Abstract
Small regulatory RNAs (sRNAs) play an important role for posttranscriptional gene regulation in bacteria. sRNAs recognize their target messenger RNAs (mRNAs) by base-pairing, which is often facilitated by interactions with the bacterial RNA-binding proteins Hfq or ProQ. The FinO/ProQ RNA-binding protein domain was first discovered in the bacterial repressor of conjugation, FinO. Since then, the functional role of FinO/ProQ-like proteins in posttranscriptional gene regulation was extensively studied in particular in the enterobacteria E. coli and Salmonella enterica and a wide range of sRNA-targets was identified for these proteins. In addition, enterobacterial ProQ homologs also recognize and protect the 3′-ends of a number of mRNAs from exonucleolytic degradation. However, the RNA-binding properties of FinO/ProQ proteins with regard to the recognition of different RNA targets are not yet fully understood. Here, we present the solution NMR structure of the so far functionally uncharacterized ProQ homolog Lpp1663 from Legionella pneumophila as a newly confirmed member and a minimal model system of the FinO/ProQ protein family. In addition, we characterize the RNA-binding preferences of Lpp1663 with high resolution NMR spectroscopy and isothermal titration calorimetry (ITC). Our results suggest a binding preference for single-stranded uridine-rich RNAs in the vicinity of stable stem–loop structures. According to chemical shift perturbation experiments, the single-stranded U-rich RNAs interact mainly with a conserved RNA-binding surface on the concave site of Lpp1663.
Keywords: Legionella, NMR structure, ProQ, RNA-binding protein, sRNA
INTRODUCTION
Posttranscriptional regulation of gene expression is an important mechanism for bacteria to quickly adapt to environmental changes. Particularly striking examples in this regard are host-activated bacterial virulence (Westermann et al. 2019), the response to oxidative stress or the regulation of iron metabolism (Altuvia et al. 1997; Massé and Gottesman 2002). Posttranscriptional gene regulation mechanisms in bacteria often depend on the action of trans acting small noncoding regulatory RNAs (sRNAs). These sRNAs are 50–300 nt in length and recognize the mRNA of a target gene via a short complementary nucleotide stretch, the so-called “seed” region (Gorski et al. 2017). In principle, one sRNA can regulate different mRNA species, while one mRNA can be targeted by several different sRNAs. Many sRNAs are negative regulators that decrease mRNA stability or inhibit translation initiation (Nitzan et al. 2017). sRNA-stability and efficient complex formation with the target mRNA often depend on the RNA-binding protein (RBP) Hfq (Updegrove et al. 2016). Hfq is a hexameric, “doughnut-shaped” RBP that belongs to the family of Sm-like proteins (Sauter et al. 2003). Coimmunoprecipitation experiments of affinity-tagged Hfq with bound RNAs coupled with next generation RNA sequencing approaches identified numerous sRNA binding-partners for Hfq in a number of different bacterial species (Holmqvist et al. 2016; Heidrich et al. 2017; Chihara et al. 2019). Thus, Hfq is apparently involved in many regulatory pathways. According to structural and biophysical studies of different Hfq proteins and Hfq–RNA complexes it acts as an RNA-interaction platform by providing three distinct RNA-binding surfaces (Schumacher et al. 2002, Link et al. 2009; Mikulecky et al. 2011; Someya et al. 2012; Panja et al. 2013; Dimastrogiovanni et al. 2014) and thereby promotes base-pairing between sRNAs and their mRNA targets.
A chaperoning activity with regard to sRNA–mRNA duplex formation and the stabilization of sRNAs against RNases was also reported as an important function for a new class of bacterial RNA chaperones, the proteins of the FinO/ProQ family (Arthur et al. 2003; Chaulk et al. 2010; Smirnov et al. 2017). In contrast to the hexameric Hfq, ProQ proteins are monomers and contain a conserved FinO/ProQ core domain. This domain comprises five characteristic α-helices that are connected by structurally well-defined loops (Ghetu et al. 2000; Chaulk et al. 2010; Gonzalez et al. 2017; Olejniczak and Storz 2017). Many FinO/ProQ family members have additional amino- or carboxy-terminal domains and/or unstructured extensions, which often contain a large number of basic amino acid residues (Fig. 1; Supplemental Fig. S1A). E. coli FinO features long α-helical extensions of the FinO/ProQ core domain at its amino as well as its carboxyl terminus (Ghetu et al. 2000). E. coli and Salmonella ProQ contain an additional carboxy-terminal domain that is structurally similar to eukaryotic Tudor domains. Tudor domains form β-barrel-like structures and in eukaryotes interact with other proteins by recognizing methylated arginine or lysine residues (Lu and Wang 2013). It is very likely that the FinO/ProQ-domains harbor the core RNA-binding activity, but the additional domains/extensions also contribute to RNA-binding or are required for duplex formation between sRNA and mRNA (Arthur et al. 2003; Attaiech et al. 2016; Gonzalez et al. 2017; Stein et al. 2020). According to recent genetic experiments a conserved positively charged area on the so-called “concave” site of the ProQ/FinO core domain (Pandey et al. 2020) is a central structural determinant for RNA-binding. However, how exactly FinO/ProQ proteins recognize their RNA targets is still not known.
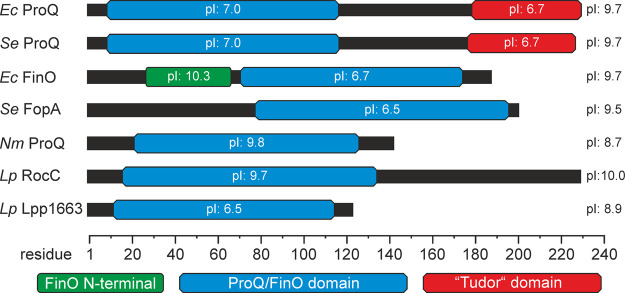
FIGURE 1.
Domain architecture of selected ProQ/FinO domain proteins. ProQ/FinO-proteins often carry additional amino- or carboxy-terminal domains or extensions. The core domain comprises ∼100 amino acids and is mainly responsible for RNA-binding. The theoretical pI (isoelectric point) values of the isolated domains as well as for the full-length proteins are indicated. (Ec) Escherichia coli, (Se) Salmonella enterica, (Nm) Neisseria meningitidis, (Lp) Legionella pneumophila.
Interestingly, some members of the FinO/ProQ family were shown to bind various RNA targets, whereas other members of the family bind only a small number of target RNAs with very high specificity. The classical example in the later regard is the E. coli FinO protein, which is part of the F-plasmid encoded conjugation repression system in enteric bacteria. This protein exclusively binds the FinP sRNA and the complementary region of the corresponding traJ mRNA (van Biesen and Frost 1994). Similarly, the very recently identified FinO/ProQ-like protein FopA, which is plasmid encoded, binds primarily to the Inc sRNA which is involved in plasmid replication in Salmonella (Gerovac et al. 2020).
The E. coli ProQ protein was initially identified as a specific regulator of the osmoregulatory transporter ProP (Kunte et al. 1999). However, decades later it was discovered to be a general sRNA-binding protein interacting with a large number of different sRNA targets (Smirnov et al. 2016). In many cases the sRNA targets of ProQ are not bound by Hfq and vice versa suggesting a functional complementarity of the two proteins. Some sRNA–mRNA pairs, however, are bound by both proteins implicating competing roles for ProQ and Hfq in certain regulatory pathways (Melamed et al. 2020).
Regarding the function of ProQ/FinO proteins, it is unclear at the moment why proteins like FinO or FopA bind specifically to only a few RNA targets whereas ProQ has a much wider target range. RNase footprinting experiments showed that both E. coli FinO and ProQ protected the GC-rich 3′-terminator stem–loops and the uridine-rich 3′-end extensions in their target RNAs upon binding (Arthur et al. 2011; Smirnov et al. 2017). Furthermore, a single-stranded 3′-tail of at least 6 nt was required for high-affinity RNA binding by E. coli FinO (Jerome and Frost 1999). Recently, it was shown that enterobacterial ProQ also associates with stem–loop structures at the 3′ end of mRNAs and protects them against ribonucleases (Holmqvist et al. 2018). Furthermore, a number of known ProQ sRNA targets have single-stranded 3′-overhangs with an average length of 4–6 nt (Olejniczak and Storz 2017). A very recent study aimed at identifying target RNAs for the N. meningitidis ProQ homolog NMB1681 by UV CLIP-seq also found binding sites for this protein at mRNA or sRNA 3′-ends including stem–loop structures and U-rich single-stranded regions (Bauriedl et al. 2020).
