Abstract
Scavenger receptors are a superfamily of membrane-bound receptors that recognize both self and nonself targets. Scavenger receptor class A (SR-A) has five known members (SCARA1 to -5 or SR-A1 to -A5), which are type II transmembrane proteins that form homotrimers on the cell surface. SR-A members recognize various ligands and are involved in multiple biological pathways. Among them, SCARA5 can function as a ferritin receptor; however, the interaction between SCARA5 and ferritin has not been fully characterized. Here, we determine the crystal structures of the C-terminal scavenger receptor cysteine-rich (SRCR) domain of both human and mouse SCARA5 at 1.7 and 2.5 Å resolution, respectively, revealing three Ca2+-binding sites on the surface. Using biochemical assays, we show that the SRCR domain of SCARA5 recognizes ferritin in a Ca2+-dependent manner, and both L- and H-ferritin can be recognized by SCARA5 through the SRCR domain. Furthermore, the potential binding region of SCARA5 on the surface of ferritin is explored by mutagenesis studies. We also examine the interactions of ferritin with other SR-A members and find that SCARA1 (SR-A1, CD204) and MARCO (SR-A2, SCARA2), which are highly expressed on macrophages, also interact with ferritin. By contrast, SCARA3 and SCARA4, the two SR-A members without the SRCR domain, have no detectable binding with ferritin. Overall, these results provide a mechanistic view regarding the interactions between the SR-A members and ferritin that may help to understand the regulation of ferritin homeostasis by scavenger receptors.
Keywords: SCARA5, SR class A, scavenger receptor, ferritin, SRCR domain, scavenger receptor, ferritin, protein structure, X-ray crystallography, cell surface receptor
Scavenger receptors (SRs) were originally identified by their abilities to recognize different forms of modified low-density lipoproteins (LDLs) with broad binding specificities (1–4). Over the past decades, SRs have been shown to be widely expressed in various tissues and play important roles in host defense, such as sensing and cleaning different pathogens, including bacteria (Escherichia coli, Staphylococcus aureus, Neisseria meningitides, etc.) and parasites (Plasmodium falciparum) (5). For example, SRs, including SCARA1, MARCO, SCARA4, SCARA5, CD36, and CD163, have been shown to be able to bind bacteria (5, 6). And they may also act as a helper for viral infection (5, 7). On the other hand, SRs also recognize and clean self-ligands such as extracellular matrix and dead cells for homeostasis (7–9). SRs have been classified into more than 10 subclasses largely based on sequence similarities (9, 10). Among them, SR class A (SR-A) has five known members, including SCARA1 (SR-A1, CD204, MSR1, SR-AI, etc.), MARCO (SR-A2, SCARA2), SCARA3 (CSR), SCARA4 (SRCL), and SCARA5 (TESR) (11, 12). They are type II transmembrane proteins with similar domain arrangement, containing an N-terminal cytoplasmic region followed by a transmembrane helix and a large C-terminal extracellular portion. The SR-A members have been predicted to form homotrimers on the cell surface, and their ectodomain may contain coiled-coil regions, a collagen-like (CL) region, and a C-terminal C-type lectin-like domain or a scavenger receptor cysteine-rich (SRCR) domain (11, 12). Despite the structural similarities, the functions of SR-A members appear to be diverse, and the mechanisms of receptor-ligand interactions are not fully understood (13–16).
In the SR-A family, SCARA5 was initially found to be expressed in embryonic and adult mouse testis (17). Further characterization showed that SCARA5 existed in various tissues restricted to epithelial cells, and the SCARA5-transfected cells could bind Escherichia coli and Staphylococcus aureus but not zymosan (18). Unlike other SR-A members, such as SCARA1 and MARCO, SCARA5 was unable to interact with acetylated or oxidized LDLs (18). Sequence analysis shows that SCARA5 has relatively high similarity with SCARA1; its ectodomain contains a spacer region, a CL region, and an SRCR domain (Fig. 1A) (18). The SRCR domains are commonly found in SRs, which usually contain about 110 residues. A number of crystal structures of the SRCR domains have been determined, for example, M2BP (19), MARCO (16), SCARA1 (20), SALSA (SRCR1, SRCR8) (21), pCD163 (SRCR5) (22), CD6 (23), CD5 (24), and hepsin (25). They adopt a relatively conserved fold with three or four disulfide bonds (26). The SRCR domains may contain cation-binding sites for Ca2+ or Mg2+, which are typically found in the crystal structures, and these cation sites along with pH and surface electrostatic potential can contribute to the ligand recognition of the domain. The ligands for the SRCR domain appear to be diverse (27). For example, the macrophage receptor CD163 is involved in the clearance of hemoglobin/haptoglobin complex via the SRCR2 and SRCR3 domain in a Ca2+- and pH-dependent manner (28–30); the lymphocyte cell surface receptor CD6 interacts with the activated leukocyte cell adhesion molecule (ALCAM/CD166) through the SRCR3 domain (23, 31). Recently, the SRCR domain of SCARA1 has been found to be able to recognize spectrin specifically on dead cells and mediates dead cell internalization (20).
Figure 1.
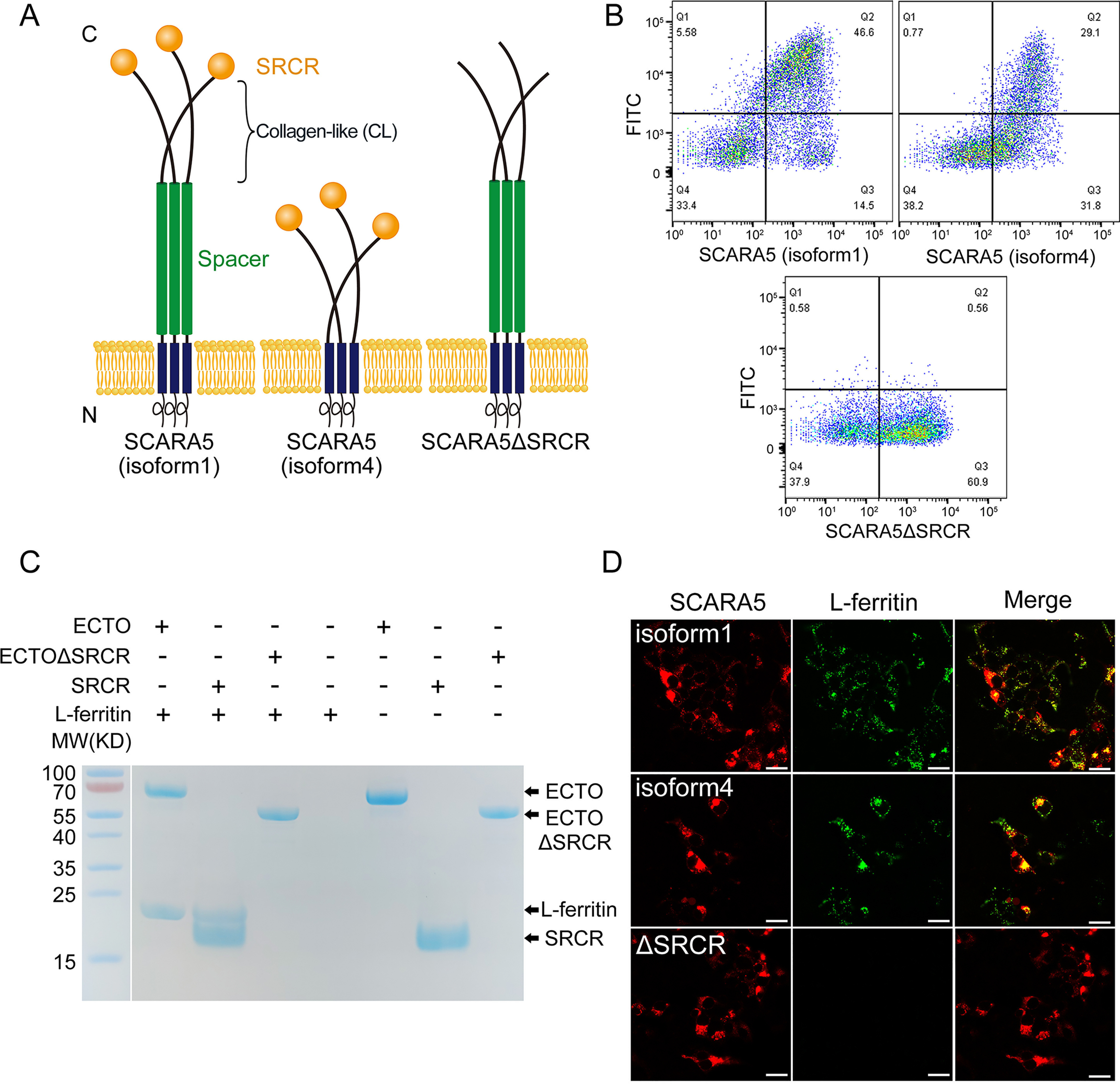
SCARA5 interacts with L-ferritin via the SRCR domain. A, schematic representation of the full-length SCARA5 (isoform 1), SCARA5 (isoform 4), and SCARA5ΔSRCR. B, FACS data showed that the FITC-labeled L-ferritin bound to the HEK293 cells transfected with hSCARA5 (isoform 1) or hSCARA5 (isoform 4) but did not bind to the SCARA5ΔSRCR-transfected cells. C, SDS-PAGE of the pulldown assays with FLAG tag showed that the ectodomain (ECTO) and the SRCR domain of SCARA5 could interact with L-ferritin, but the ectodomain without the SRCR domain (ECTOΔSRCR) had no interaction with L-ferritin. Empty beads had no interaction with L-ferritin either. D, fluorescent images showed that the hSCARA5 (isoform 1)– or the hSCARA5 (isoform 4)–transfected cells could internalize the FITC-labeled L-ferritin, but the SCARA5ΔSRCR-transfected cells had no binding to L-ferritin (bar, 25 μm).
Ferritins are present in most of the tissues as cytosolic proteins for iron storage (32). Human ferritins are assembled by two subunits, ferritin heavy chain (FTH, ∼21 kDa) and ferritin light chain (FTL, ∼19 kDa), with ratios ranging from 2:22 to 20:4 in different tissues (33, 34). FTH catalyzes the oxidation of Fe(II), whereas FTL promotes the nucleation and storage of Fe(III). Ferritin is also circulated outside cells as serum ferritin that is relatively iron-poor (35–37) and mainly composed of FTL (38–41). Although many aspects of serum ferritin remain unclear, evidence has shown that it might be involved in iron delivery (15, 42, 43), angiogenesis (44–46), inflammation, immunity, and cancer (47). It could act as an inflammatory marker and is associated with a variety of diseases, including acute respiratory distress syndrome (48), ALS (49), and atherosclerosis (50). Serum ferritin might be considered as a leakage product from damaged cells (51), and the iron-loaded ferritin within serum and tissue needs to be cleaned properly as it might be toxic to cells (52, 53). However, the innate mechanism of ferritin scavenging remains unclear.
