Abstract
The ferric uptake regulator (Fur) is a global transcription factor that regulates intracellular iron homeostasis in bacteria. The current hypothesis states that when the intracellular “free” iron concentration is elevated, Fur binds ferrous iron, and the iron-bound Fur represses the genes encoding for iron uptake systems and stimulates the genes encoding for iron storage proteins. However, the “iron-bound” Fur has never been isolated from any bacteria. Here we report that the Escherichia coli Fur has a bright red color when expressed in E. coli mutant cells containing an elevated intracellular free iron content because of deletion of the iron–sulfur cluster assembly proteins IscA and SufA. The acid-labile iron and sulfide content analyses in conjunction with the EPR and Mössbauer spectroscopy measurements and the site-directed mutagenesis studies show that the red Fur protein binds a [2Fe-2S] cluster via conserved cysteine residues. The occupancy of the [2Fe-2S] cluster in Fur protein is ∼31% in the E. coli iscA/sufA mutant cells and is decreased to ∼4% in WT E. coli cells. Depletion of the intracellular free iron content using the membrane-permeable iron chelator 2,2´-dipyridyl effectively removes the [2Fe-2S] cluster from Fur in E. coli cells, suggesting that Fur senses the intracellular free iron content via reversible binding of a [2Fe-2S] cluster. The binding of the [2Fe-2S] cluster in Fur appears to be highly conserved, because the Fur homolog from Hemophilus influenzae expressed in E. coli cells also reversibly binds a [2Fe-2S] cluster to sense intracellular iron homeostasis.
Keywords: iron homeostasis, iron–sulfur cluster, ferric uptake regulator (Fur), iron–sulfur protein, iron metabolism, repressor protein, Escherichia coli (E. coli), Mossbauer spectroscopy, electron paramagnetic resonance (EPR), intracellular iron homeostasis, IscA
The bacterial intracellular “free” iron concentration is primarily regulated by a global transcription factor, Ferric uptake regulator (Fur) (1–4). It has been generally assumed that when the intracellular free iron concentration is elevated, Fur binds free ferrous iron, and the iron-bound Fur represses the genes encoding for iron uptake systems and stimulates the genes encoding for iron storage proteins (5–9). The crystallographic studies of the Fur proteins from Escherichia coli (10), Mycobacterium tuberculosis (11), Vibrio cholerae (12), Helicobacter pylori (13), Campylobacter jejuni (14), and Francisella tularensis (15) have revealed that Fur protein exists as a homodimer or tetramer (8) with each monomer containing three putative metal-binding sites. The first metal-binding site (site 1) is coordinated by His-87, Asp-89, Glu-108, and His-125 (the residue numbers are based on the E. coli Fur), whereas the second site (site 2) is coordinated by His-33, Glu-81, His-88, and His-90 (12). The third metal-binding site (site 3) is formed by three conserved cysteine residues (Cys-93, Cys-96, and Cys-133) (11–15). However, the metal-binding sites in purified Fur proteins are often occupied by zinc or other metal ions, and the “iron-bound” Fur has never been isolated from any bacteria.
Iron–sulfur proteins are the major iron-containing proteins in cells (16). Recent studies have demonstrated that iron–sulfur clusters in proteins are assembled by a group of dedicated proteins (17, 18). Among the iron–sulfur cluster assembly proteins in E. coli, IscA has been characterized as an alternative scaffold (19) or iron chaperone to recruit the intracellular free iron for iron–sulfur cluster assembly (20, 21). Depletion of IscA and its homologs inhibits the [4Fe-4S] cluster assembly without affecting the [2Fe-2S] cluster assembly in E. coli (22), Saccharomyces cerevisiae (23), and human (24) cells, indicating that the [2Fe-2S] clusters and [4Fe-4S] clusters have distinct biogenesis pathways (25). Furthermore, deletion of IscA and its homologs significantly increases the intracellular free iron content in E. coli (22), S. cerevisiae (26), and human (24) cells. Inspired by these observations, we reasoned that the global iron regulator Fur may become iron-bound in the E. coli mutant cells in which IscA and its paralog SufA are deleted. Here, we find that recombinant E. coli Fur protein indeed has a bright red color when expressed in the E. coli iscA/sufA mutant cells under aerobic growth conditions. The iron and sulfide content analyses and the EPR and Mössbauer spectroscopy measurements show that the red Fur protein binds a novel [2Fe-2S] cluster. Site-directed mutagenesis studies further indicate that the conserved cysteine residues (Cys-93, Cys-96, and Cys-133) in the E. coli Fur are required for the binding of the [2Fe-2S] cluster. The occupancy of the [2Fe-2S] cluster in Fur protein is ∼31% when expressed in the E. coli iscA/sufA mutant cells and is decreased to ∼4% in WT E. coli cells. Moreover, when the intracellular free iron content is depleted using the membrane-permeable iron chelator 2,2´-dipyridyl (200 μm), the [2Fe-2S] cluster in Fur is effectively removed in both WT and iscA/sufA mutant E. coli cells. Because the addition of 2,2´-dipyridyl (200 μm) to E. coli cells switches on the expression of the Fur-repressed targeted genes in E. coli cells (27), we propose that the E. coli Fur may sense the intracellular free iron content via reversible binding of a [2Fe-2S] cluster. Importantly, binding of the [2Fe-2S] cluster in Fur appears to be highly conserved as the Fur homolog from Hemophilus influenzae can also bind a [2Fe-2S] cluster via the conserved cysteine residues when expressed in E. coli cells.
Results
Deletion of the iron–sulfur cluster assembly protein IscA and its paralog SufA leads to accumulation of the intracellular free iron content in E. coli cells
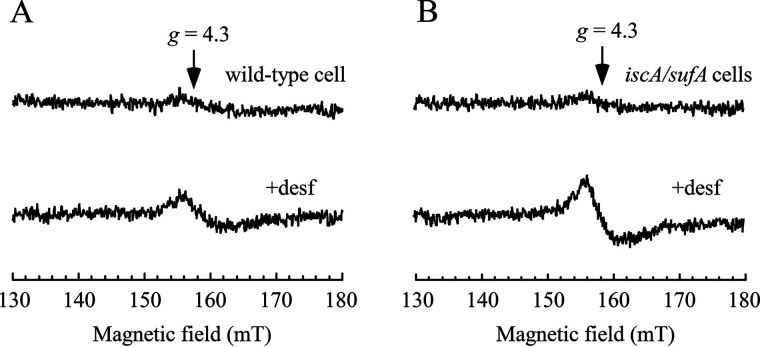
In S. cerevisiae (26) and human (24) cells, depletion of the iron–sulfur cluster assembly protein IscA homologs results in substantial iron accumulation in mitochondria. To evaluate the intracellular free iron content in the E. coli mutant cells in which IscA and its paralog SufA were deleted (22), we used the whole-cell EPR measurements following the procedures described in Ref. 28. Briefly, exponentially growing E. coli cells were treated with the membrane-permeable iron chelator desferrioxamine. The cells were then washed with the membrane-impermeable iron chelator diethylenetriaminepentaacetic acid to remove the extracellular free iron. Because the desferrioxamine-ferric iron complex has an EPR signal at g = 4.3, the amplitude of the EPR signal represents the relative concentration of the intracellular “chelatable” iron (28). The detected intracellular chelatable iron pool has been defined as the free iron associated with metabolites (e.g. GSH, citrate, and phosphorylated sugar intermediates) (6, 28–30). As shown in Fig. 1, the intracellular chelatable iron content in the iscA/sufA mutant cells is approximately two times that of WT cells grown in LB medium under aerobic conditions. This assay is of limited utility because it cannot observe ferrous iron and ferric nanoparticles (both of which have been identified in cells (31, 32)); therefore, it would be difficult to determine the exact concentration of the intracellular free iron in E. coli cells. Nevertheless, the results in Fig. 1 clearly suggested that deletion of IscA and its homolog SufA in E. coli cells increases the intracellular chelatable iron content, consistent with the previous observations made in S. cerevisiae (26) and human (24) cells.
Figure 1.
Deletion of IscA and SufA results in accumulation of the intracellular free iron content in E. coli cells. A, EPR spectra of E. coli WT cells treated with or without desferrioxamine (desf). B, EPR spectra of the E. coli iscA/sufA mutant cells treated with or without desferrioxamine. The experimental conditions are described under “Experimental procedures.” The data are representative of three independent experiments.