Interestingly, Legionella pneumophila contains two ProQ-homologs (Attaiech et al. 2016). RocC (Lpp0148—regulator of competence chaperone) specifically binds the RocR sRNA and its plasmid-encoded homolog—the RocRp sRNA—as its main targets (Attaiech et al. 2016; Durieux et al. 2019). The RocR sRNA recognizes mRNAs encoding the proteins of the DNA-uptake system and regulates their expression. The major determinant of RocR sRNA binding to RocC is its 3′-terminal stem–loop which is extended by a 6-nt-long single-stranded uridine-rich tail while the first two stem–loops of the RocR sRNA are involved in interactions with its target mRNAs (Attaiech et al. 2016).
The second ProQ homolog in L. pneumophila Lpp1663 (121 amino acids) only consists of a ProQ-domain and very short but rather basic amino- and carboxy-terminal extensions (Fig. 1; Supplemental Fig. S1A). It therefore represents a minimal version of a FinO/ProQ family member protein. So far, this protein has not been structurally and functionally characterized, but the presence of a FinO/ProQ domain suggests a likely function as an RNA-binding protein. Using NMR-spectroscopy, we determined a high-resolution structure of this protein in solution and characterized its general RNA-binding properties. We identified a potential RNA-binding site and could observe a clear preference for binding U-rich single-stranded RNAs. Furthermore, we show that a stem–loop structure in the vicinity of the U-rich region increases the affinity of Lpp1663 to RNA. With this study, we provide new insights into the structural determinants for RNA-binding of ProQ/FinO proteins that will help to understand the functional role of this protein domain in additional bacterial species.
RESULTS AND DISCUSSION
Lpp1663 adopts a ProQ/FinO fold
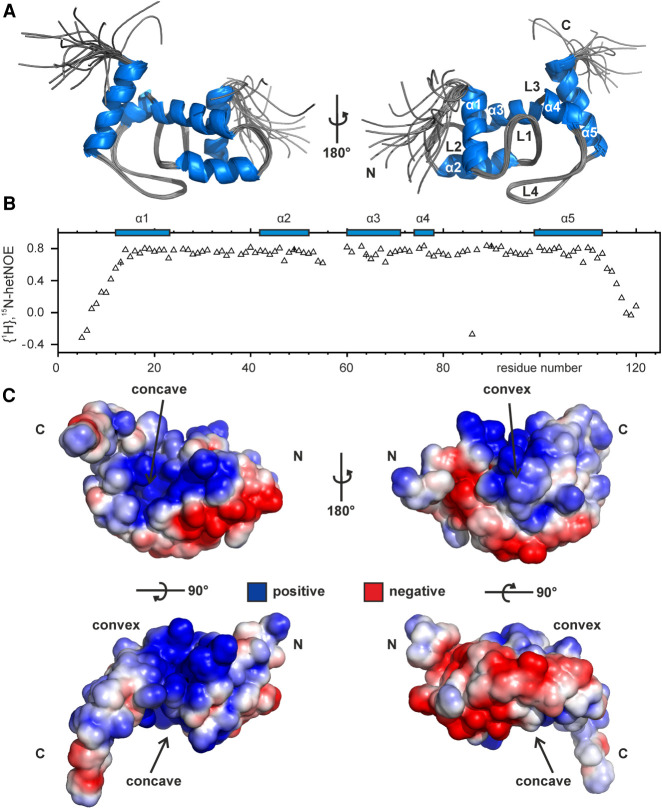
The NMR solution structure of Lpp1663 was solved using 1892 NOE-based distance restraints and 172 chemical shift derived torsion angle restraints using protocols described previously (Christ et al. 2012; Hacker et al. 2015). The calculated 20 structures with the lowest energy have an rmsd for the backbone heavy atoms in the structurally ordered regions (residues 12 to 113) of 0.4 Å (Table 1; Fig. 2A). According to 1H,15N-HetNOE-data both the basic amino (residues 1 to 12) and carboxyl terminus (residues 113 to 120) are flexible (Fig. 2B). In addition, there is no evidence for even a transient interaction of the basic termini with the core of the protein since a deletion of the two termini (Lpp1663tr—aa 10 to 117) causes only minimal chemical shift perturbations for residues in the ProQ core domain (Supplemental Fig. S1B). The fold of Lpp1663 features 5 α-helices (α1–α5) in agreement with previous predictions based on chemical shifts (Immer et al. 2018) which are connected by four structurally very well defined rigid loops (L1–L4) that lack additional regular secondary structure elements. Similar to the other reported ProQ/FinO structures, the surface of Lpp1663 shows two positively charged patches, a larger one on the concave site and a smaller one on the convex site of the protein that might play a role in RNA-binding (Fig. 2C). Furthermore, the flexible amino and carboxyl termini are also rich in positively charged basic residues.
TABLE 1.
NMR and refinement statistics of the Lpp1663 solution structure
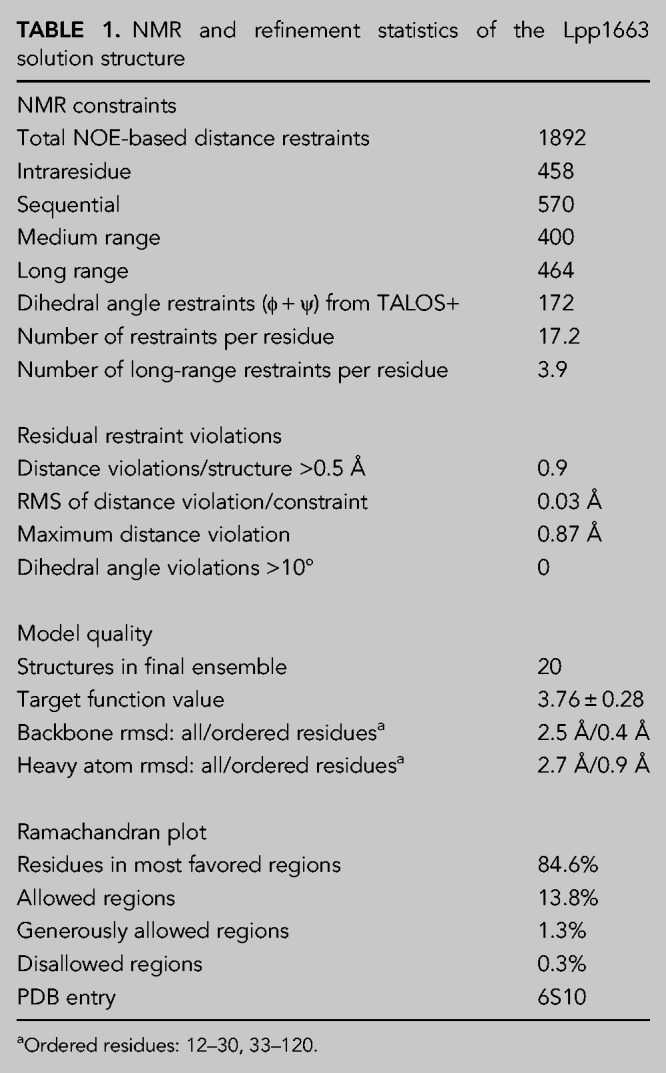
FIGURE 2.
NMR solution structure and electrostatic surface features of Lpp1663. (A) Lpp1663 has a ProQ/FinO fold with five central helices that are connected by four structurally well-defined loops. Shown is an overlay of the 10 final structures with an rmsd of 0.4 Å (ordered residues) that were deposited in the PDB under accession number 6S10. (B) The {1H}, 15N heteronuclear NOE of 15N-labeled Lpp1663 indicates the flexible amino- and carboxyl termini of the protein. Secondary structure elements are depicted by blue boxes. (C) The electrostatic surface potential of the lowest energy structure reveals two positively charged patches on either side of the protein, the concave and the convex site. Positive and negative charges are colored in blue and red. Amino- and carboxyl termini are indicated as N and C, respectively.