Although SCARA5 does not bind modified LDLs, it has been shown that it could be a ferritin receptor mediating ferritin uptake in embryonic cells (15) and might also help ferritin crossing the blood retinal barrier (54), but the interaction between SCARA5 and ferritin has not been characterized. Here we investigate the binding of SCARA5 with ferritin and determine the crystal structure of the SRCR domain of SCARA5 and also explore the interactions of ferritin with the SR-A members by biochemical assays, which may help to understand the mechanisms of ferritin recognition via the SR-A family.
Results
SCARA5 can recognize and internalize ferritin through the C-terminal SRCR domain
SCARA5 has been reported to recognize and internalize ferritin (15), but the mechanism of the interaction between SCARA5 and ferritin was unclear. Here, we transfected HEK293 cells with the mCherry-tagged full-length human SCARA5 (isoform 1) and an SRCR domain deletion mutant of SCARA5 (SCARA5ΔSRCR), respectively (Fig. 1A). The transfected cells were fed with the FITC-labeled L-ferritin. The FACS results showed that the SCARA5-transfected cells could bind to L-ferritin, but SCARA5ΔSRCR had no binding to L-ferritin under similar conditions (Fig. 1B). The fluorescent images also showed that L-ferritin co-localized with SCARA5 on the transfected cells, but no L-ferritin was found on the SCARA5ΔSRCR-transfected cells (Fig. 1D), suggesting that the SRCR domain could mediate the uptake of L-ferritin. Moreover, we expressed and purified a series of truncation mutants of SCARA5 using insect cells, including the intact ectodomain, the ectodomain without the SRCR domain (ΔSRCR), and the SRCR domain alone (Fig. 1A), all fused with a C-terminal His6 tag and a FLAG tag (DYKDDDK). The FLAG tag pulldown results showed that the intact ectodomain and the SRCR domain alone could pull down L-ferritin directly, but no L-ferritin was detected using the ectodomain without the SRCR portion (Fig. 1C), confirming that the SRCR domain is the binding domain of L-ferritin on SCARA5.
In addition, we also tested the binding of ferritin with the SCARA5 isoform 4, which does not have the spacer region of isoform 1 (Fig. 1A), and found that isoform 1 had higher ferritin binding affinity or internalization efficiency than isoform 4 (Fig. 1, B and D), which may be caused by the different lengths of the two isoforms as longer tethers may reduce steric hindrance from cell membrane (55), thus making the SRCR domain more accessible to the ligands.
Crystal structure of the SRCR domain of SCARA5
The expression of the SRCR domain of SCARA5 alone in insect cells usually gave low yields. By contrast, the fragment including the CL region (Lys305–Met392) and the SRCR domain (CL-SRCR; Lys305–His495 for hSCARA5) could be expressed at high levels, and the CL portion of the CL-SRCR fragment could be degraded quickly (Fig. 2A). Interestingly, unlike the CL-SRCR fragment of SCARA1 that could form homotrimers during expression (Fig. 2A) (20), the potential trimeric form of the CL-SRCR fragment of SCARA5 was not identified during expression and purification (Fig. 2A). However, the SEC profile of the intact ectodomain of SCARA5 had an elution volume similar to that of the ectodomain of SCARA1 (Fig. S1A), which has been shown to form trimers (20), thereby suggesting that the intact ectodomain of SCARA5 also trimerized. Indeed, the cross-linked ectodomain as well as the ectodomain without the SRCR domain of SCARA5 run as trimers on SDS-PAGE (Fig. S1B), which was consistent with the previous reports showing that SCARA5 formed homotrimers on the cell surface (18). Therefore, the monomeric state of the CL-SRCR fragment expressed in vitro implied that the trimerization of SCARA5 might depend on the spacer region of this receptor (Fig. 1A).
Figure 2.
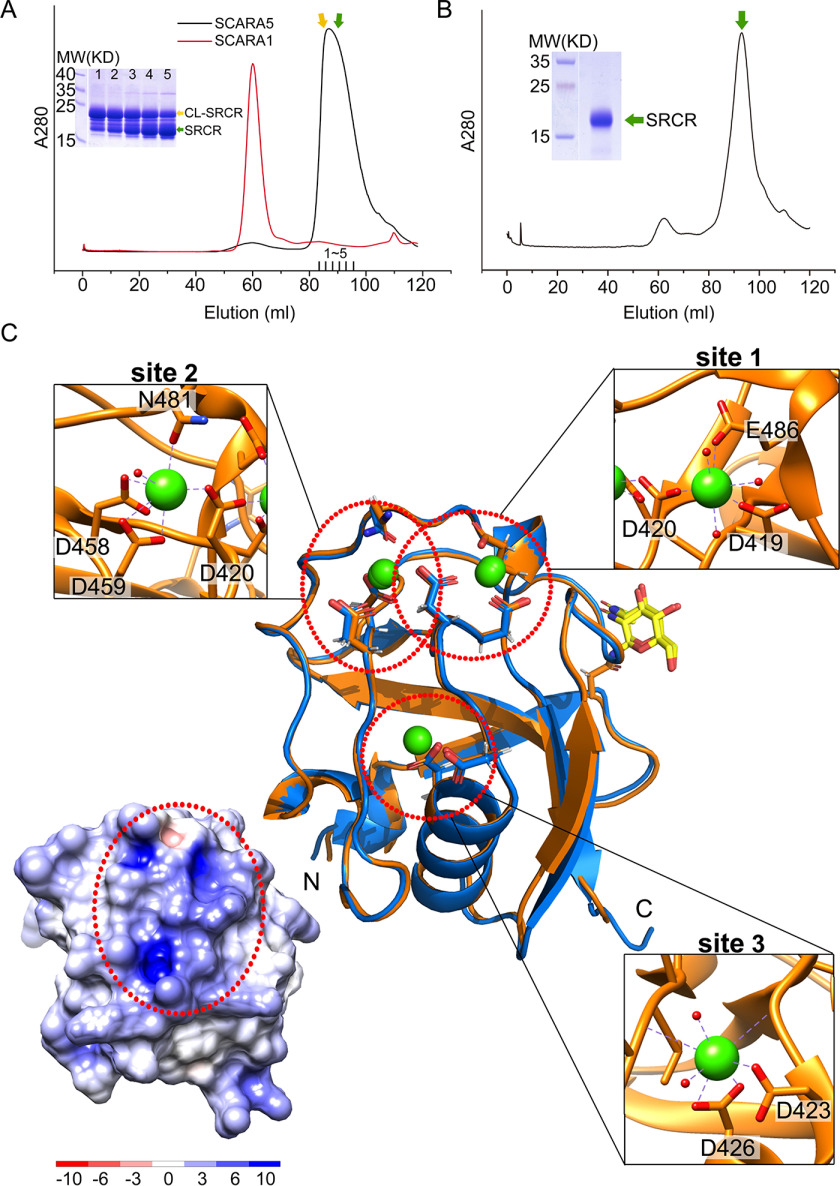
Crystal structures of the SRCR domain of SCARA5. A, SEC profiles of the CL-SRCR fragments of mSCARA1 and hSCARA5 and the SDS-PAGE of the CL-SRCR fragment of hSCARA5 from the SEC elution peak (fractions 1–5), showing the CL-SRCR fragment (orange arrow) as well as the SRCR fragment resulting from degradation (green arrow). B, SEC profile and SDS-PAGE of the CL-SRCR fragment of hSCARA5 after 36 h of degradation, resulting in an SRCR fragment for crystallization (green arrow). C, superposition of the crystal structures of human (orange) and mouse (blue) SRCR domain of SCARA5. Three Ca2+-binding sites (dashed red circles) are zoomed in at insets (Ca2+ are shown as green spheres; water molecules are shown as small red spheres), respectively. The glycans (N-acetylglucosamine, NAG) are colored in yellow. The surface electrostatic potential of the SRCR domain of hSCARA5 is shown (bottom left) with the same orientation as the crystal structure; the positively charged region is indicated by the dashed red oval.
To prepare the sample for crystallization, the CL-SRCR fragment of SCARA5 was expressed in insect cells as secreted proteins, and then the supernatant was dialyzed for 36 h, resulting in a stable fragment containing the SRCR domain (Fig. 2B), which was used for crystal screening and functional characterizations. The crystals of the SRCR domains from both human and mouse SCARA5 (hSCARA5 or mSCARA5) were obtained, and the structures were solved by molecular replacement and refined to 1.7 and 2.5 Å resolution, respectively (Tables 1 and 2). In the crystal structures, the SRCR domain and a short N-terminal α-helix are identified (Fig. 2C). The SRCR domain adopts a typical SRCR fold (Fig. 2C), similar to the SRCR domains of SCARA1 (20) and MARCO (16). Nevertheless, the structural alignments of the SRCR domains from SCARA5, SCARA1, and MARCO revealed some differences, especially in the loop regions (Fig. S4). The glycans associated with Asn397 (for hSCARA5) or Asn393 (for mSCARA5) can be visualized in the electron density (Fig. 2C). The crystal structures of human and mouse SRCR domains are quite similar with the r.m.s. deviation of the Cα atoms of 0.5 Å, consistent with the high sequence identity (about 91%) between the two domains (Fig. 2C and Fig. S4).
Table 1.
X-ray data collection and processing
| Protein | SRCR (hSCARA5) | SRCR (mSCARA5) |
|---|---|---|
| Diffraction source | BL18U1, SSRF | BL18U1, SSRF |
| Wavelength (Å) | 0.98 | 0.98 |
| Detector | Dectris PILATUS 6M | Dectris PILATUS 6M |
| Temperature (K) | 100 | 100 |
| Space group | P 212121 | P31 |
| a, b, c (Å) | 38.2, 48.8, 70.8 | 39.8, 39.8, 132.2 |
| α, β, γ (degrees) | 90, 90, 90 | 90, 90, 120 |
| Resolution range (Å) | 28.65–1.65 (1.71–1.65)a | 27.14–2.30 (2.38–2.30) |
| Rmerge | 0.082 (0.393) | 0.095 (0.326) |
| Rpim | 0.026 (0.125) | 0.043 (0.117) |
| No. of unique reflections | 15,075 (1465) | 10,380 (1040) |
| Completeness (%) | 99.59 (98.80) | 99.43 (99.11) |
| Multiplicity | 12.7 (12.7) | 10.3 (9.9) |
| Mean I/σ(I) | 29.03 (6.88) | 26.22 (7.00) |
| Overall B factor from Wilson plot (Å2) | 13.69 | 38.26 |
aValues in parentheses are for the highest-resolution shell.
Table 2.
