Significance
Despite widespread applications, the origins of functional connectivity remain elusive. Here we analyze human functional neuroimaging data. We decompose resting-state functional connectivity across time to assess the contributions of moment-to-moment activity cofluctuations to the overall connectivity pattern. We show that functional connectivity is driven by a small number of high-amplitude frames. We show that these frames are underpinned by a specific mode of brain activity; that the topography of this mode gets modulated during in-scanner tasks; and that high-amplitude frames encode personalized, subject-specific information. In summary, our parameter-free method provides an exact mathematical link between functional connectivity and frame-wise cofluctuations, creating opportunities for studying both static and time-varying functional brain networks.
Keywords: functional connectivity, dynamics, time-varying connectivity, naturalistic stimuli
Abstract
Resting-state functional connectivity is used throughout neuroscience to study brain organization and to generate biomarkers of development, disease, and cognition. The processes that give rise to correlated activity are, however, poorly understood. Here we decompose resting-state functional connectivity using a temporal unwrapping procedure to assess the contributions of moment-to-moment activity cofluctuations to the overall connectivity pattern. This approach temporally resolves functional connectivity at a timescale of single frames, which enables us to make direct comparisons of cofluctuations of network organization with fluctuations in the blood oxygen level-dependent (BOLD) time series. We show that surprisingly, only a small fraction of frames exhibiting the strongest cofluctuation amplitude are required to explain a significant fraction of variance in the overall pattern of connection weights as well as the network’s modular structure. These frames coincide with frames of high BOLD activity amplitude, corresponding to activity patterns that are remarkably consistent across individuals and identify fluctuations in default mode and control network activity as the primary driver of resting-state functional connectivity. Finally, we demonstrate that cofluctuation amplitude synchronizes across subjects during movie watching and that high-amplitude frames carry detailed information about individual subjects (whereas low-amplitude frames carry little). Our approach reveals fine-scale temporal structure of resting-state functional connectivity and discloses that frame-wise contributions vary across time. These observations illuminate the relation of brain activity to functional connectivity and open a number of directions for future research.
Resting-state functional connectivity (rsFC) refers to the correlation structure of functional magnetic resonance imaging (fMRI) blood oxygen level-dependent (BOLD) activity, usually estimated over the course of an entire scan session (1, 2). Interindividual differences in rsFC have been linked to variation in biological age (3, 4), cognitive state (5), and clinical status (6). Other studies have emphasized the dynamic nature of rsFC, using sliding window techniques to generate temporally blurred estimates of rsFC across time (7–9) and linking changes in network architecture to behavior (10, 11) and phenotypes (12, 13).
Despite intense interest and widespread application, the processes that underpin and shape rsFC are not fully understood. For instance, how do moment-to-moment fluctuations in connectivity contribute to the pattern of rsFC estimated over longer timescales? How are changes in connectivity supported by instantaneous fluctuations in brain activity?
In principle, these questions can be addressed using sliding-window approaches to track fluctuations in rsFC across time. However, the windowing procedure induces a blurring effect, making it impossible to localize time-varying connectivity in time and assess the contributions made by individual frames (14). On the other hand, methods like coactivation patterns (CAPs) allow for brain dynamics to be characterized at the resolution of single frames (15–19). Using these types of methods, previous studies have shown that coactivity patterns fluctuate across time, with the brain’s system-level organization expressed only during a select set of time points (20). However, these approaches generally require the specification of a seed region or a threshold for determining what constitutes high-amplitude activity. Consequently, a comprehensive and mathematically precise explanation for how these coactivity patterns combine to give rise to rsFC over longer timescales remains elusive (21).
Here we address these questions using a mathematically exact decomposition of rsFC into its frame-wise contributions, explicitly linking instantaneous patterns of cofluctuation to rsFC over longer timescales (22, 23). We find that at rest, cofluctuations are bursty and occur intermittently as part of whole-brain cofluctuation events that are uncorrelated with respiration, cardiac cycle, and in-scanner motion. We then show that rsFC estimated using only high-amplitude frames is highly correlated with rsFC estimated over the entire scan session, indicating that rsFC and its system-level organization are driven by cofluctuations during relatively few frames. We then show that high-amplitude cofluctuations are underpinned by the activation of a particular spatial mode of brain activity in which default mode and control networks are anticorrelated with sensorimotor and attentional systems. We then present two careful examinations of high-amplitude cofluctuations. First, we demonstrate that time series of cofluctuation amplitude synchronize across subjects during movie watching, and second, we show that subjects’ “fingerprints” are enhanced during high-amplitude frames compared to low-amplitude frames.
Results
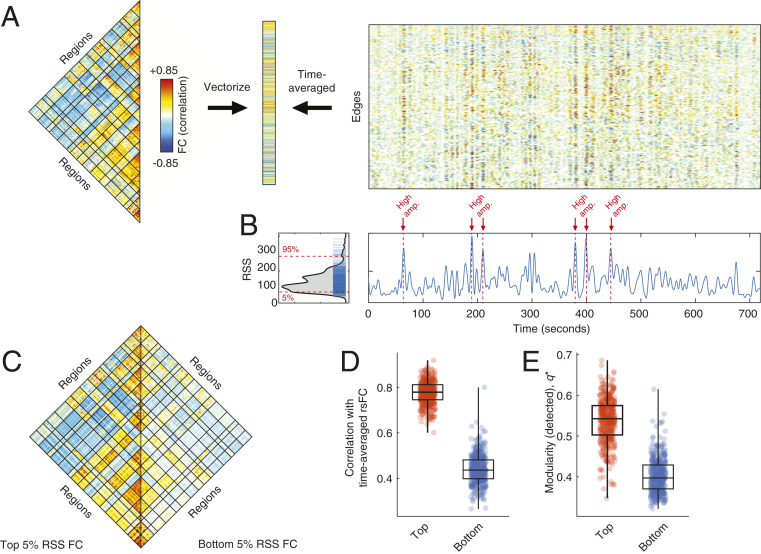
The strength of rsFC between two brain regions can be quantified as the Pearson correlation of their fMRI BOLD time series, which is calculated (after z-scoring) as the mean value of their element-wise product (24). By omitting the averaging step, we can temporally unwrap the correlation measure, which results in a new set of time series—one for every pair of brain regions (network edges)—whose elements represent the magnitude of cofluctuation between those regions resolved at every moment in time (Fig. 1A). These edge time series can be analyzed directly to pinpoint both the magnitude and timing of cofluctuations between pairs of brain regions.
Fig. 1.
Cofluctuation time series reveal bursty structure of resting-state functional connectivity. (A) We use a temporal unwrapping of the Pearson correlation to generate cofluctuation time series for every pair of brain regions (edges). The elements of the cofluctuation time series are the element-wise products of z-scored regional BOLD time series that, when averaged across time, yield vectors that are exactly equal to the Pearson correlation coefficient and can be rearranged to create a resting-state functional connectivity matrix. (B) We find that the cofluctuation time series contains moments in time where many edges collectively cofluctuate. We can identify these moments by calculating the RSS across all cofluctuation time series and plotting this value as a function of time. In B we label high- and low-amplitude frames. The distribution of edge cofluctuation amplitude is heavy tailed. We wanted to assess the contribution of high- and low-amplitude frames to the overall pattern of functional connectivity. To do this, we extracted the top and bottom 5% of all time points (ordered by cofluctuation amplitude) and estimated functional connectivity from those points alone. (C) Average functional connectivity across 100 subjects using top 5% (Left) and bottom 5% (Right). (D) In general, the networks estimated using the top 5% of time points were much more similar to traditional functional connectivity than those estimated using the bottom 5% of time points. (E) We performed a similar comparison of network modularity using networks reconstructed using top and bottom 5% frames.