The ferric uptake regulator (Fur) has a bright red color when expressed in the E. coli iscA/sufA mutant cells
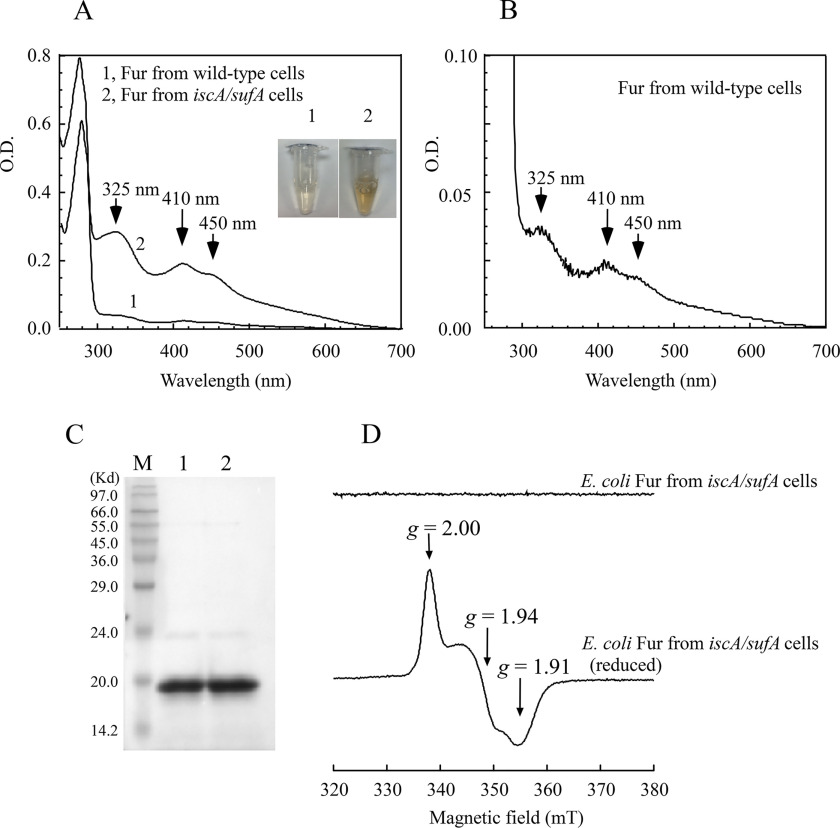
Here, we took advantage of the E. coli iscA/sufA mutant, which has an elevated intracellular free iron content (Fig. 1), to explore the possible iron binding of the global iron regulator Fur in vivo. In the experiments, we introduced a plasmid (pBAD) expressing the E. coli Fur into E. coli WT and the iscA/sufA mutant cells grown in LB medium under aerobic conditions. Recombinant Fur protein was purified from both cells. As reported previously by other research groups (33–35), the Fur protein purified from WT E. coli cells is essentially colorless. In contrast, the Fur protein purified from the E. coli iscA/sufA mutant cells has a bright red color (inset in Fig. 2A). The UV-visible absorption measurements showed that the red Fur protein has three major absorption peaks at 325, 410, and 450 nm, in addition to the protein peak at 280 nm (Fig. 2A), suggesting that the red Fur protein may bind a mononuclear iron or iron–sulfur cluster.
Figure 2.
The E. coli Fur has a bright red color when expressed in the E. coli iscA/sufA mutant cells. Recombinant E. coli Fur was expressed in the E. coli WT and the E. coli iscA/sufA mutant cells grown in LB medium under aerobic conditions. A, UV-visible absorption spectra of purified Fur proteins. Spectrum 1, Fur protein purified from WT E. coli cells. Spectrum 2, Fur protein purified from the iscA/sufA mutant cells. The protein (100 μm) was dissolved in buffer containing 20 mm Tris (pH 8.0) and 500 mm NaCl. Inset, photographs of purified Fur proteins from WT (panel 1) and the iscA/sufA mutant (panel 2) cells. B, UV-visible absorption spectrum of the Fur protein purified from WT E. coli cells. Same as in A, except the y axis is expanded by 8-fold. C, SDS-PAGE gel of purified Fur proteins from WT E. coli cells (lane 1) and from the iscA/sufA mutant cells (lane 2). Lane M, molecular mass ladder. D, EPR spectra of the E. coli Fur protein purified from the iscA/sufA mutant cells. The Fur protein (500 μm) was reduced with freshly prepared sodium dithionite (10 mm) and immediately frozen in liquid nitrogen until the EPR measurement.
Careful examination of the UV-visible spectra in Fig. 2A revealed that the Fur protein purified from WT E. coli cells also has the absorption peaks at 325, 410, and 450 nm, although their amplitudes are only approximately one-eighth of those of the red Fur protein purified from the E. coli iscA/sufA mutant cells (Fig. 2B). The small amplitudes of the absorption peaks at 325, 410, and 450 nm of the Fur protein purified from WT E. coli cells might have been overlooked previously, especially when the concentration of purified Fur protein was low. Thus, the red Fur protein is present not only in the iscA/sufA mutant cells but also in WT E. coli cells, and the relative concentration of the red Fur protein in the iscA/sufA mutant cells is approximately eight times that in WT cells.
The red Fur protein contains a [2Fe-2S] cluster
To determine whether the red Fur protein binds a mononuclear iron or iron–sulfur cluster, we first analyzed the acid-labile iron and sulfide contents of the protein. From three independent experiments, we found 0.6 ± 0.2 iron and 0.4 ± 0.2 sulfide atoms per each Fur monomer in the purified red Fur protein, suggesting that the red Fur protein likely contains an iron–sulfur cluster. Because the E. coli Fur has previously been characterized as a zinc-binding protein (33), we also analyzed the zinc content of purified Fur and found that the zinc content is decreased from 1.8 ± 0.3 atoms per Fur monomer when purified from WT E. coli cells to 1.4 ± 0.2 atoms per Fur monomer when purified from the iscA/sufA mutant cells (n = 3), indicating that binding of iron–sulfur cluster affects zinc binding in Fur.
The iron–sulfur cluster in the red Fur protein was further investigated by the EPR spectroscopy. Although as-purified red Fur protein has no EPR signals, the dithionite-reduced red Fur protein has an EPR signal at gx = 1.91, gy = 1.94, and gz = 2.00 (Fig. 2D), which is similar to that of other iron–sulfur proteins (36, 37), thus confirming that the red Fur protein contains an iron–sulfur cluster. Interestingly, unlike other iron–sulfur proteins, the reduced [2Fe-2S] cluster in Fur is not stable and quickly decomposes, thus preventing spin quantification of the [2Fe-2S] cluster in Fur using the EPR spectroscopy.
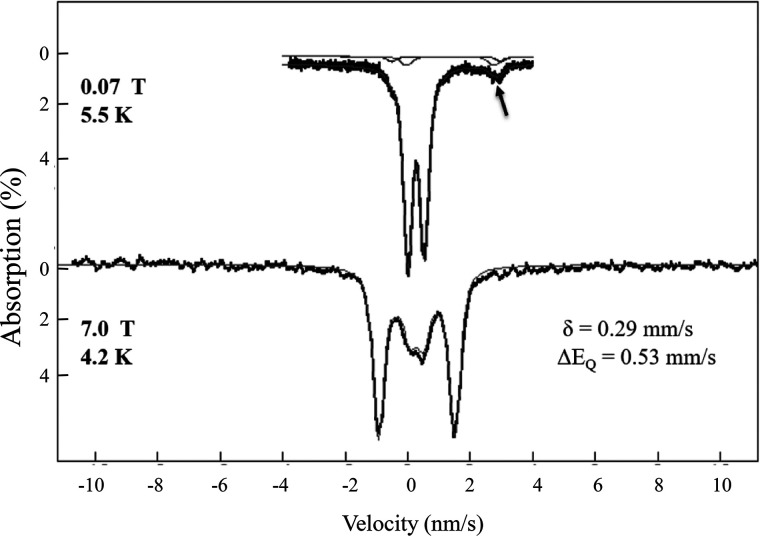
To explore the nature of the iron–sulfur cluster in the red Fur protein, we have utilized Mössbauer spectroscopy (38). The 57Fe-enriched Fur protein was prepared from the E. coli iscA/sufA mutant cells grown in 57Fe-enriched M9 minimum medium. The Mössbauer spectroscopy was conducted at variable temperatures and in applied magnetic fields (38, 39). As shown in Fig. 3, the 5.5 K Mössbauer spectrum in a 70-mT applied field exhibits a well-defined quadrupole doublet with isomer shift δ = 0.29(2) mm/s, ΔEQ of 0.53(1) mm/s and line widths of 0.33 mm/s. These parameters are typical for [2Fe-2S]2+ clusters with distorted tetrahedral thiolate coordination (39, 40) and are very different from the Mössbauer spectrum of the in vitro ferrous iron-reconstituted E. coli Fur which has an isomer shift of δ = 1.19 (1) mm/s and a quadrupole splitting of ΔEQ = 3.47 (2) mm/s (41). The spectrum in 7.0-T applied magnetic field exhibits magnetic splitting that could be attributed solely to the applied field. The absence of internal fields in the 7.0-T spectrum indicates that the cluster in the sample is diamagnetic, consistent with two iron Fe3+ ions, which are antiferromagnetically coupled. Together with the observed values of the δ and ΔEQ, which are similar to values seen for the protein-bound [2Fe-2S] clusters (39, 40), the diamagnetism of the sample provides unambiguous evidence for the presence of an oxidized [2Fe-2S]2+ cluster, which is also consistent with the UV-visible absorption spectrum (Fig. 2A). The spectra in low applied magnetic fields show a minor spectral component, indicated by the small arrow in Fig. 3 (top spectrum). Collecting spectra at 180 K, at which the fast spin fluctuation limit is a reasonable assumption, affords the quantification of all iron in the sample in terms of two components: the major [2Fe-2S] cluster component and the minor ferrous iron component. Based on these spectra, the ferrous iron component contributes ∼8% of the total iron in the sample, and the rest of the iron (∼92% of the total iron) exists as a [2Fe-2S] cluster in the Fur protein. At low temperature (6 K or below), the ferrous component can further be simulated with two distinct doublets with equal contributions (δ = 1.23(1)/ΔEQ = 3.50 mm/s and δ = 1.36/ΔEQ = 2.80 mm/s), which correspond to high-spin ferrous iron with pseudo-octahedral N/O coordination (42). The ferrous components appear as two small quadrupole doublets in the 0.07-T spectrum, and they are in the background, broadened by hyperfine in interactions in the high-field spectrum. Thus, the Mössbauer spectroscopy studies demonstrated that the red Fur protein primarily binds a [2Fe-2S] cluster likely with cysteine coordination.