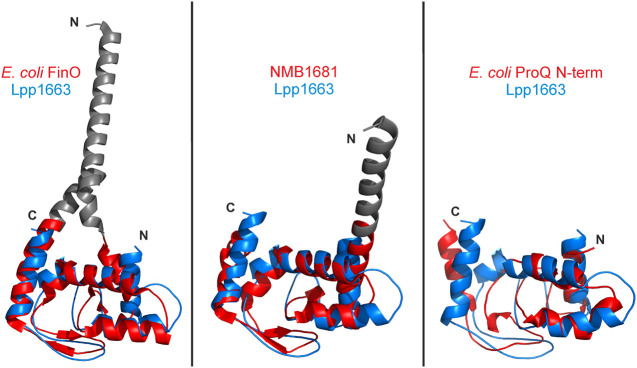
The high resolution structure of Lpp1663 as shown in Figure 2A strongly resembles the prototypic ProQ/FinO domain fold with a Cα- rmsd of 1.8 Å (80 residues) and 1.9 Å (91 residues) to the E. coli FinO and N. meningitidis NMB1681 structures, respectively (Fig. 3). The sequence identity of Lpp1663 is ∼28% to E. coli FinO and ∼25% to NMB1681. The E. coli ProQ amino-terminal domain (lowest energy structure of the NMR-determined structural bundle—Gonzalez et al. 2017) aligned with our Lpp1663 structure with an rmsd of 4.2 Å for 75 residue pairs, despite a higher sequence identity of the two proteins of 34%. The reason for the rather large rmsd between the E. coli ProQ amino-terminal domain and Lpp1663 are conformational differences in the loops connecting the core helices of the ProQ domain, as well as differences in the orientation of helices α2 and α5. It should be noted, however, that the structure of the E. coli ProQ NTD shows also rather high rmsd values in alignments with the X-ray structures of E. coli FinO (4.5 Å, 92 residues) and NMB1681 (5.6 Å, 93 residues).
FIGURE 3.
The ProQ/FinO fold is well conserved. The lowest energy Lpp1663 NMR solution structure (blue) was superposed with the FinO/ProQ domains of E. coli FinO (PDB:1DVO), N. meningitidis NMB1681 (PDB:3MW6), and E. coli ProQ (PDB:5NB9) colored in red. The Cα-rmsd between the structures are 1.8 Å (80 atom pairs), 1.9 Å (91 atom pairs), and 4.2 Å (75 atom pairs), respectively, which shows the high degree of conservation of the core fold for the ProQ/FinO domain. The rmsd was calculated with the alignment tool in PyMol (The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC.). Flexible amino- and carboxyl termini are not shown and loops were smoothed for clarity. Amino- and carboxyl termini are indicated as N and C, respectively.
The similarity of the structure and surface properties of Lpp1663 and the proteins of the ProQ/FinO family lends additional support to the notion that Lpp1663 is also a functional homolog of these proteins. Due to the absence of additional domains or extended amino- and carboxy-terminal extension segments, it represents a minimalistic model for further investigations of the RNA-binding properties of the ProQ protein family.
Lpp1663 has a preference for U-rich single-stranded RNAs
Since Lpp1663 has not yet been functionally characterized there are no known target RNAs for this protein. RocC—the other ProQ-like protein in L. pneumophila—interacts specifically with the 3′-terminal stem–loop III of the RocR sRNA and its 3′ single-stranded extension (Supplemental Fig. S2A; Attaiech et al. 2016). An RNA containing stem–loop III and the 3′ single-stranded extension of RocR bound to Lpp1663 with a KD of 28 µM in ITC experiments (Supplemental Fig. S2B). Thus, the affinity of Lpp1663 for this RNA is ∼40-fold lower in comparison to the affinity of the FinO/ProQ core domain of RocC for which this RNA is a specific target (Attaiech et al. 2016). Interestingly, a short single-stranded oligomer with the sequence 5′-CCUUUCU-3′ which corresponds to the isolated single-stranded 3′-end of the RocR sRNA bound to Lpp1663 with a similar KD of 41 µM (Supplemental Fig. S2C). In contrast, the RocR stem–loop III without the 3′-terminal single-stranded extension did not bind to Lpp1663 (Supplemental Fig. S2D). Thus, the single-stranded 3′-tail of RocR is the major contributor to this interaction.
Based on these results as well as on the previously reported RNA-binding preferences for other members of the ProQ/FinO protein family we hypothesized that short single-stranded U-rich oligomers might be suitable model systems to derive initial insights into the RNA-binding mechanism of Lpp1663.
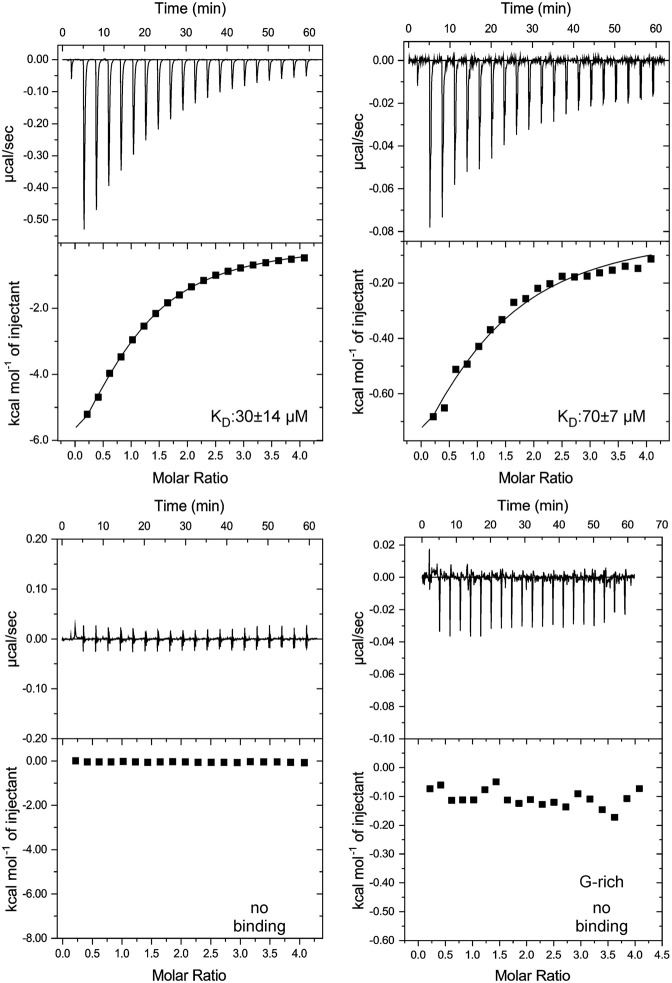
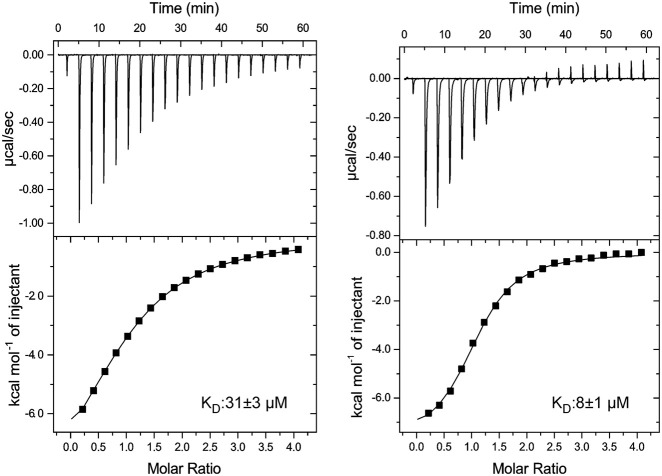
Initially, we tested the binding of a single-stranded oligo U6 RNA to Lpp1663 using isothermal titration calorimetry (ITC). This experiment yielded a dissociation constant (KD) of ∼30 µM and a complex stoichiometry of 1:1 for oligo U6 RNA (Fig. 4). Usually, ProQ/FinO proteins bind their target RNAs with a KD in the nanomolar range (Jerome and Frost 1999; Attaiech et al. 2016; Smirnov et al. 2017), indicating that a short single-stranded U-rich sequence is not the sole element required for a biologically relevant RNA-binding activity. Due to the presence of positively charged patches on the surface of Lpp1663, the interaction of oligo U6 with Lpp1663 might just be an effect of electrostatic complementarity with the negatively charged RNA backbone. To test if there is any sequence preference in single-stranded RNA-binding we titrated Lpp1663 with oligo A6, C6 and a G-rich hexameric RNA (sequence GGAGGA) (Fig. 4). We avoided using an oligo G6 RNA since such a sequence could form G-quadruplex structures. The affinity of Lpp1663 to oligo A6 was decreased 2.5-fold (∼70 µM) compared to oligo U6. The interaction of oligo C6 and the G-rich RNA with Lpp1663 was either very weak or absent and therefore yielded no interpretable binding curves in the corresponding ITC experiments (Fig. 4). Since Lpp1663 has a clear preference for oligo U, and Lpp1663 seems to be able to discriminate between the two pyrimidine bases uridine and cytosine, the interaction of the two binding partners is not only due to electrostatic interactions between the positively charged residues of the protein and the negatively charged RNA phosphate backbone.
FIGURE 4.