In rsFC Is Driven by Short-Lived and High-Amplitude Cofluctuation Events and in High-Amplitude Frames Are Driven by Fluctuations of Task-Positive/Task-Negative Mode of Brain Activity, we analyze cofluctuation time series constructed from functional imaging data acquired as part of the Human Connnectome Project (25, 26) (see Materials and Methods for details). All results reported in those sections generated using these data; we replicate these findings using a second dataset (27), with results reported in SI Appendix. In Intersubject Synchrony of Whole-Brain Cofluctuation Amplitude during Passive Movie Watching and in High-Amplitude Cofluctuations Enhance Identifiability, we analyze an independently acquired movie-watching dataset (28, 29) and data from the Midnight Scan Club (MSC) (27, 30), respectively.
rsFC Is Driven by Short-Lived and High-Amplitude Cofluctuation Events.
While past studies have used sliding window methods to generate estimates of moment-to-moment fluctuations in rsFC (8, 9), the use of a windowing procedure results in a temporally blurred estimate of rsFC. This restricts the timescale of observations regarding dynamic changes in functional connectivity to the width of the time window, generally on the order of dozens of frames (approximately 1 min of real time, although we note that recently, several groups have explored methods for estimating time-varying FC at a frame-wise timescale, e.g., refs. 31 and 32). Here we address this limitation using cofluctuation times series.
When analyzed across the whole brain, we find that edge time series exhibit bursty behavior, such that the amplitude of cofluctuations (quantified by computing the root sum square [RSS]) moves around a mean value but is punctuated by brief, intermittent, and disproportionately large fluctuations (Fig. 1B). These high-amplitude frames are not directly related to cardiac and respiratory cycles, in-scanner head motion (SI Appendix, Fig. S1), and spectral properties of fMRI BOLD time series (SI Appendix, Fig. S2) and appear aperiodic with heavy-tailed distributions of size, duration, and intervals (SI Appendix, Fig. S3).
To better understand how instantaneous cofluctuations contribute to whole-brain rsFC, we isolated high-amplitude frames and compared them with low-amplitude episodes (top and bottom 5% in terms of cofluctuation amplitude; 60 frames for HCP; see SI Appendix, Fig. S4, for comparisons at other percentiles). We then estimated rsFC separately for each category, using only fMRI BOLD data corresponding to those time points, and compared the resulting networks. First, we found that connection weights were significantly stronger during high-amplitude frames than low-amplitude (within-sample t test, ; Fig. 1C). Next, we calculated the similarity of rsFC estimated during high- and low-amplitude episodes with respect to time-averaged rsFC estimated using the full time series. We found that the high-amplitude networks were highly correlated with rsFC () while the low-amplitude networks were much less correlated () and that these differences were highly significant (t test, ; Fig. 1D). We also performed an analogous comparison of network modularity (33), an index that can be interpreted as a measure of how segregated a network’s systems are from one another. As before, we found that modularity was greater in the high-amplitude networks () compared to the networks estimated from low-amplitude frames () (t test, ; Fig. 1E).
In SI Appendix we show similar results in a second dataset (SI Appendix, Fig. S5). We also demonstrate that these effects persist with highly conservative motion censoring (SI Appendix, Fig. S6), when using an alternative strategy for estimating networks from the top and bottom 5% time points (SI Appendix, Fig. S7), and when comparing against a null model that preserves the temporal structure of cofluctuation amplitude while sampling frames randomly from the entire time series (SI Appendix, Fig. S8).
Collectively, these results suggest that rsFC, estimated over long timescales, is driven by a small number of brief, intermittent, and high-amplitude cofluctuations. The network structure over these points in time contributes disproportionately to the overall modularity and system-level organization of cerebral cortex, as estimated from long-time averages of rsFC. In contrast, low-amplitude cofluctuations are only weakly related to the overall pattern of rsFC and correspond to less modular architectures.
High-Amplitude Frames Are Driven by Fluctuations of Task-Positive/Task-Negative Mode of Brain Activity.
In the previous section we demonstrated that time-averaged rsFC can be explained by high-amplitude cofluctuations that occur during a relatively small number of frames. It remains unclear, however, whether high-amplitude frames are underpinned by a specific pattern of brain activity or whether they reflect contributions from multiple distinct patterns. Here we address this question directly, by investigating the patterns of brain activity that occur at the same time as high-amplitude frames.
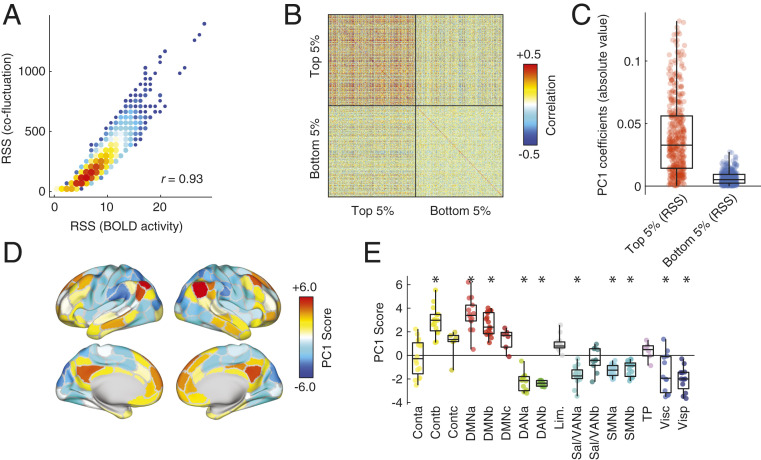
As a first point of comparison, we calculated the RSS of both the cofluctuation time series as well as the z-scored fMRI BOLD time series. We found that across subjects, these time series were highly correlated (), indicating that high-amplitude frames have an almost one-to-one correspondence with high-amplitude BOLD fluctuations (Fig. 2A). This relationship is expected; because cofluctuations are calculated as products of z-scored regional activity, their amplitudes will necessarily be correlated with one another.
Fig. 2.
Relationship of network cofluctuations with BOLD fluctuations. In rsFC Is Driven by Short-Lived and High-Amplitude Cofluctuation Events we demonstrated that resting-state functional connectivity could be explained on the basis of relatively few frames during which high-amplitude cofluctuations occurred. Here we relate those cofluctuation frames to BOLD activity fluctuations. We first calculate the RSS amplitude of BOLD activity at each time point and compare that to the amplitude of cofluctuations. (A) Pooling data from across subjects, we find that these two variables are highly correlated. (B) To investigate this relationship further, we extract mean activity patterns for each subject and for each scan during the top and bottom 5% time points, indexed according to cofluctuation amplitude. Here we show the correlation matrix of those activity vectors. (C) We then performed a principal component analysis of this correlation matrix and found that absolute value of coefficients for the first component (PC1) were greater for the top 5% than the bottom 5%, and (D and E) the PC1 score corresponded to activity patterns that emphasized correlated fluctuations of default mode and control networks that were weakly or anticorrelated with fluctuations elsewhere in the brain. Asterisks indicate systems whose mean PC1 score was significantly greater (more positive or negative) than expected by chance (permutation test; FDR fixed at 5%; ). These observations suggest that high-amplitude cofluctuations, which drive resting-state functional connectivity, are underpinned by instantaneous activation and deactivation of default mode and control network areas.