Crystallographic statistics of the SRCR domain of SCARA5
| Protein | SRCR (hSCARA5) | SRCR (mSCARA5) |
|---|---|---|
| Resolution range (Å) | 27.67–1.70 (1.76–1.70)a | 23.85–2.50 (2.59–2.50) |
| Completeness (%) | 99.66 (98.92) | 99.43 (99.11) |
| No. of reflections, working set | 15,075 (1465) | 8067 (784) |
| No. of reflections, test set | 1508 (147) | 804 (80) |
| Final Rcryst | 0.159 (0.138) | 0.199 (0.241) |
| Final Rfree | 0.176 (0.144) | 0.242 (0.272) |
| Rpim | 0.026 (0.098) | 0.043 (0.082) |
| CC½ | 0.997 (0.980) | 0.978 (0.994) |
| No. of non-H atoms | ||
| Protein | 870 | 1774 |
| Solvent | 112 | 38 |
| Ligands | 17 | 34 |
| NAG | 14 | 28 |
| Ca2+ | 3 | 6 |
| Total | 999 | 1702 |
| r.m.s. deviations | ||
| Bonds (Å) | 0.013 | 0.010 |
| Angles (degrees) | 1.32 | 1.10 |
| Average B factors (Å2) | 19.61 | 50.81 |
| Protein | 17.40 | 50.39 |
| Water | 34.69 | 50.83 |
| Ca2+ | 11.95 | 46.81 |
| Ramachandran plot | ||
| Favored regions (%) | 96.23 | 92.06 |
| Additionally allowed (%) | 3.77 | 7.94 |
| Outliers (%) | 0 | 0 |
aValues in parentheses are for the highest-resolution shell.
In the crystal structure of the SRCR domain of SCARA5, three Ca2+-binding sites are identified, including site 1 (Asp419, Asp420, and Glu486), site 2 (Asp458, Asp459, and Asn481), and site 3 (Asp423 and Asp426) (Fig. 2C). The sequence alignments show that the SRCR domain of SCARA5 has ∼90% sequence identity among mammalians, and the residues forming the Ca2+-binding sites are well-conserved (Fig. S5). The three Ca2+ sites generate a positively charged region on the surface (Fig. 2C), which may correlate with the binding of ferritin or other ligands.
The interaction between the SRCR domain of SCARA5 and ferritin is Ca2+-dependent
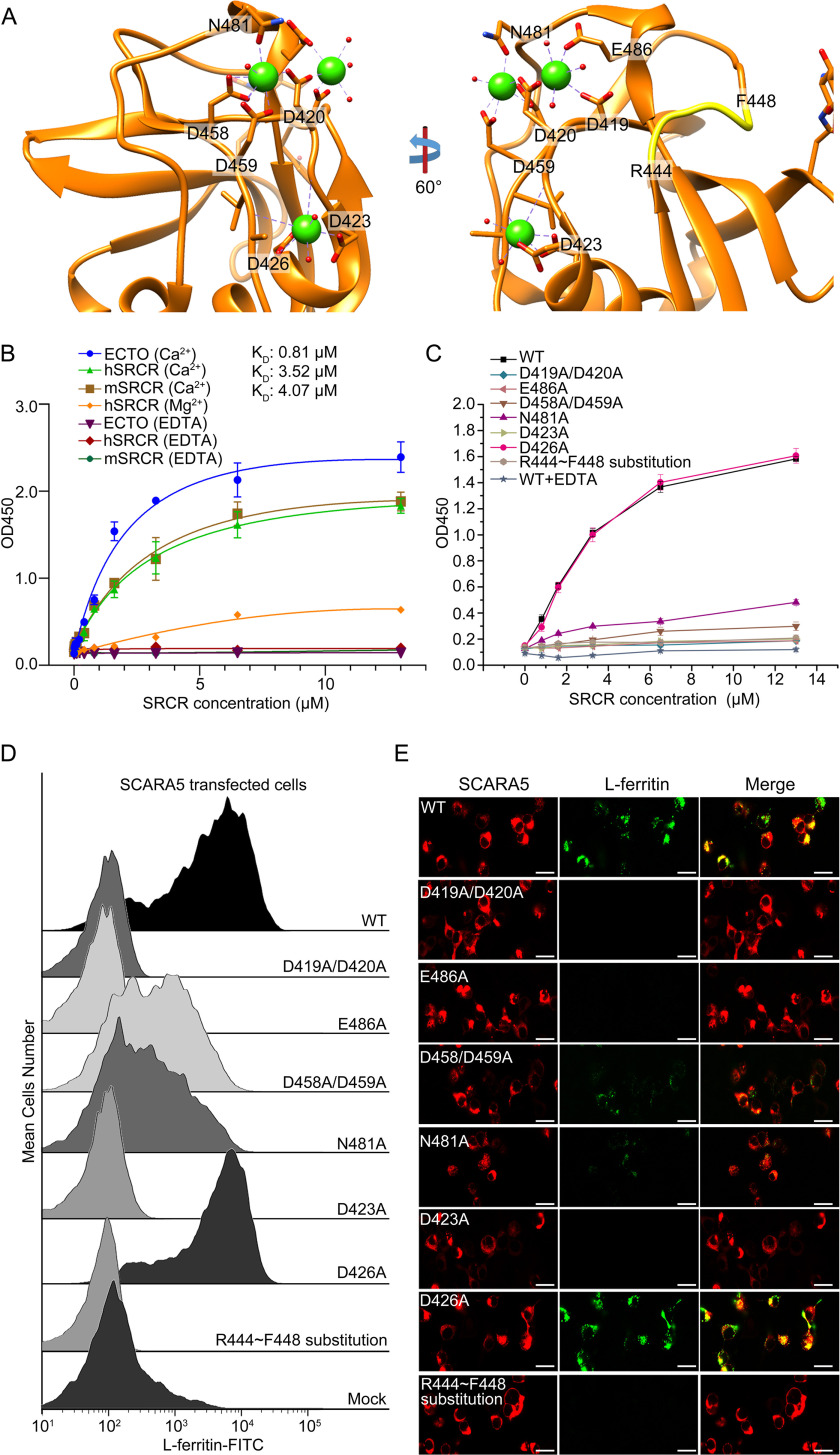
Previous reports have shown that the Ca2+ was involved in the ligand binding of the SRCR domains from different SRs (16, 20, 28). For SCARA5, ELISA results showed that both the ectodomain of SCARA5 and the SRCR domain alone bound to L-ferritin similarly, and the binding could be eliminated by the addition of EDTA (Fig. 3B), confirming the importance of Ca2+ for the binding. To verify the roles of the three Ca2+ sites, several single or double mutants involving the Ca2+-coordinating residues were constructed for hSCARA5, including D419A/D420A and E486A for site 1, D458A/D459A and N481A for site 2, and D423A and D426A for site 3 (Fig. 3A). These mutants were applied for both cell assays and recombinant protein expression. ELISA results showed that all the mutants except D426A could reduce the interactions between L-ferritin and the SRCR domain of SCARA5 (Fig. 3C). Both FACS and fluorescent imaging showed that mutants D419A/D420A and E486A from site 1 could eliminate L-ferritin binding or internalization almost completely (Fig. 3, D and E), suggesting that this site might be important for the recognition of L-ferritin. Meanwhile, mutants D458A/D459A and N481A from site 2 revealed attenuated binding with L-ferritin (Fig. 3, D and E), suggesting that this site might also contribute to ferritin recognition. The mutagenesis data for site 3 showed that mutant D423A could abolish the binding with L-ferritin, but mutant D426A had almost no effect on the interaction (Fig. 3, D and E); therefore, it was possible that residue Asp423, rather than the Ca2+ at site 3, was involved in ferritin recognition.
Figure 3.
Mutagenesis studies of the interaction between the SRCR domain of SCARA5 and L-ferritin. A, residues around the Ca2+-binding sites and the loop region (yellow) involved in the mutagenesis studies on the SRCR domain of hSCARA5. B, ELISA data showed that the ectodomain of hSCARA5 (ECTO) and the SRCR domains of human and mouse SCARA5 bound to L-ferritin in the presence of Ca2+ (5 mm), and the binding affinities (KD) were calculated based on the curve fitting, respectively. By contrast, Mg2+ reduced the binding affinity with L-ferritin, and EDTA could eliminate the binding completely. C, ELISA data showed that all of the mutants except D426A could reduce the binding of the SRCR domain with L-ferritin. D, FACS data showed all of the mutants of SCARA5 except D426A could reduce the binding of L-ferritin with the mutant-transfected cells. Mutants D458A/D459A and N481A from the Ca2+-binding site 2 retained some binding affinities with L-ferritin. Mock, nontransfected cells. E, fluorescent images showed that all of the mutants of SCARA5 except D426A could reduce the internalization of L-ferritin with the mutant-transfected cells (bar, 25 μm). Error bars, S.D.
In addition, Mg2+ was also applied in the assays to replace Ca2+, and it showed that Mg2+ could reduce the binding activities with L-ferritin significantly (Fig. 3B). This was similar to the previous reports showing that Mg2+ could reduce the binding affinities of SCARA1 and MARCO to the ligands (16, 20).
Both human L-ferritin and H-ferritin interact with the SRCR domain of SCARA5
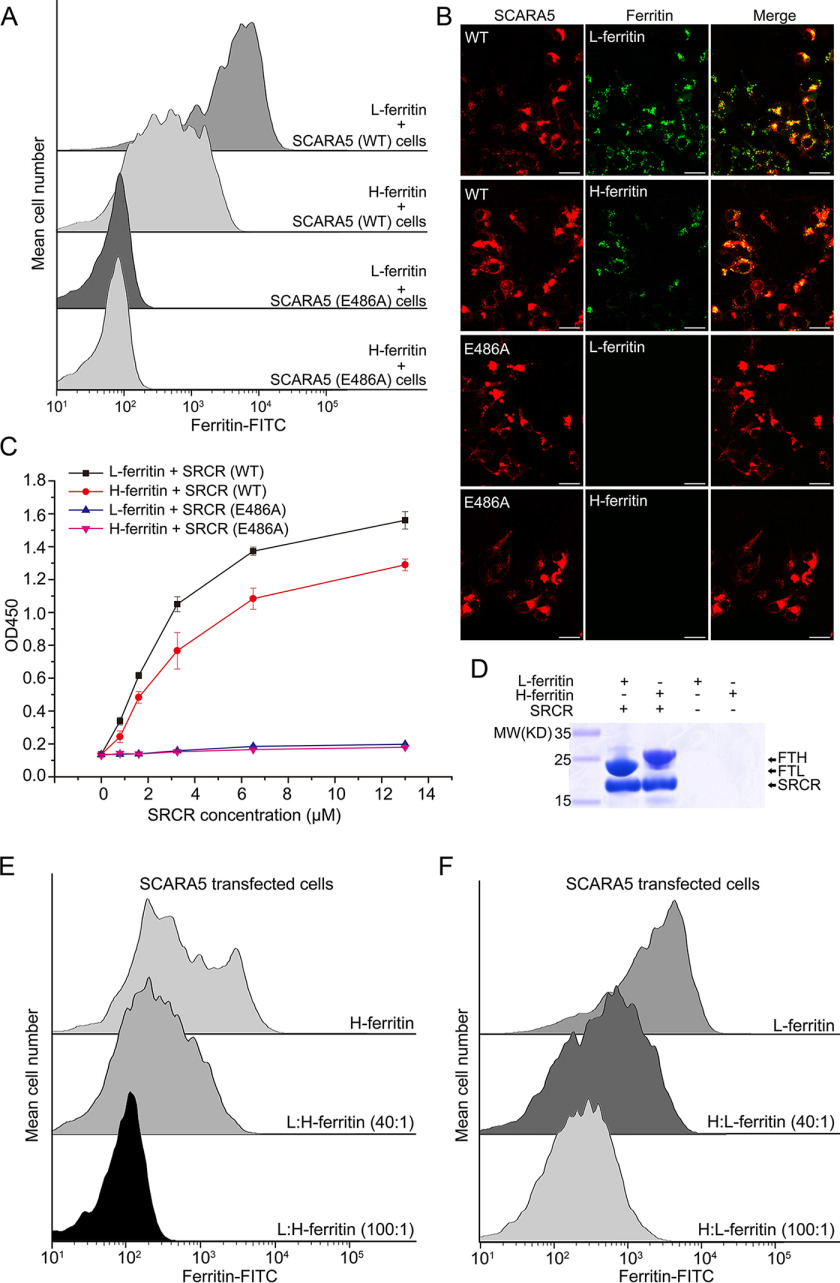
Previous reports suggested that the SCARA5-expressing cells could internalize L-ferritin, but not H-ferritin (15). In our results, both FACS data and fluorescent images showed that the FITC-labeled H-ferritin could bind to or be internalized by the SCARA5-transfected cells, but with lower affinity than L-ferritin (Fig. 4, A and B). To validate this result, both pulldown assays and ELISA were applied to monitor the interaction between SCARA5 and H-ferritin, and the results showed that the SRCR domain of SCARA5 could also pull down H-ferritin, but the binding affinity was weaker than L-ferritin (Fig. 4, C and D).
