitin hydrolase, UBP-1, modulate in vivo susceptibility to artemisinin and chloroquine in Plasmodium berghei. Antimicrob Agents Chemother 64:e02484-19. doi: 10.1128/AAC.02484-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hunt P, Afonso A, Creasey A, Culleton R, Sidhu AB, Logan J, Valderramos SG, McNae I, Cheesman S, do Rosario V, Carter R, Fidock DA, Cravo P. 2007. Gene encoding a deubiquitinating enzyme is mutated in artesunate- and chloroquine-resistant rodent malaria parasites. Mol Microbiol 65:27–40. doi: 10.1111/j.1365-2958.2007.05753.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Henrici RC, van Schalkwyk DA, Sutherland CJ. 2019. Modification of pfap2mu and pfubp1 markedly reduces ring-stage susceptibility of Plasmodium falciparum to artemisinin in vitro. Antimicrob Agents Chemother 64:e01542-19. doi: 10.1128/AAC.01542-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dogovski C, Xie SC, Burgio G, Bridgford J, Mok S, McCaw JM, Chotivanich K, Kenny S, Gnadig N, Straimer J, Bozdech Z, Fidock DA, Simpson JA, Dondorp AM, Foote S, Klonis N, Tilley L. 2015. Targeting the cell stress response of Plasmodium falciparum to overcome artemisinin resistance. PLoS Biol 13:e1002132. doi: 10.1371/journal.pbio.1002132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mok S, Ashley EA, Ferreira PE, Zhu L, Lin Z, Yeo T, Chotivanich K, Imwong M, Pukrittayakamee S, Dhorda M, Nguon C, Lim P, Amaratunga C, Suon S, Hien TT, Htut Y, Faiz MA, Onyamboko MA, Mayxay M, Newton PN, Tripura R, Woodrow CJ, Miotto O, Kwiatkowski DP, Nosten F, Day NPJ, Preiser PR, White NJ, Dondorp AM, Fairhurst RM, Bozdech Z. 2015. Drug resistance. Population transcriptomics of human malaria parasites reveals the mechanism of artemisinin resistance. Science 347:431–435. doi: 10.1126/science.1260403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee AH, Symington LS, Fidock DA. 2014. DNA repair mechanisms and their biological roles in the malaria parasite Plasmodium falciparum. Microbiol Mol Biol Rev 78:469–486. doi: 10.1128/MMBR.00059-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Franke-Fayard B, Djokovic D, Dooren MW, Ramesar J, Waters AP, Falade MO, Kranendonk M, Martinelli A, Cravo P, Janse CJ. 2008. Simple and sensitive antimalarial drug screening in vitro and in vivo using transgenic luciferase expressing Plasmodium berghei parasites. Int J Parasitol 38:1651–1662. doi: 10.1016/j.ijpara.2008.05.012. [DOI] [PubMed] [Google Scholar]
- 53.Janse CJ, Waters AP, Kos J, Lugt CB. 1994. Comparison of in vivo and in vitro antimalarial activity of artemisinin, dihydroartemisinin and sodium artesunate in the Plasmodium berghei-rodent model. Int J Parasitol 24:589–594. doi: 10.1016/0020-7519(94)90150-3. [DOI] [PubMed] [Google Scholar]
- 54.Lee RS, Waters AP, Brewer JM. 2018. A cryptic cycle in haematopoietic niches promotes initiation of malaria transmission and evasion of chemotherapy. Nat Commun 9:1689. doi: 10.1038/s41467-018-04108-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bridgford JL, Xie SC, Cobbold SA, Pasaje CFA, Herrmann S, Yang T, Gillett DL, Dick LR, Ralph SA, Dogovski C, Spillman NJ, Tilley L. 2018. Artemisinin kills malaria parasites by damaging proteins and inhibiting the proteasome. Nat Commun 9:3801. doi: 10.1038/s41467-018-06221-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li H, O'Donoghue AJ, van der Linden WA, Xie SC, Yoo E, Foe IT, Tilley L, Craik CS, da Fonseca PCA, Bogyo M. 2016. Structure- and function-based design of Plasmodium-selective proteasome inhibitors. Nature 530:233–236. doi: 10.1038/nature16936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yoo E, Stokes BH, de Jong H, Vanaerschot M, Kumar T, Lawrence N, Njoroge M, Garcia A, Van der Westhuyzen R, Momper JD, Ng CL, Fidock DA, Bogyo M. 2018. Defining the determinants of specificity of Plasmodium proteasome inhibitors. J Am Chem Soc 140:11424–11437. doi: 10.1021/jacs.8b06656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fidock DA, Rosenthal PJ, Croft SL, Brun R, Nwaka S. 2004. Antimalarial drug discovery: efficacy models for compound screening. Nat Rev Drug Discov 3:509–520. doi: 10.1038/nrd1416. [DOI] [PubMed] [Google Scholar]