Given that fluctuations in BOLD activity are greater during high-amplitude frames compared to low-amplitude frames, we asked whether they formed a consistent and recognizable pattern of activity. To address this question, we calculated the mean activity pattern for each subject during high- and low-amplitude frames and computed between-subject and between-scan similarity (Fig. 2B). In general, activity during high-amplitude frames was more correlated across subjects compared to the activity patterns during low-amplitude frames (t test, ). To better understand what was driving these correlations, we performed a principal components analysis of the activity patterns during high- and low-amplitude frames, aggregated over all subjects and scans. We focused on the first principal component (PC1), which explained 26% of total variance. The coefficients for PC1 were, on average, much greater for high-amplitude frames compared to low-amplitude (t test, ; Fig. 2C), indicating that PC1 was descriptive of activity patterns during high-amplitude frames but less so for low-amplitude frames. We then mapped component scores for PC1 onto the cortical surface and found that PC1 corresponded to a mode of activity that delineates regions in default mode and control networks from sensorimotor and attentional networks (Fig. 2 D and E). We replicated these results in a second dataset (SI Appendix, Fig. S9).
These results suggest that underlying high-amplitude frames is a mode of brain activity whose spatial pattern resembles the traditional task-positive/task-negative division of the brain (34). This pattern of activity is similar across individuals, suggesting a conserved mechanism by which rsFC emerges from brain activity. These observations suggest a fundamental link between distinct patterns of brain activity and connectivity while further clarifying the origins of high-amplitude frames.
Intersubject Synchrony of Whole-Brain Cofluctuation Amplitude during Passive Movie Watching.
In the previous sections we showed that rsFC can be viewed as an average of time-varying cofluctuations. We also showed that time-averaged rsFC is disproportionately impacted by high-amplitude frames that are, themselves, underpinned by a specific mode of brain activity and were not clearly related to motion or physiological artifacts. What, then, is the purpose of high-amplitude frames? Are they random cofluctuations, or are they related to fluctuations in an individual’s brain/cognitive state? To address these questions, we explored the cofluctuation time series for a cohort of 29 subjects that were scanned multiple times at rest and while passively viewing complex, naturalistic stimuli (movies) (28).
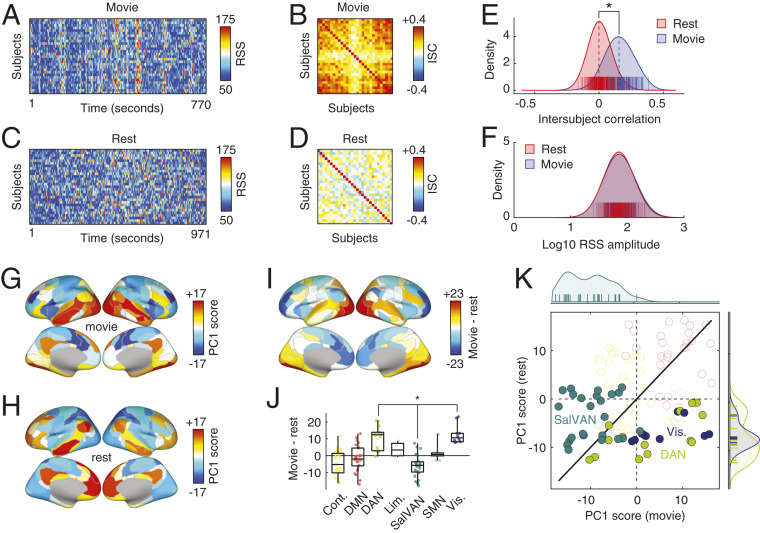
Specifically, we computed edge time series for all subjects and scans for both conditions. From these edge time series, we estimated the cofluctuation amplitude across all node pairs. For a given scan, this procedure results in 29 time series (1 per subject) of identical length. We found that cofluctuation time series were correlated across subjects during movie watching (Fig. 3 A and B) but uncorrelated during rest (Fig. 3 C and D). We directly compared the distributions of intersubject correlations between conditions (all scans from the same condition pooled together), discovering that as expected, the mean intersubject correlation was greater during movie watching than at rest (permutation test, ; Fig. 3E). Importantly, we found no difference between conditions for the overall amplitude of RSS values (permutation test, ; Fig. 3F).
Fig. 3.
Whole-brain cofluctuation amplitude synchronizes during passive movie watching. We compared cofluctuation time series during resting state and movie watching. For both conditions, we computed cofluctuation time series for 29 subjects. We show those time series in A (movie) and C (rest). We find that when subjects watch movies, their cofluctuation time series are synchronous, presumably due to the shared audiovisual stimulus. At rest, cofluctuation time series are asynchronous. We demonstrate this synchrony by computing the intersubject correlation matrix of subjects’ cofluctuation time series. We show matrices for movie watching and rest in B and D, respectively. By comparing the elements of these matrices, we demonstrate statistically that movie watching leads to increased intersubject correlations. We show the distributions in E. We find, however, that the overall amplitude of fluctuations (RSS) is not statistically different from one condition to the other (F). To further contrast these two conditions, we repeated the analysis from High-Amplitude Frames Are Driven by Fluctuations of Task-Positive/Task-Negative Mode of Brain Activity to identify modes of brain activity that underpin high-amplitude frames. We find that the resting mode recapitulates the topographic distribution reported in the previous section (H), emphasizing a task-positive/task-negative division. During movie watching, however, the mode of activity emphasizes contributions of visual and dorsal attention networks (G). In I–K, we compare rest and movie-watching modes of activity more directly. I depicts the region-wise differences in modes, J groups those differences by system, and K presents them as a scatterplot, highlighting differences associated with visual, dorsal attention, and salience/ventral attention networks.
Next, we explored differences between movie watching and rest in terms of brain activity patterns during high- and low-amplitude frames. Our exploration consisted of two analyses. First, and as in the previous section, we extracted activity patterns during high- and low-amplitude frames (top and bottom 5% by cofluctuation amplitude) separately for the movie-watching and resting conditions. We then performed principal component analysis (PCA) on these matrices and retained the top PC score for each condition. Interestingly, these PC scores exhibited distinct topography; the movie-watching PC (Fig. 3G) emphasized activity in visual and dorsal attention networks, whereas the resting PC (Fig. 3H) recapitulated the pattern shown in the previous section, emphasizing a task-positive/task-negative mode of activity. To directly compare these two patterns, we computed their element-wise (region-wise) difference and grouped these differences by system (Fig. 3 I and J). As expected, we found statistically significant differences in the dorsal attention and visual systems (movie rest; false discovery rate fixed at ) and salience/ventral attention system (movie rest). These differences are further evident when we plot the PCs against one another, revealing that these systems deviate from the identity line (Fig. 3K).
We note that another strategy for comparing movie-watching and resting-state conditions is to analyze them simultaneously by concatenating high-amplitude activity patterns from both conditions into a single matrix and jointly decomposing that matrix using PCA. This procedure results in modes of activity that are shared across both conditions. Here we retain both the first and second PCs (SI Appendix, Fig. S10 A and C), whose spatial topography is similar to what we show in Fig. 3 G and H. As expected, we find differences between the two maps in terms of their PC coefficients, with movie-watching frames loading more strongly onto the first map (permutation test, ; SI Appendix, Fig. S10B) and resting frames loading more strongly onto the second (permutation test, ; SI Appendix, Fig. S10D).
Viewed collectively, these results complement our previous findings that cofluctuation time series are not clearly related to motion or physiological artifacts. Importantly, we demonstrate that subjects’ cofluctuation time series synchronize when jointly presented with complex, time-varying, and naturalistic stimuli. This observation, combined with the topographic differences between movie-watching and resting-state activity during high-amplitude frames, strongly suggests that cofluctuation amplitude is at least in part modulated by subjects’ cognitive states.