Lpp1663 has a preference for U-rich single-stranded RNAs. The ITC thermograms show Lpp1663 titrated with an oligo U6, A6, C6, or a 6mer G-rich RNA. Lpp1663 has the highest affinity to oligo U6 (top, left). In comparison, the affinity to oligo A6 is ∼2.5-fold lower (top, right). Lpp1663 does not bind oligo C6 or the G-rich 6mer single-stranded RNA (bottom) in ITC-experiments.
The concave site of Lpp1663 harbors a potential RNA binding region
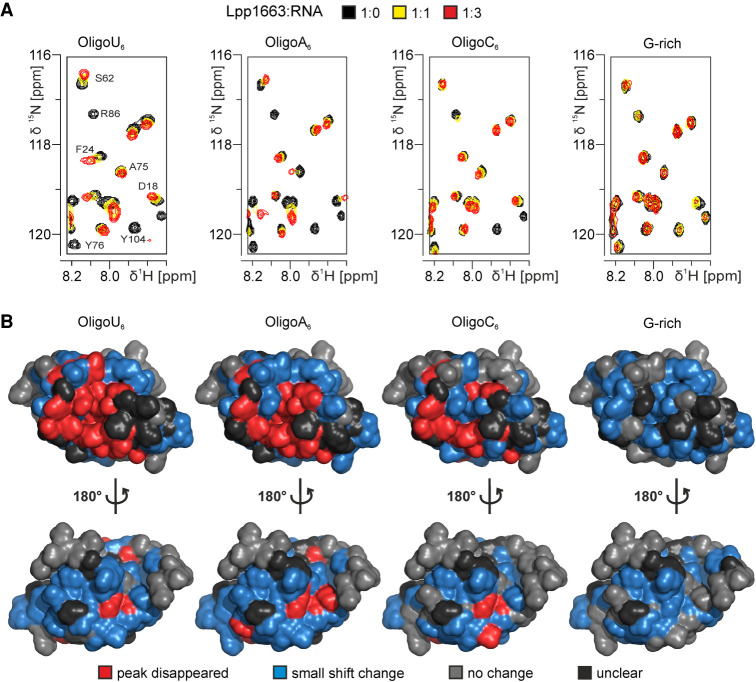
To investigate which parts of the protein are involved in binding RNA by NMR-spectroscopy we titrated 15N labeled Lpp1663 with up to five equivalents of the unlabeled oligo N6 RNAs and recorded changes in the chemical shifts of the amide group signals in 1H, 15N HSQC spectra. (Fig. 5; Supplemental Fig. S3A). NMR titration experiments are carried out at significantly higher protein concentrations compared to our ITC experiments. Thus, they also yield information about interactions that were too weak to be analyzed reliably by ITC and consequently, spectral changes were observed in titrations with all four oligo N6 RNAs.
FIGURE 5.
Lpp1663 interacts with RNA through its core domain and mainly with the concave face of the protein. (A) Sections of 1H, 15N HSQC spectra of 15N-labeled Lpp1663 titrated with oligo U6, A6, C6, or a G-rich hexameric RNA. NMR signal assignments are given for residues undergoing substantial changes in signal intensity or chemical shift in the oligo U6 titration. (B) Results of the NMR-titrations (three equivalents of RNA) were plotted on the surface of one of the lowest energy structures of Lpp1663. Changes in the chemical shift are indicated as the following: (red) peak disappeared, (blue) small shift, (gray) no change, (black) unclear. The most affected region is the concave face of Lp1663 as can be seen for oligo U6 and oligo A6. For oligo C6 and the G-rich RNA, mostly minor chemical shift changes can be observed. The amino- and carboxyl termini are not involved in RNA-binding. Amino- and carboxyl termini are indicated as N and C, respectively.
In all titration experiments, a number of Lpp1663 amide group signals gradually changed their chemical shifts upon titration with the RNAs. For these signals, the difference in chemical shifts of the apo and the RNA-bound state is small compared to the rate of interconversion between the RNA-bound and the RNA-free state of the protein—they are in fast exchange on the NMR time scale. Other signals disappeared in the course of the titration (Fig. 5A). For these signals, the difference of their chemical shift in the apo and the RNA-bound state is larger and similar to the rate of interconversion between the RNA-bound and RNA-free state of the protein—they are in intermediate exchange on the NMR time scale. In order to map the observed NMR spectral changes on the surface of Lpp1663 we colored residues with disappearing signals red, signals with shift changes blue and those with signals unaffected by RNA addition gray. Residues with signals that could not be analyzed due to overlap with other signals and proline residues were colored black (Fig. 5B).
The most dramatic effects on the 1H, 15N HSQC spectrum of Lpp1663 were observed upon titration with oligo U6 as expected based on the dissociation constants measured by ITC. For many amino acid residues on a continuous surface at the concave site of the protein, the signals of the amide protons gradually weakened and disappeared upon increasing the RNA concentration indicating binding in intermediate exchange. This surface includes the amino-terminal half of helix α2, helices α3, α4, and α5, as well as loop L1 (Fig. 5B) and is surrounded by additional residues showing gradual chemical shift changes. Gradual chemical shift changes in agreement with fast exchange are also observed for the convex site of the protein upon titration with oligo U6. However, since the concave and the convex surface of Lpp1663 are only separated by a single layer of α-helices it is unclear if these changes report on weak and probably unspecific binding to this surface or if they are due to indirect allosteric effects of RNA-binding to the concave surface. The titration experiments with oligo A6 also reveal the concave surface of the protein as a hotspot for RNA-induced spectral changes. In contrast, the effects induced by addition of the G-rich oligomer and oligo C6 are much more limited (Fig. 5B). In particular, for the G-rich oligomer the induced spectral changes are very minor in agreement with a very weak and unspecific RNA–protein interaction.
Interestingly, the concave surface region containing the amino acid residues with the most pronounced spectral changes upon oligo U6 titration contains many highly conserved residues. This includes L38, G41 and the partially conserved positively charged residue K39 in loop L1, Y76 and L77 in helix α4, and R86 in loop L4 (Olejniczak and Storz 2017) reinforcing the notion that this area represents the primary RNA-binding site of Lpp1663 (Supplemental Fig. S4A). Another residue in loop L1 with a signal disappearing upon titration with oligo U6 RNA is the partially conserved K36. In contrast to the other affected residues in loop L1, the backbone amide group and the sidechain of this residue point toward the convex surface of ProQ. However, as mentioned above, this might indicate an allosteric conformational change in loop L1 in response to RNA binding rather than a direct interaction. An RNA binding surface on the concave site of the E. coli ProQ domain very similar to our observations was recently proposed by Berry and coworkers based on results from a bacterial three-hybrid mutational screen aimed at identifying amino acids important for RNA-binding in vivo (Pandey et al. 2020). Substitution of conserved residues with alanine as well as random mutagenesis revealed an essential role in RNA-binding for, e.g., G37, K54, R58, Y70, L71, R80, and D82 (E. coli ProQ numbering). These amino acids correspond to G41, K60, R64, Y76, L77, R86, and D88 in Lpp1663, which are all affected by oligo U6 binding in the NMR titration experiments, respectively. Residue R80 (E. coli ProQ numbering)/R86 (Lpp1663 numbering) from this list warrants a more detailed discussion. In the NMR solution structure of E. coli ProQ, the side chain of R80 points toward the convex site of the protein (Gonzalez et al. 2017). Since its mutation to A lead to a decrease in RNA-binding in the bacterial three-hybrid experiments Berry and coworkers assigned this residue to an RNA-binding site on the convex surface of the protein (Pandey et al. 2020). In our structural bundle representing the NMR solution structure of Lpp1663, the equivalent R86 side chain points toward the concave face of the protein in all structures (Supplemental Fig. S4B,C). Furthermore, in the two crystal structures of E. coli FinO and N. meningitidis NMB1681, the arginine side chain points toward the concave site as well, suggesting that this residue is part of the RNA binding surface on the concave face in the majority of ProQ-like proteins (Supplemental Fig. S4D).
We also tested the influence of oligonucleotide length on the binding affinity and the affected binding surface. Therefore, we titrated Lpp1663 with uridine-5′-monophosphate (UMP), oligo U4 and oligo U8 in ITC and/or NMR experiments. NMR titrations did not show evidence for even a transient interaction of UMP with Lpp1663 (Supplemental Fig. S3B). On the other hand, according to ITC experiments oligo U4 with a KD of 31 ± 3 µM has almost the same affinity for Lpp1663 as oligo U6 (Fig. 6). In contrast, extending the RNA by two additional uridine nucleotides to oligo U8 resulted in a 3.5-fold increase in the affinity (KD = 8 ± 1.0 µM, Fig. 6). However, NMR-based titration experiments with the oligo U4 and oligo U8 RNAs and 15N- labeled Lpp1663 (Supplemental Fig. S3B) identified the same surface area on the concave site of the protein as the major RNA binding site (Supplemental Fig. S5).