- 59.Ménard D, Khim N, Beghain J, Adegnika AA, Shafiul-Alam M, Amodu O, Rahim-Awab G, Barnadas C, Berry A, Boum Y, Bustos MD, Cao J, Chen J-H, Collet L, Cui L, Thakur G-D, Dieye A, Djallé D, Dorkenoo MA, Eboumbou-Moukoko CE, Espino F-E-CJ, Fandeur T, Ferreira-da-Cruz M-F, Fola AA, Fuehrer H-P, Hassan AM, Herrera S, Hongvanthong B, Houzé S, Ibrahim ML, Jahirul-Karim M, Jiang L, Kano S, Ali-Khan W, Khanthavong M, Kremsner PG, Lacerda M, Leang R, Leelawong M, Li M, Lin K, Mazarati J-B, Ménard S, Morlais I, Muhindo-Mavoko H, Musset L, Na-Bangchang K, Nambozi M, Niaré K, Noedl H, KARMA Consortium, et al. 2016. A worldwide map of Plasmodium falciparum K13-propeller polymorphisms. N Engl J Med 374:2453–2464. doi: 10.1056/NEJMoa1513137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Anderson TJ, Nair S, McDew-White M, Cheeseman IH, Nkhoma S, Bilgic F, McGready R, Ashley E, Pyae Phyo A, White NJ, Nosten F. 2017. Population parameters underlying an ongoing soft sweep in Southeast Asian malaria parasites. Mol Biol Evol 34:131–144. doi: 10.1093/molbev/msw228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Amaratunga C, Witkowski B, Dek D, Try V, Khim N, Miotto O, Ménard D, Fairhurst RM. 2014. Plasmodium falciparum founder populations in western Cambodia have reduced artemisinin sensitivity in vitro. Antimicrob Agents Chemother 58:4935–4937. doi: 10.1128/AAC.03055-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Siddiqui FA, Boonhok R, Cabrera M, Mbenda HGN, Wang M, Min H, Liang X, Qin J, Zhu X, Miao J, Cao Y, Cui L. 2020. Role of Plasmodium falciparum Kelch13 protein mutations in P falciparum populations from northeastern Myanmar in mediating artemisinin resistance. mBio 11:e01134-19. doi: 10.1128/mBio.01134-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang J, Huang Y, Zhao Y, Ye R, Zhang D, Pan W. 2018. Introduction of F446I mutation in the K13 propeller gene leads to increased ring survival rates in Plasmodium falciparum isolates. Malar J 17:248. doi: 10.1186/s12936-018-2396-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang Z, Wang Y, Cabrera M, Zhang Y, Gupta B, Wu Y, Kemirembe K, Hu Y, Liang X, Brashear A, Shrestha S, Li X, Miao J, Sun X, Yang Z, Cui L. 2015. Artemisinin resistance at the China-Myanmar border and association with mutations in the K13 propeller gene. Antimicrob Agents Chemother 59:6952–6959. doi: 10.1128/AAC.01255-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huang F, Takala-Harrison S, Jacob CG, Liu H, Sun X, Yang H, Nyunt MM, Adams M, Zhou S, Xia Z, Ringwald P, Bustos MD, Tang L, Plowe CV. 2015. A single mutation in K13 predominates in southern China and is associated with delayed clearance of Plasmodium falciparum following artemisinin treatment. J Infect Dis 212:1629–1635. doi: 10.1093/infdis/jiv249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tun KM, Jeeyapant A, Imwong M, Thein M, Aung SS, Hlaing TM, Yuentrakul P, Promnarate C, Dhorda M, Woodrow CJ, Dondorp AM, Ashley EA, Smithuis FM, White NJ, Day NP. 2016. Parasite clearance rates in upper Myanmar indicate a distinctive artemisinin resistance phenotype: a therapeutic efficacy study. Malar J 15:185. doi: 10.1186/s12936-016-1240-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tun KM, Imwong M, Lwin KM, Win AA, Hlaing TM, Hlaing T, Lin K, Kyaw MP, Plewes K, Faiz MA, Dhorda M, Cheah PY, Pukrittayakamee S, Ashley EA, Anderson TJC, Nair S, McDew-White M, Flegg JA, Grist EPM, Guerin P, Maude RJ, Smithuis F, Dondorp AM, Day NPJ, Nosten F, White NJ, Woodrow CJ. 2015. Spread of artemisinin-resistant Plasmodium falciparum in Myanmar: a cross-sectional survey of the K13 molecular marker. Lancet Infect Dis 15:415–421. doi: 10.1016/S1473-3099(15)70032-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nyunt MH, Hlaing T, Oo HW, Tin-Oo L-LK, Phway HP, Wang B, Zaw NN, Han SS, Tun T, San KK, Kyaw MP, Han E-T. 2015. Molecular assessment of artemisinin resistance markers, polymorphisms in the K13 propeller, and a multidrug-resistance gene in the eastern and western border areas of Myanmar. Clin Infect Dis 60:1208–1215. doi: 10.1093/cid/ciu1160. [DOI] [PubMed] [Google Scholar]