Figure 4.
SCARA5 recognizes both L-ferritin and H-ferritin. A, FACS data showed that both FITC-labeled L-ferritin and H-ferritin bound to the hSCARA5 transfected cells, but not the hSCARA5 (E486A)-transfected cells. B, fluorescent images showed that both L-ferritin and H-ferritin could be internalized by the hSCARA5-transfected cells, but not the hSCARA5 (E486A)-transfected cells (bar, 25 μm). C, ELISA data showed that both L-ferritin and H-ferritin could bind to the SRCR domain of hSCARA5, but not to the SRCR (E486A) mutant. D, FLAG tag pulldown assays showed that both L-ferritin and H-ferritin could interact with the SRCR domain of hSCARA5. E, FACS data showed that the unlabeled L-ferritin could block the binding of the FITC-labeled H-ferritin by the SCARA5-transfected cells almost completely. F, FACS data showed that unlabeled H-ferritin could partially block the binding of the FITC-labeled L-ferritin by the SCARA5-transfected cells. Error bars, S.D.
In the meantime, the unlabeled L-ferritin or H-ferritin was added to the SCARA5-transfected cells fed with the FITC-labeled H-ferritin or L-ferritin at different molar ratios to see whether L-ferritin and H-ferritin could inhibit each other for binding with SCARA5. Indeed, the FACS data showed that L-ferritin and H-ferritin could block each other for binding, and L-ferritin could inhibit H-ferritin more efficiently than H-ferritin inhibiting L-ferritin (Fig. 4, E and F). This was consistent with the pulldown and ELISA data shown above and suggested that L-ferritin and H-ferritin might share a similar binding region on the SRCR domain of SCARA5.
Mutagenesis studies reveal the potential binding region of SCARA5 on ferritin surface
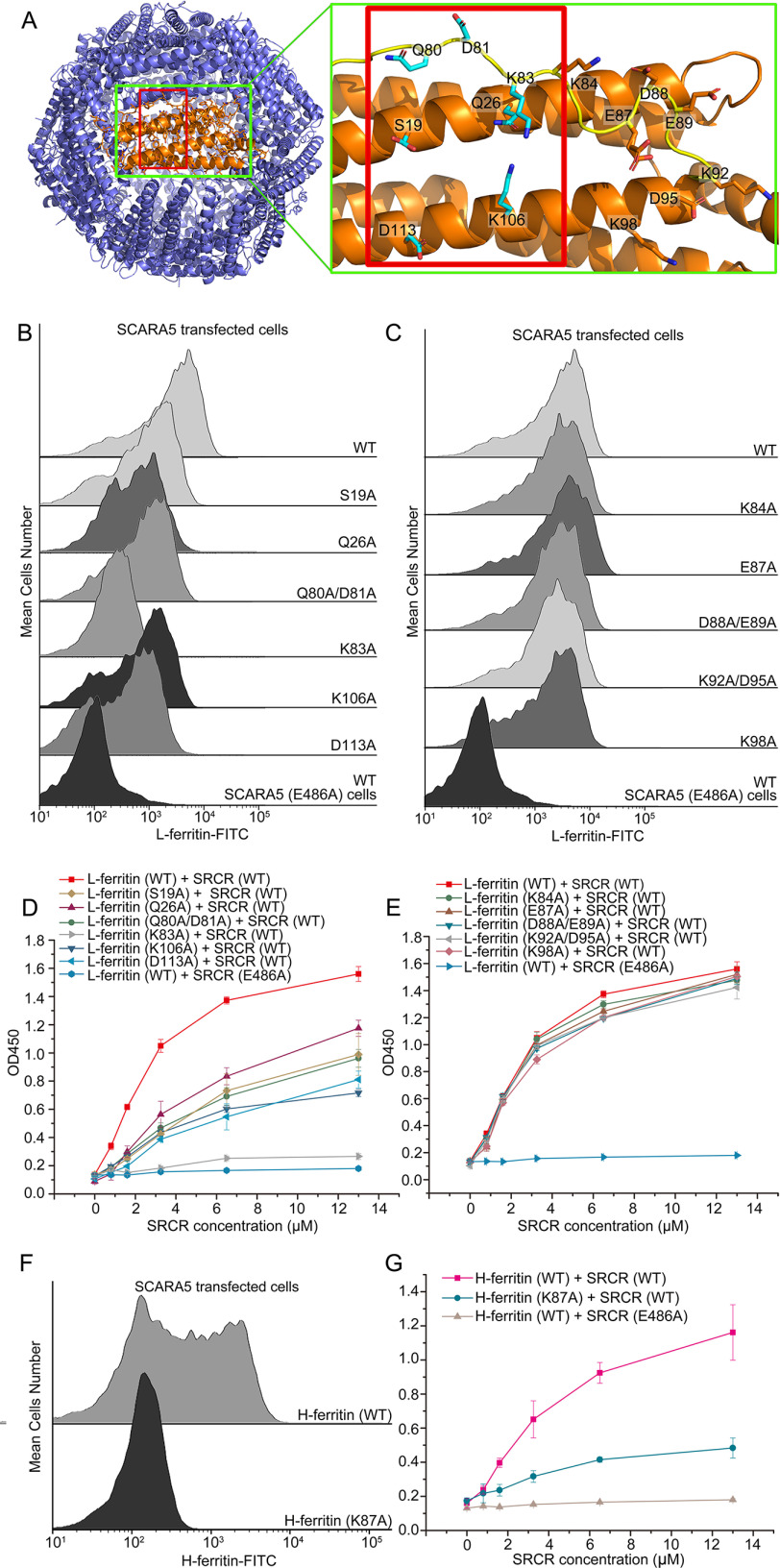
To identify the SCARA5-binding region on ferritin, a series of mutants of the surface residues of human FTL were constructed, including D12A, S19A, Q26A, Q80A/D81A, K83A, K84A, E87A, D88A/E89A, K92A/D95A, K98A, K106A, and D113A (Fig. 5A). The ferritin mutants were expressed and purified similarly as the WT (Fig. S3), and all of the mutants showed reasonable solubility except the mutant D12A, which aggregated easily during concentration. The FACS data showed that the mutants K84A, E87A, D88A/E89A, K92A/D95A, and K98A had almost no effect on the interaction between L-ferritin and SCARA5 (Fig. 5C). By contrast, mutants S19A, Q26A, Q80A/D81A, K83A, K106A, and D113A showed weaker binding with SCARA5 (Fig. 5B). ELISAs were also applied to examine the binding affinities, and the results showed that mutants S19A, Q26A, Q80A/D81A, K83A, K106A, and D113A had lower binding affinities with the SRCR domain of SCARA5 than the WT (Fig. 5D), whereas mutants K84A, E87A, D88A/E89A, K92A/D95A and K98A retained similar affinities with the WT (Fig. 5E). Among them, residue Lys83 appeared to have an important role, as the mutant K83A almost eliminated binding between L-ferritin and SCARA5 (Fig. 5, B and D). These data roughly defined the binding region of SCARA5 on the surface of L-ferritin (Fig. 5A).
Figure 5.
Mutagenesis studies of the SCARA5 binding region on ferritin. A, residues on the surface of L-ferritin (left; Protein Data Bank entry 2FFX) involved in the mutagenesis studies are labeled in the green rectangles. The potential binding region of SCARA5 on ferritin is indicated by the red rectangles. B, FACS data showed that mutants S19A, Q26A, Q80A/D81A, K83A, K106A, and D113A of L-ferritin reduced binding affinities with SCARA5. C, FACS data showed that mutants K84A, E87A, D88A/E89A, K92A/D95A, and K98A of L-ferritin retained similar binding affinities with SCARA5 as the WT. D, ELISA data showed that mutants S19A, Q26A, Q80A/D81A, K83A, K106A, and D113A of L-ferritin reduced the binding affinities with the SRCR domain SCARA5. E, ELISA data showed that mutants K84A, E87A, D88A/E89A, K92A/D95A, and K98A of L-ferritin retained similar binding affinities with the SRCR domain SCARA5. F, FACS data showed that the mutant K87A of H-ferritin reduced the binding with SCARA5 significantly. G, ELISA data showed that the mutant K87A of H-ferritin reduced the binding with the SRCR domain of SCARA5 significantly. Error bars, S.D.
The sequence identity between human FTL and FTH is about 56%. The structural alignment suggested that residue Lys83 of FTL corresponded to Lys87 of FTH. The FACS results showed that the mutant K87A of FTH could reduce the binding of H-ferritin to the SCARA5-transfected cells significantly (Fig. 5F), and the ELISA data also showed weaker binding between the SRCR domain of SCARA5 and the mutant K87A (Fig. 5G). This is consistent with the inhibition assay results described above, supporting that L-ferritin and H-ferritin might share a similar binding region on their surfaces with SCARA5 (Fig. 5A).