High-Amplitude Cofluctuations Enhance Identifiability.
In the previous sections, we showed that high-amplitude frames contribute disproportionately to the brain’s static FC patterns, shape its modular structure, are underpinned by a population-level mode of brain activity, and synchronize during viewing of naturalistic stimuli. Do they also enhance the identifiability of individual subjects? That is, does FC estimated during high-amplitude frames bear a stronger signature of a subject than FC estimated during low-amplitude frames?
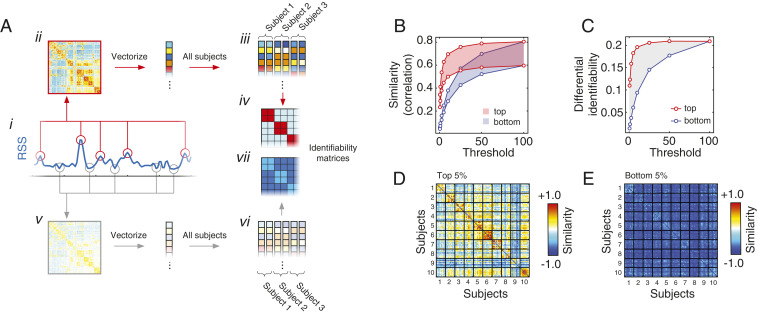
To test this question, we calculated the cofluctuation amplitude at each time point (Fig. 4A, i). We then isolated the frames with the highest amplitude and estimated FC using only these frames (Fig. 4A, ii). Repeating this procedure for all subjects and scans resulted in a set of 100 feature vectors (10 subjects 10 scans) that encode a subjects’ FC patterns (Fig. 4A, iii). We then compute the similarity matrix—otherwise known as the identifiability matrix—and compare the mean within-subject similarity to the mean between-subject similarity (Fig. 4A, iv). This measure—the differential identifiability—indicates how much more similar subjects’ FC patterns are to themselves than to those of other subjects. We repeat this entire procedure using only low-amplitude frames (Fig. 4A, v–vii) and compare low- and high-amplitude differential identifiability.
Fig. 4.
Connectome fingerprints are strong during high-amplitude frames and weaker during low-amplitude frames. We investigated whether subject-specific features of rsFC were more prevalent during high- or low-amplitude cofluctuations. To address this question, we identified frames (time points) with the highest and lowest cofluctuation amplitude and estimated subjects FC using these data only. We then calculated the intersubject similarity matrix, i.e., the identifiability matrix. (A) Illustration of this general procedure, beginning by isolating high-amplitude time points (i), estimation of FC (ii), repeating this procedure for all subjects (iii), and estimating the intersubject similarity matrix (iv). An identical procedure was carried out for low-amplitude frames and is illustrated in v–vii. (B) We calculated the mean within- and between-subject similarity using both the top (red) and bottom (blue) frames, ordered by cofluctuation amplitude. For each set of frames, we produce two separate curves, one for the within-subject similarity and another for the between-subject similarity. The area between the curves is the differential identifiability, or the extent to which subjects’ FC patterns are more similar to themselves than to FC estimated from other subjects. (C) We found that differential identifiability was always greater when FC was estimated using the top frames, ordered by amplitude. For the sake of visualization, we show identifiability matrices estimated using (D) high- and (E) low-amplitude frames.
In general, we find that intersubject similarity is greater during high-amplitude frames than compared to low-amplitude frames and over a range of thresholds (Fig. 4B). Importantly, we also find that the gap between within- and between-subject similarity, differential identifiability, is also greater when estimated using high-amplitude frames (Fig. 4 C–E). This observation suggests that high-amplitude frames may carry more individualized and distinguishable information about subjects than low-amplitude frames.
Discussion
Here we presented a general approach for temporally unwrapping Pearson correlations to generate time series of interregional cofluctuations along network edges. This simple procedure enables us to parse the contributions made by individual frames to rsFC. We find that in general, we can accurately estimate whole-brain rsFC and its system-level organization using data from a relatively small number of frames. Importantly, we link these frames to a high-modularity brain state and to a specific mode of brain activity, in which default mode and control networks fluctuate in opposition to sensorimotor and attention systems. Our results also suggest that the cofluctuation patterns, at a coarse scale, capture cognitively relevant fluctuations in brain state and that high-amplitude frames encode signatures of an individual.
Decomposing Static FC into Cofluctuation Snapshots.
Central to this paper is the observation that static FC can be nonparametrically decomposed into a series of time-varying snapshots, each of which expresses an instantaneous pattern of interregional cofluctuation. Critically, the average of these patterns across time is exactly equal to whole-brain static FC. This mathematical truism allows us to neatly assess the contributions of momentary cofluctuations to the overall FC pattern and to establish a clear link between FC and fluctuations in brain activity.
Our findings, which complement previous work (15–18, 20, 32), leverage a mathematically exact decomposition of FC into its frame-wise contributions to suggest that static FC is driven by contributions from relatively few time points, namely, those with the highest levels of cofluctuation amplitude. Frames with low levels of cofluctuation, on the other hand, contribute little. Because the cofluctuation time series are estimated at a temporal resolution of single frames, we directly compared high-amplitude frames with coincident patterns of brain activity. We established that at rest, high-amplitude cofluctuations occur in tandem with a specific mode of brain activity that emphasizes oppositional activation of sensorimotor and association cortex. Notably, similar patterns of activity have been reported in other state-based analyses of brain dynamics (39, 40), linking this pattern of activity to mentation during wakeful rest and working memory performance.
The observations reported here both clarify and challenge core assumptions concerning static FC and brain network dynamics (41). Specifically, our findings suggest that whole-brain FC follows a bursty trajectory through a high-dimensional state space, with extended periods of quietude punctuated by brief and intermittent events, whose timing is not clearly related to motion or physiological artifacts (42). This observation leads to several questions, the most important of which concerns the origins of high-amplitude frames. Are they spontaneous occurrences? Are they relevant, in any way, to ongoing cognitive processes? How individualized are high-amplitude frames?
Linking High-Amplitude Cofluctuations to Cognition and Individual Differences.
We performed two separate analyses in order to help clarify the origins of high-amplitude frames. First, we performed a comparison of their structure at rest and during movie watching (28). While the amplitude of cofluctuations was not statistically different between conditions, we found that cofluctuations were correlated during movie watching, suggesting that high-amplitude frames may be driven by audiovisual features in movies. This finding supports the hypothesis that the timing of high-amplitude frames is linked to perception and processing of sensory information and further suggests that high-amplitude frames are not simply spontaneous occurrences. These observations open up possibilities for future studies that leverage the temporal structure of cofluctuation amplitudes to track changes in an individual’s cognitive state across time.
Importantly, we also discover differences in the mode of brain activity underpinning high-amplitude cofluctuations in movie watching compared to rest. In particular, we found stronger expression of visual and dorsal attention networks, brain systems that one might hypothesize to play an important role in processing visual information and redirecting attentional resources while viewing complex naturalistic stimuli (43, 44). This finding also demonstrates that although high-amplitude cofluctuations can occur spontaneously in correlated, modular systems, their character and timing are modulated by time-varying sensory input, presenting an opportunity for future studies to comprehensively map the task-evoked topography of high-amplitude cofluctuations (45).