FIGURE 6.
Variation of the length of oligo U does alter the binding affinity of Lpp1663. ITC thermograms of Lpp1663 titrated with oligo U8 and oligo U4. Compared to oligo U6 (29 µM) oligo U4 binds to Lpp1663 with a very similar affinity while oligo U8 binds with an approximately fourfold increased affinity.
To further support the importance of the positively charged surface patch on the concave face of the protein for single-stranded oligo U binding we mutated residues Y76 or R86 (Lpp1663 numbering) to A and characterized the affinity of the two mutant proteins for oligo U8 by ITC (Supplemental Fig. S6A). Remarkably, both single point mutations Y76A and R86A completely abolished RNA binding. Importantly, both mutant proteins are folded properly according to their 1D 1H NMR spectra (Supplemental Fig. S6B). Our results agree well with the in vivo results of Berry and coworkers (Pandey et al. 2020) for E. coli ProQ, where the equivalent point mutations also abrogated RNA binding.
In all NMR-based titration experiments, we noticed that the chemical shifts of the amino acids in the positively charged flexible amino and carboxyl termini were not affected upon RNA addition and are therefore apparently not important for interactions with single-stranded RNAs. ITC titration experiments with the amino- and carboxy-terminally truncated protein variant (Lpp1663tr aa 10–117) confirmed that the affinity for oligo U8 is virtually the same as for the full-length protein (Supplemental Fig. S7; KD = 10 µM). These results also suggest that the unstructured termini do not contribute with unspecific electrostatic interactions to the binding of single-stranded RNA oligonucleotides.
The influence of RNA stem–loops on Lpp1663 RNA-binding
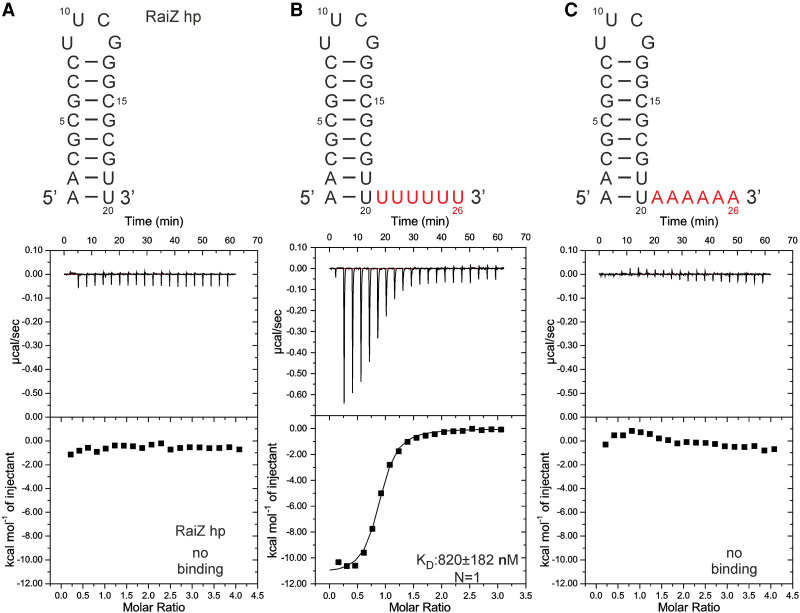
While Lpp1663 preferentially binds to single-stranded oligo U rather than to oligo A, C or a G-rich RNA and oligo U8 binds Lpp1663 with a KD in the low µM range this KD is still ∼10-fold higher than the KD for the RocC/RocR stem–loop III interaction and significantly higher than the KDs reported for other ProQ/FinO like proteins for their targets (Attaiech et al. 2016; Smirnov et al. 2016). This suggests that in biologically relevant Lpp1663–RNA interactions additional RNA elements might be needed to further contribute to the affinity. Thus, we tested the influence of adding a 3′ terminator stem–loop to oligo U on the affinity of the protein–RNA interaction. As there is no known biological target of Lpp1663, we chose the 3′-terminator hairpin of the RaiZ sRNA which is a verified target of Salmonella ProQ (Smirnov et al. 2017). We tested an RNA construct containing only the isolated RaiZ 3′ terminator hairpin (RaiZ hp) and hairpins with either a single-stranded 3′ oligo U6 (RaiZ hp U6) or A6 tail (RaiZ hp A6). The native RaiZ terminator hairpin stem is predicted to feature two terminal A:U Watson–Crick base pairs and is succeeded by an oligo U4 3′ single-stranded tail. The RNAs were folded properly according to their 1D 1H NMR spectra (Supplemental Fig. S8). In ITC experiments the isolated RaiZ hairpin (RaiZ hp) did not show binding to Lpp1663 (Fig. 7A). However, the RaiZ hp U6 RNA bound Lpp1663 with a KD of ∼800 nM (Fig. 7B) which is ∼10-fold lower than the KD for single-stranded oligo U8 and similar to the KD observed for the biologically relevant RocC/RocR interaction (Attaiech et al. 2016). In contrast, the RaiZ hp A6 RNA did not show binding in ITC titrations (Fig. 7C). Thus, both the presence of the structured stem–loop element and the sequence of the 3′-terminal single-stranded tail are important determinants of RNA-binding by Lpp1663. In agreement with our results the importance of a structured hairpin element for RNA-binding to enterobacterial ProQ was postulated recently (Westermann et al. 2019). For the E. coli FinO protein which binds specifically to its RNA-target—stem–loop II of the FinP sRNA—the presence of a 3′ single-stranded tail improved the affinity of this interaction ∼10-fold compared to an RNA construct consisting only of stem–loop II (Jerome and Frost 1999). Interestingly, however, in this case the relative importance of the two RNA structural elements—the stem–loop and the single-stranded 3′-tail—for the binding affinity seems to be reversed compared to the interaction between Lpp1663 and RaiZ hp U6. This might be due to the presence of the long amino-terminal α-helical extension of the ProQ core fold in FinO and its postulated interaction with the stem of the FinP sRNA stem–loop II (Arthur et al. 2011).
FIGURE 7.
A hairpin containing RNA binds to Lpp1663 with enhanced affinity. (A–C) Secondary structure of the RaiZ hairpin variants used in this study and ITC thermograms of Lpp1663 titrated with these constructs. The six U and A residues that are not present in the RaiZ hp construct are labeled in red.
The results of the NMR titrations for RaiZ hp U6 are very similar in comparison to the titrations with oligo U6 (Supplemental Fig. S3C). The previously identified RNA binding surface on the concave face of Lpp1663 was again affected strongly in the titration with the RaiZ hp U6 RNA. However, the surface area on the concave face of Lpp1663 affected by the presence of the RaiZ hp U6 RNA (Supplemental Fig. S9) is slightly increased compared with the binding surface for oligo U6. This suggests that the single-stranded oligo U stretch of RaiZ hp U6 is bound similar to the isolated single-stranded oligo U RNA. The stem–loop region of the RaiZ hp U6 RNA apparently only supplements the interactions with the single-stranded 3′-tail of the RNA. A computational model for the complex of FinO and FinP stem–loop II RNA based on SAXS, enzymatic probing and mutational data also suggested that the single-stranded 3′-tail of the FinP RNA is recognized by the concave face of the core domain, whereas the stem forms additional contacts to the long amino-terminal helical extension of FinO (Arthur et al. 2011). For Lpp1663 the absence of chemical shift perturbations for residues in the flexible amino- and carboxy-terminal extensions in titration experiments with the RaiZ hp U6 RNA suggest that here the short extensions do not form additional interactions with the stem part of the RNA. This is borne out by the results of an ITC titration experiment where the RaiZ hp U6 RNA is added to the amino- and carboxy-terminally truncated variant Lpp1663tr (amino acids 10–117). The KD for this interaction is essentially the same as the one involving the WT protein (KD = 1.2 µM, Supplemental Fig. S10). Thus, the flexible basic tails of Lpp1663 might be important for chaperoning RNA duplex formation as shown, for example, for the amino-terminal helix of FinO or the carboxy-terminal domain of E. coli ProQ (Arthur et al. 2003; Chaulk et al. 2011).