Other SR-A members, SCARA1 and MARCO, also interact with ferritin
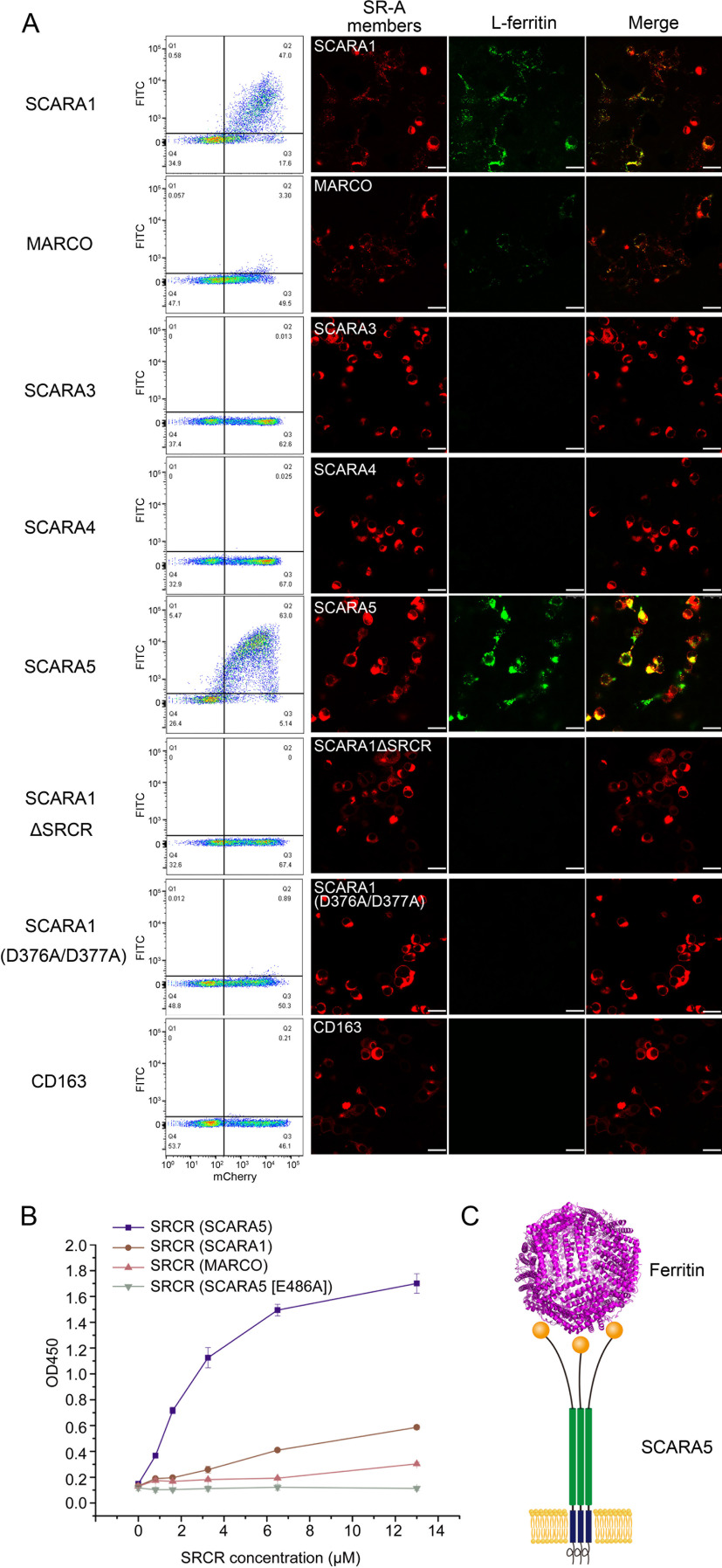
Because SR-A members share similar structural features, we explored the binding properties of the SR-A members with ferritin. HEK293 cells were transfected with the mCherry-tagged human SR-A members, respectively, and the FITC-labeled human ferritin was applied for binding assays. The FACS data showed that besides SCARA5, SCARA1 also had binding signals with both L-ferritin and H-ferritin, but weaker than that of SCARA5 (Fig. 6A and Fig. S2). Meanwhile, MARCO showed much weaker but detectable binding with both L- and H-ferritin (Fig. 6A and Fig. S2). By contrast, SCARA3 and SCARA4 had no detectable binding with either L-ferritin or H-ferritin (Fig. 6A and Fig. S2). The fluorescent images of the corresponding samples also showed that L-ferritin could be co-localized with SCARA5, SCARA1, and MARCO reasonably well (Fig. 6A), although the signal for MARCO was much weaker than that of SCARA5 or SCARA1. In addition, the SRCR domain deletion mutant (ΔSRCR) of SCARA1 was also constructed and applied for the binding assays, showing that SCARA1 could not bind to L-ferritin in the absence of the SRCR domain (Fig. 6A). The binding of the SRCR domains of SCARA5, SCARA1, and MARCO with L-ferritin was also tested using ELISA, and the data showed that the SRCR domain of SCARA5 had the strongest binding affinity among the three SRCR domains (Fig. 6B). In parallel, mouse SCARA5, SCARA1, and MARCO were also applied for the assays, and similar results were obtained (Fig. S2). Furthermore, we also tested the interaction of L-ferritin with human CD163, which contains nine SRCR domains, but no binding was detected (Fig. 6A), suggesting that the SRCR domains from the SR-A family might have binding specificities for ferritin.
Figure 6.
Interactions of L-ferritin with the SR-A members. A, FACS data (left) and fluorescent images (right) showed that human SCARA1, MARCO, and SCARA5 could bind to L-ferritin, whereas SCARA3, SCARA4, SCARA1ΔSRCR, SCARA1 (D376A/D377A), and CD163 had no binding to L-ferritin (bar, 25 μm). B, ELISA data showed that the SRCR domain of human SCARA5 had higher binding affinity with L-ferritin than the SRCR domains of human SCARA1 and MARCO. C, schematic model for the recognition of ferritin by SCARA5 on the cell surface. Error bars, S.D.
The structural and sequence alignments between the SRCR domains of SCARA5 and SCARA1 showed an r.m.s. deviation of the Cα atoms of 0.7 Å and a sequence identity of 59%. The crystal structure of the SRCR domain of SCARA1 has only one Ca2+-binding site (20), which corresponds to the Ca2+ site 1 for SCARA5 and MARCO (Fig. S4). The mutations of this site (D376A/D377A) on SCARA1 eliminated the binding with L-ferritin (Fig. 6A), similar to the results for SCARA5 shown above, suggesting that this site was also important for ferritin recognition.
The structural and sequence alignments showed that the SRCR domain of MARCO had slightly less similarity to SCARA5 than SCARA1 with an r.m.s. deviation of the Cα atoms of 1.1 Å and a sequence identity of 48% (Fig. S4B). In fact, all three Ca2+– or divalent cation–binding sites on the SRCR domain of SCARA5 were retained similarly for MARCO (Fig. S4B). The superposition of the SRCR domains showed that a loop region of SCARA5 (Arg444–Phe448) that located near the Ca2+ site 1 was shorter in MARCO (Ser471–Tyr473) (16), and they adopted different conformations in the two molecules (Fig. S4B), which might affect the interaction with ferritin. Indeed, when the loop region of SCARA5 was replaced by the loop from MARCO, the mutant lost the binding affinity with L-ferritin (Fig. 3, C–E). Meanwhile, the MARCO mutant containing the loop from SCARA5 revealed enhanced interaction with L-ferritin (Fig. S2C). These results suggest that although the Ca2+ sites are important for ferritin binding, the surface profile or other residues may also affect the accessibility or interaction of ferritin with the receptors. In addition, we also replaced the N-terminal SRCR domain of CD163 with the SRCR domain of hSCARA5, but the chimeric molecule did not show any detectable binding with ferritin (Fig. S2D). This is not surprising; as CD163 is a type I transmembrane protein, the SRCR domain might have different orientations that may also be important for ligand recognition.
Discussion
The SRCR domains are commonly found in SRs and are able to recognize various ligands (27). The ligand recognition by the SRCR domain usually needs divalent cations, such as Ca2+ (56). In the SR-A family, it has been shown that Ca2+ or Mg2+ is required for ligand binding for SCARA1 and MARCO (16, 20); therefore, it is not surprising that the SRCR domain of SCARA5 binds ferritin in a Ca2+-dependent manner. Moreover, the binding of ferritin is not limited to SCARA5; it is shared by other SR-A members, including SCARA1 and MARCO, although their binding affinities with ferritin are weaker than SCARA5. Among the three Ca2+ sites found in the SRCR domain of SCARA5, the Ca2+ site 1 is conserved in SCARA1 and also important for ferritin recognition. It has been reported that this Ca2+ site on SCARA1 is critical for recognizing spectrin from dead cells (20), suggesting that this site might be involved in multiple-ligand recognition. By contrast, MARCO also has three divalent cation–binding sites, similar to SCARA5, but its affinity to ferritin is very low, probably due to the hindrance from other residues and the surface profile of the molecule. In addition, CD163, a scavenger receptor with several SRCR domains, does not show binding activities with ferritin, suggesting that the binding of ferritin may be limited to a specific set of SRCR domains, such as those in the SR-A family.
A distinct feature of SR-A family members is that they form homotrimers on the cell surface (11, 12, 57–59). It has been shown that the trimeric form of SCARA1 increases its binding affinity with spectrin significantly (20). Unlike SCARA1, the in vitro expressed CL-SRCR fragment of SCARA5 is monomeric, and the CL region can be degraded rapidly, suggesting that the CL-SRCR portion of SCARA5 might adopt an open conformation rather than a compact trimerized state. Because ferritin particles are relatively large, ∼12 nm in diameter, the open conformation of SCARA5 may facilitate the binding and internalization of ferritin particles (Fig. 6C).
It has been shown that L- and H-ferritin have different tissue distributions (33, 47). In fact, most of the ferritin particles isolated from tissues are heteropolymers assembled by both H and L subunits with different ratios (34). The recognition of ferritin by SCARA5 and other SR-A members suggests that they may act as general receptors for ferritin particles.
Serum ferritin has been widely used in clinical tests for a number of diseases as it is often associated with inflammation, immunity, and cancer (47). It has been proposed that serum ferritin might be a leakage product from damaged cells (51) and could be toxic to cells (52, 53). The recognition of ferritin by the SR-A member suggests that these molecules could act as scavenging receptors for ferritin. Moreover, SCARA1 and MARCO are highly expressed on macrophages, and they might be able to monitor ferritin levels and induce inflammatory responses, thereby providing a clue for the correlation between ferritin and inflammation. Meanwhile, SCARA5 has relatively wide tissue distribution and could internalize ferritin either for ferritin removal or iron delivery.
Recently, it has been shown that degradation of ferritin by autophagy could promote ferroptosis (60–63). Therefore, the SR-A members with ferritin recognizing and uptake functions might be involved in regulating ferritin homeostasis and cell death, but the exact roles of the receptors in these pathways need to be elucidated in the future. In addition, the recombinant ferritins have long been used as nanocages for drug delivery (64–66); therefore, the SR-A members could be the potential targets for the therapeutic strategies against cancer and other diseases.
Materials and methods
Protein expression and purification
The cDNA of hSCARA1 to -5 (GenBankTM numbers NM_138715.2, NM_006770, NM_016240.2, NM_130386, and NM_028903.2), mSCARA1, -2, and -5 (L04274.1, NM_010766.2, and NM_173833.5), and human ferritin (FTL, NM_000146.3; FTH, NM_002032.2) were purchased from Sino Biological lnc. or synthesized by Sangon Biotech.