In our second analysis, we asked whether high-amplitude cofluctuations were personalized and idiosyncratic (27, 46, 47). To address this question, we estimated subjects’ FC separately using high- and low-amplitude frames and compared these networks in terms of their differential identifiability—the extent to which the similarity of FC patterns was stronger within subjects than between (48). Surprisingly, we found that identifiability was significantly stronger during high-amplitude cofluctuations, suggesting that subject-specific information is expressed more strongly during those frames.
Collectively, these findings suggest that the structure of high-amplitude cofluctuations is highly organized. It tracks time-varying fluctuations in cognitive state and is deeply personalized. These are key observations with clear implications for the study of brain–behavior associations, clinical neuroscience, and phenotype discovery, where the ability to make inferences is limited by the amount of data available. Our results suggest that by capitalizing on the fact that high-amplitude cofluctuations carry more subject-specific information than low-amplitude cofluctuations, it may be possible to generate robust network-level biomarkers using a relatively small number of frames and reducing the amount of required data (49) [we note this concept is being explored with other imaging modalities (50)]. This approach may be especially useful in clinical and developmental neuroscience, which study populations with characteristics that generally prohibit the extended scan durations necessary for stably estimating FC (51). For instance, because high-amplitude cofluctuations carry more information about static FC than low-amplitude cofluctuations and because they encode identifiable features of subjects, implementing experimental paradigms that elicit large cofluctuations at greater frequency may obviate the need for long scan sessions and large quantities of data and yield superior estimates of FC.
System-Level Organization Emergences from the Structure of High-Amplitude Cofluctuations.
Last, our findings hint at a crucial link between instantaneous fluctuations in activity and the organization of rsFC (31, 52). Many studies have found that the community structure of rsFC resembles known coactivation patterns, including task-evoked activity (53, 54). Here we proposed a strategy that enabled us to tease apart the precise contribution of instantaneous BOLD fluctuations (and their topography) to rsFC.
We demonstrated that a particular pattern of activity involving default mode and control regions is primarily responsible for driving high-amplitude frames and, in turn, whole-brain rsFC. While this mode made the greatest contribution, it is likely that other modes make nontrivial contributions as well. By extending the definition to include lower-amplitude fluctuations, we expect to find patterns of activity that correspond to other, well-known brain systems (16). Moreover, we speculate that these patterns likely recombine in different proportions as a function of task complexity and domain (53, 55) and across individuals (47).
We note that other studies have shown, using a wide range of approaches, that time-varying and static rsFC are related to one another and that this relationship depends, to a large extent, on high-amplitude network states. For instance, ref. 18 identified time points of high-amplitude activity using sliding-window methods and seed-based analyses to show that activity coincides with increased correlation (rsFC). Indeed, comparable results have been reported using similar methods (15, 17, 20, 36, 38, 42, 56–58). The principal finding of these studies is that high-amplitude activity is somehow related to stronger FC or the expression of particular brain systems.
While illuminating, these papers have some limitations. Notably, they describe relationships between FC and high-amplitude states but lack a mathematical mechanism for why this relationship exists. In other cases, the observations require user-defined parameters to determine what constitutes high-amplitude activity, to specify a seed voxel or region for computing FC, or the width of a sliding window. In contrast, our approach addresses these limitations directly. Our first finding is in agreement with the above-referenced papers but is precipitated from a mathematically precise decomposition of static FC into its exact frame-wise contributions. With this decomposition, we can determine how individual time points combine to give rise to patterns of time-averaged, static FC. This recombination requires no additional parameterization and no sliding window and is naturally compared for all pairwise connections (not just seeds). In summary, the framework we present here serves to unify these previous observations by providing a mathematical framework to explicitly link instantaneous patterns of cofluctuation to static FC.
In future work, the proportion of variance explained by different patterns and other statistics related to high-amplitude frames, including the frequency with which they occur, may serve as potent correlates of cognitive and disease state. Because high-amplitude frames appear to drive the overall configuration of rsFC, we further speculate that their statistics may serve as important complements to traditional measures of rsFC.
Future Work.
The approach developed here presents several exciting opportunities for future studies. These include investigating time-varying FC using cofluctuation patterns, which provide frame-wise estimates of network structure and circumvent limitations of sliding window approaches (7, 14, 59). Other possibilities include mapping the relationship of structural connectivity to regional fluctuations or interregional cofluctuations during high- and low-amplitude frames (60) and studying individual differences in cognitive, development, and disease state based on features extracted from high-amplitude frames, which we show provide more reliable estimates of subject-level networks.
A remaining open question concerns the neurobiology that shapes high-amplitude cofluctuations. On one hand, their infrequent occurrence could reflect a dynamic strategy for limiting the consumption of metabolic resources (9, 61). This theory is supported by previous reports demonstrating that metabolic activity is elevated within the default mode network (62–64), a system that overlaps closely with the dominant mode of activity we find underpins high-amplitude frames. On the other hand, high-amplitude frames are, to some extent, a mathematical necessity emerging in correlated, modular systems. For instance, a group of brain regions form a mutually correlated functional module, then by definition their activity (and coactivity) will follow similar time courses, with a tendency to fluctuate together at the same instants in time. However, the observation that high-amplitude cofluctuations synchronize during movie watching suggests that they are also related to some underlying psychological process (with a presumed neurobiological correlate). Future work should investigate the neurobiological underpinnings of high-amplitude cofluctuations in greater detail.
Importantly, the entire cofluctuation time series enterprise could be extended in several important ways, including by applying it to other imaging modalities, e.g., electrophysiological recordings (65–68) or fluorescence imaging data (69, 70). Additionally, it would be straightforward to calculate cofluctuation time series after partialing out the effects of activity from other regions in the brain (24) or to investigate temporal dependencies and lags between brain regions (71, 72).
Finally, the approach developed here decomposes functional connections into their exact frame-wise contributions. We speculate that this decomposition might offer a selective means of addressing in-scanner motion, e.g., by identifying and censoring time points impacted by motion on an edge-wise basis (73, 74).
Relationship with Existing Approaches.
We note that the analysis of cofluctuation time series is conceptually similar to several existing methods (20, 36–38) or in some cases even builds upon shared mathematical machinery (75). For instance, multiplication of temporal derivatives (MTDs) (76) calculates the element-wise products using differenced activity time series for all pairs of nodes. These time series are then convolved with a kernel to generate smooth estimates of time-varying nFC. Although similar, our approach relies on untransformed activity to estimate edge time series, thereby preserving the relationship between static nFC and the mean value of each edge time series. Furthermore, our approach omits the smoothing step, making it, in principle, capable of detecting fluctuations in network structure over shorter timescales compared to MTDs. Another related method is CAPs (15, 16), which extracts and clusters voxel- or vertex-level activity during high-activity frames. Because a voxel can be coactive under different contexts, the cluster centroids spatially overlap with one another.
Although these approaches arrive at similar conclusions, they possess distinct advantages and disadvantages that make some methods uniquely well suited for testing specific hypotheses and research questions. For instance, MTDs and the analysis of cofluctuation time series presented here are appropriate for tracking patterns of connectivity across time. In the case of cofluctuation time series, which are mathematically related to the static FC pattern, our approach is especially well suited for assessing the contributions of frame-wise cofluctuation patterns to the brain’s overall FC (we note that this relationship, to our knowledge, has not been previously discussed in the extant literature). CAPS and innovation-driven CAPS on the other hand, are better suited for studying activity patterns and tracking their cooccurrences across time. In principle, a systematic and careful comparison of these methods could be carried out in future work.