- 69.Li H, Ponder EL, Verdoes M, Asbjornsdottir KH, Deu E, Edgington LE, Lee JT, Kirk CJ, Demo SD, Williamson KC, Bogyo M. 2012. Validation of the proteasome as a therapeutic target in Plasmodium using an epoxyketone inhibitor with parasite-specific toxicity. Chem Biol 19:1535–1545. doi: 10.1016/j.chembiol.2012.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gantt SM, Myung JM, Briones MR, Li WD, Corey EJ, Omura S, Nussenzweig V, Sinnis P. 1998. Proteasome inhibitors block development of Plasmodium spp. Antimicrob Agents Chemother 42:2731–2738. doi: 10.1128/AAC.42.10.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stokes BH, Yoo E, Murithi JM, Luth MR, Afanasyev P, da Fonseca PCA, Winzeler EA, Ng CL, Bogyo M, Fidock DA. 2019. Covalent Plasmodium falciparum-selective proteasome inhibitors exhibit a low propensity for generating resistance in vitro and synergize with multiple antimalarial agents. PLoS Pathog 15:e1007722. doi: 10.1371/journal.ppat.1007722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mukherjee A, Bopp S, Magistrado P, Wong W, Daniels R, Demas A, Schaffner S, Amaratunga C, Lim P, Dhorda M, Miotto O, Woodrow C, Ashley EA, Dondorp AM, White NJ, Wirth D, Fairhurst R, Volkman SK. 2017. Artemisinin resistance without PfKelch13 mutations in Plasmodium falciparum isolates from Cambodia. Malar J 16:195. doi: 10.1186/s12936-017-1845-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal omega. Mol Syst Biol 7:539–539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Heckman KL, Pease LR. 2007. Gene splicing and mutagenesis by PCR-driven overlap extension. Nat Protoc 2:924–932. doi: 10.1038/nprot.2007.132. [DOI] [PubMed] [Google Scholar]
- 75.Burda P-C, Roelli MA, Schaffner M, Khan SM, Janse CJ, Heussler VT. 2015. A Plasmodium phospholipase is involved in disruption of the liver stage parasitophorous vacuole membrane. PLoS Pathog 11:e1004760. doi: 10.1371/journal.ppat.1004760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Philip N, Orr R, Waters AP. 2013. Transfection of rodent malaria parasites. Methods Mol Biol 923:99–125. doi: 10.1007/978-1-62703-026-7_7. [DOI] [PubMed] [Google Scholar]
- 77.Rueden CT, Schindelin J, Hiner MC, DeZonia BE, Walter AE, Arena ET, Eliceiri KW. 2017. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics 18:529. doi: 10.1186/s12859-017-1934-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fivelman QL, Adagu IS, Warhurst DC. 2004. Modified fixed-ratio isobologram method for studying in vitro interactions between atovaquone and proguanil or dihydroartemisinin against drug-resistant strains of Plasmodium falciparum. Antimicrob Agents Chemother 48:4097–4102. doi: 10.1128/AAC.48.11.4097-4102.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Boampong JN, Ameyaw EO, Aboagye B, Asare K, Kyei S, Donfack JH, Woode E. 2013. The curative and prophylactic effects of xylopic acid on plasmodium berghei infection in mice. J Parasitol Res 2013:356107. doi: 10.1155/2013/356107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vega-Rodríguez J, Pastrana-Mena R, Crespo-Lladó KN, Ortiz JG, Ferrer-Rodríguez I, Serrano AE. 2015. Implications of glutathione levels in the Plasmodium berghei response to chloroquine and artemisinin. PLoS One 10:e0128212. doi: 10.1371/journal.pone.0128212. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Schematic and amino acid alignment of P. falciparum and P. berghei K13. (A) P. falciparum K13 protein showing amino acid positions and the protein domains. Positions of K13 mutations that have been investigated in this study are indicated. Equivalent amino acid positions for P. berghei are indicated in parallel at the bottom (in blue). (B) Protein alignment of P. falciparum and P. berghei K13 showing conservation at the mutation sites. Alignments were carried out using Clustal Omega protein alignment tool. Conserved residues are indicated by “*.” Download FIG S1, TIF file, 2.9 MB (2.9MB, tif) .
Copyright © 2020 Simwela et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
CRISPR/Cas9 editing strategy and RFLP analysis of P. berghei K13 mutant lines. (A) Schematic of CRISPR/Cas9 strategy used to introduce K13 mutations into P. berghei asexual blood-stage parasites. For this, 20-bp sgRNA targeting regions within 0 to 30 bp of the mutation site (see Tables S1 and S2) were designed to contain Esp3I digestion overhangs and were cloned into the Cas9 plasmid ABR099, as previously described (46). Donor templates were generated by overlapping extension PCR as described in Materials and Methods and were subsequently cloned into the ABR099 plasmids carrying the appropriate sgRNA (Tables S1 and S2) at the HincII linker site. The donor templates carried the mutation of interest as well as silent mutations to introduce a restriction site for RFLP analysis and to inactivate the PAM site recognition sequence (as illustrated in the schematic). Details of plasmids, sgRNA, and lines generated are in Table S1. (B) RFLP analysis of transfected parasite populations before and after challenge with artesunate (AS) at 20 or 64 mg/kg for the G2022 (C592Y.1), G2023 (C592Y.2), G2024 (I555T), and G2025 (R551T) lines. RFLP analysis was carried out using the indicated restriction enzymes on PCR fragments amplified from genomic DNA of transfected parasites. PCRs used primers that bound on either side of the donor DNA (Table S1 and S2). (C) RFLP analysis of parasite bulk populations for the G1957 (F458I), G1979 (Y505H), and G1989 (M488I) transfected lines. RFLP analysis was carried out using the indicated restriction enzymes. In both RFLP analyses, “*” indicates that these lines were uncloned. Download FIG S2, TIF file, 2.8 MB (2.7MB, tif) .
Copyright © 2020 Simwela et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DNA sequence and further RFLP analysis of P. berghei K13 mutant lines. (A) Sequencing analysis of the G2025 line showing the presence of traces for both the silent mutations and the R551T substation in the original transfection. DNA sequencing showing the absence of the C592Y and I555T nucleotide substitutions and the presence of minor traces of the silent mutations in the G2023 (B) and G2024 (C) transfected lines; “*” on the transfectant parasite lines indicates that the line was not cloned. (D) RFLP analysis of the G2042 (C592Y, sgRNA 2, TAT codon), G2043 (C592Y, sgRNA 2, TAC codon), and G2044 (C592Y, sgRNA 1, TAC codon) transfected lines (Table S1), showing further unsuccessful attempts to introduce the C592Y in P. berghei. RFLP analysis of the G2045 control line (C592C, sgRNA 1, silent mutations control) where editing was readily achieved is shown for comparison. (E) Sequencing analysis of the G2045 line showing successful editing with high efficiency to introduce silent mutations without the C592Y substitution. DNA sequence analysis showing high efficiency editing to introduce silent mutations and mutations of interest in the G1957 (F458I) (F), G1989 (M488I) (G), and G1979 (Y505H) (H) lines. (I) RFLP analysis with indicated restriction enzymes for the cloned parasite lines G1957 (F458I), G1979 (Y505H), G1989 (M488I), and G2025 (R551T, AS 64 mg/kg). Download FIG S3, TIF file, 2.8 MB (2.8MB, tif) .