In conclusion, we show here that Lpp1663 structurally belongs to the ProQ/FinO family and is a genuine RNA-binding protein with a preference for RNA-substrates containing single-stranded U-rich regions in the vicinity of stem–loop elements. The primary RNA-binding surface is located on the concave face of the protein and includes a number of highly conserved amino acid residues such as Y76 and R86. Apparently, this RNA-binding site is present in other members of the FinO/ProQ protein family (Ghetu et al. 2002; Chaulk et al. 2010; Pandey et al. 2020). In particular, it was suggested to be the binding site for the 3′ single-stranded terminal tail in the interaction between FinO and the stem–loop II of the FinP sRNA (Jerome and Frost 1999). On the other hand, the comparison of the available structures and sequences for functionally characterized FinO/ProQ family members suggest that the presence of large structured or unstructured amino- or carboxy-terminal extensions of the ProQ core domain as observed in FinO, RocC or FopA is related to a high substrate specificity in these proteins. These extensions could either directly contribute to the recognition of specific RNA targets as suggested for FinO (Arthur et al. 2011) or they could sterically restrict the access to the core binding site by transiently folding back on the concave surface of the core domain. However, since no high-resolution structural information for an RNA/protein complex nor data about their intrinsic dynamics are available for FinO/ProQ-family members with specific RNA-substrate recognition, this currently remains speculation.
The genome of L. pneumophila harbors two proteins that are members of the FinO/ProQ family—RocC and Lpp1663. The function of RocC was demonstrated to be rather specific (Attaiech et al. 2016) and here we show that the specific sRNA target of RocC—the sRNA RocR—is bound only weakly by Lpp1663. On the other hand, Lpp1663 is apparently able to bind with a biologically significant affinity to RNAs containing single-stranded U-rich elements next to structured stem–loops. Thus, Lpp1663 might therefore functionally resemble those FinO/ProQ family members with a broad substrate binding ability and a more general mRNA-protection and RNA chaperone function. However, a final assessment of the function of Lpp1663 will have to await the identification of its native RNA targets in vivo.
MATERIALS AND METHODS
Cloning of Lpp1663 plasmids
The Lpp1663 coding sequence was cloned into a pET11a vector with an amino-terminal hexahistidine tag and a TEV cleavage site as a synthetic gene (GenScript) with the codon usage optimized for E. coli expression.
Lpp1663 point mutations Y76A and R86A were obtained by site-directed mutagenesis as described previously (Liu and Naismith 2008). For the PCR reactions the following primer pairs were used: R86A_forward (GCGGCGGTGGATATTTATGGTAACGAGGTTGATGTGGTT) and R86A_reverse (TCCACCGCCGCGGTGTCCGGCTTCTGG); Y76A_forward (GCGGCGCTGAGCTGCCAGAAGCCGGAC) and Y76A_reverse (CTCAGCGCGCCGGG CTCGCGCTATAGT). The introduced point mutation is underlined in the complementary region, the nonoverlapping primer pairs are highlighted in italic.
The PCR products were treated with DpnI (1 U/1 ng DNA) in cut smart buffer for 2 h at 37°C followed by 20 min of heat inactivation at 80°C. A total of 10 µL of the reaction mix was used for transformation of chemically competent E. coli DH5α cells. Plasmids were isolated from grown colonies and verified by sequencing.
Protein expression and purification
The full-length Lpp1663 protein, Lpp1663tr (residues 10 to 117), and the amino acid point mutants Lpp1663 Y76A and R86A, were expressed and purified as described in detail previously (Immer et al. 2018).
Briefly, protein expression was induced at OD600 ∼ 0.8 with 1 mM IPTG at 20°C for ∼16 h in E. coli BL21(DE3) Gold (Agilent Technologies/Stratagene). Media were supplemented with ampicillin (100 µg/mL). For uniformly 15N- or 15N,13C-labeling, the protein was expressed in M9 minimal medium containing 1 g/L 15NH4Cl or 1 g/L 15NH4Cl and 2.5 g/L 13C6-D-glucose (Cambridge Isotope Laboratories). Amino acid–specific labeled protein was expressed in M9 medium as described in Muchmore et al. (1989) with the following labeling schemes: α-15N lysine (0.225 g/L); α-15N tyrosine (0.17 g/L); α-15N phenylalanine (0.13 g/L); and U-13C proline (0.1 g/L).
Escherichia coli cells were lysed by sonication in lysis buffer (50 mM Tris/HCl pH 7.5, 500 mM NaCl, 10 mM imidazole, 5 mM β-mercaptoethanol, RNase and DNase (Roche) and cOmplete Protease Inhibitor (Roche). The lysate was cleared by centrifugation (8000g, 4°C, 30 min) and loaded on a HisTrap HP column (GE Healthcare). The hexahistidine tag was cleaved off using a recombinantly produced His-tag containing TEV protease. The tag and the protease were removed by a second purification step on a HisTrap HP column, followed by cation exchange chromatography (HiPrep 16/10 SP column, GE Healthcare). The protein was further purified by gel filtration (HiPrep 16/60 Sephacryl S-100 High resolution column, GE Healthcare) equilibrated with NMR buffer (50 mM sodium phosphate, pH 6.5, 100 mM NaCl, 2 mM β-mercaptoethanol).
NMR measurements and structure calculation
NMR measurements were performed as described previously (Immer et al. 2018). All NMR spectra were recorded on Bruker AVANCE III HD 600, 700, and 800 MHz spectrometers equipped with cryogenic triple resonance probes at 298 K. 1H chemical shifts were internally referenced with 50 µM DSS (4,4-dimethyl-4-silapentane-1-sulfonic acid). The heteronuclear 13C and 15N chemical shifts were referenced indirectly with the appropriate conversion factors (Markley et al. 1998). All protein samples (400–500 µM protein concentration) were supplemented with 10% (v/v) D2O in NMR buffer (50 mM sodium phosphate buffer pH 6.5, 100 mM NaCl, 2 mM β-mercaptoethanol).
The backbone assignment was obtained using the following BEST-TROSY-based triple resonance 3D-experiments with 13C,15N uniformly labeled Lpp1663: HNCO, HN(CA)CO, HNCACB (Favier and Brutscher 2011). 1H,15N-HSQC and 1H,13C-HSQC spectra were recorded for α-15N-lysine, α-15N-tyrosine, α-15N-phenylalanine, and 13C-proline-labeled protein to verify the assigned backbone amide signals. Side chain signals were assigned using 3D HBHA(CO)NH, (H)CCH-TOCSY, H(C)CH-TOCSY; (H)C(CO)NH and H(CCO)NH. Distance restraints for the structure calculation were obtained by analyzing cross peaks in 13C-NOESY-HSQC (with the 13C offset and delays optimized for aliphatic and aromatic carbon nuclei in two separate experiments, mixing time 150 msec) and 15N-NOESY-HSQC (mixing time 150 msec) experiments (Sattler et al. 1999). A {1H}, 15N heteronuclear NOE experiment of the amide resonances was recorded to identify flexible regions of the protein (Farrow et al. 1994).
Spectra were processed using the Bruker TOPSPIN 3.2 software and analyzed with CARA (Keller 2004). Torsion angle restraints were calculated with TALOS+ (Shen and Bax 2013) based on the chemical shift assignments. NOE distance restraints were automatically picked with the ATNOS/CANDID module from UNIO (Herrmann et al. 2002) and manually curated with CcpNmr Analysis (Vranken et al. 2005) The structure was calculated with CYANA (Güntert 2009), performing seven iterations with 100 initial structures and 10 final structures. Restrained energy refinement of the NMR structure was performed with the AMBER force field (Ponder and Case 2003) using the OPAL module (Luginbühl et al. 1996). The structure quality was assessed with the Protein Structure Validation Software (PSVS) (Bhattacharya et al. 2007). The electrostatic surface potential was calculated with the PDB2PQR web server (Dolinsky et al. 2004) and visualized with the APBS plug-in for PyMOL (Baker et al. 2001). All figures of the structures were prepared with PyMOL (The PyMOL Molecular Graphics System, Version 2.0, Schrödinger, LLC).
The NMR assignments and structure are deposited in the BMRB and the PDB under accession numbers 27453 and 6S10, respectively.
Preparation of RNA
The following RNA sequences were obtained commercially from Dharmacon: RocR 3′ single-stranded tail (5′-CCUUUCU-3′), oligo U4, U6, and U8, oligo A6, oligo C6, G-rich (5′-GGAGGA-3′), RaiZ hp (5′-AACGCGCCUUCGGGCGCGUU-3′), RaiZ hp U6 (5′-AACGCGCCUUCGGGC GCGUUUUUUUU-3′), RaiZ hp A6 (5′-AACGCGCCUUCGGGCGCGUUAAAAAA-3′). RNAs were deprotected and lyophilized twice as suggested by the manufacturer. Afterward, the samples were resuspended in NMR buffer in adequate concentrations. The secondary structure of the RaiZ constructs was tested using 1D 1H NMR spectra (Supplemental Fig. S8; Fürtig et al. 2003). The RocR stem–loop III RNAs with (5′-GGGUCAAUUGGCGACACACUGAUUGGCCCUUUCU-3′) and without (5′-GGGUCAAUUGGCGACACACUGAUUGGCCC-3′) the 3′ single-stranded tail were prepared by in vitro transcription with T7-RNA polymerase and purified as described in detail previously (Duchardt-Ferner et al. 2016).