For the crystallization of the SRCR domain of hSCARA5, the cDNAs of hCL-hSRCR (Lys305–His495) and a chimeric fragment containing the mCL region and hSRCR (mCL-hSRCR: Lys305–Met388 (mSCARA5) + Ile393–His495 (hSCARA5)) were generated and fused with an N-terminal melittin signal peptide and a C-terminal His6 tag. The SRCR domain of mSCARA5 (mCL-mSRCR, Lys305–Thr489) was constructed similarly. For pulldown assays and ELISA, the ectodomain (Ser82–His495), ECTOΔSRCR (Ser82–Met392), and CL-SRCR (Lys305–His495) of hSCARA5 were constructed and fused with an N-terminal melittin signal peptide and a C-terminal His6 tag coupled with a FLAG tag. The SRCR domain of hSCARA1 was expressed and purified as described previously (20). The SRCR domain of hMARCO was constructed with an N-terminal GFP coupled with a His6 tag. All of the constructs were subcloned into the pFastBac vector (Invitrogen) using NovoRec recombinant enzyme (Novoprotein).
For protein expression, recombinant baculoviruses were generated following the Bac-to-Bac baculovirus expression protocol (Invitrogen); High-5 cells (Invitrogen) were used for protein expression using ESF921 medium (Expression Systems). The supernatants were collected after 72 h of infection and then buffer-exchanged with 25 mm Tris, 150 mm NaCl, pH 8.0, by dialysis, and then applied to nickel-nitrilotriacetic acid chromatography (Ni-NTA Superflow, Qiagen) or directly applied to Ni Smart beads (Smart-Lifesciences, Changzhou, China) for purification following the manufacturer's protocol. The eluates were loaded onto HiLoad Superdex 200 preparation grade column (GE Healthcare) with Tris-NaCl buffer (25 mm Tris, 150 mm NaCl, 1.5 mm Ca2+, pH 7.5) for further purification. The purified proteins were loaded onto SDS-PAGE for detection. Human ferritins, including both L-ferritin and H-ferritin, were expressed in E. coli BL21 DE3 cells (Novagen) using the pET28a expression vector and purified according to the procedures reported previously (67).
Crystallization and structural determination
The CL-SRCR fragments of SCARA5 were expressed as described above. After 36 h of dialysis, the CL-SRCR fragment was fully degraded into a stable fragment and used for crystallization. Proteins were concentrated to ∼8.5 mg/ml (measured by UV absorption at 280 nm) using Amicon Ultra-15 (3000 molecular weight cutoff; Merck) in buffer containing 150 mm NaCl, 10 mm Tris, pH 7.4, 1.5 mm Ca2+. Crystal screening was done at 18 °C by the sitting-drop vapor-diffusion method using 96-well plates (Swissci) with commercial screening kits (Hampton Research). A Mosquito nanoliter robot (TTP Labtech) was used to set up 400-nl sitting drops. The crystals for the SRCR domain of hSCARA5 were obtained using the mCL-hSRCR fragment as this construct gave better crystals (40% (w/v) PEG 400, 0.2 m sodium chloride, 0.1 m Tacsimate, pH 8.0). The crystals of the SRCR domain of mSCARA5 were grown in a solution containing 20% (w/v) PEG 3350, 0.2 m ammonium sulfate (pH 6.0). Diffraction data were collected using a PILATUS 6M detector at the BL18U beamline of the National Facility for Protein Science Shanghai (NFPS) at the Shanghai Synchrotron Radiation Facility (SSRF) and processed using the HKL-3000 package (68). The diffraction data statistics are summarized in Table 1.
The crystal structure of the SRCR domain of mSCARA5 was solved by molecular replacement using the structure of the SRCR domain of mSCARA1 (Protein Data Bank entry 6J02) as a search model. The crystal structure of the SRCR domain of hSCARA5 was solved by molecular replacement using the solved SRCR domain of mSCARA5 as a search model. Molecular replacement, model building, and refinement were performed using the PHENIX suite (69) and Coot (70). The refinement statistics are summarized in Table 2.
Mutagenesis of the SR-A members and ferritins
Mutants for hSCARA5 include D419A/D420A, E486A, D458A/D459A, N481A, D423A, D426A, the loop substitution mutant (Arg444–Phe448 of hSCARA5 was replaced by Ser471–Tyr473 of mMARCO), and the truncation mutant hSCARA5ΔSRCR (deletion of Ile393–His495). The loop substitution mutant of mMARCO was constructed by replacing Ser471–Tyr473 of mMARCO with Arg444–Phe448 of hSCARA5. Mutants of hSCARA1 include D376A/D377A and the truncation mutant hSCARA1ΔSRCR (deletion of Val350–Leu451). The domain substitution mutant of hCD163 was constructed by replacing the SRCR1 domain (Leu51–Ser151) of hCD163 with the SRCR domain (Ile393–Asn493) of hSCARA5. Mutants of human L-ferritin include D12A, S19A, Q26A, Q80A/D81A, K83A, K84A, E87A, D88A/E89A, K92A/D95A, K98A, K106A, and D113A. The mutant of human H-ferritin is K87A.
Mutations were introduced into the corresponding plasmids during PCR using KOD DNA polymerase (Sparkjade Science Co., Ltd.). The template plasmids were digested by DpnI (Thermo Fisher Scientific). The digested PCR products were ligated by ligation high (TOYOBO). The mutants were expressed similarly as the WT as described above.
Pulldown assay
Anti-FLAG G1 affinity resin beads (GeneScript) were mixed with the purified FLAG-tagged proteins, respectively, including the ectodomain (ECTO), the ectodomain without the SRCR domain (ECTOΔSRCR), the SRCR domain of hSCARA5 (∼200 µg for each sample), and human L-ferritin or H-ferritin (∼400 µg) in 800 µl of binding buffer (25 mm Tris, 150 mm NaCl, pH 7.4) supplemented with either 5 mm Ca2+ or 5 mm EDTA. After incubation for 1 h at room temperature, the beads were washed three times with the binding buffer. The proteins bound to the anti-FLAG G1 affinity resin were eluted using 100 µl of 3xFLAG peptide (400 µg/ml) and loaded onto 10% SDS-polyacrylamide gels (GeneScript) running with MES running buffer (50 mm MES, 50 mm Tris, 0.1% SDS, 1 mm EDTA, pH 7.3) for detection.
ELISA experiments
Ferritins, including WT L-ferritin, H-ferritin, and the mutants, were coated onto 96-well MaxiSorp plates (Nunc) with 2–4 µg of protein/well at 4 °C overnight in the coating buffer (3.5 mm NaHCO3, 1.5 mm Na2CO3, pH 9.6). The plates were blocked with the TBS buffer (25 mm Tris, 150 mm NaCl, pH 7.4) containing 5% (w/v) BSA for 3 h. The purified SRCR domain or ectodomain was serially diluted and added to each well with corresponding binding buffer (25 mm Tris, 150 mm NaCl, pH 7.4, 1% BSA), supplemented either with 5 mm Ca2+, 5 mm EDTA, or 5 mm MgSO4. Then the plates were incubated at room temperature for 1 h. After incubation, plates were washed with the corresponding binding buffer (without 1% BSA) five times. Mouse anti-FLAG M2 antibody (Sigma) was premixed with horseradish peroxidase–conjugated goat anti-mouse IgG antibody (Signalway Antibody) in a concentration ratio of 1:3. For His6 detection, horseradish peroxidase–conjugated anti-His6 antibody (Proteintech) was used. The antibodies were added to each well at 1:1000 dilution, and the plates were incubated at room temperature for 1 h. After washing five times with the corresponding binding buffer (without 1% BSA) buffer, 100 µl of chromogenic substrate (1 µg/ml tetramethylbenzidine, 0.006% H2O2 in 0.05 m phosphate citrate buffer, pH 5.0) was added to each well and incubated for 10–20 min at 37 °C. Then 50 µl of H2SO4 (2.0 m) was added to each well to stop the reactions. The plates were read at 450 nm on a Synergy Neo machine (BioTek Instruments). The binding affinities with L-ferritin (KD values) for the ectodomain (trimer) and the SRCR domain of SCARA5 were calculated based on the fitting of the sigmoidal curves of ELISA using software GraphPad Prism version 6 (71, 72).
Cross-linking experiment
The purified N-terminal GFP-tagged ectodomain of SCARA5 (ECTO) and the ectodomain without the SRCR domain (ECTOΔSRCR) (25 mm Hepes, 150 mm NaCl, pH 7.4) were treated with 1% (w/v) glutaraldehyde for 10 min at room temperature, and then 2 mm glycine was added to terminate the cross-linking reaction. The samples were suspended in 25 µl of SDS-loading buffer and loaded onto SDS-PAGE for separation, and a fluorescent image was taken with a gel-imaging system (Bio-Rad).
Flow cytometry and confocal microscopy
For flow cytometry and confocal microscopy, ferritins were labeled with FITC (Sigma) according to the manufacturer's protocol. Briefly, the purified protein (5 mg/ml) was added with a 10-fold molar excess of FITC in protein storage buffer, incubated for 2 h at room temperature. The nonreacted dye was desalted by Amicon Ultra-15 (100,000 molecular weight cutoff; Merck), and the fluorescent dye/protein ratio was determined by UV spectroscopy.
HEK293T cells were transfected with the pTT5 vectors containing the mCherry-tagged WT proteins or the mutants. After 24 h of transfection, the FITC-labeled ferritins were added to media with a normalized amount according to different fluorescent dye/protein ratios and incubated for 12 h at 37 °C. For competition assays, unlabeled ferritins were added at the same time in a different ratio with the FITC-labeled ferritins.
For flow cytometry, trypsin-digested cells were resuspended in 1 ml of cell culture medium (90% Dulbecco's modified Eagle's medium, 10% fetal bovine serum) and analyzed by an LSR Fortessa flow cytometer (BD Biosciences). Data analysis was performed using FlowJo software (Tree Star).
For confocal microscopy, cells grown on a coverslip were fixed by 4% paraformaldehyde. After washing twice with PBS, cells were mounted onto slides and observed using a Leica SP8 microscope.
Data availability
The structures of the SRCR domains of human and mouse SCARA5 have been deposited in the Protein Data Bank with accession numbers 7C00 (for hSCARA5) and 7BZZ (for mSCARA5), respectively.
Supplementary Material
Acknowledgments
We thank the National Center for Protein Science Shanghai (the Integrated Laser Microscopy system and the Protein Expression and Purification system) for instrumental support and technical assistance. We also thank beamline BL18U1 of the National Facility for Protein Science Shanghai (NFPS) at Shanghai Synchrotron Radiation Facility for assistance in X-ray diffraction data collection.
This article contains supporting information.
Author contributions—B. Y. data curation; B. Y. and Y. H. formal analysis; B. Y. and Y. H. validation; B. Y., C. C., Y. Wu, L. G., D. K., Z. Z., Y. Wang, E. Z., and Y. L. investigation; B. Y. and Y. H. methodology; B. Y. and Y. H. writing-original draft; B. Y. and Y. H. writing-review and editing; Y. L. and Y. H. resources; Y. H. conceptualization; Y. H. supervision; Y. H. funding acquisition; Y. H. project administration.
Funding and additional information—This work is supported by National Natural Science Foundation of China Grant 91957102 and the Chinese Academy of Sciences Facility-based Open Research Program (to Y. H.).
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- SR
- scavenger receptor
- CL
- collagen-like
- SRCR
- scavenger receptor cysteine-rich
- FTH
- ferritin heavy chain
- FTL
- ferritin light chain
- r.m.s.