Figure 3.
Variable-field Mössbauer spectra of the 57Fe-enriched Fur protein purified from the E. coli iscA/sufA mutant cells. The 57Fe-labeled E. coli Fur protein was purified from the E. coli iscA/sufA mutant cells growing in M9 minimum medium supplemented with 57Fe (10 μm). The protein concentration of 57Fe-labeled Fur was ∼1.0 mm. Mössbauer spectra were collected at cryogenic temperatures at 5.5 K and 70 mT (top spectrum) and 4.2 K and 7.0 T (bottom spectrum). The magnetic field was applied parallel to the observed γ-radiation. Hash marks are raw data, and lines are spectral simulations with the parameters shown and discussed in the text. The line widths (full width at half-maximum) are 0.33 mm/s. The arrow marks the high-energy line of the ferrous components discussed in the text.
The cysteine residues in Fur are the likely ligands for the [2Fe-2S] cluster
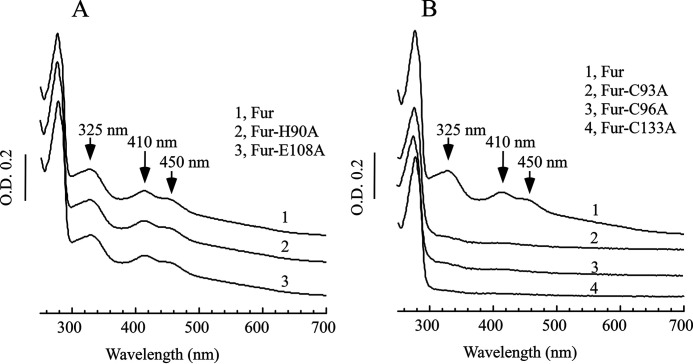
The E. coli Fur protein has three distinct metal-binding sites (10). Sites 1 and 2 are coordinated by His, Asp, and Glu, and binding of zinc in these sites has been attributed to stabilization of the Fur structure (10). To test whether site 1 or site 2 is involved in binding the [2Fe-2S] cluster, we constructed the Fur mutants in which the selected amino acid residues in site 1 or site 2 were replaced with alanine. The Fur mutants were then expressed in the E. coli iscA/sufA mutant cells and purified. Fig. 4A shows the results of two E. coli Fur mutants (Glu-108 to Ala of site 1 and His-90 to Ala of site 2). Both mutations have very little or no effect on the [2Fe-2S] cluster binding in the Fur protein when expressed in the E. coli iscA/sufA mutant cells. Similar results were also observed when His-87 (site 1) or His-88 (site 2) was replaced with Ala (data not shown). Thus, sites 1 and 2 of the E. coli Fur protein are likely not involved in binding the [2Fe-2S] cluster.
Figure 4.
The conserved cysteine residues in the E. coli Fur are the ligands for the [2Fe-2S] cluster. The Fur mutant proteins were constructed by the site-directed mutagenesis and purified from the E. coli iscA/sufA mutant cells. A, UV-visible absorption spectra of the WT Fur (spectrum 1), the mutant H190A (spectrum 2), and the mutant E108A (spectrum 3). B, UV-visible absorption spectra of the WT Fur (spectrum 1), the mutant C93A (spectrum 2), the mutant C96A (spectrum 3), and the mutant C133A (spectrum 4). Each spectrum was offset for clarity. The protein concentrations were ∼100 μm. The results are representative of three independent experiments.
The third metal-binding site in the E. coli Fur protein consists of three conserved cysteine residues (Cys-93, Cys-96, and Cys-133) (10). We thus constructed Fur mutants in which each of the cysteine residues was replaced with alanine. Fig. 4B shows that when Cys-93, Cys-96, or Cys-133 was replaced with Ala, the E. coli Fur mutant protein failed to bind the [2Fe-2S] cluster when expressed in the E. coli iscA/sufA mutant cells, suggesting that these cysteine residues are required for the E. coli Fur to bind the [2Fe-2S] cluster. The fourth cysteine residue (Cys-138) in the E. coli Fur is not conserved. Attempts to replace Cys-138 with Ala in the E. coli Fur protein were not successful. We instead replaced Cys-138 with Ser and found that mutation of Cys-138 to Ser did not abolish the [2Fe-2S] cluster binding in the E. coli Fur but significantly changed the UV-visible absorption spectrum of the [2Fe-2S] cluster in Fur (Fig. S1), indicating that Cys-138 could provide the fourth ligand for the [2Fe-2S] cluster in Fur. Regardless, the results clearly suggested that the E. coli Fur protein likely binds the [2Fe-2S] cluster via the conserved cysteine residues (Fig. 4B), which is consistent with the Mössbauer spectroscopy data (Fig. 3).
Depletion of the intracellular free iron content removes the [2Fe-2S] cluster from the Fur in E. coli cells
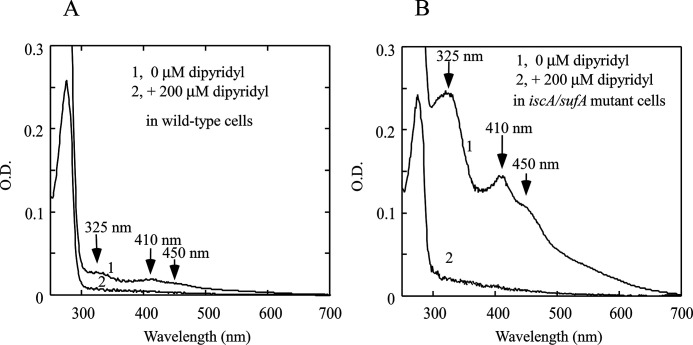
If the [2Fe-2S] cluster-bound Fur protein represents an active form of the Fur repressor in E. coli cells when the intracellular free iron content is elevated, it is expected that depletion of the intracellular free iron content will remove the [2Fe-2S] cluster from Fur. Because the membrane-permeable iron chelator 2,2´-dipyridyl has often been used to deplete the intracellular free iron content in E. coli cells (27), we treated WT E. coli cells expressing recombinant Fur protein with 2,2´-dipyridyl. Fur protein was then purified from the cells. Fig. 5A shows that addition of 200 μm of 2,2´-dipyridyl effectively removes the [2Fe-2S] cluster from the Fur protein in WT E. coli cells. We also treated the E. coli iscA/sufA mutant cells expressing recombinant Fur with 200 μm of 2,2´-dipyridyl and found that the [2Fe-2S] cluster is also removed from the Fur protein in the E. coli iscA/sufA mutant cells by 2,2´-dipyridyl (Fig. 5B). Furthermore, the binding of the [2Fe-2S] cluster in Fur is reversible in E. coli cells, because addition of ferrous ammonium sulfate (300 μm) to the 2,2´-dipyridyl–treated E. coli cells restores the [2Fe-S] cluster binding in Fur (data not shown). Because addition of 2,2´-dipyridyl to E. coli cells switches on the expression of the Fur-repressed targeted genes in E. coli cells (27), we propose that removal of the [2Fe-2S] cluster from Fur in E. coli cells by 2,2´-dipyridyl may represent deactivation of Fur in response to depletion of the intracellular free iron content and that Fur senses the intracellular free iron content via reversible binding of the [2Fe-2S] cluster in E. coli cells.