NMR titrations
For NMR titrations, 1H,15N-HSQCs of 80 µM uniformly 15N-labeled Lpp1663 were recorded in NMR buffer and titrated with up to three or five equivalents of RNA at 298 K. Chemical shift perturbations were analyzed manually with Cara (Keller 2004). Shift changes were categorized as the following: peak disappeared (intermediate exchange regime), peak shifted (fast exchange regime), no change, no analysis possibly due to signal overlap or residue is a proline and cannot be identified in a 1H,15N-HSQC spectrum. Residues that showed the same type of chemical shift change were highlighted on the surface of Lpp1663 in the same color.
ITC measurements
ITC measurements were performed at 298 K in NMR buffer using a MicroCal iTC200 calorimeter (Malvern Panalytical). Twenty-five, 50, or 80 µM protein samples were titrated with 0.375, 0.5, 1, or 2 mM RNA (obtained from Dharmacon as described above) by 19 serial injections of 2 µL at a stirring speed of 750 rpm. Data processing and determination of binding constants were performed with an Origin7 (OriginLab) based software provided commercially (Malvern Panalytical) using a one-site binding model. Experiments were performed in triplicates. The results are summarized in Supplemental Table 1.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Center of Biomolecular Magnetic Resonance (BMRZ) and the Deutsche Forschungsgemeinschaft in the framework of the CRC 902 “Molecular mechanisms of RNA-based regulation” project B17 (to J.W.). We are grateful to Marcel Horn for preparing the RocR stem–loop RNAs and the members of the Wöhnert group for helpful discussions.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.077354.120.
REFERENCES
- Altuvia S, Weinstein-Fischer D, Zhang A, Postow L, Storz G. 1997. A small, stable RNA induced by oxidative stress: role as a pleiotropic regulator and antimutator. Cell 90: 43–53. 10.1016/S0092-8674(00)80312-8 [DOI] [PubMed] [Google Scholar]
- Arthur DC, Ghetu AF, Gubbins MJ, Edwards RA, Frost LS, Glover JNM. 2003. FinO is an RNA chaperone that facilitates sense–antisense RNA interactions. EMBO J 22: 6346–6355. 10.1093/emboj/cdg607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur DC, Edwards RA, Tsutakawa S, Tainer JA, Frost LS, Glover JNM. 2011. Mapping interactions between the RNA chaperone FinO and its RNA targets. Nucleic Acids Res 39: 4450–4463. 10.1093/nar/gkr025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attaiech L, Boughammoura A, Brochier-Armanet C, Allatif O, Peillard-Fiorente F, Edwards RA, Omar AR, MacMillan AM, Glover M, Charpentier X. 2016. Silencing of natural transformation by an RNA chaperone and a multitarget small RNA. Proc Natl Acad Sci 113: 8813–8818. 10.1073/pnas.1601626113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. 2001. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci 98: 10037–10041. 10.1073/pnas.181342398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauriedl S, Gerovac M, Heidrich N, Bischler T, Vogel J, Schoen C. 2020. The minimal meningococcal ProQ protein has an intrinsic capacity for structure-based global RNA recognition. Nat Commun 11: 2823 10.1038/s41467-020-16650-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A, Tejero R, Montelione GT. 2007. Evaluating protein structures determined by structural genomics consortia: tools for structure quality evaluation. Proteins 795: 778–795. [DOI] [PubMed] [Google Scholar]
- Chaulk S, Lu J, Tan K, Arthur DC, Edwards RA, Frost LS, Joachimiak A, Glover JNM. 2010. N. meningitidis 1681 is a member of the FinO family of RNA chaperones. RNA Biol 7: 812–819. 10.4161/rna.7.6.13688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaulk SG, Smith-Frieday MN, Arthur DC, Culham DE, Edwards RA, Soo P, Frost LS, Keates RAB, Glover JNM, Wood JM. 2011. ProQ is an RNA chaperone that controls ProP levels in Escherichia coli. Biochemistry 50: 3095–3106. 10.1021/bi101683a [DOI] [PubMed] [Google Scholar]
- Chihara K, Bischler T, Barquist L, Monzon VA, Noda N, Vogel J, Tsuneda S. 2019. Conditional Hfq association with small noncoding RNAs in Pseudomonas aeruginosa revealed through comparative UV cross-linking immunoprecipitation followed by high-throughput sequencing. MSystems 4: 1–15. 10.1128/mSystems.00590-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christ NA, Bochmann S, Gottstein D, Duchardt-Ferner E, Hellmich UA, Düsterhus S, Kötter P, Güntert P, Entian KD, Wöhnert J. 2012. The first structure of a lantibiotic immunity protein, SpaI from Bacillus subtilis, reveals a novel fold. J Biol Chem 287: 35286–35298. 10.1074/jbc.M112.401620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimastrogiovanni D, Fröhlich KS, Bandyra KJ, Bruce HA, Hohensee S, Vogel J, Luisi BF. 2014. Recognition of the small regulatory RNA RydC by the bacterial Hfq protein. Elife 3: e05375 10.7554/eLife.05375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA. 2004. PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res 32: W665–W667. 10.1093/nar/gkh381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchardt-Ferner E, Gottstein-Schmidtke SR, Weigand JE, Ohlenschläger O, Wurm JP, Hammann C, Suess B, Wöhnert J. 2016. What a difference an OH makes: conformational dynamics as the basis for the ligand specificity of the neomycin-sensing riboswitch. Angew Chem Int Ed Engl 55: 1527–1530. 10.1002/anie.201507365 [DOI] [PubMed] [Google Scholar]
- Durieux I, Ginevra C, Attaiech L, Picq K, Juan PA, Jarraud S, Charpentier X. 2019. Diverse conjugative elements silence natural transformation in Legionella species. Proc Natl Acad Sci 116: 18613–18618. 10.1073/pnas.1909374116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrow NA, Muhandiram R, Pascal SM, Kay LE, Singer AU, Forman-Kay JD, Kay CM, Gish G, Pawson T, Shoelson SE. 1994. Backbone dynamics of a free and a phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry 33: 5984–6003. 10.1021/bi00185a040 [DOI] [PubMed] [Google Scholar]
- Favier A, Brutscher B. 2011. Recovering lost magnetization: polarization enhancement in biomolecular NMR. J Biomol NMR 49: 9–15. 10.1007/s10858-010-9461-5 [DOI] [PubMed] [Google Scholar]
- Fürtig B, Richter C, Wöhnert J, Schwalbe H. 2003. NMR spectroscopy of RNA. ChemBioChem 4: 936–962. 10.1002/cbic.200300700 [DOI] [PubMed] [Google Scholar]
- Gerovac M, El Mouali Y, Kuper J, Kisker C, Barquist L, Vogel J. 2020. Global discovery of bacterial RNA-binding proteins by RNase-sensitive gradient profiles reports a new FinO domain protein. RNA 10.1261/rna.076992.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghetu AF, Gubbins MJ, Frost LS, Glover JNM. 2000. Crystal structure of the bacterial conjugation repressor FinO. Nat Struct Biol 7: 565–569. 10.1038/76790 [DOI] [PubMed] [Google Scholar]
- Ghetu AF, Arthur DC, Kerppola TK, Glover JNM. 2002. Probing FinO-FinP RNA interactions by site-directed protein-RNA crosslinking and gelFRET. RNA 8: 816–823. 10.1017/S1355838202026730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez GM, Hardwick SW, Maslen SL, Mark Skehel J, Holmqvist E, Vogel J, Bateman A, Luisi BF, William Broadhurst R. 2017. Structure of the Escherichia coli ProQ RNA-binding protein. RNA 23: 696–711. 10.1261/rna.060343.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorski SA, Vogel J, Doudna JA. 2017. RNA-based recognition and targeting: sowing the seeds of specificity. Nat Rev Mol Cell Biol 18: 215–228. 10.1038/nrm.2016.174 [DOI] [PubMed] [Google Scholar]
- Güntert P. 2009. Automated structure determination from NMR spectra. Eur Biophys J 38: 129–143. 10.1007/s00249-008-0367-z [DOI] [PubMed] [Google Scholar]