Finally, we note that our approach incorporates elements of both CAPs and MTDs. Like MTDs and other sliding-window methods, our approach yields a time series of node-by-node matrices, each of which encodes pairwise relationships among brain regions across time. In the case of sliding-window methods, the elements in each matrix represent estimates of instantaneous correlations, which requires first estimating variances (a second moment) from a limited set of observations, which can lead to inaccurate and noisy FC inference, especially when the number of observations is small (narrow windows) (14, 77). Instead, the matrices generated using our approach encode instantaneous cofluctuation magnitudes, like CAPs. That is, each element indicates the magnitude with which the activity of two brain regions is instantaneously deflecting in the same direction. Importantly, deflections are calculated with respect to means and variances estimated using the full set of observations (the entire scan). Consequently, the time-varying estimates of cofluctuations reported here may be less sensitive to noise than sliding-window methods.
Additionally, it is important to note that while our approach and CAPs are similar in some ways, e.g., they both operate on single-frame timescales, our method is nonetheless distinct and has unique advantages. Most importantly, our method is built upon a mathematically exact decomposition of static rsFC into its frame-wise contributions. This decomposition enables us to quantify, precisely, how individual time points impact static rsFC. Here we use this property to demonstrate that a relatively small number of frames are necessary to explain rsFC [a finding that has been reported elsewhere (15, 17, 36)], to show that cofluctuations become synchronized during movie watching, and that high-amplitude frames can be used to enhance subjects’ connectome-based fingerprints. Second, our method is parameter-free; the decomposition does not depend on a specification of ad hoc thresholds for high-amplitude activity nor does it require that we select a seed region or brain system in advance. Rather, our decomposition method considers all activity levels and the entire network simultaneously. As noted earlier, we envision this approach being useful for future studies of time-varying FC, for generating more sensitive and subject-specific biomarkers, and in conjunction with non-fMRI imaging modalities.
Limitations.
One of the most key limitations concerns the calculation and interpretation of cofluctuation time series. The procedure for calculating edge time series begins by z-scoring each brain region’s activity time series. This procedure, however, is only appropriate if the sample mean and SD are temporally invariant (78). If there is a sustained increase or decrease in activity, e.g., the effect of a blocked task, then the z-scoring procedure can result in a biased mean and SD resulting in poor estimates of fluctuations in activity. To minimize the likelihood of this occurring, we focused on resting-state and movie-watching data rather than blocked tasks. In future work, investigation of task-evoked cofluctuations could be investigated by employing already common preprocessing steps, e.g., constructing task regressors to remove the first-order effect of tasks on activity (79).
Conclusion.
In conclusion, our study discloses a link between cortical activity and rsFC, facilitating a statistical explanation of the brain’s system-level architecture in terms of intermittent, short-lived, high-amplitude fluctuations in activity and coactivity. Our methodological framework is readily applicable to other imaging datasets and recording modalities, including observations at neuronal scales, enabling the study of neural coactivity at unprecedented temporal resolution.
Materials and Methods
Datasets.
We analyzed three separate datasets. Specifically, we focused on resting-state data from both The Human Connectome Project (HCP) and MSC. These data were processed similarly, the details of which are described in this section. The third dataset, which has been analyzed elsewhere (28, 49, 80), includes both resting-state and movie-watching data from a cohort of 29 individuals. This dataset was processed separately using a different procedure and is described in its own section. The processing pipelines resulted in parcellations of cortex into parcels (nodes) (81) in the case of the HCP and MSC datasets and parcels (52) in the case of the Indiana University dataset. The processing pipelines are described in detail in SI Appendix.
Cofluctuation Time Series.
Constructing networks from fMRI data (or any neural time series data) requires estimating the statistical dependency between every pair of time series. The magnitude of that dependency is usually interpreted as a measure of how strongly (or weakly) those voxels or parcels are functionally connected to each other. By far the most common measure of statistic dependence is the Pearson correlation coefficient. Let and be the time series recorded from voxels or parcels and , respectively. We can calculate the correlation of and by first z-scoring each time series, such that , where and are the time-averaged mean and SD. Then, the correlation of with can be calculated as . Repeating this procedure for all pairs of parcels results in a node-by-node correlation matrix, i.e., an estimate of FC. If there are nodes, this matrix has dimensions .
To estimate edge-centric networks, we need to modify the above approach in one small but crucial way. Suppose we have two z-scored parcel time series, and . To estimate their correlation we calculate the mean of their element-wise product (not exactly the average, because we divide by rather than ). Suppose, instead, that we never calculate the mean and simply stop after calculating the element-wise product. This operation would result in a vector of length whose elements encode the moment-by-moment cofluctuation magnitude of parcels and . For instance, suppose at time , parcels and simultaneously increased their activity relative to baseline. These increases are encoded in and as positive entries in the th position, so their product is also positive. The same would be true if and decreased their activity simultaneously (because the product of negatives is a positive). On the other hand, if increased while decreased (or vice versa), this would manifest as a negative entry. Similarly, if either or increased or decreased while the activity of the other was close to baseline, the corresponding entry would be close to zero.
Accordingly, the vector resulting from the element-wise product of and can be viewed as encoding the magnitude of moment-to-moment cofluctuations between and . An analogous vector can easily be calculated for every pair of parcels (network nodes), resulting in a set of cofluctuation (edge) time series. With parcels, this results in pairs, each of length .
Modularity Maximization.
Modularity maximization is a heuristic for detecting communities in networks (82). Intuitively, it attempts to decompose a network into nonoverlapping subnetworks such that the observed density of connections within subnetworks maximally exceeds what would be expected by chance, where chance is determined by the user. The actual process of detecting communities is accomplished by choosing community assignments that maximize a modularity quality function, , defined as
| [1] |
where is the element of the modularity matrix, which represents the observed weight of the connection between nodes and minus the expected weight. The variable is the community assignment of node , and is the Kronecker delta function, whose value is 1 when and 0 otherwise. The modularity, , is effectively a sum over all edges that fall within communities and is optimized when the observed weights of connections is maximally greater than the expected. In general, larger values of are thought to reflect superior community partitions.
Signed and Correlation Matrices.
In this manuscript, we used the following variant of modularity, , which has been shown to be especially well suited for use with correlation matrices (33):
| [2] |
where . In this expression, represents either the positive or negative elements of the correlation matrix, , and .
Differential Identifiability.
Let be an FC matrix. We can represent this matrix as an -dimensional vector by extracting its upper triangle elements. We can assess the similarity of two matrices, and , by computing the similarity of their vector representations. Suppose we had multiple scans from multiple individuals. Let and be the average within- and between-subject similarity. Differential identifiability, then, is simply (48). Intuitively, the larger the value of , the stronger the population-level fingerprint (46).
Code Availability.
Code and example data for generating and analyzing edge time series are available in Github (https://github.com/brain-networks/edge-ts).
Supplementary Material
Acknowledgments
R.F.B. and F.Z.E. acknowledge support from Indiana University Office of the Vice President for Research Emerging Area of Research Initiative, Learning: Brains, Machines and Children. This work was supported by the NIH (grants R01MH110630 and R00MH094409 to D.P.K. and T32HD007475 Postdoctoral Traineeship to L.B.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2005531117/-/DCSupplemental.
Data Availability.
Two of the imaging datasets come from publicly available, open-access repositories. HCP data can be accessed freely via https://db.humanconnectome.org/ after signing a data use agreement. MSC data can be accessed via OpenfMRI at https://openneuro.org/datasets/ds000224/versions/1.0.3. Postprocessed data from the Indiana University study data and code have been deposited in Figshare and GitHub (postprocessed data are available at https://doi.org/10.6084/m9.figshare.12971162).