Figure 5.
Depletion of intracellular free iron content removes the [2Fe-2S] cluster from Fur protein in E. coli cells. A, UV-visible absorption spectra of the E. coli Fur protein. Fur proteins were purified from the WT E. coli cells treated with 0 μm (spectrum 1) or 200 μm (spectrum 2) of 2,2´-dipyridyl, respectively. B, UV-visible absorption spectra of the E. coli Fur proteins. Fur proteins were purified from the E. coli iscA/sufA mutant cells treated with 0 μm (spectrum 1) or 200 μm (spectrum 2) of 2,2´-dipyridyl, respectively. The concentrations of purified proteins were ∼60 μm. The results are representative of three independent experiments.
The H. influenzae Fur also binds a [2Fe-2S] cluster
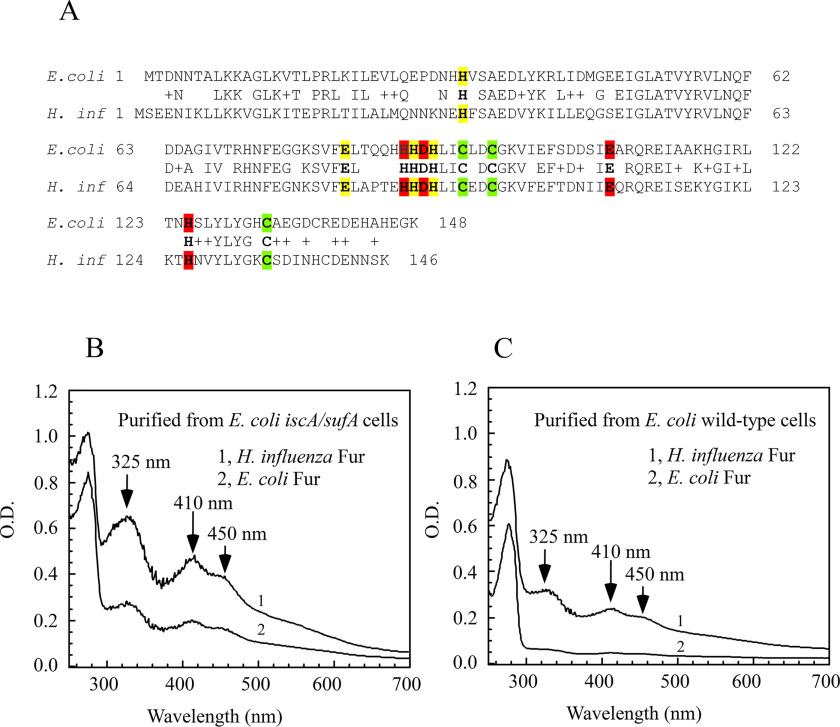
The Fur protein is highly conserved among bacteria (10–15). To test whether other Fur proteins can also bind a [2Fe-2S] cluster, we synthesized a gene encoding the Fur homolog from H. influenzae, a Gram-negative facultatively anaerobic pathogenic bacterium. The H. influenzae Fur has 65% identity and 77% similarity with the E. coli Fur (Fig. 6A). The H. influenzae Fur protein was expressed in the E. coli iscA/sufA mutant cells and purified from the cells. As shown in Fig. 6B, the purified H. influenzae Fur has the same absorption peaks at 325, 410, and 450 nm as the E. coli red Fur protein. Furthermore, mutations of the conserved cysteine residues (Cys-94, Cys-97, and Cys-134) to Ala in the H. influenzae Fur eliminated the [2Fe-2S] cluster binding (data not shown), suggesting that like the E. coli Fur, the H. influenzae Fur binds the [2Fe-2S] cluster via the conserved cysteine residues.
Figure 6.
The H. influenzae Fur also binds a [2Fe-2S] cluster when expressed in E. coli cells. A, the sequence alignment of the H. influenzae Fur and the E. coli Fur. The three metal-binding sites in Fur proteins are highlighted in red (site 1), yellow (site 2), and green (site 3), respectively. B, UV-visible absorption spectra of the H. influenzae Fur (spectrum 1) and the E. coli Fur (spectrum 2) purified from the E. coli iscA/sufA mutant cells. C, UV-visible absorption spectra of the H. influenzae Fur (spectrum 1) and the E. coli Fur (spectrum 2) purified from the E. coli WT cells. The protein concentrations were 100 μm. The results are representative of three independent experiments.
Interestingly, although the protein yield of the H. influenzae Fur was similar to that of the E. coli Fur when expressed in the E. coli iscA/sufA mutant cells, the amplitudes of the absorption peaks at 325, 410, and 450 nm of the purified H. influenzae Fur were much higher than those of the red E. coli Fur (Fig. 6B). Assuming that the extinction coefficient of the [2Fe-2S] cluster in Fur proteins at 450 nm is 10 mm−1 cm−1 (36, 37), we estimated that the occupancy of the [2Fe-2S] cluster in the H. influenzae Fur expressed in the E. coli iscA/sufA mutant cells is ∼68%, whereas that of the E. coli Fur protein is ∼31% (Fig. 6B). This result implies that the H. influenzae Fur has a higher binding affinity for the [2Fe-2S] cluster than the E. coli Fur in the E. coli iscA/sufA mutant cells. To test this idea further, we expressed the H. influenzae Fur and the E. coli Fur in WT E. coli cells and purified both proteins. As shown in Fig. 6C, the occupancy of the [2Fe-2S] cluster in the H. influenzae Fur (∼30%) is again much higher than that of the E. coli Fur (∼ 4%) when expressed in WT E. coli cells. We also carried out the in vitro reconstitution experiments. When the apo-form E. coli Fur was incubated with 4-fold excess of ferrous iron and sulfide in the presence of DTT, only ∼5% of the apo-form E. coli Fur was reconstituted with a [2Fe-2S] cluster. In contrast, when the apo-form H. influenzae Fur was incubated with 4-fold excess of ferrous iron and sulfide under the same experimental conditions, ∼50% of the apo-form H. influenzae Fur was reconstituted with a [2Fe-2S] cluster (Fig. S2). Thus, whereas both the E. coli Fur and the H. influenzae Fur can bind a [2Fe-2S] cluster via conserved cysteine residues, the H. influenzae Fur has a much higher binding affinity for the [2Fe-2S] cluster than the E. coli Fur, suggesting that E. coli and H. influenzae may have distinct genetic responses to intracellular iron homeostasis via Fur.
Discussion
In the past decades, it has been well-established that when the intracellular free iron content is elevated in bacteria, the global transcription factor Fur binds free ferrous iron to repress the genes encoding for iron uptake systems and to stimulate the genes encoding for iron storage proteins in bacteria (5–9). Although the purified E. coli Fur has been reconstituted with ferrous iron in vitro (41, 43), the iron-bound Fur has never been isolated from E. coli or any other bacteria. This could be because the intracellular free iron content is mainly regulated by Fur (5–9), and substantially increasing the intracellular free iron concentration could be challenging without deleting Fur in bacteria. The E. coli iscA/sufA mutant (22) provides a unique opportunity to explore the possible iron binding of Fur in vivo, because deficiency of iron–sulfur cluster biogenesis caused by deletion of IscA and its homologs increases the intracellular free iron content (22, 24, 26) (Fig. 1). Furthermore, deletion of IscA and its homologs only inhibits [4Fe-4S] cluster assembly without affecting [2Fe-2S] cluster assembly in E. coli (22), S. cerevisiae (23), and human (24) cells. Here, we took advantage of the E. coli iscA/sufA mutant cells (22) and found that the Fur protein expressed in the E. coli iscA/sufA mutant cells has a bright red color. The iron and sulfide content analyses in conjunction with the UV-visible absorption, EPR, and Mössbauer measurements suggest that the red Fur protein primarily binds a [2Fe-2S] cluster, and only a minor fraction of the mononuclear iron coordination (∼8% of total iron content) is associated with Fur (possibly in site 1 or 2). Additional studies reveal that the [2Fe-2S] cluster-bound Fur is present not only in the iscA/sufA mutant cells but also in WT E. coli cells (Fig. 2). The occupancy of the [2Fe-2S] cluster in the Fur protein is ∼31% in the E. coli iscA/sufA mutant cells and is decreased to ∼4% in WT E. coli cells. Furthermore, depletion of the intracellular free iron content using the membrane-permeable iron chelator 2,2´-dipyridyl (200 μm) effectively removes the [2Fe-2S] cluster from the Fur protein in both WT and iscA/sufA mutant cells (Fig. 5), suggesting that the E. coli Fur senses the intracellular free iron content via reversible binding of a [2Fe-2S] cluster in cells.