- Hacker C, Christ NA, Duchardt-Ferner E, Korn S, Göbl C, Berninger L, Düsterhus S, Hellmich UA, Madl T, Kötter P, et al. 2015. The solution structure of the lantibiotic immunity protein NisI and its interactions with nisin. J Biol Chem 290: 28869–28886. 10.1074/jbc.M115.679969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidrich N, Bauriedl S, Barquist L, Li L, Schoen C, Vogel J. 2017. The primary transcriptome of Neisseria meningitidis and its interaction with the RNA chaperone Hfq. Nucleic Acids Res 45: 6147–6167. 10.1093/nar/gkx168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann T, Güntert P, Wüthrich K. 2002. Protein NMR structure determination with automated NOE-identification in the NOESY spectra using the new software ATNOS. J Biomol NMR 24: 171–189. 10.1023/A:1021614115432 [DOI] [PubMed] [Google Scholar]
- Holmqvist E, Wright PR, Li L, Bischler T, Barquist L, Reinhardt R, Backofen R, Vogel J. 2016. Global RNA recognition patterns of post-transcriptional regulators Hfq and CsrA revealed by UV crosslinking in vivo. EMBO J 35: 991–1011. 10.15252/embj.201593360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmqvist E, Li L, Bischler T, Barquist L, Vogel J. 2018. Global maps of ProQ binding in vivo reveal target recognition via RNA structure and stability control at mRNA 3′ ends. Mol Cell 70: 971–982. 10.1016/j.molcel.2018.04.017 [DOI] [PubMed] [Google Scholar]
- Immer C, Hacker C, Wöhnert J. 2018. NMR resonance assignments for a ProQ homolog from Legionella pneumophila. Biomol NMR Assign 12: 319–322. 10.1007/s12104-018-9831-6 [DOI] [PubMed] [Google Scholar]
- Jerome LJ, Frost LS. 1999. In vitro analysis of the interaction between the FinO protein and FinP antisense RNA of F-like conjugative plasmids. J Biol Chem 274: 10356–10362. 10.1074/jbc.274.15.10356 [DOI] [PubMed] [Google Scholar]
- Keller RLJ. 2004. The computer aided resonance assignment tutorial. Cantina Verlag, Goldau, Switzerland. [Google Scholar]
- Kunte HJ, Crane RA, Culham DE, Richmond D, Wood JM. 1999. Protein ProQ influences osmotic activation of compatible solute transporter ProP in Escherichia coli K-12. J Bacteriol 181: 1537–1543. 10.1128/JB.181.5.1537-1543.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link TM, Valentin-Hansen P, Brennana RG. 2009. Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. Proc Natl Acad Sci 106: 19292–19297. 10.1073/pnas.0908744106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Naismith JH. 2008. An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol 8: 91 10.1186/1472-6750-8-91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Wang GG. 2013. Tudor: a versatile family of histone methylation “readers.” Trends Biochem Sci 38: 1–18. 10.1016/j.tibs.2012.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luginbühl P, Güntert P, Billeter M, Wüthrich K. 1996. The new program OPAL for molecular dynamics simulations and energy refinements of biological macromolecules. J Biomol NMR 8: 136–146. 10.1007/BF00211160 [DOI] [PubMed] [Google Scholar]
- Markley JL, Bax A, Arata Y, Hilbers CW, Kaptein R, Sykes BD, Wright PE, Wüthrich K. 1998. Recommendations for the presentation of NMR structures of proteins and nucleic acids. IUPAC-IUBMB-IUPAB Inter-Union Task Group on the standardization of data bases of protein and nucleic acid structures determined by NMR spectroscopy. J Biomol NMR 12: 1–23. 10.1023/A:1008290618449 [DOI] [PubMed] [Google Scholar]
- Massé E, Gottesman S. 2002. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc Natl Acad Sci 99: 4620–4625. 10.1073/pnas.032066599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melamed S, Adams PP, Zhang A, Zhang H, Storz G. 2020. RNA-RNA interactomes of ProQ and Hfq reveal overlapping and competing roles. Mol Cell 77: 411–425. 10.1016/j.molcel.2019.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikulecky PJ, Kaw MK, Brescia CC, Takach JC, Darren D, Feig AL. 2011. Escherichia coli Hfq has distinct interaction surfaces for DsrA, rpoS and poly(A) RNAs. Nat Stuct Mol Biol 11: 1206–1214. 10.1038/nsmb858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchmore DC, McIntosh LP, Russell CB, Anderson DE, Dahlquist FW. 1989. Expression and nitrogen-15 labeling of proteins for proton and nitrogen-15 nuclear magnetic resonance. Methods Enzymol 177: 44–73. 10.1016/0076-6879(89)77005-1 [DOI] [PubMed] [Google Scholar]
- Nitzan M, Rehani R, Margalit H. 2017. Integration of bacterial small RNAs in regulatory networks. Annu Rev Biophys 46: 131–148. 10.1146/annurev-biophys-070816-034058 [DOI] [PubMed] [Google Scholar]
- Olejniczak M, Storz G. 2017. ProQ/FinO-domain proteins: another ubiquitous family of RNA matchmakers? Mol Microbiol 104: 905–915. 10.1111/mmi.13679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey S, Gravel CM, Stockert OM, Wang CD, Hegner CL, LeBlanc H, Berry KE. 2020. Genetic identification of the functional surface for RNA binding by Escherichia coli ProQ. Nucleic Acids Res 48: 4507–4520. 10.1093/nar/gkaa144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panja S, Schu DJ, Woodson SA. 2013. Conserved arginines on the rim of Hfq catalyze base pair formation and exchange. Nucleic Acids Res 41: 7536–7546. 10.1093/nar/gkt521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponder JW, Case DA. 2003. Force fields for protein simulations. Adv Protein Chem 66: 27–85. 10.1016/S0065-3233(03)66002-X [DOI] [PubMed] [Google Scholar]
- Sattler M, Schleucher J, Griesinger C. 1999. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog Nucl Magn Reson Spectrosc 34: 93–158. 10.1016/S0079-6565(98)00025-9 [DOI] [Google Scholar]
- Sauter C, Basquin J, Suck D. 2003. Sm-like proteins in Eubacteria: the crystal structure of the Hfq protein from Escherichia coli. Nucleic Acids Res 31: 4091–4098. 10.1093/nar/gkg480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher MA, Pearson RF, Møller T, Valentin-Hansen P, Brennan RG. 2002. Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: a bacterial Sm-like protein. EMBO J 21: 3546–3556. 10.1093/emboj/cdf322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Bax A. 2013. Protein backbone and sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks. J Biomol NMR 56: 227–241. 10.1007/s10858-013-9741-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnov A, Förstner KU, Holmqvist E, Otto A, Günster R, Becher D, Reinhardt R, Vogel J. 2016. Grad-seq guides the discovery of ProQ as a major small RNA-binding protein. Proc Natl Acad Sci 113: 11591–11596. 10.1073/pnas.1609981113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnov A, Wang C, Drewry LL, Vogel J. 2017. Molecular mechanism of mRNA repression in trans by a ProQ-dependent small RNA. EMBO J 36: 1029–1045. 10.15252/embj.201696127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Someya T, Baba S, Fujimoto M, Kawai G, Kumasaka T, Nakamura K. 2012. Crystal structure of Hfq from Bacillus subtilis in complex with SELEX-derived RNA aptamer: insight into RNA-binding properties of bacterial Hfq. Nucleic Acids Res 40: 1856–1867. 10.1093/nar/gkr892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein EM, Kwiatkowska J, Basczok MM, Gravel CM, Berry KE, Olejniczak M. 2020. Determinants of RNA recognition by the FinO domain of the Escherichia coli ProQ protein. Nucleic Acids Res 48: 7502–7519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updegrove TB, Zhang A, Storz G. 2016. Hfq: the flexible RNA matchmaker. Curr Opin Microbiol 30: 133–138. 10.1016/j.mib.2016.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Biesen T, Frost LS. 1994. The FinO protein of IncF plasmids binds FinP antisense RNA and its target, traJ mRNA, and promotes duplex formation. Mol Microbiol 14: 427–436. 10.1111/j.1365-2958.1994.tb02177.x [DOI] [PubMed] [Google Scholar]
- Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, Laue ED. 2005. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins Struct Funct Genet 59: 687–696. 10.1002/prot.20449 [DOI] [PubMed] [Google Scholar]
- Westermann AJ, Venturini E, Sellin ME, Förstner KU, Hardt WD, Vogel J. 2019. The major RNA-binding protein ProQ impacts virulence gene expression in Salmonella enterica serovar typhimurium. MBio 10: 1–21. 10.1128/mBio.02504-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.