References
- 1.Craddock R. C., et al. , Imaging human connectomes at the macroscale. Nat. Methods 10, 524–539 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rogers B. P., Morgan V. L., Newton A. T., Gore J. C., Assessing functional connectivity in the human brain by fMRI. Magn. Reson. Imaging 25, 1347–1357 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gu S., et al. , Emergence of system roles in normative neurodevelopment. Proc. Natl. Acad. Sci. U.S.A. 112, 13681–13686 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Betzel R. F., et al. , Changes in structural and functional connectivity among resting-state networks across the human lifespan. Neuroimage 102, 345–357 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Cole M. W., Bassett D. S., Power J. D., Braver T. S., Petersen S. E., Intrinsic and task-evoked network architectures of the human brain. Neuron 83, 238–251 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fornito A., Zalesky A., Breakspear M., The connectomics of brain disorders. Nat. Rev. Neurosci. 16, 159–172 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Hutchison R. M., et al. , Dynamic functional connectivity: Promise, issues, and interpretations. Neuroimage 80, 360–378 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allen E. A., et al. , Tracking whole-brain connectivity dynamics in the resting state. Cereb. Cortex 24, 663–676 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zalesky A., Fornito A., Cocchi L., Gollo L. L., Breakspear M., Time-resolved resting-state brain networks. Proc. Natl. Acad. Sci. U.S.A. 111, 10341–10346 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kucyi A., Davis K. D., Dynamic functional connectivity of the default mode network tracks daydreaming. Neuroimage 100, 471–480 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Barttfeld P., et al. , Signature of consciousness in the dynamics of resting-state brain activity. Proc. Natl. Acad. Sci. U.S.A. 112, 887–892 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Damaraju E., et al. , Dynamic functional connectivity analysis reveals transient states of dysconnectivity in schizophrenia. Neuroimage Clin. 5, 298–308 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sakoğlu Ü., et al. , A method for evaluating dynamic functional network connectivity and task-modulation: Application to schizophrenia. MAGMA. 23, 351–366 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hindriks R., et al. , Can sliding-window correlations reveal dynamic functional connectivity in resting-state fMRI?. Neuroimage 127, 242–256 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu X., Duyn J. H., Time-varying functional network information extracted from brief instances of spontaneous brain activity. Proc. Natl. Acad. Sci. U.S.A. 110, 4392–4397 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karahanoğlu F. I., Van De Ville D., Transient brain activity disentangles fMRI resting-state dynamics in terms of spatially and temporally overlapping networks. Nat. Commun. 6, 7751 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tagliazucchi E., Balenzuela P., Fraiman D., Chialvo D. R., Criticality in large-scale brain fMRI dynamics unveiled by a novel point process analysis. Front. Physiol. 3, 15 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petridou N., Gaudes C. C., Dryden I. L., Francis S. T., Gowland P. A., Periods of rest in fMRI contain individual spontaneous events which are related to slowly fluctuating spontaneous activity. Hum. Brain Mapp. 34, 1319–1329 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaudes C. C., Petridou N., Francis S. T., Dryden I. L., Gowland P. A., Paradigm free mapping with sparse regression automatically detects single-trial functional magnetic resonance imaging blood oxygenation level dependent responses. Hum. Brain Mapp. 34, 501–518 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allan T. W., et al. , Functional connectivity in MRI is driven by spontaneous bold events. PloS One 10, e0124577 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Preti M. G., Bolton T. A. W., Van De Ville D., The dynamic functional connectome: State-of-the-art and perspectives. Neuroimage 160, 41–54 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Faskowitz J., Zamani Esfahlani F., Jo Y., Sporns O., Betzel R. F., Edge-centric functional network representations of human cerebral cortex reveal overlapping system-level architecture. bioRxiv:799924 (13 October 2019). [DOI] [PubMed] [Google Scholar]
- 23.Jo Y., et al. , The diversity and multiplexity of edge communities within and between brain systems. 10.1101/2020.05.05.067777 (6 May 2020). [DOI] [PubMed] [Google Scholar]
- 24.Smith S. M., et al. , Network modeling methods for fMRI. Neuroimage 54, 875–891 (2011). [DOI] [PubMed] [Google Scholar]
- 25.Van Essen D. C., et al. , The Wu-minn human connectome project: An overview. Neuroimage 80, 62–79 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Essen D. C., et al. ; Wu-Minn HCP Consortium, Data from “Human connectome project resting-state data.” ConnectomeDB. https://db.humanconnectome.org/ Accessed 1 September 2019. [Google Scholar]
- 27.Gordon E. M., et al. , Precision functional mapping of individual human brains. Neuron, 95, 791–807 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Betzel R. F., Byrge L., Esfahlani F. Z., Kennedy D. P.. Temporal fluctuations in the brain’s modular architecture during movie-watching. Neuroimage 213, 116687 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Betzel R., Kennedy D., Byrge L., Data from “Resting-state and movie-watching data.” Figshare. 10.6084/m9.figshare.12971162 Deposited 17 September 2020. [DOI] [Google Scholar]
- 30.Gordon E. M., et al. , Data from “The midnight scan club (MSC) dataset.” OpenNeuro. https://openneuro.org/datasets/ds000224/versions/1.0.3 Accessed 1 September 2019. [Google Scholar]
- 31.Pedersen M., Omidvarnia A., Zalesky A., Jackson G. D.. On the relationship between instantaneous phase synchrony and correlation-based sliding windows for time-resolved fMRI connectivity analysis. Neuroimage 181, 85–94 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Cabral J., et al. , Exploring mechanisms of spontaneous functional connectivity in MEG: How delayed network interactions lead to structured amplitude envelopes of band-pass filtered oscillations. Neuroimage 90, 423–435 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Rubinov M., Sporns O., Weight-conserving characterization of complex functional brain networks. Neuroimage 56, 2068–2079 (2011). [DOI] [PubMed] [Google Scholar]
- 34.Mennes M., et al. , Inter-individual differences in resting-state functional connectivity predict task-induced bold activity. Neuroimage 50, 1690–1701 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tagliazucchi E., Siniatchkin M., Laufs H., Chialvo D. R., The voxel-wise functional connectome can be efficiently derived from co-activations in a sparse spatio-temporal point-process. Front. Neurosci. 10, 381 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cifre I., Zarepour M., Horovitz S. G., Cannas S., Chialvo D. R., On why a few points suffice to describe spatiotemporal large-scale brain dynamics. arXiv:1707.00759 (3 July 2017). [Google Scholar]
- 37.Gutierrez-Barragan D., Basson M. A., Panzeri S., Gozzi A., Infraslow state fluctuations govern spontaneous fMRI network dynamics. Curr. Biol. 29, 2295–2306 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang X., Pan W.-J., Keilholz S. D., The relationship between bold and neural activity arises from temporally sparse events. Neuroimage 207, 116390 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cornblath E. J., et al. , Temporal sequences of brain activity at rest are constrained by white matter structure and modulated by cognitive demands. Commun Biol. 3, 261 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karapanagiotidis T., et al. , The psychological correlates of distinct neural states occurring during wakeful rest. bioRxiv:2019.12 (12 March 2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deco G., Jirsa V. K., McIntosh A. R., Emerging concepts for the dynamical organization of resting-state activity in the brain. Nat. Rev. Neurosci. 12, 43–56 (2011). [DOI] [PubMed] [Google Scholar]