Copyright © 2020 Simwela et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Plasmids, generated lines, transfection efficiencies, and outcome genotypes. RFLP analysis of the bulk transfected parasites was carried out on PCR fragments amplified using diagnostic PCR primers exterior of the donor template: GU5300 plus GU5301 (1,111 bp) for K13 and GU5186 plus GU4895 (946 bp) for UBP-1. Download Table S1, XLSX file, 0.1 MB (11KB, xlsx) .
Copyright © 2020 Simwela et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Clearance kinetics of P. berghei K13 mutants upon AS treatment. Parasite clearance curves in mice with established parasitemias of K13 mutant lines following treatment with 64 mg/kg artesunate (AS) at the indicated times (see arrows) for the three cohorts. See Materials and Methods and Fig. 3A. Download FIG S4, TIF file, 2.7 MB (3.3MB, tif) .
Copyright © 2020 Simwela et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Microscopic analysis of P. berghei K13 mutants upon AS treatment. Microscopic analysis of Giemsa-stained thin blood smears showing preferential survival of UBP-1 (B and H) and K13 mutant parasites G1957F458I (C and I), G1979Y505H (D and J), G1989M488I (E and K), and G2025R551T (F and L) compared to wild-type parasites (A and G) upon treatment with AS. Cohorts 1 and 2 are shown in panels A to F, and cohorts 3 and 4 are shown in panels G to L. Smears were taken at time points corresponding to those shown in the clearance plots in Fig. 3A and in Fig. S4. Second and third dose treatment days are indicated by black arrows. Red arrows indicate viable parasites. Viability was deemed significant if at least 4 viable parasites were observed upon observation of at least 10 microscopic fields. Download FIG S5, TIF file, 2.8 MB (2.7MB, tif) .
Copyright © 2020 Simwela et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Growth competition of the parent 1804WT and UBP-1 V2721F mutant line compared to the G159WT in the presence or absence of AS or CQ. Growth competition assays with the wild-type parent 1804WT or the UBP-1 G1980V2721F line compared to wild-type GFP-expressing G159WT line in the presence or absence of artesunate (AS) or chloroquine (CQ) drug pressure. Parasites were mixed at a 1:1 ratio, injected into mice, and left treated or untreated with AS at 50 mg/kg or CQ at 15 mg/kg as described in Materials and Methods and Fig. 4. (A) Percentage population changes of the 1804WT and wild-type G159WT in the absence of drug and on the day of recrudescence on day 9 for AS or CQ. (B) Proportion representations of the G159WT line in mixture with G1980V2721F, in the absence of drug or upon AS or CQ treatment, on the days of recrudescence. Download FIG S6, TIF file, 2.7 MB (2.7MB, tif) .
Copyright © 2020 Simwela et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
List of primers used in the study and their descriptions. Download Table S2, XLSX file, 0.1 MB (11.9KB, xlsx) .
Copyright © 2020 Simwela et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Recrudescence of P. berghei K13 and UBP-1 mutants compared to that of the wild type in infected mice treated with ART at 80 mg/kg as described for Fig. 3B. Groups of three or four mice (M0 to M4) were infected with ∼106 parasites on day 0 and treated from 3 h with ART at 80 mg/kg as indicated by arrows. A recrudescent event was recorded as “−” for negative smears or “+” with associated parasitemia (% infected erythrocytes). Refers to Fig. 3B. Download Table S3, XLSX file, 0.1 MB (15KB, xlsx) .
Copyright © 2020 Simwela et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ΣFIC50 values for DHA and EY5-125 drug combination ratios in the 1804WT, G1989M488I, and G2025R551T lines. ΣFIC50 values are derived from FIC50 data as plotted in Fig. 5C. Download Table S4, XLSX file, 0.1 MB (9.9KB, xlsx) .
Copyright © 2020 Simwela et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.