The UV-visible absorption and EPR spectra of the E. coli red Fur protein are reminiscent of [2Fe-2S] cluster-containing proteins (36, 37). The unambiguous evidence for the presence of a [2Fe-2S] cluster in the E. coli red Fur protein comes from the Mössbauer spectroscopic studies. In the literature, the Mössbauer isomer shifts (δ) for diamagnetic iron–sulfur clusters with tetrahedral cysteine ligation have been documented to be in the range of ∼0.27 mm/s for the [2Fe-2S]2+ and ∼0.45 mm/s for the [4Fe-4S]2+ (38) (Table S1). The Mössbauer parameters observed in the E. coli red Fur protein (δ = 0.29(2) mm/s and ΔEQ = 0.53(1)) (Fig. 3) represent a typical [2Fe-2S] cluster and are virtually identical to the [2Fe-2S] cluster associated with the oxygen-exposed FNR protein (δ = 0.28(1) mm/s and ΔEQ = 0.58(2) mm/s) (39) and the [2Fe-2S] cluster of the human mitochondrial glutaredoxin 2 (Grx2) (δ = 0.27 mm/s and ΔEQ = 0.60 mm/s) (40). It should be pointed out that the Mössbauer spectrum of the E. coli red Fur protein (Fig. 3) is very different from the Mössbauer spectrum of the in vitro ferrous iron-reconstituted E. coli Fur, which has an isomer shift of δ = 1.19 (1) mm/s and a quadrupole splitting of ΔEQ = 3.47(2) mm/s (41, 44), representing the binding of ferrous iron at site 2 via His-33, Glu-81, His-88, and His-90 in Fur protein (44). Although a small fraction (∼8%) of the total iron content in the red Fur protein is found to be the mononuclear iron component (some of the iron component could be generated during protein purification), ∼92% of the total iron content in the red Fur protein is assigned to the [2Fe-2S] cluster in the protein (Fig. 3). Thus, against all previous ideas, the E. coli Fur is a novel [2Fe-2S] cluster-binding protein.
Iron–sulfur clusters are the major group of iron-containing co-factors in cells. It has been reported that biogenesis of iron–sulfur clusters is regulated not only by the iron–sulfur cluster assembly transcription factor IscR (45) but also by the global iron regulator Fur (3, 46). Our finding that Fur senses the intracellular free iron content via binding of a [2Fe-2S] cluster provides a new aspect for the physiological link between intracellular free iron homeostasis and iron–sulfur cluster biogenesis. Use of an iron–sulfur cluster to sense the intracellular free iron content is not unprecedented. In mammalian cells, IRP-1 (iron regulatory protein 1) regulates the intracellular free iron content by reversible binding of a [4Fe-4S] cluster in response to an elevated intracellular free iron content (47). It is appealing to suggest that the E. coli Fur, like IRP-1, may also bind a [4Fe-4S] cluster in response to elevation of the intracellular free iron content. However, this is not likely, because (a) the E. coli iscA/sufA mutant cells cannot assemble [4Fe-4S] clusters in proteins under aerobic growth conditions (22), eliminating the possibility of the [4Fe-4S] cluster binding in Fur protein in the iscA/sufA mutant cells; (b) purification of recombinant Fur protein from the E. coli iscA/sufA cells under argon atmosphere does not significantly change the content of the [2Fe-2S] cluster in Fur protein (data not shown); (c) the H. influenzae Fur proteins expressed in both WT and iscA/sufA mutant E. coli cells contain the [2Fe-2S] cluster (Fig. 6); and (d) in yeast cells, the cellular iron sensors Yap5 of S. cerevisiae (48, 49) and Fep1 of Pichia pastoris (50) also bind a [2Fe-2S] cluster in response to an elevated intracellular free iron content. Thus, it is most likely that Fur binds a [2Fe-2S] cluster (not a [4Fe-4S] cluster or a mononuclear iron) when the intracellular free iron content is elevated in the E. coli iscA/sufA mutant cells. Use of a [2Fe-2S] cluster in Fur to sense the intracellular free iron content may represent physiological connections between intracellular iron homeostasis and regulation of acid tolerance, oxidative stress response, and bacterial virulence (8), because assembly of the [2Fe-2S] cluster in Fur requires not only the intracellular free iron but also sulfide that is derived from l-cysteine by cysteine desulfurase IscS (17). In this context, we propose that while elevation of the intracellular free iron content together with available sulfide leads to assembly of a [2Fe-2S] cluster in Fur and formation of an active Fur repressor in cells, depletion of the intracellular free iron content results in disassembly of the [2Fe-2S] cluster in Fur and inactivates Fur as a repressor. In WT E. coli cells, only ∼4% of Fur protein binds a [2Fe-2S] cluster, indicating that majority of Fur will be in an inactive form under normal growth conditions. In the E. coli iscA/sufA mutant cells, an elevated intracellular free iron content increases the occupancy of the [2Fe-2S] cluster in Fur protein to ∼31% (Fig. 2), which would shift a significant amount of inactive Fur to an active Fur repressor. Thus, reversible binding of the [2Fe-2S] cluster in Fur may reflect the intracellular free iron content and define the activity of Fur as a global transcription regulator in E. coli cells.
Fur is highly conserved among bacteria (11–15). With a few exceptions, the conserved three cysteine residues in Fur proteins from both Gram-negative and Gram-positive bacteria are arranged in a CX2CX37C motif. The CX2C sequence has often been associated with proteins that bind iron–sulfur clusters (51). Using the site-directed mutagenesis, we have identified three conserved cysteine residues in the E. coli Fur as the likely ligands for the [2Fe-2S] cluster. The fourth cysteine residue (Cys-138) is not conserved, and mutation of Cys-138 to Ser seemed to change the [2Fe-2S] cluster binding in the E. coli Fur. Thus, the fourth ligand for the [2Fe-2S] cluster could be Cys-138 in the E. coli Fur. It is worth mentioning that among 16 mutations of the metal-binding sites in the E. coli Fur protein, only Cys-93 and Cys-96 were found to be important for the repressor activity of Fur in E. coli cells (52), indicating that the [2Fe-2S] cluster binding could be crucial for the physiological function of Fur in E.coli cells. Interestingly, although both the E. coli Fur and the H. influenzae Fur can bind a [2Fe-2S] cluster via the conserved cysteine residues, the binding affinity of the H. influenzae Fur for the [2Fe-2S] cluster is significantly higher than that of the E. coli Fur in E. coli cells (Fig. 6, B and C). The higher binding affinity for the [2Fe-2S] cluster implies that the H. influenzae Fur will become an active repressor at a lower intracellular free iron content than the E. coli Fur. Thus, H. influenzae and E. coli likely have distinct genetic responses to intracellular iron homeostasis via Fur, and the higher binding affinity of the H. influenzae Fur for the [2Fe-2S] cluster could be vital for regulating intracellular iron homeostasis in H. influenzae. The specific amino acid residues that contribute to the higher binding affinity of the H. influenzae Fur for the [2Fe-2S] cluster and their physiological significance remain to be further investigated.