- 42.Thompson W. H., Fransson P., Bursty properties revealed in large-scale brain networks with a point-based method for dynamic functional connectivity. Sci. Rep. 6, 39156 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sonkusare S., Breakspear M., Guo C., Naturalistic stimuli in neuroscience: Critically acclaimed. Trends Cogn. Sci. 23, 699–714 (2019). [DOI] [PubMed] [Google Scholar]
- 44.Hasson U., Nir Y., Levy I., Fuhrmann G., Malach R., Intersubject synchronization of cortical activity during natural vision. Science 303, 1634–1640 (2004). [DOI] [PubMed] [Google Scholar]
- 45.Cole M. W., et al. , Multi-task connectivity reveals flexible hubs for adaptive task control. Nat. Neurosci. 16, 1348–1355 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Finn E. S., et al. , Functional connectome fingerprinting: Identifying individuals using patterns of brain connectivity. Nat. Neurosci. 18, 1664–1671 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gratton C., et al. , Functional brain networks are dominated by stable group and individual factors, not cognitive or daily variation. Neuron 98, 439–452 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Amico E., Goñi J., The quest for identifiability in human functional connectomes. Sci. Rep. 8, 8254 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Byrge L., Kennedy D. P., High-accuracy individual identification using a “thin slice” of the functional connectome. Netw. Neurosci. 3, 363–383 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nielsen J. A., Mair R. W., Baker J. T., Buckner R. L., Precision brain morphometry: Feasibility and opportunities of extreme rapid scans. bioRxiv:530436 (26 January 2019). [Google Scholar]
- 51.Laumann T. O., et al. , Functional system and areal organization of a highly sampled individual human brain. Neuron 87, 657–670 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yeo B. T., et al. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J. Neurophysiol. 106, 1125–1165 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith S. M., et al. , Correspondence of the brain’s functional architecture during activation and rest. Proc. Natl. Acad. Sci. U.S.A. 106, 13040–13045 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Crossley N. A., et al. Cognitive relevance of the community structure of the human brain functional coactivation network. Proc. Natl. Acad. Sci. U.S.A. 110 11583–11588 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yarkoni T., Poldrack R. A., Van. Essen D. C., Nichols T. E., Tor D. W., Large-scale automated synthesis of human functional neuroimaging data. Nat. Methods. 8, 665–670 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Betzel R. F., Fukushima M., Ye H., Zuo X.-N., Sporns O., Dynamic fluctuations coincide with periods of high and low modularity in resting-state functional brain networks. Neuroimage 127, 287–297 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thompson W. H., Peter F., The mean–variance relationship reveals two possible strategies for dynamic brain connectivity analysis in fMRI. Front. Hum. Neurosci. 9, 398 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cabral J., et al. , Cognitive performance in healthy older adults relates to spontaneous switching between states of functional connectivity during rest. Sci. Rep. 7, 5135 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lurie D. J., et al. , Questions and controversies in the study of time-varying functional connectivity in resting fMRI. Netw. Neurosci. 4, 30–69 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Suárez L. E., Markello D. R., Betzel R. F., Misic B., Linking structure and function in macroscale brain networks. Trends Cogn. Sci. 24, 302–315 (2020). [DOI] [PubMed] [Google Scholar]
- 61.Laughlin S. B., Sejnowski J. T., Communication in neuronal networks. Science 301, 1870–1874 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Raichle M. E., et al. , A default mode of brain function. Proc. Natl. Acad. Sci. U.S.A. 98, 676–682 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Raichle M. E., Snyder A. Z., A default mode of brain function: A brief history of an evolving idea. Neuroimage 37, 1083–1090 (2007). [DOI] [PubMed] [Google Scholar]
- 64.Passow S., et al. , Default-mode network functional connectivity is closely related to metabolic activity. Hum. Brain Mapp. 36, 2027–2038 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Betzel R. F., et al. , Structural, geometric and genetic factors predict interregional brain connectivity patterns probed by electrocorticography. Nat. Biomed. Eng. 3, 902–916 (2019). [DOI] [PubMed] [Google Scholar]
- 66.Khambhati A. N., Davis K. A., Lucas T. H., Litt B., Bassett D. S.. Virtual cortical resection reveals push-pull network control preceding seizure evolution. Neuron 91, 1170–1182 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Burns S. P., et al. , Network dynamics of the brain and influence of the epileptic seizure onset zone. Proc. Natl. Acad. Sci. U.S.A., 111, E5321–E5330 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.He B. J., Snyder A. Z., Zempel J. M., Smyth M. D., Raichle M. E.. Electrophysiological correlates of the brain’s intrinsic large-scale functional architecture. Proc. Natl. Acad. Sci. U.S.A. 105, 16039–16044 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mann K., Gallen C. L., Clandinin T. R., Whole-brain calcium imaging reveals an intrinsic functional network in drosophila. Curr. Biol. 27, 2389–2396 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Betzel R., Organizing principles of whole-brain functional connectivity in zebrafish larvae. bioRxiv:496414 (15 December 2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mitra A., Raichle M. E., How networks communicate: Propagation patterns in spontaneous brain activity. Philos. Trans. R. Soc. Lond. B Biol. Sci. 371, 20150546 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mitra A., Snyder A. Z., Tyler B., Raichle M. E., Lag threads organize the brain’s intrinsic activity. Proc. Natl. Acad. Sci. U.S.A. 112, E2235–E2244 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Power J. D., Barnes K. A., Snyder A. Z., Schlaggar B. L., Petersen S. E., Spurious but systematic correlations in functional connectivity mri networks arise from subject motion. Neuroimage 59, 2142–2154 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Power J. D., et al. , Methods to detect, characterize, and remove motion artifact in resting state fMRI. Neuroimage, 84, 320–341 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.van Oort E. S. B., et al. , Functional parcellation using time courses of instantaneous connectivity. Neuroimage, 170, 31–40 (2018). [DOI] [PubMed] [Google Scholar]
- 76.Shine J. M., et al. , Estimation of dynamic functional connectivity using multiplication of temporal derivatives. Neuroimage 122, 399–407 (2015). [DOI] [PubMed] [Google Scholar]
- 77.Leonardi N., Van De Ville D., On spurious and real fluctuations of dynamic functional connectivity during rest. Neuroimage 104, 430–436 (2015). [DOI] [PubMed] [Google Scholar]
- 78.Liegeois R., Laumann T. O., Snyder A. Z., Zhou J., Yeo B. T. T., Interpreting temporal fluctuations in resting-state functional connectivity MRI. Neuroimage 163, 437–455 (2017). [DOI] [PubMed] [Google Scholar]
- 79.Cole Ml. W., et al. , Task activations produce spurious but systematic inflation of task functional connectivity estimates. Neuroimage 189, 1–18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Byrge L., Kennedy D. P., Accurate prediction of individual subject identity and task, but not autism diagnosis, from functional connectomes. Hum Brain Mapp. 41, 2249–2262 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schaefer A., et al. , Local-global parcellation of the human cerebral cortex from intrinsic functional connectivity MRI. Cereb. Cortex 28, 3095–3114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Newman M. E. J., Girvan M., Finding and evaluating community structure in networks. Phys. Rev. E 69, 026113 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Two of the imaging datasets come from publicly available, open-access repositories. HCP data can be accessed freely via https://db.humanconnectome.org/ after signing a data use agreement. MSC data can be accessed via OpenfMRI at https://openneuro.org/datasets/ds000224/versions/1.0.3. Postprocessed data from the Indiana University study data and code have been deposited in Figshare and GitHub (postprocessed data are available at https://doi.org/10.6084/m9.figshare.12971162).