- root mean square.
References
- 1. Brown M. S., and Goldstein J. L. (1979) Receptor-mediated endocytosis: insights from the lipoprotein receptor system. Proc. Natl. Acad. Sci. U. S. A. 76, 3330–3337 10.1073/pnas.76.7.3330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brown M. S., Goldstein J. L., Krieger M., Ho Y. K., and Anderson R. G. (1979) Reversible accumulation of cholesteryl esters in macrophages incubated with acetylated lipoproteins. J. Cell Biol. 82, 597–613 10.1083/jcb.82.3.597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Russell D. W., Yamamoto T., Schneider W. J., Slaughter C. J., Brown M. S., and Goldstein J. L. (1983) cDNA cloning of the bovine low density lipoprotein receptor: feedback regulation of a receptor mRNA. Proc. Natl. Acad. Sci. U. S. A. 80, 7501–7505 10.1073/pnas.80.24.7501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sege R. D., Kozarsky K., Nelson D. L., and Krieger M. (1984) Expression and regulation of human low-density lipoprotein receptors in Chinese hamster ovary cells. Nature 307, 742–745 10.1038/307742a0 [DOI] [PubMed] [Google Scholar]
- 5. Pombinho R., Sousa S., and Cabanes D. (2018) Scavenger receptors: promiscuous players during microbial pathogenesis. Crit. Rev. Microbiol. 44, 685–700 10.1080/1040841X.2018.1493716 [DOI] [PubMed] [Google Scholar]
- 6. Peiser L., Gough P. J., Kodama T., and Gordon S. (2000) Macrophage class A scavenger receptor-mediated phagocytosis of Escherichia coli: role of cell heterogeneity, microbial strain, and culture conditions in vitro. Infect. Immun. 68, 1953–1963 10.1128/iai.68.4.1953-1963.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Canton J., Neculai D., and Grinstein S. (2013) Scavenger receptors in homeostasis and immunity. Nat. Rev. Immunol. 13, 621–634 10.1038/nri3515 [DOI] [PubMed] [Google Scholar]
- 8. Plüddemann A., Neyen C., and Gordon S. (2007) Macrophage scavenger receptors and host-derived ligands. Methods 43, 207–217 10.1016/j.ymeth.2007.06.004 [DOI] [PubMed] [Google Scholar]
- 9. PrabhuDas M. R., Baldwin C. L., Bollyky P. L., Bowdish D. M. E., Drickamer K., Febbraio M., Herz J., Kobzik L., Krieger M., Loike J., McVicker B., Means T. K., Moestrup S. K., Post S. R., Sawamura T., et al. (2017) A consensus definitive classification of scavenger receptors and their roles in health and disease. J. Immunol. 198, 3775–3789 10.4049/jimmunol.1700373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Prabhudas M., Bowdish D., Drickamer K., Febbraio M., Herz J., Kobzik L., Krieger M., Loike J., Means T. K., Moestrup S. K., Post S., Sawamura T., Silverstein S., Wang X. Y., and El Khoury J. (2014) Standardizing scavenger receptor nomenclature. J. Immunol. 192, 1997–2006 10.4049/jimmunol.1490003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Whelan F. J., Meehan C. J., Golding G. B., McConkey B. J., and Bowdish D. M. (2012) The evolution of the class A scavenger receptors. BMC Evol. Biol. 12, 227 10.1186/1471-2148-12-227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bowdish D. M., and Gordon S. (2009) Conserved domains of the class A scavenger receptors: evolution and function. Immunol. Rev. 227, 19–31 10.1111/j.1600-065X.2008.00728.x [DOI] [PubMed] [Google Scholar]
- 13. Gowen B. B., Borg T. K., Ghaffar A., and Mayer E. P. (2001) The collagenous domain of class A scavenger receptors is involved in macrophage adhesion to collagens. J. Leukocyte Biol. 69, 575–582 10.1189/jlb.69.4.575 [DOI] [PubMed] [Google Scholar]
- 14. Feinberg H., Taylor M. E., and Weis W. I. (2007) Scavenger receptor C-type lectin binds to the leukocyte cell surface glycan Lewisx by a novel mechanism. J. Biol. Chem. 282, 17250–17258 10.1074/jbc.M701624200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li J. Y., Paragas N., Ned R. M., Qiu A., Viltard M., Leete T., Drexler I. R., Chen X., Sanna-Cherchi S., Mohammed F., Williams D., Lin C. S., Schmidt-Ott K. M., Andrews N. C., and Barasch J. (2009) Scara5 is a ferritin receptor mediating non-transferrin iron delivery. Dev. Cell 16, 35–46 10.1016/j.devcel.2008.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ojala J. R., Pikkarainen T., Tuuttila A., Sandalova T., and Tryggvason K. (2007) Crystal structure of the cysteine-rich domain of scavenger receptor MARCO reveals the presence of a basic and an acidic cluster that both contribute to ligand recognition. J. Biol. Chem. 282, 16654–16666 10.1074/jbc.M701750200 [DOI] [PubMed] [Google Scholar]
- 17. Sarraj M. A., McClive P. J., Wilmore H. P., Loveland K. L., and Sinclair A. H. (2005) Novel scavenger receptor gene is differentially expressed in the embryonic and adult mouse testis. Dev. Dyn. 234, 1026–1033 10.1002/dvdy.20594 [DOI] [PubMed] [Google Scholar]
- 18. Jiang Y., Oliver P., Davies K. E., and Platt N. (2006) Identification and characterization of murine SCARA5, a novel class A scavenger receptor that is expressed by populations of epithelial cells. J. Biol. Chem. 281, 11834–11845 10.1074/jbc.M507599200 [DOI] [PubMed] [Google Scholar]
- 19. Hohenester E., Sasaki T., and Timpl R. (1999) Crystal structure of a scavenger receptor cysteine-rich domain sheds light on an ancient superfamily. Nat. Struct. Biol. 6, 228–232 10.1038/6669 [DOI] [PubMed] [Google Scholar]
- 20. Cheng C., Hu Z., Cao L., Peng C., and He Y. (2019) The scavenger receptor SCARA1 (CD204) recognizes dead cells through spectrin. J. Biol. Chem. 294, 18881–18897 10.1074/jbc.RA119.010110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Reichhardt M. P., Loimaranta V., Lea S. M., and Johnson S. (2020) Structures of SALSA/DMBT1 SRCR domains reveal the conserved ligand-binding mechanism of the ancient SRCR fold. Life Sci. Alliance 3, e201900502 10.26508/lsa.201900502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ma H., Jiang L., Qiao S., Zhi Y., Chen X. X., Yang Y., Huang X., Huang M., Li R., and Zhang G. P. (2017) The crystal structure of the fifth scavenger receptor cysteine-rich domain of porcine CD163 reveals an important residue involved in porcine reproductive and respiratory syndrome virus infection. J. Virol. 91, e01897 10.1128/jvi.01897-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chappell P. E., Garner L. I., Yan J., Metcalfe C., Hatherley D., Johnson S., Robinson C. V., Lea S. M., and Brown M. H. (2015) Structures of CD6 and its ligand CD166 give insight into their interaction. Structure 23, 1426–1436 10.1016/j.str.2015.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rodamilans B., Muñoz I. G., Bragado-Nilsson E., Sarrias M. R., Padilla O., Blanco F. J., Lozano F., and Montoya G. (2007) Crystal structure of the third extracellular domain of CD5 reveals the fold of a group B scavenger cysteine-rich receptor domain. J. Biol. Chem. 282, 12669–12677 10.1074/jbc.M611699200 [DOI] [PubMed] [Google Scholar]
- 25. Somoza J. R., Ho J. D., Luong C., Ghate M., Sprengeler P. A., Mortara K., Shrader W. D., Sperandio D., Chan H., McGrath M. E., and Katz B. A. (2003) The structure of the extracellular region of human hepsin reveals a serine protease domain and a novel scavenger receptor cysteine-rich (SRCR) domain. Structure 11, 1123–1131 10.1016/s0969-2126(03)00148-5 [DOI] [PubMed] [Google Scholar]
- 26. Resnick D., Pearson A., and Krieger M. (1994) The SRCR superfamily: a family reminiscent of the Ig superfamily. Trends Biochem. Sci. 19, 5–8 10.1016/0968-0004(94)90165-1 [DOI] [PubMed] [Google Scholar]
- 27. Sarrias M. R., Grønlund J., Padilla O., Madsen J., Holmskov U., and Lozano F. (2004) The scavenger receptor cysteine-rich (SRCR) domain: an ancient and highly conserved protein module of the innate immune system. Crit. Rev. Immunol. 24, 1–37 10.1615/critrevimmunol.v24.i1.10 [DOI] [PubMed] [Google Scholar]
- 28. Nielsen M. J., Andersen C. B., and Moestrup S. K. (2013) CD163 binding to haptoglobin-hemoglobin complexes involves a dual-point electrostatic receptor-ligand pairing. J. Biol. Chem. 288, 18834–18841 10.1074/jbc.m113.471060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Madsen M., Møller H. J., Nielsen M. J., Jacobsen C., Graversen J. H., van den Berg T., and Moestrup S. K. (2004) Molecular characterization of the haptoglobin.hemoglobin receptor CD163: ligand binding properties of the scavenger receptor cysteine-rich domain region. J. Biol. Chem. 279, 51561–51567 10.1074/jbc.M409629200 [DOI] [PubMed] [Google Scholar]
- 30. Kristiansen M., Graversen J. H., Jacobsen C., Sonne O., Hoffman H. J., Law S. K., and Moestrup S. K. (2001) Identification of the haemoglobin scavenger receptor. Nature 409, 198–201 10.1038/35051594 [DOI] [PubMed] [Google Scholar]
- 31. Patel D. D., Wee S. F., Whichard L. P., Bowen M. A., Pesando J. M., Aruffo A., and Haynes B. F. (1995) Identification and characterization of a 100-kD ligand for CD6 on human thymic epithelial cells. J. Exp. Med. 181, 1563–1568 10.1084/jem.181.4.1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Arosio P., Ingrassia R., and Cavadini P. (2009) Ferritins: a family of molecules for iron storage, antioxidation and more. Biochim. Biophys. Acta 1790, 589–599 10.1016/j.bbagen.2008.09.004 [DOI] [PubMed] [Google Scholar]
- 33. Harrison P. M., and Arosio P. (1996) The ferritins: molecular properties, iron storage function and cellular regulation. Biochim. Biophys. Acta 1275, 161–203 10.1016/0005-2728(96)00022-9 [DOI] [PubMed] [Google Scholar]
- 34. Arosio P., Adelman T. G., and Drysdale J. W. (1978) On ferritin heterogeneity: further evidence for heteropolymers. J. Biol. Chem. 253, 4451–4458 [PubMed] [Google Scholar]