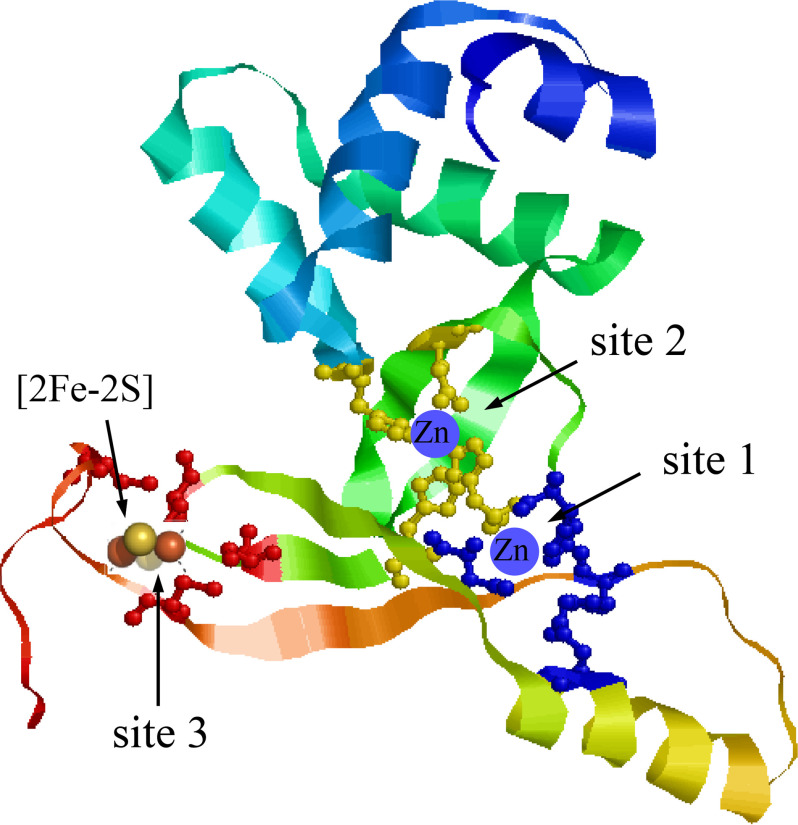
In summary, the E. coli Fur is a novel [2Fe-2S] protein, and occupancy of the [2Fe-2S] cluster in Fur is regulated by the intracellular free iron content. When the intracellular free iron content is elevated, Fur reversibly binds a [2Fe-2S] cluster via the conserved cysteine residues in E. coli cells. Because only the crystal structure of a truncated E. coli Fur (residues 1-82) is available (10), we have modeled a full-length E. coli Fur protein (Fig. 7) using the RaptorX structure prediction software (53). Overall, the predicted structure of the full-length E. coli Fur is similar to the crystal structures of Fur proteins from other bacteria (11–15). In the predicted model, the conserved cysteine residues (Cys-93, Cys-96, and Cys-113) in the E. coli Fur are closely positioned for hosting a [2Fe-2S] cluster (Fig. 7). We envision that binding of the [2Fe-2S] cluster will change the protein conformation of Fur in response to an elevated intracellular free iron content and switch an inactive Fur to an active [2Fe-2S]-bound Fur repressor in bacteria.
Figure 7.
A structure model of the full-length E. coli Fur. Full-length sequence of the E. coli Fur was modeled as described in Ref. 53. Site 1 is coordinated by His-87, Asp-89, Glu-108, and His-125. Site 2 is coordinated by His-33, Glu-81, His-88, and His-90. Site 3 is formed by Cys-93, Cys-96, and Cys-133. The structure model of the E. coli Fur was visualized using RasMol (57). The zinc-binding sites and the [2Fe-2S] cluster binding site are indicated.
Experimental procedures
E. coli strains
The E. coli iscA/sufA mutant was previously constructed from WT E. coli strain (MC4100) as described in Ref. 22. With exception of the Mössbauer sample preparation, E. coli WT and the iscA/sufA mutant strains were grown in LB medium at 37 °C under aerobic conditions.
Protein purification
Genes encoding the E. coli Fur and the H. influenzae Fur were synthesized (Genscript Co.) and cloned to plasmid pBAD for protein expression in E. coli cells. The plasmid with the cloned gene was introduced into the E. coli WT (MC4100) or the iscA/sufA mutant cells. Fur protein was overproduced in the E. coli cells by adding 0.2% l-arabinose for 4 h and purified following the procedure described in Ref. 33. In some experiments, an N-terminal His tag was used for quick purification of Fur protein from E. coli cells. The N-terminal His tag has no contribution to the [2Fe-2S] cluster binding in Fur protein, because the Fur protein with or without His tag purified from the E. coli iscA/sufA mutant cells have the same red color and same UV-visible absorption spectrum. The purity of all purified proteins was greater than 95% as judged by electrophoresis analysis on a 15% polyacrylamide gel containing SDS followed by staining with Coomassie Blue. The UV-visible absorption spectra of purified proteins were recorded in a Beckman DU640 UV-visible absorption spectrometer. The extinction coefficients for the E. coli apo-Fur and the H. influenzae apo-Fur at 280 nm are 6.2 and 6.9 mm−1 cm−1, respectively.
Site-directed mutagenesis studies
Site-directed mutagenesis was carried out using the QuikChange kit (Agilent Co.). The mutations were confirmed by direct sequencing (Eurofins Genomics Co.). The mutated Fur proteins were expressed in the E. coli iscA/sufA mutant cells and purified as described for the WT Fur protein.
Iron, sulfide, and zinc content analyses
Total iron content in protein samples was determined using the iron indicator ferrozine following the procedures described in Ref. 54. The absorption peak at 562 nm of the Fe(II)–ferrozine complex was used for quantifying the iron content using an extinction coefficient of 27.9 mm−1 cm−1. The sulfide content in protein samples was determined following the procedures described by Siegel (55). The zinc content in protein samples was determined using 4-(2-pyridylazo)-resorcinol as described in Ref. 56.
EPR measurements of purified fur
The X-band EPR spectra were recorded using a Bruker model ESR-300 spectrometer equipped with an Oxford Instruments 910 continuous flow cryostat. Routine EPR conditions were as follows: microwave frequency, 9.47 GHz; microwave power, 1.0 mW; modulation frequency, 100 kHz; modulation amplitude, 1.2 mT; temperature, 20 K; and receiver gain, 2 × 105.
Intracellular chelatable iron content analyses using the whole-cell EPR
The intracellular free iron content in E. coli cells were measured following the procedures described in Ref. 28. Overnight bacterial cultures were diluted 100-fold into 500 ml of fresh LB medium at 37 °C with agitation. The cultures were grown to an A600 of 0.2. The cells were harvested by centrifugation at 8000 rpm for 5 min, and the pellets were resuspended in LB containing 20 mm desferrioxamine to an A600 of 5.0. The cells were then incubated at 37 °C for 15 min and washed with 10 mm diethylenetriaminepentaacetic acid once. The suspension was further washed twice with 20 mm cold Tris-Cl (pH 7.4) and resuspended in 20 mm cold Tris-Cl (pH 7.4) containing 10% glycerol. The cell suspension was transferred to an EPR tube and frozen in liquid nitrogen until EPR measurements.
Mössbauer spectroscopy
For the Mössbauer experiments, the 57Fe-labeled Fur was prepared by expressing the protein in the E. coli iscA/sufA mutant cells grown in M9 minimum medium supplemented with 20 amino acids (each amino acid at 40 μg/ml), thiamine (0.5 μg/ml), glycerol (0.2%), and 57Fe (10 μm) under aerobic growth conditions. The 57Fe-labeled Fur purified from the E. coli iscA/sufA mutant cells had the same UV-visible absorption spectrum with the absorption peaks at 325, 410, and 450 nm. The Mössbauer spectra were recorded on a closed-cycle refrigerator spectrometer (model CCR4K; SeeCo, Edina, MN, USA) equipped with a 0.07-T permanent magnet, maintaining temperatures between 5 and 300 K. High-field (7.0 T) spectra were collected in Dr. Yisong (Alex) Guo's laboratory (Carnegie Mellon University) on a constant-acceleration spectrometer housed in a cryostat equipped with a superconducting magnet at 4.2 K. The samples consisted of buffered solutions of protein in Delrin 1.0-ml cups, frozen in liquid nitrogen. The isomer shifts are quoted at 5 K with respect to iron metal standard at 298 K. The Mössbauer spectra were analyzed using the software SpinCount (Michael Hendrich, Ph.D., Carnegie Mellon University and WMOSS4 (Ion Prisecaru).
Data availability
All data generated during this study are included in this published article and its supporting information files.
Supplementary Material
Acknowledgments
We gratefully acknowledge Dr. Yisong (Alex) Guo (Carnegie Mellon University) and his laboratory for collecting high-field Mössbauer spectra.
Footnotes
This article contains supporting information.
Author contributions—C. R. F., H. T., K. A. V., C. V. P., and H. D. data curation; C. R. F., H. T., K. A. V., C. V. P., and H. D. formal analysis; C. R. F., H. T., K. A. V., C. V. P., and H. D. validation; C. R. F., H. T., K. A. V., C. V. P., and H. D. investigation; C. R. F., H. T., K. A. V., C. V. P., and H. D. methodology; C. V. P. and H. D. resources; C. V. P. software; C. V. P. and H. D. supervision; C. V. P. and H. D. visualization; C. V. P. and H. D. project administration; C. V. P. and H. D. writing-review and editing; H. D. conceptualization; H. D. funding acquisition; H. D. writing-original draft.
Funding and additional information—This work was supported by National Institutes of Health Grant R15GM129564. C. R. F. was supported by The Louis Stokes Alliance for Minority Participation Bridge to Doctorate program at Louisiana State University from NSF. The Mössbauer spectroscopy work was funded by start-up funds from The College of Arts and Sciences at University of Saint Thomas (MN) to C. V. P. and a Collaborative Research Grant to K. A. V. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
References
- 1. Andrews S., Norton I., Salunkhe A. S., Goodluck H., Aly W. S., Mourad-Agha H., and Cornelis P. (2013) Control of iron metabolism in bacteria. Met. Ions Life Sci. 12, 203–239 10.1007/978-94-007-5561-1_7 [DOI] [PubMed] [Google Scholar]
- 2. Seo S. W., Kim D., Latif H., O'Brien E. J., Szubin R., and Palsson B. O. (2014) Deciphering Fur transcriptional regulatory network highlights its complex role beyond iron metabolism in Escherichia coli. Nat. Commun. 5, 4910 10.1038/ncomms5910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Roncarati D., Pelliciari S., Doniselli N., Maggi S., Vannini A., Valzania L., Mazzei L., Zambelli B., Rivetti C., and Danielli A. (2016) Metal-responsive promoter DNA compaction by the ferric uptake regulator. Nat. Commun. 7, 12593 10.1038/ncomms12593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wofford J. D., Bolaji N., Dziuba N., Outten F. W., and Lindahl P. A. (2019) Evidence that a respiratory shield in Escherichia coli protects a low-molecular-mass Fe(II) pool from O2-dependent oxidation. J. Biol. Chem. 294, 50–62 10.1074/jbc.RA118.005233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fillat M. F. (2014) The FUR (ferric uptake regulator) superfamily: diversity and versatility of key transcriptional regulators. Arch. Biochem. Biophys. 546, 41–52 10.1016/j.abb.2014.01.029 [DOI] [PubMed] [Google Scholar]
- 6. Beauchene N. A., Mettert E. L., Moore L. J., Keleş S., Willey E. R., and Kiley P. J. (2017) O2 availability impacts iron homeostasis in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 114, 12261–12266 10.1073/pnas.1707189114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pinochet-Barros A., and Helmann J. D. (2018) Redox sensing by Fe2+ in bacterial Fur family metalloregulators. Antioxid. Redox Signal. 29, 1858–1871 10.1089/ars.2017.7359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nader S., Pérard J., Carpentier P., Arnaud L., Crouzy S., and Michaud-Soret I. (2019) New insights into the tetrameric family of the Fur metalloregulators. Biometals 32, 501–519 10.1007/s10534-019-00201-8 [DOI] [PubMed] [Google Scholar]
- 9. Pi H., and Helmann J. D. (2017) Sequential induction of Fur-regulated genes in response to iron limitation in Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 114, 12785–12790 10.1073/pnas.1713008114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pecqueur L., D'Autreaux B., Dupuy J., Nicolet Y., Jacquamet L., Brutscher B., Michaud-Soret I., and Bersch B. (2006) Structural changes of Escherichia coli ferric uptake regulator during metal-dependent dimerization and activation explored by NMR and X-ray crystallography. J. Biol. Chem. 281, 21286–21295 10.1074/jbc.M601278200 [DOI] [PubMed] [Google Scholar]
- 11. Lucarelli D., Russo S., Garman E., Milano A., Meyer-Klaucke W., and Pohl E. (2007) Crystal structure and function of the zinc uptake regulator FurB from Mycobacterium tuberculosis. J. Biol. Chem. 282, 9914–9922 10.1074/jbc.M609974200 [DOI] [PubMed] [Google Scholar]
- 12. Sheikh M. A., and Taylor G. L. (2009) Crystal structure of the Vibrio cholerae ferric uptake regulator (Fur) reveals insights into metal co-ordination. Mol. Microbiol. 72, 1208–1220 10.1111/j.1365-2958.2009.06718.x [DOI] [PubMed] [Google Scholar]
- 13. Dian C., Vitale S., Leonard G. A., Bahlawane C., Fauquant C., Leduc D., Muller C., de Reuse H., Michaud-Soret I., and Terradot L. (2011) The structure of the Helicobacter pylori ferric uptake regulator Fur reveals three functional metal binding sites. Mol. Microbiol. 79, 1260–1275 10.1111/j.1365-2958.2010.07517.x [DOI] [PubMed] [Google Scholar]
- 14. Butcher J., Sarvan S., Brunzelle J. S., Couture J.-F., and Stintzi A. (2012) Structure and regulon of Campylobacter jejuni ferric uptake regulator Fur define apo-Fur regulation. Proc. Natl. Acad. Sci. U.S.A. 109, 10047–10052 10.1073/pnas.1118321109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pérard J., Nader S., Levert M., Arnaud L., Carpentier P., Siebert C., Blanquet F., Cavazza C., Renesto P., Schneider D., Maurin M., Coves J., Crouzy S., and Michaud-Soret I. (2018) Structural and functional studies of the metalloregulator Fur identify a promoter-binding mechanism and its role in Francisella tularensis virulence. Commun. Biol. 1, 93 10.1038/s42003-018-0095-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Johnson D. C., Dean D. R., Smith A. D., and Johnson M. K. (2005) Structure, function, and formation of biological iron–sulfur clusters. Annu. Rev. Biochem. 74, 247–281 10.1146/annurev.biochem.74.082803.133518 [DOI] [PubMed] [Google Scholar]
- 17. Zheng L., Cash V. L., Flint D. H., and Dean D. R. (1998) Assembly of iron–sulfur clusters: identification of an iscSUA-hscBA-fdx gene cluster from Azotobacter vinelandii. J. Biol. Chem. 273, 13264–13272 10.1074/jbc.273.21.13264 [DOI] [PubMed] [Google Scholar]
- 18. Braymer J. J., and Lill R. (2017) Iron–sulfur cluster biogenesis and trafficking in mitochondria. J. Biol. Chem. 292, 12754–12763 10.1074/jbc.R117.787101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ollagnier-De-Choudens S., Sanakis Y., and Fontecave M. (2004) SufA/IscA: reactivity studies of a class of scaffold proteins involved in [Fe-S] cluster assembly. J. Biol. Inorg. Chem. 9, 828–838 10.1007/s00775-004-0581-9 [DOI] [PubMed] [Google Scholar]
- 20. Yang J., Tan G., Zhang T., White R. H., Lu J., and Ding H. (2015) Deletion of the proposed iron chaperones IscA/SufA results in accumulation of a red intermediate cysteine desulfurase IscS in Escherichia coli. J. Biol. Chem. 290, 14226–14234 10.1074/jbc.M115.654269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yang J., Bitoun J. P., and Ding H. (2006) Interplay of IscA and IscU in biogenesis of iron–sulfur clusters. J. Biol. Chem. 281, 27956–27963 10.1074/jbc.M601356200 [DOI] [PubMed] [Google Scholar]
- 22. Tan G., Lu J., Bitoun J. P., Huang H., and Ding H. (2009) IscA/SufA paralogues are required for the 4Fe-4S cluster assembly in enzymes of multiple physiological pathways in Escherichia coli under aerobic growth conditions. Biochem. J. 420, 463–472 10.1042/BJ20090206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mühlenhoff U., Richter N., Pines O., Pierik A. J., and Lill R. (2011) Specialized function of yeast Isa1 and Isa2 in the maturation of mitochondrial [4Fe-4S] proteins. J. Biol. Chem. 286, 41205–41216 10.1074/jbc.M111.296152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sheftel A. D., Wilbrecht C., Stehling O., Niggemeyer B., Elsässer H. P., Mühlenhoff U., and Lill R. (2012) The human mitochondrial ISCA1, ISCA2, and IBA57 proteins are required for [4Fe-4S] protein maturation. Mol. Biol. Cell 23, 1157–1166 10.1091/mbc.E11-09-0772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Landry A. P., Cheng Z., and Ding H. (2013) Iron binding activity is essential for the function of IscA in iron–sulphur cluster biogenesis. Dalton Transactions 42, 3100–3106 10.1039/c2dt32000b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jensen L. T., and Culotta V. C. (2000) Role of Saccharomyces cerevisiae ISA1 and ISA2 in iron homeostasis. Mol. Cell Biol. 20, 3918–3927 10.1128/mcb.20.11.3918-3927.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McHugh J. P., Rodríguez-Quiñones F., Abdul-Tehrani H., Svistunenko D. A., Poole R. K., Cooper C. E., and Andrews S. C. (2003) Global iron-dependent gene regulation in Escherichia coli: a new mechanism for iron homeostasis. J. Biol. Chem. 278, 29478–29486 10.1074/jbc.M303381200 [DOI] [PubMed] [Google Scholar]