- 35. Arosio P., Yokota M., and Drysdale J. W. (1977) Characterization of serum ferritin in iron overload: possible identity to natural apoferritin. Br. J. Haematol. 36, 199–207 10.1111/j.1365-2141.1977.tb00640.x [DOI] [PubMed] [Google Scholar]
- 36. Worwood M., Dawkins S., Wagstaff M., and Jacobs A. (1976) The purification and properties of ferritin from human serum. Biochem. J. 157, 97–103 10.1042/bj1570097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Addison G. M., Beamish M. R., Hales C. N., Hodgkins M., Jacobs A., and Llewellin P. (1972) An immunoradiometric assay for ferritin in the serum of normal subjects and patients with iron deficiency and iron overload. J. Clin. Pathol. 25, 326–329 10.1136/jcp.25.4.326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cazzola M., Arosio P., Bellotti V., Bergamaschi G., Dezza L., Iacobello C., Ruggeri G., Zappone E., Albertini A., and Ascari E. (1985) Immunological reactivity of serum ferritin in patients with malignancy. Tumori 71, 547–554 [DOI] [PubMed] [Google Scholar]
- 39. Ghosh S., Hevi S., and Chuck S. L. (2004) Regulated secretion of glycosylated human ferritin from hepatocytes. Blood 103, 2369–2376 10.1182/blood-2003-09-3050 [DOI] [PubMed] [Google Scholar]
- 40. Lukina E. A., Levina A. A., Mokeeva R. A., and Tokarev Y. N. (1993) The diagnostic significance of serum ferritin indices in patients with malignant and reactive histiocytosis. Br. J. Haematol. 83, 326–329 10.1111/j.1365-2141.1993.tb08289.x [DOI] [PubMed] [Google Scholar]
- 41. Santambrogio P., Cozzi A., Levi S., and Arosio P. (1987) Human serum ferritin G-peptide is recognized by anti-L ferritin subunit antibodies and concanavalin-A. Br. J. Haematol. 65, 235–237 10.1111/j.1365-2141.1987.tb02271.x [DOI] [PubMed] [Google Scholar]
- 42. Leimberg M. J., Prus E., Konijn A. M., and Fibach E. (2008) Macrophages function as a ferritin iron source for cultured human erythroid precursors. J. Cell. Biochem. 103, 1211–1218 10.1002/jcb.21499 [DOI] [PubMed] [Google Scholar]
- 43. Sibille J. C., Kondo H., and Aisen P. (1988) Interactions between isolated hepatocytes and Kupffer cells in iron metabolism: a possible role for ferritin as an iron carrier protein. Hepatology 8, 296–301 10.1002/hep.1840080218 [DOI] [PubMed] [Google Scholar]
- 44. Coffman L. G., Brown J. C., Johnson D. A., Parthasarathy N., D'Agostino R. B. Jr., Lively M. O., Hua X., Tilley S. L., Muller-Esterl W., Willingham M. C., Torti F. M., and Torti S. V. (2008) Cleavage of high-molecular-weight kininogen by elastase and tryptase is inhibited by ferritin. Am. J. Physiol. Lung Cell. Mol. Physiol. 294, L505–L515 10.1152/ajplung.00347.2007 [DOI] [PubMed] [Google Scholar]
- 45. Coffman L. G., Parsonage D., D'Agostino R. Jr., Torti F. M., and Torti S. V. (2009) Regulatory effects of ferritin on angiogenesis. Proc. Natl. Acad. Sci. U. S. A. 106, 570–575 10.1073/pnas.0812010106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Parthasarathy N., Torti S. V., and Torti F. M. (2002) Ferritin binds to light chain of human H-kininogen and inhibits kallikrein-mediated bradykinin release. Biochem. J. 365, 279–286 10.1042/BJ20011637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang W., Knovich M. A., Coffman L. G., Torti F. M., and Torti S. V. (2010) Serum ferritin: past, present and future. Biochim. Biophys. Acta 1800, 760–769 10.1016/j.bbagen.2010.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Connelly K. G., Moss M., Parsons P. E., Moore E. E., Moore F. A., Giclas P. C., Seligman P. A., and Repine J. E. (1997) Serum ferritin as a predictor of the acute respiratory distress syndrome. Am. J. Respir. Crit. Care Med. 155, 21–25 10.1164/ajrccm.155.1.9001283 [DOI] [PubMed] [Google Scholar]
- 49. Goodall E. F., Haque M. S., and Morrison K. E. (2008) Increased serum ferritin levels in amyotrophic lateral sclerosis (ALS) patients. J. Neurol. 255, 1652–1656 10.1007/s00415-008-0945-0 [DOI] [PubMed] [Google Scholar]
- 50. Kiechl S., Willeit J., Egger G., Poewe W., and Oberhollenzer F. (1997) Body iron stores and the risk of carotid atherosclerosis: prospective results from the Bruneck study. Circulation 96, 3300–3307 10.1161/01.cir.96.10.3300 [DOI] [PubMed] [Google Scholar]
- 51. Kell D. B., and Pretorius E. (2014) Serum ferritin is an important inflammatory disease marker, as it is mainly a leakage product from damaged cells. Metallomics 6, 748–773 10.1039/c3mt00347g [DOI] [PubMed] [Google Scholar]
- 52. Kurz T., Gustafsson B., and Brunk U. T. (2011) Cell sensitivity to oxidative stress is influenced by ferritin autophagy. Free Radic. Biol. Med. 50, 1647–1658 10.1016/j.freeradbiomed.2011.03.014 [DOI] [PubMed] [Google Scholar]
- 53. Bresgen N., Jaksch H., Lacher H., Ohlenschläger I., Uchida K., and Eckl P. M. (2010) Iron-mediated oxidative stress plays an essential role in ferritin-induced cell death. Free Radic. Biol. Med. 48, 1347–1357 10.1016/j.freeradbiomed.2010.02.019 [DOI] [PubMed] [Google Scholar]
- 54. Mendes-Jorge L., Ramos D., Valença A., López-Luppo M., Pires V. M., Catita J., Nacher V., Navarro M., Carretero A., Rodriguez-Baeza A., and Ruberte J. (2014) L-ferritin binding to scara5: a new iron traffic pathway potentially implicated in retinopathy. PLoS One 9, e106974 10.1371/journal.pone.0106974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. McMahon C., Baier A. S., Pascolutti R., Wegrecki M., Zheng S., Ong J. X., Erlandson S. C., Hilger D., Rasmussen S. G. F., Ring A. M., Manglik A., and Kruse A. C. (2018) Yeast surface display platform for rapid discovery of conformationally selective nanobodies. Nat. Struct. Mol. Biol. 25, 289–296 10.1038/s41594-018-0028-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Andersen C. B., and Moestrup S. K. (2014) How calcium makes endocytic receptors attractive. Trends Biochem. Sci. 39, 82–90 10.1016/j.tibs.2013.12.003 [DOI] [PubMed] [Google Scholar]
- 57. Doi T., Higashino K., Kurihara Y., Wada Y., Miyazaki T., Nakamura H., Uesugi S., Imanishi T., Kawabe Y., and Itakura H. (1993) Charged collagen structure mediates the recognition of negatively charged macromolecules by macrophage scavenger receptors. J. Biol. Chem. 268, 2126–2133 [PubMed] [Google Scholar]
- 58. Krieger M. (1992) Molecular flypaper and atherosclerosis: structure of the macrophage scavenger receptor. Trends Biochem. Sci. 17, 141–146 10.1016/0968-0004(92)90322-z [DOI] [PubMed] [Google Scholar]
- 59. Krieger M., and Herz J. (1994) Structures and functions of multiligand lipoprotein receptors: macrophage scavenger receptors and LDL receptor-related protein (LRP). Annu. Rev. Biochem. 63, 601–637 10.1146/annurev.bi.63.070194.003125 [DOI] [PubMed] [Google Scholar]
- 60. Gao M., Monian P., Pan Q., Zhang W., Xiang J., and Jiang X. (2016) Ferroptosis is an autophagic cell death process. Cell Res. 26, 1021–1032 10.1038/cr.2016.95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hou W., Xie Y., Song X., Sun X., Lotze M. T., Zeh H. J. 3rd, Kang R., and Tang D. (2016) Autophagy promotes ferroptosis by degradation of ferritin. Autophagy 12, 1425–1428 10.1080/15548627.2016.1187366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Latunde-Dada G. O. (2017) Ferroptosis: Role of lipid peroxidation, iron and ferritinophagy. Biochim. Biophys. Acta 1861, 1893–1900 10.1016/j.bbagen.2017.05.019 [DOI] [PubMed] [Google Scholar]
- 63. Tang M., Chen Z., Wu D., and Chen L. (2018) Ferritinophagy/ferroptosis: Iron-related newcomers in human diseases. J. Cell. Physiol. 233, 9179–9190 10.1002/jcp.26954 [DOI] [PubMed] [Google Scholar]
- 64. Sun C., Yang H., Yuan Y., Tian X., Wang L., Guo Y., Xu L., Lei J., Gao N., Anderson G. J., Liang X. J., Chen C., Zhao Y., and Nie G. (2011) Controlling assembly of paired gold clusters within apoferritin nanoreactor for in vivo kidney targeting and biomedical imaging. J. Am. Chem. Soc. 133, 8617–8624 10.1021/ja200746p [DOI] [PubMed] [Google Scholar]
- 65. Truffi M., Fiandra L., Sorrentino L., Monieri M., Corsi F., and Mazzucchelli S. (2016) Ferritin nanocages: a biological platform for drug delivery, imaging and theranostics in cancer. Pharmacol. Res. 107, 57–65 10.1016/j.phrs.2016.03.002 [DOI] [PubMed] [Google Scholar]
- 66. Zhen Z., Tang W., Guo C., Chen H., Lin X., Liu G., Fei B., Chen X., Xu B., and Xie J. (2013) Ferritin nanocages to encapsulate and deliver photosensitizers for efficient photodynamic therapy against cancer. ACS Nano 7, 6988–6996 10.1021/nn402199g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wang Z., Li C., Ellenburg M., Soistman E., Ruble J., Wright B., Ho J. X., and Carter D. C. (2006) Structure of human ferritin L chain. Acta Crystallogr. D Biol. Crystallogr. 62, 800–806 10.1107/S0907444906018294 [DOI] [PubMed] [Google Scholar]
- 68. Minor W., Cymborowski M., Otwinowski Z., and Chruszcz M. (2006) HKL-3000: the integration of data reduction and structure solution–from diffraction images to an initial model in minutes. Acta Crystallogr. D Biol. Crystallogr. 62, 859–866 10.1107/S0907444906019949 [DOI] [PubMed] [Google Scholar]
- 69. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 66, 213–221 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 71. Cerny L. C., Stasiw D. M., and Zuk W. (1981) The logistic curve for the fitting of sigmoidal data. Physiol. Chem. Phys. 13, 221–230 [PubMed] [Google Scholar]
- 72. Raghava G. P., and Agrewala J. N. (1994) Method for determining the affinity of monoclonal antibody using non-competitive ELISA: a computer program. J. Immunoassay 15, 115–128 10.1080/15321819408013942 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The structures of the SRCR domains of human and mouse SCARA5 have been deposited in the Protein Data Bank with accession numbers 7C00 (for hSCARA5) and 7BZZ (for mSCARA5), respectively.