- 28. Woodmansee A. N., and Imlay J. A. (2002) Quantitation of intracellular free iron by electron paramagnetic resonance spectroscopy. Methods Enzymol. 349, 3–9 10.1016/S0076-6879(02)49316-0 [DOI] [PubMed] [Google Scholar]
- 29. Hider R. C., and Kong X. (2013) Iron speciation in the cytosol: an overview. Dalton Trans. 42, 3220–3229 10.1039/c2dt32149a [DOI] [PubMed] [Google Scholar]
- 30. Varghese S., Wu A., Park S., Imlay K. R., and Imlay J. A. (2007) Submicromolar hydrogen peroxide disrupts the ability of Fur protein to control free-iron levels in Escherichia coli. Mol. Microbiol. 64, 822–830 10.1111/j.1365-2958.2007.05701.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Miao R., Martinho M., Morales J. G., Kim H., Ellis E. A., Lill R., Hendrich M. P., Münck E., and Lindahl P. A. (2008) EPR and Mössbauer spectroscopy of intact mitochondria isolated from Yah1p-depleted Saccharomyces cerevisiae. Biochemistry 47, 9888–9899 10.1021/bi801047q [DOI] [PubMed] [Google Scholar]
- 32. Hudder B. N., Morales J. G., Stubna A., Münck E., Hendrich M. P., and Lindahl P. A. (2007) Electron paramagnetic resonance and Mössbauer spectroscopy of intact mitochondria from respiring Saccharomyces cerevisiae. J. Biol. Inorg. Chem. 12, 1029–1053 10.1007/s00775-007-0275-1 [DOI] [PubMed] [Google Scholar]
- 33. Althaus E. W., Outten C. E., Olson K. E., Cao H., and O'Halloran T. V. (1999) The ferric uptake regulation (Fur) repressor is a zinc metalloprotein. Biochemistry 38, 6559–6569 10.1021/bi982788s [DOI] [PubMed] [Google Scholar]
- 34. Mills S. A., and Marletta M. A. (2005) Metal binding characteristics and role of iron oxidation in the ferric uptake regulator from Escherichia coli. Biochemistry 44, 13553–13559 10.1021/bi0507579 [DOI] [PubMed] [Google Scholar]
- 35. Hohle T. H., and O'Brian M. R. (2016) Metal-specific control of gene expression mediated by Bradyrhizobium japonicum Mur and Escherichia coli Fur is determined by the cellular context. Mol. Microbiol. 101, 152–166 10.1111/mmi.13381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li H., Mapolelo D. T., Dingra N. N., Keller G., Riggs-Gelasco P. J., Winge D. R., Johnson M. K., and Outten C. E. (2011) Histidine 103 in Fra2 is an iron–sulfur cluster ligand in the [2Fe-2S] Fra2–Grx3 complex and is required for in vivo iron signaling in yeast. J. Biol. Chem. 286, 867–876 10.1074/jbc.M110.184176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cheng Z., Landry A. P., Wang Y., and Ding H. (2017) Binding of nitric oxide in CDGSH-type [2Fe-2S] clusters of the human mitochondrial protein Miner2. J. Biol. Chem. 292, 3146–3153 10.1074/jbc.M116.766774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pandelia M. E., Lanz N. D., Booker S. J., and Krebs C. (2015) Mössbauer spectroscopy of Fe/S proteins. Biochim. Biophys. Acta 1853, 1395–1405 10.1016/j.bbamcr.2014.12.005 [DOI] [PubMed] [Google Scholar]
- 39. Popescu C. V., Bates D. M., Beinert H., Münck E., and Kiley P. J. (1998) Mössbauer spectroscopy as a tool for the study of activation/inactivation of the transcription regulator FNR in whole cells of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 95, 13431–13435 10.1073/pnas.95.23.13431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lillig C. H., Berndt C., Vergnolle O., Lönn M. E., Hudemann C., Bill E., and Holmgren A. (2005) Characterization of human glutaredoxin 2 as iron–sulfur protein: a possible role as redox sensor. Proc. Natl. Acad. Sci. U.S.A. 102, 8168–8173 10.1073/pnas.0500735102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jacquamet L., Dole F., Jeandey C., Oddou J.-L., Perret E., Le Pape L., Aberdam D., Hazemann J.-L., Michaud-Soret I., and Latour J.-M. (2000) First spectroscopic characterization of Fe(II)–Fur, the physiological active form of the Fur protein. J. Am. Chem. Soc. 122, 394–395 10.1021/ja991932+ [DOI] [Google Scholar]
- 42. Gutlich P., Bill E., and Trautwein A. X. (2011) Mössbauer Spectroscopy and Transition Metal Chemistry: Fundamentals and Applications, Springer-Verlag, Berlin [Google Scholar]
- 43. Hamed M. Y., Neilands J. B., and Huynh V. (1993) Binding of the ferric uptake regulation repressor protein (Fur) to Mn(II), Fe(II), Co(II), and Cu(II) ions as co-repressors: electronic absorption, equilibrium, and 57Fe Mössbauer studies. J. Inorg. Biochem. 50, 193–210 10.1016/0162-0134(93)80025-5 [DOI] [PubMed] [Google Scholar]
- 44. Katigbak J., and Zhang Y. (2012) Iron binding site in a global regulator in bacteria: ferric uptake regulator (Fur) protein: structure, Mössbauer properties, and functional implication. J. Phys. Chem. Lett. 2012, 3503–3508 10.1021/jz301689b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mettert E. L., and Kiley P. J. (2014) Coordinate regulation of the Suf and Isc Fe-S cluster biogenesis pathways by IscR is essential for viability of Escherichia coli. J. Bacteriol. 196, 4315–4323 10.1128/JB.01975-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lee K. C., Yeo W. S., and Roe J. H. (2008) Oxidant-responsive induction of the suf operon, encoding a Fe-S assembly system, through Fur and IscR in Escherichia coli. J. Bacteriol. 190, 8244–8247 10.1128/JB.01161-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rouault T. A., and Maio N. (2017) Biogenesis and functions of mammalian iron–sulfur proteins in the regulation of iron homeostasis and pivotal metabolic pathways. J. Biol. Chem. 292, 12744–12753 10.1074/jbc.R117.789537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Li L., Miao R., Bertram S., Jia X., Ward D. M., and Kaplan J. (2012) A role for iron–sulfur clusters in the regulation of transcription factor Yap5-dependent high iron transcriptional responses in yeast. J. Biol. Chem. 287, 35709–35721 10.1074/jbc.M112.395533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rietzschel N., Pierik A. J., Bill E., Lill R., and Mühlenhoff U. (2015) The basic leucine zipper stress response regulator Yap5 senses high-iron conditions by coordination of [2Fe-2S] clusters. Mol. Cell Biol. 35, 370–378 10.1128/MCB.01033-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cutone A., Howes B. D., Miele A. E., Miele R., Giorgi A., Battistoni A., Smulevich G., Musci G., and di Patti M. C. B. (2016) Pichia pastoris Fep1 is a [2Fe-2S] protein with a Zn finger that displays an unusual oxygen-dependent role in cluster binding. Sci. Rep. 6, 31872 10.1038/srep31872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Belmonte L., and Mansy S. S. (2017) Patterns of ligands coordinated to metallocofactors extracted from the Protein Data Bank. J. Chem. Inf. Model. 57, 3162–3171 10.1021/acs.jcim.7b00468 [DOI] [PubMed] [Google Scholar]
- 52. Coy M., Doyle C., Besser J., and Neilands J. B. (1994) Site-directed mutagenesis of the ferric uptake regulation gene of Escherichia coli. Biometals 7, 292–298 10.1007/BF00144124 [DOI] [PubMed] [Google Scholar]
- 53. Zhu J., Wang S., Bu D., and Xu J. (2018) Protein threading using residue co-variation and deep learning. Bioinformatics 34, i263–i273 10.1093/bioinformatics/bty278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cowart R. E., Singleton F. L., and Hind J. S. (1993) A comparison of bathophenanthrolinedisulfonic acid and ferrozine as chelators of iron(II) in reduction reactions. Anal. Biochem. 211, 151–155 10.1006/abio.1993.1246 [DOI] [PubMed] [Google Scholar]
- 55. Siegel L. M. (1965) A direct microdetermination of sulfide. Anal. Biochem. 11, 126–132 10.1016/0003-2697(65)90051-5 [DOI] [PubMed] [Google Scholar]
- 56. Tan G., Landry A. P., Dai R., Wang L., Lu J., and Ding H. (2012) Competition of zinc ion for the 2Fe-2S cluster binding site in the diabetes drug target protein mitoNEET. Biometals 25, 1177–1184 10.1007/s10534-012-9580-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sayle R. A., and Milner-White E. J. (1995) RASMOL: biomolecular graphics for all. Trends Biochem. Sci. 20, 374 10.1016/S0968-0004(00)89080-5 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated during this study are included in this published article and its supporting information files.